Abstract
Phylogenetic, developmental, and brain-imaging studies suggest that human personality is the integrated expression of three major systems of learning and memory that regulate (1) associative conditioning, (2) intentionality, and (3) self-awareness. We have uncovered largely disjoint sets of genes regulating these dissociable learning processes in different clusters of people with (1) unregulated temperament profiles (i.e., associatively conditioned habits and emotional reactivity), (2) organized character profiles (i.e., intentional self-control of emotional conflicts and goals), and (3) creative character profiles (i.e., self-aware appraisal of values and theories), respectively. However, little is known about how these temperament and character components of personality are jointly organized and develop in an integrated manner. In three large independent genome-wide association studies from Finland, Germany, and Korea, we used a data-driven machine learning method to uncover joint phenotypic networks of temperament and character and also the genetic networks with which they are associated. We found three clusters of similar numbers of people with distinct combinations of temperament and character profiles. Their associated genetic and environmental networks were largely disjoint, and differentially related to distinct forms of learning and memory. Of the 972 genes that mapped to the three phenotypic networks, 72% were unique to a single network. The findings in the Finnish discovery sample were blindly and independently replicated in samples of Germans and Koreans. We conclude that temperament and character are integrated within three disjoint networks that regulate healthy longevity and dissociable systems of learning and memory by nearly disjoint sets of genetic and environmental influences.
Subject terms: Genetics, Neuroscience, Psychology
Introduction
An individual’s unique pattern of behaviors, feelings, and thoughts is the expression of his or her personality, which is a strong predictor of the physical, mental, and social aspects of current and future health across the lifespan [1–3]. Personality is defined briefly as the way a person learns to adapt to experience, or, more specifically, as the dynamic organization within the individual of the psychobiological systems by which a person both shapes and adapts uniquely to an ever-changing internal and external environment [4].
Different genetic and neurobiological systems are involved in regulating distinct aspects of personality, which are traditionally described as temperament and character [5–8]. Temperament refers to innate biological predispositions that influence automatic emotional reactivity and habits; it is moderately stable throughout the lifespan, but can develop with aging and behavioral conditioning [1, 9, 10]. Put another way, temperament refer to the form or style of automatic behavior: How do you act and express yourself spontaneously [11, 12]? In contrast, character refers to mental self-government (i.e., what people make of themselves intentionally and creatively) [13], which develops in a saltatory manner throughout life [6, 14]. Learning to self-govern can be further partitioned by the basic questions that distinguish self-control and self-actualization. For self-control, we must answer the question “What do you intend to do?”, which involves the executive (intrapersonal) and legislative (interpersonal) functions of intentional self-control based on more-or-less logical calculation and analysis of personal goals, facts, beliefs, and social conventions. For self-actualization, we must answer a question about meaning in a transpersonal context: “Why, Where, When are you going to do it?”, which involves the judicial (transpersonal) functions of self-aware evaluation and intuitive appraisal of values and relationships as a theory or narrative with contextual insight into its meaning (why) at a particular place (where) and time (when) [4, 15]. Thus the architecture of human personality may correspond to the structure of human learning with its distinct systems for procedural, semantic, and self-aware learning and memory [16, 17]. Unfortunately, remarkably little is known about how the underlying genetic and environmental influences for these dissociable systems are integrated to express the complex phenotypes that we recognize as the self-organized profiles that describe personality and learning.
Human personality provides a highly instructive example of the challenges that must be faced in efforts to identify the molecular mechanisms involved in the causes and development of complex phenotypes [18–20]. In previous work, we found that the genetic variants associated with personality do not operate independently; rather they are organized as clusters of particular single-nucleotide polymorphisms (SNPs) that co-occur in subgroups of subjects, which we call SNP sets [18–20]. The SNP sets were each comprised of SNPs in many coding and noncoding genes that are distributed throughout the genome [21, 22], presumably due to the need for multiple genes to act in concert, not independently [20]. In general, geneticists must expect that each gene affects many traits and many genes affect each trait because evolutionary selection operates on whole organisms, not individual genes or traits [23]. Our specific findings confirmed consistent prior evidence from studies of twins and their families and from genome-wide association studies [24] that many genes act in concert with one another to influence human personality [25–28]. Thus the high heritability of personality expected from twin studies [25–28] was not missing, but was distributed into several disjoint components in which the subjects had distinct genotypic and phenotypic features [18–20].
Specifically, we found three clusters of people who were distinguished by heritable configurations of the temperament traits related to individual differences in procedural learning of habits and skills [19]. The three clusters were also characterized by individual differences in automatic behavioral activity and emotional expression, and corresponded closely to temperament clusters described as “easy”, “difficult”, and “slow to warm-up” [8, 11, 29] (Supplementary Information, see Tables S1, S2, and reference [8] for review). People in our “reliable” cluster resembled children with an “easy temperament” and adults who were conscientious extraverts because they were well controlled in activity and were warm and calm emotionally; put another way, they were high in Reward Dependence (i.e., sentimental, friendly, and approval seeking), low in Novelty Seeking (i.e., deliberate, thrifty, and orderly), low in Harm Avoidance (i.e., optimistic, confident, outgoing, and vigorous), and high in Persistence (i.e., determined). People in our “sensitive” temperament cluster resembled children with a “difficult temperament” and adults who are neurotic and unstable because they were under-controlled in activity and emotionally hypersensitive: in other words, they were high in Harm Avoidance (i.e., pessimistic, fearful, shy, and fatigable), high in Novelty Seeking (i.e., impulsive and extravagant), and high in Reward Dependence (i.e., sentimental and friendly), so they frequently had approach-avoidance conflicts, rejection sensitivity, and disorganized attachments. People in our “antisocial” temperament cluster resembled children with a “slow to warm-up” temperament and adults who are socially detached, careless, and impulsive: that is, they were low in Reward Dependence (i.e., cold, detached, and independent), low in Persistence (i.e., easily discouraged), and high in Novelty Seeking (i.e., extravagant, rule-breaking but not inquisitive), which is frequently associated with maladaptive antisocial conduct. Furthermore, the genes associated with each of these three temperament profiles were largely unique to that profile.
Likewise, we identified five heritable clusters of people with distinct profiles of character traits involving various combinations of high versus low scores on Self-directedness, Cooperativeness, and Self-transcendence [18]. Three of these profiles were usually associated with physical, mental, and social well-being, and were described as resourceful (i.e., high in Self-directedness only), organized (i.e., high in both Self-directedness and Cooperativeness, but not Self-transcendence), or creative (i.e., high in all three character traits). The other two were usually associated with poor physical, mental, and social functioning, and were described as “dependent” (high in Cooperativeness but low in Self-directedness and Self-transcendence) or “apathetic” (low in all three character traits). As with temperament, the genes associated with each of the five character profiles were largely different.
We found that personality depends on sets of genes that regulate and coordinate the dynamic functions required for people to learn to adapt to changing circumstances, including molecular processes for neurodevelopment, neuroplasticity, neurogenesis, neurotransmission, stress reactivity, energy metabolism, neuroprotection, resilience, and healthy longevity [18, 19]. The genes we found to be associated with personality were nearly always expressed in the brain, but these brain functions depended on interactions with variability in genes regulating pathways that are particularly important in brain but involve general housekeeping functions that occur in most or all cell types, such as the regulation of energy metabolism, circadian rhythmicity, and cellular repair [30, 31].
Furthermore, we found that the genes that encode variability in human temperament are enriched in highly conserved molecular pathways, the Ras-MEK-ERK and PI3K-AKT-mTOR pathways, which are activated in experimental animals by stress reactivity and associative conditioning (i.e., psychobiological system 1 for classical and operant conditioning) [32–35]. For example, the responses of these pathways to patterns of reward and punishment regulate neuroplasticity in the striatum, thereby modulating the integration of frontocortical and mesolimbic signaling during associative conditioning in humans and other amniotes (i.e., reptiles, birds, and mammals) (see Supplementary Information, Vignette 1) [36–38].
In contrast, the genes we found encoding human character are associated with two brain systems for higher cognitive processes involving intentional self-control or self-awareness [16, 17, 39]. Specifically, a second network (psychobiological system 2) involves specialized bipolar neurons in the anterior insular cortex, frontal operculum, and anterior cingulate cortex that are present in great apes and humans, but not in other primates [40, 41]. These Von Economo neurons are functionally connected to temporal and parietal neocortical regions in brain circuits that support saliency detection, resolution of emotional conflicts, and social cooperation for mutual benefit in great apes and humans [40–43]. This system also supports intentional self-control of voluntary behavior and purposeful use of symbols with further development of the inferior parietal cortex as a convergence area for touch, hearing, and vision in humans (see Supplementary Information, Vignette 2) [40, 44]. Another network (psychobiological system 3) involves regions of late-myelinating neocortex in frontal, parietal, and temporal regions found only in humans, and is associated with the emergence of human capacities for self-awareness, insight (i.e., immediate, accurate, and deep intuitive understanding), creative imagination, altruism, and autobiographical memory (see Supplementary Information, Vignette 3) [40, 44–46]. These three brain networks normally interact in a coordinated manner [47–49], but they are dissociable developmentally [17, 46, 50] and functionally [17, 48, 49, 51–54].
Our previous findings also suggest the hypothesis that different molecular processes may regulate associative learning, intentionality, and self-awareness. Specifically, we uncovered largely disjoint sets of genes regulating these three distinct learning processes in different clusters of people with unhealthy temperament profiles (network 1 with associative conditioning), organized character profiles (network 2 with intentionality), and creative character profiles (network 3 with self-awareness), respectively [18, 19]. However, little is known about the molecular processes by which temperament and character are organized into integrated networks, how such integrated networks are associated with specific genetic and environmental influences, or how crucial such integration is for health and well-being.
Our computational approach to genome-wide association study (GWAS) is an extension of the efforts of many geneticists to analyze GWAS in terms of sets of multiple markers that are significantly and reproducibly related to complex phenotypes, rather than single markers that neglect the complex interactions among multiple variables [18, 19, 21], or polygenic risk scores that depend on multiple markers neither significantly nor consistently related to the phenotype [55–58] (See Supplementary Information for a review). There are a wide range of possible models that may account for the missing heritability in GWAS by consideration of interactions among multiple genetic, cultural, and environmental variables [59–62]. At one extreme, polygenic risk scores are computed as the sum of the effects of many genes acting independently in a linear regression model [20]. In contrast, the omnigenic model assumes a more structured architecture in which there are core genes with large effects on phenotype [63, 64], as well as peripheral genes distributed throughout the genome with weak individual effects [65].
Rather than assume any particular model, in this study we have used unsupervised learning to uncover the genetic, environmental, and phenotypic architecture of complex personality clusters and then replicate our findings in independent samples from different cultures. As in our prior work [18, 19], we use data-driven methods to uncover the joint organization of deep networks of temperament and character profiles, as well as the genetic and environmental architecture of their joint relationship. We hypothesize that the genetic networks associated with the joint relations of temperament and character networks may reflect the three different systems of learning and memory that emerged separately in the long phylogeny of human beings as incrementally more conscious and flexible ways of adapting to changing external environmental conditions and internal aspirations [40]. We test this hypothesis in a data-driven analysis so that our hypotheses do not bias or restrict our results.
Subjects and methods
Description of the samples
Our discovery sample was the Young Finns Study, an epidemiological study of 2149 healthy Finnish subjects. All subjects had thorough standardized genotypic, environmental, and phenotypic assessments, including administration of the Temperament and Character Inventory (TCI) [10, 66]. We replicated the results in two independent samples of 902 healthy adults from Germany [67] and 1052 from Korea [68, 69] in which comparable genotypic and phenotypic features were available [18, 19].
Personality assessment
All subjects completed the TCI to assess seven heritable dimensions of personality [13] (see Supplementary Information, Table S1).
Personality health indices
People at risk of poor adaptive functioning were identified as the bottom decile of the sum of TCI Self-directedness and Cooperativeness [3]. In contrast, people with strong adaptive functioning were identified as the top decile of the product of all three TCI character traits, as in prior work [3, 70]. We confirmed the validity of these indicators of health in our discovery sample with independent measures of positive affect balance, perceived social support, physical behaviors (exercise, smoking, and diet), and objective laboratory findings for ideal health recommended by the American Heart Association [71–73] (Supplementary Tables S3A, B). Our indices provided a consistent measure of health status in all three samples.
Environmental assessments
The environmental variables measured for the Finns included reports by the main caretaker (usually the mother) of parental tolerance (i.e., acceptance of the child), emotional warmth (i.e., nurturance of the child), strict disciplinary style, as well as the parent’s education, income, and the family’s urban/rural residency during childhood in 1980 and 1983 [18, 19], and stressful life events and urban versus rural residency during adulthood in 2001 [18, 19]. Comparable environmental data about the German and Korean subjects were not available.
Computational procedures
Our machine-learning approach [74, 75] uses the nonnegative matrix factorization (NMF) method, which identifies multidimensional patterns within different types of data. Our deep unsupervised NMF process uncovers naturally occurring associations between patterns across different types of data. This algorithm is described in Supplementary Fig. S1 and elsewhere [18, 19].
Our web server application for phenotype–genotype many-to-many relations analysis (PGMRA) in GWAS is published [76] and online at http://phop.ugr.es/fenogeno.
Replications
PGMRA was used to uncover the complex genotypic–phenotypic associations in two replication samples (Germans and Koreans) independent of information about the discovery sample. The process used in the discovery sample was blindly and independently repeated in each replication sample without assuming homogeneity within or across samples [20]. We accounted for ethnicity in each sample by using the first three principal components for ancestral stratification of SNP genotypes, as in prior work [18, 19]. Then matching of genotypic–phenotypic associations across samples was identified using parsimonious models that balance accuracy with model complexity, thereby avoiding overfitting [77]. Models were learned independently in diverse samples to provide a stringent test of reproducibility despite complexity that might result from possible genetic, ethnic, cultural, and environmental heterogeneity [18–20].
Results
Identifying phenotypic networks of temperament and character
We identified 44 fine-grained sets of subjects with distinct configurations of TCI character subscales regardless of their temperament (i.e., character sets or biclusters) using PGMRA, as we have previously reported [18]. The character sets varied in personality features (TCI subscales), numbers of subjects, and health status (well- or ill-being, Supplementary Table S4). The character sets were grouped into five deep clusters (i.e., character profiles) by recurrently applying PGMRA for convolutive NMF, minimizing the Cophenetic correlation coefficient [18]. These five clusters were labeled as Resourceful (i.e., self-directed only), Organized (i.e., self-directed and cooperative), Creative (i.e. self-directed, cooperative, and self-transcendent), Dependent (i.e., cooperative only), or Apathetic (low in all three character scales) using traditional labels [18] (Supplementary Table S1).
Likewise we identified 55 fine-grained sets of subjects with distinct configurations of TCI temperament subscales (i.e., temperament sets) using PGMRA, as we have previously reported [19] (Supplementary Table S4). The temperament sets were grouped in three deep clusters (i.e., temperament profiles) by recurrently applying PGMRA for convolutive NMF, minimizing the Cophenetic correlation coefficient [19]. These distinct temperament profiles were labeled as Reliable (i.e., low in Novelty Seeking and Harm Avoidance, high in Reward Dependence and Persistence), Sensitive (high in Harm Avoidance, Novelty Seeking, and Reward Dependence), or Antisocial (High in Novelty Seeking, and low in Reward Dependence and Persistence) using traditional labels [19].
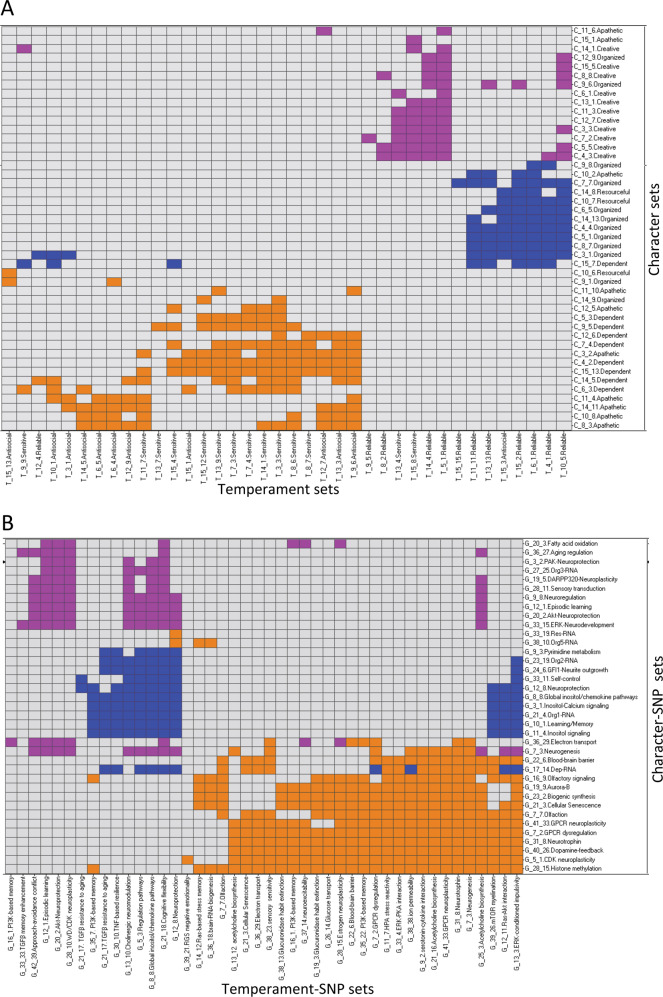
Next we tested for associations among the temperament and character sets sharing the same individuals. We found 265 deep phenotypic associations among 39 temperament sets and 46 character sets by recurrently applying PGMRA for convolutive learning. These associations were statistically significant by Fisher’s exact test (5E−14 < p < 1E−05, Supplementary Table S4 and Fig. S1) and by a permutation test (empirical p < 5E−04). To assess the phenotypic architecture of these relationships, we identified subgroups of subjects with distinct joint temperament–character features using PGMRA. These associations were organized as three deep phenotypic networks that were nearly disjoint (i.e., shared few subjects between different temperament–character associations), as shown in Fig. 1a.
Fig. 1.
The phenotypic architecture of personality. a Relationships among Temperament and Character Sets are naturally partitioned into three subnetworks using NMF (bidirectional-clustering) techniques: Creative-Reliable (violet), Organized-Reliable (blue), and Emotional-Unreliable (orange). b Relationships among SNP sets associated with Temperament and Character Sets composing the three networks shown in a: self-awareness (violet), self-control (blue), and emotional reactivity (orange)
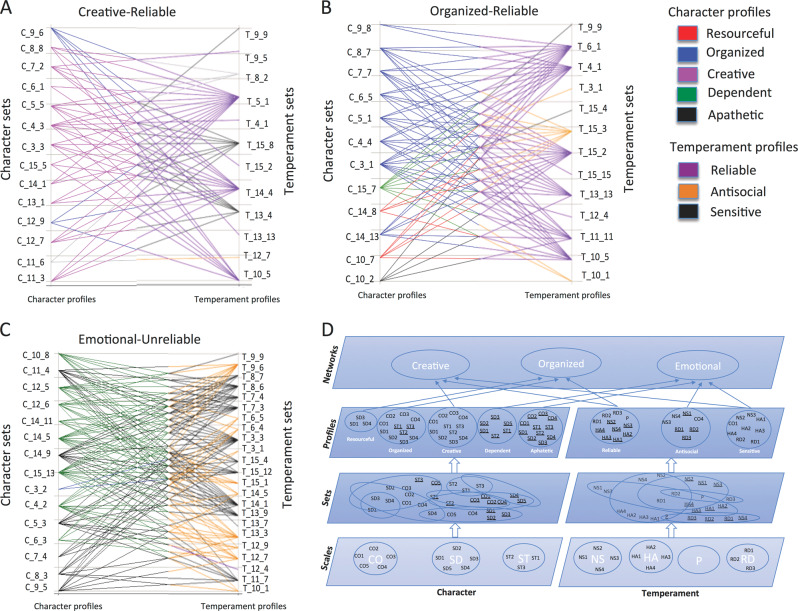
Despite the three phenotypic networks being nearly disjoint from one another, the associations among the temperament and character sets within each network were highly complex. In other words, within each network the same character set was associated with multiple temperament sets, and different character sets could be associated with the same temperament set (Fig. 2). The networks are represented at different levels of granularity in Fig. 2d. In order to make terminology clear, the deep organization of the personality networks is displayed in Fig. 2d as a hierarchy of deep descriptive complexity ascending from (i) separate scales and subscales of temperament and of character, (ii) sets of multiple subscales of temperament and of character, (iii) profiles of temperament sets and of character sets, and (iv) joint networks of temperament and character profiles.
Fig. 2.
Relationships among Temperament and Character Sets composing the three networks shown in Fig. 1. a The Creative-Reliable subnetwork (light violet, violet) contains primarily Character Sets with creative subjects, who also display a reliable temperament. b The Organized-Reliable subnetwork (blue, violet) primarily contains Character Sets with organized subjects, who also display reliable temperament. c The Emotional-Unreliable subnetwork (green, black, and orange) contains Character Sets with dependent and apathetic subjects, who also display sensitive and antisocial temperaments. d The hierarchical organization of personality: temperament and character scales and subscales, sets of subscales (see Table S3), profiles, and networks
Specifically, the healthiest phenotypic network is primarily comprised of subjects with a Creative character profile associated with a Reliable temperament profile (66% of 56 character–temperament set combinations) (see Figs. 1a and 2a, Supplementary Fig. S2A, D). Therefore, we called it the Creative-Reliable network, but it also includes some subjects with Organized characters (2 of 14 character sets in network) and/or Sensitive temperaments (3 of 12 temperament sets in network) as a result of the complex relations within the network (see Fig. 2a). The second phenotypic network was largely comprised of individuals with an Organized character profile associated with a Reliable temperament profile (72% of 72 character–temperament set combinations) (see Figs. 1a and 2b, Supplementary Fig. S2B, D). It was named the Organized-Reliable network but included some individuals with a Resourceful character (2 of 12 character sets in network) and/or antisocial temperament (3 of 13 temperament sets in network) (see Fig. 2b). The least healthy phenotypic network was named the Emotional-Unreliable network because it was comprised of emotionally reactive, injudicious, and maladapted individuals with Dependent characters (8 of 15 character sets in network) or Apathetic characters (6 of 15 characters sets in network) associated with Sensitive temperaments (12 of 23 temperament sets in network) or Antisocial temperaments (11 of 23 temperament sets in network) (see Figs. 1a and 2c, Supplementary Fig. S2C, D). Both the dependent and apathetic character profiles were usually combined with the sensitive temperament: 82% of 72 combinations for the dependent profile, and 58% of 45 combinations for the apathetic character profile (see Fig. 2c).
The three networks were similar in terms of numbers of constituent sets and subjects: there were 674, 801, and 603 subjects for networks 1 (emotional-unreliable), 2 (organized-reliable), and 3 (creative-reliable), respectively (Supplementary Table S5). In total 4.6% of subjects were not assigned to a network because they did not reach the threshold for significant association with a particular network. A prototypical vignette for each network is presented in Supplementary Information.
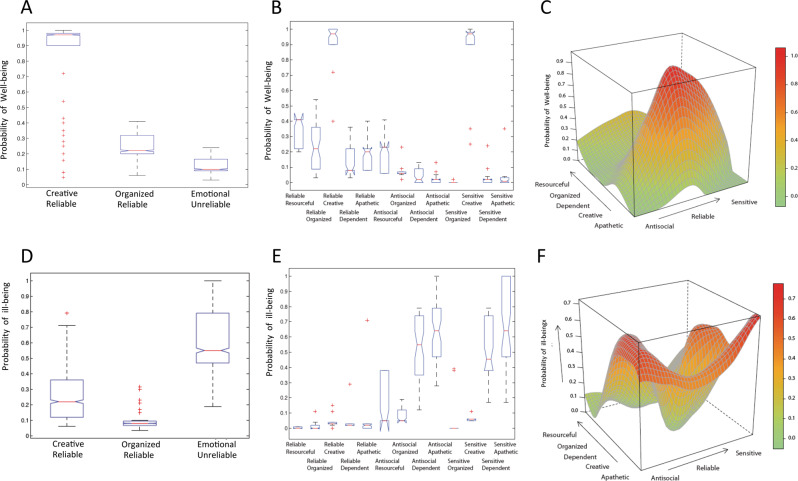
The health status of subjects also strongly distinguished the networks. The Creative-Reliable network had the highest levels of well-being, but also had a slightly higher risk of ill-being than the Organized-Reliable network (Fig. 3a–d, Supplementary Fig. S3). As expected, the Emotional-Unreliable network had the highest level of ill-being and the lowest level of well-being (p < 1.06E−23, ANOVA, Fig. 3a, d–f). Overall, the three networks differed significantly from each other in the well-being and ill-being indices (ANOVA, p < 2.29E−26).
Fig. 3.
Evaluation of the probability of health measured for Temperament and Character associations in the three phenotypic subnetworks using ANOVA statistics (p value < 1E−20), and T-test among the three subnetworks. The three networks, Creative-Reliable, Organized-Reliable, and Emotional-Unreliable, significantly differ from each other (p value < 1E−04) in their probability of well-being (a) and ill-being (d). b, e Evaluation of probability of health in Temperament and Character Sets and their relationships with SNP Sets using ANOVA statistics. b Well-being and e Ill-being evaluated for Temperament and Character Sets with respect to their profiles. c, f Surfaces representing the health function of the uncovered relationships between Temperament and Character Sets. The probability of health (z-axis; red high; green: low) was calculated based on the distribution of the status of subjects within each relationship, and the surface was plotted interpolating the relation domains. The order adopted for plotting relationships are calculated based on clustering shared subjects in Character (x-axis) and in Temperament (y-axis) Sets using Hypergeometric statistics (see “Method” section). (Close-located sets in an edge share more subjects than those located far away.) c Well-being surface. f Ill-being surface
We observed that the effect size on the well-being of subjects in different temperament–character configurations did not depend on the strength of the temperament–character association or number of subjects in their intersection. We also estimated the effect size of differences between the sets in each of the three networks by comparing their means and standard deviations for well-being and ill-being. The differences in the means of well-being for the sets in the creative-reliable (0.87 ± 0.21), organized-reliable (0.23 ± 0.012), and emotional-unreliable (0.022 ± 0.030) networks were highly significant (One-Way ANOVA, F = 7388.04, df = 2, p < 0.0001). The differences in the means for ill-being for the sets in the creative-reliable (0.63 ± 0.14), organized-reliable (0.018 ± 0.039), and emotional-unreliable (0.62 ± 0.21) networks were also highly significant (one-way ANOVA, F = 2651.63, df = 2, p < 0.0001). For well-being, the differences between the means of the sets in each pair of networks were two to four times their standard deviations, which was significant (p < 0.01) for each pair using Tukey’s range HSD test. For ill-being, the differences between the means of the sets that made up the creative-reliable and emotional-unreliable networks was four standard deviations, larger (15 standard deviations) for the differences between the means of organized-reliable and emotional-unreliable networks, and smaller (0.33 standard deviation) for the differences between the means of creative-reliable and organized-reliable networks. Although the differences in mean levels of ill-being between networks varied in size, they were all significant (p < 0.01) by Turkey’s range HSD test. In brief, there were large differences in the size of the differences in means of each pair of networks for both well-being and ill-being, except that the greater risk of ill-being in the creative-reliable network compared with the organized-reliable network was small, as illustrated in Fig. 3.
Identifying genetic networks associated with the phenotypic networks
We identified 66 SNP sets associated with at least one joint temperament–character association (Fig. 1b and Supplementary Tables S6 and S7). Fourteen SNP sets were previously deeply related to both character and temperament sets [18, 19]. Another 24 SNP sets were previously directly associated with character sets and indirectly with other temperament sets. Another 28 SNP sets were directly associated with temperament sets and indirectly with other character sets. Thus there were 66 SNP sets associated with at least one joint temperament–character relationship.
Nearly disjoint clusters of these 66 SNP sets distinguished the three phenotypic networks, as shown in Fig. 1b. In other words, the SNP sets associated with joint character–temperament relations were organized as three genotypic networks (Figs. 1b and 4a). Based on their distinguishing molecular processes (Supplementary Tables S6 and S7), the genotypic networks were labeled “emotional reactivity”, “self-control”, and “self-awareness” to describe the basic functional process of each briefly.
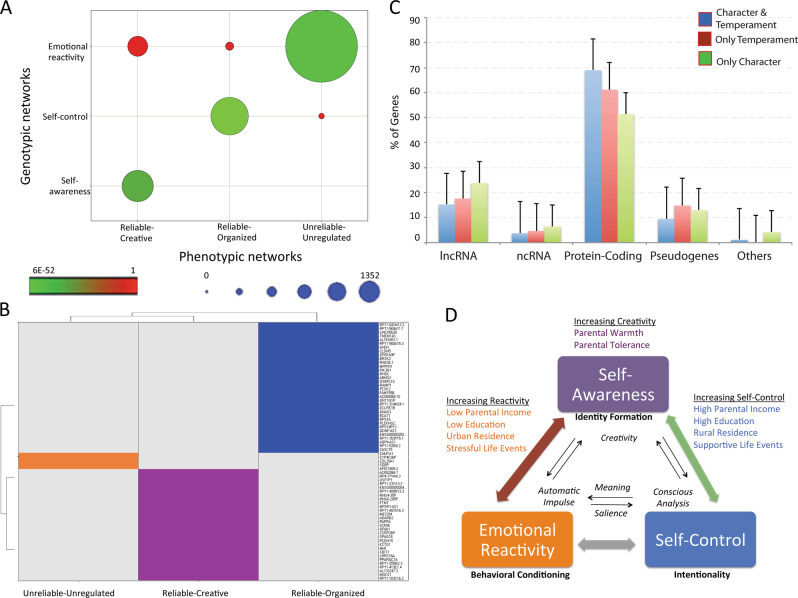
Fig. 4.
a Correlation between the phenotypic (Fig. 1a) and the genotypic (Fig. 1b) networks (p < 6E−52, Hypergeometric statistics). Color codes indicate low (red) to high (green) statistical significance. The size of the circles indicates the number of coincident phenotypic–genotypic relationships. b Relationships among key genes associated with Temperament and Character Sets that discriminate the three networks shown in Fig. 1a, b: self-awareness (violet), self-control (blue), and emotional reactivity (orange). (See AND/OR relationships in Fig. S5.) c Types of genetic variants mapped by SNP sets associated with character: specific molecular consequences [genes related only to character sets (green) were less often protein coding and more often RNA genes than those also associated with temperament sets (red color), or genes related to both character and temperament exhibit higher proportion of protein-coding genes. See subtypes in Fig. S4B]. d Relationships among environmental sets associated with Genotypic subnetworks (Fig. 1). Environmental sets can belong to one or more networks
Each of the phenotypic networks was strongly associated with only one of the genotypic networks, as shown in Fig. 4a: the Creative-Reliable network with the genotypic network for self-awareness, the Organized-Reliable network with the genotypic network for intentional self-control, and the Emotional-Unreliable network with the genotypic network for emotional reactivity and associative conditioning. There was little genotypic overlap among the three phenotypic networks: that is, few of the SNP sets associated with character (0–3%) or temperament (4.6–10.6%) were shared by any of the possible combinations of the three phenotypic networks (Supplementary Table S8).
In addition to the SNP sets associated with the integrated phenotypic network, we identified 759 genes that were associated with one or more of the three phenotypic sets in a robust way (i.e., the gene was present in multiple network-associated SNP sets). Another 213 genes were associated with the phenotypic networks, but were recognized in only one SNP set. All 972 genes are listed in Supplementary Tables S9 and S10 along with the gene’s name, type, known functions, and associated phenotypic network. The types of genes and their chromosomal locations are shown in Fig. 4b, c, Supplementary Figs. S4 and S5, Tables S9 and S10.
Among the 759 genes robustly associated with one or more of the three phenotypic networks, we found that 67.2% were unique to a single-phenotypic network: 265 for the Creative-Reliable network, 211 for the Organized-Reliable network, and 34 for the Emotional-Unreliable network. Among all 972 genes associated with one or more of the three phenotypic networks, we found that 72.4% were unique to a single-phenotypic network: 327 for the Creative-Reliable network, 273 for the Organized-Reliable network, and 106 for the Emotional-Unreliable network. Examples of the various types of genes that are unique to each of the phenotypic networks are displayed with their names in Fig. 4b. The full list of genes and their functions is available in Supplementary Fig. S5, Supplementary Tables S9 and S10.
Identifying environments that distinguished the phenotypic networks
We also tested sets of environmental variables for their ability to distinguish the three phenotypic networks. The Environmental sets are clusters of subjects with particular measured environmental experiences. There were 22 Environmental sets directly and deeply associated with the temperament and/or character components of the three networks (Table 1 and Supplementary Table S11). Another 24 Environmental sets were indirectly related with the networks as a result of gene–environment interactions (i.e., with network-associated SNP sets as mediators) (Table 1). Eight environmental sets were both directly and indirectly related to the three networks (Table 1, Supplementary Fig. S6 and Table S11).
Table 1.
Environmental sets related to the three phenotypic networks. Environmental sets are described by their features, and direct and indirect associations with the Temperament and Character phenotypes within each network. *see Supplementary Table S11 for additional description of the variables
| Direct associations | Description | Temperament associations | Character associations | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Reliable-creative | Reliable-organized | Emotional-unregulated | Reliable-creative | Reliable-organized | Emotional-unregulated | |||||
| Environment sets | ||||||||||
| E_10_1 | High parental education 1983 | High parental education 1980 | Yes | |||||||
| E_14_7 | High parental education 1983 | High parental education 1980 | Yes | |||||||
| E_5_2 | Rural residency 1980 | Rural residency 1983 | Yes | |||||||
| E_6_2 | Rural residency 1980 | Rural residency 1983 | Yes | |||||||
| E_6_4 | High parental income 1980 | High parental income 1983 | Yes | |||||||
| E_7_5 | Rural residency 2101 | Yes | Yes | Yes | ||||||
| E_14_14 | High income 1980 | Yes | Yes | |||||||
| E_11_4 | Rural residency 2101 | Yes | ||||||||
| E_13_13 | Rural residency 2101 | Yes | ||||||||
| E_8_1 | Rural residency 1980 | Rural residency 1983 | Yes | |||||||
| E_15_15 | Stressful life events | Yes | ||||||||
| E_13_6 | Rural residency 1980 | High parental income 1983 | High parental income 80 | Yes | Yes | |||||
| E_15_14 | Rural residency 1980 | High parental income 1983 | High income 80 | Yes | Yes | |||||
| E_8_6 | Rural residency 2101 | Yes | Yes | |||||||
| E_10_2 | Stressful life events | Yes | ||||||||
| E_11_7 | Stressful life events | Yes | ||||||||
| E_9_5 | Stressful life events | Yes | ||||||||
| E_9_7 | High parental income 1980 | High parental income 1983 | Yes | |||||||
| E_13_4 | High Parental Income 1980 | Yes | ||||||||
| E_14_10 | Urban residency 1980 | High parental income 1983 | Yes | |||||||
| E_3_2 | Stressful life events | Low parental income 1980 | Rural residency 2101 | Yes | ||||||
| E_8_8 | Parental tolerance 1983 | Parental tolerance 1980 | Parental emotional warmth 83 | Yes | ||||||
| Indirect associations | ||||||||||
| E_13_9 | Low parental income 1980 | Low parental income 1983 | Yes | Yes | Yes | |||||
| E_14_12 | Low parental income 1980 | Low parental income 1983 | Yes | Yes | ||||||
| E_8_5 | Low parental income 1980 | Low parental income 1983 | Yes | Yes | Yes | |||||
| E_10_2 | Stressful life events | Yes | Yes | |||||||
| E_13_10 | High parental education 1983 | High parental education 1980 | Yes | Yes | ||||||
| E_13_11 | Stressful life events | Yes | ||||||||
| E_14_1 | Parental tolerance 1980 | Yes | Yes | |||||||
| E_14_5 | Stressful life events | Yes | Yes | |||||||
| E_15_15 | Stressful life events | Yes | Yes | |||||||
| E_15_5 | High parental emotional warmth 1983 | High parental emotional warmth 1980 | Yes | Yes | ||||||
| E_6_1 | Rural residency 2101 | Yes | Yes | |||||||
| E_6_2 | Rural residency 1980 | Rural residency 1983 | Yes | Yes | ||||||
| E_10_6 | Parental tolerance 1980 | Parental tolerance 1983 | High parental emotional warmth 1983 | Yes | ||||||
| E_11_7 | Stressful life events | Yes | ||||||||
| E_15_4 | Low parental income 1980 | Yes | ||||||||
| E_9_5 | Stressful life events | Yes | ||||||||
| E_15_2 | High parental education 1983 | High parental education 1980 | Yes | Yes | Yes | Yes | ||||
| E_7_2 | Stressful life events | Yes | Yes | Yes | ||||||
| E_14_3 | High parental income 1983 | Yes | Yes | |||||||
| E_14_2 | Rural residency 2101 | Yes | ||||||||
| E_15_8 | Rural residency 2101 | Yes | ||||||||
| E_4_3 | Urban residency 1980 | Urban residency 1983 | Yes | |||||||
| E_5_1 | Urban residency 1980 | Urban residency 1983 | Yes | |||||||
| E_7_5 | Rural residency 2101 | Yes | ||||||||
| E_11_11 | Low parental income 1980 | Low parental income 1983 | Yes | |||||||
| E_14_10 | Urban residency 1980 | High parental income 1983 | Yes | |||||||
| E_3_2 | Stressful Life Events | Low Parental Income 1980 | Rural Residency 2101 | Yes | ||||||
| E_5_4 | Urban residency 2101 | Low parental education 1980 | Low parental education 1983 | Low parental income 1980 | Yes | |||||
| E_5_5 | Rural residency 2101 | Yes | ||||||||
| E_7_4 | Low parental income 1980 | Low parental income 1983 | Yes | |||||||
| E_8_7 | Stressful life events | Yes | ||||||||
| E_9_6 | Low parental income 1980 | Low parental income 1983 | Yes | |||||||
Environmental sets were associated with 18.5% of the 265 temperament–character relationships. They were directly associated with 7.6% of 265 temperament–character relationships on average in any network, but most direct relations occurred with only the Organized-reliable network (26%, Supplementary Table S12). Environmental sets were, on average, indirectly associated with 10.9% of all temperament–character relationships, and there were substantial numbers of associations with each phenotypic network: 5.3% with the creative-reliable network, 11.0% with the organized-reliable network, and 13.3% with the emotional-unreliable network (Supplementary Table S12).
We found the environmental sets distinguished the three phenotypic sets in a nearly disjoint fashion, as shown in Fig. 4d and Supplementary Fig. S6. The features of the environmental sets are detailed in Table 1, Fig. 4d, and Supplementary Table S11.
Genotypic–phenotypic relationships influence health
The probabilities of well-being and ill-being were evaluated in each of the phenotypic networks using phenotypic information alone (Fig. 3, Supplementary Fig. S3) and together with genotypic information (Supplementary Fig. S7). Combining genotypic and phenotypic information provided more information than the phenotypes alone for both well-being (Fig. 3b, c vs Supplementary Figs. S3A, S7A) and ill-being (Fig. 3e, f vs. Supplementary Figs. S3B and S7B). When health indices were based on the joint genotypic–phenotypic relationship instead of only the phenotype, all possible combinations of temperament and character profiles were strongly distinguished by their probabilities of well-being (ANOVA, p value < 4.75E−31) and ill-being (ANOVA, p value < 1.35E−49). Therefore health status provided a general characteristic of the phenotypic networks that was shared by all subjects and component sets. Consequently, health status provided a continuous metric by which to compare the effects of genotypic and environmental sets on the phenotypic networks.
We evaluated the effects of genotypic sets and environmental sets on the three networks to provide rough benchmarks of their relative contributions from a linear modeling perspective despite the limitations of such approaches. We estimated the effects of genotypic sets alone, environmental sets alone, and their joint effects in separate linear regressions on the phenotype specified as three phenotypic networks ordered by the values 1, 2, and 3 in correspondence to their mean levels of well-being. To standardize the unit of measurement for the different types of sets (genotypic, phenotypic, and environmental), we used the average well-being and/or ill-being status of their constituent sets of individuals. This provided a general and continuous characteristic of personality that could be compared for all subjects in each network (Supplementary Information: Section 10 and Tables S13 and S14). We used the R2 to compare the results obtained by the regressions, which were estimated by the average of tenfold cross-validation (Supplementary Information, Section 10). For ill-being, genotypic effects alone produced an R2 of 0.58, environmental effects alone an R2 of 0.27, and their joint effects an R2 of 0.68, showing the importance of both genotypic and environmental influences on ill-being. In contrast, for well-being, genotypic effects alone produced an R2 of 0.81, environmental effects alone an R2 of 0.23, and together an R2 of 0.82, so environmental influences did not add substantially to the explanatory effects of the genotypes for well-being. The details about specifying the phenotypic, genotypic, and environmental variables in terms of health status of genotypic sets, environmental sets, and phenotypic sets (temperament sets, character sets) is provided in the Supplementary Information (Section 10, Tables S13 and S14). Significance testing of the full model is detailed in Table S14.
Replication of results in two independent samples
We tested the replicability of our findings in the Finnish sample by carrying out the same analyses of the genotypic and phenotypic architecture blindly in the German and Korean samples, thereby allowing for possible heterogeneity within and across independent samples from different cultures (Supplementary Information, Section 9). We evaluated the matching between each replication sample and the discovery sample of all aspects of the complex genotypic–phenotypic architecture (genotypic sets, phenotypic sets, and their relations) by a permutation test. Of the associations between genotypic sets and the phenotypic character sets identified in the Finnish sample, 84% were identified in the German sample and 94% in the Korean sample [18]. The associations between genotypic sets and the phenotypic temperament sets identified in the Finnish sample closely matched those observed in the Korean sample (89%) and in the German (76%) sample [19]. Finally, the replication between genotypic–phenotypic relationships when both temperament and character were considered were also replicated in the Korean (80%) and German samples (64%) (Supplementary Tables S15 and S16). The replication of the genotypic–phenotypic relations was reduced in the Germans (Supplementary Fig. S8), as expected because they were screened to exclude individuals with psychopathology, including personality disorders, as shown elsewhere [18, 19].
The strong replication across samples of all aspects of the complex phenotypic architecture of the networks (i.e., the matching of the temperament sets, character sets, and their relations in each replication sample to that of the discovery sample) was confirmed by a permutation test in both Koreans (1.0E−14 < p < 1.0E−02, Fisher’s exact test, Supplementary Table S17) and Germans (1.0E−11 p < 1.0E−02, Fisher’s exact test, Supplementary Table S18).
Discussion
We have uncovered three robust findings about human personality for the first time. First, human personality is organized as a hierarchy of deep descriptive complexity ascending from (i) separate scales and subscales of temperament and of character, (ii) sets of multiple subscales of temperament and of character, (iii) profiles of temperament sets and of character sets, and finally to (iv) joint networks of temperament and character profiles. The genetic building blocks of the phenotypic hierarchy encode the temperament profiles and character profiles, not their constituent sets of personality subscales and not the three learning networks. Second, the three temperament–character networks are nearly disjoint phenotypes despite the marked complexity of temperament–character relationships within each of the three networks. Third, nearly disjoint sets of genetic and environmental variables strongly distinguish the three phenotypic networks. The strong association of both genetic and environmental variables with these integrated networks is surprising because the measured impact of the same environmental variables on the constituents of the integrated networks is weak, as we previously reported [18, 19]. Overall, we found that genes encode temperament profiles and character profiles separately, and then these are integrated by genetic–environmental interactions into complex adaptive networks.
These findings together suggest that the major role of environmental influences on personality development is the organization of the relations among temperament and character, not on the temperament and character profiles themselves that are largely genetically independent. Despite the novelty and surprising nature of these results, they are robust because we have strongly replicated the phenotypic and genotypic findings regardless of cultural and environmental differences among three independent samples of Finns, Germans, and Koreans. The replication was done in an unbiased fashion and avoided overfitting by balancing accuracy with model complexity while allowing for heterogeneity within and across independent samples, which is not necessarily the case for other GWAS methods [20] (see Supplementary Information, Polygenic Risk Scores). These findings have several important implications for both research and clinical practice.
Importance of both genetic and environmental influences
Although genetic influences on temperament and character are substantial and robust, the impact of measured environmental influences has been weak [78–80]. The heritability of temperament and of character in the Finnish sample was 48% and 57%, respectively, whereas the influence of the environmental covariates did not exceed 3% when temperament and character were considered separately, as previously reported [18, 19]. Here we explored wider four-dimensional sets of interactions among temperament, character, genes, and environments that allowed for both direct associations and gene–environmental interactions associated with the phenotypic networks. This data-driven approach allowed us to identify many gene–environment interactions associated with the phenotypic networks, which would not have been detected in linear models of environmental influences on individual personality traits [20]. In contrast, prior work focused on individual traits rather than integrated profiles that characterize the organization of self-regulatory processes within a well-adapted person. This difference suggests that the major role of environmental influences is to interact with genetic predisposition in the activation of learning processes that result in integrated adaptive networks. For example, parental warmth and tolerance usually nurtures creativity in children (Fig. 4d), particularly those with genotypic sets for self-awareness and reliable temperaments (e.g., E_10_6 in Table 1), but it can also enable dependence in individuals with genotypic sets for emotional reactivity (e.g., E_8_8 in Table 1).
Specifically, we found that environmental variables had substantial influence on the risk of ill-being in particular; specifically, environmental and genotypic variables together explained an additional 10% of the phenotypic variance than did genotypic variables alone for ill-being (Supplementary Table S13). In contrast, genotypic variables had even larger effects on both ill-being (58%) and well-being (81%). Likewise well-being was most often high in people in the creative-reliable phenotypic network (see Fig. 3), which had the fewest proportion of associations with environmental sets (directly 1.8%, indirectly 5.3%) (Supplementary Table S12). Ill-being was frequently high in the other two networks; the organized-reliable network had the most environmental associations (directly 26%, indirectly 11%) and the emotional-unreliable network was intermediate (directly 0%, indirectly 13.3%) (Supplementary Table S12). These findings indicate people with creative-reliable profiles are usually resilient and healthy regardless of external conditions (Supplementary Table S3), whereas others are more vulnerable to stressful life events and social influences on disease and mortality, such as limited education and socioeconomic opportunity (Table 1) [18, 81, 82].
Both our work on personality [18, 19] and other work on the human exposome [83] converge on the idea that both environmental perturbations and the emotional reactivity of unregulated temperament networks lead to ill-health with multiple chronic noncommunicable disorders as a result of impaired regulation of gene co-expression, which has been called regulatory decoherence [84]. That is, environmental perturbations and the unhealthy character profiles found in the emotional-unreliable network impair the orchestration of gene co-expression, as has been most directly supported in other independent research by reduced correlations among sets of particular mRNA transcripts [84] and related psychophysiological processes that promote health, such as heart rate variability [85], rather than by variation in the presence or absence of particular genetic markers.
What are the molecular mechanisms by which genes related to personality may regulate the co-expression of genes that are far apart in the genome in coherent ways that are adaptive and beneficial for health? An important clue is that most of the genes associated with temperament are protein coding whereas those for character have a preponderance of noncoding RNA genes, particularly long noncoding (lnc) RNAs [18, 19]. LncRNAs have important functions that influence complex patterns of adaptive functioning and health by transcriptional and posttranscriptional regulation of gene expression, coordination of the co-expression of sets of genes, and chromatin remodeling [86–89]. A reasonable next step for further investigation is to examine patterns of co-expression of the personality related genes (both coding and noncoding) we have identified in blood and other tissues [84]. Then putative patterns of coherence versus decoherence can be tested for association with indices of health versus ill-being under a variety of conditions of physical, psychological and social stress, or other environmental perturbations. The indices of health may involve metabolic states, psychophysiological processes like heart rate variability, or other indices of the physical, mental, and social aspects of health. The indices of ill-being may involve disorders of various types (physical, mental, or social) with various patterns of comorbidity. Such investigations may clarify the roles of coding and noncoding genes in health and disease, as well as the mechanisms of associations of personality with ill-being and well-being via direct effects on gene expression and co-expression versus indirect effects via life style choice or environmental perturbation.
Implications of three systems of learning for health promotion
We hypothesized that the disjoint sets of genes and environments associated with integrated temperament–character networks may correspond to the stepwise emergence of three different systems of learning and memory in the phylogeny of human beings [40, 44]. In other words, the integrated phenotypic networks are made up of people who express the prototypical features of each of three major systems of learning and memory present in modern human beings: associative conditioning for emotional reactivity in the Emotional-Unreliable network, intentional self-regulation in the organized-reliable network, and self-awareness of autobiographical memory in the creative-reliable network. Put another way, the three phenotypic networks are nearly disjoint prototypes of major systems of learning and memory, which are known to be neurologically and developmentally dissociable [17, 46].
Our findings that people with the creative-reliable profile were healthier than others confirm earlier work about how to describe a healthy human personality [70, 90–93]. We found that people with creative characters and reliable temperaments have greater well-being, including objective indicators of healthy longevity, such as optimal cardiovascular health, when compared with others. We also found that the genes for personality are expressed in most organ systems, not only the brain (Supplementary Fig. S9), so the physical, mental, and social aspects of health are expected to be strongly interdependent. Consequently, norms for healthy functioning need to consider the importance of self-transcendent functions, such as spontaneous creativity [94], altruism [95], and generosity [96], which are sometimes neglected [70, 97].
Our finding that healthy human functioning involves the integration of three systems of learning with qualitatively distinct properties [5] has strong implications for the modeling and use of artificial intelligence (AI) [98]. There is growing evidence that effective promotion of health and well-being depends on coordinated change in all three networks for self-awareness, intentional self-control, and behavioral conditioning, rather than any of these alone [99–102]. Optimizing the utility of AI for health promotion is likely to require it to recognize and address all three human learning systems in an integrative manner that facilitates its interaction with human beings in ways that are personalized to be satisfying, meaningful, and harmonious.
Strengths and limitations
The strengths of this research are the use of unrestrictive data-driven methods, and replication of genotypic and phenotypic findings independently in samples that vary in their cultural and environmental features. A potential weakness of all clustering methods is that the number of clusters and their content are uncertain, but the methods adopted here addressed this carefully, as shown by the robust replication in independent samples [18–20]. We have shown that our methods are valid even in the presence of epistasis, pleiotropy, and heterogeneity, which cannot be accounted for by polygenic models that assume the independence of genes and environmental influences [20] (Supplementary Fig. S10). Our findings document the robustness of unsupervised machine-learning methods that allow the deconstruction and reintegration of the complex architecture of human personality.
Nevertheless, there are limitations to our study. We did not have comparable environmental data in the Korean and German samples, so only the genotypic and phenotypic findings were replicated here. The estimates of effect size used linear regressions on sets of variables, which captured interactions within the sets, but not between them. Also our findings are based on cross-sectional associations, so no causal inferences are justified.
Conclusions and recommendations
Our data-driven findings support the hypothesis that human temperament and character are integrated as three nearly disjoint phenotypic networks regulated by complex interactions among nearly disjoint sets of genetic and environmental influences. Furthermore, these phenotypic networks are comprised of people who may express the prototypical features of three major systems of learning and memory that have previously been distinguished by their stepwise emergence in phylogeny and by their dissociable brain circuitry.
Our results show that the well-being of modern human beings depends on the maturation and integration of functions that interact to support healthy longevity, creative self-awareness, and self-transcendent functions such as moderation and altruism. In other words, physical, mental, social, and spiritual aspects of health cannot be separated because of the reciprocal interactions among the functions that support the well-being of the whole person. Mental or physical health cannot be adequately assessed, treated, or promoted as a set of separate diseases or traits.
Supplementary information
Acknowledgements
The Young Finns Study has been financially supported by the Academy of Finland: grants 286284, 322098, 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), 41071 (Skidi), and 308676; the Social Insurance Institution of Finland; Competitive State Research Financing of the Expert Responsibility area of Kuopio, Tampere and Turku University Hospitals (grant X51001); Juho Vainio Foundation; Paavo Nurmi Foundation; Finnish Foundation for Cardiovascular Research; Finnish Cultural Foundation; Tampere Tuberculosis Foundation; Emil Aaltonen Foundation; Yrjö Jahnsson Foundation; Signe and Ane Gyllenberg Foundation; Diabetes Research Foundation of Finnish Diabetes Association: and EU Horizon 2020 (grant 755320 for TAXINOMISIS); and Tampere University Hospital Supporting Foundation. The American Foundation for Suicide Prevention supported the study of healthy Germans. The national Healthy Twin Family Register of Korea supported the study of healthy Koreans. The Anthropedia Foundation and the Spanish Ministry of Science and Technology TIN2012-38805 and DPI2015-69585-R supported this collaboration.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Igor Zwir, Coral Del-Val
Supplementary information
The online version of this article (10.1038/s41380-019-0579-x) contains supplementary material, which is available to authorized users.
References
- 1.Zohar AH, Zwir I, Wang J, Cloninger CR, Anokhin AP. The development of temperament and character during adolescence: the processes and phases of change. Dev Psychopathol. 2019;31:601–17. [DOI] [PubMed]
- 2.Kern ML, Della Porta SS, Friedman HS. Lifelong pathways to longevity: personality, relationships, flourishing, and health. J Pers. 2014;82:472–84. doi: 10.1111/jopy.12062. [DOI] [PubMed] [Google Scholar]
- 3.Cloninger CR, Zohar AH. Personality and the perception of health and happiness. J Affect Disord. 2011;128:24–32. doi: 10.1016/j.jad.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Cloninger CR. Feeling good: the science of well-being. New York: Oxford University Press; 2004. p. 374.
- 5.Cloninger CR, Abou-Saleh MT, Mrazek DA, Moller H-J. Biological perspective on psychiatry for the person. Int J Pers Cent Med. 2011;1:137–9. doi: 10.5750/ijpcm.v1i1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloninger CR, Svrakic NM, Svrakic DM. Role of personality self-organization in development of mental order and disorder. Dev Psychopathol. 1997;9:881–906. doi: 10.1017/s095457949700148x. [DOI] [PubMed] [Google Scholar]
- 7.Shiner RL, Buss KA, McClowry SG, Putnam SP, Saudino KJ, Zentner M. What is Temperament Now? Assessing progress in temperament research on the twenty-fifth anniversary of Goldsmith et al. (1987) Child Dev Perspect. 2012;6:436–44. [Google Scholar]
- 8.Cloninger CR, Cloninger KM, Zwir I, Keltigangas-Jarvinen L. The complex genetics and biology of human temperament: a review of traditional concepts in relation to new molecular findings. Trans Psychiatry. (In press). [DOI] [PMC free article] [PubMed]
- 9.Cloninger CR. A systematic method for clinical description and classification of personality variants. A proposal. Arch Gen Psychiatry. 1987;44:573–88. doi: 10.1001/archpsyc.1987.01800180093014. [DOI] [PubMed] [Google Scholar]
- 10.Josefsson K, Jokela M, Cloninger CR, Hintsanen M, Salo J, Hintsa T, et al. Maturity and change in personality: developmental trends of temperament and character in adulthood. Dev Psychopathol. 2013;25:713–27. doi: 10.1017/S0954579413000126. [DOI] [PubMed] [Google Scholar]
- 11.Thomas A, Chess S, Birch HG. Temperament and behavior disorders in children. New York: New York University Press; 1968. [Google Scholar]
- 12.Strelau J. The regulative theory of temperament: current status. Personal Individ Differ. 1996;20:131–42. [Google Scholar]
- 13.Cloninger CR, Svrakic DM, Przybeck TR. A psychobiological model of temperament and character. Arch Gen Psychiatry. 1993;50:975–90. doi: 10.1001/archpsyc.1993.01820240059008. [DOI] [PubMed] [Google Scholar]
- 14.Vaillant GE, Vaillant CO. Natural history of male psychological health, XII: a 45-year study of predictors of successful aging at age 65. Am J Psychiatry. 1990;147:31–37. doi: 10.1176/ajp.147.1.31. [DOI] [PubMed] [Google Scholar]
- 15.Campanella F, Crescentini C, Urgesi C, Fabbro F. Mindfulness-oriented meditation improves self-related character scales in healthy individuals. Compr Psychiatry. 2014;55:1269–78. doi: 10.1016/j.comppsych.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Tulving E. Episodic memory: from mind to brain. Annu Rev Psychol. 2002;53:1–25. doi: 10.1146/annurev.psych.53.100901.135114. [DOI] [PubMed] [Google Scholar]
- 17.Tulving E. Multiple memory systems and consciousness. Hum Neurobiol. 1987;6:67–80. [PubMed] [Google Scholar]
- 18.Zwir I, Arnedo J, Del-Val C, Pulkki-Raback L, Konte B, Yang SS, et al. Uncovering the complex genetics of human character. Mol Psychiatry. 2018. 10.1038/s41380-018-0263-6. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 19.Zwir I, Arnedo J, Del-Val C, Pulkki-Raback L, Konte B, Yang SS, et al. Uncovering the complex genetics of human temperament. Mol Psychiatry. 2018. 10.1038/s41380-018-0264-5. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 20.Zwir I, Mishra P, Del-Val C, Gu CC, de Erausquin G, Lehtimäki T, et al. Uncovering the complex genetics of human personality: response from authors on the PGMRA Model. Mol Psychiatry. 2019. 10.1038/s41380-019-0399-z. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 21.Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, Hunter DJ, et al. Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet. 2010;86:929–42. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill WG, Zhang XS. On the pleiotropic structure of the genotype-phenotype map and the evolvability of complex organisms. Genetics. 2012;190:1131–7. doi: 10.1534/genetics.111.135681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verweij KJ, Yang J, Lahti J, Veijola J, Hintsanen M, Pulkki-Raback L, et al. Maintenance of genetic variation in human personality: testing evolutionary models by estimating heritability due to common causal variants and investigating the effect of distant inbreeding. Evolution. 2012;66:3238–51. doi: 10.1111/j.1558-5646.2012.01679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eaves LJ, Heath AC, Neale MC, Hewitt JK, Martin NG. Sex differences and non-additivity in the effects of genes on personality. Twin Res. 1998;1:131–7. doi: 10.1375/136905298320566267. [DOI] [PubMed] [Google Scholar]
- 26.Eaves LJ, Heath AC, Martin NG, Maes HH, Neale MC, Kendler KS, et al. Comparing the biological and cultural inheritance of personality and social attitudes in the Virginia 30,000 study of twins and their relatives. Twin Res. 1999;2:62–80. doi: 10.1375/136905299320565933. [DOI] [PubMed] [Google Scholar]
- 27.Finkel D, McGue M. Sex differences and nonadditivity in heritability of the Multidimensional Personality Questionnaire Scales. J Pers Soc Psychol. 1997;72:929–38. doi: 10.1037//0022-3514.72.4.929. [DOI] [PubMed] [Google Scholar]
- 28.Keller MC, Coventry WL, Heath AC, Martin NG. Widespread evidence for non-additive genetic variation in Cloninger’s and Eysenck’s Personality Dimensions using a Twin Plus Sibling Design. Behav Genet. 2005;35:707–21. doi: 10.1007/s10519-005-6041-7. [DOI] [PubMed] [Google Scholar]
- 29.Carey WB, McDevitt SC. Stability and change in individual temperament diagnoses from infancy to early childhood. J Am Acad Child Psychiatry. 1978;17:331–7. doi: 10.1016/s0002-7138(10)60096-9. [DOI] [PubMed] [Google Scholar]
- 30.Faraone SV. The omnigenic model: implications for psychiatric genetics. J Psychiatry Brain Sci. 2017;2:S1. [Google Scholar]
- 31.Hess JL, Akutagava-Martins GC, Patak JD, Glatt SJ, Faraone SV. Why is there selective subcortical vulnerability in ADHD? Clues from postmortem brain gene expression data. Mol Psychiatry. 2018;23:1787–93. doi: 10.1038/mp.2017.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng S, Zhang Y, Zhang J, Wang H, Ren B. ERK in learning and memory: a review of recent research. Int J Mol Sci. 2010;11:222–32. doi: 10.3390/ijms11010222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tronson NC, Corcoran KA, Jovasevic V, Radulovic J. Fear conditioning and extinction: emotional states encoded by distinct signaling pathways. Trends Neurosci. 2012;35:145–55. doi: 10.1016/j.tins.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rapanelli M, Frick LR, Zanutto BS. Differential gene expression in the rat hippocampus during learning of an operant conditioning task. Neuroscience. 2009;163:1031–8. doi: 10.1016/j.neuroscience.2009.07.037. [DOI] [PubMed] [Google Scholar]
- 35.Atkins CM, Selcher JC, Petraitis JJ, Trzaskos JM, Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nat Neurosci. 1998;1:602–9. doi: 10.1038/2836. [DOI] [PubMed] [Google Scholar]
- 36.Cerovic M, d’Isa R, Tonini R, Brambilla R. Molecular and cellular mechanisms of dopamine-mediated behavioral plasticity in the striatum. Neurobiol Learn Mem. 2013;105:63–80. doi: 10.1016/j.nlm.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 37.Reiner A, Medina L, Veenman CL. Structural and functional evolution of the basal ganglia in vertebrates. Brain Res Brain Res Rev. 1998;28:235–85. doi: 10.1016/s0165-0173(98)00016-2. [DOI] [PubMed] [Google Scholar]
- 38.Reiner A. Functional circuitry of the avian basal ganglia: implications for basal ganglia organization in stem amniotes. Brain Res Bull. 2002;57:513–28. doi: 10.1016/s0361-9230(01)00667-0. [DOI] [PubMed] [Google Scholar]
- 39.Van Schuerbeek P, Baeken C, De Raedt R, De Mey J, Luypaert R. Individual differences in local gray and white matter volumes reflect differences in temperament and character: a voxel-based morphometry study in healthy young females. Brain Res. 2011;1371:32–42. doi: 10.1016/j.brainres.2010.11.073. [DOI] [PubMed] [Google Scholar]
- 40.Cloninger CR. The evolution of human brain functions: the functional structure of human consciousness. Aust NZ J Psychiatry. 2009;43:994–1006. doi: 10.3109/00048670903270506. [DOI] [PubMed] [Google Scholar]
- 41.Allman JM, Tetreault NA, Hakeem AY, Manaye KF, Semendeferi K, Erwin JM, et al. The von Economo neurons in the frontoinsular and anterior cingulate cortex. Ann N Y Acad Sci. 2011;1225:59–71. doi: 10.1111/j.1749-6632.2011.06011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Posner MI, Rothbart MK. Research on attention networks as a model for the integration of psychological science. Annu Rev Psychol. 2007;58:1–23. doi: 10.1146/annurev.psych.58.110405.085516. [DOI] [PubMed] [Google Scholar]
- 43.Cauda F, Torta DM, Sacco K, D’Agata F, Geda E, Duca S, et al. Functional anatomy of cortical areas characterized by Von Economo neurons. Brain Struct Funct. 2013;218:1–20. doi: 10.1007/s00429-012-0382-9. [DOI] [PubMed] [Google Scholar]
- 44.Cloninger CR, Kedia S. The phylogenesis of human personality: identifying the precursors of cooperation, altruism, & well-being. In: Sussman RW, Cloninger CR, editors. The origins of cooperation and altruism, 1st ed. New York: Springer; 2011. pp. 63–110.
- 45.Tulving E. Episodic memory and common sense: how far apart? Philos Trans R Soc Lond B Biol Sci. 2001;356:1505–15. doi: 10.1098/rstb.2001.0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levine B. Autobiographical memory and the self in time: brain lesion effects, functional neuroanatomy, and lifespan development. Brain Cogn. 2004;55:54–68. doi: 10.1016/S0278-2626(03)00280-X. [DOI] [PubMed] [Google Scholar]
- 47.Ullman MT. A neurocognitive perspective on language: the declarative/procedural model. Nat Rev Neurosci. 2001;2:717–26. doi: 10.1038/35094573. [DOI] [PubMed] [Google Scholar]
- 48.Lee AC, Robbins TW, Graham KS, Owen AM. “Pray or Prey?” dissociation of semantic memory retrieval from episodic memory processes using positron emission tomography and a novel homophone task. Neuroimage. 2002;16:724–35. doi: 10.1006/nimg.2002.1101. [DOI] [PubMed] [Google Scholar]
- 49.Levine B, Turner GR, Tisserand D, Hevenor SJ, Graham SJ, McIntosh AR. The functional neuroanatomy of episodic and semantic autobiographical remembering: a prospective functional MRI study. J Cogn Neurosci. 2004;16:1633–46. doi: 10.1162/0898929042568587. [DOI] [PubMed] [Google Scholar]
- 50.Finn AS, Kalra PB, Goetz C, Leonard JA, Sheridan MA, Gabrieli JD. Developmental dissociation between the maturation of procedural memory and declarative memory. J Exp Child Psychol. 2016;142:212–20. doi: 10.1016/j.jecp.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nissen MJ, Knopman DS, Schacter DL. Neurochemical dissociation of memory systems. Neurology. 1987;37:789–94. doi: 10.1212/wnl.37.5.789. [DOI] [PubMed] [Google Scholar]
- 52.Graham KS, Simons JS, Pratt KH, Patterson K, Hodges JR. Insights from semantic dementia on the relationship between episodic and semantic memory. Neuropsychologia. 2000;38:313–24. doi: 10.1016/s0028-3932(99)00073-1. [DOI] [PubMed] [Google Scholar]
- 53.Svoboda E, McKinnon MC, Levine B. The functional neuroanatomy of autobiographical memory: a meta-analysis. Neuropsychologia. 2006;44:2189–208. doi: 10.1016/j.neuropsychologia.2006.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fink A, Benedek M, Koschutnig K, Papousek I, Weiss EM, Bagga D, et al. Modulation of resting-state network connectivity by verbal divergent thinking training. Brain Cogn. 2018;128:1–6. doi: 10.1016/j.bandc.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 55.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krapohl E, Patel H, Newhouse S, Curtis CJ, von Stumm S, Dale PS, et al. Multi-polygenic score approach to trait prediction. Mol Psychiatry. 2018;23:1368–74. doi: 10.1038/mp.2017.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Machiela MJ, Chen CY, Chen C, Chanock SJ, Hunter DJ, Kraft P. Evaluation of polygenic risk scores for predicting breast and prostate cancer risk. Genet Epidemiol. 2011;35:506–14. doi: 10.1002/gepi.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cloninger CR, Rice J, Reich T. Multifactorial inheritance with cultural transmission and assortative mating. II. a general model of combined polygenic and cultural inheritance. Am J Hum Genet. 1979;31:176–98. [PMC free article] [PubMed] [Google Scholar]
- 60.Feldman MW, Ramachandran S. Missing compared to what? Revisiting heritability, genes and culture. Philos Trans R Soc Lond B Biol Sci. 2018;373:1–8. doi: 10.1098/rstb.2017.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arnedo J, Svrakic DM, Del Val C, Romero-Zaliz R, Hernandez-Cuervo H, Fanous AH, et al. Uncovering the hidden risk architecture of the schizophrenias: confirmation in three independent genome-wide association studies. Am J Psychiatry. 2015;172:139–53. doi: 10.1176/appi.ajp.2014.14040435. [DOI] [PubMed] [Google Scholar]
- 62.Selzam S, Krapohl E, von Stumm S, O’Reilly PF, Rimfeld K, Kovas Y, et al. Predicting educational achievement from DNA. Mol Psychiatry. 2017;22:267–72. doi: 10.1038/mp.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–86. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boyle EA, Li Y, Pritchard JK. The omnigenic model: response from the authors. J Psychiatry Brain Sci. 2017;2:S8. [Google Scholar]
- 65.Võsa U, Claringbould A, Westra H-J, Bonder MJ, Deelen P, Zeng B, et al. Unraveling the polygenic architecture of complex traits using blood eQTL meta-analysis. 2018. https://www.biorxiv.org/content/10.1101/447367v1.
- 66.Raitakari OT, Juonala M, Ronnemaa T, Keltikangas-Jarvinen L, Rasanen L, Pietikainen M, et al. Cohort profile: the cardiovascular risk in Young Finns Study. Int J Epidemiol. 2008;37:1220–6. doi: 10.1093/ije/dym225. [DOI] [PubMed] [Google Scholar]
- 67.Cook TB, Brenner LA, Cloninger CR, Langenberg P, Igbide A, Giegling I, et al. Latent infection with toxoplasma gondii: association with trait aggression and impulsivity in healthy adults. J Psychiatr Res. 2015;60:87–94. doi: 10.1016/j.jpsychires.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 68.Gombojav B, Song YM, Lee K, Yang S, Kho M, Hwang YC, et al. The Healthy Twin Study, Korea updates: resources for omics and genome epidemiology studies. Twin Res Hum Genet. 2013;16:241–5. doi: 10.1017/thg.2012.130. [DOI] [PubMed] [Google Scholar]
- 69.Sung J, Cho SI, Lee K, Ha M, Choi EY, Choi JS, et al. Healthy Twin: a twin-family study of Korea-protocols and current status. Twin Res Hum Genet. 2006;9:844–8. doi: 10.1375/183242706779462822. [DOI] [PubMed] [Google Scholar]
- 70.Cloninger CR. What makes people healthy, happy, and fulfilled in the face of current world challenges? Mens Sana Monogr. 2013;11:16–24. doi: 10.4103/0973-1229.109288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Josefsson K, Cloninger CR, Hintsanen M, Jokela M, Pulkki-Raback L, Keltikangas-Jarvinen L. Associations of personality profiles with various aspects of well-being: a population-based study. J Affect Disord. 2011;133:265–73. doi: 10.1016/j.jad.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 72.Pulkki-Raback L, Elovainio M, Hakulinen C, Lipsanen J, Hintsanen M, Jokela M, et al. Cumulative effect of psychosocial factors in youth on ideal cardiovascular health in adulthood: the Cardiovascular Risk in Young Finns Study. Circulation. 2015;131:245–53. doi: 10.1161/CIRCULATIONAHA.113.007104. [DOI] [PubMed] [Google Scholar]
- 73.Serlachius A, Elovainio M, Juonala M, Shea S, Sabin M, Lehtimaki T, et al. High perceived social support protects against the intergenerational transmission of obesity: the Cardiovascular Risk in Young Finns Study. Prev Med. 2016;90:79–85. doi: 10.1016/j.ypmed.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 74.Zwir I, Shin D, Kato A, Nishino K, Latifi T, Solomon F, et al. Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA. 2005;102:2862–7. doi: 10.1073/pnas.0408238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zwir I, Huang H, Groisman EA. Analysis of differentially-regulated genes within a regulatory network by GPS genome navigation. Bioinformatics. 2005;21:4073–83. doi: 10.1093/bioinformatics/bti672. [DOI] [PubMed] [Google Scholar]
- 76.Arnedo J, del Val C, de Erausquin GA, Romero-Zaliz R, Svrakic D, Cloninger CR, et al. PGMRA: a web server for (phenotype x genotype) many-to-many relation analysis in GWAS. Nucleic Acids Res. 2013;41:W142–149. doi: 10.1093/nar/gkt496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deb K. Multi-objective optimization using evolutionary algorithms. 1st ed. New York: John Wiley & Sons; 2001.
- 78.Gillespie NA, Cloninger CR, Heath AC, Martin NG. The genetic and environmental relationship between Cloninger’s dimensions of temperament and character. Pers Individ Differ. 2003;35:1931–46. doi: 10.1016/S0191-8869(03)00042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heath AC, Cloninger CR, Martin NG. Testing a model for the genetic structure of personality: a comparison of the personality systems of Cloninger and Eysenck. J Pers Soc Psychol. 1994;66:762–75. doi: 10.1037//0022-3514.66.4.762. [DOI] [PubMed] [Google Scholar]
- 80.Lester N, Garcia D, Lundstrom S, Brandstrom S, Rastam M, Kerekes N, et al. The genetic and environmental structure of the character sub-scales of the temperament and character inventory in adolescence. Ann Gen Psychiatry. 2016;15:10. doi: 10.1186/s12991-016-0094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilkinson R, Marmot M. Social determinants of health: the solid facts. In: WHO, editor. 2nd edn. Copenhagen: World Health Organization; 2003.
- 82.Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–45. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 83.Dennis KK, Marder E, Balshaw DM, Cui Y, Lynes MA, Patti GJ, et al. Biomonitoring in the era of the exposome. Environ Health Perspect. 2017;125:502–10. doi: 10.1289/EHP474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lea A, Subramaniam M, Ko A, Lehtimäki T, Raitoharju E, Kahonen M, et al. Genetic and environmental perturbations lead to regulatory decoherence. eLife. 2019;8:e40538. doi: 10.7554/eLife.40538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zohar AH, Cloninger CR, McCraty R. Personality and heart rate variability: exploring pathways from personality to cardiac coherence and health. J Soc Sci. 2013;1:32–39. [Google Scholar]
- 86.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteom Bioinforma. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 88.Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. 2016;17:601–14. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- 89.Yang Y, Zhou R, Li W, Liu Y, Zhang Y, Ao H, et al. Dynamic transcriptome analysis reveals potential long non-coding RNAs governing postnatal pineal development in pig. Front Genet. 2019;10:409. doi: 10.3389/fgene.2019.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bae HT, Sebastiani P, Sun JX, Andersen SL, Daw EW, Terracciano A, et al. Genome-wide association study of personality traits in the long life family study. Front Genet. 2013;4:65. doi: 10.3389/fgene.2013.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Subramaniam K, Kounios J, Parrish TB, Jung-Beeman M. A brain mechanism for facilitation of insight by positive affect. J Cogn Neurosci. 2009;21:415–32. doi: 10.1162/jocn.2009.21057. [DOI] [PubMed] [Google Scholar]
- 92.Vaillant GE. Positive mental health: is there a cross-cultural definition? World Psychiatry. 2012;11:93–99. doi: 10.1016/j.wpsyc.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jeste DV, Palmer BW. Positive psychiatry: a clinical handbook. Virginia, United States: American Psychiatric Association Publishing; 2015. [Google Scholar]
- 94.Limb CJ, Braun AR. Neural substrates of spontaneous musical performance: an FMRI study of jazz improvisation. PLoS One. 2008;3:e1679. doi: 10.1371/journal.pone.0001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moll J, Krueger F, Zahn R, Pardini M, de Oliveira-Souza R, Grafman J. Human fronto-mesolimbic networks guide decisions about charitable donation. Proc Natl Acad Sci USA. 2006;103:15623–8. doi: 10.1073/pnas.0604475103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Daw ND, O’Doherty JP, Dayan P, Seymour B, Dolan RJ. Cortical substrates for exploratory decisions in humans. Nature. 2006;441:876–9. doi: 10.1038/nature04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.APA. Diagnostic and statistical manual (DSM-5). 5th ed. Washington, D.C.: American Psychiatric Association; 2013.
- 98.Tegmark M. Life 3.0: being human in the age of artificial intelligence. Reprint edn. Vintage; New York, NY, 2018.
- 99.Mirowsky J, Ross CE. Education, personal control, lifestyle and health: a human capital hypothesis. Res Aging. 1998;20:415–49. [Google Scholar]
- 100.Hayes SA, Orsillo SM, Roemer L. Changes in proposed mechanisms of action during an acceptance-based behavior therapy for generalized anxiety disorder. Behav Res Ther. 2010;48:238–45. doi: 10.1016/j.brat.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cloninger CR, Cloninger KM. People create health: effective health promotion is a creative process. Int J Pers-centered Med. 2013;3:114–22. doi: 10.5750/ijpcm.v3i2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nigg CR, Burbank PM, Padula C, Dufresne R, Rossi JS, V WF, et al. Stages of change across ten health risk behaviors for older adults. Gerontologist. 1999;39:473–82. doi: 10.1093/geront/39.4.473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.