Abstract
Background
Mitochondria have been shown to play vital roles during severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and coronavirus disease 2019 (COVID-19) development. Currently, it is unclear whether mitochondrial DNA (mtDNA) variants, which define mtDNA haplogroups and determine oxidative phosphorylation performance and reactive oxygen species production, are associated with COVID-19 risk.
Methods
A population-based case–control study was conducted to compare the distribution of mtDNA variations defining mtDNA haplogroups between healthy controls (n = 615) and COVID-19 patients (n = 536). COVID-19 patients were diagnosed based on molecular diagnostics of the viral genome by qPCR and chest X-ray or computed tomography scanning. The exclusion criteria for the healthy controls were any history of disease in the month preceding the study assessment. MtDNA variants defining mtDNA haplogroups were identified by PCR-RFLPs and HVS-I sequencing and determined based on mtDNA phylogenetic analysis using Mitomap Phylogeny. Student’s t-test was used for continuous variables, and Pearson’s chi-squared test or Fisher’s exact test was used for categorical variables. To assess the independent effect of each mtDNA variant defining mtDNA haplogroups, multivariate logistic regression analyses were performed to calculate the odds ratios (ORs) and 95% confidence intervals (CIs) with adjustments for possible confounding factors of age, sex, smoking and diseases (including cardiopulmonary diseases, diabetes, obesity and hypertension) as determined through clinical and radiographic examinations.
Results
Multivariate logistic regression analyses revealed that the most common investigated mtDNA variations (> 10% in the control population) at C5178a (in NADH dehydrogenase subunit 2 gene, ND2) and A249d (in the displacement loop region, D-loop)/T6392C (in cytochrome c oxidase I gene, CO1)/G10310A (in ND3) were associated with a reduced risk of severe COVID-19 (OR = 0.590, 95% CI 0.428–0.814, P = 0.001; and OR = 0.654, 95% CI 0.457–0.936, P = 0.020, respectively), while A4833G (ND2), A4715G (ND2), T3394C (ND1) and G5417A (ND2)/C16257a (D-loop)/C16261T (D-loop) were related to an increased risk of severe COVID-19 (OR = 2.336, 95% CI 1.179–4.608, P = 0.015; OR = 2.033, 95% CI 1.242–3.322, P = 0.005; OR = 3.040, 95% CI 1.522–6.061, P = 0.002; and OR = 2.890, 95% CI 1.199–6.993, P = 0.018, respectively).
Conclusions
This is the first study to explore the association of mtDNA variants with individual’s risk of developing severe COVID-19. Based on the case–control study, we concluded that the common mtDNA variants at C5178a and A249d/T6392C/G10310A might contribute to an individual’s resistance to developing severe COVID-19, whereas A4833G, A4715G, T3394C and G5417A/C16257a/C16261T might increase an individual’s risk of developing severe COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40779-021-00351-2.
Keywords: MtDNA variations, SARS-CoV-2, COVID-19, Risk, Han Chinese
Background
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in a worldwide crisis of formidable morbidity and mortality. The epidemiology, diagnosis, risk factors and treatments of COVID-19 have been explored intensively since the outbreak in Wuhan (Hubei Province, China) in December 2019. Many studies have shown that the clinical features of COVID-19 range from an asymptomatic state to acute respiratory distress syndrome (ARDS) and multiorgan dysfunction. Most COVID-19 patients develop a respiratory tract infection with common symptoms of cough, fever and shortness of breath. Other reported symptoms are weakness, malaise, respiratory distress, muscle pain, sore throat and loss of taste and/or smell. A number of patients develop severe fatal consequences resulting from a surge of inflammatory events (also known as the cytokine storm) [1, 2]. These clinical characteristics hint that although SARS-CoV-2 infection is the causative factor of COVID-19, not all individuals exposed to SARS-CoV-2 will develop COVID-19, especially severe COVID-19, strongly suggesting that the gene-environment interactions exist in COVID-19 progression. An individual’s hereditary susceptibility and innate capacities of antioxidant and immune responses to SARS-CoV-2 might contribute to this process. Currently, known factors related to an individual’s susceptibility to severe COVID-19 include: advanced age; male sex; blood group A in Europeans; comorbidities of cardiopulmonary diseases, diabetes, obesity and hypertension [3]; a genomic segment of ~ 50 kb inherited from Neanderthals currently carried by ~ 50% of people in South Asia and ~ 16% of people in Europe [4]; a 3p21.31 gene cluster [5]; mutations in 13 protein-coding genes of the interferon pathway [6]; and other loci [7]. Whether there are other specific molecular markers to predict the risk of COVID-19 remains unclear.
Recently, mitochondria have been inferred to be interrelated and interacted with oxidative stress and inflammation during SARS-CoV-2 infection and COVID-19 progression [8, 9]. Furthermore, mitochondria have been shown to be indispensable regulators of innate and adaptive immune responses [10] and the activation, development, maintenance and survival of immune cells [11]. These findings provide clues that mitochondria might be associated with an individual’s susceptibility to COVID-19.
As the hub of cellular oxidative homeostasis, mitochondria generate approximately 85% reactive oxygen species (ROS) when they produce usable energy through oxidative phosphorylation (OXPHOS). In contrast to other cellular organelles, mitochondria have their own DNA (mtDNA). Interestingly, the common “nonpathological” mtDNA variation defining mtDNA haplogroups determines OXPHOS performance and ROS production in humans and mice [12]. Additionally, these mtDNA variations exert a considerable influence on longevity [13], help human beings adapt to different environments [14, 15], and are associated with susceptibility to human diseases in conditions where ROS generated by mitochondria play a part [16–19]. Biological functional analysis by introducing human-associated point mutations into yeast mtDNA has revealed that certain human mtDNA variations in the key catalytic domains of mtDNA-CYB significantly changed the complex III activity or drug sensitivity of yeast [20]. Here, we hypothesized that certain mtDNA variants defining mtDNA haplogroups might be related to an individual’s susceptibility to COVID-19. To test this hypothesis, we performed a population-based case–control study for the first time to compare the distribution of mtDNA variants defining mtDNA haplogroups between COVID-19 patients and healthy controls in a Han Chinese population from Central China.
Methods
Study population
This population-based case–control study was approved by the Ethics Committee of Hubei University of Medicine (Hubei, China) (2020-TH-063 and 2020-TH-064). COVID-19 patients (n = 536) were recruited from Taihe Hospital (the First Affiliated Hospital of Hubei University of Medicine, Shiyan, Hubei Province, China) and the People’s Hospital of Hubei Province (the Affiliated Hospital of Wuhan University, Wuhan, Hubei Province, China) from February 2020 to March 2020. COVID-19 patients were diagnosed based on molecular diagnostics of the viral genome by qPCR and chest X-ray or computed tomography scanning, and stratified into cases with moderate (non-ICU) and severe (ICU) disease (patients with ARDS, multiple organ dysfunction, or metabolic acidosis). Age- and sex-matched healthy volunteers (n = 615) were individuals who underwent physical examinations at the two hospitals. The exclusion criterion for the healthy controls was any history of disease in the one-month preceding the study assessment. All subjects were unrelated for at least three generations. After explaining the purpose and procedures of the study, all the participants signed a written informed consent form and completed a detailed questionnaire on their smoking habits.
Genomic DNA extraction
Three millilitres of peripheral blood from each subject were drawn into Na-EDTA tubes. After incubation at 55 °C for 30 min to inactivate the potential SARS-CoV-2, the blood samples were stored at -80 °C prior to genomic DNA extraction. Genomic DNA was extracted from peripheral blood using the Ezup Column Blood Genomic DNA Purification Kit (Lot#: B518253-0100, Sangon Biotech Co., Ltd, Shanghai, China).
Detection of mtDNA variations defining mtDNA haplogroups
MtDNA variants defining mtDNA haplogroups were identified using PCR-restriction fragment length polymorphism (PCR–RFLP) and replenished by hypervariable segment I (HVS-I) sequencing as previously described [15, 16, 18, 19]. Briefly, after the entire mtDNA was amplified into 22 overlapping PCR fragments, the PCR fragments were digested with different restriction endonucleases and replenished by sequencing HVS-I. Two × Taq Plus Master Mix II was used for PCR–RFLP and HVS-I sequencing (Lot#: P213-01, Vazyme Co., Ltd, Nanjing, China). The restriction endonucleases AluI, AvaII, BamHI, BstNI, DdeI, HaeII, HaeIII, HhaI, HincII and HinfI were used in this study (Takara Co., Ltd, Dalian, China). The primers for PCR-RFLPs and HVS-I sequencing were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China), and the primer sequences are presented in Additional file 1: Table S1. The mtDNA polymorphisms defining mtDNA haplogroups were determined based on mtDNA phylogenetic analysis using Mitomap Phylogeny [14]. MtDNA variants were given in the format [ancestral base][position number][derived base]. The Human Genome Variation Society (HGVS) validation of the mtDNA variants is presented in Additional file 1: Table S2.
Comparison of the mtDNA variants defining mtDNA haplogroups between cases and controls
To explore whether the mtDNA variants defining mtDNA haplogroups were associated with individual’s susceptibility to COVID-19, the distribution of mtDNA variants defining mtDNA haplogroups were compared between the pooled cases and controls after the mtDNA variants defining mtDNA haplogroups were identified using PCR–RFLP replenished by HVS-I sequencing for all of the subjects. Additionally, the distribution of mtDNA variants defining mtDNA haplogroups were analysed between controls and moderate cases or severe cases.
Data analysis
Student’s t-test was used for continuous variables, and Pearson’s chi-squared test or Fisher’s exact test was used for categorical variables. For multiple comparisons of mtDNA variants defining mtDNA haplogroups, Bonferroni correction was applied (the required significance level was P = 0.05/number of comparisons). To assess the independent effect of each mtDNA variant defining mtDNA haplogroups, multivariate logistic regression analyses were performed to calculate the odds ratios (ORs) and 95% confidence intervals (CIs) with adjustments for the possible confounding factors of age, sex, smoking and diseases (including cardiopulmonary diseases, diabetes, obesity and hypertension) as determined through clinical and radiographic examinations. All statistical analyses were performed using SPSS Statistics 25 for Mac (SPSS Inc., Chicago, IL, USA).
Results
MtDNA variations defining mtDNA haplogroups in controls and pooled cases
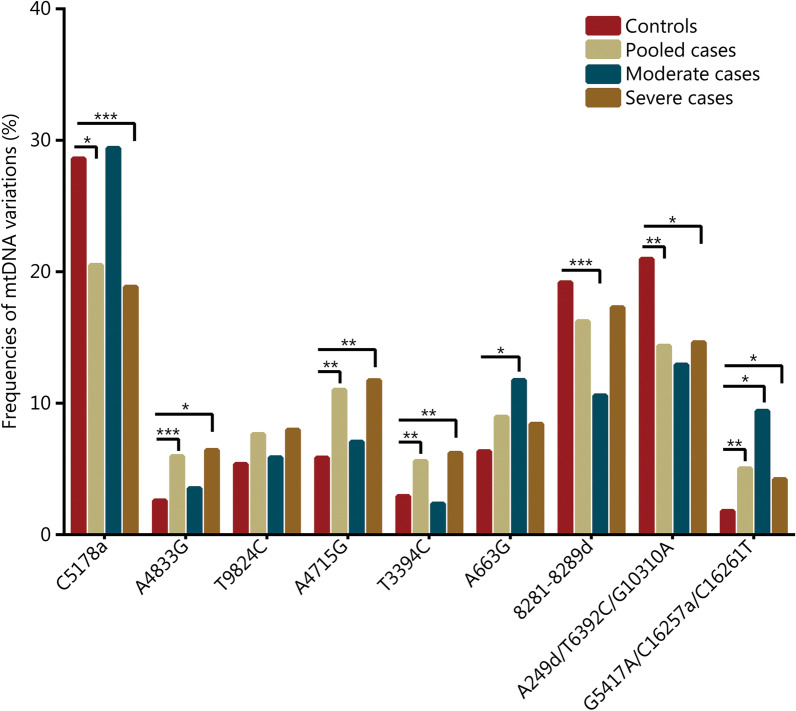
In total, 536 unrelated COVID-19 patients and 615 healthy controls were recruited in this study. As shown in Table 1, the COVID-19 patients smoked more cigarettes than controls (P < 0.001). After the cases were stratified into moderate and severe cases, the severe patients were found to be older than controls (P < 0.001). MtDNA variants defining mtDNA haplogroups were detected for all subjects. Pearson’s chi-squared test or Fisher’s exact test showed that mtDNA variants A4833G (ND2, defining mtDNA haplogroup G), A4715G (ND2, defining mtDNA haplogroup M8), T3394C (ND1, defining mtDNA haplogroup M9), and G5417A (ND2)/C16257a (D-loop)/C16261T (D-loop) (defining mtDNA haplogroup N9a) were significantly higher (P = 0.005, 0.002, 0.027 and 0.003, respectively), while mtDNA variants C5178a (ND2, defining haplogroup D; the letter “a” indicates nucleotide transversion) and A249d (D-loop)/T6392C (cytochrome c oxidase I gene, CO1)/G10310A (ND3) (defining haplogroup F; the letter “d” indicates nucleotide deletion) were significantly lower in COVID-19 patients than in controls (P = 0.002 and 0.004, respectively). When Bonferroni correction was applied, A4715G, G5417A/C16257a/C16261T and C5178a reached the required P value of < 0.0033 (0.05/15). Multivariate logistic regression analyses with adjustments for age, sex, smoking and diseases revealed that, based on a P value of < 0.05, A4833G, A4715G, T3394C and G5417A/C16257a/C16261T were associated with an increased risk of COVID-19 (OR = 4.384, 95% CI 2.103–9.137, P < 0.001; OR = 1.876, 95% CI 1.167–3.021, P = 0.009; OR = 2.618, 95% CI 1.339–5.128, P = 0.005; and OR = 3.401, 95% CI 1.486–7.752, P = 0.004, respectively). In contrast, C5178a and A249d (D-loop)/T6392C (CO1)/G10310A (ND3) variants were associated with a reduced risk of COVID-19 (OR = 0.681, 95% CI 0.504–0.919, P = 0.012; and OR = 0.639, 95% CI 0.483–0.847, P = 0.002, respectively) (Table 2 and Fig. 1).
Table 1.
Clinical characteristics of the study population
| Characteristic | Controls (n = 615) | COVID-19 patients | ||||||
|---|---|---|---|---|---|---|---|---|
| Pooled (n = 536) | P value | Moderate (n = 85) | P value | Severe (n = 451) | P value* | P value# | ||
| Gender [n(%)] | 0.590a | 0.559a | 0.413a | 0.280a | ||||
| Male | 356 (57.89) | 319 (59.51) | 46 (54.11) | 273 (60.53) | ||||
| Female | 259 (42.11) | 217 (40.49) | 39 (45.88) | 178 (39.47) | ||||
| Age (years, ± SD) | 58.29 ± 13.87 | 60.83 ± 14.46 | 0.052b | 59.96 ± 14.10 | 0.300b | 61.68 ± 14.83 | < 0.001b | 0.324b |
| Age [n (%)] | 0.689a | 0.977a | 0.458a | 0.634a | ||||
| ≤ 40 | 48 (7.87) | 46 (8.58) | 6 (7.09) | 40 (8.87) | ||||
| 40–50 | 106 (17.24) | 82 (15.30) | 15 (17.65) | 67 (14.86) | ||||
| 50–60 | 112 (18.21) | 91 (16.98) | 17 (20.00) | 74 (16.41) | ||||
| 60–69 | 217 (35.28) | 186 (34.70) | 31 (36.47) | 155 (34.37) | ||||
| > 70 | 132 (21.46) | 131 (24.44) | 16 (18.82) | 115 (25.50) | ||||
| Pack-years of smoking [n(%)]c | 0.098a | 0.522a | 0.858a | 0.509a | ||||
| 0–20 | 543 (88.29) | 455 (84.89) | 70 (82.35) | 385 (85.37) | ||||
| > 20 | 72 (11.71) | 81 (15.11) | 15 (17.65) | 66 (14.63) | ||||
| Mean pack-years (year, ± SD) | 6.35 ± 10.86 | 9.27 ± 12.54 | < 0.001b | 8.76 ± 10.24 | 0.054b | 10.87 ± 15.67 | < 0.001b | 0.233b |
aχ2-test or Fisher's exact test
bt-test
cThe median number of pack-years of cigarette smoking of the pooled COVID-19 patients and healthy controls was utilized as the cut-off point to stratify the subjects (1 pack-year = 20 cigarettes per day for 1 year)
*Severe cases versus controls
#Severe cases versus moderate cases
Table 2.
Frequencies of mtDNA variants defining mtDNA haplogroups among controls and pooled cases
| MtDNA variations | mtDNA haplogroups | Controls (n = 615) [n(%)] | Pooled cases (n = 536) [n(%)] | P value (χ2)a | Adjusted P valueb | OR (95% CI)b |
|---|---|---|---|---|---|---|
| A663G | A | 39 (6.34) | 48 (8.96) | 0.117 | 0.060 | 1.567 (0.980–2.500) |
| 8281-8289d c | B | 118 (19.19) | 87 (16.23) | 0.217 | 0.150 | 0.781 (0.557–1.093) |
| C5178a c | D | 176 (28.62) | 110 (20.52) | 0.002 | 0.012 | 0.681 (0.504–0.919) |
| A249d/T6392C/G10310A | F (F1 + F3) | 129 (20.98) | 77 (14.37) | 0.004 | 0.002 | 0.639 (0.483–0.847) |
| A4833G | G | 16 (2.60) | 32 (5.97) | 0.005 | < 0.001 | 4.384 (2.103–9.137) |
| T9824C | M7 | 33 (5.37) | 41 (7.65) | 0.119 | 0.519 | 1.181 (0.712–1.957) |
| A4715G | M8 (M8a + C + Z) | 36 (5.85) | 59 (11.01) | 0.002 | 0.009 | 1.876 (1.167–3.021) |
| T3394C | M9 | 18 (2.93) | 30 (5.60) | 0.027 | 0.005 | 2.618 (1.339–5.128) |
| G16274A | M2 | 8 (1.30) | 5 (0.93) | 0.399 | 0.896 | 0.899 (0.180–4.484) |
| T16126C | M3 | 10 (1.63) | 7 (1.30) | 0.616 | 0.466 | 0.572 (0.128–2.567) |
| T16311C | M10 | 9 (1.46) | 6 (1.12) | 0.592 | 0.418 | 0.509 (0.099–2.610) |
| G6023A | M13 | 4 (0.65) | 4 (0.75) | 1.000 | 0.514 | 0.566 (0.102–3.137) |
| T489C/C10400T/T14783C/G15043A | M* | 1 (0.16) | 1 (0.19) | 1.000 | NA | |
| G5417A/C16257a/C16261T | N9a | 11 (1.79) | 27 (5.04) | 0.003 | 0.004 | 3.401 (1.486–7.752) |
| T1391C/T16311C | R1 | 7 (1.14) | 2 (0.37) | 0.187 | 0.219 | 0.353 (0.067–1.859) |
aχ2-test or Fisher's exact test. Bonferroni corrected P < 0.05/n (n = 15)
bP value and ORs (95% CIs) determined by multivariate logistic regression analysis, adjusted for age, gender, smoking and diseases (including cardiopulmonary diseases, diabetes, obesity and hypertension determined through clinical and radiographic examinations)
cThe letter “d” indicates nucleotide deletion, the letter "a" indicates nucleotide transversion
NA not available
Fig. 1.
Frequencies of mtDNA variants defining mtDNA haplogroups in controls and COVID-19 patients. Significant differences between the two groups are labelled as follows: *P < 0.05, **P < 0.01, ***P < 0.001 [adjusted P value was determined by multivariate logistic regression analysis, adjusted for age, sex, smoking and diseases (including cardiopulmonary diseases, diabetes, obesity and hypertension)]
Frequencies of mtDNA variants defining mtDNA haplogroups in controls and moderate cases
When COVID-19 patients were stratified into cases with moderate (non-ICU) and severe (ICU) disease (patients with ARDS, multiple organ dysfunction, or metabolic acidosis), Pearson’s chi-squared test or Fisher’s exact test showed that the mtDNA variants A663G (12S rRNA, defining mtDNA haplogroup A) and G5417A/C16257a/C16261T were significantly higher (P ≤ 0.001), while the 8281–8289d variant (specific to mtDNA haplogroup B, located in a non-coding region) was significantly lower in moderate COVID-19 patients than in controls (P < 0.001). All of these mtDNA variants (A663G, G5417A/C16257a/C16261T, and 8281–8289d) reached the required P value of < 0.0033 when Bonferroni correction was applied. Multivariate logistic regression analyses with adjustments for age, sex, smoking and diseases showed that 8281–8289d was associated with a reduced risk of moderate COVID-19 (OR = 0.034, 95% CI 0.016–0.068, P < 0.001), whereas A663G and G5417A/C16257a/C16261T variants were related to an increased risk of moderate COVID-19 (OR = 2.445, 95% CI 1.116–5.348, P = 0.026 and OR = 3.984, 95% CI 1.325–11.904, P = 0.014, respectively) (Table 3 and Fig. 1).
Table 3.
Distribution of mtDNA variants defining mtDNA haplogroups among controls and moderate cases
| MtDNA variations | MtDNA haplogroups | Controls (n = 615) [n(%)] | Moderate cases (n = 85)[n(%)] | P value (χ2)a | Adjusted P valueb | OR (95% CI)b |
|---|---|---|---|---|---|---|
| A663G | A | 39 (6.34) | 10 (11.76) | < 0.001 | 0.026 | 2.445 (1.116–5.348) |
| 8281–8289d c | B | 118 (19.19) | 9 (10.59) | < 0.001 | < 0.001 | 0.034 (0.016–0.068) |
| C5178a c | D | 176 (28.62) | 25 (29.41) | 0.898 | 0.362 | 1.289 (0.747–2.222) |
| A249d/T6392C/G10310A | F | 129 (20.98) | 11 (12.94) | 0.110 | 0.092 | 1.969 (1.896–4.326) |
| A4833G | G | 16 (2.60) | 3 (3.53) | 0.494 | 0.712 | 1.182 (0.486–2.875) |
| T9824C | M7 | 33 (5.37) | 5 (5.88) | 1.000 | 0.765 | 1.185 (0.389–3.610) |
| A4715G | M8 (M8a + C + Z) | 36 (5.85) | 6 (7.06) | 0.808 | 0.910 | 1.449 (0.700–3.003) |
| T3394C | M9 | 18 (2.93) | 2 (2.35) | 1.000 | 0.903 | 0.910 (0.199–4.167) |
| T489C/C10400T/T14783C/G15043A |
M* (M2 + M3 + M10 + M13 + M) |
32 (5.20) | 6 (7.06) | 0.446 | NA | |
| G5417A/C16257a/C16261T | N9a | 11 (1.79) | 8 (9.41) | 0.001 | 0.014 | 3.984 (1.325–11.904) |
| T1391C/T16311C | R1 | 7 (1.14) | 0 | 0.607 | 0.999 | 0 |
aχ2-test or Fisher's exact test. Bonferroni corrected P < 0.05/n (n = 15)
bAdjusted P value and ORs (95% CIs) determined by multivariate logistic regression analysis, adjusted for age, gender, smoking and diseases (including cardiopulmonary diseases, diabetes, obesity and hypertension determined through clinical and radiographic examinations)
cThe letter “d” indicates nucleotide deletion, the letter “a” indicates nucleotide transversion
NA not available
Distribution of mtDNA variants defining mtDNA haplogroups in controls and severe cases
Pearson’s chi-squared test or Fisher’s exact test demonstrated that the variants A4833G, A4715G, T3394C and G5417A/C16257a/C16261T were significantly higher (P = 0.003, 0.001, 0.010 and 0.023, respectively), while C5178a and A249d/T6392C/G10310A were significantly lower in severe COVID-19 patients than in controls (P < 0.001 and P = 0.008, respectively). When Bonferroni correction was applied, A4833G, A4715G and C5178a remained significant. Multivariate logistic regression analyses with adjustments for covariates showed that C5178a and A249d/T6392C/G10310A variants were associated with a reduced risk of severe COVID-19 (OR = 0.590, 95% CI 0.428–0.814, P = 0.001 and OR = 0.654, 95% CI 0.457–0.936, P = 0.020), while A4833G, A4715G, T3394C and G5417A/C16257a/C16261T variants were related to an increased risk of severe COVID-19 (OR = 2.336, 95% CI 1.179–4.608, P = 0.015; OR = 2.033, 95% CI 1.242–3.322, P = 0.005; OR = 3.040, 95% CI 1.522–6.061, P = 0.002; and OR = 2.890, 95% CI 1.199–6.993, P = 0.018, respectively) (Table 4 and Fig. 1).
Table 4.
Distribution of mtDNA variants defining mtDNA haplogroups among controls and severe cases
| MtDNA variations | MtDNA haplogroups | Controls (n = 615) [n(%)] | Severe cases (n = 451) [n(%)] | P value (χ2)a | Adjusted P valueb | OR (95% CI) b |
|---|---|---|---|---|---|---|
| A663G | A | 39 (6.34) | 38 (8.43) | 0.231 | 0.141 | 1.451 (0.884–2.381) |
| 8281–8289d c | B | 118 (19.19) | 78 (17.29) | 0.472 | 0.202 | 0.796 (0.561–1.130) |
| C5178a c | D | 176 (28.62) | 85 (18.85) | < 0.001 | 0.001 | 0.590 (0.428–0.814) |
| A249d/T6392C/G10310A | F | 129 (20.98) | 66 (14.63) | 0.008 | 0.020 | 0.654 (0.457–0.936) |
| A4833G | G | 16 (2.60) | 29 (6.43) | 0.003 | 0.015 | 2.336 (1.179–4.608) |
| T9824C | M7 | 33 (5.37) | 36 (7.98) | 0.101 | 0.390 | 0.795 (0.472–1.341) |
| A4715G | M8 (M8a + C + Z) | 36 (5.85) | 53 (11.75) | 0.001 | 0.005 | 2.033 (1.242–3.322) |
| T3394C | M9 | 18 (2.93) | 28 (6.21) | 0.010 | 0.002 | 3.040 (1.522–6.061) |
| T489C/C10400T/T14783C/G15043A |
M* (M2 + M10 + M11 + M13a + M) |
32 (5.20) | 17 (3.77) | NA | ||
| G5417A/C16257a/C16261T | N9a | 11 (1.79) | 19 (4.21) | 0.023 | 0.018 | 2.890 (1.199–6.993) |
| T1391C/T16311C | R1 | 7 (1.14) | 2 (0.44) | 0.316 | 0.358 | 0.460 (0.088–2.410) |
aχ2-test or Fisher's exact test. Bonferroni corrected P < 0.05/n (n = 15)
bAdjusted P value and ORs (95% CIs) determined by multivariate logistic regression analysis, adjusted for age, gender, smoking and diseases (including cardiopulmonary diseases, diabetes, obesity and hypertension determined through clinical and radiographic examinations)
cThe letter “d” indicates nucleotide deletion, the letter “a” indicates nucleotide transversion
NA not available
Discussion
To provide insight into COVID-19 risk in terms of mtDNA variation, we conducted a case–control study in a Han Chinese population from Central China. Our data demonstrated that the most common investigated mtDNA variants (> 10% in the control population) at C5178a (ND2) and A249d (D-loop)/T6392C (CO1)/G10310A (ND3) contributed to an individual’s resistance to developing severe COVID-19, whereas A4833G (ND2), A4715G (ND2), T3394C (ND1) and G5417A (ND2)/C16257a (D-loop)/C16261T (D-loop) polymorphisms increased the risk of severe COVID-19 in this population. Additionally, the mtDNA variants A663G (12S rRNA) and G5417A/C16257a/C16261T increased an individual’s risk of developing moderate COVID-19, while the 8281–8289d variant (located in a non-coding region) decreased an individual’s risk of developing moderate COVID-19. The mtDNA variants G5417A/C16257a/C16261T were risk factors for both moderate and severe COVID-19.
As the hub of cellular oxidative homeostasis, mitochondria not only play a role in oxidative stress and inflammation in SARS-CoV-2 infection and COVID-19 development [8, 9, 21–24], but are also indispensable regulators of the innate and adaptive immune responses in the process of SARS-CoV-2 infection and COVID-19 development [10, 11, 24–26]. Moreover, mitochondrial residency of SARS-CoV-2 with a stronger signal compared to its coronavirus relatives further implied that mitochondria are the major cellular organelle affected by oxidative stress and inflammation caused by SARS-CoV-2 infection [27]. In comparison with nuclear DNA, mtDNA is particularly susceptible to oxidative damage due to its direct exposure to ROS, limited DNA repair capacity and absence of protection by histones. The decline in mitochondrial function with aging might explain the phenomena of high mortality rate in elderly COVID-19 patients to a certain extent [1, 13, 17]. Thus, when SARS-CoV-2 infects cells, the common “nonpathological” mtDNA variants, which define mtDNA haplogroups and determine OXPHOS performance and ROS production, contribute to an individual’s capacity for antioxidant and immune responses to protect cells from SARS-CoV-2 infection and COVID-19 development or can aggravate the process. Consistent with this, the mtDNA variation C5178a (Leu237Met) in ND2, defining mtDNA haplogroup D and proposed to be an efficient oxidant scavenger [28], was significantly lower in both the total cohort of COVID-19 patients and severe COVID-19 patients compared to controls in this study. The protective effect of the C5178a mtDNA variant has been reported to increase human longevity [13], to be beneficial for diabetic patients against atherosclerotic and myocardial infarction [29], and to decrease an individual’s risk of developing acute mountain sickness, lung cancer, chronic obstructive pulmonary disease (COPD) and other diseases [16–18, 29]. Therefore, the protective effect of C5178a against oxidative damage as an efficient oxidant scavenger might protect cells from the oxidative destruction caused by SARS-CoV-2 infection and decrease an individual’s risk of developing COVID-19, especially for severe COVID-19.
The A249d (D-loop), T6392C (CO1, synonymous mutation) and G10310A (ND3, synonymous mutation) variants are common variants of mtDNA sub-haplogroups F1–4 [30]. In our study, G12406A (Val24Ile in ND5, defining mtDNA sub-haplogroup F1) and T16298C/C16304T/T16362C (D-loop, defining mtDNA sub-haplogroup F3) were detected, and both had significantly lower frequencies in COVID-19 patients (P = 0.043 and 0.018, respectively). As expected, the combined sub-haplogroups F1 and F3 (representing haplogroup F) were associated with a decreased risk of COVID-19 and severe COVID-19. In Asian populations, haplogroup F is a positive factor associated with a long life-span [31], confers beneficial effects on the resistance of metabolic syndrome (MetS) [32], and improves the physical performance of athletes [33]. A249d occurs within the H-strand replication origin and mitochondrial transcription factor A (mtTF1) binding site, suggesting that A249d may have an impact on mtDNA replication and transcription. Recently, synonymous mutations have been reported to alter translation speed through codon optimality and protein folding in the nuclear genome, which may impact cell fitness [34]. Thus, we deduced that the variants A249d (D-loop)/T6392C (CO1)/G10310A (ND3) might have protective functions against COVID-19 by regulating the replication and transcription of mtDNA and/or the translation speed through codon optimization, protein folding or other mechanisms.
The mtDNA variants A4833G (Thr122Ala in NADH dehydrogenase subunit 2, ND2), A4715G (synonymous mutation in ND2), T3394C (Tyr30His in ND1) and G5417A (synonymous mutation in ND2)/C16257a (D-loop)/C16261T (D-loop) were found to increase an individual’s risk of developing severe COVID-19 in our study. Of note, A4715G, T3394C and G5417A/C16257a/C16261T are reported to be associated with an increased risk of type II diabetes mellitus (T2DM) in the Chinese population [35]. A4715G is a risk factor for moderate and severe non-alcoholic fatty liver disease [36]. Meanwhile, T3394C helps native Tibetans adapt to hypoxic environments because it has higher complex I activity [15]. However, in low-altitude areas, T3394C increases an individual’s risk of many diseases, including Leber’s hereditary optic neuropathy [37], hypertension [38] and T2DM [39]. Additionally, T3394C is a candidate variant that counteracts longevity [40]. Similar to T3394C, A4833G is significantly more frequent in native Tibetans residing at high altitudes than in Han Chinese individuals living in low-altitude areas (32/289 vs. 39/1605, P = 0.0001) [15], which might help native Tibetans adapt to hypoxic environments. Similarly, A4833G is a risk factor for lung cancer [18], COPD [19] and recurrent oral ulceration in low-altitude areas [41]. Therefore, T3394C and A4833G might be risk factors for severe COVID-19 through the same mechanism as in other human diseases occurring in the low-altitude areas.
The mtDNA variant G5417A in ND2 (synonymous mutation, specific for mtDNA haplogroup N9) confers a higher risk of MetS development in HIV-infected patients [42]. In the Chinese population, G5417A/C16257a/C16261T is a risk factor for diabetic nephropathy due to more ROS and fragmented mitochondria [35], which might account for it being a risk factor for both moderate and severe COVID-19 in this Han Chinese population.
Interestingly, certain variants in human mtDNA significantly change specific OXPHOS enzyme activity and response to OXPHOS targeting agents in yeast models [20], indicating that the mtDNA variants detected in this study might alter COVID-19 severity by affecting specific OXPHOS enzyme activity and the response to certain drugs/treatments. Therapeutic regimens based on the variant status of mtDNA may improve the outcomes of patients with COVID-19.
Of note, a mtDNA deletion of approximately 800 bp was detected during the PCR–RFLP analysis in this study. In our previous studies, an 822 bp mtDNA deletion was identified and demonstrated to be positively associated with cigarette smoking and mtDNA haplogroups [18, 19]. Because the blood samples used in this study were incubated at 55 °C for 30 min to inactivate any potential SARS-CoV-2, the mtDNA deletion was not further analysed to avoid possible mtDNA breakage caused by incubation at a higher temperature.
This study had some limitations. First, during the extreme clinical circumstances of the pandemic, especially at the beginning of the epidemic, we were unable to recruit asymptomatic patients and collect detailed clinical data (for example, levels of inflammatory cytokines, immune factors and disease outcome) in a very short period of time, which will be important to investigate in follow-up studies. Second, PCR–RFLP and HVS-I sequencing approaches were used to identify mtDNA variants in the study. Although they can detect known variants defining mtDNA haplogroups, these methods are unable to detect other mtDNA variants and heteroplasmy information on all variants, as other novel techniques such as the next generation sequencing (NGS) of mtDNA do. In future projects, NGS and other novel methods will be utilized to provide more comprehensive information on mtDNA variation. Third, two mtDNA macrohaplogroups M and N, emerging from African-specific mtDNA L3 in Northeast Africa, left Africa successfully and colonized the rest of the world. The mtDNA macrohaplogroup N gave rise to multiple European, Asian and Native American mtDNA lineages (including N1, N2, N9, N*, A, X, R0, JT, R9, R*, B and U, while mtDNA macrohaplogroup M gave rise to only Asian and Native American haplogroups (consisting of M7, M8, M9, G, D and M*). Of all the Asian mtDNA lineages, only haplogroups A, C, D and X became enriched in Northeast Siberia and crossed the Bering Land Bridge into the Americas and haplogroup B colonized the Pacific Islands, demonstrating that haplogroups A, C, D and X have been subjected to much greater cold stress than haplogroup B or the African macro-haplogroup L [43, 44]. These findings show that human mtDNA exhibits dramatic and region-specific sequence variation in geographically localized indigenous populations. In China, with economic development and population flow, the frequencies of mtDNA variations defining mtDNA haplogroups vary across populations. Because we only analysed mtDNA variations in the Han Chinese population from Central China, large-scale studies are needed in other populations.
Conclusions
Our findings revealed for the first time that the common mtDNA variants at C5178a (ND2) and A249d (D-loop)/T6392C (CO1)/G10310A (ND3) contributed to resistance to COVID-19 development, whereas A4833G (ND2), A4715G (ND2), T3394C (ND1) and G5417A (ND2)/C16257a (D-loop)/C16261T (D-loop) variants may be risk factors, providing evidence that the gene-environment interactions exist in COVID-19 progression.
Supplementary Information
Additional file 1: Table S1. The primers for PCR-RFLPs and HVS-I sequencing. Table S2. The Human Genome Variation Society (HGVS) validation of mtDNA variations mentioned in the study
Acknowledgements
The authors thank all the participants for their willingness to contribute to the understanding of COVID-19. Many thanks to Dr. Bin Qiao for his kind gift of DNA samples from COVID-19 patients (the People’s Hospital of Hubei Province, Wuhan University).
Abbreviations
- ARDS
Acute respiratory distress syndrome
- CIs
Confidence intervals
- CO1
Cytochrome c oxidase I
- COVID-19
Coronavirus disease 2019
- COPD
Chronic obstructive pulmonary disease
- HGVS
The Human Genome Variation Society
- HVS-I
Hypervariable segment I
- HVS-II
Hypervariable segment II
- mtDNA
Mitochondrial DNA
- MetS
Metabolic syndrome
- ND1
NADH dehydrogenase subunit 1
- ND2
NADH dehydrogenase subunit 2
- ND5
NADH dehydrogenase subunit 5
- NGS
The next generation sequencing
- OXPHOS
Oxidative phosphorylation
- ORs
Odds ratios
- PCR–RFLP
PCR-restriction fragment length polymorphism
- ROS
Reactive oxygen species
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- T2DM
Type II diabetes mellitus
Authors' contributions
FYJ and YFW contributed to study conception and design; YW, XHW and XHL performed the experiments; ZCF, CYP, YQL, LYY, LYS, SYZ, WC, LFW and HCM participated in the sample collection, storage and DNA isolation; XHW, XHL, FYJ, and SLY participated in data collection and analysis; FYJ and YFW contributed reagents/materials/analysis tools; FYJ, YFW and JMT wrote the manuscript. All the authors read and approved the final manuscript.
Funding
This work was supported by grants from the Special Project of Contingency Research for COVID-19 (No: 2020XGFYZR11), the Cultivating Project for Young Scholar at Hubei University of Medicine (No: 2018QDJZR01) awarded to Dr. Fuyun Ji, and the Special Project of Contingency Research for COVID-19 at Hubei University of Medicine (No: 2020XGFYZR03) awarded to Dr. Yunfu Wang. The funders had no role in study design, collection, analysis and interpretation of data, writing of the manuscript and in decision to submit the article for publication.
Availability of data and materials
The data that supported the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Hubei University of Medicine (2020-TH-063 and 2020-TH-064). All the participants signed a written informed consent form to participate the project.
Consent for publication
All the authors have reviewed the final version of manuscript and signed a written consent form for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Yi Wu, Xian-Hui Wang and Xi-Hua Li contributed equally to this work
Contributor Information
Yi Wu, Email: wuyihb@126.com.
Xian-Hui Wang, Email: 174409895@qq.com.
Xi-Hua Li, Email: lixh87@126.com.
Li-Yuan Song, Email: 547405688@qq.com.
Shi-Long Yu, Email: 18223171269@163.com.
Zhi-Cheng Fang, Email: 13593751009@163.com.
Yu-Quan Liu, Email: lyqliuyuquan123456@163.com.
Le-Yong Yuan, Email: leyongyuan@whu.edu.cn.
Chun-Yan Peng, Email: chunyan_peng@taihehospital.com.
Shen-Yi Zhang, Email: shenyizhang@foxmail.com.
Wang Cheng, Email: wang_cheng1997@163.com.
Hong-Chao Ma, Email: 179291159@qq.com.
Li-Feng Wang, Email: 2078816836@qq.com.
Jun-Ming Tang, Email: tangjm416@163.com.
Yun-Fu Wang, Email: wangyunfu@hbmu.edu.cn.
Fu-Yun Ji, Email: jifuyun@263.net, Email: jfuyun2018@hbmu.edu.cn.
References
- 1.Huang CL, Wang YM, Li XW, Ren LL, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang CF, Xie J, Zhao L, Fei XC, Zhang H, Tan Y, et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine. 2020;57:102833. doi: 10.1016/j.ebiom.2020.102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou F, Yu T, Du RH, Fan GH, Liu Y, Liu ZB, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeberg H, Paabo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587(7835):610–612. doi: 10.1038/s41586-020-2818-3. [DOI] [PubMed] [Google Scholar]
- 5.Severe Covid-19 GWAS Group. Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, et al. Genomewide association study of severe COVID-19 with respiratory failure. N Engl J Med. 2020;383(16):1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q, Bastard P, Liu Z, Pen JL, Moncada-Velez M, Chen J, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(6515):eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novelli G, Biancolella M, Mehrian-Shai R, Erickson C, Pollitt KJG, Vasiliou V, et al. COVID-19 update: the first 6 months of the pandemic. Hum Genom. 2020;14(1):48. doi: 10.1186/s40246-020-00298-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jumana S, Peyssonnaux C, Singh KK, Edeas M. Mitochondria and microbiota dysfunction in COVID-19 pathogenesis. Mitochondrion. 2020;54:1–7. doi: 10.1016/j.mito.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F, Nie JY, Wang HZ, Zhao Q, Zhang YX. Characteristics of peripheral lymphocyte subset alteration in COVID-19 Pneumonia. J Infect Dis. 2020;221(11):1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42(3):406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angajala A, Lim S, Phillips JB, Kim JH, Clayton Y, You ZB, et al. Diverse roles of mitochondria in immune responses: novel insights into immuno-metabolism. Front Immunol. 2018;9:1605. doi: 10.3389/fimmu.2018.01605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreno-Loshuertos R, Acín-Pérez R, Fernández-Silva P, Movilla N, Pérez-Martos A, Rodriguez de Cordoba S, et al. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet. 2006;38(11):1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka M, Gong JS, Zhang J, Yoneda M, Yagi K. Mitochondrial genotype associated with longevity. Lancet. 1998;351(9097):185–186. doi: 10.1016/S0140-6736(05)78211-8. [DOI] [PubMed] [Google Scholar]
- 14.Mishmar D, Eduardo RP, Golik P, Macaulay V, Clark AG, Hosseini S, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100(1):171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji F, Sharpley MS, Derbeneva O, Alves LS, Qian P, Wang Y, et al. Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans. Proc Natl Acad Sci USA. 2012;109(19):7391–7396. doi: 10.1073/pnas.1202484109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li FX, Ji FY, Zheng SZ, Yao W, Qian GS. MtDNA haplogroups M7 and B in southwestern Han Chinese at risk for acute mountain sickness. Mitochondrion. 2011;11(4):553–558. doi: 10.1016/j.mito.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng SZ, Qian P, Li FX, Qian GS, Wang CZ, Wu GM, et al. Association of mitochondrial DNA variations with lung cancer risk in a Han Chinese population from Southwestern China. PLoS ONE. 2012;7(2):e31322. doi: 10.1371/journal.pone.0031322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng SZ, Wang CZ, Qian GS, Wu GM, Guo RL, Li Q, et al. Role of mtDNA haplogroups in COPD susceptibility in a southwestern Han Chinese population. Free Radic Biol Med. 2012;53(3):473–481. doi: 10.1016/j.freeradbiomed.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 20.Song Z, Laleve A, Vallières C, McGeehan JE, Lloyd RE, Meunier B. Human mitochondrial cytochrome b variants studied in yeast: Not all are silent polymorphisms. Hum Mutat. 2016;37(9):933–941. doi: 10.1002/humu.23024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markus H, Hannah KW, Simon S, Nadine K, Stefan P. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian SF, Xiong Y, Liu H, Niu L, Guo JC, Liao MY, et al. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod Pathol. 2020;33(6):1007–1014. doi: 10.1038/s41379-020-0536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao HL, Zhao LX, Yang J, Tong N, An L, Liu QT, et al. Association between ACE2/ACE balance and pneumocyte apoptosis in a porcine model of acute pulmonary thromboembolism with cardiac arrest. Mol Med Rep. 2018;17(3):4221–4228. doi: 10.3892/mmr.2018.8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D, Guo R, Lei L, Liu HJ, Wang YW, Wang YL, et al. COVID-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes. J Leukoc Biol. 2021;109(1):13–22. doi: 10.1002/JLB.4HI0720-470R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann RF, Zarrintan S, Brandenburg SM, Kol A, de Bruin HG, Jafari S, et al. Prolonged cigarette smoke exposure alters mitochondrial structure and function in airway epithelial cells. Respir Res. 2013;14(1):97. doi: 10.1186/1465-9921-14-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann RF, Jonker MR, Brandenburg SM, de Bruin HG, Ten Hacken NHT, van Oosterhout AJM, et al. Mitochondrial dysfunction increases pro-inflammatory cytokine production and impairs repair and corticosteroid responsiveness in lung epithelium. Sci Rep. 2019;9(1):15047. doi: 10.1038/s41598-019-51517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu KE, Zou J, Chang HY. RNA-GPS Predicts SARS-CoV-2 RNA Localization to host mitochondria and nucleolus. Cell Syst. 2020;11(1):102–108.e3. doi: 10.1016/j.cels.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA. 1996;93(26):15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takagi K, Yamada YJ, Gong JS, Sone T, Yokota M, Tanaka M. Association of a 5178C→A (Leu237Met) polymorphism in the mitochondrial DNA with a low prevalence of myocardial infarction in Japanese individuals. Atherosclerosis. 2004;175(2):281–286. doi: 10.1016/j.atherosclerosis.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Yao YG, Kong QP, Wang CY, Zhu CL, Zhang YP. Different matrilineal contributions to genetic structure of ethnic groups in the Silk Road region in China. Mol Biol Evol. 2004;21(12):2265–2280. doi: 10.1093/molbev/msh238. [DOI] [PubMed] [Google Scholar]
- 31.Feng J, Zhang JY, Liu M, Wan G, Qi KY, Zheng CG, et al. Association of mtDNA haplogroup F with healthy longevity in the female Chuang population. China Exp Gerontol. 2011;46(12):987–993. doi: 10.1016/j.exger.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Hu C, He X, Li X, Sun L, Zheng C, Liang Q, et al. Comparative study for the association of mitochondrial haplogroup F+ and metabolic syndrome between longevity and control population in Guangxi Zhuang Autonomous Region. China J Nutr Health Aging. 2018;22(2):302–307. doi: 10.1007/s12603-017-0915-2. [DOI] [PubMed] [Google Scholar]
- 33.InWook H, Kim K, Choi EJ, Han J. Association of mitochondrial haplogroup F with physical performance in Korean population. Genom Inform. 2019;17(1):e11. doi: 10.5808/GI.2019.17.1.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh IM, Bowman MA, Soto Santarriaga IF, Rodriguez A, Clark PL. Synonymous codon substitutions perturb cotranslational protein folding in vivo and impair cell fitness. Proc Natl Acad Sci USA. 2020;117(7):3528–3534. doi: 10.1073/pnas.1907126117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang HZ, Hu NQ, Zhao QY, Wang BQ, Zhou HB. MtDNA haplogroup N9a increases the risk of type 2 diabetes by altering mitochondrial function and intracellular mitochondrial signals. Diabetes. 2018;67(7):1441–1453. doi: 10.2337/db17-0974. [DOI] [PubMed] [Google Scholar]
- 36.Lu MY, Huang JF, Liao YC, Bai RK, Trieu RB, Chuang WL, et al. Mitochondrial polymorphism 12361A>G is associated with nonalcoholic fatty liver disease. Transl Res. 2012;159(1):58–59. doi: 10.1016/j.trsl.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 37.Nakaso K, Adachi Y, Fusayasu E, Doi K, Imamura K, Yasui K, et al. Leber's hereditary optic neuropathy with olivocerebellar degeneration due to G11778A and T3394C mutations in the mitochondrial DNA. J Clin Neurol. 2012;8(3):230–234. doi: 10.3988/jcn.2012.8.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao JZ, Chen CG, Lin WH, Dong ZB, Xu JZ. Clinical and molecular features of a Han Chinese family with maternally transmitted hypertension. Int J Clin Exp Pathol. 2017;10(7):7384–7389. [PMC free article] [PubMed] [Google Scholar]
- 39.Tang DL, Zhou X, Li X, Zhao L, Liu F. Variation of mitochondrial gene and the association with type 2 diabetes mellitus in a Chinese population. Diabetes Res Clin Pract. 2006;73(1):77–82. doi: 10.1016/j.diabres.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Zheng HX, Liu ZY, Qin ZD, Chen F, Qian DG, et al. Mitochondrial genomes and exceptional longevity in a Chinese population: the Rugao longevity study. Age (Dordr) 2015;37(1):9750. doi: 10.1007/s11357-015-9750-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun M, Fu SM, Wang LF, Dong GY, Wu D. Hypervariable region polymorphism of mtDNA of recurrent oral ulceration in Chinese. PLoS ONE. 2012;7(9):e45359. doi: 10.1371/journal.pone.0045359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao D, Ding YY, Lin HJ, Chen XX, Shen WW. Mitochondrial haplogroups N9 and G are associated with metabolic syndrome among human immunodeficiency virus-infected patients in China. AIDS Res Hum Retroviruses. 2019;35(6):536–543. doi: 10.1089/aid.2018.0151. [DOI] [PubMed] [Google Scholar]
- 43.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303(5655):223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 44.Wallace DC. Mitochondrial DNA variation in human radiation and disease. Cell. 2015;163(1):33–38. doi: 10.1016/j.cell.2015.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The primers for PCR-RFLPs and HVS-I sequencing. Table S2. The Human Genome Variation Society (HGVS) validation of mtDNA variations mentioned in the study
Data Availability Statement
The data that supported the findings of this study are available from the corresponding author upon reasonable request.