Abstract
Major depressive disorder (MDD) is a serious mental illness, characterized by high morbidity, which has increased in recent decades. However, the molecular mechanisms underlying MDD remain unclear. Previous studies have identified altered metabolic profiles in peripheral tissues associated with MDD. Using curated metabolic characterization data from a large sample of MDD patients, we meta-analyzed the results of metabolites in peripheral blood. Pathway and network analyses were then performed to elucidate the biological themes within these altered metabolites. We identified 23 differentially expressed metabolites between MDD patients and controls from 46 studies. MDD patients were characterized by higher levels of asymmetric dimethylarginine, tyramine, 2-hydroxybutyric acid, phosphatidylcholine (32:1), and taurochenodesoxycholic acid and lower levels of l-acetylcarnitine, creatinine, l-asparagine, l-glutamine, linoleic acid, pyruvic acid, palmitoleic acid, l-serine, oleic acid, myo-inositol, dodecanoic acid, l-methionine, hypoxanthine, palmitic acid, l-tryptophan, kynurenic acid, taurine, and 25-hydroxyvitamin D compared with controls. l-tryptophan and kynurenic acid were consistently downregulated in MDD patients, regardless of antidepressant exposure. Depression rating scores were negatively associated with decreased levels of l-tryptophan. Pathway and network analyses revealed altered amino acid metabolism and lipid metabolism, especially for the tryptophan–kynurenine pathway and fatty acid metabolism, in the peripheral system of MDD patients. Taken together, our integrated results revealed that metabolic changes in the peripheral blood were associated with MDD, particularly decreased l-tryptophan and kynurenic acid levels, and alterations in the tryptophan–kynurenine and fatty acid metabolism pathways. Our findings may facilitate biomarker development and the elucidation of the molecular mechanisms that underly MDD.
Subject terms: Biochemistry, Diagnostic markers, Depression
Introduction
Major depressive disorder (MDD) is a mental disorder with symptoms that include low mood for at least 2 weeks, loss of interest, fatigue, and feelings of guilt [1]. MDD is a serious mental illness, characterized by high morbidity and a high suicide rate [2], and was the leading cause of disability in 2016 [3]. The lifetime prevalence of MDD has been reported to be ~20%, with increasing morbidity during recent decades [4, 5]. However, the clinical diagnosis of MDD remains underestimated because of high diagnostic error rates in primary care [6]. As a complex and heterogeneous mental disease, no robust peripheral biomarkers currently exist for MDD, and the molecular mechanisms underlying this disease remain unclear, which has impeded objective diagnoses and clinical therapy [7, 8].
Among the various omic techniques, metabolomics, which can be used to characterize the metabolic profiles of biological samples, is the best tool for determining phenotypes and can be utilized to identify disease-specific biomarkers and mechanisms [9, 10]. Mass spectrometry (MS)- and nuclear magnetic resonance (NMR)-based techniques have been widely used during biomarker development and to examine the molecular mechanisms that underly neuropsychiatric diseases [11, 12]. A range of studies has revealed perturbed metabolomes in the peripheral tissues of MDD patients. For example, we previously reported the use of plasma metabolic profiles as potential biomarkers among adult and adolescent patients with MDD [13, 14]. Further, we found that the dysbiosis of the gut microbiome may play a causal role in the induction of depressive-like behaviors in rodents via effects on metabolism [15]. Other studies have also reported that metabolic changes in the periphery may represent potential therapeutic targets for depression [16, 17]. These findings imply that metabolites may play important roles during the brain–body interactions involved in depression, and peripheral metabolite levels have been associated with hippocampal subfield volumes [18], hypothalamic pituitary adrenal axis activity [19], and neurocognitive function [20].
Despite advances in metabolomics research examining MDD, the majority of these nontargeted or targeted metabolomic studies have had small sample sizes and have reported inconsistent findings, limiting their clinical applicability. To date, a range of meta-analyses has reported decreased levels of tryptophan, kynurenic acid, and kynurenine and increased glutamate levels, in MDD patients [21–23]. Despite these studies, which have examined the levels of one or more metabolites, no meta-analyses have been performed examining the metabolomic profiles of MDD. A comprehensive bioinformatics analysis, based on the differential metabolites identified at the metabolomics level, may provide important insights into the pathological molecular mechanisms underlying MDD.
Thus, the aim of the present study was to identify metabolic changes in the peripheral blood of MDD patients. We first generated a curated list of metabolites in the peripheral blood of MDD patients, according to the results of preexisting studies, and performed a meta-analysis to identify differential metabolites, which may serve as robust biomarkers for the clinical diagnosis of MDD. We then performed comprehensive pathway and network analyses to examine the biological themes associated with these metabolic changes.
Materials and methods
Identification of relevant studies
A flowchart describing the process used to identify relevant studies is shown in Supplementary Fig. 1. Detailed methods describing the identification of relevant studies are provided in Supplementary Materials. In brief, clinical studies that compared metabolic changes between MDD and control groups were identified in MENDA (http://menda.cqmu.edu.cn:8080/index.php), our online database of existing metabolic characterization studies associated with depression. We updated our search up to January 2019, using our previously reported search terms [24]. In this study, we narrowed our selection according to the following steps. We included studies that compared metabolite levels in serum or plasma samples between MDD patients and controls and detected these changes using MS-based or NMR-based techniques.
Data curation
We extracted study information and metabolite data from the selected studies. Recorded study information included methodological and demographic information, including the biological samples used, the recruiting area, sample size, mean age, percent of females, antidepressant-free patients, disease severity measure, and the analytical platform used. Curated metabolite data included each metabolite examined, with accompanying statistic data (mean, standard deviation, and sample size). Other data, such as standard errors, p values, or interquartile ranges, were transformed, as previously reported [25].
Identification of differential metabolites in MDD
Meta-analyses were performed using statistical software (Stata v14.0; Stata Co., College Station, TX, USA), as follows. Only metabolites reported in at least three different datasets were selected for analysis [26]. Standardized effect sizes across studies were combined, based on the statistics reported for each metabolite (mean, standard deviation, and sample size) by the original reports. The standardized mean difference (SMD) and the 95% confidence interval (CI) were estimated. A random-effects model was used to determine the expected high degree of heterogeneity across studies, as reported in previous molecular studies [27, 28]. Positive and negative SMD values indicated higher and lower levels of metabolites in the MDD group, relative to the control group, respectively. Statistical significance was set at p < 0.05.
Publication bias was assessed using the Egger’s test, with a p value < 0.10 indicating the presence of publication bias [29]. The Duval and Tweedie’s trim and fill method was also used to reduce bias among the pooled estimates [30]. To test the sources of potential heterogeneity, subgroup analyses were performed, according to antidepressant use (yes or no) and the biological sample used (plasma or serum). Sensitivity analyses were performed for studies that recruited patients with a mean age of >18 years, by excluding studies that recruited pediatric patients, and for studies using MS analytic platforms, by excluding NMR studies. Meta-regression analyses were performed to investigate the influence of each variable (sample size, the proportion of females, mean age, and disease severity) on the SMD. Details describing disease severity assessment are provided in Supplementary Materials. Meta-regression analyses were only conducted for metabolites that were reported in at least ten datasets, as results using fewer datasets can lead to increased risk of spurious findings [31].
Bioinformatics analysis
To reveal the biological functions of the differential metabolites and their interactions, we performed pathway and network analyses. Detailed methods are available in Supplementary Materials. In brief, altered metabolic pathways were identified, using metabolic pathway analysis in MetaboAnalyst 4.0 [32], which performed an enrichment analysis based on predefined KEGG pathways and differential metabolites [33]. Ingenuity pathways analysis (IPA, http://www.ingenuity.com) was used for further analysis. Canonical pathway analysis was performed to identify altered pathways within the Ingenuity Pathway Knowledge Base. For both metabolic pathway analysis and canonical pathway analysis, a pathway with a p value < 0.05 was considered to be significantly enriched. Network analysis was also performed in IPA to construct molecular networks, based on interactions among input metabolites and other biological molecules.
Results
Characteristics of the included studies
Lists of included and excluded studies are provided in Supplementary Data. A total of 49 comparisons (48 two-arm comparisons and 1 three-arm comparison) were included. The characteristics of the included studies are summarized in Supplementary Table 1. Of these comparisons, 47 used the MS platform, 33 measured metabolites in plasma, and 24 recruited antidepressant-free patients. All included studies examined the associations between metabolites and MDD using cross-sectional data. The sample sizes ranged from 16 to 2812 participants per study, with a median study size of 89 participants. The mean age of participants ranged from 14.0 to 72.2 years, with a median age of 37.1 years. The median proportion of females was 58.1%.
Differential blood metabolites associated with MDD
A total of 83 metabolites, which were reported in at least three different datasets, were used for the differential metabolite identification (Table 1). A total of 23 metabolites were found to be significantly differentially regulated between the MDD and control groups (Supplementary Fig. 2). MDD patients were characterized by higher levels of asymmetric dimethylarginine, tyramine, 2-hydroxybutyric acid, phosphatidylcholine (32:1), and taurochenodesoxycholic acid and lower levels of l-acetylcarnitine, creatinine, l-asparagine, l-glutamine, linoleic acid, pyruvic acid, palmitoleic acid, l-serine, oleic acid, myo-inositol, dodecanoic acid, l-methionine, hypoxanthine, palmitic acid, l-tryptophan, kynurenic acid, taurine, and 25-hydroxyvitamin D. Forest plots for these differential metabolites are shown in Supplementary Fig. 3. The heterogeneity among effect sizes was significant for 15 differential metabolites (p for heterogeneity <0.05). The Egger test indicated potential publication bias for l-tryptophan (p = 0.004), l-serine (p = 0.034), 25-hydroxyvitamin D (p = 0.060), l-glutamine (p = 0.067), l-asparagine (p = 0.079), and tyramine (p = 0.091). After quantifying the potential effects of small-study bias, using the trim and fill method, the imputations resulted in larger summary SMDs (−0.70 for l-tryptophan; −1.80 for l-serine; −0.30 for 25-hydroxyvitamin D; −2.63 for l-glutamine; −2.20 for l-asparagine; and 1.12 for tyramine).
Table 1.
Effect sizes of blood metabolites between patients with major depressive disorder and controls.
| Metabolites | N Comparisons | N MDD/CON | SMD (95% CI) | p valuea (overall) | I2 | p valueb (heterogeneity) | p valuec (Egger) |
|---|---|---|---|---|---|---|---|
| Kynurenic acid | 13 | 797/653 | −0.42 (−0.52 to −0.31) | <0.001 | 1.2% | 0.434 | 0.647 |
| 2-Hydroxybutyric acid | 3 | 76/83 | 1.02 (0.69 to 1.36) | <0.001 | 0.0% | 0.652 | 0.990 |
| Dodecanoic acid | 4 | 151/114 | −0.73 (−0.99 to −0.48) | <0.001 | 0.0% | 0.544 | 0.495 |
| 25-Hydroxyvitamin D | 3 | 1130/521 | −0.29 (−0.39 to −0.18) | <0.001 | 0.0% | 0.811 | 0.060 |
| Tyramine | 3 | 133/121 | 1.12 (0.70 to 1.55) | <0.001 | 58.8% | 0.089 | 0.091 |
| Phosphatidylcholine (32:1) | 3 | 150/130 | 0.56 (0.32 to 0.80) | <0.001 | 0.0% | 0.482 | 0.117 |
| l-Tryptophan | 26 | 2390/2928 | −0.46 (−0.66 to −0.26) | <0.001 | 87.8% | <0.001 | 0.004 |
| Myo-inositol | 3 | 113/77 | −0.79 (−1.17 to −0.41) | <0.001 | 28.3% | 0.248 | 0.619 |
| Linoleic acid | 5 | 148/104 | −0.99 (−1.46 to −0.51) | <0.001 | 63.4% | 0.027 | 0.348 |
| Oleic acid | 5 | 148/104 | −0.84 (−1.31 to −0.36) | 0.001 | 63.6% | 0.027 | 0.368 |
| Taurochenodesoxycholic acid | 4 | 176/180 | 0.33 (0.11 to 0.54) | 0.003 | 0.0% | 0.446 | 0.320 |
| Creatinine | 5 | 186/175 | −1.79 (−3.02 to −0.56) | 0.004 | 95.7% | <0.001 | 0.321 |
| l-Glutamine | 8 | 329/287 | −1.28 (−2.19 to −0.37) | 0.006 | 95.8% | <0.001 | 0.067 |
| Asymmetric dimethylarginine | 4 | 191/342 | 2.52 (0.68 to 4.36) | 0.007 | 97.9% | <0.001 | 0.202 |
| l-Acetylcarnitine | 3 | 138/109 | −2.06 (−3.58 to −0.54) | 0.008 | 95.4% | <0.001 | 0.106 |
| Hypoxanthine | 6 | 259/386 | −0.70 (−1.21 to −0.18) | 0.009 | 85.9% | <0.001 | 0.391 |
| Palmitic acid | 4 | 139/94 | −0.69 (−1.23 to −0.16) | 0.011 | 71.2% | 0.015 | 0.288 |
| Pyruvic acid | 3 | 101/92 | −0.92 (−1.68 to −0.17) | 0.017 | 80.7% | 0.006 | 0.370 |
| l-Asparagine | 6 | 221/195 | −1.33 (−2.45 to −0.21) | 0.020 | 95.7% | <0.001 | 0.079 |
| Palmitoleic acid | 3 | 93/69 | −0.90 (−1.70 to −0.10) | 0.027 | 79.8% | 0.007 | 0.228 |
| l-Methionine | 8 | 356/306 | −0.73 (−1.40 to −0.06) | 0.032 | 93.4% | <0.001 | 0.299 |
| l-Serine | 8 | 296/245 | −0.90 (−1.73 to −0.07) | 0.034 | 94.5% | <0.001 | 0.034 |
| Taurine | 6 | 270/218 | −0.33 (−0.65 to −0.01) | 0.043 | 65.0% | 0.014 | 0.399 |
| Glycine | 8 | 325/262 | −0.91 (−1.82 to 0.01) | 0.051 | 95.7% | <0.001 | 0.096 |
| Capric acid | 4 | 151/114 | −1.31 (−2.65 to 0.03) | 0.056 | 95.6% | <0.001 | 0.033 |
| l-Lysine | 6 | 221/195 | −1.03 (−2.10 to 0.04) | 0.059 | 95.4% | <0.001 | 0.053 |
| Citric acid | 5 | 143/135 | −0.96 (−1.97 to 0.05) | 0.063 | 92.5% | <0.001 | 0.571 |
| l-Valine | 8 | 325/262 | −0.71 (−1.48 to 0.06) | 0.071 | 94.3% | <0.001 | 0.049 |
| l-Kynurenine | 18 | 2020/2595 | −0.17 (−0.36 to 0.02) | 0.077 | 81.9% | <0.001 | 0.948 |
| l-Phenylalanine | 7 | 271/245 | 0.68 (−0.08 to 1.44) | 0.079 | 93.4% | <0.001 | 0.133 |
| l-Threonine | 8 | 296/245 | −0.64 (−1.36 to 0.09) | 0.085 | 93.1% | <0.001 | 0.088 |
| Urea | 3 | 113/89 | −2.11 (−4.59 to 0.38) | 0.097 | 97.9% | <0.001 | 0.018 |
| Gluconic acid | 3 | 89/75 | −0.27 (−0.58 to 0.05) | 0.099 | 0.0% | 0.793 | 0.412 |
| l-Tyrosine | 8 | 329/287 | −0.31 (−0.69 to 0.06) | 0.099 | 79.4% | <0.001 | 0.954 |
| Aminoadipic acid | 5 | 190/136 | 1.15 (−0.22 to 2.51) | 0.100 | 96.4% | <0.001 | 0.032 |
| Gamma-aminobutyric acid | 7 | 246/184 | 0.81 (−0.17 to 1.79) | 0.104 | 94.5% | <0.001 | 0.775 |
| Creatine | 5 | 186/175 | −0.72 (−1.61 to 0.16) | 0.108 | 93.2% | <0.001 | 0.385 |
| l-Alanine | 8 | 325/262 | −0.67 (−1.48 to 0.15) | 0.109 | 94.8% | <0.001 | 0.060 |
| Phosphatidylethanolamine (34:2) | 3 | 150/130 | 0.38 (−0.12 to 0.87) | 0.136 | 73.0% | 0.025 | 0.146 |
| Stearic acid | 3 | 64/54 | −0.42 (−1.00 to 0.16) | 0.152 | 47.7% | 0.148 | 0.938 |
| l-Isoleucine | 7 | 279/237 | −0.58 (−1.40 to 0.25) | 0.171 | 94.4% | <0.001 | 0.113 |
| Succinic acid | 4 | 163/139 | 0.69 (−0.35 to 1.74) | 0.194 | 94.4% | <0.001 | 0.001 |
| Ornithine | 8 | 370/498 | −0.31 (−0.78 to 0.17) | 0.205 | 89.1% | <0.001 | 0.236 |
| Homovanillic acid | 3 | 117/114 | 1.36 (−0.80 to 3.52) | 0.216 | 97.8% | <0.001 | 0.316 |
| Glyceric acid | 4 | 122/99 | 1.29 (−0.76 to 3.35) | 0.217 | 97.1% | <0.001 | 0.613 |
| l-Leucine | 8 | 325/262 | −0.14 (−0.37 to 0.08) | 0.217 | 41.5% | 0.102 | 0.870 |
| Adenosine diphosphate | 3 | 76/83 | 2.01 (−1.20 to 5.22) | 0.220 | 98.0% | <0.001 | 0.448 |
| l-Malic acid | 3 | 76/83 | 0.87 (−0.57 to 2.31) | 0.235 | 93.7% | <0.001 | 0.908 |
| Indoleacetic acid | 3 | 82/83 | 1.49 (−1.03 to 4.00) | 0.247 | 97.6% | <0.001 | 0.300 |
| Symmetric dimethylarginine | 3 | 166/317 | 1.05 (−0.75 to 2.84) | 0.253 | 97.7% | <0.001 | 0.384 |
| Arachidonic acid | 3 | 93/69 | −0.52 (−1.42 to 0.37) | 0.253 | 84.4% | 0.002 | 0.435 |
| Pyroglutamic acid | 5 | 174/149 | −0.64 (−1.76 to 0.49) | 0.267 | 95.1% | <0.001 | 0.680 |
| Serotonin | 5 | 144/184 | −0.26 (−0.76 to 0.23) | 0.301 | 77.9% | 0.001 | 0.575 |
| Dimethylglycine | 3 | 76/83 | 1.18 (−1.08 to 3.44) | 0.305 | 97.0% | <0.001 | 0.543 |
| l-Carnitine | 3 | 138/109 | −0.40 (−1.18 to 0.37) | 0.308 | 88.2% | <0.001 | 0.587 |
| Isocitric acid | 4 | 122/108 | 1.06 (−0.98 to 3.10) | 0.308 | 97.4% | <0.001 | 0.186 |
| Choline | 4 | 134/125 | 0.75 (−0.72 to 2.22) | 0.316 | 96.2% | <0.001 | 0.253 |
| Glycoursodeoxycholic acid | 3 | 144/130 | 0.19 (−0.19 to 0.58) | 0.327 | 53.5% | 0.116 | 0.800 |
| Ethanolamine | 5 | 208/170 | −0.94 (−2.84 to 0.97) | 0.336 | 98.1% | <0.001 | 0.613 |
| cis-Aconitic acid | 3 | 76/83 | 1.42 (−1.50 to 4.35) | 0.341 | 97.9% | <0.001 | 0.304 |
| 1-Methylhistidine | 4 | 161/111 | −0.33 (−1.01 to 0.35) | 0.342 | 85.7% | <0.001 | 0.540 |
| l-Lactic acid | 5 | 180/150 | −0.37 (−1.23 to 0.49) | 0.400 | 92.3% | <0.001 | 0.731 |
| Deoxycholic acid | 3 | 144/130 | −0.17 (−0.60 to 0.25) | 0.427 | 61.1% | 0.076 | 0.985 |
| O-Phosphoethanolamine | 4 | 161/111 | −0.21 (−0.75 to 0.33) | 0.448 | 78.3% | 0.003 | 0.814 |
| 5-Hydroxylysine | 3 | 135/86 | 0.73 (−1.22 to 2.68) | 0.464 | 97.3% | <0.001 | 0.107 |
| Citrulline | 8 | 349/473 | 0.18 (−0.31 to 0.68) | 0.466 | 89.3% | <0.001 | 0.533 |
| l-Proline | 6 | 216/155 | −0.35 (−1.40 to 0.70) | 0.510 | 94.9% | <0.001 | 0.068 |
| l-alpha-aminobutyric acid | 3 | 145/112 | 0.17 (−0.40 to 0.73) | 0.563 | 77.4% | 0.012 | 0.416 |
| Sarcosine | 4 | 144/105 | −0.20 (−0.95 to 0.55) | 0.602 | 86.5% | <0.001 | 0.794 |
| Quinolinic acid | 10 | 453/415 | −0.06 (−0.32 to 0.20) | 0.660 | 69.7% | <0.001 | 0.500 |
| 3-Hydroxybutyric acid | 4 | 134/125 | 0.11 (−0.41 to 0.63) | 0.670 | 74.6% | 0.008 | 0.821 |
| Betaine | 4 | 128/133 | 0.26 (−0.99 to 1.51) | 0.679 | 95.3% | <0.001 | 0.410 |
| 4-Hydroxyproline | 5 | 170/130 | −0.16 (−0.97 to 0.65) | 0.701 | 90.3% | <0.001 | 0.794 |
| l-Histidine | 7 | 279/237 | 0.09 (−0.51 to 0.70) | 0.763 | 90.2% | <0.001 | 0.479 |
| l-Aspartic acid | 7 | 262/211 | 0.09 (−0.53 to 0.72) | 0.771 | 90.0% | <0.001 | 0.991 |
| Phosphatidylcholine (32:0) | 3 | 150/130 | 0.08 (−0.51 to 0.66) | 0.799 | 80.6% | 0.006 | 0.694 |
| Hydroxykynurenine | 6 | 261/300 | 0.03 (−0.19 to 0.25) | 0.804 | 37.7% | 0.155 | 0.615 |
| l-Cystine | 3 | 135/86 | 0.25 (−2.70 to 3.20) | 0.870 | 98.5% | <0.001 | 0.435 |
| l-Glutamic acid | 7 | 271/245 | 0.06 (−0.73 to 0.86) | 0.873 | 94.1% | <0.001 | 0.492 |
| 3-Aminoisobutanoic acid | 4 | 161/111 | −0.06 (−1.03 to 0.92) | 0.911 | 93.0% | <0.001 | 0.932 |
| beta-Alanine | 5 | 170/130 | −0.02 (−0.47 to 0.43) | 0.944 | 70.5% | 0.009 | 0.697 |
| l-Arginine | 9 | 404/498 | −0.01 (−0.73 to 0.70) | 0.970 | 95.2% | <0.001 | 0.578 |
| Cholesterol | 3 | 113/86 | 0.00 (−1.17 to 1.16) | 0.997 | 92.3% | <0.001 | 0.867 |
CI confidence interval, CON control, MDD major depressive disorder, N number, SMD standardized mean difference
ap value for between-group effect sizes
bp value for heterogeneity calculated using a chi-square analysis
cp value for Egger’s test. A p < 0.1 is indicative of this asymmetry
Subgroup analysis based on antidepressant exposure revealed that antidepressant-free MDD (AF-MDD) patients had higher levels of l-proline and 3-hydroxybutyric acid and lower levels of creatinine, l-tryptophan, kynurenic acid, l-leucine, l-kynurenine, taurine, and l-histidine compared with controls. Antidepressant-treated MDD (AT-MDD) patients had decreased levels of glycine, hypoxanthine, kynurenic acid, l-asparagine, l-glutamine, l-serine, and l-tryptophan compared with controls (all p < 0.05; Supplementary Table 2). Interestingly, six of the seven differential metabolites identified in AT-MDD patients were shared among all MDD patients, whereas five of the nine differential metabolites identified in AF-MDD patients were unique to AF-MDD (Supplementary Fig. 4a). Furthermore, l-tryptophan and kynurenic acid were consistently downregulated, regardless of antidepressant exposure. There were no significant differences in the SMDs between biological samples (plasma versus serum; Supplementary Fig. 4b, Supplementary Table 3).
Sensitivity analysis stratified by mean age found 16 differential metabolites between adult MDD patients and controls aged > 18 years (Supplementary Table 4), two of which, phosphatidylcholine (32:1) and myo-inositol, were only significantly dysregulated in adult patients (Supplementary Fig. 4c). Sensitivity analysis stratified by analytic technique identified 20 differential metabolites in studies that used MS platforms, all of which were also identified using all analytic techniques (Supplementary Fig. 4d, Supplementary Table 5). In the meta-regression analysis, no significant associations between factors (sample size, proportion of females, mean age, and disease severity) and effect sizes (all meta-regression p > 0.05; Supplementary Table 6) were identified, except for a significant negative correlation between disease severity and effect size for l-tryptophan [slope = −0.040, 95% CI (−0.076, −0.005), p = 0.029; Supplementary Fig. 5], suggesting that patients with higher depression rating scores may have lower levels of l-tryptophan.
Bioinformatics analysis of blood metabolites in MDD
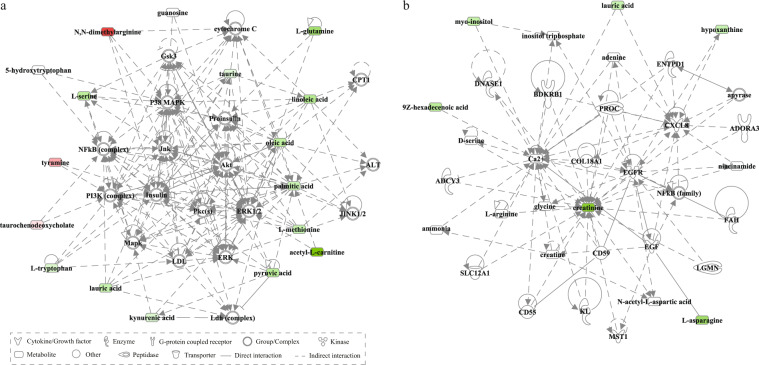
Bioinformatics analysis was performed separately for three groups of differential metabolites (all MDD, AF-MDD, and AT-MDD patients). For all patients, metabolic pathway analysis using MetaboAnalyst identified 10 altered metabolic pathways; the top-ranked metabolic pathways were “nitrogen metabolism,” “aminoacyl-tRNA biosynthesis,” and “fatty acid biosynthesis.” Canonical pathway analysis in IPA identified 20 significantly disturbed canonical pathways; the top-ranked pathways were “tRNA charging,” “glycine betaine degradation,” and “l-serine degradation” (Table 2). To better understand the interactions between pathways, we summarized these pathways in a brief plot (Fig. 1). Network analysis in IPA revealed a significantly altered network for “developmental disorder, hereditary disorder, metabolic disease” (score 42), which was associated with 15 differential metabolites (Fig. 2a), and for “small molecule biochemistry, increased levels of creatinine, cell signaling” (score 13), which was associated with six differential metabolites (Fig. 2b).
Table 2.
Altered pathways in the blood of patients with major depressive disorder.
| Pathways | p valuea |
|---|---|
| Metabolic pathways identified by MetaboAnalyst | |
| Nitrogen metabolism | <0.001 |
| Aminoacyl-tRNA biosynthesis | <0.001 |
| Fatty acid biosynthesis | 0.001 |
| Alanine, aspartate and glutamate metabolism | 0.001 |
| Linoleic acid metabolism | 0.008 |
| Glycine, serine and threonine metabolism | 0.009 |
| Cyanoamino acid metabolism | 0.009 |
| Cysteine and methionine metabolism | 0.013 |
| Taurine and hypotaurine metabolism | 0.014 |
| Arginine and proline metabolism | 0.031 |
| Canonical pathways identified by IPA | |
| tRNA charging | <0.001 |
| Glycine betaine degradation | <0.001 |
| l-Serine degradation | <0.001 |
| Asparagine biosynthesis I | 0.003 |
| Palmitate biosynthesis I (animals) | 0.006 |
| Superpathway of methionine degradation | 0.007 |
| Cysteine biosynthesis III (mammalia) | 0.010 |
| NAD biosynthesis II (from tryptophan) | 0.021 |
| Folate transformations I | 0.021 |
| HIF1α signaling | 0.024 |
| Asparagine degradation I | 0.035 |
| Glutamine degradation I | 0.035 |
| Pyruvate fermentation to lactate | 0.047 |
| Glycine biosynthesis III | 0.047 |
| Alanine degradation III | 0.047 |
| Alanine biosynthesis II | 0.047 |
| Myo-inositol biosynthesis | 0.047 |
| l-Cysteine degradation II | 0.047 |
| Phosphatidylethanolamine biosynthesis III | 0.047 |
| Glycine biosynthesis I | 0.047 |
ap values were calculated from hypergeometric tests in MetaboAnalyst, and from Fisher’s exact tests in Ingenuity pathways analysis
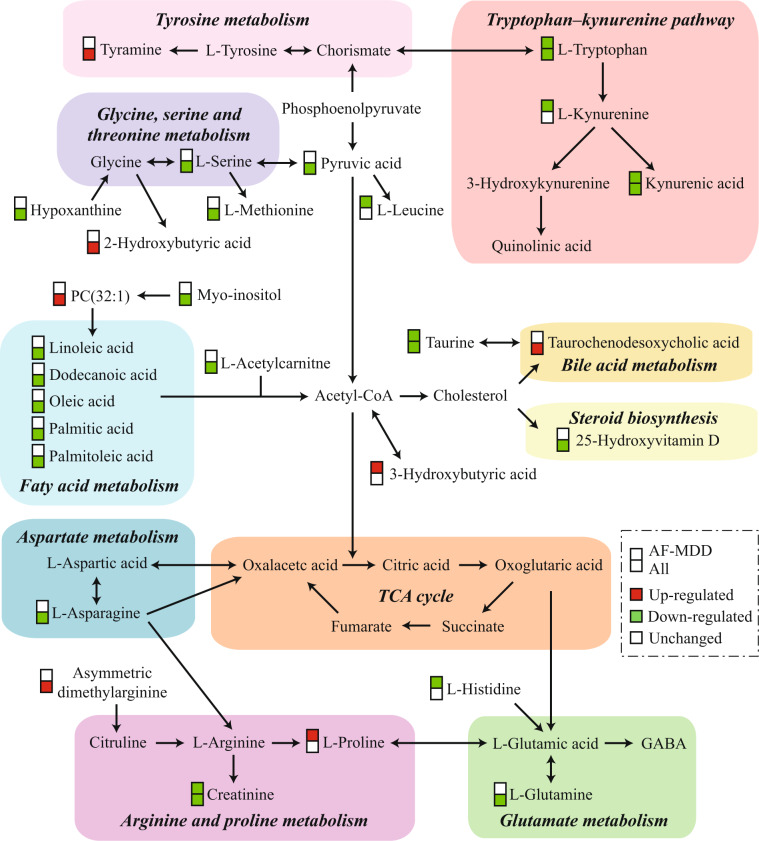
Fig. 1. A simplified schematic diagram of the altered metabolic pathways in the blood of patients with MDD.
For differential metabolites, boxes in red represent increased levels, boxes in green represent decreased levels, and boxes in white represent no significant change when compared with controls. Upper and lower boxes represent antidepressant-free major depressive disorder (AF-MDD) and all patients, respectively. Acetyl-CoA acetyl coenzyme A; GABA gamma-aminobutyric acid; MDD major depressive disorder; PC phosphatidylcholine; TCA tricarboxylic acid cycle.
Fig. 2. Altered networks associated with the differential metabolites in the blood of patients with MDD.
a This network is associated with “developmental disorder, hereditary disorder, and metabolic disease” (score 42, 15 differential metabolites). b This network is associated with “small molecule biochemistry, increased levels of creatinine, and cell signaling” (score 13, six differential metabolites). Differential metabolites highlighted in red represent increased levels, whereas metabolites highlighted in green represent decreased levels when compared with controls.
We then investigated the altered pathways and networks associated with antidepressant exposure. Respectively, five and nine metabolic pathways were significantly altered in AF-MDD and AT-MDD patients (Supplementary Table 7). “Aminoacyl-tRNA biosynthesis,” “nitrogen metabolism,” and “tryptophan metabolism” were shared pathways among both groups of patients. The canonical pathway analysis identified 4 and 45 significantly disturbed pathways for AF-MDD and AT-MDD patients, respectively (Supplementary Table 8), “tRNA charging” was a shared pathway. Network analysis revealed that “organismal injury and abnormalities, increased levels of creatinine, small molecule biochemistry” (score 27, nine differential metabolites) was a significantly altered network for AF-MDD (Supplementary Fig. 6a), and that “amino acid metabolism, cell-to-cell signaling and interaction, molecular transport” (score 21, seven differential metabolites) was significantly altered for AT-MDD (Supplementary Fig. 6b).
Discussion
In the present study, we integrated the peripheral blood metabolic profiles from a large sample of MDD patients and controls and found that the levels of five metabolites were significantly elevated in the peripheral blood of MDD patients compared with controls, whereas the levels of 18 metabolites were reduced in MDD patients compared with controls. l-tryptophan and kynurenic acid were consistently downregulated in MDD patients, regardless of antidepressant exposure. Moreover, we explored the biological themes associated with these metabolic changes, based on pathway and network analyses, which suggested that several pathways involved in amino acid metabolism and lipid metabolism, especially the tryptophan–kynurenine pathway and fatty acid metabolism, were significantly perturbed in the peripheral blood of MDD patients.
Increasing investigations have sought to identify potential blood-based biomarkers of psychiatric diseases [34, 35], and our findings may facilitate further biomarker development for MDD. Among the altered metabolites, only tryptophan, kynurenic acid, and 25-hydroxyvitamin D were identified in studies that examined relatively large samples (>1000 participants), which may provide more credible estimates than smaller samples. Furthermore, we observed heterogeneity for the metabolic changes among the included studies, which could partially be explained by antidepressant exposure. Thus, antidepressant exposure should be considered in future metabolic biomarker studies. Subgroup analyses of antidepressant exposure found that only tryptophan and kynurenic acid were consistently downregulated in MDD patients, regardless of antidepressant exposure, and meta-regression analysis revealed that patients with higher depression scores might have lower levels of tryptophan, which is consistent with previous meta-analyses [21, 23]. These findings suggest that certain metabolic markers may be used to distinguish the MDD disease state and to monitor the therapeutic response [36, 37], with the most convincing evidence existing for l-tryptophan, followed by kynurenic acid.
Despite these findings, whether any of these metabolites can be used as biomarkers for MDD remains unresolved. Biomarkers in peripheral blood might not, in theory, reflect metabolic changes in the brain, which requires further direct evidence. Moreover, the utility of any individual metabolite biomarker for MDD practically remains unclear. For example, we reported that the area under the curve for tryptophan associated with MDD was 0.74, which remains inadequate for clinical practice, suggesting that diagnostic systems that include panels of differential metabolites are likely to result in better diagnostic efficacy than individual metabolites [38]. Therefore, more metabolic profiling studies are necessary to develop promising diagnostic systems. Given the heterogeneity of depression, such as concomitant somatic diseases, clinical variances should be addressed to improve the diagnostic performance. Because metabolomics studies face challenges from methodological sources of variance, the application of more rigorous experimental designs and processes will be necessary for future progress, as described in methodological reviews [39, 40].
Our integrated results also provided clues to the potential biological mechanisms that underly MDD. We found significantly disturbed amino acid metabolism in the peripheral blood of MDD patients. The levels of five amino acids (serine, methionine, asparagine, glutamine, and tryptophan) were decreased in the blood samples from all patients, and the levels of four amino acids (histidine, leucine, taurine, and tryptophan) were decreased in AF-MDD patients compared with controls. Pathway analyses revealed nitrogen metabolism and tRNA charging to be among the top-ranked pathways, reflecting perturbations in amino acid metabolism [41, 42]. Moreover, we found that the tryptophan–kynurenine pathway was the most enriched amino acid metabolism pathway. This pathway produces both neuroprotective (kynurenic acid) and neurotoxic (3-hydroxykynurenine and quinolinic acid) metabolites [43], and decreased levels of kynurenic acid have been reported to indicate increased neurotoxic burdens during in the course of depression, which could be reversed by physical exercise and electroconvulsive therapy [16, 44]. Furthermore, pro-inflammatory cytokines (e.g., interferon) have been demonstrated to mediate the enzymatic activity of the kynurenine pathway [45], resulting in decreased neuroprotective effects for kynurenic acid. This result is consistent with the findings that interferon-induced depression in ~50% of patients receiving interferon treatment [46] and that interferon treatment resulted in decreased levels of tryptophan and increased levels of kynurenine and quinolinic acid in the rodent brain [47, 48]. Finally, anti-inflammatory treatments have been reported to decrease depressive symptoms in patients [49]. Overall, these data suggest that the tryptophan–kynurenine pathway may be involved in depression by mediating inflammatory responses.
Other amino acid metabolism pathways, as summarized in Fig. 1, were also found to be enriched. One interpretation of these findings is that these pathways are involved in neurotransmission. Glutamine is a precursor of glutamate and gamma-aminobutyric acid [50], and altered levels of circulating glutamine may affect the gamma-aminobutyric acid levels in the brain [51]. Proline is synthesized from glutamic acid, and chronic hyperprolinemia can lead to reduced glutamate uptake, increased adenosine triphosphate catabolism, and increased pro-inflammatory cytokine levels [52–54]. Moreover, monoamine neurotransmitters may be involved in MDD, as tryptophan is the precursor for serotonin and tyrosine is the precursor for catecholamines. In addition to neurotransmission, these disturbed pathways have also been associated with energy metabolism. In addition to the downregulated amino acids, being glucogenic or ketogenic, other downregulated metabolites, including creatinine, hypoxanthine, and pyruvic acid, were associated with periphery energy dyshomeostasis [55, 56]. Thus, these results suggested that disturbed amino acid metabolism may contribute to depression by modulating neurotransmission and energy metabolism.
Our integrated results also suggested that lipid metabolism was dysregulated during MDD. Fatty acid biosynthesis was among the top-ranked metabolic pathways, and the levels of five fatty acids and l-acetylcarnitine were significantly decreased in the blood of MDD patients compared with controls. Fatty acid alterations may contribute to depression through several mechanisms, such as affecting cell membrane structure, biological stress, and inflammatory responses [57]. l-Acetylcarnitine plays a pivotal role in the transport of fatty acids into the mitochondria for beta-oxidation, and l-acetylcarnitine supplementation was reported to have antidepressive effects [58]. We also found increased levels of taurochenodesoxycholic acid, a bile acid formed from taurine in the liver, in the blood of MDD patients. Animal studies of depression have reported increased taurochenodesoxycholic acid levels and decreased taurine levels in the liver [59, 60], indicating that primary bile acid biosynthesis may be associated with MDD. Furthermore, we observed decreased levels of 25-hydroxyvitamin D in the present study, which was consistent with a previous meta-analysis [61]. 25-Hydroxyvitamin D is the primary form of vitamin D in the human body, and a deficiency in vitamin D has been associated with higher rates of suicide and the elevation of pro-inflammatory cytokines [62, 63]. Taken together, these findings suggested that alterations in lipid metabolism may play a key role in the pathogenesis of MDD.
Our network analysis also revealed that the mitogen-activated protein kinase (MAPK) signaling pathway, which includes Mapk, extracellular signal-regulated kinase 1/2, c-Jun N-terminal kinase (Jnk), P38 MAPK, and protein kinase C [64], and the phosphoinositide 3-kinase/protein kinase B (PI3K/AKT) signaling pathway, which includes PI3K, Akt, glycogen synthase kinase 3, and insulin [65], were both linked to the identified altered networks, indicating cross-talk between signaling pathways and differentially expressed metabolites. We also reported decreased peripheral levels of myo-inositol, which forms the structural basis for secondary messengers in the phosphoinositol system [66]. Overall, our data supported the potential involvement of signaling pathways in the peripheral metabolic changes observed in MDD patients.
This study has several limitations. First, without metabolic and clinical data from individual patients, the adjustment of potential confounders was not possible during our analysis. The integration of individual patient data is required in future studies. Second, the sample size was relatively small for certain metabolites in our analysis, resulting in low statistical power for those metabolites. Further studies with more patients are required to validate our findings. Third, we only included studies that reported MS-based or NMR-based techniques. This decision was made a priori because these techniques have higher accuracies and wider detection ranges than other techniques, which has revolutionized metabolite measurement [67]. Although this strategy is also used in other meta-analyses [26, 68], however, this limitation may also lead to selection bias. For example, previous studies utilizing high-performance liquid chromatography with fluorescence detection reported decreased or unchanged tryptophan levels in MDD patients compared with controls [69, 70]. Fourth, confidence in metabolite annotation and quantification remains a primary challenge for metabolomics studies [71, 72]; therefore, plausible metabolite candidates from the included studies may limit the generalization of our findings. Finally, during the process of metabolite identification and standardization across studies, a small number of metabolites were lost due to the use of non-standardized metabolite nomenclature.
Summary
We identified differential metabolites in the peripheral blood of MDD patients, using a meta-analysis of curated metabolic characterization data from a large sample of patients. Subgroup analyses showed that antidepressant exposure was the most important source of heterogeneity. Pathway and network analyses revealed disturbances of amino acid and lipid metabolism, especially the tryptophan–kynurenine pathway and fatty acid metabolism, in the peripheral systems of MDD patients. This integrated approach may facilitate the development of biomarkers for MDD and help to determine the underlying molecular mechanisms associated with MDD. Future replication studies using larger sample sizes are necessary to confirm our findings.
Supplementary information
Acknowledgements
This work was supported by the National Key Research and Development Program of China (Grant no. 2017YFA0505700), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (Grant no. 2019PT320002300), and the Natural Science Foundation Project of China (Grant no. 81820108015).
Author contributions
Concept, design, and supervision of the study: PX; data acquisition/analysis/interpretation: JP, YL, HZ, LT, SG, YY, XC, YC, LY, YR, XZ, SX, XS, LL, PZ, and HW; drafting manuscript: JP and YL; revised the manuscript: HZ and JP, with the help of YL; drafting display items: JP and HZ. All authors read and approved the final version of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Juncai Pu, Yiyun Liu, Hanping Zhang
Supplementary information
The online version of this article (10.1038/s41380-020-0645-4) contains supplementary material, which is available to authorized users.
References
- 1.Mann JJ. The medical management of depression. N Engl J Med. 2005;353:1819–34. doi: 10.1056/NEJMra050730. [DOI] [PubMed] [Google Scholar]
- 2.Nanayakkara S, Misch D, Chang L, Henry D. Depression and exposure to suicide predict suicide attempt. Depress Anxiety. 2013;30:991–6. doi: 10.1002/da.22143. [DOI] [PubMed] [Google Scholar]
- 3.Vos T, Abajobir AA, Abate KH, Abbafati C, Abbas KM, Abd-Allah F, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1211–59. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasin DS, Sarvet AL, Meyers JL, Saha TD, Ruan WJ, Stohl M, et al. Epidemiology of adult DSM-5 major depressive disorder and its specifiers in the United States. JAMA Psychiatry. 2018;75:336–46. doi: 10.1001/jamapsychiatry.2017.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberger AH, Gbedemah M, Martinez AM, Nash D, Galea S, Goodwin RD. Trends in depression prevalence in the USA from 2005 to 2015: widening disparities in vulnerable groups. Psychol Med. 2018;48:1308–15. doi: 10.1017/S0033291717002781. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell AJ, Vaze A, Rao S. Clinical diagnosis of depression in primary care: a meta-analysis. Lancet. 2009;374:609–19. doi: 10.1016/S0140-6736(09)60879-5. [DOI] [PubMed] [Google Scholar]
- 7.Guest PC, Guest FL, Martins-de Souza D. Making sense of blood-based proteomics and metabolomics in psychiatric research. Int J Neuropsychopharmacol. 2016;19:pyv138. doi: 10.1093/ijnp/pyv138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan V, Nestler EJ. The molecular neurobiology of depression. Nature. 2008;455:894–902. doi: 10.1038/nature07455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guijas C, Montenegro-Burke JR, Warth B, Spilker ME, Siuzdak G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat Biotechnol. 2018;36:316–20. doi: 10.1038/nbt.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17:451–9. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sethi S, Brietzke E. Omics-based biomarkers: application of metabolomics in neuropsychiatric disorders. Int J Neuropsychopharmacol. 2015;19:pyv096. doi: 10.1093/ijnp/pyv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinones MP, Kaddurah-Daouk R. Metabolomics tools for identifying biomarkers for neuropsychiatric diseases. Neurobiol Dis. 2009;35:165–76. doi: 10.1016/j.nbd.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Zheng P, Gao HC, Li Q, Shao WH, Zhang ML, Cheng K, et al. Plasma metabonomics as a novel diagnostic approach for major depressive disorder. J Proteome Res. 2012;11:1741–8. doi: 10.1021/pr2010082. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Liu L, Lan X, Cohen D, Zhang Y, Ravindran AV, et al. Polyunsaturated fatty acids metabolism, purine metabolism and inosine as potential independent diagnostic biomarkers for major depressive disorder in children and adolescents. Mol Psychiatry. 2019;24:1478–88. doi: 10.1038/s41380-018-0047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry. 2016;21:786–96. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 16.Agudelo LZ, Femenía T, Orhan F, Porsmyr-Palmertz M, Goiny M, Martinez-Redondo V, et al. Skeletal muscle PGC-1α1 modulates kynurenine metabolism and mediates resilience to stress-induced depression. Cell. 2014;159:33–45. doi: 10.1016/j.cell.2014.07.051. [DOI] [PubMed] [Google Scholar]
- 17.Nasca C, Xenos D, Barone Y, Caruso A, Scaccianoce S, Matrisciano F, et al. L-acetylcarnitine causes rapid antidepressant effects through the epigenetic induction of mGlu2 receptors. Proc Natl Acad Sci USA. 2013;110:4804–9. doi: 10.1073/pnas.1216100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doolin K, Allers KA, Pleiner S, Liesener A, Farrell C, Tozzi L, et al. Altered tryptophan catabolite concentrations in major depressive disorder and associated changes in hippocampal subfield volumes. Psychoneuroendocrinology. 2018;95:8–17. doi: 10.1016/j.psyneuen.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 19.Sorgdrager FJH, Doornbos B, Penninx BWJH, de Jonge P, Kema IP. The association between the hypothalamic pituitary adrenal axis and tryptophan metabolism in persons with recurrent major depressive disorder and healthy controls. J Affect Disord. 2017;222:32–9. doi: 10.1016/j.jad.2017.06.052. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Zheng W, Liu W, Wang C, Zhan Y, Li H, et al. Cross-sectional relationship between kynurenine pathway metabolites and cognitive function in major depressive disorder. Psychoneuroendocrinology. 2019;101:72–9. doi: 10.1016/j.psyneuen.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Ogawa S, Fujii T, Koga N, Hori H, Teraishi T, Hattori K, et al. Plasma L-tryptophan concentration in major depressive disorder: new data and meta-analysis. J Clin Psychiatry. 2014;75:e906–15. doi: 10.4088/JCP.13r08908. [DOI] [PubMed] [Google Scholar]
- 22.Inoshita M, Umehara H, Watanabe SY, Nakataki M, Kinoshita M, Tomioka Y, et al. Elevated peripheral blood glutamate levels in major depressive disorder. Neuropsychiatr Dis Treat. 2018;14:945–53. doi: 10.2147/NDT.S159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogyu K, Kubo K, Noda Y, Iwata Y, Tsugawa S, Omura Y, et al. Kynurenine pathway in depression: a systematic review and meta-analysis. Neurosci Biobehav Rev. 2018;90:16–25. doi: 10.1016/j.neubiorev.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 24.Pu J, Yu Y, Liu Y, Tian L, Gui S, Zhong X, et al. MENDA: a comprehensive curated resource of metabolic characterization in depression. Brief Bioinform. 2019. 10.1093/bib/bbz055. [DOI] [PMC free article] [PubMed]
- 25.Higgins JPT, Green S. Cochrane handbook for systematic reviews of interventions version 5.1.0. Oxford: The Cochrane Collaboration; 2011. [Google Scholar]
- 26.Guasch-Ferré M, Hruby A, Toledo E, Clish CB, Martínez-González MA, Salas-Salvadó J, et al. Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care. 2016;39:833–46. doi: 10.2337/dc15-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green MJ, Matheson SL, Shepherd A, Weickert CS, Carr VJ. Brain-derived neurotrophic factor levels in schizophrenia: a systematic review with meta-analysis. Mol Psychiatry. 2011;16:960–72. doi: 10.1038/mp.2010.88. [DOI] [PubMed] [Google Scholar]
- 28.Dowlati Y, Herrmann N, Swardfager W, Liu H, Sham L, Reim EK, et al. A meta-analysis of cytokines in major depression. Biol Psychiatry. 2010;67:446–57. doi: 10.1016/j.biopsych.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 29.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–63. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 31.Köhler CA, Freitas TH, Maes M, de Andrade NQ, Liu CS, Fernandes BS, et al. Peripheral cytokine and chemokine alterations in depression: a meta-analysis of 82 studies. Acta Psychiatr Scand. 2017;135:373–87.. doi: 10.1111/acps.12698. [DOI] [PubMed] [Google Scholar]
- 32.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–94. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia J, Wishart DS. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26:2342–4. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- 34.Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 2013;11:126. doi: 10.1186/1741-7015-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan MK, Gottschalk MG, Haenisch F, Tomasik J, Ruland T, Rahmoune H, et al. Applications of blood-based protein biomarker strategies in the study of psychiatric disorders. Prog Neurobiol. 2014;122:45–72. doi: 10.1016/j.pneurobio.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Perlis RH. Abandoning personalization to get to precision in the pharmacotherapy of depression. World Psychiatry. 2016;15:228–35. doi: 10.1002/wps.20345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beger RD, Dunn W, Schmidt MA, Gross SS, Kirwan JA, Cascante M, et al. Metabolomics enables precision medicine: “a white paper, community perspective”. Metabolomics. 2016;12:149. doi: 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan J, Xia J, Deng F, Liang W, Wu J, Yin B, et al. Diagnosis of major depressive disorder based on changes in multiple plasma neurotransmitters: a targeted metabolomics study. Transl Psychiatry. 2018;8:130. doi: 10.1038/s41398-018-0183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andreazza AC, Laksono I, Fernandes BS, Toben C, Lewczuk P, Riederer P, et al. Guidelines for the standardized collection of blood-based biomarkers in psychiatry: steps for laboratory validity - a consensus of the Biomarkers Task Force from the WFSBP. World J Biol Psychiatry. 2019;20:340–51.. doi: 10.1080/15622975.2019.1574024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulaszewska MM, Weinert CH, Trimigno A, Portmann R, Andres Lacueva C, Badertscher R, et al. Nutrimetabolomics: an integrative action for metabolomic analyses in human nutritional studies. Mol Nutr Food Res. 2019;63:e1800384. doi: 10.1002/mnfr.201800384. [DOI] [PubMed] [Google Scholar]
- 41.Wu G. Amino acids: metabolism, functions, and nutrition. Amino Acids. 2009;37:1–17. doi: 10.1007/s00726-009-0269-0. [DOI] [PubMed] [Google Scholar]
- 42.Francklyn CS, Mullen P. Progress and challenges in aminoacyl-tRNA synthetase-based therapeutics. J Biol Chem. 2019;294:5365–85.. doi: 10.1074/jbc.REV118.002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lovelace MD, Varney B, Sundaram G, Lennon MJ, Lim CK, Jacobs K, et al. Recent evidence for an expanded role of the kynurenine pathway of tryptophan metabolism in neurological diseases. Neuropharmacology. 2017;112:373–88. doi: 10.1016/j.neuropharm.2016.03.024. [DOI] [PubMed] [Google Scholar]
- 44.Guloksuz S, Arts B, Walter S, Drukker M, Rodriguez L, Myint AM, et al. The impact of electroconvulsive therapy on the tryptophan-kynurenine metabolic pathway. Brain Behav Immun. 2015;48:48–52. doi: 10.1016/j.bbi.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 45.Yasui H, Takai K, Yoshida R, Hayaishi O. Interferon enhances tryptophan metabolism by inducing pulmonary indoleamine 2,3-dioxygenase: its possible occurrence in cancer patients. Proc Natl Acad Sci USA. 1986;83:6622–6. doi: 10.1073/pnas.83.17.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baranyi A, Meinitzer A, Stepan A, Putz-Bankuti C, Breitenecker RJ, Stauber R, et al. A biopsychosocial model of interferon-alpha-induced depression in patients with chronic hepatitis C infection. Psychother Psychosom. 2013;82:332–40. doi: 10.1159/000348587. [DOI] [PubMed] [Google Scholar]
- 47.Fischer CW, Eskelund A, Budac DP, Tillmann S, Liebenberg N, Elfving B, et al. Interferon-alpha treatment induces depression-like behaviour accompanied by elevated hippocampal quinolinic acid levels in rats. Behav Brain Res. 2015;293:166–72. doi: 10.1016/j.bbr.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 48.Saito K, Markey SP, Heyes MP. Effects of immune activation on quinolinic acid and neuroactive kynurenines in the mouse. Neuroscience. 1992;51:25–39. doi: 10.1016/0306-4522(92)90467-g. [DOI] [PubMed] [Google Scholar]
- 49.Köhler O, Benros ME, Nordentoft M, Farkouh ME, Iyengar RL, Mors O, et al. Effect of anti-inflammatory treatment on depression, depressive symptoms, and adverse effects: a systematic review and meta-analysis of randomized clinical trials. JAMA Psychiatry. 2014;71:1381–91. doi: 10.1001/jamapsychiatry.2014.1611. [DOI] [PubMed] [Google Scholar]
- 50.Bak LK, Schousboe A, Waagepetersen HS. The glutamate/GABA-glutamine cycle: aspects of transport, neurotransmitter homeostasis and ammonia transfer. J Neurochem. 2006;98:641–53. doi: 10.1111/j.1471-4159.2006.03913.x. [DOI] [PubMed] [Google Scholar]
- 51.Wang L, Maher TJ, Wurtman RJ. Oral L-glutamine increases GABA levels in striatal tissue and extracellular fluid. FASEB J. 2007;21:1227–32. doi: 10.1096/fj.06-7495com. [DOI] [PubMed] [Google Scholar]
- 52.Ferreira AG, da Cunha AA, Scherer EB, Machado FR, da Cunha MJ, Braga A, et al. Evidence that hyperprolinemia alters glutamatergic homeostasis in rat brain: neuroprotector effect of guanosine. Neurochem Res. 2012;37:205–13. doi: 10.1007/s11064-011-0604-1. [DOI] [PubMed] [Google Scholar]
- 53.Savio LE, Vuaden FC, Rosemberg DB, Bogo MR, Bonan CD, Wyse AT. Long-term proline exposure alters nucleotide catabolism and ectonucleotidase gene expression in zebrafish brain. Metab Brain Dis. 2012;27:541–9. doi: 10.1007/s11011-012-9321-y. [DOI] [PubMed] [Google Scholar]
- 54.Loureiro SO, Sidegum DS, Biasibetti H, Pereira MS, de Oliveira DL, Pessoa-Pureur R, et al. Crosstalk among disrupted glutamatergic and cholinergic homeostasis and inflammatory response in mechanisms elicited by proline in astrocytes. Mol Neurobiol. 2016;53:1065–79.. doi: 10.1007/s12035-014-9067-0. [DOI] [PubMed] [Google Scholar]
- 55.Brosnan JT, Brosnan ME. Creatine: endogenous metabolite, dietary, and therapeutic supplement. Annu Rev Nutr. 2007;27:241–61. doi: 10.1146/annurev.nutr.27.061406.093621. [DOI] [PubMed] [Google Scholar]
- 56.Harkness RA. Hypoxanthine, xanthine and uridine in body fluids, indicators of ATP depletion. J Chromatogr. 1988;429:255–78. doi: 10.1016/s0378-4347(00)83873-6. [DOI] [PubMed] [Google Scholar]
- 57.Mocking RJT, Assies J, Ruhé HG, Schene AH. Focus on fatty acids in the neurometabolic pathophysiology of psychiatric disorders. J Inherit Metab Dis. 2018;41:597–611. doi: 10.1007/s10545-018-0158-3. [DOI] [PubMed] [Google Scholar]
- 58.Wang SM, Han C, Lee SJ, Patkar AA, Masand PS, Pae CU. A review of current evidence for acetyl-l-carnitine in the treatment of depression. J Psychiatr Res. 2014;53:30–7. doi: 10.1016/j.jpsychires.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 59.Jia HM, Li Q, Zhou C, Yu M, Yang Y, Zhang HW, et al. Chronic unpredictive mild stress leads to altered hepatic metabolic profile and gene expression. Sci Rep. 2016;6:23441. doi: 10.1038/srep23441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lei T, Wang Y, Li M, Zhang X, Lv C, Jia L, et al. A comparative study of the main constituents and antidepressant effects of raw and vinegar-baked Bupleuri Radix in rats subjected to chronic unpredictable mild stress. RSC Adv. 2017;7:32652–63.. [Google Scholar]
- 61.Ju SY, Lee YJ, Jeong SN. Serum 25-hydroxyvitamin D levels and the risk of depression: a systematic review and meta-analysis. J Nutr Health Aging. 2013;17:447–55. doi: 10.1007/s12603-012-0418-0. [DOI] [PubMed] [Google Scholar]
- 62.Grudet C, Malm J, Westrin A, Brundin L. Suicidal patients are deficient in vitamin D, associated with a pro-inflammatory status in the blood. Psychoneuroendocrinology. 2014;50:210–9. doi: 10.1016/j.psyneuen.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 63.Banerjee A, Khemka VK, Roy D, Dhar A, Sinha Roy TK, Biswas A, et al. Role of pro-inflammatory cytokines and vitamin D in probable Alzheimer’s Disease with depression. Aging Dis. 2017;8:267–76. doi: 10.14336/AD.2016.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 65.Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Croze ML, Soulage CO. Potential role and therapeutic interests of myo-inositol in metabolic diseases. Biochimie. 2013;95:1811–27. doi: 10.1016/j.biochi.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 67.Lu W, Su X, Klein MS, Lewis IA, Fiehn O, Rabinowitz JD. Metabolite measurement: pitfalls to avoid and practices to follow. Annu Rev Biochem. 2017;86:277–304. doi: 10.1146/annurev-biochem-061516-044952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goveia J, Pircher A, Conradi LC, Kalucka J, Lagani V, Dewerchin M, et al. Meta-analysis of clinical metabolic profiling studies in cancer: challenges and opportunities. EMBO Mol Med. 2016;8:1134–42. doi: 10.15252/emmm.201606798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Messaoud A, Mensi R, Douki W, Neffati F, Najjar MF, Gobbi G, et al. Reduced peripheral availability of tryptophan and increased activation of the kynurenine pathway and cortisol correlate with major depression and suicide. World J Biol Psychiatry. 2018. 10.1080/15622975.2018.1468031. [DOI] [PubMed]
- 70.Hennings A, Schwarz MJ, Riemer S, Stapf TM, Selberdinger VB, Rief W. Exercise affects symptom severity but not biological measures in depression and somatization - results on IL-6, neopterin, tryptophan, kynurenine and 5-HIAA. Psychiatry Res. 2013;210:925–33. doi: 10.1016/j.psychres.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 71.Chaleckis R, Meister I, Zhang P, Wheelock CE. Challenges, progress and promises of metabolite annotation for LC-MS-based metabolomics. Curr Opin Biotechnol. 2019;55:44–50. doi: 10.1016/j.copbio.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 72.Emwas AH, Roy R, McKay RT, Tenori L, Saccenti E, Gowda GAN, et al. NMR spectroscopy for metabolomics research. Metabolites. 2019;9:E123. doi: 10.3390/metabo9070123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.