Abstract
Psychological disorders are associated with increased risk of severe inflammatory bowel disease (IBD) by causing gut microbiota dysbiosis and colonic mucosal barrier damage. However, the interaction between chronic restraint stress (CRS), gut microbiota composition, and colonic mucus remains unclear. We demonstrated that mice under CRS conditions exhibited alterations in microbiota composition, disruption of colonic mucus, and aggravation of colitis. In addition, the abundance of Akkermansia muciniphila was significantly decreased in mice under CRS and UC patients with depression, and positively associated with the expression of MUC2. After antibiotic treatment, the recipient mice colonized with CRS microbiota showed barrier defects and severe colitis. Administration of Akkermansia muciniphila was found to restore colonic mucus and modify the gut microbiota. We confirm that CRS-mediated gut microbiota dysbiosis results in colonic mucosal barrier damage and aggravation of colitis. Our results suggest that A. muciniphila is expected to be a potential probiotic to protect and treat colonic mucus that is involved in IBD with psychological disorders.
Keywords: chronic restraint stress, colitis, colonic mucus, fecal microbiota transplantation, Akkermansia muciniphila
Introduction
Inflammatory bowel disease (IBD), comprising ulcerative colitis (UC) and Crohn’s disease (CD), has become a global burden with rapidly increasing morbidity in the past 20 years. Epidemiological surveys indicate that established prevalence populations of IBD occur in 1.5 million people in American and 2.2 million in Europe, and affected populations are gradually growing worldwide (Cosnes et al., 2011; Molodecky et al., 2012). Numerous studies have also demonstrated that a high frequency of psychological disorder, such as depression or anxiety, is observed in IBD patients. Depression affects an estimated over 25% persons with IBD, and two to three times higher than healthy individuals (Walker et al., 2008). It is noteworthy that depression triggers severity of IBD, which included lower quality of life, hospitalization, risk of surgery, and IBD flare (Panara et al., 2014; Mikocka-Walus et al., 2016; Kochar et al., 2018). However, mechanisms behind the role of psychological disorders or depression in the aggravation of IBD are underexplored.
Recent evidence suggests that the risk factors of depression in aggravating IBD are closely related to the brain-gut axis, which is considered a bidirectional link for interactions between the brain and the gut (Al Omran and Aziz, 2014). The gut is considered to be among the biological factors that significantly influence the function and structure of the brain. In turn, the brain modulates the gut microbiota and microenvironment. Previous data from animal models have demonstrated that the induction of depression is involved in the development of experimental colitis by immune system dysfunction and gut microbiota dysbiosis (Ghia et al., 2009; Gao et al., 2018). It has been suggested that gut microbiota, as an independent component, significantly modulates the brain-gut axis (Rhee et al., 2009). Therefore, screening specific gut microbes plays an important role in exploring a novel therapeutic option.
Colonic mucus has been recognized as the first physical barrier that effectively protects the colon against toxins and pathogenic microorganism invasion. It consists of two layers, an outer layer attached to the gut microbiota and an inner layer attached to the epithelium. Colonic mucus is produced and maintained by an extensively glycosylated mucin-2 (MUC2), which is secreted by goblet cells and renews the inner mucus layer approximately every hour (Johansson et al., 2011; Ambort et al., 2012). The damage of colonic mucus leads to the aggravation of colitis in mice and humans (Johansson et al., 2008; Fu et al., 2011; Johansson et al., 2014). Enhanced intestinal permeability and gut microbiota dysbiosis caused by chronic stress has also been reported to result in mucosal immune reactions and intestinal inflammation (Kiliaan et al., 1998; Wei et al., 2019). However, the relationship between the gut microbiota and its influence on colonic mucus within the brain-gut axis warrants further investigation.
Our results reveal that chronic restraint stress (CRS), a credible procedure for establishing a model of depression in mice, induced alterations in gut microbiota and dysfunction in the colonic mucosal barrier and prompted the development of experimental colitis. Furthermore, abundance of Akkermansia muciniphila (A. muciniphila) was significantly reduced in mice under CRS and UC patients with depression. Subsequently, gut microbiota transplantation experiments were performed in recipient mice to investigate the causal relationship between the gut microbiota and colonic mucosal barrier. A. muciniphila supplementation could relieve depression-like symptoms and aggravation of colitis in recipient mice.
Materials and Methods
Animals
Male C57BL/6N mice weighing 18–20 g were purchased from Charles River (Nanjing, China). All animals were group-housed (three to four mice per cage) in a specific pathogen-free (SPF) condition under 12 h light-dark cycle with diet and water ad libitum. The mice were adapted to the laboratory environment for 7 days prior to the start of the experiments. The experimental protocols were approved by the Ethics Committee for Animal Experiments of the Jiangsu Provincial Hospital of Traditional Chinese Medicine. All efforts were made to minimize the suffering and number of mice used during the experiments.
Experiment 1: Chronic Restraint Stress and Dextran Sodium Sulfate–Induced Colitis Protocol
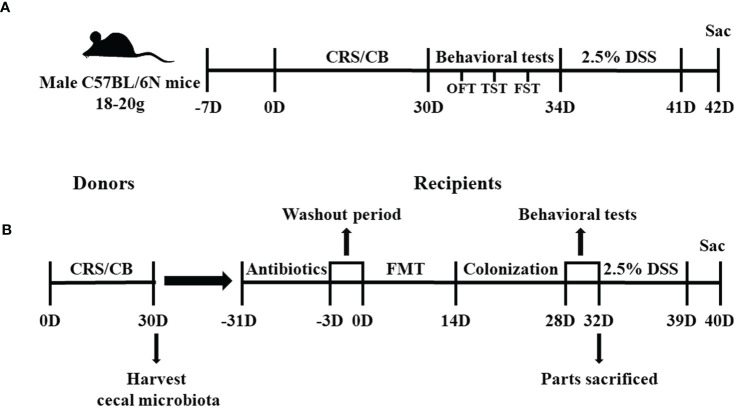
In Experiment 1 ( Figure 1A ), 32 mice were randomly assigned to four groups: conventional breeding (CB; n = 8), CRS (n = 8), dextran sodium sulfate (DSS; n = 8), and DSS+CRS (n = 8). The CRS procedure was conducted in accordance with previously reported methods (Gao et al., 2018; Son et al., 2019). Briefly, mice in the CRS and DSS+CRS groups were limited to a 50 ml centrifuge tube with good ventilation for 3 h/day (9:00 a.m. to 12:00 a.m.) for 30 consecutive days, during which they were not allowed to move back or forth during the procedure. Behavioral tests were performed to assess depressive-related behaviors under CRS. Then, mice in DSS and DSS+CRS group were subjected to drink 2.5% (w/v) DSS (36–50 kDa, MP Biomedical) for 7 days. On day 8 after DSS administration, mice in all experimental groups were euthanized to assess the entire colon length and histological score. Two mice in the DSS+CRS group were removed from the experiment due to severe colitis that induced unexpected death during DSS administration.
Figure 1.
Study timelines of Experiments 1 and 2. (A) Graphical timeline of Experiment 1 including CRS and DSS-induced colitis. (B) Graphical timeline of Experiment 2 including antibiotic treatment and FMT. CRS, chronic restraint stress; DSS, dextran sodium sulfate; FMT, Fecal microbiota transplants.
Experiment 2: Gut Microbiota Transplants and Colonization
In Experiment 2 ( Figure 1B ), 72 mice were randomly assigned to eight groups: donor CB (N = 10), donor CRS (N = 20), recipient-CB (RE-CB, n = 6), recipient-CRS (RE-CRS, N = 6), recipient CRS- Akkermansia muciniphila (RE-CRS-AKK, n = 6), recipient-CB-DSS (RE-CB-DSS, N = 8), recipient-CRS-DSS (RE-CRS-DSS, n = 8), recipient CRS–Akkermansia muciniphila–DSS (RE-CRS-AKK-DSS, N = 8). The mice in the donor CRS group were restrained in centrifuge tubes for 3 h daily for 30 consecutive days, as described above. All mice in the donor group were euthanized on day 31 to harvest the cecal microbial contents for fecal microbiota transplantation (FMT). Cecal contents from the donor group were mixed together in a 50 ml centrifuge tube based on the activity group (donor CB mice or donor CRS mice) and rapidly suspended in phosphate-buffered saline (PBS) with 0.1 g contents in 1 ml PBS. Cecal microbial suspensions were kept at −20°C and thawed overnight at 25°C prior to gavage. FMT was performed as previously described (Jimeno et al., 2018). Mice in the recipient group were given the following antibiotic cocktail ad libitum for 30 days to deplete all gut microbiota before FMT: ampicillin, neomycin, metronidazole (all at 1 g/ml), and vancomycin (0.5 g/ml). All antibiotics were obtained from Sigma-Aldrich. After 3 days washout period, 100 μl of frozen-and-thawed suspension was then administered by oral gavage into each recipient mouse for 14 days (Samuelson et al., 2017). In addition, two recipient groups treated with CRS suspension were orally administered with another 100 μl of A. muciniphila containing 1×108 bacteria in parallel with groups given an equal volume of PBS as control (Routy et al., 2018). Behavioral tests were performed after gut microbiota colonization. At the end of the behavioral tests, some recipient mice were euthanized, and others received 2.5% DSS in drinking water for 7 days.
Behavioral Tests
Open Field Test
The open field test (OFT) was conducted using an empty square apparatus (40 cm × 40 cm × 40 cm). Each mouse was gently placed in the corner of the open field arena, and its movements were recorded using a camera. The experimental arena was cleaned with 70% ethanol for each test. The total distance was analyzed using ANY-maze software to measure spontaneous activity.
Tail Suspension Test
The tail suspension test (TST) was performed to assess depression-like behavior and antidepressant efficacy, as described previously (Opal et al., 2014). Mice were individually suspended 50 cm from the floor by placing the tape 1 cm away from the tip of the tail. The test lasted for 6 min, and activity during the last 4 min was evaluated using ANY-maze software to quantify immobility time.
Forced Swim Test
The day after the TST, the mice were subjected to the forced swim test (FST) to assess depression-like behavior (Liu et al., 2016). Each mouse was individually placed in a glass cylinder (20 cm diameter, 40 cm height) containing water (23–25°C) for a duration of 6 min. Mice were kept immobile for the final 4 min and analyzed using the ANY-maze software. After the test, the mice were immediately placed in a clean and dry cage.
A. muciniphila Culture
A. muciniphila strain MucT (DSM 22959) was kindly given by Prof. Li Liu (Glycomics and Glycan Bioengineering Research Center, College of Food Science and Technology, Nanjing Agricultural University, China). A. muciniphila was grown in brain heart infusion broth for 48 h at 37°C in an anaerobic incubator (Sheldon Manufacturing, USA) containing a N2/CO2/H2 (86:7:7) gas phase (Derrien et al., 2004; Shin et al., 2019). The final suspensions of A. muciniphila with 1×109 colony-forming units (CFU)/ml were determined by measuring the optical density in PBS under 600 nm (Routy et al., 2018).
Patient Characteristics
We obtained 35 human fecal samples from patients with active UC from the Jiangsu Province Hospital of Chinese Medicine. The study was performed according to the principles of the Helsinki Declaration, and approval was obtained from the Institutional Review Board of Jiangsu Provincial Hospital of Traditional Chinese Medicine, China on October 12, 2018 (approval code 2018NL-093-02). Depression was assessed using the Patient Health questionnaire-9 (PHQ-9) scales (Kroenke et al., 2010). Patients with scores ≥10 were defined as moderate to severe depression and included (Kroenke et al., 2001). The exclusion criteria were as follows: systemic infectious diseases, autoimmune diseases, gastrointestinal surgery, and history of other mental disorders. The patient characteristics are shown in Supplementary Table 1 .
A. muciniphila Quantification
The abundance of A. muciniphila in 35 fecal samples from patients with active UC was quantified using quantitative real-time PCR analysis. Bacterial DNA isolation was conducted using the Stool DNA Isolation Kit (TIANGEN, China) according to the manufacturer’s instructions. The quantity and purity of the extracted DNA were assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). Previously reported gene-targeted primers were used for the detection of A. muciniphila species (AM1-F: 5’-CAGCACGTGAAGGTGGGGAC-3’; AM2-R: 5’-CCTTGCGGTTGGCTTCAGAT -3’) (Collado et al., 2007). The A. muciniphila primers were purchased from Invitrogen (USA). Amplification reactions were performed in a total volume of 20 μl, including 2 μl of DNA template, 10 μl SYBR Green PCR Master Mix 2× (TaKaRa, China), and 0.5 μl of each primer and probe. The ABI 7500 real-time PCR system (Applied Biosystems, USA) was used to conduct the assays. The thermal profiles were constructed based on this protocol. In summary, the samples were held at 55°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and annealing at 60°C for 1 min. Standard curves were obtained by comparing the cycle threshold (CT) with 10-fold serial dilutions of standard DNA templates from the culture of A. muciniphila strain MucT (DSM 22959) as previously reported (Song et al., 2004). Each analysis was run in duplicate on the same plate. The assay results were expressed as Log10 CFU/g of fecal stool.
RNA Sequencing and Function Analysis
Total RNA was extracted from 12 frozen colonic tissue samples from mice [three tissues per group × four groups (CB, CRS, DSS, and DSS+CRS)] using a total RNA Extractor kit (Sangon Biotech, China) according to the manufacturer’s protocol. The quality and quantity of the isolated RNA samples were assessed using a Qubit 2.0 Fluorometer (Thermo Scientific). Libraries were established using the VAHTS mRNA-seq V2 Library Prep Kit for Illumina (Vazyme Biotech, China) according to the manufacturer’s instructions. Subsequently, libraries were sequenced on the Illumina Hiseq TM 2500 platform, generating an average of 150 bp paired-end reads (Reuter et al., 2015). To evaluate differentially expressed transcripts (DEGs) between groups, we annotated and quantified the transcripts using DESeq (Anders and Huber, 2010). Finally, the absolute value of log2 fold change was calculated to extract the DEGs (greater than 0.50 and p < 0.05). Volcano maps or heat maps were used to visualize the differential genes. To evaluate the function of DEGs, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis using KOBAS 3.0. DAVID (v6.8) was used to perform the gene ontology (GO) functions enriched based on the DEGs. Pathways containing more than two differentially expressed genes were screened. The RNA-sequence read data were deposited in the Sequence Read Archive (SRA) at the NCBI database (accession number: PRJNA758283).
Quantitative Polymerase Chain Reaction
Quantitative polymerase chain reaction (qPCR) was used to confirm the differentially expressed genes of MUC2 in Experiments 1 and 2. Total RNA from colonic samples was isolated using TRIzoI Reagent (Invitrogen). After reverse transcription of RNA into cDNA using the Prime Script RT reagent Kit (TaKaRa, China), qPCR was conducted using the ABI 7500 real-time PCR system (Applied Biosystems) with SYBR Green PCR Master Mix 2× (TaKaRa, China). Gene changes were calculated using the 2−ΔΔCt method by normalizing to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Merrick et al., 2012). Primer details are shown in Supplementary Table 2 .
Histology and Immunohistochemistry
Colons were removed from the mice, flushed with PBS, fixed in 10% formaldehyde overnight at room temperature, and embedded in paraffin. The specimens were subsequently sectioned at 5 μm thick and stained with hematoxylin and eosin (H&E). Three slides in H&E staining of each colon were selected and analyzed by a blinded pathologist. Histopathological scores were assessed based on a previously described method (Sann et al., 2013).
Immunohistochemistry analysis was carried out as follows: the prepared sections were treated with primary antibodies using horseradish peroxidase-conjugated goat anti-rabbit IgG (Servicebio, China) overnight at 4°C. After washing in PBS, the slices were treated with the secondary antibody rabbit anti-MUC2 (Servicebio, China) for 50 min and then stained with diaminobenzidine. The expression of MUC2 were expressed as the number of positive cells per villus using ImageJ software.
Periodic Acid-Schiff and Alcian Blue Staining
After deparaffinization and rehydration, the sections were stained with periodic acid-Schiff and Alcian blue (PAS/AB). The goblet cells are blue. The number of goblet cells was counted using Image J software and expressed as positive cells per villus.
Gut Microbiota Analysis
Microbial DNA from cecal specimens was extracted using a DNA extraction kit (Tiangen, China) from samples of approximately 100 mg, following the manufacturer’s instructions. The 16S rRNA gene sequencing was conducted using Novogene. Briefly, the V3–V4 region of the microbial 16S rRNA gene was amplified with specific primers 341F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’). After the PCR product was purified, amplicon libraries were generated using the Illumina TruSeq DNA PCR-Free Library Preparation Kit (Illumina, USA) and then sequenced on the Illumina HiSeq platform for paired-end 250 bp raw reads. Collected raw data from the gut microbiota were uploaded to the NCBI database with the accession number PRJNA756202. To obtain high-quality data, reads containing more than 10% unknown nucleotides and less than 80% of bases with quality (Q-value) >20 were discarded (Ding et al., 2020). Subsequently, operational taxonomic units (OTUs) were clustered using UPARSE (v7.1) with a 0.97 threshold (Edgar, 2013). Each OTU was annotated using taxonomic information analysis according to the RDP database (Wang et al., 2007). All the data were analyzed by I-Sanger platform (http://www.i-sanger.com). We used the Shannon index as a metric to analyze differences in alpha diversity between the groups. Additionally, principal coordinate analysis (PCoA) was performed using unweighted UniFrac distances at the OTU level, and an analysis of similarity test (ANOSIM) was performed to assess significant differences in the bacterial community composition. Spearman correlation was used to analyze the relationships between the relative abundance of A. muciniphila and stress-associated gene counts, and the results were visualized using Cytoscape (v3.5.1).
Statistical Analysis
All results are shown as mean ± SEM. Data were analyzed by one-way analysis of variance (ANOVA) with Tukey–Kramer test between two groups and two-way ANOVA with Bonferroni’s post-hoc test for multiple comparisons. Differences were considered statistically significant at p < 0.05.
Results
The study timeline of the two independent experiments is depicted in Figure 1 .
Experiment 1: CRS Caused Adverse Psychological Effects, Colitis Aggravation, and Microbiota Dysbiosis
CRS Induced Depressive-Like Behaviors and Severe Colitis
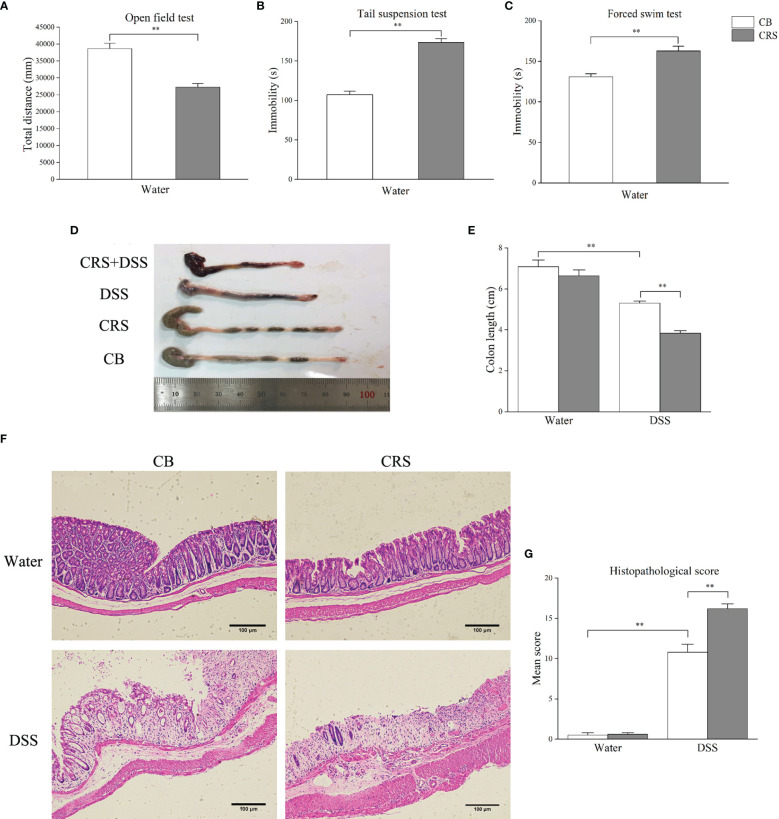
The CRS animal model exhibited depression-like behavior prior to DSS administration. Compared with the CB group, CRS mice showed significantly less movement distance in the OFT ( Figure 2A ). In addition, increased immobility and reduced swimming durations were observed in mice under CRS conditions compared to those in the CB group ( Figures 2B, C ). Following DSS exposure for 7 days, CRS mice developed severe colitis characterized by bloody diarrhea, severe infiltration of inflammatory cells, extensive villus injury of the bowel wall, and hyperplasia of reactive epithelial cells. Colon length was shorter in the DSS+CRS group than in the DSS group ( Figures 2D, E ). In addition, the histopathological scores were higher in the DSS+CRS group than in the DSS group ( Figures 2F, G ).
Figure 2.
CRS aggravated DSS-induced colon injury. (A–C) Behavioral tests were performed before DSS, including OFT, TST, FST (n = 8). (D) Representative pictures of colon were selected between groups; (E) Colon length was recorded from each group (n = 6–8). (F) Representative pathological pictures of colonic injury were selected between groups (magnification ×40, scale bar: 100 μm). (G) Colonic injury score was assessed by histopathological score based on H&E staining (n = 5). *p < 0.05, **p < 0.01.
A. muciniphila Abundance Was Reduced in CRS Mice
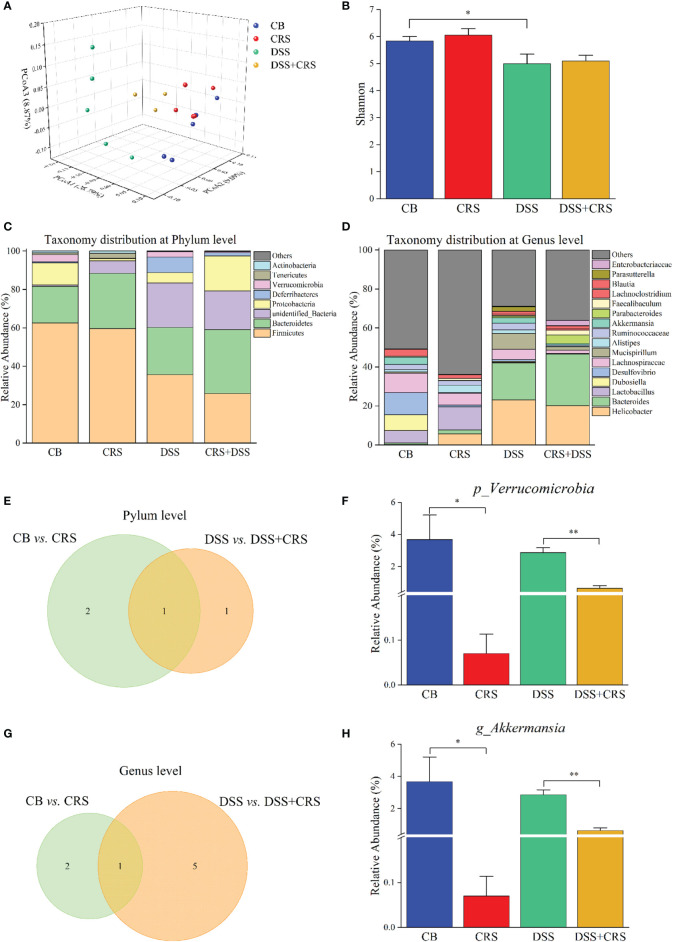
We utilized 16S rRNA sequencing to quantify and characterize the gut microbiota of mice under CRS conditions. We observed a decrease in the Shannon index in the DSS group compared to that in the CB group (p < 0.05), whereas no difference in alpha diversity was observed from the comparisons of CB versus CRS, and DSS versus DSS+CRS, respectively ( Figure 3B ). Unweighted UniFrac PCoA analysis indicated a distinct difference in beta-diversity at the OTU level among the four groups ( Figure 3A ). In contrast to the CB group, the microbiomes of mice in the CRS group exhibited significant changes based on the ANOSIM values (R = 0.761, p = 0.01). However, when comparing the DSS group with the DSS+CRS group, we found no differences (R = 0.323, p = 0.13). Next, we analyzed the microbiota community structure of each group. At the phylum and genus levels, the composition of microbiota showed dramatic changes in the CRS group or DSS+CRS group ( Figures 3C, D ). At the phylum level, three and two dominant phyla were identified from the comparisons of CB versus CRS, and DSS versus DSS+CRS, respectively ( Figure 3E ). In comparison to the CB group, the relative abundance of Proteobacteria and Verrucomicrobia was decreased and that of Melainabacteria was increased in the CRS group ( Figures S1A, B ). The relative abundance of Proteobacteria in the CRS+DSS group was higher than that in the DSS group, while the relative abundance of Verrucomicrobia was decreased. The relative abundance of Verrucomicrobia was significantly decreased in both comparisons at the phylum level ( Figure 3F ). At the genus level, the Venn diagram revealed three and six alternations of dominant genera from the comparisons of CB versus CRS, and DSS versus DSS+CRS, respectively ( Figure 3G ). When compared with the CB group, the relative abundance of Odoribacter was increased and that of Akkermansia and Desulfovibrio was decreased in the CRS group ( Figures S1C, D ). Moreover, there was a marked increase in the proportion of Erysipelatoclostridium, Enterococcus, and Parabacteroides and a decrease in Romboutsia, Dubosiella, and Akkermansia in the DSS+CRS group compared to those observed in the DSS group ( Figures S1E–I ). The mice subjected to CRS exhibited decreased Akkermansia abundance in the two comparisons at the genus level ( Figure 3H ).
Figure 3.
Reduction of A. muciniphila was observed in CRS mice. (A) PCoA according to unweighted Unifrac presented a clear site-specific clustering (n = 3–5). (B) Shannon index values among four experimental groups. (C) Microbiota composition at the phylum level. (D) Microbiota composition at the genus level. (E) Venn diagram showing only one differentially abundant species at the phylum level altered under CRS. (F) Relative abundance of Verrucomicrobia significantly decreased between groups. (G) Venn diagram showing only one differentially abundant species at the genus level altered under CRS. (H) Relative abundance of A muciniphila had significantly decreased between different groups. *p < 0.05, **p < 0.01.
Prevalence of A. muciniphila in UC Patients With Depression
To assess prevalence of A. muciniphila in UC patients with depression, a total of 35 fecal samples were collected from 16 UC patients with depression and 19 UC patients as control. Real-time PCR data further affirmed that the abundance of A. muciniphila was reduced in UC patients with depression ( Figure 4 ). Above all, these results demonstrated that a lower abundance of A. muciniphila under CRS might be associated with aggravation of colitis.
Figure 4.
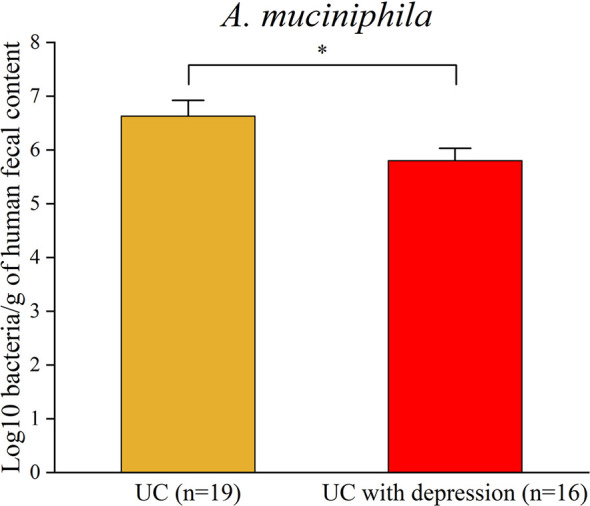
Real-time PCR analysis validated the reduction in A. muciniphila in UC patients with depression. *p < 0.05.
Interactions Between Differentially Expressed Genes Regulated by CRS and A. muciniphila in Mice
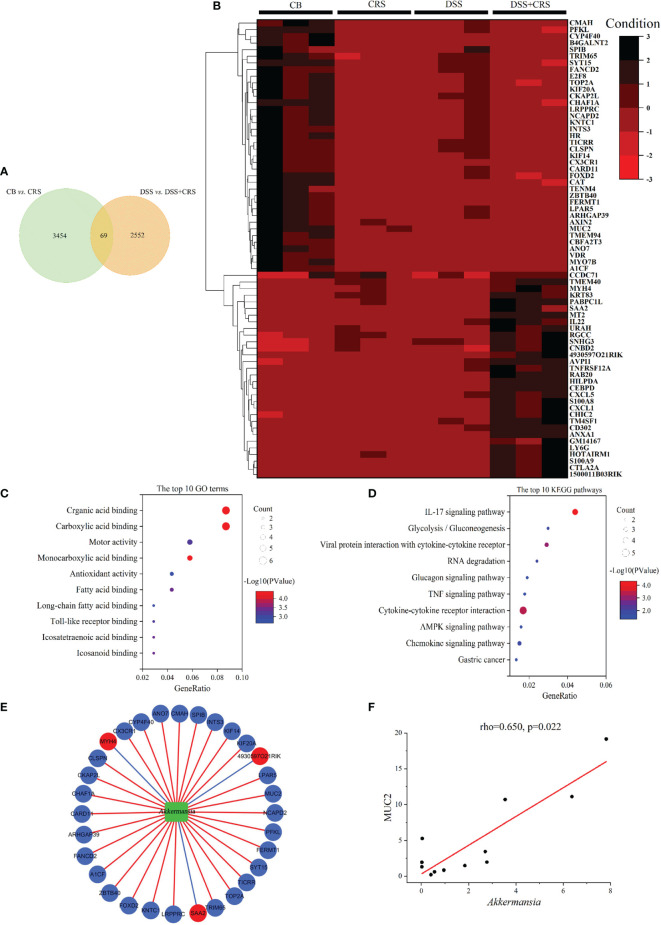
To determine how CRS exerted a harmful effect in DSS-induced colitis, we conducted DEG profiles in the colonic tissue of mice from the comparisons of CB versus CRS, and DSS versus DSS+CRS. Hence, 3,523 and 2,621 DEGs were identified respectively ( Figures S2A, D ). Notably, 69 DEGs were significantly altered in the two comparisons ( Figure 5A ). Expression levels of these 69 DEGs are shown as a heat map ( Figure 5B ). The 15 upregulated and downregulated genes most related to the two comparisons are presented in Supplementary Table 3 and Supplementary Table 4 . GO and KEGG analyses have been widely used to explore biological functions and large-scale pathway information based on the high-throughput sequencing data. When comparing the CB with CRS mice, the 10 most significant GO terms revealed that CRS mainly controlled the structural constituents of ribosomes, transcriptional coregulator activity, and enzyme activator activity ( Figure S2B ). KEGG pathway analysis showed that CRS mostly regulated Alzheimer’s, Parkinson’s, and Huntington’s diseases ( Figure S2C ). When comparing the DSS with DSS+CRS mice, the 10 most significant GO terms indicated that CRS was related to receptor ligand, cytokine, and cytokine receptor activity ( Figure S2E ). Pathway analysis revealed that the 10 most enriched pathways included the IL-17 signaling pathway, TNF signaling pathway, and Th17 cell differentiation pathway ( Figure S2F ).
Figure 5.
DEGs under CRS were analyzed by high-throughput RNA-Seq. (A) Venn diagram showing 69 DEGs changed under CRS from the two comparisons. (B) Heat-map presented 69 DEG in the two comparisons (n = 3). (C) The 10 most disordered GO terms were identified from the 69 DEG in the two comparisons. (D) The top 10 KEGG pathways were identified from the 69 DEG in the two comparisons; (E) Network visualized the correlations between A. muciniphila and genes. Green node represented A muciniphila. Red and Blue nodes represented the raising and lowering of DEGs. Blue lines represented negative correlation, and red lines represented positive correlation; (F) Scatterplot showing that MUC2 was positively correlated with A. muciniphila.
Subsequently, 69 DEGs were screened from the above two comparisons and subjected to GO and KEGG pathway analyses. GO analysis showed that the identified DEGs were associated with fatty acid binding, long-chain fatty acid binding, and Toll-like receptor binding ( Figure 5C ). KEGG pathway analysis revealed that the DEGs were related to the IL-17, TNF, and AMPK signaling pathways ( Figure 5D ).
To verify the relationship between host genes and altered microbial communities and to identify their potential roles in worsening the severity of colitis, we performed a correlation analysis between 69 DEGs regulated by CRS and A. muciniphila. The correlation analysis revealed that 34 DEGs were significantly correlated with A. muciniphila ( Figure 5E ). Notably, MUC2 expression was positively correlated with A. muciniphila ( Figure 5F ).
CRS Inhibited Mucus Production and Goblet Cell Numbers in the Colon
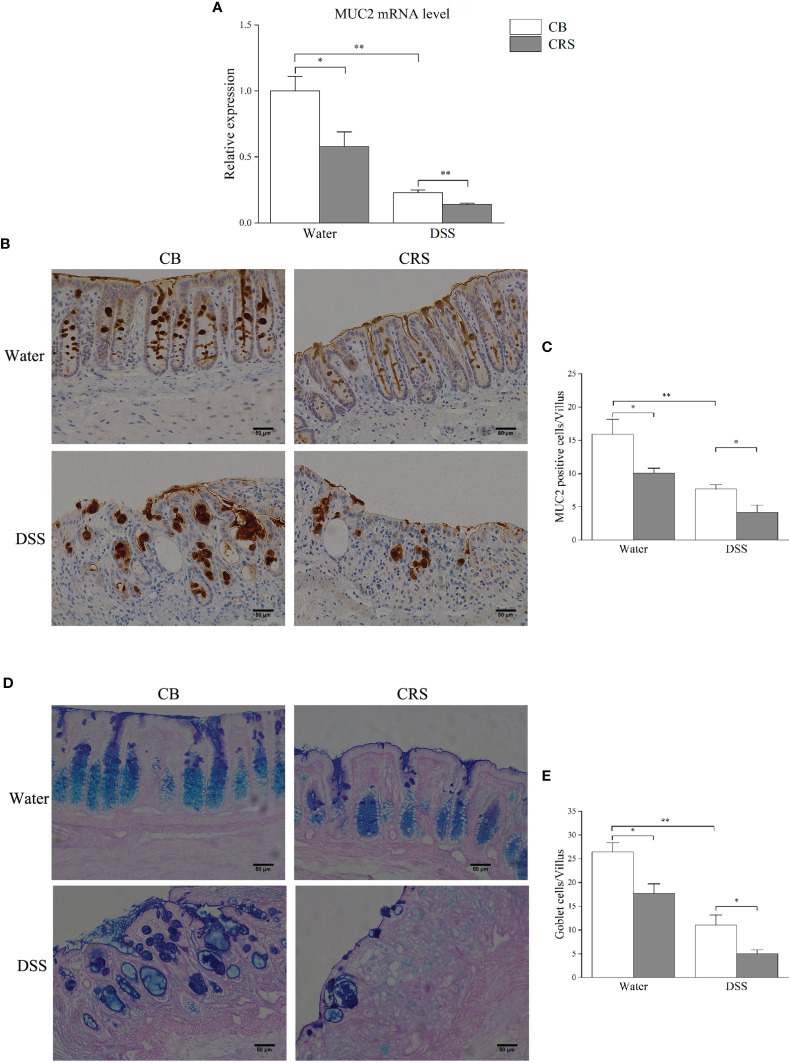
qRT-PCR was performed to validate the expression of MUC2, which was selected from candidate DEGs, and correlation analysis. Our results demonstrate that CRS decreased MUC2 expression ( Figure 6A ), and MUC2-positive cells in each villus were significantly decreased in mice under CRS ( Figures 6B, C ). Due to mucin produced by goblet cells, we next examined the colon in mice from different groups and counted the number of goblet cells by PAS/AB staining. Our results revealed that the number of goblet cells was significantly decreased in all CRS mice compared to that in non-CRS mice ( Figures 6D, E ).
Figure 6.
Chronic restraint stress prompted colonic mucosal barrier damage. (A) Relative expression of MUC2 was validated by qPCR. (B) Representative pictures shown immunostaining of MUC2 in colon (magnification ×200, scale bar: 50 μm). (C) The numbers of MUC2-positive cells in each villus (n = 4–5); (D) Representative pictures showing that colonic specimens stained with PAS/AB (magnification ×200, scale bar: 50 μm); (E) Numbers of goblet cells in each villus (n = 4–5). *p < 0.05, **p < 0.01.
Experiment 2. A. muciniphila Supplementation Prevented Mucosal Barrier Defects and Aggravation of Colitis
A. muciniphila Supplementation Alleviated Depression-Like Behaviors and Aggravated Colitis in Recipient Mice
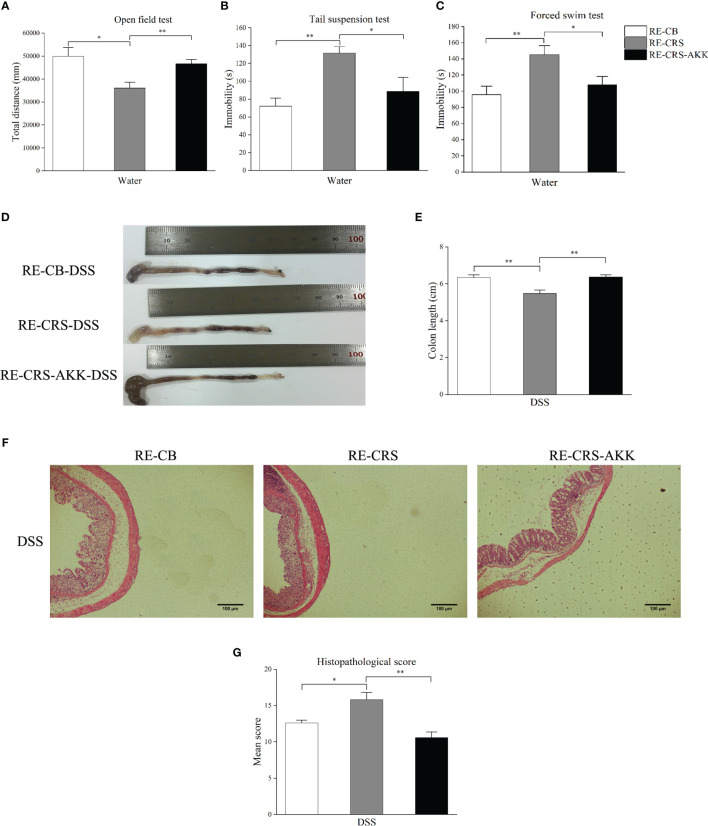
To further evaluate the role of CRS-driven dysbiosis in the aggravation of colitis associated with A. muciniphila, we performed FMT in recipient mice. Compared to the recipient mice colonized with the CB microbiota, the recipient mice colonized with the CRS microbiota showed increased depression-like behaviors in the OFT, TST, and FST. When compared with recipient mice colonized with CRS microbiota, A. muciniphila supplementation alleviated depression-like behaviors, including decreased immobility time in the TST and FST, and increasing movement distance in the OFT ( Figures 7A –C ). After DSS administration for 7 days, significant severe colitis was observed in recipient mice colonized with CRS microbiota compared to the CB microbiota. Conversely, A. muciniphila supplementation increased colon length and reduced the histopathological scores compared to the recipient mice colonized with CRS microbiota ( Figures 7D –G ).
Figure 7.
A. muciniphila supplementation alleviated depression-like behavior and colitis aggravation in recipient mice. (A–C) Depression-like behavior in recipient mice was alleviated by oral A. muciniphila supplementation by behavioral tests (n = 6). (D) Representative colon photographs from mice receiving DSS treatment. (E) Colon length recorded at time of sacrifice (n = 8). (F) Representative pathological photographs of colonic injury in mice receiving DSS treatment (magnification ×40, scale bar: 100 μm). (G) Colonic injury score assessed by histopathological score based on H&E staining in recipient mice (n = 4–5). *p < 0.05, **p < 0.01.
A. muciniphila Supplementation Inhibited Colonic Mucosal Barrier Damage in Recipient Mice
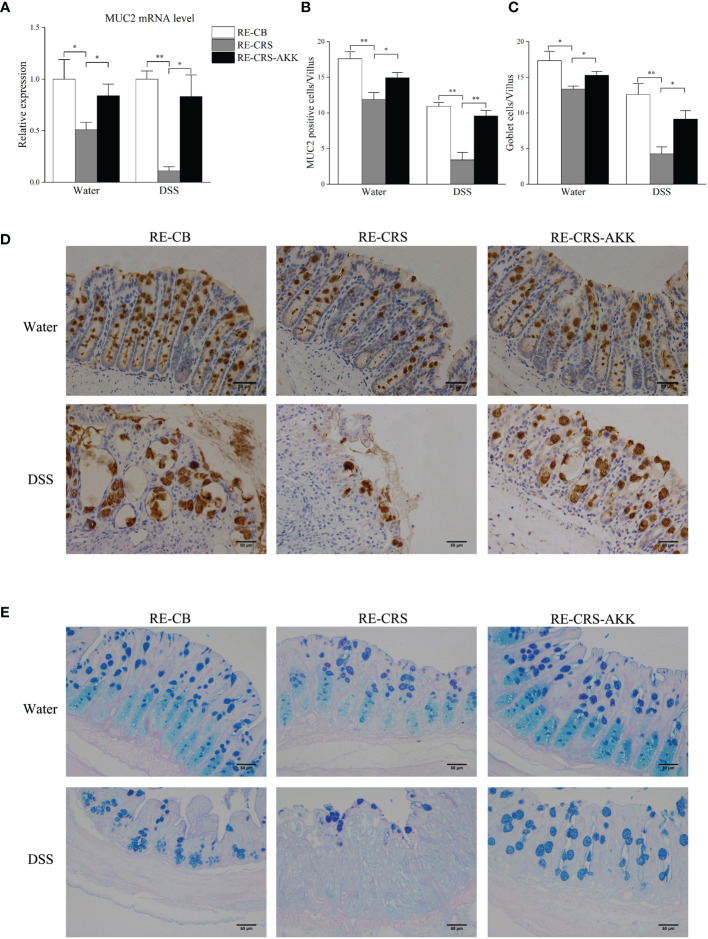
To elucidate and understand whether dysbiosis of gut microbiota is causal for colonic mucosal barrier defects, we analyzed mucosal barrier function in recipient mice. Mice colonized with the CRS microbiota exhibited disrupted colonic mucosal barrier before and after DSS exposure, while A. muciniphila supplementation significantly enhanced the expression of MUC2 ( Figure 8A ) and increased the number of goblet cells and MUC2-positive cells in each villus ( Figures 8B –E ).
Figure 8.
A. muciniphila supplementation protected colonic mucosal defects in recipient mice. (A) A. muciniphila supplementation affected relative expression of MUC2 in mice. (B) Numbers of MUC2-positive cells in each villus (n = 4–5). (C) Numbers of goblet cells in each villus (n = 4–5). (D) Representative photographs of colonic MUC2 immunostaining (magnification ×200, scale bar: 50 μm). (E) Representative photographs of colonic specimens stained with PAS/AB (magnification ×200, scale bar: 50 μm). *p < 0.05, **p < 0.01.
A. muciniphila Supplementation Remodeled the Gut Microbiota in Recipient Mice
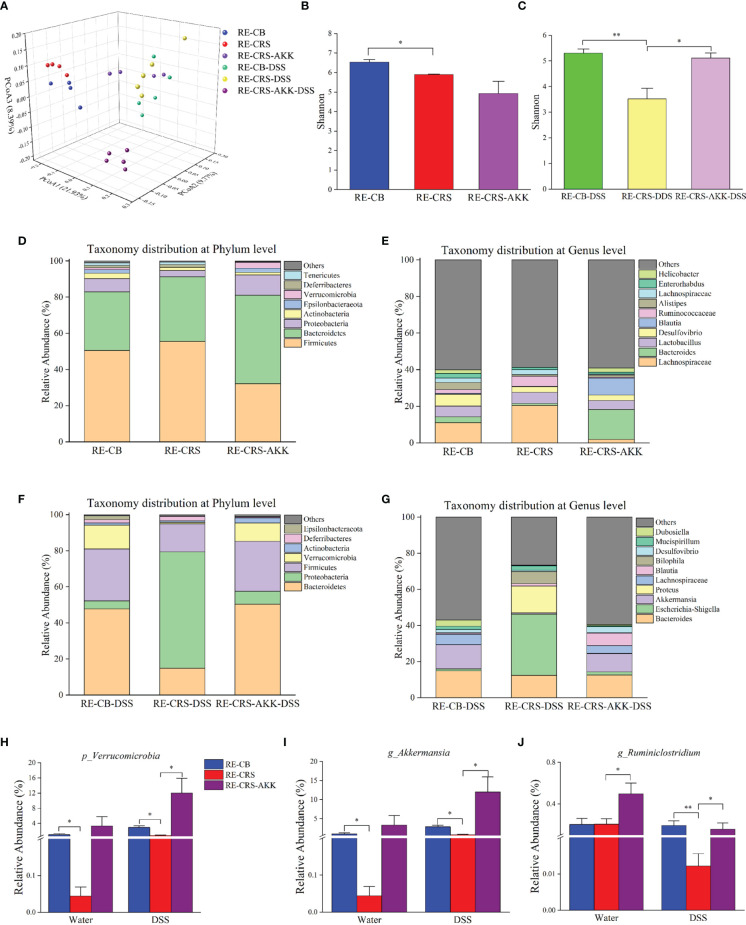
Based on the 16S rRNA gene-based profile, the Shannon diversity index revealed that A. muciniphila supplementation increased alpha diversity after DSS exposure compared to recipient mice colonized with the CRS microbiota ( Figure 9C ). Moreover, A. muciniphila supplementation revealed no significant difference in the Shannon diversity index before and after DSS administration compared with recipient mice colonized with the CB microbiota, indicating that alpha diversity was partly recovered after A. muciniphila treatment ( Figures 9B, C ). PCoA on unweighted UniFrac distances was performed to characterize differences among these groups ( Figure 9A ). ANOSIM analysis showed that A. muciniphila supplementation resulted in distinct gut microbial communities before and after DSS administration compared with the mice colonized with the CRS microbiota (RE-CB with RE-CRS-AKK: R = 0.82, p = 0.03; RE-CB-DSS with RE-CRS-AKK-DSS: R = 0.89, p = 0.01). The microbial composition at the phylum and genus levels in each recipient group is shown in Figures 9D –G . In agreement with Experiment 1, the relative abundance of Verrucomicrobia and Akkermansia was decreased in mice colonized with the CRS microbiota before and after DSS administration than in mice colonized with the CB microbiota. Following DSS exposure, A. muciniphila supplementation significantly increased the relative abundance of Verrucomicrobia and Akkermansia ( Figures 9H, I ). Notably, A. muciniphila supplementation significantly enhanced the relative abundance of Ruminiclostridium ( Figure 9J ).
Figure 9.
A. muciniphila supplementation remodeled the gut microbiota in recipient mice. (A) PCoA according to unweighted Unifrac revealed site-specific clustering (n = 4–5). (B, C) The alpha diversity, Shannon index were shown in each recipient group; (D–G) relative abundance compositions of microbiota at the phylum and genus level presented in each recipient group. (H) The representative phylum of Verrucomicrobia was differentially represented after A. muciniphila treatment in recipient mice. (I–J) Two representative genera were differentially represented after A. muciniphila. *p < 0.05, **p < 0.01.
Discussion
A growing body of evidence has revealed that depression has negative effects on the course of IBD, but the influence of these psychological disorders on IBD severity remains obscure. In this study, we demonstrated that CRS aggravates DSS-induced colitis with shorter colon length, higher histopathological scores, colonic mucus damage, and gut microbiota dysbiosis. The associations between psychological disorders and IBD through the brain-gut axis, including the hypothalamus-pituitary axis, activation of the sympathetic nervous system, increased proinflammatory cytokines, and decreased intestinal permeability (Bonaz and Bernstein, 2013; Brzozowski et al., 2016; Labanski et al., 2020). However, the exact mechanisms underlying the interaction between gut microbiota and colonic mucus require further exploration.
In the last decade, the effects of the gut microbiota on IBD have received much attention. It is widely reported that gut microbiota disturbance or dysbiosis influences the progress of IBD (Ananthakrishnan, 2015; Ananthakrishnan et al., 2018). Numerous studies from animal models and clinical studies have indicated that stress clearly causes gut microbiota dysbiosis (Galley and Bailey, 2014; Bharwani et al., 2016; Osadchiy et al., 2019). In this study, we also found that dysbiosis induced by CRS is characterized by a dramatic alteration in the gut microbiota construction and composition. Notably, a reduced abundance of A. muciniphila was observed in mice subjected to CRS. A large-scale microbiome population cohort from patients with depression demonstrated that A. muciniphila could promote the secretion of serotonin, which indicated that A. muciniphila plays an important role in depression (Valles-Colomer et al., 2019). In our study, we found that A. muciniphila was reduced in UC patients with depression. This discovery has great potential for the search of new biomarkers and treatment for IBD with psychological disorders.
IBD is also associated with damage of colonic mucosal barriers, or so-called “leaky gut.” The colonic mucus largely relies on the release of MUC2. A hydrated glycosylated protein, MUC2, is produced by goblet cells, adhering to the surface of the colon to prevent luminal microbes and pathogen invasion (Hansson and Johansson, 2010; Johansson et al., 2011). Furthermore, multiple studies have indicated that colonic barrier loss occurs in mice under stress conditions (Söderholm et al., 2002; Yu et al., 2010; Oliver et al., 2012). Although our methods were different, we also observed a decrease in MUC2-positive cells and goblet cells in each villus after 30 days of CRS. The mRNA expression of MUC2 in colon tissue was also significantly decreased in mice under CRS. Previous studies have shown that MUC2−/− mice exhibit severe colitis, which is accompanied by a remarkable downregulation of goblet cells (Morampudi et al., 2016). These results suggest that CRS-associated colonic mucus defects may lead to the development of colitis.
Culminating from the changes of gut microbiota paralleled the damage of colonic mucus in Experiment 1, we aim to determine whether gut microbiota is causal for the colonic mucosal defects. Therefore, we designed an experiment using a gut microbiota-depletion mouse model followed by engraftment by either CRS mice or control CB donor mice to indicate the critical role of gut microbiota in pathogenesis. In Experiment 2, we observed that recipient mice with transplantation of CRS microbiota exhibited severe colitis. Meanwhile, reduced numbers of MUC2-positive cells and goblet cells, together with a reduction of MUC2 mRNA levels, were observed in the colons of recipient mice colonized with the CRS microbiota, indicating that CRS-mediated gut microbiota changes are responsible for colonic mucus damage and colitis aggravation. Interestingly, depression-like symptoms were observed in recipient mice colonized with microbiota obtained from CRS mice. This finding supports the potential that the “brain-gut-microbiota” axis may exist as a bidirectional interaction and communication (Rhee et al., 2009; Al Omran and Aziz, 2014).
16S rRNA high-throughput sequencing showed that the microbiota composition of the CRS donor and recipient mice was reduced in A. muciniphila. Furthermore, MUC2 levels were positively correlated with the abundance of A. muciniphila. Considering this relationship, we investigated whether A. muciniphila is causally involved in colonic mucus repair. Consistently, A. muciniphila supplementation prevented colitis aggravation and strengthened colonic mucosal barriers in recipient mice by improving the expression of MUC2 and increased the number of goblet cells and MUC2-positive cells. A. muciniphila has been identified as a probiotic and has many reported beneficial effects, including promoting the metabolism of the mucus layer, thereby promoting a healthy microenvironment of epithelial cells and gut barrier integrity (Derrien et al., 2004; Derrien et al., 2010; Everard et al., 2013). Moreover, A. muciniphila has been found to restore a damaged gut barrier in vitro (Reunanen et al., 2015). Our findings are in agreement with previous results.
Our previous studies demonstrated that FMT or probiotic therapy was identified as novel interventions for gastrointestinal disease, which is attributed to the normalization of gut microbial dysbiosis (Chen et al., 2018; Bu et al., 2020). The present study indicated that A. muciniphila supplementation increased the relative abundance of Ruminiclostridium in recipient mice. Ruminiclostridium is an anaerobic bacterium that degrades cellulose (Xu et al., 2013; Xu et al., 2015). A recent study has shown that the levels of Ruminiclostridium correlate with an increased number of microglia, suggesting that Ruminiclostridium plays an important role in the brain-gut axis (Loman et al., 2019). These results revealed that A. muciniphila not only prevented damage to colonic mucus but also modified gut microbiota.
This study has certain limitations. First, A. muciniphila supplementation alleviated depression-like behaviors in recipient mice before DSS administration. Accordingly, we detected the colonization of A. muciniphila by 16S rRNA sequencing. The relative abundance of A. muciniphila supplementation tended to increase, but no statistically significant difference was observed. The failure of colonization suggested that A. muciniphila should undergo repeated transfers, probably due to its sensitivity to oxygen (Derrien et al., 2004). Further studies on the mechanisms of A. muciniphila on antidepressant phenotypes are needed. Additionally, A. muciniphila inhibits the aggravation of colitis in mice under CRS, and clinical studies on the efficacy and safety of A. muciniphila in IBD patients with depression are warranted.
Conclusion
This study demonstrated that CRS-mediated gut microbiota dysbiosis resulted in damage to colonic mucus and the development of colitis. A. muciniphila supplementation protected colonic mucus and inhibited aggravation of colitis. Our findings may open new probiotic options for treating IBD patients with psychological disorders, which are involved in the damage of colonic mucus.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA756202 and PRJNA758283.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of Jiangsu Provincial Hospital of Traditional Chinese Medicine. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Ethics Committee for Animal Experiments of Jiangsu Provincial Hospital of Traditional Chinese Medicine. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
TC and RW designed the experiments and wrote the original manuscript. ZD, XY, YD, ZF, and FB performed the experiments. QW, JZ, and LL analyzed the data. YC, GS, and QN edited the original manuscript. All of authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions and the leading talent project of Chinese medicine of Jiangsu Province (Grant No. SLJ0203).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.723856/full#supplementary-material
References
- Al Omran Y., Aziz Q. (2014). The Brain-Gut Axis in Health and Disease. Adv. Exp. Med. Biol. 817, 135–153. doi: 10.1007/978-1-4939-0897-4_6 [DOI] [PubMed] [Google Scholar]
- Ambort D., Johansson M. E., Gustafsson J. K., Nilsson H. E., Ermund A., Johansson B. R., et al. (2012). Calcium and pH-Dependent Packing and Release of the Gel-Forming MUC2 Mucin. Proc. Natl. Acad. Sci. U.S.A. 109 (15), 5645–5650. doi: 10.1073/pnas.1120269109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthakrishnan A. N. (2015). Epidemiology and Risk Factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 12 (4), 205–217. doi: 10.1038/nrgastro.2015.34 [DOI] [PubMed] [Google Scholar]
- Ananthakrishnan A. N., Bernstein C. N., Iliopoulos D., Macpherson A., Neurath M. F., Ali R. A. R., et al. (2018). Environmental Triggers in IBD: A Review of Progress and Evidence. Nat. Rev. Gastroenterol. Hepatol. 15 (1), 39–49. doi: 10.1038/nrgastro.2017.136 [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential Expression Analysis for Sequence Count Data. Genome Biol. 11 (10), R106. doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharwani A., Mian M. F., Foster J. A., Surette M. G., Bienenstock J., Forsythe P. (2016). Structural & Functional Consequences of Chronic Psychosocial Stress on the Microbiome & Host. Psychoneuroendocrinology 63, 217–227. doi: 10.1016/j.psyneuen.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Bonaz B. L., Bernstein C. N. (2013). Brain-Gut Interactions in Inflammatory Bowel Disease. Gastroenterology 144 (1), 36–49. doi: 10.1053/j.gastro.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Brzozowski B., Mazur-Bialy A., Pajdo R., Kwiecien S., Bilski J., Zwolinska-Wcislo M., et al. (2016). Mechanisms by Which Stress Affects the Experimental and Clinical Inflammatory Bowel Disease (IBD): Role of Brain-Gut Axis. Curr. Neuropharmacol 14 (8), 892–900. doi: 10.2174/1570159x14666160404124127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu F., Zhang S., Duan Z., Ding Y., Chen T., Wang R., et al. (2020). A Critical Review on the Relationship of Herbal Medicine, Akkermansia Muciniphila, and Human Health. BioMed. Pharmacother. 128, 110352. doi: 10.1016/j.biopha.2020.110352 [DOI] [PubMed] [Google Scholar]
- Chen T., Zhou Q., Zhang D., Jiang F., Wu J., Zhou J. Y., et al. (2018). Effect of Faecal Microbiota Transplantation for Treatment of Clostridium Difficile Infection in Patients With Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis of Cohort Studies. J. Crohns Colitis 12 (6), 710–717. doi: 10.1093/ecco-jcc/jjy031 [DOI] [PubMed] [Google Scholar]
- Collado M. C., Derrien M., Isolauri E., de Vos W. M., Salminen S. (2007). Intestinal Integrity and Akkermansia Muciniphila, a Mucin-Degrading Member of the Intestinal Microbiota Present in Infants, Adults, and the Elderly. Appl. Environ. Microbiol. 73 (23), 7767–7770. doi: 10.1128/aem.01477-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosnes J., Gower-Rousseau C., Seksik P., Cortot A. (2011). Epidemiology and Natural History of Inflammatory Bowel Diseases. Gastroenterology 140 (6), 1785–1794. doi: 10.1053/j.gastro.2011.01.055 [DOI] [PubMed] [Google Scholar]
- Derrien M., van Passel M. W., van de Bovenkamp J. H., Schipper R. G., de Vos W. M., Dekker J. (2010). Mucin-Bacterial Interactions in the Human Oral Cavity and Digestive Tract. Gut Microbes 1 (4), 254–268. doi: 10.4161/gmic.1.4.12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., Vaughan E. E., Plugge C. M., de Vos W. M. (2004). Akkermansia Muciniphila Gen. Nov., Sp. Nov., a Human Intestinal Mucin-Degrading Bacterium. Int. J. Syst. Evol. Microbiol. 54 (Pt 5), 1469–1476. doi: 10.1099/ijs.0.02873-0 [DOI] [PubMed] [Google Scholar]
- Ding N., Zhang X., Zhang X. D., Jing J., Liu S. S., Mu Y. P., et al. (2020). Impairment of Spermatogenesis and Sperm Motility by the High-Fat Diet-Induced Dysbiosis of Gut Microbes. Gut 69 (9), 1608–1619. doi: 10.1136/gutjnl-2019-319127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: Highly Accurate OTU Sequences From Microbial Amplicon Reads. Nat. Methods 10 (10), 996–998. doi: 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Everard A., Belzer C., Geurts L., Ouwerkerk J. P., Druart C., Bindels L. B., et al. (2013). Cross-Talk Between Akkermansia Muciniphila and Intestinal Epithelium Controls Diet-Induced Obesity. Proc. Natl. Acad. Sci. U.S.A. 110 (22), 9066–9071. doi: 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Wei B., Wen T., Johansson M. E., Liu X., Bradford E., et al. (2011). Loss of Intestinal Core 1-Derived O-Glycans Causes Spontaneous Colitis in Mice. J. Clin. Invest. 121 (4), 1657–1666. doi: 10.1172/jci45538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galley J. D., Bailey M. T. (2014). Impact of Stressor Exposure on the Interplay Between Commensal Microbiota and Host Inflammation. Gut Microbes 5 (3), 390–396. doi: 10.4161/gmic.28683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Cao Q., Cheng Y., Zhao D., Wang Z., Yang H., et al. (2018). Chronic Stress Promotes Colitis by Disturbing the Gut Microbiota and Triggering Immune System Response. Proc. Natl. Acad. Sci. U.S.A. 115 (13), E2960–E2969. doi: 10.1073/pnas.1720696115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghia J. E., Blennerhassett P., Deng Y., Verdu E. F., Khan W. I., Collins S. M. (2009). Reactivation of Inflammatory Bowel Disease in a Mouse Model of Depression. Gastroenterology 136 (7), 2280–2288.e2281–2284. doi: 10.1053/j.gastro.2009.02.069 [DOI] [PubMed] [Google Scholar]
- Hansson G. C., Johansson M. E. (2010). The Inner of the Two Muc2 Mucin-Dependent Mucus Layers in Colon Is Devoid of Bacteria. Gut Microbes 1 (1), 51–54. doi: 10.4161/gmic.1.1.10470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno R., Brailey P. M., Barral P. (2018). Quantitative Polymerase Chain Reaction-Based Analyses of Murine Intestinal Microbiota After Oral Antibiotic Treatment. J. Vis. Exp. 141. doi: 10.3791/58481 [DOI] [PubMed] [Google Scholar]
- Johansson M. E., Gustafsson J. K., Holmén-Larsson J., Jabbar K. S., Xia L., Xu H., et al. (2014). Bacteria Penetrate the Normally Impenetrable Inner Colon Mucus Layer in Both Murine Colitis Models and Patients With Ulcerative Colitis. Gut 63 (2), 281–291. doi: 10.1136/gutjnl-2012-303207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. E., Larsson J. M., Hansson G. C. (2011). The Two Mucus Layers of Colon Are Organized by the MUC2 Mucin, Whereas the Outer Layer Is a Legislator of Host-Microbial Interactions. Proc. Natl. Acad. Sci. U.S.A. 108 Suppl;1 (Suppl 1), 4659–4665. doi: 10.1073/pnas.1006451107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. E., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G. C. (2008). The Inner of the Two Muc2 Mucin-Dependent Mucus Layers in Colon Ss Devoid of Bacteria. Proc. Natl. Acad. Sci. U.S.A. 105 (39), 15064–15069. doi: 10.1073/pnas.0803124105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiliaan A. J., Saunders P. R., Bijlsma P. B., Berin M. C., Taminiau J. A., Groot J. A., et al. (1998). Stress Stimulates Transepithelial Macromolecular Uptake in Rat Jejunum. Am. J. Physiol. 275 (5), G1037–G1044. doi: 10.1152/ajpgi.1998.275.5.G1037 [DOI] [PubMed] [Google Scholar]
- Kochar B., Barnes E. L., Long M. D., Cushing K. C., Galanko J., Martin C. F., et al. (2018). Depression Is Associated With More Aggressive Inflammatory Bowel Disease. Am. J. Gastroenterol. 113 (1), 80–85. doi: 10.1038/ajg.2017.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke K., Spitzer R. L., Williams J. B. (2001). The PHQ-9: Validity of a Brief Depression Severity Measure. J. Gen. Intern. Med. 16 (9), 606–613. doi: 10.1046/j.1525-1497.2001.016009606.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke K., Spitzer R. L., Williams J. B., Löwe B. (2010). The Patient Health Questionnaire Somatic, Anxiety, and Depressive Symptom Scales: A Systematic Review. Gen. Hosp Psychiatry 32 (4), 345–359. doi: 10.1016/j.genhosppsych.2010.03.006 [DOI] [PubMed] [Google Scholar]
- Labanski A., Langhorst J., Engler H., Elsenbruch S. (2020). Stress and the Brain-Gut Axis in Functional and Chronic-Inflammatory Gastrointestinal Diseases: A Transdisciplinary Challenge. Psychoneuroendocrinology 111:104501. doi: 10.1016/j.psyneuen.2019.104501 [DOI] [PubMed] [Google Scholar]
- Liu Z., Qi Y., Cheng Z., Zhu X., Fan C., Yu S. Y. (2016). The Effects of Ginsenoside Rg1 on Chronic Stress Induced Depression-Like Behaviors, BDNF Expression and the Phosphorylation of PKA and CREB in Rats. Neuroscience 322, 358–369. doi: 10.1016/j.neuroscience.2016.02.050 [DOI] [PubMed] [Google Scholar]
- Loman B. R., Jordan K. R., Haynes B., Bailey M. T., Pyter L. M. (2019). Chemotherapy-Induced Neuroinflammation Is Associated With Disrupted Colonic and Bacterial Homeostasis in Female Mice. Sci. Rep. 9 (1), 16490. doi: 10.1038/s41598-019-52893-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick B. A., Auerbach S. S., Stockton P. S., Foley J. F., Malarkey D. E., Sills R. C., et al. (2012). Testing an Aflatoxin B1 Gene Signature in Rat Archival Tissues. Chem. Res. Toxicol. 25 (5), 1132–1144. doi: 10.1021/tx3000945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikocka-Walus A., Pittet V., Rossel J. B., von Känel R. (2016). Symptoms of Depression and Anxiety Are Independently Associated With Clinical Recurrence of Inflammatory Bowel Disease. Clin. Gastroenterol. Hepatol. 14 (6), 829–835.e821. doi: 10.1016/j.cgh.2015.12.045 [DOI] [PubMed] [Google Scholar]
- Molodecky N. A., Soon I. S., Rabi D. M., Ghali W. A., Ferris M., Chernoff G., et al. (2012). Increasing Incidence and Prevalence of the Inflammatory Bowel Diseases With Time, Based on Systematic Review. Gastroenterology 142 (1), 46–54.e42; quiz e30. doi: 10.1053/j.gastro.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Morampudi V., Dalwadi U., Bhinder G., Sham H. P., Gill S. K., Chan J., et al. (2016). The Goblet Cell-Derived Mediator RELM-β Drives Spontaneous Colitis in Muc2-Deficient Mice by Promoting Commensal Microbial Dysbiosis. Mucosal Immunol. 9 (5), 1218–1233. doi: 10.1038/mi.2015.140 [DOI] [PubMed] [Google Scholar]
- Oliver S. R., Phillips N. A., Novosad V. L., Bakos M. P., Talbert E. E., Clanton T. L. (2012). Hyperthermia Induces Injury to the Intestinal Mucosa in the Mouse: Evidence for an Oxidative Stress Mechanism. Am. J. Physiol. Regul. Integr. Comp. Physiol. 302 (7), R845–R853. doi: 10.1152/ajpregu.00595.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opal M. D., Klenotich S. C., Morais M., Bessa J., Winkle J., Doukas D., et al. (2014). Serotonin 2C Receptor Antagonists Induce Fast-Onset Antidepressant Effects. Mol. Psychiatry 19 (10), 1106–1114. doi: 10.1038/mp.2013.144 [DOI] [PubMed] [Google Scholar]
- Osadchiy V., Martin C. R., Mayer E. A. (2019). The Gut-Brain Axis and the Microbiome: Mechanisms and Clinical Implications. Clin. Gastroenterol. Hepatol. 17 (2), 322–332. doi: 10.1016/j.cgh.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panara A. J., Yarur A. J., Rieders B., Proksell S., Deshpande A. R., Abreu M. T., et al. (2014). The Incidence and Risk Factors for Developing Depression After Being Diagnosed With Inflammatory Bowel Disease: A Cohort Study. Aliment Pharmacol. Ther. 39 (8), 802–810. doi: 10.1111/apt.12669 [DOI] [PubMed] [Google Scholar]
- Reunanen J., Kainulainen V., Huuskonen L., Ottman N., Belzer C., Huhtinen H., et al. (2015). Akkermansia Muciniphila Adheres to Enterocytes and Strengthens the Integrity of the Epithelial Cell Layer. Appl. Environ. Microbiol. 81 (11), 3655–3662. doi: 10.1128/aem.04050-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter J. A., Spacek D. V., Snyder M. P. (2015). High-Throughput Sequencing Technologies. Mol. Cell 58 (4), 586–597. doi: 10.1016/j.molcel.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S. H., Pothoulakis C., Mayer E. A. (2009). Principles and Clinical Implications of the Brain-Gut-Enteric Microbiota Axis. Nat. Rev. Gastroenterol. Hepatol. 6 (5), 306–314. doi: 10.1038/nrgastro.2009.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B., Le Chatelier E., Derosa L., Duong C. P. M., Alou M. T. (2018). Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy Against Epithelial Tumors. Science 359 (6371), 91–97. doi: 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- Samuelson D. R., Shellito J. E., Maffei V. J., Tague E. D., Campagna S. R., Blanchard E. E., et al. (2017). Alcohol-Associated Intestinal Dysbiosis Impairs Pulmonary Host Defense Against Klebsiella Pneumoniae. PloS Pathog. 13 (6), e1006426. doi: 10.1371/journal.ppat.1006426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sann H., Erichsen J., Hessmann M., Pahl A., Hoffmeyer A. (2013). Efficacy of Drugs Used in the Treatment of IBD and Combinations Thereof in Acute DSS-Induced Colitis in Mice. Life Sci. 92 (12), 708–718. doi: 10.1016/j.lfs.2013.01.028 [DOI] [PubMed] [Google Scholar]
- Shin J., Noh J. R., Chang D. H., Kim Y. H., Kim M. H., Lee E. S., et al. (2019). Elucidation of Akkermansia Muciniphila Probiotic Traits Driven by Mucin Depletion. Front. Microbiol. 10, 1137. doi: 10.3389/fmicb.2019.01137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderholm J. D., Yang P. C., Ceponis P., Vohra A., Riddell R., Sherman P. M., et al. (2002). Chronic Stress Induces Mast Cell-Dependent Bacterial Adherence and Initiates Mucosal Inflammation in Rat Intestine. Gastroenterology 123 (4), 1099–1108. doi: 10.1053/gast.2002.36019 [DOI] [PubMed] [Google Scholar]
- Song Y., Liu C., Finegold S. M. (2004). Real-Time PCR Quantitation of Clostridia in Feces of Autistic Children. Appl. Environ. Microbiol. 70 (11), 6459–6465. doi: 10.1128/aem.70.11.6459-6465.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H., Yang J. H., Kim H. J., Lee D. K. (2019). A Chronic Immobilization Stress Protocol for Inducing Depression-Like Behavior in Mice. J. Vis. Exp. 147). doi: 10.3791/59546 [DOI] [PubMed] [Google Scholar]
- Valles-Colomer M., Falony G., Darzi Y. (2019). The Neuroactive Potential of the Human Gut Microbiota in Quality of Life and Depression. Nat. Microbiol. 4 (4), 623–632. doi: 10.1038/s41564-018-0337-x [DOI] [PubMed] [Google Scholar]
- Walker J. R., Ediger J. P., Graff L. A., Greenfeld J. M., Clara I., Lix L., et al. (2008). The Manitoba IBD Cohort Study: A Population-Based Study of the Prevalence of Lifetime and 12-Month Anxiety and Mood Disorders. Am. J. Gastroenterol. 103 (8), 1989–1997. doi: 10.1111/j.1572-0241.2008.01980.x [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences Into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 73 (16), 5261–5267. doi: 10.1128/aem.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Li Y., Tang W., Sun Q., Chen L., Wang X., et al. (2019). Chronic Unpredictable Mild Stress in Rats Induces Colonic Inflammation. Front. Physiol. 10, 1228. doi: 10.3389/fphys.2019.01228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Huang R., Teng L., Jing X., Hu J., Cui G., et al. (2015). Cellulosome Stoichiometry in Clostridium Cellulolyticum Is Regulated by Selective RNA Processing and Stabilization. Nat. Commun. 6, 6900. doi: 10.1038/ncomms7900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Huang R., Teng L., Wang D., Hemme C. L., Borovok I., et al. (2013). Structure and Regulation of the Cellulose Degradome in Clostridium Cellulolyticum. Biotechnol. Biofuels 6 (1), 73. doi: 10.1186/1754-6834-6-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Yin P., Liu F., Cheng G., Guo K., Lu A., et al. (2010). Effect of Heat Stress on the Porcine Small Intestine: A Morphological and Gene Expression Study. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 156 (1), 119–128. doi: 10.1016/j.cbpa.2010.01.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA756202 and PRJNA758283.