Abstract
Background
Adult childhood cancer survivors are at risk for frailty, including low muscle mass and weakness (sarcopenia). Using peripheral blood mitochondrial DNA copy number (mtDNAcn) as a proxy for functional mitochondria, this study describes cross-sectional associations between mtDNAcn and sarcopenia among survivors.
Methods
Among 1762 adult childhood cancer survivors (51.6% male; median age = 29.4 years, interquartile range [IQR] = 23.3-36.8), with a median of 20.6 years from diagnosis (IQR = 15.2-28.2), mtDNAcn estimates were derived from whole-genome sequencing. A subset was validated by quantitative polymerase chain reaction and evaluated cross-sectionally using multivariable logistic regression for their association with sarcopenia, defined by race-, age-, and sex-specific low lean muscle mass or weak grip strength. All statistical tests were 2-sided.
Results
The prevalence of sarcopenia was 27.0%, higher among female than male survivors (31.5% vs 22.9%; P < .001) and associated with age at diagnosis; 51.7% of survivors with sarcopenia were diagnosed ages 4-13 years (P = .01). Sarcopenia was most prevalent (39.0%) among central nervous system tumor survivors. Cranial radiation (odds ratio [OR] = 1.84, 95% confidence interval [CI] = 1.32 to 2.59) and alkylating agents (OR = 1.34, 95% CI = 1.04 to 1.72) increased, whereas glucocorticoids decreased odds (OR = 0.72, 95% CI = 0.56 to 0.93) of sarcopenia. mtDNAcn decreased with age (β = −0.81, P = .002) and was higher among female survivors (β = 9.23, P = .01) and among survivors with a C allele at mt.204 (β = −17.9, P = .02). In adjusted models, every standard deviation decrease in mtDNAcn increased the odds of sarcopenia 20% (OR = 1.20, 95% CI = 1.07 to 1.34).
Conclusions
A growing body of evidence supports peripheral blood mtDNAcn as a biomarker for adverse health outcomes; however, this study is the first to report an association between mtDNAcn and sarcopenia among childhood cancer survivors.
Five-year survival rates for pediatric cancers approach 85% (1). The estimated 500 000 childhood cancer survivors living in the United States today are at increased risk for an aging phenotype (2-4), with abnormal body composition and reduced strength conferring disproportionate risk for chronic disease and mortality (2,3,5,6). Reductions in lean muscle mass and strength observed among survivors (2,5,7) are similar to age-related loss of muscle mass and function characteristic of sarcopenia, a determinant of frailty in geriatric populations (5,6,8). Although primary cancer diagnosis and therapy are implicated as risk factors (2,4,9‐11), molecular mechanisms involved in premature development of sarcopenia or frailty in childhood cancer survivors are not yet elucidated (9).
Dysfunctional mitochondria are implicated in sarcopenia pathogenesis (12‐14). As the primary source of cellular energy, mitochondria produce adenosine triphosphate via oxidative phosphorylation (OXPHOS). Multiple copies of a circular, double-stranded, 16.5 kb mitochondrial genome encode 13 polypeptides instrumental for cellular respiration and RNA machinery (2 rRNAs and 22 tRNAs) for translation (15,16). When mitochondrial DNA (mtDNA) integrity is compromised, either through environmental damage or replication error, OXPHOS capacity is impeded (17,18), and impaired cellular function and pathological states ensue. Skeletal muscles, highly metabolic and densely packed with mitochondria (19‐21), are particularly vulnerable; neuromuscular dysfunction is a common feature of diseases caused by mtDNA mutations (22,23).
Age-related decline in mitochondrial function is described, with mitochondrial DNA copy number (mtDNAcn) used as a proxy for mitochondrial function and OXPHOS capacity (21,24), in skeletal muscle and peripheral blood (PB) samples from healthy individuals (25‐28). Some data indicate that copy number losses begin in middle age (29,30), and associations between PB mtDNAcn and frailty are apparent among older adults (25,30,31).
To advance understanding about the relationship between mitochondrial function and physiological decline, we examined the association between mtDNAcn and sarcopenia among adult survivors of childhood cancer.
Methods
Study Population
Participants were enrolled onto the institutional review board–approved St Jude Lifetime Cohort (SJLIFE) and assessed September 7, 2007, and June 30, 2015. SJLIFE assesses health outcomes among survivors of childhood cancer (32,33). Survivors aged 18 years and older at follow-up, and 10 or more years fromprimary diagnosis were eligible. Assessments were all completed 10 or more years from diagnosis. Informed consent was obtained prior to protocol-driven uniform clinical evaluation, physical performance testing, and collection of biological specimens. Of 1848 germline samples sequenced, analysis was restricted to 1762 (95.3%) survivorswith available muscle mass and grip strength measures who were not treated with allogeneic stem-cell transplantation.
Outcome
Sarcopenia, defined by low lean muscle mass or handgrip weakness (at least 1 component), was the primary outcome. Relative appendicular lean muscle mass was determined from dual x-ray absorptiometry and height; the sum of extremity lean mass divided by height in meters squared. Isometric handgrip strength (kilograms) was measured in sitting using a Jamar handheld dynamometer, with the shoulder at 0°-10°, elbow at 90° flexion, and forearm neutral (34,35). Individuals were classified with low mass when relative lean mass was less than −1.5 standard deviations below age-, sex-, and race-specific values (36). Muscle weakness was determined from body mass index–specific cut-points for strength (8).
Whole-Genome Sequencing (WGS) mtDNAcn Estimates
During clinical evaluation, PB samples were obtained and cryopreserved. Germline DNA was extracted using the DNA Blood Mini kit (Qiagen, Valencia, CA); 1 ug of fluorometrically quantified material was subjected to whole-genome pair-end sequencing on a HiSeq X Ten System (Illumina Inc, San Diego, CA) at HudsonAlpha Institute for Biotechnology Genomic Services Laboratory (Huntsville, AL). Using Burrows-Wheeler Aligner v.0.7.12 (37), sequencing reads were aligned to human reference genome build 38 (GRCh38) and the revised human mitochondrial reference (NC_012920) (37‐39). Average coverages for genomic and mitochondrial DNA were called using the depth command in SAMtools v.1.2 (Source Forge, San Diego, CA; http://samtools.sourceforge.net/) (40) and were considered proportional to copy number. To validate the pipeline, DNA from a 143B osteosarcoma cell line lacking mitochondrial DNA (ρ0) was sequenced as a negative control. Mitochondrial DNA copy number (mtDNAcn) was estimated by multiplying the ratio of mitochondrial to genomic coverage by 2 to account for the diploid nature of autosomal DNA (31).
Quantitative Polymerase Chain Reaction (qPCR) mtDNAcn Estimates
Although WGS is considered a precise method of determining mtDNAcn (28), qPCR is often used to validate next-generation sequencing (30,31). WGS estimates of mtDNAcn were validated for a subset (n = 95) of survivors by qPCR using SsoAdvanced Universal SYBR Green Supermix reagent (Bio-Rad, Hercules, CA) and 10 ng of germline DNA from survivors and the (ρ0) negative control. Primers against the mitochondrial-encoded NADH dehydrogenase 2 (MT-ND2) gene were designed using National Center for Biotechnology Information Primer Blast (Bethesda, MD) (forward: 5′-ACCAAACCCAGCTACGCAAA-3′; reverse: 5′-AGTAGTAGGGTCGTGGTGCT-3′), and primers used against the nuclear-encoded β2-microglobulin (β2M) gene (forward: 5′-TGCTGTCTCCATGTTTGATGTATCT-3′; reverse: 5′-TCTCTGCTCCCCACCTCTAAGT-3′) were published previously (41). qPCR reaction and thermocycling conditions were conducted according to manufacturer protocol. The assay was performed on a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA). Relative mtDNAcn was determined by taking the difference in CT values between β2M and MT-ND2 genes and accounting for diploid nuclear DNA (2 x 2ΔCT) (41).
Independent Variables
Variables included sex; race and ethnicity; cancer diagnosis (hematological, central nervous system [CNS], solid [non-CNS] tumors, and other malignancies); smoking status; physical activity status; average daily dietary protein intake; age at diagnosis; age at assessment; time since diagnosis (in quartiles); type of chemotherapeutic agent (alkylating, anthracyclines, glucocorticoids); and radiation site exposures (any, chest, cranial, abdominal). Demographic characteristics and treatment exposures were extracted from medical records; smoking, physical activity status (42), and dietary intake (43) were self-reported. Single nucleotide polymorphisms (associated gene) and 1 mtDNA sequence transition associated with sarcopenia in other populations were identified by WGS and included rs464553 (NUDT3), rs1028883 (KLF5), rs3129753 (HLA-DQB1-AS1) (44), rs9991501 (HSD17B11), rs2287926 (VCAN), rs4842924 (ADAMTSL3), rs9936385 (FTO), rs2943656, (IRS1) (45), and mt.204C (46). For mtDNA haplogroup assignment, sequencing data was submitted through HaploFind (47) and categorized as “Ancestral,” “Asian,” “European,” or “Reference,” based on human mtDNA migratory patterns (48).
Statistical Analysis
Descriptive statistics characterized the study population. Survivors without sarcopenia were compared with survivors with 1 sarcopenia component and both sarcopenia components with χ2 and Fisher exact tests for categorical measures and Wilcoxon rank-sum tests for median age at assessment and mtDNAcn. The association between WGS and qPCR was evaluated using Pearson correlation. Associations between sex, ancestry, age, treatment exposures, mt.204C, and mtDNAcn were evaluated in general linear models. Multivariable logistic regression was used to assess risk factors for sarcopenia with estimates reported as odds ratios (ORs) with 95% confidence intervals (CIs). Variable selection for model inclusion used 2 methods. Biologically important factors (sex, age at diagnosis, age at assessment, corticosteroid treatment, and radiation exposures) (2,49‐52) along with factors identified from bivariate analyses (P < .05) were included in a multivariable model with mtDNAcn and 2- and 3-way interactions between treatment exposures. Elastic net was then used to select covariates for model inclusion to assess prediction accuracy and reduce prediction error (Supplementary Table 1, available online) (53). Variables from both methods were put into the final model and retained if statistically significant for 1 or both component outcomes. To determine if mtDNAcn mediated the association between host and treatment-related risk factors and sarcopenia, a separate model that retained all factors except mtDNAcn was constructed to evaluate potential changes in parameter estimates (54). Analyses were conducted with SAS version 9.4 (SAS Institute, Inc, Cary, NC). All statistical tests were 2-sided, and a P value less than .05 was considered statistically significant.
Results
Population
Of 1848 survivors sequenced, 95.3% (n = 1762) had performance data available. Participants and nonparticipants did not differ by sex or age at assessment. Nonparticipants were less likely to have exposures to chest and/or abdominal radiation, anthracyclines, or corticosteroids and more likely to be diagnosed at ages 9-13 years, have solid tumors, and be Black.
Participant characteristics are in Table 1. The prevalence of sarcopenia was 27.0%, with median mtDNAcn of 301 and 310 among survivors with and without sarcopenia, respectively (P = .004). Differences in haplogroup, race, and ethnicity were not statistically significant. Nearly half of participants survived 10-19 years, with no differences in median age between those with and without sarcopenia. The prevalence of sarcopenia varied by sex (female 31.5%, male 22.9%; P < .001) and was most prevalent (39.0%) among survivors of CNS malignancies and those diagnosed at ages 4-13 years (51.7%; P = .01).
Table 1.
Characteristics of 1762 childhood cancer survivors with and without sarcopeniaa
| Characteristic | Sarcopenia (both components) A | Sarcopenia (at least one component) B | No sarcopenia (no components) C | P b | P b |
|---|---|---|---|---|---|
| No. (%) | No. (%) | No. (%) | A vs C | B vs C | |
| Total | 62 (3.5) | 476 (27.0) | 1286 (73.0) | ||
| Median age at assessment (IQR), y | 26.4 (21.4-34.7) | 28.9 (22.7-37.3) | 29.6 (23.5-36.6) | .04 | .45 |
| Median mtDNAcn (IQR) | 284.2 (235.6-342.1) | 301.0 (256.0-354.4) | 309.8 (260.5-378.9) | .008 | .004 |
| mtDNAcn standard deviation (IQR) | 3.2 (2.7-3.8) | 3.4 (2.9-4.0) | 3.5 (2.9-4.3) | .008 | .004 |
| Sex | |||||
| Female | 41 (66.1) | 268 (56.3) | 584 (45.4) | .001 | <.001 |
| Male | 21 (33.9) | 208 (43.7) | 702 (54.6) | ||
| Race | |||||
| White | 57 (91.9) | 394 (82.8) | 1088 (84.6) | .28 | .62 |
| Black | 5 (8.1) | 79 (16.6) | 192 (14.9) | ||
| Other | 0 (0.0) | 3 (0.6) | 6 (0.5) | ||
| Ethnicity | |||||
| Non-Hispanic | 59 (95.2) | 468 (98.3) | 1262 (98.1) | .10 | .80 |
| Hispanic | 3 (4.8) | 8 (1.7) | 24 (1.9) | ||
| Age at diagnosis, y | |||||
| 0-3 | 15 (24.2) | 137 (28.8) | 386 (30.0) | .02 | .04 |
| 4-8 | 26 (41.9) | 136 (28.6) | 312 (24.3) | ||
| 9-13 | 11 (17.7) | 110 (23.1) | 265 (20.6) | ||
| 14-24 | 10 (16.1) | 93 (19.5) | 323 (25.1) | ||
| Age at assessment, y | |||||
| 18-24 | 28 (45.2) | 167 (35.1) | 404 (31.4) | .16 | .18 |
| 25-30 | 12 (19.4) | 107 (22.5) | 303 (23.6) | ||
| 31-37 | 12 (19.4) | 93 (19.5) | 306 (23.8) | ||
| 38-64 | 10 (16.1) | 109 (22.9) | 273 (21.2) | ||
| Time since diagnosis, y | |||||
| 10-19 | 36 (58.1) | 234 (49.2) | 604 (47.0) | .13 | .08 |
| 20-29 | 19 (30.7) | 130 (27.3) | 420 (32.7) | ||
| 30-49 | 7 (11.3) | 112 (23.5) | 262 (20.4) | ||
| Diagnostic group | |||||
| Hematological | 24 (38.7) | 206 (43.3) | 720 (56.0) | <.001 | <.001 |
| CNS | 20 (32.3) | 90 (18.9) | 141 (11.0) | ||
| Solid, non-CNS | 16 (25.8) | 178 (37.4) | 417 (32.4) | ||
| Other malignancies | 2 (3.2) | 2 (0.4) | 8 (0.6) | ||
| Any radiation | |||||
| Yes | 42 (67.7) | 313 (65.8) | 693 (53.9) | .03 | <.001 |
| No | 20 (32.3) | 163 (34.2) | 593 (46.1) | ||
| Abdominal radiation | |||||
| Yes | 20 (32.3) | 145 (30.5) | 258 (20.1) | .02 | <.001 |
| No | 42 (67.7) | 331 (69.5) | 1028 (79.9) | ||
| Cranial radiation | |||||
| Yes | 33 (53.2) | 178 (37.4) | 349 (27.1) | <.001 | <.001 |
| No | 29 (46.8) | 298 (62.6) | 937 (72.9) | ||
| Chest radiation | |||||
| Yes | 23 (37.1) | 183 (38.5) | 371 (28.9) | .16 | .001 |
| No | 39 (62.9) | 293 (61.6) | 915 (71.2) | ||
| Anthracycline | |||||
| Yes | 31 (50.0) | 237 (49.8) | 811 (63.1) | .04 | <.001 |
| No | 31 (50.0) | 239 (50.2) | 475 (36.9) | ||
| Alkylating agents | |||||
| Yes | 45 (72.6) | 285 (59.9) | 753 (58.6) | .03 | .62 |
| No | 17 (27.4) | 191 (40.1) | 533 (41.4) | ||
| Glucocorticoids | |||||
| Yes | 24 (38.7) | 174 (36.6) | 604 (47.0) | .20 | <.001 |
| No | 38 (61.3) | 302 (63.4) | 682 (53.0) | ||
| Smoking | |||||
| Never | 45 (73.8) | 313 (67.0) | 848 (66.7) | .48 | .44 |
| Current | 10 (16.4) | 97 (20.8) | 242 (19.0) | ||
| Former | 6 (9.8) | 57 (12.2) | 182 (14.3) | ||
| Physical activity | |||||
| ≥150 min MVPA/wk | 25 (40.3) | 203 (42.6) | 746 (58.0) | .006 | <.001 |
| <150 min MVPA/wk | 37 (59.7) | 273 (57.4) | 540 (42.0) | ||
| Dietary protein intake | |||||
| <15.9% male/<15.5% female | 37 (59.7) | 276 (58.0) | 685 (53.3) | .32 | .15 |
| ≥15.9% male/≥15.5% female | 25 (40.3) | 200 (42.0) | 601 (46.7) | ||
| Haplogroup | |||||
| Ancestral | 6 (9.7) | 85 (17.9) | 210 (16.4) | .05 | .38 |
| Asian | 5 (8.1) | 10 (2.1) | 36 (2.8) | ||
| European | 28 (45.2) | 200 (42.1) | 503 (39.2) | ||
| Reference | 23 (37.1) | 180 (37.9) | 534 (41.6) | ||
| Genotypes | |||||
| rs464553 | |||||
| GG | 43 (69.4) | 264 (55.8) | 713 (55.8) | .10 | .96 |
| GT | 15 (24.2) | 172(36.4) | 461 (36.0) | ||
| TT | 4 (6.4) | 37(7.8) | 105 (8.2) | ||
| rs1028883 | |||||
| GG | 24 (38.7) | 166 (35.1) | 412 (32.2) | .41 | .36 |
| GT | 25 (40.3) | 229 (48.4) | 624 (48.8) | ||
| TT | 13 (21.0) | 78 (16.5) | 243 (19.0) | ||
| rs3129753 | |||||
| CC | 2 (3.2) | 20 (4.2) | 37 (2.9) | .56 | .37 |
| CG | 21 (33.9) | 132 (27.9) | 355 (27.8) | ||
| GG | 39 (62.9) | 321 (67.9) | 887 (69.4) | ||
| rs9991501 | |||||
| CC | 55 (88.7) | 440 (93.0) | 1214 (94.9) | .10 | .05 |
| CT | 7 (11.3) | 30 (6.4) | 64 (5.0) | ||
| TT | 0 (0.0) | 3 (0.6) | 1 (0.1) | ||
| rs2287926 | |||||
| AA | 0 (0.0) | 13 (2.8) | 29 (2.3) | .48 | .52 |
| AG | 16 (25.8) | 103 (21.8) | 308 (24.1) | ||
| GG | 46 (74.2) | 357 (75.5) | 942 (73.6) | ||
| rs4842924 | |||||
| CC | 19 (30.7) | 107 (22.6) | 248 (19.4) | .006 | .25 |
| CT | 34 (54.8) | 211 (44.6) | 618 (48.3) | ||
| TT | 9 (14.5) | 155 (32.8) | 413 (32.3) | ||
| rs9936385 | |||||
| CC | 5 (8.1) | 74 (15.6) | 220 (17.2) | .13 | .51 |
| CT | 31 (50.0) | 227 (48.0) | 629 (49.2) | ||
| TT | 26 (41.9) | 172 (36.4) | 430 (33.6) | ||
| rs2943656 | |||||
| AA | 11 (17.7) | 82 (17.3) | 159 (12.4) | .63 | .03 |
| AG | 28 (45.1) | 212 (44.8) | 594 (44.9) | ||
| GG | 23 (37.1) | 179 (37.9) | 526 (41.1) | ||
| mt204.C transition | |||||
| Yes | 7 (11.3) | 47 (9.9) | 104 (8.1) | .38 | .24 |
| No | 55 (88.7) | 427 (90.1) | 1174 (92.9) |
Missing: haplogroup data for 1 survivor with sarcopenia and 3 survivors without sarcopenia, smoking data for 9 survivors with sarcopenia, and 14 survivors without sarcopenia, genotype data for 3 survivors with and 7 survivors without sarcopenia, mt204 data for 2 survivors with sarcopenia and 8 without sarcopenia. CNS = central nervous system; IQR = interquartile range; mtDNAcn = mitochondrial DNA copy number; MVPA = moderate and vigorous physical activity.
χ2 test used to calculate 2-sided P values.
mtDNAcn Validation
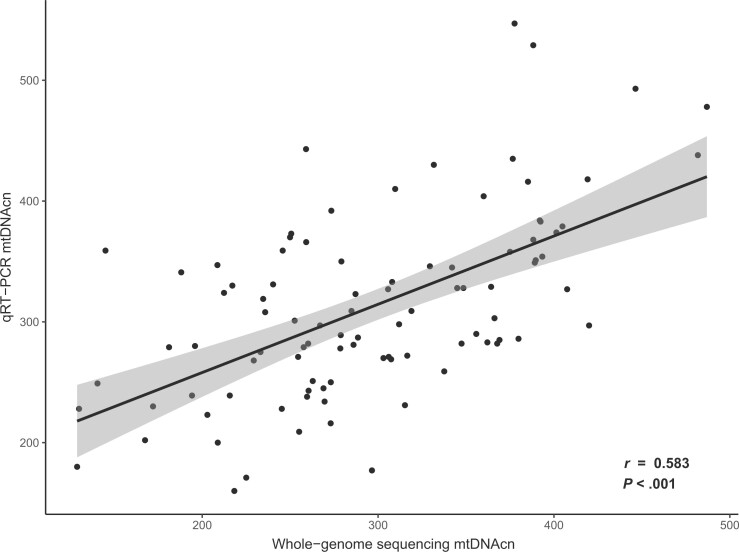
We found a positive, statistically significant linear relationship between WGS and qPCR estimates (Pearson correlation, r = 0.583; P < .001) (Figure 1). In adjusted models, mtDNAcn was associated with female sex (β = 9.23; P = .04), age (β = −0.81; P = .002), and with a C vs T allele at mt.204 (β = −17.9; P = .02). The only treatment-related risk factor associated with mtDNAcn was age at diagnosis of 4-13 years (β = −14.2; P = .01).
Figure 1.
qPCR validation of whole-genome sequencing mtDNAcn estimates. MtDNAcn estimates from 95 randomly selected whole-genome sequenced samples are plotted along the x-axis vs mtDNAcn estimates derived from qPCR of the same samples plotted along the y-axis. Pearson correlation test P value is 2-sided. mtDNAcn = mitochondrial DNA copy number; qPCR = quantitative polymerase chain reaction; qRT-PCR = real-time quantitative PCR.
Bivariate Models
In bivariate models (Table 2), mtDNAcn, female sex, age at diagnosis, and radiation exposure were positively associated with having at least 1 component of sarcopenia. Using blood malignancies as the reference, CNS (OR = 2.23, 95% CI = 1.64 to 3.03) and solid tumor (OR = 1.49, 95% CI = 1.18 to 1.89) survivors had higher odds of sarcopenia. Glucocorticoids were associated with decreased odds (OR = 0.65, 95% CI = 0.52 to 0.81). Additionally, although anthracyclines appeared to be associated with reduced odds of sarcopenia, the association was dependent on radiation exposure; survivors who received radiation were less likely to receive anthracyclines. Among survivors (n = 756) who never received radiation, anthracyclines were not associated with sarcopenia (OR = 0.81, 95% CI = 0.60 to 1.11). Inactivity (<150 minutes of moderate or vigorous physical activity per week) (OR = 1.86, 95% CI = 1.50 to 2.30) and AA genotype at rs2943656 (IRS1) (OR = 1.52, 95% CI = 1.11 to 2.08) were also associated with sarcopenia. Alternately, secondary analysis (both components) indicated that mtDNAcn, female sex, age at diagnosis, time since diagnosis, tumor type, cranial radiation, alkylating agent exposures, inactivity, Asian haplogroup, and presence of a T allele at rs9991501 (HSD17B11) were associated with sarcopenia. A TT genotype at rs4842924 (ADAMTSL3) appeared protective.
Table 2.
Potential risk factors for sarcopenia—bivariate modelsa
| Risk factor | Sarcopenia (both components) (n = 62) |
Sarcopenia (at least one component) (n = 476) |
||
|---|---|---|---|---|
| No. (%) | OR (95% CI) | No. (%) | OR (95% CI) | |
| mtDNAcn, 1 SD (91.44) decrease | 1.44 (1.08 to 1.92) | 1.19 (1.06 to 1.32) | ||
| Sex | ||||
| Female | 41 (66.1) | 2.14 (1.25 to 3.66) | 268 (56.3) | 1.55 (1.25 to 1.91) |
| Male | 21 (33.9) | 1.00 (Referent) | 208 (43.7) | 1.00 (Referent) |
| Race | ||||
| White | 57 (91.9) | 1.00 (Referent) | 394 (82.8) | 1.00 (Referent) |
| Black | 5 (8.1) | NE | 79 (16.6) | 1.13 (0.58 to 1.51) |
| Other | 0 (0.0) | NE | 3 (0.6) | 1.38 (0.34 to 5.55) |
| Ethnicity | ||||
| Non-Hispanic | 59 (95.2) | 1.00 (Referent) | 468 (98.3) | 1.00 (Referent) |
| Hispanic | 3 (4.8) | 2.93 (0.87 to 9.89) | 8 (1.7) | 0.90 (0.40 to 2.01) |
| Age at diagnosis, y | ||||
| 0-3 | 15 (24.2) | 1.20 (0.53 to 2.70) | 137 (28.8) | 1.23 (0.91 to 1.67) |
| 4-8 | 26 (41.9) | 2.50 (1.19 to 5.25) | 136 (28.6) | 1.51 (1.12 to 2.06) |
| 9-13 | 11 (17.7) | 1.23 (0.51 to 2.92) | 110 (23.1) | 1.44 (1.05 to 1.99) |
| 14-24 | 10 (16.1) | 1.00 (Referent) | 93 (19.5) | 1.00 (Referent) |
| Age at assessment, y | ||||
| 18-24 | 28 (45.2) | 1.00 (Referent) | 167 (35.1) | 1.00 (Referent) |
| 25-30 | 12 (19.4) | 0.59 (0.29 to 1.16) | 107 (22.5) | 0.85 (0.64 to 1.14) |
| 31-37 | 12 (19.4) | 0.60 (0.30 to 1.20) | 93 (19.5) | 0.74 (0.55 to 0.99) |
| 38-64 | 10 (16.1) | 0.52 (0.25 to 1.09) | 109 (22.9) | 0.97 (0.73 to 1.29) |
| Time since diagnosis, y | ||||
| 10-19 | 36 (58.1) | 1.00 (Referent) | 234 (49.2) | 1.00 (Referent) |
| 20-29 | 19 (30.7) | 0.80 (0.45 to 1.40) | 130 (27.3) | 0.80 (0.62 to 1.02) |
| 30-49 | 7 (11.3) | 0.42 (0.19 to 0.96) | 112 (23.5) | 1.10 (0.84 to 1.44) |
| Diagnostic group | ||||
| Hematological | 24 (38.7) | 1.00 (Referent) | 206 (43.3) | 1.00 (Referent) |
| Central nervous system (CNS) | 20 (32.3) | 3.56 (1.93 to 6.57) | 90 (18.9) | 2.23 (1.64 to 3.03) |
| Solid, non-CNS | 16 (25.8) | 1.04 (0.55 to 1.97) | 178 (37.4) | 1.49 (1.18 to 1.89) |
| Other malignancies | 2 (3.2) | 9.40 (1.89 to 46.61) | 2 (0.4) | 0.87 (0.18 to 4.15) |
| Any radiation | ||||
| Yes | 42 (67.7) | 1.60 (0.93 to 2.75) | 313 (65.8) | 1.64 (1.32 to 2.05) |
| No | 20 (32.3) | 1.00 (Referent) | 163 (34.2) | 1.00 (Referent) |
| Abdominal radiation | ||||
| Yes | 20 (32.3) | 1.64 (0.95 to 2.82) | 145 (30.5) | 1.75 (1.38 to 2.21) |
| No | 42 (67.7) | 1.00 (Referent) | 331 (69.5) | 1.00 (Referent) |
| Cranial radiation | ||||
| Yes | 33 (53.2) | 2.78 (1.67 to 4.63) | 178 (37.4) | 1.60 (1.28 to 2.00) |
| No | 29 (46.8) | 1.00 (Referent) | 298 (62.6) | 1.00 (Referent) |
| Chest radiation | ||||
| Yes | 23 (37.1) | 1.30 (0.77 to 2.20) | 183 (38.5) | 1.54 (1.24 to 1.92) |
| No | 39 (62.9) | 1.00 (Referent) | 293 (61.6) | 1.00 (Referent) |
| Anthracycline | ||||
| Yes | 31 (50.0) | 0.67 (0.40 to 1.12) | 237 (49.8) | 0.58 (0.47 to 0.72) |
| No | 31 (50.0) | 1.00 (Referent) | 239 (50.2) | 1.00 (Referent) |
| Alkylating agents | ||||
| Yes | 45 (72.6) | 1.88 (1.07 to 3.32) | 285 (59.9) | 1.06 (0.85 to 1.31) |
| No | 17 (27.4) | 1.00 (Referent) | 191 (40.1) | 1.00 (Referent) |
| Glucocorticoids | ||||
| Yes | 24 (38.7) | 0.79 (0.47 to 1.33) | 174 (36.6) | 0.65 (0.52 to 0.81) |
| No | 38 (61.3) | 1.00 (Referent) | 302 (63.4) | 1.00 (Referent) |
| Smoking | ||||
| Never | 45 (73.8) | 1.00 (Referent) | 313 (67.0) | 1.00 (Referent) |
| Current | 10 (16.4) | 0.75 (0.38 to 1.51) | 97 (20.8) | 1.09 (0.83 to 1.42) |
| Former | 6 (9.8) | 0.64 (0.27 to 1.51) | 57 (12.2) | 0.85 (0.61 to 1.17) |
| Physical activity | ||||
| ≥150 min MVPA/wk | 25 (40.3) | 1.00 (Referent) | 203 (42.6) | 1.00 (Referent) |
| <150 min MVPA/wk | 37 (59.7) | 1.76 (1.05 to 2.95) | 273 (57.4) | 1.86 (1.50 to 2.30) |
| Dietary protein intake | ||||
| <15.9% male/<15.5% female | 37 (58.1) | 1.24 (0.74 to 2.08) | 276 (56.9) | 1.21 (0.98 to 1.50) |
| ≥15.9% male/≥15.5% female | 25 (41.9) | 1.00 (Referent) | 200 (43.1) | 1.00 (Referent) |
| Haplogroup | ||||
| Ancestral | 6 (9.7) | 0.62 (0.25 to 1.55) | 85 (17.9) | 1.20 (0.89 to 1.63) |
| Asian | 5 (8.1) | 3.66 (1.33 to 10.13) | 10 (2.1) | 0.82 (0.40 to 1.69) |
| European | 28 (45.2) | 1.25 (0.71 to 2.19) | 200 (42.1) | 1.18 (0.93 to 1.49) |
| Reference | 23 (37.1) | 1.00 (Referent) | 180 (37.9) | 1.00 (Referent) |
| Genotype | ||||
| rs464553 | ||||
| GG | 43 (69.4) | 1.00 (Referent) | 264 (55.8) | 1.00 (Referent) |
| GT | 15 (24.2) | 0.53 (0.29 to 0.96) | 172(36.4) | 1.01 (0.81 to 1.26) |
| TT | 4 (6.4) | 0.63 (0.22 to 1.78) | 37(7.8) | 0.95 (0.64 to 1.42) |
| rs1028883 | ||||
| GG | 24 (38.7) | 1.00 (Referent) | 166 (35.1) | 1.00 (Referent) |
| GT | 25 (40.3) | 0.70 (0.39 to 1.23) | 229 (48.4) | 0.91 (0.72 to 1.15) |
| TT | 13 (21.0) | 0.97 (0.49 to 1.94) | 78 (16.5) | 0.80 (0.58 to 1.09) |
| rs3129753 | ||||
| CC | 2 (3.2) | 1.09 (0.26 to 4.63) | 20 (4.2) | 1.50 (0.85 to 2.61) |
| CG | 21 (33.9) | 1.35 (0.79 to 2.32) | 132 (27.9) | 1.03 (0.81 to 1.30) |
| GG | 39 (62.9) | 1.00 (Referent) | 321 (67.9) | 1.00 (Referent) |
| rs9991501 | ||||
| CC | 55 (88.7) | 1.00 (Referent) | 440 (93.0) | 1.00 (Referent) |
| CT | 7 (11.3) | 2.34 (1.04 to 5.29) | 30 (6.4) | 1.29 (0.83 to 2.02) |
| TT | 0 (0.0) | NE | 3 (0.6) | 8.27 (0.86 to 79.74) |
| rs2287926 | ||||
| AA | 0 (0.0) | NE | 13 (2.8) | 1.18 (0.61 to 2.30) |
| AG | 16 (25.8) | 1.10 (0.62 to 1.97) | 103 (21.8) | 0.88 (0.68 to 1.13) |
| GG | 46 (74.2) | 1.00 (Referent) | 357 (75.5) | 1.00 (Referent) |
| rs4842924 | ||||
| CC | 19 (30.7) | 1.00 (Referent) | 107 (22.6) | 1.00 (Referent) |
| CT | 34 (54.8) | 0.76 (0.43 to 1.34) | 211 (44.6) | 0.79 (0.60 to 1.04) |
| TT | 9 (14.5) | 0.29 (0.13 to 0.64) | 155 (32.8) | 0.87 (0.65 to 1.17) |
| rs9936385 | ||||
| CC | 5 (8.1) | 0.38 (0.15 to 1.01) | 74 (15.6) | 0.84 (0.61 to 1.16) |
| CT | 31 (50.0) | 0.83 (0.49 to 1.41) | 227 (48.0) | 0.90 (0.72 to 1.14) |
| TT | 26 (41.9) | 1.00 (Referent) | 172 (36.4) | 1.00 (Referent) |
| rs2943656 | ||||
| AA | 11 (17.7) | 1.42 (0.68 to 2.95) | 82 (17.3) | 1.52 (1.11 to 2.08) |
| AG | 28 (45.1) | 1.07 (0.61 to 1.87) | 212 (44.8) | 1.05 (0.83 to 1.32) |
| GG | 23 (37.1) | 1.00 (Referent) | 179 (37.9) | 1.00 (Referent) |
| mt.204C | ||||
| Yes | 7 (11.3) | 1.36 (0.61 to 3.06) | 47 (9.9) | 1.24 (0.87 to 1.78) |
| No | 55 (88.7) | 1.00 (Referent) | 427 (90.1) | 1.00 (Referent) |
Missing: haplogroup data for 1 survivor with sarcopenia and 3 survivors without sarcopenia, smoking data for 9 survivors with sarcopenia and 14 survivors without sarcopenia. CI = confidence interval; mtDNAcn = mitochondrial DNA copy number; MVPA = moderate or vigorous physical activity; NE = not estimable; OR = odds ratio.
Multivariable Models
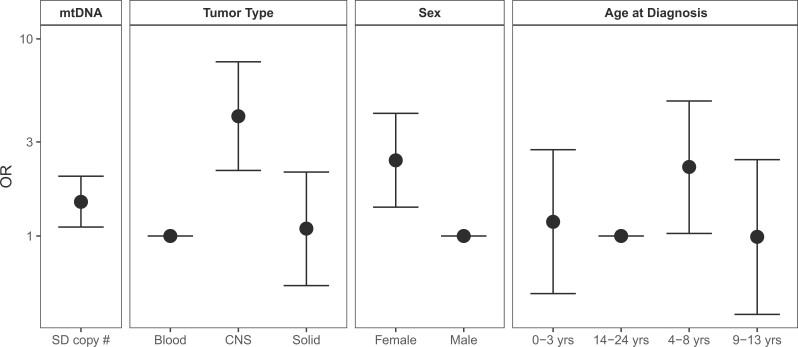
In multivariable models (Table 3), for every standard deviation decrease in mtDNAcn, the odds of having 1 component of sarcopenia increased by 20% (OR = 1.20, 95% CI = 1.07 to 1.34). Odds of sarcopenia were higher among female survivors (OR = 1.58, 95% CI = 1.27 to 1.99) and survivors treated with cranial radiation and not exposedto anthracyclines (OR = 1.84, 95% CI = 1.32 to 2.59). Exposure to glucocorticoids was associated with reduced odds (OR = 0.72, 95% CI = 0.56 to 0.93), whereas alkylators were associated with increased odds (OR = 1.34, 95% CI = 1.04 to 1.72) of sarcopenia. Inactivity (OR = 1.68, 95% CI = 1.34 to 2.10) and AA genotype at rs2943656 (IRS1) (OR = 1.48, 95% CI = 1.10 to 2.01) also remained associated with increased odds of sarcopenia. In analysis adjusting for tumor type instead of treatment exposure (Figure 2), a 1 standard deviation decrease in mtDNAcn was associated with 1.49 (95% CI = 1.11 to 2.01) increased odds, with survivors of CNS tumors (OR = 4.05, 95% CI = 2.15 to 7.65) having highest odds of sarcopenia. There was no evidence of mediation between treatment and odds of sarcopenia. Parameter estimates did not change appreciably when mtDNAcn was removed from models.
Table 3.
Multivariable associations between treatment and presence of sarcopeniaa
| Risk factor | Sarcopenia (both components) (n = 62) |
Sarcopenia (at least one component) (n = 476) |
||
|---|---|---|---|---|
| Row % | OR (95% CI) | Row % | OR (95% CI) | |
| mtDNAcn, 1 SD decrease | 1.61 (1.18 to 2.20) | 1.20 (1.07 to 1.34) | ||
| Sex | ||||
| Female | 4.8 | 3.13 (1.74 to 5.62) | 31.5 | 1.58 (1.27 to 1.99) |
| Male | 2.3 | 1.00 (Referent) | 22.9 | 1.00 (Referent) |
| Age at diagnosis, y | ||||
| 0-3 | 2.9 | 0.83 (0.35 to 1.95) | 26.2 | 0.88 (0.65 to 1.21) |
| 4-8 | 5.8 | 1.00 (0.39 to 2.57) | 30.4 | 0.70 (0.50 to 1.00) |
| 9-13 | 2.9 | 1.96 (0.90 to 4.27) | 29.3 | 1.04 (0.75 to 1.42) |
| 14-24 | 2.4 | 1.00 (Referent) | 22.4 | 1.00 (Referent) |
| Chest radiation | ||||
| Yes | 4.2 | 1.57 (0.85 to 2.89) | 33.0 | 1.61 (1.26 to 2.04) |
| No | 3.2 | 1.00 (Referent) | 24.3 | 1.00 (Referent) |
| Cranial radiation/anthracyclines | ||||
| Neither | 2.2 | 1.00 (Referent) | 27.2 | 1.00 (Referent) |
| Anthracyclines/no cranial radiation | 2.4 | 0.61 (0.24 to 1.54) | 22.5 | 0.77 (0.56 to 1.07) |
| Cranial radiation/no anthracyclines | 7.1 | 3.29 (1.38 to 7.83) | 41.8 | 1.84(1.32 to 2.59) |
| Both cranial radiation and anthracyclines | 5.1 | 1.55 (0.53 to 4.55) | 22.7 | 0.89 (0.57 to 1.39) |
| Alkylating agents | ||||
| Yes | 4.3 | 2.90 (1.46 to 5.77) | 27.5 | 1.34 (1.04 to 1.72) |
| No | 2.4 | 1.00 (Referent) | 26.4 | 1.00 (Referent) |
| Glucocorticoids | ||||
| Yes | 3.1 | 0.68 (0.36 to 1.30) | 22.4 | 0.72 (0.56 to 0.93) |
| No | 3.9 | 1.00 (Referent) | 30.7 | 1.00 (Referent) |
| Physical Activity | ||||
| ≥150 min MVPA/wk | 2.6 | 1.00 (Referent) | 21.4 | 1.00 (Referent) |
| <150 min MVPA/wk | 4.6 | 1.71 (0.97 to 3.01) | 33.6 | 1.68 (1.34 to 2.10) |
| Haplogroup | ||||
| Ancestral | 2.0 | 0.70 (0.26 to 1.90) | 28.8 | 1.07 (0.77 to 1.49) |
| Asian | 10.9 | 4.21 (1.33 to 13.30) | 21.7 | 1.00 (0.47 to 2.11) |
| European | 4.0 | 1.32 (0.72 to 2.41) | 28.5 | 1.25 (0.98 to 1.60) |
| Reference | 3.2 | 1.00 (Referent) | 25.2 | 1.00 (Referent) |
| Genotype | ||||
| rs2943656 | ||||
| AA | 4.6 | 1.40 (0.67 to 2.93) | 34.0 | 1.48 (1.10 to 2.01) |
| AG/GG | 3.4 | 1.00 (Referent) | 25.9 | 1.00 (Referent) |
| rs4842924 | ||||
| TT | 1.6 | 0.35 (0.16 to 0.76) | 27.3 | 1.00 (0.78 to 1.28) |
| CT/CC | 4.6 | 1.00 (Referent) | 26.9 | 1.00 (Referent) |
Missing: Haplogroup data for 1 survivor with sarcopenia and 3 survivors without sarcopenia. Models are adjusted for age at assessment. CI = confidence interval; mtCN = mitochondrial DNA copy number; MVPA = moderate or vigorous physical activity; OR = odds ratio.
Figure 2.
Relative odds of sarcopenia by mtDNAcn, tumor type, sex, and age at diagnosis. Models were additionally adjusted for age at assessment and haplotype. The error bars represent the 95% confidence intervals for the odds ratios (ORs). CNS = central nervous system; mtDNA = mitochondrial DNA.
In secondary analysis (both low lean mass and weakness), sarcopenia prevalence was 3.5% (Table 1). In this context, the association between mtDNAcn and sarcopenia was stronger (OR = 1.61, 95% CI = 1.18 to 2.20) and emphasized the influence of cranial radiation (OR = 3.29, 95% CI = 1.38 to 7.83) and female sex (OR = 3.13, 95% CI = 1.74 to 5.62) (Table 3). Exposure to alkylators remained positively associated with sarcopenia (OR = 2.90, 95% CI = 1.46 to 5.77); treatment with corticosteroids did not (OR = 0.68, 95% CI = 0.36 to 1.30). Survivors of Asian ancestry (OR = 4.21, 95% CI = 1.33 to 13.30) had increased odds for both low lean mass and weakness.
Discussion
Adult survivors of childhood cancer are at increased risk for physiological frailty, with concomitant reduction of muscle mass or function (3). Our study found a prevalence of sarcopenia among survivors of 27.0%, with female survivors disproportionately affected. By comparison, an evidence-based evaluation of 9 studies comprising 10 063 general population participants reported that 21.3% of men and 28.0% of women aged 65 years and older were sarcopenic (55). Given the median age of participants was 29.4 years, our data highlight early appearance of sarcopenia among survivors.
Loss of mitochondrial capacity occurs with aging, where mtDNA oxidative damage and impaired adenosine triphosphate production are associated with decreased mtDNA content and reduced skeletal muscle aerobic performance and function (27,56). In addition, decreases in PB mtDNAcn are associated with poor health (30) and frailty (25). We hypothesized that mitochondrial alterations could account for sarcopenia in our population with changes manifesting in reduced PB mtDNAcn. Indeed, we found that the odds of having sarcopenia increased 20.0% for each standard deviation decrease in PB mtDNAcn.
By contrast, prior studies in the general population have not detected associations between mtDNAcn and components of frailty, including grip strength (31). Thus, the association between mtDNAcn and sarcopenia among adult survivors of childhood cancer suggests potential differences between the pathobiology of sarcopenia in the geriatric population and among childhood cancer survivors.
Survivors in SJLIFE were exposed to antineoplastic and or cytotoxicmodalities capable of damaging mitochondria (57) and skeletal muscle. Although other data suggest that anthracyclines cause dose-related cardiac/skeletal muscle dysfunction through reactive oxygen species-mediated impairment or deletion of mtDNA (58‐62), our study failed to find an association, likely because our cohort received fairly low anthracycline doses. Treatment with alkylating agents, however, was associated with sarcopenia and is congruent with evidence of disruption of the mitochondrial electron transport system by acrolein, an end product of cyclophosphamide metabolism (63,64). We also report an association between sarcopenia and radiation exposure, a modality capable of generating unrepairable DNA double strand breaks (DSBs) (38). Radiation is exquisitely detrimental for the mitochondrial genome, which cannot avail itself of protective histones and repair mechanisms afforded to nuclear DNA (65,66). In mice, mitochondrial damage persists for months after radiation exposure (67,68).
Whereas glucocorticoids (GC) are implicated in deterioration of muscle mass and function during processes intended to counter stress or inflammation (69), counterintuitively, we found GC protective against early appearance of sarcopenia. One possible hypothesis is that peroxisome proliferator-activated γ coactivator 1-a, a master regulator of mitochondrial biogenesis and dynamic sensor of physiological and metabolic signaling (70,71), responds to conformational changes in GC receptors (72), suppressing transcription factors involved in catabolic responses. Promotion of fast-to-slow muscle fiber conversion may protect skeletal muscle from atrophic processes (73,74). Functional studies are required to confirm this hypothesis.
Evidence that mtDNA damage is causative in the pathobiology of sarcopenia comes from 2 murine models: 1) mtDNA-mutator mice, which harbor a mutation that impairs the proofreading function of mitochondrial polymerase PolG (75‐77); and 2) mito-Pst1 mice, which express a mitochondria-targeted form of endonuclease Pst1 under an inducible promoter (78). The proofreading defect in mtDNA mutator mice leads to accumulation of somatic mtDNA mutations, disruption of mitochondrial respiratory complexes, and reductions in mitochondrial respiratory capacity (75‐77). These mice have an aging phenotype that includes sarcopenia and premature death (75‐77). Similarly, the short-term induction of DSBs that exclusively target mitochondria in mito-Pst1 mice results in mtDNA depletion, loss of muscle mass, and reduction of myosatellite cell populations (78). Thus, even transient mitochondrial damage may be sufficient to provoke sarcopenia.
The prospect of delayed skeletal muscle dysfunction from short-term mitochondrial damage to muscle satellite cells (MSCs) is intriguing, as 51.7% of survivors with sarcopenia in our cohort were treated for cancer at ages 4-13 years, a time when MSCs are actively proliferating and contributing to skeletal muscle mass (79). During postnatal development, MSCs proliferate and contribute to a burgeoning population of myonuclei supporting muscle fiber growth (80). Activity stops during puberty, where the total number of fibers is maintained until middle age (11,81). After puberty, MSCs are relegated to quiescent niches until muscle injury elicits reentry into the cell cycle to repair muscle fibers or self-renew and return to the niche (82). Because MSCs are sensitive to chemotherapy-induced oxidative stress or mitochondrial DSBs (83), the finite supply of MSCs may become prematurely exhausted or undergo early senescence, perpetuating an early sarcopenic phenotype in susceptible survivors.
The primary aim of this study was to evaluate the association between PB mtDNAcn and sarcopenia. However, we did investigate associations between several sequence variants and sarcopenia. First, mt.204C, a mtDNA single nucleotide variant present in 6.3% of the population and previously associated with reduced grip strength in elderly individuals (46), showed an association with mtDNAcn in our study. We also found an association between an AA genotype in rs2943656 (IRS1) and sarcopenia. This SNP is intergenic but located within histone mark–identified promoters or enhancers in skeletal muscle samples and thought to alter regulatory motifs associated with body composition and longevity (45). Because all but 13 of roughly 800-1500 gene products required to sustain mitochondria function are encoded by the nuclear genome (84‐86), additional studies investigating associations between nuclear and mtDNA variants and sarcopenia are warranted. Such variants may better explain the observed reduction in mtDNAcn and identify at-risk survivors for interventions.
We found moderate correlation between qPCR and WGS estimation of mtDNAcn, which suggests the possibility of measurement error. However, a recent study of the Atherosclerosis Risk in Communities study and the Multi-Ethnic Study of Atherosclerosis found that mtDNAcn measured from WGS is more statistically significantly associated with mtDNAcn correlates (age, sex, white blood cell count, Duffy locus genotype, incident cardiovascular disease) compared with all other methods, including qPCR (87). Another potential limitation of our study is sampling bias. Although only 5.0% of eligible survivors did not have performance data, they did differ from those with performance data by several diagnostic and treatment variables. This may have biased our results. A third limitation is the study’s cross-sectional design; our primary independent variable, mtDNAcn, and our phenotype, sarcopenia, were measured concomitantly, limiting our ability to infer causality. We did not have measures prior to or immediately following cancer therapy; our findings provide support for prospective studies to determine if there is a causal relationship. Finally, despite widespread use of PB mtDNAcn as a biomarker in health outcome studies, there is discordance as to degree of correlation in mtDNAcn among tissues (eg, PB and muscle) within an individual (28). Additional studies are required to determine if decreased PB mtDNAcn observed in childhood cancer survivors with sarcopenia is also associated with a reduction in mtDNAcn in muscle. It is also possible that PB mtDNAcn, rather than being a surrogate for mtDNAcn in other tissues, is an indicator of global oxidative stress–induced damage (88).
In conclusion, we demonstrate that decreased mtDNAcn in childhood cancer survivors is associated with sarcopenia. This association is statistically significant even when both reduced mass and function are required criteria for sarcopenia. Our data identify potential roles for cranial radiotherapy, alkylators, sex, and genetics in the pathobiology of sarcopenia and suggest a compelling role for mitochondrial dysfunction and/or oxidative damage as agents of pathological change among childhood cancer survivors.
Funding
This work was supported by the National Cancer Institute at the National Institutes of Health (CA174851, Ness; CA195547, Hudson; CA21765, Roberts; GM132231, Kundu) and the American Lebanese Syrian Associated Charities.
Notes
Role of the funder: The funder had no role in design of the study; the collection, analysis or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Disclosures: The authors have nothing to disclose.
Author contributions: KM, CEW, ZW, JZ, MK, KKN conceptualized the study. KM, CLW, HLM, JE, KKN curated the data and did the experiments. KM, CRH, CEW, ZW, and KKN analyzed the data. MMH, KKN, and MK obtained funding, YY, AM, MMH, LLR, JZ supervised the project, KM, CRH, and KKN wrote the original draft of the manuscript. All authors reviewed and edited the manuscript.
Acknowledgements: The authors would like to acknowledge Tracie Gatewood for her assistance preparing the manuscript.
Data Availability
The data are available from the corresponding author upon request.
Supplementary Material
References
- 1. Howlader N, Noone AM, Krapcho M, et al. (eds). National Cancer Institute Surveillance, Epidemiology, and End Results Program Nov 2016 https://seer.cancer.gov/csr/1975_2014/. Published April 2017. Accessed January 20, 2021.
- 2. Ness KK, Krull KR, Jones KE, et al. Physiologic frailty as a sign of accelerated aging among adult survivors of childhood cancer: a report from the St Jude Lifetime cohort study. J Clin Oncol. 2013;31(36):4496–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ness KK, Baker KS, Dengel DR, et al. Body composition, muscle strength deficits and mobility limitations in adult survivors of childhood acute lymphoblastic leukemia. Pediatr Blood Cancer. 2007;49(7):975-981. [DOI] [PubMed] [Google Scholar]
- 4. van Brussel M, Takken T, van der Net J, et al. Physical function and fitness in long-term survivors of childhood leukaemia. Pediatr Rehabil. 2006;9(3):267–274. [DOI] [PubMed] [Google Scholar]
- 5. Meacham LR, Gurney JG, Mertens AC, et al. Body mass index in long-term adult survivors of childhood cancer: a report of the Childhood Cancer Survivor Study. Cancer. 2005;103(8):1730–1739. [DOI] [PubMed] [Google Scholar]
- 6. Fried LP, Ferrucci L, Darer J, et al. Untangling the concepts of disability, frailty, and comorbidity: implications for improved targeting and care. J Gerontol A Biol Sci Med Sci. 2004;59(3):255–263. [DOI] [PubMed] [Google Scholar]
- 7. Boland AM, Gibson TM, Lu L, et al. Dietary protein intake and lean muscle mass in survivors of childhood acute lymphoblastic leukemia: report from the St. Jude Lifetime Cohort study. Phys Ther. 2016;96(7):1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fried LP, Tangen CM, Walston J, et al. Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci. 2001;56(3):M146–M156. [DOI] [PubMed] [Google Scholar]
- 9. Ness KK, Armstrong GT, Kundu M, et al. Frailty in childhood cancer survivors. Cancer. 2015;121(10):1540–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hudson MM, Oeffinger KC, Jones K, et al. Age-dependent changes in health status in the Childhood Cancer Survivor Cohort. J Clin Oncol. 2015;33(5):479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scheede-Bergdahl C, Jagoe RT.. After the chemotherapy: potential mechanisms for chemotherapy-induced delayed skeletal muscle dysfunction in survivors of acute lymphoblastic leukaemia in childhood. Front Pharmacol. 2013;4:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sousa-Victor P, Munoz-Canoves P.. Regenerative decline of stem cells in sarcopenia. Mol Aspects Med. 2016;50:109–117. [DOI] [PubMed] [Google Scholar]
- 13. Ibebunjo C, Chick JM, Kendall T, et al. Genomic and proteomic profiling reveals reduced mitochondrial function and disruption of the neuromuscular junction driving rat sarcopenia. Mol Cell Biol. 2013;33(2):194–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rygiel KA, Picard M, Turnbull DM.. The ageing neuromuscular system and sarcopenia: a mitochondrial perspective. J Physiol. 2016;594(16):4499–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andrews RM, Kubacka I, Chinnery PF, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. [DOI] [PubMed] [Google Scholar]
- 16. Vafai SB, Mootha VK.. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491(7424):374–383. [DOI] [PubMed] [Google Scholar]
- 17. Greaves LC, Nooteboom M, Elson JL, et al. Clonal expansion of early to mid-life mitochondrial DNA point mutations drives mitochondrial dysfunction during human ageing. PLoS Genet. 2014;10(9):e1004620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin YF, Schulz AM, Pellegrino MW, et al. Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature. 2016;533(7603):416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Porter C, Wall BT.. Skeletal muscle mitochondrial function: Is it quality or quantity that makes the difference in insulin resistance? J Physiol. 2012;590(23):5935–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Romanello V, Sandri M.. Mitochondrial quality control and muscle mass maintenance. Front Physiol. 2015;6:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. D’Erchia AM, Atlante A, Gadaleta G, et al. Tissue-specific mtDNA abundance from exome data and its correlation with mitochondrial transcription, mass and respiratory activity. Mitochondrion. 2015;20:13–21. [DOI] [PubMed] [Google Scholar]
- 22. Taylor RW, Turnbull DM.. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mkaouar-Rebai E, Felhi R, Tabebi M, et al. Mitochondrial DNA triplication and punctual mutations in patients with mitochondrial neuromuscular disorders. Biochem Biophys Res Commun. 2016;473(2):578–585. [DOI] [PubMed] [Google Scholar]
- 24. Clay Montier LL, Deng JJ, Bai Y.. Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genomics. 2009;36(3):125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ashar FN, Moes A, Moore AZ, et al. Association of mitochondrial DNA levels with frailty and all-cause mortality. J Mol Med (Berl). 2015;93(2):177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Menshikova EV, Ritov VB, Fairfull L, et al. Effects of exercise on mitochondrial content and function in aging human skeletal muscle. J Gerontol A Biol Sci Med Sci. 2006;61(6):534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Short KR, Bigelow ML, Kahl J, et al. Decline in skeletal muscle mitochondrial function with aging in humans. Proc Natl Acad Sci USA. 2005;102(15):5618–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wachsmuth M, Hubner A, Li M, et al. Age-related and heteroplasmy-related variation in human mtDNA copy number. PLoS Genet. 2016;12(3):e1005939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knez J, Marrachelli VG, Cauwenberghs N, et al. Peripheral blood mitochondrial DNA content in relation to circulating metabolites and inflammatory markers: a population study. PLoS One. 2017;12(7):e0181036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mengel-From J, Thinggaard M, Dalgard C, et al. Mitochondrial DNA copy number in peripheral blood cells declines with age and is associated with general health among elderly. Hum Genet. 2014;133(9):1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ding J, Sidore C, Butler TJ, et al. Assessing mitochondrial DNA variation and copy number in lymphocytes of ∼2,000 Sardinians using tailored sequencing analysis tools. PLoS Genet. 2015;11(7):e1005306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hudson MM, Ness KK, Nolan VG, et al. Prospective medical assessment of adults surviving childhood cancer: study design, cohort characteristics, and feasibility of the St. Jude Lifetime Cohort study. Pediatr Blood Cancer. 2011;56(5):825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ojha RP, Oancea SC, Ness KK, et al. Assessment of potential bias from non-participation in a dynamic clinical cohort of long-term childhood cancer survivors: results from the St. Jude Lifetime Cohort Study. Pediatr Blood Cancer. 2013;60(5):856–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mathiowetz V, Kashman N, Volland G, et al. Grip and pinch strength: normative data for adults. Arch Phys Med Rehabil. 1985;66(2):69–74. [PubMed] [Google Scholar]
- 35. Mathiowetz V, Rennells C, Donahoe L.. Effect of elbow position on grip and key pinch strength. J Hand Surg Am. 1985;10(5):694–697. [DOI] [PubMed] [Google Scholar]
- 36. Kelly TL, Wilson KE, Heymsfield SB.. Dual energy X-ray absorptiometry body composition reference values from NHANES. PLoS One. 2009;4(9):e7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Noda A. Radiation-induced unrepairable DSBs: their role in the late effects of radiation and possible applications to biodosimetry. J Radiat Res. 2018;59(suppl 2):ii114–ii120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.National Center for Biotechnology Information, US National Library of Medicine. NCBI > DNA & RNA > Nucleotide Database. Bethesda, MD; 2018. https://www.ncbi.nlm.nih.gov/nuccore/NC_012920. Accessed January 15, 2021. [Google Scholar]
- 40. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Venegas V, Wang J, Dimmock D, et al. Real-time quantitative PCR analysis of mitochondrial DNA content. Curr Protoc Hum Genet 2011;Chapter 19:Unit 19.7. [DOI] [PubMed] [Google Scholar]
- 42. Campbell KL, Winters-Stone KM, Wiskemann J, et al. Exercise guidelines for cancer survivors: consensus statement from international multidisciplinary roundtable. Med Sci Sports Exerc. 2019;51(11):2375–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wright JD, Wang CY. Trends in intake of energy and macronutrients in adults from 1999-2000 through 2007-2008. NCHS Data Brief. 2010;(49):1-8. https://www.ncbi.nlm.nih.gov/pubmed/21211164. Accessed January 15, 2021. [PubMed]
- 44. Singh AN, Gasman B.. Disentangling the genetics of sarcopenia: prioritization of NUDT3 and KLF5 as genes for lean mass & HLA-DQB1-AS1 for hand grip strength with the associated enhancing SNPs & a scoring system. BMC Med Genet. 2020;21(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zillikens MC, Demissie S, Hsu YH, et al. Large meta-analysis of genome-wide association studies identifies five loci for lean body mass. Nat Commun. 2017;8(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moore AZ, Biggs ML, Matteini A, et al. Polymorphisms in the mitochondrial DNA control region and frailty in older adults. PLoS One. 2010;5(6):e11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vianello D, Sevini F, Castellani G, et al. HAPLOFIND: a new method for high-throughput mtDNA haplogroup assignment. Hum Mutat. 2013;34(9):1189–1194. [DOI] [PubMed] [Google Scholar]
- 48.Center for Mitochondrial & Epigenomic Medicine, Children’s Hospital of Philadelphia, MITOMAP: a human mitochondrial genome database; 2018. https://www.mitomap.org/MITOMAP. Accessed January 15, 2021.
- 49. Trajanoska K, Schoufour JD, Darweesh SK, et al. Sarcopenia and its clinical correlates in the general population: the Rotterdam study. J Bone Miner Res. 2018;33(7):1209–1218. [DOI] [PubMed] [Google Scholar]
- 50. Klein GL. The effect of glucocorticoids on bone and muscle. Osteoporos Sarcopenia. 2015;1(1):39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mitsui T, Azuma H, Nagasawa M, et al. Chronic corticosteroid administration causes mitochondrial dysfunction in skeletal muscle. J Neurol. 2002;249(8):1004–1009. [DOI] [PubMed] [Google Scholar]
- 52. Gawade PL, Hudson MM, Kaste SC, et al. A systematic review of selected musculoskeletal late effects in survivors of childhood cancer. Curr Pediatr Rev. 2014;10(4):249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zou H, Hastie T.. Regularization and variable selection via the elastic net. J Roy Stat Soc B. 2005;67(5):768–768. [Google Scholar]
- 54. Baron RM, Kenny DA.. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51(6):1173–1182. [DOI] [PubMed] [Google Scholar]
- 55. Dam TT, Peters KW, Fragala M, et al. An evidence-based comparison of operational criteria for the presence of sarcopenia. J Gerontol A Biol Sci Med Sci. 2014;69(5):584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Conley KE, Jubrias SA, Esselman PC.. Oxidative capacity and ageing in human muscle. J Physiol. 2000;526(pt 1):203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sorensen JC, Cheregi BD, Timpani CA, et al. Mitochondria: Inadvertent targets in chemotherapy-induced skeletal muscle toxicity and wasting? Cancer Chemother Pharmacol. 2016;78(4):673–683. [DOI] [PubMed] [Google Scholar]
- 58. Lebrecht D, Kokkori A, Ketelsen UP, et al. Tissue-specific mtDNA lesions and radical-associated mitochondrial dysfunction in human hearts exposed to doxorubicin. J Pathol. 2005;207(4):436–444. [DOI] [PubMed] [Google Scholar]
- 59. Pui CH, Evans WE.. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354(2):166–178. [DOI] [PubMed] [Google Scholar]
- 60. Lipshultz SE, Lipsitz SR, Sallan SE, et al. Chronic progressive cardiac dysfunction years after doxorubicin therapy for childhood acute lymphoblastic leukemia. J Clin Oncol. 2005;23(12):2629–2636. [DOI] [PubMed] [Google Scholar]
- 61. Gouspillou G, Scheede-Bergdahl C, Spendiff S, et al. Anthracycline-containing chemotherapy causes long-term impairment of mitochondrial respiration and increased reactive oxygen species release in skeletal muscle. Sci Rep. 2015;5:8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Octavia Y, Tocchetti CG, Gabrielson KL, et al. Doxorubicin-induced cardiomyopathy: from molecular mechanisms to therapeutic strategies. J Mol Cell Cardiol. 2012;52(6):1213–1225. [DOI] [PubMed] [Google Scholar]
- 63. Luo J, Shi R.. Acrolein induces oxidative stress in brain mitochondria. Neurochem Int. 2005;46(3):243–252. [DOI] [PubMed] [Google Scholar]
- 64. Crouch ML, Knowels G, Stuppard R, et al. Cyclophosphamide leads to persistent deficits in physical performance and in vivo mitochondria function in a mouse model of chemotherapy late effects. PLoS One. 2017;12(7):e0181086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Prithivirajsingh S, Story MD, Bergh SA, et al. Accumulation of the common mitochondrial DNA deletion induced by ionizing radiation. FEBS Lett. 2004;571(1-3):227–232. [DOI] [PubMed] [Google Scholar]
- 66. Wang L, Kuwahara Y, Li L, et al. Analysis of Common Deletion (CD) and a novel deletion of mitochondrial DNA induced by ionizing radiation. Int J Radiat Biol. 2007;83(7):433–442. [DOI] [PubMed] [Google Scholar]
- 67. Antipova VN, Lomaeva MG, Zyrina NV.. Mitochondrial DNA deletions in tissues of mice after ionizing radiation exposure. Int J Radiat Biol. 2018;94(3):282–288. [DOI] [PubMed] [Google Scholar]
- 68. Barjaktarovic Z, Schmaltz D, Shyla A, et al. Radiation-induced signaling results in mitochondrial impairment in mouse heart at 4 weeks after exposure to X-rays. PLoS One. 2011;6(12):e27811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Braun TP, Marks DL.. The regulation of muscle mass by endogenous glucocorticoids. Front Physiol. 2015;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lin J, Handschin C, Spiegelman BM.. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1(6):361–370. [DOI] [PubMed] [Google Scholar]
- 71. Puigserver P, Wu Z, Park CW, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829-. [DOI] [PubMed] [Google Scholar]
- 72. Liu X, Wang Y, Ortlund EA.. First high-resolution crystal structures of the glucocorticoid receptor ligand-binding domain-peroxisome proliferator-activated gamma coactivator 1-alpha complex with endogenous and synthetic glucocorticoids. Mol Pharmacol. 2019;96(4):408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sandri M, Lin J, Handschin C, et al. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA. 2006;103(44):16260–16265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang L, Zhou Y, Wu W, et al. Skeletal muscle-specific overexpression of PGC-1alpha induces fiber-type conversion through enhanced mitochondrial respiration and fatty acid oxidation in mice and pigs. Int J Biol Sci. 2017;13(9):1152–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hiona A, Sanz A, Kujoth GC, et al. Mitochondrial DNA mutations induce mitochondrial dysfunction, apoptosis and sarcopenia in skeletal muscle of mitochondrial DNA mutator mice. PLoS One. 2010;5(7):e11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kujoth GC, Hiona A, Pugh TD, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481–484. [DOI] [PubMed] [Google Scholar]
- 77. Li-Harms X, Milasta S, Lynch J, et al. Mito-protective autophagy is impaired in erythroid cells of aged mtDNA-mutator mice. Blood. 2015;125(1):162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang X, Pickrell AM, Rossi SG, et al. Transient systemic mtDNA damage leads to muscle wasting by reducing the satellite cell pool. Hum Mol Genet. 2013;22(19):3976–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Xia CY, Liu Y, Yang HR, et al. Reference intervals of mitochondrial DNA copy number in peripheral blood for Chinese minors and adults. Chin Med J (Engl). 2017;130(20):2435–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dhawan J, Rando TA.. Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 2005;15(12):666–673. [DOI] [PubMed] [Google Scholar]
- 81. Lexell J, Sjostrom M, Nordlund AS, et al. Growth and development of human muscle: a quantitative morphological study of whole vastus lateralis from childhood to adult age. Muscle Nerve. 1992;15(3):404–409. [DOI] [PubMed] [Google Scholar]
- 82. . Bentzinger CF, Wang YX, Rudnicki MA.. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4(2):a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen Y, Jungsuwadee P, Vore M, et al. Collateral damage in cancer chemotherapy: oxidative stress in nontargeted tissues. Mol Interv. 2007;7(3):147–156. [DOI] [PubMed] [Google Scholar]
- 84. Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 85. Pagliarini DJ, Calvo SE, Chang B, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Calvo SE, Mootha VK.. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010;11:25–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Longchamps RJ, Castellani CA, Yang SY, et al. Evaluation of mitochondrial DNA copy number estimation techniques. PLoS One. 2020;15(1):e0228166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kuznetsova T, Knez J.. Peripheral blood mitochondrial DNA and myocardial function. Adv Exp Med Biol. 2017;982:347–358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available from the corresponding author upon request.