Abstract
India, with around 15 million COVID-19 cases, recently became the second worst-hit nation by the SARS-CoV-2 pandemic. In this study, we analyzed the mutation and selection landscape of 516 unique and complete genomes of SARS-CoV-2 isolates from India in a 12-month span (from Jan to Dec 2020) to understand how the virus is evolving in this geographical region. We identified 953 genome-wide loci displaying single nucleotide polymorphism (SNP) and the Principal Component Analysis and mutation plots of the datasets indicate an increase in genetic variance with time. The 42% of the polymorphic sites display substitutions in the third nucleotide position of codons indicating that non-synonymous substitutions are more prevalent. These isolates displayed strong evidence of purifying selection in ORF1ab, spike, nucleocapsid, and membrane glycoprotein. We also find some evidence of localized positive selections ORF1ab, spike glycoprotein, and nucleocapsid. The CDSs for ORF3a, ORF8, nucleocapsid phosphoprotein, and spike glycoprotein were found to evolve at rapid rate. This study will be helpful in understanding the dynamics of rapidly evolving SARS-CoV-2.
Keywords: SARS-CoV-2, Genome, India, SNP, CDS, Codon, Genetic distance, Evolution, Selection
Graphical Abstract
1. Introduction
The outbreak of coronavirus disease (COVID-19) was reported from Wuhan, China on 31 December 2019 and by March 11, 2020, it was declared as a global pandemic. The COVID-19 is caused by a novel human pathogen which was initially named 2019-novel coronavirus (2019-nCoV) and later officially named Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by the Coronaviridae Study Group (CSG) of the International Committee on Taxonomy of Viruses (Gorbalenya et al., 2020). The complete genome of the SARS-CoV-2 was first obtained and submitted by Wang et al. (2020). Phylogenetic analysis of the submitted sequence revealed 89.1% nucleotide similarity between the SARS-CoV-2 and SARS-like coronaviruses (genus Betacoronavirus, subgenus Sarbecovirus) previously found in bats (Hu et al., 2018), indicating viral spill-over from animals to humans (Wu et al., 2020).
The genome of SARS-CoV-2 includes an orderly arrangement of 5′-untranslated region (UTR), replicase complex (ORF1ab), spike glycoprotein, ORF3a, envelope, membrane glycoprotein, ORF6, ORF7a, ORF7b, ORF8, nucleocapsid phosphoprotein, ORF10, and 3′-UTR (Jungreis et al., 2021, Wang et al., 2020). The ORF1ab refers to the two ORFs, ORF1a and ORF1b, combined via programmed -1 frameshift four codons before the end of ORF1a. ORFs 1a and 1b are broadly responsible for control of genome expression and viral replication, respectively (Jungreis et al., 2021). The SARS-CoV-2 is an enveloped virus, which belongs to the family of coronaviruses and carries ~ 30 kb positive-sense single-stranded RNA as its genetic material (Laamarti et al., 2020). SARS-CoV-2 has been estimated to origin somewhere between Oct and Dec 2019 (van Dorp et al., 2020) and has been reported to evolve relatively slowly (Singh and Yi, 2021). However, it has witnessed explosive population growth by circulation in millions of humans since its outbreak in Dec 2019. Therefore, the dynamics of its mutation and selection is an exceptionally active area of ongoing research.
According to the World Health Organization (WHO), COVID-19 has infected nearly 148 million individuals globally and claimed 3.12 million lives until April 27, 2021. The exponential spread of the SARS-CoV-2 virus across the globe has steered substantial interest in its genome structure, evolution, and mutations. The first case of COVID-19 in India was reported from the southern state Kerala on January 27, 2020 (Andrews et al., 2020) and since then the nation has witnessed ~ 17.6 million COVID-19 cases with ~ 198 thousand mortality by April 27, 2021. With such a large number of active cases and mortality, India was declared as the second worst hit nation in April 2021. This made us more interested in studying the mutation and selection in SARS-CoV-2 isolates from India.
The virus's fitness requirements, including the host’s immune response, transmission, and colonization of new host species, changes rapidly. In addition, the virus also needs to maintain its essential functions such as its ability to infect and replicate within the host cell. On account of the antagonistic interplay between quickly changing fitness requirements of the virus and maintenance of its essential functions, viruses undergo strong and diverse selective forces (Spielman et al., 2019). Considering that the genetic makeup and thereby the immune system of the host greatly varies in different geographical regions, it is conceivable that the SARS-CoV-2 would experience varying immune responses geographically and consequently virus substrains in India may undergo selection differently. This led us to our questions (i) which sites in SARS-CoV-2 isolates from India are evolving at higher rates, and (ii) which of the coding sequences (CDSs) are under selection pressure and are evolving rapidly.
In this study, we characterize the single nucleotide polymorphism in 516 complete genomes of SARS-CoV-2 sub strains isolated in India in 2020 (Jan 2020–Dec 2020). We studied the entire genomes and also the individual coding sequences (CDSs) of these complete genomes and analyzed the single nucleotide polymorphism (SNP), genetic diversity, and selection pressure on individual CDSs over one year of time. This study would help us understand the dynamics of SARS-CoV-2 mutation and selection in Indian population. This study could be insightful for understanding various aspects of the virus including its pathogenesis, which is critical in combating COVID-19.
2. Methods
2.1. Acquisition of accession number for complete genomes of SARS-CoV-2 isolated in India
The csv record file containing information on the complete and partial genome of SARS-CoV-2 was obtained from NCBI on March 6, 2021. This record consisted of information about both the complete and the partial genomes of SARS-CoV-2 from different geographical locations. The csv record file includes accession, release date, species, length, nucleotide completeness, geographical location, host, isolation source, and collection date (Supplementary File S1). The rows containing information only for complete genome sequences were extracted and the record of partial genomes was excluded (Supplementary File S2). The record of complete genomes was further subset based on rows containing the string “India” under the column ‘Geo_Location’. The record was sorted on the basis of ‘Collection_Date’ year and month of isolation and the rows with no month information were removed. A new column was added for Year and Month for time-wise analysis of SARS-CoV-2 genomes (Supplementary File S3).
2.2. Sequence retrieval and processing
The accession numbers for complete genomes of SARS-CoV-2 (collected in India) and one reference genome (Accession NC_045512) were retrieved from the record file (Supplementary File S3) and the fasta sequences were obtained from NCBI using the read.GenBank function of ape package (Paradis et al., 2004, Paradis and Schliep, 2019). The resultant fasta file (Supplementary File S4) was then converted into a dataframe and a new column of ‘Year_Month’ was added for the date of sample collection.
The dataframe was screened for the sequences containing gaps, designated by the character “N”, which were then excluded to create a clean dataframe of complete sequences with no “N”. The redundant sequences from the clean dataframe were then excluded to create a clean-unique dataframe (Supplementary File S5). The Biostrings package (Pages et al., 2013) was used for the efficient manipulation of sequences. As the name of each sequence was large, a new column, namely SN ‘for a short sequence name’ was added with the purpose of adding a small header (reflecting their order of collection) to each sequence. Prior to adding the new name, the dataframe was sorted on the basis of the date of collection so that the new name was given in an order of date of collection (Supplementary File S6). This was then converted into fasta format. The resultant fasta file (Supplementary File S7) of unique sequences was opened in BioEdit software (Hall et al., 2011, Hall, 1999) and the poly-A tail, wherever present, were trimmed to avoid any interference in multiple sequence alignment. The redundant sequences were removed, and only unique sequences were used for the analysis, unless otherwise stated.
The number of unique sequences in genomes and also in individual sequences were also used as a raw measure of variation in the corresponding sequences. Considering the fact that (i) all the different CDS sequences are of different length, and (ii) with the increase in the length it is expected to observe increased mutations, it is conceivable that CDS of large size would display larger numbers of unique sequence compared to the CDS of smaller sizes. Therefore, in order to avoid the sequence size-dependent bias in determining the number of unique sequences for each CDS, the numbers of unique sequences were normalized with their sequence length using the following expression: , where U and l represent the number of unique sequences for a CDS and the length of CDS, respectively among 516 unique SARS-CoV-2 genomes. The dataframes, wherever needed, were converted into fasta files using R script as previously described (Negi et al., 2020, Pal and Negi, 2019). The computational analysis of data was performed using R programming language (Gentleman and Ihaka, 2000, Ihaka and Gentleman, 1996) in the platform-independent IDE RStudio (Racine, 2012) unless otherwise stated.
2.3. Multiple sequence alignment
The multiple sequence alignment of the clean unique genomes of SARS-CoV-2 collected from India was performed as described previously (Negi et al., 2020, Pal and Negi, 2019). The msa package (Bodenhofer et al., 2015), which provides an interface to the multiple sequence alignment in R was used for obtaining an alignment of 517 genome sequences. The muscle algorithm (Edgar, 2004a, Edgar, 2004b) was utilized for multiple sequence alignment. The conversion of alignment into matrix and other downstream processing was performed by using the seqinr package (Charif and Lobry, 2007). The coordinates of individual CDSs of the reference genome were obtained from NCBI and used for extraction of corresponding CDSs from all the SARS-CoV-2 genomes used in this study. The multiple sequence alignment of individual CDSs were used for obtaining the distance matrices and genetic distances using ape (Paradis et al., 2004, Paradis and Schliep, 2019), phangorn (Schliep, 2011), stats (R Core Team, 2020), dplyr (Wickham et al., 2021), readr (Wickham and Hester, 2020), and ggplot2 (Ginestet, 2011) packages.
2.4. Single nucleotide polymorphism, and principal component analysis
The analysis of single nucleotide polymorphism distribution was performed using the ape, ggplot2, and adegenet package (Jombart, 2008, Jombart and Ahmed, 2011). The ‘DNAbin2genind’ function was used to convert the alignment to genind objects, which then were used as input for determining the SNP loci and alleles from entire genomes and each CDSs. The SNP density and position along the alignment were plotted using the ‘snpposi.plot’ function. The statistical analysis of the SNPs distribution was performed using the Monte Carlo simulation, which tests if the distribution of SNPs is random. The alternative hypothesis, in this case, would be that SNPs are clustered. The Monte Carlo simulation is based on the distances of each SNP to their closest SNP, providing a measure of clustering for each SNP. The nucleotide positions of the SNPs in codons (C1, C2, C3) for each CDSs were also determined and plotted. Mutations in the entire genome and the individual CDSs were determined and plotted using ‘findMutations’, graphMutations, and ‘gengraph’ functions. The tables contained in the genind objects were subjected to a principal component analysis to determine the summary of the genetic diversity among the sampled individuals.
2.5. Selection pressure and position-by position evolutionary rate analysis
We studied both the positive and negative selections in the SARS-CoV-2 isolates obtained from India in the year 2020. First, we studied the alignments to determine if any of the CDSs is/are undergoing positive/diversifying selection at any site. For this, all the alignments of different CDSs were subjected to ‘Branch-site Unrestricted Statistical Test for Episodic Diversification’ (BUSTED) model (Murrell et al., 2015), which is a gene-wide model to test for positive selection either throughout the evolutionary tree (pervasive) or only on some lineages (episodic). The ORF1ab is a polyprotein with some internal stop codons in its CDS. In the CDS for ORF1ab, there is no stop codon from nsp1 to nsp10 (coordinate 266–13441 of the reference genome NC_045512) containing 4392 codons and from RdRp to nsp16 (coordinate 13468–21552 of the reference genome NC_045512) containing 2695 codons. Therefore, two different CDSs corresponding to nsp1 to nsp10 (nsp1_10) and RdRp to nsp16 (RdRp_nsp16) with no stop codons were used separately in the analysis. In addition to the gene-wide analysis of selection pressure, we also performed site-wide analysis using the ‘Mixed Effects Model of Evolution’ (MEME) method (Murrell et al., 2012) and ‘Single-Likelihood Ancestor Counting’ (SLAC) method (Kosakovsky Pond and Frost, 2005). All the selection analyses were performed on the Datamonkey server (Pond and Frost, 2005, Weaver et al., 2018). The MEME model identifies the site(s) in a gene subjected to positive/diversifying selection, whereas the SLAC method tests both positive/diversifying and negative/purifying selection at each site by employing an integration of maximum-likelihood (ML) and counting approaches. Based on the recommendations in previous studies (Spielman et al., 2019), the P-value threshold of P ≤ 0.05 for gene-wide method BUSTED and P ≤ 0.1 for the site-level methods MEME and SLAC were used in this study.
The neutrality test, to determine if the SARS-CoV-2 sequences are evolving neutrally or non-randomly, was performed using Tajima's Neutrality Test (Tajima, 1989) in MEGA X (Kumar et al., 2018). The coding data was translated using the standard genetic code table and all the ambiguous positions were removed for each sequence pair. The position-by-position evolutionary rates of the deduced amino acid sequences of all the CDSs were determined using MEGA X (Kumar et al., 2018). The sequences were translated assuming a standard genetic code table and the substitution pattern and relative rates were estimated under the Jones-Taylor-Thornton (JTT) model (Jones et al., 1992).
3. Results
3.1. SARS-CoV-2 complete genome isolates from India
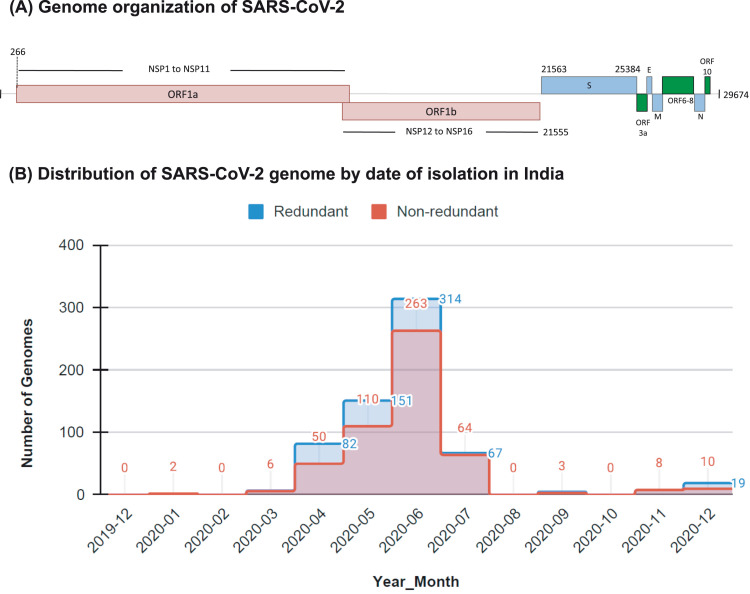
The SARS-CoV-2 csv record obtained on March 6 2021 comprised a total of 91705 accessions from different geographical regions across the world. The subsetting of data for removing partial genomes decreased the number of accession records to 53365 complete genomes of SARS-CoV-2. Further subsetting based on Geo_Location to extract complete sequence records from India resulted in only 655 accession records. The record for reference genome (Accession number NC_045512) was added and the file was converted into fasta format. Screening of gaps in the data of 656 sequences identified 75 sequences with 'N', which upon removal from the dataset resulted in 581 sequences. The redundant sequences were removed for further analysis resulting in only 516 unique SARS-CoV-2 genomes. The distribution of 655 redundant and 516 non-redundant SARS-CoV-2 genomes by date of sample collection in India shows a large number of complete genomes from April 2020 to July 2020 in the NCBI database ( Fig. 1).
Fig. 1.
Time-wise distribution of the SARS-CoV-2 genomes isolated in India. The numbers of complete genomes of SARS-CoV-2 were plotted against the time of their collection over a period of 12 months from January 2020 to December 2020. The distributions of 516 unique, non-redundant genomes and 655 redundant genomes are shown in red and blue bars, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Number of unique sequences corresponding to the sequence length
With reference to the 29868 nucleotides long reference genome (Accession NC_045512), a total of 516 unique SARS-CoV-2 genomes were identified from India within a 12-month span (Jan. 2020–Dec. 2020). This is a raw indication of rapid ongoing mutation in the viral genome. Besides the entire genome, the individual coding sequences (CDS) were also analyzed for the number of unique sequences for each CDS among 516 unique genomes.
The normalized numbers of unique sequences for the entire genome and for the longest ORF (ORF1ab) were nearly similar and were found to be 17.30 and 18.97 respectively. The normalized numbers of unique sequences for all the other CDS were notably higher than that of the entire genome/ORF1ab ranging from 26.31 to 70.30. The maximum numbers of normalized unique sequences (70.30) were displayed by ORF3a. This was followed by ORF8, nucleocapsid phosphoprotein, ORF6, ORF7a, membrane glycoprotein, spike glycoprotein, ORF7b, envelope, and ORF10 with normalized unique sequences 60.60, 58.87, 43.71, 41.32, 39.03, 37.18, 31.00, 26.66, and 26.31, respectively ( Fig. 2, and Supplementary Materials S8) indicating the number of unique sequences as a raw measure of mutation in the viral genome.
Fig. 2.
Unique sequences and sequence length for each CDS in SARS-CoV-2 genomes isolated in India. The numbers of unique sequences for each CDS were normalized based on the length of the respective CDS to avoid the sequence size-dependent bias. The sequence length does not include the last three nucleotides of the stop codon. The CDS in the y-axis are arranged after the entire genome (ent_genome) in an order of their increasing size where ORF1ab is the longest CDS and ORF10 is the smallest CDS.
3.3. ORF3a, ORF8, and nucleocapsid phosphoprotein CDSs exhibit higher percentages of polymorphic sites compared to that in entire genome sequences
Multiple sequence alignment of 517 sequences was performed using the muscle algorithm of msa package and the resultant alignment was converted to fasta format using in-house R script. The fasta file of the muscle alignment of 517 unique genomes was analyzed for polymorphism using the adegenet package (Jombart, 2008, Jombart and Ahmed, 2011). In the genome-wide analysis of SNPs, 3.19% sites of the entire genome i.e., 953 loci displayed SNP, of which 936 loci comprised 2 alleles, whereas 17 loci were identified with three alleles making altogether 1923 alleles in the entire genome ( Fig. 3, Table 1).
Fig. 3.
SNP in the entire genome and the corresponding CDS of 517 SARS-CoV-2 genomes isolated in India. The numbers of sites (y-axis) were plotted against the number of alleles (x-axis).
Table 1.
SNPs in 517 complete genomes collected in India.
| Genome/CDS | Coordinates |
SNP |
Number of loci with |
|||||
|---|---|---|---|---|---|---|---|---|
| Start | End | Length | Number of Loci | % loci (relative to sequence length) | Total Alleles | 2 alleles | 3 alleles | |
| ent_genome | 1 | 29871 | 29871 | 953 | 3.190 | 1923 | 936 | 17 |
| Envelope | 26245 | 26472 | 228 | 5 | 2.193 | 10 | 5 | 0 |
| membrane glycoprotein | 26523 | 27191 | 669 | 21 | 3.139 | 43 | 20 | 1 |
| nucleocapsid phosphoprotein | 28274 | 29533 | 1260 | 77 | 6.111 | 159 | 72 | 5 |
| ORF10 | 29558 | 29674 | 117 | 2 | 1.709 | 4 | 2 | 0 |
| ORF1ab | 266 | 21555 | 21290 | 583 | 2.738 | 1171 | 578 | 5 |
| ORF3a | 25393 | 26220 | 828 | 55 | 6.643 | 111 | 54 | 1 |
| ORF6 | 27202 | 27387 | 186 | 7 | 3.763 | 14 | 7 | 0 |
| ORF7a | 27394 | 27759 | 366 | 13 | 3.552 | 27 | 12 | 1 |
| ORF7b | 27756 | 27887 | 132 | 2 | 1.515 | 4 | 2 | 0 |
| ORF8 | 27894 | 28259 | 366 | 23 | 6.284 | 46 | 23 | 0 |
| spike glycoprotein | 21563 | 25384 | 3822 | 126 | 3.297 | 254 | 124 | 2 |
| Overall CDS | NA | 29264 | 914 | 3.123 | 1843 | 899 | 15 | |
| Overall non-coding regions | NA | 607 | 39 | 6.425 | 80 | 37 | 2 | |
After analyzing genome-wide SNP, we studied 11 coding regions of the genome including envelope, membrane glycoprotein, nucleocapsid phosphoprotein, ORF1ab, ORF10, ORF3a, ORF6, ORF7a, ORF7b, ORF8, and spike glycoprotein. The CDSs for ORF1ab, spike glycoprotein, nucleocapsid phosphoprotein, and ORF3a displayed more than 50 polymorphic sites with 583, 126, 77, and 55 polymorphic loci (Table 1). However, all the CDSs are of varying length and it is plausible that with the increase in length of CDS, the degree of polymorphism will also increase. Therefore, we analyzed the percentage of polymorphic loci relative to the length of the respective CDS.
Three of the CDSs including, ORF3a (828 nt), ORF8 (366 nt), and nucleocapsid phosphoprotein (1260 nt) displayed 6.64%, 6.28%, and 6.11% polymorphic sites, respectively. The ORF3a displayed 72 loci with 2 alleles and 5 loci with 3 alleles, whereas 77 polymorphic loci of nucleocapsid phosphoprotein consisted of 72 loci with 2 alleles and 5 loci with 3 alleles. All the 23 polymorphic sites of ORF8 displayed 2 alleles per loci. The percentage of polymorphic sites in these three CDSs are ~ 2-fold compared to that in the entire genome. This suggests that ORF3a, ORF8, and nucleocapsid phosphoprotein display a higher rate of mutation compared to that of the entire genome or other CDSs. The CDSs corresponding to ORF6 (186 nt), ORF7a (366 nt), and spike (3822) displayed 3.76%, 3.55%, and 3.29% of their total nucleotides as polymorphic sites. The ORF6 CDS comprised 7 polymorphic sites with 2 alleles per site; the ORF7a exhibited 13 polymorphic sites including 12 loci with 2 alleles per site and 1 locus with 3 alleles. The CDS for spike comprised 126 polymorphic sites including 124 loci with 2 alleles each and 2 loci with 3 alleles per site. The CDS for membrane glycoprotein (669 nt) displayed 3.13% (21 loci) of their total nucleotides as polymorphic sites including 20 loci and 1 locus with 2 and 3 alleles per site, respectively. The CDS for ORF1ab exhibited 2.73% of its sites as the polymorphic sites, which are represented by 583 loci including 578 with 2 alleles per site and 5 with 3 alleles per site. The percentage of polymorphic sites in these CDSs (ORF6, ORF7a, spike glycoprotein, membrane glycoprotein, and ORF1ab) are slightly higher/lower but nearly equal to that in the entire genome indicating that the mutation rate in these CDSs is similar to that of the entire genome.
The percent polymorphic sites in CDSs corresponding to envelope (228 nt), ORF10 (117 nt), and ORF7b (132 nt) were found to be 2.19%, 1.70%, and 1.51%. The envelope, ORF10, and ORF7b displayed 5, 2, and 2 polymorphic sites, respectively. All these polymorphic sites exhibit 2 alleles per site. The notably lower percent polymorphic sites in envelope, ORF10, and ORF7b compared to that of the entire genome suggest that these CDSs are relatively more stable than other CDSs of the SARS-CoV-2 genome (Fig. 3 and Table 1). All the CDSs altogether (29264 nt) consisted of 3.12% polymorphic sites relative to its overall length. In contrast, the non-coding regions of the genome exhibited 6.42% polymorphic sites (relative to its length), which is twice the percentage of polymorphic sites of CDS (Table 1).
3.4. Distribution of single nucleotide polymorphism (SNP) across the genome
The densities of polymorphic sites of genomes or individual CDSs were plotted against their nucleotide position to visualize the distribution of SNPs across the length of the genome ( Fig. 4). The genome-wide SNP distribution appeared random in the graph. The Monte Carlo simulation was performed to analyze whether SNPs are clustered or randomly distributed in the genome. Based on 999 replicates with a simulated p-value of 0.03, the values for standard observation, expectation value, and variance were found to be − 2.5537026, 10.5835836, and 0.3845399, respectively, indicating that the alternative hypothesis is not true and the distribution of SNPs in the 517 complete genomes of SARS-CoV-2 genome is random. After analyzing genome-wide SNP distribution, we analyzed the SNP distribution of 11 coding regions of the genome including envelope, membrane glycoprotein, nucleocapsid phosphoprotein, ORF1ab, ORF3a, ORF6, ORF7a, ORF7b, ORF8, ORF10, and spike glycoprotein (Fig. 4). The CDS for envelope exhibited SNP at 5 loci, 4 of which were clustered together from 136th to 184th nucleotide position, while one SNP was found at 12th nucleotide (Fig. 4). Similarly, the SNP loci in membrane glycoprotein, ORF6, ORF7a, and ORF7b also displayed SNP loci in small clusters (Fig. 4).
Fig. 4.
Distribution of polymorphic sites on 517 SARS-CoV-2 genome and their corresponding CDSs. The nucleotide positions are shown in x-axis and the density of polymorphism is represented at y-axis. The three different codon positions of the polymorphic loci in the CDS are shown in three different colors. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. Majority of SNP loci are at codon position 3
The SNP loci of various CDS were also analyzed for the presence of SNP at nucleotide positions C1, C2 and C3 in the codons. The CDSs for envelope, ORF7a, and ORF8 displayed the majority of SNP loci at codon position C1 with 80%, 46%, and 48% of their respective total SNP loci, respectively ( Fig. 5a and Table 2). The CDSs corresponding to ORF6, membrane glycoprotein, spike glycoprotein, ORF1ab, and nucleocapsid phosphoprotein displayed the majority of SNPs loci at codon position C3. The percent abundance of C3 SNP loci in ORF6, membrane glycoprotein, spike glycoprotein, ORF1ab, and nucleocapsid phosphoprotein were found to be 71%, 67%, 45%, 43%, and 38% of their respective total SNP loci, respectively (Fig. 5a). The ORF3a displayed a nearly similar abundance of SNP loci at codon positions C1, C2, and C3 with 35% C1, 31% C2, and 35% C3. The C2 codon position was not found to be the most abundant SNP loci position in any of the CDSs. However, it was found to be the second most abundant codon position for the SNP loci in the CDS of ORF1ab, ORF8, and spike glycoprotein.
Fig. 5.
Codon positions of SNP loci. (a) The codon positions of SNP loci for different CDS in 517 non-redundant complete genomes (isolated in India) were identified and their percentage relative to the total SNP loci was plotted as a stacked bar chart. The CDS with less than 5 SNP loci were not included in the analysis. (b) The codon positions of SNP loci from the CDS region of the entire genome were determined and their % abundance relative to the number of SNP loci was plotted as a doughnut chart.
Table 2.
Codon positions of CDS expressing SNP.
| CDS | SNP |
||||||
|---|---|---|---|---|---|---|---|
| Total loci | C1 | C2 | C3 | %C1 | %C2 | %C3 | |
| Envelope | 5 | 4 | 0 | 1 | 80 | 0 | 20 |
| membrane glycoprotein | 21 | 4 | 3 | 14 | 19 | 14 | 67 |
| nucleocapsid phosphoprotein | 77 | 26 | 22 | 29 | 34 | 29 | 38 |
| ORF1ab | 583 | 135 | 199 | 249 | 23 | 34 | 43 |
| ORF3a | 55 | 19 | 17 | 19 | 35 | 31 | 35 |
| ORF6 | 7 | 2 | 0 | 5 | 29 | 0 | 71 |
| ORF7a | 13 | 6 | 3 | 4 | 46 | 23 | 31 |
| ORF7b | 2 | 1 | 1 | 0 | 50 | 50 | 0 |
| ORF8 | 23 | 11 | 9 | 3 | 48 | 39 | 13 |
| ORF10 | 2 | 1 | 0 | 1 | 50 | 0 | 50 |
| spike glycoprotein | 126 | 28 | 41 | 57 | 22 | 33 | 45 |
| TOTAL | 914 | 237 | 295 | 382 | 26 | 32 | 42 |
Interestingly, 42% of the total SNP loci in the CDS of the entire genome were found at codon position C3, whereas 26% and 32% of total SNP loci were identified at codon position C1 and C2, respectively (Fig. 5b). Unlike C1, the C2 was not found as the most abundant SNP loci codon position in any of the CDSs. However, the C2 represents the second most abundant codon position for SNP loci with 32% of the total SNP loci in the CDS.
3.6. Principal component analysis of SNP data suggests an increase in genetic diversity with time
To discover any potential pattern in SNP data of 517 non-redundant complete SARS-CoV-2 genomes, the genind object of the entire genomes was subjected to the principal component analysis (PCA). The missing data (NAs) in the genind object were replaced by the mean allele frequency. The eigenvalues, which indicate the amount of variance represented by each principal component, were also calculated and a scatter plot was generated. Although the SNP data of the majority of genomes were found to cluster together with that of the reference genome, the SNP data of some of the genomes represented by 'F' or 'I' (followed by some numbers) were found to have separate clusters ( Fig. 6). Interestingly, the genomes with ID starting with “F” or “I” represent genomes of SARS-CoV-2 isolated in July 2020 and afterward suggesting that the genetic diversity in the virus population is increasing with time.
Fig. 6.
Principal component analysis (PCA) of the SNP data extracted from entire genomes isolated from 517 non-redundant complete SARS-CoV-2 genomes.
3.7. The genetic distance of genomes and their CDS revealed time-dependent variation
Multiple sequence alignment of 517 sequences was performed using muscle algorithm in msa package and the resultant alignment was converted to fasta format using in-house R script. The distance matrix for individual CDSs were obtained using their respective alignment file with ape and phangorn packages. The TN93 nucleotide substitution model (Tamura and Nei, 1993) was selected for creating a distance matrix. The TN93 model accounts for the difference between transitions and transversions and differentiates the two kinds of transitions i.e. purine to purine and pyrimidine to pyrimidine. The genetic distance plotted against time for entire genomes as well as individual coding regions demonstrates a gradual increase in the genetic distance with time ( Fig. 7).
Fig. 7.
Genetic distance of the SARS-CoV-2 genomes (isolated in India) and their individual coding regions. The genetic distance of all the 517 complete genomes (ent_genome) or individual coding sequences (envelope, membrane glycoprotein, nucleocapsid phosphoprotein, ORF10, ORF1ab, ORF3a ORF6, ORF7a, ORF7b, ORF8, spike glycoprotein) from 517 genomes were plotted against their time of collection.
The visualization of the patterns of genetic distances among 517 complete genomes and among the various CDSs was also obtained from the multiple sequence alignment using the adegenet package. The mutation data, which includes details of mutations from one sequence to another, for the entire genome and also for the CDSs were obtained using the findMutations function. The genegraph function was then used for constructing graphs based on genetic distances ( Fig. 8). In these graphs the genetically close individuals are connected and clustered near each other and vice versa. These graphs indicate that the viral isolates, which were obtained at the later stage of the 12-month time duration, appeared distantly and as separate cluster(s) for the entire genome and for the largest CDS, ORF1ab. Interestingly a similar pattern was observed in CDSs corresponding to nucleocapsid phosphoprotein and spike glycoprotein indicating that among all the CDSs, spike glycoprotein and nucleocapsid phosphoprotein display high divergence from reference sequence isolated in December 2019.
Fig. 8.
Mutation plot in 517 non-redundant complete genomes isolated in India. The genetically close individuals are connected and clustered near each other.
3.8. Tajima’s D indicates abundance of non-random evolution in all the CDSs of SARS-CoV-2
Tajima neutrality test was performed on the SARS-CoV-2 CDSs to determine if the virus is evolving randomly or through selection pressure. Tajima’s test statistics, D, represent the difference between the two measures of genetic diversity- the mean number of pairwise differences and the number of segregating sites. In the neutrally evolving populations, these two measures of genetic diversity are expected to be the same and therefore, the expected value for D is zero. The positive value of D indicates lack of rare alleles, which signifies the balancing selection and decrease in population size, while the negative value of D is obtained due to the abundance of rare alleles relative to expectation, which manifests population size expansion due to selective sweep. Interestingly, the Tajima’s D for all the CDSs were found to be negative ( Table 3), indicating that SARS-CoV-2 viral population in India is evolving under selective sweep in response to host immunity ( Table 4).
Table 3.
Results from Tajima's Neutrality Test. Evolutionary analyses were conducted in MEGA X.
| CDSs | m | N | S | Ps | Ө | π | D |
|---|---|---|---|---|---|---|---|
| nsp1_10 | 301 | 4391 | 186 | 0.042359371 | 0.006742263 | 0.000537337 | - 2.822767096 |
| RdRp_nsp16 | 214 | 2694 | 127 | 0.047141797 | 0.007935189 | 0.000827848 | - 2.783749436 |
| spike glycoprotein | 142 | 1273 | 80 | 0.062843676 | 0.011365128 | 0.001933301 | - 2.618418883 |
| nucleocapsid phosphoprotein | 74 | 419 | 50 | 0.119331742 | 0.024480773 | 0.006228578 | - 2.449889264 |
| membrane glycoprotein | 26 | 222 | 7 | 0.031531532 | 0.008263071 | 0.003686764 | - 1.695285254 |
| envelope | 6 | 75 | 4 | 0.053333333 | 0.023357664 | 0.017777778 | - 1.295030853 |
| ORF3a | 58 | 273 | 39 | 0.142857143 | 0.030861252 | 0.006973668 | - 2.577232862 |
| ORF6 | 8 | 61 | 3 | 0.049180328 | 0.01896762 | 0.012295082 | - 1.447514187 |
| ORF7a | 15 | 119 | 7 | 0.058823529 | 0.018090851 | 0.007843137 | - 2.039958116 |
| ORF7b | 4 | 43 | 2 | 0.046511628 | 0.025369979 | 0.023255814 | - 0.709896168 |
| OTF8 | 22 | 117 | 15 | 0.128205128 | 0.035169414 | 0.011655012 | - 2.41070917 |
| ORF10 | 3 | 38 | 1 | 0.026315789 | 0.01754386 | 0.01754386 | n/c |
Abbreviations: m = number of sequences, n = total number of sites, S = Number of segregating sites, Ps = S/n, Ө = Ps /a1, π = nucleotide diversity, and D is the Tajima test statistic.
Table 4.
Sites of SARS-CoV-2 genome under positive/diversifying or negative/purifying selection.
| SARS-CoV-2 genomes |
BUSTED (gene-wide) | MEME (positive/diversifying selection) |
SLAC (positive & negative selection) |
||||
|---|---|---|---|---|---|---|---|
| CDSs | Codons | Positive/diversifying selection | Total sites | Codon positions | Total sites | Codon position (Positive selection) | Codon position (Negative selection) |
| nsp1_10 | 4392 | No | 3 | 676, 3405, 3606a | 18 | 3606a | 16, 857, 924, 1273, 1345, 1868, 1925, 2352, 2560, 2638, 2677, 2839, 3291, 3568, 3785, 4117, 4218 |
| RdRp_nsp16 | 2695 | No | 4 | 314a, 871, 1176, 1701 | 6 | 314a | 619, 942, 952, 1036, 1804, 2509 |
| spike glycoprotein | 1273 | No | 1 | 627 | 3 | NA | 206, 294, 789, 856 |
| nucleocapsid phosphoprotein | 419 | Yes | 2 | 3, 204 | 4 | NA | 110, 157, 234, 298 |
| membrane glycoprotein | 222 | No | 0 | NA | 1 | NA | 71 |
| envelope | 75 | No | 0 | NA | 0 | NA | NA |
| ORF3a | 275 | No | 0 | NA | 0 | NA | NA |
| ORF6 | 61 | No | 0 | NA | 0 | NA | NA |
| ORF7a | 121 | No | 0 | NA | 0 | NA | NA |
| ORF7b | 43 | No | 0 | NA | 0 | NA | NA |
| ORF8 | 121 | No | 0 | NA | 0 | NA | NA |
| ORF10 | 38 | No | 0 | NA | 0 | NA | NA |
Sites identified in more than one method.
Tajima test identified 186, 127, 80, 50, 7, 4, 39, 3, 7, 2, 15, and 1 segregating sites in the CDSs corresponding to nsp1_10, RdRp_nsp1, spike glycoprotein, nucleocapsid phosphoprotein, membrane glycoprotein, envelope, ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10, respectively. The Ps (ratio of the number of segregating sites and the total number of sites in each CDS) values suggest that ORF3a, ORF8, nucleocapsid phosphoprotein, and spike glycoprotein of the viral genome are undergoing rapid non-random evolution.
3.9. The ORF1ab, spike glycoprotein, and nucleocapsid phosphoprotein displayed evidence of positive/diversifying selection
We performed BUSTED, MEME, and SLAC analysis to study gene-wide and site-wide selection analysis of the SARS-CoV-2 CDSs. The BUSTED analysis with synonymous rate variation found no evidence of gene-wide episodic diversifying selection in CDSs for nsp1_10, RdRp_nsp16, spike glycoprotein, membrane glycoprotein, envelope, ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10. However, BUSTED found evidence of gene-wide episodic diversifying selection in the CDSs corresponding to nucleocapsid phosphoprotein, indicating that at least one site on at least one test branch has experienced diversifying selection pressure (Supplementary material, S9 and Table 3). Being a gene-wide model, BUSTED identifies if a gene displays positive/diversifying selection at any of its sites, however, it does not provide the statistically valid identification of specific sites undergoing positive/diversifying selection. Therefore, we further analyzed all the CDSs using the Mixed Effects Model of Evolution (MEME) model, which identifies the site(s) in a gene subjected to diversifying selection. The MEME analysis of CDSs corresponding to membrane glycoprotein, envelope, ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10 did not display episodic positive/diversifying selection in any of its sites. The MEME analysis of nsp1_10, RdRp_nsp16, spike glycoprotein, and nucleocapsid phosphoprotein identified 3, 4, 1, and 2 sites, respectively under episodic diversifying selection at p < = 0.1 (Table 3). The three sites of nsp1_10 include codon positions 676, 3405, and 3606, whereas the four sites of RdRp_nsp16 consist of codon positions 314, 871, 1176, 1701. The spike glycoprotein displayed diversifying selection at codon position 627, while nucleocapsid phosphoprotein CDS exhibited positive diversifying selection at codon positions 3 and 204 (Supplementary material, S10 and S11).
3.10. The ORF1ab, spike glycoprotein, nucleocapsid phosphoprotein, and membrane glycoprotein exhibited negative/purifying selection
After a gene-wide test for positive selection using BUSTED followed by site-wide positive/diversifying selection using the MEME, we studied the site-level positive/diversifying or negative/purifying selection using the SLAC method. The CDSs corresponding to envelope, ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10 displayed neither positive nor negative selection in any of their residues. The CDS for nsp1_10 and RdRp_nsp16 of ORF1ab displayed positive/diversifying selection at codon positions 3606 (in nsp1_10), and 314 (in RdRp_nsp16), respectively. Apart from these two sites of ORF1ab, none of the sites in any of the CDSs exhibited positive/diversifying selection in the SLAC method.
The nsp1_10, RdRp_nsp16, spike glycoprotein, membrane protein, and nucleocapsid phosphoprotein displayed negative/purifying selection at 17, 6, 4, 1, and 4 sites, respectively (Table 3). In nsp1_10 CDS of ORF1ab polyprotein, codon positions 16 (in nsp1), 857 (in nsp3), 924 (in nsp3), 1273 (in nsp3), 1345 (in nsp3), 1868 (in nsp3), 1925 (in nsp3), 2352 (in nsp3), 2560 (in nsp3), 2638 (in nsp3), 2677 (in nsp3), 2839 (in nsp4), 3291 (in nsp5), 3568 (in nsp5), 3606 (in nsp6), 3785 (in nsp6), 4117 (in nsp8), and 4218 (in nsp9) displayed evidence of negative/purifying selection. The next highest number of sites displaying negative/purifying selection were observed in RdRp_nsp16 CDS of ORF1ab polyprotein. The six sites in RdRp_nsp16 including the codon positions 619 (in RdRp), 942 (in helicase), 952 (in helicase), 1036 (in helicase), 1804 (exonuclease), and 2509 (in nsp16) displayed the evidence of negative/purifying selection. The CDS for spike glycoprotein, nucleocapsid phosphoprotein and membrane glycoprotein exhibited the signs of negative/purifying selections at codon positions 206 (in spike glycoprotein), 294 (in spike glycoprotein), 789 (in spike glycoprotein), 856 (in spike glycoprotein), 110 (in nucleocapsid phosphoprotein), 157 (in nucleocapsid phosphoprotein), 234 (in nucleocapsid phosphoprotein), 298 (in nucleocapsid phosphoprotein), and 71 (in membrane glycoprotein).
3.11. Spike glycoprotein glycoprotein, nucleocapsid phosphoprotein phosphoprotein, ORF3a, and ORF8 are evolving at higher rate than the rest of the CDSs
The site-wise evolutionary rate analysis of sequence data was performed using MEGA X under the Jones-Taylor-Thornton (JTT) model. The evolutionary rates were scaled such that the average evolutionary rate across all sites is 1 implying that sites with a rate less than 1 are evolving slower than average, while sites with a rate more than 1 are evolving faster than average. For the overall CDSs of SARS-CoV-2, a total of 5% amino acid sites were observed to be evolving faster than the average (Supplementary material S12, S13, and S14). The CDSs corresponding to ORF3a, nucleocapsid phosphoprotein, ORF8, and spike glycoprotein displayed 14%, 12%, 12%, and 6% amino acid sites evolving faster than the average, while the rest of the CDSs exhibited 5% or less amino acid sites evolving faster than the average (Supplementary material S12–S16). This indicates that the ORF3a, nucleocapsid phosphoprotein, ORF8, and spike glycoprotein are evolving at a higher rate than other CDSs in SARS-CoV-2.
4. Conclusions
Within one year of the outbreak of SARS-CoV-2 in China, we identified 516 unique and complete genomes that were isolated from India. The majority of the isolates were obtained from April to July 2020. After July 2020, we observed a sharp decrease in the virus genome isolates probably because of the decrease in the COVID-19 cases in India from August to December 2020. The CDS for ORF1ab is the largest CDS of the SARS-CoV-2 genome and encodes for the polyprotein. With 21.29 kb in size, the ORF1ab covers nearly 71% of the genome; the rest of the genome is primarily covered by other CDSs corresponding to spike glycoprotein, ORF3a, envelope, membrane glycoprotein, ORF6, ORF7a, ORF7b, ORF8, nucleocapsid phosphoprotein, and ORF10. The rapid accumulation of mutations is the fundamental basis of virus evolution and variability in its genome. RNA viruses such as SARS-CoV-2 undergo rapid evolution because of high mutation rates (Jenkins et al., 2002). In addition, the exponential spread of SARS-CoV-2 in pandemic since the first outbreak in Dec 2019, provides the virus favorable conditions to evolve rapidly. In addition, the genetic makeup and the resultant immune response of the host also plays a major role in virus evolution. In this study, we screened the CDSs of 516 genomes of SARS-CoV-2 from Indian population to study the impact of the immune challenge to SARS-CoV-2 in Indian population on virus evolution.
The SNP analysis identified 953 loci displaying SNP genome-wide consisting of 914 in the CDS and 39 in the non-coding region. The 914 and 39 polymorphic sites represent 3.12% of the CDS, and 6.42% of the non-coding region, respectively. Such a high percentage of polymorphic sites in the non-coding region is conceivable considering that the mutations in non-coding regions may not compromise the pathogenicity and survival of the virus. The majority of SNP loci (42% of the total polymorphic sites) were found to be at the 3rd nucleotide position of the codons, suggesting that the majority of substitutions in the SARS-CoV-2 genome are of synonymous type due to redundancy of the codon. Our results, including the PCA of the genome dataset and analysis of genetic distances, suggest an increase in the genetic diversity in the SARS-CoV-2 isolates with time. Particularly, the viruses isolated from India in July 2020 and afterward displayed high variance. The mutation plot of individual CDSs indicates high divergence with time in ORF1ab, spike glycoprotein, and nucleocapsid phosphoprotein.
The selection process in which deleterious alleles are selectively removed is known as purifying (or negative) selection. This is a means of stabilizing selection by getting rid of detrimental genetic polymorphism. In contrast, positive selection is a process in which advantageous genetic variants are selected and increases in the population over time. Both weak deleterious selection and rare positive diversifying selection are now widely identified as important evolutionary forces (Fay et al., 2002).
The adaptive evolution of viruses including their adaptive immune escape is characterized by the positive selection in their genes. The BUSTED alternative model for the gene-wide method identified a very small proportion i.e., only 0.027% of sites in nucleocapsid phosphoprotein evolved under a very large ω of over 100 (obtained value 4001.526). Apart from nucleocapsid phosphoprotein, none of the genes of SARS-CoV-2 displayed evidence of positive/diversifying selection in BUSTED. This does not rule out the possible evidence of positive selection in other genes of SARS-CoV-2. It could be because of a lack of statistical power wherein the datasets for other genes did not contain statistically significant selection.
The site-wide study of the alignments of all the CDSs using MEME model identified one site each in nsp2 (676 of nsp1_10 CDS), nsp5 (3405 of nsp1_10 CDS), nsp6 (3606 of nsp1_10 CDS), helicase (1176 of RdRp_nsp16 CDS), exonuclease (1701 of RdRp_nsp16 CDS) and spike glycoprotein (627), and 2 sites each in RdRp (314 and 871 of RdRp_nsp16 CDS) and nucleocapsid phosphoprotein (3 and 204). The SLAC method identified only two sites under positive selection - codon position 3606 of nsp6 (in nsp1_10 CDS), and codon position 314 of RdRp (in RdRp_nsp16 CDS). Identification of a lesser number of sites under positive selection in the SLAC method is conceivable as unlike MEME, SLAC needs a larger number of substitutions to achieve significance (Spielman et al., 2019).
The SLAC method also identified statistically significant evidence of negative/purifying selection at 32 sites - 1 in nsp1, 10 in nsp3, 1 in nsp4, 2 in nsp5, 2 in nsp6, 1 in nsp8, 1 nsp9, 2 RdRp, 3 in helicase, 1 in exonuclease, 1 in nsp16, 4 in spike glycoprotein, 4 in nucleocapsid phosphoprotein, and 1 in membrane glycoprotein. We did not observe any statistically significant evidence of either positive/diversifying or negative/purifying selection in the CDSs for envelope, ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10.in the isolates of the year 2020 from India. Previous studies on SARS-CoV-2 with other coronaviruses reported strong and global purifying selection along with some site-specific (localized) positive selections (Singh and Yi, 2021).
Our results also suggest that SARS-CoV-2 isolates from India are experiencing strong purifying selection to remove deleterious new mutations particularly in the CDSs corresponding to nsp1, nsp3, nsp4, nsp5, nsp6, nsp8, nsp9, RdRp, helicase, exonuclease, nsp16, spike glycoprotein, nucleocapsid phosphoprotein, and membrane glycoprotein. Also, the localized sites displaying positive selection indicate adaptive selection facilitating the maintenance of nonsynonymous substitution in these sites of nsp2, nsp5, nsp6, helicase, exonuclease, RdRp, spike glycoprotein, and nucleocapsid phosphoprotein. It is likely that these substitutions provide viruses some sort of advantages in terms of their pathogenicity and immune escape. Our analyses, including Tajima neutrality test, mutation analysis, and site-wise evolutionary rate analysis indicate that ORF3a, ORF8, nucleocapsid phosphoprotein, and spike glycoprotein of the viral genome are undergoing rapid non-random evolution. The ORF3a encoded protein is the largest accessory protein with a TRAF binding motif, which promotes the activation of NLRP3 and NFκB inflammasome (Majumdar and Niyogi, 2020). ORF8 also encodes for an accessory protein, which interferes with the host immune response, induces IL17 signaling pathway, and induces overexpression of the proinflammatory factors (Lin et al., 2021). Considering the interplay of these viral proteins with the host immune response, it is conceivable that they are evolving at a rapid rate due to host immune challenge. These findings on mutations, selection pressure, and evolutionary rates of SARS-CoV-2 isolates from India populations are important for an improved understanding of the rapidly evolving virus and would be helpful in combating COVID-19.
CRediT authorship contribution statement
Archana Pal: Conceptualization, Experimental design, Data acquisition. Shefali Dobhal: Experimental design, Writing – review & editing. Kishore Kumar Dey: Experimental design, Writing – review & editing. Anish Kumar Sharma: Experimental design, Writing – review & editing. Vivek Savani: Data acquisition, Writing – review & editing. Vishal Singh Negi: Conceptualization, Experimental design, Data processing and analysis, Coding, Writing – original draft, Writing – review & editing. All authors reviewed the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors are thankful to Dr. Ratnesh Singh (Texas A&M Agrilife) for valuable discussion and suggestions.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.compbiolchem.2021.107594.
Appendix A. Supplementary material
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Code Availability
The code for this study is available at the GitHub repository (https://github.com/vishalsnegi/SARS-CoV-2_Geo_India).
References
- Andrews M.A., Areekal B., Rajesh K.R., Krishnan J., Suryakala R., Krishnan B., Muraly C.P., Santhosh P.V. First confirmed case of COVID-19 infection in India: a case report. Indian J. Med. Res. 2020;151:490–492. doi: 10.4103/ijmr.IJMR_2131_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhofer U., Bonatesta E., Horejš-Kainrath C., Hochreiter S. msa: an R package for multiple sequence alignment. Bioinformatics. 2015;31:3997–3999. doi: 10.1093/bioinformatics/btv494. [DOI] [PubMed] [Google Scholar]
- Charif D., Lobry J.R. In: Structural Approaches to Sequence Evolution: Molecules, Networks, Populations, Biological and Medical Physics, Biomedical Engineering. Bastolla U., Porto M., Roman H.E., Vendruscolo M., editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2007. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis; pp. 207–232. [DOI] [Google Scholar]
- Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:1–19. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J.C., Wyckoff G.J., Wu C.-I. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- Gentleman R., Ihaka R. Lexical scope and statistical computing. J. Comput. Graph. Stat. 2000;9:491–508. [Google Scholar]
- Ginestet C. ggplot2: Elegant graphics for data analysis. J. R. Stat. Soc.: Ser. A (Stat. Soc.) 2011;174:245–246. doi: 10.1111/j.1467-985X.2010.00676_9.x. [DOI] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J., Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T., Biosciences I., Carlsbad C. BioEdit: an important software for molecular biology. GERF Bull. Biosci. 2011;2:60–61. [Google Scholar]
- Hall T.A. c1979-c2000. Information Retrieval Ltd; London: 1999. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT; pp. 95–98. (Nucleic Acids Symposium Series). [Google Scholar]
- Ihaka R., Gentleman R. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. doi: 10.1080/10618600.1996.10474713. [DOI] [Google Scholar]
- Jenkins G.M., Rambaut A., Pybus O.G., Holmes E.C. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 2002;54:156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Jombart T., Ahmed I. Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jungreis I., Sealfon R., Kellis M. SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes. Nat. Commun. 2021;12:2642. doi: 10.1038/s41467-021-22905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S.L., Frost S.D.W. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Fu B., Yin S., Li Z., Liu H., Zhang H., Xing N., Wang Y., Xue W., Xiong Y., Zhang S. ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. Iscience. 2021;24(4) doi: 10.1016/j.isci.2021.102293. Apr 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar P., Niyogi S. ORF3a mutation associated with higher mortality rate in SARS-CoV-2 infection. Epidemiol. Infect. 2020;148:148. doi: 10.1017/S0950268820002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B., Wertheim J.O., Moola S., Weighill T., Scheffler K., Pond S.L.K. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B., Weaver S., Smith M.D., Wertheim J.O., Murrell S., Aylward A., Eren K., Pollner T., Martin D.P., Smith D.M., Scheffler K., Kosakovsky Pond S.L. Gene-wide identification of episodic selection. Mol. Biol. Evol. 2015;32:1365–1371. doi: 10.1093/molbev/msv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi A.P., Singh R., Sharma A., Negi V.S. Insights into high mobility group A (HMGA) proteins from Poaceae family: an in silico approach for studying homologs. Comput. Biol. Chem. 2020;87 doi: 10.1016/j.compbiolchem.2020.107306. [DOI] [PubMed] [Google Scholar]
- Pages, H., Aboyoun, P., Gentleman, R., DebRoy, S., Pages, M.H., IRanges, L., BSgenome, S., XStringSet-class, R., MaskedXString-class, R., XStringSet-io, R., 2013. Package ‘Biostrings.
- Pal A., Negi V.S. Plant Cenh3 evolution is congruent with the phylogeny of plant species. Int. J. Sci. Technol. Res. 2019;8:1473–1476. [Google Scholar]
- Paradis E., Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pond S.L.K., Frost S.D.W. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Racine J.S. RStudio: a platform-independent IDE for R and sweave. J. Appl. Econ. 2012;27:167–172. doi: 10.1002/jae.1278. [DOI] [Google Scholar]
- Schliep K.P. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D., Yi S.V. On the origin and evolution of SARS-CoV-2. Exp. Mol. Med. 2021;53:1–11. doi: 10.1038/s12276-021-00604-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman S.J., Weaver S., Shank S.D., Magalis B.R., Li M., Kosakovsky Pond S.L. In: Evolutionary Genomics: Statistical and Computational Methods, Methods in Molecular Biology. Anisimova M., editor. Springer; New York, NY: 2019. Evolution of viral genomes: interplay between selection, recombination, and other forces; pp. 427–468. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Wang C., Liu Z., Chen Z., Huang X., Xu M., He T., Zhang Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020;92:667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S., Shank S.D., Spielman S.J., Li M., Muse S.V., Kosakovsky Pond S.L. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018;35:773–777. doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H., Hester, J., 2020. readr: Read Rectangular Text Data.
- Wickham, H., François, R., Henry, L., Müller, K., 2021. dplyr: A Grammar of Data Manipulation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Data Availability Statement
The code for this study is available at the GitHub repository (https://github.com/vishalsnegi/SARS-CoV-2_Geo_India).