Abstract
Obesity is a growing human health concern word wide and imposes adverse effects on many cell types and organ systems, including the kidneys. Obesity interferes with various cellular processes by increasing lipid accumulation and oxidation, insulin resistance, and inflammation. Autophagy is an important cellular process to maintain hemostasis and preserve resources but might be altered in obesity. Interestingly experimental studies have shown either an increase or decrease in the rate of autophagy and accumulation of by-products and mediators of this cascade in kidneys of obese individuals. Hence, whether autophagy is beneficial or detrimental under these conditions remains to be unresolved. This review aims to summarize emerging evidence linking superfluous fat accumulation to alterations in autophagy. Elucidating the role of autophagy in the pathogenesis and complications of obesity in the kidney might help in identification of therapeutic targets to prevent or delay development of chronic kidney disease in obese subjects.
Keywords: Autophagy, kidney, obesity, lipids
Introduction
Nowadays obesity is the growing global health concern of the modern world and developing countries and may increase morbidity and mortality. Data from the U.S. Centers for Disease Control and Prevention (CDC) show that a rising number of Americans have obesity, a risk factor for several disease including severe COVID-19 complications. Obesity can increase the risk of severe illness, hospitalization, and even death secondary to COVID-19, with notable racial and ethnic disparities [1]. In turn, in the COVID-19 era, working from home, lack of daily physical activity, increase in stress, inconsistent access to fresh food, and falling economic status may impact on worldwide diet and personal life span and obesity related diseases. For example, the pandemic will probably increase out-of-school time for many children in the US and aggravate the risk for weight gain [2].
Obesity is defined by excess body weight secondary to excessive fat accumulation in fat cells and is often assessed by body mass index (BMI). This expansion of the adipose tissue is a result of exceeding energy input expenditure. [3]. The hyper-energy availability despite low energy consumption, causes an elevation in fatty acid triglyceride, low density lipoproteins (LDL), and total cholesterol [4]. These changes disturb neurohormonal and metabolic regulation, resulting in an increase in oxidative stress, inflammation, apoptosis, lipotoxicity, catecholamines overflow, and interrupted cellular processes. [3, 5]. Obese patients also have a higher risk of developing chronic kidney disease (CKD) a propensity that is influenced by sex and race [6, 7].
Obesity may damage cellular targets by inducing inflammation, lipotoxicity, and by dysregulating local and circulating levels of adipokines and cytokines. Recent evidence implicates altered autophagy, a homeostatic mechanism responsible for recycling of damaged cells and organelles[8]. However, the role of altered autophagy in kidney injury in obesity remains obscure. This review article aims to summarize recent data on the involvement of autophagy in structural and functional impairment in the kidney of obese subjects.
Obesity: definition, consequences and impact
Excess obesity that might in turn cause metabolic syndrome is a growing epidemic. In 2014, over 600 million individuals were reported to be obese worldwide. The age adjusted prevalence of obesity in the US was reported to be over 35% in men and 40.4% among women[9, 10]. It is estimated that by the year 2030 57.8% (3.3 billion people) of the world adult population will have a BMI higher than 25[11].
Yet, the definition of obesity might be variable, given that BMI does not account for variable patterns of fat distribution in the body[12] and the common classification of obesity differs between adults and children[7]. Alternative effective methods to determine obesity include dual-energy x-ray absorptiometry[13], waist-circumference[14] or Waist stature ratio[15].
The roots of obesity are multifactorial, and include the interaction among genetic, environmental, hormonal and psychological factors[16]. The list of clinical outcomes of obesity is extensive, but its severity depends on additional presence of excess body fat in adipose deposits and around normally lean tissues. This “ectopic fat deposition” around heart, liver and kidney causes damage to these organs[17] by facilitating access of cytokines and adiponekines. In addition, excess nutrients can overwhelm endogenous cellular energy production and utilization. For example, obesity inhibits mitochondrial fatty acid ß-oxidation in tissue[18], leading to loss of mitochondrial cristae membrane and matrix density and in turn H2o2 release from the mitochondria that causes further damage to the cell [19]. Lipotoxcity has also been implicated in regulation of cell survival and demise, and may interfere with cellular processes responsible for elimination or recycling of damaged proteins [20], lipid droplets and waste products [21].
Among these processes, autophagy mechanisms are often activated in animal models[22] that are genetically predisposed to obesity or in obese human subjects. Such alternations have been documented in various cell types, including the-kidneys[23].
The characteristic glomerular damage during obesity is histologically defined as glomerulomegaly in the presence or absence of glomerulosclerosis [24], and may lead to focal segmental glomerulosclerosis (FSGS). Glomerular and tubular hypertrophy results in increased kidney size, with renal tubular overload and increased proximal tubular sodium and water reabsorption. Activation of the intrarenal renin-angiotensin-aldosterone system and renal sympathetic nerves may further compound renal metabolic imbalance [25]. Initially, glomerular filtration rate rises and induces hyperfiltration, but ultimately proceeds to proteinuria and CKD.
Autophagy dysregulation may play role in these histologic alterations. In the kidney, the podocytes appear to be primary victims that are damaged during adiposity and are also the cells showing the highest basal rate of autophagy.
Nevertheless, the precise role of autophagy and its inhibition or reinforcement in the context of obesity is controversial and incompletely understood. A number of proteins participate in autophagic flux, and several different autophagic pathways have been described so that an increase in one autophagic element does not necessarily indicate upregulation of an entire pathway. This presents a unique challenge for pinpointing the precise direction of the autophagic cascade and its regulation in obesity.
Mechanism of autophagy
Autophagy is an evolutionarily-conserved cellular mechanism used to engulf injured cells and organelles and retain their energy by degradation and recycling of intracellular materials to maintain homeostasis. However, its most fundamental role is adaptation to metabolic demands [26]. This process was first discovered in the early 50s and was boosted by the discovery of two mechanisms by cancer biologists, identification of AuTophaGy-related (ATG) genes in yeast [27] and its mamelian ATG homolog ATG6/BECN1 as a tumor suppressor gene[28, 29]. The main protein complex is composed of ATG, which serves as a functional coordinator of the cascade [30]. Autophagy is now considered as the primary process to recycle damaged organelles in order to curb cellular energy requirement for maintaining the structure and function, and for preventing further cellular damage[31].
This multi-step process involves several defined steps ,including initiation, nucleation, expansion fusion and ultimately degradation. Each step can get perturbed during obesity.
The initiation of autophagy in mammalian cells is mediated by Unc-51-like kinase (ULK)-1 and -2 [32], the activation of which is controlled by two nutrient regulated proteins: 5' adenosine monophosphate-activated kinase (AMPK) and the mammalian target of rapamycin (mTOR) complex [26]. AMPK is activated by cellular energy depletion while mTOR is activated upon nutrient abundance. In addition, microtubule-associated protein 1A/1B-light chain-3 (LC3) is a key player during autophagy. LC3-I conjugates to phosphatidylethanolamine forming LC3-II which is then recruited to the autophagosomal membrane [33]. The exact regulation of this fusion is not yet well understood, but it is believed that the cytoskeleton component and the motor proteins play the major role, which is then followed by soluble N-ethylmalemid-sensitive factor attachment protein receptor (SNARE)-mediated fusion[34].
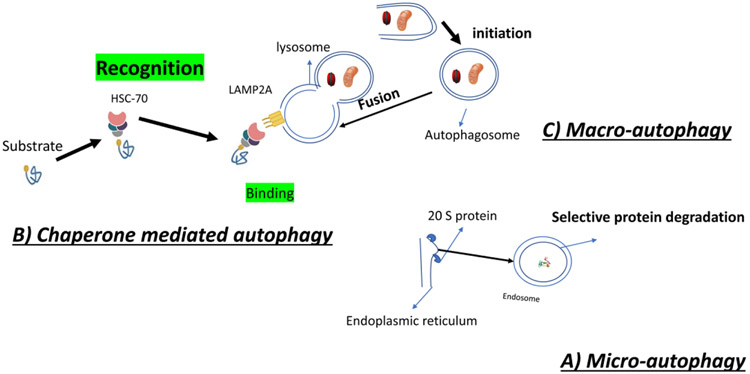
Three major types of autophagy have been described: micro-autophagy, chaperone mediated autophagy and macro-autophagy (Figure 1).
Figure 1:
Autophagy has three sub types: A) Micro-autophagy which is executed by 2 subtype proteins called core protein(20 s proteins) and is formation of microsome from endoplasmic reticulum in order to degrade useless protein or small cell obstacle. B) Chaperone mediated autophagy, which is performed by recognition of the targeted substrate(for degradation)by Heat Shock Cognate 70 (HSC70), which is further phagocytosed in to the lysosome by binding to Lysosome associated membrane protein 2A (LAMP2A).C) Macro-autophagy is performed by formation of autophagosome and its fusion to lysosome.
Micro-autophagy is described as an extension of lysosomal membrane to take in intracellular contents. Its molecular pathway is dependent on Ca2+/Calmodulin, but independent of components involved in vesicle fusion[35, 36]. The endoplasmic reticulum (ER) expands and triggers formation of whorls, which are further selectively taken up into microsomes to form autophagosomes[36].In contrast to macro-autophagy, studies have shown that micro-autophagy decreases during starvation and protein degradation [37]. In mammalian cells the heat-shock cognate 70 (HSC70) conjugation with endosomal acidic phospholipids plays a pivotal role in this pathway [38]. In contrast to other pathways, micro-autophagy is accomplished by assembly of two major subcomplexes called the 20S proteasome or core particle to form the microsome (small lysosome)[39].
Chaperon-mediated autophagy is instigated by recognition of proteins carrying the pentapeptide KFERQ sequence by HSC70, which then unfolds the protein and bring it to lysosome-associated membrane protein (LAMP)-2A receptor, which is in turn transported into the lysosome in order to get degraded[40]. It is estimated that about 30% of all cytosolic proteins carry this sequence [41].Regulation of this process is controlled by substrate binding to LAMP2A. Changing LAMP2A level modulate chaperone-mediated autophagy and its upregulation can occur in response to mild oxidative stress, protein-damaging toxins, and extended periods of nutrient deprivation, although the signaling process requires additional studies[42].
Macro-autophagy is the chief regulated form of autophagy that responds to environmental and physiological cues[43]. This high-capacity process initially takes place by encircling targeted proteins and organelles and formation of an autophagosome. Then these autophagosomes fuse with lysosomes and their content gets degraded [29]. It is completely distinct from the two other pathways, because the initial site of sequestration occurs away from the limiting membrane of the lysosomes, and involves formation of cytosolic vesicles transporting cargo to the organelle [44]. In addition, the autophagosome is formed anew, whereas in the other pathways it is formed by expansion and budding of preexisting organelles [45].
An alternative is another classification defines autophagy based on specific organelles removed from the cytoplasm. This taxonomy includes mitophagy (removal of damaged mitochondria) which is predominantly mediated by LC3-associated autophagy receptors[42], lysophagy (lysosome), aggrephagy(protein aggregates), and lipophagy {to remove lipid droplets).
Autophagy in obesity
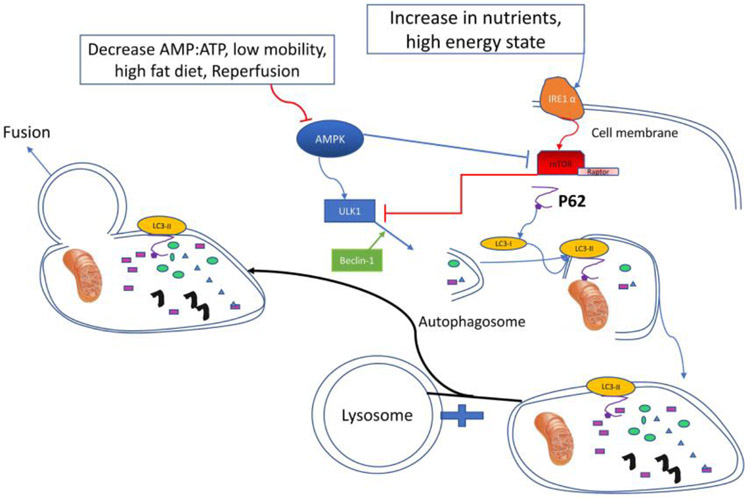
Obesity often involves a high energy state and can activate several different cellular cascades. Obesity-induced ER stress is a main mediator of cellular damage, especially in adipose tissue, and triggers an increase in inflammatory elements [46]. A central mediator of this link to ER stress is inositol-requiring enzyme (IRE)-1,α conserved transmembrane ER sensor that normally activates the unfolded protein response that monitors protein-folding to maintain the ER and cellular function. IRE-1 responds to metabolic cues and nutrient stress conditions, and its dysregulation favors pro-inflammatory M1 macrophage polarization and contributes to progression of obesity and metabolic disorders[47]. During these situations, autophagy can play a pivotal role to eliminate the insult, by removing the malfunctioning parts of the cell like ER (ER-phagy)[48]or lipid droplets[49]. This high-energy state can also increase mTOR activity (associated with increased anabolic metabolism), especially in the liver[50] (Figure 2).
Figure 2:
Inositol Requiring Enzyme-1α(IRE-1α) senses the high energy state which amplify activity of mammalian target of rapamycin (mTOR) which inhibit unc-51 like autophagy kinase 1 (ULK-1) and inhibits further Autophagy process. The most important proteins in formation and fusion of autophagosome and lysosome are microtubule-associated light chain 3(LC3) and P62. Beclin-1 can also reinforce Autophagy in high demand conditions. Low adenosine mono phosphate (AMP)portion to adenosine three phosphate(ATP) which is an indicator of low energy state can inhibit activated protein kinase(AMPK) and inhibit the circuit at its first stages.
Reliance on BMI to define obesity may be responsible for the “obesity paradox”, [51] For example, obesity has been shown to be beneficial in patients with end-stage renal disease (ESRD) undergoing dialysis[52]. Possibly higher adiposity of non-visceral fat may be associated with better outcomes. Furthermore, the J-shaped association between BMI and mortality may be offset by muscle wasting that characterize advanced CKD. Furthermore, survival studies indicate a marked contrast between protective short-term effects and adverse long-term effects of obesity in ESRD[52]. A comparable paradox may involve autophagy that has been proposed to be either inhibited or magnified in obesity. Accumulating evidence illustrates potential opposing effects of nutritional status on autophagy. The stress insult associated with superfluous nutrition in obesity can stimulate autophagy in certain tissues, whereas lipotoxicity can interfere with autophagy by inhibiting autophagosome formation or acidification[53].
For instance, ER stress can induce autophagy via multiple mechanisms but can inhibit it by others[54, 55].This apparent discrepancy might be related to differential effects of autophagy-linked gene knock-out and its molecular pattern in different tissues of different obese models fed a high-fat diet, as shown in Table 1. These molecular patterns demonstrate diverse roles of different genes and their protein in the autophagy cascade[56, 57], although additional studies are needed to reveal whether these genes also have non-autophagy-related functions, such as anti-or pro-inflammatory.
Table 1:
Hyper activated or knocked out genes in model animals kept on high fat diet, caused different results on expression of autophagy proteins
| Gene | Effected Organ | Change in autophagy |
Model animal |
|---|---|---|---|
| Becn2−/− | Whole body | Suppressed | Mice [91] |
| ATG5 overexpression | Whole body | enhanced | Mice [92] |
| ATG7+/− | Whole body | Suppressed | ob/ob mice [93] |
| Lamp2+/− | Whole body | Suppressed | Mice [94] |
| Bif1−/− | Whole body | Suppressed | Mice [95] |
| Adenoviral Tfeb overexpression | Liver | Enhanced | Mice [96] |
| Tfeb−/− | Liver | Suppressed | Mice [97] |
| Atg7−/− | Liver | Suppressed | Mice [98] |
| Adenoviral Atg7 knockdown | Liver | Enhanced | ob/ob mice [99] |
| Fip200−/− | Liver | Suppressed | Mice [100] |
| Vps34−/− | Liver | Suppressed | Mice [101] |
| Atg7−/− | Pancreas | Suppressed | Mice [102] |
| Atg7−/− | Pancreas | Suppressed | ob/ob mice [103] |
| ATG5−/− | Kidney | Suppressed | ob/ob mice [104] |
| ULK1−/− | Kidney | suppressed | Swine [105] |
In adipose tissue, especially omental, obesity up-regulates autophagy that correlates with the degree of obesity, visceral fat distribution, and adipocyte hypertrophy[58], possibly in response to ER stress. This elevated autophagy in adipose tissue in turn might contribute to insulin resistance by raising insulin receptors degradation. In contrast, decreased hepatocyte autophagy in murine obesity has also been implicated in defective insulin signaling and insulin resistance[59], and in ß-cells induces autophagic failure associated with accumulation of damaged mitochondria and increased oxidative stress[60]. Clearly, autophagy plays different roles in regulating metabolic homeostasis in different organs. Presumably, for example while autophagy can directly increase during ER stress in pancreatic ß-cells or kidney cells, obesity can also cause a decrease in degradation of autophagosomes, which in turn causes compensatory upregulation of relevant proteins, and as a result decreased autophagy in these states[61].
Furthermore, the efficiency of autophagy might vary and obscure elucidation of its role. Robust autophagy should decrease accumulation of substrate such as lipid droplets and ubiquitin adaptor protein p62/SQSTM1, whereas in many studies these substrates were found to accumulate during high energy state [50, 62, 63], suggesting inefficient autophagy. These compensatory processes complicate interpretation of the role of autophagy in metabolic regulation. A decrease of autophagy in obesity might also be mediated by inhibition of autophagosome and lysosome fusion or by a defect in lysosomal acidification[64, 65]. Although the autophagy cycle might be defective in high-energy state, this inhibition can lead to upregulation of compensatory pathways that are beneficial against consequences of obesity [66].
Regulation of kidney autophagy during obesity
In the adult kidney, basal autophagy serves as homeostatic mechanism in parenchymal cells, including podocytes, proximal tubular epithelial cells, glomerular mesangial cells and glomerular endothelial cells, are responsible for keeping renal integrity [67]. LC3 staining of the mouse kidney shows a sizeable distribution in podocytes, an indicator of constitutive autophagy in these cells [68]. This process might be required to suppress stress in high-functioning podocytes and enable recycling and degradation of cellular material in these cells with limited proliferation and turnover capacity, akin to its role in long-living neurons.
Indeed, in high-fat-diet fed mice, ectopic glomerular lipid accumulation is accompanied by increased insulin resistance, fusion of podocyte foot processes, and excessive autophagy[69]. Several studies demonstrated the role of mTOR and AMPK in podocyte’s autophagy[66, 70], although its inhibition in obesity might involve LC3 dysregulation. Proximal tubular cells show low level of basal autophagy under physiologic conditions[71].
The exact mechanism of how renal cellular autophagy is disrupted in obesity is still under investigation. Activation of compensatory autophagy in the kidneys of patients with superfluous nutrient supply may be complicated by competing process that offset this ostensible defense mechanism, such as aging[72] or diabetes. Diabetic nephropathy induces down-regulation of AMPK phosphorylation and ULK1 phosphorylation and upregulating of mTOR phosphorylation [73], and in rats sucrose-feeding suppresses beclin-1 and LC3-B [74]. In murine diabetic kidney disease, inhibition of autophagy by ATG7 ablation in proximal tubular cells aggravates renal hypertrophy, tubular injury, inflammation, fibrosis, and albuminuria, implying a protective role for autophagy (Table 1) [75]. Hence, dysregulation of nutrient and energy-sensing pathways and intracellular stress signals, including reactive oxygen species, have all been implicated in kidneys of animal models[73]. Also, mutation of ATG5 in the renal epithelium might bear a direct effect on decreasing amount of autophagy. Moreover, differences among organs and tissues may further complicate the understanding of its role in the kidney. For example, obesity upregulates mRNA levels of atg5, atg7, and lc3b in adipose tissue from people with obesity but its association with diabetes as fat depot-related and observed in visceral but not subcutaneous fat[76].
Autophagy also controls lipid metabolism in the kidney by degrading lipid droplets [77] and activating peroxisome proliferator-activated receptor (PPAR)-α. Increasing level of cholesterol and fatty acid elicits accumulation of harmful oxidized and ubiquitinated proteins and organelles in podocytes. Exerting high stress that requires high rates of autophagy for podocyte maintenance. Attenuation of the autophagy cascade accelerates damage to these cells, and in the absence of a compensatory mechanism, might lead to development of FSGS. Thus, deficient autophagy and high-mTORC1 characterize many diseases, including diabetes, aging, and obesity[24]. This impaired cycle causes further accumulation of damaged particles which increase the cellular stress level (e.g., ER stress) and will lead to further podocyte injury, and eventually, kidney malfunction and CKD [69]. Indeed in mice with highly-active mTOR gene, pharmacological suppression with rapamycin can inhibit further kidney damage[70].
Interestingly, lipophagy plays a major role not only during nutrient surplus, but also during dearth. For example, starvation in proximal tubular cells uptake fatty acid and hydrolyze lipid droplets for use. Impaired lipid-droplet degradation in autophagy-deficiant proximal tubular cells blunt ATP production and leads to cell death[61]. By the same token, excessive autophagy may also be detrimental. Upregulation of ATG5 and beclin-1 secondary to cellular stress may increase autophagy, leading to higher rate of apoptosis, and in turn kidney cellular loss. For example, cisplatin phosphorylates p38 MAPK (mitogen-activated protein kinase) and ERK, magnifying autophagy of tubular epithelial cells and thus exacerbating kidney damage [78]. Evidently, the balance of autophagy needs to remain tightly controlled to sustain cellular viability.
Therapeutic Opportunities
Several therapeutic strategies targeting deficient autophagy have been attempted, although few are available clinically, and usually for other indications. In Danon disease, a rare vacuolar myopathy associated with mutations in the LAMP2 gene, transfer of a recombinant adeno-associated virus-9 carrying human LAMP2B gene abrogated impaired autophagic flux and improved cardiac and hepatic function [79]. In neurodegenerative diseases and other conditions, enhancement of autophagy using interventions such as rapamycin [80], hypoxia-inducible factor prolyl hydroxylases inhibitors [81], or ubiquitin-specific protease deubiquitinates that are critical for protein quality control [82] have all been shown to lead to metabolic and physiologic improvements. In the kidney, pharmacologic inhibition of mTOR enhances autophagy after cisplatin treatment, and the PKC-delta inhibitor rottlerin suppresses cisplatin-induced phosphorylation of AKT, mTOR, p70S6 kinase, and ULK1, resulting in upregulation of autophagy and protection against cisplatin nephrotoxicity [83]. Furthermore, the reno-protective effect of glucagon-like peptide-1 might be achieved via an AMPK-mTOR-autophagy-reactive oxygen species signaling axis [84]. However, less is known about the effects of specific autophagy-targeting on the kidney in obesity. Future studies and clinical trials are needed to determine the suitability of autophagy as a therapeutic target in obesity.
At the same time, such development would need to be undertaken cautiously. Given that cancer cells rely on autophagy for their own metabolism and aberrant activation of autophagy may foster tumorigenesis [85], modulating autophagy might be favorable for cancer development. In addition, autophagy might favor cyst formation, suggesting that drugs related to autophagy regulation should be considered with caution for treating patients with polycystic kidney disease [86].
Summary
Obesity is a growing concern in the modern era healthcare system. Fast pace of modern life and low-nutritional benefit fast foods accelerate development of obesity, especially in developed, but also in developing countries. Obesity can initially cause glomerulomegaly which might further progress to FSGS and kidney failure. An emerging mechanism that mediates renal cellular damage involves and thereby disruption of cellular hemostasis. Autophagy has been reported to either increase or decrease in obesity and its related conditions, but most consistent signals suggest that this process is alternated prominent molecular pathways such as ATG5, mTOR and AMPK are dysregulated during obesity. In particular, in the setting of the setting of oxidative stress due to lipid oxidation and ER stress, loss of kidney integrity might be accelerated.
Conclusion
Obesity may dysregulate autophagy in divergent ways so that either a decrease or increase might be dependent to the cell type. In the kidney. Most evidence supports the notion that impaired autophagy interferes with cellular homeostasis and vitality in obesity. Additional studies are needed to establish the role of autophagy mediators as therapeutic targets to prevent kidney injury in obesity.
Table 2:
Autophagy alternation in different organs of different animals. The direction of autophagy is measured by concentration of autophagic cascade proteins
| Animal Model | Metabolic Manipulation |
Organ | Autophagy Alternation |
Reference |
|---|---|---|---|---|
| Minipigs | High fat diet | Heart | Decrease | [87] |
| Rat | Metabolic syndrome | Adipose tissue | Increase | [88] |
| Mice | High fat diet | Heart | Decrease | [89] |
| Mice | Metabolic syndrome | Liver | Increase | [90] |
| Rat | High fat diet | Peripheral Nerve | Increase | [91] |
| Rat | High fat diet | Kidney | Decrease | [92] |
Acknowledgments
This study was partly supported by NIH grant numbers DK120292, DK122734, and AG062104.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: Dr. Lerman is an advisor to AstraZeneca. The remaining author declare no conflict of interest.
References
- 1.<obesity-race-ethnicity-and-COVID-19.pdf>.
- 2.Rundle AG, et al. , COVID-19-Related School Closings and Risk of Weight Gain Among Children. Obesity (Silver Spring), 2020. 28(6): p. 1008–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, et al. , beta-Arrestins promote podocyte injury by inhibition of autophagy in diabetic nephropathy. Cell Death Dis, 2016. 7: p. e2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortega FB, Lavie CJ, and Blair SN, Obesity and Cardiovascular Disease. Circ Res, 2016. 118(11): p. 1752–70. [DOI] [PubMed] [Google Scholar]
- 5.Farahani RA, et al. , Metabolic Syndrome Impairs 3D Mitochondrial Structure, Dynamics, and Function in Swine Mesenchymal Stem Cells. Stem Cell Rev Rep, 2020. 16(5): p. 933–945. [DOI] [PubMed] [Google Scholar]
- 6.Grubbs V, et al. , Body mass index and early kidney function decline in young adults: a longitudinal analysis of the CARDIA (Coronary Artery Risk Development in Young Adults) study. Am J Kidney Dis, 2014. 63(4): p. 590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hruby A and Hu FB, The Epidemiology of Obesity: A Big Picture. Pharmacoeconomics, 2015. 33(7): p. 673–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anand SK, et al. , Entrenching role of cell cycle checkpoints and autophagy for maintenance of genomic integrity. DNA Repair (Amst), 2020. 86: p. 102748. [DOI] [PubMed] [Google Scholar]
- 9.Olaya B, et al. , Country-level and individual correlates of overweight and obesity among primary school children: a cross-sectional study in seven European countries. BMC Public Health, 2015. 15: p. 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cattaneo A, et al. , Overweight and obesity in infants and pre-school children in the European Union: a review of existing data. Obes Rev, 2010. 11(5): p. 389–98. [DOI] [PubMed] [Google Scholar]
- 11.Kawakami T, et al. , Deficient Autophagy Results in Mitochondrial Dysfunction and FSGS. J Am Soc Nephrol, 2015. 26(5): p. 1040–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith KB and Smith MS, Obesity Statistics. Prim Care, 2016. 43(1): p. 121–35, ix. [DOI] [PubMed] [Google Scholar]
- 13.Moore ML, et al. , Segmental body composition evaluation by bioelectrical impedance analysis and dual-energy X-ray absorptiometry: Quantifying agreement between methods. Clin Nutr, 2020. 39(9): p. 2802–2810. [DOI] [PubMed] [Google Scholar]
- 14.Janssen I, Katzmarzyk PT, and Ross R, Waist circumference and not body mass index explains obesity-related health risk. Am J Clin Nutr, 2004. 79(3): p. 379–84. [DOI] [PubMed] [Google Scholar]
- 15.Ashwell M and Hsieh SD, Six reasons why the waist-to-height ratio is a rapid and effective global indicator for health risks of obesity and how its use could simplify the international public health message on obesity. Int J Food Sci Nutr, 2005. 56(5): p. 303–7. [DOI] [PubMed] [Google Scholar]
- 16.van der Klaauw AA and Farooqi IS, The hunger genes: pathways to obesity. Cell, 2015. 161(1): p. 119–132. [DOI] [PubMed] [Google Scholar]
- 17.Shulman GI, Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N Engl J Med, 2014. 371(12): p. 1131–41. [DOI] [PubMed] [Google Scholar]
- 18.Coughlan MT, et al. , Mapping time-course mitochondrial adaptations in the kidney in experimental diabetes. Clin Sci (Lond), 2016. 130(9): p. 711–20. [DOI] [PubMed] [Google Scholar]
- 19.Munusamy S, et al. , Obesity-induced changes in kidney mitochondria and endoplasmic reticulum in the presence or absence of leptin. Am J Physiol Renal Physiol, 2015. 309(8): p. F731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chade AR, et al. , Effects of proteasome inhibition on the kidney in experimental hypercholesterolemia. J Am Soc Nephrol, 2005. 16(4): p. 1005–12. [DOI] [PubMed] [Google Scholar]
- 21.Conley SM, et al. , Metabolic Syndrome Induces Release of Smaller Extracellular Vesicles from Porcine Mesenchymal Stem Cells. Cell Transplant, 2019. 28(9-10): p. 1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Vries AP, et al. , Fatty kidney: emerging role of ectopic lipid in obesity-related renal disease. Lancet Diabetes Endocrinol, 2014. 2(5): p. 417–26. [DOI] [PubMed] [Google Scholar]
- 23.Yamahara K, et al. , Obesity-mediated autophagy insufficiency exacerbates proteinuria-induced tubulointerstitial lesions. J Am Soc Nephrol, 2013. 24(11): p. 1769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Agati VD, et al. , Obesity-related glomerulopathy: clinical and pathologic characteristics and pathogenesis. Nat Rev Nephrol, 2016. 12(8): p. 453–71. [DOI] [PubMed] [Google Scholar]
- 25.Tsuboi N, et al. , The Renal Pathology of Obesity. Kidney Int Rep, 2017. 2(2): p. 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collaborators, G.B.D.R.F., et al. , Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet, 2015. 386(10010): p. 2287–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeshige K, et al. , Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol, 1992. 119(2): p. 301–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang XH, et al. , Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature, 1999. 402(6762): p. 672–6. [DOI] [PubMed] [Google Scholar]
- 29.Klionsky DJ, Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol, 2007. 8(11): p. 931–7. [DOI] [PubMed] [Google Scholar]
- 30.Park JM, et al. , ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy, 2018. 14(4): p. 584–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia HZ, et al. , Hyperbranched-hyperbranched polymeric nanoassembly to mediate controllable co-delivery of siRNA and drug for synergistic tumor therapy. J Control Release, 2015. 216: p. 9–17. [DOI] [PubMed] [Google Scholar]
- 32.Alers S, et al. , The incredible ULKs. Cell Commun Signal, 2012. 10(1): p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walczak M and Martens S, Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy, 2013. 9(3): p. 424–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorincz P and Juhasz G, Autophagosome-Lysosome Fusion. J Mol Biol, 2020. 432(8): p. 2462–2482. [DOI] [PubMed] [Google Scholar]
- 35.Sattler T and Mayer A, Cell-free reconstitution of microautophagic vacuole invagination and vesicle formation. J Cell Biol, 2000. 151(3): p. 529–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uttenweiler A, Schwarz H, and Mayer A, Microautophagic vacuole invagination requires calmodulin in a Ca2+-independent function. J Biol Chem, 2005. 280(39): p. 33289–97. [DOI] [PubMed] [Google Scholar]
- 37.Mortimore GE, Hutson NJ, and Surmacz CA, Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci U S A, 1983. 80(8): p. 2179–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahu R, et al. , Microautophagy of cytosolic proteins by late endosomes. Dev Cell, 2011. 20(1): p. 131–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karmon O and Ben Aroya S, Spatial Organization of Proteasome Aggregates in the Regulation of Proteasome Homeostasis. Front Mol Biosci, 2019. 6: p. 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaushik S, et al. , Chaperone-mediated autophagy at a glance. J Cell Sci, 2011. 124(Pt 4): p. 495–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiang HL and Dice JF, Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J Biol Chem, 1988. 263(14): p. 6797–805. [PubMed] [Google Scholar]
- 42.Gatica D, Lahiri V, and Klionsky DJ, Cargo recognition and degradation by selective autophagy. Nat Cell Biol, 2018. 20(3): p. 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizushima N and Levine B, Autophagy in Human Diseases. N Engl J Med, 2020. 383(16): p. 1564–1576. [DOI] [PubMed] [Google Scholar]
- 44.Parzych KR and Klionsky DJ, An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal, 2014. 20(3): p. 460–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z and Klionsky DJ, Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol, 2010. 22(2): p. 124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Odegaard JI and Chawla A, The immune system as a sensor of the metabolic state. Immunity, 2013. 38(4): p. 644–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bujisic B and Martinon F, IRE1 gives weight to obesity-associated inflammation. Nat Immunol, 2017. 18(5): p. 479–480. [DOI] [PubMed] [Google Scholar]
- 48.Bernales S, Schuck S, and Walter P, ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy, 2007. 3(3): p. 285–7. [DOI] [PubMed] [Google Scholar]
- 49.Singh R and Cuervo AM, Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol, 2012. 2012: p. 282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ju L, et al. , Obesity-associated inflammation triggers an autophagy-lysosomal response in adipocytes and causes degradation of perilipin 1. Cell Death Dis, 2019. 10(2): p. 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gallagher D, et al. , Healthy percentage body fat ranges: an approach for developing guidelines based on body mass index. Am J Clin Nutr, 2000. 72(3): p. 694–701. [DOI] [PubMed] [Google Scholar]
- 52.Kovesdy CP, et al. , Body mass index, waist circumference and mortality in kidney transplant recipients. Am J Transplant, 2010. 10(12): p. 2644–51. [DOI] [PubMed] [Google Scholar]
- 53.Toledo M, et al. , Autophagy Regulates the Liver Clock and Glucose Metabolism by Degrading CRY1. Cell Metab, 2018. 28(2): p. 268–281 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rashid HO, et al. , ER stress: Autophagy induction, inhibition and selection. Autophagy, 2015. 11(11): p. 1956–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zahid MDK, et al. , CCAAT/enhancer-binding protein beta (C/EBPbeta) knockdown reduces inflammation, ER stress, and apoptosis, and promotes autophagy in oxLDL-treated RAW264.7 macrophage cells. Mol Cell Biochem, 2020. 463(1-2): p. 211–223. [DOI] [PubMed] [Google Scholar]
- 56.Hsu HC, et al. , Time-dependent cellular response in the liver and heart in a dietary-induced obese mouse model: the potential role of ER stress and autophagy. Eur J Nutr, 2016. 55(6): p. 2031–43. [DOI] [PubMed] [Google Scholar]
- 57.Chen Y, et al. , omega-3 Fatty acids reverse lipotoxity through induction of autophagy in nonalcoholic fatty liver disease. Nutrition, 2015. 31(11-12): p. 1423–1429 e2. [DOI] [PubMed] [Google Scholar]
- 58.Kovsan J, et al. , Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab, 2011. 96(2): p. E268–77. [DOI] [PubMed] [Google Scholar]
- 59.Yang L, et al. , Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab, 2010. 11(6): p. 467–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu H, et al. , Intermittent fasting preserves beta-cell mass in obesity-induced diabetes via the autophagy-lysosome pathway. Autophagy, 2017. 13(11): p. 1952–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Minami S, et al. , Lipophagy maintains energy homeostasis in the kidney proximal tubule during prolonged starvation. Autophagy, 2017. 13(10): p. 1629–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gonzalez-Rodriguez A, et al. , Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis, 2014. 5: p. e1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jeong SJ, et al. , p62/SQSTM1 and Selective Autophagy in Cardiometabolic Diseases. Antioxid Redox Signal, 2019. 31(6): p. 458–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mauvezin C, et al. , Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun, 2015. 6: p. 7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodriguez-Navarro JA and Cuervo AM, Dietary lipids and aging compromise chaperone-mediated autophagy by similar mechanisms. Autophagy, 2012. 8(7): p. 1152–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bork T, et al. , Podocytes maintain high basal levels of autophagy independent of mtor signaling. Autophagy, 2020. 16(11): p. 1932–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto T, et al. , Time-dependent dysregulation of autophagy: Implications in aging and mitochondrial homeostasis in the kidney proximal tubule. Autophagy, 2016. 12(5): p. 801–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Orhon I, et al. , Primary-cilium-dependent autophagy controls epithelial cell volume in response to fluid flow. Nat Cell Biol, 2016. 18(6): p. 657–67. [DOI] [PubMed] [Google Scholar]
- 69.Guo H, et al. , Glucagon-like peptide-1 analog prevents obesity-related glomerulopathy by inhibiting excessive autophagy in podocytes. Am J Physiol Renal Physiol, 2018. 314(2): p. F181–F189. [DOI] [PubMed] [Google Scholar]
- 70.Cina DP, et al. , Inhibition of MTOR disrupts autophagic flux in podocytes. J Am Soc Nephrol, 2012. 23(3): p. 412–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakai S, et al. , Proximal Tubule Autophagy Differs in Type 1 and 2 Diabetes. J Am Soc Nephrol, 2019. 30(6): p. 929–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pyo JO, et al. , Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun, 2013. 4: p. 2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X, et al. , Mangiferin prevents diabetic nephropathy progression and protects podocyte function via autophagy in diabetic rat glomeruli. Eur J Pharmacol, 2018. 824: p. 170–178. [DOI] [PubMed] [Google Scholar]
- 74.Ruiz-Ramirez A, et al. , Kidney dysfunction induced by a sucrose-rich diet in rat involves mitochondria ROS generation, cardiolipin changes, and the decline of autophagy protein markers. Am J Physiol Renal Physiol, 2020. 318(1): p. F53–F66. [DOI] [PubMed] [Google Scholar]
- 75.Ma Z, et al. , p53/microRNA-214/ULK1 axis impairs renal tubular autophagy in diabetic kidney disease. J Clin Invest, 2020. 130(9): p. 5011–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kosacka J, et al. , Autophagy in adipose tissue of patients with obesity and type 2 diabetes. Mol Cell Endocrinol, 2015. 409: p. 21–32. [DOI] [PubMed] [Google Scholar]
- 77.Schulze RJ, et al. , Direct lysosome-based autophagy of lipid droplets in hepatocytes. Proc Natl Acad Sci U S A, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lassen KG, et al. , Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A, 2014. 111(21): p. 7741–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manso AM, et al. , Systemic AAV9.LAMP2B injection reverses metabolic and physiologic multiorgan dysfunction in a murine model of Danon disease. Sci Transl Med, 2020. 12(535). [DOI] [PubMed] [Google Scholar]
- 80.Gao G, et al. , Rapamycin regulates the balance between cardiomyocyte apoptosis and autophagy in chronic heart failure by inhibiting mTOR signaling. Int J Mol Med, 2020. 45(1): p. 195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singh A, et al. , Hypoxia-inducible factor (HIF) prolyl hydroxylase inhibitors induce autophagy and have a protective effect in an in-vitro ischaemia model. Sci Rep, 2020. 10(1): p. 6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aron R, et al. , Deubiquitinase Usp12 functions noncatalytically to induce autophagy and confer neuroprotection in models of Huntington's disease. Nat Commun, 2020. 11(1): p. 2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang D, et al. , Protein Kinase Cdelta Suppresses Autophagy to Induce Kidney Cell Apoptosis in Cisplatin Nephrotoxicity. J Am Soc Nephrol, 2017. 28(4): p. 1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang S, et al. , Glucagon-like peptide-1 alleviates diabetic kidney disease through activation of autophagy by regulating AMP-activated protein kinase-mammalian target of rapamycin pathway. Am J Physiol Endocrinol Metab, 2020. 319(6): p. E1019–E1030. [DOI] [PubMed] [Google Scholar]
- 85.Chen X, et al. , Ethanol Extract of Brucea javanica Seed Inhibit Triple-Negative Breast Cancer by Restraining Autophagy via PI3K/Akt/mTOR Pathway. Front Pharmacol, 2020. 11: p. 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee EJ, et al. , Autophagy induction promotes renal cyst growth in polycystic kidney disease. EBioMedicine, 2020. 60: p. 102986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li SJ, et al. , The high-fat diet induces myocardial fibrosis in the metabolically healthy obese minipigs-The role of ER stress and oxidative stress. Clin Nutr, 2017. 36(3): p. 760–767. [DOI] [PubMed] [Google Scholar]
- 88.Kosacka J, et al. , Up-regulated autophagy: as a protective factor in adipose tissue of WOKW rats with metabolic syndrome. Diabetol Metab Syndr, 2018. 10: p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Andres AM, et al. , Discordant signaling and autophagy response to fasting in hearts of obese mice: Implications for ischemia tolerance. Am J Physiol Heart Circ Physiol, 2016. 311(1): p. H219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu TY, et al. , FNDC5 Alleviates Hepatosteatosis by Restoring AMPK/mTOR-Mediated Autophagy, Fatty Acid Oxidation, and Lipogenesis in Mice. Diabetes, 2016. 65(11): p. 3262–3275. [DOI] [PubMed] [Google Scholar]
- 91.Kosacka J, et al. , Increased autophagy in peripheral nerves may protect Wistar Ottawa Karlsburg W rats against neuropathy. Exp Neurol, 2013. 250: p. 125–35. [DOI] [PubMed] [Google Scholar]
- 92.Ruiz-Ramirez A, et al. , Kidney dysfunction induced by a sucrose-rich diet in rat involves mitochondria ROS generation, cardiolipin changes, and the decline of autophagy protein markers. Am J Physiol Renal Physiol, 2020. 318(1): p. F53–F66. [DOI] [PubMed] [Google Scholar]
- 93.Kuramoto K, et al. , Autophagy activation by novel inducers prevents BECN2-mediated drug tolerance to cannabinoids. Autophagy, 2016. 12(9): p. 1460–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peng X, et al. , ATG5-mediated autophagy suppresses NF-kappaB signaling to limit epithelial inflammatory response to kidney injury. Cell Death Dis, 2019. 10(4): p. 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lim YM, et al. , Systemic autophagy insufficiency compromises adaptation to metabolic stress and facilitates progression from obesity to diabetes. Nat Commun, 2014. 5: p. 4934. [DOI] [PubMed] [Google Scholar]
- 96.Terasawa K, et al. , Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes. Biochem Biophys Res Commun, 2016. 479(3): p. 489–495. [DOI] [PubMed] [Google Scholar]
- 97.Frangez Z, et al. , BIF-1 inhibits both mitochondrial and glycolytic ATP production: its downregulation promotes melanoma growth. Oncogene, 2020. 39(26): p. 4944–4955. [DOI] [PubMed] [Google Scholar]
- 98.Wang S, et al. , Tumor necrosis factor-inducible gene 6 protein ameliorates chronic liver damage by promoting autophagy formation in mice. Exp Mol Med, 2017. 49(9): p. e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim JS, et al. , Impaired autophagy: A mechanism of mitochondrial dysfunction in anoxic rat hepatocytes. Hepatology, 2008. 47(5): p. 1725–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kongara S and Karantza V, The interplay between autophagy and ROS in tumorigenesis. Front Oncol, 2012. 2: p. 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lodder J, et al. , Macrophage autophagy protects against liver fibrosis in mice. Autophagy, 2015. 11(8): p. 1280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iwahashi K, et al. , Autophagy impairment in pancreatic acinar cells causes zymogen granule accumulation and pancreatitis. Biochem Biophys Res Commun, 2018. 503(4): p. 2576–2582. [DOI] [PubMed] [Google Scholar]
- 103.Quan W, Jung HS, and Lee MS, Role of autophagy in the progression from obesity to diabetes and in the control of energy balance. Arch Pharm Res, 2013. 36(2): p. 223–9. [DOI] [PubMed] [Google Scholar]
- 104.Li H, et al. , Atg5-mediated autophagy deficiency in proximal tubules promotes cell cycle G2/M arrest and renal fibrosis. Autophagy, 2016. 12(9): p. 1472–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu B, et al. , Hyperoside attenuates renal aging and injury induced by D-galactose via inhibiting AMPK-ULK1 signaling-mediated autophagy. Aging (Albany NY), 2018. 10(12): p. 4197–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]