Abstract
Respiratory tract infections are a major cause of morbidity and mortality worldwide in young children. Concepts such as the gut-lung axis have highlighted the impact of microbial communities at distal sites in mediating disease locally. However, little is known about the extent to which microbial communities from multiple body sites are linked, and how this relates to disease susceptibility. Here, we combine 16S-based rRNA sequencing data from 112 healthy, term born infants, spanning three body sites (oral cavity, nasopharynx, gut) and the first six months of life. Using a cross-niche microbial network approach, we show that, already from the first week of life on, there is a strong association between both network structure and species essential to these structures (hub species), and consecutive susceptibility to respiratory tract infections in this cohort. Our findings underline the crucial role of cross-niche microbial connections in respiratory health.
Subject terms: Microbiome, Microbial ecology
In order to assess the extent by which microbial communities from multiple body sites are linked and related to disease susceptibility, Reyman and Clerc et al. combined 16S-based rRNA sequencing data from 112 healthy, term born infants, spanning three body sites (oral cavity, nasopharynx, gut) over the first six months of life. They demonstrate a strong association between network structure and species and susceptibility to respiratory tract infections, suggesting a crucial role of cross-niche microbial connections in respiratory health.
Introduction
The human microbiome is widely recognised as an important mediator of health and disease, making it the subject of extensive study. The microbiome is highly variable between individuals but also within one individual when studied over time or across body sites1. Exposures to a wide range of environmental factors, such as delivery mode, antibiotics, and diet, have been shown to contribute to this variation2. In general, microbiome research has focused on the relation between the microbial community composition of a single anatomical niche and health or disease parameters.
However, it is becoming increasingly clear that direct or indirect effects of microbial communities at distal body sites exist and can also have major implications for both development, as well as severity of a number of diseases locally3. One well-established example of cross-niche microbial interaction is the gut-lung axis. It has been shown that microbial components and metabolites in both the gut and lung are capable of modulating immunity not only locally but also systemically4. Additionally, specific taxa in both the gastrointestinal and respiratory tract are associated with lung diseases such as asthma, chronic obstructive pulmonary disease, and respiratory tract infections (RTIs)4.
Next to variation in species presence/absence and abundance within niches, other features such as the microbial network structure may also contribute to susceptibility to RTIs. Commonly, studies of microbiome networks have been restricted to microbial networks within a single niche5,6. For instance, a study performed in patients suffering from inflammatory bowel disease (IBD) showed that the community structure of the microbial network within the gut was distorted in IBD patients7. However, it has been hypothesised that the structure of the network formed by the microbes within a specific niche might not only affect local disease development and/or severity but might also have more systemic effects related to disease8. Even though examples for such effects are emerging from the literature, we currently lack a conceptual understanding if and to what extent variation in the structure of cross-niche microbial networks might also affect disease susceptibility locally. Understanding these relationships would substantially increase our ability to formulate holistic hypotheses about the development and improvement of integrated diagnostic and treatment approaches for a wide range of diseases.
Here, we, therefore, investigate the structure of microbial networks across different body sites in order to identify signatures in the overall microbial community network in infancy that can be associated with susceptibility to RTIs in the first year of life. To do so, we used 16S rRNA sequencing data from samples of a Dutch cohort of 112 healthy, term born infants collected longitudinally over the first 6 months of life and spanning three body sites (oral cavity, nasopharynx, and gastrointestinal tract). We aimed to (1) compare the development of the microbial communities per niche over time, (2) build cross-niche microbial networks, (3) identify hub species within the networks, and (4) study network structure and hub species in relation to RTI susceptibility during the first year of life. Using this stepwise approach, we found a strong association between cross-niche network structure and hub species and consecutive susceptibility to respiratory tract infections.
Results
Niche comparison
Of the 1,250 available samples obtained from the three niches at 1 week, 2, 4, and 6 months of life of 112 healthy infants, 1,248 samples fulfilled our quality threshold (433 faecal samples, 430 nasopharyngeal samples, and 385 saliva samples). Sequencing of these samples resulted in 58,608,834 high quality reads with a minimum Good’s coverage of 99.47% (median 99.97%). The overall Operational Taxonomical Unit (OTU)-table, including the samples of the three niches collected at the four time points, contained 1,148 bacterial OTUs distributed over 18 bacterial phyla, with Firmicutes being the most abundant phylum and Streptococcus the most abundant genus.
In the oral cavity, the most abundant genus over the first 6 months of life was Streptococcus, while this was Moraxella for the nasopharynx and Bifidobacterium for the gut. Observed species richness was highest in the nasopharyngeal samples, with 895 OTUs identified, compared to 746 OTUs in the saliva and 595 OTUs in the faecal samples. In addition, the nasopharynx also contained the highest number of unique OTUs (defined as observed in a single niche only), namely 232, followed by 121 in the gut and only 38 in the oral cavity. In other words, the oral niche contained a relatively high number of observed species but few unique OTUs, and so it seems to be a reservoir sharing many OTUs with the nasopharynx and gut. Although highly variable in abundance, a total of 331 overlapping OTUs were observed in all three niches, including the top 10 most abundant OTUs in the overall dataset of the combined niches: Streptococcus (1), Bifidobacterium (2), Moraxella (3), Staphylococcus (4), Corynebacterium propinquum (5), Streptococcus salivarius (6), Dolosigranulum (7), Escherichia coli (9), Veillonella (10), and Haemophilus (8).
Supplementary Fig. 1 shows the succession patterns of the 15 most abundant OTUs in each niche, highlighting the gradual increase of Bifidobacterium (2) over time in the gut (Supplementary Fig. 1a). In the nasopharynx, the initial high abundance of Staphylococcus (4) at 1 week of life was gradually replaced by an increasing abundance of C. propinquum (5), Dolosigranulum (7), Moraxella (3), and Haemophilus (8) (Supplementary Fig. 1b). Lastly, the figure highlights an overall dominance of Streptococcus (1) in the oral cavity (Supplementary Fig. 1c). Observed species richness increased over time in both faecal and saliva samples, though this effect was most pronounced in saliva (linear mixed effect model including age and diversity per niche: p < 0.0001 for both niches; Supplementary Fig. 2).
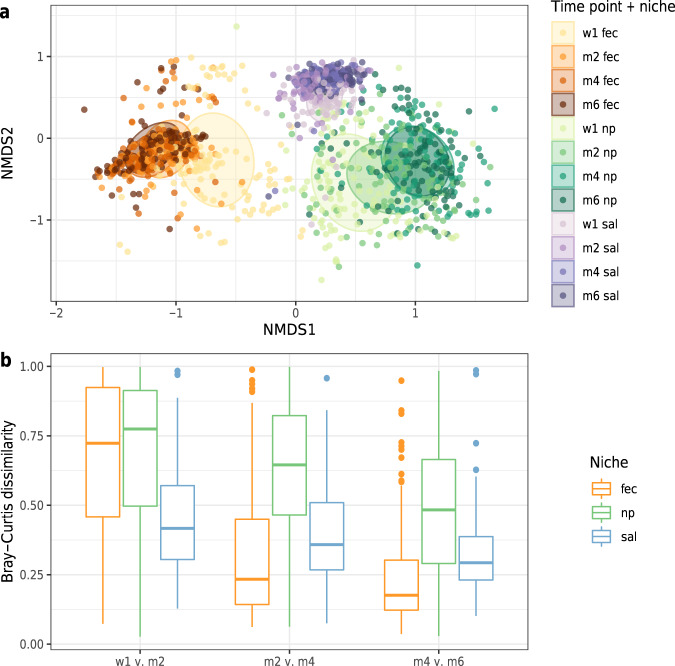
Microbial community development over time (shown in Fig. 1a) was most stable for the oral cavity in early-life, although this was overtaken by higher stability in the gut at the later time points (median Bray–Curtis [BC] dissimilarity between month 4 and 6 for faecal samples 0.18 versus 0.29 for saliva, Wilcoxon test, p < 0.0001; Fig. 1b). The microbial community composition in the nasopharynx showed the lowest temporal stability compared to the other two niches over the first 6 months of life. As expected, the community composition differed significantly between the three niches at all time points, although niche composition was most similar at the earliest time point, in line with the mutual origin of initial microbial seeding at birth. This was followed by a gradual deviation into niche-specific communities over time (calculating the association of niche with composition using PERMANOVA, week 1: R2 31.8%, month 2: R2 47.8%, month 4; R2 57.5%, month 6: R2 56.3%, all p-values < 0.0001).
Fig. 1. Development of the microbiota community composition for individual niches over time.
Non-metric multidimensional scaling (nMDS) plot (a), based on Bray–Curtis (BC) dissimilarity between samples, visualising the overall gut microbial community composition stratified for each niche per time point. Each data point represents the microbial community composition of one sample. The ellipses represent the standard deviation of data points belonging to each group, with the centre points of the ellipses calculated using the mean of the coordinates per group. The stress of the ordination was 0.18. In panel b the temporal community stability for each niche is shown. As a measure of microbiota stability, we calculated the Bray–Curtis distance between consecutive sample pairs belonging to each individual per time interval, i.e., between week 1 and month 2, between month 2 and month 4, and between month 4 and month 6. Boxplots with medians are shown; the lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles); the upper and lower whiskers extend from the hinge to the largest and smallest value no further than 1.5*IQR from the hinge; outliers are plotted individually. Fec faeces, np nasopharynx, sal saliva. Temporal stability was highest for the saliva samples in early-life, which was later overtaken by a higher stability in consecutive faecal samples (median BC dissimilarity between months 4 and 6 for faecal samples 0.18 versus 0.29 for saliva, Wilcoxon test, p < 0.0001).
Characteristics explaining the overall community composition across niches and time points were in order of importance, niche itself (R2 42.4%, adjusted p-value 0.00079), followed by age with a much smaller effect size (R2 1.8%, adjusted p-value 0.00079), and presence of siblings <5 years, pets, breastfeeding at time of sampling, season of birth, mode of delivery, exposure to antibiotics in the month prior to sampling, day care attendance, pacifier use, and duration of hospital stay after birth (Supplementary Table 1). When studying the niches separately and cross-sectionally per time point, we observed different effect sizes of associations between environmental variables and microbial community composition (Supplementary Table 2). In total, we identified eight covariates to be associated with the nasopharyngeal microbiome (among others presence of siblings <5 years, breastfeeding at sampling and season of birth), seven covariates for the faecal microbiome (among others mode of delivery, breastfeeding at sampling and day care attendance), and only three covariates for the salivary microbiome composition (breastfeeding at sampling, antibiotic use 1 month prior to sampling and season of birth).
Network analysis
We identified a total of 107 niche indicator OTUs (defined as characteristic, but not exclusive, for a particular niche), with 18 niche indicator OTUs specific for the nasopharynx, 37 for the gut and 52 for the oral cavity. Participants were divided into three groups based on the distribution of the number of RTIs they experienced over the first year of life (0-2 RTIs, 3-4 RTIs, 5–7 RTIs). After selecting the 100 most abundant OTUs per niche and per RTI group, we constructed for each of the three RTI groups a cross-niche microbial network per time point using the SpiecEasi pipeline9. We studied the formation of bacterial clusters within these cross-niche networks, and whether clusters were indicative of a specific niche or not, based on the proportion of niche indicator OTUs the clusters contained. In the results below, we will further refer to the cross-niche networks as networks.
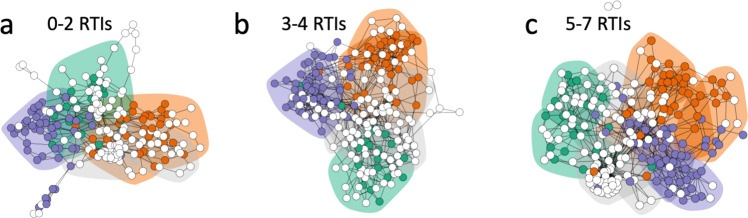
We found that at 1 week of age, the network of the least susceptible group (0–2 RTIs) was structured into six clusters (one indicative of nasopharynx, one saliva, two faeces, and two mixed), while the network of the average susceptible group (3–4 RTIs) contained seven clusters (one nasopharynx, one saliva, two faeces, and three mixed) and the network of the most susceptible group (5–7 RTIs) contained nine clusters (two nasopharynx, two saliva, three faeces, and two mixed; Supplementary Fig. 3). The number of clusters per network generally decreased over time with cluster sizes increasing (Fig. 2a, b). However, cluster sizes in the networks from the 5–7 RTI group showed a trend towards less increase over time compared to clusters from networks from the 0–2 RTI group (negative binomial GLM: 5–7 RTI group estimate = − 0.3, p = 0.071), which coincided with a higher number of clusters retained in the 5–7 RTI networks. This resulted in the networks of the lowest and average susceptible RTI groups each defined by four clusters each, whereas the network from the most susceptible group still contained six clusters at the age of 6 months, suggesting more fragmented networks. The two extra clusters identified in the most susceptible group had a saliva and faecal origin, thereby split the oral and gut communities into two communities each (Supplementary Fig. 3). Figure 3 depicts the networks of each RTI group at time point week 1.
Fig. 2. Cluster number and size of the cross-niche microbial networks built per RTI group.
Number of main clusters (a) and mean size of main clusters (b) over time. Number of clusters at week 1 were n = 22 (n =6 for 0–2 RTIs, n = 7 for 3–4 RTIs and n = 9 for 5–7 RTIs), n = 20 at month 2 (n = 6 for 0–2 RTIs, n = 7 for 3–4 RTIs and n = 7 for 5–7 RTIs), n = 16 at month 4 (n = 6 for 0–2 RTIs, n = 4 for 3–4 RTIs and n = 6 for 5–7 RTIs) and n = 14 at month 6 (n = 4 for 0–2 RTIs, n = 4 for 3–4 RTIs and n = 6 for 5–7 RTIs). Boxplots with medians are shown; the lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles); the upper and lower whiskers extend from the hinge to the largest and smallest value no further than 1.5 *IQR from the hinge. RTI stands for number of respiratory tract infections experienced over the first year of life.
Fig. 3. Cross-niche networks for each RTI group at week 1.
Microbial cross-niche networks using data from the nasopharynx, gut, and oral cavity (a–c). Nodes represent individual OTUs and were coloured depending on their indicator species identity (orange = faeces-specific, green = nasopharynx-specific, blue = saliva specific, white = not niche-specific). Shaded areas around groups of nodes represent clusters defined by walktrap community analysis and are shaded depending on their niche identity (orange = faeces-specific, green = nasopharynx-specific, blue = saliva specific, grey = mixed cluster/not niche-specific).
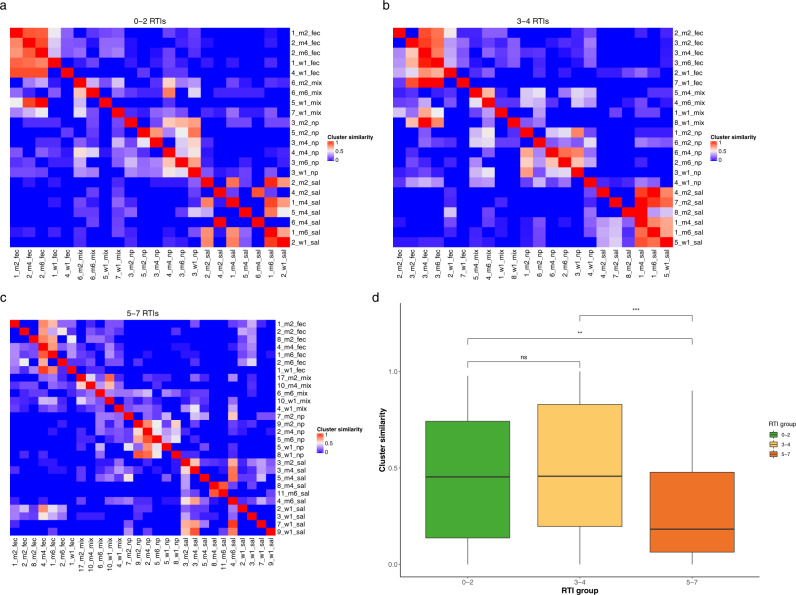
We then studied the similarity of clusters over time as a measure of temporal network stability (Fig. 4). The mean similarity over time differed between RTI groups, with 45.6% ± 4.9 SE similarity over time for the least susceptible group, 51.8% ± 5.1 SE for the average susceptible group and 29.0% ± 3.2 SE for the most susceptible group (Kruskal–Wallis test: X2 = 15.7, p = 0.0004, Fig. 4d). Specifically, the similarity of clusters in the networks of the most susceptible group was lower compared to the other two RTI groups (Dunn post-hoc test, 0–2 vs 3–4 RTIs: p = 0.328, 0–2 vs 5–7 RTIs: p = 0.012, 3–4 vs 5–7 RTIs: p = 0.0006; Fig. 4d). In stratified analyses per niche, we found significant differences in cluster similarity between RTI groups for saliva clusters (similarity over time 44.0% ± 9.9 SE in the 0–2 RTI group, 78.9% ± 6.7 SE in the 3–4 RTI group, and 19.6% ± 4.5 SE in the 5–7 RTI group, Kruskal–Wallis test X2 = 19.1, p < 0.0001, Supplementary Fig. 4). In addition, we found non-significant trends for the association between RTI susceptibility and loss in cluster similarity for faecal clusters (similarity over time 49.9% ± 11.1 SE in the 0–2 RTI group, 49.4% ± 9.3 SE in the 3–4 RTI group, and 41.0% ± 6.9 SE in the 5–7 RTI group, Kruskal–Wallis test X2 = 1.00, p = 0.612, Supplementary Fig. 4) and nasopharyngeal clusters (similarity over time 54.5% ± 5.8 SE in the 0–2 RTI group, 42.7 % ± 7.4 SE in the 3-4 RTI group, and 35.3% ± 6.2 SE in the 5–7 RTI group, Kruskal–Wallis test X2 = 4.28, p = 0.118). Overall, this suggests reduced cluster stability over time in networks from children from the 5–7 RTI group.
Fig. 4. Stability and similarity of individual network clusters.
Heatmaps (a–c) displaying the similarity between the cluster compositions of all clusters at all time points for networks for the 0–2 RTI group, 3–4 RTI group and 5–7 RTI group, respectively. Cluster similarity was calculated as the proportion of OTUs shared between two clusters (1 represents complete overlap between all OTUs of two clusters, whereas 0 represents no OTUs being shared between two clusters). Each cluster is labelled with a unique number to distinguish between clusters with the same niche-identity, the time point of the network from which the cluster originates (w1 = week 1, m2 = month 2, m4 = month 4, m6 = month 6) and the niche-identity of the cluster (fec = faecal cluster, np = nasopharyngeal cluster, sal = saliva cluster, mix = no clear niche-identity of cluster). d Mean cluster similarity for all clusters per RTI group across time. The number of clusters that are compared to each other is n = 22 for 0–2 RTIs, n = 22 for 3–4 RTIs, and n = 28 for 5–7 RTIs. Boxplots with medians are shown; the lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles); the upper and lower whiskers extend from the hinge to the largest and smallest value no further than 1.5 *IQR from the hinge. In order to assess the extent by which microbial communities from multiple body sites are linked and related to disease susceptibility, Reyman and Clerc et al. combined 16S-based rRNA sequencing data from 112 healthy, term born infants, spanning three body sites (oral cavity, nasopharynx, and gut) over the first 6 months of life. They demonstrate a strong association between network structure and species and susceptibility to respiratory tract infections, suggesting a crucial role of cross-niche microbial connections in respiratory health.
Hub species
To identify OTUs with an important role for the network structure, we calculated the degree, betweenness centrality and closeness centrality for each of the OTUs in each network (only considering main clusters). Table 1 lists the 134 hub species found, also reporting whether they were niche indicator species or not, and for which RTI network (least, average, or most susceptible) they were identified as being important. We identified only 5 OTUs (Akkermansia (280), Moraxella (190), Actinomyces (58), Veillonella (10) and Prevotella melaninogenica (22)) as hub species in all three RTI groups, indicating that different OTUs are central to the network structure of the different RTI groups.
Table 1.
Hub species for all RTI cohorts.
| OTU | Hub ID | Niche indicator | No. thresholds met |
|---|---|---|---|
| Actinomyces_58 | All | Sal | 1 |
| Akkermansia_280 | All | None | 1 |
| Moraxella_190 | All | None | 1 |
| Prevotella_melaninogenica_22 | All | Sal | 1 |
| Veillonella_10 | All | Sal | 1 |
| 7B_8_687 | 0–2 | None | 2 |
| Acinetobacter_calcoaceticus_146 | 0–2 | None | 1 |
| Aggregatibacter_354 | 0–2 | None | 1 |
| Alloscardovia_316 | 0–2 | None | 2 |
| Altererythrobacter_748 | 0–2 | None | 1 |
| Anaerostipes_142 | 0–2 | None | 1 |
| Blastococcus_281 | 0–2 | None | 2 |
| Blastococcus_707 | 0–2 | None | 1 |
| Blautia_212 | 0–2 | None | 1 |
| Chroococcidiopsis_338 | 0–2 | None | 1 |
| Chroococcidiopsis_593 | 0–2 | None | 3 |
| Chryseobacterium_299 | 0–2 | None | 1 |
| Corynebacterium_111 | 0–2 | None | 2 |
| Corynebacterium_162 | 0–2 | None | 1 |
| Craurococcus_sp_HM28_1_726 | 0–2 | None | 1 |
| Cupriavidus_31 | 0–2 | None | 1 |
| Curvibacter_286 | 0–2 | None | 1 |
| Dorea_86 | 0–2 | None | 1 |
| Erysipelotrichaceae_149 | 0–2 | None | 2 |
| Lachnospiraceae_176 | 0–2 | None | 2 |
| Limnobacter_245 | 0–2 | None | 2 |
| Megasphaera_133 | 0–2 | None | 2 |
| Megasphaera_sp_TrE9262_383 | 0–2 | None | 1 |
| Moraxella_131 | 0–2 | None | 1 |
| Neisseria_lactamica_47 | 0–2 | None | 1 |
| Pseudobutyrivibrio_195 | 0–2 | None | 2 |
| Rubellimicrobium_596 | 0–2 | None | 1 |
| Ruminococcus_bromii_L2_63_112 | 0–2 | None | 2 |
| Sphingomonas_573 | 0–2 | None | 1 |
| Subdoligranulum_71 | 0–2 | None | 1 |
| Varibaculum_199 | 0–2 | None | 1 |
| Escherichia_Shigella_267 | 0–2 | Fec | 1 |
| Comamonadaceae_118 | 0–2 | Np | 1 |
| Dolosigranulum_pigrum_ATCC_51524_147 | 0–2 | Np | 1 |
| Enhydrobacter_138 | 0–2 | Np | 1 |
| Pseudomonas_158 | 0–2 | Np | 1 |
| Haemophilus_211 | 0–2 | Sal | 1 |
| Helcococcus_179 | 0–2 and 3–4 | None | 2 |
| Scardovia_wiggsiae_F0424_127 | 0–2 and 3–4 | None | 2 |
| Bifidobacterium_2 | 0–2 and 3–4 | Fec | 2 |
| Rothia_259 | 0–2 and 3–4 | Sal | 1 |
| Streptococcus_1 | 0–2 and 3–4 | Sal | 1 |
| Actinomyces_100 | 0–2 and 5–7 | None | 1 |
| Blautia_308 | 0–2 and 5–7 | None | 1 |
| Moryella_119 | 0–2 and 5–7 | None | 1 |
| Gallibacterium_Salpingitidis_141 | 0–2 and 5–7 | Sal | 1 |
| Akkermansia_210 | 3–4 | None | 1 |
| Akkermansia_40 | 3–4 | None | 1 |
| boneC3G7_70 | 3–4 | None | 2 |
| Clostridium_colinum_487 | 3–4 | None | 1 |
| Clostridium_nexile_166 | 3–4 | None | 2 |
| Corynebacterium_500 | 3–4 | None | 2 |
| Corynebacterium_aurimucosum_ATCC_700975_260 | 3–4 | None | 2 |
| Haemophilus_226 | 3–4 | None | 1 |
| Lachnoanaerobaculum_115 | 3–4 | None | 1 |
| Lactobacillus_plantarum_337 | 3–4 | None | 1 |
| Modestobacter_323 | 3–4 | None | 1 |
| Moraxella_204 | 3–4 | None | 1 |
| Morganella_294 | 3–4 | None | 1 |
| Neisseria_345 | 3–4 | None | 2 |
| Peptococcus_like_sp_oral_clone_I070_371 | 3–4 | None | 2 |
| Propionibacterium_312 | 3–4 | None | 2 |
| Sphingomonas_228 | 3–4 | None | 1 |
| Turicella_264 | 3–4 | None | 2 |
| Zymomonas_335 | 3–4 | None | 2 |
| Bifidobacteriaceae_213 | 3–4 | Fec | 1 |
| Clostridium_sensu_stricto_1_33 | 3–4 | Fec | 1 |
| Collinsella_220 | 3–4 | Fec | 1 |
| Eggerthella_189 | 3–4 | Fec | 2 |
| Klebsiella_13 | 3–4 | Fec | 1 |
| Corynebacterium_350 | 3–4 | Np | 1 |
| Moraxella_3 | 3–4 | Np | 1 |
| Alloprevotella_23 | 3–4 | Sal | 1 |
| Bergeyella_108 | 3–4 | Sal | 1 |
| Lachnoanaerobaculum_128 | 3–4 | Sal | 1 |
| Lactobacillales_63 | 3–4 | Sal | 1 |
| Rothia_126 | 3–4 | Sal | 1 |
| Rothia_16 | 3–4 | Sal | 1 |
| Rothia_192 | 3–4 | Sal | 1 |
| Rothia_287 | 3–4 | Sal | 1 |
| Solobacterium_moorei_232 | 3–4 | Sal | 1 |
| Streptococcus_20 | 3–4 | Sal | 1 |
| Veillonella_66 | 3–4 | Sal | 1 |
| Collinsella_110 | 3–4 and 5–7 | None | 1 |
| Lachnospiraceae_98 | 3–4 and 5–7 | None | 2 |
| Campylobacter_123 | 3–4 and 5–7 | Sal | 1 |
| Veillonella_sp_DNF00869_15 | 3–4 and 5–7 | Sal | 1 |
| Actinobaculum_schaalii_FB123_CNA_2_408 | 5–7 | None | 1 |
| Bacteroides_84 | 5–7 | None | 2 |
| Capnocytophaga_196 | 5–7 | None | 2 |
| Erysipelotrichaceae_102 | 5–7 | None | 1 |
| Erysipelotrichaceae_231 | 5–7 | None | 2 |
| Flavonifractor_200 | 5–7 | None | 1 |
| Fusicatenibacter_saccharivorans_44 | 5–7 | None | 1 |
| Haemophilus_haemolyticus_663 | 5–7 | None | 1 |
| Lachnospiraceae_144 | 5–7 | None | 2 |
| Lachnospiraceae_230 | 5–7 | None | 2 |
| Lachnospiraceae_302 | 5–7 | None | 1 |
| Lactobacillus_fermentum_91 | 5–7 | None | 1 |
| Moraxella_153 | 5–7 | None | 1 |
| Moraxella_412 | 5–7 | None | 1 |
| Moraxella_494 | 5–7 | None | 1 |
| Neisseria_1000 | 5–7 | None | 1 |
| Neisseria_469 | 5–7 | None | 2 |
| Neisseria_527 | 5–7 | None | 1 |
| Neisseria_949 | 5–7 | None | 1 |
| Neisseria_meningitidis_830 | 5–7 | None | 1 |
| Parascardovia_denticolens_F0305_486 | 5–7 | None | 1 |
| Peptoniphilus_sp_S7MS8_148 | 5–7 | None | 1 |
| Peptostreptococcaceae_117 | 5–7 | None | 1 |
| Peptostreptococcaceae_75 | 5–7 | None | 1 |
| Prevotella_sp_oral_clone_ID019_94 | 5–7 | None | 1 |
| Ruminococcaceae_160 | 5–7 | None | 2 |
| Ruminococcaceae_358 | 5–7 | None | 1 |
| Streptococcus_Salivarius_subsp_thermophilus_364 | 5–7 | None | 1 |
| Subdoligranulum_249 | 5–7 | None | 1 |
| Bacteroides_74 | 5–7 | Fec | 1 |
| Bifidobacterium_dentium_Bd1_28 | 5–7 | Fec | 1 |
| Blautia_19 | 5–7 | Fec | 1 |
| Collinsella_26 | 5–7 | Fec | 1 |
| Erysipelotrichaceae_78 | 5–7 | Fec | 1 |
| Parabacteroides_distasonis_82 | 5–7 | Fec | 1 |
| ratAN060301C_27 | 5–7 | Fec | 1 |
| Ruminococcus_gnavus_CC55_001C_24 | 5–7 | Fec | 1 |
| Haemophilus_8 | 5–7 | Np | 1 |
| Actinomyces_sp_oral_clone_DR002_81 | 5–7 | Sal | 1 |
| Haemophilus_215 | 5–7 | Sal | 1 |
| Haemophilus_413 | 5–7 | Sal | 1 |
| Veillonella_156 | 5–7 | Sal | 2 |
Np nasopharynx, sal saliva, Fec faeces, None no niche indicator OTU.
The column niche indicator shows whether an OTU is also a niche indicator OTU. Hub ID indicates for which RTI group (or combination of RTI groups) an OTU is a hub species. Rows are ordered firstly by hub ID, then niche indicator (None, faeces, np or saliva), then on OTU in alphabetical order.
To assess whether the identified hub species were high or low abundant species, we calculated summary statistics for all hub species split by the RTI group(s) for which they were identified as hub species (Supplementary Table 3). The mean abundance of hub species in the 0–2 RTI group networks was 0.0150% ± 0.54 SD (range 0–55.6%), 0.34% ± 3.8 SD for the 3–4 RTI group (range 0–99.5%) and 0.12% ± 1.8 SD (range 0–96.5%) for the 5–7 RTI group underlining hub OTUs are often low abundant species. Furthermore, most hub species were not identified as niche indicator OTUs. This suggests that niche indicator OTUs are less relevant for the microbial network structure and vice versa, that hub species are less niche-specific.
The hub species unique to the 0–2 RTI networks included previously reported respiratory health-associated OTUs such as Neisseria lactamica (47), a low abundant lactic acid-producing Dolosigranulum pigrum (147), and Corynebacterium (162 and 111) OTUs. Also, OTUs associated with the production of butyrates, such as Ruminococcus bromii (112), Megasphaera (133 and 383), and Anaerostipes (142), were found to be hub species in the 0–2 RTI networks. Alternatively, hub species in the 5–7 RTI networks included OTUs previously associated with poorer respiratory health, such as the proteobacterial OTUs Haemophilus (8, 215, 413), Haemophilus haemolyticus (663), and Neisseria (469, 527, 949, 1,000), and the anaerobic Lachnospiraceae (144, 230, 302). Additionally, Actinobaculum schaalii (408) is often associated with (invasive) infections, the proinflammatory Ruminococcus gnavus (24) and the faecal pH increasing Peptostreptococcaceae (75 and 117) were hub species for the networks of infants who experienced 5–7 RTIs. Also, cariogenic species such as Bifidobacterium dentium (28) and Parascardovia denticolens (486) were identified as hub species in the 5–7 RTI networks.
Discussion
Previous studies from this healthy Dutch birth cohort showed already that the nasopharyngeal, oral, and gut microbiota individually develops along specific trajectories, and that various environmental factors are associated with the microbial community composition and development of these three niches in the first months of life. Also, associations between the microbiome development of each of these separate niches and respiratory health have been observed, with an accelerated maturation of the nasopharyngeal microbiome being associated with the number of RTIs experienced in the first year of life, a loss of topography of the upper respiratory microbiome (oropharynx and nasopharynx) being shown to precede RTIs and an association is found between the faecal microbiota composition at one week of life with the number of RTIs experienced in the first year of life10–12. Although this suggests an interplay between the microbial communities across these niches, this has previously not been studied. Here, we investigated cross-niche infant microbiota composition and development and its relationship with susceptibility to RTIs within the first year of life. We show multiple pieces of evidence that support a link between network structure and RTI susceptibility, which are (1) the increase in network fragmentation (increased number of clusters) with increase in RTI susceptibility, (2) the decrease in network stability (decrease in cluster similarity) with an increased RTI susceptibility, and (3) the identification of hub species associated with respiratory health in the least susceptible RTI group vs. identification of hub species associated with (respiratory) dysbiosis in the most susceptible RTI group. To our knowledge, this is the first study using a cross-niche microbial network strategy to study the composition of the wider infant microbiome in relation to (respiratory) health.
In the rapidly developing early-life microbiome, we observed that the anatomical niche was by itself the most important explanatory variable for the microbial community composition, suggesting that the niche environment is the main driver of the composition of the local microbiome. This has already been found in other studies8, but we also identified additional environmental drivers of microbiota composition and development, among others mode of delivery, feeding type and the presence of siblings <5 years in the household11–13.
Building cross-niche microbial networks allowed us to additionally investigate the connections between local and distal microbial communities across the body and how this related to respiratory health. In doing so, we were not only able to identify clusters of OTUs in the cross-niche networks that were niche-specific but also clusters of bacteria that were niche-independent. The network clusters found to be dominated by niche-specific bacteria are in line with previous results from human microbiome network studies8.
When analysing the cross-niche network structure in relation to RTI susceptibility, we observed that the number of clusters within a network was highest for the most susceptible group, while the temporal stability of clusters was lowest. This demonstrates that increased susceptibility to RTIs might (in part) be driven by an increase in network fragmentation, which is further characterised by a decrease in similarity across clusters with the same niche-specificity. Of note is the fact that we were unable to statistically test the difference in cluster numbers across time and RTI group, as for each of these categories, only one network could be built given the available data. However, our finding of fragmentation is in line with a study focusing on the structure of (niche-specific) gut microbial co-occurrence networks in IBD patients, which was more distorted when compared to networks in healthy individuals7. Further, a theoretical study by Ma et al. supports our observation by showing that properties of critical network structures were associated with microbiome-associated diseases14.
When studying hub OTUs central to the respective cross-niche networks15,16, we observed little overlap between RTI groups. In order to place the identity of the hub OTUs that we identified into a disease/health-related context, we performed a non-systematic literature search for associations between hub species and (respiratory) health. We found OTUs previously associated with low susceptibility to, and low severity of, respiratory infections, such as Corynebacterium, D. pigrum and N. lactamica, as hub species in networks from our least susceptible group. Both Corynebacterium and D. pigrum have been consistently associated with a health-associated microbiota maturation and a decreased risk of developing RTIs in later life11,17,18. Furthermore, N. lactamica has been shown in human challenge studies to inhibit colonization by the pathogenic Neisseria meningitidis, which supports that N. lactamica plays a role in a resilient microbial community network19. Inversely, we observed hub species that were previously associated with recurrent respiratory infections, such as Haemophilus and Lachnospiraceae, in the networks of the group experiencing the most RTIs, but generally not in the group with few RTI episodes11,20–22. Therefore, our data suggest that the presence of these bacteria is not incidental but potentially central to a less beneficial bacterial community structure associated with more RTI episodes.
Not only did we identify hub species that were previously associated with respiratory health, but we also identified OTUs that were previously associated with gut health, oral health, immunological diseases, and infections as hub species in the networks of infants experiencing either 0–2 or 5–7 RTIs. With respect to gut health, beneficial OTUs known to produce or enhance the production of butyrates, such as R. bromii (112), Megasphaera (133 and 383), and Anaerostipes (142), were found to be hub species in the networks of the least susceptible group23–25. The short-chain fatty acid butyrate is a microbial end-product of the human gut fermentation process and an essential metabolite in the gut environment, being the preferred energy source for colon epithelial cells. It has anti-inflammatory properties and lowers the pH of the gut environment, in this way inhibiting growth of pathogens23. Conversely, Peptostreptococcaceae spp. (75 and 117), previously associated with increased faecal pH, were identified as hub species for the networks from the most susceptible group26. Additionally, in this group, the mucin degrader R. gnavus (24) was identified as a hub species. This OTU has previously been associated with a broad range of immunological disorders, such as paediatric allergy, IBD and psoriatic arthritis, and also with failure of faecal microbiota transplantation27–32. Also, in the most susceptible RTI networks, we observed A. schaalii (408), involved in urinary tract infections, as hub species, as well as the cariogenic B. dentium (28) and P. denticolens (486)33–35. Altogether, these observations suggest that cross-niche bacterial networks may stand at the basis of overall systemic susceptibility to inflammation-driven health conditions. They further imply that the overall construction of microbial networks, in addition to merely the presence/absence or abundance of specific commensals or pathogens, can act as a driving force behind inflammation-mediated disease.
Of note, although the more predominant OTUs were generally niche-specific, most hub species were actually lower abundant taxa and not associated with a specific niche. This supports the “rare taxa” concept, which postulates that the abundance of a species is not necessarily the best determinant for its importance within the microbial community structure36. Studying the human microbiome in a more system-centric context, therefore, might provide insight into the importance and roles of lesser-known microbes. In addition, it is important to highlight the fact that we were able to shed some light on the degree of cross-niche connectivity between microbial communities within the body.
Relationships between microbial communities across body sites have already been pointed out, for example, by Madan et al.37, who showed that the introduction of solid foods in children with cystic fibrosis not only changed the microbiome composition within the gut but also that of the respiratory tract. These connections are likely a result of indirectly connected microbe-host interactions, such as immunological pathways triggered by one microbe that affect distally located microbes or metabolic products used by distally located microbes38,39, rather than direct microbe–microbe interactions, which is due to their physical separation, less likely. Examples for such effects of distally located microbes affecting susceptibility to RTI’s have already been presented in the literature. Using a gain-of-function genetic mouse model, Steed et al. were able to show that desaminotyrosine produced by the human gut commensal Clostridium orbiscindens augmented IFN-I signalling, thereby rendering protection from influenza morbidity and mortality in infected mice40. In addition, Olsen and Yamazaki present a mechanism by which systemic diffusion of inflammatory mediators from periodontal lesions can lead to or contribute to the development of a multitude of gastrointestinal conditions such as non-fatty liver disease, rheumatoid arthritis, or cancer41.
One key strength of our study is the integrated use of microbiota data across different body sites to build cross-niche microbial networks in a longitudinal fashion, using samples collected from a prospectively followed birth cohort of children recruited at a single hospital in the Netherlands. By using a cross-niche network approach with an algorithm based on conditional independence rather than correlation, we showed that besides the known associations between niche-specific microbiota and respiratory health, there is likely also an association between the overall infant microbiome network features and respiratory health. Furthermore, we were able to identify hub species that were important for the network structure, which were often niche-independent, suggesting these microbes might be missed or their value might be underestimated when the microbiota is studied within a single niche. With respect to future studies, our results demonstrate that careful consideration of the composition and connectivity of local and distal microbial communities can enhance our understanding of the magnitude of systemic dysbiosis potentially underlying health and disease. However, we are aware that experimental limitations might not always allow the study of the composition of the infant microbiome on a systemic scale, although studies like ours may still provide a framework to formulate testable hypotheses that incorporate more than one niche community.
Limitations of our study are mainly inherent to the experimental design, including only children recruited at a single hospital in the Netherlands, warranting the need for similar studies covering a broader range of geographical variation. In addition, future in vitro/in vivo studies are needed to understand the biological mechanisms underpinning these findings. Furthermore, we potentially lack statistical power when comparing network cluster similarities across RTI groups within niches, as there were only between one and four niche-specific clusters in any one network. Lastly, we only considered univariate relationships between environmental factors and infant microbiome composition as a means to pre-select factors to be combined in a multivariate model. This approach allowed us to pre-screen a large number of covariates for their potential effect on the infant microbiome without the risk of overfitting. However, it does not take into account potential interactions/correlations between individual covariates.
In conclusion, we observed cohesive and stable microbial networks across body sites already from early life on, which in turn were associated with a lower susceptibility for RTIs in the first year of life. In contrast, we found more fragmented and unstable cross-niche networks over the first 6 months of life in infants with higher susceptibility to RTIs. Finally, we identified bacteria that were central to each of the cross-niche microbial network structures, though these differed between RTI groups. These bacteria were often not indicative for a specific niche within the body, and represented low abundant species, underlining the potential importance of low abundant bacteria for microbiome function.
Methods
Data collection
A detailed description of the study design and inclusion criteria of the birth cohort can be found elsewhere13. In short, we used microbiota data from samples obtained from 112 healthy infants born at term (≥37 weeks gestational age) recruited at a single Dutch hospital at four different time points (week 1, month 2, month 4, and month 6) and from three different niches, namely the gastrointestinal tract (faecal samples), the upper respiratory tract (nasopharyngeal swabs), and the oral cavity (saliva samples). Metadata was available for the first year of life, but matched samples collected in parallel were only available until month 6. Written informed consent was obtained from parents of all children and the study was approved by METC Noord-Holland (M012-015, NTR3986). Infants were divided into three groups based on the distribution of the number of RTIs they experienced over the first year of life (0–2 RTIs, 3–4 RTIs, 5–7 RTIs), and as described in previous publications using data from this birth cohort Supplementary Fig. 5)10.
16S rRNA gene sequencing
DNA extraction and library preparation of the bacterial 16S rRNA gene V4 region was performed as described previously13. Samples were sequenced in 27 individual library pools, each containing DNA extraction and qPCR blanks as negative control; and a mock community (positive control). All pools were sequenced on the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA). The raw sequencing reads were trimmed using Sickle v1.33 (with the quality threshold set to q = 30 and the length threshold set to 150 nucleotides)42; error corrected using BayesHammer (SPAdes v3.8.1)43 and assembled using PANDAseq v2.1044. Following removal of chimeric sequences, reads were grouped into Operational Taxonomic Units (OTUs) using VSEARCH v2.0.3 with a 97% similarity threshold45. Taxonomic annotation was performed using QIIME v1.9.1 based on the SILVA database v11946,47. We removed spurious OTUs by filtering at a minimum relative abundance of 0.1% and presence in at least two samples48. We combined each OTU name with a number representing its rank in the OTU table, based on the relative abundance in the overarching dataset to discriminate OTUs with the same taxonomic annotation.
Statistical analysis
Analyses regarding the microbiota composition have been performed in R version 3.4.3, while network analyses have been performed in R versions and 3.6.249.
Niche comparison
Results of analyses of observed species richness were based on raw read counts (Supplementary Fig. 6 for rarefaction curves). Similar results were obtained when rarefying read counts to a sequencing depth of 3,000 reads. We used the lmer function (lme4 package)50 to study the temporal changes of species richness within and across niches, with participants added as random effect to take repeated measures into account. To compare compositions of microbial communities across niches, we used non-metric multidimensional scaling (nMDS) plots based on ordinations that used the Bray–Curtis (BC) dissimilarity matrix of relative abundance data as input (function ordinate with parameter trymax 10,000; vegan package)51. Stability of the microbiota composition in the first 6 months of life within each niche was visualised by measuring the BC dissimilarities between consecutive samples of each participant, and we tested differences across niches using Wilcoxon’s signed rank test.
We used permutational multivariate analysis of variance (PERMANOVA) with 1,999 permutations to investigate the associations between microbiota composition (response variable) and environmental covariates using the function adonis2 (vegan package)51. The covariates tested were mode of delivery (vaginal vs. caesarean section), season of birth (summer, autumn, winter, spring), hospital stay duration after birth in day parts, presence of siblings <5 years of age (yes vs. no), presence of pets in the household (none, cat(s), dog(s), cat(s) and dog(s), other), breastfeeding at sampling moment (yes vs. no), attendance of day care at sampling moment (yes vs. no), use of pacifier at sampling moment (yes vs. no) and use of antibiotics in the month prior to sampling moment (yes vs. no). Covariates that were significantly associated in univariate models (per time point per niche, Supplementary Figs. 7–12) with microbiota community composition at a minimum of one-time point were included in a multivariate model that included niche as well as age and subject to control for repeated measures. P-values were adjusted for multiple testing using the Benjamini–Hochberg method52.
Network construction and cluster definition
Prior to network construction, we split the data by RTI group and filtered the OTU tables to only include the 100 most abundant OTUs per niche, and within each RTI group in order to avoid overestimation of the impact of very rare taxa on the overall network structure. Because of overlapping OTUs between niches, this resulted in a final dataset that included 220 OTUs for the 0–2 RTI group, 214 OTUs for the 3–4 RTI group and 228 OTUs for the 5–7 RTI group. All combined, we identified 315 non-redundant OTUs, of which 149 OTUs were present in all three groups (Supplementary Fig. 13). Using the top 100 prevalent OTUs per niche instead of the 100 most abundant OTUs resulted in a smaller dataset of only 251 OTUs in total. Seventy-four percent of those 251 OTUs identified using a prevalence filter were also identified using an abundance filter, and since the overlap between the two methods was large, we decided to continue our analysis using the abundance filter since this resulted in a bigger number of OTUs to be available for network construction.
We defined niche indicator OTUs as OTUs that were characteristic for a particular niche. To identify such OTUs, we ran a niche indicator species analysis using the function multipatt (indicspec package)53. This function calculates an indicator value for each OTU at each niche, taking its total abundance per niche into account. An OTU has deemed a niche indicator when it had an association value of >0.5 with a p-value < 0.05 for one specific niche.
For network construction, we used the SpiecEasi pipeline with Meinshausen–Buhlmann estimation (number of StARS repetitions = 50, lambda scaling factor = 1e-2)9. This method is based on the concept of conditional independence rather than correlation, making it less likely to detect spurious connections between taxa that are indirectly connected but not directly connected. Further, it uses centred log-ratio transformation of the data to overcome the compositionality of microbiome data. In total, we constructed 12 networks (3 RTI groups × 4 time points) using the methods described above, with the number of OTUs remaining constant in each network. We used random walks (number of steps = 5) to group OTUs into clusters that were closely associated using the function walktrap.community (igraph package)54. We chose a step length of five since the number of clusters plateaued after this value (Supplementary Fig. 14). We considered clusters with ten or more OTUs as “main clusters”, as smaller clusters usually contained only one or two OTUs (Supplementary Fig. 15). The OTUs within a cluster can be either niche indicator OTUs (saliva, faeces, or nasopharynx) or non-niche indicator OTUs. Therefore, if the composition of a cluster were random, we would expect an equal proportion of any of these four OTU classes within a cluster, i.e. each OTU class would have a prevalence of 25%. We considered a cluster niche-specific if one of the OTU classes exceeded that threshold value of 25%. This is a very conservative threshold as none of the classes of niche indicator OTUs actually made up 25% of OTUs (between 5.7% and 11.5% of OTUs within an RTI group). The use of this conservative threshold allowed us to unambiguously assign cluster identity. If there were two classes of niche indicator OTUs that met this threshold within a single cluster, then the largest set determined the niche annotation. If there were not enough niche indicator OTUs that met this threshold, the cluster was defined as a “mixed (niche) cluster”.
Cluster stability analysis
We compared the composition of all clusters from each time-specific network per RTI group using an n × n matrix, where cluster size was not taken into account. We then calculated the difference in cluster composition from a scale from 0 to 1, where 1 indicates complete similarity (i.e., two clusters share 100% of their OTUs). We used ANOVA to assess whether cluster similarity was different across niches.
Hub species analysis
To identify hub species (i.e., OTUs playing a significant role in determining the structure of a community network), we calculated three important network metrics for each OTU within a network at each time point: (1) degree (the number of connections each OTU has within a network), (2) betweenness centrality (number of shortest paths that pass through a specific OTU within a network), and (3) closeness centrality (the reciprocal of the sum of the length of the shortest paths between an OTU and all other OTUs in a network). The correlation data between network metrics is shown in Supplementary Fig. 16. We followed the definitions from Banerjee et al. and Berry and Widder by defining hub species as those OTUs with a high degree, high closeness centrality and low betweenness centrality15,16. We defined high degree or closeness centrality as the top 10% of the distribution of each of those metrics and low betweenness centrality as the bottom 10% of the distribution for betweenness. By doing so, we ensured a selection of OTUs that were at the extremes of the distributions for all three values (see Supplementary Fig. 17). We selected OTUs within RTI groups that met at least one of those conditions as hub species.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors are most grateful for the participation of all the children and their families in the Microbiome Utrecht Infant study (MUIS study). We would like to acknowledge all the members of our research team at the Spaarne Gasthuis Academy and the UMC Utrecht, the Department of Obstetrics and Gynaecologist of the Spaarne Gasthuis (Hoofddorp and Haarlem, the Netherlands), and participating midwifery clinics. This work was supported in part by the Netherlands Organisation for Scientific Research (NWO-VIDI; grant 91715359) and CSO/NRS Scottish Senior Clinical Fellowship award (SCAF/16/03). The study was co-sponsored by the Spaarne Hospital Hoofddorp and Haarlem and the University of Edinburgh.
Author contributions
K.A., M.L.J.N.C. and R.H. performed the laboratory work. M.R. and M.C. analysed the data. M.R., M.C. and D.B. wrote the manuscript. M.A.v.H and E.A.M.S. provided critical comments on the manuscript.
Data availability
Raw sequence data underlying the findings of this study have been previously deposited in the NCBI Sequence Read Archive (SRA) database as part of other publications with BioProject IDs PRJNA353336, PRJNA450937, and PRJNA481243. The results presented in this manuscript are based on a subset of the sequences deposited under these BioProjects. The source data underlying the tables and figures presented in this manuscript are provided in Supplementary Data 1.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Karli Montague-Cardoso.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Marta Reyman, Melanie Clerc.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02755-1.
References
- 1.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bokulich NA, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016;8:343ra82–343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang D, et al. The cross-talk between gut microbiota and lungs in common lung diseases. Front. Microbiol. 2020;11:301. doi: 10.3389/fmicb.2020.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budden KF, et al. Emerging pathogenic links between microbiota and the gut–lung axis. Nat. Rev. Microbiol. 2016;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 5.Hall CV, et al. Co-existence of network architectures supporting the human gut. Microbiome. iScience. 2019;22:380–391. doi: 10.1016/j.isci.2019.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soret P, et al. Respiratory mycobiome and suggestion of inter-kingdom network during acute pulmonary exacerbation in cystic fibrosis. Sci. Rep. 2020;10:3589. doi: 10.1038/s41598-020-60015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldassano SN, Bassett DS. Topological distortion and reorganized modular structure of gut microbial co-occurrence networks in inflammatory bowel disease. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep26087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faust K, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurtz ZD, et al. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 2015;11:5. doi: 10.1371/journal.pcbi.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosch AATM, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. a prospective cohort study. Am. J. Respir. Crit. Care Med. 2017;196:1582–1590. doi: 10.1164/rccm.201703-0554OC. [DOI] [PubMed] [Google Scholar]
- 11.Man WH, et al. Loss of microbial topography between oral and nasopharyngeal microbiota and development of respiratory infections early in life. Am. J. Respir. Crit. Care Med. 2019;200:760–770. doi: 10.1164/rccm.201810-1993OC. [DOI] [PubMed] [Google Scholar]
- 12.Reyman M, et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 2019;10:4997. doi: 10.1038/s41467-019-13014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosch AATM, et al. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine. 2016;9:336–345. doi: 10.1016/j.ebiom.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma, Z. (Sam) Critical network structures and medical ecology mechanisms underlying human microbiome-associated diseases. Science23, 6 (2020). [DOI] [PMC free article] [PubMed]
- 15.Banerjee S, Schlaeppi K, van der Heijden MGA. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018;16:567–576. doi: 10.1038/s41579-018-0024-1. [DOI] [PubMed] [Google Scholar]
- 16.Berry D, Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014;5:219. doi: 10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biesbroek G, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 2014;190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 18.Teo SM, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deasy AM, et al. Nasal inoculation of the commensal Neisseria lactamica inhibits carriage of Neisseria meningitidis by young adults: a controlled human infection study. Clin. Infect. Dis. 2015;60:1512–1520. doi: 10.1093/cid/civ098. [DOI] [PubMed] [Google Scholar]
- 20.Man WH, et al. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir. Med. 2019;7:417–426. doi: 10.1016/S2213-2600(18)30449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickson RP, et al. Lung microbiota predict clinical outcomes in critically ill patients. Am. J. Respir. Crit. Care Med. 2020;201:555–563. doi: 10.1164/rccm.201907-1487OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langevin S, et al. Early nasopharyngeal microbial signature associated with severe influenza in children: a retrospective pilot study. J. Gen. Virol. 2017;98:2425–2437. doi: 10.1099/jgv.0.000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baxter NT, et al. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. MBio. 2019;10:e02566–18. doi: 10.1128/mBio.02566-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuille S, Reichardt N, Panda S, Dunbar H, Mulder IE. Human gut bacteria as potent class I histone deacetylase inhibitors in vitro through production of butyric acid and valeric acid. PLoS One. 2018;13:e0201073. doi: 10.1371/journal.pone.0201073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front. Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henrick BM, et al. Elevated Fecal pH Indicates a Profound change in the breastfed infant gut microbiome due to reduction of Bifidobacterium over the past century. mSphere. 2018;3:e00041–18. doi: 10.1128/mSphere.00041-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chua HH, et al. Intestinal dysbiosis featuring abundance of Ruminococcus gnavus associates with allergic diseases in infants. Gastroenterology. 2018;154:154–167. doi: 10.1053/j.gastro.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Png CW, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 29.Tailford LE, et al. Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat. Commun. 2015;6:1–12. doi: 10.1038/ncomms8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henke MT, et al. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl Acad. Sci. U. S. A. 2019;116:12672–12677. doi: 10.1073/pnas.1904099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scher JU, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67:128–139. doi: 10.1002/art.38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuentes S, et al. Microbial shifts and signatures of long-term remission in ulcerative colitis after faecal microbiota transplantation. ISME J. 2017;11:1877–1889. doi: 10.1038/ismej.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lotte R, Lotte L, Ruimy R. Actinotignum schaalii (formerly Actinobaculum schaalii): a newly recognized pathogen-review of the literature. Clin. Microbiol. Infect. 2016;22:28–36. doi: 10.1016/j.cmi.2015.10.038. [DOI] [PubMed] [Google Scholar]
- 34.Ventura M, et al. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 2009;5:e1000785. doi: 10.1371/journal.pgen.1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gueimonde M, et al. Genome sequence of Parascardovia denticolens IPLA 20019, isolated from human breast milk. J. Bacteriol. 2012;194:4776–4777. doi: 10.1128/JB.01035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynch MDJ, Neufeld JD. Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 2015;13:217–229. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]
- 37.Madan JC, et al. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: Interaction between intestinal and respiratory tracts and impact of nutritional exposures. MBio. 2012;3:e00251–12. doi: 10.1128/mBio.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grainger, J., Daw, R. & Wemyss, K. Systemic instruction of cell-mediated immunity by the intestinal microbiome [version 1; referees: 2 approved]. F1000Research10.12688/f1000research.14633.1 (2018). [DOI] [PMC free article] [PubMed]
- 39.Fahimipour AK, Gross T. Mapping the bacterial metabolic niche space. Nat. Commun. 2020;11:1–8. doi: 10.1038/s41467-020-18695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steed AL, et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science. 2017;357:498–502. doi: 10.1126/science.aam5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olsen I, Yamazaki K. Can oral bacteria affect the microbiome of the gut? J. Oral. Microbiol. 2019;11:1586422. doi: 10.1080/20002297.2019.1586422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joshi, N. A. & Fass, J. N. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files. https://github.com/najoshi/sickle (2011).
- 43.Nikolenko SI, Korobeynikov AI, Alekseyev MA. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics. 2013;14:S7. doi: 10.1186/1471-2164-14-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinforma. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian S, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R Core Team. R: A language and environment for statistical computing. https://www.eea.europa.eu/data-and-maps/indicators/oxygen-consuming-substances-in-rivers/r-development-core-team-2006 (2017).
- 50.Bates D, Mächler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015;67:1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 51.Oksanen, J. et al. vegan: Community ecology package. R package version 2.5–7. https://cran.r-project.org/web/packages/vegan/index.html (2020).
- 52.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple. Test. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 53.De Cáceres M, Legendre P, Moretti M. Improving indicator species analysis by combining groups of sites. Oikos. 2010;119:1674–1684. doi: 10.1111/j.1600-0706.2010.18334.x. [DOI] [Google Scholar]
- 54.Csárdi G, Nepusz T. The igraph software package for complex network research. InterJournal, Complex Syst. 2006;1695:1–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Raw sequence data underlying the findings of this study have been previously deposited in the NCBI Sequence Read Archive (SRA) database as part of other publications with BioProject IDs PRJNA353336, PRJNA450937, and PRJNA481243. The results presented in this manuscript are based on a subset of the sequences deposited under these BioProjects. The source data underlying the tables and figures presented in this manuscript are provided in Supplementary Data 1.