CONSPECTUS:
Any chemist studying the interaction of molecules with lipid assemblies will eventually be confronted by the topic of membrane bilayer heterogeneity and may ultimately encounter the heterogeneity of natural membranes. In artificial bilayers, heterogeneity is defined by phase segregation that can be in the nano- and micrometer range. In biological bilayers, heterogeneity is considered in the context of small (10–200 nm) sterol and sphingolipid-enriched heterogeneous and highly dynamic domains. Several techniques can be used to assess membrane heterogeneity in living systems. Our approach is to use a fluorescent reporter molecule immersed in the bilayer, which, by changes in its spectroscopic properties, senses physical-chemistry aspects of the membrane. This dye in combination with microscopy and fluctuation techniques can give information about membrane heterogeneity at different temporal and spatial levels: going from average fluidity to number and diffusion coefficient of nanodomains. LAURDAN (6-dodecanoyl-2-(dimethylamino) naphthalene), is a fluorescent probe designed and synthesized in 1979 by Gregorio Weber with the purpose to study the phenomenon of dipolar relaxation. The spectral displacement observed when LAURDAN is either in fluid or gel phase permitted the use of the technique in the field of membrane dynamics. The quantitation of the spectral displacement was first addressed by the generalized polarization (GP) function in the cuvette, a ratio of the difference in intensity at two wavelengths divided by their sum. In 1997, GP measurements were done for the first time in the microscope, adding to the technique the spatial resolution and allowing the visualization of lipid segregation both in liposomes and cells. A new prospective to the membrane heterogeneity was obtained when LAURDAN fluorescent lifetime measurements were done in the microscope. Two channel lifetime imaging provides information on membrane polarity and dipole relaxation (the two parameters responsible for the spectral shift of LAURDAN), and the application of phasor analysis allows pixel by pixel understanding of these two parameters in the membrane. To increase temporal resolution, LAURDAN GP was combined with fluctuation correlation spectroscopy (FCS) and the motility of nanometric highly packed structures in biological membranes was registered. Lately the application of phasor analysis to spectral images from membranes labeled with LAURDAN allows us to study the full spectra pixel by pixel in an image. All these methodologies, using LAURDAN, offer the possibility to address different properties of membranes depending on the question being asked. In this Account, we will focus on the principles, advantages, and limitations of different approaches to orient the reader to select the most appropriate technique for their research.
Graphical Abstract
INTRODUCTION
Biological membranes are inherently heterogeneous due to the asymmetry of composition of the two monolayers.5-7 In addition to differences in the protein content, the actual lipid composition of the monolayers also differs. The thermodynamic barriers to lipids transferring between bilayers are significant and such translocation is usually only accomplished by enzymes known as flippases. In addition to this bilayer asymmetry, heterogeneity also exists in the lateral organization of membranes and is susceptible to thermodynamic variables such as temperature, pressure and chemical potential. The lateral organization of phospholipids is defined in the context of a supramolecular organization, for example, in terms of solid order (gel, Lβ), liquid disorder (liquid crystalline, ld or Lα) or liquid order (lo). Lipid phase segregation in the nano to micrometer range is fundamentally important for synthetic membranes. In biological membranes, the situation is more complicated due to the compositional heterogeneity and physical characteristics. Lateral organization of membranes in living cells is manifested by small (10–200 nm) sterol and sphingolipid-enriched heterogeneous and highly dynamic domains, as discussed by Pike.8,9
Several techniques can be used to assess membrane heterogeneity in living systems.1,10-12 Our approach was to use a fluorescent reporter molecule immersed in the bilayer. The changes in the spectroscopic properties of this probe provide information on membrane physical properties without perturbing the system under study. Since Gregorio Weber synthesized this molecule in 1979,13 LAURDAN (6-dodecanoyl-2-(dimethylamino) naphthalene), used in combination with several biophysical methods, has provided information on membrane heterogeneity at different scales. These diverse approaches will be described in the following sections.
LAURDAN, THE FLUORESCENT EXPLORER FOR MEMBRANE HETEROGENEITY
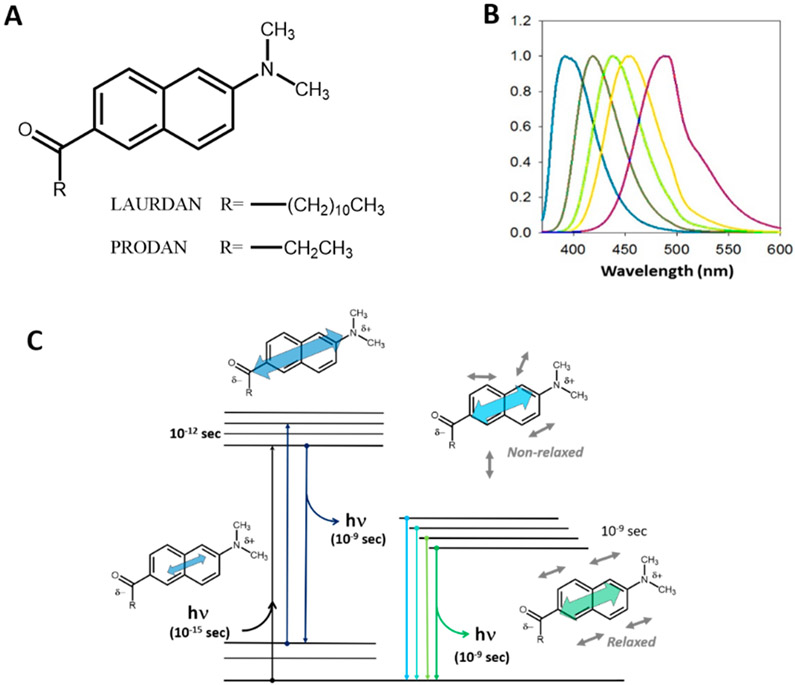
LAURDAN belongs to the family of probes having a naphthalene moiety modified on the 2,6 ring positions (Figure 1A), with electron donor (amino) and acceptor (carbonyl) groups, which can stabilize charge transfer and engender large excited state dipole moments.13 These excited state dipoles respond dramatically to the polarity of their surroundings, as shown in Figure 1B, which illustrates the change in the emission spectra of LAURDAN in solvents of varying dielectric constants. Although Weber never used LAURDAN himself in any published studies, when he synthesized it he clearly had in mind its potential for membrane studies, such as he had carried out earlier.14
Figure 1.
(A) Chemical structure of LAURDAN and PRODAN. (B) Emission spectra of LAURDAN in different solvents with increasing polarities (from left to right: hexane, benzene, chloroform, acetonitrile, and ethanol). (C) Perrin–Jabłoński diagram for 6-alkyl-2-dimethyl amino naphthalene, showing the ground state and the first singlet excited state. Excitation results in an increase in charge separation and hence an increase in the magnitude of the fluorophore’s dipole moment, which then induces relaxation of the solvent dipoles.
The rationale behind the synthesis of the LAURDAN family was their sensitivity to two different physicochemical properties characterizing their environment: the dielectric environment associated with the solvent surrounding the probes (Figure 1C) and the relaxation of solvent molecules (carrying dipoles) around the fluorophore dipole during the excited state. Furthermore, the selection of the alkyl chain size allows for modulating the lipophilic/hydrophilic balance of the molecule and, therefore, permits the probe to be preferentially located in specific regions of microheterogeneous systems, such as cells. LAURDAN has been proven to be especially useful in studies on lipid systems in general and on biological membranes in particular.
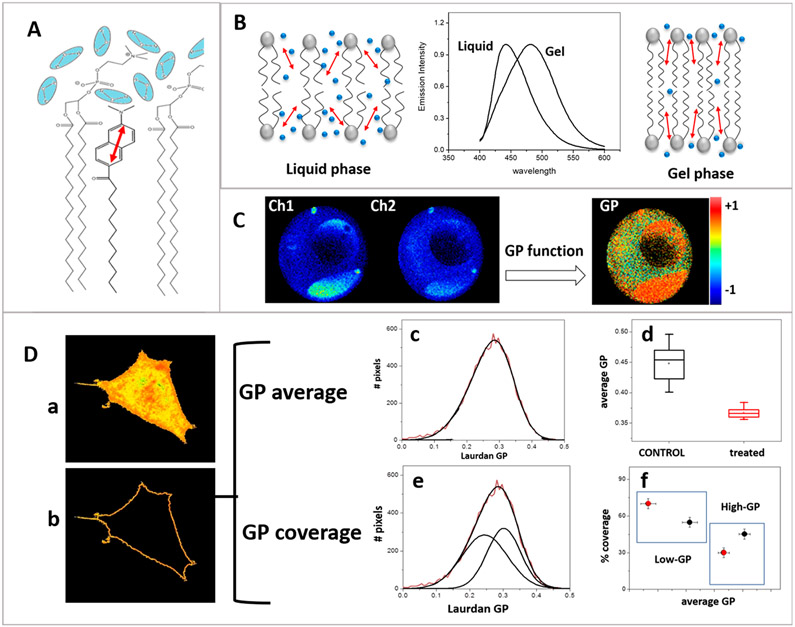
At the membrane, the naphthalene moiety of LAURDAN localizes at the bilayer interphase below the carbonyl group of the phospholipid, while the lauric acid tail embeds in the bilayer (Figure 2A). At this location LAURDAN can sense the presence of water molecules having rotational times which are in the range of LAURDAN’s lifetime (nanoseconds).15
Figure 2.
LAURDAN generalized polarization. (A) LAURDAN location at the membrane interphase. (B) LAURDAN spectra in gel (~440 nm) and liquid (~490 nm) phases. Red arrows represent LAURDAN in the membrane, and blue spheres represent the water molecules. (C) GP image obtained using two channels observed through bandpass filters (Ch1 and Ch2). DOPC/DPPC/CHOL 1:1:1 GUV showing coexistence of liquid ordered (orange) and liquid disordered (yellow) phase separation at 15.7°C. (D) GP analysis of HeLa cells incubated with LAURDAN at 37°C: (a) GP image for a HeLa cell, (b) pixels corresponding to the plasma membrane, and (c) GP histogram of the pixels from the membrane. (d) GP average analysis used to follow cholesterol removal.34 The GP center is used to compare control and treated cells. (e, f) Same data analyzed by GP coverage. The GP histogram is resolved into the two best Gaussians; center and area are used to create the coverage plot (f).
Both the number of confined water molecules and their relaxation rates are modified by the physical properties of the membrane. As the membrane becomes less organized (more fluid), more mobile water molecules can be found near the LAURDAN ring, experiencing dipolar relaxation (DR) around the excited LAURDAN, which causes a shift in the emission spectrum to lower energy (Figure 2B).
At this point, we would like to define some terminology for the readers: membrane polarity sensed by LAURDAN relates to the number of water molecules present in the bilayer, while dipolar relaxation (DR) relates to reorientation of their dipoles by rotation, in response to the change in LAURDAN’s dipole moment upon excitation. Both terms depend on the lipid order. Polarity is a discrete parameter while DR is continuous (time dependent). The term “fluidity” is a qualitative general term used to describe the viscosity in the membrane (capability of the lipids to flow or rotate), and is determined by the water content, the water relaxation and the lipid order of the bilayer. We will see in the following sections that depending on the technique used, information on polarity and DR can be obtained independently (FLIM) or mixed (GP and spectral phasor). In the latest case, the information obtained is related with the general term “fluidity”.
THE GP APPROACH TO MEASURE MEMBRANE HETEROGENEITY
The spectral displacement shown by LAURDAN in synthetic membranes was first studied in 1986 following the time evolution of its emission spectrum.16 Quantification of LAURDAN’s spectral displacement in cuvette studies was addressed by the generalized polarization (GP) function, defined in 1990 by Parassasi et al.17 The GP function is defined as
| (1) |
where IB and IR are the emission intensities observed at wavelengths on the blue and red sides of the spectrum, respectively (for example, near 440 nm (blue) and 490 nm (red)).
In 1997, imaging GP on cells was done for the first time using two-photon excitation to minimize photobleaching.18 Using two bandpass filters (Ch1 (blue) and Ch2 (red)) the GP function at each pixel in the image was calculated (Figure 2C). This method considers a model with two extreme molecular environments (gel at 440 nm and liquid at 490 nm) and the determination of different environments will be restricted to these two values and the linear properties of the GP function.
Using this approach lipid domain segregation in giant unilamellar vesicles (GUVs) made of pure lipid mixtures were visualized,19-24 a phenomenon that was predicted but which could not be seen in cuvette measurements. For example, Figure 2C shows GUVs made of the raft mixture DOPC/DPPC/CHOL 1:1:1 presenting a coexistence of liquid ordered (orange) and liquid disordered (yellow) phases at 15.7 °C. In live cells (Figure 2D), LAURDAN GP imaging shows differences in the fluidity of plasma and internal membranes. To study the internal membranes, for instance, the nuclear membrane, a second fluorescent marker must be used to identify the organelle and use it as a template to isolate the corresponding pixels in the GP image.25 To study the plasma membrane, the corresponding pixels may be isolated (Figure 2Db) and the pixel distribution histogram (GP histogram) is obtained. The width of the GP histogram corresponding to the membrane demonstrates that the membrane is rather heterogeneous in nature (i.e., a range of GP values are found in the membrane) but the resolution of the GP images is not sufficient to see the structural membrane segregation observed in GUVs. GP imaging and the average GP value (the center of the histogram of GP values) provide information about changes in membrane fluidity. This technique is easy to implement, and the analysis can be done using software such as SimFCS26 and available ImageJ routines.27 The GP method allows for rapid data acquisition and analysis of several cells, providing good statistics for biological studies with cells in culture.28-30 The use of GP histograms to account for heterogeneity in terms of raft domains was done using the “coverage analysis”.31 In this approach (Figure 2D), the GP histogram is resolved into two Gaussians and the two areas with their respective average GP values are interpreted as the lo/ld phases. Considering that lo/ld are defined in pure lipid systems in equilibrium, a situation not applicable to biological membranes, we interpret this analysis as subpopulations of domains with different fluidities (high and low GP populations). The coverage approach can be a good complementary analysis to follow changes in fluidity domain distributions, due to changes in membrane composition30 or cell interactions.32 Although there are data from GP experiments in the literature obtained on fixed samples, the validity of this approach is limited due to the extreme sensitivity of LAURDAN to the membrane physical state and water activity.33
LAURDAN GP AND FLUORESCENCE CORRELATION SPECTROSCOPY: ADDITION OF TIME RESOLUTION
LAURDAN and GP imaging can differentiate areas having different fluidities, but the spatial resolution is still determined by the optics of the confocal microscope (~300 nm, diameter of the point spread function, PSF). To detect nanometric and mobile domains in live cell membranes, spatial and temporal resolution must be increased. Our approach was to combine the LAURDAN GP technique and the temporal resolution of fluorescence correlation spectroscopy (FCS).35,36 Our aim was to detect fluctuations in LAURDAN’s GP, which can be related to the mobility of highly packed membrane structures, smaller than the PSF in the membrane of live cells.1
FCS37-39 is based on analysis of fluctuations within the PSF volume generated in a conventional confocal microscope or by using two-photon excitation.40 The fluorescence fluctuations generated by molecules diffusing in and out of the PSF is used to obtain information related to the diffusion of the molecules. Autocorrelation analysis41 yields the diffusion coefficient (Dcoef) and the concentration of particles in the observation volume, and photon counting histogram analysis42,43 provides the number of fluorophores in the observation volume and their intrinsic molecular brightness.44
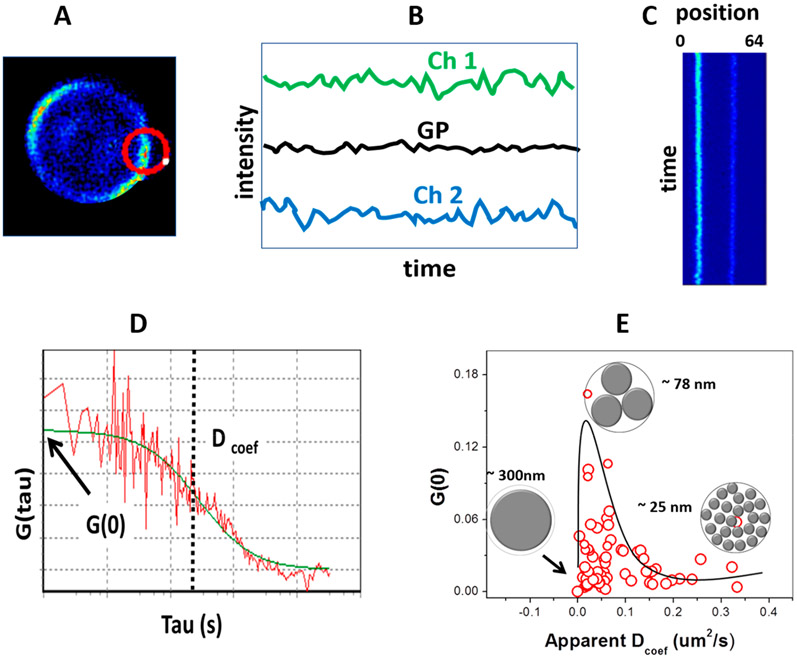
In the point-FCS approach, the PSF is kept immobile and the information obtained is restricted to the volume being illuminated/observed. This configuration, however, may be problematic when used with live cells, since cell movement and photobleaching can occur. In order to avoid such problems, we used circular scanning-FCS (csFCS). In the csFCS method,45 the intensity fluctuations are acquired along a 2D-orbit and they are used to create a map or “carpet” (x/y vs time). The csFCS method is useful when the laser beam illuminates a precise region such as a biological membrane.45 GP-FCS is done by performing circular scanning FCS, using two channels in the detection (Ch1 and Ch2) having the traditional GP filters (Figure 3A). The intensity traces from Ch1 and Ch2 are used to calculate a GP trace (Figure 3B) and a location versus time map (Figure 3C). In this representation, the autocorrelation function is applied to the pixels corresponding to the membrane and the diffusion and number of objects passing through the PSF is determined. Finally, the experimental autocorrelation function is fitted using the mathematical expression for csFCS for diffusion1,35 and the Dcoef together with the number of particles for the structures producing the fluctuation are determined (Figure 3D).36,38,46
Figure 3.
LAURDAN GP and scanning FCS. (A) Scanning FCS in rabbit red blood cells stained with LAURDAN at 37 °C with laser scanning in a circular pattern. (B) Two channels (440 and 490 nm) were acquired to obtain the GP trace. (C) GP trace data were transformed in an x–y representation (each orbit over time). Locations corresponding to the membrane were analyzed by autocorrelation giving G(0) and diffusion coefficient Dcoef (μm2/s) for several red blood cells. (D) Autocorrelation analysis. (E) Plot of G(0) versus Dcoef, showing the structures responsible for the fluctuations with sizes of 300, 75, and 25 nm.
Two parameters were determined for GP fluctuations in the membranes of erythrocytes and CHO cells1 and presented in an xy plot (Figure 3E). Our interpretation of this data was based on the question of how big the fluctuating structures should be to accommodate the experimental N value (number of particles) inside the PSF of 300 nm, assuming the structures as well as the PSF are circular. These fluctuations could be explained by the existence of tightly packed microdomains moving in a more fluid background with sizes ranging from 20 to 300 nm.1 Several features of these structures were common to the two cell types studied: (i) domains of different sizes coexisted in the same cell; (ii) the map of Dcoef vs number of structures in both cells types was similar; (iii) incubation of the cells with cholesterol acceptor (rHDL) produced the coalescence of small domains into larger ones. All these observations support theories stating that assembling/reassembling of these structures occurs in live cell membranes and may play a role in biological processes. This technique is a very delicate method used to study lipid domains in vivo and addresses the dynamics in terms of mobility and supramolecular organization; it requires a robust implementation of both data acquisition and data analysis.
FLUORESCENCE LIFETIME IMAGING AND PHASOR ANALYSIS: MORE DETAILS ON THE MEMBRANE PROPERTIES (BEYOND THE GP)
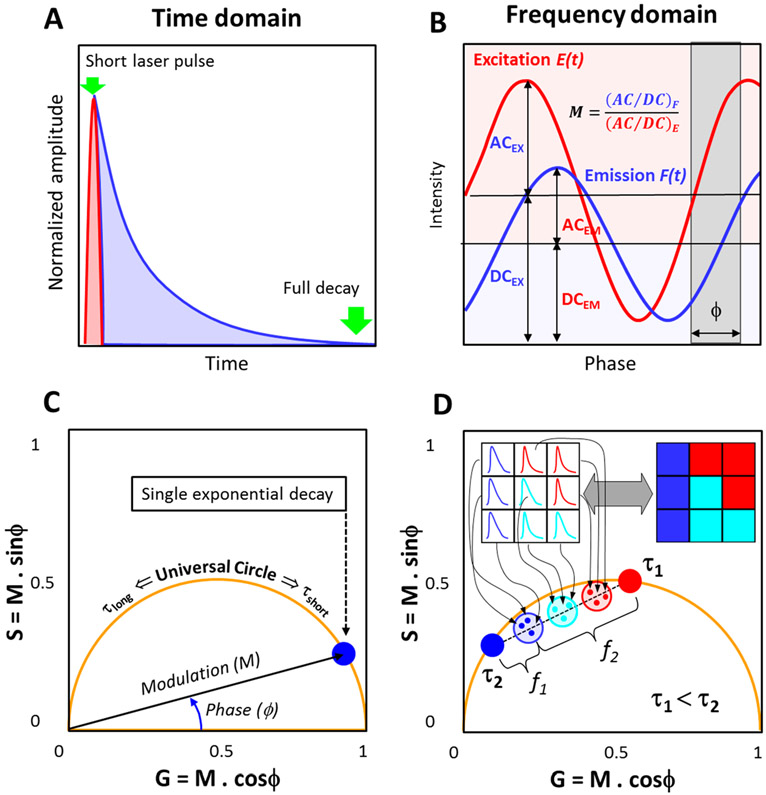
Excited state lifetimes have traditionally been measured by either “time-domain” or “frequency domain” methods30 (Figure 4A and B). The phasor plot, which can be implemented using either time or frequency domain data, was first introduced by Jameson et al. as a geometrical representation to illustrate the existence of any single exponential decay independently of the lifetime or light modulation frequency.47 The difficulty of traditional analyses of lifetimes increases with the number of decay components and the complexity of the analysis is further amplified when one seeks to analyze a lifetime image from a biological system, which may be highly heterogeneous.
Figure 4.
Schematic representation of the phasor FLIM analysis: (A, B) Lifetime measurements using the time and frequency domain method, respectively. (C) For the phasor transformation, the phase shift, ϕ, and relative modulation, M, are used to obtain the x-axis (G) and y-axis (S) of the phasor plot. Single exponential decays fall on the universal circle (orange semicircle). (D) In a FLIM image (represented as a 3 × 3 image in the inset), the decay from each pixel maps to a single point in the phasor plot. Multiexponential decay phasors are located inside the universal circle (represented by the shaded circles). The linear combination property allows one to obtain the fraction of each component (as represented in figure D). The reciprocity principle (gray arrow) enables one to connect the intensity image and the phasor plot data. The cursors (blue, cyan, and red) are valuable tools to select a region of interest in phasor plot and then trace back to where these pixels, selected by the cursors, are located in the image.
The use of phasors in fluorescence was relatively dormant until the early part of the twenty-first century when phasors began to be applied to fluorescence lifetime imaging microscopy (FLIM). The first to suggest “phasor-type plots” for use in FLIM were Clayton et al.,48 whose point was to resolve two lifetime components in a FLIM image. Other laboratories also made use of “phasor-type plots”, naming them AB plots49 or polar plots,50 but the first use of the modern phasor-FLIM approach for cell work was from the Laboratory for Fluorescence Dynamics.51,52
For phasor analysis, experimental data (acquired by either frequency or time domain) are transformed into coordinates “x” (G component) and “y” (S component) of the phasor plot. The equations for this transformation were first presented by Weber.53 In the frequency domain, the phasor plot transformation can be done using the following notations.
| (2) |
| (3) |
For time domain data the following transformations are used:
| (4) |
| (5) |
where n and ω correspond to the harmonic number and the angular modulation frequency of the excitation, respectively. With a pulsed excitation source, ω is equal to 2π/T, where T is the period of the pulses.
Using eqs 2, 3 or 4, 5, the fluorescence decay at each pixel of a FLIM image is phasor transformed (Figure 4C) and placed in a phasor plot (Figure 4D). In this type of plot, the perimeter of the semicircle is known as the “universal circle” and all single exponential lifetimes must fall somewhere on this semicircle. As the lifetime increases, the position on the “universal circle” moves anticlockwise from τ = 0 at (1,0) to τ ~ ∞ at (0,0). The position of the phasor on the universal circle depends on the modulation frequency of the excitation light. Perhaps the most relevant properties of the phasor transformation are the principles of linear combination and reciprocity. The linear combination refers to the possibility to geometrically resolve the proportions of two fluorescence species (in the simplest case) by the lever rule of vector additions.54 By noting the position of each of the two components or by obtaining higher harmonics and resolving the equations, it is possible to elucidate the fraction of both components, Figure 4D. The reciprocity principle is due to the connection between the real and imaginary space, in which it is possible to locate regions of interest (using the cursor) in the phasor plot and highlighting those pixels in the image, and vice versa. This is exemplified in Figure 4C.
This graphical representation of the FLIM images is a useful framework to interpret and analyze any possible fluorescence decay without previous assumptions, fittings or models, and is a powerful tool to visualize the enormous amount of data obtained in FLIM. The phasor plot approach to FLIM has been used in numerous different applications such as studies of ions in cells,55,56 proteins conformation in vitro57 or quantitative biology58 and a detailed discussion of the method and modern software was recently published.59
Application of the FLIM phasor approach to LAURDAN fluorescence in membranes of live cells was introduced by Golfetto, Hinde, and Gratton in 2013.60 As stated earlier, the fluorescent properties of LAURDAN may change due to the polarity of the environment and also due to dipolar relaxation of water molecules around the LAURDAN dipole during the emission lifetime.61-64 Independent phasor analysis of two simultaneous FLIM images, obtained through different bandpass filters, allows the evaluation of these two properties separately. The blue channel is used to evaluate the polarity, and the green channel to observe dipolar relaxation (DR).60,62,65,66 By monitoring the decay through the two bandpass filters (the GP filters), we can isolate LAURDAN information from different environments: the blue channel refers to LAURDAN where no relaxation occurs and the green channel where DR take place.
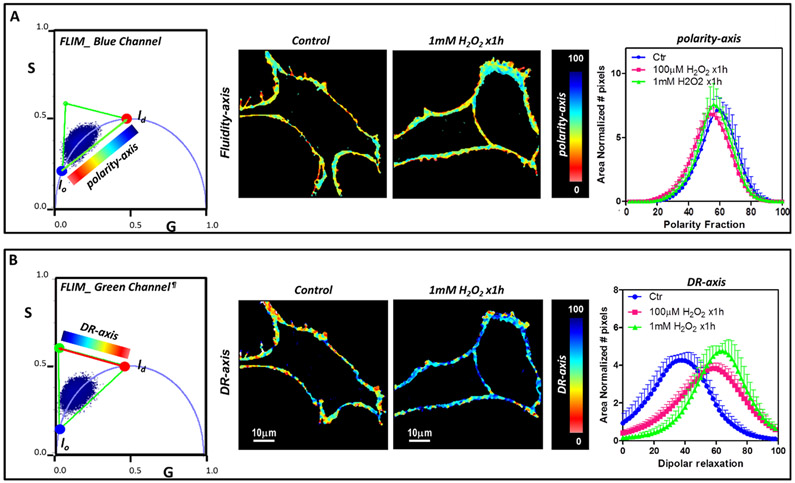
Figure 5 shows an example of this methodology to study the effect of H2O2 in NIH-3T3 cells.3 The phasor plot for the blue and green channels (left panel) shows a cluster of phasors originating from the plasma membrane of control and treated NIH-3T3 cells. Treated cells showed decreased polarity and higher dipolar relaxation as compared with the control. By analyzing the fractional histograms for the blue and green channels one may quantify the results (right panel). In this analysis, we used a three components approach developed to obtain information independently from each of the dimensions (polarity and DR). FLIM phasors may provide valuable information on membrane heterogeneity at the molecular level. FLIM is available in some commercial microscopes but like all precise techniques, it needs a robust implementation of data acquisition and analysis.
Figure 5.
FLIM phasor three component analysis for the effect of H2O2 in NIH-3T3 cells. H2O2 treated versus nontreated cells were compared. (A) Analysis for the blue channel. A color palette for the polarity-axis was used to obtain the corresponding figures (middle panel) and histogram of polarity fraction (right panel). (B) Analysis for the green channel. Images were generated using the coloring scheme shown in the phasor plot for the dipolar relaxation axis and the histogram for the dipolar relaxation enables quantification of the effect of H2O2 (right image). For both analysis cell images were masked to analyze pixels from the plasma membrane. lo and ld refer to liquid order and liquid disorder environments, respectively. We note that dipolar relaxation may bring phasor points outside of the universal circle due to the additional phase shift.
THE SPECTRAL PHASOR SPACE FOR LAURDAN ANALYSIS2,67,68
With the advent of hyperspectral imaging, new avenues opened for analysis of the spectra of solvatochromic probes like LAURDAN. A spectral image is composed of a series of images obtained simultaneously at different wavelength ranges in the emission spectra. The resultant spectral image contains the full spectrum in each pixel. Spectral phasor analysis is the transformation of the spectral image, pixel-by-pixel, into Fourier space, by calculating the real and imaginary components called G and S according to the following equations
| (6) |
| (7) |
I(λ) represents the intensity at every wavelength, n is the harmonic number, and λi is the initial wavelength. After this transformation, the (G, S) pair is plotted in a polar plot, the spectral phasor67 (Figure 6A). Phasor analysis applied to spectra shares analysis tools with phasor analysis applied to lifetimes: linear combinations, additivity, reciprocity, etc. However, a very important difference from the lifetime phasor plot is that the universal circle in the spectral phasor plot does not have any physical meaning. In the spectral phasor, the four quarters and the circle with 1 unit radii enclose all the possible positions for the phasors. It is important to mention that to compare different spectra, using spectral phasor analysis, the same wavelength range must be used.
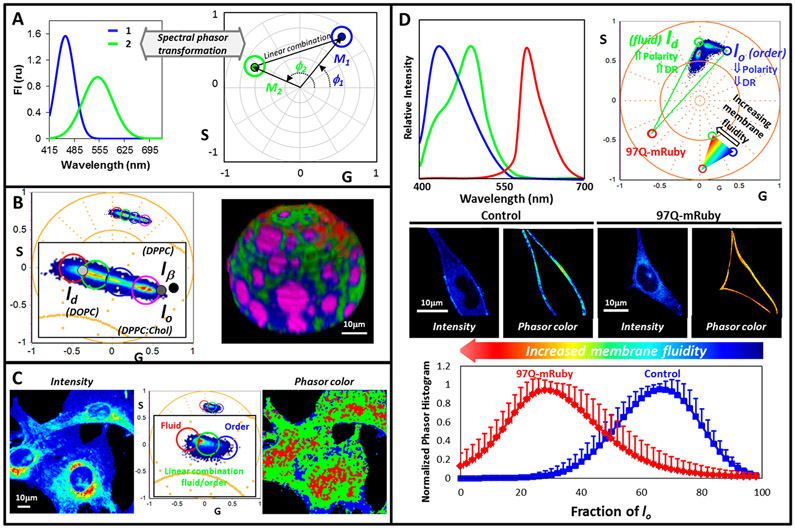
Figure 6.
Spectral phasor analysis. (A) Spectral phasor represents spectra as vectors of length (M) and with phase (ϕ), which are related to the spectrum λmax and width. As the spectrum λmax increases, the phasor moves counterclockwise from position (1,0) 2π (phase increases). Increase on spectrum width moves the phasor closer to the center (modulation decreases). (B) 3D Spectral phasor analysis of a GUV (DOPC/SM/cholesterol; 1:1:1 molar) labeled with LAURDAN. The black and grays dots correspond to the position obtained for solid order (lβ), liquid order (lo), and liquid disordered (ld) phases. Using the cursor selection, the GUV is colored. (C) Phasor plot analysis for live NIH-3T3 cells labeled with LAURDAN. Cursor selection generates the phasor color image on the left. (D) Three-component analysis. LAURDAN and mRuby protein spectra define the LAURDAN trajectory in a three component triangle. The inserted color palette was used to color the intensity images of Hek-293 control and expressing 97Q-mRuby according to the phasor distribution.
Spectral phasor analysis applied to LAURDAN has a very important difference compared to LAURDAN GP analysis. The GP analysis considers a linear GP range to describe the heterogeneities of a system and any component outside of this linear GP range will not be considered. The spectral phasor analysis overcomes this issue by transforming the full spectrum into polar coordinates. The method is model-free and allows for the analysis of different spectral signals and the relationship among them, for instance LAURDAN and autofluorescence.
Figure 6B-D shows the use of this methodology in three selected examples of increasing complexity. The first (Figure 6B) shows a tridimensional spectral phasor analysis of a GUV composed of dioleoylphospatidylcholine, DOPC; sphingomyelin, SM; and cholesterol; 1:1:1 molar, labeled with LAURDAN. Visual examination of the corresponding phasor plot shows a phasor cluster aligned between the straight line defined by the reference phasors corresponding to lo (liquid ordered) and ld (liquid disordered) pure phases. This observation indicates the presence of two molecular environments (the lo and ld) in the GUV and regions where the two components are mixed with different fractions. In a 3D view, it is possible to identify micrometer size liquid order domains in the GUV surrounded by a liquid disorder membrane.69
In natural membranes, the scenario is more complex. First, the concept of lo/ld can be used as a reference point for the phasor location; however, the complexity of cellular membrane is far from the conditions where lo/ld are defined in pure lipids. Second, the spatial resolution of the spectral phasor analysis (which depends on the confocal microscope) does not allow us to observe the nanometric lipid domains postulated for natural membranes (below the PSF). However, phasors from areas in the membrane where nanodomains and fluid regions coexist will locate, in the phasor plot, between the linear combination of “pure” nanodomains and fluid regions and that location in the plot is related to the fractional contribution of each component. The example in Figure 6C shows the spectral image analyzed for NIH-3T3 cells labeled with LAURDAN. Visual inspection of the data, without assuming a model, shows a cluster of phasors organized along a line, which indicates the linear combination between two surrounding environments for LAURDAN (red and blue cursors). Phasor points in the middle of this distribution correspond to pixels with different fractions of order and fluid (green cursor) components. By using the cursor selection tool, it is possible to identify regions with different physical characteristics, for instance blue pixels are associated with ordered regions in the plasma membrane. Quantification of the number of pixels having different order allow to follow the dynamics of the lipid domain under certain stimulus.29
Finally, one very powerful application of the spectral phasor analysis is the three-component analysis, designed to study membrane dynamics and membrane proteins simultaneously. As an example, Figure 6D shows the analysis of LAURDAN in Hek-293 cells expressing the human full-length mutant huntingtin protein (mHTT) with 97 polyQ genetically encoded with mRuby (97polyQ-mRuby).70 In this case, three references were used: two for fluidity, lo–ld trajectory for LAURDAN (first and second component), and one for mRuby protein fluorescence (third component). Using the three-component analysis,71 the fraction of each component can be obtained separately even if two signals are present in a single pixel. In the phasor plot corresponding to the membrane of cells expressing the 97polyQ protein, one can observe a shift in the LAURDAN fluorescence toward the ld component, indicating a relation between the more fluid areas in the membrane and the protein. This experimental design can be applied to relate specific membrane molecular markers with membrane fluidity domains (Figure 6D).
The advantages of spectral phasors to study live cells rely on the versatility of the method which allows the design of unique analyses depending on the complexity of the system.67,72 From the technical point of view, several commercial instruments allow for acquisition of spectral images, and there are different options for software analysis (SimFCS26) and ImageJ routines.27,73
FINAL REMARKS
Having known Gregorio Weber, we are sure that when he synthesized LAURDAN, he visualized its potential for membrane studies. This Account describes advances in the interpretation and use of LAURDAN for detecting membrane fluidity or, more properly, water dipolar relaxation rates around the excited LAURDAN dipole. Using the phasor method, we can build continuous parameters (indices) in a more complete form than offered by the two-state model implicit with the GP concept, which is critical for understanding complex microenvironments such as cells experiencing different conditions or in animals.
ACKNOWLEDGMENTS
G.G. is supported by Agencia Nacional de Investigación y Desarrollo (ANID formerly FONDECYT) through Grants 1160705 and 1200597. L.M. is supported by Comision Sectiorial de Investigación Científica (CSIC) and Agencia Nacional de Investigación e Innovación (ANII), grant I+D–2018 #85 and FCE_3_2018_1_149047 and the Chan Zuckerberg Initiative. D.M.J. is supported in part by NIH Grant R21MH119516. S.A.S. is supported by Grant 1201028 from ANID. E.G. is supported in part by NIH Grant P41-GM103540.
Biography
German Gunther, born in Chile, received his doctorate in Chemistry from the Universidad de Chile. He joined the Facultad de Ciencias Químicas y Farmacéuticas in 1995 and teaches Physical Chemistry. He is presently Associate Professor. His research is focused on characterization of microheterogeneous systems (particularly membranes modified with glycolipids) and on the design and synthesis of fluorescent probes and glycosensitizers.
Leonel Malacrida, born in Uruguay, earned a PhD in Biology at the PEDECIBA; his thesis was recognized by IUPAB for the Latinoamerican Postdegree in Biophysics. He was a postdoc with Enrico Gratton in the Laboratory for Fluorescence Dynamics from 2015 to 2019. Today, Dr. Malacrida is the head of the Advanced Bioimaging Unit in a shared position at the Hospital de Clínicas, Universidad de la República-Uruguay and the Institut Pasteur of Montevideo. His research is related to novel instrumentation and methods in the Biophotonics field to address biomedical questions.
David M. Jameson, born in the United States, received his doctorate in Biochemistry at the University of Illinois at Urbana—Champaign under the supervision of Gregorio Weber. He carried out a postdoctoral period in Paris at the Universidad de Paris-Sud before returning to Gregorio Weber’s laboratory for more postdoctoral research. His first faculty appointment was at UT Southwestern Medical Center. In 1988, he joined the University of Hawaii and is presently Full Professor in the Department of Cell and Molecular Biology. His current research interests concern protein interactions, in vitro and in cells, and he has been working recently on the large GTPase dynamin and on membrane remodeling by the protein Arc/Arg3.1.
Enrico Gratton, born in Italy, received his doctorate in physics from the University of Rome. In 1976, he arrived as a postdoctoral fellow with Gregorio Weber at the University of Illinois at Urbana–Champaign. In 1979, he joined the Physics Department at UIUC and continued a collaboration with Gregorio Weber in fluorescence instrumentation design. In 1986, he was awarded a grant from the NIH to establish the first national facility dedicated to fluorescence spectroscopy: The Laboratory for Fluorescence Dynamics. He moved to the University of California, Irvine in 2006 where he is Full Professor in the Bioengineering Department. His research interests include design of new fluorescence instruments, protein dynamics, and I.R. spectroscopy of biological substances.
Susana Sánchez, born in Chile, received her doctorate in Chemistry from the Universidad Católica de Valparaiso. She came to the University of Illinois at Urbana–Champaign (UIUC) in 1996 to work with Gregorio Weber, and in 1997 she became a postdoctoral fellow in the Laboratory for Fluorescence Dynamics (LFD) directed by Enrico Gratton. In 2001, she started as a Research Scientist and User Coordinator of the LFD. In 2013, she went back to Chile to the Universidad de Concepcion, Facultad de Ciencias Químicas and became an Assistant Professor in 2015. Her research focuses on the topic of membrane heterogeneity.
Footnotes
The authors declare no competing financial interest.
Contributor Information
German Gunther, Facultad de Ciencias Químicas y Farmacéuticas, Universidad de Chile, Santiago 8380492, Chile.
Leonel Malacrida, Advanced Microscopy and Biophotonics Unit, Hospital de Clínicas, Universidad de la República, Montevideo-Uruguay. Advanced Bioimaging Unit, Institut Pasteur Montevideo, 90600 Montevideo, Uruguay.
David M. Jameson, Department of Cell and Molecular Biology, John A. Burns School of Medicine, University of Hawaii, Honolulu, Hawaii 96813, United States
Enrico Gratton, Laboratory for Fluorescence Dynamics, 3210 Natural Sciences II, University of California, Irvine, Irvine, California 92697-2725, United States.
Susana A. Sánchez, Departamento de Polímeros, Facultad de Ciencias Químicas, Universidad de Concepción, Concepción 4070371, Chile.
REFERENCES
- (1). Sanchez SA; Tricerri MA; Gratton E Laurdan generalized polarization fluctuations measures membrane packing micro-heterogeneity in vivo. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 7314–7319. Combination of LAURDAN generalized polarization and fluorescence correlation spectroscopy demonstrates the existence in the membrane of mobile nanometer size higlly packed structures.
- (2). Malacrida L; Gratton E; Jameson DM Model-free methods to study membrane environmental probes: a comparison of the spectral phasor and generalized polarization approaches. Methods Appl. Fluoresc 2015, 3, 047001. A critical comparison of the GP and spectral phasor methods used in conjunction with Laurdan to investigate membrane systems.
- (3). Malacrida L; Gratton E LAURDAN fluorescence and phasor plots reveal the effects of a H2O2 bolus in NIH-3T3 fibroblast membranes dynamics and hydration. Free Radical Biol. Med 2018, 128, 144–156. Presents an application of Laurdan and phasor plots in live cells and demonstrates application to studies of membrane dynamics.
- (4). Castro-Castillo V; Gajardo J; Sandoval-Altamirano C; Gratton E; Sanchez S; Malacrida L; Gunther G CAPRYDAA, an anthracene dye analog to LAURDAN: a comparative study using cuvette and microscopy. J. Mater. Chem. B 2020, 8, 88–99. Presents a new fluorophore, CAPRYDAA, which possesses the environmental sensitivity properties of LAURDAN, but which has the advantage of longer wavelength excitation and higher photostability.
- (5).Israelachvili JN; Marcelja S; Horn RG Physical Principles of Membrane Organization. Q. Rev. Biophys 1980, 13, 121–200. [DOI] [PubMed] [Google Scholar]
- (6).Lombard J Once upon a time the cell membranes: 175 years of cell boundary research. Biol. Direct 2014, 9, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Morigaki K; Tanimoto Y Evolution and development of model membranes for physicochemical and functional studies of the membrane lateral heterogeneity. Biochim. Biophys. Acta, Biomembr 2018, 1860, 2012–2017. [DOI] [PubMed] [Google Scholar]
- (8).Pike LJ Lipid rafts: bringing order to chaos. J. Lipid Res 2003, 44, 655–667. [DOI] [PubMed] [Google Scholar]
- (9).Pike LJ Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res 2006, 47, 1597–1598. [DOI] [PubMed] [Google Scholar]
- (10).Gaus K; Gratton E; Kable EP; Jones AS; Gelissen I; Kritharides L; Jessup W Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 15554–15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Jacobson K; Mouritsen OG; Anderson RG Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol 2007, 9, 7–14. [DOI] [PubMed] [Google Scholar]
- (12).He HT; Marguet D Detecting nanodomains in living cell membrane by fluorescence correlation spectroscopy. Annu. Rev. Phys. Chem 2011, 62, 417–436. [DOI] [PubMed] [Google Scholar]
- (13).Weber G; Farris FJ Synthesis and Spectral Properties of a Hydrophobic Fluorescent Probe: 6-Propionyl-2-(dimethylamino)-naphthalene. Biochemistry 1979, 18, 3075–3078. [DOI] [PubMed] [Google Scholar]
- (14).Shinitzky M; Dianoux AC; Gitler C; Weber G Microviscosity and order in the hydrocarbon region of micelles and membranes determined with fluorescent probes. I. Synthetic micelles. Biochemistry 1971, 10, 2106–2113. [DOI] [PubMed] [Google Scholar]
- (15).Parasassi T; Krasnowska EK; Bagatolli L; Gratton E Laurdan and Prodan as polarity-sensitive fluorescent membrane probes. J. Fluoresc 1998, 8, 365–373. [Google Scholar]
- (16).Parasassi T; Conti F; Gratton E Time-Resolved Fluorescence Emission-Spectra of Laurdan in Phospholipid-Vesicles by Multifrequency Phase and Modulation Fluorometry. Cell Mol. Biol 1986, 32, 103–108. [PubMed] [Google Scholar]
- (17).Parasassi T; De Stasio G; d'Ubaldo A; Gratton E Phase Fluctuation in Phospholipid-membranes Revealed by LAURDAN fluorescence. Biophys. J 1990, 57, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Parasassi T; Gratton E; Yu WM; Wilson P; Levi M Two-photon fluorescence microscopy of Laurdan generalized polarization domains in model and natural membranes. Biophys. J 1997, 72, 2413–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sanchez SA; Bagatolli LA; Gratton E; Hazlett TL A two-photon view of an enzyme at work: Crotalus atrox venom PLA(2) interaction with single-lipid and mixed-lipid giant unilamellar vesicles. Biophys. J 2002, 82, 2232–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Tricerri MA; Toledo JD; Sanchez SA; Hazlett TL; Gratton E; Jonas A; Garda HA Visualization and analysis of apolipoprotein A-I interaction with binary phospholipid bilayers. J. Lipid Res 2005, 46, 669–678. [DOI] [PubMed] [Google Scholar]
- (21).Nicolini C; Celli A; Gratton E; Winter R Pressure tuning of the morphology of heterogeneous lipid vesicles: A two-photon-excitation fluorescence microscopy study. Biophys. J 2006, 91, 2936–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Bagatolli LA; Gratton E Two-photon fluorescence microscopy observation of shape changes at the phase transition in phospholipid giant unilamellar vesicles. Biophys. J 1999, 77, 2090–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bagatolli LA; Sanchez SA; Hazlett T; Gratton E Giant vesicles, laurdan, and two-photon fluorescence microscopy: Evidence of lipid lateral separation in bilayers. Methods Enzymol. 2003, 360, 481–500. [DOI] [PubMed] [Google Scholar]
- (24).Dietrich C; Bagatolli LA; Volovyk ZN; Thompson NL; Levi M; Jacobson K; Gratton E Lipid rafts reconstituted in model membranes. Biophys. J 2001, 80, 1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Navarro-Lerida I; Pellinen T; Sanchez SA; Guadamillas MC; Wang Y; Mirtti T; Calvo E; Del Pozo MA Rac1 nucleocytoplasmic shuttling drives nuclear shape changes and tumor invasion. Dev. Cell 2015, 32, 318–334. [DOI] [PubMed] [Google Scholar]
- (26). www.lfd.uci.edu/globals/. [Google Scholar]
- (27). www.optinav.info/Generalized_Polarization_Analysis.htm. [Google Scholar]
- (28).Sanchez SA; Tricerri MA; Ossato G; Gratton E Lipid packing determines protein-membrane interactions: Challenges for apolipoprotein A-I and high density lipoproteins. Biochim. Biophys. Acta, Biomembr 2010, 1798, 1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Gunther G; Herlax V; Lillo MP; Sandoval-Altamirano C; Belmar LN; Sanchez SA Study of rabbit erythrocytes membrane solubilization by sucrose monomyristate using laurdan and phasor analysis. Colloids Surf., B 2018, 161, 375–385. [DOI] [PubMed] [Google Scholar]
- (30).Jaureguiberry MS; Tricerri MA; Sanchez SA; Finarelli GS; Montanaro MA; Prieto ED; Rimoldi OJ Role of plasma membrane lipid composition on cellular homeostasis: learning from cell line models expressing fatty acid desaturases. Acta Biochim. Biophys. Sin 2014, 46, 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Navarro-Lerida I; Sanchez-Perales S; Calvo M; Rentero C; Zheng Y; Enrich C; Del Pozo MA A palmitoylation switch mechanism regulates Rac1 function and membrane organization. EMBO J. 2012, 31, 534–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Fernandez-Perez EJ; Sepulveda FJ; Peters C; Bascunan D; Riffo-Lepe NO; Gonzalez-Sanmiguel J; Sanchez SA; Peoples RW; Vicente B; Aguayo LG Effect of Cholesterol on Membrane Fluidity and Association of Abeta Oligomers and Subsequent Neuronal Damage: A Double-Edged Sword. Front. Aging Neurosci 2018, 10, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Owen DM; Gaus K Optimized Time-Gated Generalized Polarization Imaging of Laurdan and di-4-ANEPPDHQ for Membrane Order Image Contrast Enhancement. Microsc. Res. Tech 2009, 73, 618–622. [DOI] [PubMed] [Google Scholar]
- (34).Sanchez SA; Tricerri MA; Gratton E Interaction of high density lipoprotein particles with membranes containing cholesterol. J. Lipid Res 2007, 48, 1689–1700. [DOI] [PubMed] [Google Scholar]
- (35).Celli A; Beretta S; Gratton E Phase fluctuations on the micron-submicron scale in GUVs composed of a binary lipid mixture. Biophys. J 2008, 94, 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Celli A; Gratton E Dynamics of lipid domain formation: fluctuation analysis. Biochim. Biophys. Acta, Biomembr 2010, 1798, 1368–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Magde D; Elson E; Webb WW Thermodynamic Fluctuations in a Reacting System-Measurement by Fluorescence Correlation Spectrsocopy. Phys. Rev. Lett 1972, 29, 705–708. [Google Scholar]
- (38).Berland KM; So PTC; Gratton E Two-Photon fluorescence correlation spectroscopy: method and applications to the intracellular environment. Biophys. J 1995, 68, 694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Denk W; Strickler JH; Webb WW Two-photon laser scanning fluorescence microscopy. Science 1990, 248, 73–76. [DOI] [PubMed] [Google Scholar]
- (40).Jameson DM Introduction to Fluorescence; CRC Press: Boca Raton, FL, 2014. [Google Scholar]
- (41).Thompson NL Fluorescence Correlation Spectroscopy; Plenum Press: New York, 1991; pp 337–378. [Google Scholar]
- (42).Chen Y; Muller JD; Berland KM; Gratton E Fluorescence fluctuation spectroscopy. Methods 1999, 19, 234–352. [DOI] [PubMed] [Google Scholar]
- (43).Muller JD; Chen Y; Gratton E Resolving heterogeneity on the single molecular level with the photon-counting histogram. Biophys. J 2000, 78, 474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Chen Y; Muller JD; Ruan Q; Gratton E Molecular brightness characterization of EGFP in vivo by fluorescence fluctuation spectroscopy. Biophys. J 2002, 82, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Gunther G; Jameson DM; Aguilar J; Sanchez SA Scanning fluorescence correlation spectroscopy comes full circle. Methods 2018, 140, 52–61. [DOI] [PubMed] [Google Scholar]
- (46).Digman MA; Brown CM; Sengupta P; Wiseman PW; Horwitz AR; Gratton E Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys. J 2005, 89, 1317–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Jameson DM; Gratton E; Hall RD The Measurement and Analysis of Heterogeneous Emissions by Multifrequency Phase and Modulation Fluorometry. Appl. Spectrosc. Rev 1984, 20, 55–106. [DOI] [PubMed] [Google Scholar]
- (48).Clayton AH; Hanley QS; Verveer PJ Graphical representation and multicomponent analysis of single-frequency fluorescence lifetime imaging microscopy data. J. Microsc 2004, 213, 1–5. [DOI] [PubMed] [Google Scholar]
- (49).Hanley QS; Clayton AH AB-plot assisted determination of fluorophore mixtures in a fluorescence lifetime microscope using spectra or quenchers. J. Microsc 2005, 218, 62–67. [DOI] [PubMed] [Google Scholar]
- (50).Redford GI; Clegg RM Polar plot representation for frequency-domain analysis of fluorescence lifetimes. J. Fluoresc 2005, 15, 805–815. [DOI] [PubMed] [Google Scholar]
- (51).Caiolfa VR; Zamai M; Malengo G; Andolfo A; Madsen CD; Sutin J; Digman MA; Gratton E; Blasi F; Sidenius N Monomer dimer dynamics and distribution of GPI-anchored uPAR are determined by cell surface protein assemblies. J. Cell Biol 2007, 179, 1067–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Digman MA; Caiolfa VR; Zamai M; Gratton E The phasor approach to fluorescence lifetime imaging analysis. Biophys. J 2008, 94, L14–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Weber G Resolution of the fluorescence lifetimes in a heterogeneous system by phase and modulation measurements. J. Phys. Chem 1981, 85, 949–953. [Google Scholar]
- (54).Ranjit S; Malacrida L; Gratton E Differences between FLIM phasor analyses for data collected with the Becker and Hickl SPC830 card and with the FLIMbox card. Microsc. Res. Tech 2018, 81, 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Celli A; Sanchez S; Behne M; Hazlett T; Gratton E; Mauro T The epidermal Ca(2+) gradient: Measurement using the phasor representation of fluorescent lifetime imaging. Biophys. J 2010, 98, 911–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Sanchez S; Bakas L; Gratton E; Herlax V Alpha hemolysin induces an increase of erythrocytes calcium: a FLIM 2-photon phasor analysis approach. PLoS One 2011, 6, e21127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).James NG; Ross JA; Stefl M; Jameson DM Applications of phasor plots to in vitro protein studies. Anal. Biochem 2011, 410, 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Chen LC; Lloyd WR 3rd; Chang CW; Sud D; Mycek MA Fluorescence lifetime imaging microscopy for quantitative biological imaging. Methods Cell Biol. 2013, 114, 457–488. [DOI] [PubMed] [Google Scholar]
- (59).Ranjit S; Malacrida L; Jameson DM; Gratton E Fit-free analysis of fluorescence lifetime imaging data using the phasor approach. Nat. Protoc 2018, 13, 1979–2004. [DOI] [PubMed] [Google Scholar]
- (60).Golfetto O; Hinde E; Gratton E Laurdan fluorescence lifetime discriminates cholesterol content from changes in fluidity in living cell membranes. Biophys. J 2013, 104, 1238–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Harris FM; Best KB; Bell JD Use of laurdan fluorescence intensity and polarization to distinguish between changes in membrane fluidity and phospholipid order. Biochim. Biophys. Acta, Biomembr 2002, 1565, 123–128. [DOI] [PubMed] [Google Scholar]
- (62).Bonaventura G; Barcellona ML; Golfetto O; Nourse JL; Flanagan LA; Gratton E Laurdan Monitors Different Lipids Content in Eukaryotic Membrane During Embryonic Neural Development. Cell Biochem. Biophys 2014, 70, 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Gaus K; Zech T; Harder T Visualizing membrane microdomains by Laurdan 2-photon microscopy (Review). Mol. Membr. Biol 2006, 23, 41–48. [DOI] [PubMed] [Google Scholar]
- (64).Malacrida L; Jameson DM; Gratton E A multidimensional phasor approach reveals LAURDAN photophysics in NIH-3T3 cell membranes. Sci. Rep 2017, 7, 9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Golfetto O; Hinde E; Gratton E The Laurdan spectral phasor method to explore membrane micro-heterogeneity and lipid domains in live cells. Methods Mol. Biol 2015, 1232, 273–290. [DOI] [PubMed] [Google Scholar]
- (66).Vest R; Wallis R; Jensen LB; Haws AC; Callister J; Brimhall B; Judd AM; Bell JD Use of steady-state laurdan fluorescence to detect changes in liquid ordered phases in human erythrocyte membranes. J. Membr. Biol 2006, 211, 15–25. [DOI] [PubMed] [Google Scholar]
- (67).Malacrida L; Astrada S; Briva A; Bollati-Fogolin M; Gratton E; Bagatolli LA Spectral phasor analysis of LAURDAN fluorescence in live A549 lung cells to study the hydration and time evolution of intracellular lamellar body-like structures. Biochim. Biophys. Acta, Biomembr 2016, 1858, 2625–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Fereidouni F; Bader AN; Gerritsen HC Spectral phasor analysis allows rapid and reliable unmixing of fluorescence microscopy spectral images. Opt. Express 2012, 20, 12729–12741. [DOI] [PubMed] [Google Scholar]
- (69).Malacrida L Surfactante pulmonar durante la lesión pulmonar aguda:desde la fisiopatología a los aspectos biofísicos de su disfunción. Ph.D. Thesis, Universidad de la República, Uruguay, 2014. [Google Scholar]
- (70).Sameni S; Malacrida L; Tan Z; Digman MA Alteration in Fluidity of Cell Plasma Membrane in Huntington Disease Revealed by Spectral Phasor Analysis. Sci. Rep 2018, 8, 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Ranjit S; Dvornikov A; Dobrinskikh E; Wang X; Luo Y; Levi M; Gratton E Measuring the effect of a Western diet on liver tissue architecture by FLIM autofluorescence and harmonic generation microscopy. Biomed. Opt. Express 2017, 8, 3143–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Sena F; Sotelo-Silveira M; Astrada S; Botella MA; Malacrida L; Borsani O Spectral phasor analysis reveals altered membrane order and function of root hair cells in Arabidopsis dry2/sqe1–5 drought hypersensitive mutant. Plant Physiol. Biochem 2017, 119, 224–231. [DOI] [PubMed] [Google Scholar]
- (73).Di Giacinto F; De Angelis C; De Spirito M; Maulucci G Quantitative imaging of membrane micropolarity in living cells and tissues by spectral phasors analysis. MethodsX 2018, 5, 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]