Abstract
On the basis of biochemical, phenotypic, and 16S rRNA analyses, Helicobacter canis was isolated from Bengal cats with and without chronic diarrhea. Because the cats were coinfected with other potential pathogens, including Campylobacter helveticus, and because H. canis was isolated from nondiarrheic cats, the causal role of H. canis in producing the diarrhea could not be proven. Histologically, the colons of the four affected cats were characterized by mild to moderate neutrophilic, plasmacytic, and histocytic infiltrates in the lamina propria. Rare crypt abscesses were also noted for three cats but were a more prominent feature of the colonic lesions noted for the fourth cat. This is the first observation of H. canis in cats and raises the possibility that H. canis, like H. hepaticus and H. bilis in mice, can cause inflammation of the colon, particularly in hosts with immune dysregulation. Further studies are needed to determine the importance of H. canis as a primary enteric pathogen in cats and the role of cats in the possible zoonotic spread of H. canis to humans.
The bacterial genus Helicobacter contains at least 18 species (15). These organisms colonize the gastrointestinal tracts of several mammalian and avian hosts. The type species, Helicobacter pylori, and the ferret gastric pathogen H. mustelae have well-documented causal roles in the development of peptic ulceration and neoplasia (9, 11, 12, 18, 29, 43). Other Helicobacter spp. have been associated with enteritis (5, 40) and inflammatory bowel disease (IBD) (3, 41). Some helicobacters, such as H. canis (2), H. pullorum (37), H. heilmannii (35), and “H. rappini” and H. cinaedi (10), may be zoonotic. The original descriptions of H. canis were from the feces of healthy and diarrheic dogs (38) and a child with enteritis (2). H. canis also has been isolated from a dog with hepatitis (14). H. canis is closely related genetically to H. hepaticus, an enteric helicobacter which can produce hepatitis, hepatic neoplasia, and IBD in some mouse strains (8, 13). Because IBD is a common clinical finding in domestic cats (39) and because Helicobacter spp. are associated with IBD in mice and rats, the possible relationship between helicobacters and IBD in cats should be explored (3, 20, 34). The purpose of this study was to ascertain whether Helicobacter spp. could be isolated from cats with and without diarrhea.
CASE REPORT
Four Bengal cats (Asian leopard cat-domestic cat hybrids) were evaluated from a cattery with 20 other cats, 75% of which had episodic severe, watery, projectile, and mucus- and blood-tinged diarrhea during the preceding 6 months. Each of the four hospitalized cats examined in this study had a previous history of diarrhea. The younger of the four cats, cats 1 and 2, were 8-month-old intact females presenting clinically with vomiting, lack of appetite, weight loss, and severe dehydration, were underweight (23), and were aware of but uninterested in their surroundings. Cat 3 was a 2-year-old intact female with vesicular faucitis, bilateral serous ocular discharge, and mild conjunctivitis consistent with calicivirus infection (31) and mild feline acne. Cat 4 was a 4-year-old intact male. Both of the adult cats were hydrated, in good condition, and normally responsive. Abdominal palpation of all four cats revealed fluid- and gas-filled small intestines and no mesenteric lymphadenopathy. Diarrhea in cats 1 and 2 was persistent, with the consistency of water, a foul odor, and more than 10 bowel movements per day. Adult cats 3 and 4 had intermittent diarrhea; their diarrheic feces were soft to liquid and malodorous. Gastrointestinal endoscopy was performed on cats 3 and 4 by standard techniques. Fresh gastrointestinal biopsy samples were obtained during endoscopy of the stomachs, proximal small intestines, and colons of the cats. Feces also were collected from nine asymptomatic cats for Helicobacter sp. isolation.
MATERIALS AND METHODS
Clinical pathology.
Serum antibodies against viruses were evaluated by an indirect immunofluorescence assay with feline infectious peritonitis virus UCD1-infected Felis catus whole fetal (fcwf-4) cells as a substrate for feline enteric coronavirus (32) and feline immunodeficiency virus (FIV) Petaluma-infected fcwf-4 cells as a substrate for FIV (45). Feline leukemia virus was evaluated by an enzyme-linked immunosorbent assay for p27 antigen (25).
Parasites and ova were evaluated by fecal flotation on 70% sodium dichromate (specific gravity, 1.34)-saturated zinc sulfate solution and direct fluorescence (Merifluor C/G; Meridian Diagnostics, Cincinnati, Ohio) for Crytosporidium spp. Giardia antigen was evaluated by an enzyme immunoassay (ProSpecT; Alexon, Sunnyvale, Calif.).
Gross pathology.
Biopsy samples from cats 3 and 4 were either rapidly frozen in OCT medium (Sakura Finetech, Torrance, Calif.) in a 2-methylbutane bath within liquid nitrogen or collected into 10% formalin, fixed overnight, embedded in paraffin, cut into 5-μm thin sections, and stained with hematoxylin and eosin and Warthin-Starry stains. Cats 1 and 2 were euthanized with an intravenous overdose of barbiturates and immediately necropsied. Necropsy tissues were processed as described for biopsy specimens.
Bacterial cultures.
Fecal specimens (two each from the four cats) were plated onto seven different agars for bacterial culturing. These were MacConkey (PML, Rancho Cordova, Calif.), CVA (cefoperazone-vancomycin-amphotericin B; Remel Laboratories, Lenexa, Kans.), fresh BHI (brain heart infusion with 2.5 mg of trimethoprim, 5 mg of vancomycin, 1.25 IU of polymyxin B, and 2 mg of amphotericin B per ml), fresh brucella (fetal calf serum, trimethoprim, vancomycin, polymyxin B, and amphotericin B; Anaerobe Systems, San Jose, Calif.), prereduced anaerobically sterilized modified agar with cefoxitin, cycloserine, and fructose (CCFA; Anaerobe Systems), egg yolk, and FDA (5,000 mg of polymyxin B per ml, 10% vancomycin in water, 10% trimethoprim in ethanol, and amphotericin B). CVA agar was incubated at 37 and 42°C under microaerobic conditions in vented jars containing N2, H2, and CO2 (90:5:5). An additional nine cats without diarrhea were surveyed from the same colony approximately 6 months after the initial evaluation of the four cats with diarrhea. These nondiarrheic cats were specifically screened for Helicobacter spp. The biochemical tests and phenotypic characterization used were based on previously described media and methods used by us and others for identifying, characterizing, and naming other Helicobacter spp. (16). Colony morphology and Gram staining of bacteria were determined by use of organisms obtained after 72 h of incubation of blood agar under microaerobic conditions. Bile tolerance was determined by growth of organisms on 1.5% bile media (1.5% desiccated ox bile [Oxoid] in 5% blood agar). A sample was also placed in selenite broth. After overnight incubation, the selenite broth was subcultured onto XLT4 (xylose-lysine-Tergitol 4) agar (PML). FDA agar was incubated under microaerobic conditions at 42°C. Brucella and CCFA agars were incubated microaerobically at 37°C. When colonies appeared on FDA agar, they were subcultured to fresh brucella agar and kept at 37°C.
The presence in feces of Clostridium difficile toxin A was evaluated with a monoclonal antibody enzyme-linked immunosorbent assay-based C. difficile toxin A kit (PET reverse passive latex agglutination [RPLA]; Unipath, Tokyo, Japan). The presence of enterotoxigenic C. perfringens was evaluated with a Pet RPLA assay.
DNA extraction for PCR analysis.
DNA was extracted from cultured organisms with a High Pure PCR template preparation kit (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) according to the manufacturer’s directions. Briefly, the samples were lysed and incubated with 40 μl of proteinase K for 1 h at 55°C. Binding buffer (200 μl) was added to each sample and incubated for 10 min at 72°C before 100 μl of isopropanol was added (16). The samples were placed in filter tubes and centrifuged at 8,000 rpm for 1 min. The flowthrough was discarded, 500 μl of wash buffer was added to the samples, and the samples were centrifuged as before. This washing step was repeated three times. Elution of the DNA was achieved by adding 200 μl of elution buffer to the filter tubes and centrifuging the samples for 1 min at 8,000 rpm.
PCR amplification of bacterial DNA.
The primer sequence chosen for PCR amplification recognizes a region of the 16S rRNA gene specific for members of the Helicobacter genus. The sets of primers used amplified a product of 1.2 kb. PCR amplification was achieved by use of a previously described method (16). Briefly, 20 μl of the DNA preparation was added to 100 μl of a reaction mixture containing 1× Taq polymerase buffer (supplied by Pharmacia, Uppsala, Sweden, but supplemented with 1 M MgCl2 to a final concentration of 2.25 mM), 0.5 μM each primer, 200 μm each deoxynucleotide, and 200 μg of bovine serum albumin per ml. Samples were heated at 94°C for 4 min, briefly centrifuged, and cooled to 61°C. At this time, 2.5 U of Taq polymerase (Pharmacia) and 1.0 U of polymerase enhancer (Perfect Match; Stratagene, La Jolla, Calif.) were added, and then 100 μl of mineral oil was laid over the samples. For amplification of the 1.2-kb fragment, the following conditions were used: 35 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 3 min, and elongation at 72°C for 3 min, followed by an elongation step of 8 min at 72°C. A 15-μl aliquot of each sample was electrophoresed through a 1% agarose gel, stained with ethidium bromide, and viewed by UV illumination.
RFLP analysis.
The 1.2-kb PCR-amplified Helicobacter DNA from the two strains of H. canis isolated from the diarrheic cats, the two strains from the nondiarrheic cats, an H. canis strain from a dog, and H. canis ATCC 51401 was subjected to restriction fragment length polymorphism (RFLP) analysis. DNA digestion was accomplished by the addition of 10 U each of the restriction endonucleases HhaI and BfaI (New England Biolabs, Beverly, Mass.) and 1 μl of restriction buffer (New England Biolabs) to 16 μl of DNA and incubation at 37°C for 3 h. The samples were separated on a 6% Visigel separation matrix (Stratagene), stained with ethidium bromide, and viewed by UV illumination.
Genomic DNA extraction for 16S rRNA gene sequencing.
Bacteria isolated from both fecal samples from one cat were cultured on blood agar plates, and the cells were harvested and washed twice with 1 ml of double-distilled H2O. The pellets were suspended in STET buffer (8% sucrose, 50 mM EDTA, 0.1% Triton X-100, 50 mM Tris-HCl [pH 8.0]), and lysozyme (hen egg white; Boehringer) was added to a final concentration of 3 mg/ml. The suspension was incubated for 12 min at 37°C and then lysed with 1% sodium dodecyl sulfate. RNase A (bovine pancreas; Boehringer) was added to a final concentration of 0.05 mg/ml, and the solution was incubated for 1 h at 37°C. Then, 0.1 volume of a 5% cetyltrimethylammonium bromide–0.5 NaCl solution (Sigma Chemical Co., St. Louis, Mo.) was added, and the solution was gently mixed and incubated at 65°C for 10 min. DNA was extracted with an equal volume of phenol-chloroform (1:1, vol/vol), precipitated overnight in 0.3 M sodium acetate with 2 volumes of absolute ethanol at −20°C, and pelleted by centrifugation at 13,000 × g for 1 h at 4°C. The ethanol was decanted, and the pellet was air dried and suspended in sterile distilled water.
16S rRNA gene sequencing.
The sequences of the 16S rRNA genes of the two H. canis strains (MIT 98-152 and MIT 98-153) isolated from two fecal samples from one cat were determined. For the amplification of 16S rRNA cistrons, 16S rRNA gene sequencing, and 16S rRNA data analysis, we used the methods described by Fox et al. (16). Briefly, primers C70 and B37 (16) were used to amplify the 16S rRNA genes. The amplicons were purified and directly sequenced by use of a TAQuence cycle sequencing kit (U.S. Biochemicals, Cleveland, Ohio). The 16S rRNA gene sequences were entered into a program for the analysis of 16S rRNA data in Microsoft Quikbasic for use with IBM PC-compatible computers and aligned as previously described (30). The database used contains approximately 100 Helicobacter, Wolinella, Arcobacter, and Campylobacter sequences and more than 900 sequences for other bacteria. Similarity matrices were constructed from the aligned sequences by use of only base positions for which 90% of the strains had data and were corrected for multiple base changes by the method of Jukes and Cantor (21). Phylogenetic trees were constructed by the neighbor-joining method (33).
Nucleotide sequence accession number.
The 16S rRNA sequences for strains MIT 98-152 and MIT 98-153 have been deposited in GenBank under one accession number, AF177475, given that the two isolates are identical.
RESULTS
Clinical pathology.
Complete blood counts and serum chemistries were within normal limits for all four cats. The cats were serologically negative for feline leukemia virus antigen and antibodies against FIV. All four were seropositive for feline enteric coronavirus at a minimum dilution of 1:100.
Large numbers of Cryptosporidium sp. oocytes were observed on fecal flotation; these oocytes were fluorescence positive when tested with the fluorescence-tagged Cryptosporidium sp. antibody in all four cats. All cats were negative for Giardia spp. and Giardia antigen.
Gross pathology.
Gross lesions were similar in both adult cats on endoscopy and included fluid distention of the large and small intestines, thickened erythematous mucosal tissue in the duodenum, and diffuse, patchy mottling in the proximal colon. The distal colon, stomach, and esophagus were all within normal limits. Gross lesions observed during necropsy of the young cats were limited to fluid and gas distention of the small and large intestines. Other abdominal organs were within normal limits.
Histopathologic findings from colonic biopsies of the two adult cats (cats 3 and 4) and colonic tissues from the two young cats (cats 1 and 2) included mild to moderate neutrophilic, plasmacytic, and histiocytic infiltrates in the lamina propria and rare crypt necrosis. Neutrophilic infiltrates were clustered in some sections, but bacteria were not evident in these lesions. Bacterial overgrowth of luminal contents was notable and included spiral bacteria adherent to the mucosal surface. Colonic sections from one of the young cats had more prominent crypt necrosis and a focal, intramural pyogranulomatous infiltrate that effaced local lymphoid tissues. Small intestinal lesions in all of the cats included mild to moderate lamina propria infiltrates of plasma cells and neutrophils, transepithelial migration of leukocytes, and mild villus blunting. Rare crypt abscesses with peripheral fibrosis also were present in one young cat. All four cats had intraglandular and intraparietal cell spiral bacteria (presumed to be gastric helicobacter-like organisms) in the stomach, and one of the adult cats had associated gastric lesions that included plasmacytic and neutrophilic gastritis, minimal gland necrosis, and mild hyperplasia with intestinal metaplasia. Livers from the two young cats had minimal plasmacytic infiltrates in portal areas and prominent Kupffer cells.
Bacterial cultures.
Gram staining of the fecal smears from cat 4 showed mixed flora, with numerous large gram-positive rods consistent with Clostridium spp. There was growth of a small number of Clostridium colonies, but no clostridial toxins were detected in toxin assays. Cat 3 had mixed flora on fecal smear Gram staining, with large gram-positive rods with spores and large numbers of small and large spiral gram-negative rods. The feces were culture and toxin A positive for C. difficile.
Growth on FDA agar after 48 h at 37°C consisted of small white to clear watery colonies with spiral gram-negative slender rods. Subcultures grew at 42°C but not at 25°C. The isolates were oxidase positive and catalase and urease negative, did not grow in 3.5% sodium chloride or 1% glycine, were sensitive to 30 mg of nalidixic acid and 30 mg of cephalothin, and did not hydrolyze hippurate. Based on the data, the bacteria were identified as Campylobacter helveticus.
Small colonies were visible on CVA medium after 3 days of incubation under microaerobic conditions. These colonies were observed in fecal cultures for three of the four diarrheic cats and five of the nine nondiarrheic cats. Direct examination of the colonies revealed the presence of curved gram-negative rods. The isolates grew at 37 and 42°C but not at 25°C. The organisms were oxidase positive and catalase, urease, and indoxyl acetate negative, did not reduce nitrate to nitrite, did not hydrolyze hippurate, but did grow in the presence of 1.5% bile. The isolates were resistant to cephalothin (30 μg) but sensitive to nalidixic acid (30 μg). The organisms isolated from the feces were provisionally identified as H. canis.
PCR.
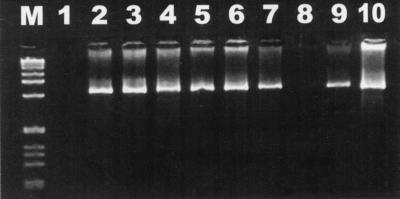
Bacteria isolated from seven of the eight fecal samples from the four diarrheic cats amplified a 1.2-kb product specific for Helicobacter spp. (Fig. 1). All nine fecal samples from the nondiarrheic cats had a similar Helicobacter-specific 1.2-kb product.
FIG. 1.
Electrophoresis of DNA amplified by Helicobacter-specific PCR on a 6% Visigel separation matrix with Helicobacter-specific primers. Lanes: M, 1-kb DNA ladder; 1, reagent control; 2, H. canis ATCC 51401; 3 to 10, DNAs extracted from feline fecal isolates.
RFLP analysis.
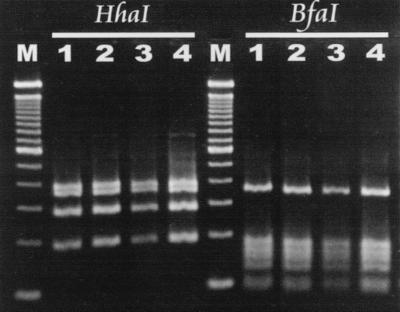
With the restriction enzymes HhaI and BfaI, the two strains from the diarrheic cats had patterns identical to those of an H. canis strain isolated from the liver of a dog (14) and H. canis ATCC 51401 (Fig. 2). Helicobacter spp. isolated from two cats without diarrhea had RFLP patterns consistent with H. canis isolated from two diarrheic cats (data not shown).
FIG. 2.
Comparison of RFLP patterns of the 1.2-kb Helicobacter-specific PCR product with restriction enzymes BfaI and HhaI. Lanes: M, 100-bp DNA ladder; 1, H. canis MIT dog isolate (14); 2, H. canis ATCC 51401; 3, MIT 98-152; 4, MIT 98-153.
16S rRNA analysis.
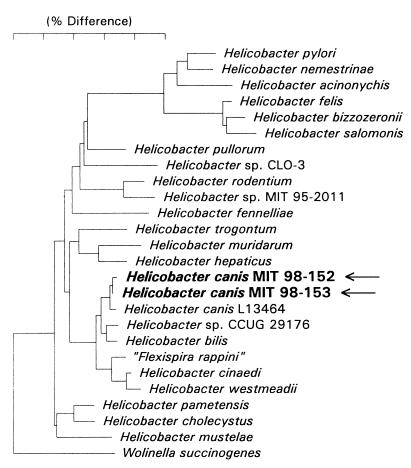
The sequence of the cat isolates (MIT 98-152 and MIT 98-153) differed from that of the type strain of H. canis (L13464) by 4 bases (Fig. 3). There is an intervening sequence (IVS) in the 16S rRNA gene in about one-third of H. canis strains (24). However, the two feline H. canis isolates did not have an IVS. Whether the H. canis strains with IVSs represent one or more distinct subspecies has not been determined.
FIG. 3.
Phylogenetic tree constructed on the basis of 16S rRNA sequence similarity values. The two H. canis strains from diarrheic cats are in bold. Scale bar, 5% difference in nucleotide sequences, as determined by measuring the lengths of the horizontal lines connecting two species.
DISCUSSION
To our knowledge, this is the first report of H. canis in cats, and it is significant for several reasons, including the possibility that the organism has zoonotic potential. This organism is related to other helicobacters previously associated with colitis and proctitis, including H. hepaticus (3, 17), H. cinaedi (40), H. fennelliae (5, 40), and H. bilis (20, 34). The murine IBD produced by H. hepaticus and H. bilis is most pronounced in immunosuppressed rodents (3, 8, 20, 34, 41). However, in immunocompetent mouse strains, H. hepaticus also can induce enterocolitis and typhlitis (17, 44). Mice also can develop hepatitis, hepatic adenomas, and hepatocellular carcinoma as a result of H. hepaticus infection (42). H. canis has been previously reported in a child with gastroenteritis (2), dogs with and without diarrhea (38), and a puppy with necrotizing hepatitis (14).
The clinically ill Bengal cats in the present report had severe diarrhea, enterocolitis, and mild portal hepatitis associated with multiple primary and opportunistic pathogens, including H. canis, Cryptosporidium spp., C. perfringens, C. difficile, and C. helveticus. A gram-negative organism similar to H. heilmannii was observed in the stomachs of these cats. Although gastric helicobacters are common in cats and are associated with mild to moderate inflammation, definite clinical signs have not been linked to helicobacter-associated gastritis in cats (27, 28). Feline enteric coronavirus antibody also was detected in the serum, but this is a common finding in cattery cats (7) and is not typically associated with clinical enteritis (31). Because each of the protozoal and bacterial organisms isolated from these affected cats has been associated with diarrhea, it is not clear which if any was the primary pathogen and how significant synergistic interactions among the intestinal flora were in causing clinical disease.
Cryptosporidium spp. cause diarrhea in humans and animals, but the condition is self-limiting unless the subject is immunosuppressed (19). Cryptosporidiosis with oocyst shedding occurs in normal and diarrheic cats, and experimental attempts to infect cats with C. parvum commonly result in shedding with no clinical signs (31). Similarly, C. perfringens is frequently detected in stools of normal cats, although toxin production may suggest that C. perfringens is at least partially responsible for some clinical signs. Outbreaks of C. perfringens enteritis have been reported for cats maintained in a cattery (6) and captive cheetahs (4). Pseudomembranous colitis due to C. difficile has been reported for humans with underlying disease or undergoing antibiotic therapy; the infection often results from nosocomial exposure (22). As with C. perfringens, the diarrhea and intestinal inflammation are due to an exotoxin produced by C. difficile. C. helveticus is a catalase-negative or weakly positive Campylobacter sp. which was first reported for normal cats but has also been isolated from the feces of diarrheic cats (26, 36). The diversity of pathogens in the cats in this study suggests that they may have been immunocompromised. However, the cats were not infected with retroviruses and appeared healthy except for the enteritis. Alternatively, one or several of the bacterial organisms may have caused primary bacterial colitis and may, under certain conditions, be considered primary pathogens.
As noted for the cats in this study and other cats screened by us (10a), coinfection with Campylobacter spp. and Helicobacter spp. is commonly observed. Such coinfection also has been documented for humans with diarrheic feces (1). This situation poses a particular diagnostic challenge in correctly identifying both campylobacters and helicobacters in one fecal specimen, given the similarities in their phenotypic and biochemical profiles. As in our study, the use of specific and sensitive PCR primers distinguishing the two genera may be required.
The cats in the present report had enteritis and periportal hepatitis. However, cats without diarrhea were also colonized with H. canis. The data indicates that H. canis is endemic in this cat colony, given that H. canis was isolated from multiple cats at two time points (6 months apart). It will be important to further evaluate H. canis for causal roles in IBD and hepatitis. Additionally, studies are needed to determine whether or not enterohepatic disease associated with H. canis infection occurs more commonly in certain hosts of a particular genotype (e.g., particular breeds of dogs or cats), in immunosuppressed hosts, and in association with certain other enteric infections. The study of H. canis in cats with IBD may also provide insight into the etiopathogenesis of IBD in humans.
ACKNOWLEDGMENTS
This work was supported by grants to Janet E. Foley from the Krade and Maddox Endowments to the Center for Companion Animal Health, the San Francisco Foundation, and the University of California at Davis School of Veterinary Medicine and in part by NIH grants R01CA67529 (to J.G.F.), R01DK52413 (to J.G.F.), RR07036 (to J.G.F.), and DE-10374 (to F.E.D.).
We thank Amy Poland for technical assistance and Dwight Hirsh, Carol Glaser, and Patrick Foley for suggestions and interpretations.
REFERENCES
- 1.Allos B M, Lastovica A J, Blaser M J. Atypical campylobacters and related microorganisms. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 849–865. [Google Scholar]
- 2.Burnens A P, Stanley J, Schaad U B, Nicolet J. Novel Campylobacter-like organism resembling Helicobacter fennelliae isolated from a boy with gastroenteritis and from dogs. J Clin Microbiol. 1993;31:1916–1917. doi: 10.1128/jcm.31.7.1916-1917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cahill R J, Foltz C J, Fox J G, Dangler C A, Powrie F, Schauer D B. Inflammatory bowel disease: an immune mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65:3126–3131. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Citino S. Chronic intermittent Clostridium perfringens enterotoxicosis in a group of cheetahs (Acinonyx jubatus jubatus) J Zool Wildl Med. 1995;26:2. [Google Scholar]
- 5.Fennell C L, Totten P A, Quinn T C, Patton D L, Holmes K K, Stamm W E. Characterization of Campylobacter-like organisms isolated from homosexual men. J Infect Dis. 1984;149:58–66. doi: 10.1093/infdis/149.1.58. [DOI] [PubMed] [Google Scholar]
- 6.Foley J E, Hirsh D C, Pedersen N C. An outbreak of Clostridium perfringens enteritis in a cattery of Bengal cats and inadvertant experimental transmission to specific pathogen free cats. Feline Pract. 1996;24:31–35. [Google Scholar]
- 7.Foley J E, Poland A, Carlson J, Pedersen D C. Patterns of feline coronavirus infection and fecal shedding from cats in multiple-cat environments. J Am Vet Med Assoc. 1997;210:1307–1312. [PubMed] [Google Scholar]
- 8.Foltz C J, Fox J G, Cahill R J, Murphy R C, Yan L, Shames B, Schauer D B. Spontaneous inflammatory bowel disease in multiple mutant mouse lines: association with colonization by Helicobacter hepaticus. Helicobacter. 1998;3:69–78. doi: 10.1046/j.1523-5378.1998.08006.x. [DOI] [PubMed] [Google Scholar]
- 9.Fox J G. In vivo models of gastric Helicobacter infections. In: Hunt R H, Tytgat G, editors. Helicobacter pylori: basic mechanisms to clinical cure. Dordrecht, The Netherlands: Kluwer Academic; 1994. pp. 3–27. [Google Scholar]
- 10.Fox J G. The expanding genus of Helicobacter: pathogenic and zoonotic potential. Semin Gastrointest Dis. 1997;8:124–141. [PubMed] [Google Scholar]
- 10a.Fox, J. G. Unpublished data.
- 11.Fox J G, Correa P, Taylor N S, Lee A, Otto G, Murphy J C, Rose R. Helicobacter mustelae associated gastritis in ferrets: an animal model of Helicobacter pylori gastritis in humans. Gastroenterology. 1990;99:352–361. doi: 10.1016/0016-5085(90)91016-y. [DOI] [PubMed] [Google Scholar]
- 12.Fox J G, Dangler C A, Sager W, Borkowski R, Gliatto J M. Helicobacter mustelae associated gastric adenocarcinoma in ferrets (Mustela putorius furo) Vet Pathol. 1997;34:225–229. doi: 10.1177/030098589703400308. [DOI] [PubMed] [Google Scholar]
- 13.Fox J G, Dewhirst F E, Tully J G, Paster B J, Yan L, Taylor N S, Collins M J, Gorelick P L, Ward J M. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox J G, Drolet R, Higgins R, Messier S, Yan L, Dewhirst F E. Helicobacter canis isolated from a dog liver with multifocal necrotizing hepatitis. J Clin Microbiol. 1996;34:2479–2482. doi: 10.1128/jcm.34.10.2479-2482.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox J G, Lee A. The role of Helicobacter species in newly recognized gastrointestinal tract diseases of animals. Lab Anim Sci. 1997;47:222–255. [PubMed] [Google Scholar]
- 16.Fox J G, Yan L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter isolated from bile, livers, and intestines of aged, inbred mouse strains. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox J G, Yan L, Shames B, Campbell J, Murphy J C, Li X. Persistent hepatitis and enterocolitis in germfree mice infected with Helicobacter hepaticus. Infect Immun. 1996;64:3673–3681. doi: 10.1128/iai.64.9.3673-3681.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham D Y. Helicobacter pylori infection in the pathogenesis of duodenal ulcer and gastric cancer: a model. Gastroenterology. 1997;113:1983–1991. doi: 10.1016/s0016-5085(97)70019-2. [DOI] [PubMed] [Google Scholar]
- 19.Guerrant R, Thielman N. Emerging enteric protozoa: Cryptosporidium, Cyclospora, and Microsporidia. In: Armstrong D, Hughes J, editors. Emerging infections. Washington, D.C: American Society for Microbiology; 1998. pp. 233–245. [Google Scholar]
- 20.Haines D C, Gorelick P L, Battles J K, Pike K M, Anderson R J, Fox J G, Taylor N S, Shen Z, Dewhirst F E, Anver M R, Ward J M. Inflammatory large bowel disease in immunodeficient rats naturally and experimentally infected with Helicobacter bilis. Vet Pathol. 1998;35:202–208. doi: 10.1177/030098589803500305. [DOI] [PubMed] [Google Scholar]
- 21.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 22.Knoop F, Owens M, Crocker I. Clostridium difficile: clinical disease and diagnosis. Clin Microbiol Rev. 1993;6:251–265. doi: 10.1128/cmr.6.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laflamme D, Kealy R, Schmidt D. Estimation of body fat by body condition score. J Vet Int Med. 1994;8:154. [Google Scholar]
- 24.Linton D, Clewley J P, Burnens A, Owen J, Stanley J. An intervening sequence (IVS) in the 16S rRNA gene of the eubacterium Helicobacter canis. Nucleic Acids Res. 1994;22:1954–1958. doi: 10.1093/nar/22.11.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutz H, Pedersen N C, Durbin R, Theilen G H. Monoclonal antibodies to three epitopic regions of feline leukemia virus P27 and their use in enzyme-linked immunosorbent assay of p27. J Immunol Methods. 1983;56:209–220. doi: 10.1016/0022-1759(83)90413-1. [DOI] [PubMed] [Google Scholar]
- 26.Moreno G, Griffiths P, Connerton I, Park R. Occurrence of campylobacters in small domestic and laboratory animals. J Appl Bacteriol. 1993;75:49–54. doi: 10.1111/j.1365-2672.1993.tb03406.x. [DOI] [PubMed] [Google Scholar]
- 27.Neiger R, Dieterich C, Burnens A, Waldvogel A, Corthesey-Theulaz I, Halter F, Lauterburg B, Schmassmann A. Detection and prevalence of Helicobacter infection in pet cats. J Clin Microbiol. 1998;36:634–637. doi: 10.1128/jcm.36.3.634-637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otto G, Hazell S H, Fox J G, Howlett C R, Murphy J C, O’Rourke J L, Lee A. Animal and public health implications of gastric colonization of cats by Helicobacter-like organisms. J Clin Microbiol. 1994;32:1043–1049. doi: 10.1128/jcm.32.4.1043-1049.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsonnet J, Friedman G D, Vandersteen D P, Chang Y, Vogelman J H, Orentreich N, Sibley R J. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 30.Paster B J, Dewhirst F E. Phylogeny of Campylobacter, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int J Syst Bacteriol. 1988;38:56–62. [Google Scholar]
- 31.Pedersen N. Feline infectious diseases. Goletta, Calif: American Veterinary Publisher; 1988. [Google Scholar]
- 32.Pedersen N C. Serologic studies of naturally occurring feline infectious peritonitis. Am J Vet Res. 1976;37:1449–1453. [PubMed] [Google Scholar]
- 33.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 34.Shomer N H, Dangler C A, Fox J G. Helicobacter bilis induced inflammatory bowel disease in defined-flora scid mice. Infect Immun. 1997;65:4858–4864. doi: 10.1128/iai.65.11.4858-4864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solnick J V, O’Rourke J, Lee A, Paster B J, Dewhirst F E, Tompkins L S. An uncultured gastric spiral organism is a newly identified Helicobacter in humans. J Infect Dis. 1993;168:379–385. doi: 10.1093/infdis/168.2.379. [DOI] [PubMed] [Google Scholar]
- 36.Stanley J, Burnens A, Linton D, On S, Costas M, Owen R. Campylobacter helveticus sp. nov., a new thermophilic species from domestic animals: characterization and cloning of a species-specific DNA probe. J Gen Microbiol. 1992;138:2293–2303. doi: 10.1099/00221287-138-11-2293. [DOI] [PubMed] [Google Scholar]
- 37.Stanley J, Linton D, Burens A P, Dewhirst F E, On S L, Porter A, Owen R J, Costas M. Helicobacter pullorum sp. nov.—genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology. 1994;140:3441–3449. doi: 10.1099/13500872-140-12-3441. [DOI] [PubMed] [Google Scholar]
- 38.Stanley J, Linton D, Burens A P, Dewhirst F E, Owen R J. Helicobacter canis sp. nov., a new species from dogs: an integrated study of phenotype and genotype. J Gen Microbiol. 1993;139:2495–2504. doi: 10.1099/00221287-139-10-2495. [DOI] [PubMed] [Google Scholar]
- 39.Tams T. Inflammatory bowel disease. In: August J, editor. Consultations in feline internal medicine. Philadelphia, Pa: The W. B. Saunders Co.; 1991. pp. 409–413. [Google Scholar]
- 40.Totten P A, Fennel C L, Tenover F C. Campylobacter cinaedi (sp. nov.) and Campylobacter fennelliae (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J Infect Dis. 1985;151:131–139. doi: 10.1093/infdis/151.1.131. [DOI] [PubMed] [Google Scholar]
- 41.Ward J M, Anver M R, Haines D C, Melhorn J M, Gorelick P, Yan L, Fox J G. Inflammatory large bowel disease in immunodeficient mice naturally infected with Helicobacter hepaticus. Lab Anim Sci. 1996;46:15–20. [PubMed] [Google Scholar]
- 42.Ward J M, Fox J G, Anver M R, Haines D C, George C V, Collins M J, Gorelick P L, Nagashima K, Gonda M A, Gilden R V, Tully J G, Russell R J, Benveniste R E, Paster B J, Dewhirst F E, Donovan J C, Anderson L M, Rice J M. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst. 1994;86:1222–1227. doi: 10.1093/jnci/86.16.1222. [DOI] [PubMed] [Google Scholar]
- 43.Warren J D, Marshall B J. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1273–1275. [PubMed] [Google Scholar]
- 44.Whary M T, Morgan T J, Dangler C A, Gaudes K J, Taylor N S, Fox J G. Chronic active hepatitis induced by Helicobacter hepaticus in the A/JCr mouse is associated with a Th1 cell-mediated immune response. Infect Immun. 1998;66:3142–3148. doi: 10.1128/iai.66.7.3142-3148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto J K, Hansen H, Ho E W, Morishita T Y, Okuda T, Sawa T R, Nakamura R M, Pedersen N C. Epidemiologic and clinical aspects of feline immunodeficiency virus infection in cats from the continental United States and Canada and possible mode of transmission. J Am Vet Med Assoc. 1989;194:213–220. [PubMed] [Google Scholar]