Abstract
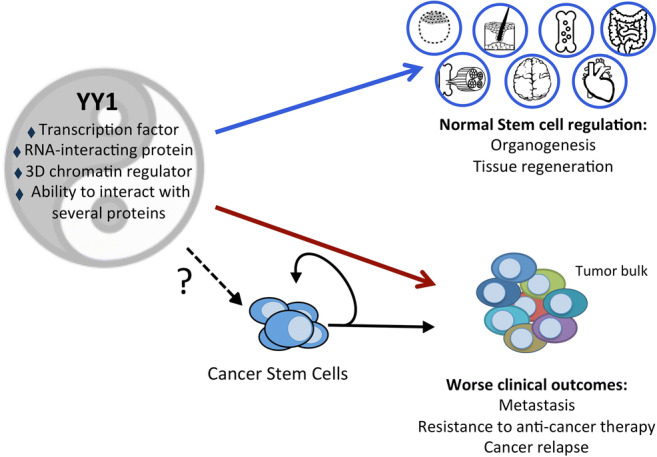
Accumulating evidence strongly indicates that the presence of cancer stem cells (CSCs) leads to the emergence of worse clinical scenarios, such as chemo- and radiotherapy resistance, metastasis, and cancer recurrence. CSCs are a highly tumorigenic population characterized by self-renewal capacity and differentiation potential. Thus, CSCs establish a hierarchical intratumor organization that enables tumor adaptation to evade the immune response and resist anticancer therapy. YY1 functions as a transcription factor, RNA-binding protein, and 3D chromatin regulator. Thus, YY1 has multiple effects and regulates several molecular processes. Emerging evidence indicates that the development of lethal YY1-mediated cancer phenotypes is associated with the presence of or enrichment in cancer stem-like cells. Therefore, it is necessary to investigate whether and to what extent YY1 regulates the CSC phenotype. Since CSCs mirror the phenotypic behavior of stem cells, we initially describe the roles played by YY1 in embryonic and adult stem cells. Next, we scrutinize evidence supporting the contributions of YY1 in CSCs from a number of various cancer types. Finally, we identify new areas for further investigation into the YY1-CSCs axis, including the participation of YY1 in the CSC niche.
Graphical Abstract
Keywords: YY1, Cancer stem cells, Self‐renewal, Differentiation potential
Introduction
Yin yang-1 (YY1) has been identified as a transcription factor with a dual function activating or repressing gene expression. It recognizes and binds to specific DNA sequences through its COOH-terminal region, which harbors four C2H2-type zinc finger motifs. YY1 also contains a REPO domain, which is an anchoring element 25 amino acids in length that recruits the polycomb group (PcG) proteins to DNA [1]. Thus, YY1 mediates a closed chromatin conformation by recruiting the polycomb repressive complex 2 (PRC2) containing histone methyltransferase EHZ2, which induces methylation of histone H3 at lysine 27 [2]. In contrast, YY1 mediates transcriptional activation by interacting with the INO80 chromatin remodeling complex in the regulatory regions of the genes [3]. Interestingly, the YY1-INO80 axis is also associated with homologous recombination-based DNA repair by potentially binding to a recombination intermediate structure [4]. Recently, it was shown that YY1 mediates the formation of genome-wide loops, contributing to the expression of cell type-specific genes [5–7]. Importantly, YY1 is able to interact with a number of RNA molecules, such as long noncoding RNAs (lncRNAs) [8–10], and proteins, including HDACs [11, 12], c-Myc [13], CTCF [14], p53 [15], Sp1 [16], and p300 [17]. Consequently, YY1 is implicated in several processes, such as transcriptional regulation, epigenetic modulation, chromatin remodeling, DNA repair, cell proliferation, cell death, differentiation, and development.
In cancer, the expression levels or functions of YY1 are commonly altered. Specifically, high expression levels of YY1 have been detected in several cancer types, including prostate [18, 19], breast [20], cervical [21, 22], glioma [23, 24], gastric [25], melanoma [26], liver [27], lung [28], etc. Notably, YY1 expression levels correlate with worse clinical outcomes, such as metastasis, invasion, resistance to anticancer therapy, and cancer recurrence [29–31]. Furthermore, YY1 is involved in multiple molecular mechanisms, enhancing all hallmarks of cancer [31]. Therefore, YY1 is conceptualized as an oncogenic protein fueling the emergence of lethal cancer phenotypes.
The cancer stem cell (CSC) model suggests the presence of two functional cell types in a tumor: CSCs and non-CSCs. The first type is a highly tumorigenic subpopulation with stem-like functional features: self-renewal capacity and differentiation potential. Similar to normal stem cells (SCs), CSCs proliferate by undergoing symmetrical cell divisions, securing their self-renewal and differentiation potentials [32, 33]. Non-CSCs are nontumorigenic heterogeneous cancer cells, hierarchically generated from CSCs through asymmetrical cell divisions [34, 35]. The frequency of CSCs is variable from 0.1 % or less up to 40 % [36–39], and non-CSCs certainly form the bulk of a tumor. Importantly, CSCs lead to the emergence of highly lethal cancer phenotypes associated with metastasis, chemo- and radiotherapy resistance, and cancer recurrence.
YY1 triggers the development of deadly cancer phenotypes with enriched presence of highly tumorigenic cells in the tumors, suggesting that YY1 may regulate the CSC phenotype. Here, we present evidence linking these two fields of cancer research that share a common denominator, to be implicated in the generation of worse clinical scenarios. CSCs mirror the stemness properties of self-renewal and differentiation potential; thus, we describe the role of YY1 in the biology of embryonic and adult SCs as a biological framework to show the role of YY1 in CSCs in various cancer types. Finally, we present possible new areas for future studies into the YY1-CSCs axis.
YY1 Regulates Early Embryonic Development
Stemness potential plays an essential role during embryonic development. Thus, evaluation of the relevance of YY1 to the regulation of stemness potential requires assessment of its role during embryonic development. Donohoe and colleagues observed a very early lethal phenotype in YY1 knockout mice because the embryos died during the peri-implantation period at the blastocyst stage [40]. The blastocyst stage is characterized by the presence of ICM, blastocoelic cavity, and trophoblasts. Importantly, YY1 ablation or downregulation decreased the expression of the master regulators of embryogenesis, Oct4 and Sox2, in the inner cell mass (ICM, also known as the embryoblast) [41]. Interestingly, trophoblasts that form the outer layer of the blastocyst express high levels of YY1 [40]. Similarly, YY1 downregulation has been associated with recurrent miscarriage in patients [42]. Therefore, YY1 is essential for early embryonic development.
Embryonic stem cells (ESCs) are isolated from the ICM during the blastocyst stage of embryonic development. Notably, YY1 is required for stemness potential of these cells. Three gene expression subsignatures, known as the Core, PRC, and Myc modules, establish the regulatory circuitry of ESC responsible for self-renewal and pluripotency [43–45]. The transcription factors Oct4, Sox2, and Nanog mediate the core gene expression module by forming specialized autoregulatory and feedforward loops via interactions with their own and target promoter sequences [44, 45]. The PRC module is defined as those gene co-occupied and regulated by Suz12, Eed, Phc1, and Rnf2 [43]. The Myc module regulates more genes than the Core or PRC modules. Notably, the Myc module is overrepresented in cancer tissue samples [43]. Importantly, YY1 is apparently associated with the three modules of stemness regulation in ESCs. Specifically, YY1 mediates transcriptional activation of the Oct4 and Sox2 genes by forming long-range interactions by looping [46]. Moreover, YY1 can interact with members of the BAF complex, such as Smarc4, in the regulatory regions of transcriptionally activated pluripotent genes Nanog, Oct4, Myc, and others [47]. Thus, YY1 is associated with the Core module by transactivating its crucial transcription factors. Additionally, YY1 is associated with the PRC and Myc modules by cointeracting with a subset of the regulatory regions of certain genes. Interestingly, YY1 polarizes its binding from these regulatory regions to ones associated with the Core module upon interaction with Smac4 [47]. Furthermore, YY1 and transcription factors of the Myc module, including Myc itself, cobind to a subset of regulatory DNA sequences to drive their transcriptional activation, highlighting that YY1 may contribute to the regulatory circuitry of ESC [48]. In agreement with the role of YY1 in the three regulatory modules of ESC, YY1 downregulation decreases pluripotency and concomitantly induces ESC differentiation [47]. Interestingly, YY1 drives active gene expression in ESCs by interacting with the INO80 remodeling complex, whereas the YY1-PRC2 complex axis mediates gene silencing [48]. Therefore, YY1 regulates the transcriptional landscape of ESCs and is thus essential for the biology of ESCs.
After the blastocyst is implanted in the uterus, the primitive streak is formed marking the initiation of gastrulation, which results in the formation of a trilaminar embryonic disc from a bilaminar embryonic disc originating from the ICM. Significantly, YY1 ablation results in failed migration of primitive streak-derived cells into the interior of the embryo. This effect results from persistent E-cadherin expression that inhibits epithelial-mesenchymal transition (EMT) and generation of three germ cell layers [49]. Thus, YY1 is essential for the establishment of three germ layers: ectoderm, mesoderm, and endoderm.
YY1 Modulates the Stemness Potential During Organogenesis
YY1 plays a critical role in the early stages of embryonic development; thus, YY1 is expected to play a significant role in organogenesis by regulating the generation of lineage-committed stem and progenitor cells and modulating their stemness potential. Generation of conditional YY1 knockout mice demonstrated the role of YY1 during organogenesis (Table 1). The following sections present and discuss the information on the role of YY1 in the stemness potential that governs organ formation and residing adult stem cells. To have a better understanding of the available information, it will be presented taking as reference the three germ layers.
Table 1.
Role of YY1 in organogenesis
| Germ layer | Tissue or organ analyzed | Mouse models | Major findings | Ref. |
|---|---|---|---|---|
| Ectoderm | Brain |
• Disruption of one allele in ES cells by homologous recombination before microinjection into C57BL/6 blastocysts generated heterozygous YY1 mice. • Conditional YY1 ablation in cerebral cortex was achieved by generating the Emx1-Cre;YY1f/f mouse line. • The Emx1-CreER(t2);YY1f/f mouse line induced time-dependent conditional YY1 KO in the cerebral cortex. • p53 ablation in cerebral cortex lacking YY1 expression was achieved by engineering the Emx1-Cre;YY1f/f Trp53f/f mouse line. • YY1 ablation in early neuroepithelial cells in the mid-hindbrain region was achieved by generating the mouse line Cre-En1;YY1f/f. • Ablation of YY1 in premigratory neural crest stem cells (NCSCs) was achieve by generating the Wnt1-Cre;YY1f/f transgenic mouse line. |
• Heterozygous YY1 KO embryos showed brain defects, such as altered brain symmetries and exencephaly. ▪ YY1 ablation in the cerebral cortex decreased the forebrain size by inducing apoptosis-mediated cell death. Notably, p53 KO partially rescued the YY1 ablation phenotype in this brain region. ▪ YY1 regulated earlier stages of neural progenitor cell differentiation by gene expression associated with mitochondrial function and protein translation. ▪ YY1 ablation in the mid-hindbrain region provoked perinatal death due to dorsal midbrain hypoplasia and cerebellar agenesis. ▪ Neuroepithelial cells lacking YY1 expression inhibited cell cycle progression and activated apoptosis. ▪ Homozygous deletion of the YY1 gene in NCSCs induced craniofacial and midbrain defects. |
[40, 50–52] |
| Skin |
• Time-dependent homozygous deletion of the YY1 gene in postmigratory neural crest was achieved by generating Sox10-CreER(T2);YY1 f/f transgenic mice. • YY1 was time-dependently ablated in the adult melanocytic lineage by generating the Tyr-CreER(T2);YY1f/f mouse line. |
• Ablation of the YY1 gene in early stages of neural crest development reduced the melanocyte population. Additionally, these embryos presented a reduction in the size of the dorsal root ganglia. • The survival and proliferation rates of adult melanocyte stem cells were compromised after YY1 ablation. |
[50, 53]. | |
| Mesoderm | Hematopoietic system |
• Time-dependent conditional YY1 ablation was achieved by generating the Mx1-Cre;YY1f/f mouse line. • Competitive congenic bone marrow transplantations were performed into lethally irradiated mice (CD45.1+) by using a mixture of CD45.2+ bone marrow progenitors (Mx1-Cre or Mx1-Cre;YY1f/f) with CD45.1 competitor cells at the 1:1 or 1:9 ratios. • Bone marrow transplantations were achieved using Mx1-Cre;YY1 f/f bone marrow progenitor cells that overexpressed either YY1 or its variant lacking the REPO domain. |
• YY1 ablation in bone marrow progenitors induced a pancytopenic phenotype. • LT-HSC, ST-HSC, multipotent progenitor cells, and myeloid linages were increased by ectopic expression of YY1. • Ectopic expression of YY1 inhibited B-cell population. • YY1 ablation exhausted HSC population by lowering c-kit singling to induce the loss of the quiescence state. • Ectopic expression of YY1 or YY1ΔREPO rescued from the YY1 ablation phenotype. |
[54] |
| Hearth |
• Ablation of the YY1 gene in mesodermal precursor cells was achieved by generating the Msp1-Cre;YY1f/f transgenic mouse line. • YY1 ablation in cardiac progenitor cells (CPCs) was achieved by generating the Nkx2.5-Cre;YY1f/f mouse line. • Y1 expression was knocked out in cardiomyocytes by generating the α-MHC-Cre;YY1 f/f mouse line. |
• Specific deletion of the YY1 gene in mesodermal cells resulted in early embryonic lethality due to the loss of the CPC population. • Conditional ablation of YY1 in the CPCs during cardiac development resulted in developmental defects. • Deletion of YY1 in cardiomyocytes provoked congenital defects in the heart. |
[55, 56]. | |
| Skeletal muscle |
• YY1 ablation in muscle stem cells was achieved by generating the Pax7-Cre;YY1f/f mouse line. • Time-dependent YY1 ablation in satellite cells was achieved by generating the Pax7-CreER(T2)/+;YY1f/f mouse line. • Conditional YY ablation in skeletal muscle cell was achieved by generating the Myo-Cre;YY1 f/f mouse line |
▪ Ablation of the YY1 gene in muscle progenitor cells induced neonatal death by suffocation due to insufficient diaphragm development. ▪ Injury-induced muscle regeneration was not correctly performed by adult satellite cells lacking YY1 expression. ▪ Loss of YY1 expression in satellite cells induced transcriptional activation of mitochondrial genes that are normally repressed by the YY1-PCR2 complex. ▪ Glycolytic HIF1α-responsive genes were inhibited in muscle stem cells lacking YY1 expression due to the absence of YY1-mediated stabilization of HIF1-α. |
[57, 58] | |
| Endoderm | Intestine |
• Time-dependent YY1 ablation in intestinal epithelium was achieved by generating the Villin-CreER(T2);YY1f/f mouse line. • Intestinal Lrg5+ stem cells lacking YY1 expression were traced by crossing Villin-CreER(T2);YY1 f/f mice with Lrg5+-EGFP-Ires-CreER(T2) mice. • YY1 ablation in the intestinal epithelium was achieved by generating the Shh-Cre;YY1f/f mouse line. |
▪ Loss of YY1 expression in the intestinal epithelium was incompatible with life. ▪ Intestinal Lgr5+ stem cells lacking YY1 expression differentiated and migrated to the villi leaving their crypt base localization. ▪ YY1 was essential for the maintenance of the stemness potential of intestinal stem cells by regulating the gene expression associated with the mitochondrial function. |
[59, 60]. |
| Lung | • YY1 ablation in the lung mesenchyme was achieved by generating Dermo-Cre;YY1 f/f or Shh-Cre;YY1f/f mice |
• The loss of YY1 expression in the lung mesenchyme compromised the lung development. Importantly, club cells and type-1 pneumocytes were dramatically decreased in the pulmonary epithelium. • Deletion of the YY1 gene in the pulmonary epithelium resulted in respiratory failures at birth. Activation of apoptosis and reduced proliferation were detected in the pulmonary epithelium lacking YY1 expression. Additionally, several cell types were lost due to YY1 ablation, such as club, ciliated, goblet, and smooth muscle cells. |
[61, 62] |
YY1 Regulates Stem Cells Derived from the Ectodermal Lineage
The role of YY1 in stem cells associated with the brain and epidermal development from the ectoderm germ cell lineages was investigated using conditional KO mice (Table 1).
Role of YY1 in Brain Development
YY1-mediated functions have been extensively characterized in the brain. The earliest description of the effects of YY1 on embryonic development was reported by Donohae et al. The authors demonstrated that a small proportion of embryos carrying heterozygous YY1 allele ablation exhibited growth and neurulation defects and structural brain alterations [40]. Conditional knockout of YY1 in premigratory neural crest cells was shown to induce embryonic death at E14.5, presenting midbrain and craniofacial defects. Additionally, a marked reduction in neural crest derivatives located in the enteric nervous system, dorsal root ganglia, and skin was observed in this model [50]. Dong and colleagues demonstrated that Cre-mediated deletion of the YY1 gene in the mid-hindbrain region induced hypoplasia and cerebellar agenesis by inhibiting cell cycle progression and inducing apoptotic death in neuroepithelial stem cells in a p53-dependent manner [51]. Another study demonstrated that knockout of the YY1 gene during cortex development decreased the forebrain size due to reduced proliferation and increased apoptotic death in cortical progenitor cells [52]. Interestingly, the ablation of YY1 affected neuronal progenitor cells (NPCs) at the earlier stages of differentiation during cortex development [52]. Similarly, Beagan et al. showed that YY1 is essential for NPC commitment by mediating the formation of specific DNA loops that attract distal and proximal regulatory elements to genomic regions demarcated by CTCF [6]. Thus, loss of YY1 compromises stemness potential of neural crest stem cells and progenitors cells, such as NPCs.
Determination of the molecular mechanisms governed by YY1 in NPCs required high-throughput technologies. Specifically, RNA-seq, YY1 ChIP-seq, and functional analyses were performed in NPCs after YY1 ablation. The lack of YY1 expression downregulated direct target genes associated with mitochondrial metabolism and protein translation [52]. On the other hand, a decrease in the cortex size induced by YY1 ablation was partially rescued by inhibiting cell death by knockout of the p53 gene [52]. YY1 negatively regulates p53 function [15, 63], and this molecular mechanism appears to be essential for brain development. Similarly, YY1 deletion in the mid-hindbrain region induced p53-dependent cell death [51]. Interestingly, YY1 knockout in this region compromised YY1-mediated Wnt1 transcriptional activation [51]. Importantly, ectopic expression demonstrated that Wnt1 is a key factor that regulates mid-hindbrain size [64]. Overall, studies using all these mouse models demonstrated that YY1 modulates brain development by regulating the proliferation, cell death, mitochondrial metabolism, and 3D chromatin configuration. Importantly, alterations in the human YY1 gene lead to craniofacial abnormalities accompanied by deficient intellectual abilities such as those observed in YY1 and Gabriele-de Vries syndromes [65, 66]. Investigations of patients diagnosed with YY1 syndrome showed that the affinity of YY1 interaction with its target DNA sites is reduced, provoking the global loss of H3K27 acetylation [65]. Thus, YY1 is a key regulator of cerebral organogenesis, and alterations in YY1 lead to several brain pathologies.
Role of YY1 in Skin Development
Neural crest (NC) cells are an embryonic subpopulation involved in the generation of diverse cell types and tissues, such as connective tissue, cartilage, bone, and endocrine and melanocyte cells. Knockout of the YY1 gene in premigratory NC cells lead to embryonic lethality at E14.5. Additionally, ablation of YY1 reduced NC derivatives, including melanocytes, at E13.5 [50]. Moreover, time-dependent conditional ablation of YY1 in postmigratory NC cells at E10.5 decreased the melanocyte subpopulation [50]. Cre-mediated YY1 ablation in the melanocytic lineage was not lethal; however, these mice exhibited lighter skin pigmentation and hair graying [50, 53]. Thus, YY1 is essential for the stemness potential of both embryonic and adult melanocyte stem cells.
YY1 Regulates Stem Cells Derived from the Mesoderm Lineage
The mesoderm is the last germ layer formed in the trilaminar embryonic disc and marks the beginning of the gastrulation stage of embryonic development. Experiments using YY1 KO embryos showed that YY1 plays an essential role in the formation of the mesoderm by regulating primitive streak formation that precedes the gastrulation stage [49]. Analysis of the organs and tissues generated from this layer demonstrated the regulatory role of YY1 in heart, muscle, and hematopoietic development.
YY1 Regulates the Hearth Development
The Mesp1 gene encodes an essential transcription factor expressed in all cardiac mesodermal cells [67]. A conditional YY1 KO mouse line with ablation of the YY1 floxed alleles under Mesp1-Cre expression was engineered to investigate the role of YY1 in early embryonic heart development. YY1 ablation in cardiac mesodermal cells resulted in embryonic lethality due to a decrease in the cardiac progenitor cell (CPC) population [55]. Additionally, conditional YY1 KO in CPCs induced embryonic lethality, which was attributed to cell cycle inhibition, impaired EMT, and downregulation of several direct YY1 targets [56]. Concordantly, ectopic YY1 overexpression in ESC-derived CPCs maintained their limited stemness potential by regulating the epigenetic state of CPC-associated genes [68]. Importantly, overexpression of YY1 in embryonic stem cells enhanced the generation of CPCs [55]. Additionally, ablation of YY1 in cardiomyocytes provoked congenital abnormalities in cardiac histology due to decreased proliferative potential and enhanced activation of apoptosis in cardiomyocytes [56]. Overall, YY1 regulates stemness potential needed for the generation of CPCs, maintains limited stemness potential of CPCs, and is required for proliferative potential of cardiac lineage-committed cells to generate cardiomyocytes.
YY1 Regulates the Muscle Development
Embryonic muscle progenitor cells express the transcription factors Pax3 and Pax7 (paired box proteins 3 and 7) necessary for the formation of fetal myofibers and skeletal muscle [69, 70]. YY1 was knocked out in Pax7+ cells to determine its role in muscle progenitor cells [57]. The data indicated that knockout mice died by suffocation after birth due to insufficient development of the diaphragm muscle evidenced by reduced levels of MyHC and troponin T proteins and concomitant loss of muscle fibers. Additionally, YY1-ablated mice had a smaller body size than that of the control animals [57]. Interestingly, ablation of YY1 in skeletal muscle cells during embryonic development did not induce significant changes at birth; however, the animals exhibited postnatal growth delay, showing a dwarf-like phenotype [58]. Additionally, these mice were characterized by exercise intolerance due to downregulation of YY1-mediated mitochondrial bioenergetics after YY1 ablation. Therefore, YY1 plays an essential role in muscle development.
YY1 Regulates Adult Muscle Stem Cells
In addition to the role of YY1 in organogenesis of skeletal muscle, YY1 also participates in muscle regeneration by regulating the metabolism of satellite cells. Since YY1 ablation in embryonic muscle stem cells is deleterious, a conditional YY1 KO mouse line was engineered for inducible YY1 ablation in Pax7+ satellite cells [57]. This mouse model did not show any abnormalities up to 1 year after YY1 ablation; however, injury-induced muscle regeneration in the animals resulted in a small fiber size with severe atrophy [57]. Similarly, the embryos harboring skeletal muscle cells lacking YY1 expression did not have detectable abnormalities [58]. Importantly, activation of YY1-ablated Pax7+ cells by injury resulted in low proliferative potential, which was rescued by lentivirus-mediated ectopic expression of YY1 [57], highlighting the role of YY1 in the expansion of activated muscle stem cells. RNA-seq and YY1 ChIP-seq assays were used to identify molecular pathways regulated by YY1, and the loss of YY1 inhibited cell cycle progression and reactivated silenced target genes of YY1 associated with mitochondrial function [57]. Interestingly, YY1 ablation in differentiated skeletal muscle cells compromised the mitochondrial gene expression profile, altering mitochondrial structure and decreasing mitochondrial function [58]. This result suggests that mitochondrial dependence on YY1 is different in activated muscle stem cells versus differentiated muscle cells. On the other hand, YY1-mediated stabilization of the HIF1α protein was lost in activated Pax7+ cells lacking the YY1 gene, provoking the loss of the expression of glycolytic HIF1-responsive genes [57]. Overall, YY1 plays a vital role during satellite cell activation by favoring the HIF1a-mediated expression program of glycolytic genes and repressing mitochondrial function.
YY1 Regulates Hematopoietic Stem Cells
Both self-renewal and differentiation potentials enable hematopoietic stem cells (HSCs) to maintain the entire hematopoietic system, which was clearly shown in bone marrow transplantation experiments performed in lethally irradiated mice. Specifically, the LSK (Lin−Scahic-kithi cells) compartment was shown to contain cells with the ability to repopulate the bone marrow. Pan X et al. demonstrated that mice undergoing transplantation of bone marrow progenitors ectopically expressing the YY1 gene increased LSK population in comparison with mice transplanted with bone marrow progenitors transduced with an empty vector [71]. Importantly, in-depth analysis of LSK population demonstrated that ectopic YY1 expression increased the LT-HSC population defined as CD48−CD150+ cells, highlighting that YY1 enhances the self-renewal potential of these cells [71]. Similarly, the levels of short-term (ST)-HSCs, multipotent progenitors, myeloid progenitors, monocytes, and neutrophils were also increased 30 weeks after bone marrow transplantation, suggesting that YY1 mediates the multipotency and differentiation potential of HSCs [54]. Additionally, YY1 ablation decreased these populations (Table 1). Thus, YY1 seems to regulate the stemness potential of HSCs.
To further assess the relationship between YY1 and self-renewal capacity, competitive congenic bone marrow transplantations were performed in lethally irradiated CD45.1+ mice by using a mixture of bone marrow progenitors from the donors harboring the CD45.1+ wild-type genotype with those carrying either the CD45.2+ Mx1-Cre YY1f/f or CD45.2+ Mx1-Cre genotypes. A very low presence of CD45.2+ cells lacking YY1 expression was detected in the reconstituted hematopoietic system compared to that obtained in the case of either CD45.1+ wild-type- or CD45.2+ Mx1-Cre-transplanted cells. Additionally, no CD45.2+ cells with absent YY1 expression were found in the secondary bone marrow transplantations [54]. This result was explained by the fact that YY1 ablation exhausted the HSC population by inducing cell cycle progression and compromising the quiescence state of the cells. On the other hand, the REPO domain of YY1 can recruit the PRC2 complex to DNA [2]; thus, Zhanping and colleagues analyzed whether the REPO domain is implicated in the pro-stemness effects of YY1 on HSCs. Ectopic expression of a YY1 variant lacking the REPO domain rescued the loss of the YY1 phenotype, similar to wild-type YY1, suggesting that YY1-mediated PRC2 recruitment to DNA is dispensable for the regulation of the stemness potential of HSCs. Similarly, the YY1-PRC2 axis was not overrepresented in gene regulation in ESCs [48]. Overall, YY1 plays a crucial role in the hematopoietic system by regulating HSC stemness.
Neither YY1 ablation nor ectopic expression was associated with the emergence of hematological malignancies; however, YY1 overexpression increased the myeloid lineage [54, 71]. Accumulated mutations [72, 73] and epigenetic changes [74] in the HSC population have been associated with some types of myeloid leukemia; thus, detection of high expression levels of YY1 in some patients with myeloid leukemia cancers suggests that YY1 may play an oncogenic role [75]. Hence, it has been demonstrated that YY1 is associated with antitumor therapy resistance of leukemia cells [76].
YY1 Regulates Stem Cells Derived from the Endoderm Lineage
The endoderm is the innermost gem layer that generates digestive and respiratory tubes. The role of YY1 in the development of the intestine and lung was investigated by using conditional mouse models (Table 1).
YY1 Regulates Gut Development
The association of YY1 with the stemness potential is evident in the gut development. Specifically, YY1 knockout in the intestinal epithelium inhibited villus growth during intestinal development, and maximal reduction of the growth was observed at E18.5 [59]. This effect was attributed to the deficient differentiation potential of enterocytes. Notably, the loss of YY1 in the intestinal epithelium inhibited gene expression associated with mitochondrial function [59]. Even though YY1 expression was ablated as early as E9.5, the YY1 ablation phenotype was discernible until E16.5, during which higher expression of mitochondria-associated genes is needed for normal development of the intestinal epithelium [59]. This finding suggested that YY1 regulates later stages of intestinal development by mediating the expression of mitochondria-associated genes. Similarly, enterocyte differentiation in the later stages of intestinal development was decreased after the deletion of the mitochondrial transcription factor TFAM [77]. Alterations in the relationship between YY1 and mitochondrial function in the intestine have clinical implications. Necrotizing enterocolitis (NEC) is a pathologic human condition characterized by gut development delay and is fatal in preterm infants. The samples from patients diagnosed with NEC showed downregulation of direct targets of YY1 associated with mitochondrial function [59]. Thus, YY1 regulates the differentiation potential of enterocytes during intestinal development.
YY1 Regulates Adult Intestinal Stem Cells
The adult intestine is another organ with high cell turnover rates, suggesting the presence of a cell population with stemness potential. Specifically, Lgr5+ intestinal stem cells generate all cell types of the gut epithelium. Perekatt and colleagues ablated YY1 expression in Lgr5+ intestinal stem cells and tracked the cells in an in vivo model to demonstrate the regulatory role of YY1. The results indicated that Lgr5+ intestinal stem cells lacking YY1 expression abandoned their crypt-base localization and migrated onto the villi to undergo differentiation and cell death [60]. The vacant stem niche was filled with intestinal stem cells expressing YY1 [60]. It was performed gene expression microarrays and YY1 ChIP-seq analyses of crypt epithelial cells lacking YY1 expression to clarify the molecular mechanisms mediated by YY1 in these cells. The loss of YY1 inhibited the expression of the genes encoding the mitochondrial complex I components and transactivated genes associated with cell cycle progression and RNA processing [60]. Consistently, the stemness potential of adult intestinal stem cells was compromised due to mitochondrial dysfunction resulting from the loss of HSP60, a mitochondrial chaperone [78], or ablation of the mitochondrial transcription factor TFAM [77]. Interestingly, in vitro assays showed that YY1 downregulation compromised in vitro organoid generation due to ROS inhibition, indicating that mitochondrial function is required for the stem cell phenotype [60]. Thus, YY1 maintains adult intestinal stemness by inducing a transcriptional program needed for mitochondrial function and cellular metabolism.
Role of YY1 in Lung Development
As previously mentioned, ablation of the YY1 gene results in peri-implantation lethality. To circumvent this, the animals harboring YY1 hypomorphic alleles (YY1flox − neo/flox−neo) were crossed with mice heterozygous for the YY1 null allele (YY1−/+). Thus, generated mouse line was genotyped as YY1flox − neo/− and displayed YY1 expression levels equal to 25 % of the normal expression. Approximately 50 % of the embryos harboring this genotype were alive at birth; however, the animals succumbed within two days postnatally due to inability to breathe attributed to the collapse of alveoli [79]. In agreement with this finding, Bérube and colleagues showed that conditional ablation of the YY1 gene in the lung mesenchyme induced structural alterations in embryonic lungs at E18.5, such as a decrease in club and pneumocyte type I cells. [61]. Interestingly, ablation of YY1 in the lung epithelium in another conditional mouse model also induced respiratory complications at birth that ended with postnatal death. Structural analyses of organogenesis of embryonic lung in these mice showed that YY1 regulated apoptosis and proliferation and is thus essential for lung branching [62]. Moreover, YY1 ablation diminished the density and diameter of the ring cartilage distributed along the trachea [62]. YY1 ablation in the pulmonary epithelium did not alter the precursors of basal cells expressing p63; however, this manipulation decreased the number of ciliated, club, and goblet cells, suggesting compromised differentiation potential [62]. Overall, lung development of various conditional KO mouse models targeting the YY1 gene demonstrated the regulatory role of YY1 in the differentiation potential of committed pulmonary progenitor cells.
Role of YY1 in the Cancer Stem Cell Phenotype
Previous sections described the role of YY1 in the regulation of the stemness potential of embryonic stem, lineage-committed progenitor, and adult stem cells (Table 1). Interestingly, in certain biological settings, YY1 modulates stemness potential by regulating the transcriptional landscape associated with mitochondrial function [50, 59, 60, 69]. In cancer, the CSC phenotype has been associated with mitochondrial function, and targeting this function is emerging as a potential anticancer therapy for specific elimination of CSCs [80, 81]. Pancreatic adenocarcinoma stem cells are sensitive to metformin treatment that inhibits mitochondrial function due to a decrease in the expression of PCG-1α, a mitochondrial transcription factor [82]. PCG-1α interacts with YY1, inducing co-binding to the promoter sequences of mitochondrial genes and activating their transcription [83]. Similarly, ectopic downregulation of either YY1 or PCG-1α compromises mitochondrial function, reducing the oxygen consumption rate and ATP production levels [83, 84]. Importantly, mTOR promoted physical interaction between YY1 and PCG-1α enhancing mitochondrial function [83]. mTOR inhibition compromised CSC traits [85, 86] by modulating mitochondrial function in several types of cancers, such as pancreatic, ovarian, and bone cancers [82, 87–89], suggesting that the YY1-mTOR-PGC-1α axis plays the key role in the biology of CSCs.
YY1 is implicated in the regulation of the Core regulatory circuitry in ESCs. Analysis of the data retrieved from an experimental protein database of cancer patient tissues enabled stratification of 17 types of cancer based on the expression patterns of YY1 and stem cell transcription factors. The expression levels of Sox2 and Oct4 were significantly associated with each other in all tested cancer types [90]. Notably, YY1 and Oct4 protein levels strongly correlated with each other in all analyzed cancer types [90]. Additionally, high expression levels of both YY1 and Sox2 were common in specific types of cancer. These findings are in agreement with the fact that YY1 mediates gene expression and stabilizes the levels of proteins encoded by the Oct-4 and Sox-2 genes [41]. Since Sox2 and Oct4 are essential for ESC biology [91]; codetection of these proteins enables experimental isolation of CSCs [92, 93], and targeting of these proteins inhibits tumor growth [94]; thus, associations of YY1 with Oct4 and Sox2 suggest a possible role for YY1 in the CSC phenotype.
Role of YY1 in Brain CSCs
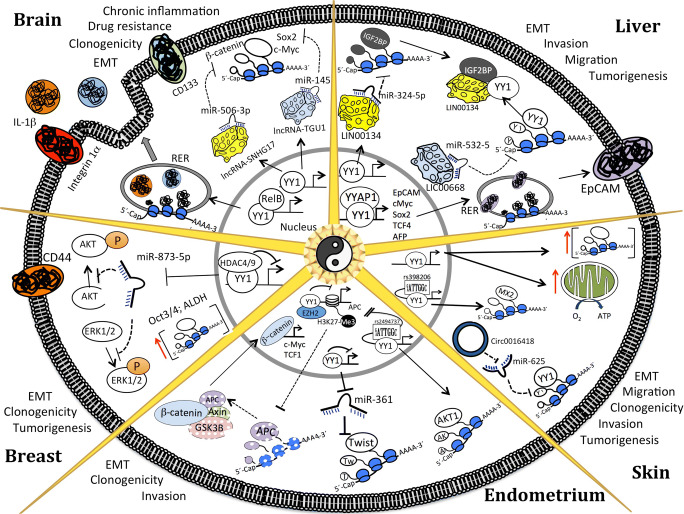
Glioma tumors are characterized by extensive infiltration of immune cells, contributing to the formation of a tumor microenvironment that triggers metastasis and resistance to anticancer treatments [95]. On the other hand, specific signaling pathways are associated with the CSC phenotype, such as NFκB, Wnt, and Notch [95]. RelB is a member of the NFκB signaling pathway and generates a specific cytokine profile responsible for myeloid cell recruitment to glioblastoma tumors (Fig. 1). Importantly, YY1 cointeracts with RelB in the promoter sequences of these cytokine genes to govern their transcriptional activation [96]. Thus, YY1 might support the CSC phenotype by contributing to the generation of an immunosuppressive microenvironment. The Wnt/CTNNB1 pathway is well known to be associated with both tumorigenesis and CSC phenotype [97, 98]. YY1 upregulates the expression lncRNA-SNHG17, which extends the half-life of CTNNB1 by sponging its direct negative regulator, miR-506-3p [99]. Significantly, the downregulation of lncRNA-SNHG17 inhibits tumor growth in vitro and in vivo [99]. Thus, YY1 regulates the activation of the Wnt1 pathway in glioma cells. The Notch pathway is another signaling pathway implicated in the CSC phenotype. lncRNA taurine upregulated gene 1 (TUG1) is a direct target gene of Notch that maintains the glioma CSC phenotype manifested as high expression of CD15, Sox2, and Myc genes and an increase in neurosphere formation [23]. Significantly, the pro-stem functions of TUG1 are mediated by sponging miRNA-145 to impede the miRNA-145-dependent degradation of Sox2 and Myc mRNAs [23]. Moreover, undifferentiated state of the cells is maintained by TUG1 through cointeraction with the YY1-PRC2 complex that induces silencing of differentiation-associated genes, such as BDNF, NGF, and NTF3 [23]. Therefore, YY1 appears to be an essential branch of several signaling pathways associated with the CSC phenotype in brain tumors (Fig. 1).
Fig. 1.
Molecular pathways modulated by YY1 in CSCs. Molecular pathways altered by YY1 in CSCs from endometrial, brain, liver, skin, and breast cancers are shown. RER, rough endoplasmic reticulum; EMT, epithelial-mesenchymal transition
Glioblastoma multiforme (GBM) is one of the most aggressive brain malignancies and is characterized by the lowest patient survival rate due to chemotherapy resistance [100]. YY1 appears to govern this outcome by enhancing the properties of CSCs. Specifically, YY1 was overexpressed in cisplatin-resistant glioblastoma cells. YY1 loss-of-function assays demonstrated that YY1 mediates the growth of tumor spheres and expression of CSC markers, such as CD133, STAT3, and integrin-α6 [101]. Importantly, the downregulation of YY1 by miR-186 was sufficient to induce cisplatin-mediated cell death [101]. On the other hand, temozolomide is used as first-line chemotherapy in newly diagnosed glioblastoma patients and in patients with refractory anaplastic astrocytomas [102]. shRNA-mediated downregulation of YY1 in temozolomide-resistant glioblastoma multiforme cells was shown to compromise the CSC population evaluated based on CD133 expression and spheroid and colony formation assays [103]. Notably, miR-7-5p is a negative regulator of YY1, and the levels of miR-7-5p expression were downregulated in temozolomide-resistant glioblastoma cells; thus, miR-7-5p overexpression reversed temozolomide resistance in vivo [103]. Overall, YY1 contributes to anticancer therapy resistance in GBM by enhancing the CSC phenotype (Fig. 1).
Role of YY1 in Hepatic CSCs
Hepatocellular carcinoma (HCC) is one of the most aggressive tumors. The levels of YY1 expression were shown to be higher in the samples of patient with HCC than that in the normal counterpart tissue samples [9, 10, 104]. Moreover, YY1 has been shown to trigger molecular mechanisms leading to HCC carcinogenesis [105] and the development of worse clinical phenotypes, such as angiogenesis [27] and antineoplastic drug resistance [104]. YY1 enhanced the migration, invasion, and epithelial-mesenchymal transition of HCCs by inducing the expression of linc01134, a lncRNA expressed at a high level in HCC [9]. linc01134 impeded miRNA-324-5p-mediated downregulation of IGF2BP1 by sponging miRNA-324-5p [9]. IGF2BP1 interacts with and extends the half-life of YY1 mRNA; thus, YY1 can regulate its own expression by regulating linc01134 expression in HCC [9]. Importantly, ectopic YY1 expression was sufficient to trigger epithelial-mesenchymal transition and invasive and migratory potential of the cells [9]. Therefore, YY1 triggers aggressive phenotypes in HCC in a linc01134-dependent and linc01134-independent manner. Another study demonstrated that the levels of YY1 expression were protected by linC00668, which sponges miR-532-5 to inhibit miR-532-5-mediated degradation of YY1 [10]. Importantly, both YY1 and linC00668 enhanced the proliferation, EMT, migration, and invasiveness of the cells [10]. Interestingly, ectopic YY1 expression rescued the inhibition of tumor growth in vivo and in vitro induced by downregulation of linc00668 [10]. Therefore, YY1 is an oncogenic regulator of HCC located upstream of linc00668 or linc01134. Overall, these studies showed that YY1 extensively regulates cellular processes strongly associated with the CSC phenotype, such as EMT, angiogenesis, migration, and invasion [106]. Thus, YY1 is expected to regulate the CSC phenotype in HCC.
Investigation of the molecular pathways in CSCs obtained from the samples of HCC patients provided new insight into the role of YY1 in the CSC phenotype. Analysis of transcriptomic datasets generated using the samples of HCC patients enriched in CSCs, which were selected based on the high expression levels of the EpCAM and AFP genes, which are well-known CSC biomarkers [107], indicated that the levels of the expression of direct transcriptional targets of YY1 were the highest [108]. One of these genes, YY1AP, was further characterized as a driver of CSC phenotype because the lowest survival rates of patient correlated with high copy number of YY1AP [108]. Similarly, YY1AP enhanced the in vitro and in vivo tumorigenic potential in YY1AP loss-of-function assays [108]. Significantly, YY1AP requires an association with transcriptional activation of the expression of the EpCAM gene induced by YY1 [108]. Thus, YY1 regulates the YY1AP-mediated CSC phenotype in HCC.
Role of YY1 in Endometrial CSCs
Endometrial cancer is the commonest gynecologic malignancy. Previous studies showed that the PRC2 complex containing EZH2 plays a crucial role in endometrial cancer by promoting the proliferation, migration, and metastasis [109, 110]. These effects are partially attributed to EZH2-mediated silencing of tumor suppressor miRNAs [111, 112]. miR-101 downregulates EZH2 and was thus overexpressed to investigate new tumor suppressor miRNAs potentially repressed by EZH2 [113]. miRNA-361 was identified out of miRNAs upregulated by miR-101 overexpression due to higher expression in normal endometrial tissues than that in their cancerous counterpart tissues [113]. The results of the miRNA-361 gain- and loss-of-function assays indicated that tumor suppressor functions miRNA-361 involve inhibition of invasiveness, spheroid formation potential, clonogenicity, EMT, and expression of CSC biomarkers [113]. YY1 is required to promote the recruitment of EZH2 to the miRNA-361 promoter [113], suggesting that YY1 governs the CSC phenotype in endometrial cancer. Additionally, YY1 was shown to silence APC gene expression in endometrial cancer cells by enhancing EZH2-mediated H3K27me3 deposition on the APC promoter [114]. YY1-mediated APC silencing relieved β-catenin repression, allowing the transactivation of its direct gene targets that enhanced tumorigenic potential both in vitro and in vivo [114]. On the other hand, YY1 regulates the oncogenic PI3K/AKT signaling pathway in endometrial tumors. The results of genome-wide association studies indicated that the minor allele of the SNP rs2494737 located at the 14q32.33 locus was associated with increased risk for endometrial tumors [115]. The results of ChIP-3 C, gene reporter, EMSA, and YY1 loss-of-function assays demonstrated that this allele generates a YY1-binding site in a distal regulatory region of the AKT1 gene that enhanced transcriptional activation [115]. Both the PI3K/AKT and Wnt/b-catenin signaling pathways modulate the CSC phenotype in endometrial cancer [116]; thus, YY1 might regulate the CSC phenotype, leading to worse clinical outcomes. Evidence supporting this hypothesis includes YY1 overexpression in tumor samples of the patients and inhibition of in vitro and in vivo tumorigenic potential by YY1 downregulation [114].
Role of YY1 in Skin CSCs
Melanoma is one of the deadliest skin cancers worldwide [117]. Apparently, YY1 plays a key role in melanoma because its expression levels correlate with advanced tumor stages [26, 118] and ectopic YY1 downregulation inhibits the proliferation, migration, invasion, and resistance to anticancer therapies [26, 118, 119]. Interestingly, the rs398206-A allele is associated with increased risk for melanoma because it generates to a YY1-binding site that results in transcriptional activation of the MX2 gene, which enhances melanomagenesis on a BRAFV600E background [120]. YY1 is essential for the generation of embryonic melanocytic lineage and adult melanocyte stem cells [53], and melanoma is strongly associated with the CSC phenotype [121–124]; thus, YY1 could be regulating the melanoma CSC phenotype.
The results of the experiments in a mouse melanoma model expressing human oncogene N-RasQ61K only in melanocytic lineage on an INK4a-deficient background [125] demonstrated that YY1 is coexpressed with a melanocytic marker in both hair follicles and skin melanomas [50]. This finding suggests a possible association between YY1 and undifferentiated cellular state. Tyr::CreERT2;YY1fl/fl or Tyr::CreERT2;YY1fl/wt inducible floxed YY1 mice were crossed with the model melanoma strain mentioned above to generate a mouse line with inducible YY1 ablation in melanoma cells aiming to investigate the role of YY1 in melanoma. Ablation of only one YY1 allele was sufficient to inhibit melanoma formation, indicating that melanogenesis is linked to YY1 expression [50]. RNA-seq and YY1 ChIP-Seq analyses were performed after RNAi-mediated knockdown of YY1 to extend these observations to human melanoma cells [50]. siRNA-mediated YY1 downregulation decreased the expression of direct YY1 target genes associated with the mitochondrial electron transfer chain, tricarboxylic acid cycle, protein synthesis, and other fundamental metabolic pathways [50]. Similarly, functional assays demonstrated that mitochondrial bioenergetics, protein synthesis, and other metabolic pathways were compromised by the loss of YY1 expression in melanoma cells [50]. Thus, YY1 apparently regulates CSC metabolism in melanoma (Fig. 1).
Role of YY1 in Breast CSCs
The CSC phenotype has been extensively studied in breast cancer [126, 127], showing its association with worse clinical scenarios, such as resistance to anticancer therapies, metastasis, angiogenesis, and cancer recurrence [128, 129]. Importantly, YY1 expression levels correlate with the presence of breast cancer [130, 131]. The results of YY1 loss- and gain-of-function assays showed that YY1 fuels the migratory, invasive, colony-forming, and in vivo tumorigenic potentials of breast cancer cells [130]. Similarly, YY1 is associated with life-threatening processes, such as the development of metastasis [130, 132, 133]. Since these results are indicators of the presence of the CSC population, YY1 may play a role in the regulation of the CSC phenotype. Interestingly, YY1 decreased p27 protein levels in in vitro and in vivo assays by enhancing Sk2-mediated p27 polyubiquitination [130]. p27 induces cell cycle inhibition by targeting the cyclin E-CDK2 complex, suggesting that YY1 contributes to oncogenic behavior by enabling cell cycle progression [134]. Interestingly, a decrease or lack of p27 expression have been associated with the stemness potential of human ESCs [135] and the generation of induced pluripotent stem cells [136], suggesting that Sk2-mediated p27 degradation induced by YY1 may favor the CSC phenotype.
Another study demonstrated that breast CSCs enriched by mammosphere culture had lower NMI expression levels than that in non-CSCs [137]. The results of the NMI loss- and gain-of-function assays demonstrated that NMI decreases several CSC features, such as the expression of stem cell transcription factors, frequency of the CD44+CD24− cell population, mammosphere-forming potential, EMT, and xenograft tumor formation [137]. These effects were attributed to NMI-mediated downregulation of the hTERT gene, a well-known pro-stemness factor in breast cancer [138, 139]. NMI lacks a DNA-interacting domain and modulates gene expression by functioning as a coactivator. NMI immunoprecipitation coupled with mass spectrometry was performed to identify transcription factors associated with NMI-mediated downregulation of hTERT; the results identified YY1 as an NMI-interacting protein. The YY1-NMI complex was able to silence the expression of the hTERT gene by binding to the hTERT promoter; however, this axis was not directly assessed in the CSC population [137]. Thus, the role of YY1 in breast CSCs was incompletely determined. Importantly, breast tumor samples were recently shown to have high levels of expression of YY1 and stem cell transcription factors, such as Oct4, Sox2, and Nanog [140]. Importantly, the ectopic modulation of YY1 demonstrated its positive regulatory role in the CSC phenotype due to an increase in several stemness traits, including the expression of stem cell transcription factors, sphere-forming potential, percentage of CD44+CD24− cells, and the ability to form tumors in vivo [140]. Additionally, YY1 positively regulated the ERK1/2 and PI3K/AKT pathways, and inhibition of these pathways by small molecules decreased the tumorigenic effects of YY1 [140]. Importantly, YY1 inhibited miR879-5p expression by interacting with its promoter sequence [140]. Since miR879-5p inhibits the YY1-mediated CSC phenotype, the repressive role of YY1 in the regulation of the miR879-5p gene is required for the maintenance of the breast CSC phenotype (Fig. 1).
Potential Role of YY1 in the CSC Niche
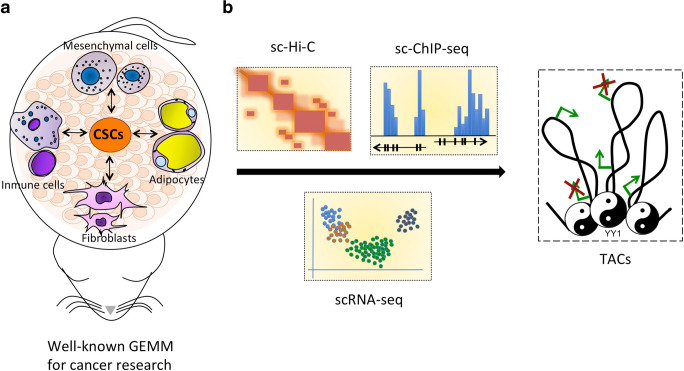
Microenvironment (ME) of the CSC niche provides correct signals to the CSC population that secure their stemness properties. This ME is sculpted by corrupted tumor-associated cells (TACs), such as fibroblasts, mesenchymal stem cells, immune cells, and adipocytes, that exchange certain molecules and establish specific interactions with CSCs [141]. Co-engraftment of CSCs with tumor-associated fibroblasts, which are one of the main populations mediating the formation of the CSC niche, enhances tumor growth [142], antitumor therapy resistance [143], invasion, and metastasis [144]. Targeting TACs or molecules secreted by TACs inhibits the CSC phenotype, and TACs are thus emerging as attractive targets for the development of anti-CSC therapies. Therefore, investigation of YY1-mediated functions in TACs that support the CSC niche is required to advance our understanding of the role of YY1 in the CSC phenotype. Some interesting results have been reported; however, the data on these relationships are lacking. Tumor-associated macrophages (TAMs) are M2-polarized macrophages that enhance tumor growth via their immunosuppressive functions. Macrophages expressing miR-125a were shown to suppress the polarization of the M2 phenotype to inhibit tumor growth in vivo [145]. Importantly, YY1 apparently promoted the development of TAMs by repressing miR-125a expression. Specifically, YY1 is able to interact with a distal regulatory element that governs miR-125a silencing in an RYBP-dependent manner [145]. Thus, the role of YY1 in various TACs supporting the CSC niche requires further investigation.
The lack of observations on the role of YY1 in the CSC niche may be a consequence of the lack of in vivo models for studies of the CSC niche. CSCs are functionally defined as the cells that can generate tumors in the limiting dilution and serial tumor transplantation assays in immunocompromised mice. Potentially, the failure of a putative CSC population to display tumorigenic behavior assessed in these in vivo assays indirectly reflects the addiction of CSCs to a correct CSC niche. Thus, generation of the in vivo models, in which TACs are assessed and modulated, such as immune cells, is required to study CSCs to gain new insight into human cancer biology. Therefore, the development of genetically engineered mice with conditional and active YY1 ablation in specific TACs is needed to circumvent this limitation. Since YY1 mediates specific 3D chromatin configurations needed for cell identity, we suggest that YY1 may govern both CSCs and the CSC niche by imposing cell type-specific chromatin configurations. Thus, single-cell high-throughput technologies, such as single-cell ChIP-seq [146, 147] and single-cell chromatin conformation capture [148, 149], may be able to promote this area of research (Fig. 2).
Fig. 2.
Potential role of YY1 in the regulation of the CSC niche. a The use of well-known genetically engineered mouse models for cancer research with conditional ablation of YY1 in specific TACs, such as mesenchymal stem cells (MSCs), immune cells, tumor-associated fibroblasts, and adipocytes, may enable to determine the role of YY1 in tumor-associated cells (TACs) regulating the CSC niche. b YY1 is currently conceptualized as a 3D chromatin regulator; thus, it will be of interest to determine whether YY1 imposes specific gene expression in TACs supporting the CSC niche by modulating their chromatin configuration
Concluding Remarks
The use of genetically engineered mouse lines coupled with high-throughput technologies of molecular biology enabled to investigate the role of YY1 in the stemness potential in limited scenarios during embryonic development and in some adult stem cells. Thus, YY1 governs the stemness potential by regulating the transcriptional landscape associated with cellular metabolism, energetics, and cell death. In cancer, in addition to the modulation of molecular processes similar to those in normal stem cells, YY1 governs the CSC phenotype via tumor-dependent molecular events (Fig. 1). This effect may be a consequence of the presence of various tissue-specific selection pressures. On the other hand, analysis of the regulatory effects of YY1 on TACs supporting the CSC niche, which is an essentially unexplored area, is needed to enrich our understanding of the role of YY1 in the CSC phenotype. Notably, most YY1-mediated actions regulating the CSC phenotype have been polarized to YY1-mediated transcriptional regulation of the target genes. However, YY1 is emerging as a dynamic regulator of the three-dimensional chromatin configuration. Since 3D genome organization dictates cell identity and its dynamic rearrangement is associated with cellular responsiveness [150, 151], we envision that YY1 may induce specific chromatin conformation states needed for TACs that support the CSC niche. Thus, in vitro and in vivo models for the investigations of the role of YY1 in the chromatin configuration in specific TACs, which support the CSC niche, may be able to identify new druggable molecular targets.
Abbreviations
- YY1
yin yang-1
- CSCs
cancer stem cells
- TACs
tumor-associated cells
- PRC2
polycomb repressive complex 2
- lncRNA
long non-coding RNA
- SC
stem cell
- ICM
inner cell mass
- EMT
epithelial-mesenchymal transition
- ESC
embryonic stem cell
- NPC
neural progenitor cell
- ChIP
chromatin immunoprecipitation
- CPC
cardiac progenitor cell
- MyHC
myosin heacy chain
- NEC
necrotizing enterocolitis
- HSC
hematopoietic stem cell
- PGC-1α
peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- NFKB
nuclear factor kappa B
- ME
microenvironment
Author Contributions
AMS and AFPH performed the literature review, data interpretation and contributed to the first draft writing. GUMR conceived and designed the study, wrote the first draft and the final version of this manuscript. All authors read and approved the final version of this manuscript.
Funding
GUMR is supported by grants from National Council of Science and Technology (CONACyT-México; A1-S-16997), National Autonomous University of México (PAPIIT-UNAM; IA204820), and Mexican Federal Founds of Children’s Hospital of Mexico (HIM/2018/055-SSA1525). AMS is supported by grants from Mexican Federal Founds of Children’s Hospital of Mexico (HIM/2017/145-SSA1469).
Data Availability
All data presented in this review are totally available and present in the text.
Code Availability
Not applicable.
Declarations
Conflict of Interest
No potential conflicts of interest were disclosed.
Ethics Approval
Not applicable.
Consent for Publication
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wilkinson, F., Pratt, H., & Atchison, M. L. (2010). PcG recruitment by the YY1 REPO domain can be mediated by Yaf2. Journal of Cellular Biochemistry, 109(3), 478–486. [DOI] [PMC free article] [PubMed]
- 2.Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(51):19296–19301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, et al. YY1 functions with INO80 to activate transcription. Nature Structural & Molecular Biology. 2007;14(9):872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- 4.Wu S, Shi Y, Mulligan P, Gay F, Landry J, Liu H, Lu J, Qi HH, Wang W, Nickoloff JA, et al. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nature Structural & Molecular Biology. 2007;14(12):1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atchison ML. Function of YY1 in long-distance DNA interactions. Frontiers in Immunology. 2014;5:45. doi: 10.3389/fimmu.2014.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beagan JA, Duong MT, Titus KR, Zhou L, Cao Z, Ma J, Lachanski CV, Gillis DR, Phillips-Cremins JE. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Research. 2017;27(7):1139–1152. doi: 10.1101/gr.215160.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, et al. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017;171(7):1573–1588 e1528. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang XC, Gu AP, Zheng CY, Li YB, Liang HF, Wang HJ, Tang XL, Bai XX, Cai J. YY1/LncRNA GAS5 complex aggravates cerebral ischemia/reperfusion injury through enhancing neuronal glycolysis. Neuropharmacology. 2019;158:107682. doi: 10.1016/j.neuropharm.2019.107682. [DOI] [PubMed] [Google Scholar]
- 9.Rong Z, Wang Z, Wang X, Qin C, Geng W. Molecular interplay between linc01134 and YY1 dictates hepatocellular carcinoma progression. Journal of Experimental & Clinical Cancer Research: CR. 2020;39(1):61. doi: 10.1186/s13046-020-01551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.You, G., Zhou, C., & Xuan, W. (2020). LncRNA LINC00668 promotes cell proliferation, migration, invasion ability and EMT process in hepatocellular carcinoma by targeting miR-532-5p/YY1 axis. Bioscience Reports 40(5), BSR20192697. [DOI] [PMC free article] [PubMed]
- 11.Yang WM, Yao YL, Sun JM, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. The Journal of Biological Chemistry. 1997;272(44):28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 12.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Journal of Molecular Cell Biology. 2001;21(17):5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrivastava A, Saleque S, Kalpana GV, Artandi S, Goff SP, Calame K. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science. 1993;262(5141):1889–1892. doi: 10.1126/science.8266081. [DOI] [PubMed] [Google Scholar]
- 14.Donohoe, M. E., Zhang, L. F., Xu, N., Shi, Y., & Lee, J. T. (2007). Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Molecular Cell,25(1), 43–56. [DOI] [PubMed]
- 15.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117(7):859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Lee, J. S., Galvin, K. M., & Shi, Y. (1993). Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proceedings of the National Academy of Sciences of the United States of America, 90(13), 6145–6149. [DOI] [PMC free article] [PubMed]
- 17.Lee, J. S., Galvin, K. M., See, R. H., Eckner, R., Livingston, D., Moran, E., & Shi, Y. (1995). Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes & Development, 9(10), 1188–1198. [DOI] [PubMed]
- 18.Kashyap V, Bonavida B. Role of YY1 in the pathogenesis of prostate cancer and correlation with bioinformatic data sets of gene expression. Genes & Cancer. 2014;5(3–4):71–83. doi: 10.18632/genesandcancer.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang N, Li X, Wu CW, Dong Y, Cai M, Mok MT, Wang H, Chen J, Ng SS, Chen M, et al. microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene. 2013;32(42):5078–5088. doi: 10.1038/onc.2012.526. [DOI] [PubMed] [Google Scholar]
- 20.Powe DG, Akhtar G, Habashy HO, Abdel-Fatah T, Rakha EA, Green AR, Ellis IO. Investigating AP-2 and YY1 protein expression as a cause of high HER2 gene transcription in breast cancers with discordant HER2 gene amplification. Breast Cancer Research: BCR. 2009;11(6):R90. doi: 10.1186/bcr2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He G, Wang Q, Zhou Y, Wu X, Wang L, Duru N, Kong X, Zhang P, Wan B, Sui L, et al. YY1 is a novel potential therapeutic target for the treatment of HPV infection-induced cervical cancer by arsenic trioxide. International Journal of Gynecological Cancer: Official Journal of the International Gynecological Cancer Society. 2011;21(6):1097–1104. doi: 10.1097/IGC.0b013e31821d2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baritaki S, Sifakis S, Huerta-Yepez S, Neonakis IK, Soufla G, Bonavida B, Spandidos DA. Overexpression of VEGF and TGF-beta1 mRNA in Pap smears correlates with progression of cervical intraepithelial neoplasia to cancer: implication of YY1 in cervical tumorigenesis and HPV infection. International Journal of Oncology. 2007;31(1):69–79. [PubMed] [Google Scholar]
- 23.Katsushima K, Natsume A, Ohka F, Shinjo K, Hatanaka A, Ichimura N, Sato S, Takahashi S, Kimura H, Totoki Y, et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nature Communications. 2016;7:13616. doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baritaki S, Chatzinikola AM, Vakis AF, Soulitzis N, Karabetsos DA, Neonakis I, Bonavida B, Spandidos DA. YY1 Over-expression in human brain gliomas and meningiomas correlates with TGF-beta1, IGF-1 and FGF-2 mRNA levels. Cancer Investigation. 2009;27(2):184–192. doi: 10.1080/07357900802210760. [DOI] [PubMed] [Google Scholar]
- 25.Kang W, Tong JH, Chan AW, Zhao J, Dong Y, Wang S, Yang W, Sin FM, Ng SS, Yu J, et al. Yin Yang 1 contributes to gastric carcinogenesis and its nuclear expression correlates with shorter survival in patients with early stage gastric adenocarcinoma. Journal of Translational Medicine. 2014;12:80. doi: 10.1186/1479-5876-12-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao G, Li Q, Wang A, Jiao J. YY1 regulates melanoma tumorigenesis through a miR-9 ~ RYBP axis. Journal of Experimental & Clinical Cancer Research: CR. 2015;34:66. doi: 10.1186/s13046-015-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W, Li Z, Qin R, Wang X, An H, Wang Y, Zhu Y, Liu Y, Cai S, Chen S, et al. YY1 promotes endothelial cell-dependent tumor angiogenesis in hepatocellular carcinoma by transcriptionally activating VEGFA. Frontiers in Oncology. 2019;9:1187. doi: 10.3389/fonc.2019.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, T., Wang, G., Yang, L., Peng, B., Wen, Y., Ding, G., & Wang, Z. (2017). Transcription factor YY1 modulates lung cancer progression by activating lncRNA-PVT1. DNA and Cell Biology, 36(11), 947–958. [DOI] [PubMed]
- 29.Wang W, Li D, Sui G. YY1 is an inducer of cancer metastasis. Critical Reviews in Oncogenesis. 2017;22(1–2):1–11. doi: 10.1615/CritRevOncog.2017021314. [DOI] [PubMed] [Google Scholar]
- 30.Sarvagalla S, Kolapalli SP, Vallabhapurapu S. The two sides of YY1 in cancer: a friend and a foe. Frontiers in Oncology. 2019;9:1230. doi: 10.3389/fonc.2019.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meliala ITS, Hosea R, Kasim V, Wu S. The biological implications of Yin Yang 1 in the hallmarks of cancer. Theranostics. 2020;10(9):4183–4200. doi: 10.7150/thno.43481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shackleton M. Normal stem cells and cancer stem cells: similar and different. Seminars in Cancer Biology. 2010;20(2):85–92. doi: 10.1016/j.semcancer.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Rycaj K, Tang DG. Cell-of-origin of cancer versus cancer stem cells: assays and interpretations. Cancer Research. 2015;75(19):4003–4011. doi: 10.1158/0008-5472.CAN-15-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciurea ME, Georgescu AM, Purcaru SO, Artene SA, Emami GH, Boldeanu MV, Tache DE, Dricu A. Cancer stem cells: biological functions and therapeutically targeting. International Journal of Molecular Sciences. 2014;15(5):8169–8185. doi: 10.3390/ijms15058169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, Zhang G, Wang X, Dong Z, Chen F, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduction and Targeted Therapy. 2020;5:8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vargaftig J, Taussig DC, Griessinger E, Anjos-Afonso F, Lister TA, Cavenagh J, Oakervee H, Gribben J, Bonnet D. Frequency of leukemic initiating cells does not depend on the xenotransplantation model used. Leukemia. 2012;26(4):858–860. doi: 10.1038/leu.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarry JE, Murphy K, Perry R, Sanchez PV, Secreto A, Keefer C, Swider CR, Strzelecki AC, Cavelier C, Recher C, et al. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in NOD/SCID/IL2Rgammac-deficient mice. The Journal of Clinical Investigation. 2011;121(1):384–395. doi: 10.1172/JCI41495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong Y, Guan K, Zhou C, Ma W, Wang D, Zhang Y, Zhang S. Cancer stem cells sustaining the growth of mouse melanoma are not rare. Cancer Letters. 2010;292(1):17–23. doi: 10.1016/j.canlet.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 39.Johnston MD, Maini PK, Jonathan Chapman S, Edwards CM, Bodmer WF. On the proportion of cancer stem cells in a tumour. Journal of Theoretical Biology. 2010;266(4):708–711. doi: 10.1016/j.jtbi.2010.07.031. [DOI] [PubMed] [Google Scholar]
- 40.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Molecular and Cellular Biology. 1999;19(10):7237–7244. doi: 10.1128/MCB.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallingford MC, Hiller J, Zhang K, Mager J. YY1 is required for posttranscriptional stability of SOX2 and OCT4 proteins. Cellular Reprogramming. 2017;19(4):263–269. doi: 10.1089/cell.2017.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian FJ, Cheng YX, Li XC, Wang F, Qin CM, Ma XL, Yang J, Lin Y. The YY1/MMP2 axis promotes trophoblast invasion at the maternal-fetal interface. The Journal of Pathology. 2016;239(1):36–47. doi: 10.1002/path.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 46.Gao F, Wei Z, An W, Wang K, Lu W. The interactomes of POU5F1 and SOX2 enhancers in human embryonic stem cells. Scientific Reports. 2013;3:1588. doi: 10.1038/srep01588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Wu X, Wei C, Huang X, Ma Q, Huang X, Faiola F, Guallar D, Fidalgo M, Huang T, et al. YY1 positively regulates transcription by targeting promoters and super-enhancers through the BAF complex in embryonic stem cells. Stem Cell Reports. 2018;10(4):1324–1339. doi: 10.1016/j.stemcr.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vella P, Barozzi I, Cuomo A, Bonaldi T, Pasini D. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Research. 2012;40(8):3403–3418. doi: 10.1093/nar/gkr1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trask MC, Tremblay KD, Mager J. Yin-Yang1 is required for epithelial-to-mesenchymal transition and regulation of Nodal signaling during mammalian gastrulation. Developmental Biology. 2012;368(2):273–282. doi: 10.1016/j.ydbio.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varum S, Baggiolini A, Zurkirchen L, Atak ZK, Cantu C, Marzorati E, Bossart R, Wouters J, Hausel J, Tuncer E, et al. Yin Yang 1 orchestrates a metabolic program required for both neural crest development and melanoma formation. Cell Stem Cell. 2019;24(4):637–653 e639. doi: 10.1016/j.stem.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 51.Dong X, Kwan KM. Yin Yang 1 is critical for mid-hindbrain neuroepithelium development and involved in cerebellar agenesis. Molecular Brain. 2020;13(1):104. doi: 10.1186/s13041-020-00643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zurkirchen L, Varum S, Giger S, Klug A, Hausel J, Bossart R, Zemke M, Cantu C, Atak ZK, Zamboni N, et al. Yin Yang 1 sustains biosynthetic demands during brain development in a stage-specific manner. Nature Communications. 2019;10(1):2192. doi: 10.1038/s41467-019-09823-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Song JS, Bell RJ, Tran TN, Haq R, Liu H, Love KT, Langer R, Anderson DG, Larue L, et al. YY1 regulates melanocyte development and function by cooperating with MITF. PLoS Genetics. 2012;8(5):e1002688. doi: 10.1371/journal.pgen.1002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu Z, Hong CC, Kong G, Assumpcao A, Ong IM, Bresnick EH, Zhang J, Pan X. Polycomb group protein YY1 is an essential regulator of hematopoietic stem cell quiescence. Cell Reports. 2018;22(6):1545–1559. doi: 10.1016/j.celrep.2018.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gregoire S, Karra R, Passer D, Deutsch MA, Krane M, Feistritzer R, Sturzu A, Domian I, Saga Y, Wu SM. Essential and unexpected role of Yin Yang 1 to promote mesodermal cardiac differentiation. Circulation Research. 2013;112(6):900–910. doi: 10.1161/CIRCRESAHA.113.259259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beketaev I, Zhang Y, Kim EY, Yu W, Qian L, Wang J. Critical role of YY1 in cardiac morphogenesis. Developmental Dynamics: an Official Publication of the American Association of Anatomists. 2015;244(5):669–680. doi: 10.1002/dvdy.24263. [DOI] [PubMed] [Google Scholar]
- 57.Chen, F., Zhou, J., Li, Y., Zhao, Y., Yuan, J., Cao, Y., Wang, L., Zhang, Z., Zhang, B., Wang, C. C., et al. (2019). YY1 regulates skeletal muscle regeneration through controlling metabolic reprogramming of satellite cells. The EMBO Journal, 38(10), 1–23. [DOI] [PMC free article] [PubMed]
- 58.Blattler, S. M., Verdeguer, F., Liesa, M., Cunningham, J. T., Vogel, R. O., Chim, H., Liu, H., Romanino, K., Shirihai, O. S., Vazquez, F., et al. (2012). Defective mitochondrial morphology and bioenergetic function in mice lacking the transcription factor Yin Yang 1 in skeletal muscle. Molecular and Cellular Biology, 32(16), 3333–3346. [DOI] [PMC free article] [PubMed]
- 59.Kumar N, Srivillibhuthur M, Joshi S, Walton KD, Zhou A, Faller WJ, Perekatt AO, Sansom OJ, Gumucio DL, Xing J, et al. A YY1-dependent increase in aerobic metabolism is indispensable for intestinal organogenesis. Development. 2016;143(20):3711–3722. doi: 10.1242/dev.137992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perekatt AO, Valdez MJ, Davila M, Hoffman A, Bonder EM, Gao N, Verzi MP. YY1 is indispensable for Lgr5 + intestinal stem cell renewal. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(21):7695–7700. doi: 10.1073/pnas.1400128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berube-Simard FA, Prudhomme C, Jeannotte L. YY1 acts as a transcriptional activator of Hoxa5 gene expression in mouse organogenesis. PLoS One. 2014;9(4):e93989. doi: 10.1371/journal.pone.0093989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boucherat O, Landry-Truchon K, Berube-Simard FA, Houde N, Beuret L, Lezmi G, Foulkes WD, Delacourt C, Charron J, Jeannotte L. Epithelial inactivation of Yy1 abrogates lung branching morphogenesis. Development. 2015;142(17):2981–2995. doi: 10.1242/dev.120469. [DOI] [PubMed] [Google Scholar]
- 63.Gronroos E, Terentiev AA, Punga T, Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(33):12165–12170. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Panhuysen M, Vogt Weisenhorn DM, Blanquet V, Brodski C, Heinzmann U, Beisker W, Wurst W. Effects of Wnt1 signaling on proliferation in the developing mid-/hindbrain region. Molecular and Cellular Neurosciences. 2004;26(1):101–111. doi: 10.1016/j.mcn.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 65.Gabriele M, Vulto-van Silfhout AT, Germain PL, Vitriolo A, Kumar R, Douglas E, Haan E, Kosaki K, Takenouchi T, Rauch A, et al. YY1 haploinsufficiency causes an intellectual disability syndrome featuring transcriptional and chromatin dysfunction. American Journal of Human Genetics. 2017;100(6):907–925. doi: 10.1016/j.ajhg.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nabais Sa, M. J., Gabriele, M., Testa, G., & de Vries, B. B. A. (1993). Gabriele-de Vries syndrome. In M. P. Adam, H. H. Ardinger, R. A. Pagon, S. E. Wallace, L. J. H. Bean, K. Stephens, & A. Amemiya (Eds.), GeneReviews((R)). Seattle (WA), 1993-2021. [PubMed]
- 67.Witman N, Sahara M. Cardiac progenitor cells in basic biology and regenerative medicine. Stem Cells International. 2018;2018:8283648. doi: 10.1155/2018/8283648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gregoire S, Li G, Sturzu AC, Schwartz RJ, Wu SM. YY1 expression is sufficient for the maintenance of cardiac progenitor cell state. Stem Cells. 2017;35(8):1913–1923. doi: 10.1002/stem.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sambasivan R, Yao R, Kissenpfennig A, Van Wittenberghe L, Paldi A, Gayraud-Morel B, Guenou H, Malissen B, Tajbakhsh S, Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138(17):3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- 70.Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435(7044):948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 71.Pan X, Jones M, Jiang J, Zaprazna K, Yu D, Pear W, Maillard I, Atchison ML. Increased expression of PcG protein YY1 negatively regulates B cell development while allowing accumulation of myeloid cells and LT-HSC cells. PLoS One. 2012;7(1):e30656. doi: 10.1371/journal.pone.0030656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li XL, Xue Y, Yang YJ, Zhang CX, Wang Y, Duan YY, Meng YN, Fu J. Hematopoietic stem cells: cancer involvement and myeloid leukemia. European Review for Medical and Pharmacological Sciences. 2015;19(10):1829–1836. [PubMed] [Google Scholar]
- 74.Bartholdy B, Christopeit M, Will B, Mo Y, Barreyro L, Yu Y, Bhagat TD, Okoye-Okafor UC, Todorova TI, Greally JM, et al. HSC commitment-associated epigenetic signature is prognostic in acute myeloid leukemia. The Journal of Clinical Investigation. 2014;124(3):1158–1167. doi: 10.1172/JCI71264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Erkeland SJ, Valkhof M, Heijmans-Antonissen C, Delwel R, Valk PJ, Hermans MH, Touw IP. The gene encoding the transcriptional regulator Yin Yang 1 (YY1) is a myeloid transforming gene interfering with neutrophilic differentiation. Blood. 2003;101(3):1111–1117. doi: 10.1182/blood-2002-04-1207. [DOI] [PubMed] [Google Scholar]
- 76.Antonio-Andres, G., Rangel-Santiago, J., Tirado-Rodriguez, B., Martinez-Ruiz, G. U., Klunder-Klunder, M., Vega, M. I., Lopez-Martinez, B., Jimenez-Hernandez, E., Torres Nava, J., Medina-Sanson, A., et al. (2018) Role of Yin Yang-1 (YY1) in the transcription regulation of the multi-drug resistance (MDR1) gene. Leukemia & Lymphoma, 59(11), 2628–2638. [DOI] [PubMed]
- 77.Srivillibhuthur M, Warder BN, Toke NH, Shah PP, Feng Q, Gao N, Bonder EM, Verzi MP. TFAM is required for maturation of the fetal and adult intestinal epithelium. Developmental Biology. 2018;439(2):92–101. doi: 10.1016/j.ydbio.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berger E, Rath E, Yuan D, Waldschmitt N, Khaloian S, Allgauer M, Staszewski O, Lobner EM, Schottl T, Giesbertz P, et al. Mitochondrial function controls intestinal epithelial stemness and proliferation. Nature Communications. 2016;7:13171. doi: 10.1038/ncomms13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, Shi Y. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Molecular and Cellular Biology. 2006;26(9):3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lamb R, Ozsvari B, Lisanti CL, Tanowitz HB, Howell A, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: treating cancer like an infectious disease. Oncotarget. 2015;6(7):4569–4584. doi: 10.18632/oncotarget.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song IS, Jeong JY, Jeong SH, Kim HK, Ko KS, Rhee BD, Kim N, Han J. Mitochondria as therapeutic targets for cancer stem cells. World Journal of Stem Cells. 2015;7(2):418–427. doi: 10.4252/wjsc.v7.i2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, Barneda D, Sellers K, Campos-Olivas R, Grana O, et al. MYC/PGC-1alpha balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metabolism. 2015;22(4):590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 83.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450(7170):736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 84.Ji, K., Zheng, J., Lv, J., Xu, J., Ji, X., Luo, Y. B., Li, W., Zhao, Y., & Yan, C. (2015). Skeletal muscle increases FGF21 expression in mitochondrial disorders to compensate for energy metabolic insufficiency by activating the mTOR-YY1-PGC1alpha pathway. Free Radical Biology & Medicine, 84, 161–170. [DOI] [PubMed]
- 85.Francipane MG, Lagasse E. Therapeutic potential of mTOR inhibitors for targeting cancer stem cells. British Journal of Clinical Pharmacology. 2016;82(5):1180–1188. doi: 10.1111/bcp.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xia P, Xu XY. PI3K/Akt/mTOR signaling pathway in cancer stem cells: from basic research to clinical application. American Journal of Cancer Research. 2015;5(5):1602–1609. [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao B, Luo J, Wang Y, Zhou L, Che J, Wang F, Peng S, Zhang G, Shang P. Metformin suppresses self-renewal ability and tumorigenicity of osteosarcoma stem cells via reactive oxygen species-mediated apoptosis and autophagy. Oxidative Medicine and Cellular Longevity. 2019;2019:9290728. doi: 10.1155/2019/9290728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Singh BN, Kumar D, Shankar S, Srivastava RK. Rottlerin induces autophagy which leads to apoptotic cell death through inhibition of PI3K/Akt/mTOR pathway in human pancreatic cancer stem cells. Biochemical Pharmacology. 2012;84(9):1154–1163. doi: 10.1016/j.bcp.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 89.Alvero AB, Montagna MK, Holmberg JC, Craveiro V, Brown D, Mor G. Targeting the mitochondria activates two independent cell death pathways in ovarian cancer stem cells. Molecular Cancer Therapeutics. 2011;10(8):1385–1393. doi: 10.1158/1535-7163.MCT-11-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaufhold S, Garban H, Bonavida B. Yin Yang 1 is associated with cancer stem cell transcription factors (SOX2, OCT4, BMI1) and clinical implication. Journal of Experimental & Clinical Cancer Research: CR. 2016;35:84. doi: 10.1186/s13046-016-0359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rizzino A, Wuebben EL. Sox2/Oct4: A delicately balanced partnership in pluripotent stem cells and embryogenesis. Biochimica et Biophysica Acta. 2016;1859(6):780–791. doi: 10.1016/j.bbagrm.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 92.Gao W, Wu D, Wang Y, Wang Z, Zou C, Dai Y, Ng CF, Teoh JY, Chan FL. Development of a novel and economical agar-based non-adherent three-dimensional culture method for enrichment of cancer stem-like cells. Current Stem Cell Research & Therapy. 2018;9(1):243. doi: 10.1186/s13287-018-0987-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang B, Raviv A, Esposito D, Flanders KC, Daniel C, Nghiem BT, Garfield S, Lim L, Mannan P, Robles AI, et al. A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Reports. 2015;4(1):155–169. doi: 10.1016/j.stemcr.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vaddi, P. K., Stamnes, M. A., Cao, H., & Chen, S. (2019). Elimination of SOX2/OCT4-associated prostate cancer stem cells blocks tumor development and enhances therapeutic response. Cancers, 11(9), 1331. [DOI] [PMC free article] [PubMed]
- 95.Ma Q, Long W, Xing C, Chu J, Luo M, Wang HY, Liu Q, Wang RF. Cancer stem cells and immunosuppressive microenvironment in glioma. Frontiers in Immunology. 2018;9:2924. doi: 10.3389/fimmu.2018.02924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Waters MR, Gupta AS, Mockenhaupt K, Brown LN, Biswas DD, Kordula T. RelB acts as a molecular switch driving chronic inflammation in glioblastoma multiforme. Oncogenesis. 2019;8(6):37. doi: 10.1038/s41389-019-0146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bengoa-Vergniory N, Kypta RM. Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. Cellular and Molecular Life Sciences: CMLS. 2015;72(21):4157–4172. doi: 10.1007/s00018-015-2028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Takahashi-Yanaga F, Kahn M. Targeting Wnt signaling: can we safely eradicate cancer stem cells? Clinical Cancer Research: an Official Journal of the American Association for Cancer Research. 2010;16(12):3153–3162. doi: 10.1158/1078-0432.CCR-09-2943. [DOI] [PubMed] [Google Scholar]
- 99.Li H, Li T, Huang D, Zhang P. Long noncoding RNA SNHG17 induced by YY1 facilitates the glioma progression through targeting miR-506-3p/CTNNB1 axis to activate Wnt/beta-catenin signaling pathway. Cancer Cell International. 2020;20:29. doi: 10.1186/s12935-019-1088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Walid MS, Smisson HF, III, Robinson JS., Jr. Long-term survival after glioblastoma multiforme. Southern Medical Journal. 2008;101(9):971–972. doi: 10.1097/SMJ.0b013e31818005e5. [DOI] [PubMed] [Google Scholar]
- 101.Li J, Song J, Guo F. miR-186 reverses cisplatin resistance and inhibits the formation of the glioblastoma-initiating cell phenotype by degrading Yin Yang 1 in glioblastoma. International Journal of Molecular Medicine. 2019;43(1):517–524. doi: 10.3892/ijmm.2018.3940. [DOI] [PubMed] [Google Scholar]
- 102.Johannessen TC, Bjerkvig R. Molecular mechanisms of temozolomide resistance in glioblastoma multiforme. Expert Review of Anticancer Therapy. 2012;12(5):635–642. doi: 10.1586/era.12.37. [DOI] [PubMed] [Google Scholar]
- 103.Jia B, Liu W, Gu J, Wang J, Lv W, Zhang W, Hao Q, Pang Z, Mu N, Zhang W, et al. MiR-7-5p suppresses stemness and enhances temozolomide sensitivity of drug-resistant glioblastoma cells by targeting Yin Yang 1. Experimental Cell Research. 2019;375(1):73–81. doi: 10.1016/j.yexcr.2018.12.016. [DOI] [PubMed] [Google Scholar]
- 104.Dong S, Ma X, Wang Z, Han B, Zou H, Wu Z, Zang Y, Zhuang L. YY1 promotes HDAC1 expression and decreases sensitivity of hepatocellular carcinoma cells to HDAC inhibitor. Oncotarget. 2017;8(25):40583–40593. doi: 10.18632/oncotarget.17196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Y, Kasim V, Yan X, Li L, Meliala ITS, Huang C, Li Z, Lei K, Song G, Zheng X, et al. Yin Yang 1 facilitates hepatocellular carcinoma cell lipid metabolism and tumor progression by inhibiting PGC-1beta-induced fatty acid oxidation. Theranostics. 2019;9(25):7599–7615. doi: 10.7150/thno.34931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu Y, Zhang J, Zhang X, Zhou H, Liu G, Li Q. Cancer stem cells: a potential breakthrough in HCC-targeted therapy. Frontiers in Pharmacology. 2020;11:198. doi: 10.3389/fphar.2020.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Research. 2008;68(5):1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 108.Zhao X, Parpart S, Takai A, Roessler S, Budhu A, Yu Z, Blank M, Zhang YE, Jia HL, Ye QH, et al. Integrative genomics identifies YY1AP1 as an oncogenic driver in EpCAM(+) AFP(+) hepatocellular carcinoma. Oncogene. 2015;34(39):5095–5104. doi: 10.1038/onc.2014.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang C, Shao S, Zhang Y, Wang L, Liu J, Fang F, Li P, Wang B. LncRNA PCAT1 promotes metastasis of endometrial carcinoma through epigenetical downregulation of E-cadherin associated with methyltransferase EZH2. Life Sciences. 2020;243:117295. doi: 10.1016/j.lfs.2020.117295. [DOI] [PubMed] [Google Scholar]
- 110.Roh JW, Choi JE, Han HD, Hu W, Matsuo K, Nishimura M, Lee JS, Kwon SY, Cho CH, Kim J, et al. Clinical and biological significance of EZH2 expression in endometrial cancer. Cancer Biology & Therapy. 2020;21(2):147–156. doi: 10.1080/15384047.2019.1672455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nature Medicine. 2016;22(2):128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cao Q, Mani RS, Ateeq B, Dhanasekaran SM, Asangani IA, Prensner JR, Kim JH, Brenner JC, Jing X, Cao X, et al. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell. 2011;20(2):187–199. doi: 10.1016/j.ccr.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ihira K, Dong P, Xiong Y, Watari H, Konno Y, Hanley SJ, Noguchi M, Hirata N, Suizu F, Yamada T, et al. EZH2 inhibition suppresses endometrial cancer progression via miR-361/Twist axis. Oncotarget. 2017;8(8):13509–13520. doi: 10.18632/oncotarget.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang Y, Zhou L, Lu L, Wang L, Li X, Jiang P, Chan LK, Zhang T, Yu J, Kwong J, et al. A novel miR-193a-5p-YY1-APC regulatory axis in human endometrioid endometrial adenocarcinoma. Oncogene. 2013;32(29):3432–3442. doi: 10.1038/onc.2012.360. [DOI] [PubMed] [Google Scholar]
- 115.Painter JN, Kaufmann S, O’Mara TA, Hillman KM, Sivakumaran H, Darabi H, Cheng THT, Pearson J, Kazakoff S, Waddell N, et al. A common variant at the 14q32 endometrial cancer risk locus activates AKT1 through YY1 binding. American Journal of Human Genetics. 2016;98(6):1159–1169. doi: 10.1016/j.ajhg.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dong P, Konno Y, Watari H, Hosaka M, Noguchi M, Sakuragi N. The impact of microRNA-mediated PI3K/AKT signaling on epithelial-mesenchymal transition and cancer stemness in endometrial cancer. Journal of Translational Medicine. 2014;12:231. doi: 10.1186/s12967-014-0231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maverakis E, Cornelius LA, Bowen GM, Phan T, Patel FB, Fitzmaurice S, He Y, Burrall B, Duong C, Kloxin AM, et al. Metastatic melanoma - a review of current and future treatment options. Acta Dermato-Venereologica. 2015;95(5):516–524. doi: 10.2340/00015555-2035. [DOI] [PubMed] [Google Scholar]
- 118.Zou Y, Wang SS, Wang J, Su HL, Xu JH. CircRNA_0016418 expedites the progression of human skin melanoma via miR-625/YY1 axis. European Review for Medical and Pharmacological Sciences. 2019;23(24):10918–10930. doi: 10.26355/eurrev_201912_19795. [DOI] [PubMed] [Google Scholar]
- 119.Du J, Ren W, Yao F, Wang H, Zhang K, Luo M, Shang Y, O’Connell D, Bei Z, Wang H, et al. YY1 cooperates with TFEB to regulate autophagy and lysosomal biogenesis in melanoma. Molecular Carcinogenesis. 2019;58(11):2149–2160. doi: 10.1002/mc.23105. [DOI] [PubMed] [Google Scholar]
- 120.Choi J, Zhang T, Vu A, Ablain J, Makowski MM, Colli LM, Xu M, Hennessey RC, Yin J, Rothschild H, et al. Massively parallel reporter assays of melanoma risk variants identify MX2 as a gene promoting melanoma. Nature Communications. 2020;11(1):2718. doi: 10.1038/s41467-020-16590-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marzagalli M, Raimondi M, Fontana F, Montagnani Marelli M, Moretti RM, Limonta P. Cellular and molecular biology of cancer stem cells in melanoma: Possible therapeutic implications. Seminars in Cancer Biology. 2019;59:221–235. doi: 10.1016/j.semcancer.2019.06.019. [DOI] [PubMed] [Google Scholar]
- 122.Bedogni B, Paus R. Hair(y) matters in melanoma biology. Trends in Molecular Medicine. 2020;26(5):441–449. doi: 10.1016/j.molmed.2020.02.005. [DOI] [PubMed] [Google Scholar]
- 123.Lang D, Mascarenhas JB, Shea CR. Melanocytes, melanocyte stem cells, and melanoma stem cells. Clinics in Dermatology. 2013;31(2):166–178. doi: 10.1016/j.clindermatol.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kohler C, Nittner D, Rambow F, Radaelli E, Stanchi F, Vandamme N, Baggiolini A, Sommer L, Berx G, van den Oord JJ, et al. Mouse cutaneous melanoma induced by mutant BRaf arises from expansion and dedifferentiation of mature pigmented melanocytes. Cell Stem Cell. 2017;21(5):679–693 e676. doi: 10.1016/j.stem.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 125.Ackermann J, Frutschi M, Kaloulis K, McKee T, Trumpp A, Beermann F. Metastasizing melanoma formation caused by expression of activated N-RasQ61K on an INK4a-deficient background. Cancer Research. 2005;65(10):4005–4011. doi: 10.1158/0008-5472.CAN-04-2970. [DOI] [PubMed] [Google Scholar]
- 126.Casarsa C, Oriana S, Coradini D. The controversial clinicobiological role of breast cancer stem cells. Journal of Oncology. 2008;2008:492643. doi: 10.1155/2008/492643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Britton KM, Kirby JA, Lennard TW, Meeson AP. Cancer stem cells and side population cells in breast cancer and metastasis. Cancers. 2011;3(2):2106–2130. doi: 10.3390/cancers3022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhou J, Chen Q, Zou Y, Chen H, Qi L, Chen Y. Stem cells and cellular origins of breast cancer: updates in the rationale, controversies, and therapeutic implications. Frontiers in Oncology. 2019;9:820. doi: 10.3389/fonc.2019.00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Butti R, Gunasekaran VP, Kumar TVS, Banerjee P, Kundu GC. Breast cancer stem cells: Biology and therapeutic implications. The International Journal of Biochemistry & Cell Biology. 2019;107:38–52. doi: 10.1016/j.biocel.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 130.Wan M, Huang W, Kute TE, Miller LD, Zhang Q, Hatcher H, Wang J, Stovall DB, Russell GB, Cao PD, et al. Yin Yang 1 plays an essential role in breast cancer and negatively regulates p27. The American Journal of Pathology. 2012;180(5):2120–2133. doi: 10.1016/j.ajpath.2012.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang, W., Feng, B., Meng, Y., Wang, J., Geng, B., Cui, Q., Zhang, H., Yang, Y., & Yang, J. (2019). FAM3C-YY1 axis is essential for TGFbeta-promoted proliferation and migration of human breast cancer MDA-MB-231 cells via the activation of HSF1. Journal of Cellular and Molecular Medicine, 23(5), 3464–3475. [DOI] [PMC free article] [PubMed]
- 132.Schiano, C., Franzese, M., Pane, K., Garbino, N., Soricelli, A., Salvatore, M., de Nigris, F., & Napoli, C. (2019). Hybrid (18)F-FDG-PET/MRI measurement of standardized uptake value coupled with Yin Yang 1 signature in metastatic breast cancer. A preliminary study. Cancers, 11(10), 1444. [DOI] [PMC free article] [PubMed]
- 133.Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies metastatic pathways and transcription factors in breast cancer. BMC Cancer. 2008;8:394. doi: 10.1186/1471-2407-8-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Abukhdeir AM, Park BH. P21 and p27: roles in carcinogenesis and drug resistance. Expert Reviews in Molecular Medicine. 2008;10:e19. doi: 10.1017/S1462399408000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Menchon C, Edel MJ, Izpisua Belmonte JC. The cell cycle inhibitor p27Kip(1) controls self-renewal and pluripotency of human embryonic stem cells by regulating the cell cycle, brachyury and twist. Cell Cycle. 2011;10(9):1435–1447. doi: 10.4161/cc.10.9.15421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhan Z, Song L, Zhang W, Gu H, Cheng H, Zhang Y, Yang Y, Ji G, Feng H, Cheng T, et al. Absence of cyclin-dependent kinase inhibitor p27 or p18 increases efficiency of iPSC generation without induction of iPSC genomic instability. Cell Death & Disease. 2019;10(4):271. doi: 10.1038/s41419-019-1502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Feng X, Xu X, Xiao X, Zou K, Yu W, Wu J, Tang R, Gao Y, Hao J, Zhao X, et al. NMI inhibits cancer stem cell traits by downregulating hTERT in breast cancer. Cell Death & Disease. 2017;8(5):e2783. doi: 10.1038/cddis.2017.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chung SS, Aroh C, Vadgama JV. Constitutive activation of STAT3 signaling regulates hTERT and promotes stem cell-like traits in human breast cancer cells. PLoS One. 2013;8(12):e83971. doi: 10.1371/journal.pone.0083971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Paranjape AN, Mandal T, Mukherjee G, Kumar MV, Sengupta K, Rangarajan A. Introduction of SV40ER and hTERT into mammospheres generates breast cancer cells with stem cell properties. Oncogene. 2012;31(15):1896–1909. doi: 10.1038/onc.2011.378. [DOI] [PubMed] [Google Scholar]
- 140.Guo Q, Wang T, Yang Y, Gao L, Zhao Q, Zhang W, Xi T, Zheng L. Transcriptional factor Yin Yang 1 promotes the stemness of breast cancer cells by suppressing miR-873-5p transcriptional activity. Molecular Therapy Nucleic Acids. 2020;21:527–541. doi: 10.1016/j.omtn.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhu Z, Zhu X, Yang S, Guo Z, Li K, Ren C, Zhou Y, Dou J. Yin-yang effect of tumour cells in breast cancer: from mechanism of crosstalk between tumour-associated macrophages and cancer-associated adipocytes. American Journal of Cancer Research. 2020;10(2):383–392. [PMC free article] [PubMed] [Google Scholar]
- 142.Hasegawa T, Yashiro M, Nishii T, Matsuoka J, Fuyuhiro Y, Morisaki T, Fukuoka T, Shimizu K, Shimizu T, Miwa A, et al. Cancer-associated fibroblasts might sustain the stemness of scirrhous gastric cancer cells via transforming growth factor-beta signaling. International Journal of Cancer. 2014;134(8):1785–1795. doi: 10.1002/ijc.28520. [DOI] [PubMed] [Google Scholar]
- 143.Sansone P, Savini C, Kurelac I, Chang Q, Amato LB, Strillacci A, Stepanova A, Iommarini L, Mastroleo C, Daly L, et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(43):E9066–E9075. doi: 10.1073/pnas.1704862114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Karagiannis GS, Poutahidis T, Erdman SE, Kirsch R, Riddell RH, Diamandis EP. Cancer-associated fibroblasts drive the progression of metastasis through both paracrine and mechanical pressure on cancer tissue. Molecular Cancer Research: MCR. 2012;10(11):1403–1418. doi: 10.1158/1541-7786.MCR-12-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao JL, Huang F, He F, Gao CC, Liang SQ, Ma PF, Dong GY, Han H, Qin HY. Forced Activation of notch in macrophages represses tumor growth by upregulating miR-125a and disabling tumor-associated macrophages. Cancer Research. 2016;76(6):1403–1415. doi: 10.1158/0008-5472.CAN-15-2019. [DOI] [PubMed] [Google Scholar]
- 146.Grosselin K, Durand A, Marsolier J, Poitou A, Marangoni E, Nemati F, Dahmani A, Lameiras S, Reyal F, Frenoy O, et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nature Genetics. 2019;51(6):1060–1066. doi: 10.1038/s41588-019-0424-9. [DOI] [PubMed] [Google Scholar]
- 147.Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nature Biotechnology. 2015;33(11):1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tan L, Xing D, Chang CH, Li H, Xie XS. Three-dimensional genome structures of single diploid human cells. Science. 2018;361(6405):924–928. doi: 10.1126/science.aat5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zheng H, Xie W. The role of 3D genome organization in development and cell differentiation. Nature Reviews. Molecular Cell Biology. 2019;20(9):535–550. doi: 10.1038/s41580-019-0132-4. [DOI] [PubMed] [Google Scholar]
- 151.Bonev B, Cavalli G. Organization and function of the 3D genome. Nature Reviews. Genetics. 2016;17(11):661–678. doi: 10.1038/nrg.2016.112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data presented in this review are totally available and present in the text.
Not applicable.