In this study, Howell et al. found that the ability of RUNX1 to promote endothelial-to-hematopoietic transition (EHT) depends on its ability to recruit the TGFβ signaling effectors AP-1 and SMAD2/3, which in turn is determined by the changing chromatin landscape in embryonic versus fetal ECs. Their work provides insight into the regulation of EndoMT and EHT that will guide reprogramming efforts for clinical applications.
Keywords: hematopoiesis, hemogenic endothelium, RUNX1, TGFβ, endothelial-to-hematopoietic transition, endothelial-to-mesenchymal transition
Abstract
Hematopoietic stem and progenitor cells (HSPCs) are generated de novo in the embryo from hemogenic endothelial cells (HECs) via an endothelial-to-hematopoietic transition (EHT) that requires the transcription factor RUNX1. Ectopic expression of RUNX1 alone can efficiently promote EHT and HSPC formation from embryonic endothelial cells (ECs), but less efficiently from fetal or adult ECs. Efficiency correlated with baseline accessibility of TGFβ-related genes associated with endothelial-to-mesenchymal transition (EndoMT) and participation of AP-1 and SMAD2/3 to initiate further chromatin remodeling along with RUNX1 at these sites. Activation of TGFβ signaling improved the efficiency with which RUNX1 specified fetal ECs as HECs. Thus, the ability of RUNX1 to promote EHT depends on its ability to recruit the TGFβ signaling effectors AP-1 and SMAD2/3, which in turn is determined by the changing chromatin landscape in embryonic versus fetal ECs. This work provides insight into regulation of EndoMT and EHT that will guide reprogramming efforts for clinical applications.
The de novo generation of hematopoietic stem and progenitor cells (HSPCs) occurs during embryogenesis, beginning at embryonic day (E)9.5 in mice, from a transient population of hemogenic endothelial cells (HECs) (Zovein et al. 2008; Lancrin et al. 2009; Boisset et al. 2010). The dorsal aorta (DA) is the best-characterized site of HSPC formation from HECs. HECs first appear as endothelial cells (ECs) in a monolayer interconnected by tight junctions, then undergo an endothelial-to-hematopoietic transition (EHT) to form HSPCs (Kissa and Herbomel 2010). HECs can be distinguished from non-HECs by expression of the transcription factor (TF) RUNX1, which is required for EHT (North et al. 1999; Kissa and Herbomel 2010). Deletion of RUNX1 in ECs completely blocks HSPC formation, indicating that RUNX1 is required intrinsically in ECs for EHT (Chen et al. 2009). RUNX1 also appears to be sufficient to induce EHT and HSPC formation from some populations of non-HECs. For example, ectopic expression of RUNX1 in non-HECs from E8.5 mouse embryos could efficiently specify them as hemogenic (Eliades et al. 2016). Similarly, ectopic expression of RUNX1 in non-HECs in E7.5–E8.5 embryos promoted the formation of HSPCs in the DA and heart (Yzaguirre et al. 2018). However, ectopic expression of RUNX1 in non-HECs after E8.5 failed to induce ectopic blood cell formation, indicating that the ability of ECs to respond to RUNX1 and differentiate into blood cells in situ is restricted to specific developmental times. The mechanism underlying the loss of an EC's competence to be specified by RUNX1 as an HEC is not known. It could be caused, for example, by cell-intrinsic epigenetic alterations in ECs that occur during development, or potentially by the location of ECs in nonpermissive environments for EHT and HSPC formation.
During endogenous HSPC formation, only the ventral wall of the DA gives rise to hematopoietic stem cells (HSCs) (Taoudi and Medvinsky 2007). This polarization of the DA is thought to arise from gradients of growth factors such as bone morphogenetic protein (BMP) and Notch ligands that are required for Runx1 expression (Robert-Moreno et al. 2005, 2008). Transforming growth factor β (TGFβ) ligands and receptors act upstream of Notch signaling during EHT and HSPC formation through transcriptional regulation of the Notch ligand jag1a and runx1 (Burns et al. 2005). TGFβ and Notch signaling regulate epithelial-to-mesenchymal transition (EMT) and endothelial-to-mesenchymal transition (EndoMT), processes very similar to EHT, although whether TGFβ signaling regulates the morphological changes of EHT is still a matter of debate (Ottersbach 2019). In multiple other systems, RUNX1 has been shown to directly regulate TGFβ signaling and EMT-related genes (VanOudenhove et al. 2016; Zhou et al. 2018; Lu et al. 2020).
RUNX1 orchestrates EHT by activating repressors of endothelial fate in HECs, such as the TFs GFI1 and GFI1B (Lancrin et al. 2012), and by initiating hematopoietic and cell migration transcriptional programs (Lie-A-Ling et al. 2014). RUNX1 is recruited by TFs such as FLI1 and SCL to sites where they are bound, but RUNX1 also recruits FLI1 and SCL to de novo sites in intergenic regions that are likely enhancers (Lichtinger et al. 2012; Gao et al. 2020). RUNX1 could bind regions of chromatin that were not enriched for the active histone mark H3K9Ac, and following RUNX1 binding H3K9Ac was strongly increased (Lichtinger et al. 2012). Therefore, RUNX1 is essential for orchestrating widespread chromatin changes that specify ECs as hemogenic.
Endogenous HECs are specified during early gestation (E8.5–E9.5), then undergo EHT by midgestation (E10.5–E11.5) and are depleted prior to birth (Kauts et al. 2016). However postnatal ECs can be reprogrammed into HECs through ectopic expression of a cocktail of TFs that includes RUNX1, FOSB, GFI1, and SPI1 (Lis et al. 2017). Both GFI1 and SPI1 are direct downstream targets of RUNX1 (Huang et al. 2008; Lancrin et al. 2012), suggesting that RUNX1 may be the primary driver of reprogramming. Direct reprogramming of patient-specific adult ECs into HSC-generating HECs has the potential to cure hematological diseases. However, to increase the efficiency of reprogramming, it is imperative to understand what defines a cell that is competent to be reprogrammed.
Here we address what makes ECs competent to be efficiently specified as HECs by RUNX1. We ectopically expressed RUNX1 in non-HECs at different developmental time points to determine when and why ECs lose their competency to be specified as hemogenic and undergo EHT. We show that ECs dramatically lose this competency over a 4-d period of embryonic development in mice. This loss of competence correlated with the decreased accessibility of EndoMT-related genes that are activated by TGFβ signaling and are targets of AP-1, SMAD2/3, and RUNX1. Activation of ECs with TGFβ ligand could improve the efficiency of HEC specification of fetal ECs where RUNX1 alone was insufficient to activate the TGFβ-related component of its HEC specification program. We propose that hemogenic specification competency is dependent on a naïve chromatin landscape that allows RUNX1 to recruit AP-1 and SMAD to TGFβ target genes that mediate the morphological changes required for EHT.
Results
RUNX1 can efficiently specify both embryonic and fetal ECs as hemogenic, although fetal ECs require a higher dose of RUNX1
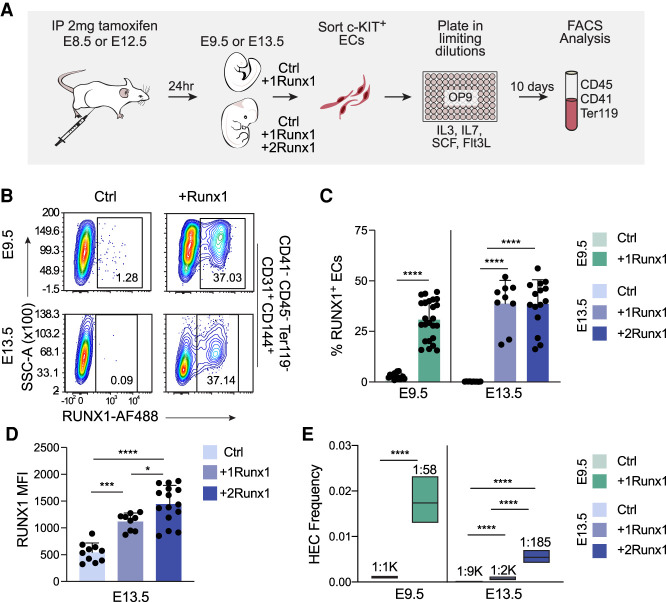
To identify the developmental window of competency for ECs to be specified by RUNX1 as HECs, we activated ectopic RUNX1 expression in ECs that would not normally express RUNX1 at E8.5 (embryonic ECs), when endogenous HECs are specified, and E12.5 (fetal ECs), after most endogenous HECs have undergone EHT. We used mice containing a Runx1 cDNA preceded by a floxed translational stop codon expressed from the Rosa26 locus, and activated RUNX1 expression using an endothelial-specific tamoxifen-inducible Cre driven from the Cdh5 (encoding vascular endothelial cadherin) regulatory sequences (Yzaguirre et al. 2018). Mice with Tg(Cdh5-Cre) and one or two conditional Rosa26Runx1 alleles are referred to as +1Runx1 and +2Runx1, respectively, and littermate controls lacking Tg(Cdh5-Cre) are referred to as Ctrl. We previously showed that Cre expression from the Cdh5-Cre transgene had no effect on HSPC formation from HECs; thus, we did not include Tg(Cdh5-Cre); Rosa26+/+ controls in the current study (Yzaguirre et al. 2018).
We initiated ectopic Runx1 expression at E8.5 and E12.5 in utero and harvested embryos or fetuses 24 h later. We sorted ECs, plated them in limiting dilutions on OP9 stromal cells in conditions that support hematopoiesis (Nakano et al. 1994), and determined the frequency of HECs that generated hematopoietic cells (CD41+, CD45+, and/or Ter119+) 10 d later by flow cytometry (Fig. 1A). Approximately 30% of ECs in E9.5 +1Runx1 embryos and ∼38% of ECs in both E13.5 +1Runx1 and +2Runx1 fetuses were RUNX1+ 24 h after tamoxifen delivery (Fig. 1B,C). Although the percentages of RUNX1+ ECs in E13.5 +1Runx1 and +2Runx1 fetuses were equivalent, RUNX1 levels were ∼1.3-fold higher in +2Runx1 ECs (Fig. 1D). All major organs at E13.5 had comparable percentages of RUNX1+ ECs (Supplemental Fig. S1A); therefore, we analyzed ECs from entire fetuses. We sorted and analyzed c-KIT+ ECs in all of our experiments (Supplemental Fig. S1B), as endogenous HECs are enriched in the c-KIT+ population (Nadin et al. 2003).
Figure 1.
RUNX1 can efficiently specify both embryonic and fetal ECs as hemogenic, although fetal ECs require a higher dose of RUNX1. (A) Schematic illustrating limiting dilution assays for quantifying HEC frequency. (B) Representative contour plots of RUNX1 expression in ECs from Ctrl and +Runx1 embryos and fetuses 24 h after tamoxifen delivery. (C) Quantification of percentage (mean ± SD) of RUNX1+ ECs. (D) Quantification of the median fluorescence intensity (MFI; ±SD) of RUNX1 expression in ECs from E13.5 Ctrl, +1Runx1, and +2Runx1 fetuses. For B–D, ECs were purified as CD41−CD45−Ter119−CD144+CD31+ cells (Supplemental Fig. S1B). Each point represents pooled embryos (E9.5) or a single fetus (E13.5). Data are from three E9.5 and six E13.5 litters/independent experiment. To determine significance, unpaired two-tailed Student's t-test was used for E9.5 and one-way ANOVA and Tukey's multiple comparison test was used for E13.5. (E) Frequency of HECs (± 95% CI) in c-KIT+ ECs. HEC frequencies are indicated above the bars. Data represent four biological replicates using pooled cells from litters of E9.5 embryos collected in four independent experiments, and three biological replicates using pooled cells from E13.5 fetuses collected in three independent experiments. Significance was determined using ELDA. In all figures: (****) P ≤ 0.0001, (***) P ≤ 0.001, (**) P ≤ 0.01, (*) P ≤ 0.05, (ns) P > 0.05.
The frequency of functional HECs significantly increased when RUNX1 was induced at E8.5, from ∼1:1000 in E9.5 Ctrl ECs to 1:58 in E9.5 +1Runx1 ECs (Fig. 1E). The frequency of hematopoietic progenitors (HPs) capable of forming colonies when directly plated in methylcellulose (1:43,600) was much lower than the frequency of HECs (Supplemental Table S1); therefore, hematopoietic cells in the cultures were primarily derived from ECs and not from contaminating HPs. RUNX1 induction at E12.5 increased the frequency of HECs at E13.5 by 4.5-fold, from ∼1:9000 in Ctrl fetuses to ∼1:2000 in +1Runx1 fetuses (Fig. 1E); however, the HEC frequency was only moderately higher (less than threefold) than the frequency of contaminating HPs (Supplemental Table S1). Therefore, E13.5 +1Runx1 ECs were only weakly reprogrammed by RUNX1 into HECs. Increased RUNX1 levels in E13.5 +2Runx1 ECs elevated the frequency of HECs to 1:185, approaching, but still 69% lower than, the 1:58 frequency in E9.5 +1Runx1 ECs. In summary, both fetal and embryonic ECs are competent to be specified as HECs by RUNX1, but efficient specification of fetal ECs required higher RUNX1 levels.
To determine whether RUNX1 expression in ECs did not simply up-regulate hematopoietic markers, but also generated functional HPs, we plated sorted c-KIT+ ECs on OP9 stromal cells, harvested cells from wells containing round cells and in which ECs had been plated in limiting doses (i.e., wells seeded with single ECs), and replated the cells in methylcellulose (Supplemental Fig. S2A). Cells harvested from wells seeded with E9.5 +1Runx1 and E13.5 +2Runx1 ECs gave rise to multilineage colonies (Supplemental Fig. S2B,C). Both E9.5 +1Runx1 and E13.5 +2Runx1 ECs gave rise to phenotypic myeloid cells, erythroid cells, B cells, and T cells when plated under conditions that support the differentiation of these lineages (Supplemental Fig. S2D; Nakano et al. 1994; Schmitt and Zúñiga-Pflücker 2002). Therefore, the HECs specified by RUNX1 gave rise to multilineage HPs.
RUNX1 induced few transcriptional changes at 24 h after induction
To determine whether the more efficient hemogenic specification of E9.5 ECs correlated with early transcriptional changes, we sorted E9.5 and E13.5 ECs 24 h after RUNX1 induction and performed RNA sequencing (RNA-seq). There were relatively few changes in the transcriptomes of any EC populations 24 h following RUNX1 induction (Supplemental Fig. S3A–C). The most transcriptional changes occurred in E13.5 +1Runx1 ECs, which were the least efficiently specified by RUNX1, suggesting that the majority of the transcriptional changes at 24 h were not important for efficient hemogenic specification. Runx1 was significantly up-regulated in all ECs, as were Itgb3 and AI467606, both known direct targets of RUNX1 (Ferreras et al. 2011; Lichtinger et al. 2012). Itgb3 encodes the cell surface receptor CD61, which marks endogenous HECs (Huang et al. 2016). Increases in both CD61+ and CD61+RUNX1+ ECs were observed in E9.5 +1Runx1 embryos and E13.5 +2Runx1 fetuses (Supplemental Fig. S3D,E).
Gross changes in the chromatin structure of ECs occur between E9.5 and E13.5
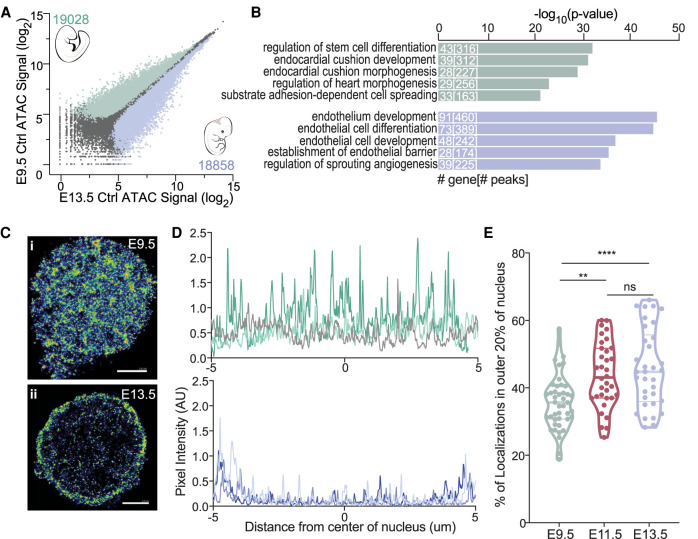
We investigated whether the chromatin landscapes played a role in efficient specification using assay for transposase-accessible chromatin using sequencing (ATAC-seq). The chromatin landscape was vastly different at baseline in E9.5 and E13.5 Ctrl ECs, with ECs at each stage containing almost 20,000 differentially accessible peaks (Fig. 2A). Gene ontology (GO) analysis of these differentially accessible regions showed that stem cell, endocardial cushion development/morphogenesis, and other heart development genes were more accessible in E9.5 Ctrl ECs, while endothelial-specific genes were more accessible in E13.5 Ctrl ECs (Fig. 2B). Several genes preferentially accessible in E9.5 ECs, including Twist1, Snai1, and Hey1/2, are required for EndoMT, a type of EMT that occurs during endocardial cushion formation (Lamouille et al. 2014). We hypothesize that the accessibility of EndoMT-related genes may be a determinant of efficient hemogenic specification.
Figure 2.
Gross changes in the chromatin structure of ECs occur between E9.5 and E13.5. (A) Scatter plot of ATAC-seq peak signals for a consensus peak set in all samples for E9.5 Ctrl (Y-axis) and E13.5 Ctrl (X-axis). Numbers and colored dots represent differentially accessible peaks calculated using EdgeR (FDR < 0.05). Pearson correlation of all ATAC-seq replicate samples are in Supplemental Figure S4. (B) GO terms for differentially accessible peaks for E9.5 Ctrl (top) and E13.5 Ctrl (bottom). The top five terms are shown. Numbers in bars represent the number of genes and peaks/regions for each GO term. (C) Purified E9.5 (panel i) and E13.5 (panel ii) ECs stained for H2b. H2b color-coded by chromatin density. (Blue) Density <0.003 nm2, (green) density between 0.003 and 0.02 nm2, (red) density >0.02 nm2. Scale bar, 2µm. (D) Signal profile of H2b signal across the nucleus (the center of the plot corresponds to center of the nucleus) of E9.5 (top) or E13.5 (bottom) ECs. Each line represents a single nucleus. Three representative cells are shown. (E) Percent of H2b localizations in the outer 20% boundaries of the nucleus. Each data point represents a single nucleus; data were collected from three independent experiments.
Chromatin also underwent gross structural changes between E9.5 and E13.5. Stochastic optical reconstruction microscopy (STORM) with an antibody to histone H2b demonstrated that E9.5 ECs had diffuse chromatin that was evenly spread throughout the nucleus, typical of the distribution seen in early embryos, whereas chromatin was concentrated in the nuclear periphery in E13.5 ECs, as seen in more differentiated cells (Fig. 2C,D; Ricci et al. 2017). The relocalization of chromatin to the nuclear periphery occurred over a several-day period, as E9.5 ECs had significantly less chromatin in the outer 20% of the nucleus than E11.5 ECs, and E13.5 ECs had the greatest amount (Fig. 2E). Peripheralization of chromatin is associated with repressive chromatin and gene silencing (Lanctôt et al. 2007), indicating that fetal ECs displayed a more restrictive global chromatin landscape, which potentially affected RUNX1's ability to impart its hemogenic specification program.
RUNX1 increased chromatin accessibility at hematopoietic genes in both embryonic and fetal ECs
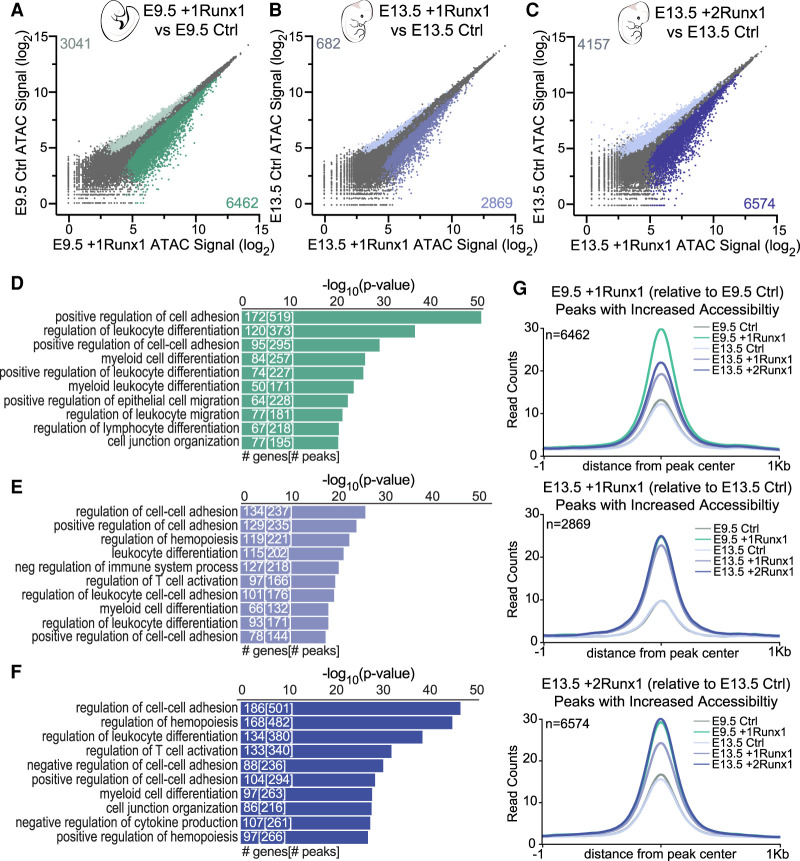
We next examined the changes in chromatin accessibility mediated by RUNX1. RUNX1 induced similar changes in chromatin accessibility in E9.5 +1Runx1 and E13.5 +2Runx1 ECs (Fig. 3A,C), and fewer changes in E13.5 +1Runx1 ECs (Fig. 3B). Fewer peaks were closed than opened by RUNX1 in all ECs (Fig. 3A–C). GO terms for genes associated with peaks closed by RUNX1 at E9.5 were related to vasculogenesis and angiogenesis (Supplemental Fig. S5A), reflecting RUNX1's role in shutting down the endothelial program. Notably, angiogenesis and vasculogenesis GO terms were not enriched in genes closed by RUNX1 in E13.5 ECs. Instead, pathways related to metabolism and Rap signaling were associated with genes closed by RUNX1 in E13.5 +1Runx1 ECs, and terms related to cell biological processes were enriched for genes closed in E13.5 +2Runx1 ECs (Supplemental Fig. S5B,C). The inefficient silencing of the angiogenesis and vasculogenesis programs may contribute to the relatively inefficient hemogenic specification of E13.5 ECs.
Figure 3.
RUNX1 increased chromatin accessibility at hematopoietic genes in both embryonic and fetal ECs. (A–C) Scatter plot of ATAC-seq peak signals for a consensus peak set following RUNX1 induction for E9.5 +1Runx1 (A), E13.5 +1Runx1 (B), and E13.5 +2Runx1 (C). Coloring and peak number are based on differential peaks calculated using EdgeR (FDR < 0.05). (D–F) GO biological process terms for peaks with increased accessibility (relative to Ctrl) for E9.5 +1Runx1 (D), E13.5 +1Runx1 (E), and E13.5 +2Runx1 (F). Peak regions were linked to genes using GREAT. The top 10 terms are shown. Numbers in bars represent the number of genes and peaks/regions for each GO term. (G) Averaged peak profile plots (normalized to reads per genomic content [RPGC]) of peaks with increased accessibility (shown in plots as n) following RUNX1 relative to Ctrl at each time point for E9.5 +1Runx1, E13 .5 +1Runx1, and E13.5 +2Runx1.
GO terms of genes in regions opened by RUNX1 in both E9.5 and E13.5 ECs included hematopoietic and EHT-related terms, such as “positive regulation of cell adhesion” and “cell junction organization,” and terms for different hematopoietic lineages such as myeloid and lymphoid differentiation (Fig. 3D–F). Genes encoding transcripts induced by RUNX1, such as Itgb3, had corresponding increases in chromatin accessibility (Supplemental Fig. S5D). Taken together, this demonstrates that RUNX1 can activate at least a subset of its hematopoietic program in both embryonic and fetal ECs.
Regions that gained accessibility in E13.5 +2Runx1 ECs did so to a greater degree than in E13.5 +1Runx1 ECs, and regions that gained accessibility in either E13.5 +1Runx1 or +2Runx1 ECs did so also in E9.5 +1Runx1 ECs (Fig. 3G; Supplemental Fig. S6A,B). Conversely, 40% of regions opened by RUNX1 in E9.5 +1Runx1 ECs were not opened in E13.5 +2Runx1 ECs (Supplemental Fig. S6C,D). The regions opened by RUNX1 only in E9.5 +1Runx1 ECs were enriched for endocardial cushion development/morphogenesis as well as hematopoietic and EHT terms (Supplemental Fig. S6E). The ability of RUNX1 to mediate changes in chromatin accessibility at these migration/EndoMT-related genes appears to be a determinant of the efficiency of hemogenic specification observed in embryonic ECs.
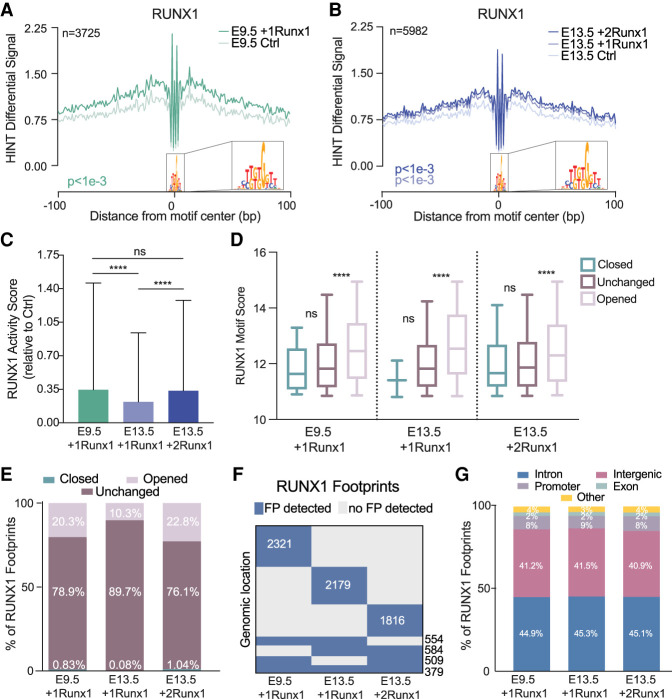
RUNX1 binds predicted high-affinity sites to open chromatin
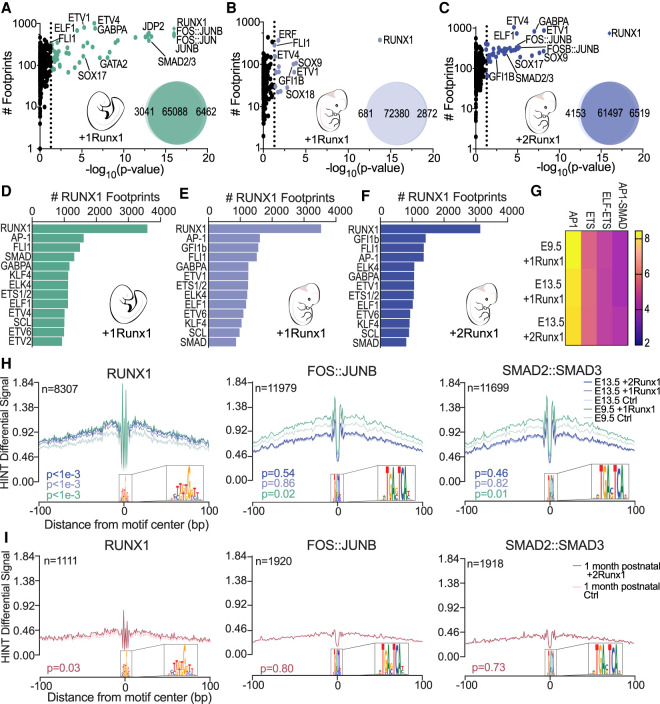
We predicted TF binding across accessible chromatin using Hmm-based identification of transcription factor footprints from ATAC-seq data (HINT-ATAC) (Li et al. 2019). Because the Tn5 cleavage enzyme used for ATAC-seq is unable to cleave where TFs are bound to DNA, there is a resulting dip in reads, or “footprint,” that can then be matched to TF motifs to infer the specific TF bound. To improve accuracy, we filtered out TFs that were not expressed based on RNA-seq data (<1.5 FPKM). RUNX1 opened chromatin in the vicinity of its predicted binding sites, as demonstrated by the overall increase in average normalized read counts surrounding RUNX1 footprints (Fig. 4A,B). We predicted the global activity of RUNX1, represented as a TF activity score, using HINT differential, which measures the depth of the footprint plus the total number of reads in the flanking region for all footprints, then determines the difference between two biological conditions (e.g., between E9.5 +1Runx1 and E9.5 Ctrl ECs) (Li et al. 2019). E9.5 +1Runx1 and E13.5 +2Runx1 ECs have comparable RUNX1 activity scores, which are significantly higher than the RUNX1 activity score in E13.5 +1Runx1 ECs, suggesting that RUNX1 bound more weakly and induced fewer changes in chromatin accessibility in E13.5 +1Runx1 ECs (Fig. 4C). Relatively weak RUNX1 binding sites were enriched under RUNX1 footprints in already accessible chromatin (closed or unchanged) compared with sites in chromatin RUNX1 opened, as predicted by the average motif scores for RUNX1 footprints in closed, unchanged, and opened chromatin (Fig. 4D). The distribution of RUNX1 footprints at opened versus unchanged chromatin correlated with the efficiency of hemogenic specification. Specifically, a significantly greater proportion (∼20%) of RUNX1 footprints in both E9.5 +1Runx1 and E13.5 +2Runx1 ECs was located in regions of chromatin with increased accessibility, compared with only ∼10% in E13.5 +1Runx1 ECs (Fig. 4E). The genomic locations of RUNX1 footprints appeared to be largely unique to each EC population, with a relatively smaller number of footprints shared by two or more EC populations (Fig. 4F), underscoring that the hemogenic specification program that RUNX1 is imparting is vastly different not only between embryonic and fetal ECs, but also for varying doses of RUNX1 in fetal ECs. There were no apparent differences in the genomic feature location of the RUNX1 footprints for any of the conditions (Fig. 4G). In summary, under suboptimal conditions, i.e., lower RUNX1 levels and a fetal chromatin landscape (E13.5 +1Runx1), a greater proportion of RUNX1 appeared to bind to weaker sites in already accessible chromatin. Increasing the levels of RUNX1 (in E13.5 +2Runx1 ECs) or the more naïve chromatin landscape in E9.5 ECs increased the proportion of RUNX1 footprints in closed chromatin, where RUNX1 could access higher-affinity sites and open the chromatin.
Figure 4.
RUNX1 binds predicted high-affinity loci to open chromatin. (A,B) RUNX1 footprint plot following RUNX1 induction for E9.5 +1Runx1 (A) and E13.5 +1Runx1 and E13.5 +2Runx1 (B). Plots show average normalized read counts and P-value calculated using HINT differential. (n) Number of nonredundant RUNX1 footprints. RUNX1 footprints detected in each condition were E9.5 +1Runx1: 3725, E13.5 +1Runx1: 3657, and E13.5 +2Runx1: 3259 (nonredundant footprints in E13.5 +1Runx1 and +2Runx1 ECs: 5982). (C) Bar plot (mean ± SD) of the TF activity score for each RUNX1 footprint called shown as average score relative to Ctrl at each time point. One-way ANOVA and Tukey's multiple comparison test were applied to determine significance. (D) RUNX1 motif score for called RUNX1 footprints shown with respect to changed regions of chromatin (opened, closed, or unchanged relative to the Ctrl at each time point). Significance between regions was determined using one-way ANOVA and Tukey's multiple comparison test. (E) Location of the RUNX1 footprints with respect to changed regions of chromatin (opened, closed, or unchanged relative to the Ctrl at each time point) shown as a percentage of total RUNX1 footprints in each sample. P-value calculated using repeated pairwise comparison of proportions Fisher's test in R; differences in all are significant (P < 0.05) except for closed regions between E9.5 +1Runx1 and E13.5 +2Runx1. (F) Clustering of RUNX1 footprints; each row of the heat map represents a genomic location, and numbers overlaid on the heat map represent the number of RUNX1 footprints. (G) Distribution of RUNX1 footprints with respect to genomic feature. (Other) 3UTR, miRNA, ncRNA, transcription termination sites (TTSs), pseudo gene, and 5UTR.
Colocalization of RUNX1, AP-1, and SMAD footprints is a determinant of efficient HEC specification
To identify cooperating TFs that could participate in driving hemogenic specification, we used bias-free transcription factor footprint enrichment test (biFET) to find enrichment of TF footprints in regions of chromatin with increased accessibility (Youn et al. 2019). RUNX1 and AP-1 (FOS:JUNB, FOS:JUN, and JUN:JUNB) were the most strongly enriched TF footprints in regions of increased accessibility following RUNX1 induction in E9.5 ECs (Fig. 5A). Footprints of SMAD2/3, GATA2, and several ETS TFs (ETV1/4, GABPA, and ELF1) were also significantly enriched (Fig. 5A), suggesting that RUNX1 directs multiple families of TFs to help drive hemogenic specification of embryonic ECs, potentially through different mechanisms. In contrast, no other TF footprints were as highly enriched as RUNX1 footprints in chromatin with increased accessibility in E13.5 +1Runx1 ECs, suggesting that RUNX1 was unable to efficiently recruit additional TFs to chromatin it was opening (Fig. 5B). RUNX1 at higher doses in E13.5 +2Runx1 ECs, however, was able to direct more TFs, including AP-1 and SMAD2/3 to regions of chromatin with increased accessibility (Fig. 5C). Approximately 40% of RUNX1 footprints were located within 200 bp of AP-1, SMAD, FLI1, and other ETS family footprints, indicating that RUNX1 was recruiting these TFs to local sites (Fig. 5D–F).
Figure 5.
Colocalization of RUNX1, AP-1, and SMAD footprints is a determinant of efficient HEC specification. (A–C) Scatter plots showing enriched TF footprints at regions of chromatin with increased accessibility (relative to Ctrl at each time point) for E9.5 +1Runx1 (A), E13.5 +1Runx1 (B), and E13.5 +2Runx1 (C). The number of footprints for each TF at regions of chromatin with increased accessibility is displayed on the Y-axis for the +Runx1 condition. (Diamonds) P-value ≪ 1 × 10−16, (colored circles) P < 0.05. P-values calculated using biFET. Venn diagrams represent the number of peaks closed (left), unchanged (middle), and opened (right) following RUNX1 induction in each sample. (D–F) TF footprints co-occurring (±200 bp) with RUNX1 footprints shown as number of RUNX1 footprints for E9.5 +1Runx1 (D), E13.5 +1Runx1 (E), and E13.5 +2Runx1 (F). (G) Top combination of motifs enriched near RUNX1 footprints; select pairs are shown. Scale is Z-score calculated using potentially collaborating transcription factor finder (PC-TrAFF). AP-1 and ETS refer to combinations of the respective TF family members. (H,I) RUNX1 (left), FOS:JUNB (middle), and SMAD2:SMAD3 (right) footprint plots following RUNX1 induction for embryonic and fetal ECs (H) and 1-mo postnatal ECs (I). Plots show average normalized read counts calculated using HINT differential around all footprints in all samples. (n) Number of nonredundant footprints. P-values shown are relative to each time point Ctrl and were calculated using HINT differential.
SMAD and AP-1 are effectors of TGFβ family signaling. SMADs are directly phosphorylated by TGFβ type I receptors, while activation of AP-1 occurs through TAK1, TRAF6, and JNKs (Moustakas and Heldin 2005). Activated AP-1 and SMAD directly interact with each other and coregulate TGFβ target genes during EMT (Liberati et al. 1999; Sundqvist et al. 2013). SMAD requires TFs such as AP-1 and RUNX (RUNX1/2/3) for effective and specific DNA binding (Jia and Meng 2021), and both SMAD and AP-1 directly interact with RUNX1 (Hanai et al. 1999; D'Alonzo et al. 2002). We searched for evidence of cooperativity between SMAD, AP-1, and RUNX1 in regulating TGFβ target genes in embryonic ECs. The percentage of RUNX1 footprints near AP-1 footprints was similar in both embryonic and fetal ECs (∼44%) (Fig. 5D–F), and the combination of AP-1 and SMAD motifs was significantly enriched (P < 1 × 10−3) near RUNX1 footprints in all conditions (Fig. 5G). However, the percentage of RUNX1 footprints near SMAD footprints was greater in embryonic ECs (∼36%) compared with fetal ECs (∼25%), suggesting that RUNX1 was better able to relocate SMAD in embryonic ECs (Fig. 5D–F). Supporting this, regions that gained accessibility only in E9.5 +1Runx1 ECs (Supplemental Fig. S6D) were significantly enriched for AP-1 and SMAD2/3 footprints relative to regions that gained accessibility in both E9.5 +1Runx1 and E13.5 +2Runx1 (Supplemental Fig. S6F), demonstrating that critical AP-1 and SMAD binding sites are not being opened in fetal ECs. Our data suggest that SMAD, AP-1, and TGFβ-related target genes are a crucial part of RUNX1's hemogenic specification program in embryonic ECs, and that the ability of RUNX1 to recruit AP-1 and SMAD to local sites contributed to the difference in the efficiency with which RUNX1 could specify embryonic and fetal ECs as hemogenic.
Regions of chromatin around RUNX1 footprints were similarly accessible in E9.5 and E13.5 ECs prior to RUNX1 induction, and RUNX1 appeared to open chromatin to the same degree (Fig. 5H). In contrast, FOS:JUNB (an AP-1 TF) and SMAD2:SMAD3 were able to locally open chromatin only in embryonic ECs even though their footprints were enriched in E13.5 +2Runx1 ECs (Fig. 5H). The chromatin around FOS:JUNB and SMAD2:SMAD3 footprints was more accessible at baseline in E9.5 Ctrl ECs than in E13.5 Ctrl ECs, and chromatin accessibility in E9.5 ECs further increased following RUNX1 induction, but did not significantly increase in E13.5 ECs (Fig. 5H). The accessibility of chromatin directly flanking (±100 bp) other TFs enriched at regions of chromatin with increased accessibility; namely, GATA2, GABPA, ETV4, FLI1, SCL, ELF1, and GFI1B, did not significantly change following RUNX1 induction in either embryonic or fetal ECs, suggesting that unlike FOS:JUNB and SMAD2:SMAD3, these TFs were not driving local chromatin remodeling (Supplemental Fig. S7A,B).
The ability of RUNX1 to open and recruit additional TFs to chromatin decreases with age and tissue specialization of ECs. RUNX1 alone was unable to efficiently specify human umbilical vein ECs (HUVECs) as HECs (Sandler et al. 2014). Similarly, ECs purified from the livers of 1-mo postnatal +2Runx1 mice contained no HECs able to produce blood cells ex vivo (data not shown). RUNX1 was unable to open chromatin to the same degree in 1-mo postnatal ECs (Fig. 5I, Supplemental Fig. S8A) compared with embryonic or fetal ECs (Fig. 4A,B). Postnatal ECs showed the largest changes in gene expression following RUNX1 induction (Supplemental Fig. S8B), indicating that the degree of gene up-regulation at 24 h does not correlate with efficient hemogenic specification. Regions of opened chromatin were enriched for hematopoietic-related terms, demonstrating that even in nonpermissive reprogramming conditions, RUNX1 was able to activate a portion of the HEC specification program (Supplemental Fig. S8C). Only RUNX1 and BHLHE22 were enriched at regions of chromatin with increased accessibility in 1-mo postnatal ECs (Supplemental Fig. S8D). AP-1 and SMAD factors were not enriched in opened chromatin and did not participate in chromatin opening (Fig. 5I), and their targets in embryonic ECs were inaccessible in postnatal ECs (Supplemental Fig. S8E). In summary, AP-1 and SMAD2/3 collaborate with RUNX1 to open chromatin in embryonic ECs. Their failure to open chromatin correlates with the reduced competence of fetal or postnatal ECs to be efficiently specified by RUNX1 as HECs.
RUNX1 primes regulatory regions in fetal ECs that are active in embryonic ECs
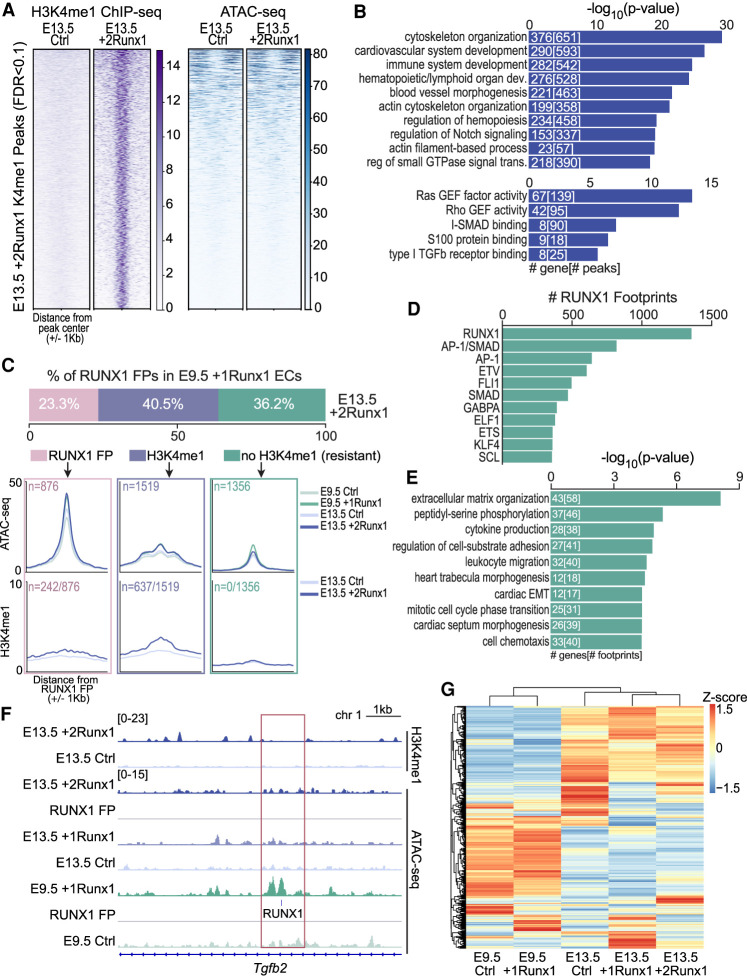
To capture early events induced by RUNX1 that precede chromatin remodeling, we performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) for the histone mark H3K4me1 in E13.5 Ctrl and +2Runx1 ECs. H3K4me1, which is associated mostly with enhancers, is typically deposited prior to chromatin opening and enhancer activation (Calo and Wysocka 2013). Based on ATAC-seq, 5398 sites in largely inaccessible regions of chromatin significantly (FDR < 0.1) gained H3K4me1 in E13.5 +2Runx1 ECs (Fig. 6A). These sites were enriched for regulatory regions of EHT-related and hematopoietic genes, as well as some TGFβ signaling-related terms (Fig. 6B). We analyzed the sites of RUNX1 footprints in E9.5 +1Runx1 ECs (Fig. 4A) to determine whether RUNX1 was able to induce H3K4me1 at these sites in E13.5 +2Runx1 ECs (Fig. 6C). Approximately 23% of the RUNX1 footprints in E9.5 +1Runx1 ECs were also present in E13.5 +2Runx1 ECs, ∼40% were absent in E13.5 +2Runx1 ECs but were marked by H3K4me1, and ∼36% had neither a RUNX1 footprint nor H3K4me1 marks in E13.5 +2Runx1 ECs. Sites that had neither a RUNX1 footprint nor a H3K4me1 mark in E13.5 +2Runx1 ECs were categorized as “resistant” to RUNX1 activity (Fig. 6C). The majority of the resistant sites (60%) were located near (±200 bp) AP-1 or SMAD footprints in E9.5 ECs (Fig. 6D), and many were associated with endocardial morphogenesis genes downstream from the TGFβ signaling pathway (Fig. 6E; Supplemental Table S2). For example, at a regulatory region of the RUNX1 target Tgfb2 (VanOudenhove et al. 2016), a RUNX1 footprint was present in newly accessible chromatin in E9.5 +1Runx1 ECs, but this site neither gains accessibility nor H3K4me1 in E13.5 +2Runx1 ECs (Fig. 6F). Expression of these TGFβ signaling pathway genes was also differentially regulated in fetal and embryonic ECs (Fig. 6G), consistent with the regulatory regions of these genes being “resistant” in fetal ECs. Taken together, these data suggest that RUNX1 is largely able to execute the hematopoietic portion of its HEC specification program in both embryonic and fetal ECs, but the morphological component of EHT mediated by TGFβ-related genes is not fully activated in fetal ECs.
Figure 6.
RUNX1 primes regulatory regions in fetal ECs that are active in embryonic ECs. (A) Heat maps of H3K4me1 (left, purple) and ATAC-seq (right, blue) signals (scales are normalized to RPGC read counts) for 5398 regions newly marked (FDR < 0.1) by H3K4me1 following RUNX1 induction in E13.5 +2Runx1 ECs. (B) The top 10 biological process terms (top graph), and top five molecular function terms (bottom graph) associated with H3K4me1 peaks gained in E13.5 +2Runx1 ECs. The numbers of genes and peaks associated with each term are overlaid on the bars. (C) Annotation of the RUNX1 footprints found in E9.5 +1Runx1 ECs and in E13.5 +2Runx1 ECs. The top bar graph shows footprints in E9.5 +1Runx1 ECs binned based on their status in E13.5 +2Runx1 ECs. Categories include RUNX1 footprints present in E13.5 +2Runx1 ECs (pink), no RUNX1 footprint in E13.5 +2Runx1 ECs but marked by H3K4me1 (±500 bp) (lavender), or no H3K4me1 (or RUNX1 footprint) in E13.5 +2Runx1 ECs (±500 bp) (green). The top metagene plots show ATAC-seq signal, and the bottom metagene plots show H3K4me1 signal at these regions. In the top plots, n represents the total number of regions plotted for both ATAC-seq and H3K4me1 plots, and in the bottom panel, n represents the number of regions that were marked by new H3K4me1 in E13.5 +2Runx1 relative to E13.5 Ctrl ECs shown as a fraction of all regions. Colored boxes around metagene plots signify same-colored regions as shown in the bar plot above. (D) The number of RUNX1 footprints with co-occurring (±200 bp) TF footprints in E9.5 +1Runx1 ECs that were not present or marked by H3K4me1 in E13.5 +2Runx1 ECs (“resistant”; boxed in green in C). (E) GO terms for “resistant” genes. The top 10 terms are shown; numbers in bars represent the number of genes and footprints/regions for each GO term. (F) Genome browser view showing normalized H3K4me1 signal in E13.5 +2Runx1 and Ctrl ECs (top), normalized ATAC-seq signal in E13.5 +2Runx1, E13.5 +1Runx1, E13.5 Ctrl, E9.5 +1Runx1, and E9.5 Ctrl ECs, and RUNX1 footprint (RUNX1; denoted by small blue box) in the Tgfb2 locus. (G) Expression of genes from E, represented as a heat map based on RNA-seq data normalized to FPKM and shown as a Z-score for each gene.
Reduced accessibility of chromatin to AP-1 and SMAD2/3 limits RUNX1-driven hemogenic specification of fetal ECs
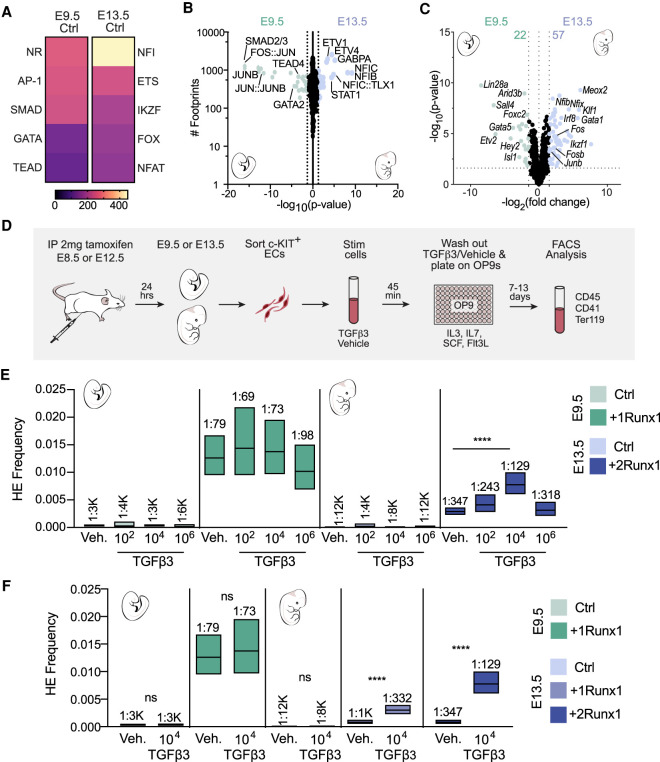
AP-1 and SMAD motifs and footprints were more strongly enriched in E9.5 than in E13.5 Ctrl ECs (Fig. 7A,B). This was not caused by increased levels of AP-1 or SMAD2/3 in E9.5 ECs; transcripts encoding the AP-1 factors Fos, Junb, and Fosb were present at significantly higher levels in E13.5 Ctrl ECs (Fig. 7C), and there were no significant differences in Smad2/3 transcript levels (data not shown). As SMAD2/3 must be phosphorylated to enter the nucleus and bind DNA, we determined whether there were differences in activated SMAD2/3 in E9.5 and E13.5 ECs. The percentage of ECs that contained phosphorylated SMAD2/3 (pSMAD2/3+) in the heart and DA was equivalent in E9.5 and E13.5 Ctrl ECs (Supplemental Fig. S9A–C), and nearly all ECs in E9.5 +1Runx1 and E13.5 +2Runx1 ECs that were RUNX1+ were also pSMAD2/3+ (Supplemental Fig. S9D,E). Therefore, TGFβ signaling is similarly active in embryonic and fetal ECs, indicating that the decreased ability of SMAD2/3 to open chromatin in E13.5 ECs following RUNX1 induction is likely due to differences in chromatin that limited the activity of pSMAD2/3 and its partner AP-1 at critical target genes for hemogenic specification.
Figure 7.
Activation of TGFβ signaling cooperates with RUNX1 to increase the efficiency of hemogenic specification of fetal ECs. (A) Heat maps of the top five enriched TF motif families in differentially accessible peaks between E9.5 and E13.5 Ctrl. Score is −log10(adjusted P-value) calculated using MEME AME. (B,C) Scatter plots of TF footprints enriched (B) and volcano plot of differentially expressed TFs (C; FDR < 0.05 and FC > |1.5|) between E9.5 Ctrl (left) and E13.5 Ctrl (right). For B: (diamonds) P-value ≪ 1 × 10−16, (colored dots) P < 0.05. P-value was calculated using biFET. (C) Colored dots and numbers represent differentially expressed TFs. (D) Schematic illustrating the isolation of ECs, ex vivo stimulation with TGFβ3, and limiting dilution assays for quantification of HEC frequency. (E,F) Frequency of HECs (±95% CI) in c-KIT+ ECs following stimulation with 102, 104, or 106 molecules of TGFβ3 per cell or vehicle (E), and 104 molecules of TGFβ3 per cell or vehicle (F). HEC frequencies are indicated above the bars. Data represent four biological replicates using pooled cells from litters of E9.5 Ctrl and +1Runx1 embryos collected in six independent experiments, and four (E13.5 +1Runx1) and five (E13.5 +2Runx1) biological replicates from E13.5 litters collected in 11 independent experiments. Significance was determined using ELDA software.
Activation of TGFβ signaling cooperates with RUNX1 to increase the efficiency of hemogenic specification of fetal ECs
We hypothesized that the failure to activate shared target genes of RUNX1, AP-1, and SMAD2/3 that are also downstream targets of TGFβ signaling was limiting the hemogenic specification of fetal ECs, and reasoned that increased TGFβ signaling would elevate the concentrations of activated AP-1 and SMAD2/3 in the nucleus (Moustakas and Heldin 2005) and help overcome this barrier. Both TGFβ1 and TGFβ3 are required for HSPC formation in zebrafish; we chose to stimulate ECs with TGFβ3 because it was shown to act after Runx1 expression and HEC specification to promote EHT (Monteiro et al. 2016). To test this hypothesis, we induced RUNX1 expression in embryos and fetuses, isolated ECs, and subjected them to a short pulse of TGFβ3 (45 min) to induce a transient increase in pSMAD2/3 and AP-1. Since TGFβ ligand stimulation is dose-dependent and cell type-specific (Zi et al. 2011), we tested three concentrations of TGFβ3 (102, 104, and 106 molecules/cell) on E9.5 +1Runx1 and E13.5 +2Runx1 ECs to determine the optimal dose for stimulating EHT. Following stimulation with TGFβ3 ex vivo, we performed a limiting dilution assay to determine the frequency of HECs (Fig. 7D). TGFβ3 did not increase the frequency of HECs in E9.5 ECs at any dose (Fig. 7E; Supplemental Table S3). Our interpretation of this negative result is that the chromatin in the regulatory regions of TGFβ signaling targets is already accessible and bound by a combination of AP-1, SMAD2/3, and RUNX1 in E9.5 +1Runx1 ECs. In contrast, stimulation of E13.5 +2Runx1 ECs with 104 molecules of TGFβ3 per cell significantly increased the frequency of HECs by approximately threefold, whereas doses of 102 or 106 molecules of TGFβ3 per cell did not significantly increase EHT (Fig. 7E). TGFβ3 could also increase the frequency of HECs in E13.5 +1Runx1 ECs when exposed to the optimal TGFβ3 dose (104 molecules/cell) (Fig. 7F). These results suggest that a short-term increase in TGFβ signaling could partially overcome the chromatin barrier to efficient HEC specification in fetal ECs, but higher amounts TGFβ3 had no effect on EHT. These data also show that RUNX1 was crucial for directing AP-1 and SMAD2/3 factors to HEC specification targets, as the frequency of HECs was not increased upon TGFβ3 treatment of E9.5 and E13.5 Ctrl ECs. Taken together, these data show that TGFβ signaling is a crucial component of RUNX1's HEC specification program, and short-term activation of TGFβ signaling can partially overcome a resistant chromatin landscape in fetal ECs.
Discussion
RUNX1 is sufficient to specify embryonic and fetal ECs as functional HECs
Our results demonstrate that RUNX1 alone is sufficient to specify embryonic and fetal ECs as HECs that can generate multilineage HPs, and that TGFβ target genes are a critical component of RUNX1's HEC specification program. A higher dose of RUNX1 is required to specify fetal ECs due to the relative inaccessibility of TGFβ target genes, but this reduced efficiency of hemogenic specification could be partially overcome by transient activation of TGFβ signaling. In contrast, postnatal ECs are not competent to be specified as HECs even when exposed to a high dose of RUNX1. These data indicate that as ECs mature and acquire organotypic functions, they lose competence to be specified as HECs due to changes in the chromatin landscape, specifically at TGFβ EndoMT-related genes that are regulated by a combination of AP-1, SMAD2/3, and RUNX1.
Striking parallels between EHT and EndoMT
EHT and EndoMT are both developmentally restricted processes that involve down-regulation of endothelial junction proteins to allow ECs to become motile (Lamouille et al. 2014). EndoMT in the heart occurs between E9.5 and E10.5 and significantly diminishes by E13 (Camenisch et al. 2002)—timing that strikingly mirrors that of HEC specification and EHT. Another parallel between EHT and EndoMT is the involvement of a RUNX protein: RUNX1 in EHT (Chen et al. 2009) and RUNX2 in EndoMT (Tavares et al. 2018). Additionally, EHT and EndoMT share a number of signaling pathways, including TGFβ and Notch (Ottersbach 2019). EHT has also been observed endogenously in the heart at E9.5–E10.5 (Nakano et al. 2013), and could be amplified by overexpressing RUNX1 (Yzaguirre et al. 2018), again highlighting the parallels between hematopoietic and cardiac development. We found that the accessibility of genes shared between the EHT and EndoMT programs was a determinant of HEC specification by RUNX1, and that TGFβ activation could improve the specification efficiency when these targets were not as accessible. As development progresses, ECs lose their ability to undergo EHT/EndoMT, likely due to chromatin compaction of these genes.
AP-1 and SMAD factors and TGFβ signaling targets play a significant role in HEC specification by RUNX1
TGFβ target genes are coregulated by AP-1 and SMAD (Liberati et al. 1999; Sundqvist et al. 2013), and both factors can interact directly with RUNX1 (Hanai et al. 1999; D'Alonzo et al. 2002). All RUNX proteins (RUNX1/2/3) have been implicated in TGFβ signaling and EMT in various systems (Voon and Thiery 2017). All three proteins—RUNX1, AP-1, and SMAD2/3—in combination are shown here to be coregulators of EHT.
In an ESC model of hematopoiesis, a dominant-negative FOS that abrogated all AP-1 binding had no effect on HEC specification but resulted in reduced expression of morphological-related genes, including TGFβ-related genes (Obier et al. 2016). This is consistent with our model that RUNX1 by itself is sufficient for implementing the hematopoietic cell fate program, but AP-1 proteins, in cooperation with RUNX1, activate TGFβ-related genes needed for establishing the morphological attributes of EHT. A previous study reported that postnatal ECs can be reprogrammed into HECs via the ectopic expression of FOSB, GFI1, RUNX1, and SPI1, and the highest reprogramming efficiency required all four factors (Sandler et al. 2014). Both GFI1 and SPI1 are direct downstream targets of RUNX1 and known regulators of embryonic hematopoiesis (Huang et al. 2008; Lancrin et al. 2012). In contrast, FOSB has no known role in the specification of either HECs or EHT, but our results suggest that FOSB may have facilitated reprogramming by inducing the expression of TGFβ morphological-related genes. The dosage of TFs, particularly in a nonpermissive chromatin state, is also likely critical to reprogramming efficiency. In this study, increasing the concentration of AP-1 and SMAD factors in the nucleus with TGFβ3 ligand addition improved reprogramming efficiency of fetal ECs, which have a nonpermissive chromatin state. This is likely due to sufficient levels of phosphorylated SMAD and AP-1 factors for RUNX1 to direct them to critical inaccessible EHT/EndoMT-related TGFβ target genes.
Several studies have shown inhibiting TGFβ signaling could augment HSPC production (Wang et al. 2012; Vargel et al. 2016; Lis et al. 2017), directly contradicting our results. One possible explanation for this discrepancy is that the common TGFβ antagonist (SB431542) used in the former studies has since been shown to increase the amount of phosphorylated SMAD2/3 in some systems (Thambyrajah et al. 2018). It is also possible that the differences in the TGFβ dose and duration of TGFβ exposure are responsible for this discrepancy. Cellular responses to TGFβ are highly sensitive to both dose and duration of exposure. Short exposures to TGFβ produce transient, graded responses, while longer exposure and higher concentrations of TGFβ produce sustained, switch-like responses (Zi et al. 2011). We found that short exposure to a TGFβ dose predicted to induce a transient, graded response (104 molecules/cell) (Zi et al. 2011) was the optimal dose for inducing EHT, whereas a higher TGFβ dose that is known to induce a sustained response (106 molecules/cell) (Zi et al. 2011) did not stimulate EHT. Based on our calculations, it appears that other groups likely used concentrations of TGFβ ligand (>106 molecules of TGFβ per cell) that failed to increase EHT in our system (Wang et al. 2012; Vargel et al. 2016). We hypothesize that transient activation of TGFβ signaling and RUNX1-dependent recruitment of SMAD2/3 along with AP-1 augments hematopoietic cell fate specification in the early phases of EHT, but excessive prolonged TGFβ signaling might derail EHT.
Defining a plastic cellular state that is competent to be respecified and establishing methods to induce competency with minimal manipulation of the cells will be imperative to enhance the efficiency, durability, and safety of cellular reprogramming. This work provides insight into not only regulation of EHT and reprogramming of ECs to HSCs, but additionally any reprogramming of an epithelial cell that must undergo EMT or EndoMT. The ability to reprogram cells into HSC-producing HECs that can be expanded in vitro will aid in disease modeling, drug discovery, and clinical applications.
Materials and methods
Data and code availability
The data generated in this study are available at Gene Expression Omnibus (GEO) under accession number GSE174591.
Mice
This study was performed in accordance with the approved institutional animal care and use committee (IACUC) protocol 803789 of the University of Pennsylvania. We obtained Tg(Cdh5-CreERT) [Tg(Cdh5-cre/ERT2)1Rha] mice from Ralf Adams (Sörensen et al. 2009), and RUNX1 conditional knock-in mice (Rosa26Runx1/Runx1) from Qiufu Ma (Qi et al. 2017). Timed matings were performed between Rosa26Runx1/Runx1, Rosa26Runx1/+, or +/+ females and Tg(Cdh5-CreERT); Rosa26Runx1/Runx1 males to obtain E13.5 fetuses. B6C3F1 females were purchased from Charles River Laboratories, superovulated, and mated to Tg(Cdh5-CreERT); Rosa26Runx1/Runx1 males to obtain E9.5 embryos. For superovulation, 3-wk-old B6C3F1 females were injected intraperitoneally with 5 IU of pregnant mare serum gonadotropin (Prospec Protein Specialists) 2 d prior to mating. Forty-eight hours later, the B6C3F1 females were injected intraperitoneally with 5 IU of human chorionic gonadotropin (Sigma-Aldrich) and placed in cages with males overnight for mating. For embryonic and fetal experiments, pregnant dams were injected intraperitoneally with 2 mg of tamoxifen (Sigma-Aldrich). One-month-old mice received 2 mg of tamoxifen via oral gavage.
Cell preparation
Embryonic and fetal cells were prepared for flow cytometry, cell sorting, and progenitor assays as follows: Embryos and fetuses were separated from yolk sacs and dissociated individually in 0.125% collagenase type I (Sigma-Aldrich) for 20–60 min at 37°C. Samples were washed with 10% FBS + PBS and filtered. Fetuses and postnatal organs were minced prior to enzyme digestion. For fetal organ flow assays, E13.5 fetuses were dissected by organ and dissociated separately as described above.
Fluorescence-activated cell sorting and flow cytometry
The Foxp3/transcription factor staining buffer set (eBioscience) was used to fix and permeabilize cells for RUNX1 intracellular staining. Cells were blocked with Fc block prior to immunostaining. All antibodies used are listed in Supplemental Table S4. Cells were analyzed on an LSR II flow cytometer (BD Biosciences), and data were analyzed with FlowJo or FCS Express. Cells were sorted on a BD Influx with a 100-µm nozzle.
Limiting dilution hemogenic endothelium assay
OP9 stromal cells were cultured in aMEM containing 10% FBS and 1× penicillin–streptomycin. One day prior to initiating the HE assays, OP9 cells were plated (4000 cells/well) in 96-well plates. On day 1, sorted ECs were plated in limiting dilutions with three to 10 replicates for each dilution on confluent OP9s. For TGFβ stimulation, ECs were washed, 102–106 molecules of TGFβ3 ligand (R&D Systems) or vehicle (4 mM HCL + 0.1% BSA) per cell were added to cells, and cells were incubated for 45 min at 37°C and 5% CO2, then washed, and plated in limiting dilutions on confluent OP9 cells. The media was supplemented with 10 ng/mL SCF, 10 ng/mL IL7, 10 ng/mL IL3, and 10 ng/mL Flt3L (PeproTech). On days 7–13, cells were then harvested and analyzed by flow cytometry for positive wells (CD41+, CD45+, and/or Ter119+). The HEC frequency and P-values were calculated with Extreme Limiting Dilution Analysis (ELDA) software (Hu and Smyth 2009).
Additional experimental details are in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank Andrea Stout and Jasmine Zhao at the Cell and Developmental Biology Microscopy Core for confocal microscopy assistance; William DeMuth, William Murphy, and Andrew Morschauser in the Flow Cytometry and Cell Sorting Resource Laboratory for cell sorting assistance; Peter Relich for assistance with STORM data processing; Qiufu Ma and Zijing Liu for the inducible Rosa26 Runx1 mice; and Ralf Adams for the Cdh5-Cre mice. This work was supported by R01HL091724 (to N.A.S.), U01HL100405 (to N.A.S.), R01HD089245 (to K.T. and N.A.S.), 1F31HL150952-01 (to E.D.H.), T32 HD083185 (to E.D.H.), 1F31HL120615 (to A.D.Y.), T32 DK07780 (to A.D.Y.) and T32CA09140 (to A.D.Y.).
Author contributions: K.T., N.A.S., and E.D.H. conceived the project and wrote the manuscript. All authors discussed the results and revised the manuscript. E.D.H., A.D.Y., and R.L. performed experiments and the functional assays. E.D.Y., P.G., and A.D.Y. generated the genomics data. E.D.H. and B.H. performed bioinformatics analyses. M.L. and S.R. provided resources and supervision.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348738.121.
Competing interest statement
The authors declare no competing interests.
References
- Boisset JC, Van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. 2010. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464: 116–120. 10.1038/nature08764 [DOI] [PubMed] [Google Scholar]
- Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. 2005. Hematopoietic stem cell fate is established by the Notch – Runx pathway. Genes Dev 19: 2331–2342. 10.1101/gad.1337005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49: 825–837. 10.1016/j.molcel.2013.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camenisch TD, Molin DGM, Person A, Runyan RB, Gittenberger-de Groot AC, McDonald JA, Klewer SE. 2002. Temporal and distinct TGFβ ligand requirements during mouse and avian endocardial cushion morphogenesis. Dev Biol 248: 170–181. 10.1006/dbio.2002.0731 [DOI] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. 2009. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457: 887–891. 10.1038/nature07619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alonzo RC, Selvamurugan N, Karsenty G, Partridge NC. 2002. Physical interaction of the activator protein-1 factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter activation. J Biol Chem 277: 816–822. 10.1074/jbc.M107082200 [DOI] [PubMed] [Google Scholar]
- Eliades A, Wareing S, Marinopoulou E, Fadlullah MZH, Patel R, Grabarek JB, Plusa B, Lacaud G, Kouskoff V. 2016. The hemogenic competence of endothelial progenitors is restricted by Runx1 silencing during embryonic development. Cell Rep 15: 2185–2199. 10.1016/j.celrep.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreras C, Lancrin C, Lie-A-Ling M, Kouskoff V, Lacaud G. 2011. Identification and characterization of a novel transcriptional target of RUNX1/AML1 at the onset of hematopoietic development. Blood 118: 594–597. 10.1182/blood-2010-06-294124 [DOI] [PubMed] [Google Scholar]
- Gao P, Chen C, Howell ED, Li Y, Tober J, Uzun Y, He B, Gao L, Zhu Q, Siekmann AF, et al. 2020. Transcriptional regulatory network controlling the ontogeny of hematopoietic stem cells. Genes Dev 34: 950–964. 10.1101/gad.338202.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai JI, Chen LF, Kanno T, Ohtani-Fujita N, Kim WY, Guo WH, Imamura T, Ishidou Y, Fukuchi M, Shi MJ, et al. 1999. Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Cα promoter. J Biol Chem 274: 31577–31582. 10.1074/jbc.274.44.31577 [DOI] [PubMed] [Google Scholar]
- Hu Y, Smyth GK. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347: 70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, et al. 2008. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet 40: 51–60. 10.1038/ng.2007.7 [DOI] [PubMed] [Google Scholar]
- Huang K, Gao J, Du J, Ma N, Zhu Y, Wu P, Zhang T, Wang W, Li Y, Chen Q, et al. 2016. Generation and analysis of GATA2w/eGFP human ESCs reveal ITGB3/CD61 as a reliable marker for defining hemogenic endothelial cells during hematopoiesis. Stem Cell Reports 7: 854–868. 10.1016/j.stemcr.2016.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Meng A. 2021. TGFβ family signaling and development. Development 148: dev188490. 10.1242/dev.188490 [DOI] [PubMed] [Google Scholar]
- Kauts ML, Vink CS, Dzierzak E. 2016. Hematopoietic (stem) cell development—how divergent are the roads taken? FEBS Lett 590: 3975–3986. 10.1002/1873-3468.12372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K, Herbomel P. 2010. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464: 112–115. 10.1038/nature08761 [DOI] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. 2014. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15: 178–196. 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. 2009. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457: 892–895. 10.1038/nature07679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancrin C, Mazan M, Stefanska M, Patel R, Lichtinger M, Costa G, Vargel Ö, Wilson NK, Möröy T, Bonifer C, et al. 2012. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood 120: 314–322. 10.1182/blood-2011-10-386094 [DOI] [PubMed] [Google Scholar]
- Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. 2007. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet 8: 104–115. 10.1038/nrg2041 [DOI] [PubMed] [Google Scholar]
- Li Z, Schulz MH, Look T, Begemann M, Zenke M, Costa IG. 2019. Identification of transcription factor binding sites using ATAC-seq. Genome Biol 20: 45. 10.1186/s13059-019-1642-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati NT, Datto MB, Frederick JP, Shen X, Wong C, Rougier-Chapman EM, Wang XF. 1999. Smads bind directly to the Jun family of AP-1 transcription factors. Proc Natl Acad Sci 96: 4844–4849. 10.1073/pnas.96.9.4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtinger M, Ingram R, Hannah R, Müller D, Clarke D, Assi SA, Lie-A-Ling M, Noailles L, Vijayabaskar MS, Wu M, et al. 2012. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J 31: 4318–4333. 10.1038/emboj.2012.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie-A-Ling M, Marinopoulou E, Li Y, Patel R, Stefanska M, Bonifer C, Miller C, Kouskoff V, Lacaud G. 2014. RUNX1 positively regulates a cell adhesion and migration program in murine hemogenic endothelium prior to blood emergence. Blood 124: e11–e20. 10.1182/blood-2014-04-572958 [DOI] [PubMed] [Google Scholar]
- Lis R, Karrasch CC, Poulos MG, Kunar B, Redmond D, Duran JGB, Badwe CR, Schachterle W, Ginsberg M, Xiang J, et al. 2017. Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature 545: 439–445. 10.1038/nature22326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Yang Z, Yu D, Lin J, Cai W. 2020. RUNX1 regulates TGF-β induced migration and EMT in colorectal cancer. Pathol Res Pract 216: 153142. 10.1016/j.prp.2020.153142 [DOI] [PubMed] [Google Scholar]
- Monteiro R, Pinheiro P, Joseph N, Peterkin T, Koth J, Repapi E, Bonkhofer F, Kirmizitas A, Patient R. 2016. Transforming growth factor B drives hemogenic endothelium programming and the transition to hematopoietic stem cells. Dev Cell 38: 358–370. 10.1016/j.devcel.2016.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. 2005. Non-Smad TGF-β signals. J Cell Sci 118: 3573–3584. 10.1242/jcs.02554 [DOI] [PubMed] [Google Scholar]
- Nadin BM, Goodell MA, Hirschi KK. 2003. Phenotype and hematopoietic potential of side population cells throughout embryonic development. Blood 102: 2436–2443. 10.1182/blood-2003-01-0118 [DOI] [PubMed] [Google Scholar]
- Nakano T, Kodama H, Honjo T. 1994. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science 265: 1098–1101. 10.1126/science.8066449 [DOI] [PubMed] [Google Scholar]
- Nakano H, Liu X, Arshi A, Nakashima Y, Van Handel B, Sasidharan R, Harmon AW, Shin JH, Schwartz RJ, Conway SJ, et al. 2013. Haemogenic endocardium contributes to transient definitive haematopoiesis. Nat Commun 4: 1564. 10.1038/ncomms2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck NA. 1999. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126: 2563–2575. 10.1242/dev.126.11.2563 [DOI] [PubMed] [Google Scholar]
- Obier N, Cauchy P, Assi SA, Gilmour J, Lie-A-Ling M, Lichtinger M, Hoogenkamp M, Noailles L, Cockerill PN, Lacaud G, et al. 2016. Cooperative binding of AP-1 and TEAD4 modulates the balance between vascular smooth muscle and hemogenic cell fate. Dev 143: 4324–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottersbach K. 2019. Endothelial-to-haematopoietic transition: an update on the process of making blood. Biochem Soc Trans 47: 591–601. 10.1042/BST20180320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Huang C, Wu X, Tao Y, Yan J, Shi T, Cao C, Han L, Qiu M, Ma Q, et al. 2017. Hierarchical specification of pruriceptors by runt-domain transcription factor runx1. J Neurosci 37: 5549–5561. 10.1523/JNEUROSCI.0094-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci MA, Cosma MP, Lakadamyali M. 2017. Super resolution imaging of chromatin in pluripotency, differentiation, and reprogramming. Curr Opin Genet Dev 46: 186–193. 10.1016/j.gde.2017.07.010 [DOI] [PubMed] [Google Scholar]
- Robert-Moreno A, Espinosa L, de la Pompa JL, Bigas A. 2005. RBPjκ-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development 132: 1117–1126. 10.1242/dev.01660 [DOI] [PubMed] [Google Scholar]
- Robert-Moreno À, Guiu J, Ruiz-Herguido C, López ME, Inglés-Esteve J, Riera L, Tipping A, Enver T, Dzierzak E, Gridley T, et al. 2008. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J 27: 1886–1895. 10.1038/emboj.2008.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler VM, Lis R, Liu Y, Kedem A, James D, Elemento O, Butler JM, Scandura JM, Rafii S. 2014. Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature 511: 312–318. 10.1038/nature13547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt TM, Zúñiga-Pflücker JC. 2002. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity 17: 749–756. 10.1016/S1074-7613(02)00474-0 [DOI] [PubMed] [Google Scholar]
- Sörensen I, Adams RH, Gossler A. 2009. DLL1-mediated notch activation regulates endothelial identity in mouse fetal arteries. Blood 113: 5680–5688. 10.1182/blood-2008-08-174508 [DOI] [PubMed] [Google Scholar]
- Sundqvist A, Zieba A, Vasilaki E, Herrera Hidalgo C, Söderberg O, Koinuma D, Miyazono K, Heldin CH, Landegren U, Ten Dijke P, et al. 2013. Specific interactions between Smad proteins and AP-1 components determine TGFβ-induced breast cancer cell invasion. Oncogene 32: 3606–3615. 10.1038/onc.2012.370 [DOI] [PubMed] [Google Scholar]
- Taoudi S, Medvinsky A. 2007. Functional identification of the hematopoietic stem cell niche in the ventral domain of the embryonic dorsal aorta. Proc Natl Acad Sci 104: 9399–9403. 10.1073/pnas.0700984104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares ALP, Brown JA, Ulrich EC, Dvorak K, Runyan RB. 2018. Runx2-I is an early regulator of epithelial–mesenchymal cell transition in the chick embryo. Dev Dyn 247: 542–554. 10.1002/dvdy.24539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambyrajah R, Fadlullah MZH, Proffitt M, Patel R, Cowley SM, Kouskoff V, Lacaud G. 2018. HDAC1 and HDAC2 modulate TGF-β signaling during endothelial-to-hematopoietic transition. Stem Cell Reports 10: 1369–1383. 10.1016/j.stemcr.2018.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanOudenhove JJ, Medina R, Ghule PN, Lian JB, Stein JL, Zaidi SK, Stein GS. 2016. Transient RUNX1 expression during early mesendodermal differentiation of hESCs promotes epithelial to mesenchymal transition through TGFB2 signaling. Stem Cell Reports 7: 884–896. 10.1016/j.stemcr.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargel Ö, Zhang Y, Kosim K, Ganter K, Foehr S, Mardenborough Y, Shvartsman M, Enright AJ, Krijgsveld J, Lancrin C. 2016. Activation of the TGFβ pathway impairs endothelial to haematopoietic transition. Sci Rep 6: 1–15. 10.1038/srep21518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voon DCC, Thiery JP. 2017. The emerging roles of RUNX transcription factors in epithelial-mesenchymal transition. Adv Exp Med Biol 962: 471–489. 10.1007/978-981-10-3233-2_28 [DOI] [PubMed] [Google Scholar]
- Wang C, Tang X, Sun X, Miao Z, Lv Y, Yang Y, Zhang H, Zhang P, Liu Y, Du L, et al. 2012. TGFβ inhibition enhances the generation of hematopoietic progenitors from human ES cell-derived hemogenic endothelial cells using a stepwise strategy. Cell Res 22: 194–207. 10.1038/cr.2011.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn A, Marquez EJ, Lawlor N, Stitzel ML, Ucar D. 2019. BiFET: sequencing bias-free transcription factor footprint enrichment test. Nucleic Acids Res 47: e11–. 10.1093/nar/gky1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yzaguirre AD, Howell ED, Li Y, Liu Z, Speck NA. 2018. Runx1 is sufficient for blood cell formation from non-hemogenic endothelial cells in vivo only during early embryogenesis. Development 145: dev158162. 10.1242/dev.158162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Luo M, Cai W, Zhou S, Feng D, Xu C, Wang H. 2018. Runt-related transcription factor 1 (RUNX1) promotes TGF-β-induced renal tubular epithelial-to-mesenchymal transition (EMT) and renal fibrosis through the PI3K subunit p110δ. EBioMedicine 31: 217–225. 10.1016/j.ebiom.2018.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi Z, Feng Z, Chapnick DA, Dahl M, Deng D, Klipp E, Moustakas A, Liu X. 2011. Quantitative analysis of transient and sustained transforming growth factor-β signaling dynamics. Mol Syst Biol 7: 492. 10.1038/msb.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, et al. 2008. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 3: 625–636. 10.1016/j.stem.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available at Gene Expression Omnibus (GEO) under accession number GSE174591.