Abstract
Starch is a vital energy source for living organisms and is a key raw material and additive in the food and non-food industries. Starch has received continuous attention in multiple research fields. The endosperm of cereals (e.g., rice, corn, wheat, and barley) is the most important site for the synthesis of storage starch. Around 2010, several excellent reviews summarized key progress in various fields of starch research, serving as important references for subsequent research. In the past 10 years, many achievements have been made in the study of starch synthesis and regulation in cereals. The present review provides an update on research progress in starch synthesis of cereal endosperms over the past decade, focusing on new enzymes and non-enzymatic proteins involved in starch synthesis, regulatory networks of starch synthesis, and the use of elite alleles of starch synthesis-related genes in cereal breeding programs. We also provide perspectives on future research directions that will further our understanding of cereal starch biosynthesis and regulation to support the rational design of ideal quality grain.
Key words: cereal, starch biosynthesis, regulation network, endosperm development, quality improvement
Over the past 10 years, great progress has been made in the field of starch biosynthesis in the cereal endosperm. This review summarizes recent findings on the molecular regulation and networks that underlie starch synthesis in the endosperm of major cereals, highlights challenges that remain to be overcome, and discusses prospects for future research.
Introduction
Starch is a natural polymer composed of glucose that is unique to plants and algae. The metabolism of starch is the hub of energy metabolism and is widely involved in almost all aspects of plant growth and development. Storage starch, synthesized in the seeds, tubers, corms, and roots of plants, is the main substance used by plants to store carbohydrates and is the most important energy source for all living organisms, as well as an important industrial raw material and additive (Zeeman et al., 2010). The degradation of storage starch can also affect seed germination and pre-harvest sprouting (Tai et al., 2021). Storage starch is a semi-crystalline, insoluble particle composed of two α-glucose polymers, linear amylose and highly branched amylopectin linked by α-1,4- and α-1,6-glycosidic linkages (Zhu et al., 2020b). The composition of amylose and amylopectin, and the hierarchical fine structures they form, determine the diversity of starch physicochemical properties, cereal grain quality, and industrial uses. Extensive research has been carried out on storage starch in fields such as plant science, food science, polymer science, and various industrial fields. For details, please refer to several excellent reviews and book chapters published previously (Jeon et al., 2010; Kötting et al., 2010; Zeeman et al., 2010; Shen et al., 2011; Tetlow et al., 2015; Wang et al., 2015; Pfister and Zeeman, 2016; Zaman and Sarbini, 2016; Crofts et al., 2017; Thalmann and Santelia, 2017; Kaur et al., 2018; Seung and Smith, 2018; Xia et al., 2018; Abt and Zeeman, 2020; Jukanti et al., 2020; Seung, 2020; Sharma et al., 2020; Zhou et al., 2020a; Tai et al., 2021).
The cereal endosperm is a major source of storage starch in plants. In the first decade of the 21st century, great progress was made in the functional analysis of key starch synthesis-related genes (SSRGs) in cereal endosperms and their correlations with the physicochemical properties of starches with different components and structures. This work has been well summarized previously (Jeon et al., 2010; Kötting et al., 2010; Zeeman et al., 2010). In the past 10 years, a series of advances have been made in the area of starch biosynthesis within the cereal endosperm. This review aims to update recent findings in cereals, especially in the endosperm of major cereals, including rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum), and barley (Hordeum vulgare).
The main text of the review is divided into five parts. In the first part, we briefly introduce the physicochemical properties of starch and its applications in food and non-food industries. In the second part, we detail the starch biosynthesis pathway in the endosperm of cereals based on progress made in the past decade and on previous reviews. In the third part, we summarize the regulatory network and mechanism of starch synthesis in cereal endosperms. In the fourth part, we discuss genetic improvement of cereal quality using accumulated knowledge on the genes involved in starch synthesis and its regulation, especially the SSRGs. Finally, we highlight the challenges that remain to be overcome in cereal endosperm starch research, and we raise prospects for future research.
Physicochemical properties and applications of cereal starch
Physicochemical properties of cereal starch
Starch physicochemical properties, including hardness, stickiness, gelatinization, retrogradation, digestibility, crystallization, and elasticity, are key factors for the manufacture and functional features of starch-based foods and industrial raw materials. To understand the relationship between the composition/structure of starch and its properties, starch characteristics are widely parameterized. Amylose content (AC) is the most classic parameter that reflects the composition of starch, whereas diverse parameters have been established for starch structure (Li et al., 2020a). Among them, chain-length distribution (CLD) is the fundamental component of starch structure and physicochemical properties. The CLD of amylose has been divided into three fractions based on the degree of polymerization (DP): short (DP 100–500), medium (DP 500–5000), and long (DP 5000–20 000) amylose chains (Li et al., 2016). Alternatively, it has been divided into two fractions: fraction 1 (DP 100–1000) and fraction 2 (DP 1000–20 000) (Li et al., 2017a). The CLD of amylopectin has been divided into diverse chain categories in different reports (Zhu, 2018). Generally, it has been divided into four fractions based on the amylopectin cluster model. These are the A (DP 6–12), B1 (DP 13–24), B2 (DP 25–36), and B3 (DP >36) chains, of which the A and B1 chains account for more than 90% (Yu et al., 2019; Jukanti et al., 2020). In addition, a series of indicators and instruments have been summarized and developed to quantify the physicochemical properties of starch. These include the gelatinization properties of starch, which are mainly indicated by gelatinization temperature (GT), and the crystallization properties, which are determined by crystallinity degree (Tan et al., 1999; Dankar et al., 2018).
After decades of exploration, the correlations among quantitative indicators, starch physicochemical properties, and cereal grain quality have been largely revealed and established. For starch composition, AC shows a negative correlation with starch stickiness, digestion rate, crystallinity degree, and transparency of polished rice (AC < 13%) and a positive correlation with starch hardness, elasticity, and retrogradation rate (Gong et al., 2019; Huang et al., 2020a; Li et al., 2020a; Zhang et al., 2020b). Therefore, AC is the most critical indicator that determines the physicochemical properties of starch and its end use. For the CLD of amylopectin, short (A and B1) amylopectin chains form amylopectin crystalline regions, and longer A and B1 chains can form longer double helices that require a higher temperature to dissociate than do shorter double helices in the crystalline region (Noda et al., 1998; Li et al., 2020a). Thus, the numbers of amylopectin A chains and B1 chains are negatively and positively correlated with GT, respectively, and longer amylopectin A and B1 chains reduce the starch digestion rate (Martinez et al., 2018). Starch that contained a higher proportion of amylopectin with DP 70–100 showed a less viscous but more elastic texture (Li et al., 2016). For the CLD of amylose, shorter amylose medium chains are relatively easily retrograded, whereas shorter amylose medium-to-long chains and the amount of short-to-medium chain amylose help to reduce the starch digestion rate (Li et al., 2020a).
Applications of cereal starch in food and non-food industries
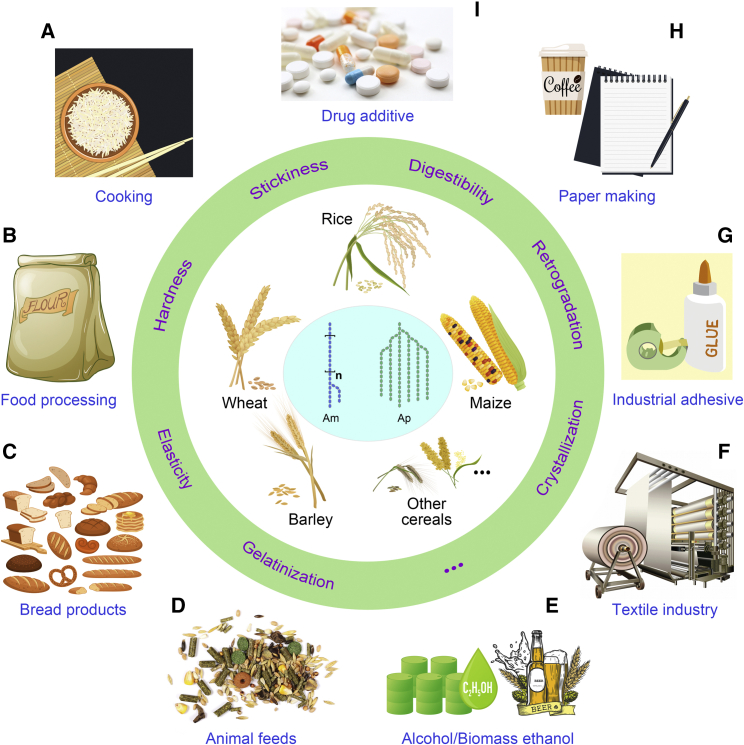
Starch from cereal endosperms is used mainly as food, providing about 50% of the daily energy needed by the human body (Figure 1A and 1C). Consumer preferences and dietary habits in different regions of the world result in diverse needs for starchy food and require cereal starches with diverse physicochemical properties (Huang et al., 2020a). For instance, cereals with a lower AC, such as waxy corn and soft rice, tend to be preferred in Southeast Asia, despite being less healthy because of their high digestibility and high glycemic index (GI) (Dong et al., 2019a; Parween et al., 2020). Cereal grains are also crucial ingredients for animal feeds (Figure 1D).
Figure 1.
Physicochemical properties and industrial applications of cereal starch.
The major cereal grains are shown within the circle, and the middle circle highlights the physicochemical properties of cereal starch, including hardness, stickiness, gelatinization, retrogradation, digestibility, crystallization, and elasticity. The pictures outside the circle (A–I) present the applications of cereal starch in food and non-food industries. Am, amylose; Ap, amylopectin.
As the second most abundant carbohydrate in nature after cellulose, starch’s plentiful sources, easy access, low cost, and natural, renewable, and biodegradable characteristics have led to its widespread use in industrial applications. Using physical, chemical, or enzymatic methods such as molecular cutting, rearrangement, oxidation, or introduction of substituents into starch molecules can produce modified starches with altered, enhanced, or new physicochemical properties. Modified starch overcomes the shortcomings of natural starch, such as poor freeze-thaw stability, low solubility, easy retrogradation, and low transparency, further expanding its range of applications in related industries (Masina et al., 2017; Wang et al., 2020c). In food processing, modified starch is widely added to food as a partial fat substitute to provide the shape, taste, thickening, gelling, adhesiveness, and stability required by the food system while avoiding the perception that high-fat meat products will cause cancer (Heinen et al., 2009; Kapelko-Żeberska et al., 2015; Kaur et al., 2018) (Figure 1B). Starch is also used in the alcoholic beverage industry and as a feedstock in bioethanol production for clean energy (Zeeman et al., 2010) (Figure 1E). In the textile industry, modified starch is an important material for warp sizing, printing paste, and silk printing paste (Hebeish et al., 2008) (Figure 1F). Low- and non-amylose starch is very sticky and is an ideal industrial adhesive (Onusseit, 1992) (Figure 1G). Starch with a molecular structure similar to that of cellulose in papermaking fiber is also widely used in the papermaking industry to improve the strength, stiffness, smoothness, gloss, and whiteness of paper (Shen et al., 2011) (Figure 1H). Starch-based biodegradable plastic is considered to be an ideal replacement for traditional plastics in packaging (Gross and Kalra, 2002) (Figure 1H). Modified starch is one of the most important excipients for various new preparations in the pharmaceutical industry (Elvira et al., 2002) (Figure 1I). Starch also has remarkable potential for the preparation of nanofibers using the electrospinning process. Electrospun starch nanofibers have applications in drug delivery, tissue engineering, and the manufacture of wound dressings, and their continued development and applications are expected to further expand the use and commercial value of starch (Liu et al., 2017).
Starch biosynthesis pathway in cereal endosperms
The pathway of starch biosynthesis in the endosperm is highly conserved among cereals. A number of key enzymes, including ADP-glucose pyrophosphorylase (AGPase), granule-bound starch synthase (GBSS), soluble starch synthase (SS), starch branching enzyme (SBE), debranching enzyme (DBE), disproportionating enzyme (DPE), and starch/α-glucan phosphorylase (PHO), have been found to participate in the synthesis of starch in the cereal endosperm (Zeeman et al., 2010; Seung and Smith, 2018). Each of the starch synthesis-related enzymes (SSREs) in cereals is present in a variety of isozymes, and these isozymes can form heterologous multi-enzyme complexes with other isozymes or homomultimers with themselves in vivo to perform their functions (Utsumi et al., 2011; Crofts et al., 2015). In addition, some novel non-enzymatic proteins, such as PROTEIN TARGETING TO STARCH (PTST), have also been found to participate in starch synthesis in the cereal endosperm (Peng et al., 2014; Seung et al., 2015). From the perspective of the whole genome, the types and functions of SSREs are highly conserved in rice, maize, wheat, and barley, despite the differences in copy number and isoenzyme number owing to variations in chromosome number among cereal species (Ohdan et al., 2005; Radchuk et al., 2009; Yan et al., 2009; Kang et al., 2013a) (Table 1). A brief depiction of the starch biosynthesis pathway in the cereal endosperm is shown in Figure 2. For specific information, please refer to previous reviews (Jeon et al., 2010; Zeeman et al., 2010; Abt and Zeeman, 2020).
Table 1.
Enzymes and non-enzymatic proteins involved in starch synthesis during cereal endosperm development.
| Enzymes/non-enzymatic proteins | Rice (Oryza sativa L.) |
Maize (Zea mays L.) |
Wheat (Triticum aestivum L.) |
Barley (Hordeum vulgare L.) |
||||
|---|---|---|---|---|---|---|---|---|
| Gene name | Acc. No./ID | Gene name | Acc. No./ID | Gene name | Acc. No./ID | Gene name | Acc. No./ID | |
| ADPG pyrophosphorylase (AGPase, EC 2.7.7.27)a | OsAGPS1 | AY028315 | ZmAGPS1 | AY032604 | TaAGPS1 | AY727927 | HvAGP-S1 | AAO16183 |
| OsAGPS2a | EF122437 | ZmAGPS2a-1/Bt2 | AF330035 | TaAGPS2-a | X66 080 | HvAGP-S2a | CAA88449 | |
| ZmAGPS2a-2 | DQ118038 | |||||||
| OsAGPS2b | AP004459 | ZmAGPS2b | AF334960 | TaAGPS2-b | EU582678 | HvAGP-S2b | CAA88450 | |
| OsAGPL1 | AY028314 | ZmAGPL1/Sh2 | BT016868 | TaAGPL1 | Z21969 | HvAGP-L1 | CAA47626 | |
| OsAGPL2/GIF2 | D50317 | ZmAGPL2 | Z38111 | TaAGPL2 | DQ406820 | HvAGP-L2 | AAC49729 | |
| OsAGPL3 | NM-001065811 | ZmAGPL3 | EF694838 | |||||
| OsAGPL4 | NM-001057719 | ZmAGPL4 | EF694839 | |||||
| ADPG transporter | OsBt1 | Os02g0202400 | ZmBt1 | M79333 | TaBt1 | BT008958.1 | HvBt1 | AY560327.2 |
| Granule-bound starch synthase (GBSS, EC 2.4.1.21) | OsGBSSI | AB425323 | ZmGBSSI | AY109531 | TaGBSSI | AF286320 | HvGBSSIa | AAM74051 |
| HvGBSSIb | AAM74054 | |||||||
| OsGBSSII | AY069940 | ZmGBSSIIa | EF471312 | TaGBSSII | AF109395 | |||
| ZmGBSSIIb | EF472248 | |||||||
| Soluble starch synthase (SS, EC 2.4.1.21) | OsSSI | AY299404 | ZmSSI | AF036891 | TaSSI | AJ292521 | HvSSI | AAF37876 |
| OsSSIIa/SSII-3/ALK | AF419099 | ZmSSIIa/Su2 | AF019296 | TaSSIIa/Sgp1 | AJ269503 | HvSSIIa/Sex6 | AAN28307 | |
| OsSSIIb/SSII-2 | AF395537 | ZmSSIIb-2 | EF472249 | TaSSIIb | EU333947 | HvSSIIb | AAN28307 | |
| ZmSSIIb-1 | AF019297 | |||||||
| OsSSIIc/SSII-3 | AF383878 | ZmSSIIc | EU284113 | TaSSIIc | EU307274 | |||
| OsSSIIIa/SSIII-2/Flo5 | AY100469 | ZmSSIIIa/du1 | AF023159 | TaSSIIIa | AF258608 | HvSSIIIa | AAF87999 | |
| OsSSIIIb/SSIII-1 | AF432915 | ZmSSIIIb-1 | EF472250 | TaSSIIIb | EU333946 | HvSSIIIb | AAL40942 | |
| ZmSSIIIb-2 | EF472251 | |||||||
| OsSSIVa/SSIV-1 | AY373257 | ZmSSIV | EU599036 | TaSSIV | AY044844 | HvSSIV | AAK97773 | |
| OsSSIVb/SSIV-2 | AY373258 | |||||||
| OsSSV | EU621837.1 | ZmSSV | NM_001 130 131.1 | |||||
| Starch branching enzyme (SBE, EC 2.4.1.18) | OsSBEI/SBE1 | EF122471 | ZmBEI | AY105679 | TaBEI | Y12320 | HvSBEI | AAP72268 |
| OsSBEIIa/SBE4 | AB023498 | ZmBEIIa | EF433557 | TaBEIIa | AF286319 | HvSBEIIa | AAC69753 | |
| OsSBEIIb/SBE3 | D16201 | ZmBEIIb/Ae | EU333945 | TaBEIIb | AY740401 | HvSBEIIb | AAC69754 | |
| OsSBEIII | AK066930 | ZmBEIII | ZMU18908 | TaBEIII | JQ346193 | |||
| Debranching enzyme (DBE, EC 3.2.1.68 and EC 3.2.1.41) | OsISA1 | AB015615 | ZmISA1/Su1 | ZMU18908 | TaISA1 | AF548380 | HvISA1 | AAM46866 |
| OsISA2 | NM-001061991 | ZmISA2 | EU976060 | TaISA2 | JX473824 | HvISA2 | BAD08581 | |
| OsISA3 | NM-001069968 | ZmISA3 | AY172634 | TaISA3 | JN412069 | HvISA3 | BAD89532 | |
| OsPUL | D50602 | ZmPUL/Zpu1 | AF080567 | TaPUL | EF137375 | HvPUL | AAD34348 | |
| Starch/α-glucan phosphorylase (PHO, EC 2.4.1.1) | OsPHOL/PH O 1 | AF327055 | ZmPHOL/Sh4/SP | EU857640 | TaPHOL | EU595762 | HvPHOL/PHS1 | KF195562 |
| OsPHOH | NM-001051358 | ZmPHOH | EU971442 | TaPHOH | AF275551 | HvPHOH/PHS2 | KF147849 | |
| Disproportionating enzyme (DPE, EC 2.4.1.25) | OsDPE1 | AB626975 | ZmDPE1 | BT061520 | TaDPE1 | DQ068045 | ||
| OsDPE2 | AK067082 | ZmDPE2 | BT055804 | TaDPE2 | BQ294920 | |||
| Protein targeting to starch (PTST) | OsGBP | LOC_Os02g04330 | GPM177 | NP_001132796.1 | HvPTST1 | HORVU6Hr1G018500 | ||
| OsFL O 6 | LOC_Os03g48170 | TaBGC1 | HvFL O 6/Fra | HORVU4Hr1G004470 | ||||
Most of the information was collected from previous reports (Ohdan et al., 2005; Radchuk et al., 2009; Yan et al., 2009; Jeon et al., 2010; Kang et al., 2013a; Ma et al., 2013; Soliman et al., 2014; Liu et al., 2015b; Cuesta-Seijo et al., 2017; Wang et al., 2019; Abt and Zeeman, 2020); some genes are predicted by sequence alignment, and their authenticity and function require further verification; Acc. No., accession number.
The naming of AGPS1 and AGPS2 is contrary to that of Yan et al. (2009), Radchuk et al. (2009) and Kang et al. (2013a), but consistent with their original citations (Johnson et al., 2003; Ohdan et al., 2005) and the NCBI website.
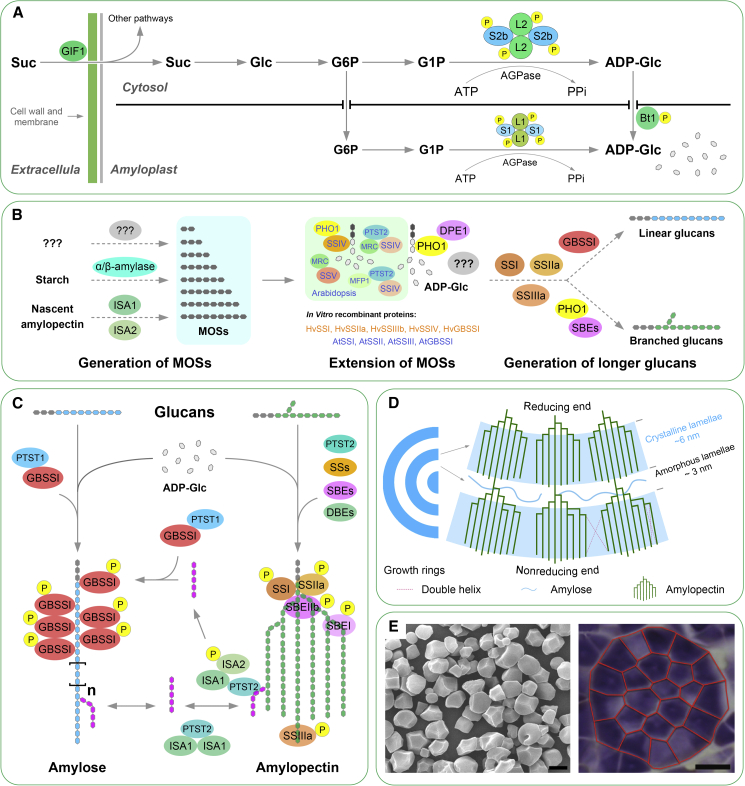
Figure 2.
Starch biosynthesis in the cereal endosperm: rice as an example.
(A) Generation and transport of ADP glucose (ADPG), the major substrate for starch synthesis. Sucrose synthesized by photosynthesis is unloaded and distributed by GRAIN INCOMPLETE FILLING 1 (GIF1) (Wang et al., 2008), then processed into ADPG in the cytosol and amyloplast by AGPase after a series of reactions and transport. In higher plants, AGPase is a heterotetramer composed of two large subunits (AGPL) and two small subunits (AGPS). ADPG synthesized in the cytosol is transported to the amyloplast through Brittle 1 (Bt1).
(B) Generation of primers for starch synthesis. The initiation of starch synthesis requires the availability of malto-oligosaccharides (MOSs). MOSs have been found to be continuously generated through trimming of nascent amylopectin by ISA1 and ISA2 in the process of starch synthesis or released by α/β-amylases during starch degradation (Myers et al., 2000; Fulton et al., 2008; Abt and Zeeman, 2020). However, the de novo synthesis of the original MOSs in cereals is still unclear. In rice, the PHO1–DPE1 complex enhanced synthesis of long MOSs in vitro, and an unknown factor(s) can compensate for the function of PHO1 at room temperature (Satoh et al., 2008; Hwang et al., 2016). SSIV and its interaction partners, including PHO1, PTST2, MFP1, and MRC, were found to participate in the extension of MOSs in Arabidopsis (Malinova et al., 2018). In addition, recombinant SSs and GBSSI from barley and Arabidopsis were demonstrated to elongate MOSs as short as maltose in vitro (Brust et al., 2013; Cuesta-Seijo et al., 2016). MOSs are further processed into linear glucans and branched glucans, which act as primers to initiate the synthesis of amylose and amylopectin, respectively. PHO1 and SBEs combine to promote the extension of MOSs and the synthesis of branched glucans by activating each other's mutual capacities in rice (Nakamura et al., 2012).
(C) Synthesis of amylose and amylopectin. Amylose in cereal endosperms is synthesized by GBSSI, whereas amylopectin is synthesized by coordinated cooperation of SSs, SBEs, and DBEs. PTST1 and PTST2, two non-enzymatic proteins, are also involved in the starch synthesis pathway. The isozymes of starch synthesis-related enzymes in cereals form heterologous multi-enzyme complexes with other isozymes or homo-multimers with themselves in vivo to perform their functions. These complexes include, but are not limited to, trimers composed of SSI, SSIIa, and SBEIIa/SBEIIb (Utsumi et al., 2011; Crofts et al., 2015). ISA1 can form homo-oligomers with itself or heterologous complexes with ISA2, and PTST2 interacts with ISA1, enabling its binding to starch granules (Utsumi et al., 2011; Peng et al., 2014). PTST1 interacts with GBSSI to help the latter’s localization to the surface of starch granules, and GBSSI then controls amylose synthesis in the form of oligomers after being phosphorylated (Liu et al., 2013; Wang et al., 2020b).
(D) Schematic diagram of the internal growth ring structure of starch granules formed by amylose and amylopectin. Adjacent α-1,4-glucan chains (DP ≥ 10) in or between amylopectin can form double helices and self-assemble to form crystalline lamellae in the starch granule matrix (Pfister and Zeeman, 2016). Amylopectin branch points and amylose form amorphous lamellae. The crystalline lamellae alternate with the amorphous lamellae containing branch points of amylopectin and amylose to form concentric growth rings with a period of 9 nm (Abt and Zeeman, 2020).
(E) Scanning electron micrograph of extracted starch granules from mature rice grains (left) and a schematic diagram of compound starch granules formed by multiple starch granules (traced as red polygons) in the amyloplast (right) (Wei et al., 2010; Matsushima et al., 2015). Scale bar, 5 μm.
Suc, sucrose; Glc, glucose; G6P, glucose 6-phosphate; G1P, glucose 1-phosphate; ATP, adenosine triphosphate; PPi, pyrophosphate; ADP-Glc, adenosine diphosphate-glucose; MOSs, malto-oligosaccharides; GBSS, granule-bound starch synthase; SS, soluble starch synthase; SBE, starch branching enzyme; DBE, debranching enzyme; ISA, isoamylase; DPE, disproportionating enzyme; PHO, starch/α-glucan phosphorylase; PTST, protein targeting to starch; MFP1, MAR BINDING FILAMENT-LIKE PROTEIN 1; MRC, MYOSIN-RESEMBLING CHLOROPLAST PROTEIN; P, phosphorylation.
Generation of substrates and primers for starch synthesis
AGPase is responsible for the synthesis of ADP glucose (ADPG), the major substrate for starch synthesis (Pfister and Zeeman, 2016). In higher plants, AGPase is a heterotetramer composed of two large subunits (AGPL) and two small subunits (AGPS). The isozymes of AGPL and AGPS can be classified into cytosolic and plastidial types according to their cellular localization (Saripalli and Gupta, 2015). The cytosolic type accounts for most of the total AGPase activity in the cereal endosperm, and mutation of its encoding genes often leads to severe grain-filling defects (Crumpton-Taylor et al., 2011; Huang et al., 2014; Wei et al., 2017). ADPG, synthesized in the cytosol of the developing endosperm, is transported to the amyloplast through the plastid ADPG transporter BT1 (Brittle 1), which is located in the amyloplast envelope (Kirchberger et al., 2007; Cakir et al., 2016; Li et al., 2017b) (Figure 2A).
Malto-oligosaccharides (MOSs) are required as primers for starch synthesis (Seung and Smith, 2018) (Figure 2B). The de novo synthesis of the original MOSs remains unclear. However, MOSs have been found to be continuously generated through trimming of nascent amylopectin by isoamylase 1 (ISA1) and ISA2 during starch synthesis or released by α/β-amylases during starch degradation (Myers et al., 2000; Fulton et al., 2008; Abt and Zeeman, 2020). Recombinant SSI-SSIV and GBSSI from barley and Arabidopsis elongated MOSs as short as maltose with low affinities in vitro (Brust et al., 2013; Cuesta-Seijo et al., 2016). PHO1 (also known as PHS1 and SP) was reported to interact with SSIV in Arabidopsis and may function in the extension of MOSs (Malinova et al., 2018). In rice, PHO1 assembles with DPE1 to use a broader range of sugars to synthesize MOSs and enhance the synthesis of long MOSs in vitro (Hwang et al., 2016). PHO1 was also demonstrated to combine with SBEs to promote the extension of MOSs and the synthesis of branched glucans in rice, and similar interactions were also observed in protein extracts from the endosperms of wheat, maize, and barley (Tetlow et al., 2004; Nakamura et al., 2012; Subasinghe et al., 2014; Cuesta-Seijo et al., 2017). However, the functions of PHO1 and DPE1 remain controversial, as only the isolated cases mentioned above have been reported.
Synthesis of amylopectin
Amylopectin is synthesized by at least SS, SBE, and DBE (Kötting et al., 2010; Crofts et al., 2015) (Figure 2C). SS is responsible for the extension of the α-1–4 glycosidic bond in amylopectin; it has the largest number of isoforms and the most complex functions among the SSREs (Ohdan et al., 2005). SSI, SSII, and SSIII are generally believed to be responsible for the elongation of α-glucan chains during amylopectin synthesis, whereas SSIV and its interaction partners, including PHO1, PTST2, MAR BINDING FILAMENT-LIKE PROTEIN 1 (MFP1), and MYOSIN-RESEMBLING CHLOROPLAST PROTEIN/PROTEIN INVOLVED IN STARCH INITIATION 1 (MRC/PII1), are involved in starch granule initiation (Jeon et al., 2010; Crofts et al., 2015; Seung et al., 2018; Abt and Zeeman, 2020). The novel starch synthase SSV was recently identified, and it shows conserved sequences and gene structures in all green plants (Liu et al., 2015b). SSV shows no enzymatic activity but interacts with MRC to manipulate starch granule initiation in Arabidopsis chloroplasts, similar to its close relative SSIV (Abt and Zeeman, 2020; Abt et al., 2020).
SBE is the only enzyme that acts on glucan to produce branches connected by α-1,6-glycoside bonds, and it generally includes three types in cereals: SBEI (SBE1), SBEII, and SBEIII (Han et al., 2007; Tian et al., 2009). SBEII, including SBEIIb (SBE3) and SBEIIa (SBE4), plays a major role during amylopectin synthesis in the cereal endosperm (Tian et al., 2009; Sawada et al., 2018). Significantly, SBEIIa mainly compensates for the function of SBEI, instead of SBEIIb, by forming amylopectin medium chains in the rice endosperm (Sawada et al., 2018). SBEIII, which shares low similarity with SBEI and SBEII, has been found in many higher plants, including rice, maize, and wheat, and only TaSBEIII was identified to be associated with the synthesis of both A and B granules in wheat grains (Han et al., 2007; Kang et al., 2013b).
DBE, comprising ISA1, ISA2, ISA3, and pullulanase (PUL), hydrolyzes α-1,6-glycosidic bonds and corrects the wrong branches in starch synthesis to ensure the orderly synthesis of amylopectin. ISA1, also known as Sugary 1 (Su1) and Pre-harvest sprouting 8 (PHS8), is critical for the orderly synthesis of amylopectin in cereal endosperms (James et al., 1995; Burton et al., 2002; Du et al., 2018). Mutation of ISA1 causes a massive accumulation of highly branched glucan and phytoglycogen in grains and also causes pre-harvest sprouting (Du et al., 2018). ISA1 can form homo-oligomers with itself or heterologous complexes with ISA2, in which ISA1 plays a catalytic role and ISA2 plays a regulatory role (Utsumi et al., 2011). Moreover, ISA1, ISA2, and SSIII can act synergistically to inhibit the accumulation of phytoglycogen (Lin et al., 2011).
In cereal endosperms, the enzymes involved in amylopectin synthesis usually act in the form of multi-enzyme complexes (Kötting et al., 2010; Crofts et al., 2017). For instance, trimers composed of SSI, SSIIa, and SBEIIa/SBEIIb have been observed in the developing endosperms of wheat, maize, and rice; within these trimers, SSIIa is located between SSI and SBEII and is responsible for binding to starch granules (Hennen-Bierwagen et al., 2008; Tetlow et al., 2008; Liu et al., 2012; Hayashi et al., 2018).
Synthesis of amylose
Amylose in cereal endosperms is synthesized by granule-bound starch synthase I (GBSSI), which is encoded by the Waxy (Wx) gene (Shure et al., 1983; Wang et al., 1990) (Figure 2C). GBSSI controls amylose synthesis in the form of oligomers after being phosphorylated on the surface of starch granules (Liu et al., 2013). All three PTST proteins are involved in the synthesis of starch, but only PTST1 is involved in the synthesis of amylose (Seung et al., 2017). PTST1 interacts with GBSSI to help the latter’s localization to the surface of starch granules (Seung et al., 2015). The C-terminal carbohydrate binding module (CBM) of PTST1 is required for correct GBSSI localization, and mutation of CBM can cause GBSSI to remain in the plastid stroma (Wang et al., 2020b). The ptst1 mutants in Arabidopsis and barley showed significantly reduced GBSS protein accumulation without amylose synthesis, consistent with the gbss mutants (Seung et al., 2015; Zhong et al., 2018). The OsGBP gene in rice is homologous to PTST1 in Arabidopsis and barley, but the reduced AC in the endosperm of the osgbp mutant is still much higher than that of glutinous rice, suggesting that other mechanisms are involved in localizing GBSSI to starch granules (Wang et al., 2020b).
Initiation and morphogenesis of starch granules
Starch in the cereal endosperm exists in the form of semi-crystalline, insoluble starch granules (Abt and Zeeman, 2020) (Figure 2D). The morphology of starch granules, including number, size, shape, and distribution in amyloplasts, is diverse among cereal endosperm cells. The amyloplast in the endosperm of maize and most members of the Triticeae (including wheat and barley) contains only one simple starch granule, and the starch granules of Triticeae are bimodal in size (Myers et al., 2011; Seung and Smith, 2018). In the rice endosperm, multiple starch granules initiate in amyloplasts and fuse into a larger compound structure during grain maturation (Matsushima et al., 2015) (Figure 2E). The compound starch granules are considered to be the ancestral form in the Gramineae, and the number of starch granules per amyloplast is determined in the early developmental stage (Matsushima et al., 2013, 2015).
The matrix of starch granules is composed mainly of amylopectin. Thus, enzymes and proteins involved in amylopectin synthesis, such as ISA1, SBEs, SSIII, SSIV, SSV, and PTST2, also function in the initiation and morphogenesis of starch granules. As mentioned above, ISA1 is vital for ensuring the correct formation of starch granules instead of highly branched glucan or phytoglycogen (Burton et al., 2002). FLOURY ENDOSPERM6 (FLO6), which is homologous to PTST2 in Arabidopsis, B-GRANULE CONTENT 1 (BGC1) in wheat, and Franubet (Fra) in barley, modulates the formation of compound starch granules in the rice endosperm through interaction with ISA1, enabling its binding to the starch granules (Peng et al., 2014; Saito et al., 2018; Chia et al., 2019). The double mutation of ISA1 and SBEIIa eliminated the sugary endosperm, resulting in normal starch granule formation (Lee et al., 2017). Gradually decreasing the expression of SBEs can lead to the formation of various heterogeneous starch granules, including polygonal, aggregate, elongated, and hollow granules (Wang et al., 2018). Studies in wheat indicated that SSIV and BGC1 are required for proper granule initiation, as TaSSIVb-D mutation led to a decrease in starch granules per chloroplast (Guo et al., 2017), and mutation of either SSIV or BGC1 resulted in multiple initiations of amyloplasts and the formation of compound granules (Hawkins et al., 2021). Mutation of SSIVb (SSIV-2) or SSIIIa did not show even a slight effect on starch granule morphology in the rice endosperm, but their combination resulted in small and spherical starch granules (Toyosawa et al., 2016). SSIII also functions in starch granule initiation in the absence of SSIV, and the absence of SSV negatively affects the number of starch granules in Arabidopsis chloroplasts, similar to that of SSIV (Szydlowski et al., 2009; Abt et al., 2020).
In cereals, starch granule morphology after mutation of the Wx gene is mostly normal, indicating that amylose is not essential for the formation of starch granules (Seung and Smith, 2018). However, mutations of PTST1 or OsGBP, which interacts with GBSSI, cause dramatic changes in starch granule morphology (Seung et al., 2015; Zhong et al., 2018; Wang et al., 2020b).
Regulatory network of starch synthesis in cereal endosperms
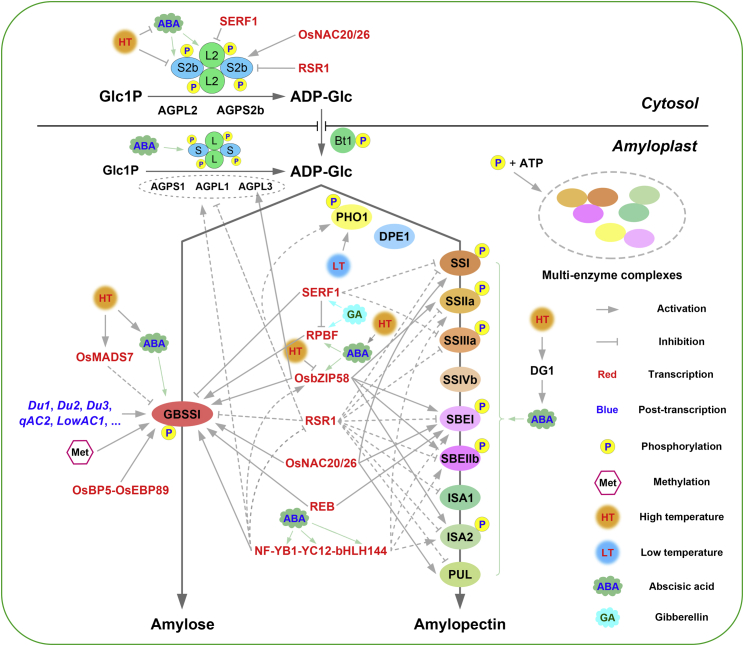
Starch metabolism is regulated by various upstream pathways. In the past decade, considerable progress has been made in revealing the upstream regulatory network of starch synthesis in the endosperms of a number of cereals, especially rice, as summarized in Figure 3.
Figure 3.
Regulatory network of starch synthesis in the cereal endosperm: rice as an example.
At the transcriptional level, several key transcription factors (red font) and methylation are involved in the stimulation or inhibition of starch synthesis-related genes (SSRGs). At the post-transcriptional level, some Du (Dull) genes (Du1, Du2, Du3, etc.) and quantitative trait loci (QTLs) (qAC2, LowAC1, etc.) have been reported to regulate amylose synthesis by manipulating the splicing efficiency of Wx mRNA. At the post-translational level, phosphorylation affects starch synthesis by promoting the formation of multi-enzyme complexes. Gibberellin (GA) modulates the expression of SERF1 and RPBF in starch synthesis to promote starch accumulation in the developing rice endosperm. Abscisic acid (ABA) synthesized in rice and maize leaves directly activates the expression of most SSRGs and multiple hub transcription factors in the rice caryopsis after long-distance leaf-to-caryopsis ABA transport. Defective grain-filling 1 (DG1)-mediated long-distance ABA transport is sensitive to high temperature (HT) during seed development. HT also downregulates the expression of SSRGs and the stability of their encoded enzymes, especially genes encoding AGPase and GBSSI, resulting in chalky grains and defective starch accumulation. Low temperature (LT) promotes the function of PHO1. The arrowed lines indicate activation, and the bar-ended lines indicate inhibition. The dotted lines indicate that the transcription factor can regulate the transcription of the target gene, but there is no direct interaction evidence. The abbreviations for the enzymes, transcription factors, and genes are the same as those listed in Tables 1 and 2.
Transcriptional regulation
At the transcription level, the expression of SSRGs is regulated by a series of transcription factors (TFs) (Table 2). The basic leucine zipper (bZIP) family of TFs plays important roles in cereal endosperm development. For example, Opaque2 (O2) not only transactivates the expression of key SSRGs and sucrose synthase (SUS)-encoding genes but also binds to the GCN4 motif in the promoters of almost all zein genes in maize, indicating its role as a hub for corn kernel development (Li et al., 2015; Zhang et al., 2016; Deng et al., 2020). The prolamin box binding factor (PBF) from the DNA binding with one finger (Dof) family interacts with O2 to additively regulate the expression of certain SSRGs and zein genes or directly activates the expression of these genes (Zhang et al., 2016). OsbZIP58 (also known as RISBZ1) and rice PBF (RPBF) are the homologs of maize O2 and PBF, respectively. OsbZIP58 and RPBF show both complementarity and functional redundancy with one another to regulate the transcription of genes involved in starch and protein synthesis during endosperm development (Yamamoto et al., 2006; Kawakatsu et al., 2009; Wang et al., 2013). OsbZIP58 and RPBF also directly regulate the expression of the gene encoding lysine ketoglutarate reductase/saccharopine dehydrogenase (LKR/SDH), a key regulator of free lysine content in plants (Kawakatsu and Takaiwa, 2010). Similar conclusions were also obtained with the O2 and PBF homologs in wheat and barley (Guo et al., 2020b; Orman-Ligeza et al., 2020).
Table 2.
Transcription factors involved in starch synthesis during cereal endosperm development.
| TF | Locus | TF family | Key references |
|---|---|---|---|
| Oryza sativa L. | |||
| OsbZIP58/RISBZ1 | LOC_Os07g08420 | bZIP | Kawakatsu et al., 2009; Wang et al., 2013; Xu et al., 2020 |
| OsbZIP33/REB/RISBZ2 | LOC_Os03g58250 | bZIP | Yang et al., 2001; Cai et al., 2002 |
| OsbZIP76 | LOC_Os09g34880 | bZIP | Niu et al., 2020 |
| OsNAC20 | LOC_Os01g01470 | NAC | Wang et al., 2020a |
| OsNAC26 | LOC_Os01g29840 | NAC | |
| ONAC127 | LOC_Os11g31340 | NAC | Ren et al., 2021 |
| ONAC129 | LOC_Os11g31380 | NAC | |
| NF-YB9 | LOC_Os06g17480 | NF-Y | Niu et al., 2021 |
| NF-YB1 | LOC_Os02g49410 | NF-Y | Zhiguo et al., 2018; Bello et al., 2019 |
| NF-YC12 | LOC_Os10g11580 | NF-Y | |
| bHLH144 | LOC_Os04g35010 | bHLH | |
| OsMADS7 | LOC_Os08g41950 | MADS-box | Zhang et al., 2018 |
| OsMADS29 | LOC_Os02g07430 | MADS-box | Yin and Xue, 2012 |
| OsMADS6 | LOC_Os02g45770 | MADS-box | Zhang et al., 2010 |
| RSR1 | LOC_Os05g03040 | AP2/EREBP | Fu and Xue, 2010 |
| OsEBP-89 | LOC_Os03g08460 | AP2/EREBP | Zhu et al., 2003 |
| RPBF | LOC_Os02g15350 | DOF | Yamamoto et al., 2006; Kawakatsu et al., 2009 |
| SERF1 | LOC_Os05g34730 | DREB | Schmidt et al., 2014 |
| OsBP-5 | LOC_Os03g43810 | MYC-like | Zhu et al., 2003 |
| Zea mays L. | |||
| ZmABI19 | Zm00001d011712 | B3 domain | Yang et al., 2020 |
| Opaque2/O2 | GRMZM2G015534 | bZIP | Zhang et al., 2016; Deng et al., 2020 |
| ZmbZIP22 | GRMZM2G043600 | bZIP | Li et al., 2018a; Dong et al., 2019b |
| ZmbZIP91 | GRMZM2G043600 | bZIP | Chen et al., 2015 |
| PBF | GRMZM2G146283 | DOF | Zhang et al., 2016 |
| ZmDOF36 | GRMZM2G137502 | DOF | Wu et al., 2019 |
| ZmNAC36 | GRMZM2G154182 | NAC | Zhang et al., 2014b |
| ZmNAC126 | Zm00001d005028 | NAC | Xiao et al., 2020 |
| ZmNAC128/ZmNAC34 | GRMZM2G062650 | NAC | Peng et al., 2019; Zhang et al., 2019d |
| ZmNAC130 | GRMZM2G154182 | NAC | |
| ZmMYB14 | GRMZM2G172327 | MYB | Xiao et al., 2017 |
| ZmEREB156 | GRMZM2G421033 | AP2/EREBP | Huang et al., 2016 |
| Opaque11/O11 | GRMZM2G147685 | bHLH | Feng et al., 2018 |
| ZmMADS1a | GRMZM2G160687 | MADS-box | Dong et al., 2019c |
| ZmABI4 | GRMZM2G093595 | AP2/ERF | Hu et al., 2012 |
| Triticum aestivum L. | |||
| TaSPA | TraesCS1B02G343500, TraesCS1D02G332200 | bZIP | Guo et al., 2020b |
| TubZIP28/TabZIP28 | TRIUR3_00 571, AML47732 | bZIP | Song et al., 2020 |
| TaNAC019-A, B, D | TraesCS3A02G077900, TraesCS3B02G092800, TraesCS3D02G078500 | NAC | Liu et al., 2020; Gao et al., 2021 |
| TaRSR1 | JX473823 | AP2/EREBP | Kang et al., 2013a |
| TaPBF/LSY3 | TraesCS5A02G155900, TraesCS5B02G154100, TraesCS5D02G161000 | DOF | Orman-Ligeza et al., 2020 |
| Hordeum vulgare L. | |||
| HvSUSIBA2 | AY323206 (GenBank) | WRKY | Sun et al., 2003 |
| PBF/LYS3 | HORVU5Hr1G048700 | DOF | Orman-Ligeza et al., 2020 |
AP2/EREBP, APETALA2/ethylene-responsive element binding protein; AP2/ERF, APETALA2/ethylene-responsive factor; bZIP, basic leucine zipper transcription factor; DOF, DNA binding with one finger; MYB, myeloblastosis; NAC, NAM/ATAF/CUC domain transcription factor; NF-Y, nuclear factor Y; PBF, prolamin box binding factor.
In addition to O2 and its homologs, several other bZIP TFs have also been shown to participate in the transcriptional regulation of SSRGs in maize, rice, wheat, and barley. ZmbZIP22 positively regulates prolamin synthesis in corn kernels by binding to the promoter of the gene encoding 27-kDa γ-zein and negatively affects starch synthesis by regulating key genes involved in sugar and starch metabolism (Li et al., 2018a; Dong et al., 2019b). ZmbZIP22 interacts with PBF but not with O2 (Li et al., 2018a). ZmbZIP91 positively regulates the expression of SSRGs in the maize leaf and endosperm by binding to ACTCAT elements in their promoters (Chen et al., 2015). Knockout or knockdown of OsbZIP76, which regulates endosperm cellularization in rice, caused precocious cellularization in the rice caryopsis and enhanced the expression of SSRGs, leading to earlier starch accumulation (Niu et al., 2020). OsbZIP33/REB/RISBZ2 interacts with GCN4 motifs in the promoters of Wx, SBE1, and α-globulin genes in rice (Yang et al., 2001; Cai et al., 2002). In the wheat endosperm, TubZIP28/TabZIP28 binds to the promoters of genes encoding cytosolic AGPase, enhancing their transcription (Song et al., 2020). In barley, more than 10 bZIP TFs were identified with expression patterns similar to those of SSRGs, two of which could bind to the promoter of HvAGPS (Zhong et al., 2021).
Recently, some NAM/ATAF/CUC (NAC) TFs involved in the regulation of starch and storage protein synthesis in developing cereal grains were cloned. Three NAC members related to starch synthesis were identified in maize: ZmNAC126, ZmNAC128/ZmNAC34, and ZmNAC130. Among these, ZmNAC126 modulates starch accumulation in corn kernels by increasing starch synthesis and inhibiting starch degradation (Xiao et al., 2020). The functions of ZmNAC128/ZmNAC34 and ZmNAC130 are redundant. They compete for a 6-bp consensus sequence (ACGCAA) in the promoters of starch and zein synthesis-related genes, such as Bt2 and 16-kDa γ-zein, to regulate the accumulation of starch and storage protein in corn kernels (Peng et al., 2019; Zhang et al., 2019d). ZmaNAC36, which is highly expressed in the maize endosperm and is co-expressed with most of the SSRGs, may also play a role in starch synthesis in the maize endosperm (Zhang et al., 2014b). In rice, OsNAC20 and OsNAC26 bind directly to key genes of starch and protein synthesis in the endosperm. However, the high similarity of their amino acid sequences has resulted in redundancy, which was verified by the fact that only the osnac20/26 double mutant showed significantly decreased starch and storage protein content in the endosperm (Wang et al., 2020a). In the linked region of rice chromosome 11, a gene cluster encoding four NAC members (ONAC025, ONAC127, ONAC128, and ONAC129) was identified (Fang et al., 2008). ONAC127 and ONAC129 are not directly involved in starch synthesis in the rice endosperm, but they do form heterodimers that participate in apoplastic transport and the heat stress response to regulate rice grain filling, including starch accumulation (Ren et al., 2021). In wheat, the endosperm-specific TaNAC019 was also found to regulate glutenin and starch accumulation by directly activating the expression of relevant genes (Liu et al., 2020; Gao et al., 2021).
A group of nuclear factor Ys (NF-Ys), which are predominantly expressed in the rice endosperm, were also reported to be regulators of rice endosperm development (Yang et al., 2017; Zhiguo et al., 2018). Recent findings showed that OsNF-YB1 is specifically expressed in the aleurone layer of the rice developing endosperm and regulates grain filling and endosperm development by interacting with other TFs, including OsbZIP76, OsERF115, OsNF-YC11, OsNF-YC12, and bHLH144 (Bai et al., 2016; Bello et al., 2019; Xiong et al., 2019; Niu et al., 2020). For instance, NF-YB1 binds with NF-YC12 and bHLH144 to form an NF-Y heterotrimer complex (NF-YB1-YC12-bHLH144), and mutation of each gene in the complex can alter starch synthesis in the rice endosperm (Bello et al., 2019). A rice LEC1-like TF known as OsNF-YB9 functions in starch synthesis by affecting SSRG expression and starch granule morphology (Niu et al., 2021).
Furthermore, the maize MYB TF ZmMYB14 binds to the promoters of multiple SSRGs, including ZmBT1, to activate their expression (Xiao et al., 2017). ZmMYB138 and ZmMYB115 are candidate TFs involved in the regulation of starch synthesis in the maize endosperm, as suggested by expression correlation analysis (Hu et al., 2021). ZmABI4, ZmEREB156, and HvSUSIBA2 are TFs related to the modulation of SSRGs such as SSIIIa, SSI, and ISA1 by abscisic acid (ABA) and/or sugar signaling (Sun et al., 2003; Hu et al., 2012; Huang et al., 2016). OsMADS7, OsBP-5, and OsEBP89 were identified in rice as being associated with Wx gene regulation and its response to high temperature (Cai et al., 2002; Zhu et al., 2003; Zhang et al., 2018). Rice Starch Regulator1 (RSR1), an APETALA2/ethylene-responsive element binding protein (AP2/EREBP), is a negative regulator of SSRGs that is expressed predominantly in seeds of rice and wheat (Fu and Xue, 2010; Kang et al., 2013a). Suppression of OsMADS29 expression inhibits starch biosynthesis and grain filling in the rice endosperm by causing defective programmed cell death (Yin and Xue, 2012). OsMADS6 is highly expressed in the florets and endosperm and regulates starch accumulation by regulating genes that encode AGPase (Zhang et al., 2010).
Upstream of these TFs, the DEHYDRATION-RESPONSE ELEMENT BINDING (DREB)-type TF SALT-RESPONSIVE ERF1 (SERF1) was reported to manipulate the synthesis and degradation of starch by timing the expression of RPBF in rice (Schmidt et al., 2014). O11 is a central hub of maize endosperm development and nutrient metabolism that directly regulates related key TFs, including O2 and PBF (Feng et al., 2018). The B3 domain-containing TF ZmABI19 can coordinate the expression of O11, O2, and other key TFs and has been defined as a hub of hubs (Yang et al., 2020; Zhan, 2020).
Methylation regulation
Previous studies have shown that genes preferentially expressed in the rice endosperm, including SSRGs, usually show reduced expression because of methylation, indicating that methylation is also involved in the regulation of starch synthesis (Zemach et al., 2010). Recently, similar overall methylation patterns were found in the SSRGs expressed in the maize endosperm (Hu et al., 2021). In the developing maize endosperm, the coding regions of low-expression SSRGs are highly methylated, and expression levels of SBE1 and Su1 correlate significantly and negatively with predicted DNA methylation marks (Hu et al., 2021). Thus, DNA methylation may act as a switch in the global regulation of starch biosynthesis. Moreover, high CpG methylation in Wx is closely related to low AC in rice grains, and two neighboring CpG islands were detected in the promoter region of Wx, indicating that DNA methylation is involved in the regulation of amylose synthesis (Anacleto et al., 2019).
Post-transcriptional regulation
At the post-transcriptional level, some Du (Dull) genes and quantitative trait loci (QTLs) were reported to regulate amylose synthesis by manipulating the splicing efficiency of Wx mRNA in the rice endosperm. Du1 and Du2 showed similar genetic interactions with Wx alleles: the level of GBSSI protein in both the du1 and du2 mutants was decreased by inefficient splicing of Wxb pre-mRNA, whereas neither mutant affected the expression of the Wxa allele. The Du1 gene encodes a member of the pre-mRNA processing (Prp1) family that is a component of the U4/U6 small nuclear ribonucleoprotein required for spliceosome assembly (Zeng et al., 2007). Du3 modulates Wx pre-mRNA splicing by encoding the rice homolog of a cap-binding protein 20-kDa subunit (CBP20), a component of the heterodimeric nuclear cap-binding complex. The novel QTL qAC2 and the novel dull gene LowAC1, which encodes an RNA recognition motif protein, were recently shown to have a function related to Wxb pre-mRNA splicing, similar to that of du1 and du2 (Takemoto-Kuno et al., 2015; Igarashi et al., 2021). The post-transcriptional regulation of Wx may also be involved in grain filling at high temperatures, which will be discussed below.
Post-translational regulation via phosphorylation
Post-translational phosphorylation is an important aspect of starch synthesis regulation: the formation of multi-enzyme complexes depends on protein phosphorylation, and broad-spectrum phosphatase treatment can prevent the formation of high-molecular-weight protein complexes (Grimaud et al., 2008; Tetlow et al., 2008; Kötting et al., 2010; Ahmed et al., 2015). 32P-labeled autoradiography and phosphorylated proteomics further confirmed the phosphorylation of enzymes involved in the formation of multi-enzyme complexes in the starch synthesis pathway (Tetlow et al., 2004; Walley et al., 2013). In the developing maize endosperm, the phosphorylation sites of SSREs, including AGPase large and small subunits, PHO1, SBEs, ISA2, and Bt1, were identified using high-throughput proteome, transcriptome, and phosphorylated proteome analysis (Walley et al., 2013). For example, a systematic study of the phosphorylation sites of maize SBEIIb demonstrated that three serine residue sites, Ser286, Ser297, and Ser649, can be phosphorylated (Makhmoudova et al., 2014). These phosphorylation sites form salt bridges to stabilize the SBEIIb protein structure, and their mutation caused a decreased phosphorylation level of recombinant SBEIIb. The three phosphorylation sites show different degrees of conservation in different crop species and SBE isoforms, indicating functional differentiation among them. The difference in the conservation of Ser286 among the SBEs may explain why phosphorylation can change the activities of SBEIIa and SBEIIb but has no effect on the activity of SBEI in wheat (Tetlow et al., 2008). Several phosphorylation sites were also found in the rice GBSSI protein, such as the S415P substitution that was shown to change the phosphorylation level of GBSSI, thereby modulating its activity to a moderate level in Wxlv (Zhang et al., 2019a). In wheat, the amount of phosphorylated AGPase increased during the progress of seed development and was positively correlated with relative increases in AGPase activity and starch accumulation (Ferrero et al., 2020).
Regulation by hormones
Hormones play vital roles in all stages of plant growth and development, including endosperm starch synthesis. In the process of starch metabolism, gibberellin (GA) functions mainly in starch degradation by regulating Ramy1A through the TF GAMyb, but it also modulates the expression of the key TFs SERF1 and RPBF in starch synthesis to promote starch accumulation in the developing rice endosperm (Schmidt et al., 2014). Previous studies showed that ABA could greatly enhance AGPL1 (ApL3) induction by sugar in cultured rice cells (Akihiro et al., 2005). In maize, the novel TF ZmEREB156 was found to mediate the regulation of ABA and sucrose in starch synthesis (Huang et al., 2016). Sugar accumulation resulting from mutation of ISA1 decreased the expression of OsABI3 and OsABI5 and reduced sensitivity to ABA (Du et al., 2018). ABA induces the accumulation of ZmSSI mRNA in maize endosperm. The ZmSSI promoter contains a CACCG motif that interacts with the ABA pathway of the TF ABI4 in vitro (Hu et al., 2012). ABI4 enhances the activity of promoters containing the CACCG motif, including ZmSSI (Hu et al., 2012). A recent study indicated that starch synthesis in developing seeds is regulated mainly by leaf-derived ABA. ABA synthesized in rice leaves directly activates the expression of most SSRGs and multiple hub TFs in the rice caryopsis following long-distance leaf-to-caryopsis ABA transport. Defective grain-filling 1 (DG1), a functionally conserved multi-drug and toxic compound extrusion transporter in cereals, regulates grain filling by manipulating long-distance leaf-to-caryopsis ABA transport (Qin et al., 2021).
Response to temperature
The environment is also a key determinant of starch synthesis in cereal endosperms, mainly through temperature fluctuations during the reproductive phase (Fan et al., 2019). The pho1 mutant showed shrunken rice grains when grown at 20°C, whereas most grains were plump when grown at 30°C, suggesting that low temperature promotes the function of PHO1 and that an unknown factor(s) can compensate for PHO1 function at room temperature (Satoh et al., 2008). The negative effect of high temperature (HT) on starch synthesis is more severe and more common during the reproductive period of cereals. At the genetic level, HT significantly downregulates the expression of SSRGs and induces the expression of genes encoding α-amylase, resulting in chalky grains and defective starch accumulation (Zhang et al., 2018). AGPase and GBSSI are among the SSREs that are severely affected by HT during grain ripening (Denyer et al., 1994; Boehlein et al., 2007; Yamakawa et al., 2007). HT significantly destabilizes AGPase heterotetramers, dramatically reducing ADPG synthesis (Saripalli and Gupta, 2015). Unlike the AGPase from potato tubers, the AGPases of rice and maize endosperms lack a conserved QTCL (Gln-Thr-Cys-Leu) motif and are therefore heat labile (Boehlein et al., 2007). An insertion of cysteine in the N terminus of AGPS2b in maize and rice enhanced the heat stability of their heterotetramers (Linebarger et al., 2005; Hwang et al., 2019). The L379F mutation of rice AGPS2b was also found to improve the heat stability of AGPase by increasing the number of hydrogen bonds between two AGPS2b subunits and the intermolecular interactions between AGPL2 and AGPS2b (Hwang et al., 2019).
HT inhibits Wx-encoded GBSSI and AC accumulation, resulting in rice grains with poor eating quality (Zhang et al., 2018). One of the mechanisms is that HT induced the alternative splicing of OsbZIP58, thereby reducing the transcription of the Wx gene (Xu et al., 2020). HT promotes the formation of the truncated OsbZIP58β protein relative to the full-length OsbZIP58α protein, and OsbZIP58β showed lower transcriptional activity than OsbZIP58α under HT conditions (Xu et al., 2020). Specific suppression of the floral organ identity gene OsMADS7 in the rice endosperm stabilized AC under HT stress without reducing spikelet fertility (Zhang et al., 2018). The post-transcriptional regulation of Wxb is a critical mechanism for maintaining a stable AC at HT. Four QTLs that can stabilize rice AC at HT by increasing the splicing efficiency of Wx pre-mRNA (qSAC3, qHAC4, qHAC8a, and qHAC8b) were detected in a genome-wide survey of chromosome segment substitution lines derived from a cross between the indica variety 9311 and the japonica variety Nipponbare (Zhang et al., 2014a, 2019b).
Furthermore, inhibition of α-amylase gene expression resulted in fewer chalky grains under HT (Hakata et al., 2012). The presence of heat-stable PGD1 or PGD2, two heat-stable isotypes of 6-phosphogluconate dehydrogenase, in the endosperm amyloplast mitigated maize yield losses caused by HT stress (Ribeiro et al., 2020). Recently, HT was also found to affect starch accumulation in the rice and maize endosperms by regulating the efficiency of DG1-mediated leaf-to-caryopsis ABA transport. HT increased DG1 expression, causing increased transport of ABA to the caryopsis and thus altering the expression of SSRGs (Qin et al., 2021).
Genetic improvement of cereal starch quality
Research on SSRGs in cereals has resulted in numerous breakthroughs in cereal breeding for grain quality improvement, especially the application of elite natural and mutagenic alleles of key SSRGs (Tian et al., 2009; Chen and Bao, 2016; Zeng et al., 2017). The genetic engineering approach has also been widely used in the genetic modification of SSRGs.
The Wx gene that controls the synthesis of amylose is the most widely used gene in cereal breeding (Huang et al., 2020a). Since the cloning of the Wx gene in the 1980s, transgenic lines with various ACs have been established using antisense RNA, homologous recombination, and overexpression, and the effects of different ACs on starch physicochemical properties have been clarified (Shimada et al., 1993; Terada et al., 2000, 2002; Itoh et al., 2003; Liu et al., 2015a). In recent years, diverse Wx alleles have been cloned in rice, maize, wheat, and barley (Asare et al., 2012; Guzmán and Alvarez, 2016; Luo et al., 2020). Cloning of numerous null wx alleles in maize has enabled the convenient breeding of waxy maize with a unique chewing texture. The combination of one, two, or three null wx (wx-A1, wx-B1, and wx-D1) alleles allows the breeding of partial waxy wheat with a reduced AC and amylose-free waxy wheat (Guzmán et al., 2012). Flours with different ACs and physicochemical properties can also be obtained by mixing waxy flour and non-waxy flour in different proportions. The Wx gene was selected preferentially during rice domestication (Anacleto et al., 2019). In rice, at least nine functional Wx alleles responsible for various ACs between 0% and 30% have been cloned, including Wxlv, Wxa, Wxin, Wxb, Wxmw/la, Wxop/hp, Wxmq, Wxmp, and wx (Zhang et al., 2019a). Significant achievements have been made through the introgression of Wxb alleles (low to medium AC) and Wxop/hp, Wxmq, and Wxmp alleles (low AC) to reduce the AC of targeted rice varieties (Huang et al., 2020a). Recently, the rare Wx allele Wxmw/Wxla (AC ∼13.5%), derived from the homologous recombination of Wxin and Wxb, was identified through genome-wide association analysis and map-based cloning (Zhang et al., 2020b; Zhou et al., 2020b). Rice carrying Wxmw/Wxla showed an excellent eating and cooking quality, similar to soft rice, but its grains remained transparent, unlike the opaque endosperm of soft rice cultivars (Zhang et al., 2020a; Zhou et al., 2020b). Moreover, a series of novel Wx alleles was created by editing the promoter and coding region of rice Wx, and similar strategies could be adopted to create more novel Wx alleles (Huang et al., 2020b; Zeng et al., 2020; Xu et al., 2021). Direct editing of the Wx gene in superior germplasm could reduce the impact of linkage drag on yield (Gao et al., 2020).
The ALK gene, also known as SSIIa and SSII-3, controls GT and is another crucial gene in rice quality improvement breeding (Gao et al., 2003). Three typical ALK alleles were identified based on three key single-nucleotide polymorphisms (SNPs) among rice cultivars (Nakamura et al., 2005; Waters et al., 2006; Gao et al., 2011). ALKa and ALKb encode inactive japonica-type SSIIaj that produces a low GT. ALKc encodes the active indica-type SSIIai that produces a high GT. ALKb induces a slightly lower GT but has a wider distribution among rice subpopulations than ALKa, and both their GTs and distributions are lower than those of ALKc (Chen et al., 2020). In addition, ALKd, caused by a G/T SNP in exon 1 of the ALKc allele, was identified from the high-GT indica rice cultivar Zhonghui 9308, which had a higher GT and improved retrogradation properties (Zhang et al., 2020a). The SSIIa-deficient mutant ssiia, with no SSIIa activity or SSIIa protein and a lower GT, was also identified following N-methyl-N-nitrosourea mutagenesis of the japonica rice cultivar Kinmaze (Miura et al., 2018). In rice breeding programs, ALK alleles that produce low and medium GTs are generally preferred.
Sweet corn is an important direction in maize breeding. The improvement of sweet corn involves multiple genes and their allelic variants, including Su1, Su2, Se1, Bt1, Bt2, and Sh2. Mutant alleles of isoamylase 1, designated su1, were first used for sweet corn breeding, and su1 hybrids were mostly planted before 1985 (Schultz and Juvik, 2004). However, the rapid conversion of sugar to starch and the moisture loss in su1 sweet corn hybrids after harvest caused a rapid decline in ear quality, restricting their storage, transport, and marketability (Schultz and Juvik, 2004). The su1 allele and the se1 mutant can be combined to breed enhanced sweet corn. The se1 gene is a recessive modification gene of su1 that is inherited independently from su1 (Zhang et al., 2019c). After the use of su1, sh2 was used extensively in commercial varieties to breed super-sweet corn; the sh2-i allele, which encodes a partially inactivated AGPase, was obtained using ethyl methanesulfonate (EMS) mutagenesis (Lal et al., 1999). The combination of sh2-i with su1 and sh2 can produce multi-gene compound sweet corn varieties that overcome the genetic defects of the single sh2 gene (Dodson-Swenson and Tracy, 2015). Knockout of Wx using genome editing also facilitated sucrose accumulation in corn kernels, and the simultaneous abolition of Wx and Sh2 created super-sweet and waxy compound corn (Dong et al., 2019a).
Slow-digestible starch and resistant starch (RS) with a low GI are beneficial for the prevention and treatment of type 2 diabetes, obesity, and other blood glucose diseases. Increasing their content is a topic of considerable interest in current cereal research and breeding (Jukanti et al., 2020). Increasing AC and extra-long chains of amylopectin in cereal grains are the keys to GI reduction because of their negative correlation with starch digestion rate and positive correlation with RS content (Xia et al., 2018; Huang et al., 2020a). As early as the 1940s and 1950s, ssii and amylose extender (ae) mutants were used for high-amylose maize breeding (Li et al., 2019). A series of ae mutants characterized by loss of SBEIIb activity were identified in maize, among which ae 1.2 lost SBEIIb catalytic activity and was unable to bind to amylopectin because it lacked a 28-amino-acid peptide (Val272–Pro299). The granule morphology and physicochemical characteristics of ae 1.2 are distinct from those of the regular frameshift ae mutant as well as the wild type (Liu et al., 2011). The effects of each SBE isoform on cereal grain AC and RS content were determined using RNA interference and antisense RNA technology (Regina et al., 2006, 2010; Carciofi et al., 2012; Zhu et al., 2012). SBEIIb mutation in rice and maize significantly increased the AC and RS content (Li et al., 2019). Enhanced AC and RS content were also observed in the SBEIIb and SBEIIa double mutant but not in the SBEIIa single mutant (Zhu et al., 2012). However, this is incongruent with reports in which the opposite effects were observed in wheat and barley (Regina et al., 2006, 2010). Consequently, sbe-rs, a natural allele of SBEIIb/SBE3 that can be used for rice breeding, was cloned through the mapping of Jiangtangdao 1, a rice cultivar with a high RS content and low GI (Yang et al., 2012). Two mutant alleles of SBEIIb that produced moderate changes in starch gelatinization and AC and a fine-tuned texture were obtained by screening a mutant population of the japonica rice variety Nipponbare, and they were named altered gelatinization 1 (age1) and age2 (Nakata et al., 2018). In addition, low GI breeding lines with different mutation combinations of SBEIIa and SBEIIb have been obtained through map-based cloning and EMS mutagenesis in wheat (Hazard et al., 2015; Regina et al., 2015; Schönhofen et al., 2016). More recently, genome editing technology has successfully enabled the development of novel rice and wheat germplasms with a low GI through mutation of SBEs (Sun et al., 2017; Guo et al., 2020a; Li et al., 2020b). In addition to SBEs, mutations in SSIIa and SSIIIa also reportedly increased the grain AC of cereals (Shimbata et al., 2012; Zhu et al., 2013; Zhou et al., 2016; Miura et al., 2018; Li et al., 2019).
Perspectives
Comprehensive analysis of the functions of SSRGs
Fully and systematically revealing SSRG functions, interactions, and effects on grain quality is the foundation of starch-related research and its applications; it is essential, albeit not highly innovative. Nevertheless, research on SSRGs has mainly focused on isoforms that are predominantly expressed in the cereal endosperm, whereas the roles of SSRGs that are expressed at low levels in the endosperm or predominantly expressed in other tissues remain unclear (Jeon et al., 2010; Pfister and Zeeman, 2016). Recent studies have found that some SSRGs that are weakly expressed in cereal endosperms nonetheless play significant roles in endosperm starch synthesis and quality formation (Tian et al., 2009; Lee et al., 2017; Zeng et al., 2017). For example, knockdown or knockout of SSIIb (SSII-2), which is expressed at a low level in the endosperm, suppressed the expression of SSIIa, SSIIb, and Wx, resulting in decreased AC, altered amylopectin CLD, and novel low-amylose rice with soft, transparent grains (Li et al., 2018b; Huang et al., 2021). Numerous mutation combinations with various starch properties can be isolated from different SSRG mutations, but only a few such combinations have been obtained, significantly limiting relevant studies (Jeon et al., 2010). The emergence of genome-editing technology provides the possibility for the systematic creation of related mutants, fully revealing SSRG functions, interactions, and effects on grain quality (Chen et al., 2019; Zhu et al., 2020a).
In-depth study of the regulatory network of starch synthesis
The cloning of ZmABI19 and O11, which directly regulate key TFs of corn kernel development such as O2 and PBF, has shed light on the further upstream regulatory network of starch synthesis in cereal endosperms (Feng et al., 2018; Yang et al., 2020). Recent research has demonstrated that some microRNAs (miRNAs) modulate starch synthesis by affecting TFs that regulate SSRG expression and that DNA methylation may function as a switch to regulate starch synthesis by manipulating SSRG and miRNA expression levels in the early stage of grain development (Cheng et al., 2021; Hu et al., 2021). Furthermore, phosphorylation is widely involved in the regulation of starch synthesis (Makhmoudova et al., 2014). However, little is known about the protein kinases and phosphorylases that regulate SSRE phosphorylation, and this topic warrants further work to parse the underlying mechanisms.
Underlying mechanism of the metabolic connection between starch and other storage substances in cereal endosperms
The reduction of starch synthesis in cereal endosperms can lead to a general increase in the accumulation of multiple primary and secondary metabolites, such as sugars, fatty acids, amino acids, and phytosterols. It can also trigger extensive metabolic changes throughout the plant via transcriptional reprogramming (Baysal et al., 2020). TFs that regulate SSRGs also directly regulate genes involved in storage protein synthesis and lysine synthesis (Kawakatsu and Takaiwa, 2010; Zhang et al., 2016). These observations outline a complex regulatory network in which TFs coordinately regulate the accumulation of storage substances in cereal endosperms through the dynamic balance and distribution of carbon and nitrogen; however, only a small portion of this network has been analyzed.
Toward the rational design of cereal starches
The physicochemical properties of starch are closely related to the grain quality of cereals. Further optimization of starch synthesis in cereal endosperms is an important direction in future cereal breeding for grain quality improvement. Nonetheless, there are still many hurdles to overcome. Substantial quality improvements that result from alterations in starch synthesis are often accompanied by negative effects on grain yield and other aspects of grain quality, such as grain chalkiness (Li et al., 2018b). An increase in grain AC may drastically reduce starch digestion rate and reduce eating and cooking quality; the inhibition of SBEIIb or SSIIIa led to an increase in AC and a reduction in grain yield (Zhu et al., 2012; Zhou et al., 2016). Compared with SSRGs predominantly expressed in the endosperm and pleiotropic TFs, SSRGs with a relatively low level of expression in the endosperm appear to be more suitable for crop breeding because their mutations tend to cause moderate changes in starch synthesis (Huang et al., 2021). Furthermore, fine-tuning of the expression or enzyme activity of SSRGs with predominant expression in the endosperm is also likely to be useful in developing elite cereal varieties with improved grain quality without compromising yield traits (Huang et al., 2020b; Xu et al., 2021).
Funding
This work was supported by grants from the National Natural Science Foundation of China (grant numbers 31825019, 32072032, and 31901517) and the PAPD Programs from Jiangsu Province Government.
Author contributions
Qiaoquan Li, L.H., Qianfeng Li, and C.Z. conceived the project. L.H. and H.T. collected the references and prepared the figures and tables. L.H. and Qiaoquan Li wrote the manuscript. All authors read and approved the final version of the manuscript.
Acknowledgments
We gratefully appreciate manuscript editing by Dr. Qing Liu (CSIRO Agriculture and Food, Australia). No conflict of interest declared.
Published: September 2, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
References
- Abt M.R., Zeeman S.C. Evolutionary innovations in starch metabolism. Curr. Opin. Plant Biol. 2020;55:109–117. doi: 10.1016/j.pbi.2020.03.001. [DOI] [PubMed] [Google Scholar]
- Abt M.R., Pfister B., Sharma M., Eicke S., Bürgy L., Neale I., Seung D., Zeeman S.C. STARCH SYNTHASE5, a noncanonical starch synthase-like protein, promotes starch granule initiation in Arabidopsis. Plant Cell. 2020;32:2543–2565. doi: 10.1105/tpc.19.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Z., Tetlow I.J., Ahmed R., Morell M.K., Emes M.J. Protein–protein interactions among enzymes of starch biosynthesis in high-amylose barley genotypes reveal differential roles of heteromeric enzyme complexes in the synthesis of A and B granules. Plant Sci. 2015;233:95–106. doi: 10.1016/j.plantsci.2014.12.016. [DOI] [PubMed] [Google Scholar]
- Akihiro T., Mizuno K., Fujimura T. Gene expression of ADP-glucose pyrophosphorylase and starch contents in rice cultured cells are cooperatively regulated by sucrose and ABA. Plant Cell Physiol. 2005;46:937–946. doi: 10.1093/pcp/pci101. [DOI] [PubMed] [Google Scholar]
- Anacleto R., Badoni S., Parween S., Butardo V.M., Jr., Misra G., Cuevas R.P., Kuhlmann M., Trinidad T.P., Mallillin A.C., Acuin C. Integrating a genome-wide association study with a large-scale transcriptome analysis to predict genetic regions influencing the glycaemic index and texture in rice. Plant Biotechnol. J. 2019;17:1261–1275. doi: 10.1111/pbi.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare E.K., Båga M., Rossnagel B.G., Chibbar R.N. Polymorphism in the barley granule bound starch synthase 1 (Gbss1) gene associated with grain starch variant amylose concentration. J. Agric. Food Chem. 2012;60:10082–10092. doi: 10.1021/jf302291t. [DOI] [PubMed] [Google Scholar]
- Bai A.N., Lu X.D., Li D.Q., Liu J.X., Liu C.M. NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res. 2016;26:384–388. doi: 10.1038/cr.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal C., He W., Drapal M., Villorbina G., Medina V., Capell T., Khush G.S., Zhu C., Fraser P.D., Christou P. Inactivation of rice starch branching enzyme IIb triggers broad and unexpected changes in metabolism by transcriptional reprogramming. Proc. Natl. Acad. Sci. U S A. 2020;117:26503–26512. doi: 10.1073/pnas.2014860117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello B.K., Hou Y., Zhao J., Jiao G., Wu Y., Li Z., Wang Y., Tong X., Wang W., Yuan W. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.) Plant Biotechnol. J. 2019;17:1222–1235. doi: 10.1111/pbi.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehlein S.K., Shaw J.R., Stewart J.D., Hannah L.C. Heat stability and allosteric properties of the maize endosperm ADP-glucose pyrophosphorylase are intimately intertwined. Plant Physiol. 2007;146:289–299. doi: 10.1104/pp.107.109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust H., Orzechowski S., Fettke J., Steup M. Starch synthesizing reactions and paths: in vitro and in vivo studies. J. Appl. Glycosci. 2013;60:3–20. doi: 10.5458/jag.jag.JAG-2012_018. [DOI] [Google Scholar]
- Burton R.A., Jenner H., Carrangis L., Fahy B., Fincher G.B., Hylton C., Laurie D.A., Parker M., Waite D., Van Wegen S. Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J. 2002;31:97–112. doi: 10.1046/j.1365-313X.2002.01339.x. [DOI] [PubMed] [Google Scholar]
- Cai Y., Xie D., Wang Z., Hong M. Interaction of rice bZIP protein REB with the 5′-upstream region of both rice SBE1 gene and Waxy gene. Chin. Sci. Bull. 2002;47:310–314. doi: 10.1360/02tb9074. [DOI] [Google Scholar]
- Cakir B., Shiraishi S., Tuncel A., Matsusaka H., Satoh R., Singh S., Crofts N., Hosaka Y., Fujita N., Hwang S.-K. Analysis of the rice ADP-glucose transporter (OsBT1) indicates the presence of regulatory processes in the amyloplast stroma that control ADP-glucose flux into starch. Plant Physiol. 2016;170:1271–1283. doi: 10.1104/pp.15.01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carciofi M., Blennow A., Jensen S.L., Shaik S.S., Henriksen A., Buléon A., Holm P.B., Hebelstrup K.H. Concerted suppression of all starch branching enzyme genes in barley produces amylose-only starch granules. BMC Plant Biol. 2012;12:223. doi: 10.1186/1471-2229-12-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Yi Q., Cao Y., Wei B., Zheng L., Xiao Q., Xie Y., Gu Y., Li Y., Huang H. ZmbZIP91 regulates expression of starch synthesis-related genes by binding to ACTCAT elements in their promoters. J. Exp. Bot. 2015;67:1327–1338. doi: 10.1093/jxb/erv527. [DOI] [PubMed] [Google Scholar]
- Chen K., Wang Y., Zhang R., Zhang H., Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- Chen Y., Bao J. Underlying mechanisms of zymographic diversity in starch synthase I and pullulanase in rice-developing Endosperm. J. Agric. Food Chem. 2016;64:2030–2037. doi: 10.1021/acs.jafc.5b06030. [DOI] [PubMed] [Google Scholar]
- Chen Z., Lu Y., Feng L., Hao W., Li C., Yang Y., Fan X., Li Q., Zhang C., Liu Q. Genetic dissection and functional differentiation of ALKa and ALKb, two natural alleles of the ALK/SSIIa gene, responding to low gelatinization temperature in rice. Rice. 2020;13:39. doi: 10.1186/s12284-020-00393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Pan M., E Z., Zhou Y., Niu B., Chen C. The maternally expressed polycomb group gene OsEMF2a is essential for endosperm cellularization and imprinting in rice. Plant Com. 2021;2:100092. doi: 10.1016/j.xplc.2020.100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia T., Chirico M., King R., Ramirez-Gonzalez R., Saccomanno B., Seung D., Simmonds J., Trick M., Uauy C., Verhoeven T. A carbohydrate-binding protein, B-GRANULE CONTENT 1, influences starch granule size distribution in a dose-dependent manner in polyploid wheat. J. Exp. Bot. 2019;71:105–115. doi: 10.1093/jxb/erz405. [DOI] [PubMed] [Google Scholar]
- Crofts N., Nakamura Y., Fujita N. Critical and speculative review of the roles of multi-protein complexes in starch biosynthesis in cereals. Plant Sci. 2017;262:1–8. doi: 10.1016/j.plantsci.2017.05.007. [DOI] [PubMed] [Google Scholar]
- Crofts N., Abe N., Oitome N.F., Matsushima R., Hayashi M., Tetlow I.J., Emes M.J., Nakamura Y., Fujita N. Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 2015;66:4469–4482. doi: 10.1093/jxb/erv212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumpton-Taylor M., Grandison S., Png K.M.Y., Bushby A.J., Smith A.M. Control of starch granule numbers in Arabidopsis chloroplasts. Plant Physiol. 2011;158:905–916. doi: 10.1104/pp.111.186957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta-Seijo J., Ruzanski C., Krucewicz K., Meier S., Hägglund P., Svensson B., Palcic M. Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development. PLoS One. 2017;12:e0175488. doi: 10.1371/journal.pone.0175488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta-Seijo J.A., Nielsen M.M., Ruzanski C., Krucewicz K., Beeren S.R., Rydhal M.G., Yoshimura Y., Striebeck A., Motawia M.S., Willats W.G.T. In vitro biochemical characterization of all barley endosperm starch synthases. Front. Plant Sci. 2016;6:1265. doi: 10.3389/fpls.2015.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankar I., Haddarah A., Omar F.E.L., Pujolà M., Sepulcre F. Characterization of food additive-potato starch complexes by FTIR and X-ray diffraction. Food Chem. 2018;260:7–12. doi: 10.1016/j.foodchem.2018.03.138. [DOI] [PubMed] [Google Scholar]
- Deng Y., Wang J., Zhang Z., Wu Y. Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize. Plant Biotechnol. J. 2020;18:1897–1907. doi: 10.1111/pbi.13349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer K., Hylton C., Smith A. The effect of high temperature on starch synthesis and the activity of starch synthase. Funct. Plant Biol. 1994;21:783–789. doi: 10.1071/PP9940783. [DOI] [Google Scholar]
- Dodson-Swenson H.G., Tracy W.F. Endosperm carbohydrate composition and kernel characteristics of shrunken2-intermediate (sh2-i/sh2-i Su1/Su1) and shrunken2-intermediate–sugary1-reference (sh2-i/sh2-i su1-r/su1-r) in sweet corn. Crop Sci. 2015;55:2647–2656. doi: 10.2135/cropsci2015.03.0188. [DOI] [Google Scholar]
- Dong L., Qi X., Zhu J., Liu C., Zhang X., Cheng B., Mao L., Xie C. Supersweet and waxy: meeting the diverse demands for specialty maize by genome editing. Plant Biotechnol. J. 2019;17:1853–1855. doi: 10.1111/pbi.13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q., Xu Q., Kong J., Peng X., Zhou W., Chen L., Wu J., Xiang Y., Jiang H., Cheng B. Overexpression of ZmbZIP22 gene alters endosperm starch content and composition in maize and rice. Plant Sci. 2019;283:407–415. doi: 10.1016/j.plantsci.2019.03.001. [DOI] [PubMed] [Google Scholar]
- Dong Q., Wang F., Kong J., Xu Q., Li T., Chen L., Chen H., Jiang H., Li C., Cheng B. Functional analysis of ZmMADS1a reveals its role in regulating starch biosynthesis in maize endosperm. Sci. Rep. 2019;9:3253. doi: 10.1038/s41598-019-39612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Xu F., Fang J., Gao S., Tang J., Fang S., Wang H., Tong H., Zhang F., Chu J. Endosperm sugar accumulation caused by mutation of PHS8/ISA1 leads to pre-harvest sprouting in rice. Plant J. 2018;95:545–556. doi: 10.1111/tpj.13970. [DOI] [PubMed] [Google Scholar]
- Elvira C., Mano J.F., San Román J., Reis R.L. Starch-based biodegradable hydrogels with potential biomedical applications as drug delivery systems. Biomaterials. 2002;23:1955–1966. doi: 10.1016/S0142-9612(01)00322-2. [DOI] [PubMed] [Google Scholar]
- Fan X., Li Y., Lu Y., Zhang C., Li E., Li Q., Tao K., Yu W., Wang J., Chen Z. The interaction between amylose and amylopectin synthesis in rice endosperm grown at high temperature. Food Chem. 2019;301:125258. doi: 10.1016/j.foodchem.2019.125258. [DOI] [PubMed] [Google Scholar]
- Fang Y., You J., Xie K., Xie W., Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics. 2008;280:547–563. doi: 10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- Feng F., Qi W., Lv Y., Yan S., Xu L., Yang W., Yuan Y., Chen Y., Zhao H., Song R. OPAQUE11 is a central hub of the regulatory network for maize endosperm development and nutrient metabolism. Plant Cell. 2018;30:375–396. doi: 10.1105/tpc.17.00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero D.M.L., Piattoni C.V., Asencion Diez M.D., Rojas B.E., Hartman M.D., Ballicora M.A., Iglesias A.A. Phosphorylation of ADP-glucose pyrophosphorylase during wheat seeds development. Front. Plant Sci. 2020;11:1058. doi: 10.3389/fpls.2020.01058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu F., Xue H. Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 2010;154:927–938. doi: 10.1104/pp.110.159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton D.C., Stettler M., Mettler T., Vaughan C.K., Li J., Francisco P., Gil M., Reinhold H., Eicke S., Messerli G.l. β-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. Plant Cell. 2008;20:1040–1058. doi: 10.1105/tpc.107.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Gadlage M.J., Lafitte H.R., Lenderts B., Yang M., Schroder M., Farrell J., Snopek K., Peterson D., Feigenbutz L. Superior field performance of waxy corn engineered using CRISPR-Cas9. Nat. Biotechnol. 2020;38:579–581. doi: 10.1038/s41587-020-0444-0. [DOI] [PubMed] [Google Scholar]
- Gao Y., An K., Guo W., Chen Y., Zhang R., Zhang X., Chang S., Rossi V., Jin F., Cao X. The endosperm-specific transcription factor TaNAC019 regulates glutenin and starch accumulation and its elite allele improves wheat grain quality. Plant Cell. 2021;33:603–622. doi: 10.1093/plcell/koaa040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Zeng D., Cui X., Zhou Y., Yan M., Huang D., Li J., Qian Q. Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci. China C Life Sci. 2003;46:661–668. doi: 10.1360/03yc0099. [DOI] [PubMed] [Google Scholar]
- Gao Z., Zeng D., Cheng F., Tian Z., Guo L., Su Y., Yan M., Jiang H., Dong G., Huang Y. ALK, the key gene for gelatinization temperature, is a modifier gene for gel consistency in rice. J. Integr. Plant Biol. 2011;53:756–765. doi: 10.1111/j.1744-7909.2011.01065.x. [DOI] [PubMed] [Google Scholar]
- Gong B., Cheng L., Gilbert R.G., Li C. Distribution of short to medium amylose chains are major controllers of in vitro digestion of retrograded rice starch. Food Hydrocolloid. 2019;96:634–643. doi: 10.1016/j.foodhyd.2019.06.003. [DOI] [Google Scholar]
- Grimaud F., Rogniaux H., James M.G., Myers A.M., Planchot V. Proteome and phosphoproteome analysis of starch granule-associated proteins from normal maize and mutants affected in starch biosynthesis. J. Exp. Bot. 2008;59:3395–3406. doi: 10.1093/jxb/ern198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross R.A., Kalra B. Biodegradable polymers for the environment. Science. 2002;297:803–807. doi: 10.1126/science.297.5582.803. [DOI] [PubMed] [Google Scholar]
- Guo D., Ling X., Zhou X., Li X., Wang J., Qiu S., Yang Y., Zhang B. Evaluation of the quality of a high-resistant starch and low-glutelin rice (Oryza sativa L.) generated through CRISPR/Cas9-mediated targeted mutagenesis. J. Agric. Food Chem. 2020;68:9733–9742. doi: 10.1021/acs.jafc.0c02995. [DOI] [PubMed] [Google Scholar]
- Guo D., Hou Q., Zhang R., Lou H., Li Y., Zhang Y., You M., Xie C., Liang R., Li B. Over-expressing TaSPA-B reduces prolamin and starch accumulation in wheat (Triticum aestivum L.) grains. Int. J. Mol. Sci. 2020;21:3257. doi: 10.3390/ijms21093257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Liu Y., Li X., Yan Z., Xie Y., Xiong H., Zhao L., Gu J., Zhao S., Liu L. Novel mutant alleles of the starch synthesis gene TaSSIVb-D result in the reduction of starch granule number per chloroplast in wheat. BMC Genomics. 2017;18:358. doi: 10.1186/s12864-017-3724-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán C., Alvarez J.B. Wheat waxy proteins: polymorphism, molecular characterization and effects on starch properties. Theor. Appl. Genet. 2016;129:1–16. doi: 10.1007/s00122-015-2595-9. [DOI] [PubMed] [Google Scholar]
- Guzmán C., Caballero L., Martín L.M., Alvarez J.B. Waxy genes from spelt wheat: new alleles for modern wheat breeding and new phylogenetic inferences about the origin of this species. Ann. Bot. 2012;110:1161–1171. doi: 10.1093/aob/mcs201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakata M., Kuroda M., Miyashita T., Yamaguchi T., Kojima M., Sakakibara H., Mitsui T., Yamakawa H. Suppression of α-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol. J. 2012;10:1110–1117. doi: 10.1111/j.1467-7652.2012.00741.x. [DOI] [PubMed] [Google Scholar]
- Han Y., Sun F.-J., Rosales-Mendoza S., Korban S.S. Three orthologs in rice, Arabidopsis, and Populus encoding starch branching enzymes (SBEs) are different from other SBE gene families in plants. Gene. 2007;401:123–130. doi: 10.1016/j.gene.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Hawkins E., Chen J., Watson-Lazowski A., Ahn-Jarvis J., Barclay J.E., Fahy B., Hartley M., Warren F.J., Seung D. STARCH SYNTHASE 4 is required for normal starch granule initiation in amyloplasts of wheat endosperm. New Phytol. 2021;230:2371–2386. doi: 10.1111/nph.17342. [DOI] [PubMed] [Google Scholar]
- Hayashi M., Crofts N., Oitome N.F., Fujita N. Analyses of starch biosynthetic protein complexes and starch properties from developing mutant rice seeds with minimal starch synthase activities. BMC Plant Biol. 2018;18:59. doi: 10.1186/s12870-018-1270-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazard B., Zhang X., Naemeh M., Hamilton M.K., Rust B., Raybould H.E., Newman J.W., Martin R., Dubcovsky J. Mutations in durum wheat SBEII genes affect grain yield components, quality, and fermentation responses in rats. Crop Sci. 2015;55:2813–2825. doi: 10.2135/cropsci2015.03.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebeish A., Aly A.A., El-Shafei A.M., Zaghloul S. Innovative starch derivatives as textile auxiliaries for application in sizing, finishing and flocculation. Starch. 2008;60:97–109. doi: 10.1002/star.200700627. [DOI] [Google Scholar]
- Heinen M.M., Verhage B.A.J., Goldbohm R.A., van den Brandt P.A. Meat and fat intake and pancreatic cancer risk in The Netherlands Cohort Study. Int. J. Cancer. 2009;125:1118–1126. doi: 10.1002/ijc.24387. [DOI] [PubMed] [Google Scholar]
- Hennen-Bierwagen T.A., Liu F., Marsh R.S., Kim S., Gan Q., Tetlow I.J., Emes M.J., James M.G., Myers A.M. Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. Plant Physiol. 2008;146:1892–1908. doi: 10.1104/pp.108.116285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Li Y., Zhang J., Liu H., Tian M., Huang Y. Binding of ABI4 to a CACCG motif mediates the ABA-induced expression of the ZmSSI gene in maize (Zea mays L.) endosperm. J. Exp. Bot. 2012;63:5979–5989. doi: 10.1093/jxb/ers246. [DOI] [PubMed] [Google Scholar]
- Hu Y., Li Y., Weng J., Liu H., Yu G., Liu Y., Xiao Q., Huang H., Wang Y., wei B. Coordinated regulation of starch synthesis in maize endosperm by microRNAs and DNA methylation. Plant J. 2021;105:108–123. doi: 10.1111/tpj.15043. [DOI] [PubMed] [Google Scholar]
- Huang B., Hennen-Bierwagen T.A., Myers A.M. Functions of multiple genes encoding ADP-glucose pyrophosphorylase subunits in maize endosperm, embryo, and leaf. Plant Physiol. 2014;164:596–611. doi: 10.1104/pp.113.231605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Xie S., Xiao Q., Wei B., Zheng L., Wang Y., Cao Y., Zhang X., Long T., Li Y. Sucrose and ABA regulate starch biosynthesis in maize through a novel transcription factor, ZmEREB156. Sci. Rep. 2016;6:27590. doi: 10.1038/srep27590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Sreenivasulu N., Liu Q. Waxy editing: old meets new. Trends Plant Sci. 2020;25:963–966. doi: 10.1016/j.tplants.2020.07.009. [DOI] [PubMed] [Google Scholar]
- Huang L., Li Q., Zhang C., Chu R., Gu Z., Tan H., Zhao D., Fan X., Liu Q. Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol. J. 2020;18:2164–2166. doi: 10.1111/pbi.13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Gu Z., Chen Z., Yu J., Chu R., Tan H., Zhao D., Fan X., Zhang C., Li Q. Improving rice eating and cooking quality by coordinated expression of the major starch synthesis-related genes, SSII and Wx, in endosperm. Plant Mol. Biol. 2021;106:419–432. doi: 10.1007/s11103-021-01162-8. [DOI] [PubMed] [Google Scholar]
- Hwang S.K., Koper K., Satoh H., Okita T.W. Rice endosperm starch phosphorylase (Pho1) assembles with disproportionating enzyme (Dpe1) to form a protein complex that enhances synthesis of malto-oligosaccharides. J. Biol. Chem. 2016;291:19994–20007. doi: 10.1074/jbc.M116.735449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S.K., Singh S., Maharana J., Kalita S., Tuncel A., Rath T., Panda D., Modi M.K., Okita T.W. Mechanism underlying heat stability of the rice endosperm cytosolic ADP-glucose pyrophosphorylase. Front. Plant Sci. 2019;10:70. doi: 10.3389/fpls.2019.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi H., Ito H., Shimada T., Kang D.-J., Hamada S. A novel rice dull gene, LowAC1, encodes an RNA recognition motif protein affecting Waxyb pre-mRNA splicing. Plant Physiol. Bioch. 2021;162:100–109. doi: 10.1016/j.plaphy.2021.02.035. [DOI] [PubMed] [Google Scholar]
- Itoh K., Ozaki H., Okada K., Hori H., Takeda Y., Mitsui T. Introduction of Wx transgene into rice wx mutants leads to both high- and low-amylose rice. Plant Cell Physiol. 2003;44:473–480. doi: 10.1093/pcp/pcg068. [DOI] [PubMed] [Google Scholar]
- James M.G., Robertson D.S., Myers A.M. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell. 1995;7:417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.S., Ryoo N., Hahn T.R., Walia H., Nakamura Y. Starch biosynthesis in cereal endosperm. Plant Physiol. Bioch. 2010;48:383–392. doi: 10.1016/j.plaphy.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Johnson P.E., Patron N.J., Bottrill A.R., Dinges J.R., Fahy B.F., Parker M.L., Waite D.N., Denyer K. A low-starch barley mutant, Risø 16, lacking the cytosolic small subunit of ADP-glucose pyrophosphorylase, reveals the importance of the cytosolic isoform and the identity of the plastidial small subunit. Plant Physiol. 2003;131:684–696. doi: 10.1104/pp.013094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukanti A.K., Pautong P.A., Liu Q., Sreenivasulu N. Low glycemic index rice—a desired trait in starchy staples. Trends Food Sci. Technol. 2020;106:132–149. doi: 10.1016/j.tifs.2020.10.006. [DOI] [Google Scholar]
- Kang G., Xu W., Liu G., Peng X., Guo T. Comprehensive analysis of the transcription of starch synthesis genes and the transcription factor RSR1 in wheat (Triticum aestivum) endosperm. Genome. 2013;56:115–122. doi: 10.1139/gen-2012-0146. [DOI] [PubMed] [Google Scholar]
- Kang G., Li S., Zhang M., Peng H., Wang C., Zhu Y., Guo T. Molecular cloning and expression analysis of the starch-branching enzyme III gene from common wheat (Triticum aestivum) Biochem. Genet. 2013;51:377–386. doi: 10.1007/s10528-013-9570-4. [DOI] [PubMed] [Google Scholar]
- Kapelko-Żeberska M., Zięba T., Singh A.V. Surface Modification of Biopolymers. Wiley; Hoboken, NJ: 2015. Physically and chemically modified starches in food and non-food industries; pp. 173–193. [DOI] [Google Scholar]
- Kaur J., Kaur G., Sharma S., Jeet K. Cereal starch nanoparticles—a prospective food additive: a review. Crit. Rev. Food Sci. 2018;58:1097–1107. doi: 10.1080/10408398.2016.1238339. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T., Takaiwa F. Differences in transcriptional regulatory mechanisms functioning for free lysine content and seed storage protein accumulation in rice grain. Plant Cell Physiol. 2010;51:1964–1974. doi: 10.1093/pcp/pcq164. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T., Yamamoto M.P., Touno S.M., Yasuda H., Takaiwa F. Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J. 2009;59:908–920. doi: 10.1111/j.1365-313X.2009.03925.x. [DOI] [PubMed] [Google Scholar]
- Kirchberger S., Leroch M., Huynen M.A., Wahl M., Neuhaus H.E., Tjaden J. Molecular and biochemical analysis of the plastidic ADP-glucose transporter (ZmBT1) from Zea mays. J. Biol. Chem. 2007;282:22481–22491. doi: 10.1074/jbc.M702484200. [DOI] [PubMed] [Google Scholar]
- Kötting O., Kossmann J., Zeeman S.C., Lloyd J.R. Regulation of starch metabolism: the age of enlightenment? Curr. Opin. Plant Biol. 2010;13:320–328. doi: 10.1016/j.pbi.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Lal S., Choi J.-H., Hannah L.C. The AG dinucleotide terminating introns is important but not always required for pre-mRNA splicing in the maize endosperm. Plant Physiol. 1999;120:65–72. doi: 10.1104/pp.120.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Choi M.S., Lee G., Jang S., Yoon M.R., Kim B., Piao R., Woo M.O., Chin J.H., Koh H.J. Sugary endosperm is modulated by starch branching enzyme IIa in rice (Oryza sativa L.) Rice. 2017;10:33. doi: 10.1186/s12284-017-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Yue Y., Chen H., Qi W., Song R. The ZmbZIP22 transcription factor regulates 27-kD γ-zein gene transcription during maize endosperm development. Plant Cell. 2018;30:2402–2424. doi: 10.1105/tpc.18.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wu A., Yu W., Hu Y., Li E., Zhang C., Liu Q. Parameterizing starch chain-length distributions for structure-property relations. Carbohyd. Polym. 2020;241:116390. doi: 10.1016/j.carbpol.2020.116390. [DOI] [PubMed] [Google Scholar]
- Li C., Qiao Z., Qi W., Wang Q., Yuan Y., Yang X., Tang Y., Mei B., Lv Y., Zhao H. Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of Opaque2 in maize. Plant Cell. 2015;27:532–545. doi: 10.1105/tpc.114.134858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Gidley M.J., Dhital S. High-amylose starches to bridge the “Fiber Gap”: development, structure, and nutritional functionality. Compr. Rev. Food Sci. F. 2019;18:362–379. doi: 10.1111/1541-4337.12416. [DOI] [PubMed] [Google Scholar]
- Li H., Prakash S., Nicholson T.M., Fitzgerald M.A., Gilbert R.G. Instrumental measurement of cooked rice texture by dynamic rheological testing and its relation to the fine structure of rice starch. Carbohydr. Polym. 2016;146:253–263. doi: 10.1016/j.carbpol.2016.03.045. [DOI] [PubMed] [Google Scholar]
- Li H., Fitzgerald M.A., Prakash S., Nicholson T.M., Gilbert R.G. The molecular structural features controlling stickiness in cooked rice, a major palatability determinant. Sci. Rep. 2017;7:43713. doi: 10.1038/srep43713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Jiao G., Sun Y., Chen J., Zhong Y., Yan L., Jiang D., Ma Y., Xia L. Modification of starch composition, structure and properties through editing of TaSBEIIa in both winter and spring wheat varieties by CRISPR/Cas9. Plant Biotechnol. J. 2020;19:937–951. doi: 10.1111/pbi.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.F., Huang L.C., Chu R., Li J., Jiang M.Y., Zhang C.Q., Fan X.L., Yu H.X., Gu M.H., Liu Q.Q. Down-regulation of SSSII-2 gene expression results in novel low-amylose rice with soft, transparent grains. J. Agric. Food Chem. 2018;66:9750–9760. doi: 10.1021/acs.jafc.8b02913. [DOI] [PubMed] [Google Scholar]
- Li S., Wei X., Ren Y., Qiu J., Jiao G., Guo X., Tang S., Wan J., Hu P. OsBT1 encodes an ADP-glucose transporter involved in starch synthesis and compound granule formation in rice endosperm. Sci. Rep. 2017;7:40124. doi: 10.1038/srep40124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Huang B., Zhang M., Zhang X., Rivenbark J., Lappe R.L., James M.G., Myers A.M., Hennen-Bierwagen T.A. Functional interactions between starch synthase III and isoamylase-type starch-debranching enzyme in maize endosperm. Plant Physiol. 2011;158:679–692. doi: 10.1104/pp.111.189704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linebarger C.R.L., Boehlein S.K., Sewell A.K., Shaw J., Hannah L.C. Heat stability of maize endosperm ADP-glucose pyrophosphorylase is enhanced by insertion of a cysteine in the N terminus of the small subunit. Plant Physiol. 2005;139:1625–1634. doi: 10.1104/pp.105.067637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.R., Huang W.X., Cai X.L. Oligomerization of rice granule-bound starch synthase 1 modulates its activity regulation. Plant Sci. 2013;210:141–150. doi: 10.1016/j.plantsci.2013.05.019. [DOI] [PubMed] [Google Scholar]
- Liu D., Wang W., Cai X. Modulation of amylose content by structure-based modification of OsGBSS1 activity in rice (Oryza sativa L.) Plant Biotechnol. J. 2015;12:1297–1307. doi: 10.1111/pbi.12228. [DOI] [PubMed] [Google Scholar]
- Liu F., Ahmed Z., Lee E.A., Donner E., Liu Q., Ahmed R., Morell M.K., Emes M.J., Tetlow I.J. Allelic variants of the amylose extender mutation of maize demonstrate phenotypic variation in starch structure resulting from modified protein-protein interactions. J. Exp. Bot. 2011;63:1167–1183. doi: 10.1093/jxb/err341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Romanova N., Lee E.A., Ahmed R., Evans M., Gilbert E.P., Morell M.K., Emes M.J., Tetlow I.J. Glucan affinity of starch synthase IIa determines binding of starch synthase I and starch-branching enzyme IIb to starch granules. Biochem. J. 2012;448:373–387. doi: 10.1042/bj20120573. [DOI] [PubMed] [Google Scholar]
- Liu G., Gu Z., Hong Y., Cheng L., Li C. Electrospun starch nanofibers: recent advances, challenges, and strategies for potential pharmaceutical applications. J. Control Release. 2017;252:95–107. doi: 10.1016/j.jconrel.2017.03.016. [DOI] [PubMed] [Google Scholar]
- Liu H., Yu G., Wei B., Wang Y., Zhang J., Hu Y., Liu Y., Yu G., Zhang H., Huang Y. Identification and phylogenetic analysis of a novel starch synthase in maize. Front. Plant Sci. 2015;6:1013. doi: 10.3389/fpls.2015.01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hou J., Wang X., Li T., Majeed U., Hao C., Zhang X. The NAC transcription factor NAC019-A1 is a negative regulator of starch synthesis in wheat developing endosperm. J. Exp. Bot. 2020;71:5794–5807. doi: 10.1093/jxb/eraa333. [DOI] [PubMed] [Google Scholar]
- Luo M., Shi Y., Yang Y., Zhao Y., Zhang Y., Shi Y., Kong M., Li C., Feng Z., Fan Y. Sequence polymorphism of the waxy gene in waxy maize accessions and characterization of a new waxy allele. Sci. Rep. 2020;10:15851. doi: 10.1038/s41598-020-72764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Jiang Q., Zhang X., Lan X., Pu Z., Wei Y., Liu C., Lu Z., Zheng Y. Structure and expression of barley starch phosphorylase genes. Planta. 2013;238:1081–1093. doi: 10.1007/s00425-013-1953-6. [DOI] [PubMed] [Google Scholar]
- Makhmoudova A., Williams D., Brewer D., Massey S., Patterson J., Silva A., Vassall K.A., Liu F., Subedi S., Harauz G. Identification of multiple phosphorylation sites on maize endosperm starch branching enzyme IIb, a key enzyme in amylopectin biosynthesis. J. Biol. Chem. 2014;289:9233–9246. doi: 10.1074/jbc.M114.551093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinova I., Qasim H.M., Brust H., Fettke J. Parameters of starch granule genesis in chloroplasts of Arabidopsis thaliana. Front. Plant Sci. 2018;9:761. doi: 10.3389/fpls.2018.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M.M., Li C., Okoniewska M., Mukherjee I., Vellucci D., Hamaker B. Slowly digestible starch in fully gelatinized material is structurally driven by molecular size and A and B1 chain lengths. Carbohyd. Polym. 2018;197:531–539. doi: 10.1016/j.carbpol.2018.06.021. [DOI] [PubMed] [Google Scholar]
- Masina N., Choonara Y.E., Kumar P., du Toit L.C., Govender M., Indermun S., Pillay V. A review of the chemical modification techniques of starch. Carbohyd. Polym. 2017;157:1226–1236. doi: 10.1016/j.carbpol.2016.09.094. [DOI] [PubMed] [Google Scholar]
- Matsushima R., Maekawa M., Sakamoto W. Geometrical formation of compound starch grains in rice implements voronoi diagram. Plant Cell Physiol. 2015;56:2150–2157. doi: 10.1093/pcp/pcv123. [DOI] [PubMed] [Google Scholar]
- Matsushima R., Yamashita J., Kariyama S., Enomoto T., Sakamoto W. A phylogenetic re-evaluation of morphological variations of starch grains among poaceae species. J. Appl. Glycosci. 2013;60:37–44. doi: 10.5458/jag.jag.JAG-2012_006. [DOI] [Google Scholar]
- Miura S., Crofts N., Saito Y., Hosaka Y., Oitome N.F., Watanabe T., Kumamaru T., Fujita N. Starch synthase IIa-deficient mutant rice line produces endosperm starch with lower gelatinization temperature than japonica rice cultivars. Front. Plant Sci. 2018;9:645. doi: 10.3389/fpls.2018.00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A.M., Morell M.K., James M.G., Ball S.G. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 2000;122:989–998. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A.M., James M.G., Lin Q., Yi G., Stinard P.S., Hennen-Bierwagen T.A., Becraft P.W. Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell. 2011;23:2331–2347. doi: 10.1105/tpc.111.087205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Ono M., Utsumi C., Steup M. Functional interaction between plastidial starch phosphorylase and starch branching enzymes from rice during the synthesis of branched maltodextrins. Plant Cell Physiol. 2012;53:869–878. doi: 10.1093/pcp/pcs030. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Francisco P.B., Hosaka Y., Sato A., Sawada T., Kubo A., Fujita N. Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol. Biol. 2005;58:213–227. doi: 10.1007/s11103-005-6507-2. [DOI] [PubMed] [Google Scholar]
- Nakata M., Miyashita T., Kimura R., Nakata Y., Takagi H., Kuroda M., Yamaguchi T., Umemoto T., Yamakawa H. MutMapPlus identified novel mutant alleles of a rice starch branching enzyme IIb gene for fine-tuning of cooked rice texture. Plant Biotechnol. J. 2018;16:111–123. doi: 10.1111/pbi.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B., Zhang Z., Zhang J., Zhou Y., Chen C. The rice LEC1-like transcription factor OsNF-YB9 interacts with SPK, an endosperm specific sucrose synthase protein kinase, and functions in seed development. Plant J. 2021;106:1233–1246. doi: 10.1111/tpj.15230. [DOI] [PubMed] [Google Scholar]
- Niu B., Deng H., Li T., Sharma S., Yun Q., Li Q., E Z., Chen C. OsbZIP76 interacts with OsNF-YBs and regulates endosperm cellularization in rice (Oryza sativa) J. Integr. Plant Biol. 2020;62:1983–1996. doi: 10.1111/jipb.12989. [DOI] [PubMed] [Google Scholar]
- Noda T., Takahata Y., Sato T., Suda I., Morishita T., Ishiguro K., Yamakawa O. Relationships between chain length distribution of amylopectin and gelatinization properties within the same botanical origin for sweet potato and buckwheat. Carbohyd. Polym. 1998;37:153–158. doi: 10.1016/S0144-8617(98)00047-2. [DOI] [Google Scholar]
- Ohdan T., Francisco P.B., Jr., Sawada T., Hirose T., Terao T., Satoh H., Nakamura Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 2005;56:3229–3244. doi: 10.1093/jxb/eri292. [DOI] [PubMed] [Google Scholar]
- Onusseit H. Starch in industrial adhesives: new developments. Ind. Crop Prod. 1992;1:141–146. doi: 10.1016/0926-6690(92)90012-K. [DOI] [Google Scholar]
- Orman-Ligeza B., Borrill P., Chia T., Chirico M., Doležel J., Drea S., Karafiátová M., Schatlowski N., Solomon C.U., Steuernagel B. LYS3 encodes a prolamin-box-binding transcription factor that controls embryo growth in barley and wheat. J. Cereal Sci. 2020;93:102965. doi: 10.1016/j.jcs.2020.102965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parween S., Anonuevo J.J., Butardo V.M., Jr., Misra G., Anacleto R., Llorente C., Kosik O., Romero M.V., Bandonill E.H., Mendioro M.S. Balancing the double-edged sword effect of increased resistant starch content and its impact on rice texture: its genetics and molecular physiological mechanisms. Plant Biotechnol. J. 2020;18:1763–1777. doi: 10.1111/pbi.13339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C., Wang Y., Liu F., Ren Y., Zhou K., Lv J., Zheng M., Zhao S., Zhang L., Wang C. FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J. 2014;77:917–930. doi: 10.1111/tpj.12444. [DOI] [PubMed] [Google Scholar]
- Peng X., Wang Q., Wang Y., Cheng B., Zhao Y., Zhu S. A maize NAC transcription factor, ZmNAC34, negatively regulates starch synthesis in rice. Plant Cell Rep. 2019;38:1473–1484. doi: 10.1007/s00299-019-02458-2. [DOI] [PubMed] [Google Scholar]
- Pfister B., Zeeman S.C. Formation of starch in plant cells. Cell. Mol. Life Sci. 2016;73:2781–2807. doi: 10.1007/s00018-016-2250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Zhang G., Hu B., Wu J., Chen W., Ren Z., Liu Y., Xie J., Yuan H., Tu B. Leaf-derived ABA regulates rice seed development via a transporter-mediated and temperature-sensitive mechanism. Sci. Adv. 2021;7:eabc8873. doi: 10.1126/sciadv.abc8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk V.V., Borisjuk L., Sreenivasulu N., Merx K., Mock H.-P., Rolletschek H., Wobus U., Weschke W. Spatiotemporal profiling of starch biosynthesis and degradation in the developing barley grain. Plant Physiol. 2009;150:190–204. doi: 10.1104/pp.108.133520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina A., Kosar-Hashemi B., Ling S., Li Z., Rahman S., Morell M. Control of starch branching in barley defined through differential RNAi suppression of starch branching enzyme IIa and IIb. J. Exp. Bot. 2010;61:1469–1482. doi: 10.1093/jxb/erq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina A., Bird A., Topping D., Bowden S., Freeman J., Barsby T., Kosar-Hashemi B., Li Z., Rahman S., Morell M. High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc. Natl. Acad. Sci. U S A. 2006;103:3546–3551. doi: 10.1073/pnas.0510737103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina A., Berbezy P., Kosar-Hashemi B., Li S., Cmiel M., Larroque O., Bird A.R., Swain S.M., Cavanagh C., Jobling S.A. A genetic strategy generating wheat with very high amylose content. Plant Biotechnol. J. 2015;13:1276–1286. doi: 10.1111/pbi.12345. [DOI] [PubMed] [Google Scholar]
- Ren Y., Huang Z., Jiang H., Wang Z., Wu F., Xiong Y., Yao J. A heat stress responsive NAC transcription factor heterodimer plays key roles in rice grain filling. J. Exp. Bot. 2021;72:2947–2964. doi: 10.1093/jxb/erab027. [DOI] [PubMed] [Google Scholar]
- Ribeiro C., Hennen-Bierwagen T.A., Myers A.M., Cline K., Settles A.M. Engineering 6-phosphogluconate dehydrogenase improves grain yield in heat-stressed maize. Proc. Natl. Acad. Sci. U S A. 2020;117:33177–33185. doi: 10.1073/pnas.2010179117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Tanaka T., Sato K., Vrinten P., Nakamura T. A single nucleotide polymorphism in the “Fra” gene results in fractured starch granules in barley. Theor. Appl. Genet. 2018;131:353–364. doi: 10.1007/s00122-017-3006-1. [DOI] [PubMed] [Google Scholar]
- Saripalli G., Gupta P.K. AGPase: its role in crop productivity with emphasis on heat tolerance in cereals. Theor. Appl. Genet. 2015;128:1893–1916. doi: 10.1007/s00122-015-2565-2. [DOI] [PubMed] [Google Scholar]
- Satoh H., Shibahara K., Tokunaga T., Nishi A., Tasaki M., Hwang S.-K., Okita T.W., Kaneko N., Fujita N., Yoshida M. Mutation of the plastidial α-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell. 2008;20:1833–1849. doi: 10.1105/tpc.107.054007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada T., Itoh M., Nakamura Y. Contributions of three starch branching enzyme isozymes to the fine structure of amylopectin in rice endosperm. Front. Plant Sci. 2018;9:1536. doi: 10.3389/fpls.2018.01536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Schippers J.H.M., Mieulet D., Watanabe M., Hoefgen R., Guiderdoni E., Mueller-Roeber B. SALT-RESPONSIVE ERF1 is a negative regulator of grain filling and gibberellin-mediated seedling establishment in rice. Mol. Plant. 2014;7:404–421. doi: 10.1093/mp/sst131. [DOI] [PubMed] [Google Scholar]
- Schönhofen A., Hazard B., Zhang X., Dubcovsky J. Registration of common wheat germplasm with mutations in SBEII genes conferring increased grain amylose and resistant starch content. J. Plant Regist. 2016;10:200–205. doi: 10.3198/jpr2015.10.0066crg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J.A., Juvik J.A. Current models for starch synthesis and the sugary enhancer1 (se1) mutation in Zea mays. Plant Physiol. Bioch. 2004;42:457–464. doi: 10.1016/j.plaphy.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Seung D. Amylose in starch: towards an understanding of biosynthesis, structure and function. New Phytol. 2020;228:1490–1504. doi: 10.1111/nph.16858. [DOI] [PubMed] [Google Scholar]
- Seung D., Smith A.M. Starch granule initiation and morphogenesis—progress in Arabidopsis and cereals. J. Exp. Bot. 2018;70:771–784. doi: 10.1093/jxb/ery412. [DOI] [PubMed] [Google Scholar]
- Seung D., Schreier T.B., Bürgy L., Eicke S., Zeeman S.C. Two plastidial coiled-coil proteins are essential for normal starch granule initiation in arabidopsis. Plant Cell. 2018;30:1523–1542. doi: 10.1105/tpc.18.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D., Soyk S., Coiro M., Maier B.A., Eicke S., Zeeman S.C. PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol. 2015;13:e1002080. doi: 10.1371/journal.pbio.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D., Boudet J., Monroe J., Schreier T.B., David L.C., Abt M., Lu K.-J., Zanella M., Zeeman S.C. Homologs of PROTEIN TARGETING TO STARCH control starch granule initiation in arabidopsis leaves. Plant Cell. 2017;29:1657–1677. doi: 10.1105/tpc.17.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Aguado R., Murtinho D., Valente A.J.M., Mendes De Sousa A.P., Ferreira P.J.T. A review on cationic starch and nanocellulose as paper coating components. Int. J. Biol. Macromol. 2020;162:578–598. doi: 10.1016/j.ijbiomac.2020.06.131. [DOI] [PubMed] [Google Scholar]
- Shen J., Song Z., Qian X., Ni Y. Carbohydrate-based fillers and pigments for papermaking: a review. Carbohydr. Polym. 2011;85:17–22. doi: 10.1016/j.carbpol.2011.02.026. [DOI] [Google Scholar]
- Shimada H., Tada Y., Kawasaki T., Fujimura T. Antisense regulation of the rice Waxy gene expression using a PCR-amplified fragment of the rice genome reduces the amylose content in grain starch. Theor. Appl. Genet. 1993;86:665–672. doi: 10.1007/BF00222654. [DOI] [PubMed] [Google Scholar]
- Shimbata T., Ai Y., Fujita M., Inokuma T., Vrinten P., Sunohara A., Saito M., Takiya T., Jane J.l., Nakamura T. Effects of homoeologous wheat starch synthase IIa genes on starch properties. J. Agric. Food Chem. 2012;60:12004–12010. doi: 10.1021/jf303623e. [DOI] [PubMed] [Google Scholar]
- Shure M., Wessler S., Fedoroff N. Molecular identification and isolation of the Waxy locus in maize. Cell. 1983;35:225–233. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- Soliman A., Ayele B., Daayf F. Biochemical and molecular characterization of barley plastidial ADP-glucose transporter (HvBT1) PLoS One. 2014;9:e98524. doi: 10.1371/journal.pone.0098524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Luo G., Shen L., Yu K., Yang W., Li X., Sun J., Zhan K., Cui D., Liu D. TubZIP28, a novel bZIP family transcription factor from Triticum urartu, and TabZIP28, its homologue from Triticum aestivum, enhance starch synthesis in wheat. New Phytol. 2020;226:1384–1398. doi: 10.1111/nph.16435. [DOI] [PubMed] [Google Scholar]
- Subasinghe R.M., Liu F., Polack U.C., Lee E.A., Emes M.J., Tetlow I.J. Multimeric states of starch phosphorylase determine protein–protein interactions with starch biosynthetic enzymes in amyloplasts. Plant Physiol. Bioch. 2014;83:168–179. doi: 10.1016/j.plaphy.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Sun C., Palmqvist S., Olsson H., Borén M., Ahlandsberg S., Jansson C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003;15:2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Jiao G., Liu Z., Zhang X., Li J., Guo X., Du W., Du J., Francis F., Zhao Y. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 2017;8:298. doi: 10.3389/fpls.2017.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szydlowski N., Ragel P., Raynaud S., Lucas M.M., Roldán I., Montero M., Muñoz F.J., Ovecka M., Bahaji A., Planchot V. Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant Cell. 2009;21:2443–2457. doi: 10.1105/tpc.109.066522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai L., Wang H.J., Xu X.J., Sun W.H., Ju L., Liu W.T., Li W.Q., Sun J., Chen K.M. Cereal pre-harvest sprouting: a global agricultural disaster regulated by complex genetic and biochemical mechanisms. J. Exp. Bot. 2021;72:2857–2876. doi: 10.1093/jxb/erab024. [DOI] [PubMed] [Google Scholar]
- Takemoto-Kuno Y., Mitsueda H., Suzuki K., Hirabayashi H., Ideta O., Aoki N., Umemoto T., Ishii T., Ando I., Kato H. qAC2, a novel QTL that interacts with Wx and controls the low amylose content in rice (Oryza sativa L.) Theor. Appl. Genet. 2015;128:563–573. doi: 10.1007/s00122-014-2432-6. [DOI] [PubMed] [Google Scholar]
- Tan Y., Yu S., Xing Y., Xu C., Zhang Q., Li J. The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor. Appl. Genet. 1999;99:642–648. doi: 10.1007/s001220051279. [DOI] [PubMed] [Google Scholar]
- Terada R., Urawa H., Inagaki Y., Tsugane K., Iida S. Efficient gene targeting by homologous recombination in rice. Nat. Biotechnol. 2002;20:1030–1034. doi: 10.1038/nbt737. [DOI] [PubMed] [Google Scholar]
- Terada R., Nakajima M., Isshiki M., Okagaki R.J., Wessler S.R., Shimamoto K. Antisense Waxy genes with highly active promoters effectively suppress Waxy gene expression in transgenic rice. Plant Cell Physiol. 2000;41:881–888. doi: 10.1093/pcp/pcd008. [DOI] [PubMed] [Google Scholar]
- Tetlow I.J., Liu F., Emes M.J. In: Starch: Metabolism and Structure. Nakamura Y., editor. Springer Japan; Tokyo: 2015. Protein-protein interactions during starch biosynthesis; pp. 291–313. [DOI] [Google Scholar]
- Tetlow I.J., Wait R., Lu Z., Akkasaeng R., Bowsher C.G., Esposito S., Kosar-Hashemi B., Morell M.K., Emes M.J. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein–protein interactions. Plant Cell. 2004;16:694–708. doi: 10.1105/tpc.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow I.J., Beisel K.G., Cameron S., Makhmoudova A., Liu F., Bresolin N.S., Wait R., Morell M.K., Emes M.J. Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol. 2008;146:1878–1891. doi: 10.1104/pp.108.116244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann M., Santelia D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017;214:943–951. doi: 10.1111/nph.14491. [DOI] [PubMed] [Google Scholar]
- Tian Z., Qian Q., Liu Q., Yan M., Liu X., Yan C., Liu G., Gao Z., Tang S., Zeng D. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. U S A. 2009;106:21760–21765. doi: 10.1073/pnas.0912396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyosawa Y., Kawagoe Y., Matsushima R., Crofts N., Ogawa M., Fukuda M., Kumamaru T., Okazaki Y., Kusano M., Saito K. Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm. Plant Physiol. 2016;170:1255–1270. doi: 10.1104/pp.15.01232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi Y., Utsumi C., Sawada T., Fujita N., Nakamura Y. Functional diversity of isoamylase oligomers: the ISA1 homo-oligomer is essential for amylopectin biosynthesis in rice endosperm. Plant Physiol. 2011;156:61–77. doi: 10.1104/pp.111.173435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley J.W., Shen Z., Sartor R., Wu K.J., Osborn J., Smith L.G., Briggs S.P. Reconstruction of protein networks from an atlas of maize seed proteotypes. Proc. Natl. Acad. Sci. U S A. 2013;110:E4808–E4817. doi: 10.1073/pnas.1319113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Wang J., Zhu X., Hao W., Wang L., Li Q., Zhang L., He W., Lu B., Lin H. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 2008;40:1370–1374. doi: 10.1038/ng.220. [DOI] [PubMed] [Google Scholar]
- Wang J., Xu H., Zhu Y., Liu Q., Cai X. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 2013;64:3453–3466. doi: 10.1093/jxb/ert187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chen Z., Zhang Q., Meng S., Wei C. The NAC transcription factors OsNAC20 and OsNAC26 regulate starch and storage protein synthesis. Plant Physiol. 2020;184:1775–1791. doi: 10.1104/pp.20.00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Hu P., Lin L., Chen Z., Liu Q., Wei C. Gradually decreasing starch branching enzyme expression is responsible for the formation of heterogeneous starch granules. Plant Physiol. 2018;176:582–595. doi: 10.1104/pp.17.01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Li C., Copeland L., Niu Q., Wang S. Starch retrogradation: a comprehensive review. Compr. Rev. Food Sci. F. 2015;14:568–585. doi: 10.1111/1541-4337.12143. [DOI] [Google Scholar]
- Wang W., Wei X., Jiao G., Chen W., Wu Y., Sheng Z., Hu S., Xie L., Wang J., Tang S. GBSS-BINDING PROTEIN, encoding a CBM48 domain-containing protein, affects rice quality and yield. J. Integr.Plant Biol. 2020;62:948–966. doi: 10.1111/jipb.12866. [DOI] [PubMed] [Google Scholar]
- Wang X., Huang L., Zhang C., Deng Y., Xie P., Liu L., Cheng J. Research advances in chemical modifications of starch for hydrophobicity and its applications: a review. Carbohyd. Polym. 2020;240:116292. doi: 10.1016/j.carbpol.2020.116292. [DOI] [PubMed] [Google Scholar]
- Wang Y., Hou J., Liu H., Li T., Wang K., Hao C., Liu H., Zhang X. TaBT1, affecting starch synthesis and thousand kernel weight, underwent strong selection during wheat improvement. J. Exp. Bot. 2019;70:1497–1511. doi: 10.1093/jxb/erz032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wu Z., Xing Y., Zheng F., Guo X., Zhang W., Hong M. Nucleotide sequence of rice Waxy gene. Nucleic Acids Res. 1990;18:5898. doi: 10.1093/nar/18.19.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters D.L.E., Henry R.J., Reinke R.F., Fitzgerald M.A. Gelatinization temperature of rice explained by polymorphisms in starch synthase. Plant Biotechnol. J. 2006;4:115–122. doi: 10.1111/j.1467-7652.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- Wei C., Qin F., Zhu L., Zhou W., Chen Y., Wang Y., Gu M., Liu Q. Microstructure and ultrastructure of high-amylose rice resistant starch granules modified by antisense RNA inhibition of starch branching enzyme. J. Agric. Food Chem. 2010;58:1224–1232. doi: 10.1021/jf9031316. [DOI] [PubMed] [Google Scholar]
- Wei X., Jiao G., Lin H., Sheng Z., Shao G., Xie L., Tang S., Xu Q., Hu P. GRAIN INCOMPLETE FILLING 2 regulates grain filling and starch synthesis during rice caryopsis development. J. Integr. Plant Biol. 2017;59:134–153. doi: 10.1111/jipb.12510. [DOI] [PubMed] [Google Scholar]
- Wu J., Chen L., Chen M., Zhou W., Dong Q., Jiang H., Cheng B. The DOF-domain transcription factor ZmDOF36 positively regulates starch synthesis in transgenic maize. Front. Plant Sci. 2019;10:465. doi: 10.3389/fpls.2019.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Zhu D., Wang R., Cui Y., Yan Y. Crop resistant starch and genetic improvement: a review of recent advances. Theor. Appl. Genet. 2018;131:2495–2511. doi: 10.1007/s00122-018-3221-4. [DOI] [PubMed] [Google Scholar]
- Xiao Q., Wang Y., Du J., Li H., Wei B., Wang Y., Li Y., Yu G., Liu H., Zhang J. ZmMYB14 is an important transcription factor involved in the regulation of the activity of the ZmBT1 promoter in starch biosynthesis in maize. FEBS J. 2017;284:3079–3099. doi: 10.1111/febs.14179. [DOI] [PubMed] [Google Scholar]
- Xiao Q., Wang Y., Li H., Zhang C., Wei B., Wang Y., Huang H., Li Y., Yu G., Liu H. Transcription factor ZmNAC126 plays an important role in transcriptional regulation of maize starch synthesis-related genes. Crop J. 2020 doi: 10.1016/j.cj.2020.04.014. [DOI] [Google Scholar]
- Xiong Y., Ren Y., Li W., Wu F., Yang W., Huang X., Yao J. NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice. J. Exp. Bot. 2019;70:3765–3780. doi: 10.1093/jxb/erz168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Li X., Zhang H., Wang L., Zhu Z., Gao J., Li C., Zhu Y. High temperature inhibits the accumulation of storage materials by inducing alternative splicing of OsbZIP58 during filling stage in rice. Plant Cell Environ. 2020;43:1879–1896. doi: 10.1111/pce.13779. [DOI] [PubMed] [Google Scholar]
- Xu Y., Lin Q., Li X., Wang F., Chen Z., Wang J., Li W., Fan F., Tao Y., Jiang Y. Fine-tuning the amylose content of rice by precise base editing of the Wx gene. Plant Biotechnol. J. 2021;19:11–13. doi: 10.1111/pbi.13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H., Hirose T., Kuroda M., Yamaguchi T. Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 2007;144:258–277. doi: 10.1104/pp.107.098665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M.P., Onodera Y., Touno S.M., Takaiwa F. Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol. 2006;141:1694–1707. doi: 10.1104/pp.106.082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Pan X., Jiang H., Wu G. Comparison of the starch synthesis genes between maize and rice: copies, chromosome location and expression divergence. Theor. Appl. Genet. 2009;119:815–825. doi: 10.1007/s00122-009-1091-5. [DOI] [PubMed] [Google Scholar]
- Yang D., Wu L., Hwang Y.-S., Chen L., Huang N. Expression of the REB transcriptional activator in rice grains improves the yield of recombinant proteins whose genes are controlled by a Reb-responsive promoter. Proc. Natl. Acad. Sci. U S A. 2001;98:11438–11443. doi: 10.1073/pnas.201411298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Sun C., Bai J., Luo Z., Shi B., Zhang J., Yan W., Piao Z. A putative gene sbe3-rs for resistant starch mutated from SBE3 for starch branching enzyme in rice (Oryza sativa L.) PLoS One. 2012;7:e43026. doi: 10.1371/journal.pone.0043026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Guo L., Ji C., Wang H., Wang J., Zheng X., Xiao Q., Wu Y. The B3 domain-containing transcription factor ZmABI19 coordinates expression of key factors required for maize seed development and grain filling. Plant Cell. 2020;33:104–128. doi: 10.1093/plcell/koaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Lu Z., Xiong Y., Yao J. Genome-wide identification and co-expression network analysis of the OsNF-Y gene family in rice. Crop J. 2017;5:21–31. doi: 10.1016/j.cj.2016.06.014. [DOI] [Google Scholar]
- Yin L., Xue H. The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development. Plant Cell. 2012;24:1049–1065. doi: 10.1105/tpc.111.094854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Li H., Zou W., Tao K., Zhu J., Gilbert R.G. Using starch molecular fine structure to understand biosynthesis-structure-property relations. Trends Food Sci. Technol. 2019;86:530–536. doi: 10.1016/j.tifs.2018.08.003. [DOI] [Google Scholar]
- Zaman S.A., Sarbini S.R. The potential of resistant starch as a prebiotic. Crit. Rev. Biotechnol. 2016;36:578–584. doi: 10.3109/07388551.2014.993590. [DOI] [PubMed] [Google Scholar]
- Zeeman S.C., Kossmann J., Smith A.M. Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 2010;61:209–234. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Silva P., Rodrigues J.A., Dotson B., Brooks M.D., Zilberman D. Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. U S A. 2010;107:18729–18734. doi: 10.1073/pnas.1009695107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D., Liu T., Ma X., Wang B., Zheng Z., Zhang Y., Xie X., Yang B., Zhao Z., Zhu Q. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5′UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 2020;18:2385–2387. doi: 10.1111/pbi.13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D., Tian Z., Rao Y., Dong G., Yang Y., Huang L., Leng Y., Xu J., Sun C., Zhang G. Rational design of high-yield and superior-quality rice. Nat. Plants. 2017;3:17031. doi: 10.1038/nplants.2017.31. [DOI] [PubMed] [Google Scholar]
- Zeng D., Yan M., Wang Y., Liu X., Qian Q., Li J. Du1, encoding a novel Prp1 protein, regulates starch biosynthesis through affecting the splicing of Wxb pre-mRNAs in rice (Oryza sativa L.) Plant Mol. Biol. 2007;65(4):501–509. doi: 10.1007/s11103-007-9186-3. [DOI] [PubMed] [Google Scholar]
- Zhan J. A hub of hubs: the central role of ZmABI19 in the regulatory network of maize grain filling. Plant Cell. 2020;33:9–10. doi: 10.1093/plcell/koaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Yang Y., Chen Z., Chen F., Pan L., Lu Y., Li Q., Fan X., Sun Z., Liu Q. Characteristics of grain physicochemical properties and the starch structure in rice carrying a mutated ALK/SSIIa gene. J. Agric. Food Chem. 2020;68:13950–13959. doi: 10.1021/acs.jafc.0c01471. [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhu J., Chen S., Fan X., Li Q., Lu Y., Wang M., Yu H., Yi C., Tang S. Wxlv, the ancestral allele of rice Waxy gene. Mol. Plant. 2019;12:1157–1166. doi: 10.1016/j.molp.2019.05.011. [DOI] [PubMed] [Google Scholar]
- Zhang C., Yang Y., Chen S., Liu X., Zhu J., Zhou L., Lu Y., Li Q., Fan X., Tang S. A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J. Integr. Plant Biol. 2020;63:889–901. doi: 10.1111/jipb.13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Xu H., Feng M., Zhu Y. Suppression of OsMADS7 in rice endosperm stabilizes amylose content under high temperature stress. Plant Biotechnol. J. 2018;16:18–26. doi: 10.1111/pbi.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Duan L., Dai J., Zhang C., Li J., Gu M., Liu Q., Zhu Y. Major QTLs reduce the deleterious effects of high temperature on rice amylose content by increasing splicing efficiency of Wx pre-mRNA. Theor. Appl. Genet. 2014;127:273–282. doi: 10.1007/s00122-013-2216-4. [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhou L., Xu H., Wang L., Liu H., Zhang C., Li Q., Gu M., Wang C., Liu Q. The qSAC3 locus from indica rice effectively increases amylose content under a variety of conditions. BMC Plant Biol. 2019;19:275. doi: 10.1186/s12870-019-1860-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nallamilli B.R., Mujahid H., Peng Z. OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa) Plant J. 2010;64:604–617. doi: 10.1111/j.1365-313X.2010.04354.x. [DOI] [PubMed] [Google Scholar]
- Zhang J., Chen J., Yi Q., Hu Y., Liu H., Liu Y., Huang Y. Novel role of ZmaNAC36 in co-expression of starch synthetic genes in maize endosperm. Plant Mol. Biol. 2014;84:359–369. doi: 10.1007/s11103-013-0153-x. [DOI] [PubMed] [Google Scholar]
- Zhang X., Mogel K.J.H.v., Lor V.S., Hirsch C.N., De Vries B., Kaeppler H.F., Tracy W.F., Kaeppler S.M. Maize sugary enhancer1 (se1) is a gene affecting endosperm starch metabolism. Proc. Natl. Acad. Sci. U S A. 2019;116:20776–20785. doi: 10.1073/pnas.1902747116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zheng X., Yang J., Messing J., Wu Y. Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc. Natl. Acad. Sci. U S A. 2016;113:10842–10847. doi: 10.1073/pnas.1613721113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Dong J., Ji C., Wu Y., Messing J. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc. Natl. Acad. Sci. U S A. 2019;116:11223–11228. doi: 10.1073/pnas.1904995116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhiguo E., Li T., Zhang H., Liu Z., Deng H., Sharma S., Wei X., Wang L., Niu B., Chen C. A group of nuclear factor Y transcription factors are sub-functionalized during endosperm development in monocots. J. Exp. Bot. 2018;69:2495–2510. doi: 10.1093/jxb/ery087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Feng X., Li Y., Guzmán C., Lin N., Xu Q., Zhang Y., Tang H., Qi P., Deng M. Genome-wide identification of bZIP transcription factor genes related to starch synthesis in barley (Hordeum vulgare L.) Genome. 2021;0:1–14. doi: 10.1139/gen-2020-0195. [DOI] [PubMed] [Google Scholar]
- Zhong Y., Blennow A., Kofoed-Enevoldsen O., Jiang D., Hebelstrup K.H. Protein Targeting to Starch 1 is essential for starchy endosperm development in barley. J. Exp. Bot. 2018;70:485–496. doi: 10.1093/jxb/ery398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Wang L., Liu G., Meng X., Jing Y., Shu X., Kong X., Sun J., Yu H., Smith S.M. Critical roles of soluble starch synthase SSIIIa and granule-bound starch synthase Waxy in synthesizing resistant starch in rice. Proc. Natl. Acad. Sci. U S A. 2016;113:12844–12849. doi: 10.1073/pnas.1615104113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Xia D., He Y. Rice grain quality—traditional traits for high quality rice and health-plus substances. Mol. Breed. 2020;40:1. doi: 10.1007/s11032-019-1080-6. [DOI] [Google Scholar]
- Zhou H., Xia D., Zhao D., Li Y., Li P., Wu B., Gao G., Zhang Q., Wang G., Xiao J. The origin of Wxla provides new insights into the improvement of grain quality in rice. J. Integr. Plant Biol. 2020;63:878–888. doi: 10.1111/jipb.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F. Relationships between amylopectin internal molecular structure and physicochemical properties of starch. Trends Food Sci. Technol. 2018;78:234–242. doi: 10.1016/j.tifs.2018.05.024. [DOI] [Google Scholar]
- Zhu F., Bertoft E., Källman A., Myers A.M., Seetharaman K. Molecular structure of starches from maize mutants deficient in starch synthase III. J. Agric. Food Chem. 2013;61:9899–9907. doi: 10.1021/jf402090f. [DOI] [PubMed] [Google Scholar]
- Zhu H., Li C., Gao C. Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020;21:661–677. doi: 10.1038/s41580-020-00288-9. [DOI] [PubMed] [Google Scholar]
- Zhu J., Yu W., Zhang C., Zhu Y., Xu J., Li E., Gilbert R.G., Liu Q. New insights into amylose and amylopectin biosynthesis in rice endosperm. Carbohydr. Polym. 2020;230:115656. doi: 10.1016/j.carbpol.2019.115656. [DOI] [PubMed] [Google Scholar]
- Zhu L., Gu M., Meng X., Cheung S.C.K., Yu H., Huang J., Sun Y., Shi Y., Liu Q. High-amylose rice improves indices of animal health in normal and diabetic rats. Plant Biotechnol. J. 2012;10:353–362. doi: 10.1111/j.1467-7652.2011.00667.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Cai X., Wang Z., Hong M. An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J. Biol. Chem. 2003;278:47803–47811. doi: 10.1074/jbc.M302806200. [DOI] [PubMed] [Google Scholar]