SUMMARY
Gram-negative bacteremia is a devastating public health threat, with high mortality in vulnerable populations and significant costs to the global economy. Concerningly, rates of both Gram-negative bacteremia and antimicrobial resistance in the causative species are increasing. Gram-negative bacteremia develops in three phases. First, bacteria invade or colonize initial sites of infection. Second, bacteria overcome host barriers, such as immune responses, and disseminate from initial body sites to the bloodstream. Third, bacteria adapt to survive in the blood and blood-filtering organs. To develop new therapies, it is critical to define species-specific and multispecies fitness factors required for bacteremia in model systems that are relevant to human infection. A small subset of species is responsible for the majority of Gram-negative bacteremia cases, including Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii. The few bacteremia fitness factors identified in these prominent Gram-negative species demonstrate shared and unique pathogenic mechanisms at each phase of bacteremia progression. Capsule production, adhesins, and metabolic flexibility are common mediators, whereas only some species utilize toxins. This review provides an overview of Gram-negative bacteremia, compares animal models for bacteremia, and discusses prevalent Gram-negative bacteremia species.
KEYWORDS: Acinetobacter, Escherichia coli, Klebsiella, Pseudomonas aeruginosa, bacteremia, bloodstream infections, Gram-negative bacteria, pathogenesis, sepsis
INTRODUCTION
Gram-negative bacteremia is a significant public health threat affecting both healthy individuals and those with underlying comorbidities but is especially dangerous in vulnerable populations like the elderly and hospitalized patients (1). Clinical presentation of bacteremia can be primary, originating from an unknown source, or secondary, originating from infections such as pneumonia. Among primary bacteremias, a significant subset are thought to arise from gut colonization. A small group of species cause the majority of Gram-negative bacteremia cases, and each species is uniquely adapted for initial infection or colonization at specific body sites. To establish bacteremia, pathogens must (i) invade initial sites of infection or colonization, (ii) disseminate to the bloodstream, and (iii) survive in the blood. Clinical studies have identified some host and bacterial factors that correlate with better patient outcome and with disease progression. Animal models have provided insights into bacterial fitness factors required for initial body site invasion and bloodstream fitness for a few species. However, our understanding of shared and species-specific factors involved in the three steps of Gram-negative bacteremia pathogenesis is incomplete. This review summarizes what has been learned from clinical studies, describes how animal models assess bacteremia fitness factors, and discusses major gaps in knowledge that limit insight into Gram-negative bacteremia. Filling these knowledge gaps could uncover potential targets for future therapies to treat these destructive infections.
BACTEREMIA OVERVIEW
Bacteremia, the presence of bacteria in the bloodstream, is an urgent public health issue (1) that can initiate devastating diseases (2, 3) and costs the global economy billions of dollars each year (4). Clinical bacteremia can result in sepsis, defined as life-threatening organ dysfunction caused by a dysregulated host response to infection (5). Annually, there are an estimated 1.7 million sepsis cases (6) in the United States and more than 30 million cases globally (7), leading the World Health Organization (WHO) to declare sepsis a global health priority (8). Sepsis can result in permanent dysfunction, including cognitive impairment or organ failure (9, 10) and is associated with variable, yet incredibly high, mortality (7, 10, 11). Further, bloodstream infections are also associated with a high mortality risk of 27% (12). The severity and high mortality for conditions associated with bacteremia underscore the importance of investigating these complex infections.
Bacteria can enter the blood by multiple routes. Bacteremia with no evident source is considered primary (13); secondary bacteremia can arise through dissemination from an infection, commonly pneumonia or urinary tract infections (UTIs), or through contaminated medical devices. Transient bacteremia, caused by minor procedures such as dental cleanings, can occur but is typically cleared from the blood shortly after introduction (14) and therefore has undetermined clinical significance.
Clinical presentation of bacteremia and sepsis can vary based on patient factors, the underlying pathogen, and the source of infection. Typically, bacteremia is associated with fever but can be asymptomatic. Sepsis can cause organ dysfunction in the respiratory, hepatic, cardiovascular, central nervous, renal, and coagulation systems. The severity of dysfunction can be assessed through laboratory testing and intensive monitoring and using the results to calculate the sequential organ failure assessment (SOFA) score (5). A score of >2 on the 4-point scale is associated with a mortality risk of 10%, greater than that of an acute myocardial infarction. Because sepsis patients need to be rapidly managed, the increased mortality risk in suspected cases can be rapidly identified by a quick score (qSOFA) defined by two or more of the following: high respiratory rate (>22/min), low systolic blood pressure (<100 mm Hg), and altered mentation. Patients with sepsis can progress to septic shock, with an in-hospital mortality of >40%.
Bacteremia and sepsis carry large economic consequences (4, 11, 15). Hospitalization due to bacteremia may be 20 days longer than other stays and can be even longer if the underlying pathogen is antimicrobial resistant (15), while hospitalizations for sepsis are ∼75% longer than stays for other conditions (1). This contributes to sepsis being the most expensive condition for hospitals, costing the United States $24 billion in 2013 (4). Generally, risk factors for bacteremia and sepsis include older age, immunosuppressive medications, and underlying comorbidities (1, 11), but risk factors vary depending on initial site of invasion and underlying pathogen. Due to longer life expectancy, rising use of immunosuppressive therapies, and increased emergence of antimicrobial-resistant pathogens, bacteremia will continue to be a costly disease. Further, cognitive and physical impairment after sepsis can require lifetime supportive care at a major expense to individuals (1, 9, 11). Thus, bacteremia conveys immense socioeconomic burdens.
Bloodstream infections can be caused by a number of microbes, including bacteria and fungi, although bacteria are responsible for over 90% of cases (10, 12, 16, 17). Bacteremia can be broadly classified into infections by Gram-positive or Gram-negative species. Gram-negative bacteremia is increasing in prevalence and antibiotic resistance, yet it is largely understudied. Thus, Gram-negative bacteremia is a public health concern and further investigation is necessary. This review highlights pathogenesis of Gram-negative bacteremia, discusses relevant pathogens, and compares bacteremia models in mice.
GRAM-NEGATIVE BACTEREMIA
Between 1997 and 2013, the number of bacteremia cases caused by Gram-negative species increased substantially, from 33% to 43% (18). This estimate accounts only for bacteremia cases caused by the 10 most prevalent species and is therefore likely an underestimate. Along with increased incidence of Gram-negative bacteremia, emergence of antimicrobial resistance among these species, which often complicates treatment and increases mortality, was repeatedly reported (16, 18–24). Rates of bacteremia due to multidrug-resistant Enterobacterales more than doubled between 1997 and 2016, from 6.3% to 15.8% (18). The relative frequencies of pathogens underlying bacteremia has shifted over the past decades. From 1997 to 2016, the top 10 species isolated from clinical bacteremia included Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Enterococcus faecalis, Staphylococcus epidermidis, Enterobacter cloacae, Streptococcus pneumoniae, Enterococcus faecium, and Acinetobacter baumannii (18). In 2005 E. coli replaced S. aureus as the most prevalent bloodstream pathogen and A. baumannii entered the top 10. K. pneumoniae is consistently the third most prevalent bloodstream pathogen, and P. aeruginosa has emerged as fourth in prevalence, emphasizing the significance of Gram-negative species in these infections. This small subset of Gram-negative species significantly contributes to disease burden. E. coli accounts for 6 to 27% of bacteremia cases, K. pneumoniae for 5 to 13%, P. aeruginosa for 4 to 9%, and A. baumannii for 1 to 13% (12, 17, 18, 20, 25). Infection rates for individual species vary between studies, likely due to patient demographics and geography, the year data were collected, and definitions of bacteremia. However, studies consistently identify members of the order Enterobacterales as highly prevalent, specifically E. coli and K. pneumoniae, as well as non-Enterobacterales species, including P. aeruginosa and A. baumannii. Although A. baumannii is the fifth most prevalent Gram-negative bacteremia pathogen, its clinical significance is marked by staggeringly high multidrug resistance rates reaching nearly 71% (18). Therefore, it is important to discuss these four problematic Gram-negative species in detail in the context of bacteremia.
Reservoirs for Gram-negative bacteremia pathogens include the environment and intestinal tract colonization. It is likely that all of these pathogens have some environmental reservoir, as none are obligate human pathogens. Colonization can occur after exposure to ubiquitous environmental reservoirs, including contaminated medical equipment, water, and companion animals (26–28). Highly contaminated environments, like wastewater treatment plants and hospital drainage, are particularly problematic and may serve as hubs of pathogen accumulation that provide a space for exchange and acquisition of genetic elements, specifically, antimicrobial resistance genes (26, 29). For Enterobacterales, particularly E. coli and K. pneumoniae (30–32), gastrointestinal colonization can persist and serve as a reservoir for transmission and colonization of another person or reintroduction into the environment. Colonization can also be an intermediate step between exposure to an environmental reservoir and infection. For the non-Enterobacterales, including P. aeruginosa, A. baumannii, and Serratia marcescens, colonization is typically transient and environmental exposure is more of a direct risk for subsequent infection. Colonization or environmental exposure can progress to a bloodstream infection, manifesting clinically as primary bacteremia. Alternatively, colonization or environmental exposure can progress to a different infection (pneumonia or urinary tract infection) and then to a secondary bacteremia.
Gram-negative bacteremia is associated with intermittent bacterial presence in the blood (33) and often arises as a secondary infection seeded from an initial source which disseminates to the bloodstream. Health care-associated infections (HAIs) and community-acquired infections (CAIs) have roughly equal contributions to Gram-negative bacteremia disease burden, although exact proportions vary at a species- and strain-specific level (17, 18). E. coli is the most common species isolated from community-onset bacteremia, underlying 26.6% of cases, and second most common within hospital onset, underlying 21.3% of cases. K. pneumoniae is the third most prevalent species for both community- and hospital-onset bacteremia, responsible for 7.2% and 8.8% of cases, respectively. P. aeruginosa is the fourth most common species isolated from hospital-onset bacteremia (7.4% of cases) and ranked fifth among species causing community-onset bacteremia (7.3% of cases). A. baumannii is a frequent cause of hospital-onset bacteremia, accounting for 3.2% of cases, but is not highly prevalent among community-onset infections (18).
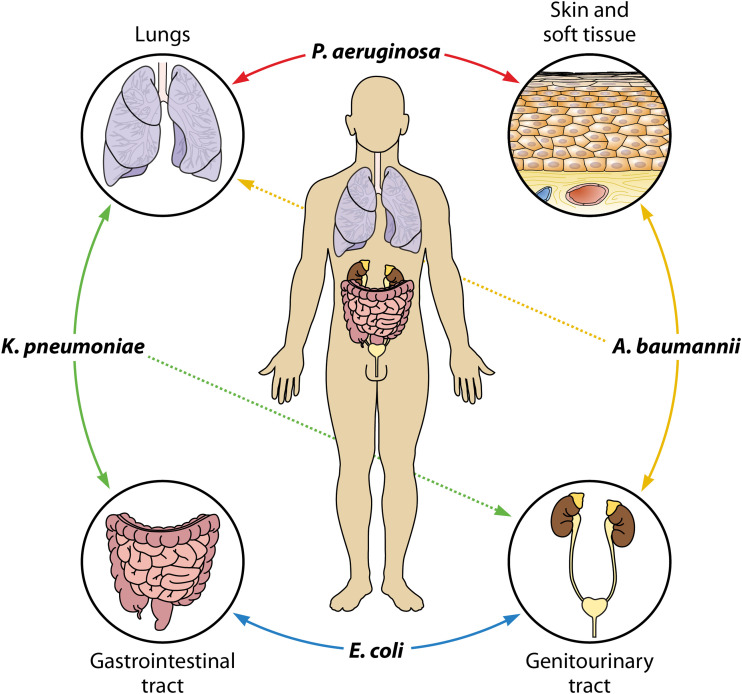
For HAIs, the length of intensive care unit (ICU) stay is about 44 days longer for Gram-negative bacteremia than other ICU stays (34). Longer length of stay for health care-associated bacteremia is likely due to the invasive nature of equipment utilized in ICUs, multiple underlying patient comorbidities, and increased surveillance. Most Gram-negative bacteremia cases are monomicrobial (10, 34, 35) and stem from preexisting infections. Primary bacteremia can originate in the gastrointestinal tract, which is responsible for about 7 to 22% of ICU-associated Gram-negative bacteremia cases (34, 35). E. coli can cross intestinal barriers, especially in immunocompromised patients, to initiate bacteremia. Concordance for K. pneumoniae between gut colonizers and bacteremia isolates has been well documented (31, 32). How bacterial species colonize and exit the gastrointestinal tract remains to be investigated. Pneumonia is the most common initial infection leading to secondary bacteremia (34, 35). Of Gram-negative species, K. pneumoniae and P. aeruginosa are often isolated from bacteremia cases secondary to pneumonia. The genitourinary tract is also a common initial infection site. UTIs are predominantly caused by E. coli, but a 17-year investigation found a significant increase in the incidence of ICU-associated UTIs by K. pneumoniae and A. baumannii (36). Bacteremia can also initiate from sources with direct access to the blood, like contaminated medical equipment and soft tissue wounds, which are particularly problematic with A. baumannii and P. aeruginosa (37, 38) (Fig. 1).
FIG 1.
Gram-negative species are adapted to colonize or infect diverse initial sites, which may progress to secondary bacteremia.
Virulence factors required for initial-site invasion have been studied in many Gram-negative species. However, factors mediating dissemination and bloodstream survival have been either minimally or only recently described. Clinical studies and animal models have identified remarkably few virulence factors required for Gram-negative bacteremia, partially due to the understudied nature of these infections. It is critical to identify mechanisms governing each phase of bacteremia for problematic species (Fig. 2). Microbial factors that have been defined are largely related to capsule production, adherence, and metabolic diversity (Table 1). Capsule production may be particularly important in aiding immune evasion during initial infection and during bloodstream survival. Adherence, mediated by fimbriae and other adhesins, may be most important in earlier phases of bacteremia when pathogens bind to specific receptors at the initial infection site and disseminate to the blood. Genes regulating metabolic diversity are likely significant throughout all phases of bacteremia but may be most significant in later phases, when bacteria must survive the transition from initial infection to survival in the bloodstream. Investigating virulence factors at each phase of infection across multiple species will identify mechanisms that are shared or unique among the Gram-negative bacteremia pathogens that could be targeted by future therapies, providing greater comprehension of the mechanisms of pathogenesis.
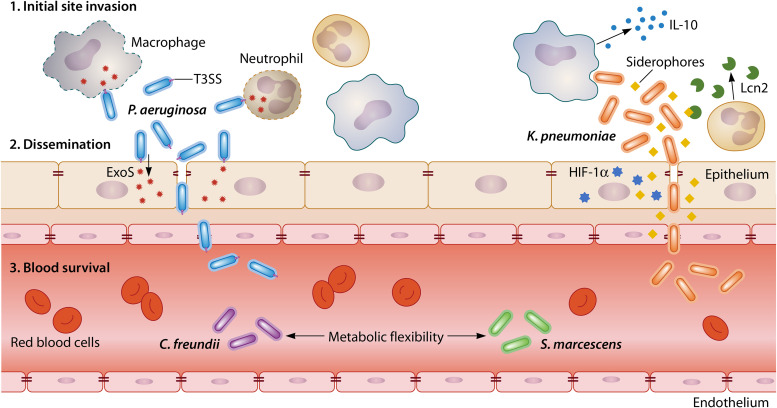
FIG 2.
Gram-negative bacteremia pathogenesis broadly involves three phases. (Step 1) Invasion. Bacteria must invade initial sites of colonization or infection and evade host immune responses using mechanisms such as capsule production (K. pneumoniae) and secretion of exotoxins (P. aeruginosa). Initial-site specificity varies by species, with certain bacteria being adapted for invasion at specific sites. (Step 2) Dissemination. After invasion, bacteria penetrate host epithelial barriers to access the blood. Dissemination requires factors such as adhesins and exotoxins (P. aeruginosa) or activation of specific pathways in epithelial cells (HIF-1α for K. pneumoniae). (Step 3) Survival. Once in the blood, species must survive a new environment through metabolic flexibility (C. freundii and S. marcescens) and evade immune clearance with capsule production. Each bacteremia phase must be investigated to better comprehend pathogenesis at both species-specific and multispecies levels.
TABLE 1.
Bacteremia factors can be shared across multiple species or conserved in only one species, and many factors have been confirmed in only one phase of bacteremiaa
| Category | Fitness gene(s) | Species | Subfunction | Initial site | Intraperitoneal | TVI | Reference(s) |
|---|---|---|---|---|---|---|---|
| Metabolism | speAB, gltB, gdhA | P. mirabilis | Amino acid | X | 44 | ||
| pqqL, c1220 | E. coli | Amino acid | X | 50 | |||
| ddc | A. baumannii | Amino acid | X | 45 | |||
| glnA | P. mirabilis | Amino acid; carbohydrate; energy | X | 44 | |||
| nanA | E. coli | Carbohydrate | X | 49 | |||
| mtlD | C. freundii | Carbohydrate | X | 43 | |||
| pfkA | C. freundii, S. marcescens | Carbohydrate | X | 42, 43 | |||
| mgtB, pgm | S. marcescens | Carbohydrate; energy | X | 42 | |||
| gltA | K. pneumoniae | Carbohydrate | X | X | 53 | ||
| pntB | A. baumannii | Cofactor and vitamin | X | 45 | |||
| cysE | C. freundii | Energy | X | 43 | |||
| tonB | E. coli, K. pneumoniae | Iron transport | X (K. pneumoniae) | X | 49, 99 | ||
| hma, chuA | E. coli | Iron transport | X | 49 | |||
| feoB | A. baumannii | Iron transport | X | 45 | |||
| Cellular maintenance | pilVS, fim, pap | E. coli | Adherence | X | X | 49, 50 | |
| pic, vat | E. coli | Autotransporter serine protease | X | 50 | |||
| pgaABCD | E. coli | Biofilm formation | X | X | 50 | ||
| ksl | E. coli | Capsule production | X | 49 | |||
| wzx | S. marcescens | Capsule production | X | 42 | |||
| sufl | C. freundii | Cell growth | X | 43 | |||
| UMH9_0939 | S. marcescens | Membrane synthesis | X | 42 | |||
| mltB | A. baumannii | Membrane synthesis | X | X | 51 | ||
| dam | K. pneumoniae | Replication and repair | X | X | 104 | ||
| ruvA | C. freundii | Replication and repair | X | 43 | |||
| Environmental response | sapABCDF, oppA | E. coli | Membrane transport | X | 50 | ||
| tatC | C. freundii, P. mirabilis | Membrane transport | X | 43, 44 | |||
| potB | P. mirabilis | Membrane transport | X | 44 | |||
| ompK36 | K. pneumoniae | Membrane transport | X | 103 | |||
| fepA | A. baumannii | Signal transduction | X | 45 | |||
| ntrB | P. mirabilis | Signal transduction | X | 44 | |||
| exoU, exoS, exoT, exlA | P. aeruginosa | Toxins | X | 55, 120, 122 | |||
| Unclassified | yddB | E. coli | Uncharacterized | X | 50 | ||
| UMH9_0544 | S. marcescens | Uncharacterized | X | 42 | |||
TVI, tail vein injection.
MODELING BACTEREMIA
Based on studies of blood culture yields for diagnostic testing, bacteremia is often characterized by low microbial abundance in patient samples (reviewed in reference 39). For example, Gram-negative bacteremia in humans often presents as intermittent shedding from an initial source. As a result, blood CFU can fluctuate over time and present with bloodstream abundance often as low as 10 CFU/ml (40, 41). Several animal models have been employed to identify virulence factors required for bacteremia but do not fully mimic human disease (Table 2). Some models use a single direct blood inoculum as high as 2 × 107 CFU/ml (42–45). Although high inocula are likely necessary to ensure reproducible experimental infections, this difference in bacterial blood density between models and human disease must be considered for influences on metabolism or host immune responses.
TABLE 2.
Summary of current bacteremia models and known advantages and limitations
| Model | Phase of bacteremia |
Advantages | Limitations | Reference(s) | ||
|---|---|---|---|---|---|---|
| Initial site | Dissemination | Bloodstream survival | ||||
| Retropharyngeal/Intratracheal infection | X | X | Identifies factors involved in lung fitness and dissemination; models secondary bacteremia | Tight bottleneck in dissemination step can limit genetic screens; difficult to distinguish factors involved in primary site infection from those involved in dissemination; frequency and quantity of dissemination may be variable | 46 – 48 | |
| Urinary tract infection | X | X | Identifies factors involved in genitourinary fitness and dissemination; models secondary bacteremia | 44 | ||
| Tail vein injection | X | Low bottleneck allows for genetic screens; monitoring of hematogenous spread to other organs | Unknown if the high inoculum typically used for infection mimics human bacteremia; few bacteria detectable in blood after infection; injection is technically challenging | 42–45, 49–52 | ||
| Intraperitoneal injection | X | X | May be ideal for studying liver colonization; less technically challenging than tail vein injection; reproducibly causes bacteremia from multiple species | Phase of bacteremia unclear (dissemination from peritoneal cavity vs. drainage in the portal vein is unknown) | 53 | |
| Serum killing | X | Can use mouse or human serum; reduction in animal use, commercially available reagents, inexpensive, scalable, no bottleneck for genetic screens; can individually examine serum growth and serum killing |
In vitro: short-term studies only; limited immune interactions Killing: complement is not a major barrier to bacteremia in some species; lacks immune cells that may interact with complement to kill bacteria Growth: provides only serum nutrients but not those from primary site; some species do not grow in 100% serum and require medium supplementation |
42, 43, 53, 57, 58 | ||
| Serum growth | X | |||||
Animal models are often used to investigate initial infections that seed secondary bacteremia. While these approaches can identify fitness factors required for initial-site invasion and blood dissemination, distinguishing roles at distinct steps of pathogenesis may be difficult. Furthermore, bloodstream fitness mechanisms may not be discernible. Modeling bacteremia from pneumonia can be accomplished through retropharyngeal and intratracheal infection and subsequent harvesting of the liver and spleen to enumerate CFU dissemination (46–48). Since pneumonia is a leading cause of secondary bacteremia for many species, including K. pneumoniae and P. aeruginosa, these models can identify host-pathogen interactions for dissemination. Bacteremia from initial UTI models have also uncovered a number of genes linked to catheter-associated UTI and bloodstream infection (44). However, both pneumonia and UTI models are limited by bottlenecks affecting subsequent bacteremia (Table 2). Bottlenecks are barriers that can cause stochastic loss of populations and may confound experimental systems unless appropriately assessed. In the lung dissemination model, there is likely a bottleneck in crossing epithelial barriers while also evading host inflammatory responses. UTI models encounter bottlenecks through the necessity of ascending the urinary tract and crossing kidney tubules to enter the blood. Another initial-site infection is that of indwelling devices. Modeling bacteremia associated with indwelling devices in mice is complex to due to ethical considerations and experimental variability, and direct blood inoculation could appropriately model device-associated infections. Although initial-site modeling requires optimization, these systems are crucial to uncovering mechanisms of pathogenesis at early phases of bacteremia. Models must also be optimized to monitor dissemination, rather than initial-site fitness alone, to fully understand early phases of bacteremia.
Models that directly introduce bacteria to the blood bypass the steps of initial infection and dissemination and can specifically identify factors required for bloodstream fitness. Tail vein injections (TVIs) are used to inoculate mice intravascularly and have been extensively used to model bacteremia (49) (Table 2). Using E. coli, TVIs have been validated as a bacteremia model system. Shortly after TVI, E. coli was found in multiple organs, including the spleen, liver, and heart, indicating hematogenous spread. Uropathogenic E. coli (UPEC), but not a nonpathogenic strain, was recovered from the spleen and liver 24 h after TVI, indicating clearance of nonpathogenic strains by murine immune responses (49). TVIs have been used to model bacteremia from many species, including E. coli (49, 50), A. baumannii (45, 51), Citrobacter freundii (43), S. marcescens (42, 52), and Proteus mirabilis (44). These models have high utility, in part, due to a low bottleneck effect. By direct introduction of bacteria to the blood, virulence and fitness factors can be monitored in the environment of interest. Hematogenous spread after TVI also allows monitoring of disease processes at secondary organs, such as the brain, which helps identify mechanisms required for pathogenesis in infections resulting from bacteremia. Compared to human bacterial burden, TVI bacteremia utilizes an incredibly high bacterial blood load. However, injection of lower numbers of CFU may prevent detection of bacteria in distal organs and reduce experimental sensitivity. Interestingly, although TVI models use large initial doses, few circulating bacteria are subsequently found in murine blood (49). It is possible that these models render similar bacterial blood density between clinical and murine bacteremia, but this remains to be determined.
Intraperitoneal (i.p.) injections are also used to model bacteremia (53) (Table 2). Here, bacteria introduced directly into the peritoneal cavity can traverse the thin peritoneal layer for blood dissemination, which could mimic barrier permeation during infection. Peritoneal fluid is drained to the liver by the portal vein; thus, species that colonize the liver may successfully establish bacteremia. i.p. injections may also establish a reservoir for intermittent shedding which could mirror human bacteremia pathology. However, there are technical considerations with i.p. injections, including appropriate placement of the inoculum into the peritoneal cavity. Needle placement must avoid internal organs, fat, and muscle and must also avoid damaging the peritoneal wall. It is also difficult to assess accurate needle placement during i.p. injection; gentle aspiration after needle placement can determine whether an organ has been punctured, but this technique does not indicate if the needle is placed into fat or muscle. i.p. injections could also differ from TVIs in having a tighter experimental bottleneck. TVIs place bacteria directly into the blood, allowing the inoculum to circulate prior to filtration in the spleen and liver. i.p. injections result in peritoneal inflammation or drainage to the liver, where bacteria that cannot survive liver filtration may be eliminated. i.p. injections are beneficial due to minimal required equipment and high reproducibility, but it may be unclear which phase of bacteremia is being modeled in this system for detecting fitness factors. However, this model could be highly informative for species predicted to colonize the liver.
Together, initial infection site models can identify mechanisms that are important during establishment of infection and also determine bacterial and host factors mediating blood dissemination. These models are helpful for investigating early phases of bacteremia but may not be helpful in identifying factors relevant in later disease after bacteria have exited original sites. To model later phases of bacteremia, namely, bloodstream survival, TVI and i.p. injections can be used. Experimental design and model rationale must be clearly defined in order to appropriately evaluate what certain models can and cannot report about bacteremia pathogenesis (Table 2). Factors required for dissemination may be dispensable in the blood, and factors governing bloodstream survival may not be necessary for dissemination, as hypothesized for P fimbriae in E. coli bacteremia (49, 54), discussed below.
Additionally, choosing an appropriate initial site of infection is a significant consideration for bacteremia modeling, as this can largely dictate disease progression. Intranasal inoculation with P. aeruginosa results in severe pneumonia and dissemination to the liver and spleen (55). In contrast, direct intravascular injection of the same strain results in minimal CFU burden in either the lung or spleen. This difference in pathogenicity is not described, but perhaps bacteria are primed for infection through physiologic adaptation in the lung, or perhaps initial infection overwhelms host responses that can otherwise clear bloodstream infection. For both models, the P. aeruginosa strain was rarely detected in the blood itself, indicating a poor microenvironment for survival (55). Similarly, from a transposon screen, arnA was suspected to encode a virulence factor required for P. mirabilis dissemination from the kidney to the bloodstream but was dispensable for P. mirabilis survival in the blood (44). This was again demonstrated for K. pneumoniae, where mutants attenuated in a model of gut to liver dissemination were as virulent as the parent strain in an i.p. bacteremia model (56). These findings indicate differences in microenvironments and interactions with host responses at various sites. Thus, when modeling bacteremia it is critical to consider how specific models address distinct questions. Direct blood injection may be appropriate for discerning bacterial blood fitness factors but may not illuminate bacterial mechanisms of dissemination from initial infection. Likewise, it may be inappropriate to model bacterial dissemination by direct injection into the bloodstream.
Another limitation of animal modeling is harvesting organs to deduce circulating bacterial burden. In some bacteremia models, species are rarely isolated from murine blood but are readily harvested from spleen and liver (55). Further, microenvironments of the blood, liver, and spleen are not equal, which may influence fitness factors at each site. As demonstrated in K. pneumoniae bacteremia, the citrate synthase gene gltA is required for liver and spleen colonization but is dispensable in the blood (53). Since spleen and liver burden is not assessed in clinical cases, it is difficult to interpret how these findings translate to human disease.
Aside from animal modeling, in vitro systems may be used to identify bacteremia factors (Table 2). Serum killing assays have been used to evaluate bacterial resistance to complement and the membrane attack complex. For example, these assays identified the importance of capsule in bloodstream infections, demonstrating that acapsular K. pneumoniae is more susceptible to complement-mediated killing (57, 58). Another in vitro approach, serum growth assays, can identify factors required for bloodstream replication and have illuminated diverse bacterial metabolic strategies in the blood (42, 43, 53, 57). Both serum killing and growth assays can identify bacterial factors involved later phases of bacteremia. Because human serum is readily available from commercial sources and residual clinical specimens, a substantial advantage of in vitro serum-based methods is the ability to evaluate whether findings from animal models may translate to human disease. However, serum is only a fraction of whole blood, and other components that influence pathogenesis, such as circulating immune cells and cytokine responses, are not accounted for in these assays. For serum growth, nutrients from the initial site and end organs from dissemination are not included. Since factors may have species-specific dependency, it is important to validate animal findings in these systems.
In summary, animal systems must be carefully considered when modeling bacteremia but provide unique insight into pathogenesis of infections from multiple sources and species. A combination of initial-site dissemination and direct bloodstream fitness modeling is appropriate for identifying factors involved in early and later bacteremia. Additionally, since bacteremia is broadly associated with underlying comorbidities, animal models of these conditions could illuminate bacterial factors unique to age, immunosuppression, and hematologic malignancy.
ESCHERICHIA COLI BACTEREMIA
Introduction and Epidemiology
E. coli is consistently the most prevalent Gram-negative bacteremia pathogen and since 2005 has been reported as the most common bacteremia pathogen overall (18). Due to the vast number of identified strains and disease associations, E. coli has been divided into several pathotypes. Extraintestinal pathogenic E. coli (ExPEC) includes uropathogenic E. coli (UPEC) and sepsis-associated E. coli (SEPEC), which can both cause E. coli bacteremia. Among all HAIs, E. coli is the fourth leading cause of infection (12, 16, 18, 25). As with other Gram-negative pathogens, E. coli bacteremia risk factors include immunosuppression and underlying comorbidities (59–61). Distribution of community and health care-associated E. coli bacteremia is approximately similar, but health care-associated cases correlate with older age, while community-acquired cases correlate with younger age (61). The gastrointestinal tract serves as a major reservoir for E. coli bacteremia (59, 60), and individuals may become colonized by interactions with the vast number of ExPEC environmental niches (reviewed in reference 30). In the environment, ExPEC has been isolated from recreational water, contaminated rainwater, and wastewater (26). Wildlife and agriculture may also serve as reservoirs through which the intestinal tract may become colonized after consumption of contaminated foods (30).Transmission of colonizing strains can occur between household members and companion animals (27, 62).
E. coli bacteremia has been monitored in selected populations, but epidemiological studies have not appropriately estimated global disease burdens. Annual population incidence of E. coli bacteremia is approximately 30.3 per 100,000 (61). Estimates of E. coli bacteremia mortality are incredibly broad, spanning from 5 to 30% (59–61), due to the high level of heterogeneity among E. coli strains capable of causing bacteremia and associations with severe underlying comorbidities. Additionally, many studies addressing E. coli bacteremia have been conducted in cohorts with hematologic malignancies, making broad mortality estimates difficult to assess.
Subtypes
Spanning E. coli pathotypes are phylogroups A, B1, B2, and D. Largely, group A is associated with commensal clones and B2 with pathogenic clones, particularly ExPEC (63). However, all phylogroups have been represented in E. coli bacteremia clinical isolates. B2 is closely connected to E. coli bacteremia, and groups B2 and D express more virulence factors that include toxins, adhesins, and siderophores, which aid in initial-site invasion, dissemination, and bloodstream survival (54, 59, 63–66).
Antimicrobial resistance in E. coli bacteremia is alarming. E. coli can carry antimicrobial resistance plasmids encoding β-lactamases (bla), extended spectrum β-lactamases (ESBLs), and carbapenemases (23, 61). Plasmid-based ESBLs can convey resistance to cephalosporins, and carbapenemases can confer resistance to nearly all β-lactams. In Western Europe and South Korea, E. coli bacteremia isolates often belong to sequence types (STs) associated with antimicrobial resistance, including ST131, which can carry plasmid-based resistance genes, including blaCTX-M (59, 65, 67, 68). Future studies should identify unique features of ST131 to understand increased sustained global spread and bacteremia pathogenicity.
Sites of Origin
E. coli bacteremia is primarily attributed to urinary tract infection (UTI) and gastrointestinal origins (59, 60). UTIs and E. coli bacteremia are tightly linked. Indeed, UTIs are a dominant source of E. coli bacteremia (59) and E. coli is the most common source of bacteremic UTI (69). Since bacteria can ascend the ureters to the kidney parenchyma and cross renal tubule epithelium and capillary endothelium to enter the bloodstream during UTI, UPEC strains and SEPEC strains often overlap pathotypes. Interestingly, mortality rates for E. coli bacteremia originating from UTI are about 17% lower than those for E. coli bacteremia from other sources (59). Since UPEC is adapted to infect the genitourinary tract, perhaps these strains can advantageously disseminate from the bladder, but fitness may not always translate once organisms are in the blood.
The gut microbiota also harbors potential E. coli bacteremia strains that colonize the gastrointestinal tract. In particular, E. coli bacteremia isolates from phylogroups A or B1 are often predicted to have disseminated from the gut. Patients with hematologic malignancies are especially vulnerable to E. coli bacteremia through increased permeability of the intestinal mucosa and dissemination of colonizing strains (60, 64). Conditions associated with immune modulation, such as organ transplant, cancer, and Crohn’s disease, strongly correlate with development of E. coli bacteremia (61). Thus, these infections may largely be established by colonizing strains not controlled by the host.
Bacteremia Factors
There is no single virulence factor predictive of E. coli fitness in bacteremia, but factors that have been identified largely contribute to immune evasion, adherence, and metabolic diversity (Table 1). Due to the high diversity of E. coli, some bacteremia factors required in one setting may be dispensable in another due to strain variation and differential expression of virulence factors, as with P fimbriae. Capsule production is also correlated with E. coli bacteremia (49, 63), likely serving as a defense mechanism against immune opsonization during initial infection and in the blood. E. coli may modulate host immune responses through pic, encoding a protein required for E. coli bacteremia fitness that reduces leukocyte activity during inflammation (50). Additional defense against innate immunity is conveyed through sapR by protection against antimicrobial peptides.
Adhesins have a clear function in E. coli bacteremia pathogenesis. Uroepithelial adhesion, critical for bacteremic UTI dissemination, is mediated by type IV pilin genes (50). In mice, fimbrial adherence contributes to systemic bacteremia modeled by TVI, as mutants lacking type I and P fimbriae are outcompeted by wild-type strains (49). In humans, P fimbriae expressed by UPEC bind to the P blood group antigen on erythrocytes, implying a potential role for erythrocyte interactions in systemic spread (49). However, mice lack this globoside receptor, and P fimbria-mediated adherence to it likely does not occur in this model. Nevertheless, P fimbria mutants are outcompeted by wild-type UPEC following murine transurethral infection, and the fitness defect can be complemented in trans, suggesting an unknown advantageous property conferred by P fimbriae in the ascending UTI mouse model (70) and perhaps the TVI model. Comparing gut-colonizing E. coli isolates that did and did not progress to E. coli bacteremia revealed that the only virulence factor genes expressed significantly more in blood-disseminating strains were afa/dr (64), encoding Afa and Dr adhesins. Of note, this study utilized clinical isolates from patients with hematologic malignancies; therefore, this virulence factor may relate to damaged mucosal barriers and may not correlate with gut-to-blood dissemination in heathy individuals. Poly-N-acetylglucosamine, an extracellular E. coli polysaccharide that aids in biofilm formation and extracellular matrix binding, is also required for E. coli bacteremia blood survival (50). This polysaccharide may assist in anchoring E. coli to kidney tubule epithelium or indwelling devices, which could have implications for initial-site invasion and dissemination, although this should be further investigated, since this factor was uncovered in a TVI bacteremia model.
Metabolic diversity is critical for transitioning between environmental niches, as nutrients at initial infection sites are often more abundant than in the blood. In murine TVI bacteremia, the E. coli genes c1220 and oppA are required for maximum bloodstream fitness (50). These genes encode proteins involved in oligopeptide uptake and shikimate pathway biosynthesis, representing a requirement to utilize diverse carbon sources in the blood. There is also evidence that E. coli metabolizes sialic acid, a component of erythrocyte outer membranes, as a carbon source; nanA mutants, which cannot metabolize sialic acid, are at a significant disadvantage in TVI bacteremia (49). In clinical isolates, siderophore expression is associated with B2 bacteremia pathotypes (59). Siderophores are chelating molecules which allow bacteria to scavenge iron in environments where this critical resource may be highly limited (71), as in the blood. Increased siderophore expression in E. coli bacteremia likely aids in iron uptake and bloodstream survival.
There are conflicting reports on the requirement of some factors. One report suggests a requirement for hemolysin in E. coli bacteremia (72), while another shows dispensability (49), suggesting strain- and model-specific differences. Since hemolysin can lyse red blood cells, perhaps certain strains downregulate this mechanism in the blood to avoid inflammation. There are also conflicting reports on the requirement of P fimbrial genes. E. coli bacteremia isolates demonstrate a protective correlation of P fimbrial gene expression with patient survival rates (59). However, mouse models show a requirement for P fimbriae in establishing bacteremia (49) and UTI (70). Perhaps P fimbrial genes are required to exit initial sites and establish infection but also trigger a host immune response for pathogen clearance. Survival in the blood may require a balance between expression of adhesion genes for dissemination and repression once in the bloodstream to minimize detection. This discrepancy may also be explained by bacteremia pathogenicity being a function of combinations of virulence factors and not reliant on single factors. One study found no prediction for hemolysin or P fimbrial expression in E. coli bacteremia clinical isolates but did find that these factors in combination were predictive for blood dissemination (54). Differential requirements for dissemination from initial sites may also explain discrepancies, along with differences in bacteremia pathogenesis between humans and mice.
KLEBSIELLA PNEUMONIAE BACTEREMIA
Introduction and Epidemiology
K. pneumoniae is a leading pathogen in Gram-negative bacteremia and the third most common cause of HAIs overall (12, 18, 20, 25). From the few studies addressing the global epidemiology of K. pneumoniae bacteremia, annual population incidence is an estimated 7.1 per 100,000 people and, as with E. coli bacteremia, is likely underreported (73). In a Canadian population, approximately 70% of cases were health care associated, while 30% were community acquired (73); in a Korean population, health care- and community-associated infections each contributed to roughly half of K. pneumoniae bacteremia cases (74). The gastrointestinal tract serves as a major reservoir for K. pneumoniae bacteremia (31, 32). It is unclear how initial gut colonization occurs, but K. pneumoniae has been isolated from many niches, including water and livestock (29, 75, 76), and may be introduced into the gut after interaction with these reservoirs.
Broadly, K. pneumoniae infections are a public health threat given high rates of antibiotic resistance (23). K. pneumoniae is inherently resistant to ampicillin through a narrow-spectrum β-lactamase (bla) (77, 78). Two widely documented K. pneumoniae carbapenemases (KPC) are the plasmid-based KPC-2 and KPC-3, and KPC composition in carbapenem-resistant (CR) K. pneumoniae bacteremia isolates geographically varies. In New York City, KPC composition within CR K. pneumoniae bacteremia isolates was 70% blaKPC-2 and 30% blaKPC-3 (79); in Italy, case distribution was 10% blaKPC-2 and 90% blaKPC-3 (80). Importantly, mortality rates are similar between KPC subtypes (79). These KPC studies reported minimal detection of other carbapenemases, but separate cohorts have reported non-KPC carbapenemases in CR K. pneumoniae bacteremia. In South and Southeast Asia, blaOXA-232 and blaNDM-1 account for 95% of CR K. pneumoniae bacteremia isolates, and blaKPC isolates were not detected (81). K. pneumoniae STs associated with bacteremia are diverse and vary geographically. In New York City, antimicrobial-susceptible K. pneumoniae bacteremia is highly diverse, represented by 127 STs among 194 isolates, and the most common STs (ST20, ST37, and ST45) account for only 13% of isolates. CR K. pneumoniae bacteremia is more clonally representative, with ST258 alone accounting for 63% of cases and, along with ST17 and ST392, representing 86% of isolates (79). In South and Southeast Asia, ST15, ST23, ST14, and ST231 each account for >6% of K. pneumoniae bacteremia isolates, and ST15 is responsible for the majority of CR cases (81). Intensive epidemiological studies of K. pneumoniae bacteremia should be performed to properly evaluate global prevalence of K. pneumoniae bacteremia and the contributing sequence types.
Subtypes
K. pneumoniae can be classified into different subtypes based on virulence and carbapenem resistance. Capsular polysaccharide production is an established K. pneumoniae virulence factor and protects from host immune defenses (reviewed in reference 82). Classic K. pneumoniae, the most common subtype in Western countries, is characterized by lack of excessive capsule, but is pathogenic and can carry antimicrobial resistance plasmids. Hypervirulent (HV) K. pneumoniae, first reported in Southeast Asia in 1980 to 1990, is linked to CAIs and liver abscess formation even in healthy individuals. Excessive capsule production is characteristic of HV K. pneumoniae (reviewed in references 83 and 84). The invasive nature of HV K. pneumoniae is partially linked to hypermucoviscosity, a phenotype indicated by excess exopolysaccharide coating which increases phagocytosis resistance partially by preventing deposition of complement on the bacterial surface. Another characteristic is the production of multiple siderophores, which scavenge iron from the host (71, 84). Therefore, in the context of bacteremia, consideration of infectious K. pneumoniae subtypes is necessary, since strains differ in capsule production and host interactions.
Prognosis of K. pneumoniae bacteremia largely depends on the infectious strain. Antimicrobial susceptible K. pneumoniae bacteremia is associated with ∼26% mortality, a risk that increases with antimicrobial resistance. The frequency of bacteremia caused by ESBL-producing K. pneumoniae varies geographically, composing 23% of isolates in Northern Italy, 36% in the United States, 37% in South Africa, 59% in Argentina, and 60% in India (80, 81, 85). CR K. pneumoniae bacteremia incidence also varies by location, accounting for 5% of K. pneumoniae bacteremia cases in Vietnam, 7% in Northern Italy, 13% in New York City, 28 to 33% in Eastern China, and 50% in India (79–81, 86, 87). Health care-associated K. pneumoniae bacteremia predicts higher mortality than CAI, likely due to increased antimicrobial resistance (74, 87). CR K. pneumoniae and HV K. pneumoniae can problematically merge into CR-HV K. pneumoniae, which is concerning due to both high antimicrobial resistance and virulence (81). As with other cases of Gram-negative bacteremia, mortality is associated with older age, immunosuppression, and underlying comorbidities.
Sites of Origin
Pneumonia, UTI, and gut colonization are leading sources of K. pneumoniae bacteremia (34, 35, 73, 79, 85). In roughly half of cases, nosocomial K. pneumoniae bacteremia is associated with primary bacteremia, likely arising from gut colonization (73). Health care-associated K. pneumoniae bacteremia arises from UTI, primary infection, or biliary tract infection in 76% of cases. Community-acquired K. pneumoniae bacteremia is linked to UTI or biliary tract infection in 66% of cases. Pneumonia is a common source of K. pneumoniae bacteremia across environments, underlying 11% of nosocomial infections, 10% of HAI, and 5% of CAI. Interestingly, K. pneumoniae bacteremia originating from catheters is associated with higher survival than secondary bacteremia from other sources (88). This reflects differences in pathophysiology, as infections from indwelling devices may involve direct introduction of pathogens to the bloodstream, unlike infections originating from sites where pathogens must overcome epithelial barriers and immune evasion. Compared to other secondary bacteremia isolates, catheter-sourced K. pneumoniae bacteremia isolates are less hypermucoviscous, potentially increasing susceptibility to opsonization (89). K. pneumoniae is responsible for 12% of hospital-acquired pneumonia (25). It is unclear what percentage of organisms progress to bacteremia, but K. pneumoniae-bacteremic pneumonia carries a mortality risk of 37% (85, 90). Secondary K. pneumoniae bacteremia from UTIs predicts a mortality risk of 12% (85, 90). Thus, secondary bacteremia from distal sites may result from bacteria that are more pathogenic than those originating from indwelling devices, but more studies on K. pneumoniae bacteremia from catheters is required.
Primary K. pneumoniae bacteremia may arise from the gastrointestinal tract after colonization. High concordance between gut-colonizing K. pneumoniae and isolates from pneumonia, UTI, and blood indicate a critical role in initial gut colonization for progression to secondary infection and bacteremia (31, 32). Gut colonization may also precede pyogenic liver abscess (PLA), an infection which generates pockets of bacterial growth in the hepatic parenchyma associated with HV K. pneumoniae and is particularly common in Southeast Asia (83, 91, 92). This unique clinical presentation should raise suspicion of a Klebsiella infection. K. pneumoniae site-specific fitness factors have been extensively studied for pneumonia but remain largely unknown for other sites.
Bacteremia Factors
By using murine pneumonia models to investigate secondary bacteremia, some K. pneumoniae factors required for lung to spleen dissemination have been identified. Siderophores are required for blood dissemination (93) through interaction with host lung epithelial HIF-1α, although the exact mechanism is unknown. In agreement with a role for inflammation in dissemination, a wecA mutant with stunted lipopolysaccharide (LPS) production had significantly lower spleen dissemination after lung infection (46). Although pneumonia models appropriately identify K. pneumoniae fitness factors at initial infection sites, current literature often reports defects in both lung fitness and spleen dissemination. Therefore, it is difficult to discern whether these factors relate to lower blood dissemination due to stunted lung fitness or an inability to adapt in the blood.
Neutrophils and alveolar macrophages are required for initial containment of K. pneumoniae lung infection and bacteremia (94). Lipocalin 2 (Lcn2), an innate immune protein, can prevent dissemination from the lung to the spleen, and mice deficient in Lcn2 have higher mortality after K. pneumoniae lung infection (95, 96). Host thrombospondin-1 (TSP-1), an extracellular matrix protein, is required for K. pneumoniae lung fitness and spleen migration (97). TSP-1 peptides can inactivate neutrophil elastase, a serine protease used to degrade pathogens, and could be advantageous for K. pneumoniae survival during lung inflammation prior to bloodstream infection. K. pneumoniae can also survive innate immune cell killing through evasion of pyroptosis via upregulation of host interleukin 10 (IL-10). The exact mechanisms are unknown, but strains that upregulate IL-10 establish higher lung and spleen K. pneumoniae burdens (98).
By modeling gut colonization and liver metastasis, factors required for dissemination from the gut to the blood have been identified (56). In one study, mutations in these factors did not alter serum resistance, implying large contributions from initial site-specific fitness in bacteremia. One identified gene cluster, mrkCDF, encoding anchoring proteins for type III fimbriae, likely aids in transmigration of intestinal epithelium. Another gene, ymdF, encodes a hypothetical protein that is linked to oxidative stress regulation and evasion of phagocytosis. In a separate investigation, iron acquisition encoded by kfu was required for infection by intragastric colonization but dispensable for i.p.-modeled bacteremia (99). K. pneumoniae factors associated with bacteremia secondary to PLA have also been investigated. Siderophores have some functional redundancy in bacteremia virulence, but tonB, required for siderophore import, is essential for maximum virulence in an i.p. model (99). By using an i.p. injection and monitoring subsequent liver invasion, HV K. pneumoniae K1/K2 serotypes were found to more readily establish PLA, confer higher lethality, and induce greater neutrophil influx than K62 or acapsular strains (100). Accordingly, there may be connections between neutrophil infiltration and necrosis with establishment of liver abscesses prior to bacteremia by hypervirulent strains.
K. pneumoniae bloodstream survival partially depends on capsular polysaccharide production, as mutants with decreased capsule have attenuated virulence in an i.p. model (101). In human serum, rfaH, lpp, and arnD are required for evasion of complement-mediated serum killing, and all encode products that contribute to capsule and outer membrane structure (57, 58, 102). aroE, involved in aromatic amino acid synthesis, is required for evasion of serum killing, although the mechanism is unknown (57). Mutations in ompK36, encoding an outer membrane porin, also decrease bloodstream virulence (103). Regulation of DNA methylation partially contributes to bloodstream fitness, as mutations in dam result in a higher lethal dose than the parent strain, although dam mutants still confer mouse lethality (104). Mechanisms of how capsule and DNA methylation influence blood fitness are not understood and should be further investigated. Metabolic flexibility for K. pneumoniae is partially conveyed through gltA, encoding a citrate synthase dispensable for fitness in the bloodstream but required for maximum fitness in the spleen and liver (53). Carbohydrate metabolism is also essential in human serum through genes such as pfkA and galE (102). Biosynthesis of uracil through pyrE is required for K. pneumoniae serum growth, indicating a potential target for future antibiotics (102).
Together, these observations indicate that a potential cycle occurs where K. pneumoniae in the lung upregulates host responses to control infection. To evade killing by immune cells, K. pneumoniae utilizes unknown mechanisms to manipulate host IL-10 or utilize TSP-1. In turn, K. pneumoniae factors like siderophores can upregulate proinflammatory cytokines, which increase angiogenesis and epithelial permeability, providing an escape mechanism for bacterial cells from initial lung infection to the bloodstream (Fig. 2). While this cycle may be relevant for pneumonia, site-specific fitness in UTI and gut colonization should be further investigated. Additionally, mechanisms governing K. pneumoniae dissemination and bloodstream survival have not been thoroughly assessed.
Overall, factors involved in K. pneumoniae bacteremia are similar to those involved in E. coli pathogenesis where capsule, adhesins, and siderophores contribute to bacteremia (Table 1). Notably, K. pneumoniae lacks defined toxins, such as hemolysin or cytotoxic necrotizing factor, which E. coli may utilize.
PSEUDOMONAS AERUGINOSA BACTEREMIA
Introduction and Epidemiology
P. aeruginosa is the third leading Gram-negative species isolated from clinical bacteremia, and estimated mortality rates span a large range, from 21 to 62% (12, 18, 20, 105–108). Mortality calculations are confounded by a high incidence of P. aeruginosa infections in cystic fibrosis patients (109, 110) and the critically ill and depend on characteristics of the infecting strain, including antimicrobial resistance and exotoxin profile. Therefore, is it difficult to comprehensively estimate the global burden of P. aeruginosa bacteremia. The primary reservoir for P. aeruginosa, including strains expressing the clinically relevant exotoxins encoded by exoS and exoU, is moist environmental niches, such as lakes, swimming pools, faucets, and sinks, rather than a human host (111, 112).
Subtypes
Multidrug-resistant P. aeruginosa has been labeled as a serious threat by the CDC (23) and a species of critical concern by the WHO (24). However, preventive efforts have deceased rates of P. aeruginosa infection by 29% since 2013 (23), an encouraging sign that this trend may continue. Like other Gram-negative species, P. aeruginosa is equipped with plentiful antimicrobial resistance mechanisms. Efflux systems like MexAB-OprM render P. aeruginosa inherently resistant to multiple antimicrobials (reviewed in reference 113), and chromosomal AmpC cephalosporinase confers inherent resistance to some β-lactams (113). Importantly, P. aeruginosa can acquire plasmids carrying genes for carbapenemases, such as blaKPC and blaVIM (23, 107, 108, 114), and carbapenem-resistant P. aeruginosa bacteremias are more lethal (105, 107).
Analysis of P. aeruginosa bacteremia isolates indicates that no single ST infects a majority of patients (115, 116). However, in Spain, all carbapenem-resistant P. aeruginosa bacteremia isolates belonged to ST235 or ST175 (108). When the same samples were analyzed for ExoU, a potent bacteremia virulence factor, all ST235 P. aeruginosa bacteremia isolates were ExoU positive, while no ST175 isolate expressed ExoU. Further, P. aeruginosa bacteremia with a ST235 isolate predicted significantly higher 30-day mortality. Therefore, special attention must be paid to P. aeruginosa bacteremia caused by ST235 isolates because of unique features contributing to pathogenicity.
Sites of Origin
The average length of hospital stay prior to the onset of bacteremia is about 14 days longer for P. aeruginosa infections than other species, emphasizing the strong link between HAIs and P. aeruginosa bacteremia (20, 105). Further, P. aeruginosa bacteremia with antimicrobial-resistant strains is more associated with HAIs than nonresistant strains (108). Community-acquired P. aeruginosa bacteremia is less prevalent, and initial sites of infection in this subset have not been extensively documented. Therefore, future studies should evaluate initial sites and features of causative strains in CAI.
Pneumonia is the most prominent initial infection for secondary P. aeruginosa bacteremia, associated both with higher mortality and antimicrobial-resistant P. aeruginosa bacteremia (105–107, 117). P. aeruginosa is also problematic in infections involving skin and soft tissue lesions, which provide a direct route for establishing P. aeruginosa bacteremia. A unique pathology of P. aeruginosa bacteremia is the formation of ecthyma gangrenosum (necrotic skin lesions), particularly in immunocompromised patients (38).
Bacteremia Factors
Factors governing P. aeruginosa lung infection and blood dissemination have been investigated. One study reported a significant increase in virulence factor protein levels for P. aeruginosa bacteremia compared to isolates from the same patient at initial sites of infection (118). Upregulated proteins included LecA and RpoN, an adhesion factor and RNA polymerase sigma factor, respectively. Although higher in bloodstream protein abundance, mRNA transcript levels were similar between initial-site and blood isolates, suggesting posttranslational regulation of virulence factors in different environments. These findings may encourage investigation of both transcriptional and translational virulence regulation mechanisms.
As P. aeruginosa is a prevalent cause of pneumonia, factors promoting dissemination from the lung to the blood have been explored (reviewed in reference 119). In particular, the type III secretion system (T3SS) has been identified as leading to increased mortality in human infection and animal models (106, 120). Bacterial T3SS machinery injects virulent effector proteins directly into host cell cytoplasm. T3SS exotoxins of P. aeruginosa include ExoU, ExoS, and ExoT (120). ExoU and ExoS contribute to higher lung bacterial burden and worse clinical outcomes but are rarely found in the same isolate (106, 111). ExoU, a phospholipase which disrupts host membrane integrity, maximizes dissemination from the lung to the bloodstream (120). Expression patterns of exotoxins in P. aeruginosa bacteremia cannot predict initial sites of infection, but pneumonia isolates trend toward higher levels of ExoU (106). Additionally, man-made environmental water reservoirs like faucets, drains, and tubs are significantly associated with exoU+ strains of P. aeruginosa compared to exoS+ strains (111). ExoU expression in P. aeruginosa bacteremia correlates with higher mortality (120), and therefore, screening for exoU+ strains could be explored as an indicator for initiation of aggressive therapy.
Innate immune cells are recruited to the lung during P. aeruginosa infection (119). Two T3SS effector proteins, ExoS and ExoT, aid directly in evasion of host immune cells by disruption of the actin cytoskeleton (119, 121). In early P. aeruginosa pneumonia, ExoS is injected specifically into neutrophils as opposed to other immune cells (122). During later infection, type I pneumocytes, which contribute to lung epithelial architecture, are injected with ExoS. Type I pneumocytes with ExoS form aggregates which increase in size and abundance of dead cells as infection progresses (119). The presence of these aggregates correlates with bacterial leakage from the lung and blood dissemination. Although injected into phagocytes, ExoT cannot form type I pneumocyte aggregates (122), highlighting distinct roles for ExoS and ExoT in P. aeruginosa bacteremia.
It is important to note T3SS-independent virulence factors. ExlA, a toxin secreted by T3SS-negative P. aeruginosa, is a potent necrotizing factor for epithelial and myeloid cells. In murine pneumonia models, lung invasion and blood dissemination are governed by ExlA, which in the absence of T3SS can disrupt pulmonary vascular barriers (55). P. aeruginosa can also employ a type II secretion system (T2SS), utilizing the virulence factor LasB. This protease can dissociate extracellular matrix proteins and cleave adherens junctions through interactions with vascular endothelial cadherin (123), providing bloodstream access. Investigations of T2SS and T3SS effector proteins have formed a model of P. aeruginosa bacteremia establishment from pneumonia (Fig. 2). Upon lung infection, strains expressing ExoU alone can directly kill host cells through disruption of the cell membrane. Strains expressing ExoS and ExoT can use these exotoxins to evade early innate immune cell killing. ExoS can then kill type I pneumocytes, disrupting epithelial barriers. Meanwhile, ExoT can cleave extracellular matrix proteins and contribute to vascular leakage (119, 123).
The pathogenicity of P. aeruginosa T3SS extends beyond pneumonia, and it is required for virulence in burn models (124), indicating broad usage of T3SS in P. aeruginosa bacteremia. Establishment of P. aeruginosa bacteremia from other initial sites must be further characterized to evaluate site-specific factors. Additionally, P. aeruginosa mechanisms for bloodstream fitness have not been addressed. As opposed to K. pneumoniae, P. aeruginosa expresses a repertoire of potent exotoxins, which more closely reflects proteins like hemolysin and cytotoxic necrotizing factor in E. coli.
ACINETOBACTER BAUMANNII BACTEREMIA
Introduction and Epidemiology
A. baumannii is a clinically problematic Gram-negative species that causes bacteremia (A. baumannii bacteremia), especially in health care settings, with an estimated mortality risk ranging from 20 to 39% (37, 87, 125). High mortality may be due to the prevalence of A. baumannii bacteremia in ICUs and associations with comorbidities, supported by the finding that community-acquired A. baumannii bacteremia cases have slightly lower mortality rates. A. baumannii infections are closely linked to hospital environments, with a longer stay increasing A. baumannii bacteremia risk (126). Hospital environments serve as abundant reservoirs for A. baumannii, which has been isolated from the surfaces of portable medical equipment, mattresses, and sinks (28, 127). A. baumannii can colonize the skin, nasal tract, and trachea, but patient isolates are often identical to hospital environmental isolates taken during their stay (28, 128). It is difficult to determine whether colonization from hospital environments is transient and could allow subsequent spread to community reservoirs (28, 127). Diverse niches of A. baumannii extend to soil and fertilizer, but it is unclear how relevant these habitats are to CAIs (129, 130).
Although A. baumannii accounts for a relatively low percentage of overall bacteremia cases, multidrug resistance is globally problematic for this species (131). Drug-resistant A. baumannii infections were classified as an urgent threat by the CDC in 2019 due to high rates of antimicrobial resistance, including strains that are nonsusceptible to all available antibiotics (23). In 2017, the WHO classified resistance in A. baumannii as a critical priority (24). In Taiwan, over 15% of A. baumannii clinical isolates are carbapenem resistant (126), and globally, over 71% are multidrug resistant (18). In the United States, 27% of mechanically ventilated patients were colonized with a multidrug-resistant strain of A. baumannii (132). Of A. baumannii bacteremia isolates, ∼1% in a multicenter study were pan-drug resistant, and colistin was the only reliable antimicrobial against these infections. A. baumannii utilizes resistance strategies, including chromosomal β-lactamases, efflux pumps, and aminoglycoside-modifying enzymes. After human serum albumin exposure, A. baumannii upregulates transcription of β-lactamases, indicating potentially inherent antimicrobial resistance mechanisms in serum (133).
Sites of Origin and Bacteremia Factors
HAIs account for 75% of A. baumannii infections and compose about 86% of antimicrobial-resistant strains (134). Pneumonia is a common source of A. baumannii bacteremia for community-acquired cases. For UTIs, rates of developing A. baumannii bacteremia are similar between community- and health care-acquired cases. Invasive devices are also sources of A. baumannii bacteremia but are primarily linked to health care-acquired infections. The ability of A. baumannii to survive on many different surfaces (13, 127, 135) is hypothesized to be one reason this species is uniquely steadfast in hospitals (18, 28, 127). Hospitalized patients are frequently colonized, which can progress to opportunistic infection after antibiotic treatment (28, 127). Nasal colonization is particularly common after long-term medical care, during which reported colonization rates are 70 to 92% in Taiwan (128) and 63% in the United States. Infection prevention strategies have reduced A. baumannii bacteremia, indicating that continued efforts could reduce disease burden (37).
A. baumannii bacteremia frequently arises as a secondary infection from contaminated health care equipment and surgical site infections (37, 126, 134). The formation of robust biofilms is an advantageous survival mechanism of A. baumannii and underlies the prevalence of infections associated with indwelling and contaminated devices. Once in the blood, A. baumannii downregulates genes involved in biofilm formation and adhesion and increases expression of motility-associated genes like pilQ, suggesting involvement of planktonic cells in dissemination (133, 136). Factors governing initial-site infection of catheters are likely highly linked to those required for biofilm establishment and maintenance, although this is understudied due to the modeling limitations discussed above. For pneumonia, mltB, which encodes a lytic transglycosylase, is required for murine respiratory tract colonization (51). How mltB and factors required for pneumonia contribute to bacterial dissemination is not clear. In this model, mice were depleted of neutrophils prior to infection, and therefore, interactions between innate immune responses and A. baumannii dissemination from the lung should be explored.
Survival of A. baumannii in the blood has been partially described. Genes regulating immune evasion and adhesion are especially important to pathogenesis. After exposure to human serum, there is minimal killing of A. baumannii, suggesting a degree of inherent serum resistance (45). Capsule has been indicated as a protective mechanism due to the upregulation of multiple capsule synthesis genes following serum exposure (133). Additionally, A. baumannii can bind plasminogen, a regulator of the complement cascade, preventing formation of terminal complement complexes on the surface of the bacteria (137). In part, serum resistance is conferred by mltB, involved in peptidoglycan remodeling and cell membrane integrity (51). Deletion of mltB renders A. baumannii more susceptible to physiologic stressors, including oxidative, acidic, and osmotic challenge. Biofilm formation and adherence to epithelial cells are also significantly impacted by deletion of mltB through disruption of pilus assembly.
A. baumannii strains defined by one study as hypervirulent, causing 100% mortality in mice when administered at a standard dose, appear to be well adapted to survival in the bloodstream. In mice, sustained circulating CFU are observed over multiple days for hypervirulent strains, while hypovirulent isolates are cleared within hours of inoculation (138). Serum fitness is largely linked to complement, with hypervirulent A. baumannii being more resistant to complement-mediated killing than other strains. Neutrophils are central in the initial immune response to A. baumannii infection, but additional activities of macrophages and complement synergistically eradicate infection (138). Antimicrobial peptide resistance is partially conferred to A. baumannii by pntB, feoB, and fepA which also contribute to intracellular macrophage survival (45). These factors increase persistence in the midst of immune responses, which varies by strain.
Toll-like receptor 4 (TLR4), a host pattern recognition receptor, detects LPS and initiates inflammation. Paradoxically, even though TLR4 initiates immune responses, TLR4−/− mice have lower mortality rates than wild-type mice during A. baumannii bacteremia (139). This is not due to differences in bacterial burden and is instead attributed to lower levels of host inflammation and sepsis-associated inflammatory markers (139). Inhibition of LPS detection diminishes bacterial blood density through macrophage phagocytosis and also minimizes release of TLR4-dependent sepsis-associated markers, which increases mouse survival. Thus, complex interactions between LPS- and TLR4-mediated inflammation should be broadly considered for Gram-negative bacteremia.
Bloodstream survival for A. baumannii may rely more heavily on survival mechanisms to persist and maintain bacterial density in the blood than traditional virulence factors (45, 51, 138). In support of this theory, in vivo transcriptome analysis revealed that majority of genes upregulated during A. baumannii bacteremia are associated with capsule biosynthesis and iron acquisition (136). Other A. baumannii virulence factors were either downregulated or unchanged, which could be strain specific. This highlights similarities between A. baumannii, K. pneumoniae, and E. coli. Survival for these species seems to be linked to the ability to adapt to diverse environmental niches and upregulation of capsule to protect from opsonization. Unlike P. aeruginosa, A. baumannii and K. pneumoniae do not appear to utilize potent toxins or virulence factors to perpetuate bacteremia.
OTHER GRAM-NEGATIVE BACTEREMIA SPECIES
A relatively small number of additional Gram-negative species not highlighted above are emerging as frequent causes of bacteremia and also contribute to the global disease burden. This secondary group of Gram-negative bacteremia species are generally less well studied, and comparatively little is known about the physiology and pathogenesis of these organisms during infection. Continued investigation of bacteremia characteristics attributed to these species will provide additional insight into unique and conserved pathogenic mechanisms among the larger group of Gram-negative bacteremia-causing organisms.
Serratia marcescens is a member of the family Yersiniaceae that engages in both pathogenic and nonpathogenic human interactions, but it also has broad distribution in other environments. S. marcescens is commonly isolated from water and soil and can be found in association with plants, animals, and insects (140, 141). Specific correlations between colonization with S. marcescens and the onset of bacteremia are lacking in the current literature; however, surveys have shown S. marcescens colonization of patients in health care settings (142–145). As an etiologic agent of bacteremia, S. marcescens is among the 10 most common causes of nosocomial bloodstream infections, responsible for 2 to 4% of all cases (12, 20). Localized S. marcescens bacteremia outbreaks have also resulted from numerous reported instances of contaminated medical equipment (146–148), and the ability of S. marcescens to thrive in diverse environments may contribute to its stable colonization of medical surfaces and solutions. Pediatric populations appear to suffer more-severe adverse outcomes associated with S. marcescens bacteremia than with non-Serratia bacteremias, as exemplified by a greater length of stay, higher likelihood of ICU admission, and higher in-hospital mortality (149). Historically, Serratia infections indicative of hematogenous spread have been linked to ocular infections such as keratitis, especially in contact lens use (13). In TVI murine bacteremia, S. marcescens capsule synthesis contributes to disease, indicated by a significant reduction in spleen and kidney dissemination for wzx mutants compared to a wild-type strain (42). Mutations in pgm, which encodes a phosphoglucomutase linked to capsule production, also decrease bloodstream fitness and kidney colonization. Other branches of glucose metabolism are significant in S. marcescens bacteremia, indicated by reduced fitness when pfkA, encoding the glycolytic enzyme phosphofructokinase, is disrupted. S. marcescens bacteremia also requires the siderophore serratiochelin (52). Clinical observations of S. marcescens antibiotic resistance among bacteremia isolates (150) are a cause for concern. The variations of plasmid-encoded resistance genes discussed above are also relevant for S. marcescens. Furthermore, S. marcescens harbors chromosomal ampC genes, which can facilitate resistance to multiple β-lactam antibiotics, including cephalosporins, when expressed at high levels (151, 152). Multiple carbapenem resistance genotypes have also been reported for S. marcescens (153, 154). Finally, S. marcescens is inherently resistant to polymyxins (155), which further reduces the available treatment options in cases of acquired resistance to more commonly used antimicrobials.
Citrobacter freundii is a colonizer of the human gastrointestinal tract and can also be found in environments such as water and soil (156). As a pathogen, C. freundii is largely opportunistic and associated with many different types of infections (157–159), including a small but increasing fraction of bacteremia cases (16). Given that C. freundii is a frequent colonizer, it is not surprising that C. freundii bacteremia is largely linked to comorbidities. Intra-abdominal cancers and surgery-associated infections particularly underlie C. freundii bacteremia, but catheter-associated and genitourinary infections have also been reported (160, 161). C. freundii bacteremia most commonly originates from the urinary tract or from gastrointestinal sources of infection (161, 162). Drug resistance for C. freundii bacteremia can be facilitated in part by chromosomally encoded ampC genes in addition to infrequent carbapenem resistance. Using a TVI model of murine bacteremia, factors involved in bloodstream fitness have been identified. Pathways relating to DNA recombination and repair, like RuvABC complexes, are essential to C. freundii bacteremia by potentially aiding in defense against damaging host responses (43). Metabolism-associated factors also aid in C. freundii bacteremia, particularly that encoded by pfkA, which supports glycolysis, and that encoded by mtlD, which supports mannitol metabolism and the regeneration of the NAD+ pool (43). Protein transport by TatC is also required for blood fitness. In support of the notion that common fitness strategies are shared among Gram-negative bacteremia species, 42 fitness factor homologs were found between C. freundii and S. marcescens during murine bacteremia (43).
Proteus mirabilis is a Gram-negative species that frequently causes catheter-associated UTI in the elderly and can progress to secondary bacteremia (163, 164). Natural reservoirs of P. mirabilis include soil, water, and decomposing animal matter (165), and the organism can be found infrequently in human fecal samples (166, 167). Bacteremia caused by P. mirabilis accounts for ∼2% of community-acquired bloodstream infections (18). P. mirabilis is known for its extensive adherence and swarming capabilities, which likely contribute to infection. Hydronephrosis and urolithiasis resulting from P. mirabilis genitourinary tract infection can increase susceptibility to bacteremia (13). In a P. mirabilis TVI bacteremia model, tatA and tatC were necessary for blood fitness through contributions from protein translocation, reflecting similar requirement observed in C. freundii. Metabolic modification is also necessary for bloodstream fitness, as indicated by requirement of nitrogen assimilation genes, including gltB, glnA, gdhA, and ntrB. Polyamine biosynthesis via speAB and potB are required for P. mirabilis bloodstream fitness, indicating a fitness advantage for metabolic diversity in the blood (44). Like Serratia, P. mirabilis also exhibits very poor susceptibility to polymyxins (155).
Individual species underlying bacteremia are discussed above. However, these species may belong to clinical complexes where other closely related members also cause bacteremia. The Klebsiella clinical complex includes K. pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae, which account for 70%, 20%, and 10% of Klebsiella bacteremia cases, respectively (168). Virulence factors associated with K. pneumoniae are also found in K. variicola, including iron-scavenging mechanisms encoded by ybtS, entB, and kfu, and the adhesin-encoding mrkD (168). Mortality rates are similar for bacteremia caused by different Klebsiella clinical complex species, indicating that species outside K. pneumoniae should be investigated (168). Similarly, the Acinetobacter calcoaceticus-A. baumannii complex (ABC) encompasses four genomospecies: A. baumannii, A. nosocomialis, A. calcoaceticus, and A. pittii (169). A. baumannii and A. nosocomialis contribute to the majority of ABC bacteremia and predict similar mortality rates (169); therefore, further study of A. nosocomialis bacteremia is warranted.
Enterobacter cloacae is actually a complex of closely related species that is connected with health care-associated bacteremia (18, 170) and underlies 2.9% of Gram-negative bacteremia, making it the fourth most common pathogen in these infections. Like many of the other species described in this section, E. cloacae can be isolated from both human and environmental sources, with human colonization correlated with underlying illness and antibiotic treatment (171). The most common sources of Enterobacter bacteremia in one multicenter study of >250 cases were vascular catheters, the gastrointestinal tract, and the urinary tract (172). This complex often has extensive ESBL production and may deploy resistance to heavy metals (13). ESBL-producing E. cloacae strains have been reported in bacteremia, including strains carrying blaSHV and blaCTX. The carriage rate of chromosomal ampC genes among Enterobacter bloodstream isolates is also very high (151, 152). Factors associated with E. cloacae bacteremia fitness have yet to be defined and must be investigated. It is important to consider both individual species and species complexes when defining virulence factors, as mechanisms may be conserved across similar species. Identifying unique characteristics making one species more virulent than a member of the same complex could indicate promising targets for future therapies.
Antimicrobial resistance in Gram-negative bacteremia is especially alarming (131). In 2013 and 2019 reports, the CDC classified carbapenem-resistant Enterobacteriaceae (CRE) as an urgent threat (22, 23) and emphasized waning options for antibiotics that treat Gram-negative infections. Multidrug-resistant infections, defined infections caused by organisms with nonsusceptibility to one agent in more than three antimicrobial categories (173), in Enterobacteriaceae increased by 10% between 1997 and 2016, highlighting the need to further investigate bacteremia by these species (18). Gram-negative antimicrobial resistance, aside from that among members of the Enterobacteriaceae, also threatens public health. A. baumannii and P. aeruginosa can harbor carbapenemases and transfer these enzymes to other species through mobile genetic elements (23, 35). Additionally, carriage of carbapenemases in species including S. marcescens, C. freundii, and E. cloacae indicates widespread mechanisms of resistance that must be urgently addressed.
CONCLUSION
The pathogenesis of Gram-negative bacteremia requires establishment of initial infection or colonization, dissemination, and bloodstream survival. Fitness at initial sites has been widely studied, but it does not always indicate factors needed to transition to bacteremia. Dissemination factors may involve adherence mechanisms or, in the case of some species, toxins which disrupt epithelial barriers. Persistence in the blood is largely linked to metabolic diversity and capsule production to evade immune responses. Among the Gram-negative bacteremia species that have been interrogated using unbiased and genome-wide approaches to fitness factor identification in the mammalian bloodstream, there has been a predominance of mechanisms that support evasion of immune defenses and promote metabolic diversity, some of which are shared across multiple species. Individual species may use unique factors for colonization and dissemination but a common set of genes for bloodstream survival. Characterizing these common factors may facilitate future therapeutic strategies that target the physiologic requirements for bacterial fitness in the bloodstream environment to disrupt the progression of bacteremia across multiple species. Importantly, antimicrobial resistance has brought new urgency to understanding the steps of bacteremia, since the absence of effective therapy significantly increases the risk of death.
ACKNOWLEDGMENTS
C.L.H. is supported by the Lung Immunopathology Training Grant (T32HL007517), M.T.A. is supported by AI148767, and M.T.A., M.A.B., and H.L.T.M. are supported by AI134731 from the National Institutes of Health.
Biographies

Caitlyn L. Holmes, Ph.D., is a postdoctoral fellow in the Department Microbiology and Immunology and the Department of Pathology at the University of Michigan Medical School. She received her doctoral degree in Cellular and Molecular Pathology from the University of Wisconsin–Madison in 2019. Her past work has focused on innate immunity and cell death via neutrophil extracellular trap release. In 2020, Dr. Holmes began her current work in the Bachman laboratory investigating Klebsiella pneumoniae pathogenesis during pneumonia and bloodstream infection.

Mark T. Anderson, Ph.D., is a Research Assistant Professor in the Department of Microbiology and Immunology at the University of Michigan Medical School. He received his doctoral degree in Microbiology, Immunology, and Cancer Biology from the University of Minnesota Medical School and completed a postdoctoral fellowship at Northwestern University Medical School. Dr. Anderson has studied the physiology and pathogenesis of multiple Gram-negative bacterial species throughout his career. In 2014, he began his current research investigating bacterial strategies for survival and replication in the mammalian bloodstream environment, focusing primarily on two opportunistic pathogens, Serratia marcescens and Citrobacter freundii.

Harry L. T. Mobley, Ph.D., is a professor in the Department of Microbiology and Immunology at the University of Michigan Medical School. Dr. Mobley received his Ph.D. at the University of Louisville School of Medicine and conducted postdoctoral work at the University of Maryland School of Medicine in biochemistry and vaccine development, after which he joined the faculty in the Division of Infectious Diseases and rose to the rank of Professor of Microbiology and Immunology. After 23 years at University of Maryland, Dr. Mobley accepted an appointment as the Frederick G. Novy Professor and Chair of the Department of Microbiology and Immunology at the University of Michigan in 2004, serving in this role until 2019. Dr. Mobley’s research focuses on uropathogenic E. coli and Proteus mirabilis, but he has also studied Helicobacter pylori and now Gram-negative bacterial species causing bacteremia, including E. coli, Klebsiella pneumoniae, Acinetobacter baumannii, Citrobacter freundii, and Serratia marcescens.

Michael A. Bachman, M.D. Ph.D., is an associate professor in the Departments of Pathology and Microbiology and Immunology at the University of Michigan Medical School (Michigan Medicine) and Associate Director of the Clinical Microbiology Laboratory. As part of the Medical Scientist Training Program at the University of Michigan, he received his M.D. and his Ph.D. in Microbiology and Immunology in 2004. He completed his clinical pathology residency and clinical microbiology fellowship at the Hospital of the University of Pennsylvania and his postdoctoral fellowship in the Department of Microbiology at the Perelman School of Medicine, University of Pennsylvania. During this time, he began to study Klebsiella as a leading and increasingly antibiotic-resistant cause of health care-associated infections. In 2011, he returned to the University of Michigan as an assistant professor. His research is focused on risk factors for Klebsiella infections and the pathogenesis of Gram-negative bacteremia.
REFERENCES
- 1.Hall MJ, Williams SN, DeFrances CJ, Golosinskiy A. 2011. Inpatient care for septicemia or sepsis: a challenge for patients and hospitals. NCHS Data Brief 2011:1–8. [PubMed] [Google Scholar]
- 2.Holland TL, Baddour LM, Bayer AS, Hoen B, Miro JM, Fowler VG. 2016. Infective endocarditis. Nat Rev Dis Primers 2:16059. 10.1038/nrdp.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kavanagh N, Ryan EJ, Widaa A, Sexton G, Fennell J, O'Rourke S, Cahill KC, Kearney CJ, O'Brien FJ, Kerrigan SW. 2018. Staphylococcal osteomyelitis: disease progression, treatment challenges, and future directions. Clin Microbiol Rev 31:e00084-17. 10.1128/CMR.00084-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torio CM, Moore BJ. 2016. National inpatient hospital costs: the most expensive conditions by payer, 2013. Healthcare Cost and Utilization Project (HCUP) Statistical Briefs no. 204. [PubMed]
- 5.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC. 2016. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 315:801–810. 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC. 2020. Sepsis data and reports: clinical information. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 7.Fleischmann C, Scherag A, Adhikari NK, Hartog CS, Tsaganos T, Schlattmann P, Angus DC, Reinhart K, Trialists I, International Forum of Acute Care Trialists. 2016. Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am J Respir Crit Care Med 193:259–272. 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- 8.Reinhart K, Daniels R, Kissoon N, Machado FR, Schachter RD, Finfer S. 2017. Recognizing sepsis as a global health priority—a WHO resolution. N Engl J Med 377:414–417. 10.1056/NEJMp1707170. [DOI] [PubMed] [Google Scholar]
- 9.Iwashyna TJ, Ely EW, Smith DM, Langa KM. 2010. Long-term cognitive impairment and functional disability among survivors of severe sepsis. JAMA 304:1787–1794. 10.1001/jama.2010.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin GS, Mannino DM, Eaton S, Moss M. 2003. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med 348:1546–1554. 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 11.Hajj J, Blaine N, Salavaci J, Jacoby D. 2018. The “centrality of sepsis”: a review on incidence, mortality, and cost of care. Healthcare (Basel) 6:90. 10.3390/healthcare6030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 13.Bennet J, Dolin R, Blaser M. 2020. Mandell, Douglas, and Bennett's principles and practice of Infectious Diseases, 9th ed. Elsevier, Philadelphia, PA. [Google Scholar]
- 14.Seifert H. 2009. The clinical importance of microbiological findings in the diagnosis and management of bloodstream infections. Clin Infect Dis 48(Suppl 4):S238–S245. 10.1086/598188. [DOI] [PubMed] [Google Scholar]
- 15.Riu M, Chiarello P, Terradas R, Sala M, Garcia-Alzorriz E, Castells X, Grau S, Cots F. 2016. Cost attributable to nosocomial bacteremia. Analysis according to microorganism and antimicrobial sensitivity in a university hospital in Barcelona. PLoS One 11:e0153076. 10.1371/journal.pone.0153076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodríguez-Créixems M, Alcalá L, Muñoz P, Cercenado E, Vicente T, Bouza E. 2008. Bloodstream infections: evolution and trends in the microbiology workload, incidence, and etiology, 1985-2006. Medicine (Baltimore) 87:234–249. 10.1097/MD.0b013e318182119b. [DOI] [PubMed] [Google Scholar]
- 17.Diekema DJ, Beekmann SE, Chapin KC, Morel KA, Munson E, Doern GV. 2003. Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J Clin Microbiol 41:3655–3660. 10.1128/JCM.41.8.3655-3660.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diekema DJ, Hsueh PR, Mendes RE, Pfaller MA, Rolston KV, Sader HS, Jones RN. 2019. The microbiology of bloodstream infection: 20-year trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother 63:e00355-19. 10.1128/AAC.00355-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albrecht SJ, Fishman NO, Kitchen J, Nachamkin I, Bilker WB, Hoegg C, Samel C, Barbagallo S, Arentzen J, Lautenbach E. 2006. Reemergence of gram-negative health care-associated bloodstream infections. Arch Intern Med 166:1289–1294. 10.1001/archinte.166.12.1289. [DOI] [PubMed] [Google Scholar]
- 20.Marra AR, Camargo LF, Pignatari AC, Sukiennik T, Behar PR, Medeiros EA, Ribeiro J, Girão E, Correa L, Guerra C, Brites C, Pereira CA, Carneiro I, Reis M, de Souza MA, Tranchesi R, Barata CU, Edmond MB, Group BSS, Brazilian SCOPE Study Group. 2011. Nosocomial bloodstream infections in Brazilian hospitals: analysis of 2,563 cases from a prospective nationwide surveillance study. J Clin Microbiol 49:1866–1871. 10.1128/JCM.00376-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Neill J. 2016. Tackling drug-resistant infections globally: final report and recommendations. Review on Antimicrobial Resistance, London, United Kingdom. [Google Scholar]
- 22.CDC. 2013. Antibiotic resistance threats in the United State, 2013. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 23.CDC. 2019. Antibiotic resistance threats in the United States, 2019. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 24.WHO. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 25.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Team Emerging Infection Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoelle J, Johnson JR, Johnston BD, Kinkle B, Boczek L, Ryu H, Hayes S. 2019. Survey of US wastewater for carbapenem-resistant Enterobacteriaceae. J Water Health 17:219–226. 10.2166/wh.2019.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson JR, Stell AL, Delavari P. 2001. Canine feces as a reservoir of extraintestinal pathogenic Escherichia coli. Infect Immun 69:1306–1314. 10.1128/IAI.69.3.1306-1314.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith AR, Vowles M, Horth RZ, Smith L, Rider L, Wagner JM, Sangster A, Young EL, Schuckel H, Stewart J, Gruninger RJ, Rossi A, Oakeson KF, Nakashima AK. 2020. Infection control response to an outbreak of OXA-23 carbapenemase-producing carbapenem-resistant Acinetobacter baumannii in a skilled nursing facility in Utah. Am J Infect Control Epub ahead of print. 10.1016/j.ajic.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SH, Wei CI, Tzou YM, An H. 2005. Multidrug-resistant Klebsiella pneumoniae isolated from farm environments and retail products in Oklahoma. J Food Prot 68:2022–2029. 10.4315/0362-028x-68.10.2022. [DOI] [PubMed] [Google Scholar]
- 30.Manges AR, Johnson JR. 2015. Reservoirs of extraintestinal pathogenic Escherichia coli. Microbiol Spectr 3:UTI-0006-2012. 10.1128/microbiolspec.UTI-0006-2012. [DOI] [PubMed] [Google Scholar]
- 31.Gorrie CL, Mirceta M, Wick RR, Edwards DJ, Thomson NR, Strugnell RA, Pratt NF, Garlick JS, Watson KM, Pilcher DV, McGloughlin SA, Spelman DW, Jenney AWJ, Holt KE. 2017. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis 65:208–215. 10.1093/cid/cix270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin RM, Cao J, Brisse S, Passet V, Wu W, Zhao L, Malani PN, Rao K, Bachman MA. 2016. Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere 1:e00261-16. 10.1128/mSphere.00261-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canzoneri CN, Akhavan BJ, Tosur Z, Andrade PEA, Aisenberg GM. 2017. Follow-up blood cultures in Gram-negative bacteremia: are they needed? Clin Infect Dis 65:1776–1779. 10.1093/cid/cix648. [DOI] [PubMed] [Google Scholar]
- 34.Sligl W, Taylor G, Brindley PG. 2006. Five years of nosocomial Gram-negative bacteremia in a general intensive care unit: epidemiology, antimicrobial susceptibility patterns, and outcomes. Int J Infect Dis 10:320–325. 10.1016/j.ijid.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Sligl WI, Dragan T, Smith SW. 2015. Nosocomial Gram-negative bacteremia in intensive care: epidemiology, antimicrobial susceptibilities, and outcomes. Int J Infect Dis 37:129–134. 10.1016/j.ijid.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 36.Weinstein RA, Gaynes R, Edwards JR, National Nosocomial Infections Surveillance System. 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis 41:848–854. 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 37.Russell DL, Uslan DZ, Rubin ZA, Grogan TR, Martin EM. 2018. Multidrug resistant Acinetobacter baumanii: a 15-year trend analysis. Infect Control Hosp Epidemiol 39:608–611. 10.1017/ice.2018.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korte AKM, Vos JM. 2017. Ecthyma gangrenosum. N Engl J Med 377:e32. 10.1056/NEJMicm1702302. [DOI] [PubMed] [Google Scholar]
- 39.Jorgensen JH, Pfaller MA (ed). 2015. Manual of clinical microbiology, 11th ed. ASM Press, Washington, DC. [Google Scholar]
- 40.Wain J, Diep TS, Ho VA, Walsh AM, Nguyen TT, Parry CM, White NJ. 1998. Quantitation of bacteria in blood of typhoid fever patients and relationship between counts and clinical features, transmissibility, and antibiotic resistance. J Clin Microbiol 36:1683–1687. 10.1128/JCM.36.6.1683-1687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henry NK, McLimans CA, Wright AJ, Thompson RL, Wilson WR, Washington JA. 1983. Microbiological and clinical evaluation of the isolator lysis-centrifugation blood culture tube. J Clin Microbiol 17:864–869. 10.1128/JCM.17.5.864-869.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson MT, Mitchell LA, Zhao L, Mobley HLT. 2017. Capsule production and glucose metabolism dictate fitness during Serratia marcescens bacteremia. mBio 8:e00740-17. 10.1128/mBio.00740-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson MT, Mitchell LA, Zhao L, Mobley HLT. 2018. Citrobacter freundii fitness during bloodstream infection. Sci Rep 8:11792. 10.1038/s41598-018-30196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armbruster CE, Forsyth VS, Johnson AO, Smith SN, White AN, Brauer AL, Learman BS, Zhao L, Wu W, Anderson MT, Bachman MA, Mobley HLT. 2019. Twin arginine translocation, ammonia incorporation, and polyamine biosynthesis are crucial for Proteus mirabilis fitness during bloodstream infection. PLoS Pathog 15:e1007653. 10.1371/journal.ppat.1007653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subashchandrabose S, Smith S, DeOrnellas V, Crepin S, Kole M, Zahdeh C, Mobley HL. 2016. Acinetobacter baumannii genes required for bacterial survival during bloodstream infection. mSphere 1:e00013-15. 10.1128/mSphere.00013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawlor MS, Hsu J, Rick PD, Miller VL. 2005. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol 58:1054–1073. 10.1111/j.1365-2958.2005.04918.x. [DOI] [PubMed] [Google Scholar]
- 47.Bachman MA, Miller VL, Weiser JN. 2009. Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog 5:e1000622. 10.1371/journal.ppat.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Twentyman J, Morffy Smith C, Nims JS, Dahler AA, Rosen DA. 2020. A murine model demonstrates capsule-independent adaptive immune protection in survivors of Klebsiella pneumoniae repiratory tract infection. Dis Model Mech 13:dmm043240. 10.1242/dmm.043240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith SN, Hagan EC, Lane MC, Mobley HL. 2010. Dissemination and systemic colonization of uropathogenic Escherichia coli in a murine model of bacteremia. mBio 1:e00262-10. 10.1128/mBio.00262-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subashchandrabose S, Smith SN, Spurbeck RR, Kole MM, Mobley HLT. 2013. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog 9:e1003788. 10.1371/journal.ppat.1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crépin S, Ottosen EN, Peters K, Smith SN, Himpsl SD, Vollmer W, Mobley HLT. 2018. The lytic transglycosylase MltB connects membrane homeostasis and in vivo fitness of Acinetobacter baumannii. Mol Microbiol 109:745–762. 10.1111/mmi.14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weakland DR, Smith SN, Bell B, Tripathi A, Mobley HLT. 2020. The Serratia marcescens siderophore serratiochelin is necessary for full virulence during bloodstream infection. Infect Immun 88:e00117-20. 10.1128/IAI.00117-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vornhagen J, Sun Y, Breen P, Forsyth V, Zhao L, Mobley HLT, Bachman MA. 2019. The Klebsiella pneumoniae citrate synthase gene, gltA, influences site specific fitness during infection. PLoS Pathog 15:e1008010. 10.1371/journal.ppat.1008010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szemiako K, Krawczyk B, Samet A, Śledzińska A, Nowicki B, Nowicki S, Kur J. 2013. A subset of two adherence systems, acute pro-inflammatory pap genes and invasion coding dra, fim, or sfa, increases the risk of Escherichia coli translocation to the bloodstream. Eur J Clin Microbiol Infect Dis 32:1579–1582. 10.1007/s10096-013-1913-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouillot S, Munro P, Gallet B, Reboud E, Cretin F, Golovkine G, Schoehn G, Attrée I, Lemichez E, Huber P. 2017. Pseudomonas aeruginosa Exolysin promotes bacterial growth in lungs, alveolar damage and bacterial dissemination. Sci Rep 7:2120. 10.1038/s41598-017-02349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tu YC, Lu MC, Chiang MK, Huang SP, Peng HL, Chang HY, Jan MS, Lai YC. 2009. Genetic requirements for Klebsiella pneumoniae-induced liver abscess in an oral infection model. Infect Immun 77:2657–2671. 10.1128/IAI.01523-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bachman MA, Breen P, Deornellas V, Mu Q, Zhao L, Wu W, Cavalcoli JD, Mobley HL. 2015. Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. mBio 6:e00775. 10.1128/mBio.00775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Short FL, Di Sario G, Reichmann NT, Kleanthous C, Parkhill J, Taylor PW. 2020. Genomic profiling reveals distinct routes to complement resistance in Klebsiella pneumoniae. Infect Immun 88:e00043-20. 10.1128/IAI.00043-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mora-Rillo M, Fernández-Romero N, Navarro-San Francisco C, Díez-Sebastián J, Romero-Gómez MP, Fernández FA, López JRA, Mingorance J. 2015. Impact of virulence genes on sepsis severity and survival in Escherichia coli bacteremia. Virulence 6:93–100. 10.4161/21505594.2014.991234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Samet A, Śledzińska A, Krawczyk B, Hellmann A, Nowicki S, Kur J, Nowicki B. 2013. Leukemia and risk of recurrent Escherichia coli bacteremia: genotyping implicates E. coli translocation from the colon to the bloodstream. Eur J Clin Microbiol Infect Dis 32:1393–1400. 10.1007/s10096-013-1886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laupland KB, Gregson DB, Church DL, Ross T, Pitout JD. 2008. Incidence, risk factors and outcomes of Escherichia coli bloodstream infections in a large Canadian region. Clin Microbiol Infect 14:1041–1047. 10.1111/j.1469-0691.2008.02089.x. [DOI] [PubMed] [Google Scholar]
- 62.Gibson JS, Cobbold RN, Trott DJ. 2010. Characterization of multidrug-resistant Escherichia coli isolated from extraintestinal clinical infections in animals. J Med Microbiol 59:592–598. 10.1099/jmm.0.018002-0. [DOI] [PubMed] [Google Scholar]
- 63.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 67:546–553. 10.1128/IAI.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krawczyk B, Śledzińska A, Szemiako K, Samet A, Nowicki B, Kur J. 2015. Characterisation of Escherichia coli isolates from the blood of haematological adult patients with bacteraemia: translocation from gut to blood requires the cooperation of multiple virulence factors. Eur J Clin Microbiol Infect Dis 34:1135–1143. 10.1007/s10096-015-2331-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodríguez-Baño J, Mingorance J, Fernández-Romero N, Serrano L, López-Cerero L, Pascual A, ESBL-REIPI Group. 2012. Virulence profiles of bacteremic extended-spectrum β-lactamase-producing Escherichia coli: association with epidemiological and clinical features. PLoS One 7:e44238. 10.1371/journal.pone.0044238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272. 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 67.Ciesielczuk H, Doumith M, Hope R, Woodford N, Wareham DW. 2015. Characterization of the extra-intestinal pathogenic Escherichia coli ST131 clone among isolates recovered from urinary and bloodstream infections in the United Kingdom. J Med Microbiol 64:1496–1503. 10.1099/jmm.0.000179. [DOI] [PubMed] [Google Scholar]
- 68.Cha MK, Kang CI, Kim SH, Cho SY, Ha YE, Wi YM, Chung DR, Peck KR, Song JH, Korean Network for Study on Infectious Diseases. 2016. Comparison of the microbiological characteristics and virulence factors of ST131 and non-ST131 clones among extended-spectrum β-lactamase-producing Escherichia coli causing bacteremia. Diagn Microbiol Infect Dis 84:102–104. 10.1016/j.diagmicrobio.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 69.Al-Hasan MN, Eckel-Passow JE, Baddour LM. 2010. Bacteremia complicating gram-negative urinary tract infections: a population-based study. J Infect 60:278–285. 10.1016/j.jinf.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buckles EL, Luterbach CL, Wang X, Lockatell CV, Johnson DE, Mobley HL, Donnenberg MS. 2015. Signature-tagged mutagenesis and co-infection studies demonstrate the importance of P fimbriae in a murine model of urinary tract infection. Pathog Dis 73:ftv014. 10.1093/femspd/ftv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holden VI, Bachman MA. 2015. Diverging roles of bacterial siderophores during infection. Metallomics 7:986–995. 10.1039/c4mt00333k. [DOI] [PubMed] [Google Scholar]
- 72.Brzuszkiewicz E, Brüggemann H, Liesegang H, Emmerth M, Olschläger T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emody L, Gottschalk G, Hacker J, Dobrindt U. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci U S A 103:12879–12884. 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meatherall BL, Gregson D, Ross T, Pitout JD, Laupland KB. 2009. Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am J Med 122:866–873. 10.1016/j.amjmed.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 74.Kang CI, Kim SH, Bang JW, Kim HB, Kim NJ, Kim EC, Oh MD, Choe KW. 2006. Community-acquired versus nosocomial Klebsiella pneumoniae bacteremia: clinical features, treatment outcomes, and clinical implication of antimicrobial resistance. J Korean Med Sci 21:816–822. 10.3346/jkms.2006.21.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Podschun R, Pietsch S, Höller C, Ullmann U. 2001. Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl Environ Microbiol 67:3325–3327. 10.1128/AEM.67.7.3325-3327.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davis GS, Waits K, Nordstrom L, Weaver B, Aziz M, Gauld L, Grande H, Bigler R, Horwinski J, Porter S, Stegger M, Johnson JR, Liu CM, Price LB. 2015. Intermingled Klebsiella pneumoniae populations between retail meats and human urinary tract infections. Clin Infect Dis 61:892–899. 10.1093/cid/civ428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Babini GS, Livermore DM. 2000. Are SHV beta-lactamases universal in Klebsiella pneumoniae? Antimicrob Agents Chemother 44:2230. 10.1128/aac.44.8.2230-2230.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard AS, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine MH, Decré D, Brisse S. 2014. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gomez-Simmonds A, Greenman M, Sullivan SB, Tanner JP, Sowash MG, Whittier S, Uhlemann AC. 2015. Population structure of Klebsiella pneumoniae causing bloodstream infections at a New York City tertiary care hospital: diversification of multidrug-resistant isolates. J Clin Microbiol 53:2060–2067. 10.1128/JCM.03455-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Girometti N, Lewis RE, Giannella M, Ambretti S, Bartoletti M, Tedeschi S, Tumietto F, Cristini F, Trapani F, Gaibani P, Viale P. 2014. Klebsiella pneumoniae bloodstream infection: epidemiology and impact of inappropriate empirical therapy. Medicine (Baltimore) 93:298–309. 10.1097/MD.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wyres KL, Nguyen TNT, Lam MMC, Judd LM, van Vinh Chau N, Dance DAB, Ip M, Karkey A, Ling CL, Miliya T, Newton PN, Lan NPH, Sengduangphachanh A, Turner P, Veeraraghavan B, Vinh PV, Vongsouvath M, Thomson NR, Baker S, Holt KE. 2020. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med 12:11. 10.1186/s13073-019-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paczosa MK, Mecsas J. 2016. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shon AS, Bajwa RP, Russo TA. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4:107–118. 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Russo TA, Marr CM. 2019. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 32:e00001-19. 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paterson DL, Ko WC, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, Bonomo RA, Rice LB, Wagener MM, McCormack JG, Yu VL. 2004. International prospective study of Klebsiella pneumoniae bacteremia: implications of extended-spectrum beta-lactamase production in nosocomial Infections. Ann Intern Med 140:26–32. 10.7326/0003-4819-140-1-200401060-00008. [DOI] [PubMed] [Google Scholar]
- 86.Xiao T, Zhu Y, Zhang S, Wang Y, Shen P, Zhou Y, Yu X, Xiao Y. 2020. A retrospective analysis of risk factors and outcomes of carbapenem-resistant Klebsiella pneumoniae bacteremia in nontransplant patients. J Infect Dis 221:S174–S183. 10.1093/infdis/jiz559. [DOI] [PubMed] [Google Scholar]
- 87.Xu M, Fu Y, Kong H, Chen X, Chen Y, Li L, Yang Q. 2018. Bloodstream infections caused by Klebsiella pneumoniae: prevalence of blaKPC, virulence factors and their impacts on clinical outcome. BMC Infect Dis 18:358. 10.1186/s12879-018-3263-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zarkotou O, Pournaras S, Tselioti P, Dragoumanos V, Pitiriga V, Ranellou K, Prekates A, Themeli-Digalaki K, Tsakris A. 2011. Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin Microbiol Infect 17:1798–1803. 10.1111/j.1469-0691.2011.03514.x. [DOI] [PubMed] [Google Scholar]
- 89.Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, Lee CC, Fung CP, Chuang YC. 2006. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis 42:1351–1358. 10.1086/503420. [DOI] [PubMed] [Google Scholar]
- 90.Qureshi ZA, Paterson DL, Potoski BA, Kilayko MC, Sandovsky G, Sordillo E, Polsky B, Adams-Haduch JM, Doi Y. 2012. Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob Agents Chemother 56:2108–2113. 10.1128/AAC.06268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee HC, Chuang YC, Yu WL, Lee NY, Chang CM, Ko NY, Wang LR, Ko WC. 2006. Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: association with invasive syndrome in patients with community-acquired bacteraemia. J Intern Med 259:606–614. 10.1111/j.1365-2796.2006.01641.x. [DOI] [PubMed] [Google Scholar]
- 92.Togawa A, Yoshimura M, Tokushige C, Matsunaga A, Takata T, Takamatsu Y. 2020. Development of risk factor-based scoring system for detection of hypervirulent Klebsiella pneumoniae bloodstream infections. Gut Pathog 12:34. 10.1186/s13099-020-00374-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holden VI, Breen P, Houle S, Dozois CM, Bachman MA. 2016. Klebsiella pneumoniae siderophores induce inflammation, bacterial dissemination, and HIF-1α stabilization during pneumonia. mBio 7:e01397-16. 10.1128/mBio.01397-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Broug-Holub E, Toews GB, van Iwaarden JF, Strieter RM, Kunkel SL, Paine R, Standiford TJ. 1997. Alveolar macrophages are required for protective pulmonary defenses in murine Klebsiella pneumonia: elimination of alveolar macrophages increases neutrophil recruitment but decreases bacterial clearance and survival. Infect Immun 65:1139–1146. 10.1128/IAI.65.4.1139-1146.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bachman MA, Oyler JE, Burns SH, Caza M, Lépine F, Dozois CM, Weiser JN. 2011. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun 79:3309–3316. 10.1128/IAI.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bachman MA, Lenio S, Schmidt L, Oyler JE, Weiser JN. 2012. Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. mBio 3:e00224-11. 10.1128/mBio.00224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao Y, Olonisakin TF, Xiong Z, Hulver M, Sayeed S, Yu MT, Gregory AD, Kochman EJ, Chen BB, Mallampalli RK, Sun M, Silverstein RL, Stolz DB, Shapiro SD, Ray A, Ray P, Lee JS. 2015. Thrombospondin-1 restrains neutrophil granule serine protease function and regulates the innate immune response during Klebsiella pneumoniae infection. Mucosal Immunol 8:896–905. 10.1038/mi.2014.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Codo AC, Saraiva AC, Dos Santos LL, Visconde MF, Gales AC, Zamboni DS, Medeiros AI. 2018. Inhibition of inflammasome activation by a clinical strain of Klebsiella pneumoniae impairs efferocytosis and leads to bacterial dissemination. Cell Death Dis 9:1182. 10.1038/s41419-018-1214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hsieh PF, Lin TL, Lee CZ, Tsai SF, Wang JT. 2008. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 197:1717–1727. 10.1086/588383. [DOI] [PubMed] [Google Scholar]
- 100.Fung CP, Chang FY, Lin JC, Ho DM, Chen CT, Chen JH, Yeh KM, Chen TL, Lin YT, Siu LK. 2011. Immune response and pathophysiological features of Klebsiella pneumoniae liver abscesses in an animal model. Lab Invest 91:1029–1039. 10.1038/labinvest.2011.52. [DOI] [PubMed] [Google Scholar]
- 101.Ehrenworth L, Baer H. 1956. The pathogenicity of Klebsiella pneumoniae for mice: the relationship to the quantity and rate of production of type-specific capsular polysaccharide. J Bacteriol 72:713–717. 10.1128/JB.72.5.713-717.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weber BS, De Jong AM, Guo ABY, Dharavath S, French S, Fiebig-Comyn AA, Coombes BK, Magolan J, Brown ED. 2020. Genetic and chemical screening in human blood serum reveals unique antibacterial targets and compounds against Klebsiella pneumoniae. Cell Rep 32:107927. 10.1016/j.celrep.2020.107927. [DOI] [PubMed] [Google Scholar]
- 103.Chen JH, Siu LK, Fung CP, Lin JC, Yeh KM, Chen TL, Tsai YK, Chang FY. 2010. Contribution of outer membrane protein K36 to antimicrobial resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 65:986–990. 10.1093/jac/dkq056. [DOI] [PubMed] [Google Scholar]
- 104.Fang CT, Yi WC, Shun CT, Tsai SF. 2017. DNA adenine methylation modulates pathogenicity of Klebsiella pneumoniae genotype K1. J Microbiol Immunol Infect 50:471–477. 10.1016/j.jmii.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 105.Tam VH, Rogers CA, Chang KT, Weston JS, Caeiro JP, Garey KW. 2010. Impact of multidrug-resistant Pseudomonas aeruginosa bacteremia on patient outcomes. Antimicrob Agents Chemother 54:3717–3722. 10.1128/AAC.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.El-Solh AA, Hattemer A, Hauser AR, Alhajhusain A, Vora H. 2012. Clinical outcomes of type III Pseudomonas aeruginosa bacteremia. Crit Care Med 40:1157–1163. 10.1097/CCM.0b013e3182377906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rossi Gonçalves I, Dantas RCC, Ferreira ML, Batistão DWDF, Gontijo-Filho PP, Ribas RM. 2017. Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation. Braz J Microbiol 48:211–217. 10.1016/j.bjm.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Recio R, Villa J, Viedma E, Orellana M, Lora-Tamayo J, Chaves F. 2018. Bacteraemia due to extensively drug-resistant Pseudomonas aeruginosa sequence type 235 high-risk clone: facing the perfect storm. Int J Antimicrob Agents 52:172–179. 10.1016/j.ijantimicag.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 109.Langan KM, Kotsimbos T, Peleg AY. 2015. Managing Pseudomonas aeruginosa respiratory infections in cystic fibrosis. Curr Opin Infect Dis 28:547–556. 10.1097/QCO.0000000000000217. [DOI] [PubMed] [Google Scholar]
- 110.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rutherford V, Yom K, Ozer EA, Pura O, Hughes A, Murphy KR, Cudzilo L, Mitchell D, Hauser AR. 2018. Environmental reservoirs for exoS+ and exoU+ strains of Pseudomonas aeruginosa. Environ Microbiol Rep 10:485–492. 10.1111/1758-2229.12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bertrand X, Thouverez M, Talon D, Boillot A, Capellier G, Floriot C, Hélias JP. 2001. Endemicity, molecular diversity and colonisation routes of Pseudomonas aeruginosa in intensive care units. Intensive Care Med 27:1263–1268. 10.1007/s001340100979. [DOI] [PubMed] [Google Scholar]
- 113.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Poirel L, Nordmann P, Lagrutta E, Cleary T, Munoz-Price LS. 2010. Emergence of KPC-producing Pseudomonas aeruginosa in the United States. Antimicrob Agents Chemother 54:3072. 10.1128/AAC.00513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McCarthy KL, Wailan AM, Jennison AV, Kidd TJ, Paterson DL. 2018. P. aeruginosa blood stream infection isolates: a “full house” of virulence genes in isolates associated with rapid patient death and patient survival. Microb Pathog 119:81–85. 10.1016/j.micpath.2018.03.062. [DOI] [PubMed] [Google Scholar]
- 116.Yin S, Chen P, You B, Zhang Y, Jiang B, Huang G, Yang Z, Chen Y, Chen J, Yuan Z, Zhao Y, Li M, Hu F, Gong Y, Peng Y. 2018. Molecular typing and carbapenem resistance mechanisms of Pseudomonas aeruginosa isolated from a Chinese burn center from 2011 to 2016. Front Microbiol 9:1135. 10.3389/fmicb.2018.01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Micek ST, Lloyd AE, Ritchie DJ, Reichley RM, Fraser VJ, Kollef MH. 2005. Pseudomonas aeruginosa bloodstream infection: importance of appropriate initial antimicrobial treatment. Antimicrob Agents Chemother 49:1306–1311. 10.1128/AAC.49.4.1306-1311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hickey C, Schaible B, Nguyen S, Hurley D, Srikumar S, Fanning S, Brown E, Crifo B, Matallanas D, McClean S, Taylor CT, Schaffer K. 2018. Increased virulence of bloodstream over peripheral isolates of P. aeruginosa identified through post-transcriptional regulation of virulence factors. Front Cell Infect Microbiol 8:357. 10.3389/fcimb.2018.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Berube BJ, Rangel SM, Hauser AR. 2016. Pseudomonas aeruginosa: breaking down barriers. Curr Genet 62:109–113. 10.1007/s00294-015-0522-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shaver CM, Hauser AR. 2004. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun 72:6969–6977. 10.1128/IAI.72.12.6969-6977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Garrity-Ryan L, Kazmierczak B, Kowal R, Comolli J, Hauser A, Engel JN. 2000. The arginine finger domain of ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect Immun 68:7100–7113. 10.1128/iai.68.12.7100-7113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rangel SM, Diaz MH, Knoten CA, Zhang A, Hauser AR. 2015. The role of ExoS in dissemination of Pseudomonas aeruginosa during pneumonia. PLoS Pathog 11:e1004945. 10.1371/journal.ppat.1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Golovkine G, Faudry E, Bouillot S, Voulhoux R, Attrée I, Huber P. 2014. VE-cadherin cleavage by LasB protease from Pseudomonas aeruginosa facilitates type III secretion system toxicity in endothelial cells. PLoS Pathog 10:e1003939. 10.1371/journal.ppat.1003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Holder IA, Neely AN, Frank DW. 2001. Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns 27:129–130. 10.1016/s0305-4179(00)00142-x. [DOI] [PubMed] [Google Scholar]
- 125.Martín-Aspas A, Guerrero-Sánchez FM, García-Colchero F, Rodríguez-Roca S, Girón-González JA. 2018. Differential characteristics of Acinetobacter baumannii colonization and infection: risk factors, clinical picture, and mortality. Infect Drug Resist 11:861–872. 10.2147/IDR.S163944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee NY, Chang TC, Wu CJ, Chang CM, Lee HC, Chen PL, Lee CC, Ko NY, Ko WC. 2010. Clinical manifestations, antimicrobial therapy, and prognostic factors of monomicrobial Acinetobacter baumannii complex bacteremia. J Infect 61:219–227. 10.1016/j.jinf.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 127.Shenoy ES, Pierce VM, Sater MRA, Pangestu FK, Herriott IC, Anahtar MN, Bramante JT, Kwon DS, Hawkins FR, Suslak D, West LR, Huntley MH, Hooper DC. 2020. Community-acquired in name only: a cluster of carbapenem-resistant Acinetobacter baumannii in a burn intensive care unit and beyond. Infect Control Hosp Epidemiol 41:531–538. 10.1017/ice.2020.15. [DOI] [PubMed] [Google Scholar]
- 128.Liou ML, Chen KH, Yeh HL, Lai CY, Chen CH. 2017. Persistent nasal carriers of Acinetobacter baumannii in long-term-care facilities. Am J Infect Control 45:723–727. 10.1016/j.ajic.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 129.Byrne-Bailey KG, Gaze WH, Kay P, Boxall AB, Hawkey PM, Wellington EM. 2009. Prevalence of sulfonamide resistance genes in bacterial isolates from manured agricultural soils and pig slurry in the United Kingdom. Antimicrob Agents Chemother 53:696–702. 10.1128/AAC.00652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sarma PM, Bhattacharya D, Krishnan S, Lal B. 2004. Assessment of intra-species diversity among strains of Acinetobacter baumannii isolated from sites contaminated with petroleum hydrocarbons. Can J Microbiol 50:405–414. 10.1139/w04-018. [DOI] [PubMed] [Google Scholar]
- 131.De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, Paterson DL, Walker MJ. 2020. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev 33:e00181-19. 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Thom KA, Maragakis LL, Richards K, Johnson JK, Roup B, Lawson P, Harris AD, Fuss EP, Pass MA, Blythe D, Perencevich EN, Wilson L, Maryland MDRO Prevention Collaborative. 2012. Assessing the burden of Acinetobacter baumannii in Maryland: a statewide cross-sectional period prevalence survey. Infect Control Hosp Epidemiol 33:883–888. 10.1017/S0195941700031489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Quinn B, Rodman N, Jara E, Fernandez JS, Martinez J, Traglia GM, Montaña S, Cantera V, Place K, Bonomo RA, Iriarte A, Ramírez MS. 2018. Human serum albumin alters specific genes that can play a role in survival and persistence in Acinetobacter baumannii. Sci Rep 8:14741. 10.1038/s41598-018-33072-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen CT, Wang YC, Kuo SC, Shih FH, Chen TL, How CK, Yang YS, Lee YT. 2018. Community-acquired bloodstream infections caused by Acinetobacter baumannii: a matched case-control study. J Microbiol Immunol Infect 51:629–635. 10.1016/j.jmii.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 135.Thom KA, Johnson JK, Lee MS, Harris AD. 2011. Environmental contamination because of multidrug-resistant Acinetobacter baumannii surrounding colonized or infected patients. Am J Infect Control 39:711–715. 10.1016/j.ajic.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Murray GL, Tsyganov K, Kostoulias XP, Bulach DM, Powell D, Creek DJ, Boyce JD, Paulsen IT, Peleg AY. 2017. Global gene expression profile of Acinetobacter baumannii during bacteremia. J Infect Dis 215:S52–S57. 10.1093/infdis/jiw529. [DOI] [PubMed] [Google Scholar]
- 137.Koenigs A, Zipfel PF, Kraiczy P. 2015. Translation elongation factor Tuf of Acinetobacter baumannii is a plasminogen-binding protein. PLoS One 10:e0134418. 10.1371/journal.pone.0134418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bruhn KW, Pantapalangkoor P, Nielsen T, Tan B, Junus J, Hujer KM, Wright MS, Bonomo RA, Adams MD, Chen W, Spellberg B. 2015. Host fate is rapidly determined by innate effector-microbial interactions during Acinetobacter baumannii bacteremia. J Infect Dis 211:1296–1305. 10.1093/infdis/jiu593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lin L, Tan B, Pantapalangkoor P, Ho T, Baquir B, Tomaras A, Montgomery JI, Reilly U, Barbacci EG, Hujer K, Bonomo RA, Fernandez L, Hancock RE, Adams MD, French SW, Buslon VS, Spellberg B. 2012. Inhibition of LpxC protects mice from resistant Acinetobacter baumannii by modulating inflammation and enhancing phagocytosis. mBio 3:e00312-12. 10.1128/mBio.00312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Grimont PA, Grimont F. 1978. The genus Serratia. Annu Rev Microbiol 32:221–248. 10.1146/annurev.mi.32.100178.001253. [DOI] [PubMed] [Google Scholar]
- 141.Mahlen SD. 2011. Serratia infections: from military experiments to current practice. Clin Microbiol Rev 24:755–791. 10.1128/CMR.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Byrne AH, Herra CM, Aucken H, Keane CT. 2001. Rate of carriage of Serratia marcescens in patients with and without evidence of infection. Scand J Infect Dis 33:822–826. 10.1080/00365540110077385. [DOI] [PubMed] [Google Scholar]
- 143.Moles L, Gómez M, Moroder E, Jiménez E, Escuder D, Bustos G, Melgar A, Villa J, Del Campo R, Chaves F, Rodríguez JM. 2019. Serratia marcescens colonization in preterm neonates during their neonatal intensive care unit stay. Antimicrob Resist Infect Control 8:135. 10.1186/s13756-019-0584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Young VM, Moody MR, Morris MJ. 1980. Distribution of Serratia marcescens serotypes in cancer patients. J Med Microbiol 13:333–339. 10.1099/00222615-13-2-333. [DOI] [PubMed] [Google Scholar]
- 145.Filius PM, Gyssens IC, Kershof IM, Roovers PJ, Ott A, Vulto AG, Verbrugh HA, Endtz HP. 2005. Colonization and resistance dynamics of gram-negative bacteria in patients during and after hospitalization. Antimicrob Agents Chemother 49:2879–2886. 10.1128/AAC.49.7.2879-2886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sunenshine RH, Tan ET, Terashita DM, Jensen BJ, Kacica MA, Sickbert-Bennett EE, Noble-Wang JA, Palmieri MJ, Bopp DJ, Jernigan DB, Kazakova S, Bresnitz EA, Tan CG, McDonald LC. 2007. A multistate outbreak of Serratia marcescens bloodstream infection associated with contaminated intravenous magnesium sulfate from a compounding pharmacy. Clin Infect Dis 45:527–533. 10.1086/520664. [DOI] [PubMed] [Google Scholar]
- 147.Blossom D, Noble-Wang J, Su J, Pur S, Chemaly R, Shams A, Jensen B, Pascoe N, Gullion J, Casey E, Hayden M, Arduino M, Budnitz DS, Raad I, Trenholme G, Srinivasan A, Serratia in Pre-filled Syringes Investigation Team Group. 2009. Multistate outbreak of Serratia marcescens bloodstream infections caused by contamination of prefilled heparin and isotonic sodium chloride solution syringes. Arch Intern Med 169:1705–1711. 10.1001/archinternmed.2009.290. [DOI] [PubMed] [Google Scholar]
- 148.Ostrowsky BE, Whitener C, Bredenberg HK, Carson LA, Holt S, Hutwagner L, Arduino MJ, Jarvis WR. 2002. Serratia marcescens bacteremia traced to an infused narcotic. N Engl J Med 346:1529–1537. 10.1056/NEJMoa012370. [DOI] [PubMed] [Google Scholar]
- 149.Johnson A, Watson D, Dreyfus J, Heaton P, Lampland A, Spaulding AB. 2020. Epidemiology of Serratia bloodstream infections among hospitalized children in the United States, 2009-2016. Pediatr Infect Dis J 39:e71–e73. 10.1097/INF.0000000000002618. [DOI] [PubMed] [Google Scholar]
- 150.Moradigaravand D, Boinett CJ, Martin V, Peacock SJ, Parkhill J. 2016. Recent independent emergence of multiple multidrug-resistant Serratia marcescens clones within the United Kingdom and Ireland. Genome Res 26:1101–1109. 10.1101/gr.205245.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cheng L, Nelson BC, Mehta M, Seval N, Park S, Giddins MJ, Shi Q, Whittier S, Gomez-Simmonds A, Uhlemann AC. 2017. Piperacillin-tazobactam versus other antibacterial agents for treatment of bloodstream infections due to AmpC β-lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother 61:e00276-17. 10.1128/AAC.00276-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chaubey VP, Pitout JD, Dalton B, Gregson DB, Ross T, Laupland KB. 2014. Clinical and microbiological characteristics of bloodstream infections due to AmpC β-lactamase producing Enterobacteriaceae: an active surveillance cohort in a large centralized Canadian region. BMC Infect Dis 14:647. 10.1186/s12879-014-0647-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bush K, Pannell M, Lock JL, Queenan AM, Jorgensen JH, Lee RM, Lewis JS, Jarrett D. 2013. Detection systems for carbapenemase gene identification should include the SME serine carbapenemase. Int J Antimicrob Agents 41:1–4. 10.1016/j.ijantimicag.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 154.Rodríguez C, Brengi S, Cáceres MA, Mochi S, Viñas MR, Merletti G, Raya RR, Centrón D. 2017. Polyclonal dissemination of KPC-2 in Serratia marcescens, including a clone with epidemic behaviour in the nosocomial niche. Int J Antimicrob Agents 49:657–658. 10.1016/j.ijantimicag.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 155.Gales AC, Jones RN, Sader HS. 2006. Global assessment of the antimicrobial activity of polymyxin B against 54 731 clinical isolates of Gram-negative bacilli: report from the SENTRY antimicrobial surveillance programme (2001–2004). Clin Microbiol Infect 12:315–321. 10.1111/j.1469-0691.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- 156.Borenshtein D, Schauer DB. 2006. The fenus Citrobacter, p 90–98. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes, vol 6. Proteobacteria: gamma subclass. Springer New York, New York, NY. 10.1007/0-387-30746-x_5. [DOI] [Google Scholar]
- 157.Brenner DJ, O'Hara CM, Grimont PA, Janda JM, Falsen E, Aldova E, Ageron E, Schindler J, Abbott SL, Steigerwalt AG. 1999. Biochemical identification of Citrobacter species defined by DNA hybridization and description of Citrobacter gillenii sp. nov. (formerly Citrobacter genomospecies 10) and Citrobacter murliniae sp. nov. (formerly Citrobacter genomospecies 11). J Clin Microbiol 37:2619–2624. 10.1128/JCM.37.8.2619-2624.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hodges GR, Degener CE, Barnes WG. 1978. Clinical significance of Citrobacter isolates. Am J Clin Pathol 70:37–40. 10.1093/ajcp/70.1.37. [DOI] [PubMed] [Google Scholar]
- 159.Samonis G, Karageorgopoulos DE, Kofteridis DP, Matthaiou DK, Sidiropoulou V, Maraki S, Falagas ME. 2009. Citrobacter infections in a general hospital: characteristics and outcomes. Eur J Clin Microbiol Infect Dis 28:61–68. 10.1007/s10096-008-0598-z. [DOI] [PubMed] [Google Scholar]
- 160.Liu LH, Wang NY, Wu AY, Lin CC, Lee CM, Liu CP. 2018. Citrobacter freundii bacteremia: risk factors of mortality and prevalence of resistance genes. J Microbiol Immunol Infect 51:565–572. 10.1016/j.jmii.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 161.Shih CC, Chen YC, Chang SC, Luh KT, Hsieh WC. 1996. Bacteremia due to Citrobacter species: significance of primary intraabdominal infection. Clin Infect Dis 23:543–549. 10.1093/clinids/23.3.543. [DOI] [PubMed] [Google Scholar]
- 162.Drelichman V, Band JD. 1985. Bacteremias due to Citrobacter diversus and Citrobacter freundii. Incidence, risk factors, and clinical outcome. Arch Intern Med 145:1808–1810. 10.1001/archinte.1985.00360100068010. [DOI] [PubMed] [Google Scholar]
- 163.Schaffer JN, Pearson MM. 2015. Proteus mirabilis and urinary tract infections. Microbiol Spectr 3:UTI-0017-2013. 10.1128/microbiolspec.UTI-0017-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jacobsen SM, Stickler DJ, Mobley HL, Shirtliff ME. 2008. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin Microbiol Rev 21:26–59. 10.1128/CMR.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wenner JJ, Rettger LF. 1919. A systematic study of the Proteus group of bacteria. J Bacteriol 4:331–353. 10.1128/JB.4.4.331-353.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Müller HE. 1986. Occurrence and pathogenic role of Morganella-Proteus-Providencia group bacteria in human feces. J Clin Microbiol 23:404–405. 10.1128/JCM.23.2.404-405.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Peerbooms PG, Verweij AM, MacLaren DM. 1985. Uropathogenic properties of Proteus mirabilis and Proteus vulgaris. J Med Microbiol 19:55–60. 10.1099/00222615-19-1-55. [DOI] [PubMed] [Google Scholar]
- 168.Imai K, Ishibashi N, Kodana M, Tarumoto N, Sakai J, Kawamura T, Takeuchi S, Taji Y, Ebihara Y, Ikebuchi K, Murakami T, Maeda T, Mitsutake K, Maesaki S. 2019. Clinical characteristics in blood stream infections caused by Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae: a comparative study, Japan, 2014–2017. BMC Infect Dis 19:946. 10.1186/s12879-019-4498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Koizumi Y, Sakanashi D, Ohno T, Yamada A, Shiota A, Kato H, Hagihara M, Watanabe H, Asai N, Watarai M, Murotani K, Yamagishi Y, Suematsu H, Mikamo H. 2019. The clinical characteristics of Acinetobacter bacteremia differ among genomospecies: a hospital-based retrospective comparative analysis of genotypically identified strains. J Microbiol Immunol Infect 52:966–972. 10.1016/j.jmii.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 170.Lee CC, Lee NY, Yan JJ, Lee HC, Chen PL, Chang CM, Wu CJ, Ko NY, Wang LR, Chi CH, Ko WC. 2010. Bacteremia due to extended-spectrum-beta-lactamase-producing Enterobacter cloacae: role of carbapenem therapy. Antimicrob Agents Chemother 54:3551–3556. 10.1128/AAC.00055-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sanders WE, Sanders CC. 1997. Enterobacter spp.: pathogens poised to flourish at the turn of the century. Clin Microbiol Rev 10:220–241. 10.1128/CMR.10.2.220-241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Álvarez-Marín R, Navarro-Amuedo D, Gasch-Blasi O, Rodríguez-Martínez JM, Calvo-Montes J, Lara-Contreras R, Lepe-Jiménez JA, Tubau-Quintano F, Cano-García ME, Rodríguez-López F, Rodríguez-Baño J, Pujol-Rojo M, Torre-Cisneros J, Martínez-Martínez L, Pascual-Hernández Á, Jiménez-Mejías ME, Spanish Network for Research in Infectious Diseases/Enterobacter spp. Bacteriemia Project Group. 2020. A prospective, multicenter case control study of risk factors for acquisition and mortality in Enterobacter species bacteremia. J Infect 80:174–181. 10.1016/j.jinf.2019.09.017. [DOI] [PubMed] [Google Scholar]
- 173.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]