Abstract
The poly(A) polymerase of the budding yeast Saccharomyces cerevisiae (Pap1) is a 64-kDa protein essential for the maturation of mRNA. We have found that a modified Pap1 of 90 kDa transiently appears in cells after release from α-factor-induced G1 arrest or from a hydroxyurea-induced S-phase arrest. While a small amount of modification occurs in hydroxyurea-arrested cells, fluorescence-activated cell sorting analysis and microscopic examination of bud formation indicate that the majority of modified enzyme is found at late S/G2 and disappears by the time cells have reached M phase. The reduction of the 90-kDa product upon phosphatase treatment indicates that the altered mobility is due to phosphorylation. A preparation containing primarily the phosphorylated Pap1 has no poly(A) addition activity, but this activity is restored by phosphatase treatment. A portion of Pap1 is also polyubiquitinated concurrent with phosphorylation. However, the bulk of the 64-kDa Pap1 is a stable protein with a half-life of 14 h. The timing, nature, and extent of Pap1 modification in comparison to the mitotic phosphorylation of mammalian poly(A) polymerase suggest an intriguing difference in the cell cycle regulation of this enzyme in yeast and mammalian systems.
Addition of a poly(A) tail to the 3′ end of a eukaryotic mRNA is an essential step in the regulation of gene expression. Polyadenylation enhances initiation of translation (45), mRNA stability (5), and transport of mRNA from the nucleus to the cytoplasm (25). The importance of this modification is reflected in the fact that all eukaryotic mRNAs, except for the replication-dependent histone mRNAs of metazoans, are polyadenylated. In the yeast Saccharomyces cerevisiae, polyadenylation occurs in the nucleus in a two-step reaction (for recent reviews, see references 39, 50, and 53). Site-specific endonucleolytic cleavage of the primary transcript by the yeast cleavage factors CF I and II is followed by addition of a poly(A) tail to the upstream fragment by a complex of poly(A) polymerase (Pap1), CF I, and polyadenylation factor I (PF I). Pap1, a monomeric polypeptide of 64 kDa, is the key enzyme which catalyzes the addition of poly(A) tail to the mRNA (32, 33). The purified enzyme has no specificity for genuine yeast mRNA 3′ ends, but specificity for the correct 3′ end is conferred by association with other mRNA processing factors (43). Furthermore, interaction with these factors also determines the length of the poly(A) tail (2, 30, 40, 55).
Among the cloned genes encoding 3′-end processing factors in yeast and mammals, PAP1 shows the greatest sequence homology to its mammalian counterpart. There is a 47% identity between the yeast and mammalian proteins throughout the 450 amino acids in the amino-terminal region, which is thought to comprise the catalytic domain of these enzymes (37). The C-terminal regions vary significantly; in particular, the yeast Pap1 lacks the serine/threonine (Ser/Thr)-rich regulatory domain found in the mammalian enzyme (10). In contrast to its mammalian homologue, the yeast Pap1 is not required for the initial mRNA 3′-end cleavage (7, 41).
Posttranslational modification by phosphorylation and ubiquitination plays a major role in regulation of various aspects of cellular physiology, including progression through the mitotic cell cycle (13), transcriptional activation, signal transduction, and receptor-mediated endocytosis (22). For several reasons, we were interested in whether the yeast Pap1 was subject to such regulatory modifications. First, very little is known about the posttranslational modifications of RNA-binding proteins and the effects on nuclear processes. Second, mammalian poly(A) polymerase (PAP) is modified by phosphorylation on the cyclin-dependent kinase (cdk) consensus sites located in the Ser/Thr-rich region. Hyperphosphorylation occurs specifically during mitosis (M), resulting in inactivation of PAP (9). Numerous proteins, including components of the transcriptional and translational machinery, are phosphorylated as a result of mitotic cdk p34cdc2 activity in mammalian cells, leading to a general repression of nuclear RNA transcription and reduced protein synthesis in mitosis (20). Phosphorylation-mediated gene repression may be needed to facilitate mitotic events, such as chromosome condensation and segregation and dissociation of the nuclear lamina that is seen in mammalian cells. Down-regulation of the mammalian PAP by phosphorylation during mitosis is perhaps a mechanism to prevent inappropriate polyadenylation and probably affects synthesis of all polyadenylated transcripts (9, 10). Phosphorylation of the yeast enzyme has not been investigated. Furthermore, we have previously identified a protein, Ufd1 (named Ufd for ubiquitin fusion degradation), which interacts specifically with Pap1 in a two-hybrid screen (12). Ufd1 was also independently identified by Johnson et al. (27) and was shown to have a role in the ubiquitination process. However, ubiquitination of PAP or mRNA processing factors has not been reported.
In this study, we show that yeast Pap1 undergoes posttranslational modifications at the S-G2 transition of the cell cycle. The modifications involve both phosphorylation and ubiquitination but do not alter the stability of the bulk of cellular Pap1. Analysis of strains harboring mutations in the gene encoding Cdc28, a homologue of the mammalian cdc2 kinase that modifies the mammalian PAP at mitosis (9), suggests that Cdc28 activity is not directly required for Pap1 phosphorylation but may be involved in the correct timing of the modification. This is consistent with the lack of cdc2-cyclin B consensus phosphorylation sites in Pap1. However, the phosphorylated species does appear to be an inactive form of Pap1. Our studies indicate that the cell cycle regulation of yeast Pap1 is different from that reported for the mammalian enzyme.
MATERIALS AND METHODS
Yeast strains and cell culture.
All cultures were grown to early log phase, with an optical density at 600 nm of 1, in yeast extract peptone medium (YPD) or synthetic medium (SM) supplemented with 2% glucose, at 30°C except where indicated. S. cerevisiae strains used in this study were gifts from D. Finley (Harvard Medical School, Boston, Mass.), M. Hochstrasser (University of Chicago, Chicago, Ill.), S. Reed (The Scripps Research Institute, La Jolla, Calif.), M. Tyers (Samuel Lunenfeld Research Institute, Toronto, Ontario, Canada), and A. Amon (The Whitehead Institute, Boston, Mass.) and are as follows: W3031-A (MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100), Fc12-18 (MATa bar1 trp1 leu2 ura3 his2 ade1), cdc28-4 cells (MATa ura3 leu2 ade1 trp1 arg4 his2 cdc28-4), cdc28-13 cells (MATa ura3 leu2 ade2 trp1 arg4 his2 cdc28-13), cdc28-1N cells (MATa ura3 leu2 ade2 trp1 arg4 his2 cdc28-1N), cdc9-1 cells (MATα ura3 leu1 ade2 trp1 can1 cdc9-1), cdc13-1 cells (MATα ura3 his7 trp1 can1 cdc13-1), cdc4-1 cells (MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 cdc4-1), and cdc34-2 cells (MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 cdc34-2). The HA-Smt3 construct was a gift from David Leggett (Harvard Medical School). The 2μm-based plasmids expressing the hemagglutinin (HA) and Myc epitope-tagged forms of ubiquitin used in this study were described previously (24). For ubiquitin overexpression from the CUP1 promoter of these plasmids, cells were induced by addition of 100 μM CuSO4 for 5 h at 30°C.
Immunoprecipitation and Western blot analysis.
Yeast cells were harvested, washed with cold water, and resuspended in cell lysis buffer (30 mM HEPES [pH 7.5], 100 mM potassium acetate, 2 mM magnesium acetate, 0.5 M sorbitol, 1 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]) supplemented with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin A per ml, 1 μg of pepstatin, 10 μg of tolysulfonyl phenylalanyl chloromethyl ketone [TPCK] per ml, 10 μg of Nα-p-tosyl-l-lysine chloromethyl ketone [TLCK] per ml, 50 mM N-ethylmaleimide) and phosphatase inhibitors (50 mM β-glycerophosphate, 1 mM sodium orthovanadate). Samples were lysed with glass beads by vortex mixing (three times for 1 min each time with 1-min cooling intervals on ice). Cell lysates were centrifuged at 15,000 × g for 20 min at 4°C. The protein concentration of the supernatant was measured by the Bradford assay.
Immunoprecipitation was performed with 5 mg of protein from total cell lysate using 100 μl of anti-Pap1 monoclonal antibody (31) or with a control monoclonal antibody against β-galactosidase. Unless indicated, the antibody specific for an epitope in the C terminus of Pap1 was used. After incubation at 4°C for 18 h, immune complexes were collected on 20 μl of protein A-Sepharose beads for 2 h. Beads were washed three times with wash buffer containing 50 mM Tris-HCl (pH 7.9), 200 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 1 mM phenylmethylsulfonyl fluoride at 4°C. For phosphatase treatment, immunoprecipitates were incubated with 200 U of lambda phosphatase or with 1 U of T-cell protein tyrosine phosphatase or 1 U of protein phosphatase 1 (New England BioLabs) in 20 μl of a solution containing 50 mM Tris-HCl (pH 8.0) and 1 mM ubiquitin aldehyde (Calbiochem) at 37°C for 30 min. Inactivation of the phosphatase was accomplished by incubating the enzyme at 65°C for 10 min prior to addition to the beads. Samples were treated with sample buffer containing 5 mM dithiothreitol, boiled for 3 min, and resolved on a 10% polyacrylamide-SDS gel. Proteins were transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 1% fetal bovine serum for 1 h and then incubated with a mouse monoclonal anti-Pap1 antibody at a 1:500 dilution, a rabbit polyclonal antibody against the HA epitope at a 1:2,000 dilution (Santa Cruz Biotechnology, Inc.), or antibody against ubiquitin (Sigma) at a 1:2,000 dilution, at 4°C for 18 h. Antibody binding was detected by chemiluminescence (Amersham).
Cell synchronization and release.
Chemical arrest was performed by treatment for 3 h with α-factor mating pheromone (2 μg/ml) for G1 phase, hydroxyurea (10 mg/ml) for S phase, or nocodazole (15 μg/ml) in M phase at 30°C, unless noted. To release, cells were washed twice and resuspended in fresh media. Arrest was confirmed by fluorescence-activated cell sorting (FACS) analysis and microscopic examination of bud formation. Samples were collected and pelleted at various time intervals. Typically, a 150-ml culture with an optical density at 600 nm of 1 was used for protein lysate, 2 ml was used for total RNA preparation, and 1 ml was used for FACS analysis and determination of the budding index.
Flow cytometry.
Flow cytometric DNA quantitation was determined by the following procedure (14). Cells were fixed in 70% ethanol for 18 h at 4°C. Samples were then washed with and resuspended in 1 ml of 50 mM sodium citrate (pH 7.5) and then treated with 25 μl of 10-mg/ml RNase A at 37°C for 1 h followed by addition of 50 μl of 20-mg/ml proteinase K and incubation at 37°C for another hour. The DNA was stained by addition of 1 ml of 50 mM sodium citrate containing 5 μg of propidium iodide per ml. Samples were incubated in the dark overnight at 4°C and analyzed on a FACScan (Becton Dickinson). For each histogram, 25,000 cells were analyzed with the ModiFIT cell cycle analysis software to calculate the percentages of cells in G1, S, and G2/M phases.
Pulse-chase analysis.
The pulse-chase analysis regimen was adapted from the regimen of Amon et al. (1) with minor modifications. Yeast cells were grown in YPD to log phase at 30°C. A 50-ml culture was then washed in synthetic medium without methionine (SM-Met) and grown in 5 ml of SM-Met medium supplemented with 0.1 mM methionine for 5 min. Cells were then labeled with 500 μCi of Trans35S-label (ICN) (85% [35S]methionine, 15% [35S]cysteine) for 30 min at 30°C. Cells were then centrifuged and resuspended at time zero in YPD medium containing 2 mM methionine, 2 mM cysteine, and when noted, 1 mg of cycloheximide per ml. Aliquots of cells were removed at different times during the chase, washed twice with cold water, and stored at −80°C before protein extraction. Protein extracts were prepared as described above by the glass bead disruption method. Incorporation of 35S was determined by counting tricholoroacetic acid-precipitable material. Aliquots of extract containing equivalent disintegrations per minute were precleared with 20 μl of protein A-Sepharose beads for 15 min. Supernatant was incubated with anti-Pap1 antibody for 2 h at 4°C, after which 20 μl of protein A-Sepharose beads was added for an additional 2 h. The beads were washed twice with 1 ml of the wash buffer and twice with 1 ml of wash buffer containing 2 M urea. Beads were resuspended in sample buffer containing dithiothreitol, boiled, and resolved on a 10% polyacrylamide-SDS gel. The gel was then stained with Coomassie blue, dried, and subjected to autoradiography or quantified with a PhosphorImager system (Molecular Dynamics, Sunnyvale, Calif.). The half-life of Pap1 was calculated from the formula t1/2 = 1n 2/degradation rate by using Fig. P software (BioSoft, Cambridge, United Kingdom) (11).
In vitro polyadenylation assays.
For processing extracts, cdc9-1 and cdc13-1 cells were grown to logarithmic phase in YPD medium at the permissive temperature of 24°C and then shifted to the restrictive temperature of 37°C for 2 h. Cell cycle arrest was confirmed by microscopic examination and FACS analysis. Cell extract for polyadenylation assays was prepared as described previously (30). Extract was prepared in the presence of phosphatase inhibitors (50 mM β-glycerophosphate and 1 mM sodium orthovanadate). For separation of Pap1 forms, 1 ml of cell extract (10 mg/ml) was loaded onto a 1-ml Q fast anion-exchange column (Pharmacia), equilibrated with buffer B, containing 50 mM Tris-HCl (pH 7.9), 80 mM potassium chloride, and 1 mM phenylmethylsulfonyl fluoride. The column was washed with 7 volumes of buffer B. The bound proteins were then eluted with steps of 150 or 500 mM potassium chloride. Fractions (3 ml each) were immunoprecipitated with Pap1 antibody. The Pap1 immunoprecipitates were divided into two parts and analyzed by immunoblotting and for Pap1 activity. The polyadenylation assay was performed by incubating the Pap1 immunoprecipitates with 20 mM Tris-HCl (pH 8.0), 2% polyethylene glycol 8000, 20 mM creatine phosphate, 1 mM MnCl2, 2 mM ATP, and an α-32P-labeled RNA substrate in a final volume of 20 μl for 20 min at 30°C. Phosphatase treatment was performed in 50 mM Tris-HCl, pH 8.0 for 5 min at 30°C. The RNA substrate contains the 161 nucleotides of the wild-type GAL7 sequence upstream of the poly(A) site (7). Resulting products were analyzed on a 6% polyacrylamide–8.3 M urea gel and visualized by autoradiography.
RESULTS
Pap1 is modified during the cell cycle.
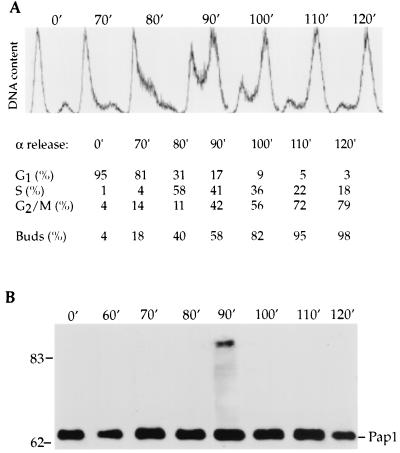
As a first step towards investigating potential regulation of the yeast Pap1, we analyzed the amount and type of Pap1 found in the cells as they progressed through the cell cycle. Wild-type cells were blocked by α-factor pheromone at G1. After release, cell samples were removed at various time intervals to determine the position in the cell cycle by measuring DNA content by FACS and by examining bud formation microscopically. α-factor treatment resulted in a G1 arrest, as evidenced by the uniform population of cells with 1 N content of DNA and the accumulation of unbudded cells (Fig. 1A). The percentage of cells at different stages of the cell cycle was calculated to better estimate the timing of Pap1 modification. After release from α-factor arrest, cells continued in a synchronized fashion to replicate their DNA and progress through M phase.
FIG. 1.
Analysis of Pap1 modification during the S. cerevisiae cell cycle. Fc12-18 wild-type cells were synchronized with α-factor mating pheromone at G1 (0 min [0′]) followed by a release into YPD medium. (A) Synchrony was assessed by measuring the DNA content of cells by FACS analysis, and the percentages of cells at different stages of the cell cycle were determined using the cellfit cell cycle program, as shown below the relevant times. At the indicated times (in minutes), the occurrence (percentage) of buds in synchronized culture was determined by microscopic examination. (B) Pap1 was immunoprecipitated from cell extract at various time intervals with anti-Pap1 antibody and resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis was performed with anti-Pap1 antibody. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel.
To examine Pap1 during the cell cycle, equal amounts of total protein from cell lysates taken at each time point were immunoprecipitated with a monoclonal antibody that recognizes an epitope in the C terminus of Pap1 (31). The immunoprecipitates were then analyzed by electrophoresis and immunoblotting. The 64-kDa Pap1 band was found at all time points (Fig. 1B). At 90 min after release from α-factor arrest, the Pap1 antibody also detected species with higher molecular masses with the major form being approximately 90 kDa. The appearance of this Pap1 band coincided with the S-G2 transition determined by FACS analysis and by budding index (Fig. 1A). At this time, an equal number of cells are in S or G2/M and thus have just finished replicating their DNA or are in the process of replication. The occurrence of the larger Pap1 was remarkably transient, appearing and disappearing within 20 min, such that it was absent at M phase, when the majority of the cells had large buds and had replicated their DNA (Fig. 1B). The total amount of Pap1 did not vary significantly in this and in the other cell cycle experiments, suggesting that Pap1 abundance does not fluctuate substantially during the cell cycle. The 90-kDa band is most likely a modified form of Pap1, since there is only one copy of the gene and no possibility of variants due to alternative splicing (33).
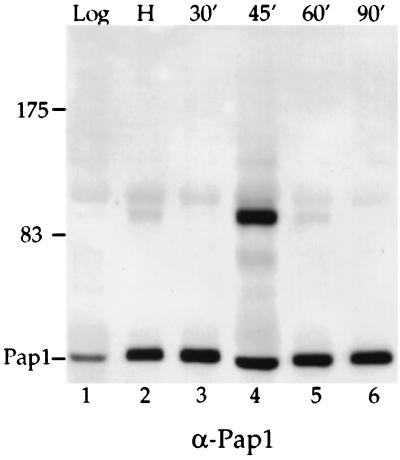
To further investigate the point at which Pap1 is modified in the cell cycle, cells were arrested with hydroxyurea, which inhibits ribonucleotide reductase and results in cell cycle arrest in S phase. For reasons discussed below, the hydroxyurea-arrested cells in this experiment also expressed a ubiquitin gene tagged with the influenza virus hemagglutinin epitope (HA-Ub) (15). After release into fresh medium lacking hydroxyurea, samples were removed at various time points for Pap1 detection. The 90-kDa Pap1 initially was present at a low level in hydroxyurea-treated cells (Fig. 2, lane 2). However, the amount of 90-kDa Pap1 greatly increased 45 min after hydroxyurea release (Fig. 2, lane 4), declined at 60 min, and disappeared at 90 min (Fig. 2, lanes 5 and 6). Consistent with these observations, we did not detect the modified Pap1 in cells treated with nocodazole, which blocks cells at the M phase of the cell cycle (data not shown). The combined data from these experiments suggest that the larger Pap1 species first appeared during S phase, accumulated during the late S-G2 transition, and disappeared at the M phase of the cell cycle.
FIG. 2.
Analysis of Pap1 modification detected in cells blocked with hydroxyurea. Cells containing the HA-Ub construct were grown in selective medium at 25°C (without copper induction) and then blocked at S phase with hydroxyurea. Cells were released into fresh medium lacking hydroxyurea. Extracts from cells taken from a log-phase yeast culture (lane 1), from cells treated with hydroxyurea (H) (lane 2), and from cells removed at various time points (in minutes) after release from the hydroxyurea block (lanes 3 to 6) were immunoprecipitated with anti-Pap1 antibody, separated by SDS-PAGE, and then probed with anti-Pap1 antibody (α-Pap1) (lanes 1 to 6). The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel.
Pap1 is phosphorylated.
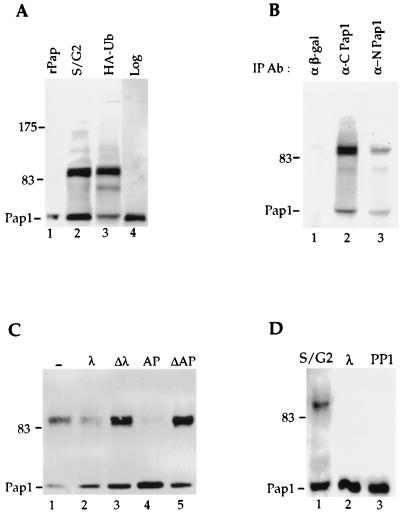
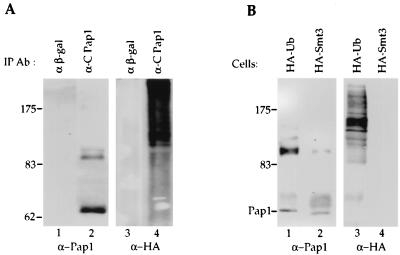
The 90-kDa form of Pap1 is not found in recombinant Pap1 (rPap1) made in Escherichia coli or cells from a log-phase yeast culture (Fig. 3A, lanes 1 and 4). However, overexpression of ubiquitin in an asynchronized cell culture of the same yeast strain resulted in accumulation of Pap1 isoforms which comigrated with those observed in the late S/G2 phase of cells synchronized by α-factor arrest (Fig. 3A, lanes 2 and 3). In these samples, additional species between 64 and 90 kDa in size and greater than 90 kDa are also visible. To verify that the 90-kDa species was indeed a form of Pap1, we tested whether it could also be immunoprecipitated with a monoclonal antibody directed against an epitope in the N terminus of Pap1 (Fig. 3B). Both Pap-specific antibodies, but not an unrelated antibody against β-galactosidase, immunoprecipitate the 90-kDa form, as well as the 64-kDa Pap1. Furthermore, the 90-kDa species is also detected when blots are probed with either the N terminus or C terminus anti-Pap1 specific antibody (data not shown). These results confirm that the 90-kDa species contains a form of Pap1 which has probably been posttranslationally modified.
FIG. 3.
Pap1 phosphorylation. (A) Pap1 was immunoprecipitated from extract prepared from synchronized Fc12-18 cells which have entered late S/G2 after release from α-factor arrest (lane 2), from unsynchronized W303 cells overexpressing HA-ubiquitin (lane 3), or from unsynchronized W303 cells without ubiquitin overexpression (lane 4). rPap is recombinant Pap1 made in E. coli (lane 1). (B) Extracts from W303 cells overexpressing ubiquitin were immunoprecipitated with anti-β-galactosidase (α β-gal) (lane 1) or with the C-terminal (lane 2) or N-terminal (lane 3) Pap1 antibodies, and the immunoblot was probed with the C-terminal Pap1 antibody. IP Ab, immunoprecipitation antibody. (C) The 90-kDa Pap1 species is sensitive to phosphatases. Immunoprecipitates were incubated without phosphatase (−) (lane 1) or with 200 U of lambda phosphatase (λ) (lane 2), 2 U of alkaline phosphatase (AP) (lane 4), or heat-inactivated lambda phosphatase (Δλ) (lane 3) on alkaline phosphatase (ΔAP) (lane 5). (D) Phosphorylation of Pap1 at S/G2 of the cell cycle. Phosphatase treatment of the Pap1 immunoprecipitate from the 90-min time point shown in Fig. 1B. Lane 1, no phosphatase; lane 2, lambda phosphatase; lane 3, protein phosphatase 1 (PP1). Unless indicated otherwise, the immunoprecipitations and immunoblotting were performed with the C-terminus-specific Pap antibody. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gels.
To determine whether this 90-kDa form represented phosphorylated species, the Pap1 immunoprecipitate from cells overexpressing HA-tagged ubiquitin was subjected to phosphatase treatment. This analysis showed that the 90-kDa band is sensitive to treatment with lambda phosphatase at a concentration which gives specific removal of phosphate groups from Ser/Thr residues (Fig. 3C, lane 2). Heat-inactivated lambda phosphatase had no effect on the higher Pap1 band (Fig. 3C, lane 3). Treatment with alkaline phosphatase also resulted in a decrease in the intensity of the 90-kDa Pap1 protein (Fig. 3C, lane 4). No reduction was observed with heat-inactivated alkaline phosphatase (Fig. 3C, lane 5) or with the inclusion of phosphatase inhibitor (data not shown). The 90-kDa band is also sensitive to a Ser/Thr-specific phosphatase, protein phosphatase 1, but not to the T-cell tyrosine phosphatase (data not shown). Thus, the phosphorylation of Pap1 is likely to be on Ser/Thr residues of the enzyme.
Treatment of the Pap1 species found in cells at S/G2 by lambda phosphatase or protein phosphatase 1 also eliminates the 90-kDa Pap1 (Fig. 3D, lanes 2 and 3), indicating that the cell-cycle-specific species also contains phosphate. While the magnitude of the mobility alteration of the phosphorylated form on SDS-polyacrylamide gels is surprising, a similar shift in size also has been observed upon hyperphosphorylation of the mammalian PAP (9).
Phosphorylation is associated with Pap1 inactivation.
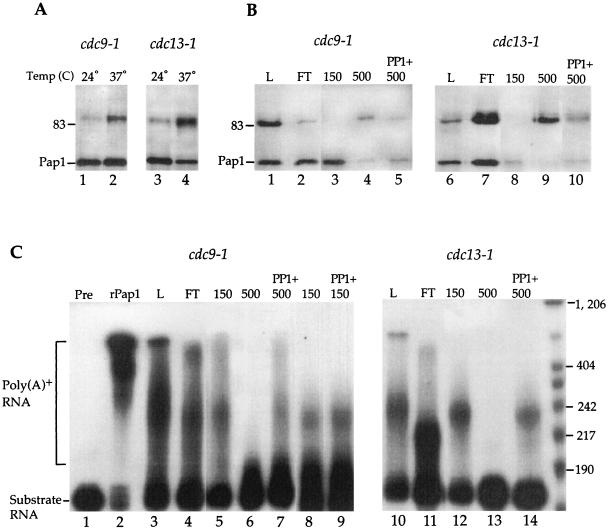
To characterize the effects of Pap1 modifications, we took advantage of two mutations, cdc9-1 and cdc13-1, that cause cell cycle arrest specifically in the S/G2 phase when cells are grown at the nonpermissive temperature. The cdc9-1 mutant encodes a temperature-sensitive DNA ligase which results in unligated Okazaki fragments at the restrictive temperature (28). The cdc13-1 mutant has defective metabolism of the telomere-associated DNA. These mutants arrest in the late S phase or in the early G2 phase as a consequence of DNA damage detected by the RAD9 checkpoint (52). In these strains, low levels of the 90-kDa Pap1 are present even at the permissive temperature (Fig. 4A, lanes 1 and 3). The 90-kDa Pap1 species increases relative to the 64-kDa form in the cdc9 and cdc13 cells at the restrictive temperature, consistent with these strains arresting at the time in the cell cycle coinciding with the Pap1 modification (Fig. 4A, lanes 2 and 4).
FIG. 4.
Chromatographic separation of phosphorylated Pap1 and recovery of poly(A) addition activity by phosphatase treatment. (A) Immunoprecipitated Pap1 from extracts made from cdc9-1 and cdc13-1 cells grown at 24°C (lanes 1 and 3) or 37°C (lanes 2 and 4). (B) Pap1 species obtained from cdc9-1 and cdc13-1 extracts after Q fast column fractionation. Immunoprecipitated Pap1 from either total cell lysate (L) at 37°C (lanes 1 and 6) or an equal proportion of different Q fast fractions (flowthrough [FT] or elutions with 150 and 500 mM KCl) as indicated over lanes 2 to 5 and 7 to 10 were separated by SDS-PAGE and then probed with anti-Pap1 antibody. Phosphatase treatment of the 500 mM fraction was performed for 5 min (lanes 5 and 10). For panels A and B, the position of the 83-kDa molecular mass marker is indicated to the left of the gels. (C) Pap1 activity of the Q fast fractions analyzed by addition of poly(A) tail onto radioactive substrate RNA. Polyadenylation assays were performed using 50 ng of the yeast rPap1 (lane 2) or immunoprecipitated Pap1 from total extract (L) (lanes 3 and 10), different Q fast fractions (lanes 4 to 6 and 11 to 13), and phosphatase-treated 150 mM fractions (lane 9) and 500 mM fractions (lanes 7 and 14). Untreated substrate RNA is shown in lane 1. The positions of the poly(A)+ and substrate RNA on the gel are indicated to the left of the gel, and the positions of size markers (in bases) are shown to the right.
Crude extracts contain a mixture of modified and unmodified Pap1. To assess the effect of the Pap1 phosphorylation, we separated the two species by column chromatography. Extracts competent for in vitro polyadenylation were prepared from cdc9-1 and cdc13-1 cells (30). These extracts were applied to a Q fast anion-exchange column, and proteins were eluted by 150 or 500 mM potassium chloride. An equal proportion of each fraction was immunoprecipitated with Pap1 antibody and assayed for Pap1 level and nonspecific polyadenylation activity. A mixture of Pap1 forms is present in the flowthrough fractions (Fig. 4B, lanes 2 and 7). Most of the 64-kDa Pap1 remaining on the column is eluted by 150 mM KCl (Fig. 4B, lanes 3 and 8). While a small amount of the 64-kDa Pap1 is present in the 500 mM KCl fraction, this fraction is greatly enriched for the 90-kDa species (Fig. 4B, lanes 4 and 9). The activity of the Pap1 is not preserved if immunoprecipitates are extensively treated with phosphatase, as shown in Fig. 3C and D. Thus, for activity assays, the Pap1 was treated with protein phosphatase 1 in 50 mM Tris for 5 min. This milder treatment still resulted in a significant decrease in the 90-kDa species in the 500 mM fraction and an increase in the 64-kDa Pap1 band (Fig. 4B, compare lanes 4 and 5 and 9 and 10). This confirms our previous result that the mobility of the 90-kDa form was due to phosphorylation.
The immunoprecipitates were then assayed for nonspecific Pap1 activity. This assay measures the ability of Pap1 to incorporate AMP onto a radioactive RNA substrate in the presence of manganese. The purified rPap1 efficiently polyadenylated the RNA, resulting in an adenosine tract of greater than 400 nucleotides (Fig. 4C, lane 2). The Pap1 present in the crude extract, flowthrough, or the 150 mM fraction is active, resulting in polyadenylated RNA seen primarily as a band about 50 to 70 nucleotides longer than the RNA substrate with some diffuse slower- and faster-migrating products (Fig. 4C, lanes 3 to 5 and 10 to 12). The shorter poly(A) tails seen in these samples in comparison to the rPap1 sample are probably due to differences in specific activity or to the presence of Pap1-associated factors which are known to regulate the length of the poly(A) tract (2, 36, 40, 55). No polyadenylation was detected with the 90-kDa Pap1 from the 500 mM KCl fraction (Fig. 4C, lanes 6 and 13). However, activity was recovered by treatment of the 500 mM KCl immunoprecipitate with protein phosphatase 1 prior to the polyadenylation assay (Fig. 4C, lanes 7 and 14). The activity of the 150 mM KCl fraction is not affected by phosphatase treatment (Fig. 4C, lanes 8 and 9). These results suggest that the 90-kDa Pap1 species is an inactive form of Pap1 which becomes active upon phosphatase treatment.
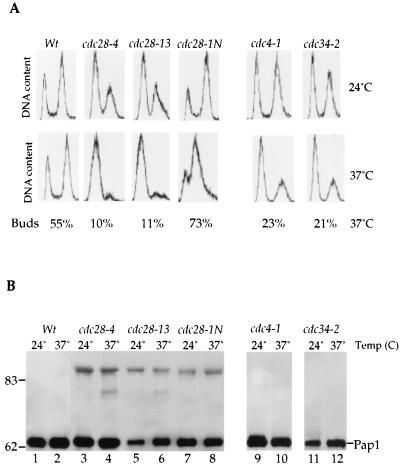
Mammalian PAP is highly phosphorylated by cdc2-CDK at multiple sites in the Ser/Thr-rich domain located in the C-terminal region of the enzyme (9). However, the yeast Pap1 lacks this Ser/Thr-rich domain and there is only one Thr-Pro site that resembles a putative phosphorylation site for Cdc28, the yeast homologue of cdc2-CDK. We examined the Pap1 species in three different cdc28 mutant strains. Cells harboring the cdc28-4 and cdc28-13 alleles arrest at G1, whereas a strain with the cdc28-1N allele arrests at G2 in the cell cycle (34, 42, 48). Cell cycle arrests of the various mutants were confirmed by cellular morphology, FACS analysis, and budding index (Fig. 5A). The 90-kDa Pap1 was present in cdc28 mutants even at 24°C and was not visible in extracts from the isogenic wild-type strain (Fig. 5B). A decrease in the amount of the 90-kDa Pap1 was not observed when these Cdc28 mutant cells were shifted to the restrictive temperature in all three cdc28 mutants (Fig. 5B). The appearance of the 90-kDa Pap1 species in the cdc28-4 and cdc28-13 mutants, which arrest at G1 in the cell cycle, was surprising, given our previous results with cells synchronized by α-factor or hydroxyurea arrest. To investigate this finding further, we examined the Pap1 species in two mutants, cdc4-1 and cdc34-2 mutants, which also arrest at G1 in the cell cycle (16, 18) (Fig. 5A). We did not detect the 90-kDa phosphorylated band in these mutants at either temperature (Fig. 5B, lanes 9 to 12). These data suggest that phosphorylation of yeast Pap1 is mediated by a protein kinase distinct from Cdc28. However, Cdc28 kinase may play an indirect role in regulation of the Pap1 phosphorylation.
FIG. 5.
Effects of cdc28 mutations on phosphorylation of Pap1. Wild-type (Wt) and cdc28-4, cdc28-13, cdc28-1N, cdc4-1, and cdc34-2 mutant cells were grown at 24°C and then shifted to 37°C for 2.5 h. (A) Cell cycle arrest was confirmed by FACS analysis and budding index. (B) Pap1 from the wild type (lanes 1 and 2) and cdc28 (lanes 3 to 8), cdc4-1 (lanes 9 and 10), and cdc34-2 (lanes 11 and 12) mutants was analyzed by immunoprecipitation followed by immunoblotting. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel.
Pap1 is ubiquitinated.
We also asked if Pap1 is modified by ubiquitination in addition to phosphorylation. To show the presence of ubiquitinated Pap1, we employed a commonly used HA-tagged ubiquitin construct, which is under control of a copper-inducible promoter (24). In the absence of copper, this construct has been reported to produce ubiquitin at a level approximately 2-fold higher than the endogenous level, while in the presence of copper, the level increases 100-fold (15, 24). When cells containing this construct are grown in the presence of copper, the 90-kDa Pap1 accumulates (Fig. 3A, lane 3; Fig. 6A, lane 2; Fig. 6B, lane 1). Neither the 64-kDa form nor the 90-kDa form of Pap1 is found in immunoprecipitations performed with anti-β-galactosidase antibody (Fig. 6A, lane 1). If the Pap1 immunoprecipitate is instead probed with anti-HA antibody, a ladder of high-molecular-weight proteins is detected in samples from cells expressing the HA-Ub fusion (Fig. 6A, lane 4; Fig. 6B, lane 3). Ubiquitination of proteins typically produces a pattern of higher-molecular-weight species such as those observed in the Pap1 immunoprecipitate (15, 38). The fact that these are not detected in an immunoprecipitation performed with antibodies against β-galactosidase (Fig. 6A, lane 3) suggests that they are specific to Pap1 and represent modification of Pap1 by ubiquitination. None of these appear to comigrate with the bands detected with the Pap1 antibody and thus are not likely to be abundant species, as one might expect for a modification which usually targets proteins for degradation. Because multiple HA-ubiquitination adducts are probably present in each band, it is easier to detect these species with the HA antibody. Overexpression of a ubiquitin-like protein, Smt3 (46) as an HA-Smt3 fusion does not result in any HA-reactive species detectable in Pap1 immunoprecipitates (Fig. 6B, lane 4), suggesting that Pap1 is not a target for this type of modification.
FIG. 6.
Pap1 is ubiquitinated in vivo. (A) Extracts from W303 cells overexpressing an HA-Ub fusion were used for immunoprecipitation (IP) with anti-β-galactosidase (α β-gal) (lanes 1 and 3) or with the C-terminal Pap1 antibody (lanes 2 and 4), and the immunoblot was probed with the C-terminal Pap1 (lanes 1 and 2) or anti-HA antibodies (lanes 3 and 4). (B) Immunoprecipitated Pap1 from W303 cells overexpressing HA-tagged ubiquitin (lanes 1 and 3) or HA-tagged Smt3 (lanes 2 and 4) was probed with anti-Pap1 (lanes 1 and 2) or anti-HA (lanes 3 and 4) antibodies. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gels.
Pap1 is ubiquitinated at late S/G2 in the cell cycle.
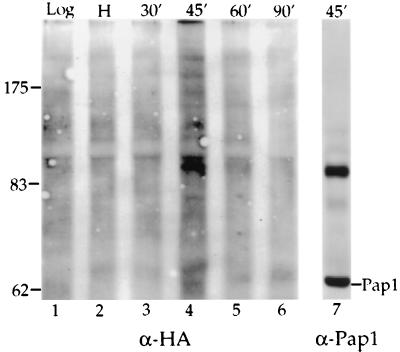
Our data show that phosphorylated Pap1 accumulates at late S/G2, and we wanted to see if the ubiquitination of Pap1 occurred at the same time. For this experiment, we used extracts prepared for the cell cycle time course shown in Fig. 2, in which W303 cells carrying the copper-inducible HA-Ub construct are released from a hydroxyurea-induced arrest in S phase. As discussed above, overexpression of ubiquitin causes the accumulation of phosphorylated Pap1 in unsynchronized cells and apparently perturbs the cell cycle regulation of this modification. To avoid this complication, copper was not used to induce high levels of the ubiquitin fusion. However, even in the absence of copper, there is sufficient expression of HA-Ub to allow detection of Pap1-ubiquitin conjugates with the HA antibody. When Pap1 immunoprecipitates were probed with the HA antibody, a ladder of ubiquitinated species was detected 45 min after release (Fig. 7, lane 4), concurrent with the appearance of the 90-kDa phosphorylated species (Fig. 2, lane 4). When ubiquitin is not overexpressed, it appears that less material is found in higher-molecular-weight conjugates such as seen in Fig. 6, and there is accumulation of a species which reproducibly migrates slightly slower than the 90-kDa phosphorylated form (Fig. 7, compare lanes 4 and 7).
FIG. 7.
Pap1 is ubiquitinated at S/G2. Pap1 immunoprecipitates prepared from cells after release from the hydroxyurea block as described in the legend to Fig. 2 were analyzed for ubiquitin-containing species by probing with HA antibody (α-HA) (lanes 1 to 6). For comparison, lane 4 of Figure 2 is shown again as lane 7.
The stability of the 64-kDa Pap1 does not change during the cell cycle.
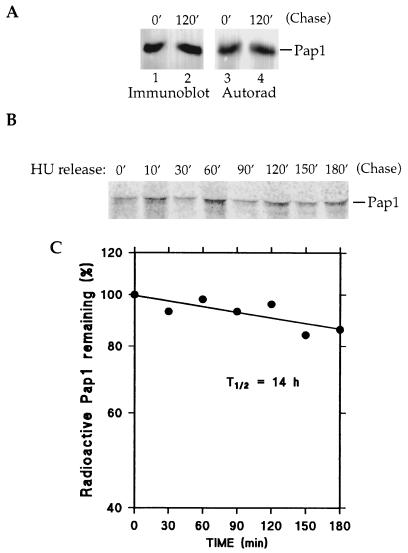
The above results demonstrate that a fraction of Pap1 becomes ubiquitinated at the S/G2 phase of the cell cycle. However, the steady-state levels of the 64-kDa Pap1 do not change after the S/G2 modification has occurred (Fig. 1B and 2). Since ubiquitination commonly targets a protein for degradation, we determined the stability of the 64-kDa protein by pulse-chase analysis with [35S]methionine/cysteine in the presence of cycloheximide. Radiolabeled extracts from asynchronous W303 cells prepared at the beginning of or 2 h into the chase period were immunoprecipitated with antibody to the C terminus of Pap1. Pap1 was first detected by immunoblotting (Fig. 8A, lanes 1 and 2), and the blot was then subjected to autoradiography (Fig. 8A, lanes 3 and 4). The amounts of Pap1 at the beginning and end of the chase period detected by blotting were identical, indicating that synthesis of the new protein had been blocked by cycloheximide. More important was the finding that the levels of Pap1 detected by radioactivity had also not changed. Thus, there was no turnover of Pap1 during this time.
FIG. 8.
The 64-kDa Pap1 is a stable protein. (A) Wild-type W303 cells were pulse-labeled with [35S]methionine/cysteine for 30 min and then chased with excess unlabeled methionine and cysteine in the presence of cysteine in the presence of cycloheximide for 120 min (120′). Radiolabeled Pap1 was immunoprecipitated, separated by SDS-PAGE, and then transferred to a polyvinylidene difluoride membrane. The 64-kDa Pap1 was detected by immunoblot analysis (lanes 1 and 2) and then subjected to autoradiography (Autorad) to show 35S-labeled Pap1 (lanes 3 and 4). (B) An experiment similar to that shown in panel A was conducted with synchronized cells. Wild-type cells were blocked at S with hydroxyurea (HU) for 120 min. Cell cycle arrest was monitored visually and determined to be complete when greater than 90% of cell showed large buds. Pulse-chase analysis was performed as described above for panel A but in the absence of cycloheximide. Samples were withdrawn at the indicated time points (in minutes), and Pap1 immunoprecipitates were detected by autoradiography. (C) 35S-labeled Pap1 from the experiment of panel B was quantified by PhosphorImager analysis. The graph shows the half-life of Pap1 calculated from the values found.
We also investigated whether Pap1 became unstable at a particular point in the cell cycle. Wild-type cells were arrested in S phase by hydroxyurea prior to the pulse. Samples were removed at different time points after the release from the hydroxyurea block and addition of excess unlabeled methionine/cysteine and in this experiment, in the absence of cycloheximide. PhosphorImager detection of immunoprecipitated Pap1 indicated that the level of 64-kDa Pap1 did not change throughout the cell cycle (Fig. 8B), confirming that it is a stable protein and that its level is not regulated in a cell cycle-dependent manner. The 64-kDa Pap1 has an estimated half-life of 14 h (Fig. 8C). We were not able to detect the 90-kDa Pap1 species under these conditions, perhaps because none of the time points sampled coincided with the narrow window of Pap1 phosphorylation.
DISCUSSION
Our results demonstrate that the yeast Pap1 is phosphorylated and ubiquitinated in a cell cycle-dependent manner. By examination of synchronized cells, we have found that Pap1 phosphorylation begins at S and persists through G2 but disappears before nuclear division. Consistent with this result, a small amount of modified Pap1 is found in cells arrested in S by hydroxyurea. Furthermore, the phosphorylation does not appear to require the activity of the Cdc28 kinase. These findings are in contrast to those for the mammalian PAP, which is phosphorylated by the Cdc28 homologue in the M phase of the cell cycle (9). The difference in the timing of modification implies an intriguing difference in cell cycle regulation of mRNA polyadenylation in the yeast and mammalian systems.
In the budding yeast S. cerevisiae, unlike mammalian cells, there is a lack of clear definition between S, G2, and M, and a partial overlap between the S and M phases (17). Events that would be restricted to late G2 or M phase in other organisms, including mitotic spindle assembly and activation of cyclin B-Cdc28 complexes, begin during the S phase (13). In addition, nuclear envelope disassembly, a distinctive hallmark of mammalian M phase, does not occur in budding yeast, and there is less dramatic chromosome condensation. These features of budding yeast are perhaps necessary to allow formation of a daughter bud, growth of the bud, and migration of the nucleus to the bud neck before mitosis.
These distinctions correlate with a difference in regulating the poly(A) polymerase. In mammals, down-regulation of PAP phosphorylation during mitosis is thought to be part of the general repression of mRNA synthesis seen at this time. Our data show that this modification in yeast occurs at S/G2 and not mitosis. This is the period of DNA replication, spindle pole formation, and active growth of the budding daughter cell, and a time when the expression of many genes involved in these events must be correctly choreographed. In mammals, the Cdc2-cyclin B complex phosphorylates PAP. While we do not yet know the kinase responsible for Pap1 phosphorylation, the continued appearance of the 90-kDa Pap1 in strains defective for Cdc28 suggests that it is not the yeast cdc2-cyclin B homologue. The three cdc28 mutations that we have examined have been studied extensively. The cdc28-4 and cdc28-13 mutants, which arrest in G1, are both thermolabile for kinase activity in vitro (44, 48). The cdc28-1N mutant is not a temperature-sensitive kinase in vitro (48), and the effect leading to a G2/M arrest is thought to involve its interaction with B-type cyclins or Cks1 (29, 48). However, compared to the protein from the wild-type strain, the protein from the cdc28-1N mutant is highly unstable at the restrictive temperature (4). Our data show that the cell cycle regulation of the Pap1 phosphorylation is perturbed in these mutant strains, indicating that Cdc28 probably modulates the activity of enzymes involved in producing or removing the phosphorylated Pap1.
Like the mammalian PAP, we have found that Pap1 phosphorylation is also associated with enzyme inactivation. It does not appear to affect the cellular localization of the enzyme during the cell cycle (data not shown). However, in contrast to the mammalian situation, only a portion of Pap1 is phosphorylated. Furthermore, the overall amount of poly(A) does not change during the cell cycle (data not shown). These observations suggest that a possible function for Pap1 modification may be to regulate the polyadenylation of a subset of yeast mRNAs whose products are needed primarily during, but not after, S phase. One class of such mRNAs might be those encoding histones (21, 35). The amount of histone mRNAs in a cell increases by as much as 20-fold during the period of DNA replication, and part of this regulation occurs at a posttranscriptional level. In higher eukaryotes, this posttranscriptional component involves both 3′-end processing and mRNA stability, but the nature of this aspect of the regulation has not been determined for yeast. In contrast to higher eukaryotes, which use a specialized cleavage machinery for histone mRNA 3′-end processing, the yeast histone pre-mRNAs undergo both cleavage and polyadenylation (21). Another interesting example is telomerase RNA, an untranslated stable nuclear RNA involved in replicating the ends of the chromosomes. This RNA is induced in S phase, and a small fraction of the S. cerevisiae telomerase RNA is polyadenylated in a Pap1-dependent manner in vivo (6). Finally, a recent genomewide analysis of the yeast mitotic cell has found that the levels of at least 74 different mRNA transcripts peaked in S phase (8). Negative regulation of polyadenylation by Pap1 modification may contribute to the decline in the levels of these transcripts as cells move out of S phase. Such a regulation could occur throughout the nucleus or be more pronounced by limitation to a subnuclear space, if, for example, the Pap1-modifying enzymes were recruited or activated only in the vicinity of certain transcriptional units.
It was surprising to find that a small amount of Pap1 was ubiquitinated. The ubiquitin system is generally associated with rapid protein degradation, yet Pap1 is a stable protein with an estimated half-life of 14 h. Ubiquitination can have regulatory functions that do not involve proteolysis. For example, the first protein identified as a ubiquitin target, histone H2A, is a stably ubiquitinated protein in vivo (19). Ubiquitin conjugation has also been implicated in protein-protein interaction (3) and in the endocytosis of membrane proteins (23). A ubiquitinated form of actin, arthrin, is a stable myofibril component involved in thin filament assembly or function of the myofibrillar protein of Drosophila flight muscle (3).
Other stable proteins also become ubiquitinated. One example is the large subunit of the yeast RNA polymerase II, Rbp1 (26). Ubiquitination of Rbp1 is mediated by the ubiquitin-protein ligase, Rsp5. However, Pap1 is not a substrate for this class of ligases (J. Huibregtse, N. Mizrahi, and C. Moore, unpublished data). Another intriguing example is the nucleotide excision repair protein Rad23 (51). This normally stable protein becomes short-lived under certain conditions (47). Rad23 degradation is thought to be mediated by an N-terminal Ub-like domain and the UFD pathway (49). This pathway is involved in the degradation of artificial fusion proteins, which have an in-frame ubiquitin moeity at their N terminus. Other physiological substrates for this pathway have not been identified (49). Interestingly, Pap1 interacts specifically with Ufd1 (12) and has two regions with approximately the same amount of homology to ubiquitin as the Rad23 Ub-like domain, suggesting that its ubiquitination may require the UFD pathway. Ubiquitination and subsequent degradation of a stable protein such as Pap1 may be important to eliminate defective protein, to tightly control its cellular level, or to remove protein not associated with polyadenylation specificity factors. In support of these possibilities, it has been shown that overexpression of PAP in chicken cells is detrimental to cell growth (54).
While phosphorylation is often a prerequisite for recognition by the ubiquitination machinery (22), the relationship between the phosphorylated and ubiquitinated forms of Pap1 is not clear. The ubiquitinated species appear to be larger than the 90-kDa phosphorylated form. In addition, overexpression of ubiquitin causes accumulation of the 90-kDa Pap1 in unsynchronized cells, but this may be an indirect effect. However, we have also observed that phosphatase treatment does not affect the ubiquitinated forms (data not shown), suggesting that these modifications may occur independently. The resolution of this issue will require mapping the sites of phosphorylation and ubiquitination and identifying the proteins involved in Pap1 modifications.
We do not yet understand the full significance of Pap1 modifications reported here but they may be critical for progression through the cell cycle in yeast. The ramifications of these multiple modifications on Pap1 activity and its interaction with the RNA substrate and other RNA processing factors during the cell cycle will be important areas of future investigation.
ACKNOWLEDGMENTS
Plasmids and strains used in this study were generously provided by Dan Finley, David Leggett, Angelika Amon, Steve Reed, Mike Tyers, Paul Ferrigno, and Mark Hochstrasser. We are grateful to Paulo Dice, Jerry Faust, and Gary Sahagian for critical comments and continuous encouragement and support during the course of this study. We thank Dorothy Fallows, Paul Ferrigno, Elizabeth Joyce, Steffen Helmling, and Debu Raychaudhuri for critically reading the manuscript. Special thanks goes to Marco Kessler for providing Pap1 antibodies, and Ana Maria Cuervo for help determining the Pap1 half-life in Fig. 8C.
Neptune Mizrahi was supported by a grant from the Lucille P. Markey Foundation to the Physiology Department. This work was also supported by ACS and NIH grants to C.L.M.
REFERENCES
- 1.Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;81:269–277. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- 2.Armani N, Minet M, LeGouar M, Lacroute F, Wyres F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball E, Karlik C C, Saville D L, Sparrow J C, Bullard B, Fyrberg E A. Arthrin, a myofibrillar protein of insect flight muscle, is an actin-ubiquitin conjugate. Cell. 1987;51:221–228. doi: 10.1016/0092-8674(87)90149-8. [DOI] [PubMed] [Google Scholar]
- 4.Betting J, Seufert W. A yeast Ubc9 mutant protein with temperature-sensitive in vivo function is subject to conditional proteolysis by a ubiquitin- and proteasome-dependent pathway. J Biol Chem. 1996;271:25790–25796. doi: 10.1074/jbc.271.42.25790. [DOI] [PubMed] [Google Scholar]
- 5.Carpousis A J, Vanzo N F, Raynal L C. mRNA degradation: a tale of poly(A) and multiprotein machines. Trends Biochem Sci. 1999;15:24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- 6.Chapon C, Cech T R, Zaug A J. Polyadenylation of telomerase RNA in budding yeast. RNA. 1997;3:1337–1351. [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Moore C L. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol Cell Biol. 1992;12:3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho R J, Campbell M J, Winzeler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, Davis R W. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;1:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 9.Colgan D, Murthy K, Prives C, Manley J. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- 10.Colgan D, Manley J. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 11.Cuervo A M, Hu W, Lim B, Dice J F. IkB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell. 1998;9:1995–2010. doi: 10.1091/mbc.9.8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Olmo M, Mizrahi N, Gross S, Moore C. The Uba2 and Ufd1 proteins of S. cerevisiae interact with poly(A) polymerase and affect the polyadenylation activity of extracts. Mol Gen Genet. 1997;255:209–218. doi: 10.1007/s004380050491. [DOI] [PubMed] [Google Scholar]
- 13.Deshaies R J. Phosphorylation and proteolysis: partners in the regulation of cell division in budding yeast. Curr Opin Genet Dev. 1997;7:7–16. doi: 10.1016/s0959-437x(97)80103-7. [DOI] [PubMed] [Google Scholar]
- 14.Dien B, Peterson M, Srienc F. Cell cycle analysis of Saccharomyces cerevisiae. Methods Cell Biol. 1994;42:457–475. doi: 10.1016/s0091-679x(08)61090-0. [DOI] [PubMed] [Google Scholar]
- 15.Ellison M J, Hochstrasser M. Epitope-tagged ubiquitin. J Biol Chem. 1991;266:21150–21157. [PubMed] [Google Scholar]
- 16.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 17.Forsburg S L, Nurse P. Cell cycle regulation in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Annu Rev Cell Biol. 1991;7:227–256. doi: 10.1146/annurev.cb.07.110191.001303. [DOI] [PubMed] [Google Scholar]
- 18.Goebl M G, Yochem J, Jentsch S, McGrath J P, Varshavsky A. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 19.Goldknopf I L, Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc Natl Acad Sci USA. 1977;74:864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottesfeld J, Forbes D. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 21.Heintz N. The regulation of histone gene expression during the cell cycle. Biochim Biophys Acta. 1991;1088:327–339. doi: 10.1016/0167-4781(91)90122-3. [DOI] [PubMed] [Google Scholar]
- 22.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 23.Hicke L, Reizman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 24.Hochstrasser M, Ellison M J, Chau V, Varshavsky A. The short-lived MATa-2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, Carmichael G G. A suboptimal 5′ splice site is a cis-acting determinant of nuclear export of polyomavirus late mRNAs. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.11.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huibregtse J, Yang J, Beaudenon S. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson E, Ma P, Ota I, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 28.Johnston L H, Nasmyth K A. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978;274:891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- 29.Kaiser P, Moncollin V, Clarke D J, Watson M H, Bertolaet B L, Reed S I, Bailly E. Cyclin-dependent kinase and Cks/Suc1 interact with the proteasome in yeast to control proteolysis of M-phase targets. Genes Dev. 1999;13:1190–1202. doi: 10.1101/gad.13.9.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessler M, Henry M, Gross S, Shen E, Zhao J, Silver P, Moore C. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kessler M M, Zhelkovsky A M, Skvorak A, Moore C L. Monoclonal antibodies to yeast poly(A) polymerase (PAP) provide evidence for association of PAP with Cleavage Factor I. Biochemistry. 1995;34:1750–1759. doi: 10.1021/bi00005a032. [DOI] [PubMed] [Google Scholar]
- 32.Lingner J, Radtke I, Wahle E, Keller W. Purification and characterization of poly(A) polymerase from S. cerevisiae. J Biol Chem. 1991;266:8741–8746. [PubMed] [Google Scholar]
- 33.Lingner J, Kellermann J, Keller W. Cloning and expression of the essential gene for poly(A) polymerase from S. cerevisiae. Nature. 1991;354:496–498. doi: 10.1038/354496a0. [DOI] [PubMed] [Google Scholar]
- 34.Lorincz A, Reed S I. Sequence analysis of temperature-sensitive mutations in the Saccharomyces cerevisiae gene CDC28. Mol Cell Biol. 1986;6:4099–4103. doi: 10.1128/mcb.6.11.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lycan D E, Osley M A, Hereford L M. Role of transcriptional and posttranscriptional regulation in expression of histone genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:614–621. doi: 10.1128/mcb.7.2.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangus D, Amrani N, Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol Cell Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 38.Mayer T U, Braun T, Jentsch S. Role of the proteasome in membrane extraction of a short-lived ER-transmembrane protein. EMBO J. 1998;17:3251–3257. doi: 10.1093/emboj/17.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minvielle-Sebastia L, Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to the transcription. Curr Opin Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- 40.Minvielle-Sebastia L, Preker P, Wiederkehr T, Strahm Y, Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc Natl Acad Sci USA. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel D, Butler J S. Conditional defect in mRNA 3′ end processing caused by mutation in the gene for poly(A) polymerase. Mol Cell Biol. 1992;12:3297–3304. doi: 10.1128/mcb.12.7.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piggott J R, Rai R, Carter B L. A bifunctional gene product involved in two phases of the yeast cell cycle. Nature. 1982;298:391–393. doi: 10.1038/298391a0. [DOI] [PubMed] [Google Scholar]
- 43.Preker P J, Lingner J, Minville-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast polyadenylation factor that interacts with poly(A) polymerase. Cell. 1995;89:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 44.Reed S I, Wittenberg C. Mitotic role for the Cdc28 protein kinase of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:5697–5701. doi: 10.1073/pnas.87.15.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sachs A, Sarnow P, Hentze M. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 46.Saitoh H, Pu R T, Dasso M. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 47.Schauber C, Chen L, Tongaonkar P, Vega I, Lamberston D, Potts W, Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 48.Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher A B, Nasmyth K. The role of CDC28 and cyclins during mitosis in budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- 49.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 50.Wahle E, Rüegsegger U. 3′-end processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;648:1–18. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 51.Watkins J F, Sung P, Prakash L, Prakash S. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol Cell Biol. 1993;13:7757–7765. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 53.Zhao J, Hyman L, Moore C L. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao W, Manley J L. Deregulation of poly(A) polymerase interferes with cell growth. Mol Cell Biol. 1998;18:5010–5020. doi: 10.1128/mcb.18.9.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhelkovsky A, Helmling S, Moore C L. Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol Cell Biol. 1998;18:5942–5951. doi: 10.1128/mcb.18.10.5942. [DOI] [PMC free article] [PubMed] [Google Scholar]