Abstract
The mechanism of conversion of the human sperm basal body to a centrosome after fertilization, and its role in supporting human early embryogenesis, has not been directly addressed so far. Using proteomics and immunofluorescence studies, we show here that the human zygote inherits a basal body enriched with centrosomal proteins from the sperm, establishing the first functional centrosome of the new organism. Injection of human sperm tails containing the basal body into human oocytes followed by parthenogenetic activation, showed that the centrosome contributes to the robustness of the early cell divisions, increasing the probability of parthenotes reaching the compaction stage. In the absence of the sperm-derived centrosome, pericentriolar material (PCM) components stored in the oocyte can form de novo structures after genome activation, suggesting a tight PCM expression control in zygotes. Our results reveal that the sperm basal body is a complex organelle which converts to a centrosome after fertilization, ensuring the early steps of embryogenesis and successful compaction. However, more experiments are needed to elucidate the exact molecular mechanisms of centrosome inheritance in humans.
Keywords: centrosome, microtubule organizing centers, fertilization, embryo early development, compaction, centriole, pericentriolar material, human
Introduction
The centrosome is the main microtubule organizing center (MTOC) of the cell. It consists of two centrioles surrounded by a mass of proteins known as pericentriolar material (PCM) (Rusan and Rogers, 2009). Centrosomes are important for intracellular organization, spindle assembly, asymmetric cell division and polarity establishment as well as for the assembly of cilia and flagella. They are therefore essential for tissue architecture and function, and the development of healthy organisms (Wu and Akhmanova, 2017). Defects in centrosome function and/or number are associated with conditions including cancer, ciliopathies and infertility. In cycling cells, the number of centrosomes is tightly controlled in a cell cycle-dependent manner. Centrioles duplicate once (and only once) during interphase, and in mitosis, the duplicated centrosomes localize each at one pole of the bipolar spindle defining spindle organization and orientation. After cell division, each daughter cell inherits one of the two centrosomes (Nigg and Stearns, 2011).
Fertilization entails the fusion of the male and the female gametes. In mammals, the regulation of the centrosome number per cell is established during fertilization. Indeed, human oocytes do not contain centrioles (Sathananthan, 1997), while the sperm has two centrioles (proximal and distal centrioles) in the midpiece that function as the basal body of the flagella (Familiari et al., 2006). After fertilization, the paternal pronucleus is associated with a centriole, suggesting that the centrosome is paternally inherited during fertilization (Sathananthan et al., 1991; Simerly et al., 1995; Van Blerkom and Davis, 1995). Studies on fertilized eggs from various species that also inherit the centrosome paternally (i.e. rhesus monkeys, drosophila and cows) showed that the sperm basal body suffers a process known as ‘centrosome reduction’ (Manandhar et al., 2000). This process consists of the elimination of the PCM and the remodeling of the centriolar microtubules, generating an atypical centrosome. The current model is that upon fertilization, the sperm atypical centrioles recruit PCM components stored in the oocyte cytoplasm to assemble a functional centrosome. A very limited number of studies have addressed this process in humans, except for a few reports showing that the human sperm shares many characteristics with those of other species (Sathananthan et al., 1996; Simerly et al., 1999; Manandhar et al., 2000; Fishman et al., 2018; Khanal et al., 2021). Indeed, the human sperm distal centriole is also remodeled, although it retains a subset of centrosomal proteins, suggesting that sperm PCM reduction is not completed in humans (Fishman et al., 2018; Khanal et al., 2021). Nevertheless, the degree of conservation or loss of PCM components at the human sperm basal body, and the functional implications of these proteins for early embryonic development are still unresolved questions.
The oocyte cytoplasm provides a large store of maternal components, including proteins and mRNAs that are essential for the first embryonic divisions, that occur in the absence of transcription (Conti and Franciosi, 2018). Little is known about the storage of maternal PCM components in oocytes and how they contribute to the transition of the sperm basal body into the centrosome of the zygote. Many of these studies have been performed in mice and, although this is an excellent model to study early embryonic development, it is not ideal to study the centrosome because these phases occur in the absence of centrioles, which form de novo at the blastocyst stage in mice (Hiraoka et al., 1989). The early embryonic cell divisions in mice are instead supported by acentriolar MTOCs (aMTOCs) (Gueth-Hallonet et al., 1993; Courtois et al., 2012; Howe and FitzHarris, 2013). This suggests that although centrosomes are essential for healthy organisms, they may not be needed during the early embryo cell divisions, which may rely on spindle assembly acentrosomal pathways acting during oocyte meiosis.
In this work, we first determined the protein composition of the human sperm basal body by proteomics and super-resolution microscopy, and then devised a functional approach to study the role of the sperm-derived centrosome in the early cell divisions of parthenotes. Our results suggest that the human sperm provides the zygote with centrioles and an important mass of centrosomal proteins, which may be important for the assembly of the first functional centrosome of the new organism. Furthermore, our data suggest that the sperm-derived centrosome increases the robustness of the first cell divisions of the parthenotes, leading to compaction. Our data also suggest that the expression of centrosomal proteins in the early parthenotes has to be tightly regulated to avoid the untimely formation of spontaneous MTOC-like aggregates in the embryonic cells. These findings not only improve our understanding of centrosome biology during fertilization and early development but also provide novel putative avenues to explore the causes of early embryo arrest and idiopathic infertility in humans.
Materials and methods
Ethics
Approval to conduct this study was obtained from the ethical committee of the Parc de Recerca Biomèdica of Barcelona (PRBB), Barcelona, Spain. All procedures performed were in accordance with the ethical standards of the institutional research committees and with the 1964 Helsinki declaration of the Ethical principles for medical research involving human subjects, as revised in 2013 in Fortaleza (World Medical Association, 2013). Written informed consent to participate was obtained from all participants prior to their inclusions in the studies. All oocyte donors are between 18 and 35 years old, have a BMI <30 kg/m2, normal karyotype and adequate ovarian reserve, and did not present systemic or reproductive conditions. Sperm samples were obtained from donors that are between 35 and 51 years old, have a BMI <30 kg/m2 and the semen analysis was normal.
Sperm thawing and swim-up
To thaw sperm samples, straws were incubated at 37°C for 5 min. To remove the sperm cryoprotectant, the sample was diluted with the same volume of Sperm Rinse (Vitrolife, Göteborg) and centrifuged for 5 min at 300 g at room temperature. Then, the pellet was washed twice with 1 ml of Sperm Rinse (Vitrolife, Göteborg) and centrifuged at 372 g for 5 min at room temperature. To perform the sperm swim-up, 0.1 ml of Sperm Rinse was carefully loaded on the top of the pellets. The tubes were oriented at 45° for 10 min at room temperature. During this time, the motile spermatozoa swam to the Sperm Rinse fraction. Finally, the Sperm Rinse fraction was carefully collected avoiding the aspiration of the pellet. For the tails-injected experiment, after thawing the sperm straw, the sample was diluted with 3 ml of Sperm Rinse and centrifuged once at 300 g for 5 min. Immediately, after this wash, the sperm swim-up was performed as previously described.
Sperm centriole enrichment
Two different methods were used to analyze sperm centrosomal composition. Only normozoospermic samples with >50% of A + B motility were used. The first approach was based on a previously published protocol (Firat-Karalar et al., 2014) but with some modifications. Frozen samples were thawed and washed twice with PBS and checked under the microscope for any defects caused by the freezing and/or thawing cycles (if any defect was detected, they were discarded). The total amount of cells used at each experiment was of ≈60 million. Washed samples were pelleted at 850 g, for 10 min and resuspended with 350 µl of PBS. Then, samples were sonicated 5 times at 70% output for 15 s, with 30 s intervals (Bioruptor). Samples (1 µl of each) were taken to check the sonication efficiency under a bright-field microscope. Sperm tails were separated from the heads through a 30% sucrose cushion (centrifugation at 200 g for 10 min at 4°C). A sample of the tails fraction was taken (1 µl) and squashed under an 18 × 18 mm coverslip to check its purity. To sequentially extract sperm tails proteins, the tails fraction was diluted with the same volume of Buffer 1 (100 mM Tris HCl pH 8, 4 mM EGTA, 4 mM EDTA, 1000 mM NaCl, 2% NP40, 0.2% β-mercaptoethanol, 2 mM DTT, protease inhibitors in ddH2O) and incubated 1 h at 4°C with gentle agitation. After this incubation, the sample was centrifuged at maximum velocity (16 000 g) for 10 min at 4°C. The pellet and the supernatant were separated in two different tubes to be treated differentially. In the supernatant, the proteins solubilized by buffer 1 are found. A total of 125 µl of 100% trichloroacetic acid was added to the supernatant and kept at 4°C for 10 min. To pellet the precipitated proteins, the sample was centrifuged at maximum velocity at 4°C, 5 min. The supernatant was removed, and the pellet was washed twice with cold acetone and then dried at 95°C. Finally, the pellet was diluted with 6 M urea and 200 mM of ABC (ammonium bicarbonate, Sigma, MO, USA) in ddH2O. The pellet fraction from the first extraction was sequentially extracted with extraction buffer 2 (50 mM Tris HCl pH 8, 600 mM KSCN, 2 mM DTT, protease inhibitors in ddH2O) and extraction buffer 3 (50 mM Tris HCl pH 8, 4 M urea, protease inhibitors in ddH2O). Three different extractions diluted in 6 M urea and 200 mM ABC were finally obtained for proteomic analysis.
For the second approach, sonicated tails were pelleted at 4°C at the maximum velocity and diluted with LB (Loading Buffer, 2% w/v sodium dodecyl sulphate (SDS), 10% Glycerol, 50 mM Tris HCl pH 6.8, 5% β-mercaptoethanol). Samples were run on a precasted gradient gel (4–15% Criterion TGX, 12 + 2 wells, 45 μl, BioRad, CA, USA) for 45 min at 60 mA. Gels were stained for 10 min using Coomassie (Coomassie Brilliant Blue R250 (Thermo Fisher Scientific, WA, USA), 10% acetic acid, 50% methanol) and destained with 10% methanol and 10% acetic acid. Gels were cut into nine bands to process for proteomic analysis.
Mass spectrometry
Soluble samples were reduced with 10 mM DTT, 37°C, 60 min and alkylated in the dark with iodoacetamide (IAM, 20 mM, 25°C, 30 min). The resulting protein extract was first diluted to 2M urea for overnight digestion with LysC (Wako, USA) at 37°C and then diluted 2-fold for 8 h digestion with trypsin (Promega, USA) at 37°C.
Gel band samples were destained with 40% acetonitrile (ACN)/100 mM ABC, reduced with 10 mM DTT at 56°C for 30 min and alkylated in the dark with IAM (55 mM, 25°C, 30 min). Gel bands were then dehydrated with ACN and digested overnight with trypsin at 37°C.
After digestion, the peptide mix was acidified with formic acid and desalted with a MicroSpin C18 column (The Nest Group, Inc, MA, USA) prior to liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis.
Samples were analyzed using an LTQ-Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific, WA, USA) coupled to an EasyLC (Thermo Fisher Scientific (Proxeon), Odense, Denmark). Peptides were loaded onto the 2-cm Nano Trap column with an inner diameter of 100 μm packed with C18 particles of 5 μm particle size (Thermo Fisher Scientific, WA, USA) and were separated by reversed-phase chromatography using a 25-cm column with an inner diameter of 75 μm, packed with 1.9 μm C18 particles (Nikkyo Technos Co., Ltd. Japan). Chromatographic gradients started at 93% buffer A and 7% buffer B with a flow rate of 250 nl/min for 5 min and gradually increased to 65% buffer A and 35% buffer B in 60 min for the in solution samples and 120 min for the gels. After each analysis, the column was washed for 15 min with 10% buffer A and 90% buffer B (Buffer A: 0.1% formic acid in water. Buffer B: 0.1% formic acid in ACN).
The mass spectrometer was operated in data-dependent acquisition (DDA) mode and full MS scans with 1 micro scans at resolution of 60 000 were used over a mass range of m/z 350-2000 with detection in the Orbitrap. Auto gain control (AGC) was set to 1E6, dynamic exclusion (60 s) and charge state filtering disqualifying singly charged peptides was activated. In each cycle of DDA analysis, following each survey scan the top 20 most intense ions with multiple charged ions above a threshold ion count of 5000 were selected for fragmentation at normalized collision energy of 35%. Fragment ion spectra produced via collision-induced dissociation were acquired in the Ion Trap: AGC was set to 5e4, isolation window of 2.0 m/z, activation time of 0.1 ms and maximum injection time of 100 ms were used. All data were acquired with Xcalibur software (v2.2, Thermo Fisher Scientific, WA, USA).
For peptide identification, proteome Discoverer software suite (v1.4, Thermo Fisher Scientific) and the Mascot search engine (v2.5, Matrix Science (Perkins et al., 1999)) were used. Samples were searched against a Swiss-Prot human database plus a list of common contaminants and all the corresponding decoy entries (20 797 entries). Trypsin was chosen as enzyme and a maximum of three miscleavages were allowed. Carbamidomethylation (C) was set as a fixed modification, whereas oxidation (M) and acetylation (N-terminal) were used as variable modifications. Searches were performed using a peptide tolerance of 7 ppm and a product ion tolerance of 0.5 Da. Resulting data files were filtered for a false discovery rate <5%.
Our proteomic datasets are available on PRIDE (PXD028348).
Oocyte warming
Oocytes were warmed according to a standard procedure (Cryotop, Kitazato, BioPharma Co., Ltd; Japan). Briefly, thawing solution (TS) medium was prewarmed at 37°C for 45 min prior to use. Diluent solution (DS) and washing solution (WS) were used at room temperature (Kitazato). The oocyte straw cap was carefully removed and then quickly immersed in a dish with 1 ml of TS medium for 1 min. Then, oocytes were incubated in DS medium for 3 min. Finally, oocytes were placed to the WS medium for 5 min and transferred to a second plate containing WS for 1 min before incubating them for 2 h at 37°C, in a 6% CO2 atmosphere to let them recover from warming.
Sperm tails injection
In an ICSI dish of 60 mm of diameter, several drops of G-MOPS plus containing one oocyte each (Vitrolife, Gothenburg, Sweden) surrounded a central PVP drop (polyvinylpyrrolidone; Vitrolife, Gothenburg, Sweden) containing the sperm swim-up sample. The plate was overlaid with mineral oil (FujiFilm, Tokyo, Japan) to prevent the evaporation of the drops and then placed on the ICSI microscope stage (Olympus IX50, Olympus, Tokyo, Japan). Two different pipettes were needed to perform tails separation and injection into oocytes. A PZD (Partial Zona Dissection, Vitrolife, Gothenburg, Sweden) pipette was used for the sperm head-tail separation, and an ICSI micropipette (Vitrolife, Gothenburg, Sweden) to perform the tail injection into the oocyte. Both pipettes were located in a double needle holder. The PZD pipette was placed at the interface between the sperm head and the midpiece. With an accurate blow, the separation of both parts was achieved. Immediately after the separation, the tails were collected. To confirm that the separated tails had the centrosome and not DNA, tails were aspirated with the ICSI micropipette and loaded onto a glass microscope slide (25 mm × 75 mm) and fixed with 4% paraformaldehyde (PFA) to immunodetect centrosomes and DNA. In all the cases, only one tail was injected per oocyte. A certain number of oocytes were sham injected as controls.
Oocyte activation
Tail-injected and control oocytes were washed four times with G1-PLUS™ medium (Vitrolife, Gothenburg, Sweden) and incubated for 30 min at 37°C, 6% CO2. The oocyte activation protocol (AOA) included three 10-min incubations in 10 µM ionomycin (calcium ionophore, MP Biomedicals, CA, USA), and three 30-min washes in G1-PLUS™ medium (Vitrolife, Gothenburg, Sweden) at 37°C, 6% CO2. All the plates were covered with OVOIL™. Finally, oocytes were transferred to a plate with SAGE medium (Origio, Malov, Denmark) at 37°C, 6% CO2 to be placed in the time-lapse system: Primovision or Embryoscope (Vitrolife, Gothenburg, Sweden). These two systems took images of each sample every 5 min, allowing for morphologic and the kinetic analyses.
DNA cloning and transfection
DNA sequences of the 10 uncharacterized proteins were obtained from the ORFeome service (Centre for Genomic Regulation). These sequences were cloned into a pDEST vector that contained either cyan fluorescent protein at the N-terminus (Kanamycin resistant) or green fluorescent protein at the C-terminus (Ampicillin resistant) (Addgene, MA, USA).
HeLa cells (CCL2; ATCC, VA, USA) were grown at 37°C in a 5% CO2 atmosphere in DMEM 4.5 g/l Glucose, supplemented with Ultraglutamine (Lonza, Basilea, Switzerland), with 10% fetal bovine serum (Invitrogen, CA, USA), 100 units/ml of penicillin and 100 μg/ml of streptomycin. HeLa cells were regularly checked for mycoplasma contamination. DNA were transfected with the same volume of X-tremeGENE (Sigma, MO, USA) and 100 µl of Opti-MEM (Thermo Fisher Scientific, WA, USA) using 500 ng DNA per well in a 12-well plate (150 000 cells/well) of HeLa cells already attached to a glass coverslip (18 mm diameter). After 24 h of protein expression (at 37°C in a 5% CO2 atmosphere), cells were collected and washed twice with 1 ml of PBS prior to fixation with cold methanol (10 min) or 4% PFA (15 min, Sigma, MO, USA).
Sperm immunofluorescence
To perform sperm immunofluorescence (IF) analysis, thawed sperm samples were washed twice with PBS and then loaded onto a poly-l-lysine coated glass coverslip (12 mm of diameter—from 50 000 to 100 000 cells). After 30 min, the supernatant was removed and the cells were fixed with either 4% PFA for 1 h, or methanol for 10 min. After several washes with PBS, samples were permeabilized with 0.5% Triton X-100 PBS for 15 min at room temperature. Samples were loaded with 5% BSA in PBS to block nonspecific interactions for 2 h. The same blocking solution was used to incubate the primary and the secondary antibody for 1 h and 45 min, respectively, and mounted in 10% Mowiol (Sigma, MO, USA) in 0.1M Tris HCl at pH 8.2, 25% glycerol. Confocal images were obtained using a TCS-SP5 microscope (Leica Microsystems, Wetzlar, Germany) with a 63× objective. Lasers and spectral detection bands were chosen for the optimal imaging of Alexa Fluor 488 and 568 signals. Two-channel colocalization analysis was performed using ImageJ (National Institutes of Health, MD, USA). Stimulated emission depletion microscopy (STED) images were taken on a TCS SP8 STED3X microscope (Leica Microsystems, Wetzlar, Germany).
Oocyte and parthenote IF
In a prewarmed plate (37°C), oocytes and parthenotes were mixed with Tyrode’s solution (Sigma, MO, USA) to remove the zona pellucida. Then, a quick wash with prewarmed PBS was performed. Samples were then fixed with prewarmed 4% PFA for 15 min. Oocytes and parthenotes were permeabilized with PBS/0.2% Triton X-100 for 15 min. After washing the samples once with PBS-T (PBS, 0.1% Tween 20) and three more times with PBS-TB (PBS, 0.1% Tween20, 2% BSA Fraction V) for 20 min each, samples were blocked with 5% normal goat serum (Vector Laboratories, CA, USA) in PBS-TB (freshly prepared) for 3 h. All primary antibodies were incubated overnight at 4°C with the blocking solution. Then, three washes of 20 min in PBS-TB were performed while shaking to eliminate the remaining primary antibody. The secondary antibody was only incubated for 1 h at room temperature in PBS-TB together with Hoechst 33342 (1 µg/ml, Invitrogen, CA, USA). Samples were mounted with Vectashield (Vector Laboratories, CA, USA) and visualized using Zeiss 780 confocal/multiphoton microscopy at 63× with 80% glycerol (Carl Zeiss, Oberkochem, Germany).
Cell culture IF
HeLa transfected cells were blocked and permeabilized at the same time with IF medium (0.1% Triton X-100, 2% BSA in PBS 1×) for 30 min. The primary and the secondary antibodies diluted in IF (0.1% Triton X-100, 2% BSA in PBS 1×) medium were placed onto the samples for 50 and 45 min, respectively. The samples mounted with 10% Mowiol (Sigma, MO, USA) were visualized at 63× or with the Leica TCS SP5 upright microscope (Leica Microsystems, Wetzlar, Germany). Two-channel colocalization analysis was performed using ImageJ (National Institutes of Health, MD, USA).
SDS-PAGE and western blot
To perform SDS-PAGE (SDS-polyacrylamide gel electrophoresis), sperm extracts (intact, heads and tails fractions) were obtained by diluting the samples with 1× LB and freezing and boiling the extracts three times. Then, samples were run on 4–20% Protean TGX Precast protein gels (Biorad, CA, USA). To transfer proteins, the iBlot dry system (Thermo Fisher Scientific, WA, USA) was used. Polyvinylidene difluoride membranes were blocked with TBS 5% milk 1 h. The primary and secondary antibodies were diluted with 2% milk in TBS and incubated overnight at 4°C and for 1 h at room temperature, respectively. Blots were developed using the Odyssey Infrared imaging system (LI-COR Biosciences, NE, USA).
Antibodies
The following primary antibodies were used: rabbit anti-centrin (Merck Millipore, MA, USA, 20H5) at 1:100, mouse anti-acetylated tubulin (Sigma, MO, USA, T7451) at 1:1000, rabbit anti-Cep63 (Merck Millipore, MA, USA, 06-1292) at 1:100, rabbit anti-Pericentrin (Abcam, Cambridge, UK, ab448) at 1:500, mouse anti-protamine (Novus Biologicals, CO, USA, H00005619) at 1:100, rabbit anti-β-tubulin (Abcam, Cambridge, UK, ab6046) at 1:1000, mouse anti-α-tubulin (Sigma, MO, USA, DM1A T6199) at 1:1000 in sperm and at 1:100 in oocytes and parthenotes, mouse anti-γ-tubulin (Sigma, MO, USA, GTU-448) at 1:100, rabbit anti-Poc1B (Thermo Fisher Scientific, MA, USA, PA5-24495) at 1:100, rabbit anti-WDR62 (Bethyl Laboratories, TX, USA, A301-560A) at 1:100, rabbit anti-Nek9 (Dr. Joan Roig gift) at 1:100, rabbit anti-Pontin (Home Made) at 1:100 and rabbit anti-Reptin (Home Made) at 1:100. Secondary antibodies anti-rabbit and anti-mouse conjugated to Alexa-488 and 568 (Invitrogen, CA, USA) were used at 1:1000 in sperm and cell culture, and 1:100 in oocyte and parthenotes for IF and 680 (Invitrogen, CA, USA) or IRdye 800 CW (LI-COR Biosciences, NE, USA) at 1:10 000 for western blots. Hoechst 33342 (1 µg/ml, Invitrogen, CA, USA) was used to visualize DNA.
Primo-vision/embryoscope analysis
To analyze the kinetics of parthenote development, the time of cell division and compaction, early pseudo-blastocyst and expanded pseudo-blastocyst was measured in minutes and transformed to hours.
Gene ontology enrichment and STRING analysis
The Gene Ontology (GO) analyses were performed using the ‘Gene Ontology Consortium’
(http://geneontology.org/page/go-enrichment-analysis). Data were confirmed using Uniprot (https://www.uniprot.org) and Protein Atlas (https://www.proteinatlas.org) databases. STRING (version 11.0) was used to perform the protein–protein interaction network (https://string-db.org).
Results
The human sperm centrioles are associated with centrosomal proteins
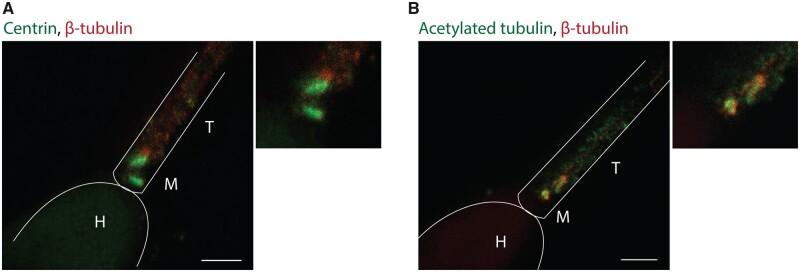
The human sperm basal body consists of a proximal centriole with a conventional microtubule-based organization and a distal centriole that has been described as a degenerated centriole (Avidor-Reiss et al., 2015). In order to get further insights on the nature of these centrioles, we determined centrin and acetylated α-tubulin localization patterns with super-resolution microscopy (STED). Centrin is a structural protein that localizes inside the centrioles and is often used as a centriolar marker (White et al., 2000). We found that centrin localizes to both centrioles (Fig. 1A), suggesting that the distal centriole still retains some basic intrinsic features of a conventional centriole. Acetylated α-tubulin is usually found in long-lived microtubules, such as the centriole microtubules, and they are considered a marker of their structural stability (Amargant et al., 2019). We found that both centrioles contain acetylated tubulin suggesting that they both are stable microtubule assemblies (Fig. 1B). Taken together, our results suggest that the centrioles of the human sperm basal body retain some of the basic features of conventional centrioles.
Figure 1.
Sperm centrioles are stable structures. (A) Super-resolution imaging (STED) of human spermatozoa stained for centrin and β-tubulin. Scale bar: 1 μm. (B) Human sperm immunofluorescence (IF) of acetylated tubulin and β-tubulin in the human sperm centrioles visualized with STED. Scale bar: 1 μm. H, head; M, midpiece; T, tail. N = 2 different sperm samples.
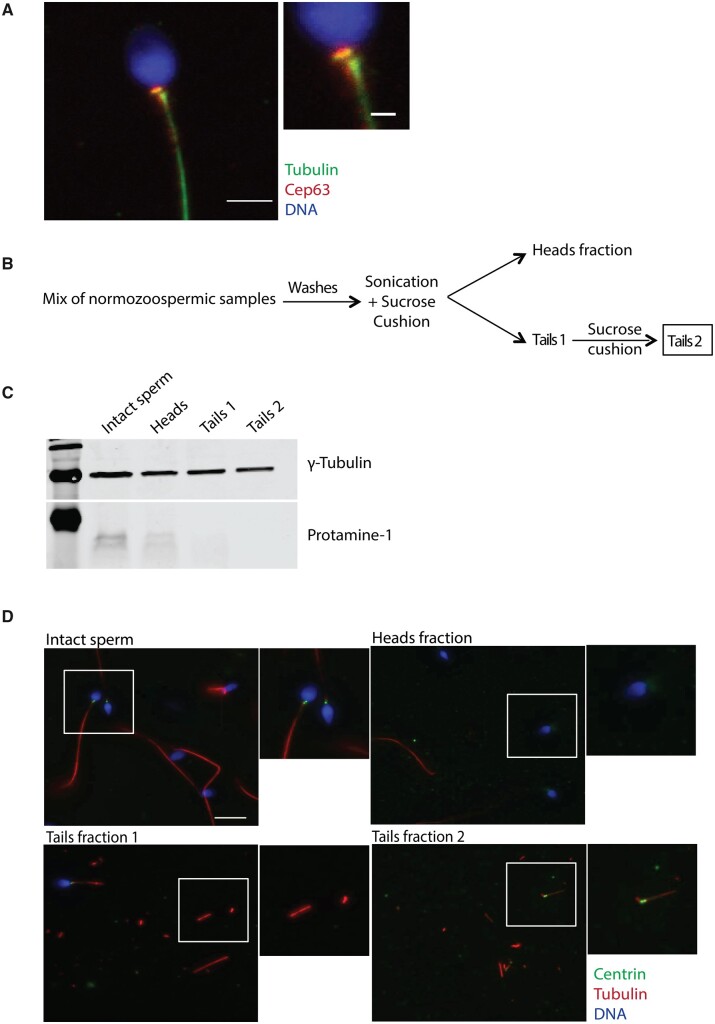
To check whether the sperm centrioles are associated with centrosomal proteins, we performed IF for Cep63, a protein involved in the centrosome duplication cycle (Brown et al., 2013; Watanabe et al., 2016). We found that Cep63 mainly localizes to the proximal centriole in the human sperm (Fig. 2A). These data, in agreement with previous reports (Fishman et al., 2018; Khanal et al., 2021), suggest that the human sperm basal body might be associated with centrosomal proteins. Altogether we conclude that the human sperm basal body includes two centrioles that show some basic features of conventional centrosomes, suggesting that it might not undergo a full process of centrosome reduction and establishing the foundation to perform a large scale approach to identify the human sperm PCM (Manandhar et al., 2000; Manandhar and Schatten, 2000).
Figure 2.
Sperm centrosome enrichment to identify centrosomal components. (A) IF images on human sperm to visualize Cep63, α-tubulin and DNA. Scale bar: 3 μm, inset: 1 μm. N = 4 different sperm samples. (B) Schematic representation of the sonication and enrichment protocol. Only normozoospermic samples with ≥50% of A + B motility were used. N = 3 different experiments. (C) Western blot analysis of cell lysates from the different sperm fractions shown in B to detect heads (protamine 1) and tails (γ-tubulin). N = 3 different experiments (see Supplementary Information for the uncropped blot). (D) Representative images of intact and sperm fractions stained for DNA (blue), centrin (green) and α-tubulin (red). Scale: 10 μm. N = 3 different experiments.
The human sperm basal body is associated with a complex set of centrosomal proteins
To define the complexity of the proteins associated with the human sperm basal body, we used mass spectrometry. Centrosomal proteins are usually in very low abundance compared with other cellular components and they may be even less represented in sperm (Bauer et al., 2016). We therefore aimed at reducing the sample complexity before mass spectrometry by separating the sperm heads from the tails and basal bodies (Amaral et al., 2013; Baker et al., 2013). Human normozoospermic samples were sonicated and centrifuged twice through a 30% sucrose cushion in order to obtain one fraction enriched in heads and another enriched in tails (Fig. 2B). The purity of the tail fraction, evaluated by bright-field microscopy, showed that less than 0.01% of the samples contained a few contaminating heads (Supplementary Fig. S1). To further test the purity of the tail fraction, we checked for the presence of protamines as a marker for chromatin and γ-tubulin as a marker of centrosomes in the head and tail fractions (Fig. 2C and Supplementary Fig. S2). Western blot analysis showed that intact sperm lysates were positive for protamines and γ-tubulin as expected. In contrast, protamines were not detected in the tail fraction whereas γ-tubulin was present. These data confirmed that the tail fraction contains very low levels of head contaminants, if any. We next examined if centrioles were present in the sperm tail fraction by IF analysis (Fig. 2D). Indeed, we could detect a positive signal for centrin at one of the ends of some tail fragments (10%) indicating that basal bodies were recovered in the tail fraction.
In order to identify as many centrosomal proteins as possible, we used different approaches for processing the tail fractions before mass spectrometry analysis. First, we aimed at differentially extracting the centrosomal proteins using a range of detergents, as previously described (Firat-Karalar et al., 2014). The resulting samples derived from three successive extractions steps from three independent experiments were independently analyzed by mass spectrometry, leading to the identification of 1545 proteins with at least two unique peptides. In addition, as a complementary approach, the sperm tails were directly solubilized in LB and the proteins resolved by SDS-PAGE. The gel was excised in nine fragments that were analyzed independently by mass spectrometry. This approach resulted in the identification of 3210 proteins with at least two unique peptides. Altogether we identified 3406 proteins with at least two unique peptides for the human sperm tail proteome with a significant enrichment for basal body and tail proteins (Supplementary Table SI).
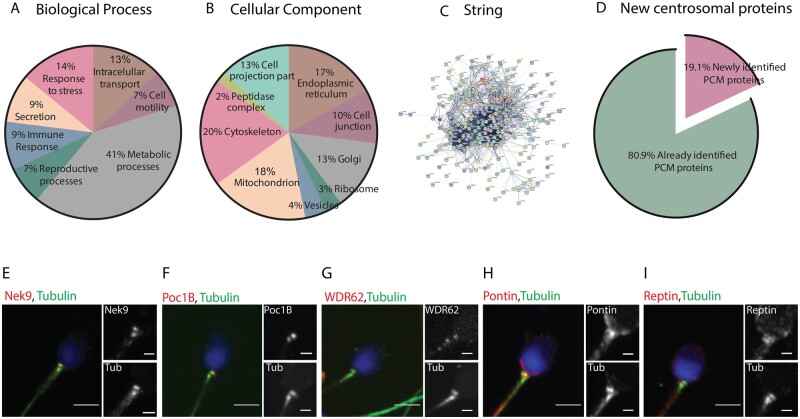
GO analysis based on biological processes and cellular localization showed that many of the 3406 proteins are involved in metabolic processes, response to stress and intracellular transport, in agreement with previous reports (Martinez-Heredia et al., 2006; Baker et al., 2007; Amaral et al., 2013; Baker et al., 2013; Wang et al., 2013; Jumeau et al., 2015). Considering their cellular localization, the majority had GO terms corresponding to mitochondria and cytoskeleton. Others had GO terms corresponding to the Golgi apparatus, the endoplasmic reticulum and vesicles (Fig. 3A and B) (Amaral et al., 2013).
Figure 3.
Classification of the human sperm tail proteins. (A) Gene Ontology of the 3406 proteins based on their biological process. (B) Gene ontology of the same 3406 proteins based on their subcellular localization. (C) String network of the 251 identified centrosomal proteins. (D) Comparison of the pericentriolar material (PCM) proteins only identified in this work (19.1%—pink fraction) to all the previously published human sperm proteomic data (green fraction). (E–I) Antibodies against five of the centrosomal proteins identified in our proteomic analysis and co-stained with tubulin to visualize the centrioles. Nek9 (E), Poc1B (F) and WDR62 (G) co-localize with the proximal and distal centriole. Pontin (H) and Reptin (I) have a diffuse localization around the basal body. Scale: 3 μm, insets: 1 μm. N = 2 different sperm samples.
Interestingly, 170 proteins were associated with the GO terms: centrosome and/or centriole. Extending our analysis to the proteins identified with only one peptide (centrosomal proteins are in very low abundance), we found an additional 81 proteins with these GO terms (Supplementary Table SII). To obtain some supporting evidence for these proteins being centrosomal, we checked them individually in the databases Uniprot and Protein Atlas. In Uniprot, 149 proteins out of the 251 total number of identified centrosomal proteins (97 identified with two unique peptides and 52 with one unique peptide) have a described centrosomal function and/or localization. In Protein Atlas, 116 were also classified as centrosomal (68 identified with two unique peptides and 48 with one unique peptide). We could not find data in these two databases for the remaining 69 proteins (Supplementary Table SII). Then, a protein-protein interaction network using STRING database and the 251 proteins was established, also showing a highly interconnected network (Fig. 3C). In addition, we validated the localization of five centrosomal proteins from our proteomic analysis (Fig. 3E–I). Nek9, Poc1B (POC1 centriolar protein homolog B) and WDR62 (WD repeat-containing protein 62) localized in both centrioles, while Pontin and Reptin, which for a complex (Torreira et al., 2008), had a more diffuse localization around the sperm basal body. Taken altogether, our comprehensive analysis shows that the human sperm basal body is associated with a complex centrosomal proteome, including at least 251 proteins, and we have demonstrated that five of these 251 localize to the basal body in the human sperm. To our knowledge, this is the first time that four of these proteins (Nek9, WDR62, Pontin and Reptin) are visualized at the human sperm.
Interestingly, the functions of the identified centrosomal proteins are very diverse including the regulation of centriole structure and length (centrin, POC1B), microtubule nucleation (tubulin γ-1, GCP2, GCP3 and GCP6), phosphorylation of multiple factors (Nek9 and Aurora C Kinases), centrosome cycle regulation and biogenesis (Cep135, Cep170) and PCM organization (ODF2, PCM1). When we compared our PCM proteins with all the proteins identified until 2018 by proteomics in the human spermatozoa, we found that some of the centrosomal proteins we identified have not been reported previously as present in human sperm (12 of the 170 centrosomal proteins with two peptides and 36 out of the 81 centrosomal proteins with one peptide; in total 48 out of the 251 proteins (19.1%)) (Castillo et al., 2018) (Fig. 3D, Supplementary Table SII).
The proteome of isolated centrosomes from KE37 cells contains a higher number of associated proteins (Bauer et al., 2016). To determine whether we only detected abundant proteins using our approach, we checked the reported abundance of the proteins we identified in the KE37 centrosome proteome database. Data were available for 20 of the proteins we identified, including some abundant ones, such as ODF2 and POC1B, others having intermediate abundance values, such as OFD1 and Cep76, and yet others with low abundance, such as Cep170 and Cep290. These data suggest that we obtained a good representation of the centrosomal proteins associated with the human sperm basal body.
Since we had successfully identified many centrosomal proteins in the human sperm samples, we decided to explore whether any of the 26 proteins identified in at least two independent experiments and currently uncharacterized could be novel centrosomal components (Supplementary Table SIII). We obtained constructs for expression of 10 of them with a fluorescent tag (Supplementary Table SIII) in HeLa cells. IF analysis showed that three of them co-localized with centrin, suggesting that they are novel human centrosomal proteins (Supplementary Fig. S3). Consistently, one of them (C7orf31) was recently identified in bovine sperm and validated through localization studies as a novel centrosomal protein (Firat-Karalar et al., 2014). In summary, we have identified 251 centrosomal proteins and three novel centrosomal components in human sperm. Our results suggest that the human sperm basal body is associated with a complex pericentriolar protein network.
The early embryonic centrosomal proteins are biparentally inherited
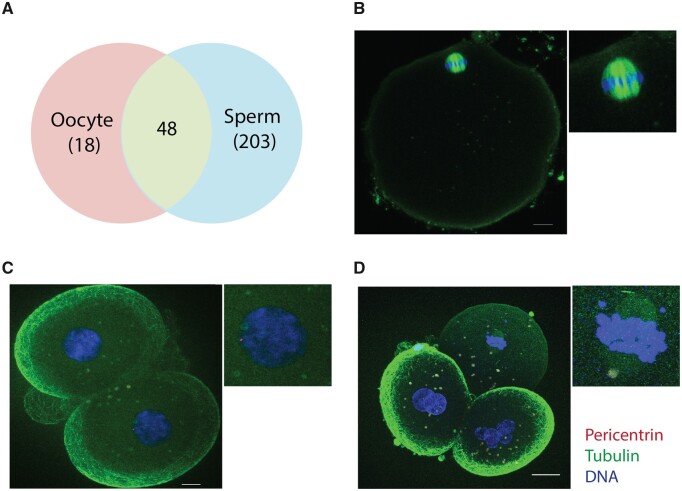
The current view is that centrosomal proteins are recruited from the oocyte cytoplasm by the sperm centrioles to assemble the first centrosome of the future organism. However, our data revealed that the sperm basal body is already associated with a complex network of centrosomal proteins. This suggested that upon fertilization the sperm not only provides half of the chromosomes to the zygote and centrioles but also a high number of centrosomal proteins, suggesting that in fact the first centrosome that assembles in the zygote has a mixed PCM derived from both the sperm basal body and the oocyte cytoplasm. To gain some insights into the centrosomal proteins stored in the human oocyte cytoplasm that may be recruited we first analyzed the 1376 proteins identified in the human oocyte (Virant-Klun et al., 2016). Only 66 of these proteins have the GO terms: centrosome and/or MTOC, including 48 that we identified in the human sperm (Fig. 4A, Supplementary Table SIV). Thus, these data suggest that the first centrosome of the zygote is of biparental origin with some centrosomal proteins coming from the sperm basal body and others being recruited from the oocyte cytoplasm.
Figure 4.
The zygote centrosomal composition is biparentally inherited during fertilization. (A) Venn diagram showing overlap of 48 centrosomal proteins in the human oocyte and sperm. (B) Representative IF image of an in vitro matured metaphase II oocyte stained for pericentrin, tubulin and DNA. Scale: 10 μm. N = 10 oocytes. (C) IF images of abnormally fertilized human embryos at D + 2 stained for pericentrin, tubulin and DNA. Scale: 10 μm. N = 13 fertilized human embryos. (D) Pericentrin, tubulin and DNA staining of a D + 2 parthenogenetically activated human oocyte. Scale: 20 μm. N = 6 activated human oocytes.
To test this idea, we focused on pericentrin, a centrosomal component involved in the recruitment of other centrosomal proteins (Kim and Rhee, 2014). We identified pericentrin in our sperm proteome although with only one unique peptide. In the oocyte, pericentrin was not detected by either IF (Fig. 4B) or proteomics (Holubcova et al., 2015; Virant-Klun et al., 2016). To test whether pericentrin was present in the human embryo, we used in vitro fertilized human oocytes that had 1 pronucleus (PN) or ≥3 PN embryos, as these embryos are discarded from IVF cycles and are easy to obtain. We found that 87% of embryos with 1 PN and 100% of embryos with ≥3 PN contained pericentrin positive foci (Fig. 4C, Supplementary Figs S4A and S4B). Interestingly, in parthenogenetically activated human oocytes, a pericentrin signal was not detected (Fig. 4D), suggesting that pericentrin may be paternally inherited or the sperm-derived centrioles are needed to recruit pericentrin from the oocyte cytoplasm. Altogether these data suggest that both the human sperm and the oocyte provide centrosomal components for the zygote centrosome. They further suggest that the contribution of the sperm goes beyond providing centrioles by also providing a complex set of proteins for the assembly of the first functional centrosome of the future organism.
The sperm-derived centrosome provides robustness to the early cell division cycles of the human parthenotes
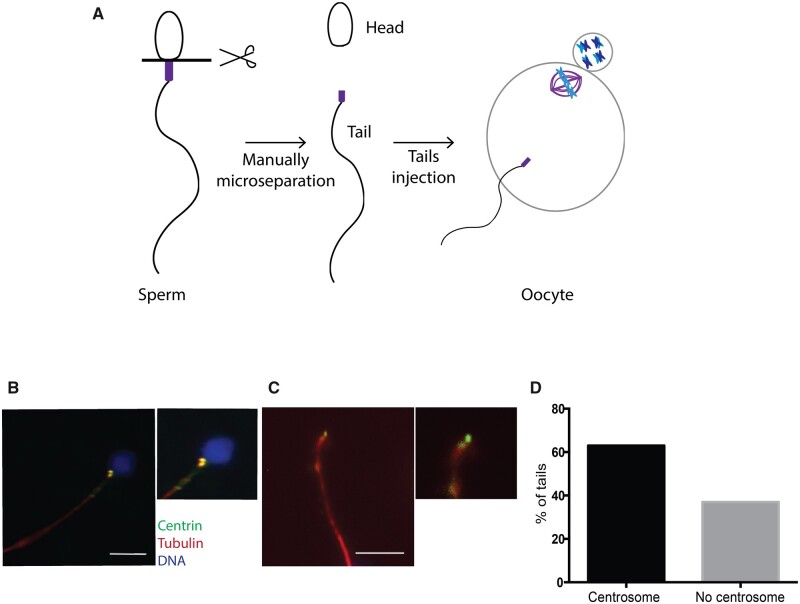
To evaluate the role and contribution of the sperm-derived centrosome in early embryonic development, we devised an experimental system to inject meiotically mature human oocytes with sperm basal bodies and follow their pseudo-development upon activation (Fig. 5A). First, we checked that 83.9% of ejaculated normozoospermic sperm showed a positive signal for centrin indicating that they do have centrioles (Fig. 5B). To isolate these centrioles, we microsurgically severed sperm tails at the transition between the flagella and the head (Methods and Fig. 5A). To confirm that the severed tails contained the centrioles we monitored the presence of centrin in these samples. We found that 63.1% of the severed sperm tails showed either one or two centrin fluorescent dots, and no traces of DNA (Fig. 5C and D). Thus, the injection of these severed tails into oocytes, followed by parthenogenetic activation, should provide a model to study the role of the sperm-derived centrosome in the initial phases of human development.
Figure 5.
The sperm basal body localizes to the microsurgically separated tails. (A) Schematic representation of our functional assay. (B) IF for centrin, tubulin and DNA on intact sperm. Scale: 7.5 μm. (C) Representative IF images of a manually separated sperm tail stained for centrin, tubulin and DNA. Scale: 7.5 μm. (D) Graph showing the percentage of isolated tails with centrosomes. N = 2 different sperm samples.
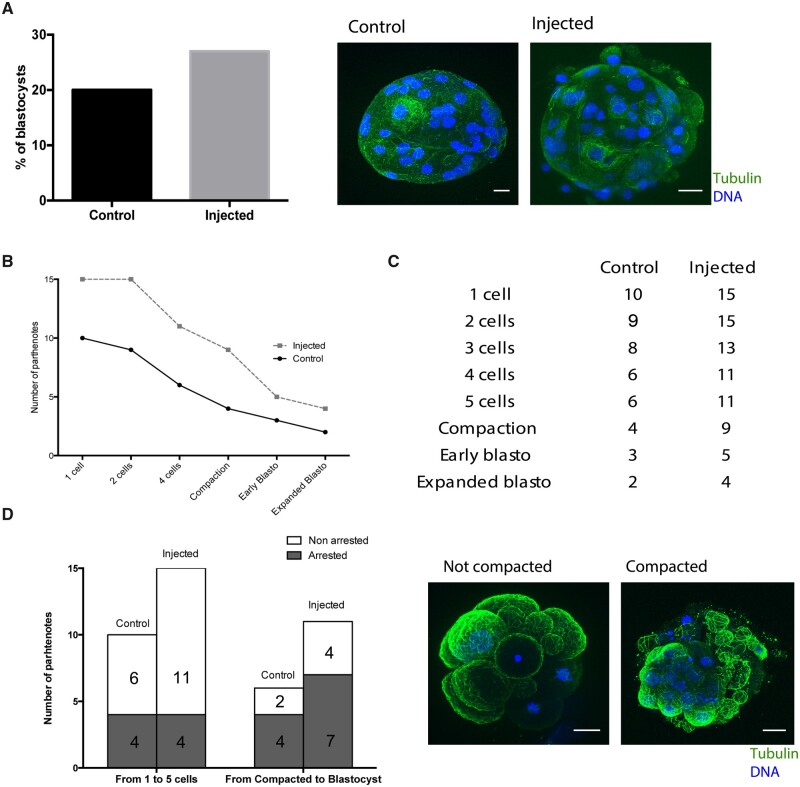
Next, we injected severed sperm tails into human oocytes and parthenogenetically activated them (Fig. 5A). Parthenotes are a good experimental model to mimic the early stages of embryo development owing to the similarity of most parameters used for evaluating in vitro development in activated or fertilized oocytes (Paffoni et al., 2007). We then monitored by time-lapse both the control (sham-injected, n = 10) and tail-injected (n = 15) parthenotes for up to 5 days of pseudo-development in two independent experiments. The percentage of control parthenotes that reached the pseudo-blastocyst stage by Day 5 (20%) was in line with published reports (Paffoni et al., 2007), and it was slightly higher in tail-injected parthenotes (27%) (Fig. 6A). However, tail-injected parthenotes had a slightly higher survival rate at all the time points examined: 1-, 2-, 3-, 4, and 5 cells, compaction, early pseudo-blastocyst and expanded pseudo-blastocyst stages (Fig. 6B and Supplementary Table SV). This was particularly evident before compaction when the large-scale embryonic transcription begins in the human species (Fig. 6B and C). In fact, 4 of the 10 control parthenotes arrested before compaction (from 1 to 5 cells), whereas only 4 of the 15 tail-injected parthenotes arrested before this stage (Fig. 6D). From the 6 control and 11 injected oocytes that did not arrest before compaction, 4 and 7 pseudoembryos respectively arrested after compaction started (Fig. 6D). Despite that the limited access to viable human oocytes restricted the use of robust statistical analysis, our data suggest that tail-injected parthenotes may have an increased probability to go through the earlier cell divisions successfully compared to control ones.
Figure 6.
Sperm centrosome inheritance during fertilization ensures parthenotes compaction. (A) The graph on the left shows the percentage of parthenotes that form a blastocyst-like structure at D + 5 in controls and injected oocytes. The images on the right are representative pseudo-blastocysts obtained in control and injected oocytes. Scale: 20 μm. (B) Developmental progress of control versus injected parthenotes. The graph represents the number of parthenotes that achieved each cellular or embryonic stage. (C) Table with the number of control and injected parthenotes in each cellular and embryonic stage. (D) The graph on the left shows the rate of control and injected oocytes that arrested before or after compaction. On the right, representative images of non-compacted and compacted parthenotes. Scale: 20 μm. N = 10 control and N = 15 injected oocytes in 2 independent experiments.
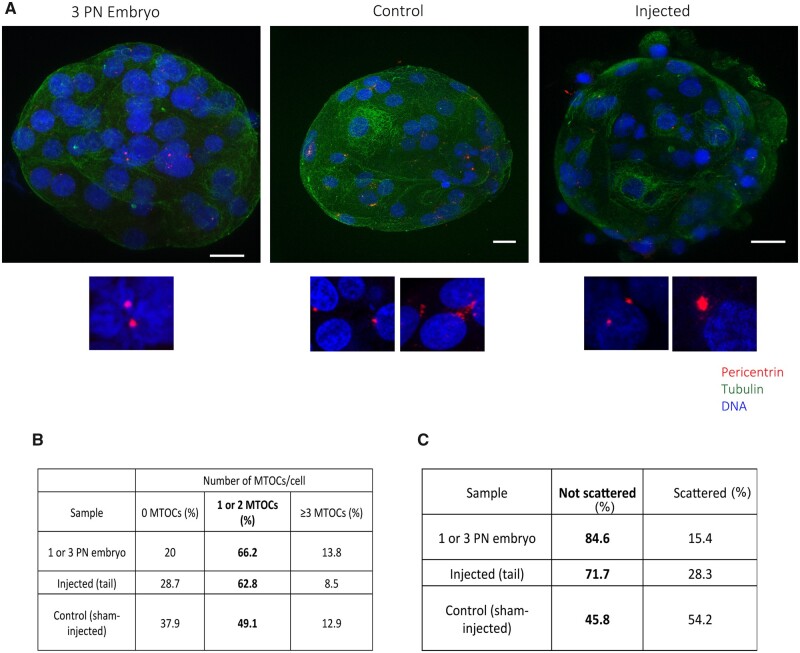
To confirm that the tail-injected oocytes contained centrosomes, we fixed the parthenotes at Day 5 and processed them for IF. We detected pericentrin in 8 out of the 15 tail-injected parthenotes. Moreover, the proportion of cells with 1 or 2 centrosomes (as expected in normal cell cycles) highly resembles the ones we quantified in discarded embryos from 1 or 3 pronuclei: 66.2% in embryos and 62.8% in tail-injected parthenotes (Fig. 7A and B). These data suggested that tail-injected parthenotes do contain centrosomes that cycle correctly. In summary, our experimental approach suggests for the first time that the human sperm-derived centrosome plays an important role in ensuring the robustness of the early human parthenote cell divisions up to compaction when additional requirements provided by a large scale embryonic transcription come into play.
Figure 7.
Microtubule organizing centers (MTOCs) can be formed de novo in control pseudo-blastocysts and after the activation of the embryonic genome. (A) Representative IF images of blastocyst and pseudo-blastocyst of abnormal fertilized oocytes, control and injected oocytes stained for pericentrin, tubulin and DNA. The lower panels are magnifications of the MTOCs for each condition. Scale: 20 μm. (B) Number of MTOCs per cell in abnormal fertilized oocytes, control and injected oocytes. (C) Table showing the percentage of scattered MTOCs per sample. N = 5 3PN embryos, N = 10 control and N = 15 injected oocytes.
Human parthenotes can form de novo MTOCs
While performing the IF studies to monitor pericentrin in parthenotes, we surprisingly found that those derived from control oocytes also contained pericentrin positive aggregates at the early pseudo-blastocyst stage after compaction (Supplementary Table SIV and Fig. 7A). These aggregates could potentially be de novo MTOCs, similar to those forming in mice embryos at blastocyst stage. When we quantified their number of MTOC structures, we found that the proportion of cells having pericentrin positive MTOC-like aggregates was lower (49.1%) than in the tail-injected parthenotes. Moreover, the morphology of these aggregates was different than those detected in tail-injected parthenotes; they had irregular shapes and some degree of scattering that was not usually observed in the tail-injected parthenotes (Fig. 7A—lower panels and C). More than half of the pericentrin aggregates (54.2%) observed in the sham-injected parthenotes showed this ‘scattered’ morphology whereas this was the case for only very few pericentrin positive MTOCs in abnormal fertilized embryos (15.4%) and injected oocytes (28.3%) (Fig. 7C).
We hypothesized that embryonic genome activation could be involved in the formation of de novo pericentrin MTOC-like aggregates. In humans, at Day 2 after fertilization, a first burst of embryonic genome activation occurs, but it is not until Days 3 and 4 that the major activation wave happens (Vassena et al., 2011). We therefore monitored pericentrin by IF in another set of sham-injected and parthenogenetically activated oocytes fixed at Day 3 (before the major wave of embryonic genome activation), and at Day 5 (after embryonic genome activation). None of the oocytes fixed at Day 3 had pericentrin signal (0 out of 6). The single parthenote out of five parthenotes that progressed until the pseudo-blastocyst stage contained pericentrin aggregates (Supplementary Table SVI).
Since the centrosome was previously proposed to define the kinetics of the first embryonic spindle bipolarization (Cavazza et al., 2016), we checked whether the presence of the sperm-derived centrosome had any influence on the early embryonic development kinetics considering only the tail-injected parthenotes that did have MTOCs detected by IF. Although these parthenotes reached each of the cellular and embryonic stages faster than non-injected controls, overall the development kinetics do not differ in control versus injected samples (Supplementary Fig. S5).
Altogether, our results indicate that the paternal inheritance of the basal body is important to establish the centrosome structure and number per cell in the developing pseudoembryo.
Discussion
The mechanism of centrosome inheritance during fertilization is an essential process to ensure the proper number of centrosomes per cell and their correct function in the new organism, yet this process has not been studied in humans. Our study supports the hypothesis that the human sperm basal body is a remodeled centrosome with an atypical structure that is associated with an extensive variety of centrosomal proteins. Moreover, we addressed for the first time the role of the human sperm-derived centrosome in the initial phases of human pseudoembryo development. Our data suggest that the human sperm-derived centrosome plays an important role during the early developmental events leading to embryo compaction.
It is currently accepted that the human sperm basal body consists of two highly remodeled centrioles and little associated PCM. After fertilization, the sperm would therefore only provide one functional centriole to the oocyte and little, if any, PCM (Fawcett and Phillips, 1969; Manandhar and Schatten, 2000; Avidor-Reiss et al., 2015). However, two functional centrosomes need to be assembled in the zygote to provide the two first cells with one centrosome each after the first division of the zygote (Palermo et al., 1997). Our study provides novel data that support an alternative mechanism. High-resolution microscopy has recently shown that the sperm distal centriole consists of an atypical, splayed microtubule structure that retains some centrosomal proteins (Fishman et al., 2018; Khanal et al., 2021). Our super resolution analysis of centrin and acetylated tubulin localization in the human sperm basal body supports these findings. It also suggests that, despite the atypical structure of the distal centriole, it is most likely a stable structure that retains some intrinsic structural characteristics shared with the proximal one. In any case, further studies are needed to comprehensively analyze the stability of both proximal and distal centrioles in the human sperm.
The identification of centrosomal proteins is usually difficult because of their low abundance in cells (Bauer et al., 2016). This issue was particularly challenging for this work because of the process of centrosome reduction in sperm (Manandhar et al., 2000; Avidor-Reiss et al., 2015). We could, however, enrich the samples by eliminating the sperm heads and identify more than 3406 sperm tail proteins, as previously performed in humans and in other species such as cows (Amaral et al., 2013; Firat-Karalar et al., 2014; Cao et al., 2018; Agarwal et al., 2020). To our knowledge, this is the most complex human sperm tail proteome and also the only one that carefully examined the sperm PCM proteins. Most of the identified proteins are related to the cytoskeleton or mitochondria, in agreement with previous results (Martinez-Heredia et al., 2006; Baker et al., 2007; Amaral et al., 2013; Baker et al., 2013; Wang et al., 2013; Jumeau et al., 2015). The proteome also includes 251 centrosomal proteins with 183 having validated localizations and/or functions at the centrosome. We confirmed the centrosomal localization of five of the centrosomal proteins identified in our proteomic analysis, including Nek9, Poc1B and WDR62, demonstrating that our proteomic approach is effective in identifying novel human sperm centrosomal components. Interestingly, these proteins have a variety of functions at various levels including structural roles, the mechanism of centriole duplication and microtubule nucleation. This is somehow surprising since these functions are in principle not required in the mature sperm. In the human oocyte, we found that some centrosomal components are stored as proteins (Virant-Klun et al., 2016) suggesting that the centrosomal proteins may be biparentally inherited. The sperm-derived centrosomal proteins may play an important role after fertilization to promote a quick transition from the sperm basal body to a functional centrosome in the zygote cytoplasm. Interestingly, a similar mechanism was recently proposed in Drosophila. In flies, the sperm centrosome retains some centrosomal components that are essential to support normal embryogenesis (Khire et al., 2016).
The localization of a subset of human sperm centrosomal proteins has been recently described using IF (Fishman et al., 2018; Khanal et al., 2021) and our proteomic analysis further identified 7 out of 20 proteins that Fishman et al. (2018) already described. Interestingly, we identified three centrosomal proteins that could not be detected by IF analysis (γ-tubulin chain 1, pericentrin and PCM 1), either because of technical limitations, such as accessibility in fixed samples, or their very low abundance, opening the possibility that many other centrosomal proteins may also be present in the basal body of the mature sperm.
The injection of microsurgically severed human sperm tails containing the basal body in parthenogenetically activated oocytes offered a unique system to test directly the role of the sperm basal body during preimplantation human development independently of other sperm components such as the paternal genome. This is in fact the first time that the assembly and role of the first functional centrosome in zygotes obtained using human gametes could be addressed following human parthenotes pseudo-development. Owing to limited access to viable human oocytes and the technical complexity of the methodology, we had to work with a small sample size. This restricted the use of robust statistical analysis of the results and hampered the possibility of addressing mechanistically the role of the centrosome in human early development. Despite these limitations, our data suggest that the sperm-derived centrosome helps parthenotes to transit successfully through the early cell divisions up to compaction. Compaction is characterized by cell internalization and embryonic reorganization (Maitre et al., 2016) that in mice is partially mediated by the orientation of the cell division orchestrated by the aMTOC (Korotkevich et al., 2017).
Surprisingly, we could detect the formation of de novo MTOCs in control pseudo-blastocysts. In humans, embryonic genome activation starts as early as the 4-cell stage (D + 2 of embryo development) but, the peak of gene expression occurs when the embryo is at the morula stage (compaction) (Vassena et al., 2011). Our results suggest that when the genome is activated, the production of centrosomal proteins increases considerably after D + 3. Centrosomal proteins may cluster and form independent identities probably through a phase separation mechanism (Woodruff et al., 2017). Their ability to accumulate tubulin triggers microtubule nucleation. The appearance of MTOCs at the pseudo-blastocyst stage is kinetically similar to de novo centrosome formation in mice (Courtois et al., 2012; Howe and FitzHarris, 2013). Although we cannot assume that in the parthenotes they contain centrioles, it suggests that MTOCs activity is important for the development of complex organisms.
Together, our results provide novel insights about the mechanism of centrosome inheritance upon fertilization in humans and its importance in supporting early embryogenesis. These results may also have an important impact in ART in which 30% of the fertilized oocytes arrest before compaction. We show here that centrosome dysfunction may not only be associated with sperm motility and morphology alterations (Jumeau et al., 2017; Amargant et al., 2018) but also it could derive from an defective conversion into a fully functional centrosome in the zygote that could explain some of the unexpected early embryo arrests. Further studies using our novel tail-injection method are likely to provide new strategies to address unexpected cases of human infertility.
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Data availability
All data are in the article or in its online supplemental material. The mass spectrometry proteomics data are available via ProteomeXchange with identifier PXD028348.
Supplementary Material
Acknowledgements
We want to thank all members of the Basic Laboratory from Clinica EUGIN as well as Vernos Lab for fruitful discussions. We also thank all the clinical embryology laboratory staff from Clínica EUGIN and CIRH for their assistance with sample collection and the proteomic facility at the Center for Genomic Regulation for their assistance with the proteomic analysis. We also thank Dr. Joan Roig (IBMB CSIC, Barcelona) for the gift of the anti-Nek9 antibody.
Authors’ roles
F.A. performed the experiments, analyzed data and wrote the paper. A.P. performed sperm tails separation and injection experiments. A.F.-V. performed oocyte warming and provided technical support with the microscope imaging. M.D. performed AOA experiments. M.M. analyzed parthenotes development. R.V. and I.V. supervised the work, designed experiments, interpreted results and critically reviewed and edited the manuscript.
Funding
F.A. was supported by a fellowship from the Agency for Management of University and Research Grants from the Government of Catalonia (AGAUR-2014 DI 065). Work in the Vernos lab was supported by the Spanish Ministry of Economy and Competitiveness (BFU2015-68726-P), the Ministry of Science, Innovation and Universities (PGC2018-096976-B-I00) and intramural funds from the Centre for Genomic Regulation. Intramural funding from Clínica EUGIN partially supported the study. We acknowledge the support of the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) to the EMBL partnership, the Spanish Ministry of Economy and Competitiveness, Centro de Excelencia ‘Severo Ochoa’ and the CERCA programme/Generalitat de Catalunya.
Conflict of interest
The authors declare no conflict of interests.
References
- Agarwal A, Panner Selvam MK, Baskaran S.. Proteomic analyses of human sperm cells: understanding the role of proteins and molecular pathways affecting male reproductive health. Int J Mol Sci 2020;21:1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral A, Castillo J, Estanyol JM, Ballesca JL, Ramalho-Santos J, Oliva R.. Human sperm tail proteome suggests new endogenous metabolic pathways. Mol Cell Proteomics 2013;12:330–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amargant F, Barragan M, Vassena R, Vernos I.. Insights of the tubulin code in gametes and embryos: from basic research to potential clinical applications in humans. Biol Reprod 2019;100:575–589. [DOI] [PubMed] [Google Scholar]
- Amargant F, Garcia D, Barragan M, Vassena R, Vernos I.. Functional analysis of human pathological semen samples in an oocyte cytoplasmic ex vivo system. Sci Rep 2018;8:15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T, Khire A, Fishman EL, Jo KH.. Atypical centrioles during sexual reproduction. Front Cell Dev Biol 2015;3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MA, Naumovski N, Hetherington L, Weinberg A, Velkov T, Aitken RJ.. Head and flagella subcompartmental proteomic analysis of human spermatozoa. Proteomics 2013;13:61–74. [DOI] [PubMed] [Google Scholar]
- Baker MA, Reeves G, Hetherington L, Muller J, Baur I, Aitken RJ.. Identification of gene products present in Triton X-100 soluble and insoluble fractions of human spermatozoa lysates using LC-MS/MS analysis. Proteomics Clin Appl 2007;1:524–532. [DOI] [PubMed] [Google Scholar]
- Bauer M, Cubizolles F, Schmidt A, Nigg EA.. Quantitative analysis of human centrosome architecture by targeted proteomics and fluorescence imaging. EMBO J 2016;35:2152–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NJ, Marjanovic M, Luders J, Stracker TH, Costanzo V.. Cep63 and cep152 cooperate to ensure centriole duplication. PLoS One 2013;8:e69986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Cui Y, Zhang X, Lou J, Zhou J, Bei H, Wei R.. Proteomic profile of human spermatozoa in healthy and asthenozoospermic individuals. Reprod Biol Endocrinol 2018;16:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo J, Jodar M, Oliva R.. The contribution of human sperm proteins to the development and epigenome of the preimplantation embryo. Hum Reprod Update 2018;24:535–555. [DOI] [PubMed] [Google Scholar]
- Cavazza T, Peset I, Vernos I.. From meiosis to mitosis—the sperm centrosome defines the kinetics of spindle assembly after fertilization in Xenopus. J Cell Sci 2016;129:2538–2547. [DOI] [PubMed] [Google Scholar]
- Conti M, Franciosi F.. Acquisition of oocyte competence to develop as an embryo: integrated nuclear and cytoplasmic events. Hum Reprod Update 2018;24:245–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois A, Schuh M, Ellenberg J, Hiiragi T.. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol 2012;198:357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Familiari G, Heyn R, Relucenti M, Nottola SA, Sathananthan AH.. Ultrastructural dynamics of human reproduction, from ovulation to fertilization and early embryo development. Int Rev Cytol 2006;249:53–141. [DOI] [PubMed] [Google Scholar]
- Fawcett DW, Phillips DM.. The fine structure and development of the neck region of the mammalian spermatozoon. Anat Rec 1969;165:153–164. [DOI] [PubMed] [Google Scholar]
- Firat-Karalar EN, Sante J, Elliott S, Stearns T.. Proteomic analysis of mammalian sperm cells identifies new components of the centrosome. J Cell Sci 2014;127:4128–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman EL, Jo K, Nguyen QPH, Kong D, Royfman R, Cekic AR, Khanal S, Miller AL, Simerly C, Schatten G. et al. A novel atypical sperm centriole is functional during human fertilization. Nat Commun 2018;9:2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueth-Hallonet C, Antony C, Aghion J, Santa-Maria A, Lajoie-Mazenc I, Wright M, Maro B.. gamma-Tubulin is present in acentriolar MTOCs during early mouse development. J Cell Sci 1993;105(Pt 1):157–166. [DOI] [PubMed] [Google Scholar]
- Hiraoka L, Golden W, Magnuson T.. Spindle-pole organization during early mouse development. Dev Biol 1989;133:24–36. [DOI] [PubMed] [Google Scholar]
- Holubcova Z, Blayney M, Elder K, Schuh M.. Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science 2015;348:1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, FitzHarris G.. A non-canonical mode of microtubule organization operates throughout pre-implantation development in mouse. Cell Cycle 2013;12:1616–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumeau F, Chalmel F, Fernandez-Gomez FJ, Carpentier C, Obriot H, Tardivel M, Caillet-Boudin ML, Rigot JM, Rives N, Buee L. et al. Defining the human sperm microtubulome: an integrated genomics approach. Biol Reprod 2017;96:93–106. [DOI] [PubMed] [Google Scholar]
- Jumeau F, Com E, Lane L, Duek P, Lagarrigue M, Lavigne R, Guillot L, Rondel K, Gateau A, Melaine N. et al. Human spermatozoa as a model for detecting missing proteins in the context of the chromosome-centric human proteome project. J Proteome Res 2015;14:3606–3620. [DOI] [PubMed] [Google Scholar]
- Khanal S, Leung MR, Royfman A, Fishman EL, Saltzman B, Bloomfield-Gadelha H, Zeev-Ben-Mordehai T, Avidor-Reiss T.. A dynamic basal complex modulates mammalian sperm movement. Nat Commun 2021;12:3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khire A, Jo KH, Kong D, Akhshi T, Blachon S, Cekic AR, Hynek S, Ha A, Loncarek J, Mennella V. et al. Centriole remodeling during spermiogenesis in Drosophila. Curr Biol 2016;26:3183–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Rhee K.. Importance of the CEP215-pericentrin interaction for centrosome maturation during mitosis. PLoS One 2014;9:e87016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich E, Niwayama R, Courtois A, Friese S, Berger N, Buchholz F, Hiiragi T.. The apical domain is required and sufficient for the first lineage segregation in the mouse embryo. Dev Cell 2017;40:235–247.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitre JL, Turlier H, Illukkumbura R, Eismann B, Niwayama R, Nedelec F, Hiiragi T.. Asymmetric division of contractile domains couples cell positioning and fate specification. Nature 2016;536:344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar G, Schatten G.. Centrosome reduction during Rhesus spermiogenesis: gamma-tubulin, centrin, and centriole degeneration. Mol Reprod Dev 2000;56:502–511. [DOI] [PubMed] [Google Scholar]
- Manandhar G, Simerly C, Schatten G.. Highly degenerated distal centrioles in rhesus and human spermatozoa. Hum Reprod 2000;15:256–263. [DOI] [PubMed] [Google Scholar]
- Martinez-Heredia J, Estanyol JM, Ballesca JL, Oliva R.. Proteomic identification of human sperm proteins. Proteomics 2006;6:4356–4369. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T.. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 2011;13:1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffoni A, Brevini TA, Somigliana E, Restelli L, Gandolfi F, Ragni G.. In vitro development of human oocytes after parthenogenetic activation or intracytoplasmic sperm injection. Fertil Steril 2007;87:77–82. [DOI] [PubMed] [Google Scholar]
- Palermo GD, Colombero LT, Rosenwaks Z.. The human sperm centrosome is responsible for normal syngamy and early embryonic development. Rev Reprod 1997;2:19–27. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS.. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999;20:3551–3567. [DOI] [PubMed] [Google Scholar]
- Rusan NM, Rogers GC.. Centrosome function: sometimes less is more. Traffic 2009;10:472–481. [DOI] [PubMed] [Google Scholar]
- Sathananthan AH. Ultrastructure of the human egg. Hum Cell 1997;10:21–38. [PubMed] [Google Scholar]
- Sathananthan AH, Kola I, Osborne J, Trounson A, Ng SC, Bongso A, Ratnam SS.. Centrioles in the beginning of human development. Proc Natl Acad Sci USA 1991;88:4806–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathananthan AH, Ratnam SS, Ng SC, Tarin JJ, Gianaroli L, Trounson A.. The sperm centriole: its inheritance, replication and perpetuation in early human embryos. Hum Reprod 1996;11:345–356. [DOI] [PubMed] [Google Scholar]
- Simerly C, Wu GJ, Zoran S, Ord T, Rawlins R, Jones J, Navara C, Gerrity M, Rinehart J, Binor Z. et al. The paternal inheritance of the centrosome, the cell's microtubule-organizing center, in humans, and the implications for infertility. Nat Med 1995;1:47–52. [DOI] [PubMed] [Google Scholar]
- Simerly C, Zoran SS, Payne C, Dominko T, Sutovsky P, Navara CS, Salisbury JL, Schatten G.. Biparental inheritance of gamma-tubulin during human fertilization: molecular reconstitution of functional zygotic centrosomes in inseminated human oocytes and in cell-free extracts nucleated by human sperm. Mol Biol Cell 1999;10:2955–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torreira E, Jha S, Lopez-Blanco JR, Arias-Palomo E, Chacon P, Canas C, Ayora S, Dutta A, Llorca O.. Architecture of the pontin/reptin complex, essential in the assembly of several macromolecular complexes. Structure 2008;16:1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Blerkom J, Davis P.. Evolution of the sperm aster after microinjection of isolated human sperm centrosomes into meiotically mature human oocytes. Hum Reprod 1995;10:2179–2182. [DOI] [PubMed] [Google Scholar]
- Vassena R, Boue S, Gonzalez-Roca E, Aran B, Auer H, Veiga A, Izpisua Belmonte JC.. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 2011;138:3699–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virant-Klun I, Leicht S, Hughes C, Krijgsveld J.. Identification of maturation-specific proteins by single-cell proteomics of human oocytes. Mol Cell Proteomics 2016;15:2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Guo Y, Zhou T, Shi X, Yu J, Yang Y, Wu Y, Wang J, Liu M, Chen X. et al. In-depth proteomic analysis of the human sperm reveals complex protein compositions. J Proteomics 2013;79:114–122. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Honda S, Konishi A, Arakawa S, Murohashi M, Yamaguchi H, Torii S, Tanabe M, Tanaka S, Warabi E. et al. Autophagy controls centrosome number by degrading Cep63. Nat Commun 2016;7:13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RA, Pan Z, Salisbury JL.. GFP-centrin as a marker for centriole dynamics in living cells. Microsc Res Tech 2000;49:451–457. [DOI] [PubMed] [Google Scholar]
- Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA.. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 2017;169:1066–1077.e10. [DOI] [PubMed] [Google Scholar]
- World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 2013;310:2191–2194. [DOI] [PubMed] [Google Scholar]
- Wu J, Akhmanova A.. Microtubule-organizing centers. Annu Rev Cell Dev Biol 2017;33:51–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are in the article or in its online supplemental material. The mass spectrometry proteomics data are available via ProteomeXchange with identifier PXD028348.