Abstract
Body composition measures derived from already available electronic medical records (computed tomography [CT] scans) can have significant value, but automation of measurements is needed for clinical implementation. We sought to use artificial intelligence to develop an automated method to measure body composition and test the algorithm on a clinical cohort to predict mortality. We constructed a deep learning algorithm using Google’s DeepLabv3+ on a cohort of de‐identified CT scans (n = 12,067). To test for the accuracy and clinical usefulness of the algorithm, we used a unique cohort of prospectively followed patients with cirrhosis (n = 238) who had CT scans performed. To assess model performance, we used the confusion matrix and calculated the mean accuracy of 0.977 ± 0.02 (0.975 ± 0.018 for the training and test sets, respectively). To assess for spatial overlap, we measured the mean intersection over union and mean boundary contour scores and found excellent overlap between the manual and automated methods with mean scores of 0.954 ± 0.030, 0.987 ± 0.009, and 0.948 ± 0.039 (0.983 ± 0.013 for the training and test set, respectively). Using these automated measurements, we found that body composition features were predictive of mortality in patients with cirrhosis. On multivariate analysis, the addition of body composition measures significantly improved prediction of mortality for patients with cirrhosis over Model for End‐Stage Liver Disease alone (P < 0.001). Conclusion: The measurement of body composition can be automated using artificial intelligence and add significant value for incidental CTs performed for other clinical indications. This is proof of concept that this methodology could allow for wider implementation into the clinical arena.
Abbreviations
- BMI
body mass index
- CI
confidence interval
- CT
computed tomography
- HR
hazard ratio
- HU
Hounsfield unit
- MELD
Model for End‐Stage Liver Disease
- NAFLD
nonalcoholic fatty liver disease
- NRI
net reclassification improvement
There is emerging interest in the use of artificial intelligence in health care, but the implementation into clinical practice is only just beginning.( 1 ) Although diagnostic codes and laboratory values represent important data sources, radiological imaging such as body computed tomography (CT), which can provide phenotypic information, has yet to be fully leveraged.
Within body CT scans are measurements of body composition and nutritional status, which have significant predictive value in multiple disease states including chronic liver disease.( 2 , 3 , 4 , 5 ) The original Child‐Turcotte‐Pugh score to assess for mortality in patients with cirrhosis included assessment for nutritional status; however, this was removed and substituted with laboratory values because there was not a method to quantitatively measure nutritional status. In recent years, visceral fat as measured by CT scans has become the gold standard and known driver for metabolic syndrome and nonalcoholic fatty liver disease (NAFLD).( 6 , 7 , 8 ) The relative quantity and quality of visceral fat are associated with prognosis and cardiovascular outcome.( 6 ) Additionally, there is now increasing recognition that muscle mass and quality are important determinants of outcome.( 9 , 10 ) In NAFLD, muscle mass and quality identify patients at risk for development of liver fibrosis and cirrhosis. Once cirrhosis has developed, muscle loss can become a surrogate for body composition and poor nutritional status in patients with cirrhosis, and is associated with worse prognosis.( 11 , 12 ) To be useful for clinical care, however, muscle loss needed to be clearly defined and be quantifiable. In patients with liver disease, advanced imaging such as CT and magnetic resonance imaging measurements of muscle mass are the most accurate and accepted modality for reliable measurement, as changes in fluid status, bone density, and ascites make the use of other measurement technologies unreliable.( 13 ) Similar to muscle size and density, fat and bone density measurements are also key predictive factors that can be extracted from routinely collected abdominal CT scans to more precisely predict outcome and provide added value for clinical decision making.( 3 , 14 , 15 , 16 , 17 , 18 ) However, the standard methods for measuring body composition in CT scans involve tedious manual delineation,( 19 ) which is time‐consuming and imprecise due to interobserver and intra‐observer variabilities.( 17 ) It is also unrealistic for implementation into clinical care. Future use of body composition features within the electronic medical records for enhanced clinical application would require more automation to improve accuracy and reproducibility of measurements.
We hypothesize that artificial intelligence technology, which has revolutionized many aspects of modern life, can now automate measurements of body composition from CT scans obtained for clinical purposes in a cohort of patients with cirrhosis. Our aim is to demonstrate that these automated measurements can provide significant clinical value and be useful for patient stratification.
Patients and Methods
Patient Training and Testing Cohorts
Training Data Set: The training cohort consists of 12,067 CT scans from 10,354 de‐identified patients in the Morphomics database at the University of Michigan, where a standard axial slice was available with “ground truth” manual segmentation of the skin, fascia, and skeletal muscle boundaries at the inferior lumbar spine vertebra, L3. The CT scans in our cohort were acquired using GE LightSpeed (GE, Boston, MA), Siemens Discovery (Erlangen, Germany, and Toshiba Acquilion (Tokyo, Japan) scanners. The tube voltage was set at 120 kV for all scans, while the tube current ranged between 100 mA and 500 mA based on body mass. Both training and test sets used a combination of enhanced and unenhanced CTs, which we have previously shown to be equivalent in cross‐sectional area measurements.( 20 )
Clinical Validation Data Set: Model performance and its clinical relevance were investigated using CT scans from a cohort of patients with cirrhosis (n = 238) from the University of Michigan hepatology clinics. Patients prospectively enrolled in a chronic disease monitoring system (Avitracks; Avicenna Medical Systems, Ann Arbor, MI) with a diagnosis of cirrhosis (confirmed by a board certified hepatologist)( 21 ) from March 1, 2010, to July 30, 2015, were used for this study if they had a CT scan with the L3 slice within 365 days. At the time of enrollment, we excluded patients who had hepatocellular carcinoma or those referred for liver transplantation. All demographic and clinical details were extracted from chart review; baseline data were obtained within 6 months of the CT, and death was confirmed with the national death index. None of the CT scans from the clinical validation data set were included in the training data set.
Ground‐Truth Segmentation
The scans in the training and test sets were first processed using a semi‐automated, high‐throughput methodology (Analytic Morphomics) as previously described by our group.( 22 , 23 , 24 , 25 ) Briefly, de‐identified DICOM (Digital Imaging and Communications in Medicine) files associated with each CT were loaded into the Analytic Morphomics spatial database. This was followed by processing each scan with a series of algorithms developed in MATLAB (MathWorks, Natick, MA), to landmark aspects of spinal vertebral bodies as well as their image axis coordinates. These coordinates were then used to load the axial image most inferior to L3. Manual segmentation of boundary geometries of skin, fascia, spine, and skeletal muscle was performed by a trained research analyst who also performed manual quality control and edited resultant boundaries by using custom‐designed graphical user interfaces. Depending on the condition, this process lasted 5‐20 minutes per scan. All research analysts underwent standard training and testing before initiating any work. Multiple trained research analysts were used for this initial process and worked on what cohort was random. The manually generated masks were then inspected and adjusted by two quality control analysts to further improve measurement consistency. The results were considered our “ground truth.” In internal testing of our quality assurance process, the concordance of results from repeated measures on the same set of scans was greater than 0.99. Body feature composition measures, such as muscle and fat areas and density and bone density, were then derived from the corrected boundary geometry using density‐based thresholding and morphological operations.
Semantic Segmentation Model Development
Image data were preprocessed by windowing between −800 and 700 Hounsfield unit (HU). The scale of each image was then normalized between the range of 0 and 1. The proposed model uses Google’s DeepLabv3+( 27 ) architecture. Briefly, the DeepLabv3 model used depth‐separable convolutions, atrous spatial pyramid pooling, which allowed a larger field of vision and the pixel‐level accuracy required for this task. This model used the Xception( 26 , 27 ) convolutional neural network, which was pretrained using the ImageNet database (http://www.image‐net.org) to classify over 1000 object categories. The DeepLabv3+ with Xception network consists of 205 layers including encoder decoders, dilated convolutions, and skip connections. To match the input image to the size and channel requirements of this architecture, the single‐channel, grayscale image was replicated to create a 3‐channel RGB (red, green, blue) image size of 400 × 400 × 3. Every pixel within each image was grouped into one of five classes: skin, fascia, muscle, spine, and background. The classes are balanced with “background” having the highest number of observations and “spine” having the least. Training data were augmented with additional synthetic data by x‐axis and y‐axis rotation, translation, and scale. The model uses an Adam optimizer with an initial learning rate of 0.00001, mini‐batch size of 8, and the maximum number of epochs was set to 8. The order of the data was randomly shuffled after each epoch. The loss function was assessed by a pixel classification layer that uses categorical cross‐entropy loss.
Statistical Analysis
The quality of the semantic segmentation results was evaluated using the following region‐based and contour‐based score metrics( 28 ) for each class: accuracy, intersection over union, boundary F1( 28 ) contour matching score, and Dice similarity coefficient. These metrics have values ranging from 0 to 1, where 0 indicates there is no similarity and 1 indicates absolute similarity. Intraclass correlation coefficient (ICC) for the entire cohort was calculated using the “single fixed raters” analysis (ICC[3, 1]) in Shrout and Fleiss convention.( 29 )
To assess the body composition features of those with NAFLD (n = 78) versus those with chronic liver disease from other etiologies (n = 160), Student t test was conducted. Prognostic models of transplant‐free survival were developed using Cox proportional hazard regression analysis. Mortality was indexed from the date of the initial CT and censored at the last documented clinical visit or liver transplantation. Only one CT was used per patient. Five Morphomic variables (total muscle area index, visceral fat area and density, and subcutaneous fat area and density) were selected a priori as the initial input of the multivariate analysis based on clinical judgment of significance. The final predictive model was then developed using forward/backward selection under the Cox regression framework with Model for End‐Stage Liver Disease (MELD) and Morphomic variables in considerations, which optimizes the Akaike information criterion.( 30 ) The statistical comparison was conducted between this final predictive model and the MELD model that serves as reference model. The performance of the models was assessed with C‐statistics using the method described by Harrell et al( 31 ). We also assessed whether the addition of Morphomic features improved prediction accuracy by using a modification of the continuous net reclassification improvement (NRI) methodology that allowed for the censored data analysis.( 32 ) The continuous NRI does not require risk stratification into categories compared with the traditional category‐based NRI. We chose to use the continuous NRI because, to our knowledge, there is no consensus categorization of mortality risk in patients with liver diseases. The continuous NRI was obtained using the Hmisc package for the R statistical program.( 33 )
Results
Deep Learning Model of Body Composition
Using incidental abdominal CT scans at the L3 level, we created masks for the skin, fascia, muscle wall, and spine (Fig. 1A). These manually delineated masks provided the ground truth to train a deep learning model, which were used for measurements of subcutaneous fat, visceral fat, skeletal muscle, and bone (Fig. 1B). The intersection and difference overlap between the model’s prediction and ground truth appeared to be comparable (Fig. 1C,D). The performance was further assessed by comparing the model’s predictions to its ground truth for the training set (n = 12,067) and hold‐out, clinical test set (n = 238).
FIG. 1.
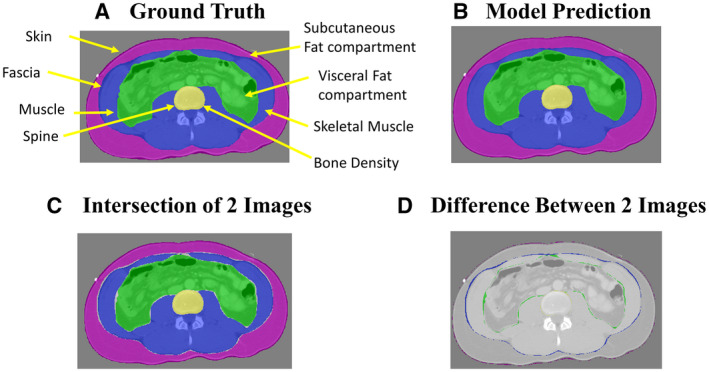
(A,B) Representative manually delineated ground truth and model prediction using artificial intelligence of body composition using abdominal CT scan at L3 level. (C,D) Representative intersection and difference between ground truth and model prediction.
We found that the model performed at a mean accuracy of 0.977 ± 0.02 and 0.975 ± 0.018 for the training and test sets, respectively (Table 1). To assess for spatial overlap, we measured the mean intersection over union and mean boundary contour scores and found excellent spatial overlap with mean scores of 0.954 ± 0.030 and 0.987 ± 0.009, and 0.948 ± 0.039 and 0.983 ± 0.013, for the training and test sets, respectively (Table 1). Additionally, we saw exceptional similarity between ground truth and prediction by the high Dice score coefficient in the test set (0.970 ± 0.021; Table 1).
TABLE 1.
Performance Accuracy of Deep Learning Model in Training and Test Sets
| Class | Training Set (n = 12,067) | Test Set (n = 238) | Dice Score Coefficient | ||||
|---|---|---|---|---|---|---|---|
| Accuracy | Intersection Over Union | BF Score | Accuracy | Intersection Over Union | BF Score | ||
| Background | 0.997 | 0.994 | 0.997 | 0.996 | 0.993 | 0.996 | 0.9964 |
| Skin | 0.979 | 0.954 | 0.991 | 0.975 | 0.953 | 0.990 | 0.9697 |
| Fascia | 0.958 | 0.920 | 0.980 | 0.949 | 0.899 | 0.975 | 0.9450 |
| Muscle | 0.985 | 0.973 | 0.975 | 0.987 | 0.975 | 0.963 | 0.9864 |
| Spine | 0.966 | 0.930 | 0.990 | 0.966 | 0.917 | 0.990 | 0.9557 |
| Mean ± SD | 0.977 ± 0.015 | 0.954 ± 0.030 | 0.987 ± 0.009 | 0.975 ± 0.018 | 0.948 ± 0.039 | 0.983 ± 0.014 | 0.970 ± 0.021 |
Model performance is assessed using accuracy, intersection over union, boundary contour score, and Dice score coefficient.
Abbreviation: BF, boundary contour.
Baseline Cohort Characteristics
To assess the clinical relevance and applicability of our deep learning model, we analyzed our study cohort of 238 patients with cirrhosis (Tables 2, 3, 4, 5). The cohort’s baseline characteristics are detailed in Table 2. In brief, our patients were 58 (51, 65) years of age, on average, with 56.72% being male. Overall, 88.24% were White, with 4.62% being Black and 2.10% being Asian and Pacific Islander. The average body mass index (BMI) was 29.0 (24.9, 33.8). In terms of the etiology of their cirrhosis, 21.43% were secondary to alcohol, 28.57% were secondary to hepatitis C infection, and 32.77% were secondary to NAFLD. Of those with alcoholic hepatitis, 68.63% were male; of those with hepatitis C, 66.18% were male; of those with NAFLD, 47.44% were male. The median MELD score was 10 (8, 13), and 47.90% were Child‐Pugh class A. Overall, 165 (69.3%) had at least one characteristic of decompensation (variceal bleed, ascites, or hepatic encephalopathy). The median follow‐up time was 1,891 days. At the end of follow‐up, 22 (9.2%) developed hepatocellular carcinoma, 105 (44%) patients died, and 1 (0.4%) had liver transplantation. Of the patients who died, 91 of 105 (86.7%) had decompensated cirrhosis before death.
TABLE 2.
Baseline Characteristics of the Total Cohort
| Characteristic | Median (Q25, Q75) (n = 238) |
|---|---|
| Age | 58 (51, 65) |
| Male (%) | 56.72 |
| Race (%) | |
| White | 88.24 |
| Black | 4.62 |
| Asian and Pacific Islander | 2.10 |
| Other or unknown | 5.04 |
| BMI | 29.0 (24.9, 33.8) |
| Etiology (%) with % male | |
| Alcoholic cirrhosis | 21.43 (68.63% male) |
| HCV | 28.57 (66.18% male) |
| NAFLD | 32.77 (47.44% male) |
| Other | 17.23 (43.90% male) |
| MELD | 10 (8, 13) |
| Child‐Pugh class (%) | |
| A | 47.90 |
| B | 38.24 |
| C | 13.87 |
| Variceal bleed (%) | 61.27 |
| Encephalopathy (%) | 28.15 |
| Ascites (%) | 50.42 |
| Platelets | 105 (75,148) |
| Albumin | 3.5 (3, 4) |
| Bilirubin | 1.2 (0.7, 2.1) |
| INR | 1.2 (1.1, 1.3) |
| Creatine | 0.8 (0.7, 1.0) |
Abbreviation: INR, international normalized ratio.
TABLE 3.
Comparing Body Composition Features of Those With NAFLD (n = 78) Versus Those With Chronic Liver Disease From Other Etiologies (n = 160)
| Body Composition | NAFLD (n = 78, mean ± SEM) | Non‐NAFLD Etiologies (n = 160, mean ± SEM) | P Value |
|---|---|---|---|
| Total muscle index* | 49.5 ± 9.9 | 47.9 ± 9.5 | 0.23 |
| Total muscle density (HU) | 33.5 ± 9.1 | 37.3 ± 9.3 | 0.003 |
| Visceral fat area (cm2) | 210.8 ± 129.8 | 133.7 ± 92.4 | <0.001 |
| Visceral fat density (HU) | −88.0 ± 10.8 | −84.7 ± 9.6 | 0.025 |
| Subcutaneous fat area (cm2) | 255.6 ± 14.7 | 211.1 ± 9.8 | 0.005 |
| Subcutaneous fat density (HU) | −98.7 ± 12.0 | −96.8 ± 12.0 | 0.24 |
| Bone mineral density (HU) | 145.2 ± 49.8 | 142.9 ± 52.6 | 0.74 |
Total muscle index = total muscle area/height2.
TABLE 4.
Univariate Cox Regression to Assess Predictors of Mortality in Patients With Cirrhosis (n = 238)
| Variable | Cox Univariate HR (95% CI) | P Value |
|---|---|---|
| Total muscle area | 0.987 (0.981, 0.994) | <0.0001 |
| Total muscle index* | 0.955 (0.944, 0.988) | 0.0002 |
| Total muscle density | 0.952 (0.932, 0.972) | <0.0001 |
| Visceral fat area | 0.998 (0.996, 0.999) | 0.02 |
| Visceral fat density | 1.043 (1.022, 1.051) | <0.0001 |
| Subcutaneous fat area | 0.998 (0.996, 0.999) | 0.02 |
| Subcutaneous fat density | 1.035 (1.019, 1.051) | <0.0001 |
| Bone mineral density | 1.001 (0.999, 1.002) | 0.57 |
Index = area/height2.
TABLE 5.
Parsimonious Multivariable Analysis for Mortality Risk Prediction in Patients With Cirrhosis
| C‐statistic | P Value | |
|---|---|---|
| MELD | 0.66 (0.55‐0.78) | REF |
| MELD + Morphomics* | 0.71 (0.61‐0.82) | <0.0001 |
Total muscle index, visceral fat area and density, and subcutaneous fat area and density were included as variables for the initial Morphomics model; the final Morphomics model includes total muscle area index and subcutaneous fat density.
Visceral and Subcutaneous Fat Areas in Those With NAFLD Are Significantly Increased
It is well‐established that patients with chronic liver disease secondary to NAFLD often present clinically with metabolic syndrome and higher percentage of body fat.( 34 , 35 , 36 , 37 ) Using our deep learning model, we obtained automated body composition measures on the incidental abdominal CT scans at L3 and compared them in patients with NAFLD versus those with chronic liver disease from other etiologies (Table 3). Within our cohort of patients with cirrhosis, 78 of 238 (33%) had a primary diagnosis of NAFLD as the cause of their liver disease. Not surprisingly, these patients were more likely to be obese, with a mean BMI of 34 ± 1.3 versus 28.5 ± 0.5 (P < 0.0001). Visceral fat area was significantly increased in those with liver disease from NAFLD (210.8 ± 129.8 cm2) as compared to those with non‐NAFLD etiologies (133.7 ± 92.4 cm2; P < 0.001). Subcutaneous fat area was also significantly higher in those with NAFLD versus non‐NAFLD cirrhosis (255.6 ± 14.7 cm2 vs. 211.1 ± 9.8 cm2; P = 0.005). Although patients with cirrhosis have higher fat areas, this was not associated with an increase in total muscle area or increased bone density. Furthermore, there was decreased muscle and visceral fat attenuation (HU) in patients with NAFLD cirrhosis (33.5 ± 9.1 vs. 37.3 ± 9.3 HU; P = 0.003; −88.0 ± 10.8 vs. −84.7 ± 9.6 HU; P = 0.025), suggestive of lower muscle quality or myosteatosis.
Body Composition Is Predictive of Mortality in Patients With Cirrhosis
Recognizing the importance of body composition as an important determinant of mortality in patients with cirrhosis,( 16 ) we used our deep learning model to extract features from clinical CT scans to predict mortality. In the univariate Cox proportional hazards regression analysis, we found that there were many components of body composition that were significantly associated with mortality (Table 4). For example, muscle mass and muscle attenuation were significant determinants of mortality in patients with cirrhosis with a hazard ratio (HR) of 0.98 per cm2 of total skeletal muscle area (P < 0.0001) and remained significant when adjusted for height with a HR of 0.96 for skeletal muscle index (P = 0.002). Muscle attenuation as measured by skeletal muscle HU showed a HR of 0.95 with higher attenuation associated with greater survival (P < 0.0001). Increased visceral and subcutaneous fat areas were somewhat associated with improved survival (HR = 0.99 and 0.99; P = 0.02) and did not remain significant when adjusted by height. But interestingly, increased visceral and subcutaneous fat densities were significantly associated with increased mortality (HR = 1.03 and 1.04: P < 0.0001).
As body composition features differ by sex in cirrhosis, we further stratified our results by gender (Supporting Table S1). In the female‐only cohort, skeletal muscle area and index were not associated with survival. Skeletal muscle attenuation was somewhat associated with greater survival (HR = 0.99; P = 0.04). Increased visceral fat area was again associated with improved survival (HR = 0.99; P = 0.04), but subcutaneous fat area was not. Increased visceral and subcutaneous fat densities both showed a HR of 1.05, therefore again associated with increased mortality (P = 0.0005 and <0.0001).
To determine whether these body composition features can be used to improve the performance of the MELD score, we used backward and forward selection to develop the best predictive model (Table 5). A priori, five clinically relevant body Morphomic features were used as initial input variables: total muscle area index, visceral fat area and density, and subcutaneous fat area and density. The best model selected included MELD, total muscle area index, and subcutaneous fat density, producing c‐statistics of 0.71 (confidence interval [CI] = 0.61‐0.82) compared with 0.66 (CI = 0.55‐0.78) using MELD alone. The NRI score showed an enhanced predictability of mortality using the new model when compared with MELD alone (P < 0.0001). Using this model and the median score as the cutoff, high‐risk and low‐risk patients (with median survival of 2.93 years vs. 7.01 years, respectively) can be easily differentiated using Kaplan‐Meier analysis (P < 0.0001; Fig. 2).
FIG. 2.
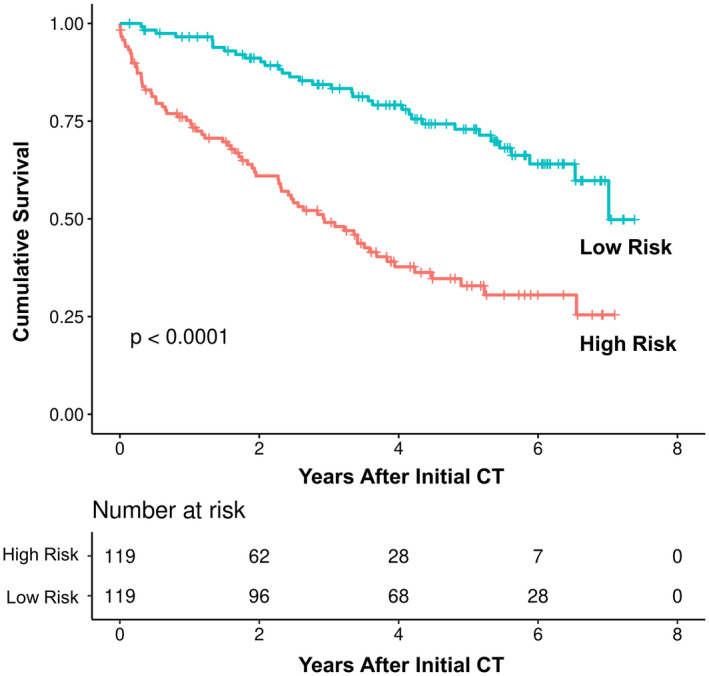
Kaplan‐Meier analysis comparing patients with high versus low risks in MELD + Morphomics model.
Discussion
CT scans are routinely used in clinical practice for diagnostic purposes, but within the image files are important measures of body composition including muscle, fat, and bone, which can provide valuable information regarding patient phenotypes. These body composition features are key predictive factors that affect clinical outcome and have significant value for clinical decision making.( 3 , 14 , 15 , 16 , 17 , 18 ) However, standard methods for measuring body composition include visual examination and time‐consuming manual delineation,( 19 ) which are not amenable for clinical implementation. In this study, we show that objective measures of body composition can be derived from incidental CT scans ordered for clinical purposes, and, more importantly, this process can be fully automated using deep‐learning methods. We demonstrate that our convolutional neural networks are able to accurately identify the key areas for measurement and show high correlation with manual measurements.
Using the automated measurements, we demonstrate how body composition features differ between those patients with cirrhosis with NAFLD as the primary diagnosis compared to those from other etiologies. Although BMI predictably was higher in patients with NAFLD, examination of the body composition showed that this increase in BMI was not proportionally reflected in all body components. Visceral and subcutaneous fat areas were significantly increased, but not muscle mass indexed to height or bone density. In fact, we saw a decreased bone density in those with NAFLD when compared to those with cirrhosis from other etiologies, although this decrease was not significant, likely secondary to our small cohort. These changes may reflect the pathophysiology of NAFLD and further compounded by the low attenuation of the muscle density in our cohort of patients with NALFD, which may be related to fat infiltration and/or myosteatosis.( 38 ) Having the ability to measure these features automatically and easily in CT scans that were performed for other purposes could have potential benefit for patients. One can envision that patients given this information could have tangible targets for behavior modifications and other interventions.
We also found that muscle mass remains a significant predictor of clinical outcome in patients with cirrhosis. In addition, muscle attenuation was predictive of survival with lower attenuation (likely lower quality muscle) and associated with worse survival. Interestingly, increased visceral and subcutaneous fat areas in this population were protective and likely reflect the overall nutritional status that has also been seen in other cohorts.( 39 ) Increased fat density, on the other hand, similar to our prior studies, was associated with worse survival.( 40 ) We hypothesize that this may represent early portal hypertension versus other changes with the fat compartments. An increase in water content within the fat compartments increases overall HU units, as water molecules within this compartment are denser than fat.( 40 ) Alternatively, a difference in the type of adipocytes could also change the density or HU within the fat compartment.( 41 ) Regardless of the cause, there is a clear association of higher fat HU with worse survival. This association of increased fat density and mortality has also been demonstrated in a recent study published on a cohort of patients with hepatocellular carcinoma.( 15 ) Compared with the entire cohort, increased muscle attenuation and visceral fat remain significantly associated with survival in a female‐only cohort. This again demonstrates the impact of the overall nutritional status on mortality in cirrhosis. The association of increased fat density on mortality remains striking in the female‐only cohort.
Given these findings, we sought to determine whether these body composition features derived from standard CT scans could provide incremental value to the commonly used MELD model, which relies on laboratory values alone. Within the electronic medical system, implementation of laboratory‐based values can easily be performed, but this does not preclude inclusion of other modalities such as imaging studies. Using features automatically extracted from clinical CT scans, we found that the addition of body composition features to MELD provided incremental discriminatory ability. Using artificial intelligence to automate the body composition measurements, we have demonstrated the first step for potential use of this technology in the electronic medical record system, allowing for a path to implementation in clinical care delivery.
It is notable that our cohort is derived from a general hepatology clinic, which excluded patients who have been referred for transplant. We think that work on this cohort, which is reflective of a nontransplant population with cirrhosis, is of significant value, as it represents results from a broader population perspective.
Although our training cohort was one of the largest examined, our study cohort was limited by the relatively small sample size within one academic health center. Future studies are necessary to validate the utility of our automated algorithm in a larger population.
The measurement of body composition can be fully automated using artificial intelligence, and it had excellent performance in a cohort of patients with cirrhosis. Incorporation of these automated measurements can add significant value for incidental CTs. This is proof of principal that this methodology could allow for wider implementation into the clinical arena, including the potential for mortality prediction in cirrhosis and behavioral modification in NAFLD.
Supporting information
Table S1
Potential conflict of interest: Dr. N. Wang is a founder of and owns equity in Prenovo, AMI, and EndoscopIQ. Dr. S. Wang is a founder of and owns equity in Applied Morphomics and Prenovo.
Supported by the Agency for Healthcare Research and Quality (R01 HS027183), National Cancer Institute (U01 CA230669), and US Department of Veteran Affairs (IIR 17‐269).
References
- 1. Smith M, Sattler A, Hong G, Lin S. From code to bedside: implementing artificial intelligence using quality improvement methods. J Gen Intern Med 2021;36:1061‐1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Waits SA, Kim EK, Terjimanian MN, Tishberg LM, Harbaugh CM, Sheetz KH, et al. Morphometric age and mortality after liver transplant. JAMA Surg 2014;149:335‐340. [DOI] [PubMed] [Google Scholar]
- 3. Sharma P, Parikh ND, Yu J, Barman P, Derstine BA, Sonnenday CJ, et al. Bone mineral density predicts posttransplant survival among hepatocellular carcinoma liver transplant recipients. Liver Transpl 2016;22:1092‐1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tapper EB, Derstine B, Baki J, Su GL. Bedside measures of frailty and cognitive function correlate with sarcopenia in patients with cirrhosis. Dig Dis Sci 2019;64:3652‐3659. [DOI] [PubMed] [Google Scholar]
- 5. Carey EJ, Lai JC, Sonnenday C, Tapper EB, Tandon P, Duarte‐Rojo A, et al. A North American expert opinion statement on sarcopenia in liver transplantation. Hepatology 2019;70:1816‐1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Therkelsen KE, Pedley A, Rosenquist KJ, Hoffmann U, Massaro JM, Murabito JM, et al. Adipose tissue attenuation as a marker of adipose tissue quality: associations with six‐year changes in body weight. Obesity 2016;24:499‐505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van der Poorten D, Milner K‐L, Hui J, Hodge A, Trenell MI, Kench JG, et al. Visceral fat: a key mediator of steatohepatitis in metabolic liver disease. Hepatology 2008;48:449‐457. [DOI] [PubMed] [Google Scholar]
- 8. Yu SJ, Kim W, Kim D, Yoon J‐H, Lee K, Kim JH, et al. Visceral obesity predicts significant fibrosis in patients with nonalcoholic fatty liver disease. Medicine (United States) 2015;94:e2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Linge J, Ekstedt M, Dahlqvist LO. Adverse muscle composition is linked to poor functional performance and metabolic comorbidities in NAFLD. JHEP Rep 2020;3:100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee Y‐H, Kim SU, Song K, Park JY, Kim DY, Ahn SH, et al. Sarcopenia is associated with significant liver fibrosis independently of obesity and insulin resistance in nonalcoholic fatty liver disease: nationwide surveys (KNHANES 2008‐2011). Hepatology 2016;63:776‐786. [DOI] [PubMed] [Google Scholar]
- 11. Thandassery RB, Aldo Montano‐Loza DJ. Role of nutrition and muscle in cirrhosis. Curr Treatment Options Gastroenterol 1938;14:257‐273. [DOI] [PubMed] [Google Scholar]
- 12. Charlton MR, Pockros PJ, Harrison SA. Impact of obesity on treatment of chronic hepatitis C. Hepatology 2006;43:1177‐1186. [DOI] [PubMed] [Google Scholar]
- 13. Giusto M, Lattanzi B, Albanese C, Galtieri A, Farcomeni A, Giannelli V, et al. Sarcopenia in liver cirrhosis: the role of computed tomography scan for the assessment of muscle mass compared with dual‐energy X‐ray absorptiometry and anthropometry. Comparative Study. Eur J Gastroenterol Hepatol 2015;27:328‐334. [DOI] [PubMed] [Google Scholar]
- 14. van Vugt JLA, Alferink LJM, Buettner S, Gaspersz MP, Bot D, Darwish Murad S, et al. A model including sarcopenia surpasses the MELD score in predicting waiting list mortality in cirrhotic liver transplant candidates: a competing risk analysis in a national cohort. J Hepatol 2018;68:707‐714. [DOI] [PubMed] [Google Scholar]
- 15. Ebadi M, Tandon P, Moctezuma‐Velazquez C, Ghosh S, Baracos VE, Mazurak VC, et al. Low subcutaneous adiposity associates with higher mortality in female patients with cirrhosis. J Hepatol 2018;69:608‐616. [DOI] [PubMed] [Google Scholar]
- 16. Tapper EB, Zhang P, Garg R, Nault T, Leary K, Krishnamurthy V, et al. Body composition predicts mortality and decompensation in compensated cirrhosis patients: a prospective cohort study. JHEP Rep 2020;2:100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Engelke K, Museyko O, Wang L, Laredo JD. Quantitative analysis of skeletal muscle by computed tomography imaging—state of the art. J Orthopaedic Translation 2018;15:91‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Durand F, Buyse S, Francoz C, Laouénan C, Bruno O, Belghiti J, et al. Prognostic value of muscle atrophy in cirrhosis using psoas muscle thickness on computed tomography. J Hepatol 2014;60:1151‐1157. [DOI] [PubMed] [Google Scholar]
- 19. Wang NC, Zhang P, Tapper EB, Saini S, Wang SC, Su GL. Automated measurements of muscle mass using deep learning can predict clinical outcomes in patients with liver disease. Am J Gastroenterol 2020;115:1210‐1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Derstine BA, Holcombe SA, Goulson RL, Ross BE, Wang NC, Sullivan JA, et al. Quantifying sarcopenia reference values using lumbar and thoracic muscle areas in a healthy population. J Nutr Health Aging 2018;22:180‐185. [DOI] [PubMed] [Google Scholar]
- 21. Aberra FB, Essenmacher M, Fisher N, Volk ML. Quality improvement measures lead to higher surveillance rates for hepatocellular carcinoma in patients with cirrhosis. Dig Dis Sci 2013;58:1157‐1160. [DOI] [PubMed] [Google Scholar]
- 22. Harbaugh CM, Terjimanian MN, Lee JS, Alawieh AZ, Kowalsky DB, Tishberg LM, et al. Abdominal aortic calcification and surgical outcomes in patients with no known cardiovascular risk factors. Ann Surg 2013;257:774‐781. [DOI] [PubMed] [Google Scholar]
- 23. Zhang P, Parenteau C, Wang LU, Holcombe S, Kohoyda‐Inglis C, Sullivan J, et al. Prediction of thoracic injury severity in frontal impacts by selected anatomical morphomic variables through model‐averaged logistic regression approach. Accid Anal Prev 2013;60:172‐180. [DOI] [PubMed] [Google Scholar]
- 24. Huhdanpaa H, Douville C, Baum K, Krishnamurthy VN, Holcombe S, Enchakalody B, et al. Development of a quantitative method for the diagnosis of cirrhosis. Scand J Gastroenterol 2011;46:1468‐1477. [DOI] [PubMed] [Google Scholar]
- 25. Englesbe MJ, Lee JS, He K, Fan L, Schaubel DE, Sheetz KH, et al. Analytic morphomics, core muscle size, and surgical outcomes. Ann Surg 2012;256:255‐261. [DOI] [PubMed] [Google Scholar]
- 26. Chollet F. Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, Hawaii, 2017. pp 1251‐1258. [Google Scholar]
- 27. Chen L‐C, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder‐decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of the European Conference on Computer Vision, Munich, Germany, 2018. [Google Scholar]
- 28. Csurka G, Larlus D, Perronnin F. What is a good evaluation measure for semantic segmentation? In: Proceedings of the British Machine Vision Conference, Bristol, United Kingdom; 2013. pp. 32.1‐32.11. [Google Scholar]
- 29. Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull 1979;86:420‐428. [DOI] [PubMed] [Google Scholar]
- 30. Venables WN, Ripley BD. Modern Applied Statistics with S‐Plus, Second Edition. Berlin, Germany: Springer; 1997. [Google Scholar]
- 31. Schmid M, Wright MN, Ziegler A. On the use of Harrell’s C for clinical risk prediction via random survival forests. Expert Syst App Int J 2016;63:450‐459. [Google Scholar]
- 32. Pencina MJ, D’Agostino RB, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med 2011;30:11‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Uno H, Tian L, Cai T, Kohane IS, Wei LJ. A unified inference procedure for a class of measures to assess improvement in risk prediction systems with survival data. Stat Med 2013;32:2430‐2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. MacHado MV, Diehl AM. Pathogenesis of nonalcoholic steatohepatitis. Gastroenterology 2016;150:1769‐1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yki‐Järvinen H. Non‐alcoholic fatty liver disease as a cause and a consequence of metabolic syndrome. Lancet Diabetes Endocrinol 2014;2:901‐910. [DOI] [PubMed] [Google Scholar]
- 36. Tilg H, Moschen AR, Roden M. NAFLD and diabetes mellitus. Nat Rev Gastroenterol Hepatol 2017;14:32‐42. [DOI] [PubMed] [Google Scholar]
- 37. Buzzetti E, Pinzani M, Tsochatzis EA. The multiple‐hit pathogenesis of non‐alcoholic fatty liver disease (NAFLD). Metab Clin Exp 2016;65:1038‐1048. [DOI] [PubMed] [Google Scholar]
- 38. Montano‐Loza AJ, Angulo P, Meza‐Junco J, Prado CMM, Sawyer MB, Beaumont C, et al. Sarcopenic obesity and myosteatosis are associated with higher mortality in patients with cirrhosis. J Cachexia Sarcopenia Muscle 2016;7:126‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Caan BJ, Feliciano EMC, Kroenke CH, Permanente K, California N. The importance of body composition in explaining the overweight paradox in cancer. AACR 2018;78:1906‐1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parikh ND, Zhang P, Singal AG, Derstine BA, Krishnamurthy V, Barman P, et al. Body composition predicts survival in patients with hepatocellular carcinoma treated with transarterial chemoembolization. Cancer Res Treatment 2018;50:530‐537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee JJ, Pedley A, Hoffmann U, Massaro JM, Keaney JF, Vasan RS, et al. Cross‐sectional associations of computed tomography (CT)‐derived adipose tissue density and adipokines: the Framingham heart study. J Am Heart Assoc 2015;5:e002545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1


