Abstract
Dpb11 is required for chromosomal DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Here, we report detection of a physical complex containing Dpb11 and DNA polymerase ɛ (Dpb11-Polɛ complex). During the S phase of the cell cycle, Dpb11 associated preferentially with DNA fragments containing autonomously replicating sequences (ARSs), at the same time as Polɛ associated with these fragments. Association of Dpb11 and Polɛ with these fragments was mutually dependent, suggesting that the Dpb11-Polɛ complex associates with the ARS. Moreover, Dpb11 was required for the association of Polα-primase with the fragments. Thus, it seems likely that association of the Dpb11-Polɛ complex with the ARS fragments is required for the association of the Polα-primase complex. Hydroxyurea inhibits late-origin firing in S. cerevisiae, and the checkpoint genes, RAD53 and MEC1, are involved in this inhibition. In the presence of hydroxyurea at temperatures permissive for cell growth, Polɛ in dpb11-1 cells associated with early- and late-origin fragments. In wild-type cells, however, it associated only with early-origin fragments. This indicates that Dpb11 may also be involved in the regulation of late-origin firing. Overall, these results suggest that Dpb11 controls the association between DNA polymerases α and ɛ and the ARS.
Eukaryotic chromosomal DNA replicates exactly once per cell cycle, in the S phase. In Saccharomyces cerevisiae, chromosomal DNA replication is initiated at a restricted region known as the autonomously replicating sequence (ARS) (reviewed in references 10 and 43). An origin recognition complex (ORC), comprising six subunits, is bound to each ARS throughout the cell cycle (6, 14). At the end of mitosis, six Mcm family proteins (Mcm2 to -7) are also loaded with the ORC onto the ARS as components of the prereplicative complex (pre-RC) (14). The Mcm proteins have a conserved amino acid sequence and form large complexes (11, 29, 48). Loading of these proteins requires both the ORC and Cdc6 (1, 15, 46).
At the onset of S phase, Cdk and Cdc7 protein kinases facilitate loading of the single-stranded DNA binding protein, RPA, onto the ARS (47). Then, the three DNA polymerases (Polα, -δ, and -ɛ), essential for chromosomal DNA replication (reviewed in reference 45), are recruited to the ARS region to initiate DNA synthesis (1, 2, 47). Association of Polα and Polɛ with the ARS region is dependent on Cdc45 (2), which associates with the ARS in the G1 and S phases of the cell cycle (1, 2) and interacts with the Mcm proteins (12, 21, 22, 24, 32, 52).
All the replication proteins described above are well conserved from yeasts through humans. The Cdc45 protein of Xenopus laevis is also required for association of Polα with chromatin DNA (31), as observed in yeast. Moreover, Cdc45 and Polα form a complex in Xenopus egg extracts (31). In in vitro simian virus 40 (SV40) DNA replication, T antigen unwinds the replication origin and RPA binds the unwound single-stranded DNA. DNA primase, tightly associated with Polα, then synthesizes an RNA primer. This RNA primer is used by Polα to synthesize a short DNA strand, followed by elongation of the DNA strand by Polδ and/or Polɛ, using the short DNA fragment as a primer (43). Although the cellular counterpart of the SV40 T antigen has not been identified, it is believed that similar reactions take place during chromosomal DNA replication.
DNA replication in eukaryotic cells initiates from multiple replication origins that fire throughout the S phase of the cell cycle; some origins fire early, others fire late (17, 25). In S. cerevisiae, hydroxyurea (HU), which inhibits ribonucleotide reductase and consequently inhibits DNA synthesis, blocks the firing of late origins. In rad53 and mec1 mutants (defective in cell cycle checkpoints), however, late-origin firing is not blocked by HU (40). Methyl methane sulfonate similarly inhibits late-origin firing, and this inhibition is not observed in rad53 or orc2 mutants (42). From these observations, it is proposed that the regulation of late-origin firing is important for the S-phase checkpoint (13, 40, 42).
The DPB11 gene was isolated as a multicopy suppressor of mutations in the POL2 and DPB2 genes, which encode the catalytic and second-largest subunits of Polɛ, respectively (5). The amino acid sequence of Dpb11 is similar to the sequence of the Cut5 (also known as Rad4) protein of Schizosaccharomyces pombe. This protein is required for the onset of the S-phase and cell cycle checkpoint in S. pombe (30, 37–39, 49). Both Dpb11 and Cut5 have four copies of the BRCA1 C-terminus (BRCT) domain, which is thought to be an interaction domain between proteins (7, 9, 51).
In thermosensitive dpb11-1 mutants, S-phase progression is delayed when the temperature is shifted up, followed by cell division with unequal chromosome segregation. In the presence of HU, dpb11-1 cells also have an elongated spindle, indicating that mitosis has started without the completion of DNA replication. Furthermore, there is a strong genetic interaction between Dpb11 and Polɛ; high-copy DPB11 suppresses the growth defects of pol2-11 and dpb2-1, and no combination of dpb11-1 with one of pol2-11, pol2-18, and dpb2-1 is obtained. This suggests that Dpb11 interacts with Polɛ and is required for DNA replication and the S-phase checkpoint (5).
To gain a broader understanding of the function of Dpb11, we tried to identify the factors that interact with Dpb11 by isolating synthetic lethal mutations with dpb11-1 (sld). So far, we have isolated five SLD genes. SLD1 is identical to the DPB3 gene that encodes the third-largest subunit of Polɛ (4), and SLD4 is identical to CDC45 (22). The SLD2 gene encodes a 52-kDa protein that forms a complex with Dpb11 that is essential for DNA replication. From this analysis, we suggested that a Dpb11-Sld2 complex is required for one of the steps close to the initiation of DNA replication (27). The SLD2 gene was independently isolated as the DRC1 (DNA replication and checkpoint 1) gene, and the drc1-8 mutant was found to be defective in the S-phase checkpoint (50).
To further elucidate the function of Dpb11, we analyzed the association between Dpb11, DNA polymerases, and chromatin DNA, using a chromatin immunoprecipitation (CHIP) assay (44). In the S phase of the cell cycle, Dpb11 and Polɛ simultaneously associated with DNA fragments containing an ARS, and their association was mutually dependent. We also detected a complex of Dpb11 and Pol2 that was most abundant during the S phase. Moreover, Dpb11 was required for blockage of late-origin firing by HU. Therefore, we suggest that Dpb11 is required for DNA polymerases to associate with the ARS and for blockage of late-origin firing.
MATERIALS AND METHODS
Plasmids.
Hemagglutinin (HA) or c-myc epitope-tagged gene fusions were constructed by PCR amplification of the coding regions of interest and insertion of the resulting PCR products into plasmid pUC18. NotI-3HA or NotI-9myc cassettes were inserted into the resulting plasmids, creating a C-terminal fusion with each coding sequence. An N-terminally deleted fragment of each gene fusion was subcloned into an integrating vector of the YIp family. The resulting plasmids, YIplac128-POL2-3HA/C and YIplac211-DPB11-9myc/C, were digested with PstI before yeast transformation. For Mcm4-3HA, p404-CDC54-HA/C (1) was digested with NruI prior to yeast transformation.
Yeast strains.
W303-1Ab (Δbar1) was constructed from W303-1A (MATa ura3-1 trp1-1 leu2-3,112 his3-11,15 ade2-1 can1-100) by replacing the endogenous BAR1 gene with a URA3 insertion mutant allele and subsequently popping out the URA3 gene. W303-1Ab (bar1) was the parent strain for most of the strains used in this study. W303-1Ab was transformed with YIplac211dpb11-1 (5), and the Ura+ transformants were grown at 25°C on plates containing 5-fluoroorotic acid. A temperature-sensitive colony (YHM1) was isolated from the plates. Strains harboring POL2-3HA, DPB11-9my, or MCM4-3HA were constructed by a homologous recombination that disrupted the wild-type allele and simultaneously created a C-terminal epitope-tagged version of the gene in W303-1Ab and YHM1. The resulting strains were YHM01 (POL2-3HA), YHM08 (MCM4-3HA), YHM011 (DPB11-9myc), YHM11 (dpb11-1 POL2-3HA), and YHM18 (dpb11-1 MCM4-3HA). YHM21 (rad53-1 POL2-3HA), YHM311 (mcm5-1 DPB11-9myc), and YHM511 (cdc17-1 DPB11-9myc) were constructed from YHY301a (MATa rad53-1; W303 background), K5581 (MATa mcm5-1; W303 background), and K6877 (MATa cdc17-1; W303 background) by disruption of BAR1 and integration of POL2-3HA or DPB11-9myc. YHM01101 (POL2-3HA DPB11-9myc) was constructed by further integration of Pol2-3HA into YHM011. YHM013 (MATa PRI1-9myc Δbar1) and YHM113 (MATa dpb11-1 PRI1-9myc Δbar1) were segregants from a cross of YHM11 and K7213 (MATα PRI1-9myc; W303 background). YHM014 (MATa Δbar1 RFA1-18myc) was a segregant from a cross of K7131 (MATα RFA1-18myc; W303 background) and W303-1Ab. YHM114 was constructed by replacing the endogenous DPB11 gene of YHM014 with the dpb11-1 mutant allele as YHM1 was constructed. Similarly, YHM611 (MATa rfa2-2 DPB11-9myc) was constructed by replacing the endogenous RFA2 gene of YHM011 with the rfa2-2 allele on pHC36 obtained from P. Plevani (University of Milan, Milan, Italy). YHA500 (MATa dpb2-1 POL2-3HA DPB11-9myc) and YHA503 (MATa dpb2-1 DPB11-9myc) were segregants from a cross of YHA422 (MATα dpb2-1 Δbar1; W303 background) and YHM01101. K5581, K6877, K7131, and K7213 were obtained from T. Tanaka (Research Institute of Molecular Pathology, Vienna, Austria) and YHY301a from K. Shirahige (Nara Institute of Science and Technology, Nara, Japan). YHA410 and YHA411 used for two-dimensional (2D) gel analysis were described previously (27).
Synchronization of cells.
For α-factor arrest and release experiments at 16°C, cells were suspended in fresh YPD medium (26) containing 30 ng of α-factor per ml and incubated for 3 h at 25°C. The α-factor was removed by centrifugation, and the cells were resuspended at 1.3 × 107 cells/ml in YPD containing 0.1 mg of actinase E (Kakenseiyaku) per ml and incubated further at 16°C. For α-factor arrest-and-release experiments at 25 and 36°C, the cells were suspended in fresh YEPR medium (2% raffinose instead of glucose in YPD) and incubated for 2 h at 25°C. The α-factor was added to a final concentration of 30 ng/ml and incubated for 3 h at 25°C. Additionally, 0.2 M HU was added and incubated for 1 h at 25°C. The α-factor was removed by centrifugation, and the cells were resuspended at 1.3 × 107 cells/ml in YEPR containing 0.1 mg of actinase E per ml and 0.2 M HU and incubated at 25°C or 36°C.
Immunoprecipitation.
Approximately 109 cells with a density of 107 cells/ml were spheroplasted according to Donovan et al. (15) with modifications. Cells were incubated at room temperature for 10 min in 12.5 ml of prespheroplasting buffer (100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] [pH 9.4] and 10 mM dithiothreitol), followed by incubation at 30°C for 15 min with agitation in 5 ml of YPD supplemented with 0.6 M sorbitol, 25 mM Tris-HCl [pH 7.5], and 100 μl of Zymolase20T (50 mg/ml) (Seikagakukogyo). The spheroplasts were incubated in 5 ml of YPD–0.7 M sorbitol–25 mM Tris-HCl (pH 7.5) at 30°C for 20 min, followed by washing the spheroplasts twice with 2 ml of cross-linking buffer (0.4 M sorbitol, 150 mM Na acetate, 2 mM Mg2+ acetate, 20 mM HEPES-KOH [pH 7.5]). The spheroplasts were suspended in 2 ml of cross-linking buffer containing the cross-linking agent dithiobis(succinimidylpropionate) (DSP; Pierce) to a final concentration of 2 mM (20, 36). After 30-min incubation at room temperature, the reaction was stopped by incubation for 5 min with 50 mM Tris-HCl (pH 7.5). The spheroplasts were pelleted in a microfuge, washed with 1 ml of cross-linking buffer, and resuspended in lysis buffer (20 mM HEPES-KOH [pH 7.5] 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1× Complete Proteinase Inhibitor Cocktail (Roche), and 1 mM phenylmethylsulfonyl fluoride). The lysates were clarified by centrifugation, and stored at −80°C. Extracts (1 mg) were incubated with anti-HA monoclonal antibody (MAb) 12CA5 at 4°C for 3 h, followed by incubation for 1 h with protein G-Sepharose beads (Pharmacia). The beads were washed three times in lysis buffer supplemented with 360 mM NaCl. In the immunoprecipitation-immunoblotting experiments, 15 μg of protein from each extract was loaded per lane. Extracts and immunoprecipitates were separated in a sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel and analyzed by Western blotting.
CHIP assay.
The CHIP assay used in this study is based on the methods described by Tanaka et al. (46) with some modifications. The soluble fraction of the whole-cell extract was discarded, and the insoluble pellet was resuspended in 500 μl of lysis buffer. This chromatin-containing suspension was sonicated to yield an average DNA size of 500 bp (range, 100 to 1,000 bp), clarified by centrifugation, and subjected to immunoprecipitation with anti-HA MAbs (12CA5) or anti-myc (9E11) MAbs conjugated to protein G-Sepharose beads (Pharmacia). PCR was carried out in 50-μl volumes containing 1/60 to 1/15 of the anti-myc or anti-HA antibody immunoprecipitates, respectively, or 1/6,000 of the cross-linked DNA samples derived from the whole-cell extract. Taq polymerase (AmpliTaq Gold; Perkin-Elmer) and the corresponding buffer system were used. PCR primer sequences and cycle conditions are available upon request. Three pairs of primers were used together in each PCR. The final concentration of each primer was set up to amplify each fragment evenly from total genomic DNA. The PCR products were separated in a 2.5% agarose gel and stained with 0.2 μg of ethidium bromide per ml. The gels were photographed using a Polaroid MP-4 Land Camera, and the negatives were scanned directly into Photoshop 3.0 (Adobe).
Other methods.
Fluorescence-activated cell sorter (FACS) and 2D gel analyses were performed as described previously (27).
RESULTS
Dpb11 physically interacts with Polɛ.
Previously, we have shown a strong genetic interaction between Dpb11 and Polɛ (5). Using a two-hybrid assay (data not shown), we have also observed an interaction between Dpb11 and Dpb2, the second-largest subunit of Polɛ (3). These results suggest that Dpb11 and Polɛ form a complex. We have subsequently tried to detect a physical interaction between Dpb11 and the catalytic subunit of Polɛ, Pol2 (33).
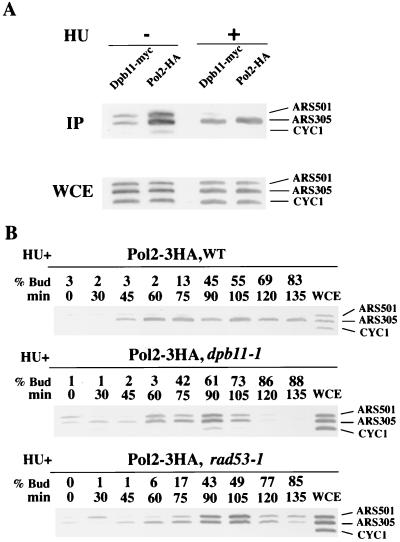
Cells harboring DPB11-9myc and POL2-3HA on their chromosomes were grown, and the cell wall was digested. The resultant spheroplasts were treated with the cross-linking agent, DSP, and Pol2 was precipitated from the cell lysates with an anti-HA antibody. The precipitated sample was subjected to SDS-polyacrylamide gel electrophoresis, and Dpb11 and Pol2 were detected on the membranes using anti-myc and anti-HA antibodies, respectively. As shown in Fig. 1A, Dpb11 coimmunoprecipitated with Pol2, although in the absence of the cross-linking agent, no coimmunoprecipitation was detected. To know the specificity of coimmunoprecipitation in the presence of the cross-linking agent, we examined whether Mcm2 and tubulin coprecipitate with Pol2 using anti-Mcm2 and antitubulin antibodies. Then, we found that neither Mcm2 nor tubulin coimmunoprecipitates with Pol2 (Fig. 1B). These results suggest that the specificity of coimmunoprecipitation remains in the presence of the cross-linking agent, and therefore the Dpb11-Pol2 complex is not an artifact of cross-linking. It is also suggested that Pol2 weakly or transiently interacts with Dpb11 because the Dpb11-Pol2 complex was only detected in the presence of the cross-linking agent.
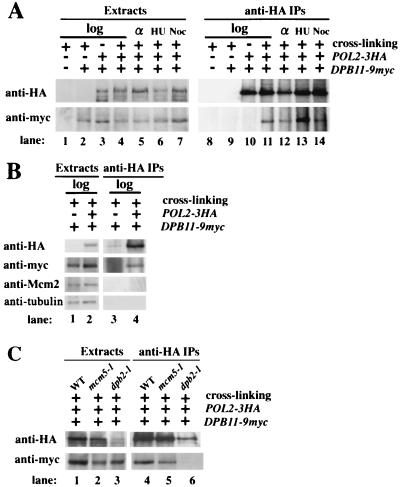
FIG. 1.
Dpb11 and Pol2 form a complex. (A) Cells of W303-1Ab, YHM011 (DPB11-9myc), and YHM01101 (POL2-3HA DPB11-9myc) were spheroplasted and treated with DSP (+) or left untreated (−). Lysates were prepared from log-phase cells (log) and cells arrested by α-factor (α), HU, and nocodazole (Noc) with the indicated genotypes: an epitope-tagged gene (+) and a wild-type allele (−). HA-tagged Pol2 was immunoprecipitated with anti-HA mouse MAb 12CA5. Extracts or immunoprecipitates were separated in a SDS–7.5% polyacrylamide gel and myc-tagged Dpb11 and HA-tagged Pol2 were detected by immunoblotting, using anti-myc rabbit polyclonal antibodies or anti-HA mouse MAb 16B12, respectively. (B) Extracts and immunoprecipitates from YHM011 (lanes 1 and 3) and YHM01101 (lanes 2 and 4) were separated in a SDS–7.5% polyacrylamide gel as described in panel A. In addition to Pol2 and Dpb11, Mcm2 and tubulin were detected by immunoblotting with anti-Mcm2 (yN-19; Santa Cruz Biotechnology) and anti-tubulin (YOL1/34) antibodies. (C) YHM01101, YHM31101 (mcm5-1), and YHA500 (dpb2-1 POL2-3HA DPB11-9myc) cells were grown at 25°C and then cultured at 36°C for 1 h. The cell lysates were subjected to immunoprecipitation using an anti-HA antibody.
We also examined the formation of a complex between Dpb11 and Polɛ during the cell cycle. Cells were arrested with α-factor (G1 phase), HU (S phase), or nocodazole (M phase) and treated with DSP, and Pol2 was immunoprecipitated. Coimmunoprecipitation of Dpb11 and Pol2 was observed in all the cells examined, and Dpb11 was most abundant in the precipitates from cells arrested by HU (Fig. 1A). Furthermore, Pol2 and Dpb11 coimmunoprecipitation was also observed in mcm5-1 mutant defective in the initiation of DNA replication at the restrictive temperature (Fig. 1C). These results suggested that formation of a complex between Dpb11 and Pol2 does not depend on DNA synthesis. Dpb11 did not associate with the ARS fragments of the mcm5-1 mutants (see below), and the Dpb11-Pol2 complex remained intact after DNase I treatment (data not shown), suggesting that the Dpb11-Pol2 complex is not bridged by DNA fragments.
We further examined the Dpb11-Pol2 complex formation in dpb2-1 cells from which Pol2 and Dpb2 are rarely copurified (3). The amount of Pol2 was reduced in the dpb2-1 cells, probably because of the unstable Polɛ complex, and coimmunoprecipitation of Pol2 and Dpb11 was not observed (Fig. 1C). Coimmunoprecipitation of these proteins could not be observed even when the amount of Pol2 was increased by using five times the number of dpb2-1 cells (data not shown). This result indicates that the amount of the Dpb11-Polɛ complex is severely reduced in dpb2-1 cells, and complex formation between Dpb11 and Polɛ requires Dpb2.
Neutral-neutral 2D gel analysis of replication intermediates formed in synchronized dpb11-1 cells.
In a previous analysis of DNA replication intermediates in asynchronous dpb11-1 cells by neutral-neutral 2D gel electrophoresis (8), we showed that DNA replication initiated from ARS306 is rapidly abolished after temperature upshift (27). As a replication fork initiating from an origin other than ARS306 has been observed in dpb11-1 cells (27), we analyzed the replication intermediates in synchronized dpb11-1 cells.
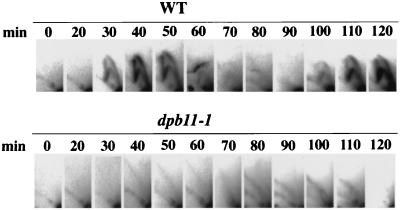
Wild-type and dpb11-1 cells were arrested in late G1 phase with α-factor and released in fresh medium at a temperature nonpermissive for cell growth. Cells were taken at 10-min intervals from the cultures, and the replication intermediates from an active origin, ARS1, were analyzed by 2D gel electrophoresis (8). Bubble arc signals, indicative of replication initiation in the ARS1 region, were observed in the wild-type cells from 30 to 50 min and 100 to 120 min after release from α-factor, suggesting the presence of two replication cycles. However, neither bubble arcs nor Y arcs that are generated from replication forks initiating from outside the ARS1 and passing through the ARS1 region were observed in the dpb11-1 cells (Fig. 2). This indicates that DNA synthesis initiating from ARS1 and other origins is reduced at temperature nonpermissive for cell growth, suggesting that Dpb11 is required for the initiation and/or the early step of elongation of DNA replication at ARS.
FIG. 2.
Neutral-neutral 2D gel analysis of the chromosomal ARS1 locus in synchronized cells. YHA410 (WT) and YHA411 (dpb11-1) cells were arrested in G1 phase by α-factor and released to YPD medium at 37°C. The same number of cells at each time point (in minutes) was subjected to 2D gel electrophoresis and probed for the 5-kb NcoI fragment containing ARS1.
Dpb11 and Polɛ associate simultaneously with ARS fragments in S phase.
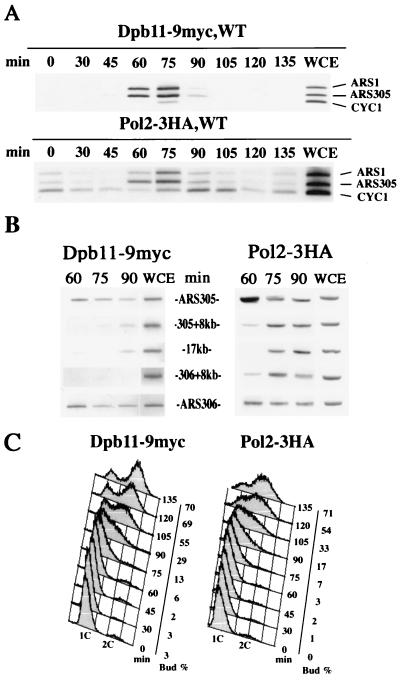
To investigate the function of Dpb11 during the cell cycle, we used a CHIP assay (44). Cells harboring Pol2-3HA or Dpb11-9myc were arrested by α-factor and released at 16°C to slow movement of the replication fork (1). Cells were withdrawn from the cultures every 15 min and fixed with formaldehyde. The chromatin fraction was obtained from these cells, sonicated, and subjected to immunoprecipitation with anti-HA or anti-myc antibodies. The whole cell extracts and the immunoprecipitates were subjected to SDS-polyacrylamide gel electrophoresis, followed by Western blotting with anti-HA or anti-myc antibody. The protein levels of Pol2-3HA and Dpb11-9myc in the whole-cell extracts and the immunoprecipitates did not fluctuate during the cell cycle. DNA was extracted from the immunoprecipitates and analyzed by PCR to determine the relative abundance of specific sequences bound to the immunoprecipitated HA- or myc-tagged proteins. We used three sets of PCR primers to amplify two ARS fragments (ARS1 and ARS305) fired early in S phase (18), and one non-ARS fragment in the CYC1 gene.
The CHIP assay showed that Pol2 associates with the ARS fragments containing ARS1 or ARS305 60 to 75 min after release from α-factor and then reassociates with non-ARS fragments as previously reported (Fig. 3A) (1). Dpb11 also preferentially associated with the ARS fragments 60 to 75 min after release and dissociated from the fragments thereafter. Thus, Dpb11 and Pol2 associate with ARS fragments at the same timing during the cell cycle. An association between Dpb11 and non-ARS fragments was not detected in this experiment (Fig. 3A). To examine whether Dpb11 associates with non-ARS fragments, we also used the primers specific to five unique sequences between the ARS305 and ARS306 that are the same as those described by Aparicio et al. (1). As shown in Fig. 3B, association signals of Pol2 with non-ARS fragments 8 kb distant from either ARS (305 plus 8 kb and 306 plus 8 kb) (Fig. 3B) were detected weakly at 60 min and strongly at 75 and 90 min. Pol2 association was also detected with the more distal fragment (17 kb) (Fig. 3B) at 75 min and 90 min. On the contrary, significant signals of Dpb11 association were detected only with ARS305 and ARS306 fragments. These results suggest that the Dpb11 association is limited to ARS fragments.
FIG. 3.
Dpb11 associates with the ARS fragments in S phase. (A and B) YHM01 (POL2-3HA, WT) and YHM011 (DPB11-9myc, WT) cells were arrested in G1 phase and released in YPD medium at 16°C. Cells were withdrawn from the culture every 15 min and fixed with formaldehyde. The chromatin fraction was sonicated and used for immunoprecipitation of HA- and myc-tagged proteins. PCR was performed either on immunoprecipitates derived from the same number of cells at each time point (in minutes) or on the 0-min chromatin fraction from the whole-cell extract (WCE). Note that faint signals of non-ARS fragments on the 90-min immunoprecipitates from YHM011 in panel B were not reproducibly obtained. (C) The DNA content was measured by FACS analysis of the samples collected in panel A. The percentage of budded cells is also shown.
Association of Dpb11 with the ARS fragments depends on Mcm, RPA, and Dpb2 but not Polα.
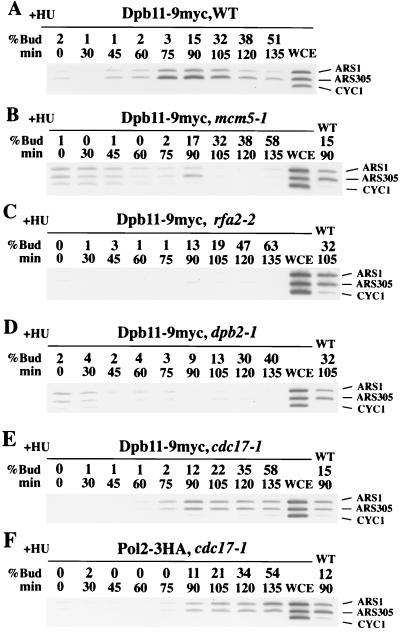
The pre-RC is assembled at ARSs from late M to G1 phase. This complex is then activated by Cdk and Cdc7 protein kinases, and DNA polymerases are recruited to the activated origins at the initiation of DNA replication (1). To determine if the association of Dpb11 with ARS fragments depends on Mcm5 (one of the pre-RC components), we performed a CHIP assay using mcm5-1 cells. We detected almost no selective signals in wild-type cells at 36°C by the CHIP assay, probably because Dpb11 moves rapidly away from the ARS fragments. To slow replication fork movement at 36°C, we inhibited ribonucleotide reductase and subsequently inhibited DNA synthesis, using HU. Under these conditions, Dpb11 was shown to be associated with ARS fragments in the wild-type cells (Fig. 4A), similar to the results obtained in the absence of HU at 16°C (Fig. 3A). However, no significant amplification of DNA fragments was detected in the mcm5-1 cells at the restrictive temperature (Fig. 4B). Thus, Mcm5 is required for Dpb11 association with the ARS fragments. Since mcm5-1 cells have a temperature-sensitive allele that causes a defect in pre-RC formation at the restrictive temperatures (46), it is likely that pre-RC formation is a prerequisite for Dpb11 association with the ARS fragments.
FIG. 4.
Association of the Dpb11 protein with the ARS fragments depends on the function of Mcm5, Rfa2, and Dpb2 but not on Cdc17 (Polα). YHM011 (DPB11-9myc) (A), YHM311 (DPB11-9myc mcm5-1) (B), YHM611 (DPB11-9myc rfa2-2) (C), YHA503 (DPB11-9myc dpb2-1) (D), YHM511 (DPB11-9myc cdc17-1) (E), and YHM501 (POL2-3HA cdc17-1) (F) cells were treated with α-factor for 3 h, 0.2 M HU was added, and the culture was incubated for another hour. The α-factor was removed, and the cells were suspended in YEPR medium containing 0.2 M HU at 36°C. PCR was performed as described in the legend to Fig. 3. In the presence of HU, the G1 cells of all strains retained 1C DNA contents throughout the progression of budding (data not shown). Note that the signals appeared after release from α factor in panels B and D were not observed reproducibly.
After the pre-RC is assembled on the ARS region, the single-stranded DNA binding protein, RPA, also associates with this region. RPA is required for association of the Polα-primase complex with the ARS region (47). To determine whether Dpb11 association depends on RPA, we performed a CHIP assay using rfa2-2 cells. These cells have a temperature-sensitive allele in the second-largest subunit of RPA (41) and do not associate Pri1, a subunit of DNA primase (19) with their ARS regions at the restrictive temperature (47). As shown in Fig. 4C, no significant amplification of DNA fragments was observed in these cells, suggesting that Dpb11 association is also dependent on the presence of functional RPA.
As Dpb11 physically interacts with Polɛ (Fig. 1), we investigated the dependency of the association between Dpb11 and the ARS fragments on DNA polymerase. The cdc17-1 mutation in the catalytic subunit of Polα (10) causes temperature-sensitive initiation and association defects between Pri1 and ARS regions (47). Using 2D agarose gel analysis, we found that dpb2-1 mutants rapidly lose their bubble arc signals after a temperature shift to the restrictive temperature (data not shown), suggesting that this mutation causes an initiation defect. Complex formation between Dpb11 and Pol2 was also defective in dpb2-1 cells (Fig. 1B). Therefore, cdc17-1 and dpb2-1 mutants were used for further analysis.
As shown in Fig. 4D, no significant signals for Dpb11 association with the ARS fragments were observed in dpb2-1 cells. In contrast, a significant signal was observed for the association between Dpb11, Pol2, and the ARS fragments in cdc17-1 cells (Fig. 4E and F). Thus, Dpb11 association with the ARS fragments depends on Polɛ but not on Polα. As no Polδ initiation defective mutants have been reported, we could not determine whether Dpb11 association depends on Polδ.
Dpb11-1 mutant cells are defective in association between DNA polymerase and ARS fragment.
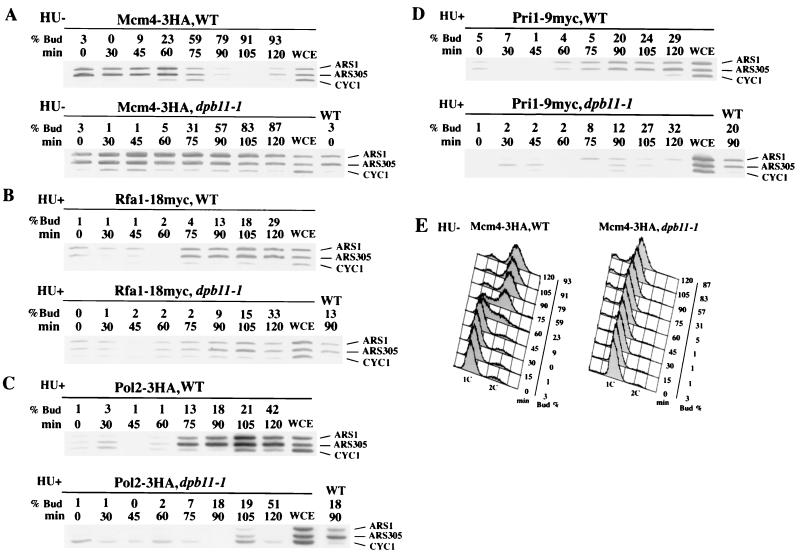
As Mcm proteins associate with the ARS fragments at the end of mitosis (14), we predicted that Mcm proteins remain associated with the ARS fragments in cells lacking Dpb11 function. To test this prediction, dpb11-1 cells harboring Mcm4-3HA were arrested in G1 phase by α-factor and released at 36°C without HU. As expected, Mcm4 remained associated with the ARS fragments throughout progress of budding in dpb11-1 cells, whereas it dissociated from the ARS fragments when DNA replication started (later than 60 min after release) in wild-type cells (Fig. 5A and E). Thus, Mcm association with ARSs does not require the presence of functional Dpb11, but dissociation of Mcm from these sequences does. This is probably because the Mcm proteins dissociate from the ARS fragments after DNA synthesis begins (1, 46), and DNA synthesis does not begin in dpb11-1 cells.
FIG. 5.
Association of Pol2 and Pri1 with ARS fragments requires Dpb11. (A to D). Association of Mcm4, Rfa1, Pol2, and Pri1 with ARS1- and ARS305-containing fragments in wild-type and dpb11-1 cells. YHM08 (MCM4-3HA) (A), YHM18 (MCM4-3HA dpb11-1) (A), YHM014 (RFA1-18myc) (B), YHM114 (RFA1-18myc dpb11-1) (B), YHM01 (POL2-3HA) (C), YHM11 (POL2-3HA dpb11-1) (C), YHM013 (PRI1-9myc) (D), and YHM113 (PRI1-9myc dpb11-1) (D) cells were arrested by α-factor and released to YEPR medium in the presence of HU at 36°C, as described in the legend to Fig. 4. PCR was performed as described in Fig. 3. (E) The DNA content of the cells were measured by FACS analysis after the cells had been cultured at 36°C without HU. In the presence of HU, the DNA content of cells remained exclusively 1C throughout the progression of budding.
After Mcm association, RPA associates with the ARS region (47). As our results indicate that Dpb11 association requires functional RPA (Fig. 4C), we used the CHIP assay to examine RPA association in dpb11-1 cells that harbor Rfa1-18myc. As shown in Fig. 5B, signals indicative of Rfa1 association with the ARS fragments were detected even in dpb11-1 cells. This result suggests that RPA association with the ARS fragments occurs independently of the Dpb11 association.
Furthermore, we investigated the association of Pol2 and Pri1 with the ARS fragments in dpb11-1 cells to determine whether Dpb11 plays a role in the association between DNA polymerases and ARS regions. Dpb11-1 cells harboring Pol2-3HA or Pri1-9myc arrested by α-factor were released at 36°C in the presence of HU. No significant association signals were detected in the dpb11-1 cells, whereas ARS association signals for both proteins were observed in wild-type cells (Fig. 5C and D). These results suggest that Dpb11 is required for association of Polɛ and Polα-primase with ARS fragments. Since Dpb11 association with the ARS fragments depended on Polɛ but not Polα, it seems likely that association of the Dpb11-Polɛ complex with ARS fragments is required for Polα association with the ARS fragments. We were not able to detect association of Polδ with the ARS fragments by CHIP assay, even in wild-type cells (data not shown). Thus, we could not determine when and how Polδ associates with the ARS fragments during the initiation of DNA replication.
DNA polymerases associate with late origins in the presence of HU in dpb11-1 cells.
Because dpb11-1 cells show elongated spindles in the presence of HU (5), they are thought to be defective not only in DNA replication but also in the S-phase checkpoint. Since late-origin firing is inhibited in the presence of HU in wild-type cells but not in mec1 and rad53 cells, it has been proposed that the checkpoint proteins, Mec1 and Rad53, control late-firing origins (40). In the presence of HU, RPA and Cdc45 have been shown to associate with late-origin regions in rad53 cells but not in wild-type cells (2, 47). Furthermore, Rad53 protein kinase is activated by HU at a lower level in dpb11-1 cells than wild-type cells (50). Therefore, we investigated whether Dpb11 is involved in the control of late-firing origins.
First, we examined the association of Pol2 and Dpb11 with late-origin regions in the presence of HU. Cells harboring the tagged one of these two genes arrested by α-factor were released into YEPR (26) containing HU at 25°C, and cells were collected 75 min later. We used two sets of PCR primers to amplify early- and late-origin fragments and one set for a nonorigin fragment in the CYC1 gene. We monitored ARS305 as an early origin and ARS501 as a late origin (18). As shown in Fig. 6A, Dpb11 and Pol2 associated with the ARS305 fragment but not with the ARS501 fragment, similar to the action of RPA (47) in wild-type cells. Thus, association of Dpb11 and Pol2 with late-firing-origin fragments is inhibited by HU in wild-type cells.
FIG. 6.
Pol2 associates with a late-firing origin-containing fragment as well as with a early-firing origin-containing fragment in dpb11-1 mutants. (A) YHM011 (DPB11-9myc) and YHM01 (POL2-3HA) cells arrested by α-factor were released to YPD medium without HU and cultured at 16°C (−HU) or to YEPR medium supplemented with HU and cultured at 25°C (+HU) (time = 0 min). The DNA from cells taken at 75 min was subjected to PCR amplification as outlined in the legend to Fig. 3. (B) YHM01 (POL2-3HA), YHM11 (POL2-3HA dpb11-1), and YHM21 (POL2-3HA rad53-1) cells were arrested by α-factor, released in YEPR medium supplemented with 0.2 M HU, and cultured at 25°C. PCR was performed as described in the legend to Fig. 3.
Association of Pol2 with the ARS305 and ARS501 fragments in α-factor-synchronized dpb11-1 cells was examined at 25°C, a temperature permissive for cell growth and for firing ARS305 and ARS501 (data not shown). Pol2 associated with the ARS305 fragment but not with the ARS501 fragment in the presence of HU in wild-type cells (Fig. 6B). Pol2 also associated with the ARS305 fragment in dpb11-1 cells. Thus, the Dpb11-1 protein is functional at 25°C for the ARS association, which is consistent with the firing of ARS305 at 25°C. Moreover, Pol2 associated with the ARS501 fragment in dpb11-1 cells as well as in rad53-1 cells (Fig. 6B). These results suggest that Dpb11 is involved in the control of late-firing origins, which may be important for the S-phase checkpoint.
DISCUSSION
Dpb11 functions for association of DNA polymerases with ARS regions.
In a previous study, we suggested that Dpb11 is required for one of the steps close to the initiation of DNA replication (27). In the present study, three lines of evidence suggest that DNA synthesis cannot initiate without functional Dpb11. First, significant signals were not observed in synchronized dpb11-1 cells by 2D gel analysis (Fig. 2). Second, Dpb11 associated with the ARS fragments after the assembly of the pre-RC (Fig. 4B), and Dpb11 was required for association of the Polα-primase complex (Fig. 4E). The Polα-primase complex is essential for initiation of DNA synthesis because it is the only enzyme known to synthesize the primer RNA (19). Third, the Mcm proteins did not dissociate from the ARS fragments without functional Dpb11 (Fig. 5A). It has been suggested that Mcm proteins dissociate from the pre-RCs on ARSs when DNA synthesis begins (1, 46). Therefore, it is likely that Dpb11 is involved in the initiation of DNA synthesis.
The CHIP assay showed that Dpb11 associates preferentially with the ARS fragments in S phase (Fig. 3A). In this assay, proteins were cross-linked to DNA in vivo, and the chromatin DNA was purified and sonicated. The sheared chromatin was immunoprecipitated with specific antibodies, and the DNA from the immunoprecipitate was then amplified with specific primers. As sonication sheared the DNA into approximately 500-bp fragments, we could not limit the association sequence of Dpb11 to less than a 500-bp sequence. This means that we cannot determine whether Dpb11 associates with the ARS sequence, as the ARS sequence only spans about 100 bp. Therefore, we conclude that Dpb11 association with ARS region (ARS sequence plus the sequence surrounding the ARS) is essential for initiation of DNA synthesis because Dpb11 association is required for Polα association.
We have shown in this study that Dpb11 and Polɛ form a complex (Fig. 1). Consistent with formation of this complex, we also found that their association with ARS fragments is mutually dependent (Fig. 4D and 5C). These observations, combined with the results that show the kinetics of Dpb11 and Pol2 association with the ARS fragments is similar (Fig. 3A), suggest that the Dpb11-Polɛ complex associates with ARS regions during initiation of DNA replication. After initiation, the Dpb11-Polɛ complex seems to disassemble because after this stage Dpb11 does not appear to be associated with a non-ARS fragment, and Pol2 moves to a non-ARS fragment (Fig. 3A and B). Therefore, it seems likely that Dpb11 plays a role in the association of Polɛ with the ARS region.
We do not know exactly what role Dpb11 plays in the association of Polα and Polɛ with the ARS. It has been shown that Cdc45, which interacts with Mcm proteins (12, 21, 22, 24, 32, 52), is required for the loading of Polα and Polɛ (2). CDC45 was isolated as SLD4 in our previous sld screening, suggesting that Cdc45 interacts with Dpb11, directly or indirectly (27). The CHIP assay has been used to show that Cdc45 associates with early-firing ARS regions in the G1 and S phases of the cell cycle, associates with late-firing ARS regions in S phase, and also dissociates like Mcm proteins in S phase (1, 2). Dpb11 associates with both early- and late-firing ARS regions in S phase (Fig. 3 and 6A). These results together suggest that Dpb11 interacts with Mcm proteins through Cdc45, which may be important for Polɛ loading onto the ARS.
S-phase progression of dpb11-1 cells after HU arrest was delayed (27) and short DNA fragments were accumulated in dpb11-10 cells at high temperature (35), suggesting that Dpb11 may also play a role in the elongation step of DNA replication. In the present study, we did not observe any association of Dpb11 with non-ARS fragments by the CHIP assay. However, we cannot neglect the possibility that Dpb11-antibody interactions are interfered with at the replication fork by other replication proteins or the replicating DNA, and therefore Dpb11-DNA association is less easily detected by the CHIP assay. It is also possible that Dpb11 is required for reassociation of Polɛ with chromatin DNA during the elongation step or after HU arrest.
Complex formation between Dpb11 and Polɛ.
In the present study, the Dpb11-Polɛ complex was detected with the cross-linking agent, DSP (Fig. 1A). As DSP cross-links proteins that face each other, all subunits of Polɛ could be cross-linked. Thus, we could not determine which subunit of Polɛ binds to Dpb11. However, detection of an interaction between Dpb11 and Dpb2 in the two-hybrid assay and the absence of a Dpb11-Polɛ complex in dpb2-1 cells (Fig. 1C) suggest that the most probable candidate is Dpb2, the second-largest subunit of Polɛ. As the pol2-11 and dpb2-1 mutations both weaken the interaction between Pol2 and Dpb2 (3, 16), an increase in the copy number of DPB11 may also improve the formation of Dpb11 and mutant Polɛ complexes and suppress the growth defects of pol2-11 and dpb2-1.
Recently, Kesti et al. (28) reported that the Pol2 C-terminal region is essential for DNA replication and cell growth, while the N-terminal polymerase domains of this protein are dispensable for cell growth. As Pol2-11 lacks 32 C-terminal amino acid residues (34) and the Pol2 C-terminal region is required for complex formation between Pol2 and Dpb2 (33), the Pol2 C-terminal region could be essential for Dpb11-Polɛ complex formation. DPB3, a gene that encodes the third-largest subunit of Polɛ, is not essential for cell growth (4). However, cells bearing the sld1-1 mutation of DPB3 and the dpb11-1 mutation are lethal (27). This lethality may be caused by a decrease in the number of Pol2-Dpb2 complexes in a cell, since less than 50% of the Polɛ activity of wild-type cells has been recovered from dpb3 cells (4). Nevertheless, we cannot eliminate the possibility that other proteins connect Dpb11 to Polɛ. If this does occur, Dpb11 and Polɛ may be present in the same complex.
Since Dpb11 has four BRCT domains that are important for protein-protein interactions (5, 7, 9, 51), Dpb11 might be a scaffold protein for assembly of DNA replication proteins including Polα and Polɛ at the initiation of DNA replication. From this point of view, the Dpb11-Polɛ complex might be a core complex in the assembly of replication proteins because the association between Polα and the ARS region depends on Dpb11. Studies of the in vitro SV40 replication system suggest that Polα is the first to associate with the replication origin because the Polα-primase complex initiates DNA synthesis (23). We therefore propose that the complex consisting of the Dpb11-Polɛ core, Polα and other replication proteins, associates with ARS region.
Since a small amount of the Dpb11-Polɛ complex was observed in G1 and M phases (Fig. 1A), this complex may also function for DNA repair, and other factors may be required for association of the Dpb11-Polɛ complex with the ARS region. We have previously shown that Dpb11 and Sld2 form a complex essential for DNA replication, and dpb11-1 cells are defective in the formation of this complex (27). Thus, Sld2 may play an important role in association of Dpb11-Polɛ complex with the ARS region and disassembly of the complex after DNA synthesis starts.
Dpb11 controls the association of DNA polymerases with ARS regions.
In the presence of HU, late-origin firing is inhibited in wild-type cells. However, in rad53 or mec1 cells defective in checkpoint, DNA synthesis initiates from both early- and late-firing origins in the presence of HU (40). In the presence of HU, Mcm proteins associate with early- and late-firing-origin regions; however, RPA and Cdc45 only associate with early-origin regions. In rad53 cells, RPA and Cdc45 associate with early and late origins in the presence of HU (2, 47).
In the present study, we have shown that in the presence of HU, Dpb11 and Pol2 behave as RPA and Cdc45 behave in wild-type cells (Fig. 6A), and Pol2 associates with both early- and late-firing origins in dpb11-1 cells (Fig. 6B). As Dpb11 is partly required for activation of Rad53 protein kinase (50), Dpb11 may monitor the initiation of DNA replication and then communicate with Rad53. Both the ARS305 and ARS501 fragments were simultaneously amplified from dpb11-1 cells after Pol2 immunoprecipitation. In rad53 cells, Pol2 associated with the ARS501 fragment at a later stage than these cells did with the ARS305 fragment (Fig. 6B). Analysis with 2D gels of dpb11-1 cells cultured in the absence of HU showed that initiation of DNA replication from the early origin, ARS1, occurred at almost the same time as initiation from ARS501 (data not shown). Similar observations have been reported for the orc2-1 mutation. This mutation is located in the second-largest subunit of the ORC and makes the ORC defective in the initiation of DNA replication. The orc2-1 mutation affects the timing of origin firing, and orc2-1 mutants are defective in inhibition of late-origin firing when cells are exposed to methyl methane sulfonate (42). Therefore, it seems likely that Dpb11 is involved in the control of activation of early origins and in blocking late-origin firing, which may be important for the S-phase checkpoint.
Overall, the results reported in this paper strongly suggest that there is a distinct regulatory step for association of DNA polymerases with replication origins. This step might be controlled by the cell cycle as well as the S-phase checkpoint, and Dpb11 is an integral part of this novel regulatory mechanism. Cut5, a probable counterpart of Dpb11 in fission yeast, also has BRCT domains and functions in both DNA replication and checkpoints (30, 37–39, 49). This suggests that a similar protein may be involved in the association of DNA polymerases with replication origins and the monitoring of this step in other eukaryotic cells.
ACKNOWLEDGMENTS
We thank T. Tanaka, K. Nasmyth, O. M. Aparicio, S. Bell, P. Plevani, and K. Shirahige for information about PCR primers, strains, and plasmids; T. Yasuda, H. Ohmori, and N. Shimamoto for information about cross-linkers; C. Wittenberg for information about Pol2 before publication; L. Johnston, Y. Kamimura, and J. Tomizawa for critical reading of the manuscript.
This study is partially supported by grants-in-aid from the Ministry of Education, Science, Sports, and Culture, Japan, to A.S. and H.A.
REFERENCES
- 1.Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio O M, Stout A M, Bell S P. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci USA. 1999;96:9130–9135. doi: 10.1073/pnas.96.16.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araki H, Hamatake R K, Johnston L H, Sugino A. DPB2, the gene encoding DNA polymerase II subunit B, is required for chromosome replication in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:4601–4605. doi: 10.1073/pnas.88.11.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araki H, Hamatake R K, Morrison A, Johnson A L, Johnston L H, Sugino A. Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19:4867–4872. doi: 10.1093/nar/19.18.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Araki H, Leem S-H, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II(ɛ) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci USA. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origin of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 7.Bork P, Hoffman K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 8.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 9.Callebaut I, Mornon J-P. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 10.Campbell J L, Newlon C S. Chromosomal DNA replication. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 41–146. [Google Scholar]
- 11.Chong J P, Thommes P, Blow J J. The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem Sci. 1996;21:102–106. [PubMed] [Google Scholar]
- 12.Dalton S, Hopwood B. Characterization of Cdc47p-minichromosome maintenance complex in Saccharomyces cerevisiae: identification of Cdc45p as a subunit. Mol Cell Biol. 1997;17:5867–5875. doi: 10.1128/mcb.17.10.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desany B A, Alcasabas A A, Bachant J B, Elledge S J. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diffley J F X, Cocker J H, Dowell S J, Harwood J, Rowley A. Stepwise assembly of initiation complexes at budding yeast replication origins during the cell cycle. J Cell Sci Suppl. 1995;19:67–72. doi: 10.1242/jcs.1995.supplement_19.9. [DOI] [PubMed] [Google Scholar]
- 15.Donovan S, Harwood J, Drury L S, Diffley J F X. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dua R, Levy D L, Campbell J L. Role of the putative Zinc finger domain of Saccharomyces cerevisiae DNA polymerase ɛ in DNA replication and the S/M checkpoint pathway. J Biol Chem. 1998;273:30046–30055. doi: 10.1074/jbc.273.45.30046. [DOI] [PubMed] [Google Scholar]
- 17.Fangman W L, Brewer B J. A question of time-replication origins of eukaryotic chromosomes. Cell. 1992;71:363–366. doi: 10.1016/0092-8674(92)90505-7. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson B M, Brewer B J, Reynolds A E, Fangman W L. A yeast origin of replication is activated late in S phase. Cell. 1991;65:507–515. doi: 10.1016/0092-8674(91)90468-e. [DOI] [PubMed] [Google Scholar]
- 19.Foiani M, Lucchini G, Plevani P. The DNA polymerase α-primase complex couples DNA replication, cell-cycle progression and DNA-damage response. Trends Biochem Sci. 1997;22:424–427. doi: 10.1016/s0968-0004(97)01109-2. [DOI] [PubMed] [Google Scholar]
- 20.Gunzburg J D, Riehl R, Weinberg R A. Identification of a protein associated with p21ras by chemical crosslinking. Proc Natl Acad Sci USA. 1989;86:4007–4011. doi: 10.1073/pnas.86.11.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardy C F J. Identification of Cdc45p, an essential factor required for DNA replication. Gene. 1997;187:239–246. doi: 10.1016/s0378-1119(96)00761-5. [DOI] [PubMed] [Google Scholar]
- 22.Hennesy K M, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 23.Heredeen D, Kelly T J. SV40 DNA replication. In: Blow J J, editor. Eukaryotic DNA replication. Oxford, United Kingdom: Oxford University Press; 1996. pp. 29–65. [Google Scholar]
- 24.Hopwood B, Dalton S. Cdc45p assembles into a complex with Cdc46/Mcm5p, is required for minichromosome maintenance, and is essential for chromosomal DNA replication. Proc Natl Acad Sci USA. 1996;93:12309–12314. doi: 10.1073/pnas.93.22.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huberman J A, Riggs A D. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968;32:327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- 26.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 27.Kamimura Y, Masumoto H, Sugino A, Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol. 1998;18:6102–6109. doi: 10.1128/mcb.18.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kesti T, Flick K, Keränen S, Syväoja J E, Wittenberg C. DNA polymerase ɛ catalytic domains are dispensable for DNA replication, repair and cell viability. Mol Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 29.Lei M, Kawasaki Y, Tye B K. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5081–5090. doi: 10.1128/mcb.16.9.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McFarlane R J, Carr A M, Price C. Characterization of the Schizosaccharomyces pombe rad4/cut5 mutant phenotypes: dissection of DNA replication and G2 checkpoint control function. Mol Gen Genet. 1997;255:332–340. doi: 10.1007/s004380050504. [DOI] [PubMed] [Google Scholar]
- 31.Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moir D, Stewart S E, Osmond B C, Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: properties and pseudoreversion studies. Genetics. 1982;100:547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrison A, Araki H, Clark A B, Hamatake R K, Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1991;62:1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- 34.Navas T A, Zhou Z, Elledge S J. DNA polymerase ɛ links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 35.Reid R D J, Fiorani P, Sugawara M, Bjornsti M-A. CDC45 and DPB11 are required for processive DNA replication and resistance to DNA topoisomerase I-mediated DNA damage. Proc Natl Acad Sci USA. 1999;96:11440–11445. doi: 10.1073/pnas.96.20.11440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth R A, Pierce S B. In vivo cross-linking of protein disulfide isomerase to immunoglobulins. Biochemistry. 1987;26:4179–4182. doi: 10.1021/bi00388a001. [DOI] [PubMed] [Google Scholar]
- 37.Saka Y, Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- 38.Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saka Y, Fantes P, Sutani T, McInerny C, Creanor J, Yanagida M. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 1994;13:5319–5329. doi: 10.1002/j.1460-2075.1994.tb06866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santocanale C, Diffley J F X. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 41.Santocanale C, Neecke H, Longhese M P, Lucchini G, Plevani P. Mutations in the gene encoding the 34 kDa subunit of yeast replication protein A cause defective S phase progression. J Mol Biol. 1995;254:595–607. doi: 10.1006/jmbi.1995.0641. [DOI] [PubMed] [Google Scholar]
- 42.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 43.Stillman B. Initiation of chromosomal DNA replication in eukaryotes: lessons from lambda. J Biol Chem. 1994;269:7047–7050. [PubMed] [Google Scholar]
- 44.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 45.Sugino A. Yeast DNA polymerases and their role at the replication fork. Trends Biochem Sci. 1995;20:319–323. doi: 10.1016/s0968-0004(00)89059-3. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka T, Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tye B-K. The MCM2-3-5 proteins: are they replication licensing factors? Trends Cell Biol. 1994;4:160–166. doi: 10.1016/0962-8924(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 49.Verkade H M, O'Connell M J. Cut5 is a component of the UV-responsive DNA damage checkpoint in fission yeast. Mol Gen Genet. 1998;260:426–433. doi: 10.1007/s004380050913. [DOI] [PubMed] [Google Scholar]
- 50.Wang H, Elledge S J. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:3824–3829. doi: 10.1073/pnas.96.7.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Morera S, Bates P A, Whitehead P C, Coffer A I, Hainbucher K, Nash R A, Sternberg M J E, Lindahl T, Freemont P S. Structure of an XRCC1 BRCT domain: a new protein-protein interaction module. EMBO J. 1998;21:6404–6411. doi: 10.1093/emboj/17.21.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zou L, Mitchell J, Stillman B. CDC45, a novel yeast gene that functions with the origin recognition complex and proteins in initiation of DNA replication. Mol Cell Biol. 1997;17:553–563. doi: 10.1128/mcb.17.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]