Abstract
Central obesity is a leading health concern with a great burden carried by ethnic minority populations, especially Hispanics/Latinos. Genetic factors contribute to the obesity burden overall and to inter-population differences. We aimed to identify the loci associated with central adiposity measured as waist-to-hip ratio (WHR), waist circumference (WC) and hip circumference (HIP) adjusted for body mass index (adjBMI) by using the Hispanic Community Health Study/Study of Latinos (HCHS/SOL); determine if differences in associations differ by background group within HCHS/SOL and determine whether previously reported associations generalize to HCHS/SOL. Our analyses included 7472 women and 5200 men of mainland (Mexican, Central and South American) and Caribbean (Puerto Rican, Cuban and Dominican) background residing in the USA. We performed genome-wide association analyses stratified and combined across sexes using linear mixed-model regression. We identified 16 variants for waist-to-hip ratio adjusted for body mass index (WHRadjBMI), 22 for waist circumference adjusted for body mass index (WCadjBMI) and 28 for hip circumference adjusted for body mass index (HIPadjBMI), which reached suggestive significance (P < 1 × 10−6). Many loci exhibited differences in strength of associations by ethnic background and sex. We brought a total of 66 variants forward for validation in cohorts (N = 34 161) with participants of Hispanic/Latino, African and European descent. We confirmed four novel loci (P < 0.05 and consistent direction of effect, and P < 5 × 10−8 after meta-analysis), including two for WHRadjBMI (rs13301996, rs79478137); one for WCadjBMI (rs3168072) and one for HIPadjBMI (rs28692724). Also, we generalized previously reported associations to HCHS/SOL, (8 for WHRadjBMI, 10 for WCadjBMI and 12 for HIPadjBMI). Our study highlights the importance of large-scale genomic studies in ancestrally diverse Hispanic/Latino populations for identifying and characterizing central obesity susceptibility that may be ancestry-specific.
Introduction
Obesity, and especially central obesity, is a leading risk factor for metabolic and cardiovascular diseases (CVDs), with the greatest burden carried by minority populations (1–4), particularly Hispanic/Latino Americans and African Americans (5). Emerging evidence suggests that genetic factors may contribute not only to the obesity burden overall, explaining 40–70% of the inter-individual variation (6), but also to population-specific differences in obesity susceptibility (7–12). For example, although a majority of the >1000 genome-wide association study (GWAS)-identified obesity [body mass index (BMI), waist-to-hip ratio (WHR), waist circumference (WC), hip circumference (HIP) and body fat percentage] loci generalize across populations (13–20), recent studies in populations of Asian (19,20) and African (16,21) ancestry have revealed a number of novel and population-specific loci. These observations highlight the importance of large-scale genomic studies in ancestrally diverse populations, including Hispanic/Latinos, to identify obesity-susceptibility loci, and more specifically, alleles that are ancestry-specific and may thus partly explain disparities. However, no large-scale GWAS for any obesity-related traits has been performed in Hispanic/Latino populations despite their increased prevalence of obesity.
While obesity is commonly assessed by BMI, measures of central adiposity, such as WHR and WC, are predictors of increased cardiometabolic risk independent of BMI (22–25). Here, we consider three measures of central obesity: WHR, WC and HIP after accounting for overall body size, measured as BMI [waist-to-hip ratio adjusted for body mass index (WHRadjBMI), waist circumference adjusted for body mass index (WCadjBMI) and hip circumference adjusted for body mass index (HIPadjBMI)]. Larger WHR indicates higher visceral fat and is associated with increased risk for type 2 diabetes (T2D) and CVD (26–28), while smaller WHR indicates a proportionately greater fat accumulation around the hips and is associated with lower risk for T2D, hypertension and dyslipidemia (29). Previous GWAS have identified WHR, WC and HIP loci, which are enriched for association with other cardiometabolic traits and suggested that different fat distribution patterns can have distinct genetic underpinnings (30–32). Identifying genetic risk variants across these traits in Hispanic/Latinos may provide insights into these mechanisms and highlight population-specific variants that increase susceptibility to obesity in specific groups.
We aimed to: (1) identify novel genetic loci associated with central obesity, measured here as WHRadjBMI, WCadjBMI and HIPadjBMI, in Hispanics/Latinos; (2) determine if differences in genetic associations by background group (mainland or Caribbean) and sex exist in Hispanic Community Health Study/Study of Latinos (HCHS/SOL) and (3) assess generalization of central adiposity-associated loci, discovered in European, African and multi-ethnic studies, to Hispanics/Latinos.
Results
Discovery
We identified 16 loci for WHRadjBMI, 22 for WCadjBMI and 28 for HIPadjBMI, which exhibited suggestive evidence of association in the HCHS/SOL (N = 12 472, 58% women, Supplementary Material, Table S1) in at least one stratum (Table 1, Figs 1–3, Supplementary Material, Tables S2–S4, Supplementary Material, Figs S1–S21). For WHRadjBMI, we identified four loci that reach suggestive significance (P < 1 × 10−6) in the combined sexes, including rs12435790 near KIAA0391, which is within a previously reported WHRadjBMI locus [+/−500 Kb from tag single nucleotide polymorphism (SNP)] (33). We also identified five loci for men only, including one reaching genome-wide significance (GWS, P < 5 × 10−8). A total of eight suggestive loci were identified in the women-only analyses, including one, rs115981023 in TAOK3, which also reached suggestive significance in the combined sexes analysis and identified rs79478137 in solute carrier family 22 (organic cation transporter), member 18 antisense (SLC22A18AS) near a previously implicated WHRadjBMI locus (34). For WCadjBMI, we identified nine loci, including one GWS locus in the combined sexes; 11 for men only, including two SNPs that reach GWS, and two for women only. Of the WCadjBMI loci identified, two were nearby previously reported WCadjBMI loci, rs6809759 near PROK2 (men-only) (14,15,17) and rs77319470 near ADAMTS3 (sexes-combined) (15,17,35). For HIPadjBMI, we identified eight loci that reach P < 1 × 10−6 for the combined sexes; nine for men only, including one in a locus that reached suggestive significance for the combined sexes as well (near ANO10), and 12 for women only, including one SNP in a locus that reached suggestive significance for the combined sexes as well (near LPPR4). Of the WCadjBMI loci, rs10818474 near MEGF9 was within 500 Kb of a recently reported WHRadjBMI association in women (14).
Table 1.
Summary of association results for all loci that passed replication criteria
| Stratum | dbSNPID | CHR | POS (GRCh38) | Nearest gene | EAF | Other allele | Stage | EAF | Beta | SE | P | N |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WHRadjBMI | ||||||||||||
| AAb-combined | rs13301996 | 9 | 120 570 806 | CDK5RAP2 | T | G | SOL | 0.8080 | 0.0050 | 0.0010 | 5.69E-07 | 12 672 |
| Replication | 0.8720 | 0.0036 | 0.0014 | 1.10E-02 | 12 496 | |||||||
| SOL + replication | 0.8295 | 0.0045 | 0.0008 | 2.88E-08 | 25 168 | |||||||
| HLc−women | rs79478137 | 11 | 2 891 739 | SLC22A18AS | T | C | SOL | 0.0150 | -0.0230 | 0.0040 | 2.03E-07 | 7472 |
| Replication | 0.0169 | -0.0116 | 0.0054 | 3.12E-02 | 6582 | |||||||
| SOL + replication | 0.0157 | -0.0189 | 0.0032 | 3.64E-09 | 14 054 | |||||||
| WCadjBMI | ||||||||||||
| EURd-combined | rs3168072 | 11 | 61 864 038 | FADS2 | A | T | SOL | 0.7250 | 0.5140 | 0.1020 | 5.28E-07 | 12 674 |
| Replication | 0.9750 | 2.0132 | 0.6323 | 1.45E-03 | 8845 | |||||||
| SOL + replication | 0.7313 | 0.5520 | 0.1007 | 4.21E-08 | 21 519 | |||||||
| HIPadjBMI | ||||||||||||
| EURd-women | rs28692724e | 14 | 77 027 445 | IRF2BPL | T | C | SOL | 0.4250 | 0.0020 | 0.0004 | 7.32E-07 | 7462 |
| Replication | 0.303 | 0.789 | 0.305 | 9.62E-03 | 4678 | |||||||
| SOL + replication | 0.3781 | 5.4900a | 4.02E-08 | 12 140 | ||||||||
EAF, estimated allele frequency; CHR, chromosome; pos, position; SE, standard error. Genome-wide significant (P<5×10−8) values are highlighted in bold.
aFor rs28692724, SOL analyses were performed on log10-transformed HIP, while replication analyses in European descent population used untransformed hip measurements. In the SOL + replication analyses, an z-score is provided instead of a beta.
bAA replication samples included: ARIC Study, Multi-Ethnic Study of Atherosclerosis (MESA) Study, Women’s Health Initiative (WHI) Study.
cHispanic Latino (HL) replication samples included: Genetics of Latinos Diabetic Retinopathy (GOLDR), HCHS/SOL, Mexican–American Hypertension Study (HTN), MACAD, MESA, Mexico-City, 1982 Pelotas Birth Cohort (PELOTAS), Starr County Health Studies (STARR), WHI.
dEuropean American (EA) replication samples included: ARIC.
ers28692724 is <500 Kb from a previously reported SNP nominally associated with WHRadjBMI (PMID: 28552196).
Figure 1 .
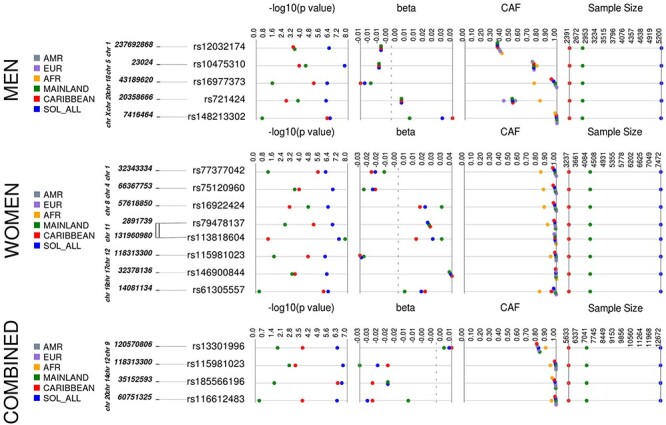
WHRadjBMI Synthesis View plot that shows −log10 P-values, beta (effect estimate), effect/coded allele frequency (CAF) and sample size across analysis samples for all loci that reached suggestive significance in one or more of our discovery strata. This chart also shows the CAF of each of our top loci by background group and by 1000 genomes reference panel. European, EUR; Latin American, AMR; African, AFR.
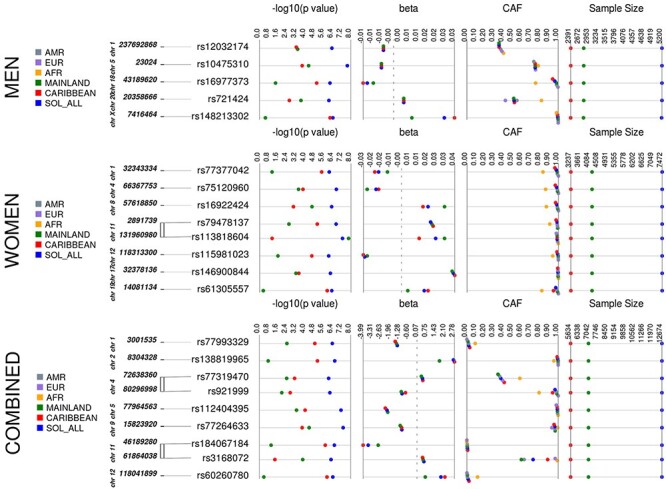
Figure 3 .
HIPadjBMI Synthesis View plot that shows the −log10 P-values, beta (effect estimate), effect/CAF and sample size across analysis samples for all loci that reached suggestive significance in one or more of our discovery strata. This chart also shows the CAF of each of our top loci by background group and by 1000 genomes reference panel. European, EUR; Latin American, AMR; African, AFR.
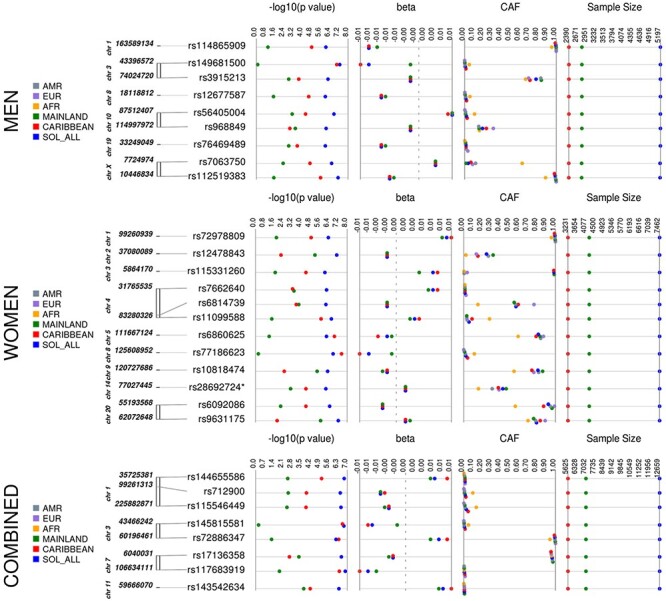
Figure 2 .
WCadjBMI Synthesis View plot that shows −log10 P-values, beta (effect estimate), effect/CAF, and sample size across analysis samples for all loci that reached suggestive significance in one or more of our discovery strata. This chart also shows the CAF of each of our top loci by background group and by 1000 genomes reference panel. European, EUR; Latin American, AMR; African, AFR.
Association differences by genetic ancestry
All of the top loci were directionally consistent in each background group, yet many of the loci exhibited effect heterogeneity by background group (Table 2, Figs 1–3, Supplementary Material, Tables S5–S7), as exhibited by moderate-to-high I2 values [I-squared heterogeneity (ISQ) > 65%) and/or significant interaction across background groups (Pdiff < 0.05). For example, rs113818604 (β = 0.0269, P = 5.47 × 10−8), I2 = 78.5%, Pdiff = 0.38) in NTM is significantly associated with WHRadjBMI in women from the mainland background groups [N = 4220, minor allele frequency (MAF) = 0.014, β = 0.0343, P = 1.63 × 10−8] but not in women from Caribbean background groups (N = 3238, MAF = 0.013, β = 0.0144, P = 0.08) (Supplementary Material, Table S5). Also, for the women-only primary analysis, the rs77186623 in LOC105375745 locus associated with HIPadjBMI (β = −0.006, P = 1.74 × 10−7) exhibited nominally significant interaction by background group (I2 = 55.3%, Pdiff = 0.042) and was GWS in the Caribbean group (N = 3231, MAF = 0.041, β = −0.0078, P = 3.05 × 10−8) but not significant in the mainland group (N = 4216, MAF = 0.008, β = −0.0015, P = 0.567, Supplementary Material, Table S7). Additional examples that cannot be explained because of power (i.e. MAF and sample size are similar) for WHRadjBMI include rs77377042 near MARCKSL1 and rs61305557 in C19orf67 for women and rs16977373 near RIT2 for men; for WCadjBMI in women-only, these include rs76842062 in MAP4K4 and rs76941364 near COBL; and for HIPadjBMI, these include rs6860625 near NREP for women and rs145815581 in ANO10 for the combined sexes.
Table 2.
Summary of top association results in SOL background group analyses
| Stratum | dnSNPID | CHR | POS (GRCh38) | Nearest gene | EAF | Other allele | Background group | EAF | Beta | SE | P | N | Pdiff | EAF | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFR | EUR | AMR | |||||||||||||||
| WHRadjBMI | |||||||||||||||||
| AA-combined | rs13301996 | 9 | 120 570 806 | CDK5RAP2 | T | G | SOL | 0.8080 | 0.0050 | 0.0010 | 5.69E-07 | 12 672 | 1.88E-01 | 0.886 | 0.809 | 0.794 | |
| Mainland | 0.8220 | 0.0034 | 0.0014 | 1.95E-02 | 7013 | ||||||||||||
| Caribbean | 0.7890 | 0.0061 | 0.0017 | 2.52E-04 | 5633 | ||||||||||||
| HL-women | rs79478137 | 11 | 2 891 739 | SLC22A18AS | T | C | SOL | 0.0150 | -0.0230 | 0.0040 | 2.03E-07 | 7472 | 3.97E-01 | 0.095 | 0.011 | 0.010 | |
| Mainland | 0.0080 | -0.0241 | 0.0080 | 2.72E-03 | 4220 | ||||||||||||
| Caribbean | 0.0250 | -0.0250 | 0.0056 | 8.87E-06 | 3238 | ||||||||||||
| WCadjBMI | |||||||||||||||||
| EUR-combined | rs3168072 | 11 | 61 864 038 | FADS2 | A | T | SOL | 0.7250 | 0.5140 | 0.1020 | 5.28E-07 | 12 674 | 3.94E-01 | 0.990 | 0.967 | 0.630 | |
| Mainland | 0.5969 | 0.4552 | 0.1204 | 1.57E-04 | 7013 | ||||||||||||
| Caribbean | 0.8846 | 0.4170 | 0.2075 | 4.45E-02 | 5635 | ||||||||||||
| HIPadjBMI | |||||||||||||||||
| EUR-Women | rs28692724a | 14 | 77 027 445 | IRF2BPL | T | C | SOL | 0.4250 | 0.0020 | 0.0004 | 7.32E-07 | 7462 | 2.31E-01 | 0.160 | 0.384 | 0.310 | |
| Mainland | 0.462 | 0.0017 | 0.001 | 8.70E-04 | 4216 | ||||||||||||
| Caribbean | 0.3765 | 0.0025 | 0.0006 | 4.02E-05 | 3232 | ||||||||||||
aEAF for reference population obtained from ExAC; all other estimated EAFs are from 1000 Genomes Project Phase 3.
For other loci, allele frequency and linkage disequilibrium (LD) differences across Hispanic/Latino populations likely contributed to observed differences in the magnitude of effect and significance levels (Supplementary Material, Table S8). For example, while the magnitude of effect for the rs115981023 TAOK3 association with WHRadjBMI in women (β = −0.029, P = 8.88x10−7, I2 = 0, Pdiff = 0.391) was similar across background groups, the P-value was far more significant in the Caribbean background group (MAF = 0.016, β = −0.030, P = 2.72 × 10−5) when compared with the mainland (MAF = 0.003, β = −0.027, P = 0.025), likely because of the higher MAF in the Caribbean group. Of note, the minor allele at this SNP is more common in the 1000 Genomes AFR compared with the EUR and AMR reference samples (Supplementary Material, Table S5), and the local ancestry of participants at this locus indicate that those with African ancestry exhibit the highest MAF (Supplementary Material, Table S8). Additional loci where the significance level differences between Caribbean and mainland background groups appear to be driven by increased MAF owing to African ancestry in Caribbean populations include the SLC22A18AS and CDH4 loci for WHRadjBMI; LOC102723448, FZD7, WSB2 and ACTRT2 loci for WCadjBMI; and COQ2, LPPR4, TMEM63A and FHIT loci for HIPadjBMI (Supplementary Material, Tables S5–S8). Rs12478843 in HEATR5B (β = −0.002, P = 8.2 × 10−8, I2 = 1.7%, Pdiff = 0.385) is more significantly associated with HIPadjBMI in mainland (MAF = 0.320, β = −0.002, P = 6.50 × 10−6) women when compared with Caribbean (MAF = 0.154, β = −0.002, P = 6.03 × 10−3), likely reflecting the higher MAF among those from mainland Latin America with greater Native American ancestry (Supplementary Material, Table S8). Similarly, differences in effect magnitude between mainland and Caribbean background groups for the TAF4 (HIPadjBMI in women) and the ESRRG (WCadjBMI in men) loci may also be owing to higher MAF in the mainland group because of a greater proportion of Native American ancestry (Supplementary Material, Tables S6–S8).
Replication
We brought 66 variants forward for replication in nine cohort studies (N up to 34 161), with participants of Hispanic/Latino, African and European descent, and for further examination of replication by ancestral background (Supplementary Material, Tables S1–S4). Our criteria for replication included both nominal evidence of an association (P < 0.05), consistent direction of effect between the replication results and the HCHS/SOL discovery results for any ancestry/sex stratum and genome-wide significance (P < 5 × 10−8) when meta-analyzed together with HCHS/SOL. Based on these criteria, we were able to replicate four novel loci (Table 1) after combining our HCHS/SOL discovery sample with specific ancestry results. For WHRadjBMI in men and women combined, rs13301996 was significant after meta-analyzing HCHS/SOL with the African American replication sample (P = 2.88 × 10−8). For WHRadjBMI in women only, rs79478137 was GWS after combining HCHS/SOL with the Hispanic/Latino replication sample (P = 3.64 × 10−9). For WCadjBMI in men and women combined, rs3168072 was significant after combining HCHS/SOL with the European American replication sample (P = 4.21 × 10−8). For HIPadjBMI in women only, rs28692724 was significant after meta-analyzing HCHS/SOL with the European American replication sample (P = 4.02 × 10−8).
Of note, for rs13301996, which only replicated in African Americans, we saw a larger effect size in the Caribbean background group compared with the mainland, although this is not a significant difference (Table 2, Supplementary Material, Table S5, Supplementary Material, Fig. S2A). This finding may provide insight into why the variants were more successful upon replication with a particular ancestry. For the remaining loci, there is little difference in effect magnitude between the Caribbean and the mainland background groups, which could explain differences in replication by ancestral group.
Generalization of previous loci
We examined previously reported association regions from the Genetic Investigation of Anthropometric Traits (GIANT) Consortium (14) to assess generalization to the HCHS/SOL (Supplementary Material, Tables S9–S11, Supplementary Material, Figs S22–S30). To account for the differences in LD between GIANT (primarily European descent populations) and HCHS/SOL (highly admixed Hispanic/Latino populations), we report generalization results based on the lead generalized SNP (the SNP with lowest r-value in the region of the previously reported variant in GIANT). In sex-combined analyses, there were a total of 12 association regions across the genome, which generalized to HCHS/SOL for WHRadjBMI (r < 0.05), including three for both women-only and sexes-combined, three for women-only and six for the sexes-combined analysis (Supplementary Material, Table S9). A total of 15 association regions generalized to HCHS/SOL for WCadjBMI, including seven sex-specific loci (two for men, five for women; Supplementary Material, Table S10), one for the sexes-combined only and seven for more than one stratum. Of note, we identified rs6809759 near PROK2, which was significantly associated with WCadjBMI in HCHS/SOL for men-only and sexes-combined and was within 500 kb (+/−) of rs12330322, as identified in Shungin et al. (14). However, this previously identified locus did not generalize to HCHS/SOL (r > 0.05) and may represent an independent association signal in a known region [i.e. all GIANT variants at this locus with P < 1 × 10−6 exhibit r > 0.05 in HCHS/SOL and rs6809759 had a P > 1 × 10−6 in Shungin et al. (14) (P = 1.4 × 10−1)]. A total of 40 regions generalized to HCHS/SOL for HIPadjBMI, including 29 for sexes-combined, three of which were significant for both women-only and sexes-combined analyses (Supplementary Material, Table S11).
Because some of the SNPs previously reported by GIANT may not have generalized owing to lack of power in HCHS/SOL, we calculated individual-level genetic scores based on trait-increasing alleles for each central adiposity phenotype (Supplementary Material, Table S12) and sex stratum (three association tests per phenotype). For genetic scores based on SNPs with P-value < 1 × 10−7 in GIANT, all association tests were significant (P < 0.05). For genetic scores calculated from GIANT SNPs with 1 × 10−7 < P < 1 × 10−6, six of the nine association tests were significant. Given that only three out of 27 analyses had a P > 0.05, there is considerable overlap in the association results of Hispanics/Latinos to those previously reported in the GIANT multi-ethnic analysis.
Biological curation
We examined the four SNPs (i.e. rs13301996, rs79478137, rs28692724 and rs3168072) in novel loci identified in the replication analyses (Table 1) for association with other phenotypes, gene expression and metabolites in publicly available data using Phenoscanner (36,37), and we assessed the potential regulatory role of these variants and those in LD using publicly available databases, including RegulomeDB (38), Haploreg (39), UCSC GenomeBrowser (40) and GTeX (41). Known associations with these variants meeting Bonferroni-corrected significance after correcting for number of reported associations in Phenoscanner for the four variants within each category (P < 0.05/7631 = 6.55 × 10−5 for GWAS; P < 0.05/88 = 5.68 × 10−4 for gene expression; P < 0.05/488 = P < 1.02 × 10−4 for metabolites) are provided in Supplementary Material, Tables S13–S15.
WHRadjBMI-associated variant, rs13301996, which is intronic to cyclin-dependent kinase 5 (CDK5) regulatory subunit-associated protein 2 (CDK5RAP2), was significantly associated with the expression of 15 genes and one lncRNA across 17 tissue types (Supplementary Material, Table S13). The most significant of these associations was with multiple epidermal growth factor-like domains 9 (MEGF9) in whole blood (P = 1.8 × 10−149), a gene that rests 30 Kb upstream of rs1330996. This SNP is also significantly associated with expression of MEGF9 in subcutaneous adipose tissue, sun-exposed skin and T-cells. Additionally, our lead variant in CDK5RAP2 is associated with the expression of MEGF9 in whole blood and the testis and with the expression of proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 (PSMD5) and/or PSMD5-AS1 in several relevant tissues, including whole blood, tibial artery, tibial nerve, lung, thyroid, esophagus muscle, skeletal muscle, liver, cerebellum and subcutaneous adipose tissues, among others. There is additional support for a regulatory role of rs13301996 and those with which it is in high LD (r2 > 0.8). For example, our lead SNP lies just outside of a DNase hypersentivitiy cluster; lies within a region with evidence of histone modification in nine tissues, including brain, skin, muscle and heart; and likely falls in a transcription factor binding site active in skeletal and lung tissue; etc. (Supplementary Material, Table S16) (38–40). While there are multiple lines of evidence for a regulatory role of this variant and multiple genes, rs13301996 has RegulomeDB score of 6, indicating little evidence of binding.
WHRadjBMI-associated SNP, rs79478137, is a low-frequency variant (MAF = 1.6%) intronic in SLC22A18AS. This region is subject to genomic imprinting (42), which has been linked with Beckwith-Wiedemann syndrome, a disease caused by an increased rate of growth in children (43–45). Our lead variant is associated with two Electronic Health Record (EHR)-derived phenotypes (cause of death: multisystem degeneration; and cause of death: tongue, unspecified) (Supplementary Material, Table S13) in Phenoscanner. There is limited evidence of a regulatory role for our lead SNP (RegulomeDB score = 4), but rs79478137 is in perfect LD with several variants with evidence of regulation (histone modification, open chromatin, DNAse hypersensitivity and transcription factor binding) in more than 50 tissues, including blood, pancreas, liver and skeletal muscle, hippocampal tissues, etc. (Supplementary Material, Table S16) (38–40).
WCadjBMI-associated SNP, rs3168072, was significantly associated with existing GWAS traits present in Phenoscanner, including ‘cause of death: other specified respiratory disorders’ (Supplementary Material, Table S13). Additionally, rs3168072 is significantly associated with the expression of several genes in whole blood but is most significantly associated with the expression of transmembrane protein 258 (TMEM258) (Supplementary Material, Table S14). Rs3168072 is ~95 Kb upstream of TMEM258. Our lead variant is likely to play a role in gene expression regulation (RegulomeDB score = 2b, ‘likely to affect binding’) (38). Additionally, our lead variant and those in high LD (R2 > 0.8) lie within known DNase hypersentivitiy regions and within active areas of histone modification, open chromatin and likely gene enhancer regions (Supplementary Material, Table S16) (38–40). Our lead SNP associated with WCadjBMI, rs3168072, is significantly associated with five lipid-related metabolites (Supplementary Material, Table S15), including ‘Other polyunsaturated fatty acids than 18:2’, ‘CH2 groups in fatty acids’, ‘Ratio of bis allylic bonds to double bonds in lipids’, ‘CH2 groups to double bonds ratio’ and ‘Ratio of bis allylic bonds to total fatty acids in lipids’.
Our lead SNP associated with HIPadjBMI in women, rs28692724 (NC_000014.9:g.77027445C>T), is a synonymous variant exonic to interferon regulatory factor 2-binding protein-like (IRF2BPL) that is significantly associated with expression of the same gene in whole blood (Supplementary Material, Table S14). Additionally, this variant lies in a known CCCTC-binding factor (CTCF)-binding site (RegulomDB Score = 2b), among other transcription factors, and a DNAse Hypersentivity cluster (Supplementary Material, Table S16) (38–40).
Discussion
We performed the first large-scale GWAS of three central adiposity traits (i.e. WHRadjBMI, WCadjBMI and HIPadjBMI) in a sample of approximately 12 672 Hispanic/Latino individuals. We identified 16 variants that were suggestively associated (P < 1 × 10−6) with WHRadjBMI, 22 for WCadjBMI and 28 for HIPadjBMI. Of these 66 variants that were suggestively associated with the three central adiposity traits, four novel loci replicated after meta-analysis with replication samples. Additionally, we demonstrated that eight previously identified GWAS loci generalized to Hispanic/Latino study participants for WHRadjBMI, 10 for WCadjBMI and 12 for HIPadjBMI in HCHS/SOL.
Discovery of four novel loci
Given the large number of published GWAS on central adiposity measures, it may seem surprising that four novel loci (rs13301996, rs79478137, rs28692724 and rs3168072) were mapped. There are a few explanations for these novel findings, including (1) previous GWAS were primarily conducted in European populations. Indeed, all four novel SNPs were absent from previous GIANT HapMap imputed analyses (14), and one (rs28692724) of the four absent from a more recent GWAS that included Europeans from the UK Biobank (33); (2) the consideration of a broad spectrum of ancestrally diverse Hispanic/Latino populations, including not just those of Mexican ancestry but also those with ancestry from the Caribbean, Central, and/or South America (46); (3) the use of the entire 1000 Genomes Phase I Reference panel, including populations with Native American ancestry: Mexico (MXL), Colombia (CLM) and Puerto Rico (PUR); (4) demonstrated differences in the patterning of body composition by ancestry (47,48). More specifically, African ancestry populations have lower body fat percentages than men and women of non-Hispanic European, Native American and East Asian ancestry at the same BMI. Additionally, non-Hispanic African ancestry men and women have greater skeletal and muscle mass than their non-Hispanic European ancestry counterparts who, in turn, have greater skeletal and muscle mass than men and women of East Asian origin (47,49–51).
Recent GWAS for coding variation of WHRadjBMI identified the importance of central adiposity genes in lipid regulation, storage and homeostasis (52). Similarly, we found a novel association of a variant in FADS2 (rs3168072) with WCadjBMI following meta-analysis of HCHS/SOL results with the results from an independent sample of European descent individuals, which further implies a role of this locus in central adiposity and lipid homeostasis. Genetic variations in the FADS2 gene has been associated with several traits related to obesity and cardiometabolic health, including fatty acid metabolism and adipose tissue inflammation, leading to an interaction between weight loss and FADS2 genes in the regulation of adipose tissue inflammation (53). A nearby variant, rs174546 (R2 = 0.3523, D′ = 0.916 in AMR), in FADS1 has previously been associated with four lipid traits (54). The A allele (MAF = 38%) for our lead SNP is associated with greater WC in our samples and is nearly fixed among sub-Saharan African populations (99% in 1000 Genomes AFR) at very high frequency in European populations (97% in EUR) and at a lower frequency in East Asian (75% in EAS) and Native American populations (63% in AMR). Rs3168072 is intronic to FADS2—a member of the fatty acid desaturase (FADS) gene family—and is involved in the endogenous conversion of short-chain polyunsaturated fatty acids to long chain fatty acids. The FADS cluster of genes appears to have been under strong selection in several human populations, which likely explains the large differences in allele frequencies across global populations (55–58) and why previous GWASs of waist traits that primarily focused on European descent populations did not detect an association signal in this region.
We identified a novel association for WHRadjBMI with rs13301996 following meta-analysis with an independent sample of African descent individuals. Rs13301996 is intronic to CDK5RAP2, which encodes a regulator of CDK5 activity (59), interacts with CDK5R1 and pericentrin (PCNT) (59), plays a role in centriole engagement and microtubule nucleation (60) and has been linked to primary microcephaly and Alzheimer’s disease (61,62). In addition, we identified a novel association for WHRadjBMI with rs79478137 (P-value = 3.64E−9) in Hispanic/Latino women. Rs79478137 is intronic to the antisense SLC22A18AS gene, which is highly expressed in the liver and kidney as well as in the gastrointestinal tract and placenta. Very little is known of the biological role of this gene (63), and SLC22A18AS has no counterpart in mice or other rodents (64). Thus, although its genomic organization is known, the regulation and function of this gene is not understood (65).
Lastly, we identified a novel association for HIPadjBMI at rs28692724 following meta-analysis with an independent sample of European women. Rs28692724 is a synonymous variant in IRF2BPL, which encodes a transcription factor that, acting within the neuroendocrine system, plays a role in regulating female reproductive function (66).
Differences in association by background group
Many of the loci mapped in this study displayed effect heterogeneity by background group. For example, the NTM locus associated with WHRadjBMI in women, displayed nearly 3-fold the effect size in the mainland background group when compared with the Caribbean background group. Also, for the women-only primary analysis, rs77186623 in the LOC105375745 locus displayed a 4-fold greater effect in the Caribbean background group compared with the mainland group. These and other loci displaying heterogeneity by background group (i.e. MARCKSL1, C19orf67, RIT2, MAP4K4, COBL, NREP and ANO10) were not validated in replication analyses, possibly due in part to heterogeneity by background group.
Limitations
A limitation of this study was the small sample size within each HCHS/SOL background group. However, the use of genetic analysis groups in our main analyses accounted for the heterogeneity of genetic effects among ethnic groups often ignored in GWAS studies. Compared with self-identified background groups, genetic analysis groups are more genetically homogeneous and lack principal component outliers in stratified analysis, which may hinder detection of and adjustment for important population structure when ignored (67). In addition, genetic analysis groups allow all individuals to be classified in a specific group, whereas many individuals in HCHS/SOL have a missing or non-specific self-identified background (67). Therefore, by using genetic analysis groups in our analysis rather than self-identified groups, we have increased our study’s power to detect novel and previously documented associations with central adiposity traits (67). Owing to the diverse background of our discovery population, another limitation was the lack of an ideal replication study. We attempted to overcome this limitation by focusing on both multi-ethnic meta-analyses, which would validate those associations that generalize across ancestries, and meta-analyses stratified by ancestry, which may allow for validation of more population-specific associations. However, it is possible that the limited Native American ancestry present across our replication cohorts may have hindered replication, and further analyses in more diverse Hispanic/Latino populations are needed to confirm the relevance of promising central adiposity associated loci identified in our study. Last, we attempted to leverage bioinformatics databases to assist in evaluating the potential functional effects of our top associations, including lookup of previous evidence of cis regulation of gene expression. However, a possible limitation of these lookups is the lack of diversity in resources like GTEx, which are derived from European ancestry populations (Supplementary Material, Table S14), and thus our tag SNP may not be well represented owing to differences in the LD structure. Future investigations into the potential regulatory function of our associated loci are needed in ancestrally relevant sample populations and available Omics data.
Conclusion
We identified four novel loci for central adiposity traits in a large population of Hispanic/Latino Americans. We also found that several previously identified central adiposity loci discovered in European American populations generalized to Hispanic/Latino Americans. Many of the loci interrogated exhibit background-group-specific effects, likely owing to population history (admixture and natural selection), that have resulted in changes in LD, or allele frequency differences or owing to variation in etiology. These observations highlight the importance of large-scale genomic studies in ancestrally diverse populations for identifying obesity-susceptibility loci that generalize and those that are ancestry-specific.
Materials and Methods
Study sample
Details on the study and sampling design of the HCHS/SOL have been previously described (68). Briefly, HCHS/SOL is a community-based prospective cohort study of 16 415 self-identified Hispanic/Latino adults who were aged 18–74 years at screening from randomly selected households in four US field centers (Chicago, IL; Miami, FL; Bronx, NY and San Diego, CA) with baseline examination (2008–2011) and yearly telephone follow-up assessment for at least 3 years. The HCHS/SOL cohort includes participants who self-identified as being Central American (n = 1732), Cuban (n = 2348), Dominican (n = 1473), Mexican (n = 6472), Puerto-Rican (n = 2728) and South American (n = 1072). The goals of the HCHS/SOL are to describe the prevalence of risk and the protective factors for chronic conditions (e.g. CVD, diabetes and pulmonary disease) and to quantify all-cause mortality, fatal and non-fatal CVD and pulmonary disease and pulmonary disease exacerbation over time. The baseline clinical examination (69) included comprehensive biological (e.g. anthropometrics, blood draw, oral glucose tolerance test, ankle brachial pressure index and electrocardiogram), behavioral (e.g. dietary intake assessed with two 24 h recalls, physical activity assessment by accelerometer and self-report, overnight sleep exam for apneic events, tobacco and alcohol assessed by self-report) and socio-demographic (e.g. socioeconomic status and migration history) assessments. This study was approved by the institutional review boards at each field center where all subjects gave written informed consent.
Participants in HCHS/SOL self-identified their background as Mexican, Central American, South American (mainland), Puerto Rican, Cuban or Dominican (Caribbean). Some participants chose ‘more than one’, ‘other’ or chose not to self-identify. We addressed the missing or inconsistent data in self-identified background groups by defining ‘genetic analysis groups’ described in Conomos et al. (67). To increase power in this analysis, we chose to stratify by the broader mainland or Caribbean categories rather than more specific groups. In this paper, we will use the term ‘background group’ to refer to a super-group of genetic analysis groups by geographic region, mainland or Caribbean. Hispanics/Latinos have admixed ancestry from three continents: Africa, America and Europe. In general, participants from the mainland group have higher proportions of American ancestry and lower African ancestry, while participants in the Caribbean group have higher proportions of African ancestry (67).
Phenotypes
All variables were taken from the baseline visit. Participants were dressed in scrub suits or light non-constricting clothing, and shoes were removed for weight and height measurements. WC and HIP were measured using Gulick II 150 and 250 cm anthropometric tape and rounded to the nearest centimeter (cm). Height was measured using a wall-mounted stadiometer and rounded to the nearest cm, and weight measured with a Tanita Body Composition Analyzer, TBF-300A, to the nearest tenth of a kg. Height and weight were used to calculate BMI (kg/m2). We applied a log10 transformation on HIP owing to its non-normal trait distribution.
Genotyping
Our analyses included 7472 women and 5200 men of mainland (Mexican, Central and South American) or Caribbean (Puerto Rican, Cuban and Dominican) ancestry residing in the USA. All participants were genotyped on the Illumina SOL Omni2.5M custom content array, which was subsequently used to impute millions of additional variants, based on the entire 1000 Genomes Phase I Reference panel, including populations with Native American ancestry: MXL, CLM and PUR. Pre-phasing was performed using SHAPEIT, followed by imputation with IMPUTE2 (70,71).
Discovery analyses
Owing to known differences in genetic effects on waist and hip traits between men and women (14,32,72), we analyzed associations stratified by sex for each trait, in addition to the entire sample. We used linear mixed-model regression, assuming an additive genetic model adjusted for age, age2, study center, sample weights, genetic analysis background group (67,73), principal components to account for ancestry, population structure using kinship coefficients and sample eigenvectors, household, census block group and sex in the combined analysis. Kinship, household and block group were treated as random effects in each model. Sample weights were incorporated in our models as a fixed effect to account for oversampling of the communities in the 45–74 age group (n = 9714, 59.2%), which was intended to facilitate the examination of HCHS/SOL target outcomes. HCHS/SOL sampling weights are the product of a ‘base weight’ (reciprocal of the probability of selection) and three adjustments: (1) non-response adjustments made relative to the sampling frame, (2) trimming to handle extreme values (to avoid a few weights with extreme values being overly influential in the analyses) and (3) calibration of weights to the 2010 US Census according to age, sex and Hispanic background. We used genetic analysis groups in our analyses that accounted for heterogeneity of genetic effects among ethnic groups. Compared with self-identified background groups, genetic analysis groups are more genetically homogeneous and lack principal component outliers in stratified analysis, which may hinder detection of and adjustment for important population structure when ignored (67). In addition, genetic analysis groups allow all individuals to be classified in a specific group, whereas many individuals in HCHS/SOL have a missing or non-specific self-identified background (67). Also, we conducted stratified analyses by region (mainland vs. Caribbean) to identify potential heterogeneity in effect by background group. We examined heterogeneity across background group using I2 statistics calculated using METAL (74) and tested for significant interaction (Pdiff < 0.05) by background group using EasyStrata (75).
To decrease the number of spurious associations, we filtered all results on MAF < 0.5%, Hardy–Weinberg Equilibrium (HWE) P < 1 × 10−7, minor allele count [MAC (effective N)] < 30 (67). Additionally, we categorized suggestive loci as those with variants reaching P < 1 × 10−6 and with at least one additional variant within 500 kb+/− with a P < 1 × 10−5. We used regional association plots produced in LocusZoom to visualize association regions using unrelated individuals from HCHS/SOL for LD (http://locuszoom.sph.umich.edu/).
Local ancestry estimation
We estimated local ancestry (African, Native American and European) using RFMix (76), which applies a conditional-random-field-based approach for estimation to inform differences by background group. We used 236 456 genotyped SNPs available in both HCHS/SOL and reference-panel datasets in the Human Genome Diversity Project (HGDP) (77), HapMap 3 (78) and 1000 Genomes phase 1 for detecting African, Native American and European ancestry. We used BEAGLE (v.4) to phase and impute sporadic missing genotypes in the HCHS/SOL and reference-panel datasets (79).
Replication and meta-analyses
An aim of our study was to identify genetic variants that associate with central adiposity, which may vary by ancestry. Therefore, we sought to replicate our association findings using 1000 Genomes imputed GWAS data available in independent cohorts, including eight studies with Hispanics/Latinos (HL: N up to 12 341), three studies with African Americans (AA; N up to 12 496) and one study with European-Americans (EUR: N up to 8845). Study design and descriptive statistics for each replication study are provided in Supplementary Material, Table S1. Each replication study excluded individuals who were pregnant or exhibited extreme values for waist or hip measures (outside of ±4 SD from the mean). Each study used measures from a single visit with the greatest sample size. We used linear regression (or linear mixed effects models if the study had related individuals) association analyses on the trait residuals after adjustment for age, age2, principal components to account for ancestry, BMI, other study specific factors (e.g. study center) and sex in the sex-combined analysis, stratified by race/ethnicity where applicable for each SNP that reached suggestive significance (P < 1 × 10−6) in the discovery analysis.
We employed a fixed-effects meta-analysis using the inverse variance-weighted method for WHRadjBMI and WCadjBMI. For HIPadjBMI, owing to trait transformations, we used sample-size-weighted meta-analysis. All meta-analyses were implemented in METAL (80). We conducted meta-analyses stratified by race/ethnicity group and combined across groups. We included SNPs with a study- and stratum-specific imputation quality (Rsq) greater than 0.4, HWE P-value greater than 1 × 10−7 and a MAC greater than five. To declare statistical significance for replicated loci, we required in each replication sample a trait and stratum-specific P < 0.05 with a consistent direction of effect with discovery and genome-wide significance (P < 5 × 10−8) when meta-analyzed together with HCHS/SOL.
Generalization
To examine whether previously reported association regions generalized to the HCHS/SOL, we downloaded the publicly available multi-ethnic (European, Asian and African ancestry) GWAS results from the GIANT consortium (14) for WHRadjBMI, WCadjBMI and HIPadjBMI (https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files#GIANT_consortium_2012-2015_GWAS_Metadata_is_Available_Here_for_Download) in men, women and sexes-combined, and then we applied the framework of Sofer et al. (2017) for generalization testing (81). We took all variant associations with P < 1 × 10−6 in GIANT and identified the matching association test in HCHS/SOL. For each such association, we calculated a directional False Discovery Rate (FDR) r-value by combining the P-values from GIANT and HCHS/SOL, while accounting for multiple testing and for the direction of estimated associations in each of the studies. An association was declared as generalized, while controlling for the FDR at the 0.05 level, if its r-value was smaller than 0.05. Multiple SNPs from the same region were tested. Therefore, in an iterative procedure, we pruned the results list by identifying the SNP with the lowest r-value in an analysis, then finding all SNPs in a 1 MB region around it and removing them from the list. Thus, the number of generalized regions is the number of generalized SNPs in the pruned list.
We also hypothesized that some regions did not generalize owing to lack of power (the HCHS/SOL sample size is much smaller than the GIANT sample size). To test this, we took all tested SNPs from the non-generalized regions and considered the GIANT multi-ethnic GWAS results. In an iterative procedure, we pruned the list by first identifying the SNP with lowest GIANT P-value in the analysis, then found all SNPs in a 1 MB region around it and removed them from the list. We repeated until no SNPs remained. All the SNPs in the pruned list were selected solely based on their GIANT P-values. Since there were many such variants, we further grouped them according to their P-values. Groups were formed by trait, sex (men, women and combined) and GIANT P-value (between 10−6 and 10−7, between 10−7 and 10−8 and smaller than 10−8). For each such group of SNPs, we created a genetic risk score (GRS) in HCHS/SOL. For each sex stratum and each group of SNPs, the value of the GRS was the sum of all trait increasing alleles in that group. We chose an unweighted GRS as effect sizes derived from primarily European ancestry GWAS are not easily transferable to admixed populations (82). We tested the GRS in the appropriate analysis group (men, women and combined). A low P-value implies that some of the SNPs in the group are likely associated with the trait in HCHS/SOL.
Biological curation
To gain further insight into the possible functional role of the identified variants and to assess the relevance of our identified variants with other phenotypes, we conducted lookups of our replicated variants in multiple publicly available databases, including PhenoScanner (36), RegulomeDB (38), Haploreg (39) and UCSC GenomeBrowser (40). Additionally, we conducted lookups of nearby genes in GTeX (41). The R package HaploR was used to query HaploReg and RegulomeDB (https://cran.r-project.org/web/packages/haploR/vignettes/haplor-vignette.html).
Supplementary Material
Acknowledgements
The authors thank the staff and participants of HCHS/SOL for their important contributions. (Investigators website—http://www.cscc.unc.edu/hchs/). The HCHS/SOL is a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (HHSN268201300001I/N01-HC-65233), University of Miami (HHSN268201300004I/N01-HC-65234), Albert Einstein College of Medicine (HHSN268201300002I/N01-HC-65235), University of Illinois at Chicago—HHSN268201300003I/N01-HC-65236 Northwestern Univ) and San Diego State University (HHSN268201300005I/N01-HC-65237). The following Institutes/Centers/Offices have contributed to the HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Neurological Disorders and Stroke and National Institute of Health (NIH) Institution-Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). The Atherosclerosis Risk in Communities Study (ARIC) has been funded in whole or in part with Federal funds from the NHLBI, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HHSN268201700005I), R01HL087641, R01HL059367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. The authors thank the staff and participants of the ARIC study for their important contributions. This research was supported by the GUARDIAN (Genetics Underlying Diabetes in Hispanics) Study, NIDDK contract DK085175; Mexican-American Coronary Artery Disease (MACAD) NHLBI, contracts R01-HL088457, R01-HL-60030; NHLBI, Hypertension and Insulin Resistance (HTN-IR) contracts R01-HL067974, R01-HL-55005 and R01-HL 067974; National Institutes of Health, Genetics of Latino Diabetic Retinopathy (GOLDR) contract EY014684. MESA and the MESA SHARe projects are conducted and supported by the NHLBI in collaboration with MESA investigators. Support for MESA is provided by contracts 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079 and UL1-TR-001420. Also supported by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881 and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The Mexico City 1 and Mexico City 2 studies were supported in Mexico by the Fondo Sectorial de Investigación en Salud y Seguridad Social (SSA/IMSS/ISSSTECONACYT, project 150352), Temas Prioritarios de Salud Instituto Mexicano del Seguro Social (2014-FIS/IMSS/PROT/PRIO/14/34) and the Fundación IMSS. We thank Miguel Alexander Vazquez Moreno, Daniel Locia and Araceli Méndez Padrón for technical support in Mexico. In Canada, this research was enabled in part by two Canadian Institutes for Health Research (CIHR) Operating grants to EJP, a CIHR New Investigator Award to EJP and by support provided by Compute Ontario (www.computeontario.ca) and Compute Canada (www.compute.canada.ca). Funding support for the ‘Epidemiology of putative genetic variants: The Women’s Health Initiative’ study is provided through the NHGRI grants HG006292 and HL129132. The WHI program is funded by the NHLBI, National Institutes of Health, US Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C and HHSC271201100004C. The authors thank the WHI investigators and staff, for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whiscience.org/publications/WHI_investigators_shortlist.pdf.
Conflict of Interest statement. None declared.
Funding
National Institute of Health (NIH), National Heart, Lung, and Blood Institutes (NHLBI) (grant K99/R00 HL 130580) and American Heart Association (AHA) (grant 13POST16500011) to A.E.J.; National Heart, Lung, and Blood Institutes (NHLBI) (R01-HL136528 to Y.C.K.); National Institute of Health National Heart, Lung, and Blood Institutes (grant T32 HL007055 to H.M.H.); National Institute of Health National Heart, Lung, and Blood Institutes (grant R01HL142302 to K.E.N., M.G., J.E.B. and L.P.); National Institute of Health (N01-HC-65233, N01-HC-65234, N01-HC-65235, N01-HC-65236, N01-HC-65237, R01HL087641, R01HL059367, R01HL086694, U01HG004402, UL1RR025005, DK085175, R01-HL088457, R01-HL-60030, R01-HL067974, R01-HL-55005, R01-HL 067974, EY014684, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1TR001881, DK063491, HG006292 and HL129132); Fondo Sectorial de Investigación en Salud y Seguridad Social (150352); Temas Prioritarios de Salud Instituto Mexicano del Seguro Social (2014-FIS/IMSS/PROT/PRIO/14/34); the Fundación IMSS; Canadian Institutes for Health Research (to E.P.); Compute Ontario; Compute Canada.
References
- 1. Flegal, K.M., Carroll, M.D., Kit, B.K. and Ogden, C.L. (2012) Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999-2010. JAMA, 307, 491–497. [DOI] [PubMed] [Google Scholar]
- 2. Ogden, C.L., Carroll, M.D., Curtin, L.R., McDowell, M.A., Tabak, C.J. and Flegal, K.M. (2006) Prevalence of overweight and obesity in the United States, 1999-2004. JAMA, 295, 1549–1555. [DOI] [PubMed] [Google Scholar]
- 3. Stevens, G.A., Singh, G.M., Lu, Y., Danaei, G., Lin, J.K., Finucane, M.M., Bahalim, A.N., McIntire, R.K., Gutierrez, H.R., Cowan, M. et al. (2012) National, regional, and global trends in adult overweight and obesity prevalences. Popul. Health Metrics, 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Finucane, M.M., Stevens, G.A., Cowan, M.J., Danaei, G., Lin, J.K., Paciorek, C.J., Singh, G.M., Gutierrez, H.R., Lu, Y., Bahalim, A.N. et al. (2011) National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet, 377, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hales C.M., Carroll M.D., Fryar C.D., Ogden C.L. (2017) Prevalence of obesity among adults and youth: United States, 2015-2016. NCHS Data Brief, 288, 1–8. [PubMed] [Google Scholar]
- 6. Elks, C.E., den Hoed, M., Zhao, J.H., Sharp, S.J., Wareham, N.J., Loos, R.J. and Ong, K.K. (2012) Variability in the heritability of body mass index: a systematic review and meta-regression. Front. Endocrinol. (Lausanne), 3, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Norden-Krichmar, T.M., Gizer, I.R., Libiger, O., Wilhelmsen, K.C., Ehlers, C.L. and Schork, N.J. (2014) Correlation analysis of genetic admixture and social identification with body mass index in a Native American community. Am. J. Hum. Biol., 26, 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ravussin, E., Valencia, M.E., Esparza, J., Bennett, P.H. and Schulz, O. (1994) Effects of a traditional lifestyle on obesity in Pima Indians. Diabetes Care, 17, 1067–1074. [DOI] [PubMed] [Google Scholar]
- 9. Bogardus, C., Lillioja, S., Ravussin, E., Abbott, W.G., Zawadzki, J.K., Young, A., Knowler, W.C., Jacobowitz, R. and Moll, P.P. (1986) Familial dependence of the resting metabolic rate. N. Engl. J. Med., 315, 96–100. [DOI] [PubMed] [Google Scholar]
- 10. Knowler, W.C., Pettitt, D.J., Saad, M.F., Charles, M.A., Nelson, R.G., Howard, B.V., Bogardus, C. and Bennett, P.H. (1991) Obesity in the Pima Indians: its magnitude and relationship with diabetes. Am. J. Clin. Nutr., 53, 1543S–1551S. [DOI] [PubMed] [Google Scholar]
- 11. Abate, N. and Chandalia, M. (2007) Ethnicity, type 2 diabetes & migrant Asian Indians. Indian J. Med. Res., 125, 251–258. [PubMed] [Google Scholar]
- 12. Nassir, R., Qi, L., Kosoy, R., Garcia, L., Allison, M., Ochs-Balcom, H.M., Tylavsky, F., Manson, J.E., Shigeta, R., Robbins, J. et al. (2012) Relationship between adiposity and admixture in African-American and Hispanic-American women. Int. J. Obes., 36, 304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Locke, A.E., Kahali, B., Berndt, S.I., Justice, A.E., Pers, T.H., Day, F.R., Powell, C., Vedantam, S., Buchkovich, M.L., Yang, J. et al. (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature, 518, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shungin, D., Winkler, T.W., Croteau-Chonka, D.C., Ferreira, T., Locke, A.E., Magi, R., Strawbridge, R.J., Pers, T.H., Fischer, K., Justice, A.E. et al. (2015) New genetic loci link adipose and insulin biology to body fat distribution. Nature, 518, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Justice, A.E., Winkler, T.W., Feitosa, M.F., Graff, M., Fisher, V.A., Young, K., Barata, L., Deng, X., Czajkowski, J., Hadley, D. et al. (2017) Genome-wide meta-analysis of 241,258 adults accounting for smoking behaviour identifies novel loci for obesity traits. Nat. Commun., 8, 14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ng, M.C.Y., Graff, M., Lu, Y., Justice, A.E., Mudgal, P., Liu, C.T., Young, K., Yanek, L.R., Feitosa, M.F., Wojczynski, M.K. et al. (2017) Discovery and fine-mapping of adiposity loci using high density imputation of genome-wide association studies in individuals of African ancestry: African Ancestry Anthropometry Genetics Consortium. PLoS Genet., 13, e1006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Graff, M., Scott, R.A., Justice, A.E., Young, K.L., Feitosa, M.F., Barata, L., Winkler, T.W., Chu, A.Y., Mahajan, A., Hadley, D. et al. (2017) Genome-wide physical activity interactions in adiposity - a meta-analysis of 200,452 adults. PLoS Genet., 13, e1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berndt, S.I., Gustafsson, S., Magi, R., Ganna, A., Wheeler, E., Feitosa, M.F., Justice, A.E., Monda, K.L., Croteau-Chonka, D.C., Day, F.R. et al. (2013) Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet., 45, 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wen, W., Cho, Y.S., Zheng, W., Dorajoo, R., Kato, N., Qi, L., Chen, C.H., Delahanty, R.J., Okada, Y., Tabara, Y. et al. (2012) Meta-analysis identifies common variants associated with body mass index in east Asians. Nat. Genet., 44, 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cho, Y.S., Go, M.J., Kim, Y.J., Heo, J.Y., Oh, J.H., Ban, H.J., Yoon, D., Lee, M.H., Kim, D.J., Park, M. et al. (2009) A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet., 41, 527–534. [DOI] [PubMed] [Google Scholar]
- 21. Monda K.L., Chen G.K., Taylor K.C., Palmer C., Edwards T.L., Lange L.A., Ng M.C., Adeyemo A.A., Allison M.A., Bielak L.F., et al. (2013) A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet., 45, 690–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Folsom, A.R., Kushi, L.H., Anderson, K.E., Mink, P.J., Olson, J.E., Hong, C.P., Sellers, T.A., Lazovich, D. and Prineas, R.J. (2000) Associations of general and abdominal obesity with multiple health outcomes in older women: the Iowa Women’s Health Study. Arch. Intern. Med., 160, 2117–2128. [DOI] [PubMed] [Google Scholar]
- 23. Lavie, C.J., Milani, R.V. and Ventura, H.O. (2009) Obesity and cardiovascular disease: risk factor, paradox, and impact of weight loss. J. Am. Coll. Cardiol., 53, 1925–1932. [DOI] [PubMed] [Google Scholar]
- 24. Huxley, R., Mendis, S., Zheleznyakov, E., Reddy, S. and Chan, J. (2010) Body mass index, waist circumference and waist:hip ratio as predictors of cardiovascular risk—a review of the literature. Eur. J. Clin. Nutr., 64, 16–22. [DOI] [PubMed] [Google Scholar]
- 25. Zyriax, B.C., Schoeffauer, M., Klipstein-Grobusch, K., Boeing, H. and Windler, E. (2011) Differential association of anthropometric parameters with coronary risk in women—data of the CORA study. Obes. Facts, 4, 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carey, V.J., Walters, E.E., Colditz, G.A., Solomon, C.G., Willett, W.C., Rosner, B.A., Speizer, F.E. and Manson, J.E. (1997) Body fat distribution and risk of non-insulin-dependent diabetes mellitus in women. The Nurses’ Health Study. Am. J. Epidemiol., 145, 614–619. [DOI] [PubMed] [Google Scholar]
- 27. Wang, Y., Rimm, E.B., Stampfer, M.J., Willett, W.C. and Hu, F.B. (2005) Comparison of abdominal adiposity and overall obesity in predicting risk of type 2 diabetes among men. Am. J. Clin. Nutr., 81, 555–563. [DOI] [PubMed] [Google Scholar]
- 28. Canoy, D. (2008) Distribution of body fat and risk of coronary heart disease in men and women. Curr. Opin. Cardiol., 23, 591–598. [DOI] [PubMed] [Google Scholar]
- 29. Snijder, M.B., Dekker, J.M., Visser, M., Yudkin, J.S., Stehouwer, C.D., Bouter, L.M., Heine, R.J., Nijpels, G. and Seidell, J.C. (2003) Larger thigh and hip circumferences are associated with better glucose tolerance: the Hoorn study. Obes. Res., 11, 104–111. [DOI] [PubMed] [Google Scholar]
- 30. Lango Allen, H., Estrada, K., Lettre, G., Berndt, S.I., Weedon, M.N., Rivadeneira, F., Willer, C.J., Jackson, A.U., Vedantam, S., Raychaudhuri, S. et al. (2010) Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature, 467, 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fox, C.S., White, C.C., Lohman, K., Heard-Costa, N., Cohen, P., Zhang, Y., Johnson, A.D., Emilsson, V., Liu, C.T., Chen, Y.D. et al. (2012) Genome-wide association of pericardial fat identifies a unique locus for ectopic fat. PLoS Genet., 8, e1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Randall, J.C., Winkler, T.W., Kutalik, Z., Berndt, S.I., Jackson, A.U., Monda, K.L., Kilpelainen, T.O., Esko, T., Magi, R., Li, S. et al. (2013) Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet., 9, e1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pulit, S.L., Stoneman, C., Morris, A.P., Wood, A.R., Glastonbury, C.A., Tyrrell, J., Yengo, L., Ferreira, T., Marouli, E., Ji, Y. et al. (2019) Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet., 28, 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tachmazidou, I., Suveges, D., Min, J.L., Ritchie, G.R.S., Steinberg, J., Walter, K., Iotchkova, V., Schwartzentruber, J., Huang, J., Memari, Y. et al. (2017) Whole-genome sequencing coupled to imputation discovers genetic signals for anthropometric traits. Am. J. Hum. Genet., 100, 865–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wen, W., Kato, N., Hwang, J.Y., Guo, X., Tabara, Y., Li, H., Dorajoo, R., Yang, X., Tsai, F.J., Li, S. et al. (2016) Genome-wide association studies in East Asians identify new loci for waist-hip ratio and waist circumference. Sci. Rep., 6, 17958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Staley, J.R., Blackshaw, J., Kamat, M.A., Ellis, S., Surendran, P., Sun, B.B., Paul, D.S., Freitag, D., Burgess, S., Danesh, J. et al. (2016) PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics, 32, 3207–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kamat, M.A., Blackshaw, J.A., Young, R., Surendran, P., Burgess, S., Danesh, J., Butterworth, A.S. and Staley, J.R. (2019) PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics, 35, 4851–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boyle, A.P., Hong, E.L., Hariharan, M., Cheng, Y., Schaub, M.A., Kasowski, M., Karczewski, K.J., Park, J., Hitz, B.C., Weng, S. et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res., 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ward, L.D. and Kellis, M. (2016) HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res., 44, D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M. and Haussler, D. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. GTEx Consortium (2013) The genotype-tissue expression (GTEx) project. Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gallagher, E., Mc Goldrick, A., Chung, W.Y., Mc Cormack, O., Harrison, M., Kerin, M., Dervan, P.A. and Mc Cann, A. (2006) Gain of imprinting of SLC22A18 sense and antisense transcripts in human breast cancer. Genomics, 88, 12–17. [DOI] [PubMed] [Google Scholar]
- 43. Gardiner, K., Chitayat, D., Choufani, S., Shuman, C., Blaser, S., Terespolsky, D., Farrell, S., Reiss, R., Wodak, S., Pu, S. et al. (2012) Brain abnormalities in patients with Beckwith-Wiedemann syndrome. Am. J. Med. Genet. A, 158A, 1388–1394. [DOI] [PubMed] [Google Scholar]
- 44. Weksberg, R., Shuman, C., Caluseriu, O., Smith, A.C., Fei, Y.L., Nishikawa, J., Stockley, T.L., Best, L., Chitayat, D., Olney, A. et al. (2002) Discordant KCNQ1OT1 imprinting in sets of monozygotic twins discordant for Beckwith-Wiedemann syndrome. Hum. Mol. Genet., 11, 1317–1325. [DOI] [PubMed] [Google Scholar]
- 45. Brioude, F., Kalish, J.M., Mussa, A., Foster, A.C., Bliek, J., Ferrero, G.B., Boonen, S.E., Cole, T., Baker, R., Bertoletti, M. et al. (2018) Expert consensus document: clinical and molecular diagnosis, screening and management of Beckwith-Wiedemann syndrome: an international consensus statement. Nat. Rev. Endocrinol., 14, 229–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abecasis, G.R., Auton, A., Brooks, L.D., DePristo, M.A., Durbin, R.M., Handsaker, R.E., Kang, H.M., Marth, G.T. and McVean, G.A. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rao, G., Powell-Wiley, T.M., Ancheta, I., Hairston, K., Kirley, K., Lear, S.A., North, K.E., Palaniappan, L. and Rosal, M.C. (2015) Identification of obesity and cardiovascular risk in ethnically and racially diverse populations: a scientific statement from the American Heart Association. Circulation, 132, 457–472. [DOI] [PubMed] [Google Scholar]
- 48. Heymsfield, S.B., Peterson, C.M., Thomas, D.M., Heo, M. and Schuna, J.M., Jr. (2016) Why are there race/ethnic differences in adult body mass index-adiposity relationships? A quantitative critical review. Obes. Rev., 17, 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wagner, D.R. and Heyward, V.H. (2000) Measures of body composition in blacks and whites: a comparative review. Am. J. Clin. Nutr., 71, 1392–1402. [DOI] [PubMed] [Google Scholar]
- 50. Rush, E.C., Goedecke, J.H., Jennings, C., Micklesfield, L., Dugas, L., Lambert, E.V. and Plank, L.D. (2007) BMI, fat and muscle differences in urban women of five ethnicities from two countries. Int. J. Obes., 31, 1232–1239. [DOI] [PubMed] [Google Scholar]
- 51. Hull, H.R., Thornton, J., Wang, J., Pierson, R.N.,.J., Kaleem, Z., Pi-Sunyer, X., Heymsfield, S., Albu, J., Fernandez, J.R., Vanitallie, T.B. et al. (2011) Fat-free mass index: changes and race/ethnic differences in adulthood. Int. J. Obes., 35, 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Justice, A.E., Karaderi, T., Highland, H.M., Young, K.L., Graff, M., Lu, Y., Turcot, V., Auer, P.L., Fine, R.S., Guo, X. et al. (2019) Protein-coding variants implicate novel genes related to lipid homeostasis contributing to body-fat distribution. Nat. Genet., 51, 452–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vaittinen, M., Walle, P., Kuosmanen, E., Mannisto, V., Kakela, P., Agren, J., Schwab, U. and Pihlajamaki, J. (2016) FADS2 genotype regulates delta-6 desaturase activity and inflammation in human adipose tissue. J. Lipid Res., 57, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Teslovich, T.M., Musunuru, K., Smith, A.V., Edmondson, A.C., Stylianou, I.M., Koseki, M., Pirruccello, J.P., Ripatti, S., Chasman, D.I., Willer, C.J. et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature, 466, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fumagalli, M., Moltke, I., Grarup, N., Racimo, F., Bjerregaard, P., Jorgensen, M.E., Korneliussen, T.S., Gerbault, P., Skotte, L., Linneberg, A. et al. (2015) Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science, 349, 1343–1347. [DOI] [PubMed] [Google Scholar]
- 56. Mathieson, I., Lazaridis, I., Rohland, N., Mallick, S., Patterson, N., Roodenberg, S.A., Harney, E., Stewardson, K., Fernandes, D., Novak, M. et al. (2015) Genome-wide patterns of selection in 230 ancient Eurasians. Nature, 528, 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mathias, R.A., Fu, W., Akey, J.M., Ainsworth, H.C., Torgerson, D.G., Ruczinski, I., Sergeant, S., Barnes, K.C. and Chilton, F.H. (2012) Adaptive evolution of the FADS gene cluster within Africa. PLoS One, 7, e44926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mathias, R.A., Sergeant, S., Ruczinski, I., Torgerson, D.G., Hugenschmidt, C.E., Kubala, M., Vaidya, D., Suktitipat, B., Ziegler, J.T., Ivester, P. et al. (2011) The impact of FADS genetic variants on omega6 polyunsaturated fatty acid metabolism in African Americans. BMC Genet., 12, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sukumaran, S.K., Stumpf, M., Salamon, S., Ahmad, I., Bhattacharya, K., Fischer, S., Müller, R., Altmüller, J., Budde, B., Thiele, H. et al. (2017) CDK5RAP2 interaction with components of the hippo signaling pathway may play a role in primary microcephaly. Mol. Genet. Genom., 292, 365–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Barrera, J.A., Kao, L.R., Hammer, R.E., Seemann, J., Fuchs, J.L. and Megraw, T.L. (2010) CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev. Cell, 18, 913–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Erten-Lyons, D., Wilmot, B., Anur, P., McWeeney, S., Westaway, S.K., Silbert, L., Kramer, P. and Kaye, J. (2011) Microcephaly genes and risk of late-onset Alzheimer disease. Alzheimer Dis. Assoc. Disord., 25, 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miron, J., Picard, C., Nilsson, N., Frappier, J., Dea, D., Theroux, L. and Poirier, J. (2018) CDK5RAP2 gene and tau pathophysiology in late-onset sporadic Alzheimer’s disease. Alzheimers Dement., 14, 787–796. [DOI] [PubMed] [Google Scholar]
- 63. Schwienbacher, C., Sabbioni, S., Campi, M., Veronese, A., Bernardi, G., Menegatti, A., Hatada, I., Mukai, T., Ohashi, H., Barbanti-Brodano, G. et al. (1998) Transcriptional map of 170-kb region at chromosome 11p15.5: identification and mutational analysis of the BWR1A gene reveals the presence of mutations in tumor samples. Proc. Natl. Acad. Sci. U. S. A., 95, 3873–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bajaj, V., Markandaya, M., Krishna, L. and Kumar, A. (2004) Paternal imprinting of the SLC22A1LS gene located in the human chromosome segment 11p15.5. BMC Genet., 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bajaj, V., Singhmar, P. and Kumar, A. (2008) Promoter characterization and regulation of expression of an imprinted gene SLC22A18AS. Gene, 424, 40–47. [DOI] [PubMed] [Google Scholar]
- 66. Heger, S., Mastronardi, C., Dissen, G.A., Lomniczi, A., Cabrera, R., Roth, C.L., Jung, H., Galimi, F., Sippell, W. and Ojeda, S.R. (2007) Enhanced at puberty 1 (EAP1) is a new transcriptional regulator of the female neuroendocrine reproductive axis. J. Clin. Invest., 117, 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Conomos, M.P., Laurie, C.A., Stilp, A.M., Gogarten, S.M., McHugh, C.P., Nelson, S.C., Sofer, T., Fernandez-Rhodes, L., Justice, A.E., Graff, M. et al. (2016) Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet., 98, 165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lavange, L.M., Kalsbeek, W.D., Sorlie, P.D., Aviles-Santa, L.M., Kaplan, R.C., Barnhart, J., Liu, K., Giachello, A., Lee, D.J., Ryan, J. et al. (2010) Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Ann. Epidemiol., 20, 642–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sorlie, P.D., Aviles-Santa, L.M., Wassertheil-Smoller, S., Kaplan, R.C., Daviglus, M.L., Giachello, A.L., Schneiderman, N., Raij, L., Talavera, G., Allison, M. et al. (2010) Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann. Epidemiol., 20, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Howie, B., Marchini, J. and Stephens, M. (2011) Genotype imputation with thousands of genomes. G3 (Bethesda), 1, 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Delaneau, O., Marchini, J. and Zagury, J.F. (2011) A linear complexity phasing method for thousands of genomes. Nat. Methods, 9, 179–181. [DOI] [PubMed] [Google Scholar]
- 72. Winkler, T.W., Justice, A.E., Graff, M., Barata, L., Feitosa, M.F., Chu, S., Czajkowski, J., Esko, T., Fall, T., Kilpelainen, T.O. et al. (2015) The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet., 11, e1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Conomos, M.P., Miller, M.B. and Thornton, T.A. (2015) Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol., 39, 276–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Willer, C.J., Li, Y. and Abecasis, G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Winkler, T.W., Kutalik, Z., Gorski, M., Lottaz, C., Kronenberg, F. and Heid, I.M. (2015) Easy strata: evaluation and visualization of stratified genome-wide association meta-analysis data. Bioinformatics, 31, 259–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Maples, B.K., Gravel, S., Kenny, E.E. and Bustamante, C.D. (2013) RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet., 93, 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cavalli-Sforza, L.L. (2005) The human genome diversity project: past, present and future. Nat. Rev. Genet., 6, 333–340. [DOI] [PubMed] [Google Scholar]
- 78. International Hap Map Consortium, Altshuler, D.M., Gibbs, R.A., Peltonen, L., Altshuler, D.M., Gibbs, R.A., Peltonen, L., Dermitzakis, E., Schaffner, S.F., Yu, F. et al. (2010) Integrating common and rare genetic variation in diverse human populations. Nature, 467, 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Browning, S.R. and Browning, B.L. (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet., 81, 1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Heid, I.M., Jackson, A.U., Randall, J.C., Winkler, T.W., Qi, L., Steinthorsdottir, V., Thorleifsson, G., Zillikens, M.C., Speliotes, E.K., Magi, R. et al. (2010) Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet., 42, 949–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sofer, T., Heller, R., Bogomolov, M., Avery, C.L., Graff, M., North, K.E., Reiner, A.P., Thornton, T.A., Rice, K., Benjamini, Y. et al. (2017) A powerful statistical framework for generalization testing in GWAS, with application to the HCHS/SOL. Genet. Epidemiol., 41, 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Martin, A.R., Kanai, M., Kamatani, Y., Okada, Y., Neale, B.M. and Daly, M.J. (2019) Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet., 51, 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


