ABSTRACT
The neonatal body provides a range of potential habitats, such as the gut, for microbes. These sites eventually harbor microbial communities (microbiotas). A “complete” (adult) gut microbiota is not acquired by the neonate immediately after birth. Rather, the exclusive, milk-based nutrition of the infant encourages the assemblage of a gut microbiota of low diversity, usually dominated by bifidobacterial species. The maternal fecal microbiota is an important source of bacterial species that colonize the gut of infants, at least in the short-term. However, development of the microbiota is influenced by the use of human milk (breast feeding), infant formula, preterm delivery of infants, caesarean delivery, antibiotic administration, family details and other environmental factors. Following the introduction of weaning (complementary) foods, the gut microbiota develops in complexity due to the availability of a diversity of plant glycans in fruits and vegetables. These glycans provide growth substrates for the bacterial families (such as members of the Ruminococcaceae and Lachnospiraceae) that, in due course, will dominate the gut microbiota of the adult. Although current data are often fragmentary and observational, it can be concluded that the nutrition that a child receives in early life is likely to impinge not only on the development of the microbiota at that time but also on the subsequent lifelong, functional relationships between the microbiota and the human host. The purpose of this review, therefore, is to discuss the importance of promoting the assemblage of functionally robust gut microbiotas at appropriate times in early life.
KEYWORDS: early life, gut microbiota, infants
INTRODUCTION
WHY IS THE GUT MICROBIOTA OF EARLY LIFE IMPORTANT?
In adult humans, the gut harbors a complex microbial community that is largely comprised of bacterial species and often referred to as the microbiota (1). The taxonomic composition of the microbiota is individualistic and remains relatively stable over a reasonable period of time (2, 3). Detailed knowledge of the composition of the gut microbiota, as revealed by the study of feces of westerners, is available due to the use of high-throughput DNA sequencing methods and bioinformatic analysis of data (1). In addition, metagenomic studies have revealed the constancy of metabolic capacity of the microbiota between individual humans despite differences in taxonomic content (4). Communities with high α-diversity (numerous kinds of bacteria) are considered desirable because they have functional resilience due to metabolic redundancies (5–8). Thus, loss of a species due to ecological perturbation need not result in a loss of function that might impair normal host-microbiota relationships; this represents a kind of “insurance policy.” Therefore, constancy (robustness) of functional characteristics of microbiotas is desirable (9, 10).
The newborn human does not acquire the gut microbiota as a complete entity. Rather, just as the child develops physically and mentally over a timespan of years, the development of the gut microbiota also proceeds in a longitudinal, more or less predictable manner, mainly associated with changes to the dietary intake of the child (11–21). Although it might seem sensible to implant a complete microbiota from healthy parents in the gut of progeny immediately after birth (hereditary acquisition of microbiota lineages), the nutritional requirements of the infant do not support the establishment of an adult (climax) microbial community. The substrates required for the growth of many members of the Lachnospiraceae and Ruminococcaceae, for example, which are major taxa comprising the adult microbiota, are absent in exclusively milk-fed babies (22, 23).
Interest in the development of the gut microbiota during early life of humans has been stimulated by the desire to ameliorate childhood conditions such as the effects of preterm birth (24–26), allergies, asthma, eczema (27–30), sleep disorder (31), autistic spectrum disorder (32–35), type 1 diabetes (36), malnourishment (37, 38), and obesity (39). Further, as well as assisting in the optimal physiological development of tissues and organs, the development of a functionally resilient microbiota established by good dietary habits in childhood might contribute to better health long term due to lessening the impacts of metabolic diseases in the fifth decade of life and beyond (40–42). Thus, research of the gut microbiota in early life has three worthy goals. These goals are to understand the ecological processes that drive the establishment of bacteria in the infant gut and the impact of the neonatal microbiota on formative processes in the infant and to define the progression that culminates in a robust assemblage of species and associated functions characteristic of the adult microbiota.
THE PIONEERS
Historical studies assumed that the infant in utero was sterile (the sterile womb paradigm) and that first contact with bacteria occurred during passage through the birth canal. Although it makes sense that the infant develops in an environment protected from potential pathogens prior to birth, some studies have challenged this concept and have described the detection of bacterial DNA in amniotic fluid and fetal tissues (skin, lung, thymus, spleen, and gut) (43–47). It has been reasoned that an “amniotic microbiota” might impact on the development of the fetus that is bathed in, and swallows, amniotic fluid in utero. A “placental microbiome” has likewise been reported (48, 49). Although receiving wide publicity, the existence of amniotic/fetal and placental microbiotas has been discounted, mainly on the basis of very low biomass in samples (suggesting contamination) and inadequately controlled PCR procedures used to search for bacterial DNA in samples (45, 50–55). It is noteworthy that bacteria are known to occur in amniotic fluid samples where prelabor rupture of membranes has happened, resulting in microbial contamination (56, 57). The historical derivation of germfree mammals by hysterectomy and hand rearing under sterile conditions provides strong evidence that the mammalian foetus in utero is sterile (58). However, this topic is likely to stimulate much further debate.
Also debatable is the biological importance of bacterial species detected in human milk (59–66). In the more extreme view, the infant might, through suckling, be inoculated with bacteria originating in the mother’s gut microbiota. These bacteria might migrate from the gut lumen, circulating to the mammary glands within blood phagocytes (67). This prospect, too, is counterintuitive to the historical concept of breast milk as a hygienic, nutritive source for baby. Through breastfeeding, the mother provides dependent, refuge-bound offspring with nutrients from maternal reserves. The selective advantage to lactating mothers, who can provide infant nutrition without having found food recently, is substantial when food supplies are uncertain (68). Nipples make possible the direct transfer of milk from the breast to the infant digestive tract. It makes sense to keep this food supply relatively free of interference by bacteria; the microbes will otherwise remove nutrients from milk, might produce metabolites toxic to the infant, produce metabolites with unpleasant taste, or be overt pathogens. Milk expressed from the breast contains bacteria, but these are mainly bacteria characteristic of cutaneous or oral habitats. Staphylococci (especially Staphylococcus epidermidis) and streptococci are consistently the most prevalent and abundant bacteria detected across studies (69–74). Moreover, microbial numbers in human milk are low (usually 1 × 103 to 1 × 105 CFU/ml), suggesting contamination of milk during collection or due to suckling (retrograde inoculation) rather than colonization of the lactiferous ducts and sinuses within the breast (67, 75). Milk obtained before the infant feeds (“foremilk”) is dominated by bacteria characteristic of the maternal skin microbiota, whereas after feeding (“hindmilk”), bacteria characteristic of the oral microbiota of the infant are the most common (76). Thus, there is strong evidence that human milk bacteria are allochthonous species in that they originate in other habitats.
The contribution of the maternal fecal microbiota to the early seeding of the infant gut is well established (77–84). Could it be that the proximity of the opening of the birth canal to the anus is not coincidental? Defecation by mother during childbirth is very common, including at the final “push”; the baby’s head emerges usually in the best fitting orientation with the face looking toward the rectum, and there is a lot of fluid (“the waters”) rinsing the area, making cross-contamination possible. Meta-analysis of fecal metagenomic data from approximately 1,900 familial subjects recruited to nine studies demonstrated the predominant influence of the maternal fecal microbiota in the acquisition of bacterial strains during the first year of life (84). Bifidobacterium, Bacteroides, Parabacteroides, Faecalibacterium, Blautia, and Ruminococcus strains were particularly involved. The fecal microbiotas of adult twins tend to be more similar in composition compared to those of their siblings (4, 85–87). This supports the view that the taxa entering the gut around the time of birth may have long-term effects on gut microbiota composition because the twins are born into the same environment, whereas the birth of siblings is distanced from them in time, space, and circumstance. Genetic influences and very long-term cohabitation effects, of course, cannot be ruled out (87, 88). Further, the fecal microbiota of infants delivered by sterile caesarean section (C-section) is more likely to lack fecal bacteria such as bifidobacteria and Bacteroides and is more likely to contain a diverse variety of oxygen-tolerant taxa, including those usually found on the skin and in the hospital environment (82, 89–98). This observation has been considered to indicate an influence of vaginal bacteria in early colonization but really points to the importance of the anal/birth canal proximity. Microbiological studies with C-section infants are somewhat confounded because the mothers are routinely administered antibiotics prior to delivery of the child (intrapartum antibiotic prophylaxis). “Vaginal seeding” involves the deliberate exposure, immediately after delivery of C-section infants, to vaginal secretions collected from the mother on a gauze swab (99). Based on the examination of a relatively small number of babies, vaginal seeding is reported to “partially restore” the fecal microbiotas of C-section infants (99) but does not produce the dramatic difference in microbiota composition obtained by oral inoculation of C-section infants with preparations of maternal feces (“maternal fecal microbiota transplant”) (100). Moreover, the influence of the vaginal microbiota on the fecal microbiota of even vaginally delivered infants is brief. Typical vaginal bacteria (Gardnerella vaginalis, Atopobium vaginae, Lactobacillus crispatus, and Lactobacillus iners) are detected (at low relative abundance) in infant feces only within the first few days after birth (84). Some clinicians have questioned the safety of vaginal seeding because they fear increased likelihood of transfer of potential pathogens from mother to child in an ill-defined system (101). The compositional differences in microbiotas detected in infants delivered vaginally or by C-section (without vaginal seeding) disappear eventually (microbiota development converges between the two groups), so catch-up in the assembly of a characteristic infant microbiota is possible (102–104). When this happens is difficult to determine accurately because few fecal sampling times prior to 3 months after birth have been included in microbiota studies. It may occur at less than 3 months or sometimes more than 12 months after birth. The importance of these early differences in microbiota composition may be relevant to development of the immune system (asthma and allergies) or metabolic disease (obesity) according to some researchers. These conclusions are based on the relative risk of the specific diseases or conditions in vaginally or C-section delivered people later in life (105, 106).
Antibiotic administration to infants is also likely to contribute to disruption of the development of the gut microbiota. Infants born preterm commonly receive antibiotic treatment as a prophylactic measure (107–113). Of interest is the work of Vatanen et al., who found a differential effect of antibiotic treatment on bifidobacterial species: B. longum and B. breve populations appeared to be relatively unscathed by antibiotic exposure compared to the other common bifidobacterial species detected in the gut microbiota of infants (109). Also of note, the size of bifidobacterial populations in the fecal microbiota is inversely related to the prevalence of antibiotic resistance determinants detected in the microbial community (108).
ASSEMBLY OF THE MICROBIOTA INVOLVES AN ECOLOGICAL SUCCESSION
Regardless of source of inoculating bacteria and other potential confounders, the establishment of a microbiota in the infant gut in the days, weeks, and months following birth is characteristically an ecological (biological) succession. An initial, heterogeneous collection of species is replaced by specific bacterial populations, consistently detected in the majority of infants, which expand or decrease in size over time in a characteristic manner (114). The metabolic activities of the early arrivals lower the oxidation-reduction potential (Eh of meconium, +175 mV; 2-day-old infants, −113 mV) of the digesta and so provide a more suitable environment for the establishment of obligate anaerobes that will eventually dominate the microbiota. The qualitative and quantitative changes characteristic of the ecological succession are regulated by factors described as autogenic (formed within the habitat, such as competition for nutrients and Eh) or allogeneic (influences from outside the habitat, such as growth substrates in the food that is consumed) (114).
Tissier reported, in 1905, the predominance and persistence of Gram-positive bacterial cells in the feces of infants fed human milk exclusively (115). A greater variety of bacterial morphotypes was present in the feces of infants fed cow’s milk. This difference between fecal microbiotas of infants fed human milk or formula (based on ruminant milk) still exists today even though modern formula resembles human milk more closely in nutritional aspects (89, 116–122). In most comparisons, members of the genus Bifidobacterium are abundant in the feces of infants fed exclusively human milk (relatively about 60% of the microbiota on average, but 100% of the microbiota in some infants) whereas the community has a more varied taxonomic composition when infants are exclusively formula-fed (about 40% bifidobacteria) (78, 118, 120, 123–130) (Table 1). Curiously, although fed human milk, some infants have very low relative abundances of bifidobacteria in the feces. Sometimes, none at all are detected even when more than one fecal sample is collected and examined. It is possible that these results are due to lack of sufficient sensitivity of detection methods (including poor choice of PCR primers and DNA extraction methods), in conjunction with fluctuating population levels of bifidobacteria (131, 132). However, there really may be infants who missed exposure to bifidobacteria at a critical time (the “window of opportunity/infectivity”). Normally, however, bifidobacteria predominate in the microbiota of infants during the exclusively milk fed period of life.
TABLE 1.
Examples of differences in fecal microbiotas of infants fed human milk or cow’s milk formula
| Bacterial family | Mean % (SEM)a |
|
|---|---|---|
| Human milk | Cow’s milk formula | |
| Bifidobacteriaceae | 61.36 (6.28) | 40.99 (5.16) |
| Lachnospiraceae | 4.22 (2.65) | 22.11 (4.52) |
| Erysipelotrichaceae | 0.21 (0.15) | 7.99 (2.34) |
| Enterobacteriaceae | 8.22 (2.40) | 4.42 (1.14) |
| Coriobacteriaceae | 6.10 (2.67) | 4.59 (2.20) |
| Streptococcaceae | 4.12 (2.81) | 4.04 (1.46) |
| Clostridiaceae | 2.67 (1.33) | 6.23 (2.80) |
| Enterococcaceae | 0.88 (0.38) | 3.80 (0.83) |
| Bacteroidaceae | 4.93 (1.99) | 0.03 (0.02) |
| Lactobacillaceae | 1.75 (0.69) | 0.07 (0.03) |
| Veillonellaceae | 1.59 (0.81) | 0.26 (0.12) |
| Peptostreptococcaceae | 0.19 (0.10) | 0.94 (0.56) |
| Ruminococcaceae | 0.35 (0.24) | 0.64 (0.42) |
Relative abundances of the 13 most highly represented bacterial families in the feces of 2-month-old infants fed human milk or formula; 30 infants per group (data from reference 120) are presented. Statistically significantly different values are indicated in boldface.
The relative abundance of bifidobacteria begins to decline once weaning (the introduction of solid foods to the diet, complementary feeding) begins, usually at 4 to 6 months of age in western countries. The intake of milk (both human and formula) declines from then on and, concomitantly, an increased amount of plant glycans (dietary fiber) is consumed (20). Thus, growth substrates for bacteria provided by milk are reduced in concentration in the gut, while those associated with “solid” foods increase. The increasing complexity of the diet between 6 and 12 months of age provides a wider range of potential growth substrates for obligately anaerobic bacteria that have degradative and fermentative capacities in relation to dietary fiber (11, 14, 17, 18, 133–135). Therefore, the diversity of bacterial types comprising the microbiota increases because, although population sizes of Bifidobacteriaceae are reduced and Bacteroidaceae remain much the same, those of Lachnospiraceae and Ruminococcaceae are increased. The relatively large Enterobacteriaceae population of early life declines in size as the obligately anaerobic taxa that produce short-chain fatty acids, inhibitory to the enterobacteria, establish and proliferate in the gut (15, 20, 136, 137) (Fig. 1). Stewart et al. (138) provided a comprehensive view of the development of the fecal microbiota of approximately 900 children, at risk of developing type 1 diabetes, which were recruited in various locations in the United States and Europe and sampled monthly from 3 months of age. Three phases of microbiota development could be recognized using 16S rRNA gene and metagenomic sequence analysis: a “developmental” phase (months 3 to 14), a “transitional” phase (months 15 to 30), and a “stable” phase (months 31 to 46). Particular taxa were identified as markers of each phase because of changes in relative abundances over time (for example, bifidobacteria in the developmental phase, Subdoligranulum and Anaerostipes in the transitional phase, and the Eubacterium rectale group in the stable phase). Based on taxonomic criteria, most studies point to the assembly of an adult-type microbiota by 2 years of age, but further research may show that the process continues for an extended time (139–144). Firm conclusions will not be drawn until gaps in the sampling of age groups 3 to 6 years, 6 to 12 years, and 12 to 18 years have been filled (141). Nevertheless, biochemical features of adult feces that reflect bacterial degradation of human secretions (decreased protease activity, increased mucin and bilirubin degradation, and cholesterol reduction to coprostanol) generally appear by age 12 to 24 months (145–150).
FIG 1.
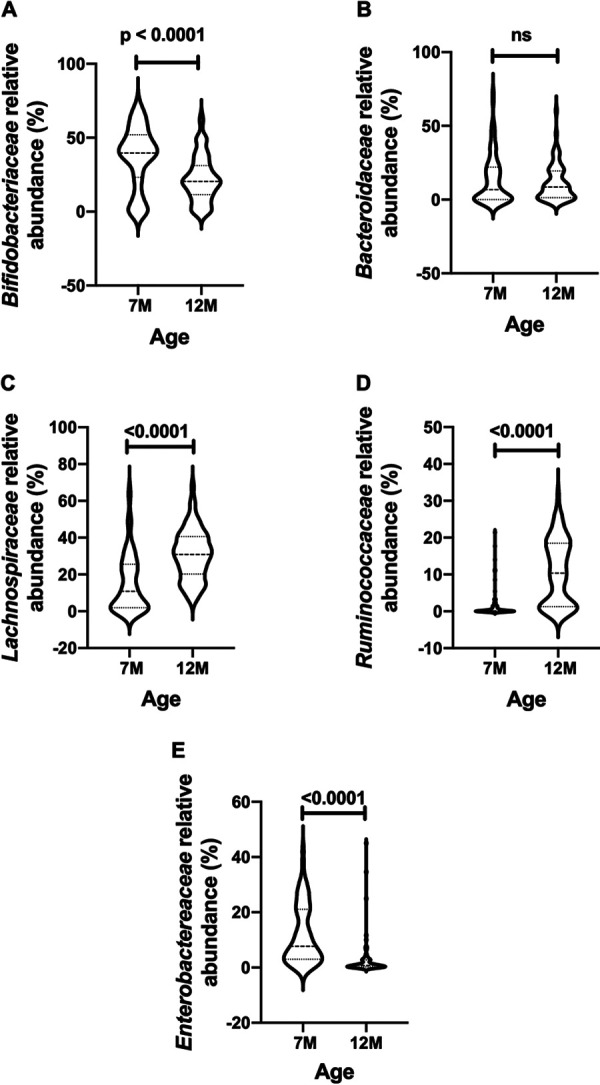
Examples of differences in the relative abundances of selected bacterial families detected in the feces of the same New Zealand infants (n = 74) sampled at 7 compared to 12 months of age. (A) Bifidobacteriaceae; (B) Bacteroidaceae; (C) Lachnospiraceae; (D) Ruminococcaceae; (E) Enterobacteriaceae. The data are from reference 20.
Although a general pattern, outlined above, of assembly of the gut microbiota of infants can be recognized, confounding factors modify the acquisition of the microbiota. For example, ethnic differences in microbiota composition can be detected even prior to the start of complementary feeding in Chinese, Malay, and Indian infants living in Singapore (103). Ethnic differences in microbiota composition point to the potential impact of human genetics, and/or fecal microbiotas of family members, and/or general household environment on the development of the gut microbiota (142, 151, 152). All things considered, the genotype of the host probably has little impact on the composition of the adult microbiota, but the situation in infants has not been investigated comprehensively (153–156). Differences in gut microbiota compositions between ethnic groups prior to weaning might also be due to environmental factors such as dietary preferences of the mother and other household members that could select for particular bacterial strains that would be dispersed to the infants (142, 157, 158). Humans sharing the same environment tend to have microbiotas that are more similar in composition, indicating greater dispersal/acquisition of gut bacteria (159–162).
Environmental ecologists support the “Baas Becking hypothesis” in which it is postulated that all types of microbes occur in all soils, no matter where (“everything is everywhere”), but that the properties of different soils in different locations favor the assemblage of microbial communities of different compositional proportions (“but the environment selects”) (163). Equally, the infant gut may be exposed to bacterial species from a variety of sources, but a characteristic community, of different complexity to that of the mother’s adult microbiota is assembled during the first 3 months of life. This infant community is simple in composition (low alpha-diversity), dominated by the family Bifidobacteriaceae, in contrast to the highly diverse microbiota of an adult. The differences in complexity provide something of a conundrum because, ecologically, high diversity equates to functional resilience. Perhaps the functional requirements of the microbiota of infants are quite simple at this stage of life and so can be provided by a simple but highly networked community? The relatively limited nutritional resources available to bacteria in the habitat could be depleted by a low diversity, yet metabolically highly specialized, community that would ensure competitive exclusion of pathogens. In contrast, in a more complex nutritional environment, a higher diversity microbiota would be required to fill all of the ecological niches (164). With these differences in view, the bowel of exclusively human milk-fed infants is likely to provide an example of environmental selection due to the provision of a “fertile soil” in which a limited collection of specialized bacteria can thrive (165).
HUMAN MILK OLIGOSACCHARIDES PROVIDE A “FERTILE SOIL” FOR SOME BACTERIAL SPECIES
Complex carbohydrates, synthesized in the mammary glands, form the third most abundant solid component of mature human milk (10 to 15 g/liter) (166–168). They are referred to as human milk oligosaccharides (HMOs), of which there are at least 150 types (169, 170). These are constructed from five building blocks: glucose, galactose, N-acetylglucosamine, fucose, and sialic acid. In general, they consist of a lactose core linked to lacto-N-biose or to N-acetyllactosamine. The lactose core can be elongated at the C-3 position of galactose with repeats of lacto-N-biose (Galβ1-3GlcNAc; referred to as type 1 chain) or N-acetyllactosamine (Galβ1-4GlcNAc; type 2 chain) (171). The terminal lactose in the chains may be linked to fucose (α1-2, α1-3, or α1-4 linkages) or sialic acid (N-acetylneuraminic acid [Neu5Ac]; α2-3 or α2-6 linkages) (172, 173). Fucosylation depends in part on the “secretor” status of the mother; the FUT2 gene encodes a fucosyltransferase and results in milk containing α1-2 fucosylated HMOs. These are absent in the milk of “nonsecretor” mothers because they have a FUT2 gene that encodes an inactive protein (174, 175). Elongation of chains can occur at the C-6 position of galactose, resulting in branched molecules. HMOs can be separated from human milk to give an acidic fraction (mostly sialylated oligosaccharides; 12 to 14% of HMOs in milk) and a neutral fraction containing oligosaccharides that may be fucosylated (35 to 50% of HMOs in milk) or nonfucosylated (42 to 55% of HMOs in milk) (176). HMOs that have a type 1 chain linkage (such as lacto-N-tetraose [LNT]) are not degraded by human β-galactosidase in the small bowel, whereas those with a type 2 linkage (such as lacto-N-neotetraose [LNnT]) are susceptible to cleavage (171). However, type 1 chains are predominant in human milk, and it is considered that much of the HMO fraction of milk is not digested in the small bowel but passes to the colon where the oligosaccharides are metabolized by bifidobacterial species (172). Bacteroides thetaiotaomicron, Bacteroides vulgatus, Bacteroides fragilis, and Akkermansia muciniphila are also able to degrade HMOs because HMO structures make them similar to those of mucin oligosaccharides (Lewis antigens) (177, 178). The Bacteroides species are more abundant in the feces of infants fed human milk than cow’s milk formula, indicating that HMOs enrich for these bacteria, too, and divert their degradative activities away from mucins in the mucus layer lining the gut (120, 179, 180). The molecular mixture of HMOs detected in human milk varies between mothers but remains constant, per individual, throughout lactation. However, quantities diminish with time (168, 181–187). Comparison of the amounts and structures of HMOs in human milk of mothers and the feces of their infants provides evidence of transit of HMOs through the gut and degradation of at least some of these structures by bacteria, as well as bacterial utilization of carbohydrates linked to proteins (N-glycans) (185–187).
The common bifidobacteria detected in global studies of infant feces fed human milk include B. longum subspecies infantis, B. longum subsp. longum, B. breve, and B. bifidum. Other species detected in some studies include Bifidobacterium animalis subsp. lactis, Bifidobacterium pseudocatenulatum, Bifidobacterium dentium, Bifidobacterium adolescentis, Bifidobacterium catenulatum, Bifidobacterium kashiwanohense, and Bifidobacterium scardovii (78, 120, 123, 126, 188–192). There is a good biochemical fit between the common bifidobacterial species and HMOs because the bifidobacteria produce fucosidases, hexosaminidase, sialidase, biosidase, and galactosidases that hydrolyze the oligosaccharides (170, 193–212). These enzymes are located at the cell surface of B. bifidum, whereas they are intracellular in B. longum subsp. infantis and subsp. longum and in B. breve (213). Thus, HMOs are internalized by the latter species before hydrolysis occurs, whereas cleavage of the oligosaccharides by B. bifidum results in accumulation of fucose and sialic acid in the extracellular environment because the bacteria do not use these substances for growth (205, 214–216). Fucose and sialic acid are, however, growth substrates for some strains of B. breve, thus providing an opportunity for syntrophy between the two species (214, 215). However, cleavage of fucose or sialic acid from simple HMOs such as 2′-O-fucosyllactose (2FL) or 6′-O-sialyllactose also liberates lactose that is utilized for growth by both B. bifidum and B. breve. Therefore, a seemingly beneficial cross-feeding interaction between the bifidobacterial species might become competitive (215). In vitro, B. bifidum has been shown to adjust to the situation because its abundance is the same in monoculture as in coculture with B. breve in medium containing 2FL. B. bifidum achieves this in coculture by increasing transcription of the fucosidase gene so that more lactose becomes available and by increasing transcription of genes associated with carbohydrate transport and with energy production and conversion (215). B. bifidum also has the capacity to use mucin as a substrate, perhaps explaining why it persists in the feces of infants even when weaned, at which time HMOs are no longer available (217–219). The biochemical pathways associated with the utilization of HMOs by bifidobacterial species has been well reviewed recently by Sakanaka et al. (211). There is more lactose in human milk than the infant can utilize (traces can be detected in infant feces), so this substance, too, is available for microbial growth (132). Much more information about the intricacies of metabolic interaction among bifidobacteria is required in order to understand nutritional regulation of these populations.
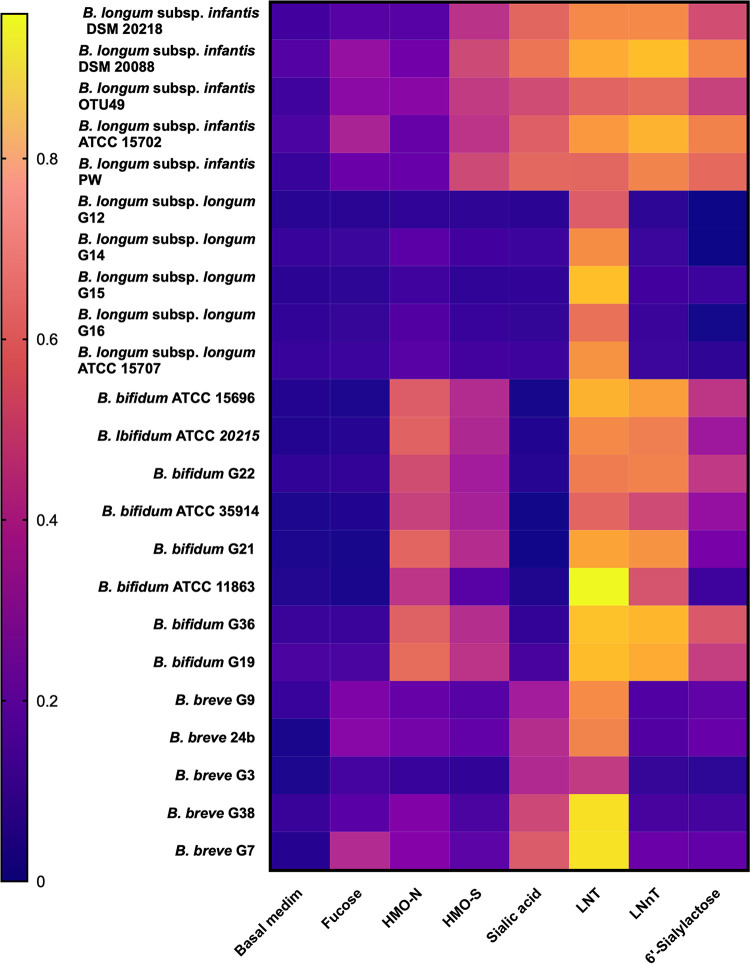
Efficiency of use of HMOs and their component parts for growth is markedly different between bifidobacterial species and can be used to differentiate between them in laboratory culture (Fig. 2). B. longum subsp. infantis has much the best growth on HMO substrates, probably because of the expression of a cluster of genes that form an HMO utilization locus in the genome of the bacteria (206). The capacity of subsp. infantis to preferentially utilize and efficiently grow on the most abundant (especially fucosylated) HMOs (220, 221) should ensure its dominance in the gut of infants fed human milk, yet this seems not always to be the case. Infants in California and in some other regions of the United States, as well as in particular countries, are reported to have low abundances of subsp. infantis. However, data are relatively sparse, and it is difficult to gain a good global perspective (95, 124, 130, 189, 192, 222–227). A paucity of subsp. infantis could be related to feeding practices, or perhaps some environments are not conducive to the transmission of subsp. infantis (224). Nevertheless, bifidobacteria in general are abundant in the gut microbiota of infants, and bacterial genes associated with HMO utilization are more abundant in fecal DNA when infants are receiving human milk than during weaning (109).
FIG 2.
Heat map showing the growth (optical density after 24 h anaerobic incubation) of bifidobacterial species (B. longum subsp. infantis, B. longum subsp. longum, B. bifidum, and B. breve) in media containing HMO fractions (0.2% [wt/vol]) or component molecules (0.2% [wt/vol]) or basal medium (no added carbohydrates). The data are from references 191 and 215.
BIFIDOBACTERIAL FUNCTIONS IN RELATION TO INFANT WELL-BEING
A preponderance of bifidobacteria in the gut may mediate competitive exclusion of bacterial pathogens by virtue of the creation of an acidic environment due to fermentation products acetate and lactate (propanediol in the case of fermentation of fucose by B. longum subsp. infantis and B. breve) (215, 228). Fecal pH of infants fed human milk is lower than that of formula-fed (cow’s milk-based) infants (229–232). Although the amount of historical data are limited, some investigators argue that fecal pH of western infants may have increased temporally due to a paucity of subsp. infantis in the gut microbiota (233). Infants fed formula have a fecal metabolome suggestive of greater metabolism of peptides and amino acids than do human milk-fed infants. This probably reflects the lower oligosaccharide content of formula, higher protein content of formula, different microbiota composition, and slower gut transit time in formula-fed infants compared to those fed human milk (234, 235).
Apart from a competitive exclusion role, bifidobacteria may be important to infant development because they produce folate (vitamin B9), which is a coenzyme or cosubstrate in single-carbon transfers in the synthesis of nucleic acids and metabolism of amino acids (13, 236). An important folate-dependent reaction is the conversion of homocysteine to methionine in the synthesis of S-adenosylmethionine, a methyl donor. Another folate-dependent reaction, the methylation of deoxyuridylate to thymidylate in the formation of DNA, is required for proper cell division (237). The enrichment of genes involved with the de novo biosynthesis of folate (known to be absorbed from the colon) in infants relative to the favored synthesis of another B vitamin, cobalamin, in adults is interesting because it provides circumstantial evidence that folate production in the infant gut is of developmental significance (13, 238).
A baby’s brain is only 15% formed at birth and in 3 years increases in weight from 300 g to 1.2 kg and exceeds the rate of growth of any other organ or body tissue (239). The infant is born with neurons already formed but the synaptic connections between these cells are mostly established and elaborated after birth, causing a large nutritional demand for the biosynthesis of gangliosides. Nutrition of the infant in early life affects developmental processes, including cognition (240–243). While long-chain fatty acids in milk (such as docosahexanoic acid) have been the focus of much of this research, there is tantalizing evidence that sialic acid is also important for optimal brain development and cognition (244, 245). Sialylated oligosaccharides are detectable in the blood of infants and the sialic acid content is higher in the brain of human milk-fed infants relative to formula-fed infants (246–248). Perhaps metabolism of HMOs by bifidobacterial species, particularly the extracellular sialidase activity of B. bifidum, makes a contribution to this phenomenon?
Many clues to the influences of the gut microbiota on the mammalian host have been obtained from comparisons of the biochemical and physiological characteristics of germfree mice with conventional animals (which have a microbiota) (249). Among the differences in these animal groups is the underdevelopment of immune features, including the lesser size of ileocecal lymph nodes and the gamma-globulin fraction of blood of germfree animals. Experiments with ex-germfree mice colonized by a gut microbiota show that the presence of the bacteria programs the immune system toward tolerance of members of the microbiota while still retaining the ability to prevent invasion and dispersion of pathogens in the body (29, 250–255). The maintenance of this homeostasis is dependent on tightly controlled immune responses as well as partitioning of functions (mucosal relative to systemic immunity) (256–258). Modulation of immune function can be achieved for the most part by inoculating adult ex-germfree mice at any age with a gut microbiota (259). However, the ability to restore some cellular defects (for example, negating the mucosal accumulation of invariant natural killer T cells in gut mucosa) that occur in the absence of the microbiota is restricted to a short time interval in early life and thus is age dependent (29, 259). Therefore, the members of the gut microbiota in very early life of humans may have important interactions with the developing immune system. In this context, a variety of molecules associated with bifidobacterial cells are reported to stimulate aspects of the immune system, and the size of bifidobacterial populations has been correlated with the amount of salivary sIgA production and B cell maturation (260–263). Bifidobacterial effects on the expression of the dendritic-cell activation marker CD83 and the production of interleukin-10 (IL-10), as well as response to vaccinations by human infants, have been reported (124, 192). Infants inoculated with the EVC001 strain of B. longum subsp. infantis harbor abundant bifidobacteria in the gut, and there is a negative correlation with fecal cytokines IL-13, IL-17A, IL-21, and IL-33 that indicates a silencing of Th2 and Th17 responses in the gut. This phenomenon may be mediated by a specific metabolite (indole-3-lactic acid) produced by bifidobacteria when they utilize HMOs (264). However, overall, research on this topic seems to lack cohesion and relation to real life. It may indeed be an intractable situation given that few babies have low or absent bifidobacterial populations for comparative purposes, that low bifidobacterial populations are counterbalanced by those of other bacterial species, and that sampling of appropriate immune features is difficult and therefore very limited in neonatal humans.
The maternal production of milk rich in substances that do not obviously contribute to the nutrition of the child is certainly curious and indicates a phenomenon of evolutionary importance. As well as providing “fertilizer” to encourage a predominance of bifidobacteria in the colon that could contribute to competitive exclusion of pathogens in the gut and assist infant nutrition as indicated above, HMOs probably also contribute to host defense through decoy functions in that they resemble receptors in the gut mucosa to which pathogens, including viruses, may bind. “Mopping up” pathogens in the gut lumen with HMOs would minimize opportunity for mucosal adherence and infection to occur. Examples of possible decoy roles apply to the bacterial pathogens Campylobacter jejuni, enteropathogenic Escherichia coli, viruses such as norovirus, and the protozoan Entamoeba histolytica (265–270). However, some caution is required because G10P[11] rotavirus infection of neonates has been reported to be facilitated by the presence of HMOs (271). In vitro studies indicate inhibitory effects of HMOs on some bacteria and promotion of epithelial cell integrity (272–277). These effects may be specific to particular HMOs (268).
THE RESEARCH PATH AHEAD
Human milk for human infants in the months following birth is clearly the desired option to satisfy the nutritional requirements of the child. Exclusive breast milk feeding during the first 6 months of life and partially breast milk-feeding during weaning, is recommended by the World Health Organization, as well as by professional medical associations and agencies (278). Maternal antibodies in milk, iron-sequestering molecules, and molecular decoys help provide protection of the newborn infant from infections (279, 280). For example, data gathered from about 20,000 mother/infant pairs at a time when antibiotics were not available in pediatrics (1930s), show that infants fed totally “artificially” demonstrated 3-fold-higher morbidity and 7-fold-higher mortality from gastrointestinal disease than infants fed human milk. Partially artificially fed infants had 2.5 times more morbidity and 3 times higher mortality than infants fed human milk (279, 281). HMOs are probably molecular decoys allied to protection from infection but collaterally encourage the development of an abundant bifidobacterial population in the gut microbiota during the period of exclusive human milk feeding. The bifidobacterial population commonly contains several species that have varying abilities with respect to utilization of HMOs. The relative efficiency of utilization of HMOs by different bifidobacterial species has been studied, but the interactive relationships between species and between bifidobacterial and other gut inhabitants has only been studied superficially. There is a need, therefore, to define how a stable (robust) microbiota is maintained during the first months of life, probably by means of cooccurrence or metabolic networks (282, 283). Once identified, the trophic networks could be established using in vitro systems, probably including simple chemostats (215) and tested for resilience when nutritional factors are altered.
The early contribution of bacterial strains from maternal feces to infants is likely to be very important, but further research is needed to establish specific effects resulting from this early seeding. Indeed, murine studies using antibiotic-treated and gnotobiotic animals suggest that, even before birth, the maternal gut microbiota influences the concentrations of numerous molecules in maternal serum and hence in fetal brain (284). Thus, the maternal microbiota metabolism may influence neural development even in utero! An exciting prospect is therefore the study of the influence of maternal microbiota pre- and postpartum to delineate a potentially critical and long-term relationship that goes beyond the usual mother-child nurture.
Modification of infant formula will continue to be made so as to increase the oligosaccharide content and provide “probiotic” bacteria that might function in the gut ecosystem. Space limitation does not allow substantial review of these two topics, but examples are provided with comments in Table 2. The nascent use of synthetic HMOs, 2′-O-fucosyllactose (2FL) and lacto-N-neotetraose (LNnT) as additives for infant formula is of considerable interest (325–328). 2FL is the major HMO in milk from secretor mothers, comprising almost 30% of all HMOs present (327). As discussed above, the enrichment of bifidobacteria in the infant gut may not be the sole evolutionary function of HMOs and galacto- and fructo-oligosaccharides (GOS/FOS) are only able to provide growth substrates for bacteria. Thus, the inclusion of synthetic HMOs in formula may be an important advance. To date, trials with infants indicate that formula supplemented with 2FL/LNnT is safe for ingestion by neonates (329, 330), but the bifidobacterium-stimulating effect is likely to be species-specific because some bifidobacteria (for example, strains of B. breve) cannot metabolize these HMOs (Fig. 2) (191, 215). The few microbiological investigations of infants fed supplemented (synthetic HMOs) relative to nonsupplemented formula are somewhat unsatisfactory because the bifidobacterial data are superficial and the numbers of infants examined are small (328). The use of LNnT in formula is curious because it is susceptible to cleavage by human β-galactosidase (171). Caution about reliance on the addition of single HMOs is suggested because different developmental effects of human milk in the infant might relate to specific HMOs. Certainly, the addition of one or two HMOs to formula does not replicate the features of human milk where a diversity of HMO structures is a characteristic feature.
TABLE 2.
Examples of probiotic, prebiotic, and synbiotic approaches to influencing the assemblage of gut microbiota in infants
| Approach | Organism | Description |
|---|---|---|
| Probiotics | L. rhamnosus | In separate studies, Lactobacillus rhamnosus strain GG or strain HN001 (DR20) was administered to mothers whose children were at high risk of developing allergies. The probiotic was taken by mothers before the baby was born and was administered postnatally to the infants for 6 months. The occurrence of atopic eczema at 2 years of age was recorded as the main point of comparison between placebo and test groups. Atopic eczema was diagnosed in 50% fewer probiotic-administered infants compared to controls (285, 286). Strain HN001 was shown to be present in about 60 to 70% of the infants in the test group (5% in the placebo group) when feces were tested on a single occasion during the course of the study (286). Thus, efficacy could be linked to the passage of the probiotic strain through the gut. Probiotic trials like this do not relate directly to the natural situation where lactobacilli, although detectable, are present at low relative abundances in the gut microbiota compared to values associated with the common bifidobacterial species (120). Nevertheless, there has been much pediatric interest in this topic, and there is an extensive review literature regarding the impact of the administration of probiotic products on the prevalence of allergies and atopy in children (287–293). In general, clinical views of the effectiveness of these products are rather mixed. |
| B. infantis (presumbaly B. infantis subsp. infantis; B. bifidum included in later versions of this product) and Lactobacillus acidophilus or B. breve M-16V | Premature, very-low-birthweight infants (<1,500 g) are highly susceptible to necrotizing enterocolitis (NEC) wherein the infants show poor feeding and spilling of gastric contents (which may be bile stained), abdominal distension, and blood in feces. Necrotic inflammation of the intestine may lead to perforation and bacterial infection and, potentially, NEC has high mortality (294). Reduction in historical rates of NEC have been reported when administration of these probiotics to infants in neonatal intensive care has been introduced as a routine practice (295–300). | |
| B. animalis subsp. lactis BB-12 | Administration of formula containing B. animalis subsp. lactis BB-12 augments bifidobacterial abundance in the feces of preterm infants, and enterobacterial proportions are reduced (302, 303). | |
| Synbiotic | B. breve M-16V, galacto- and fructo-oligosaccharides | Feeding formula containing B. breve M-16V in combination with short-chain galacto-oligosaccharide and long-chain fructo-oligosaccharide augments bifidobacterial proportions in the microbiota of C-section infants. Both the probiotic strain and naturally colonizing B. breve strains were boosted in the infant gut. Fecal pH was lowered compared to controls due to increased acetate production by the microbiota, and there was a reduction in the proportion of Enterobacteriaceae (301). |
| Prebiotics | Galacto- and fructo-oligosaccharides | Efforts have been made to increase the oligosaccharide content of formula by the addition of naturally occurring or synthetic oligosaccharides. Galacto- and fructo-oligosaccharides (GOS and FOS, respectively) are common prebiotics used for this purpose (304–308). Commercially available GOS contain oligosaccharides composed of galactose residues that are variously linked β-D-1,2, β-D-1,3, β-D-1,4, and β-D-1,6 and range in degrees of polymerization (DP) from 2 to 5. Each oligosaccharide is terminated with a glucose residue. FOS contain two series of β-D-2,1-linked fructo-oligosaccharides, one of which contains a terminal glucose (GF series, with DPs of 2 to 6) and one which does not (F series, with DPs of 2 to 6) (309–311). As such, GOS and FOS do not chemically resemble HMOs, but various bifidobacterial species are known to utilize GOS and FOS for growth in laboratory experiments (312–314). Inclusion of the substrates in formula tends to result in somewhat augmented total bifidobacterial populations in infant feces relative to controls (315–322). In coculture experiments, B. breve has demonstrated the greatest trophic potential with respect to GOS/FOS to outcompete the subspecies of B. longum (323). Unfortunately, the in vivo effect of GOS/FOS dietary supplementation on the proportions of the different species within bifidobacterial populations has not been included in trials in which the outcome of feeding supplemented formula has been compared to that of nonsupplemented formula. This may be due to past difficulties in differentiating between bifidobacterial species using DNA-based methodologies (224, 324). |
In-depth knowledge of the immunological influences of bifidobacterial species in very early life requires more research. Specifically, we need to know whether they are the critical bacterial species that affect the aspects of immune development that, as mentioned above, cannot be replicated in older gnotobiotic animals. Since most human infants are naturally exposed to bifidobacteria, much of this work will probably require comparisons of germfree and gnotobiotic animals exposed to bifidobacteria and fed human milk. However, comparisons of the effects of infant formulas with or without bifidobacterial components may also be useful in this research. The success of this approach will depend on which bifidobacterial species are included in the formula and must also take into consideration the biochemical differences that exist between human milk and formulas. In other words, the background nutritional environment may confound extrapolation of results from formula-fed infants to those fed human milk.
Although the bifidobacteria have been a major focus of the gut microbiota in early life throughout microbiological history, the predominance of these bacteria in the gut is a transient phenomenon and is largely over by just a few months postpartum. The commencement of weaning and the development of a highly diverse microbiota that provides a platform for the rest of life is equally important. Knowledge of how microbial communities with high functional resilience are constructed is required, with a particular need to understand how complex microbiotas promote health through involvement in molecular signaling in human physiology (331–334). It is relevant to this topic that there is a reduced prevalence of allergies in children whose microbiota is characterized by the end of the first year of life by an enrichment of propionate- and butyrate-producing bacteria (335, 336). The main drivers of the development of the gut microbiota after weaning and throughout the rest of life are the complex plant glycans contained in the diet (135). Therefore, encouraging the development of a functionally resilient microbiota in the first year or two of life requires that the infant be exposed to a wide spectrum of tastes and textures of the diverse foods provided by Earth’s bounty. An intake of a variety of whole foods, especially those containing complex plant polysaccharides, will promote microbiotas of high functional resilience (135). Traditionally, parents have been encouraged to start spoon-feeding their infant puréed foods from around 6 months of age, progressing to mashed and then chopped foods in the hope that they will be eating family foods by about 12 months of age. An alternative method of complementary feeding, known as baby-led weaning, encourages babies to feed themselves whole pieces of food from the family meal from 6 months of age, instead of being offered “baby food” (337–341). As a result, infants following this procedure are more likely to eat the same foods as the rest of the family than their baby-food counterparts. Because the introduction of “solid” foods is a major determinant of the development of the gut microbiota, it is possible that the more rapid transition to the family diet observed in baby-led approaches could beneficially influence the assembly of the gut microbiota in these children. However, recent microbiota research in baby-led weaning, using mediation analysis, indicates that adequate intakes of “fruit and vegetables” and “dietary fiber” are critical for development of a diverse microbiota and that this may not be sustained by family foods if they are not soundly based on good nutrition for the whole family (20). Care must also be taken that the infants do not bite off more than they can chew; choking is not a desirable outcome (342).
Despite the large volume of research that has been directed to studying the bacterial taxa comprising gut microbiotas in early life, there is yet little detail about the molecular biology of microbe-human interactions. It seems quite likely that future breakthroughs in understanding the early life relationships between the gut microbiota and humans will arise from genetical (including gene transcription), biochemical, neurological, and physiological (including immunology) studies rather than predominantly taxonomic, compositional investigations. However, since much of this work will probably have to be accomplished using murine models, clear translations to human biology may be difficult to perceive given the different nutritional, gut microbiota, and developmental trajectories that occur between humans and mice. It is a difficult task worth pursuing because the early life events associated with the assemblage of the gut microbiota may impact the human host throughout its remaining life span!
ACKNOWLEDGMENT
Gerald W. Tannock is Professor Emeritus of the University of Otago, Dunedin, New Zealand, and is hosted by the Department of Microbiology and Immunology.
Contributor Information
Gerald W. Tannock, Email: gerald.tannock@otago.ac.nz.
Danilo Ercolini, University of Naples Federico II.
REFERENCES
- 1.Franzosa EA, Hsu T, Sirota-Madi A, Shafquat A, Abu-Ali G, Morgan XC, Huttenhower C. 2015. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol 13:360–372. 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konopka A. 2009. What is microbial community ecology? ISME J 3:1223–1230. 10.1038/ismej.2009.88. [DOI] [PubMed] [Google Scholar]
- 6.Mori AS, Furukawa T, Sasaki T. 2013. Response diversity determines the resilience of ecosystems to environmental change. Biol Rev Camb Philos Soc 88:349–364. 10.1111/brv.12004. [DOI] [PubMed] [Google Scholar]
- 7.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcón Ó, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G, Dominguez-Bello MG. 2015. The microbiome of uncontacted Amerindians. Sci Adv 1:e1500183. 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyte KZ, Schluter J, Foster KR. 2015. The ecology of the microbiome: networks, competition, and stability. Science 350:663–666. 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 9.Willis KJ, Jeffers ES, Tovar C. 2018. What makes a terrestrial ecosystem resilient? Science 359:988–989. 10.1126/science.aar5439. [DOI] [PubMed] [Google Scholar]
- 10.Sheykhali S, Fernández-Gracia J, Traveset A, Ziegler M, Voolstra CR, Duarte CM, Eguíluz VM. 2020. Robustness to extinction and plasticity derived from mutualistic bipartite ecological networks. Sci Rep 10:9783. 10.1038/s41598-020-66131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fallani M, Amarri S, Uusijarvi A, Adam R, Khanna S, Aguilera M, Gil A, Vieites JM, Norin E, Young D, Scott JA, Doré J, Edwards CA, The Infabio Team. 2011. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology (Reading) 157:1385–1392. 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 12.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA 108(Suppl):4578–4585. 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergström A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, Mølgaard C, Michaelsen KF, Licht TR. 2014. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol 80:2889–2900. 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J, Jun W. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:690–703. 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Laursen MF, Andersen LB, Michaelsen KF, Mølgaard C, Trolle E, Bahl MI, Licht TR. 2016. Infant gut microbiota development is driven by transition to family foods independent of maternal obesity. mSphere 1:e00069-15. 10.1128/mSphere.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laursen MF, Bahl MI, Michaelsen KF, Licht TR. 2017. First foods and gut microbes. Front Microbiol 8:356. 10.3389/fmicb.2017.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qasem W, Azad MB, Hossain Z, Azad E, Jorgensen S, Castillo San Juan S, Cai C, Khafipour E, Beta T, Roberts LJ, 2nd, Friel J. 2017. Assessment of complementary feeding of Canadian infants: effects on microbiome and oxidative stress, a randomized controlled trial. BMC Pediatr 17:54. 10.1186/s12887-017-0805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galazzo G, van Best N, Bervoets L, Dapaah IO, Savelkoul PH, Hornef MW, Lau S, Hamelmann E, Penders J, GI-MDH Consortium. 2020. Development of the microbiota and associations with birth mode, diet, and atopic disorders in a longitudinal analysis of stool samples, collected from infancy through early childhood. Gastroenterology 158:1584–1596. 10.1053/j.gastro.2020.01.024. [DOI] [PubMed] [Google Scholar]
- 20.Leong C, Haszard JJ, Lawley B, Otal A, Taylor RW, Szymlek-Gay EA, Fleming EA, Daniels L, Fangupo LJ, Tannock GW, Heath AM. 2018. Mediation analysis as a means of identifying dietary components that differentially affect the fecal microbiota of infants weaned by modified baby-led and traditional approaches. Appl Environ Microbiol 84:e00914-18. 10.1128/AEM.00914-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leong C, Haszard JJ, Heath AM, Tannock GW, Lawley B, Cameron SL, Szymlek-Gay EA, Gray AR, Taylor BJ, Galland BC, Lawrence JA, Otal A, Hughes A, Taylor RW. 2020. Using compositional principal component analysis to describe children’s gut microbiota in relation to diet and body composition. Am J Clin Nutr 111:70–78. 10.1093/ajcn/nqz270.. [DOI] [PubMed] [Google Scholar]
- 22.Biddle A, Stewart L, Blanchard J, Leschine S. 2013. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 5:627–640. 10.3390/d5030627. [DOI] [Google Scholar]
- 23.Stackebrandt E. 2014. The family Lachnospiraceae, p 197–201. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes. Springer, Berlin, Germany. [Google Scholar]
- 24.Butel MJ, Suau A, Campeotto F, Magne F, Aires J, Ferraris L, Kalach N, Leroux B, Dupont C. 2007. Conditions of bifidobacterial colonization in preterm infants: a prospective analysis. J Pediatr Gastroenterol Nutr 44:577–582. 10.1097/MPG.0b013e3180406b20. [DOI] [PubMed] [Google Scholar]
- 25.Stewart CJ, Embleton ND, Marrs EC, Smith DP, Nelson A, Abdulkadir B, Skeath T, Petrosino JF, Perry JD, Berrington JE, Cummings SP. 2016. Temporal bacterial and metabolic development of the preterm gut reveals specific signatures in health and disease. Microbiome 4:67. 10.1186/s40168-016-0216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jayasinghe TN, Vatanen T, Chiavaroli V, Jayan S, McKenzie EJ, Adriaenssens E, Derraik JGB, Ekblad C, Schierding W, Battin MR, Thorstensen EB, Cameron-Smith D, Forbes-Blom E, Hofman PL, Roy NC, Tannock GW, Vickers MH, Cutfield WS, O’Sullivan JM. 2020. Differences in compositions of gut bacterial populations and bacteriophages in 5 to 11 year olds born preterm compared to full term. Front Cell Infect Microbiol 10:276. 10.3389/fcimb.2020.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stsepetova J, Sepp E, Julge K, Vaughan E, Mikelsaar M, de Vos WM. 2007. Molecularly assessed shifts of Bifidobacterium ssp. and less diverse microbial communities are characteristic of 5-year-old allergic children. FEMS Immunol Med Microbiol 51:260–269. 10.1111/j.1574-695X.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- 28.Storrø O, Øien T, Langsrud Ø, Rudi K, Dotterud C, Johnsen R. 2011. Temporal variations in early gut microbial colonization are associated with allergen-specific immunoglobulin E but not atopic eczema at 2 years of age. Clin Exp Allergy 41:1545–1554. 10.1111/j.1365-2222.2011.03817.x. [DOI] [PubMed] [Google Scholar]
- 29.El Aidy S, Hooiveld G, Tremaroli V, Bäckhed F, Kleerebezem M. 2013. The gut microbiota and mucosal homeostasis: colonized at birth or at adulthood, does it matter? Gut Microbes 4:118–124. 10.4161/gmic.23362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, Mohn WW, Turvey SE, Finlay BB, CHILD Study Investigators. 2015. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 7:307ra152. 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 31.Heath AM, Haszard JJ, Galland BC, Lawley B, Rehrer NJ, Drummond LN, Sims IM, Taylor RW, Otal A, Taylor B, Tannock GW. 2020. Association between the faecal short-chain fatty acid propionate and infant sleep. Eur J Clin Nutr 74:1362–1365. 10.1038/s41430-019-0556-0. [DOI] [PubMed] [Google Scholar]
- 32.Finegold SM, Downes J, Summanen PH. 2012. Microbiology of regressive autism. Anaerobe 18:260–262. 10.1016/j.anaerobe.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 33.Borre YE, O’Keeffe GW, Clarke G, Stanton C, Dinan TG, Cryan JF. 2014. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol Med 20:509–518. 10.1016/j.molmed.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Xu M, Xu X, Li J, Li F. 2019. Association between gut microbiota and autism spectrum disorder: a systematic review and meta-analysis. Front Psychiatry 10:473. 10.3389/fpsyt.2019.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bezawada N, Phang TH, Hold GL, Hansen R. 2020. Autism spectrum disorder and the gut microbiota in children: a systematic review. Ann Nutr Metab 76:16–29. 10.1159/000505363. [DOI] [PubMed] [Google Scholar]
- 36.Lee HS, Burkhardt BR, McLeod W, Smith S, Eberhard C, Lynch K, Hadley D, Rewers M, Simell O, She JX, Hagopian B, Lernmark A, Akolkar B, Ziegler AG, Krischer JP, TEDDY study group. 2014. Biomarker discovery study design for type 1 diabetes in the environmental determinants of diabetes in the young (TEDDY) study. Diabetes Metab Res Rev 30:424–434. 10.1002/dmrr.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA, Jr, Ahmed T, Gordon JI. 2014. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510:417–421. 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, Fan YM, Henrissat B, Leyn SA, Rodionov DA, Osterman AL, Maleta KM, Newgard CB, Ashorn P, Dewey KG, Gordon JI. 2016. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351. 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanislawski MA, Dabelea D, Wagner BD, Iszatt N, Dahl C, Sontag MK, Knight R, Lozupone CA, Eggesbø M. 2018. Gut microbiota in the first 2 years of life and the association with body mass index at age 12 in a Norwegian birth cohort. mBio 9:e01751-18. 10.1128/mBio.01751-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker WA. 2017. The importance of appropriate initial bacterial colonization of the intestine in newborn, child, and adult health. Pediatr Res 82:387–395. 10.1038/pr.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonnenburg ED, Sonnenburg JL. 2019. The ancestral and industrialized gut microbiota and implications for human health. Nat Rev Microbiol 17:383–390. 10.1038/s41579-019-0191-8. [DOI] [PubMed] [Google Scholar]
- 42.Fan Y, Pedersen O. 2021. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol 19:55–71. 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 43.Rautava S, Luoto R, Salminen S, Isolauri E. 2012. Microbial contact during pregnancy, intestinal colonization and human disease. Nat Rev Gastroenterol Hepatol 9:565–576. 10.1038/nrgastro.2012.144. [DOI] [PubMed] [Google Scholar]
- 44.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. 2016. Human gut colonization may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep 6:23129. 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim ES, Rodriguez C, Holtz LR. 2018. Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community. Microbiome 6:87. 10.1186/s40168-018-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rackaityte E, Halkias J, Fukui EM, Mendoza VF, Hayzelden C, Crawford ED, Fujimura KE, Burt TD, Lynch SV. 2020. Viable bacterial colonization is highly limited in the human intestine in utero. Nat Med 26:599–607. 10.1038/s41591-020-0761-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishra A, Lai GC, Yao LJ, Aung TT, Shental N, Rotter-Maskowitz A, Shepherdson E, Singh GSN, Pai R, Shanti A, Wong RMM, Lee A, Khyriem C, Dutertre CA, Chakarov S, Srinivasan KG, Shadan NB, Zhang XM, Khalilnezhad S, Cottier F, Tan ASM, Low G, Chen P, Fan Y, Hor PX, Lee AKM, Choolani M, Vermijlen D, Sharma A, Fuks G, Straussman R, Pavelka N, Malleret B, McGovern N, Albani S, Chan JKY, Ginhoux F. 2021. Microbial exposure during early human development primes fetal immune cells. Cell 184:3394–3409. 10.1016/j.cell.2021.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. 2014. The placenta harbors a unique microbiome. Sci Transl Med 6:237ra65. 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fricke WF, Ravel J. 2021. Microbiome or no microbiome: are we looking at the prenatal environment through the right lens? Microbiome 9:9. 10.1186/s40168-020-00947-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez-Muñoz ME, Arrieta MC, Ramer-Tait AE, Walter J. 2017. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome 5:48. 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leiby JS, McCormick K, Sherrill-Mix S, Clarke EL, Kessler LR, Taylor LJ, Hofstaedter CE, Roche AM, Mattei LM, Bittinger K, Elovitz MA, Leite R, Parry S, Bushman FD. 2018. Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome 6:196. 10.1186/s40168-018-0575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, Parkhill J, Charnock-Jones DS, Smith GCS. 2019. Human placenta has no microbiome but can contain potential pathogens. Nature 572:329–334. 10.1038/s41586-019-1451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Theis KR, Romero R, Winters AD, Greenberg JM, Gomez-Lopez N, Alhousseini A, Bieda J, Maymon E, Pacora P, Fettweis JM, Buck GA, Jefferson KK, Strauss JF, III, Erez O, Hassan SS. 2019. Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomics. Am J Obstet Gynecol 220:267.e1–267.e39. 10.1016/j.ajog.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gschwind R, Fournier T, Kennedy S, Tsatsaris V, Cordier A-G, Barbut F, Butel M-J, Wydau-Dematteis S. 2020. Evidence for contamination as the origin for bacteria found in human placenta rather than a microbiota. PLoS One 15:e0237232. 10.1371/journal.pone.0237232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walter J, Hornef MW. 2021. A philosophical perspective on the prenatal in utero microbiome debate. Microbiome 9:5. 10.1186/s40168-020-00979-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, Kim CJ, Erez O, Edwin S, Relman DA. 2008. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One 3:e3056. 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DiGiulio DB, Romero R, Kusanovic JP, Gómez R, Kim CJ, Seok KS, Gotsch F, Mazaki-Tovi S, Vaisbuch E, Sanders K, Bik EM, Chaiworapongsa T, Oyarzún E, Relman DA. 2010. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am J Reprod Immunol 64:38–57. 10.1111/j.1600-0897.2010.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luckey TD. 1963. Germfree life and gnotobiology. Academic Press, New York, NY. [Google Scholar]
- 59.Martín R, Jiménez E, Heilig H, Fernández L, Marín ML, Zoetendal EG, Rodríguez JM. 2009. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol 75:965–969. 10.1128/AEM.02063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. 2014. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol 16:2891–2904. 10.1111/1462-2920.12238. [DOI] [PubMed] [Google Scholar]
- 61.Biagi E, Quercia S, Aceti A, Beghetti I, Rampelli S, Turroni S, Faldella G, Candela M, Brigidi P, Corvaglia L. 2017. The bacterial ecosystem of mother’s milk and infant’s mouth and gut. Front Microbiol 8:1214. 10.3389/fmicb.2017.01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams JE, Carrothers JM, Lackey KA, Beatty NF, Brooker SL, Peterson HK, Steinkamp KM, York MA, Shafii B, Price WJ, McGuire MA, McGuire MK. 2019. Strong multivariate relations exist among milk, oral, and fecal microbiomes in mother-infant dyads during the first six months postpartum. J Nutr 149:902–914. 10.1093/jn/nxy299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lackey KA, Williams JE, Meehan CL, Zachek JA, Benda ED, Price WJ, Foster JA, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SE, Prentice A,K, Dg Kvist LJ, Otoo GE, García-Carral C, Jiménez E, Ruiz L, Rodríguez JM, Pareja RG, Bode L, McGuire MA, McGuire MK. 2019. What’s normal? microbiomes in human milk and infant feces are related to each other but vary geographically: the INSPIRE study. Front Nutr 6:45. (Erratum, 7:12, 2020.) . 10.3389/fnut.2019.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Differding MK, Mueller NT. 2020. Human milk bacteria: seeding the infant gut? Cell Host Microbe 28:151–153. 10.1016/j.chom.2020.07.017. [DOI] [PubMed] [Google Scholar]
- 65.Fehr K, Moossavi S, Sbihi H, Boutin RCT, Bode L, Robertson B, Yonemitsu C, Field CJ, Becker AB, Mandhane PJ, Sears MR, Khafipour E, Moraes TJ, Subbarao P, Finlay BB, Turvey SE, Azad MB. 2020. Breastmilk feeding practices are associated with the co-occurrence of bacteria in mothers’ milk and the infant gut: the child cohort study. Cell Host Microbe 28:285–297. 10.1016/j.chom.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 66.Lugli GA, Duranti S, Milani C, Mancabelli L, Turroni F, Alessandri G, Longhi G, Anzalone R, Viappinai A, Tarracchini C, Bernasconi S, Yonemitsu C, Bode L, Goran MI, Ossiprandi MC, van Sinderen D, Ventura M. 2020. Investigating bifidobacteria and human milk oligosaccharide composition of lactating mothers. FEMS Microbiol Ecol 96:fiaa049. 10.1093/femsec/fiaa049. [DOI] [PubMed] [Google Scholar]
- 67.Perez PF, Doré J, Leclerc M, Levenez F, Benyacoub J, Serrant P, Segura-Roggero I, Schiffrin EJ, Donnet-Hughes A. 2007. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 119:e724–e732. 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- 68.Dall SR, Boyd IL. 2004. Evolution of mammals: lactation helps mothers to cope with unreliable food supplies. Proc Biol Sci 271:2049–2057. 10.1098/rspb.2004.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. 2012. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr 96:544–551. 10.3945/ajcn.112.037382. [DOI] [PubMed] [Google Scholar]
- 70.González R, Maldonado A, Martín V, Mandomando I, Fumadó V, Metzner KJ, Sacoor C, Fernández L, Macete E, Alonso PL, Rodríguez JM, Menendez C. 2013. Breast milk and gut microbiota in African mothers and infants from an area of high HIV prevalence. PLoS One 8:e80299. 10.1371/journal.pone.0080299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, Adisetiyo H, Zabih S, Lincez PJ, Bittinger K, Bailey A, Bushman FD, Sleasman JW, Aldrovandi GM. 2017. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr 171:647–654. 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwab C, Voney E, Ramirez GA, Vischer M, Lacroix C. 2019. Characterization of the cultivable microbiota in fresh and stored mature human breast milk. Front Microbiol 10:2666. 10.3389/fmicb.2019.02666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Treven P, Mahnič A, Rupnik M, Golob M, Pirš T, Matijašić BB, Lorbeg PM. 2019. Evaluation of human milk microbiota by 16S rRNA gene next-generation sequencing (NGS) and cultivation/MALDI-TOF mass spectrometry identification. Front Microbiol 10:2612. (Erratum, 11:31, 2020.) 10.3389/fmicb.2019.02612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Asbury MR, Butcher J, Copeland JK, Unger S, Bando N, Comelli EM, Forte V, Kiss A, LeMay-Nedjelski L, Sherman PM, Stintzi A, Tomlinson C, Wang PW, O’Connor DL. 2020. Mothers of preterm infants have individualized breast milk microbiota that changes temporally based on maternal characteristics. Cell Host Microbe 28:669–682. 10.1016/j.chom.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 75.Grönlund MM, Gueimonde M, Laitinen K, Kociubinski G, Grönroos T, Salminen S, Isolauri E. 2007. Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin Exp Allergy 37:1764–1772. 10.1111/j.1365-2222.2007.02849.x. [DOI] [PubMed] [Google Scholar]
- 76.Laursen MF, Pekmez CT, Larsson MW, Lind MV, Yonemitsu C, Larnkjær A, Mølgaard C, Bode L, Dragsted LO, Michaelsen KF, Licht TR, Bahl MI. 2021. Maternal milk microbiota and oligosaccharides contribute to the infant gut microbiota assembly. ISME Commun 1:21. 10.1038/s43705-021-00021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tannock GW, Fuller R, Smith SL, Hall MA. 1990. Plasmid profiling of members of the family Enterobacteriaceae, lactobacilli, and bifidobacteria to study the transmission of bacteria from mother to infant. J Clin Microbiol 28:1225–1228. 10.1128/jcm.28.6.1225-1228.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grönlund MM, Grześkowiak Ł, Isolauri E, Salminen S. 2011. Influence of mother’s intestinal microbiota on gut colonization in the infant. Gut Microbes 2:227–233. 10.4161/gmic.2.4.16799. [DOI] [PubMed] [Google Scholar]
- 79.Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. 2016. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res 26:1612–1625. 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, Armanini F, Truong DT, Manara S, Zolfo M, Beghini F, Bertorelli R, De Sanctis V, Bariletti I, Canto R, Clementi R, Cologna M, Crifò T, Cusumano G, Gottardi S, Innamorati C, Masè C, Postai D, Savoi D, Duranti S, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Milani C, Mangifesta M, Anzalone R, Viappiani A, Yassour M, Vlamakis H, Xavier R, Collado CM, Koren O, Tateo S, Soffiati M, Pedrotti A, Ventura M, Huttenhower C, Bork P, Segata N. 2018. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24:133–145.e5. 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yassour M, Jason E, Hogstrom LJ, Arthur TD, Tripathi S, Siljander H, Selvenius J, Oikarinen S, Hyöty H, Virtanen SM, Ilonen J, Ferretti P, Pasolli E, Tett A, Asnicar F, Segata N, Vlamakis H, Lander ES, Huttenhower C, Knip M, Xavier RJ. 2018. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe 24:146–154. 10.1016/j.chom.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mitchell CM, Mazzoni C, Hogstrom L, Bryant A, Bergerat A, Cher A, Pochan S, Herman P, Carrigan M, Sharp K, Huttenhower C, Lander ES, Vlamakis H, Xavier RJ, Yassour M. 2020. Delivery mode affects stability of early infant gut microbiota. Cell Rep Med 1:100156. 10.1016/j.xcrm.2020.100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nilsen M, Lokmic A, Angell IL, Lødrup Carlsen KC, Carlsen KH, Haugen G, Hedlin G, Jonassen CM, Marsland BJ, Nordlund B, Rehbinder EM, Saunders CM, Skjerven HO, Snipen L, Staff AC, Söderhäll C, Vettukattil R, Rudi K. 2021. Fecal microbiota nutrient utilization potential suggests mucins as drivers for initial gut colonization of mother-child-shared bacteria. Appl Environ Microbiol 87:e02201-20. 10.1128/AEM.02201-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Podlesny D, Fricke WF. 2021. Strain inheritance and neonatal gut microbiota development: a meta-analysis. Int J Med Microbiol 311:151483. 10.1016/j.ijmm.2021.151483. [DOI] [PubMed] [Google Scholar]
- 85.Zoetendal EG, Akkermans ADL, Akkermans-van Vliet VM, de Visser AGM, de Vos WM. 2001. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb Ecol Health Dis 13:129–134. 10.1080/089106001750462669.. [DOI] [Google Scholar]
- 86.Tims S, Derom C, Jonkers DM, Vlietinck R, Saris WH, Kleerebezem M, de Vos WM, Zoetendal EG. 2013. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J 7:707–717. 10.1038/ismej.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koo H, Hakim JA, Crossman DK, Lefkowitz EJ, Morrow CD. 2019. Sharing of gut microbial strains between selected individual sets of twins cohabitating for decades. PLoS One 14:e0226111. 10.1371/journal.pone.0226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, Spector TD, Keinan A, Ley RE, Gevers D, Clark AG. 2015. Host genetic variation impacts microbiome composition across human body sites. Genome Biol 16:191. 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. 2006. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–521. 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 90.Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. 2010. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev 86(Suppl 1):13–15. 10.1016/j.earlhumdev.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 91.Dogra S, Sakwinska O, Soh SE, Ngom-Bru C, Brück WM, Berger B, Brüssow H, Lee YS, Yap F, Chong YS, Godfrey KM, Holbrook JD, GUSTO Study Group. 2015. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio 6:e02419-14. 10.1128/mBio.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stokholm J, Thorsen J, Chawes BL, Schjorring S, Krogfelt KA, Bonnelykke K, Bisgaard H. 2016. Cesarean section changes neonatal gut colonization. J Allergy Clin Immunol 138:881–889. 10.1016/j.jaci.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 93.Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, Sears MR, Becker AB, Scott JA, Kozyrskyj AL. 2013. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ 185:385–394. 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Bjorksten B, Engstrand L, Andersson AF. 2014. Decreased gut microbiota diversity, delayed Bacteroidetes colonization and reduced Th1 responses in infants delivered by caesarean section. Gut 63:559–566. 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 95.Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, Kubota H, Swinkels S, Sakai T, Oishi K, Kushiro A, Knol J. 2016. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 11:e0158498. 10.1371/journal.pone.0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, Kumar N, Stares MD, Rodger A, Brocklehurst P, Field N, Lawley TD. 2019. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574:117–121. 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chin N, Méndez-Lagares G, Taft DH, Laleau V, Kieu H, Narayan NR, Roberts SB, Mills DA, Hartigan-O’Connor DJ, Flaherman VJ. 2021. Transient effect of infant formula supplementation on the intestinal microbiota. Nutrients 13:807. 10.3390/nu13030807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lay C, Chu CW, Purbojati RW, Acerbi E, Drautz-Moses DI, de Sessions PF, Jie S, Ho E, Kok YJ, Bi X, Chen S, Mak SY, Chua MC, Goh AEN, Chiang WC, Rao R, Chaithongwongwatthana S, Khemapech N, Chongsrisawat V, Martin R, Roeselers G, Ho YS, Hibberd ML, Schuster SC, Knol J, JULIUS Study Group. 2021. A synbiotic intervention modulates meta-omics signatures of gut redox potential and acidity in elective caesarean born infants. BMC Microbiol 21:191. 10.1186/s12866-021-02230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, Bokulich NA, Song SJ, Hoashi M, Rivera-Vinas JI, Mendez K, Knight R, Clemente JC. 2016. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med 22:250–253. 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Korpela K, Helve O, Kolho KL, Saisto T, Skogberg K, Dikareva E, Stefanovic V, Salonen A, Andersson S, de Vos WM. 2020. Maternal fecal microbiota transplantation in cesarean-born infants rapidly restores normal gut microbial development: a proof-of-concept study. Cell 183:324–334. 10.1016/j.cell.2020.08.047. [DOI] [PubMed] [Google Scholar]
- 101.Cunnington AJ, Sim K, Deierl A, Kroll JS, Brannigan E, Darby J. 2016. “Vaginal seeding” of infants born by caesarean section. BMJ 352:i227. 10.1136/bmj.i227. [DOI] [PubMed] [Google Scholar]
- 102.Murphy R, Morgan XC, Wang XY, Wickens K, Purdie G, Fitzharris P, Otal A, Lawley B, Stanley T, Barthow C, Crane J, Mitchell EA, Tannock GW. 2019. Eczema-protective probiotic alters infant gut microbiome functional capacity but not composition: subsample analysis from a RCT. Benef Microbes 10:5–17. 10.3920/BM2017.0191. [DOI] [PubMed] [Google Scholar]
- 103.Xu J, Lawley B, Wong G, Otal A, Chen L, Ying TJ, Lin X, Pang WW, Yap F, Chong YS, Gluckman PD, Lee YS, Chong MF, Tannock GW, Karnani N. 2020. Ethnic diversity in infant gut microbiota is apparent before the introduction of complementary diets. Gut Microbes 11:1362–1373. 10.1080/19490976.2020.1756150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Busi SB, de Nies L, Habier J, Wampach L, Fritz JV, Heintz-Buschart A, May P, Halder R, de Beaufort C, Wilmes P. 2021. Persistence of birth mode-dependent effects on gut microbiome composition, immune system stimulation and antimicrobial resistance during the first year of life. ISME Commun 1:8. 10.1038/s43705-021-00003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sandall J, Tribe RM, Avery L, Mola G, Visser GH, Homer CS, Gibbons D, Kelly NM, Kennedy HP, Kidanto H, Taylor P, Temmerman M. 2018. Short-term and long-term effects of caesarean section on the health of women and children. Lancet 392:1349–1357. 10.1016/S0140-6736(18)31930-5. [DOI] [PubMed] [Google Scholar]
- 106.Stokholm J, Thorsen J, Blaser MJ, Rasmussen MA, Hjelmsø M, Shah S, Christensen ED, Chawes BL, Bønnelykke K, Brix S, Mortensen MS, Brejnrod A, Vestergaard G, Trivedi U, Sørensen SJ, Bisgaard H. 2020. Delivery mode and gut microbial changes correlate with an increased risk of childhood asthma. Sci Transl Med 12:eaax9929. 10.1126/scitranslmed.aax9929. [DOI] [PubMed] [Google Scholar]
- 107.Yassour M, Vatanen T, Siljander H, Hämäläinen AM, Härkönen T, Ryhänen SJ, Franzosa EA, Vlamakis H, Huttenhower C, Gevers D, Lander ES, Knip M, Xavier RJ, DIABIMMUNE Study Group. 2016. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med 8:343ra81. 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Taft DH, Liu J, Maldonado-Gomez MX, Akre S, Huda MN, Ahmad SM, Stephensen CB, Mills DA. 2018. Bifidobacterial dominance of the gut in early life and acquisition of antimicrobial resistance. mSphere 3:e00441-18. 10.1128/mSphere.00441-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, Lernmark Å, Hagopian WA, Rewers MJ, She JX, Toppari J, Ziegler AG, Akolkar B, Krischer JP, Stewart CJ, Ajami NJ, Petrosino JF, Gevers D, Lähdesmäki H, Vlamakis H, Huttenhower C, Xavier RJ. 2018. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562:589–594. 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Casaburi G, Duar RM, Vance DP, Mitchell R, Contreras L, Frese SA, Smilowitz JT, Underwood MA. 2019. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob Resist Infect Control 8:131. 10.1186/s13756-019-0583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Berger B, Porta N, Foata F, Grathwohl D, Delley M, Moine D, Charpagne A, Siegwald L, Descombes P, Alliet P, Puccio G, Steenhout P, Mercenier A, Sprenger N. 2020. Linking human milk oligosaccharides, infant fecal community types, and later risk to require antibiotics. mBio 11:e03196-19. 10.1128/mBio.03196-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Korpela K, Salonen A, Saxen H, Nikkonen A, Peltola V, Jaakkola T, de Vos W, Kolho KL. 2020. Antibiotics in early life associate with specific gut microbiota signatures in a prospective longitudinal infant cohort. Pediatr Res 88:438–443. 10.1038/s41390-020-0761-5. [DOI] [PubMed] [Google Scholar]
- 113.Uzan-Yulzari A, Turta O, Belogolovski A, Ziv O, Kunz C, Perschbacher S, Neuman H, Pasolli E, Oz A, Ben-Amram H, Kumar H, Ollila H, Kaljonen A, Isolauri E, Salminen S, Lagström H, Segata N, Sharon I, Louzoun Y, Ensenauer R, Rautava S, Koren O. 2021. Neonatal antibiotic exposure impairs child growth during the first six years of life by perturbing intestinal microbial colonization. Nat Commun 12:443. 10.1038/s41467-020-20495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Alexander M. 1971. Microbial ecology. Wiley, New York, NY. [Google Scholar]
- 115.Tissier H. 1905. Repartition des microbes dans l’intestin du nourisson. Ann Institut Pasteur (Paris) 19:109–123. [Google Scholar]
- 116.Cooperstock MS, Zedd AJ. 1983. Intestinal flora of infants, p 79–99. In Hentges DJ (ed), Human intestinal microflora in health and disease. Academic Press, New York, NY. [Google Scholar]
- 117.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr 30:61–67. 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 118.Favier CF, Vaughan EE, De Vos WM, Akkermans AD. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol 68:219–226. 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bezirtzoglou E, Tsiotsias A, Welling GW. 2011. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 17:478–482. 10.1016/j.anaerobe.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 120.Tannock GW, Lawley B, Munro K, Gowri Pathmanathan S, Zhou SJ, Makrides M, Gibson RA, Sullivan T, Prosser CG, Lowry D, Hodgkinson AJ. 2013. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl Environ Microbiol 79:3040–3048. 10.1128/AEM.03910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Azad MB, Konya T, Persaud RR, Guttman DS, Chari RS, Field CJ, Sears MR, Mandhane PJ, Turvey SE, Subbarao P, Becker AB, Scott JA, Kozyrskyj AL, CHILD Study Investigators. 2016. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG 123:983–993. 10.1111/1471-0528.13601. [DOI] [PubMed] [Google Scholar]
- 122.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, D Lieber A, Wu F, Perez-Perez GI, Chen Y, Schweizer W, Zheng X, Contreras M, Dominguez-Bello MG, Blaser MJ. 2016. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 8:343ra82. 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Biavati B, Castagnoli P, Crociani F, Trovatelli LD. 1984. Species of the Bifidobacterium in the feces of infants. Microbiologica 7:341–345. [PubMed] [Google Scholar]
- 124.Young SL, Simon MA, Baird MA, Tannock GW, Bibiloni R, Spencely K, Lane JM, Fitzharris P, Crane J, Town I, Addo-Yobo E, Murray CS, Woodcock A. 2004. Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested from cord blood. Clin Diagn Lab Immunol 11:686–690. 10.1128/CDLI.11.4.686-690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. 2010. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology (Reading) 156:3329–3341. 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- 126.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O’Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Makino H, Kushiro A, Ishikawa E, Kubota H, Gawad A, Sakai T, Oishi K, Martin R, Ben-Amor K, Knol J, Tanaka R. 2013. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS One 8:e78331. 10.1371/journal.pone.0078331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Grześkowiak Ł, Collado MC, Mangani C, Maleta K, Laitinen K, Ashorn P, Isolauri E, Salminen S. 2012. Distinct gut microbiota in southeastern African and northern European infants. J Pediatr Gastroenterol Nutr 54:812–816. 10.1097/MPG.0b013e318249039c. [DOI] [PubMed] [Google Scholar]
- 129.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Gorfer V, Tett A, Segata N, van Sinderen D, Ventura M. 2015. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 81:7078–7087. 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lewis ZT, Sidamonidze K, Tsaturyan V, Tsereteli D, Khachidze N, Pepoyan A, Zhgenti E, Tevzadze L, Manvelyan A, Balayan M, Imnadze P, Torok T, Lemay DG, Mills DA. 2017. The fecal microbial community of breast-fed infants from Armenia and Georgia. Sci Rep 7:40932. 10.1038/srep40932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Walker AW, Martin JC, Scott P, Parkhill J, Flint HJ, Scott KP. 2015. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 3:26. 10.1186/s40168-015-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tannock GW, Lee PS, Wong KH, Lawley B. 2016. Why don’t all infants have bifidobacteria in their stool? Front Microbiol 7:834. 10.3389/fmicb.2016.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Scott KP, Duncan SH, Flint HJ. 2008. Dietary fibre and the gut microbiota. Br Nutr Found Nutr Bull 33:201–211. 10.1111/j.1467-3010.2008.00706.x. [DOI] [Google Scholar]
- 134.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tannock GW. 2021. Modulating the gut microbiota of humans by dietary intervention with plant glycans. Appl Environ Microbiol 87:e02757-20. 10.1128/AEM.02757-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Avershina E, Storrø O, Øien T, Johnsen R, Pope P, Rudi K. 2014. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol Ecol 87:280–290. 10.1111/1574-6941.12223. [DOI] [PubMed] [Google Scholar]
- 137.Vallès Y, Artacho A, Pascual-García A, Ferrús ML, Gosalbes MJ, Abellán JJ, Francino MP. 2014. Microbial succession in the gut: directional trends of taxonomic and functional change in a birth cohort of Spanish infants. PLoS Genet 10:e1004406. 10.1371/journal.pgen.1004406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni H, Metcalf GA, Muzny D, Gibbs RA, Vatanen T, Huttenhower C, Xavier RJ, Rewers M, Hagopian W, Toppari J, Ziegler AG, She JX, Akolkar B, Lernmark A, Hyoty H, Vehik K, Krischer JP, Petrosino JF. 2018. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562:583–588. 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Agans R, Rigsbee L, Kenche H, Michail S, Khamis HJ, Paliy O. 2011. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol 77:404–412. 10.1111/j.1574-6941.2011.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Berding K, Holscher HD, Arthur AE, Donovan SM. 2018. Fecal microbiome composition and stability in 4- to 8-year-old children is associated with dietary patterns and nutrient intake. J Nutr Biochem 56:165–174. 10.1016/j.jnutbio.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 141.Derrien M, Alvarez AS, de Vos WM. 2019. The gut microbiota in the first decade of life. Trends Microbiol 27:997–1010. 10.1016/j.tim.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 142.Nakayama J, Watanabe K, Jiang J, Matsuda K, Chao SH, Haryono P, La-Ongkham O, Sarwoko MA, Sujaya IN, Zhao L, Chen KT, Chen YP, Chiu HH, Hidaka T, Huang NX, Kiyohara C, Kurakawa T, Sakamoto N, Sonomoto K, Tashiro K, Tsuji H, Chen MJ, Leelavatcharamas V, Liao CC, Nitisinprasert S, Rahayu ES, Ren FZ, Tsai YC, Lee YK. 2019. Author correction: diversity in gut bacterial community of school-age children in Asia. Sci Rep 9:6530. 10.1038/s41598-019-42780-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhong H, Penders J, Shi Z, Ren H, Cai K, Fang C, Ding Q, Thijs C, Blaak EE, Stehouwer CDA, Xu X, Yang H, Wang J, Wang J, Jonkers DMAE, Masclee AAM, Brix S, Li J, Arts ICW, Kristiansen K. 2019. Impact of early events and lifestyle on the gut microbiota and metabolic phenotypes in young school-age children. Microbiome 7:2. 10.1186/s40168-018-0608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Roswall J, Olsson LM, Kovatcheva-Datchary P, Nilsson S, Tremaroli V, Simon MC, Kiilerich P, Akrami R, Krämer M, Uhlén M, Gummesson A, Kristiansen K, Dahlgren J, Bäckhed F. 2021. Developmental trajectory of the healthy human gut microbiota during the first 5 years of life. Cell Host Microbe 10.1016/j.chom.2021.02.021.. [DOI] [PubMed] [Google Scholar]
- 145.Norin KE, Gustafsson BE, Lindblad BS, Midtvedt T. 1985. The establishment of some microflora associated biochemical characteristics in feces from children during the first years of life. Acta Paediatr Scand 74:207–212. 10.1111/j.1651-2227.1985.tb10951.x. [DOI] [PubMed] [Google Scholar]
- 146.Midtvedt AC, Carlstedt-Duke B, Norin KE, Saxerholt H, Midtvedt T. 1988. Development of five metabolic activities associated with the intestinal microflora of healthy infants. J Pediatr Gastroenterol Nutr 7:559–567. 10.1097/00005176-198807000-00014. [DOI] [PubMed] [Google Scholar]
- 147.Midtvedt AC, Midtvedt T. 1992. Production of short chain fatty acids by the intestinal microflora during the first 2 years of human life. J Pediatr Gastroenterol Nutr 15:395–403. 10.1097/00005176-199211000-00005. [DOI] [PubMed] [Google Scholar]
- 148.Midtvedt AC, Midtvedt T. 1993. Conversion of cholesterol to coprostanol by the intestinal microflora during the first two years of human life. J Pediatr Gastroenterol Nutr 17:161–168. 10.1097/00005176-199308000-00006. [DOI] [PubMed] [Google Scholar]
- 149.Midtvedt AC, Carlstedt-Duke B, Midtvedt T. 1994. Establishment of a mucin-degrading intestinal microflora during the first two years of human life. J Pediatr Gastroenterol Nutr 18:321–326. 10.1097/00005176-199404000-00012. [DOI] [PubMed] [Google Scholar]
- 150.Tannock GW. 1995. Normal microflora: an introduction to microbes inhabiting the human body. Chapman and Hall, London, United Kingdom. [Google Scholar]
- 151.Zhang J, Guo Z, Xue Z, Sun Z, Zhang M, Wang L, Wang G, Wang F, Xu J, Cao H, Xu H, Lv Q, Zhong Z, Chen Y, Qimuge S, Menghe B, Zheng Y, Zhao L, Chen W, Zhang H. 2015. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J 9:1979–1990. 10.1038/ismej.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Quin C, Gibson DL. 2020. Human behavior, not race or geography, is the strongest predictor of microbial succession in the gut bacteriome of infants. Gut Microbes 11:1143–1171. 10.1080/19490976.2020.1736973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, MetaHIT Consortium, et al. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Goodrich JK, Davenport ER, Clark AG, Ley RE. 2017. The relationship between the human genome and microbiome comes into view. Annu Rev Genet 51:413–433. 10.1146/annurev-genet-110711-155532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Deschasaux M, Bouter KE, Prodan A, Levin E, Groen AK, Herrema H, Tremaroli V, Bakker GJ, Attaye I, Pinto-Sietsma SJ, van Raalte DH, Snijder MB, Nicolaou M, Peters R, Zwinderman AH, Bäckhed F, Nieuwdorp M. 2018. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat Med 24:1526–1531. 10.1038/s41591-018-0160-1. [DOI] [PubMed] [Google Scholar]
- 156.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu J, Wijmenga C, Zhernakova A, Elinav E, Segal E. 2018. Environment dominates over host genetics in shaping human gut microbiota. Nature 555:210–215. 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 157.Gómez-Gallego C, Morales JM, Monleón D, Du Toit E, Kumar H, Linderborg KM, Zhang Y, Yang B, Isolauri E, Salminen S, Collado MC. 2018. Human breast milk NMR metabolomic profile across specific geographical locations and its association with the milk microbiota. Nutrients 10:1355. 10.3390/nu10101355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.van Leeuwen SS, Stoutjesdijk E, Ten Kate GA, Schaafsma A, Dijck-Brouwer J, Muskiet FAJ, Dijkhuizen L. 2018. Regional variations in human milk oligosaccharides in Vietnam suggest FucTx activity besides FucT2 and FucT3. Sci Rep 8:16790. 10.1038/s41598-018-34882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Caporaso JG, Knights D, Clemente JC, Nakielny S, Gordon JI, Fierer N, Knight R. 2013. Cohabiting family members share microbiota with one another and with their dogs. Elife 2:e00458. 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Laursen MF, Zachariassen G, Bahl MI, Bergström A, Høst A, Michaelsen KF, Licht TR. 2015. Having older siblings is associated with gut microbiota development during early childhood. BMC Microbiol 15:154. 10.1186/s12866-015-0477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Martinez I, Stegen JC, Maldonado GM, Eren AM, Siba PM, Greenhill AR, Walter J. 2015. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep 11:527–538. 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 162.Gaulke CA, Sharpton TJ. 2018. The influence of ethnicity and geography on human gut microbiome composition. Nat Med 24:1495–1496. 10.1038/s41591-018-0210-8. [DOI] [PubMed] [Google Scholar]
- 163.de Wit R, Bouvier T. 2006. “Everything is everywhere, but, the environment selects”: what did Baas Becking and Beijerinck really say? Environ Microbiol 8:755–758. 10.1111/j.1462-2920.2006.01017.x. [DOI] [PubMed] [Google Scholar]
- 164.Duar RM, Henrick BM, Casaburi G, Frese SA. 2020. Integrating the ecosystem services framework to define dysbiosis of the breastfed infant gut: the role of B. infantis and human milk oligosaccharides. Front Nutr 7:33. 10.3389/fnut.2020.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Winogradsky S, Winogradsky H. 1933. Etudes sur la microbiologie du sol. Ann Inst Pasteur 50:350–432. [Google Scholar]
- 166.Zivkovic AM, German JB, Lebrilla CB, Mills DA. 2011. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci USA 108(Suppl 1):4653–4658. 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bode L. 2012. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22:1147–1162. 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xu G, Davis JC, Goonatilleke E, Smilowitz JT, German JB, Lebrilla CB. 2017. Absolute quantitation of human milk oligosaccharides reveals phenotypic variations during lactation. J Nutr 147:117–124. 10.3945/jn.116.238279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.van Niekerk E, Autran CA, Nel DG, Kirsten GF, Blaauw R, Bode L. 2014. Human milk oligosaccharides differ between HIV-infected and HIV-uninfected mothers and are related to necrotizing enterocolitis incidence in their preterm very-low-birth-weight infants. J Nutr 144:1227–1233. 10.3945/jn.113.187799. [DOI] [PubMed] [Google Scholar]
- 170.Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. 2014. Breast milk oligosaccharides: structure-function relationships in the neonate. Annu Rev Nutr 34:143–169. 10.1146/annurev-nutr-071813-105721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H. 2012. Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology 22:361–368. 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- 172.German JB, Freeman SL, Lebrilla CB, Mills DA. 2008. Human milk oligosaccharides: evolution, structures and bioselectivity as the substrates for intestinal bacteria. Nestle Nutr Workshop Ser Pediatr Program 62:205–222. 10.1159/000146322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kobata A. 2010. Structures and application of oligosaccharides in human milk. Proc Jpn Acad Ser B Phys Biol Sci 86:731–747. 10.2183/pjab.86.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kumazaki T, Yoshida A. 1984. Biochemical evidence that secretor gene, Se, is a structural gene encoding a specific fucosyltransferase. Proc Natl Acad Sci USA 81:4193–4197. 10.1073/pnas.81.13.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, Lemay DG, Van Tassell ML, Miller MJ, Jin YS, German JB, Lebrilla CB, Mills DA. 2015. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 3:13. 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Plaza-Díaz J, Fontana L, Gil A. 2018. Human milk oligosaccharides and immune system development. Nutrients 10:1038. 10.3390/nu10081038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL. 2011. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10:507–514. 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kostopoulos I, Elzinga J, Ottman N, Klievink JT, Blijenberg B, Aalvink S, Boeren S, Mank M, Knol J, de Vos WM, Belzer C. 2020. Akkermansia muciniphila uses human milk oligosaccharides to thrive in the early life conditions in vitro. Sci Rep 10:14330. 10.1038/s41598-020-71113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Karav S, Casaburi G, Frese SA. 2018. Reduced colonic mucin degradation in breastfed infants colonized by Bifidobacterium longum subsp. infantis EVC001. FEBS Open Bio 8:1649–1657. 10.1002/2211-5463.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ma J, Li Z, Zhang W, Zhang C, Zhang Y, Mei H, Zhuo N, Wang H, Wang L, Wu D. 2020. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci Rep 10:15792. 10.1038/s41598-020-72635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Samuel TM, Binia A, de Castro CA, Thakkar SK, Billeaud C, Agosti M, Al-Jashi I, Costeira MJ, Marchini G, Martínez-Costa C, Picaud J-C, Stiris T, Stoicescu S-M, Vanpeé M, Domellöf M, Austin S, Sprenger N. 2019. N. Impact of maternal characteristics on human milk oligosaccharide composition over the first 4 months of lactation in a cohort of healthy European mothers. Sci Rep 9:11767. 10.1038/s41598-019-48337-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Borewicz K, Gu F, Saccenti E, Hechler C, Beijers R, de Weerth C, van Leeuwen SS, Schols HA, Smidt H. 2020. The association between breastmilk oligosaccharides and faecal microbiota in healthy breastfed infants at two, six, and twelve weeks of age. Sci Rep 10:4270. 10.1038/s41598-020-61024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Shenker NS, Perdones-Montero A, Burke A, Stickland S, McDonald JAK, Alexander-Hardiman K, Flanagan J, Takats Z, Cameron SJS. 2020. Metabolomic and metataxonomic fingerprinting of human milk suggests compositional stability over a natural term of breastfeeding to 24 months. Nutrients 12:3450. 10.3390/nu12113450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sprenger N, Lee LY, De Castro CA, Steenhout P, Thakkar SK. 2017. Longitudinal change of selected human milk oligosaccharides and association to infants’ growth, an observatory, single center, longitudinal cohort study. PLoS One 12:e0171814. 10.1371/journal.pone.0171814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Davis JC, Totten SM, Huang JO, Nagshbandi S, Kirmiz N, Garrido DA, Lewis ZT, Wu LD, Smilowitz JT, German JB, Mills DA, Lebrilla CB. 2016. Identification of oligosaccharides in feces of breast-fed infants and their correlation with the gut microbial community. Mol Cell Proteomics 15:2987–3002. 10.1074/mcp.M116.060665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Karav S, Le Parc A, Leite Nobrega de Moura Bell JM, Frese SA, Kirmiz N, Block DE, Barile D, Mills DA. 2016. Oligosaccharides released from milk glycoproteins are selective growth substrates for infant-associated bifidobacteria. Appl Environ Microbiol 82:3622–3630. 10.1128/AEM.00547-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Borewicz K, Gu F, Saccenti E, Arts ICW, Penders J, Thijs C, Leeuwen SS, Lindner C, Nauta A, Leusen E, Schols HA, Smidt H. 2019. Correlating infant faecal microbiota composition and human milk oligosaccharide consumption by microbiota of one-month-old breastfed infants. Mol Nutr Food Res 63:e1801214. 10.1002/mnfr.201801214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol 65:4506–4512. 10.1128/AEM.65.10.4506-4512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Youn SY, Seo JM, Ji GE. 2008. Evaluation of the PCR method for identification of Bifidobacterium species. Lett Appl Microbiol 46:7–13. 10.1111/j.1472-765X.2007.02263.x. [DOI] [PubMed] [Google Scholar]
- 190.Sheu SJ, Hwang WZ, Chiang YC, Lin WH, Chen HC, Tsen HY. 2010. Use of tuf gene-based primers for the PCR detection of probiotic Bifidobacterium species and enumeration of bifidobacteria in fermented milk by cultural and quantitative real-time PCR methods. J Food Sci 75:M521–M527. 10.1111/j.1750-3841.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- 191.Lawley B, Centanni M, Watanabe J, Sims I, Carnachan S, Broadbent R, Lee PS, Wong KH, Tannock GW. 2018. Tuf gene sequence variation in Bifidobacterium longum subsp. infantis detected in the fecal microbiota of Chinese infants. Appl Environ Microbiol 84:e00336-18. 10.1128/AEM.00336-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Huda MN, Ahmad SM, Alam MJ, Khanam A, Kalanetra KM, Taft DH, Raqib R, Underwood MA, Mills DA, Stephensen CB. 2019. Bifidobacterium abundance in early infancy and vaccine response at 2 years of age. Pediatrics 143:e20181489. 10.1542/peds.2018-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr 20:699–722. 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 194.Ward RE, Niñonuevo M, Mills DA, Lebrilla CB, German JB. 2007. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res 51:1398–1405. 10.1002/mnfr.200700150. [DOI] [PubMed] [Google Scholar]
- 195.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA 105:18964–18969. 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Garrido D, Kim JH, German JB, Raybould HE, Mills DA. 2011. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One 6:e17315. 10.1371/journal.pone.0017315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. 2010. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol 76:7373–7381. 10.1128/AEM.00675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sela DA, Mills DA. 2010. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol 18:298–307. 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, German JB, Chen X, Lebrilla CB, Mills DA. 2011. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem 286:11909–11918. 10.1074/jbc.M110.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom HJ, Joachimiak A, Lebrilla CB, Mills DA. 2012. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol 78:795–803. 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Turroni F, Strati F, Foroni E, Serafini F, Duranti S, van Sinderen D, Ventura M. 2012. Analysis of predicted carbohydrate transport systems encoded by Bifidobacterium bifidum PRL2010. Appl Environ Microbiol 78:5002–5012. 10.1128/AEM.00629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Garrido D, Ruiz-Moyano S, Mills DA. 2012. Release and utilization of N-acetyl-d-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp. infantis. Anaerobe 18:430–435. 10.1016/j.anaerobe.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Garrido D, Dallas DC, Mills DA. 2013. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology (Reading) 159:649–664. 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Egan M, O’Connell Motherway M, Ventura M, van Sinderen D. 2014. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol 80:4414–4426. 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Garrido D, Ruiz-Moyano S, Lemay DG, Sela DA, German JB, Mills DA. 2015. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci Rep 5:13517. 10.1038/srep13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Garrido D, Ruiz-Moyano S, Kirmiz N, Davis JC, Totten SM, Lemay DG, Ugalde JA, German JB, Lebrilla CB, Mills DA. 2016. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596. Sci Rep 6:35045. 10.1038/srep35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S, Ogawa E, Kodama H, Yamamoto K, Yamada T, Matsumoto S, Kurokawa K. 2016. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun 7:11939. 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bottacini F, Morrissey R, Esteban-Torres M, James K, van Breen J, Dikareva E, Egan M, Lambert J, van Limpt K, Knol J, O’Connell Motherway M, van Sinderen D. 2018. Comparative genomics and genotype-phenotype associations in Bifidobacterium breve. Sci Rep 8:10633. 10.1038/s41598-018-28919-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.James K, O’Connell Motherway M, Penno C, O’Brien RL, van Sinderen D. 2018. Bifidobacterium breve UCC2003 employs multiple transcriptional regulators to control metabolism of particular human milk oligosaccharides. Appl Environ Microbiol 84:e02774-17. 10.1128/AEM.02774-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lugli GA, Mancino W, Milani C, Duranti S, Turroni F, van Sinderen D, Ventura M. 2018. Reconstruction of the bifidobacterial pan-secretome reveals the network of extracellular interactions between bifidobacteria and the infant gut. Appl Environ Microbiol 84:e00796-18. 10.1128/AEM.00796-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sakanaka M, Gotoh A, Yoshida K, Odamaki T, Koguchi H, Xiao JZ, Kitaoka M, Katayama T. 2019. Varied pathways of infant gut-associated bifidobacterium to assimilate human milk oligosaccharides: prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients 12:71. 10.3390/nu12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lawson MAE, O’Neill IJ, Kujawska M, Gowrinadh Javvadi S, Wijeyesekera A, Flegg Z, Chalklen L, Hall LJ. 2020. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J 14:635–648. 10.1038/s41396-019-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. 2011. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem 286:34583–34592. 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gotoh A, Katoh T, Sakanaka M, Ling Y, Yamada C, Asakuma S, Urashima T, Tomabechi Y, Katayama-Ikegami A, Kurihara S, Yamamoto K, Harata G, He F, Hirose J, Kitaoka M, Okuda S, Katayama T. 2018. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci Rep 8:13958. 10.1038/s41598-018-32080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Centanni M, Ferguson SA, Sims IM, Biswas A, Tannock GW. 2019. Bifidobacterium bifidum ATCC 15696 and Bifidobacterium breve 24b metabolic interaction based on 2′-O-fucosyl-lactose studied in steady-state cultures in a Freter-style chemostat. Appl Environ Microbiol 85:e02783-18. 10.1128/AEM.02783-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tsukuda N, Yahagi K, Hara T, Watanabe Y, Matsumoto H, Mori H, Higashi K, Tsuji H, Matsumoto S, Kurokawa K, Matsuki T. 2021. Key bacterial taxa and metabolic pathways affecting gut short-chain fatty acid profiles in early life. ISME J 15:2574–2590. 10.1038/s41396-021-00937-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sánchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci USA 107:19514–19519. 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Turroni F, Milani C, van Sinderen D, Ventura M. 2011. Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human-microbe co-evolution. Gut Microbes 2:183–189. 10.4161/gmic.2.3.16105. [DOI] [PubMed] [Google Scholar]
- 219.Egan M, Motherway MO, Kilcoyne M, Kane M, Joshi L, Ventura M, van Sinderen D. 2014. Cross-feeding by Bifidobacterium breve UCC2003 during cocultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol 14:282. 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. 2007. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem 55:8914–8919. 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 221.Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, Mills DA. 2010. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem 58:5334–5340. 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Balamurugan R, Magne F, Balakrishnan D, Suau A, Ramani S, Kang G, Ramakrishna BS. 2010. Faecal bifidobacteria in Indian neonates and the effect of asymptomatic rotavirus infection during the first month of life. Indian J Med Res 132:721–727. [PMC free article] [PubMed] [Google Scholar]
- 223.Tsuji H, Oozeer R, Matsuda K, Matsuki T, Ohta T, Nomoto K, Tanaka R, Kawashima M, Kawashima K, Nagata S, Yamashiro Y. 2012. Molecular monitoring of the development of intestinal microbiota in Japanese infants. Benef Microbes 3:113–125. 10.3920/BM2011.0038. [DOI] [PubMed] [Google Scholar]
- 224.Lawley B, Otal A, Moloney-Geany K, Diana A, Houghton L, Heath AM, Taylor RW, Tannock GW. 2019. Fecal microbiotas of Indonesian and New Zealand children differ in complexity and bifidobacterial taxa during the first year of life. Appl Environ Microbiol 85:e01105-19. 10.1128/AEM.01105-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Vatanen T, Plichta DR, Somani J, Münch PC, Arthur TD, Hall AB, Rudolf S, Oakeley EJ, Ke X, Young RA, Haiser HJ, Kolde R, Yassour M, Luopajärvi K, Siljander H, Virtanen SM, Ilonen J, Uibo R, Tillmann V, Mokurov S, Dorshakova N, Porter JA, McHardy AC, Lähdesmäki H, Vlamakis H, Huttenhower C, Knip M, Xavier RJ. 2019. Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat Microbiol 4:470–479. 10.1038/s41564-018-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Casaburi G, Duar RM, Brown H, Mitchell RD, Kazi S, Chew S, Cagney O, Flannery RL, Sylvester KG, Frese SA, Henrick BM, Freeman SL. 2021. Metagenomic insights of the infant microbiome community structure and function across multiple sites in the United States. Sci Rep 11:1472. 10.1038/s41598-020-80583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.O’Brien CE, Meier AK, Cernioglo K. 2021. Early probiotic supplementation with B. infantis in breastfed infants leads to persistent colonization at 1 year. Pediatr Res 10.1038/s41390-020-01350-0.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Schwab C, Ruscheweyh HJ, Bunesova V, Pham VT, Beerenwinkel N, Lacroix C. 2017. Trophic interactions of infant bifidobacteria and Eubacterium hallii during l-fucose and fucosyllactose degradation. Front Microbiol 8:95. 10.3389/fmicb.2017.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Barbero GJ, Runge G, Fischer D, Crawford MN, Torres FE, Gyorgy P. 1952. Investigations on the bacterial flora, pH, and sugar content in the intestinal tract of infants. J Pediatr 40:152–163. 10.1016/s0022-3476(52)80176-3. [DOI] [PubMed] [Google Scholar]
- 230.Pratt AG, Read WT, Jr.. 1955. Influence of type of feeding on pH of stool, pH of skin, and incidence of perianal dermatitis in the newborn infant. J Pediatr 46:539–543. 10.1016/s0022-3476(55)80259-4. [DOI] [PubMed] [Google Scholar]
- 231.Bullen CL, Willis AT. 1971. Resistance of the breast-fed infant to gastroenteritis. Br Med J 3:338–343. 10.1136/bmj.3.5770.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ogawa K, Ben RA, Pons S, de Paolo MI, Bustos Fernández L. 1992. Volatile fatty acids, lactic acid, and pH in the stools of breast-fed and bottle-fed infants. J Pediatr Gastroenterol Nutr 15:248–252. 10.1097/00005176-199210000-00004. [DOI] [PubMed] [Google Scholar]
- 233.Henrick BM, Hutton AA, Palumbo MC, Casaburi G, Mitchell RD, Underwood MA, Smilowitz JT, Frese SA. 2018. elevated fecal pH indicates a profound change in the breastfed infant gut microbiome due to reduction of Bifidobacterium over the past century. mSphere 3:e00041-18. 10.1128/mSphere.00041-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chow J, Panasevich MR, Alexander D, Vester Boler BM, Rossoni Serao MC, Faber TA, Bauer LL, Fahey GC. 2014. Fecal metabolomics of healthy breast-fed versus formula-fed infants before and during in vitro batch culture fermentation. J Proteome Res 13:2534–2542. 10.1021/pr500011w. [DOI] [PubMed] [Google Scholar]
- 235.He X, Parenti M, Grip T, Lönnerdal B, Timby N, Domellöf M, Hernell O, Slupsky CM. 2019. Fecal microbiome and metabolome of infants fed bovine MFGM supplemented formula or standard formula with breast-fed infants as reference: a randomized controlled trial. Sci Rep 9:11589. 10.1038/s41598-019-47953-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.D’Aimmo MR, Mattarelli P, Biavati B, Carlsson NG, Andlid T. 2012. The potential of bifidobacteria as a source of natural folate. J Appl Microbiol 112:975–984. 10.1111/j.1365-2672.2012.05261.x. [DOI] [PubMed] [Google Scholar]
- 237.Crider KS, Yang TP, Berry RJ, Bailey LB. 2012. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv Nutr 3:21–38. 10.3945/an.111.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Aufreiter S, Gregory JF, III, Pfeiffer CM, Fazili Z, Kim YI, Marcon N, Kamalaporn P, Pencharz PB, O’Connor DL. 2009. Folate is absorbed across the colon of adults: evidence from cecal infusion of 13C-labeled [6S]-5-formyltetrahydrofolic acid. Am J Clin Nutr 90:116–123. 10.3945/ajcn.2008.27345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Stiles J, Jernigan TL. 2010. The basics of brain development. Neuropsychol Rev 20:327–348. 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Svennerholm L, Boström K, Fredman P, Månsson JE, Rosengren B, Rynmark BM. 1989. Human brain gangliosides: developmental changes from early fetal stage to advanced age. Biochim Biophys Acta 1005:109–117. 10.1016/0005-2760(89)90175-6. [DOI] [PubMed] [Google Scholar]
- 241.Uauy R, Peirano P. 1999. Breast is best: human milk is the optimal food for brain development. Am J Clin Nutr 70:433–434. 10.1093/ajcn/70.4.433. [DOI] [PubMed] [Google Scholar]
- 242.Uauy R, Mena P, Peirano P. 2001. Mechanisms for nutrient effects on brain development and cognition. Nestle Nutr Workshop Ser Clin Perform Programme 5:41–72. 10.1159/000061845. [DOI] [PubMed] [Google Scholar]
- 243.Goyal MS, Venkatesh S, Milbrandt J, Gordon JI, Raichle ME. 2015. Feeding the brain and nurturing the mind: linking nutrition and the gut microbiota to brain development. Proc Natl Acad Sci USA 112:14105–14112. 10.1073/pnas.1511465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wang B, Miller JB, McNeil Y, McVeagh P. 1998. Sialic acid concentration of brain gangliosides: variation among eight mammalian species. Comp Biochem Physiol A Mol Integr Physiol 119:435–439. 10.1016/S1095-6433(97)00445-5. [DOI] [PubMed] [Google Scholar]
- 245.Wang B. 2012. Molecular mechanism underlying sialic acid as an essential nutrient for brain development and cognition. Adv Nutr 3:465S–472S. 10.3945/an.112.001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wang B, McVeagh P, Petocz P, Brand-Miller J. 2003. Brain ganglioside and glycoprotein sialic acid in breastfed compared with formula-fed infants. Am J Clin Nutr 78:1024–1029. 10.1093/ajcn/78.5.1024. [DOI] [PubMed] [Google Scholar]
- 247.Goehring KC, Kennedy AD, Prieto PA, Buck RH. 2014. Direct evidence for the presence of human milk oligosaccharides in the circulation of breastfed infants. PLoS One 9:e101692. 10.1371/journal.pone.0101692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ruhaak LR, Stroble C, Underwood MA, Lebrilla CB. 2014. Detection of milk oligosaccharides in plasma of infants. Anal Bioanal Chem 406:5775–5784. 10.1007/s00216-014-8025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Gordon HA, Pesti L. 1971. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol Rev 35:390–429. 10.1128/MMBR.35.4.390-429.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Hooper LV, Gordon JI. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115–1118. 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 251.Round JL, Mazmanian SK. 2010. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA 107:12204–12209. 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Sims IM, Frese SA, Walter J, Loach D, Wilson M, Appleyard K, Eason J, Livingston M, Baird M, Cook G, Tannock GW. 2011. Structure and functions of exopolysaccharide produced by gut commensal Lactobacillus reuteri 100–23. ISME J 5:1115–1124. 10.1038/ismej.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. 2012. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336:489–493. 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. 2016. How colonization by microbiota in early life shapes the immune system. Science 352:539–544. 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Honda K, Littman DR. 2016. The microbiota in adaptive immune homeostasis and disease. Nature 535:75–84. 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 256.Macpherson AJ, Geuking MB, McCoy KD. 2005. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology 115:153–162. 10.1111/j.1365-2567.2005.02159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273. 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ganal-Vonarburg SC, Hornef MW, Macpherson AJ. 2020. Microbial-host molecular exchange and its functional consequences in early mammalian life. Science 368:604–607. 10.1126/science.aba0478. [DOI] [PubMed] [Google Scholar]
- 259.Gensollen T, Blumberg RS. 2017. Correlation between early-life regulation of the immune system by microbiota and allergy development. J Allergy Clin Immunol 139:1084–1091. 10.1016/j.jaci.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Sjögren YM, Tomicic S, Lundberg A, Böttcher MF, Björkstén B, Sverremark-Ekström E, Jenmalm MC. 2009. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin Exp Allergy 39:1842–1851. 10.1111/j.1365-2222.2009.03326.x. [DOI] [PubMed] [Google Scholar]
- 261.Lundell AC, Björnsson V, Ljung A, Ceder M, Johansen S, Lindhagen G, Törnhage CJ, Adlerberth I, Wold AE, Rudin A. 2012. Infant B cell memory differentiation and early gut bacterial colonization. J Immunol 188:4315–4322. 10.4049/jimmunol.1103223. [DOI] [PubMed] [Google Scholar]
- 262.Alessandri G, Ossiprandi MC, MacSharry J, van Sinderen D, Ventura M. 2019. Bifidobacterial dialogue with its human host and consequent modulation of the immune system. Front Immunol 10:2348. 10.3389/fimmu.2019.02348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Li M, Bai Y, Zhou J, Huang W, Yan J, Tao J, Fan Q, Liu Y, Mei D, Yan Q, Yuan J, Malard P, Wang Z, Gu J, Tanigchi N, Li W. 2019. Core fucosylation of maternal milk N-glycan evokes B cell activation by selectively promoting the l-fucose metabolism of gut Bifidobacterium spp. and Lactobacillus spp. mBio 10:e00128-19. 10.1128/mBio.00128-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Henrick BM, Rodriguez L, Lakshmikanth T, Pou C, Henckel E, Arzoomand A, Olin A, Wang J, Mikes J, Tan Z, Chen Y, Ehrlich AM, Bernhardsson AK, Mugabo CH, Ambrosiani Y, Gustafsson A, Chew S, Brown HK, Prambs J, Bohlin K, Mitchell RD, Underwood MA, Smilowitz JT, German JB, Frese SA, Brodin P. 2021. Bifidobacterium-mediated immune system imprinting early in life. Cell 184:3884–3898. 10.1016/j.cell.2021.05.030. [DOI] [PubMed] [Google Scholar]
- 265.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. 2003. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem 278:14112–14120. 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- 266.Newburg DS, Ruiz-Palacios GM, Morrow AL. 2005. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr 25:37–58. 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 267.Jantscher-Krenn E, Lauwaet T, Bliss LA, Reed SL, Gillin FD, Bode L. 2012. Human milk oligosaccharides reduce Entamoeba histolytica attachment and cytotoxicity in vitro. Br J Nutr 108:1839–1846. 10.1017/S0007114511007392. [DOI] [PubMed] [Google Scholar]
- 268.Manthey CF, Autran CA, Eckmann L, Bode L. 2014. Human milk oligosaccharides protect against enteropathogenic Escherichia coli attachment in vitro and EPEC colonization in suckling mice. J Pediatr Gastroenterol Nutr 58:165–168. 10.1097/MPG.0000000000000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yu ZT, Nanthakumar NN, Newburg DS. 2016. The human milk oligosaccharide 2’-fucosyllactose quenches Campylobacter jejuni-induced inflammation in human epithelial cells HEP-2 and HT-29 and in mouse intestinal mucosa. J Nutr 146:1980–1990. 10.3945/jn.116.230706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Facinelli B, Marini E, Magi G, Zampini L, Santoro L, Catassi C, Monachesi C, Gabrielli O, Coppa GV. 2019. Breast milk oligosaccharides: effects of 2′-fucosyllactose and 6′-sialyllactose on the adhesion of Escherichia coli and Salmonella fyris to Caco-2 cells. J Matern Fetal Neonatal Med 32:2950–2952. 10.1080/14767058.2018.1450864. [DOI] [PubMed] [Google Scholar]
- 271.Ramani S, Stewart CJ, Laucirica DR, Ajami NJ, Robertson B, Autran CA, Shinge D, Rani S, Anandan S, Hu L, Ferreon JC, Kuruvilla KA, Petrosino JF, Venkataram Prasad BV, Bode L, Kang G, Estes MK. 2018. Human milk oligosaccharides, milk microbiome and infant gut microbiome modulate neonatal rotavirus infection. Nat Commun 9:5010. 10.1038/s41467-018-07476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Morrow AL, Ruiz-Palacios GM, Altaye M, Jiang X, Guerrero ML, Meinzen-Derr JK, Farkas T, Chaturvedi P, Pickering LK, Newburg DS. 2004. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J Pediatr 145:297–303. 10.1016/j.jpeds.2004.04.054. [DOI] [PubMed] [Google Scholar]
- 273.Lin AE, Autran CA, Szyszka A, Escajadillo T, Huang M, Godula K, Prudden AR, Boons GJ, Lewis AL, Doran KS, Nizet V, Bode L. 2017. Human milk oligosaccharides inhibit growth of group B Streptococcus. J Biol Chem 292:11243–11249. 10.1074/jbc.M117.789974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Kuntz S, Kunz C, Rudloff S. 2009. Oligosaccharides from human milk induce growth arrest via G2/M by influencing growth-related cell cycle genes in intestinal epithelial cells. Br J Nutr 101:1306–1315. 10.1017/S0007114508079622. [DOI] [PubMed] [Google Scholar]
- 275.Srutkova D, Schwarzer M, Hudcovic T, Zakostelska Z, Drab V, Spanova A, Rittich B, Kozakova H, Schabussova I. 2015. Bifidobacterium longum CCM 7952 promotes epithelial barrier function and prevents acute DSS-induced colitis in strictly strain-specific manner. PLoS One 10:e0134050. 10.1371/journal.pone.0134050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Holscher HD, Bode L, Tappenden KA. 2017. Human milk oligosaccharides influence intestinal epithelial cell maturation in vitro. J Pediatr Gastroenterol Nutr 64:296–301. 10.1097/MPG.0000000000001274. [DOI] [PubMed] [Google Scholar]
- 277.Bienenstock J, Buck RH, Linke H, Forsythe P, Stanisz AM, Kunze WA. 2013. Fucosylated but not sialylated milk oligosaccharides diminish colon motor contractions. PLoS One 8:e76236. 10.1371/journal.pone.0076236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Victora CG, Bahl R, Barros AJ, França GV, Horton S, Krasevec J, Murch S, Sankar MJ, Walker N, Rollins NC, Lancet Breastfeeding Series Group. 2016. Breastfeeding in the 21st century: epidemiology, mechanisms, and lifelong effect. Lancet 387:475–490. 10.1016/S0140-6736(15)01024-7. [DOI] [PubMed] [Google Scholar]
- 279.Institute of Medicine (US) Committee on the Evaluation of the Addition of Ingredients New to Infant Formula. 2004. Infant formula: evaluating the safety of new ingredients. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 280.Newburg DS. 2005. Innate immunity and human milk. J Nutr 135:1308–1312. 10.1093/jn/135.5.1308. [DOI] [PubMed] [Google Scholar]
- 281.Grulee CG, Sanford HN, Herron PH. 1934. Breast and artificial feeding. Influence on morbidity and mortality of twenty thousand infants. JAMA 103:735–739. 10.1001/jama.1934.02750360011006. [DOI] [Google Scholar]
- 282.Bauer E, Thiele I. 2018. From network analysis to functional metabolic modeling of the human gut microbiota. mSystems 3:e00209-17. 10.1128/mSystems.00209-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Röttjers L, Faust K. 2018. From hairballs to hypotheses-biological insights from microbial networks. FEMS Microbiol Rev 42:761–780. 10.1093/femsre/fuy030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Vuong HE, Pronovost GN, Williams DW, Coley EJL, Siegler EL, Qiu A, Kazantsev M, Wilson CJ, Rendon T, Hsiao EY. 2020. The maternal microbiome modulates fetal neurodevelopment in mice. Nature 586:281–286. 10.1038/s41586-020-2745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Kalliomäki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. 2001. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357:1076–1079. 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 286.Wickens K, Black PN, Stanley TV, Mitchell E, Fitzharris P, Tannock GW, Purdie G, Crane J. 2008. Probiotic Study Group. A differential effect of 2 probiotics in the prevention of eczema and atopy: a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol 122:788–794. 10.1016/j.jaci.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 287.Markowitz JE, Bengmark S. 2002. Probiotics in health and disease in the pediatric patient. Pediatr Clin North Am 49:127–141. 10.1016/s0031-3955(03)00112-3. [DOI] [PubMed] [Google Scholar]
- 288.Savilahti E, Kukkonen K, Kuitunen M. 2009. Probiotics in the treatment and prevention of allergy in children. World Allergy Organ J 2:69–76. 10.1097/WOX.0b013e3181a45ee5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Tang ML, Lahtinen SJ, Boyle RJ. 2010. Probiotics and prebiotics: clinical effects in allergic disease. Curr Opin Pediatr 22:626–634. 10.1097/MOP.0b013e32833d9728. [DOI] [PubMed] [Google Scholar]
- 290.Elazab N, Mendy A, Gasana J, Vieira ER, Quizon A, Forno E. 2013. Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics 132:e666–e676. 10.1542/peds.2013-0246. [DOI] [PubMed] [Google Scholar]
- 291.Fiocchi A, Burks W, Bahna SL, Bielory L, Boyle RJ, Cocco R, Dreborg S, Goodman R, Kuitunen M, Haahtela T, Heine RG, Lack G, Osborn DA, Sampson H, Tannock GW, Lee BW, WAO Special Committee on Food Allergy and Nutrition. 2012. Clinical use of probiotics in pediatric allergy (CUPPA): a world allergy organization position paper. World Allergy Organ J 5:148–167. 10.1097/WOX.0b013e3182784ee0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Fooladi AA, Khani S, Hosseini HM, Mousavi SF, Aghdam EM, Nourani MR. 2013. Impact of altered early infant gut microbiota following breastfeeding and delivery mode on allergic diseases. Inflamm Allergy Drug Targets 12:410–418. 10.2174/1871528112666131205113129. [DOI] [PubMed] [Google Scholar]
- 293.Zuccotti G, Meneghin F, Aceti A, Barone G, Callegari ML, Di Mauro A, Fantini MP, Gori D, Indrio F, Maggio L, Morelli L, Corvaglia L, Italian Society of Neonatology. 2015. Probiotics for prevention of atopic diseases in infants: systematic review and meta-analysis. Allergy 70:1356–1371. 10.1111/all.12700. [DOI] [PubMed] [Google Scholar]
- 294.Grave GD, Nelson SA, Walker WA, Moss RL, Dvorak B, Hamilton FA, Higgins R, Raju TN. 2007. New therapies and preventive approaches for necrotizing enterocolitis: report of a research planning workshop. Pediatr Res 62:510–514. 10.1203/PDR.0b013e318142580a. [DOI] [PubMed] [Google Scholar]
- 295.Lin HC, Su BH, Chen AC, Lin TW, Tsai CH, Yeh TF, Oh W. 2005. Oral probiotics reduce the incidence and severity of necrotizing enterocolitis in very low birth weight infants. Pediatrics 115:1–4. 10.1542/peds.2004-1463. [DOI] [PubMed] [Google Scholar]
- 296.Wong CB, Iwabuchi N, Xiao JZ. 2019. Exploring the science behind Bifidobacterium breve M-16V in infant health. Nutrients 11:1724. 10.3390/nu11081724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Meyer MP, Chow SSW, Alsweiler J, Bourchier D, Broadbent R, Knight D, Lynn AM, Patel H. 2020. Probiotics for prevention of severe necrotizing enterocolitis: experience of New Zealand neonatal intensive care units. Front Pediatr 8:119. 10.3389/fped.2020.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Robertson C, Savva GM, Clapuci R, Jones J, Maimouni H, Brown E, Minocha A, Hall LJ, Clarke P. 2020. Incidence of necrotising enterocolitis before and after introducing routine prophylactic Lactobacillus and Bifidobacterium probiotics. Arch Dis Child Fetal Neonatal Ed 105:380–386. 10.1136/archdischild-2019-317346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Frese SA, Hutton AA, Contreras LN, Shaw CA, Palumbo MC, Casaburi G, Xu G, Davis JCC, Lebrilla CB, Henrick BM, Freeman SL, Barile D, German JB, Mills DA, Smilowitz JT, Underwood MA. 2017. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere 2:e00501-17. 10.1128/mSphere.00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Henrick BM, Chew S, Casaburi G, Brown HK, Frese SA, Zhou Y, Underwood MA, Smilowitz JT. 2019. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr Res 86:749–757. 10.1038/s41390-019-0533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Chua MC, Ben-Amor K, Lay C, Goh AEN, Chiang WC, Rao R, Chew C, Chaithongwongwatthana S, Khemapech N, Knol J, Chongsrisawat V. 2017. Effect of synbiotic on the gut microbiota of cesarean delivered infants: a randomized, double-blind, multicenter study. J Pediatr Gastroenterol Nutr 65:102–106. 10.1097/MPG.0000000000001623. [DOI] [PubMed] [Google Scholar]
- 302.Mohan R, Koebnick C, Schildt J, Schmidt S, Mueller M, Possner M, Radke M, Blaut M. 2006. Effects of Bifidobacterium lactis Bb12 supplementation on intestinal microbiota of preterm infants: a double-blind, placebo-controlled, randomized study. J Clin Microbiol 44:4025–4031. 10.1128/JCM.00767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Mohan R, Koebnick C, Schildt J, Mueller M, Radke M, Blaut M. 2008. Effects of Bifidobacterium lactis Bb12 supplementation on body weight, fecal pH, acetate, lactate, calprotectin, and IgA in preterm infants. Pediatr Res 64:418–422. 10.1203/PDR.0b013e318181b7fa. [DOI] [PubMed] [Google Scholar]
- 304.Palframan RJ, Gibson GR, Rastall RA. 2003. Carbohydrate preferences of Bifidobacterium species isolated from the human gut. Curr Issues Intest Microbiol 4:71–75. [PubMed] [Google Scholar]
- 305.Boehm G, Moro G. 2008. Structural and functional aspects of prebiotics used in infant nutrition. J Nutr 138:1818S–1828S. 10.1093/jn/138.9.1818S. [DOI] [PubMed] [Google Scholar]
- 306.Macfarlane GT, Steed H, Macfarlane S. 2008. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J Appl Microbiol 104:305–344. 10.1111/j.1365-2672.2007.03520.x. [DOI] [PubMed] [Google Scholar]
- 307.Barboza M, Sela DA, Pirim C, Locascio RG, Freeman SL, German JB, Mills DA, Lebrilla CB. 2009. Glycoprofiling bifidobacterial consumption of galacto-oligosaccharides by mass spectrometry reveals strain-specific, preferential consumption of glycans. Appl Environ Microbiol 75:7319–7325. 10.1128/AEM.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Oozeer R, van Limpt K, Ludwig T, Ben Amor K, Martin R, Wind RD, Boehm G, Knol J. 2013. Intestinal microbiology in early life: specific prebiotics can have similar functionalities as human-milk oligosaccharides. Am J Clin Nutr 98:561S–571S. 10.3945/ajcn.112.038893. [DOI] [PubMed] [Google Scholar]
- 309.Torres DPM, Gonçalves MDPF, Teixeira JA, Rodrigues LR. 2010. Galacto-Oligosaccharides: production, properties, applications, and significance as prebiotics. Compr Rev Food Sci Food Saf 9:438–454. 10.1111/j.1541-4337.2010.00119.x. [DOI] [PubMed] [Google Scholar]
- 310.Flores-Maltos DA, Mussatto SI, Contreras-Esquivel JC, Rodríguez-Herrera R, Teixeira JA, Aguilar CN. 2016. Biotechnological production and application of fructooligosaccharides. Crit Rev Biotechnol 36:259–267. 10.3109/07388551.2014.953443. [DOI] [PubMed] [Google Scholar]
- 311.van Leeuwen SS, Kuipers BJH, Dijkhuizen L, Kamerling JP. 2016. Comparative structural characterization of 7 commercial galacto-oligosaccharide (GOS) products. Carbohydr Res 425:48–58. 10.1016/j.carres.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 312.Watson D, O’Connell MM, Schoterman MHC, van Neerven RJJ, Nauta A, van Sinderen D. 2013. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J Appl Microbiol 114:1132–1146. 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- 313.Sotoya H, Shigehisa A, Hara T, Matsumoto H, Hatano H, Matsuki T. 2017. Identification of genes involved in galactooligosaccharide utilization in Bifidobacterium breve strain YIT 4014T. Microbiology (Reading) 163:1420–1428. 10.1099/mic.0.000517. [DOI] [PubMed] [Google Scholar]
- 314.Valdés-Varela L, Ruas-Madiedo P, Gueimonde M. 2017. In vitro fermentation of different fructo-oligosaccharides by Bifidobacterium strains for the selection of synbiotic combinations. Int J Food Microbiol 242:19–23. 10.1016/j.ijfoodmicro.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 315.Moro G, Minoli I, Mosca M, Fanaro S, Jelinek J, Stahl B, Boehm G. 2002. Dosage-related bifidogenic effects of galacto- and fructooligosaccharides in formula-fed term infants. J Pediatr Gastroenterol Nutr 34:291–295. 10.1097/00005176-200203000-00014. [DOI] [PubMed] [Google Scholar]
- 316.Bakker-Zierikzee AM, Alles MS, Knol J, Kok FJ, Tolboom JJ, Bindels JG. 2005. Effects of infant formula containing a mixture of galacto- and fructo-oligosaccharides or viable Bifidobacterium animalis on the intestinal microflora during the first 4 months of life. Br J Nutr 94:783–790. 10.1079/BJN20051451. [DOI] [PubMed] [Google Scholar]
- 317.Knol J, Scholtens P, Kafka C, Steenbakkers J, Gro S, Helm K, Klarczyk M, Schöpfer H, Böckler HM, Wells J. 2005. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J Pediatr Gastroenterol Nutr 40:36–42. 10.1097/00005176-200501000-00007. [DOI] [PubMed] [Google Scholar]
- 318.Holscher HD, Faust KL, Czerkies LA, Litov R, Ziegler EE, Lessin H, Hatch T, Sun S, Tappenden KA. 2012. Effects of prebiotic-containing infant formula on gastrointestinal tolerance and fecal microbiota in a randomized controlled trial. JPEN J Parenter Enteral Nutr 36:95S–105S. 10.1177/0148607111430087. [DOI] [PubMed] [Google Scholar]
- 319.Vandenplas Y, De Greef E, Veereman G. 2014. Prebiotics in infant formula. Gut Microbes 5:681–687. 10.4161/19490976.2014.972237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Matsuki T, Tajima S, Hara T, Yahagi K, Ogawa E, Kodama H. 2016. Infant formula with galacto-oligosaccharides (OM55N) stimulates the growth of indigenous bifidobacteria in healthy term infants. Benef Microbes 7:453–461. 10.3920/BM2015.0168. [DOI] [PubMed] [Google Scholar]
- 321.Akkerman R, Faas MM, de Vos P. 2019. Non-digestible carbohydrates in infant formula as substitution for human milk oligosaccharide functions: effects on microbiota and gut maturation. Crit Rev Food Sci Nutr 59:1486–1497. 10.1080/10408398.2017.1414030. [DOI] [PubMed] [Google Scholar]
- 322.Borewicz K, Suarez-Diez M, Hechler C, Beijers R, de Weerth C, Arts I, Penders J, Thijs C, Nauta A, Lindner C, Van Leusen E, Vaughan EE, Smidt H. 2019. The effect of prebiotic fortified infant formulas on microbiota composition and dynamics in early life. Sci Rep 9:2434. 10.1038/s41598-018-38268-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Sims IM, Tannock GW. 2020. Galacto- and fructo-oligosaccharides utilized for growth by cocultures of bifidobacterial species characteristic of the infant gut. Appl Environ Microbiol 86:e00214-20. 10.1128/AEM.00214-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Lawley B, Munro K, Hughes A, Hodgkinson AJ, Prosser CG, Lowry D, Zhou SJ, Makrides M, Gibson RA, Lay C, Chew C, Lee PS, Wong KH, Tannock GW. 2017. Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. PeerJ 5:e3375. 10.7717/peerj.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Marriage BJ, Buck RH, Goehring KC, Oliver JS, Williams JA. 2015. Infants fed a lower calorie formula with 2′FL show growth and 2′FL uptake like breast-fed infants. J Pediatr Gastroenterol Nutr 61:649–658. 10.1097/MPG.0000000000000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Puccio G, Alliet P, Cajozzo C, Janssens E, Corsello G, Sprenger N, Wernimont S, Egli D, Gosoniu L, Steenhout P. 2017. Effects of infant formula with human milk oligosaccharides on growth and morbidity: a randomized multicenter trial. J Pediatr Gastroenterol Nutr 64:624–631. 10.1097/MPG.0000000000001520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Vandenplas Y, Berger B, Carnielli VP, Ksiazyk J, Lagström H, Sanchez LM, Migacheva N, Mosselmans JM, Picaud JC, Possner M, Singhal A, Wabitsch M. 2018. Human milk oligosaccharides: 2′-fucosyllactose (2′-fl) and lacto-N-neotetraose (LNnT) in infant formula. Nutrients 10:1161. 10.3390/nu10091161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Berger PK, Plows JF, Jones RB, Alderete TL, Yonemitsu C, Poulsen M, Ryoo JH, Peterson BS, Bode L, Goran MI. 2020. Human milk oligosaccharide 2′-fucosyllactose links feedings at 1 month to cognitive development at 24 months in infants of normal and overweight mothers. PLoS One 15:e0228323. 10.1371/journal.pone.0228323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.EFSA NDA Panel (EFSA Panel on Dietetic Products, Nutrition, and Allergies). 2015. Scientific opinion on the safety of lacto-N-neotetraose as a novel food ingredient pursuant to regulation (EC) no 258/97. EFSA J 13:4183. [Google Scholar]
- 330.EFSA NDA Panel (EFSA Panel on Dietetic Products, Nutrition, and Allergies). 2015. Scientific opinion on the safety of 2′-O-fucosyllactose as a novel food ingredient pursuant to regulation (EC) no 258/97. EFSA J 13:4184. 10.2903/j.efsa.2015.4184. [DOI] [Google Scholar]
- 331.Fischbach MA, Segre JA. 2016. Signaling in host-associated microbial communities. Cell 164:1288–1300. 10.1016/j.cell.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Cox LM, Weiner HL. 2018. Microbiota signaling pathways that influence neurologic disease. Neurotherapeutics 15:135–145. 10.1007/s13311-017-0598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.He J, Zhang P, Shen L, Niu L, Tan Y, Chen L, Zhao Y, Bai L, Hao X, Li X, Zhang S, Zhu L. 2020. Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int J Mol Sci 21:6356. 10.3390/ijms21176356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Deleu S, Machiels K, Raes J, Verbeke K, Vermeire S. 2021. Short chain fatty acids and its producing organisms: an overlooked therapy for IBD? EBioMedicine 66:103293. 10.1016/j.ebiom.2021.103293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Cait A, Cardenas E, Dimitriu PA, Amenyogbe N, Dai D, Cait J, Sbihi H, Stiemsma L, Subbarao P, Mandhane PJ, Becker AB, Moraes TJ, Sears MR, Lefebvre DL, Azad MB, Kollmann T, Turvey SE, Mohn WW. 2019. Reduced genetic potential for butyrate fermentation in the gut microbiome of infants who develop allergic sensitization. J Allergy Clin Immunol 144:1638–1647. 10.1016/j.jaci.2019.06.029. [DOI] [PubMed] [Google Scholar]
- 336.Roduit C, Frei R, Ferstl R, Loeliger S, Westermann P, Rhyner C, Schiavi E, Barcik W, Rodriguez-Perez N, Wawrzyniak M, Chassard C, Lacroix C, Schmausser-Hechfellner E, Depner M, von Mutius E, Braun-Fahrländer C, Karvonen AM, Kirjavainen PV, Pekkanen J, Dalphin JC, Riedler J, Akdis C, Lauener R, O’Mahony L, PASTURE/EFRAIM study group. 2019. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 74:799–809. 10.1111/all.13660. [DOI] [PubMed] [Google Scholar]
- 337.Rapley G, Murkett T. 2008. Baby-led weaning: helping your baby love good food. Random House, London, United Kingdom. [Google Scholar]
- 338.Brown A, Lee M. 2011. A descriptive study investigating the use and nature of baby-led weaning in a UK sample of mothers. Matern Child Nutr 7:34–47. 10.1111/j.1740-8709.2010.00243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Beal JA. 2016. Baby-led weaning. MCN Am J Matern Child Nurs 41:373. 10.1097/NMC.0000000000000283. [DOI] [PubMed] [Google Scholar]
- 340.D’Andrea E, Jenkins K, Mathews M, Roebothan B. 2016. Baby-led weaning: a preliminary investigation. Can J Diet Pract Res 77:72–77. 10.3148/cjdpr-2015-045. [DOI] [PubMed] [Google Scholar]
- 341.Morison BJ, Taylor RW, Haszard JJ, Schramm CJ, Williams EL, Fangupo LJ, Fleming EA, Luciano A, Heath AL. 2016. How different are baby-led weaning and conventional complementary feeding? A cross-sectional study of infants aged 6–8 months. BMJ Open 6:e010665. 10.1136/bmjopen-2015-010665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Fangupo LJ, Heath AM, Williams SM, Erickson Williams LW, Morison BJ, Fleming EA, Taylor BJ, Wheeler BJ, Taylor RW. 2016. A baby-led approach to eating solids and risk of choking. Pediatrics 138:e20160772. 10.1542/peds.2016-0772. [DOI] [PubMed] [Google Scholar]