Abstract
Influenza viruses hijack host cell factors at each stage of the viral life cycle. After host cell entry and endosomal escape, the influenza viral ribonucleoproteins (vRNPs) are released into the cytoplasm where the classical cellular nuclear import pathway is usurped for nuclear translocation of the vRNPs. Transcription takes place inside the nucleus at active host transcription sites, and cellular mRNA export pathways are subverted for export of viral mRNAs. Newly synthesized RNP components cycle back into the nucleus using various cellular nuclear import pathways and host-encoded chaperones. Replication of the negative-sense viral RNA (vRNA) into complementary RNA (cRNA) and back into vRNA requires complex interplay between viral and host factors. Progeny vRNPs assemble at the host chromatin and subsequently exit from the nucleus—processes orchestrated by sets of host and viral proteins. Finally, several host pathways appear to play a role in vRNP trafficking from the nuclear envelope to the plasma membrane for egress.
All viruses usurp the host machinery to assist their replication and influenza virus is no exception. There are four distinct influenza virus types—namely, A, B, C, and D. Respiratory disease in humans is caused predominantly by influenza A and B viruses (IAV and IBV), whereas influenza C virus (ICV) is known to cause sporadic disease in children. Influenza D virus infects cattle. Two IAV subtypes (H3N2 and H1N1, based on antigenic diversity of the hemagglutinin [HA] and neuraminidase [NA] glycoproteins), and two IBV lineages (Victoria and Yamagata) currently circulate in humans, causing seasonal epidemics that affect millions (Krammer et al. 2018). The single-stranded negative-sense RNA genome of IAV (vRNA) consists of eight segments encoding 10 core polyproteins: the RNA-dependent RNA polymerase (RdRp) subunits polymerase basic 1 (PB1), polymerase basic 2 (PB2), and polymerase acidic (PA), the glycoproteins HA and NA, nucleoprotein (NP), the matrix protein M1, the M2 ion channel, the nonstructural protein NS1, and, finally, the nuclear export protein (NEP) (previously known as NS2). An additional suite of virus strain–dependent, nonessential accessory proteins has been described (Vasin et al. 2014).
Eighteen HA subtypes and 11 NA subtypes have been identified in IAV to date, all of which present in avian species except H17N10 and H18N11, which infect bats (Tong et al. 2012, 2013). A large and antigenically diverse reservoir of IAV subtypes is present in wild waterfowl from whence viruses transfer into mammalian host species including pigs, horses, dogs, and humans (Lycett et al. 2019). Although most of these zoonoses will be dead-end infections, zoonotic events can lead to pandemics when the virus accumulates sufficient mutations to allow for effective transmission among members of the new host species. Recent species jumps of H5 and H7 subtypes, lethal in poultry, have led to substantial worry among global health experts. Influenza pandemics occur every few decades, most recently in 2009 (H1N1 swine flu) but also in 1968 (H3N2 Hong Kong flu), 1957 (H2N2 Asian flu), and, most notoriously, in 1918 (H1N1 Spanish flu), the latter killing at least 50 million people.
Mammalian adaptive mutations are required by avian influenza strains before they can cause a pandemic. These include changes in HA, NA, and the heterotrimeric RdRp and NP (for review, see Long et al. 2019b). The PB2 subunit of the polymerase in particular is well known to undergo mammalian adaptions. Substitution of the avian-signature glutamate at position 627 and aspartate 701 to mammalian-signature 627K and 701N are common in zoonotic and human-adapted strains (Subbarao et al. 1993; Czudai-Matwich et al. 2014) and adapt polymerase to using mammalian host factors as discussed in this review. Additional adaptations are believed to counteract mammalian antiviral immune pathways. The retinoic acid–inducible gene 1 protein (RIG-I) recognizes influenza vRNA on the basis of a 5′-terminal triphosphate moiety and an adjacent patch of double-stranded RNA (dsRNA) that is the viral promoter binding the RdRp (for review, see Fodor and te Velthuis 2019; Wandzik et al. 2020). During infection, the viral NS1 protein counteracts RIG-I, which would otherwise result in the induction of interferon (IFN) and IFN-stimulated gene (ISG) expression, such as MX1, protein kinase R (PKR), and IFITM3 (Pichlmair et al. 2006; Rehwinkel et al. 2010; Rajsbaum et al. 2012). It has also been suggested that the PB2 host-adapting mutation E627K masks the pathogen-associated molecular pattern (PAMP) on the incoming genome from detection by RIG-I (Weber et al. 2015).
Antivirals against influenza virus are in limited supply. The M2 ion channel inhibitors amantadine and rimantadine have been discontinued as all circulating human influenza viruses have developed resistance in the form of an S31N substitution in the RNA segment encoding M2. Oseltamivir (Tamiflu) is an NA inhibitor targeting the sialidase activity of NA, but NA inhibitors are also compromised by resistance-conferring mutations such as NA-H275Y that spread through the seasonal H1N1 viral population. Recently, a variety of compounds that target the replication machinery has come to the fore (for review, see Takashita 2020). Active sites for RNA binding, cleavage, and elongation are highly conserved among influenza viruses, so the RdRp is a potentially promising target for antiviral agents. The PB1-targeting purine analog favipiravir acts as a chain terminator and has been approved for use in Japan. Resistance mutations have, however, been described, in particular, a K229R substitution in the PB1 subunit alongside a fitness-compensating PA-P653L mutation (Goldhill et al. 2018). Pimodivir occupies the N7-methyl guanosine (m7G) cap-binding domain of the PB2 subunit, which is required for cap snatching and, therefore, transcription of the vRNA. Pimodivir-resistant H1N1 and H3N2 viruses are routinely detected in clinical trials with this compound (Trevejo et al. 2018; Finberg et al. 2019). Finally, baloxavir marboxil binds to the PA endonuclease domain, also required for cap snatching (Omoto et al. 2018). Approved in Japan and in the United States, an I38T substitution in PA seems to confer resistance against the drug in circulating H1N1 and H3N2 viruses (Hayden et al. 2018; Noshi et al. 2018; Hirotsu et al. 2020; Uehara et al. 2020).
Given that influenza virus acquires resistance to antivirals targeting virus-encoded proteins rapidly and consistently, an alternative strategy may be called for. Host-directed therapy targets host-encoded rather than virus-encoded factors and as such aims to sidestep rapid accumulation of resistance mutations on the premise that viruses cannot directly control the physical appearance of host proteins. The strategy is to interfere with host cell factors that are required by a pathogen for replication or persistence (for review, see Kaufmann et al. 2018).
The viral life cycle involves the following steps, all of which require host factors (for review, see Eisfeld et al. 2015; Peacock et al. 2019). Upon virus binding and endocytosis, the viral ribonucleoproteins (vRNPs) are released into the cytoplasm and imported into the host cell nucleus. Once inside the nucleus, the negative-sense vRNA is transcribed into mRNA, which is transferred to the cytoplasm for translation (for review, see Wandzik et al. 2020). Newly generated HA, NA, M1, and M2 traffic to lipid rafts in the plasma membrane, whereas PB1, PB2, PA, NP, NEP, and a subset of M1 are recycled into the nucleus where they are required for genome replication and nuclear export of progeny vRNPs. Replication takes place in two steps—namely, synthesis of an intermediate positive-sense complementary RNA (cRNA), which in turn is used as a template for the synthesis of additional vRNA for secondary transcription and assembly into vRNPs for packaging in progeny virions (for review, see Fodor and te Velthuis 2019). The final steps of the life cycle are vRNP transport from the perinuclear region to the plasma membrane, followed by particle assembly.
In this review, we will examine the host factors required for nuclear import of the vRNPs and vRNP subunits, transcription and replication of the genome, nuclear export of progeny vRNPs, and trafficking of vRNPs for egress.
NUCLEAR IMPORT
Unlike most RNA viruses that infect vertebrates, influenza virus transcribes and replicates its genome in the host cell nucleus. The advantages of nuclear over cytoplasmic replication are at least fourfold (Hutchinson and Fodor 2012). First, access to the nucleus facilitates cap snatching, the process by which the viral RdRp obtains 5′ cap structures from cellular RNA species to act as primers for transcription of the viral mRNA. Although cap snatching is found also in some cytoplasmic replicating viruses, including the Bunyaviridae and Arenaviridae families (Mir et al. 2008), nuclear cap snatching is deemed more efficient and provides direct access to cellular pathways of cotranscriptional mRNA processing and nuclear export (Engelhardt and Fodor 2006). Second, nuclear replication allows access to the splicing machinery, enabling expansion of the coding capacity of the small genome (∼13.5 kb). Indeed, spliced segments 7 and 8 encode the ion channel M2 and the NEP, respectively, whereas the full-length transcripts encode the M1 matrix protein and the multifunctional innate immune antagonist NS1. Third, direct interaction with the carboxy-terminal domain (CTD) of the cellular RNA polymerase II (RNA Pol II) via the PA subunit of the RdRp leads to inhibition of RNA Pol II elongation and causes it to be degraded, leading to efficient host shutoff (Rodriguez et al. 2007; Vreede and Fodor 2010; Vreede et al. 2010). Fourth, one of the main mechanisms by which influenza virus is detected by the host is via the cytoplasmic pathogen recognition receptor (PRR) RIG-I. Encapsidating newly synthesized RNA within RNPs in the nucleus may partially mask the RNA molecular patterns capable of triggering an antiviral response (Rehwinkel et al. 2010).
Prior to Nuclear Import
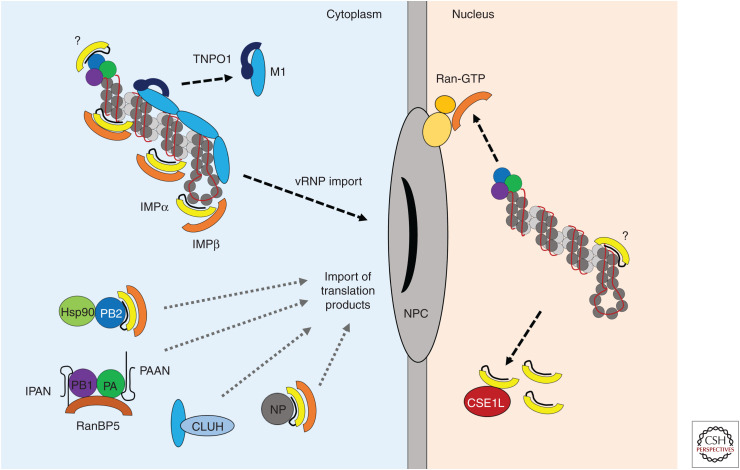
Upon entry into the target cell, vRNPs are dissociated from the viral matrix protein M1 in the low-pH environment of the endocytosed virion when M2 ion channels open within the acidified endosome (Martin and Heleniust 1991a; Bui et al. 1996). The vRNPs are then actively transported into the nucleus where transcription and replication take place (Fig. 1; Martin and Helenius 1991b; Kemler et al. 1994). Recently, TNPO1 (transportin 1), a member of the importin β (IMPβ) family, was implicated in the removal of M1 matrix protein and debundling of vRNPs just after release of the viral cores into the cytoplasm (Miyake et al. 2019). During virus entry, acidification triggers a conformational change in M1 that exposes a noncanonical proline–tyrosine nuclear localization signal (PY-NLS) close to the amino terminus (Lee et al. 2006). TNPO1 then associates with the PY-NLS exposed in the primed M1, stripping M1 from vRNPs and allowing dissociation of the eight vRNPs from each other. vRNPs are subsequently transferred to α importins for nuclear import. IAVs thus use different α and β importins for distinct steps in host cell entry.
Figure 1.
Nuclear import of viral ribonucleoproteins (vRNPs) and translation products. Following fusion of the viral and endosomal membranes, incoming vRNPs associated with M1 matrix protein enter the cytoplasm. The importin β (IMPβ) family member TNPO1/transportin 1 strips M1 from the vRNPs, thus allowing dissociation of the eight vRNPs from each other and releasing them for interactions with the classical nuclear import machinery. vRNPs interact with importin α (IMPα) adaptor proteins that recognize a nonclassical nuclear localization signal (NLS) at the surface-exposed amino terminus of the nucleoprotein (NP). Direct interaction between IMPα and the polymerase basic 2 (PB2) subunit of the viral RNA–dependent RNA polymerase (RdRp) has also been described, but the biological relevance of this is not known. IMPα is complexed with the IMPβ transport receptor, which mediates translocation into the nucleus by interactions with the nuclear pore complex (NPC). IMPβ is dissociated from the cargo in the nucleus by Ran-GTP associated with the NPC. IMPα is removed separately by CSE1L, although some IMPα may remain associated with the vRNP and support viral replication via unknown mechanisms. The importins subsequently cycle back into the cytoplasm. After primary transcription and translation of viral proteins, NP and RdRp components are transported back into the nucleus to support replication and synthesis of progeny vRNPs. PB2 and NP are imported on their own via classical nuclear import, the former in conjunction with its chaperone Hsp90. PB1 and PA are imported as heterodimers in a nonclassical manner through direct interaction with the β importin RanBP5. PB1 and PA are maintained in a stable conformation and protected against degradation by the nonproteinaceous chaperones long noncoding RNA IPAN (lncRNA-IPAN) and PA-associated noncoding RNA (lncRNA-PAAN), respectively. A subset of newly synthesized M1, required for the vRNP nuclear export complex later in the viral life cycle, is believed to associate with the mitochondrial host protein CLUH for nuclear translocation.
The Classical Nuclear Import Pathway
The classical pathway of cellular protein nuclear import is mediated by IMPα adaptor proteins, which recognize basic NLSs on cargo proteins. IMPα isoforms, of which there are seven in humans, are in turn recognized by the importin β (IMPβ) transport receptor via the amino terminal importin β-binding (IBB) domain, which is a functional NLS in itself and autoinhibits IMPα so that recognition of NLSs occurs only when IMPβ is available. The ternary IMPα–IMPβ-cargo complex subsequently traverses the nuclear pore complex (NPC) through interaction of IMPβ with phenylalanine–glycine (FG) repeats on the nucleoporins that compose the NPC. Once inside the nucleus, IMPβ is dissociated by Ran-GTP associated with the NPC. IMPα is dissociated from the cargo by a separate mechanism involving the cellular CSE1L protein (Kutay et al. 1997). Subtle differences in NLS affinity, conformational flexibility, and IBB autoinhibition between IMPα isoforms lead to a flexible and functionally redundant transport machinery. It has been proposed that IMPα subfamilies α2 and α3 provide a fast track for nuclear import of important cellular cargos, and that these subfamilies are specifically targeted by influenza vRNPs (Pumroy et al. 2015).
Nuclear Import of vRNPs
Influenza vRNPs are transported into the nucleus by the interaction between the NP subunits associated with the genome and IMPα (O'Neill et al. 1995). IAV NP carries a “nonclassical” NLS (ncNLS) at the amino terminus, which is surface-exposed and thus available for IMPα binding (Arranz et al. 2012; Moeller et al. 2012). This motif binds IMPα isoforms and is vital for vRNP nuclear import in digitonin-permeabilized cells and for influenza virus replication (Neumann et al. 1997; Wang et al. 1997; Cros et al. 2005). Ultrastructural analysis of the surface availability of the ncNLS on purified vRNPs revealed multiple exposed motifs along the vRNP filament (Wu et al. 2007). IMPα 5 binding prevents the oligomerization of NP in vitro (Boulo et al. 2011), suggesting an additional role for α importins as NP chaperones. It is highly likely that the same IMPα isoform also imports newly synthesized NP into the nucleus during replication.
Once the vRNP has entered the nucleus and primary transcription given rise to new viral proteins, newly synthesized NP and polymerase subunits must be imported into the nucleus to support replication of cRNA and of new vRNA genomes. PB2 has been shown to interact with multiple IMPα isoforms (Tarendeau et al. 2007; Gabriel et al. 2008, 2011), but NP is sufficient to traffic vRNA into the nucleus. Thus, the PB2–IMPα interaction is likely more important for trafficking newly synthesized PB2 into the nucleus to support and transactivate replication (Cros et al. 2005). The mammalian-adaptive D701N substitution in the PB2 subunit has been suggested to specifically bind IMPα isoforms in different host species (Gabriel et al. 2008, 2011; Boivin and Hart 2011), and was shown to promote nuclear import of vRNPs in mammalian cells (Sediri et al. 2015). The latter observation implies some role for PB2 in import of vRNPs but this has not been entirely resolved.
The differential use of different IMPα isoforms has been associated with host range restriction of avian origin polymerases in mammalian cells. Mammalian-adapted viruses are affected most by silencing of importins α1 and α7, whereas avian-origin viruses require importins α1 and α3 (Gabriel et al. 2011). IMPα 7 knockout (KO) mice are less susceptible to influenza infection, and specifically show reduced tropism of virus in the lung (Resa-Infante et al. 2014). Viral mutants that can regain infectivity in this system acquire mutations allowing binding of NP to different isoforms (Resa-Infante et al. 2015, 2019). Adaptation of an avian-signature H7N7 virus by serial lung passage in mice include key adaptations in PB2 (D701N) and NP (N319K) that increase affinity to mammalian IMPα 1.
Noncanonical Roles of α Importins
An additional role for IMPα isoforms 1 and 7 as positive regulators of mammalian-signature but not avian-signature viral polymerase activity in mammalian cells has been suggested independent of nuclear import functions (Resa-Infante et al. 2008; Hudjetz and Gabriel 2012). Importins may remain associated with the viral polymerase even after nuclear import suggesting they play a role in polymerase function. Mutation of the PB2 NLS resulted in reduced RdRp activity in a minireplicon assay despite proper formation of polymerase complexes (Resa-Infante et al. 2008). Moreover, replacing the PB2 NLS with an ectopic SV40 NLS enabled PB2 nuclear import but abrogated polymerase function, again implying that specific IMPα association with the PB2 subunit in the nucleus is required for RdRp activity. RNAi-mediated silencing of IMPα 1 and 7 in human 293 T cells resulted in reduced activity of human-adapted PB2-627K polymerase as measured by minireplicon assay, but the avian-signature 627E polymerase was unaffected (Hudjetz and Gabriel 2012). The exact role of this interaction of PB2 with specific IMPα isoforms is not yet clear.
IMPα-Independent Import of PB1 and PA
Whereas newly synthesized PB2 and NP are imported into the nucleus on their own via the classical import pathway, PA-PB1 is imported as a dimeric subcomplex (Loucaides et al. 2009; Huet et al. 2010; Hutchinson et al. 2011). PB1 contains a bipartite NLS that directly interacts with the β importin-binding protein 5 (RanBP5) (Deng et al. 2006). PA contains NLSs between residues 124–139 and 186–247. PA-PB1 forms a stable heterodimeric submodule that strongly interacts with 5′ vRNA, and moreover forms a stable and stoichiometric 1:1:1 complex with RanBP5 as modeled by small-angle X-ray scattering (SAXS) (Swale et al. 2016). Interestingly, when bound to RanBP5, the PA-PB1 dimer loses its capacity to bind the viral RNA promoter, suggesting that PA-PB1 will pick up the vRNA once it dissociates from RanBP5 after nuclear entry. A recent X-ray crystallography study identified the residues of RanBP5 required for the interaction with influenza virus PA-PB1 heterodimer (Swale et al. 2020). The classical IMPα–IMPβ import pathway is thus not required for nuclear accumulation of PB1 and PA.
TRANSCRIPTION
Upon entry into the nucleus, the incoming viral RdRp associated with the vRNPs transcribes the negative-sense genomic RNA segments into positive-strand mRNA in cis (Fig. 2). mRNA molecules are capped at the 5′ end by an m7G cap, derived from capped nascent host RNA Pol II transcripts, in particular small nucleolar RNAs (snoRNAs) and small nuclear RNAs (snRNAs) (Gu et al. 2015; Koppstein et al. 2015). Cap snatching is carried out by the RdRp utilizing the PB2 cap-binding domain (residues 320–485) and the PA endonuclease (residues 1–196). The 10- to 15-nt capped host RNA fragments are then used as primers for transcription of viral mRNA (for review, see Fodor and te Velthuis 2019; Wandzik et al. 2020).
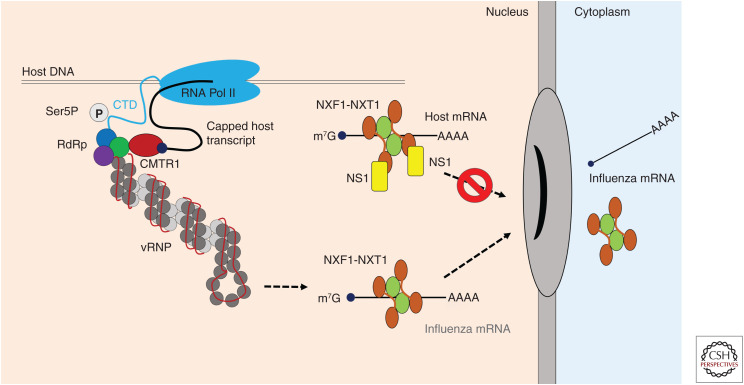
Figure 2.
Transcription and export of viral mRNA. Influenza virus acquires 5′ N7-methyl guanosine (m7G)-capped host transcripts from actively transcribing host RNA polymerase II (RNA Pol II), which it subsequently uses to prime viral transcription. To facilitate cap snatching, the viral RdRp associates with the carboxy-terminal domain (CTD) of the large subunit of RNA Pol II through interaction between the PA subunit and the serine 5 phosphorylated (Ser5P) CTD. The methyltransferase CMTR1 appears to have a role in cap snatching but the mechanism is not known. Influenza mRNAs are exported via the NXF1/TAP pathway through interaction with the mRNA export receptor NXF1-NXT1, which mediates translocation across the nuclear pore complex (NPC) via interaction with nucleoporins. Later in infection, as viral proteins accumulate, NXF1-NXT1 complexes are targeted by the virus-encoded virulence factor nonstructural protein 1 (NS1) to block host mRNA export, in particular, antiviral transcripts. (vRNP) Viral ribonucleoprotein.
The Role of Host RNA Polymerase II
Cap snatching is enabled by direct interaction of the large PA CTD (residues 258–713) with the CTD of host RNA Pol II (for review, see Walker and Fodor 2019). Human cellular RNA Pol II is a complex composed of 12 core subunits; the large subunit RPB1 has an unstructured flexible CTD consisting of 52 heptad repeats with the consensus sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 (Y1S2P3T4S5P6S7). Posttranslational modification of the CTD regulates transcription and cotranscriptional processing of host RNAs (Zaborowska et al. 2016). Early in cellular transcription, Ser5 on the CTD is phosphorylated as a trigger for mRNA-capping enzymes to associate with the CTD. Throughout elongation, CTD Ser5 is gradually dephosphorylated and Ser2 becomes phosphorylated instead. The switch between Ser5P and Ser2P is coupled with the release of 5′ end processing factors and recruitment of 3′ end processing factors. Influenza RdRp interacts directly with Pol II by binding to SerP5 CTD at transcription start sites, which puts it in close proximity to the capped host transcripts it requires for initiating its own transcription (Engelhardt et al. 2005). Fluorescence recovery after photobleaching (FRAP) analysis was harnessed to analyze the intranuclear dynamics of GFP-tagged RdRp in living cells. Treatment of the cells with inhibitors of RNA Pol II actinomycin D and α-amanitin led to mobility shifts of the RdRp, confirming the influence that the cellular Pol II transcription complex has on the viral RNP (Loucaides et al. 2009).
RdRp does not associate with promoter regions of Pol I or Pol III genes, and furthermore shows no affinity with Ser2P CTD associated with elongating Pol II. Pol II activity is required for viral mRNA synthesis but not cRNA or vRNA synthesis. Moreover, viral infection leads to inhibition of polymerase elongation; fewer Pol II units were found associated with internal regions of host genes in infected cells (Chan et al. 2006). This elongation inhibition is likely a result of competitive binding of the viral RdRp to the CTD, blocking its normal interface with host transcriptional machinery. Alternatively or in addition, the removal of 5′ caps from nascent host transcripts by the RdRp may lead to degradation of the remaining transcript by Xrn2 exonuclease, leading to early termination (Brannan et al. 2012).
Interaction of influenza virus with RNA Pol II results in profound changes to the host cell transcripts. A recent study using mammalian native elongating transcript sequencing (mNET-seq) confirmed there is decreased RNA Pol II occupancy downstream of transcription start sites in influenza virus–infected cells. The authors also detected generation of polyadenylated downstream-of-gene transcripts (DoGs) that extend beyond normal poly(A) sites as a result of catastrophically altered termination (CAT) reminiscent of a cellular reaction to severe stress like osmotic shock or heat shock (Bauer et al. 2018).
Recent structural work has provided insight in the molecular details of RdRp PA subunit interaction with the RNA Pol II CTD and how influenza A, B, and C polymerases compare to one another (Lukarska et al. 2017; Serna Martin et al. 2018). Ser5P Pol II CTD binding, in combination with vRNA promoter binding to the RdRp, is believed to promote a transcription preinitiation conformation in the RdRp to facilitate cap snatching; in other words, association with RNA Pol II activates the transcriptase conformation of the viral polymerase. As a result of RdRp interacting with host RNA Pol II, it has been estimated that 8 hr postinfection more than one-half of all the mRNA in an influenza virus–infected cell is viral mRNA (Bercovich-Kinori et al. 2016).
A Role for CMTR1 in Cap Snatching
CMTR1 is the human 2′-O-ribose cap methyltransferase that adds a methyl group to the 5′-7-methylguanosine cap of eukaryotic mRNA to form the cap1 structure (Smietanski et al. 2014). As influenza virus requires 2′-O-methylation of the mRNA cap for efficient recognition and cleavage by PA (Bouloy et al. 1980; Wakai et al. 2011), it was interesting to find CMTR1 as a major hit in a pooled genome-wide CRISPR–Cas9 screen (Li et al. 2020). Infection of CMRT1 KO A549 cells showed that loss of this protein conferred protection against a variety of H1N1 and H3N2 virus strains. Immunoprecipitation of capped viral and host mRNA using an anti-eIF4E antibody revealed that there was less cap1-associated viral mRNA in CMTR1 KO cells, presumably because cap snatching was impaired. Furthermore, loss of CMTR1 resulted in an increased antiviral IFN response in infected cells and conferred synergistic protection against IAV infection with the endonuclease inhibitor baloxavir marboxil. How exactly CMTR1 facilitates cap snatching remains to be elucidated.
Influenza mRNA Nuclear Export
Cellular mRNAs are predominantly exported to the cytoplasm via the NXF1/TAP pathway through acquisition of the principal mRNA export receptor NXF1-NXT1. NXF1-NXT1 interacts with the FG repeats of nucleoporins, which is required for translocation of mRNAs through the NPC (Stewart 2010). The IAV nonstructural protein NS1 is known to inhibit nuclear export of host mRNA, thus contributing to NS1-mediated inhibition of host immune responses (Satterly et al. 2007). Recently, the structural basis for this block was elucidated by cocrystallization of NS1 in complex with NXF1-NXT1, revealing that NS1 acts by preventing binding of NXF1-NXT1 to nucleoporins by occupying the nucleoporin-binding domain of NXF1 (Zhang et al. 2019b). Transcripts of IFN- or immune-regulated genes were among the top transcripts retained in the host nucleus following IAV infection. A mutant virus unable to inhibit NXF1-NXT1 was attenuated.
On the other hand, NXF1 has been routinely detected as an important host factor for influenza replication in genome-wide screens (Hao et al. 2008; Shapira et al. 2009; Karlas et al. 2010; Watanabe et al. 2010) and was also shown to interact with RdRp subunits using a split luciferase complementation screen (Munier et al. 2013). Influenza mRNAs are transported from the nucleus by different host factors including the NXF1 and CRM1 pathways (Wang et al. 2008; Carmody and Wente 2009). RNAi-mediated knockdown of NXF1 showed that segment 4 (HA), 7 (M), and 8 (NS1) mRNAs were the most reliant on this pathway (Read and Digard 2010), and this explains the attenuation of virus replication in cells lacking this host factor. Thus, there is an interesting and unresolved dichotomy between viral reliance on the NXF1 pathway for nuclear export of viral mRNAs and its shutoff via NS1 to avoid expression of antiviral genes.
REPLICATION
Replication of the influenza virus genome occurs in two steps (for review, see Fodor and te Velthuis 2019; Wandzik et al. 2020). First, the RdRp synthesizes cRNA, a process that requires newly synthesized RdRp components and NP (Jorba et al. 2009; York et al. 2013). These cRNPs are then used as templates for the synthesis of additional vRNPs for secondary transcription and later for export across the nuclear membrane and to the plasma membrane for incorporation into progeny virions. cRNA synthesis on the vRNA template is subtly distinct from vRNA synthesis on a cRNA template, which requires primer realignment (for review, see te Velthuis and Fodor 2016). Host factors have been postulated to play supportive roles in all steps of the replication cycle (Fig. 3). Oligomerization of trimeric RdRp molecules into dimers, trimers, or even higher-order complexes is now believed to be required for efficient replication (Chang et al. 2015; Chen et al. 2019; Fan et al. 2019; Peng et al. 2019).
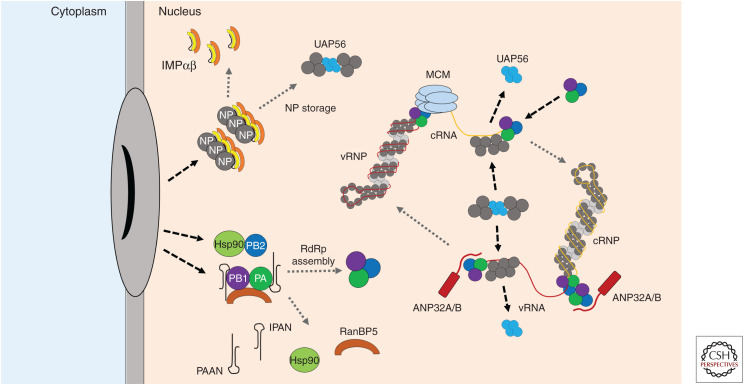
Figure 3.
Replication and ribonucleoprotein (RNP) assembly. As newly synthesized nucleoprotein (NP) and RNA-dependent RNA polymerase (RdRp) components accumulate in the nucleus, the genome is replicated into a positive-sense complementary genome (cRNA), which is packaged into a cRNP by newly synthesized RdRp and NP. cRNA synthesis from a vRNA template is believed to require the minichromosome maintenance (MCM) complex, which stabilizes the transition from initiation to elongation. Functional RdRp complexes are assembled from PA-PB1 dimers and PB2 as their respective chaperones PA-associated noncoding RNA (lncRNA-PAAN), long noncoding RNA IPAN (lncRNA-IPAN), and Hsp90 dissociate. NP assembles with UAP56 after nuclear import, which shields it from aggregation and stores it as trimeric units. UAP56 is subsequently competed off the NP as the latter encapsidates newly synthesized viral RNA. ANP32 proteins are shown as stabilizing a trans-activating polymerase required for cRNA-to-vRNA replication or stabilizing a trimeric RdRp associating with the 3′ end of nascent vRNA. (IMP) Importin.
MCM
The minichromosome maintenance (MCM) complex was identified as a nuclear factor supporting de novo replication initiation of cRNA on a vRNA template (Kawaguchi and Nagata 2007). MCM was shown to interact with the PA subunit of the RdRp, and siRNA-mediated knockdown of subunits MCM2 and MCM3 led to reduced viral RNA accumulation in infected human cells. The authors propose a model in which MCM stabilizes replication elongation complexes during the transition from replication initiation to elongation, preventing abortive replication and allowing the synthesis of full-length cRNA.
Hsp90 and UAP56
Host factors stimulating influenza virus polymerase in vitro were isolated from nuclear extract of uninfected HeLa cells (Shimizu et al. 1994; Momose et al. 1996). Initially called RNA polymerase activating factor (RAF) 1 and RAF2, these distinct fractions were subsequently identified as Hsp90 and UAP56, respectively (Momose et al. 2001, 2002).
Hsp90 is an abundant heat shock protein that functions as a molecular chaperone and contributes to the correct folding, activation, and assembly of a wide variety of transcription factors, steroid receptors, and protein kinases (Picard 2002; Pearl and Prodromou 2006). Hsp90 was found to interact with PB2 in the absence or presence of vRNA, but its RdRp stimulatory activity was mapped to the acidic middle region of Hsp90, not the amino terminal chaperone domain. Normally exclusively cytoplasmic, Hsp90 relocalized to the nucleus in influenza virus–infected cells. Exogenous expression of PB2 or the heterotrimeric RdRp in HeLa cells also resulted in nuclear relocalization of Hsp90 (Naito et al. 2007). Hsp90 was also found to interact with PB1. Treatment of cells with Hsp90 inhibitors resulted in impaired viral growth and reduced the nuclear levels of RNPs, presumably owing to selective degradation of PB1 and PB2 in the absence of functional Hsp90 (Chase et al. 2008).
RAF2/UAP56 is a member of the DExD/H-box RNA helicase family and plays critical roles in healthy cells in pre-mRNA splicing and mRNA nuclear export (Luo et al. 2001; Shen 2009). The ATPase and unwinding activities of UAP56 are required for spliceosome assembly and maturation (Shen et al. 2007, 2008). Furthermore, UAP56 forms part of the transcript export (TREX) complex for efficient export of spliced mRNAs to the cytoplasm (MacMorris et al. 2003).
UAP56 was shown to interact with H1N1 PR8 NP by yeast two-hybrid assay. UAP56 stimulated vRNA synthesis in in vitro assays; interestingly, the helicase activity was not required for this. Furthermore, although interaction of UAP56 with free NP was demonstrated, it did not bind RNA-associated NP. The amino terminal 20 amino acids of NP were found to be sufficient for UAP56 binding. As this is also where the NLS of NP is located, it is possible that IMPα and UAP56 bind competitively to the same motif on NP. The proposal is that UAP56 acts as a chaperone for NP, facilitating formation of NP–RNA complexes. Indeed, chaperones are important for nucleic acid binding of basic proteins such as histones, which tend to aggregate and become inactive in the absence of appropriate substrates (Momose et al. 2001).
UAP56 stimulates the elongation activity of the viral polymerase, possibly by facilitating the encapsidation of nascent cRNA, which is degraded by host cellular nucleases unless it is stabilized by newly synthesized RdRp and NP (Vreede et al. 2004; Kawaguchi et al. 2011); this might work in conjunction with MCM (Kawaguchi and Nagata 2007). Accumulation of vRNA and cRNA was reduced and delayed in UAP56 knockdown cells. Thus, UAP56 facilitates replication reaction–coupled encapsidation of the nascent viral RNA as an NP molecular chaperone (Kawaguchi et al. 2011). Again, the ATP-dependent RNA helicase activity of UAP56 was not required for the encapsidating function.
Binding stoichiometry analysis showed that the NP–UAP56 complex consists of NP and UAP56 at a 3:1 molar ratio, indicating that UAP56 interacts predominantly with trimeric NP. The molecular weight of the complex on the gel filtration column—∼440 kDa—suggests that the NP–UAP56 complex consists of six molecules of NP and two molecules of UAP56 (Hu et al. 2017). This suggests a model in which a UAP56 dimer binds to two trimeric NP complexes, stimulating transfer of NP trimers onto viral RNA as a molecular chaperone. Indeed, trimeric NP has higher RNA-binding activity than monomeric NP (Tarus et al. 2012). Atomic force microscopy (AFM) analysis of NP–UAP56 shows a dumbbell-shaped complex considered as two trimeric NP units connected by dimeric UAP56.
Although not required for its chaperone function, the helicase function of UAP56 (and its paralog URH49) may serve the purpose of preventing dsRNA formation in the cell during influenza virus infection, which would be favorable for the virus to avoid innate immune sensing by PRRs. Indeed, virus infection of UAP56-depleted cells leads to the rapid accumulation of dsRNA in the perinuclear region as well as activation of the dsRNA-dependent PKR, which is part of the antiviral response. Although it cannot be ruled out that accumulation of viral dsRNA was caused by aggregation of NP in the absence of its chaperone UAP56, UAP56 may thus be utilized directly by influenza virus to prevent the formation of dsRNA (Wisskirchen et al. 2011). UAP56 was also found to interact with NS1 but the biological significance of this association remains unclear (Chiba et al. 2018). URH49 shows an identity of 90% at the amino acid level with UAP56 and also interacts with NP. Interestingly, depletion of either URH49 or UAP56 by RNAi led to increased expression levels of the paralog, and URH49, like UAP56, is involved in the nuclear export of viral mRNA (Wisskirchen et al. 2011).
ANP32 Proteins
Acidic (leucine-rich) nuclear phosphoproteins of 32 kDa, a small family comprising ANP32A, ANP32B, and ANP32E, fulfil many cellular functions including the regulation of gene expression, cell death, and intracellular transport (for review, see Reilly et al. 2014). All of the ANP32 proteins associate with histones, albeit in different ways (Tochio et al. 2010; Obri et al. 2014; Saavedra et al. 2017; Kleiner et al. 2018) and they have been linked to the replication cycles of a variety of viruses including paramyxoviruses and retroviruses (Bodem et al. 2011; Wang et al. 2019b; Günther et al. 2020). ANP32 proteins have an amino-terminal leucine-rich repeat (LRR) domain and a low-complexity acidic region (LCAR) (Huyton and Wolberger 2007; Tochio et al. 2010) and are believed to have histone acetyl transferase (HAT) inhibitory activity, placing them at the chromatin within the nucleus.
ANP32A and ANP32B proteins have been identified as host factors for influenza replication in proteomics, RNAi, and CRISPR screens (Bradel-Tretheway et al. 2011; Watanabe et al. 2014; Li et al. 2020). ANP32A (pp32) and ANP32B (APRIL) were purified from nuclear extract of uninfected HeLa cells and were shown to support in vitro synthesis of vRNA on a cRNA template (Sugiyama et al. 2015).
ANP32 proteins have been implicated as key host factors that determine host range restriction of avian influenza viruses in mammalian cells. A functional screen led to the discovery that avian ANP32A, which carries an additional 33 amino acids between the LRR and the LCAR domains, is able to rescue activity of avian-signature (PB2-627E) RdRp in human cells (Long et al. 2016). This finding addressed a long-standing debate about whether restriction of avian influenza polymerase in mammalian cells was due to a positive factor in avian cells (Moncorgé et al. 2010) or a restriction factor in mammalian cells (Mehle and Doudna 2008; Weber et al. 2015). The E627K mutation often seen in adaptation of avian influenza viruses to replication in mammalian cells allows the RdRp to co-opt the shorter mammalian ANP32 proteins, which lack the 33-amino-acid insertion. Artificial insertion of the avian ANP32A-specific 33 amino acids into human ANP32A or ANP32B enabled avian RdRp activity when coexpressed in human cells. ANP32A has since been shown to interact directly with influenza RdRp and localize to the cell nucleus (Domingues and Hale 2017; Baker et al. 2018; Long et al. 2019a; Mistry et al. 2020). In vitro binding assays with purified RdRp components showed that this interaction maps to the 627 domain of PB2 but is independent of the nature of the amino acid at position 627; thus, interaction alone does not explain the host specificity phenotype (Baker et al. 2018).
Twenty-seven of the 33 additional amino acid residues in chicken ANP32A derive from a gene duplication event in the flighted bird ancestor, but the six remaining amino acids are unique to avian ANP32A and contain a SUMO interaction motif-like sequence (SIM) (Domingues and Hale 2017). The SIM confers increased binding to the RdRp complex and is key for the ability to complement avian RdRp. Different bird species encode one or more splice variants of ANP32A, containing either 33, 29, or no additional amino acids. ANP32A lacking the SIM (ANP32A29) was shown to support avian RdRp but to a lesser extent than the variant containing the full 33 amino acids (Baker et al. 2018). Interestingly, ratites like the ostrich lack the gene duplication and thus the additional 33-amino-acid sequence. In support of the notion that the PB2-E627K adaptation facilitates use of shorter mammalian ANP32 proteins, viral replication in ostriches selected for this or other humanizing PB2 mutations (Shinya et al. 2009).
The ratios of ANP32A splice variants (full-length including the 33-amino-acid insertion, 29 amino acids lacking the putative SIM, or mammalian-like lacking the whole 33 amino acids) vary among avian species with passerine species like swallows, blackbirds, and magpies encoding a much larger proportion of the shorter (mammalian-like) splice variants than the migratory reservoir species like ducks and geese normally considered IAV carriers. Competitive replication assays in the presence of mixtures of ANP32A splice variants suggested different splice variants can drive mammalian adaptations in the PB2 gene, and mathematical modeling supported the concept that ANP32A splice ratios in birds could be harnessed to track and predict acquisition and maintenance of mammalian RdRp adaptations (Domingues et al. 2019).
A split luciferase complementation assay was used to demonstrate robust interaction between chicken (chA) or human ANP32A (huA) with IAV RdRp and it was confirmed that chA interacts much more strongly with the RdRp than huA (Mistry et al. 2020). Bimolecular fluorescence complementation (BiFC) assays showed the interaction between the RdRp and ANP32A occurs in the cell nucleus, placing the host factor at the site of influenza replication. Removal of the PB2-627 domain resulted in decreased binding of chA but not huA, suggesting the sequences within the 33-amino-acid insertion in chA are responsible for the increased binding to the RdRp. Indeed, removal of the 33 amino acids, as well as removal of the first four amino acids containing the putative SIM, strongly reduced the interaction and likewise abolished avian-signature RdRp activity. Binding of ANP32A to the RdRp was affected by the presence of viral-like RNA molecules of various lengths, binding of which by the RdRp is known to result in major conformational shifts (Thierry et al. 2016). Binding affinity to ANP32A was increased if the RdRp was inactive, but binding of active RdRp to ANP32A decreased in the presence of the RNA template, suggesting that ANP32A does not interact with actively synthesizing RdRp. Mutations in the vRNA promoter have been shown to enable avian-origin RdRp activity in human cells in the absence of mammalian adaptations in PB2 (Neumann and Hobom 1995; Crescenzo-Chaigne et al. 2002). ANP32 proteins were still essential for replication of this altered RNA however (Mistry et al. 2020).
Solution-state NMR spectroscopy combined with quantitative ensemble analysis was used to analyze complexes formed between the avian and human ANP32A LCAR domain and the avian (PB2-627E) or human-adapted (PB2-627K) IAV polymerase. It was found that the unique avian hexapeptide containing the SIM-like motif likely interacts specifically with the linker between the 627 and NLS domains of PB2. In this study, the E627K mutation did confer tighter binding with the negatively charged human ANP32A LCAR, as it completes a continuous ridge of solvent-exposed positively charged residues available for interaction. Bearing in mind that in other studies binding differences between 627E and 627K RdRp to human ANP32 proteins were not observed, it is still not clear whether interaction alone explains the host range mutation or whether the differences in binding can only be observed using certain methodologies. Interestingly, the authors observed an electrostatic interaction between the known proviral aspartate at position 130 of ANP32A and the basic arginine residue R646 in the 627 domain of PB2 (Camacho-Zarco et al. 2020). Overall, the interactions were shown to be highly flexible and polyvalent. Any role of the LRR domain in binding the RdRp remains unclear.
CRISPR–Cas9-mediated KO of human ANP32A and ANP32B carried out in 293 T cells or low-ploidy eHAP cells demonstrated that ANP32A and ANP32B are functionally redundant essential host factors for influenza virus polymerase activity and replication in human cells (Staller et al. 2019; Zhang et al. 2019a). Deletion of either ANP32A or ANP32B had negligible effects on virus replication, but replication was severely restricted in cells lacking both paralogs (double KO). Residues 129 and 130 at the carboxy-terminal end of the LRR have been heavily implicated in proinfluenza virus function (Long et al. 2019a; Staller et al. 2019; Zhang et al. 2019a). Chicken ANP32B, mouse ANP32A, and human ANP32E lack proinfluenza viral function owing to amino acid substitutions in the proviral dyad 129N-130D. Thus, the redundancy for proviral activity of ANP32 proteins seen in human cells is not present in mice or chickens. Indeed, a conditional ANP32B KO mouse model was shown to resist IAV-induced morbidity and mortality (Beck et al. 2020), despite the presence of wild-type ANP32A in the animals. A recent preprint identified both homozygous and heterozygous individuals for the single-nucleotide variant (SNV) ANP32B-D130A in the Latino population of the gnomAD database (Staller et al. 2020). This variant might exert a dominant-negative effect over both the wild-type ANP32B allele and the functionally redundant wild-type ANP32A alleles, which would mean carriers of the SNV have some protection against influenza virus infection.
It has recently been shown that swine ANP32A (swA), uniquely among mammalian ANP32 proteins, is somewhat capable of supporting avian-signature polymerases in mammalian cells (Peacock et al. 2020; Zhang et al. 2020). This observation supports the notion of pigs as a “mixing vessel” or intermediate host in the generation of pandemic viruses: avian influenza viruses may be capable of just enough replication in the pig host to obtain the necessary mutations for successful zoonosis into other mammalian species. Overexpression experiments in human cells lacking ANP32A and ANP32B expression (dKO) with ANP32A and ANP32B proteins from a variety of natural mammalian host species including human, swine, dog, horse, seal, and bat showed that swA had a unique capacity to rescue polymerase activity and viral replication of a wide variety of avian IAV subtypes (H7N9, H7N2, H5N1, H1N1) as well as swine-origin 2009 pH1N1 and Eurasian avian-like isolates from pigs (Peacock et al. 2020; Zhang et al. 2020). Reciprocal mutations in human and swine ANP32A showed that proavian polymerase activity mapped to residues 106 and 156, which are valine and serine in swA but isoleucine and proline in other mammalian ANP32A proteins. These substitutions result in increased binding of swA to avian-signature influenza RdRp (Peacock et al. 2020; Zhang et al. 2020). Although swine ANP32B has the ability to support RdRp activity, it would appear that in swine cells ANP32A is the dominant proviral factor. CRISPR–Cas9-mediated ablation of swA in pig epithelial PK15 cells (AKO) resulted in a sharp reduction in IAV H7N9 and H9N2 polymerase activity as well as reduced H9N2 titers. Complementation in PK15 AKO cells with huA-I106V/P156S double-mutant increased, whereas swA-V106I/S156P double-mutant reduced H9N2 titers (Zhang et al. 2020). The 2009 pandemic virus had its immediate precursor in swine. A PA-N321K substitution found in second- and third-wave 2009 pH1N1 isolates was likely selected as a direct adaptation to human ANP32 proteins. Direct comparison of the activity of polymerase constellations from a pair of first- (PA-321N) and third-wave (PA-321K) pH1N1 isolates in human and pig cells revealed that human ANP32A has a greater capacity to boost activity of the latter constellation, compared with swine ANP32A (Peacock et al. 2020).
Chicken cells lacking ANP32A expression (chA KO) or expressing a mammalian-like ANP32A protein lacking the 33-amino-acid insertion resulted in restriction of 627E but not 627K polymerase (Long et al. 2019a; Park et al. 2020). As chicken ANP32B and ANP32E transcripts were readily detected in these cells, it is clear that these family members could not support IAV RdRp activity (Long et al. 2019a). The chicken Anp32B gene (chB) encodes isoleucine and asparagine at positions 129 and 130, and this is the reason why it lacks proinfluenza virus function. Split luciferase complementation and coimmunoprecipitation assays show that binding of chB to the viral polymerase is abrogated due to this sequence difference (Long et al. 2019a; Zhang et al. 2019a). In fact, the N129I mutation alone is sufficient to break interaction and proviral activity of ANP32 proteins from a variety of relevant species including humans and pigs (Peacock et al. 2020).
Many avian H7N9 and H9N2 viruses acquire the PB2-E627K substitution when passaged in MDCK cells or mice, but some H9N2 isolates do not. Recombinant viruses in an H7N9 background sensitive to 627K acquisition were generated with single-genome segments from an H9N2 virus that did not evolve 627K. The gene driving the emergence of PB2-E627K in mice was found to be PA; the H7N9 virus with H9N2 PA did not acquire 627K in a mouse passage experiment, whereas the H7N9 PA conferred low RdRp activity in human cells. This phenomenon was mapped to four highly conserved residues in the amino-terminal domain of PA (Liang et al. 2019). H7N9 viruses were further found to replicate poorly in human ANP32A KO cells and mice, which is surprising as KO of both ANP32A and ANP32B is normally required in human cells to see reduced H1N1, H3N2, and humanized H5N1 titers (Staller et al. 2019; Zhang et al. 2019a), whereas murine ANP32A has been shown to lack proinfluenza viral activity (Staller et al. 2019; Zhang et al. 2019a).
To sum up, ANP32 proteins are essential proviral factors that support the replication of influenza virus RNAs by the RdRp. Differences in ANP32 sequences in different species play important roles in host range restriction, and these proteins represent perhaps the most promising host targets for future influenza control strategies.
Nonproteinaceous Host Factors Support Replication
Long noncoding RNAs (lncRNAs) are transcripts of >200 nt that lack peptide-coding capacity but nonetheless fulfil important functions in diverse cellular processes. In addition, many lncRNAs play a role in virus–host interactions (Meng et al. 2017). Hundreds of lncRNAs are up- or down-regulated upon IAV infection, the majority of which are associated with immune responses that contribute to antiviral defense (Peng et al. 2010; Winterling et al. 2014). Some IFN-independent lncRNAs promote rather than counteract influenza virus infection (for review, see Wang and Cen 2020).
In one study, RNA sequencing (RNA-seq) and real-time PCR were used to identify up-regulated lncRNAs in influenza virus–infected A549 cells. Repression of the lncRNA PSMB8-AS1 by CRISPR interference (CRISPRi) led to reduced viral protein accumulation in infected cells and a slight but significant drop in virus titers (More et al. 2019). Another IAV-induced IFN-independent lncRNA, lncRNA-ACOD1 (aconitate decarboxylase 1), was shown to promote viral replication indirectly through its effects on cellular metabolism (Wang et al. 2017). Knockdown of lncRNA-ACOD1 by RNAi resulted in lower H1N1 PR8 viral load in A549 cells. The authors identified glutamic-oxaloacetic transaminase (GOT2), a key metabolic enzyme, as an lncRNA-ACOD1-binding cellular protein. Virus-induced metabolic changes in wild-type cells were abolished when lncRNA-ACOD1 was depleted. The authors propose a model in which lncRNA-ACOD1 facilitates virus replication through stimulating GOT2 activity and production of its metabolites, which can subsequently be harnessed by the virus to support its energy-dependent activities.
Using microarray analysis, several lncRNAs were identified that were differentially expressed during H1N1 WSN infection of A549 cells (Winterling et al. 2014). One of these, the IFN-independent large intergenic ncRNA (lincRNA) VIN (virus-inducible lincRNA), was up-regulated by IAVs of subtypes H7N7, H1N1, and H3N2, but not by IBV. VIN is localized in the cell nucleus and knockdown by RNAi resulted in a tenfold decrease in WSN titers in A549 cells. However, the mechanism through which VIN supports IAV replication has not been elucidated.
A loss-of-function screen using an endoribonuclease-prepared short interfering RNA (esiRNA) library targeting human lncRNAs was carried out in 293 T cells that express Gaussia luciferase (Gluc) upon infection with IAV. This approach yielded two lncRNAs that are hijacked by the viral replication machinery to support efficient viral RNA synthesis (Wang et al. 2018, 2019a). The first of these, PA-associated noncoding RNA (lncRNA-PAAN), was specifically induced by IAV infection in multiple human cell types. Knockdown of lncRNA-PAAN resulted in attenuated viral replication and RdRp activity while also impairing association of the PB1 and PB2 subunits. Native RNA immunoprecipitation (RIP) analysis showed that lncRNA-PAAN interacted with the PA subunit of the polymerase, likely acting as its chaperone while promoting the assembly of the viral polymerase (Wang et al. 2018).
The other major hit lncRNA-IPAN (influenza virus PB1-associated noncoding RNA) is also induced by IAV infection and believed to promote viral replication through its association with PB1, preventing its degradation (Wang et al. 2019a). IAV infection did not only increase the level of lncRNA-IPAN but also recruited it into the nucleus. lncRNA-IPAN silencing led to a strong reduction of NP in knockdown cells, whereas progeny virus titers diminished. CRISPR–Cas9-mediated depletion of lncRNA-IPAN led to a significant reduction in viral protein and RNA levels in KO cells infected with IAV. Reduced RdRp activity in a minireplicon system suggested that lncRNA-IPAN specifically supported IAV RNA replication. Accelerated turnover of PB1 but not PB2 or PA in the absence of lncRNA-IPAN suggested that it plays a role in stabilizing PB1 and indeed PB1 coprecipitates with lncRNA-IPAN. The authors suggest a model in which lncRNA-IPAN associates with PB1 in the cytoplasm where it stabilizes and protects the viral protein from degradation by the host machinery. PB1 then brings lncRNA-IPAN into the nucleus where it dissociates upon formation of the heterotrimeric RdRp complex.
NUCLEAR EXPORT AND BEYOND
Once sufficient progeny vRNPs have been generated, these must be exported out of the nucleus and toward the plasma membrane for budding into new virus particles. Host factors are heavily involved in both processes (Figs. 4 and 5).
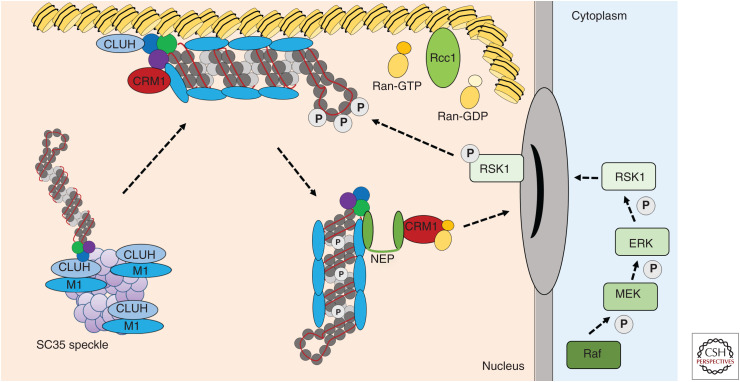
Figure 4.
Nuclear export of progeny viral ribonucleoproteins (vRNPs). Nuclear export of progeny vRNPs is triggered by the Raf/MEK/ERK phosphorylation cascade leading to S269 and S392 phosphorylation of nucleoprotein (NP) by the ERK effector RSK1. Progeny vRNPs are trafficked to the chromatin via SC35 nuclear speckles where they pick up M1, which is held there by the clustered mitochondria protein homolog CLUH. At the chromatin, the export complex is assembled proximal to the Ran guanine exchange factor Rcc1, which regenerates Ran-GTP from Ran-GDP. The soluble export receptor CRM1, also known as exportin 1, also localizes to the chromatin. The vRNP export complex consists of the vRNP coated with M1, which is bound in turn by the viral nuclear export protein (NEP). NEP, in possession of a nuclear export signal (NES), is believed to form a bridge between the vRNP and the CRM1–Ran–GTP complex, which mediates nuclear export through interaction with the nuclear pore complex (NPC).
Figure 5.
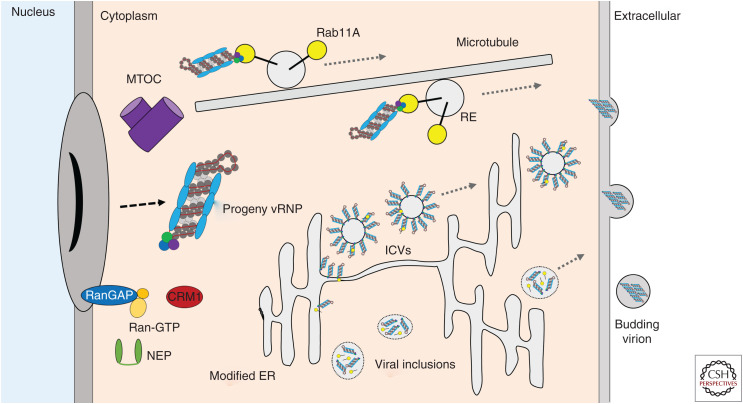
Cytoplasmic trafficking of progeny viral ribonucleoproteins (vRNPs). The nuclear export complex disassembles at the perinuclear region upon Ran-GTP hydrolysis to Ran-GDP by the Ran GTPase-activating protein RanGAP, which is associated with the nuclear pore complex (NPC). The preferred model for what happens next used to be the association of the vRNPs with recycling endosomes (REs) through interaction with Ras-related protein Rab11A at the microtubule organizing center (MTOC). Rab11A-positive RE traffic along microtubules upon recruitment of molecular motors. More recently, involvement of modified endoplasmic reticulum (ER) structures rather than RE has been proposed as driving vRNP trafficking. This model comes in various incarnations, including irregular coated vesicles (ICVs) adorned with vRNPs and viral inclusions that resemble liquid organelles containing the vRNPs. In each of these cases, Rab11A is also associated with the structures, which would explain the colocalization between vRNPs and Rab11A that led to the initial model. All models result in accumulation of vRNPs at the plasma membrane where they are packaged in novel virions for budding.
Involvement of the Raf/MEK/ERK Signaling Pathway
Nuclear export of progeny vRNPs does not occur constitutively but is regulated by cellular signaling pathways to ensure temporal control over vRNP migration to the cytoplasm in the later stages of the viral life cycle. (Pleschka et al. 2001; Nencioni et al. 2009; Eisfeld et al. 2011; Ehrhardt et al. 2013). The Ras-dependent Raf/MEK/ERK mitogen-activated protein (MAP) kinase signaling pathway regulates many cellular functions including proliferation, differentiation, cell metabolism, and immune response. Downstream targets of the pathway may be phosphorylated directly by ERK or alternatively by ERK-activated kinases like the p90 ribosomal S6 kinases (RSK1/2) (Katz et al. 2007; Cargnello and Roux 2011). HA accumulation in the plasma membrane at the late stage of the influenza viral cycle activates the Raf/MEK/ERK pathway, triggering nuclear export of vRNPs (Ludwig et al. 2004; Marjuki et al. 2006). This ensures timely regulation of vRNP export late in the viral life cycle when sufficient glycoproteins have accumulated for budding of progeny virions. MEK inhibitors may be promising antivirals against IAV and IBV; no escape mutants were detected after multipassage use (Ludwig et al. 2004). MEK and ERK were identified separately as host factors required for influenza virus replication in a genome-wide RNAi screen (König et al. 2010).
MEK inhibition also alters the interaction of M1 with vRNPs, thus resulting in a block in the assembly of the export complex at the dense chromatin (Schreiber et al. 2020). Two phosphorylation sites on the viral NP, serine 269 and serine 392, remained unphosphorylated in the presence of the MEK inhibitor CI-1040. Phosphorylation of these residues is carried out by RSK1, whose activation is mediated by virus-induced Raf/MEK/ERK pathway activity. Knockdown by siRNA of the RSK1 isoform, as well as inhibition of both RSK isoforms by the compound BI-D1870, led to nuclear retention of vRNPs and reduced virus titers in A549 cells. Knockdown of RSK2, a related kinase activated in the same way, increased virus replication, which was not surprising as RSK2 is involved in antiviral response (Kakugawa et al. 2009). Because the RSK inhibitors block both isoforms, the proviral function of RSK1 seems dominant over the antiviral function of RSK2.
Involvement of the CRM1 Pathway
CRM1 is a soluble adaptor molecule that binds to leucine-rich nuclear export signals (NESs) on cargo proteins in the nucleoplasm. It then forms a trimeric complex with Ran-GTP that mediates export of the substrate (Petosa et al. 2004). Leptomycin B (LMB) specifically inhibits CRM1/exportin 1, a member of the IMPβ family, by covalently modifying a specific cysteine residue, which is thought to interfere with the normal formation of a stable complex with the export substrate (Kudo et al. 1999).
CRM1 was identified as a host factor required for influenza virus replication in a combined Co-IP/LC/MS and RNAi screen (Watanabe et al. 2010). NP and CRM1 were found to interact in vivo and nuclear retention of both RNA-free NP and RNPs is seen in infected cells treated with LMB. Conversely, cotransfection of exogenous CRM1 resulted in a significant increase in cytoplasmic NP. The inhibitory effect of LMB on RNP nuclear export was seen with a variety of IAV strains and in multiple cell lines including primary chicken embryo fibroblasts (CEFs), so use of the CRM1 export pathway is a general feature of influenza viruses (Elton et al. 2001; Watanabe et al. 2001).
vRNPs are exported in conjunction with M1 and NEP. Indeed, NEP interacted directly with CRM1 in a mammalian two-hybrid system, and NEP was crucial for nuclear export of vRNP complexes (Neumann et al. 2000). Although the amino terminal NES of NEP was not required for NEP binding to CRM1, mutations in the NES did lead to nuclear retention of NP, so the NES functions critically in the nuclear export of NP and, by extension, of vRNPs (Neumann et al. 2000). NEP was also the only viral protein to interact with nucleoporins in a yeast two-hybrid system, and antibodies directed against NEP, injected directly into the cell nucleus, inhibited the cytoplasmic accumulation of free NP and RNPs. The NEP amino-terminal effector domain was recognized as a bona fide NES comparable with known NESs of viral and cellular origin, and mapped to amino acid residues 11–23 (O'Neill et al. 1998). A second NES has also been proposed (Huang et al. 2013).
The daisy-chain model of nuclear export of vRNPs postulates that M1 binds directly to vRNPs with NEP acting as a bridge between M1 and CRM1 to facilitate translocation across the NPC. It is believed that the vRNP export complexes assemble at the host chromatin, bringing them in close contact to the Ran guanine exchange factor Rcc1, which is responsible for generating Ran-GTP and driving CRM1-dependent nuclear export (Chase et al. 2011). Recycled cellular CRM1–Ran-GTP complexes are thus usurped by influenza vRNPs after nucleotide exchange by Rcc1 on the chromatin. In this way, influenza virus gains preferential access to host cell export machinery by locating vRNP cargo proximal to the sites of Ran-GTP regeneration (Chase et al. 2011). vRNA but not cRNA was found in the relevant chromatin fraction, alongside proteinaceous RNP components, M1, small amounts of NEP, Rcc1, and core histone H3. CRM1 was found to relocalize to the sites on dense chromatin where vRNP export complexes also accumulate (Chase et al. 2011).
A subset of newly synthesized M1 is imported into the nucleus via its NLSs where it interacts with the newly formed vRNPs, in particular NP (Martin and Heleniust 1991a; Whittaker et al. 1996). However, H5N1 virus M1 alone appeared incapable of interacting with the vRNP in the nucleus. Rather, vRNP–M1 interaction was found to require the CTD of NEP (Brunotte et al. 2014). This leads to a slightly different model in which the CTD of NEP enhances the binding affinity of M1 to the vRNP. All in all, it appears that progeny vRNPs associated with viral M1 and NEP are exported to the cytoplasm through the CRM1-dependent nuclear export pathway as vRNP–M1–NEP–CRM1 complexes.
The Involvement of CLUH
CLUH is a PB2-binding host protein with a role in subcellular transport of vRNPs (Watanabe et al. 2010). CLUH plays a key role in the subnuclear transport of vRNPs to the assembly site of viral nuclear export complexes via nuclear speckles (Ando et al. 2016). Pandemic 2009 H1N1, seasonal H3N2, and IBV titers were reduced in cells depleted of CLUH by siRNA and nuclear retention of M1, NEP, and vRNPs was observed in the CLUH knockdown cells. CLUH normally functions as a mitochondrial protein, but in infected cells it translocates to the nucleoplasm and nuclear matrix where it colocalizes with PB2 and M1. CLUH and M1 accumulate in SC32-positive nuclear speckles where they are joined by progeny vRNPs via interaction with PB2. They are then translocated to the chromatin-bound region where the vRNP export complexes are assembled in the presence of NEP and CRM1. Thus, CLUH is involved in the subnuclear transport of progeny vRNPs to the assembly site of viral nuclear export complexes at the dense chromatin via SC35-positive speckles.
Progeny vRNP Transport to the Plasma Membrane
Until recently, it seemed clear-cut that vRNPs are transported from the nucleus to the plasma membrane on microtubule networks in association with Rab11A-positive recycling endosomes (REs) (Bruce et al. 2010; Amorim 2019). Rab11A specifically marks REs, which sort and transport cargo to the apical cell membrane. Rab11A recruits molecular motors to RE through interaction with the Rab11 family interacting proteins (FIPs). Rab11-FIPs can associate with both actin and microtubules; therefore, Rab11A-RE can use multiple cytoskeletal networks for transport.
After nuclear export, progeny vRNPs accumulate near the microtubule organizing center (MTOC) adjacent to the nuclear membrane (Momose et al. 2007; Amorim et al. 2011). Live-cell imaging approaches using fluorescently tagged vRNP components have shown vRNP movement along microtubules with a characteristic intermittent saltatory motion (Amorim et al. 2011; Avilov et al. 2012). Cells treated with microtubule depolymerizing agents had altered vRNP distribution and reduced viral growth. Depletion of Rab11A by RNAi and overexpression of a dominant-negative Rab11A mutant (Rab11A-GDP) impaired the association of vRNPs with Rab11A-positive vesicles, disrupted accumulation of vRNPs at the plasma membrane, and sharply reduced the output of infectious progeny virus (Eisfeld et al. 2011). Several recent reports, however, have put the question to the accepted model of vRNP trafficking along microtubules in association with Rab11A.
Discrepant results included the modest reduction in viral titers seen on depolymerization of microtubules and decrease in Rab11A binding of FIP adaptors upon infection, suggesting that association between Rab11A and microtubules might be compromised (Vale-Costa et al. 2016). Ultrastructural inspection of cytoplasmic sites positive for Rab11A and vRNPs, using correlative light and electron microscopy (CLEM) showed clustered vesicles of heterogeneous sizes from where coiled-coil structures resembling vRNPs protruded. Similar vesicles, renamed irregular coated vesicles (ICVs), were observed more recently by EM, which seemed to extend from an extensively swollen and tubulated endoplasmic reticulum (ER) (de Castro Martin et al. 2017). This suggests that it may be extensions of the ER and not the REs that regulate vRNP transport. Rab11A was found associated with the modified ER and with irregular coated vesicles as well. So this alternative model postulates that once progeny vRNPs reach the cytoplasm, they first bind to a modified ER from where Rab11A-coated vesicles loaded with vRNPs are subsequently released and directed to the plasma membrane (Amorim 2019).
Another report also suggested the existence of a Rab11A-RE independent pathway for cytoplasmic transport of vRNPs, based on the observation that vRNP subcellular location could be uncoupled from Rab11A in the presence of the microtubule depolymerization agent nocodazole (Nturibi et al. 2017). Further supporting data for involvement of the ER rather than microtubules comes from the detection of viral inclusions forming in the vicinity of ER exit sites (ERES) (Alenquer et al. 2019). These inclusions are not membrane-bound, rather they have properties of liquid organelles and contain both vRNPs and Rab11A. Thus, accumulating evidence suggests that Rab11 subcellular localization is redirected and its function is impaired during IAV infection.
Three-dimensional movement of fluorescently tagged Rab11A-RE and IAV vRNPs was tracked in infected A549 cells using dual-view inverted selective-plane illumination microscopy (diSPIM). Although Rab11A motion was dependent on microtubules in A549 cells, depletion of microtubule filaments by nocodazole treatment had little impact on vRNP movement (Bhagwat et al. 2020). In addition, a large reduction in the amount of dynein, the minus-end-directed microtubule motor, associated with Rab11A was observed in IAV-infected cells. This might be the mechanism behind altered Rab11A movement seen during IAV infection. As more than one-half of the IAV vRNP puncta moved independently of Rab11A spots and vice versa, this study provides yet more evidence for a microtubule and Rab11A-independent mechanism of cytoplasmic vRNP transport.
A unifying explanation would be that influenza virus has evolved several mechanisms of cytoplasmic vRNP trafficking involving either Rab11A-RE and the microtubule network or the ER and alternative vesicles or even liquid-phase organelles.
DISCUSSION
Cellular factors are involved in all stages of the influenza life cycle. As details of these essential virus–host interactions are unveiled, we may be given the opportunity to target them specifically, leading to novel means of intervention. Understanding how viral pathogens usurp the cell machinery is key for the development of antiviral agents. At the same time, studying how and why viruses subvert cellular proteins and pathways will give us valuable insights in the workings of the host cell itself. As RNA viruses are prone to rapid evolution, compounds aimed at virus-encoded proteins will inexorably result in the rapid emergence of drug resistance. A focus on host-directed therapy may lead to novel longer-lasting therapeutics and antiviral strategies.
ACKNOWLEDGMENT
This article has been made freely available online courtesy of TAUNS Laboratories.
Footnotes
Editors: Gabriele Neumann and Yoshihiro Kawaoka
Additional Perspectives on Influenza: The Cutting Edge available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this subject collection.
- Alenquer M, Vale-Costa S, Etibor TA, Ferreira F, Sousa AL, Amorim MJ. 2019. Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat Commun 10: 1629. 10.1038/s41467-019-09549-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim MJ. 2019. A comprehensive review on the interaction between the host GTPase Rab11 and influenza A virus. Front Cell Dev Biol 6: 176. 10.3389/fcell.2018.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim MJ, Bruce EA, Read EK, Foeglein A, Mahen R, Stuart AD, Digard P. 2011. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J Virol 85: 4143–4156. 10.1128/JVI.02606-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Yamayoshi S, Tomita Y, Watanabe S, Watanabe T, Kawaoka Y. 2016. The host protein CLUH participates in the subnuclear transport of influenza virus ribonucleoprotein complexes. Nat Microbiol 1: 16062. 10.1038/nmicrobiol.2016.62 [DOI] [PubMed] [Google Scholar]
- Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J. 2012. The structure of native influenza virion ribonucleoproteins. Science 338: 1634–1637. 10.1126/science.1228172 [DOI] [PubMed] [Google Scholar]
- Avilov SV, Moisy D, Naffakh N, Cusack S. 2012. Influenza A virus progeny vRNP trafficking in live infected cells studied with the virus-encoded fluorescently tagged PB2 protein. Vaccine 30: 7411–7417. 10.1016/j.vaccine.2012.09.077 [DOI] [PubMed] [Google Scholar]
- Baker SF, Ledwith MP, Mehle A. 2018. Differential splicing of ANP32A in birds alters its ability to stimulate RNA synthesis by restricted influenza polymerase. Cell Rep 24: 2581–2588.e4. 10.1016/j.celrep.2018.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DLV, Tellier M, Martínez-Alonso M, Nojima T, Proudfoot NJ, Murphy S, Fodor E. 2018. Influenza virus mounts a two-pronged attack on host RNA polymerase II transcription. Cell Rep 23: 2119–2129.e3. 10.1016/j.celrep.2018.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S, Zickler M, Pinho dos Reis V, Günther T, Grundhoff A, Reilly PT, Mak TW, Stanelle-Bertram S, Gabriel G. 2020. ANP32B deficiency protects mice from lethal influenza A virus challenge by dampening the host immune response. Front Immunol 11: 450. 10.3389/fimmu.2020.00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercovich-Kinori A, Tai J, Gelbart IA, Shitrit A, Ben-Moshe S, Drori Y, Itzkovitz S, Mandelboim M, Stern-Ginossar N. 2016. A systematic view on influenza induced host shutoff. eLife 5: e18311. 10.7554/eLife.18311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat AR, Le Sage V, Nturibi E, Kulej K, Jones J, Guo M, Tae Kim E, Garcia BA, Weitzman MD, Shroff H, et al. 2020. Quantitative live cell imaging reveals influenza virus manipulation of Rab11A transport through reduced dynein association. Nat Commun 11: 23. 10.1038/s41467-019-13838-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodem J, Schied T, Gabriel R, Rammling M, Rethwilm A. 2011. Foamy virus nuclear RNA export is distinct from that of other retroviruses. J Virol 85: 2333–2341. 10.1128/JVI.01518-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin S, Hart DJ. 2011. Interaction of the influenza A virus polymerase PB2 C-terminal region with importin α isoforms provides insights into host adaptation and polymerase assembly. J Biol Chem 286: 10439–10448. 10.1074/jbc.M110.182964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulo S, Akarsu H, Lotteau V, Müller CW, Ruigrok RW, Baudin F. 2011. Human importin α and RNA do not compete for binding to influenza A virus nucleoprotein. Virology 409: 84–90. 10.1016/j.virol.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Bouloy M, Plotch SJ, Krug RM. 1980. Both the 7-methyl and the 2′-O-methyl groups in the cap of mRNA strongly influence its ability to act as primer for influenza virus RNA transcription. Proc Natl Acad Sci 77: 3952–3956. 10.1073/pnas.77.7.3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradel-Tretheway BG, Mattiacio JL, Krasnoselsky A, Stevenson C, Purdy D, Dewhurst S, Katze MG. 2011. Comprehensive proteomic analysis of influenza virus polymerase complex reveals a novel association with mitochondrial proteins and RNA polymerase accessory factors. J Virol 85: 8569–8581. 10.1128/JVI.00496-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, et al. 2012. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell 46: 311–324. 10.1016/j.molcel.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce EA, Digard P, Stuart AD. 2010. The Rab11 pathway is required for influenza A virus budding and filament formation. J Virol 84: 5848–5859. 10.1128/JVI.00307-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunotte L, Flies J, Bolte H, Reuther P, Vreede F, Schwemmle M. 2014. The nuclear export protein of H5N1 influenza A viruses recruits matrix 1 (M1) protein to the viral ribonucleoprotein to mediate nuclear export. J Biol Chem 289: 20067–20077. 10.1074/jbc.M114.569178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M, Whittaker G, Helenius A. 1996. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol 70: 8391–8401. 10.1128/JVI.70.12.8391-8401.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Zarco AR, Kalayil S, Maurin D, Salvi N, Delaforge E, Milles S, Jensen MR, Hart DJ, Cusack S, Blackledge M. 2020. Molecular basis of host-adaptation interactions between influenza virus polymerase PB2 subunit and ANP32A. Nat Commun 11: 3656. 10.1038/s41467-020-17407-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargnello M, Roux PP. 2011. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev 75: 50–83. 10.1128/MMBR.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody SR, Wente SR. 2009. mRNA nuclear export at a glance. J Cell Sci 122: 1933–1937. 10.1242/jcs.041236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AY, Vreede FT, Smith M, Engelhardt OG, Fodor E. 2006. Influenza virus inhibits RNA polymerase II elongation. Virology 351: 210–217. 10.1016/j.virol.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Chang S, Sun D, Liang H, Wang J, Li J, Guo L, Wang X, Guan C, Boruah BM, Yuan L, et al. 2015. Cryo-EM structure of influenza virus RNA polymerase complex at 4.3 Å resolution. Mol Cell 57: 925–935. 10.1016/j.molcel.2014.12.031 [DOI] [PubMed] [Google Scholar]
- Chase G, Deng T, Fodor E, Leung BW, Mayer D, Schwemmle M, Brownlee G. 2008. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology 377: 431–439. 10.1016/j.virol.2008.04.040 [DOI] [PubMed] [Google Scholar]
- Chase GP, Rameix-Welti MA, Zvirbliene A, Zvirblis G, Götz V, Wolff T, Naffakh N, Schwemmle M. 2011. Influenza virus ribonucleoprotein complexes gain preferential access to cellular export machinery through chromatin targeting. PLoS Pathog 7: e1002187. 10.1371/journal.ppat.1002187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KY, Santos Afonso ED, Enouf V, Isel C, Naffakh N. 2019. Influenza virus polymerase subunits co-evolve to ensure proper levels of dimerization of the heterotrimer. PLoS Pathog 15: e1008034. 10.1371/journal.ppat.1008034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Hill-Batorski L, Neumann G, Kawaoka Y. 2018. The cellular DExD/H-box RNA helicase UAP56 co-localizes with the influenza A virus NS1 protein. Front Microbiol 9: 2192. 10.3389/fmicb.2018.02192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescenzo-Chaigne B, van der Werf S, Naffakh N. 2002. Differential effect of nucleotide substitutions in the 3′ arm of the influenza A virus vRNA promoter on transcription/replication by avian and human polymerase complexes is related to the nature of PB2 amino acid 627. Virology 303: 240–252. 10.1006/viro.2002.1637 [DOI] [PubMed] [Google Scholar]
- Cros JF, García-Sastre A, Palese P. 2005. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 6: 205–213. 10.1111/j.1600-0854.2005.00263.x [DOI] [PubMed] [Google Scholar]
- Czudai-Matwich V, Otte A, Matrosovich M, Gabriel G, Klenk HD. 2014. PB2 mutations D701N and S714R promote adaptation of an influenza H5N1 virus to a mammalian host. J Virol 88: 8735–8742. 10.1128/JVI.00422-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro Martin IF, Fournier G, Sachse M, Pizarro-Cerda J, Risco C, Naffakh N. 2017. Influenza virus genome reaches the plasma membrane via a modified endoplasmic reticulum and Rab11-dependent vesicles. Nat Commun 8: 1396. 10.1038/s41467-017-01557-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Engelhardt OG, Thomas B, Akoulitchev AV, Brownlee GG, Fodor E. 2006. Role of Ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. J Virol 80: 11911–11919. 10.1128/JVI.01565-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues P, Hale BG. 2017. Functional insights into ANP32A-dependent influenza A virus polymerase host restriction. Cell Rep 20: 2538–2546. 10.1016/j.celrep.2017.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues P, Eletto D, Magnus C, Turkington HL, Schmutz S, Zagordi O, Lenk M, Beer M, Stertz S, Hale BG. 2019. Profiling host ANP32A splicing landscapes to predict influenza A virus polymerase adaptation. Nat Commun 10: 3396. 10.1038/s41467-019-11388-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt C, Rückle A, Hrincius ER, Haasbach E, Anhlan D, Ahmann K, Banning C, Reiling SJ, Kühn J, Strobl S, et al. 2013. The NF-κB inhibitor SC75741 efficiently blocks influenza virus propagation and confers a high barrier for development of viral resistance. Cell Microbiol 15: 1198–1211. 10.1111/cmi.12108 [DOI] [PubMed] [Google Scholar]
- Eisfeld AJ, Kawakami E, Watanabe T, Neumann G, Kawaoka Y. 2011. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J Virol 85: 6117–6126. 10.1128/JVI.00378-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeld AJ, Neumann G, Kawaoka Y. 2015. At the centre: influenza A virus ribonucleoproteins. Nat Rev Microbiol 13: 28–41. 10.1038/nrmicro3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton D, Simpson-Holley M, Archer K, Medcalf L, Hallam R, McCauley J, Digard P. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J Virol 75: 408–419. 10.1128/JVI.75.1.408-419.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt OG, Fodor E. 2006. Functional association between viral and cellular transcription during influenza virus infection. Rev Med Virol 16: 329–345. 10.1002/rmv.512 [DOI] [PubMed] [Google Scholar]
- Engelhardt OG, Smith M, Fodor E. 2005. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J Virol 79: 5812–5818. 10.1128/JVI.79.9.5812-5818.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Walker AP, Carrique L, Keown JR, Serna Martin I, Karia D, Sharps J, Hengrung N, Pardon E, Steyaert J, et al. 2019. Structures of influenza A virus RNA polymerase offer insight into viral genome replication. Nature 573: 287–290. 10.1038/s41586-019-1530-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finberg RW, Lanno R, Anderson D, Fleischhackl R, van Duijnhoven W, Kauffman RS, Kosoglou T, Vingerhoets J, Leopold L. 2019. Phase 2b study of pimodivir (JNJ-63623872) as monotherapy or in combination with oseltamivir for treatment of acute uncomplicated seasonal influenza A: TOPAZ trial. J Infect Dis 219: 1026–1034. 10.1093/infdis/jiy547 [DOI] [PubMed] [Google Scholar]
- *.Fodor E, te Velthuis AJW. 2019. Structure and function of the influenza virus transcription and replication machinery. Cold Spring Harb Perspect Med 10: a038398. 10.1101/cshperspect.a038398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Herwig A, Klenk HD. 2008. Interaction of polymerase subunit PB2 and NP with importin α1 is a determinant of host range of influenza A virus. PLoS Pathog 4: e11. 10.1371/journal.ppat.0040011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Klingel K, Otte A, Thiele S, Hudjetz B, Arman-Kalcek G, Sauter M, Shmidt T, Rother F, Baumgarte S, et al. 2011. Differential use of importin-α isoforms governs cell tropism and host adaptation of influenza virus. Nat Commun 2: 156. 10.1038/ncomms1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhill DH, te Velthuis AJW, Fletcher RA, Langat P, Zambon M, Lackenby A, Barclay WS. 2018. The mechanism of resistance to favipiravir in influenza. Proc Natl Acad Sci 115: 11613–11618. 10.1073/pnas.1811345115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Gallagher GR, Dai W, Liu P, Li R, Trombly MI, Gammon DB, Mello CC, Wang JP, Finberg RW. 2015. Influenza A virus preferentially snatches noncoding RNA caps. RNA 21: 2067–2075. 10.1261/rna.054221.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther M, Bauer A, Müller M, Zaeck L, Finke S. 2020. Interaction of host cellular factor ANP32B with matrix proteins of different paramyxoviruses. J Gen Virol 101: 44–58. [DOI] [PubMed] [Google Scholar]
- Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454: 890–893. 10.1038/nature07151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden FG, Sugaya N, Hirotsu N, Lee N, de Jong MD, Hurt AC, Ishida T, Sekino H, Yamada K, Portsmouth S, et al. 2018. Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N Engl J Med 379: 913–923. 10.1056/NEJMoa1716197 [DOI] [PubMed] [Google Scholar]
- Hirotsu N, Sakaguchi H, Sato C, Ishibashi T, Baba K, Omoto S, Shishido T, Tsuchiya K, Hayden FG, Uehara T, et al. 2020. Baloxavir marboxil in Japanese pediatric patients with influenza: safety and clinical and virologic outcomes. Clin Infect Dis 71: 971–981. 10.1093/cid/ciz908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Gor V, Morikawa K, Nagata K, Kawaguchi A. 2017. Cellular splicing factor UAP56 stimulates trimeric NP formation for assembly of functional influenza viral ribonucleoprotein complexes. Sci Rep 7: 14053. 10.1038/s41598-017-13784-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Chen J, Chen Q, Wang H, Yao Y, Chen J, Chen Z. 2013. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. J Virol 87: 767–778. 10.1128/JVI.06519-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudjetz B, Gabriel G. 2012. Human-like PB2 627K influenza virus polymerase activity is regulated by importin-α1 and -α7. PLoS Pathog 8: e1002488. 10.1371/journal.ppat.1002488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet S, Avilov SV, Ferbitz L, Daigle N, Cusack S, Ellenberg J. 2010. Nuclear import and assembly of influenza A virus RNA polymerase studied in live cells by fluorescence cross-correlation spectroscopy. J Virol 84: 1254–1264. 10.1128/JVI.01533-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson EC, Fodor E. 2012. Nuclear import of the influenza A virus transcriptional machinery. Vaccine 30: 7353–7358. 10.1016/j.vaccine.2012.04.085 [DOI] [PubMed] [Google Scholar]
- Hutchinson EC, Orr OE, Man Liu S, Engelhardt OG, Fodor E. 2011. Characterization of the interaction between the influenza A virus polymerase subunit PB1 and the host nuclear import factor Ran-binding protein 5. J Gen Virol 92: 1859–1869. 10.1099/vir.0.032813-0 [DOI] [PubMed] [Google Scholar]
- Huyton T, Wolberger C. 2007. The crystal structure of the tumor suppressor protein pp32 (Anp32a): structural insights into Anp32 family of proteins. Protein Sci 16: 1308–1315. 10.1110/ps.072803507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorba N, Coloma R, Ortín J. 2009. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog 5: e1000462. 10.1371/journal.ppat.1000462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakugawa S, Shimojima M, Goto H, Horimoto T, Oshimori N, Neumann G, Yamamoto T, Kawaoka Y. 2009. Mitogen-activated protein kinase-activated kinase RSK2 plays a role in innate immune responses to influenza virus infection. J Virol 83: 2510–2517. 10.1128/JVI.02416-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, et al. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463: 818–822. 10.1038/nature08760 [DOI] [PubMed] [Google Scholar]
- Katz M, Amit I, Yarden Y. 2007. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim Biophys Acta 1773: 1161–1176. 10.1016/j.bbamcr.2007.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann SHE, Dorhoi A, Hotchkiss RS, Bartenschlager R. 2018. Host-directed therapies for bacterial and viral infections. Nat Rev Drug Discov 17: 35–56. 10.1038/nrd.2017.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Nagata K. 2007. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. EMBO J 26: 4566–4575. 10.1038/sj.emboj.7601881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Momose F, Nagata K. 2011. Replication-coupled and host factor-mediated encapsidation of the influenza virus genome by viral nucleoprotein. J Virol 85: 6197–6204. 10.1128/JVI.00277-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler I, Whittaker G, Helenius A. 1994. Nuclear import of microinjected influenza virus ribonucleoproteins. Virology 202: 1028–1033. 10.1006/viro.1994.1432 [DOI] [PubMed] [Google Scholar]
- Kleiner RE, Hang LE, Molloy KR, Chait BT, Kapoor TM. 2018. A chemical proteomics approach to reveal direct protein–protein interactions in living cells. Cell Chem Biol 25: 110–120.e3. 10.1016/j.chembiol.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, et al. 2010. Human host factors required for influenza virus replication. Nature 463: 813–817. 10.1038/nature08699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppstein D, Ashour J, Bartel DP. 2015. Sequencing the cap-snatching repertoire of H1N1 influenza provides insight into the mechanism of viral transcription initiation. Nucleic Acids Res 43: 5052–5064. 10.1093/nar/gkv333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG, et al. 2018. Influenza. Nat Rev Dis Primers 4: 3. 10.1038/s41572-018-0002-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci 96: 9112–9117. 10.1073/pnas.96.16.9112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. 1997. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 90: 1061–1071. 10.1016/S0092-8674(00)80372-4 [DOI] [PubMed] [Google Scholar]
- Lee BJ, Cansizoglu AE, Süel KE, Louis TH, Zhang Z, Chook YM. 2006. Rules for nuclear localization sequence recognition by karyopherin β2. Cell 126: 543–558. 10.1016/j.cell.2006.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Clohisey SM, Chia BS, Wang B, Cui A, Eisenhaure T, Schweitzer LD, Hoover P, Parkinson NJ, Nachshon A, et al. 2020. Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection. Nat Commun 11: 164. 10.1038/s41467-019-13965-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Jiang L, Li J, Zhao Q, Wang J, He X, Huang S, Wang Q, Zhao Y, Wang G, et al. 2019. Low polymerase activity attributed to PA drives the acquisition of the PB2 E627K mutation of H7N9 avian influenza virus in mammals. MBio 10: e01162.. 10.1128/mBio.01162-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JS, Giotis ES, Moncorgé O, Frise R, Mistry B, James J, Morisson M, Iqbal M, Vignal A, Skinner MA, et al. 2016. Species difference in ANP32A underlies influenza A virus polymerase host restriction. Nature 529: 101–104. 10.1038/nature16474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JS, Idoko-Akoh A, Mistry B, Goldhill D, Staller E, Schreyer J, Ross C, Goodbourn S, Shelton H, Skinner MA, et al. 2019a. Species specific differences in use of ANP32 proteins by influenza A virus. eLife 8: e45066. 10.7554/eLife.45066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JS, Mistry B, Haslam SM, Barclay WS. 2019b. Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol 17: 67–81. 10.1038/s41579-018-0115-z [DOI] [PubMed] [Google Scholar]
- Loucaides EM, von Kirchbach JC, Foeglein A, Sharps J, Fodor E, Digard P. 2009. Nuclear dynamics of influenza A virus ribonucleoproteins revealed by live-cell imaging studies. Virology 394: 154–163. 10.1016/j.virol.2009.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig S, Wolff T, Ehrhardt C, Wurzer WJ, Reinhardt J, Planz O, Pleschka S. 2004. MEK inhibition impairs influenza B virus propagation without emergence of resistant variants. FEBS Lett 561: 37–43. 10.1016/S0014-5793(04)00108-5 [DOI] [PubMed] [Google Scholar]
- Lukarska M, Fournier G, Pflug A, Resa-Infante P, Reich S, Naffakh N, Cusack S. 2017. Structural basis of an essential interaction between influenza polymerase and Pol II CTD. Nature 541: 117–121. 10.1038/nature20594 [DOI] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644–647. 10.1038/35098106 [DOI] [PubMed] [Google Scholar]
- Lycett SJ, Duchatel F, Digard P. 2019. A brief history of bird flu. Philos Trans R Soc Lond B Biol Sci 374: 20180257. 10.1098/rstb.2018.0257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMorris M, Brocker C, Blumenthal T. 2003. UAP56 levels affect viability and mRNA export in Caenorhabditis elegans. RNA 9: 847–857. 10.1261/rna.5480803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjuki H, Alam MI, Ehrhardt C, Wagner R, Planz O, Klenk HD, Ludwig S, Pleschka S. 2006. Membrane accumulation of influenza A virus hemagglutinin triggers nuclear export of the viral genome via protein kinase Cα-mediated activation of ERK signaling. J Biol Chem 281: 16707–16715. 10.1074/jbc.M510233200 [DOI] [PubMed] [Google Scholar]
- Martin K, Helenius A. 1991a. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67: 117–130. 10.1016/0092-8674(91)90576-K [DOI] [PubMed] [Google Scholar]
- Martin K, Helenius A. 1991b. Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol 65: 232–244. 10.1128/JVI.65.1.232-244.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Doudna JA. 2008. An inhibitory activity in human cells restricts the function of an avian-like influenza virus polymerase. Cell Host Microbe 4: 111–122. 10.1016/j.chom.2008.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XY, Luo Y, Anwar MN, Sun Y, Gao Y, Zhang H, Munir M, Qiu HJ. 2017. Long non-coding RNAs: emerging and versatile regulators in host-virus interactions. Front Immunol 8: 1663. 10.3389/fimmu.2017.01663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MA, Duran WA, Hjelle BL, Ye C, Panganiban AT. 2008. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc Natl Acad Sci 105: 19294–19299. 10.1073/pnas.0807211105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry B, Long JS, Schreyer J, Staller E, Sanchez-David RY, Barclay WS. 2020. Elucidating the interactions between influenza virus polymerase and host factor ANP32A. J Virol 94: e01353-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Keusch JJ, Decamps L, Ho-Xuan H, Iketani S, Gut H, Kutay U, Helenius A, Yamauchi Y. 2019. Influenza virus uses transportin 1 for vRNP debundling during cell entry. Nat Microbiol 4: 578–586. 10.1038/s41564-018-0332-2 [DOI] [PubMed] [Google Scholar]
- Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. 2012. Organization of the influenza virus replication machinery. Science 338: 1631–1634. 10.1126/science.1227270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose F, Handa H, Nagata K. 1996. Identification of host factors that regulate the influenza virus RNA polymerase activity. Biochimie 78: 1103–1108. 10.1016/S0300-9084(97)86736-3 [DOI] [PubMed] [Google Scholar]
- Momose F, Basler CF, O'Neill RE, Iwamatsu A, Palese P, Nagata K. 2001. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J Virol 75: 1899–1908. 10.1128/JVI.75.4.1899-1908.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose F, Naito T, Yano K, Sugimoto S, Morikawa Y, Nagata K. 2002. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J Biol Chem 277: 45306–45314. 10.1074/jbc.M206822200 [DOI] [PubMed] [Google Scholar]
- Momose F, Kikuchi Y, Komase K, Morikawa Y. 2007. Visualization of microtubule-mediated transport of influenza viral progeny ribonucleoprotein. Microbes Infect 9: 1422–1433. 10.1016/j.micinf.2007.07.007 [DOI] [PubMed] [Google Scholar]
- Moncorgé O, Mura M, Barclay WS. 2010. Evidence for avian and human host cell factors that affect the activity of influenza virus polymerase. J Virol 84: 9978–9986. 10.1128/JVI.01134-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- More S, Zhu Z, Lin K, Huang C, Pushparaj S, Liang Y, Sathiaseelan R, Yang X, Liu L. 2019. Long non-coding RNA PSMB8-AS1 regulates influenza virus replication. RNA Biol 16: 340–353. 10.1080/15476286.2019.1572448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munier S, Rolland T, Diot C, Jacob Y, Naffakh N. 2013. Exploration of binary virus–host interactions using an infectious protein complementation assay. Mol Cell Proteomics 12: 2845–2855. 10.1074/mcp.M113.028688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Momose F, Kawaguchi A, Nagata K. 2007. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J Virol 81: 1339–1349. 10.1128/JVI.01917-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nencioni L, De Chiara G, Sgarbanti R, Amatore D, Aquilano K, Marcocci ME, Serafino A, Torcia M, Cozzolino F, Ciriolo MR, et al. 2009. Bcl-2 expression and p38MAPK activity in cells infected with influenza A virus: impact on virally induced apoptosis and viral replication. J Biol Chem 284: 16004–16015. 10.1074/jbc.M900146200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Hobom G. 1995. Mutational analysis of influenza virus promoter elements in vivo. J Gen Virol 76: 1709–1717. 10.1099/0022-1317-76-7-1709 [DOI] [PubMed] [Google Scholar]
- Neumann G, Castrucci MR, Kawaoka Y. 1997. Nuclear import and export of influenza virus nucleoprotein. J Virol 71: 9690–9700. 10.1128/JVI.71.12.9690-9700.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Hughes MT, Kawaoka Y. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J 19: 6751–6758. 10.1093/emboj/19.24.6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noshi T, Kitano M, Taniguchi K, Yamamoto A, Omoto S, Baba K, Hashimoto T, Ishida K, Kushima Y, Hattori K, et al. 2018. In vitro characterization of baloxavir acid, a first-in-class cap-dependent endonuclease inhibitor of the influenza virus polymerase PA subunit. Antiviral Res 160: 109–117. 10.1016/j.antiviral.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Nturibi E, Bhagwat AR, Coburn S, Myerburg MM, Lakdawala SS. 2017. Intracellular colocalization of influenza viral RNA and Rab11A is dependent upon microtubule filaments. J Virol 91: e01179. 10.1128/JVI.01179-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obri A, Ouararhni K, Papin C, Diebold ML, Padmanabhan K, Marek M, Stoll I, Roy L, Reilly PT, Mak TW, et al. 2014. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 505: 648–653. 10.1038/nature12922 [DOI] [PubMed] [Google Scholar]
- Omoto S, Speranzini V, Hashimoto T, Noshi T, Yamaguchi H, Kawai M, Kawaguchi K, Uehara T, Shishido T, Naito A, et al. 2018. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci Rep 8: 9633. 10.1038/s41598-018-27890-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill RE, Jaskunas R, Blobel G, Palese P, Moroianu J. 1995. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J Biol Chem 270: 22701–22704. 10.1074/jbc.270.39.22701 [DOI] [PubMed] [Google Scholar]
- O'Neill RE, Talon J, Palese P. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J 17: 288–296. 10.1093/emboj/17.1.288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YH, Chungu K, Lee SB, Woo SJ, Cho HY, Lee HJ, Rengaraj D, Lee JH, Song CS, Lim JM, et al. 2020. Host-specific restriction of avian influenza virus caused by differential dynamics of ANP32 family members. J Infect Dis 221: 71–80. 10.1093/infdis/jiz506 [DOI] [PubMed] [Google Scholar]
- Peacock TP, Sheppard CM, Staller E, Barclay WS. 2019. Host determinants of influenza RNA synthesis. Annu Rev Virol 6: 215–233. 10.1146/annurev-virology-092917-043339 [DOI] [PubMed] [Google Scholar]
- Peacock TP, Swann OC, Salvesen HA, Staller E, Leung PB, Goldhill DH, Zhou H, Lillico SG, Whitelaw CBA, Long JS, et al. 2020. Swine ANP32A supports avian influenza virus polymerase. J Virol 94: e00132. 10.1128/JVI.00132-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. 2006. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem 75: 271–294. 10.1146/annurev.biochem.75.103004.142738 [DOI] [PubMed] [Google Scholar]
- Peng Q, Liu Y, Peng R, Wang M, Yang W, Song H, Chen Y, Liu S, Han M, Zhang X, et al. 2019. Structural insight into RNA synthesis by influenza D polymerase. Nat Microbiol 4: 1750–1759. 10.1038/s41564-019-0487-5 [DOI] [PubMed] [Google Scholar]
- Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, Bradel-Tretheway BG, Korth MJ, Castle JC, Biery MC, et al. 2010. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio 1: e00206. 10.1128/mBio.00206-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petosa C, Schoehn G, Askjaer P, Bauer U, Moulin M, Steuerwald U, Soler-López M, Baudin F, Mattaj IW, Müller CW. 2004. Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol Cell 16: 761–775. 10.1016/j.molcel.2004.11.018 [DOI] [PubMed] [Google Scholar]
- Picard D. 2002. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci 59: 1640–1648. 10.1007/PL00012491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314: 997–1001. 10.1126/science.1132998 [DOI] [PubMed] [Google Scholar]
- Pleschka S, Wolff T, Ehrhardt C, Hobom G, Planz O, Rapp UR, Ludwig S. 2001. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat Cell Biol 3: 301–305. 10.1038/35060098 [DOI] [PubMed] [Google Scholar]
- Pumroy RA, Ke S, Hart DJ, Zachariae U, Cingolani G. 2015. Molecular determinants for nuclear import of influenza A PB2 by importin α isoforms 3 and 7. Structure 23: 374–384. 10.1016/j.str.2014.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villán E, García-Sastre A, Gack MU. 2012. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog 8: e1003059. 10.1371/journal.ppat.1003059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read EK, Digard P. 2010. Individual influenza A virus mRNAs show differential dependence on cellular NXF1/TAP for their nuclear export. J Gen Virol 91: 1290–1301. 10.1099/vir.0.018564-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, et al. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140: 397–408. 10.1016/j.cell.2010.01.020 [DOI] [PubMed] [Google Scholar]
- Reilly PT, Yu Y, Hamiche A, Wang L. 2014. Cracking the ANP32 whips: important functions, unequal requirement, and hints at disease implications. Bioessays 36: 1062–1071. 10.1002/bies.201400058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resa-Infante P, Jorba N, Zamarreño N, Fernández Y, Juárez S, Ortín J. 2008. The host-dependent interaction of α-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS One 3: e3904. 10.1371/journal.pone.0003904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resa-Infante P, Thieme R, Ernst T, Arck PC, Ittrich H, Reimer R, Gabriel G. 2014. Importin-α7 is required for enhanced influenza A virus replication in the alveolar epithelium and severe lung damage in mice. J Virol 88: 8166–8179. 10.1128/JVI.00270-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resa-Infante P, Paterson D, Bonet J, Otte A, Oliva B, Fodor E, Gabriel G. 2015. Targeting importin-α7 as a therapeutic approach against pandemic influenza viruses. J Virol 89: 9010–9020. 10.1128/JVI.00583-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resa-Infante P, Bonet J, Thiele S, Alawi M, Stanelle-Bertram S, Tuku B, Beck S, Oliva B, Gabriel G. 2019. Alternative interaction sites in the influenza A virus nucleoprotein mediate viral escape from the importin-α7 mediated nuclear import pathway. FEBS J 286: 3374–3388. 10.1111/febs.14868 [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Pérez-González A, Nieto A. 2007. Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II. J Virol 81: 5315–5324. 10.1128/JVI.02129-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra F, Rivera C, Rivas E, Merino P, Garrido D, Hernández S, Forné I, Vassias I, Gurard-Levin ZA, Alfaro IE, et al. 2017. PP32 and SET/TAF-Iβ proteins regulate the acetylation of newly synthesized histone H4. Nucleic Acids Res 45: 11700–11710. 10.1093/nar/gkx775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci 104: 1853–1858. 10.1073/pnas.0610977104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A, Boff L, Anhlan D, Krischuns T, Brunotte L, Schuberth C, Wedlich-Söldner R, Drexler H, Ludwig S. 2020. Dissecting the mechanism of signaling-triggered nuclear export of newly synthesized influenza virus ribonucleoprotein complexes. Proc Natl Acad Sci 117: 16557–16566. 10.1073/pnas.2002828117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sediri H, Schwalm F, Gabriel G, Klenk HD. 2015. Adaptive mutation PB2 D701N promotes nuclear import of influenza vRNPs in mammalian cells. Eur J Cell Biol 94: 368–374. 10.1016/j.ejcb.2015.05.012 [DOI] [PubMed] [Google Scholar]
- Serna Martin I, Hengrung N, Renner M, Sharps J, Martínez-Alonso M, Masiulis S, Grimes JM, Fodor E. 2018. A mechanism for the activation of the influenza virus transcriptase. Mol Cell 70: 1101–1110.e4. 10.1016/j.molcel.2018.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, et al. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139: 1255–1267. 10.1016/j.cell.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H. 2009. UAP56- a key player with surprisingly diverse roles in pre-mRNA splicing and nuclear export. BMB Rep 42: 185–188. 10.5483/BMBRep.2009.42.4.185 [DOI] [PubMed] [Google Scholar]
- Shen J, Zhang L, Zhao R. 2007. Biochemical characterization of the ATPase and helicase activity of UAP56, an essential pre-mRNA splicing and mRNA export factor. J Biol Chem 282: 22544–22550. 10.1074/jbc.M702304200 [DOI] [PubMed] [Google Scholar]
- Shen H, Zheng X, Shen J, Zhang L, Zhao R, Green MR. 2008. Distinct activities of the DExD/H-box splicing factor hUAP56 facilitate stepwise assembly of the spliceosome. Genes Dev 22: 1796–1803. 10.1101/gad.1657308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Handa H, Nakada S, Nagata K. 1994. Regulation of influenza virus RNA polymerase activity by cellular and viral factors. Nucleic Acids Res 22: 5047–5053. 10.1093/nar/22.23.5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya K, Makino A, Ozawa M, Kim JH, Sakai-Tagawa Y, Ito M, Le QM, Kawaoka Y. 2009. Ostrich involvement in the selection of H5N1 influenza virus possessing mammalian-type amino acids in the PB2 protein. J Virol 83: 13015–13018. 10.1128/JVI.01714-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smietanski M, Werner M, Purta E, Kaminska KH, Stepinski J, Darzynkiewicz E, Nowotny M, Bujnicki JM. 2014. Structural analysis of human 2′-O-ribose methyltransferases involved in mRNA cap structure formation. Nat Commun 5: 3004. 10.1038/ncomms4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller E, Sheppard CM, Neasham PJ, Mistry B, Peacock TP, Goldhill DH, Long JS, Barclay WS. 2019. ANP32 proteins are essential for influenza virus replication in human cells. J Virol 93: e00217. 10.1128/JVI.00217-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller E, Baillon L, Frise R, Peacock TP, Sheppard CM, Sancho-Shimizu V, Barclay WS. 2020. A rare variant in Anp32B impairs influenza virus replication in human cells. bioRxiv 10.1101/2020.04.06.027482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. 2010. Nuclear export of mRNA. Trends Biochem Sci 35: 609–617. 10.1016/j.tibs.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Subbarao EK, London W, Murphy BR. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol 67: 1761–1764. 10.1128/JVI.67.4.1761-1764.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama K, Kawaguchi A, Okuwaki M, Nagata K. 2015. Pp32 and APRIL are host cell-derived regulators of influenza virus RNA synthesis from cRNA. eLife 4: e08939. 10.7554/eLife.08939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swale C, Monod A, Tengo L, Labaronne A, Garzoni F, Bourhis JM, Cusack S, Schoehn G, Berger I, Ruigrok RW, et al. 2016. Structural characterization of recombinant IAV polymerase reveals a stable complex between viral PA-PB1 heterodimer and host RanBP5. Sci Rep 6: 24727. 10.1038/srep24727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swale C, Da Costa B, Sedano L, Garzoni F, McCarthy AA, Berger I, Bieniossek C, Ruigrok RWH, Delmas B, Crépin T. 2020. X-ray structure of the human karyopherin RanBP5: an essential factor for influenza polymerase nuclear trafficking. J Mol Biol 432: 3353–3359. 10.1016/j.jmb.2020.03.021 [DOI] [PubMed] [Google Scholar]
- *.Takashita E. 2020. Influenza polymerase inhibitors: mechanisms of action and resistance. Cold Spring Harb Perspect Med 10.1101/cshperspect.a038687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, et al. 2007. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol 14: 229–233. 10.1038/nsmb1212 [DOI] [PubMed] [Google Scholar]
- Tarus B, Bakowiez O, Chenavas S, Duchemin L, Estrozi LF, Bourdieu C, Lejal N, Bernard J, Moudjou M, Chevalier C, et al. 2012. Oligomerization paths of the nucleoprotein of influenza A virus. Biochimie 94: 776–785. 10.1016/j.biochi.2011.11.009 [DOI] [PubMed] [Google Scholar]
- te Velthuis AJ, Fodor E. 2016. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat Rev Microbiol 14: 479–493. 10.1038/nrmicro.2016.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry E, Guilligay D, Kosinski J, Bock T, Gaudon S, Round A, Pflug A, Hengrung N, El Omari K, Baudin F, et al. 2016. Influenza polymerase can adopt an alternative configuration involving a radical repacking of PB2 domains. Mol Cell 61: 125–137. 10.1016/j.molcel.2015.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochio N, Umehara T, Munemasa Y, Suzuki T, Sato S, Tsuda K, Koshiba S, Kigawa T, Nagai R, Yokoyama S. 2010. Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain. J Mol Biol 401: 97–114. 10.1016/j.jmb.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, et al. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci 109: 4269–4274. 10.1073/pnas.1116200109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9: e1003657. 10.1371/journal.ppat.1003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevejo JM, Asmal M, Vingerhoets J, Polo R, Robertson S, Jiang Y, Kieffer TL, Leopold L. 2018. Pimodivir treatment in adult volunteers experimentally inoculated with live influenza virus: a Phase IIa, randomized, double-blind, placebo-controlled study. Antiviral Ther 23: 335–344. 10.3851/IMP3212 [DOI] [PubMed] [Google Scholar]
- Uehara T, Hayden FG, Kawaguchi K, Omoto S, Hurt AC, De Jong MD, Hirotsu N, Sugaya N, Lee N, Baba K, et al. 2020. Treatment-emergent influenza variant viruses with reduced baloxavir susceptibility: impact on clinical and virologic outcomes in uncomplicated influenza. J Infect Dis 221: 346–355. [DOI] [PubMed] [Google Scholar]
- Vale-Costa S, Alenquer M, Sousa AL, Kellen B, Ramalho J, Tranfield EM, Amorim MJ. 2016. Influenza A virus ribonucleoproteins modulate host recycling by competing with Rab11 effectors. J Cell Sci 129: 1697–1710. 10.1242/jcs.188409 [DOI] [PubMed] [Google Scholar]
- Vasin AV, Temkina OA, Egorov VV, Klotchenko SA, Plotnikova MA, Kiselev OI. 2014. Molecular mechanisms enhancing the proteome of influenza A viruses: an overview of recently discovered proteins. Virus Res 185: 53–63. 10.1016/j.virusres.2014.03.015 [DOI] [PubMed] [Google Scholar]
- Vreede FT, Fodor E. 2010. The role of the influenza virus RNA polymerase in host shut-off. Virulence 1: 436–439. 10.4161/viru.1.5.12967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreede FT, Jung TE, Brownlee GG. 2004. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J Virol 78: 9568–9572. 10.1128/JVI.78.17.9568-9572.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreede FT, Chan AY, Sharps J, Fodor E. 2010. Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology 396: 125–134. 10.1016/j.virol.2009.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakai C, Iwama M, Mizumoto K, Nagata K. 2011. Recognition of cap structure by influenza B virus RNA polymerase is less dependent on the methyl residue than recognition by influenza A virus polymerase. J Virol 85: 7504–7512. 10.1128/JVI.02375-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AP, Fodor E. 2019. Interplay between influenza virus and the host RNA polymerase II transcriptional machinery. Trends Microbiol 27: 398–407. 10.1016/j.tim.2018.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandzik JM, Kouba T, Cusack S. 2020. Structure and function of influenza polymerase. Cold Spring Harb Perspect Med 10.1101/cshperspect.a038372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cen S. 2020. Roles of lncRNAs in influenza virus infection. Emerg Microbes Infect 9: 1407–1414. 10.1080/22221751.2020.1778429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Palese P, O'Neill RE. 1997. The NPI-1/NPI-3 (karyopherin α) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol 71: 1850–1856. 10.1128/JVI.71.3.1850-1856.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cui ZQ, Han H, Zhang ZP, Wei HP, Zhou YF, Chen Z, Zhang XE. 2008. Imaging and characterizing influenza A virus mRNA transport in living cells. Nucleic Acids Res 36: 4913–4928. 10.1093/nar/gkn475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xu J, Wang Y, Cao X. 2017. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 358: 1051–1055. 10.1126/science.aao0409 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang Y, Zhou R, Zhao J, Zhang Y, Yi D, Li Q, Zhou J, Guo F, Liang C, et al. 2018. Host long noncoding RNA lncRNA-PAAN regulates the replication of influenza A virus. Viruses 10: 330. 10.3390/v10060330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang Y, Li Q, Zhao J, Yi D, Ding J, Zhao F, Hu S, Zhou J, Deng T, et al. 2019a. Influenza virus exploits an interferon-independent lncRNA to preserve viral RNA synthesis through stabilizing viral RNA polymerase PB1. Cell Rep 27: 3295–3304.e4. 10.1016/j.celrep.2019.05.036 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Na L, Du C, Zhang Z, Zheng YH, Wang X. 2019b. ANP32A and ANP32B are key factors in the Rev-dependent CRM1 pathway for nuclear export of HIV-1 unspliced mRNA. J Biol Chem 294: 15346–15357. 10.1074/jbc.RA119.008450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Takizawa N, Katoh M, Hoshida K, Kobayashi N, Nagata K. 2001. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res 77: 31–42. 10.1016/S0168-1702(01)00263-5 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Watanabe S, Kawaoka Y. 2010. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7: 427–439. 10.1016/j.chom.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kawakami E, Shoemaker JE, Lopes TJ, Matsuoka Y, Tomita Y, Kozuka-Hata H, Gorai T, Kuwahara T, Takeda E, et al. 2014. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe 16: 795–805. 10.1016/j.chom.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Sediri H, Felgenhauer U, Binzen I, Bänfer S, Jacob R, Brunotte L, García-Sastre A, Schmid-Burgk JL, Schmidt T, et al. 2015. Influenza virus adaptation PB2-627 K modulates nucleocapsid inhibition by the pathogen sensor RIG-I. Cell Host Microbe 17: 309–319. 10.1016/j.chom.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker G, Bui M, Helenius A. 1996. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J Virol 70: 2743–2756. 10.1128/JVI.70.5.2743-2756.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterling C, Koch M, Koeppel M, Garcia-Alcalde F, Karlas A, Meyer TF. 2014. Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA Biol 11: 66–75. 10.4161/rna.27504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisskirchen C, Ludersdorfer TH, Müller DA, Moritz E, Pavlovic J. 2011. The cellular RNA helicase UAP56 is required for prevention of double-stranded RNA formation during influenza A virus infection. J Virol 85: 8646–8655. 10.1128/JVI.02559-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WW, Weaver LL, Panté N. 2007. Ultrastructural analysis of the nuclear localization sequences on influenza A ribonucleoprotein complexes. J Mol Biol 374: 910–916. 10.1016/j.jmb.2007.10.022 [DOI] [PubMed] [Google Scholar]
- York A, Hengrung N, Vreede FT, Huiskonen JT, Fodor E. 2013. Isolation and characterization of the positive-sense replicative intermediate of a negative-strand RNA virus. Proc Natl Acad Sci 110: E4238–E4245. 10.1073/pnas.1315068110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborowska J, Egloff S, Murphy S. 2016. The pol II CTD: new twists in the tail. Nat Struct Mol Biol 23: 771–777. 10.1038/nsmb.3285 [DOI] [PubMed] [Google Scholar]
- Zhang H, Li H, Wang W, Wang Y, Han GZ, Chen H, Wang X. 2020. A unique feature of swine ANP32A provides susceptibility to avian influenza virus infection in pigs. PLoS Pathog 16: e1008330. 10.1371/journal.ppat.1008330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang Z, Wang Y, Wang M, Wang X, Zhang X, Ji S, Du C, Chen H, Wang X. 2019a. Fundamental contribution and host range determination of ANP32A and ANP32B in influenza A virus polymerase activity. J Virol 93: e00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Xie Y, Muñoz-Moreno R, Wang J, Zhang L, Esparza M, García-Sastre A, Fontoura BMA, Ren Y. 2019b. Structural basis for influenza virus NS1 protein block of mRNA nuclear export. Nat Microbiol 4: 1671–1679. 10.1038/s41564-019-0482-x [DOI] [PMC free article] [PubMed] [Google Scholar]