Abstract
Melanin is a dark color pigment biosynthesized naturally in most living organisms. Fungal melanin is a major putative virulence factor of Mucorales fungi that allows intracellular persistence by inducing phagosome maturation arrest. Recently, it has been shown that the black pigments of Rhizopus delemar is of eumelanin type, that requires the involvement of tyrosinase (a copper-dependent enzyme) in its biosynthesis. Herein, we have developed a series of compounds (UOSC-1–14) to selectively target Rhizopus melanin and explored this mechanism therapeutically. The compounds were designed based on the scaffold of the natural product, cuminaldehyde, identified from plant sources and has been shown to develop non-selective inhibition of melanin production. While all synthesized compounds showed significant inhibition of Rhizopus melanin production and limited toxicity to mammalian cells, only four compounds (UOSC-1, 2, 13, and 14) were selected as promising candidates based on their selective inhibition to fungal melanin. The activity of compound UOSC-2 was comparable to the positive control kojic acid. The selected candidates showed significant inhibition of Rhizopus melanin but not human melanin by targeting the fungal tyrosinase, and with an IC50 that are 9 times lower than the reference standard, kojic acid. Furthermore, the produced white spores were phagocytized easily and cleared faster from the lungs of infected immunocompetent mice and from the human macrophages when compared with wild-type spores. Collectively, the results suggested that the newly designed derivatives, particularly UOSC-2 can serve as promising candidate to overcome persistence mechanisms of fungal melanin production and hence make them accessible to host defenses.
Introduction
Melanin is predominantly dark-colored polymer widely distributed in bacteria, fungi, plants, and animals [1]. Several biological functions of melanin have been reported including its role as free radical scavenger, as cation-binding material, and as protection from UV radiation [1]. In fungi, melanin is more likely correlated with spore formation, virulence of pathogenic fungi, and evasion from host defense mechanisms or stressful environmental conditions [2,3]. In mammals, melanin is produced in specialized pigment-producing cells known as melanocytes [4]. In mammals, melanin pigments play several diverse and important roles, including thermoregulation, camouflage, and sexual attraction [5].
It has been reported that melanin can provide the pathogenic fungi including Rhizopus delemar with a protective shield from host defensive mechanisms and hence allow their persistence in the human body through antigen mimicry mechanism [6]. The presence of pathogenic fungi in the human body in a dormant state facilitates their pathogenesis once the conditions are appropriate.
Melanin can be classified into two major classes, eumelanins, and pheomelanins. R. delemar melanin has been identified as of eumelanin type [6]. Eumelanin is a black/brown colored cross-linked polymer of the monomer 5,6-dihydroxyindole (DHI) and 5, 6-dihydroxyindole-2-carboxylic acid (DHICA). The rate-limiting step in eumelanin biosynthesis is the enzymatic oxidation of tyrosine or l-3,4-dihydroxyphenylalanine (l-DOPA) to its corresponding O-dopaquinone [7]. It is the only step in eumelanin biosynthesis that is controlled by an enzyme, named tyrosinase [8,9]. Although the crystal structures of most tyrosinases are similar, there are key differences among tyrosinases from different sources. For instance, both fungal and human tyrosinases showed key differences [10–12]. Specifically, the mushroom tyrosinase is a cytosolic enzyme while the human tyrosinase is a membrane bound [13]. Furthermore, mushroom tyrosinase is a tetramer contains binuclear copper-binding site which is located in the bottom of the active pocket. Each copper ion co-ordinated by three histidine residues. The ligands of the first Cu-A are His 61, His85, and His94, while the second Cu-B surrounded with His259, His263, and His296 [14]. In contrast, the human tyrosinase is a monomer that is highly glycosylated during its complex maturation process. Human tyrosinase contains binuclear zinc site instead of the copper ions in the case of mushroom tyrosinase. Zn-A is co-ordinated with His192, His215, and His224, while Zn-B is co-ordinated with his377, His404, and His381 [15].
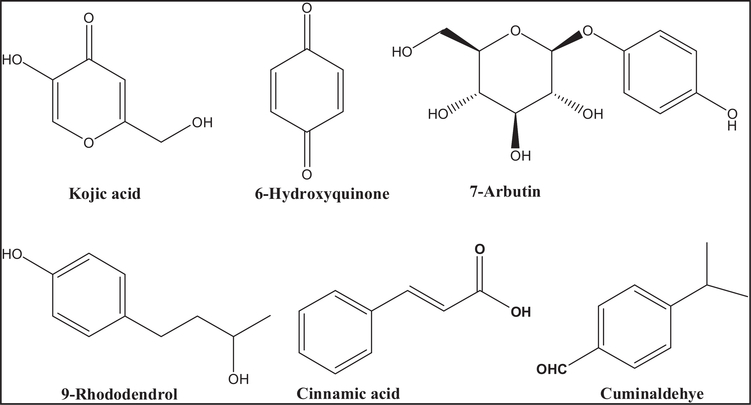
Numerous compounds were identified as tyrosinase inhibitor from natural and synthetic sources, such as kojic acid, hydroquinone, arbutin, 4-methoxycinnamic acid, and rhododendrol (Figure 1). However, they show instability and undesirable side effects including cytotoxicity, dermatitis, skin cancer, and neurodegenerative disorders because of interaction with human cells [16–18]. Furthermore, most identified inhibitors lack clinical efficacy since they were evaluated using mushroom tyrosinase as a target. On the other hand, despite the potent fungal tyrosinase inhibition activities of the naturally isolated aldehyde, cuminaldehyde (Figure 1), it shows significant toxic activities [19,20].
Figure 1.
Structures of synthetic and natural compounds show the ability to inhibit melanin production from variable organisms.
The aim of this research study is to design and synthesis safe and selective compounds with promising inhibitory activity on fungal melanin biosynthesis in particular R. delemar which is the main cause of the lethal infection, mucormycosis [6]. To achieve this goal, cuminaldehyde was employed as a lead structure to develop optimized inhibitors for fungal melanin biosynthesis with high activity and enhanced physicochemical properties and limited toxicity.
Materials and methods
General chemistry procedures
Most chemicals and solvents were of analytical grade and, when necessary, were purified and dried by standard methods. Reactions were monitored by thin-layer chromatography (TLC) using pre-coated silica gel plates (kiesel gel 60 F254, BDH), and spots were visualized under UV light (254 nm). Melting points were determined using a Gallenkamp melting point apparatus and are uncorrected. Column chromatography was performed with Merck silica gel 60 (40–60 μM). 1H-NMR and 13C-NMR spectra were recorded on a Bruker spectrometer at 500 MHz. Chemical shifts were expressed in parts per million (ppm) and coupling constant J values were represented in Hz. Mass spectroscopic data were obtained through electrospray ionization (ESI) mass spectrum. Detailed synthesis and spectroscopic data of UOSC-1–14 are described in Supplementary Information.
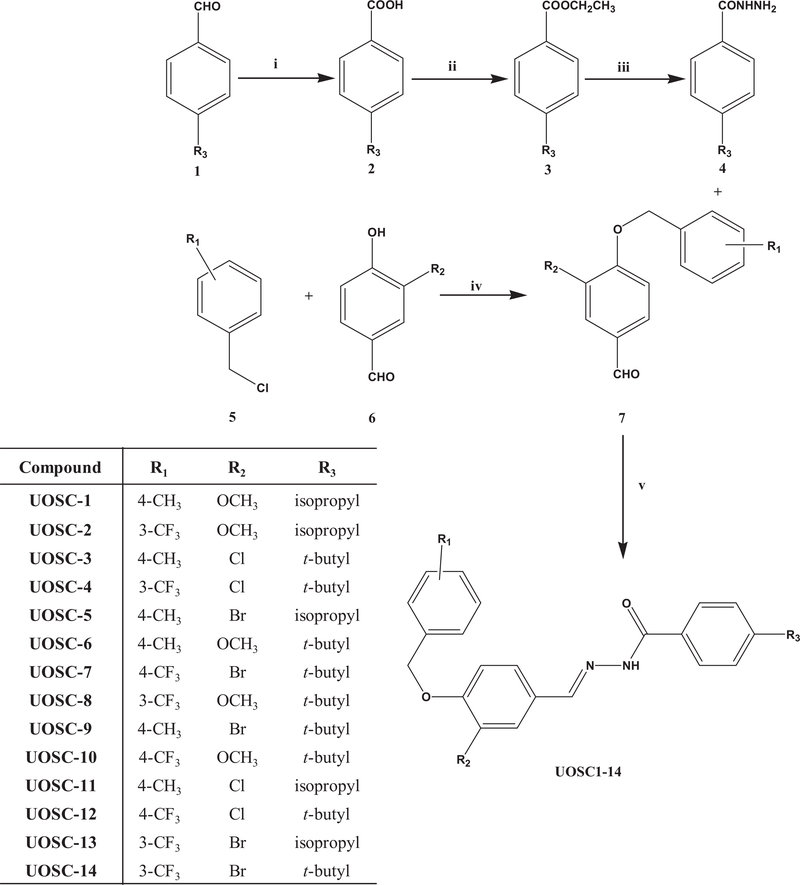
Cuminic acid was prepared from oxidation of cuminaldehyde following a reported procedure [21]. For the synthesis of ethyl 4-substituted benzoate (3), a mixture of appropriate 4-substituted benzoic acid 2 (10 mmol), absolute ethanol (20 ml), and concentrated sulfuric acid (2 ml) was refluxed for 3 h. Excess ethanol was distilled under reduced pressure and the resulting oil was rendered alkaline with aqueous sodium bicarbonate and extracted with methylene chloride (2 × 50 ml). The combined organic extract was dried over anhydrous sodium sulfate and distilled under reduced pressure to give the corresponding ester in a yield of 68%. For the synthesis of 4-substituted benzoic acid hydrazide (4), to a solution of ethyl 4-substituted benzoate (3) (10 mmol) in ethanol (10 ml), excess hydrazine monohydrate (5 ml) was added. The reaction mixture was refluxed for 24 h, and left to cool to room temperature. The formed precipitate was collected by filtration, washed with water followed by cold ethanol to remove excess hydrazine and left to dry to give the corresponding acid hydrazide. For the synthesis of 3-substituted-4-(4 substituted-benzyloxy) benzaldehyde (7), to a mixture of substituted benzyl chloride (5) (10 mmol), K2CO3 (12 mmol) and KI (trace amount) in 20 ml acetonitrile, 3-substituted-4-hydroxybenzaldehde (6) was added dropwise under inert nitrogen and stirred overnight, and then evaporate under reduced pressure. The crude mixture was quenched with water and the resulting un-dissolved solid was collected by filtration, washed with water, dried, and re-crystallized from aqueous ethanol to give the titled compound. For the synthesis of 4-substituted benzoic acid [3-substituted-4-(4-substituted-benzyloxy)-benzylidene]-hydrazide UOSC-1–14, a mixture of the acid hydrazide (4) (10 mmol) and appropriate aromatic aldehydes (10 mmol) (7) in glacial acetic acid (6 ml) was heated at reflux for 3–4 h until the reaction was completed. The reaction mixture was concentrated under reduced pressure, cooled and the solid obtained was filtered and crystallized.
Biological evaluation
Organism and growth conditions
The spore-forming fungus, Rhizopus delemar-9980, was maintained on potato dextrose agar (PDA) plates [33]. To test the effect of UOSC compounds on fungal melanin production, a fungal inoculum of 2 × 105 colony-forming unit (CFU)/ml was suspended in PD broth and streaked evenly on a PDA plates. The agar plates were cut into 1 cm discs using cork borer. The discs were suspended separately into a 12-well plate containing 1 ml PD broth mixed with different concentrations of UOSC-1–14 compounds. The plates were incubated for 3 days at 37°C in the dark. Kojic acid and cuminaldehyde were employed as positive controls, while DMSO (the solvent used to dissolve UOSC compounds) was used as negative control. Kojic acid is the common reference used to inhibit melanin production. Cuminaldehyde is the lead compound used as scaffold to synthesis UOSC compounds [20]. The fungal growth was monitored visually until the negative control wells showed the production of fully grown black spores. For melanin quantifications, clear pictures were taken for all wells at the same magnification power. The black color of Rhizopus spores were assessed by setting a ‘threshold’ of the black color using the tool followed by measuring the intensity of the black color using ImageJ (1.52n for Windows) [22]. In parallel, the fungal hyphae were collected, dried by cheese cloth followed by taking the weight.
To identify the melanin inhibitory concentration of the newly synthesized compounds, a pilot experiment was conducted. R. delemar spores were incubated in liquid media containing different concentrations (6.3, 12.5, 25, 50, and 100 μg/ml) of UOSC-1–14 compounds and kojic acid. The experiment was repeated at 5, 10, and 15 μg/ml for UOSC or 12.5, 25, and 50 μg/ml for kojic acid. The lowest concentration (10 μg/ml) of UOSC compounds that showed significant inhibition in melanin production and limited toxicity on normal fibroblasts was selected for subsequent experiments. However, 50 μg/ml kojic acid was selected since it is the only concentration caused persistent melanin inhibition activity.
Inhibition of tyrosinase activity and calculation of IC50
The inhibitory activity of UOSC-1–14 compounds on the diphenolase activity of mushroom tyrosinase was investigated using l-DOPA as a substrate. Mushroom tyrosinase is commercially available from Agaricus bisporus (Cat. # T3824–25KU, Sigma–Aldrich). The spectrophotometric activity assay for tyrosinase was performed according to the previously reported method [23] with modifications. Briefly, 30 units (0.01 mg) of tyrosinase enzyme were pre-incubated with the compounds (dissolved in DMSO) in phosphate buffer (pH 6.8) at concentrations 1, 5, 10, 25, 50, 75, and 100 μg/ml for 20 min at 25°C. l-DOPA solution (0.5 mM) was added to the mixture and the reaction was monitored with the change in absorbance at 470 nm due to DOPA chrome formation. All measurements were in triplicate for each concentration. Kojic acid and DMSO were employed as positive and negative controls, respectively. The inhibition percentage was calculated from the below equation:
Inhibition ratio (%) = [(B − S)/B] × 100, where B and S are the absorbance for the blank and samples. The IC50 was calculated according to Ismaya et al. [23]. The IC50 values were calculated using GraphPad Prism 5.0 software.
To evaluate the activity of the developed compounds on R. delemar melanin production, fungal spores were treated with UOSC-1, UOSC-2, UOSC-13, UOSC-14, kojic acid (as positive control), cuminaldehyde (the lead compound), and DMSO as negative control for 3 days at 10 μg/ml. The same weight of the grown hyphae (0.9 g) was grinded using the mortar and pestle method. The total protein of fungal hyphae was extracted using 300 μl triton lysis buffer containing 30 μl protease inhibitor (50 mM Tris, 150 mM NaCl, 5 mM EGTA, 1% Triton-X100) followed by centrifugation at 14 000 rpm at 4°C for 10 min. The supernatant was separated and the extracted protein was quantified using Pierce BCA protein assay kit (Cat# 23227, Thermo Scientific). A pilot experiment was performed using protein extracted from untreated-fungal hyphae in order to determine the optimal concentration of protein and l-DOPA and incubation time. The same protein weight (50 μg) of treated-, control-treated, and non-treated fungi was employed in tyrosinase activity assay as mentioned before. Briefly, fungal protein extracts were mixed with 10 mM l-DOPA in 0.5 M sodium phosphate buffer (pH 6.8) and incubated in dark at 37°C for 3 h. The developed dark color due to melanin production was measured using microplate reader at 475 nm. Data obtained were expressed as mean ± standard error of the mean of three independent experiments.
Kinetic studies
A series of experiments were performed to determine the inhibition kinetics of compound UOSC-2. The inhibitor concentrations were 1, 2.5, and 5 μM, while the substrate (l-DOPA) concentrations were set at 0.625, 1.25, 2.5, and 5 mM in all kinetic studies. Pre-incubation and measurement time was as the same as mushroom tyrosinase inhibition assay. The tyrosinase inhibition rate was then calculated using Lineweaver-Burk plot using GraphPad prism. The Michaelis–Menten constant (Km) and the maximum reaction velocity (Vmax) were calculated at different concentrations of l-DOPA for 10 min [24].
Melanin staining of melanoma cell line
Melanoma cells line (B16 cells) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, and 100 mg/ml streptomycin. Cells were passaged routinely and maintained at 37°C and 5% CO2. For each experiment, 5000 cells were seeded into each well of a clear flat-bottom 96-well plate and allowed to adhere for 24 h. The cells were then incubated with the tested compounds at a concentration of 10 μg/ml in triplicate. Kojic acid and DMSO were employed as positive and negative controls, respectively. The plates were then incubated for 24 h, followed by treatment with staining solution (1% aqueous ferric chloride (30 ml) and 1% aqueous potassium ferricyanide (10 ml), combine and mix well) by adding 0.1 ml to each well and stand for 10 min [25]. The excess solution was removed and washed with distilled water twice. An equal volume of phosphate buffer was added and photos using inverted microscope were taken prior to analysis.
MTT cell viability assay
The reduction of yellow tetrazolium salt 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was used to measure cellular metabolic activity as a proxy for cell viability [26]. To measure the toxicity of UOSC compounds, fibroblasts (HFF-1) cell lines were cultured until reached 80% confluency. A 96-well plate was seeded with 4000 cells per 100 μl media and incubated at 37°C for 24 h. All compounds at 100 μg/ml were added onto the cells and incubated for further 24 h. Freshly prepared MTT solution (5 mg/ml) was added to each well (20 μl per well) and followed by incubation for 2 h at 37°C. The supernatants were then removed and 100 μl DMSO was added and incubated until formazan violet crystals were developed and the OD540 were measured.
In vivo virulence studies of white versus wild (black) types Rhizopus spores
Immunocompetent mice were infected intratracheal with 106 fungal spores according to Andrianaki et al. [6]. The mice were anesthetized by intraperitoneal injection of 0.2 ml mixture of ketamine (82.5 mg/kg, Phoenix, St. Joseph, MO) and xylazine (6 mg/kg, Lloyd Laboratories, Shenandoah, IA). The intratracheal injection of the fungal spores was performed according to Luo et al. [27]. The mice were then euthanized by cervical dislocation at different time points including 4, 24, and 72 h, the lungs were homogenized, and fungal CFU counts were assessed. All animal studies have been taken place at the Lundquist Institute for Biomedical Innovations, Torrance, CA, U.S.A. Animal studies were approved by the IACUC of the Lundquist Institute for Biomedical Innovations at Harbor-UCLA Medical Center and according to the NIH guidelines for animal housing and care.
De-melanization of Rhizopus spores enhanced the phagocytosis and clearance activities of macrophages
Peripheral blood was obtained in EDTA vacutainer collection tubes from healthy male and female individuals after signing an informed consent form approved by the Institutional Review Board (IRB) of the Lundquist Institute for Biomedical Innovations at Harbor-UCLA Medical Center. The blood was pooled in a 50 ml falcon tube. Peripheral blood mononuclear cells (PBMCs) were isolated from the whole blood by density gradient centrifugation through over layering 12.5 ml blood over 10 ml Histopaque-1077 (Sigma) followed by centrifugation at RCF 400 for 25 min at room temperature without brakes. Interfaces containing PBMCs were collected, washed in RPMI-1640 (Sigma), counted and seeded into 96-well tissue culture plates at a density of 10 000 cells per 200 μl of RPMI-1640 media supplemented with 10% fetal bovine serum (Sigma) and 1% penicillin/streptomycin (Sigma). At least six wells were employed for each treatment per each experiment. Plates were incubated for 24 h at 37°C and 5% CO2. Supernatants including floating non-monocytic cells were removed by gentle wash with pre-warmed PBS. Spores were added at a final ratio of 2 : 1 phagocyte. Co-cultures were incubated for 2, 5, and 16 h. At the end of each time point, non-phagocytosed spores were gently rinsed away with PBS and the remaining macrophages were incubated with cold water for 30 min to allow the lysis of phagocytes. The obtained lysates from each well were diluted and cultured on PDA (HiMedia) for 24 h at 37°C.
Procedures of molecular modeling
Molecular modeling for the binding of UOSC compounds with either the mushroom or the human tyrosinases was performed according to Hamdy et al. [28] and Debnath et al. [29].
Preparation of the target enzymes.
The crystal structures of both mushroom (2Y9X) and human (5M8M) tyrosinases were downloaded from the Protein Data Bank at http://www.rcsb.org. The energy of the system was minimized using MOE 2008.10 (Molecular Operating Environment) available at http://www.chemcomp.com.
Preparation of the ligands.
The compounds were prepared with MOE 2008.10. The structures of the compounds and the energy minimization were carried out using MOE 2008.10. The placement criteria were adjusted to be MMFF94× force field until RMSD gradient of 0.05 kcal mol−1 Å was reached.
Molecular docking.
Compounds UOSC-1, 2, 13, and 14 were docked within the binding site of mushroom and human tyrosinases. The placement criteria were adjusted to be Triangle Matcher. Rescoring 1 was selected to be London Gand Retain 10 poses. In our study, we prefer to make refinement with Force field and rescoring 2 was chosen to be London G.
Calculation of binding energy.
The free binding energies of the ligands with the active sites of tyrosinase enzymes were calculated using PlayMolecule software available at https://www.playmolecule.org [30].
Molecular dynamics simulation.
MD was carried out using GROMACS 5.1.2 software package [31]. The topology files of mushroom tyrosinase protein (PDB 2Y9X) were directly created by GROMACS, whereas Acpype software [32] was used to generate topology files of the compounds. Compounds UOSC-1, 2, 13, and 14 complexes and the enzyme alone were immersed in the center of the cubic box with margin of 1 nm, and then the boxes were filled with TIP3P water model. Sodium and chloride ions were added to neutralize the systems, then the complexes were relaxed via energy minimization using the steepest descent minimization algorithm until the maximum force is under1000 kcal/mol/nm. The complexes were equilibrated through two steps, first one was NVT ensemble (constant number of particles, volume, and temperature) for 100 picoseconds ( ps) to stabilize the system at 300 K, and the second step was NPT (constant number of particles, pressure, and temperature) running for 100 ps. Finally, after the equilibration of each system, 10 ns MD simulation was performed.
Study approval
All procedures involving the use of mice and human were performed in accordance with the relevant guidelines and regulations. The use of mice was approved by the IACUC of The Lundquist Institute for Biomedical Innovations at Harbor-UCLA Medical Center, according to the NIH guidelines for animal housing and care. Human peripheral blood collection was approved by the IRB of The Lundquist Institute for Biomedical Innovations at Harbor-UCLA Medical Center under protocol #11671.
Statistical methods
The data were graphed using Graph Pad 5.0 for Windows (GraphPad Software, La Jolla, CA, U.S.A.). The statistical significance was analyzed using one-way analysis of variance (ANOVA) using either Bonferroni’s multiple comparisons test or Dunnett’s multiple comparison test. P-value < 0.05 was considered significant.
Results
Rational design of UOSC compounds
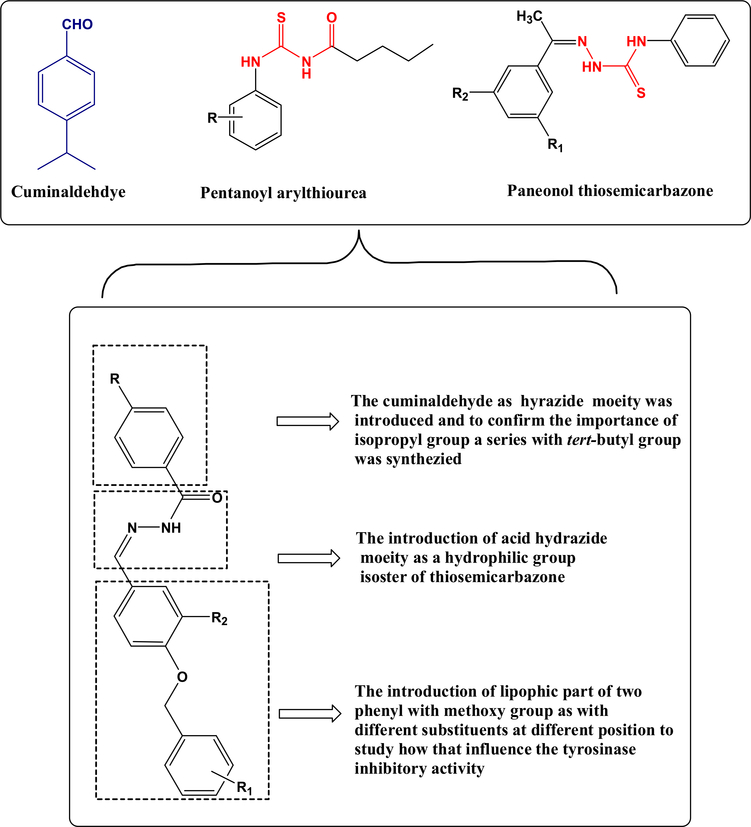
Cuminaldehyde, a natural volatile oil extracted from a plant source, causes reduction of melanin production from R. delemar [20]. Cuminaldehyde has been identified as a non-specific tyrosinase inhibitor [16,19,33]. We have used cuminaldehyde as a scaffold to synthesize a library of compounds (UOSC-1–14) with specific potent and safe fungal tyrosinase inhibition activities, particularly against R. delemar, without affecting the human tyrosinase.
The newly developed compounds were designed based on the structural comparison of cuminaldehyde to ligands (Figure 1) known to inhibit fungal tyrosinase enzyme. The presence of 3D crystal structure of fungal tyrosinase enzyme with various inhibitory ligands and the help of modeling techniques provided an excellent opportunity for structure-based design of potential candidates to rationalize the structural elements responsible for the selective inhibitory activity of the enzyme. The toxic aldehyde group of cuminaldehyde was replaced with polar hydrophilic acid hydrazide moiety. The lone pair of electrons on N and carbonyl oxygen provided better hydrogen bond formation with the binuclear copper active site of the fungal tyrosinase enzyme. Furthermore, the introduction of the proper lipophilic group with different substitutions influenced the orientation and interaction of the compounds with the hydrophobic residues at the active site of fungal tyrosinase enzyme (Figure 2). The aforementioned designs resulted in the synthesis of novel UOSC-1–14 compounds (Figure 3) with enhanced physiochemical properties and selectively to inhibit R. delemar fungal tyrosinase.
Figure 2.
The rational design of melanin biosynthesis inhibitors based on cuminaldehyde scaffold compared with reported anti-melanin compounds.
Figure 3. Synthesis scheme of UOSC compounds.
Reagents and conditions: i. I2-NaOH/TBHP, 70°C, 10–16 h; ii. Conc H2SO4, EtOH/3 h; iii. NH2NH2/EtOH, 24 h; iv. K2CO3, KI, acetontrile, overnight stirring under nitrogen; v. EtOH-glacial acetic acid, 4 h; R1: 4-CH3, 3-CF3, or 4-CF3; R2: Cl, Br, or OCH3; R3: isopropyl (cuminaldeyde) or t-butyl.
The newly designed compounds caused effective inhibition of R. delemar melanin production
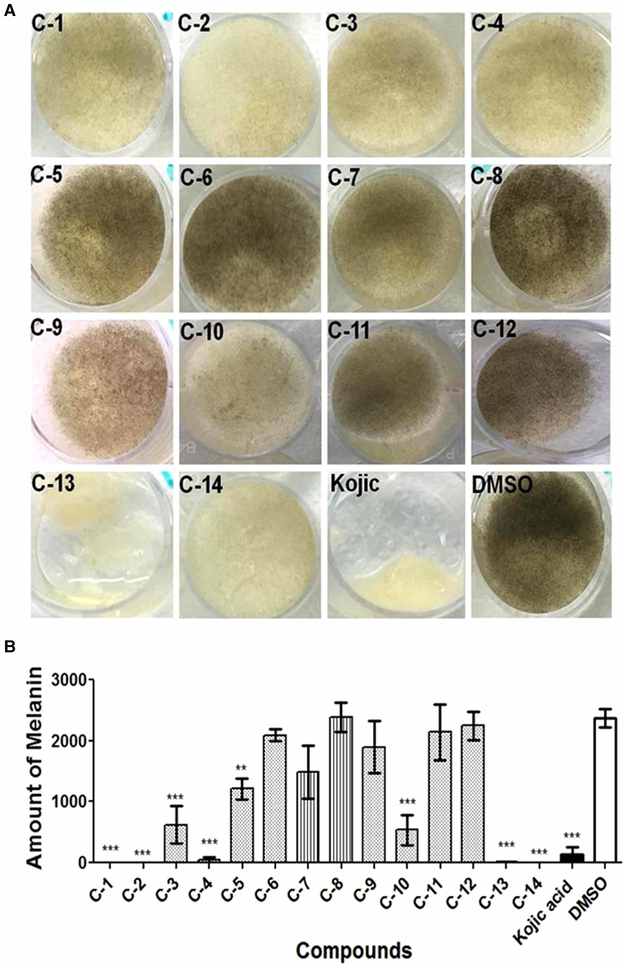
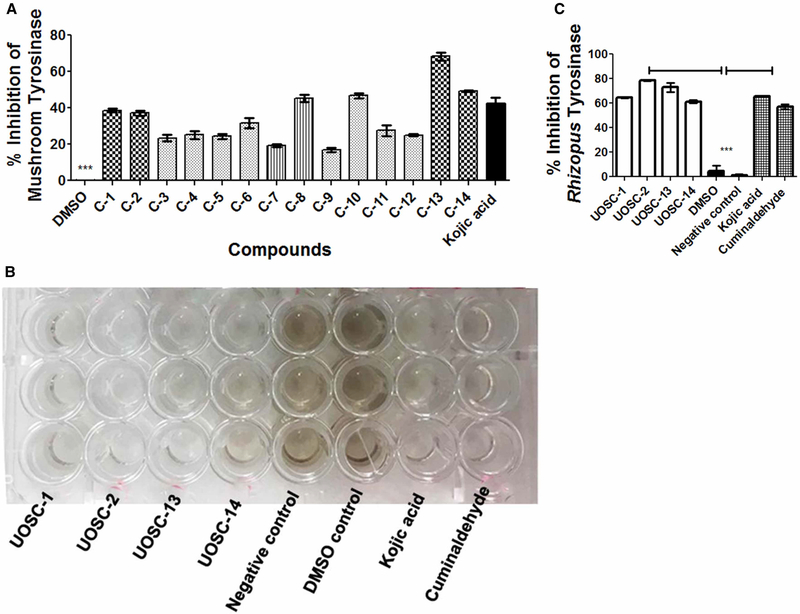
To test for the melanin inhibition activity of the newly synthesized compounds, R. delemar spores were incubated for 3 days in liquid media containing 10 μg/ml of UOSC-1–14 compounds in comparison with 50 μg/ml kojic acid, a general de-pigmentation compound [34]. Out of the 14 compounds, UOSC-1, 2, 3, 4, 10, 13, and 14 showed significant inhibition (One-way ANOVA, P < 0.0001) of R. delemar melanin production (Figure 4A,B). While UOSC-1, 2, 3, 4, 10, and 14 did not reduce the growth of fungal hyphae (weight range = ~33.1 ± 0.6 − 33.5 ± 0.9 mg), UOSC-13 caused significant reduction in the growth of fungal hyphae (weight = 20.5 ± 2.5 mg) similar to kojic acid (Figure 4A). Thus, UOSC-1, 2, 3, 4, 10, and 14 were selected as potent anti-melanin candidates for the R. delemar fungus.
Figure 4. Effect of UOSC compounds on the melanin production of R. delemar.
(A) Direct effect of UOSC compounds on melanin biosynthesis when incubated for 3 days with R. delemar spores in liquid culture at 10 μg/ml in comparison with DMSO and kojic acid as negative and positive controls, respectively. (B) Measurement of melanin production from R. delemar after treatment was performed using ImageJ. The data was analyzed using one-way ANOVA and statistical significance was calculated with Dunnett’s multiple comparisons test and significance level indicated by asterisks (*, P < 0.05: **, P < 0.001; ***, P < 0.0001). The data display the mean ± SEM of three biological replicas.
UOSC-1, 2, 13, and 14 are potent candidates to inhibit R. delemar melanin production
The inhibitory activities of all tested compounds on fungal tyrosinase enzyme were tested in comparison with kojic acid using a mushroom tyrosinase purified from A. bisporus and a fresh total cell protein extract of R. delemar. Although all compounds showed significant inhibition of mushroom tyrosinase activity when compared with kojic acid (One-way ANOVA, P < 0.0001), UOSC-1, 2, 8, 10, 13, and 14 showed the highest inhibition activity (Figure 5A). On the other hand, the inhibitory effect of the developed derivatives on the melanin biosynthesis of R. delemar was the highest with UOSC-1, 2, 13, and 14, kojic acid and cuminaldehyde (One-way ANOVA, P < 0.0001) (Figure 5B,C). Compound UOSC-2 showed the highest inhibition activity (Figure 5C).
Figure 5. Inhibition effect of UOSC compounds at 10 μg/ml on fungal melanin.
(A) Inhibition effect of UOSC compounds on mushroom tyrosinase using ELISA kits. (B) Inhibition effect of UOSC compounds on R. delemar tyrosinase enzyme activity compared with kojic acid and cuminaldehyde. (C) Graphical representation for the inhibition of UOSC compounds on Rhizopus tyrosinase enzyme activity compared with kojic acid and cuminaldehyde. The data were analyzed using one-way ANOVA and statistical significance was calculated using Dunnet’s multiple comparisons test and significance level indicated by asterisks (*, P < 0.05: **, P < 0.001; ***, P < 0.0001). The data display the mean ± SEM of at least three biological replicas.
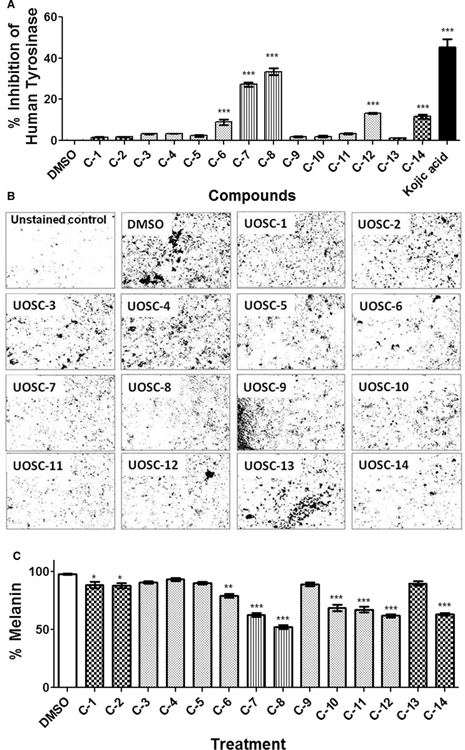
Mushroom tyrosinase shows limited homology to mammalian ones (~30% similarity based on Smith–Waterman sequence alignment, Data not shown) and this renders it as a well-suited model for studies on melanogenesis [23,35]. Thus, any variation in the activities of the tested compounds between both mushroom and human tyrosinases will support the potent inhibition selectivity of UOSC compounds against fungal melanin biosynthesis but not the human, and hence offers minimal toxicity. Consequently, the inhibitory activities of UOSC compounds were tested on human tyrosinase (Cat. # T8455–1MG, Fragment 369–3771 Sigma–Aldrich) and melanoma cells (employed as melanin-producing human cells). Compared with kojic acid, UOSC-6, 7, 8, 12, and 14 showed significant inhibition of human tyrosinase (P < 0.0001) (Figure 6A). However, UOSC-7, 8, 10, 11, 12, and 14 caused significant inhibition of melanin production from melanoma cells (Figure 6B,C). Collectively, UOSC-1, 2, 13, and 14 can be considered as excellent candidates against R. delemar melanin production. Because UOSC-8 showed similar inhibitory activities on both fungal and human tyrosinase, the compound cannot be selected as candidate (Figures 5A and 6A). Although UOSC-10 showed significant inhibition to fungal tyrosinase but not human tyrosinase, the compound caused significant inhibition (P < 0.005) of melanin production from melanoma cells; thus the compound also excluded from the selection. On the other hand, UOSC-14 showed anti-melanin activities against both fungus and human (Figures 5C and 6C).
Figure 6. Inhibition effects of UOSC compounds at 10 μg/ml on human melanin.
(A) Inhibition effect of UOSC compounds on human tyrosinase using ELISA kits. (B,C) Effect of UOSC compounds on the melanoma cell line. The melanin was stained according to Schmorl Melanin Stain [36] as indicated in the methodology section. (B) Melanin staining (C) Representative measurement of melanin from melanoma cells by densitometry. The data were analyzed using one-way ANOVA and statistical significance was calculated using Dunnet’s multiple comparisons test and significance level indicated by asterisks (*, P<0.05: **, P<0.001; ***, P<0.0001). The data display the mean ± SEM of three biological replicas.
UOSC-1, 2, 13, and 14 showed high tyrosinase-inhibitory activities with limited toxicity
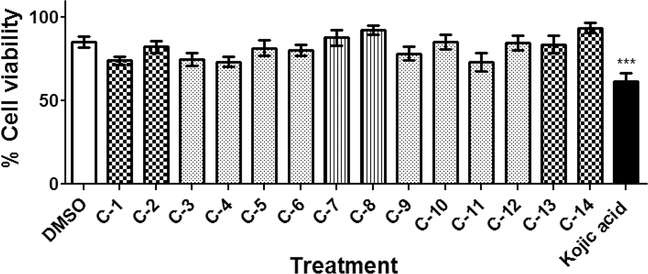
The toxicity of all developed compounds was tested on mammalian cells using the normal fibroblasts HFF-1 cell line. All tested compounds showed limited toxicity when compared with kojic acid toxicity (Figure 7, P < 0.005). In contrast, the inhibitory activities of all tested compounds to mushroom tyrosinase were tested at seven different concentrations (1, 5, 10, 25, 50, 75, and 100 μg/ml). Although all tested compounds showed variable inhibitory activities to mushroom tyrosinase, UOSC-1, 2, 13, and 14 showed the lowest IC50 of 0.01, 0.0074, 0.0086, and 0.009 μM, respectively (Table 1).
Figure 7. Toxicity of UOSC compounds.
The effect of compounds on mammalian fibroblast cell line using MTT assay. Mammalian cells were seeded in 96-well plate until confluency followed by incubation with the compounds overnight prior to MTT assay. The data display the mean of the percentage of the survival rate of mammalian cells ± SEM of six experimental replicas. A P-value < 0.05 was considered as significant.
Table 1.
Calculated IC50 (molar concentration) of the UOSC compounds on mushroom tyrosinase
| Compounds | IC50 (μM) |
|---|---|
|
| |
| UOSC-1 | 0.01 |
| UOSC-2 | 0.0074 |
| UOSC-3 | 0.0195 |
| UOSC-4 | 0.0147 |
| UOSC-5 | 0.034 |
| UOSC-6 | 0.0837 |
| UOSC-7 | >0.1875 |
| UOSC-8 | 0.0807 |
| UOSC-9 | >0.207 |
| UOSC-10 | 0.0287 |
| UOSC-11 | 0.102 |
| UOSC-12 | 0.043 |
| UOSC-13 | 0.0086 |
| UOSC-14 | 0.0099 |
Computational modeling predicted that UOSC-1, 2, 13, and 14 are selective inhibitors of fungal tyrosinase
To confirm the selectivity of UOSC-1, 2, 13, and 14 to the fungal (represented by mushroom), but not the human tyrosinase, in-depth docking studies for the interaction of UOSC-1, 2, 13, and 14 within the hydrophobic binding pocket of mushroom tyrosinase (PDB:2Y9X) was performed in comparison with their interactions within human tyrosinase (PDB:5M8M) (Figure 8, Supplementary Figures S1–S3 and Supplementary Information) [37]. These binding interactions were further compared with the interactions of the enzyme with its native ligands, tropolone, and kojic acid. Furthermore, the binding mode and binding energies were investigated using Molecular Operating Environment (MOE) [38] and PlayMolecule [30], respectively.
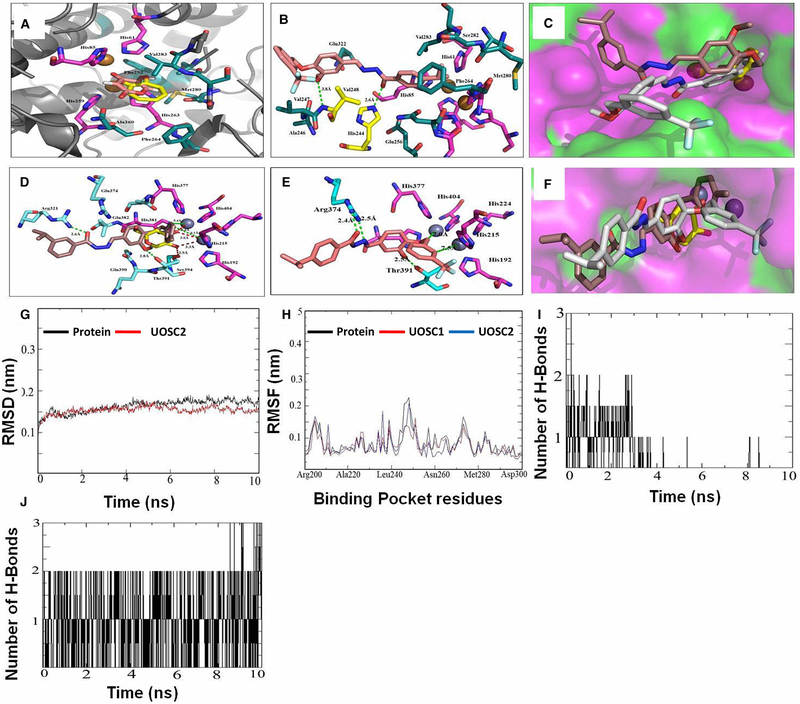
Figure 8. Computational modeling of UOSC-1, 2, 13, and 14 with fungal tyrosinase compared with human tyrosinase.
(A–C) Interaction of the native ligands (tropolone and kojic acid) and compounds UOSC1 or UOSC-2 within the active site of mushroom tyrosinase. (A) Illustrated the interaction of native ligand tropolone (yellow stick) and kojic acid (light pink stick) within the active site of mushroom tyrosinase (PDB:2Y9X). Copper ions represented as copper sphere surrounded with histidine residues (pink stick). (B) Illustrated the interactions of UOSC-2 (light brown stick) with residues (dark green stick). UOSC-2 showed H-bonds (yellow stick) with His244 and Val248. (C) Binding mode of native ligand, tropolone (yellow stick), UOSC-2 (white stick), and UOSC-1 (brown stick). UOSC-2 occupied the active center similar to native ligand. UOSC-2 is so near to copper ions at a distance of 3.8 Å. The remaining structure of UOSC-2 extended in the binding pocket, the surface represented as hydrophilic (pink), hydrophobic (green), and neutral (white). (D–F) Interactions of UOSC-1 or UOSC-2 and the native ligand, kojic acid within the active site of human tyrosinase. (D) Illustrated the interactions of kojic acid (light yellow stick) and UOSC-1 (brown stick) within the human tyrosinase active site (PDB:5M8M). (E) Illustrated the interactions of UOSC-2 (light brown stick) within human tyrosinase active site. (F) Illustrated the binding mode of UOSC-2 (white stick), UOSC-1 (brown stick), and kojic acid (yellow stick). Human tyrosinase surface represented as hydrophilic (pink), hydrophobic (green), and neutral (white). Zinc ions represented as violet spheres, histidines ligands represented as pink stick and pocket residues are represented as cyan stick. Kojic acid, the native ligand represented as yellow stick. H-bonds between UOSC-2 and the active site are green dots, while H-bonds between kojic acid and the pocket site are red dots. H-bonds between kojic acids and Zn ions are shorter than the H-bonds between UOSC-2 and Zn ions. The H-bonds between UOSC-2 and Zn ions are not in accepted H-bond length. (G–J) Molecular Dynamics (MD) simulation of the newly designed tyrosinase inhibitors. (G) Illustrated the RMSD values over 10 ns simulations of the protein (black) and UOSC-2 (red). (H) Illustrated the RMSF values in nanometer over 10 ns simulations of the protein (black) and UOSC-2 (blue). (I–J) H-bonds interaction over 10 ns MD simulation of (I) kojic acid and (J) UOSC-2.
Compared with the mushroom tyrosinase enzyme’s binding modes with tropolone and kojic acid (Figure 8A), the enzyme’s binding mode with UOSC-1, 2, 13, and 14 are buried beside the copper ions active site, while UOSC-13 and 14 are being displaced away by a small distance (Supplementary Figure S1 and Supplementary Information). The four compounds showed π-p stacking with copper ions ligands (His61, His 85, His259, and His263) which may lead to a change in the geometry of copper ions center and a disturbance in the redox reactions of tyrosinase. The four compounds illustrated strong interactions with the key and reserved residues of the hydrophobic pocket site including Hiss244, Val248, and Phe264. Moreover, the binding mode of UOSC-2 (Figure 8B) showed that it can occupy the active center similar to the native ligand. The compound was very close to the copper ions, while the remaining of the compound’s structure was extended in the binding pocket, making UOSC-2 as the most promising mushroom tyrosinase inhibitors (Figure 8C) and may explain its superiority as anti-melanin when compared with kojic acid.
Co-crystallization of kojic acid with human tyrosinase shows a close contact to Zn atom active site in a distance of 3.3–3.6 Å, and a H-bond formed between O-atom of C=O group and the OH of the key residue Ser394 at distance 2.9 Å (Figure 8D). Compared with kojic acid, UOSC compounds including in particular UOSC-2 moved far away from the Zn binuclear active site of the human tyrosinase (Figure 8E,F, Supplementary Figure S2 and Supplementary Information).
To further predict the possible binding mode for each compound within the pocket site of mushroom tyrosinase, the binding free energy (ΔGBind) and the binding affinity pKd [39,40] were calculated (Table 2). It was found that the four compounds showed binding energies less than the reference compound kojic acid = −5.78 and higher binding affinity than kojic acid (pKd = 4.28). UOSC-2, which showed the best IC50 = 0.0074 μM, illustrated the lowest free binding energy (−9.13 Kcal/mol) and the best binding affinity (pKd = 6.76) (Table 2). This result is in agreement with the concept that a compound with the best IC50 should exhibit the lowest binding energy [40]. The results obtained confirmed that the newly designed derivatives exhibited good binding mode which conformed to their lowest binding energies and high binding affinity than kojic acid. Furthermore, the orientation of the four compounds within human tyrosinase was far away from the active site and this is conformed to the higher binding energies; explaining the selectively of the newly designed UOSC-1, 2, 13, and 14 compounds to the mushroom tyrosinase.
Table 2.
Free binding energy (ΔG) and binding affinity (Pkd) of compounds 1, 2, 13, and 14 and selected known anti-melanin compounds with mushroom versus human tyrosinase
| Mushroom tyrosinase |
Human tyrosinase |
||||
|---|---|---|---|---|---|
| Cp. no. | IC50 (μM) | ΔG (Kcal/mol) | Pkd | ΔG (Kcal/mol) | Pkd |
|
| |||||
| UOSC-1 | 0.01 | −8.10 | 6.0 | −7.73 | 5.73 |
| UOSC-2 | 0.0074 | −9.13 | 6.76 | −7.82 | 5.79 |
| UOSC-13 | 0.0086 | −8.07 | 5.98 | −7.36 | 5.45 |
| UOSC-14 | 0.0099 | −8.13 | 6.02 | −7.73 | 5.72 |
| Kojic acid | 0.23 | −5.78 | 4.28 | – | – |
| Cinnamic acid | – | −7.84 | – | – | – |
| Thiosemicarbazone | – | −7.40 | – | – | – |
| Umbeliferone | – | −7.52 | – | – | – |
| Reservatrol | – | −7.40 | – | – | – |
| Chalcone | – | −8.61 | – | −8.96 | – |
Molecular dynamics (MD) simulations was carried out to further confirm the accuracy of docking results and to obtain a more accurate ligand–protein binding model in a case close to the natural conditions [41]. Docking of UOSC compounds with the enzyme was tested using MD simulation for 10 ns. The MD simulations of the produced complexes with the mushroom tyrosinase were checked for root mean square deviation (RMSD) to confirm the stability of the protein–inhibitors in the solvent system. The produced complexes showed low RMSD values (0.15 nm) (Figure 8, Supplementary Figure S3 and Supplementary Information). Furthermore, the complex produced from UOSC-2 showed smooth RMSD curves (Figure 8G). The compound reached the equilibrium at 1 ns with ~0.14 nm RMSD which is less than the enzyme alone (Figure 8G). These results indicated that the most stable form of mushroom tyrosinase occurred in the presence of the aforementioned inhibitors in particular compound UOSC-2. Furthermore, the root mean square fluctuation (RMSF) of UOSC compounds–tyrosinase complexes were high (~0.4 nm) with the first 100 residues, indicating the high flexibility of this part. The remaining residues showed low RMSF value (~0.2 nm), particularly the binding pocket residues 200–300 (Figure 8, Supplementary Figure S3 and Supporting Information). The low flexibility of the binding pocket residues indicated the stability of the enzyme due to the presence of inhibitors [29]. Furthermore, the RMSF of UOSC-2 with residues between 240 and 250 in the binding pocket was lower than the protein alone (Figure 8H), which may be attributed to the H-bond between UOSC-2 and His244 and Val248.
The compactness of inhibitor–enzyme complexes was measured by the Radius of gyration (Rg). The Rg of the four inhibitor–protein complexes was ~2.055 nm and remained steady over the dynamics measurements (Figure 8, Supplementary Figure S3 and Supplementary Information). Studying the H-bonds over 10 ns indicated that UOSC-2, 13, and 14 showed stable and strong H-bonds with the tyrosinase enzyme much better than kojic acid (Figure 8, Supplementary Figure S3 and Supplementary Information). Compared with kojic acid (Figure 8I), UOSC-2 illustrated the most stable and maintained H-bonds (Figure 8J), supporting its superiority against fungal melanin.
UOSC-2 showed competitive reversible inhibition activity against fungal tyrosinase
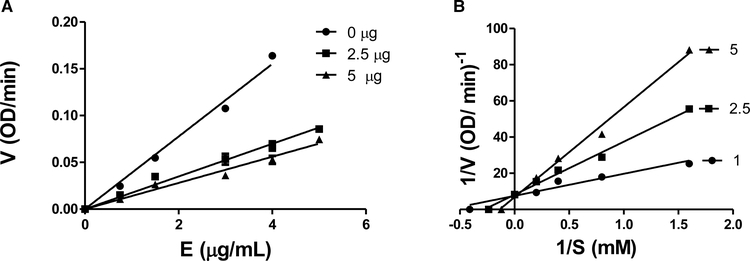
The inhibition mechanism of UOSC-2 on mushroom tyrosinase enzyme was determined using l-DOPA as substrate. The relationship between enzyme activity and its concentration in the presence of different concentrations of UOSC-2 showed straight lines pass through the origin. The enzyme inhibition activity due to UOSC-2 is dose-dependent. As the concentration of UOSC-2 increased, the enzyme activity is reduced, while the amount of enzyme is not affected (Figure 9A). Moreover, Lineweaver–Burk plots were generated in order to determine the inhibition type. The plots of the enzyme activity (1/V) versus the concentration of substrate (1/[S]) at different inhibitor concentrations gave straight lines, which all passed through one point. The results indicated that compound UOSC-2 inhibited the diphenolase activity of tyrosinase in a dose-dependent manner [24]. With increasing the concentrations of the compound, Km value increased and Vmax value remained the same, while the enzyme activity was inhibited; confirming that the compound is a competitive reversible inhibitor (Figure 9B).
Figure 9. Activity of mushroom tyrosinase enzyme in the presence of different concentrations of L-DOPA as substrate and different concentrations of UOSC-2 inhibitor.
(A) A plot of enzyme concentration (S) versus the enzyme activities (V) at different concentrations of the inhibitor (UOSC-2). The presence of inhibitor did not reduce the amount of enzyme, but resulted in an inhibition of the enzyme activity. The UOSC-2 inhibited the diphenolase activity of tyrosinase in a dose-dependent manner. (B) Lineweaver−Burk plot of mushroom tyrosinase enzyme inhibition by different concentrations of UOSC-2 (1, 2.5, and 5 μM) in the presence of different concentrations of l-DOPA as a substrate (0.675, 1.25, 2.5, and 5 mM). The reciprocal tyrosinase inhibitory activity was plotted against the reciprocal substrate concentration (double reciprocal plot). The data were obtained as mean values of at least three biological replica of 1/V, the inverse of the absorbance increases at a wavelength of 492 nm per min of three independent experiments.
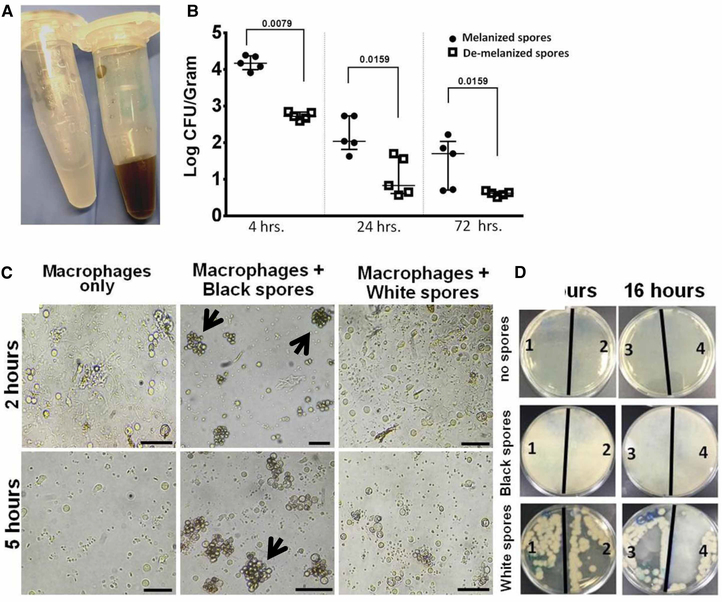
De-melanized R. delemar (white) spores were phagocytized and cleared faster than the wild-type (black) spores
To determine the potential incorporation of anti-melanin activity of the inhibitors in fungal pathogenesis, we treated R. delemar spores with UOSC-2 at 10 μg/ml for 3 days. The generated white spores were compared for their infectivity with wild-type spores using the intratracheally infected immunocompetent mouse model as previously described [6]. Infection of mice with de-melanized R. delemar spores (Figure 10A) resulted in rapid fungal clearance from the lungs, when compared with infection with wild-type melanized fungal spores (Figure 10B). To discern if the white spores become prone to phagocytosis, peripheral human blood-enriched macrophages were co-cultured with white and wild-type spores (Figure 10A) and evaluated at 2, 5, and 16 h time points. The efficiency of fungal clearance by macrophages was also compared by culturing the macrophages lysates on fungal growth media (PDA) at the end of each time point. In contrast with wild-type black spores which persisted in the macrophages without being killed, white spores were phagocytized more efficiently and cleared faster within ~5 h. No spores clumping observed in association with macrophages co-cultured with white spores (Figure 10C). The growth of intracellular white (albino) conidia following macrophage lysis was significantly inhibited as compared with wild-type conidia (Figure 10D).
Figure 10. De-melanization of Rhizopus (white) spores enhanced the phagocytic ability of macrophages.
(A) White Rhizopus spores developed after treatment with UOSC-2 for 3 days compared with wild-type black spores. (B) Fungal burden from the lungs of infected immunocompetent mice (n = 5 per experimental group) with 106 fungal spores. The mice were sacrificed at the indicated time points, lungs were collected and the lungs fungal burden were assessed by CFU plating on PDA containing 0.1% triton. The data were analyzed using Student’s t-test and statistical significance was calculated. The data display the mean ± SEM of five different mice. (C) Representative photomicrographs of macrophages obtained from peripheral blood monocytes incubated with Rhizopus spores at a ratio of 1 macrophage to 2 spores at different time points. Developed white spores failed to cluster and were eliminated rapidly as compared with non-treated wild-type spores (indicated by black arrows). Scale bar = 50 μm. (D) Viability of spores after phagocytosis events. At the end of each time point, excess spores were washed with pre-warmed PBS and the remaining macrophages were lysed. The lysates were further diluted and cultured overnight on PDA. White spores viable numbers capable of growing on media were reduced after phagocytosis when compared with wild-type spores. The numbers on the plates (1–4) indicated the number of replicas per each treatment.
Collectively, the newly designed compounds, particularly UOSC-2, are promising candidates to overcome the persistence mechanism of the pathogenic R. delemar fungus due to melanin.
Discussion
Fungal melanin exhibits efficient resistance mechanisms against most stressful conditions including human host defensive mechanisms [1]; thus allowing the persistence of the pathogenic fungi and development of pathogenesis mechanisms. Similarly, R. delemar which is the most common cause of mucormycosis (a life-threatening infection), demonstrates unique persistence mechanisms in animal models causing phagosome maturation arrest [6]. Furthermore, it has been reported that fungal melanin can bind to antifungal compounds including amphotericin B and caspofungin, thereby reducing their fungicidal activities [42].
Cuminaldehyde showed significant inhibition of fungal melanin biosynthesis including R. delemar melanin [20], and has been reported as potent tyrosinase inhibitor [33], but as any aldehyde, it has cytotoxic activities on mammalian cells and cannot be used for clinical applications [43]. Thus, the compound was employed in this study as scaffold to build potent and selective inhibitors against R. delemar melanin biosynthesis. We have successfully generated a series of compounds that showed potent inhibition of fungal melanin biosynthesis and significant inhibition of the rate-limiting melanin biosynthesis enzyme, tyrosinase, and limited toxicity on both melanoma and fibroblasts cell lines. Only compounds UOSC-1, 2, 13, and 14 were selected as promising candidates inhibitors for fungal melanin biosynthesis, since they showed limited effects on human melanin. In contrast with naturally identified anti-melanin compounds including cuminaldehyde and synthetic anti-melanin compounds, the newly designed compounds showed significant selectivity against fungal melanin, particularly R. delemar melanin. This activity may be attributed to the perfect binding of the aforementioned compounds with the fungal tyrosinase active site but not the human one.
UOSC-1, 2, 13, and 14 exhibited excellent binding with mushroom tyrosinase, as a representative to fungal tyrosinase particularly R. delemar, due to (i) the good binding mode of the compounds within the mushroom tyrosinase active site, similar to the enzyme’s native ligand, (ii) the vicinity of the compounds to the catalytic copper ions of the active site and (iii) the interactions of the compounds with the key residues of the binding pocket as the four compounds illustrated strong H-bonds profile and hydrophobic interactions with the hydrophobic residues (Hiss244, Val248, and Phe264). These interactions stabilized the compounds within the active site of fungal tyrosinase. Additionally, the hydrophobic nature of the four compounds facilitates their hydrophobic interactions with the hydrophobic residues in the binding site of mushroom tyrosinase, whereas the active site in human tyrosinase were lining mostly with the hydrophilic residues of the compounds. Therefore, they are displaced away from the binuclear Zn binding site in the case of human tyrosinase.
The newly designed compounds differ in structures from other reported synthetic and natural anti-melanin compounds including cinnamic acid, thiosemicarbazone, umbeliferone, and resveratrol derivatives (Supplementary Figure S4). The free binding energy (ΔG) of UOSC compounds particularly compound UOSC-1, 2, 13, and 14 with mushroom tyrosinase was lower when compared with the aforementioned compounds except chalcone (Table 1). The results indicated that UOSC compounds and chalcone can strongly and efficiently bind with the pocket site of fungal tyrosinase. In contrast and compared with UOSC compounds, chalcone derivatives showed lower ΔG with human tyrosinase (Table 1), indicating in-selectivity of chalcone derivatives. Therefore, we can conclude that UOSC compounds showed efficient and selective binding capacities to fungal tyrosinase.
In conclusion, UOSC-1, 2, 13, and 14 showed significant inhibition activities against fungal melanin biosynthesis particularly against R. delemar melanin. The aforementioned compounds showed excellent binding capabilities within the fungal tyrosinase binuclear copper ions active site but not with the human tyrosinase binuclear zinc-binding site. This binding capability was attributed to the hydrophobic feature of the new designs, the substituent that facilitate the orientation of the compounds within the fungal tyrosinase active site, and the formation of hydrogen bond between the compounds and the fungal tyrosinase active site. Our new designs are considered as the first report for compounds that can distinguish between fungal (R. delemar) and human melanin biosynthesis. We propose that the newly designed compounds can be used effectively either alone or in combination therapy to ameliorate fungal infections in particular against R. delemar. Moreover, the newly designed compounds in particular compounds UOSC-1, 2, 13, and 14 can serve as promising lead drugs for not only Rhizopus infection but also other melanin-dependent serious fungal infections such as Cryptococcus.
Other examined inhibitors such as UOSC-6, 7, 8, and 14 significantly affecting human melanin production, thereby can be developed against diseases with hyper-pigmentation disorders such as actinic and senile lentigines, melasma, and post-inflammatory hyperpigmentation, that are considered as major cosmetic problems.
Supplementary Material
Acknowledgements
This work was supported by grants from the University of Sharjah, U.A.E. to S.S.M.S., partially to M.M. and by the Public Health Service grant R01 AI063503 to A.S.I.
Funding
UOS (1701110120-P) grant to SS and Public Health Service (AI063503) grant to A.S.I.
Abbreviations
- CFU
colony forming unit
- IRB
Institutional Review Board
- l-DOPA
l-3,4-dihydroxyphenylalanine
- PBMCs
peripheral blood mononuclear cells
- PDA
potato dextrose agar
- PDA
potato dextrose agar
- RMSF
root mean square fluctuation
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Riley PA (1997) Melanin. Int. J. Biochem. Cell Biol 29, 1235–1239 10.1016/S1357-2725(97)00013-7 [DOI] [PubMed] [Google Scholar]
- 2.Bell AA and Wheeler MH (1986) Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol 24, 411–451 10.1146/annurev.py.24.090186.002211 [DOI] [Google Scholar]
- 3.Gadd GM (1980) Melanin production and differentiation in batch cultures of the polymorphic fungus aureobasidium pullulans. FEMS Microbiol. Lett 9, 237–240 10.1111/j.1574-6968.1980.tb05644.x [DOI] [Google Scholar]
- 4.Iozumi K, Hoganson GE, Pennella R, Everett MA and Fuller BB (1993) Role of tyrosinase as the determinant of pigmentation in cultured human melanocytes. J. Investig. Dermatol 100, 806–811 10.1111/1523-1747.ep12476630 [DOI] [PubMed] [Google Scholar]
- 5.Tadokoro T, Kobayashi N, Zmudzka BZ, Ito S, Wakamatsu K, Yamaguchi Y et al. (2003) UV-induced DNA damage and melanin content in human skin differing in racial/ethnic origin. FASEB J. 17, 1177–1179 10.1096/fj.02-0865fje [DOI] [PubMed] [Google Scholar]
- 6.Andrianaki AM, Kyrmizi I, Thanopoulou K, Baldin C, Drakos E, Soliman SSM et al. (2018) Iron restriction inside macrophages regulates pulmonary host defense against rhizopus species. Nat. Commun 9, 3333 10.1038/s41467-018-05820-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ando H, Kondoh H, Ichihashi M and Hearing VJ (2007) Approaches to identify inhibitors of melanin biosynthesis via the quality control of tyrosinase. J. Investig. Dermatol 127, 751–761 10.1038/sj.jid.5700683 [DOI] [PubMed] [Google Scholar]
- 8.Korner A and Pawelek J (1982) Mammalian tyrosinase catalyzes three reactions in the biosynthesis of melanin. Science 217, 1163–1165 10.1126/science.6810464 [DOI] [PubMed] [Google Scholar]
- 9.Kameyama K, Takemura T, Hamada Y, Sakai C, Kondoh S, Nishiyama S et al. (1993) Pigment production in murine melanoma cells is regulated by tyrosinase, tyrosinase-related protein 1 (trp1), dopachrome tautomerase (trp2), and a melanogenic inhibitor. J. Investig. Dermatol 100, 126–131 10.1111/1523-1747.ep12462778 [DOI] [PubMed] [Google Scholar]
- 10.Kupper U, Niedermann DM, Travaglini G and Lerch K (1989) Isolation and characterization of the tyrosinase gene from neurospora crassa. J. Biol. Chem 264, 17250–17258 [PubMed] [Google Scholar]
- 11.Lerch K (1982) Primary structure of tyrosinase from neurospora crassa. II. complete amino acid sequence and chemical structure of a tripeptide containing an unusual thioether. J. Biol. Chem 257, 6414–6419 [PubMed] [Google Scholar]
- 12.Kwon BS, Haq AK, Pomerantz SH and Halaban R (1987) Isolation and sequence of a cDNA clone for human tyrosinase that maps at the mouse c-albino locus. Proc. Natl. Acad. Sci. U.S.A 84, 7473–7477 10.1073/pnas.84.21.7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishioka K (1978) Particulate tyrosinase of human malignant melanoma. Eur. J. Biochem 85, 137–146 10.1111/j.1432-1033.1978.tb12221.x [DOI] [PubMed] [Google Scholar]
- 14.Bagherzadeh K, Shirgahi Talari F, Sharifi A, Ganjali MR, Saboury AA and Amanlou M (2015) A new insight into mushroom tyrosinase inhibitors: docking, pharmacophore-based virtual screening, and molecular modeling studies. J. Biomol. Struct. Dyn 33, 487–501 10.1080/07391102.2014.893203 [DOI] [PubMed] [Google Scholar]
- 15.Lai X, Wichers HJ, Soler-Lopez M and Dijkstra BW (2017) Structure of human tyrosinase related protein 1 reveals a binuclear zinc active site important for melanogenesis. Angew. Chem 56, 9812–9815 10.1002/anie.201704616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y-J and Uyama H (2005) Tyrosinase inhibitors from natural and synthetic sources: structure, inhibition mechanism and perspective for the future. Cell Mol. Life Sci 62, 1707–1723 10.1007/s00018-005-5054-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ennes SBP, Paschoalick RC and Alchorne MMDA (2000) A double-blind, comparative, placebo-controlled study of the efficacy and tolerability of 4% hydroquinone as a depigmenting agent in melasma. J. Dermatol. Treat 11, 173–179 10.1080/09546630050517333 [DOI] [Google Scholar]
- 18.Romaguera C and Grimalt F (1985) Leukoderma from hydroquinone. Contact Dermat. 12, 183–183 10.1111/j.1600-0536.1985.tb01101.x [DOI] [PubMed] [Google Scholar]
- 19.Jiménez M, Chazarra S, Escribano J, Cabanes J and García-Carmona F (2001) Competitive inhibition of mushroom tyrosinase by 4-substituted benzaldehydes. J. Agric. Food Chem 49, 4060–4063 10.1021/jf010194h [DOI] [PubMed] [Google Scholar]
- 20.Soliman SSM, Alsaadi AI, Youssef EG Khitrov G, Noreddin AM, Husseiny MI et al. (2017) Calli essential oils synergize with lawsone against multidrug resistant pathogens. Molecules 22, 2223 10.3390/molecules22122223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazra S, Deb M and Elias AJ (2017) Iodine catalyzed oxidation of alcohols and aldehydes to carboxylic acids in water: a metal-free route to the synthesis of furandicarboxylic acid and terephthalic acid. Green Chem. 19, 5548–5552 10.1039/C7GC02802D [DOI] [Google Scholar]
- 22.Jensen EC (2013) Quantitative analysis of histological staining and fluorescence using image J. Anat. Rec 296, 378–381 10.1002/ar.22641 [DOI] [PubMed] [Google Scholar]
- 23.Ismaya WT, Rozeboom HJ, Weijn A, Mes JJ, Fusetti F, Wichers HJ et al. (2011) Crystal structure of Agaricus bisporus mushroom tyrosinase: identity of the tetramer subunits and interaction with tropolone. Biochemistry 50, 5477–5486 10.1021/bi200395t [DOI] [PubMed] [Google Scholar]
- 24.Uchida R, Ishikawa S and Tomoda H (2014) Inhibition of tyrosinase activity and melanine pigmentation by 2-hydroxytyrosol. Acta Pharm. Sin. B 4, 141–145 10.1016/j.apsb.2013.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orchard GE (2019) 14 - Pigments and minerals. In Bancroft’s Theory and Practice of Histological Techniques, 8th edn (Suvarna SK, Layton C and Bancroft JD, eds), pp. 198–230, Churchill Livingstone Elsevier, London, UK [Google Scholar]
- 26.Soliman S, Mohammad MG, El-Keblawy AA, Omar H, Abouleish M, Madkour M et al. (2018) Mechanical and phytochemical protection mechanisms of calligonum comosum in arid deserts. PLoS One 13, e0192576–e0192576 10.1371/journal.pone.0192576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo G, Gebremariam T, Lee H, French SW, Wiederhold NP, Patterson TF et al. (2013) Efficacy of liposomal amphotericin B and posaconazole in intratracheal models of murine mucormycosis. Antimicrob. Agents Chemother 57, 3340–3347 10.1128/AAC.00313-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamdy R, Ziedan N, Ali S, El-Sadek M, Lashin E, Brancale A et al. (2013) Synthesis and evaluation of 3-(benzylthio)-5-(1H-indol-3-yl)-1,2,4-triazol-4-amines as Bcl-2 inhibitory anticancer agents. Bioorg. Med. Chem. Lett 23, 2391–2394 10.1016/j.bmcl.2013.02.029 [DOI] [PubMed] [Google Scholar]
- 29.Debnath U, Verma S, Singh P, Rawat K, Gupta SK, Tripathi RK et al. (2015) Synthesis, biological evaluation and molecular modeling studies of new 2,3-diheteroaryl thiazolidin-4-ones as NNRTIs. Chem. Biol. Drug. Des 86, 1285–1291 10.1111/cbdd.12591 [DOI] [PubMed] [Google Scholar]
- 30. https://www.playmolecule.org/
- 31.Abraham MJ, der Spoel DV, Lindahl E and Hess B (2016) The GROMACS DevelopmentTeam. GROMACS User Manual version 5.1.2
- 32.da Silva S, and Vranken AW and F, W. (2012) ACPYPE - AnteChamber PYthon parser interfacE. BMC Research Notes. 5, 367–367 10.1186/1756-0500-5-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubo I and Kinst-Hori I (1998) Tyrosinase inhibitors from cumin. J. Agric. Food Chem 46, 5338–5341 10.1021/jf980226+ [DOI] [Google Scholar]
- 34.Saruno R, Kato F and Ikeno T (1979) Kojic acid, a tyrosinase inhibitor from aspergillus albus. Agric. Biol. Chem 43, 1337–1338 10.1080/00021369.1979.10863620 [DOI] [Google Scholar]
- 35.Khan MTH (2007) Heterocyclic compounds against the enzyme tyrosinase essential for melanin production: Biochemical features of inhibition. In Bioactive Heterocycles III (Khan MTH, ed.), pp. 119–138, Springer, Berlin, Heidelberg [Google Scholar]
- 36.Bancroft JD and Gamble M (2008) Theory and Practice of Histological Techniques, Elsevier Health Sciences, Philadelphia, PA, USA [Google Scholar]
- 37. https://www.rcsb.org/
- 38. http://www.chemcomp.com.
- 39.Cavalheiro M and Teixeira MC (2018) Candida biofilms: Threats, challenges, and promising strategies. Front. Med 5, 28 10.3389/fmed.2018.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kassem AF, Batran RZ, Abbas EMH, Elseginy SA, Shaheen MNF and Elmahdy EM (2019) New 4-phenylcoumarin derivatives as potent 3C protease inhibitors: Design, synthesis, anti-HAV effect and molecular modeling. Eur. J. Med. Chem 168, 447–460 10.1016/j.ejmech.2019.02.048 [DOI] [PubMed] [Google Scholar]
- 41.Mohamed TK, Batran RZ, Elseginy SA, Ali MM and Mahmoud AE (2019) Synthesis, anticancer effect and molecular modeling of new thiazolylpyrazolyl coumarin derivatives targeting VEGFR-2 kinase and inducing cell cycle arrest and apoptosis. Bioorg. Chem 85, 253–273 10.1016/j.bioorg.2018.12.040 [DOI] [PubMed] [Google Scholar]
- 42.Gómez BL and Nosanchuk JD (2003) Melanin and fungi. Curr. Opin. Infect. Dis 16, 91–96 10.1097/00001432-200304000-00005 [DOI] [PubMed] [Google Scholar]
- 43.LoPachin RM and Gavin T (2014) Molecular mechanisms of aldehyde toxicity: a chemical perspective. Chem. Res. Toxicol 27, 1081–1091 10.1021/tx5001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.