Abstract
Coenzyme F430, the prosthetic group of methyl coenzyme M reductase (MCR), is a key compound in methane metabolism. We applied coenzyme F430 as a function-specific biomarker of methanogenesis to subsurface marine sediments collected below the sulfate reduction zone to investigate the distribution and activity of methanogens. In addition, we examined the kinetics of the epimerization of coenzyme F430, which is the first stage of the degradation process after cell death, at various temperatures (4, 15, 34, 60 °C) and pH (5, 7, 9) conditions, which cover in situ conditions of drilled sediments used in this study. The degradation experiments revealed that the kinetics of the epimerization well follow the thermodynamic laws, and the half-life of coenzyme F430 is decreasing from 304 days to 11 h with increasing the in situ temperature. It indicates that the native F430 detected in the sediments is derived from living methanogens, because the abiotic degradation of F430 is much faster than the sedimentation rate and will not be fossilized. Based on coenzyme F430 analysis and degradation experiments, the native form of F430 detected in subseafloor sediments off the Shimokita Peninsula originates from living methanogen cells, which is protected from degradation in cells but disappears soon after cell death. The biomass of methanogens calculated from in situ F430 concentration and F430 contents in cultivable methanogen species decreases by 2 orders of magnitude up to a sediment depth of 2.5 km, with a maximum value at ∼70 m below the seafloor (mbsf), while the proportion of methanogens to the total prokaryotic cell abundance increases with the depth, which is 1 to 2 orders of magnitude higher than expected previously. Our results indicate the presence of undetectable methanogens using conventional techniques.
Keywords: methanogenesis, Archaea, function-specific compound analysis, coenzyme F430, biological methanogenesis, marine sediment
1. Introduction
In continental margin sediments, a vast quantity of methane is accumulated as free gas and gas hydrates,1 which has attracted increasing interest in terms of both greenhouse gas emissions and energy resources. Several biogeochemical and microbiological studies have suggested that microbial methanogenesis notably contributes to methanogenesis in subseafloor sediments.1−4
Microbial methanogenesis is one of the major metabolic pathways in an aerobic subsurface sediment including subseafloor sediments, which is mostly driven by methanogenic archaea.5−7 In general, methanogenic archaea are strict anaerobes and drive the last stage of organic matter degradation. Their activities have been observed from near the surface to a few hundred meters below the seafloor (mbsf) and potentially occur to a few km deep in continental margin sediments.8,9 Recent updates in environmental microbiology have proposed the occurrence of previously uncultivated lineages in addition to known previously cultivated methanogenic lineages in subseafloor environments.10 In contrast, the abundance and activity of methanogens and environmental factors controlling methanogenesis remain ambiguous due to the lack of a direct technique to detect ongoing microbial activities in subsurface marine sediments. Therefore, we cannot differentiate in situ and ex situ microbial methanogenesis, although it is crucial to understand not only the microbially mediated carbon cycle but also the mechanisms of gas reservoirs and gas hydrate formations (occurrence, transport, and accumulation).
Source-specific compound (biomarker) analysis has been widely applied in the investigation of biogeochemical processes and reconstruction of paleoenvironments. One of the advantages of this approach is the quantitative detection of in situ organic molecules reflecting source organisms and environmental factors when they are synthesized. Intact polar lipids of membrane lipids, for example, have been considered to investigate the distributions of archaea and bacteria in subseafloor sediments.11,12 In regard to function-specific compounds for methane metabolism, coenzyme factor F430 analysis has recently been developed and applied as a biomarker for methane-metabolizing archaea. Coenzyme F430 was originally identified as a prosthetic compound of methyl coenzyme M reductase (MCR), which catalyzes the last step of microbial methanogenesis.13,14 Coenzyme F430 is a key compound for anaerobic methanogenesis, because the reaction step associated with the MCR–coenzyme F430 complex commonly occurs in all known methanogenic pathways, including hydrogenotrophic, aceticlastic, methylotrophic methanogenesis, and newly identified methoxydotrophic pathways.13−15 Methanogenic archaea without MCR have yet to be discovered. Anaerobic methanotrophic archaea (ANMEs) undergoing reverse methanogenesis, referred to as the anaerobic oxidation of methane (AOM), are other source organisms possessing the MCR–F430 complex.16−18 The AOM process is performed by bacterial partners such as sulfate-reducing bacteria. Given that the AOM process requires electron acceptors, such as sulfate,19,20 the application of coenzyme F430 analysis in subseafloor sediments below the sulfate reduction zone should clarify in situ methanogenesis and its quantity.21−23
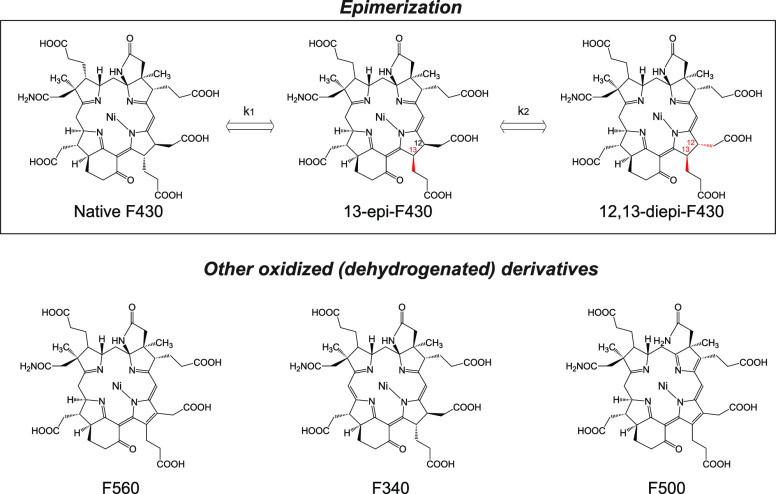
A major concern regarding the application of coenzyme F430 analysis in the assessment of ongoing modern methanogenesis in subsurface marine sediments is the accumulation of F430 as a relic of past methanogenesis. If F430 is stable and persists in subsurface sediments, it does not simply reflect modern methanogenesis, but if it immediately decomposes after cell death, it may be adopted as a function-specific biomarker of methanogenesis. Similar arguments have been made in studies on microbial membrane lipid biomarkers as well as DNA to extract modern and past biological signals from environmental samples.12,24,25 It is predictable that F430 does not survive in sediments for a long time based on observation of derivatives through epimerization and oxidation under laboratory conditions (Figure 1).26,27
Figure 1.
Structures of native coenzyme F430 and its derivatives.
Epimerization of coenzyme F430 could be a major initial degradation process in subseafloor methanogenic environments where oxidants are limited, and the kinetics should be a function of the temperature and time. However, the kinetics of the epimerization reaction has not been quantitatively investigated, except under heated conditions.26
Here, we present the kinetics of epimerization of coenzyme F430 at various temperatures and pH values to examine the stability and potential of coenzyme accumulation in subseafloor environments. Additionally, we report the quantitative distribution of coenzyme F430 in marine sediments off the Shimokita Peninsula and interpret the origin of coenzyme F430 and modern methanogenesis.
2. Results and Discussion
2.1. Rate of Epimerization and Implications for the Stability of Coenzyme F430 in Marine Sediments
To evaluate the stability of coenzyme F430 under subseafloor conditions, a crude fraction of coenzyme F430 extracted from methanogenic granules21,22 was dissolved in pH-adjusted solutions (pH values of 5, 7, 9) and incubated at temperatures of 4, 15, 34, and 60 °C in anaerobic closed vials. Native coenzyme F430 and its epimers were monitored to investigate the kinetics of the epimerization of coenzyme F430, because other known derivatives, including 12,13-didehydro-F430 (F560), 19,20-didehydro-F430 (F340), and 6-deamino-6,8,12,13-tetrahydro-F430–71-carbonic acid-72-amide (F500), are major degradation products in the presence of oxygen.27
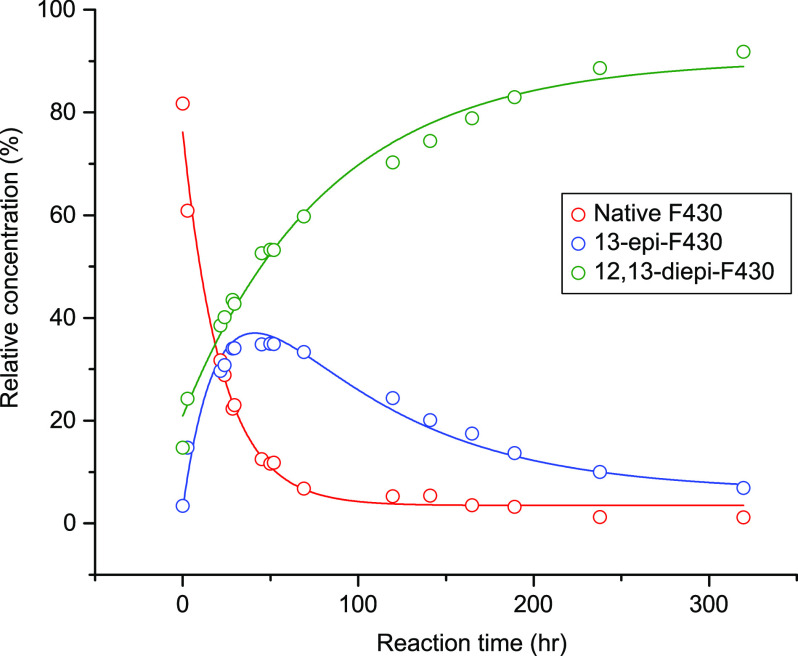
During epimerization, the conformation of native coenzyme F430 is altered into a thermodynamically stable form, 12,13-diepi-F430, via 13-epi-F430. For example, in the degradation experiment at 60 °C and pH 7, the relative abundance of native F430 exponentially decreased over time, while 12,13-diepi-F430 exponentially increased (Figure 2). Since 13-epi-F430 is an intermediate of the epimerization of coenzyme F430, an upward trend changed at approximately 20 h. Similar trends were observed at the different temperatures and pH values, although the trends became less clear at the lower temperatures due to the relatively low epimerization rate (in particular, no notable change was observed at 4 °C over 2000 h).
Figure 2.
Time-series changes in the fractions of native and epimerized F430 during the degradation experiment at 60 °C and pH 7.
The degradation and growth curves of F430 and 12,13-diepi-F430 can be fitted with the following first-order equation (R2 > 0.94)
| 1 |
where k is a rate constant and t is the reaction time. Based on the fitting curves, the rate constants for the decrease in native F430 and the increase in 12,13-diepi-F430 at several temperatures and pH values were calculated, as summarized in Table 1. The rate constants for the decrease in native F430 increased with the temperature: 4.54–6.19 × 10–2, 0.60–1.27 × 10–2, and 0.49–1.0 × 10–3 at 60, 34, and 15 °C, respectively. The constants for the increase in the above diepimer were 1.22–2.69 × 10–2, 0.22–1.04 × 10–2, and 0.38–1.07 × 10–3 at 60, 34, and 15 °C, respectively, which are slightly lower than those for the decrease in native F430. Both reactions seemed to be accelerated under both acidic and alkaline conditions. Based on the degradation experiments, the curves exhibited intercepts, which correlate with reaction temperature. The intercept would reflect the reversibility of epimerization and temperature dependence of its equilibria.
Table 1. Experimentally Determined Rate Constants of the Epimerization and Half-Lives of Native F430 and 12,13-Diepi-F430.
| pH 5 | pH 7 | pH 9 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Experimental temperature (°C) | knative | Standard (Std.) error | t1/2 (h) | knative | Std. error | t1/2 (h) | knative | Std. error | t1/2 (h) |
| 60 | 4.61 × 10–02 | 3.80 × 10–03 | 15 | 4.54 × 10–02 | 4.51 × 10–03 | 15 | 6.19 × 10–02 | 2.13 × 10–03 | 11 |
| 34 | 1.27 × 10–02 | 4.07 × 10–04 | 55 | 5.95 × 10–03 | 2.53 × 10–04 | 116 | 9.02 × 10–03 | 4.95 × 10–04 | 77 |
| 15 | 1.00 × 10–03 | 1.86 × 10–04 | 693 | 4.85 × 10–04 | 2.53 × 10–04 | 1429 | 4.52 × 10–04 | 1.62 × 10–04 | 1533 |
| 2 | 3.03 × 10–04 | 2291 | 9.49 × 10–05 | 7306 | 1.07 × 10–04 | 6464 | |||
| Experimental temperature (°C) | kdiepi | Std. error | t1/2 (h) | kdiepi | Std. error | t1/2 (h) | kdiepi | Std. error | t1/2 (h) |
|---|---|---|---|---|---|---|---|---|---|
| 60 | 2.65 × 10–02 | 2.82 × 10–03 | 26 | 1.22 × 10–02 | 1.20 × 10–03 | 57 | 2.69 × 10–02 | 8.52 × 10–04 | 26 |
| 34 | 1.04 × 10–02 | 7.07 × 10–04 | 67 | 2.20 × 10–03 | 2.51 × 10–04 | 315 | 3.70 × 10–03 | 7.74 × 10–04 | 187 |
| 15 | 1.07 × 10–03 | 2.20 × 10–04 | 648 | 3.77 × 10–04 | 2.59 × 10–04 | 1836 | 4.74 × 10–04 | 1.69 × 10–04 | 1463 |
| 2 | 4.73 × 10–04 | 1467 | 1.20 × 10–04 | 5791 | 1.25 × 10–04 | 5558 |
With the use of the calculated rate constants, the fraction of epimerized F430 relative to the total F430 (F) can be expressed with the following equation
| 2 |
The half-lives of native F430 ranged from 11 to 1533 h (Table 1), which consistently increased with decreasing temperature.
2.2. Degradation Rate of Coenzyme F430 in Marine Sediments
To our knowledge, based on field studies and laboratory experiments, the habitats of methanogens may occur in deep subsurface sediments up to 2 km below the seafloor.2,4,7,9,29,30 Just below the seafloor where sulfate remains, hydrogenotrophic methanogenesis may be excluded, but methylotrophic methanogenesis with noncompetitive carbon substrates may occur.31−34 In addition, AOM is known to be an important process in the sulfate reduction zone just beneath the sulfate–methane transition zone (SMTZ). Thus, coenzyme F430 near the SMTZ could be derived from both methanogens and ANMEs. Although methanogenesis is a major metabolic process below the sulfate reduction zone, it remains ambiguous how deep methanogenesis persists and potentially persists down to several km below the seafloor.9,35
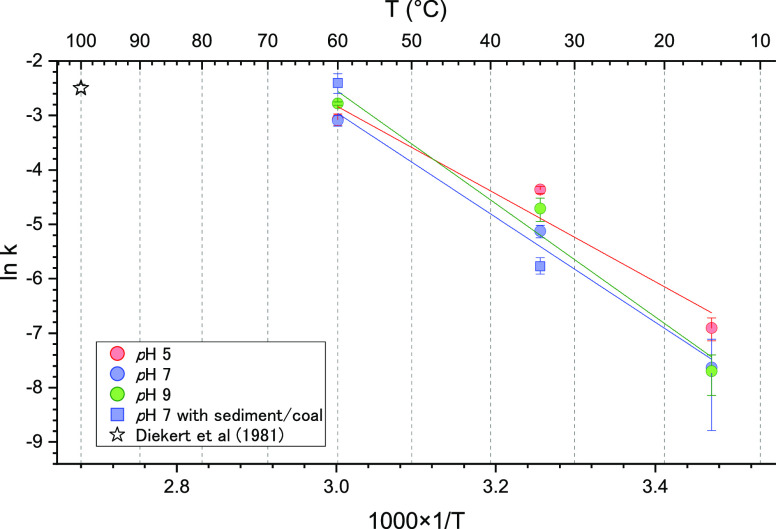
The degradation rates directly determined in this study indicate that free coenzyme F430 is readily degradable and that 99% of its native structure is epimerized within a year at temperatures higher than 15 °C when assuming an open system or an irreversible epimerization reaction. In contrast, direct determination of the degradation rate at lower temperatures, for example, at 2–4 °C, which is the general temperature at the sediment–water interface, is difficult because of the low degradation rate. However, as displayed in Figures 3 and S1, the Arrhenius plot shows a negative correlation between the rate constant and reciprocal of the temperature. The rate constants at 2 °C, therefore, can be estimated via extrapolation of the regression lines, which reveal half-lives ranging from 95 to 305 days at pH values ranging from 5 to 9. Considering a general sedimentation rate at the continental margin (10–30 mm y–1), it is considered that the degradation rate of coenzyme F430 is high enough to investigate in situ signals of microbial methanogenesis.
Figure 3.
Arrhenius plot of the epimerization of native F430.
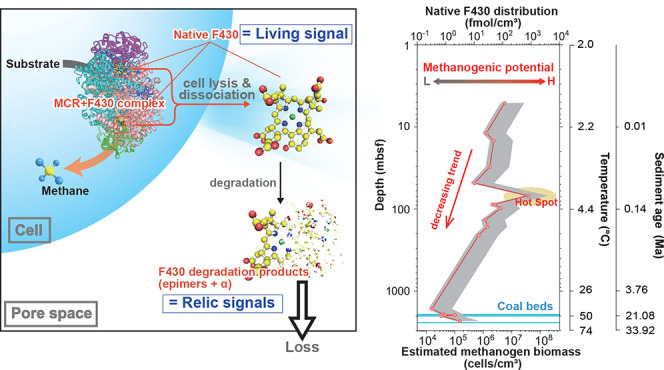
The resistance of F430 to degradation after death of methanogen cells should not be notable at this time scale, although it is bound to the MCR protein. In general, coenzyme F430 is extracted from intact cells using heat extraction in an aqueous solution, ultrasonic extraction with an acid solution, or suspension of cell extracts in aqueous ethanol,17,26,36 which indicates that coenzyme F430 is excreted from MCR via destruction of the protein structure. Coenzyme F430 bound to proteins in living cells is not epimerized even at high growth temperatures (∼65 °C) due to steric effects. However, soon after cell death, coenzyme F430 likely dissociates from proteins and is released in a free form. Thus, the native form of coenzyme F430 could be mostly degraded within a few years after cell death in natural environments (99% degraded in 6 years at 2 °C and 1 year at 15 °C) and could be a biomarker for living methanogenic archaea in deep subseafloor methanogenic sediments.
2.3. Distribution of Coenzyme F430 and Its Origin in the Subseafloor Sediments off the Shimokita Peninsula
Scientific drilling was performed during the JAMSTEC CK06–06 cruise and International Ocean Discovery Program (IODP) Expedition 337 by D/V Chikyu at Site C9001 located off the Shimokita Peninsula. The sediments were drilled down to 2466 mbsf, which is correlated with the late Oligocene (∼28 Ma), and the in situ temperature reached up to 60 °C.9,37,38 Several geochemical and microbiological studies have indicated the presence of methanogenic archaea in not only the upper part of sediments (∼200 mbsf) but also in sediments below 2000 mbsf where lignite coal is embedded.39,40 Hence, this drilling site is suitable for application of the analysis and findings from our degradation experiments of coenzyme F430. Indeed, coenzyme F430 has been detected in previous studies in certain sediment horizons.9,21,22 Here, we show the vertical distribution of coenzyme F430 in the drilled sediments and interpret its significance to the subseafloor biosphere.
The concentration of coenzyme F430 ranged from 0.34 to 795 fmol cm–3 throughout the sediment core. Overall, the concentration decreased with increasing depth; the concentration ranged from 27 to 116 fmol cm–3 just below the sulfate–methane interface (∼4 mbsf)40 and from 0.34 to 3.11 fmol cm–3 near the coalbed layer, located at approximately 2000 mbsf. This trend is similar to that of the cell abundance determined via the improved method using SYBR Green but is not correlated with the corresponding methane concentration (Figures 4 and S2).9,37,40,41
Figure 4.
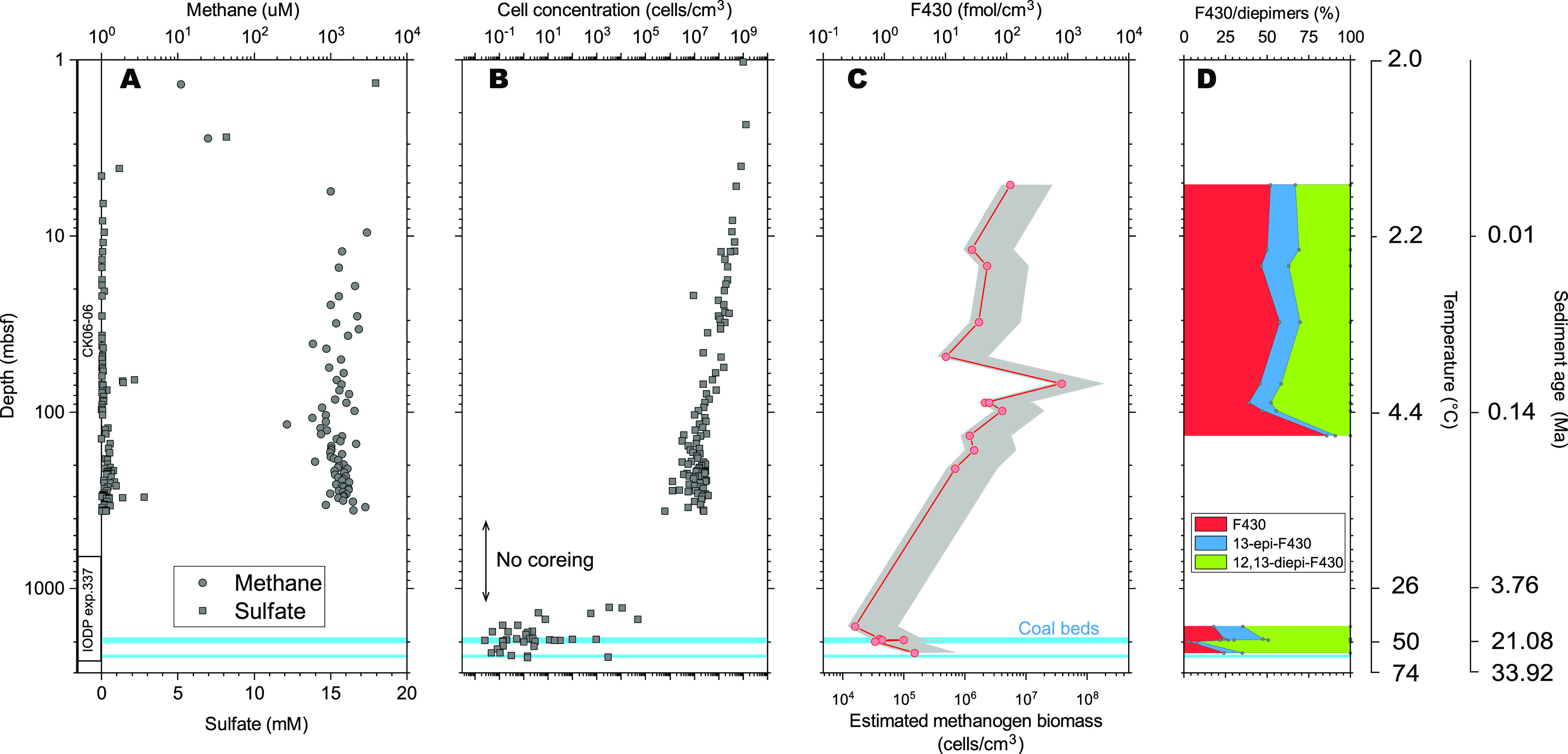
Comparison of the coenzyme F430 concentration and relative abundance of its epimers to other biogeochemical data. Concentrations of methane and sulfate (A) as well as prokaryotic cell abundance (B).9,37,40,41 Concentration of F430 and estimated methanogen biomass by eq 7. The gray shade is a possible range of the biomass given maximal and minimal F430 content in a cell of cultivable methanogens. Relative abundance of native F430 and its epimers (D). The light blue bars indicate locations of coalbeds.
To clarify the representativeness of coenzyme F430 as a biomarker tool, the source of coenzyme F430 in the sediments must be considered. The source may be characterized by source organisms and their production timing (modern vs past). Potential source organisms for coenzyme F430 are methanogens and ANMEs. However, a possible contribution originating from modern ANMEs should be excluded, because the coenzyme F430 detected in this study occurs deeper than 5 mbsf, where the sulfate required for the AOM process has almost been entirely consumed. In addition, methylthio-F430, which is a derivative of coenzyme F430 specifically detected in ANME-2, was not detected.17 Thus, the source organism of coenzyme F430 detected in sediments of this study can be derived from methanogens.
It can be examined whether F430 detected from sediments reflects relic signals by application of above determined kinetics of epimerization. However, the determined kinetics did not consider the effects of mineral matrices in sediment, which can both accelerate and inhibit degradation of organic matter. Hence, additional degradation experiments with actual sediment used in this study were performed to realistically examine the effects of mineral matrices. As the results of the epimerization experiments with sediment (collected from 30 mbsf) at 34 °C and coal (1998 mbsf) at 60 °C, rate constants k were 0.00312 and 0.091, which are quite similar to those determined by experiments without sediment (Figures 3 and S3). Major mineral components were quartz, feldspar, calcite, halite, and pyrite for sediment as well as lignite, quartz, and kaolinite for coal based on The powder X-ray diffraction (XRD) analysis (Figure S4). Thus, the effects of mineral matrices on the kinetics of epimerization of native F430 are small, and determined k values without sediment in more detail can be applicable for consideration of degradation of F430 in the sediments.
To formulate the above determined kinetics of the epimerization of coenzyme F430 and determine the potential contribution of relic signals in the sediments off Shimokita, we applied a simple box model, similar to the case of sedimentary organic matter and intact polar lipids, based on the equations below.12,42
| 3 |
| 4 |
| 5 |
where Finput is the flux of the newly synthesized coenzyme F430 in the sediments and Fdegraded is the degradation flux estimated with eq 2. The rate constant of coenzyme F430 degradation k at a given temperature was calculated based on the regression lines of the Arrhenius plot (Figure 3) as follows
| 6 |
The sediment age and temperature were estimated based on a depth-age model and the geothermal gradient measured during previous expeditions.37,38 To assess how fast coenzyme F430 is degraded in the sediments after methanogen cell death, we assumed that the initial F430 concentration is 116 fmol cm–3 at 5 mbsf (Figure 4) and that coenzyme F430 is not newly synthesized in the sediments (i.e., Finput = 0). We also assumed that coenzyme F430 is released from MCR immediately after cell death. In this simple box model, the concentration of coenzyme F430 decreased to 0.04 fmol cm–3 over the next 1 cm of burial and to 1 × 10–5 fmol cm–3 at a depth of 2 cm and beyond. Thus, coenzyme F430 does not persist in sediments, and the contributions of relic signals of methanogenesis and AOM to the native coenzyme F430 distribution should be limited. In contrast, native coenzyme F430 occurs in deep subseafloor environments, suggesting that it is newly synthesized or maintained by living methanogenic archaea.
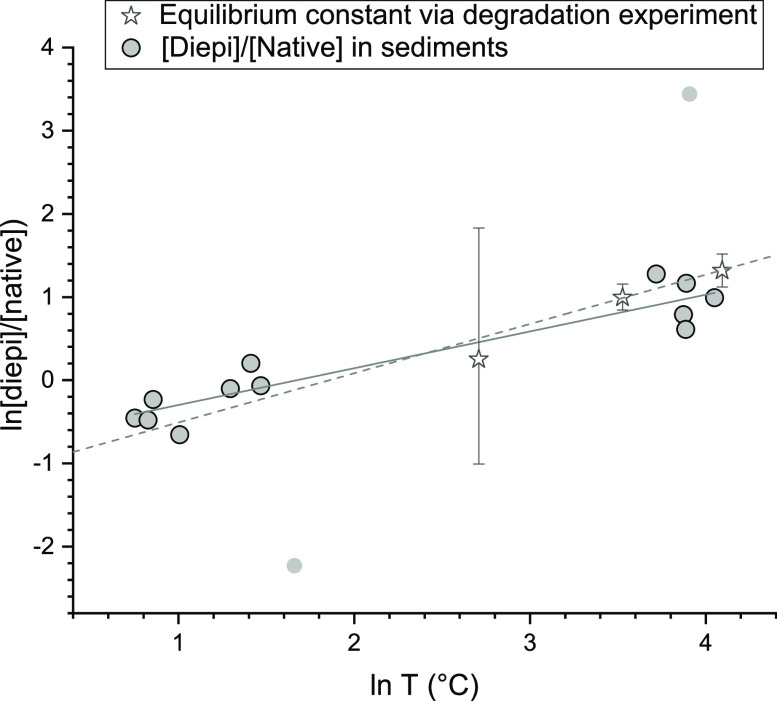
The [12,13-diepi-F430]/[native F430] ratio (diepi/native) observed in the cored sediments increased with increasing in situ temperature. Surprisingly, this trend agreed well with that of the coenzyme F430 degradation experiment at pH 7, although the complexity of both systems is quite different (Figure 5). This simply indicates that the in situ diepi/native F430 ratio is predominantly controlled by the kinetics of epimerization due to the notably higher reaction rate than that of other biotic, physical, or chemical processes.
Figure 5.
Relationship between the [12,13-diepi-F430]/[native F430] ratio and temperature in the sediments and degradation experiments. The light gray dots are treated as outliers in the calculation of the regression line.
The concentration of coenzyme F430 seemed to decrease with an apparent half-life of 0.1 million years based on the depth profile of F430 and sediment age (Figure 4). If the concentration of native F430 is only determined by new biosynthesis of coenzyme F430 (eq 5), a biosynthetic rate comparable to that of epimerization is required. However, the metabolic activity in subseafloor environments should be much lower than that under cultivation conditions, as observed in 14C tracer experiments.2,4,43,44 A model to explain in situ native F430 and its epimer distributions satisfying (1) the decrease in the F430 concentration based on a half-life of 0.1 million years, (2) abiotic degradation of native F430 and production of its epimers on a day-to-hour scale, and (3) low metabolic activities is shown in Figure 6.
Figure 6.
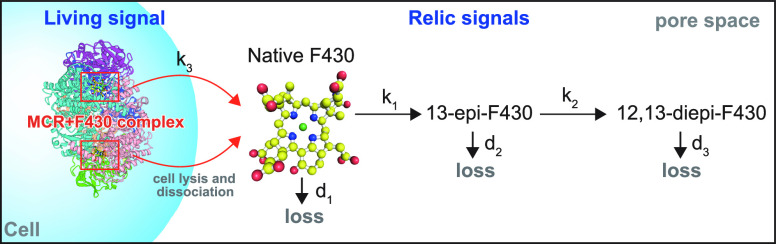
Proposed model for the protection/degradation of coenzyme F430 in the sediment pore space. 3D images of MCR and native F430 were drawn using data from Protein Data Bank (PDB ID: 1E6Y).
In this model, protein-bound coenzyme F430 biosynthesized and maintained in methanogen cells is not epimerized. After cell death, coenzyme F430 dissociates to form free native F430, and epimerization starts. Assuming an open system where free native F430 and its epimers exit the system via physical and chemical means (diffusion, migration, and further degradation), 12,13-diepi-F430 does not infinitely accumulate. This model is not definitive. For example, if assuming a salvage pathway of coenzyme F430 or its epimers, similar to the case of cobalamine (vitamin B12), other models could be possible. The important aspect of this model is the provision of a coenzyme F430 pool in cells (MCR-bound F430) to prevent coenzyme F430 from epimerizing, which satisfies the above three requirements. According to this model, the measured coenzyme F430 concentration in the sediments represents the sum of coenzyme F430 in methanogen cells and free native F430, although the latter (reaction intermediate) should be a small portion.
Thus, source characterization of coenzyme F430 based on the in situ concentration of native F430 and its derivatives as well as degradation experiments and modeling indicate that F430 detected in the sediments stems from living methanogenic archaea rather than ANMEs and relic signals.
Given that coenzyme F430 detected in the sediments originates from living methanogens, the amount of coenzyme F430 may reflect the methanogen biomass. Based on the coenzyme F430 contents in methanogen cultures (580 ± 240 nmol g–1 dry cells),26 the biomass of methanogens (n) can be calculated from the in situ coenzyme F430 concentration.23
| 7 |
where C is the concentration of coenzyme F430 in the sample and culture, and mcell is the average weight of methanogen cells in the marine sediments (referred from prokaryotic cell weight, 36 × 10–15 g).45 This conversion suitably agreed with the in situ biomass of methane-metabolizing archaea estimated via other biological techniques in natural environments, including paddy soils, groundwater, and microbial mats dominated by ANMEs.22,23
The calculated methanogen cell abundance was between 1.4 × 104 and 3.8 × 107 cells cm–3 and decreased with the depth (with increasing age and temperature) according to an entire trend of F430 concentration (Figure 4). The decreasing trend in both the prokaryotic biomass and estimated methanogen biomass could be attributed to the decreasing available resources (substrates, nutrients, pore space, etc.). Our data indicate the highest methanogen biomass at ∼70 mbsf. In contrast, the proportion of methanogen cells estimated via coenzyme F430 analysis and prokaryotic cell counting ranged from 0.2 to 164% from near-surface sediments to a depth of 208 mbsf. In deeper sediments (below 1277 mbsf), the proportion ranged at least from 34 to 313%, although total cell abundance was highly scattered.41 As an overall trend, the decreasing rate of the coenzyme F430 concentration with increasing depth was lower than that of the prokaryotic cell abundance, resulting in an increasing trend of the methanogen proportion in the deeper part of the sediments.
Since coenzyme F430 directly catalyzes methanogenic reactions, it could be adopted as a biomarker representing the in situ methanogenic reaction rate rather than the biomass. In an anaerobic bioreactor system, it has been observed that the F430 concentration is highly correlated with the methane production rate.46 Based on the specific activity of MCR determined via cell suspension and activated MCR (Min: 10–15 U mg–1, Max: 100 U mg–1),47,48 the reactivity of coenzyme F430 for methanogenesis ranges from 2.2 to 22 nmol d–1 fmol–1. When the activity of coenzyme F430 was applied to the concentration in our sediments, the calculated methanogenic activity ranged from 7 to 17000 nmol d–1 cm–3 or a tenth of the range.
2.4. Comparison to Microbiological Aspects
In general, the methanogen population in subseafloor sediments has been estimated to be as low as 0.1% or lower based on DNA-dependent molecular biological analysis of the 16S rRNA gene and methanogen- and ANME-specific mcrA genes.3,4,7 At Site C9001, the methanogen and ANME abundance levels were estimated to vary between 0.07 and 0.97%, and they decreased with increasing depth above 116 mbsf based on DNA-dependent analysis. Genes of mcrA were amplified for all sediment samples above 360 mbsf, while quantification of mcrA was unsuccessful below 78 mbsf. Thus, the decreasing trend of the methanogen abundance with increasing depth in DNA-dependent analysis is consistent with that determined in our study, but the estimated abundance seems to be much lower than that estimated via coenzyme F430 analysis. As a local observation, the highest abundance of methanogen and ANME-related DNA (Methanocellalles, Methanosarcinales, and ANME-1) has been detected, and methanogens belonging to Methanosarcina, Methanobacterium, Methanococcoides, and Methanobrevibactor have been isolated at ∼70 mbsf,39,40 which is consistent with the highest methanogen abundance estimated via coenzyme F430 analysis at similar depths.
While the decreasing trend of the methanogen biomass is consistent between both the DNA-dependent and coenzyme F430 analysis methods, there exists a large gap in the estimated biomass between these approaches. In principle, polymerase chain reaction (PCR)-dependent molecular ecological analyses are always affected by primer mismatching and other factors during PCR amplification. Recent PCR-independent techniques and metagenomic analysis methods have discovered methanogenic and methanotrophic groups possessing MCR genes based on novel euryarchaeal and noneuryachaeal lineages in various environments,10 including the subseafloor biosphere. In addition, some of these novel lineages of methane-metabolizing archaea cannot be detected by the current primer set, which might result in underestimation of the methanogenic biomass via DNA analysis.
If the biomass estimation results based on coenzyme F430 are fitted to those obtained via DNA analysis, the F430 content in cells must be ten to several hundred times higher than that under optimum growth conditions. Excess biosynthesis of coenzyme F430 has been observed only under optimal growth conditions with a sufficiently high Ni concentration.49 Our estimation already took into account the biosnthysis of excess F430, which suitably agrees with the methanogenic and ANME populations detected in natural environments such as paddy soils, microbial communities, and a deep aquifer in a previous study.22 Further biosynthesis of excess F430 unlikely occurs in subsurface marine environments where the energy and nutrient supplies are limited. The dissolved Ni concentration in pore water of marine sediment is thought to be lower than the optimum growth condition of methanogens.50,51 Assuming the Ni-limited condition of marine sediment, the excess F430 in cells become less, resulting in higher estimated methanogen biomass and proportion (Figure 4). The estimation of methanogen biomass can be optimized by further constraining the F430 content in a cell and cell weight in subseafloor environments.
Estimation of the methanogenic activity via F430 analysis also indicated a large gap compared with estimated methanogenic activity in sediment from various oceanic regions based on the 14C tracer experiments, which are generally pmol level (CO2 reduction: 0.005 to 3.69 nmol d–1 cm–3; acetate fermentation: 0.00001 to 1240 nmol d–1 cm–3).2,4,43,44 Although it is difficult to directly compare the results obtained with these technically different approaches (in situ vs ex situ methods), the activity calculated with the available specific activity of MCR in cultivated cells may cause overestimation. The activity of MCR is controlled by the oxidation state of the Ni center of coenzyme F430. In addition to the in situ F430 concentration, the turnover rate of the Ni(I) ⇄ Ni(II) cycle of coenzyme F430 in subseafloor environments should also be considered as a constraint to obtain better estimation results.
3. Experimental Section
3.1. Extraction of Coenzyme F430 from Marine Sediments
Inner parts of sediment cores were used for extraction. Samples were extracted with 1% formic acid by ultrasonication for 30 min on ice. The supernatant was recovered by centrifugation at 4 °C. This step was repeated three times. The combined supernatant was introduced to a Q Sepharose fast flow column, which is pre-equilibrated with 50 mm of Tris/HCl (pH 7.5) and washed with deionized water. The recovered eluent was introduced to a C18 SPE column, which was washed with methanol and conditioned with 1% formic acid prior to use. An absorbed F430 fraction on the column was eluted with 100% methanol. The recovered F430 fraction was dried and stored at −20 °C prior to further treatment. The dried F430 fraction was reacted with BF3/methanol in a closed vial at 40 °C for 3.5 h to convert F430 to its pentamethyl ester (F430M). Water was added to the vial, and the aqueous phase was extracted three times with dichloromethane (DCM). The organic phase was recovered after centrifugation and dried under N2 stream. Silica gel column chromatography was performed for the F430 M fraction to remove organic matrices in the sample if required.23
3.2. LC and LC/MS Analyses for F430
Method of high performance liquid chromatography (HPLC) for F430M determination was modified after Mayr et al.17 The samples were dissolved in H2O/acetonitrile (85/15, v/v). Compounds were separated using an Agilent 1100 or 1200 series HPLC equipped with a ZORBAX Eclipse XDB-C18 (4.6 × 250 mm; 5 μm p.s.; Agilent Technologies) and a guard column with 100 mM NaClO4 and acetonitrile as mobile phases, at 0.5 mL min–1 of flow rate. The gradient condition was started at 0% B followed by 30% B after 3 min and then 90% B after 90 min. For both HPLC analyses, a signal of 430 nm was monitored using a photodiode array detector (DAD).
The method of online mass spectrometry (LC/MS/MS) for F430M determination was previously reported by Kaneko et al.22 Briefly, the analysis was performed using an Agilent HPLC 1260 Infinity coupled to a 6460 Triple Quadrupole (QQQ) LC/MS system or HPLC 1260 Infinity coupled to a 6490 QQQ-LC/MS. F430 M was analyzed in positive ion mode by electrospray ionization (ESI) and an Agilent jet stream. Source and sheath gas temperatures were set at 300 and 250 °C, respectively. Source and sheath gas flow rates were set to 5 and 11 L min–1, respectively. Capillary and nozzle voltages were set at 3500 and 500 V, respectively. For MRM analysis, the fragmentor voltage was 180 V, and the collision energy was 0 V. Both precursor and product ions of F430 were set to m/z 975.4. Compound separation by HPLC was conducted using a ZORBAX Eclipse XDB-C18 (4.6 × 250 mm; 5 μm p.s.) or a ZORBAX Eclipse SB-C18 (0.5 × 250 mm; 5 μm p.s.). Mobile phases were 10 mM ammonium acetate and acetonitrile. The flow rate was 0.5 mL min–1 or 16 μL min–1. The gradient condition was started at 0% B followed by 30% B after 3 min and then 90% B after 90 min.
3.3. Degradation Experiments for Native Coenzyme F430
The F430 fraction extracted from methanogenic granules21,22 was used for the degradation experiments. The experiments were conducted in anaerobic glass vials capped with butyl rubber stoppers. The pH of solutions was adjusted to pH 5, 7, and 9 with potassium hydrogen phthalate/NaOH, KH2PO4/NaOH, and sodium tetraborate/HCl, respectively, and then deoxygenated by N2 bubbling. The dried F430 fraction was dissolved in each pH-adjusted anaerobic solution with cooling on ice in an anaerobic chamber. After the vials were capped, vials were transferred to incubators, which were set at 60, 34, 15, and 4 °C. Each solution was periodically sampled followed by methyl esterification as described above. The derivatized samples were analyzed by HPLC. To obtain precise peak area especially for epimers that are partially coeluted, peaks on a chromatogram were deconvoluted by Gaussian peak fitting using Origin graph (Figure S5).
Freeze-dried and grinded sediment (30 mbsf) and coal (1998 mbsf) were partitioned into 2 and 1 g, respectively, and transferred to 10 and 7 glass tubes. Coenzyme F430 (1 ng) dissolved in methanol (0.1 nmol/μL) and 3 mL of phosphate buffer solution 0.5 ng were injected to sediment and coal, respectively. Aliquots of 3 and 1.5 mL of pH 7 solution degassed prior to use were added to the glass tubes containing sediment. A 0.5 ng sample of coenzyme F430 and 1.5 mL of phosphate buffer solution were added to the glass tubes containing coal. The glass tubes were sealed with butyl rubber stoppers. The above treatments were performed in an anaerobic chamber. The sediment samples were incubated at 34 °C, and coal samples were incubated at 60 °C. The tubes were periodically sampled followed by extraction of F430 and methyl esterification and analyzed by LC–MS as described above. The XRD patterns of ground samples were measured by using a powder diffractometer (RINT-2500, Rigaku in GSJ-Lab, AIST) with graphite monochromatized Cu Kα radiation (λ = 0.1541 nm) at 40 kV and 100 mA in the 2–70° (2θ) range with a scanning rate of 1° min–1.
4. Conclusions
In this study, we determine the depth profile of coenzyme F430 and its degradation products in subseafloor sediments down to a depth of 2.5 km off the Shimokita Peninsula to investigate the distribution and activity of methanogens based on a newly developed function-specific biomarker for methane-metabolizing archaea. Since coenzyme F430 is a key compound directly determining the in situ methane metabolic activity via catalysis, it is theoretically adopted as a powerful function-specific biomarker. The detection approach via mcrA gene analysis relies on the same basic concept as does this method. Coenzyme F430 does not contain phylogenetic information, but in contrast, it targets all methane-metabolizing archaea, including unknown archaea. The quantitative detection of coenzyme F430, which is directly associated with enzymatic reaction of methanogenesis, more precisely represents the in situ biomass and activity of methane-metabolizing archaea living in subsurface environments than other techniques including gene analysis. The high rate of abiotic degradation also increases the utility of this method. The rate increases with increasing depth and temperature (i.e., sediment depth), which is an advantage in the study of microbial activity in subsurface environments where the activity is extremely low.
The high sensitivity of coenzyme F430 analysis is another advantage, as it quantifies the concentration of F430 in deep sediments, where quantification via DNA analysis is difficult due to the low biomass. Direct comparison to other microbial and geochemical techniques is difficult at this stage. RNA-based detection of methanogens and PCR-independent detection, such as metagenomic analysis and in situ cultivation, will enable a comparison of these fields and help to refine the precise conversion of the coenzyme F430 concentration to the in situ biomass and activity of methane-metabolizing archaea. Nevertheless, the distribution of coenzyme F430 in this study should better reflect the relative importance of the in situ methanogen biomass and activity.
In this study, we determine the kinetics of the epimerization of coenzyme F430, which represents the early stage of abiotic degradation after cell death in anaerobic subsurface environments. The half-life of coenzyme F430 become shorter with increasing temperature from a year at 2 °C of seafloor temperature to 11 h at 60 °C corresponding to the in situ temperature of 2.5 km coalbeds, indicating that the native form of coenzyme F430 disappears soon after cell death if biosynthesis or in vivo protection does not occur in sediments. In the actual distribution of coenzyme F430 in the marine sediments at Site C9001, coenzyme F430 persists down to 2.5 km below the seafloor, while the relationship between the native/diepi F430 ratio and in situ temperature suitably agrees with that determined in the F430 degradation experiments. To explain the conflicting findings whereby native coenzyme F430 persists while the ratio well reflects the thermodynamical behavior of epimerization, we suggest a model in which native F430 is protected by the host protein MCR in living methanogens cells.
Based on coenzyme F430 analysis, the methanogenic potential decreases with increasing depth, and the potential in the coalbed layer at a depth of 2.5 km is 2 orders of magnitude lower than that in near-surface sulfate-depleted sediments. A notably high potential is determined at approximately 70 mbsf, indicating a hot spot of methanogenesis. Biomass calculation based on available reference data, such as the F430 content in cultured methanogens and estimated cell weight in subsurface environments, results in values ranging from 1.4 × 104 to 3.8 × 107 cells cm–3. The methanogen population with respect to total prokaryotic biomass is 1 to 2 orders of magnitude higher than that expected by other microbial techniques (0.1 to 1%). The explored methanogenic potential based on coenzyme F430 analysis indicates that the presence of unexplored methanogens that cannot be detected by conventional techniques in subsurface marine sediments.
Acknowledgments
This work was supported by JSPS Kakenhi Grants JP25610166, JP15H05332, and 16H04083. This research used samples provided by the International Ocean Discovery Program (IODP). We thank the scientific parties of JAMSTEC CK06-06 and IODP Expedition 337. We thank two reviewers (anonymous and Dr. Marcus Elvert) for their careful and constructive comments and suggestions during a peer-review process of this manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.1c00307.
Supporting experimental data, Figures S1–S5 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Milkov A. V. Molecular and stable isotope compositions of natural gas hydrates: A revised global dataset and basic interpretations in the context of geological settings. Org. Geochem. 2005, 36, 681–702. 10.1016/j.orggeochem.2005.01.010. [DOI] [Google Scholar]
- Parkes R. J.; Webster G.; Cragg B. A.; Weightman A. J.; Newberry C. J.; Ferdelman T. G.; Kallmeyer J.; Jørgensen B. B.; Aiello I. W.; Fry J. C. Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature 2005, 436, 390–394. 10.1038/nature03796. [DOI] [PubMed] [Google Scholar]
- Biddle J. F.; Fitz-Gibbon S.; Schuster S. C.; Brenchley J. E.; House C. H. Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 10583–10588. 10.1073/pnas.0709942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry J. C.; Parkes R. J.; Cragg B. A.; Weightman A. J.; Webster G. Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiol. Ecol. 2008, 66, 181–196. 10.1111/j.1574-6941.2008.00566.x. [DOI] [PubMed] [Google Scholar]
- Reeburgh W. S. Oceanic methane biogeochemistry. Chem. Rev. 2007, 107, 486–513. 10.1021/cr050362v. [DOI] [PubMed] [Google Scholar]
- Thauer R. K.; Kaster A. K.; Seedorf H.; Buckel W.; Hedderich R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008, 6, 579–591. 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- Valentine D. L. Emerging topics in marine methane biogeochemistry. Annual Review of Marine Science 2011, 3, 147–171. 10.1146/annurev-marine-120709-142734. [DOI] [PubMed] [Google Scholar]
- Colwell F. S.; Boyd S.; Delwiche M. E.; Reed D. W.; Phelps T. J.; Newby D. T. Estimates of biogenic methane production rates in deep marine sediments at Hydrate Ridge, Cascadia margin. Appl. Environ. Microbiol. 2008, 74, 3444–3452. 10.1128/AEM.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki F.; Hinrichs K. U.; Kubo Y.; Bowles M. W.; Heuer V. B.; Hong W. L.; Hoshino T.; Ijiri A.; Imachi H.; Ito M.; Kaneko M.; Lever M. A.; Lin Y. S.; Methe B. A.; Morita S.; Morono Y.; Tanikawa W.; Bihan M.; Bowden S. A.; Elvert M.; Glombitza C.; Gross D.; Harrington G. J.; Hori T.; Li K.; Limmer D.; Liu C. H.; Murayama M.; Ohkouchi N.; Ono S.; Park Y. S.; Phillips S. C.; Prieto-Mollar X.; Purkey M.; Riedinger N.; Sanada Y.; Sauvage J.; Snyder G.; Susilawati R.; Takano Y.; Tasumi E.; Terada T.; Tomaru H.; Trembath-Reichert E.; Wang D. T.; Yamada Y. Exploring deep microbial life in coal-bearing sediment down to 2.5 km below the ocean floor. Science 2015, 349, 420–424. 10.1126/science.aaa6882. [DOI] [PubMed] [Google Scholar]
- Evans P. N.; Boyd J. A.; Leu A. O.; Woodcroft B. J.; Parks D. H.; Hugenholtz P.; Tyson G. W. An evolving view of methane metabolism in the Archaea. Nat. Rev. Microbiol. 2019, 17, 219–232. 10.1038/s41579-018-0136-7. [DOI] [PubMed] [Google Scholar]
- Lipp J. S.; Morono Y.; Inagaki F.; Hinrichs K. U. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 2008, 454, 991–994. 10.1038/nature07174. [DOI] [PubMed] [Google Scholar]
- Lipp J. S.; Hinrichs K. U. Structural diversity and fate of intact polar lipids in marine sediments. Geochim. Cosmochim. Acta 2009, 73, 6816–6833. 10.1016/j.gca.2009.08.003. [DOI] [Google Scholar]
- Thauer R. K. Biochemistry of methanogenesis: A tribute to Marjory Stephenson:1998 Marjory Stephenson prize lecture. Microbiology 1998, 144, 2377–2406. 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- Thauer R. K. Methyl (Alkyl)-coenzyme M reductases: Nickel F-430-containing enzymes involved in anaerobic methane formation and in anaerobic oxidation of methane or of short chain alkanes. Biochemistry 2019, 58, 5198–5220. 10.1021/acs.biochem.9b00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayumi D.; Mochimaru H.; Tamaki H.; Yamamoto K.; Yoshioka H.; Suzuki Y.; Kamagata Y.; Sakata S. Methane production from coal by a single methanogen. Science 2016, 354, 222–225. 10.1126/science.aaf8821. [DOI] [PubMed] [Google Scholar]
- Scheller S.; Goenrich M.; Boecher R.; Thauer R. K.; Jaun B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 2010, 465, 606–608. 10.1038/nature09015. [DOI] [PubMed] [Google Scholar]
- Mayr S.; Latkoczy C.; Krüger M.; Günther D.; Shima S.; Thauer R. K.; Widdel F.; Jaun B. Structure of an F430 variant from archaea associated with anaerobic oxidation of methane. J. Am. Chem. Soc. 2008, 130, 10758–10767. 10.1021/ja802929z. [DOI] [PubMed] [Google Scholar]
- Krüger M.; Meyerdierks A.; Glöckner F. O.; Amann R.; Widdel F.; Kube M.; Reinhardt R.; Kahnt J.; Böcher R.; Thauer R. K.; Shima S. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 2003, 426, 878–881. 10.1038/nature02207. [DOI] [PubMed] [Google Scholar]
- Haroon M. F.; Hu S.; Shi Y.; Imelfort M.; Keller J.; Hugenholtz P.; Yuan Z.; Tyson G. W. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 2013, 500, 567–570. 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- Boetius A.; Ravenschlag K.; Schubert C. J.; Rickert D.; Widdel F.; Gieseke A.; Amann R.; Jørgensen B. B.; Witte U.; Pfannkuche O. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 2000, 407, 623–626. 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- Takano Y.; Kaneko M.; Kahnt J.; Imachi H.; Shima S.; Ohkouchi N. Detection of coenzyme F430 in deep sea sediments: A key molecule for biological methanogenesis. Org. Geochem. 2013, 58, 137–140. 10.1016/j.orggeochem.2013.01.012. [DOI] [Google Scholar]
- Kaneko M.; Takano Y.; Chikaraishi Y.; Ogawa N. O.; Asakawa S.; Watanabe T.; Shima S.; Krüger M.; Matsushita M.; Kimura H.; Ohkouchi N. Quantitative analysis of coenzyme F430 in environmental samples: A new diagnostic tool for methanogenesis and anaerobic methane oxidation. Anal. Chem. 2014, 86, 3633–3638. 10.1021/ac500305j. [DOI] [PubMed] [Google Scholar]
- Kaneko M.; Takano Y.; Ogawa N. O.; Sato Y.; Yoshida N.; Ohkouchi N. Estimation of methanogenesis by quantification of coenzyme F430 in marine sediments. Geochem. J. 2016, 50, 453–460. 10.2343/geochemj.2.0410. [DOI] [Google Scholar]
- Lengger S. K.; Lipsewers Y. A.; de Haas H.; Sinninghe Damsté J. S.; Schouten S. Lack of 13C-label incorporation suggests low turnover rates of thaumarchaeal intact polar tetraether lipids in sediments from the Iceland shelf. Biogeosciences 2014, 11, 201–216. 10.5194/bg-11-201-2014. [DOI] [Google Scholar]
- Kirkpatrick J. B.; Walsh E. A.; D’Hondt S. Fossil DNA persistence and decay in marine sediment over hundred-thousand-year to million-year time scales. Geology 2016, 44, 615–618. 10.1130/G37933.1. [DOI] [Google Scholar]
- Diekert G.; Konheiser U.; Piechulla K.; Thauer R. K. Nickel requirement and factor F430 content of methanogenic bacteria. J. Bacteriol. 1981, 148, 459–464. 10.1128/jb.148.2.459-464.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor L.The Last Step of Methanogenesis in Archaea: Mechanistic and Spectroscopic Investigations of the Reactivity and Coordination Chemistry Thioether-Thiol/Phenol Model Substrates with Coen-Zyme F430 and its Nickel Model Complexes. Doctoral Thesis, Naturwissenschaften ETH Zürich, Zürich, 2001. [Google Scholar]
- Wellsbury P.; Goodman K.; Barth T.; Cragg B. A.; Barnes S. P.; Parkes R. J. Deep marine biosphere fuelled by increasing organic matter availability during burial and heating. Nature 1997, 388, 573–576. 10.1038/41544. [DOI] [Google Scholar]
- Parkes R. J.; Wellsbury P.; Mather I. D.; Cobb S. J.; Cragg B. A.; Hornibrook E. R. C.; Horsfield B. Temperature activation of organic matter and minerals during burial has the potential to sustain the deep biosphere over geological timescales. Org. Geochem. 2007, 38, 845–852. 10.1016/j.orggeochem.2006.12.011. [DOI] [Google Scholar]
- Oremland R. S.; Polcin S. Methanogenesis and sulfate reduction: Competitive and noncompetitive substrates in estuarine sediments. Appl. Environ. Microbiol. 1982, 44, 1270–1276. 10.1128/aem.44.6.1270-1276.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R. Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol. Ecol. 1999, 28, 193–202. 10.1111/j.1574-6941.1999.tb00575.x. [DOI] [Google Scholar]
- Parkes R. J.; Brock F.; Banning N.; Hornibrook E. R. C.; Roussel E. G.; Weightman A. J.; Fry J. C. Changes in methanogenic substrate utilization and communities with depth in a salt-marsh, creek sediment in southern England. Estuarine, Coastal Shelf Sci. 2012, 96, 170–178. 10.1016/j.ecss.2011.10.025. [DOI] [Google Scholar]
- Zhuang G. C.; Heuer V. B.; Lazar C. S.; Goldhammer T.; Wendt J.; Samarkin V. A.; Elvert M.; Teske A. P.; Joye S. B.; Hinrichs K. U. Relative importance of methylotrophic methanogenesis in sediments of the Western Mediterranean Sea. Geochim. Cosmochim. Acta 2018, 224, 171–186. 10.1016/j.gca.2017.12.024. [DOI] [Google Scholar]
- Takai K.; Nakamura K.; Toki T.; Tsunogai U.; Miyazaki M.; Miyazaki J.; Hirayama H.; Nakagawa S.; Nunoura T.; Horikoshi K. Cell proliferation at 122 degrees C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 10949–10954. 10.1073/pnas.0712334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiemke A. K.; Shelnutt J. A.; Scott R. A. Coordination chemistry of F430. J. Biol. Chem. 1989, 264, 11236–11245. 10.1016/S0021-9258(18)60454-5. [DOI] [PubMed] [Google Scholar]
- Aoike K.CK06–06 D/V Chikyu Shakedown Cruise Offshore Shimokita. Laboratory Operation Report; Yokohama: Tokyo, Japan, 2007.
- Inagaki F.; Hinrichs K. U.; Kubo Y.. Deep coalbed biosphere off Shimokita: Microbial processes and hydrocarbon system associated with deeply buried coalbed in the ocean; IODP Expedition 337 Preliminary Report; 2012. http://publications.iodp.org/preliminary_report/337/index.html.
- Imachi H.; Aoi K.; Tasumi E.; Saito Y.; Yamanaka Y.; Saito Y.; Yamaguchi T.; Tomaru H.; Takeuchi R.; Morono Y.; Inagaki F.; Takai K. Cultivation of methanogenic community from subseafloor sediments using a continuous-flow bioreactor. ISME J. 2011, 5, 1913–1925. 10.1038/ismej.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunoura T.; Takaki Y.; Shimamura S.; Kakuta J.; Kazama H.; Hirai M.; Masui N.; Tomaru H.; Morono Y.; Imachi H.; Inagaki F.; Takai K. Variance and potential niche separation of microbial communities in subseafloor sediments off Shimokita Peninsula, Japan. Environ. Microbiol. 2016, 18, 1889–1906. 10.1111/1462-2920.13096. [DOI] [PubMed] [Google Scholar]
- Morono Y.; Terada T.; Masui N.; Inagaki F. Discriminative detection and enumeration of microbial life in marine subsurface sediments. ISME J. 2009, 3, 503–511. 10.1038/ismej.2009.1. [DOI] [PubMed] [Google Scholar]
- Westrich J. T.; Berner R. A. The role of sedimentary organic matter in bacterial sulfate reduction: The G model tested. Limnol. Oceanogr. 1984, 29, 236–249. 10.4319/lo.1984.29.2.0236. [DOI] [Google Scholar]
- Parkes R. J.; Cragg B. A.; Wellsbury P. Recent studies on bacterial populations and processes in subseafloor sediments: A review. Hydrogeol. J. 2000, 8, 11–28. 10.1007/PL00010971. [DOI] [Google Scholar]
- Yoshioka H.; Maruyama A.; Nakamura T.; Higashi Y.; Fuse H.; Sakata S.; Bartlett D. H. Activities and distribution of methanogenic and methane-oxidizing microbes in marine sediments from the Cascadia Margin. Geobiology 2010, 8, 223–233. 10.1111/j.1472-4669.2009.00231.x. [DOI] [PubMed] [Google Scholar]
- Whitman W. B.; Coleman D. C.; Wiebe W. J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 6578–6583. 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passaris I.; Van Gaelen P.; Cornelissen R.; Simoens K.; Grauwels D.; Vanhaecke L.; Springael D.; Smets I. Cofactor F430 as a biomarker for methanogenic activity: Application to an anaerobic bioreactor system. Appl. Microbiol. Biotechnol. 2018, 102, 1191–1201. 10.1007/s00253-017-8681-y. [DOI] [PubMed] [Google Scholar]
- Goubeaud M.; Schreiner G.; Thauer R. K. Purified methyl-coenzyme-M reductase is activated when the enzyme-bound coenzyme F430 is reduced to the nickel(I) oxidation state by titanium(III) citrate. Eur. J. Biochem. 1997, 243, 110–114. 10.1111/j.1432-1033.1997.00110.x. [DOI] [PubMed] [Google Scholar]
- Becker D. F.; Ragsdale S. W. Activation of methyl-SCoM reductase to high specific activity after treatment of whole cells with sodium sulfide. Biochemistry 1998, 37, 2639–2647. 10.1021/bi972145x. [DOI] [PubMed] [Google Scholar]
- Ankel-Fuchs D.; Jaenchen R.; Gebhardt N. A.; Thauer R. K. Functional relationship between protein-bound and free factor F430 in Methanobacterium. Arch. Microbiol. 1984, 139, 332–337. 10.1007/BF00408375. [DOI] [Google Scholar]
- Shaw T. J.; Gieskes J. M.; Jahnke R. A. Early diagenesis in differing depositional environments: the response of transition metals in pore water. Geochim. Cosmochim. Acta 1990, 54, 1233–1246. 10.1016/0016-7037(90)90149-F. [DOI] [Google Scholar]
- Glass J. B.; Orphan V. J. Trace metal requirements for microbial enzymes involved in the production and consumption of methane and nitrous oxide. Front. Microbiol. 2012, 3, 61. 10.3389/fmicb.2012.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.