In this review, Alendar and Berns discuss emerging evidence that the chromodomain helicase DNA-binding (CHD) family of enzymes is subjected to frequent DNA copy number alterations or mutations and shows aberrant expression in malignancies and other human diseases.
Keywords: cancer, chromatin remodeling, chromodomain helicase DNA-binding proteins, development and disease
Abstract
Chromatin is highly dynamic, undergoing continuous global changes in its structure and type of histone and DNA modifications governed by processes such as transcription, repair, replication, and recombination. Members of the chromodomain helicase DNA-binding (CHD) family of enzymes are ATP-dependent chromatin remodelers that are intimately involved in the regulation of chromatin dynamics, altering nucleosomal structure and DNA accessibility. Genetic studies in yeast, fruit flies, zebrafish, and mice underscore essential roles of CHD enzymes in regulating cellular fate and identity, as well as proper embryonic development. With the advent of next-generation sequencing, evidence is emerging that these enzymes are subjected to frequent DNA copy number alterations or mutations and show aberrant expression in malignancies and other human diseases. As such, they might prove to be valuable biomarkers or targets for therapeutic intervention.
All cells in our body face the challenge of accommodating ∼2 m of DNA in a 5-µm nucleus. The fundamental unit of compaction is the nucleosome that, together with linker histone H1, other structural nonhistone DNA-binding proteins, and RNA, facilitates DNA assembly into higher-order chromatin (Yadav et al. 2018). Nevertheless, the chromatin-packaged DNA must be accessible to the myriad of factors that catalyze transcription, repair, replication, and recombination (Radman-Livaja and Rando 2010). Transcriptional activity of genes is mechanistically coupled to their local chromatin environment, shaped by the actions of enzymes that either catalyze the deposition or removal of covalent modifications on histones and DNA or use ATP hydrolysis to mobilize nucleosomes (Voss and Hager 2014).
ATP-dependent chromatin remodelers have a SNF2 helicase-like ATPase domain (Ryan and Owen-Hughes 2011) and, based on the presence of other functional domains, can be broadly divided into four main protein families: SWI/SNF, ISWI, CHD, and INO80 complexes (Clapier et al. 2017).
Here, we highlight the major findings of research on the CHD family of proteins and discuss challenges that lie ahead. Other families of chromatin remodelers have been extensively reviewed elsewhere (Narlikar et al. 2013; Kadoch and Crabtree 2015; Clapier et al. 2017).
Domain organization, structural features, and regulatory principles
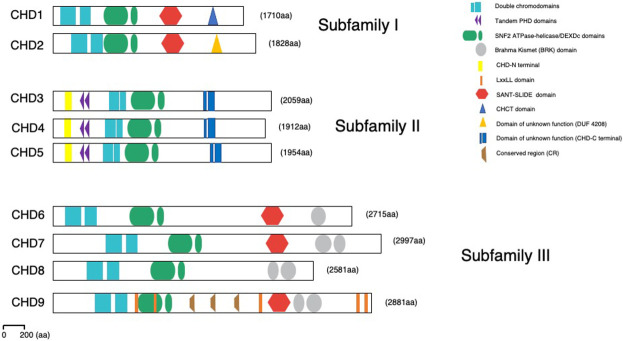
Almost three decades ago, the group of the late Robert Perry (Delmas et al. 1993) cloned the first member of this remarkable family, murine Chd1, and coined the term “chromodomain–helicase–DNA-binding protein.” CHD proteins are highly conserved in evolution and play pivotal roles in organismal development and tumorigenesis. Based on their constituent domains, CHD remodelers are classified into three subfamilies: subfamily I (CHD1 and CHD2), subfamily II (CHD3–5), and subfamily III (CHD6–9) (Fig. 1; Clapier et al. 2017).
Figure 1.
Structural domain organization of the CHD enzymes. Domain representation of all three CHD subfamilies as indicated in the text. All CHDs have double chromodomains (blue squares) and SNF2-like ATPase/DEXDc helicase domains (green ovals). Subfamily II members contain tandem PHD Zn finger-like domains (purple triangles). Subfamily III members have additional Brahma Kismet (BRK) domains at the C terminus (gray oval); enzymes of both subfamilies I and III contain SANT-SLIDE domains at C terminus (red hexagon). These domains facilitate engagement and stabilization of the interaction with the chromatin substrate and allow efficient ATP hydrolysis and DNA translocation. Recently, a number of novel structural domains have been identified: CHD-N-terminal, CHD-C-terminal, CHCT, DUF4208, and conserved region (CR). CHD9 associates with nuclear receptors through LxxLL recognition motifs (orange stripes).
CHDs are a family of large proteins that exist either as monomers or constituents of multimeric complexes, exerting specific functions in different cell types or developmental stages (Supplemental Table S1; Micucci et al. 2015). Moreover, alternative promoter usage or splicing can result in several protein isoforms, generating additional functional complexity (Kita et al. 2012; Nishiyama et al. 2012; Kunkel et al. 2018, 2020). They are subjected to a range of post-translational modifications (Schmidt and Schreiber 1999; Choudhary et al. 2009; Urquhart et al. 2011; Kessler et al. 2012; Piatti et al. 2015; Salomon-Kent et al. 2015; Yu et al. 2015; Luijsterburg et al. 2016; Zhao et al. 2017) and reside in the nucleus and nucleolus (Zentner et al. 2010; Kita et al. 2012; Xie et al. 2012; Salomon-Kent et al. 2015; Hoffmeister et al. 2017), as well as in the cytoplasm (Stokes and Perry 1995; Salomon-Kent et al. 2015), where they exert context-dependent functions.
The hallmark features of all CHD remodelers are the presence of double chromodomains at their N-terminal region and a central SNF2 helicase-like ATPase domain (Fig. 1). The double chromodomains mediate association with chromatin by directly binding to methylated lysines on histone H3 tails (Flanagan et al. 2005, 2007; Sims et al. 2005), nucleosomal DNA (Nodelman et al. 2017), poly(ADP-ribose) (Murawska and Brehm 2011), or RNA (Akhtar et al. 2000), serving as nucleosome recognition surface and allosteric regulators of the enzymatic activity of the ATPase motor (Hauk et al. 2010). Notably, the chromodomains of subfamily II members lack key residues required for binding to methylated histone tails but have evolved the ability to bind DNA substrates (Flanagan et al. 2007). The ATPase motor of CHD enzymes is related to the catalytic subunit of superfamily 2 (SF2) helicases (Flaus et al. 2006), and uses ATP hydrolysis to disrupt histone–DNA contacts within the nucleosome to alter chromatin packaging (Hauk and Bowman 2011). Furthermore, CHD enzymes harbor several auxiliary domains that facilitate substrate recognition and chromatin binding, ATPase activity, and coupling of ATP hydrolysis to productive DNA translocation (Fig. 1; Marfella and Imbalzano 2007; Ramírez et al. 2012; Silva et al. 2015; Luijsterburg et al. 2016; Mohanty et al. 2016).
The subfamily II members CHD3/CHD4 (Mi-2α/β) and CHD5 are core components of the multimeric enzymatic complex with both chromatin remodeling and histone deacetylase activity, termed the nucleosome remodeling and histone deacetylase complex (NuRD) (Supplemental Table S1 and references therein). They contain additional tandem N-terminal zinc finger-like, paired plant homeodomains (tPHD) (Mansfield et al. 2011), which appear to be structurally independent modules that can bind separate unmodified or K9-methylated H3 tails on a single nucleosome (Supplemental Table S1; Musselman et al. 2012).
The third subfamily (CHD6–9) includes recently identified homologs of Drosophila Kismet protein (KIS-L). They are evolutionarily conserved and harbor additional structural features such as the Brahma and Kismet domain (BRK), the conserved region (CR) domains (Schuster and Stöger 2002; Shur and Benayahu 2005), and the SANT-SLIDE-like domain (Fig. 1; Boyer et al. 2004). The BRK domains appear to mediate CHD7/8 interaction with CCCTC-binding factor (CTCF) (Ishihara et al. 2006), and between CHD7 and CHD8 themselves (Batsukh et al. 2010). The SANT-SLIDE-like DNA-binding domain (DBD) is also present at the C terminus of CHD1 and CHD2 and binds linker DNA to direct nucleosomal sliding (McKnight et al. 2011; Ryan et al. 2011; Liu et al. 2014a).
Chromatin recruitment and allosteric regulation of the ATPase activity
Since CHD proteins have limited DNA sequence specificity (Stokes and Perry 1995), their chromatin association is mediated by interactions with transcription factors, modified histones, RNA, poly(ADP-ribose), and methylated DNA (Supplemental Table S1). Current evidence supports a model in which CHD association with chromatin takes place in a stepwise fashion. Interactions with specific transcription factors (TF) or subunits of the multimeric protein complexes in which CHDs reside govern recruitment to target chromatin sites (Längst and Manelyte 2015), as exemplified by the chromatin recruitment of the NuRD complex through interactions with a panoply of developmental and tissue-specific transcription factors (Supplemental Table S1 and references therein); the NuRD complex also binds methylated DNA, acting as an important nexus between gene silencing and DNA methylation (Zhang et al. 1999).
Moreover, the presence of several distinct histone and RNA- and DNA-binding modules facilitates CHD chromatin association (Fig. 1; Clapier and Cairns 2009; Längst and Manelyte 2015; Hendrickson et al. 2016). In general, double chromodomains of CHD enzymes (CHD1/2 and CHD7–9) bind methylated histone tails associated with active chromatin, such as H3K4me1/2/3 and H3K36me3, or marks of repressive chromatin states (CHD3/4/5/9), such as H3K9me2/3 and H3K27me3 (Supplemental Table S1 and references therein). Moreover, it appears that poly(ADP-ribose)-dependent recruitment of CHD2–4 and CHD6/7 to chromatin plays an important role in DNA damage response (Luijsterburg et al. 2016; Smith et al. 2018; Moore et al. 2019; Rother et al. 2020).
Intradomain allosteric regulation
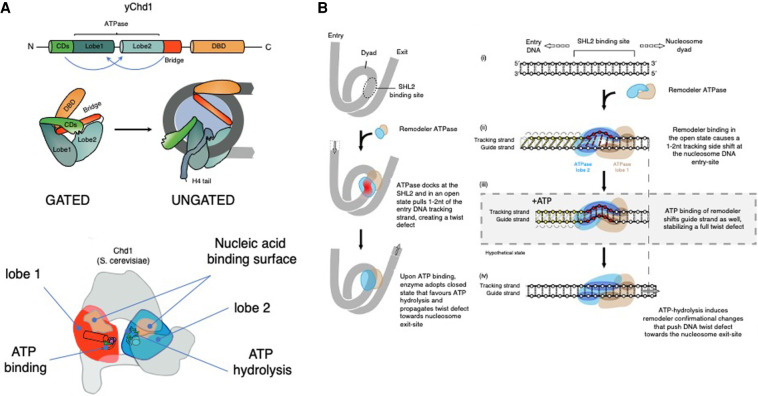
A great number of studies have addressed the biochemical and structural properties of CHD functional domains in isolation; however, their function in the context of the full-length proteins has remained ill defined. Recent studies, largely spearheaded by Bowman's group (Hauk et al. 2010; Hauk and Bowman 2011; McKnight et al. 2011; Nodelman and Bowman 2013; Nodelman et al. 2016, 2017, 2021; Tokuda et al. 2018; Winger et al. 2018), provide insight into the long-standing question of how the ATPase motor integrates information from the auxiliary domains to orchestrate engagement of the nucleosome substrates and catalyze accurate nucleosome sliding. An overarching model is emerging in which N-terminal chromodomains (yChd1) and PHD domains (CHD4) fold onto the bilobed ATPase motor, preventing nucleic acid binding and ATP hydrolysis (Fig. 2; Hauk et al. 2010; Ryan et al. 2011; Morra et al. 2012; Watson et al. 2012; Nodelman and Bowman 2013; Liu et al. 2014a; Nodelman et al. 2017, 2021). Consequently, in the absence of nucleosome substrates, key highly conserved residues across the central cleft of the ATPase motor are kept in an open conformation, thus preventing ATP hydrolysis (Hauk and Bowman 2011; Watson et al. 2012; Morra et al. 2012; Nodelman and Bowman 2013; Nodelman et al. 2017, 2021). Additionally, interactions with extranucleosomal elements such as linker DNA and protruding histone H4 tails trigger major conformational changes and dislodge the autoinhibitory domains, allowing the ATPase motor to adopt an active conformation (Fig. 2; Brehm et al. 2000; Hauk et al. 2010; Bouazoune and Kingston 2012; Farnung et al. 2017, 2020; Ludwigsen et al. 2017; Sundaramoorthy et al. 2017; Tokuda et al. 2018; Yan and Chen 2020; Nodelman et al. 2021).
Figure 2.
Autoinhibitory regulation and nucleosome sliding mechanism of SF2-type chromatin remodelers. (A) The ATPase motor of CHD enzymes is comprised of two RecA-like lobes ([red] lobe1, [blue] lobe2) that adopt a closed confirmation to form a contiguous surface that allows nucleic acid binding and efficient ATP hydrolysis. Biophysical and structural studies of yChd1, CHD2, and CHD4 revealed autoinhibitory domain organization whereby flanking domains fold onto the ATPase motor to prevent activation by improper substrates (Hauk et al. 2010; Ryan et al. 2011; Morra et al. 2012; Watson et al. 2012; Liu et al. 2014a). The N-terminal tandem chromodomains (CD; green) of yChd1 contain a linker helix with several conserved acidic residues that stacks onto the DNA-binding surface of lobe2 and negatively regulates yChd1 activity (Hauk and Bowman 2011; Narlikar et al. 2013). Full inhibition is achieved through folding of the “bridge” inhibitory domain adjacent to the ATPase motor (Bridge; orange). Moreover, autoinhibitory interaction of the ATPase motor with the C-terminal domains uncouples ATP hydrolysis from productive DNA translocation of Chd1p and CHD4, respectively (Hauk et al. 2010; Morra et al. 2012; Watson et al. 2012). Upon chromatin binding, interactions of CHD domains with linker DNA and protruding histone H4 tails (H4; gray) trigger major conformational change and dislodging of the autoinhibitory domains, allowing the ATPase motor to adopt an active conformation (Hauk and Bowman 2011; Nodelman et al. 2021). The interplay between the ATPase motor and flanking domains not only provides means for autoinhibition but also can promote substrate recognition and ATPase activity. In the case of CHD4, the N-terminal tandem chromodomains and PHD domains stimulate ATPase motor activity and DNA translocation (Morra et al. 2012; Watson et al. 2012), whereas the respective C-terminal DNA-binding regions of CHD1, CHD2, and CHD7 stimulate activity of the ATPase motor (Ryan and Owen-Hughes 2011; Bouazoune and Kingston 2012; Liu et al. 2014a). Thus, intradomain allosteric regulation keeps the enzyme in an inactive conformation to ensure substrate specificity, while on the other hand facilitating ATPase motor activity and DNA translocation. (GATED) yChd1 closed confirmation, (UNGATED) open confirmation, (light blue) nucleosome core, (gray) wrapped DNA. Features required for the efficient ATP binding and hydrolysis in lobe1 and lobe2 are indicated: ATP binding (magenta circles), ATP hydrolysis (green lines), and nucleic acid binding surfaces on each lobe (opaque areas) (Hauk and Bowman 2011). Top panel cartoon adapted with permission from Springer Nature from Mueller-Planitz et al. (2013), © 2013. Bottom panel republished with permission of Elsevier Science and Technology Journals from Hauk and Bowman (2011); permission conveyed through Copyright Clearance Center, Inc. (B) Historically, nucleosome crystal structure and earlier models of nucleosome sliding revealed that nucleosomal DNA can accommodate an extra base pair at the internal DNA location termed superhelix location 2 (SHL2) (Luger et al. 1997), which is a docking site for many ATP-dependent chromatin remodelers. Recent evidence shows that CHD, ISWI, and Snf2-type remodelers shift DNA discontinuously around the nucleosome (Farnung et al. 2017, 2020; Winger et al. 2018; Sabantsev et al. 2019; Yan and Chen 2020; Zhong et al. 2020; Nodelman et al. 2021). Remarkably, in an open conformation, the ATPase motor bound at the nucleosomal SHL2 position pulls 1–2 bp of DNA at the nucleosome entry site, creating a shift of only one DNA strand (tracking strand) with respect to the other (guide strand), resulting in a twist defect. Subsequent ATP binding induces closed confirmation of the ATPase motor, pulling the entire base pair of the DNA at SHL2 and forcing the DNA twist defect toward the nucleosomal exit site. Upon ATP hydrolysis, the motor readopts the open confirmation again, priming the nucleosome for the next translocation cycle. This sequential, ATP-driven conformational transition between open and closed states propels the DNA around the nucleosome histone core (Liu et al. 2017; Farnung et al. 2017, 2020; Armache et al. 2019; Nodelman et al. 2021; Yan and Chen 2020). Republished with permission of Elsevier Science and Technology Journals from Bowman (2019); permission conveyed through Copyright Clearance Center, Inc.
Intriguingly, the interplay between the ATPase motor and flanking domains not only permits autoinhibition, but also promotes substrate recognition and ATPase activity (Morra et al. 2012; Watson et al. 2012) (Ryan et al. 2011; Bouazoune and Kingston 2012; Liu et al. 2014a). Thus, intradomain allosteric regulation keeps the enzyme in an inactive conformation to ensure substrate specificity, while facilitating ATPase motor activity and DNA translocation once specific interactions with chromatin substrate are consolidated.
Nucleosome binding and translocase activity
Like other members of the SF2 helicase family, CHD remodelers are intrinsically active DNA translocases that harness the energy of ATP hydrolysis to propel DNA around the octamer and mobilize nucleosomes (Hauk et al. 2010; Mueller-Planitz et al. 2013; Nodelman and Bowman 2013; Nodelman et al. 2016; Farnung et al. 2017; Yan and Chen 2020). Recent evidence shows that SNF2-type remodelers shift DNA discontinuously, as DNA movement at the nucleosome entry site precedes that of the exit site (Farnung et al. 2017; Sabantsev et al. 2019; Zhong et al. 2020), inducing a “twist defect” of the nucleosomal DNA (Fig. 2; Winger et al. 2018; Farnung et al. 2020; Yan and Chen 2020; Nodelman et al. 2021).
Upon chromatin association, ATPase motor anchoring on the nucleosome and interactions of diverse CHD auxiliary domains with chromatin features (histone H4 tails, linker DNA, or other chromatin regulators) stabilize binding, prime the nucleosome for remodeling, and promote ATPase activity (Farnung et al. 2017, 2020; Liu et al. 2017; Armache et al. 2019). The concomitant cycles of ATP binding, hydrolysis, and dissociation of the hydrolysis product trigger a succession of conformational changes of the enzyme, promoting DNA translocation and nucleosome remodeling (Fig. 2; Mueller-Planitz et al. 2013; Yan and Chen 2020). The process is driven by the binding affinity of the remodeler for the nucleosome substrates, which, upon remodeling, decreases, leading to the release of the enzyme, thus marking the end point of the remodeling reaction (Längst and Manelyte 2015).
Functionally, chromatin remodelers alter histone–DNA interactions by promoting assembly, disruption, and repositioning of nucleosomes and incorporation of histone variants (Mueller-Planitz et al. 2013). The majority of CHDs (1/2/3/4/7/8/9) appear to predominantly slide nucleosomes (Lusser et al. 2005; Liu et al. 2014a; Salomon-Kent et al. 2015; Hoffmeister et al. 2017; Manning and Yusufzai 2017), whereas CHD5 and CHD6 disrupt nucleosomes without causing sliding (Quan and Yusufzai 2014; Manning and Yusufzai 2017). Notably, CHD1, CHD2, and ISWI-type remodelers are the only members of the SF2 helicases able to assemble periodic nucleosome arrays in an ATP-dependent manner (Lusser et al. 2005; Stockdale et al. 2006; Liu et al. 2014a). Importantly, CHD enzymes can also exhibit distinct remodeling activities in the absence of accessory subunits of the complexes in which they reside (CHD3/4 and NuRD) (Hoffmeister et al. 2017).
Collectively, biophysical and structural studies reveal that CHD remodelers display intricate conformational intradomain allosteric regulation and seem to share a common translocation mechanism with other SF2 ATPases, yet individual CHDs exhibit largely distinct nucleosome binding and remodeling activities (Quan and Yusufzai 2014; Liu et al. 2014a; Manning and Yusufzai 2017). This aligns with the nonredundant functions they fulfil in chromatin organization and transcription, resulting in distinct phenotypes in animal models and human disease upon their disruption.
Multimeric protein complexes
CHDs function either as monomers or constituents of multimeric enzymatic complexes (Micucci et al. 2015) that remodel chromatin and catalyze deposition or removal of histone modifications (Supplemental Table S1 and references therein). The nucleosome remodeling activity of CHD proteins facilitates interaction of histone methyltransferases (KMTs) and deacetylases (HDACs) with histone tail substrates. The combinatorial assembly of the noncatalytic subunits confers functional specificity to the CHD remodelers (Wang and Zhang 2001; Hoffmeister et al. 2017). The subunit composition is governed by the cell type and developmental stage (Micucci et al. 2015; Nitarska et al. 2016), and, in response to cellular cues, CHD enzymes are tethered to particular genomic loci where they exert specific functions (Lai and Wade 2011).
Biochemically the NuRD complex is characterized best. It harbors CHD3/4/5 chromatin remodelers and HDAC1 and HDAC2 catalytic subunits, next to several auxiliary subunits (Supplemental Table S1; Tong et al. 1998; Wade et al. 1998, 1999; Xue et al. 1998; Zhang et al. 1998, 1999; Kim et al. 1999; Hoffmeister et al. 2017; Tencer et al. 2017). The concerted action of CHD3/4 (Mi2-α/β) and HDAC subunits of the NuRD complex results in histone deacetylation, chromatin compaction, and transcriptional repression (Tong et al. 1998; Xue et al. 1998). Notably, the core CHD3/4/5 remodelers can form distinct NuRD complexes with specific functions (Nitarska et al. 2016). On the other hand, activators of transcription such as CHD8 can also regulate transcription as part of the histone methyltransferase subcomplex termed WAR (WDR5, ASH2L, and RBBP5) (Supplemental Table S1; Yates et al. 2010). Curiously, experimental evidence suggests that CHD family proteins also interact with each other (CHD7–CHD8 [Batsukh et al. 2010] and CHD1–CHD2 [Luijsterburg et al. 2016]) raising an interesting possibility of direct functional cooperativity in regulating chromatin-templated processes.
Chromatin organization and transcriptional regulation
Every cell type is characterized by a distinct set of genes that can be transcriptionally on, off, poised, or oscillating, resulting from the complex interplay between genomic regulatory elements and cognate binding factors (Signolet and Hendrich 2014). Access of specific TFs and chromatin regulators to underlying DNA is determined by the nucleosome positioning and an array of histone and DNA modifications (Lai and Pugh 2017).
Association with promoters and enhancers
RNA polymerase II (RNAPII) transcription typically initiates at the transcription start site (TSS) embedded within the core promoter sequence that is usually devoid of nucleosomes, termed the nucleosome-free region (NFR), which is demarcated by the +1 nucleosome that represents a major obstacle for RNAPII productive elongation (Fig. 3). The CHDs’ activity enables RNAPII to overcome nucleosomal barriers during transcription (Li et al. 2007).
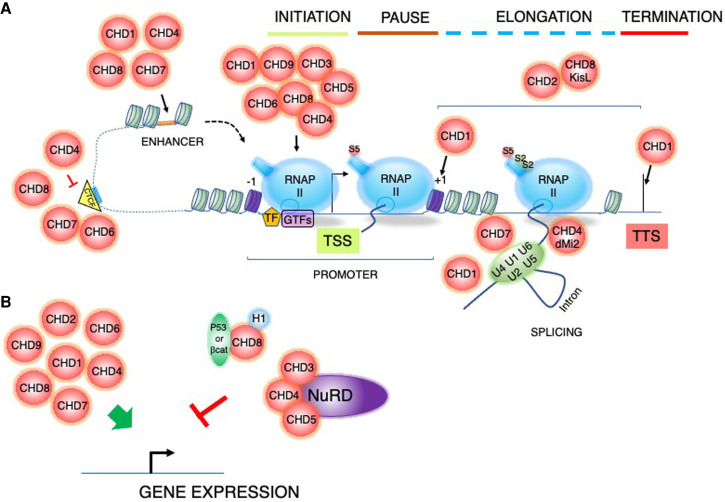
Figure 3.
Regulation of the transcription cycle. (A) CHD enzymes are involved at different stages of transcription: initiation, promoter escape, elongation, splicing, and termination, as well as higher-order chromatin organization. The promoter nucleosome-free region (NFR) is demarcated by the ±1 nucleosomes (purple). CHDs also regulate nucleosome accessibility at enhancer elements and modulate CTCF-mediated chromatin organization. (TSS) Transcription start site, (TTS) transcription termination site, (TF) transcription factors, (GTFs) general transcription factors, (U1/2/4/5/6) spliceosome, (orange rectangle) enhancer elements, (blue rectangle) CTCF-binding site. RNA polymerase II (RNAPII) phosphorylation status of the C-terminal domain is represented by the red (serine 5 phosphorylation-paused RNAPII) and green (serine 2 phosphorylation-elongating RNAPII) circles. (B) CHD enzymes can both promote and inhibit transcription of different subsets of genes in a context-dependent manner. CHD8 and linker histone H1 (blue circle) form a trimeric repressive complex with either P53 or β-catenin (green oval) to repress target gene expression.
The landmark study by the group of Matthieu Gérard (Dieuleveult et al. 2016) comprehensively charted chromatin binding of CHD proteins and their role in the gene expression regulation in mouse ES cells. It revealed a dynamic interplay between remodelers at promoter-proximal nucleosomes and a variable correlation between promoter enrichment of remodelers and transcriptional activity of the corresponding genes (Fig. 3). Moreover, it demonstrated dynamic CHD binding at gene promoters with different NFR regions, serving different functional purposes. Specifically, promoter occupancy of CHD1/2/9 directly correlates with H3K4me3 levels, CHD8 binds promoters with intermediate levels of H3K4me3, whereas CHD4/6 bind promoters irrespective of the H3K4me3 status. The observed binding profile of CHD4 aligns with reports that the NuRD complex displays a broader chromatin association in ES cells and contributes to both transcriptional activation and repression (Hu and Wade 2012; Bornelöv et al. 2018). Additional studies in primary and cancer cells show enrichment of CHD5 at the promoter-proximal regions largely devoid of H3K4me3 or H3K27me3 (Egan et al. 2013; Paul et al. 2013), and CHD3/NuRD enrichment at H3K9 acetylated (H3K9ac) target genes (Tencer et al. 2017). In contrast, CHD2 correlates with the H3K36me3 mark and is distributed throughout the gene body of transcribed genes and at transcription termination sites (TTSs) (Dieuleveult et al. 2016; Rom et al. 2019), likely modulating mRNA processing and RNAPII elongation and termination, as previously shown for yChd1 (Fig. 3; Murawska and Brehm 2011). This seminal study highlights several important general concepts. First, that CHD chromatin remodelers exhibit position-specific nucleosome binding tailored to the local chromatin architecture and DNA composition (CpG content). Second, that individual CHD remodelers can function as transcriptional activators of one subset of genes and as repressors of another.
Besides binding to promoter-proximal regions, CHDs remodel nucleosomes at enhancers to modulate enhancer–promoter communication and transcription initiation (Fig. 3). For instance, the CHD7 association with active and/or poised enhancers is important for RNAPII transcription in diverse cell lineages (Bajpai et al. 2010; Schnetz et al. 2010; Marie et al. 2018). Moreover, several lines of evidence show that CHD4/NuRD enhancer occupancy is critical for the transcriptional regulation in mouse ES cells (Bornelöv et al. 2018) and during T-cell development (Williams et al. 2004). Notably, nuclear receptors mediate CHD enhancer recruitment to regulate expression of androgen receptor (AR)-responsive genes (CHD1 and CHD8) (Menon et al. 2010; Metzger et al. 2016), progesterone (PR)-responsive genes (CHD8) (Ceballos-Chávez et al. 2015), and PPARγ- and C/EBPβ-regulated genes (CHD7) (Kita et al. 2018). These studies highlight different means of CHD recruitment to enhancer elements, eliciting distinct transcriptional outcomes governed by cellular and developmental contexts.
Transcriptional elongation and splicing
Current evidence supports a model in which chromatin structure (Schwartz et al. 2009) and RNAPII elongation rate (Batsché et al. 2006) play a critical role in splicing. The CHDs appear to act as an interface between chromatin epigenetic information and the RNAPII and pre-mRNA processing machinery (CHD1 [Sims et al. 2007; Lee et al. 2017] and CHD7 [Bélanger et al. 2018]), modulating both the RNAPII elongation (CHD8 [Rodríguez-Paredes et al. 2009] and dKisL [Srinivasan et al. 2008]) and splicing efficiency (CHD1 [Sims et al. 2007; Lee et al. 2017]). Furthermore, direct association with nascent RNA is demonstrated for dMi-2 (Murawska et al. 2011) and CHD1/2/4/7 (Hendrickson et al. 2016), providing additional means by which CHD enzymes may affect cotranscriptional processing. However, it remains unclear whether any of the CHD–RNA interactions affect biological functions of RNA molecules or CHD recruitment to specific genomic locations.
Higher-order chromatin regulation
In addition to shaping the local chromatin landscape and modulating transcription of individual genes, CHD proteins act globally to regulate higher-order chromatin structure, thereby regulating genome stability, transcriptional regulation, and DNA damage repair.
The role of CHD1 in maintaining high-order chromatin organization and genome stability has become evident in prostate cancer, where CHD1-null tumors display distinctive patterns of chromoplexy (Stephens et al. 2011; Baca et al. 2013). Moreover, several studies show that CHD1 (Boginya et al. 2019), CHD4 (Fasulo et al. 2012), and CHD6 (Sancho et al. 2015) appear to be required for the loading of cohesin, a critical mediator of transcription regulation and genome stability that is frequently mutated in cancer.
The CTCF insulator protein modulates transcription and orchestrates long-distance chromatin interactions (Ling et al. 2006), facilitating genome partitioning into functionally distinct regions called topologically associated domains (TADs) (Dixon et al. 2012). Biochemical studies reveal CHDs’ association with CTCF (CHD7 and CHD8) (Ishihara et al. 2006; Allen et al. 2007), and with the human CTCF paralog BORIS (brother of the regulator of imprinted sites) (CHD8) (Nguyen et al. 2008). Moreover, CHDs occupy many CTCF-binding sites (CHD8) and modulate insulation and topological organization of CTCF target loci (CHD8 [Ishihara et al. 2006] and CHD6 [Sancho et al. 2015]) (Fig. 3). Curiously, in mouse ES cells, the CHD4/ChAHP complex prevents CTCF binding to sequences within the transposable elements and thereby safeguards the evolutionarily conserved spatial chromatin organization (Kaaij et al. 2019). Given that CHD9 appears to play a role in regulation of chromatin organization in oocytes (Ooga et al. 2018), research into the role of the CHD family in higher-order chromatin organization is warranted.
Transcriptional outcomes
As demonstrated in diverse model organisms, CHD enzymes regulate nucleosome density and chromatin organization at promoter and enhancer elements, which can lead to different transcriptional outcomes (Fig. 3). For instance, some, such as CHD1, are required for RNAPII transcription (Krogan et al. 2002; Simic 2003; Srinivasan et al. 2005; Warner et al. 2007; Skene et al. 2014; Dieuleveult et al. 2016; Baumgart et al. 2017) and play a role in transcription initiation (Lin et al. 2011) and in RNAPII-directed turnover of promoter-proximal nucleosomes, promoter escape, and subsequent elongation and splicing (Sims et al. 2007; Skene et al. 2014). Similarly, mounting evidence shows that mammalian CHD2 (Rom et al. 2019), CHD6 (Lutz et al. 2006), CHD8 (Rodríguez-Paredes et al. 2009; Ceballos-Chávez et al. 2015), and CHD9 (Shur et al. 2006a; Lee and Stallcup 2017; Alendar et al. 2020; Newton and Pask 2020; Yoo et al. 2020) potentiate transcription of distinct target genes in diverse cell types. CHD enzymes also contribute to transcriptional repression, best exemplified by the concerted action of chromatin remodeling and histone deacetylase catalytic subunits of the NuRD complex, which generally leads to the generation of hypoacetylated, densely packed chromatin (Lai and Wade 2011) and facilitates PRC2-mediated silencing of target genes in ESCs (Reynolds et al. 2012).
The capacity of CHD enzymes to function as either transcriptional activators or repressors is likely caused by the mode of CHD recruitment to target loci, the local chromatin landscape, and the subunit composition of the multimeric protein complexes in which they reside (Fig. 3). For instance, CHD4 alone promotes transcription in T cells (Williams et al. 2004) and ES cells (Dieuleveult et al. 2016; Bornelöv et al. 2018), whereas a great number of studies reviewed here show that in a NuRD complex it contributes to transcriptional repression. Similarly, in addition to its role in promoting transcription (Yates et al. 2010; Ceballos-Chávez et al. 2015), CHD8 inhibits β-catenin (Nishiyama et al. 2004) and P53 (Nishiyama et al. 2009) transactivation by forming a repressive trimeric complex with linker histone H1.
In some instances, different CHD remodelers are required for distinct stages of transcription—e.g., to regulate initiation (dBRM), promoter clearance (dKisL), and elongation (dCHD1) (Srinivasan et al. 2005)—or act synergistically with other chromatin regulators to modulate transcription in a given cellular context (CHD7 and PBAF [Bajpai et al. 2010], as well as CHD4 and Polycomb repressive complex 2 [PRC2] [Sparmann et al. 2013]). Moreover, different chromatin remodelers with opposing functions can engage in a “tug-of-war” action to fine-tune transcription in response to extracellular cues—SWI/SNF and NuRD/Mi-2 (Ramirez-Carrozzi et al. 2006; Gao et al. 2009), esBAF and NuRD (Yildirim et al. 2011), and Cockayne syndrome protein B (CSB) and NuRD (Xie et al. 2012).
As discussed in the following section, this mode of action may be particularly relevant in embryonic development, where cells rapidly switch transcriptional programs during lineage commitment and differentiation.
Stem cell maintenance, lineage commitment, differentiation, and development
Accurate transcriptional control is paramount for organismal development and requires cross-talk between myriad transcription factors and chromatin regulators that act in concert to control expression of the right genes at the right time. A great body of work discussed below highlights the important roles CHD enzymes play in stem cell self-renewal and pluripotency, as well as in directing cell fate decisions in development (Fig. 4).
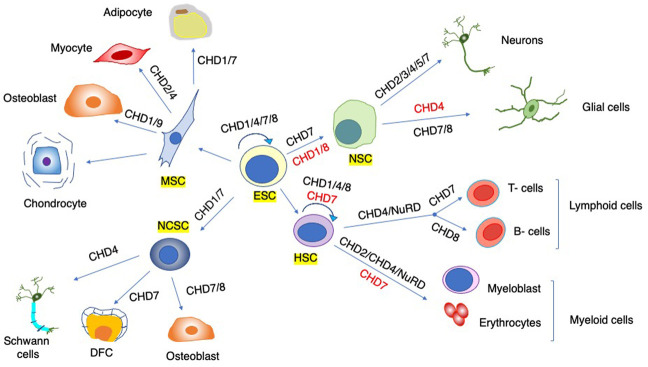
Figure 4.
Cell fate specification and lineage commitment. CHD functions are important in diverse stem cell populations (annotated by yellow rectangles). Some CHDs are important for the emergence and self-renewal of stem cells (circular arrow), and others are vital for lineage commitment and differentiation (straight arrows). Distinct CHDs can promote (black) or inhibit (red) differentiation. For simplicity, not all intermediate precursors and terminally differentiated cells are depicted. (ESC) Embryonic stem cell, (HSC) hematopoietic stem cell, (MSC) mesenchymal stem cell, (NSC) neural stem cell, (NCSC) neural crest stem cell, (DFC) dental follicle cells.
The majority of the Chd germline knockouts in animal models display severe early developmental phenotypes, pointing to their nonredundant functions (Supplemental Table S1 and references therein). For instance, Chd7 and Chd8 germline knockout mice are embryonic-lethal due to the widespread P53-mediated apoptosis (Nishiyama et al. 2009; Nostrand et al. 2014). Notably, Chd5 and Chd9 knockouts are viable and provide an exception to the rule, with Chd9−/− animals having no overt phenotype (Alendar et al. 2020), and Chd5−/− displaying impaired spermatogenesis and fertility (Li et al. 2014). There are contrasting reports of phenotypes observed in Chd2 knockout mice, ranging from perinatal lethality (Marfella et al. 2006, 2008; Kulkarni et al. 2008) to brain-specific developmental defects (Meganathan et al. 2017), or absence of any overt phenotypes (Supplemental Table S1; Rom et al. 2019). Single-exon deletion Chd6 mice are viable but display motor coordination impairment (Lathrop et al. 2010). Although no germline knockout mice are reported to date, both Chd1 (Gaspar-Maia et al. 2009) and Chd4 (O'Shaughnessy-Kirwan et al. 2015) clearly play critical roles in development, as their depletion leads to impaired blastocyst formation, preimplantation defects, and embryonic lethality.
As discussed below, most of our insight into the roles of CHDs in guiding cell fate decisions and development comes from the studies of primary cell cultures and tissue-specific conditional knockouts (Supplemental Table S1). Gene dosage seems to be important, and haploinsufficiency can cause pathogenesis in both mice and humans, indicating that functions of chromatin remodelers in specific cellular processes are rate-limiting.
CHDs in stem and progenitor cells
ES cells are self-renewing, pluripotent cells characterized by high expression of chromatin remodeling enzymes (Efroni et al. 2008), pervasive global transcription, and relatively open chromatin organization (Orkin and Hochedlinger 2011; Percharde et al. 2017). The gene expression heterogeneity of ESCs creates clonal diversification and transcriptional plasticity, securing accurate response to differentiation cues while maintaining the capacity for self-renewal (Ram and Meshorer 2009). These features of pluripotent ESCs are at least in part regulated by the ATP-dependent chromatin remodeling enzymes (Landry et al. 2008; Ho et al. 2009, 2011).
Importantly, CHD enzymes form intricate regulatory networks with core pluripotency factors, forming complexes with SOX2, OCT4, and NANOG (CHD1, CHD2, CHD7, and CHD3/4/5/NuRD) (Schnetz et al. 2009; Pardo et al. 2010; Reynolds et al. 2012; Gagliardi et al. 2013; Semba et al. 2017; Bornelöv et al. 2018). They also associate with promoters and enhancers of Sox2, Pou5f1, and Nanog to modulate their expression (CHD1, CHD2, CHD4, and CHD7) (Gaspar-Maia et al. 2009; Pardo et al. 2010; Schnetz et al. 2010; Reynolds et al. 2012; Gagliardi et al. 2013; Suzuki et al. 2015; Semba et al. 2017). Moreover, expression of Chd1, Chd9, and several subunits of the NURD complex is directly regulated by the core pluripotency factors (Kim et al. 2008; Orkin et al. 2008; Orkin and Hochedlinger 2011). As such, CHD enzymes clearly play an important role in modulating the ESCs’ core pluripotency regulatory circuitry.
In ESCs, CHDs appear to occupy distinct enhancers, as well as promoters of both active and bivalent genes marked by H3K4m3 and H3K4me3/H3K27me3, respectively (Gaspar-Maia et al. 2009; Schnetz et al. 2009; Lin et al. 2011; Dieuleveult et al. 2016). Bivalent developmental genes are largely kept silent in ESCs but are poised for activation upon differentiation (Bernstein et al. 2006; Voigt et al. 2013). The CHD1-mediated (Konev et al. 2007; Gaspar-Maia et al. 2009; Baumgart et al. 2017) and CHD2-mediated incorporation of histone variants (H2A.Z and H3.3) (Harada et al. 2012; Shen et al. 2015b; Semba et al. 2017) facilitates establishment of bivalency and bookmarking of key developmental genes (Harikumar and Meshorer 2015). In turn, this promotes decommissioning of the pluripotency enhancers by CHD4/NuRD/LSD1 (Gehre et al. 2020) and concomitant shutdown of the ESC-specific transcription during differentiation (Reynolds et al. 2012; Whyte et al. 2012).
Transcriptional heterogeneity and plasticity of ESCs allow prompt cellular response to developmental cues. In this regard, CHD4 and CHD7 act as transcriptional rheostats to delimit expression of the lineage- and ES-specific genes, facilitating cell fate changes (Schnetz et al. 2010; Reynolds et al. 2012; O'Shaughnessy-Kirwan et al. 2015; Bornelöv et al. 2018). Consequently, in the absence of CHD4/NuRD, ESCs are unable to down-regulate pluripotency genes and accurately engage critical lineage-specific genes (Kaji et al. 2006; Reynolds et al. 2012).
Thus, the net effect of intricate CHD transcriptional regulatory mechanisms that operate in ESCs is twofold: curtailing expression of ESC-specific genes and core pluripotency factors while maintaining tight regulation of expression of key instructive factors required for rapid transcriptional response upon lineage commitment and differentiation.
Reprogramming
Chromatin remodelers also play critical roles during reprograming of somatic cells to induced pluripotency (iPS) by overexpression of Oct4, Sox2, Klf4, and cMyc (OSKM) factors (Orkin and Hochedlinger 2011). Notably, in both mouse and human somatic cells, the NuRD complex appears to modulate iPS reprogramming, although further studies are required to clarify its exact role, as current literature provides contrasting experimental evidence suggesting that it can both inhibit (Luo et al. 2013; Rais et al. 2013) and promote (Santos et al. 2014) reprogramming. The contribution of other CHD proteins to iPS reprograming is less understood; while CHD1 is critical for efficient reprogramming of mouse fibroblasts into iPS cells, CHD2 and CHD7 appear to be dispensable (Gaspar-Maia et al. 2009; Schnetz et al. 2009; Semba et al. 2017). Recently, iPS cells were successfully derived from patients carrying heterozygous mutations in CHD7 (CHARGE syndrome) and CHD8 (autism spectrum disorders [ASDs]), indicating that, while their heterozygosity appears to drive human neurodevelopmental pathologies, gene dosage is not critical for somatic cell reprogramming (Chai et al. 2018; Deneault et al. 2018). This holds great promise as a model system for studying underlying molecular perturbations.
Lineage commitment and differentiation
The iconic concept of developmental specification as an epigenetic landscape introduced by Waddington (1957) likens differentiation of a pluripotent cell to a ball on the top of the hill going down the cascade of branching ridges and valleys, representing specific developmental trajectories leading to different cell fates. As discussed below, CHDs alter chromatin landscape and modulate barriers between distinct cell fates, either fortifying commitment toward a specified developmental trajectory (increasing the barrier) or lowering the threshold for differentiation and permitting random sampling of different cellular fates (reducing the barrier).
The association of CHDs with chromatin is dynamic during differentiation. Some family members (CHD1 and CHD7) appear important for both stemness and differentiation of ESCs, whereupon their chromatin occupancy alters, tracking the transcriptional activation and H3K4me3 redistribution to differentiation-specific genes (Gaspar-Maia et al. 2009; Schnetz et al. 2009, 2010). In contrast, CHD2 remains on a subset of bivalent genes throughout differentiation, likely preserving resolution of their poised chromatin state for later developmental stages (Semba et al. 2017).
Beyond their importance in ESC biology, the roles of CHDs extend to other embryonic and adult stem and progenitor cells (Fig. 4). Some, such as CHD1, have a general role in the expansion of stem and progenitor populations during developmental transitions (Percharde et al. 2017), or fulfil critical roles in lineage commitment and differentiation of the diverse stem cells and early progenitors (CHD4/NuRD) (Fig. 4; Williams et al. 2004; Kaji et al. 2006; Kashiwagi et al. 2007; Yoshida et al. 2008; Hung et al. 2012; Reynolds et al. 2012; Luo et al. 2013; O'Shaughnessy-Kirwan et al. 2015; Bornelöv et al. 2018; Hirota et al. 2019). Additionally, individual CHD enzymes can also contribute to the development of specific germ layers and lineages. For instance, CHD1/2/9 are important for mesoderm formation and lineage commitment of mesenchymal stem cells (MSCs) toward osteoblasts (CHD9 [Shur and Benayahu 2005; Marom et al. 2006; Shur et al. 2006a,b] and CHD1 [Baumgart et al. 2017]) and adipocytes (CHD1) (Baumgart et al. 2017). Similarly, CHD7 and CHD8 appear to be important for the osteogenic differentiation of neural crest cells (NCCs) (Liu et al. 2020; Fan et al. 2021), whereas CHD7 appears to be required for both oligodendrocyte maturation and osteoblast bone formation (He et al. 2016), indicating that CHD enzymes may act as developmental hubs of diverse lineages.
Several CHDs are also implicated in myogenic differentiation and muscle organogenesis (Fig. 4), as exemplified by the critical role of CHD2 in differentiation of myogenic progenitors in mice (Harada et al. 2012). Evidence is emerging that the CHD4/NuRD complex is an important regulator of cardiac and skeletal muscle progenitor cell identity (Mammen et al. 2009; Arco et al. 2016; Sreenivasan et al. 2020), as evident from Chd4-null cardiomyocytes that form a hybrid muscle tissue in mice (Wilczewski et al. 2018), and the notion that mutations in both CHD4 and CHD7 are implicated in congenital heart disease (Homsy et al. 2015). Similarly, CHD4/NuRD is critical for the self-renewal and multilineage differentiation potential of hematopoietic stem cells (Wade et al. 1999; Fujita et al. 2004; Williams et al. 2004; Yoshida et al. 2008) and fulfils important roles in guiding lymphocyte development (B cell [Fujita et al. 2004; Arends et al. 2019; Yoshida et al. 2019] and T-cell [Yasui et al. 2002; Williams et al. 2004; Naito et al. 2007]), governing the postnatal switch in β-globin expression (Xu et al. 2010) and establishment of immune tolerance (Tomofuji et al. 2020).
Other CHDs play important roles as well (Fig. 4). For instance, while some are vital for the emergence (CHD1 [Koh et al. 2015] and CHD8 [Shooshtarizadeh et al. 2019; Nita et al. 2021; Tu et al. 2021]) and differentiation of hematopoietic stem and progenitor cells (HSPCs) toward erythroid and myeloid precursors (CHD2) (Nagarajan et al. 2009; Varnoosfaderani et al. 2020), others appear to have an opposing role (CHD7) (Fig. 4; Hsu et al. 2013, 2020). Importantly, patient genomic data and studies in animal models point to driver roles of CHD2 (Berquam-Vrieze et al. 2011; Quesada et al. 2012; Rodríguez et al. 2015; Kim et al. 2016), CHD7 (Zhen et al. 2017), CHD8 (Shingleton and Hemann 2015; Shen et al. 2015a; Grande et al. 2019; Newell et al. 2019), and CHD9 (Mikkers et al. 2002; Shou et al. 2006) of various hematopoietic malignancies (Supplemental Table S1).
Neuronal development and pathologies
During neurodevelopment, CHD remodelers regulate chromatin accessibility and gene expression of key transcriptional networks converging on the SHH (Feng et al. 2017), Notch (Jones et al. 2015), FGF (Hurd et al. 2010; Engelen et al. 2011), TGF-β (Fueyo et al. 2018), WNT/β-catenin (Sakamoto et al. 2000; Nishiyama et al. 2004), and PI3K (Micucci et al. 2015) signaling pathways and regulating P53 transactivation (Nishiyama et al. 2009; Nostrand et al. 2014), thereby controlling many aspects of brain development and functionality. Importantly, in the last decade, a great number of whole-genome sequencing studies identified mutations of CHD enzymes in neurodevelopmental disorders, with haploinsufficiency as a mechanism driving these pathologies (Supplemental Table S1 and references therein). Phenotypic characterization of patient-derived cell lines and CHD animal models provides valuable insight into the underlying molecular mechanisms that govern development of these pathologies.
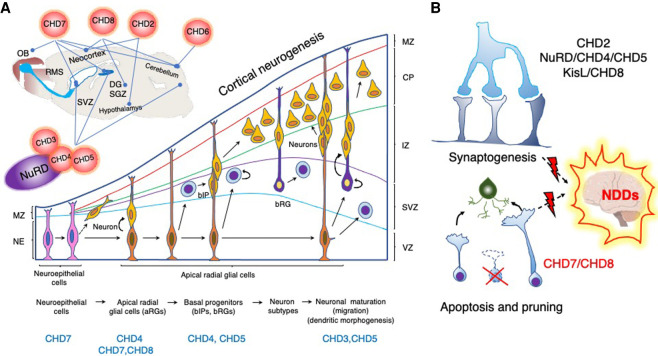
CHDs have distinct yet overlapping roles in neural stem cell niches in the cerebrum (subventricular zone [SVZ]) and hippocampus (subgranular zone of the dentate gyrus [SGZ/DG]; CHD3/4/5/NuRD) (Egan et al. 2013; Sparmann et al. 2013; Nitarska et al. 2016), where they regulate neurogenesis and neuronal differentiation (CHD2 [Shen et al. 2015b; Kim et al. 2018; Lalli et al. 2020], CHD4 [Sparmann et al. 2013], and CHD5 [Egan et al. 2013]) and neuronal migration and maturation (CHD3 and CHD5) (Fig. 5; Egan et al. 2013; Nitarska et al. 2016). Likewise, both CHD7 (Engelen et al. 2011; Feng et al. 2013, 2017; Micucci et al. 2014; Jones et al. 2015; He et al. 2016; Yao et al. 2020) and CHD8 (Sakamoto et al. 2000; Nishiyama et al. 2004; Ronan et al. 2013; Sugathan et al. 2014; Durak et al. 2016; Katayama et al. 2016; Kawamura et al. 2021) are vital for brain development and regulate functions of stem cells in diverse brain regions. Thus, CHD enzymes play vital roles both in brain development (CHD2, CHD3/4/5, CHD7, and CHD8) and adult neurogenesis (CHD2, NuRD, and CHD7). It is becoming increasingly evident that some of the CHD7/8-mediated neurodevelopmental disorders develop, at least in part, as a result of dysregulation of P53 transactivation during brain development (Nostrand et al. 2014; Marie et al. 2018; Wade et al. 2019; Hurley et al. 2021).
Figure 5.
CHDs regulate different aspects of neurodevelopment. (A, top) CHD enzymes are vital for the development and functionality of different parts of the brain: the cerebral neocortex (CHD2, CHD3/4/5/NuRD, CHD7, and CHD8), cerebellum (CHD4/NuRD, CHD6, CHD7, and CHD8), hypothalamus (CHD2), thalamus (CHD8), and hippocampus (CHD4/5/NuRD, CHD7, and CHD8). They regulate neural stem cell niches in the cerebrum (subventricular zone [SVZ]) and hippocampus (subgranular zone of the dentate gyrus [SGZ/DG]) and are essential for proper neuronal differentiation and migration. CHD7 is important for the development of the inner ear and olfactory system and regulates migration of the neuroblasts that emerge from the neural stem cells in the SVZ and proceed through the rostral migratory stream (RMS) to the olfactory bulb (OB). The cartoon represents the sagittal section of the rodent brain and highlights CHD functions in the respective brain regions (lines). (Bottom) CHDs are required at various stages of cortical neurogenesis. During the early stages of cortical neurogenesis, symmetrical division of the neuroepithelial cells (NEs) in the ventricular zone (VZ) expands their pool, whereas asymmetrical division gives rise to apical neural precursor cells (NPCs), such as apical radial glial cells (aRGs), and pioneer neurons. Apical progenitor cells give rise to neurons through basal intermediate precursors (bIP) and radial glial cells (bRG) that reside in the subventricular zone (SVZ). Neurons use aRG and bRG fibers to migrate to the specified layer in the cortical plate (CP), where they establish synaptic connections with subplate layer neurons during maturation. CHD remodelers involved at distinct stages are highlighted bellow. (MZ) Marginal zone, (IZ) intermediate zone. Cortical neurogenesis cartoon representation adapted with permission from Sokpor et al. (2018). (B, top) Synaptogenesis between neurons initiates during embryogenesis, but synaptic plasticity proceeds throughout life. Synaptic plasticity is the property of synapses to strengthen or weaken in response to changes in both the amplitude and the temporal dynamics of neuronal activity. Sensory inputs and intrinsic brain activity can effect long-term changes in synaptic efficacy and eventually increase or decrease neuronal connectivity by modulating the number of synapses (Bourgeron 2015). Some CHD enzymes regulate neuronal connectivity (CHD4, CHD5, and CHD8) and excitability (CHD2 and CHD8), whereas others regulate synaptic vesicle recycling (KisL) and expression of genes essential for synaptic homeostasis and plasticity (CHD8). (Bottom) Apoptosis and pruning are required for the maintenance and refinement of the neural circuitries, and CHD7/8 inhibition of P53-mediated apoptosis plays an active role in this process. Dysregulation of CHD enzyme function affects synaptogenesis and apoptosis and leads to aberrant neurogenesis and connectivity, which results in the emergence of diverse neurodevelopmental disorders (NDDs).
Moreover, CHD enzymes are also critical for the maturation of glial cells and myelination of the central nervous system (CNS; CHD7/8) (Feng et al. 2013; He et al. 2016; Doi et al. 2017; Marie et al. 2018; Zhao et al. 2018a) and peripheral nervous system (CHD4) (Fig. 5; Srinivasan et al. 2006; Hung et al. 2012). Intriguingly, CHD8 can functionally compensate for the CHD7 loss at least partially during differentiation of oligodendrocyte precursor cells (OPCs) (Marie et al. 2018). These findings, together with evidence of CHD7–CHD8 association (Batsukh et al. 2010), suggest a requirement for concerted action of these remodelers.
Beyond their role in regulating cell fate decisions during neurogenesis, CHD enzymes are also important for neuronal excitability (CHD2 [Meganathan et al. 2017] and CHD8 [Platt et al. 2017; Jung et al. 2018; Ellingford et al. 2021]), and for synaptic connectivity and strength (CHD4 [Yamada et al. 2014], CHD5 [Egan et al. 2013; Pisansky et al. 2017], and CHD8 [Platt et al. 2017; Jung et al. 2018; Ellingford et al. 2021]). The CHD8 regulation of synaptic homeostasis and plasticity is vital for neocortical (Kweon et al. 2021) and cerebellar (Kawamura et al. 2021) development and is dysregulated in ASDs, as discussed below (Fig. 5).
Neurodevelopmental disorders
As highlighted above, CHD enzymes are critical for the development of the cerebrum (CHD2, CHD3/4/5/NuRD, CHD7, and CHD8), cerebellum (CHD4/NuRD, CHD6, CHD7, and CHD8), hypothalamus (CHD2), thalamus (CHD8), and hippocampus (CHD4/5/NuRD, CHD7, and CHD8) (Fig. 5). Disruptions of CHD functions can result in different numbers of mature neurons and neuroglia, mislocalization of neurons, and altered synaptic activity in various parts of the brain, thereby affecting cognitive functions, the integration of sensory information, memory, learning, and motor coordination.
Therefore, it comes as no surprise that a large number of recent whole-genome sequencing studies of patients with NDDs (ASD, CHARGE, schizophrenia, intellectual disability, developmental disorders, and epilepsy) identified de novo heterozygous mutations, SNVs, small deletions, and copy number variations in CHD1, CHD2, CHD3, CHD4, CHD6, CHD7, and CHD8 (Supplemental Table S1 and references therein; O'Roak et al. 2012a,b, 2014; Sanders et al. 2012; Bernier et al. 2014; Iossifov et al. 2014; Krumm et al. 2014; Study 2017; Coe et al. 2019).
The de novo loss-of-function mutations in CHD8 are recurrent in ASD patients, and Chd8 heterozygous mice faithfully recapitulate a variety of symptoms found in these patients (Durak et al. 2016; Katayama et al. 2016; Gompers et al. 2017; Platt et al. 2017; Jung et al. 2018; Suetterlin et al. 2018). Likewise, mounting evidence indicates that dysregulation of CHD7/SOX2 gene networks is driving human Alagille, Pallister-Hall, and Feingold syndromes (Engelen et al. 2011). Moreover, CHD7 regulates development of diverse tissues of neuronal origin that are affected in the autosomal dominant congenital CHARGE syndrome (Micucci et al. 2015), and 90% of the CHARGE patients harbor CHD7 heterozygous mutations (Supplemental Table S1 and reference therein; Ravenswaaij-Arts and Martin 2017). Importantly, Chd7 germline heterozygous and tissue-specific knockout mice display most of the features observed in CHARGE individuals (Supplemental Table S1). Biochemical studies reveal that patient-specific mutations of CHD7 chromodomains modulate ATP-dependent remodeling activity in a mutation-dependent manner (Bouazoune and Kingston 2012; Yan et al. 2020). Moreover, as reviewed earlier, dysregulation of the CHD7–P53 axis plays an important role in CHARGE etiology, and P53 mutations are reported in 10%–30% of CHARGE individuals (Nostrand et al. 2014).
Similarly, several recent studies reveal that haploinsufficiency of CHD1 (Pilarowski et al. 2018), CHD2 (Pinto et al. 2010; Capelli et al. 2012; Rauch et al. 2012; Carvill et al. 2013; Suls et al. 2013; Chénier et al. 2014; Lund et al. 2014; Thomas et al. 2015; Wilson et al. 2021), and CHD5 (Pisansky et al. 2017; Network et al. 2021) contributes to the emergence of NDDs with diverse phenotypic features, whereas recurrent de novo CHD3 and CHD4 missense mutations are drivers of novel human neurodevelopmental syndromes termed Snijders Blok-Campeu syndrome (Blok et al. 2019; Drivas et al. 2020) and Sifrim–Hitz–Weiss syndrome (SIHIWES) (Weiss et al. 2016, 2020), respectively. These CHD mutations are likely gene-disrupting mutations (LGD) that result in alteration or complete loss of protein function. Notably, gene dosage seems to play a role in the etiology of pathologies, as both inactivation and duplication of CHD3 (Drivas et al. 2020) and CHD4 (Coe et al. 2019) can result in NDDs.
Importantly, as discussed in the following section, it is increasingly appreciated that similar signaling and metabolic pathways are dysregulated in both NDDs and human cancers. This holds promise for the repurposing of anticancer drugs for the treatment of some NDD symptoms.
CHDs in cancer
The genome sequencing endeavor of human cancers has identified numerous somatic mutations, copy number alterations, and chromosomal rearrangements of chromatin proteins (Flavahan et al. 2017), including CHD family members (Supplemental Table S1 and references therein). These mutations affect mostly only one allele, likely causing protein inactivation, providing support for the notion that haploinsufficiency drives pathogenesis both in human cancers and neurodevelopmental disorders. Additionally, epigenetic silencing of CHDs by promoter hypermethylation is documented across various types of cancers (Supplemental Table S1). Notably, complete inactivation is occasionally observed in prostate cancer (CHD1; homozygous deletion) and neuroblastoma (CHD5; inactivation of both alleles) (Supplemental Table S1). Given that intradomain allosteric regulation appears to be a general mechanism controlling chromatin association and enzymatic activity of CHD enzymes, it is plausible that CHD translocations, mutations or copy number alterations modulate folding and allosteric regulation or alter assembly and subunit composition of the CHD protein complexes, resulting in aberrant enzymatic activity and pathogenicity. Functional diversification of CHD enzymes can be further expanded through association with cancer-specific chimeric TFs generated by chromosomal translocations, as illustrated by PML-RARα that repurposes the NuRD complex to drive leukemogenic transcriptional programs (Morey et al. 2008). To what extent these genetic CHD perturbations are clear drivers or mere passengers in tumorigenesis is not clear, but as discussed below, for some (CHD1, CHD4, CHD5, and CHD7), a causal role in human malignancies has been clearly established.
One of the cardinal features of cancer cells is the ability to sustain proliferation through persistent activation of mitogenic signaling pathways (Fig. 6; Hanahan and Weinberg 2011). The CHD enzymes act downstream from key developmental signaling pathways that are frequently perturbed in tumorigenesis, such as WNT/β-catenin (CHD5 and CHD8) (Nishiyama et al. 2004; Fatemi et al. 2014), LIF/STAT3 (CHD8) (Yamashina et al. 2006), SHH (CHD7) (Engelen et al. 2011), NOTCH (CHD7) (Engelen et al. 2011), SMAD/BMP (CHD7) (Jiang et al. 2012; Liu et al. 2014b), and TGF-β/SMAD (CHD4 and CHD8) (Wang et al. 2009; Fueyo et al. 2018). Thus, chromatin remodeling activity of CHD enzymes appears to be critical for translating information from ligand-mediated signaling pathways to the transcriptional machinery—e.g., ERK/CHD7 (Badodi et al. 2017), WNT/β-catenin/CHD8 (SAWADA et al. 2013), PTEN/CHDl (Zhao et al. 2017), and K-RASG13D/CHD1 (Yu et al. 2015)—and it is likely that the CHD modulation of developmental pathways also contributes to tumorigenesis.
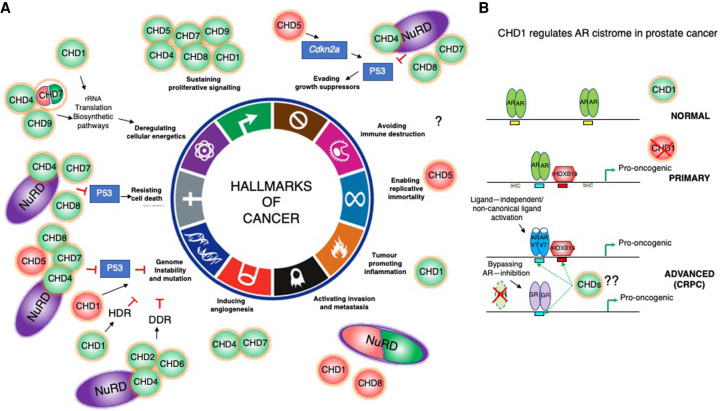
Figure 6.
The role of CHDs in tumorigenesis. (A) CHD enzymes are context-dependent tumor suppressors (TS; red) and oncogenes (green) that contribute to acquisition of different hallmarks of cancer. They can drive tumorigenesis by directly promoting transcription of oncogenes and repression of TS genes (CDKN2A and TP53; blue rectangles). Some enzymes can exert both oncogenic and TS function (CHD1, CHD4/NuRD, CHD7, and CHD8). For simplicity, not all players that are mentioned in the text are indicated in the cartoon. CHD functions in human malignancies are still largely unexplored, and it is likely that all members are dysregulated and mutated in diverse cancers and contribute to different aspects of tumorigenesis. Cartoon representation adapted from Hanahan and Weinberg (2011) with permission from Elsevier. (B, top) CHD1 nucleosome remodeling activity restricts binding of AR (green ovals) to prostetic lineage enhancers (yellow rectangles) in normal prostates. (Middle) During prostate tumorigenesis, in primary tumors, CHD1 loss leads to AR cistrome redistribution to a different subset of enhancers (blue rectangles), with HOXB13 (red hexagon) binding motifs (red squares) to drive expression of pro-oncogenic gene networks. (Bottom) More advanced, castration-resistant prostate cancers (CRPCs) are characterized by amplifications or mutations in the AR ligand-binding domain (LBD; AR-V7 as an example; blue ovals) (Jeselsohn et al. 2015; Watson et al. 2015). The LBD mutations allow promiscuous activation by noncanonical ligands (adrenal androgens, estrogen, progesterone, and glucocorticoids) to secure sustained expression of a subset of critical AR target genes (Watson et al. 2015). Additionally, antiandrogen-resistant prostate cancers can fully bypass the requirement for AR signaling by activating GR-mediated (violet ovals) transcriptional regulation of the AR cistrome (Arora et al. 2013). The role of other CHDs in the regulation of alternative transcriptional pathways that sustain expression of the AR-driven oncogenic gene networks in CRPC prostate cancers is unknown.
Importantly, CHD chromatin remodelers promote acquisition of different hallmarks of cancer by directly activating expression of pro-oncogenic transcriptional networks or inhibiting expression of tumor suppressor genes (Fig. 6). For instance, several examples of the pro-oncogenic function of CHD enzymes have been documented: regulating cell cycle and proliferation (CHD8 [Caldon et al. 2009; Rodríguez-Paredes et al. 2009; Subtil-Rodríguez et al. 2014; Dingar et al. 2015; Shen et al. 2015a], CHD4 [D'Alesio et al. 2016; 2019], CHD1 [Yu et al. 2015], and CHD9 [Yoo et al. 2020]), modulating angiogenesis (CHD4 [Yoo et al. 2006; Wang et al. 2020] and CHD7 [Boyd et al. 2019]), affecting the tumor microenvironment (CHD1 [Zhao et al. 2020]), impairing P53 transactivation (CHD4 [Luo et al. 2000], CHD7 [Nostrand et al. 2014], and CHD8 [Tu et al. 2021]), regulating expression of rRNA genes (CHD7 [Zentner et al. 2010], CHD4 [Zhao et al. 2018b], and CHD9 [Salomon-Kent et al. 2015]), and biosynthetic pathways (CHD1 [Bulut-Karslioglu et al. 2018] and CHD8 [Subtil-Rodríguez et al. 2014; Ceballos-Chávez et al. 2015]) (Fig. 6). Moreover, some, like the multifaceted CHD4/NuRD complex, promote tumorigenesis by epigenetic silencing of key tumor suppressor genes in colorectal cancer (Cai et al. 2014) (Xia et al. 2017).
As noted earlier, haploinsufficiency of chromatin remodelers drives tumorigenesis, and down-regulation of CHD expression is observed in several human cancers (Supplemental Table S1). CHD5 is best characterized as a tumor suppressor, and its inactivation drives progression of various human cancers (Supplemental Table S1 and references therein; Kolla et al. 2014). CHD5 is a potent regulator of cellular proliferation, senescence, and apoptosis owing to transcriptional regulation of the Cdkn2a locus, and its compromised expression in various human cancers blunts induction of key cellular gatekeeper pathways p16INK4a/RB and p14ARF/p53, setting the stage for cancer development (Mills 2017).
These studies, together with the roles of CHDs in the brain (CHD7) and hormone-dependent cancers (CHD1, CHD4/NuRD, and CHD8) discussed below, highlight the role of individual CHD enzymes as context-dependent oncogenes or tumor suppressors in human malignancies.
Brain cancers
CHD enzymes are vital for the proper development and functionality of the nervous system, and their reduced activity can give rise to neuronal tumors of variable origin. For instance, in neuroblastoma, loss of CHD5 (Kolla et al. 2014) and CHD9 (Lasorsa et al. 2016) correlates with poor prognosis. Notably, it appears that CHD7 inactivation drives human gliomas (Boyd et al. 2019), whereas both overexpression (Machado et al. 2019) and inactivation of CHD7 promotes medulloblastoma (MB) progression (Badodi et al. 2017, 2021). Indeed, CHD7 inactivation drives a MB subtype with increased activation of the ERK and mTOR signaling pathways (Badodi et al. 2017, 2021). Importantly, this metabolic adaptation creates a clinically relevant cancer vulnerability, as combined treatment with inositol and cisplatin greatly improves survival of MB xenografts (Badodi et al. 2021).
Hormone-driven cancers
There is ample evidence that CHD enzymes govern transcription in response to ligand-mediated activation of nuclear receptors in human cancers, either through direct association with nuclear receptors (NRs; CHD6, CHD8, and CHD9) (Surapureddi et al. 2002; Shur and Benayahu 2005; Marom et al. 2006; Surapureddi et al. 2006; Shur et al. 2006a; Menon et al. 2010), or indirectly through modulating NR-mediated transcriptional activity (CHD1, CHD4, and CHD7) (Augello et al. 2019; Zhang et al. 2020; Zhao et al. 2020; Paakinaho et al. 2021). These regulatory interactions are particularly important for prostate and breast cancers (CHD1 [Berger et al. 2011; Grasso et al. 2012; Huang et al. 2012; Liu et al. 2012; Burkhardt et al. 2013] and CHD8 (Caldon et al. 2009; Menon et al. 2010; Ceballos-Chávez et al. 2015; Metzger et al. 2016]) discussed below.
Prostate cancer
The progression of prostate cancer is mainly driven by PTEN loss, AR amplification or mutation, and ETS gene fusion (TMRPSS2-ERG) (Jamaspishvili et al. 2018). Recent genomic studies of prostate cancers also identified recurrent mutations (CHD1/3), amplifications (CHD7), or promoter methylation (CHD5/8) of CHD family members (Supplemental Table S1). Intriguingly, a CHD8 functional dichotomy is observed, whereby decreased expression through promoter methylation drives early stages of prostate cancer tumorigenesis, whereas higher levels are associated with advanced clinical features (Damaschke et al. 2014).
Next to PTEN loss, CHD1 inactivation is the most frequent event in localized prostate cancer and advanced castration-resistant prostate cancer (CRPC) samples (Supplemental Table S1 and references therein). Importantly, a large body of work in cellular and animal models established CHD1 as a bone fide tumor suppressor in prostate cancer (Berger et al. 2011; Grasso et al. 2012; Huang et al. 2012; Liu et al. 2012; Baca et al. 2013; Burkhardt et al. 2013; Rodrigues et al. 2015; Shenoy et al. 2017; Zhao et al. 2017, 2020; Augello et al. 2019; Li et al. 2020; Zhang et al. 2020). The CHD1-null tumors define a specific subset of prostate tumors with high genome instability, characterized by intrachromosomal rearrangements, gene deletions, and high frequency of recurrent small copy number amplifications (Baca et al. 2013).
However, CHD1 can also serve as an oncogenic driver of distinct PTEN− and TMPRSS2-ERG+ subtypes of prostate cancers. Indeed, high CHD1 levels positively correlate with the Gleason grade in patients (Zhao et al. 2017). Strikingly, PTEN and CHD1 deletions are mutually exclusive, and CHD1 appears to be a synthetic-essential gene required for the growth of PTEN-deficient tumors by promoting transcriptional activation of the prosurvival TNF–NFκB pathway (Zhao et al. 2017). Additionally, CHD1 regulation of IL-6 expression fosters an immunosuppressive tumor microenvironment and promotes progression of PTEN-deficient prostate cancers (Zhao et al. 2020). This newly defined PTEN–CHD1 paradigm appears to be equally important for the growth of breast and colorectal cancers as well, and CHD1 targeting in PTEN-negative breast and prostate cancers could be a viable strategy to impair tumor growth (Zhao et al. 2017).
Importantly, it appears that CHD1 nucleosome remodeling activity commands AR chromatin distribution and prostate tumorigeneses, as it favors either occupancy of canonical prostatic lineage-specific enhancers or pro-oncogenic genes enriched for HOXB13 binding motif (Fig. 6; Burkhardt et al. 2013; Pomerantz et al. 2015; Augello et al. 2019). Specifically, upon CHD1 loss during prostate epithelial transformation, altered chromatin accessibility leads to extensive redistribution of the AR cistrome to HOBX13 target regions (Pomerantz et al. 2015), resulting in transcriptional activation of pro-oncogenic pathways (Augello et al. 2019).
Together, the data suggest that CHD1, similar to CHD8, exerts dichotomous functions, having context-dependent tumor-suppressive or oncogenic roles in prostate cancer progression. Moreover, the CHD1 pleiotropic role modulates AR cistrome transactivation and the tumor microenvironment, thereby promoting prostate cancer progression. As such, the CHD1 status in patients may serve as an indicator of chromosomal instability and a predictor of metastatic potential.
Breast cancer
Owing to its multisubunit composition and context-dependent functionality, the NuRD complex can have opposing roles in breast cancer progression (Lai and Wade 2011), both inhibiting (Fujita et al. 2003) and promoting (Fu et al. 2011) EMT and enhancing insensitivity to endocrine therapy (Fu et al. 2011). Additionally, evidence suggests that both CHD1 and CHD8 might be involved in ER+ breast cancer progression (Tan et al. 2014; Ceballos-Chávez et al. 2015). Notably, similar to CHD1 modulation of the AR cistrome in prostate cancer, NuRD chromatin occupancy regulates the binding potential and genomic distribution of ER (Serandour et al. 2018) and is critical for the growth of ER+ breast cancers (Farcas et al. 2021). These studies are indicative of complex roles of CHD family members in breast cancer progression, warranting further studies of the underlying mechanisms.
DNA damage repair signaling
Although not a focus of this review, it is noteworthy highlighting critical roles of CHD enzymes in modulating double-stranded break (DSB) DNA damage repair (DDR) pathway signaling (for further reading, see Stanley et al. 2013; Rother and Attikum 2017). Current evidence indicates that CHD2 (Luijsterburg et al. 2016), CHD3/4 (Rother and Attikum 2017), and CHD7 (Rother et al. 2020) recruitment to DSBs orchestrates nonhomologous end joining (NHEJ), whereas CHD1 (Kari et al. 2016) and CHD3/4 (Pan et al. 2012) are important for homology-directed repair (HDR). Moreover, displacement of CHD3 appears vital for the chromatin relaxation and repair of heterochromatic DSBs by either NHEJ or HDR (Goodarzi et al. 2011; Klement et al. 2014). In all of these instances, upon DNA damage, CHD remodeling activity is required either to promote transcription of inhibitors of oxidative damage (CHD6) (Moore et al. 2019), or to shape local chromatin structure and facilitate recruitment of the DNA repair machinery to sites of damage (CHD1 [Kari et al. 2016; Shenoy et al. 2017; Zhou et al. 2018], CHD2 [Luijsterburg et al. 2016], CHD3 [Goodarzi et al. 2011; Klement et al. 2014], CHD4 [Rother and Attikum 2017], and CHD7 [Colbert et al. 2014; Rother et al. 2020]).
Clinical relevance as potential biomarkers and actionable therapeutic targets
Current cancer treatment modalities rely on the use of standard cytotoxic chemotherapeutic agents and relatively recent approaches, such as targeted drugs, endocrine therapies, and immunotherapy. Therapeutic effects of anticancer drugs are largely determined by the landscape of genetic and epigenetic events acquired during tumor evolution. Emerging evidence indicates that CHDs may hold promise as actionable targets and serve as biomarkers and predictors of metastatic potential, patient survival, and therapeutic response, as well as chemoresistance (Wagner et al. 2018), in several cancers. As such, they could help inform patient stratification and response to therapies.
As CHDs are dysregulated across various cancers, they may serve as novel stratification biomarkers (CHD1: prostate cancer), and predictors of patient survival (CHD4: colorectal cancer and hepatocellular carcinoma [Xia et al. 2017] and nonsmall lung cancer [Xu et al. 2016], CHD8: gastric cancer [SAWADA et al. 2013], CHD5: various cancer types [Mills 2017], and CHD7: pancreatic cancer [Colbert et al. 2014] and glioblastoma [Boyd et al. 2019]) and therapeutic outcomes (CHD2: colorectal cancer [Bandrés et al. 2007]) (Supplemental Table S1).
It is increasingly apparent that some cancers rely on the tumor-promoting function of CHD enzymes, phenomena known as “oncogene addition,” as their depletion elicits a strong antitumor effect and could be exploited as vulnerabilities in at least some cancer types (CHD1: PTEN-negative prostate cancer [Zhao et al. 2017], CHD4: AML [Sperlazza et al. 2015; Heshmati et al. 2016, 2018] and glioblastoma [Chudnovsky et al. 2014; McKenzie et al. 2019], and CHD7: pancreatic cancer [Colbert et al. 2014] and glioblastoma [Machado et al. 2019]). Additionally, studies in animal models provide evidence that CHD7 (Zhen et al. 2017) and CHD8 (Shingleton and Hemann 2015; Shen et al. 2015a) are required for the growth of hematopoietic cancers and therefore might represent vulnerabilities in human hematopoietic malignancies. Alternatively, given the promoter silencing by hypermethylation in CHD nullizygous tumors (Supplemental Table S1), reactivation of the wild-type locus using demethylating agents might be a viable therapeutic strategy (CHD5) (Fatemi et al. 2014).
Importantly, from a clinical perspective, loss of CHD function renders cells sensitive to treatment with irradiation (CHD1 [Kari et al. 2016; Shenoy et al. 2017] and CHD2 [Rajagopalan et al. 2012]), PARP inhibitors (CHD1 [Kari et al. 2016; Shenoy et al. 2017] and CHD4 [Pan et al. 2012; Nio et al. 2015]), chemotherapeutic drugs (CHD1 [Kari et al. 2016; Shenoy et al. 2017] and CHD7 [Colbert et al. 2014]), genotoxic agents (CHD4 [Sperlazza et al. 2015]), monoclonal antibodies (CHD4 [D'Alesio et al. 2019]), and combined inositol hexaphosphate (IP6) and cisplatin treatment (CHD7 [Badodi et al. 2021]). Conversely, loss of CHDs can also confer resistance to therapeutic treatments, as evident from enzalutamide resistance in prostate cancer (CHD1) (Zhang et al. 2020) and the relapse of SCLC (CHD8) (Supplemental Table S1; Wagner et al. 2018).
Although showing great promise in some malignancies (Cheng et al. 2019), the field of epigenetic targeted therapy is still in its infancy. Given the CHDs’ domain conservation and structure, functional dichotomy, protein isoforms, and multimeric complexes they reside in, as well as the severity of phenotypes in homozygous animal models, there are considerable challenges regarding drug design and selectivity.
An interesting approach could be the use of synthetic histone mimic compounds, such as influenza virus nonstructural protein 1 (NS1), which, when dimethylated and trimethylated, is able to target CHD1 chromodomains faithfully mimicking the CHD1–H3K4me2/3 interaction (Marazzi et al. 2012). This might enable selective inhibition of CHD activity in various pathological conditions.
Additional concern when considering CHDs as actionable targets is the combinatorial assembly of protein subunits that governs chromatin association and functional specificity of CHD complexes. Interrogation of functional and compositional differences between physiological and tumor-specific CHD complexes could provide an opportunity for development of new therapeutic strategies. The emergence of novel biochemical and structural approaches, such as cryo-EM, will facilitate determination of the structure of tumor-specific CHD protein complexes, and may allow development of selective small molecule inhibitors aimed at disrupting key interfaces that sustain tumor-specific CHD transcriptional programs.
Concluding remarks and future prospects
Chromodomain helicase DNA-binding proteins are an evolutionarily conserved family of ATP-dependent chromatin remodeling enzymes that play pivotal roles in regulation of the DNA-templated processes. The sheer number of CHD remodelers, their functional nonredundancy, and the unique domain composition of the different classes point to more specific and programmatic functions. Curiously, many CHD genes are expressed in the nervous system, and it is conceivable that the evolutionary development of the nervous system might have prompted CHD diversification and specialization of their functions, or has been the result of the rewiring of the regulatory circuitry that adapted to the increased complexity of the non-protein-coding complement of the genome (Mattick 2007).
Given their functional complexity, it is still unclear whether the primary function of CHD proteins is to locally remodel chromatin to regulate gene expression and DNA-repair or to maintain higher-order chromatin structure, and how the enzymes have been repurposed for each function within a given cellular context. Recent efforts in mapping CHD chromatin occupancy provide insight into the mechanism of transcriptional regulation. However, the role of CHDs in higher-order chromatin organization is still poorly understood, and it is tempting to speculate that distinct CHD enzymes might, each in their own right, safeguard particular or overlapping spatially organized “chromatin territories.” Given the dynamic nature of CHD chromatin association, a systematic approach using ChIP-seq, RNA-seq, and chromosome capture conformation assays in a defined model system (differentiation: ES vs. NPC; pathology: normal vs. tumor/NDD samples) might permit integrative modeling of their function in chromatin organization and start addressing this important question.
Recently, the role of liquid–liquid phase separation (LLPS) in chromatin compartmentalization and organization has received much attention (Hnisz et al. 2017; Gibson et al. 2019). Importantly, this process is at least in part driven by nucleosome spacing, linker length, and histone H1 binding (Hnisz et al. 2017; Gibson et al. 2019), and CHD enzymatic activity is instrumental in modulating these chromatin features. Additionally, as CHDs are organized in subnuclear compartments (CHD7 and CHD9), as well as occupy enhancers (CHD1, CHD4, and CHD7) and interact with nascent RNAs (CHD4 and CHD7), both of which are shown to affect chromatin compaction, exploring the involvement of CHDs in chromatin organization by LLPS is an attractive strategy.
The CHD proteins instruct cells to maintain their identity, divide, differentiate, or die, thereby orchestrating organismal development or malignant transformation. Disruption of CHD remodelers in mice is often embryonic-lethal. Phenotypic discrepancies of individual Chd knockouts between different studies highlight the importance of careful selection of the targeting strategy (homologous recombination or CRISPR) when manipulating proteins of such domain complexity, as removal of one critical exon or functional domain can elicit phenotypically different outcomes compared with the complete loss-of-function protein.
Genetic alterations of several CHD family members are identified in human cancers and neurodevelopmental syndromes, and mouse models carrying mutated Chd7/8 are valuable to further our understanding of underlying NDD molecular mechanisms and identification of viable therapeutic strategies. So far, only patient-specific mutations of CHD3, CHD4, and CHD7 are shown to alter nucleosome remodeling activity (Bouazoune and Kingston 2012; Kovač et al. 2018; Blok et al. 2019; Yan et al. 2020). Systematic analysis of patient-specific CHD mutants in defined cell-based or animal model systems might provide insight into their mechanism of action and relevance for pathogenesis. In addition, biochemical purification and structural characterization of CHD protein complexes specific for particular developmental (Nitarska et al. 2016; Hoffmeister et al. 2017) or pathological (i.e., normal vs. tumor/NDD sample) conditions would inform about the specific functional contexts in which CHDs operate. This would potentially allow the designing of drugs specifically aimed at disrupting key interfaces that sustain tumor-specific CHD transcriptional programs.
Three decades of the intense biochemical and genetic analysis of the CHD family members in different model organisms has provided insight into the mechanistic principles that govern the function of CHD remodelers in vitro and in vivo. However, it is becoming evident that we have barely exposed the tip of the iceberg, and that their complex roles in regulating gene expression, chromatin organization, development, and disease are yet to be fully appreciated.
Supplementary Material
Acknowledgments
We apologize to our colleagues whose work we did not cite due to space limitations. We thank anonymous reviewers for their helpful comments and suggestions.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.348897.121.
Competing interest statement
The authors declare no competing interests.
References
- Akhtar A, Zink D, Becker PB. 2000. Chromodomains are protein-RNA interaction modules. Nature 407: 405–409. 10.1038/35030169 [DOI] [PubMed] [Google Scholar]
- Alendar A, Lambooij J-P, Bhaskaran R, Lancini C, Song J-Y, Vugt Hv, Snoek M, Berns A. 2020. Gene expression regulation by the chromodomain helicase DNA-binding protein 9 (CHD9) chromatin remodeler is dispensable for murine development. PLoS One 15: e0233394. 10.1371/journal.pone.0233394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MD, Religa TL, Freund SMV, Bycroft M. 2007. Solution structure of the BRK domains from CHD7. J Mol Biol 371: 1135–1140. 10.1016/j.jmb.2007.06.007 [DOI] [PubMed] [Google Scholar]
- Arco PG-D, Perdiguero E, Yunes-Leites PS, Acín-Pérez R, Zeini M, Garcia-Gomez A, Sreenivasan K, Jiménez-Alcázar M, Segalés J, López-Maderuelo D, et al. 2016. The chromatin remodeling complex Chd4/NuRD controls striated muscle identity and metabolic homeostasis. Cell Metab 23: 881–892. 10.1016/j.cmet.2016.04.008 [DOI] [PubMed] [Google Scholar]
- Arends T, Dege C, Bortnick A, Danhorn T, Knapp JR, Jia H, Harmacek L, Fleenor CJ, Straign D, Walton K, et al. 2019. CHD4 is essential for transcriptional repression and lineage progression in B lymphopoiesis. Proc Natl Acad Sci 116: 10927–10936. 10.1073/pnas.1821301116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache JP, Gamarra N, Johnson SL, Leonard JD, Wu S, Narlikar GJ, Cheng Y. 2019. Cryo-EM structures of remodeler–nucleosome intermediates suggest allosteric control through the nucleosome. Elife 8: e46057. 10.7554/eLife.46057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, Shah N, Cai L, Efstathiou E, Logothetis C, et al. 2013. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 155: 1309–1322. 10.1016/j.cell.2013.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augello MA, Liu D, Deonarine LD, Robinson BD, Huang D, Stelloo S, Blattner M, Doane AS, Wong EWP, Chen Y, et al. 2019. CHD1 loss alters AR binding at lineage-specific enhancers and modulates distinct transcriptional programs to drive prostate tumorigenesis. Cancer Cell 35: 603–617.e8. 10.1016/j.ccell.2019.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, et al. 2013. Punctuated evolution of prostate cancer genomes. Cell 153: 666–677. 10.1016/j.cell.2013.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badodi S, Dubuc A, Zhang X, Rosser G, Jaeger MDC, Kameda-Smith MM, Morrissy AS, Guilhamon P, Suetterlin P, Li X-N, et al. 2017. Convergence of BMI1 and CHD7 on ERK signaling in medulloblastoma. Cell Rep 21: 2772–2784. 10.1016/j.celrep.2017.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badodi S, Pomella N, Zhang X, Rosser G, Whittingham J, Niklison-Chirou MV, Lim YM, Brandner S, Morrison G, Pollard SM, et al. 2021. Inositol treatment inhibits medulloblastoma through suppression of epigenetic-driven metabolic adaptation. Nat Commun 12: 2148. 10.1038/s41467-021-22379-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang C-P, Zhao Y, Swigut T, Wysocka J. 2010. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463: 958–962. 10.1038/nature08733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandrés E, Malumbres R, Cubedo E, Honorato B, Zarate R, Labarga A, Gabisu U, Sola J, García-Foncillas J. 2007. A gene signature of 8 genes could identify the risk of recurrence and progression in Dukes’ B colon cancer patients. Oncol Rep 17: 1089–1094. [PubMed] [Google Scholar]
- Batsché E, Yaniv M, Muchardt C. 2006. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biology 13: 22–29. 10.1038/nsmb1030 [DOI] [PubMed] [Google Scholar]
- Batsukh T, Pieper L, Koszucka AM, Velsen Nv, Hoyer-Fender S, Elbracht M, Bergman JEH, Hoefsloot LH, Pauli S. 2010. CHD8 interacts with CHD7, a protein which is mutated in CHARGE syndrome. Hum Mol Genet 19: 2858–2866. 10.1093/hmg/ddq189 [DOI] [PubMed] [Google Scholar]
- Baumgart SJ, Najafova Z, Hossan T, Xie W, Nagarajan S, Kari V, Ditzel N, Kassem M, Johnsen SA. 2017. CHD1 regulates cell fate determination by activation of differentiation-induced genes. Nucleic Acids Res 45: 7722–7735. 10.1093/nar/gkx377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bélanger C, Bérubé-Simard F-A, Leduc E, Bernas G, Campeau PM, Lalani SR, Martin DM, Bielas S, Moccia A, Srivastava A, et al. 2018. Dysregulation of cotranscriptional alternative splicing underlies CHARGE syndrome. Proc Natl Acad Sci 115: E620–E629. 10.1073/pnas.1715378115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. 2011. The genomic complexity of primary human prostate cancer. Nature 470: 214–220. 10.1038/nature09744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Silfhout ATV, et al. 2014. Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158: 263–276. 10.1016/j.cell.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Berquam-Vrieze KE, Nannapaneni K, Brett BT, Holmfeldt L, Ma J, Zagorodna O, Jenkins NA, Copeland NG, Meyerholz DK, Knudson CM, et al. 2011. Cell of origin strongly influences genetic selection in a mouse model of T-ALL. Blood 118: 4646–4656. 10.1182/blood-2011-03-343947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok LS, Rousseau J, Twist J, Ehresmann S, Takaku M, Venselaar H, Rodan LH, Nowak CB, Douglas J, Swoboda KJ, et al. 2019. CHD3 helicase domain mutations cause a neurodevelopmental syndrome with macrocephaly and impaired speech and language. Nat Commun 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boginya A, Detroja R, Matityahu A, Frenkel-Morgenstern M, Onn I. 2019. The chromatin remodeler Chd1 regulates cohesin in budding yeast and humans. Sci Rep 9: 8929. 10.1038/s41598-019-45263-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornelöv S, Reynolds N, Xenophontos M, Gharbi S, Johnstone E, Floyd R, Ralser M, Signolet J, Loos R, Dietmann S, et al. 2018. The nucleosome remodeling and deacetylation complex modulates chromatin structure at sites of active transcription to fine-tune gene expression. Mol Cell 71: 56–72.e4. 10.1016/j.molcel.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouazoune K, Kingston RE. 2012. Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc Natl Acad Sci 109: 19238–19243. 10.1073/pnas.1213825109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron T. 2015. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci 16: 551–563. 10.1038/nrn3992 [DOI] [PubMed] [Google Scholar]
- Bowman GD. 2019. Uncovering a new step in sliding nucleosomes. Trends Biochem Sci 44: 643–645. 10.1016/j.tibs.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd NH, Walker K, Ayokanmbi A, Gordon ER, Whetsel J, Smith CM, Sanchez RG, Lubin FD, Chakraborty A, Tran AN, et al. 2019. Chromodomain helicase DNA-binding protein 7 is suppressed in the perinecrotic/ischemic microenvironment and is a novel regulator of glioblastoma angiogenesis. Stem Cells Dayt Ohio 37: 453–462. 10.1002/stem.2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Latek RR, Peterson CL. 2004. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol 5: 158–163. 10.1038/nrm1314 [DOI] [PubMed] [Google Scholar]
- Brehm A, Längst G, Kehle J, Clapier CR, Imhof A, Eberharter A, Müller J, Becker PB. 2000. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. Embo J 19: 4332–4341. 10.1093/emboj/19.16.4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, Macrae TA, Oses-Prieto JA, Covarrubias S, Percharde M, Ku G, Diaz A, McManus MT, Burlingame AL, Ramalho-Santos M. 2018. The transcriptionally permissive chromatin state of embryonic stem cells is acutely tuned to translational output. Cell Stem Cell 22: 369–383.e8. 10.1016/j.stem.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt L, Fuchs S, Krohn A, Masser S, Mader M, Kluth M, Bachmann F, Huland H, Steuber T, Graefen M, et al. 2013. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res 73: 2795–2805. 10.1158/0008-5472.CAN-12-1342 [DOI] [PubMed] [Google Scholar]
- Cai Y, Geutjes E-J, Lint Kd, Roepman P, Bruurs L, Yu L-R, Wang W, Blijswijk Jv, Mohammad H, Rink Id, et al. 2014. The NuRD complex cooperates with DNMTs to maintain silencing of key colorectal tumor suppressor genes. Oncogene 33: 2157–2168. 10.1038/onc.2013.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldon CE, Sergio CM, Schütte J, Boersma MN, Sutherland RL, Carroll JS, Musgrove EA. 2009. Estrogen regulation of cyclin E2 requires cyclin D1 but not c-Myc. Mol Cell Biol 29: 4623–4639. 10.1128/MCB.00269-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelli LP, Krepischi ACV, Gurgel-Giannetti J, Mendes MFS, Rodrigues T, Varela MC, Koiffmann CP, Rosenberg C. 2012. Deletion of the RMGA and CHD2 genes in a child with epilepsy and mental deficiency. Eur J Med Genet 55: 132–134. 10.1016/j.ejmg.2011.10.004 [DOI] [PubMed] [Google Scholar]
- Carvill GL, Heavin SB, Yendle SC, McMahon JM, O'Roak BJ, Cook J, Khan A, Dorschner MO, Weaver M, Calvert S, et al. 2013. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet 45: 825–830. 10.1038/ng.2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos-Chávez M, Subtil-Rodríguez A, Giannopoulou EG, Soronellas D, Vázquez-Chávez E, Vicent GP, Elemento O, Beato M, Reyes JC. 2015. The chromatin remodeler CHD8 is required for activation of progesterone receptor-dependent enhancers. Plos Genet 11: e1005174. 10.1371/journal.pgen.1005174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai M, Sanosaka T, Okuno H, Zhou Z, Koya I, Banno S, Andoh-Noda T, Tabata Y, Shimamura R, Hayashi T, et al. 2018. Chromatin remodeler CHD7 regulates the stem cell identity of human neural progenitors. Gene Dev 32: 165–180. 10.1101/gad.301887.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, Han J, Wei X. 2019. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther 4: 62. 10.1038/s41392-019-0095-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chénier S, Yoon G, Argiropoulos B, Lauzon J, Laframboise R, Ahn JW, Ogilvie CM, Lionel AC, Marshall CR, Vaags AK, et al. 2014. CHD2 haploinsufficiency is associated with developmental delay, intellectual disability, epilepsy and neurobehavioural problems. J Neurodev Disord 6: 9. 10.1186/1866-1955-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840. [DOI] [PubMed] [Google Scholar]
- Chudnovsky Y, Kim D, Zheng S, Whyte WA, Bansal M, Bray M-A, Gopal S, Theisen MA, Bilodeau S, Thiru P, et al. 2014. ZFHX4 interacts with the NuRD core member CHD4 and regulates the glioblastoma tumor-initiating cell state. Cell Rep 6: 313–324. 10.1016/j.celrep.2013.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Iwasa J, Cairns BR, Peterson CL. 2017. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Bio 18: 407–422. 10.1038/nrm.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe BP, Stessman HAF, Sulovari A, Geisheker MR, Bakken TE, Lake AM, Dougherty JD, Lein ES, Hormozdiari F, Bernier RA, et al. 2019. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat Genet 51: 106–116. 10.1038/s41588-018-0288-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert LE, Petrova AV, Fisher SB, Pantazides BG, Madden MZ, Hardy CW, Warren MD, Pan Y, Nagaraju GP, Liu EA, et al. 2014. CHD7 expression predicts survival outcomes in patients with resected pancreatic cancer. Cancer Res 74: 2677–2687. 10.1158/0008-5472.CAN-13-1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alesio C, Punzi S, Cicalese A, Fornasari L, Furia L, Riva L, Carugo A, Curigliano G, Criscitiello C, Pruneri G, et al. 2016. RNAi screens identify CHD4 as an essential gene in breast cancer growth. Oncotarget 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alesio C, Bellese G, Gagliani MC, Lechiara A, Dameri M, Grasselli E, Lanfrancone L, Cortese K, Castagnola P. 2019. The chromodomain helicase CHD4 regulates ERBB2 signaling pathway and autophagy in ERBB2+ breast cancer cells. Biol Open 8: bio038323. 10.1242/bio.038323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaschke NA, Yang B, Blute ML, Lin CP, Huang W, Jarrard DF. 2014. Frequent disruption of chromodomain helicase DNA-binding protein 8 (CHD8) and functionally associated chromatin regulators in prostate cancer. Neoplasia 16: 1018–1027. 10.1016/j.neo.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V, Stokes DG, Perry RP. 1993. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci 90: 2414–2418. 10.1073/pnas.90.6.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneault E, White SH, Rodrigues DC, Ross PJ, Faheem M, Zaslavsky K, Wang Z, Alexandrova R, Pellecchia G, Wei W, et al. 2018. Complete disruption of autism-susceptibility genes by gene editing predominantly reduces functional connectivity of isogenic human neurons. Stem Cell Rep 11: 1211–1225. 10.1016/j.stemcr.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieuleveult Md, Yen K, Hmitou I, Depaux A, Boussouar F, Dargham DB, Jounier S, Humbertclaude H, Ribierre F, Baulard C, et al. 2016. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature 530: 113–116. 10.1038/nature16505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingar D, Kalkat M, Chan P-K, Srikumar T, Bailey SD, Tu WB, Coyaud E, Ponzielli R, Kolyar M, Jurisica I, et al. 2015. BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J Proteomics 118: 95–111. 10.1016/j.jprot.2014.09.029 [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi T, Ogata T, Yamauchi J, Sawada Y, Tanaka S, Nagao M. 2017. Chd7 collaborates with Sox2 to regulate activation of oligodendrocyte precursor cells after spinal cord injury. J Neurosci 37: 10290–10309. 10.1523/JNEUROSCI.1109-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drivas TG, Li D, Nair D, Alaimo JT, Alders M, Altmüller J, Barakat TS, Bebin EM, Bertsch NL, Blackburn PR, et al. 2020. A second cohort of CHD3 patients expands the molecular mechanisms known to cause Snijders Blok-Campeau syndrome. Eur J Hum Genet 28: 1422–1431. 10.1038/s41431-020-0654-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durak O, Gao F, Kaeser-Woo YJ, Rueda R, Martorell AJ, Nott A, Liu CY, Watson LA, Tsai L-H. 2016. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat Neurosci 19: 1477–1488. 10.1038/nn.4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Grice SL, McKay RDG, Buetow KH, et al. 2008. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2: 437–447. 10.1016/j.stem.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan CM, Nyman U, Skotte J, Streubel G, Turner S, O'Connell DJ, Rraklli V, Dolan MJ, Chadderton N, Hansen K, et al. 2013. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev Cell 26: 223–236. 10.1016/j.devcel.2013.07.008 [DOI] [PubMed] [Google Scholar]
- Ellingford RA, Panasiuk MJ, Meritens Ed, Shaunak R, Naybour L, Browne L, Basson MA, Andreae LC. 2021. Cell-type-specific synaptic imbalance and disrupted homeostatic plasticity in cortical circuits of ASD-associated Chd8 haploinsufficient mice. Mol Psychiatr 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen E, Akinci U, Bryne JC, Hou J, Gontan C, Moen M, Szumska D, Kockx C, Ijcken Wv, Dekkers DHW, et al. 2011. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet 43: 607–611. 10.1038/ng.825 [DOI] [PubMed] [Google Scholar]
- Fan X, Masamsetti VP, Sun JQ, Engholm-Keller K, Osteil P, Studdert J, Graham ME, Fossat N, Tam PP. 2021. TWIST1 and chromatin regulatory proteins interact to guide neural crest cell differentiation. Elife 10: e62873. 10.7554/eLife.62873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas AM, Nagarajan S, Cosulich S, Carroll JS. 2021. Genome-wide estrogen receptor activity in breast cancer. Endocrinology 162: bqaa224-. 10.1210/endocr/bqaa224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnung L, Vos SM, Wigge C, Cramer P. 2017. Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature 550: 539–542. 10.1038/nature24046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnung L, Ochmann M, Cramer P. 2020. Nucleosome-CHD4 chromatin remodeler structure maps human disease mutations. Elife 9: e56178. 10.7554/eLife.56178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasulo B, Deuring R, Murawska M, Gause M, Dorighi KM, Schaaf CA, Dorsett D, Brehm A, Tamkun JW. 2012. The Drosophila MI-2 chromatin-remodeling factor regulates higher-order chromatin structure and cohesin dynamics in vivo. Plos Genet 8: e1002878. 10.1371/journal.pgen.1002878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi M, Paul TA, Brodeur GM, Shokrani B, Brim H, Ashktorab H. 2014. Epigenetic silencing of CHD5, a novel tumor-suppressor gene, occurs in early colorectal cancer stages. Cancer 120: 172–180. 10.1002/cncr.28316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Khan MA, Bellvis P, Zhu Z, Bernhardt O, Herold-Mende C, Liu H-K. 2013. The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell 13: 62–72. 10.1016/j.stem.2013.05.002 [DOI] [PubMed] [Google Scholar]
- Feng W, Kawauchi D, Körkel-Qu H, Deng H, Serger E, Sieber L, Lieberman JA, Jimeno-González S, Lambo S, Hanna BS, et al. 2017. Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme. Nat Commun 8: 14758. 10.1038/ncomms14758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JF, Mi L-Z, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. 2005. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438: 1181–1185. 10.1038/nature04290 [DOI] [PubMed] [Google Scholar]
- Flanagan JF, Blus BJ, Kim D, Clines KL, Rastinejad F, Khorasanizadeh S. 2007. Molecular implications of evolutionary differences in CHD double chromodomains. J Mol Biol 369: 334–342. 10.1016/j.jmb.2007.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Martin DMA, Barton GJ, Owen-Hughes T. 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34: 2887–2905. 10.1093/nar/gkl295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Gaskell E, Bernstein BE. 2017. Epigenetic plasticity and the hallmarks of cancer. Science 357: eaal2380. 10.1126/science.aal2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Qin L, He T, Qin J, Hong J, Wong J, Liao L, Xu J. 2011. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res 21: 275–289. 10.1038/cr.2010.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fueyo R, Iacobucci S, Pappa S, Estarás C, Lois S, Vicioso-Mantis M, Navarro C, Cruz-Molina S, Reyes JC, Rada-Iglesias A, et al. 2018. Lineage specific transcription factors and epigenetic regulators mediate TGFβ-dependent enhancer activation. Nucleic Acids Res 414: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA. 2003. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell 113: 207–219. 10.1016/S0092-8674(03)00234-4 [DOI] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA. 2004. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell 119: 75–86. 10.1016/j.cell.2004.09.014 [DOI] [PubMed] [Google Scholar]
- Gagliardi A, Mullin NP, Tan ZY, Colby D, Kousa AI, Halbritter F, Weiss JT, Felker A, Bezstarosti K, Favaro R, et al. 2013. A direct physical interaction between Nanog and Sox2 regulates embryonic stem cell self-renewal. EMBO J 32: 2231–2247. 10.1038/emboj.2013.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Lukin K, Ramírez J, Fields S, Lopez D, Hagman J. 2009. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci 106: 11258–11263. 10.1073/pnas.0809485106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. 2009. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460: 863–868 . 10.1038/nature08212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehre M, Bunina D, Sidoli S, Lübke MJ, Diaz N, Trovato M, Garcia BA, Zaugg JB, Noh K-M. 2020. Lysine 4 of histone H3.3 is required for embryonic stem cell differentiation, histone enrichment at regulatory regions and transcription accuracy. Nat Genet 52: 273–282. 10.1038/s41588-020-0586-5 [DOI] [PubMed] [Google Scholar]
- Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK. 2019. Organization of chromatin by intrinsic and regulated phase separation. Cell 179: 470–484.e21. 10.1016/j.cell.2019.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompers AL, Su-Feher L, Ellegood J, Copping NA, Riyadh MA, Stradleigh TW, Pride MC, Schaffler MD, Wade AA, Catta-Preta R, et al. 2017. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat Neurosci 20: 1062–1073. 10.1038/nn.4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Kurka T, Jeggo PA. 2011. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 18: 831–839. 10.1038/nsmb.2077 [DOI] [PubMed] [Google Scholar]
- Grande BM, Gerhard DS, Jiang A, Griner NB, Abramson JS, Alexander TB, Allen H, Ayers LW, Bethony JM, Bhatia K, et al. 2019. Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood 133: 1313–1324. 10.1182/blood-2018-09-871418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. 2012. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487: 239–243. 10.1038/nature11125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Harada A, Okada S, Konno D, Odawara J, Yoshimi T, Yoshimura S, Kumamaru H, Saiwai H, Tsubota T, Kurumizaka H, et al. 2012. Chd2 interacts with H3.3 to determine myogenic cell fate. Embo J 31: 2994–3007. 10.1038/emboj.2012.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar A, Meshorer E. 2015. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep 16: 1609–1619. 10.15252/embr.201541011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauk G, Bowman GD. 2011. Structural insights into regulation and action of SWI2/SNF2 ATPases. Curr Opin Struc Biol 21: 719–727. 10.1016/j.sbi.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauk G, McKnight JN, Nodelman IM, Bowman GD. 2010. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell 39: 711–723. 10.1016/j.molcel.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, Marie C, Zhao C, Kim B, Wang J, Deng Y, Clavairoly A, Frah M, Wang H, He X, et al. 2016. Chd7 cooperates with Sox10 and regulates the onset of CNS myelination and remyelination. Nat Neurosci 19: 678–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson D, Kelley DR, Tenen D, Bernstein B, Rinn JL. 2016. Widespread RNA binding by chromatin-associated proteins. Genome Biol 17: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heshmati Y, Turköz G, Harisankar A, Linnarsson S, Dimitriou M, Sinha I, Lehmann S, Qian H, Walfridsson J. 2016. Identification of CHD4 as a potential therapeutic target of acute myeloid leukemia. Blood 128: 1648. 10.1182/blood.V128.22.1648.1648 [DOI] [Google Scholar]
- Heshmati Y, Türköz G, Harisankar A, Kharazi S, Boström J, Dolatabadi EK, Krstic A, Chang D, Månsson R, Altun M, et al. 2018. The chromatin-remodeling factor CHD4 is required for maintenance of childhood acute myeloid leukemia. Haematologica 103: 1169–1181. 10.3324/haematol.2017.183970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota A, Nakajima-Koyama M, Ashida Y, Nishida E. 2019. The nucleosome remodeling and deacetylase complex protein CHD4 regulates neural differentiation of mouse embryonic stem cells by down-regulating p53. J Biol Chem 294: 195–209. 10.1074/jbc.RA118.004086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. 2017. A phase separation model for transcriptional control. Cell 169: 13–23. 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. 2009. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci 106: 5181–5186. 10.1073/pnas.0812889106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. 2011. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol 13: 903–913. 10.1038/ncb2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister H, Fuchs A, Erdel F, Pinz S, Gröbner-Ferreira R, Bruckmann A, Deutzmann R, Schwartz U, Maldonado R, Huber C, et al. 2017. CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Nucleic Acids Res 45: 10534–10554. 10.1093/nar/gkx711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. 2015. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350: 1262–1266. 10.1126/science.aac9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J, Huang H-T, Lee C-T, Yu S, Zon LI, Speck NA. 2013. Chromatin remodeling enzyme CHD7 negatively regulate hematopoietic stem cell function. Blood 122: 2413–2413. 10.1182/blood.V122.21.2413.2413 [DOI] [Google Scholar]
- Hsu J, Huang H-T, Lee C-T, Choudhuri A, Wilson NK, Abraham BJ, Moignard V, Kucinski I, Yu S, Hyde RK, et al. 2020. CHD7 and Runx1 interaction provides a braking mechanism for hematopoietic differentiation. Proc Natl Acad Sci 117: 23626–23635. 10.1073/pnas.2003228117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Wade PA. 2012. NuRD and pluripotency: a complex balancing act. Cell Stem Cell 10: 497–503. 10.1016/j.stem.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Gulzar ZG, Salari K, Lapointe J, Brooks JD, Pollack JR. 2012. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene 31: 4164–4170. 10.1038/onc.2011.590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H, Kohnken R, Svaren J. 2012. The nucleosome remodeling and deacetylase chromatin remodeling (NuRD) complex is required for peripheral nerve myelination. J Neurosci 32: 1517–1527. 10.1523/JNEUROSCI.2895-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd EA, Poucher HK, Cheng K, Raphael Y, Martin DM. 2010. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development 137: 3139–3150. 10.1242/dev.047894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley S, Mohan C, Suetterlin P, Ellingford R, Riegman KLH, Ellegood J, Caruso A, Michetti C, Brock O, Evans R, et al. 2021. Distinct, dosage-sensitive requirements for the autism-associated factor CHD8 during cortical development. Mol Autism 12: 16. 10.1186/s13229-020-00409-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. 2014. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515: 216–221. 10.1038/nature13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Oshimura M, Nakao M. 2006. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell 23: 733–742. 10.1016/j.molcel.2006.08.008 [DOI] [PubMed] [Google Scholar]
- Jamaspishvili T, Berman DM, Ross AE, Scher HI, Marzo AMD, Squire JA, Lotan TL. 2018. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol 15: 222–234. 10.1038/nrurol.2018.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeselsohn R, Buchwalter G, Angelis CD, Brown M, Schiff R. 2015. ESR1 mutations—a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol 12: 573–583. 10.1038/nrclinonc.2015.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Zhou Y, Xian L, Chen W, Wu H, Gao X. 2012. The mutation in Chd7 causes misexpression of Bmp4 and developmental defects in telencephalic midline. Am J Pathol 181: 626–641. 10.1016/j.ajpath.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Jones KM, Sarić N, Russell JP, Andoniadou CL, Scambler PJ, Basson MA. 2015. CHD7 maintains neural stem cell quiescence and prevents premature stem cell depletion in the adult hippocampus. Stem Cells 33: 196–210. 10.1002/stem.1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Park H, Choi Y, Kang H, Lee E, Kweon H, Roh JD, Ellegood J, Choi W, Kang J, et al. 2018. Sexually dimorphic behavior, neuronal activity, and gene expression in Chd8-mutant mice. Nat Neurosci 5: 1. [DOI] [PubMed] [Google Scholar]
- Kaaij LJT, Mohn F, Weide Rvd, Wit Ed, Bühler M. 2019. The ChAHP complex counteracts chromatin looping at CTCF sites that emerged from SINE expansions in mouse. Cell 178: 1437–1451.e14. 10.1016/j.cell.2019.08.007 [DOI] [PubMed] [Google Scholar]
- Kadoch C, Crabtree GR. 2015. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci Adv 1: e1500447. 10.1126/sciadv.1500447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. 2006. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol 8: 285–292. 10.1038/ncb1372 [DOI] [PubMed] [Google Scholar]
- Kari V, Mansour WY, Raul SK, Baumgart SJ, Mund A, Grade M, Sirma H, Simon R, Will H, Dobbelstein M, et al. 2016. Loss of CHD1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness. EMBO Rep 17: 1609–1623. 10.15252/embr.201642352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi M, Morgan BA, Georgopoulos K. 2007. The chromatin remodeler Mi-2β is required for establishment of the basal epidermis and normal differentiation of its progeny. Development 134: 1571–1582. 10.1242/dev.001750 [DOI] [PubMed] [Google Scholar]
- Katayama Y, Nishiyama M, Shoji H, Ohkawa Y, Kawamura A, Sato T, Suyama M, Takumi T, Miyakawa T, Nakayama KI. 2016. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature 537: 675–679. 10.1038/nature19357 [DOI] [PubMed] [Google Scholar]
- Kawamura A, Katayama Y, Kakegawa W, Ino D, Nishiyama M, Yuzaki M, Nakayama KI. 2021. The autism-associated protein CHD8 is required for cerebellar development and motor function. Cell Rep 35: 108932. 10.1016/j.celrep.2021.108932 [DOI] [PubMed] [Google Scholar]
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. 2012. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 335: 348–353. 10.1126/science.1212728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10: 345–355. 10.1016/S1074-7613(00)80034-5 [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 133: 1290. 10.1016/j.cell.2008.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-A, Hwang B, Park SN, Huh S, Im K, Choi S, Chung HY, Huh J, Seo E-J, Lee J-H, et al. 2016. Genomic profile of chronic lymphocytic leukemia in Korea identified by targeted sequencing. PLoS One 11: e0167641. 10.1371/journal.pone.0167641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Khoshkhoo S, Frankowski JC, Zhu B, Abbasi S, Lee S, Wu YE, Hunt RF. 2018. Chd2 Is necessary for neural circuit development and long-term memory. Neuron 100: 1180–1193.e6. 10.1016/j.neuron.2018.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita Y, Nishiyama M, Nakayama KI. 2012. Identification of CHD7S as a novel splicing variant of CHD7 with functions similar and antagonistic to those of the full-length CHD7L. Genes Cells 17: 536–547. 10.1111/j.1365-2443.2012.01606.x [DOI] [PubMed] [Google Scholar]
- Kita Y, Katayama Y, Shiraishi T, Oka T, Sato T, Suyama M, Ohkawa Y, Miyata K, Oike Y, Shirane M, et al. 2018. The autism-related protein CHD8 cooperates with C/EBPβ to regulate adipogenesis. Cell Rep 23: 1988–2000. 10.1016/j.celrep.2018.04.050 [DOI] [PubMed] [Google Scholar]
- Klement K, Luijsterburg MS, Pinder JB, Cena CS, Nero VD, Wintersinger CM, Dellaire G, Attikum Hv, Goodarzi AA. 2014. Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J Cell Biol 207: 717–733. 10.1083/jcb.201405077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh FM, Lizama CO, Wong P, Hawkins JS, Zovein AC, Ramalho-Santos M. 2015. Emergence of hematopoietic stem and progenitor cells involves a Chd1-dependent increase in total nascent transcription. Proc Natl Acad Sci 112: E1734–E1743. 10.1073/pnas.1424850112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolla V, Zhuang T, Higashi M, Naraparaju K, Brodeur GM. 2014. Role of CHD5 in human cancers: 10 years later. Cancer Res 74: 652–658. 10.1158/0008-5472.CAN-13-3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, et al. 2007. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science 317: 1087–1090. 10.1126/science.1145339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovač K, Sauer A, Mačinković I, Awe S, Finkernagel F, Hoffmeister H, Fuchs A, Müller R, Rathke C, Längst G, et al. 2018. Tumour-associated missense mutations in the dMi-2 ATPase alters nucleosome remodelling properties in a mutation-specific manner. Nat Commun 9: 2112. 10.1038/s41467-018-04503-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22: 6979–6992. 10.1128/MCB.22.20.6979-6992.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, O'Roak BJ, Shendure J, Eichler EE. 2014. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci 37: 95–105. 10.1016/j.tins.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Nagarajan P, Wall J, Donovan DJ, Donell RL, Ligon AH, Venkatachalam S, Quade BJ. 2008. Disruption of chromodomain helicase DNA binding protein 2 (CHD2) causes scoliosis. Am J Med Genet A 146A: 1117–1127. 10.1002/ajmg.a.32178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel GR, Tracy JA, Jalufka FL, Lekven AC. 2018. CHD8short, a naturally-occurring truncated form of a chromatin remodeler lacking the helicase domain, is a potent transcriptional coregulator. Gene 641: 303–309. 10.1016/j.gene.2017.10.058 [DOI] [PubMed] [Google Scholar]
- Kunkel GR, Lisciandro HG, Winter HL. 2020. The human chd8 gene is transcribed from two distant upstream promoters. Biochem Bioph Res Co 532: 190–194. 10.1016/j.bbrc.2020.08.051 [DOI] [PubMed] [Google Scholar]
- Kweon H, Jung WB, Im GH, Ryoo J, Lee J-H, Do H, Choi Y, Song Y-H, Jung H, Park H, et al. 2021. Excitatory neuronal CHD8 in the regulation of neocortical development and sensory-motor behaviors. Cell Rep 34: 108780. 10.1016/j.celrep.2021.108780 [DOI] [PubMed] [Google Scholar]
- Lai WKM, Pugh BF. 2017. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat Rev Mol Cell Bio 18: 548–562. 10.1038/nrm.2017.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AY, Wade PA. 2011. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer 11: 588–596. 10.1038/nrc3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli MA, Avey D, Dougherty JD, Milbrandt J, Mitra RD. 2020. High-throughput single-cell functional elucidation of neurodevelopmental disease–associated genes reveals convergent mechanisms altering neuronal differentiation. Genome Res 30: 1317–1331. 10.1101/gr.262295.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E, Matta J, Tessarollo L, Zhang YE, Ko MSH, et al. 2008. Essential role of chromatin remodeling protein bptf in early mouse embryos and embryonic stem cells. Plos Genet 4: e1000241. 10.1371/journal.pgen.1000241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längst G, Manelyte L. 2015. Chromatin remodelers: from function to dysfunction. Genes (Basel) 6: 299–324. 10.3390/genes6020299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorsa VA, Formicola D, Pignataro P, Cimmino F, Calabrese FM, Mora J, Esposito MR, Pantile M, Zanon C, Mariano MD, et al. 2016. Exome and deep sequencing of clinically aggressive neuroblastoma reveal somatic mutations that affect key pathways involved in cancer progression. Oncotarget 7: 21840–21852 . 10.18632/oncotarget.8187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop MJ, Chakrabarti L, Eng J, Rhodes CH, Lutz T, Nieto A, Liggitt HD, Warner S, Fields J, Stöger R, et al. 2010. Deletion of the Chd6 exon 12 affects motor coordination. Mamm Genome 21: 130–142. 10.1007/s00335-010-9248-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Stallcup MR. 2017. Glucocorticoid receptor binding to chromatin is selectively controlled by coregulator Hic-5 and chromatin remodeling enzymes. J Biol Chem 292: 9320–9334. jbc.M117.782607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Park D, Iyer VR. 2017. The ATP-dependent chromatin remodeler Chd1 is recruited by transcription elongation factors and maintains H3K4me3/H3K36me3 domains at actively transcribed and spliced genes. Nucleic Acids Res 45: 8646. 10.1093/nar/gkx636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. 2007. The role of chromatin during transcription. Cell 128: 707–719. 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- Li W, Wu J, Kim S-Y, Zhao M, Hearn SA, Zhang MQ, Meistrich ML, Mills AA. 2014. Chd5 orchestrates chromatin remodelling during sperm development. Nat Commun 5: 3812. 10.1038/ncomms4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Xu C, Lee HJ, Ren S, Zi X, Zhang Z, Wang H, Yu Y, Yang C, Gao X, et al. 2020. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature 580: 93–99. 10.1038/s41586-020-2135-x [DOI] [PubMed] [Google Scholar]
- Lin JJ, Lehmann LW, Bonora G, Sridharan R, Vashisht AA, Tran N, Plath K, Wohlschlegel JA, Carey M. 2011. Mediator coordinates PIC assembly with recruitment of CHD1. Gene Dev 25: 2198–2209. 10.1101/gad.17554711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312: 269–272. 10.1126/science.1123191 [DOI] [PubMed] [Google Scholar]
- Liu W, Lindberg J, Sui G, Luo J, Egevad L, Li T, Xie C, Wan M, Kim S-T, Wang Z, et al. 2012. Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene 31: 3939–3948. 10.1038/onc.2011.554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JC, Ferreira CG, Yusufzai T. 2014a. Human CHD2 Is a chromatin assembly ATPase regulated by its chromo- and DNA-binding domains. J Biol Chem 290: 25–34. 10.1074/jbc.M114.609156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Harmelink C, Peng Y, Chen Y, Wang Q, Jiao K. 2014b. CHD7 interacts with BMP R-SMADs to epigenetically regulate cardiogenesis in mice. Hum Mol Genet 23: 2145–2156. 10.1093/hmg/ddt610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li M, Xia X, Li X, Chen Z. 2017. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 544: 440–445. 10.1038/nature22036 [DOI] [PubMed] [Google Scholar]
- Liu C, Li Q, Xiao Q, Gong P, Kang N. 2020. CHD7 regulates osteogenic differentiation of human dental follicle cells via PTH1R signaling. Stem Cells Int 2020: 8882857. 10.1155/2020/8882857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwigsen J, Pfennig S, Singh AK, Schindler C, Harrer N, Forné I, Zacharias M, Mueller-Planitz F. 2017. Concerted regulation of ISWI by an autoinhibitory domain and the H4 N-terminal tail. Elife 6: 1240. 10.7554/eLife.21477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, Krijger Id, Wiegant WW, Shah RG, Smeenk G, Groot Ad, Pines A, Vertegaal ACO, Jacobs JJL, Shah GM, et al. 2016. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol Cell 61: 547–562. 10.1016/j.molcel.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund C, Brodtkorb E, Øye A-M, Røsby O, Selmer KK. 2014. CHD2 mutations in Lennox-Gastaut syndrome. Epilepsy Behav 33: 18–21. 10.1016/j.yebeh.2014.02.005 [DOI] [PubMed] [Google Scholar]
- Luo J, Su F, Chen D, Shiloh A, Gu W. 2000. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408: 377–381. 10.1038/35042612 [DOI] [PubMed] [Google Scholar]
- Luo M, Ling T, Xie W, Sun H, Zhou Y, Zhu Q, Shen M, Zong L, Lyu G, Zhao Y, et al. 2013. NuRD blocks reprogramming of mouse somatic cells into pluripotent stem cells. Stem Cells 31: 1278–1286. 10.1002/stem.1374 [DOI] [PubMed] [Google Scholar]
- Lusser A, Urwin DL, Kadonaga JT. 2005. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 12: 160–166. 10.1038/nsmb884 [DOI] [PubMed] [Google Scholar]
- Lutz T, Stöger R, Nieto A. 2006. CHD6 is a DNA-dependent ATPase and localizes at nuclear sites of mRNA synthesis. Febs Lett 580: 5851–5857. 10.1016/j.febslet.2006.09.049 [DOI] [PubMed] [Google Scholar]
- Machado RAC, Schneider H, Pereira CD, Lichtenstein F, Andrade F, Fujita A, Trombetta-Lima M, Weller M, Bowman-Colin C, Sogayar MC. 2019. CHD7 promotes glioblastoma cell motility and invasiveness through transcriptional modulation of an invasion signature. Sci Rep 9: 3952. 10.1038/s41598-019-39564-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammen AL, Casciola-Rosen LA, Hall JC, Christopher-Stine L, Corse AM, Rosen A. 2009. Expression of the dermatomyositis autoantigen Mi-2 in regenerating muscle. Arthritis Rheum 60: 3784–3793. 10.1002/art.24977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BJ, Yusufzai T. 2017. The ATP-dependent chromatin remodeling enzymes CHD6, CHD7, and CHD8 exhibit distinct nucleosome binding and remodeling activities. J Biol Chem 292: 11927–11936. 10.1074/jbc.M117.779470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield RE, Musselman CA, Kwan AH, Oliver SS, Garske AL, Davrazou F, Denu JM, Kutateladze TG, Mackay JP. 2011. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J Biol Chem 286: 11779–11791. 10.1074/jbc.M110.208207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazzi I, Ho JSY, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, et al. 2012. Suppression of the antiviral response by an influenza histone mimic. Nature 483: 428–433. 10.1038/nature10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella CGA, Imbalzano AN. 2007. The Chd family of chromatin remodelers. Mutat Res Fundam Mol Mech Mutagen 618: 30–40. 10.1016/j.mrfmmm.2006.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella CGA, Ohkawa Y, Coles AH, Garlick DS, Jones SN, Imbalzano AN. 2006. Mutation of the SNF2 family member Chd2 affects mouse development and survival. J Cell Physiol 209: 162–171. 10.1002/jcp.20718 [DOI] [PubMed] [Google Scholar]
- Marfella CGA, Henninger N, LeBlanc SE, Krishnan N, Garlick DS, Holzman LB, Imbalzano AN. 2008. A mutation in the mouse Chd2 chromatin remodeling enzyme results in a complex renal phenotype. Kidney Blood Press Res 31: 421–432. 10.1159/000190788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie C, Clavairoly A, Frah M, Hmidan H, Yan J, Zhao C, Steenwinckel JV, Daveau R, Zalc B, Hassan B, et al. 2018. Oligodendrocyte precursor survival and differentiation requires chromatin remodeling by Chd7 and Chd8. Proc Natl Acad Sci 95: 201802620–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marom R, Shur I, Hager GL, Benayahu D. 2006. Expression and regulation of CReMM, a chromodomain helicase-DNA-binding (CHD), in marrow stroma derived osteoprogenitors. J Cell Physiol 207: 628–635. 10.1002/jcp.20611 [DOI] [PubMed] [Google Scholar]
- Mattick JS. 2007. A new paradigm for developmental biology. J Exp Biol 210: 1526–1547. 10.1242/jeb.005017 [DOI] [PubMed] [Google Scholar]
- McKenzie LD, LeClair JW, Miller KN, Strong AD, Chan HL, Oates EL, Ligon KL, Brennan CW, Chheda MG. 2019. CHD4 regulates the DNA damage response and RAD51 expression in glioblastoma. Sci Rep 9: 4444. 10.1038/s41598-019-40327-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight JN, Jenkins KR, Nodelman IM, Escobar T, Bowman GD. 2011. Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol Cell Biol 31: 4746–4759. 10.1128/MCB.05735-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meganathan K, Lewis EMA, Gontarz P, Liu S, Stanley EG, Elefanty AG, Huettner JE, Zhang B, Kroll KL. 2017. Regulatory networks specifying cortical interneurons from human embryonic stem cells reveal roles for CHD2 in interneuron development. Proc Natl Acad Sci 114: E11180–E11189. 10.1073/pnas.1712365115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon T, Yates JA, Bochar DA. 2010. Regulation of androgen-responsive transcription by the chromatin remodeling factor CHD8. Mol Endocrinol 24: 1165–1174. 10.1210/me.2009-0421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Willmann D, McMillan J, Forne I, Metzger P, Gerhardt S, Petroll K, Maessenhausen Av, Urban S, Schott A-K, et al. 2016. Assembly of methylated KDM1A and CHD1 drives androgen receptor-dependent transcription and translocation. Nat Struct Mol Biol 23: 132–139. 10.1038/nsmb.3153 [DOI] [PubMed] [Google Scholar]
- Micucci JA, Layman WS, Hurd EA, Sperry ED, Frank SF, Durham MA, Swiderski DL, Skidmore JM, Scacheri PC, Raphael Y, et al. 2014. CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum Mol Genet 23: 434–448. 10.1093/hmg/ddt435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micucci J, Sperry ED, Martin DM. 2015. Chromodomain helicase DNA binding proteins in stem cells and human developmental diseases. Stem Cells Dev 24: 917–926. 10.1089/scd.2014.0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E, Berns A, Romeyn L. 2002. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet 32: 153–159. 10.1038/ng950 [DOI] [PubMed] [Google Scholar]
- Mills AA. 2017. The chromodomain helicase DNA-binding chromatin remodelers: family traits that protect from and promote cancer. Csh Perspect Med 7: a026450. 10.1101/cshperspect.a026450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B, Helder S, Silva APG, Mackay JP, Ryan DP. 2016. The chromatin remodelling protein CHD1 contains a previously unrecognised C-terminal helical domain. J Mol Biol 428: 4298–4314. 10.1016/j.jmb.2016.08.028 [DOI] [PubMed] [Google Scholar]
- Moore S, Berger ND, Luijsterburg MS, Piett CG, Stanley FKT, Schräder CU, Fang S, Chan JA, Schriemer DC, Nagel ZD, et al. 2019. The CHD6 chromatin remodeler is an oxidative DNA damage response factor. Nat Commun 10: 241. 10.1038/s41467-018-08111-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L, Brenner C, Fazi F, Villa R, Gutierrez A, Buschbeck M, Nervi C, Minucci S, Fuks F, Croce LD. 2008. MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Mol Cell Biol 28: 5912–5923. 10.1128/MCB.00467-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra R, Lee BM, Shaw H, Tuma R, Mancini EJ. 2012. Concerted action of the PHD, chromo and motor domains regulates the human chromatin remodelling ATPase CHD4. Febs Lett 586: 2513–2521. 10.1016/j.febslet.2012.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Planitz F, Klinker H, Becker PB. 2013. Nucleosome sliding mechanisms: new twists in a looped history. Nat Struct Mol Biol 20: 1026–1032. 10.1038/nsmb.2648 [DOI] [PubMed] [Google Scholar]
- Murawska M, Brehm A. 2011. CHD chromatin remodelers and the transcription cycle. Biochem Soc Symp 2: 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawska M, Hassler M, Renkawitz-Pohl R, Ladurner A, Brehm A. 2011. Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. Plos Genet 7: e1002206. 10.1371/journal.pgen.1002206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Ramírez J, Sims JK, Mansfield RE, Oliver SS, Denu JM, Mackay JP, Wade PA, Hagman J, Kutateladze TG. 2012. Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression. Proc Natl Acad Sci 109: 787–792. 10.1073/pnas.1113655109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan P, Onami TM, Rajagopalan S, Kania S, Donnell R, Venkatachalam S. 2009. Role of chromodomain helicase DNA-binding protein 2 in DNA damage response signaling and tumorigenesis. Oncogene 28: 1053–1062. 10.1038/onc.2008.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Arco PG, Williams CJ, Georgopoulos K. 2007. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2β determine silencer activity and Cd4 gene expression. Immunity 27: 723–734. 10.1016/j.immuni.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. 2013. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154: 490–503. 10.1016/j.cell.2013.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network UD, Parenti I, Lehalle D, Nava C, Torti E, Leitão E, Person R, Mizuguchi T, Matsumoto N, Kato M, et al. 2021. Missense and truncating variants in CHD5 in a dominant neurodevelopmental disorder with intellectual disability, behavioral disturbances, and epilepsy. Hum Genet 140: 1109–1120. 10.1007/s00439-021-02283-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell F, Kong Y, Wilmott JS, Johansson PA, Ferguson PM, Cui C, Li Z, Kazakoff SH, Burke H, Dodds TJ, et al. 2019. Whole-genome landscape of mucosal melanoma reveals diverse drivers and therapeutic targets. Nat Commun 10: 3163. 10.1038/s41467-019-11107-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AH, Pask AJ. 2020. CHD9 upregulates RUNX2 and has a potential role in skeletal evolution. Bmc Mol Cell Biology 21: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P, Bar-Sela G, Sun L, Bisht KS, Cui H, Kohn E, Feinberg AP, Gius D. 2008. BAT3 and SET1A form a complex with CTCFL/BORIS to modulate H3K4 histone dimethylation and gene expression. Mol Cell Biol 28: 6720–6729. 10.1128/MCB.00568-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nio K, Yamashita T, Okada H, Kondo M, Hayashi T, Hara Y, Nomura Y, Zeng SS, Yoshida M, Hayashi T, et al. 2015. Defeating EpCAM(+) liver cancer stem cells by targeting chromatin remodeling enzyme CHD4 in human hepatocellular carcinoma. J Hepatol 63: 1164–72 10.1016/j.jhep.2015.06.009 [DOI] [PubMed] [Google Scholar]
- Nishiyama M, Nakayama K, Tsunematsu R, Tsukiyama T, Kikuchi A, Nakayama KI. 2004. Early embryonic death in mice lacking the β-catenin-binding protein Duplin. Mol Cell Biol 24: 8386–8394. 10.1128/MCB.24.19.8386-8394.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Oshikawa K, Tsukada Y, Nakagawa T, Iemura S, Natsume T, Fan Y, Kikuchi A, Skoultchi AI, Nakayama KI. 2009. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat Cell Biol 11: 172–182. 10.1038/ncb1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Skoultchi AI, Nakayama KI. 2012. Histone H1 recruitment by CHD8 is essential for suppression of the Wnt-β-catenin signaling pathway. Mol Cell Biol 32: 501–512. 10.1128/MCB.06409-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nita A, Muto Y, Katayama Y, Matsumoto A, Nishiyama M, Nakayama KI. 2021. The autism-related protein CHD8 contributes to the stemness and differentiation of mouse hematopoietic stem cells. Cell Rep 34: 108688. 10.1016/j.celrep.2021.108688 [DOI] [PubMed] [Google Scholar]
- Nitarska J, Smith JG, Sherlock WT, Hillege MMG, Nott A, Barshop WD, Vashisht AA, Wohlschlegel JA, Mitter R, Riccio A. 2016. A functional switch of NuRD chromatin remodeling complex subunits regulates mouse cortical development. Cell Rep 17: 1683–1698. 10.1016/j.celrep.2016.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodelman IM, Bowman GD. 2013. Nucleosome sliding by Chd1 does not require rigid coupling between DNA-binding and ATPase domains. EMBO Rep 14: 1098–1103. 10.1038/embor.2013.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodelman IM, Horvath KC, Levendosky RF, Winger J, Ren R, Patel A, Li M, Wang MD, Roberts E, Bowman GD. 2016. The Chd1 chromatin remodeler can sense both entry and exit sides of the nucleosome. Nucleic Acids Res 44: 7580–91. 10.1093/nar/gkw406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodelman IM, Bleichert F, Patel A, Ren R, Horvath KC, Berger JM, Bowman GD. 2017. Interdomain communication of the Chd1 chromatin remodeler across the DNA gyres of the nucleosome. Mol Cell 65: 447–459.e6. 10.1016/j.molcel.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodelman IM, Shen Z, Levendosky RF, Bowman GD. 2021. Autoinhibitory elements of the Chd1 remodeler block initiation of twist defects by destabilizing the ATPase motor on the nucleosome. Proc Natl Acad Sci 118: e2014498118. 10.1073/pnas.2014498118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nostrand JLV, Brady CA, Jung H, Fuentes DR, Kozak MM, Johnson TM, Lin C-Y, Lin C-J, Swiderski DL, Vogel H, et al. 2014. Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature 514: 228–232. 10.1038/nature13585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooga M, Funaya S, Hashioka Y, Fujii W, Naito K, Suzuki MG, Aoki F. 2018. Chd9 mediates highly loosened chromatin structure in growing mouse oocytes. Biochem Bioph Res Co 500: 583–588. 10.1016/j.bbrc.2018.04.105 [DOI] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. 2011. Chromatin connections to pluripotency and cellular reprogramming. Cell 145: 835–850. 10.1016/j.cell.2011.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Wang J, Kim J, Chu J, Rao S, Theunissen TW, Shen X, Levasseur DN. 2008. The transcriptional network controlling pluripotency in ES cells. Cold Spring Harb Sym 73: 195–202. 10.1101/sqb.2008.72.001 [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. 2012a. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338: 1619–1622. 10.1126/science.1227764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. 2012b. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485: 246–250. 10.1038/nature10989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Stessman HA, Boyle EA, Witherspoon KT, Martin B, Lee C, Vives L, Baker C, Hiatt JB, Nickerson DA, et al. 2014. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun 5: 5595. 10.1038/ncomms6595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shaughnessy-Kirwan A, Signolet J, Costello I, Gharbi S, Hendrich B. 2015. Constraint of gene expression by the chromatin remodelling protein CHD4 facilitates lineage specification. Development 142: 2586–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paakinaho V, Lempiäinen JK, Sigismondo G, Niskanen EA, Malinen M, Jääskeläinen T, Varjosalo M, Krijgsveld J, Palvimo JJ. 2021. SUMOylation regulates the protein network and chromatin accessibility at glucocorticoid receptor-binding sites. Nucleic Acids Res 49: 1951–1971. 10.1093/nar/gkab032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M-R, Hsieh H-J, Dai H, Hung W-C, Li K, Peng G, Lin S-Y. 2012. Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and Its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment. J Biol Chem 287: 6764–6772. 10.1074/jbc.M111.287037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J. 2010. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 6: 382–395. 10.1016/j.stem.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Kuo A, Schalch T, Vogel H, Joshua-Tor L, McCombie WR, Gozani O, Hammell M, Mills AA. 2013. Chd5 requires PHD-mediated histone 3 binding for tumor suppression. Cell Rep 3: 92–102. 10.1016/j.celrep.2012.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Bulut-Karslioglu A, Ramalho-Santos M. 2017. Hypertranscription in development, stem cells, and regeneration. Dev Cell 40: 9–21. 10.1016/j.devcel.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti P, Lim CY, Nat R, Villunger A, Geley S, Shue YT, Soratroi C, Moser M, Lusser A. 2015. Embryonic stem cell differentiation requires full length Chd1. Sci Rep 5: 8007. 10.1038/srep08007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilarowski GO, Vernon HJ, Applegate CD, Boukas L, Cho MT, Gurnett CA, Benke PJ, Beaver E, Heeley JM, Medne L, et al. 2018. Missense variants in the chromatin remodeler CHD1 are associated with neurodevelopmental disability. J Med Genet 55: 561–566. 10.1136/jmedgenet-2017-104759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466: 368–372. 10.1038/nature09146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisansky MT, Young AE, O'Connor MB, Gottesman II, Bagchi A, Gewirtz JC. 2017. Mice lacking the chromodomain helicase DNA-binding 5 chromatin remodeler display autism-like characteristics. Transl Psychiat 7: e1152. 10.1038/tp.2017.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Zhou Y, Slaymaker IM, Shetty AS, Weisbach NR, Kim J-A, Sharma J, Desai M, Sood S, Kempton HR, et al. 2017. Chd8 mutation leads to autistic-like behaviors and impaired striatal circuits. Cell Rep 19: 335–350. 10.1016/j.celrep.2017.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, Cejas P, Vazquez F, Cook J, Shivdasani RA, et al. 2015. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet 47: 1346–1351. 10.1038/ng.3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J, Yusufzai T. 2014. The tumor suppressor chromodomain helicase DNA-binding protein 5 (CHD5) remodels nucleosomes by unwrapping. J Biol Chem 289: 20717–20726. 10.1074/jbc.M114.568568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez-Trillos A, et al. 2012. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 44: 47–52. 10.1038/ng.1032 [DOI] [PubMed] [Google Scholar]
- Radman-Livaja M, Rando OJ. 2010. Nucleosome positioning: how is it established, and why does it matter? Dev Biol 339: 258–266. 10.1016/j.ydbio.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. 2013. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502: 65–70. 10.1038/nature12587 [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Nepa J, Venkatachalam S. 2012. Chromodomain helicase DNA-binding protein 2 affects the repair of X-ray and UV-induced DNA damage. Environ Mol Mutagen 53: 44–50. 10.1002/em.20674 [DOI] [PubMed] [Google Scholar]
- Ram EVSR, Meshorer E. 2009. Transcriptional competence in pluripotency. Gene Dev 23: 2793–2798. 10.1101/gad.1881609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez J, Dege C, Kutateladze TG, Hagman J. 2012. MBD2 and multiple domains of CHD4 are required for transcriptional repression by Mi-2/NuRD complexes. Mol Cell Biol 32: 5078–5088. 10.1128/MCB.00819-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. 2006. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Gene Dev 20: 282–296. 10.1101/gad.1383206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Donato ND, et al. 2012. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 380: 1674–1682. 10.1016/S0140-6736(12)61480-9 [DOI] [PubMed] [Google Scholar]
- Ravenswaaij-Arts Cv, Martin DM. 2017. New insights and advances in CHARGE syndrome: diagnosis, etiologies, treatments, and research discoveries. Am J Medical Genetics Part C Seminars Medical Genetics 175: 397–406. 10.1002/ajmg.c.31592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O'Shaughnessy A, Mosaku O, Signolet J, Brennecke P, Kalkan T, et al. 2012. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell 10: 583–594. 10.1016/j.stem.2012.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues LU, Rider L, Nieto C, Romero L, Karimpour-Fard A, Loda M, Lucia MS, Wu M, Shi L, Cimic A, et al. 2015. Coordinate loss of MAP3K7 and CHD1 promotes aggressive prostate cancer. Cancer Res 75: 1021–1034. 10.1158/0008-5472.CAN-14-1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez D, Bretones G, Quesada V, Villamor N, Arango JR, López-Guillermo A, Ramsay AJ, Baumann T, Quirós PM, Navarro A, et al. 2015. Mutations in CHD2 cause defective association with active chromatin in chronic lymphocytic leukemia. Blood 126: 195–202. 10.1182/blood-2014-10-604959 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Paredes M, Ceballos-Chávez M, Esteller M, García-Domínguez M, Reyes JC. 2009. The chromatin remodeling factor CHD8 interacts with elongating RNA polymerase II and controls expression of the cyclin E2 gene. Nucleic Acids Res 37: 2449–2460. 10.1093/nar/gkp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom A, Melamed L, Gil N, Goldrich MJ, Kadir R, Golan M, Biton I, Perry RB-T, Ulitsky I. 2019. Regulation of CHD2 expression by the chaserr long noncoding RNA gene is essential for viability. Nat Commun 10: 5092. 10.1038/s41467-019-13075-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. 2013. From neural development to cognition: unexpected roles for chromatin. Nature Reviews Genetics 14: 347–359. 10.1038/nrg3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother MB, Attikum Hv. 2017. DNA repair goes hip-hop: SMARCA and CHD chromatin remodellers join the break dance. Philosophical Transactions Royal Soc B Biological Sci 372: 20160285. 10.1098/rstb.2016.0285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother MB, Pellegrino S, Smith R, Gatti M, Meisenberg C, Wiegant WW, Luijsterburg MS, Imhof R, Downs JA, Vertegaal ACO, et al. 2020. CHD7 and 53BP1 regulate distinct pathways for the re-ligation of DNA double-strand breaks. Nat Commun 11: 5775. 10.1038/s41467-020-19502-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DP, Owen-Hughes T. 2011. Snf2-family proteins: chromatin remodellers for any occasion. Curr Opin Chem Biol 15: 649–656. 10.1016/j.cbpa.2011.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T. 2011. The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. Embo J 30: 2596–2609. 10.1038/emboj.2011.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabantsev A, Levendosky RF, Zhuang X, Bowman GD, Deindl S. 2019. Direct observation of coordinated DNA movements on the nucleosome during chromatin remodelling. Nat Commun 10: 1720. 10.1038/s41467-019-09657-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto I, Kishida S, Fukui A, Kishida M, Yamamoto H, Hino S, Michiue T, Takada S, Asashima M, Kikuchi A. 2000. A novel β-catenin-binding protein inhibits β-catenin-dependent Tcf activation and axis formation. J Biol Chem 275: 32871–32878. 10.1074/jbc.M004089200 [DOI] [PubMed] [Google Scholar]
- Salomon-Kent R, Marom R, John S, Dundr M, Schiltz LR, Gutierrez J, Workman J, Benayahu D, Hager GL. 2015. New face for chromatin-related mesenchymal modulator: n-CHD9 localizes to nucleoli and interacts With ribosomal genes. J Cell Physiol 230: 2270–2280. 10.1002/jcp.24960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho A, Li S, Paul T, Zhang F, Aguilo F, Vashisht A, Balasubramaniyan N, Leleiko NS, Suchy FJ, Wohlschlegel JA, et al. 2015. CHD6 regulates the topological arrangement of the CFTR locus. Hum Mol Genet 24: 2724–2732. 10.1093/hmg/ddv032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. 2012. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485: 237–241. 10.1038/nature10945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Rd, Tosti L, Radzisheuskaya A, Caballero IM, Kaji K, Hendrich B, Silva JCR. 2014. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell 15: 102–110. 10.1016/j.stem.2014.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada G, Ueo H, Matsumura T, Uchi R, Ishibashi M, Mima K, Kurashige J, Takahashi Y, Akiyoshi S, Sudo T, et al. 2013. CHD8 is an independent prognostic indicator that regulates Wnt/β-catenin signaling and the cell cycle in gastric cancer. Oncol Rep 30: 1137–1142. 10.3892/or.2013.2597 [DOI] [PubMed] [Google Scholar]
- Schmidt DR, Schreiber SL. 1999. Molecular association between ATR and Two components of the nucleosome remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry 38: 14711–14717. 10.1021/bi991614n [DOI] [PubMed] [Google Scholar]
- Schnetz MP, Bartels CF, Shastri K, Balasubramanian D, Zentner GE, Balaji R, Zhang X, Song L, Wang Z, Laframboise T, et al. 2009. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res 19: 590–601. 10.1101/gr.086983.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG, Adams DJ, Flicek P, Crawford GE, Laframboise T, et al. 2010. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet 6: e1001023. 10.1371/journal.pgen.1001023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster EF, Stöger R. 2002. CHD5 defines a new subfamily of chromodomain-SWI2/SNF2-like helicases. Mamm Genome 13: 117–119. 10.1007/s00335-001-3042-6 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G. 2009. Chromatin organization marks exon-intron structure. Nat Struct Mol Biology 16: 990–995. 10.1038/nsmb.1659 [DOI] [PubMed] [Google Scholar]
- Semba Y, Harada A, Maehara K, Oki S, Meno C, Ueda J, Yamagata K, Suzuki A, Onimaru M, Nogami J, et al. 2017. Chd2 regulates chromatin for proper gene expression toward differentiation in mouse embryonic stem cells. Nucleic Acids Res 45: 8758–8772 10.1093/nar/gkx475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serandour AA, Mohammed H, Miremadi A, Mulder KW, Carroll JS. 2018. TRPS1 regulates oestrogen receptor binding and histone acetylation at enhancers. Oncogene 37: 5281–5291. 10.1038/s41388-018-0312-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe J-S, Suzuki Y, Pappin DJ, Joshua-Tor L, Vakoc CR. 2015a. NSD3-Short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell 60: 847–859. 10.1016/j.molcel.2015.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T, Ji F, Yuan Z, Jiao J. 2015b. CHD2 is required for embryonic neurogenesis in the developing cerebral cortex. Stem Cells 33: 1794–1806. 10.1002/stem.2001 [DOI] [PubMed] [Google Scholar]
- Shenoy TR, Boysen G, Wang MY, Xu QZ, Guo W, Koh FM, Wang C, Zhang LZ, Wang Y, Gil V, et al. 2017. CHD1 loss sensitizes prostate cancer to DNA damaging therapy by promoting error-prone double-strand break repair. Ann Oncol 28: 1495–1507. 10.1093/annonc/mdx165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingleton JR, Hemann MT. 2015. The chromatin regulator CHD8 Is a context-dependent mediator of cell survival in murine hematopoietic malignancies. PLoS One 10: e0143275. 10.1371/journal.pone.0143275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shooshtarizadeh P, Helness A, Vadnais C, Brouwer N, Beauchemin H, Chen R, Bagci H, Staal FJT, Coté J-F, Möröy T. 2019. Gfi1b regulates the level of Wnt/β-catenin signaling in hematopoietic stem cells and megakaryocytes. Nat Commun 10: 1270. 10.1038/s41467-019-09273-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou Y, Ma Z, Lu T, Sorrentino BP. 2006. Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc Natl Acad Sci 103: 11730–11735. 10.1073/pnas.0603635103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shur I, Benayahu D. 2005. Characterization and functional analysis of CReMM, a novel chromodomain helicase DNA-binding protein. J Mol Biol 352: 646–655. 10.1016/j.jmb.2005.06.049 [DOI] [PubMed] [Google Scholar]
- Shur I, Socher R, Benayahu D. 2006a. In vivo association of CReMM/CHD9 with promoters in osteogenic cells. J Cell Physiol 207: 374–378. 10.1002/jcp.20586 [DOI] [PubMed] [Google Scholar]
- Shur I, Solomon R, Benayahu D. 2006b. Dynamic interactions of chromatin-related mesenchymal modulator, a chromodomain helicase-DNA-binding protein, with promoters in osteoprogenitors. Stem Cells 24: 1288–1293. 10.1634/stemcells.2005-0300 [DOI] [PubMed] [Google Scholar]
- Signolet J, Hendrich B. 2014. The function of chromatin modifiers in lineage commitment and cell fate specification. FEBS J 282: 1692–1702. 10.1111/febs.13132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva APG, Ryan DP, Galanty Y, Low JKK, Vandevenne M, Jackson SP, Mackay JP. 2015. The N-terminal region of CHD4 is essential for activity and contains a HMG-box-like-domain that can bind poly(ADP-ribose). J Biol Chem 291: 924–938. 10.1074/jbc.M115.683227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. Embo J 22: 1846–1856. 10.1093/emboj/cdg179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. 2005. Human but Not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via Its tandem chromodomains. J Biol Chem 280: 41789–41792. 10.1074/jbc.C500395200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, Millhouse S, Chen C-F, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676. 10.1016/j.molcel.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Hernandez AE, Groudine M, Henikoff S. 2014. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. Elife 3: e02042. 10.7554/eLife.02042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Sellou H, Chapuis C, Huet S, Timinszky G. 2018. CHD3 and CHD4 recruitment and chromatin remodeling activity at DNA breaks is promoted by early poly(ADP-ribose)-dependent chromatin relaxation. Nucleic Acids Res 46: 6087–6098. 10.1093/nar/gky334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokpor G, Castro-Hernandez R, Rosenbusch J, Staiger JF, Tuoc T. 2018. ATP-dependent chromatin remodeling during cortical neurogenesis. Front Neurosci 12: 226. 10.3389/fnins.2018.00226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, Xie Y, Verhoeven E, Vermeulen M, Lancini C, Gargiulo G, Hulsman D, Mann M, Knoblich JA, Lohuizen Mv. 2013. The chromodomain helicase Chd4 is required for polycomb-mediated inhibition of astroglial differentiation. Embo J 32: 1598–1612. 10.1038/emboj.2013.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperlazza J, Rahmani M, Beckta J, Aust M, Hawkins E, Wang SZ, Zhu SZ, Podder S, Dumur C, Archer K, et al. 2015. Depletion of the chromatin remodeler CHD4 sensitizes AML blasts to genotoxic agents and reduces tumor formation. Blood 126: 1462–1472. 10.1182/blood-2015-03-631606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan K, Ianni A, Künne C, Strilic B, Günther S, Perdiguero E, Krüger M, Spuler S, Offermanns S, Arco PG, et al. 2020. Attenuated epigenetic suppression of muscle stem cell necroptosis Is required for efficient regeneration of dystrophic muscles. Cell Rep 31: 107652. 10.1016/j.celrep.2020.107652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Armstrong JA, Deuring R, Dahlsveen IK, McNeill H, Tamkun JW. 2005. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA polymerase II. Development 132: 1623–1635. 10.1242/dev.01713 [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Mager GM, Ward RM, Mayer J, Svaren J. 2006. NAB2 represses transcription by interacting with the CHD4 subunit of the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem 281: 15129–15137. 10.1074/jbc.M600775200 [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Dorighi KM, Tamkun JW. 2008. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. Plos Genet 4: e1000217. 10.1371/journal.pgen.1000217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley FKT, Moore S, Goodarzi AA. 2013. CHD chromatin remodelling enzymes and the DNA damage response. Mutat Res 750: 31–44. 10.1016/j.mrfmmm.2013.07.008 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40. 10.1016/j.cell.2010.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. 2006. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem 281: 16279–16288. 10.1074/jbc.M600682200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes DG, Perry RP. 1995. DNA-binding and chromatin localization properties of CHD1. Mol Cell Biol 15: 2745–2753. 10.1128/MCB.15.5.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Study DDD. 2017. Prevalence and architecture of de novo mutations in developmental disorders. Nature 542: 433–438. 10.1038/nature21062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtil-Rodríguez A, Vázquez-Chávez E, Ceballos-Chávez M, Rodríguez-Paredes M, Martín-Subero JI, Esteller M, Reyes JC. 2014. The chromatin remodeller CHD8 is required for E2F-dependent transcription activation of S-phase genes. Nucleic Acids Res 42: 2185–2196. 10.1093/nar/gkt1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetterlin P, Hurley S, Mohan C, Riegman KLH, Pagani M, Caruso A, Ellegood J, Galbusera A, Crespo-Enriquez I, Michetti C, et al. 2018. Altered neocortical gene expression, brain overgrowth and functional over-connectivity in Chd8 haploinsufficient mice. Cereb Cortex 28: 2192–2206. 10.1093/cercor/bhy058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugathan A, Biagioli M, Golzio C, Erdin S, Blumenthal I, Manavalan P, Ragavendran A, Brand H, Lucente D, Miles J, et al. 2014. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci 111: E4468–E4477. 10.1073/pnas.1405266111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suls A, Jaehn JA, Kecskés A, Weber Y, Weckhuysen S, Craiu DC, Siekierska A, Djémié T, Afrikanova T, Gormley P, et al. 2013. De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am J Hum Genet 93: 967–975. 10.1016/j.ajhg.2013.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy R, Hughes AL, Singh V, Wiechens N, Ryan DP, El-Mkami H, Petoukhov M, Svergun DI, Treutlein B, Quack S, et al. 2017. Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes. Elife 6: e22510. 10.7554/eLife.22510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surapureddi S, Yu S, Bu H, Hashimoto T, Yeldandi AV, Kashireddy P, Cherkaoui-Malki M, Qi C, Zhu Y-J, Rao MS, et al. 2002. Identification of a transcriptionally active peroxisome proliferator-activated receptor α-interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. PNAS 99: 11836–11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surapureddi S, Viswakarma N, Yu S, Guo D, Rao MS, Reddy JK. 2006. PRIC320, a transcription coactivator, isolated from peroxisome proliferator-binding protein complex. Biochem Biophys Res Commun 343: 535–543. 10.1016/j.bbrc.2006.02.160 [DOI] [PubMed] [Google Scholar]
- Suzuki S, Nozawa Y, Tsukamoto S, Kaneko T, Manabe I, Imai H, Minami N. 2015. CHD1 acts via the Hmgpi pathway to regulate mouse early embryogenesis. Development 142: 2375–2384. 10.1242/dev.120493 [DOI] [PubMed] [Google Scholar]
- Tan S, Ding K, Li R, Zhang W, Li G, Kong X, Qian P, Lobie PE, Zhu T. 2014. Identification of miR-26 as a key mediator of estrogen stimulated cell proliferation by targeting CHD1, GREB1 and KPNA2. Breast Cancer Res Bcr 16: R40. 10.1186/bcr3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tencer AH, Cox KL, Di L, Bridgers JB, Lyu J, Wang X, Sims JK, Weaver TM, Allen HF, Zhang Y, et al. 2017. Covalent modifications of histone H3K9 promote binding of CHD3. Cell Rep 21: 455–466. 10.1016/j.celrep.2017.09.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RH, Zhang LM, Carvill GL, Archer JS, Heavin SB, Mandelstam SA, Craiu D, Berkovic SF, Gill DS, Mefford HC, et al. 2015. CHD2 myoclonic encephalopathy is frequently associated with self-induced seizures. Neurology 84: 951–958. 10.1212/WNL.0000000000001305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD. 2018. The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome. Nucleic Acids Res 46: 4978–4990. 10.1093/nar/gky206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomofuji Y, Takaba H, Suzuki HI, Benlaribi R, Martinez CDP, Abe Y, Morishita Y, Okamura T, Taguchi A, Kodama T, et al. 2020. Chd4 choreographs self-antigen expression for central immune tolerance. Nat Immunol 21: 892–901. 10.1038/s41590-020-0717-2 [DOI] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. 1998. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395: 917–921. 10.1038/27699 [DOI] [PubMed] [Google Scholar]
- Tu Z, Wang C, Davis AK, Hu M, Zhao C, Xin M, Lu R, Zheng Y. 2021. Chromatin remodeler CHD8 governs hematopoietic stem/progenitor survival by regulating ATM-mediated P53 protein stability. Blood 138: 221–233. 10.1182/blood.2020009997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart AJ, Gatei M, Richard DJ, Khanna KK. 2011. ATM mediated phosphorylation of CHD4 contributes to genome maintenance. Genome Integr 2: 1. 10.1186/2041-9414-2-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnoosfaderani FS, Palau A, Dong W, Persson J, Durand-Dubief M, Svensson JP, Lennartsson A. 2020. A regulatory role for CHD2 in myelopoiesis. Epigenetics 15: 702–714. 10.1080/15592294.2019.1710913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee W-W, Reinberg D. 2013. A double take on bivalent promoters. Genes Dev 27: 1318–1338. 10.1101/gad.219626.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Hager GL. 2014. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet 15: 69–81. 10.1038/nrg3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. 1957. The strategy of the genes. George Allen & Unwin, London. [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. 1998. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol 8: 843–848. 10.1016/S0960-9822(98)70328-8 [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. 1999. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet 23: 62–66. 10.1038/12664 [DOI] [PubMed] [Google Scholar]
- Wade AA, Lim K, Catta-Preta R, Nord AS. 2019. Common CHD8 genomic targets contrast With model-specific transcriptional impacts of CHD8 haploinsufficiency. Front Mol Neurosci 11: 481. 10.3389/fnmol.2018.00481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AH, Devarakonda S, Skidmore ZL, Krysiak K, Ramu A, Trani L, Kunisaki J, Masood A, Waqar SN, Spies NC, et al. 2018. Recurrent WNT pathway alterations are frequent in relapsed small cell lung cancer. Nat Commun 9: 3787. 10.1038/s41467-018-06162-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-B, Zhang Y. 2001. Mi2, an auto-antigen for dermatomyositis, is an ATP-dependent nucleosome remodeling factor. Nucleic Acids Res 29: 2517–2521. 10.1093/nar/29.12.2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, et al. 2009. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138: 660–672. 10.1016/j.cell.2009.05.050 [DOI] [PubMed] [Google Scholar]
- Wang Y, Chen Y, Bao L, Zhang B, Wang JE, Kumar A, Xing C, Wang Y, Luo W. 2020. CHD4 promotes breast cancer progression as a coactivator of hypoxia-inducible factors. Cancer Res 80: 3880–3891. 10.1158/0008-5472.CAN-20-1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner MH, Roinick KL, Arndt KM. 2007. Rtf1 Is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol 27: 6103–6115. 10.1128/MCB.00772-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AA, Mahajan P, Mertens HDT, Deery MJ, Zhang W, Pham P, Du X, Bartke T, Zhang W, Edlich C, et al. 2012. The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4. J Mol Biol 422: 3–17. 10.1016/j.jmb.2012.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PA, Arora VK, Sawyers CL. 2015. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 15: 701–711. 10.1038/nrc4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Terhal PA, Cohen L, Bruccoleri M, Irving M, Martinez AF, Rosenfeld JA, Machol K, Yang Y, Liu P, et al. 2016. De novo mutations in CHD4, an ATP-dependent chromatin remodeler gene, cause an intellectual disability syndrome with distinctive dysmorphisms. Am J Hum Genet 99: 934–941. 10.1016/j.ajhg.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Lazar HP, Kurolap A, Martinez AF, Paperna T, Cohen L, Smeland MF, Wallen S, Solveig H, Keren B, et al. 2020. The CHD4-related syndrome: a comprehensive investigation of the clinical spectrum, genotype-phenotype correlations, and molecular basis. Genetics Medicine Official J Am Coll Medical Genetics 22: 389–397. 10.1038/s41436-019-0612-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. 2012. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482: 221–225. 10.1038/nature10805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczewski CM, Hepperla AJ, Shimbo T, Wasson L, Robbe ZL, Davis IJ, Wade PA, Conlon FL. 2018. CHD4 and the NuRD complex directly control cardiac sarcomere formation. Proc Natl Acad Sci 115: 6727–6732. 10.1073/pnas.1722219115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Naito T, Arco PG-D, Seavitt JR, Cashman SM, Souza BD, Qi X, Keables P, Andrian UHV, Georgopoulos K. 2004. The chromatin remodeler Mi-2β is required for CD4 expression and T cell development. Immunity 20: 719–733. 10.1016/j.immuni.2004.05.005 [DOI] [PubMed] [Google Scholar]
- Wilson M-M, Henshall DC, Byrne SM, Brennan GP. 2021. CHD2-Related CNS pathologies. Int J Mol Sci 22: 588. 10.3390/ijms22020588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger J, Nodelman IM, Levendosky RF, Bowman GD. 2018. A twist defect mechanism for ATP-dependent translocation of nucleosomal DNA. Elife 7: 391. 10.7554/eLife.34100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Huang W, Bellani M, Seidman MM, Wu K, Fan D, Nie Y, Cai Y, Zhang YW, Yu L-R, et al. 2017. CHD4 has oncogenic functions in initiating and maintaining epigenetic suppression of multiple tumor suppressor genes. Cancer Cell 31: 653–668.e7. 10.1016/j.ccell.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Ling T, Zhou Y, Feng W, Zhu Q, Stunnenberg HG, Grummt I, Tao W. 2012. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc Natl Acad Sci 109: 8161–8166. 10.1073/pnas.1201262109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. 2010. Transcriptional silencing of γ-globin by BCL11A involves long-range interactions and cooperation with SOX6. Gene Dev 24: 783–798. 10.1101/gad.1897310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Liu F, Zhou J, Bai C. 2016. CHD4 is associated with poor prognosis of non-small cell lung cancer patients through promoting tumor cell proliferation. Eur Respir J 48: PA2862. 10.1183/13993003.congress-2016.PA2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 2: 851–861. 10.1016/S1097-2765(00)80299-3 [DOI] [PubMed] [Google Scholar]
- Yadav T, Quivy J-P, Almouzni G. 2018. Chromatin plasticity: a versatile landscape that underlies cell fate and identity. Sci New York N Y 361: 1332–1336. 10.1126/science.aat8950 [DOI] [PubMed] [Google Scholar]
- Yamada T, Yang Y, Hemberg M, Yoshida T, Cho HY, Murphy JP, Fioravante D, Regehr WG, Gygi SP, Georgopoulos K, et al. 2014. Promoter decommissioning by the NuRD chromatin remodeling complex triggers synaptic connectivity in the mammalian brain. Neuron 83: 122–134. 10.1016/j.neuron.2014.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashina K, Yamamoto H, Chayama K, Nakajima K, Kikuchi A. 2006. Suppression of STAT3 activity by Duplin, which is a negative regulator of the Wnt signal. J Biochem 139: 305–314. [DOI] [PubMed] [Google Scholar]
- Yan L, Chen Z. 2020. A unifying mechanism of DNA translocation underlying chromatin remodeling. Trends Biochem Sci 45: 217–227. 10.1016/j.tibs.2019.09.002 [DOI] [PubMed] [Google Scholar]
- Yan S, Thienthanasit R, Chen D, Engelen E, Brühl J, Crossman DK, Kesterson R, Wang Q, Bouazoune K, Jiao K. 2020. CHD7 regulates cardiovascular development through ATP-dependent and -independent activities. Proc Natl Acad Sci 117: 28847–28858. 10.1073/pnas.2005222117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Hannum DF, Zhai Y, Hill SF, Albanus RD, Oliveira, Lou W, Skidmore JM, Sanchez G, Saiakhova A, et al. 2020. CHD7 promotes neural progenitor differentiation in embryonic stem cells via altered chromatin accessibility and nascent gene expression. Sci Rep 10: 17445. 10.1038/s41598-020-74537-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. 2002. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419: 641–645. 10.1038/nature01084 [DOI] [PubMed] [Google Scholar]
- Yates JA, Menon T, Thompson BA, Bochar DA. 2010. Regulation of HOXA2 gene expression by the ATP-dependent chromatin remodeling enzyme CHD8. Febs Lett 584: 689–693. 10.1016/j.febslet.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung J-H, Chen PB, Dong X, Ee L-S, Weng Z, Rando OJ, Fazzio TG. 2011. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147: 1498–1510. 10.1016/j.cell.2011.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo Y-G, Kong G, Lee M-O. 2006. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1α protein by recruiting histone deacetylase 1. Embo J 25: 1231–1241. 10.1038/sj.emboj.7601025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, La H, Lee EJ, Choi H-J, Oh J, Thang NX, Hong K. 2020. ATP-Dependent Chromatin remodeler CHD9 controls the proliferation of embryonic stem cells in a cell culture condition-dependent manner. Biology (Basel) 9: 428. 10.3390/biology9120428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, et al. 2008. The role of the chromatin remodeler Mi-2β in hematopoietic stem cell self-renewal and multilineage differentiation. Gene Dev 22: 1174–1189. 10.1101/gad.1642808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Hu Y, Zhang Z, Emmanuel AO, Galani K, Muhire B, Snippert HJ, Williams CJ, Tolstorukov MY, Gounari F, et al. 2019. Chromatin restriction by the nucleosome remodeler Mi-2β and functional interplay with lineage-specific transcription regulators control B-cell differentiation. Gene Dev 33: 763–781. 10.1101/gad.321901.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Swatkoski S, Holly A, Lee LC, Giroux V, Lee C-S, Hsu D, Smith JL, Yuen G, Yue J, et al. 2015. Oncogenesis driven by the Ras/Raf pathway requires the SUMO E2 ligase Ubc9. Proc Natl Acad Sci 112: E1724–E1733. 10.1073/pnas.1415569112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Hurd EA, Schnetz MP, Handoko L, Wang C, Wang Z, Wei C, Tesar PJ, Hatzoglou M, Martin DM, et al. 2010. CHD7 functions in the nucleolus as a positive regulator of ribosomal RNA biogenesis. Hum Mol Genet 19: 3491–3501. 10.1093/hmg/ddq265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig H-P, Lane WS, Reinberg D. 1998. The dermatomyositis-specific autoantigen Mi2 Is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95: 279–289. 10.1016/S0092-8674(00)81758-4 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev 13: 1924–1935. 10.1101/gad.13.15.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhou C, Li X, Barnes SD, Deng S, Hoover E, Chen C-C, Lee YS, Zhang Y, Wang C, et al. 2020. Loss of CHD1 promotes heterogeneous mechanisms of resistance to AR-targeted therapy via chromatin dysregulation. Cancer Cell 37: 584–598.e11. 10.1016/j.ccell.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Lu X, Wang G, Lan Z, Liao W, Li J, Liang X, Chen JR, Shah S, Shang X, et al. 2017. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature 542: 484–488 10.1038/nature21357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Dong C, Frah M, Deng Y, Marie C, Zhang F, Xu L, Ma Z, Dong X, Lin Y, et al. 2018a. Dual requirement of CHD8 for chromatin landscape establishment and histone methyltransferase recruitment to promote CNS myelination and repair. Dev Cell 45: 753–768.e8. 10.1016/j.devcel.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Sentürk N, Song C, Grummt I. 2018b. lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Gene Dev 32: 836–848. 10.1101/gad.311688.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Cai L, Lu X, Liang X, Li J, Chen P, Ittmann M, Shang X, Jiang S, Li H, et al. 2020. Chromatin regulator, CHD1, remodels the immunosuppressive tumor microenvironment in PTEN-deficient prostate cancer. Cancer Discov 10: CD-19-1352. 10.1158/2159-8290.CD-19-1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen T, Kwon EM, Zhao L, Hsu J, Hyde RK, Lu Y, Alemu L, Speck NA, Liu PP. 2017. Chd7 deficiency delays leukemogenesis in mice induced by Cbfb-MYH11. Blood 130: 2431–2442. 10.1182/blood-2017-04-780106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Paudel BP, Ryan DP, Low JKK, Franck C, Patel K, Bedward MJ, Torrado M, Payne RJ, Oijen Av, et al. 2020. CHD4 slides nucleosomes by decoupling entry- and exit-side DNA translocation. Nat Commun 11: 1519. 10.1038/s41467-020-15183-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li J, Serafim RB, Ketchum S, Ferreira CG, Liu JC, Coe KA, Price BD, Yusufzai T. 2018. Human CHD1 is required for early DNA-damage signaling and is uniquely regulated by its N terminus. Nucleic Acids Res 290: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.