DNA sensor ABCF1 links genome surveillance to pluripotency gene transcription in embryonic stem cells.
Abstract
OCT4 and SOX2 confer pluripotency by recruiting coactivators to activate stem cell–specific transcription. However, the composition of coactivator complexes and their roles in maintaining stem cell fidelity remain unclear. Here, we report the ATP-binding cassette subfamily F member 1 (ABCF1) as a coactivator for OCT4/SOX2 critical for stem cell self-renewal. The intrinsically disordered low-complexity domain (LCD) of ABCF1 contributes to phase separation in vitro and transcriptional activation of pluripotency genes by mediating multivalent interactions with SOX2 and co-dependent coactivators XPC and DKC1. These LCD-driven transcription factor–coactivator interactions critical for pluripotency gene expression are disrupted by DNA damage, likely due to LCD-dependent binding of ABCF1 to damage-generated intracellular DNA fragments instead of SOX2. This study identifies a transcriptional coactivator that uses its LCD to form selective multivalent interactions to regulate stem cell self-renewal and exit from pluripotency when genome integrity is compromised.
INTRODUCTION
Stem cell pluripotency is largely driven by core transcription factors including OCT4 and SOX2 (1). Genome-wide studies demonstrated extensive co-binding of OCT4 and SOX2 at key pluripotency genes and across the embryonic stem (ES) cell genome (2, 3). It has also been shown that proper reactivation of this pluripotency gene network by OCT4 and SOX2 in somatic cells is a critical barrier to cellular reprogramming (4, 5). Therefore, OCT4 and SOX2 are essential for both maintenance and reacquisition of stem cell pluripotency.
Transcriptional activation of pluripotency genes by OCT4 and SOX2 requires transcriptional coactivators (6). Recent studies showed that multivalent interactions between intrinsically disordered low-complexity domains (LCDs) found in coactivators such as Mediator and transcriptional activators OCT4 and SOX2 form protein-rich hubs at genomic loci, thereby increasing the local concentration of these factors to promote transactivation (7–9). Mediator is required for transcription of most RNA polymerase II (Pol II) genes in somatic and pluripotent cells by virtue of its ability to interact with Pol II and diverse transcription factors (10). However, we and others have previously shown that OCT4 and SOX2 require nuclear factors enriched in pluripotent stem cells to robustly activate transcription (11, 12). Therefore, LCD-driven interactions by Mediator and other cell-ubiquitous coactivators are unlikely sufficient to account for the cell type– and gene-specific transactivation by OCT4 and SOX2 in ES cells. We surmised that additional coactivators may be involved to nucleate the assembly of stem cell–specific transcriptional hubs at pluripotency gene loci.
We took an unbiased in vitro approach to identify coactivators that can reconstitute transcriptional activation by OCT4 and SOX2 (11). By biochemical fractionation of human pluripotent cell nuclear extracts, we uncovered three coactivators that are enriched in ES cells and specifically required by OCT4 and SOX2 to activate transcription of the NANOG promoter in vitro (11). We previously reported that the first two stem cell coactivators (SCCs) are the XPC nucleotide excision repair complex and the dyskerin (DKC1) ribonucleoprotein complex (RNP) (11, 13–15). However, we found that robust activation of the NANOG gene by XPC and DKC1 requires an additional coactivator activity (SCC-B) (15). Therefore, revealing the identity and the transcriptional mechanisms by which SCC-B coordinates with XPC and DKC1 to promote the stem cell fate is fundamental to understanding the molecular basis of pluripotency.
Here, we identify SCC-B as adenosine 5′-triphosphate (ATP)–binding cassette subfamily F member 1 (ABCF1). We demonstrate that ABCF1 has an N-terminal LCD that can undergo liquid-liquid phase separation (LLPS) to form droplets in vitro. Using biochemical approaches and chromatin immunoprecipitation (ChIP) assays, we show that ABCF1 potently stimulates OCT4/SOX2-dependent transcriptional activation through LCD-mediated selective multivalent interactions with XPC, DKC1, SOX2, and Pol II, thereby forming a stem cell–specific transcriptional ensemble at pluripotency gene promoters. Consistent with its role as a critical SCC, disruption of ABCF1 compromises stem cell maintenance, pluripotency gene expression, and somatic cell reprogramming.
In somatic cells, ABCF1 has been implicated in the detection of aberrant intracellular DNA and ubiquitin conjugation in the innate immune pathway (16, 17). We provide evidence that the LCD of ABCF1 is also required for binding of these DNAs and that they can modulate ABCF1’s ability to form multivalent interactions with SOX2, suggesting potential mechanisms for regulating ABCF1 assembly in cells that may influence transcriptional activity. We show that, when ES cells are challenged with DNA damage or pathogen-derived DNAs, ABCF1 binds these aberrant DNAs accumulated in cells, concomitant with loss of interaction with SOX2 and dissociation of ABCF1 from gene promoters targeted by SOX2 and OCT4. This contributes to disruption of pluripotency gene expression and elimination of compromised ES cells through spontaneous differentiation. Our findings reveal that ABCF1 functions as a cell-specific transcriptional coactivator and regulates stem cell pluripotency in response to genome instability in an LCD-dependent manner.
RESULTS
An SCC essential for SCC-dependent transcriptional activation by OCT4 and SOX2
We have previously shown that robust activation of the human NANOG promoter by OCT4 and SOX2 in vitro requires three distinct SCCs present in a human pluripotent cell nuclear extract (11, 15). The first SCC, the XPC complex, separated from the other two at the Poros-HQ anion exchange chromatographic step, while the second SCC, the DKC1 complex (SCC-A), segregated from the remaining unknown coactivator (SCC-B) at the Poros-Heparin (Poros-HE) step (Fig. 1A). Starting with nuclear extracts prepared from 400 liters of a pluripotent human embryonal carcinoma cell line N-TERA2 (NT2), we serially fractionated the nuclear extracts over seven chromatographic columns. We tracked SCC-B activity in various protein fractions by assessing their ability to restore SCC-dependent transcriptional activation by OCT4 and SOX2 (schematic diagram of the in vitro transcription and biochemical complementation assay is shown in fig. S1A). In the final Mono S chromatographic step, salt-eluted fractions were assayed in in vitro transcription reactions. We found that adding fractions 14 and 15 to reactions containing purified XPC and DKC1 complexes potently stimulated transcription of the NANOG promoter template (Fig. 1B). These results demonstrated an important role of SCC-B in mediating cooperative coactivation by SCCs. Furthermore, our results suggested that the bulk of SCC-B likely resided in these fractions. Accordingly, SDS–polyacrylamide gel electrophoresis (SDS-PAGE) of these Mono S fractions revealed that fractions 14 and 15 were highly enriched with a polypeptide at ~110 kDa along with multiple apparent breakdown products (Fig. 1C).
Fig. 1. Purification and identification of SCC-B as ATP-binding cassette subfamily F member 1 (ABCF1).
(A) Chromatography scheme for purification of SCC-B from NT2 nuclear extracts (NT2 NE). NT2 NE is first subjected to ammonium sulfate precipitation (55% saturation) followed by a series of chromatographic columns including nickel affinity agarose (Ni-NTA), cation exchangers phosphocellulose (P11), heparin (Poros-HE), Mono S, anion exchanger Poros-HQ, hydroxyapatite (HAP), and gel filtration medium Superose 6. (B) Input fraction containing SCC-B activity from the Poros-HE step (IN), flow-through (FT), and various salt-eluted Mono S fractions was assayed for their ability to stimulate OCT4/SOX2-dependent transcription from the human NANOG promoter template engineered with four extra copies of the oct-sox composite binding element (bottom). All reactions contain purified general transcription factors (GTFs), Pol II, OCT4, SOX2, and recombinant XPC and DKC1 complexes. Transcribed RNA products are subjected to primer extension and visualized by autoradiography. (C) Mono S fractions assayed in in vitro transcription are separated on a polyacrylamide gel and stained with silver. Filled arrowhead indicates the ~110-kDa polypeptide that comigrates with SCC-B transcriptional activity. (D) Coomassie staining of Mono S fraction 14 demonstrates purification to homogeneity. (E) ABCF1 is enriched in human ES cells. Down-regulation of ABCF1 in human ES cell line H9 upon exit from pluripotency. (F) ABCF1 is enriched in mouse ES cells. mRNA and protein levels of ABCF1 in pluripotent D3 mouse ES cells carrying V5 epitope–tagged Abcf1 alleles (V5-ABCF1 knock-in) are compared to their differentiated counterparts.
To identify these polypeptides that co-migrate with SCC-B activity, we pooled and separated peak Mono S fractions using SDS-PAGE. Tryptic digestion of the gel slice containing the 110-kDa protein band followed by mass spectrometry identified SCC-B to be ABCF1 (Fig. 1D). Identification of ABCF1 as the active constituent of SCC-B activity was unexpected because it has not been previously associated with transcriptional regulation or any cellular processes in the nucleus. To corroborate the mass spectrometry results, we showed by Western blotting that ABCF1 resides in nuclear and cytoplasmic fractions of human and mouse ES cells (fig. S1B), and is enriched exclusively in the phosphocellulose 1 M KCl (P1M) nuclear fraction that contains SCC activities, but not in the transcriptionally inactive P0.3 and P0.5 fractions (Fig. 1A and fig. S1C) (11). Furthermore, we found that the mRNA and protein level of ABCF1 in human ES cells and a CRISPR-Cas9–mediated V5-tagged ABCF1 knock-in mouse ES cell line decreased sharply when these cells were induced to undergo differentiation (Fig. 1, E and F, and fig. S1, D and E). Here, we identified ABCF1 as a stem cell–enriched coactivator and a key player in mediating the synergistic transcriptional activation by OCT4 and SOX2 (11).
ABCF1 is required for pluripotency gene expression and stem cell pluripotency
We next set out to determine whether ABCF1 is required for stem cell maintenance and pluripotency by performing loss-of-function studies using lentiviruses expressing short hairpin RNAs (shRNAs) that target ABCF1 (Fig. 2A and fig. S2A). Compared to control D3 mouse ES cells, ABCF1 knockdown resulted in rapid collapse of the tightly packed ES cell colonies and the appearance of flattened and elongated cells with reduced alkaline phosphatase (AP) activity, a marker for pluripotent cells (Fig. 2B). We also found that depletion of ABCF1 in both mouse and human ES cells reduced proliferation and viability (fig. S2B). These observations indicated that loss of ABCF1 in ES cells compromises self-renewal capacity and promotes spontaneous differentiation. Consistent with the morphological changes indicative of compromised stem cell identity, depletion of ABCF1 in mouse ES cells resulted in significant decrease in mRNA levels of key pluripotency–associated genes, some of which are known direct targets of OCT4 and SOX2 (Fig. 2C) (2, 3). Concomitant with the down-regulation of these pluripotency genes, we observed increased expression of lineage-specific genes related to the ectoderm, mesoderm, and trophectoderm at the expense of endoderm marker induction (Fig. 2D). These data suggest that loss of ABCF1 destabilizes the pluripotency gene network and restricts the ability of ES cells to efficiently differentiate into all three embryonic germ layers. Our finding is consistent with a previously unexplained observation that Abcf1 knockout mouse embryos die at 3.5 days post-coital, a developmental stage that coincides with the emergence of pluripotent cells in the inner cell mass of the blastocyst (18) and from which ES cells are derived (19).
Fig. 2. ABCF1 is required for stem cell pluripotency and recruited to regulatory regions of key pluripotency genes targeted by OCT4 and SOX2 in mouse ES cells.
(A) shRNA-mediated knockdown of ABCF1 in mouse ES cells. Asterisk denotes a nonspecific band. (B) Depletion of ABCF1 in mouse ES cells leads to colony collapse with flattened cell morphology and reduced alkaline phosphatase (AP) staining, indicating spontaneous differentiation. (C) Loss of ABCF1 in mouse ES cells compromises pluripotency gene expression. Quantification of mRNA levels of pluripotency genes is analyzed by qPCR and normalized to Actb. (D) Depletion of ABCF1 induces expression of genes associated with the three embryonic germ layers and the trophectoderm, analyzed by qPCR as in (C). (E) Depletion of ABCF1 blocks somatic cell reprogramming. AP+ colonies are counted after 14 days (11 days with doxycycline followed by 3 days of dox withdrawal) post-induction (dpi). (F) Single-cell suspensions of 14 dpi reprogrammed CF-1 MEFs as described in (E) are stained with anti-mouse SSEA-1 and analyzed by flow cytometry. (G) Micrococcal nuclease (MNase) ChIP analysis of ABCF1 occupancy on control and enhancer regions of Oct4, Sox2, and Nanog gene loci in V5-ABCF1 knock-in D3 mouse ES cells. Representative data showing the enrichment of V5-ABCF1 (gray bars) compared to control IgGs (white bars) are analyzed by qPCR and expressed as percentage of input chromatin. Schematic diagrams of OCT4/SOX2 binding sites of each gene and the relative positions of the amplicons are shown at the bottom. (H) ABCF1 is not recruited to the promoter of housekeeping gene Actb.
Given the importance of ABCF1 in stem cell maintenance and expression of genes that are also known to promote somatic cell reprogramming (e.g., Nanog, Klf4, Esrrb, Prdm14, and Nr5a2) (Fig. 2C) (20, 21), we next assessed whether ABCF1 is required for the generation of induced pluripotent stem (iPS) cells from mouse embryonic fibroblasts (MEFs). To this end, we transduced MEFs with lentiviruses expressing nontargeting control shRNA or two independent shRNAs specific for ABCF1 and initiated reprogramming by doxycycline-induced expression of OCT4, KLF4, SOX2, and c-MYC (22). ABCF1 knockdown resulted in a marked decrease in the number of AP-positive iPS cell colonies formed (Fig. 2E and fig. S2C). Flow cytometry analysis further demonstrated that loss of ABCF1 led to a decrease in cells expressing SSEA-1+, a cell surface marker that indicates early stage of reprogramming (Fig. 2F). Our results are consistent with a role of ABCF1 in overcoming early barriers to reacquisition of pluripotency during cellular reprogramming.
We reasoned that if ABCF1 functions as a transcriptional coactivator for OCT4 and SOX2 in ES cells, ABCF1 ought to occupy the genome at cis-regulatory regions that are bound by OCT4 and SOX2. Therefore, we performed ChIP assays to investigate the binding of ABCF1 at known pluripotency gene loci targeted by OCT4 and SOX2 in V5-ABCF1 knock-in mouse ES cells using an anti-V5 antibody. We performed micrococcal nuclease–ChIP (MNase-ChIP) using formaldehyde and the protein-protein cross-linker ethylene glycol bis(succinimidylsuccinate) (EGS) and found that ABCF1 was enriched at OCT4/SOX2 co-bound regions including core pluripotency genes Oct4 (Pou5f1), Sox2, and Nanog (Fig. 2G and fig. S2, D and E). By contrast, we did not observe any significant enrichment of ABCF1 at housekeeping genes β-actin (Actb) and Dhfr (Fig. 2H and fig. S2F), consistent with ABCF1 acting as a selective coactivator for OCT4 and SOX2. Together, the data presented thus far suggest a classical coactivator function of ABCF1 both in vitro with naked DNA and in the context of chromatin in ES cells.
LCD in ABCF1 mediates LLPS in vitro
ABCF1 belongs to a large class of transporters that couples ATP hydrolysis to the active transport of substrates across cell membranes (23). While most ABC proteins contain transmembrane domains (TMDs) as expected, ABCF1 lacks TMD and is therefore not predicted to function as a transporter. In somatic cells, ABCF1 has been implicated as a regulator of mRNA translation (24, 25), an E2 ubiquitin–conjugating enzyme (16), and a sensor for intracellular double-stranded DNAs involved in innate immune responses (17). Note that these reported activities of ABCF1 all reside in the cytoplasm. Our findings here thus point to a hitherto unknown nuclear function of ABCF1 in gene regulation in ES cells.
ABCF1 contains two conserved nucleotide-binding domains (NBDs) and a ~300–amino acid N-terminal domain that is predicted to be highly disordered (26) (Fig. 3A, top). This is in agreement with a recent structural study on ABCF1, which indicated that the N-terminal domain cannot be crystallized (27). The N-terminal domain of ABCF1 is a low-complexity region that contains a polyglutamine (polyQ) tract and is unusually rich in charged amino acids, of which ~40% are divided between lysine (K) and glutamic acid (E) residues (Fig. 3A, bottom) (28). This unique amino acid composition is conserved among vertebrates (fig. S3A) and consistent with one of the largest classes of intrinsically disordered region called polyampholytes (29). These clusters of positively and negatively charged amino acid, interspersed with hydrophobic residues such as phenylalanine, are known to promote LLPS (30, 31). Given the recent appreciation that the ability of LCDs in transcription factors and coactivators to form LLPS droplets in vitro often correlates with their transactivation activities (7–9, 32), we next investigated whether ABCF1 proteins are also capable of forming liquid droplets in vitro and in cells.
Fig. 3. The LCD of ABCF1 phase separates in vitro.
(A) Top: Unstructured regions in human ABCF1. X axis indicates position of the amino acids, and Y axis shows the probability of disordered sequences. Regions that are above the value of 0.5 are predicted to be unstructured. The schematic diagram denotes protein domains of ABCF1: intrinsically disordered low-complexity domain (LCD; yellow, amino acids 1 to 302) and two nucleotide-binding domains (NBDs; blue). Bottom: Amino acid composition of human ABCF1. Each of the 20 amino acids is counted and marked as a black bar at that position in ABCF1. One-letter abbreviations for amino acids are used. Q, E, and K residues in the LCD are highlighted. (B) Schematic diagrams of recombinant GFP fusion ABCF1 proteins used in vitro droplet assays. (C) Representative images of droplet formation with FL ABCF1, NBDs, LCD, or GFP. Proteins (13 μM) are added to droplet formation buffer containing 200 mM NaCl. The average number and size (arbitrary unit) of droplets in each image are indicated (n = 10). (D) Representative images of droplet formation with FL (n = 10) and LCD (n = 5) in the presence of buffer (Ctrl) or 10% 1,6-hexanediol (1,6-Hex). Relative droplet size of FL and LCD is indicated. (E) Fluorescence images of stable mouse D3 ES cell lines expressing GFP, FL ABCF1, or NBDs. Scale bars, 20 μm. (F) The percentage of cells having nuclear puncta per image field in the presence of buffer (Ctrl) or 3% 1,6-Hex is analyzed (n = 9 to 10).
Recombinant green fluorescent protein (GFP) fusion proteins of full-length (FL) ABCF1 were purified and added to reaction buffers containing a crowding agent [10% polyethylene glycol, molecular weight 800 (PEG-8000)] to mimic the intracellular crowding environment in vitro (Fig. 3B and fig. S3B). Fluorescence microscopy of the protein mixture revealed that ABCF1 readily formed micrometer-sized liquid droplets at or above a physiological salt concentration (200 mM NaCl) (Fig. 3C) and at a lower protein concentration range used in in vitro transcription assays (~100 nM) (fig S3C). We also observed droplet formation with other inert crowding agents [e.g., Ficoll and bovine serum albumin (BSA)] and in more complex mixtures such as mouse ES cell nuclear extracts (fig. S3D). Deletion of the LCD (i.e., NBDs-GFP) markedly reduced the number of droplets, while LCD alone (amino acids 1 to 302) forms even bigger droplets that are sensitive to salt concentration (fig. S3E). Furthermore, addition of the alcohol 1,6-hexanediol (1,6-Hex), which is known to disrupt LLPS, significantly suppressed droplet formation (Fig. 3D) (32). These results indicate that the LCD is essential for ABCF1 to phase separate into liquid-like bodies in vitro, but the structured NBDs may function to regulate its tendency.
In ES cells stably expressing FL-GFP or NBD-GFP, we found that FL ABCF1 resides predominantly in the cytoplasm, consistent with previous observations (Fig. 3E and fig. S3F) (28). In the nucleus, FL-GFP forms discrete puncta in a subpopulation of stable cells, likely due to heterogeneity of FL-GFP expression in these cells (Fig. 3F). By contrast, although NBDs-GFP proteins were more uniformly distributed in the cells and accumulated in the nucleus at higher levels than FL ABCF1, we failed to observe puncta structures. 1,6-Hex treatment significantly reduced the number of ES cells with FL-GFP puncta (Fig. 3F). Together, these results demonstrate that LCD-dependent multivalent interactions by ABCF1 can occur in ES cells and drive puncta formation in the nucleus, at least in the context of overexpression.
Transcriptional coactivation mediated by the LCD in ABCF1
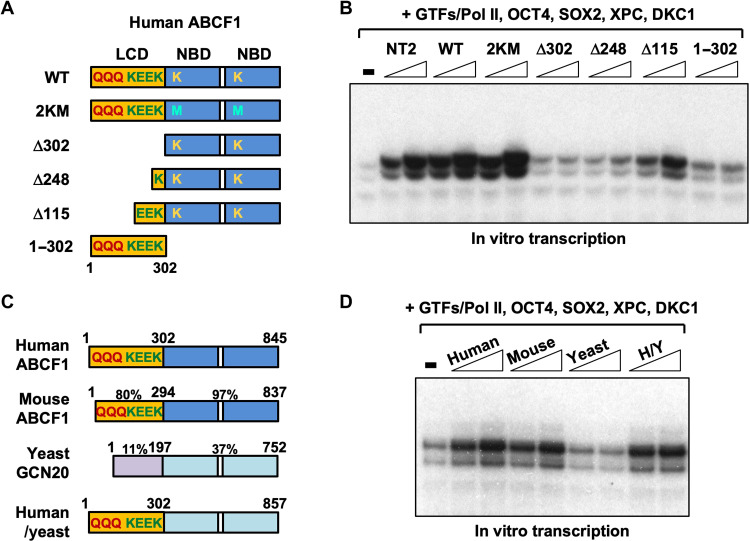
We next asked whether the ability of ABCF1 to undergo phase separation in vitro correlates with transcriptional activities, as has been observed in other LCD-containing transcription factors (7–9). We generated recombinant FL, wild-type (WT) ABCF1, the LCD fragment (1–302), and a series of N-terminal truncations of the LCD in FL ABCF1. To address whether its catalytic activity is required for transcriptional coactivation, we also purified an adenosine triphosphatase (ATPase)–defective mutant ABCF1 protein where the two conserved ATP-binding lysine residues (K324 and K664) in the Walker A motif of both NBDs were substituted with methionine (2KM) (Fig. 4A and fig. S4A) (28). When added to in vitro transcription assay, the FL WT and 2KM mutant proteins were as active as endogenous ABCF1 purified from NT2 cells (Fig. 4B). However, deletion of polyQ alone (Δ115) or together with the K/E-rich domain (Δ248) led to a progressive loss of transcriptional activity, and the two NBDs by themselves (Δ302) were completely inactive. Unexpectedly, the LCD fragment (1–302) also lacked transcriptional activity. These results provide several insights. First, ABCF1 is unambiguously shown to be the sole constituent of SCC-B. Second, ATP binding and hydrolysis by ABCF1 are dispensable for transcription, unlike for ABCF1’s role in translation (27, 28). Third, while the entire LCD is essential for full coactivator activity, the LCD by itself is not sufficient to activate transcription. These observations suggest that the NBDs also contribute to the full transcriptional activity of ABCF1.
Fig. 4. Transcription coactivation by ABCF1 requires its LCD.
(A) Schematic diagram of FL wild-type (WT) ABCF1 protein depicting the N-terminal LCD (yellow) containing a polyglutamine (polyQ) tract and lysine (K)/glutamic acid (E)–rich regions. The two conserved lysine residues (K324 and K664) in the Walker A motif of each of the two NBDs (blue) in ABCF1 are highlighted. FL WT ABCF1, ATP-binding defective lysine-to-methionine mutant (2KM), various truncated ABCF1 proteins lacking part (Δ248 and Δ115), or all of the LCD (Δ302) as well as the LCD by itself (1–302) are purified from E. coli. (B) Transcriptional activities of the various recombinant ABCF1 proteins shown in (A) are assayed (over a twofold concentration range) together with recombinant XPC and DKC1 complexes in in vitro transcription as described in Fig. 1. (C) Schematic representation of the human and mouse ABCF1 and yeast homolog GCN20. The percentage of sequence similarity among human, mouse, and yeast homolog is indicated. Domain-swapped hybrid protein between the human LCD and yeast NBDs (H/Y) is generated and purified from E. coli. (D) Titration over a twofold concentration range of human and mouse ABCF1, yeast GCN20, and human-yeast hybrid (H/Y) proteins is assayed in in vitro transcription reactions.
The yeast homolog of ABCF1, GCN20, shares with its human and mouse counterpart a conserved NBD (33). However, the residues outside of NBD (amino acids 1 to 197) are highly divergent from mammalian ABCF1. This domain lacks the polyQ tract and is not K/E-rich (Fig. 4C and fig. S4B). This region is predicted to be structured, unlike the LCDs found in human and mouse ABCF1 proteins (fig. S4C). As predicted based on a requirement for the LCD, we found that purified GCN20 exhibited no coactivator activity in the in vitro transcription assay (Fig. 4D and fig. S4D). However, replacing the GCN20 N-terminal domain with the human ABCF1 LCD fully conferred transcriptional stimulatory capacity to the hybrid protein in vitro. These observations provide strong biochemical evidence that the mammalian-specific LCD confers coactivator activity.
Specificity of LCD-mediated interactions by ABCF1
Having identified the LCD as a “transactivation domain” in ABCF1, we next investigated the mechanisms by which the LCD potentiates stem cell–specific transcription. We have previously shown that transcriptional activation by OCT4 and SOX2 selectively requires SCCs but not the prototypical coactivator Mediator at least in vitro (11). To gain mechanistic insights into the specificity of the LCD in ABCF1, we tested whether the LCD can associate with its two co-dependent coactivators, XPC and DKC1, as well as Pol II by performing glutathione S-transferase (GST) pull-down assay. We incubated NT2 cell nuclear extract with immobilized LCD from human ABCF1 (1–302) and the transcriptionally inactive N-terminal domain from yeast GCN20 (1–197) as GST fusion proteins. Western blot analysis revealed that only LCD/ABCF1 was able to bind XPC, DKC1, and Pol II (Fig. 5A and fig. S5A). To further demonstrate the specificity of LCD/ABCF1 for the XPC and DKC1 complexes, we also tested the transactivation domain from human transcription factor SREBP1a, which harbors an LCD and is known to bind Mediator (34). No detectable binding of XPC and DKC1 was observed, but the fusion protein was able to pull down Pol II, likely indirectly through a Mediator–Pol II interaction (10). Using transient transfection and coimmunoprecipitation experiments in 293T cells, we showed that ABCF1 interacted preferentially with SOX2 but not OCT4 (Fig. 5B). We further confirmed the interaction between ABCF1 and SOX2 in mouse ES cell extracts (Fig. 5C). Deletion of the LCD from ABCF1 (NBDs), which abrogates its transcriptional activity in vitro, completely abolished its ability to bind SOX2 (Fig. 5D). We next showed that the high-mobility group (HMG) box DNA binding domain of SOX2, but not its activation domain (AD), is required for binding ABCF1 (fig. S5, B and C), consistent with studies indicating that interactions of SOX family transcription factors with cofactors are often mediated by their conserved HMG domain (35). There are 20 SOX genes in human and mice that are divided into different groups (A to H) (36). Although these SOX proteins share an HMG box with more than 80% sequence identity, we found that ABCF1/LCD showed a strong preference for SOX2 and SOX1 from group B1, but not SOX11 (group C) or SOX10 (group E) (fig. S5D). These results highlight the ability of ABCF1/LCD to discriminate small differences in protein sequences of the HMG boxes, despite high similarity in their overall structure. Together, we conclude that the transcriptional defect observed with LCD-truncated ABCF1 is likely due to its failure to establish multivalent interactions with SOX2, SCCs, and the Pol II machinery.
Fig. 5. Transcriptional coactivation by ABCF1 is mediated by LCD-dependent selective interactions.
(A) GST fusion proteins containing the LCD of human ABCF1 (1–302), the N-terminal domain of yeast GCN20 (1–197), and the transactivation domain of human transcription factor SREBP1a (1–50) are incubated with buffer only (−) or NT2 NEs (+). Input (IN) indicated. (B) Whole-cell extracts (WCEs) from 293T cells cotransfected with plasmid expressing V5-tagged ABCF1 together with either empty plasmid (−) or plasmids expressing FLAG-tagged SOX2 (S) or OCT4 (O) are immunoprecipitated with anti-FLAG antibody. (C) Input V5-ABCF1 KI mouse ES cell WCEs (IN) and IPs by IgG and anti-SOX2 antibodies are analyzed by Western blotting. (D) HA IPs from 293T cells overexpressing HA-tagged SOX2 (SOX2-HA) with V5-tagged FL or LCD-truncated human ABCF1 (NBDs). (E) ABCF1 knockdown rescue assay. mRNAs from control mouse ES cells (shNT) overexpressing RFP, and ABCF1 knockdown ES cells (sh1) overexpressing RFP, V5-tagged FL, LCD-truncated human ABCF1 (NBDs), or mouse NANOG are analyzed for Nanog, Fgf4, and Klf4 mRNA levels by qPCR. (F) Colony formation assays of cells described in (E). Differentiation status is evaluated on the basis of AP staining intensity and colony morphology. Representative images of AP staining of control and rescued ES cells are shown (right).
To provide in vivo evidence that the ABCF1 LCD is critical for OCT4/SOX2-dependent transcription, we performed knockdown rescue experiments in mouse ES cells. Simultaneous knockdown of mouse ABCF1 and ectopic expression of FL human ABCF1, but not LCD-truncated ABCF1 (NBDs) or control red fluorescent protein (RFP), restored expression of several ABCF1-dependent pluripotency genes such as Nanog, Fgf4, and Klf4 in mouse ES cells (Fig. 5E and fig. S5E). These results demonstrate that the LCD in ABCF1 is required for pluripotency gene expression in vivo.
We next showed that ectopic expression of NANOG alone was sufficient to restore pluripotency gene expressions (Fgf4 and Klf4) in ABCF1-deficient ES cells (Fig. 5E). This result indicates that Nanog is likely a critical downstream target of ABCF1, consistent with our in vitro transcription results. Unlike ABCF1, transcription factor NANOG has no other known roles beyond transcription in ES cells. The fact that NANOG can bypass the requirement of ABCF1 strongly suggests that the transcriptional defect in ABCF1 knockdown ES cells is unlikely due to disruption of other cellular processes that may be carried out by ABCF1. Rather, our data thus far suggest that ABCF1 directly controls pluripotency gene expression in vitro and in vivo. Consistent with these results, we found that defect of ABCF1-depleted ES cells in forming AP-positive pluripotent colonies in limited dilution assays can be largely overcome by ectopic expression of FL ABCF1 and NANOG but not NBDs or control RFP (Fig. 5F). Therefore, we conclude that ABCF1 supports self-renewal by activating stem cell–specific transcription through LCD-dependent assembly of select activator and coactivators with the Pol II transcription machinery.
ABCF1 senses aberrant intracellular DNAs that arise from genome instability
DNA sensing by the innate immune system underpins many physiological responses to DNA (37), including immunity to DNA viruses and bacteria (38) and inflammatory responses to intracellular self-DNAs that arise from genome instability (39–41). Because ABCF1, an essential SCC shown here, has also been reported to act as an important sensor for intracellular DNAs in the innate immune system in somatic cells (17), we next asked whether ABCF1 can also recognize these DNAs in ES cells and how intracellular DNAs may influence transcriptional coactivation by ABCF1.
We took two independent approaches to test whether ABCF1 can bind intracellular DNAs in ES cells. As a first approach, we incubated mouse ES cell extracts containing ectopically expressed FL and LCD-truncated (NBDs) human ABCF1 proteins with a 5′ biotinylated single-stranded (ss) or two different double-stranded (ds) DNA oligonucleotides containing sequences derived from Listeria monocytogenes. ds-matched (ds-M) contains a consensus sox motif, while ds-unmatched (ds-UM) does not. In neutrophils, SOX2 has been shown to act as a sequence-specific innate immune sensor for sox motif–containing DNAs such as ds-M (42). Given that ABCF1 binds SOX2 in ES cells, we asked whether ABCF1 bound DNA, and if so, whether binding was direct or indirect through SOX2. Western blot analyses of protein-DNA complexes captured by streptavidin beads revealed that both ds-M and ds-UM efficiently pulled down FL ABCF1 but not NBDs, while neither binds ssDNA (Fig. 6A). Similar results were also obtained with endogenous ABCF1 (fig. S6A). These results indicate that ABCF1 binds to short dsDNAs in an LCD-dependent, but sequence-independent, and therefore SOX2-independent manner. Because the LCD important for LLPS is also required for DNA binding, we next investigated how DNA may influence ABCF1 droplet formation. Consistent with our DNA pull-down results, we found that fluorescently labeled (Cy5) ds-UM, but not ssDNAs, was efficiently incorporated into FL ABCF1 droplets. Addition of dsDNAs markedly increased both the number and size of the ABCF1 droplets that are 1,6-Hex sensitive (Fig. 6B). Furthermore, dsDNAs promoted droplet formation at lower protein concentrations (Fig. 6C). Together, these results demonstrate that dsDNA promotes homotypic LCD-LCD interactions by ABCF1 and condensate formation in vitro.
Fig. 6. dsDNAs modulate phase separation of ABCF1 in vitro and pluripotency gene expressions in ES cells.
(A) WCEs from D3 mouse ES cells stably expressing V5-tagged FL ABCF1 or NBDs are incubated with three different 5′ biotinylated 98-mer oligonucleotides: single-stranded (ss), double-stranded (ds) with SOX2-binding motif (matched, ds-M), or ds lacking the sox-motif (unmatched, ds-UM). (B) Representative images of droplet formation with FL ABCF1 (9 μM) in the presence of 12.5 nM of Cy5-labeled (magenta) ss or ds-UM DNAs. Number and size of droplets are analyzed. Scale bar, 20 μm. (C) Phase separation diagram of FL ABCF1 with or without ds-UM at indicated concentrations. Blue and red dots indicate the presence and absence of droplets, respectively. Bottom: Fluorescence microscopy images showing that dsDNA (ds-UM) stimulates droplet formation of FL ABCF1 at low protein concentration (0.025 μM). (D) DNA copurified with ABCF1 IP from WCEs prepared from ETO-treated (20 μM) V5-ABCF1 knock-in mouse ES cells, denoted by vertical bar. (E) The percentage of stable ES cells (GFP, FL, or NBDs) containing puncta per image field upon treatment with DMSO or ETO (2 and 20 μM) is analyzed (n = 9 to 10). Representative images of cells expressing GFP, FL, or NBDs treated with ETO (20 μM) are indicated (right). Scale bar, 20 μm.
For the second approach, we investigated whether ABCF1 can recognize short DNAs accumulated in cells following DNA damage–induced stress. We treated V5-ABCF1 knock-in mouse ES cells with etoposide (ETO), a DNA damaging agent that is known to induce DNA fragmentation by damage-activated endonucleases (40). The resulting self-DNA fragments were readily detected in the cell nucleus 6 hours after treatment, while ABCF1 and SOX2 protein levels remained unchanged (fig. S6, B and C). We showed that immunoprecipitation of ABCF1 from ETO-treated ES cell extracts coprecipitated small DNA fragments that correspond to the size of a mononucleosome (Fig. 6D). Using the ABCF1-GFP stable ES cells, we found that ETO treatment at higher concentration (20 μM) significantly increased the number of ES cells with FL ABCF1 puncta at 6 hours (Fig. 6E), coinciding with accumulation of DNA fragments in cells (fig. S6B). ETO treatment did not promote NBDs-GFP puncta formation, indicating that DNA damage–induced ABCF1 puncta formation remained LCD dependent. Furthermore, transfection of Cy5-labeled dsDNAs into ES cells led to a similar increase in FL ABCF1 puncta, consistent with a role of dsDNA in promoting LCD-dependent puncta formation (fig. S6D). However, we did not detect significant colocalization of Cy5 signals with ABCF1-GFP puncta. Because we showed that a substoichiometric level of dsDNA relative to ABCF1 (~700-fold lower in concentration) can potently stimulate droplet formation in vitro (Fig. 6B), it is likely that a punctum that contains hundreds of ABCF1 protein may carry only a few Cy5-dsDNAs. Therefore, detection of single or a few Cy5-labeled dsDNA molecules is likely below the detection threshold of confocal microscopy. It remains possible that there are alternative mechanisms by which dsDNA induces ABCF1 puncta formation in ES cells. Nonetheless, using multiple approaches, we provided evidence that ABCF1 and dsDNA interact in ES cells and that dsDNA can promote LCD-dependent condensate formation by ABCF1-GFP in vitro and in cells.
Aberrant intracellular DNAs disrupt ABCF1-SOX2 interactions and pluripotency gene expression
To understand the effect of intracellular DNA on the coactivator function of ABCF1, we showed that transfection of dsDNAs into ES cells compromised an interaction between endogenous ABCF1 and SOX2 (Fig. 7A). To assess the consequence of a compromised ABCF1-SOX2 interaction by dsDNA on pluripotency gene expression, we transfected 5′ 6-carboxyfluorescein (6-FAM)–labeled ss, ds-M, or ds-UM oligonucleotides into mouse ES cells. Transfected, 6-FAM–positive cells were enriched by flow cytometry (fig. S7A). Gene expression analyses by quantitative polymerase chain reaction (qPCR) revealed that transfection of ds-M or ds-UM down-regulated pluripotency genes and up-regulated differentiation genes compared to ssDNAs (Fig. 7B and fig. S7B). These data are consistent with our observation that ABCF1 selectively binds dsDNA but not ssDNA. Furthermore, we found that interaction of ABCF1 with SOX2 was also disrupted in ES cells treated with high concentration of ETO (20 μM), where accumulation of intracellular DNAs was detected (Fig. 7A and figs. S6B and S7C). Similar to the negative effect of dsDNA transfection on pluripotency gene expression (Fig. 7B), significant down-regulation of pluripotency-associated genes and up-regulation of lineage-specific genes were observed only at higher ETO concentrations (e.g., 20 μM), consistent with the notion that a stable SOX2-ABCF1 interaction is required for robust pluripotency gene transcription (fig. S7, D and E). To probe the mechanism by which dsDNA may interfere with ABCF1’s ability to bind SOX2, we performed in vitro droplet assays as proxy for studying how dsDNA modulates LCD-driven multivalent interactions. ABCF1-GFP droplets efficiently incorporated SOX2-mCherry but not control mCherry (Fig. 7C and fig. S7, F and G). Likewise, we did not observe significant enrichment of control GFP by SOX2-mCherry, indicating that interaction between ABCF1 and SOX2 in droplet formation buffer was specific (fig. S7G). However, in the presence of fluorescently labeled (Cy5) dsDNA (ds-UM), we found that its incorporation compromised the ability of ABCF1 droplets to recruit SOX2, resulting in significant reduction in SOX2-mCherry signal intensity and size that overlapped with ABCF1-GFP (Fig. 7C). For example, areas within an ABCF1 droplet that were enriched with dsDNAs incorporated lower levels of SOX2. Conversely, droplets that contained lower levels of DNA tend to display stronger SOX2 signals (Fig. 7D). Together, these results suggest that interaction of ABCF1 with dsDNAs may compete with SOX2. Consistent with our observations that dsDNAs and DNA damage can disrupt ABCF1-SOX2 interaction and pluripotency gene expression in ES cells, we observed loss of ABCF1 enrichment at its target genes such as Oct4, Sox2, and Nanog in ETO-treated ES cells by ChIP-qPCR, while histone H3 levels at these genomic loci remained unchanged (Fig. 7E and fig. S7, H to J).
Fig. 7. dsDNAs disrupt interaction between ABCF1 and SOX2, target gene expression, and stem cell maintenance.
(A) dsDNA transfection (left) or ETO treatment (20 μM; right) disrupts ABCF1-SOX2 interaction. Input (IN) and IgG or SOX2 IPs from WCEs of V5-ABCF1 knock-in mouse ES cells are analyzed by Western blotting. (B) Mouse ES cells transfected with 5′ 6-carboxyfluorescein (6-FAM)–labeled ss, ds-M, or ds-UM are enriched by FACS. Expression levels of pluripotency are analyzed by qPCR. (C) Representative images of droplet formation with FL ABCF1-GFP (green) and SOX2-mCherry (red) in the presence or absence of Cy5-labeled dsDNA (ds-UM, magenta). Scale bars, 20 μm. Fluorescence intensity levels of FL and SOX2 that colocalize in droplets are obtained as described in Materials and Methods. Bottom: Relative fluorescence intensity and size of SOX2 signals that overlap with FL in droplets are quantified from independent images (n = 5). (D) Line-scan profiles of fluorescence intensity of FL, SOX2, and dsDNA in indicated droplets. (E) MNase-ChIP of ABCF1 in DMSO- and ETO-treated V5-ABCF1 knock-in mouse ES cells. Enrichment of ABCF1 is analyzed by qPCR as in Fig. 2G. (F) MNase-ChIP of ABCF1 in DMSO- and 1,6-Hex (1.5%)–treated V5-ABCF1 knock-in mouse ES cells. Enrichment of ABCF1 (left) and Pol II (right) is analyzed by qPCR as in (E). (G) Colony formation assays in control and ABCF1 gain-of-function D3 mouse ES cells. AP-positive colonies are counted and indicated as a relative unit. Representative images of each AP-stained cells are indicated (right).
To provide further evidence that the transcription factor–coactivator hubs assembled at the SOX2/OCT4 enhancers are mediated by LCD/ABCF1-driven multivalent interactions, we showed that treatment of ES cells with 1,6-Hex significantly reduced the enrichment of ABCF1 and Pol II at the Oct4 and Nanog enhancers, while H3K4me3 level remained unchanged (Fig. 7F and fig. S7K). Our results suggest that LCD-dependent interaction between ABCF1 and intracellular dsDNAs may come at the cost of transcription, indicating that ABCF1 may be limiting in ES cells. If this were the case, increasing cellular concentration of ABCF1 would enhance DNA damage tolerance in ES cells by increasing the pool of ABCF1 proteins available for transcriptional activation. We found that, compared to RFP, ectopic expression of FL ABCF1 significantly enhanced the self-renewal capacity of ETO-treated ES cells as shown by an increase in number of AP-positive colonies formed in limiting dilution assays (Fig. 7G and fig. S7, L and M). By contrast, deletion of the LCD (i.e., NBDs) compromised the ability of ABCF1 to overcome ETO-induced stresses. We further demonstrated that transfection of dsDNAs alone in the absence of DNA damage was sufficient to recapitulate the negative effect of ETO treatment on stem cell maintenance, consistent with disruption of ABCF1-SOX2 interaction and decreased pluripotency gene expressions observed in dsDNA-transfected ES cells (Fig. 7, A and B). ABCF1 overexpression again enhanced the resistance of ES cells to transfected DNAs (fig. S7N). Note that we did not observe significant differences between RFP and ABCF1 gain-of-function ES cells in the absence of DNA damage or dsDNA transfection, thus suggesting a specific protective effect of ABCF1 overexpression on self-renewal capacity when ES cells encounter genome instability or high levels of intracellular DNAs. Our studies thus reveal a new function of ABCF1 and its LCD in stem cell–specific transcriptional control and a potential mechanism through which stem cell pluripotency can be regulated in response to genome instability by modulating multivalent interactions.
DISCUSSION
Role of LCD in mediating stem cell–specific transcriptional activation
In this study, we identified ABCF1 as a critical transcriptional coactivator for stem cell maintenance and showed that the LCD of ABCF1 is indispensable for both physical interaction with SOX2, XPC, DKC1, and Pol II and functional reconstitution of ABCF1 coactivator activity both in vitro and in vivo. The flexible nature of LCDs is thought to facilitate the dynamic interaction with multiple protein partners by virtue of their ability to rapidly adopt an ensemble of conformations (43). Note that LCDs are not simply unstructured sequences that bind promiscuously to any proteins; instead, they can be selective for binding partners (8, 44). In this regard, the conformationally flexible XPC protein also contains several highly disordered regions that we found, however, to be dispensable for transcriptional activation (11, 13, 14). These observations reveal the unique ability of ABCF1 LCD to integrate multiple lines of information encoded by SOX2, SCCs, and the Pol II machinery, likely by forming a hub of these factors at target gene promoters through selective multivalent interactions (8). Consistent with this hypothesis, we find that these ABCF1-mediated interactions at pluripotency gene loci are sensitive to 1,6-Hex, a chemical that interferes with LLPS of many LCDs (32). Our findings here thus provide a clear example of an LCD that can impart activator preference and target gene specificity to a transcriptional coactivator and activate cell-specific transcription by a protein:protein–driven local high-concentration mechanism.
Paradoxically, we find that the LCD by itself is inactive in transcription (Fig. 4B). This observation points to a potential critical regulatory function of the NBDs in transcriptional coactivation. We show that, in the absence of the NBDs, the LCD becomes “hyperactive” in forming significantly bigger LLPS droplets (up to 10 μm in diameter) in vitro (Fig. 3C). We speculate that the higher propensity of the LCD to self-associate could interfere with its ability to interact with transcription factors, coactivators, and the transcription machinery. It remains possible that the NBDs provide additional contact points critical for transactivation.
Stable expression of ABCF1 in ES cells resulted in fewer puncta structures than anticipated, given its propensity to undergo phase separation in vitro. This may reflect the heterogenous expression levels of ABCF1 and the fact that not all LCD-mediated multivalent interactions by transcription factors appear as large condensates in cells. It has been shown that transcriptional activation by LCD-containing transcription factors can occur in the form of dynamic multivalent interaction hubs rather than large phase-separated bodies in cells (8). Large puncta structures could mark a subset of genomic loci such as super-enhancers that are highly enriched with transcription factors, where the abundance of LCD-driven multivalent interactions is thought to promote phase separation at these loci (32). It is unclear whether ABCF1 is recruited to the majority or a subset of SOX2-bound super-enhancers. It remains possible that there are differences in phase separation behavior of ABCF1 in vitro and in ES cells. Future experiments will be required to determine whether endogenous ABCF1 can form condensates and how they may contribute to stem cell–specific transcriptional activation by ABCF1 in ES cells. However, we showed that the number of ABCF1-GFP puncta increases in response to DNA damage (Fig. 6E), suggesting that these LCD interaction–driven puncta display a capacity to respond to changing cellular environment. While it is unclear whether endogenous ABCF1 also forms puncta upon DNA damage, we suggest that the underlying LCD-mediated interactions that drive ABCF1-GFP puncta formation may still play important biological roles and could inform the mechanisms by which ABCF1 regulates pluripotency gene transcription in ES cells.
Function of ABCF1/LCD-mediated multivalent interactions in stem cell maintenance
Because LCD-mediated interactions are highly dynamic (8, 45), targeting these transient and multivalent interactions between transcription factors and SCCs could provide an effective means of modulating the pluripotency gene network in a rapid manner in response to changing cellular environment. It has been shown that molecular crowding by LCD is particularly sensitive to changes in concentration, where a less than twofold decrease can be sufficient to cause an LCD-mediated phase-separated body to be disrupted (46, 47). Therefore, we suggest that a decrease in ABCF1 protein concentration, such as during stem cell differentiation (Fig. 1, E and F), could lead to rapid dissolution of the transcriptional apparatus, pluripotency gene expression, and the pluripotent state as a result.
While we showed that ABCF1 can directly bind short intracellular dsDNA that arise from genome instability, we would like to stress that this is likely distinct from the mechanism by which ABCF1 is recruited to specific genomic loci such as the SOX2 enhancers in ES cells. We find that detection of ABCF1 at SOX2-bound genomic regions requires a protein:protein cross-linker (EGS). This observation is consistent with the notion that ABCF1 does not bind genomic DNA directly. Rather, transcriptional coactivators are often recruited to specific genomic regions through direct interactions with sequence-specific transcription factors; such is the case here between ABCF1 and SOX2. Under optimal growth conditions, there are likely multiple mechanisms to return ES cells experiencing low level of DNA damage to homeostasis, such as enhanced DNA repair (48). We surmise that when these cellular insults turn catastrophic, accumulation of aberrant short dsDNAs in damaged ES cells could compromise ABCF1-SOX2 interaction, in part, due to the ability of these dsDNAs to interfere with multivalent interactions between ABCF1 and SOX2 (Fig. 7A). Dissociation of ABCF1 from SOX2 destabilizes the SOX2 transcriptional complexes at target pluripotency gene promoters anchored by ABCF1 LCD, leading to down-regulation of pluripotency gene expression and spontaneous differentiation of compromised ES cells. Future studies will be required to identify features of DNA damage–induced short dsDNA (e.g., length and structure such as the presence of free DNA ends) that are recognized by ABCF1. We propose that the LCD-mediated ABCF1-SOX2 interaction may serve as an important checkpoint for self-renewal by modulating pluripotency gene expressions in homeostasis and in response to genome instability in ES cells.
MATERIALS AND METHODS
DNA constructs and antibodies
Complementary DNAs (cDNAs) for human and mouse ABCF1 were obtained from cDNA libraries generated from total RNAs isolated from human embryonic carcinoma cell line NTERA-2 (NT2) and mouse ES cell line D3. Intronless human SOX1 and SOX11 cDNAs were cloned from human genomic DNA. Human SOX10 was subcloned from an FL cDNA clone purchased from National Institutes of Health (NIH) Mammalian Gene Collection (MGC). Yeast GCN20 cDNA was amplified from purified Saccharomyces cerevisiae genomic DNA. For expression of GCN20, FL human and mouse ABCF1, as well as various truncations of human ABCF1 in Escherichia coli, corresponding cDNAs were cloned into a modified pMtac-His6 vector containing a His6 tag at the N terminus and a FLAG tag at the C terminus. Mutations of the two Walker A motifs in human ABCF1 (K324M and K664M) were created by using the QuikChange II Site-Directed Mutagenesis Kit (Agilent). Human ABCF1 and yeast GCN20 domain fusion cDNAs were created by PCR-mediated ligation and cloned into modified pMtac-His6 vector. For in vitro droplet assays, C-terminal enhanced GFP (eGFP) fusion proteins containing FL, LCD (1–302), and LCD-truncated human ABCF1 (NBDs) were inserted into pMtac vector in frame with a flexible linker (GGSGGGSG) followed by eGFP. For in vivo expression of FL ABCF1-eGFP and LCD-eGFP fusion proteins, corresponding human ABCF1 cDNAs were inserted into pFLAG-CMV5a in frame with a flexible linker (GGSGGGSG) followed by eGFP. mCherry cDNA was subcloned into pFLAG-CMV5a in frame with a flexible linker (GGSGGGSG) and a C-terminal FLAG tag. mCherry-GGSGGGSG-SOX2-FLAG was generated by subcloning human SOX2 cDNA in frame between the flexible linker and FLAG tag. GST fusion proteins containing the human ABCF1 LCD (1–302), yeast GCN20 (amino acids 1 to 197), and transcription factor SREBP1a (amino acids 1 to 50) were generated by inserting the corresponding cDNA fragments into pGEX4T-3 vector (Sigma-Aldrich). V5-tagged FL and NBDs of human ABCF1 and untagged mouse Nanog were cloned into lentiviral overexpression vector pHAGE-IRES-Neo (11). cDNAs for human OCT4 and SOX2 were cloned into the pFLAG-CMV5a mammalian expression vector (Sigma-Aldrich).
Polyclonal antibodies against ABCF1 (13950-1-AP) were purchased from ProteinTech; XPC (A301-122A) and mouse Nanog (A300-397A) from Bethyl Laboratories; DKC1 (H-300), OCT4 (N-19), and Pol II (N-20) from Santa Cruz Biotechnology; SOX2 (AB5603) from EMD Millipore; V5 ChIP grade (ab15828) from Abcam; and histone H3 (ChIP formulated, no. 2650) from Cell Signaling Technology (CST). Monoclonal antibodies against ACTB (66009-1) were purchased from ProteinTech; hemagglutinin (HA) tag (C29F4) and Rbp1 (Pol II) N-terminal domain (NTD) (D8L4Y) from CST; FLAG tag (M2), α-tubulin (T5168), and trimethyl histone H3 (Lys4) (H3K4me3) (05-745R-S) from Sigma-Aldrich; RFP (600-401-379) from Rockland; and V5 tag (R96025) from Life Technologies.
Cell culture
NT2 cell line was obtained from the American Type Culture Collection (ATCC). NT2, 293T, and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) high glucose with GlutaMAX (Gibco) supplemented with 10% fetal bovine serum (FBS). Large-scale culture of NT2 cells was described (11). Feeder-independent human ES cell line H9 was purchased from WiCell Research Institute. H9 ES cells were cultured in mTeSR1 (STEMCELL Technologies) with normocin (50 μg/ml; InvivoGen) on Matrigel-coated tissue culture plates (Corning). D3 mouse ES cell line was purchased from ATCC and adapted to feeder-free condition as described (11). Medium was changed daily. Cell cultures were passaged by StemPro Accutase (Gibco) for human ES cells and trypsin for mouse ES cells.
For ChIP experiments, mouse ES cells were adapted to 2i/LIF culture condition. Mouse ES cells were passaged in 2i/LIF, serum-free medium composed of 1:1 mix of Advanced DMEM/F12 and Neurobasal medium (Gibco) supplemented with N2 (Gibco), B27 (Gibco), l-glutamine (GlutaMAX, Gibco), β-mercaptoethanol (0.1 mM; Sigma-Aldrich), BSA (50 μg/ml; Sigma-Aldrich), PD0325901 (1 μM; Sigma-Aldrich), CHIR99021 (3 μM; EMD Millipore), and LIF (103 U/ml) for at least four passages before the cells were used for ChIP and reverse transcription (RT)–qPCR analyses.
Differentiation of H9 ES cells was induced by exchanging human ES cell growth medium with DMEM/F12 (Gibco) containing 2 mM l-glutamine, 12.5% FBS, and normocin (50 μg/ml) for up to 14 days. V5-ABCF1 knock-in D3 mouse ES cells were induced to differentiate by maintaining cells in regular medium containing 5 μM all-trans retinoic acid (Sigma-Aldrich) for 7 days.
In vitro transcription assay
In vitro transcription reactions, the human NANOG transcription template, purification of activators OCT4 and SOX2, general transcription factors, RNA Pol II, and recombinant XPC complex were described (11). Recombinant XPC complex purified from Sf9 and DKC1 complex reconstituted and purified from Sf9 cells or E. coli (15) were supplemented in the in vitro transcription reactions to assay for SCC-B activity.
Purification of SCC-B/ABCF1
All steps were performed at 4°C. Nuclear extracts were prepared from 400 liters of NT2 cells. Partially purified P11-P1M and Ni-NTA flowthrough (Ni-FT) fractions were prepared as described (11). The Ni-FT fraction was dialyzed against buffer D [20 mM Hepes (pH 7.9) and 2 mM MgCl2] at 0.2 M KCl with 0.0025% NP-40 and 10% glycerol (all buffers from then on contained 0.0025% NP-40 and 10% glycerol unless otherwise stated). This Ni-FT fraction was applied to a Poros 20 HQ column (Applied Biosystems), subjected to a four–column volume (CV) linear gradient from 0.2 to 0.4 M KCl (Q0.3), washed at 0.52 M KCl, and developed with a 13-CV linear gradient from 0.52 to 1.0 M KCl. Transcriptionally active Q0.3 fraction (0.32 to 0.4 M) was pooled and applied directly to hydroxyapatite (HAP) type II ceramic resin (Bio-Rad), washed first at 0.38 M, and then lowered to 0.1 M KCl in 3 CV. HAP column buffer was then exchanged and washed extensively with buffer D at 0.03 M KPi (pH 6.8) without KCl and NP-40. The HAP column was subjected to a 20-CV linear gradient from 0.03 to 0.6 M KPi. Active HAP fractions eluting from 0.2 to 0.3 M KPi were pooled and separated on a Superose 6 XK 16/70 gel filtration column (130 ml; GE Healthcare) equilibrated with buffer D + 0.1 mM EDTA at 0.15 M KCl. Active Superose 6 fractions with an apparent molecular mass of 400 to 600 kDa were pooled and supplemented with insulin (0.25 mg/ml; Roche). Pooled fractions were applied to a Poros-HE column (Applied Biosystems), equilibrated in buffer D + 0.1 mM EDTA at 0.15 M KCl, and subjected to a 34-CV linear gradient from 0.15 to 1 M KCl. SCC-B containing HE fractions eluted from 0.35 to 0.43 M KCl. Active HE fractions eluting from 0.35 to 0.43 M KCl were supplemented with insulin (0.3 mg/ml) and dialyzed to 0.15 M KCl in buffer D + 0.1 mM EDTA + 0.01% NP-40. The dialyzed HE fraction was applied to a Mono S PC 1.6/5 column (GE Healthcare), washed, and developed with a 20-CV linear gradient from 0.15 to 0.65 M KCl. Transcriptionally active SCC-B/ABCF1 fractions eluted from 0.29 to 0.31 M KCl.
Mass spectrometry analysis
Peak Mono S fractions were pooled, concentrated using a Spin-X centrifugal concentrator, separated by SDS-PAGE, and stained with PageBlue (Thermo Fisher Scientific), and protein bands were excised, digested with trypsin, and extracted. Peptide pools from each gel slice were analyzed by matrix-assisted laser desorption time-of-flight mass spectrometry (Bruker Reflex III). Selected mass values were used to search protein databases linked to PROWL (Rockefeller University) using ProFound and protein databases linked to Expasy (Swiss Institute of Bioinformatics, Geneva) using PeptIdent.
Generation of endogenously V5-tagged ABCF1 knock-in mouse ES cell line
Single-guide RNA (sgRNA) targeting genomic region immediately downstream of the ATG translation start codon of ABCF1 was cloned into LentiCRISPRv2 vector (Addgene). An ss donor oligonucleotide containing a V5 tag followed by a flexible linker GSSG sequence in frame with the second amino acid of ABCF1, which is flanked by left and right homology arms of about 70 base pairs (bp), was synthesized [Integrated DNA Technologies (IDT)]. Both the LentiCRISPRv2-sgRNA plasmid and the ss donor oligonucleotides were transfected into D3 mouse ES cell line using Lipofectamine 3000 (Invitrogen). Transfected cells were selected with puromycin (1.5 μg/ml) for 3 days. Cells were then expanded in the absence of puromycin to avoid integration of the LentiCRISPRv2-sgRNA plasmid into the genome. Single clones were plated into 96-well plates. Positive clones were identified by PCR and confirmed by sequencing and Western blotting. Clones selected for further analysis were confirmed to be puromycin-sensitive and express similar levels of key pluripotency genes such as Nanog, Oct4, and Sox2. See table S1 for sgRNA and ss donor oligonucleotide sequences.
Purification of recombinant proteins
pMtac expression plasmids were transformed into BL21-CodonPlus-RIPL competent cells (Agilent). Expression of His6-tagged proteins was induced at 30°C with 0.5 mM isopropyl-β-D-thiogalactopyranoside for 4 hours. Cell pellets were lysed in high-salt lysis buffer HSLB [50 mM tris-HCl (pH 7.9), 0.5 M NaCl, 0.6% Triton X-100, 0.05% NP-40, and 10% glycerol] with imidazole (10 mM) and lysozyme (0.5 mg/ml). Sonicated lysates were cleared by ultracentrifugation and incubated with Ni–nitrilotriacetic acid (NTA) resin for 1.5 hours at 4°C. Bound proteins were washed extensively with HSLB with 20 mM imidazole, equilibrated with 0.3 M NaCl HGN [25 mM Hepes (pH 7.9), 10% glycerol, and 0.05% NP-40] with 20 mM imidazole, and eluted with 0.25 M imidazole in 0.3 M NaCl HGN. Peak fractions were pooled and incubated with anti-FLAG M2 agarose (Sigma-Aldrich) for 3 hours at 4°C. Immunoprecipitated proteins were washed extensively with 0.7 M NaCl HEMG [25 mM Hepes (pH 7.9), 0.1 mM EDTA, 12.5 mM MgCl2, and 10% glycerol] with 0.1% NP-40 and equilibrated with 0.25 M NaCl HEMG with 0.1% NP-40.
pFLAG-CMV5a plasmids expressing C-terminal FLAG-tagged mCherry or mCherry-SOX2 were transfected into 293T cells with polyethyleneimine. Purification of FLAG-tagged proteins using anti-FLAG M2 agarose beads was described previously (11).
For recombinant proteins used in in vitro transcription assays, bound proteins were eluted in the same buffer containing FLAG peptides (0.4 mg/ml). For in vitro LLPS droplet assays, bound proteins were extensively washed with 0.25 M NaCl HEMG without NP-40 before the proteins were eluted with FLAG peptides in the same buffer without detergent. Eluted proteins were filtered through a 0.22-μm filter. Proteins used in droplet assays were concentrated by using a Spin-X UF concentrator (10,000 molecular weight cutoff) (Corning). Protein concentrations were determined by Bradford assays. Purified proteins were snap-frozen in liquid nitrogen and stored at −80°C.
pGEX4T-3 expression plasmids were transformed into BL21-CodonPlus-RIPL competent cells (Agilent). Protein expression was induced at 20°C for 3.5 hours. Bacterial pellets were resuspended in 1× phosphate-buffered saline (PBS) containing 3 M dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and complete protease inhibitor cocktail (Sigma-Aldrich) and snap-frozen in liquid nitrogen. Sonicated lysates were cleared by ultracentrifugation. Cleared lysates were then incubated with Glutathione Sepharose 4 Fast Flow (GE Healthcare) to immobilize GST fusion proteins.
In vitro droplet assay and imaging analysis
Recombinant eGFP fusion proteins were diluted into indicated protein and NaCl concentration with 10% PEG-8000 (Sigma-Aldrich) with or without 10% 1,6-Hex and incubated for 30 min at room temperature. For ABCF1-SOX2-dsDNA droplet reactions, eGFP or ABCF1-eGFP (6.4 μM), mCherry or mCherry-SOX2 (3.2 μM), and Cy5-labeled dsDNA (ds-UM, 6.4 μM) or water were mixed in buffer containing 10% PEG-8000 and incubated for 30 min to 1 hour at room temperature. The solutions were loaded into an in-house chamber and imaged with a confocal microscope with a 63× objective. For analyzing colocalization of GFP, mCherry, and Cy5 signals in droplets, custom ImageJ macro code was written to identify green/red channel mask size/intensity and overlay area. Each channel first goes through a Gaussian blur filter (radius 1 pixel) to reduce noise, then “Yen dark” threshold algorithm is applied to green channel, and “Otsu dark” threshold algorithm is applied to red channel to achieve green/red positive mask image. (“Yen” and “Otsu” are standard ImageJ build-in threshold algorithms). The overlay region (both green/red positive) is generated by binary “AND” function on green and red mask. All intensity values on green/red/overlay region were measured on four to five raw images for each condition. Droplet number, area, and line-scan profiles of intensity for each fluorescence channel were analyzed using ImageJ.
Immunofluorescence microscopy
D3 mouse ES cells were transduced with lentiviruses [multiplicity of infection (MOI) of 10] expressing untagged GFP, FLAG-tagged, GFP fusion FL, or LCD-truncated (NBDs) human ABCF1 and selected with neomycin (500 μg/ml). For examining subcellular localization of ABCF1, transduced ES cells were plated on cover glass and fixed 24 hours later in 4% paraformaldehyde for 10 min at room temperature and permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature. After three washes in PBS for 5 min each, cells were blocked with 5% BSA and 5% goat serum for 1 hour at room temperature and incubated with primary antibodies (1:200; anti-GFP; Abcam) overnight at 4°C. Following three washes, cells were incubated with secondary antibodies [1:500; goat anti-rabbit immunoglobulin G (IgG) Alexa Fluor 594; Life Technologies] for 1 hour at room temperature. Cover glasses were mounted on slides with Mountant with 4′,6-diamidino-2-phenylindole (DAPI) (Life Technologies) and imaged with a confocal microscope.
GST pull-down assay
Nuclear extracts from NT2 cells were prepared as described (49). Proteins were precipitated with ammonium sulfate (55% saturation) and resuspended in buffer D [20 mM Hepes (pH 7.9), 2 mM MgCl2, and 20% glycerol] containing 20 mM KCl and 0.01% NP-40 to about a fourth of the starting volume of nuclear extracts. Soluble extracts were dialyzed against buffer D containing 0.2 M KCl, 0.2 mM EDTA, 10% glycerol, and 0.01% NP-40 (buffer HEGN). Dialyzed nuclear extracts were cleared by centrifugation.
Bacterial lysates containing GST fusion proteins were immobilized onto Glutathione Sepharose 4 Fast Flow in 1× PBS and BSA (30 μg/ml). Bound proteins were washed extensively with STE buffer [20 mM tris (pH 8.0), 1 mM EDTA, 1% Triton X-100, 0.5% NP-40, 10% glycerol] containing 1 M NaCl. Approximately 20 μg of GST-ABCF1 (1–302), equimolar of GST-GCN20 (1–197), and GST-SREBP1a (1–50) were immobilized onto the Sepharose beads. Bound GST fusion proteins were equilibrated to 0.2 M NaCl HEGN.
Approximately 3 mg of NT2 nuclear extracts was incubated with the immobilized GST fusion proteins overnight at 4°C. Bound proteins were washed extensively with 0.3 M NaCl HEGN and then with 0.1 M NaCl HEGN. Proteins bound to GST fusion proteins were eluted by incubating the Sepharose slurry twice with 0.1 M NaCl HEGN containing 0.2% sarkosyl.
Coimmunoprecipitation assay
pHAGE-V5-ABCF1 (FL and NBDs), pFLAG-CMV5a-OCT4, and pFLAG-CMV5a-SOX1, 2 (FL, HMG, and AD), 10, and 11 expression plasmids were cotransfected in various combinations into 293T cells using Lipofectamine 2000 (Invitrogen). Transfected cells on 10-cm dishes were lysed with 1 ml of lysis buffer [0.25 M NaCl, 20 mM Hepes (pH 7.9), 2 mM MgCl2, 0.5% NP-40, and 10% glycerol] 40 hours after transfection. Cell lysates were collected and homogenized by passing through a 25-gauge needle five times. Lysates were cleared by centrifugation at 15,000 rpm for 25 min at 4°C. Cleared lysates were then incubated with anti-FLAG M2 agarose preblocked with BSA (5 mg/ml) for 3 to 4 hours at 4°C. Bound proteins were washed extensively with lysis buffer followed by FLAG peptide elution (0.4 mg/ml) in buffer HEMG containing 0.3 M NaCl and 0.1% NP-40.
To detect interaction between endogenous ABCF1 and SOX2 under normal and DNA damage conditions, V5-ABCF1 knock-in D3 mouse ES cells were treated with dimethyl sulfoxide (DMSO) or ETO (20 μM; Sigma-Aldrich) for 6 hours. Whole-cell extracts were prepared using lysis buffer [20 mM Hepes (pH 7.9), 0.12 M NaCl, 1 mM EDTA, 1% NP-40, 1 mM DTT, 1 mM benzamidine, 0.5 mM PMSF, and complete protease inhibitors (Sigma-Aldrich)]. To assess the effect of dsDNA on ABCF1-SOX2 interaction, pHAGE-RFP-IRES-Neo plasmids were digested to completion with Eco RV, Hind III, and Xho I to generate DNA fragments of varying sizes ranging from 79 bp to 2.8 kb. Digested DNAs were purified by phenol/chloroform extraction and ethanol precipitation. Purified DNAs were transfected into mouse ES cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol. Cells were harvested 4 hours after transfection. Cell lysates were incubated on ice for 10 min, and lysates were cleared by centrifugation. Cleared lysates were incubated with IgG or anti-SOX2 antibodies at 4°C overnight. Lysates were incubated with Protein A Sepharose (GE Healthcare) for 1 to 2 hours at 4°C, and bound proteins were washed extensively with lysis buffer and eluted with SDS sample buffer with boiling.
Micrococcal nuclease–ChIP
V5-ABCF1 knock-in D3 mouse ES cells were first adapted to 2i/LIF conditions. Cells were treated with DMSO/ETO (80 μM, 8.5 hours) or DMSO/1,6-Hex (1.5%, 30 min) before proceeding with ChIP. Cells were cross-linked with EGS for 20 min and then with formaldehyde for 5 min. Nuclei were prepared as described (15). Nuclei were equilibrated with MNase buffer [10 mM tris-HCl (pH 7.5), 15 mM NaCl, 60 mM KCl, 1 mM CaCl2, 1 mM PMSF, complete protease inhibitors (Sigma)] before nuclei were digested with MNase in the same buffer as described (50). Briefly, nuclei from 4 × 107 of cells were resuspended in 0.5 ml of MNase digestion buffer and digested with 2400 U of MNase (New England Biolabs) in the presence of 3.3 mM CaCl2 and 0.2% Triton X-100 at 30°C for 12 min with vigorous shaking (1200 rpm). Digestion was terminated by adding 12.5 μl of MNase stop buffer (250 mM EDTA, 250 mM EGTA, 0.5% SDS, and 125 mM NaCl). MNase-digested nuclei slurry was sonicated using Bioruptor 300 (Diagenode) to disrupt the nuclear membrane. Released chromatin was clarified by centrifugation. Input chromatin and immunoprecipitated DNA were reverse cross-linked at 65°C overnight. DNA was purified by phenol/chloroform extraction followed by chloroform extraction. DNA was precipitated by adding 1/10 volume of 3 M sodium acetate (pH 5.5) and three volumes of ethanol. Precipitated DNA was washed once with 70% ethanol and resuspended in 1× TE buffer [10 mM tris-HCl (pH 8.0) and 0.1 mM EDTA]. Purified DNA was quantified by real-time PCR with KAPA SYBR FAST qPCR Master Mix (KAPA Biosystems) and gene-specific primers using the CFX Touch Real-Time PCR Detection System (Bio-Rad). Primer sequences are shown in table S2.
shRNA-mediated knockdown and rescue by lentiviral infection
For lentivirus production, nontargeting control and pLKO plasmids targeting mouse ABCF1 (Sigma-Aldrich), or pHAGE plasmids for overexpression were cotransfected with packaging vectors (psPAX2 and pMD2.G; Addgene) into 293T cells using Lipofectamine 2000 (Invitrogen). Supernatants were collected at 48 hours and again at 72 hours. Virus preparation was performed as described (11). Functional viral titer was determined by transduction of limited dilution of concentrated viruses into HeLa or NIH 3T3 cells followed by counting antibiotic-resistant colonies to obtain colony-forming unit per milliliter. Cells were infected with viruses in the presence of polybrene (8 μg/ml). For knockdown experiments in mouse ES cells, lentiviruses expressing control nontargeting shRNA or two independent shRNAs targeting ABCF1 were used to infect cells. Infected cells were selected with puromycin (1.5 μg/ml). Pluripotency status of control and ABCF1 knockdown cells was analyzed using an AP detection kit (EMD Millipore) or by RT-qPCR analysis of mRNAs purified using TRIzol reagent (Life Technologies).
For rescue experiments in ABCF1 knockdown mouse ES cells, mouse ES cells were coinfected with viruses expressing shABCF1 (pLKO-shABCF1-1) at MOI of 15 and viruses expressing either control RFP at MOI of 6, mouse Nanog at MOI of 6, FL hABCF1 (FL), or N-terminally truncated ABCF1 (NBDs) at MOI of 6 and 1, respectively. We used a lower MOI for NBDs because it expresses at a substantially higher level than FL ABCF1 in mouse ES cells even at MOI of 1 (see fig. S5E). Coinfected cells were selected with puromycin (1 μg/ml) and neomycin (750 μg/ml).
Small interfering RNA–mediated knockdown in human ES cells
For knockdown of ABCF1 in human ES cells, H9 cells were transfected with 3.3 μM of si-nontargeting (siNT) and siABCF1 (siGENOME SMARTpool, Dharmacon) using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. Forty-eight hours after transfection, cell viability and morphology were documented before RNAs were collected by TRIzol reagent (Life Technologies) and processed for RT-qPCR analysis.
Somatic cell reprogramming and flow cytometry
CF-1 MEFs (Charles River Laboratories) were transduced with inducible STEMCCA and rtTA lentivirus–containing supernatants overnight in polybrene (8 μg/ml; Sigma-Aldrich). Doxycycline (2 μg/ml; Sigma-Aldrich) was supplemented to complete mouse ES cell medium to induce expression of OCT4, KLF4, SOX2, and c-MYC. Reprogramming was assayed by AP staining (EMD Millipore) or by flow cytometry analysis using anti–SSEA-1 (BioLegend) on BD LSRFortessa, performed according to the manufacturers’ protocols.
RNA isolation, RT, and real-time PCR analysis
Cells were rinsed once with 1× PBS. Total RNA was extracted and purified using TRIzol reagent (Life Technologies) followed by deoxyribonuclease I treatment (Invitrogen). cDNA synthesis was performed with 1 μg of total RNA using the iScript cDNA Synthesis Kit (Bio-Rad) and diluted 10-fold. Real-time PCR analysis was carried out with iTaq Universal SYBR Green (Bio-Rad) and gene-specific primers using the CFX Touch Real-Time PCR Detection System (Bio-Rad). Results were normalized to β-actin. Primer sequences are shown in table S3.
Nucleofection of oligonucleotides and flow cytometry
D3 mouse ES cells were detached and washed once with 1× PBS. For each nucleofection, 3 × 106 cells were resuspended in 80 μl of mouse ES cell nucleofection solution (Lonza). Equimolar (0.32 nmol) of 6-FAM–labeled ss, ds-M, or ds-UM DNA oligonucleotides (IDT) (table S4) was nucleofected into mouse ES cells using an Amaxa nucleofector (Lonza) according to the manufacturer’s instructions. Cells mock-nucleofected with water were used as a negative control. Nucleofected cells were recovered in 1 ml of prewarmed ES cell medium at 37°C for 10 min before cells were transferred to 10-cm culture plates. Twenty-four hours after plating, cells were detached, rinsed with 1× PBS, resuspended in 0.2 ml of ice-cold PBS, and filtered into BD filter cap tubes for fluorescence-activated cell sorting (FACS). Fluorescein isothiocyanate (FITC)–positive cells were sorted on a BD FACSAria II instrument using the following settings: 70 μm nozzle, 70 psi pressure, frequency of 85, amplitude of 5, and 70 sheath pressures. Mock-nucleofected negative control cells were used for setting the gate for FITC+ cells. Once the gate is adjusted, FITC+ cells were collected directly into TRIzol (Life Technologies) solution and immediately frozen in dry ice.
Cytoplasmic and nuclear fractionation
DMSO- or ETO-treated mouse ES cells were washed with PBS twice and lysed with buffer A [10 mM Hepes (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.4% NP-40, 1 mM DTT, and complete protease inhibitors (Sigma-Aldrich)] on ice for 10 min. Lysates were cleared by spinning down at 15,000g for 3 min at 4°C. Supernatants were kept as cytoplasmic fractions. Nuclei were washed with buffer A three times and lysed with buffer B [20 mM Hepes (pH 7.9), 0.4 M NaCl, 1 mM EDTA, 10% glycerol, 1 mM DTT, and complete protease inhibitors (Sigma-Aldrich)] with vigorous shaking for 2 hours or overnight at 4°C. Lysates were cleared by centrifugation at 15,000g for 5 min at 4°C. Supernatants were kept as nuclear fractions.
ABCF1-DNA interaction analysis
For DNA pull-down assay, approximately 5 × 106 V5-ABCF1 knock-in D3 mouse ES cells were lysed in 0.2 ml of lysis buffer [0.14 M NaCl, 50 mM Hepes (pH 7.9), 1 mM EDTA, 2 mM MgCl2, 0.5% NP-40, and 10% glycerol]. Cell lysates were homogenized by passing through a 25-gauge needle five times. Cell lysates were cleared by centrifugation. 5′ Biotinylated ss or ds oligonucleotides labeled with biotin at the 5′ end of the sense strand were synthesized. All oligos contain modified, exonuclease-resistant nucleotides (phosphorothioate bonds) in the last five nucleotides at the 5′ and 3′ ends of both sense and antisense strands (IDT). These oligonucleotides were incubated with cell lysates for 1.5 hours at 4°C. For each pull-down reaction, 100 μl of Dynabeads MyOne Streptavidin T1 bead slurry was first preblocked in lysis buffer containing BSA (50 mg/ml) for 1 hour at 4°C. The preblocked beads were then incubated with cell lysate containing oligonucleotides for 1 hour at 4°C. Biotinylated DNA oligonucleotides and associated protein factors captured onto the beads were washed extensively with lysis buffer. Bound proteins were eluted by boiling the beads in sample buffer. Sequences of the 5′ biotinylated oligonucleotides are available in table S4.
To analyze binding of ABCF1 to endogenous dsDNA fragments, V5-ABCF1 knock-in D3 mouse ES cells were treated with DMSO or ETO (20 μM) for 6 hours to induce DNA fragmentation. Whole-cell extracts were prepared by using the same lysis buffer as for DNA pull-down assay. Cell lysates were homogenized and cleared. Cleared lysates were incubated with IgG or anti-V5 overnight at 4°C. Protein A Sepharose was added to lysates and incubated for 1 to 2 hours at 4°C. Bound proteins were washed extensively with lysis buffer. Bound DNAs were eluted by ribonuclease and proteinase K treatment. Eluted DNAs were extracted by phenol-chloroform extraction and purified by ethanol precipitation. Purified DNAs were run on a 6% urea-denaturing gel, stained with SYBR Gold (Invitrogen), and analyzed.
Colony formation assay
To examine colony-forming ability of ABCF1-overexpressing cells under DNA damaging condition, D3 mouse ES cells were transduced with lentiviruses expressing untagged RFP, V5-tagged FL human ABCF1, or V5-tagged LCD-truncated (NBDs) ABCF1. Mouse ES cells were transduced with lentiviruses at MOI of 3 and selected with neomycin (500 μg/ml). RFP- or ABCF1-overexpressing mouse ES cells (200 or 1000) were plated on 24-well plates. Cells were allowed to recover for 24 hours before treatment with DMSO or ETO (1 μM) for 1, 2, or 4 hours, after which fresh medium without ETO was replaced. After 6 days, cells were fixed and stained for AP activity (EMD Millipore). AP-positive cells were counted and analyzed. For transient transfection of dsDNA oligonucleotides into D3 mouse ES cells stably expressing GFP, FLAG-tagged GFP fusion FL, and NBDs ABCF1, 100 cells were plated on 24-well plates 24 hours before transfection with water or ds-UM (0.2 μg) using Lipofectamine 3000 (Invitrogen). Cells were replaced with fresh medium the next day and were fixed and stained for AP activity (EMD Millipore) 3 days later. AP-positive colonies were counted.
Amino acid composition analysis
The amino acid composition of a protein was analyzed in R with custom scripts. The occurrence of each amino acid is counted by using the package “stringr” and plotted with the package “plot. matrix.”
Statistical analysis
To determine statistical significance, P values were calculated by using unpaired two-sided Student’s t test. P values less than 0.05 (<0.05) were considered as statistically significant, and they were indicated with * (*P < 0.05). All data represent the mean ± SEM (error bars) except for Fig. 6I. For Fig. 6I, box and whisker plot was used. The central mark represents median, and edges represent 25th and 75th percentiles. The whiskers indicate 5th to 95th percentiles.
Acknowledgments
We thank G. Dailey for help with expression constructs; S. Zheng for tissue culture assistance; and S. Zhou and D. King for mass spectrometry analysis, E. Guo for advice on in vitro droplet assay, and A. Hansen at Massachusetts Institute of Technology for discussion on microscopy. We thank R. Tjian, S. Agarwal, Y.-C. Hsu, Y. Isogai, Z. Zhang, A. Hansen, B. Zhang, E. Guo, S. Chong, and C. Cattoglio for valuable discussion.
Funding: We acknowledge the following funding sources: NIH grant R01HL125527, Harvard Stem Cell Institute, Boston Biomedical Innovation Center, Charles H. Hood Foundation, Brigham Research Institute, and Brigham and Women’s Hospital HVC Junior Faculty Research Awards to Y.W.F.
Author contributions: Y.W.F. conceived and supervised this study. Y.W.F., E.-B.C., M.V., and M.Z. designed the experiments. E.-B.C. and Y.W.F. conducted biochemical purification. E.-B.C., M.V., and Y.W.F. performed in vitro assays. C.I. purified general transcription factors for in vitro transcription assays. J.W. assisted in purification of recombinant proteins. E.-B.C., M.V., and M.Z. performed cell-based assays and gene expression analyses. M.V. and M.Z. performed ChIP and FACS analyses. J.J.H. performed somatic cell reprogramming experiments and FACS analysis. L.D. wrote code for in vitro droplet imaging analysis. Y.W.F., E.-B.C., and M.V. wrote the manuscript. All authors edited the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S4
REFERENCES AND NOTES
- 1.Dunn S.-J., Martello G., Yordanov B., Emmott S., Smith A. G., Defining an essential transcription factor program for naïve pluripotency. Science 344, 1156–1160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marson A., Levine S. S., Cole M. F., Frampton G. M., Brambrink T., Johnstone S., Guenther M. G., Johnston W. K., Wernig M., Newman J., Calabrese J. M., Dennis L. M., Volkert T. L., Gupta S., Love J., Hannett N., Sharp P. A., Bartel D. P., Jaenisch R., Young R. A., Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134, 521–533 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim J., Chu J., Shen X., Wang J., Orkin S. H., An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K., Yamanaka S., A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 17, 183–193 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Brumbaugh J., Di Stefano B., Hochedlinger K., Reprogramming: Identifying the mechanisms that safeguard cell identity. Development 146, dev182170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong Y. W., Cattoglio C., Yamaguchi T., Tjian R., Transcriptional regulation by coactivators in embryonic stem cells. Trends Cell Biol. 22, 292–298 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boija A., Klein I. A., Sabari B. R., Dall’Agnese A., Coffey E. L., Zamudio A. V., Li C. H., Shrinivas K., Manteiga J. C., Hannett N. M., Abraham B. J., Afeyan L. K., Guo Y. E., Rimel J. K., Fant C. B., Schuijers J., Lee T. I., Taatjes D. J., Young R. A., Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G. M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., Tjian R., Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J. L., McKnight S. L., Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harper T. M., Taatjes D. J., The complex structure and function of Mediator. J. Biol. Chem. 293, 13778–13785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fong Y. W., Inouye C., Yamaguchi T., Cattoglio C., Grubisic I., Tjian R., A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell 147, 120–131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodda D. J., Chew J. L., Lim L. H., Loh Y. H., Wang B., Ng H. H., Robson P., Transcriptional regulation of Nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731–24737 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Cattoglio C., Zhang E. T., Grubisic I., Chiba K., Fong Y. W., Tjian R., Functional and mechanistic studies of XPC DNA-repair complex as transcriptional coactivator in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 112, E2317–E2326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang E. T., He Y., Grob P., Fong Y. W., Nogales E., Tjian R., Architecture of the human XPC DNA repair and stem cell coactivator complex. Proc. Natl. Acad. Sci. U.S.A. 112, 14817–14822 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong Y. W., Ho J. J., Inouye C., Tjian R., The dyskerin ribonucleoprotein complex as an OCT4/SOX2 coactivator in embryonic stem cells. eLife 3, e03573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arora H., Wilcox S. M., Johnson L. A., Munro L., Eyford B. A., Pfeifer C. G., Welch I., Jefferies W. A., The ATP-binding cassette gene ABCF1 functions as an E2 ubiquitin-conjugating enzyme controlling macrophage polarization to dampen lethal septic shock. Immunity 50, 418–431.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Lee M. N., Roy M., Ong S. E., Mertins P., Villani A. C., Li W., Dotiwala F., Sen J., Doench J. G., Orzalli M. H., Kramnik I., Knipe D. M., Lieberman J., Carr S. A., Hacohen N., Identification of regulators of the innate immune response to cytosolic DNA and retroviral infection by an integrative approach. Nat. Immunol. 14, 179–185 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilcox S. M., Arora H., Munro L., Xin J., Fenninger F., Johnson L. A., Pfeifer C. G., Choi K. B., Hou J., Hoodless P. A., Jefferies W. A., The role of the innate immune response regulatory gene ABCF1 in mammalian embryogenesis and development. PLOS ONE 12, e0175918 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans M. J., Kaufman M. H., Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981). [DOI] [PubMed] [Google Scholar]
- 20.Iseki H., Nakachi Y., Hishida T., Yamashita-Sugahara Y., Hirasaki M., Ueda A., Tanimoto Y., Iijima S., Sugiyama F., Yagami K. I., Takahashi S., Okuda A., Okazaki Y., Combined overexpression of JARID2, PRDM14, ESRRB, and SALL4A dramatically improves efficiency and kinetics of reprogramming to induced pluripotent stem cells. Stem Cells 34, 322–333 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Xu Y., Zhang M., Li W., Zhu X., Bao X., Qin B., Hutchins A. P., Esteban M. A., Transcriptional control of somatic cell reprogramming. Trends Cell Biol. 26, 272–288 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Sommer C. A., Stadtfeld M., Murphy G. J., Hochedlinger K., Kotton D. N., Mostoslavsky G., Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 27, 543–549 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasiliou V., Vasiliou K., Nebert D. W., Human ATP-binding cassette (ABC) transporter family. Hum. Genomics 3, 281–290 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coots R. A., Liu X.-M., Mao Y., Dong L., Zhou J., Wan J., Zhang X., Qian S.-B., m6A facilitates eIF4F-independent mRNA translation. Mol. Cell 68, 504–514.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paytubi S., Morrice N. A., Boudeau J., Proud C. G., The N-terminal region of ABC50 interacts with eukaryotic initiation factor eIF2 and is a target for regulatory phosphorylation by CK2. Biochem. J. 409, 223–231 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Mészáros B., Erdos G., Dosztányi Z., IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 46, W329–W337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu L., Jiang Y., Cheng C., Wu D., Meng B., Chen Z., Zhu Y., Shaw N., Ouyang S., Liu Z. J., Crystal structure of ATP-bound human ABCF1 demonstrates a unique conformation of ABC proteins. Structure 26, 1259–1265.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Paytubi S., Wang X., Lam Y. W., Izquierdo L., Hunter M. J., Jan E., Hundal H. S., Proud C. G., ABC50 promotes translation initiation in mammalian cells. J. Biol. Chem. 284, 24061–24073 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Der Lee R., Buljan M., Lang B., Weatheritt R. J., Daughdrill G. W., Dunker A. K., Fuxreiter M., Gough J., Gsponer J., Jones D. T., Kim P. M., Kriwacki R. W., Oldfield C. J., Pappu R. V., Tompa P., Uversky V. N., Wright P. E., Babu M. M., Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pak C. W., Kosno M., Holehouse A. S., Padrick S. B., Mittal A., Ali R., Yunus A. A., Liu D. R., Pappu R. V., Rosen M. K., Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell 63, 72–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vodnala M., Choi E. B., Fong Y. W., Low complexity domains, condensates, and stem cell pluripotency. World J. Stem Cells 13, 416–438 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabari B. R., Dall’Agnese A., Boija A., Klein I. A., Coffey E. L., Shrinivas K., Abraham B. J., Hannett N. M., Zamudio A. V., Manteiga J. C., Li C. H., Guo Y. E., Day D. S., Schuijers J., Vasile E., Malik S., Hnisz D., Lee T. I., Cisse I. I., Roeder R. G., Sharp P. A., Chakraborty A. K., Young R. A., Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vazquez de Aldana C. R., Marton M. J., Hinnebusch A. G., GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2 alpha kinase GCN2 in amino acid-starved cells. EMBO J. 14, 3184–3199 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Näär A. M., Beaurang P. A., Zhou S., Abraham S., Solomon W., Tjian R., Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398, 828–832 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Wilson M., Koopman P., Matching SOX: Partner proteins and co-factors of the SOX family of transcriptional regulators. Curr. Opin. Genet. Dev. 12, 441–446 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Sarkar A., Hochedlinger K., The Sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell 12, 15–30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paludan S. R., Bowie A. G., Immune sensing of DNA. Immunity 38, 870–880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikawa H., Ma Z., Barber G. N., STING regulates intracellular DNA-mediated, type i interferon-dependent innate immunity. Nature 461, 788–792 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKenzie K. J., Carroll P., Martin C. A., Murina O., Fluteau A., Simpson D. J., Olova N., Sutcliffe H., Rainger J. K., Leitch A., Osborn R. T., Wheeler A. P., Nowotny M., Gilbert N., Chandra T., Reijns M. A. M., Jackson A. P., CGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunphy G., Flannery S. M., Almine J. F., Connolly D. J., Paulus C., Jønsson K. L., Jakobsen M. R., Nevels M. M., Bowie A. G., Unterholzner L., Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol. Cell 71, 745–760.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harding S. M., Benci J. L., Irianto J., Discher D. E., Minn A. J., Greenberg R. A., Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia P., Wang S., Ye B., Du Y., Huang G., Zhu P., Fan Z., Sox2 functions as a sequence-specific DNA sensor in neutrophils to initiate innate immunity against microbial infection. Nat. Immunol. 16, 366–375 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Choi U. B., Sanabria H., Smirnova T., Bowen M. E., Weninger K. R., Spontaneous switching among conformational ensembles in intrinsically disordered proteins. Biomolecules 9, 114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Y. E., Manteiga J. C., Henninger J. E., Sabari B. R., Dall’Agnese A., Hannett N. M., Spille J. H., Afeyan L. K., Zamudio A. V., Shrinivas K., Abraham B. J., Boija A., Decker T. M., Rimel J. K., Fant C. B., Lee T. I., Cisse I. I., Sharp P. A., Taatjes D. J., Young R. A., Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weng J., Wang W., Dynamic multivalent interactions of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 62, 9–13 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Wang J., Choi J. M., Holehouse A. S., Lee H. O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D., Poser I., Pappu R. V., Alberti S., Hyman A. A., A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., Sawaya M. R., Eisenberg D., Tycko R., McKnight S. L., Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maynard S., Swistowska A. M., Lee J. W., Liu Y., Liu S.-T., Bettencourt A., Cruz D. A., Rao M., De Souza-Pinto N. C., Zeng X., Bohr V. A., Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cells 26, 2266–2274 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dignam J. D., Lebovitz R. M., Roeder R. G., Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mieczkowski J., Cook A., Bowman S. K., Mueller B., Alver B. H., Kundu S., Deaton A. M., Urban J. A., Larschan E., Park P. J., Kingston R. E., Tolstorukov M. Y., MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 7, 11485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S4