Introduction
Machine learning approaches have many applications in radiology. Over the last several years, the majority of attention has been garnered by applications focused on assisted interpretation of medical images. However, a significant component of radiology encompasses the generation of medical images prior to interpretation, the upstream component. These elements include modality selection for an exam, hardware design, protocol selection for an exam, data acquisition, image reconstruction, and image processing. Here, we give the reader insight into these aspects of upstream Artificial Intelligence (AI), conveying the breadth of the emerging field, some of the techniques, and the potential impact of the applications.
Discussion
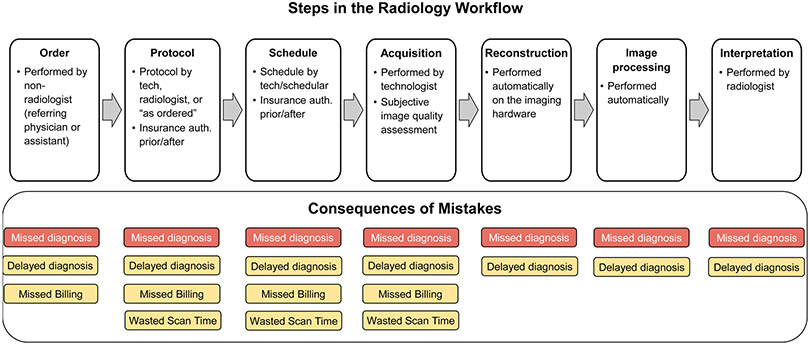
The standard radiology workflow can undergo seven steps; 1 - conversion of a patient’s clinical question into a radiology exam order, usually performed by a non-radiologist physician, 2 – conversion of an exam order into an exam protocol, usually performed by a radiologist or radiology technologist based on institution-specific guidelines, 3 – scheduling of an exam onto a specific scanner, usually performed by a radiology schedular or technologist, 4 – adaption of a protocol to a specific device, usually performed by a radiology technologist, 5 – acquisition and reconstruction of acquired imaging data into images, usually performed automatically with software algorithms directly on the imaging hardware, 6 – image processing, to retrospectively improve image quality and produce multiplanar reformations and quantitative parameter maps, typically performed directly on the imaging hardware, and 7 – evaluation of images to render a diagnosis related to the patient’s clinical question, usually performed by a radiologist (Fig. 1). Applications and research areas for AI algorithms in radiology have typically focused on the final 2 steps in the radiology workflow.1 Several AI techniques exist to perform automated classification, detection, or segmentation of medical images, many of which are even approved for clinical use by the Food and Drug Administration (FDA). Furthermore, the acquisition of raw data for generating diagnostic quality images is also being used for reducing scan time or radiation dosage of medical images, or retrospectively improving resolution or signal to noise ratio (SNR) of images.
Figure 1:
A summary of the radiology workflow starting from the ordering of an examination and ending with a radiologist interpretation of the acquired imaging. Errors during each of the seven stages of this workflow can lead to delays or the rendering of an incorrect final diagnosis decision.
Despite a dearth of research studies, there exist substantial challenges for all steps in the ‘pre-image-acquisition’ workflow (steps 1-4) that may be alleviated using data-driven benefits that AI techniques can offer. For example, a patient’s initial exam order may not always be correctly defined by a radiology specialist, which can lead to delays in insurance authorizations and the schedule of the correct examination. Next, conversion of an order to a protocol is commonly performed by radiology fellows at many academic institutions, while many private practice organizations may not have such a luxury. Furthermore, with ‘protocol creep’ of new protocols being aggregated in an unorganized manner, choosing the appropriate protocol for the patient becomes a more challenging task.2 The scheduling of patients to different scanners also requires considerations for complex cases that may require a radiologist presence. Following successful protocoling and scheduling, tuning the scanner parameters to cater to the specific exam order needs and patient requirements (such as image resolution, coils, and breath holds for MRI, dose optimization and contrast needs for CT, etc.) can directly affect image quality, and consequently, downstream utility for eventual image interpretation.
Errors can occur during any part of the radiology pre-image-acquisition workflow and can lead to delayed diagnoses, or, in the worst case, missed diagnoses. Each of these four steps is repetitive, but because of the innate variability between patients, these tasks have not lent themselves to automated techniques. Using manual techniques to complete such tasks is prone to user error, leads to delays in the workflow, and is an inefficient use of human capital particularly when these tasks require radiologist oversight. The application of pre-image-acquisition AI can lead to potential gains in efficiency and patient care. There are several factors that these tasks ripe for automation with AI techniques. First, all the tasks are already performed on computer systems, entailing that all inputs to and output of the tasks are already digitized, recorded, and can be retrospectively extracted. Second, as large volumes (hundreds to thousands) of most exam types are performed every year, even at a single academic center, large institution-specific data sets needed to train AI algorithms are already available. Finally, a stepped pathway for incorporating these algorithms into the radiology workflow can be achievable. Initial versions of the ML algorithms can suggest or pre-select choices for use as a clinical decision support system with continued human oversight. Following this, continual-learning strategies can be used to improve such algorithms to have higher reliability than humans for eventual transition to full automation.
Recent advances in natural language processing (NLP) have been utilized to perform automated assignment for CT and MRI protocols using procedure names, modality, section, indications, and patient history as inputs to guide a clinical decision support model.3 Similar NLP systems have also been previously used to use unstructured data for performing automated protocoling for musculoskeletal MRI examinations4,5 as well as brain MRI examinations.6 Newer AI techniques for NLP that utilize the Bidirectional Encoder Representations from Transformers (BERT) model7 have also been used in the context of automated protocoling.8 Advances in multi-modality data integration presents an exciting opportunity to improve the upstream workflow by further combining data from a patient’s medical record, along with AI models that rely on conventional computer vision and NLP techniques.9
Although the techniques and applications of upstream AI are broad, below we discuss some representative examples across various commonly utilized medical imaging modalities.
Improved Image Formation in Ultrasound
In ultrasound image reconstruction, raw echo data is received by an array of transducer sensors. This “channel data” is processed to produce an image of the underlying anatomy and function. Traditional ultrasound image reconstruction can include various noise artifacts that degrade signal and image quality. Rather than attempting to correct these using already-reconstructed images, upstream ultrasound research utilizes the raw channel data to address the artifacts at their source. AI has found a wide range of early uses in upstream ultrasound, a few of which we highlight here.
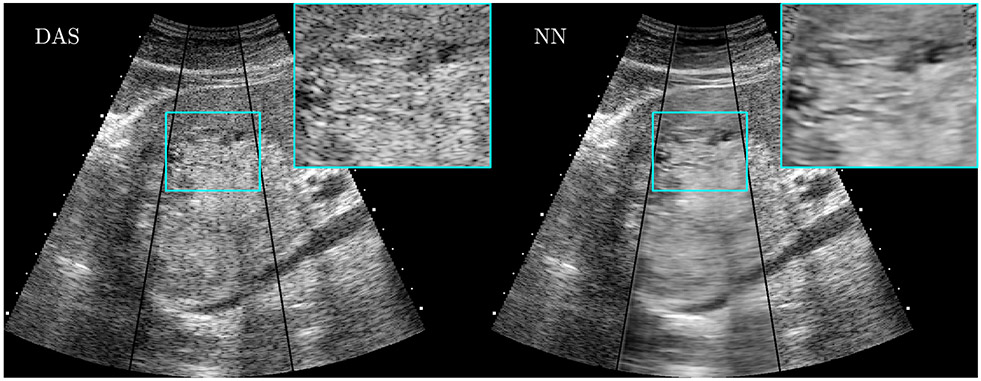
A major source of ultrasound image degradation is reverberation clutter.10 Clutter arises from shallow reverberations that are incorrectly superimposed upon echoes from deeper tissue, resulting in decorrelated channel data. This decorrelation manifests in ordinary B-mode images as a dense haze and also degrades the ability to perform Doppler and other advanced imaging techniques. Several non-AI techniques have been proposed to remove clutter from channel data directly, improving channel data correlations and B-mode image contrast, albeit at great computational cost.11-13 Recently, AI has been used to achieve the same goal using a data-driven approach.14,15 In our work, we used matched simulations of ultrasound sensor data with reverberation clutter (input) and without reverberation clutter (output) to train a convolutional neural network (CNN).15 Figure 2 demonstrates how the CNN removes high-frequency clutter from simulated channel data. In Figure 3, the CNN removes clutter from in vivo channel data, resulting in B-mode images with improved structure visualization and contrast. The CNN restores correlations in the raw channel data and thus enables subsequent advanced imaging techniques, such as sound speed estimation and phase aberration correction.
Figure 2:
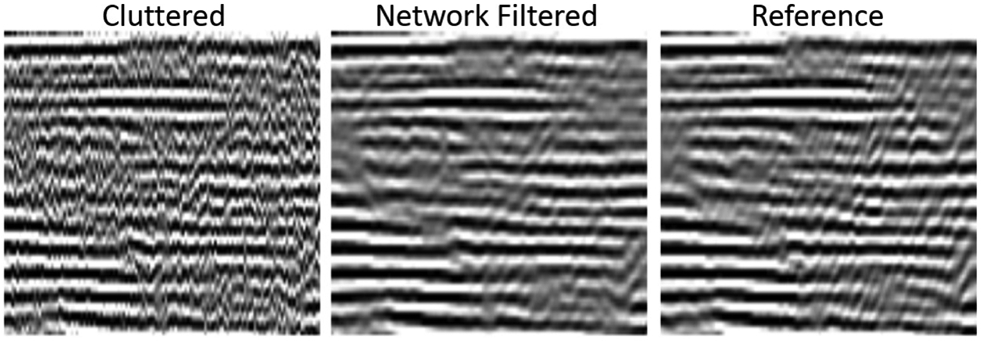
Images of simulated channel data comparing cluttered, CNN-filtered, and reference uncluttered images. Reverberation clutter, which appears as a high frequency noise across the channels, is removed by the CNN while preserving the structure of reflections from true targets. Data from Brickson LL, Hyun D, Jakovljevic M, Dahl JJ. Reverberation Noise Suppression in Ultrasound Channel Signals Using a 3D Fully Convolutional Neural Network. IEEE Trans Med Imaging. 2021;PP.
Figure 3:
B-mode images of a longitudinal cross-section of a carotid artery and thyroid. The CNN-filtered image visualizes several hypoechoic and anechoic targets that were originally obscured by clutter. Data from Brickson LL, Hyun D, Jakovljevic M, Dahl JJ. Reverberation Noise Suppression in Ultrasound Channel Signals Using a 3D Fully Convolutional Neural Network. IEEE Trans Med Imaging. 2021;PP.
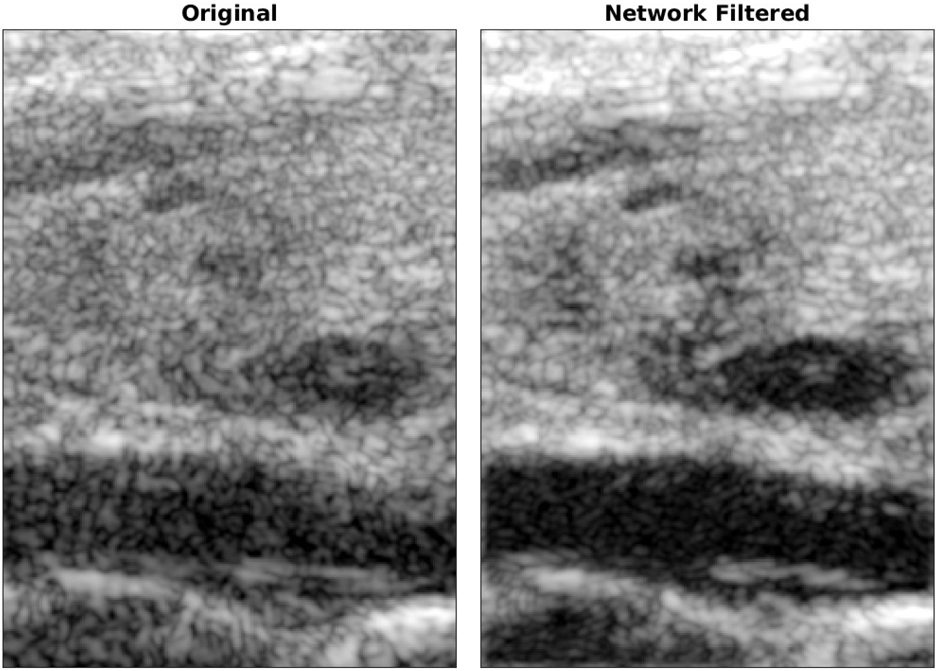
Another major noise source in ultrasound images is a pervasive grainy texture called speckle. Speckle is an unavoidable artifact arising from the finite resolution of ultrasound imaging systems. While speckle can be useful for certain applications, it is largely treated as an undesirable noise that degrades B-mode images of the underlying tissue echogenicity. Rather than filtering speckle from already-reconstructed B-mode images, we trained a convolutional neural network (CNN) to estimate the tissue echogenicity directly from raw channel data using simulations of ultrasound speckle and known ground truth echogenicity.16 In simulations, the trained CNN accurately and precisely estimated the true echogenicity. In phantom and in vivo experiments, where the ground truth echogenicity is unknown, the CNN outperformed traditional signal processing and image processing speckle reduction techniques according to standard image quality metrics such as contrast, contrast-to-noise ratio, and signal-to-noise ratio. Figure 4 shows an in vivo example of a liver lesion acquired using a clinical scanner. This CNN has been further demonstrated in real-time on a prototype scanner.17
Figure 4:
CNN-based ultrasound speckle reduction and detail preservation. (left) A traditional B-mode image of a complex focal liver lesion. (right) The CNN output when provided with the same raw data. Data from Hyun D, Brickson LL, Looby KT, Dahl JJ. Beamforming and Speckle Reduction Using Neural Networks. IEEE Trans Ultrason Ferroelectr Freq Control. 2019;66(5):898-910.
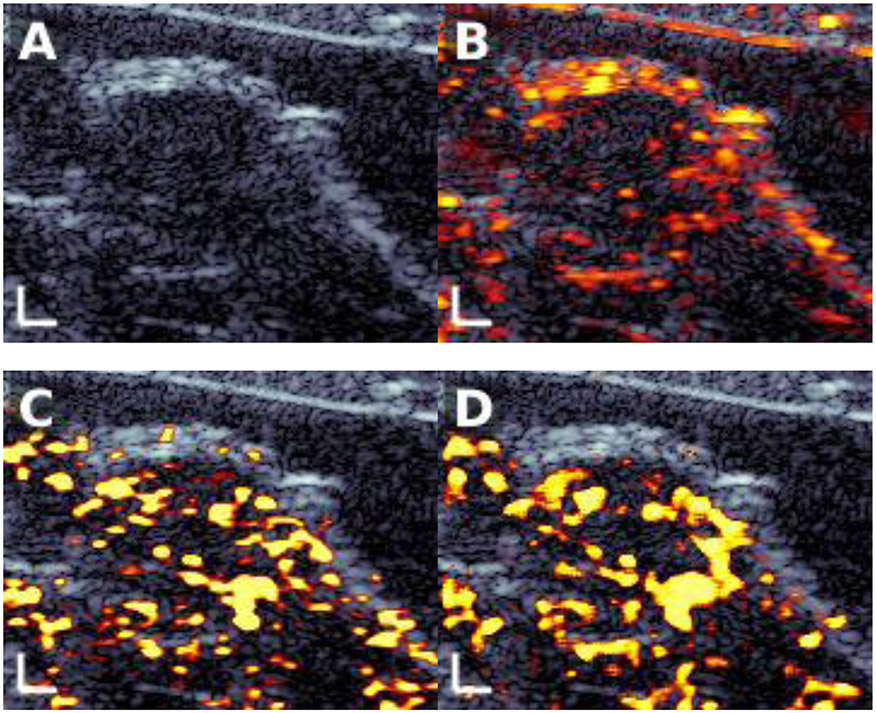
Another emerging application in ultrasound is contrast-enhanced imaging, including with targeted contrast agents, to detect the presence of disease. Contrast-enhanced images are presented alongside ordinary B-mode images to highlight any disease-bound contrast agents in the field of view. A key challenge in contrast-enhanced imaging, particularly molecular imaging, is to detect echoes from contrast agents while suppressing echoes from background tissue. Current state-of-the-art imaging identifies molecular signals retrospectively after a strong destructive ultrasound pulse eliminates them from the field of view, and thus cannot be used for real-time imaging. We recently used CNNs to nondestructively emulate the performance of destructive imaging, enabling real-time molecular imaging for early cancer detection.18 Rather than treating B-mode and contrast-enhanced modes as separate images, the CNN combines raw data from both to improve the final molecular image. As shown in Fig. 5, the trained CNN (panel D) was able to mimic the performance of the state-of-the-art destructive approach (panel C), enabling high quality real-time ultrasound molecular imaging.
Figure 5:
Ultrasound molecular imaging of a breast cancer tumor in a transgenic mouse. (A) B-mode image, with the tumor in the center. (B) Contrast-enhanced image overlaid on the B-mode image. (C) Destructive state-of-the-art molecular image. (D) Output of the trained CNN using only nondestructive input data. The nondestructive CNN closely matched the destructive image, showing the potential for AI-enabled real-time molecular imaging.
In addition to these, AI has found numerous other upstream ultrasound applications, including adaptive beamforming,19 ultrasound localization microscopy,20 surgical guidance,21 vector flow Doppler imaging, 22,23 as well as many others.24 The range and variety of applications highlight the extraordinary adaptability of AI techniques to ultrasound signal and image processing.
CT Dose Optimization
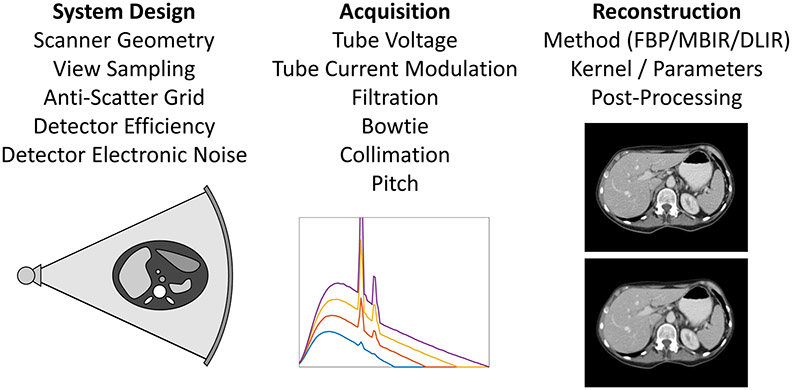
Over 75 million computed tomography (CT) scans are performed annually in the United States, with the number expected to continue increasing.25 Despite clear clinical value, ionizing radiation dose has been a major concern for CT. In addition, there are significant variations in radiation dose and image quality across institutions, protocols, scanners, and patients. While CT has evolved from the manual adjustment of dose control parameters such as tube current, to automatic tube current modulation, CT dose optimization still has major challenges in achieving the most efficient use of radiation dose for each patient and clinical task.26,27 An overview of existing technologies for controlling dose and image quality is shown in Fig. 6.
Figure 6:
Various technologies for controlling CT dose and image quality are listed under the categories of system design, acquisition, and reconstruction. Sub-figures show (left to right): overall scanner geometry, x-ray spectra from different tube voltages, and two reconstructions with different kernels showing sharper image but higher noise (top) and lower noise but smoother image (bottom).
Reconstructed CT images contain quantum noise when the ALARA (as low as reasonably achievable) principle is followed to reduce radiation exposure. As a result, important details in the image could be hidden by the noise, leading to degradation in the diagnostic value of CT. It is therefore crucial to assess the quality of reconstructed CT images at reduced dose. Dose and image noise have a complex dependency on patient size (ranging from pediatrics to obese adults), patient anatomy, CT acquisition, and reconstruction. As can be seen in Fig. 7, CT images at lower doses relative to the routine dose induce higher noise. Unfortunately, there is no consistent image quality assessment measure that has been established to objectively quantify images from different imaging protocols, vendors, sites, etc.28 Having such capabilities would help balance image quality and radiation dose. At every step of the CT scan, from system design to acquisition to reconstruction, efforts could then be made to lower radiation dose.29 As a result, the quality of the acquired CT images could be varied and assessed to guide dose reduction. In addition, effective dose to the patient needs to account for the different radiosensitivity of different organs (for example using the ICRP tissue weights).30 Therefore, an accurate, reliable, and region-focused assessment of dose and image quality is extremely important.
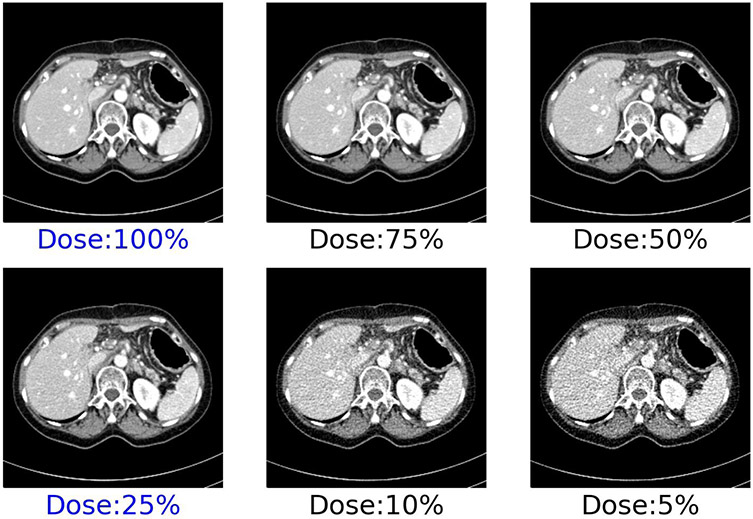
Figure 7:
CT slice of an abdomen scan showing higher noise at lower dose levels. Mayo patient CT projection data library93 provides routine (100%) dose and low-dose (25%) after inserting noise in the projection data. The additional four dose levels are synthesized from these two, assuming only quantum noise.
In an effort to combat increased noise in low-dose settings, CT image reconstruction has been going through a paradigm shift, from conventional filtered backprojection (FBP) to model-based iterative reconstruction (MBIR), and now to deep learning image reconstruction (DLIR).31 FBP, although fast and computationally efficient, incurs higher noise and streak artifacts at low dose. Towards improving image quality at low dose, iterative reconstruction has emerged which preserves diagnostic information, although low-contrast detail remains a challenge and can impact detectability of subtle pathologies. With the advent of machine learning and deep learning in particular, high diagnostic quality is created with reduced noise at low dose, with the additional benefit of faster reconstruction than iterative reconstruction.32 For example, 1) generating standard-dose images from low-dose images in deep learning-based CT denoising33; 2) through incorporating Deep CNN in the complex iterative reconstruction process, less noisy images can be obtained at low dose34; 3) improved image quality of dual energy CT, particularly for large patients, low-energy virtual monoenergetic images, and material decomposition35; 4) reduction of metal artifacts through deep learning techniques.36,37 Deep learning-based image reconstruction enables significantly lower image noise and higher image quality (IQ score and better CNR) over FBP and MBIR, which can be traded off for dose reduction.38 However, dose reduction needs to be balanced against an acceptable level of diagnostic accuracy. Therefore, not only phantom data but also patient data are needed to understand the impacts of dose reduction on image quality (using objective metrics) as well as the task-specific diagnostic performance (such as detecting liver lesions).39
Conventionally, medical images are assessed subjectively requiring manual labor, long times, and observer variations, which is impractical for use in a clinical workflow. It is therefore desirable to perform fast, automatic, and objective image quality assessment. Objective assessment can be of two types: reference-based and non-reference-based. A number of reference-based image assessment metrics have been widely used in medical imaging. For example, PSNR, MSE, CNR, SSIM, and RMSE are often used. However, these metrics rely on the availability of high-quality reference images. Since our goal is to acquire CT scans at reduced dose, we cannot assume to have access to high-quality (and high-dose) images of the same patient.40 Therefore, non-reference assessments are preferred so that we can quantify any CT image without requiring the corresponding reference images at high dose. In addition to the established image quality assessment metrics, a few non-reference assessment methods are also available, such as a blind/referenceless image spatial quality evaluator based on natural scene statistics that operates in the spatial domain.41 A growing body of research has recently emerged focusing on no-reference and automated image quality assessment using deep learning. For example, Patwari et al. used a regression CNN to learn mapping noisy CT images to GSSIM scores, as an assessment of the reconstructed image quality.42 Kim et al. proposed a no-reference image quality assessment framework via two-stage training: regression into objective error map and subjective score.43 Another deep learning-based non-reference assessment has recently been proposed leveraging self-supervised prediction of noise levels.44
In summary, reduction of CT dose directly impacts image quality. Preserving image quality at low dose has been an active area of research. Deep learning-based methods have achieved some promising results in CT image reconstruction, denoising, artifact reduction, and image quality assessment. However, to bring dose optimization to CT acquisition, prediction of the expected dose and image quality has to be real time, patient specific, organ specific, and task specific. This is a complex and challenging problem for which deep learning algorithms are likely to excel.
Optimized Data Acquisition in MRI via Deep Learning
In MRI, data is acquired in the Fourier domain, or k-space, which represents spatial and temporal frequency information in the object being imaged. Sampling of k-space is necessarily finite. Therefore, optimizing sampling patterns and designing k-space readout trajectories that enable collecting the most informative k-space samples for image reconstruction can improve the resulting diagnostic information in images. Besides improved image quality, the design of optimized sampling trajectories can lead to shortened data acquisition for improved efficiency. Recently, several deep learning methods have been proposed for learning k-space sampling patterns jointly with image reconstruction algorithmic parameters. These methods have shown improved reconstructed image quality compared to reconstructions obtained with predetermined trajectories such as variable density sampling. Deep learning methods for data sampling can be separated into two categories: active and non-active (fixed) strategies. These two strategies differ primarily in whether the learned sampling schemes are fixed or not at inference time.
Given a target k-space undersampling or acceleration factor, the non-active (fixed) strategies produce undersampling masks or sampling trajectories using a set of fully-sampled images. After the training procedure is completed, the learned sampling pattern is fixed, and new scans can be acquired using the learned trajectory (Figs 8A&B). Image reconstruction can then be performed using the reconstruction network learned as a part of the training process. Several studies45,46 focus on the Cartesian sampling case and model binary sampling masks probabilistically. These methods can be applied to either 2D or 3D Cartesian sampling to determine the optimal set of phase encodes. As an extension to more general parameterized, or non-Cartesian, k-space trajectories,47 one can directly optimize for k-space coordinates by restricting the optimization to a set of separable variables such as horizontal and vertical directions in 2-D plane. The framework presented in Weiss et al. additionally incorporates gradient system constraints into the learning process.48 This enables finding the optimal gradient waveforms in a data-driven manner.
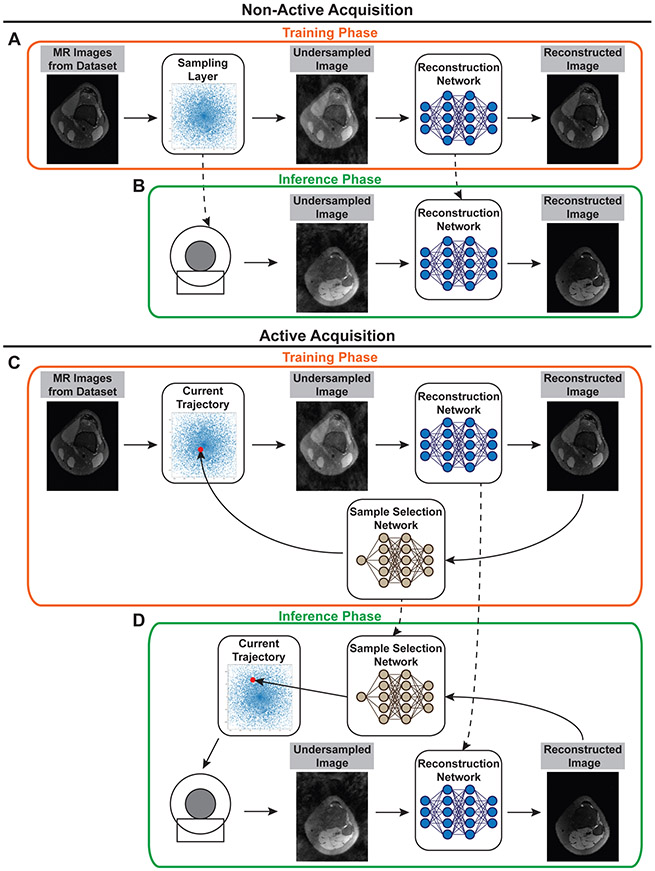
Figure 8:
Non-active and active data strategies for deep learning based data sampling methods. Non-active (fixed) strategies optimize sampling trajectories and reconstruction networks at training time (A). At inference time (B), the sampling trajectory is fixed, and the corresponding gradient waveforms are programmed in the scanner for acquisition. The optimized reconstruction network is then used for reconstructing images from undersampled measurements. Active strategies (C&D) use an additional neural network that suggests the next sample to collect using the reconstruction obtained from existing samples. The process is repeated until a desired metric or uncertainty threshold is met. Due to their sequential nature, active strategies require an additional mechanism that generates gradient waveforms on-the-fly for acquiring the samples proposed by the sample selection network.
Active acquisition strategies,49-52 on the other hand, attempt to predict in real-time the next k-space samples to be acquired using information from existing samples from the same acquisition, i.e. the same patient/scan (Figs 8 C&D). These methods employ an additional neural network that suggests the next sample to collect by measuring the reconstruction quality or uncertainty during the acquisition. Active techniques therefore have the benefit of obtaining subject-specific data acquisition strategies. In other words, active sampling strategies tailor the sampling pattern to new scans and patients.
In general, the non-active acquisitions are easier to implement in scanner hardware. However, such acquisitions can be suboptimal on new types of data or scans. For example, a sampling pattern that is optimal for brain imaging might not be the best trajectory for abdominal imaging. On the other hand, active acquisitions could adapt to new kinds of data at the expense of added complexity in determining and generating gradient waveforms on-the-fly.
Optimization of data sampling using deep learning is an active area of research and many open questions exist for further investigation. The effects of imaging non-idealities such as off-resonance, relaxation effects and motion on the optimized sampling pattern have not yet been fully investigated. In addition, the findings on deep learning-based data sampling approaches in MR literature typically rely on retrospective studies and simulations. Their applicability to prospective studies and clinical settings should be investigated to understand the full potential and applicability of learned sampling patterns.
Image Reconstruction in MRI: Supervised Deep Learning
To reconstruct rapidly acquired k-space data into high-quality images, advanced image reconstruction methods are necessary. Previously, parallel imaging and compressed sensing (PI-CS) methods53,54 have been used to iteratively reconstruct highly undersampled data, thus enabling rapid MRI scans. However, due to their iterative nature, PI-CS methods suffer from impractically long reconstruction times which limit their clinical applicability. More recently, deep learning-based reconstruction approaches have demonstrated much faster reconstruction times, and in some cases, better image quality than PI-CS methods. These approaches are based on machine learning models which must first be trained to learn the process of reconstructing raw data into images.
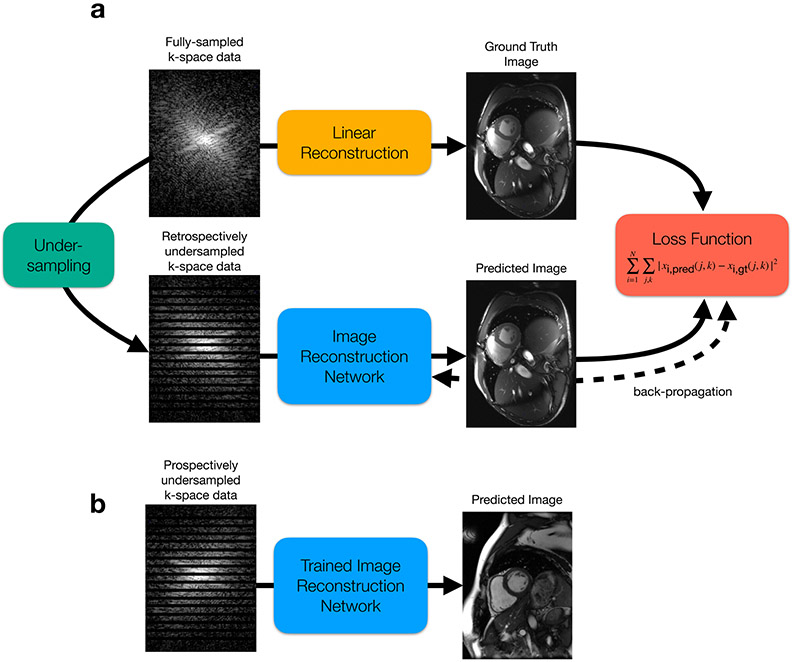
The most common way of training a model to reconstruct MR images is through supervised learning. In the case of reconstruction, the inputs are undersampled k-space data, which the model uses to output a reconstructed image. Supervision here is enabled by utilizing fully sampled k-space data, which may have either been directly acquired through a lengthy scan or synthesized from under-sampled data via more conventional image reconstruction techniques. The model itself is learned by updating the model parameters such that the difference between the model output and the fully sampled ground truth is minimized. This process, known as stochastic gradient descent, is done repeatedly for each example in the training dataset until the model has fully converged (Fig. 9).
Figure 9:
(a) MRI reconstruction model training pipeline. If fully-sampled k-space data is available, a linear reconstruction is performed to generate the high-quality, ground truth image. The fully-sampled k-space data is then retrospectively undersampled by throwing away k-space lines, simulating how the scanner would undersample data in a true, accelerated acquisition. This undersampled data is given to the network for reconstruction. The network is then trained to output a predicted image, which is enforced to be “close” to the ground truth image via a loss function. A simple mean-squared-error loss is shown here. The model parameters are then iteratively updated by a stochastic gradient descent (SGD) algorithm, which intends to minimize the loss function, thereby minimizing the difference between the predicted image and the ground truth image. (b) Once the model is fully trained, it can be used to “infer” or reconstruct images from prospectively undersampled data in an efficient manner.
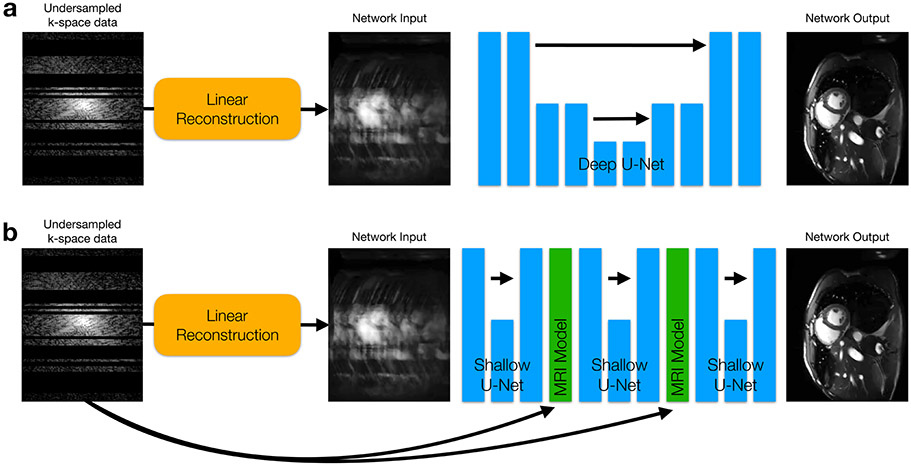
Choosing the type of model to use for reconstruction tasks remains an active area of research. Similar to image classification approaches, the most common type of model used to learn MR image reconstruction is a deep convolutional neural network (CNN).55-57 CNNs consist of convolutions, cascaded with non-linear activation functions, which are able to extract features from images at various hierarchical levels of detail and use them for reconstruction (Fig. 10A). As in any other application of deep learning, deeper networks (i.e. more cascades) are capable of learning better, more accurate reconstruction models. However, deeper networks also consist of more learnable parameters, and are therefore more likely to overfit to the training dataset. In other words, if not provided with enough training data to support the number of network parameters, the network will begin to memorize the data, degrading performance on data which was not seen during training.
Figure 10:
(a) An example of a deep convolutional neural network (CNN) for MRI reconstruction. There are many degrees of freedom in designing CNNs. One popular architecture is called a U-Net, which repeatedly applies convolutions and down-sampling layers to extract both low-resolution and high-resolution features. (b) Unrolled neural networks instead apply shallow CNNs (again U-Nets are shown) cascaded with simulated MRI model projections which ensure that the output of each CNN does not deviate from the raw, undersampled k-space data. These projections also make use of Fourier transforms and coil sensitivity information to convert intermediate network outputs back to the original multi-channel k-space domain where the projection is performed.
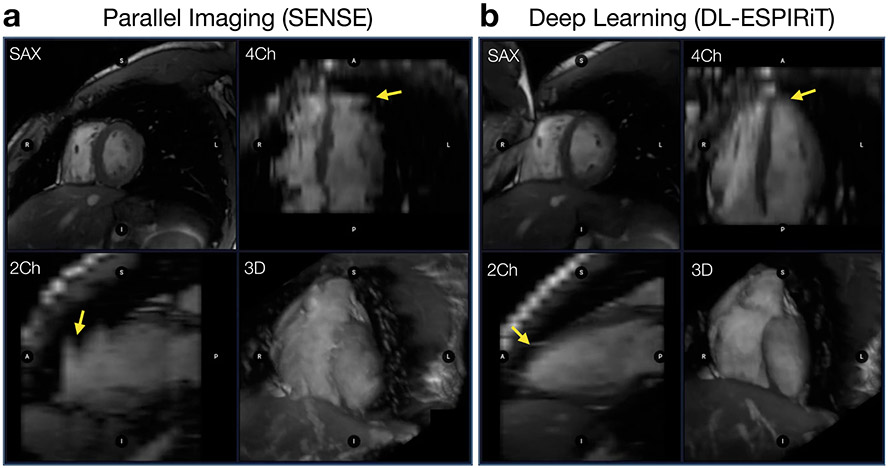
Recently, it has been demonstrated that deep CNN methods benefit from incorporating additional information about the MRI acquisition as an aid for image reconstruction. This is done by interleaving shallow CNNs with projections of a simulated MRI scanning model (Fig. 10B).58-60 This interleaved model, known as an unrolled neural network (UNN), is treated as one deep model and trained end-to-end in the same way as a conventional CNN. By incorporating additional acquisition information, the network is never allowed to produce images that are relatively inconsistent with the raw k-space data. As a result, UNN reconstruction methods reduce the overfitting problem, and allow for better generalizability to unseen data. Initial proof-of-concept studies have demonstrated that UNNs outperform conventional reconstruction methods across a wide variety of clinical applications, such as 2D cardiac cine MRI scans61 for example (Fig. 11). However, further research is necessary to demonstrate their robustness and effectiveness across large clinical populations.
Figure 11:
Two 2D cardiac cine scans are performed on a pediatric patient with 17 short-axis view (SAX) slices covering the heart and scan parameters: TE=1.4ms, TR=3.3ms, matrix size=200x180. Reformats are shown to visualize 4-chamber (4Ch), 2-chamber (2Ch) views along with a 3D rendering. (a) The first scan is performed with 2X undersampling and reconstructed using a standard parallel imaging technique. (b) The second scan is performed with 12X undersampling and reconstructed using a deep learning approach. With deep learning-powered acceleration, the scan time is shortened from 6 breath-holds down to a single breath-hold. This not only has important implications for patient comfort, but also for the accuracy of volumetric assessments from these images, since the inevitable variations between breath-holds is significantly reduced in the DL images (yellow arrows).
Besides undersampled MRI reconstruction that utilizes raw k-space data, an additional class of techniques perform image-to-image translations (for example, low-resolution to high-resolution, low SNR to high SNR, etc) in order to overcome the challenge that raw k-space data is not always archived from clinical MRI scans. Such techniques have previously been utilized to transform low-resolution images into higher-resolution images, under the constraints of maintaining segmentation, quantitative parameter mapping, and diagnostic accuracy.62-64 These methods have been demonstrated across varying anatomies, including neuro, cardiac, and musculoskeletal MRI.65-68 Deep learning based approaches have also been utilized to alleviate the constraints of long scan times for accelerating quantitative MRI69-71 scans.
MR Image reconstruction: Unsupervised Deep Learning
As described above, supervised DL reconstruction methods56,58,59,72-78 may provide more robustness, higher quality, and faster reconstruction speed than conventional image reconstruction approaches. However, the need for fully-sampled data for supervised training poses a problem for applications such as dynamic contrast enhancement (DCE), 3D cardiac cine, 4D flow, low-dose contrast agent imaging, and real-time imaging, where collecting fully-sampled datasets is time-consuming, difficult, or impossible. As a result, supervised DL-based methods often cannot be used in these applications. There are two main possible ways to address this problem. First, PI-CS reconstructions can be used as ground truth for DL frameworks.75 However, reconstructions of the DL model are unlikely to surpass PI-CS reconstruction methods with respect to image quality. Another way is to re-formulate DL training in a way that only leverages undersampled datasets for training, otherwise known as unsupervised learning.79-86
In unsupervised learning, the goal is to train a network to reconstruct an image that is consistent with a distribution of images. Therefore, paired inputs and outputs are no longer necessary like they were for supervised learning. This can be achieved using generative adversarial networks (GANs),87 which in the past, have been used to learn and model complex distributions of images.88,89 GAN training involves jointly training two networks: a generator network whose goal is to reconstruct an image from undersampled k-space data, and a discriminator whose goal is to discern whether or not the generator output came from the same underlying distribution as the training data. During training, these two networks are pitted against each other such that the generator continuously learns to reconstruct higher quality images, while the discriminator becomes better at discerning outputs from the generator.
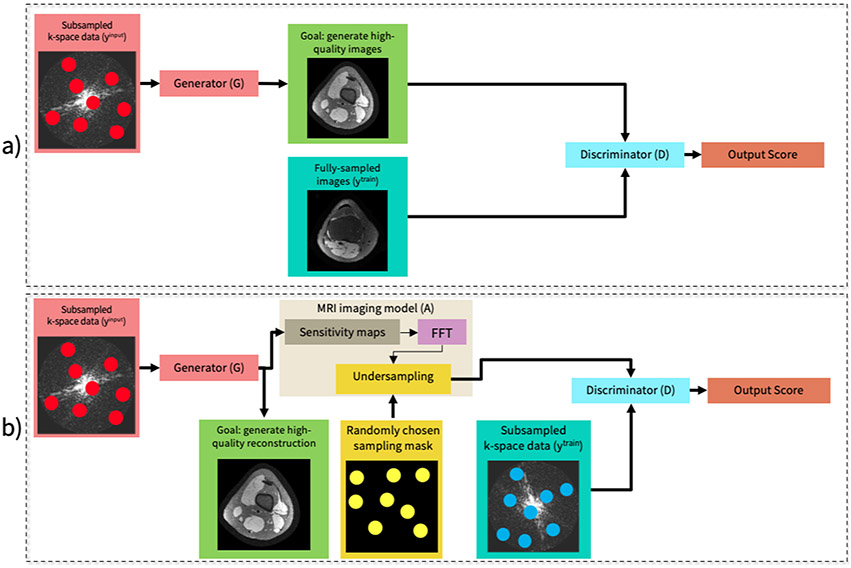
GANs can be used to train MRI reconstruction models in an unsupervised fashion in two situations. If fully-sampled data is available, the generator can be trained to output images that match the distribution of a training dataset comprised of fully-sampled images90 (Figure 12a). If only undersampled data is available, a slightly different framework known as AmbientGAN91 can be used (Figure 12b). In AmbientGAN, the generator output is converted back to k-space and is undersampled through simulation. Then the discriminator is tasked with determining if the undersampled output is consistent with the undersampled training data distribution. This approach has been demonstrated to be effective for unsupervised DL reconstruction, when the undersampling is sufficiently varied throughout the training dataset.92
Figure 12:
A conventional supervised learning system (a) and an unsupervised system (b). (a) Framework overview in a supervised setting with a conditional GAN when fully-sampled datasets are available. (b) Framework overview in an unsupervised setting. The input to the generator network is an undersampled complex-valued k-space data and the output is a reconstructed two-dimensional complex-valued image. Next, a sensing matrix comprised of coil sensitivity maps, an FFT and a randomized undersampling mask (drawn independently from the input k-space measurements) is applied to the generated image to simulate the imaging process. The discriminator takes simulated and observed measurements as inputs and tries to differentiate between them.
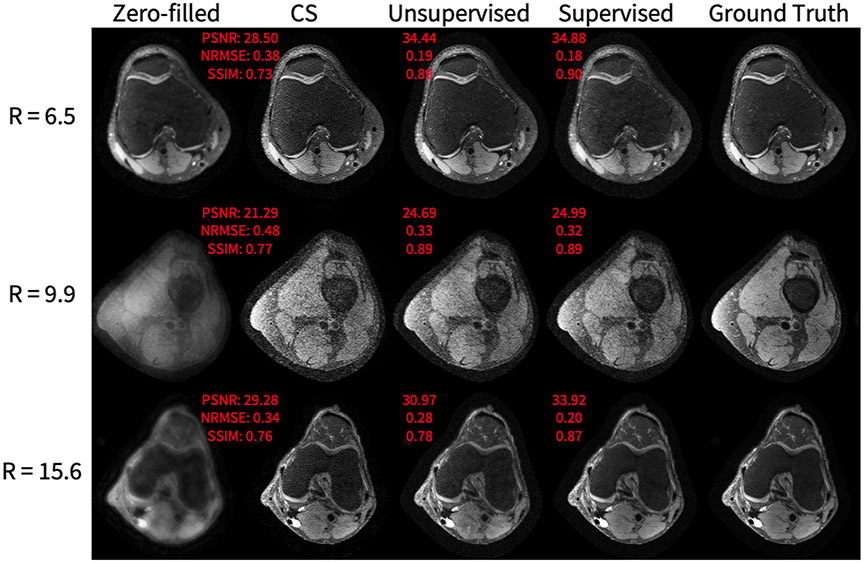
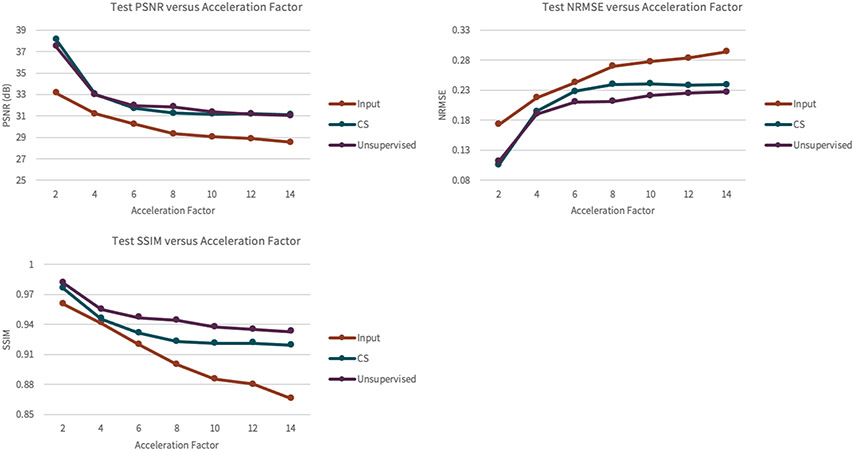
As shown in Figure 13, the unsupervised method achieves superior SSIM, PSNR, and NRMSE on a knee dataset compared to compressed sensing reconstruction. However, the supervised GAN is superior in comparison to the proposed unsupervised GAN, which is expected because the supervised GAN has access to fully-sampled data, giving the network a stronger prior. Of note, the superior performance of the unsupervised GAN compared to compressed sensing is fairly negligible for lower accelerations, but becomes more significant at accelerations of 4-6 (Figure 14), highlighting the potential of achieving imaging speed ups greater than can be obtained conventionally. Representative results from a DCE dataset are shown in Figure 15. The generator greatly improves input image quality by recovering sharpness and adding more structure to input images. Additionally, the proposed method produces a sharper reconstruction compared to CS. In the first row, the anatomical right kidney of the unsupervised GAN is visibly much sharper than input and CS.
Figure 13.
Knee application representative results, showing, from left to right: the input undersampled complex image to the generator, the output of the unsupervised generator, the output of the supervised generator, and the fully-sampled image. The acceleration factors of the input image are 6.5, 9.9, and 15.6, from top to bottom. The quantitative metrics that are plotted next to the images are for the slice that is shown. In all rows, the unsupervised GAN has superior PSNR, NRMSE, and SSIM compared to CS. In the first row, the unsupervised GAN has metrics that are notably worse than the supervised GAN. In the middle row and last rows, the unsupervised GAN has metrics that come close to the performance of the supervised GAN.
Figure 14.
The results of the reconstruction performance on the set of knee scans of the unsupervised GAN as a function of the acceleration factor of the training datasets. The y-axis represents PSNR, NRMSE, or SSIM, depending on the plot. The x-axis represents the acceleration factor of the datasets. The gap between PSNR of CS and the unsupervised model is fairly negligible over the range of accelerations. The gap between NRMSE of CS and the unsupervised model is fairly negligible at first, for low accelerations, but becomes more significant at an acceleration of 6 and beyond. The gap between SSIM of CS and the unsupervised model is fairly negligible at first, for an acceleration of 2, but becomes more significant at an acceleration of 4 and beyond.
Figure 15.
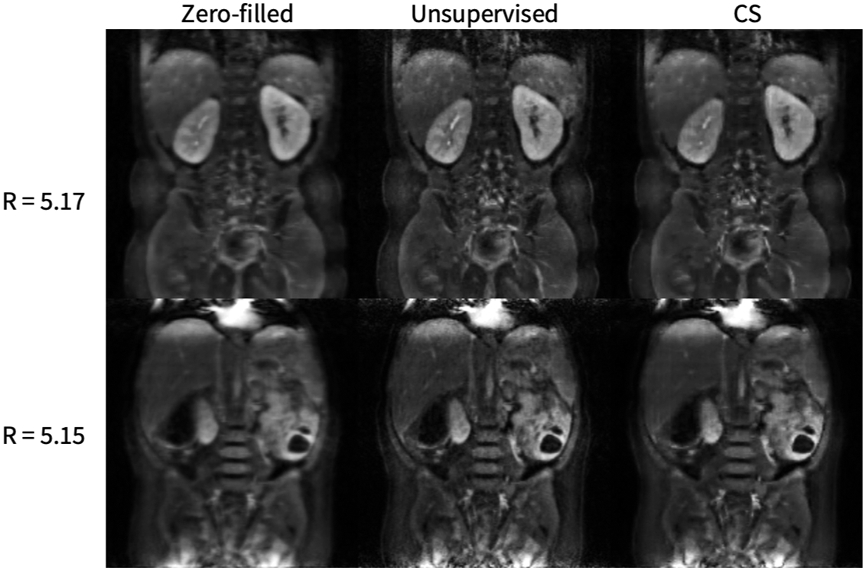
2D DCE application representative results, where the left slice is the magnitude of one input undersampled complex image to the generator, the middle slice is the output of the generator and the right slice is a compressed sensing L1-wavelet regularization. The generator greatly improves the input image quality by recovering sharpness and adding more structure to the input images. Additionally, the proposed method produces a sharper reconstruction compared to CS. In the first row, the kidneys of the unsupervised GAN are visibly much sharper than that of the input and CS.
The main advantage of this method over existing DL reconstruction methods is eliminating the need for fully-sampled data. Another benefit is other additional datasets are not needed to use as ground-truth, as in some other works on semi-supervised training.90 Additionally, the method produces better quality reconstruction compared to baseline compressed sensing methods.
One significant challenge in the implementation of unsupervised methods is ensuring adequate clinical validation. In supervised methods, a ground truth image can be obtained for comparison. In that circumstance, comparison can be by quantitative image quality metrics, such as SNR. However, an open question in the field is whether conventional image quality metrics are adequate to ensure diagnostic performance. Thus, clinical assessment by expert radiologists is critical. The challenge of clinical validation in unsupervised methods is often harder, since the big applications are for cases where a gold standard is impossible to obtain, such as high spatiotemporal resolution DCE or perfusion MRI. In this case, validation would require comparison against either another modality, such as CT, or by assessing clinical outcomes.
Summary
The field of upstream AI for medical imaging is broad, covering the steps in the medical imaging process from selection of an imaging modality to image processing. Potential benefits include a more streamlined clinical practice, faster and more accurate exam scheduling and protocoling, faster exams, and improved image quality. In this article, we have highlighted only a few representative applications, giving the reader insight into directions in this rapidly evolving field.
KEY POINTS.
This article reviews multiple applications of artificial intelligence (AI) in which it has been used to vastly improve upstream components of the medical imaging pipeline.
All medical imaging examinations can be accelerated by using AI to help manage exam schedules and imaging protocols.
In ultrasound, AI has been used to form high-quality images with enhanced robustness to speckle noise.
In computed tomography, AI-based image reconstruction approaches have been used to reconstruct high-quality images from low-dose acquisitions.
In magnetic resonance imaging, AI has been used to design optimal data sampling schemes, and reconstruction approaches to accelerate scan time
SYNOPSIS.
Artificial intelligence (AI) has the potential to drastically improve workflows in radiology at multiple stages of the imaging pipeline. However, most of the attention has been garnered by applications focused on improving the end of the pipeline – image interpretation. This article reviews how AI can be applied to improve upstream aspects of the imaging pipeline as well including modality selection for an exam, hardware design, protocol selection for an exam, data acquisition, image reconstruction and image processing. A breadth of applications and their potential for impact is shown across multiple imaging modalities including ultrasound, computed tomography, and magnetic resonance imaging.
Clinical Care Points.
As of today, there has been little research on validating upstream AI approaches in large clinical cohorts for deep learning-based image reconstruction approaches.
Although these methods demonstrate great potential for accelerating clinical workflows in radiology, further studies are necessary to understand and determine the robustness of such approaches across a variety of patient populations.
Future studies should not only validate upstream AI approaches based on standard image quality metrics, but also based on quantitative measurements which serve as clinical endpoints to ensure that the algorithms are truly improving the standard-of-care.
DISCLOSURE STATEMENT
Dr. Dahl receives technical and in-kind support from Siemens Healthcare and serves as a technical advisor to Cephasonics Ultrasound and Vortex Imaging, Inc. Dr. Chaudhari has provided consulting services to Skope MR, Subtle Medical, Culvert Engineering, Edge Analytics, Image Analysis Group, ICM, and Chondrometrics GmbH; and is a shareholder in Subtle Medical, LVIS Corp. and Brain Key; and received research support from GE Healthcare and Philips. Dr. Hyun has a consulting relationship with Exo Imaging, Inc. Dr. Imran receives research support from GE Healthcare. Dr. Wang receives research support from GE Healthcare, Siemens Healthineers, and Varex Imaging. Elizabeth Cole receives support from GE Healthcare. Drs. Loening, Vasanawala, and Chaudhari receive research support from GE Healthcare. Dr. Vasanawala has a consulting relationship with Arterys and HeartVista.
This research was supported by NIBIB grants R01 EB013661, R01 EB015506, P41 EB015891, R01 EB002524, R01 EB009690, R01 EB026136, and R01 EB027100, by NIAMS grant R01 AR063643, by NCI grant R01-CA218204, by NICHD grant R01-HD086252, by a seed grant from the Stanford Cancer Institute, and a NCI-designated Comprehensive Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Christopher M. Sandino, Department of Electrical Engineering, Stanford University, Stanford, CA.
Elizabeth K. Cole, Department of Electrical Engineering, Stanford University, Stanford, CA.
Cagan Alkan, Department of Electrical Engineering, Stanford University, Stanford, CA.
Akshay S. Chaudhari, Departments of Radiology and Biomedical Data Science, Stanford, CA.
Andreas M. Loening, Department of Radiology, Stanford University, Stanford, CA.
Dongwoon Hyun, Department of Radiology, Stanford University, Stanford, CA.
Jeremy Dahl, Department of Radiology, Stanford University, Stanford, CA.
Abdullah-Al-Zubaer Imran, Radiological Sciences Laboratory, Stanford University, Stanford, CA.
Adam S. Wang, Radiology and Electrical Engineering, PhD, Stanford University, Stanford, CA.
Shreyas S. Vasanawala, Department of Radiology, Stanford University, Stanford, CA.
References
- 1.Chaudhari AS, Sandino CM, Cole EK, et al. Prospective Deployment of Deep Learning in MRI: A Framework for Important Considerations, Challenges, and Recommendations for Best Practices. J Magn Reson Imaging. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sachs PB, Hunt K, Mansoubi F, Borgstede J. CT and MR Protocol Standardization Across a Large Health System: Providing a Consistent Radiologist, Patient, and Referring Provider Experience. J Digit Imaging. 2017;30(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalra A, Chakraborty A, Fine B, Reicher J. Machine Learning for Automation of Radiology Protocols for Quality and Efficiency Improvement. J Am Coll Radiol. 2020;17(9):1149–1158. [DOI] [PubMed] [Google Scholar]
- 4.Lee YH. Efficiency Improvement in a Busy Radiology Practice: Determination of Musculoskeletal Magnetic Resonance Imaging Protocol Using Deep-Learning Convolutional Neural Networks. J Digit Imaging. 2018;31(5):604–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trivedi H, Mesterhazy J, Laguna B, Vu T, Sohn JH. Automatic Determination of the Need for Intravenous Contrast in Musculoskeletal MRI Examinations Using IBM Watson's Natural Language Processing Algorithm. J Digit Imaging. 2018;31(2):245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown AD, Marotta TR. Using machine learning for sequence-level automated MRI protocol selection in neuroradiology. J Am Med Inform Assoc. 2018;25(5):568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devlin J, Chang MW, Lee K, Toutanova K. BERT: Pre-training of deep bidirectional transformers for language understanding. Paper presented at: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies2019; Minneapolis, Minnesota. [Google Scholar]
- 8.Lau W, Aaltonen L, Gunn M, Yetisgen-Yildiz M. Automatic Assignment of Radiology Examination Protocols Using Pre-trained Language Models with Knowledge Distillation. ArXiv. 2020;abs/2009.00694. [PMC free article] [PubMed] [Google Scholar]
- 9.Huang SC, Pareek A, Seyyedi S, Banerjee I, Lungren MP. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. NPJ Digit Med. 2020;3:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinton GF, Trahey GE, Dahl JJ. Sources of image degradation in fundamental and harmonic ultrasound imaging using nonlinear, full-wave simulations. IEEE Trans Ultrason Ferroelectr Freq Control. 2011;58(4):754–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byram B, Dei K, Tierney J, Dumont D. A model and regularization scheme for ultrasonic beamforming clutter reduction. IEEE Trans Ultrason Ferroelectr Freq Control. 2015;62(11):1913–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin J, Huang L. Spatial Prediction Filtering of Acoustic Clutter and Random Noise in Medical Ultrasound Imaging. IEEE Trans Med Imaging. 2017;36(2):396–406. [DOI] [PubMed] [Google Scholar]
- 13.Jennings J, Jakovljevic M, Biondi E, Dahl J, Biondi B. Estimating signal and structured noise in ultrasound data using prediction-error filters. Paper presented at: 2019 SPIE Medical Imaging2019; San Diego, California. [Google Scholar]
- 14.Luchies AC, Byram BC. Deep Neural Networks for Ultrasound Beamforming. IEEE Trans Med Imaging. 2018;37(9):2010–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brickson LL, Hyun D, Jakovljevic M, Dahl JJ. Reverberation Noise Suppression in Ultrasound Channel Signals Using a 3D Fully Convolutional Neural Network. IEEE Trans Med Imaging. 2021;PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyun D, Brickson LL, Looby KT, Dahl JJ. Beamforming and Speckle Reduction Using Neural Networks. IEEE Trans Ultrason Ferroelectr Freq Control. 2019;66(5):898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyun D, Li L, Steinberg I, Jakovljevic M, Klap T, Dahl JJ. An Open Source GPU-Based Beamformer for Real-Time Ultrasound Imaging and Applications. Paper presented at: 2019 IEEE International Ultrasonics Symposium (IUS) 2019; Glasgow, United Kingdom. [Google Scholar]
- 18.Hyun D, Abou-Elkacem L, Bam R, Brickson LL, Herickhoff CD, Dahl JJ. Nondestructive Detection of Targeted Microbubbles Using Dual-Mode Data and Deep Learning for Real-Time Ultrasound Molecular Imaging. IEEE Trans Med Imaging. 2020;39(10):3079–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luijten B, Cohen R, de Bruijn FJ, et al. Adaptive Ultrasound Beamforming Using Deep Learning. IEEE Trans Med Imaging. 2020;39(12):3967–3978. [DOI] [PubMed] [Google Scholar]
- 20.van Sloun RJ, Solomon O, Bruce M, et al. Super-resolution Ultrasound Localization Microscopy through Deep Learning. IEEE Trans Med Imaging. 2020;PP. [DOI] [PubMed] [Google Scholar]
- 21.Nair AA, Washington KN, Tran TD, Reiter A, Lediju Bell MA. Deep Learning to Obtain Simultaneous Image and Segmentation Outputs From a Single Input of Raw Ultrasound Channel Data. IEEE Trans Ultrason Ferroelectr Freq Control. 2020;67(12):2493–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanziola A, Robins T, Riemer K, M. T. A Deep Learning Approach to Synthetic Aperture Vector Flow Imaging. Paper presented at: 2018 IEEE International Ultrasonics Symposium (IUS)2018; Kobe, Japan. [Google Scholar]
- 23.Li Y, Hyun D, Dahl JJ. Vector Flow Velocity Estimation from Beamsummed Data Using Deep Neural Networks. Paper presented at: 2019 IEEE International Ultrasonics Symposium (IUS)2019; Glasgow, United Kingdom. [Google Scholar]
- 24.van Sloun RJ, Cohen R, Eldar YC. Deep Learning in Ultrasound Imaging. in Proceedings of the IEEE. 2020;108(1):11–29. [Google Scholar]
- 25.Over 75 Million CT Scans Are Performed Each Year and Growing Despite Radiation Concerns. https://idataresearch.com/over-75-million-ct-scans-are-performed-each-year-and-growing-despite-radiation-concerns. Published 2018. Accessed January 4, 2021.
- 26.Larson DB, Boland GW. Imaging Quality Control in the Era of Artificial Intelligence. J Am Coll Radiol. 2019;16(9 Pt B):1259–1266. [DOI] [PubMed] [Google Scholar]
- 27.Lell MM, Kachelriess M. Recent and Upcoming Technological Developments in Computed Tomography: High Speed, Low Dose, Deep Learning, Multienergy. Invest Radiol. 2020;55(1):8–19. [DOI] [PubMed] [Google Scholar]
- 28.Meineke A, Rubbert C, Sawicki LM, et al. Potential of a machine-learning model for dose optimization in CT quality assurance. Eur Radiol. 2019;29(7):3705–3713. [DOI] [PubMed] [Google Scholar]
- 29.McCollough CH, Bruesewitz MR, Kofler JM Jr., CT dose reduction and dose management tools: overview of available options. Radiographics. 2006;26(2):503–512. [DOI] [PubMed] [Google Scholar]
- 30.ICRP. The 2007 Recommendations of the International Commission on Radiological Protection. 2007.
- 31.Willemink MJ, Noel PB. The evolution of image reconstruction for CT-from filtered back projection to artificial intelligence. Eur Radiol. 2019;29(5):2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eberhard M, Alkadhi H. Machine Learning and Deep Neural Networks: Applications in Patient and Scan Preparation, Contrast Medium, and Radiation Dose Optimization. J Thorac Imaging. 2020;35 Suppl 1:S17–S20. [DOI] [PubMed] [Google Scholar]
- 33.Tian C, Fei L, Zheng W, Xu Y, Zuo W, Lin CW. Deep learning on image denoising: An overview. Neural Netw. 2020;131:251–275. [DOI] [PubMed] [Google Scholar]
- 34.Kambadakone A Artificial Intelligence and CT Image Reconstruction: Potential of a New Era in Radiation Dose Reduction. J Am Coll Radiol. 2020;17(5):649–651. [DOI] [PubMed] [Google Scholar]
- 35.Liu SZ, Cao Q, Tivnan M, et al. Model-based dual-energy tomographic image reconstruction of objects containing known metal components. Phys Med Biol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katsura M, Sato J, Akahane M, Kunimatsu A, Abe O. Current and Novel Techniques for Metal Artifact Reduction at CT: Practical Guide for Radiologists. Radiographics. 2018;38(2):450–461. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Yu H. Convolutional Neural Network Based Metal Artifact Reduction in X-Ray Computed Tomography. IEEE Trans Med Imaging. 2018;37(6):1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akagi M, Nakamura Y, Higaki T, et al. Deep learning reconstruction improves image quality of abdominal ultra-high-resolution CT. Eur Radiol. 2019;29(11):6163–6171. [DOI] [PubMed] [Google Scholar]
- 39.Greffier J, Hamard A, Pereira F, et al. Image quality and dose reduction opportunity of deep learning image reconstruction algorithm for CT: a phantom study. Eur Radiol. 2020;30(7):3951–3959. [DOI] [PubMed] [Google Scholar]
- 40.Kaza RK, Platt JF, Goodsitt MM, et al. Emerging techniques for dose optimization in abdominal CT. Radiographics. 2014;34(1):4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mittal A, Moorthy AK, Bovik AC. No-reference image quality assessment in the spatial domain. IEEE Trans Image Process. 2012;21(12):4695–4708. [DOI] [PubMed] [Google Scholar]
- 42.Patwari M, Gutjahr R, Raupach R, Maier A. Measuring CT Reconstruction Quality with Deep Convolutional Neural Networks. 2019; Cham. [Google Scholar]
- 43.Kim J, Nguyen AD, Lee S. Deep CNN-Based Blind Image Quality Predictor. IEEE Trans Neural Netw Learn Syst. 2019;30(1):11–24. [DOI] [PubMed] [Google Scholar]
- 44.Imran AAZ, Pal D, Patel B, Wang A. SSIQA: Multi-task learning for non-reference CT image quality assessment with self-supervised noise level prediction. Paper presented at: International Symposium on Biomedical Imaging (ISBI)2021. [Google Scholar]
- 45.Bahadir CD, Dalca AV, Sabuncu MR. Learning-Based Optimization of the Under-Sampling Pattern in MRI. Paper presented at: International Conference on Information Processing in Medical Imaging2019; Hong Kong, China. [Google Scholar]
- 46.Huijben IAM, Veeling BS, van Sloun RJ. Learning Sampling and Model-Based Signal Recovery for Compressed Sensing MRI. Paper presented at: 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) 2020; Barcelona, Spain. [Google Scholar]
- 47.Aggarwal HK, Jacob M. J-MoDL: Joint Model-Based Deep Learning for Optimized Sampling and Reconstruction. IEEE Journal of Selected Topics in Signal Processing. 2020;14(6):1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss T, Senouf O, Vedula S, Michailovich O, Zibulevsky M, Bronstein A. PILOT: Physics-Informed Learned Optimal Trajectories for Accelerated MRI. ArXiv. 2019;abs/1909.05773. [Google Scholar]
- 49.Bakker T, Hoof HV, Welling M. Experimental design for MRI by greedy policy search. ArXiv. 2020;abs/2010.16262. [Google Scholar]
- 50.Jin K, Unser M, Yi K. Self-Supervised Deep Active Accelerated MRI. ArXiv. 2019;abs/1901.04547. [Google Scholar]
- 51.Pineda L, Basu S, Romero A, Calandra R, Drozdzal M. Active MR k-space Sampling with Reinforcement Learning. Paper presented at: Medical Image Computing and Computer-Assisted Intervention2020; Lima, Peru. [Google Scholar]
- 52.Zhang Z, Romero A, Muckley M, Vincent P, Yang L, Drozdzal M. Reducing Uncertainty in Undersampled MRI Reconstruction With Active Acquisition. Paper presented at: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)2019; Long Beach, CA. [Google Scholar]
- 53.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–1195. [DOI] [PubMed] [Google Scholar]
- 54.Vasanawala SS, Alley MT, Hargreaves BA, Barth RA, Pauly JM, Lustig M. Improved pediatric MR imaging with compressed sensing. Radiology. 2010;256(2):607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin KH, McCann MT, Froustey E, Unser M. Deep Convolutional Neural Network for Inverse Problems in Imaging. IEEE Trans Image Process. 2017;26(9):4509–4522. [DOI] [PubMed] [Google Scholar]
- 56.Mardani M, Gong E, Cheng JY, et al. Deep Generative Adversarial Neural Networks for Compressive Sensing MRI. IEEE Trans Med Imaging. 2019;38(1):167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee D, Yoo J, Tak S, Ye JC. Deep Residual Learning for Accelerated MRI Using Magnitude and Phase Networks. IEEE Trans Biomed Eng. 2018;65(9):1985–1995. [DOI] [PubMed] [Google Scholar]
- 58.Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med. 2018;79(6):3055–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aggarwal HK, Mani MP, Jacob M. MoDL: Model-Based Deep Learning Architecture for Inverse Problems. IEEE Trans Med Imaging. 2019;38(2):394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sandino CM, Cheng JY, Chen F, Mardani M, Pauly JM, Vasanawala SS. Compressed Sensing: From Research to Clinical Practice with Deep Neural Networks. IEEE Signal Process Mag. 2020;37(1):111–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandino CM, Lai P, Vasanawala SS, Cheng JY. Accelerating cardiac cine MRI using a deep learning-based ESPIRiT reconstruction. Magn Reson Med. 2021;85(1):152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chaudhari AS, Stevens KJ, Wood JP, et al. Utility of deep learning super-resolution in the context of osteoarthritis MRI biomarkers. J Magn Reson Imaging. 2020;51(3):768–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chaudhari AS, Fang Z, Hyung Lee J, Gold GE, Hargreaves BA. Deep Learning Super-Resolution Enables Rapid Simultaneous Morphological and Quantitative Magnetic Resonance Imaging. Paper presented at: Machine Learning for Medical Image Reconstruction2018; Granada, Spain. [Google Scholar]
- 64.Chaudhari AS, Grissom MJ, Fang Z, et al. Diagnostic Accuracy of Quantitative Multi-Contrast 5-Minute Knee MRI Using Prospective Artificial Intelligence Image Quality Enhancement. AJR Am J Roentgenol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian Q, Bilgic B, Fan Q, et al. Improving in vivo human cerebral cortical surface reconstruction using data-driven super-resolution. Cereb Cortex. 2021;31(1):463–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Y, Xie Y, Zhou Z, Shi F, Christodoulou A, Li D. Brain MRI super resolution using 3D deep densely connected neural networks. Paper presented at: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) 2018; Washington, DC. [Google Scholar]
- 67.Oktay O, Bai W, Lee M, et al. Multi-input Cardiac Image Super-Resolution Using Convolutional Neural Networks. Paper presented at: International Conference on Medical Image Computing and Computer-Assisted Intervention2016; Athens, Greece. [Google Scholar]
- 68.Chaudhari AS, Fang Z, Kogan F, et al. Super-resolution musculoskeletal MRI using deep learning. Magn Reson Med. 2018;80(5):2139–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cai C, Wang C, Zeng Y, et al. Single-shot T2 mapping using overlapping-echo detachment planar imaging and a deep convolutional neural network. Magn Reson Med. 2018;80(5):2202–2214. [DOI] [PubMed] [Google Scholar]
- 70.Bollmann S, Rasmussen KGB, Kristensen M, et al. DeepQSM - using deep learning to solve the dipole inversion for quantitative susceptibility mapping. Neuroimage. 2019;195:373–383. [DOI] [PubMed] [Google Scholar]
- 71.Gibbons EK, Hodgson KK, Chaudhari AS, et al. Simultaneous NODDI and GFA parameter map generation from subsampled q-space imaging using deep learning. Magn Reson Med. 2019;81(4):2399–2411. [DOI] [PubMed] [Google Scholar]
- 72.Chen F, Taviani V, Malkiel I, et al. Variable-Density Single-Shot Fast Spin-Echo MRI with Deep Learning Reconstruction by Using Variational Networks. Radiology. 2018;289(2):366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang G, Yu S, Dong H, et al. DAGAN: Deep De-Aliasing Generative Adversarial Networks for Fast Compressed Sensing MRI Reconstruction. IEEE Trans Med Imaging. 2018;37(6):1310–1321. [DOI] [PubMed] [Google Scholar]
- 74.Diamond S, Sitzmann V, Heide F, Wetzstein G. Unrolled Optimization with Deep Priors. ArXiv. 2017;abs/1705.08041. [Google Scholar]
- 75.Cheng JY, Chen F, Alley M, Pauly J, Vasanawala S. Highly Scalable Image Reconstruction using Deep Neural Networks with Bandpass Filtering. ArXiv. 2018;abs/1805.03300. [Google Scholar]
- 76.Souza R, Lebel RM, Frayne R. A Hybrid, Dual Domain, Cascade of Convolutional Neural Networks for Magnetic Resonance Image Reconstruction. Proceedings of The 2nd International Conference on Medical Imaging with Deep Learning; 2019; London, England. [Google Scholar]
- 77.Eo T, Jun Y, Kim T, Jang J, Lee HJ, Hwang D. KIKI-net: cross-domain convolutional neural networks for reconstructing undersampled magnetic resonance images. Magn Reson Med. 2018;80(5):2188–2201. [DOI] [PubMed] [Google Scholar]
- 78.Cole E, Cheng JY, Pauly J, Vasanawala S. Analysis of Complex-Valued Convolutional Neural Networks for MRI Reconstruction. ArXiv. 2020;abs/2004.01738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tamir JI, Yu S, Lustig M. Unsupervised Deep Basis Pursuit: Learning inverse problems without ground-truth data. Paper presented at: International Society for Magnetic Resonance in Medicine 27th Annual Meeting & Exhibition 2019; Montreal, Quebec, Canada. [Google Scholar]
- 80.Zhussip M, Soltanayev S, Chun S. Training Deep Learning Based Image Denoisers From Undersampled Measurements Without Ground Truth and Without Image Prior. Paper presented at: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2019; Long Beach, CA. [Google Scholar]
- 81.Lehtinen J, Munkberg J, Hasselgren J, et al. Noise2Noise: Learning Image Restoration without Clean Data. Proceedings of the 35th International Conference on Machine Learning; 2018. [Google Scholar]
- 82.Soltanayev S, Chun SY. Training deep learning based denoisers without ground truth data. 32nd Conference on Neural Information Processing Systems(NeurIPS); 2018; Montreal, Canada. [Google Scholar]
- 83.Yaman B, Hosseini SAH, Moeller S, Ellermann J, Ugurbil K, AkÁakaya M. Self-Supervised Physics-Based Deep Learning MRI Reconstruction Without Fully-Sampled Data. Paper presented at: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) 2020; Iowa City, IA, USA. [Google Scholar]
- 84.Chen F, Cheng JY, Pauly JM, Vasanawala SS. Semi-Supervised Learning for Reconstructing Under-Sampled MR Scans. Paper presented at: International Society for Magnetic Resonance in Medicine 27th Annual Meeting & Exhibition 2019; Montreal, Quebec, Canada. [Google Scholar]
- 85.Wu Z, Xiong Y, Yu S, Lin D. Unsupervised Feature Learning via Non-Parametric Instance-level Discrimination. Paper presented at: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition2018; Salt Lake City, UT, USA. [Google Scholar]
- 86.Sim B, Oh G, Ye JC. Optimal Transport Structure of CycleGAN for Unsupervised Learning for Inverse Problems. Paper presented at: 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP)2020; Barcelona, Spain. [Google Scholar]
- 87.Goodfellow IJ, Pouget-Abadie J, Mirza M, et al. Generative Adversarial Nets. 28th Conference on Neural Information Processing Systems; 2014; Montreal, Canada. [Google Scholar]
- 88.Zhu JY, Krähenbühl P, Shechtman E, Efros AA. Generative Visual Manipulation on the Natural Image Manifold. Paper presented at: Computer Vision – ECCV 20162016; Amsterdam, The Netherlands. [Google Scholar]
- 89.Radford A, Metz L, Chintala S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. 4th International Conference on Learning Representations; 2016; San Juan, Puerto Rico. [Google Scholar]
- 90.Lei K, Mardani M, Pauly J, Vasanawala S. Wasserstein GANs for MR Imaging: From Paired to Unpaired Training. IEEE Transactions on Medical Imaging. 2021;40(1):105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bora A, Price E, Dimakis A. AmbientGAN: Generative models from lossy measurements. Paper presented at: 6th International Conference on Learning Representations2018; Vancouver, Canada. [Google Scholar]
- 92.Cole EK, Pauly J, Vasanawala S, Ong F. Unsupervised MRI Reconstruction with Generative Adversarial Networks. ArXiv. 2020;abs/2008.13065. [Google Scholar]
- 93.Chen B, Duan X, Yu Z, Leng S, Yu L, McCollough C. Technical Note: Development and validation of an open data format for CT projection data. Med Phys. 2015;42(12):6964–6972. [DOI] [PMC free article] [PubMed] [Google Scholar]