Abstract
Background
EGFR amplification occurs in about 1% of metastatic colorectal cancers (mCRCs) but is not routinely tested as a prognostic or predictive biomarker for patients treated with anti-EGFR monoclonal antibodies. Herein, we aimed to characterize the clinical and molecular landscape of EGFR-amplified mCRC.
Methods
In this multinational cohort study, we compared clinical data of 62 patients with EGFR-amplified vs 1459 EGFR nonamplified mCRC, as well as comprehensive genomic data of 35 EGFR-amplified vs 439 EGFR nonamplified RAS/BRAF wild-type and microsatellite stable (MSS) tumor samples. All statistical tests were 2-sided.
Results
EGFR amplification was statistically significantly associated with left primary tumor sidedness and RAS/BRAF wild-type status. All EGFR-amplified tumors were MSS and HER2 nonamplified. Overall, EGFR-amplified samples had higher median fraction of genome altered compared with EGFR-nonamplified, RAS/BRAF wild-type MSS cohort. Patients with EGFR-amplified tumors reported longer overall survival (OS) (median OS = 71.3 months, 95% confidence interval [CI] = 50.7 to not available [NA]) vs EGFR-nonamplified ones (24.0 months; 95% CI = 22.8 to 25.6; hazard ratio [HR] = 0.30, 95% CI = 0.20 to 0.44; P < .001; adjusted HR = 0.46, 95% CI = 0.30 to 0.69; P < .001). In the subgroup of patients with RAS/BRAF wild-type mCRC exposed to anti-EGFR-based therapy, EGFR amplification was again associated with better OS (median OS = 54.0 months, 95% CI = 35.2 to NA, vs 29.1 months, 95% CI = 27.0 to 31.9, respectively; HR = 0.46, 95% CI = 0.28 to 0.76; P = .002).
Conclusion
Patients with EGFR-amplified mCRC represent a biologically defined subgroup and merit dedicated clinical trials with novel and more potent EGFR-targeting strategies beyond single-agent monoclonal antibodies.
The anti-EGFR monoclonal antibodies (mAbs) cetuximab and panitumumab are used in patients with RAS/BRAF wild-type metastatic colorectal cancer (mCRC). Both primary tumor sidedness and molecular selection beyond RAS and BRAF mutational status help refine the identification of patients with higher chance of benefit from EGFR inhibition, for example, those with left-sided primary tumors and lack of rare primary resistance alterations (HER2/MET amplification, gene fusions, PIK3CA/PTEN pathway deregulation, and microsatellite instability [MSI-high]) (1,2). Even after initial tumor responses, the emergence of secondary resistance almost invariably limits the long-term efficacy of EGFR blockade. The clinical experience suggests that an extremely limited subset of patients shows exceptional and long-lasting responses to anti-EGFR-based therapies even across multiple lines. Therefore, although the implementation of biomarkers has proceeded over time according to the paradigm of negative selection, preclinical evidence suggested that some biomarkers, such as EGFR amplification and IRS2 amplification or activating mutations, may have a positive predictive value (3).
EGFR dependency in colorectal cancer relies on the functional activation of the receptor by high levels of endogenous ligands, such as AREG and EREG, and accounts for the therapeutic efficacy of anti-EGFR mAbs in most of well-selected patients (4, 5). However, EGFR constitutive activation secondary to gene amplification might define a small subset of oncogene-addicted colorectal tumors for which cetuximab or panitumumab might be used as the matched targeted therapy.
Considering the rarity of patients with EGFR-amplified mCRC, the association between EGFR amplification and specific clinical and molecular features and the outcomes with anti-EGFR mAbs have not been addressed yet. Moreover, it is not clear whether this molecular subgroup may deserve the personalized development of new anti-EGFR strategies. Based on these considerations, we conducted a multinational effort aimed at investigating the landscape of EGFR amplification in patients with mCRC.
Methods
Patient Population
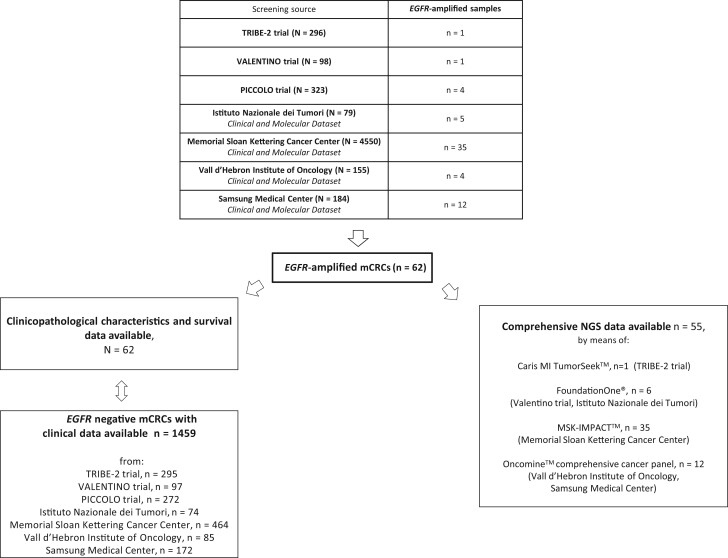
As shown in Figure 1, 62 patients with EGFR-amplified mCRC were retrieved from 7 screening sources: TRIBE-2 trial (6), VALENTINO trial (7), PICCOLO trial (8), and prospective datasets established at Fondazione IRCCS Istituto Nazionale dei Tumori di Milano, Memorial Sloan Kettering Cancer Center, Vall d’Hebron Institute of Oncology, and Samsung Medical Center. We also retrieved clinical data from 1459 patients bearing EGFR-nonamplified mCRC that served as a retrospective control cohort (see Figure 1). The assays used to test for EGFR amplification for each screening platform are detailed in Supplementary Table 1 (available online). Briefly, all cases were assessed by next-generation sequencing (NGS) and required at least 6 EGFR copies. In cases with available tumor tissue, in situ hybridization was also performed. In situ hybridization criteria to identify EGFR amplification were EGFR/CEP7 ratio of at least 2 and the presence of EGFR gene clusters or at least 15 gene copies in at least 10% of cells (9). The study was approved by the Fondazione IRCCS Istituto Nazionale dei Tumori di Milano institutional review board (INT 117/15) and was conducted in accordance with the ethical principles for medical research involving human subjects adopted in the Declaration of Helsinki.
Figure 1.
Study flow diagram. mCRC = metastatic colorectal cancer; NGS = next-generation sequencing.
Molecular Analyses
Comprehensive NGS data were available for 55 out of 62 EGFR-amplified tumors. NGS assays for every screening platform are detailed in Figure 1 and Supplementary Table 1 (available online). Genes present in all gene panels were kept for the analyses. Frequency of genomic alterations including mutations, copy number variations (CNVs), and gene fusions were compared with those reported in a subgroup of RAS/BRAF wild-type, microsatellite stable (MSS) mCRC (10). CNV data were acquired as .seg files from cBioportal (10) for both EGFR-amplified and -nonamplified samples and processed with the copynumber package (11).
For patients with available plasma samples in concomitance with the emergence of secondary resistance to anti-EGFR-based therapy, digital droplet polymerase chain reaction was performed to detect KRAS, NRAS, BRAF and EGFR extracellular domain (ECD) mutations in circulating tumor DNA, as previously described (12).
Statistical Analyses
Overall survival (OS) was calculated as the time from diagnosis of metastatic disease until death or last follow-up for alive patients. Association between EGFR amplification status and patients or/and disease characteristics was assessed by means of Kruskal-Wallis, χ2, or Fisher exact test, as appropriate. The Kaplan-Meier estimator and Cox proportional hazards regression were used for survival analysis with the survival, survminer, and survMisc packages. Follow-up time was estimated using the reverse Kaplan-Meier method. In Cox proportional hazards regression models, all the covariates statistically significantly associated with OS at the univariable analyses were included in the multivariable model. The assumption of proportionality was verified by means of the Grønnesby and Borgan overall goodness-of-fit test (13). EGFR CNV was modeled by means of 3-knots natural cubic splines (using the splines package) to assess flexible fit and to check for nonlinearity (14). Furthermore, a maximally selected log-rank statistic method for OS was used to find an optimal cutoff value of EGFR CNV using the maxstat package (15). All tests were 2-sided at α equals 5%. Statistical analyses were performed using the R (version 3.5.0) and R Studio (version 1.1.447).
Results
Patient Population
The overall prevalence of EGFR amplification in the screened population was 1.1% (62 out of 5685 screened samples). Baseline features and clinical outcomes of patients with EGFR-amplified mCRC were compared with a cohort of 1459 patients without EGFR amplification (Figure 1).
Clinical and Pathological Characteristics of EGFR-Amplified mCRC
As shown in Table 1, EGFR-amplified mCRCs were located more frequently in the rectum (48.4% vs 23.3%), and only 5 cases were right-sided. Regarding mutational profile, EGFR-amplified cancers were more frequently RAS/BRAF wild type (95.2% vs 48.7%), and all of them were MSS. As expected, history of exposure to anti-EGFR therapy was more frequent in patients with EGFR-amplified vs -nonamplified cancers (56.5% vs 36.0%). Primary tumor resection (74.2% vs 50.7%) and 1 single site of metastases (71.0% vs 45.5%) were also statistically significantly associated with EGFR amplification. Of note, RAS/BRAF wild-type mCRCs arising from the left colon or rectum had a 10.70-fold higher chance of bearing EGFR amplification compared with other cases (odds ratio [OR] = 10.70, 95% confidence interval [CI] = 5.34 to 24.66; P < .001).
Table 1.
Patients and disease characteristics in the entire study population and according to EGFR status
| Characteristics | Total (N = 1521) |
EGFR-non-amplified (n = 1459) |
EGFR-amplified (n = 62) |
P a |
|---|---|---|---|---|
| Age, y | .07 | |||
| Median (IQR) | 60 (51-67) | 60 (51-68) | 56 (50-62) | |
| Sex, No. (%) | .15 | |||
| Female | 710 (46.7) | 687 (47.1) | 23 (37.1) | |
| Male | 811 (53.3) | 772 (52.9) | 39 (62.9) | |
| ECOG PS, No. (%) | .30 | |||
| 0 | 807 (54.6) | 769 (54.3) | 38 (61.3) | |
| 1-2 | 670 (45.4) | 646 (45.7) | 24 (38.7) | |
| Primary tumor location, No. (%) | <.001 | |||
| Rectum | 368 (24.3) | 338 (23.3) | 30 (48.4) | |
| Left colon | 675 (44.7) | 648 (44.7) | 27 (43.5) | |
| Right colon | 468 (31.0) | 463 (32.0) | 5 (8.1) | |
| Primary tumor resection, No. (%) | <.001 | |||
| Yes | 647 (51.9) | 601 (50.7) | 46 (74.2) | |
| No | 600 (48.1) | 584 (49.3) | 16 (25.8) | |
| Synchronous metastases, No. (%) | .07 | |||
| No | 313 (25.1) | 291 (24.5) | 22 (35.5) | |
| Yes | 936 (74.9) | 896 (75.5) | 40 (64.5) | |
| No. of metastatic sites, No. (%) | <.001 | |||
| 1 | 702 (46.5) | 658 (45.5) | 44 (71.0) | |
| >1 | 807 (53.5) | 789 (54.5) | 18 (29.0) | |
| RAS/BRAF status, No. (%) | .002 | |||
| All wild type | 768 (50.6) | 709 (48.7) | 59 (95.2) | |
| RAS mutated | 623 (41.0) | 621 (42.7) | 2 (3.2) | |
| BRAF mutated | 127 (8.4) | 126 (8.6) | 1 (1.6) | |
| MSI status, No. (%) | .19 | |||
| MSS | 1240 (96.0) | 1178 (95.8) | 62 (100) | |
| MSI-high | 52 (4.0) | 52 (4.2) | 0 (0) | |
| Anti-EGFR therapy, No. (%) | .002 | |||
| Yes | 555 (63.1) | 520 (36.0) | 35 (56.5) | |
| No | 950 (36.9) | 923 (64.0) | 27 (43.5) |
P values were based on Fisher exact test, χ2, or Kruskal-Wallis test, whenever appropriate. All statistical tests were 2-sided. ECOG PS = Eastern Cooperative Oncology Group performance status; IQR = interquartile range; MSI-high = microsatellite instability high; MSS = microsatellite stable.
Molecular Characteristics of EGFR-Amplified mCRC
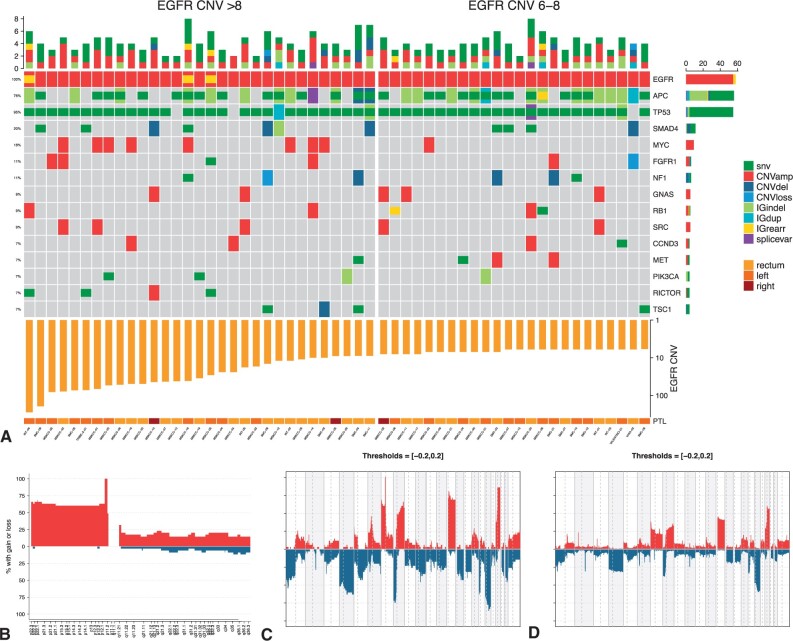
Comprehensive NGS data were available for 55 EGFR-amplified tumors, with 134 genes sequenced in all samples (Figure 1). The alterations profiles are depicted in the heatmap in Figure 2, A. Notably, TP53 was the most frequently mutated gene, with alterations occurring in almost all samples (52 of 55, 94.5%). Among the others, APC and SMAD4 were found altered in at least 20% of the samples.
Figure 2.
EGFR-amplified samples alterations profile. A) Heatmap showing the genomic profiles of patients with EGFR amplification. B) Copy number variation frequency for chromosome 7 of EGFR-amplified samples (EGFR is located in the p arm of chromosome 7). Gains are shown in red and losses are shown in blue; the threshold for both gains and losses is set at 0.5. C) and (D) show copy number variation frequencies for, respectively, patients with and without EGFR amplification. amp = amplification; CNV = copy number variation; del = deletion; dup = duplication; IG = intragenic; rear = rearrangement; splicevar = splice variant.
We then investigated whether EGFR amplification was associated with particular patterns of either gene amplifications or chromosomal alterations. To reduce potential biases related to the heterogeneity of sequencing platforms, we restricted our analysis to the Memorial Sloan Kettering Cancer Center cohort including 439 EGFR-nonamplified, RAS/BRAF wild type, MSS controls as described in Yaeger et al. (10). HER2 amplification was found in 26 of 439 EGFR nonamplified cases, whereas none of the 35 EGFR-amplified carried this alteration (5.9% vs 0%; P = .24), possibly suggesting their mutual exclusivity as drivers of oncogene addiction. Notably, samples with EGFR amplification were enriched in the amplification of the p arm of chromosome 7 (P < .001; Figure 2, B). The complete copy number alteration profiles of both EGFR-amplified and -nonamplified tumors are depicted in Figure 2, C and D; overall, EGFR-amplified samples were enriched in chromosomal gains and losses, and they showed a higher median fraction of genome-altered compared with nonamplified samples (35.2% vs 21.8%; P < .001; Supplementary Figure 1, available online).
Prognostic Impact of EGFR Amplification in mCRC
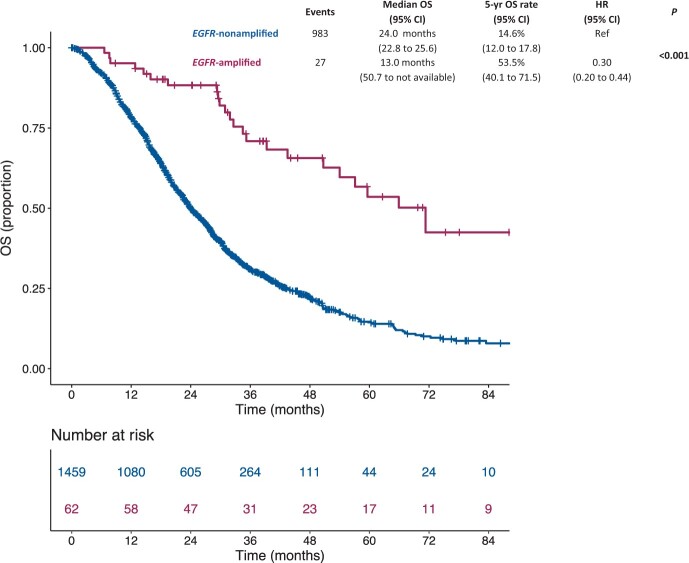
Median follow-up time was 40.3 (interquartile range [IQR] = 28.7-58.9) months. Patients with EGFR-amplified mCRC showed a better OS (median OS = 71.3 months [95% CI = 50.7 to NA; 5-year OS rate = 53.5% [95% CI = 40.1% to 71.5%]) compared with patients with EGFR-nonamplified cancers (median OS = 24.0 months [95% CI = 22.8 to 25.6]; 5-year OS rate = 14.6% [95% CI = 12.0% to 17.8%]; hazard ratio [HR] = 0.30, 95% CI = 0.20 to 0.44, P < .001) (Figure 3). Similar results were observed in the subgroup of patients with RAS/BRAF wild-type status (Supplementary Figure 2, available online). In the multivariable model including the other characteristics statistically significantly associated with OS (ie, age, sex, Eastern Cooperative Oncology Group performance status, primary tumor location, primary tumor resection, presence of synchronous metastases, number of metastatic sites, and RAS/BRAF status), the presence of EGFR amplification had an independent positive prognostic impact (adjusted HR = 0.46, 95% CI = 0.30 to 0.69; P < .001), whereas the exposure to anti-EGFR therapy was no longer statistically significant (adjusted HR = 0.99, 95% CI = 0.80 to 1.22; P = .90; Table 2).
Figure 3.
Kaplan-Meier estimates of overall survival according to the presence of EGFR amplification in the entire study population (N = 1521). Blue lines indicate patients with EGFR nonamplified mCRC (n = 1459), whereas violet lines indicate patients with EGFR-amplified mCRC (n = 62). Patients with EGFR-amplified mCRC showed a better overall survival compared with patients with EGFR-nonamplified mCRC. CI = confidence interval; HR = hazard ratio; mCRC = metastatic colorectal cancer; OS = overall survival.
Table 2.
Cox proportional hazards regression models for overall survival
| Characteristics | Univariable analysis |
Multivariable model |
||
|---|---|---|---|---|
| HR (95% CI) | P a | HR (95% CI) | P a | |
| Age, 10 years increase | 1.09 (1.03 to 1.15) | .001 | 1.08 (1.01 to 1.15) | .02 |
| Sex | .03 | .06 | ||
| Female | Referent | Referent | ||
| Male | 0.87 (0.77 to 0.98) | 0.86 (0.74 to 1.01) | ||
| ECOG PS | <.001 | <.001 | ||
| 0 | Referent | Referent | ||
| 1-2 | 1.51 (1.33 to 1.72) | 1.49 (1.28 to 1.74) | ||
| Primary tumor location | <.001 | .23 | ||
| Rectum | Referent | Referent | ||
| Left colon | 0.90 (0.77 to 1.05) | 0.84 (0.69 to 1.02) | ||
| Right colon | 1.20 (1.02 to 1.42) | 0.92 (0.74 to 1.14) | ||
| Primary tumor resection | <.001 | <.001 | ||
| Yes | Referent | Referent | ||
| No | 1.70 (1.47 to 1.96) | 1.47 (1.23 to 1.75) | ||
| Synchronous metastases | <.001 | .03 | ||
| No | Referent | Referent | ||
| Yes | 1.42 (1.19 to 1.69) | 1.25 (1.02 to 1.52) | ||
| No. of metastatic sites | <.001 | <.001 | ||
| 1 | Referent | Referent | ||
| >1 | 1.67 (1.47 to 1.90) | 1.34 (1.15 to 1.56) | ||
| RAS/BRAF status | <.001 | <.001 | ||
| All wild type | Referent | Referent | ||
| RAS mutated | 1.23 (1.07 to 1.40) | 1.36 (1.10 to 1.68) | ||
| BRAF mutated | 2.57 (2.08 to 3.17) | 2.41 (1.78 to 3.27) | ||
| MSI status | .62 | |||
| MSS | Referent | |||
| MSI | 1.10 (0.75 to 1.60) | |||
| EGFR status | <.001 | <.001 | ||
| Not amplified | Referent | Referent | ||
| Amplified | 0.30 (0.20 to 0.44) | 0.46 (0.30 to 0.69) | ||
| Anti-EGFR therapy | .002 | .90 | ||
| No | Referent | Referent | ||
| Yes | 0.81 (0.71 to 0.92) | 0.99 (0.80 to 1.22) | ||
P values are based on the Likelihood ratio test, 2-sided, in Cox proportional hazard regression analyses. ECOG PS = Eastern Cooperative Oncology Group performance status; CI = confidence interval; HR = hazard ratio; MSI = microsatellite instability high; MSS = microsatellite stable.
Implications of EGFR Amplification in Patients With RAS/BRAF Wild-Type mCRC Treated With anti-EGFR Agents
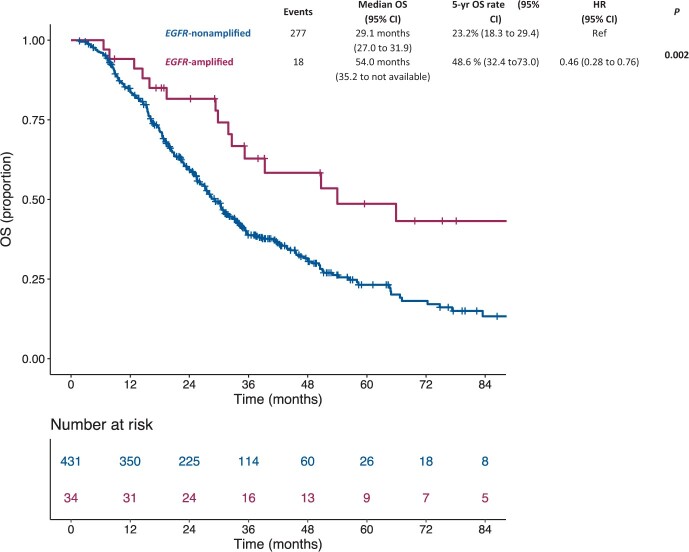
In the subgroup of 465 patients with RAS/BRAF wild-type mCRC who received an anti-EGFR-containing regimen during their disease history, 34 had EGFR amplification, and 431 were EGFR nonamplified. EGFR amplification was associated with a statistically significantly better OS compared with EGFR-nonamplified status (median OS = 54.0 months [95% CI = 35.2 to NA] vs 29.1 months [95% CI = 27.0 to 31.9]; 5-year OS rate = 48.6% [95% CI = 32.4% to 73.0%] vs 23.2% [95% CI = 18.3% to 29.4%]; HR = 0.46, 95% CI = 0.28 to 0.76, P = .002) (Figure 4). Notably, among patients with RAS/BRAF wild-type and EGFR-amplified tumors, there was no statistically significant OS difference according to exposure to anti-EGFR therapy or not (P = .21; data not shown). Treatment data and clinical outcomes in individual patients with EGFR amplification are shown in Supplementary Table 2 (available online).
Figure 4.
Kaplan-Meier estimates of overall survival according to the presence of EGFR amplification in the subgroup of patients with RAS/BRAF wild-type mCRC treated with anti-EGFR agents (n = 465). Blue lines indicate patients with EGFR-nonamplified mCRC (n = 431), whereas violet lines indicate patients with EGFR-amplified mCRC (n = 34). Patients with EGFR-amplified mCRC showed a better overall survival compared with patients with EGFR nonamplified mCRC. P values were calculated by means of the likelihood ratio test, 2-sided, in a Cox univariable regression model. CI = confidence interval; HR = hazard ratio; mCRC = metastatic colorectal cancer; OS = overall survival.
In the subgroup of patients with EGFR-amplified, RAS/BRAF wild-type mCRC who received an anti-EGFR-based option during their disease history, EGFR CNV status was available for 34 patients (Supplementary Table 2, available online). Regarding CNV as a continuous variable, we observed a nonlinear effect on the log-hazard function (Supplementary Figure 3, A, available online). Starting from this observation, we then modeled a cutoff for EGFR CNV and found that patients with 6-8 EGFR copies had better OS compared with those with EGFR CNV of more than 8 (5-year OS rate = 90.0% [95% CI = 73.2% to 100%] vs 24.5% [95% CI = 9.8% to 61.2%]; HR = 5.28, 95% CI = 1.47 to 18.97, P = .01) (Supplementary Figure 3, C, available online). We sought to investigate whether EGFR-amplified mCRCs with higher EGFR CNV are enriched with on-target genomic co-alterations, which may impair single-agent EGFR targeting with mAbs (Supplementary Table 2, available online). We discovered the novel EGFR-LANCL2 fusion in 1 sample bearing 250 EGFR copies (Supplementary Figure 4, available online), and we found 2 EGFR deletions: one involving exons 25-28 in one sample with 41 EGFR copies as already described in Cho et al. (16) and one involving exons 5-7 in one sample with 29 gene copies. EGFR pathogenic mutations were never detected in EGFR-amplified subgroup. Finally, liquid biopsies obtained at the time of secondary resistance to anti-EGFR-based therapy were available for a small subgroup of 5 patients and were analyzed for the presence of RAS, BRAF, and EGFR ECD mutations. Notably, although no alterations were detected in 2 samples possibly because of low amounts of circulating tumor DNA, only EGFR ECD mutations but no RAS/BRAF-acquired mutations were detected in the 3 analyzed cases (Supplementary Table 2 and Supplementary Figure 5, available online).
Discussion
Anti-EGFR agents cetuximab or panitumumab are recommended in eligible patients with RAS/BRAF wild-type, left-sided mCRC, independently from the presence of other molecular alterations, especially positive predictors such as EGFR amplification (17). Moreover, investigating the role of EGFR amplification as a driver of oncogene addiction is challenging because of its low prevalence (about 1%). In recent years, HER2 amplification (found in 3% of all mCRCs) has been increasingly recognized as a relevant therapeutic target for several anti-HER2 targeted strategies (18). Here, we showed that, as for HER2 overexpression or amplification, EGFR amplification is highly enriched in left-sided, RAS/BRAF wild-type, and MSS tumors. These associations are even stronger than those reported in HER2-amplified tumors: in fact, RAS/BRAF co-mutations or right-sided primary site are extremely uncommon in EGFR-amplified subgroup. Finally, HER2 and EGFR amplifications are mutually exclusive in our dataset.
Although EGFR amplification was associated with positive prognostic features such as RAS/BRAF wild-type status and left-sided primary tumor location, it was independently associated with better OS: notably, the 5-year OS rate of patients with EGFR-amplified mCRC was 53.5%. Several explanations may be hypothesized for such a robust impact. In fact, EGFR amplification may be associated with enhanced chemosensitivity, as described for glioblastoma (19), and may be theoretically predictive of exceptional efficacy of anti-EGFR mAbs. In our dataset, the overall outcome of the EGFR-amplified population was excellent, and the anti-EGFR-based regimens and treatment lines were highly heterogeneous. Because of these reasons, we could not demonstrate a positive predictive role of EGFR amplification. Moreover, exposure to anti-EGFR therapy was not statistically significantly associated with OS, suggesting that EGFR amplification may be just a prognostic factor or, if anything, a mixed prognostic-predictive one.
A relevant matter of discussion is whether monotherapy with anti-EGFR agents achieve sufficient target inhibition in EGFR-amplified CRCs. The lesson learned from HER2-amplified CRC is based on the lack of effectiveness of either treatment with mAbs or tyrosine-kinase inhibitors alone and the role of dual HER2 inhibition as the treatment mainstay. In patients-derived xenografts of EGFR-amplified gastroesophageal cancer, we previously showed that dual EGFR blockade with a mAb plus a tyrosine-kinase inhibitor had superior efficacy compared with single agent anti-EGFR treatments (20). Consistently, a recent post hoc analysis of the REAL-3 trial showed that the presence of EGFR amplification was not a positive predictor of greater efficacy from the addition of panitumumab to initial triplet chemotherapy (21). Based on all of these considerations, optimized EGFR blockade strategies are warranted in the small but relevant subgroup of patients with EGFR-amplified mCRC.
Although acknowledging the lack of CRC-specific data on differential efficacy of the approved mAbs according to the presence of EGFR amplification or not, we attempted to investigate further biomarkers that may stratify outcomes in the subgroup of patients with EGFR-amplified mCRC and exposed to anti-EGFR-based therapy. Considering that the typical alterations of primary resistance to anti-EGFR agents beyond RAS and BRAF (1) are anectodical in EGFR-amplified subtype, we focused on EGFR CNV as an immediately available variable. Paradoxically, we noted that patients with EGFR hyperamplification had inferior outcomes compared with those with relatively lower CNV. Such results must be interpreted with extreme caution because of technical reproducibility, lack of external validation of CNV cutoff, and small sample size. However, the development of more efficient EGFR-targeting strategies other than single-agent mAbs could be even more important for patients with higher EGFR CNV. Indeed, in these cancers with EGFR hyperamplification, we found the presence of complex on-target EGFR rearrangements, such as large deletions and the newly described EGFR-LANCL2 fusion. Liquid biopsies obtained in selected clinical cases at the time of secondary resistance to anti-EGFR mAbs showed the emergence of on-target EGFR ECD mutations, but not RAS/BRAF mutations, consistent with the evidence previously reported for patients with longer responses (22) and for clinical case of EGFR-amplified gastric cancer (23). Based on these considerations on the importance of EGFR as a CRC driver gene, we assume that patients with EGFR-amplified mCRC are the optimal candidates for more potent EGFR inhibition strategies and potential chemo-free options, such as second-generation oligoclonal antibodies targeting multiple epitopes of EGFR or anti-EGFR antibody drug conjugates (24). Because of the low frequency of these alterations, we therefore advocate the design of umbrella studies of novel EGFR inhibition strategies in multiple tumor types including mCRC and gastroesophageal cancers, thus potentially leading to agnostic trials.
Our study has several limitations, including the retrospective data collection and the major reliance of the overall dataset on institutional databases—particularly the Memorial Sloan Kettering Cancer Center one—rather than clinical trials. Second, the heterogeneity of anti-EGFR-based regimens and treatment lines prevented a reliable analysis on the potential predictive role of EGFR amplification. Third, the NGS platforms used in the several screening sources were clearly different. Because NGS analyzes both tumor and stromal DNA, this technique may underestimate the true prevalence of EGFR amplification by excluding cases with relatively lower gene copy number and higher grade of genomic heterogeneity, especially when the tumor cellularity of the sample is low. On the other hand, the adoption of 6 CNV cutoff to identify EGFR amplification reasonably reflected the presence of a homogeneous pattern of EGFR amplification and thus prevented the well-known reproducibility issues of in situ hydridization assay alone (25). Finally, interlesion heterogeneity of EGFR status may have influenced the outcomes of patients included in this cohort, and liquid biopsy data are warranted.
In conclusion, the spread availability of comprehensive genomic profiling for patients with mCRC allows the concomitant assessment of guideline-recommended biomarkers and further actionable drivers such as EGFR status, favoring patients’ inclusion in clinical trials with innovative drugs (26). Noteworthy, we highlighted the importance of testing for EGFR amplification in patients with mCRC, because EGFR-amplified mCRC represents a specific subtype with a favorable prognosis and is worthy of dedicated clinical trials with novel EGFR-targeting drugs.
Funding
AIRC under IG 2019 - ID. 23624 project—P.I. Pietrantonio Filippo (FP). National Institutes of Health R01 CA233736—Yaeger Rona (RY). FONDAZIONE AIRC under 5 per Mille 2018 - ID. 21091 program—P.I. Bardelli Alberto (AB); AIRC under IG 2018 - ID. 21923 project—P.I. Bardelli Alberto (AB); AIRC-CRUK-FC AECC Accelerator Award contract 22795 (AB).
Notes
Role of the funders: The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Disclosures: Filippo Pietrantonio (FP) reports personal fees from Amgen, Roche, Sanofi, Bayer, Servier, Merck-Serono, Lilly; advisory role with Amgen, Bayer, Servier, Merk-Serono and research grants from BMS. Rona Yaeger (RY) has received research funding from Array BioPharma/Pfizer and Boehringer Ingelheim and has consulted for Array BioPharma and Natera. Elena Élez (EE) reports personal fees from Hoffman La—Roche, Bristol Myers Squibb, Servier, Amgen, Merck Serono, ArrayBiopharma, Sanofi. Chiara Cremolini (CC) reports personal fees from Roche, Amgen, Bayer, and Servier; research funding from Merck Serono; and a consulting or advisory role with Roche, Bayer, Amgen. Daniele Rossini (DR) reports personal fees from Takeda.
Author contributions: Conceptualization—FP (Filippo Pietrantonio), GR. Data curation—GR, RY, JFH, JL, EE, JS, HW (Henry Walch), FP (Filippo Pagani), MMG, MA, DR, MR, FS, SDR, HW (Henry Wood), GN, AG. Formal Analysis—GR, RY, GH, PM, GF, JL, EE, JS, AG, MM, AB, FdB, FM, CC, FP. Funding acquisition—FP, RY, AB. Investigation—All the authors. Methodology—FP, PM, HW, GF. Project administration—FP. Resources FP, RY, JL, EE, JS, MR, AB, CC. Software—PM, HW, GF. Supervision—FP, CC, FdB. Validation—FP. Visualization—FP. Writing—original draft—FP, GR, RY, JFH. Writing—review & editing—FP, GR.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author.
Supplementary Material
References
- 1. Morano F, Corallo S, Lonardi S, et al. Negative hyperselection of patients with RAS and BRAF wild-type metastatic colorectal cancer who received panitumumab-based maintenance therapy. J Clin Oncol. 2019;37(33):3099–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cremolini C, Morano F, Moretto R, et al. Negative hyper-selection of metastatic colorectal cancer patients for anti-EGFR monoclonal antibodies: the PRESSING case-control study. Ann Oncol. 2017;28(12):3009–3014. [DOI] [PubMed] [Google Scholar]
- 3. Bertotti A, Papp E, Jones S, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526(7572):263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seligman JF, Elliott F, Richman SD, et al. Combined epiregulin and amphiregulin expression levels as a predictive biomarker for panitumumab therapy benefit or lack of benefit in patients with RAS wild-type advanced colorectal cancer. JAMA Oncol. 2016;2(5):633–642. [DOI] [PubMed] [Google Scholar]
- 5. Stahler A, Stintzing S, Modest DP, et al. Amphiregulin expression is a predictive biomarker for EGFR inhibition in metastatic colorectal cancer: combined analysis of three randomized trials. Clin Cancer Res. 2020;26(24):6559–6567. [DOI] [PubMed] [Google Scholar]
- 6. Cremolini C, Antoniotti C, Rossini D, et al. ; GONO Foundation Investigators. Upfront FOLFOXIRI plus bevacizumab and reintroduction after progression versus mFOLFOX6 plus bevacizumab followed by FOLFIRI plus bevacizumab in the treatment of patients with metastatic colorectal cancer (TRIBE2): a multicentre, open-label, phase 3, randomised, controlled trial. Lancet Oncol. 2020;21(4):497–507. [DOI] [PubMed] [Google Scholar]
- 7. Pietrantonio F, Morano F, Corallo S, et al. Maintenance therapy with panitumumab alone vs panitumumab plus fluorouracil-leucovorin in patients with RAS wild-type metastatic colorectal cancer: a phase 2 randomized clinical trial. JAMA Oncol. 2019;5(9):1268–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seymour MT, Brown SR, Middleton G, et al. Panitumumab and irinotecan versus irinotecan alone for patients with KRAS wild-type, fluorouracil-resistant advanced colorectal cancer (PICCOLO): a prospectively stratified randomised trial. Lancet Oncol. 2013;14(8):749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97(9):643–655. [DOI] [PubMed] [Google Scholar]
- 10. Yaeger R, Chatila WK, Lipsyc MD, et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell. 2018;33(1):125–136. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nilsen G, Liestøl K, Van Loo P, et al. Copynumber: efficient algorithms for single- and multi-track copy number segmentation. BMC Genomics. 2012;13:591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Siravegna G, Bardelli A.. Blood circulating tumor DNA for non-invasive genotyping of colon cancer patients. Mol Oncol. 2016;10(3):475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. May S, Hosmer DW.. A simplified method of calculating an overall goodness-of-fit test for the Cox proportional hazards model. Lifetime Data Anal. 1998;4(2):109–120. [DOI] [PubMed] [Google Scholar]
- 14. Durrleman S, Simon R.. Flexible regression models with cubic splines. Stat Med. 1989;8(5):551–561. [DOI] [PubMed] [Google Scholar]
- 15. Lausen B, Schumacher M.. Maximally selected rank statistics. Biometrics. 1992;48(1):73–85. [Google Scholar]
- 16. Cho J, Pastorino S, Zeng Q, et al. Glioblastoma-derived epidermal growth factor receptor carboxyl-terminal deletion mutants are transforming and are sensitive to EGFR-directed therapies. Cancer Res. 2011;71(24):7587–7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoshino T, Arnold D, Taniguchi H, et al. Pan-Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: a JSMO-ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann Oncol. 2018;29(1):44–70. [DOI] [PubMed] [Google Scholar]
- 18. Siena S, Sartore-Bianchi A, Marsoni S, et al. Targeting the human epidermal growth factor receptor 2 (HER2) oncogene in colorectal cancer. Ann Oncol. 2018;29(5):1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu S, Gao F, Zheng S, et al. Amplification induces increased DNA damage response and renders selective sensitivity to talazoparib (PARP Inhibitor) in glioblastoma. Clin Cancer Res. 2020;26(6):1395–1407. [DOI] [PubMed] [Google Scholar]
- 20. Corso S, Pietrantonio F, Apicella M, et al. Optimized EGFR blockade strategies in EGFR addicted gastroesophageal adenocarcinomas. Clin Cancer Res. 2021. doi: 10.1158/1078-0432.CCR-20-0121 [DOI] [PubMed] [Google Scholar]
- 21. Smyth EC, Vlachogiannis G, Hedayat S, et al. EGFR amplification and outcome in a randomised phase III trial of chemotherapy alone or chemotherapy plus panitumumab for advanced gastro-oesophageal cancers. Gut. 2020. doi: 10.1136/gutjnl-2020-322658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Emburgh BO, Arena S, Siravegna G, et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun. 2016;7:13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura Y, Sasaki A, Yukami H, et al. Emergence of concurrent multiple EGFR mutations and MET amplification in a patient with EGFR-amplified advanced gastric cancer treated with cetuximab. J Clin Oncol Precis Oncol. 2020;4:1407–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arena S, Bellosillo B, Siravegna G, et al. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin Cancer Res. 2015;21(9):2157–2166. [DOI] [PubMed] [Google Scholar]
- 25. Cappuzzo F, Finocchiaro G, Rossi E, et al. EGFR FISH assay predicts for response to cetuximab in chemotherapy refractory colorectal cancer patients. Ann Oncol. 2008;19(4):717–723. [DOI] [PubMed] [Google Scholar]
- 26. Mosele F, Remon J, Mateo J, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31(11):1491–1505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author.