Abstract
Breast and colorectal cancers are two primary malignancies on which most of the research done worldwide investigates the potential genetic and environmental risk factors and thereby tries to develop therapeutic methods to improve prognosis. Breast cancer is the most diagnosed cancer type in women, while colorectal cancer is diagnosed in males as the third most and females as the second most cancer type. Though these two cancer types are predominantly seen in adult patients worldwide, in the current context, these malignancies are diagnosed at a younger age with a significant rate of incidents than previous. Such early-onset cancers are generally present at an advanced stage of the most aggressive type with a poor prognosis. In the past, the focus of the research was mainly on studying possible candidate genes to understand the onset. However, it is now recognized that genetics, epigenetics, and other environmental factors play a pivotal role in cancer susceptibility. Thus, most studies were diversified to study the behavior of host microRNAs, and the involvement of gut microbiota and good communication between them surfaced in the occurrence and state of the disease. It is understood that the impact of these factors affects the outcome of the disease. Out of the adverse outcomes identified relating to the disease, immunosuppression is one of the most concerning outcomes in the current world, where such individuals remain vulnerable to infections. Recent studies revealed that microbiome and microRNA could create a considerable impact on immunosuppression. This review focused on the behavior of host microRNAs and gut microbiome for the onset of the disease and progression, thereby influencing an individual’s immunosuppression. Understanding the interactions among microRNA, microbiome, presentation of the disease, and impact on the immune system will be immensely useful for developing future therapeutic strategies based on targeting host microRNA and the patient’s gut microbiome. Therapies such as inhibitory-miRNA therapies, miRNA mimic-based therapeutics, immune checkpoint blockade therapies, and bacteria‐assisted tumor‐targeted therapies help modulate cancer. At the same time, it paid equal attention to potential noninvasive biomarkers in diagnosis, prognosis, and therapeutics in both cancers.
Keywords: breast cancer, colorectal cancer, miRNA, microbiota, immunosuppression
Introduction
Breast Cancer
Breast cancer (BC) is the most common malignancy among females, and the leading cause of cancer death accounted for estimated 685,000 deaths in 2020. Worldwide, there were estimated 2.3 million cases diagnosed with female breast cancer in 2020, representing 11.7% of all cancer cases, accounting for almost 1 in 4 cancer cases among women.1,2 There are four molecular subtypes of breast cancer, which are highly influential by age, identified as luminal A (ER+ and/or PR+, HER2-), luminal B (ER+ and/or PR+, HER2+), human epidermal growth factor receptor 2- enriched (ER-, PR-, HER2+), and basal-like (ER-, PR-, HER2-), where these types show differences in their incidence, level of response for treatments, progression of the disease, and survival.3 Breast cancer risk increases with age, but a significant rate of incidents occur in younger patients worldwide in the current context.
Although early-onset breast cancer is rare, it is diagnosed among younger women between 15 and 39 years of age accounts for 7% of all breast cancer incidence and more than 40% of all young cancer cases.4 Positive family history of cancer is considered the most significant personal risk factor in this type of cancer with a strong genetic predisposition. Germline pathogenic mutations of BRCA1 and BRCA2 with family history play a significant role in the occurrence of early-onset where such BRCA carriers with unilateral breast cancer are at an elevated risk (16–35%) of developing contralateral breast cancer as well.5 Young premenopausal breast cancer generally presents at an advanced stage with distinct clinicopathological features related to the most aggressive subtypes with poor prognosis. Triple-negative (ER-, PR-, HER2-) and HER2-positive (HER2+) breast cancers with high-grade proliferation are the most lethal subtypes of breast cancer presented at a younger age. The survival rate of women with early-onset breast cancer is relatively lower when compared to older women, contributing to an increased breast cancer mortality rate.6 Furthermore, Luminal A, the least aggressive subtype, is much more common among breast cancer patients older than 50 years, which is much more favorable in prognosis, while luminal B is an aggressive subtype present in older patients than 70 years with high-grade proliferation.7
However, about 90% of breast cancers are sporadic with no apparent genetic predisposition. Thus, other factors play a pivotal role in sporadic breast cancers in young non-BRCA carriers without a family history or other genetic predisposition. Hormonal risk factors, obesity, and dietary habits are some of the risk factors having a pronounced impact on the disease.8 Nevertheless, recent research has discovered the involvement of the gut microbiome and host microRNA (miRNA) individually or in conjunction with some underlying mechanisms operating in breast cancer cases.
Colorectal Cancer
Worldwide, colorectal cancer (CRC) is identified as the third most common cancer diagnosed in both sexes, wherein males are the third most and females are the second most cancer type.9 In 2020, more than 1.9 million new colorectal cancer cases and about 935,000 deaths were estimated, accounting for one in 10 cancer cases and deaths worldwide.1
CRC incidence continues to be an increasingly predominant issue in Asia10 and Eastern Asian countries.11 Both CRC incidence and mortality have increased in countries with medium Human Development Index (HDI), showing rapid social and economic changes by adopting westernized lifestyles compared to countries with high HDI. Countries with high HDI have increased and improved early detection and prevention through polypectomy, improving perioperative care, thus lowering the CRC incidence and mortality rate.12,13
According to cancer statistics in the United States, there is an increased level of incidence of young-onset colorectal cancer (aged 20–49 years), where more than one-tenth of all CRC cases are young-onset comprising 11% and 18%, respectively, in the colon and rectal cancers when compared to the age group of 50 years and above.14 In the Asia-Pacific region, with a westernized urban lifestyle and a higher population of younger people, the incidence rate of young colorectal cancer has also increased, similar to the West.15 Obesity, sedentary behavior, and a high-meat, high-calorie, fat-rich, fiber-deficient diet are some factors associated with the increased risk of colorectal cancer.16 Alternatively, several differences are identified concerning tumor aggressiveness, staging, and clinical outcome in young-onset colorectal cancer compared with late-onset.17 The early detection of CRC can enhance the diagnosis procedure and thereby improve the patient’s prognosis. Early identification would increase the patient’s survival rate by administering an appropriate surgical approach before the disease metastasized. The 5-year survival rate of colorectal cancer patients with advanced-stage IV drops to 14% compared to early-stage/localized stage CRC patients with a 90% survival rate where such low survival rates can be improved by timely administration of surgical procedures.18
Most colorectal cancers are sporadic, accounting for 90% of all cases without a clear family history or proven genetic involvement, where the other 10% are considered familial types with an identified genetic predisposition.19 CRC with early-onset is a feature of the hereditary cancer syndromes, which are recognized in a young individual in advance with a family history or other risk factors, such as inflammatory bowel disease or a hereditary syndrome (Lynch syndrome/familial adenomatous polyposis). Such identification may lead to the recommendation of risk-reducing processes, such as weight reduction, increased physical activities, consumption of lots of vegetables, fruits, and whole grains, avoiding alcohol and smoking, and increasing vitamin D uptake to avoid low levels.14 Alternatively, sporadic CRC can occur by involving the dysregulation of numerous signal transduction pathways.20 Chromosomal instability (CIN), CpG island methylator phenotype (CIMP), and microsatellite instability (MSI) are the recognized pathways of carcinogenesis in CRC via somatic genetic mutations.21,22 The most common pathway is the chromosome instability pathway (CIN) that accounts for 70% of sporadic CRC incidence, and such tumors occur due to accumulation of mutations in specific oncogenes and tumor suppressor genes, such as APC, KRAS, PIK3CA, BRAF, SMAD4, TP53.23
In recent studies, the interaction of host miRNA and gut microbiome was identified in colorectal cancer. Such factors may significantly contribute to CRC in cases when genetic predisposition is not apparent and may be involved in the occurrence of sporadic colorectal cancer at a younger age.
Inter-Domain Communications Between the Gut Microbiome and Host miRNAs in Cancer
miRNAs are small, non-protein-coding endogenous RNA molecules that regulate gene expression via complementary base pairing with the 3ʹ- untranslated region of the respective mRNA results with alteration of protein expression through targeted mRNA degradation, translation repression, or gene silencing. Single miRNA can regulate several mRNAs that mediate biological functions, such as proliferation, cell signaling, DNA repair, differentiation, and stress responses.24 Due to aberrant expression of miRNAs alters the possible gene expression contributing to the development of several diseases, including cancer.25 Cancer-related miRNAs can be categorized based on their expression and processing as oncomiRs (function as oncogenes) and TS-miRs (function as tumor suppressors). Such miRNAs play critical roles in oncogenesis, metastasis, as well as when showing resistance to several therapies.26,27 In addition, there are other types of miRNAs, such as metastamiRs (metastasis-associated miRNA), regulating metastasis and metastasis-suppressor miRNAs, suppressing metastasis.28,29
The microbiome in humans is where a symbiotic relationship with microbiota confers benefits in many vital aspects of life. In addition, microbiota will be shaped up by age, lifestyle, diet/nutrition, environmental factors, host hormonal changes, underlying disease conditions, and host genetics.30 Especially, most of the members of gut microbiota are beneficial as they mediate different functions within the host, such as bioconversion of nutrients, protection against pathogenic microbes, regulation and maintenance of several host physiological functions, such as metabolism, immune homeostasis, and the nervous system.31,32
However, gut microbiota dysbiosis is a process where imbalance or perturbation of the content of the normal microbiota in the gut leading to the disruption of normal microbial equilibrium results in detrimental consequences, such as provoking different disease conditions, such as inflammatory, immune, and infectious diseases, as well as malignancies. Such modified microbiota leads to the onset and progression of cancers. Dysbiosis of the microbiome may alter biological equilibrium by regulating the host’s metabolites, genes, and proteins.31 Such events can create an impact on the immune system and the onset of obesity. Modified microbiota can cause alterations in the expression of miRNA related to lipid metabolism and thereby cause obesity and cancer.33
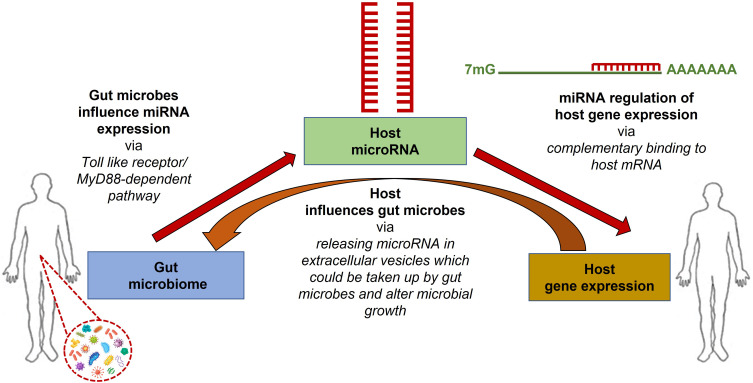
Recent studies have identified a close interaction between the gut microbiome and host miRNAs. miRNAs regulate gene expression via complementary binding to host mRNAs, while the gut microbiota can influence host miRNA expression via MyD88-dependent pathway or microbiome-derived metabolites that alter host gene expression in the colon.34 In turn, the host influences the gut microbiota via releasing host miRNAs into extracellular vesicles, where microbes can up-take them (Figure 1). Thus, bidirectional interaction of host miRNAs and gut microbiome will regulate the host gene expression.35 Alternatively, some studies have shown that miRNA expression can be altered by several dietary agents taken by the host36 and in turn, such regulated miRNA can modulate the gut microbiome, and dietary agents may directly influence the microbiome as well.37 Deregulation of miRNA expression and dysbiosis of the gut intestinal microbiota are strongly interconnected, leading to the occurrence of several diseases in the host, including cancers.
Figure 1.
Inter-domain communications occur among host-gene expression, host miRNA expression, and host gut microbiome. Different miRNA species regulate host gene expression via complementary binding to host mRNA, and the gut microbiome regulates host miRNA expression via MyD88-dependent pathway or microbiome-derived metabolites. Alternatively, the host influences the gut microbiota by releasing host miRNAs in extracellular vesicles, taking up microbes, and altering the gut microbiome.
Expression-based communication between the gut microbiome and host miRNAs can help to design druggable targets for cancer therapy. The main druggable target in the current trend is miRNA levels in cancer metabolism. Manipulating miRNA levels will help develop miRNA-based therapies in cancer treatments by suppressing the expression of oncomiRs that are otherwise overexpressed in human cancers and/or restoring the expression of TS-miRs inactivated normally in the tumor. miRNA inhibitor-based therapeutics inhibit levels of oncomiRs by several methods, such as antagomiRs (anti-miRs), antisense anti-miR oligonucleotides (AMOs), locked nucleic acid (LNA), and miRNA sponge. While miRNA mimic-based therapeutics are based on synthetic miRNA sequences, they will mimic the natural TS-miRs that can be recognized by Dicer and Ago2 proteins.38,39
Interaction of Gut Microbiome and Host miRNAs in Breast Cancer
miRNAs are highly stable regulatory RNAs found in serum and plasma at different levels.40 These miRNAs can be potential biomarkers for evaluating cancer, and many circulating miRNAs indicative and unique to breast cancer have been identified.41,42 Screening of such circulating miRNAs on cancers can be helpful to develop noninvasive and cost-effective novel biomarkers to identify patients with a poor prognosis, such as triple-negative breast cancer (TNBC) patients with early-onset, thereby improving diagnostic, prognostic values, and treatment options.43,44 Alternatively, these miRNA markers significantly correlate with tumor recurrence, survival rate, and tumor grade and are also identified as subtype-specific miRNAs acting as biomarkers for the respective cancer subtypes.
One study showed that some of the miRNA species are highly overexpressed in tumor samples when compared to non-tumor samples, such as miR-21, miR-106a, and miR-155, while some of them are comparatively under-expressed, such as miR-126, miR-199a, and miR-335 in a tumor sample, showing a correlation between breast tumor tissue types and levels of circulating miRNA. In addition, expression levels of some miRNAs such as miR-21, miR-126, miR-155, miR-199a, and miR-335 show association with clinicopathological characteristics of breast cancer such as histologic tumor grades as well as the expression of sex hormone receptors.45,46 Another study revealed that the concentration of circulating miR-21 is helpful to distinguish breast cancer patients from healthy women and recognize distant metastatic patients from patients with locoregional recurrence.47,48 It has also been shown that overexpression of miR-21 on breast cancer occurrence and progression can be used as a biomarker in the diagnosis and prognosis of the disease.49 miR-21 is identified as an oncogene from its tumor growth-promoting ability, targeting tumor suppressor genes, TPM1 and PDCD4, and inhibiting their expression.50 Circulating miRNAs can potentially be used for early detection and monitoring of the prognosis of cancer. A study has shown that plasma levels of miR-21, miR-155, and miR-10b (oncomiRs) were significantly elevated in breast cancer patients comparatively, while plasma expression of the Let-7a (tumor suppressor miRNA) was reduced when compared with those levels in the controls. Alternatively, plasma levels of miR-21, miR-155, and miR-10b were reduced after surgery, chemotherapy, or radiotherapy, whereas Let-7a was increased after treatment where miR155 showed a significant level of differences in pre- and post-operative stages, suggesting a potential biomarker and an indicator for treatment responses in the Luminal A subtype.51,52 Additionally, metastatic luminal A breast cancer can be distinguished by locally confined type based on the expression level of some specific miRNA species. miR-331 shows an elevated expression level, while miR-195 level shows downregulation in metastatic type when combined, can be used as biomarkers to distinguish metastatic from local.53 Alternatively, the specific characteristic feature of luminal A from luminal B is the upregulation of miRNA cluster miR99a/let-7c/miR-125b, which improves the overall survival of a patient with luminal A cancer, especially with high miR-99a expression, linked to favorable clinical outcome. This miRNA cluster can be considered a possible biomarker to distinguish luminal A from B and can be used as a prognostic marker within the luminal A group where some patients with luminal A show the low level of these miRNA associated with poor overall survival.54 Furthermore, pre-operative patients with luminal B subtype show an elevated level of miR-195 in the blood, which can be used as an early biomarker.51,55 Concerning HER2 enriched subtype, several miRNAs identified have tumor-suppressive function on the EGFR1 pathway while inhibiting the proliferation of cancer, such as miR-147, miR-124, and miR-193-3p. There is a loss of expression of miR-342-5p and miR-744 in HER2-positive BC when compared with HER2-negative breast cancers. Let-7f, Let-7g, miR-107, miR-10b, miR-126, miR-154, and miR-195 have been identified as HER2-positive BC-specific miRNAs. miR-4734 and miR-150-5p are identified with prognostic values and can be used as prognostic biomarkers in HER2-positive BC.56
In addition, Zeng et al revealed that the plasma level of miR-30a can be used as a diagnostic biomarker with improved sensitivity and specificity over conventional circulating markers, such as carcinoembryonic antigen (CEA) and cancer antigen 15-3 (CA15-3), showing reduced levels in breast cancer patients. In addition, miR-30a levels also show a significant association with breast cancers having ER and triple-negative status.57 Furthermore, some studies done on the behavior of miRNA were identified that miR-155 as a potential prognostic biomarker in TNBC patients, where elevated levels of miR-155 showed a protective action by reducing RAD51 expression and improving better clinical outcome for IR-based therapies in TNBC patients.43,58 Whereas miR-18b, miR-103, miR-107, and miR-652 levels interact with recurrence and reduced survival in TNBC.59 In addition, miR-376c, miR-155 and miR-17 are identified as biomarkers at the early-stage, while miR-10b is a late-stage biomarker in TNBC,60 while up-regulated miR-532-5p can be used as a potential biomarker for prognosis.50 In addition, elevated levels of miR-138 can be seen in TNBC types with the worst prognosis, highlighting its use as a diagnostic and prognostic marker where miR-138 promotes cell proliferation and inhibits apoptosis while targeting TUSC2, a tumor suppressor, and silences its action.61 Some miRNA signatures are correlated with larger tumor size such as miR-374, where some (miR-105 and miR-93-3p) with elevated levels are associated with poor survival.62 Furthermore, aberrant levels of some miRNAs in TNBC patients are associated with chemoresistance. Elevated levels of miR-181a are associated with non-responsive to neo-adjuvant chemotherapy, while low levels of miR-200c show resistance to chemotherapy and poor response to radiotherapy.63 Several studies revealed that miRNA signatures could be used as diagnostic, prognostic, and predictive biomarkers in several cancer types, especially TNBC, mainly in patients with early-onset.64
Studies were done to identify potential oncogenic miRNAs on breast cancer, promoting cell growth and proliferation, cell migrations, invasion, and metastasis.65 Several diagnostic miRNAs for early diagnosis, breast cancer molecular subtypes, and histological subtypes; predictive miRNAs associated with hormone therapies, targeted therapies, chemotherapeutic agents, radiotherapy, and prognostic miRNAs for positive and negative prognosis have been identified for breast cancer.66,67
Alternatively, a study on the association of microbiome with different breast cancer types identified that distinct microbial signatures were associated with the triple-negative and triple-positive samples but shared similar microbial patterns in the ER-positive and HER2-positive samples, which can be used as a diagnostic tool as well as treatments for different cancers.68 It was identified that the ER-positive subtype has a very diverse microbiome, while triple-negative breast cancer has the least diverse microbiome.69 Various bacterial genes in the gut microbiome can produce estrogen-metabolizing enzymes. Thus, higher levels of endogenous estrogens and their differences in metabolism are directly related to BC risk in postmenopausal patients. Since endogenous estrogen is a significant risk factor, especially for postmenopausal women, 70% of breast cancers that occur in them are estrogen receptor-positive.70
Hence, it was shown that the effect of the gut microbiome on breast cancer is modulated via circulating estrogen and its metabolites.71 The composition of microbiome present between postmenopausal and premenopausal women is different, resulting in the production of distinct metabolites. Additionally, the microbes that show synergistic action during the premenopausal stage would be competing with each other when it comes to the postmenopausal stage. According to Zhao et al, Bacteroidetes and Roseburia sp. show high abundance, while Firmicutes and Parabacteroides show lower abundance in premenopausal women. Alternatively, a higher number of Escherichia coli and Bacteroides were seen in postmenopausal women showing a low ratio of Firmicutes to Bacteroidetes than that of premenopausal women.72 Since there are differences in endogenous estrogen levels and gut microbiome composition involving the regulation of estrogen levels in pre and postmenopausal women, dysbiosis of microbiota in the gut will interact with elevated levels of circulating estrogen in postmenopausal breast cancer. Such microbiome involvement in the development and progression of breast cancer can be a future trend in identifying novel treatment methods.73 In addition, studies revealed that the composition of the gut microbiome could be varied among BC women based on their body mass, where obese women with BC have low levels of Firmicutes, Faecalibacterium prausnitzii, and Blautia sp. compared to patients with normal weight. Furthermore, the availability of some bacterial groups, Clostridium leptum cluster, Clostridium coccoides cluster, Faecalibacterium prausnitzii, and Blautia sp. are associated with the clinical stage of the patient where there is an elevated level shown in BC patients with stage II/III when compared to patients with stage 0/I.74,75
In addition to the gut microbiome, a study on the mammary microbiome showed that bacterial communities present in mammary tissue do not show any difference between tumor tissue and its adjacent normal tissue,76 but it showed cancer-promoting activity by Escherichia coli due to its high abundance in breast cancer patients when compared to healthy controls.77 Several possible mechanisms by which microbial influence can be exerted on the occurrence of breast cancer have been identified. These include regulation of chronic inflammation and immunity,78 genomic stability and DNA damage76 and metabolic function.79
Most of the studies on microbiome detection have relied on the sequencing technique of a specific region of the bacterial16S rRNA gene, while several studies were based on either qPCR or a DNA array. Circulation of digestive and microbial products in the gut can involve the development of breast cancer as such products interfere in several pathways leading to modification of gene expression or signal transduction within the host.73 Additionally, dietary uptake/lifestyle of the host can shape the gut microbiota population, then there could be a negative impact from the diet for the composition of microbiota, which leads to the progression of cancers including breast cancer. The nature of total dietary fiber (soluble fiber and insoluble fiber) uptake would matter for the existence of different microbiota in the gut, which creates an imbalance among different phylae. In the context of breast cancer, a possible dysbiosis occurs among Bacteroidetes and Firmicutes, which can be positively adjusted by taking probiotics and a healthy diet.80
Triple-negative breast carcinoma that occurs more often in young breast cancer patients is the highly aggressive type among BC. Endocrine therapies and HER2-targeted therapies are ineffective in TNBC, and chemoresistance often develops after chemotherapy. Thus, identifying effective targeted therapies for TNBC based on manipulating miRNA levels and/or microbiome status is likely to improve prognosis.81
Interaction of Gut Microbiome and Host miRNAs in Colorectal Cancer
Several miRNAs have been identified with potential biological and clinical relevance in CRC.82 Aberrant expression of miRNAs is associated with CRC progression, survival, and disease outcome of the patient. Studies have proven a differential expression of miRNA levels between colorectal cancer tissue and normal colorectal epithelium; thus, some miRNAs enhance prognostic and predictive values as biomarkers in colorectal cancer. Where such miRNAs can predict the outcome of the disease, as well as the responses to chemotherapy and radiotherapy in patients.83 Dysregulation of miRNA, will alter its involvement in different cellular pathways leading to cell proliferation, differentiation, apoptosis, and development, such as WNT/β-catenin pathway, EGFR pathway, TGF-β signaling pathway, and epithelial-to-mesenchymal transition. Thus, identifying the involvement of miRNAs in the above pathways could be beneficial in selecting possible biomarkers and treatment options for CRC.84
miRNA expression varies in different stages of CRC, where some are expressed in the late stage of CRC, such as miR-141, which is identified as a differential diagnostic biomarker, and higher levels of miR-141 in plasma are associated with poor survival of CRC cases.85 Alternatively, serum miR-21 has been identified as a promising biomarker for the early diagnosis and prognosis of CRC. miR-21 is up-regulated in many cancers, including colorectal cancer, suggesting this to be a promising diagnostic biomarker for CRC.86 Expression of miR-21 is associated with TNM stage where high miRNA expression occurs in later stages of CRC.87 Overexpression of miR-21 elevates the tumor progression associated with poor survival and response to chemotherapy in patients. It acts as an oncogenic miRNA (oncomiR) capable of regulating the expression levels of some cancer-related genes PTEN, TPM1, and PDCD.88 Also, miRNA signatures can be used as diagnostic, predictive, and prognostic biomarkers for CRC. miR-31 is associated with BRAF mutation in CRCs, where it appears to be strongly associated with the aggressive phenotype, showing poor prognosis in patients, which demonstrated that elevated levels of miR-31 expression are associated with BRAF V600E mutation in stage IV CRC patients.89 Furthermore, elevated level of miR-31 expression has a significant association with poor mortality.90 Moreover, there is a significant elevation of miR-21 and 31 expressions can be seen even in precancerous colorectal adenoma, which can be used as the target for CRC screening.88
Alternatively, host miRNA can influence the growth and the composition of bacteria in the gut microbiome in an individual. Several studies have investigated the association of miRNA expression with the microbiome in human CRC tumors and normal tissues.34 Studies using new technologies such as metagenomic sequencing have confirmed the association of the gut microbiome with colorectal carcinogenesis, where different microbes such as Fusobacterium nucleatum, Peptostreptococcus stomatis, and Parvimonas micra are found towards the development of CRC. Such CRC susceptibility and progression can be influenced by the composition of the gut microbiome via regulating processes, such as inflammation and DNA damage in the host by producing metabolites that can either develop or suppress tumors. Gut microbes are present in different sites, such as the ascending colon, distal colon, proximal ileum, and jejunum in the gut, fulfilling crucial functions such as producing vitamins and metabolizing dietary compounds.91 Out of different bacterial species present in the microbiome, the Firmicutes, and Bacteroidetes phyla were shown to be in high abundance in the gut. Such abundances of Firmicutes and Bacteroidetes and Proteobacteria were correlated with differently expressed miRNA in colorectal cancer.34 Some studies have revealed that Bacteroidetes and Firmicutes phyla are significantly correlated with miR-141-3p levels, while Actinobacteria, Bacteroidetes, Cyanobacteria, and Firmicutes show correlation with miR-200a-3p levels.32
Dysbiosis/imbalance in the microbe community in the gut is associated with the development of CRC. CRC patients show an increase in several gut microbes, such as Fusobacterium nucleatum, Bacteroides fragilis, Escherichia coli, Enterococcus faecalis while a decrease in microbes like Faecalibacterium, Blautia, Clostridium, Bifidobacterium, and Roseburia sp.91,92 Such imbalance shown in gut microbiome in CRC patients from healthy individuals tends to increase detrimental/pro-inflammatory opportunistic pathogens while decreasing commensal bacterial population such as butyrate-producing bacteria. Butyrate-producing microbes are associated with protection against CRC and colitis formation by inhibiting tumor formation, inducing apoptosis, reducing oxidative damage, and minimizing co-carcinogenic enzymes’ activity. However, with the reduction of such beneficial bacterial population in the gut, it facilitates the progression of CRC.92,93
Fusobacterium nucleatum, enterotoxigenic Bacteroides fragilis, and colibactin-producing Escherichia coli are bacterial species associated with tumorigenesis. It was found that a significant increase in Fusobacterium nucleatum in early-stage CRC patients shows a worse prognosis.94 In addition, recent studies have identified Enterotoxigenic Bacteroides Fragilis (ETBF), a subtype of B. fragilis strain producing Bacteroides fragilis toxin (BFT) that activates certain cancer-promoting pathways in epithelial cells of the colon, causing colon inflammation associated with colorectal cancer.95 Dysbiosis of the colon microbiome results in increased interleukin-17 driven inflammation, which contributes to the carcinogenesis of colorectal cancer in humans. Chronic inflammation is linked to carcinogenesis, where 20% of patients identified with ulcerative colitis, leading to CRC development within 30 years of onset.96
Association Between Breast and Colorectal Cancers
Some of the studies supported the existence of a connection between breast cancer and colorectal cancer in women, where a possible association is found between the level of sex hormones and the occurrence of colorectal cancer. Possible exposure of breast cancer patients to the elevated levels of endogenous/exogenous sex hormones due to parity, hormone/estrogen replacement therapies, and breast cancer treatments (tamoxifen) may increase the possible risk of having colorectal cancer, which is yet controversial. Furthermore, the research identified that patients diagnosed with breast cancer have a 60% increased risk of getting colorectal cancer.97 Another research work carried out by Abu-Sbeih et al identified that patients with a history of breast cancer could have a high incidence rate for adenomatous polyps occurrence irrespective of the patient’s age and having a 5% chance of getting invasive CRC. In addition, it was highlighted that a recommendation of colonoscopy decided upon the patient’s age of diagnosis of breast cancer. For example, appropriate colonoscopy screening is recommended within one year of breast cancer being diagnosed in a patient older than 40 years.98
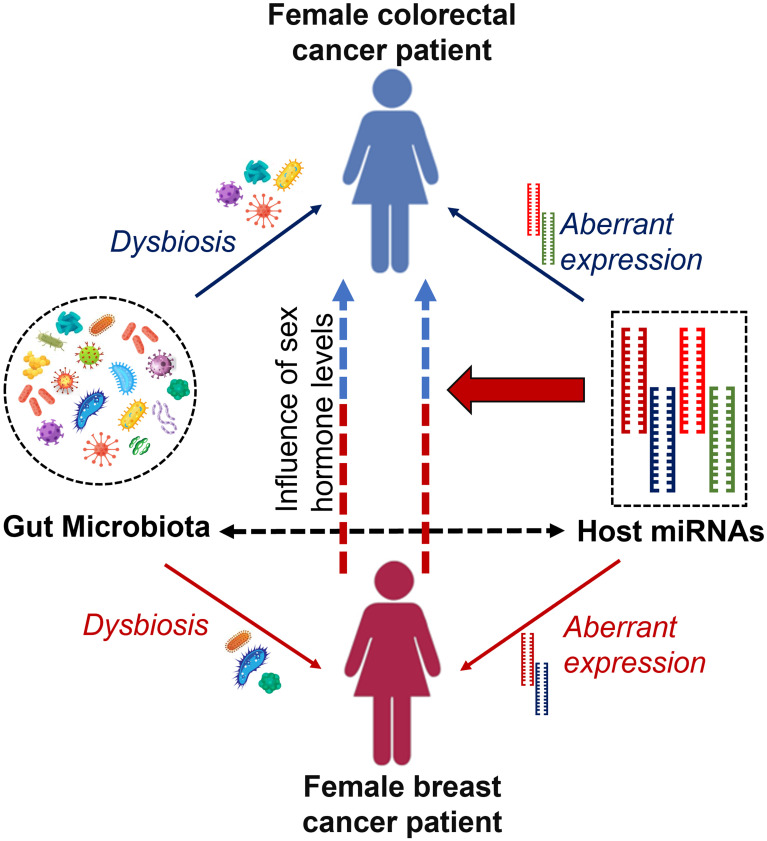
Furthermore, a link between these two cancers is established by involving a common set of miRNAs and their behavior during disease progression. As mentioned previously, miR-21 is overexpressed in breast cancer patients promoting cell proliferation, migration, and invasion and thereby acts as oncomiR,49 while miR-31, miR-143, and miR-145 are known to be under-expressed in breast cancer patients inhibiting cell proliferation.99,100 However, in colorectal cancers, the expression of miR-21 and miR-31 shows different behavior where overexpression of those miRNAs can be seen in colorectal patients promoting inflammation-associated tumorigenesis.88 In such a situation, it is worth counting the synergistic effects of miRNAs in such cancer types that can occur as second de novo malignancy in cancer survivors.100 Figure 2 illustrates the possible connection to the occurrence of colorectal cancer in females with a history of breast cancer.
Figure 2.
Occurrence of colorectal cancer in female breast cancer patients/survivors. Well-coordinated interaction between the gut microbiome and host miRNAs is maintained within the host, but dysbiosis of gut microbiota and aberrant expression of several host miRNAs can cause malignancies such as breast and colorectal cancers. Alternatively, female breast cancer patients and/or survivors show a possible risk of having colorectal cancer. Levels of endogenous and/or exogenous sex hormones and differential behavior of some common set of miRNAs cause colorectal cancer in females with a history of breast cancer.
Involvement of Host miRNAs and the Microbiota in Immune Regulation of Cancer
miRNA is a crucial player in human physiological activities, such as differentiation, cellular proliferation, development, and apoptosis, but dysregulation of miRNA expression, biogenesis, and alteration of epigenetic regulation of miRNA genes may lead to tumor development progression and response failure to therapies.101,102 Such cancer-derived miRNAs can modulate immune responses by creating an immunosuppressive tumor microenvironment while downregulating cancer immunogenicity, thereby protecting cancer cells from immune clearance.103 Immunomodulatory miRNAs (im-miRNAs) can regulate cancer immune surveillance leading to immune escape of tumors.104 Such im-miRNAs are involved in manipulating the immune response through regulating cancer antigen processing and presentation. For example, MHC class I cell surface exposure is repressed by miR-27 in colorectal cancer.105 miRNAs are also capable of regulating PD-1 expression and modulating NK cells, which facilitate cancers to escape the immune surveillance.106
Further, commensal gut microbes are essential for the maturation of the host’s innate and adaptive immune systems, maintaining gut homeostasis, thereby developing recognition and tolerance against opportunistic pathogen attacks and preventing infection.107 However, alteration of microbial community influences immune checkpoint blockers leading to immune dysregulation, thereby provoking inflammation-inducing bacteria, which promotes chronic inflammation directing to tumor development.108 Several studies revealed that gut microbes could regulate the function of the immune system by regulating miRNA expression.30
Alterations of the microbiome, miRNA transcriptome, and other factors, such as chronic stress and some treatments (chemotherapy), lead to immune dysregulation in cancer patients, leading to immunodeficiency/immunosuppression.104,109 Such an immunosuppression makes them prone to infectious diseases caused by opportunistic pathogens, such as viruses, which has been a tremendous challenge throughout the decades for cancer patients and oncologists.
In the current context, with a pandemic spreading globally, starting from the end of 2019, the novel coronavirus, COVID-19 (coronavirus disease of 2019) outbreak is a type of severe acute respiratory syndrome caused by SARS-CoV-2 virus (Severe Acute Respiratory Syndrome-Coronavirus-2) which cause severe lower respiratory disease in immunocompromised individuals compared to immunocompetent individuals. Such incidence is one of the highlighted pathogenic attacks for all individuals, especially for cancer patients. Virus virulence is more critical in disease outcomes; apart from that, several other host-related factors such as age, gender, obesity, smoking, and comorbidities such as cardiovascular disease, diabetes mellitus, and cancer matter a lot concerning the severe outcome of the disease.110 Cancer patients are known to be immunosuppressed individuals, and the situation is aggravated by certain forms of therapies, making them vulnerable to infections like COVID-19, which might be opportunistic, leading to severe consequences in such patients. Studies in China showed a higher mortality rate of 6% in cancer patients with COVID-19 infection than 1% in non-cancer patients. Susceptibility of cancer patients was evaluated to be three times more for COVID-19 infections with a poor prognosis due to their immunosuppression caused by the disease and therapies administered compared with individuals without cancer.111
Different cancer types have different susceptibilities towards viral infections, including COVID-19. As an example, colorectal cancer has a greater susceptibility towards COVID-19. Angiotensin-converting enzyme 2 (ACE2) receptor helps the coronavirus spike protein bind to the target cell, while transmembrane serine protease 2 (TMPRSS2) enables the activation of spike protein, thereby increasing the entry of the virus to the target cell. Higher expression of these two proteins in the lung epithelium increases the risk of SARS-CoV-2 virus infection. In addition to the respiratory tract, the human gastrointestinal tract expresses ACE2 and TMPRSS2. Moreover, the expression is elevated in colorectal cancer. Thus, the COVID-19 virus may infect the intestinal epithelium leading to an increased infection rate among CRC patients.112,113
However, some miRNAs can bind to viral RNA and inhibit its translation, creating an adverse impact on the viral genome. One research identified six different miRNAs, namely, miR-21-3p, miR-195-5p, miR-16-5p, miR-3065-5p, miR-424-5p, and miR-421 potential regulators of human coronaviruses.114 Alternatively, some miRNA like miR-27b can regulate ACE2 receptor, which is important for the entry of the coronavirus.115 Another research reported that miRNAs 200b-3p, 200 c-3p and 429 can regulate ACE2, while TMPRSS2 can be regulated by let-7c-5p, miRNA 98-5p, let-7 f-5p, let-7a-5p, let-7 g-5p, let-7b-5p, miR-4458, let-7e-5p, let-7i-5p, let-7d-5p and miRNA 4500.116 Such miRNAs could be promising therapeutic methods for controlling proteins promoting COVID-19 entry, especially in the gastrointestinal tract as ACE2 and TMPRSS2 are highly expressed in colorectal cancers. However, the expression levels of miRNA against the SARS-CoV-2 genome are downregulated inversely with age, so the elderly population is unlikely to benefit from controlling COVID-19 by miRNA-based methods compared to younger populations.117
In breast cancer, due to inflammatory reactions in the microenvironment, there is a risk of the reawakening of dormant cancer cells (DCC), which have entered a quiescent state upon effective treatment of primary cancer. DCC may be reactivated by escaping from the metastatic dormancy, and in instances such as SARS-CoV-2 infection, damages that occur in the respiratory tract provoke sequential immune activities, leading to pro-inflammatory responses. Such responses regulate inflammatory reactions and may contribute to the reawakening of DCCs, which promotes cancer cell proliferation.118
Promising Therapeutic Targets of Cancer
miRNAs are modulators of the cell responsible for many biological processes, such as proliferation, cell signaling, differentiation, stress responses, and DNA repair. In addition, miRNAs play a pivotal role in the differentiation, activation, and effector function of immune cells in innate and adaptive immunity. The innate immune system will provide an initial defense against pathogens, while adaptive immunity will lead to the propagation of such a response. miRNAs can regulate leading players in the innate immune systems, such as natural killer cells, macrophages, and inflammatory cytokines and chemokines.119 For example, miR-21 and miR-155 are known to be oncomiRs (oncogenic miRNAs), and both play a pivotal role in the immune regulatory process concerning breast cancer. It was revealed that miR-21 shows a bi-directional role during tumorigenesis where it also mediates an antitumor immune response,120,121 and elevated levels of miR-115 in immune cells show an antitumor immune response as well.122 Such miRNAs can be identified as a potential immunotherapeutic target for cancers, such as developing inhibitory-miRNA therapies based on antisense antimiRs.
Alternatively, gut microbiota show a high contribution towards immunotherapy of cancer, where such microbiota regulate immune functions by modulating immune checkpoint inhibitors. Most cancer immunotherapies work by reactivating the functions of the immune cells by blocking immune checkpoints and restoring the antitumor response of the immune cells.123 Immune checkpoint blockade therapy (ICB) is a promising immunotherapeutic method for certain types of cancers where the gut microbiome plays a vital role in the execution of such therapies. The composition of the gut microbiome determines the level of effectiveness of PD-1 and CTLA-4 blockades. Bacterial species such as Enterococcus, Ruminococcaceae, Akkermansia, and Bifidobacterium are increasing the effectiveness of PD-1 blockade therapy. Meanwhile, genus Bacteroides shows a biphasic effect where some strains facilitate CTLA-4 blockade therapy, while others negatively impact therapy efficacy.124
Dysbiosis of gut microbiota can induce chronic inflammatory diseases, such as cancer, which facilitate unfavorable microbes in the gut, negatively impacting immunotherapy. Either administering antibiotics or using prebiotics and synbiotics can enhance the colonization of commensal microbes in the gut by excluding harmful microbes, which is beneficial for developing antitumor immunity activities.125
Several new trends are developing in the current world concerning cancer therapies, such as using living or attenuated and/or genetically modified bacteria to treat cancer. Bacteria‐assisted tumor‐targeted therapy is a promising method of using bacteria as gene or drug delivery agents to treat cancers where bacteria alone act as an effective antitumor agent. Some of the selected bacterial species such as Clostridia, Bifidobacteria, and Salmonellae are used in animal models to express tumor suppressor genes, anti‐angiogenic genes, suicide genes, or tumor‐associated antigens in a particular tumor. In addition, bacteria are used as immunotherapeutic agents and their toxins/enzymes in cancer therapy, which are potent therapies in the future.126
Conclusion
After considering all the crucial areas discussed so far identified the relevant signatures concerning gut microbiomes and host miRNAs, highlighting their involvement in the cancer occurrence via dysregulation, awareness of such mechanisms of dysregulation of microbiomes and miRNAs in different cancers is essential for modulating cancer at its initiation and progression. Beyond that, both the gut microbiome and host miRNA signatures can be used as biomarkers to monitor the disease’s diagnosis, prognosis, prediction, and recurrence. In addition, the possible association of two cancers (breast and colorectal) concerning their occurrence in a patient is vital to consider when determining the timely diagnosis at an early-stage and after that appropriate administering treatment by considering the synergistic effects of those biomarkers for both cancers. Furthermore, dysregulation of miRNA and microbiome modulates the immune response in a cancer patient directing for immunosuppression, thereby becoming immunocompromised upon infections. Thus, in-depth knowledge of the behavior of the gut microbiome and miRNA, in particular cancer, will be very advantageous in developing possible noninvasive biomarkers for disease identification at an early-stage to minimize the possible risks. Alternatively, understanding the expression-based communication between the gut microbiome and host miRNAs can help design druggable targets in cancer therapies, such as miRNA-based therapies, to suppress the expression of oncomiRs while restoring expression of TS-miRs. In addition, therapeutic targets can be designed to increase commensal microbes in the gut while decreasing pathogenic microbes, thereby improving the gut health to develop antitumor immune activities. Usage of antibiotics, prebiotics, and synbiotics can boost the colonization of commensal microbes in the gut.
Furthermore, genetically engineered bacteria alone or with a combination of conventional methods experiment as future therapeutic targets known as bacteria‐assisted tumor‐targeted therapy to enhance the antitumor environment within cancer. In addition, immune checkpoint blockade therapies are favorable immunotherapeutic methods that can be used to treat cancer patients with the knowledge of the microbiome composition. In concluding remarks, it is worth understanding the combination and behavior of host miRNAs and gut microbiome, particularly in cancer diagnosis, therapeutics, and prognosis, while gathering essential facts for administering efficient and effective therapeutic methods.
Abbreviations
BC, breast cancer; ER+, estrogen-receptor-positive; ER-, estrogen-receptor-negative; PR+, progesterone-receptor-positive; PR-, progesterone-receptor-negative; HER2+, human epidermal growth factor receptor 2-positive; HER2-, human epidermal growth factor receptor 2-negative; miRNA, microRNA; CRC, colorectal cancer; HDI, Human Development Index; CIN, chromosomal instability; CIMP, CpG island methylator phenotype; MSI, microsatellite instability; oncomiR, oncogenic miRNAs; TS-miRs, tumor-suppressive miRNAs; metastamiRs, metastasis-associated miRNA; antagomirs, anti-miRs; AMOs, antisense anti-miR oligonucleotides; LNA, locked nucleic acid; TNBC, triple-negative breast cancer, TPM1, tropomyosin; CEA, carcinoembryonic antigen; CA15-3, cancer antigen, TUSC2, tumor suppressor candidate 2; EGFR, epidermal growth factor receptor, TGF-β, transforming growth factor beta; PDCD4, programmed cell death 4; BFT, Bacteroides fragilis toxin; ETBF, enterotoxigenic Bacteroides fragilis; im-miRNAs, immunomodulatory miRNAs, PD-1, programmed cell death protein 1; NK, natural killer; ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane serine protease 2, DCC, dormant cancer cells; ICB, immune checkpoint blockade; CTLA-4, cytotoxic T-lymphocyte-associated protein 4.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Breast cancer - WHO | World Health Organization [homepage on the Internet]. Available from: https://www.who.int/news-room/fact-sheets/detail/breast-cancer. Accessed June 14, 2021.
- 3.Johnson KS, Conant EF, Soo MS. Molecular subtypes of breast cancer: a review for breast radiologists. J Breast Imaging. 2021;3(1):12–24. doi: 10.1093/jbi/wbaa110 [DOI] [PubMed] [Google Scholar]
- 4.Anders CK, Johnson R, Litton J, Phillips M, Bleyer A. Breast cancer before age 40 years. Semin Oncol. 2009;36(3):237–249. doi: 10.1053/j.seminoncol.2009.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mavaddat N, Peock S, Frost D, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst. 2013;105(11):812–822. doi: 10.1093/jnci/djt095 [DOI] [PubMed] [Google Scholar]
- 6.Assi HA, Khoury KE, Dbouk H, Khalil LE, Mouhieddine TH, El Saghir NS. Epidemiology and prognosis of breast cancer in young women. J Thorac Dis. 2013;5(Suppl 1):S2–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durbecq V, Ameye L, Veys I, et al. A significant proportion of elderly patients develop hormone-dependent “luminal-B” tumors associated with aggressive characteristics. Crit Rev Oncol Hematol. 2008;67(1):80–92. doi: 10.1016/j.critrevonc.2007.12.008 [DOI] [PubMed] [Google Scholar]
- 8.Seiler A, Chen MA, Brown RL, Fagundes CP. Obesity, dietary factors, nutrition, and breast cancer risk. Curr Breast Cancer Rep. 2018;10(1):14–27. doi: 10.1007/s12609-018-0264-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 10.Sung JJ, Ng SC, Chan FK, et al. An updated Asia Pacific consensus recommendations on colorectal cancer screening. Gut. 2015;64(1):121–132. doi: 10.1136/gutjnl-2013-306503 [DOI] [PubMed] [Google Scholar]
- 11.Pourhoseingholi MA. Epidemiology and burden of colorectal cancer in Asia-Pacific region: what shall we do now? Transl Gastrointest Cancer. 2014;3(4):169–173. doi: 10.3978/j.issn.2224-4778.2014.08.10 [DOI] [Google Scholar]
- 12.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912 [DOI] [PubMed] [Google Scholar]
- 13.Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14(2):89–103. doi: 10.5114/pg.2018.81072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahnen DJ, Wade SW, Jones WF, et al. The increasing incidence of young-onset colorectal cancer: a call to action. Mayo Clin Proc. 2014;89(2):216–224. doi: 10.1016/j.mayocp.2013.09.006 [DOI] [PubMed] [Google Scholar]
- 15.Kyaw M, Sung JJ. Young-onset colorectal cancer in the Asia-Pacific region. Med J Aust. 2016;205(10):450–451. doi: 10.5694/mja16.00957 [DOI] [PubMed] [Google Scholar]
- 16.Marley AR, Nan H. Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet. 2016;7(3):105–114. [PMC free article] [PubMed] [Google Scholar]
- 17.Campos FG. Colorectal cancer in young adults: a difficult challenge. World J Gastroenterol. 2017;23(28):5041–5044. doi: 10.3748/wjg.v23.i28.5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong L, Ren H. Blood-based DNA methylation biomarkers for early detection of colorectal cancer. J Proteomics Bioinform. 2018;11(6):120–126. doi: 10.4172/jpb.1000477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogaert J, Prenen H. Molecular genetics of colorectal cancer. Ann Gastroenterol. 2014;27(1):9–14. [PMC free article] [PubMed] [Google Scholar]
- 20.Jass JR. Colorectal cancer: a multi-pathway disease. Crit Rev Oncog. 2006;12(3–4):273–287. doi: 10.1615/CritRevOncog.v12.i3-4.50 [DOI] [PubMed] [Google Scholar]
- 21.Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res. 2012;5(1):19–27. [PMC free article] [PubMed] [Google Scholar]
- 22.Tariq K, Ghias K. Colorectal cancer carcinogenesis: a review of mechanisms. Cancer Biol Med. 2016;13(1):120–135. doi: 10.20892/j.issn.2095-3941.2015.0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386(6625):623–627. doi: 10.1038/386623a0 [DOI] [PubMed] [Google Scholar]
- 24.Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, Galandiuk S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer. 2017;116(6):762–774. doi: 10.1038/bjc.2017.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11(12):1753–1761. doi: 10.1261/rna.2248605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frixa T, Donzelli S, Blandino G. Oncogenic microRNAs: key players in malignant transformation. Cancers. 2015;7(4):2466–2485. doi: 10.3390/cancers7040904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grammatikakis I, Gorospe M, Abdelmohsen K. Modulation of cancer traits by tumor suppressor microRNAs. Int J Mol Sci. 2013;14(1):1822–1842. doi: 10.3390/ijms14011822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Camarillo C, Marchat LA, Arechaga-Ocampo E, et al. MetastamiRs: non-coding microRNAs driving cancer invasion and metastasis. Int J Mol Sci. 2012;13(2):1347–1379. doi: 10.3390/ijms13021347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouyssou JM, Manier S, Huynh D, Issa S, Roccaro AM, Ghobrial IM. Regulation of microRNAs in cancer metastasis. Biochim Biophys Acta. 2014;1845(2):255–265. doi: 10.1016/j.bbcan.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Chen WD, Wang YD. The roles of the gut microbiota-miRNA interaction in the host pathophysiology. Mol Med. 2020;26(1):101. doi: 10.1186/s10020-020-00234-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kho ZY, Lal SK. The human gut microbiome - a potential controller of wellness and disease. Front Microbiol. 2018;9:1835. doi: 10.3389/fmicb.2018.01835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Zeng Y, Zeng D, et al. Probiotics and microRNA: their roles in the host-microbe interactions. Front Microbiol. 2021;11:604462. doi: 10.3389/fmicb.2020.604462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allegra A, Musolino C, Tonacci A, Pioggia G, Gangemi S. Interactions between the microRNAs and microbiota in cancer development: roles and therapeutic opportunities. Cancers. 2020;12(4):805. doi: 10.3390/cancers12040805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan C, Burns MB, Subramanian S, Blekhman R. Interaction between host microRNAs and the gut microbiota in colorectal cancer. mSystems. 2018;3(3):e00205–e00217. doi: 10.1128/mSystems.00205-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams MR, Stedtfeld RD, Tiedje JM, Hashsham SA. MicroRNAs-based inter-domain communication between the host and members of the gut microbiome. Front Microbiol. 2017;8:1896. doi: 10.3389/fmicb.2017.01896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karius T, Schnekenburger M, Dicato M, Diederich M. MicroRNAs in cancer management and their modulation by dietary agents. Biochem Pharmacol. 2012;83(12):1591–1601. doi: 10.1016/j.bcp.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 37.Bi K, Zhang X, Chen W, Diao H. MicroRNAs regulate intestinal immunity and gut microbiota for gastrointestinal health: a comprehensive review. Genes. 2020;11(9):1075. doi: 10.3390/genes11091075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishida M, Selaru FM. miRNA-based therapeutic strategies. Curr Anesthesiol Rep. 2013;1(1):63–70. doi: 10.1007/s40139-012-0004-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedroza-Torres A, Romero-Córdoba SL, Justo-Garrido M, et al. MicroRNAs in tumor cell metabolism: roles and therapeutic opportunities. Front Oncol. 2019;9:1404. doi: 10.3389/fonc.2019.01404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimomura A, Shiino S, Kawauchi J, et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107(3):326–334. doi: 10.1111/cas.12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng EK, Li R, Shin VY, et al. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8(1):e53141. doi: 10.1371/journal.pone.0053141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin VY, Siu JM, Cheuk I, Ng EK, Kwong A. Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. Br J Cancer. 2015;112(11):1751–1759. doi: 10.1038/bjc.2015.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peña-Chilet M, Martínez MT, Pérez-Fidalgo JA, et al. MicroRNA profile in very young women with breast cancer. BMC Cancer. 2014;14:529. doi: 10.1186/1471-2407-14-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang F, Zheng Z, Guo J, Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119(3):586–593. doi: 10.1016/j.ygyno.2010.07.021 [DOI] [PubMed] [Google Scholar]
- 46.Bhat SA, Majid S, Hassan T. MicroRNAs and its emerging role as breast cancer diagnostic marker - A review. Adv Biomark Sci Technol. 2019;1:1–8. doi: 10.1016/j.abst.2019.05.001 [DOI] [Google Scholar]
- 47.Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57(1):84–91. doi: 10.1373/clinchem.2010.151845 [DOI] [PubMed] [Google Scholar]
- 48.Cortez MA, Welsh JW, Calin GA. Circulating microRNAs as noninvasive biomarkers in breast cancer. Recent Results Cancer Res. 2012;195:151–161. doi: 10.1007/978-3-642-28160-0_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niu J, Shi Y, Tan G, et al. DNA damage induces NF-κB-dependent microRNA-21 up-regulation and promotes breast cancer cell invasion. J Biol Chem. 2012;287(26):21783–21795. doi: 10.1074/jbc.M112.355495 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Tsai HP, Huang SF, Li CF, Chien HT, Chen SC. Differential microRNA expression in breast cancer with different onset age. PLoS One. 2018;13(1):e0191195. doi: 10.1371/journal.pone.0191195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zelli V, Compagnoni C, Capelli R, et al. Circulating microRNAs as prognostic and therapeutic biomarkers in breast cancer molecular subtypes. J Pers Med. 2020;10(3):98. doi: 10.3390/jpm10030098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khalighfard S, Alizadeh AM, Irani S, Omranipour R. Plasma miR-21, miR-155, miR-10b, and Let-7a as the potential biomarkers for the monitoring of breast cancer patients. Sci Rep. 2018;8(1):17981. doi: 10.1038/s41598-018-36321-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McAnena P, Tanriverdi K, Curran C, et al. Circulating microRNAs miR-331 and miR-195 differentiate local Luminal A from metastatic breast cancer. BMC Cancer. 2019;19:436. doi: 10.1186/s12885-019-5636-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Søkilde R, Persson H, Ehinger A, et al. Refinement of breast cancer molecular classification by miRNA expression profiles. BMC Genomics. 2019;20(1):503. doi: 10.1186/s12864-019-5887-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cecene G, Ak S, Eskiler GG, et al. Circulating miR-195 as a therapeutic biomarker in Turkish breast cancer patients. Asian Pac J Cancer Prev. 2016;17(9):4241–4246. [PubMed] [Google Scholar]
- 56.Sareyeldin RM, Gupta I, Al-Hashimi I, et al. Gene expression and miRNAs profiling: function and regulation in human epidermal growth factor receptor 2 (HER2)-positive breast cancer. Cancers. 2019;11(5):646. doi: 10.3390/cancers11050646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng RC, Zhang W, Yan X, et al. Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer. Med Oncol. 2013;30(1):477. doi: 10.1007/s12032-013-0477-z [DOI] [PubMed] [Google Scholar]
- 58.Gasparini P, Lovat F, Fassan M, et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci U S A. 2014;111(12):4536–4541. doi: 10.1073/pnas.1402604111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kleivi Sahlberg K, Bottai G, Naume B, et al. A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin Cancer Res. 2015;21(5):1207–1214. doi: 10.1158/1078-0432.CCR-14-2011 [DOI] [PubMed] [Google Scholar]
- 60.Shaheen J, Shahid S, Shahzadi S, Akhtar MW, Sadaf S. Identification of circulating miRNAs as noninvasive biomarkers of triple negative breast cancer in the population of Pakistan. Pakistan J Zool. 2019;51(3):1113–1121. doi: 10.17582/journal.pjz/2019.51.3.1113.1121 [DOI] [Google Scholar]
- 61.Nama S, Muhuri M, Di Pascale F, et al. MicroRNA-138 is a prognostic biomarker for triple-negative breast cancer and promotes tumorigenesis via TUSC2 repression. Sci Rep. 2019;9(1):12718. doi: 10.1038/s41598-019-49155-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li HY, Liang JL, Kuo YL, et al. miR-105/93-3p promotes chemoresistance and circulating miR-105/93-3p acts as a diagnostic biomarker for triple negative breast cancer. Breast Cancer Res. 2017;19(1):133. doi: 10.1186/s13058-017-0918-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Piasecka D, Braun M, Kordek R, Sadej R, Romanska H. MicroRNAs in regulation of triple-negative breast cancer progression. J Cancer Res Clin Oncol. 2018;144(8):1401–1411. doi: 10.1007/s00432-018-2689-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta I, Rizeq B, Vranic S, Moustafa AA, Al Farsi H. Circulating miRNAs in HER2-positive and triple negative breast cancers: potential biomarkers and therapeutic targets. Int J Mol Sci. 2020;21(18):6750. doi: 10.3390/ijms21186750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang W, Luo YP. MicroRNAs in breast cancer: oncogene and tumor suppressors with clinical potential. J Zhejiang Univ Sci B. 2015;16(1):18–31. doi: 10.1631/jzus.B1400184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5(10):1122–1143. doi: 10.7150/thno.11543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Schooneveld E, Wildiers H, Vergote I, Vermeulen PB, Dirix LY, Van Laere SJ. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res. 2015;17:21. doi: 10.1186/s13058-015-0526-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banerjee S, Tian T, Wei Z, et al. Distinct microbial signatures associated with different breast cancer types. Front Microbiol. 2018;9:951. doi: 10.3389/fmicb.2018.00951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Banerjee S, Wei Z, Tian T, et al. Prognostic correlations with the microbiome of breast cancer subtypes. Cell Death Dis. 2021;12(9):831. doi: 10.1038/s41419-021-04092-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parida S, Sharma D. The microbiome-estrogen connection and breast cancer risk. Cells. 2019;8(12):1642. doi: 10.3390/cells8121642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fernández MF, Reina-Pérez I, Astorga JM, Rodríguez-Carrillo A, Plaza-Díaz J, Fontana L. Breast cancer and its relationship with the microbiota. Int J Environ Res Public Health. 2018;15(8):1747. doi: 10.3390/ijerph15081747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao H, Chen J, Li X, Sun Q, Qin P, Wang Q. Compositional and functional features of the female premenopausal and postmenopausal gut microbiota. FEBS Lett. 2019;593(18):2655–2664. doi: 10.1002/1873-3468.13527 [DOI] [PubMed] [Google Scholar]
- 73.Zhang J, Xia Y, Sun J. Breast and gut microbiome in health and cancer. Genes Dis. 2020;8(5):581–589. doi: 10.1016/j.gendis.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luu TH, Michel C, Bard JM, Dravet F, Nazih H, Bobin-Dubigeon C. Intestinal proportion of Blautia sp. is associated with clinical stage and histoprognostic grade in patients with early-stage breast cancer. Nutr Cancer. 2017;69(2):267–275. doi: 10.1080/01635581.2017.1263750 [DOI] [PubMed] [Google Scholar]
- 75.Laborda-Illanes A, Sanchez-Alcoholado L, Dominguez-Recio ME, et al. Breast and gut microbiota action mechanisms in breast cancer pathogenesis and treatment. Cancers. 2020;12(9):2465. doi: 10.3390/cancers12092465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Urbaniak C, Gloor GB, Brackstone M, et al. The Microbiota of breast tissue and its association with breast cancer. Appl Environ Microbiol. 2016;82(16):5039–5048. doi: 10.1128/AEM.01235-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Urbaniak C, Cummins J, Brackstone M, et al. Microbiota of human breast tissue. Appl Environ Microbiol. 2014;80(10):3007–3014. doi: 10.1128/AEM.00242-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwabe R, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13(11):800–812. doi: 10.1038/nrc3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fuhrman BJ, Feigelson HS, Flores R, et al. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J Clin Endocrinol Metab. 2014;99(12):4632–4640. doi: 10.1210/jc.2014-2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zengul AG, Demark-Wahnefried W, Barnes S, et al. Associations between dietary fiber, the fecal microbiota and estrogen metabolism in postmenopausal women with breast cancer. Nutr Cancer. 2021;73(7):1108–1117. doi: 10.1080/01635581.2020.1784444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lehmann BD, Pietenpol JA. Clinical implications of molecular heterogeneity in triple negative breast cancer. Breast. 2015;24(02):S36–40. doi: 10.1016/j.breast.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schee K, Fodstad Ø, Flatmark K. MicroRNAs as biomarkers in colorectal cancer. Am J Pathol. 2010;177(4):1592–1599. doi: 10.2353/ajpath.2010.100024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reid JF, Sokolova V, Zoni E, et al. miRNA profiling in colorectal cancer highlights miR-1. Involvement in MET-dependent proliferation. Mol Cancer Res. 2012;10(4):504–515. doi: 10.1158/1541-7786.MCR-11-0342 [DOI] [PubMed] [Google Scholar]
- 84.To KK, Tong CW, Wu M, Cho WC. MicroRNAs in the prognosis and therapy of colorectal cancer: from bench to bedside. World J Gastroenterol. 2018;24(27):2949–2973. doi: 10.3748/wjg.v24.i27.2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheng H, Zhang L, Cogdell DE, et al. Circulating plasma miR141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6(3):e17745. doi: 10.1371/journal.pone.0017745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu W, Wang Z, Shen LI, Wei Q. Circulating microRNA-21 as a potential diagnostic marker for colorectal cancer: a meta-analysis. Mol Clin Oncol. 2016;4(2):237–244. doi: 10.3892/mco.2015.702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Du B, Wu D, Yang X, et al. The expression and significance of microRNA in different stages of colorectal cancer. Medicine. 2018;97(5):e9635. doi: 10.1097/MD.0000000000009635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105(12):849–859. doi: 10.1093/jnci/djt101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kubota N, Taniguchi F, Nyuya A, et al. Upregulation of microRNA-31 is associated with poor prognosis in patients with advanced colorectal cancer. Oncol Lett. 2020;19(4):2685–2694. doi: 10.3892/ol.2020.11365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nosho K, Igarashi H, Nojima M, et al. Association of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathway. Carcinogenesis. 2014;35(4):776–783. doi: 10.1093/carcin/bgt374 [DOI] [PubMed] [Google Scholar]
- 91.Zou S, Fang L, Lee MH. Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol Rep. 2018;6(1):1–12. doi: 10.1093/gastro/gox031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang T, Cai G, Qiu Y, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6(2):320–329. doi: 10.1038/ismej.2011.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sánchez-Alcoholado L, Ramos-Molina B, Otero A, et al. The role of the gut microbiome in colorectal cancer development and therapy response. Cancers. 2020;12(6):1406. doi: 10.3390/cancers12061406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cheng Y, Ling Z, Li L. The intestinal microbiota and colorectal cancer. Front Immunol. 2020;11:615056. doi: 10.3389/fimmu.2020.615056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dai Z, Zhang J, Wu Q, et al. The role of microbiota in the development of colorectal cancer. Int J Cancer. 2019;145(8):2032–2041. doi: 10.1002/ijc.32017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Raskov H, Burcharth J, Pommergaard HC. Linking gut microbiota to colorectal cancer. J Cancer. 2017;8(17):3378–3395. doi: 10.7150/jca.20497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu Y, Segelman J, Nordgren A, Lindström L, Frisell J, Martling A. Increased risk of colorectal cancer in patients diagnosed with breast cancer in women. Cancer Epidemiol. 2016;41:57–62. doi: 10.1016/j.canep.2016.01.006 [DOI] [PubMed] [Google Scholar]
- 98.Abu-Sbeih H, Ali FS, Ge PS, et al. Patients with breast cancer may be at higher risk of colorectal neoplasia. Ann Gastroenterol. 2019;32(4):400–406. doi: 10.20524/aog.2019.0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yan X, Chen X, Liang H, et al. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer. 2014;13:220. doi: 10.1186/1476-4598-13-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kundaktepe BP, Sozer V, Papila C, et al. Associations between miRNAs and two different cancers: breast and colon. Cancer Manag Res. 2020;12:871–879. doi: 10.2147/CMAR.S227628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring, and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4(3):143–159. doi: 10.1002/emmm.201100209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cho S, Tai JW, Lu LF. MicroRNAs and their targetomes in tumor-immune communication. Cancers. 2020;12(8):2025. doi: 10.3390/cancers12082025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yi M, Xu L, Jiao Y, Luo S, Li A, Wu K. The role of cancer-derived microRNAs in cancer immune escape. J Hematol Oncol. 2020;13(1):25. doi: 10.1186/s13045-020-00848-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Omar HA, El-Serafi AT, Hersi F, et al. Immunomodulatory microRNAs in cancer: targeting immune checkpoints and the tumor microenvironment. FEBS J. 2019;286(18):3540–3557. doi: 10.1111/febs.15000 [DOI] [PubMed] [Google Scholar]
- 105.Colangelo T, Polcaro G, Ziccardi P, et al. Proteomic screening identifies calreticulin as a miR-27a direct target repressing MHC class I cell surface exposure in colorectal cancer. Cell Death Dis. 2016;7(2):e2120. doi: 10.1038/cddis.2016.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pesce S, Greppi M, Ferretti E, et al. miRNAs in NK cell-based immune responses and cancer immunotherapy. Front Cell Dev Biol. 2020;8:119. doi: 10.3389/fcell.2020.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bose M, Mukherjee P. Role of microbiome in modulating immune responses in cancer. Mediators Inflamm. 2019;2019:4107917. doi: 10.1155/2019/4107917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ge Y, Wang X, Guo Y, et al. Gut microbiota influence tumor development and alter interactions with the human immune system. J Exp Clin Cancer Res. 2021;40(1):42. doi: 10.1186/s13046-021-01845-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang L, Pan J, Chen W, Jiang J, Huang J. Chronic stress-induced immune dysregulation in cancer: implications for initiation, progression, metastasis, and treatment. Am J Cancer Res. 2020;10(5):1294–1307. [PMC free article] [PubMed] [Google Scholar]
- 110.Tahaghoghi-Hajghorbani S, Zafari P, Masoumi E, et al. The role of dysregulated immune responses in COVID-19 pathogenesis. Virus Res. 2020;290:198197. doi: 10.1016/j.virusres.2020.198197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jyotsana N, King MR. The Impact of COVID-19 on cancer risk and treatment. Cell Mol Bioeng. 2020;13(4):285–291. doi: 10.1007/s12195-020-00630-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Niu P, Lei F, Gu J. Colorectal cancer and COVID-19: do we need to raise awareness and vigilance? Cancer. 2021;127(6):979–980. doi: 10.1002/cncr.33217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu C, Wang K, Zhang M, et al. High expression of ACE2 and TMPRSS2 and clinical characteristics of COVID-19 in colorectal cancer patients. NPJ Precis Oncol. 2021;5(1):1. doi: 10.1038/s41698-020-00139-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nersisyan S, Engibaryan N, Gorbonos A, Kirdey K, Makhonin A, Tonevitsky A. Potential role of cellular miRNAs in coronavirus-host interplay. PeerJ. 2020;8:e9994. doi: 10.7717/peerj.9994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sardar R, Satish D, Birla S, Gupta D. Comparative analyses of SAR-CoV2 genomes from different geographical locations and other coronavirus family genomes reveals unique features potentially consequential to host-virus interaction and pathogenesis. bioRxiv. 2020. doi: 10.1101/2020.03.21.001586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chauhan N, Jaggi M, Chauhan SC, Yallapu MM. COVID-19: fighting the invisible enemy with microRNAs. Expert Rev Anti Infect Ther. 2021;19(2):137–145. doi: 10.1080/14787210.2020.1812385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fani M, Zandi M, Ebrahimi S, Soltani S, Abbasi S. The role of miRNAs in COVID-19 disease. Future Virol. 2021;16:301–306. doi: 10.2217/fvl-2020-0389 [DOI] [Google Scholar]
- 118.Francescangeli F, De Angelis ML, Zeuner A. COVID-19: a potential driver of immune-mediated breast cancer recurrence? Breast Cancer Res. 2020;22(1):117. doi: 10.1186/s13058-020-01360-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang Y, Alderman C, Sehlaoui A, Xiao Y, Wang W. MicroRNAs as immunotherapy targets for treating gastroenterological cancers. Can J Gastroenterol Hepatol. 2018;2018:9740357. doi: 10.1155/2018/9740357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Paladini L, Fabris L, Bottai G, Raschioni C, Calin GA, Santarpia L. Targeting microRNAs as key modulators of tumor immune response. J Exp Clin Cancer Res. 2016;35:103. doi: 10.1186/s13046-016-0375-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.He W, Wang C, Mu R, et al. MiR-21 is required for antitumor immune response in mice: an implication for its bi-directional roles. Oncogene. 2017;36(29):4212–4223. doi: 10.1038/onc.2017.62 [DOI] [PubMed] [Google Scholar]
- 122.Michaille JJ, Awad H, Fortman EC, Efanov AA, Tili E. miR-155 expression in antitumor immunity: the higher, the better? Genes Chromosomes Cancer. 2019;58(4):208–218. doi: 10.1002/gcc.22698 [DOI] [PubMed] [Google Scholar]
- 123.Dai Z, Zhang J, Wu Q, et al. Intestinal microbiota: a new force in cancer immunotherapy. Cell Commun Signal. 2020;18(1):90. doi: 10.1186/s12964-020-00599-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim E, Ahn H, Park H. A review on the role of gut microbiota in immune checkpoint blockade therapy for cancer. Mamm Genome. 2021;32:223–231. doi: 10.1007/s00335-021-09867-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li W, Deng Y, Chu Q, Zhang P. Gut microbiome and cancer immunotherapy. Cancer Lett. 2019;447:41–47. doi: 10.1016/j.canlet.2019.01.015 [DOI] [PubMed] [Google Scholar]
- 126.Sedighi M, Zahedi Bialvaei A, Hamblin MR, et al. Therapeutic bacteria to combat cancer; current advances, challenges, and opportunities. Cancer Med. 2019;8(6):3167–3181. doi: 10.1002/cam4.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]