Abstract
μ-Crystallin, encoded by the CRYM gene, binds the thyroid hormones, T3 and T4. Because T3 and T4 are potent regulators of metabolism and gene expression, and CRYM levels in human skeletal muscle can vary widely, we investigated the effects of overexpression of Crym. We generated transgenic mice, Crym tg, that expressed Crym protein specifically in skeletal muscle at levels 2.6–147.5 fold higher than in controls. Muscular functions, Ca2+ transients, contractile force, fatigue, running on treadmills or wheels, were not significantly altered, although T3 levels in tibialis anterior (TA) muscle were elevated ~190-fold and serum T4 was decreased 1.2-fold. Serum T3 and thyroid stimulating hormone (TSH) levels were unaffected. Crym transgenic mice studied in metabolic chambers showed a significant decrease in the respiratory exchange ratio (RER) corresponding to a 13.7% increase in fat utilization as an energy source compared to controls. Female but not male Crym tg mice gained weight more rapidly than controls when fed high fat or high simple carbohydrate diets. Although labeling for myosin heavy chains showed no fiber type differences in TA or soleus muscles, application of machine learning algorithms revealed small but significant morphological differences between Crym tg and control soleus fibers. RNA-seq and gene ontology enrichment analysis showed a significant shift towards genes associated with slower muscle function and its metabolic correlate, β-oxidation. Protein expression showed a similar shift, though with little overlap. Our study shows that μ-crystallin plays an important role in determining substrate utilization in mammalian muscle and that high levels of μ-crystallin are associated with a shift toward greater fat metabolism.
Keywords: Glycolysis, β-oxidation, RER, RNA-seq, Proteomics, Thyroid hormone
Graphical abstract
Highlights
-
•
μ-Crystallin (Crym) is expressed specifically in transgenic skeletal muscle at highly elevated levels.
-
•
T3 is increased ~190 fold in the Tibialis anterior muscle of Crym tg mice.
-
•
Small but significant changes in gene and protein expression in tg muscle towards a slow twitch, oxidative phenotype.
-
•
Metabolic studies show that Crym tg mice increase their use of fat as an energy source.
-
•
Female Crym tg mice gain weight faster on high fat or simple carbohydrate diets than controls.
1. Introduction
μ-Crystallin was first discovered in 1957 and called cellular thyroxine-binding protein (Tata, 1958). It was subsequently shown to bind both T3 and T4 in an NADPH-dependent manner (Hashizume et al., 1989; Tata, 1958), and to have ketimine reductase activity (Hallen et al., 2011). In 1991, μ-crystallin was characterized as a 38,000 Da polypeptide that readily forms dimers (Kobayashi et al., 1991) and shares structural homology with bacterial ornithine cyclodeaminase (Kim et al., 1992). Its structure has since been solved to 1.75 Å (Borel et al., 2014).
CRYM is highly expressed in the cerebral cortex, heart, skeletal muscle, and kidney (Kim et al., 1992), and has been linked to Deafness, Autosomal Dominant 40 (Abe et al., 2003). Importantly, μ-crystallin binds T3 with a KD = 0.3 nM (Beslin et al., 1995) while T3 binds thyroid hormone receptors (TR) α and β with a KD = 0.06 nM; T4 binds TRs at a KD = 2 nM (Sandler et al., 2004). μ−Crystallin's activity as a ketimine reductase is inhibited by T3 and T4 at subnanomolar levels, KI = 0.60 nM and KI = 0.75 nM, respectively (Hallen et al., 2015). T3 and T4 are strong regulators of metabolism and thermogenic homeostasis (Larsen et al., 1981). Because of this, proteins that interact with and regulate thyroid hormones may have a broad influence on integrative functions like gene expression and metabolism.
Consistent with its possible role in regulating metabolism associated with thyroid hormones, Crym knockout mice (Crym KO) fed a high fat diet (HFD) show increased fat mass by computer tomography and increased body weight compared to control mice (Ohkubo et al., 2019). Furthermore, Crym KO mice show significant hypertrophy of glycolytic fast twitch type IIb muscle fibers (Seko et al., 2015).
Here we address the effects of high levels of Crym expression in mammalian muscle. We initially observed high levels of μ-crystallin in several muscle biopsies of patients with facioscapulohumeral muscular dystrophy (FSHD) (Reed et al., 2007), a muscle wasting disease in which patients progressively lose muscle. At the time, the cause of FSHD was unknown and we speculated that CRYM might play a role in the disease. Later studies showed that mRNA and protein both varied widely in expression in both healthy and diseased muscle (Klooster et al., 2009). It is now widely accepted that the primary pathogen in FSHD is DUX4 (Tawil et al., 2014), a transcription factor (Dixit et al., 2007) that promotes CRYM expression (Vanderplanck et al., 2011). Thus, increases in CRYM may perturb muscle metabolism and contribute to pathology.
To examine the differences that result from the high level of expression of Crym in skeletal muscle, we developed a transgenic mouse (Crym tg) that overexpresses μ-crystallin under the control of the human skeletal actin promoter (ACTA1) and the human slow troponin I enhancer (TNNI1), to drive skeletal muscle-specific expression (Ebashi et al., 1969; Gunning et al., 1983). Here we explore the effects of overexpression of μ-crystallin on the structure and function of muscle.
2. Materials and methods
2.1. Creation of the Crym tg mouse
Mouse Crym was cloned downstream of the human skeletal actin promoter (ACTA1) and human slow troponin I enhancer element (TNNI1), (kindly provided by Dr. J. Molkentin, Cincinnati Children's Hospital Medical Center), to limit expression to differentiated skeletal muscle. The expression construct was linearized and injected into C57BL/6J mouse embryos at the Genome Modification Facility, Harvard University (Cambridge, MA). Mice received from the Harvard facility were genotyped by PCR and then rederived by artificial insemination of C57BL/6J mice, due to pinworm infestation. This was performed by Veterinary Resources, University of Maryland School of Medicine. The rederived offspring were bred; all mice were tail snipped at weaning. Genomic DNA was purified from mouse tail snips using the Nucleon Genomic DNA extraction kit (Tepnel Life Sciences, Scotland). PCR of genomic DNA was used to identify mice positive for the transgene, with the following primers: forward: TGGCCACGCGTCGACTAGTACG; reverse: AATTCGTACTAGTCGACGCGTGGCC.
As it was initially difficult to differentiate between heterozygotes and homozygotes with PCR methods, qPCR was done on several of the Crym-positive genomic DNA samples. Mice with higher levels of Crym were then bred to controls and the F1 and F2 offspring were crossed and screened to generate probable homozygotes. Mice were identified as tg/tg homozygotes if after crossing with controls they gave at least 12 Crym-positive offspring and no Crym-negative offspring by PCR. Homozygotes were then bred together to establish the line of Crym tg/tg mice.
All histologic and physiologic experiments used approximately three-month-old, male control and Crym tg mice that were anesthetized under isoflurane (2.5%). Euthanasia was by cervical dislocation under anesthesia.
All procedures were approved by the Institutional Animal Care and Use Committee, University of Maryland School of Medicine.
2.1.1. Back-crossing and genotyping
Crym tg mice were backcrossed with control C57BL/6J mice. Non-littermate F1 heterozygotes were then bred together to generate F2 mice. Tail snips of Crym tg, C57BL/6J, F1 heterozygotes, and F2 mice were taken at 6 weeks of age or older. Genomic DNA was extracted from the tail snips by following the manufacturers protocol from the PureLink Genomic DNA Mini Kit (K182001; Invitrogen) modified only by substituting DirectPCR Lysis Reagent (Tail) (102-T; Viagen Biotech, Los Angeles, CA) in place of PureLink Genomic Digestion Buffer. Crym tg, F1 heterozygotes, and C57BL/6J mice were genotyped using a multiplexed probe-based assay from IDT for Crym and Tert. The Crym assay was designed such that it would produce the same amplicon from the native Crym gene as well as the inserted transgene. A synthetic Crym amplicon was used as an interplate control and Tert was used as the control for copy number. RT-qPCR was used to determine copy number and average normalized Cq for each mouse. The amplification protocol followed the manufacturer's instructions (IDT). Mice were genotyped to one of the three genotypes when the calculated copy number and average normalized Cq was closest to a known genotype (i.e. Crym tg, C57BL/6J, or F1 heterozygotes).
2.1.2. Sequencing the transgene insertion site
Genomic DNA obtained from Crym tg mice was digested with EcoRI. We designed and had synthesized a double stranded short oligonucleotide with overhanging sequences corresponding to an EcoRI digestion that we called Crym-Adaptor. Crym-Adaptor was ligated to the digested genomic DNA. The resulting ligation fragments were amplified using a forward primer in the multiple cloning site of the transgene and a reverse primer in the Crym-Adaptor sequence. Nested primers were used to specifically amplify the sequence. The products of this last reaction were purified by gel purification and sequenced from both ends. The sequence was then evaluated with NCBI BLASTn.
2.2. Staining of longitudinal and cross sections for μ-crystallin
Mice were perfusion fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS). TA and soleus muscles were collected, snap frozen in a liquid nitrogen slush, mounted in O.C.T. (Fisher Healthcare, Hampton, NH) and sectioned at 10–20 μm with a Reichert Jung cryostat (Leica, Buffalo Grove, IL). Sections were stained using the Mouse-On-Mouse (M.O.M.) Basic Kit (BMK-2202; Vector labs, Burlingame, CA). Sections were incubated for at least 1 h at room temperature in M.O.M. blocking reagent followed by a 10 min incubation in M.O.M. diluent. Sections were incubated overnight at 4 °C in mouse monoclonal anti-μ-crystallin antibody (GTX84654; GeneTex Inc., Irvine, CA) diluted 1:100. Some sections were also incubated with rabbit anti-desmin (PA5-16705; Thermo Fisher, Waltham, MA), diluted 1:100. Sections were washed in PBS and stained for 1 h at room temperature with Alexa Fluor 568 goat anti-mouse secondary antibody (A11031) and Alexa Fluor 488 goat anti-rabbit (A32731), both from Alexa Molecular Probes (Invitrogen, Carlsbad, CA), diluted 1:200. All antibodies were made up in M.O.M. diluent. Samples were washed with PBS, mounted in Vectashield + DAPI (H-1500; Vector Laboratory), and imaged with a Nikon W-1 spinning disc confocal microscope (Nikon USA, Melville, NY). We used identical laser power and exposure settings to compare cross sections of TA muscle, of soleus muscle, and of longitudinal sections of TA muscle, but we adjusted them for each set of comparisons to optimize image clarity.
2.2.1. Fiber type staining
We used a slightly modified version of Kammoun et al.’s fiber typing protocol (Kammoun et al., 2014). Primary murine monoclonal antibodies specific for the myosin isoforms Myh7 (type I; BA-D5, isotype IgG2b), Myh2 (type IIa; SC-71, isotype IgG1), and Myh4 (type IIb; BF-F3, isotype IgM) were from the Developmental Studies Hybridoma Bank, (Iowa City, IA). Alexa Fluor 647 conjugated wheat germ agglutinin (WGA) was used to stain the myofiber surface (W32466, Thermo Fisher). The 4 reagents were used on 10 μm thick cross sections of snap frozen soleus and TA muscles (n = 5). The following secondary antibodies were used: goat anti-mouse IgG2b Cross-Adsorbed Secondary Antibody Alexa Fluor 568 (A-21144, Thermo Fisher), goat anti-mouse IgG1 Cross-Adsorbed Secondary Antibody Alexa Fluor 488 (A-21121, Thermo Fisher), and goat anti-mouse IgM mu chain Alexa Fluor 405 (ab175662, Abcam). Slides were imaged on a Nikon W-1 spinning disc microscope. Laser power and exposure were identical for comparisons of sections of control and tg TA muscles, and for soleus muscles, but were different between these tissues. We used 5 Crym tg and 4 control mice to study soleus fiber type. Four stacked images were taken at approximately 2 μm intervals and stacked to generate a single image, which were flattened using Aguet et al.’s Model-Based 2.5-D Deconvolution for Extended Depth of Field algorithm (Aguet et al., 2008). We determined that a η0 = 0.2 and a η1 = 1.3 were optimal in our images to generate an in-focus image of the flattened z stacks. For soleus muscle sections we used Myosoft (Encarnacion-Rivera et al., 2020), a Fiji (Schindelin et al., 2012) macro, to categorize fibers by their myosin heavy chain composition and measure several traits (count, area, perimeter, circularity, minimal Feret's diameter, roundness, and solidity). Statistical analysis used Real Statistics Resource Pack software (Release 6.8) (Zaiontz, 2020) as a plugin in Microsoft Excel with α = 0.05. Normality of the 7 measured metrics for each fiber type (I, IIa, IIb, IIx, I/IIa, I/IIb, IIa/IIb, and I/IIa/IIb) were determined with the Shapiro-Wilks and d'Agostino-Pearson tests, while scedasticity was determined with the mean, median, and trimmed method in Levene's test. If a dataset failed any individual test for normality or homoscedasticity it was considered not normal and heteroscedastic. Measurements that were both normal and homoscedastic were tested for significance using Student's t-test, while measurements that were normal but heteroscedastic were tested for significance using either Welch's t-test or the Brown-Forsythe test. The Kruskal-Wallis test for significance was used on measurements that failed the tests for normality but were homoscedastic. Either the Yuen-Welch or the Mann-Whitney U test were used on measurements that failed tests for both normality and homoscedasticity.
2.2.2. Fat staining
Cryosections of 5 snap frozen soleus and TA muscles from control and Crym tg mice were stained with BODIPY (493/503) according to Spangenburg et al. (2011) and imaged with a Nikon spinning disk microscope (see above). Laser power and exposure settings were maintained for each section regardless of mouse strain, but these were altered between TA and soleus tissues. Accordingly, look up table values were also different from TA to soleus samples but the same within a tissue regardless of mouse strain.
2.3. μ-Crystallin staining of isolated FDB fibers
FDB muscles were harvested bilaterally and digested in Dulbecco's modified Eagle's medium with 4 mg/mL type II collagenase (Gibco, Thermo Fisher Scientific, Waltham, MA) for 3 h at 37 °C. Tissue was transferred to FDB medium (Dulbecco's modified Eagle's medium with 2% BSA, 1 μL/mL gentamicin, and 1 μL/mL fungizone). Single myofibers were mechanically separated by trituration and allowed to incubate overnight. Isolated fibers were plated down on coverslips coated with Geltrex (A1413201; Thermo Fisher Scientific) for 2 h. Coverslips were fixed in 2% paraformaldehyde at room temperature for 15 min. They were then permeabilized with 0.25% TritonX-100 in PBS for 10 min and stained with antibody to μ-crystallin (H00001428-M03; Abnova, Taiwan), diluted 1:100, followed by Alexa Fluor 488, goat anti-mouse secondary antibody (A11029; Alexa Molecular Probes, Invitrogen), diluted 1:200, with the M.O.M. kit, as described above. Each incubation was for 1 h at room temperature. Samples were imaged on a Zeiss 510 Duo microscope (Carl Zeiss, Thornwood, NY).
2.3.1. CNFs and minimal Feret's diameter of TA cross sections
Cross sections of TA muscles were used to measure centrally nucleated fibers (CNFs) and minimal Feret's diameter (Briguet et al., 2004). Sections were stained as above but with rabbit anti-dystrophin (PAS-16734; Thermo Fisher Scientific), mounted in Vectashield + DAPI as above and imaged by confocal microscopy with a Zeiss 510 Duo microscope (Carl Zeiss). DAPI labeling was evaluated with ImageJ (NIH, Bethesda, MD), for determination of centrally nucleated fibers. Measurements of minimal Feret's diameter were obtained with Zeiss LSM Image Browser (Carl Zeiss). A total of 312 myofibers from 5 Crym tg mice and 722 fibers from 5 control mice were analyzed.
2.4. Measurements of contractile force
Nerve-evoked contractile function of gastrocnemius, extensor digitorum longus, or soleus muscles in vivo was evaluated as described (Burks et al., 2011; Olojo et al., 2011). In brief, isofluorane-anesthetized mice were placed supine on a warming pad (37 °C) of an Aurora 1300A system with knee position fixed and paw secured to the foot-plate of the 300C-FP. Percutaneous nerve stimulation was with brief (100 μsec) pulses with current adjusted to achieve maximal isometric force. The force vs frequency relationship was determined with 250 msec trains of pulses between 1 and 150 Hz and normalized to muscle mass or cross-sectional area (CSA). Fatigability was determined by delivering tetanic trains (250 ms at 80 Hz) every 2 s for 10 min for soleus or for 5 min for gastrocnemius. Data were analyzed with DMA-HT analysis software (Aurora Scientific, Ontario, Canada) and evaluated for statistical difference via the Holm-Šídák test with GraphPad Prism version 8.2.0 for Mac (GraphPad Software, La Jolla, CA).
2.5. Treadmill and ad libidum running
Mice were conditioned to the treadmill over 3 days, as follows. Mice were placed in the treadmill with no belt movement for 10 min. The next day they were made to walk at 5 m/min for 10 min; this was increased to 10 m/min the following day. For testing, mice were placed in the treadmill at a 7° incline. The speed was set at 10 m/min and was increased by 1.5 m/min every 2 min. Mice were considered exhausted when they were unable to run for 30 s consecutively. Each mouse was run 3 times with a minimum of 2 d rest between each run.
Additional Crym tg and control mice were housed in cages equipped with running wheels for 7 d. The number of times the wheel spun per minute was recorded every 6 min and analyzed to determine total distance run in different time periods, day and night.
2.6. Protein extraction
Tissues of interest (gastrocnemius, TA, soleus, diaphragm, heart, kidney, cerebral cortex and liver) were collected from 3-month old Crym tg and control mice, snap frozen in liquid nitrogen and stored at −80 °C. Protein was extracted in RIPA Buffer (R0278-50 ML, Sigma-Aldrich), prepared with cOmplete Mini, EDTA-free protease inhibitor tablets (11836170001, Sigma-Aldrich; 10 mL of RIPA buffer to every protease inhibitor tablet). Tissue was weighed and 100 μL per 10 mg of tissue of RIPA/protease inhibitor solution was added along with two 5 mm steel beads (69989; Qiagen, Hilden, Germany). Samples were placed in a TissueLyser LT (Qiagen) at 50 oscillations/sec for 3 min, briefly vortexed, put on ice for 2 min, followed by a second round of 50 oscillations/sec for 3 min in the TissueLyser. Samples were sonicated for 10 s and subjected to centrifugation at 12,470 g for 30 min at 4 °C. The supernatant was transferred to a new microfuge tube and protein concentration was determined with Bio-Rad's Protein Assay Dye Reagent Concentrate (Bradford Assay) (5000006, Bio-Rad).
2.6.1. Immunoblotting protocols
Traditional Western blot: Muscle tissue was combined with an equal volume of sample homogenate and Laemmli sample buffer (Laemmli, 1970), containing 5% 2-mercaptoethanol, then further diluted in sample buffer to a final concentration of 1 mg/mL. Samples of 10 μg of Crym tg TA homogenate and 40 μg of control TA homogenate were loaded onto 4–12% Bis-Tris gels (NuPAGE NP0321PK2, Thermo Fisher), electrophoresed for 1 h at 170 V, and transferred to nitrocellulose for 2 h at 22 V. Blots were blocked for 3 h in blocking solution (3% milk in TBS containing 0.1% Tween-20 and 10 mM azide). Monoclonal mouse anti-μ-crystallin (SC-376687, Santa Cruz Biotechnology, Inc., Dallas, TX) and monoclonal rabbit anti-lysyl-tRNA synthetase antibodies (#129080, Abcam, Cambridge, United Kingdom), used as a loading control, were diluted 1:1000 in blocking buffer and incubated overnight at room temperature. Lysyl-tRNA synthetase, encoded by the Kars gene, was used as the protein loading control because the expression of its mRNA showed minimal variance among mice and across genotypes, unlike many other common control proteins (Supplemental Document 1). (At the time we did these studies, we did not have proteomic data.) After extensive washing, the membranes were incubated for 2 h with HRP conjugated goat anti-rabbit (3-035-144, Jackson ImmunoResearch, West Grove, PA) or goat anti-mouse (47-035-146, Jackson ImmunoResearch) secondary antibody diluted 1:10,000 in blocking solution. After extensive washing, chemiluminescence was visualized with the Super Signal West Femto Maximum Sensitivity Kit (#34095, Thermo Fisher) and imaged with a BioRad ChemiDoc XRS and image lab software (#1708265, BioRad).
Capillary-based Western blot (Wes): Wes analysis was performed according to the manufacturer's instructions for the 12–230 kDa protein separation module (#SM-W004; Protein Simple, San Jose, CA) to provide an automated alternative to the traditional Western Blot. Instrument settings were as follows; stacking and separation time, 30 min; separation voltage, 375 V; time for antibody blocking, 30 min; incubation with primary antibody time, 60 min; incubation with secondary antibody time, 30 min; luminol/peroxide chemiluminescence detection time, 15 min (HDR exposure). Protein extracted from whole tissue lysates (gastrocnemius, TA, soleus, diaphragm, heart, kidney, cerebral cortex, quadriceps, liver) from 3 male Crym tg and 3 control were prepared with 2x Laemmli Sample Buffer (#1610737; Bio-Rad, Hercules, CA) to generate an overall sample protein concentration of 1.0 mg/mL for each . The primary antibody to μ-crystallin (see above) was diluted 1:25 in Protein Simple's Antibody Diluent 2. A primary antibody to a control protein, aconitase-2 (ab129069; Abcam, United Kingdom), was used at 1:200. We chose aconitase-2 as the protein loading control because this protein is abundantly expressed in many tissues, showed very little variability in expression in mice of the same genotype and between Crym tg and controls, as assayed by whole proteome LC.MS/MS (data not shown), and because the molecular mass of this protein is distinct from that of μ-crystallin. The primary and control antibodies were multiplexed. Samples were run as technical duplicates. Compass software (Compass for SW 4.0 Mac Beta; Protein Simple) was used to visualize the electropherograms and to analyze peaks of interest for area under the curve (AUC). The peak signal-to-noise ratio was set at ≥ 10 automatically by the software. Due to slight capillary-to-capillary variations in molecular weight readout, identified peaks at a molecular weight of interest were allowed a 10% range in molecular weight (as automatically set by the Wes system). AUC was analyzed with individual t-tests per tissue following the Benjamini, Krieger and Yekutieli FDR approach at Q = 1%.
2.6.2. RNA and cDNA preparation
Tissues from 3-month old Crym tg and control mice were removed, snap frozen in liquid nitrogen and stored at −80 °C.
RNA was extracted using the following protocol. Tissue was placed in a 2 mL tube with 1 mL of TRIzol Reagent (15596026; Thermo Fisher Scientific) and two 5 mm steel beads (69989; Qiagen, Hilden, Germany), loaded into a TissueLyser LT (Qiagen) and run at 50 oscillations/sec for 2 min. Samples were checked for complete homogenization after 2 min and, if incompletely homogenized, run again in 30 s cycles with checks for complete homogenization after each cycle. Samples were vortexed briefly, inverted to mix at room temperature for 5 min, and subjected to centrifugation at 12,000 g for 10 min at 4 °C. Supernatant was removed into a new 1.5 mL RNase-free tube and 200 μL of chloroform was added. After vortexing for 15 s, the sample remained at room temperature for 3 min. After a second centrifugation, at 12,000 g for 15 min at 4 °C, the top (clear) layer was removed into a new 1.5 mL RNase free tube and 500 μL of ice-cold isopropanol was added. Tubes were vortexed and inverted several times to mix, and then left at room temperature for 10 min. Centrifugation at 12,000 g for 10 min at 4 °C generated a white pellet of RNA, which was washed with 1 mL of ice-cold 75% ethanol and collected again by centrifugation (7500 g for 5 min at 4 °C). After drying at room temperature, the pellet was dissolved in 25–35 μL of RNase-, DNase-free water at 60 °C for 10 min. RNA samples were stored at −80 °C until use.
cDNA was generated with the QuantiTect Reverse Transcription Kit (205313; Qiagen). RT-qPCR was performed on a CFX Connect thermal cycler (Bio-Rad) using the cDNA and PrimeTime Gene Expression Master Mix (1055772; IDT, Coralville, IA) in 20 μL reaction volumes in a BioRad CFX Connect thermal cycler following the manufacturer's protocol for the PrimeTime qPCR Probe Assays (IDT) except for the extension of the number of amplification cycles to 60. Primers are listed below.
IDT PrimeTime RT-qPCR Probe-Based Assay.
Crym: Forward: 5′-GAGATGTTCGGGTCTGTTCAT-3'.
Probe: 5'-/FAM/TCATCACAG/ZEN/TCACCATGGCAACAGA/3IABkFQ/-3'.
Reverse: 5′-GGCTTTACCCATTCACCAAATAA-3'.
Nom1: Forward: 5′-TAAACCCAGAGTTCACTTCCTAC-3'.
Probe: 5'-/5HEX/TCGTCTTCA/ZEN/GTTTCCATCAACAGTGCA/3IABkFQ/-3'.
Reverse: 5′-CCTTCTCGCAACATTCCCA-3'.
Tert: Forward: 5′-CTCCTTTCCTCTAGGGCTATCT-3'.
Probe: 5'-/HEX/TCTCTGTCT/ZEN/CCCTTACCCACAGCT/3IABkFQ/-3'.
Reverse: 5′-AGTGCTGACATCTCATTCCTTC-3'.
Relative tissue specific expression of Crym, standard deviations, and p-values were calculated according to Taylor et al. (2019). Nom1 was used as the control gene to measure Crym expression by qPCR.
2.7. Proteomic comparison of skeletal muscle of Crym tg and control mice
Skeletal muscle samples were harvested from six 3-month-old, male, Crym tg and control littermate mice. All of the mice were perfused with cold PBS prior to muscle collection except for one of the 6 control samples. Samples, comprising half of the left TA muscle, divided longitudinally, were homogenized by bead beating (see above). Sample preparation for proteomics and nano ultra-performance liquid chromatography-tandem mass spectrometry were performed as described (Kim et al., 2019). Spectra were searched against a UniProt mouse reference proteome using Sequest HT algorithm described by Eng et al. (2008) and MS Amanda algorithm developed by Dorfer et al. (2014). Search configuration, false discovery control and quantitation were as described (Kim et al., 2019).
2.8. RNA-seq
RNA extraction and RNA-seq was performed by Genewiz (South Plainfield, NJ) on 20 mg samples of TA muscles from 3-month-old mice from 3 Crym tg littermate and 3 control littermate mice. Genewiz generated FASTQ files of the raw RNA-seq data, after removing transcripts that mapped to introns and transcripts that mapped to multiple genes. p-Values for comparisons of genes expressed in Crym tg and control mice were also from Genewiz.
2.9. Ca2+ transients
The amplitude of Ca2+ transients was recorded as described (Lukyanenko et al., 2017). In brief, isolated FDB fibers were loaded with Rhod-2AM (Thermo Fisher Scientific, Waltham, MA). Trains of voltage-induced Ca2+ transients were induced by field stimulation (A-M Systems, Carlsborg, WA). Rhod-2 was visualized with the Zeiss 510 Duo confocal microscope (Carl Zeiss). Image J 1.31v (NIH) was used for image analysis. The values reported were measured as the difference between maximal fluorescence intensity (Fmax) and background fluorescence (Fo), normalized to Fo. Quantitative data are shown as mean ± SE. A Student's t-test was used to compare the data with p < 0.05 considered statistically significant.
2.9.1. Metabolic chambers
Data were from two different cohorts of 4 Crym tg and 4 C57BL/6J control mice housed in an Oxymax/CLAMS (Columbus Instruments, Columbus, OH) open circuit indirect calorimeter, in two separate experiments. Measurements for RER, Accumulated CO2, Delta CO2, CO2 Out, Heat, Delta O2, Feed Weight, Volume CO2, Accumulated O2, Feed Accumulation, Z Total, Volume O2, O2 Out, X Total, X Ambulatory, Flow, O2 In, and CO2 In were taken directly or calculated every 18 min for approximately 92 h. Z Total and X Total refer to the total number of times an animal disrupted the infrared (IR) beam in the Z and X axis, respectively, while X Ambulatory refers to the number of times an animal disrupted at least two consecutive IR beams. Complete measurements were compiled and 24 h worth of measurements were analyzed, starting at the first dark cycle after a 24 h period of acclimation. Values were normalized to total body weight and analyzed separately for light and dark periods. Outliers were identified with the ROUT method (Motulsky and Brown, 2006) at Q = 1%, with GraphPad Prism version 6.0e for Mac (GraphPad Software, La Jolla, CA). Averages were then calculated for all measurements except for Feed Weight, Feed Accumulation, Z Total, X Total, and X Ambulatory, for which the sums were determined. Each average or sum for each mouse was tested for significance, grouped by genotype, with an unpaired, parametric t-test with Welch's Correction.
2.9.2. Diet studies
Crym tg and control male and female mice at approximately 14 weeks old were placed on one of five diets (Supplemental Document 5). The number of mice on a particular diet, assayed over time, varied from 5 to 16, due to the occasional death of a mouse, unrelated to the experiment, or to our inability to weigh them on a particular date. The diets were “normal” diet (2018SX, Envigo Madison, WI), high fat diet (D12492), low fat diet (D12450J), high simple carbohydrate diet (D15040205), or high complex carbohydrate diet (D12450K), all from Research Diets Inc. (New Brunswick, NJ) unless otherwise specified. Mouse weight (anesthetized with 2.5% isoflurane) and food weight were recorded every Monday, Wednesday, and Friday for approximately 60 d. Water was provided ad libitum.
Mouse weights were normalized to their starting body weight. Using the Real Statistics Resource Pack software (Release 6.8) (Zaiontz, 2020) to run the Mauchly and John-Nagao-Sugiura tests, we found that all of the groups violated at least one test for sphericity (α = 0.05). We therefore used GraphPad Prism 8.2.0 for Mac to apply the Greenhouse and Geisser epsilon correction factor to either a two-way repeated measures (RM) ANOVA or a mixed-effects model, specifically a compound symmetry covariance matrix fit with Restricted Maximum Likelihood (REML) if data were missing for certain dates (α = 0.05). When we observed significance with the two-way RM ANOVA or mixed-effects model (REML), we performed multiple post-hoc unpaired, two-tailed t tests assuming heteroscedasticity at α = 0.05 to determine which data points showed significant differences between Crym tg and control mice.
2.9.3. T3, T4, TSH levels
Homogenates of TA muscles and serum from four Crym tg and four control mice were sent to the clinical diagnostic service at Vanderbilt University (Nashville, TN) for analysis of T3, T4 and TSH. T3, T4 and TSH levels were analyzed by a two-tailed t-test with Welch's correction. The thyroid hormone mass (in ng/mL) was divided by total protein (in ng/mL) and used to determine the percent of total thyroid hormone per total protein in a milliliter of serum or homogenized muscle. The negative log base 10 of the percent of total protein for each value was determined and used to calculate the average and standard deviation.
2.9.4. Materials
Unless otherwise stated, all biologics were from Sigma-Aldrich and all salts were from Thermo Fisher.
3. Results
We created a transgenic mouse in which murine Crym is expressed at high levels in skeletal muscle, comparable to those seen in some human muscle biopsies, and assessed the effects an abundance of μ-crystallin would have on muscle structure and function, and on metabolism. The Crym transgene was encoded in a plasmid under the control of the human slow troponin I enhancer and the human skeletal actin promoter with the polyadenylation sequence of bovine growth hormone mRNA (Supplemental Fig. 1A). After oocyte injection, the plasmid inserted randomly into intron 12 of the Cntn6 gene on murine chromosome 6 (Supplemental Fig. 1B), as determined by sequencing (Supplemental Fig. 1C). The mice were bred to homozygosity and genotyping of the mice showed that there was a single insertion in the genome resulting in two copies of the Crym transgene in the diploid mouse genome (Supplemental Fig. 2). Back-crossing Crym tg with control mice yielded the expected Mendelian inheritance pattern (Supplementary Table 1). Cntn6 was not significantly differentially expressed in control and Crym tg mice and showed little to no expression as assayed by RNA-seq in both. This indicates that the gene was not disrupted by the transgene's insertion.
3.1. RT-qPCR, western blot, and immunofluorescence
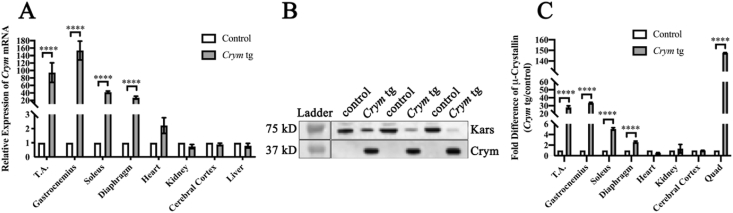
RT-qPCR of several skeletal muscles showed moderate to large increases in Crym mRNA in male Crym tg mice compared to male controls (Fig. 1A). Because many canonical control genes were significantly differentially expressed (DEG), we compared RNA-seq and LC.MS/MS data (Supplemental Document 1 and 3 respectively) to identify gene products with the smallest coefficient of variation. We used RefFinder (Xie et al., 2012) to evaluate the most promising control genes among biological and technical replicates of various tissues (data not shown). We found that Nom1 was optimal, as it showed the most stability. Using Nom1 mRNA as a standard, we observed large increases in the Crym mRNA levels in skeletal muscles of the Crym tg mice, compared both to controls and to heart muscle and non-muscle tissues. Notably, different skeletal muscles in the tg mice differed significantly in their relative expression of Crym. We found no significant differences in Crym mRNA levels in non-skeletal muscle tissues in tg and controls (Fig. 1A).
Fig. 1.
Increased Crym mRNA and μ-crystallin protein levels in Crym tg vs. control mice. A. Crym expression in several skeletal muscles and other tissues was analyzed from 3 Crym tg and 3 control mice. All analyses were in technical triplicate, except for soleus and diaphragm. B. Western blot of μ-crystallin in Crym tg and control TA muscle extracts. Control mice had 40 μg of TA homogenate per lane, and Crym tg lanes had 10 μg of TA homogenates per lane. We were unable to detect μ-crystallin reliably in controls in these blots. C. μ-Crystallin was quantitated with Protein Simple's Wes instrument. Areas under the curve, generated from electropherograms of μ-crystallin expression, were normalized to aconitase-2 expression (n = 3, in technical duplicate) in several muscles and other tissues. Multiple individual t-tests using the Benjamini, Krieger and Yekutieli FDR approach at Q = 1% determined statistical significance (∗∗∗∗ = p < 0.0001). The results show that Crym tg skeletal muscle express high levels of Crym mRNA and protein compared to controls and to other tissues analyzed.
Immunoblotting confirmed that μ-crystallin (Crym) was present in elevated to highly elevated amounts in transgenic skeletal muscles compared to controls (Fig. 1B). We used capillary-based immunoblotting in a Wes apparatus (Fig. 1C) to quantitate the differences, with aconitase 2 as a loading control, and determined that transgenic skeletal muscles contain 2.6–147.5-fold more μ-crystallin than controls, depending on the muscle. Remarkably, although all skeletal muscles we assayed showed increased levels of μ-crystallin, their relative amounts varied considerably (Fig. 1C), consistent with our qRT-PCR results. These differences were highly significant (p < 0.0001).
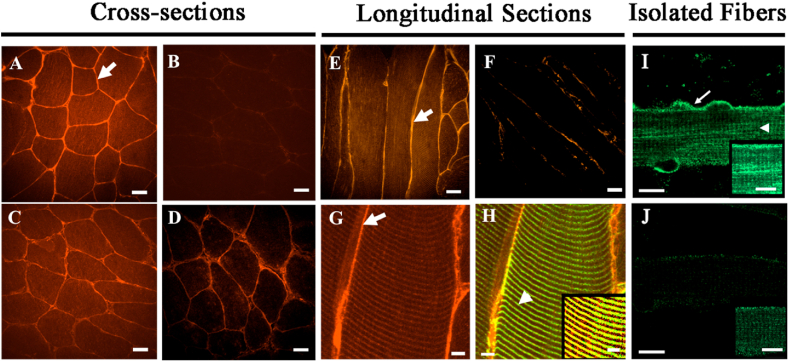
We also examined the distribution of μ-crystallin in transgenic muscle. In cross-sections of TA and flexor digitorum brevis (FDB) muscle, immunolabeling for μ-crystallin was concentrated near the sarcolemma (Fig. 2, arrows) but was also present in the myoplasm. Soleus cross sections did not show a similar concentration of μ-crystallin near the sarcolemma compared to controls (Fig. 2C and D), perhaps because they express lower amounts of the protein. Longitudinal sections of TA muscle and isolated FDB myofibers also showed high levels of μ-crystallin at or near the sarcolemma (Fig. 2E, G, I), but in addition revealed a faint striated distribution in the myoplasm (Fig. 2H and I), which in TA muscles colocalized with desmin at the level of the Z-disk (Fig. 2H, arrowhead). We did not observe immunolabeling for μ-crystallin in the capillaries or connective tissue surrounding myofibers. These results as well as results from qPCR and Wes data (Fig. 1A and C respectively) indicate that μ-crystallin is expressed at elevated levels in myofibers of the transgenic mice but not in other cell types in muscle or other tissues.
Fig. 2.
Immunolabeling of μ-crystallin in control and Crym tg skeletal muscles. A-D, Cross (A–D), longitudinal sections (E–H), of Crym tg (A, C, E, G, H) and control (B, D, F) TA (A, B, E-H) and soleus muscles (C, D) were stained with anti-μ-crystallin antibody and Alexa Fluor-568-conjugated secondary antibody. In G-H, longitudinal sections of Crym tg TA muscle (G, μ-crystallin only) were colabeled with anti-desmin and Alexa Fluor-488-conjugated secondary antibody (H, yellow color shows colabeled structures). I, J, Flexor digitorum brevis (FDB) myofibers in culture from Crym tg (I) and control mice (J) were labeled with anti-μ-crystallin antibody and Alexa-Fluor 488-conjugated secondary antibody. The results show that μ-crystallin was detected at higher levels in tg muscles than in controls. In TA muscle and FDB myofibers it was enriched at the levels of the sarcolemma (arrows, A,E,G, I) and Z-disks, colabeled with desmin (arrowhead, H) or shown without desmin colabel (arrowhead, I). Inset panels (H–J) are brightened and magnified. Crym tg (I) and control (J) FDB images were brightened and magnified equivalently. A-F, scale bars = 20 μm; G, H, scale bars = 5 μm; I, J, scale bars = 10 μm.
3.2. Hormone levels
We compared the levels of different hormones involved in thyroid hormone signaling in Crym tg and control mice. We found significantly more T3 in the TA muscle (~190 fold more) of Crym tg mice and significantly less T4 in the serum (~1.2 fold less) compared to controls (p < 0.001 and p < 0.05, respectively). Thyroid stimulating hormone (TSH) was not significantly different in serum (Table 1) of Crym tg and control mice and, as expected, was not detected in muscle. Intramuscular T4 and serum T3 also did not differ significantly between Crym tg and controls. Thus, the large increase in μ-crystallin in murine skeletal muscle is associated with an even larger increase in muscle T3.
Table 1.
T3, T4, and TSH in TA muscle and serum in Crym tg and control mice. Hormone levels were determined by a clinical diagnostic laboratory. Muscle T3 was significantly higher in Crym tg compared to controls while serum T4 was significantly lower in Crym tg compared to controls. TSH was not significantly different in serum and was not detected in muscle (n = 4). Significance was calculated using a two-tailed Student's t-test with Welch's Correction (α = 0.05).
| TA Muscle | |||||||||||
| Control | Crym tg | ||||||||||
| Percent Mean (ng/mg protein) | -Log (Percent Mean) | Std. Dev. | Std. Dev.(-Log) | n | Percent Mean (ng/mg protein) | -Log (Percent Mean) | Std. Dev. | Std. Dev.(-Log) | n | p Values | |
| T3 | 5.801E-09 | 8.252 | 1.940E-09 | 0.130 | 4 | 1.115E-06 | 6.070 | 1.065E-06 | 0.341 | 4 | 0.0003 |
| T4 | 1.008E-07 | 5.294 | 1.084E-07 | 0.701 | 4 | 2.392E-08 | 5.939 | 2.610E-08 | 0.670 | 4 | 0.2314 |
| TSH |
N.D. |
N.D. |
N/A |
N/A |
4 |
N.D. |
N.D. |
N/A |
N/A |
4 |
N/A |
| Serum |
|||||||||||
| Control |
Crym tg |
||||||||||
| Percent Mean (ng/mg protein) |
-Log (Percent Mean) |
Std. Dev. |
Std. Dev.(-Log) |
n |
Percent Mean (ng/mg protein) |
-Log (Percent Mean) |
Std. Dev. |
Std. Dev.(-Log) |
n |
p Values |
|
| T3 | 2.672E-06 | 5.577 | 3.826E-07 | 0.063 | 4 | 2.325E-06 | 5.635 | 1.865E-07 | 0.036 | 4 | 0.1732 |
| T4 | 1.146E-04 | 3.941 | 5.530E-06 | 0.021 | 4 | 9.417E-05 | 4.028 | 1.076E-05 | 0.051 | 4 | 0.0339 |
| TSH | 1.546E-03 | 2.816 | 2.652E-04 | 0.076 | 4 | 1.376E-03 | 2.885 | 5.664E-04 | 0.157 | 4 | 0.6149 |
3.2.1. Morphology and physiology
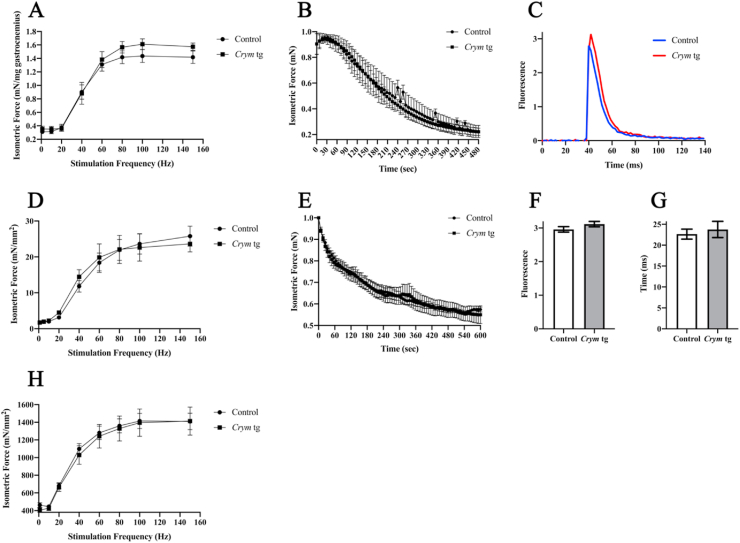
When we performed different morphological and physiological assays of the structure and function of the transgenic mice, we found no significant differences between Crym tgs and controls. The fiber sizes, distribution of fiber sizes, fiber types by immunohistochemistry of myosin heavy chains, weights, and the frequency of centrally nucleated fibers (CNFs) were not significantly altered in the TA muscles of Crym tgs (Supplemental Figs. 4 and 5C, 9C-D). Specific isometric force of contraction, maximal rate of twitch force contraction/relaxation, grip strength, maximum treadmill running speed, and distance run were also indistinguishable between Crym tg and controls (Fig. 3A and B; Supplemental Fig. 6; Supplemental Fig. 10). Fatiguability in the Crym tg mice showed a trend towards being slower than in controls, but this did not rise to the level of significance (Fig. 3B). At the single myofiber level, voltage-induced Ca2+ transients, maximal amplitudes of transients and transient decay rates, all measured with the Ca2+-sensitive fluorescent indicator, Rhod2, under conditions that measure changes in cytoplasmic and not mitochondrial Ca2+ (Giger et al., 2009), were identical in the two strains of mice (Fig. 3C, F, G). Fat staining with BODIPY (493/503) was also the same (Supplemental Fig. 7). However, we did find that Crym tg gastrocnemius muscles and mesenteric fat pads weighed slightly more than their control counterparts (p < 0.05; Supplemental Fig. 3). By these measures, the overexpression of Crym and the resulting accumulation of T3 in myofibers has minimal effects on the structure or function of murine skeletal muscle.
Fig. 3.
Contractile properties of Crym tg muscle. Force frequency curves (A, B, D, E, H) of gastrocnemius (A, B), soleus (D, E), and extensor digitorum longus (EDL) (H). Force was normalized to gastrocnemius muscle weight (A) or to soleus or EDL cross-sectional area (D, H). For fatigue, force was normalized to peak force (B, E). Ca2+ transients in isolated myofibers (C, F, G). C. Representative Ca2+ transient evoked by a voltage pulse, visualized with Rhod2. Blue, control; red, Crym tg. F. Maximal amplitudes of the Ca2+ transients. (n = 100). G. Mean time constants for the decay of the transients (n = 100). Statistics utilized the Student's t-test for Ca2+ measurements (F, G) or the Holm-Šídák test on the gastrocnemius (n = 5; A, B), soleus (control n = 4, Crym tg n = 5; D, E), or EDL (control n = 6, Crym tg n = 5; H) at α = 0.05. All error bars show standard error. The results show no statistically significant differences in any of these measurements.
3.2.2. Fiber types
We applied a machine learning approach to obtain more quantitative information about the soleus muscles in the Crym tg mouse. We did not find any significant differences in the fiber type populations or total number of fibers of Crym tg soleus muscles (782 ± 219 fibers) compared to controls (933 ± 442). Furthermore, our results obtained by determining fiber type visually closely matched the results we determined computationally (Supplemental Figs. 4C and 5C, 9A-B; Supplemental Document 4). We did discover significant differences in a few metrics, however. In particular, the minimal Feret's diameter of Crym tg Type I, I/IIb, I/IIb/IIa, IIa, IIa/IIb, IIb, and all fiber types in aggregate were significantly smaller than control fibers of the same types while Type I/IIa and IIx fibers were not significantly altered in minimal Feret's diameter. Crym tg Type I, IIb, and IIa/IIb fibers were significantly more circular than controls, whereas Type I/IIb, IIx, and all fiber types in the Crym tg in aggregate were less circular than controls (Supplemental Document 4). Using visual evaluation only, we did not observe any significant differences in myosin-specific fiber type labeling in TA or gastrocnemius muscle cross sections between Crym tg mice and controls (Supplemental Figs. 5 and 9; the sizes of these muscles were too large to analyze by machine learning with our equipment). Our results indicate that, although the fiber types in Crym tg and control soleus muscles are indistinguishable, several properties differ significantly between the two genotypes.
3.3. Metabolism
As T3 can have a profound effect on metabolism, we subjected Crym tg and control mice to metabolic studies in Comprehensive Lab Monitoring System (CLAMS) cages. We determined 18 metabolic traits and found that 6 and 4 traits (Supplemental Table 2) were significantly different between Crym tg and control mice during the light and dark cycles, respectively. The respiratory exchange ratio (RER), a measure of the oxidative preference for carbohydrates vs fats, was significantly different during both the light and dark cycles, with essentially no change in the overall energy expenditure (Table 2). Table 2 shows the mean RER of the Crym tg and control mice, compared to standardized values (Lusk, 1924). The results show that the oxidative metabolism in the transgenic mice has partially shifted away from the use of carbohydrates as an energy source and towards fats. The difference from controls is 13.7%. Other traits that also differed significantly between Crym tg and control mice in both light and dark periods were Delta CO2, and Accumulated CO2 (Supplemental Table 2). Thus, overexpression of Crym in skeletal muscle has a significant effect on murine preference for fat as a fuel source for energy metabolism.
Table 2.
Mean RER and corresponding carbohydrate and fat usage. Mean RER was determined by averaging calculated RER values for each genotype (n = 8). The closest standard RER value (taken from Lusk, G (Laemmli, 1970).) was determined and carbohydrate vs fat utilization as energy sources from the standard RER value are listed.
| Percentage of total oxygen consumed by: |
Percentage of total heat produced by: |
Calories per liter of O2 |
||||||
|---|---|---|---|---|---|---|---|---|
| Mean RER | Closest Standard RER Value | Carbohydrate | Fat | Carbohydrate | Fat | Number | Log10 | |
| Control | 0.916 | 0.92 | 72.7 | 27.3 | 74.1 | 25.9 | 4.948 | 0.69447 |
| Crym tg | 0.879 | 0.88 | 59 | 41 | 60.8 | 39.2 | 4.899 | 0.69012 |
3.3.1. Diet study
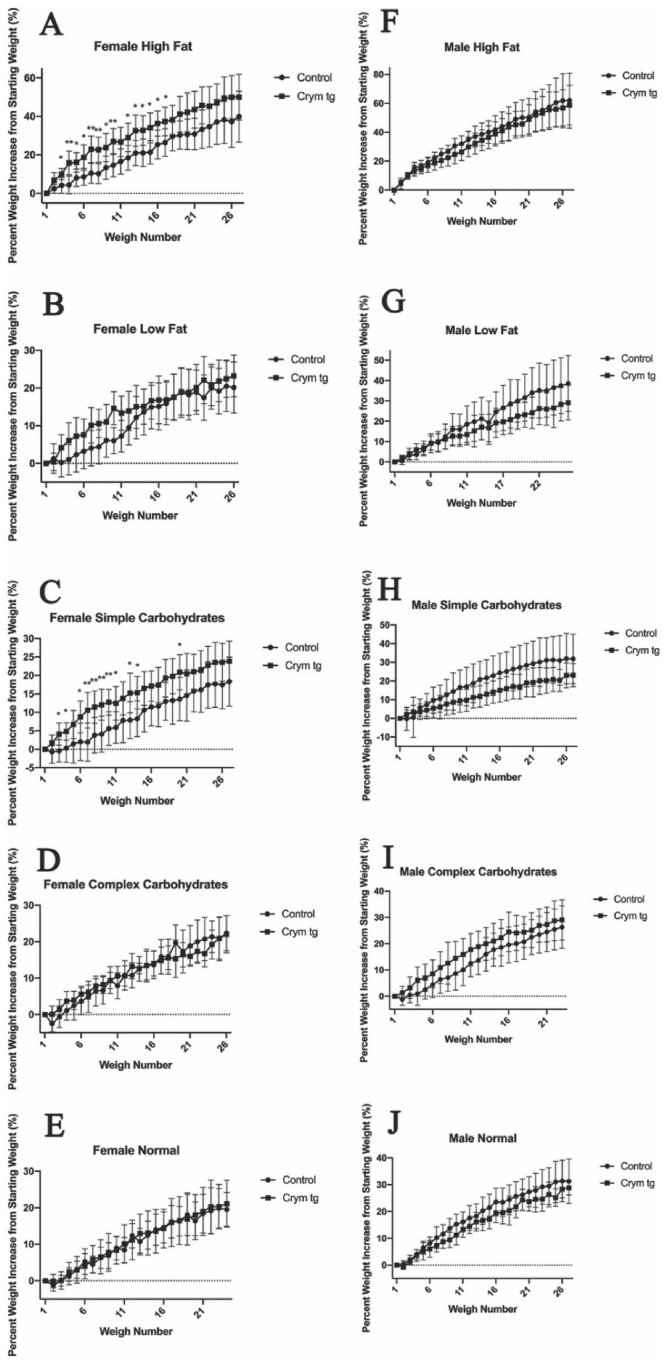
As the Crym tg mice prefer fats to carbohydrates as an energy source, we fed Crym tg and control mice high fat, low fat, high simple carbohydrate, high complex carbohydrate and normal control diets and measured their weight gain over a period of 2 months. We observed significant increases in normalized body weight of female Crym tg mice on the high simple carbohydrate and high fat diets, compared to controls. Female Crym tg mice on the other diets and male Crym tg mice on all 5 of the diets did not gain significantly more or less weight than controls (Fig. 4).
Fig. 4.
Weight gain of Crym tg and control mice on various diets.Crym tg and control mice were placed on different diets and weighed over the course of 60 days. Female (A–D) and male (F-I) Crym tg and control mice were placed on diet containing high fat (A, F), low fat (B, G), high simple carbohydrates (C, H), high complex carbohydrates (D, I), or on a normal diet (E, J) for 60 days. Mice were weighed three times a week and the percent weight increase from their starting weight was determined. Female Crym tg mice on high fat or high simple carbohydrate diets gained weight significantly more rapidly than controls. We did not observe any other significant results using a two-way repeated measures ANOVA or mixed-effects model (∗ = p < 0.05, ∗∗ = p < 0.001, n = 5–16; see Supplemental Document 5 also).
3.3.2. Global gene expression (RNA-seq)
The expression of a large number of genes can be affected by T3 (Supplemental Document 2), which is thought to act largely through thyroid responsive elements (TREs). As T3 is present in much higher levels in Crym tg mice, we had RNA-seq performed on TA muscle samples from 3 Crym tg and 3 control mice and measured more than 13,000 transcripts from approximately 8.4 × 108 reads. All samples had an average Q score >37 with approximately 87% of bases having a Q score greater than 30. After removing transcripts that aligned to introns or that mapped to multiple genes, and after Benjamini-Hochberg False Discovery Rate (FDR) correction, we found that 566 genes were significantly different between Crym tg and controls at α = 0.05 (Supplemental Document 1). Gene ontology (GO) analysis revealed a relative decrease in the expression of genes encoding proteins involved in glycolysis and glycogen metabolism and in fast contractile speeds, and an increase in the expression of genes associated with oxidative metabolism and slower contraction (Table 3, Supplemental Table 6). STRING network analysis also showed clusters of genes involved in ubiquitination, filament associated genes, and the innate immune system (Supplemental Fig. 8). After Benjamini-Hochberg FDR correction, DAVID chromosome annotation showed significantly more (p = 5.7E-7) differentially expressed transcripts arising from chromosome 6 than would be expected. Notably, only 7 of the 566 DEGs contained putative TREs (Supplemental Table 3).
Table 3.
Changes in gene expression related to glycolytic and oxidative pathways and fiber type. Genes related to the glycolytic and oxidative metabolic pathways were determined by assessing root ontological terms for DEGs (n = 566) processed using PANTHER. Enrichment of ontological terms was determined by p < 0.05 after Bonferroni correction of Fisher's exact p-values. Fiber contraction speed genes were determined by cross referencing a putative fiber type-specific list of genes (Drexler et al., 2012). Genes that shared both glycolytic and β-oxidative ontologies were removed from the analysis. The results show that Crym tg muscle exhibit changes in gene expression consistent with a shift to a slower contractile, more oxidative state.
| Glycolytic Pathway Genes | Oxidative Pathway Genes | Fast Contractile Genes | Slow Contractile Genes | ||||
| # increased | # decreased | # increased | # decreased | # increased | # decreased | # increased | # decreased |
| 5 | 11 | 29 | 20 | 10 | 43 | 54 | 8 |
In addition to its ability to bind T3, μ-crystallin is a ketimine reductase, with its activity inhibited by T3 binding (Hallen et al., 2011). Although this activity is likely inhibited by the high intracellular levels of T3 in Crym tg muscle, five genes involved in lysine degradation (a pathway including substrates that μ-crystallin acts on in its capacity as a ketimine reductase), compared to controls, with Aldh2, Ogdh and Dlst increasing in expression and Sccpdh and Setd7 decreasing in expression.
3.3.3. Global protein expression (LC.MS/MS)
Proteomic analysis used liquid chromatography-tandem mass spectrometry (LC.MS/MS) followed by bioinformatic assessment (Supplemental Document 3). GO analysis was performed on the 85 significantly differentially expressed proteins using the PANTHER Classification System which resulted in 7 statistically significant ontological classes. Four of the terms related to muscle while one was for oxidation-reduction processes (Table 4). Eleven proteins that were significantly differentially expressed in Crym tg mice compared to controls were associated with the “oxidation-reduction process” gene ontology. Five of those proteins increased in expression in Crym tg mice compared to controls while 6 decreased (Supplemental Table 5). Although the gene products revealed by LC.MS/MS and RNA-seq showed minimal overlap, the significant ontological terms in the proteomic study were similar to those found in the transcriptomic study.
Table 4.
Significant biological GO terms. Ontological terms were derived from a list of 85 differentially expressed proteins via LC.MS/MS proteomics comparing Crym tg mice to controls.
| Term | Count | p-value | Fold Enrichment |
|---|---|---|---|
| oxidation-reduction process | 11 | 3.700E-04 | 3.976 |
| cardiac muscle contraction | 4 | 9.580E-04 | 20.363 |
| cardiac myofibril assembly | 3 | 1.034E-03 | 61.088 |
| detection of muscle stretch | 2 | 1.605E-02 | 122.176 |
| protein refolding | 2 | 3.576E-02 | 54.300 |
| cardiac muscle hypertrophy | 2 | 3.576E-02 | 54.300 |
| regulation of mitotic spindle assembly | 2 | 4.740E-02 | 40.725 |
The results are consistent with the Crym tg skeletal muscle undergoing a shift towards a slower contractile, more oxidative state.
4. Discussion
As imbalances in T3, such as hypothyroidism or hyperthyroidism, can result in a variety of maladies, thyroid hormone levels are carefully controlled under normal conditions. Regulation of thyroid hormone levels are affected by the feedback loop of thyrotropin releasing hormone (TRH), TSH, T4, somatostatin (Pirahanchi and Jialal, 2019; Siler et al., 1974), monocarboxylate thyroid hormone transporters (especially MCT8/10) (van Mullem et al., 2016), and deiodinases (DIO1-3) that convert T4 transported into the cell into T3 and other metabolites (Bianco and Kim, 2006). μ-Crystallin binds T3 (Hashizume et al., 1989; Tata, 1958) and its absence has been shown to increase efflux of T3 from the cell significantly (Suzuki et al., 2007). This information, paired with the fact that CRYM levels can vary greatly in human muscle (Klooster et al., 2009), raises the question: what effects does μ-crystallin have in those individuals where it is highly expressed? To address this, we generated a skeletal muscle specific transgenic mouse, Crym tg, in which Crym mRNA and protein are expressed in skeletal muscle at high levels comparable to those seen in some humans (Homma et al., 2012; Klooster et al., 2009; Reed et al., 2007). We find that when Crym is overexpressed, metabolism shifts away from carbohydrate use toward fat use, with concomitant shifts in the expression of genes from glycolysis toward β-oxidation and from fast contractile toward slow contractile gene products.
Expression of CRYM in human skeletal muscle from 430 individuals in the GTEx database show about 20% of individuals expressing CRYM at relatively high levels while the majority of people express little to no CRYM. RT-qPCR and Western blot data [ (Klooster et al., 2009); see also (Homma et al., 2012)] show a similar percentage of individuals expressing high levels of CRYM, 30–40 times higher than low expressers. Though a marked variability in CRYM expression has been observed, to the best of our knowledge no data so far correlates human CRYM expression to energy expenditure, locomotive ability, or body composition. We have confirmed this in our own laboratory (Reed et al., unpublished). Crym tg mice produce approximately 94-fold more Crym mRNA as assayed by RT-qPCR and ~28 times more μ-crystallin protein in their TA muscles compared to control C57BL/6J mice. Thus, the Crym tg mouse model reproduces key features of human high expressers, producing Crym at levels that vastly outstrip control mice or human low expressers.
The enhancer from the TNNI1 gene that we used to increase expression of the Crym transgene is a slow skeletal muscle specific gene (Sheng and Jin, 2016) while the promoter, taken from the human skeletal actin gene (ACTA1), shows expression in both fast and slow type fibers (Ciciliot et al., 2013). To our knowledge, muscle-specific differences in gene expression driven by the skeletal actin promoter have not been documented, though it is regulated by a number of factors, including adrenergic signaling (Bishopric and Kedes, 1991). Indeed, thyroid hormone itself can reduce skeletal actin expression, but only when it is introduced exogenously at levels several orders of magnitude greater than those reached in either control or Crym tg TA muscles (Collie and Muscat, 1992; Gustafson et al., 1986). Studies of skeletal actin protein do not show differences in expression that depend on the muscle or fiber type composition, however (Giger et al., 2009; Pette and Staron, 1997), suggesting that differences in the activity of the actin promoter may not account for our results. Less is known about the activity of the TNNI1 enhancer element, but, paradoxically, soleus and diaphragm, which are more slowly contracting in mice than other skeletal muscles (Luff, 1981) have lower levels of Crym expression at the mRNA and protein level compared to other skeletal muscles in Crym tg mice, although they are elevated compared to controls. Thus, although unexpected, our results clearly show that the same promoter/enhancer pair can drive different levels of transgene expression in different muscle groups.
As expected from the high levels of Crym expressed in the muscles of the transgenic mice, we observed a large accumulation of intracellular T3 (~190-fold) in the TA muscle compared to controls. μ-Crystallin promotes accumulation of T3 in the cytoplasm in part by sequestering the hormone. Levels of total T3 in control mouse serum are unchanged in the transgenic mouse. At these levels, μ-crystallin in the muscle should be close to saturated with bound T3. The same should be true for the C57BL/6J control muscle. Although we cannot explain the 5-fold higher enrichment in the T3 level compared to μ-crystallin protein (the stoichiometry of binding is 1:1 (Hallen and Cooper, 2017)), we propose that μ-crystallin may promote the influx and slow the efflux of thyroid hormones by interacting directly with the T3 transporters at the cell surface (Friesema et al., 2008; Mori et al., 2002). This would be consistent with our finding μ-crystallin at the sarcolemma of Crym tg muscle (Fig. 2A, E, G, I).
It is also not clear why serum T4 is decreased in Crym tg mice compared to controls. TSH levels are unchanged between Crym tg and control mice, so T4 production should be unimpaired. Lower serum T4 may result from its increased intramuscular conversion into T3 and its intracellular storage once it is bound to μ-crystallin in the myoplasm. Thus, the higher levels of μ-crystallin are likely to create a “sink” for T3 within the cell that is filled by mass action of T4 transported from the serum into the myoplasm, followed by its intracellular deiodination. Additional studies will be needed to determine why T4 decreases in the serum, why T3 accumulates to such high levels in the muscles of the transgenic mice, and whether the levels of intracellular T3 accumulation vary with the levels of expression of μ-crystallin in different muscles.
We did not see any significant differences in the numbers of any fiber type or in the total number of fibers when we compared soleus muscles of Crym tg mice to controls. Similarly, we also saw no differences in fiber type between control and Crym tg TA and gastrocnemius muscles. Staining for myosin heavy chains is only one metric used to quantify fiber type as slow twitch or fast twitch, however. Future studies using ATPase staining (Brooke and Kaiser, 1970), succinic dehydrogenase staining (Bancroft and Gamble, 2008), or α-glycerophosphate dehydrogenase staining (Dubowitz et al., 2013) may present a more complete picture of any, potentially subtle, fiber type changes that occur due to the overexpression of μ-crystallin in skeletal muscle. Despite this superficial similarity, we did find differences in the minimal Feret's diameter and the circularity of particular fiber types, among other significantly different morphological characteristics, although we cannot ascribe any physiological significance to these differences. Because we see a shift towards β-oxidation as determined by RER, and slow twitch fibers preferentially utilize β-oxidation compared to fast twitch fibers (Kalmar et al., 2012) while also having a smaller size (Schiaffino and Reggiani, 2011), soleus myofibers may be shifting towards a slower twitch phenotype. Remarkably, notwithstanding the large changes in the levels of thyroid hormones that we have measured, we see very few other changes in the structure or function of the hindlimb muscles in mice. Like the fiber types, central nucleation and fat deposition are essentially unchanged. Physiologically, overall muscle function and the function of individual muscles and muscle fibers are indistinguishable between transgenic and control samples. Thus μ-crystallin's effects on muscle structure and function are not readily apparent, despite its massive effect on the distribution of T3. This is consistent with our observation that most changes in gene expression that we measure in transgenic muscle are quite modest – usually less than 50% higher or lower than controls. Whether the similarities between control and tg muscle are sustained when mice are stressed by changes in diet, or by injury or disease, must still be determined.
Because T3 has profound effects on metabolism, we also studied the Crym tg mice in metabolic cages. We found that, under normal resting conditions, RER, a measure of preference for sources of metabolic energy, was significantly different between the Crym tg mice and controls. The difference in RER corresponds to a 13.7% increase in utilization of fat as an energy source over carbohydrates in Crym tg mice compared to controls.
Consistent with the changes we observed in RER, GO analysis of RNA-seq data on Crym tg and controls showed an increase in expression of genes associated with β-oxidation and a decrease in genes associated with glycolysis. As the murine TA muscle is comprised almost entirely of fast twitch fibers, which are primarily glycolytic (Kalmar et al., 2012), we compared our 566 significantly differentially expressed genes to a list of 1343 fast and slow fiber type genes generated by Drexler et al. (2012). We found that 106 or ~19% of the genes altered in expression in the Crym tg mice encode contractile or other proteins related to fiber type (Table 3), and that the expression of genes associated with fast fiber types significantly decreased while that of genes associated with slow fiber types significantly increased (Table 3). By contrast, proteomic studies results revealed 26 significantly differentially expressed, fiber type-specific proteins, of which 7 shifted towards a slower phenotype while 19 shifted towards a faster phenotype (Table 5). As reported (Fitts et al., 1984), high levels of TH have little effect on fast twitch muscles but cause slow twitch muscles to exhibit faster twitch characteristics. Because mice have very few slow twitch fibers and the Crym tg mouse is subject to far less than the thyrotoxic levels of TH used in previous rodent studies, it is perhaps not surprising that we see minimal effects of Crym overexpression. However, we have shown for the first time that high levels of T3 in myocytes may cause curious transcriptional/translational shifts in which slow fiber type genes increase in expression while fast fiber type proteins increase in expression. Future studies evaluating the regulatory steps concerning transcription and translation may determine how TH can decrease the mRNA content of genes expressed primarily in fast fiber types but increase the protein levels of fast fiber type proteins, and vice versa. Coupled with the shift towards a preference for fat utilization and increased expression of genes involved in β-oxidative metabolism, 11 proteins related to the metabolic oxidation-reduction process, and decreased expression of genes involved in carbohydrate metabolism, it seems likely that the accumulation of T3 in muscle is associated with a shift towards a slower twitch, more β-oxidative phenotype.
Table 5.
Proteins differentially expressed in Crym tg mice compared to controls by LC.MS/MS associated with muscle twitch speed. A total of 26 of the 85 differentially expressed proteins identified via LC.MS/MS proteomics are associated with fast or slow twitch speeds. The fourth column from the left lists the fiber types these proteins are normally associated with. The fifth column indicates the nature of the shift we observed in the Crym tg. The results show changes indicating changes in proteins associated with both slow and fast twitch fibers.
| Gene | Abundance Ratio: (Crym tg)/(Control) | padj | Type of Gene | Shifted Towards |
|---|---|---|---|---|
| Ptgr2 | 0.001 | 5.06E-16 | slow | fast |
| Bdh1 | 0.314 | 5.06E-16 | slow | fast |
| Rab14 | 0.352 | 5.06E-16 | slow | fast |
| Ankrd2 | 0.388 | 3.09E-12 | slow | fast |
| Paics | 0.427 | 1.69E-07 | fast | slow |
| Csrp3 | 0.435 | 5.06E-16 | slow | fast |
| Tmem65 | 0.563 | 5.03E-04 | slow | fast |
| Cryab | 0.576 | 3.48E-07 | slow | fast |
| Dnaja4 | 0.605 | 2.78E-03 | slow | fast |
| Gbe1 | 0.639 | 1.10E-04 | slow | fast |
| Fhl1 | 0.672 | 1.10E-03 | slow | fast |
| Palmd | 0.676 | 1.36E-03 | slow | fast |
| Sdhc | 0.705 | 6.11E-03 | slow | fast |
| Hspb6 | 0.711 | 8.19E-03 | slow | fast |
| Hspb1 | 0.732 | 1.96E-02 | slow | fast |
| Pfn1 | 0.747 | 3.07E-02 | slow | fast |
| Gstm2 | 1.351 | 1.08E-02 | fast | fast |
| Casq2 | 1.363 | 8.62E-03 | slow | slow |
| Myh7b | 1.381 | 5.68E-03 | slow | slow |
| Idh2 | 1.389 | 4.59E-03 | slow | slow |
| Selenbp2 | 1.403 | 3.13E-03 | slow | slow |
| Kera | 1.445 | 1.12E-03 | fast | fast |
| Fbp2 | 1.724 | 9.86E-08 | fast | fast |
| Myl2 | 2.672 | 5.06E-16 | slow | slow |
| Gpt | 1000 | 5.06E-16 | slow | slow |
| Lta4h | 1000 | 5.06E-16 | fast | fast |
Earlier studies used thyrotoxic doses of T3 and T4 in rats to alter skeletal muscles. These doses induced a shift in slow twitch soleus muscle from slow to faster characteristics, but without accompanying changes in contractile velocity. They had no effect on fast twitch extensor digitorum longus (EDL) muscles (Fitts et al., 1984). Consistent with our observations, these results suggest that the contractile properties of muscle may not always correspond to its histochemical and biochemical properties, at least when thyroid hormones are elevated. The levels reached in Crym tg muscle are considerably below thyrotoxic levels, however. A later study examined the properties of muscle in mice lacking thyroid hormone receptors and, again, found changes in soleus but not EDL (Johansson et al., 2003). In this case, soleus muscles in the knockouts showed even slower twitch characteristics than in controls, consistent with changes seen in hypothyroid mice. These results suggest that any elevation in T3 related to transgenic expression of Crym is more likely to appear in slow twitch than in fast twitch muscles. Given the fact that only ~60% of murine soleus muscles are slow and the effects of Crym overexpression are subtler than either thyrotoxicosis or the complete ablation of the thyroid hormone receptor, significant differences in the physiological properties of Crym tg muscles might not be expected. This is congruent with our observation that the changes in gene expression of the myosin heavy and light chains in TA muscle are relatively small and inconsistent (Supplemental Table 4). While we do not see any changes in fiber type as measured by myosin heavy chain presence in soleus muscle, we do see smaller Type I/IIb and IIa fibers consistent with a shift towards a slower muscle twitch phenotype.
We saw significantly greater rates of increase in body weight of female Crym tg compared to control mice on high fat or high simple carbohydrate diets (Fig. 4A, C). There may be a sexually dimorphic effect that higher levels of μ-crystallin has on metabolism because we did not observe a similar significant increase in body weight of Crym tg males on these diets (Fig. 4F–J). Indeed Chen et al. found that mice with two X chromosomes had “accelerated weight gain on a high fat diet” and up to a 2-fold increase in adiposity compared to XY mice (Chen et al., 2012) showing a clear sexual dimorphism for fat storage. Thus, μ-crystallin may affect metabolism in a sexually dimorphic way.
The changes that we have observed are novel. To the best of our knowledge, the only comprehensive studies of thyroid hormone-dependent gene expression in mice to date were performed in liver tissue, though an older study has been reported on skeletal muscle in men (Clément et al., 2002). There are 5129 significantly differentially expressed genes shared across three whole genome microarrays comparing hyperthyroid mouse liver to euthyroid or hypothyroid mouse liver (GEO DataSets ID: 200068803, 200065947, 200021307). Of the 566 significant DEGs we identified by RNA-seq, 211 showed changes in these studies of liver. 107 were significantly increased in expression while 104 were significantly decreased in expression (Supplemental Document 2). Notably, however, only a small subset of these DEGs are known to contain TREs (Supplemental Table 3). Despite the fact that T3 is highly elevated in the TA muscles of Crym tg mice, the changes in mRNA and protein levels are modest. This is in keeping with earlier studies of the relatively modest effects of more extreme changes in thyroid hormone signaling in rodent muscle (Fitts et al., 1984; Johansson et al., 2003).
The shift in gene expression and metabolism in Crym tg mice may also be relevant to human physiology and perhaps to the pathophysiology of FSHD. CRYM levels in human muscles vary widely (Klooster et al., 2009), but no distinctive physiological differences have been linked to this variability. Such differences may appear in muscle stressed by disease, however. In particular, CRYM is likely to be linked directly or indirectly to FSHD (Reed et al., 2007; Vanderplanck et al., 2011). DUX4 expressed in FSHD muscle increases CRYM expression (Vanderplanck et al., 2011), and μ-crystallin in mice promotes a shift in RER and gene expression consistent with a shift toward a slower fiber type, which if pronounced enough would generate less force upon contraction. Notably, force generated by fast twitch muscle fibers is significantly reduced in FSHD (Lassche et al., 2013). Although Crym tg mice do not show a decrease in force, the changes we document in TA muscle could be associated with such a decrease if it manifests over decades, the time course of the disease in man.
In summary, we found that the expression of high levels of μ-crystallin in murine skeletal muscle greatly increases T3 levels in muscle, shifts metabolism to favor the use of fat over carbohydrates as an energy source, and enhances the expression of genes typical of slow skeletal muscle. Remarkably, the morphological and physiological characteristics of the muscle are not significantly altered. We therefore propose that the higher levels of μ-crystallin seen in some humans regulate gene expression and metabolism in similar, subtle ways. We speculate that individuals showing high levels of μ-crystallin expression in muscle will be more resistant to diabetes and obesity, and that mechanisms that up-regulate μ-crystallin will be beneficial in treating these conditions.
CRediT authorship contribution statement
Christian J. Kinney: Data curation, Writing - original draft. Andrea O'Neill: Data curation, Supervision. Kaila Noland: Data curation. Weiliang Huang: Data curation. Joaquin Muriel: Data curation. Valeriy Lukyanenko: Data curation. Maureen A. Kane: Data curation. Christopher W. Ward: Data curation. Alyssa F. Collier: Data curation. Joseph A. Roche: Data curation. John C. McLenithan: Data curation. Patrick W. Reed: Data curation. Robert J. Bloch: Writing - original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful to Dr. J. Molkentin (Children's Hospital, University of Cincinnati) for his gift of a plasmid, and the Mid-Atlantic NORC (Dr. S. Taylor, P.I.) for its assistance with measurements of RER. W-LH and MAK are affiliated with the University of Maryland School of Pharmacy Mass Spectrometry Center (SOP1841-IQB2014).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crphys.2021.02.003.
Funding
This work was initially supported by a subcontract to RJB from 5 U54 HD060848 (Dr. C. Emerson, Jr., P.I.), and more recently by NIH grants to RJB (2RO1 AR064268 minority supplement for CK) and to PWR (1R21 AR057519). We also acknowledge the generous support of the Kahlert Foundation (Sykesville, MD) for our research, and funding from the Graduate School, University of Maryland, Baltimore, for KN's stipend.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abe S., Katagiri T., Saito-Hisaminato A., Usami S-i, Inoue Y., Tsunoda T., Nakamura Y. Identification of CRYM as a candidate responsible for nonsyndromic deafness, through cDNA microarray analysis of human cochlear and vestibular tissues. Am. J. Hum. Genet. 2003;72:73–82. doi: 10.1086/345398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguet F., Van De Ville D., Unser M. Model-based 2.5-D deconvolution for extended depth of field in brightfield microscopy. IEEE Trans. Image Process. 2008;17:1144–1153. doi: 10.1109/TIP.2008.924393. [DOI] [PubMed] [Google Scholar]
- Bancroft J.D., Gamble M. Elsevier health sciences; 2008. Theory and Practice of Histological Techniques. [Google Scholar]
- Beslin A., Vié M.-P., Blondeau J.-P., Francon J. Identification by photoaffinity labelling of a pyridine nucleotide-dependent tri-iodothyronine-binding protein in the cytosol of cultured astroglial cells. Biochem. J. 1995;305:729–737. doi: 10.1042/bj3050729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A.C., Kim B.W. Deiodinases: implications of the local control of thyroid hormone action. J. Clin. Invest. 2006;116:2571–2579. doi: 10.1172/JCI29812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopric N.H., Kedes L. Adrenergic regulation of the skeletal alpha-actin gene promoter during myocardial cell hypertrophy. Proc. Natl. Acad. Sci. Unit. States Am. 1991;88:2132–2136. doi: 10.1073/pnas.88.6.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel F., Hachi I., Palencia A., Gaillard M.C., Ferrer J.L. Crystal structure of mouse mu-crystallin complexed with NADPH and the T3 thyroid hormone. FEBS J. 2014;281:1598–1612. doi: 10.1111/febs.12726. [DOI] [PubMed] [Google Scholar]
- Briguet A., Courdier-Fruh I., Foster M., Meier T., Magyar J.P. Histological parameters for the quantitative assessment of muscular dystrophy in the mdx-mouse. Neuromuscul. Disord. 2004;14:675–682. doi: 10.1016/j.nmd.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Brooke M.H., Kaiser K.K. Three" myosin adenosine triphosphatase" systems: the nature of their pH lability and sulfhydryl dependence. J. Histochem. Cytochem. 1970;18:670–672. doi: 10.1177/18.9.670. [DOI] [PubMed] [Google Scholar]
- Burks T.N., Andres-Mateos E., Marx R., Mejias R., Van Erp C., Simmers J.L., Walston J.D., Ward C.W., Cohn R.D. Losartan restores skeletal muscle remodeling and protects against disuse atrophy in sarcopenia. Sci. Transl. Med. 2011;3 doi: 10.1126/scitranslmed.3002227. 82ra37-82ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., McClusky R., Chen J., Beaven S.W., Tontonoz P., Arnold A.P., Reue K. The number of x chromosomes causes sex differences in adiposity in mice. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciciliot S., Rossi A.C., Dyar K.A., Blaauw B., Schiaffino S. Muscle type and fiber type specificity in muscle wasting. Int. J. Biochem. Cell Biol. 2013;45:2191–2199. doi: 10.1016/j.biocel.2013.05.016. [DOI] [PubMed] [Google Scholar]
- Clément K., Viguerie N., Diehn M., Alizadeh A., Barbe P., Thalamas C., Storey J.D., Brown P.O., Barsh G.S., Langin D. In vivo regulation of human skeletal muscle gene expression by thyroid hormone. Genome Res. 2002;12:281–291. doi: 10.1101/gr.207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collie E., Muscat G.E. The human skeletal alpha-actin promoter is regulated by thyroid hormone: identification of a thyroid hormone response element. Cell Growth Differ.: Mol. Biol. J. Am. Assoc. Canc. Res. 1992;3:31–42. [PubMed] [Google Scholar]
- Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Mattéotti C., van Acker A.M., Leo O. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. Unit. States Am. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfer V., Pichler P., Stranzl T., Stadlmann J., Taus T., Winkler S., Mechtler K. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J. Proteome Res. 2014;13:3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler H.C., Ruhs A., Konzer A., Mendler L., Bruckskotten M., Looso M., Günther S., Boettger T., Krüger M., Braun T. On marathons and Sprints: an integrated quantitative proteomics and transcriptomics analysis of differences between slow and fast muscle fibers. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.010801. M111. 010801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubowitz V., Sewry C.A., Oldfors A. Elsevier Health Sciences; 2013. Muscle Biopsy: a Practical Approach: Expert Consult; Online and Print. [Google Scholar]
- Ebashi S., Endo M., Ohtsuki I. Control of muscle contraction. Q. Rev. Biophys. 1969;2:351–384. doi: 10.1017/s0033583500001190. [DOI] [PubMed] [Google Scholar]
- Encarnacion-Rivera L., Foltz S., Hartzell H.C., Myosoft Choo H. An automated muscle histology analysis tool using machine learning algorithm utilizing Fiji/ImageJ software. PloS One. 2020;15 doi: 10.1371/journal.pone.0229041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J.K., Fischer B., Grossmann J., MacCoss M.J. A fast SEQUEST cross correlation algorithm. J. Proteome Res. 2008;7:4598–4602. doi: 10.1021/pr800420s. [DOI] [PubMed] [Google Scholar]
- Fitts R.H., Brimmer C.J., Troup J.P., Unsworth B.R. Contractile and fatigue properties of thyrotoxic rat skeletal muscle. Muscle Nerve: Offl. J. Am. Assoc. Electrodiagnost. Med. 1984;7:470–477. doi: 10.1002/mus.880070609. [DOI] [PubMed] [Google Scholar]
- Friesema E.C., Jansen J., Jachtenberg J-w, Visser W.E., Kester M.H., Visser T.J. Effective cellular uptake and efflux of thyroid hormone by human monocarboxylate transporter 10. Mol. Endocrinol. 2008;22:1357–1369. doi: 10.1210/me.2007-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giger J.M., Bodell P.W., Zeng M., Baldwin K.M., Haddad F. Rapid muscle atrophy response to unloading: pretranslational processes involving MHC and actin. J. Appl. Physiol. 2009;107:1204–1212. doi: 10.1152/japplphysiol.00344.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P., Ponte P., Blau H., Kedes L. alpha-skeletal and alpha-cardiac actin genes are coexpressed in adult human skeletal muscle and heart. Mol. Cell Biol. 1983;3:1985–1995. doi: 10.1128/mcb.3.11.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson T.A., Markham B.E., Morkin E. Effects of thyroid hormone on alpha-actin and myosin heavy chain gene expression in cardiac and skeletal muscles of the rat: measurement of mRNA content using synthetic oligonucleotide probes. Circ. Res. 1986;59:194–201. doi: 10.1161/01.res.59.2.194. [DOI] [PubMed] [Google Scholar]
- Hallen A., Cooper A.J. Reciprocal control of thyroid binding and the pipecolate pathway in the brain. Neurochem. Res. 2017;42:217–243. doi: 10.1007/s11064-016-2015-9. [DOI] [PubMed] [Google Scholar]
- Hallen A., Cooper A.J., Jamie J.F., Haynes P.A., Willows R.D. Mammalian forebrain ketimine reductase identified as μ-crystallin; potential regulation by thyroid hormones. J. Neurochem. 2011;118:379–387. doi: 10.1111/j.1471-4159.2011.07220.x. [DOI] [PubMed] [Google Scholar]
- Hallen A., Cooper A.J., Jamie J.F., Karuso P. Insights into enzyme catalysis and thyroid hormone regulation of cerebral ketimine reductase/μ-crystallin under physiological conditions. Neurochem. Res. 2015;40:1252–1266. doi: 10.1007/s11064-015-1590-5. [DOI] [PubMed] [Google Scholar]
- Hashizume K., Miyamoto T., Ichikawa K., Yamauchi K., Sakurai A., Ohtsuka H., Kobayashi M., Nishii Y., Yamada T. Evidence for the presence of two active forms of cytosolic 3, 5, 3'-triiodo-L-thyronine (T3)-binding protein (CTBP) in rat kidney. Specialized functions of two CTBPs in intracellular T3 translocation. J. Biol. Chem. 1989;264:4864–4871. [PubMed] [Google Scholar]
- Homma S., Chen J.C., Rahimov F., Beermann M.L., Hanger K., Bibat G.M., Wagner K.R., Kunkel L.M., Emerson C.P., Jr., Miller J.B. A unique library of myogenic cells from facioscapulohumeral muscular dystrophy subjects and unaffected relatives: family, disease and cell function. Eur. J. Hum. Genet. 2012;20:404–410. doi: 10.1038/ejhg.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson C., Lunde P.K., Göthe S., Lännergren J., Westerblad H. Isometric force and endurance in skeletal muscle of mice devoid of all known thyroid hormone receptors. J. Physiol. 2003;547:789–796. doi: 10.1113/jphysiol.2002.032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar B., Blanco G., Greensmith L. Determination of muscle fiber type in rodents. Curr. Protocols Mouse Biol. 2012;2:231–243. doi: 10.1002/9780470942390.mo110229. [DOI] [PubMed] [Google Scholar]
- Kammoun M., Cassar-Malek I., Meunier B., Picard B. A simplified immunohistochemical classification of skeletal muscle fibres in mouse. Eur. J. Histochem.: EJH. 2014;58 doi: 10.4081/ejh.2014.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Chen R., Sheu M., Kim N., Kim S., Islam N., Wier E.M., Wang G., Li A., Park A. Noncoding dsRNA induces retinoic acid synthesis to stimulate hair follicle regeneration via TLR3. Nat. Commun. 2019;10:2811. doi: 10.1038/s41467-019-10811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R.Y., Gasser R., Wistow G.J. mu-crystallin is a mammalian homologue of Agrobacterium ornithine cyclodeaminase and is expressed in human retina. Proc. Natl. Acad. Sci. Unit. States Am. 1992;89:9292–9296. doi: 10.1073/pnas.89.19.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klooster R., Straasheijm K., Shah B., Sowden J., Frants R., Thornton C., Tawil R., Van Der Maarel S. Comprehensive expression analysis of FSHD candidate genes at the mRNA and protein level. Eur. J. Hum. Genet. 2009;17:1615. doi: 10.1038/ejhg.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Hashizume K., Suzuki S., Ichikawa K., Takeda T. A novel NADPH-dependent cytosolic 3, 5, 3′-triiodo-LThyronine-binding protein (CTBP; 5. IS) in rat liver: a comparison with 4.7 S NADPH-dependent CTBP. Endocrinology. 1991;129:1701–1708. doi: 10.1210/endo-129-4-1701. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsen P.R., Silva J.E., Kaplan M.M. Relationships between circulating and intracellular thyroid hormones: physiological and clinical implications. Endocr. Rev. 1981;2:87–102. doi: 10.1210/edrv-2-1-87. [DOI] [PubMed] [Google Scholar]
- Lassche S., Stienen G.J., Irving T.C., Van Der Maarel S.M., Voermans N.C., Padberg G.W., Granzier H., van Engelen B.G., Ottenheijm C.A. Sarcomeric dysfunction contributes to muscle weakness in facioscapulohumeral muscular dystrophy. Neurology. 2013;80:733–737. doi: 10.1212/WNL.0b013e318282513b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luff A. Dynamic properties of the inferior rectus, extensor digitorum longus, diaphragm and soleus muscles of the mouse. J. Physiol. 1981;313:161–171. doi: 10.1113/jphysiol.1981.sp013656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukyanenko V., Muriel J.M., Bloch R.J. Coupling of excitation to Ca2+ release is modulated by dysferlin. J. Physiol. 2017;595:5191–5207. doi: 10.1113/JP274515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk G. Animal calorimetry twenty-fourth paper. Analysis of the oxidation of mixtures of carbohydrate and fat. J. Biol. Chem. 1924;59:41–42. [Google Scholar]
- Mori J-i, Suzuki S., Kobayashi M., Inagaki T., Komatsu A., Takeda T., Miyamoto T., Ichikawa K., Hashizume K. Nicotinamide adenine dinucleotide phosphate-dependent cytosolic T3 binding protein as a regulator for T3-mediated transactivation. Endocrinology. 2002;143:1538–1544. doi: 10.1210/endo.143.4.8736. [DOI] [PubMed] [Google Scholar]
- Motulsky H.J., Brown R.E. Detecting outliers when fitting data with nonlinear regression–a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinf. 2006;7:123. doi: 10.1186/1471-2105-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo Y., Sekido T., Nishio S-i, Sekido K., Kitahara J., Suzuki S., Komatsu M. Loss of μ-crystallin causes PPARγ activation and obesity in high-fat diet-fed mice. Biochem. Biophys. Res. Commun. 2019;508:914–920. doi: 10.1016/j.bbrc.2018.12.038. [DOI] [PubMed] [Google Scholar]
- Olojo R.O., Ziman A.P., Hernández-Ochoa E.O., Allen P.D., Schneider M.F., Ward C.W. Mice null for calsequestrin 1 exhibit deficits in functional performance and sarcoplasmic reticulum calcium handling. PloS One. 2011;6 doi: 10.1371/journal.pone.0027036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pette D., Staron R.S. International Review of Cytology. Elsevier; 1997. Mammalian skeletal muscle fiber type transitions; pp. 143–223. [DOI] [PubMed] [Google Scholar]
- Pirahanchi Y., Jialal I. StatPearls. Treasure Island (FL) StatPearls Publishing StatPearls Publishing LLC.; 2019. Physiology, thyroid stimulating hormone (TSH) [PubMed] [Google Scholar]
- Reed P.W., Corse A.M., Porter N.C., Flanigan K.M., Bloch R.J. Abnormal expression of mu-crystallin in facioscapulohumeral muscular dystrophy. Exp. Neurol. 2007;205:583–586. doi: 10.1016/j.expneurol.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Sandler B., Webb P., Apriletti J.W., Huber B.R., Togashi M., Lima S.T.C., Juric S., Nilsson S., Wagner R., Fletterick R.J. Thyroxine-thyroid hormone receptor interactions. J. Biol. Chem. 2004;279:55801–55808. doi: 10.1074/jbc.M410124200. [DOI] [PubMed] [Google Scholar]
- Schiaffino S., Reggiani C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seko D., Ogawa S., Li T.-S., Taimura A., Ono Y. μ-Crystallin controls muscle function through thyroid hormone action. Faseb. J. 2015;30:1733–1740. doi: 10.1096/fj.15-280933. [DOI] [PubMed] [Google Scholar]
- Sheng J.-J., Jin J.-P. TNNI1, TNNI2 and TNNI3: evolution, regulation, and protein structure–function relationships. Gene. 2016;576:385–394. doi: 10.1016/j.gene.2015.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siler T., Yen S., Vale W., Guillemin R. Inhibition by somatostatin on the release of TSH induced in man by thyrotropin-releasing factor. J. Clin. Endocrinol. Metab. 1974;38:742–745. doi: 10.1210/jcem-38-5-742. [DOI] [PubMed] [Google Scholar]
- Spangenburg E.E., Pratt S.J., Wohlers L.M., Lovering R.M. Use of BODIPY (493/503) to visualize intramuscular lipid droplets in skeletal muscle. J. Biomed. Biotechnol. 2011:2011. doi: 10.1155/2011/598358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Suzuki N., Mori J-i, Oshima A., Usami S., Hashizume K. μ-Crystallin as an intracellular 3, 5, 3′-triiodothyronine holder in vivo. Mol. Endocrinol. 2007;21:885–894. doi: 10.1210/me.2006-0403. [DOI] [PubMed] [Google Scholar]
- Tata J.R. A cellular thyroxine-binding protein fraction. Biochim. Biophys. Acta. 1958;28:91–94. doi: 10.1016/0006-3002(58)90432-3. [DOI] [PubMed] [Google Scholar]
- Tawil R., Van Der Maarel S.M., Tapscott S.J. Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skeletal Muscle. 2014;4(12) doi: 10.1186/2044-5040-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.C., Nadeau K., Abbasi M., Lachance C., Nguyen M., Fenrich J. The ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends Biotechnol. 2019;37:761–774. doi: 10.1016/j.tibtech.2018.12.002. [DOI] [PubMed] [Google Scholar]
- van Mullem A.A., van Gucht A.L., Visser W.E., Meima M.E., Peeters R.P., Visser T.J. Effects of thyroid hormone transporters MCT8 and MCT10 on nuclear activity of T3. Mol. Cell. Endocrinol. 2016;437:252–260. doi: 10.1016/j.mce.2016.07.037. [DOI] [PubMed] [Google Scholar]
- Vanderplanck C., Ansseau E., Charron S., Stricwant N., Tassin A., Laoudj-Chenivesse D., Wilton S.D., Coppée F., Belayew A. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PloS One. 2011;6 doi: 10.1371/journal.pone.0026820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Xiao P., Chen D., Xu L., Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- Zaiontz C. Real Statistics Resource Pack software. 2020. www.real-statistics.com (Release 6.8)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.