Abstract
Mouse models have made innumerable contributions to understanding the genetic basis of neurological disease and pathogenic mechanisms and to therapy development. Here we consider the current state of mouse genetic models of Developmental and Epileptic Encephalopathy (DEE), representing a set of rare but devastating and largely intractable childhood epilepsies. By examining the range of mouse lines available in this rapidly moving field and by detailing both expected and unusual features in representative examples, we highlight lessons learned in an effort to maximize the full potential of this powerful resource for preclinical studies.
Keywords: Mouse models, Epileptic encephalopathy, Disease modeling, Seizure
1. Human DEE and mouse genetic models: a tally
The past three decades witnessed the unfolding realization that epilepsy has a significant genetic component, culminating with the striking discovery that a sizeable fraction of the most devastating form of disease – a mixed set of childhood epilepsies that are together termed Developmental and Epileptic Encephalopathy (DEE) – results from single gene de novo mutations (Epi4K et al., 2013). While currently over one-quarter of all DEE cases are genetically diagnosed with high confidence by whole exome sequencing, the underlying genetic heterogeneity, early age of disease onset, and severity present major challenges to effective treatment.
Through technological advances in genome editing, the past 5 years has produced a burst of new mouse models of epilepsy, primarily for DEE, which have been examined using transcriptomics, neuroimaging, neurophysiology and behavior testing. Despite a high degree of genetic, and to a lesser extent neurophysiologic similarity between human and mice, and while many construct-valid mouse models display some form of seizure susceptibility and abnormal neurodevelopment, navigating species differences proves to be no small matter. As DEE families and clinicians have begun to engage with researchers to seek diagnosis and therapy, there is a further need for clarity on mouse strains that model specific genetic DEEs and to reflect upon their potential for preclinical research.
Towards this end, we aligned human variant data with equivalent mouse mutations in the same genes. We began by surveying the NCBI ClinVar database (further guided by OMIM database annotation) for intragenic variants classified as pathogenic or likely pathogenic in the Molecular Consequence field, and containing terms in the Conditions field such as early infantile (or early onset) epileptic encephalopathy (EIEE, EOEE), severe myoclonic epilepsy in infancy (SMEI), mental retardation (intellectual disability) or similar conditions of neurodevelopmental origin where seizures are a prominent feature. After excluding genes that had only a single DEE variant, this effort resulted in 85 genes that collectively carried over 7200 DEE variants. These genes were then sub-grouped by apparent molecular mode of action (Table 1): A. Missense only (genes for which there were no predicted loss-of-function (LOF) DEE variants, i.e. no frameshifts, deletions, critical splice site or nonsense mutations), B. Loss-of-function only (genes for which no missense variants were listed), and C. DEE conditions that included both LOF and missense variants in the same gene.
Table 1.
Seizure susceptibility in mouse models of human DEE.
| Human DEE variant type | Mutant mouse strain with DEE variant |
Other mutant mouse strain (no DEE variant) |
No mutant mouse reported | Total (unique) | |||
|---|---|---|---|---|---|---|---|
| Seizure susceptibility |
Seizure susceptibility |
||||||
| Yes | None reporteda | Yes | None reporteda | ||||
| A. Missense only | DNM1b, GNB1c, GRIN2A, KCNT1, GABRB3c, SCN1B, SCN8A | ALG13d | ALG13, ARV1, GABBR2, GABRB3, GABRG2c, KCNQ2, KCNQ3, NEUROD2, SCN1B, SCN2Ac, SCN8Ac, SLC1A2, SLC25A12, SYNGAP1e | CACNA1E, CUX2, CYFIP2, DNM1, EEF1A2, FGF12, GABRA2, GABRA5, GABRB1, GABRB2, GRIN2B, GRIN2D, KCNT1, KCNT2, MEF2C, RNF13, SETD1A, ST3GAL3, USP19 | RHOBTB2 | 36 | |
| B. Loss-of-function only | AP3B2, CPLX1, PLCB1, SZT2 f | DMXL2, DOCK7, FRRS1L, PIGQ, TRAK1 | 9 | ||||
| LOF mouse | In LOF and missense | ||||||
| C. Both LOF & missense | CACNA1Ac, HNRNPU, KCNA2e, PCDH19c, STXBP1c, SYNJ1, TBC1D24 | ARXc, CACNA2D2c, CDKL5, GABRA1c,e, GNAO1, IQSEC2, MECP2, SCN1A, SLC12A5 | AARSe, ACTL6B, ADAM22e, ARHGEF9, CAD, CHD2, DENND5A, GLS, GOT2e, HCN1e, ITPA, KCNB1, MDH2e, PIGPe, PNKP, PPP3CA, SCN3A, SIK1, SLC13A5, SMC1A, SPTAN1, UGDH, WWOX, YWHAG | 40 | |||
Excluding seizures described without documentation in manuscript text.
The missense mutation in Dnm1Ftfl mice is not based on a human DEE variant but phenocopies multiple variants in the same domain.
Mouse mutants that exhibit frequent spike-wave discharges (SWD).
C57BL/6NJ-Alg13N107S mice exhibit no spontaneous seizures behaviorally or by conventional EEG and have normal electroconvulsive and PTZ seizure threshold (W. Frankel, unpublished results).
Only one human loss of function DEE variant described.
SZT2 individuals are compound heterozygous, whereby one of the two allelic variants is always loss-of-function.
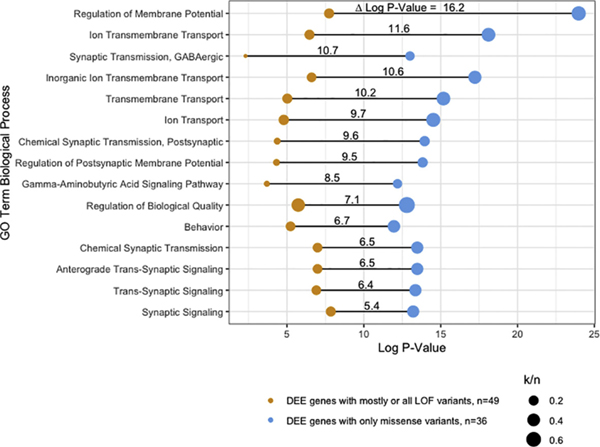
These groupings are not intended as formal, but rather to guide comparative studies. Indeed, some genes have only a few pathogenic variants. Others may be exceptions, for example, while genes in Group C contain many loss-of-function variants and it is reasonable to assume that most missense variants in these genes for the same disease are also null or hypomorphic, some may have gain-of-function (GOF) properties. Nevertheless, any division between GOF (emergence of new functional properties) and LOF (deficiency in essential properties) begs the question of whether the genes align with different biological processes. To test this idea at a basic level, we performed in silico assessment using Gene Ontology (GO) functional annotation clustering (Fig. 1), from which two conclusions emerge: i) Across the most significant GO terms, Group A clusters were almost 9 orders of magnitude more significant than Groups B/C, despite having one-third fewer genes. This may suggest that genes with missense-only DEE variants are more likely to converge on common biological processes; ii) In fact, one o GO term (“Regulation of Membrane Potential”) was 16.2 orders of magnitude more significant in the missense-only group. Although, ultimately specific genes and variants need to be tested in the lab, such clues that suggest higher-level convergence is good news for the potential for future treatments to span multiple DEEs.
Fig. 1.
Functional annotation clustering of DEE genes by variant type. Visualization of enriched biological process Gene Ontology (GO) terms in two categories of genes – DEE genes with mostly or all LOF variants and DEE genes with only missense variants. Only GO terms with at least 5 genes in either category were included. Δ Log P-Value: difference in log p-value for clustering between the two gene categories; n: total number of genes in each category; k: number of genes in the total gene set annotated to the GO term.
We then cross-tabulated DEE genetic variant types by whether a mouse model exists for a human variant using the Mouse Genome Informatics database as a guide (http://www.informatics.jax.org), confirmed by review of the primary literature or supplemented with knowledge of recent studies or those pending publication. From this straightforward comparison we can make some general observations regarding the representation of mouse models for DEE genetic variants. First, it is very clear that only a small fraction (22%, or 8 of 36) from the missense-only type (Table 1, Row A) have been modeled in mice, whereas 100% of genes that have a LOF DEE is represented by mouse mutants, mostly knockouts (Table 1, Rows B and C). This contrast is not surprising given that knockout mice (whether studied as homozygous or heterozygous) have been mainstays in the mouse genetics community for many years, whereas facile generation of missense mutations in mice (e.g. by CRISPR/Cas9) has come about only recently.
We next examined whether seizure susceptibility was observed in a corresponding mouse mutant, whether or not that mutation was equivalent to the DEE variant. The results were very promising for genes with only missense human variants: 88% of mutant mouse strains with a DEE variant (7 of 8) were reported with obvious spontaneous seizures (Table 1, Row A; Table 2; DNM1, GNB1, etc). Interestingly, 14 DEE genes that have mouse strains with LOF variants or missense variants not orthologous to human DEE variants also showed seizure susceptibility (Table 1, Row A; ALG13, ARV1, etc.). This is a likely source of confusion to the uninitiated as these mouse lines might appear to be suitable construct-valid DEE models when they may model a different type of epilepsy entirely.
Table 2.
Mouse models carrying orthologous variants (or equivalent) to human DEE and exhibiting seizure or comorbidity phenotypes.
One or more knockout or hypomorphic mouse mutation has been described.
Observed in one or more missense mutation.
Observed in one or more LOF or hypomorphic mutation.
Spike-wave discharges (SWD) only.
Blank cells mean either not tested or not susceptible.
May include pentylenetetrazole, kainic acid, electroconvulsive, audiogenic, fluorothyl or febrile stimuli.
May include growth delay, reduced body weight, small stature.
May include ataxia, dystonia, hyperactivity, hypoactivity.
Similar examination of seizure susceptibility in models based on putative loss-of-function human DEE variants gave more mixed results. Seizure susceptibility was demonstrated in only 41% (20 of 49) mutant mouse strains (Table 1, Rows B, C). Certainly the lower rate in this group is affected by negative ascertainment bias, namely a) many knockout mice were not developed by epilepsy researchers who might not recognize subtle seizure behaviors, b) some knockouts are lethal prior to seizure onset or detection, and heterozygotes are often not examined to the same degree, c) as we will discuss later, non-convulsive seizures are common in mouse models but require dedicated in vivo electrophysiology for detection, d) focal seizures can easily be missed with conventional rodent EEG, and e) detection of seizures in pre-weaning age mice is not yet commonplace even among epilepsy researchers despite the fact that this developmental period corresponds best with children. In contrast, the relatively high seizure susceptibility yield of the missense-only group almost certainly reflects a degree of positive ascertainment bias – these models were likely developed for the explicit purpose of studying the respective human epilepsy.
Many further subtleties and important details influence comparison of DEE mouse models to the respective human genetic disease, including phenotypic heterogeneity, genetic background and various technical challenges to detecting and defining seizure types or predisposition in small rodents, not to mention other neurological or neurosensory abnormalities, present in one form or another in all mouse DEE models (Table 2). We will discuss some of these issues in examples below. However, even our straightforward assessment shows that while there are already over 80 mouse mutants in genes for human DEE, seemingly a large number available to study, there remain both significant gaps (e.g. only 22% of potential GOF models have been generated in mice), and sources of overlap and confusion (e.g. GOF and LOF mouse mutants in the same gene can cause seizures).
Gaps, overlaps and complexities aside, several studies from the mouse literature provide useful, state-of-the-art examples of studying genetic construct-valid models for DEE. The following examples illustrate both desirable features in mouse models, plus some quirks that need to be reckoned with as the field moves forward.
2. SCN8A epileptic encephalopathy: strong models, paradox included
Genetic variants in genes encoding sodium channels were among the earlier channelopathies recognized in human epilepsy; first SCN1B, then SCN1A (the primary cause of Dravet Syndrome, the most common genetic DEE), SCN2A (Meisler et al., 2001), and more recently SCN8A encoding the Nav1.6 sodium channel, first described for a DEE in 2012 (Veeramah et al., 2012). As Meisler described in a recent review (Meisler, 2019), heterozygosity for gain- or altered-function variants in SCN8A are associated with the most severe DEE whereas haploinsufficiency is associated with significant but less severe disease, notably, intellectual disability accompanied by mild epilepsies. Currently in ClinVar, there are 110 SCN8A missense variants, with 60% (66/110) being annotated with epileptic encephalopathy, while only 9 loss-of-function variants are documented.
Mice carrying Scn8a loss-of-function variants were first reported in the 1960’s and 1970’s as recessive spontaneous neurological mutants with motor phenotypes and premature lethality (Dickie, 1965; Lane, 1976). It was not until 2009 that loss-of-function Scn8a heterozygotes were shown to display spontaneous spike-wave discharges, considered to model absence or petit-mal seizures (Papale et al., 2009). Unusually, these same mice confer relative resistance to convulsive seizures whether induced chemically, electrically, or by genetic mutation (Hawkins et al., 2011; Martin et al., 2007; Sun et al., 2013) – a feature that was recently exploited to show the effectiveness of Scn8a antisense gene therapy in reducing or eliminating seizures in the Scn1a mouse model of Dravet Syndrome (Lenk et al., 2020). This apparent paradox of seizure sensitivity and resistance in the same genetic model provided a glimpse into the allelic heterogeneity and complex cellular etiology of epilepsy, complexities that are becoming increasingly appreciated for SCN8A and other genetic DEEs.
Two separate SCN8A DEE gain-of-function (GOF) mutations have now been introduced into mice – N1768D (constitutive) and R1872W (conditional) – respectively based on the heterozygous variant N1768D first identified in a patient with sudden unexpected death in epilepsy (SUDEP) (Veeramah et al., 2012), and R1872W first identified in a patient with prolonged tonic-clonic seizures from 3 months of age (Ohba et al., 2014). Although just two models, the severity of the in vivo phenotype in mice and the corresponding channel kinetics in vitro track with the respective disease severity (Meisler, 2019). Both heterozygous models exhibit spontaneous seizures resulting in lethality, although the seizure phenotype of R1872W of heterozygotes is fully penetrant and mice die at a much earlier age (2–3 weeks) than in N1768D (9 weeks). Electroencephalographic evidence of epileptiform activity was observed in both models including typical high amplitude polyspike activity followed by post-ictal depression, and at least one of the two exhibited myoclonic jerks prior to development of generalized tonic-clonic seizures (GTCS). N1768D heterozygous mice were tested for various neurological and behavioral deficiencies but found to be relatively normal compared with control, except for modest deficits motor coordination and social discrimination (Wagnon et al., 2015). The R1872W conditional model was activated using cre-recombinase expression in either inhibitory or excitatory neurons, which led to the discovery that expression of the variant in excitatory neurons is necessary for seizure activity (Bunton-Stasyshyn et al., 2019).
Overall Scn8a knockin mice appear to be a satisfying model for genetic DEE, displaying features representative of some other mouse models of DEE including lethal spontaneous seizures, allelic heterogeneity, and mild-to-moderate behavioral deficits that together make them suitable models for study. The R1872W conditional mutation adds further insight into the cellular etiology of disease. Scn8a heterozygous loss-of-function mutants also show the value of studying a range of milder forms of disease with seizures over a range of severities (Inglis et al., 2020). Although the coexistence of spontaneous absence-like seizures and resistance to convulsive seizures in the same mice appears at first a paradox, it provides an object lesson about the biological complexity of developmental epilepsies, while underscoring how crucial correct Nav1.6 ion channel expression levels are for normal neuronal function. Using Scn8a conditional knockout mice, Makinson et al. (2017) reported that absence seizures are due to thalamic deficit while convulsive seizure resistance results from loss of Scn8a from cortical excitatory cells, providing an example of how seemingly paradoxical findings in Scn8a LOF mutants could be understood by considering how mutations influence distinct regions and networks differently.
3. KCNT1 DEE: multiple seizure types, brain state-dependence
Inherited variants in potassium channels were among the first well-defined albeit relatively mild monogenetic epilepsies, now termed Benign Neonatal Familial Epilepsy (Wang et al., 1998). It was not for another ten years when variants in KCNT1, the gene encoding the Na+ activated K+ channel (Slack, KNA 1.1) were associated with a range of more severe epilepsies, including EIEE, epilepsy of infancy with migrating focal seizures (EIMFS), and autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE).
Two KCNT1 GOF variants, Y796H (Y777H in mouse) and R474H (R455H in mouse), were described in mice recently, and both confer significant spontaneous seizure activity (Quraishi et al., 2020; Shore et al., 2020). The Y796H variant (c.2386 T>C) was identified as a heterozygous mutation causing both inherited and de novo cases of a severe, early-onset form of ADNFLE (Heron et al., 2012). The R474H variant (c.1421G>A) was reported in individuals with EIMFS or EOEE (Ohba et al., 2015). Both Y796H and R474H patients had intractable seizures with high frequency.
R455H homozygotes are embryonically lethal, and heterozygotes all have persistent interictal epileptiform discharges in the EEG (IED; average 16/h) and susceptibility to PTZ-induced convulsive seizures. Despite the fully penetrant IED, only half (3/7) showed spontaneous seizures in the EEG. Consistent with the respective human disease, Y777H mice have milder phenotypes and no selective lethality was observed in homozygotes. While seizures in heterozygotes are too rare to quantify, 93% of homozygotes (26/28 total) experienced seizures at an average of about one every 3.2 ± 0.4 h. In agreement with the epilepsy phenotype reported in Y796H patients [17], seizure episodes consisted of at least two types: GTCS typically lasting 30–60 s, and tonic seizures (TS) lasting 1–2 s (also observed in R455H but termed as myoclonic seizures). Frequent postictal and interictal epileptiform discharges (IED) were also observed. Interestingly, 90% of seizure episodes occurred during NREM sleep, which provides meaningful parallel with the sleep-associated seizure pattern of ADFNLE patients. Both GTCS and TS were examined further using calcium imaging, revealing large amplitude calcium events localized primarily to the secondary motor cortex region. Epileptiform activity was also detected in pre-adolescent mouse pups by performing acute unanaesthetized in vivo electrocorticography with a novel neural interface device called NeuroGrid, which makes Y777H mice especially valuable for studying childhood genetic disease. Further examination of biophysical properties and neuronal connectivity in primary neuron culture and acute slices also provide a first glimpse into the cellular basis of KCNT1 DEE, with non-fast spiking GABAergic interneurons displaying a strong impairment in action potential generation due to increased K+ current at subthreshold membrane voltages.
Taken together, the KCNT1 DEE models are satisfying as they cover different disease severities and forms consistent with the range of human patients. Although epileptic phenotypes were only tested in R455H adult mice, the model may phenocopy many key aspects of human EIMFS. For the Y777H model, while it is odd and a perhaps species-specific quirk that homozygosity is required to confer frequent seizures in mice, the relationship between seizures and activity state and the topographical mapping is surprisingly consistent with that of the ADNFLE patient.
4. GABRB3: Lennox-Gastaut Syndrome in a mouse model?
Type A GABA receptors (GABAARs), formed by pentameric assemblies of different subunit subtypes, are ligand-gated chloride channels that mediate fast inhibitory synaptic transmission. Because of the crucial inhibitory role of GABAARs, variants in many GABAAR subunit genes (e. g. GABRA1, GABRA2, GABRA5, GABRB1, GABRB2, GABRB3, GABRG2) are associated with DEE. GABRB3 variants specifically are implicated in several types of epileptic encephalopathies including infantile spasms and Lennox-Gastaut Syndrome (LGS; (Epi4K et al., 2013)). In ClinVar, 12 out of 29 missense variants are annotated with epileptic encephalopathy, and all others with annotated conditions, including 4 putative loss-of-function, are annotated with other epilepsy types. Children with missense mutations in GABRB3 have common symptoms including infantile seizure onset of multiple seizure types, and associated comorbidities such as neurodevelopmental delay, autistic behavior, and intellectual disability (Epi4K et al., 2013; Papandreou et al., 2016).
Gabrb3 knockout mice were first reported in 1998 (DeLorey et al., 1998) as a model for Angelman Syndrome since GABRB3 is within the Angelman Syndrome locus (15q11–13) together with UBE3A, but it was not until 2020 that a Gabrb3 D120N knock-in mouse was reported as the first genetic model for Lennox-Gastaut Syndrome (Qu et al., 2020). Although other missense mutations in mouse Gabrb3 (N265M and S408/9A) were described previously (Vien et al., 2015; Zeller et al., 2007), only D120N is based on a human DEE. The de novo missense mutation in GABRB3 - D120N (c.358G>A) – has been identified in two individuals with different symptoms (Epi4K, 2016) one diagnosed with LGS and another one with milder febrile and tonic-clonic seizures. The LGS patient had infantile spasms (IS) at 10 months of age (Epi4K, 2016) and later developed additional seizure types including generalized tonic-clonic, atypical absence, myoclonic and atonic seizures. EEG showed bursts of 2 Hz sharp spike-wave complexes. EEG from the other patient showed 2.5 Hz–3 Hz generalized spike-waves and diffuse slowing. It is notable that a phenotypic range arises in patients with the exact same GABRB3 variant.
While homozygous Gabrb3 D120N/D120N mouse mutants are neonatal lethal, heterozygotes do have frequent spontaneous spike-wave discharges that appear to model atypical absence-like seizures (3.0 Hz – 5.5 Hz), and less frequent tonic, myoclonic, atonic and GTCS. Spontaneous epileptic spasms were observed in young heterozygotes, which aligns with the observation that IS precedes LGS in one D120N patient. Other behavioral comorbidities seen in LGS patients such as impaired learning and memory, hyperactivity, and impaired social interactions appear to be mimicked in Gabrb3 D120N/+ heterozygotes as well. The authors assert that the heterozygotes are a convincing model of LGS because they recapitulate both the occurrence and semiology of LGS seizures.
Gabrb3 D120N/+ mice, like Kcnt1 Y777H/Y777H mice, are early examples of how specific mouse model seizure phenotypes may line-up with associated features of the human disease, while many of the accompanying behavioral features of these mice are quite mild. Moving forward, it will be very helpful to develop milder GABRB3 missense models to begin exploring the phenotypic heterogeneity and range and thus how precisely the mouse model phenotype aligns with a specific human DEE subtype – LGS.
5. CDKL5 Deficiency Disorder: nominal seizure susceptibility in an epilepsy model
Loss-of-function variants in the Chromosome X-linked gene CDKL5, encoding a serine-threonine kinase, were first shown to cause a DEE in the early 2000’s (Evans et al., 2005). Now termed CDKL5 Deficiency Disorder (CDD), disease associated mostly with loss-of-function CDKL5 variants provides a good example of early-onset intractable neurodevelopmental epilepsies with a wide range of overlapping clinical diagnoses such as autism, intellectual disability and atypical Rett’s Syndrome, usually accompanied by seizures.
A conventional Cdkl5 knockout mouse model was first reported in 2012 (Wang et al., 2012), and more recently an independent nonsense mutation, R59X, was described (Tang et al., 2019; Yennawar et al., 2019). While hemizygous males in these studies were described as having autistic-like behaviors and. Hyperactivity and reduced pain sensitivity (Gao et al., 2020b; Montanara et al., 2020), no seizure susceptibility was observed. Lately, evidence for excessive NMDAR-mediated neurotransmission was described in these models (Okuda et al., 2017; Tang et al., 2019). Although Cdkl5 knockout mice are not sensitive to the standard pro-convulsant kainic acid, one study observed increased seizure susceptibility induced by NMDA (Okuda et al., 2017). Interestingly, following anecdotal observations of behavioral seizures, it is now known that aged Cdkl5 heterozygous females (> 300 days) of both the R59X and the original knockout line actually do exhibit brief epileptic spasms and interictal spiking (Mulcahey et al., 2020). As in CDD, spasms in mice tended to occur in clusters along with increased power in the gamma and theta frequency band during quiet wake and non-REM sleep. The presence of epileptiform features in aged hemizygous males was not examined.
From a disease modeling perspective, the lack of readily-apparent seizure activity in genetic construct-valid mice might reasonably be argued as less than ideal. And, as we highlighted above, mouse knockouts published for over half of the genes representing loss-of-function DEE do not have any demonstrated seizure susceptibility (Table 1, Rows B, C). While most of those studies apparently did not include systematic efforts to detect seizures beyond passive observation, the Cdkl5 example shows that even dedicated epilepsy research labs sometimes need to dig deep to detect seizures in their mouse models. It also reinforces the reality that while some mouse models, such as Scn8a, Kcnt1 and Gabrb3 above have readily observed seizures - some with features that are strikingly similar to human - it is unrealistic to expect precise phenotypic matching in an animal model where brain maturation occurs earlier and more accelerated in real time compared with human. Notwithstanding our desire for better alignment, the Cdkl5 story and others emerging like it nevertheless show that much may be learned mechanistically at the basic cellular and network levels from studying construct-valid models, and the excitability features that may reflect key aspects of seizure predisposition.
6. Prominence of spike-wave discharges and the “absence” of characteristic DEE electrographic features in mouse models
Spike-wave discharges (SWD), the electrographic correlate of seizures in rodents characterized by behavioral arrest, are thought to be analogous to the generalized spike and wave discharges observed in human absence seizures. SWD are promiscuous in laboratory rodents having been demonstrated in many mutant mouse strains (Maheshwari and Noebels, 2014; Noebels, 1995), and they are also observed at lower incidence in several so-called normal inbred strains, although they are not typically observed in the three most commonly used strains for genetic engineering, C57BL/6J, FVB/NJ, 129/Sv (Letts et al., 2014). The reason for this predisposition is unknown – perhaps it is endemic to all mice, or alternatively it may have evolved in the ancestors of laboratory mice as a quirk due to relaxation of selective pressure over the centuries of commensality culminating with their use in biomedical research. Regardless, like Gabrb3 and Scn8a models noted above, several DEE mouse mutants exhibit spontaneous SWD (including Cacna1a (Fletcher et al., 1996; Jun et al., 1999; Noebels and Sidman, 1979), Cacna2d2 (Barclay et al., 2001), Gabra1 (Arain et al., 2012), Gabrg2 (Tan et al., 2007), Gnb1 (Colombo et al., 2019), Pcdh19 (Pederick et al., 2018), Scn2a (Ogiwara et al., 2018) and Stxbp1 (Chen et al., 2020)) well above the genetic background rate, indicating that the DEE variant is the primary determinant. Among DEE mouse models, Stxbp1 is a notable because SWD is the most prominent seizure phenotype in the otherwise fairly mildly impaired Stxbp1 haploinsufficiency mouse model (Chen et al., 2020; Miyamoto et al., 2017). STXBP1 is one of the most prevalent DEEs (Hamdan et al., 2011; Saitsu et al., 2008) but absence seizures are only one of several types of seizure observed in patients (Carvill et al., 2014).
The study of SWD in mouse models of DEE is a kind of double-edged sword. If absence seizures are not prominent in human DEE, and if laboratory mice are particularly SWD-sensitive, the corresponding mouse model might not be viewed by purists as suitable if SWD is the predominant or only seizure phenotype. On the other hand, if the defective gene is indeed the initiating cause of excitability phenotypes, then study of SWD in these models nevertheless provides a convenient means of uncovering pathogenic mechanisms at the cellular and network level which, in turn, could be generalized to other neurological systems.
Conversely, several electrographic seizure types that are characteristic of DEE have not yet been described in mouse models. The most notable include: hypsarrhythmia –EEG is characterized by chaotic background with nearly continuous random asynchronous high-voltage slow wave and spikes arising from multiple foci; paroxysmal fast activity, i.e. as seen in LGS patients; and abnormal sleep architecture. Although it is easy to conclude that the absence of these features so far, along with the dominance of SWD may be inevitable limitations of mouse models, the jury is still out because the vast majority of EEG studies in freely-moving mice is done in adults, and with a nominal number of electrodes. The novel recording device NeuroGrid noted in the KCNT1 section above, designed specifically to cover the dorsal cortical surface of a pup brain, allows recording from multiple areas simultaneously without damaging the brain in the same way that implanted electrodes would. Further development and implementation of advanced in vivo array recording technologies will be valuable for determining the full spectrum of normal and pathogenic electroencephalographic patterns in mouse pups.
7. Genetic background: expected and unexpected players
Genetic background is one of many potential sources of diverse clinical presentation for neurodevelopmental disorders and epilepsies. Although background is well known to impact seizure threshold in mouse strains and some mutant lines, including models that exhibit SWD (Frankel et al., 2001; Loscher et al., 2017), there are not yet enough data to know the degree to which it plays a role in spontaneous seizure incidence or form in some of the newer DEE models. Certainly, background genetics is known to have a very significant impact in Scn1a heterozygous knockout mice, the most widely studied model for Dravet Syndrome (Hawkins et al., 2016; Kang et al., 2019). While homozygotes are early lethal, heterozygotes have spontaneous tonic-clonic seizures with an onset, incidence and severity that varies considerably with mouse strain background. Heterozygotes on the 129/Sv mouse strain background do not have any spontaneous seizures, whereas those on either of the major C57BL/6 substrains (or F1 hybrid background between 129/Sv and C57BL/6), have lethal seizures as young adults with varying degrees of penetrance. Efforts to use Scn1a haploinsufficiency as a driver for identifying modifier genes have begun, and candidates have begun to emerge, such as that encoding a hypomorphic variant of Gabra2 in C57BL/6 J mice (Hawkins et al., 2016). However, nomination of genetic modifiers based on apparent, sensical candidate genes, such as Gabra2, may be self-limiting and come with ascertainment bias. For example, very recently it was determined that all C57BL/6 J mice – historically the most widely-used C57BL/6 substrain - have a natural LOF mutation in a neuronal tRNA gene, initially discovered as a genetic modifier of neurodegeneration (Ishimura et al., 2014). This mutation also significantly increases electroconvulsive and chemoconvulsive threshold compared with C57BL/6N substrains and even mitigates the incidence of SWD due to the Gabrg2 R43Q DEE variant (Kapur et al., 2020). Thus, while it is appealing to nominate existing candidate genes that account for some genetic background effects, this latest study suggests that fresh, unbiased approaches to identify unexpected players may also have significant value in shedding new light on genes and pathways that modify disease severity.
8. Genetic-based therapies and future expectations
So far at least three mouse DEE models (Cdkl5−/Y, Scn1a−/+, Dnm1Ftfl/Ftfl) have been deployed to illustrate the power of gene therapy for both loss-of-function and missense childhood epilepsies (Aimiuwu et al., 2020; Colasante et al., 2020; Gao et al., 2020a; Lenk et al., 2020; Yamagata et al., 2020), an approach that could supplement if not eventually replace conventional drug therapies to mitigate symptoms and improve quality of life. Although there are significant differences in timing of onset and progression of disease in rodents compared to human, conventional mouse genetic tools, such as conditional mutations, and somatic tools, such as antisense oligonucleotides and virally-delivered RNAi vectors, can be deployed to model both the genetic specificity as well as effective therapeutic windows for mouse DEE models. A recent review has discussed in detail the uses and limitations of utilizing mouse models to screen for genetic-based therapy strategies (Turner et al., 2020). It is potentially feasible to direct genetic therapies at the defective gene or RNA at or before the onset of symptoms to mitigate most, if not all, clinical features of some DEEs.
Each of the 85 DEE genes recognized in this simple survey have corroborative evidence for a causative role in DEE, ranging from nominal to overwhelming. With the considerable genetic heterogeneity that underlies DEE, the number should rise steadily with increasing genetic diagnosis. While it is very unlikely that new genes will overtake the major contributors SCN1A, CDKL5, STXBP1, SCN8A in terms of population incidence, we expect that the continued identification and laboratory modeling of rare players will contribute, perhaps significantly, to understanding and convergence of pathogenic mechanisms, in turn providing new opportunities for effective therapy that may cut across multiple genetic DEEs. Despite the complexities of modeling a neurodevelopmental human disease in a rodent, in parallel with alternatives such as the currently popular human iPSC-derived neuron culture approach (Ardhanareeswaran et al., 2017; Di Lullo and Kriegstein, 2017) and continued use of other animal models ranging from flies, to nematodes, to fish (Bessa et al., 2013; Gatto and Broadie, 2011; Sakai et al., 2018), laboratory mouse models are likely to remain prominent tools for in vivo research on DEE.
Acknowledgements and funding
The authors thank Matthew Weston, Christopher Makinson, Tristan Sands and Megha Sah for input on a draft version of this article. This work was supported by NINDS grant R37 NS013148.
References
- Aimiuwu OV, et al. , 2020. RNAi-based gene therapy rescues developmental and epileptic encephalopathy in a genetic mouse model. Mol. Ther 28, 1706–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador A, et al. , 2020. Modelling and treating GRIN2A developmental and epileptic encephalopathy in mice. Brain 143, 2039–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola E, et al. , 2014. Mapping pathological phenotypes in a mouse model of CDKL5 disorder. PLoS One 9, e91613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arain FM, et al. , 2012. Decreased viability and absence-like epilepsy in mice lacking or deficient in the GABAA receptor alpha1 subunit. Epilepsia 53, e161–e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardhanareeswaran K, et al. , 2017. Human induced pluripotent stem cells for modelling neurodevelopmental disorders. Nat. Rev. Neurol 13, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asinof SK, et al. , 2015. Independent neuronal origin of seizures and behavioral comorbidities in an animal model of a severe childhood genetic epileptic encephalopathy. PLoS Genet. 11, e1005347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay J, et al. , 2001. Ducky mouse phenotype of epilepsy and ataxia is associated with mutations in the Cacna2d2 gene and decreased calcium channel current in cerebellar Purkinje cells. J. Neurosci 21, 6095–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa C, et al. , 2013. Using C. elegans to decipher the cellular and molecular mechanisms underlying neurodevelopmental disorders. Mol. Neurobiol 48, 465–489. [DOI] [PubMed] [Google Scholar]
- Brew HM, et al. , 2007. Seizures and reduced life span in mice lacking the potassium channel subunit Kv1.2, but hypoexcitability and enlarged Kv1 currents in auditory neurons. J. Neurophysiol 98, 1501–1525. [DOI] [PubMed] [Google Scholar]
- Bunton-Stasyshyn RKA, et al. , 2019. Prominent role of forebrain excitatory neurons in SCN8A encephalopathy. Brain 142, 362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvill GL, et al. , 2014. GABRA1 and STXBP1: novel genetic causes of Dravet syndrome. Neurology 82, 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, et al. , 2020. Stxbp1/Munc18–1 haploinsufficiency impairs inhibition and mediates key neurological features of STXBP1 encephalopathy. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante G, et al. , 2020. dCas9-based Scn1a gene activation restores inhibitory interneuron excitability and attenuates seizures in Dravet syndrome mice. Mol. Ther 28, 235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, et al. , 2003. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 17, 2591–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S, et al. , 2019. G protein-coupled potassium channels implicated in mouse and cellular models of GNB1 Encephalopathy. bioRxiv 697235. [Google Scholar]
- Cremona O, et al. , 1999. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell 99, 179–188. [DOI] [PubMed] [Google Scholar]
- DeLorey TM, et al. , 1998. Mice lacking the beta3 subunit of the GABAA receptor have the epilepsy phenotype and many of the behavioral characteristics of Angelman syndrome. J. Neurosci 18, 8505–8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lullo E, Kriegstein AR, 2017. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci 18, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickie MM, 1965. Jolting. Mouse News Lett. 32, 44. [Google Scholar]
- Dong H-W, et al. , 2020. Detection of neurophysiological features in female R255X MeCP2 mutation mice. Neurobiol. Dis 145, 105083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugger SA, et al. , 2020. Neurodevelopmental deficits and cell-type-specific transcriptomic perturbations in a mouse model of HNRNPU haploinsufficiency. bioRxiv. 10.1101/2020.05.01.072512(2020.05.01.072512). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epi4K C, et al. , 2013. De novo mutations in epileptic encephalopathies. Nature 501, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epi4K C, 2016. De novo mutations in SLC1A2 and CACNA1A are important causes of epileptic encephalopathies. Am. J. Hum. Genet 99, 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JC, et al. , 2005. Early onset seizures and Rett-like features associated with mutations in CDKL5. Eur. J. Human Genet 13, 1113–1120. [DOI] [PubMed] [Google Scholar]
- Feng H, et al. , 2019. Mouse models of GNAO1-associated movement disorder: allele-and sex-specific differences in phenotypes. PLoS One 14, e0211066. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fletcher CF, et al. , 1996. Absence epilepsy in tottering mutant mice is associated with calcium channel defects. Cell 87, 607–617. [DOI] [PubMed] [Google Scholar]
- Frankel WN, et al. , 2001. Electroconvulsive thresholds of inbred mouse strains. Genomics 74, 306–312. [DOI] [PubMed] [Google Scholar]
- Frankel WN, et al. , 2009. Szt2, a novel gene for seizure threshold in mice. Genes Brain Behav. 8, 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandaglia A, et al. , 2019. A novel Mecp2Y120D knock-in model displays similar behavioral traits but distinct molecular features compared to the Mecp2-null mouse implying precision medicine for the treatment of Rett syndrome. Mol. Neurobiol 56, 4838–4854. [DOI] [PubMed] [Google Scholar]
- Gao Y, et al. , 2020a. Gene replacement ameliorates deficits in mouse and human models of cyclin-dependent kinase-like 5 disorder. Brain 143, 811–832. [DOI] [PubMed] [Google Scholar]
- Gao Y, et al. , 2020b. Gene replacement ameliorates deficits in mouse and human models of cyclin-dependent kinase-like 5 disorder. Brain J. Neurol 143, 811–832. [DOI] [PubMed] [Google Scholar]
- Gatto CL, Broadie K, 2011. Drosophila modeling of heritable neurodevelopmental disorders. Curr. Opin. Neurobiol 21, 834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, et al. , 2011. Intellectual disability without epilepsy associated with STXBP1 disruption. Eur. J. Human Genet 19, 607–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins NA, et al. , 2011. Neuronal voltage-gated ion channels are genetic modifiers of generalized epilepsy with febrile seizures plus. Neurobiol. Dis 41, 655–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins NA, et al. , 2016. Fine mapping of a Dravet syndrome modifier locus on mouse chromosome 5 and candidate gene analysis by RNA-Seq. PLoS Genet. 12, e1006398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron SE, et al. , 2012. Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat. Genet 44, 1188–1190. [DOI] [PubMed] [Google Scholar]
- Inglis GAS, et al. , 2020. Mutations in the Scn8a DIIS4 voltage sensor reveal new distinctions among hypomorphic and null Nav 1.6 sodium channels. Genes Brain Behav. 19, e12612. [DOI] [PubMed] [Google Scholar]
- Ishimura R, et al. , 2014. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov SV, et al. , 2004. Cerebellar ataxia, seizures, premature death, and cardiac abnormalities in mice with targeted disruption of the Cacna2d2 gene. Am. J. Pathol 165, 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, et al. , 1998. Multiple neurological abnormalities in mice deficient in the G protein Go. Proc. Natl. Acad. Sci. U. S. A 95, 3269–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Meisler MH, 2014. Modeling human epilepsy by TALEN targeting of mouse sodium channel Scn8a. Genesis 52, 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun K, et al. , 1999. Ablation of P/Q-type Ca(2+) channel currents, altered synaptic transmission, and progressive ataxia in mice lacking the alpha(1A)-subunit. Proc. Natl. Acad. Sci. U. S. A 96, 15245–15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SK, et al. , 2019. C57BL/6J and C57BL/6N substrains differentially influence phenotype severity in the Scn1a (+/− ) mouse model of Dravet syndrome. Epilepsia Open 4, 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur M, et al. , 2020. Expression of the neuronal tRNA n-Tr20 regulates synaptic transmission and seizure susceptibility. Neuron. 10.1016/j.neuron.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, et al. , 1997. Phospholipase C isozymes selectively couple to specific neurotransmitter receptors. Nature 389, 290–293. [DOI] [PubMed] [Google Scholar]
- Kitamura K, et al. , 2009. Three human ARX mutations cause the lissencephaly-like and mental retardation with epilepsy-like pleiotropic phenotypes in mice. Hum. Mol. Genet 18, 3708–3724. [DOI] [PubMed] [Google Scholar]
- Kruger LC, et al. , 2016. β1-C121W is down but not out: epilepsy-associated Scn1b-C121W results in a deleterious gain-of-function. J. Neurosci 36, 6213–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo FS, et al. , 2019. Disordered breathing in a mouse model of Dravet syndrome. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane PW, 1976. Scn8a - motor end-plate disease-Jackson. Mouse News Lett. 54, 40. [Google Scholar]
- Lenk GM, et al. , 2020. Scn8a antisense oligonucleotide is protective in mouse models of SCN8A encephalopathy and Dravet syndrome. Ann. Neurol 87, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts VA, et al. , 2014. Hidden in plain sight: spike-wave discharges in mouse inbred strains. Genes Brain Behav. 13, 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher W, et al. , 2017. The relevance of inter- and intrastrain differences in mice and rats and their implications for models of seizures and epilepsy. Epilepsy Behav. 73, 214–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari A, Noebels JL, 2014. Monogenic models of absence epilepsy: windows into the complex balance between inhibition and excitation in thalamocortical microcircuits. Prog. Brain Res 213, 223–252. [DOI] [PubMed] [Google Scholar]
- Makinson CD, et al. , 2017. Regulation of thalamic and cortical network synchrony by Scn8a. Neuron 93, 1165–1179 (e6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark MD, et al. , 2011. Delayed postnatal loss of P/Q-type calcium channels recapitulates the absence epilepsy, dyskinesia, and ataxia phenotypes of genomic Cacna1a mutations. J. Neurosci 31, 4311–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MS, et al. , 2007. The voltage-gated sodium channel Scn8a is a genetic modifier of severe myoclonic epilepsy of infancy. Hum. Mol. Genet 16, 2892–2899. [DOI] [PubMed] [Google Scholar]
- Martin MS, et al. , 2010. Altered function of the SCN1A voltage-gated sodium channel leads to gamma-aminobutyric acid-ergic (GABAergic) interneuron abnormalities. J. Biol. Chem 285, 9823–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisler MH, 2019. SCN8A encephalopathy: mechanisms and models. Epilepsia 60 (Suppl. 3), S86–S91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisler MH, et al. , 2001. Identification of epilepsy genes in human and mouse. Annu. Rev. Genet 35, 567–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, et al. , 2008. Two novel alleles of tottering with distinct Ca(v)2.1 calcium channel neuropathologies. Neuroscience 155, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto H, et al. , 2017. Potentiation of excitatory synaptic transmission ameliorates aggression in mice with Stxbp1 haploinsufficiency. Hum. Mol. Genet 26, 4961–4974. [DOI] [PubMed] [Google Scholar]
- Montanara PL, et al. , 2020. Cyclin-dependent–like kinase 5 is required for pain signaling in human sensory neurons and mouse models. Sci. Transl. Med 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahey PJ, et al. , 2020. Aged heterozygous Cdkl5 mutant mice exhibit spontaneous epileptic spasms. Exp. Neurol 332, 113388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noebels JL, 1995. Single locus mutations in mice expressing generalized spike-wave absence epilepsies. Ital. J. Neurol. Sci 16, 107–111. [DOI] [PubMed] [Google Scholar]
- Noebels JL, Sidman RL, 1979. Inherited epilepsy: spike-wave and focal motor seizures in the mutant mouse tottering. Science 204, 1334–1336. [DOI] [PubMed] [Google Scholar]
- Ogiwara I, et al. , 2018. Nav1.2 haplodeficiency in excitatory neurons causes absence-like seizures in mice. Commun. Biol 1, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba C, et al. , 2014. Early onset epileptic encephalopathy caused by de novo SCN8A mutations. Epilepsia 55, 994–1000. [DOI] [PubMed] [Google Scholar]
- Ohba C, et al. , 2015. De novo KCNT1 mutations in early-onset epileptic encephalopathy. Epilepsia 56, e121–e128. [DOI] [PubMed] [Google Scholar]
- Okuda K, et al. , 2017. CDKL5 controls postsynaptic localization of GluN2B-containing NMDA receptors in the hippocampus and regulates seizure susceptibility. Neurobiol. Dis 106, 158–170. [DOI] [PubMed] [Google Scholar]
- Papale LA, et al. , 2009. Heterozygous mutations of the voltage-gated sodium channel SCN8A are associated with spike-wave discharges and absence epilepsy in mice. Hum. Mol. Genet 18, 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou A, et al. , 2016. GABRB3 mutations: a new and emerging cause of early infantile epileptic encephalopathy. Dev. Med. Child Neurol 58, 416–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederick DT, et al. , 2018. Abnormal cell sorting underlies the unique X-linked inheritance of PCDH19 epilepsy. Neuron 97, 59–66 (e5). [DOI] [PubMed] [Google Scholar]
- Qu S, et al. , 2020. GABA(A) receptor β3 subunit mutation D120N causes Lennox-Gastaut syndrome in knock-in mice. Brain Commun. 2, fcaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quraishi IH, et al. , 2020. Impaired motor skill learning and altered seizure susceptibility in mice with loss or gain of function of the Kcnt1 gene encoding Slack (K(Na)1.1) Na(+)-activated K(+) channels. Sci. Rep 10, 3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim K, et al. , 2001. Complexins regulate a late step in Ca2+− dependent neurotransmitter release. Cell 104, 71–81. [DOI] [PubMed] [Google Scholar]
- Rogers EJ, et al. , 2019. An IQSEC2 mutation associated with intellectual disability and autism results in decreased surface AMPA receptors. Front. Mol. Neurosci 12, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose SJ, et al. , 2014. The first knockin mouse model of episodic ataxia type 2. Exp. Neurol 261, 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah M, et al. , 2020. Altered excitatory transmission onto hippocampal interneurons in the IQSEC2 mouse model of X-linked neurodevelopmental disease. Neurobiol. Dis 137, 104758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitsu H, et al. , 2008. De novo mutations in the gene encoding STXBP1 (MUNC18–1) cause early infantile epileptic encephalopathy. Nat. Genet 40, 782–788. [DOI] [PubMed] [Google Scholar]
- Sakai C, et al. , 2018. Zebrafish models of neurodevelopmental disorders: past, present, and future. Front. Mol. Neurosci 11, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong E, et al. , 2005. Genetic analysis of the neuronal and ubiquitous AP-3 adaptor complexes reveals divergent functions in brain. Mol. Biol. Cell 16, 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian M, et al. , 2002. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron 35, 243–254. [DOI] [PubMed] [Google Scholar]
- Shore AN, et al. , 2020. Reduced GABAergic neuron excitability, altered synaptic connectivity, and seizures in a KCNT1 gain-of-function mouse model of childhood epilepsy. Cell Rep. 33, 108303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silayeva L, et al. , 2015. KCC2 activity is critical in limiting the onset and severity of status epilepticus. Proc. Natl. Acad. Sci. U. S. A 112, 3523–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, et al. , 2013. Aberrant sodium channel activity in the complex seizure disorder of Celf4 mutant mice. J. Physiol 591, 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TJ, Zourray C, Schorge S, Lignani G, 2020. Aberrant sodium channel activity in the complex seizure disorder of Celf4 mutant mice. J. Neurochem 10.1111/jnc.15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan HO, et al. , 2007. Reduced cortical inhibition in a mouse model of familial childhood absence epilepsy. Proc. Natl. Acad. Sci. U. S. A 104, 17536–17541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, et al. , 2019. Altered NMDAR signaling underlies autistic-like features in mouse models of CDKL5 deficiency disorder. Nat. Commun 10, 2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tona R, et al. , 2019. The phenotypic landscape of a Tbc1d24 mutant mouse includes convulsive seizures resembling human early infantile epileptic encephalopathy. Hum. Mol. Genet 28, 1530–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M-S, et al. , 2015. Functional and structural deficits of the dentate gyrus network coincide with emerging spontaneous seizures in an Scn1a mutant Dravet Syndrome model during development. Neurobiol. Dis 77, 35–48. [DOI] [PubMed] [Google Scholar]
- Uemura M, et al. , 2007. OL-Protocadherin is essential for growth of striatal axons and thalamocortical projections. Nat. Neurosci 10, 1151–1159. [DOI] [PubMed] [Google Scholar]
- Veeramah KR, et al. , 2012. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am. J. Hum. Genet 90, 502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicini S, et al. , 2001. GABA(A) receptor alpha1 subunit deletion prevents developmental changes of inhibitory synaptic currents in cerebellar neurons. J. Neurosci 21, 3009–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vien TN, et al. , 2015. Compromising the phosphodependent regulation of the GABAAR β3 subunit reproduces the core phenotypes of autism spectrum disorders. Proc. Natl. Acad. Sci. U. S. A 112, 14805–14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, et al. , 2015. Convulsive seizures and SUDEP in a mouse model of SCN8A epileptic encephalopathy. Hum. Mol. Genet 24, 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HS, et al. , 1998. KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science (New York, N.Y.) 282, 1890–1893. [DOI] [PubMed] [Google Scholar]
- Wang IT, et al. , 2012. Loss of CDKL5 disrupts kinome profile and event-related potentials leading to autistic-like phenotypes in mice. Proc. Natl. Acad. Sci. U. S. A 109, 21516–21521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer VC, et al. , 2010. Axon initial segment dysfunction in a mouse model of genetic epilepsy with febrile seizures plus. J. Clin. Invest 120, 2661–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo N-S, et al. , 2002. Hyperexcitability and epilepsy associated with disruption of the mouse neuronal-specific K–Cl cotransporter gene. Hippocampus 12, 258–268. [DOI] [PubMed] [Google Scholar]
- Yamagata T, et al. , 2020. CRISPR/dCas9-based Scn1a gene activation in inhibitory neurons ameliorates epileptic and behavioral phenotypes of Dravet syndrome model mice. Neurobiol. Dis 141, 104954. [DOI] [PubMed] [Google Scholar]
- Ye J, et al. , 2015. hnRNP U protein is required for normal pre-mRNA splicing and postnatal heart development and function. Proc. Natl. Acad. Sci. U. S. A 112, E3020–E3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yennawar M, et al. , 2019. AMPA receptor dysregulation and therapeutic interventions in a mouse model of CDKL5 deficiency disorder. J. Neurosci 39, 4814–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller A, et al. , 2007. Mapping the contribution of β3-containing GABAA receptors to volatile and intravenous general anesthetic actions. BMC Pharmacol. 7, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, et al. , 2014. Loss of MeCP2 from forebrain excitatory neurons leads to cortical hyperexcitation and seizures. J. Neurosci 34, 2754–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]