Abstract
Despite that obesity is associated with many metabolic diseases, a significant proportion (10–30 %) of obese individuals is recognized as ‘metabolically healthy obeses’ (MHOs). The aim of the current study is to characterize the gut microbiome for MHOs as compared to ‘metabolically unhealthy obeses’ (MUOs). We compared the gut microbiome of 172 MHO and 138 MUO individuals from Chongqing (China) (inclined to eat red meat and food with a spicy taste), and performed validation with selected biomarkers in 40 MHOs and 33 MUOs from Quanzhou (China) (inclined to eat seafood and food with a light/bland taste). The genera Alistipes , Faecalibacterium and Odoribacter had increased abundance in both Chongqing and Quanzhou MHOs. We also observed different microbial functions in MUOs compared to MHOs, including an increased abundance of genes associated with glycan biosynthesis and metabolism. In addition, the microbial gene markers identified from the Chongqing cohort bear a moderate accuracy [AUC (area under the operating characteristic curve)=0.69] for classifying MHOs distinct from MUOs in the Quanzhou cohort. These findings indicate that gut microbiome is significantly distinct between MHOs and MUOs, implicating the potential of the gut microbiome in stratification and refined management of obesity.
Keywords: clinical indicators, gut microbiome, metabolic abnormality, obesity, two cohorts
Data Summary
Sequencing data are accessible in the National Center for Biotechnology Information (NCBI) database under BioProject accession number PRJNA539 850 (https://www.ncbi.nlm.nih.gov/bioproject/?term=539850).
Impact Statement.
A growing number of studies demonstrate the potential of gut microbiome interventions in obesity management, which needs optimization due to the different metabolic status in the obese population. Our study stratified the obese individuals to two groups: with (metabolically unhealthy obeses; MUOs) and without (metabolically healthy obeses; MHOs) metabolic dysbiosis, and identified a remarkably different gut microbiome between MUOs and MHOs. This suggests the necessity of stratifying obese individuals when applying the gut microbiome to manage obesity. Microbial genes, functions and markers in this study also provide additional clues for studies exploring distinct responses of obese subjects to gut-microbiome-targeted interventions.
Introduction
Obesity, which affects over 20–30 % of adults worldwide [1], has various metabolic co-morbidities including dyslipidaemia, hypertension and hyperglycaemia. The metabolic co-morbidities together define the metabolic syndrome that poses a high risk for type two diabetes and cardiovascular diseases [2]. Despite the common metabolic syndrome identified in obesity, about 10–30 % of obese individuals present no metabolic abnormalities [3, 4] (metabolically healthy obeses; MHOs). In comparison with obese individuals with metabolic syndrome (metabolically unhealthy obeses; MUOs), MHOs generally possess higher levels of insulin sensitivity, normal blood pressure and lower inflammatory profiles. Although those metabolic differences could be partly determined by visceral fat content, early weight gain and the development of adipose cells, other factors associated with the favourable metabolic profiles of MHOs remain poorly explored.
Recent studies indicated that the balance of host metabolic interactions is largely associated with the gut microbiome [5]. Alterations in host biological status, such as body weight, correlated with many changes in the gut microbiome [6, 7]. In addition, the gut microbiome affects host energy consumption and storage, the gut barrier and the host immune system [8, 9]. Specifically, the gut microbiome produces vitamins and short-chain fatty acids (SCFAs) that are essential to host metabolism and the immune system [10]. MUOs generally display an increasing abundance of bacterial membrane-derived lipopolysaccharides (LPSs) [11], which impair intestinal barrier integrity; hence, triggering host inflammatory responses and endotoxaemia.
To date, although multiple studies have highlighted the role of the gut microbiome in the pathogenesis of obesity and metabolic disorders, there are still many discrepant findings on the gut-microbiome patterns in the obese individuals with various metabolic disorders [12]. For instance, two important indexes of the gut microbiome, microbial diversity and the ratio of Firmicutes / Bacteroidetes (F/B), both showed contrary results in different obese cohorts [13]. Furthermore, a systematic review concluded that the associations between obesity and microbiome diversity were not found in studies with different populations characterized by weight or age [14]. The inconsistencies between different studies could be attributed to huge variability among individual gut microbiomes, which are sensitive to differences in environmental factors, dietary habits and even host genetics [7, 15]. Therefore, the association of the gut microbiome with health, identified in one cohort, requires further validation in another independent cohort.
In the present study, we applied metagenomic sequencing to compare the gut microbiome of 172 MHOs and 138 MUOs from Chongqing (China) (inclined to eat red meat and food with a spicy taste) and validated the main findings in 40 MHOs and 33 MUOs in Quanzhou (China) (inclined to eat seafood and food with a light/bland taste). The objectives of this study were to characterize the universal gut microbiome of these two groups, and identify microbial gene markers discriminating MHOs from MUOs in different populations. Our findings provide extensive insights into obesity management via gut-microbiome-targeted interventions.
Methods
Recruitment and physical examination of individuals
Two cohorts with a divergent characteristic of diet (Chongqing and Quanzhou are inclined to eat a spicy and a bland diet, respectively) were included in the study. Individuals were randomly recruited in the 180th Hospital of the People’s Liberation Army of China (Quanzhou, China) and Southwest Hospital (Chongqing, China), with the following inclusion criteria: (i) older than 18 years of age and younger than 75 years of age; (ii) not exposed to antibiotics, probiotics or proton pump inhibitors within 1 month before physical examination; (iii) not suffering from diarrhoea, constipation, haematochezia or other gastrointestinal infectious diseases within 1 month before physical examination; (iv) not undergoing an enema or other gastroenterology operation within 1 month before physical examination; (v) not suffering from mental disorders, autoimmune diseases or psychological imbalance; (vi) without a history of drug abuse; (vii) without regular usage of medicine, including anti-inflammatory and antidepressant drugs.
The recruited individuals were then subjected to physical measurements, including height, weight, waistline and blood pressure. Glucose, total cholesterol, triglyceride, low density lipoprotein, high density lipoprotein, uric acid (UA) and epidermal growth factor receptor (eGFR) were measured using a blood auto-analyser (Beckman Coulter AU5800).
Definition of MUO and MHO individuals
Considering the population in this study was Asian, a body mass index (BMI) of ≥25.0 kg m−2 was defined as a cut-off for obesity [16]. Among obese individuals, MUO individuals were selected based on the presence of both elevated level UAs (>400 μmol l−1) and metabolic syndrome [17]. Metabolic syndrome was defined following the Joint Committee for Developing Chinese Guidelines on the Prevention and Treatment of Dyslipidemia (JCDCG 2007) in Adults (≥3 of the 6 criteria): (i) waist >90 cm (male) or waist >85 cm (female), (ii) glucose ≥6.1 mmol l−1 (110 mg dl−1), (iii) triglyceride ≥1.7 mmol l−1 (150 mg dl−1), (iv) high density lipoprotein <1.04 mmol l−1 (40 mg dl−1), (v) blood pressure ≥130/85 mmHg, and (vi) eGFR <90 ml min−1 per 1.73 m2 [18]. In contrast, MHOs were defined as the obese individuals who presented normal level of UAs (≤400 μmol l−1) and did not meet at least four criteria of metabolic syndrome described above. Finally, 212 MHO (Chongqing n=172; Quanzhou n=40) and 171 MUO (Chongqing n=138; Quanzhou n=33) individuals were selected for the following analyses.
Stool sample collection
During physical examination, fresh stools were collected from individuals using sterile stool containers. Approximately 5 g stool from each individual was obtained using swabs (Huachenyang Technology). The stool samples were preserved in stool collection tubes (Axygen), and then transferred to a −80 °C refrigerator (DW-86L626; Haier) within half an hour.
Library construction and sequencing
DNA was extracted following the protocol of the QIAamp DNA stool mini kit (Qiagen) [19]. The quality and quantity of the extracted DNA were measured via both NanoDrop spectrophotometer (Thermo Scientific) and Qubit fluorometer (Life Technologies). The DNA fragment size was evaluated by agarose gel electrophoresis. The extracted DNA was used to construct short-insert libraries (350 bp), which were then paired-end sequenced on the Illumina HiSeq platform.
Quality control and taxonomic profiling
The sequenced paired-end reads were removed via FastQC when one read contained more than 10 % of ambiguous or 50 % of low quality (Phred quality score <5) bases [20]. The filtered reads were then mapped to the human genome (hg19) to eliminate host genomic contamination using Burrows–Wheeler Aligner (BWA) v. 0.7.13 with default parameters [21]. Species-level taxonomy and relative abundance were assessed using Metagenomic Phylogenetic Analysis (MetaPhlAn2) v. 2.6 based on the read alignment against a reference database containing approximately one million clade-specific gene markers derived from ~17 000 reference genomes [22, 23].
Gene profiling
The high-quality reads were mapped to an updated Integrated Gene Catalog (IGC) using BWA v. 0.7.13 with default parameters [21, 24]. The number of mapped reads for each gene was calculated following three alignment scenarios: (i) if a gene was aligned by a pair of reads, the two mapped reads were both counted for the gene; (ii) if more than one gene were aligned by a pair of reads, the two mapped reads were only counted for the gene with the highest alignment confidence; or (iii) if a gene was aligned by only one of the paired-end reads, the one mapped read was counted for the gene.
Based on the number of mapped reads, the relative abundance [Ab(g) relative ] of a gene (g) was calculated by measuring the proportion of the abundance [Ab(g)] of g over the sum of abundance of all the genes using the formula below:
Ab(g) relative =Ab(g)×100/Ab(G)
Ab(g)=U/L
The abundance of a gene [Ab(g)] was calculated by dividing the number of mapped reads in the gene (U) by the length the gene (L) [24]. The genes with zero abundance in more than 20 % of individuals were then removed.
Functional annotation
The relative abundance of KEGG orthology (KO) was calculated by summing up the relative abundance of corresponding genes based on the annotated results in the IGC database. Differentially enriched KO modules were predicted using the Wilcoxon rank sum test with P values (P value <0.05) adjusted with the Benjamini–Hochberg method [22].
Co-abundance gene groups (CAGs)
To compare the gut microbiome in MUOs and MHOs, genes with significantly different relative abundance between the two groups were identified using the Wilcoxon rank sum test (Benjamini–Hochberg P value <0.05). These associated genes were then clustered according to their abundance correlation across all samples [25]. Clusters with more than ten genes were identified as CAGs and used for further analysis. The relative abundance of each CAG was determined by the median relative abundance of genes contained within that CAG, and the taxonomic classification of the CAG referred to previous research [26]. Briefly, the CAG was annotated at the species level if 90 % of the genes in the CAG belonged to the same species. If a CAG could not be assigned as a species, the CAG was annotated at genus level when 80 % of the genes contained in the CAG were assigned as the same genus.
Construction of random forest models and selection of microbial gene markers
To predict metabolic status based on gut microbiome, random forest classification was performed among individuals from the Chongqing cohort. Firstly, the individuals of the Chongqing cohort were randomized into a training set (80 % of the individuals) and a validation set (20 % of the individuals). Secondly, random forest models were constructed by the training sets using the randomForest package in R. The models were then used to classify the validation sets. Area under the operating characteristic curve (AUC) was measured and visualized using the R package pROC, to assess microbial gene biomarkers. Since the classification was repeated 50 times independently, candidate biomarkers that were present in more than 40 models were selected as final microbial biomarkers to discriminate the two groups.
Statistics
All statistical analyses were performed in R (version 3.4.1). The principal coordinate analysis (PCoA) based on R package vegan was applied to reduce dimensionality among the two cohorts. Shannon diversity, gene count, the abundances of microbial species and CAG were compared between MHOs and MUOs using the Wilcoxon rank sum test (Wilcox.test) in R. Spearman correlation was applied to evaluate the association between microbial features and clinical indicators (using cor in R). P values were adjusted with the Benjamini–Hochberg method (False Discovery Rate, FDR <0.05) using the function p.adjust, and were plotted using the package ggplot2 in R [27]. The correlation of metabolic status with gut microbiome was assessed using the function adonis2 within the R package vegan. The dissimilarity distance among all individuals was calculated based on annotated microbial species and human metabolic status using Bray–Curtis and Euclidean dissimilarities, respectively.
Results
Physical characteristics and sequencing data
To compare the gut microbial composition of MHOs and MUOs, we carried out metagenomic shotgun sequencing on the faecal samples of 310 individuals from Chongqing (172 MHOs and 138 MUOs) and 73 from Quanzhou (40 MHOs and 33 MUOs) (Tables S1 and S2, available with the online version of this article). The high-quality sequencing reads (4.54 Gb per sample on average; Table S3) were mapped to microbial-taxonomy-specific marker genes for analysing taxonomic components in the gut microbiome.
Alterations of microbial composition
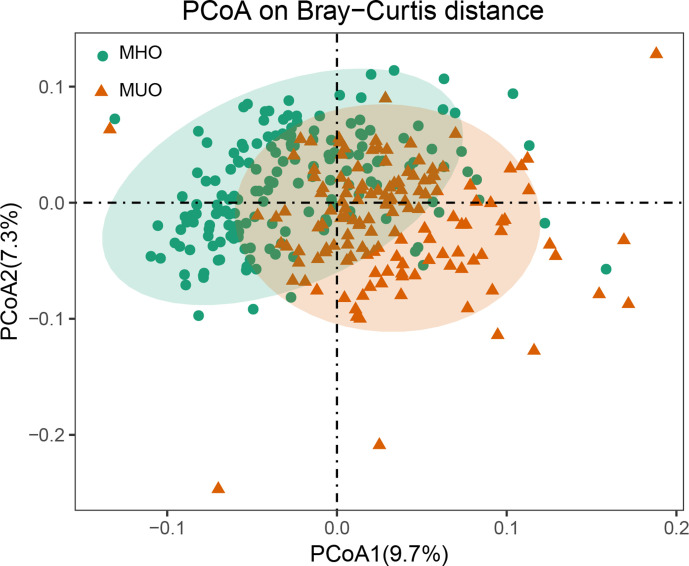
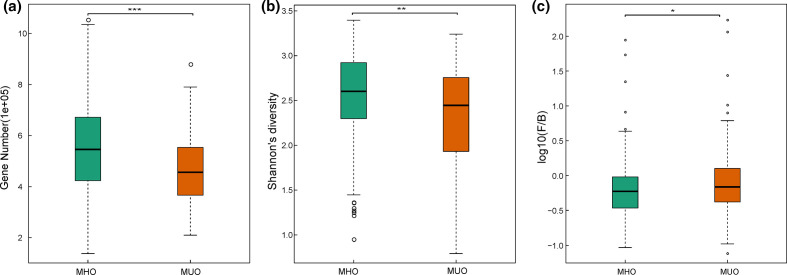
We firstly estimated the distribution of microbial samples in MHOs and MUOs from the Chongqing cohort based on microbial composition. Using PCoA, the two most significant coordinates of taxonomic profiling showed that the MHOs and MUOs formed two distinct clusters (Fig. 1). We then examined the gut microbial features, indicating that MHOs had a higher level of Shannon diversity and marginally lower ratio of F/B compared to MUOs (Fig. 2).
Fig. 1.
PCoA of microbial taxonomic profiling in MHOs and MUOs from Chongqing. Green circles represent the 172 MHOs and the brown triangles represent the 138 MUOs.
Fig. 2.
Comparison between the gut microbiome in MHO (n=172) and MUO individuals (n=138) from Chongqing. (a) Box plot of the gene count in MHO and MUO individuals; (b) Shannon diversity; and (c) F/B ratio. For (a–c), two-tailed Wilcoxon rank sum test was used to determine significance. Boxes represent the interquartile ranges (IQRs) between the first and third quartiles, and the line inside the box represents the median; whiskers represent the lowest or highest values within 1.5 times IQR from the first or third quartiles. *, **, *** represent the p-value < 0.05, < 0.01, < 0.001 respectively.
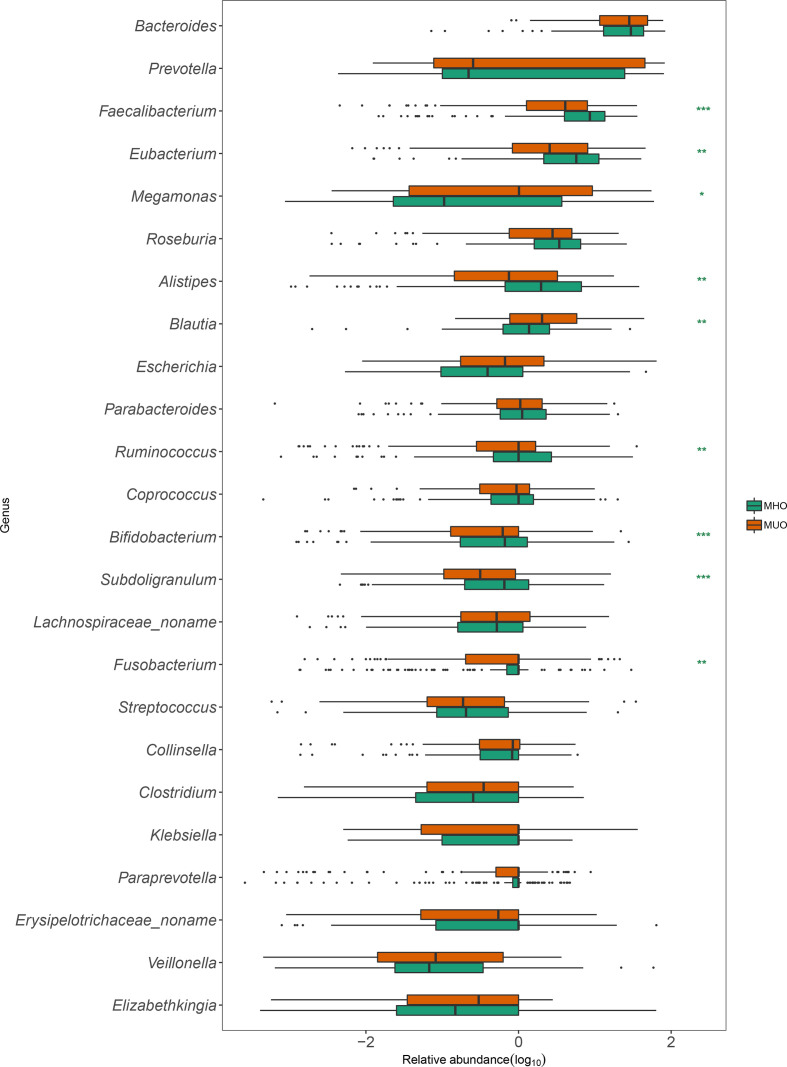
At the phylum level, MHOs had a lower proportion of Fusobacteria (P <0.01) compared to MUOs (Table S4). At the genus level, Alistipes , Bifidobacterium , Eubacterium , Faecalibacterium , Ruminococcus and Subdoligranulum showed higher relative abundance in MHOs compared to MUOs, while Escherichia , Clostridium , Fusobacterium and Megamonas were found with higher abundance in MUOs (Fig. 3).
Fig. 3.
Differentiated bacterial genera between MHOs and MUOs from Chongqing. Green boxes and brown boxes represent the relative abundance of genus enriched in MHOs and MUOs, respectively.
Different microbial genes and functions in the gut microbiome between MHOs and MUOs
We mapped sequencing reads to the IGC and identified 761 272 genes that were present in at least 20 % of the Chongqing cohort. Similar to the Shannon diversity, MHOs showed a higher gene count than the MUOs in the Chongqing cohort (Wilcoxon rank sum test, Benjamini–Hochberg P value <0.05) (Fig. 2a).
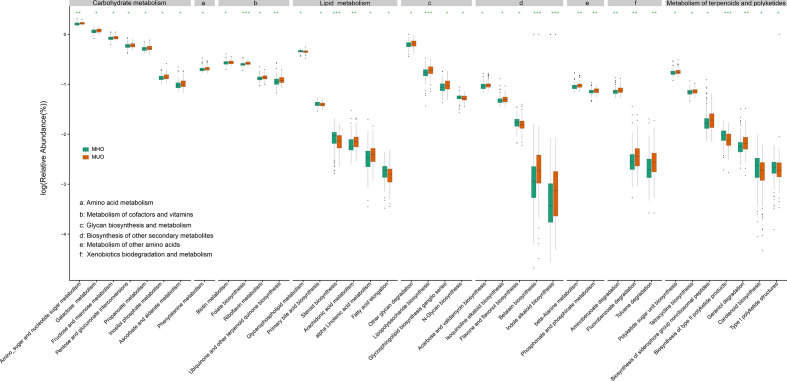
To understand the association of these different microbial genes with metabolic status, we analysed the differences of microbial functions as well as microbial gene markers between MHOs and MUOs. Genes associated with glycan biosynthesis and metabolism had a significantly different abundance between MHOs and MUOs (Fig. 4, Table S5). The overall abundance of KO groups involved in LPS biosynthesis, glycosphingolipid biosynthesis and other glycan degradation increased in MUOs, while the abundance of N-glycan biosynthesis increased in MHOs. In addition, MUOs had higher levels of microbial genes associated with carbohydrate metabolism, metabolism of cofactors and vitamins, and xenobiotics biodegradation and metabolism, when compared to MHOs.
Fig. 4.
Differentiated metabolic functional categories between MHOs and MUOs from Chongqing. Green boxes and brown boxes represent the relative abundance of metabolic functional categories enriched in MHOs and MUOs, respectively. *, **, *** represent p-value < 0.05, < 0.01, < 0.001 respectively.
Identification of microbial gene markers to discriminate MHOs from MUOs
We clustered microbial genes into 43 CAGs according to their abundance correlation among individuals (Table S6). To demonstrate the association of gut microbiome and metabolic status, we constructed random forest models to classify MHOs and MUOs using the abundance of CAGs in the Chongqing cohort (172 MHOs and 138 MUOs). Cross-validation (ten repeats of fivefold cross validation) showed that the constructed models confidently classify (mean AUC=0.75) the Chongqing individuals into the correct category (Table S7). Overall, 17 CAGs showed sufficient classification in at least 40 iterations, including 10 CAGs enriched in MUOs and 7 in MHOs. Consistent with the genus level observations, the genera of CAGs enriched in MHOs include Alistipes , Bifidobacterium (Bifidobacterium longum) and Eubacterium, whereas only Megamonas ( Megamonas hypermegale ) was found enriched in MUOs (Table 1).
Table 1.
Co-abundance groups enriched in MHOs and MUOs
|
ID |
Genus |
Species |
Mean abundance in: |
Enriched group |
Adjusted P value |
Mean Gini value |
|
|---|---|---|---|---|---|---|---|
|
MUOs |
MHOs |
||||||
|
CAG004 |
– |
– |
6.07E−06 |
5.43E−05 |
MHO |
0.000009 |
3.76 |
|
CAG007 |
– |
– |
8.18E−05 |
7.29E−05 |
MHO |
0.000062 |
3.43 |
|
CAG008 |
2.05E−05 |
7.48E−05 |
MHO |
0.00018 |
3.17 |
||
|
CAG009 |
– |
– |
0.0002 |
4.30E−05 |
MUO |
0.000031 |
3.99 |
|
CAG012 |
– |
– |
3.92E−05 |
6.84E−05 |
MUO |
0.1813 |
3.14 |
|
CAG013 |
0.00015 |
9.16E−05 |
MUO |
0.00083 |
4.14 |
||
|
CAG017 |
– |
– |
0.00048 |
0.0004 |
MUO |
0.00063 |
4.74 |
|
CAG018 |
– |
– |
0.00015 |
7.47E−05 |
MUO |
0.003154 |
3.38 |
|
CAG019 |
– |
– |
0.00014 |
9.37E−05 |
MUO |
0.001291 |
4.84 |
|
CAG023 |
– |
– |
0.00023 |
0.0002 |
MUO |
0.000031 |
4.78 |
|
CAG025 |
1.89E−05 |
7.51E−05 |
MHO |
0.000031 |
3.51 |
||
|
CAG026 |
– |
– |
0.00031 |
0.0003 |
MUO |
0.000062 |
4.42 |
|
CAG027 |
2.77E−05 |
7.56E−05 |
MHO |
0.000031 |
4.19 |
||
|
CAG033 |
– |
– |
0.00025 |
0.0002 |
MUO |
0.000105 |
4.28 |
|
CAG035 |
2.68E−05 |
7.97E−05 |
MHO |
0.000031 |
3.55 |
||
|
CAG038 |
3.03E−05 |
8.74E−05 |
MHO |
0.000171 |
3.38 |
||
|
CAG042 |
– |
– |
0.00031 |
0.0003 |
MUO |
0.000062 |
4.49 |
Validation in an independent cohort
We further applied the model trained by the Chongqing data in the Quanzhou cohort (AUC=0.69). Similar to the Chongqing cohort, higher gene count (P <0.05) and Shannon microbial diversity were detected in the gut microbiome of MHOs in Quanzhou (Fig. S1). Nevertheless, the F/B ratio in the MHOs of Quanzhou is slightly higher than that in MUOs (Fig. S1). No CAGs showed significant differences between the two groups with adjusted P values.
Discussion
Substantial evidence has shown that the gut microbiome is closely related to the development of obesity [28–30]. Nevertheless, the gut microbiome differences between obese individuals with and without abnormalities remains to be fully explored. This study observed higher gene count and microbial diversity in MHOs when compared with MUOs. Other studies upon obesity and metabolic syndrome also show a higher gut microbiome diversity in healthy individuals, suggesting a potential association between microbial richness and the host metabolic status [26, 28, 31]. In contrast, individuals with lower microbial gene count are generally characterized by higher fat mass, insulin resistance and dyslipidaemia [32]. Interestingly, the obese individuals with higher microbial gene count generally gained more improvement in decreasing body weight, insulin resistance and inflammation level through the same dietary restriction [33]. Those microbial genes could be identified through the comparison between the gut microbiome of MHOs and MUOs, given the significant difference of microbial richness between the two groups.
We then identified several microbial species associated with metabolic health status in obese individuals. Most of the associated microbial taxa identified in the Chongqing cohort were highly consistent with the findings in previous studies on obesity, metabolic syndrome and diabetes. In particular, we confirmed higher abundances of the genera Alistipes [26], Bifidobacterium [31, 34], Eubacterium [35], Faecalibacterium [29] and Subdoligranulum [26] in MHOs, and lower abundances of the genera Fusobacterium [29] and Megamonas [29] in MUOs. Our findings suggest that many microbial taxa may have a similar association with host metabolism across studies. The taxonomic annotation of CAGs was consistent with the above-mentioned gut microbiome structure analysis results, showing a reliable taxonomic profiling. Gut microbiome differences between MUOs and MHOs were also detected and validated in the Quanzhou cohort with distinct dietary habits. In addition, we detected a higher abundance of Adlercreutzia equolifaciens in Quanzhou MHOs. Recent evidence showed that A. equolifaciens is able to metabolize resveratrol and produce dihydroresveratrol, which exhibits significant antioxidant activity and, hence, could be involved in anti-inflammatory responses and protect the host from developing metabolic abnormalities [36]. These findings on microbial components warrant further investigation of the interaction between microbial components and host metabolism.
Furthermore, the differentiated microbial functions between MHOs and MUOs suggest the functional alteration during development of metabolic abnormalities in the obese individuals. We found that MUOs had increased levels of genes associated with biosynthesis pathways of LPS, which is a common product of Gram-negative bacteria. This can be partly explained by demonstrated roles of LPS in causing inflammatory responses and obesity, such as triggering the secretion of proinflammatory cytokines via binding to Toll-like receptor 4 of intestinal immune cells [37].
Though our findings suggest the potential of the gut microbiome in stratifying obese populations, classifying the obese individuals from Quanzhou with the model trained by the Chongqing data showed a reduced AUC. This is consistent with prior reports applying the gut microbiome to distinguish microbial samples in a validated cohort, and may be partly explained by differences in living environments and dietary habits that impact the gut microbiome significantly [38]. Our previous similar studies can also explain the contrary association between gut microbiome and obesity in different cohorts [14].
We acknowledge that there are some limitations in the present research. Firstly, the sample size of our study is relatively small. Secondly, although we used several metabolic traits to characterize and define MHO and MUO individuals, a widely agreed definition of the MHO is still in debate [39]. In future, more phenotypes associated with host metabolism should be included to define MHOs and MUOs. Nonetheless, the gut microbial markers reported in the present research could also serve as biomarkers for those metabolic statuses, and findings in the present study should be validated in larger cohorts with more comprehensive clinical measurements.
Taken together, we have revealed different gut microbiomes between MUOs and MHOs based on two geographically distinct cohorts in China. Furthermore, we identified microbial components and genes associated with obese individuals without metabolic abnormalities. These findings provide extensive insights into management of obesity via gut-microbiome-targeted interventions, and exploration of host/microbe–microbe interactions in obesity with distinct metabolic status.
Supplementary Data
Funding information
This work was supported by the Natural Science Foundation of China (grant no. 81 602 854, no. 81872920 and no. 81561128020), Military Healthcare programmes (grant no.15BJZ48 and no. 16BJZ40), the Shenzhen High-Level Hospital Construction Fund (YBH2019-260), the Shenzhen Key Medical Discipline Construction Fund (no. SZXK027) and the Sanming Project of Medicine in Shenzhen (no. SZSM202011016).
Acknowledgements
We thank all the nurses who helped with physical examination, clinical recording and faecal collection at the180th Hospital of the People’s Liberation Army of China and Southwest Hospital.
Author contributions
Q. Z.: Conceptualisation, Data curation, Writing‐original draft, Project administration. Z.Y.: Conceptualization, Data curation, Writing‐original draft, Writing‐review and editing. F.W.: Formal analysis. D.W: Writing‐original draft. Ya.L.: Data curation, Formal analysis, Visualization. X.Z.: Data curation, Resources. W.D.: Writing‐original draft, Resources. Yi.L.: Data curation, Formal analysis. Y.W.: Data curation, Resources. X.F.: Data curation, Formal analysis. J.C.: Methodology, Formal analysis. Yo.L.: Data curation, Resources. Y.Z.: Resources. T.K.: Formal analysis, Methodology. H.G.: Formal analysis, Methodology. S.F.: Methodology. S.L.: Writing‐review and editing. Y.H.: Data curation, Resources. X.X.: Methodology, Writing‐review and editing. D.L.: Conceptualization, Writing‐original draft, Writing‐review and editing.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This study was approved by the Ethics Committee of the General Hospital of the People’s Liberation Army (PLAGH) under registration number S2016-068-01, and the research was carried out according to the Code of Ethics of the World Medical Association. Informed consents were signed by all the participants.
Footnotes
Abbreviations: AUC, area under the operating characteristic curve; CAG, co-abundance gene group; F/B, Firmicutes/Bacteroidetes; IGC, Integrated Gene Catalog; KO, KEGG orthology; LPS, lipopolysaccharide; MHO, metabolically healthy obese; MUO, metabolically unhealthy obese; PCoA, principal coordinate analysis; UA, uric acid.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary figure and seven supplementary tables are available with the online version of this article.
References
- 1.Tilg H, Kaser A. Gut microbiome, obesity, and metabolic dysfunction. J Clin Invest. 2011;121:2126–2132. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Despres JP, Lemieux I, Bergeron J, Pibarot P, Mathieu P, et al. Abdominal obesity and the metabolic syndrome: contribution to global cardiometabolic risk. Arterioscler Thromb Vasc Biol. 2008;28:1039–1049. doi: 10.1161/ATVBAHA.107.159228. [DOI] [PubMed] [Google Scholar]
- 3.Pindjakova J, Sartini C, Lo Re O, Rappa F, Coupe B, et al. Gut dysbiosis and adaptive immune response in diet-induced obesity vs systemic inflammation. Front Microbiol. 2017;8:1157. doi: 10.3389/fmicb.2017.01157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muñoz-Garach A, Cornejo-Pareja I, Tinahones FJ. Does metabolically healthy obesity exist? Nutrients. 2016;8:320. doi: 10.3390/nu8060320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castaner O, Goday A, Park YM, Lee SH, Magkos F, et al. The gut microbiome profile in obesity: A systematic review. Int J Endocrinol. 2018;2018:4095789. doi: 10.1155/2018/4095789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap Adv Gastroenterol. 2013;6:295–308. doi: 10.1177/1756283X13482996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, et al. Minireview: gut microbiota: the neglected endocrine organ. Mol Endocrinol. 2014;28:1221–1238. doi: 10.1210/me.2014-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 11.Gérard P. Gut microbiome and obesity. How to prove causality? Ann Am Thorac Soc. 2017;14:S354–S356. doi: 10.1513/AnnalsATS.201702-117AW. [DOI] [PubMed] [Google Scholar]
- 12.Vallianou N, Stratigou T, Christodoulatos GS, Dalamaga M. Understanding the role of the gut microbiome and microbial metabolites in obesity and obesity-associated metabolic disorders: current evidence and perspectives. Curr Obes Rep. 2019;8:317–332. doi: 10.1007/s13679-019-00352-2. [DOI] [PubMed] [Google Scholar]
- 13.Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015;15:100. doi: 10.1186/s12876-015-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran-Ramos S, López-Contreras BE, Canizales-Quinteros S. Gut microbiota in obesity and metabolic abnormalities: a matter of composition or functionality? Arch Med Res. 2017;48:735–753. doi: 10.1016/j.arcmed.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Schloss PD, Iverson KD, Petrosino JF, Schloss SJ. The dynamics of a family’s gut microbiota reveal variations on a theme. Microbiome. 2014;2:25. doi: 10.1186/2049-2618-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen CP, Cheng TYD, Tsai SP, Chan HT, Hsu HL, et al. Are Asians at greater mortality risks for being overweight than Caucasians? Redefining obesity for Asians. Public Health Nutr. 2009;12:497–506. doi: 10.1017/S1368980008002802. [DOI] [PubMed] [Google Scholar]
- 17.Chizyński K, Rózycka M. Hyperuricemia. Pol Merkur Lekarski. 2005;19:693–696. [PubMed] [Google Scholar]
- 18.Goolsby MJ. National Kidney Foundation Guidelines for chronic kidney disease: evaluation, classification, and stratification. J Am Acad Nurse Pract. 2002;14:238–242. doi: 10.1111/j.1745-7599.2002.tb00119.x. [DOI] [PubMed] [Google Scholar]
- 19.Wagner Mackenzie B, Waite DW, Taylor MW. Evaluating variation in human gut microbiota profiles due to DNA extraction method and inter-subject differences. Front Microbiol. 2015;6:130. doi: 10.3389/fmicb.2015.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews S. Babraham Institute, UK; 2014. FastQC: a quality control tool for high throughput sequence data.www.bioinformatics.babraham.ac.uk/projects/fastqc [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 24.Xie H, Guo R, Zhong H, Feng Q, Lan Z, et al. Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Syst. 2016;3:572–584. doi: 10.1016/j.cels.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014;32:822–828. doi: 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- 26.Qin J, Li Y, Cai Z, Li S, Zhu J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. doi: 10.2307/2346101. [DOI] [Google Scholar]
- 28.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu R, Hong J, Xu X, Feng Q, Zhang D, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23:859–868. doi: 10.1038/nm.4358. [DOI] [PubMed] [Google Scholar]
- 30.John GK, Mullin GE. The gut microbiome and obesity. Curr Oncol Rep. 2016;18:45. doi: 10.1007/s11912-016-0528-7. [DOI] [PubMed] [Google Scholar]
- 31.Lim MY, You HJ, Yoon HS, Kwon B, Lee JY, et al. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut. 2017;66:1031–1038. doi: 10.1136/gutjnl-2015-311326. [DOI] [PubMed] [Google Scholar]
- 32.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 33.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 34.Thingholm LB, Rühlemann MC, Koch M, Fuqua B, Laucke G, et al. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe. 2019;26:252–264. doi: 10.1016/j.chom.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haro C, Garcia-Carpintero S, Alcala-Diaz JF, Gomez-Delgado F, Delgado-Lista J. The gut microbial community in metabolic syndrome patients is modified by diet. J Nutr Biochem. 2016;27:27–31. doi: 10.1016/j.jnutbio.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 36.Chaplin A, Carpéné C, Mercader J. Resveratrol, metabolic syndrome, and gut microbiota. Nutrients. 2018;10:1651. doi: 10.3390/nu10111651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerard P. Gut microbiota and obesity. Cell Mol Life Sci. 2016;73:147–162. doi: 10.1007/s00018-015-2061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Y, Wu W, Zheng HM, Li P, McDonald D, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med. 2018;24:1532–1535. doi: 10.1038/s41591-018-0164-x. [DOI] [PubMed] [Google Scholar]
- 39.Rial SA, Karelis AD, Bergeron KF, Mounier C. Gut microbiota and metabolic health: the potential beneficial effects of a medium chain triglyceride diet in obese individuals. Nutrients. 2016;8:281. doi: 10.3390/nu8050281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.