Abstract
Background
Recent comprehensive studies have revealed several molecular alterations that are frequently found in meningiomas. However, effective treatment reagents targeting specific molecular alterations have not yet been identified because of the limited number of representative research models of meningiomas.
Methods
We performed organoid cultures using meningioma cells and meningioma tumor tissues. Using immunohistochemistry and molecular analyses consisting of whole-exome sequencing, RNA-seq, and DNA methylation analyses, we compared the histological findings and molecular profiling of organoid models with those of parental tumors. Further, using these organoid models together with a public database of meningiomas, we explored molecular alterations, which are a potent treatment target for meningioma.
Results
We established 18 organoid models comprising of two malignant meningioma cells (HKBMM and IOMM-Lee), 10 benign meningiomas, four malignant meningiomas, and two solitary fibrous tumors (SFTs). The organoids exhibited consistent histological features and molecular profiles with those of the parental tumors. Using a public database, we identified that upregulated forkhead box M1 (FOXM1) was correlated with increased tumor proliferation. Overexpression of FOXM1 in benign meningioma organoids increased organoid proliferation; depletion of FOXM1 in malignant organoids decreased proliferation. Additionally, thiostrepton, a FOXM1 inhibitor combined with radiation therapy, significantly inhibited the proliferation of malignant meningioma organoid models.
Conclusions
An organoid model for meningioma enabled us to elucidate the tumor biology of meningioma along with potent treatment targets for meningioma.
Keywords: FOXM1, meningioma, organoid model
Key Points.
We established 14 organoid models of meningiomas with a 100% success rate.
Organoids maintained histological and molecular features of parental meningiomas.
In vitro studies revealed that FOXM1 contributed to the tumor growth of meningioma.
Importance of the Study.
Organoid models have recently emerged as relevant in vitro experimental models of various neoplasms. Basic and translational meningioma research has, however, been highly restricted owing to the limited number of experimental tools available. Biological experiments using organoid models of meningiomas have also not yet been established. Here, we efficiently established patient-derived meningioma organoids that recapitulated the morphological and molecular features of parental tumors. Genetic engineering experiments using organoid models and administration of FOXM1 inhibitor combined with radiation therapy for organoid models revealed that FOXM1 upregulation contributed to the proliferation of meningiomas. Our organoid models will thus provide more insight into the tumor biology of meningiomas.
Meningioma is the most common primary tumor of the central nervous system (CNS). Based on histological features, meningiomas are categorized into three grades according to the World Health Organization: Grades I, II, and III. Approximately 80% of meningiomas are benign tumors (Grade I)1 and most Grade I meningioma cases are favorably managed by surgical removal with a 5%–10% recurrence rate at 5 years.2 However, some cases of benign meningiomas located at technically challenging locations for extended total removal (eg skull base meningiomas or parasagittal meningiomas) exhibit higher recurrence rates, despite the frequent use of adjuvant radiotherapy. Effective treatment reagents for recurrent meningiomas have not yet been identified. Recent comprehensive genetic analyses revealed several alterations frequently found in meningioma tissues such as NF2, POLR2A, TRAF7, KLF4, AKT1, SMARCB1, SMARCE1, and SMO.3–6 Additionally, comprehensive transcriptional analyses revealed that forkhead box M1 (FOXM1) was one of the most important transcription factors for meningioma proliferation.7 Although the data suggested that these altered molecules contributed to the initiation or proliferation of meningiomas, the functional analyses that could reveal that these alterations were optimal treatment targets were insufficient. One of the obstacles to these analyses is the limited number of established experimental materials, such as meningioma cell lines or animal models.8,9 Reliable models for fundamental research are not only essential for the investigation of tumor biology but also for the evaluation of the efficacy of potential targeted reagents.
Recently, organoid culture technologies have been developed that represent phenotypic and molecular features in various organs, including cerebral tissues.10–12 Organoid culture technologies have been applied to model various cancers including pancreatic,13 liver,14 and gastrointestinal cancers.15 The efficiency of the organoid establishment was reported to be higher than that of tumor cell lines or patient-derived xenograft (PDX) models in various cancers.11 Additionally, patient-derived organoids were considered to better recapitulate native tumors than tumor cell lines and they might be optimal models for identifying potential treatment reagents.16 Organoid models of meningiomas could be a reliable platform for basic and clinical research on meningiomas.
In this study, we established 16 organoid models, including 10 Grade I meningiomas, three Grade II meningiomas, one Grade III meningioma, and two solitary fibrous tumors (SFTs), using 16 patient-derived tumor tissues, with a 100% success rate. These organoid models revealed morphological features and molecular profiles similar to those of parental tumors. Using these organoid models, we revealed that overexpression of the FOXM1 gene increased Grade I meningioma organoid proliferation. In contrast, knockdown of FOXM1 in malignant meningioma organoids inhibited organoid growth. Thiostrepton, a FOXM1 inhibitor,17 combined with radiation therapy significantly inhibited the proliferation of meningioma organoid models. In vitro experiments using this organoid model of benign or malignant meningioma demonstrated that aberrantly upregulated FOXM1 might be a potent treatment target.
Materials and Methods
Ethics Approval and Consent to Participate
The present study was approved by the institutional review board at Nagoya University Hospital (approval number: 2012–0067) and complied with all provisions of the World Medical Association Declaration of Helsinki. Tumor samples were collected intraoperatively upon receiving informed consent from the patients.
Generation of Organoids of Meningioma and SFT
Fresh surgically resected tissues were immediately minced into 1 mm3 piece in organoid culture medium (Dulbecco's Modified Eagle's Medium [DMEM; Sigma–Aldrich] supplemented with 10% fetal bovine serum [FBS] and 1% penicillin–streptomycin [PS; Thermo Fisher Scientific]) or serum-free culture medium (Neurobasal Medium, N-2, B-27, PS [Thermo Fisher Scientific], recombinant human FGF Protein [50 ng/mL], and EGF Human Recombinant Protein [50 ng/mL; R&D Systems]). After removal of red blood cells and debris, dissociated cells were resuspended in Matrigel Basement Membrane Matrix (Corning Inc.), which was diluted with the same amount of culture medium. Then, the Matrigel was incubated till solidification and subsequently overlaid with a culture medium. The culture medium was refreshed every few days. These organoids were passaged every 2–4 weeks. More detailed methods were described in Supplementary Methods.
For organoid cryopreservation, organoids were resuspended in BAMBANKER (GC LYMPHOTEC Inc.) and stored at −80°C. After frozen storage for more than two weeks, the organoids were quickly thawed and cultured. Histological analysis of postcryopreservation organoids was performed one week after thawing.
Malignant Meningioma and Hepatocellular Carcinoma Cells
HKBMM (RIKEN BioResource Research Center), IOMM-Lee (American Type Culture Collection [ATCC]), and HepG2 (ATCC) were cultured in DMEM with 10% FBS and 1% PS.
Organoid Fixation for Hematoxylin and Eosin (HE) Staining
After the Matrigel was removed using dispase, organoids were fixed in 4% paraformaldehyde (Wako). The organoids were enclosed with 3D Ready Atelocollagen (Koken) and embedded in paraffin.
Immunohistochemistry (IHC)
Formalin-fixed paraffin-embedded tissues were sectioned and deparaffinized accordingly. Primary antibodies were anti-Ki-67 antibody (Dako), recombinant antisomatostatin receptor 2 antibody (UMB1)-C-terminal, recombinant anti-STAT6 antibody, and recombinant anti-FOXM1 antibody (Abcam). Secondary antibodies were goat anti-mouse IgG H&L (HRP polymer; Abcam) or goat anti-rabbit IgG H&L (HRP polymer; Abcam). For image analyses of IHC staining or tumor sections, all images were obtained under appropriate conditions using a BZ-X710 microscope (KEYENCE). The Ki-67 positive index was obtained by calculating the average percentage of positively stained nuclei in three fields. More detailed methods were described in Supplementary Methods.
Western Blot Analysis
Cell lysates were extracted from organoids using radioimmunoprecipitation assay (RIPA) buffer. Primary antibodies were recombinant anti-FOXM1 antibody (Abcam) and β-Actin Mouse mAb (Cell Signaling Technology). Secondary antibodies were anti-rabbit IgG-HRP and m-IgGκ BP-HRP (Santa Cruz Biotechnology). More detailed methods were described in Supplementary Methods.
DNA and RNA Extraction From Patient Tumors, Blood, and Organoids
DNA and RNA were extracted using a QIAamp DNA Mini Kit (Qiagen) and RNeasy Mini Kit (Qiagen) following the manufacturer's instructions.
Whole-exome Sequencing (WES) and Variant Calling
In this study, candidate variants that met all the following criteria were adopted: (i) variants annotated as exonic/splicing and nonsynonymous; (ii) variants with <1% allele frequency in the Exome Aggregation Consortium and not registered in NCBI dbSNP build 138 or avsnp150; (iii) both tumor and normal sample depth ≥20; (iv) number of variant reads in tumor ≥10; (v) variant allele frequency (VAF) in tumors >0.1; and (vi) VAF in normal samples <0.05. The number of protein-altering somatic variants (normalized per megabase pair of sequencing data) identified with the criteria was 0.19, which was within the previously reported range (0.17–0.23).18 More detailed methods were described in Supplementary Methods.
Targeted Resequencing and Variant Calling
The filtered candidate variants in the parental tumor (PT) and those reported in a previous study4 were validated by PCR amplification of the corresponding reads, followed by sequencing on the MiSeq platform (Illumina) with a 75-bp paired-end mode (Supplementary Table S1). The primers used for validation are summarized in Supplementary Table S2. More detailed methods were described in Supplementary Methods.
Structural Variation (SV) Analyses
Copy number variation (CNV) analyses were performed using the CNVkit software19 with default parameters by comparing the parental tumor or organoid with its corresponding normal control (Supplementary Table S3). A heatmap was created using the heatmap command of the CNVkit.
DNA Methylation Analysis
DNA extracted from frozen tumors and matched organoids was treated with sodium bisulfite using the Zymo EZ DNA Methylation kit (Zymo Research), and then converted DNA was analyzed on an Illumina Infinium Methylation EPIC 850k BeadChIP (Illumina). More detailed methods were described in Supplementary Methods.
RNA-Seq Data Analyses
cDNA libraries were prepared using the NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs) and the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs). We also analyzed publicly available RNA-seq data of meningiomas (n = 160, Grade I: 121, Grade II: 32, Grade III: 7)20 using iDEP (‘limma-voom’ option).21 Selection of 15 protein-coding genes with reading counts of more than 25 was carried out among the 76 differentially expressed genes in Grade II or III meningiomas compared with Grade I tumors (log2 fold change > 1.5, FDR < 10–5). More detailed methods were described in Supplementary Methods.
RNA Interference
To downregulate gene expression with small interfering RNA (siRNA), two independent siRNAs targeting the FOXM1 gene (HSS103713 and HSS177135, Thermo Fisher Scientific) were used.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
RNA was reverse-transcribed with ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO Co. Ltd.) according to the manufacturer's instructions. The primer sequences used to evaluate FOXM1 and LMNA gene were described in Supplementary Methods.
Construction of the FOXM1 Expression Vector
The amplified PCR product of FOXM1 was digested with BamHI and HindIII and ligated into the multi-cloning site of the pcDNA3.0 vector. The primer sequences were described in Supplementary Methods.
Cell Proliferation Assay
Cell proliferation was assessed using a CellTiter-Glo®️ 3D Cell Viability Assay (Promega Corporation) according to the manufacturer's instructions in triplicate. After 72 h of transfection of siRNAs for FOXM1, negative control siRNA, FOXM1 expression vector, or pcDNA3.0, organoid size, and the number of generated organoids were counted in triplicate.
The organoids were treated with Thiostrepton (Santa Cruz Biotechnology) at the respective concentrations or 0.1% DMSO for 72 h with or without radiation (4 Gy) via MBR-1520R-3 (Hitachi) after 24 h of treatment.
Statistical Analyses
Differences between groups were analyzed using paired Student's t-tests, Wilcoxon signed-rank tests, and Wilcoxon rank-sum tests with R Statistical Software (version 3.6.1; R Foundation for Statistical Computing). All reported P values were two-sided, and statistical significance was set at P < .05.
Results
Organoid Culture for Malignant Meningioma Cells and Patient-Derived Tumors
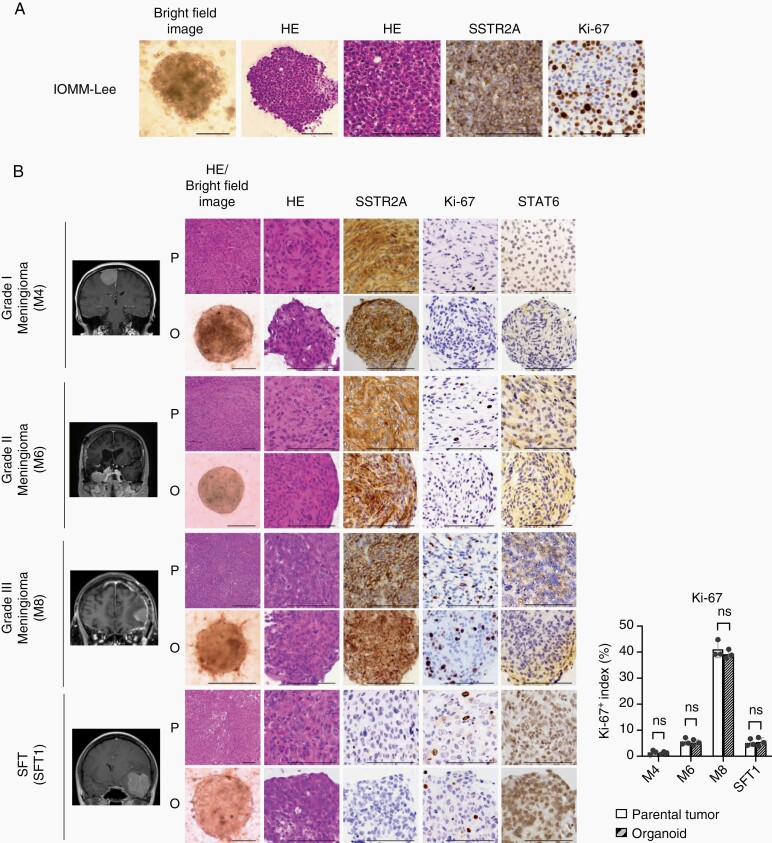
To establish optimal organoid culture conditions for meningiomas, we performed organoid culture using malignant meningioma cells (IOMM-Lee and HKBMM). Dissociated cells were resuspended in Matrigel, and these mixtures were placed on a cell culture plate. They were then overlaid with a modified organoid culture medium. Remarkably, we obtained rounded structures exhibiting histological findings similar to those of malignant meningioma tissues. Because somatostatin receptor 2A (SSTR2A) is a highly sensitive and specific marker of meningioma,22 we performed IHC using anti-SSTR2A and anti-Ki-67 antibodies. IHC revealed that these organoid models exhibited molecular features similar to those of Grade III meningiomas (Figure 1A and Supplementary Figure S1A). Using these methods, we generated patient-derived organoid models for 10 Grade I meningiomas, three Grade II meningiomas, one Grade III meningioma, and two SFTs (Table 1). Surgically resected specimens were immediately dissociated mechanically and enzymatically. Dissociated cells were plated on cell culture plates using the same method as that used for the generation of organoid models of IOMM-Lee or HKBMM cells. Dissociated cells proliferated and formed spherical organoids within one week. We defined a successful organoid generation as an event in which organoid models could survive, develop a spherical morphology, and continuously grow in culture for two weeks, as described previously.12 We confirmed that meningioma organoids could be established with a 100% succession rate. Remarkably, all Grade I meningioma organoids exhibited histological and morphological features that were compatible with those of the parental tumors. Additionally, Grades II and III meningioma and SFT organoids revealed higher expression of Ki-67 or nuclear STAT6 than Grade I meningioma organoids (Figure 1B). Overexpression of nuclear STAT6, which is induced by NAB2-STAT6 fusion, is a specific and sensitive finding of SFTs, whereas expression of STAT6 was detected only in the cytoplasm of meningioma.23 These data indicated that the established meningioma organoids maintained the histological and molecular features of corresponding parental tumors along with SFT organoids (Figure 1B and Supplementary Figure S1B). A previous study reported that successful culture of meningioma cell spheres using serum-free medium.24 DMEM with 10% FBS medium did not increase organoid size but increased the number of generated organoids of meningioma compared with serum-free medium (Supplementary Figure S1C). We continued to culture organoid models with a maximum number of passages of 26 (M1: Grade I meningioma; now we continue to culture M1 organoids). The number of passages for all organoid models is described in Table 1. Organoid models cultured after cryopreservation exhibited similar histological features and comparable Ki-67 positive index to those of organoid models before cryopreservation (Supplementary Figure S1D). Additionally, morphological features, Ki-67 positive index, and expression status of SSTR2A or nuclear STAT6 were maintained in organoids cultured over various passages (Supplementary Figure S2).
Fig. 1.
Established meningioma and SFT organoids recapitulate the histologic features of corresponding parental tumors. (A) Bright-field images, HE staining and IHC for anti-SSTR2A and anti-Ki-67 antibodies in the organoids (IOMM-Lee). Scale bars indicate 100 μm. (B) Magnetic resonance images, bright-field images, HE staining, and IHC using anti-SSTR2A, anti-Ki-67, and anti-STAT6 antibodies with Grades I (M4), II (M6), and III (M8) meningiomas and an SFT (SFT1) as parental tumors (P) and the organoids (O). Scale bars indicate 100 μm. Bar graph (right) indicating Ki-67 expression status. Ki-67 positive index was calculated and averaged in three fields. Average cell counts in a field were 130 (P) and 143 (O). The y-axis indicates Ki-67 positive index. Error bars indicate SD (ns: not significant).
Table 1.
Clinical Characteristics of 16 Parental Tumors
| ID | Patient Age/Sex | Tumor Type | Status | Tumor Location | Grade | Subtype | Passage |
|---|---|---|---|---|---|---|---|
| M1 | 52/Female | Meningioma | Primary | Petroclival | I | Meningothelial | 26 |
| M2 | 63/Female | Meningioma | Primary | Falx | I | Meningothelial | 23 |
| M3 | 58/Male | Meningioma | Primary | Olfactory groove | I | Meningothelial | 25 |
| M4 | 56/Female | Meningioma | Primary | Parasagittal | I | Meningothelial | 24 |
| M5 | 65/Female | Meningioma | Primary | Convexity | I | Meningothelial | 21 |
| M6 | 83/Female | Meningioma | Recurrent | Convexity | II | Atypical | 11 |
| M7 | 71/Female | Meningioma | Recurrent | Middle cranial fossa | II | Atypical | 13 |
| M8 | 77/Female | Meningioma | Recurrent | Convexity | III | Anaplastic | 20 |
| M9 | 64/Female | Meningioma | Primary | Cavernous sinus | I | Secretory | 4 |
| M10 | 57/Male | Meningioma | Primary | Parasagittal | I | Meningothelial | 3 |
| M11 | 37/Female | Meningioma | Primary | Tuberculum sellae | I | Meningothelial | 2 |
| M12 | 47/Female | Meningioma | Recurrent | Middle cranial fossa | I | Meningothelial | 2 |
| M13 | 59/Female | Meningioma | Primary | Falx | I | Meningothelial | 1 |
| M14 | 71/Male | Meningioma | Primary | Falx | II | Atypical | 4 |
| SFT1 | 34/Female | SFT | Primary | Middle cranial fossa | II | 23 | |
| SFT2 | 40/Male | SFT | Primary | Falx | I | 12 |
Molecular Profiling of Organoid Models and Their Corresponding Parental Tumors
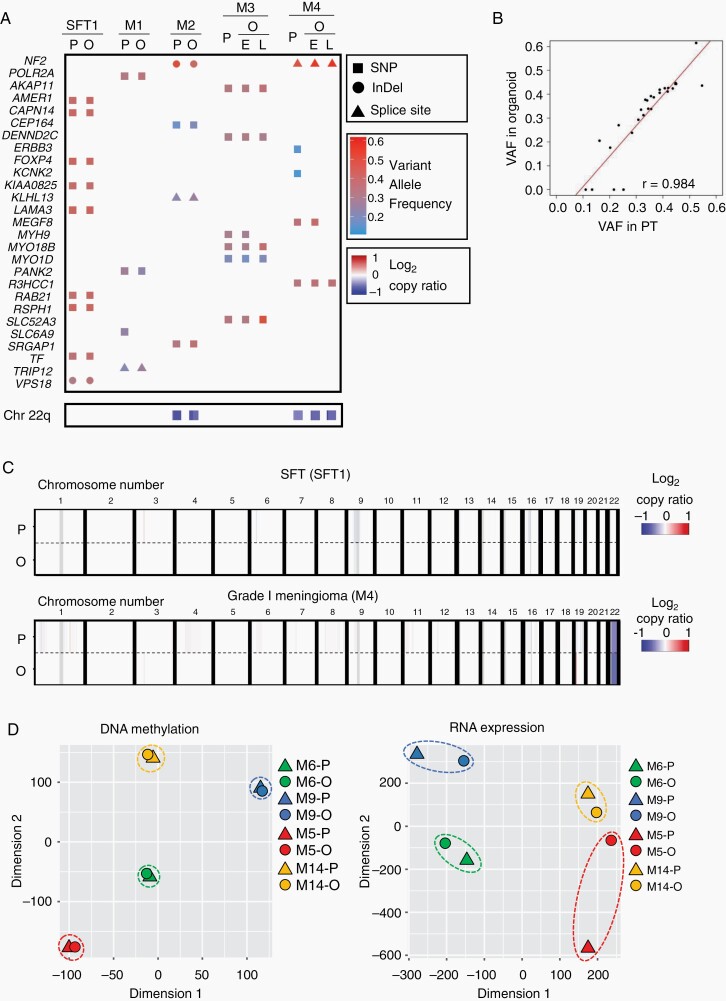
To compare the genetic profiling of organoid models with those of parental tumors, we performed WES and deep sequencing using DNA extracted from five organoids that were successfully cultured for at least 10 passages. We extracted DNA from these organoids, 2–8 weeks (M1, four passages; M2, 3, 4, and SFT1, one passage) after initiation of organoid culture, together with their corresponding parental tumor tissues and matched blood samples. WES and deep sequencing revealed that organoid models exhibited a consistent genetic mutation with those of parental tumor tissues in genes whose alterations were not only reported to be frequently found in meningiomas but also specifically found in each case (Figure 2A). Additionally, organoid models exhibited VAFs that were quite similar to those found in parental tumors (r = 0.984; Figure 2B). Structural variant analysis also revealed that loss of chromosome 22q, which is one of the most characteristic genetic alterations in meningiomas, was detected in both organoid models and parental tumor tissues with consistent copy number values (Figure 2A). In SFT and meningioma cases, other gains or losses of the whole chromosome arm were not identified in either parental tumors or organoids (Figure 2C and Supplementary Figure S3A). Based on the gene analyses of long-term cultured organoid models (M3 and M4), we found that important genetic alterations such as NF2 gene mutation and loss of chromosome 22q were retained after long-term culture, although a few gene alterations were altered (Figure 2A). The number of somatic variants (normalized per megabase pair of sequencing data) identified in long-term cultured organoid models were consistent with those of parental tumors (M3: 0.10 [long-term cultured organoid] and 0.10 [parental tumor] and M4; 0.06 [long-term cultured organoid] and 0.09 [parental tumor]). In the SFT case (SFT1), we detected NAB2-STAT6 fusion, which was the most characteristic structural variant of SFT, in both the organoid model and parental tumor tissue with the same breakpoint of these genes (Supplementary Figure S3B).
Fig. 2.
Meningioma and SFT organoids maintain genetic profiles of corresponding parental tumors. (A) Heatmap indicating somatic variants in SFT and meningioma-associated genes and a copy number variation in chromosome 22q identified by WES of the organoids (O) and corresponding parental tumors (P). In M3 and M4, E and L indicate data of organoids in their early-passages (M3; 1 passage and M4; 1 passage) or late-passages (M3; 23 passages and M4; 22 passages), respectively. Rectangles, circles, and triangles in the top panels represent single nucleotide polymorphisms (SNP), insertions or deletions (InDel), and mutations in splice site (Splice site), respectively, with the variant allele frequencies (VAFs). Blue colored rectangles in the bottom represent the relative copy numbers of chromosome 22q (Chr 22q) as indicated in log2-copy ratio. (B) Scatter plot indicating correlation of the VAFs of genetic mutations between the organoids and corresponding parental tumors. The x- or y-axis axis indicate VAF in parental tumors (PT) or organoids, respectively. (C) Structural variants in autosomal chromosomes of parental tumors (P) or organoids (O) of SFT1 and M4 (Grade I). (D) t-SNE plot indicating DNA methylation and RNA expression status of M5 (red), M9 (blue; Grade I), M6 (green), and M14 (yellow; Grade II). Circles or triangles indicate organoids (O) or parental tumors (P).
Next, we compared the DNA methylation and RNA expression status of organoid models with those of parental tumors. We extracted DNA and RNA from these organoids 2 weeks after the initiation of organoid culture, together with their corresponding parental tumor tissues. The t-distributed stochastic neighbor embedding (t-SNE) analyses revealed that all organoid models exhibited similar profiling of DNA methylation and RNA expression as those of parental tumors (M5 and M9; Grade I, M6, and M14; Grade II: Figure 2D). These data revealed that the organoid model maintained the molecular profile of these parental tumors.
Correlation of Upregulated FOXM1 Expression Level With Increased Proliferation of Meningioma
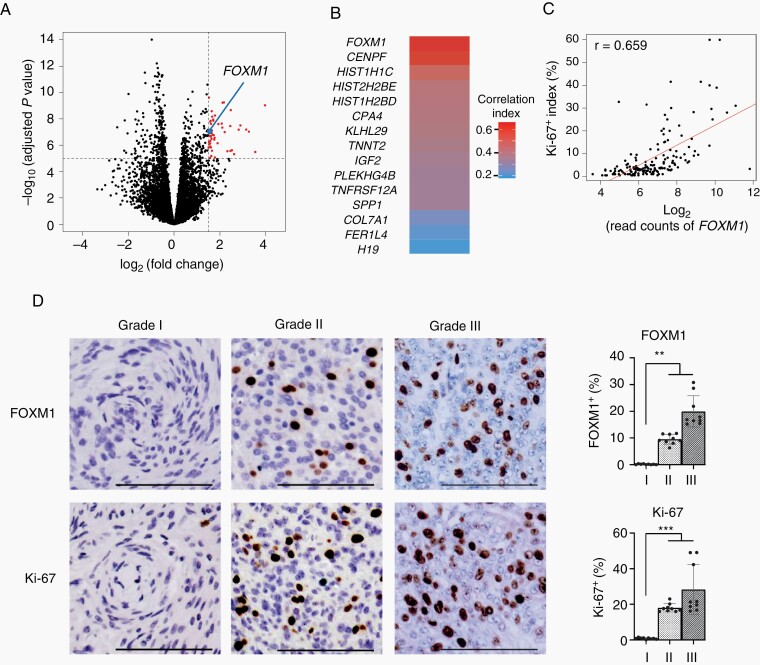
To identify the characteristic molecular alterations that contributed to meningioma proliferation, we analyzed published gene expression data for Grades I, II, and III meningiomas (GSE136661). First, we compared the gene expression profiles of Grade I and Grade II or III meningiomas. We identified 76 genes that were significantly differentially upregulated in Grade II or III meningiomas compared to those in Grade I tumors (Figure 3A). Among these 76 differentially expressed genes, we selected 15 genes that exhibited high expression levels in Grade II and III meningiomas. Among these 15 genes, we found that FOXM1 expression levels were the most correlated with Ki-67 positive indexes in meningioma tissues (r = 0.659; Figure 3B and C and Supplementary Table S4). Consistent with these data, IHC revealed higher expression levels of FOXM1 in Grade II or III meningiomas (n = 3 or 3) than in Grade I meningiomas (n = 3) (Figure 3D). Another report also demonstrated that FOXM1 contributed to the proliferation of malignant meningiomas.7,25 However, no studies have demonstrated that upregulated FOXM1 contributes to the proliferation of meningioma using in vitro experiments because of the lack of widely accepted benign meningioma cell lines. Therefore, we analyzed the overexpression or knockdown effects of FOXM1 on meningioma proliferation using established meningioma organoids.
Fig. 3.
An upregulated expression of FOXM1 is associated with an increased meningioma proliferation. (A) Volcano plot illustrating the difference in gene expression between Grade I and Grades II and III meningiomas. Log2-fold change in gene expression values is plotted on the x-axis and calculated adjusted log10P values are plotted on the y-axis. Horizontal dotted line indicates −log10 (adjusted P value) = 5; vertical dotted line indicates log2 (fold change) = 1.5. A blue dot indicates FOXM1. (B) Heatmap indicating correlation index between Ki-67-positive scores and expression levels of 15 genes upregulated in malignant meningiomas. (C) Scatter plot illustrating the correlation between Ki-67 positive index and FOXM1 expression in meningioma tissues. The x-axis indicates log2-read counts of FOXM1. The y-axis indicates the Ki-67 positive index (%). (D) IHC images (left) using either anti-FOXM1 antibody (upper) or anti-Ki-67 antibody (lower) in Grades I, II, and III meningioma tissues. Bar graphs (right) indicating either FOXM1 or Ki-67 expression status. FOXM1 or Ki-67 positive indexes were calculated and averaged in three fields of IHC samples from three various cases of each grade. The average number of counted cells were 546 (FOXM1), or 592 (Ki-67) per field. The y-axis indicates either FOXM1 or Ki-67 positive index. Scale bars indicated 100 μm. Error bars indicate SD (**P < .05 or ***P < .01).
Overexpression or Knockdown Effects of FOXM1 Gene in Meningioma Organoids
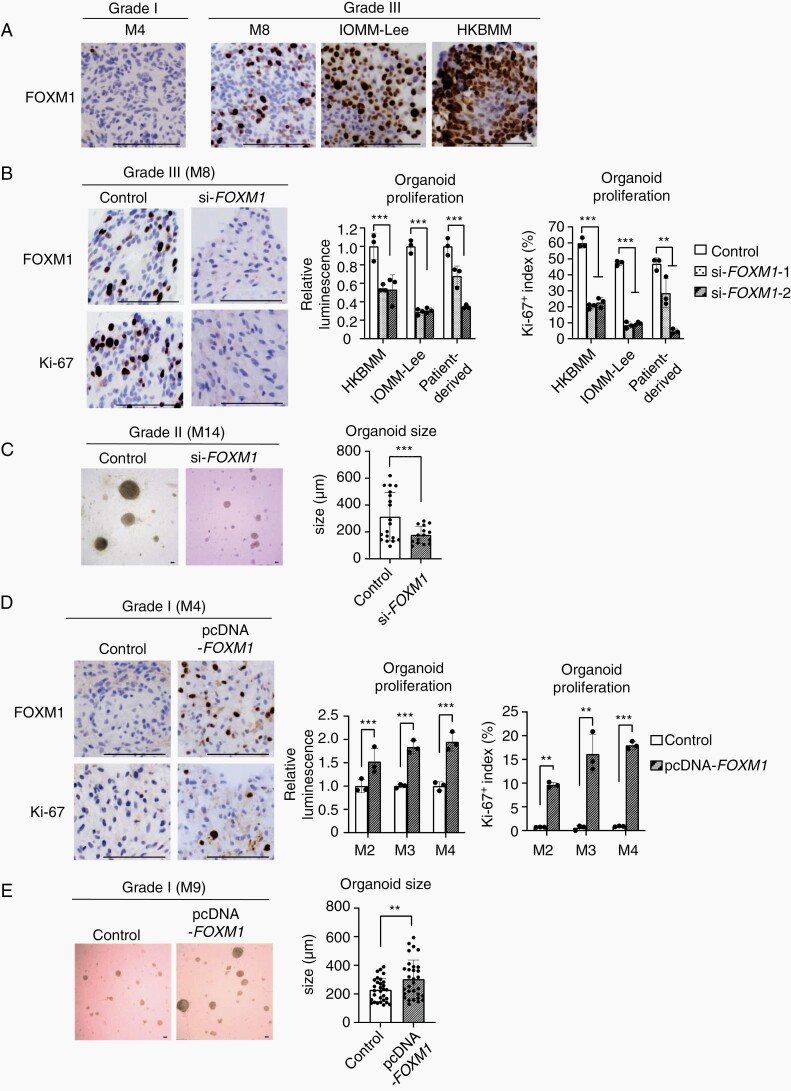
Consistent with the expression data in meningioma tissues, IHC revealed that FOXM1 expression levels were highly upregulated in Grade III meningioma organoids together with organoids generated from malignant meningioma cells (IOMM-Lee and HKBMM), while Grade I meningioma organoids exhibited low expression levels of FOXM1 (Figure 4A). We analyzed organoid proliferation after knockdown or overexpression of the FOXM1 gene using Grade I, II, or III meningioma organoids. Protein expression levels of FOXM1 were significantly downregulated or upregulated after knockdown or overexpression of FOXM1 (Supplementary Figure S4A). First, we knocked down FOXM1 using siRNA in Grade II or III meningioma organoids consisting of patient-derived, IOMM-Lee, and HKBMM. Depletion of FOXM1 significantly decreased the proliferation and size of these organoids, together with downregulation of Ki-67 expression levels (Figure 4B and C). In contrast, overexpression of FOXM1 significantly increased the proliferation and size of Grade I meningioma organoids, together with upregulation of Ki-67 expression levels (Figure 4D and E). Histological findings of Grade I meningioma organoids with overexpression of FOXM1 for 7 days were consistent with those of Grade I meningioma, but not with those of Grade II or III meningiomas (Supplementary Figure S4B). Collectively, these findings hence suggest that FOXM1 expression promotes the progression of benign meningiomas.
Fig. 4.
Altered FOXM1 expression status affected the proliferation of meningioma organoids. (A) IHC images using anti-FOXM1 antibody with the organoids of Grades I (M4) and III (M8), IOMM-Lee cells, or HKBMM cells. Scale bars indicate 100 μm. (B) IHC images (left) using anti-FOXM1 and anti-Ki-67 antibodies with Grade III organoid (M8) treated either with negative control siRNA or si-FOXM1. Scale bars indicate 100 μm. Bar graphs (right) indicating the cell viabilities or Ki-67 positive indexes. The Ki-67 positive indexes were averaged in three fields. The average number of counted cells were 200 per field. The y-axis indicates either relative luminescence to those of control or Ki-67 positive index (%). Error bars indicate SD (**P < .05 or ***P < .01). (C) Bright-field image (left) of organoid (M14; Grade II) treated either with negative control siRNA or si-FOXM1-2. Scale bars indicated 100 μm. Bar graphs (right) indicating average sizes of organoids treated with negative control siRNA or si-FOXM1-2. The organoid sizes were averaged in three fields. The y-axis indicates organoid sizes (μm). Error bars indicate SD (***P < .01). (D) IHC images (left) using anti-FOXM1 and anti-Ki-67 antibodies with Grade I organoid (M4) after transfection of either pcDNA3.0 or pcDNA3.0-FOXM1. Scale bars indicated 100 μm. Bar graphs (right) indicating the cell viabilities or Ki-67 positive indexes with these organoids. The Ki-67 positive indexes were averaged in three fields of IHC images. The y-axis indicates either relative luminescence to those of control or Ki-67 positive index (%). Error bars indicate SD (**P < .05 or ***P < .01). (E) Bright-field image (left) of organoid (M9; Grade I) after transfection of either pcDNA3.0 (Control) or pcDNA3.0-FOXM1. Scale bars indicate 100 μm. Bar graphs (right) indicating average sizes of organoids. The organoid sizes were averaged in three fields. The y-axis indicates organoid sizes (μm). Error bars indicate SD (**P < .05).
Combination Therapy of Radiation Therapy and a FOXM1 Inhibitor for Meningioma Organoids
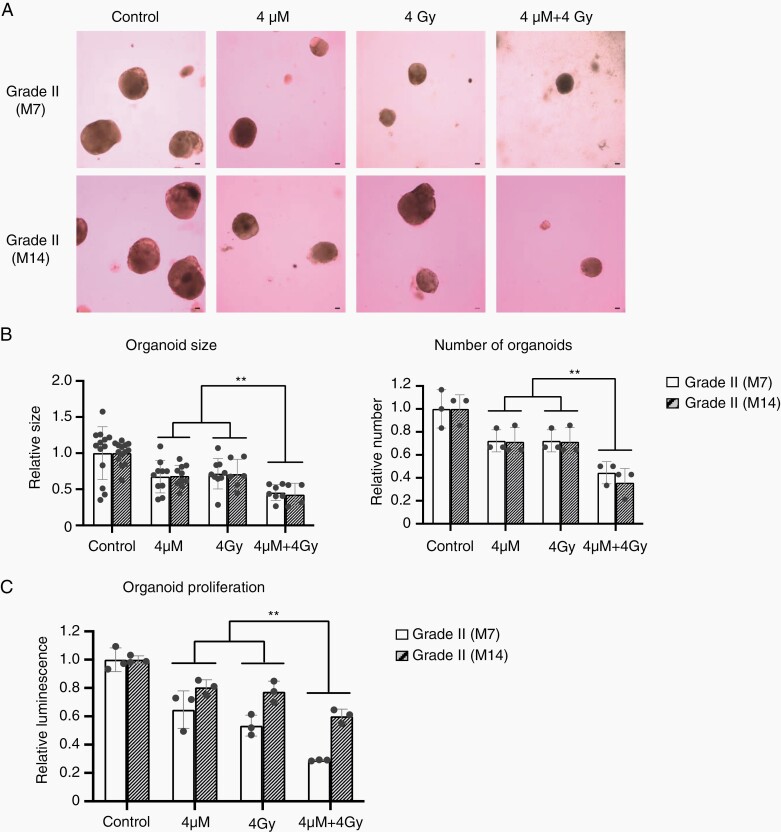
Using meningioma organoid models, we evaluated the treatment effect of radiation therapy. More than 4 Gy radiation therapy significantly inhibited proliferation of organoid models of IOMM-Lee cells, as a previous study reported that these radiation therapies inhibited cell proliferation of IOMM-Lee cells cultured with the conventional two-dimensional (2D) culture method26 (Supplementary Figure S5A). Thiostrepton, which reduces the transcriptional activity of FOXM1, is a widely used FOXM1 inhibitor.17,27 More than 4 μM thiostrepton significantly inhibited the proliferation of organoid models of IOMM-Lee cells (Supplementary Figure S5B). Therefore, we treated organoid models of Grade II meningiomas (M7 and M14; n = 2) using a combination therapy of 4 μM thiostrepton and 4 Gy radiation therapy. Combination therapy significantly decreased organoid size and the number of organoids (Figure 5A and B) along with proliferation (Figure 5C) of Grade II meningioma organoids (M7 and M14) more strongly than those treated with a single treatment or as a combination therapy for organoids of IOMM-Lee and HKBMM (Supplementary Figure S5C and D). These data revealed that combination therapy with radiation therapy and a FOXM1 inhibitor may be effective for malignant meningioma cases.
Fig. 5.
Radiation therapy and/or treatment with thiostrepton for meningioma organoid models. (A) Bright-field image of M7 (Grade II) and M14 (Grade II) organoids treated with radiation therapy (4 Gy) and/or thiostrepton (4 μM). Scale bars indicate 100 μm. (B) Bar graphs indicating average sizes or number of organoids (M7 and M14) treated with radiation therapy (4 Gy) and/or thiostrepton (4 μM). These data were averaged in three fields. The y-axis indicates relative organoid sizes or number to those of control. Error bars indicate SD (**P < .05). (C) Bar graphs indicating the cell viabilities of organoids (M7 and M14) treated with radiation therapy (4 Gy) and/or thiostrepton (4 μM). The y-axis indicates relative luminescence to those of control. Error bars indicate SD (**P < .05).
Discussion
In this study, we established a method for patient-derived organoid culture of various grade meningiomas, including Grade I meningiomas with high efficiency of the establishment. In vitro experiments using these patient-derived organoids revealed that FOXM1 directly contributed to meningioma growth. Additionally, a FOXM1 inhibitor, especially combined with radiation therapy, significantly inhibited the proliferation of meningioma organoid models. These findings suggest that patient-derived organoid models for benign or malignant meningiomas are quite effective for the biological analyses of meningiomas via in vitro experiments.
In oncological research, the most frequently used research materials are tumor cells in 2D cultures or animal models, including xenograft models. Although successful establishment of several tumor cells of meningiomas in 2D culture has been reported, most of these cells are derived from high-grade meningiomas. Although a limited number of Grade I meningioma cells in 2D culture have been established, these cells are frequently artificially immortalized by the introduction of the human telomerase reverse transcriptase (hTERT) gene.18 Artificial immortalization might be one of the most critical obstacles to understanding the biology of the original benign meningioma.8 In various cancers, during the establishment of stable cells, a subpopulation of cells among heterogeneous tumor tissues can be dominated clonally. A recent study reported that organoid models could recapitulate the cellular diversity of tumor tissues. Additionally, GBM organoid models have been reported to recapitulate some elements of the tumor microenvironment, such as the microvasculature.12 Therefore, organoid models could be utilized as useful research materials for further biological studies and for testing the efficiency of therapeutic reagents for meningiomas. Recently, PDX models, which are generated by transplantation of patient-derived cancer cells into immunodeficient mice, have emerged as efficient research models. Several studies have reported successful establishment of PDX models for meningioma; however, these studies revealed that even in malignant meningiomas, the generation of PDX models of meningiomas was quite inefficient.9 Although PDX approaches might be time- and resource-consuming, organoid models could be established using a small piece of tumor tissue for 2 weeks with a 100% success rate. Additionally, organoid models could be more optimal for high-throughput drug screening tests than PDX models.
Using recently developed 3D culture technologies, tissue-derived adult stem cells can be grown into organoids with high efficiency.11 Organoid models for various normal human tissue types have already been established, for example, among brain tumors, organoids of glioblastoma, and medulloblastoma respectively.28,29 These organoids are regarded as superior models for identifying and testing anticancer reagents.30 Additionally, genome editing experiments using the CRISPR-Cas9 system and RNA interference experiments using siRNA could be performed with organoids.20 To date, the successful establishment of organoid models of meningiomas has not been reported. In this study, we have revealed that newly established organoid models of meningiomas are available for genetic engineering experiments.
In meningiomas, several molecular alterations (eg Kruppel-like factor 4 [KLF4] and kinesin family member 11 [KIF11]) have been reported as potent treatment targets.21,31 Targeted therapies for altered AKT1 and SMO are currently being investigated in a subset of meningiomas (ClinicalTrials.gov: NCT02523014). However, clinical trials targeting specific molecular alterations in meningiomas are limited. In this study, we revealed that FOXM1, which contributes to meningioma proliferation, could be a potent therapeutic target in in vitro experiments. FOXM1, a pro-mitotic transcription factor, contributes to cell proliferation. Several studies have reported that upregulated FOXM1 plays a pivotal role in meningioma proliferation as a master transcription factor, resulting in a potent treatment target.7,25 Upregulated FOXM1 aberrantly activates the expression levels of various important genes, including β-catenin, cyclin D1, p21, interleukin 8 (IL-8), and vascular endothelial growth factor A (VEGF-A)25 thus promoting tumor progression in various cancers including glioblastoma, liver cancer, and breast cancer.32–34 Although no reagents inhibiting FOXM1 have been approved for clinical use in cancer treatment, several potent inhibitors of FOXM1 such as siomycin A or thiostrepton have been reported.17,27,35 Further investigation to unravel the molecular mechanisms by which FOXM1 contributes to meningioma proliferation using increased numbers of patient-derived organoid models might result in the establishment of a treatment strategy targeting the aberrantly activated FOXM1 pathway for refractory meningioma.
In this study, one of the intrinsic limitations of meningioma organoid culture was the lack of surrounding normal tissue. Meningiomas are formed from the meninges and malignant meningiomas frequently exhibit brain invasion. In order to unravel the tumor biology of meningioma, we may need to analyze the interaction between meningioma organoids and these normal cells. A previous study reported successful 2D culture of normal leptomeningeal cells.36 The successful establishment of cerebral organoids using human pluripotent stem cells has also been reported.37 Therefore, we could analyze the interaction between meningioma and normal cells cocultured with normal cells or organoids.
This is the first study of patient-derived meningioma organoid models that recapitulated the histological and molecular features of their parental tumors. Our model is feasible for performing genetic engineering experiments. It could also be useful for broad applications in basic and clinical research on meningiomas.
Supplementary Material
Acknowledgments
Computation was partially performed on the National Institute of Genetics (NIG) supercomputer at the Research Organization of Information and Systems (ROIS), NIG.
Conflict of interest statement. The authors have no conflicts of interest to declare.
Authorship statement. Conception and design: F.O., Y.K., A.N.; Development of methodology: S.Y., F.O., M.H., Y.S., K.S., Y.M., A.E.; acquisition of data: S.Y., M.H., K.M., K.T., T.T., A.M.; analysis and interpretation of data: S.Y., F.O., M.H., K.A., S.M., A.K., H.S., J.Y., A.A.; writing of the manuscript: S.Y., F.O., A.N.
Funding
This study was performed as a research program of the Grant-in-Aid for Scientific Research, Japan Society for the Promotion of Science [grant number 19KK0228 to A.N., and F.O.].
References
- 1. Wiemels J, Wrensch M, Claus EB. Epidemiology and etiology of meningioma. J Neurooncol. 2010; 99(3):307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Domingues PH, Sousa P, Otero Á, et al. . Proposal for a new risk stratification classification for meningioma based on patient age, WHO tumor grade, size, localization, and karyotype. Neuro Oncol. 2014; 16(5):735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Riemenschneider MJ, Perry A, Reifenberger G. Histological classification and molecular genetics of meningiomas. Lancet Neurol. 2006; 5(12):1045–1054. [DOI] [PubMed] [Google Scholar]
- 4. Clark VE, Harmancı AS, Bai H, et al. . Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. 2016; 48(10):1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clark VE, Erson-Omay EZ, Serin A, et al. . Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013; 339(6123):1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brastianos PK, Horowitz PM, Santagata S, et al. . Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013; 45(3):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vasudevan HN, Braunstein SE, Phillips JJ, et al. . Comprehensive molecular profiling identifies FOXM1 as a key transcription factor for meningioma proliferation. Cell Rep. 2018; 22(13):3672–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mei Y, Bi WL, Greenwald NF, et al. . Genomic profile of human meningioma cell lines. PLoS One. 2017; 12(5):e0178322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang H, Qi L, Du Y, et al. . Patient-derived orthotopic xenograft (PDOX) mouse models of primary and recurrent Meningioma. Cancers (Basel). 2020; 12(6):1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clevers H. Modeling development and disease with organoids. Cell. 2016; 165(7):1586–1597. [DOI] [PubMed] [Google Scholar]
- 11. Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018; 18(7):407–418. [DOI] [PubMed] [Google Scholar]
- 12. Jacob F, Salinas RD, Zhang DY, et al. . A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020; 180(1):188–204 e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boj SF, Hwang CI, Baker LA, et al. . Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015; 160(1-2):324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Broutier L, Mastrogiovanni G, Verstegen MM, et al. . Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017; 23(12):1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gjorevski N, Sachs N, Manfrin A, et al. . Designer matrices for intestinal stem cell and organoid culture. Nature. 2016; 539(7630):560–564. [DOI] [PubMed] [Google Scholar]
- 16. Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007; 130(4):601–610. [DOI] [PubMed] [Google Scholar]
- 17. Hegde NS, Sanders DA, Rodriguez R, Balasubramanian S. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat Chem. 2011; 3(9):725–731. [DOI] [PubMed] [Google Scholar]
- 18. Püttmann S, Senner V, Braune S, et al. . Establishment of a benign meningioma cell line by hTERT-mediated immortalization. Lab Invest. 2005; 85(9):1163–1171. [DOI] [PubMed] [Google Scholar]
- 19. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016; 12(4):e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patel A J, Wan YW, Al-Ouran R, et al. . Molecular profiling predicts meningioma recurrence and reveals loss of DREAM complex repression in aggressive tumors. Proc Natl Acad Sci USA. 2019; 116(43):21715–21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ge SX, Son EW, Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics. 2018; 19(1):534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schulz S, Pauli SU, Schulz S, et al. . Immunohistochemical determination of five somatostatin receptors in meningioma reveals frequent overexpression of somatostatin receptor subtype sst2A. Clin Cancer Res. 2000; 6(5):1865–1874. [PubMed] [Google Scholar]
- 23. Schweizer L, Koelsche C, Sahm F, et al. . Meningeal hemangiopericytoma and solitary fibrous tumors carry the NAB2-STAT6 fusion and can be diagnosed by nuclear expression of STAT6 protein. Acta Neuropathol. 2013; 125(5):651–658. [DOI] [PubMed] [Google Scholar]
- 24. Nigim F, Esaki S, Hood M, et al. . A new patient-derived orthotopic malignant meningioma model treated with oncolytic herpes simplex virus. Neuro Oncol. 2016; 18(9):1278–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim H, Park KJ, Ryu BK, et al. . Forkhead box M1 (FOXM1) transcription factor is a key oncogenic driver of aggressive human meningioma progression. Neuropathol Appl Neurobiol. 2020; 46(2):125–141. [DOI] [PubMed] [Google Scholar]
- 26. Skibinski CG, Williamson T, Riggins GJ. Mebendazole and radiation in combination increase survival through anticancer mechanisms in an intracranial rodent model of malignant meningioma. J Neurooncol. 2018; 140(3):529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhat UG, Halasi M, Gartel AL. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS One. 2009; 4(5):e5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hubert CG, Rivera M, Spangler LC, et al. . A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016; 76(8):2465–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ballabio C, Anderle M, Gianesello M, et al. . Modeling medulloblastoma in vivo and with human cerebellar organoids. Nat Commun. 2020; 11(1):583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van de Wetering M, Francies HE, Francis JM, et al. . Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015; 161(4):933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. von Spreckelsen N, Waldt N, Poetschke R, et al. . KLF4K409Q-mutated meningiomas show enhanced hypoxia signaling and respond to mTORC1 inhibitor treatment. Acta Neuropathol Commun. 2020; 8(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tao W, Zhang A, Zhai K, et al. . SATB2 drives glioblastoma growth by recruiting CBP to promote FOXM1 expression in glioma stem cells. EMBO Mol Med. 2020; 12(12):e12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hu G, Yan Z, Zhang C, et al. . FOXM1 promotes hepatocellular carcinoma progression by regulating KIF4A expression. J Exp Clin Cancer Res. 2019; 38(1):188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang C, Chen H, Yu L, et al. . Inhibition of FOXM1 transcription factor suppresses cell proliferation and tumor growth of breast cancer. Cancer Gene Ther. 2013; 20(2):117–124. [DOI] [PubMed] [Google Scholar]
- 35. Halasi M, Gartel AL. Suppression of FOXM1 sensitizes human cancer cells to cell death induced by DNA-damage. PLoS One. 2012; 7(2):e31761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rutka JT, Giblin J, Dougherty DV, McCulloch JR, DeArmond SJ, Rosenblum ML. An ultrastructural and immunocytochemical analysis of leptomeningeal and meningioma cultures. J Neuropathol Exp Neurol. 1986; 45(3):285–303. [DOI] [PubMed] [Google Scholar]
- 37. Lancaster MA, Renner M, Martin CA, et al. . Cerebral organoids model human brain development and microcephaly. Nature. 2013; 501(7467):373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.