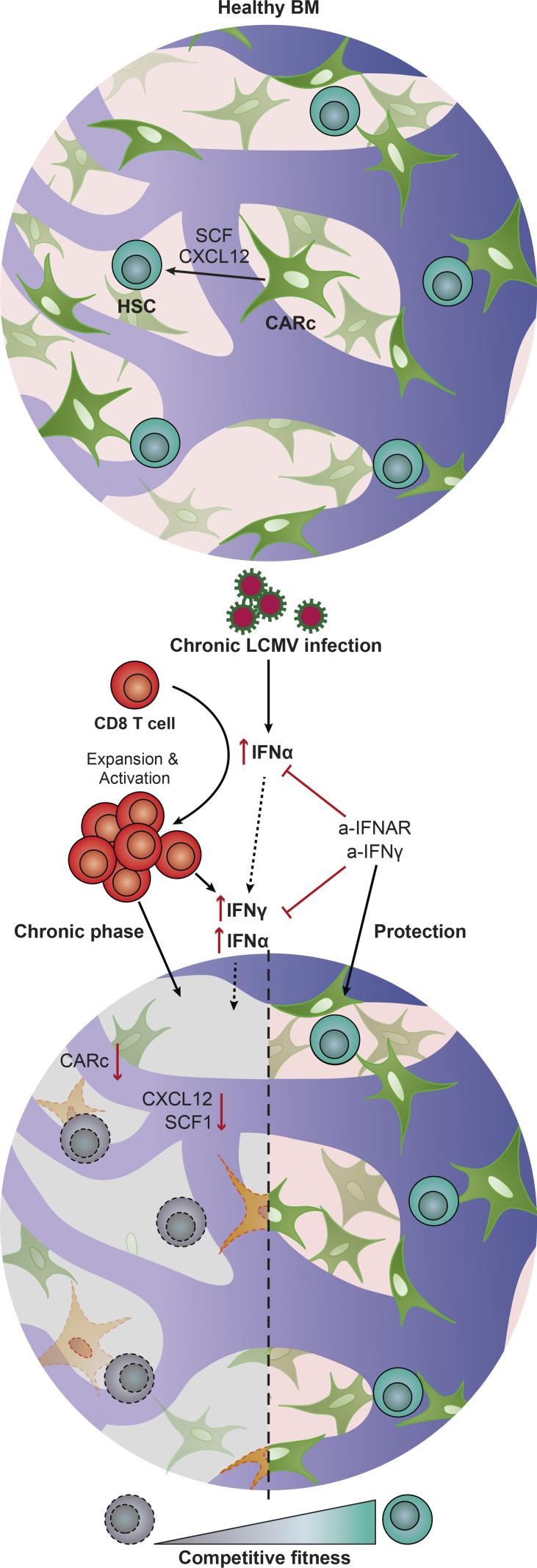
Isringhausen et al. demonstrate that chronic viral infection causes durable destruction of BM mesenchymal stromal networks and leads to long-lasting impairment of the competitive fitness of HSCs. In vivo blockage of IFN pathways protects BM and HSC function from viral-induced damage.
Abstract
Chronic viral infections are associated with hematopoietic suppression, bone marrow (BM) failure, and hematopoietic stem cell (HSC) exhaustion. However, how persistent viral challenge and inflammatory responses target BM tissues and perturb hematopoietic competence remains poorly understood. Here, we combine functional analyses with advanced 3D microscopy to demonstrate that chronic infection with lymphocytic choriomeningitis virus leads to (1) long-lasting decimation of the BM stromal network of mesenchymal CXCL12-abundant reticular cells, (2) proinflammatory transcriptional remodeling of remaining components of this key niche subset, and (3) durable functional defects and decreased competitive fitness in HSCs. Mechanistically, BM immunopathology is elicited by virus-specific, activated CD8 T cells, which accumulate in the BM via interferon-dependent mechanisms. Combined antibody-mediated inhibition of type I and II IFN pathways completely preempts degeneration of CARc and protects HSCs from chronic dysfunction. Hence, viral infections and ensuing immune reactions durably impact BM homeostasis by persistently decreasing the competitive fitness of HSCs and disrupting essential stromal-derived, hematopoietic-supporting cues.
Graphical Abstract
Introduction
The hematopoietic system has evolved to generate billions of mature cells on a daily basis (Kaushansky, 2006; Nombela-Arrieta and Manz, 2017). This unparalleled output is achieved through the organization of the system as a cellular hierarchy initiated by hematopoietic stem cells (HSCs), which self-renew while sequentially differentiating along lineage-restricted pathways, giving rise to mature blood cell derivatives (Kondo et al., 2003). During adulthood, this complex developmental process takes place in bone marrow (BM) within a specialized tissue microenvironment provided by a constellation of nonhematopoietic stromal cells (Crane et al., 2017; Morrison and Scadden, 2014). The stromal infrastructure is absolutely essential for proper hematopoietic and bone remodeling functions and encompasses a remarkable cellular and functional heterogeneity (Crane et al., 2017; Mercier et al., 2011).
Integral components of BM stroma are fibroblastic Leptin receptor (LepR+), so-called CXCL12-abundant reticular cells (CARcs), which make up the largest fraction of mesenchymal stroma and comprise a variety of multifunctional and heterogeneous adipogenic and osteogenic progenitor cells (Omatsu et al., 2010; Zhou et al., 2014; Baccin et al., 2019; Baryawno et al., 2019; Tikhonova et al., 2019; Wolock et al., 2019 Preprint). CARcs assemble as dense fibrous networks that permeate the entire extravascular space. The majority of CARc bodies are directly associated to the extraluminal side of sinusoidal microvessels and are physically connected through numerous thin cytoplasmic extensions (Gomariz et al., 2018). Through production of extracellular matrix (ECM) and the abundant secretion of a wide variety of key factors, CARcs crucially govern hematopoietic development and are best studied for their contribution to the maintenance of HSCs and progenitors (Crane et al., 2017). For instance, CARc-specific genetic deletion of stem cell factor (SCF) expression results in an almost complete depletion of phenotypic and functional hematopoietic stem and progenitor cell (HSPC) subsets (Ding et al., 2012). CARcs are also the main cellular players involved in attraction to, retention of, and survival of HSCs and multipotent progenitor cells within supportive BM tissues, through the production of the chemoattractant CXCL12 (Greenbaum et al., 2013; Ding and Morrison, 2013). The endothelial compartment is another key functional component of BM stroma. Beyond their role as major routes of cellular trafficking and nutrient delivery, sinusoidal vessels are the source of relevant cytokines involved in hematopoietic control (Ding et al., 2012). Sinusoidal endothelial cells (SECs) additionally secrete SCF, which is needed for homeostatic HSC maintenance, and growth factors that promote hematopoietic regeneration after myeloablative damage (Hooper et al., 2009; Butler et al., 2010; Greenbaum et al., 2013; Poulos et al., 2013; Itkin et al., 2016; Kusumbe et al., 2016). Thus, the preservation of functional and structural integrity of perisinusoidal stromal BM niches is absolutely indispensable for HSC and BM function.
Pathogenic infections result in molecular adaptation of hematopoietic progenitors to inflammatory stress, profound rewiring of hematopoietic developmental circuits, and qualitative and quantitative shifts in mature cell production (Pietras, 2017; Chavakis et al., 2019). In humans, viral infections are also frequently associated with BM alterations, which, depending on the viral agent and duration of infection, range in severity from transient pancytopenias to aplastic anemia or failure (Rosenfeld and Young, 1991; Pascutti et al., 2016). Despite the major health burden imposed by chronic infectious diseases worldwide, surprisingly little is known about the mechanisms by which persistent viral inflammatory states alter the hematopoietic process. Recent work indicates that viral-induced perturbations of hematopoiesis could at least in part result from the direct effects of proinflammatory cytokines and viral-derived molecules on HSPCs (Essers et al., 2009; Baldridge et al., 2010; Pietras et al., 2016). Best studied are the effects of type I and type II IFNs, which are produced early in immune responses and critically promote antiviral states. Type I IFN (IFNα), or its inducer polycytidylic-polyinosinic acid, have been shown to promote cell cycle entry and increase HSC contribution to mature cellular output (Essers et al., 2009; Sato et al., 2009). IFNα-driven proliferation induces DNA damage, and repeated stimulation results in defective HSC function in transplantation assays (Walter et al., 2015). Yet, upon exposure to IFNα, HSCs have also been shown to reenter quiescence as a protective mechanism from proliferation-driven exhaustion (Pietras et al., 2014) Therefore, the effects of type I IFN on HSC properties remain controversial, depend on the duration and timing of IFN production, and need to be explored in the context of life infections (Demerdash et al., 2021). Similarly, type II IFN (IFNγ) enhances HSC proliferation and drives a bias toward myeloid output, which is thought to contribute to rapid responses to infections (Schürch et al., 2014; Matatall et al., 2014; de Bruin et al., 2013). Sustained production of abnormally high levels of IFNγ leads to ablation of the HSC pool in experimental models of recurrent bacterial infections (Morales-Mantilla and King, 2018; Matatall et al., 2016; Baldridge et al., 2010).
Despite recent progress, global and long-term alterations of BM and HSC function have not been thoroughly evaluated using relevant models of life-persistent viral infections, in which complex dynamics of immune cell activation and migration to the BM, as well as the timed modulation of local cytokine levels, are accounted for. As a consequence, potential mechanisms of viral-mediated hematopoietic dysfunction, including the direct and indirect contribution of IFNs, remain unclear. Most importantly, how viral processes alter organization and function of the stromal cell microenvironment, and to what extent this damage may impact BM functional integrity, remain to be investigated (Nombela-Arrieta and Isringhausen, 2017). We employed the paradigmatic pathogenic model of lymphocytic choriomeningitis virus clone-13 strain (LCMV-cl13) infection (Zhou et al., 2012) to comprehensively and kinetically study BM tissue dynamics, assess the damage inflicted on the stromal infrastructure and HSC functionality, and determine the potential roles of immune cell activation and IFN signaling in hematopoietic dysfunction during persistent viral infections.
Results
LCMV infection causes transient BM hypoplasia and prolonged impairment of HSC function
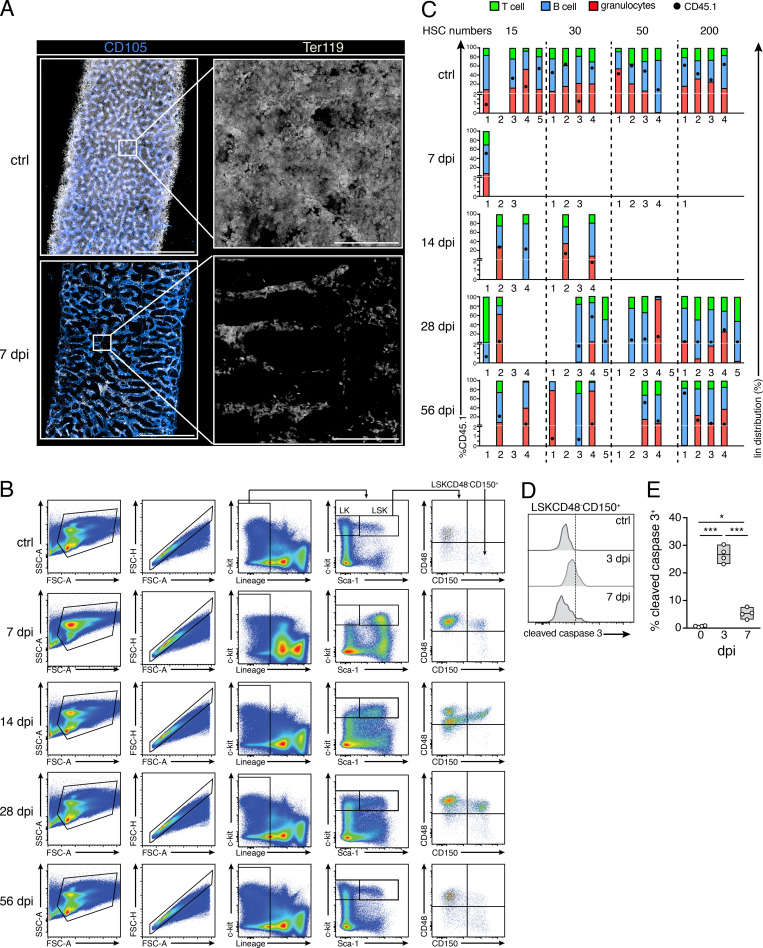
Acute LCMV infection leads to transient and reversible alterations in BM hematopoiesis, peripheral blood (PB) content, and HSC properties (Binder et al., 1997; Matatall et al., 2014). Using flow cytometry (FC) and three-dimentsional quantitative microscopy (3D QM), we performed an in-depth kinetic analysis of the population dynamics of major hematopoietic subsets upon inoculation of high doses of LCMV-cl13 (2 × 106 focus-forming units [ffu]), which leads to establishment of chronic disease. In the initial stages of infection, we detected strong and rapid drops in BM cellularity, largely caused by virtual depletion of progenitors of erythroid and B cell lineages (Fig. 1, A, B, and F). 3D microscopy revealed that the densely populating Ter-119+ erythroid population was almost totally absent from the BM 7 d post infection (dpi). At this point, few erythroid cells, most likely circulating erythrocytes, could be visualized inside sinusoids (Fig. S1 A and Video 1), which caused bones to appear conspicuously pale in these early phases (Fig. 1 C). In contrast, BM-resident CD8 T cells increased in a sustained fashion (up to 56 dpi; Fig. 1 D), while CD4 T cell frequency was reduced during initial infection phases and rose to abnormally high levels from 14 dpi until the end of the observation period (Fig. 1 E).
Figure 1.
Chronic infection with LCMV-cl13 disrupts BM hematopoiesis and leads to long-term impairment of HSC function. (A and B) FC-based quantification of total BM cellularity (A) and Ter-119+ BM erythroid progenitors (B) after infections with 2 × 106 ffu LCMV-cl13. (C) Representative pictures of thick femoral slices of uninfected control (ctrl) or chronic LCMV-cl13 infection (7 dpi). (D–I) FC-based quantification of BM CD8 T cells (D), CD4 T cells (E), and B220+ B cells (F). (G) LK (Lin−c-kit+) progenitors. (H) LSK (Lin−Sca-1+c-kit+) progenitors. (I) HSCs (LSKCD150+CD48−) at different time points after infection. (J) Quantification of Ki67/DAPI cell cycle analysis of HSCs (n = ≥4 mice per group) during the course of infections with LCMV-cl13. Statistical significance was analyzed by two-tailed Mann–Whitney U test for A–J: *, P < 0.05; **, P < 0.01; ***, P < 0.001. (K) Focus-forming assay for LCMV-cl13 in the BM during the course of infections (n = 3–5 mice per time point). (L) Top: Schematic experimental layout for ELDA transplantation performed in at least four replicates per condition. Bottom: Linear regression analyses for the transplantation, with indicated numbers representing ELDA estimates for HSC functionality (n = 4–5 mice per group) and HSC dose. Numbers of transplanted mice were lower for 7 and 14 dpi, given the scarcity of HSCs (see Fig. S1).
Figure S1.
Effects of chronic infections in BM hematopoietic and HSC function. (A) Representative image showing maximum-intensity projection of femoral diaphyseal regions of BM from uninfected control (ctrl) or 7 dpi with LCMV-cl13. Blue, CD105 (BM sinusoids); white, Ter119 (erythroid progenitors). Scale bars, 500 µm (left); 50 µm (right). (B) Gating scheme used to phenotypically define HSC and HSPC populations in the BM of control (CTRL) and infected mice. FSC, forward scatter; SSC, side scatter. (C) Analysis showing CD45.1 donor engraftment (black circle, scale on left y axis) and donor lineage distribution in T cell (green), B cell (blue), and granulocyte (red) compartments as percentage of total CD45.1 engraftment (scale on right y axis). Mice were considered engrafted when the percentage of CD45.1+ cells in PB was >0.5% 4 mo after transplantation in all three lineages. Empty columns indicate total CD45.1 donor engraftment <0.5%. (D and E) Representative histograms and quantification of the fraction of cleaved caspase 3+ HSCs in uninfected mice and 3 and 7 dpi. Statistical significance was analyzed by Mann–Whitney U test. *, P < 0.05; ***, P < 0.001.
Video 1.
LCMV infections result in acute and transient depletion of the BM erythroid cellular compartment. 3D confocal microscopy of thick femoral slices from uninfected control mice (ctrl) and mice 7 dpi with LCMV-cl13. Immunostaining against erythroid progenitors (Ter119, white) and BM sinusoidal vessels (CD105, blue). Tissue thickness was 65 µm in whole diaphysis images and 44 µm for zoomed-in images, respectively.
Infections strongly affected HSPCs. Total numbers of lineage−c-kit+ (LK) hematopoietic progenitors dropped sharply and gradually recovered over a period of 4 wk after infection (Fig. 1 G). As previously described, up-regulation of Sca-1 in response to inflammation led to minor increases in the number of Lin−c-kit+Sca-1+ (LSK) cells (Fig. 1 H). Despite this effect, phenotypic HSCs (defined hereafter as LSKCD48−CD150+; Fig. S1 B) were almost completely depleted from BM 7–14 dpi, only recovering to normal levels 2 wk later (28 dpi; Fig. 1 I). Increases in the fraction of apoptotic HSPCs, as determined by cleaved caspase-3 staining, were evident as early as 3 dpi (Fig. S1, D and E), indicating that HSC depletion was not exclusively due to the induction of rapid differentiation. However, the contraction of the HSC pool also correlated with entry into cell cycle and a pronounced reduction of quiescent (G0) HSCs (Fig. 1 J). Increased proliferation persisted until 28 dpi, when HSC numbers and hematopoietic parameters were restored. These effects correlated with the presence of virus in the marrow, which could be detected until 28 dpi (Fig. 1 K). Using extreme limiting dilution (ELDA) transplantation assays, we next assessed whether HSC functionality was altered by infections. Limiting numbers of sorted CD45.1 HSCs (15, 30, 50, or 200 cells) isolated from mice infected at different time points (7, 14, 28, and 56 dpi) were transplanted together with 3.5 × 105 total BM cells from control mice into lethally irradiated CD45.2 mice. PB CD45.1 donor engraftment was monitored for 4 mo after transplantation, when terminal analysis of BM and PB chimerism was performed. These experiments uncovered a drastic functional impairment of HSCs early after LCMV-cl13 inoculation (Fig. 1 L). Strikingly, the repopulation capacity of HSCs remained strongly diminished even at the longest time point analyzed (56 dpi), when the estimated frequency of functional repopulating cells in the phenotypic HSC pool was still decreased approximately fourfold compared with that of control HSCs (Fig. 1 L). Notably, infections did not substantially alter the lineage distribution output of engrafted HSCs (Fig. S1 C). Altogether, these data demonstrate that chronic LCMV-cl13 infection induces strong, reversible responses throughout all hematopoietic lineages, with transient loss of phenotypic HSPCs, prominent induction of proliferation, and long-lasting impairment of HSPC function.
Vascular remodeling and persistent destruction of CARc networks during LCMV-cl13 infection
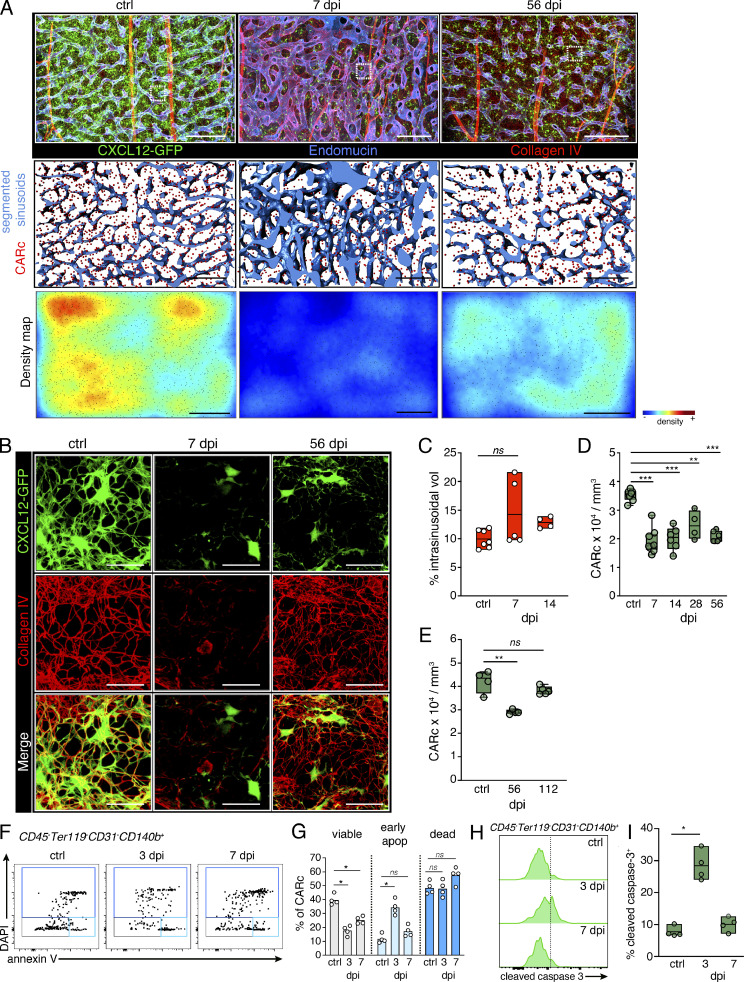
Infections alter lymphoid organ fibroblastic reticular cell (FRC) networks, thereby impairing the mounting of immune responses (Scandella et al., 2008; Mueller et al., 2007). We hypothesized that the hematopoietic defects induced by LCMV-cl13 could at least partially result from the degeneration of stromal support signals. Since stromal cells are largely neglected by FC analyses (Gomariz et al., 2018), we employed 3D QM to study the effects of LCMV-cl13 on this compartment. Early after infection, we observed variable levels of vasodilation of the sinusoidal vessel network and concomitant contraction of the extravascular space. This phenomenon peaked 3–5 dpi and in some cases was still pronounced by 7 dpi (Fig. 2, A and C; Video 2; and not depicted). Notably, high-resolution imaging revealed a marked destruction of the dense network of BM ECM (Fig. 2 B and Video 3). From 7 to 14 dpi, large tissue regions appeared entirely devoid of the otherwise dense meshwork of collagen IV+ fibers, which were completely rebuilt by 56 dpi (Fig. 2 B).
Figure 2.
Chronic LCMV infection induces transient remodeling of BM vascular and ECM networks and persistent destruction of CARc infrastructure. (A) Maximum-intensity projection of confocal image stacks from representative femoral regions of uninfected controls (ctrl), 7-, or 56-dpi Cxcl12-GFP transgenic mice. Segmented signal of sinusoidal vessels stained for CD105 is shown in the middle panel; green, CXCL12-GFP+ CARc; lower panel, tissue maps of CARc densities. Scale bars for all panels, 200 µm. (B) Zoomed-in images from regions demarcated with white dotted rectangles in low-magnification images in A. The CXCL12-GFP signal is shown in green, and collagen IV–specific signal is shown in red (ECM fibers). (C) Quantification of the fraction of intrasinusoidal volumes (vol) from total BM tissue using segmentation of sinusoidal lumina (n = 4–7 femurs from at least three independent experiments). (D) CARc densities measured by 3D QM at indicated time points after infection (n = 4–9 femurs from at least three independent experiments). (E) Quantification of CARc densities by 3D QM at late time points after infections (n = 4 mice, one long-term infection experiment). (F and G) FC analysis of apoptosis (apop) of CARc, staining with DAPI and anti-annexin V. Representative dot plots (F) and quantification (G) of viable (DAPI−annexin V−), early apoptotic (DAPI−annexin V+), and dead (DAPI+) fractions after infection with LCMV-cl13 (n = 4 mice from one representative of two independent experiments). (H and I) Cleaved caspase-3 staining of CARcs; histograms (H) and quantification (I) of positive CARc fractions (n = 4 mice, one representative of two experiments). Statistical significance was analyzed using two-tailed t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Video 2.
Acute and chronic effects of LCMV-cl13 infections on BM stromal infrastructure. 3D confocal microscopy of thick femoral slices from naive mice (ctrl) and mice 7 or 56 dpi with LCMV-cl13. CXCL12-GFP shown in green, segmented CARc spots in red, collagen IV antibody staining in red, endomucin (BM sinusoids) in blue, and c-Kit in white. The tissue thickness imaged in all cases was 80 µm.
Video 3.
Destruction and remodeling of BM ECM scaffolds during chronic LCMV infections. 3D confocal microscopy reconstruction of thick femur slices from naive mice (ctrl), or mice 7 or 56 dpi after chronic LCMV-cl13. CXCL12-GFP shown in green and anti-collagen IV staining in red. The tissue thickness imaged in all slices was 40 µm.
The functional and structural link between sinusoids, ECM, and mesenchymal CARcs raised the possibility that CARc networks could also be disrupted. 3D imaging of Cxcl12-GFP transgenic reporter mice revealed a constant decimation of the CARc population over the course of infection. At initial time points, CARc numbers were strongly diminished, and the characteristic cellular projections that form the CARc continuum appeared retracted or largely absent (Fig. 2, A and B). We confirmed that the reduction of CARc was not due to a down-regulation of Cxcl12 and GFP transgene expression in these cells, since it was also evident when CARc densities were analyzed in infected LepR-Cre/td-Tomato+ mice (Fig. S2 A). Significant increases in the fraction of apoptotic CARcs could be detected as early as 3 dpi (Fig. 2, F–I). In contrast to the effects observed in vasculature, ECM, and hematopoietic compartment, the partial ablation of CARcs was not transient but persisted at least throughout the 8 wk after infection (Fig. 2 D). Nonetheless, long-term damage was reversible, and CARc densities were almost restored by 4 mo after infection (112 dpi), thereby indicating the activation of mechanisms that repair stromal injury once inflammation subsides (Fig. 2 E). LCMV-cl13 reportedly infects monocytic cells, dendritic cells, and megakaryocytes. FC and 3D QM analyses of BM immunostained against LCMV-specific nucleoprotein uncovered that, beyond hematopoietic cells, multiple CARcs and SECs were directly infected by LCMV-cl13 (Fig. S2, C and D). HSCs were spared from infection, thus allowing us to discard direct effects of LCMV infection on their function.
Figure S2.
Chronic LCMV-cl13 directly infects endothelial cells, CARcs, and mature hematopoietic cells. (A and B) Quantification of the cellular density of LepR-Cre/tdTomato+ CARcs in the BM of uninfected mice (CTRL) and 7 dpi with LCMV-cl13 (n = 6 mice per group). Statistics were analyzed by two-tailed Mann–Whitney U test with **, P < 0.005. Representative images in which cells quantified are marked by yellow spheres are shown in B; scale bars, 100 µm. (C) Maximum-intensity projection of a representative femoral cavity 7 dpi with LCMV-cl13. White signal shows transmitted light (bone outline); blue, CD105 (BM sinusoids); red, VL4 (actively replicating LCMV-cl13 particles), and green, CXCL12-GFP (CARc). Scale bars, 2 mm (left, whole femur); 400 µm (middle, whole diaphysis); 50 µm (right, zoomed image). Arrows indicate infected GFP+ CARcs. (D) Representative dot plots for intracellular staining for viral nucleoprotein using VL4. Percentages of LCMV-cl13–infected cells in the BM 7 dpi are shown for LSKCD48−CD150+ HSCs (orange), erythroid progenitors (red), BM endothelial cells (blue), and BM CARcs (green), respectively. Levels of specific isotype control stainings for each population are shown in overlaid dark plots. FSC, forward scatter; MEP, megakaryocyte-erythroid progenitor cell.
Infection persistently affects transcriptional status and functionality of CARcs
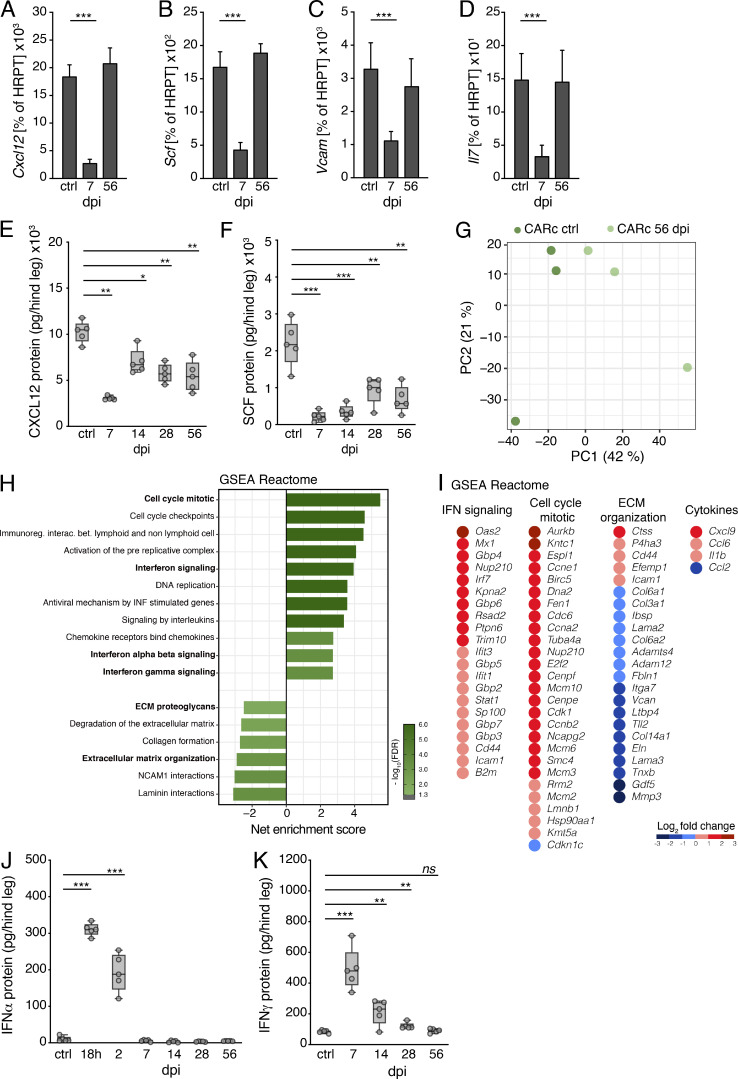
Beyond the quantitative changes, we also observed alterations in the functionality of CARcs surviving the infectious challenge. Expression of Cxcl12, Scf, Vcam-1, and Il-7 dropped in initial phases, but gradually recovered to normal levels after 8 wk (Fig. 3, A–D). Since CARcs produce the highest levels of these cytokines, the qualitative and quantitative effects observed in fibroblastic networks translated into a very pronounced decline in the overall protein concentrations of CXCL12 and SCF in BM lysates, which remained strongly reduced throughout infections (Fig. 3, E and F). To better assess the long-term impact on gene expression, we performed RNA sequencing (RNA-seq) on highly purified CARc and SEC populations sorted from mice 56 dpi. Principal component analysis revealed evident alterations in transcriptional programs at this late time point, which were more pronounced in CARcs than SECs (Fig. 3 G and not depicted). Up-regulated gene expression pathways in both cell types were fundamentally associated with inflammatory programs, especially those related to IFNα- and IFNγ-induced responses, as evidenced by gene set enrichment analysis (GSEA; Fig. 3, H and I; and Fig. S3 A). These effects were detected even though by 56 dpi, IFN levels in both plasma and BM lysates had long declined to baseline after initial bursts and were not significantly elevated (Fig. 3, J and K; and Fig. S3, B and C). Abnormally high expression levels of classic inflammatory mediators such as Il1b, which are known to impact HSC maintenance and hematopoiesis (Pietras et al., 2016; Ergen et al., 2012), were observed in CARcs (Fig. 3 I). We further observed enriched expression of cell cycle–related genes, which could be linked to the activation of regeneration programs of the CARc network at late stages after infection. Nonetheless, at this point ECM-related genes, including matrisomic proteins, degradation enzymes, and adhesion molecules, were strongly down-regulated (Fig. 3, H and I). Collectively, our data demonstrate that CARcs and, to a lower extent, SECs display inflammation-driven, functional sequelae and a sustained, abnormal, IFN-imprinted gene expression profile for prolonged periods after systemic inflammation has largely receded.
Figure 3.
Durable functional impairment of CARc after chronic LCMV-cl13 infection. (A–D) Expression of key genes involved in HSC maintenance measured by RT-qPCR in sorted BM CARcs of uninfected control (ctrl) or 7 and 56 dpi with LCMV-cl13. Expression of Cxcl12 (A), Scf (B), Vcam (C), and Il-7 (D) are shown as percentage of HRPT expression (n = 3–5 samples from at least two independent experiments). (E–H) Concentrations of CXCL12 (E) and SCF (F) in BM extracts from one hind leg (femur and tibia) at different time points after LCMV-cl13 infection as measured by ELISA. (G) Principal component (PC) analysis plot for RNA-seq analyses of CARcs from uninfected control mice and 56 dpi. (H) GSEA graph for REACTOME gene sets overrepresented and underrepresented in CARcs isolated from the BM of mice 56 dpi compared with uninfected control BM. The net enrichment score (NES) and false discovery rate (FDR) for different gene sets are represented in the graph. Immunoreg. interac. bet., immunoregulation interaction between. (I) Integrated dot plot/heatmaps depicting expression levels from RNA-seq analysis of individual genes from selected REACTOME gene sets in H that are significantly up- or down-regulated in CARc 56 dpi compared with uninfected controls. (J and K) Concentrations of IFNα and IFNγ in BM extracts from one hind leg (femur and tibia) at different time points after LCMV-cl13 infection as measured by ELISA (n = 5 mice from at least two independent experiments). Statistical significance was analyzed by two-tailed Mann–Whitney U test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05..
Figure S3.
Chronic LCMV-cl13 infection results in long-lasting transcriptomic alterations in BM stromal cells. (A) GSEA graph for REACTOME gene sets overrepresented and underrepresented in SECs isolated from the BM of mice 56 dpi, compared with uninfected control BM. The net enrichment score (NES) and false discovery rate (FDR) for different gene sets are represented in the graph. (B and C) Serum protein levels for IFNα (B) and IFNγ (C) at indicated time points after infection with LCMV-cl13 (n = 5 mice per time point), as determined by ELISA. Statistics were analyzed by two-tailed Mann–Whitney U test with *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05.
CD8 T cell infiltration of BM mediates hematopoietic alterations and CARc elimination during infection
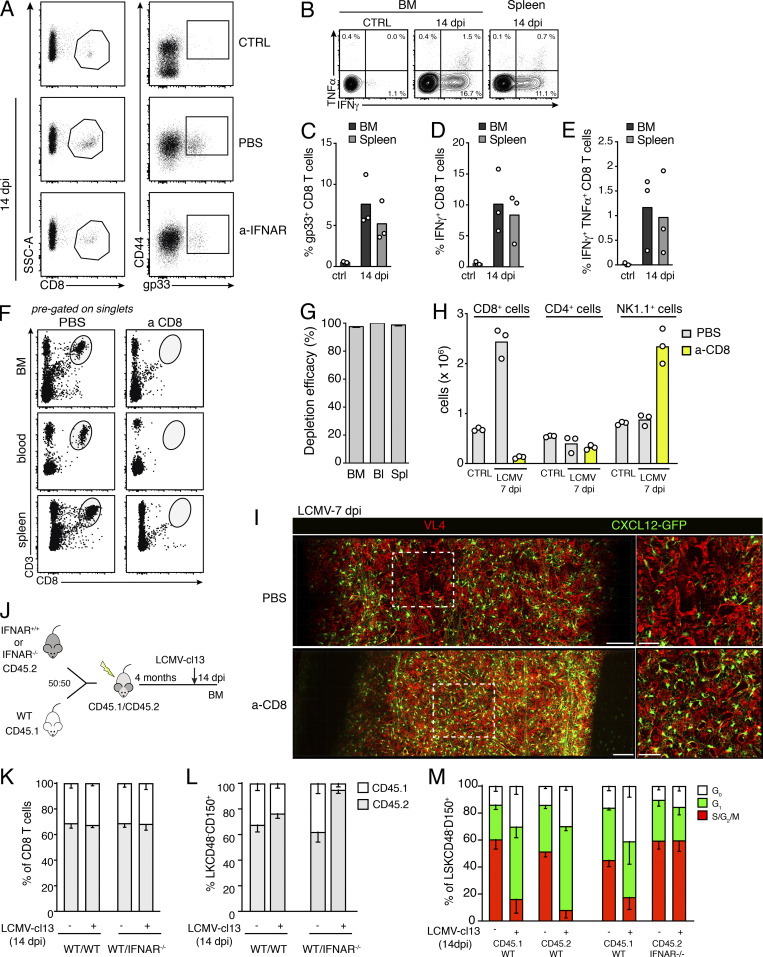
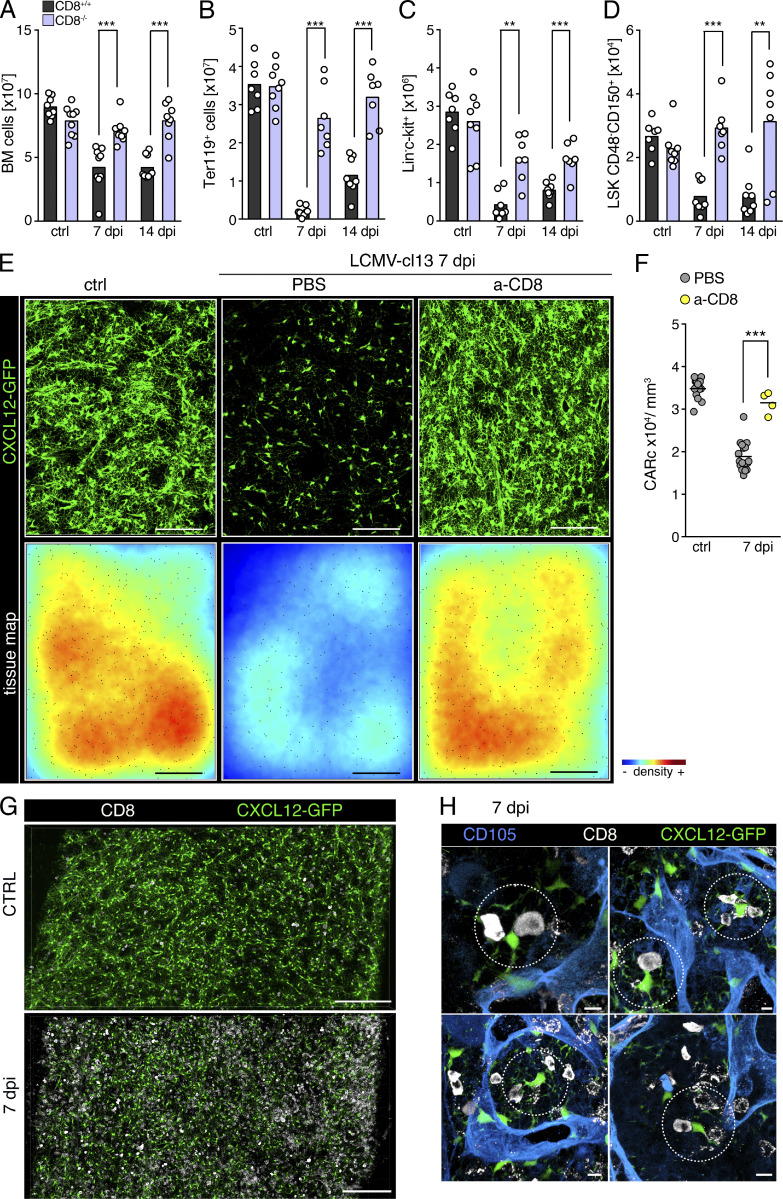
CD8+ T lymphocytes play fundamental roles in antiviral immune responses and infection clearance, but can also cause severe damage to infected tissues (Rouse and Sehrawat, 2010). Functional and phenotypic analyses showed that activated, gp33+ LCMV-specific T cells producing high levels of IFNγ and/or TNFα accumulated in the BM at early time points of immune responses (Fig. S4, A and B). At 14 dpi, the frequencies and phenotype of BM CD8 T cells were comparable to those found in the spleen (Fig. S4, C–E). Thus, we hypothesized that stromal and hematopoietic perturbations could be caused by immunopathologic cytotoxic T cell responses in the BM. To assess this, we analyzed postinfection BM dynamics in WT and CD8α-deficient mice (CD8−/−). Lack of CD8 lymphocytes completely prevented virus-induced BM hypoplasia and erythroid cell depletion (Fig. 4, A and B). Although a mild reduction in hematopoietic progenitor cells was still detectable in CD8−/− mice, the absolute numbers of phenotypic HSCs were not significantly altered (Fig. 4, C and D). To precisely quantify the contribution of CD8+ cells to viral-induced CARc ablation, we employed 3D QM in Cxcl12-GFP reporter mice in which CD8 T cells were depleted through administration of anti-CD8 (a-CD8) antibodies. Antibody-mediated depletion was highly efficient and specific for CD8 T cells before and after infection (7 dpi; Fig. S4, F–H). As observed for the hematopoietic compartment, absence of CD8 T cells largely prevented destruction of CARc networks triggered by viral challenge (Fig. 4, E and F; and Video 4). Stromal protection was almost complete, despite the BM being profusely infiltrated by the virus, as demonstrated by staining with the viral nucleoprotein-specific VL4 antibody (Fig. S4 I). Although we did observe a minor decrease in CARc abundance in CD8 T cell–depleted mice, this was attributed to the incomplete efficiency of antibody-mediated depletion and the presence of a residual CD8 T cell pool in LCMV-infected mice (Fig. S4 H). Microscopic examination revealed that during the early phases of infection, CARcs were mostly found in direct spatial association with CD8 T cells (Fig. 4, G and H). Together with the high abundance of both cell types and the relatively motile nature of CD8 cells in BM (Khan et al., 2018), these data suggest that CARcs are directly exposed to the cytotoxic action and cytokine secretion of CD8 lymphocytes. In summary, CD8 T cells are crucial in driving both the immunopathologic effects on BM stroma and the initial reductions in HSC numbers upon LCMV-cl13 infection.
Figure S4.
CD8 T cells mediate viral-induced effects in the BM. (A) Gating of gp33+ tetramer-specific CD8 T cells in the BM of control mice (CTRL) and mice treated with a-IFNAR 14 dpi with LCMV-cl13. SSC, side scatter. (B) Contour plot showing intracellular immunostaining for IFNγ and TNFα of CD44+CD8+ activated T lymphocytes in uninfected mice (CTRL) or 14 dpi with LCMV-cl13. (C–E) Percentages of CD8 gp33+ T cells (C), IFNγ+CD8+ T cells (D), and IFNγ+TNFα+ CD8+ T lymphocytes (E) in the BM and spleen 14 dpi with LCMV-cl13 (n = 3 mice per time point). (F–H) Anti-CD8 antibody administration efficiently depletes CD8+ T cells from BM and peripheral organs. (F) Dot plots showing representative gating for CD8 T lymphocytes (CD3+CD8+, pregated on singlets) in BM, blood, and spleen before and after antibody-mediated depletion. (G) Quantification of depletion efficacy, n = 4 mice. Depletion was calculated as 100% abundance in depleted mice, with abundance calculated as percentage of normal CD8+ T cell counts. Bl, blood; Spl, spleen. (H) Absolute numbers of CD8+, CD4+, and NK1.1+ cells in the BM of mice treated with PBS or anti-CD8 7 dpi with LCMV-cl13. (I) Maximum-intensity projection of a representative femoral cavity BM of mice treated with PBS or anti-CD8, 7 dpi with LCMV-cl13. Red, VL4 (actively replicating LCMV particles); green, CXCL12-GFP (BM CARcs). Scale bars, 100 µm (left, whole diaphysis); 50 µm (right, zoomed-in image). (J–M) Experimental layout depicting infection of chimeric mice and results of CD8 and HSC ratios and cell cycle status 14 dpi (J). Quantification of the ratio of CD8 T cells (K) and HSPCs (Lin−c-kit+CD48−CD150+ cells; L), in the BM of WT/WT and WT/IFNAR chimeras before and after infection. (M) Cell cycle status of WT and IFNAR−/− HSCs in chimeric mice, before infection and 14 dpi with LCMV-cl13 (n = 2–5 mice per group from two independent experiments).
Figure 4.
CD8 T lymphocytes mediate infection-induced hematopoietic effects and destruction of BM CARc upon chronic infection. (A–D) Total cell counts for BM cellularity (A), BM erythroid progenitors (B), LK hematopoietic progenitors (C), and BM HSCs (D) in CD8+/+ (black bars) and CD8−/− (light purple) mice at different time points after infection with LCMV-cl13 (n = 7–9 samples from at least three independent experiments). (E) Maximum-intensity projection of representative images from large femoral volumes of uninfected mice (ctrl) or mice subjected to LCMV-cl13 infections and treated with PBS or anti-CD8 (a-CD8)-depleting antibody and analyzed 7 dpi. Top: Green, CXCL12-GFP (CARc). Bottom: Tissue maps depicting CARc density levels according to a color-coded scale. Scale bar, 200 µm. (F) CARc densities assessed by 3D QM at indicated time points after infection (n = 4–9 samples from at least two independent experiments) in control and CD8 T cell–depleted mice. (G and H) Representative images of BM CD8+ T cells and CARc in mice before infection and 7 dpi with LCMV-cl13. Low-magnification images of global distribution and frequencies of both cell types are shown in G. (H) Examples of closely interacting CD8 T cell and CARc frequently found throughout BM tissues (dotted circles). Statistics were analyzed by two-tailed t test with **, P < 0.01; ***, P < 0.001; ns, P > 0.05.
Video 4.
Destruction of CARc networks is mediated by CD8 T cells. 3D confocal microscopy of thick femoral slices from control uninfected mice (ctrl), or mice 7 dpi with LCMV-cl13 that were pretreated with PBS or CD8-depleting antibody (a-CD8). CXCL12-GFP shown in green, segmented CARc spots shown in red, and color-coded CARc density map shown at the end of the video. Tissue thickness of all images slices was 50 µm.
Type I IFN signals drive accumulation of activated, antigen-specific CD8 T cells in the BM
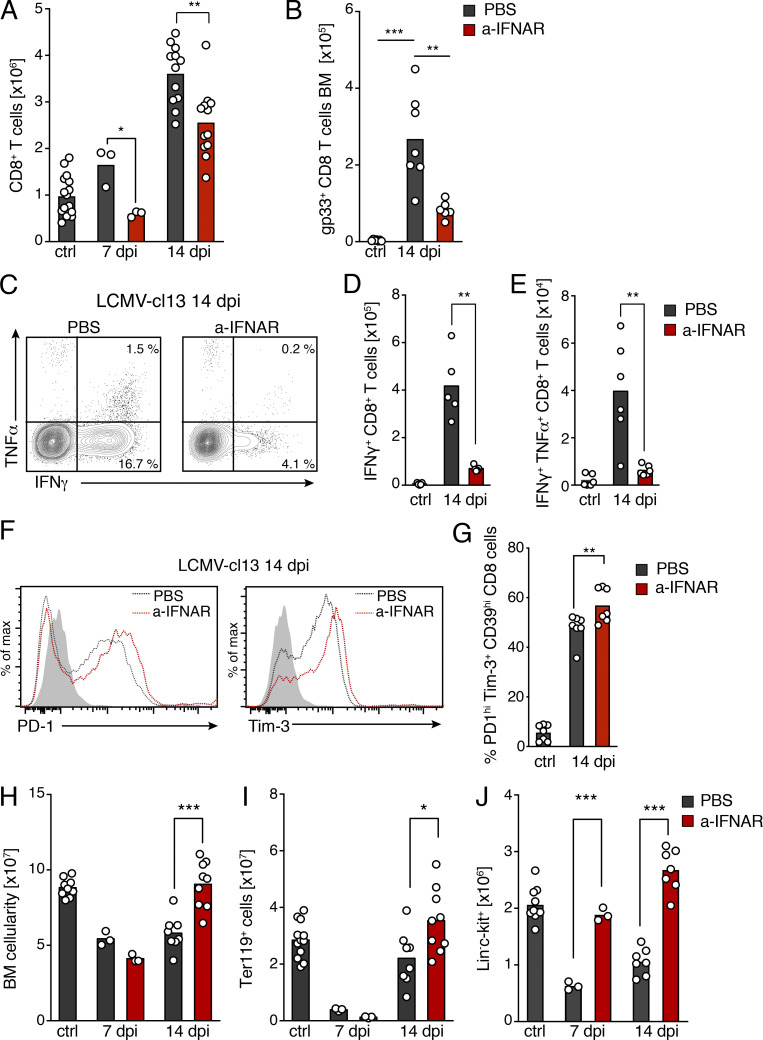
Type I IFNs are central mediators of antiviral immune responses, which control the early expansion of antigen-specific CD8 T cells, promote cytolytic activity, and regulate cytokine secretion (Crouse et al., 2015). Nonetheless, recent work has shown that in the context of persistent viral infections, exacerbated IFN production contributes to CD8 exhaustion, establishment of chronic states of immune activation, and destruction of lymphoid tissue microarchitecture (Teijaro et al., 2013; Wilson et al., 2013). Therefore, we next investigated whether blockade of type I IFN α/β receptor (IFNAR) signaling was sufficient to prevent injury to BM stroma. Because IFNAR-deficient mice display strong phenotypes in immature hematopoietic compartments and HSC cycling behavior (Essers et al., 2009; Baldridge et al., 2010), which can be confounding, we used antibody-mediated inhibition of type I IFN receptor signaling in WT mice. Blockage of IFNAR signaling partially mitigated accumulation of CD8 T cells in BM 7 and 14 dpi (Fig. 5 A). Nonetheless, in IFNAR−/−/IFNAR+/+ chimeric mice, infection led to similar recruitment of both IFNAR-deficient and -sufficient CD8 T cells, as their ratios were maintained in the blood and BM after infection (Fig. S4, J and K). Thus, IFNAR signaling is most likely not required for CD8 T cell homing or recruitment to the BM. Notably, IFNAR blockage specifically led to a very pronounced reduction of gp33+ antigen-specific and cytokine-producing activated CD8 T cells, and an increased expression of prototypical markers of cellular exhaustion upon infection (Fig. 5, B–G). These observations are therefore consistent with the major role of IFNAR as a key third signal during antiviral responses for priming and differentiation of CD8 T cells endowed with antiviral effector functions (Kolumam et al., 2005; Crouse et al., 2015). Of major importance, impairment of CD8 T cell responses in the BM correlated with a partial mitigation of infection-driven alterations in the cellular composition of BM tissues. For instance, although transient BM hypoplasia was observed at initial time points, restoration of BM cellularity was accelerated and complete within 2 wk of infection when IFNAR signaling was inhibited (Fig. 5 H). Similarly, effects on hematopoietic progenitors were detectable, although they were milder and followed faster kinetics of recovery (Fig. 5, I and J).
Figure 5.
Type I IFN signaling drives BM accumulation of activated, antigen-specific CD8 T cells. (A and B) Total cell counts of BM CD8 T lymphocytes (A) and gp33+ LCMV-specific BM CD8 T cells (B). Black bars, control mice treated with PBS before or after infection; red bars, data from mice treated with a-IFNAR blocking antibody during infection (see Materials and methods for dosage and regimens). (C) Contour plot of intracellular immunostaining for IFNγ and TNFα in BM CD8 T cells 14 dpi with or without a-IFNAR treatment. (D) Histograms showing PD-1 (left) and Tim-3 (right) expression of BM CD8 T cells 14 dpi in control and a-IFNAR–treated mice. (E) Total number of IFNγ+ CD8 T cells. (F) IFNγ+TNF-α+ CD8 T cells. (G) Percentage of PD-1hiTim-3+ T cells in total BM CD8 T cells at 14 dpi. (H) Total BM cellularity. (I and J) BM erythroid progenitors (I) and Lin−c-kit+ progenitor cells (J; n = 5–7 mice from two or three independent experiments). Statistics were analyzed using two-tailed Mann–Whitney U test with *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05.
Combined blockage of type I and II IFN signaling abrogates viral-induced injury of BM stroma
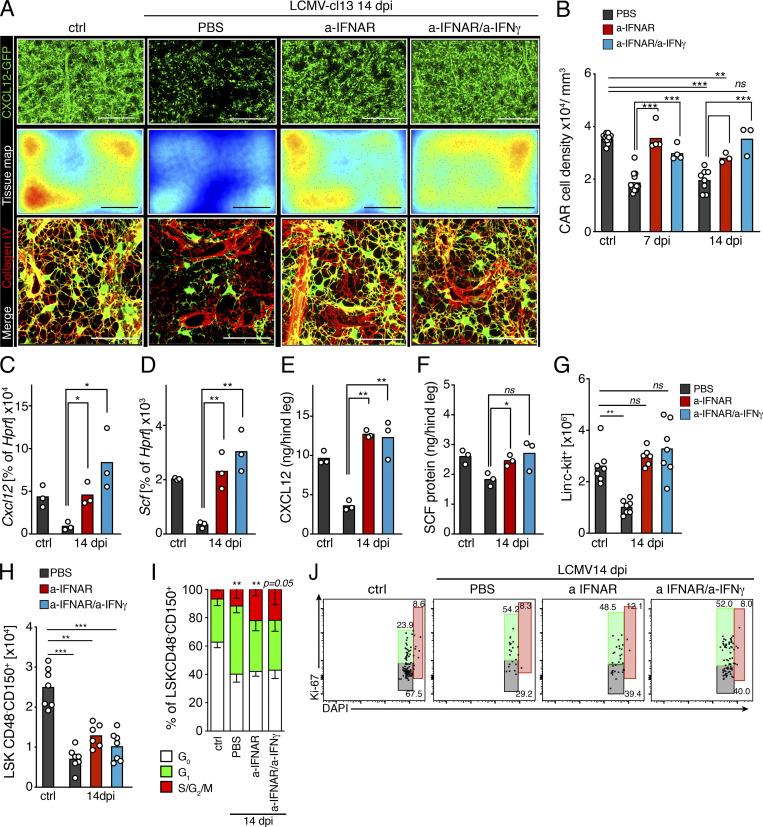
To test the potential role of IFN signaling on stromal alterations, we next investigated postinfection BM dynamics in mice receiving IFNAR blocking treatment. Administration of anti-IFNAR (a-IFNAR) had a drastic protective effect in BM stroma, strongly blocking depletion of CARcs and preserving gene expression levels of prohematopoietic factors and the integrity of ECM networks (Fig. 6, A–D; and Video 5). Nonetheless, detailed quantification of 3D images uncovered a mild, yet significant, reduction of CARc densities 14 dpi in a-IFNAR–treated mice, compared with control counterparts, which correlated with residual presence of IFNγ-producing CD8 T cells in the BM. We hypothesized that remaining loss of CARc could be mediated by local IFNγ production, which can be further exacerbated in the context of type I IFN blockade (Teijaro et al., 2013). While blockage with anti-IFNγ alone did not result in detectable preservation of CARc or progenitor numbers after infection (Fig. S5, A–F), simultaneous blockage of both IFNAR and IFNγ fully preserved CARc densities and intact networks of ECM fibers at all time points after infection analyzed (Fig. 6, A and B). This correlated with a complete restoration of protein concentrations of CXCL12 and SCF in whole BM extracts in mice undergoing combined treatment. In fact, expression levels of both factors tended to be elevated compared with controls when both type I and II IFN signaling were blocked, thereby indicating that under homeostatic conditions, tonic levels of IFN signaling may negatively regulate CARc function (Fig. 6, E and F). In summary, blockade of IFNAR signaling early in infection strongly reduces the frequencies of activated CD8 T cells and restores the supportive capacity of remaining CARc subsets. Additional inhibition of IFNγ signaling completely blunts infection-mediated damage to the CARc supportive network. Nonetheless, inhibition of IFNAR, IFNγ, or both pathways simultaneously failed to prevent the initial drop in numbers of phenotypically defined HSCs and did not block infection-induced entry into cell cycle of the HSC pool (Fig. 6, H–J; and Fig. S5 E). Of note, in chimeric WT/IFNAR−/− chimeric mice, in which CD8 T cell function was partially mitigated (Fig. S4 K), loss of HSCs and cell cycle activation mostly affected IFNAR-competent HSCs, while largely sparing those lacking the IFNAR receptor (Fig. S4, L and M). These results suggest that IFN-mediated effects on HSCs early after infection are complex and the result of both direct and possibly indirect mechanisms. Nevertheless, while antibody-mediated blockage of IFN signaling is sufficient to preclude stromal damage, it fails to prevent the initial quantitative alterations of the HSC pool.
Figure 6.
Type I and II IFN signaling mediates structural and functional damage to BM CARc networks. (A) Maximum-intensity projection of representative images of femoral bones of mice 14 dpi with LCMV-cl13 treated with PBS, a-IFNAR alone, or a-IFNAR and anti-IFNγ. Top: Green, CXCL12-GFP (CARc); scale bar, 200 µm. Middle: Color-coded tissue maps of CARc densities. Scale bar, 200 µm. Bottom: High-resolution images of zoomed-in regions depicting CARc in green and collagen IV ECM fibers in red. Scale bar, 50 µm. (B) 3D QM–based quantification of CARc density in BM tissues from uninfected (ctrl) and LCMV-cl13–infected mice (black bars) treated with a-IFNAR (red bars) or a combination of a-IFNAR and a-IFNγ (blue bars) 7 and 14 dpi (n = 3–9 samples from at least two independent experiments). (C and D) RT-qPCR of sorted BM CARc for Cxcl12 (C) and Scf (D) gene expression as percentage of HRPT expression (n = 3) for the same experimental groups as in B. (E and F) Protein concentrations of CXCL12 (E) and SCF (F) in BM extracts from one hind leg (femur + tibia; n = 3). (G and H) Absolute numbers of Lin−c-kit+ cells (G) and HSCs (H) in mice in all experimental groups 14 dpi (six mice, two independent experiments). (I and J) Quantification and representative dot plots of cell cycle analyses of HSCs as measured by staining for DAPI and Ki67 after infection with LCMV-cl13 of mice treated with IFN-blocking antibodies. Statistics were analyzed using two-tailed Mann–Whitney U test with *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05.
Video 5.
Inhibition of type I IFN signaling prevents infection-induced damage to CARc and ECM networks. 3D confocal microscopy of thick femur slices either from uninfected control mice (ctrl) or mice 14 dpi treated with PBS or type I IFN-blocking antibody (a-IFNαR). CXCL12-GFP shown in green, segmented CARc spots (whole diaphysis image) and collagen IV fibers (zoomed-in image) shown in red, color-coded CARc density map shown at the end of the video. Thickness of tissue slices was 60 µm for whole diaphysis images and 30 µm for zoomed-in images.
Figure S5.
Effects of IFN blockage on the BM and HSCs of infected and non-infected mice. (A–E) Quantification of total BM cells (A), Ter119+ erythroid progenitors (B), B220+ cells (C), Lin−c-kit+ progenitor cells (D), and LSKCD48−CD150+ cells (E) in mice after infection and blockage with either a-IFNAR or a-IFNγ (n = 5–7 mice from two independent experiments). Statistics were analyzed using Mann–Whitney U test with *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05. (F) Image-based quantification of CARc densities in the BM of mice 7 and 14 dpi with LCMV-cl13 in untreated and a-IFNγ–treated mice (n = 3–4 mice, two experiments). (G) Blockade of IFNAR does not impact cell cycle entry of HSCs without LCMV-cl13 infection. Dot plots showing representative cell cycle analyses of HSCs (LSKCD48−CD150+) using DNA labeling (DAPI) and immunostaining against Ki-67. HSCs were isolated 7 d after injection with either PBS or a-IFNAR antibody. (H) Quantification of cell cycle analyses, n = 3 mice. Statistics were analyzed by Mann–Whitney test. (I and J) Limiting-dilution transplantations of 10, 20, 50, and 100 HSCs (LSKCD48−CD150+) from control untreated mice (CTRL) and mice treated with a-IFNAR and a-IFNγ for 56 d. (I and J) Calculation of functional repopulating HSCs (I) and engraftment levels and lineage distribution (J) of donor-derived CD45-1+ cells in transplanted mice (n = 4–5 mice per group). (K) Quantification of divisional (div) history of CFSE-labeled IFNγR−/− LSKCD48−CD150+ HSCs after transplantation into control and LCMV-cl13–infected (56 dpi) mice (n = 5–6 recipient mice from two independent experiments). Statistics were analyzed by two-tailed Mann–Whitney U test with *, P < 0.05; ns, P > 0.05.
Combined blockage of IFNs protects the repopulating activity of HSCs
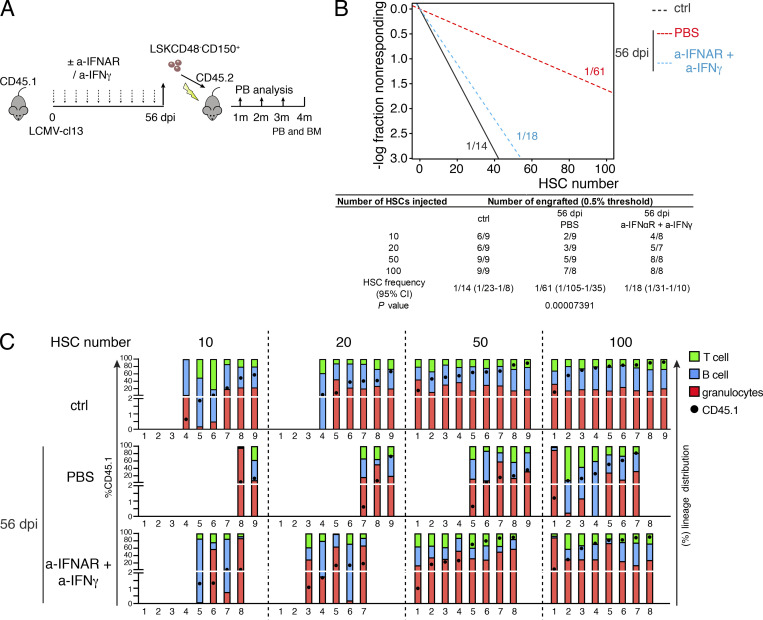
Based on its protective effect on BM stroma, we next asked whether IFN blockage would prevent the long-term loss of HSPC functionality observed in chronic phases of infection. To test this, we sorted HSCs 56 dpi from mice that had been initially treated with either PBS or combined IFNAR/IFNγ blocking therapy. HSCs were transplanted at limiting numbers (10, 20, 50, and 100 cells) together with 3.5 × 105 uninfected CD45.2 BM competitor cells into lethally irradiated recipient mice (Fig. 7 A). Confirming previous results, infections led to a strong and chronic reduction in the number of functional repopulating HSCs (Fig. 7, B and C). Most importantly, blockage of type I and II IFN signaling almost completely shielded HSCs from the appearance of chronic defects in their repopulating activity in these late phases and preserved the frequency of functional repopulating units in the HSC fraction (Fig. 7, B and C). Of note, IFNAR blockage did not increase HSC quiescence, and 56 d of combined a-IFNAR/IFNγ treatment did not result in improved repopulating activity of HSCs from control, noninfected mice, thereby indicating that IFN blockage specifically protects HSC function against the deleterious effects of infection (Fig. S5, G–J).
Figure 7.
Combined blockage of type I and II IFN signaling prevents persistent decline in HSC functionality induced by infection. (A) Schematic experimental layout for ELDA repopulation assays. m, months. (B) Linear regression analysis for the transplantation, with indicated numbers representing ELDA estimates for HSC functionality. Table indicates values for 95% confidence intervals for each experimental group. (C) Individual engraftment analysis showing overall CD45.1 donor engraftment (black circle), as well as donor lineage distribution in T cell (green), B cell (blue), and granulocyte (red) engraftment as percentage of total CD45.1 engraftment. Mice were considered engrafted when PB CD45.1 engraftment was >0.5% in myeloid (Gr1+) and lymphoid lineages (T and B). Empty columns indicate total CD45.1 donor engraftment <0.5%. Transplantation was performed in seven to nine transplanted mice per condition (two independent experiments). Statistics were analyzed using Pearson’s χ2 test, and P values are shown.
Infections increase engraftment of transplanted HSCs by eroding competitive fitness of endogenous HSCs
Next, we attempted to uncouple and independently assess the niche-derived contribution to HSC dysfunction by evaluating the impact of prior chronic infections on the ability of the BM microenvironment to maintain the functionality of unchallenged, naïve HSCs. For this, we first conducted previously established CFSE dilution assays (Takizawa et al., 2011) in which CD45.2 LSK cells were labeled with CFSE and transferred into CD45.1/CD45.2, nonmyeloablated mice that had been either injected with PBS or infected with LCMV-cl13 56 d earlier. 3 wk after transplantation, the divisional history of transplanted HSPCs was assessed by measuring the dilution of the CFSE label using FC (Fig. 8 A). As previously shown, when transplanted into untreated nonirradiated recipient mice, a relatively rare but consistent fraction of HSPCs (8.4 ± 1.9%) did not divide during the 3-wk chase period, displaying undiluted levels of CFSE (Takizawa et al., 2011; Kovtonyuk et al., 2016). In stark contrast, the quiescent fraction of HSPCs transplanted into LCMV-cl13–infected hosts was virtually undetectable (0.6 ± 0.9%), and the overall divisional dynamics were accelerated (44.3 ± 11.0% >4 divisions) compared with those transferred into unchallenged control mice (27.0 ± 5.5% >4 divisions; Fig. 8, B and C). Notably, enhanced divisional rates were also found when IFNγR−/− HSCs were transplanted into previously infected recipients, indicating that increases in proliferation are for the most part independent of the presence of residual levels of IFNγ in the BM of mice 56 dpi (Fig. S5 K).
Figure 8.
LCMV infection results in persistent functional impairment of the BM microenvironment to maintain HSC quiescence. (A) Experimental layout for CFSE label dilution analysis. (B) Representative dot plots for CFSE label dilution 21 d after transplantation of LSK cells into control (ctrl) uninfected or LCMV-cl13–infected mice, 56 dpi. CFSE dilution is shown for HSPC LSKCD48− (left) and HSC LSKCD48−CD150+ (right) subsets. Numbers above dot plots indicate divisions demarcated by gates on the CFSE axis. (C) Quantification of the percentage of transplanted LSKCD48−CD150+ cells undergoing different numbers of divisions (div) based on dilution levels of CFSE (n = 7 recipient mice per time point from two independent experiments are shown in recipient control and LCMV-cl13–infected mice, 56 dpi). Statistics were analyzed by two-tailed Mann–Whitney U test with *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ns, P > 0.05. (D) Experimental layout for long-term transplantation experiments. LSK cells from CD45.2 mice were transplanted into CD45.2/CD45.1 mice uninfected (CTRL) or infected with LCMV-cl13 (56 dpi) treated with PBS or a combination of a-IFNAR and anti-IFNγ. (E–I) Monthly values of CD45.2 chimerism in PB (E) myeloid Gr1+ (F), B220+ (G), CD4+ (H), and CD8+ (I) compartments. (J–L) CD45.2 chimerism in HSCs (LSKCD48−CD150+), multipotential progenitors (MPPs; LSKCD48+CD150−), common myeloid progenitors (CMPs; Lin−Sca-1−ckit+CD16/32−CD34+), granulocyte-monocyte progenitors (GMPs; Lin−Sca-1−ckit+CD16/32+CD34+), and common lymphoid progenitors (CLPs; Lin−Sca-1−ckit+/loCD127+) in the BM at terminal analysis 4 mo after transplantation (n = 3 mice per group, one experiment). Statistical significance was analyzed by two-tailed ANOVA with Tukey’s multiple comparison test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05.
We next reasoned that enhanced HSC proliferation in infection-damaged tissue could be explained by an inability of the BM microenvironment to support HSC quiescence, in which case increased divisional rates would lead to the rapid exhaustion of the transplanted HSC pool. To test this possibility, we transplanted large numbers (3 × 106) of unlabeled LSK cells from uninfected mice into groups of congenic, noninfected mice or recipients exposed to LCMV-cl13 (56 dpi). Engraftment was then tracked in blood and BM for a period of 16 wk (Fig. 8 D). As expected, CD45.2+ HSCs transplanted into noninfected CD45.2+/CD45.1+ hosts under nonmyeloablative conditions gave rise to low-grade chimerism in PB that ranged from 1 to 4% in different lineages (Fig. 8, E–I). Contributions to Gr1+ and B220+ cells continued to increase for 3 mo and then stabilized, while terminal analysis revealed a chimerism of ∼6–8% within HSPC subsets in BM. Unexpectedly, when transplanted into previously infected hosts (56 dpi), the levels of chimerism were significantly higher in all blood lineages and continuously increased, reaching significantly higher levels than those found in control recipients by 16 wk after transplantation (Fig. 8, E–I). Consistently, at terminal analysis, HSC chimerism virtually doubled in infected compared with noninfected recipient mice, and the numbers of donor-derived HSPC subsets were increased, while no differences in the global absolute numbers of BM cells or HSCs were detected in recipient mice from both groups (Fig. 8, J–N; and not depicted). Most importantly, augmented engraftment was completely erased by treatment of recipients with a-IFNAR/IFNγ antibodies from the beginning of the infection until the time of transplantation (56 d; Fig. 8, E–N). Taken together, these results suggest that (1) despite major structural damage, BM tissues support long-term engraftment of functional HSCs after infection, and (2) via direct and/or indirect mechanisms mediated by IFN signaling, infections imprint a long-lasting loss of competitive fitness in HSCs, which are gradually displaced by infection-naive HSCs when forced to compete under the same microenvironmental conditions.
Discussion
The frequent association of viral infections with a variety of hematologic syndromes, including BM failure, has long been noted (Rosenfeld and Young, 1991). However, the specific mechanisms by which viral processes perturb BM function remain poorly defined. Using a mouse model of chronic viral infection, we show that (1) chronic infection results in a strong impairment of HSC functionality and competitive fitness, which persists long after the inflammatory responses to the virus have receded, viral presence has been cleared, and apparent BM homeostasis has been regained; (2) long-lasting hematopoietic defects correlate with the prolonged viral-induced depletion of the supportive BM mesenchymal CARc network; (3) both injury to CARc networks and loss of HSCs are immune mediated and triggered by BM-resident virus-activated CD8 T cells; (4) the observed immunopathological alterations are mediated by type I and type II IFN signaling; and (5) combined blockage of both IFN pathways restricts BM accumulation of cytotoxic T cells, completely prevents the damage to the CARc stromal compartment, and largely protects HSCs from their functional decline, maintaining their competitive status throughout the entire course of chronic infections.
We show for the first time that key components of the BM microenvironment are profoundly altered during chronic viral infections, leading to the disruption of indispensable structural and molecular cues that make up HSPC niches. Our data thus point to this stromal damage—and specifically, suppression of CARc-derived supportive signals—as a potential contributing cause to the chronic functional defects observed in HSCs. Studies to date have for the most part addressed how viruses or ensuing inflammatory mediators directly target hematopoietic cells, with a specific focus on effects at the level of HSCs. For instance, studies using pathogenic models and infection-mimicking agents have suggested that direct activation of HSC cycling and enhanced proliferative history by IFNα and IFNγ could mechanistically underlie the loss of HSC repopulating capacity. Indeed, repeated in vivo stimulation of IFNα production eventually leads to reduced HSC repopulation capacity (Walter et al., 2015). Pathogen-driven IFNγ production also triggers HSC proliferation and can lead to virtual ablation of the HSC pool if prolonged owing to repeated infectious challenge (Matatall et al., 2016; MacNamara et al., 2011). Nonetheless, the kinetics and duration of local HSC exposure to IFNα and IFNγ shown in these previous studies do not recapitulate those naturally elicited by life-chronic viral infections. In fact, during LCMV-cl13 infection, IFNα production is strictly limited to very early and transient phases after challenge (0–2 dpi), and increased IFNγ levels gradually phase out within 4 wk after infection. Moreover, despite displaying IFNα production during infection, mice lacking CD8 T cells were entirely protected from most hematologic effects and HSC loss after LCMV-cl13 infection. Most importantly, we found that combined blockage of type I and II IFN signaling protects stroma from damage, preserves functionality, and restores competitive fitness in the chronic phases of infection, despite not preventing entry into cell cycle or initial losses of HSC numbers. Altogether, our findings argue that in LCMV-cl13 infections, (1) IFNα is an essential mediator of hematological symptoms, not only via its direct action on HSCs, but most actively through its function as a key activation signal for virus-specific CD8 T cells, which subsequently drive immunopathological damage in HSCs and stroma; (2) long-term deficiency in HSC function is largely uncoupled from the initial proliferative burst, and could arise from direct cytokine exposure or, at least partially, as a consequence of the crippled HSC niche support of the BM microenvironment; and (3) IFNs are not the sole inducers of infection-reactive HSC proliferation, which is in line with previous reports (Hirche et al., 2017).
CARcs are central to BM hematopoiesis through production of essential prohematopoietic factors. Nevertheless, it is possible that other stromal components with putative roles in HSC niches are targeted during infections and contribute to suboptimal HSC activity. Arterial endothelial cells and periarterial Ng2-CreER+ mesenchymal subsets provide functionally relevant fractions of SCF and CXCL12 and have been proposed to regulate HSC localization, cell cycle, and function (Kunisaki et al., 2013; Asada et al., 2017). Although we did not observe conspicuous remodeling or destruction within periarterial neighborhoods, further work is needed to determine whether potential aberrant transcriptomic programs or functional alterations are imprinted on these or other stromal structures with long-term consequences on BM homeostasis. Recent analyses suggest that the CARc pool is made up of a mixed group of at least four to five different subpopulations (Baccin et al., 2019; Baryawno et al., 2019; Tikhonova et al., 2019; Wolock et al., 2019 Preprint). Whether damage is inflicted with the same magnitude in all CARc subsets or, alternatively, selective dwindling or expansion of specific subpopulations occurs remains to be assessed.
The exact mechanisms by which BM-infiltrating CD8 T cells target CARcs are yet unclear. LCMV-cl13 infects both SECs and CARcs. Thus, direct cytotoxic CD8 killing, cytokine-mediated death, or a combination thereof could account for the destruction of these stromal networks. Unfortunately, LCMV-cl13 infection of perforin-deficient mice leads to supraphysiological cytokine secretion and fatal BM syndromes, making it hard to precisely determine the relative contribution of T cell–mediated killing to the demise of the stromal infrastructure (Jordan et al., 2004). Viral agents such as HIV and CMV, which are associated with strong hematologic manifestations, directly infect BM stromal cells and impair their function, thereby suggesting that cytotoxic killing could be a potential mechanism contributing to BM suppression in clinical settings (Moses et al., 1996; Scadden et al., 1990; Bahner et al., 1997; Apperley et al., 1989; Simmons et al., 1990). Nonetheless, immunopathologic loss of secondary lymphoid organ FRCs in acute LCMV infection is mostly driven by cytokines (Scandella et al., 2008), and abnormally high IFNγ production has been shown to impair and partially deplete BM mesenchymal stromal cells in transgenic mouse models (Goedhart et al., 2018). Our analysis indicates that apoptosis of CARcs is evident as early as 3 dpi, when adaptive immune responses and cytotoxic killing have not yet peaked. It seems therefore reasonable to speculate that cytokine-mediated mechanisms could directly operate to destroy the CARc network, alter its function, or prevent its regeneration upon damage.
Most importantly, despite the twofold reduction in CARc abundance, their eradication from relatively large tissue regions, and the profound decrease in hematopoietic cytokine levels, we observed that BM homeostasis is restored, with hematopoietic cellularity and output going back to normal levels, by 4 wk after infection. Furthermore, BM tissues with a twofold reduction in CARcs were competent in supporting long-term engraftment of transplanted HSCs. This suggests that, in homeostatic conditions, CARcs are present in numbers that exceed those needed to maintain basal hematopoietic activity of the BM. These findings are in line with elegant studies showing that secondary lymphoid organ function is only conspicuously impaired when >50% of the mesenchymal FRC population is destroyed (Novkovic et al., 2016).
Nonetheless, while a damaged CARc network suffices to support basal HSC and progenitor function, it may fall short in maintaining full HSC fitness required during stress. We show that within a tissue environment shaped by chronic infections, infection-experienced HSCs display a disadvantage that promotes the expansion of unchallenged competitors. Whether and to what extent such decreased fitness is imprinted by the direct but temporally limited action of inflammatory mediators on HSCs at early stages of infection, or is partially conferred by the prolonged residence in a dysfunctional tissue microenvironment, remains to be clarified. Irrespective of this, our observations support the notion that chronic inflammation can act as a trigger for the selective expansion of HSC clones, by exacerbating differences in competitive fitness that would otherwise not manifest in noninflammatory settings. Along these lines, recent studies demonstrate that inflammation or infections accelerate the competitive expansion of HSCs and multipotent progenitors harboring specific mutations, thereby reinforcing the hypothesis that inflammatory events underlie the development of age-related clonal hematopoiesis and leukemias (Higa et al., 2021; Hormaechea-Agulla et al., 2021; Caiado et al., 2021). Our findings now suggest that gradual proinflammatory remodeling of the tissue microenvironment could be an additional contributing factor and key event in hematopoietic disease, through selective dampening of the competitive status of HSCs. Interestingly, recent investigations have shown that a rapid loss of osteoblasts follows infections with Plasmodium berghei, and that protection of this stromal subset partially prevents impairment of HSC function (Haltalli et al., 2020). Together with our study, these observations clearly show that alterations in hematopoietic and structural compartments will differ qualitatively and mechanistically, depending on the specific pathogens and the nature of the immune responses elicited.
Beyond this, our work may have broader clinical implications. Latent chronic CMV infections are reactivated during myeloablative conditioning regimens in BM transplant patients, and are a common cause of engraftment failure (Simmons et al., 1990; Reddehase et al., 1992; Mayer et al., 1997). Since CMV infects stromal cells (Simmons et al., 1990; Mayer et al., 1997), infections with chronic LCMV-cl13 may represent a valid model to shed light on how underlying stromal deficiencies contribute to this clinical burden. In summary, the results here presented emphasize the importance of studying disruptions in the dynamic interplay between immune, parenchymal, and stromal compartments to uncover how pathologic challenges compromise organ function.
Materials and methods
Mice
C57BL/6J were purchased from Charles River. Cxcl12-GFP mice have been previously described (Sugiyama et al., 2006). Unless stated otherwise, experiments were performed in female mice, which were infected and analyzed between 10 and 20 wk of age. LepR-Cre (B6.129-Leprtm3(cre)Mgmj/J; 032457), Rosa26-CAG-loxp-stop-loxp-tdTomato (B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J; 007908), CD8−/− (B6.129S2-Cd8atm1Mak/J; 002665), and IFNAR−/− (B6(Cg)-Ifnar1tm1.2Ees/J) mice were from Jackson Laboratory. Animals were maintained in the animal facility of the University Hospital Zurich and treated in accordance with guidelines of the Swiss Federal Veterinary Office. Experiments and procedures were approved by the Veterinäramt des Kantons Zurich, Switzerland.
Reagents and antibodies
A detailed list of all the reagents used in this study, including antibodies for FC and 3D QM, is provided in Table S1.
Chronic LCMV infection
The LCMV-cl13 strain was propagated on baby hamster kidney 21 cells. In all cases, infections were performed by a single intravenous injection of high doses (2 × 106 ffu) of the LCMV strain clone 13.
FC analysis and cell sorting
Bones were prepared as previously described (Gomariz et al., 2018). In brief, for BM cell analyses, murine bones were isolated and thoroughly cleaned. The marrow content was flushed with a syringe directly into 5 ml of medium (DMEM GlutaMAX, 10 mM Hepes, and 10% FBS). Remaining bone fragments were carefully minced into small fragments. Tissue suspensions were thoroughly homogenized and enzymatic digestion was performed by incubation in DNase (0.2 mg/ml) and collagenase type 2 (0.04 g/ml) at 37°C for 45 min under gentle rocking. Cells were subsequently filtered through a 70-µm cell strainer, washed in PBS, blocked for 15 min using TruStain fcX, and successively stained with fluorescently labeled antibodies for 30 min at 4°C. Information on all antibodies employed in FC studies can be found in Table S1. Cells were then washed twice with PBS and resuspended in DAPI (0.5 µg/ml in PBS) before being analyzed using an LSR II Fortessa (BD Biosciences) or FACS Aria II sorter (BD Biosciences). Data analysis was performed using the FlowJo 10 software package (BD Biosciences).
RNA isolation, library preparation, and sequencing
CARcs and SECs were identified and sorted based on defined phenotypic signatures (CD45−Ter119−CD31−CD140b+Sca-1− and CD45−Ter119−CD31+Sca-1+, respectively) directly into RLT buffer (Qiagen). RNA was extracted using the RNeasy Plus Micro Kit (Qiagen). The quality of the isolated RNA was measured using a Bioanalyzer 2100 (Agilent). Library preparation, cluster generation, and sequencing were performed at the Functional Genomics Center Zurich. The libraries were prepared using the SMARTer Stranded Total RNA-Seq Pico Input Mammalian kit (Takara Bio) following the manufacturer’s instructions. For each sample, 600 pg of input total RNA was used. The TruSeq SR Cluster Kit v4-cBot-HS (Illumina) was employed for cluster generation using 8 pM of pooled normalized libraries on the cBOT system. Sequencing was performed on the Illumina HiSeq 4000 single-end 125-bp using the TruSeq SBS Kit v4-HS (Illumina).
RNA-seq of stromal populations
Bioinformatic analysis was performed using the R package ezRun (https://github.com/uzh/ezRun) within the data analysis framework SUSHI (Hatakeyama et al., 2016). In brief, raw reads were quality checked using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and FastQ Screen (https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/). Quality-controlled reads (adaptor trimmed, minimum average quality Q20, minimum tail quality Q20, and minimum read length 20 nt) were aligned to the reference genome (Ensembl GRCm38.p5) using the STAR aligner. Expression counts were computed using featureCounts in the Bioconductor package Subread. Differential expression analysis was performed using the Bioconductor DESeq2 package, where raw read counts were normalized using the quantile method, and differential expression was computed using the quasi-likelihood F test. Gene ontology enrichment analysis was performed using Bioconductor packages goseq and GOStats. Quality checkpoints, such as quality control of the alignment and count results, were implemented in ezRun and applied throughout the analysis workflow to ensure correct data interpretation.
Limiting dilution transplantation
For competitive transplantation, 6–8-wk-old CD45.1 female mice were infected with chronic dose LCMV-cl13 (2 × 106 ffu). Femoral bones were collected at predefined time points after infection, and distinct numbers of LSKCD48−CD150+ CD45.1 phenotypic HSCs were sorted on a FACS Aria II sorter (BD Biosciences) in 4-way-purity sorting mode to ensure highest purity and accuracy. Sorting was performed into FCS-coated 1.5-ml Eppendorf tubes at the numbers indicated in each experiment. After validating viability of sorted cells by microscopy, predefined doses of HSCs were transplanted together with 3.5 × 105 of total BM CD45.2 competitors into lethally irradiated CD45.2 recipient mice. Lethal irradiation consisted of two doses of 6.5 Gy in a 4-h interval. PB analysis was performed monthly for a total of 4 mo, at which time the experimental endpoint was reached and terminal BM analysis was performed. Recipients with >0.5% CD45.1 donor engraftment in all three major blood lineages (Gr1+ myeloid cells, B220+ B cells, and CD3+ T cells) were considered engrafted.
Steady-state transplantations and in vivo CFSE dilution assays
Analysis of divisional history using CFSE labeling was performed as described in Takizawa et al. (2011). The BM from 6–8-wk-old CD45.2 mice was harvested, and Lin−Sca-1+c-kit+ (LSK) cells were immunostained, sorted using a FACS Aria II sorter (BD Biosciences), and labeled for 7 min at 37°C with 2 µM CFSE in PBS and 2% FBS. The staining reaction was stopped by addition of ice-cold PBS and 10% FBS. After one wash in PBS, 3 × 105 LSK cells per recipient mouse were transplanted intravenously into 6–8-wk-old CD45.1/CD45.2 mice that had been infected 56 d before with 2 × 106 ffu of LCMV-cl13. At terminal analysis (3 wk after transplantation), recipient mice were sacrificed, BM was harvested, and cells were isolated as described above but without digestion. After immunostaining of HSC markers, cells were run on a BD Fortessa for analysis. For long-term transplantations, the same experimental scheme was used, but 3 × 106 unlabeled LSK cells were transplanted per mouse. Mice were then bled monthly through sublingual incision to follow chimerism of CD45.2+ cells in the different blood cell lineages. At terminal analysis (4 mo after transplantation), mice were sacrificed, and hematopoietic CD45.2+ chimerism was measured in all major hematopoietic lineages, HSCs, and progenitor subsets.
ELISA
For ELISA, BM supernatant was generated by flushing the BM plug from bones from one hind leg (femur + tibia) in 300 µl containing equal volumes of RayBio 2x Cell Lysis Buffer (Raybiotech/Lucerna Chem) and 2× complete ULTRA Protease Inhibitor Cocktail (Sigma-Aldrich). Levels of IFNα and IFNγ in serum and BM supernatants were tested using the VeriKine kit (PBL Assay Sciences) or Ready-SET-Go! Kit (eBioscience), respectively. SCF levels were determined using Mouse SCF ELISA (Raybiotech), and CXCL12 protein concentration was assayed with the Mouse CXCL12/SDF-1 α Quantikine ELISA Kit (R&D Systems) according to manufacturer’s instructions.
Focus-forming assays
For virus titer determination, organs were harvested and homogenized, and virus titer was calculated as previously described (Battegay et al., 1991). Briefly, blood or homogenized organs from infected mice were plated in serial dilutions and at least duplicates into wells containing MEM with 2% FCS and penicillin/streptomycin/glutamine together with MC57G cell suspension. After mixing cells, the culture plate was incubated for 2–4 h at 37°C until MC57G cells were adherent, at which a 1:1 mixture of 2% methylcellulose and 2× DMEM (10% FCS and penicillin/streptomycin) was added dropwise. After incubation for 2 d at 37°C, plaques were fixed with 4% paraformaldehyde and permeabilized in 1% Triton X-100 solution, at which time staining with primary antibody VL-4 rat anti-LCMV was performed. Secondary antibody staining was done with peroxidase-conjugated goat anti-rat IgG antibody, and color reaction was performed for 15–20 min in 0.1 M MNa2HPO4x2H2O and 0.1 M citric acid buffer containing ortho-phenylenediamine. Serial dilutions of plaque titers were counted at least in duplicate.
Real-time quantitative PCR (RT-qPCR)
RNA was isolated as previously described using RNeasy Plus Micro Kit (Qiagen; Gomariz et al., 2018). cDNA was generated with High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Gene expression was measured on a 7500 Fase Realtime PCR System with SYBR Green.
Blocking and depleting antibodies
Blocking of IFNAR and IFNγ signaling was performed as previously described (Teijaro et al., 2013; Wilson et al., 2013). Briefly, for IFNAR signaling blockage, 500 µg of InvivoMab anti-mouse IFNAR-1 monoclonal antibody (clone MAR1-5A3; BioXCell) was injected i.p. on days −1, 0, and 2 of infection with LCMV-cl13, after which 250 µg blocking antibody was injected i.p. every other day to ensure efficient blockage. For IFNγ signaling blockage, InvivoMab anti-mouse IFNγ antibody was used (clone XMG1.2, BioXCell). 500 µg blocking antibody was injected i.p. on days −1 and 0, after which 500 µg was injected i.p. every third day. For CD8 T cell depletion, mice were treated on days −3, −1, 2, and 5 after infection with 200 µg of the depleting antibody YTS 169.4 (BioXCell). Depletion of CD8 T cells was determined to be ∼90% in the BM on day 7 after infection.
CD8 T cell stimulation and cytokine production
Viral gp33 peptide (gp33-41, KAVYNFATM) was purchased from IBA Lifesciences. 1 µg/ml was used for restimulation of BM CD8+ T lymphocytes along with PMA, ionomycin, and Brefeldin A for 6 h at 37°C before surface antibody staining for 20 min, which was followed by fixation/permeabilization using the BD Fixation/Permeabilization kit (BD Biosciences). After washing, intracellular staining for IFNγ, TNF-α, and IL-2 was performed for 30 min at RT. For cytokine restimulation, single-cell suspensions of BM or spleen were generated as described above in FACS buffer containing EDTA. Surface staining of the single-cell solution was performed for t = 30 min at 4°C. Before intracellular staining and stimulation, cells were fixed in 2% paraformaldehyde (diluted in PBS) for 20 min at 4°C, followed by a washing step. For the stimulation, 1 µg/ml of LCMV-specific gp33-41 was added for 6 h at 37°C in the presence of Brefeldin A to stimulate LCMV-specific CD8 T cells. Cells were then permeabilized for t = 10 min at RT. Intracellular staining for cytokines IFNγ and TNF-α was performed for 30 min at RT. Multiparametric FC analysis was performed using LSR Fortessa, and data were analyzed using FlowJo software.
BM slice preparation, immunostaining, and optical clearing
Methods for 3D imaging of BM were adapted from previously published protocols (Gomariz et al., 2018). Mouse femurs were isolated, cleaned, and immersed in PBS/2% paraformaldehyde for 6 h at 4°C, followed by a dehydration step in 30% sucrose for 72 h at 4°C. Femurs were then embedded in cryopreserving medium (OCT) and snap frozen in liquid nitrogen. Bone specimens were iteratively sectioned using a cryostat until the BM cavity was fully exposed along the longitudinal axis. The OCT block containing the bone was then reversed and the procedure was repeated on the opposite face until a thick bone slice with bilaterally and evenly exposed BM content was obtained. Once BM slices were generated, the remaining OCT medium was removed by incubation and washing of the bone slices in PBS three times for 5 min. For immunostaining, slices were incubated in blocking solution (0.2% Triton X-100, 1% BSA, and 10% donkey serum in PBS) overnight at 4°C. Primary antibody immunostainings were performed in blocking solution for 3 d at 4°C, followed by overnight washing in PBS. Secondary antibody staining was performed for another 3 d at 4°C in blocking solution but in the absence of BSA to avoid cross-absorption. Immunostained thick femoral slices were successively washed overnight in PBS and incubated in RapiClear 1.52 for a minimum of 6 h. For observation under the confocal microscope, BM slices were mounted on glass slides using vacuum grease. Confocal microscopy was performed with 10× (HCX PL FLUOTAR), 20× (HC PL APO CS2). and 63× (HCX PL APO CS2) in SP5 and SP8 Leica confocal microscopes equipped with hybrid detectors.
Image processing and visualization of confocal microscopy data
Imaris software (Bitplane) was used for rendering of confocal image stacks into 3D reconstructions and visualizations. Individual CARcs were detected, and their coordinates were mapped with the Imaris Spots function using a predefined diameter of 7.5 µm. For calculation of CARc densities, ImarisXT was used as described previously (Gomariz et al., 2018). Gamma correction was applied exclusively for visualization purposes. For BM tissue maps, empirical probability densities were used. A custom algorithm adapted the kernel-based density estimation method to account for the different boundaries of the BM as previously described (Gomariz et al., 2018). The tissue map f(u) was calculated at every position u as defined in
where Kσ(u) is a 3D Gaussian kernel with standard deviation σ and a length of 8σ for each dimension. The value of σ was calculated as the average nearest neighbor distance between the represented cells. The result was a 3D probability distribution function of the distribution of cells. To generate 2D density maps as depicted in Figs. 2 A, 4 E, and 6 A, the 3D map was averaged along the axial direction in the volume defined by the segmented DAPI mask. Segmentation of sinusoidal volumes was performed using a deep learning convolutional network trained using 3D images of BM stained with anti-endomucin and/or CD105, as reported in Gomariz et al. (2021).
Statistical analysis of data
If not indicated otherwise, results are displayed as means ± SEM. Statistical tests employed for data analysis, significance, and P values are indicated in figure legends. Limiting dilution analysis was performed as previously described (Hu and Smyth, 2009). In brief, pairwise differences in functional HSC frequencies between groups are compared using χ2 test. The fitted models for different datasets are compared using likelihood ratio tests and asymptotic χ2 approximations to the log ratio. P values are calculated using an overall test for differences between the population groups, analogous to a one-way ANOVA test.
Online supplementary information
Fig. S1 shows effects on HSC function and apoptosis after infection with LCMV-cl13. Fig. S2 shows alterations of CARcs after infection. Fig. S3 depicts transcriptomic changes in SECs 56 dpi and levels of IFNα and IFNγ in blood during the course of infections. Fig. S4 shows the dynamics of CD8 T cells in tissues after infection, the efficiency of depletion, and the effects of infections on CD8 T cells and HSCs in WT/IFNAR−/− chimeric mice. Fig. S5 shows hematopoietic and CARc dynamics during infection and antibody-mediated blockage of IFNAR and IFNγ alone, the effects on cell cycle and engraftment capacity of HSCs in noninfected mice treated with anti IFNAR and IFNγ, and the proliferative history of CFSE-labeled IFNγR−/− HSCs transplanted into recipient mice. Table S1 lists reagents, commercial sources, and catalog numbers. Video 1 shows depletion of erythroid progenitor cells in the BM 7 dpi. Video 2 shows the structural damage to CARc networks and sinusoidal vessels throughout the course of infections with LCMV-cl13. Video 3 focuses on the alterations observed in the ECM networks and their remodeling after LCMV-cl13 infection. Video 4 shows how ablation of CD8 T cells prevents damage to CARc networks after infection. Video 5 shows that combined treatment with anti-IFNAR and anti-IFNγ protects the BM CARc infrastructure from destruction after viral infection.
Supplementary Material
lists reagents, commercial sources, and catalog numbers, if available.
Acknowledgments
We thank Urs Ziegler and José María Mateos of the Center for Microscopy and Image Analysis of the University of Zurich for assistance with confocal microscopy.
This work was supported by grants from the Heubergstiftung (to N.J. Kraeutler and A. Oxenius); the Swiss National Science Foundation to A. Oxenius (310030_166078), M.G. Manz (310030B_166673/1), and C. Nombela-Arrieta (31003A_159597/1 and 310030_185171); and the Seventh Framework Programme Marie Curie Career Integration Program (PCIG13-GA-2013-618633), the Vontobel-Stiftung (Zurich, Switzerland), the Monique Dornonville de la Cour Foundation, and the Helmut Horten Stiftung (Lugano, Switzerland) to C. Nombela-Arrieta.
Author contributions: S. Isringhausen and Y. Mun designed and performed experiments, analyzed data, wrote the manuscript, and contributed equally. L. Kovtonyuk, U. Suessbier, and N.J. Kraeutler performed experiments. H.C. Wong, G. Spaltro, and P.M. Helbling performed experiments. A. Gomariz implemented tools for image analysis and spatial statistics. T. Nagasawa contributed the Cxcl12-Gfp mouse line and discussions. A. Oxenius and M.G. Manz designed and supervised experiments. C. Nombela-Arrieta designed and directed the study and wrote the manuscript.
Data availability
The accession number of the RNA-seq analysis is GEO ID: GSE184341. Raw microscopy data can be made available by the corresponding author upon request.
References
- Apperley, J.F., Dowding C., Hibbin J., Buiter J., Matutes E., Sissons P.J., Gordon M., and Goldman J.M.. 1989. The effect of cytomegalovirus on hemopoiesis: in vitro evidence for selective infection of marrow stromal cells. Exp. Hematol. 17:38–45. [PubMed] [Google Scholar]
- Asada, N., Kunisaki Y., Pierce H., Wang Z., Fernandez N.F., Birbrair A., Ma’ayan A., and Frenette P.S.. 2017. Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nat. Cell Biol. 19:214–223. 10.1038/ncb3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccin, C., Al-Sabah J., Velten L., Helbling P.M., Grünschläger F., Hernández-Malmierca P., Nombela-Arrieta C., Steinmetz L.M., Trumpp A., and Haas S.. 2019. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol. 22:38–48. 10.1038/s41556-019-0439-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahner, I., Kearns K., Coutinho S., Leonard E.H., and Kohn D.B.. 1997. Infection of human marrow stroma by human immunodeficiency virus-1 (HIV-1) is both required and sufficient for HIV-1-induced hematopoietic suppression in vitro: demonstration by gene modification of primary human stroma. Blood. 90:1787–1798. 10.1182/blood.V90.5.1787 [DOI] [PubMed] [Google Scholar]
- Baldridge, M.T., King K.Y., Boles N.C., Weksberg D.C., and Goodell M.A.. 2010. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 465:793–797. 10.1038/nature09135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryawno, N., Przybylski D., Kowalczyk M.S., Kfoury Y., Severe N., Gustafsson K., Kokkaliaris K.D., Mercier F., Tabaka M., Hofree M., et al. 2019. A Cellular Taxonomy of the Bone Marrow Stroma in Homeostasis and Leukemia. Cell. 177:1915–1932.e16. 10.1016/j.cell.2019.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battegay, M., Cooper S., Althage A., Bänziger J., Hengartner H., and Zinkernagel R.M.. 1991. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24- or 96-well plates. J. Virol. Methods. 33:191–198. 10.1016/0166-0934(91)90018-U [DOI] [PubMed] [Google Scholar]
- Binder, D., Fehr J., Hengartner H., and Zinkernagel R.M.. 1997. Virus-induced transient bone marrow aplasia: major role of interferon-alpha/beta during acute infection with the noncytopathic lymphocytic choriomeningitis virus. J. Exp. Med. 185:517–530. 10.1084/jem.185.3.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, J.M., Nolan D.J., Vertes E.L., Varnum-Finney B., Kobayashi H., Hooper A.T., Seandel M., Shido K., White I.A., Kobayashi M., et al. 2010. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 6:251–264. 10.1016/j.stem.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiado, F., Pietras E.M., and Manz M.G.. 2021. Inflammation as a regulator of hematopoietic stem cell function in disease, aging, and clonal selection. J. Exp. Med. 218:e20201541. 10.1084/jem.20201541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavakis, T., Mitroulis I., and Hajishengallis G.. 2019. Hematopoietic progenitor cells as integrative hubs for adaptation to and fine-tuning of inflammation. Nat. Immunol. 20:802–811. 10.1038/s41590-019-0402-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane, G.M., Jeffery E., and Morrison S.J.. 2017. Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 17:573–590. 10.1038/nri.2017.53 [DOI] [PubMed] [Google Scholar]
- Crouse, J., Kalinke U., and Oxenius A.. 2015. Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol. 15:231–242. 10.1038/nri3806 [DOI] [PubMed] [Google Scholar]
- de Bruin, A.M., Demirel Ö., Hooibrink B., Brandts C.H., and Nolte M.A.. 2013. Interferon-γ impairs proliferation of hematopoietic stem cells in mice. Blood. 121:3578–3585. 10.1182/blood-2012-05-432906 [DOI] [PubMed] [Google Scholar]
- Demerdash, Y., Kain B., Essers M.A.G., and King K.Y.. 2021. Yin and Yang: The dual effects of interferons on hematopoiesis. Exp. Hematol. 96:1–12. 10.1016/j.exphem.2021.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L., and Morrison S.J.. 2013. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 495:231–235. 10.1038/nature11885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L., Saunders T.L., Enikolopov G., and Morrison S.J.. 2012. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 481:457–462. 10.1038/nature10783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergen, A.V., Boles N.C., and Goodell M.A.. 2012. Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood. 119:2500–2509. 10.1182/blood-2011-11-391730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers, M.A.G., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., and Trumpp A.. 2009. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908. 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Goedhart, M., Cornelissen A.S., Kuijk C., Geerman S., Kleijer M., van Buul J.D., Huveneers S., Raaijmakers M.H.G.P., Young H.A., Wolkers M.C., et al. 2018. Interferon-Gamma Impairs Maintenance and Alters Hematopoietic Support of Bone Marrow Mesenchymal Stromal Cells. Stem Cells Dev. 27:579–589. 10.1089/scd.2017.0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomariz, A., Helbling P.M., Isringhausen S., Suessbier U., Becker A., Boss A., Nagasawa T., Paul G., Goksel O., Székely G., et al. 2018. Quantitative spatial analysis of haematopoiesis-regulating stromal cells in the bone marrow microenvironment by 3D microscopy. Nat. Commun. 9:2532. 10.1038/s41467-018-04770-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomariz, A., Portenier T., Helbling P.M., Isringhausen S., Suessbier U., Nombela-Arrieta C., and Goksel O.. 2021. Modality attention and sampling enables deep learning with heterogeneous marker combinations in fluorescence microscopy. Nat. Mach. Intell. 3:799–811. 10.1038/s42256-021-00379-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum, A., Hsu Y.-M.S., Day R.B., Schuettpelz L.G., Christopher M.J., Borgerding J.N., Nagasawa T., and Link D.C.. 2013. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 495:227–230. 10.1038/nature11926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltalli, M.L.R., Watcham S., Wilson N.K., Eilers K., Lipien A., Ang H., Birch F., Anton S.G., Pirillo C., Ruivo N., et al. 2020. Manipulating niche composition limits damage to haematopoietic stem cells during Plasmodium infection. Nat. Cell Biol. 22:1399–1410. 10.1038/s41556-020-00601-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama, M., Opitz L., Russo G., Qi W., Schlapbach R., and Rehrauer H.. 2016. SUSHI: an exquisite recipe for fully documented, reproducible and reusable NGS data analysis. BMC Bioinformatics. 17:228. 10.1186/s12859-016-1104-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa, K.C., Goodspeed A., Chavez J.S., De Dominici M., Danis E., Zaberezhnyy V., Rabe J.L., Tenen D.G., Pietras E.M., and DeGregori J.. 2021. Chronic interleukin-1 exposure triggers selection for Cebpa-knockout multipotent hematopoietic progenitors. J. Exp. Med. 218:e20200560. 10.1084/jem.20200560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirche, C., Frenz T., Haas S.F., Döring M., Borst K., Tegtmeyer P.-K., Brizic I., Jordan S., Keyser K., Chhatbar C., et al. 2017. Systemic Virus Infections Differentially Modulate Cell Cycle State and Functionality of Long-Term Hematopoietic Stem Cells In Vivo. Cell Rep. 19:2345–2356. 10.1016/j.celrep.2017.05.063 [DOI] [PubMed] [Google Scholar]
- Hooper, A.T., Butler J.M., Nolan D.J., Kranz A., Iida K., Kobayashi M., Kopp H.-G., Shido K., Petit I., Yanger K., et al. 2009. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell. 4:263–274. 10.1016/j.stem.2009.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormaechea-Agulla, D., Matatall K.A., Le D.T., Kain B., Long X., Kus P., Jaksik R., Challen G.A., Kimmel M., and King K.Y.. 2021. Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNγ signaling. Cell Stem Cell. 28:1428–1442.e6. 10.1016/j.stem.2021.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., and Smyth G.K.. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 347:70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Itkin, T., Gur-Cohen S., Spencer J.A., Schajnovitz A., Ramasamy S.K., Kusumbe A.P., Ledergor G., Jung Y., Milo I., Poulos M.G., et al. 2016. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature. 532:323–328. 10.1038/nature17624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, M.B., Hildeman D., Kappler J., and Marrack P.. 2004. An animal model of hemophagocytic lymphohistiocytosis (HLH): CD8+ T cells and interferon gamma are essential for the disorder. Blood. 104:735–743. 10.1182/blood-2003-10-3413 [DOI] [PubMed] [Google Scholar]
- Kaushansky, K. 2006. Lineage-specific hematopoietic growth factors. N. Engl. J. Med. 354:2034–2045. 10.1056/NEJMra052706 [DOI] [PubMed] [Google Scholar]
- Khan, A.B., Carpenter B., Santos E Sousa P., Pospori C., Khorshed R., Griffin J., Velica P., Zech M., Ghorashian S., Forrest C., et al. 2018. Redirection to the bone marrow improves T cell persistence and antitumor functions. J. Clin. Invest. 128:2010–2024. 10.1172/JCI97454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolumam, G.A., Thomas S., Thompson L.J., Sprent J., and Murali-Krishna K.. 2005. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 202:637–650. 10.1084/jem.20050821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, M., Wagers A.J., Manz M.G., Prohaska S.S., Scherer D.C., Beilhack G.F., Shizuru J.A., and Weissman I.L.. 2003. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu. Rev. Immunol. 21:759–806. 10.1146/annurev.immunol.21.120601.141007 [DOI] [PubMed] [Google Scholar]
- Kovtonyuk, L.V., Manz M.G., and Takizawa H.. 2016. Enhanced thrombopoietin but not G-CSF receptor stimulation induces self-renewing hematopoietic stem cell divisions in vivo. Blood. 127:3175–3179. 10.1182/blood-2015-09-669929 [DOI] [PubMed] [Google Scholar]
- Kunisaki, Y., Bruns I., Scheiermann C., Ahmed J., Pinho S., Zhang D., Mizoguchi T., Wei Q., Lucas D., Ito K., et al. 2013. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 502:637–643. 10.1038/nature12612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumbe, A.P., Ramasamy S.K., Itkin T., Mäe M.A., Langen U.H., Betsholtz C., Lapidot T., and Adams R.H.. 2016. Age-dependent modulation of vascular niches for haematopoietic stem cells. Nature. 532:380–384. 10.1038/nature17638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNamara, K.C., Jones M., Martin O., and Winslow G.M.. 2011. Transient activation of hematopoietic stem and progenitor cells by IFNγ during acute bacterial infection. PLoS One. 6:e28669. 10.1371/journal.pone.0028669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matatall, K.A., Shen C.-C., Challen G.A., and King K.Y.. 2014. Type II interferon promotes differentiation of myeloid-biased hematopoietic stem cells. Stem Cells. 32:3023–3030. 10.1002/stem.1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matatall, K.A., Jeong M., Chen S., Sun D., Chen F., Mo Q., Kimmel M., and King K.Y.. 2016. Chronic Infection Depletes Hematopoietic Stem Cells through Stress-Induced Terminal Differentiation. Cell Rep. 17:2584–2595. 10.1016/j.celrep.2016.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, A., Podlech J., Kurz S., Steffens H.P., Maiberger S., Thalmeier K., Angele P., Dreher L., and Reddehase M.J.. 1997. Bone marrow failure by cytomegalovirus is associated with an in vivo deficiency in the expression of essential stromal hemopoietin genes. J. Virol. 71:4589–4598. 10.1128/jvi.71.6.4589-4598.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier, F.E., Ragu C., and Scadden D.T.. 2011. The bone marrow at the crossroads of blood and immunity. Nat. Rev. Immunol. 12:49–60. 10.1038/nri3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Mantilla, D.E., and King K.Y.. 2018. The Role of Interferon-Gamma in Hematopoietic Stem Cell Development, Homeostasis, and Disease. Curr. Stem Cell Rep. 4:264–271. 10.1007/s40778-018-0139-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, S.J., and Scadden D.T.. 2014. The bone marrow niche for haematopoietic stem cells. Nature. 505:327–334. 10.1038/nature12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses, A.V., Williams S., Heneveld M.L., Strussenberg J., Rarick M., Loveless M., Bagby G., and Nelson J.A.. 1996. Human immunodeficiency virus infection of bone marrow endothelium reduces induction of stromal hematopoietic growth factors. Blood. 87:919–925. 10.1182/blood.V87.3.919.bloodjournal873919 [DOI] [PubMed] [Google Scholar]
- Mueller, S.N., Matloubian M., Clemens D.M., Sharpe A.H., Freeman G.J., Gangappa S., Larsen C.P., and Ahmed R.. 2007. Viral targeting of fibroblastic reticular cells contributes to immunosuppression and persistence during chronic infection. Proc. Natl. Acad. Sci. USA. 104:15430–15435. 10.1073/pnas.0702579104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nombela-Arrieta, C., and Isringhausen S.. 2017. The Role of the Bone Marrow Stromal Compartment in the Hematopoietic Response to Microbial Infections. Front. Immunol. 7:689. 10.3389/fimmu.2016.00689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nombela-Arrieta, C., and Manz M.G.. 2017. Quantification and three-dimensional microanatomical organization of the bone marrow. Blood Adv. 1:407–416. 10.1182/bloodadvances.2016003194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novkovic, M., Onder L., Cupovic J., Abe J., Bomze D., Cremasco V., Scandella E., Stein J.V., Bocharov G., Turley S.J., and Ludewig B.. 2016. Topological Small-World Organization of the Fibroblastic Reticular Cell Network Determines Lymph Node Functionality. PLoS Biol. 14:e1002515. 10.1371/journal.pbio.1002515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omatsu, Y., Sugiyama T., Kohara H., Kondoh G., Fujii N., Kohno K., and Nagasawa T.. 2010. The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity. 33:387–399. 10.1016/j.immuni.2010.08.017 [DOI] [PubMed] [Google Scholar]
- Pascutti, M.F., Erkelens M.N., and Nolte M.A.. 2016. Impact of Viral Infections on Hematopoiesis: From Beneficial to Detrimental Effects on Bone Marrow Output. Front. Immunol. 7:364. 10.3389/fimmu.2016.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M. 2017. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood. 130:1693–1698. 10.1182/blood-2017-06-780882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M., Lakshminarasimhan R., Techner J.M., Fong S., Flach J., Binnewies M., and Passegué E.. 2014. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 211:245–262. 10.1084/jem.20131043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M., Mirantes-Barbeito C., Fong S., Loeffler D., Kovtonyuk L.V., Zhang S., Lakshminarasimhan R., Chin C.P., Techner J.-M., Will B., et al. 2016. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18:607–618. 10.1038/ncb3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos, M.G., Guo P., Kofler N.M., Pinho S., Gutkin M.C., Tikhonova A., Aifantis I., Frenette P.S., Kitajewski J., Rafii S., and Butler J.M.. 2013. Endothelial Jagged-1 is necessary for homeostatic and regenerative hematopoiesis. Cell Rep. 4:1022–1034. 10.1016/j.celrep.2013.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddehase, M.J., Dreher-Stumpp L., Angele P., Balthesen M., and Susa M.. 1992. Hematopoietic stem cell deficiency resulting from cytomegalovirus infection of bone marrow stroma. Ann. Hematol. 64(S1, Suppl):A125–A127. 10.1007/BF01715364 [DOI] [PubMed] [Google Scholar]
- Rosenfeld, S.J., and Young N.S.. 1991. Viruses and bone marrow failure. Blood Rev. 5:71–77. 10.1016/0268-960X(91)90037-D [DOI] [PubMed] [Google Scholar]
- Rouse, B.T., and Sehrawat S.. 2010. Immunity and immunopathology to viruses: what decides the outcome? Nat. Rev. Immunol. 10:514–526. 10.1038/nri2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T., Onai N., Yoshihara H., Arai F., Suda T., and Ohteki T.. 2009. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat. Med. 15:696–700. 10.1038/nm.1973 [DOI] [PubMed] [Google Scholar]
- Scadden, D.T., Zeira M., Woon A., Wang Z., Schieve L., Ikeuchi K., Lim B., and Groopman J.E.. 1990. Human immunodeficiency virus infection of human bone marrow stromal fibroblasts. Blood. 76:317–322. 10.1182/blood.V76.2.317.317 [DOI] [PubMed] [Google Scholar]
- Scandella, E., Bolinger B., Lattmann E., Miller S., Favre S., Littman D.R., Finke D., Luther S.A., Junt T., and Ludewig B.. 2008. Restoration of lymphoid organ integrity through the interaction of lymphoid tissue-inducer cells with stroma of the T cell zone. Nat. Immunol. 9:667–675. 10.1038/ni.1605 [DOI] [PubMed] [Google Scholar]
- Schürch, C.M., Riether C., and Ochsenbein A.F.. 2014. Cytotoxic CD8+ T cells stimulate hematopoietic progenitors by promoting cytokine release from bone marrow mesenchymal stromal cells. Cell Stem Cell. 14:460–472. 10.1016/j.stem.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Simmons, P., Kaushansky K., and Torok-Storb B.. 1990. Mechanisms of cytomegalovirus-mediated myelosuppression: perturbation of stromal cell function versus direct infection of myeloid cells. Proc. Natl. Acad. Sci. USA. 87:1386–1390. 10.1073/pnas.87.4.1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, T., Kohara H., Noda M., and Nagasawa T.. 2006. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 25:977–988. 10.1016/j.immuni.2006.10.016 [DOI] [PubMed] [Google Scholar]
- Takizawa, H., Regoes R.R., Boddupalli C.S., Bonhoeffer S., and Manz M.G.. 2011. Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J. Exp. Med. 208:273–284. 10.1084/jem.20101643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro, J.R., Ng C., Lee A.M., Sullivan B.M., Sheehan K.C.F., Welch M., Schreiber R.D., de la Torre J.C., and Oldstone M.B.A.. 2013. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 340:207–211. 10.1126/science.1235214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova, A.N., Dolgalev I., Hu H., Sivaraj K.K., Hoxha E., Cuesta-Domínguez Á., Pinho S., Akhmetzyanova I., Gao J., Witkowski M., et al. 2019. The bone marrow microenvironment at single-cell resolution. Nature. 569:222–228. 10.1038/s41586-019-1104-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, D., Lier A., Geiselhart A., Thalheimer F.B., Huntscha S., Sobotta M.C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P., et al. 2015. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 520:549–552. 10.1038/nature14131 [DOI] [PubMed] [Google Scholar]
- Wilson, E.B., Yamada D.H., Elsaesser H., Herskovitz J., Deng J., Cheng G., Aronow B.J., Karp C.L., and Brooks D.G.. 2013. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 340:202–207. 10.1126/science.1235208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock, S.L., Krishnan I., Tenen D.E., Matkins V., Camacho V., Patel S., Agarwal P., Bhatia R., Tenen D.G., Klein A.M., and Welner R.S.. 2019. Mapping Distinct Bone Marrow Niche Populations and Their Differentiation Paths. Cell Rep. 28:302–311.e5. 10.1016/j.celrep.2019.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., Ramachandran S., Mann M., and Popkin D.L.. 2012. Role of lymphocytic choriomeningitis virus (LCMV) in understanding viral immunology: past, present and future. Viruses. 4:2650–2669. 10.3390/v4112650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B.O., Yue R., Murphy M.M., Peyer J.G., and Morrison S.J.. 2014. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 15:154–168. 10.1016/j.stem.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists reagents, commercial sources, and catalog numbers, if available.
Data Availability Statement
The accession number of the RNA-seq analysis is GEO ID: GSE184341. Raw microscopy data can be made available by the corresponding author upon request.