Abstract
Background
The detection of environmental cues and signals via the sensory system directs behavioral choices in diverse organisms. Insect larvae rely on input from the chemosensory system, mainly olfaction, for locating food sources. In several lepidopteran species, foraging behavior and food preferences change across larval instars; however, the molecular mechanisms underlying such behavioral plasticity during larval development are not fully understood. Here, we hypothesize that expression patterns of odorant receptors (ORs) change during development, as a possible mechanism influencing instar-specific olfactory-guided behavior and food preferences.
Results
We investigated the expression patterns of ORs in larvae of the cotton leafworm Spodoptera littoralis between the first and fourth instar and revealed that some of the ORs show instar-specific expression. We functionally characterized one OR expressed in the first instar, SlitOR40, as responding to the plant volatile, β-caryophyllene and its isomer α-humulene. In agreement with the proposed hypothesis, we showed that first but not fourth instar larvae responded behaviorally to β-caryophyllene and α-humulene. Moreover, knocking out this odorant receptor via CRISPR-Cas9, we confirmed that instar-specific responses towards its cognate ligands rely on the expression of SlitOR40.
Conclusion
Our results provide evidence that larvae of S. littoralis change their peripheral olfactory system during development. Furthermore, our data demonstrate an unprecedented instar-specific behavioral plasticity mediated by an OR, and knocking out this OR disrupts larval behavioral plasticity. The ecological relevance of such behavioral plasticity for S. littoralis remains to be elucidated, but our results demonstrate an olfactory mechanism underlying this plasticity in foraging behavior during larval development.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12915-021-01159-1.
Keywords: α-Humulene, β-Caryophyllene, CRISPR-Cas9, Electrophysiology, Larval transcriptome, Modulation, Odorant receptor, Olfactometer
Background
Many animals, including insects, exhibit changes in foraging behavior and food preferences during their life [1–4]. For example, in the insect model Drosophila melanogaster, larvae are attracted to and prefer unsaturated fatty acids over saturated fatty acids, while adults prefer saturated fatty acids [3]. In Lepidoptera, larval and adult food resources differ, and within larval instars, food requirements may also change according to the instar. For instance, the older larvae of the buckeye butterfly Junonia coenia (Lepidoptera: Nymphalidae) show marked preference for young plants over flowering and fruiting mature plants, contrary to the first instar larvae which are less selective [5]. In the fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae), the early instar larvae feed predominantly on cotton leaves and progressively change their preferences to fruiting structures (squares and bolls) in late instars [6]. These preference shifts are attributed to the substantial increase of the larval body size, changes in digestive physiology, dietary needs across instars, plant phenology, and competition avoidance at late stages [5, 7–9]. However, the molecular mechanisms underlying behavioral plasticity during larval development are not fully understood.
The detection of relevant cues or signals via the sensory system is an initial step towards the display of behavior in insects. In larvae as in adults, the chemosensory system encompassing gustation and olfaction mediates the detection of chemicals emitted by a variety of sources, including food [10–12]. In particular, the olfactory system is of prime importance as it detects at distance food cues, such as specific plant odors for herbivorous insects [13]. It could be thus hypothesized that changes in the foraging behavior and food preference during larval development would be mediated by modifications in olfactory abilities, through changes either in sensing chemical signals in the peripheral olfactory organs or in the processing of chemical signals at higher brain centers. Changes in odor detection at the peripheral olfactory system during larval development have not been explored.
The peripheral olfactory organs of Lepidoptera larvae consist of antennae and maxillary palps [14]. Together, they house olfactory sensory neurons (OSNs) that express the chemosensory receptors responsible for olfaction: odorant receptors (ORs) and ionotropic receptors (IRs) [13]. The axons from these OSNs innervate one or several glomeruli in the antennal lobe, before the signals are projected into the higher brain center [15, 16]. Lepidoptera larvae generally have fewer sensilla and OSNs on their chemosensory organs than adults and a smaller repertoire of expressed ORs [17–21]. In vivo, ORs form a heteromeric complex with a universal co-receptor Orco, a prerequisite for signal transduction [22, 23]. Despite the relatively simple olfactory system of larvae, several examples show that it detects numerous host plant volatiles [21, 24, 25]. For example, Di et al. [21] showed that larvae of the cotton bollworm Helicoverpa armigera express only 17 ORs in the antennae and maxillary palps, but detect a wide range of odorants such as green leaf volatiles, terpenoids, and various other aromatic and aliphatic compounds.
We hypothesize that expression patterns of ORs change during development, as a possible mechanism influencing instar-specific olfactory-guided behavior and food preferences. To date, studies of OR differential expression in insects have mainly focused on the comparison between species, sexes, and physiological conditions (feeding/mating status) and comparison between larvae and adults [19, 20, 26–28]. Such studies revealed, for instance, male-biased pheromone receptors in adults and female-specific ORs related to host finding. For example, in Bombyx mori, females specifically express some ORs that respond to volatiles likely involved in oviposition site selection [29]. Differential expression of ORs between larval stages has not been investigated yet. To address this, we examined whether larvae of the Egyptian cotton leafworm, Spodoptera littoralis (Lepidoptera: Noctuidae), express different sets of ORs during development, and propose a novel mechanism of olfactory-guided behavioral modulation in insect larvae.
Spodoptera littoralis is a polyphagous pest across Africa and the Middle East whose larvae can feed and successfully complete their life cycle on plants belonging to more than 40 families [30]. In this species, large OR repertoires have already been described in both adults and larvae [19, 31] and odor-guided behavioral traits are well characterized [25, 32–34], providing a good basis to investigate differential expression of ORs during larval development. We selected two larval stages, first and fourth instars, to cover the developmental period where larvae actively search for food.
We first used Illumina-based RNA sequencing (RNA-seq) to compare the transcriptomes of heads of first and fourth instar larvae and revealed that some ORs were differentially expressed between the two larval stages, further confirmed by PCR analyses. We functionally characterized one of these ORs as responding to β-caryophyllene and α-humulene, two volatiles commonly found in S. littoralis host plants. In agreement with the proposed hypothesis, we showed that β-caryophyllene and α-humulene triggered different behavioral responses in first and fourth instar larvae. Knocking out this OR via CRISPR-Cas9, we confirmed that instar-specific responses towards its cognate ligands did rely on this OR expression. In adults, knocking out this OR impaired electrophysiological responses to β-caryophyllene and α-humulene, comparing the function of this receptor in vivo.
Results
Transcriptome sequencing and identification of odorant receptors in S. littoralis larvae
First, we used RNA-seq to identify ORs from the transcriptomes of first and fourth instar larval heads. A total of 15,763,159 and 30,022,191 raw sequence reads were obtained from the first and fourth instar larvae, respectively. After trimming and removing low-quality reads, we obtained 95.97% (first instar) and 94.88% (fourth instar) paired reads. Using Trinity, we assembled one transcriptome with first instar larval reads, one with fourth instars, and one with both instars + previously published adult antennae reads [31]. This last assembly yielded 50,680 sequences, with a N50 of 1954 nt, mean sequence length of 744.79 nt, and 24,264 contigs greater than 1000 nt. Benchmarking Universal Single-Copy Orthologs (BUSCO, [35]) analysis of the assembled transcriptome (first and fourth instar + adult) using the insecta_obd9 reference database showed 96.4% complete BUSCO, 75.1% complete and single copy, 21.3% complete and duplicate BUSCO, and 2.8 and 0.8% fragmented and missing BUSCOs, respectively (Additional file 1: Table S1).
Mapping first and fourth instar reads to the assembled transcriptome and estimation of expression levels revealed 34 ORs expressed in the first instar and 18 ORs in the fourth instar (Additional file 2: Table S2). Most of these ORs corresponded to previously identified receptors in S. littoralis. In addition, we identified one new OR, named SlitOR70 (amino acid sequence in Additional file 3: Table S3), expressed in both first and fourth instar transcriptomes, and RT-PCR quantification confirmed the expression in both the larval stages and adult antennae (Additional file 4: Fig. S1). We followed the same OR nomenclature previously described for S. littoralis [19, 31].
A previous study on the same species retrieved 22 ORs expressed in the fourth instar [19]. The differences in OR numbers between our study and the already published one may have resulted from tissue collection methodology or other unknown reasons. Overall expression levels of ORs were low in our larval transcriptomes. Hence, we performed an additional exhaustive OR screening in both larval instars via RT-PCR using primer pairs designed to all the 60 previously described S. littoralis ORs [19, 31]. We retrieved a set of 36 ORs expressed in larvae including OR co-receptor (Additional file 2: Table S2 and Additional file 4: Fig. S1). The comparison between our transcriptome analysis and the RT-PCR analysis revealed several false positives (first instar = 16.39 %; fourth instar = 8.19 %) (Additional file 2: Table S2, values indicated with hashtag) and false negatives (first instar = 21.3 %; fourth instar = 34.42 %) (Additional file 2: Table S2, values indicated with asterisk) of OR expressions in our larval transcriptomes.
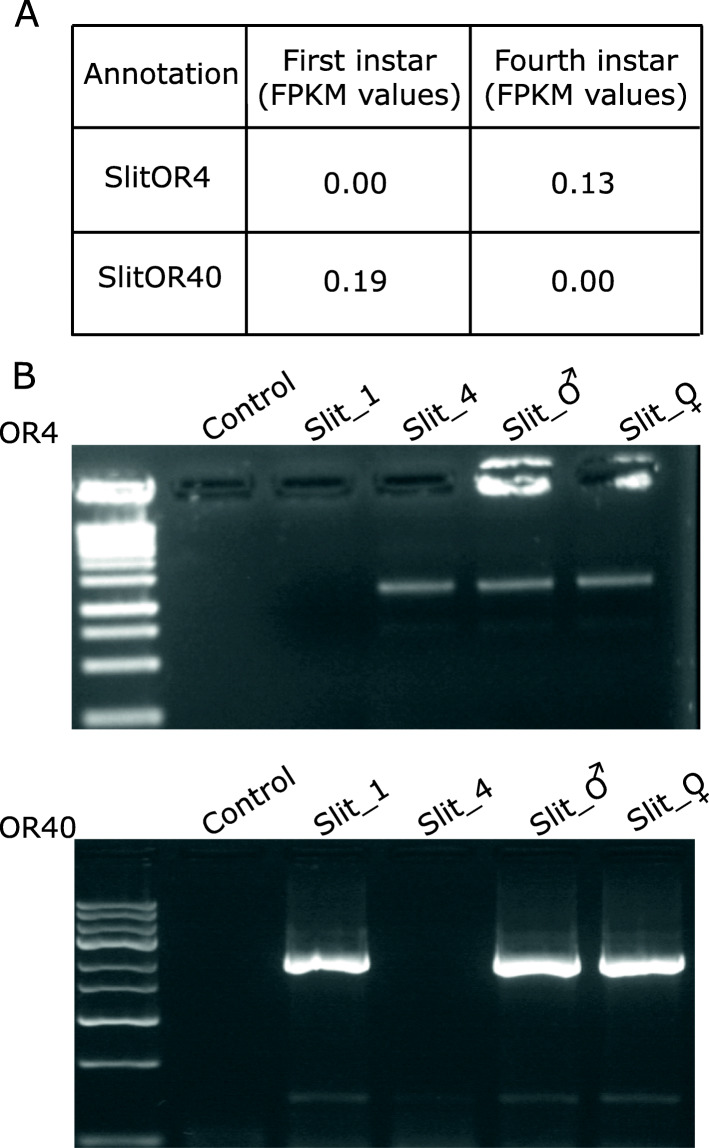
RT-PCR revealed that both stages expressed a large overlapping set of ORs, but evidenced that two receptors were stage-specific: SlitOR40 was expressed only in the first instar, while SlitOR4 was expressed only in the fourth instar, which was in agreement with the transcriptome data (Fig. 1 and Additional file 2: Table S2).
Fig. 1.
Expression of SlitOR4 and SlitOR40 in Spodoptera littoralis larvae: A Differential expression levels of two ORs in first and fourth instars measured using fragment reads per kb per million criteria. B Expression profile of SlitOR4 and SlitOR40 in RT-PCR assays performed using specific primer pairs, and comparison included negative control (control) and cDNAs from different tissues: first and fourth instar larval heads, and male (Slit_♂) and female antennae (Slit_♀) with 1kb gene ruler ladder (Thermo Fischer Scientific)
RT-PCR and RT-qPCR assays on SlitOR40 expression
We focused our investigation on SlitOR40 since SlitOR4 function was already elucidated [36]. SlitOR40 expression, as well as that of SlitOrco as a positive control, was followed in detail from first to fourth larval instars by RT-PCR (Additional file 5: Fig. S2A). Fifth and sixth larval stages were not assayed in our RT-PCR analysis, and the SlitOR40 expression in these stages remains unknown. Whereas SlitOrco was detected as expected in all samples, SlitOR40 expression was detected in first, second, and third instars, as well as in adult antennae, but it was not detected in fourth instar larval tissue (Additional file 5: Fig. S2A).
To further quantify the expression levels of SlitOR40, we performed RT-qPCR. First, Orco expression was quantified in first and fourth larval instar heads and adult male antennae using reference genes β-actin, L13a, and Ef1a [37]. Since Orco forms a heteromeric complex with ORs in nearly all OSNs [22, 38], Orco expression in the chemosensory tissue reflects the overall expression of ORs. We found that the relative Orco expression in the male antennae was significantly higher compared to first instar (187.09-fold) and fourth instar (128.38-fold) larval head tissues (Df = 28, t = −22.622; P < 0.001; Df = 28, t = 50.441, P < 0.001, respectively) (Additional file 5: Fig. S2B), probably because OSNs are enriched in adult antennae tissues compared to whole heads of larvae. Moreover, in the fourth instar larvae, Orco expression was 2.11-fold higher compared to first instar (t = 9.33; Df = 28; P < 0.001). As Orco level should correlate with the number of OSNs, we further used SlitOrco as a reference gene for SlitOR40 quantification to ensure tissue normalization according to OSN numbers. Doing this, we revealed that SlitOR40 relative expression in first instar larvae was significantly higher (19.97-fold) compared to adult male antennae (t = 26.532; Df = 28; P < 0.001), while there was no expression detected (t = 27.693, Df = 28; P < 0.001) in the fourth larval instar (Additional file 5: Fig. S2C), confirming RNA-seq and RT-PCR observations.
SlitOR40 functional study
As SlitOR40 presented a biased expression to first instars, we assayed for its ligands as they may represent important cues for this stage. To identify these ligands, SlitOR40 was heterologously expressed in the Drosophila empty neuron system [39]. The transformed Drosophila neurons were stimulated with a panel of 54 odorants and responses were measured via single sensillum recordings. At a 10-μg dose of stimuli, SlitOR40 showed significant responses to β-caryophyllene and its isomer α-humulene (Fig. 2), two plant volatiles commonly found in S. littoralis host plants [11, 40, 41]. SlitOR40 did not show a significant response to any other chemical in the panel tested even at higher doses of 100 μg.
Fig. 2.
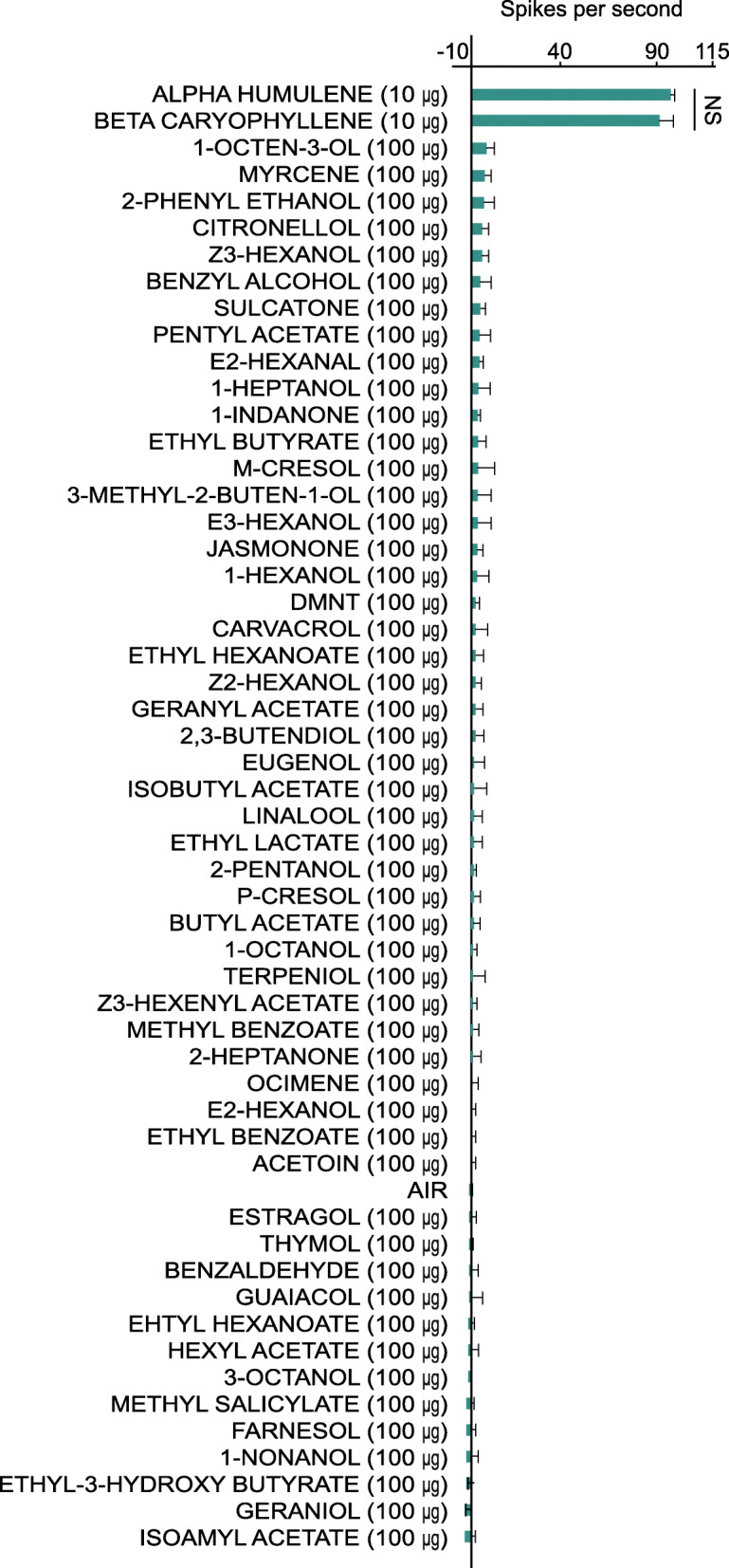
Single sensillum recordings from neurons expressing Spodoptera littoralis SlitOR40 in Drosophila melanogaster antennae. Values correspond to spikes per second produced during odor stimulation. The odor panel consisted of 54 compounds, individually loaded in disposable Pasteur glass pipettes at a dose of 100 μg. Differences in the electrophysiological responses from the empty neuron flies to β-caryophyllene (10 μg) and α-humulene (10 μg) were tested using the Student t-test. None of the remaining compounds in the panel responded significantly. The compounds are listed in decreasing order of signal strength. Error bars indicate standard deviations. The analyses were based on recordings from six flies; P = 0.76. NS not significant
Generation and characterization of SlitOR40-knockout lines
In order to test the behavioral importance of SlitOR40, we used CRISPR-Cas9 to knock out this gene in the S. littoralis genome. We tested two single guide RNAs (sgRNA) designed to target the predicted first and the second exons of the SlitOR40 gene (Additional file 6: Fig. S3A). Injection of the Cas9-sgRNA complex in eggs resulted in the generation of three different mutations in the gene, all leading to a premature termination codon (Additional file 6: Fig. S3B and S3C). One of the mutations resulted in a deletion (−) of 445 base pairs (bp) (line-10), while two other mutations resulted in an insertion (+) and a deletion (−) (line-38 = − 440 bp and + 20 bp; line-2 = − 446 bp and + 5 bp). The line-10 was chosen for crossing to homozygotes and subsequent phenotyping using behavioral and electrophysiological assays.
Larval behavioral responses to β-caryophyllene and α-humulene
To investigate if larval behavioral response to the SlitOR40 ligands would differ between larval stages and according to expression (or not) of SlitOR40, we conducted behavioral assays in a two arm olfactometer (Y-tube). We first ran a positive control looking at the behavior of first and fourth instar larvae of wild type (WT) and SlitOR40-knockout (KO) genotypes to one of the preferred host plants, cotton [42], which was provided as leaf discs (Additional file 7: Fig. S4). In four independent experiments, first and fourth instar larvae from both genotypes showed equal preference when treatments included cotton leaf discs in each arm (first instar (WT): X2 = 0.27; P = 0.87; fourth instar (WT): X2 = 0.043; P = 0.84; first instar (KO): X2 = 1.04; P = 0.3; fourth instar (KO): X2 = 0.024; P = 0.88). In another control experiment, both instars across genotypes (WT or KO) preferred cotton to a blank control (empty arm) (first instar (WT): X2 = 19.56; P < 0.001; fourth instar (WT): X2 = 16.95; P < 0.001; first instar (KO): X2 = 14.22; P < 0.001; fourth instar (KO): X2 = 23.52; P < 0.001). These results showed that both instars were similarly attracted to cotton leaf discs and confirmed that there was no behavioral difference in attraction to the discs between WT and KO larvae.
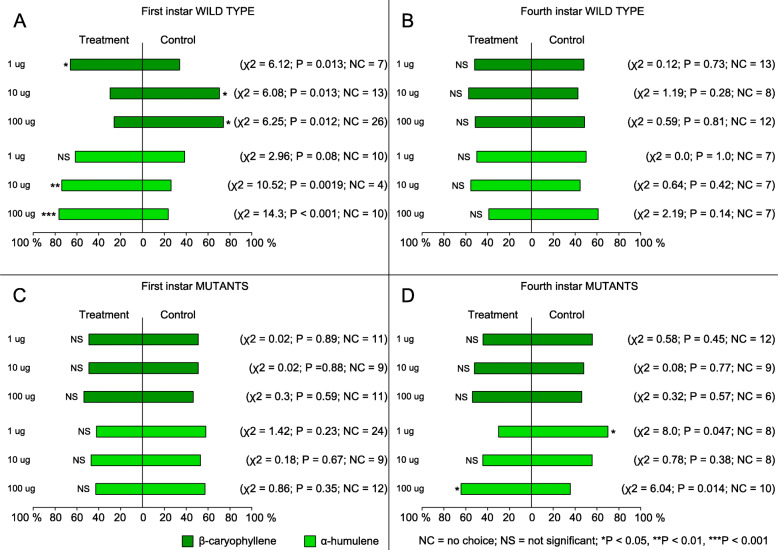
Subsequently, we compared the behavior of both instars of WT and KO genotypes to leaf discs of cotton plants + filter paper treated with solvent (paraffin oil) versus cotton leaf discs + filter paper treated with test odorants: β-caryophyllene or α-humulene (treatments). In the experiments with first instar WT larvae, cotton leaf discs with 1-μg dose of β-caryophyllene induced preference compared to control (X2 = 6.12; P = 0.013) (Fig. 3A). However, with increasing doses of β-caryophyllene (10 and 100 μg), larvae preferred the control arm (X2 = 6.08; P = 0.013; X2 = 6.25; P = 0.012, respectively). In the experiments with α-humulene, first instar larvae preferred the two higher doses of α-humulene (10 and 100 μg) (X2 = 10.52; P = 0.0019; X2 = 14.13; P < 0.001, respectively). At the 1-μg dose, first instar larvae tended towards humulene but without statistical significance (X2 = 2.96; P = 0.08). Collectively, first instar larvae responded to both β-caryophyllene and α-humulene but dose-response patterns towards the two isomers differed. In KO behavioral tests, first instar larvae showed no preference for β-caryophyllene (1 μg: X2 = 0.02; P = 0.89; 10 μg: X2 = 0.02; P = 0.88; 100 μg: X2 = 0.3; P = 0.59) nor for α-humulene over solvent (1 μg: X2 = 1.42; P = 0.23; 10 μg: X2 = 0.18; P = 0.67; 100 μg: X2 = 0.86; P = 0.35) at any of the doses tested (Fig. 3C).
Fig. 3.
Preference of first and fourth larval instar of Spodoptera littoralis wild type and SlitOR40-knockout: The data show the percentage of larvae choosing the control or one of the two odors tested at three doses in a Y-tube assay: A Behavioral responses of first instar (WT) to β-caryophyllene (1/10/100 μg doses) + cotton leaf discs vs cotton leaf discs (n = 66, 50, and 53, respectively) and α-humulene (1/10/100 μg doses) + cotton leaf discs vs cotton leaf discs (n = 67, 50, and 61, respectively). B Behavioral responses of fourth instar (WT) to β-caryophyllene (1/10/100 μg doses) + cotton leaf discs vs cotton leaf discs (n = 88, 62, and 80, respectively) and α-humulene (1/10/100 μg doses) + cotton leaf discs vs cotton leaf discs (n = 63, 63, and 43, respectively). C Behavioral responses of knockout first instars to β-caryophyllene (1/10/100 μg doses) + cotton leaf discs vs cotton leaf discs (n = 60, 58, and 65, respectively) and α-humulene (1/10/100 μg doses) + cotton leaf discs vs cotton leaf discs (n = 81, 58, and 54, respectively). D Behavioral responses of knockout fourth instars to β-caryophyllene (1/10/100 μg doses) + cotton leaf discs vs cotton leaf discs (n = 55, 57, and 56, respectively) and α-humulene (1/10/100 μg doses) + cotton leaf discs vs cotton leaf discs (n = 58, 71, and 83, respectively). Asterisks indicate statistically significant differences following a chi-square test. (***P < 0.001; **P < 0.01; *P < 0.05). NS not significant, NC number of insects that exhibited no choice
Fourth instar WT larvae chose equally control leaf discs and leaf discs treated with β-caryophyllene (1 μg: X2 = 0.12; P = 0.73; 10 μg: X2 = 1.19; P = 0.28; 100 μg: X2 = 0.59; P = 0.81) or α-humulene (1 μg: X2 = 0.0; P = 1.0; 10 μg: X2 = 0.64; P = 0.42; 100 μg: X2 = 2.19; P = 0.14) (Fig. 3B), contrary to first instar larvae.
Thus, behavioral responses in the two larval stages corroborate with SlitOR40 expression. Knockout of SlitOR40 had no effect on fourth instar larvae behavior to β-caryophyllene (Fig. 2D). Interestingly, KO fourth instar larvae showed preference to α-humulene at the highest dose tested (100 μg: X2 = 6.04; P = 0.014) (Fig. 3D). However, larval preferences to α-humulene changed with decreasing doses; a 10-μg dose induced no preference (X2 = 0.78; P = 0.38), while larvae preferred the control arm compared to the one with 1 μg of α-humulene (X2 = 8.0; P = 0.047) (Fig. 3D).
Electroantennography (EAG) responses
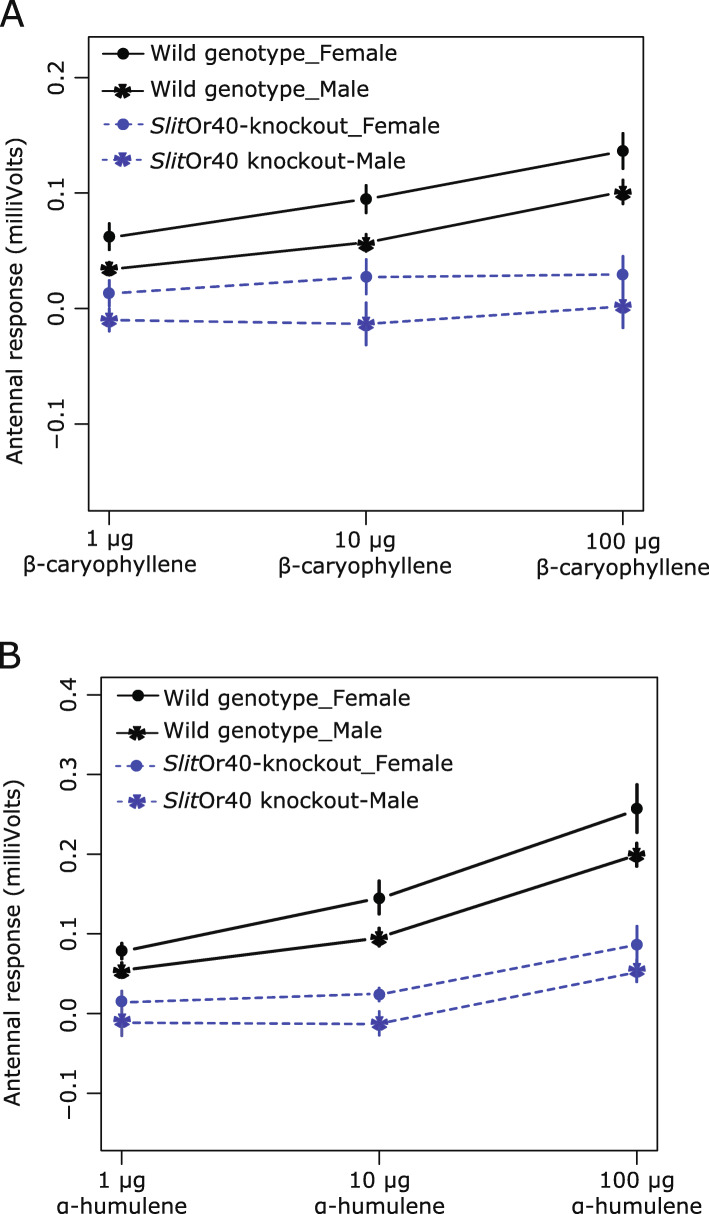
To confirm the effectiveness of knocking out SlitOR40, we recorded EAG responses from KO adult antennae in comparison to WT adult antennal responses. The EAG recordings showed no statistical difference in responses to positive control stimuli: guaiacol and (±)-linalool in both genotypes (Additional file 8: Fig. S5). However, at stimulation with SlitOR40 ligands, sex, genotype, and compound dose all significantly affected response intensity to β-caryophyllene and α-humulene (Fig. 4A, B; ANCOVA analysis, Table 1). There was a global tendency that female moth antennae responded stronger than male antennae, irrespective of doses applied and genotype. However, the slope of the dose-response to either compound did not depend on sex (dose*sex interaction non-significant). On the contrary, KO of SlitOR40 significantly modified the slope of the dose-response to β-caryophyllene and to α-humulene (dose*genotype interaction significant). In order to refine these results, we performed linear regression on response intensity as a function of compound dose, separately for each genotype and without taking into account the sex parameter. The WT moths showed a significant dose-response to both compounds (Fig. 4A, B; ANCOVA analysis, Table 2). In KO moth, the dose-response to β-caryophyllene was completely abolished (slope of the regression not significantly different from zero), while the dose-response to α-humulene was reduced but still significant (slope of the regression significantly different from zero). Therefore, as expected, SlitOR40 knockout affected the moth’s ability to perceive its cognate ligands: KO moths completely lost the ability to detect β-caryophyllene and are less sensitive than WT to α-humulene although they could still detect it.
Fig. 4.
EAG responses of Spodoptera littoralis wild type and SlitOr40-knockout moth antennae to β-caryophyllene and α-humulene: Three doses for both compounds were tested: 1 μg, 10 μg, and 100 μg (n = 10 per sex). The slope of dose-response to A β-caryophyllene and B α-humulene are sex-independent. In KO moths, the slope of dose-response is significantly different from WT (dose*genotype interaction). The response to β-caryophyllene was completely abolished in KO moths, while the dose-response to α-humulene was reduced but still significantly different from WT
Table 1.
Statistical analysis of EAG response intensity to β-caryophyllene and α-humulene. Results of the ANCOVA analyses testing the effect of compound dose, moth sex, and moth genotype on response intensity. Text in bold indicates statistical significance
| Factor | Df | β-caryophyllene | α-humulene | ||||
|---|---|---|---|---|---|---|---|
| Sum of squares | F value | P-value | Sum of squares | F value | P-value | ||
| Dose | 1 | 0.0531 | 29.2 | 2.62E−7 | 0.364 | 109 | <2E−16 |
| Sex | 1 | 0.0259 | 14.2 | 0.000235 | 0.0313 | 9.41 | 0.00257 |
| Genotype | 1 | 0.197 | 108 | <2E−16 | 0.464 | 139 | <2E−16 |
| Dose*sex | 1 | 2.00E−5 | 1.00E−2 | 0.922 | 0.00130 | 0.404 | 0.526 |
| Dose*genotype | 1 | 0.0196 | 10.8 | 0.00129 | 0.0535 | 16.1 | 9.73E−5 |
| Sex*genotype | 1 | 8.00E−5 | 0.0460 | 0.831 | 0.00140 | 0.521 | 0.521 |
| Triple interaction | 1 | 1.00E−5 | 7.00E−3 | 0.935 | 0.00130 | 0.315 | 0.576 |
| Residual | 145 | 0.264 | 0.483 | ||||
Table 2.
Statistical analysis of EAG response intensity to β-caryophyllene and α-humulene. Estimation of dose-response slopes separately for each moth genotype, irrespective of sex (linear regression). Text in bold indicates statistical significance
| β-caryophyllene | α-humulene | ||||
|---|---|---|---|---|---|
| OR40 knockout | Wild type | OR40 knockout | Wild type | ||
| Regression statistics | F-statistic | 0.866 | 41.5 | 17.4 | 87.0 |
| DF | 1;64 | 1;85 | 1;64 | 1;85 | |
| P-value | 0.356 | 6.83E−9 | 9.41E−5 | 1.17E−14 | |
| R2 | 0.0134 | 0.328 | 0.214 | 0.506 | |
| Regression slope | Estimate | 0.00700 ± 0.00752 | 0.0348 ± 0.00541 | 0.0339 ± 0.00814 | 0.0794 ± 0.00851 |
| T-value | 0.930 | 0.00541 | 4.17 | 9.33 | |
Phylogenetic analysis
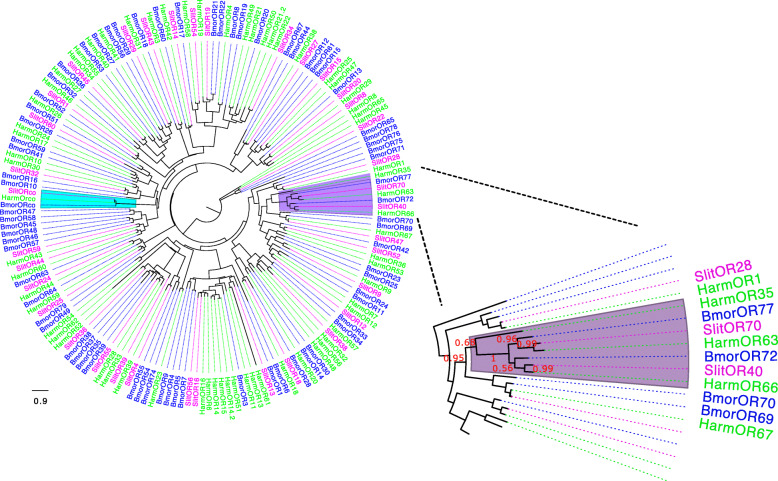
We constructed a phylogenetic tree based on the maximum likelihood method with the Bayesian information criterion (BIC) [43, 44] built using the online tool PhyML 3.0 (Fig. 5 and Additional file 9: Fig. S6). Smart Model Selection (SMS) method was used to choose the substitution model. Based on this, the “JTT+G+F” model was selected. We used amino acid sequences of ORs from larvae of S. littoralis along with ORs from larva and adult of B. mori and H. armigera [45–47]. The phylogenetic clustering of S. littoralis larval ORs (except newly identified SlitOR70) corroborates with previous reports [19, 31]. The OR of interest, SlitOR40, shared the same clade with SlitOR70. This well-supported clade (SH-aLRT = 1) contained four other ORs, two from B. mori (BmorOR72 and BmorOR77) and two from H. armigera (HarmOR63 and HarmOR66). These two last ORs from H. armigera have recently been functionally characterized, and HarmOR63 is tuned to β-caryophyllene and caryophyllene oxide [48]. Clustering of the SlitOR40, SlitOR70, BmorOR72, BmorOR77, HarmOR63, and HarmOR66 ORs’ topology did not change when all known ORs from S. littoralis [19, 31] were included in the phylogenetic analysis (Additional file 9: Fig. S6). Whether SlitOR70 is electrophysiologically “tuned” to β-caryophyllene and/or α-humulene remains to be investigated.
Fig. 5.
Maximum likelihood phylogenetic tree of ORs expressed in larvae of Spodoptera littoralis: The analyses included ORs from S. littoralis (purple), B. mori (blue), and H. armigera (green). Highlighted clades: Orco (cadet blue); SlitOR40 and SlitOR70 along with ORs from B. mori and H. armigera (lilac). Zoomed-in image: clade grouping SlitOR40, the newly identified OR SlitOR70, and four other ORs, belonging to B. mori: BmorOr72 and BmorOr77, and H. armigera: HarmOr63 and HarmOr66. The node support value (SH-aLRT) > 0.9 is considered well supported, 0.8–0.9 node values are considered weakly supported, and values < 0.8 indicate the node values are unsupported. The unrooted phylogenetic tree was built using the online tool PhyML 3.0
Discussion
In our study, we demonstrated for the first time that some ORs are differentially expressed according to larval stages in S. littoralis. The transcriptomic analysis conducted in this study revealed 36 ORs expressed in the first and fourth instar of S. littoralis. Interestingly, we did not identify any larval-specific ORs in S. littoralis, consistent with the data from the previous report [19], whereas larval-specific ORs have been identified in other Lepidoptera species such as B. mori [18].
We focused on SlitOR40, which we found expressed in the first, second, and third larval stages and not in the fourth larval stage. Through directed genome mutation, we knocked out the SlitOR40 gene and revealed that larval behavior to SlitOR40 ligands is differentially disrupted dependent on the larval stages. Precisely, CRISPR-Cas9-guided SlitOR40-KO disrupted specific odor detection abilities in the first instar larvae and affected foraging behavior. Our study confirms that modulation in expression of SlitOR40 during two developmental stages influences instar-specific attraction behavior.
SlitOR40 was deorphanized and shown to respond to the terpene β-caryophyllene and its isomer α-humulene, two volatile molecules that are commonly produced by plants, including host plants of S. littoralis, as reaction to herbivory [40, 41]. The response spectrum of SlitOR40 perfectly matched with the response spectrum of one OSN profile we previously identified via single sensillum screening in S. littoralis female moth antenna [49–51]. This OSN type was the only one found to respond to both β-caryophyllene and α-humulene. Taken together, these data suggest that SlitOR40 is the OR expressed in this OSN type.
Behavior studies revealed that the cognate ligands, β-caryophyllene and α-humulene, had a significant effect on the WT first instar larval choice, while these compounds had no significant effect on the fourth instar choice. These results clearly correlate with one major finding of our study, the absence of SlitOR40 in the fourth instar. This provides experimental evidence for differential OR expression as a molecular basis for behavioral plasticity such as changing food preference in larvae.
Interestingly, the cognate ligands elicited opposite behavioral responses (avoidance versus attraction) in the first instar WT larvae. Our results suggest that S. littoralis first instar larvae could distinguish between the isomers. As one would expect that the different ligands of one OR would trigger the same behavioral response, this finding also suggests a possible dual-channel for the detection of β-caryophyllene and/or α-humulene. Accordingly, previous studies on adults have shown that the S. littoralis female olfactory system contains two separate olfactory channels: one pathway tuned to β-caryophyllene and α-humulene, and the other pathway to α-humulene alone, and the signals innervate two different glomeruli in the antennal lobe [11, 49, 52]. This suggests that SlitOR40 is not the only receptor involved in α-humulene detection in S. littoralis female and supports the existence of a similar dual olfactory channel in larvae. Interestingly, the phylogenetic analysis of larval ORs clustered SlitOR40 in the same clade with the newly identified SlitOR70 and with HarmOR63, a receptor tuned to β-caryophyllene and caryophyllene oxide [48]. Such functional conservation of two ORs in this clade (SlitOR40 and HarmOR63) suggests that SlitOR70 constitutes a good candidate for the second channel of α-humulene detection.
Our EAG analyses of WT and SlitOR40-KO adults also support the existence of a second OR tuned to α-humulene in S. littoralis since the responses to α-humulene were not totally abolished in KO mutants, as they were for β-caryophyllene. In larvae, the scheme was more complex: if SlitOR40-KO abolished larval behavior to β-caryophyllene in the first instar, it also abolished larval behavior to α-humulene. Thus, the contribution of the other OR to the α-humulene response appeared as negligible at this developmental stage. However, the fourth instar SlitOR40-KO larvae showed a significant odor-guided behavior to α-humulene that was not evidenced in WT. A plausible explanation is that knocking out SlitOR40 may have a side effect on other OR(s), regulating or deregulating their expression, leading to a gained response to α-humulene at specific stages.
Conclusions
Our data, grouped altogether, provide the first empirical evidence that different larval stages use different ORs, bringing molecular explanation of dynamic changes in larval olfactory behavior. Specifically, our results on SlitOR40 clearly linked its expression to β-caryophyllene- and α-humulene-associated behavior in first instar larvae, but not in fourth instars. The discovery that larvae are able to respond differently to the isomers, β-caryophyllene and α-humulene (avoidance vs attraction), and that the associated behavior is instar-specific, adds a new dimension to our understanding of foraging behavior during larval development.
The volatiles β-caryophyllene and α-humulene are constitutive terpenes emitted by plants, including cotton, one of the preferred hosts of S. littoralis [53, 54]. While α-humulene is emitted at a low level compared to β-caryophyllene in undamaged plants, its emission increases during herbivory. It has been hypothesized that increasing rates of α-humulene most certainly signals herbivore-induced plant damage and therefore competition for food [55]. Both compounds have been associated with plant volatile blends that affect host plant selection for oviposition in S. littoralis and other moth species [56–60]. However, in some lepidopteran pests, β-caryophyllene and α-humulene induce different behavioral responses in female moths. For example, α-humulene stimulated oviposition in the European corn borer Ostrinia nubilalis (Pyralidae) when tested individually, unlike β-caryophyllene, which neither stimulated nor deterred it [56]. Similarly, in B. mori and Cydia strobilella (Lepidoptera; Tortricidae), α-humulene stimulated oviposition behavior, when tested individually and as a blend component [61, 62]. The behavioral responses of caterpillars to β-caryophyllene and α-humulene have been less investigated. One study reports that the neonate larvae of the congeneric species S. frugiperda are attracted to volatile blends including α-humulene induced by conspecifics’ foraging [63]. In our study, S. littoralis neonates avoided β-caryophyllene and preferred α-humulene. The ecological relevance of such differential behavior according to species, and according to instar stages as shown here, remains to be elucidated.
The previously deorphanized SlitOR4 that is tuned to (±)-linalool [36] was found in fourth but not in first instar larvae. Expression in other larval instars remains unknown and future studies are needed to clarify a possible instar-specificity and the role of (±)-linalool in larval development and behavior. Nonetheless, our results provide evidence that the molecular changes in the peripheral chemosensory system would serve changing developmental needs, reflected by larval behavioral changes. In the future, this may open new pest management strategies based on interference at adequate developmental stages for larval behavioral control.
Methods
Insects
S. littoralis lines used in this study were reared in the laboratory on a potato-based artificial diet [33, 64]. Adults and larvae were maintained at 24 ± 2 °C and 65 ± 5% relative humidity (RH) under 16:8 h light:dark (L:D) photoperiod. For the larval transcriptome, behavioral experiments, RT-PCR and RT-qPCR assays, neonate larvae (1–12 h old) and fourth instar larvae (10–11 days old) were used. For the SitOR40 and SlitOrco detailed RT-PCR analysis, RNA extractions were performed on second (3–4 days old) and third (7–8 days old) instar larvae. The whole larval period lasts 16–18 days, six instars in total.
Plant material
The cotton plants (Gossypium hirsutum L., Malavacae) (DPL90 variety) were grown under controlled conditions at 23 ± 2 °C and 70 ± 5% RH. Sodium lamps were used as supplement to natural light with 16:8 h L:D photoperiod. Leaves from 5–6-week-old plants were used for the olfactometer experiments (leaf disc of approximately 1-cm diameter).
Dissection/RNA extraction
Fourth instar larvae were starved for 6 h prior to dissection for consistency with behavioral experiments. For first and fourth instars, approximately 600 larval heads were collected. For adults, 50 pairs of male and female antennae were collected 3–4 days after eclosion from the pupae. For transcriptomics, first and fourth instar RNA extracts were used (one biological replicate). For RT-PCR and RT-qPCR, first to fourth instar and adult RNA extracts were used and five biological replicates were generated and used for cDNA synthesis for each of the four larval instars and adults. The TRIZOL (Invitrogen) extraction method was used to isolate the RNA from the target tissues, as previously described [31].
Illumina sequencing, processing, assembly, and annotation
Approximately 2 μg of total RNAs from first and fourth instar larvae were used for Illumina sequencing at BGI-Tech (China). Paired-end Illumina TruSeq libraries were generated from purified RNA using Illumina’s sample preparation kit (Illumina Inc., San Diego, CA) according to the manufacturer’s instructions and sequenced at a HiSeq 2000 platform. All raw data from this study have been deposited at NCBI (SRA accession: PRJNA507834). Reads were de novo assembled [65] in one transcriptome that contained first and fourth instar larval reads along with published reads data from adult antennae [31] (NCBI Accession: PRJNA312160, Sample Accessions: SRX2750986-SRX2750991) to provide a more robust reference for OR expression. Following the procedure described in Walker et al. [31], we removed low-quality reads and adapter contamination from the raw reads using trimmomatic tool (version 0.32) with the following parameters: ILLUMINACLIP (sequencing adapters file: TruSeq3-PE. fa:2:30:10) and TRAILING:20. Following this, Trinity pipeline (version 2.0.6) was used on the filtered data with default parameters [66]. The resulting Trinity.fasta files were then processed using the cd-hit-est tool (version 4.6.1) [67] to reduce transcript redundancy; in transcripts sharing sequence identity ≥ 95%, the longest sequence was retained and used for further analysis. Individual reads from first and fourth instar larvae were used for read mapping on the assembled full transcriptome using bowtie (version 0.12.6) [68]. The completeness of the assembled transcriptomes was assessed using the BUSCO tool (Benchmarking Universal Single-Copy Orthologs, version 3.0.2) (https://busco.ezlab.org/), against the insecta_obd9 dataset that included 1658 reference genes [35]. The transcripts’ expression level abundances were estimated using RSEM [69], and the mapping statistics were calculated using Samtool [70] for downstream analyses.
To identify the expressed ORs, we conducted a tBlastn search on the assembled transcriptome using an input file containing protein sequences of ORs previously identified in S. littoralis [19, 31], with an e-value cut-off of 1e−05. The putative OR transcripts were translated into protein sequences with the ExPASy portal [71] and were then aligned to reference transcripts to identify similarity or divergence using Clustal [72]. Newly identified SlitORs were added to the query dataset to conduct an additional tBlastn query.
RT-PCR and quantitative real-time PCR (RT-qPCR)
To prepare cDNAs from larval heads and adult antennae, total RNAs (1 μg) were reverse transcribed using the Invitrogen; ThermoScriptTM Reverse Transcriptase kit as described in the manufacturer’s protocol.
RT-PCR was performed using gene-specific primers (Additional file 10: Table S4). For all RT-PCR expression assays, 1 μL of cDNA was used with 1 μL of 10 mM of forward and reverse primers along with Dreamtaq buffer, dNTPs, and Dreamtaq enzyme making up the final volume of 25 μL. The RT-PCR amplification was as follows: 95 °C for 2’; 36 cycles X (95 °C for 30”, 53.5–65 °C (depending on primers) for 30”, 72°C for 1’ 30”, 72°C for 10’). The PCR-amplified products were run on a 1.5% agarose gel for verification.
RT-qPCR was also performed using gene-specific primers (Additional file 10: Table S4) and the amplicon length was between 100 and 200 bp. The amplification peak and the subsequent melting curve were checked to ensure primer specificity. Each RT-qPCR reaction was run in triplicates on five independent biological replicates and contained 10 μL of SYBR Green reagent (Invitrogen), 1 μL of 10 mM forward and reverse primers, 7 μL of RNase-free water, and 2μL of cDNA (5 ng/μL concentration cDNA), making the final volume of 20 μL. The reactions were performed in a 96-well plate (iQ5 qPCR System; plate type-white, Bio-Rad), and the amplification was as follows: initial denaturing step at 95 °C for 5’; 40 cycles X (95 °C 15”, annealing step 59–61 °C 25” (based on primer pairs)) 95 °C 10”, 50 to 95 °C (increment 0.5 °C) for 5”. We considered Orco as a relative reference to compare SlitOR40 expression in the larvae (heads) and male antennae, and β-actin, L13A, and Ef1A to compare Orco expression in first and fourth larval instars. For all the runs, relative expression was calculated using the 2−ΔΔCt (cycle threshold) method [73], and the geometric mean of expression values of the three reference genes were considered.
TOPO/gateway cloning of SlitOR40 and heterologous expression
The open reading frame (ORF) was amplified from larval cDNA using forward (F = GGAAGAGCTGCCTGAAATTTCAAAAGA) and reverse (R = CGACCAAGTTGTGCTCAGTAC) primer sequences and cloned into PCR8/GW/TOPO plasmids (Thermo Fisher Scientific), according to the manufacturer’s protocol. SlitOR40 was further cloned into the pUAST.attB plasmid using Clonase II enzyme mix kit (Thermo Fisher, Scientific, USA). The OR insert was sequenced using UAS1 and UAS2 sequencing primers [74] to confirm desired sequence and insertion orientation. The UAS-SlitOR40 Drosophila melanogaster lines were generated by BestGene, Inc (Chino Hills, CA, USA), using the PhiC31-mediated integration approach. Briefly, upon sequence confirmation, the recombinant pUAST-HA.attB-SlitOR40 plasmid containing a attB site was injected into attP docking site with PhiC31 activity within the third chromosome (genotype y1 M{vas-int.Dm}ZH- 2A w∗; M{3xP3-RFP.attP}ZH-86Fb) of the embryos of a fly. The UAS fly lines were then crossed with Or22a-GAL4 promoter flies that lack the endogenous OR genes OR22a and OR22b, following the crossing scheme in Gonzalez et al. [74]. The flies generated with genotype ∆halo; Or22a-GAL4/UAS-SlitOR40 were used for downstream electrophysiological recordings.
CRISPR/Cas9 egg injection
Two RNA guides were designed against exon 1 (gRNA1 sequence: TAGAGGAGAATTACAACTTG AGG) and exon 2 (gRNA2 sequence: TGCAGCTGGGAGGTATGTGG AGG) using the CRISPOR gRNA design tool (cripsor.tefor.net) [75] and the SlitOR40 genomic DNA sequence as target. Guide syntheses and Cas9 protein production were performed as previously described in Koutroumpa et al. [76]. The two gRNAs were injected together along with the Cas9 protein, in order to create more aggressive mutations with a higher probability to knockout (KO) the gene due to large deletions. Another advantage of these large deletions is that they can be visible on agarose gel with no need of further genotyping. Concentrations of the gRNAs were 45 μM and the Cas9 was 30 μM (Sp-Cas9-NLS-GFP-NLS; fusion of Streptococcus pyogenes-derived Cas9 protein and green fluorescent protein between nuclear localization signal on both N and C terminal of the protein). The gRNA:Cas9 ratio in the complex was 1.5:1. Aliquots of gRNA were denatured at 80°C for 2 min and then left on ice for 2 min before mixing them with the necessary amount of Cas9. Each complex was formed at room temperature during 10 min. Mix of the two gRNA:Cas9 complexes was done afterwards in order to avoid eventual Cas9 binding preferentially to one gRNA. The mix was then placed on ice until use. S. littoralis eggs were prepared for injection as described in Koutroumpa et al. [76]. Injections took place within one half to 1 h after oviposition in order to target the first steps in embryogenesis. We used an Eppendorf FemtoJet 4i injector.
Crude genomic DNA was extracted from one larval pseudopod (Wizard Genomic DNA Purification Kit, Promega, Madison, WI). Amplification of the gDNA was performed using the Pfu thermostable polymerase (Promega, Madison, WI) and specific primers for SlitOR40. The forward primer (TCGAACCGATATTACCATGTCTG) sitting on exon 1 and the reverse primer (ACTTTAGTCTCCTCAGTAACGT) on exon 2 amplifies a PCR product of 684 bp and includes the target sequence. First, the gDNA was denatured at 95°C for 2 min. The amplification program was 45 s at 95°C, 30 s at 62°C, and 60 s at 72°C, and this was repeated 40 times. Agarose gel analysis gave size polymorphism differences among individual amplifications, and sequencing (Biofidal, Vaulx-en-Velin, France) confirmed the gel results. Sequences were aligned and manually curated using SEQUENCHER™ 4.7 (Gene Codes Corporation, Inc.).
Injected eggs were kept at the same conditions as the regular rearing. G0 adults carrying mutagenic events were backcrossed with adults from the regular rearing (called here wild type, WT). G1 heterozygote males and females carrying the same mutation were crossed to obtain homozygote G2 SlitOR40 knockouts (KO). One G2 homozygote CRISPR line was chosen for further analysis. Adults were tested for their response in EAG and larval behavior was tested in olfactometer assays.
Olfactometer assays
Y-tube olfactometer tests were performed to examine the behavior of WT and SlitOR40-KO larvae. Olfactometers of different dimensions were selected according to different sizes of tested larvae. For fourth instar larvae, the size of the olfactometer that we designed was arm length = 14 cm, stem = 12.5 cm, and inner diameter = 2.2 cm, while for the neonate larvae, the olfactometer size was arm length = 4 cm, stem = 5 cm, and inner diameter = 0.8 cm (built at Humiglas, Södra Sandby, Sweden). For both systems, control experiments confirmed adequate settings with larvae showing equal upwind walk towards cotton leaf discs on both sides as well as discrimination between the arms providing leaf discs alone and leaf discs with control stimulus (Additional file 7: Fig. S4). Experiments were performed under diffused light (53 lux) from the top of the olfactometer. A charcoal-filtered airstream was pumped (0.1 L/min for first instar and 0.2 L/min for fourth instar) through a wash bottle containing 50 mL distilled water for humidification and then split into each arm of the olfactometer.
To achieve context-based attraction, each arm of the olfactometer had two cotton leaf discs (each 1-cm diameter) on a wet filter paper (Grade 1002, Munktell Filter AB, Munktell) as a background. Then, filter paper in one arm of the olfactometer was treated with treatment solution (β-caryophyllene or α-humulene) diluted in paraffin oil on a 1-cm2 filter paper (10 μL), versus control (10 μL paraffin oil). The behavior of WT and SlitOR40-KO larvae was tested with three different doses (1, 10, and 100 μg) of β-caryophyllene or α-humulene.
The fourth instar larvae were starved for 6–8 h prior to the experiment. Larvae were tested individually for 10 min. Larval responses were recorded as “treatment” or “control” based on their choices, and the larvae that did not enter the olfactometer arms within 10 min were treated as “no-choice.” To avoid any positional effect, treatment position was swapped between the arms of the olfactometer every fifth larva, and 10–15 larvae were tested per day. An adjusted protocol was followed for the first instar larvae. After hatching, larvae feed on the egg residues, and after approximately 45 min to 1 h, start moving towards food sources. Those larvae were selected for the behavioral assays. First instar larvae required a longer “activation time” to start moving in the olfactometer compared to fourth instar larvae. Therefore, first instar larvae were tested for 20 min.
The percentage attraction was calculated as behavioral response = (number of larvae in the arm with treatment/total number of larvae that made a choice) multiplied by 100.
Electrophysiological recordings
Single sensillum recording (SSR) on D. melanogaster neurons expressing SlitOR40
We functionally characterized the SlitOR40 receptor using the Drosophila empty neuron system [39, 77]. Four- to 5-day-old D. melanogaster female flies carrying SlitOR40 receptors expressed in ab3 sensillum “A” neurons were used for electrophysiological recordings. Recordings were repeated with six different flies. Prior to recording, a fly was immobilized in a 100-μL pipette tip with its antennae protruding outside the narrow tip. This tip was mounted on a glass slide positioning the fly ventrally, and the right antenna was gently pulled flat over the glass coverslip. The antenna was held by a micro-capillary at the flagellum to avoid antennal movement. The glass slide was gently placed under the microscope (Olympus BX51W1) across a glass tube (positioned approximately 20 mm from the fly antenna) connected to charcoal-filtered humidified main airflow (1 L/min). At the lateral side of the glass tube, a hole allowed the introduction of a disposable glass Pasteur pipette (150 mm, Assistent, # 567/1, Germany) containing the odorant on the filter paper. A reference electrode was gently inserted into the eye using a motor-controlled piezo micromanipulator (Märzhauser DC-3K, Wetzlar, Germany), while a recording microelectrode was inserted into the base of the ab3 sensilla. The extracellular action potentials from the sensory neuron were recorded using tungsten microelectrodes, sharpened using KNO2-solution. The signal was then amplified (INR-02A and AC/DC UN-06, respectively) and transferred to a computer through IDAC4 (Intelligent Data Acquisition Controller-4) for visualization. The analysis was performed using Autospike (version 3.4) software (Syntech, Kirchzarten, Germany).
Odorant compounds were diluted in paraffin oil (Merck) except 1-indanone, carvacrol, and 4,8-dimethyl-1,3,7-nonatriene (DMNT), which were diluted in hexane (LabScan) to a concentration of 10 μg/μL (Additional file 11: Table S5). A disposable glass Pasteur pipette containing 1.5 × 1 cm filter paper (size Ø 90mm; grade 1002; Munktell) was then loaded with 100 μg (diluted in 10 μL of paraffin oil or hexane) of the odorant compound to deliver the stimulus (since there was a very low electrophysiological response to most of the odorants, lower doses were not tested). β-Caryophyllene (chemical purity = 98.5%) and α-humulene (chemical purity = 96%) were tested at the dose of 10 μg (diluted in 10 μl of paraffin oil). Disposable Pasteur pipettes loaded with individual odorants were used in order to deliver odors using stimulus controller (Syntech SFC-1/b; 2.5 ml air flux) onto the fly antenna, from which responses were recorded for 0.5 s post-stimulation.
Electroantennography (EAG)
We performed EAG measurements on adult antennae to determine olfactory responses from WT and KO moths to SlitOR40 cognate ligands. Two other compounds, guaiacol and (±)-linalool, were included as positive control stimuli, as they are known to be ligands of SlitORs expressed in adult antennae [36, 78]. All odorants were diluted in hexane. Three doses of β-caryophyllene and α-humulene (100 μg, 10 μg, and 1 μg) and one dose (10 μg) of each positive control stimulus were tested.
The excised moth antennal base was placed at the tip of a glass electrode (filled with Beadle-Ephrussi Ringer solution) [79] connected to a 10x preamplifier probe that was linked to an IDAC-2 box (Syntech, Kirchzarten, Germany), while the distal end of the antenna was connected to another glass electrode for grounding. The electrodes were placed 1 cm away from an odor-delivery glass tube connected to a humidified charcoal-filtered airstream (1 L/min). A disposable Pasteur pipette (VWR International, Stockholm, Sweden) containing filter paper strip (1.5 × 0.5 cm; Whatman®) was loaded with 10 μL of odorant, which was delivered (0.5 s) on the moth antenna. The signal from the antenna was amplified and transferred to a computer for visualization using the Autospike program (version 1.2.5, Syntech).
Phylogenetic analysis
Two phylogenetic trees were built from S. littoralis ORs: one with larval expressed ORs, and the other with all described ORs [19, 31], along with OR sequences from B. mori (71 ORs from larva and adults) [45, 47] and H. armigera (66 ORs from larva and adults) [46]. All amino acid sequences were aligned using MAFFT (version 7) (https://mafft.cbrc.jp/alignment/server/) [80, 81], and an unrooted phylogenetic tree was built using PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/) [43, 82] using BioNJ algorithm and Maximum Likelihood Tree with Smart Model Selection (SMS) method [44]. This software tool integrated into the PhyML web server automatically selects the best substitution model. This method chose the “JTT+G+F” substitution model. PhyML uses both NNI (Nearest Neighbor Interchanges) and SPR (Subtree pruning and regrafting) method to rearrange and optimize the tree structure. Clade support for maximum likelihood analysis was assessed using Shimodiara-Hasegawa-approximate likelihood ratio test (SH-aLRT) [43, 83]. The nodes with support value SH-aLRT > 0.9 were considered well supported, nodes with value ranging from 0.8 to 0.9 were considered weakly supported, and node values < 0.8 were considered unsupported [43]. Consensus Newick format tree was visualized and processed in FigTree software (version 1.4.4) (http://tree.bio.ed.ac.uk/software/figtree/).
Statistical analysis
All statistical analyses were performed in R [84]. Behavioral responses of first and fourth instar larvae to different doses of β-caryophyllene and α-humulene were analyzed with binomial GLM to test the factors (instar, genotype, concentration of doses) influencing larval behavior. To analyze RT-qPCR data from larvae and adults, we checked for normal distribution and performed paired t-test on ΔCt values. EAG responses of WT and SlitOR40-KO adult moths to (±)-linalool and to guaiacol followed a normal distribution and were analyzed with ANOVA. To analyze EAG responses of moth antennae to β-caryophyllene and α-humulene (three doses of each compound), we performed ANCOVA to test the influence of doses, sex, and genotype.
Supplementary Information
Additional file 1: Table S1. BUSCO scores for transcriptomes assembly completeness: Assessment of Spodoptera littoralis larval transcriptomes assembly completeness using the Benchmarking BUSCO tool performed against insecta_obd9 database consisting of 1,658 BUSCOs.
Additional file 2: Table S2. Expression profile of Spodoptera littoralis ORs (SlitORs) in the first and fourth instar larval heads. Expression levels of ORs are displayed as FPKM values.
Additional file 3: Table S3. Newly identified SlitOR70 amino acid sequence.
Additional file 4: Figure S1. RT-PCR gel images of PCR-amplified ORs at different developmental stages of Spodoptera littoralis. Comparison included negative-control (control), first instar heads (Slit_1), fourth instar heads (Slit_4), male antennae (Slit_♂) and female antennae (Slit_♀) with 1kb gene ruler ladder (Thermo Fischer Scientific). Red rectangles around SlitOR4 and SlitOR40 represent differential expression in the first and fourth instar, respectively, supported by RT-PCR and transcriptomic analysis.
Additional file 5: Figure S2. Expression pattern (RT-PCR) and relative expression levels (RT-qPCR) of Orco and SlitOR40 across different instars. (A) RT-PCR products obtained from SlitOrco and SlitOR40 amplification from the cDNA of first, second, third and fourth instar larval heads and adult antennae. (B) Quantitative RT-qPCR expression levels (relative fold-change) of Orco in first instar larval heads, fourth instar larval heads and adult male antennae using β-actin, L13A and Ef1A as reference genes. Male antennae samples expressed significantly higher levels of Orco compared to samples from first and fourth instar larvae (Df = 28, t= -22.622, P<0.001; Df = 28, t= 50.441, P < 0.001, respectively). Similarly, in the fourth instar larvae, Orco expression was significantly higher compared to first instar (Df = 28, t = 9.33, P < 0.001). (C) SlitOR40 expression (using Orco as reference gene) in the first instar larval heads, fourth instar larval heads and adult male antennae. Relative expression of SlitOR40 in first instar larvae was significantly higher compared to adult male antennae (Df = 28, t = 26.532, P < 0.001), while in the fourth larval instar, SlitOR40 expression was not detected (Df = 28, t = 27.693, P < 0.001). The analyses are based on five biological samples. The bars indicate standard errors. Statistical differences between treatments were calculated using a non-paired Student’s t-test. *: P < 0.05.
Additional file 6: Figure S3. SlitOR40 CRISPR/Cas9 induced indels (insertion and deletion). (A) Wild Type (WT) cDNA sequence including exon1 and exon2. An asterisk separates the two exons’ sequences. Blue letters indicate the primer sequences used for genotyping. Orange and purple boxes surround sequence regions where the Cas9 cleaved SlitOR40 and correspond to the same color boxes in B. (B) The three most prominent double mutational events obtained are shown around each of the two CRISPR guides. Indels’ produced between the two guides are described (NA means not applied) and insertion sequences are indicated in red letters. The number of nucleotide differences (Δ) between the WT line and the three mutant obtained lines are given between brackets. In both A and B, green letters show the CRISPR guide sequences with their PAM highlighted in yellow. (C) Electrophoregram traces for each of the three main double mutations obtained (BioEdit). Line 10 mutants were chosen for further crossing and phenotypic analysis.
Additional file 7: Figure S4. Attraction of Spodoptera littoralis wild type and SlitOR40-knockout larvae to positive control odors: Attraction of first and fourth instar WT and KO genotypes to positive control odors (CLD, cotton leaf discs) in both arms of the olfactometer (first instar WT: n = 44; fourth instar WT: n = 24; first instar KO: n = 59; fourth instar KO: n = 43) (top bar in Fig. S4A, S4B, S4C and S4D, respectively). Similarly, behavioral responses of WT and KO genotypes of S. littoralis first and fourth instar to the odor of cotton leaf discs versus clean air (control blank) (first instar WT: n = 50; fourth instar WT: n = 48; first instar KO: n = 21; fourth instar KO: n = 32) (bottom bar in Fig. S4A, S4B, S4C and S4D, respectively).
Additional file 8: Figure S5. EAG responses of Spodoptera littoralis wild type and SlitOr40-knockout moth antennae to positive control odors: The EAG responses of WT and KO line adult moth antennae to positive control stimuli (guaiacol and (±)-linalool) were tested at 10 μg dose (minimum n = 9 per sex). There was no statistical difference in responses to positive control stimuli, guaiacol (ANOVA: Sex: Df = 1; F = 1.63; P = 0.2; Genotype: Df = 1; F = 3.55; P = 0.066; Interaction: Df = 1; F = 0.13; P = 0.72; Residual Df = 47) and (±)-linalool (ANOVA: Sex: Df = 1; F = 0.66; P = 0.42; Genotype: Df = 1; F = 0.812; P = 0.372; Interaction: Df = 1; F = 1.68; P = 0.202; Residual Df = 47) in the antennae from both genotypes. The boxplots represent mean values along with the minimum and maximum values.
Additional file 9: Figure S6. Maximum-likelihood phylogenetic tree of ORs expressed in larvae and adults of Spodoptera littoralis: The analyses included all described ORs from S. littoralis (purple), B. mori (blue) and H. armigera (green). Highlighted clades: Orco (cadet blue); SlitOR40 and SlitOR70 along with ORs from B. mori and H. armigera (lilac). The unrooted phylogenetic tree was built using online tool PhyML 3.0.
Additional file 10: Table S4. Primers used in the study: List of all primers used to confirm the expression of SlitORs, qRT-PCR assays in first and fourth larval instars, and primers used for genotyping.
Additional file 11: Table S5. List of chemical compounds used in the electrophysiological recordings.
Acknowledgements
We thank Elisabeth Marling for her help in insect rearing, Dr. Ramesh Vetukuri for technical advice on RT-qPCR analysis, and Dr. Sandeep Kushwaha for his help to submit RNA-sequence files to NCBI. The computations were performed on the resources provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX.
Abbreviations
- ORs
Odorant receptors
- IRs
Ionotropic receptors
- CRISPR
Clustered regularly interspaced short palindromic repeats
- Cas9
CRISPR-associated protein 9
- S. littoralis
Spodoptera littoralis
- S. frugiperda
Spodoptera frugiperda
- B. mori
Bombyx mori
- H. armigera
Helicoverpa armigera
- D. melanogaster
Drosophila melanogaster
- OSNs
Olfactory sensory neurons
- ORCO
Odorant receptor co-receptor
- RNA
Ribonucleic acid
- DNA
Deoxyribonucleic acid
- RNA-Seq
RNA sequencing
- RT-PCR
Real-time polymerase chain reaction
- qRT-PCR
Quantitative real-time polymerase chain reaction
- BUSCO
Benchmarking Universal Single-Copy Orthologs
- sgRNA
Single guide RNA
- Bp
Base pairs
- WT
Wild type
- KO
Knockout
- CLD
Cotton leaf disc
- μg
Microgram
- ANOVA
Analysis of variance
- ANCOVA
Analysis of co-variance
- EAG
Electroantennogram
- SSR
Single sensillum recording
- BIC
Bayesian information criterion
- L:D
Light:dark
- RH
Relative humidity
- Sp-Cas9-NLS-GFP-NLS
Streptococcus pyogenes-derived Cas9 protein and green fluorescent protein between nuclear localization signal on both N and C terminal of the protein
- DMNT
4,8-Dimethyl-1,3,7-nonatriene
- IDAC
Intelligent Data Acquisition Controller
- KNO2
Potassium nitrite
Authors’ contributions
S.V.R., E-J.J., and P.G.B. conceived the study. S.V.R. and W.B.W. collected RNA-Seq data and generated empty neuron fly lines; S.V.R. performed electrophysiological recordings, RT-PCR, and RT-qPCR; S.V.R., G.M.H., V.A.G., V.R., E.C., and G.R. did larval behavior assays; S.V.R., E-J.J., and F.K. generated SlitOR40-knockout lines and performed genotyping. S.V.R., L.C., W.W.B., F.K., P.A., E-J.J, and P.G.B. were involved in study design, data analysis, and interpretation; S.V.R. wrote the manuscript; all authors contributed to revisions. All authors have read and approved the final version of the manuscript.
Funding
The study was supported by the Linnaeus grant “Insect Chemical Ecology, Ethology and Evolution (IC-E3)” by The Swedish Research Council Formas, an Early Career Mobility Grant by Formas to S.R.V. (2016-01434), the SLU Centre for Biological Control (CBC), the French National Research Agency (ANR-16-CE21-0002-01), and the “Erasmus Student Mobility for Traineeship program.” Open Access funding provided by Swedish University of Agricultural Sciences.
Availability of data and materials
Transcriptomics data are deposited in the public biomedical database, NCBI under the following accession number: PRJNA507834. All of the data on which conclusions rely in this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fotini Koutroumpa and Paul G. Becher contributed equally to this work.
References
- 1.Lind AJ, Welsh HH. Ontogenetic changes in foraging behaviour and habitat use by the Oregon garter snake, Thamnophis atratus hydrophilus. Anim Behav. 1994;48(6):1261–1273. doi: 10.1006/anbe.1994.1362. [DOI] [Google Scholar]
- 2.Shtonda BB, Avery L. Dietary choice behavior in Caenorhabditis elegans. J Exp Biol. 2006;209(1):89–102. doi: 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fougeron AS, Farine JP, Flaven-Pouchon J, Everaerts C, Ferveur JF. Fatty-acid preference changes during development in Drosophila melanogaster. PLoS One. 2011;6(10):e26899. doi: 10.1371/journal.pone.0026899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clay TA, Pearmain EJ, McGill RAR, Manica A, Phillips RA. Age-related variation in non-breeding foraging behaviour and carry-over effects on fitness in an extremely long-lived bird. Funct Ecol. 2018;32(7):1832–1846. doi: 10.1111/1365-2435.13120. [DOI] [Google Scholar]
- 5.Quintero C, Bowers MD. Plant and herbivore ontogeny interact to shape the preference, performance and chemical defense of a specialist herbivore. Oecologia. 2018;187(2):401–412. doi: 10.1007/s00442-018-4068-8. [DOI] [PubMed] [Google Scholar]
- 6.Ali A, Luttrell RG, Pitre HN. Feeding sites and distribution of fall armyworm (Lepidoptera: Noctuidae) larvae on cotton. Environ Entomol. 1990;19(4):1060–1067. doi: 10.1093/ee/19.4.1060. [DOI] [Google Scholar]
- 7.Stockhoff BA. Ontogenetic change in dietary selection for protein and lipid by gypsy moth larvae. J Insect Physiol. 1993;39(8):677–686. doi: 10.1016/0022-1910(93)90073-Z. [DOI] [Google Scholar]
- 8.Chapman JW, Williams T, Escribano A, Caballero P, Cave RD, Goulson D. Fitness consequences of cannibalism in the fall armyworm, Spodoptera frugiperda. Behav Ecol. 1999;10(3):298–303. doi: 10.1093/beheco/10.3.298. [DOI] [Google Scholar]
- 9.Van Dam NM, Hermenau U, Baldwin IT. Instar-specific sensitivity of specialist Manduca sexta larvae to induced defences in their host plant Nicotiana attenuata. Ecol Entomol. 2001;26(6):578–586. doi: 10.1046/j.1365-2311.2001.00366.x. [DOI] [Google Scholar]
- 10.Hanson FE, Dethier VG. Role of gustation and olfaction in food plant discrimination in the tobacco hornworm, Manduca sexta. J Insect Physiol. 1973;19(5):1019–1031. doi: 10.1016/0022-1910(73)90028-0. [DOI] [PubMed] [Google Scholar]
- 11.Anderson P, Hansson BS, Löfqvist J. Plant-odour-specific receptor neurons on the antennae of female and male Spodoptera littoralis. Physiol Entomol. 1995;20(3):189–198. doi: 10.1111/j.1365-3032.1995.tb00001.x. [DOI] [Google Scholar]
- 12.Landolt PJ, Brumley JA, Smithhisler CL, Biddick LL, Hofstetter RW. Apple fruit infested with codling moth are more attractive to neonate codling moth larvae and possess increased amounts of (E,E)-α-farnesene. J Chem Ecol. 2000;26(7):1685–1699. doi: 10.1023/A:1005595014589. [DOI] [Google Scholar]
- 13.Hansson BS, Stensmyr MC. Evolution of insect olfaction. Neuron. 2011;72(5):698–711. doi: 10.1016/j.neuron.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Itagaki H, Hildebrand JG. Olfactory interneurons in the brain of the larval sphinx moth Manduca sexta. J Comp Physiol A. 1990;167(3):309–320. doi: 10.1007/BF00192566. [DOI] [PubMed] [Google Scholar]
- 15.Kuebler LS, Schubert M, Kárpáti Z, Hansson BS, Olsson SB. Antennal lobe processing correlates to moth olfactory behavior. J Neurosci. 2012;32(17):5772–5782. doi: 10.1523/JNEUROSCI.6225-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kymre JH, Liu XL, Ian E, Berge CN, Wang GR, Berg BG, et al. Distinct protocerebral neuropils associated with attractive and aversive female-produced odorants in the male moth brain. eLife. 2021;10:1–27. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laue M. Immunolocalization of general odorant-binding protein in antennal sensilla of moth caterpillars. Arthropod Struct Dev. 2000;29(1):57–73. doi: 10.1016/S1467-8039(00)00013-X. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, Yamaoka R, Touhara K. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol. 2009;19(11):881–890. doi: 10.1016/j.cub.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 19.Poivet E, Gallot A, Montagné N, Glaser N, Legeai F, Jacquin-Joly E. A comparison of the olfactory gene repertoires of adults and larvae in the noctuid moth Spodoptera littoralis. PLoS One. 2013;8(4):e60263. doi: 10.1371/journal.pone.0060263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker WB, Gonzalez F, Garczynski SF, Witzgall P. The chemosensory receptors of codling moth Cydia pomonella expression in larvae and adults. Sci Rep. 2016;6(1):23518. doi: 10.1038/srep23518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di C, Ning C, Huang LQ, Wang CZ. Design of larval chemical attractants based on odorant response spectra of odorant receptors in the cotton bollworm. Insect Biochem Mol Biol. 2017;84:48–62. doi: 10.1016/j.ibmb.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43(5):703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 23.Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452(7190):1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 24.Becher PG, Guerin PM. Oriented responses of grapevine moth larvae Lobesia botrana to volatiles from host plants and an artificial diet on a locomotion compensator. J Insect Physiol. 2009;55(4):384–393. doi: 10.1016/j.jinsphys.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 25.de Fouchier A, Sun X, Caballero-Vidal G, Travaillard S, Jacquin-Joly E, Montagné N. Behavioral effect of plant volatiles binding to Spodoptera littoralis larval odorant receptors. Front Behav Neurosci. 2018;12:264. doi: 10.3389/fnbeh.2018.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang XJ, Guo H, Di C, Yu S, Zhu L, Huang LQ, et al. Sequence similarity and functional comparisons of pheromone receptor orthologs in two closely related Helicoverpa species. Insect Biochem Mol Biol. 2014;48:63–74. doi: 10.1016/j.ibmb.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 27.Taparia T, Ignell R, Hill SR. Blood meal induced regulation of the chemosensory gene repertoire in the southern house mosquito. BMC Genomics. 2017;18(1):393. doi: 10.1186/s12864-017-3779-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crava CM, Sassù F, Tait G, Becher PG, Anfora G. Functional transcriptome analyses of Drosophila suzukii antennae reveal mating-dependent olfaction plasticity in females. Insect Biochem Mol Biol. 2018;105:51–59. doi: 10.1016/j.ibmb.2018.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Anderson AR, Wanner KW, Trowell SC, Warr CG, Jaquin-Joly E, Zagatti P, Robertson H, Newcomb RD. Molecular basis of female-specific odorant responses in Bombyx mori. Insect Biochem Mol Biol. 2009;39(3):189–197. doi: 10.1016/j.ibmb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Brown ES, Dewhurst CF. The genus Spodoptera (Lepidoptera: Noctuidae) in Africa and the Near East. Bull Entomol Res. 1975;65(2):221–262. doi: 10.1017/S0007485300005939. [DOI] [Google Scholar]
- 31.Walker WB, Roy A, Anderson P, Schlyter F, Hansson BS, Larsson MC. Transcriptome analysis of gene families involved in chemosensory function in Spodoptera littoralis (Lepidoptera: Noctuidae) BMC Genomics. 2019;20(1):428. doi: 10.1186/s12864-019-5815-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlsson MA, Anderson P, Hartlieb E, Hansson BS. Experience-dependent modification of orientational response to olfactory cues in larvae of Spodoptera littoralis. J Chem Ecol. 1999;25(11):2445–2454. doi: 10.1023/A:1020865922827. [DOI] [Google Scholar]
- 33.Saveer AM, Kromann SH, Birgersson G, Bengtsson M, Lindblom T, Balkenius A, Hansson BS, Witzgall P, Becher PG, Ignell R. Floral to green: mating switches moth olfactory coding and preference. Proc R Soc B Biol Sci. 2012;279(1737):2314–2322. doi: 10.1098/rspb.2011.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borrero-Echeverry F, Becher PG, Birgersson G, Bengtsson M, Witzgall P, Saveer AM. Flight attraction of Spodoptera littoralis (Lepidoptera, Noctuidae) to cotton headspace and synthetic volatile blends. Front Ecol Evol. 2015;3:56. doi: 10.3389/fevo.2015.00056. [DOI] [Google Scholar]
- 35.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 36.de Fouchier A, Walker WB, Montagne N, Steiner C, Schlyter F, Chertemps T, et al. Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat Commun. 2017;8(1):15709. doi: 10.1038/ncomms15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-shui S, Hai-kuo Y, Jing-hua D, Zi-ge X, Wan-qiang Q, Jin-tian L. Stability evaluation of reference genes for real-time quantitative PCR normalization in Spodoptera frugiperda (Lepidoptera : Noctuidae) J Integr Agric. 2020;19(9):2–13. doi: 10.1016/S2095-3119(20)63298-1. [DOI] [Google Scholar]
- 38.Butterwick JA, del Mármol J, Kim KH, Kahlson MA, Rogow JA, Walz T, Ruta V. Cryo-EM structure of the insect olfactory receptor Orco. Nature. 2018;560(7719):447–452. doi: 10.1038/s41586-018-0420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobritsa AA, Van Der Goes Van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37(5):827–841. doi: 10.1016/S0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- 40.Hoballah ME, Turlings TCJ. The role of fresh versus old leaf damage in the attraction of parasitic wasps to herbivore-induced maize volatiles. J Chem Ecol. 2005;31(9):2003–2018. doi: 10.1007/s10886-005-6074-7. [DOI] [PubMed] [Google Scholar]
- 41.Zakir A, Sadek MM, Bengtsson M, Hansson BS, Witzgall P, Anderson P. Herbivore-induced plant volatiles provide associational resistance against an ovipositing herbivore. J Ecol. 2013;101(2):410–417. doi: 10.1111/1365-2745.12041. [DOI] [Google Scholar]
- 42.Thöming G, Larsson MC, Hansson BS, Anderson P. Comparison of plant preference hierarchies of male and female moths and the impact of larval rearing hosts. Ecology. 2013;94(8):1744–1752. doi: 10.1890/12-0907.1. [DOI] [PubMed] [Google Scholar]
- 43.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 44.Lefort V, Longueville JE, Gascuel O. SMS: Smart Model Selection in PhyML. Mol Biol Evol. 2017;34(9):2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia Q, Wang J, Zhou Z, Li R, Fan W, Cheng D, et al. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38(12):1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Liu NY, Xu W, Papanicolaou A, Dong SL, Anderson A. Identification and characterization of three chemosensory receptor families in the cotton bollworm Helicoverpa armigera. BMC Genomics. 2014;15(1):597. doi: 10.1186/1471-2164-15-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu CZ, Zhou QZ, Liu TT, Fang SM, Wang YW, Fang X, Huang CL, Yu QY, Chen CH, Zhang Z. Evidence of peripheral olfactory impairment in the domestic silkworms: insight from the comparative transcriptome and population genetics. BMC Genomics. 2018;19(1):788. doi: 10.1186/s12864-018-5172-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo M, Du L, Chen Q, Feng Y, Zhang J, Zhang X, et al. Odorant receptors for detecting flowering plant cues are functionally conserved across moths and butterflies. Mol Biol Evol. 2021;38(4):1413–1427. doi: 10.1093/molbev/msaa300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jönsson M, Anderson P. Electrophysiological response to herbivore-induced host plant volatiles in the moth Spodoptera littoralis. Physiol Entomol. 1999;24(4):377–385. doi: 10.1046/j.1365-3032.1999.00154.x. [DOI] [Google Scholar]
- 50.Binyameen M, Anderson P, Ignell R, Seada MA, Hansson BS, Schlyter F. Spatial organization of antennal olfactory sensory neurons in the female Spodoptera littoralis moth: differences in sensitivity and temporal characteristics. Chem Senses. 2012;37(7):613–629. doi: 10.1093/chemse/bjs043. [DOI] [PubMed] [Google Scholar]
- 51.Binyameen M, Anderson P, Ignell R, Birgersson G, Razaq M, Shad SA, Hansson BS, Schlyter F. Identification of plant semiochemicals and characterization of new olfactory sensory neuron types in a polyphagous pest moth, Spodoptera littoralis. Chem Senses. 2014;39(8):719–733. doi: 10.1093/chemse/bju046. [DOI] [PubMed] [Google Scholar]
- 52.Carlsson MA, Galizia CG, Hansson BS. Spatial representation of odours in the antennal lobe of the moth Spodoptera littoralis (Lepidoptera: Noctuidae) Chem Senses. 2002;27(3):231–244. doi: 10.1093/chemse/27.3.231. [DOI] [PubMed] [Google Scholar]
- 53.Loughrin JH, Manukian A, Heath RR, Tumlinson JH. Volatiles emitted by different cotton varieties damaged by feeding beet armyworm larvae. J Chem Ecol. 1995;21(8):1217–1227. doi: 10.1007/BF02228321. [DOI] [PubMed] [Google Scholar]
- 54.Mcauslane HJ, Alborn HT. Systemic induction of allelochemicals in glanded and glandless isogenic cotton by Spodoptera exigua feeding. J Chem Ecol. 1998;24(2):399–416. doi: 10.1023/A:1022596812049. [DOI] [Google Scholar]
- 55.McCall PJ, Turlings TCJ, Loughrin J, Proveaux AT, Tumlinson JH. Herbivore-induced volatile emissions from cotton (Gossypium hirsutum L.) seedlings. J Chem Ecol. 1994;20:3039–3050. doi: 10.1007/BF02033709. [DOI] [PubMed] [Google Scholar]
- 56.Binder BF, Robbins JC. Effect of terpenoids and related compounds on the oviposition behavior of the European corn borer, Ostrinia nubilalis (Lepidoptera: Pyralidae) J Agric Food Chem. 1997;45(3):980–984. doi: 10.1021/jf960400z. [DOI] [Google Scholar]
- 57.De Moraes CM, Mescher MC, Tumlinson JH. Caterpillar-induced nocturnal plant volatiles repel conspecific females. Nature. 2001;410(6828):577–580. doi: 10.1038/35069058. [DOI] [PubMed] [Google Scholar]
- 58.Konstantopoulou MA, Krokos FD, Mazomenos BE. Chemical composition of corn leaf essential oils and their role in the oviposition behavior of Sesamia nonagrioides females. J Chem Ecol. 2004;30(11):2243–2256. doi: 10.1023/B:JOEC.0000048786.12136.40. [DOI] [PubMed] [Google Scholar]
- 59.Witzgall P, Ansebo L, Yang Z, Angeli G, Sauphanor B, Bengtsson M. Plant volatiles affect oviposition by codling moths. Chemoecology. 2005;15(2):77–83. doi: 10.1007/s00049-005-0295-7. [DOI] [Google Scholar]
- 60.Zakir A, Bengtsson M, Sadek MM, Hansson BS, Witzgall P, Anderson P. Specific response to herbivore-induced de novo synthesized plant volatiles provides reliable information for host plant selection in a moth. J Exp Biol. 2013;216:3257–3263. doi: 10.1242/jeb.083188. [DOI] [PubMed] [Google Scholar]
- 61.Bédard C, Gries R, Gries G, Bennett R. Cydia strobilella (Lepidoptera: Tortricidae): antennal and behavioral responses to host and nonhost volatiles. Can Entomol. 2002;134(6):793–804. doi: 10.4039/Ent134793-6. [DOI] [Google Scholar]
- 62.Damodaram KJP, Kempraj V, Aurade RM, Rajasekhar SB, Venkataramanappa RK, Nandagopal B, Verghese A. Centuries of domestication has not impaired oviposition site-selection function in the silkmoth, Bombyx mori. Sci Rep. 2014;4(1):7472. doi: 10.1038/srep07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carroll MJ, Schmelz EA, Meagher RL, Teal PEA. Attraction of Spodoptera frugiperda larvae to volatiles from herbivore-damaged maize seedlings. J Chem Ecol. 2006;32(9):1911–1924. doi: 10.1007/s10886-006-9117-9. [DOI] [PubMed] [Google Scholar]
- 64.Hinks CF, Byers JR. Biosystematics of the genus Euxoa (Lepidoptera: Noctuidae) v. rearing procedures, and life cycles of 36 species. Can Entomol. 1976;108(12):1345–1357. doi: 10.4039/Ent1081345-12. [DOI] [Google Scholar]
- 65.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W. Fast program for clustering and comparing large sets of protein or nucleotide sequences. In: Nelson KE, editor. Encyclopedia of metagenomics. Boston: Springer; 2015. pp. 173–177. [Google Scholar]
- 68.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7(1):539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ CT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 74.Gonzalez F, Witzgall P, Walker WB. Protocol for heterologous expression of insect odourant receptors in Drosophila. Front Ecol Evol. 2016;4:24. doi: 10.3389/fevo.2016.00024. [DOI] [Google Scholar]
- 75.Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly JS, Concordet JP. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17(1):148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koutroumpa FA, Monsempes C, François MC, De Cian A, Royer C, Concordet JP, et al. Heritable genome editing with CRISPR/Cas9 induces anosmia in a crop pest moth. Sci Rep. 2016;6(1):29620. doi: 10.1038/srep29620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hallem EA, Nicole Fox A, Zwiebel LJ, Carlson JR. Olfaction: mosquito receptor for human-sweat odorant. Nature. 2004;427(6971):212–213. doi: 10.1038/427212a. [DOI] [PubMed] [Google Scholar]
- 78.Revadi SV, Giannuzzi AV, Vetukuri R, Walker WB, Becher PG. Larval response to frass and guaiacol : detection of an attractant produced by bacteria from Spodoptera littoralis frass. J Pest Sci. 2021;94(4):1105–1118. doi: 10.1007/s10340-021-01352-9. [DOI] [Google Scholar]
- 79.Ljungberg H, Anderson P, Hansson BS. Physiology and morphology of pheromone-specific sensilla on the antennae of male and female Spodoptera littoralis (Lepidoptera: Noctuidae) J Insect Physiol. 1993;39(3):253–260. doi: 10.1016/0022-1910(93)90096-A. [DOI] [Google Scholar]
- 80.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20(4):1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 83.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55(4):539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 84.R Development Core Team R. R: A language and environment for statistical computing. Vienna; 2018. https://www.R-project.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. BUSCO scores for transcriptomes assembly completeness: Assessment of Spodoptera littoralis larval transcriptomes assembly completeness using the Benchmarking BUSCO tool performed against insecta_obd9 database consisting of 1,658 BUSCOs.
Additional file 2: Table S2. Expression profile of Spodoptera littoralis ORs (SlitORs) in the first and fourth instar larval heads. Expression levels of ORs are displayed as FPKM values.
Additional file 3: Table S3. Newly identified SlitOR70 amino acid sequence.
Additional file 4: Figure S1. RT-PCR gel images of PCR-amplified ORs at different developmental stages of Spodoptera littoralis. Comparison included negative-control (control), first instar heads (Slit_1), fourth instar heads (Slit_4), male antennae (Slit_♂) and female antennae (Slit_♀) with 1kb gene ruler ladder (Thermo Fischer Scientific). Red rectangles around SlitOR4 and SlitOR40 represent differential expression in the first and fourth instar, respectively, supported by RT-PCR and transcriptomic analysis.
Additional file 5: Figure S2. Expression pattern (RT-PCR) and relative expression levels (RT-qPCR) of Orco and SlitOR40 across different instars. (A) RT-PCR products obtained from SlitOrco and SlitOR40 amplification from the cDNA of first, second, third and fourth instar larval heads and adult antennae. (B) Quantitative RT-qPCR expression levels (relative fold-change) of Orco in first instar larval heads, fourth instar larval heads and adult male antennae using β-actin, L13A and Ef1A as reference genes. Male antennae samples expressed significantly higher levels of Orco compared to samples from first and fourth instar larvae (Df = 28, t= -22.622, P<0.001; Df = 28, t= 50.441, P < 0.001, respectively). Similarly, in the fourth instar larvae, Orco expression was significantly higher compared to first instar (Df = 28, t = 9.33, P < 0.001). (C) SlitOR40 expression (using Orco as reference gene) in the first instar larval heads, fourth instar larval heads and adult male antennae. Relative expression of SlitOR40 in first instar larvae was significantly higher compared to adult male antennae (Df = 28, t = 26.532, P < 0.001), while in the fourth larval instar, SlitOR40 expression was not detected (Df = 28, t = 27.693, P < 0.001). The analyses are based on five biological samples. The bars indicate standard errors. Statistical differences between treatments were calculated using a non-paired Student’s t-test. *: P < 0.05.
Additional file 6: Figure S3. SlitOR40 CRISPR/Cas9 induced indels (insertion and deletion). (A) Wild Type (WT) cDNA sequence including exon1 and exon2. An asterisk separates the two exons’ sequences. Blue letters indicate the primer sequences used for genotyping. Orange and purple boxes surround sequence regions where the Cas9 cleaved SlitOR40 and correspond to the same color boxes in B. (B) The three most prominent double mutational events obtained are shown around each of the two CRISPR guides. Indels’ produced between the two guides are described (NA means not applied) and insertion sequences are indicated in red letters. The number of nucleotide differences (Δ) between the WT line and the three mutant obtained lines are given between brackets. In both A and B, green letters show the CRISPR guide sequences with their PAM highlighted in yellow. (C) Electrophoregram traces for each of the three main double mutations obtained (BioEdit). Line 10 mutants were chosen for further crossing and phenotypic analysis.
Additional file 7: Figure S4. Attraction of Spodoptera littoralis wild type and SlitOR40-knockout larvae to positive control odors: Attraction of first and fourth instar WT and KO genotypes to positive control odors (CLD, cotton leaf discs) in both arms of the olfactometer (first instar WT: n = 44; fourth instar WT: n = 24; first instar KO: n = 59; fourth instar KO: n = 43) (top bar in Fig. S4A, S4B, S4C and S4D, respectively). Similarly, behavioral responses of WT and KO genotypes of S. littoralis first and fourth instar to the odor of cotton leaf discs versus clean air (control blank) (first instar WT: n = 50; fourth instar WT: n = 48; first instar KO: n = 21; fourth instar KO: n = 32) (bottom bar in Fig. S4A, S4B, S4C and S4D, respectively).
Additional file 8: Figure S5. EAG responses of Spodoptera littoralis wild type and SlitOr40-knockout moth antennae to positive control odors: The EAG responses of WT and KO line adult moth antennae to positive control stimuli (guaiacol and (±)-linalool) were tested at 10 μg dose (minimum n = 9 per sex). There was no statistical difference in responses to positive control stimuli, guaiacol (ANOVA: Sex: Df = 1; F = 1.63; P = 0.2; Genotype: Df = 1; F = 3.55; P = 0.066; Interaction: Df = 1; F = 0.13; P = 0.72; Residual Df = 47) and (±)-linalool (ANOVA: Sex: Df = 1; F = 0.66; P = 0.42; Genotype: Df = 1; F = 0.812; P = 0.372; Interaction: Df = 1; F = 1.68; P = 0.202; Residual Df = 47) in the antennae from both genotypes. The boxplots represent mean values along with the minimum and maximum values.
Additional file 9: Figure S6. Maximum-likelihood phylogenetic tree of ORs expressed in larvae and adults of Spodoptera littoralis: The analyses included all described ORs from S. littoralis (purple), B. mori (blue) and H. armigera (green). Highlighted clades: Orco (cadet blue); SlitOR40 and SlitOR70 along with ORs from B. mori and H. armigera (lilac). The unrooted phylogenetic tree was built using online tool PhyML 3.0.
Additional file 10: Table S4. Primers used in the study: List of all primers used to confirm the expression of SlitORs, qRT-PCR assays in first and fourth larval instars, and primers used for genotyping.
Additional file 11: Table S5. List of chemical compounds used in the electrophysiological recordings.
Data Availability Statement
Transcriptomics data are deposited in the public biomedical database, NCBI under the following accession number: PRJNA507834. All of the data on which conclusions rely in this study are included in this published article and its supplementary information files.