Abstract
Viral mutations are an emerging concern in reducing SARS-CoV-2 vaccination efficacy. Second-generation vaccines will need to elicit neutralizing antibodies against sites that are evolutionarily conserved across the sarbecovirus subgenus. Here, we immunized mice containing a human antibody repertoire with diverse sarbecovirus receptor-binding domains (RBDs) to identify antibodies targeting conserved sites of vulnerability. Antibodies with broad reactivity against diverse clade B RBDs targeting the conserved class 4 epitope, with recurring IGHV/IGKV pairs, were readily elicited but were non-neutralizing. However, rare class 4 antibodies binding this conserved RBD supersite showed potent neutralization of SARS-CoV-2 and all variants of concern. Structural analysis revealed that the neutralizing ability of cross-reactive antibodies was reserved only for those with an elongated CDRH3 that extends the antiparallel beta-sheet RBD core and orients the antibody light chain to obstruct ACE2-RBD interactions. These results identify a structurally defined pathway for vaccine strategies eliciting escape-resistant SARS-CoV-2 neutralizing antibodies.
Keywords: SARS-CoV-2, epitope conservation, class 4 epitope site, cross-reactivity, neutralization, variants of concern, antigenic variation, escape mutations, next generation vaccines, human antibodies, mouse model
Graphical abstract
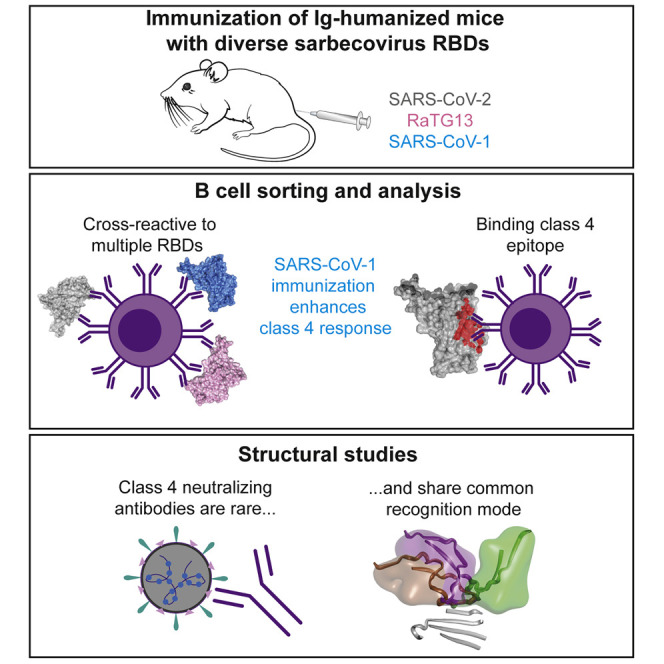
Viral mutations are an emerging concern in reducing SARS-CoV-2 vaccination efficacy. Burnett et al. immunized humanized mice with different diverse sarbecovirus RBDs to elicit antibodies targeting conserved sites. Non-neutralizing cross-reactive antibodies targeting the conserved class 4 epitope were readily elicited. Neutralizing ability was reserved only for antibodies binding this conserved supersite through an elongated CDRH3 that obstructed ACE2-RBD interactions.
Introduction
The current generation of COVID-19 vaccines provide strong protection against severe disease, but the lack of broad cross-neutralization elicited by these vaccines is a source of concern, as future variants are likely to emerge. Only a small percentage of antibodies against the spike protein on the SARS-CoV-2 (abbreviated here as CoV2) envelope are virus-neutralizing, the majority of which are directed against the receptor-binding domain (RBD) (Yuan et al., 2021). Among these, class 1 and 2 antibodies predominate, focused against epitopes in the ACE2 receptor-binding site (RBS) (Barnes et al., 2020). The RBS is highly variable while preserving ACE2 binding, and only 48% conserved between CoV2 and CoV1 compared to 84% amino acid conservation for non-RBS regions of the RBD (Cohen et al., 2021). K417N/T, E484K, and N501Y mutations in the RBS epitope have independently arisen in rapidly spreading CoV2 variants of concern in South Africa (B.1.351/beta) and Brazil/Japan (P.1/gamma) (Greaney et al., 2021; Starr et al., 2021). These mutations decrease the serum neutralization titer of mRNA-vaccinated people by up to 70% (Garcia-Beltran et al., 2021; Ikegame et al., 2021; Liu et al., 2021; Wang et al., 2021; Wu et al., 2021). This has been shown to have clinical impacts limiting vaccine efficacy (Hacisuleyman et al., 2021; Madhi et al., 2021). The ongoing risk of new zoonotic spillover events from sarbecoviruses circulating widely in bats also remains a major concern (Banerjee et al., 2021).
In order to address this problem, second-generation vaccines will need to elicit neutralizing antibodies directed against sites that are evolutionarily conserved across the sarbecovirus subgenus (clade B) including CoV2, the related bat virus RaTG13, and the more distant CoV1, which differ at key residues characterizing variants of concern such as K417, E484, and N501 (Boni et al., 2020) (Table S1B). Here, we analyzed broadly reactive antibody responses elicited by RBD-focused immunization of mice that carry human antibody gene segments (Asensio et al., 2019; Peter et al., 2021), employing multi-color flow-cytometric staining of B cells with RBDs from CoV1, CoV2, and RaTG13, coupled with single-cell RNA sequencing and antibody characterization. We found that for immunization strategies utilizing diverse sarbecovirus RBDs, class 4 antibodies dominated the broadly reactive subset and used recurring human IGHV and IGKV elements. However, such antibodies were generally not neutralizing, as previously observed for CR3022 (ter Meulen et al., 2006). In contrast, potent neutralizing antibodies binding the highly conserved class 4 epitope that sterically blocks the RBS from accessing ACE2, employing a long CDRH3 and less common IGHV and IGKV pairs, could be expanded by multiple immunizations with the CoV1 RBD. These results provide a guide for developing second-generation COVID-19 vaccine strategies.
Results
Immunization with diverse sarbecovirus antigens induced a cross-reactive response in wild-type mice
The current CoV2 vaccines utilize the whole spike protein to induce clonal selection of antibody-forming B cells. Given the goal of eliciting antibodies specifically to highly conserved regions of the RBD, we explored the capacity of the isolated CoV2 RBD compared to full trimeric spike, enhancing immunogenicity by conjugation to sheep red blood cells (SRBCs) (Figure S1). Previous experience has shown that small foreign proteins elicit more reproducible germinal center (GC) B cell populations when covalently linked to SRBCs without adjuvant than when given in adjuvant as isolated proteins or covalently linked to other immunogenic carrier proteins. Mice from C57BL/6, BALB/c, and FVB/NJ strains with distinct Ighv and Igkv loci (Collins et al., 2015) were immunized with an equivalent density of the conjugated CoV2 RBD or spike (Figure S1A) and spleen cells analyzed 7 days later by staining with CoV2 RBD fluorescent tetramers, which selectively bound to individual B cells bearing surface immunoglobulin (Ig) with an affinity for CoV2 RBD epitopes. Immunization of mice with the RBD or spike resulted in the development of RBD-specific B cells in the GC, IgG1 memory, and plasmablast compartments (Figure S1B). Full trimeric spike resulted in greater recruitment of RBD-specific cells in the GC compartment, while both antigens were equally efficient at recruiting these cells into the memory and plasmablast compartments.
To enumerate B cells with broadly reactive surface Ig within the elicited RBD response, these analyses were extended by flow cytometric staining with a panel of distinguishable fluorescent RBD tetramers from CoV2, Pangolin, RaTG13, and CoV1, representing progressively more distant RBD sequences (Table S1A). 50% to 70% of CoV2 RBD-binding B cells in the CoV2-RBD-elicited GC and memory repertoires were cross-reactive to pangolin-derived RBD and 30% to 60% were cross-reactive to RaTG13 but only 5% to 15% displayed cross-reactivity to CoV1 (Figures 1 A, 1B, and S2A). For comparison, the same flow cytometric analysis was performed following intranasal infection with 1 × 104 plaque-forming units (PFU) CoV2 in K18-hACE2-C57BL/6 mice with humanized ACE2 receptors (Johansen et al., 2020). This revealed analogous results with 60% of CoV2 RBD-binding GC B cells cross-reacting to pangolin RBD, 19% cross-reacting RaTG12, and 1.8% to CoV1 (Figures S1B and S1C).
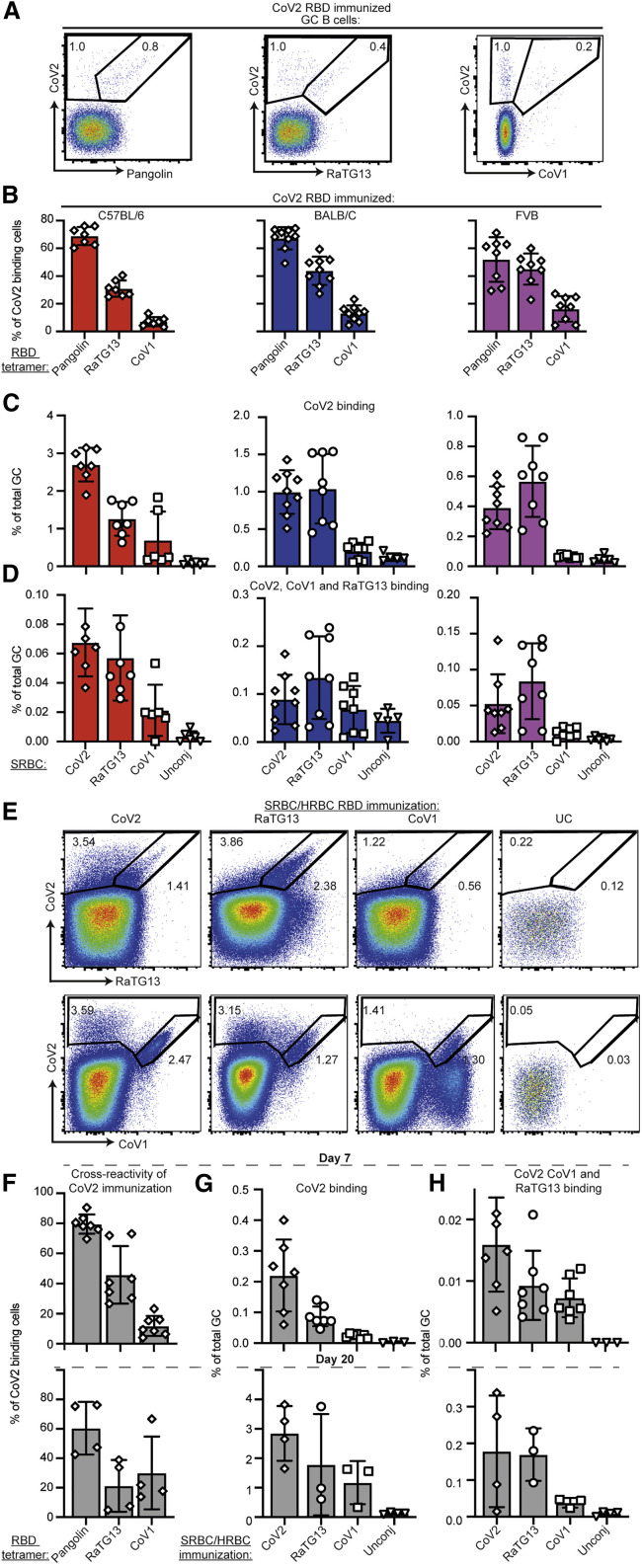
Figure 1.
B cell response elicited by RBD-focused immunization included broadly cross-reactive cells
(A) Representative flow cytometric plots showing total GC B cells (TCRB–, CD11b–, B220+, Fas+, CD38–) simultaneously stained with four different fluorescent tetramers of the indicated sarbecovirus RBDs in the spleen of BALB/C mice 7 days following immunization with CoV2-RBD conjugated to SRBCs. Percentage of total CoV2-binding GC B cells and cross-reactive B cells are indicated on each plot.
(B) Percentage of CoV2 binding GC B cells also binding to the indicated sarbecovirus RBD tetramer in individual mice (symbols) and mean ± SEM from three different inbred strains.
(C and D) Percentage of all GC B cells binding the CoV2 RBD tetramer (C) or cross-reactively binding the CoV2, RaTG13, and CoV1 RBD tetramers (D) 7 days after immunization with the indicated RBD conjugated to SRBCs or unconjugated SRBCs as a negative control.
(E) Representative flow cytometric plots of GC B cells simultaneously stained with three different fluorescent tetramers of the indicated sarbecovirus RBDs in the spleen of Ig-humanized mice with a human antibody repertoire on day 20 following four immunizations with the indicated RBD conjugated first to SRBCs and then to HRBCs.
(F) Percentage of CoV2 binding GC B cells also binding the indicated sarbecovirus RBD tetramer in individual human antibody repertoire mice (symbols) after one immunization (day 7) or four immunizations (day 20) with CoV2 RBD.
(G and H) Percentage of all GC B cells binding the CoV2 RBD tetramer (G) or cross-reactively binding the CoV2, RaTG13, and CoV1 tetramers (H) in individual humanized antibody mice (symbols).
Data pooled from two independent experiments. Columns show mean ± SEM.
See also Figures S1–S3.
Given that CoV2 RBD immunization elicited cross-reactive GC and memory B cells binding to other sarbecovirus RBDs, we next tested with the inverse also applied (Figure 1C). Immunization with the RaTG13 RBD elicited a strong CoV2 RBD-binding response. Immunization with the more distant CoV1 RBD elicited RBD-binding B cells that were mostly specific to the CoV1 RBD; however, a clear subset of the CoV1-binding cells cross-reacted with all three sarbecovirus RBDs. All three immunizations induced a subset of B cells with surface Ig that is triple-reactive: binding CoV2, RaTG13, and CoV1 RBDs (Figure 1D).
Overall, we found that both spike and the RBD induced a CoV2 specific response. A proportion of the response following CoV2 RBD immunization or infection included B cells with broad cross-reactivity. These broadly reactive B cells were also induced by immunization with CoV1 or RaTG13 RBDs.
Immunization with diverse sarbecovirus RBDs induced a cross-reactive response in human antibody VDJ transgenic mice
We next extended this analysis to C57BL/6 TRIANNI transgenic mice where the human antibody variable, diversity, and joining (V ( D ) J) element repertoire replaces the mouse V ( D ) J repertoire at the heavy chain and kappa light chain loci (Asensio et al., 2019; Peter et al., 2021). Immunization of the Ig-humanized mice with CoV2 RBD conjugated to SRBCs elicited a GC response with 79% of CoV2-binding GC B cells cross-reactive to the closely related pangolin RBD, 42% to RaTG13, and 10% to the more distant CoV1 RBD (Figure 1F). Again, 0.005% to 0.015% of all GC B cells displayed antibodies that cross-reacted to all three RBDs (Figure 1H). To prime-boost the response, the Ig-humanized mice were immunized with the RBD conjugated to SRBCs on days 0 and 6 and then with the same RBD conjugated to a different adjuvant-free immunogenic carrier, horse RBCs (HRBCs), on days 10 and 15, and the effects were explored on day 20 (Figures 1E–1H). Prime-boosting increased by 10-fold the percentage of GC B cells binding the CoV2 RBD (Figure 1G) and cross-reacting with CoV2, RaTG13, and CoV1 (Figure 1H).
We extended the flow cytometric approach to analyze which RBD epitope is recognized by each B cell responding to the different sarbecovirus RBD immunizations. Spleen cells were first incubated with equimolar monomeric CoV2, CoV1, or RaTG13 RBDs to identify B cells with surface Ig capable of binding each antigen. Bound RBD was revealed by staining the cells with fluorescent S309 binding the class 3 epitope (Pinto et al., 2020), fluorescent EY6A recognizing the class 4 epitope (Zhou et al., 2020), and fluorescent ACE2 (Figure 2 ). B cells bearing surface Ig against the class 1 or 2 epitopes would be revealed as having bound the RBD that was still recognizable by S309 and EY6A (Figure 2A). Indeed, cells with these characteristics were the most frequently elicited RBD-binding B cells in Ig-humanized mice immunized with the CoV2 RBD (Figures 2B and 2F). By contrast, the RBD bound to B cells bearing surface Ig against the canonical conserved class 4 epitope should be available for binding fluorescently labeled S309 but not EY6A (orange gate in Figures 2A and 2B) and retain availability for binding fluorescent ACE2. This was validated in spleen B cells from transgenic mice expressing the rearranged V(D)J segments of the prototypic class 4 antibody CR3022 (ter Meulen et al., 2006) (Figure 2A). Immunization of Ig-humanized mice with the RaTG13 or CoV1 RBD elicited a significantly higher proportion of their RBD-binding B cells recognizing the class 4 epitope compared to mice immunized with CoV2 RBD (Figures 2D–2F). 70% to 90% of B cells bearing class 4 surface Ig bound RBD monomers that retained availability to bind ACE2 (Figures 2F–2H), but a consistent subset bound the RBD in a way that blocked ACE2 binding.
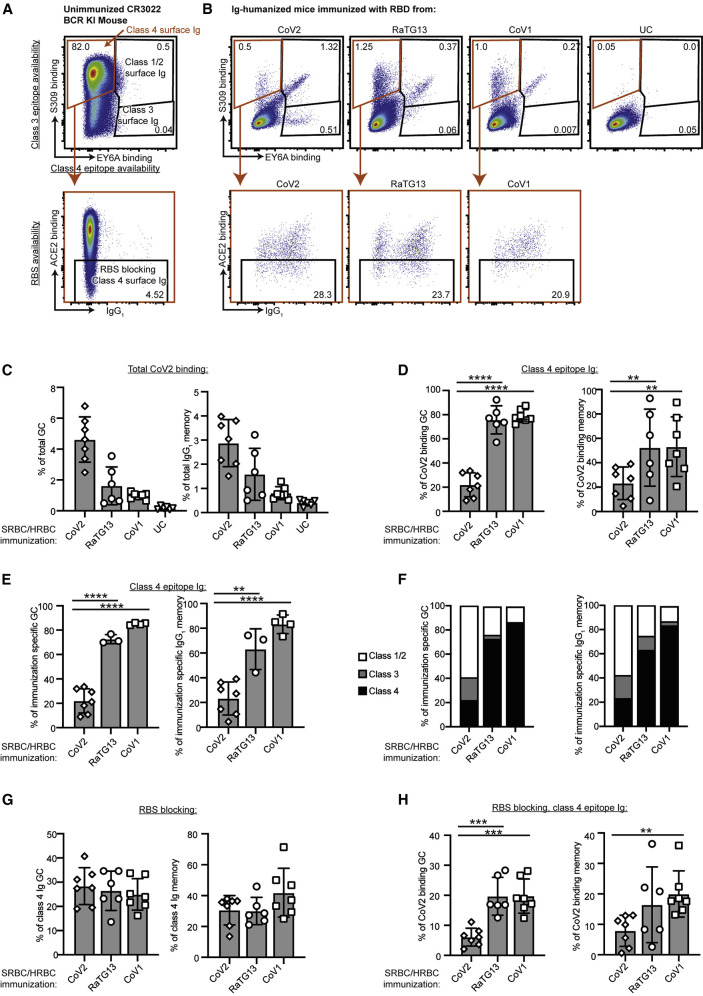
Figure 2.
Immunization with diverse RBDs shift epitope predominance of the B cell response
(A) Flow cytometric plot of spleen B cells in an unimmunized mouse with knocked in CR3022 V(D)J IGH and IGK exons encoding the prototypic class 4 epitope-specific membrane Ig, illustrating the strategy used to identify B cells recognizing different RBD epitopes based on competition for RBD binding between the membrane Ig on each cell and fluorescent ligands for class 4 (EY6A), class 3 (S309), and class 1/2 (ACE2) epitopes. B cells were first stained with CoV2 RBD, then with fluorescent anti-RBD antibodies S309 and EY6A, and finally with fluorescent ACE2.
(B) Representative flow cytometric plots of GC B cells in Ig-humanized mice on day 20 following immunization with the indicated RBDs coupled to SRBCs/HRBCs or unconjugated SRBCs/HRBCs (UC): (top panels) the proportion of B cells with membrane Ig-binding RBD and blocking either the class 4 (orange gate) or class 3 epitopes (S309– EY6A+) or class 1/2 epitopes (S309+ EY6A+); (bottom panels) the proportion of cells with membrane Ig binding the class 4 epitope but also precluding ACE2 binding.
(C) Percentage of total GC (B220+, Fas+, CD38–) B cells or IgG1 memory (B220+, IgG1+, Fas–) B cells binding to 200 ng/mL CoV2 RBD in individual Ig-humanized mice immunized with the indicated RBDs.
(D) Percentage of CoV2 RBD binding GC or IgG1 memory B cells binding to the class 4 epitope.
(E) Of GC or IgG1 memory B cells that bind the same RBD as was used for immunization, percentage that bind the class 4 epitope.
(F) Percentage of CoV2, CoV1, or RaTG13 binding GC or IgG1 memory B cells elicited by the indicated immunogens that bind to each of the indicated epitopes.
(G) Percentage of B cells with class 4 epitope binding Ig that precludes ACE2 binding.
(H) Percentage of CoV2 binding GC or IgG1 memory B cells binding to the class 4 epitope and blocking the RBS.
Data points represent individual mice. Columns show mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Data pooled from two independent experiments.
To analyze broadly cross-reactive RBD-binding B cells, we performed single-cell V(D)J mRNA sequencing of GC B cells that bound single or multiple RBD tetramers flow-cytometry sorted from the humanized mice immunized with CoV2, RaTG13, or CoV1 RBDs with the highest percentage of cross-reactive B cells (Figure 1E). Paired IGHV/IGKV sequences were obtained from 7,481 dual or single RBD tetramer-binding B cells (Figure 3 ; Tables S2A–S2C) and from an additional set of 52 triple RBD tetramer-binding B cells sorted from separate mice (Figure S4A; Table S2D). Across these immunization strategies, several IGHV/IGKV pairs were frequently used by GC B cells that cross-reacted with all three sarbecovirus RBDs: VH1-18 paired with VK6-21 or VK6D-21, VH1-46 paired with VK1-9 or VK1-6, and VH3-33 paired with VK1D-13 or VK1-9 (Figures 3B–3D and S4A).
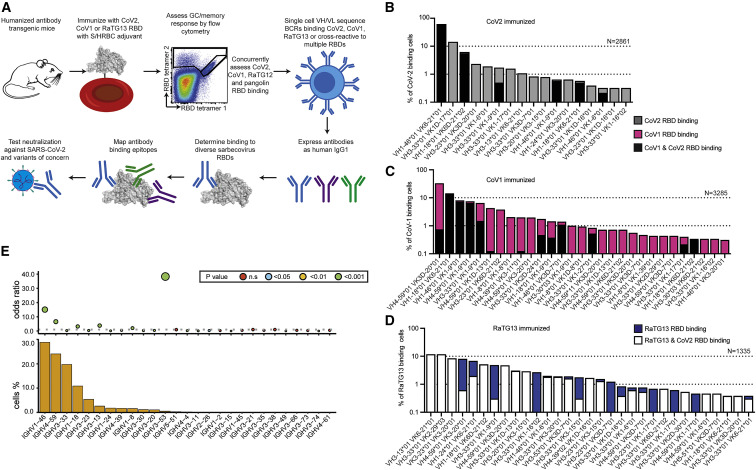
Figure 3.
B cell response to coronavirus RBDs in Ig-humanized mice included recurrent IGHV/IGKV pairs
(A) Experimental workflow.
(B–D) Percentage of sequenced RBD-binding B cells using the indicated IGHV/IGKV pairs, sorted for binding two different RBD tetramers or specific for one, from mice with human antibody repertoire immunized four times with RBD from CoV2 (B), CoV1 (C), or RaTG13 (D). Dashed lines indicate a frequency 1 or 10% of sequenced B cells.
(E) Frequency of different human IGHV elements used by RBD-binding antibodies sorted and sequenced from germinal center B cells of RBD-immunized Ig humanized mice, and the odds ratio of using each IGHV element in the RBD-elicited set compared to its frequency from sequencing single naive IgM B cells from unimmunized Ig humanized mice both analyzed by paired heavy and light chain sequencing. p value calculated by Fisher’s exact test.
See also Figure S4.
For comparison with the RBD-elicited B cells, we obtained paired IGHV/IGKV sequences from 6,000 naive B cells from the humanized mice and analyzed deep cDNA sequencing of the expressed IgM repertoires of 106 cells from two additional humanized mice and in naive B cells from 100 human blood samples (Figure S4C). This confirmed that Ig-humanized mice express diverse IGHVs. Although some IGHV elements like IGHV1-69 showed an altered frequency compared to humans, 78% of IGHV elements were used at comparable frequency to the naive repertoire in human blood, including many of those frequently used in RBD-binding antibodies: IGHV1-18, IGHV1-46, IGHV4-39, IGHV3-30.3, IGHV3-53, and IGHV3-66 (Figure S4C). IGHV3-53, which accounts for frequent germline-encoded antibodies against the hypervariable RBS region of CoV2 (Andreano and Rappuoli, 2021; Yuan et al., 2020), was strongly selected among the total set of sequenced RBD-binding B cells when compared to its frequency in the naive repertoire but was rarely used among the highly cross-reactive triple-RBD binders from the humanized mice (Figures 3B–3E and S4A). The latter result is consistent with RBD-binding antibodies isolated from CoV2-infected people and deposited in CoV-AbDab (Raybould et al., 2021), where IGHV3-53 is frequent among CoV2-specific antibodies but rarely used among antibodies that also bind CoV1 (Figure S4D).
For comparison to the Ig-humanized mice, rare triple RBD tetramer-binding memory B cells were sorted from the blood of CoV2 convalescent human patients (Figures 4A and 4B). Single-cell sequencing of 121 CoV2, CoV1, and RaTG13 triple-binders revealed that many used IGHV regions observed in triple binders from RBD-immunized Ig-humanized mice, including the recurring IGHV1-46, IGHV4-59, IGHV1-18, IGHV3-33, IGHV4-39, and IGHV3-13 (Figure 4C; Table S2H). Indeed, 92% of the IGHVs identified from the 53 triple-binding cells sorted from Ig-humanized mice were utilized among the 121 human B cells sorted with the same strategy. These IGHV elements are also used by CoV1/CoV2 RBD cross-reactive antibodies isolated from CoV2-infected people and deposited in CoV-AbDab, confirming cross-reactive B cells selected in the humanized mouse repertoire to be representative of the human repertoire (Figure S4D).
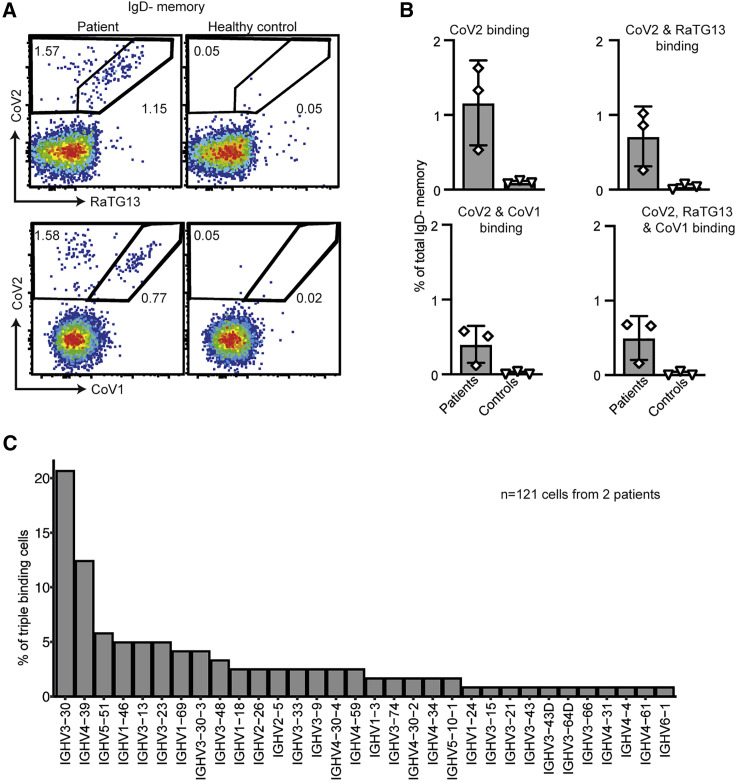
Figure 4.
Cross-reactive B cell response in human convalescent patients was comparable to Ig-humanized mice
(A) Representative flow cytometric plots of circulating IgD– CD27+ memory B cells in a convalescent patient four months after CoV2 infection and in a healthy uninfected control, stained with fluorescent tetramers of RBDs from CoV2, RaTG13, and CoV1.
(B) Percentage of memory B cells binding one, two, or all three RBD tetramers from three convalescent CoV2 patients four months post-infection and three uninfected healthy controls. Data points represent one individual. Data pooled from two independent experiments.
(C) IGHV usage among antibodies from 121 triple RBD-binding memory B cells from CoV2 convalescent patients.
See also Figure S4.
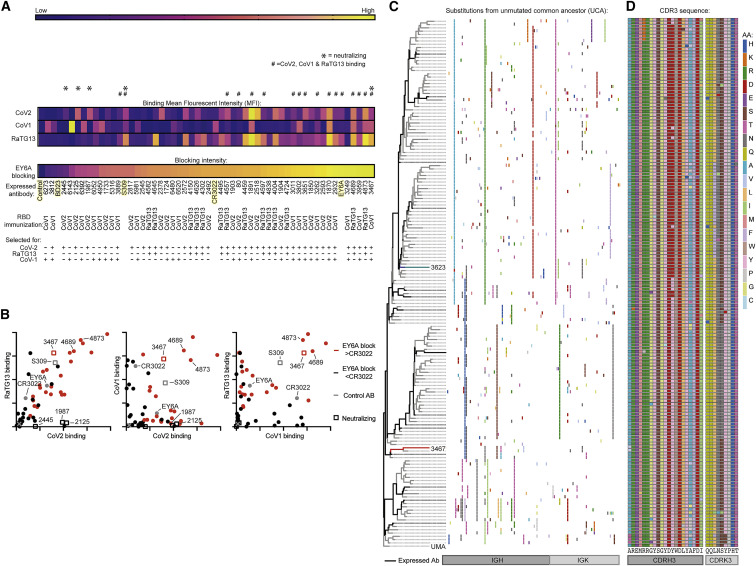
From the 7,533 RBD-binding antibody sequences obtained from RBD-immunized Ig-humanized mice, a total of 56 antibodies were selected for expression as human IgG1, representing IGHV/IGKV pairs found in >1% of sarbecovirus RBD-binding B cells or pairs recurrently selected between different immunization regimes (Table S3A). Antibodies were tested for binding to different sarbecovirus RBDs by flow cytometry against RBD-conjugated erythrocytes (Figures 5A and 5B; Table S2A). Previously described class 4 antibodies, CR3022 (ter Meulen et al., 2006) and EY6A (Zhou et al., 2020), and the class 3 antibody S309 (Pinto et al., 2020), were expressed and tested in parallel as controls. Overall, 15 antibodies bound all three RBDs (Figures 5A and 5B). Most used the recurring IGHV/IGKV pairs noted above except for AB-3467, which used VH4-59 paired with VK1-9 and was part of a very large, heavily mutated clonal expansion of cells binding CoV1 and CoV2 RBDs that comprised 7.9% of the RBD-binding GC cells sorted from a CoV1-RBD-immunized mouse (Figures 5C and 5D). Of the IGHV regions that accounted for recurrently selected class 4 sequences, IGHV1-18, IGHV1-46, IGHV3-13, IGHV3-20, and IGHV4-39, which together accounted for 76% of class 4 antibodies, were equally represented in the human and mouse naive repertoires. IGHV3-33, which accounted for 16% of the class 4 sequences, was overrepresented in the mouse naive repertoire. IGHV4-59, which accounted for 8% of class 4 antibodies and AB-3467, was underrepresented in the mouse naive repertoire (Figure S4D).
Figure 5.
Cross-reactive human antibodies expressed from sequenced RBD-binding B cells showed common properties
Representative RBD-binding antibodies from Figure 2 and from published broadly reactive antibodies CR3022, EY6A, and S309 were expressed as human IgG1s and tested in parallel.
(A) Heatmap showing binding of each antibody to the indicated sarbecovirus RBD, measured flow cytometrically as mean fluorescence intensity (MFI) of IgK staining of RBD-conjugated erythrocytes. Also shown is the percentage of inhibition of fluorescent EY6A IgG1 binding to CoV2 RBD-conjugated erythrocytes, measured flow cytometrically for each antibody in parallel. Hash symbol (#) denotes antibodies binding all three RBDs with an MFI > CR3022. Asterisk (∗) denotes CoV2-neutralizing antibodies. Control antibodies highlighted in yellow. RBD HRBCs/SRBCs immunizations and tetramer sorting strategy to elicit each antibody are indicated below.
(B) IgK MFI for binding of each expressed antibody to erythrocytes conjugated with the indicated RBD. Red symbols indicate antibodies blocking EY6A more strongly than the class 4 antibody CR3022. Published control antibodies shown by gray symbols. Squares indicate CoV2-neutralizing antibodies. Each symbol is data for one antibody. Binding control with no antibody is indicated with an X.
(A and B) Data pooled from two independent experiments.
(C) AB-3467 clonal tree. Each row summarizes nonsynonymous somatic mutations in one clonally related, tetramer-sorted, and V(D)J-sequenced B cell. Antibodies expressed for further analysis are indicated in black. AB-3467 is highlighted in red and AB-3623 in blue.
(D) CDRH3 and CDRK3 mutations across the indicated AB-3467 clonal lineage indicated in (C).
See also Figure S5.
Together, this revealed that antibodies cross-reactive to multiple RBDs predominantly used a subset of recurrently selected IGHV/IGKV pairs.
Sarbecovirus cross-reactive antibodies bound the class 4 epitope site
To identify expressed antibodies recognizing the class 4 epitope, we tested each expressed antibody for competitive inhibition of fluorescently conjugated class 4 antibody EY6A binding to the CoV2 RBD. Among 15 antibodies cross-reacting between CoV2, RaTG13, and CoV1 RBDs, 14 (93%) blocked EY6A more effectively than the positive control class 4 antibody CR3022 (Figures 5A and 5B; Table S2A). 12 of these 14 putative class 4 antibodies used the IGHV/IGKV pairs found frequently in triple-tetramer-binding B cells noted above (Figure S4B).
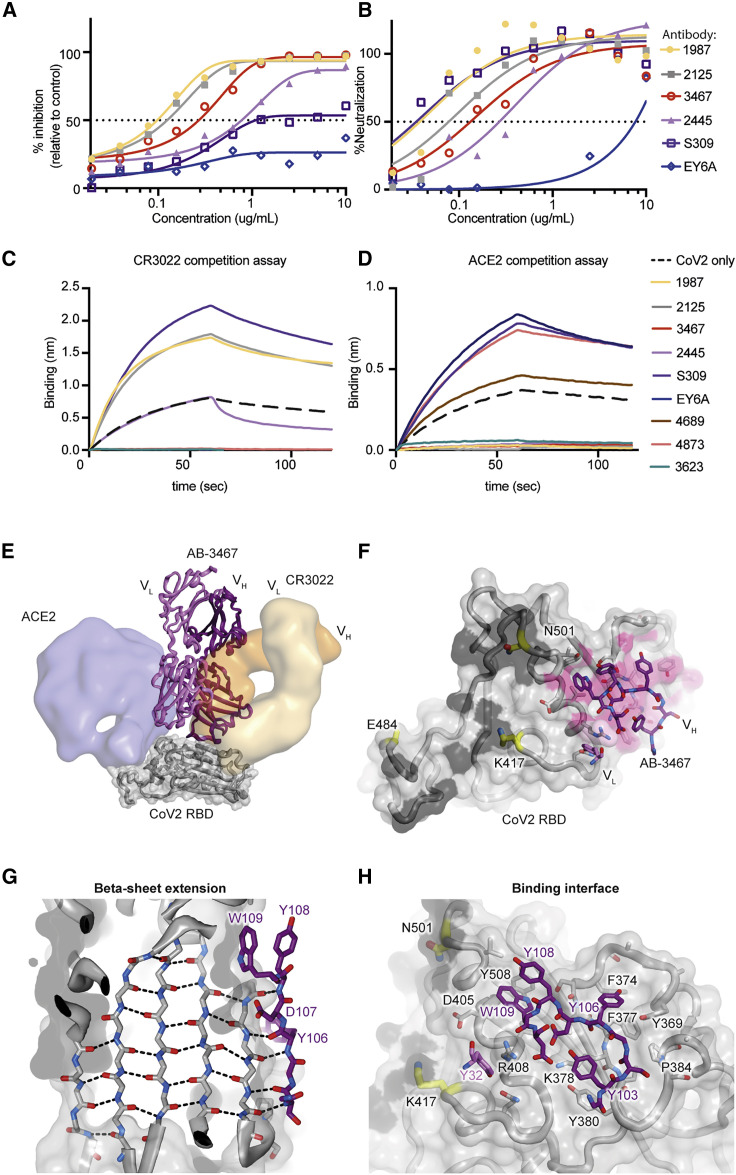
The total 36 antibodies binding strongly to CoV2 were tested at four concentrations for neutralization of lentivirus particles pseudotyped with the CoV2 spike alongside EY6A, S309, and CR3022. Only four were potent neutralizers (half maximal inhibitory concentration [IC50] < 5 μg/mL): AB-1987, IC50 = 0.097 μg/mL; AB-2126, IC50 = 0.107 μg/mL; AB-3467, IC50 = 0.247 μg/mL; and AB-2445, IC50 = 0.753 μg/mL (Figure 6 A). All of the neutralizing antibodies were more potent than S309 (IC50 = 1.2 μg/mL) and much more potent than EY6A (IC50 > 10 μg/mL). In a Vero-E6-based neutralization assay, the same four antibodies potently neutralized live CoV2 virus: AB-1987, IC50 = 0.054 μg/mL; AB-2126, IC50 = 0.106 μg/mL; AB-3467, IC50 = 0.179 μg/mL; and AB-2445, IC50 = 0.370 μg/mL (Figure 6B). Again, all four neutralized much more potently than EY6A (IC50 > 10 μg/mL), and CR3022 had no measurable CoV2 neutralizing activity. Notably, of the 14 CoV2/RaTG13/CoV1 broadly cross-reactive antibodies that competed with EY6A, only AB-3467 neutralized CoV2 virus (Figures 5A and 5B). Thus, the most consistently used IGHV/IGKV pairs in class 4 antibodies encoded antibodies that bound strongly but did not neutralize.
Figure 6.
Structural basis of neutralizing class 4 antibodies
(A and B) CoV2 spike envelope pseudotyped lentivirus neutralization (A) or neutralization of live CoV2 in Vero E6 cells (B) for the indicated antibodies. Data representative of two independent experiments.
(C and D) Biolayer interferometry competition assay showing binding of CR3022 (C) or ACE2-Fc (D) to the CoV2 RBD alone or following incubation with the indicated antibodies.
(E) Crystal structure of AB-3467 (purple ribbon) with the CoV2 RBD (gray surface and ribbon). Surfaces for CR3022 (orange) and hACE2 (blue) are overlaid for reference. The light chain of AB-3467 (light purple) would sterically hinder hACE2 binding.
(F) The CoV2 RBD surface showing the ACE2 binding interface (black); escape mutation locations are highlighted in yellow and AB-3467 contact residues on the RBD are shaded in pink. The binding region of the AB-3467 is shown as purple sticks and is distal to the known escape mutations.
(G) View of the CoV2 RBD beta sheet (gray) showing the antiparallel extension by the AB-3467 heavy chain. Key residues on AB-3467 labeled in purple (edged by CDRH3 residues Y108 [top] to S104 [bottom]).
(H) Binding interface of AB-3467 (purple) with the CoV2 RBD (gray). ACE2-binding residues are shaded black, escape mutation locations are in yellow, and key RBD residues are labeled in black text and key antibody residues in purple.
See also Figure S6.
The affinities of selected antibodies to the different sarbecovirus RBDs were measured using biolayer interferometry. AB-3467 strongly bound CoV2 (KD = 4 nM), pangolin (KD = 3.5 nM), mink (KD = 2 nM), RaTG13 (KD = 9.8 nM), and CoV1 (KD = 1.5nM) (Figure S5D). Two comparably potent EY6A blocking and broadly cross-reactive antibodies (Figures 5A and 5B) that lacked neutralizing activity had comparable affinity: AB-4873, employing the recurrent VH1-46 / VK1-9 pair, and AB-4689, employing the recurrent VH1-18 / VK6D-21 pair. The affinity of these two antibodies to RBDs from CoV2 (AB-4873 KD = 4.1 nM; AB-4689 KD = 0.24 nM), pangolin, RaTG13, and CoV1 was similar or higher than AB-3467 (Figure S5). Given that affinity was not predictive of neutralizing activity, we next sought to explore if this difference could be explained by other binding characteristics. Biolayer interferometry competition assays confirmed that AB-3467 as well as the non-neutralizing class 4 antibodies AB-4873 and AB-4689 were equally potent at blocking the binding of the class 4 antibody CR3022 to the CoV2 RBD (Figure 6C). However, among the class 4 antibodies, only antibody AB-3467 competed for binding of ACE2 by the CoV2 RBD (Figure 6D). AB-3467 thus represented the minority of class 4 broadly reactive antibodies elicited by immunization with RBDs from CoV1, RaTG13, or CoV2 that possessed a capacity to block availability of the RBS to ACE2 (Figure 2F).
By contrast, among the non-class 4 antibodies elicited by RBD immunization, the three potent CoV2 neutralizing antibodies—AB-1987, AB-2125, and AB-2445 (Figures 6A and 6B)—all competed with ACE2 for binding to the RBD (Figure 6D). These three antibodies were isolated from GC B cells with narrow specificity for CoV2 and lacking binding to RaTG13 or CoV1 RBDs (Figures 5A and 5B), consistent with the vulnerability of class 1/2 antibodies to hypervariability around the ACE2 binding site.
This highlighted that the class 4 antibodies were frequently non-neutralizing, and that neutralizing potency was associated with the ability to interfere with ACE2 binding.
Neutralizing class 4 antibodies shared common structural features
To understand how AB-3467 simultaneously competed with ACE2 and with CR3022/EY6A, we solved the crystal structure of the AB-3467 Fab bound to the CoV2 RBD (Figures 6E and 6F). The structure revealed that >90% of the RBD-binding interface was provided by the antibody heavy chain, which projected a long CDRH3 loop containing three tyrosine (Y) residues and a tryptophan (W) residue whose side chains contacted hydrophobic components of the RBD surface (Figure 6G). Much of this surface (∼80%) was also contacted by the heavy chain of CR3022 (Figure 6E; Table S3B). However, unlike CR3022, the backbone of AB-3467’s long CDRH3 loop extended the antiparallel beta sheet running through the center of the RBD (Figures 6G and 6H). The sole light chain contribution was mediated by side chain interactions of CDRL1 residue Y32 with the highly conserved RBD residue R408.
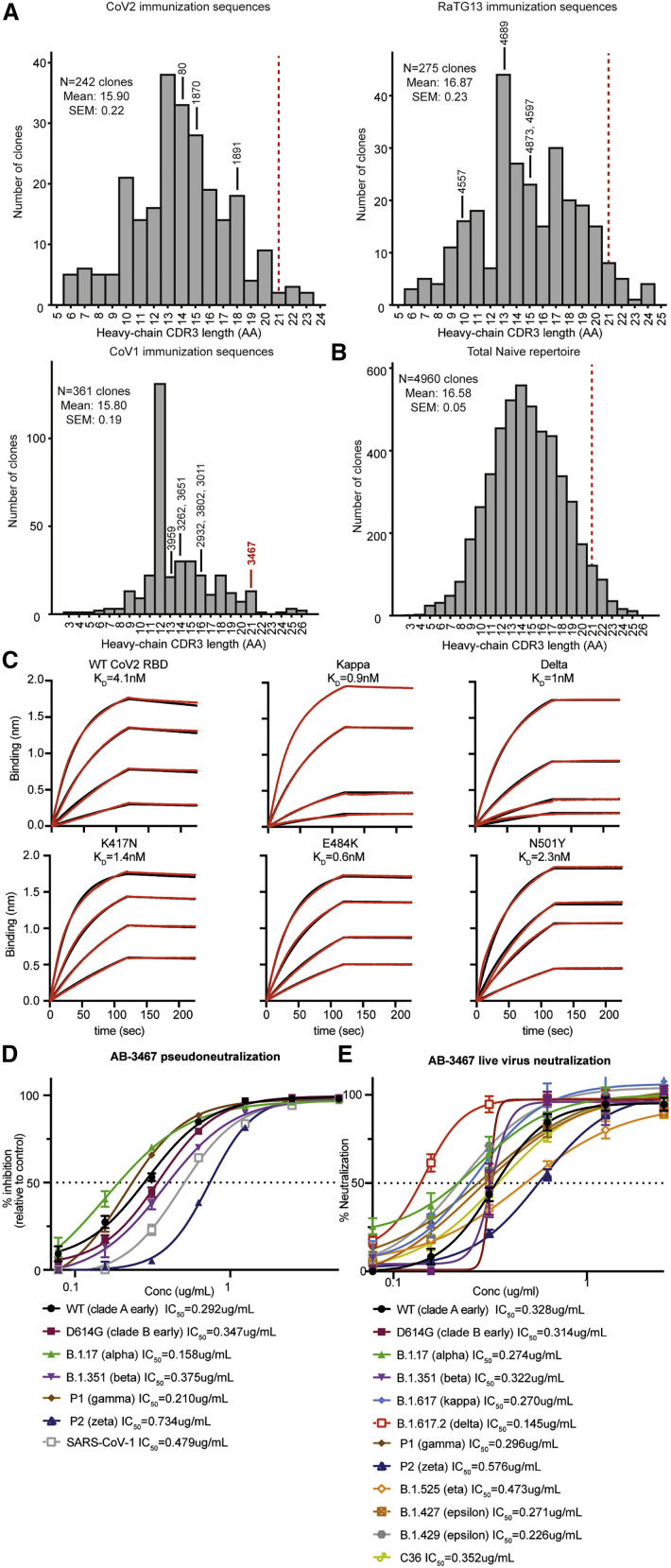
As a consequence of CDRH3 adding an antiparallel beta sheet to the RBD core, the spatial orientation of the AB-3467 heavy and light chains, with respect to the RBD surface, was rotated ∼180 degrees relative to that of CR3022 (Figure 6E). Consequently, the largely non-contacting AB-3467 light chain was projected toward the ACE2 epitope (rather than away from it), thus facilitating blockage of ACE2 binding due to steric hindrance. Inspecting deposited datasets, this rotated orientation has been seen in five other class 4 antibodies, all of which show potent CoV2 neutralization, likely through similar ACE2 steric hindrance by the light chain, and each employing a long CDRH3 loop (Jette et al., 2021; Liu et al., 2020; Saunders et al., 2021; Wrapp et al., 2020). For the previously published VHH-72 (Wrapp et al., 2020), COVA1-6 (Liu et al., 2020), and DH1047 (Saunders et al., 2021) class 4 antibodies, the long CDRH3s also engaged the RBD through beta-sheet extension (Figures S6E and S6F). Recently, Jette et al. (2021) have shown the same antiparallel beta-sheet extension in human antibodies C022 and C118. As such, antiparallel beta-sheet extension at the RBD surface, mediated through long heavy chain CDR3s, appeared to be a common mechanism of potent neutralizing class 4 antibodies. In contrast, shorter CDRH3s were present in all 14 of the class 4 antibodies with recurrent IGHV/IGKV pairs that we had expressed and confirmed broad sarbecovirus RBD reactivity but no neutralizing activity (Figure 7 A). The CDRH3 length of AB-3467 was also considerably longer than the mean length of the total sequenced human or mouse cross-reactive sequences (Figures S6H and S6I).
Figure 7.
Key features of AB-3467 facilitated its resistance to escape mutations
(A) Distribution of heavy-chain CDR3 lengths from single B cells sequenced on day 20 following immunization with each of the indicated sarbecovirus RBDs. The CDRH3 length of broadly reactive class 4 antibodies that were expressed are shown. The relative position AB-3467 would lie on each graph is indicated by a red dashed line.
(B) Distribution of heavy-chain CDR3 lengths from single B cells sequenced from a naive Ig humanized mouse with human antibody repertoire.
(C) Biolayer interferometry measurement of affinity of AB-3467 against the wild-type (WT; clade A early) CoV2 RBD and the indicated variants.
(D) Neutralization by AB-3467 of infection of ACE2-HEK293T cells by lentiviral particles pseudotyped with spike envelope from CoV1 or from each of the indicated CoV2 variants.
(E) Neutralization by AB-3467 of HEK293T cell cytopathic effect by live CoV2 virus isolates of the indicated variants of concern.
See also Figure S6
As noted above, AB-3467 was one of 260 CoV1/CoV2 RBD-binding and sequenced B cells comprising a large, highly mutated clone induced by repeated CoV1 RBD immunization (Figure 5C). The observed VH4-59/VK1-9 pairing was infrequent in the RBD response of other mice (Table S2). VH4-59 paired with VK1-9 accounted for 0.5% of the total naive repertoire in Ig-humanized mice; however, only a minority of naive B cells had CDRH3 as long as AB-3467 (Figure 7B), including naive B cells expressing the VH4-59 heavy chain (Figure S6K). To further explore the VH4-59 VK1-9 antibodies, an additional 17 antibodies were expressed from the same clonal lineage as AB-3467 as well as the unmutated common ancestor (Figure 5C). All 17 of these antibodies and the unmutated common ancestor bound to CoV2, RaTG13, CoV1, and mink RBDs and blocked EY6A, confirming that the diverse patterns of somatic mutations preserved a broad reactivity to the conserved class 4 epitope that had been established during VDJ-recombination (Table S3B). The unmutated ancestor nevertheless had 100-fold lower affinity for the CoV2 RBD (KD = 499nM) than its hypermutated descendants AB-3467 (KD = 4.1 nM) and AB-3623 (KD = 2.3 nM) (Figure S5). In addition, we expressed five clonally unrelated VH4-59 VK1-9 antibodies with different D and J segments and shorter CDRH3s but matched CDRH1/2s and CDRL1/2s. These came from B cells sorted from CoV1 RBD-immunized mice on days 7 or 20 that did not cross-react with CoV2 RBD tetramers and were much less effective at cross-reactive binding or blocking EY6A than the unmutated ancestor of AB-3467 (Table S3B). Compared to AB-3467, the highest spike binder expressed from the AB-3467 clonal lineage, AB-3623, had a divergent mutational profile outside of CDRH3 (Figures 5C and 5D) but showed similar affinity (Figure S5G) and competition with CR3022 and ACE2 (Figures 6C and 6D) and had comparable neutralization potency against live CoV2 (Figure S6K). Collectively, these data indicate that the long CDRH3 sequence generated by VDJ recombination and preserved without mutation in AB-3467 and most of the 259 clonally related B cells is a dominant feature conferring binding to the conserved class 4 site in diverse sarbecovirus RBDs.
Class 4 antibodies neutralized CoV2 variants of concern
The binding footprint of AB-3467 was highly conserved across 192,000 sarbecovirus RBD genomes (Table S1A) and separate from the mutations on the rim of the RBS, including K417N, E484K, or N501Y, that provide an escape from class 1 and class 2 antibody binding (Figure 6F). In biolayer interferometry, the affinity of AB-3467 was neither hindered by K417N, E484K, or N501Y mutations (Figure 7C). AB-3467 potently neutralized lentiviral particles bearing SARS CoV1 or CoV2 spike proteins (Figure 7D). Likewise, AB-3467 retained live CoV2 virus neutralization potency against each of the current CoV2 variants of concern: D614G, B.1.17 (alpha), B.1.351 (beta), B.1.617 (kappa), B.1.617.2 (delta), P1 (gamma), P2 (zeta), B.1.525 (eta), B.1.427 (epsilon), B.1.429 (epsilon), and C36 variants (Figure 7E), with a maximum of a 2-fold drop in IC50 against any of these variants. By contrast, class 1, class 2, and class 3 antibodies show 10-fold (Chen et al., 2021; Greaney et al., 2021) or 4-to-6-fold (Chen et al., 2021) drops in potency to these lineages.
Discussion
The importance of vaccines that target conserved epitopes has become apparent with the increasing predominance of viral mutations that decrease the neutralizing efficiency of antibodies against CoV2, particularly toward epitopes around the RBS (Greaney et al., 2021; Liu et al., 2021). Given the ongoing risk of CoV2 evolution in animal hosts (Oude Munnink et al., 2021) and the potential for the emergence of related viruses (Wahl et al., 2021; Wang et al., 2018), elicited antibodies should target not only the current CoV2 lineages but display resistance to novel variants of concern.
Of the cross-neutralizing epitopes on the RBD, antibodies targeting the class 4 epitope (Barnes et al., 2020) hold considerable hope at fulfilling this goal. This epitope is highly conserved, possibly because of its interaction with the S2 subunit in the pre-fusion spike conformation when the RBD is in the “down” state. Antibodies to this epitope show considerable affinity for both CoV1 and CoV2 and the bat precursor virus RaTG13 (Liu et al., 2020), highlighting their potential resistance to future mutational escape.
Here, we showed that immunization strategies with divergent sarbecovirus RBDs induced antibodies to the conserved class 4 epitope with a range of recurring IGHV/IGKV combinations. These antibodies were not only stimulated by CoV2 RBD immunizations but also by immunization with diverse sarbecovirus RBDs, extending recent work showing that nanoparticles containing WIV1, Rf1, RmYN02, and Pang17 RBDs could result in CoV2-specific antibodies (Cohen et al., 2021). In this study, we showed that immunization with CoV1 and RaTG13 RBDs actually resulted in a higher proportion of the response targeting this class 4 epitope compared to immunization with the CoV2 RBD. The potently neutralizing class 4 AB-3467 antibody lineage described here, similar to previously described cross-reactive antibodies (Pinto et al., 2020; Rappazzo et al., 2021; ter Meulen et al., 2006; Wec et al., 2020; Wrapp et al., 2020), was induced by CoV1 RBD exposure. Optimal RBD immunization strategies could therefore require vaccinations with non-CoV2 RBDs. Our results addressed the nature of CoV1 RBD-cross-neutralizing polyclonal antibodies detected in the sera of macaques immunized with the CoV2 RBD conjugated to ferritin in protein nanoparticles or with an isolated CoV2 RBD given as an mRNA vaccine (Saunders et al., 2021). The serum titers are 20-fold lower than against CoV2, indicating that a minority of the response recognizes conserved epitopes, but these antisera did measurably block binding of a class 4 antibody DH1047 more than convalescent patient serum. Later manuscripts have emerged suggesting that the macaque repertoire is pre-disposed toward the generation of CoV1/CoV2 cross-reactive responses compared to humans and mice and suggest caution interpreting macaque models of cross-reactive neutralizing antibodies compared with data elicited from human antibody repertoire mice (He et al., 2021). In contrast, we found that the IGHV regions accounting for 76% of class 4 antibodies were equally represented in the naive repertoire of humans and Ig-humanized mice, suggesting these are a more appropriate model of cross-reactive responses. While these previous studies provide limited analysis of the nature of class 4 antibodies elicited by RBD immunization and are limited to CoV2 RBD immunization, their results complement the findings here that second-generation immunization strategies employing diverse RBDs elicit a small but consistent expansion of class 4 antibodies with long heavy chain CDR3s targeting the RBD beta-sheet backbone to confer broad reactivity and orient light chains for the steric blockade of the ACE2 binding site.
Although we were able to identify and express numerous antibodies to the class 4 epitope, with most bearing high affinity to diverse sarbecovirus RBDs, the great majority were not neutralizing. This corresponds to the characteristics of antibodies isolated from convalescent patients in the published antibody database (CoV-AbDab) (Raybould et al., 2021) wherein 17% (428/2523) of antibodies described to bind CoV2 also bind to CoV1, but only 6% (57/899) of neutralizing CoV2 antibodies show any binding to CoV1. Taken together, these results highlight the challenges to generating neutralizing antibodies to conserved sarbecovirus epitopes. AB-3467 and four neutralizing class 4 antibodies that have been isolated from convalescent humans reveal a shared mechanism: concurrently binding the class 4 epitope and blocking ACE2 interactions with the RBS by steric hindrance (Jette et al., 2021; Liu et al., 2020; Saunders et al., 2021). In this study we found and characterized two antibodies, AB-3467 and AB-3623, induced by immunization with these unusual properties and similarly broad and potent neutralizing activity from the same expanded clone but with highly divergent CDR1 and CDR2 mutational profiles. By contrast, antibodies with the same IGHV and IGKV but different CDRH3 sequences lacked these properties. The other described neutralizing class 4 antibodies show a wide range of IGHV/IGKV/IGLV pairings (Jette et al., 2021; Liu et al., 2020; Wrapp et al., 2020), indicating that this unusual mechanism for virus neutralization can be conveyed by a wide variety of V-regions and mutational profiles.
Despite their divergent CDR1 and CDR2 sequences, AB-3467 and the other potently neutralizing class 4 antibodies so far described possess an extended CDRH3 which forms a fifth antiparallel beta sheet extending the four antiparallel beta sheets at the core of the coronavirus RBD. Antiparallel binding of CDRH3 orientates the antibody H-chain so that the light chain is projected toward the ACE2 RBC, whereupon it sterically blocks ACE2 binding without depending on contacts with the variable class 1 and class 2 antibody epitopes. AB-3467 was distinct in this manner because its binding interface consisted almost exclusively of the antibody heavy chain. As such, AB-3467 was able to function similarly to camelid single-domain antibodies (nanobodies) (Wrapp et al., 2020) while still maintaining the advantages of being a fully human antibody.
A focus of next-generation vaccination strategies should be the induction of class 4 antibodies that prevent ACE2 interactions through steric hindrance, similar to the AB-3467 clonal lineage induced here. The focus on highly conserved spike protein structural elements provides these antibodies with resistance to viral mutations, superior to that of antibodies binding the class 1, class 2, and class 3 epitopes, which show significant reductions in potency to emerging strains (Chen et al., 2021; Greaney et al., 2021). However, the requirement for a long CDRH3 makes these antibodies less common in the circulating naive B cell repertoire. Indeed, our flow-cytometric analyses indicated that a small percentage of heterologous RBD-elicited class 4 antibodies block ACE2 binding, and we found only a single expanded clone of 260 cells with these properties out of >7,500 sequenced RBD-binding GC B cells, in contrast to non-neutralizing class 4 antibodies, which were frequently induced by a range of immunization regimes and employed more common CDRH3 lengths and IGHV/IGKV pairs. Long CDRH3s increase the likelihood of self-reactivity (Mouquet et al., 2010) and downregulation of surface IgM on naive B cells (Zhang et al., 2021), so uncommon B cell precursors of neutralizing class 4 antibodies may need especially potent stimulation to overcome tolerance checkpoints and undergo clonal redemption from self-reactivity by somatic hypermutation (Burnett et al., 2018). The findings here in humanized mice set out a structurally defined pathway for these successful next-generation COVID immunization strategies.
Limitations of the study
Sheep erythrocyte immunizations represent a well-characterized, adjuvant-free method for eliciting potent, reproducible antibody responses in rodents (Paus et al., 2006; Yi et al., 2015) and have been shown to elicit potent antibody responses in humans (Leikola and Aho, 1969). However, the antigenic complexity of xenogeneic erythrocytes and their cross-reactivity with human blood cell antigens may preclude their use for large-scale vaccination (Hoffman et al., 1973; Villa and De Biasi, 1983; Yi et al., 2015). Given the evidence that the CoV2 RBD mRNA immunization of macaques elicits comparable titers of CoV1-neutralizing serum antibodies to stabilized spike mRNA vaccination (Saunders et al., 2021), the class 4 antibody responses demonstrated here may be more potently elicited by second-generation mRNA vaccines encoding CoV1 RBDs tethered to the plasma membrane.
STAR★Methods
Key resources table
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Anti-human CD21-BV421 (Clone B-ly4) | BD Bioscience | Cat#562966; RRID: AB_2737921 |
| Anti-human IgD-BV510 (Clone IA6-2) | BD Bioscience | Cat#563034; RRID: AB_2737966 |
| Anti-human CD10-BV605 (Clone HI10a) | BD Bioscience | Cat#562978; RRID: AB_2737929 |
| Anti-human CD19-BV711 (Clone SJ25C1) | BD Bioscience | Cat#563038; RRID: AB_2737970 |
| Anti-human CD20-APC-H7 (Clone H27) | BD Bioscience | Cat#560853; RRID: AB_10561681 |
| Anti-human IgG-BV786 (Clone G18-145) | BD Bioscience | Cat#564230; RRID: AB_2738684 |
| Anti-human CD27-PE CF594 (Clone M-T271) | BD Bioscience | Cat#562297; RRID: AB_11154596 |
| Anti-human CD3-BB700 (Clone HIT3a) | BD Bioscience | Cat#742207; RRID: AB_2871428 |
| anti-human IgK FITC (Clone MHK-49) | Biolegend | Cat#316506; RRID: AB_493611 |
| Anti-mouse IgK-biotin (Clone 187.1) | BD Biosciences | Cat#559750; RRID: AB_397314 |
| Anti-mouse IgG1-BUV395 (Clone 10.9) | BD Biosciences | Cat#743265; RRID: AB_2741388 |
| Anti-mouse Fas-PeCy7 (Clone Jo2) | BD Biosciences | Cat#557653; RRID: AB_396768 |
| Anti-mouse CD38-BV510 (Clone 90/CD38) | BD Biosciences | Cat#740129; RRID: AB_2739886 |
| Anti-mouse CD4 -AF700 (Clone RM4-5) | BD Biosciences | Cat#557956; RRID: AB_396956 |
| Anti-mouse B220-BUV737 (Clone RA3-6B2) | BD Biosciences | Cat#612839; RRID: AB_2870161 |
| Anti-mouse CD11b-PE (Clone M1/70) | BD Biosciences | Cat#553311; RRID: AB_394775 |
| Anti-mouse TCRB-BV711 (H57-597) | BD Bioscience | Cat#109243; RRID: AB_2629564 |
| Anti-mouse IgD APCCy7 (Clone 11-26c.2a) | Biolegend | Cat#405716; RRID: AB_10662544 |
| unlabelled anti-CD16/32 | ebioscience | Cat#14016186; RRID: AB_467135 |
| unlabelled anti-CD16/32 | BD Biosciences | Cat#553142; RRID: AB_394657 |
| Biological samples | ||
| Convalescent donor blood samples | The Kirby Institute, UNSW | https://kirby.unsw.edu.au/project/natural-history-cohort-following-sars-cov-2-infection |
| Chemicals, peptides, and recombinant proteins | ||
| Streptavidin- alkaline phosphatase | Sigma | Cat#S2890 |
| Sigmafast P-Nitrophenl Phosphate Tablets | Sigma | Cat#N1891 |
| Fixable Viability Stain 700 | BD Bioscience | Cat#564997 |
| 7AAD | Biolegend | Cat#420403 |
| N-(3-Dimethylaminopropyl)-N-ethylcarbodimide hydrochloride | Sigma | Cat#E7750 |
| Horse Blood in Alsevers | Applied Biological Products | Cat#HBBX0050 |
| Sheep Blood in Alsevers | Applied Biological Products | Cat#SHBA0050 |
| Bovine Serum Albumin | Bovogen | Cat#BSAS-NZ |
| SA-BV605 | Biolegend | Cat#405229 |
| protein G Sepharose | Genscript | Cat# L00209 |
| pCEP4 mammalian expression vector | Thermo Fisher Scientific | Cat#V04450 |
| EZ-Link NHS-PEG4-Biotinylation reagent | Thermo Fisher Scientific | Cat#21330 |
| Critical commercial assays | ||
| Chromium Single Cell 5′ Gel Bead and Library Kit v2 | 10x Genomics | Cat#1000263 |
| AF488 or AF647 conjugation kits | Invitrogen | Cat#A20181 |
| High-binding plates | Corning | Cat#3700 |
| 70 μm cell strainer | Falcon, Corning, NY, USA | Cat#352350 |
| ZebaSpin columns | Thermo Fisher Scientific | Cat#89890 |
| Amicon Ultracel centrifugal columns | Merck Millipore Ltd | Cat#UFC501096 |
| Deposited data | ||
| Crystallography | PDB entry | Accession # 7msq |
| SC-RNaseq data | Mendeley data | https://doi.org/10.17632/kxcmhj7p27.1 |
| Experimental models: Cell lines | ||
| Expi293 cells | Thermo Fisher Scientific | Cat#A14527 |
| Lenti-X 293T | Clontech Laboratories | Cat#632180 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6JAusb | Australian BioResources | https://www.abr.org.au/animals/inbred-mice |
| Mouse: Balb/C | Australian BioResources | https://www.abr.org.au/animals/inbred-mice |
| Mouse: FVB | Australian BioResources | https://www.abr.org.au/animals/inbred-mice |
| Mouse: Trianni | Dr Hans Martin-Jack | https://trianni.com/ |
| Mouse: B6.Cg-Tg(K18-hACE2)2Prlmn/J | JAX Laboratories | Cat#034860 |
| Mouse: CR3022 Knock in | This paper | N/A |
| Recombinant DNA | ||
| Spike protein plasmids | BEI Resources | Amanat et al., 2020 |
| Software and algorithms | ||
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo version 10.7.1 | Tree Star, Inc | https://www.flowjo.com/ |
| R 4.0.2 | The R Foundation | https://cran.r-project.org/bin/windows/base/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Deborah L. Burnett (d.burnett@garvan.org.au).
Material availability
CR3022 knock in mice generated in this study are available upon request and completion of a routine MTA.
Experimental model and subject details
Mice
All mice used were housed at Australian BioResources and held at the Garvan Institute of Medical Research in specific pathogen-free environments. TRIANNI mice (San Francisco, CA, USA), gained with permission from Hans-Martin Jack, expressing a fully humanized variable antibody repertoire (Peter et al., 2021), were crossed with mice with a mutation in the α1,3 galactosyltransferase (α1,3GT) gene and lacked α-Gal epitopes on glycosylated proteins normally present in non-primate mammals (Saleh et al., 2020). This ensured that they could form a more accurate model of a human antibody repertoire which is not tolerant to this epitope. Hemizygous male and female K18-hACE2 mice (B6.Cg-Tg(K18-hACE2)2Prlmn/J, JAX stock #034860) were obtained from Jackson Laboratory. Transgenic mice expressing the rearranged heavy and light chain genes of CR3022 were produced by the Mouse Engineering Garvan/ABR (MEGA) Facility using CRISPR/Cas9-mediated gene targeting in C57BL/6J embryos. Rearranged CR3022 L-VDJ heavy and L-VJ kappa exons were linked to 5′ mouse Ighv and Igkv promoters and inserted 3′ of the endogenous Ighj4 and Igkj5 segments, respectively. Mice hemizygous for both targeted loci were utilized for analysis. Mice were used between the ages of 7-12 weeks with both male and female mice included. Mice were excluded if they showed any evidence of disease prior to recruitment.
Human samples
Convalescent COVID-19 donors The COSIN (Collection of COVID-19 Outbreak Samples in NSW) study is an ongoing prospective cohort study evaluating the natural history of SARS-CoV-2 infection in New South Wales, Australia. Individuals with SARS-CoV-2 infection (confirmed by NAT) were eligible for enrolment, irrespective of disease severity. Participants were enrolled through seven hospital in- and outpatient departments and referring microbiology laboratories in New South Wales between 6th March 2020 and 17th September 2020. In this study two female and one male patient with mild disease between the ages of 55 to 78 were used. Samples were collected at follow up visits 4 months following infection in August 2020.
Blood from two male and one female healthy control, between the ages of 25-48, were collected in July 2020 in Sydney, Australia, where local transmission was very low at the time. None of these healthy controls had a history of COVID-19, were not close contacts of cases of COVID-19 and were not health care workers. None had been given any COVID-19 vaccines.
Ethics statement
The Garvan Animal Ethics or Sydney Local Health District Animal Welfare Committees approved all mice protocols and procedures.
The protocol was approved by the Human Research Ethics Committees of the Northern Sydney Local Health District and the University of New South Wales, NSW Australia (ETH00520) and was conducted according to the Declaration of Helsinki and International Conference on Harmonization Good Clinical Practice (ICH/GCP) guidelines and local regulatory requirements. Written informed consent was obtained from all participants before study procedures.
Method details
Production of novel GM mouse lines using CRISPR/Cas9
CR3022 knock in GM mouse lines were produced by the Mouse Engineering Garvan/ABR (MEGA) Facility (Moss Vale and Sydney, Australia) by CRISPR/Cas9 gene targeting in C57BL/6J mouse embryos following established molecular and animal husbandry techniques (Yang et al., 2014). Embryos for microinjection were produced by mating stud males with super-ovulated C57BL/6J females. Stud males were wild-type C57BL/6J mice. All embryos were microinjected with in vitro transcribed and polyadenylated mRNA encoding S.pyogenes Cas9 and either one or two in vitro transcribed sgRNAs. Microinjected embryos were cultured overnight and introduced into pseudo-pregnant foster mothers. Pups were screened by PCR and Sanger sequencing of ear-punch DNA to identify founder mice which were then crossed to C57BL/6J mice to establish each line.
Recombinant proteins
CoV sequences were taken from the literature in the field (Amanat et al., 2020; Zhang et al., 2020). Open reading frame of CoV proteins were cloned into the pCEP4 mammalian expression vector (Thermo Fisher Scientific) with a N-terminal IgG leader sequence and C-terminal Avitag and His tag for purification and transiently expressed in Expi293 cells (Thermo Fisher Scientific). Transfections were performed as per the manufacturer’s instructions and the protein expressed for 7 days at 37°C, 5% CO2. Recombinant CoV proteins were purified from cell culture supernatant using Talon resin (Takara). Fractions were further gel-filtration purified a S200 26/60 column plumbed with 25 mM Tris (pH 8.0), 150 mM NaCl as the running buffer. Finally, peak fractions were pooled and concentrated with spin filters (Amicon Ultracel 10KMWCO, Merck). The plasmid encoding the spike protein with C-terminal trimerization domain and His tag was a gift from the Krammer lab (BEI Resources) (Amanat et al., 2020). The protein was further purified on a Sepharose 6 gel filtration column (GE Healthcare) using an AKTA Pure FPLC instrument (GE Healthcare) to isolate the trimeric protein.
Proteins were biotinylated by incubating for 30 min at room temperature with EZ-Link NHS-PEG4-Biotinylation reagent (Thermo Fisher Scientific) at a 10:1 biotin-to-protein ratio. Free biotin was removed from the samples by repeating the buffer exchange step in a second ZebaSpin column equilibrated with PBS. Following biotinylation proteins were conjugated to the relevant streptavidin fluorophores at a 4:1 molar ratio for 1h at 4 degrees. Excess biotinylated protein that was not coupled to streptavidin was removed by size exclusion through 30KMWCO Amicon Ultracel centrifugal columns (Merck Millipore Ltd). All recombinant protein fluorophores were titrated prior to use. Tetramers were stored for a maximum of 4 weeks prior to use.
Antibodies were transiently expressed as human IgG1 in HEK293 cells using standard plate transfection and the Expi system (LifeTechnologies) and purified with protein G Sepharose (Genscript) according to the manufacturers’ recommendations. Buffer exchange was performed using Genescript desalting preparation columns. Quality control of proteins included SDS-Page and western blots. Samples that failed to meet QC or were unable to be expressed at high concentrations (< 0.01mg/mL) were removed from the analysis. Samples were frozen at −80 degrees prior to use.
Affinity measurements using biolayer interferometry (BLI)
Purified monoclonal IgG antibodies were buffer exchanged into PBS using equilibrated ZebaSpin columns (Thermo Fisher Scientific). Affinity of interactions between biotinylated antibodies and purified soluble RBD proteins were measured Biolayer Interferometry (BLItz, ForteBio). Streptavidin biosensors were rehydrated in PBS containing 0.1% w/v BSA for 1 h at room temperature. Biotinylated antibody was loaded onto the sensors “on-line” and global fits were obtained for the binding kinetics by running associations and dissociations of RBD proteins at a suitable range of molar concentrations. The global dissociation constant (KD) for each 1:1 binding interaction was determined using the BlitzPro 1.2.1.3 software. For competition assays, biotinylated ACE2-Fc or CR3022 antibody was loaded onto the streptavidin sensors on-line, and the binding kinetics determined using either 500 nM of soluble RBD, or 500nM of soluble RBD pre-incubated with 1μM of IgG for 5min, using advanced kinetics protocol.
Structural studies
Crystals were grown in a vapor-diffusion hanging-drop format by combining equal volumes (2 uL) of protein complex (∼5.5 mg/mL) with well solution (100 mM BisTrisPropane (pH 6.9), 800 mM KSCN, 10% (v/v) glycerol, 18% (w/v) PEG8000). Initial crystallization hits were obtained with a sparse-matrix commercial screen (PACT-premier (Molecular Dimensions), conditions E4, F4, G4 and H4), dispensed with a Mosquito liquid handling robot (TTP Labtech), before optimizations were performed in 24 well plates. No explicit cryoprotection protocol was employed, and crystals were directly flash frozen in liquid nitrogen. X-ray diffraction data were recorded at the Australian Synchrotron on beamline MX2 using a Dectris Eiger X16M detector. A 360 degree sweep of data was recorded then deconvolution into 3600 images (0.1° degree each). Reflections were indexed and integrated using XDS (Kabsch, 2010). Space groups were determined with POINTLESS (Evans, 2011) and scaling and merging were performed with AIMLESS (Evans and Murshudov, 2013), both part of the CCP4 suit of software (Winn et al., 2011). Data collection statistics are shown in Table S4. Structures were determined by molecular replacement using PHASER (McCoy et al., 2007). The search model for the RBD component was derived from PDB entry 7kzb (Rouet et al., 2021), while the Fab components were derived from PDB entry 7czx where the heavy and light chains were split into variable (VH + VL) and constant (CH1 + CL) domain pairings. Two RBD-Fab complexes were found in the asymmetric unit, consistent with a solvent content of ∼54%. The model was iteratively improved via rounds of refinement performed with REFMAC5 (Murshudov et al., 2011) and manual real-space inspection and adjustment performed with COOT (Emsley et al., 2010). Model and refinement statistics are shown in Table S4. The final model comprises two essentially identical RBD-Fab complexes.
Sheep red blood cells (SRBCs) conjugation and binding assays
Conjugation was performed as described previously (Burnett et al., 2018). Sheep Red Blood Cells (SRBCs) were washed in of Phosphate Buffered Saline (PBS) three times by centrifugation at 2,300 rpm (1,111 g) for 5 min at 4°C and then once in conjugation buffer. SRBCs were then resuspended in a final volume of 1 mL conjugation buffer containing 10-30μg/mL of protein for conjugation. The solution was mixed on a platform rocker on ice for 10 minutes. 10 mg N-(3-Dimethylaminopropyl)-N-ethylcarbodimide hydrochloride (EDCI) (Sigma) was then added and the solution was mixed for a further 30 minutes on ice. SRBCs were then washed four times in PBS. Confirmation of successful conjugation was performed by flow-cytometric analysis of SRBCs and shown to be proportional to the concentration of conjugated antigen. Mice were immunized with 200,000 SRBCs per mouse intravenous via retroorbital injection. For day 7 analyses mice were immunized once on day 0 and harvested on day 7. For day 20 analyses mice were immunized RBD conjugated SRBCs on days 0 and 6, and RBD conjugated horse red blood cells (HRBC) on days 10 and 15.
For assays to assess the binding of expressed antibodies to CoV protein conjugated SRBCs, 10,000 SRBCs were aliquoted into individual wells of a 96 well plate. 50uL of 10ug/mL of the relevant antibodies were added to each well and incubated for 30 mins on ice. Samples were then washed by the addition of 100uL PBS and centrifuged at 2,300 rpm (1,111 g) for 1 min at 4°C. A further incubation was then performed by adding 50uL per well of a 1:50 dilution of FITC anti-human IgK (Biolegend, Clone MHK-49), followed by a further wash step prior to transfer to tubes for flow cytometry.
For antibody competition assays by flow cytometry, 100ug of EY6A or S309 IgG1 antibody was conjugated to AF488 or AF647 conjugation kits (Invitrogen), ACE2-FC dimers were biotinylated and incubated for 30 minutes with SA-BV605 (Biolegend). Antibodies and ACE2 were titrated against CoV2 RBD-conjugated SRBCs to determine a sub-saturating dilution of the fluorescent antibody to be used to measure competition with other unconjugated antibodies. The mean fluorescent intensity of binding of these fluorescent antibodies to SARS-CoV-2 RBD conjugated SRBCs was measured after incubating the SRBCs with competing unconjugated antibodies at 10ug/mL for 30 minutes on ice, compared to the MFI with no competing antibody measured in parallel. % inhibition = MFI test/MFI no competitor x 100.
For assessment of the effects of EDCI on RBD proteins CoV2 RBD was resuspended at 10ug/mL in 1mL conjugation buffer. 10mg of EDCI was added (or sample left as a control) and the sample was mixed for a further 30 minutes on ice. Following this incubation 2ug/mL EDCI conjugated or unconjugated CoV2 RBD was added to 1 million splenocytes from a mouse with a knocked in CR3022 heavy and light chain B cell receptor and sample was incubated for a further 30 minutes. Samples were then washed by the addition of 100uL PBS and centrifuged at 2,300 rpm (1,111 g) for 1 min at 4°C. Samples were then incubated with S309- AF647 or ACE2FC-BV605 generated as described above for 30 minutes, followed by a further wash step and transfer to flow tubes.
Flow cytometry
On the day of harvest organs were collected into PBS with 1% Bovine Serum Albumin (BSA) (Bovogen), cell suspensions passed through a 70 μm cell strainer (Falcon, Corning, NY, USA) and centrifuged 1,500 rpm (440 g) for 5 min at 4°C. Fc receptors were blocked with unlabeled anti-CD16/32 (ebioscience or BD) before staining.
Single cell suspensions were labeled with the following anti-mouse antibodies: IgG1-BUV395 (10.9, BD Biosciences), Fas-PeCy7 (Jo2, BD Biosciences), CD38-BV510 (90/CD38, BD Biosciences) CD4 -AF700 (RM4-5, BD Biosciences), B220-BUV737 (RA3-6B2, BD Biosciences), CD11b-PE (M1/70, BD Biosciences), TCRB-BV711 (H57-597, BD Bioscience), IgD APCCy7 (11-26c.2a, Biolegend). Live dead discrimination was performed with 7AAD (Biolegend). B cells were determined as TCRB-, CD4-, CD11b-, B220+ cells. Germinal centers were identified as Fas+, CD38-, IgD-.
Cells were filtered using 35μm filter round-bottom FACS tubes (BD PharMingen) immediately before data acquisition on a LSR II analyzer (BD PharMingen) sorted samples were analyzed on a FACS ARIA II or III (BD PharMingen). Forward- and side-scatter threshold gates were applied to remove red blood cells and debris and approximately 2-5 × 106 events were collected per sample. Cytometer files were analyzed with FlowJo software (FlowJo LLC, Ashland, Oregon, USA).
For flow cytometric epitope binding assays spleens from immunized day 20 TRIANNI mice were incubated with SARS-CoV-2 RBD at 200ng/mL in 1% BSA. Unimmunized mice with knocked in CR3022 heavy and light chain B cell receptor were used as a staining control as affinity maturation of the CR3002 antibody has been shown to be able to mature to bind different epitopes (Rouet et al., 2021). IgG1-BUV395 (10.9, BD Biosciences), Fas-PeCy7 (Jo2, BD Biosciences), CD38-BV510 (90/CD38, BD Biosciences) CD4 -AF700 (RM4-5, BD Biosciences), B220-BUV737 (RA3-6B2, BD Biosciences), CD11b-PE (M1/70, BD Biosciences), TCRB-BV711 (H57-597, BD Bioscience), IgD APCCy7 (11-26c.2a, Biolegend) as well as S309 conjugated to AF647 and EY6A conjugated to AF488. Live dead discrimination was performed with 7AAD (Biolegend). Samples were then incubated with ACE2FC-BV605 generated as described above for 30 minutes. Between stains samples were washed by the addition of 100uL PBS and centrifuged at 2,300 rpm (1,111 g) for 1 min at 4°C. Gating of germinal center and memory B cells was performed as above.
For analysis of human samples, cryopreserved PBMCs were thawed rapidly in a 37-degree waterbath and washed with pre-warmed RPMI media supplemented with 2 mM L-glutamine, IU/mL penicillin, 50 μg/mL streptomycin and 10% heat inactivated fetal calf serum (Sigma). The cells were resuspended in PBS and counted. Single cell suspensions were labeled with the following anti-human antibodies at the indicated dilutions 1:10 CD21-BV421 (B-ly4, BD Bioscience), 1:10 IgD-BV510 (IA6-2, BD Bioscience), 1:10 CD10-BV605 (HI10a, BD Bioscience), 1:20 CD19-BV711 (SJ25C1, BD Bioscience), 1:10 CD20-APC-H7 (H27, BD Bioscience), 1:5 IgG-BV786 (G18-145, BD Bioscience), 1:25 CD27-PE CF594 (M-T271, BD Bioscience), 1:50 CD3-BB700 (HIT3a, BD Bioscience). Live dead discrimination was performed with Fixable Viability Stain 700 (BD Bioscience). Memory B cells were identified as CD3-, CD19+, CD20+, CD10+ IgD-.
Serum ELISA
High-binding plates (Corning, Corning, NY, USA) were coated with the relevant CoV recombinant proteins at 5ug/mL in a sodium Biocarbonate buffer (0.3% NaHCO3, 0.2% NaCO3, 0.01% MgCl2) overnight at 4 degrees. Wells were then blocked with 1% BSA in PBS for 1h at 35 degrees, followed by incubation with mouse serum. Bound serum antibody quantified using IgK-biotin (187.1, BD Biosciences) followed by Streptavidin- alkaline phosphatase (Sigma) and Sigmafast P-Nitrophenl Phosphate tablets (Sigma).
10x Genomics sequencing and analysis
For 10x experiments FACS-sorted single-cell germinal center B cell suspensions were used to generate barcoded single-cell 5′ cDNA libraries for each sample pool with the Chromium Single Cell 5′ Gel Bead and Library Kit v2 (10x Genomics). Libraries were assessed with an Agilent BioAnalyzer High Sensitivity DNA chip, pooled, and quantified with qPCR (KAPA Library Quantification Kit). Denatured libraries were loaded onto an Illumina NovaSeq 6000 and sequenced using a 150-cycle High-Output Kit. To process the sequencing data, we used the 10x Genomics cellranger pipeline (v2.1.0), comprising the mkfastq stage. Using cellranger mkfastq, raw base call files were demultiplexed into sample-specific FASTQ files. FASTQs were then processed with 10x Genomics cellranger vdj (v4.0.0) using a custom reference that included the human IGH and IGK variable (V/D/J) genes and the mouse Igh and Igk constant region genes. The resulting VDJ contigs were post-processed using stand-alone IgBLAST (v1.14) (Ye et al., 2013) to generate additional alignment details. Clonal lineages were defined by independent clustering of the IGH and IGK using cd-hit (Li and Godzik, 2006). IGH and IGK were subset by V gene, J gene and CDR3 length and CDR3 nucleotide sequences were clustered at 90% identity. Clonal trees were generated using the linearham package (Dhar et al., 2020). Trees were visualized using the ggtree package (Yu, 2020).
Plate-based VDJ single cell sequencing and analysis
Following flow cytometry sorting of single cells into 96-well plates, Smart-Seq2 was performed by following the protocol of Picelli et al. (Picelli et al., 2014) with the following modifications. Reactions were performed at half volumes, the IS PCR primer was reduced to 50nM final concentration and the number of PCR cycles increased to 28. Sequencing libraries were prepared using the Nextera XT Library Preparation Kit (Illumina) at one quarter of the recommended volume. Sequencing was performed using the Illumina NextSeq 500 instrumet with 150 bp paired-end reads to a median depth of ∼1 million reads per cell. Paired heavy and light chain sequences were assembled with mixcr. Paired fastqs for each cell were processed using mixcr (v3.0.9) using the analyze command with the shotgun option (Bolotin et al., 2017). The resulting contigs were then processed with IgBLAST as for the 10x VDJ datasets.
Bulk repertoire sequencing
Follicular B cells (CD19+ CD23+ CD21+) of 12-14 week old female naive TRIANNI mice were FACS-sorted and total RNA was extracted from 106 cells per sample. First-strand cDNA was prepared with mouse C-region specific primers containing unique molecular identifiers (UMI) and VDJ regions were amplified using pooled primers targeting human VH leader sequences (Kreer et al., 2020) containing UMIs.
NGS was performed on the Illumina MiSeq platform with a MiSeq Reagent Kit V32 × 300 bp paired-end (Illumina). Read Quality was ensured by FASTQC quality analysis. Quality filtering and Paired-end read merging was achieved with the pRESTO software package (Vander Heiden et al., 2014).
Error correction was achieved with UMIs as part of the cDNA synthesis (10pb) and multiplex PCR Primers (4bp). PCR and sequencing errors were corrected by UMI pattern matching and selecting reads containing valid UMIs. Reads with matching UMIS were corrected to the most abundant read per UMI (1704 - 1954 unique UMIs retained per sample). To exclude quantitative bias introduced during multiplex PCR reads were normalized to unique mRNA sequences utilizing 10bp UMIs. VDJ annotation was conducted with IMGT/HighV-QUEST (Alamyar et al., 2012) as well as the Change-O (Gupta et al., 2015) wrapper for IgBlast. The sequences were further analyzed with the ARGalaxy immune receptor pipeline(Moorhouse et al., 2014).
Analysis of IGHV gene usage
IGHV gene usage, as defined by IgBLAST, was compared to relevant naive repertoires. For humans, IGH repertoire sequencing from 114 healthy human controls was obtained from SRA (BioProject: PRJNA491287) and processed as previously reported (Nielsen et al., 2020). The naive compartment was defined as unmutated (< 0.5% median SHM) IgM clones and donors with at least 1000 unique IgM clones were retained (n = 100). For TRIANNI mice, naive B cells were sorted from an unimmunised mouse and sequences and analyzed using the 10x Genomics platform. To test if an IGHV gene’s usage differed between a response and the underlying naive repertoire, the odds ratio and P value were determined by a Fisher’s exact test using the fisher.test function from the stats package in R and plotted using the ggplot package with RStudio.
The IGHV usage among patient derived mAbs that bind RBD were extracted from the CoV-AbDab (release 16th June 2021) (Raybould et al., 2021). The csv file was downloaded and subset for mAbs meeting the following criteria: Binds to = SARS-CoV2 or SARS-CoV1, Protein + Epitope = RBD, Heavy V Gene = human IGHV, Origin = B cells from COV2 Patient. IGHV gene usage was quantified using the reported ‘Heavy V Gene’ and neutralization information was collected from the ‘Neutralizing Vs’ field. Data manipulation was performed in R using the tidyverse package.
SARS-CoV-2 infections in K18-hACE2 mice
Hemizygous male and female K18-hACE2 mice (B6.Cg-Tg(K18-hACE2)2Prlmn/J, JAX stock #034860) were obtained from Jackson Laboratory. Mice were housed in groups and fed normal rodent chow. At 6-8 weeks age, mice were intranasally inoculated with 1 × 104 PFU SARS-CoV-2 (Isolate AUS/VIC01/2020) in a 30 μL volume. Mice were weighed and monitored twice daily. Once mice lost 20% body weight, or showed any severe clinical disease, they were humanely euthanized and tissues collected for downstream processing.
SARS-CoV-2 viral-cell fusion assay
For SARS-CoV-2 viral-cell fusion assays, stable ACE2-expressing Hek293T cells were generated by lentiviral transductions and lentiviral particles pseudotyped with SARS-CoV-2 Spike envelope were produced by co-transfection with a GFP encoding lentiviral plasmids. Neutralization activity of sera was measured using a single round infection of ACE2-HEK293T with Spike-pseudotyped lentiviral particles. Virus particles were incubated with serially diluted antibodies for 1 hour at 37°C and then added onto ACE2-HEK293T cells. Following spinoculation at 1200 g for 1 hour at 18°C, the cells were moved to 37°C for 72 hours. Entry of Spike particles was imaged by GFP-positive cells (InCell Analyzer) followed by enumeration with InCarta software (Cytiva, USA). Neutralization was measured by reduction in GFP expression relative to control group infected with the virus particles without antibody treatment.
High content live SARS-CoV-2 neutralization assay
Antibodies were serially diluted and mixed in duplicate with an equal volume of virus solution at 1.25x104 TCID50/mL. After 1 hour of virus-antibody coincubation at 37°C, 40 μL were added to an equal volume of Vero E6 or HEK293T cells (5x103 cells in suspension) in 384-well plates for a final MOI = 0.05. After 72h, cells were stained with NucBlue (Invitrogen, USA) and the entire well was imaged with InCell Analyzer microscopy system (Cytiva). Nuclei counts were obtained for each well with InCarta software (Cytiva) as a proxy for measuring cytopathic effect. Counts were compared between test antibody, mock controls (defined as 100% neutralization), and infected controls (defined as 0% neutralization). Sample-mediated neutralization was calculated using the formula; % viral neutralization = (D-(1-Q))x100/D, where Q = nuclei count normalized to average of mock controls, and D = 1-Q for average of infection controls.
Quantification and statistical analysis
GraphPad Prism 8 (GraphPad Software, San Diego, USA) was used for data analysis. When the data were normally distributed, an unpaired Student’s t test was performed for analysis. When data was not normally distributed Welsh’s correction was applied. For all tests, p < 0.05 was considered as being statistically significant. Unless otherwise stated error bars represent arithmetic mean. For all figures, data points indicate individual mice. ∗ represents p < 0.05, ∗∗ represents p < 0.01, ∗∗∗ represents p < 0.001, ∗∗∗∗ represents p < 0.0001.
Acknowledgments
We thank the Garvan Institute Australian BioResources, Garvan Molecular Genetics, Flow Cytometry, and Garvan-Weizmann Single Cell facilities. We thank the Krammer lab for the plasmid-encoding CoV2 spike. Part of this work was performed on the MX2 beamline at the Australian Synchrotron, part of ANSTO. We thank L. Burnett for his comments on the manuscript. The authors would like to thank the study participants for their contribution to the research. They would like to acknowledge members of the study group. Protocol steering committee: Andrew R. Lloyd (The Kirby Institute), John Kaldor (The Kirby Institute), Greg Dore (The Kirby Institute), Tania Sorrell (Marie Bashir Institute, University of Sydney, Sydney, Australia), Jeffrey J. Post (Prince of Wales Hospital [POWH]), Bernard Hudson (Royal North Shore Hospital [RNSH], Sydney, Australia), Dominic Dwyer (NSWHP), Adam Bartlett (Sydney Children’s Hospital [SCH], Sydney, Australia), Sarah C. Sasson (UNSW), Nick Di Girolamo (UNSW), and Daniel Lemberg (SCH). Coordinating center: Marianne Byrne (clinical trials manager), Mohammed Hammoud (post-doctoral fellow and data manager), Andrew R. Lloyd (investigator), and Roshana Sultan (study coordinator). Site principal investigators: Jeffrey J. Post (POWH), Michael Mina (Northern Beaches Hospital [NBH], Sydney, Australia), Bernard Hudson (RNSH), Nicky Gilroy (Westmead Hospital, Sydney, Australia), William Rawlinson (NSWHP); Pam Konecny (St George Hospital [SGH], Sydney, Australia), Marianne Martinello (Blacktown Hospital, Blacktown, Australia), Adam Bartlett (SCH), and Gail Matthews (St Vincent’s Hospital [SVH], Sydney, Australia). Site coordinators: Dmitrii Shek and Susan Holdaway (Blacktown Hospital); Katerina Mitsakos (RNSH); Dianne How-Chow and Renier Lagunday (POWH); Sharon Robinson (SGH); Lenae Terrill (NBH); Neela Joshi (Westmead Hospital), Ying Li (Westmead Hospital), and Satinder Gill (Westmead Hospital); and Alison Sevehon (SVH).
This work received funding from National Health and Medical Research Council (NHMRC) grants 585490, 1157744, 190774, 1176351, 1176134, 1081858, 1016953, and 1113904; Australian Research Council Discovery grants 160104915 and 140103465; Garvan COVID-19 Catalytic Funding; Bill and Patricia Ritchie Family Foundation; University of Technology Sydney and Rainbow Foundation; the Snow Foundation; Bundesmininisterium für Bildung und Forschung (BMBF, 01KI2043); DFG (TRR130); the Bayersiche Forschungsstiftung (CORAdc); The Bavarian State Ministry for Science; and the BMBF-funded COVIM project (NaFoUniMedCovid19)
Author contributions
Conceptualization, D.L.B., K.J.L.J., D.B.L., R.R., R.B., R.A.B., T.H.W., P.M.H., H.-M.J., S.T., D.C., and C.C.G.; methodology, D.L.B., K.J.L.J., D.B.L., S.R.S., R.R., R.A.B., R.B., P.M.H., R.K., S.S., S.T., D.C., and C.C.G.; investigation, D.L.B., K.J.L.J., D.B.L., A.A., A.O.S., M.D.J., H.B., H.L., R.R., G.W., S.R.S., R.K., S.S., E.R., F.W., B.M.S., M.S., A.G.S., and P.S.; funding acquisition, D.L.B., K.J.L.J., R.R., R.B., R.A.B., P.M.H., T.H.W., H.-M.J., S.T., D.C., and C.C.G.; supervision, R.B., R.A.B., P.M.H., H.-M.J., T.H.W., S.T., D.C., and C.C.G.; writing – original draft, D.L.B., D.C., and C.C.G.; writing – review & editing, D.L.B., K.J.L.J., D.B.L., R.B., S.R.S., P.M.H., H.-M.J., S.T., D.C., and C.C.G.
Declaration of interests
All affiliations are listed on the title page of the manuscript. The authors and their immediate family members have no financial interests to declare. The authors have lodged a patent (Australian provisional patent application no. 2021901716) based on this work.
Published: October 29, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.10.019.
Supplemental information
Data and code availability
Crystallography for this project is uploaded to the PDB. Accession numbers are listed in the KRT.
The published article includes all datasets generated or analyzed during this study, including full scRNA-seq data in the accompanying tables, figures and supplementary material.
This study did not generate new code.
References
- Alamyar E., Duroux P., Lefranc M.P., Giudicelli V. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Rappuoli R. Immunodominant antibody germlines in COVID-19. J. Exp. Med. 2021;218:e20210281. doi: 10.1084/jem.20210281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asensio M.A., Lim Y.W., Wayham N., Stadtmiller K., Edgar R.C., Leong J., Leong R., Mizrahi R.A., Adams M.S., Simons J.F., et al. Antibody repertoire analysis of mouse immunization protocols using microfluidics and molecular genomics. MAbs. 2019;11:870–883. doi: 10.1080/19420862.2019.1583995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Mossman K., Baker M.L. Zooanthroponotic potential of SARS-CoV-2 and implications of reintroduction into human populations. Cell Host Microbe. 2021;29:160–164. doi: 10.1016/j.chom.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin D.A., Poslavsky S., Davydov A.N., Frenkel F.E., Fanchi L., Zolotareva O.I., Hemmers S., Putintseva E.V., Obraztsova A.S., Shugay M., et al. Antigen receptor repertoire profiling from RNA-seq data. Nat. Biotechnol. 2017;35:908–911. doi: 10.1038/nbt.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- Burnett D.L., Langley D.B., Schofield P., Hermes J.R., Chan T.D., Jackson J., Bourne K., Reed J.H., Patterson K., Porebski B.T., et al. Germinal center antibody mutation trajectories are determined by rapid self/foreign discrimination. Science. 2018;360:223–226. doi: 10.1126/science.aao3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.A., Gnanapragasam P.N.P., Lee Y.E., Hoffman P.R., Ou S., Kakutani L.M., Keeffe J.R., Wu H.J., Howarth M., West A.P., et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735–741. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A.M., Wang Y., Roskin K.M., Marquis C.P., Jackson K.J. The mouse antibody heavy chain repertoire is germline-focused and highly variable between inbred strains. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20140236. doi: 10.1098/rstb.2014.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar A., Ralph D.K., Minin V.N., Matsen F.A., 4th A Bayesian phylogenetic hidden Markov model for B cell receptor sequence analysis. PLoS Comput. Biol. 2020;16:e1008030. doi: 10.1371/journal.pcbi.1008030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2523. doi: 10.1016/j.cell.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N.T., Vander Heiden J.A., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S.H. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E., Hale C., Saito Y., Blachere N.E., Bergh M., Conlon E.G., Schaefer-Babajew D.J., DaSilva J., Muecksch F., Gaebler C., et al. Vaccine Breakthrough Infections with SARS-CoV-2 Variants. N. Engl. J. Med. 2021;384:2212–2218. doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W.-t., Yuan M., Callaghan S., Musharrafieh R., Song G., Silva M., Beutler N., Lee W., Yong P., Torres J., et al. Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques. bioRxiv. 2021 doi: 10.1126/scitranslmed.abl9605. 2021.2007.2005.451222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M.K., Schmidt D., Oettgen H.F. Production of antibody to sheep red blood cells by human tonsil cells in vitro. Nature. 1973;243:408–410. doi: 10.1038/243408a0. [DOI] [PubMed] [Google Scholar]
- Ikegame S., Siddiquey M.N.A., Hung C.T., Haas G., Brambilla L., Oguntuyo K.Y., Kowdle S., Chiu H.P., Stevens C.S., Vilardo A.E., et al. Neutralizing activity of Sputnik V vaccine sera against SARS-CoV-2 variants. Nat. Commun. 2021;12:4598. doi: 10.1038/s41467-021-24909-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jette C.A., Cohen A.A., Gnanapragasam P.N.P., Muecksch F., Lee Y.E., Huey-Tubman K.E., Schmidt F., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C., et al. Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies. Cell Rep. 2021;36:109760. doi: 10.1016/j.celrep.2021.109760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen M.D., Irving A., Montagutelli X., Tate M.D., Rudloff I., Nold M.F., Hansbro N.G., Kim R.Y., Donovan C., Liu G., et al. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. 2020;13:877–891. doi: 10.1038/s41385-020-00340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreer C., Döring M., Lehnen N., Ercanoglu M.S., Gieselmann L., Luca D., Jain K., Schommers P., Pfeifer N., Klein F. openPrimeR for multiplex amplification of highly diverse templates. J. Immunol. Methods. 2020;480:112752. doi: 10.1016/j.jim.2020.112752. [DOI] [PubMed] [Google Scholar]
- Leikola J., Aho K. Experimentally induced mononucleosis-like heterophile antibodies in man. Clin. Exp. Immunol. 1969;5:67–73. [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liu H., Wu N.C., Yuan M., Bangaru S., Torres J.L., Caniels T.G., van Schooten J., Zhu X., Lee C.D., Brouwer P.J.M., et al. Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. Immunity. 2020;53:1272–1280.e5. doi: 10.1016/j.immuni.2020.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A., Cai H., Sarkar R., Chen W., Cutler M., et al. Neutralizing Activity of BNT162b2-Elicited Serum - Preliminary Report. N. Engl. J. Med. 2021;384:1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhouse M.J., van Zessen D., IJspeert H., Hiltemann S., Horsman S., van der Spek P.J., van der Burg M., Stubbs A.P. ImmunoGlobulin galaxy (IGGalaxy) for simple determination and quantitation of immunoglobulin heavy chain rearrangements from NGS. BMC Immunol. 2014;15:59. doi: 10.1186/s12865-014-0059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H., Scheid J.F., Zoller M.J., Krogsgaard M., Ott R.G., Shukair S., Artyomov M.N., Pietzsch J., Connors M., Pereyra F., et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S.C.A., Yang F., Jackson K.J.L., Hoh R.A., Röltgen K., Jean G.H., Stevens B.A., Lee J.Y., Rustagi A., Rogers A.J., et al. Human B Cell Clonal Expansion and Convergent Antibody Responses to SARS-CoV-2. Cell Host Microbe. 2020;28:516–525.e5. doi: 10.1016/j.chom.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus D., Phan T.G., Chan T.D., Gardam S., Basten A., Brink R. Antigen recognition strength regulates the choice between extrafollicular plasma cell and germinal center B cell differentiation. J. Exp. Med. 2006;203:1081–1091. doi: 10.1084/jem.20060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter A.S., Roth E., Schulz S.R., Fraedrich K., Steinmetz T., Damm D., Hauke M., Richel E., Mueller-Schmucker S., Habenicht K., et al. A pair of noncompeting neutralizing human monoclonal antibodies protecting from disease in a SARS-CoV-2 infection model. Eur. J. Immunol. 2021 doi: 10.1002/eji.202149374. Published onlineAugust 6, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybould M.I.J., Kovaltsuk A., Marks C., Deane C.M. CoV-AbDab: the coronavirus antibody database. Bioinformatics. 2021;37:734–735. doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet R., Mazigi O., Walker G.J., Langley D.B., Sobti M., Schofield P., Lenthall H., Jackson J., Ubiparipovic S., Henry J.Y., et al. Potent SARS-CoV-2 binding and neutralization through maturation of iconic SARS-CoV-1 antibodies. MAbs. 2021;13:1922134. doi: 10.1080/19420862.2021.1922134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh F.M., Chandra P.K., Lin D., Robinson J.E., Izadpanah R., Mondal D., Bollensdorff C., Alt E.U., Zhu Q., Marasco W.A., et al. A New Humanized Mouse Model Mimics Humans in Lacking α-Gal Epitopes and Secreting Anti-Gal Antibodies. J. Immunol. 2020;204:1998–2005. doi: 10.4049/jimmunol.1901385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K.O., Lee E., Parks R., Martinez D.R., Li D., Chen H., Edwards R.J., Gobeil S., Barr M., Mansouri K., et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594:553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden J.A., Yaari G., Uduman M., Stern J.N.H., O’Connor K.C., Hafler D.A., Vigneault F., Kleinstein S.H. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 2014;30:1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa M.L., De Biasi S. Antigen-dependent colonies of human peripheral blood lymphocytes: an immunomorphologic study. Cell. Immunol. 1983;81:323–332. doi: 10.1016/0008-8749(83)90240-x. [DOI] [PubMed] [Google Scholar]
- Wahl A., Gralinski L.E., Johnson C.E., Yao W., Kovarova M., Dinnon K.H., 3rd, Liu H., Madden V.J., Krzystek H.M., De C., et al. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591:451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Li S.Y., Yang X.L., Huang H.M., Zhang Y.J., Guo H., Luo C.M., Miller M., Zhu G., Chmura A.A., et al. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol. Sin. 2018;33:104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., et al. VIB-CMB COVID-19 Response Team Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell. 2020;181:1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang H., Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc. 2014;9:1956–1968. doi: 10.1038/nprot.2014.134. [DOI] [PubMed] [Google Scholar]
- Ye J., Ma N., Madden T.L., Ostell J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T., Li J., Chen H., Wu J., An J., Xu Y., Hu Y., Lowell C.A., Cyster J.G. Splenic Dendritic Cells Survey Red Blood Cells for Missing Self-CD47 to Trigger Adaptive Immune Responses. Immunity. 2015;43:764–775. doi: 10.1016/j.immuni.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinformatics. 2020;69:e96. doi: 10.1002/cpbi.96. [DOI] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Wilson I.A. Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem. Biophys. Res. Commun. 2021;538:192–203. doi: 10.1016/j.bbrc.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020;30:1346–1351.e2. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Jara C.J., Singh M., Xu H., Goodnow C.C., Jackson K.J., Reed J.H. Human transitional and IgM(low) mature naive B cells preserve permissive B-cell receptors. Immunol. Cell Biol. 2021;99:865–878. doi: 10.1111/imcb.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Duyvesteyn H.M.E., Chen C.P., Huang C.G., Chen T.H., Shih S.R., Lin Y.C., Cheng C.Y., Cheng S.H., Huang Y.C., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystallography for this project is uploaded to the PDB. Accession numbers are listed in the KRT.
The published article includes all datasets generated or analyzed during this study, including full scRNA-seq data in the accompanying tables, figures and supplementary material.
This study did not generate new code.