Abstract
Whole-genome sequencing is becoming the de facto standard for bacterial outbreak surveillance and infection prevention. This is accompanied by a variety of bioinformatic tools and needs bioinformatics expertise for implementation. However, little is known about the concordance of reported outbreaks when using different bioinformatic workflows. In this multi-centre proficiency testing among 13 major Dutch healthcare-affiliated centres, bacterial whole-genome outbreak analysis was assessed. Centres who participated obtained two randomized bacterial datasets of Illumina sequences, a Klebsiella pneumoniae and a Vancomycin-resistant Enterococcus faecium, and were asked to apply their bioinformatic workflows. Centres reported back on antimicrobial resistance, multi-locus sequence typing (MLST), and outbreak clusters. The reported clusters were analysed using a method to compare landscapes of phylogenetic trees and calculating Kendall–Colijn distances. Furthermore, fasta files were analysed by state-of-the-art single nucleotide polymorphism (SNP) analysis to mitigate the differences introduced by each centre and determine standardized SNP cut-offs. Thirteen centres participated in this study. The reported outbreak clusters revealed discrepancies between centres, even when almost identical bioinformatic workflows were used. Due to stringent filtering, some centres failed to detect extended-spectrum beta-lactamase genes and MLST loci. Applying a standardized method to determine outbreak clusters on the reported de novo assemblies, did not result in uniformity of outbreak-cluster composition among centres.
Keywords: bacterial typing, Bioinformatics, Infection Prevention Control, Outbreak analysis, Proficiency test, Whole genome sequencing
Data Summary
K. pneumoniae and E. faecium Illumina sequence data is available via BioProject PRJEB15226 and PRJEB25424, respectively. For a full list of the accession numbers, see Table S1 (available in the online version of this article). Proficiency test template sheets and associated code are available at ‘https://github.com/MUMC-MEDMIC/SIGBIO-proficiencytest’ under an MIT license.
Impact Statement.
Bacterial typing and outbreak analyses are essential for performing appropriate infection prevention control. Whole-genome sequencing (WGS) is quickly becoming the gold standard in the field, notwithstanding the bioinformatic tools used to process the data and interpret the phylogenetic relation between the bacterial pathogens are currently not standardized. To date, it remains unclear what impact the use of these different tools has on the typing outcome and interpretation of outbreaks between different centres. In this study, we performed a proficiency test that focuses on the impact of different bioinformatic tools applied by centres on interpretation and possible infection-prevention decision making. The results of this study contribute to the community by: (i) exposing the extend of variations in WGS analysis resulting from usage of different bioinformatics tools, parameters and interpretation thresholds; (ii) highlighting the shortcomings of certain bioinformatic tools and decisions; (iii) providing insights on how to improve bacterial typing. We bring to light that it is essential to apply identical bioinformatic workflows to make it possible to implement inter-laboratory surveillance on regional or national level and thus improve future outbreak analysis.
Introduction
Dissemination of pathogenic bacteria is a significant contributor to healthcare-associated infections (HAI) and a global problem. For intensive care (IC) admitted patients, 11 787 (8.3 %) patients acquired a HAI in Europe in 2017 alone [1]. Infections by antimicrobial-resistant bacteria are an increased risk for mortality [2].
Of significant interest are the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter sp.), as they are associated with a burden on the economy and adverse outcomes for hospitalized patients [2, 3]. Therefore, it is essential to curb the dissemination and infections of these nosocomial pathogens by employing proper infection-prevention measurements and typing strategies to strengthen surveillance in and around healthcare facilities.
Conventional typing methods such as PFGE [4], multi-locus sequence typing (MLST) [5] and amplification fragment length polymorphism (AFLP) [6] have been used for many years to perform outbreak analysis and made bacterial epidemiology possible. These methods are robust and have well-defined guidelines [7]. Nowadays, whole-genome sequencing (WGS) has become common, as an increased number of laboratories have adopted it. The versatility, backward compatibility and ability to measure at a detailed genomic level are significant contributors to its increased implementation [8–13], thereby phasing out conventional typing methods.
WGS provides genomic data, which can be used to find genetic sample-to-sample relations [14–16]. WGS outbreak analysis is more and more applied by hospital Infection Prevention Control (IPC) teams to trace and monitor pathogenic infections [17–20] but also to traceback the source of transmission [21, 22]. Additionally, with WGS, one can detect antimicrobial resistance (AMR) genes and virulence factors, which is a beneficial add-on for clinicians and IPC [11]. To perform bacterial whole-genome-based outbreak analysis, WGS for data needs pre-processing using either one of three strategies or a combination. (i) Reference-based: by mapping sequence reads to a reference genome and detect single-nucleotide polymorphisms (SNPs). (ii) Allelic-based: by determining the allelic content and comparing these alleles between strains, commonly referred to as core genome (cg) or whole genome (wg) MLST. (iii) k-mer based: genomic data is grouped into smaller sequences of length k, and the composition of those shorter sequences is used to detect SNPs. To accompany these strategies, a vast amount of bioinformatic tools are available [10]. To date, guidelines or quality markers for WGS outbreak analysis in nosocomial settings are still in its infancy. However, minimal sequencing quality requirements and well-defined quality markers are needed to harmonize laboratories and make inter-laboratory comparisons possible [23].
Previous studies have provided insights into the inter-laboratory comparison of WGS data. A study that assessed the reproducibility of WGS-based typing by performing a ring trial with multiple centres concluded that WGS-based typing is reproducible for Staphylococcus aureus [24]. Studies show that the identification of AMR genes is reproducible [25]. However, the translation to phenotype is inconsistent [26].
A third initiative is ongoing and initiated by The Swiss Institute of Bioinformatics. They are performing a nationwide quality assessment ring trial focusing on bacterial phylogeny to eventually start a nationwide WGS outbreak surveillance platform [23].
The variety in bioinformatic workflows for outbreak analyses applied by these studies only reflects a small portion of the total diversity of procedures used among centres. However, little is known about the congruence of identifying bacterial outbreaks among these various bioinformatic workflows.
This study assessed the comparability of bacterial outbreak analyses and outcomes performed by multiple centres in the Netherlands. We aim to (i) expose the differences in bioinformatic workflows applied by centres and their effect on cluster composition, (ii) present a strategy to assess performance between centres by using an advance analysis methodology that is easy to implement and interpret, and (iii) provide guidelines for bioinformatic workflows to perform outbreak analyses.
Methods
Sequence datasets
Illumina paired-end sequencing data was obtained from the Sequence Read Archive (SRA) and extracted using fasterq-dump (-F -S) (https://github.com/ncbi/sra-tools/tree/master/tools/fasterq-dump). For both K. pneumoniae and E. faecium, 40 random datasets were selected from BioProject PRJEB15226 [27] and PRJEB25424, respectively. To the best of our knowledge, no publicly available outbreak analysis was conducted previously on these samples. File names and FASTQ headers were anonymized before distribution to the centres. For a full list of the accession numbers, sample details and metadata, see Table S1.
Standardisation of reporting
A secure data transferring service (www.surffilesender.nl) was used to provide each participating centre three standardized excel report files including an instruction manual. The first excel file is a pipeline report file in which the participants describe their pipeline(s), QC rejection parameters, and cluster cut-offs applied on the datasets. The second and third files are sheets for KP and VRE, respectively, in which the participants report genome coverage, MLST and presence of AMR genes for each sample in the dataset, as well as the sample-to-sample relation based on clonal relatedness. Participants used their routine methods and thresholds for analysis. Participating centres were asked to fill in their analysis results in the report sheets and fill out their contact details. All excel sheets were parsed using python (version 3.7.6) and jupyter (version 4.6.1) using pandas (version 0.25.3) and NumPy (version 1.17.3). When necessary, manual inspection of assemblies was done using ABRicate (version 0.9.8) [28] or mlst (version 2.19.0) [29], and inspection of reads was done using KMA (version 1.2.26) [30] and the Resfinder database (accessed 18 June 2020). These template sheets and associated code are available at https://github.com/MUMC-MEDMIC/SIGBIO-proficiencytest.
Reporting of outbreak clusters
Participants registered the outbreak clusters by inserting values in the lower triangle of a similarity matrix by placing either 0, 0.5 or 1, which indicates for ‘not related’, ‘probably related’ or ‘related’, respectively. The lower triangle similarity matrix was converted to a square similarity matrix using python. The instruction manual explicitly stated that all strains in a cluster should be related to each other to be part of a cluster. A custom python script (available at https://github.com/MUMC-MEDMIC/SIGBIO-proficiencytest), implementing networkx (version 2.4), was used to correct these missing relations. With this script, sample-to-sample relations were represented in a network graph, and missing edges were restored between samples to complete the outbreak clusters. For example, if sample A is clonally related to sample B, and sample B clonally related to sample C, sample A and C are also part of the cluster. This missing edge was added in the graph between samples A and C to complete all edges within a cluster. The resulting network graph was converted into a dissimilarity matrix for subsequent analyses. This process was manually inspected before applying to all reported results. Sample-to-sample relations, as reported by all centres, are aggregated and visualized using Cytoscape (version 3.7.2) using Prefuse Force Directed OpenCL Layout [31].
Creating additional matrixes
A summed distance matrix (SDM) was calculated by summing all the reported dissimilarity matrices per species. A majority distance matrix (MDM) was constructed by selecting values in the SDM that scored higher than half of the number of participating centres (>6.5). Thereby, maintaining only the sample-to-sample relations which represent the majority vote.
Compare outbreak clusters among centres
Dissimilarity matrices per centre and MDM were imported in R (version 3.6.3). Dissimilarity trees were inferred by using UPGMA with hclust (version 3.6.3). A geometric median of all dissimilarity trees, according to the Kendall–Colijn metric, was calculated by using the function medTree of the R package treespace (version 1.1.3.2) [32, 33]. Additionally, all trees, including the MDM tree, were compared using the multiDist function of R package treespace per species. This resulted in a pairwise distance matrix of all trees calculated using the Kendall–Colijn metric [32]. The pairwise distance matrix was used as input for hclust to create a UPGMA tree-of-centres. Visualization of the trees and metadata was done using iTOL (version 5.5.1).
Perform SNP-cut-off sweep
Pairwise core- and whole-genome SNPs (cgSNPs, wgSNPs) was used to determine if standardized cut-offs mitigate cluster composition variation. The fasta files of all de novo assemblies provided by each centre were used as input. The pairwise SNPs were calculated by split k-mer analyses as implemented by SKA (version 1.0) [34]. In short, split k-mer files (.skf) were generated for each assembly (ska fasta, default parameters). For cgSNPs we only maintained split k-mers that were present in 90 % of all assemblies per dataset. Pairwise alignments were made (‘ska align -p 0.9’), and the SNP distance was determined using snp-dists (version 0.7.0) [35]. For wgSNP analysis, pairwise SNP distance was determined directly from the .skf files (‘ska distance’). The pairwise cg- and wgSNPs were imported into R (version 3.6.3), and a sweep cut-off was applied by setting the cut-off to a range from 0 to 150 SNPs. Samples equal to or within this cut-off were set to be part of an outbreak cluster. Additionally, all strains in an outbreak cluster were related to each other to be consistent with the proficiency test method using R package igraph (version 1.2.5) [36]. Centres were compared to each other by calculating the Kendall–Colijn distance metric using the multiDist function of R package treespace.
Results
Thirteen centres who are members of the Special Interest Group Bioinformatics in Medical Microbiology (SIG-BIMM) NL Consortium participated in this study.
Sequence types
Participating centres were asked to report on conventional MLST. All 13 centres reported on sequence types (ST). Good concordance among centres on the reported STs was observed for both the KP and the VRE dataset, and for 35/40 (87.5 %) and 38/40 (95 %) samples, no discrepant STs were reported for KP and VRE, respectively.
For the KP dataset, centre 3 may have switched sequence data of KP12 with KP13 and KP23 with KP24 (Table S2). Centre 5 reported on the least number of STs for K. pneumoniae 32/40 (80 %) and was the only centre using BioNumerics (Applied Maths, Belgium). This centre mentioned that for some of the isolates no ST could be identified because not all seven required alleles were called.
Centres 2, 5, 7, 11 and 13 mentioned sample KP23 not belonging to the K. pneumoniae species but to Klebsiella variicola , a different species in the Klebsiella pneumoniae complex [37]. Interestingly, of the seven centres (2, 3, 4, 6, 8, 9, 10 and 12) using Ridom SeqSphere+, only centre 2 identified sample KP23 as K. variicola . For KP33, two centres (2, 11) reported it as ST33, and five centres (1, 2, 9, 10 and 13) appointed it to a novel sequence type.
For the VRE set, only centre 9 reported on a discordant ST for VRE18 and VRE33 (Table S3). Manual inspection of the assembled contigs for these two datasets from centre 9 revealed for VRE18, an incomplete pstS gene (548 bp /583 bp) at the end of a contig. For VRE33, no pstS was identified. The absence of this allele leads to an entirely new ST [38].
AMR reporting
We focus on beta-lactamase and vancomycin resistance genes as they are most clinically relevant. Eleven out of thirteen centres reported the presence of AMR genes. For AMR reporting in the KP dataset, the bla CTX-M genes were in concordance among all centres for 30 out of the 40 isolates (Table S4). For KP07 and KP09 centre 9, and for KP23 centre 11 did not report a bla CTX-M gene. For KP34, only seven out of eleven centres managed to detect a bla CTX-M-14. The presence of bla CTX-M-14 in KP34 using KMA [30] was confirmed. In addition, manual inspection using ABRicate confirmed this gene was absent in the de novo assembly of the centres, which did not report bla CTX-M-14. These centres filtered out contigs smaller than 1 kb from the de novo assembly (data not shown). For bla OXA genes, all centres were in complete agreement except for strain KP09, for which centre 9 did not report a bla OXA-1 gene. For bla TEM genes, mainly bla TEM-1 was reported, and centre 1 reported on bla TEM-30 instead of bla TEM-1. Centres reported a high heterogeneity on bla SHV gene variants, as only in six out of forty samples, a single variant was reported. Centre 5 was the only centre indicating the presence of blaSHV-38, a beta-lactamase with carbapenemase activity for strain KP12 and KP30. Two centres (2 and 9) reported multiple bla SHV genes per strain for most of the K. pneumoniae strains in this study. This could be reproduced using the web service of ResFinder, for which multiple bla SHV genes were reported on the same genomic location.
For the VRE dataset, 11 out of 13 centres reported on AMR genes. Here, although variation on reporting the vanA or vanB cassette, all centres agreed on the presence of vancomycin resistance gene variant A or B per strain (Table S5). Seven out of eleven centres reported directly on the vanHAX or vanHBX cassette, and the remaining four centres reported on all separate van genes present on this cassette, including vanS, vanR, vanY and vanZ genes.
Pipeline descriptions
Ten out of thirteen centres used an allele calling method for detecting outbreak clusters, of which eight centres used Ridom SeqSphere+varying from version 4.1.9 to version 6.0.2, centre 5 used the BioNumerics (version 7.6.3) software suite for outbreak analysis, and centre 1 used Pathogenwatch (https://pathogen.watch). For allele calling, six and four centres used cgMLST, wgMLST, respectively. The remaining three centres used an SNP approach (centre 7, 11 and 13). The tools used for SNP-based outbreak analysis are either SKA or kSNP3 (Fig. 1).
Fig. 1.
UPGMA tree-of-centres for both the KP as the VRE dataset. The trees indicate the relation of reported outbreak outcome of all 13 centres. Majority and geometric median calculations are added to the UPGMA trees. The data next to the UPGMA trees show the bioinformatic workflow used per centre divided in readcleaning, assembly, and outbreak analysis tools. Furthermore, cluster definitions applied per centre are plotted in barplots and the outcome of the centres is indicated in the barplots with cluster composition. Legends are integrated in the figure.
Reported sample-to-sample relations and outbreak clusters
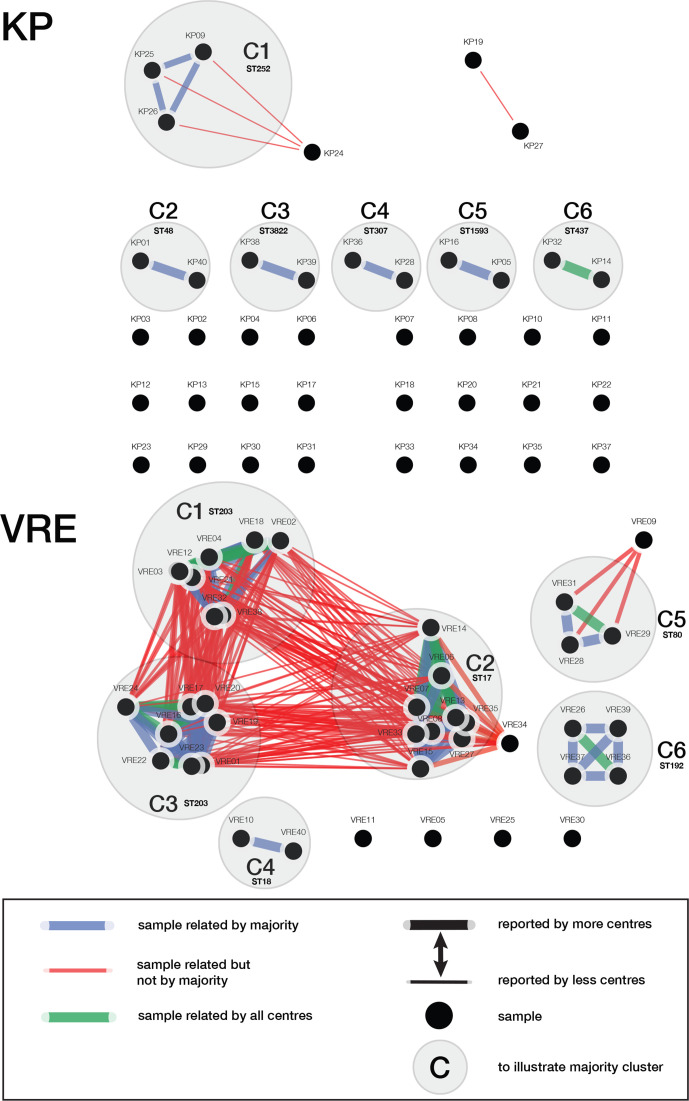
All reported sample-to-sample relations were aggregated to assess if centres reported the same outbreak clusters. In Fig. 2, the sum of all sample-to-sample relations are illustrated. For the KP dataset, we identified six independent clusters as defined by the majority of the centres. Contrarily, centres 1 and 11 reported a link between KP19 and KP27. Furthermore, only centre 5 reported sample KP24 as being part of cluster C1. Of all six majority clusters, only cluster 6 (C6) was reported by all centres.
Fig. 2.
Sample-to-sample relations as reported by the 13 participating centres. The figure is divided in the K. pneumoniae (KP) outcome and in the VRE outcome. All samples are named according the naming that was provided throughout the study. Legend of the figure can be found in the right corner of the figure. The ST that was reported by the majority of the centres was added at each cluster.
For the VRE dataset, six independent clusters were identified when using the majority vote. Firstly, centre 9 reported sample-to-sample relations between clusters that other centres identified as separate clusters: C1, C2 and C3. Centres 4 and 10 reported a maybe relation between two clusters: C1 and C3. Multiple centres linked sample VRE34 to C2, reported as related (centres 2, 3, 4 and 8), may be related (centres 6, 9 and 10) and not related (centres 1, 5,7, 11, 12 and 13). All but centre 4 reported C4. In C5, the majority did not report sample VRE09 as being part of this cluster. Nevertheless, it was reported by five centres. Lastly, C6 is well supported by 12 out of the 13 centres for containing sample VRE26, VRE36, VRE37 and VRE39. Notwithstanding, centre 7 only reported sample VRE26 with VRE36 as being linked.
KP UPGMA tree-of-centres outcome
A UPGMA tree-of-centres (Fig. 1) was used to visualize the comparison of each centre’s reported outcome, including pipeline description, cluster definition and cluster composition. Furthermore, trees identified as being the geometric median are noted in bold (Fig. 1). For KP, three groups of centres reported identical outbreak cluster content, centres 7 and 12, centres 1 and 11, and centres 2, 4, 6, 8 and 13. The latter group of centres reported identical clusters as the majority vote. Centres 3, 5, 9 and 10 reported unique cluster compositions from any other centre. Centre 5 reported the most dissimilar cluster compositions to any other centre in this study.
VRE UPGMA tree-of-centres outcome
The VRE tree-of-centres shows more dissimilarity (median 64, range 0–283) compared to the KP tree-of-centres (median 39, range 0 to 68). Centres 2, 3 and 8 reported identical outbreak clusters and content. All other centres reported unique outbreak cluster compositions. The VRE tree-of-centres clearly shows a large branch for centre 9, suggesting that centre 9 reported a very different outbreak cluster composition. Centre 9 reported on only four clusters, which included 35 out of the 40 strains. Additionally, this centre reported on the biggest individual cluster and included 26 strains, which was a composite of C1 (ST203), C2 (ST17) and C3 (ST203) (Fig. 2). However, the majority of the centres identified three separate cluster: C1, C3 (ST203) and C2 (ST17) (Fig. 2, Table S3).
Overall outbreak analysis performance
The majority vote and geometric median were calculated to evaluate centres’ outbreak analyses outcome. The KP UPGMA tree-of-centres reported an identical majority vote and the geometric median (Fig. 1). However, there is a difference between the majority vote and the geometric median in the VRE dataset, highlighting the vast diversity in reported clusters.
Centres 2 and 8 reported identical clusters for both the KP dataset as well as the VRE dataset. Centre 6 reported comparable clusters to centres 2 and 8. Centre 3 was also reporting similar to centres 2, 6 and 8 in both datasets.
Another observation is the type of cluster definitions and its wide distribution among centres. For instance, when using cg/wgMLST schemes, this varied from 7 to 150 alleles difference (Fig. 1).
Eight centres used Ridom SeqSphere+, but still, these centres reported different outbreak clusters. For the KP and VRE dataset, only four and three out of eight centres reported identical outcomes, respectively. Moreover, centres that used different bioinformatic workflows still were able to report identical outbreak clusters.
SNP-cut-off sweep
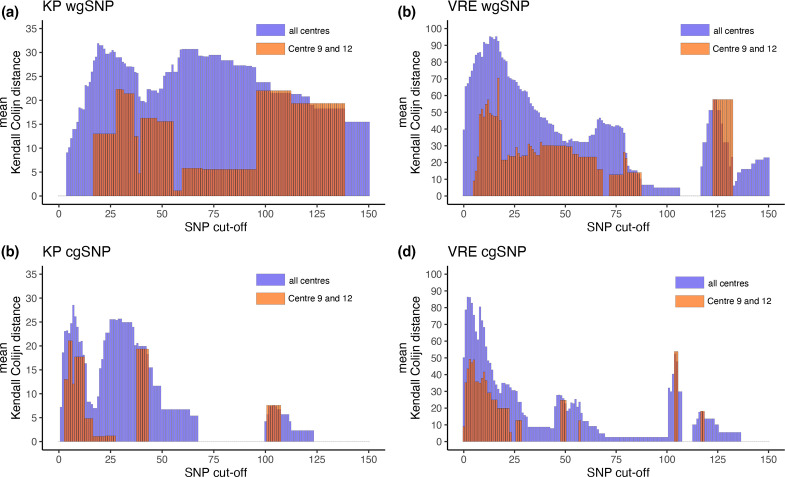
We standardized the cluster cut-offs to a range of 0 to 150 SNPs and used a single outbreak analysis tool (SKA) to remove bias that could be introduced by the various different cut-offs used by each centre. Hence, the results would give us insights into the influence of pre-processing on the outcome of each centre's cluster composition. In Fig. 3, the results of this analysis are visualized for all centres except for centre 5, who submitted faulty formatted fasta files that could not be analysed. The blue bar indicates the mean Kendall–Colijn distances calculated for all centres. The red bar indicated the distances between centres 9 and 12. A Kendall–Colijn distance of 0 would indicate no difference between the cluster composition between centres. Fig. 3(a, c) show that the blue bar plots start with 0 mean Kendall–Colijn distance due to the absence of any clusters (data not shown). The lowest cut-off to result in a full agreement of cluster composition among all centres is 68 cgSNPs in the KP dataset. Overall, the cgSNP method (Fig. 3c, d) results in lower mean Kendall–Colijn distances and shows a better agreement in cluster composition among centres compared to the wgSNP method (Fig. 3a, b).
Fig. 3.
Sweep cut-off analysis results. The barplots in this figure illustrate the mean differences between the outbreak clusters reported among centres. For example, a distance of 0 means that centres reported identical outbreak clusters. The mean distance is calculated using the Kendall–Colijn distances metric. (a) Sweep cut-off analysis of the KP samples using the wgSNP method. (b) Sweep cut-off analysis of the VRE samples using the wgSNP method. (c) Sweep cut-off analysis of the KP samples using the cgSNP method. (d) sweep cut-off analysis of the VRE samples using the cgSNP method.
Centres 9 and 12 use identical tools and near identical settings (Fig. 1), Kendall–Colijn distances are indicated by the red bars (Fig. 3). The results in Fig. 3 (indicated by the red bars) clearly show that the Kendall–Colijn distance is lower between centres 9 and 12 compared to the distance between all centres (blue bars). The lowest SNP cut-off resulting in identical reporting between centre 9 and 12 is 24 cgSNPs for the VRE dataset (Fig. 3d) and 28 cgSNPs for the KP dataset (Fig. 3c).
Impact on IPC measures
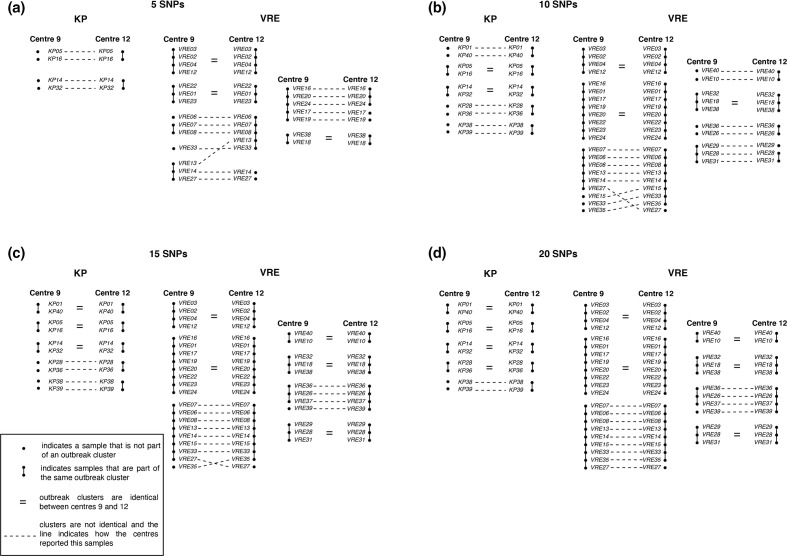
To study these differences in more detail and see the effect on potential IPC measures, cut-offs 5, 10, 15 and 20 cgSNPs were used to illustrate the differences between the sample-to-sample trees of centres 9 and 12 (see Fig. 4). Fig. 4(a) illustrates the sample-to-sample trees with a cut-off of five SNPs. Centre 9 does not have samples clustered for the KP dataset, whereas centre 12 already has two clusters of two samples each. For the VRE dataset, both centres have samples clustered. However, the composition of the clusters is not always identical. For instance, centre 12 has a cluster of five samples, of which centre 9 reported VRE33 not being part of a cluster and VRE13 being part of a different cluster. Furthermore, with 10, 15 and 20 SNPs (Fig. 4b–d), we also observe differences between centres 9 and 12 in outbreak cluster composition for both the KP and the VRE datasets.
Fig. 4.
Illustration of differences in sample-to-sample relations between centre 9 and centre 12. This figure illustrates for a sweep cut-off of 5, 10, 15 and 20 SNPs using the cgSNP method the differences in outbreak cluster composition between centre 9 and centre 12. Both given for the KP as well as the VRE samples.
Discussion
This study aimed to assess the reproducibility of WGS-based bacterial outbreak analysis and interpretation in the medical microbiology laboratory. Thirteen Dutch hospitals and university medical centres participated and entered this study with the same WGS datasets and reported results for outbreak clusters, AMR genes and ST. Hence, any form of variation or bias introduced during the sample preparation was mitigated.
Results presented here demonstrated an evident lack of reproducibility among centres, caused by differences in outbreak cluster definitions, bioinformatic workflow and quality control. The four most important findings were: (i) the large variety in cluster definitions leading to a large diversity in reported outbreak clusters, which, in the current situation, makes it impossible to compare outbreak clusters across centres. (ii) In light of the current situation, it is unachievable to obtain identical clusters when using a standardized cut-off because data processing introduces bias. (iii) The failure of detecting specific loci, such as ESBL and housekeeping genes, due to mis-assembly and too stringent post-processing. (iv) Imprecise data entry leads to erroneous conclusions. In a real-world scenario, all these issues will affect outbreak management, which impacts patient and healthcare worker safety.
To move the field of clinical bacterial typing and outbreak detection forward, we provide guidelines and recommendations based on our findings. These guidelines help to establish a workflow that has reproducible outcome, thereby minimizing the discrepancies between centres. Yet, we are aware this list is far from absolute.
Tools: All tools used in the bioinformatic workflows should be deterministic, if possible, to guarantee fully reproducible results.
Verification of species: perform identification of species, to ensure proper sample handling.
Contamination: perform identification of sample composition using a metagenomic tool [39], as contamination will affect analyses.
AMR detection and MLST typing: perform gene detection preferably using a de novo assembly-free method such as KMA [30]. This method can detect AMR and housekeeping genes using raw sequence reads as input and measure these targets’ sequence depth.
Automation of pipelines and reporting: the use of a bioinformatic management system will assist to create reproducible data analyses and facilitates standardize reporting. Furthermore, automation will limit manual intervention, which is known to be error prone.
Harmonize workflows: identical workflows ought to be used to be able to compare, share and integrate data.
Outbreak cluster comparison
The differences in reported outbreak cluster composition among centres cannot be strictly appointed to the use of specific tools. No clear relation between reported cluster outcome and use of tool or methodology was observed (Fig. 1). Three groups of centres (centres 2, 4, 6 and 8; centres 1 and 11; centres 7 and 12) used different tools for outbreak analyses yet reported identical cluster compositions. On the contrary, not all centres using Ridom Seqsphere+ (eight out of thirteen centres) reported identical cluster composition. Based on these contradictory results, we cannot appoint the effect of a particular tool on cluster composition.
To exclude the possibility that all bias was introduced using different thresholds or different outbreak analysis tools, we used a single tool and a range of thresholds to determine the cluster composition of the assemblies generated in each centre. This analysis clearly illustrated that using a single outbreak analysis tool and defining standardized SNP cut-offs is not sufficient to obtain identical cluster compositions, since the impact of pre-processing already heavily impacts the cluster outcome (Fig. 3). Even when comparing the two most closely related centres in terms of methodology and tools used, we still observe differences in outcome, leading to significant implications for outbreak management and IPC. Fig. 4 highlights the differences in outcome in a sample-to-sample comparison. These findings support the need for a more standardized way of bacterial outbreak analysis to circumvent most of these short-comings.
In our final analysis, we focused on SNP analysis and determining outbreak clusters using SNP cut-offs. These cut-offs are often calculated ad hoc [15, 40] and differ significantly among studies [41]. Combined with our findings, we can conclude that using these cut-offs when using non-identical bioinformatic outbreak analysis workflows is futile. Other analysis strategies have been proposed, for example, a method that uses a probabilistic method to infer transmissions to help solving these [42].
ST
All centres were in excellent concordance on the STs of the strains used in this study, however reporting-errors were detected for two VREs strains (VRE18 and VRE33), potentially impacting the final epidemiological assessment. All but one centre reported these two strains as ST17 and ST203, common nosocomial VRE [43–45]. One centre classified these two strains to an ST with an absent pstS, one of the seven genes in the MLST scheme for E. faecium . This would indicate the presence of rare types of VRE. However, no pstS-null vanB VRE has been reported, and only recently, the first non-typeable VRE isolates associated with a pstS-null genotype carrying a vanA cassette have been described in Australia, Korea and Scotland [38, 46, 47]. The misinterpretation was introduced by mis-assembly or too stringent post-processing and may lead to different interpretations when reporting on routine surveillance or bacterial outbreaks.
AMR
Not all centres reported on the presence of specific beta-lactamase genes. Also, high variation was observed in the reported SHV genes. Many, but not all, of these bla SHV genes result in an ESBL phenotype [48]. In addition, not all bla CTX-M genes were recovered by some centres. In a study investigating the reproducibility of AMR gene reporting, Doyle et al. reported similar discordance. However, the discordance in the reported gene variant was only minor in the genotypic resistance prediction [26]. Contradictory, in our study, both discordance in the gene variant reporting and false absence of ESBL genes was observed. Although we did not request genotypic resistance prediction reporting, failure to detect ESBL genes will influence resistance prediction. This can be of major impact as international guidelines advise contact isolation for patients carrying ESBL Enterobacteriaceae [49]. This problem is minor in practice, as strains are commonly phenotypically characterized for their AMR profile in a clinical microbiology laboratory. Analysis of how the false absence of the ESBL genes occurred demonstrated that centres that missed ESBL genes (bla CTX-M-14) removed all small contigs of up to 1 kb during post-processing. Resistance genes are often located on transposons or plasmids, which are difficult to assemble using short-read sequencing data. These hard to assemble regions can then be assembled into small contigs, sometimes of <1 kb, which would be removed by stringent post-processing. Normally, small contigs are removed as they are often associated with contamination. To overcome the failure of detecting AMR genes, one could use an assembly-free method such as KMA [30] or ARIBA [50] or simply retain these small contigs.
Data entry
This study evaluates the reporting of AMR, ST and outbreak clusters, performed by molecular trained staff, to IPC teams, thereby mimicking a crucial procedure in outbreak management. However, in this study, we found multiple incidences of inaccurate or incorrect reporting of results, such as, (i) swapping of samples KP12 with KP13 and KP23 with KP24 by centre 3, and, (ii) incomplete reporting of sample-to-sample relations, which mainly occurred by centre 9 in the VRE dataset (data not shown). These flaws in data entry can have significant consequences for IPC. It may result in extra costs and could potentially miss or identify new faulty outbreaks, leading to further transmission and follows into the closure of hospital wards, lack of patient safety and even loss of human lives [51]. When implementing WGS procedures, medical microbiology laboratories should carefully follow international norms and guidelines relating to data management [52].
Limitations
We are aware that this study is focused on the dry-lab part of outbreak analysis, thereby not taking into account the wet-lab. Assessing the combination of wet- and dry-lab will result in even larger discrepancies than observed in our study. To date, little effort has been conducted to assess the reproducibility of outbreak analyses in a clinical context. Wet-lab reproducibility has been previously evaluated but all used a single bioinformatic analysis method [24, 53]. Notwithstanding, the current situation is that centres in the Netherlands that adopted WGS-based outbreak analyses use a plethora of bioinformatic workflows. As a result, centres may communicate outcomes to each other without knowing if these results are interchangeable and may not be reproducible. Moreover, communicating outbreaks to national reference laboratories for surveillance and monitoring purposes is essential to mitigate nationwide outbreaks and prevent further spread.
Conclusion
To conclude, our study demonstrates limited reproducibility among centres applying WGS for bacterial outbreaks and AMR detection in the Netherlands. This will inevitably negatively impact IPC, healthcare workers’ and patients’ health and safety. Therefore, we advise the need for more collaboration among centres to better assess outbreaks and AMR detection through optimization and harmonization of bioinformatic tools. This would include extensive proficiency testing, open-source data sharing and formulation of guidelines [54]. Eventually, leading to harmonization of protocols and guidelines to minimize centre-to-centre variability. Furthermore, we provided guidelines for bioinformatic workflow setup, which would address most of the issues detected in this study.
Supplementary Data
Funding information
The authors received no specific grant from any funding agency.
Acknowledgements
All members of the SIG Bioinformatics in Medical Microbiology NL Consortium for supporting this study initiative. In particular the following who actively contributed to this study: W.A. van der Reijden (Streeklab Haarlem); S.D. Pas, A. Vrolijk and S. Oome (Microvida); Y. Bisselink, F. Bosma and S. Rosema (UMCG); L.M. Schouls, A. Hendrickx, M. van Santen, S. Witteveen (RIVM); S. Hermann and W. Bekers (UMCU); B.C.L. van der Putten (AIGHD); J.J. Verweij and J.J.J.M. Stohr (ETZ); J.M. Fonville and P. van Alphen (PAMM); E.C.J. Claas, M.E.M. Kraakman and S. Nooij (LUMC); A. Burggraaf and P.W. Smit (Maasstad Hospital). The Special Interest Group Bioinformatics in Medical Microbiology (SIG-BIMM) NL Consortium is comprised of: AmsterdamUMC, Amsterdam, The Netherlands; Amsterdam Institute for Global Health and Development, Amsterdam, The Netherlands; ETZ, Tilburg, The Netherlands; LUMC, Leiden, The Netherlands; Maasstad Ziekenhuis, Rotterdam, The Netherlands; Microvida, Breda, The Netherlands; MUMC+, Maastricht, The Netherlands; PAMM, Veldhoven, The Netherlands; RadboudUMC, Nijmegen, The Netherlands; RIVM, Bilthoven, The Netherlands; Streeklab Haarlem, Haarlem, The Netherlands; UMCG, Groningen, The Netherlands; UMCU, Utrecht, The Netherlands. *The Special Interest Group Bioinformatics in Medical Microbiology (SIG-BIMM) NL Consortium is comprised of: AmsterdamUMC, Amsterdam, The Netherlands; Amsterdam Institute for Global Health and Development, Amsterdam, The Netherlands; ETZ, Tilburg, The Netherlands; LUMC, Leiden, The Netherlands; Maasstad Ziekenhuis, Rotterdam, The Netherlands; Microvida, Breda, The Netherlands; MUMC+, Maastricht, The Netherlands; PAMM, Veldhoven, The Netherlands; RadboudUMC, Nijmegen, The Netherlands; RIVM, Bilthoven, The Netherlands; Streeklab Haarlem, Haarlem, The Netherlands; UMCG, Groningen, The Netherlands; UMCU, Utrecht, The Netherlands.
Author contributions
S.P.M., H.F.L.W. and L.B.A. conceived and supervised the study. J.P.M.C. and C.J. designed and performed the meta-analysis, data analysis, created figures and wrote manuscript. J.W. A.R. performed data interpretation and assisted in writing. Members of the SIG Bioinformatics in Medical Microbiology N. L., Consortium co-designed the study and participated in the study. All authors read and approved the final manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AFLP, amplification fragment length polymorphism; AMR, antimicrobial resistance; cgMLST, core-genome MLST; cgSNPs, core-genome SNPs; ESBL, extended spectrum beta-lactamase; HAI, healthcare-associated infections; IC, intensive care; IPC, Infection Prevention Control; KMA, k-mer alignment; KP, K. pneumoniae; MDM, majority distance matrix; MLST, multi-locus sequence typing; PFGE, Pulsed-field Gel Electrophoresis; SDM, summed distance matrix; SKA, Split Kmer Analysis; SNP, single-nucleotide polymorphisms; SRA, Sequence Read Archive; ST, sequence type; UPGMA, unweighted pair group method with arithmetic mean; VRE, Vancomycin-resistant Enterococcus; wgMLST, whole-genome MLST; WGS, whole-genome sequencing; wgSNPs, whole-genome SNPs.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Five supplementary tables are available with the online version of this article.
References
- 1.European Centre for Disease Prevention and Control AER for 2017: Healthcare-associated infections acquired in intensive care units. 2017.
- 2.Barrasa-Villar JI, Aibar-Remón C, Prieto-Andrés P, Mareca-Doñate R, Moliner-Lahoz J. Impact on morbidity, mortality, and length of stay of hospital-acquired infections by resistant microorganisms. Clin Infect Dis. 2017;65:644–652. doi: 10.1093/cid/cix411. [DOI] [PubMed] [Google Scholar]
- 3.Founou RC, Founou LL, Essack SY. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS One. 2017;12:e0189621. doi: 10.1371/journal.pone.0189621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannerman TL, Hancock GA, Tenover FC, Miller JM. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus . J Clin Microbiol. 1995;33:551–555. doi: 10.1128/JCM.33.3.551-555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urwin R, Maiden MCJ. Multi-locus sequence typing: A tool for global epidemiology. Trends Microbiol. 2003;11:479–487. doi: 10.1016/j.tim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect. 2007;13:1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 8.Bletz S, Mellmann A, Rothgänger J, Harmsen D. Ensuring backwards compatibility: Traditional genotyping efforts in the era of whole genome sequencing. Clin Microbiol Infect. 2015;21:347. doi: 10.1016/j.cmi.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Graham RMA, Doyle CJ, Jennison A. Real-time investigation of a Legionella pneumophila outbreak using whole genome sequencing. Epidemiol Infect. 2014;142:2347–2351. doi: 10.1017/S0950268814000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quainoo S, Coolen JPM, van Hijum S, Huynen MA, Melchers WJG, et al. Whole-genome sequencing of bacterial pathogens: The future of nosocomial outbreak analysis. Clin Microbiol Rev. 2017;30:1015–1063. doi: 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schürch AC, van Schaik W. Challenges and opportunities for whole-genome sequencing–based surveillance of antibiotic resistance. Ann N Y Acad Sci. 2017;1388:108–120. doi: 10.1111/nyas.13310. [DOI] [PubMed] [Google Scholar]
- 12.Gilchrist CA, Turner SD, Riley MF, Petri WA, Hewlett EL. Whole-genome sequencing in outbreak analysis. Clin Microbiol Rev. 2015;28:541–563. doi: 10.1128/CMR.00075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: A longitudinal molecular epidemiological study. PLoS Med. 2013;10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray RR, Tatem AJ, Johnson JA, Alekseyenko A, Pybus OG, et al. Testing spatiotemporal hypothesis of bacterial evolution using methicillin-resistant Staphylococcus aureus ST239 genome-wide data within a bayesian framework. Mol Biol Evol. 2011;28:1593–1603. doi: 10.1093/molbev/msq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris SR, Cartwright EJP, Török ME, Holden MTG, Brown NM, et al. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: A descriptive study. Lancet Infect Dis. 2013;13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program Group. et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bastiaens GJH, Cremers AJH, Coolen JPM, Nillesen MT, Boeree MJ, et al. Nosocomial outbreak of multi-resistant Streptococcus pneumoniae serotype 15A in a centre for chronic pulmonary diseases. Antimicrob Resist Infect Control. 2018;7:158. doi: 10.1186/s13756-018-0457-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes A, Ballard S, Sullivan S, Marshall C. An outbreak of vanA vancomycin-resistant Enterococcus faecium in a hospital with endemic vanB VRE. Infect Dis Heal. 2019;24:82–91. doi: 10.1016/j.idh.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Cremers AJH, Coolen JPM, Bleeker-Rovers CP, van der Geest-Blankert ADJ, Haverkate D, et al. Surveillance-embedded genomic outbreak resolution of methicillin-susceptible Staphylococcus aureus in a neonatal intensive care unit. Sci Rep. 2020;10:1–10. doi: 10.1038/s41598-020-59015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Protonotariou E, Poulou A, Politi L, Sgouropoulos I, Metallidis S, et al. Hospital outbreak due to a Klebsiella pneumoniae ST147 clonal strain co-producing KPC-2 and VIM-1 carbapenemases in a tertiary teaching hospital in Northern Greece. Int J Antimicrob Agents. 2018;52:331–337. doi: 10.1016/j.ijantimicag.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 21.van Ingen J, Kohl TA, Kranzer K, Hasse B, Keller PM, et al. Global outbreak of severe Mycobacterium chimaera disease after cardiac surgery: a molecular epidemiological study. Lancet Infect Dis. 2017;17:1033–1041. doi: 10.1016/S1473-3099(17)30324-9. [DOI] [PubMed] [Google Scholar]
- 22.Hopman J, Meijer C, Kenters N, Coolen JPM, Ghamati MR, et al. Risk assessment after a severe hospital-acquired infection associated with carbapenemase-producing Pseudomonas aeruginosa . JAMA Netw Open. 2019;2:e187665. doi: 10.1001/jamanetworkopen.2018.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adrian E, Blanc Dominique S, Gilbert G, Keller Peter M, Vladimir L, et al. Improving the quality and workflow of bacterial genome sequencing and analysis: Paving the way for a switzerland-wide molecular epidemiological surveillance platform. Swiss Med Wkly. 2018;148:w14693. doi: 10.4414/smw.2018.14693. [DOI] [PubMed] [Google Scholar]
- 24.Mellmann A, Andersen PS, Bletz S, Friedrich AW, Kohl TA, et al. High interlaboratory reproducibility and accuracy of next-generation-sequencing-based bacterial genotyping in a ring trial. J Clin Microbiol. 2017;55:908–913. doi: 10.1128/JCM.02242-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jamin C, de Koster S, van Koeveringe S, de Coninck D, Mensaert K, et al. Harmonization of whole genome sequencing for outbreak surveillance of Enterobacteriaceae and Enterococci . bioRxiv. 2020 doi: 10.1099/mgen.0.000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyle RM, O’Sullivan DM, Aller SD, Bruchmann S, Clark T, et al. Discordant bioinformatic predictions of antimicrobial resistance from whole-genome sequencing data of bacterial isolates: An inter-laboratory study. Microb Genom. 2020;6 doi: 10.1099/mgen.0.000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kluytmans-van den Bergh MFQ, Bruijning-Verhagen PCJ, Vandenbroucke-Grauls C, de Brauwer E, Buiting AGM, et al. Contact precautions in single-bed or multiple-bed rooms for patients with extended-spectrum β-lactamase-producing Enterobacteriaceae in Dutch hospitals: a cluster-randomised, crossover, non-inferiority study. Lancet Infect Dis. 2019;19:1069–1079. doi: 10.1016/S1473-3099(19)30262-2. [DOI] [PubMed] [Google Scholar]
- 28.Seemann T. ABRICATE. 2020. [ Jan 24; 2020 ]. https://github.com/tseemann/abricate accessed.
- 29.Seemann T. Mlst. 2020. [ Jun 15; 2020 ]. https://github.com/tseemann/mlst accessed.
- 30.Clausen P, Aarestrup FM, Lund O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics. 2018;19:307. doi: 10.1186/s12859-018-2336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kendall M, Colijn C. Mapping phylogenetic trees to reveal distinct patterns of evolution. Mol Biol Evol. 2016;33:2735–2743. doi: 10.1093/molbev/msw124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jombart T, Kendall M, Almagro-Garcia J, treespace CC. Statistical exploration of landscapes of phylogenetic trees. Mol Ecol Resour. 2017;17:1385–1392. doi: 10.1111/1755-0998.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris SR. SKA: Split KMER analysis toolkit for bacterial genomic epidemiology. bioRxiv. 2018:453142 [Google Scholar]
- 35.Seemann T, Klötzl F, Page AJ. snp-dists: Pairwise SNP Distance Matrix from a Fasta Sequence Alignment. 2018. [Google Scholar]
- 36.Csardi G, Nepusz T. The Igraph software package for complex network researchthe igraph software package for complex network research. InterJournal Complex Syst n.d. [Google Scholar]
- 37.Rodríguez-Medina N, Barrios-Camacho H, Duran-Bedolla J, Garza-Ramos U. Klebsiella variicola: an emerging pathogen in humans. Emerg Microbes Infect. 2019;8:973–988. doi: 10.1080/22221751.2019.1634981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemonidis K, Salih TS, Dancer SJ, Hunter IS, Tucker NP. Emergence of an Australian-like pstS-null vancomycin resistant Enterococcus faecium clone in Scotland. PLoS One. 2019;14:e0218185. doi: 10.1371/journal.pone.0218185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye SH, Siddle KJ, Park DJ, Sabeti PC. Benchmarking metagenomics tools for taxonomic classification. Cell. 2019;178:779–794. doi: 10.1016/j.cell.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goyal M, Javerliat F, Palmieri M, Mirande C, Van Wamel W, et al. Genomic evolution of Staphylococcus aureus during artificial and natural colonization of the human nose. Front Microbiol. 2019;10:1525. doi: 10.3389/fmicb.2019.01525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miro E, Rossen JWA, Chlebowicz MA, Harmsen D, Brisse S, et al. Core/whole Genome Multilocus sequence typing and core genome snp-based typing of oxa-48-producing Klebsiella pneumoniae clinical isolates from Spain. Front Microbiol. 2019;10:2961. doi: 10.3389/fmicb.2019.02961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stimson J, Gardy J, Mathema B, Crudu V, Cohen T, et al. Beyond the SNP threshold: Identifying outbreak clusters using inferred transmissions. Mol Biol Evol. 2019;36:587–603. doi: 10.1093/molbev/msy242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam MMC, Seemann T, Tobias NJ, Chen H, Haring V, et al. Comparative analysis of the complete genome of an epidemic hospital sequence type 203 clone of vancomycin-resistant Enterococcus faecium . BMC Genomics. 2013 doi: 10.1186/1471-2164-14-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo AJ, Shu JC, Liu TP, Lu J-J, Lee MH, et al. Vancomycin-resistant Enterococcus faecium at a university hospital in Taiwan, 2002–2015: Fluctuation of genetic populations and emergence of a new structure type of the Tn1546-like element. J Microbiol Immunol Infect Epub ahead of print. 2018 doi: 10.1016/j.jmii.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Johnson PDR, Ballard SA, Grabsch EA, Stinear TP, Seemann T, et al. A sustained hospital outbreak of vancomycin‐resistant Eenterococcus faecium bacteremia due to emergence of vanB E. Faecium sequence type 203. J Infect Dis Epub ahead of print. 2010 doi: 10.1086/656319. [DOI] [PubMed] [Google Scholar]
- 46.Kim HM, Chung DR, Cho SY, Huh K, Kang CI, et al. Emergence of vancomycin-resistant Enterococcus faecium ST1421 lacking the pstS gene in Korea. Eur J Clin Microbiol Infect Dis. 2020;39:1349–1356. doi: 10.1007/s10096-020-03853-4. [DOI] [PubMed] [Google Scholar]
- 47.Leong KWC, Kalukottege R, Cooley LA, Anderson TL, Wells A, et al. State-Wide genomic and epidemiological analyses of Vancomycin-Resistant Enterococcus faecium in Tasmania’s public hospitals. Front Microbiol. 2019;10:2940. doi: 10.3389/fmicb.2019.02940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liakopoulos A, Mevius D, Ceccarelli D. A review of SHV extended-spectrum β-lactamases: Neglected yet ubiquitous. Front Microbiol. 2016;7:1374. doi: 10.3389/fmicb.2016.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tacconelli E, Cataldo MA, Dancer SJ, De Angelis G, Falcone M, et al. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin Microbiol Infect. 2014;20 Suppl 1:1–55. doi: 10.1111/1469-0691.12427. [DOI] [PubMed] [Google Scholar]
- 50.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, et al. ARIBA: Rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.WHO Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report. 2020. [Google Scholar]
- 52.Turner P, Fox-Lewis A, Shrestha P, Dance DAB, Wangrangsimakul T, et al. Microbiology investigation criteria for reporting objectively (micro): A framework for the reporting and interpretation of clinical microbiology data. BMC Med. 2019;17:70. doi: 10.1186/s12916-019-1301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nouws S, Bogaerts B, Verhaegen B, Denayer S, Piérard D, et al. Impact of DNA extraction on whole genome sequencing analysis for characterization and relatedness of Shiga toxin-producing Escherichia coli isolates. Sci Rep. 2020;10:14649. doi: 10.1038/s41598-020-71207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savelkoul PHM, Koopmans MPG, Schouls L, van Rhee-Luderer R, Ossewaarde JM, et al. Richtlijn Moleculaire Typering in Het Kader Van Infectiepreventie. 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.