Abstract
All bacteria produce secreted vesicles that carry out a variety of important biological functions. These extracellular vesicles can improve adaptation and survival by relieving bacterial stress and eliminating toxic compounds, as well as by facilitating membrane remodeling and ameliorating inhospitable environments. However, vesicle production comes with a price. It is energetically costly and, in the case of colonizing pathogens, it elicits host immune responses, which reduce bacterial viability. This raises an interesting paradox regarding why bacteria produce vesicles and begs the question as to whether the benefits of producing vesicles outweigh their costs. In this review, we discuss the various advantages and disadvantages associated with Gram‐negative and Gram‐positive bacterial vesicle production and offer perspective on the ultimate score. We also highlight questions needed to advance the field in determining the role for vesicles in bacterial survival, interkingdom communication, and virulence.
Keywords: bacterial pathogenesis, bacterial secretion system, immunomodulation, interkingdom communication, membrane vesicle
Subject Categories: Membranes & Trafficking; Microbiology, Virology & Host Pathogen Interaction
Why do bacteria produce vesicles? They facilitate nutrient acquisition, survival, and adaptation, but also elicit antagonistic host responses. Close examination reveals how vesicle production yields a net positive for bacteria.

Introduction: a paradox
All bacteria produce extracellular vesicles, but why do they do this? Do the advantages overcome the disadvantages? Evidence of non‐lytic biogenesis, genetic regulation, and selective cargo incorporation help define extracellular vesicle production as a specific secretion mechanism. The diverse and complex composition of bacterial extracellular vesicles includes components that promote as well as antagonize the relationship of bacteria with their environment. As evident from the fast‐growing literature describing characteristics, activities, and reactivities of bacterial vesicles, these extracellular secreted products have been found to be both beneficial and detrimental to bacterial survival in a variety of environments (Table 1, Fig 1A–I). From an evolutionary perspective, vesicle production must ultimately benefit bacteria in some way, but how do bacteria keep the advantage in these seemingly detrimental scenarios? Here, we consider the enigmatic aspects of this secretory process.
Table 1.
Advantages and disadvantages inherent in bacterial vesicle production.
| Pros | Cons |
|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Figure 1. A delicate balance: pros and cons of bacterial vesicle production.
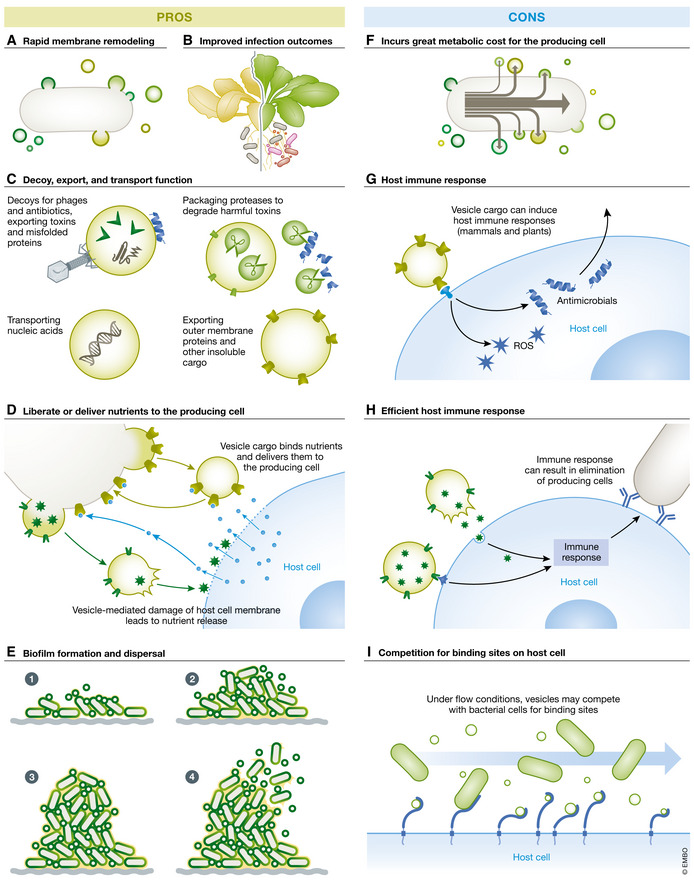
Pros: (A) Vesicle production facilitates rapid membrane remodeling in response to environmental conditions. (B) Vesicle treatment elicits plant innate immune responses that lead to improved infection outcomes. Vesicle‐mediated plant immune activation is similar to that described for induced systemic resistance and could indicate a role for vesicles in immune priming and other beneficial‐bacteria‐associated traits. (C) Vesicles play a variety of roles including (top) serving as decoys for phage and antibiotics, and exporting toxins and misfolded proteins, (right) transporting nucleic acid, (left) packaging proteases to degrade toxins that may harm the bacterial cell, (left and bottom) exporting outer membrane proteins and other insoluble cargo. Importantly, not all cargo may be contained in the same vesicle; different populations of vesicles with various cargo and distinct functionality may result from diverse production pathways. (D) Vesicles can function to liberate or deliver nutrients to the producing cell. Vesicle cargo may induce host cell nutrient release through damage to the host cell membrane. Alternatively, vesicle cargo may bind nutrients and deliver them to the producing cell. (E) Vesicles play many roles in biofilm formation and dispersal. They are critical components of the extracellular matrix and have also been shown to facilitate dispersal dependent on packaged cargo. Cons: (F) Export of macromolecules through vesicle production incurs a great metabolic cost for the producing cell. (G) Vesicle cargo induces host immune responses designed to contain and/or eliminate bacterial cells in both plant and mammalian systems. H) Under flow conditions, vesicles may compete with bacterial cells for binding sites, inhibiting bacterial attachment and colonization. (I) Vesicles activate host adaptive immune responses that, in some instances, could result in elimination of the producing cells.
Biogenesis of bacterial extracellular vesicles
Gram‐negative vesicle biogenesis
Outer membrane vesicles (OMVs), which bud from the outer membrane (OM) of Gram‐negative bacteria, are capsules of periplasmic, cell wall, and OM components (lipids, integral membrane proteins, and membrane‐associated proteins), as well as small molecules (Zhou et al, 1998; Beveridge, 1999; Kulp & Kuehn, 2010; Schwechheimer et al, 2013) (Fig 2A–C). They are heterogeneous in size, but typically range in diameter from 50 to 200 nm. The amount of OMVs released per bacterium in a given environment is both species‐ and strain‐dependent (Kulp et al, 2015; Schwechheimer & Kuehn, 2015; Orench‐Rivera & Kuehn, 2016).
Figure 2. Diverse vesicle biogenesis mechanisms and packaging could result in a variety of vesicle‐associated functions.
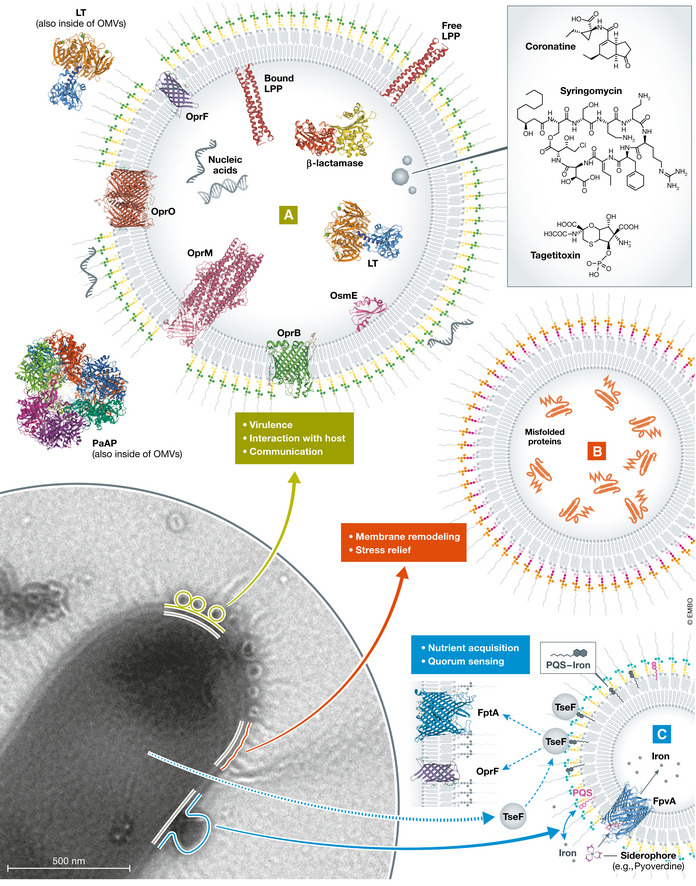
(A) Some populations of vesicles may be packaged with virulence factors, quorum‐sensing molecules, and molecules with the ability to bind host factors. (B) Bacterial stress response pathways may result in vesicles that contain misfolded proteins and lipid species to be discarded. (C) Vesicles released for the purpose of nutrient acquisition may contain siderophores or other nutrient‐binding proteins. Proteins and cargo: (A) OprO (Van den Berg, 2014; Modi et al, 2015); OprF (Zahn et al, 2014, 2015); OprB (Van den Berg, 2012a, 2012b); OprM (Akama et al, 2004a, 2004b); β‐lactamase (Golemi et al, 2001a, 2001b); LT (Merritt et al, 1993, 1994); PaAP (Nguyen et al, 2013, 2014); LPP (Shu et al, 2000a, 2000b); OsmE (Mamelli et al, 2012); Syringomycin (Duke & Dayan, 2011); Coronatine (Duke & Dayan, 2011); Tagetitoxin (Duke & Dayan, 2011); additional nucleic acids within and on the surface. (B) Misfolded proteins. (C) FpvA (Cobessi et al, 2004, 2005); PQS (Yu et al, 2007, 2009); iron. Phospholipids and LPS depicted in all vesicles.
Diverse genetic and biochemical research findings have validated that bacterial vesicles are a bona fide secretion system. Decades ago, researchers first speculated that OMVs were artefactual remnants of dead cells based on observations of increased amounts of membrane blebs in the culture media of bacteria in conditions leading to nutrient starvation (e.g., Lys‐limiting growth) (Bishop & Work, 1965; Chatterjee & Das, 1967). However, subsequent studies determined explicitly that OMV production is not necessarily linked to the loss of cellular viability or membrane integrity (Yaganza et al, 2004; McBroom et al, 2006; Schertzer & Whiteley, 2012). Further, several studies have highlighted a genetic basis for the mechanism and regulation of vesicle production by Gram‐negative bacteria (McBroom et al, 2006; McBroom & Kuehn, 2007; Schwechheimer et al, 2014, 2015; Kulp et al, 2015). Notably, mutations in stress response pathways and outer membrane constituents alter vesiculation rates (McBroom et al, 2006; Kulp et al, 2015).
These findings and subsequent studies have led to several models for vesicle formation (Fig 3A–I). First, vesicle production serves to relieve membrane stress, for example, by eliminating misfolded proteins from the periplasmic space (Fig 3A) (McBroom & Kuehn, 2007; Macdonald & Kuehn, 2013; Schwechheimer & Kuehn, 2013). Vesicle release has also been shown to relieve membrane stress resulting from accumulation of peptidoglycan fragments and lipopolysaccharide (LPS) in the periplasm (Fig 3A) (Uehara & Park, 2007; Chen et al, 2011; Haurat et al, 2011; Klein et al, 2014; Mahalakshmi et al, 2014; Schwechheimer et al, 2014). More generally, vesicle production helps resolve oxidative stress (Li et al, 1996; Berry et al, 2009; Maredia et al, 2012; Macdonald & Kuehn, 2013). In these scenarios, vesicle formation and release of the molecular pressure in the envelope is sometimes, though not always, coincident with activation of stress response pathways.
Figure 3. Mechanisms of vesicle biogenesis.
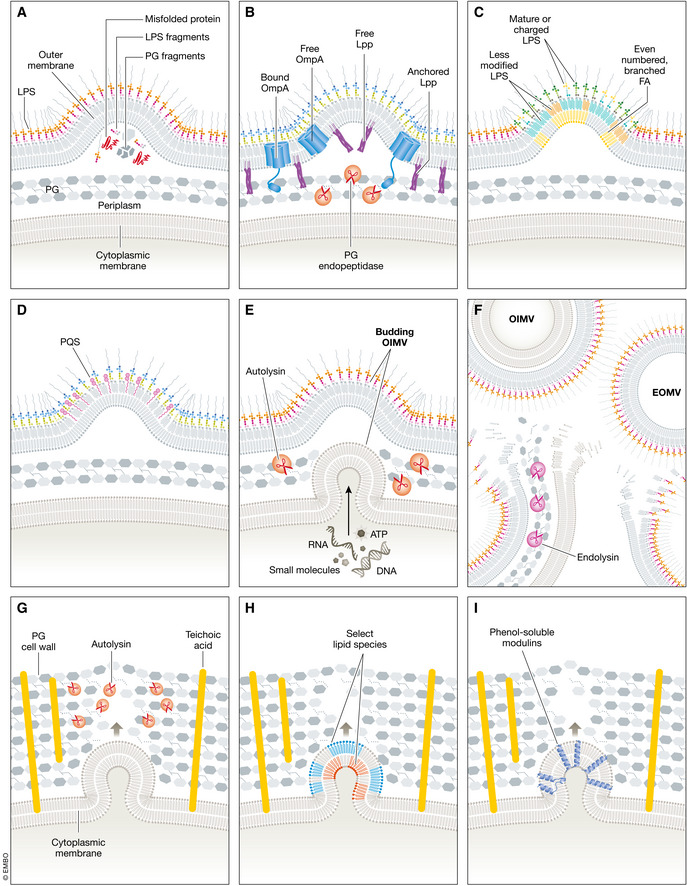
Several mechanisms of vesicle biogenesis have been proposed with notable similarities between those for Gram‐negative and Gram‐positive species. (A) An accumulation of misfolded proteins, immature LPS, and peptidoglycan fragments could result in a build‐up of pressure within the envelope. Vesicle formation and release provides one way to relieve this pressure and eliminate potentially toxic accumulation of molecules resulting from stress. (B) Vesicles bud from regions with fewer linkages to the peptidoglycan. These include linkages formed by proteins and lipoproteins, such as OmpA and Lpp, respectively. Peptidoglycan endopeptidases also transiently degrade peptidoglycan cross‐linkages to facilitate vesicle budding. (C) Specific lipids, less‐modified LPS species, and branched‐chain fatty acids are selectively exported in vesicles. Exporting this cargo in vesicles could facilitate rapid membrane remodeling during environmental shifts. Further, the precise geometry of these molecules could increase membrane bending and aid vesicle formation. (D) Insertion of small molecules, such as PQS, into the outer leaflet of the outer membrane could also increase membrane bending thus enabling vesicle formation. (E) Naturally produced OIMVs could form when the peptidoglycan is transiently degraded by autolysins. These vesicles have been shown to carry many inner membrane and cytoplasmic cargo components, including RNA, DNA, ATP, and other small molecules. (F) OIMVs and EOMVs also result from explosive cell lysis, in part due to endolysin activity. Importantly, these vesicles likely carry different cargo than naturally produced vesicles and, therefore, may have different functions. (G) Gram‐positive membrane vesicles form where the thick peptidoglycan cell wall is degraded. Autolysin activity can facilitate this degradation, creating a space through which vesicles can be released. (H) Membrane vesicles from Gram‐positive species have also been shown to contain specific types of lipids. These lipids could increase membrane fluidity or contribute to increased membrane curvature. (I) Small molecules, such as PSMs from S. aureus, can insert into cytoplasmic membrane and facilitate vesicle formation.
Vesicle production can also be the result of decreased linkages between the outer membrane and peptidoglycan (Fig 3B) (Schwechheimer & Kuehn, 2015). Numerous reports show that mutants lacking the outer membrane protein OmpA, which contains a peptidoglycan binding site, have increased vesicle production (Fig 3B) (Sonntag et al, 1978; Song et al, 2008; Deatherage et al, 2009; Moon et al, 2012; Park et al, 2012). Similar increases in vesicle production are observed when covalent linkages between Braun’s lipoprotein, Lpp, and the peptidoglycan are disrupted (Fig 3B) (McBroom et al, 2006; Deatherage et al, 2009; Kulp & Kuehn, 2010; Schwechheimer & Kuehn, 2013; Wessel et al, 2013). In fact, even subtle differences in the number of linkages between the outer membrane and peptidoglycan can have noticeable effects on vesicle production, including cases where increased numbers of linkages were found in hypovesiculating strains (Lappann et al, 2013; Schwechheimer et al, 2014, 2015).
Vesicle production is influenced by membrane fluidity and microdomains, which in turn are often modulated by temperature and lipid composition (Fig 3C). Several species including E. coli, Shewanella livingstonensis, Serratia marcescens, and Bartonella henselae display increased or decreased vesicle production in response to high or low temperatures, respectively (McBroom & Kuehn, 2007; Frias et al, 2010; McMahon et al, 2012; Roden et al, 2012). However, vesiculation by different organisms appears to have varying dependence on membrane fluidity, as Pseudomonas aeruginosa does not display altered vesicle release in response to temperature (Macdonald & Kuehn, 2013). Evidence implicating lipid species in vesicle production comes primarily from studies showing selective lipid packaging in vesicles (Fig 3C). For example, P. syringae preferentially exports even‐numbered carbon chain fatty acids, unsaturated and branched‐chain fatty acids in vesicles, all of which could increase membrane fluidity (Fig 3C) (Chowdhury & Jagannadham, 2013; Kulkarni et al, 2014). Here again, P. aeruginosa displays differing dependence on membrane fluidity as it exports phospholipids in vesicles that are typically associated with increased membrane rigidity (Tashiro et al, 2011). Subsequent studies have revealed that Salmonella enterica ssp typhimurium and Vibrio cholerae selectively export particular LPS species in vesicles (Fig 3C) (Bonnington & Kuehn, 2016; Elhenawy et al, 2016; Zingl et al, 2020). In these cases, modifications alter LPS geometry, which could facilitate membrane bending and vesicle formation (Bonnington & Kuehn, 2016; Elhenawy et al, 2016). Interestingly, vesicle formation in P. aeruginosa is also influenced by LPS composition, though these findings were based on differences in the O‐antigen rather than the Lipid A component of the LPS (Li et al, 1996; Murphy et al, 2014).
Small molecules and proteins influence vesicle formation as well. For example, in P. aeruginosa, the quorum‐sensing molecule Pseudomonas quinolone signal (PQS) stimulates vesicle production according to the bilayer‐couple model (Fig 3D) (Florez et al, 2017). In this model, PQS accumulates in the outer leaflet of the outer membrane, expanding this leaflet compared to the inner leaflet and creating tension between the two leaflets (Fig 3D). The shape of PQS further facilitates membrane curvature needed to begin the vesicle budding process. Notably, this proposed mechanism of vesicle production does not involve activation of stress response pathways. To date, this model is limited to gammaproteobacteria (Horspool & Schertzer, 2018); however, quorum‐sensing molecules have been shown to influence formation of phospholipid liposomes (Mashburn‐Warren et al, 2008) and vesicle production in Gram‐positive species (Tashiro et al, 2010). It is important to note that in bacteria normally producing PQS, vesicle formation can still occur in the absence of the PQS producing genes (Tashiro et al, 2009; Macdonald & Kuehn, 2013; Toyofuku et al, 2014), revealing the redundancy of vesicle biogenesis mechanisms in bacteria.
In a distinct process and yielding a distinct type of extracellular vesicles, Gram‐negative bacteria can also produce outer‐inner membrane vesicles (OIMVs) (Fig 3E) (Toyofuku et al, 2019; Nagakubo et al, 2020). Notably, these vesicles contain inner membrane components, nucleic acid, ATP, and other cytoplasmic content (Fig 3E) (Kadurugamuwa & Beveridge, 1995; Pérez‐Cruz et al, 2013, 2015; Clarke, 2018). Although a full biogenesis mechanism has yet to be revealed, OIMVs may form as a result of autolysin activity that transiently breaks down peptidoglycan to allow OIMV release (Kadurugamuwa & Beveridge, 1995; Pérez‐Cruz et al, 2013, 2015; Clarke, 2018). Naturally produced OIMVs have been observed in many different species including Shewanella vesiculosa, Neisseria gonorrhoeae, P. aeruginosa, Acinetobacter baumannii, V. shilonii, Pseudoalteromonas marina, Arhensia kielensis, and P. syringae, among others (Hagemann et al, 2014; Pérez‐Cruz et al, 2015; Li et al, 2016; preprint: Janda et al, 2021). In P. aeruginosa, vesicle production can also result from explosive cell lysis triggered by endolysins, yielding so‐called explosive OMVs (EOMVs) (Fig 3F) (Turnbull et al, 2016; Toyofuku et al, 2019). While naturally produced OIMVs occur independent of bacterial stress responses, EOMVs resulting from explosive cell lysis can also be the result of DNA damage and, therefore, may have different functions (Toyofuku et al, 2014; Florez et al, 2017; Cooke et al, 2019).
Gram‐positive vesicle biogenesis
Much less is known about vesicle biogenesis in Gram‐positive bacteria, though similarities to Gram‐negative vesicle biogenesis are emerging. Mounting evidence suggests that vesicles from Gram‐positive species form where the thick peptidoglycan cell wall is selectively degraded (Fig 3G) (Brown et al, 2015; Toyofuku et al, 2017, 2019; Wang et al, 2018; Andreoni et al, 2019; Nagakubo et al, 2020). Despite these disruptions to the cell wall, bacterial cell morphology remains intact during vesicle formation and release (Toyofuku et al, 2017; Wang et al, 2018; Andreoni et al, 2019). This is similar to the mechanisms of OMV formation in Gram‐negative species where membrane integrity is not compromised (Chutkan et al, 2013).
Also similar to Gram‐negative bacteria, genetic studies have revealed that Gram‐positive vesicle production may rely on global regulators that drive expression in complex genetic networks (Briaud & Carroll, 2020). For example, vesicle production in Listeria monocytogenes relies on SigB, a major stress response regulator that controls expression of many virulence factors and contributes to host cell invasion (Kim et al, 2005; Lee et al, 2013b; Liu et al, 2019). SigB is conserved in related Gram‐positive bacteria (Hecker et al, 2007); therefore, this regulator may similarly modulate vesicle biogenesis in other species.
Gram‐positive vesicle production may also depend on transient reductions in peptidoglycan cross‐linking to allow vesicles to pass through the cell wall (Wang et al, 2018). In Staphylococcus aureus, treatment with penicillin, which is known to reduce peptidoglycan cross‐linking, or mutations that disrupt peptidoglycan cross‐linking in the cell wall led to an increase in vesicle production (Wang et al, 2018). Additional studies from a number of Gram‐positive species and mycobacteria reveal that vesicles contain transpeptidases and autolysins, which are involved in peptidoglycan remodeling (Fig 3G). This result demonstrates that these molecules are at least present at the site of vesicle release and could actively contribute to vesicle biogenesis (Briaud & Carroll, 2020).
Similar to vesicle production in Gram‐negative species, Gram‐positive vesicle production may also be dictated by lipid geometry (Fig 3H). Recent lipid profiling studies from Streptococcus pyogenes and Propionibacterium acnes revealed that vesicles are enriched in phosphatidylglycerol and triacylglycerol and depleted in cardiolipin, a composition that could facilitate membrane curvature and vesicle formation (Resch et al, 2016; Jeon et al, 2018; Nagakubo et al, 2020). Similarly, the lipid composition of Listeria monocytogenes vesicles differed significantly from the bacterial cell membrane, showing enrichment in phosphatidylethanolamine, sphingolipids, and triacylglycerols (Coelho et al, 2019; Briaud & Carroll, 2020). These results suggest that vesicles are specifically packaged with distinct lipids that facility budding and release.
Just as PQS and small molecules can influence vesicle formation in Gram‐negative species, surfactant‐like phenol‐soluble modulins (PSMs) may induce Gram‐positive vesicle budding (Fig 3I) (Drin & Antonny, 2010; Nazari et al, 2012; Wang et al, 2018). Although these molecules are unique to S. aureus (Cheung et al, 2014), similar molecules may induce membrane curvature in other species to facilitate vesicle release.
Vesicle cargo selection
As the need for vesicle purification has been recognized and the purification techniques have improved (for recent reviews, see (Prados‐Rosales et al, 2014a; Klimentová & Stulík, 2015; Tulkens et al, 2020a; Liangsupree et al, 2021; Nasukawa et al, 2021)), analysis of vesicle cargo has revealed export selectivity, a hallmark of a bona fide secretory process. Numerous quantitative proteomic and lipidomic analyses have demonstrated that the composition of OMVs (McMahon et al, 2012; Bonnington & Kuehn, 2014, 2016; Elhenawy et al, 2016; Orench‐Rivera & Kuehn, 2021) does not mimic the composition of the envelope from which they are derived. Indeed, molecular “rules” have been validated that show mechanisms for specific inclusion and exclusion of soluble and membrane‐associated OMV cargo based on polypeptide sequence or physical parameters (McBroom & Kuehn, 2007; Orench‐Rivera & Kuehn, 2021). Similarly, in Gram‐positive bacteria the protein, lipid, and nucleic acid content of vesicles reveals selective packaging rather than a bulk‐flow mechanism (Resch et al, 2016; Jeon et al, 2018; Briaud & Carroll, 2020; Nagakubo et al, 2020; Tartaglia et al, 2020; Luz et al, 2021). The precise mechanisms by which each of these types of cargo are selectively packaged in vesicles remain an area of active investigation. In addition, it is widely recognized that the vesicle preparations characterized to date likely consist of a mixture of subtypes of vesicles with distinct compositions. More refined separation and sensitive analytic techniques will be necessary in the future to assign specific properties to specific subtypes, but it should be mentioned that in a natural setting, a similar variety of vesicle populations are likely to be generated. Thus, a complex mixture reflects what could be distributed into the bacterial environment.
Packaging of vesicle cargo that contributes to bacterial viability
Bacterial vesicles are energetically costly to produce, as they are composed of complex macromolecules that are metabolically expensive to synthesize (Figs 1F and 2). In addition, vesicle budding and release must overcome the energetic stability of the cell’s outermost membrane barrier that enables survival in diverse, often harsh environments. It would, therefore, seem biologically counterproductive for bacteria to release cellular resources in a process that also potentially harms the envelope’s barrier function. However, benefits of bulk export and the selective removal and retention of envelope components (i.e., envelope “remodeling”) by vesicle production would be a strong evolutionary driver, as bacterial viability would certainly outweigh energetic costs.
The first insight into the benefit of vesiculation to the cells came from analyzing vesiculation levels within a library of genetic mutants, which pointed to a link with envelope stress responses as mentioned above (McBroom et al, 2006). In a follow‐up study, it was revealed that the ability to produce vesicles benefited viability by not only increasing bulk export, but also the selective export of envelope components that would otherwise be toxic to the cell (Figs 1C and 2B) (McBroom & Kuehn, 2007).
Subsequently, studies examined whether bacteria may also benefit from using selective vesicle export to remodel their OM (Fig 1A) (Cahill et al, 2015; Bonnington & Kuehn, 2016; Elhenawy et al, 2016; Eberlein et al, 2018; Valguarnera et al, 2018; Orench‐Rivera & Kuehn, 2021). Recently, it was determined that levels of oxidizable residues and oxidized OM‐associated proteins are increased in E. coli OMV cargo in response to oxidative stress, presumably because the oxidized proteins are detrimental to the cells (Orench‐Rivera & Kuehn, 2021). Further, the OMV packaging differential between full‐length (cell‐wall bound) and truncated (unbound) OmpA increased upon oxidative stress, which may be related to the beneficial role cell‐wall associated OmpA plays in oxidative response (van der Heijden et al, 2016).
Remodeling mechanisms are especially relevant for membrane lipids because they are essential for maintaining the cell barrier and, consequently, viability in rapidly changing environmental conditions. While phospholipases including PldA degrade OM inner leaflet phospholipids, which are then recycled, to date there are no known mechanisms for degradation and recycling of LPS, the major component of the OM extracellular leaflet. Bacteria modify LPS in several ways including increasing or decreasing the O‐antigen length, attaching to lipid A positively charged and/or zwitterionic groups such as 4‐amino‐4‐deoxy‐L‐arabinose and phosphoethanolamine, palmitoylating lipid A, and modifying LPS core to improve barrier function in particular environments (Raetz et al, 2007; Capra & Laub, 2012; Chen & Groisman, 2013; Bonnington & Kuehn, 2016). However, these modifications occur in the cytoplasm, and thus without an efflux mechanism at the OM, bacteria would be forced to rely solely on entry and diffusion of these new modified LPS species throughout the membrane to adapt to environmental shifts. This would happen too slowly for bacteria to successfully generate an appropriate membrane for the new environment and would ultimately lead to bacterial death before the benefits of the LPS modifications could take effect. Exporting select lipid and LPS species in vesicles provides a fast restructuring mechanism that allows bacteria to create and maintain an appropriately adapted barrier against an otherwise stressful environment (Cahill et al, 2015; Bonnington & Kuehn, 2016; Elhenawy et al, 2016; Eberlein et al, 2018).
An example of the benefits of selectively including particular lipid cargo in vesicles is evident from studies of S. typhimurium cultures where the shift from a host extracellular to intracellular environment was mimicked by altering pH and magnesium concentration of the media (Bonnington & Kuehn, 2016). Upon shift, and even when shifting back to “extracellular conditions”, S. typhimurium preferentially exported less‐modified LPS species in vesicles (Bonnington & Kuehn, 2016). As the less‐modified LPS is exported, the overall composition of the outer membrane changes, tending toward retention of more modified LPS species. These new species benefit the bacterium and help it adapt to the new, stressful environment in several ways. For example, LPS modifications mask negatively charged phosphate moieties, helping strengthen membrane integrity when divalent ion concentrations are low (Bonnington & Kuehn, 2016). Releasing vesicles with a different lipid composition than the outer membrane could also serve to trick the host immune system upon bacterial invasion by eliciting an immune response to the vesicles instead of the bacterial cell (Bonnington & Kuehn, 2016; Zingl et al, 2020). While this secretion pathway may be energetically costly in the sense that macromolecules are “discarded” instead of recycled into components the cell may reuse, the benefit can outweigh the cost by expeditiously eliminating compounds in bulk that have become toxic or useless before they cause harm to the cell (Fig 1A, Table 1).
Characterization studies suggest that vesicles from Gram‐positive species are also packaged to benefit survival, though functional confirmation is often missing. Coagulation factors exported in vesicles could help Gram‐positive species cope with stressful host environments by facilitating biofilm formation (Lee et al, 2009). While peptidoglycan remodeling enzymes could be a result of vesicle biogenesis, this cargo could also help restructure the cell wall to help the bacterium adapt to new environments (Lee et al, 2009). Furthermore, Gram‐positive vesicles are enriched with nutrient scavenging molecules, such as siderophores (Lee et al, 2009; Schrempf et al, 2011; Brown et al, 2015; Liu et al, 2018). During transitions to environments where nutrients are limited, these molecules likely improve survival by sequestering nutrients and delivering them to the producing cell or another cell in the population (Lee et al, 2009; Schrempf et al, 2011; Brown et al, 2015; Liu et al, 2018).
In sum, at this basic physiological level the ability for bacteria to export particular cargo in vesicles can specifically improve the viability of the bacteria in stressful environments. For pathogens, improved viability due to vesicle secretion is especially beneficial in antagonistic host environments. However, vesicles play additional roles as well during host–pathogen interactions, including their use as vehicles to transport cargo used to promote microbial attack.
Vesicle‐mediated toxin and virulence factor dissemination during host–microbe interactions
Selective export of toxins and other virulence factors via vesicles and their functional delivery into host cells has been described in many cases (Figs 1C, D, G and I, and 2) (Kolling & Matthews, 1999; Horstman & Kuehn, 2000; Kuehn & Kesty, 2005; Kulp & Kuehn, 2010; Chatterjee & Chaudhuri, 2011; Rompikuntal et al, 2012; Kunsmann et al, 2015; Schwechheimer & Kuehn, 2015; Jan, 2017; Zakharzhevskaya et al, 2017; Liu et al, 2018; Briaud & Carroll, 2020; Nagakubo et al, 2020). There are several advantages to packaging such cargo in vesicles. For example, vesicles protect cargo from host defenses such as proteases and nucleases, allowing the toxins and virulence factors to successfully reach their target (Dorward & Garon, 1990; Kolling & Matthews, 1999; Horstman & Kuehn, 2000; Yaron et al, 2000; Renelli et al, 2004; Bonnington & Kuehn, 2014; Bitto et al, 2017). Vesicles also allow the simultaneous delivery and potentially synergistic interactions of a cocktail of virulence factors that can be targeted to particular types of host cells by specific, tissue tropic ligand/receptor interactions (Ellis & Kuehn, 2010; Ellis et al, 2010; Kaparakis‐Liaskos & Ferrero, 2015; Kunsmann et al, 2015; Bielaszewska et al, 2017). Importantly, packaging groups of molecules in vesicles enables delivery of virulence factors with a variety of mechanisms of action, allowing one vesicle unit to target many aspects of host cell function. For example, some cargo may modulate host cell membrane function, while other packaged material may be trafficked to the nucleus to impact host transcriptional responses (Cecil et al, 2019; Le et al, 2021). Taken together, vesicles allow specific and potent transport of toxic cocktails over long distances within the host (Dorward & Garon, 1990; Bomberger et al, 2009; Bonnington & Kuehn, 2014).
Several studies have identified detailed pathways by which functional, vesicle‐associated toxins are transported into host cells. For example, enterotoxigenic Escherichia coli (ETEC) produces heat‐labile enterotoxin (LT), which is secreted via the general secretory pathway and is associated inside and on the surface of the vesicles (Fig 2A) (Horstman et al, 2004). Once secreted, LT‐containing vesicles enter host cells where the toxin is trafficked through the Golgi and ER (Kesty et al, 2004). Inside the cell, LT acts similarly to cholera toxin, also associated with vesicles, as it modifies the adenylate cyclase pathway to increase cAMP levels, which leads to a net efflux of electrolytes and water (Kesty et al, 2004; Chatterjee & Chaudhuri, 2011). Trafficking of the toxin and the consequent responses in host cells are dependent on its association with vesicles, as soluble toxin leads to a distinct outcome (Chutkan & Kuehn, 2011).
Shiga toxin 2a from enterohemorrhagic E. coli (EHEC) is also released in association with vesicles (Bauwens et al, 2017). In conditions mimicking the human intestinal tract environment, EHEC increases vesicle production, including vesicles containing Shiga toxin 2a, leading to an increase in cytotoxicity. Vesicles from EHEC are also the exclusive secretion pathway for cytolethal distending toxin V (Bielaszewska et al, 2017). In addition to toxin packaging, E. coli vesicles have been shown recently to deliver their toxic components to host intestinal epithelial cells, where they cause DNA damage and lead to increased disease pathology (Ling et al, 2019). Toxin packaging and delivery are not limited to vesicles from E. coli. Vesicles from N. gonorrhoeae are packaged with PorB, which targets mitochondrial membranes and leads to loss of mitochondrial integrity, cytochrome C release, and activation of apoptotic caspases in macrophages (Deo et al, 2018). In Bacteroides fragilis, vesicles contain the B. fragilis toxin, which cleaves E‐cadherin and contributes to virulence in inflammatory bowel disease (Zakharzhevskaya et al, 2017).
Vesicles from Gram‐positive species are also packaged with toxins. S. aureus vesicles are packaged with alpha toxin and leucocidin, which lead directly to inflammasome activation in macrophages (Wang et al, 2020). Vesicles from Listeria monocytogenes carry toxins listeriolysin O and hemolysis, which are required for bacterial escape from the pathogen containing vacuole (Lee et al, 2013b; Brown et al, 2015). In Bacillus anthracis, components of the anthrax toxin have been found in vesicles as well as additional cytolysins (Rivera et al, 2010; Brown et al, 2015).
In addition to toxins, vesicles have been shown to contain nucleic acids that elicit host responses (Figs 1C and 2A). DNA is found both on the surface and in the lumen of bacterial vesicles from at least five Gram‐negative species and encodes virulence‐related products as well as gene products linked to pathogenesis regulation and survival in stressful conditions (Bitto et al, 2017). These vesicles are trafficked to the nucleus of mammalian cells, although it is unclear whether vesicle‐associated DNA integrates into the host genome or is ultimately translated into protein (Bitto et al, 2017). In another instance, DNA from bacterial vesicles was shown to activate host immune responses via TLR9 (Perez Vidakovics et al, 2010). As this immune response was directed toward vesicles, bacteria were ultimately able to evade the host response (Perez Vidakovics et al, 2010).
Additional studies extend such activities to vesicle‐associated RNA and Gram‐positive bacteria. For S. aureus, the data show mammalian cell immune activation in response to vesicle‐associated RNA and DNA (Rodriguez & Kuehn, 2020; Bitto et al, 2021), which could play a role in the bacterial strategy of immune evasion during infection. Recent studies characterizing the specific RNA cargo in S. aureus vesicles revealed numerous mRNA transcripts that encode virulence‐associated factors, including hld, agrBCD, psmβ1, sbi, spa, and isaB, as well as sRNAs, including RsaC, that could directly contribute to virulence in a host setting (Luz et al, 2021). Intriguingly, RNA packaging in S. aureus vesicles also depends on environmental conditions such as the presence of vancomycin, which carries implications for vesicle function during infection scenarios (Luz et al, 2021). Similarly, vesicles from Clostridium perfringens contain DNA that codes for perfringolysin O and alpha toxin and have also been shown to activate host immune responses (Jiang et al, 2014; Brown et al, 2015).
In each of these cases, it seems that vesicle‐mediated secretion of virulence determinants benefits the bacterial cell, often by contributing directly to pathogenesis. This strategy both protects virulence‐associated molecules from degradation and allows for action at a distance from the producing cell. However, vesicle contributions to virulence, described here, and viability, described earlier, must be carefully balanced with host immune activation elicited by reactive vesicle components.
Vesicle‐triggered immune activation during infection
Numerous studies of a wide variety of bacterial pathogens and host models reveal a strong mammalian immune response to bacterial vesicles, a body of literature that has been reviewed in detail (Fig 1I) (Kaparakis‐Liaskos & Ferrero, 2015; Johnston et al, 2020). In vitro and in vivo experiments reveal that the specific mammalian immune response to vesicles depends on the vesicle cargo and composition and on the originating bacterial species (Figs 1I and 2). Vesicles have been shown to activate many of the canonical innate immune responses such as interleukin (IL)‐1β and interferon expression, with many of these responses apparently dependent on nucleotide‐binding oligomerization domain (NOD) factor activation (Allison et al, 2009; Kaparakis et al, 2010; Johnston et al, 2021). By design, these host immune responses target and eliminate invading bacteria; therefore, their activation appears disadvantageous for bacterial survival.
The inflammasome response is one example of a critically important mammalian defense against pathogens (Rathinam et al, 2012; He et al, 2016; Antushevich, 2020). Upon bacterial activation of the inflammasome, an innate immune signaling cascade induces a variety of host immune responses that target and eliminate the bacterial pathogen. Activating this pathway would seem counterproductive to infection by bacterial pathogens, yet bacterial vesicles secreted by pathogens appear particularly adept to do just that. For example, although vesicles may aid in bacterial escape from vacuoles, Finethy et al showed that LPS delivered by E. coli K12 vesicles activated the non‐canonical inflammasome response in a guanylate‐binding protein 1 (GBP1)‐dependent manner (Finethy et al, 2017, 2020). Recognition of vesicle‐associated LPS is also sufficient to activate pyroptosis via caspase‐11 activation and interleukin‐1 (IL‐1) maturation (Vanaja et al, 2016). Similarly, P. aeruginosa vesicles activate the non‐canonical inflammasome dependent on the caspase‐11 pathway and caspase‐5 (Bitto et al, 2018). In contrast, free P. aeruginosa LPS activated the inflammasome via caspase‐4 (Bitto et al, 2018). In yet another example, Campylobacter jejuni packages more virulence factors in vesicles in response to a shift to human body temperature, leading to strong inflammasome activation (Taheri et al, 2019). Extending this phenomenon to Gram‐positive species, S. aureus vesicles activate the NLRP3 inflammasome in a cargo‐dependent manner, and vesicles from Streptococcus pneumoniae are internalized by immune cells and elicit immune responses that protect against pneumococcal infections (Olaya‐Abril et al, 2014; Mehanny et al, 2020; Wang et al, 2020).
Additional studies of vesicle‐mediated mammalian host immune activation have shown that Gram‐positive Clostridium difficile secretes vesicles that activate pro‐inflammatory immune responses in several types of immune cells, independent of the well‐known toxins TcdA and TcdB (Nicholas et al, 2017). Notably, these responses include induced expression of IL‐1β, IL‐6, IL‐8, and monocyte chemoattractant protein‐1 (MCP‐1) (Nicholas et al, 2017). Similar pro‐inflammatory immune activation has been characterized in response to Gram‐negative Bacteroides thetaiotaomicron and Acinetobacter nosocomialis vesicles, where B. theta vesicles trigger fulminant colitis through their associated sulfatase activity, and A. nosocomialis vesicles carry a variety of virulence factors that result in host cytotoxicity and immune responses (Hickey et al, 2015; Nho et al, 2015). In‐depth studies of an outbreak strain of E. coli also showed that dynamin‐dependent endocytosis of vesicles activated caspase‐9‐mediated apoptosis and IL‐8 secretion through delivery of a cocktail of virulence factors (Kunsmann et al, 2015). Follow‐up studies identified specific components required for entry, virulence factor delivery, and immune activation (Bielaszewska et al, 2017).
In sum, despite the substantial benefits as a stress response and mechanism for effective virulence factor dissemination, vesicle secretion can come at a heavy cost to bacterial pathogens (Table 1, Fig 1). Host immune cells react to vesicles produced by bacterial pathogens in a way that would lead to the restriction of pathogen viability and disease, presumably providing a strong evolutionary disincentive for bacteria to secrete vesicles. But it must be considered that while host immune activation creates barriers to infection or colonization success, bacteria have evolved many mechanisms to withstand and evade the immune response, and, unsurprisingly, bacterial vesicles play a role here as well.
Countering mammalian responses with vesicles
Bacteria can activate immune responses via vesicle release; however, vesicles can also be utilized either actively or passively to mitigate and overcome the host response at a distance. Additionally, rapid, vesicle‐mediated bacterial membrane remodeling and maintenance can create an effective barrier to host insults and thereby improve bacterial survival during infection (Figs 1A and 2).
Cargo to actively degrade host antimicrobial compounds
One example of vesicle‐mediated, anti‐host factor activity is induced by the host immune factor itself. LL‐37 is a cationic antimicrobial peptide and the only human cathelicidin. In the presence of butyrate and LL‐37 in the intestinal tract, enterohemorrhagic E. coli (EHEC) upregulates vesicle production and specifically packages vesicles with OmpT (Urashima et al, 2017). OmpT is a protease that breaks down LL‐37 and prevents its antimicrobial activity; however, OmpT activity is dependent on binding to LPS. Therefore, by packaging OmpT in vesicles in the gut environment, EHEC improves its survival outcome and successfully adapts to the host environment (Urashima et al, 2017). The precise mechanism by which OmpT is selectively packaged in vesicles remains unknown.
Vesicles as decoys for passive defense
Vesicles can also defeat the antimicrobial action of membrane‐active peptides by absorbing them, thereby acting to reduce the antimicrobial concentration and prevent killing of sensitive bacteria. This decoy effect has been characterized for both man‐made antimicrobials such as polymyxin, colistin, and amoxicillin as well as LL‐37 in a variety of bacterial species (Loeb & Kilner, 1978; Thompson et al, 1985; Ciofu et al, 2000; Manning & Kuehn, 2011; Schaar et al, 2011; Yun et al, 2018). It is notable that vesicles also bind and inactivate other membrane binding‐dependent bacterial antagonists, such as bacteriophage (Fig 1C) (Loeb & Kilner, 1978; Manning & Kuehn, 2011; Biller et al, 2014; Reyes‐Robles et al, 2018). Gram‐positive bacterial vesicles also play roles in evading antimicrobial responses, as they have been shown to contain biologically active β‐lactamase (Lee et al, 2013a) and absorb surface‐acting antibiotics such as daptomycin (Andreoni et al, 2019). These benefits of vesicles to improve bacterial viability in a hostile environment are likely an early evolutionary trait, as antimicrobials and phage are extremely abundant in all terrestrial and aquatic habitats.
Further, vesicles can be used as decoys to evade adaptive immune responses. Moraxella catarrhalis, for example, packages vesicles with the superantigen Moraxella IgD‐binding protein (MID), which facilitates vesicle internalization by B cells (Perez Vidakovics et al, 2010). Upon internalization, vesicle‐associated DNA leads to an immune signaling cascade that results in polyclonal IgM antibody production (Perez Vidakovics et al, 2010). Importantly, this antibody response is not directed toward the vesicle‐producing bacteria, allowing the pathogen to evade adaptive immune responses (Perez Vidakovics et al, 2010).
Cargo to directly manipulate host response
Vesicles are also used to influence host responses with highly specific action in both pathogenic and mutualistic settings. Helicobacter pylori packages vesicles with small non‐coding RNAs (sncRNAs) that target host mRNAs and reduce IL‐8 secretion upon vesicle recognition (Zhang et al, 2020). Vesicles from Vibrio fischeri are packaged with OmpU in response to acidic host pH, helping to establish mutualistic interaction with the Hawaiian bobtail squid (Lynch et al, 2019). Importantly, however, an OmpU homolog from the pathogenic V. cholerae also facilitates mutualism in this context despite functioning as a virulence factor in its native environment, linking vesicle packaging in mutualistic bacteria to that in pathogenic species (Lynch et al, 2019).
Packaging to improve survival during stress and host‐induced damage
A different type of beneficial role for vesicles that directly impacts bacterial survival in a challenging environment was uncovered in studies focused on the critical ability of bacteria to generate and maintain their OM as a barrier. Membrane remodeling of lipids and protein can be essential for bacterial survival in host environments that are designed to destroy disease‐causing bacteria. As mentioned above, vesicles can be used to remove unfavorable lipid species and quickly remodel the membrane during the transition from neutral to acidic and oxidizing environments (Cahill et al, 2015; Bonnington & Kuehn, 2016; Elhenawy et al, 2016; Eberlein et al, 2018). In host cells, bacteria encounter these conditions upon internalization by macrophages, for example, which use acidification and oxidation of intracellular compartments to kill invading bacteria. Vesicles are also used to rapidly remove otherwise detrimental proteins from the OM. For example, OmpT removal from the OM has been shown to confer resistance to bile acids during V. cholerae infection (Provenzano & Klose, 2000). During the transition to the murine gut, V.cholerae packages vesicles with OmpT, resulting in faster adaptation to the host environment (Zingl et al, 2020).
As we gain an understanding of the various contributions of vesicle components to virulence, survival, and host response, the cost/benefit analysis of vesicle production increases in complexity, particularly in the case of pathogens in the context of a mammalian host environment (Fig 1, Table 1). Secretion and delivery of virulence factors via vesicles in the host certainly benefits bacterial virulence by enabling action at a distance and may additionally include moderate tissue damage to generate nutrients for the pathogen. While vesicle activation of a robust mammalian immune response directed at the pathogen is likely detrimental to bacterial survival, this may not outweigh the benefits of vesicle production. Indeed, to counter such inhospitable host responses, vesicles can be used as decoys to absorb host antimicrobial compounds or to enable changes in the bacterial membrane and improve viability (Figs 1 and 2). As in many cases of bacterial virulence factor‐host response stand‐offs, pathogens must find the right balance in order to successfully infect a mammalian host.
Vesicle trade‐offs in the plant‐microbe system
Additional and broader biological insights into the cost/benefit equation can be found in the often overlooked but equally relevant and revealing situation in plants. Bacteria must also strike a balance in plant systems, and while plant bacterial vesicles and plant host cell–vesicle interactions have yet to be extensively interrogated, several studies of the roles bacterial vesicles play in plant environments have already proven to be rewarding.
Phytobacterial vesicle cargo
Together with data from mammalian bacterial pathogens, initial findings regarding the proteomic composition of plant bacterial vesicles from pathogenic, commensal, and environmental species reveal a common theme of using vesicles to transport virulence‐associated material. Studies show that plant bacterial vesicles contain type III secreted effectors, plant cell wall‐degrading enzymes, flagellin, EF‐Tu, and many other virulence‐associated molecules (Sidhu et al, 2008; Chowdhury & Jagannadham, 2013; Kulkarni et al, 2015; Solé et al, 2015). By extrapolating from studies in mammalian systems, these data suggest that vesicles from plant bacteria could play critical roles in bacterial virulence programs. However, studies of bacterial vesicles in plant and environmental contexts are relatively limited, and it largely remains to be determined whether these virulence‐associated factors are functionally active after delivery via vesicles and how bacteria could use vesicles to promote virulence in plant systems. Additionally, the mechanism behind plant recognition of vesicles and how plant immune responses impact ultimate vesicle function are yet unknown. Thus, we must be cautious in interpreting functional relevance from compositional data.
Vesicles to aid in virulence and the bacterial life cycle in plants
As in mammalian systems, studies in plant systems are already beginning to demonstrate that bacterial vesicles may promote the virulence of plant pathogens. For example, Xylella fastidiosa (Xf) vesicles can aid in distribution of the bacterium throughout grapevine xylem by adhering to the xylem cells and creating a coating (Ionescu et al, 2014). This vesicle coating inhibits attachment of the bacteria to the xylem, thereby allowing bacterial cells to travel further and spread more widely through the plant (Ionescu et al, 2014). In this sense, vesicles act as virulence factors that facilitate bacterial spread and colonization and lend a survival advantage to the bacteria (Table 1).
In addition to contributing to bacterial spread, Ionescu et al (2014) also propose that Xf vesicles could facilitate transitions between the insect vector and plant host during the Xf life cycle. Xf appears to control vesicle production, at least in part, through diffusible signal factor‐mediated signaling (Ionescu et al, 2014, 2016; Feitosa‐Junior et al, 2019). This regulation could allow the bacteria to produce more vesicles in the plant environment and prevent bacterial attachment to xylem cell walls, while limiting vesicle production in the insect vector, allowing bacteria to adhere to the insect mouth parts under high flow conditions and facilitating spread to other host plants (Ionescu et al, 2014, 2016; Feitosa‐Junior et al, 2019). This proposed vesicle function is similarly beneficial for bacteria as it increases colonization of the host plant and improves transmission to additional hosts.
Roles for vesicles in nutrient acquisition and survival in the plant apoplast
Recent proteomics results suggest a different beneficial function for vesicles from the plant pathogen Pseudomonas syringae pv tomato DC3000 (Pst) (preprint: Janda et al, 2021). Compared to the bacterial cell, Pst vesicles were enriched in proteins involved in siderophore transport, revealing a potential role for vesicles in iron acquisition (preprint: Janda et al, 2021). Transcription of the corresponding genes for these enriched proteins is notably upregulated during plant pathogen‐associated molecular pattern (PAMP)‐triggered innate immune responses (preprint: Janda et al, 2021), which could suggest that vesicles are released upon exposure to the inhospitable plant apoplast to sequester and deliver nutrients critical to bacterial survival (Fig 1D). These and other studies in plants have begun to reveal common beneficial themes inherent in bacterial vesicle production.
Vesicle‐mediated plant immune activation and protection
Revealing instead common disadvantages of vesicle production, Bahar et al (2016) show that vesicles from Xanthomonas campestris pv campestris elicit PAMP‐triggered immune responses in A. thaliana (Fig 1G, Table 1). In response to vesicle treatment, A. thaliana leaves trigger a reactive oxygen species (ROS) burst and upregulate defense marker genes FRK1 and At5g57220. Bahar et al (2016) also found that even with PAMP receptors mutated or genetically removed, plants were still able to mount an immune response as measured by transcription of defense marker genes. Interestingly, the only way to dampen this response was to eliminate either of two co‐receptors, SOBIR1 or BAK1.
These data suggest that plants are able to detect PAMPs on the vesicles and initiate an immune response program, but while known PAMP‐triggered immune responses may be partially responsible for the immune activation seen by Bahar et al, vesicles may also be activating novel PAMP immune pathways or even activating pathways attributed to effector‐triggered immune responses. More broadly, these data reveal the existence of plant mechanisms to detect bacterial vesicles and mount an appropriate immune response designed to contain and eliminate the invading bacteria (Fig 1G). In addition to illuminating a disadvantage of vesicle production that is conserved in plant and mammalian systems, these data highlight an interesting new use for vesicles in probing host immune systems. Given that eliminating well‐characterized plant PAMP receptor pathways failed to eliminate plant immune responses, studying plant responses to vesicles will likely reveal novel aspects of plant recognition of and interaction with bacteria.
Our recent study investigated the nature and breadth of vesicle‐mediated plant immune activation in greater depth (McMillan et al, 2021b). Using vesicles from the pathogenic bacterium P. syringae pv tomato DC3000 (Pst) and the commensal P. fluorescens Migula ATCC 13525 (Pf), we discovered that pre‐treatment with vesicles triggered an immune program that protected against bacterial and oomycete challenge (Fig 4) (McMillan et al, 2021b). This result seems to stem in part from vesicle‐mediated activation of PAMP‐triggered immune responses, including phosphorylation of MAPK (McMillan et al, 2021b). However, the duration of MAPK activation exceeds that of a strictly PAMP‐triggered response, which could suggest multiple vesicle‐associated elicitors or controlled release of immunogenic cargo (Tsuda et al, 2013; Stael et al, 2015). Intriguingly, immune activation by pathogenic and commensal plant bacterial vesicles was different. In plants, isochorismate synthase 1 (ICS1) leads to production and accumulation of salicylic acid, which is a major component of local and systemic immune responses (Wildermuth et al, 2001; Glazebrook, 2005; Jones & Dangl, 2006; Spoel & Dong, 2012; Seyfferth & Tsuda, 2014; Liu et al, 2016). Pst vesicles led to induced ICS1 expression and salicylic acid accumulation while Pf vesicles did not, despite both types of vesicles leading to similar protective effects (McMillan et al, 2021b). Indeed, even some mammalian pathogens, including EHEC, P. aeruginosa, and S. aureus, can lead to immune activation and, occasionally, plant protection independent of salicylic acid pathways. This salicylic acid‐independent protection resembles induced systemic resistance responses and could reveal a use for vesicles in plant immune priming (Zamioudis & Pieterse, 2011; Pieterse et al, 2014; Vlot et al, 2021).
Figure 4. Vesicle function depends on timing of activity and host response.
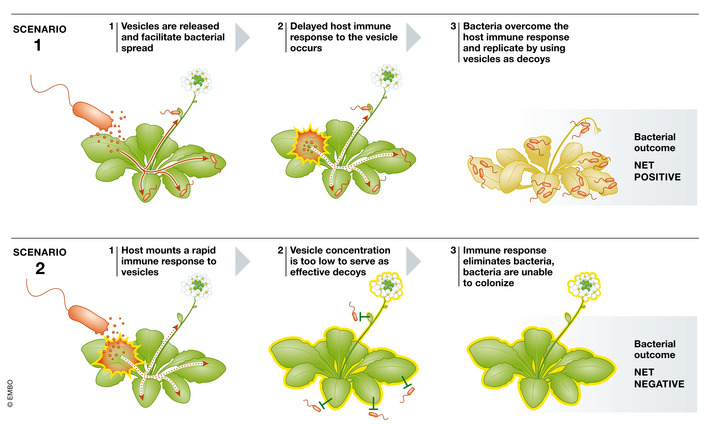
Whether vesicle production benefits or harms the producing cell ultimately depends on timing of the various vesicle‐associated functionalities and elicited host responses. Top: Potential event sequence that results in an overall benefit to the producing bacterial cell. Bottom: Event sequence that could result in a disadvantage to the bacterial cell.
Another recent study supports our findings as well as those from Bahar et al, showing that Pst vesicles lead to protection against bacterial pathogens and induce FRK1 expression in plants (preprint: Janda et al, 2021). Importantly, these results also show that Pst produces vesicles in planta that have similar biophysical properties to those isolated through traditional purification techniques (preprint: Janda et al, 2021). While current vesicle purification techniques limit the ability to test the function of vesicles produced in planta, these results provide critical evidence that vesicles are a physiologically relevant player in plant–microbe interactions.
Vesicle cargo that leads to plant immune activation
In an effort to determine which vesicle cargo were responsible for plant immune activation, we tested protection using Pst type three secretion system (T3SS) mutants. T3SS effectors are well‐studied elicitors of potent plant immune responses that can result in local and systemic protection against invading pathogens (Vlot et al, 2009, 2021; Spoel & Dong, 2012; Conrath et al, 2015). T3SS effectors have also been found in vesicles isolated from plant bacteria, including Pst (Sidhu et al, 2008; Chowdhury & Jagannadham, 2013; preprint: Janda et al, 2021). Surprisingly, pre‐treatment with vesicles from these Pst mutants resulted in the same level of protection as vesicles from wild‐type Pst, revealing that vesicle‐mediated plant protection is T3SS‐independent (McMillan et al, 2021b).
Roles for vesicles in plant growth‐defense trade‐offs
Plant immune activation that results in protection often leads to stunted plant growth as a result of growth‐defense trade‐offs (Huot et al, 2014; Pieterse et al, 2014). Accordingly, treatment with bacterial vesicles results in stunted seedling growth in A. thaliana (McMillan et al, 2021b). Using this effect as a high‐throughput assay to measure the plant immune response, we probed further the role of various vesicle cargo in plant immune activation. Strikingly, denaturing or degrading vesicle‐associated protein completely eliminated the growth‐inhibition phenotype (McMillan et al, 2021b). However, upon testing protein‐free vesicles in pathogen protection assays, we discovered that vesicle‐associated protein was not responsible for vesicle‐mediated protection against pathogens in plants (McMillan et al, 2021b).
In contrast to our results, Janda et al (preprint: Janda et al, 2021) show that Pst vesicle treatment does not lead to stunted seedling growth. This difference in function is likely due to slight variation in isolation and purification techniques and highlights an important consideration for vesicle studies. Namely, vesicle biogenesis and cargo packaging, and subsequently vesicle function, are largely dependent on the culture conditions from which the vesicles are isolated. With respect to these two studies, differences in vesicle preparation including bacterial culture time, culture density, buffer components, and density gradient medium could all contribute to the varied effect on seedling growth inhibition. Future studies should continue to carefully consider differences in preparation conditions when drawing comparative conclusions about vesicle function.
These results suggest that vesicle‐mediated plant immune activation and its resulting outcomes are complex and likely involve diverse mixtures of highly stable immune‐active molecules, potentially including lipids and small molecules. Our results also suggest that vesicle‐mediated protection against pathogens could occur in many different plant species, as protection against the oomycete Phytophthora infestans was observed in tomato in addition to the protective responses characterized in A. thaliana (McMillan et al, 2021b). Furthermore, our findings reveal a novel use for vesicles as tools to probe growth‐defense trade‐offs in plants as well as salicylic acid‐independent immune response pathways. Ultimately, vesicles may prove useful in developing agricultural treatments that lead to durable resistance in crop species.
Plant cell wall‐degrading effects of bacterial vesicles
Several additional studies have shown that vesicles or molecules contained in vesicles elicit a wide range of plant immune responses that would be disadvantageous for bacterial survival. For example, vesicles containing plant cell wall‐degrading enzymes elicit immune responses such as callose deposition and programmed cell death in plant cells, both mechanisms designed to exclude or eliminate pathogens and contain infection (Chowdhury & Jagannadham, 2013; Solé et al, 2015; Tayi et al, 2016). Intriguingly, these responses are dependent on the presence of the cell wall‐degrading enzymes in the vesicles, specifically cellulase and xylanase (Tayi et al, 2016), which parallels the situation for vesicle‐associated microbe‐associated molecular patterns (MAMPs)/PAMPs that stimulate a host immune response directed against bacteria during mammalian infections.
These examples clearly demonstrate that vesicles are both beneficial and detrimental to bacterial survival in plant systems through their roles contributing to bacterial virulence by facilitating spread and colonization and eliciting plant immune responses that limit the spread of infection, respectively. Future studies are needed to explore these trade‐offs. For instance, it would be important to determine whether there is temporal overlap or sequential timing for the vesicle‐mediated benefits and immune activation (Figs 4 and 5). The advantage of Xf spreading further may outweigh the costs of Xf vesicles activating plant immune responses if the benefit occurs before the reactive response is elicited (Fig 4). Similarly, vesicles released at different stages of bacterial growth may contain distinct cargo and, therefore, have unique functionality (Fig 5). Ultimately, whether vesicles can be deemed advantageous or disadvantageous to the bacterial cells depends on environmental conditions, the plant and bacterial genotypes, coincident infections, and other confounding factors.
Figure 5. Bacterially controlled timing of vesicle packaging and release could result in functionally distinct vesicle populations.
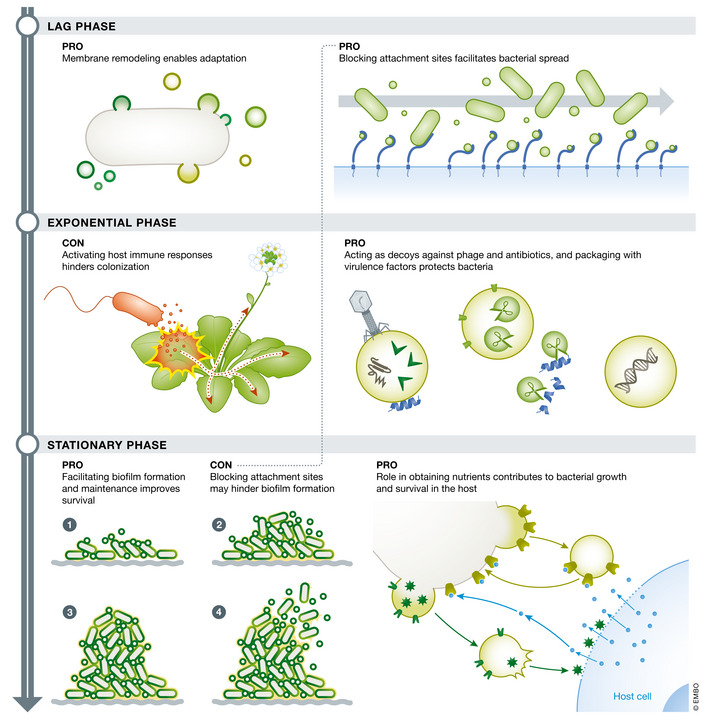
Early in bacterial growth, vesicles may be packaged with molecules that facilitate attachment and spread, while vesicles released in later growth stages may contribute to bacterial virulence or detachment. Controlled release of different vesicle populations might ensure that timing of release does not negatively interact with host immune responses, resulting in an overall benefit to the producing cell.
Limits of a reductionist approach
In evaluating the costs and benefits of vesicle production on bacterial survival and interaction with their environment, it is important to consider that vesicles are commonly studied as purified entities, effectively separated from the vesicle‐producing bacteria. Despite the strengths of such reductionist approaches to pinpoint specific effectors using thoroughly purified vesicles, it should be mentioned that the natural context of vesicles is complex, often including the vesicle‐producing cells as well as other bacteria, other types of cells, abiotic substrates, and solvents. Therefore, by extrapolating physiological conclusions from experiments using only purified components many advantages and disadvantages of vesicle production may be overlooked.
Biofilms are a well‐studied example of a complex environment that harbors vesicles. Bacterial vesicles substantially contribute to the extracellular matrix structure of a polymicrobial biofilm, which helps mediate antibiotic resistance and colonization of surfaces even during flow conditions (Fig 1E, Table 1) (Schooling & Beveridge, 2006; Yonezawa et al, 2011, 2017; Grande et al, 2015; Park et al, 2015; Gui et al, 2016; Cooke et al, 2019). In addition, the substrate of the biofilm can add complexity to the physiological situation. For instance, in the context of a coculture model using bacterial biofilms grown on lung epithelial cells, an aminopeptidase associated with P. aeruginosa vesicles was shown to facilitate bacterial cell detachment from the biofilm (Esoda & Kuehn, 2019). This ability to modulate the biofilm would benefit the bacteria by allowing it to relocate if nutrients were depleted or conditions became unfavorable. Examining the effect of purified vesicles in an environment without the bacteria or host cells present would have overlooked this critical function.
These studies reveal critical aspects of vesicle function over time under flow conditions. Modulation of aminopeptidase expression and packaging, for example, could be used to control biofilm formation or dispersal dependent on available nutrients at a given location and other environmental factors. Similarly, while X. fastidiosa vesicles facilitate bacterial spread in a plant system by coating xylem cell walls (Ionescu et al, 2014), one could imagine a situation where vesicles compete with bacteria for a limited number of binding sites (Figs 1H and 5, Table 1). In some contexts, inhibiting bacterial attachment may be detrimental for colonization success, especially under flow conditions where lack of attachment may inhibit biofilm formation. These considerations add to the importance of examining vesicle function for each producing species in its natural context.
Also overlooked in typical monoculture experiments, natural, complex bacterial environments such as microbiomes and biofilms present an opportunity for vesicles to enable interspecies behavior. To survive in mixed bacterial communities, bacteria use a variety of mechanisms to outcompete other bacterial strains. Recent studies show that vesicles play a role in these competitive interactions. In one such study, it was discovered that Chromobacterium violaceum, a Gram‐negative opportunistic pathogen common in soil and water, selectively packages vesicles with violacein, a hydrophobic antibiotic (Choi et al, 2020). These vesicles are then used to attack S. aureus and reduce S. aureus growth, presumably to confer a competitive advantage for C. violaceum (Choi et al, 2020). In fact, antagonistic vesicle function even spans kingdoms. For example, A. thaliana vesicles are selectively packaged with small RNAs that target and silence virulence genes in the fungal pathogen Botrytis cinerea and likely also in the oomycete Phytophthora infestans (Cai et al, 2018, 2020; Baldrich et al, 2019; Hou et al, 2019; He et al, 2021).
Notably, vesicles can also function in cooperative behavior. For example, Moraxella vesicles help Haemophilus influenzae evade the complement response and can improve survival of S. pneumoniae and H. influenzae by inactivating amoxicillin (Thuan Tong et al, 2007; Schaar et al, 2011). In three similar cooperative interactions, H. influenzae vesicles carrying β‐lactamase can protect group A streptococci from amoxicillin‐mediated killing, vesicles from β‐lactam‐resistant E. coli can improve survival of β‐lactam‐susceptible E. coli in the presence of ampicillin, cefoperazone, or cefotaxime, and B. thetaiotaomicron vesicles carrying surface‐associated β‐lactamases can protect S. typhimurium and gut commensals from cefotaxime (Schaar et al, 2014; Stentz et al, 2015; Kim et al, 2018). Nevertheless, such studies of vesicle contributions to cooperative and antagonistic behavior are scarce and the roles of vesicles in complex native environments remain virtually unknown. Without investigating vesicles in combination with mixed bacterial communities, many of their functions might be overlooked.
Even studies with only one bacterial species in complex media have been critical in the discovery of novel vesicle functions, such as nutrient acquisition (Figs 1D and 2C). Both vesicle‐mediated nutrient uptake and OMV production in P. aeruginosa are linked to Pseudomonas quinolone signal (PQS), a hydrophobic molecule secreted by P. aeruginosa. As mentioned above, PQS promotes OMV production when it inserts into the outermost membrane leaflet, induces membrane curvature, and thereby leads to vesicle budding (Florez et al, 2017; Horspool & Schertzer, 2018). Additionally, PQS binds iron, which is a necessary nutrient for the bacterial cell (Bredenbruch et al, 2006; Diggle et al, 2007). Lin et al (2017) showed that P. aeruginosa secretes a T6SS effector, TseF, that binds PQS and helps deliver iron‐containing vesicles to the cell via FptA and OprF (Fig 2C). Vesicles from several other species including Francisella novicida, Bacillus subtilis, S. typhimurium, and P. syringae have also been implicated in nutrient acquisition (Dubey & Ben‐Yehuda, 2011; Galkina et al, 2011; McCaig et al, 2013; Sampath et al, 2018; preprint: Janda et al, 2021). Interestingly, some of these vesicles remain attached to the parent cell in a membrane tubule, perhaps to enable delivery of nutrients more efficiently to the cell (Dubey & Ben‐Yehuda, 2011; Galkina et al, 2011; McCaig et al, 2013; Sampath et al, 2018). Importantly, the role vesicles play in nutrient acquisition could be easily missed in the absence of the producing cell.
In another instance of vesicle‐mediated nutrient acquisition, Pseudomonas putida, an environmental bacterium, was shown recently to package vesicles with enzymes to break down lignin, an abundant component of plant cell walls (Salvachúa et al, 2020). At first glance, it may seem that using vesicles to break down cell wall components would aid in virulence by activating damage associated molecular pattern‐triggered immune responses in plants. However, adding vesicles to bacterial cultures revealed that this mechanism actually allowed the bacteria to use lignin as an alternative nutrient source (Fig 1D) (Salvachúa et al, 2020). Without testing their effect on the parent bacteria and instead testing only purified vesicles in a plant system, researchers may have falsely concluded that vesicle‐mediated lignin digestion was a virulence strategy rather than a nutrient acquisition mechanism.
Important to consider in examining bacterial vesicle function in complex microbial environments is the concept of social “cheating”. In this scenario, non‐vesicle‐producing bacteria take advantage of resources packaged in vesicles from other strains or the resulting function of those vesicles and avoid the associated metabolic cost of vesicle production (Table 1). For example, in the gut environment, vesicles from Bacteroidales contain glycoside hydrolases that break down polysaccharides (Rakoff‐Nahoum et al, 2014). Non‐vesicle‐producing strains in the gut take advantage of this function by using the polysaccharide breakdown products as nutrients, which can even lead to outgrowth by the “cheater” of the producing strain (Rakoff‐Nahoum et al, 2014). P. aeruginosa strains that lack the global stress response regulator RpoS similarly take advantage of secreted aminopeptidase, which is known to associate with vesicles (Esoda & Kuehn, 2019), for its proteolytic activity to enable utilization of protein as a nutrient source (Robinson et al, 2020). This concept applies also to Gram‐positive species. Dietzia sp. DQ12‐45‐1b has been shown recently to package heme‐binding proteins in vesicles that participate in iron acquisition (Wang et al, 2021). Bacteria from a variety of related species, but not more distant species, are able to use the vesicle‐associated iron as a nutrient for their own growth (Wang et al, 2021). Vesicle production in the context of “cheaters” may pose an exacerbated metabolic burden for the producing strain, resulting in a net‐negative score in the cost‐benefit analysis. These studies reveal the importance of studying vesicle function in the context of the producing strain and also complex multi‐species environments.
There are also, of course, numerous instances where vesicles do exhibit important functions independent of the producing cell, especially in the context of long‐distance signaling. This phenomenon is exemplified in studies of gut microbiota interactions with intestinal epithelial cells. Vesicles from B. thetaiotaomicron enter gut epithelial cells via dynamin‐dependent endocytosis and are also able to transmigrate across the epithelial barrier via a paracellular route (Jones et al, 2020). Importantly, these vesicles then disseminate systemically to tissues, including the liver in mice, and are implicated in balanced gut immune function, which supports vesicle function independent of bacterial cells (Durant et al, 2020; Jones et al, 2020; preprint: Gul et al, 2021). When studied in humans, the vesicles traversed the gut epithelium into the bloodstream more readily in individuals with intestinal barrier dysfunction (Tulkens et al, 2020a, 2020b). In fact, some vesicles have even been shown to alter epithelial permeability, such as those from C. jejuni (Elmi et al, 2016). Ability to cross the mucus barrier and intestinal epithelium and interact with host immune systems has also been observed in other resident gut microbiota and pathogens, including E. coli Nissle 1917, ECOR63, and Bacteroides vulgatus (Alvarez et al, 2016; Maerz et al, 2018). By crossing mucus and epithelial barriers, vesicles effectively separate from the producing cell. While many vesicle functions may occur synergistically with bacterial cell functions, these findings support independent roles for vesicles as well.
Action at a distance is not limited to gut microbes. P. aeruginosa vesicles are able to diffuse through the mucus layer in airway epithelial cells and deliver cocktails of virulence factors in the absence of bacterial cells (Bomberger et al, 2009). Vesicles from the periodontal pathogen Aggregatibacter actinomycetemcomitans cross the blood–brain barrier and deliver small RNAs that lead to TNF‐α production (Han et al, 2019). Indeed, many studies have revealed that vesicles from diverse species can cross epithelial barriers and travel throughout an organism to exert their function (Jang et al, 2015; Stentz et al, 2018; Cuesta et al, 2021). It is important to note, however, that while vesicles may function at a distance from the bacterial cell, this may also serve to weaken the host barrier and facilitate bacterial invasion of host tissues, similar to the well‐studied functions of soluble toxins. Understanding how vesicle function differs from bacterial processes and soluble factors and predicting how far vesicles can travel through complex environments will benefit from convergence of many fields (McMillan et al, 2021a).
Beyond mammalian systems, vesicles in plant and environmental systems presumably also exhibit some functions independent of the producing cell. Vesicles produced by marine bacteria, for example, are instantly diluted in seawater and likely carried far from the producing cell by ocean currents (Biller et al, 2014). These vesicle populations have been shown to contain nucleic acids, and those from the cyanobacterium Prochlorococcus can support the growth of other bacterial species, suggesting a role in carbon cycling (Biller et al, 2014, 2017). Additional support for their role in biogeochemical cycles in marine environments comes from a recent study that shows Prochlorococcus vesicles associate with diverse bacteria and contain active enzymes that could participate in energy metabolism and extracellular biochemical reactions (preprint: Biller et al, 2020). Roles for vesicles in carbon, nutrient, and metal cycling have been reported in several other environments (Matlakowska et al, 2012; Prados‐Rosales et al, 2014b; Shao et al, 2014; Lin et al, 2017; Liu et al, 2020); however, these studies all show a direct benefit of vesicle function for the bacterial cell.
While characterization of purified vesicle cargo and function may reveal the presence or absence of molecules as well as specific vesicle‐mediated effects, only studies integrating vesicles in their larger context will reveal the potential impact they have on bacterial survival, virulence, and overall function. Parallels to this concept exist across biology and can be summed up in the basic principle of emergent properties. Time and time again scientists have realized that despite a detailed understanding of the individual components of a system, their combined function remains difficult to predict. For example, in ecological studies of species diversity, keystone species perform critical functions despite often miniscule abundances in the community (Paine, 1966, 1969, 1992; Mills et al, 1993; Davic, 2003; Smith et al, 2008; Smith & Bangs, 2009; Hale & Koprowski, 2018). In studies of the gut microbiome, researchers have found that often one species will not colonize or will have dramatically different function without the presence of certain other species (Chung et al, 2012; Surana & Kasper, 2014, 2017; Mosca et al, 2016; Turroni et al, 2019). Despite repeatedly showing the importance of emergent properties in understanding interactions and overall function in numerous contexts, we still resist its incorporation into molecular and host–pathogen studies. Of course, the reason for this hesitation is in large part due to the inevitable increase in complexity that will occur after adding even one additional element to the study. However, to move the field forward will ultimately require combining the fine details from reductionist approaches with systems level data analysis to create a holistic understanding of how these organisms and environments function on every level of complexity.
Conclusion and future directions: settling the score for vesicles
How do we settle the score? Is vesicle production overall beneficial for the bacterial cell, or does the energetic cost outweigh their usefulness? Are bacteria intentionally releasing these cargo‐filled packages or are they simply a bioproduct of stress? The studies reviewed here suggest that vesicles from both Gram‐negative and Gram‐positive species are loaded with specific cargo and ultimately benefit the bacterial cell. Despite their energetic costliness, vesicles provide a fast and selective membrane remodeling mechanism that aids bacterial survival, especially during transitions to stressful environments. Furthermore, in both mammalian and plant systems, vesicles appear to play a critical role in bacterial virulence and allow action at a distance. One could argue that their benefits to virulence are outweighed by host immune activation, which could be detrimental to bacterial viability. However, we must consider that many other virulence mechanisms result in host immune activation and yet are considered ultimately beneficial for bacterial colonization and survival. Until we are able to analyze vesicle production, transport, activities, and consequences in the environment or in animals during colonization and infection, we can only guess whether the overall outcome helps or harms the bacterial cell.
To finally tally the results, future work should incorporate findings from reductionist studies to interpret studies of vesicle function in complex bacterial communities and environmental conditions. We must begin working toward studying vesicles in their natural context, which will require development of new tools to visualize, isolate, and characterize bacterial vesicle composition, trafficking, and function. Only then can we work toward a complete understanding of why bacteria produce vesicles and determine their ultimate roles in complex environments.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by the National Science Foundation under Grant Number 1931309 and the Duke University Medical Center. We apologize to colleagues whose work has not been cited due to space constraints.
The EMBO Journal (2021) 40: e108174.
References
- Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwagi T, Narita S, Nakagawa A, Nakae T (2004a) Crystal structure of the drug‐discharge outer membrane protein. OprM. 10.2210/pdb1WP1/pdb [DOI] [PubMed]
- Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwagi T, Yoneyama H, Narita S‐I, Nakagawa A, Nakae T (2004b) Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end*. J Biol Chem 279: 52816–52819 [DOI] [PubMed] [Google Scholar]
- Allison CC, Kufer TA, Kremmer E, Kaparakis M, Ferrero RL (2009) Helicobacter pylori induces MAPK phosphorylation and AP‐1 activation via a NOD1‐dependent mechanism. J Immunol 183: 8099 [DOI] [PubMed] [Google Scholar]
- Alvarez C‐S, Badia J, Bosch M, Giménez R, Baldomà L (2016) Outer membrane vesicles and soluble factors released by probiotic Escherichia coli Nissle 1917 and commensal ECOR63 enhance barrier function by regulating expression of tight junction proteins in intestinal epithelial cells. Front Microbiol 7: 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoni F, Toyofuku M, Menzi C, Kalawong R, Mairpady Shambat S, François P, Zinkernagel AS, Eberl L (2019). Antibiotics stimulate formation of vesicles in Staphylococcus aureus in both phage‐dependent and ‐independent fashions and via different routes. Antimicrob Agents Chemother 63: e01439–e1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antushevich H (2020) Interplays between inflammasomes and viruses, bacteria (pathogenic and probiotic), yeasts and parasites. Immunol Lett 228: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar O, Mordukhovich G, Luu DD, Schwessinger B, Daudi A, Jehle AK, Felix G, Ronald PC (2016) Bacterial outer membrane vesicles induce plant immune responses. Mol Plant Microbe Interact 29: 374–384 [DOI] [PubMed] [Google Scholar]
- Baldrich P, Rutter BD, Karimi HZ, Podicheti R, Meyers BC, Innes RW (2019) Plant extracellular vesicles contain diverse small RNA species and are enriched in 10‐ to 17‐nucleotide “Tiny” RNAs. Plant Cell 31: 315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwens A, Kunsmann L, Marejková M, Zhang W, Karch H, Bielaszewska M, Mellmann A (2017) Intrahost milieu modulates production of outer membrane vesicles, vesicle‐associated Shiga toxin 2a and cytotoxicity in Escherichia coli O157:H7 and O104:H4. Environ Microbiol Rep 9: 626–634 [DOI] [PubMed] [Google Scholar]
- Berry MC, McGhee GC, Zhao Y, Sundin GW (2009) Effect of a waaL mutation on lipopolysaccharide composition, oxidative stress survival, and virulence in Erwinia amylovora . FEMS Microbiol Lett 291: 80–87 [DOI] [PubMed] [Google Scholar]
- Beveridge TJ (1999) Structures of gram‐negative cell walls and their derived membrane vesicles. J Bacteriol 181: 4725–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielaszewska M, Rüter C, Bauwens A, Greune L, Jarosch K‐A, Steil D, Zhang W, He X, Lloubes R, Fruth A et al (2017) Host cell interactions of outer membrane vesicle‐associated virulence factors of enterohemorrhagic Escherichia coli O157: Intracellular delivery, trafficking and mechanisms of cell injury. PLoS Pathog 13: e1006159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Lundeen RA, Hmelo LR, Becker KW, Arellano AA, Dooley K, Heal KR, Carlson LT, van Mooy BAS , Ingalls AE et al (2020) Prochlorococcus extracellular vesicles: molecular composition and adsorption to diverse microbes. bioRxiv 10.1101/2020.12.18.423521 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, McDaniel LD, Breitbart M, Rogers E, Paul JH, Chisholm SW (2017) Membrane vesicles in sea water: heterogeneous DNA content and implications for viral abundance estimates. ISME J 11: 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW (2014) Bacterial vesicles in marine ecosystems. Science 343: 183–186 [DOI] [PubMed] [Google Scholar]
- Bishop DG, Work E (1965) An extracellular glycolipid produced by Escherichia coli grown under lysine‐limiting conditions. Biochem J 96: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto NJ, Baker PJ, Dowling JK, Wray‐Mccann G, de Paoli A, Tran LS, Leung PL, Stacey KJ, Mansell A, Masters SL et al (2018) Membrane vesicles from Pseudomonas aeruginosa activate the noncanonical inflammasome through caspase‐5 in human monocytes. Immunol Cell Biol 96: 1120–1130. [DOI] [PubMed] [Google Scholar]
- Bitto NJ, Chapman R, Pidot S, Costin A, Lo C, Choi J, D'Cruze T, Reynolds EC, Dashper SG, Turnbull L et al (2017) Bacterial membrane vesicles transport their DNA cargo into host cells. Sci Rep 7: 7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto NJ, Cheng L, Johnston EL, Pathirana R, Phan TK, Poon IKH, O'Brien‐simpson NM, Hill AF, Stinear TP, Kaparakis‐Liaskos M (2021) Staphylococcus aureus membrane vesicles contain immunostimulatory DNA, RNA and peptidoglycan that activate innate immune receptors and induce autophagy. J Extracell Vesicles 10: e12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O'Toole GA, Stanton BA (2009) Long‐distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog 5: e1000382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnington KE, Kuehn MJ (2014) Protein selection and export via outer membrane vesicles. Biochim Biophys Acta Mol Cell Res 1843: 1612–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnington KE, Kuehn MJ (2016) Outer membrane vesicle production facilitates LPS remodeling and outer membrane maintenance in salmonella during environmental transitions. Mbio 7: e01532‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbruch F, Geffers R, Nimtz M, Buer J, Häussler S (2006) The Pseudomonas aeruginosa quinolone signal (PQS) has an iron‐chelating activity. Environ Microbiol 8: 1318–1329 [DOI] [PubMed] [Google Scholar]
- Briaud P, Carroll RK (2020) Extracellular vesicle biogenesis and functions in gram‐positive bacteria. Infect Immun 88: e00433–e520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Wolf JM, Prados‐Rosales R, Casadevall A (2015) Through the wall: extracellular vesicles in Gram‐positive bacteria, mycobacteria and fungi. Nat Rev Microbiol 13: 620–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill BK, Seeley KW, Gutel D, Ellis TN (2015) Klebsiella pneumoniae O antigen loss alters the outer membrane protein composition and the selective packaging of proteins into secreted outer membrane vesicles. Microbiol Res 180: 1–10 [DOI] [PubMed] [Google Scholar]
- Cai Q, He B, Weiberg A, Buck AH, Jin H (2020) Small RNAs and extracellular vesicles: new mechanisms of cross‐species communication and innovative tools for disease control. PLoS Pathog 15: e1008090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Qiao L, Wang M, He B, Lin F‐M, Palmquist J, Huang S‐D, Jin H (2018) Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360: 1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra EJ, Laub MT (2012) Evolution of two‐component signal transduction systems. Annu Rev Microbiol 66: 325–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil JD, Sirisaengtaksin N, O'Brien‐simpson NM, Krachler AM (2019) Outer membrane vesicle‐host cell interactions. Microbiol Spectr 7: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D, Chaudhuri K (2011) Association of cholera toxin with Vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett 585: 1357–1362 [DOI] [PubMed] [Google Scholar]
- Chatterjee SN, Das J (1967) Electron microscopic observations on the excretion of cell‐wall material by Vibrio cholerae . Microbiology 49: 1–11 [DOI] [PubMed] [Google Scholar]
- Chen HD, Groisman EA (2013) The biology of the PmrA/PmrB two‐component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67: 83–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y‐Y, Peng B, Yang Q, Glew MD, Veith PD, Cross KJ, Goldie KN, Chen D, O'Brien‐simpson N, Dashper SG et al (2011) The outer membrane protein LptO is essential for the O‐deacylation of LPS and the co‐ordinated secretion and attachment of A‐LPS and CTD proteins in Porphyromonas gingivalis . Mol Microbiol 79: 1380–1401. [DOI] [PubMed] [Google Scholar]
- Cheung GYC, Joo H‐S, Chatterjee SS, Otto M (2014) Phenol‐soluble modulins – critical determinants of staphylococcal virulence. FEMS Microbiol Rev 38: 698–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SY, Lim S, Cho G, Kwon J, Mun W, Im H, Mitchell RJ (2020) Chromobacterium violaceum delivers violacein, a hydrophobic antibiotic, to other microbes in membrane vesicles. Environ Microbiol 22: 705–713 [DOI] [PubMed] [Google Scholar]
- Chowdhury C, Jagannadham MV (2013) Virulence factors are released in association with outer membrane vesicles of Pseudomonas syringae pv. tomato T1 during normal growth. Biochim Biophys Acta 1834: 231–239 [DOI] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR et al (2012) Gut immune maturation depends on colonization with a host‐specific microbiota. Cell 149: 1578–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutkan H, Kuehn MJ (2011) Context‐dependent activation kinetics elicited by soluble versus outer membrane vesicle‐associated heat‐labile enterotoxin. Infect Immun 79: 3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutkan H, Macdonald I, Manning A, Kuehn MJ (2013) Quantitative and qualitative preparations of bacterial outer membrane vesicles. Methods Mol Biol 966: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther‐Rasmussen J, Høiby N (2000) Chromosomal β‐lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa . J Antimicrob Chemother 45: 9–13 [DOI] [PubMed] [Google Scholar]
- Clarke AJ (2018) The “hole” story of predatory outer‐membrane vesicles. Can J Microbiol 64: 589–599 [DOI] [PubMed] [Google Scholar]
- Cobessi D, Celia H, Folschweiller N, Schalk IJ, Abdallah MA, Pattus F (2004) Pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa PAO1 bound to pyoverdine. 10.2210/pdb1XKH/pdb [DOI] [PubMed]
- Cobessi D, Celia H, Folschweiller N, Schalk IJ, Abdallah MA, Pattus F (2005) The crystal structure of the pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6Å resolution. J Mol Biol 347: 121–134 [DOI] [PubMed] [Google Scholar]
- Coelho C, Brown L, Maryam M, Vij R, Smith DFQ, Burnet MC, Kyle JE, Heyman HM, Ramirez J, Prados‐Rosales R et al (2019) Listeria monocytogenes virulence factors, including listeriolysin O, are secreted in biologically active extracellular vesicles. J Biol Chem 294: 1202–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U, Beckers GJM, Langenbach CJG, Jaskiewicz MR (2015) Priming for enhanced defense. Annu Rev Phytopathol 53: 97–119 [DOI] [PubMed] [Google Scholar]
- Cooke AC, Nello AV, Ernst RK, Schertzer JW (2019) Analysis of Pseudomonas aeruginosa biofilm membrane vesicles supports multiple mechanisms of biogenesis. PLoS One 14: e0212275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta CM, Guerri C, Ureña J, Pascual M (2021) Role of microbiota‐derived extracellular vesicles in gut‐brain communication. Int J Mol Sci 22: 4235–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davic RD (2003) Linking keystone species and functional groups: a new operational definition of the keystone species concept. Conserv Ecol 7: 1–11 [Google Scholar]
- Deatherage BL, Lara JC, Bergsbaken T, Rassoulian Barrett SL, Lara S, Cookson BT (2009) Biogenesis of bacterial membrane vesicles. Mol Microbiol 72: 1395–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo P, Chow SH, Hay ID, Kleifeld O, Costin A, Elgass KD, Jiang JH, Ramm G, Gabriel K, Dougan G et al (2018) Outer membrane vesicles from Neisseria gonorrhoeae target PorB to mitochondria and induce apoptosis. PLoS Pathog 14: e1006945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Cámara M et al (2007) The Pseudomonas aeruginosa 4‐quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol 14: 87–96 [DOI] [PubMed] [Google Scholar]
- Dorward DW, Garon CF (1990) DNA is packaged within membrane‐derived vesicles of gram‐negative but not gram‐positive bacteria. Appl Environ Microbiol 56: 1960–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Antonny B (2010) Amphipathic helices and membrane curvature. FEBS Lett 584: 1840–1847 [DOI] [PubMed] [Google Scholar]
- Dubey GP, Ben‐Yehuda S (2011) Intercellular nanotubes mediate bacterial communication. Cell 144: 590–600 [DOI] [PubMed] [Google Scholar]
- Duke SO, Dayan FE (2011) Modes of action of microbially‐produced phytotoxins. Toxins 3: 1038–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant L, Stentz R, Noble A, Brooks J, Gicheva N, Reddi D, O’Connor MJ, Hoyles L, McCartney AL, Man R et al (2020) Bacteroides thetaiotaomicron‐derived outer membrane vesicles promote regulatory dendritic cell responses in health but not in inflammatory bowel disease. Microbiome 8: 88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlein C, Baumgarten T, Starke S, Heipieper HJ (2018) Immediate response mechanisms of Gram‐negative solvent‐tolerant bacteria to cope with environmental stress: cis‐trans isomerization of unsaturated fatty acids and outer membrane vesicle secretion. Appl Microbiol Biotechnol 102: 2583–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhenawy W, Bording‐Jorgensen M, Valguarnera E, Haurat MF, Wine E, Feldman MF (2016) LPS remodeling triggers formation of outer membrane vesicles in Salmonella. Mbio 7: e00940‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis TN, Kuehn MJ (2010) Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74: 81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis TN, Leiman SA, Kuehn MJ (2010) Naturally produced outer membrane vesicles from Pseudomonas aeruginosa elicit a potent innate immune response via combined sensing of both lipopolysaccharide and protein components. Infect Immun 78: 3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmi A, Nasher F, Jagatia H, Gundogdu O, Bajaj‐Elliott M, Wren B, Dorrell N (2016) Campylobacter jejuni outer membrane vesicle‐associated proteolytic activity promotes bacterial invasion by mediating cleavage of intestinal epithelial cell E‐cadherin and occludin. Cell Microbiol 18: 561–572 [DOI] [PubMed] [Google Scholar]
- Esoda CN, Kuehn MJ (2019) Pseudomonas aeruginosa leucine aminopeptidase influences early biofilm composition and structure via vesicle‐associated antibiofilm activity. Mbio 10: e02548‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitosa‐Junior OR, Stefanello E, Zaini PA, Nascimento R, Pierry PM, Dandekar AM, Lindow SE, da Silva AM (2019) Proteomic and metabolomic analyses of Xylella fastidiosa OMV‐enriched fractions reveal association with virulence factors and signaling molecules of the DSF family. Phytopathology 109: 1344–1353 [DOI] [PubMed] [Google Scholar]
- Finethy R, Dockterman J, Kutsch M, Orench‐Rivera N, Wallace GD, Piro AS, Luoma S, Haldar AK, Hwang S, Martinez J et al (2020) Dynamin‐related Irgm proteins modulate LPS‐induced caspase‐11 activation and septic shock. EMBO Rep 21: e50830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finethy R, Luoma S, Orench‐Rivera N, Feeley EM, Haldar AK, Yamamoto M, Kanneganti TD, Kuehn MJ, Coers J (2017) Inflammasome activation by bacterial outer membrane vesicles requires guanylate binding proteins. Mbio 8: e01188‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez C, Raab JE, Cooke AC, Schertzer JW (2017) Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa . Mbio 8: e1034–e1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias A, Manresa A, de Oliveira E, López‐Iglesias C, Mercade E (2010) Membrane vesicles: a common feature in the extracellular matter of cold‐adapted antarctic bacteria. Microb Ecol 59: 476–486 [DOI] [PubMed] [Google Scholar]
- Galkina SI, Romanova JM, Bragina EE, Tiganova IG, Stadnichuk VI, Alekseeva NV, Polyakov VY, Klein T (2011) Membrane tubules attach Salmonella Typhimurium to eukaryotic cells and bacteria. FEMS Immunol Med Microbiol 61: 114–124 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Golemi D, Maveyraud L, Vakulenko S, Samama J‐P, Mobashery S (2001a) Critical involvement of a carbamylated lysine in catalytic function of class D β‐lactamases. Proc Natl Acad Sci USA 98: 14280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S (2001b) OXA 10 class D beta‐lactamase at pH 6.0. 10.2210/pdb1K57/pdb [DOI] [PMC free article] [PubMed]
- Grande R, Di Marcantonio MC, Robuffo I, Pompilio A, Celia C, Di Marzio L, Paolino D, Codagnone M, Muraro R, Stoodley P et al (2015) Helicobacter pylori ATCC 43629/NCTC 11639 outer membrane vesicles (OMVs) from biofilm and planktonic phase associated with extracellular DNA (eDNA). Front Microbiol 6: 1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui MJ, Dashper SG, Slakeski N, Chen YY, Reynolds EC (2016) Spheres of influence: Porphyromonas gingivalis outer membrane vesicles. Mol Oral Microbiol 31: 365–378 [DOI] [PubMed] [Google Scholar]
- Gul L, Modos D, Fonseca S, Madgwick M, Thomas JP, Sudhakar P, Stentz R, Carding SR, Korcsmaros T (2021) Extracellular vesicles produced by the human commensal gut bacterium Bacteroides thetaiotaomicron affect host immune pathways in a cell‐type specific manner that are altered in inflammatory bowel disease. bioRxiv 10.1101/2021.03.20.436262 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann S, Stöger L, Kappelmann M, Hassl I, Ellinger A, Velimirov B (2014) DNA‐bearing membrane vesicles produced by Ahrensia kielensis and Pseudoalteromonas marina . J Basic Microbiol 54: 1062–1072 [DOI] [PubMed] [Google Scholar]
- Hale SL, Koprowski JL (2018) Ecosystem‐level effects of keystone species reintroduction: a literature review. Restor Ecol 26: 439–445 [Google Scholar]
- Han E‐C, Choi S‐Y, Lee Y, Park J‐W, Hong S‐H, Lee H‐J (2019) Extracellular RNAs in periodontopathogenic outer membrane vesicles promote TNF‐α production in human macrophages and cross the blood‐brain barrier in mice. FASEB J 33: 13412–13422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurat MF, Aduse‐Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, Feldman MF (2011) Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem 286: 1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Cai Q, Qiao L, Huang C‐Y, Wang S, Miao W, Ha T, Wang Y, Jin H (2021) RNA‐binding proteins contribute to small RNA loading in plant extracellular vesicles. Nature Plants 7: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Hara H, Núñez G (2016) Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci 41: 1012–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M, Pané‐Farré J, Uwe V (2007) SigB‐dependent general stress response in bacillus subtilis and related gram‐positive bacteria. Annu Rev Microbiol 61: 215–236 [DOI] [PubMed] [Google Scholar]
- Hickey C, Kuhn K, Donermeyer D, Porter N, Jin C, Cameron E, Jung H, Kaiko G, Wegorzewska M, Malvin N et al (2015) Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase‐dependent manner via outer membrane vesicles. Cell Host Microbe 17: 672–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horspool AM, Schertzer JW (2018) Reciprocal cross‐species induction of outer membrane vesicle biogenesis via secreted factors. Sci Rep 8: 9873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstman AL, Bauman SJ, Kuehn MJ (2004) Lipopolysaccharide 3‐deoxy‐D‐manno‐octulosonic acid (Kdo) core determines bacterial association of secreted toxins. J Biol Chem 279: 8070–8075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstman AL, Kuehn MJ (2000) Enterotoxigenic Escherichia coli secretes active heat‐labile enterotoxin via outer membrane vesicles. J Biol Chem 275: 12489–12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Zhai Y, Feng L, Karimi HZ, Rutter BD, Zeng L, Choi DS, Zhang B, Gu W, Chen X et al (2019) A phytophthora effector suppresses trans‐kingdom RNAi to promote disease susceptibility. Cell Host Microbe 25: 153–165.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY (2014) Growth‐defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant 7: 1267–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu M, Yokota K, Antonova E, Garcia A, Beaulieu E, Hayes T, Iavarone AT, Lindow SE (2016) Promiscuous diffusible signal factor production and responsiveness of the Xylella fastidiosa Rpf system. Mbio 7: e1054–e1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu M, Zaini PA, Baccari C, Tran S, da Silva AM, Lindow SE (2014) Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc Natl Acad Sci USA 111: E3910–E3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan AT (2017) Outer membrane vesicles (OMVs) of gram‐negative bacteria: a perspective update. Front Microbiol 8: 1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda M, Ludwig C, Rybak K, Meng C, Stigliano E, Botzenhardt L, Szulc B, Sklenar J, Menke FLH & Malone JG et al (2021) Biophysical and proteomic analyses suggest functions of Pseudomonas syringae pv tomato DC3000 extracellular vesicles in bacterial growth during plant infection. bioRxiv 10.1101/2021.02.08.430144 [PREPRINT] [DOI] [Google Scholar]
- Jang SC, Kim SR, Yoon YJ, Park K‐S, Kim JH, Lee J, Kim OY, Choi E‐J, Kim D‐K, Choi D‐S et al (2015) In vivo kinetic biodistribution of nano‐sized outer membrane vesicles derived from bacteria. Small 11: 456–461 [DOI] [PubMed] [Google Scholar]
- Jeon J, Park SC, Her J, Lee JW, Han J‐K, Kim Y‐K, Kim KP, Ban C (2018) Comparative lipidomic profiling of the human commensal bacterium Propionibacterium acnes and its extracellular vesicles. RSC Adv 8: 15241–15247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Kong Q, Roland KL, Curtiss R (2014) Membrane vesicles of Clostridium perfringens type A strains induce innate and adaptive immunity. Int J Med Microbiol 304: 431–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston EL, Heras B, Kufer TA, Kaparakis‐Liaskos M (2021) Detection of bacterial membrane vesicles by NOD‐like receptors. Int J Mol Sci 22: 1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston EL, Kufer TA, Kaparakis‐Liaskos M (2020) Immunodetection and pathogenesis mediated by bacterial membrane vesicles. In Bacterial membrane vesicles: biogenesis, functions and applications, Kaparakis‐Liaskos M, Kufer TA (eds), pp 159–188. Cham: Springer International Publishing; [Google Scholar]
- Jones EJ, Booth C, Fonseca S, Parker A, Cross K, Miquel‐Clopés A, Hautefort I, Mayer U, Wileman T, Stentz R et al (2020) The uptake, trafficking, and biodistribution of Bacteroides thetaiotaomicron generated outer membrane vesicles. Front Microbiol 11: 57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ (1995) Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol 177: 3998–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaparakis M, Turnbull L, Carneiro L, Firth S, Coleman HA, Parkington HC, le Bourhis L, Karrar A, Viala J, Mak J et al (2010) Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol 12: 372–385 [DOI] [PubMed] [Google Scholar]
- Kaparakis‐Liaskos M, Ferrero RL (2015) Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol 15: 375–387 [DOI] [PubMed] [Google Scholar]
- Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ (2004) Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J 23: 4538–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Marquis H, Boor KJ (2005) SigmaB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 151: 3215–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Park SB, Im SP, Lee JS, Jung JW, Gong TW, Lazarte JMS, Kim J, Seo J‐S, Kim J‐H et al (2018) Outer membrane vesicles from β‐lactam‐resistant Escherichia coli enable the survival of β‐lactam‐susceptible E. coli in the presence of β‐lactam antibiotics. Sci Rep 8: 5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G, Kobylak N, Lindner B, Stupak A, Raina S (2014) Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J Biol Chem 289: 14829–14853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimentová J, Stulík J (2015) Methods of isolation and purification of outer membrane vesicles from gram‐negative bacteria. Microbiol Res 170: 1–9 [DOI] [PubMed] [Google Scholar]
- Kolling GL, Matthews KR (1999) Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol 65: 1843–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn MJ, Kesty NC (2005) Bacterial outer membrane vesicles and the host‐pathogen interaction. Genes Dev 19: 2645–2655 [DOI] [PubMed] [Google Scholar]
- Kulkarni HM, Swamy CV, Jagannadham MV (2014) Molecular characterization and functional analysis of outer membrane vesicles from the antarctic bacterium Pseudomonas syringae suggest a possible response to environmental conditions. J Proteome Res 13: 1345–1358 [DOI] [PubMed] [Google Scholar]
- Kulkarni HM, Swamy CV, Jagannadham MV (2015) The proteome of the outer membrane vesicles of an antarctic bacterium Pseudomonas syringae Lz4W. Data Brief 4: 406–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp A, Kuehn MJ (2010) Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64: 163–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp AJ, Sun B, Ai T, Manning AJ, Orench‐Rivera N, Schmid AK, Kuehn MJ (2015) Genome‐wide assessment of outer membrane vesicle production in Escherichia coli . PLoS One 10: e0139200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunsmann L, Rüter C, Bauwens A, Greune L, Glüder M, Kemper B, Fruth A, Wai SN, He X, Lloubes R et al (2015) Virulence from vesicles: novel mechanisms of host cell injury by Escherichia coli O104:H4 outbreak strain. Sci Rep 5: 13252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappann M, Otto A, Becher D, Vogel U (2013) Comparative proteome analysis of spontaneous outer membrane vesicles and purified outer membranes of Neisseria meningitidis . J Bacteriol 195: 4425–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le LHM, Ying L, Ferrero RL (2021) Nuclear trafficking of bacterial effector proteins. Cell Microbiol 23: e13320 [DOI] [PubMed] [Google Scholar]
- Lee E‐Y, Choi D‐Y, Kim D‐K, Kim J‐W, Park JO, Kim S, Kim S‐H, Desiderio DM, Kim Y‐K, Kim K‐P et al (2009) Gram‐positive bacteria produce membrane vesicles: proteomics‐based characterization of Staphylococcus aureus‐derived membrane vesicles. Proteomics 9: 5425–5436 [DOI] [PubMed] [Google Scholar]
- Lee JH, Choi C‐W, Lee T, Kim SI, Lee J‐C, Shin J‐H (2013b) Transcription factor σB plays an important role in the production of extracellular membrane‐derived vesicles in Listeria monocytogenes . PLoS One 8: e73196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee E‐Y, Kim S‐H, Kim D‐K, Park K‐S, Kim KP, Kim Y‐K, Roh T‐Y, Gho YS (2013a) Staphylococcus aureus Extracellular vesicles carry biologically active β‐lactamase. Antimicrob Agents Chemother 57: 2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Azam F, Zhang S (2016) Outer membrane vesicles containing signalling molecules and active hydrolytic enzymes released by a coral pathogen Vibrio shilonii AK1. Environ Microbiol 18: 3850–3866 [DOI] [PubMed] [Google Scholar]
- Li Z, Clarke AJ, Beveridge TJ (1996) A major autolysin of Pseudomonas aeruginosa: subcellular distribution, potential role in cell growth and division and secretion in surface membrane vesicles. J Bacteriol 178: 2479–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liangsupree T, Multia E, Riekkola M‐L (2021) Modern isolation and separation techniques for extracellular vesicles. J Chromatogr A 1636: 461773 [DOI] [PubMed] [Google Scholar]
- Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, Wei G, Qian P‐Y, Luo Z‐Q, Shen X (2017) A Pseudomonas T6SS effector recruits PQS‐containing outer membrane vesicles for iron acquisition. Nat Commun 8: 14888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Z, Dayong C, Denggao Y, Yiting W, Liaoqiong F, Zhibiao W (2019) Escherichia coli outer membrane vesicles induced DNA double‐strand breaks in intestinal epithelial Caco‐2 cells. Med Sci Monit Basic Res 25: 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Sonbol F‐M, Huot B, Gu Y, Withers J, Mwimba M, Yao J, He SY, Dong X (2016) Salicylic acid receptors activate jasmonic acid signalling through a non‐canonical pathway to promote effector‐triggered immunity. Nat Commun 7: 13099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jing X, Ye Y, Zhan J, Ye J, Zhou S (2020) Bacterial vesicles mediate extracellular electron transfer. Environ Sci Technol Lett 7: 27–34 [Google Scholar]
- Liu Y, Defourny KAY, Smid EJ, Abee T (2018) Gram‐positive bacterial extracellular vesicles and their impact on health and disease. Front Microbiol 9: 1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Orsi RH, Gaballa A, Wiedmann M, Boor KJ, Guariglia‐Oropeza V (2019) Systematic review of the Listeria monocytogenes σB regulon supports a role in stress response, virulence and metabolism. Fut Microbiol 14: 801–828 [DOI] [PubMed] [Google Scholar]
- Loeb MR, Kilner J (1978) Release of a special fraction of the outer membrane from both growing and phage T4‐infected Escherichia coli B. Biochim Biophys Acta Biomembr 514: 117–127 [DOI] [PubMed] [Google Scholar]
- Luz BSRD, Nicolas A, Chabelskaya S, Rodovalho VDR, le Loir Y, Azevedo VADC, Felden B, Guédon E (2021) Environmental plasticity of the RNA content of Staphylococcus aureus extracellular vesicles. Front Microbiol 12: 634226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JB, Schwartzman JA, Bennett BD, McAnulty SJ, Knop M, Nyholm SV, Ruby EG (2019) Ambient pH alters the protein content of outer membrane vesicles, driving host development in a beneficial symbiosis. J Bacteriol 201: e00319–e419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald IA, Kuehn MJ (2013) Stress‐induced outer membrane vesicle production by Pseudomonas aeruginosa . J Bacteriol 195: 2971–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerz JK, Steimle A, Lange A, Bender A, Fehrenbacher B, Frick J‐S (2018) Outer membrane vesicles blebbing contributes to B. vulgatus mpk‐mediated immune response silencing. Gut Microbes 9: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalakshmi S, Sunayana MR, Saisree L, Reddy M (2014) yciM is an essential gene required for regulation of lipopolysaccharide synthesis in Escherichia coli . Mol Microbiol 91: 145–157 [DOI] [PubMed] [Google Scholar]
- Mamelli L, Spinelli S, Goemaere E, Vuillard L (2012) Putative osmotically inducible lipoprotein OsmE characterization by xray crystallography. 10.2210/pdb4DM5/pdb [DOI]
- Manning AJ, Kuehn MJ (2011) Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol 11: 258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maredia R, Devineni N, Lentz P, Dallo SF, Yu J, Guentzel N, Chambers J, Arulanandam B, Haskins WE, Weitao T (2012) Vesiculation from Pseudomonas aeruginosa under SOS. TheScientificWorldJournal 2012: 402919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn‐Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, Brandenburg K, Whiteley M (2008) Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol 69: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlakowska R, Skłodowska A, Nejbert K (2012) Bioweathering of Kupferschiefer black shale (Fore‐Sudetic Monocline, SW Poland) by indigenous bacteria: implication for dissolution and precipitation of minerals in deep underground mine. FEMS Microbiol Ecol 81: 99–110 [DOI] [PubMed] [Google Scholar]
- McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ (2006) Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol 188: 5385–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroom AJ, Kuehn MJ (2007) Release of outer membrane vesicles by Gram‐negative bacteria is a novel envelope stress response. Mol Microbiol 63: 545–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig WD, Koller A, Thanassi DG (2013) Production of outer membrane vesicles and outer membrane tubes by Francisella novicida . J Bacteriol 195: 1120–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KJ, Castelli ME, Vescovi EG, Feldman MF (2012) Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can be induced by activation of the Rcs phosphorelay system. J Bacteriol 194: 3241–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan HM, Rogers N, Wadle A, Hsu‐Kim H, Wiesner MR, Kuehn MJ, Hendren CO (2021a) Microbial vesicle‐mediated communication: convergence to understand interactions within and between domains of life. Environ Sci Process Impacts 23: 664–677 [DOI] [PubMed] [Google Scholar]
- McMillan HM, Zebell SG, Ristaino JB, Dong X, Kuehn MJ (2021b) Protective plant immune responses are elicited by bacterial outer membrane vesicles. Cell Rep 34: 108645–108666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehanny M, Koch M, Lehr C‐M, Fuhrmann G (2020) Streptococcal extracellular membrane vesicles are rapidly internalized by immune cells and alter their cytokine release. Front Immunol 11: 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt EA, Sixma TK, Kalk KH, van Zanten BAM, Hol WGJ (1993) 2.2 Angstroms crystal structure of E. coli heat‐labile enterotoxin (LT) with bound galactose. 10.2210/pdb1LTA/pdb [DOI]
- Merritt EA, Sixma TK, Kalk KH, van Zanten BAM, Hol WGJ (1994) Galactose‐binding site in Escherichia coli heat‐labile enterotoxin (LT) and cholera toxin (CT). Mol Microbiol 13: 745–753 [DOI] [PubMed] [Google Scholar]
- Mills LS, Soul XEEM, Doak DF (1993) The keystone‐species concept in ecology and conservation. Bioscience 43: 219–224 [Google Scholar]
- Modi N, Ganguly S, Bárcena‐Uribarri I, Benz R, van den Berg B, Kleinekathöfer U (2015) Structure, dynamics, and substrate specificity of the OprO Porin from Pseudomonas aeruginosa . Biophys J 109: 1429–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon DC, Choi CH, Lee JH, Choi C‐W, Kim H‐Y, Park JS, Kim SI, Lee JC (2012) Acinetobacter baumannii outer membrane protein a modulates the biogenesis of outer membrane vesicles. J Microbiol 50: 155–160 [DOI] [PubMed] [Google Scholar]
- Mosca A, Leclerc M, Hugot JP (2016) Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front Microbiol 7: 455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Park AJ, Hao Y, Brewer D, Lam JS, Khursigara CM (2014) Influence of O polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J Bacteriol 196: 1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagakubo T, Nomura N, Toyofuku M (2020) Cracking open bacterial membrane vesicles. Front Microbiol 10: 3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasukawa T, Sugimoto R, Uchiyama J, Takemura‐Uchiyama I, Murakami H, Fukuda K, Matsuzaki S, Sakaguchi M (2021) Purification of membrane vesicles from Gram‐positive bacteria using flow cytometry, after iodixanol density‐gradient ultracentrifugation. Res Microbiol 172: 103792 [DOI] [PubMed] [Google Scholar]
- Nazari M, Kurdi M, Heerklotz H (2012) Classifying surfactants with respect to their effect on lipid membrane order. Biophys J 102: 498–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DD, Pandian R, Kim DY, Ha SC, Yun KH, Kim KS, Kim JH, Kim KK (2013) Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa . 10.2210/pdb4NJQ/pdb [DOI] [PubMed]
- Nguyen DD, Pandian R, Kim D, Ha SC, Yoon H‐J, Kim KS, Yun KH, Kim J‐H, Kim KK (2014) Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa . Biochem Biophys Res Comm 447: 101–107 [DOI] [PubMed] [Google Scholar]
- Nho JS, Jun SH, Oh MH, Park TI, Choi CW, Kim SI, Choi CH, Lee JC (2015) Acinetobacter nosocomialis secretes outer membrane vesicles that induce epithelial cell death and host inflammatory responses. Microb Pathog 81: 39–45 [DOI] [PubMed] [Google Scholar]
- Nicholas A, Jeon H, Selasi GN, Na SH, Kwon HI, Kim YJ, Choi CW, Kim SI, Lee JC (2017) Clostridium difficile‐derived membrane vesicles induce the expression of pro‐inflammatory cytokine genes and cytotoxicity in colonic epithelial cells in vitro . Microb Pathog 107: 6–11 [DOI] [PubMed] [Google Scholar]
- Olaya‐Abril A, Prados‐Rosales R, McConnell MJ, Martín‐Peña R, González‐Reyes JA, Jiménez‐Munguía I, Gómez‐Gascón L, Fernández J, Luque‐García JL, García‐Lidón C et al (2014) Characterization of protective extracellular membrane‐derived vesicles produced by Streptococcus pneumoniae . J Proteomics 106: 46–60 [DOI] [PubMed] [Google Scholar]
- Orench‐Rivera N, Kuehn MJ (2016) Environmentally controlled bacterial vesicle‐mediated export. Cell Microbiol 18: 1525–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orench‐Rivera N, Kuehn MJ (2021) Differential packaging into outer membrane vesicles upon oxidative stress reveals a general mechanism for cargo selectivity. Front Microbiol 12: 1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine RT (1966) Food web complexity and species diversity. Am Nat 100: 65–75 [Google Scholar]
- Paine RT (1969) A note on trophic complexity and community stability. Am Nat 103: 91–93 [Google Scholar]
- Paine RT (1992) Food‐web analysis through field measurement of per capita interaction strength. Nature 355: 73–75 [Google Scholar]
- Park AJ, Murphy K, Surette MD, Bandoro C, Krieger JR, Taylor P, Khursigara CM (2015) Tracking the dynamic relationship between cellular systems and extracellular subproteomes in Pseudomonas aeruginosa biofilms. J Proteome Res 14: 4524–4537 [DOI] [PubMed] [Google Scholar]
- Park JS, Lee WC, Yeo KJ, Ryu K‐S, Kumarasiri M, Hesek D, Lee M, Mobashery S, Song JH, Kim SI et al (2012) Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram‐negative bacterial outer membrane. FASEB J 26: 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Vidakovics MLA, Jendholm J, Mörgelin M, Månsson A, Larsson C, Cardell L‐O, Riesbeck K (2010) B cell activation by outer membrane vesicles—a novel virulence mechanism. PLoS Pathog 6: e1000724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Cruz C, Carrión O, Delgado L, Martinez G, López‐Iglesias C, Mercade E (2013) New type of outer membrane vesicle produced by the Gram‐negative bacterium Shewanella vesiculosa M7T: implications for DNA content. Appl Environ Microbiol 79: 1874–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Cruz C, Delgado L, López‐Iglesias C, Mercade E (2015) Outer‐inner membrane vesicles naturally secreted by gram‐negative pathogenic bacteria. PLoS One 10: e0116896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, van Wees SCM, Bakker PAHM (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52: 347–375 [DOI] [PubMed] [Google Scholar]
- Prados‐Rosales R, Brown L, Casadevall A, Montalvo‐Quirós S, Luque‐Garcia JL (2014a) Isolation and identification of membrane vesicle‐associated proteins in Gram‐positive bacteria and mycobacteria. MethodsX 1: 124–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados‐Rosales R, Weinrick BC, Piqué DG, Jacobs WR, Casadevall A, Rodriguez GM (2014b) Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J Bacteriol 196: 1250–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano D, Klose KE (2000) Altered expression of the ToxR‐regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc Natl Acad Sci USA 97: 10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CR, Reynolds CM, Trent MS, Bishop RE (2007) Lipid A modification systems in gram‐negative bacteria. Annu Rev Biochem 76: 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff‐Nahoum S, Coyne MJ, Comstock LE (2014) An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol 24: 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VAK, Vanaja SK, Fitzgerald KA (2012) Regulation of inflammasome signaling. Nat Immunol 13: 333–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renelli M, Matias V, Lo RY, Beveridge TJ (2004) DNA‐containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150: 2161–2169 [DOI] [PubMed] [Google Scholar]
- Resch U, Tsatsaronis JA, le Rhun A, Stübiger G, Rohde M, Kasvandik S, Holzmeister S, Tinnefeld P, Wai SN, Charpentier E (2016) A two‐component regulatory system impacts extracellular membrane‐derived vesicle production in Group A Streptococcus. Mbio 7: e00207–e216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes‐Robles T, Dillard RS, Cairns LS, Silva‐Valenzuela CA, Housman M, Ali A, Wright ER, Camilli A (2018) Vibrio cholerae outer membrane vesicles inhibit bacteriophage infection. J Bacteriol 200: e00792–e817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera J, Cordero RJB, Nakouzi AS, Frases S, Nicola A, Casadevall A (2010) Bacillus anthracis produces membrane‐derived vesicles containing biologically active toxins. Proc Natl Acad Sci USA 107: 19002–19007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson T, Smith P, Alberts ER, Colussi‐Pelaez M, Schuster M (2020) Cooperation and cheating through a secreted aminopeptidase in the Pseudomonas aeruginosa RpoS response. Mbio 11: e03090‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden JA, Wells DH, Chomel BB, Kasten RW, Koehler JE (2012) Hemin binding protein C is found in outer membrane vesicles and protects Bartonella henselae against toxic concentrations of hemin. Infect Immun 80: 929–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez BV, Kuehn MJ (2020) Staphylococcus aureus secretes immunomodulatory RNA and DNA via membrane vesicles. Sci Rep 10: 18293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompikuntal PK, Thay B, Khan MK, Alanko J, Penttinen A‐M, Asikainen S, Wai SN, Oscarsson J (2012) Perinuclear localization of internalized outer membrane vesicles carrying active cytolethal distending toxin from Aggregatibacter actinomycetemcomitans . Infect Immun 80: 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvachúa D, Werner AZ, Pardo I, Michalska M, Black BA, Donohoe BS, Haugen SJ, Katahira R, Notonier S, Ramirez KJ et al (2020) Outer membrane vesicles catabolize lignin‐derived aromatic compounds in Pseudomonas putida KT2440. Proc Natl Acad Sci USA 117: 9302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath V, McCaig WD, Thanassi DG (2018) Amino acid deprivation and central carbon metabolism regulate the production of outer membrane vesicles and tubes by Francisella. Mol Microbiol 107: 523–541 [DOI] [PubMed] [Google Scholar]
- Schaar V, Nordström T, Mörgelin M, Riesbeck K (2011) Moraxella catarrhalis outer membrane vesicles carry β‐lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob Agents Chemother 55: 3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar V, Uddbäck I, Nordström T, Riesbeck K (2014) Group A streptococci are protected from amoxicillin‐mediated killing by vesicles containing β‐lactamase derived from Haemophilus influenzae . J Antimicrob Chemother 69: 117–120 [DOI] [PubMed] [Google Scholar]
- Schertzer JW, Whiteley M (2012) A bilayer‐couple model of bacterial outer membrane vesicle biogenesis. Mbio 3: e00297–e311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooling SR, Beveridge TJ (2006) Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol 188: 5945–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrempf H, Koebsch I, Walter S, Engelhardt H, Meschke H (2011) Extracellular streptomyces vesicles: amphorae for survival and defence. Microb Biotechnol 4: 286–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Kuehn MJ (2013) Synthetic effect between envelope stress and lack of outer membrane vesicle production in Escherichia coli . J Bacteriol 195: 4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Kuehn MJ (2015) Outer‐membrane vesicles from Gram‐negative bacteria: biogenesis and functions. Nat Rev Microbiol 13: 605–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Kulp A, Kuehn MJ (2014) Modulation of bacterial outer membrane vesicle production by envelope structure and content. BMC Microbiol 14: 324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Rodriguez DL, Kuehn MJ (2015) NlpI‐mediated modulation of outer membrane vesicle production through peptidoglycan dynamics in Escherichia coli . Microbiologyopen 4: 375–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Sullivan CJ, Kuehn MJ (2013) Envelope control of outer membrane vesicle production in Gram‐negative bacteria. Biochemistry 52: 3031–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfferth C, Tsuda K (2014) Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Front Plant Sci 5: 697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao PP, Comolli LR, Bernier‐Latmani R (2014) Membrane vesicles as a novel strategy for shedding encrusted cell surfaces. Minerals 4: 74–88 [Google Scholar]
- Shu W, Liu J, Ji H, Lu M (2000a) Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 å resolution. J Mol Biol 299: 1101–1112 [DOI] [PubMed] [Google Scholar]
- Shu W, Liu J, Ji H, Lu M (2000b) Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 angstrom resolution. 10.2210/pdb1EQ7/pdb [DOI] [PubMed]
- Sidhu VK, Vorhölter FJ, Niehaus K, Watt SA (2008) Analysis of outer membrane vesicle associated proteins isolated from the plant pathogenic bacterium Xanthomonas campestris pv. campestris. BMC Microbiol 8: 87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DW, Bangs EE (2009) Reintroduction of wolves to yellowstone national park: history, values and ecosystem restoration. Reintroduction of Top‐Order Predators 92–125
- Smith DW, Stahler DR, Becker MS (2008) Wolf recolonization of the Madison headwaters area in Yellowstone. In Terrestrial ecology, Garrott RA, White PJ, Watson FGR (eds), pp 283–303. Australia: Elsevier; [Google Scholar]
- Solé M, Scheibner F, Hoffmeister AK, Hartmann N, Hause G, Rother A, Jordan M, Lautier M, Arlat M, Büttner D (2015) Xanthomonas campestris pv. vesicatoria secretes proteases and Xylanases via the Xps type II secretion system and outer membrane vesicles. J Bacteriol 197: 2879–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J et al (2008) A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol 70: 100–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag I, Schwarz H, Hirota Y, Henning U (1978) Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol 136: 280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Stael S, Kmiecik P, Willems P, van der Kelen K, Coll NS, Teige M, van Breusegem F (2015) Plant innate immunity–sunny side up? Trends Plant Sci 20: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stentz R, Carvalho AL, Jones EJ, Carding SR (2018) Fantastic voyage: the journey of intestinal microbiota‐derived microvesicles through the body. Biochem Soc Trans 46: 1021–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stentz R, Horn N, Cross K, Salt L, Brearley C, Livermore DM, Carding SR (2015) Cephalosporinases associated with outer membrane vesicles released by Bacteroides spp. protect gut pathogens and commensals against β‐lactam antibiotics. J Antimicrob Chemother 70: 701–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana NK, Kasper DL (2014) Deciphering the tête‐à‐tête between the microbiota and the immune system. J Clin Invest 124: 4197–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana NK, Kasper DL (2017) Moving beyond microbiome‐wide associations to causal microbe identification. Nature 552: 244–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri N, Fällman M, Wai SN, Fahlgren A (2019) Accumulation of virulence‐associated proteins in Campylobacter jejuni outer membrane vesicles at human body temperature. J Proteomics 195: 33–40 [DOI] [PubMed] [Google Scholar]
- Tartaglia NR, Nicolas A, Rodovalho VDR, Luz BSRD, Briard‐Bion V, Krupova Z, Thierry A, Coste F, Burel A, Martin P et al (2020) Extracellular vesicles produced by human and animal Staphylococcus aureus strains share a highly conserved core proteome. Sci Rep 10: 8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y, Ichikawa S, Nakajima‐Kambe T, Uchiyama H, Nomura N (2010) Pseudomonas quinolone signal affects membrane vesicle production in not only gram‐negative but also gram‐positive bacteria. Microbes Environ 25: 120–125 [DOI] [PubMed] [Google Scholar]
- Tashiro Y, Inagaki A, Shimizu M, Ichikawa S, Takaya N, Nakajima‐Kambe T, Uchiyama H, Nomura N (2011) Characterization of phospholipids in membrane vesicles derived from Pseudomonas aeruginosa . Biosci Biotechnol Biochem 75: 605–607 [DOI] [PubMed] [Google Scholar]
- Tashiro Y, Sakai R, Toyofuku M, Sawada I, Nakajima‐Kambe T, Uchiyama H, Nomura N (2009) Outer membrane machinery and alginate synthesis regulators control membrane vesicle production in Pseudomonas aeruginosa . J Bacteriol 191: 7509–7519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayi L, Maku R, Patel HK, Sonti RV (2016) Action of multiple cell wall‐degrading enzymes is required for elicitation of innate immune responses during Xanthomonas oryzae pv. oryzae infection in rice. Mol Plant Microbe Interact 29: 599–608 [DOI] [PubMed] [Google Scholar]
- Thompson SS, Naidu YM, Pestka JJ (1985) Ultrastructural localization of an extracellular protease in Pseudomonas fragi by using the peroxidase‐antiperoxidase reaction. Appl Environ Microbiol 50: 1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuan Tong T, Mörgelin M, Forsgren A, Riesbeck K (2007) Haemophilus influenzae survival during complement‐mediated attacks is promoted by Moraxella catarrhalis outer membrane vesicles. J Infect Dis 195: 1661–1670 [DOI] [PubMed] [Google Scholar]
- Toyofuku M, Cárcamo‐Oyarce G, Yamamoto T, Eisenstein F, Hsiao C‐C, Kurosawa M, Gademann K, Pilhofer M, Nomura N, Eberl L (2017) Prophage‐triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis . Nat Commun 8: 481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M, Nomura N, Eberl L (2019) Types and origins of bacterial membrane vesicles. Nat Rev Microbiol 17: 13–24 [DOI] [PubMed] [Google Scholar]
- Toyofuku M, Zhou S, Sawada I, Takaya N, Uchiyama H, Nomura N (2014) Membrane vesicle formation is associated with pyocin production under denitrifying conditions in Pseudomonas aeruginosa PAO1. Environ Microbiol 16: 2927–2938 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Mine A, Bethke G, Igarashi D, Botanga CJ, Tsuda Y, Glazebrook J, Sato M, Katagiri F (2013) Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana . PLoS Genet 9: e1004015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulkens J, Vergauwen G, van Deun J, Geeurickx E, Dhondt B, Lippens L, de Scheerder M‐A, Miinalainen I, Rappu P, de Geest BG et al (2020b) Increased levels of systemic LPS‐positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut 69: 191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulkens J, de Wever O, Hendrix A (2020a) Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization. Nat Protoc 15: 40–67 [DOI] [PubMed] [Google Scholar]
- Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Cárcamo‐Oyarce G, Gloag ES, Shimoni R et al (2016) Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 7: 11220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Duranti S, Milani C, Lugli GA, van Sinderen D, Ventura M (2019) Bifidobacterium bifidum: a key member of the early human gut microbiota. Microorganisms 7: 544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Park JT (2007) An anhydro‐N‐acetylmuramyl‐L‐alanine amidase with broad specificity tethered to the outer membrane of Escherichia coli . J Bacteriol 189: 5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urashima A, Sanou A, Yen H, Tobe T (2017) Enterohaemorrhagic Escherichia coli produces outer membrane vesicles as an active defence system against antimicrobial peptide LL‐37. Cell Microbiol 19 [DOI] [PubMed] [Google Scholar]
- Valguarnera E, Scott NE, Azimzadeh P, Feldman MF (2018) Surface exposure and packing of lipoproteins into outer membrane vesicles are coupled processes in bacteroides. mSphere 3: e00559–e618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B (2012a) High pH structure of Pseudomonas putida OprB. 10.2210/pdb4GEY/pdb [DOI]
- Van den Berg B (2012b) Structural basis for outer membrane sugar uptake in pseudomonads*. J Biol Chem 287: 41044–41052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B (2014) Crystal structure of the OprO mutant protein F62Y/D114Y, 10.2210/pdb4RJX/pdb [DOI]
- Van der Heijden J, Reynolds LA, Deng W, Mills A, Scholz R, Imami K, Foster LJ, Duong F, Finlay BB, Miller SI et al (2016) Salmonella rapidly regulates membrane permeability to survive oxidative stress. Mbio 7: e01238–e1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD, Rathinam VAK (2016) Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase‐11 activation. Cell 165: 1106–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DMA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Sales JH, Lenk M, Bauer K, Brambilla A, Sommer A, Chen Y, Wenig M, Nayem S (2021) Systemic propagation of immunity in plants. New Phytol 229: 1234–1250 [DOI] [PubMed] [Google Scholar]
- Wang M, Nie Y, Wu X‐L (2021) Extracellular heme recycling and sharing across species by novel mycomembrane vesicles of a Gram‐positive bacterium. ISME J 15: 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Eagen WJ, Lee JC (2020) Orchestration of human macrophage NLRP3 inflammasome activation by Staphylococcus aureus extracellular vesicles. Proc Natl Acad Sci USA 117: 3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Thompson CD, Weidenmaier C, Lee JC (2018) Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nat Commun 9: 1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel AK, Liew J, Kwon T, Marcotte EM, Whiteley M (2013) Role of Pseudomonas aeruginosa peptidoglycan‐associated outer membrane proteins in vesicle formation. J Bacteriol 195: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Yaganza E‐S, Rioux D, Simard M, Arul J, Tweddell RJ (2004) Ultrastructural alterations of Erwinia carotovora subsp. atroseptica caused by treatment with aluminum chloride and sodium metabisulfite. Appl Environ Microbiol 70: 6800–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron S, Kolling GL, Simon L, Matthews KR (2000) Vesicle‐mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl Environ Microbiol 66: 4414–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa H, Osaki T, Woo T, Kurata S, Zaman C, Hojo F, Hanawa T, Kato S, Kamiya S (2011) Analysis of outer membrane vesicle protein involved in biofilm formation of Helicobacter pylori . Anaerobe 17: 388–390 [DOI] [PubMed] [Google Scholar]
- Yonezawa H, Osaki T, Fukutomi T, Hanawa T, Kurata S, Zaman C, Hojo F, Kamiya S (2017) Diversification of the AlpB outer membrane protein of Helicobacter pylori affects biofilm formation and cellular adhesion. J Bacteriol 199: e00729–e816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Jensen V, Feldmann I, Haussler S, Blankenfeldt W (2007) Structure of Pseudomonas quinolone signal response protein PqsE. 10.2210/pdb2Q0I/pdb [DOI] [PubMed]
- Yu S, Jensen V, Seeliger J, Feldmann I, Weber S, Schleicher E, Häussler S, Blankenfeldt W (2009) Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein. Biochemistry 48: 10298–10307 [DOI] [PubMed] [Google Scholar]
- Yun SH, Park EC, Lee S‐Y, Lee H, Choi C‐W, Yi Y‐S, Ro H‐J, Lee JC, Jun S, Kim H‐Y et al (2018) Antibiotic treatment modulates protein components of cytotoxic outer membrane vesicles of multidrug‐resistant clinical strain, Acinetobacter baumannii DU202. Clin Proteomics 15: 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn M, Basle A, van den Berg B (2014) Crystal structure of the N‐terminal beta‐barrel domain of Pseudomonas aeruginosa OprF. 10.2210/pdb4RLC/pdb [DOI]
- Zahn M, D'Agostino T, Eren E, Baslé A, Ceccarelli M, van den Berg B (2015) Small‐molecule transport by CarO, an abundant eight‐stranded β‐barrel outer membrane protein from Acinetobacter baumannii . J Mol Biol 427: 2329–2339 [DOI] [PubMed] [Google Scholar]
- Zakharzhevskaya NB, Tsvetkov VB, Vanyushkina AA, Varizhuk AM, Rakitina DV, Podgorsky VV, Vishnyakov IE, Kharlampieva DD, Manuvera VA, Lisitsyn FV et al (2017) Interaction of Bacteroides fragilis toxin with outer membrane vesicles reveals new mechanism of its secretion and delivery. Front Cell Infect Microbiol 7: 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamioudis C, Pieterse CMJ (2011) Modulation of host immunity by beneficial microbes. Mol Plant Microbe Interact 25: 139–150 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang Y, Song Z, Li R, Ruan H, Liu Q, Huang X (2020) sncRNAs packaged by Helicobacter pylori outer membrane vesicles attenuate IL‐8 secretion in human cells. Int J Med Microbiol 310: 151356 [DOI] [PubMed] [Google Scholar]
- Zhou L, Srisatjaluk R, Justus DE, Doyle RJ (1998) On the origin of membrane vesicles in gram‐negative bacteria. FEMS Microbiol Lett 163: 223–228 [DOI] [PubMed] [Google Scholar]
- Zingl FG, Kohl P, Cakar F, Leitner DR, Mitterer F, Bonnington KE, Rechberger GN, Kuehn MJ, Guan Z, Reidl J et al (2020) Outer membrane vesiculation facilitates surface exchange and in vivo adaptation of Vibrio cholerae . Cell Host Microbe 27: 225–237.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]


