ABSTRACT
Conidiation is a pivotal strategy for fungi to resist adverse environments and disperse to new habitats, which is especially important for entomopathogenic fungi whose conidia are infective as fungal pesticide propagules. However, the molecular mechanism for regulating conidiation in entomopathogenic fungi is not fully understood. Here, we characterized the regulatory mechanism of the key developmental transcription factor Mr-AbaA. Bioinformatic analysis, transcriptional profiles, and subcellular localization of Mr-abaA indicated that AbaA functioned as a transcription factor in the conidiophore development and conidium stages. Microscopic examination showed that the null mutant of Mr-abaA differentiated into defective phialides to produce an abacus structure instead of conidia. Loss of Mr-abaA resulted in the inhibition of submerged blastospore separation in vitro. Moreover, yeast (Saccharomyces cerevisiae) one-hybrid assays of interactions between genes and deletion of Mr-veA showed that Mr-AbaA regulates conidiation by interacting with the promoter regions of Mr-veA and Mr-wetA. These results demonstrate that Mr-AbaA positively regulates conidiation in Metarhizium robertsii by regulating the velvet family ortholog gene Mr-veA and contributes to the separation of blastospores in submerged culture.
IMPORTANCE Metarhizium robertsii is an emerging model entomopathogenic fungus for developing biopesticides; therefore, a comprehensive understanding of its conidiation is very important for its application. In this study, we revealed that the transcription factor Mr-AbaA is involved in the control of aerial conidiation and blastospore separation in submerged culture. Further yeast one-hybrid assays demonstrated that Mr-AbaA interacts with the promoter regions of Mr-veA and Mr-wetA, which code for proteins involved in the control of conidiation. This finding provides new insight into the regulation of the conidiation of this important entomopathogenic fungi.
KEYWORDS: conidiation, Metarhizium robertsii, transcription factor, regulation mechanism
INTRODUCTION
Many species of entomopathogenic fungi play crucial roles in worldwide agroforestry pest management. As an emerging model of entomopathogenic fungi, Metarhizium spp. have been developed for biopesticides instead of chemical insecticides because of the absence of detrimental environmental effects and the ease of mass production (1). Conidia serve as the main units of environmental dispersal, invasion, and proliferation for Metarhizium robertsii. Moreover, conidia are the main components of fungal pesticides. However, the low yield of conidia and their sensitivity to environmental conditions have limited the large-scale application of M. robertsii (2). Understanding the molecular mechanisms controlling conidium production and increasing conidium yield and resistance to stress for M. robertsii by genetic manipulation are essential for commercial development.
Asexual sporulation is the most common reproductive strategy in filamentous fungi. Conidiogenesis is genetically programmed, and distinct gene sets are responsible for the progression of each phase (3). In the genetic regulation of asexual development in filamentous fungi, many researchers have extensively studied the model fungi Neurospora crassa and Aspergillus nidulans (4, 5). Many regulatory genes, including central regulators, negative regulators, upstream activators, velvet regulators, and light-responsive genes, are involved in conidiogenesis, but a central regulatory pathway comprising the three key regulators brlA, abaA, and wetA plays a crucial role in asexual development (6, 7). Overall, the key regulators abaA and wetA are well conserved among most filamentous fungi (6, 8, 9). The C2H2 zinc finger transcription factor brlA governs the initiation of conidiophore development and subsequently activates abaA during the middle stages of conidiophore development (10). Then, WetA, the expression of which is induced by AbaA in the late stage of conidiation, activates the expression of proteins or enzymes involved in the synthesis of conidium wall components, which is required for conidial maturation (11, 12).
The abaA gene encodes a developmental transcription factor with an ATTS/TEA DNA-binding domain that is required for the differentiation of phialides during the middle stages of A. nidulans conidiation (3, 13–15). In A. nidulans and Aspergillus fumigatus, loss of abaA resulted in the formation of abnormal metulae and phialides that produce long chains of cells that appear like beads on a string, as in an abacus (16, 17). Similarly, deletion of abaA in Talaromyces (formerly Penicillium) marneffei blocks asexual development and results in aberrant conidiophores with reiterated terminal cells (18). Similar phenotypes were seen in other filamentous fungi, such as Fusarium graminearum, Penicillium digitatum, and Beauveria bassiana (19–21). In addition, abaA was reported to govern dimorphic growth in T. marneffei and B. bassiana (18, 21). As mentioned above, wetA is activated by AbaA to complete conidiation. Aside from wetA expression, AbaA also positively regulates the transcript levels of two velvet family genes, velB and vosA, during conidiogenesis and directly binds to the promoter regions of those genes in A. nidulans (22). The velB and vosA genes not only are involved in asexual development and conidiogenesis but also play interdependent roles in trehalose biogenesis, conidial viability, and controlled conidial germination (23–25).
The process of asexual reproduction in M. robertsii is divided into the vegetative growth phase and the development phase. The formation of conidia takes place in the development phase and starts with the formation of conidiophores branching repeatedly at broad angles. Afterward, the tip of the conidiophore gives rise to clavate or cylindrical phialides in dense hymenia. Finally, repeated mitotic divisions occur in phialides to produce conidia in long chains. In M. robertsii, a conserved central regulatory pathway consisting of Mr-BrlA, Mr-AbaA, and Mr-WetA was identified; Mr-BlrA regulates Mr-abaA, which in turn activates Mr-wetA during conidiation (26). Deletion of Mr-brlA and Mr-abaA resulted in inhibition of conidium production, while deletion of Mr-wetA resulted in reduced conidial yields (26). Similar phenotypes were seen in B. bassiana. Loss of brlA or abaA resulted in inhibition of aerial conidiation, while knockout mutants of wetA and vosA lost most of their conidiation capacities (21, 27). Based on a previous framework from the study of M. robertsii conidiation, we primarily focused on the characterization and regulatory mechanism of the key developmental transcription factor Mr-AbaA.
RESULTS
Characteristics and deletion of Mr-abaA.
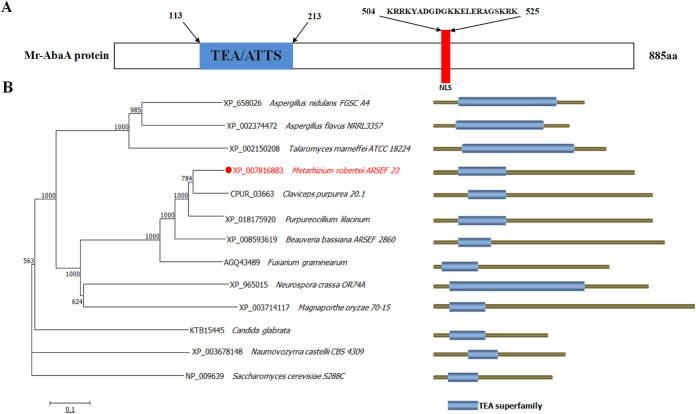
A previous analysis identified M. robertsii MAA-00694 (Mr-abaA) as a homolog of A. nidulans abaA by BLASTP (26). The open reading frame (ORF) of this gene consists of 2,658 nucleotides, contains two introns and three exons, and encodes a protein of 885 amino acids. Conserved functional domain analysis showed that it conserves an ATTS/TEA family domain (NCBI accession number pfam01285: https://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=pfam01285). A nuclear localization signal (NLS) motif was predicted in the C terminus of Mr-AbaA (residues 504 to 525) at NLStradamus (http://www.moseslab.csb.utoronto.ca/NLStradamus) (Fig. 1A). Sequence alignment analysis revealed a much higher sequence identity of Mr-abaA to the orthologs of Claviceps purpurea (82%) and Purpureocillium lilacinum (79%) than to the orthologs of these species. Further phylogenetic tree analysis indicated that Mr-abaA is also relatively closer to C. purpurea and P. lilacinum than to other fungi (Fig. 1B). Moreover, M. robertsii, C. purpurea, and P. lilacinum all belong to the family Clavicipitaceae.
FIG 1.
Bioinformatic analysis of Mr-abaA. (A) Structure domain analysis of the Mr-abaA protein. TEA, ATTS/TEA domain family. (B) Phylogenetic tree analysis of abaA orthologs from several fungi. The labels on the right display the NCBI accession numbers and the fungal species.
To assess the biological functions of Mr-abaA in M. robertsii, the targeted gene knockout vector (pDHt-SK-bar-Mr-abaA) was inserted into the wild-type (WT) strain to construct the ΔMr-abaA deletion mutant via agrobacterium-mediated homologous recombination. The confirmation of gene deletion by PCR and reverse transcription (RT)-PCR is presented in Fig. S1 in the supplemental material.
Transcriptional profiles and subcellular localization of Mr-AbaA.
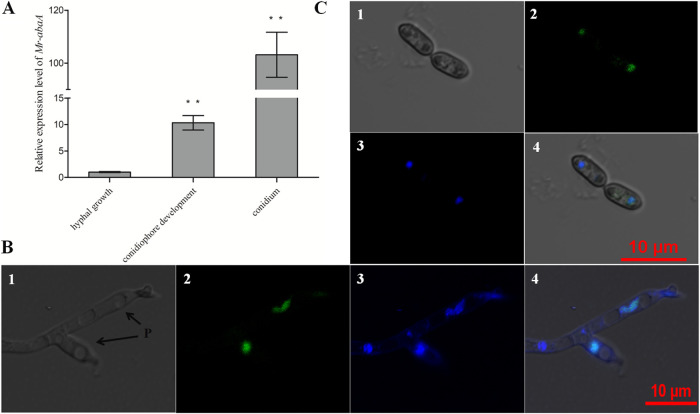
Transcriptional profiles of Mr-abaA were monitored in three different developmental stages, including hyphal growth, conidiophore development, and the conidium stage (Fig. 2A). Compared with the standard level in hyphal growth, the Mr-abaA transcript level was sharply increased in conidiophore development and the conidium stage (Tukey’s honestly significant difference [HSD] tests, P < 0.01 [n = 3]). Notably, a significant elevation to approximately 100-fold greater Mr-abaA transcript levels was detected in the conidium stage, compared with the hyphal growth stage. These data suggest that Mr-abaA may function in the conidium and conidiophore development stages.
FIG 2.
Transcriptional profiles and subcellular localization of Mr-AbaA. (A) Relative transcript levels of Mr-abaA in the WT cultures in three different developmental stages, compared with the standard level during hyphal growth. **, P < 0.01, Tukey’s HSD tests. (B and C) Subcellular localization of AbaA::GFP fusion protein expressed in the conidiophore development (B) and conidium (C) stages. Nuclei were stained with DAPI. Brightfield, expressed (green), DAPI-stained (blue), and merged views of the same field are numbered 1, 2, 3, and 4, respectively. P, phialides. Error bars in panel A indicate standard deviations of the means from three independent replicates.
The fungal cells in three different developmental stages were visualized for subcellular localization of enhanced green fluorescent protein (EGFP)-tagged Mr-AbaA fusion protein expressed in the WT strain, stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). The merged image of EGFP and DAPI staining showed that Mr-AbaA localized to the nucleus of phialides in the conidiophore development stage (Fig. 2B) and the nucleus of 10-day-old conidium (Fig. 2C). However, the green signal was not detected in the hyphal growth stage. Thus, this observation from subcellular localization analysis is consistent with the transcriptional profiles of Mr-abaA. In addition, these results imply the possibility that Mr-abaA acts as a transcription factor that functions in conidium and conidiophore development stages.
Mr-abaA is indispensable for aerial conidiation but does not affect hyphal growth.
For radial growth, ΔMr-abaA showed similar colony sizes in potato-dextrose agar (PDA), Sabouraud dextrose agar with yeast (SDAY) medium, and one-quarter-strength SDAY (1/4SDAY) medium, compared with the WT strain. These data indicated that colony growth was not affected by Mr-abaA deletion (see Fig. S2A).
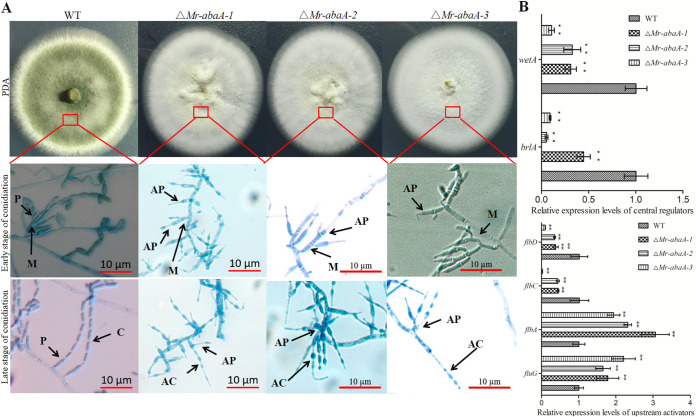
Deletion of Mr-abaA resulted in inhibition of aerial conidiation, and we were not able to obtain a Mr-abaA complementation strain using conidia as recipients. Therefore, we present assay data from three independent mutants. The gene disruption mutants and the WT strain were cultivated on different media, and their phenotypes were observed and compared. WT colonies were initially white, usually became yellow during the early development of conidia, and then became greenish as the conidia matured on PDA and 1/4 SDAY medium. However, the colony pigmentation of the ΔMr-abaA mutant was always white (Fig. 3A). Microscopically, the abacus aberrant conidia from the ΔMr-abaA mutant are responsible for the changed colony color, compared with the WT strain. Microscopic observation showed that the ΔMr-abaA mutant produces morphologically WT metulae. However, its scattered phialides produce short abacus aberrant conidia rather than cylindrical conidia (Fig. 3A). Thus, the loss of Mr-abaA interrupted the differentiation of phialides.
FIG 3.
Phenotypic analysis of the WT and ΔMr-abaA strains. (A) Three independent ΔMr-abaA strains showed a change in colony color. To observe the conidiophores on the aerial hyphae, WT and ΔMr-abaA cells were grown on PDA plates and sampled at 2.5 dpi (early stage of conidiation) and 10 dpi (late stage of conidiation). WT conidiophores had metulae (M), phialides (P), and conidia (C), whereas ΔMr-abaA conidiophores had metulae (M), abnormal phialides (AP), and abacus aberrant conidia (AC). (B) qRT-PCR analysis of the expression levels of conidiation-related genes among WT and null mutant clones. **, P < 0.01, Tukey’s HSD tests. Error bars in panel B indicate standard deviations of the means from three independent replicates.
Mr-abaA was indispensable for completing conidiation under aerial conditions. To investigate the functions of Mr-abaA in conidiation, the expression levels of conidiation-related genes in filamentous fungi were determined by quantitative RT-PCR (qRT-PCR). The results showed that the relative expression levels of genes such as upstream activators flbC and flbD or central regulators brlA and wetA were significantly downregulated, while the relative expression of others such as fluG and flbA were significantly upregulated in the ΔMr-abaA mutant, compared with the WT strain (Tukey’s HSD tests, P < 0.01 [n = 3]) (Fig. 3B) (6, 28–31). Thus, deletion of Mr-abaA resulted in significantly altered expression levels for the conidiation-related genes analyzed.
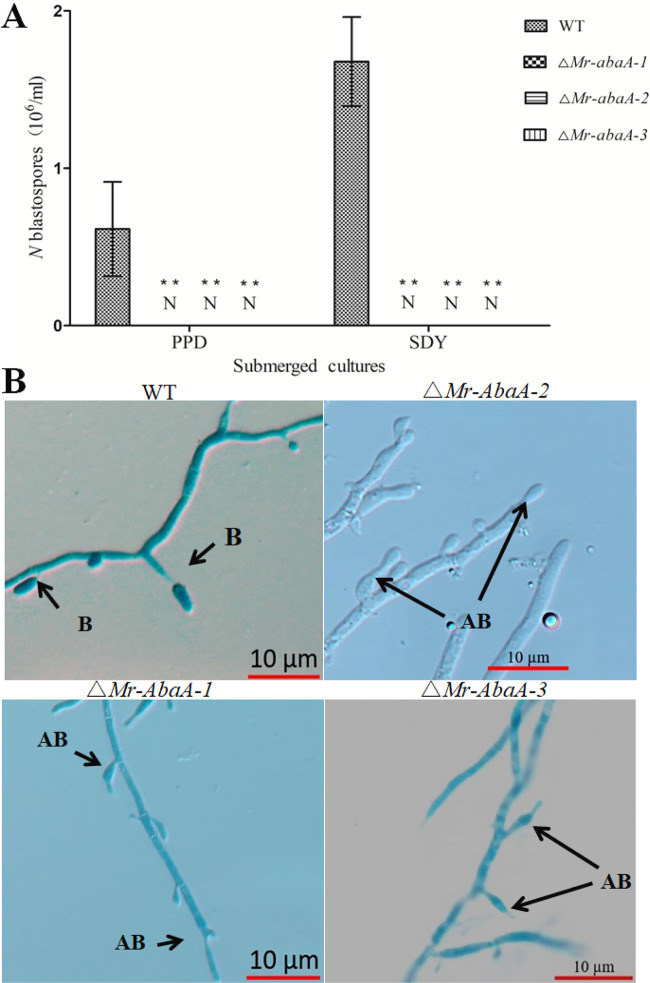
Knockout of Mr-abaA resulted in complete interruption of blastospore separation in submerged cultures. After 3 days of culture in Sabouraud dextrose broth (SDB) and potato-peptone-dextrose (PPD) broths, the WT strain generated 0.61(±0.3; n = 6) × 106 spores/ml and 1.68(±0.28; n = 6) × 106 spores/ml, respectively (Fig. 4A). However, microscopic examination demonstrated that the ΔMr-abaA mutant generated normal conidiogenous cells but blastospores were tightly connected to conidiogenous cells and did not separate from those in SDB (Fig. 4B).
FIG 4.
Indispensable roles of Mr-abaA in blastospore separation. (A) Blastospore yields were quantified from 4-day-old submerged cultures of WT and ΔMr-abaA strains in SDB medium and PPD medium, respectively. No detectable (N) blastospores were observed for the ΔMr-abaA strain. **, P < 0.01, Tukey’s HSD tests. (B) In submerged broth in vitro, the WT strain formed blastospores (B), while the ΔMr-abaA strain generated abnormal blastospores (AB). Error bars in panel A indicate standard deviations of the means from three independent replicates.
Mr-abaA is important for heat tolerance.
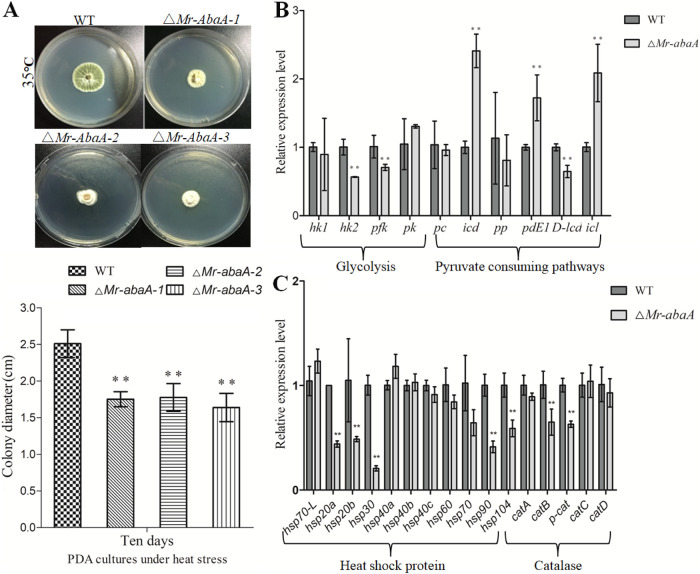
To further investigate the role of Mr-abaA in heat tolerance, the growth of WT and mutant colonies was analyzed under heat stress because of the absence of conidia in the ΔMr-abaA mutant. Intriguingly, compared with the WT strain, the mean colony diameters of three ΔMr-abaA isolates were reduced by 30% ± 3%, 29% ± 3%, and 35% ± 4% under 35°C heat stress (Tukey’s HSD tests, P < 0.01 [n = 3]) (Fig. 5A). Therefore, the ΔMr-abaA mutant showed significantly increased sensitivity to heat stress. Furthermore, some key heat-stress-responsive genes were assessed for their transcript levels in the ΔMr-abaA mutant, relative to the WT strain (32, 33). qRT-PCR results indicated that the heat stress significantly downregulated 2 of 4 genes involved in the glycolytic pathway, 5 of 11 genes encoding heat shock proteins, and 2 of 5 catalase genes but remarkably upregulated 3 of 6 genes involved in the pyruvate-consuming pathway in the ΔMr-abaA mutant (Fig. 5B and 5C).
FIG 5.
Mr-abaA is involved in heat tolerance. (A) Colony diameters of the WT strain and three Mr-abaA mutant hyphae on PDA plates with heat stress (35°C). (B and C) qRT-PCR analysis of the expression levels of heat-stress-responsive genes between WT and ΔMr-abaA strains. **, P < 0.01, Tukey’s HSD tests. Error bars in panels A and B indicate standard deviations of the means from three independent replicates.
The rate of ΔMr-abaA growth inhibitions in the presence of H2O2, Congo red, and NaCl was not different from that of the WT strain (see Fig. S2B). These results suggested that Mr-abaA is important for hyphal heat tolerance but is not involved in fungal antioxidant capacity, cell wall integrity, or osmotic stress.
Mr-AbaA regulates Mr-veA expression by directly binding to its promoters.
The An-AbaA transcription factor recognizes specific DNA motifs called abaA-response elements (AREs), which are characterized by a CATTCY sequence (15). Additionally, it was reported that abaA interacts not only with the AREs in the wetA promoter region but also with AREs in the velB and vosA promoter regions (22). A previous study showed that MAA-05862 was named velvet family gene vosA and has already been knocked out in M. robertsii (26). In this study, a BLAST search was conducted to identify genes potentially coding for transcriptional regulators of the velvet family in the M. robertsii genome database, with the A. nidulans velvet family gene veA as a query. The results showed that four velvet orthologs were found in the fungal genome. Further phylogenetic analysis indicated that MAA-01811, MAA-00244, MAA-01976, and MAA-05862 were designated Mr-veA, Mr-velB, Mr-vosA, and Mr-velC, respectively (see Fig. S3).
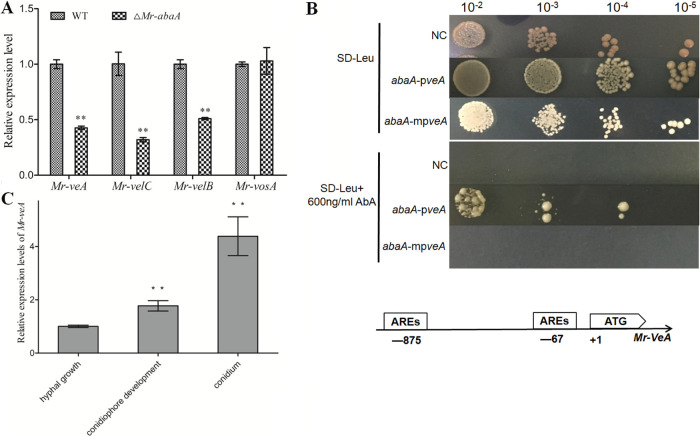
To investigate whether Mr-AbaA binds to the promoter region of velvet orthologs, bioinformatic analysis was performed, expression levels of these genes were determined, and yeast (Saccharomyces cerevisiae) one-hybrid analyses were carried out. First, the qRT-PCR analysis showed that the expression levels of Mr-veA, Mr-velB, and Mr-velC but not the expression of Mr-vosA were significantly reduced in the ΔMr-abaA mutant at the conidiophore development stage (Tukey’s HSD tests, P < 0.01 [n = 3]) (Fig. 6A). Thus, we selected Mr-veA, Mr-velB, and Mr-velC as candidates for yeast one-hybrid analysis. The positive clones showed that Mr-AbaA could physically bind to the promoter region of Mr-veA (Fig. 6B). However, recognition of the Mr-velC promoter region by endogenous yeast transcription factors resulted in unsuccessful yeast one-hybrid analysis, while Mr-AbaA could not interact with the Mr-velB promoter region (see Fig. S4). Further analysis of the promoter regions of Mr-veA, Mr-velB, and Mr-velC showed that only the Mr-veA promoter region contained two CATTCY AREs (Fig. 6B). Consistent with data from the qRT-PCR analysis and promoter sequence analysis, the yeast one-hybrid assay showed that Mr-AbaA can interact with the Mr-veA promoter region.
FIG 6.
Interaction of Mr-AbaA with the promoter region of Mr-veA. (A) qRT-PCR analysis of the expression levels of velvet family genes in the WT strain and the ΔMr-abaA strain. (B) Yeast one-hybrid assay to test the interactions of Mr-AbaA with the Mr-veA promoter regions. Yeast cells were transformed with both the pGADT7 AD vector containing the sequence of Mr-AbaA and plasmid pAbAi containing the Mr-veA promoter regions and mutated Mr-veA promoter regions. Transformed yeast cells were grown on SD-Leu medium with 600 ng/ml AbA, showing the interaction between the protein and the promoter region. NC, negative control; abaA-pveA, interaction between Mr-AbaA and the Mr-veA promoter region; abaA-mpveA, interaction between Mr-AbaA and the mutated Mr-veA promoter region; AbA, aureobasidin A. Putative ARE binding motifs are in the promoter regions of Mr-veA and Mr-wetA. One thousand-base pair portions of the Mr-veA and Mr-wetA promoter regions were analyzed. (C) Relative transcript levels of Mr-veA in the WT cultures in three different developmental stages. **, P < 0.01, Tukey’s HSD tests. Error bars in panels A and C indicate standard deviations of the means from three independent replicates.
Roles of Mr-veA in conidiation.
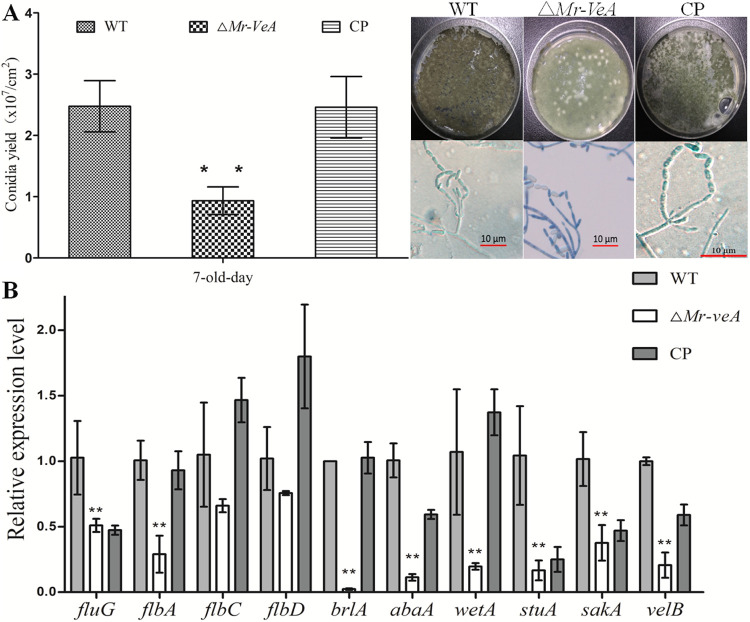
To verify the roles of Mr-veA in conidiation, the transcriptional profiles of Mr-veA were monitored in the hyphal growth, conidiophore development, and conidium stages. The results showed that the transcript level of Mr-veA was increased in the conidiophore development and conidium stages, compared with the hyphal growth stage (Fig. 6C). For further study, a null Mr-veA mutant was constructed (see Fig. S1C). Conidial yields from 7-day-old cultures of the WT strain, the ΔMr-veA mutant, and the complementation strain were quantified as 2.47(±0.42; n = 9) × 107 conidia/cm2, 0.93(±0.23; n = 9) × 107 conidia/cm2, and 2.46(±0.5; n = 9) × 107 conidia/cm2, respectively, indicating a remarkable 62% reduction of conidial yield in the absence of Mr-veA (Tukey’s HSD tests, P < 0.01 [n = 3]) (Fig. 7A). Nevertheless, microscopic examination showed that the null mutant of Mr-veA did not result in a distinctive defect in phialide formation during conidiation (Fig. 7A). Thus, deletion of Mr-veA repressed conidiation but did not alter the morphological pattern of asexual development, compared with the WT strain.
FIG 7.
Functional evaluation of Mr-veA in conidiation. (A) Conidial yield evaluated by culturing the WT, ΔMr-veA, and complementation (CP) strains on PDA plates at 7 dpi. Conidiophores were observed at the initial conidiation stage. (B) qRT-PCR analysis of the expression levels of conidiation-related genes in the WT, ΔMr-veA, and complementation strains. **, P < 0.01, Tukey’s HSD tests. Error bars in panels A and B indicate standard deviations of the means from three independent replicates.
To further study the roles of Mr-veA in conidiation, the expression levels of conidiation-related genes in filamentous fungi were assessed in the null Mr-veA mutant. The qRT-PCR results demonstrated that the relative expression levels of genes, including upstream activators flbG and flbA, central regulators brlA, abaA, and wetA, or other conidiation-related genes (stuA, sakA, and velB) were significantly downregulated in the ΔMr-veA mutant, compared with the WT strain and the complementation strain (Tukey’s HSD tests, P < 0.01 [n = 3]) (Fig. 7B).
Mr-AbaA regulates conidiation by interacting with the promoter regions of both Mr-veA and Mr-wetA.
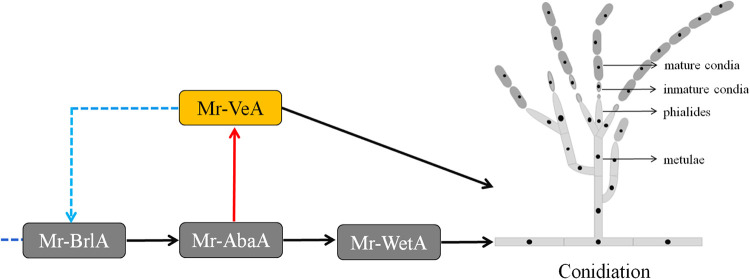
In our study, the data from the ARE search in the promoter region of Mr-wetA, the expression level of Mr-wetA in the ΔMr-abaA mutant, and the yeast one-hybrid assay results indicated that Mr-AbaA physically binds to the promoter region of Mr-wetA (see Fig. S5), which is the same as in a previous report (26). In combination with the data from analysis of the interaction of Mr-AbaA with the Mr-veA promoter region and conidial characteristics in the Mr-veA-deleted strain, we concluded that Mr-AbaA regulates conidiation by interacting with the promoter regions of both Mr-veA and Mr-wetA (Fig. 8).
FIG 8.
Putative regulatory model of the Mr-abaA-mediated regulation of conidiation in M. robertsii. Mr-AbaA positively regulated conidiation via Mr-wetA and Mr-veA, and the expression of Mr-abaA was activated by Mr-BrlA. The model shows hypothetical positive feedback control of conidiation involving Mr-AbaA, Mr-VeA, and Mr-BrlA. Solid arrows indicate positive regulation, and imaginary lines indicate uncharted regulation.
DISCUSSION
The differentiation of functional phialides is critical for the conidiogenesis of filamentous fungi. After undergoing a period of vegetative growth, M. robertsii develops into functional phialides to produce conidia. Our study showed that Mr-AbaA is localized to the nuclei of phialides and that deletion of Mr-abaA results in abnormal phialides that produce aberrant abacus conidia. The null ability of the ΔMr-abaA mutant to produce dark green conidia is responsible for the significant change in colony pigmentation. In another entomogenous fungus, B. bassiana, the ΔabaA mutant fails to generate clustered zigzag rachises (phialides) but cell clusters such as conidiation structures are infrequently present in old ΔabaA cultures (21). The abaA null mutant of A. nidulans forms aberrant conidiophores that fail to produce conidia (13). Similarly, deletion of abaA results in a defective phenotype similar to those of conidiophores in A. fumigatus, T. marneffei, P. digitatum, and F. graminearum (18–21). These findings suggest that abaA has conserved functions in the differentiation of conidiogenous structures in filamentous fungi, although the conidiation patterns in those fungi are different from each other.
A conidium is a pivotal unit for fungal survival, dispersal, and infection in the environment. The transcript level of Mr-abaA in mature conidia was significantly higher than that in other phases. Laser scanning confocal microscopy (LSCM) analysis also showed that Mr-AbaA is localized in the nucleus of mature conidia. Similar transcriptional profiles and subcellular localization of abaA were observed in F. graminearum (19). Therefore, the high level of expression of abaA in mature conidia indicated that abaA may play roles in conidial maturation in these filamentous fungi. However, the ΔMr-abaA mutant failed to generate normal conidia and, as a result, we were not able to study conidial maturation. Therefore, further studies that apply RNA interference technology to repress the transcript level of Mr-abaA in conidia will contribute to understanding the function of Mr-abaA in conidial maturation.
The regulatory pathway of abaA has been extensively studied in A. nidulans. An-AbaA directly activates not only wetA in the late stage of conidiation but also vosA and velB in conidial maturation (3, 22). A previous study also proved that Mr-BlrA can upregulate Mr-abaA, which in turn regulates Mr-wetA during conidiation in M. robertsii (26). Transcriptional profiles showed that the transcript level of Mr-wetA was increased in the conidiophore development and conidium stages, compared with the hyphal growth stage (see Fig. S6 in the supplemental material). However, our results indicated that Mr-abaA regulated conidiation via the velvet family gene Mr-veA. Compared with the hyphal growth stage, Mr-veA is highly expressed in the conidiophore development and conidium stages, compared with the hyphal growth stage. Deletion of Mr-veA also significantly repressed conidiation. Therefore, we found that a velvet family gene, veA, was directly activated by Mr-AbaA to regulate conidiation.
In this study, deletion of Mr-abaA resulted in significant downregulation of conidiation-related genes, including Mr-brlA and Mr-veA. We further found that deletion of Mr-veA also resulted in downregulation of Mr-brlA and Mr-abaA. In Aspergillus niger, it has been demonstrated that veA affects conidiation by regulating brlA expression levels (34). Therefore, there is a possibility of positive feedback control of conidiation on Mr-abaA mediated by Mr-veA and Mr-brlA. Unfortunately, Mr-VeA could not directly bind to the promoter regions of Mr-brlA, Mr-abaA, and Mr-wetA in our yeast one-hybrid assays (see Fig. S7). In Aspergillus flavus, VeA, VelB, and LaeA form a heterotrimeric complex, and FluG, which is a gene upstream of brlA, is probably an interacting partner of VelB (35). Therefore, VeA may generate a velvet complex interacting with FluG to regulate brlA expression, and the detailed mechanism by which Mr-VeA regulates Mr-brlA remains to be studied in future investigations.
The dimorphic transition between hypha and hyphal body (also called blastospore) forms is an important phenomenon in dimorphic fungi (18, 21, 36). The process of dimorphic transition has been well studied in M. robertsii; however, the molecular mechanism involved remains poorly understood. Previous studies showed that MAD1 is an adhesion protein whose mutant suppressed blastospore formation in M. robertsii (37). Our results show that Mr-abaA is indispensable for the separation of blastospores in submerged culture, but the deletion of Mr-abaA did not affect the expression level of MAD1 (see Fig. S8). T. marneffei with deletion of abaA fails to switch correctly from filamentous to yeast-like cells, and the B. bassiana ΔabaA mutant does not produce blastospores (18, 21). These results indicate that the AbaA function is conserved in these dimorphic fungi.
In conclusion, bioinformatic analysis and data on the transcriptional profiles and subcellular localization of Mr-AbaA indicated that AbaA functioned in the conidiophore development and conidium stages as a transcription factor. Microscopic examination showed that the Mr-abaA mutant differentiated into defective phialides to produce abacus structures instead of conidia. In addition, Mr-abaA is required for both aerial conidiation and submerged blastospore separation in vitro. Moreover, Mr-AbaA regulates conidiation by interacting with the promoter regions of Mr-veA and Mr-wetA. This finding provides new insight into the regulation of conidiation of this important entomopathogenic fungus.
MATERIALS AND METHODS
Strains and culture conditions.
The WT M. robertsii strain ARSEF 23 (ATCC number MYA-3075) was cultured on PDA (20% potato, 2% dextrose, and 2% agar [wt/vol]) in the dark at 25°C for 10 days to produce conidia. For liquid incubation, fungal strains were grown in PPD medium (20% potato, 2% dextrose, and 1% peptone [wt/vol]) and SDY medium (4% glucose, 1% peptone, and 1% yeast extract) at 25°C on a rotary shaker. The Y1H strain was used for yeast one-hybrid tests. Yeast cells were grown on yeast-peptone-dextrose agar (YPDA) (1% yeast extract, 2% peptone, 2% dextrose, adenine hemisulfate, and 1.5% agar), yeast-peptone-dextrose (YPD) medium (1% yeast extract, 2% peptone, and 2% dextrose), or synthetic dropout (SD) agar medium. Agrobacterium tumefaciens strain AGL-1 was cultured on solid yeast extract-beef (YEB) medium (0.5% sucrose, 1% tryptone, 0.1% yeast extract, 0.05% MgSO4·7H2O, and 1.5% agar [wt/vol]) at 28°C.
Transcriptional profiling of Mr-abaA and protein localization.
The WT strain was cultured on PDA for 10 days at 25°C in the dark and spread with 100-μl aliquots of a suspension of 107 conidia/ml. Total RNAs were extracted from samples that had been separately collected at time points of 36 h (hyphal growth), 72 h (conidiophore development), and 240 h (conidium stage) after inoculation using TRIzon reagent (Cwbio, Hefei, China). Then, RNA was reverse transcribed into cDNA using a ReverTra Ace qPCR RT master mix with genomic DNA (gDNA) remover kit (Toyobo, Japan). Three of the cDNA samples were used to assess the transcript levels of Mr-abaA via qRT-PCR with the CFX96 RT-PCR system (Bio-Rad, USA). The fungal glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used as a standard gene. The 2−ΔΔCT method was used to calculate the relative gene transcript levels (38).
To construct the AbaA::EGFP fusion protein, the full sequence of Mr-abaA with an upstream ∼1,000-bp fragment was amplified and cloned with the full sequence of EGFP into the pDHt-SK-bar vector, with which WT cells were transformed. Each transgenic strain was cultured on PDA for initial and full conidiation at 25°C in the dark. Mature conidia and hyphal cells were stained with the nucleus-specific dye DAPI and were then observed for subcellular localization under LSCM.
Phylogenetic analysis of abaA and veA in different fungi.
The Aspergillus nidulans FGSC A4 AbaA (GenBank accession number XP_658026) and VeA (GenBank accession number XP_658656.1) proteins were used as queries to search the M. robertsii genome available in the NCBI database via online BLASTP analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Amino acid sequences of abaA homologs in the genomic databases for M. robertsii, A. nidulans, A. flavus, Talaromyces marneffei, Claviceps purpurea, P. lilacinum, B. bassiana, F. graminearum, N. crassa, Magnaporthe oryzae, Candidaglabrata, Naumovozyma castellii, and Saccharomyces cerevisiae were downloaded from the NCBI database (http://ncbi.nlm.nih.gov). Phylogenetic analysis was conducted using MEGA6 software (http://www.megasoftware.net). The NLS motif of Mr-AbaA was predicted online at NLStradamus (http://www.moseslab.csb.utoronto.ca/NLStradamus).
Generation of Mr-abaA and Mr-veA mutants.
Deletions of Mr-abaA and Mr-veA were performed based on homologous recombination, as we described previously (39). Briefly, the 5′-flanking region (BamHI) and 3′-flanking region (XbaI) of the genes were amplified from gDNA by PCR and cloned onto the binary vector pDHt-SK-bar (conferring resistance to glufosinate ammonium) to construct the deletion mutant using Agrobacterium tumefaciens-mediated transformation. The integration event was verified by PCR and RT-PCR. The primers used in this study are listed in Table S1 in the supplemental material.
Mutant phenotype assays.
For the growth assay, hyphal blocks (4-mm diameter) were obtained from the WT and ΔMr-abaA strains grown on cellophane-overlaid SDAY medium for 4 days and attached centrally to PDA, SDAY, and 1/4 SDAY (amended with one-quarter of the nutrients of SDAY) plates. At 10 days postinoculation (dpi), all colony diameters were measured as indices of radial growth rates, using the cross-crossing method (40).
For chemical stress tolerance assays, hyphal blocks (4-mm diameter) of WT and three ΔMr-abaA strains were attached in the center of PDA plates with supplementary chemical reagents, including the cell wall-disturbing compound Congo red (2 mg/ml), H2O2 (5 mM) as an inducer of oxidative stress, and NaCl (0.5 M) as an inducer of osmotic stress. To investigate the fungal hypha responses to heat stress, hyphal blocks (4-mm diameter) were attached centrally to PDA plates and cultured at 35°C for 10 days, and all colony diameters were measured (32). The rate of growth inhibition (RGI) was calculated as (C − S)/C × 100, where C is the growth rate of the control and S is the growth rate under stress conditions (39).
To assay the conidiation capacity of the WT strain and each mutant, 30 μl of a suspension of 106 conidia/ml was evenly spread on PDA plates (6-mm diameter) and cultured in the dark at 25°C for 7 and 14 days. On 7 and 14 dpi, fresh conidia from the WT strain and each mutant were separately collected into 30 ml of 0.05% Tween 80, and conidia were dispersed by vibration. The concentration of conidial suspensions was measured using a hemocytometer and then converted to the number of conidia produced per unit area (square centimeter) of plate culture. Because the ΔMr-abaA mutant failed to produce conidia, its conidiation capacity was completely lost. During the culture period, the sporulation states of each strain were observed under a microscope.
For qRT-PCR analysis, each strain was cultured on PDA plates for 2.5 days, and the hyphae were collected for total RNA extraction to conduct conidiation-related gene expression analysis. qRT-PCR analysis was performed using the qPCR SYBR green master mix (Vazyme, China). Primers for qRT-PCR are listed in Table S2 in the supplemental material.
Assessment of blastospore formation was performed as described previously (21). Briefly, submerged cultures of the ΔMr-abaA and WT strains were initiated with hyphal blocks (4-mm diameter) cultured in SDY broth. After 3 days of culture, collected hyphae were rinsed twice with sterile water and filtered through lens-cleaning tissues to remove resuspended blastospores. All of the aliquots in flasks were standardized to a final concentration of fresh hyphal mass of 1 mg/ml and were incubated in SDY broth and PPD broth for 4 days with shaking (130 rpm). The blastospore concentration was assessed from each sample using a hemocytometer and was used to compute the absolute blastospore yield (number of blastospores per milliliter) in each submerged culture.
The aforementioned data from the experiments with three replicates were subjected to one-way analysis of variance, followed by Tukey’s HSD test for phenotypic changes among the tested fungal strains.
Yeast one-hybrid assay.
The Mr-veA target promoter region with an ∼615-bp DNA fragment, the mutated AREs (CttaCC) in the Mr-veA target promoter region with an ∼615-bp DNA fragment, and the Mr-wetA target promoter region with an ∼645-bp DNA fragment were amplified and cloned into the linearized pAbAi vector (Clontech, USA). The plasmids (pAbAi-pveA, pAbAi-mpveA, and pAbAi-pwetA) were linearized and cloned into Saccharomyces cerevisiae Y1HGold cells (Clontech). Transformed strains were grown on SD-Ura agar medium. Subsequently, the Mr-AbaA coding region was amplified from cDNA and cloned into the linearized pGADT7-AD vector (Clontech). The recombinant plasmid pGADT7-abaA was further transformed into the Y1H+baitGold (pAbAi-pveA, pAbAi-mpveA, and pAbAi-pwetA) strain. The transformed cells were plated on an SD-Leu agar medium with 600 ng/ml aureobasidin A to identify the interactions of Mr-AbaA with the Mr-veA and Mr-wetA promoters. Y1HGold (pAbAi-pveA, pAbAi-mpveA, and pAbAi-pwetA plus pGADT7-AD) cells were used as a negative control.
ACKNOWLEDGMENTS
This work was supported by the National Key R&D Program of China (grant 2017YFD0200400) and the National Science Foundation of China (grants 31772226, 31471821, and 31272096).
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Ting Ding, Email: dingting98@126.com.
Bo Huang, Email: bhuang@ahau.edu.cn.
Giuseppe Ianiri, University of Molise.
REFERENCES
- 1.Lacey LA, Grzywacz D, Shapiro-Ilan DI, Frutos R, Brownbridge M, Goettel MS. 2015. Insect pathogens as biological control agents: back to the future. J Invertebr Pathol 132:1–41. doi: 10.1016/j.jip.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Wang C, Wang S. 2017. Insect pathogenic fungi: genomics, molecular interactions, and genetic improvements. Annu Rev Entomol 62:73–90. doi: 10.1146/annurev-ento-031616-035509. [DOI] [PubMed] [Google Scholar]
- 3.Park HS, Yu JH. 2012. Genetic control of asexual sporulation in filamentous fungi. Curr Opin Microbiol 15:669–677. doi: 10.1016/j.mib.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Ruger-Herreros C, Corrochano LM. 2020. Conidiation in Neurospora crassa: vegetative reproduction by a model fungus. Int Microbiol 23:97–105. doi: 10.1007/s10123-019-00085-1. [DOI] [PubMed] [Google Scholar]
- 5.Ojeda-López M, Chen W, Eagle CE, Gutiérrez G, Jia WL, Swilaiman SS, Huang Z, Park HS, Yu JH, Cánovas D, Dyer PS. 2018. Evolution of asexual and sexual reproduction in the aspergilli. Stud Mycol 91:37–59. doi: 10.1016/j.simyco.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vries RP, Riley R, Wiebenga A, Aguilar-Osorio G, Amillis S, Uchima CA, Anderluh G, Asadollahi M, Askin M, Barry K, Battaglia E, Bayram Ö, Benocci T, Braus-Stromeyer SA, Caldana C, Cánovas D, Cerqueira GC, Chen F, Chen W, Choi C, Clum A, Dos Santos RAC, Damásio AR, Diallinas G, Emri T, Fekete E, Flipphi M, Freyberg S, Gallo A, Gournas C, Habgood R, Hainaut M, Harispe ML, Henrissat B, Hildén KS, Hope R, Hossain A, Karabika E, Karaffa L, Karányi Z, Kraševec N, Kuo A, Kusch H, LaButti K, Lagendijk EL, Lapidus A, Levasseur A, Lindquist E, Lipzen A, Logrieco AF, MacCabe A, Mäkelä MR, Malavazi I, Melin P, Meyer V, Mielnichuk N, Miskei M, Molnár ÁP, Mulé G, Ngan CY, Orejas M, Orosz E, Ouedraogo JP, Overkamp KM, Park HS, Perrone G, Piumi F, Punt PJ, Ram AF, Ramón A, Rauscher S, Record E, Riaño-Pachón DM, Robert V, Röhrig J, Ruller R, Salamov A, Salih NS, Samson RA, Sándor E, Sanguinetti M, Schütze T, Sepčić K, Shelest E, Sherlock G, Sophianopoulou V, Squina FM, Sun H, Susca A, Todd RB, Tsang A, Unkles SE, van de Wiele N, van Rossen-Uffink D, Oliveira JV, Vesth TC, Visser J, Yu JH, Zhou M, Andersen MR, Archer DB, Baker SE, Benoit I, Brakhage AA, Braus GH, Fischer R, Frisvad JC, Goldman GH, Houbraken J, Oakley B, Pócsi I, Scazzocchio C, Seiboth B, vanKuyk PA, Wortman J, Dyer PS, Grigoriev IV. 2017. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol 18:28. doi: 10.1186/s13059-017-1151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alkhayyat F, Chang Kim S, Yu JH. 2015. Genetic control of asexual development in Aspergillus fumigatus. Adv Appl Microbiol 90:93–107. doi: 10.1016/bs.aambs.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Etxebeste O, Otamendi A, Garzia A, Espeso EA, Cortese MS. 2019. Rewiring of transcriptional networks as a major event leading to the diversity of asexual multicellularity in fungi. Crit Rev Microbiol 45:548–563. doi: 10.1080/1040841X.2019.1630359. [DOI] [PubMed] [Google Scholar]
- 9.Mead ME, Borowsky AT, Joehnk B, Steenwyk JL, Shen XX, Sil A, Rokas A. 2020. Recurrent loss of abaA, a master regulator of asexual development in filamentous fungi, correlates with changes in genomic and morphological traits. Genome Biol Evol 12:1119–1130. doi: 10.1093/gbe/evaa107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams TH, Boylan MT, Timberlake WE. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353–362. doi: 10.1016/0092-8674(88)90198-5. [DOI] [PubMed] [Google Scholar]
- 11.Sewall TC, Mims CW, Timberlake WE. 1990. Conidium differentiation in Aspergillus nidulans wild-type and wet-white (wetA) mutant strains. Dev Biol 138:499–508. doi: 10.1016/0012-1606(90)90215-5. [DOI] [PubMed] [Google Scholar]
- 12.Marshall MA, Timberlake WE. 1991. Aspergillus nidulans wetA activates spore-specific gene expression. Mol Cell Biol 11:55–62. doi: 10.1128/mcb.11.1.55-62.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams TH, Wieser JK, Yu JH. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol Mol Biol Rev 62:35–54. doi: 10.1128/MMBR.62.1.35-54.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boylan MT, Mirabito PM, Willett CE, Zimmerman CR, Timberlake WE. 1987. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol Cell Biol 7:3113–3118. doi: 10.1128/mcb.7.9.3113-3118.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrianopoulos A, Timberlake WE. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol Cell Biol 14:2503–2515. doi: 10.1128/mcb.14.4.2503-2515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sewall TC, Mims CW, Timberlake WE. 1990. abaA controls phialide differentiation in Aspergillus nidulans. Plant Cell 2:731–739. doi: 10.1105/tpc.2.8.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao L, Yu JH. 2011. AbaA and WetA govern distinct stages of Aspergillus fumigatus development. Microbiology (Reading) 157:313–326. doi: 10.1099/mic.0.044271-0. [DOI] [PubMed] [Google Scholar]
- 18.Borneman AR, Hynes MJ, Andrianopoulos A. 2000. The abaA homologue of Penicillium marneffei participates in two developmental programmes: conidiation and dimorphic growth. Mol Microbiol 38:1034–1047. doi: 10.1046/j.1365-2958.2000.02202.x. [DOI] [PubMed] [Google Scholar]
- 19.Son H, Kim MG, Min K, Seo YS, Lim JY, Choi GJ, Kim JC, Chae SK, Lee YW. 2013. AbaA regulates conidiogenesis in the ascomycete fungus Fusarium graminearum. PLoS One 8:e72915. doi: 10.1371/journal.pone.0072915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang M, Sun X, Zhu C, Xu Q, Ruan R, Yu D, Li H. 2015. PdbrlA, PdabaA and PdwetA control distinct stages of conidiogenesis in Penicillium digitatum. Res Microbiol 166:56–65. doi: 10.1016/j.resmic.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Zhang AX, Mouhoumed AZ, Tong SM, Ying SH, Feng MG. 2019. BrlA and AbaA govern virulence-required dimorphic switch, conidiation, and pathogenicity in a fungal insect pathogen. mSystems 4:e00140-19. doi: 10.1128/mSystems.00140-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park HS, Ni M, Jeong KC, Kim YH, Yu JH. 2012. The role, interaction and regulation of the velvet regulator VelB in Aspergillus nidulans. PLoS One 7:e45935. doi: 10.1371/journal.pone.0045935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baltussen TJH, Zoll J, Verweij PE, Melchers WJG. 2020. Molecular mechanisms of conidial germination in Aspergillus spp. Microbiol Mol Biol Rev 84:e00049-19. doi: 10.1128/MMBR.00049-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park HS, Bayram O, Braus GH, Kim SC, Yu JH. 2012. Characterization of the velvet regulators in Aspergillus fumigatus. Mol Microbiol 86:937–953. doi: 10.1111/mmi.12032. [DOI] [PubMed] [Google Scholar]
- 25.Park H-S, Yu J-H. 2016. Velvet regulators in Aspergillus spp. Microbiol Biotechnol Lett 44:409–419. doi: 10.4014/mbl.1607.07007. [DOI] [Google Scholar]
- 26.Zeng G, Chen X, Zhang X, Zhang Q, Xu C, Mi W, Guo N, Zhao H, You Y, Dryburgh FJ, Bidochka MJ, St Leger RJ, Zhang L, Fang W. 2017. Genome-wide identification of pathogenicity, conidiation and colony sectorization genes in Metarhizium robertsii. Environ Microbiol 19:3896–3908. doi: 10.1111/1462-2920.13777. [DOI] [PubMed] [Google Scholar]
- 27.Li F, Shi HQ, Ying SH, Feng MG. 2015. WetA and VosA are distinct regulators of conidiation capacity, conidial quality, and biological control potential of a fungal insect pathogen. Appl Microbiol Biotechnol 99:10069–10081. doi: 10.1007/s00253-015-6823-7. [DOI] [PubMed] [Google Scholar]
- 28.Seo JA, Guan Y, Yu JH. 2006. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 172:1535–1544. doi: 10.1534/genetics.105.052258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu JH, Wieser J, Adams TH. 1996. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J 15:5184–5190. doi: 10.1002/j.1460-2075.1996.tb00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon NJ, Garzia A, Espeso EA, Ugalde U, Yu JH. 2010. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol Microbiol 77:1203–1219. doi: 10.1111/j.1365-2958.2010.07282.x. [DOI] [PubMed] [Google Scholar]
- 31.Arratia-Quijada J, Sánchez O, Scazzocchio C, Aguirre J. 2012. FlbD, a Myb transcription factor of Aspergillus nidulans, is uniquely involved in both asexual and sexual differentiation. Eukaryot Cell 11:1132–1142. doi: 10.1128/EC.00101-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, St Leger RJ, Fang W. 2017. Pyruvate accumulation is the first line of cell defense against heat stress in a fungus. mBio 8:e01284-17. doi: 10.1128/mBio.01284-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Zhu H, Cheng Y, Jiang Y, Li Y, Huang B. 2019. The polyubiquitin gene MrUBI4 is required for conidiation, conidial germination, and stress tolerance in the filamentous fungus Metarhizium robertsii. Genes 10:412. doi: 10.3390/genes10060412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Chen H, Sumarah MW, Gao Q, Wang D, Zhang Y. 2018. veA gene acts as a positive regulator of conidia production, ochratoxin A biosynthesis, and oxidative stress tolerance in Aspergillus niger. J Agric Food Chem 66:13199–13208. doi: 10.1021/acs.jafc.8b04523. [DOI] [PubMed] [Google Scholar]
- 35.Chang PK, Scharfenstein LL, Li P, Ehrlich KC. 2013. Aspergillus flavus VelB acts distinctly from VeA in conidiation and may coordinate with FluG to modulate sclerotial production. Fungal Genet Biol 58–59:71–79. doi: 10.1016/j.fgb.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Gao Q, Jin K, Ying SH, Zhang Y, Xiao G, Shang Y, Duan Z, Hu X, Xie XQ, Zhou G, Peng G, Luo Z, Huang W, Wang B, Fang W, Wang S, Zhong Y, Ma LJ, St Leger RJ, Zhao GP, Pei Y, Feng MG, Xia Y, Wang C. 2011. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet 7:e1001264. doi: 10.1371/journal.pgen.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, St Leger RJ. 2007. The MAD1 adhesin of Metarhizium anisopliae links adhesion with blastospore production and virulence to insects, and the MAD2 adhesin enables attachment to plants. Eukaryot Cell 6:808–816. doi: 10.1128/EC.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Wang T, Qiao L, Zhu J, Fan J, Zhang T, Wang ZX, Li W, Chen A, Huang B. 2017. DNA methyltransferases contribute to the fungal development, stress tolerance and virulence of the entomopathogenic fungus Metarhizium robertsii. Appl Microbiol Biotechnol 101:4215–4226. doi: 10.1007/s00253-017-8197-5. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Chen J, Hu Y, Ying SH, Feng MG. 2020. Roles of six Hsp70 genes in virulence, cell wall integrity, antioxidant activity and multiple stress tolerance of Beauveria bassiana. Fungal Genet Biol 144:103437. doi: 10.1016/j.fgb.2020.103437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00823-21_Supp_1_seq12.pdf, PDF file, 0.7 MB (689.7KB, pdf) .