Abstract
Loss of retinoblastoma (Rb) tumor suppressor function, as occurs in many cancers, leads to uncontrolled proliferation, an increased propensity to undergo apoptosis, and tumorigenesis. Rb negatively regulates multiple E2F transcription factors, but the role of the different E2F family members in manifesting the cellular response to Rb inactivation is unclear. To study the effect of deregulated E2F4 activity on cell growth control and tumorigenesis, transgenic mouse lines expressing the E2F4 gene under the control of a keratin 5 (K5) promoter were developed, and their phenotypes were compared to those of previously generated K5 E2F1 transgenic mice. In contrast to what has been observed in vitro, ectopically expressed E2F4 was found to localize to the nucleus and induce proliferation to an extent similar to that induced by E2F1 in transgenic tissue. Unlike E2F1, E2F4 does not induce apoptosis, and this correlates with the differential abilities of these two E2F species to stimulate p19ARF expression in vivo. To examine the role of E2F4 in tumor development, the mouse skin two-stage carcinogenesis model was utilized. Unlike E2F1 transgenic mice, E2F4 transgenic mice developed skin tumors with a decreased latency and increased incidence compared to those characteristics in wild-type controls. These findings demonstrate that while the effects of E2F1 and E2F4 on cell proliferation in vivo are similar, their apoptotic and oncogenic properties are quite different.
The E2F family consists of six distinct genes, E2F1 through E2F6, that encode structurally related proteins (for a review, see references 6 and 18). The DNA-binding domain located in the amino terminus represents the area of greatest homology between the six E2F species (28). Adjacent to the DNA-binding domain of each E2F is a domain involved in dimerization with the more distantly related DP1 and DP2 proteins. Each E2F species can heterodimerize with either DP1 or DP2 to generate a functional E2F factor capable of binding classical E2F sites (TTTSSSCGC [S is C or G]) with high affinity. With the exception of E2F6, the carboxy terminus of each E2F protein contains the defined transcriptional activation domain. Embedded within the transactivation domain is a region of homology involved in binding to proteins of the retinoblastoma (Rb) tumor suppressor family (Rb, p107, and p130). Binding of Rb and related proteins to E2F factors inhibits their ability to activate transcription and, in some cases, converts E2F factors from activators to repressors of transcription (10, 29, 35). E2F6 lacks transcriptional activation and Rb-binding domains and is believed to function as an inhibitor of E2F-dependent transcription independently of Rb family proteins (16, 32).
Several findings suggest that E2F4 may play an important and perhaps unique role in regulating E2F-dependent transcription and cell growth. E2F4 is the most abundant E2F species, making up the majority of the total E2F in most cells (9, 15). Unlike the other E2F genes, E2F4 is constitutively expressed throughout the cell cycle and is expressed even in quiescent cells (2, 7). Another difference between E2F4 and the other E2F proteins is that E2F4 complexes with all three members of the Rb family in a cell cycle-regulated manner (9, 15). E2F1, E2F2, and E2F3 associate exclusively with Rb, while E2F5 associates exclusively with p130. Furthermore, E2F4 appears to be regulated at the level of subcellular localization and lacks the nuclear localization signal that is present in E2Fs 1, 2, and 3 (12, 14, 17, 33). E2F4 also lacks the cyclin A-binding domain found in E2Fs 1, 2, and 3 and so is resistant to negative regulation by cyclin A-associated kinases (5). Finally, E2F4 contains a unique serine repeat domain not found in the other E2F family members. Although the function of this serine domain is unknown, it has been shown to be a target for mutation in replication error-positive colorectal cancers (8).
A number of studies have also demonstrated that the biological activity of E2F4 differs from those of the other E2F species. E2F4 is a less potent activator of transcription than E2F1 but a more potent activator than E2F5 (13, 22). This difference in transactivation potential appears to be related to both the relative strengths of the transcriptional activation domains in these E2F proteins and the presence of a nuclear localization signal found in E2F1 but not in E2F4 or E2F5. In addition, overexpression of E2F4 activates only a subset of the target genes that are activated by other E2F family members (3). Overexpression of E2F4 induces serum-starved rat embryo fibroblasts to enter S phase but not as efficiently as can E2Fs 1, 2, and 3 (3, 13). E2F5 is unable to induce S phase under these conditions, consistent with its weaker ability to activate E2F-dependent transcription. Unlike E2F1, E2F4 does not induce apoptosis when it is overexpressed in serum-starved rat embryo cells (3). Adding a nuclear localization signal to E2F4 enhances its ability to induce S phase (17). The ability of nuclear E2F4 to induce apoptosis has not been examined.
Deregulation of E2F-dependent transcription through impairment of Rb function occurs in most human cancers. This deregulation can occur through mutation of the RB1 gene, overexpression of cyclin D1, or inactivation of the p16INK4a cyclin-dependent kinase inhibitor (30). The end result of each of these events is the release of E2F factors from Rb control and the activation of E2F-dependent transcription. These processes in turn lead to uncontrolled cell proliferation and an increased propensity to undergo apoptosis. Since E2Fs 1, 2, 3, and 4 all associate with Rb, each would be expected to be deregulated in cancer cells that lack functional Rb. How the different E2F family members contribute to the loss of cell growth control and tumorigenesis as a result of Rb inactivation is unclear.
As a model to study the role of E2F in cell growth control and cancer in vivo, we previously developed transgenic mouse lines in which expression of the E2F1 gene was targeted to stratified epithelial tissue by a keratin 5 (K5) promoter (19). Deregulated expression of E2F1 results in hyperplasia, hyperproliferation, and p53-dependent apoptosis in the epidermis of K5 E2F1 transgenic mice (19, 20). Moreover, K5 E2F1 transgenic mice are predisposed to developing tumors in several epithelial tissues expressing the transgene, including the skin and odontogenic epithelium (21). In addition, the K5 E2F1 transgene can cooperate with either a v-Ha-ras transgene to induce benign skin papillomas or p53 deficiency to induce spontaneous skin carcinomas (19, 20). In sharp contrast to these oncogenic effects of E2F1, overexpression of E2F1 can suppress tumor development under some experimental conditions. K5 E2F1 transgenic mice were found to be resistant to tumor development in a two-stage chemical carcinogenesis model (21). Experiments demonstrate that tumor suppression by E2F1 occurs at the promotion stage and may involve the induction of apoptosis. Thus, E2F1 has both oncogenic and tumor-suppressive properties when it is expressed in a deregulated manner.
In this study, the phenotype of transgenic mice expressing E2F4 under the control of the same K5 promoter is examined. We find that several properties ascribed to E2F4 from in vitro studies differ when they are examined in this in vivo model. We also find that E2F4 and E2F1 are similar in their abilities to induce proliferation but differ in their abilities to induce apoptosis. The difference in apoptosis-promoting activity correlates with differential abilities to activate the expression of the p19ARF gene. Finally, E2F4 is found to have an oncogenic potential different from that of E2F1.
MATERIALS AND METHODS
Transgenic mice.
The K5 E2F4 transgene was made by cloning the full-length human E2F4 cDNA (2) into a plasmid containing the bovine K5 promoter (26), the rabbit β-globin intron 2, and the simian virus 40 polyadenylation signal (19). Founder transgenic mice were made by microinjecting the purified transgene into the pronuclei of zygotes and then by implanting the zygotes into pseudopregnant female mice. Lines were established and maintained by backcrossing the founders to the SENCAR outbreed strain (for the 4.3 line) or the SENCAR inbred strain SSIN (for the 4.0 line). K5 E2F1 transgenic mice have previously been described (19) and were in the SSIN background.
Northern and Western blot analyses.
Northern blot analysis was performed using RNA isolated from primary keratinocytes. The method for isolating primary keratinocytes from newborn pups has been described (19). Total RNA was isolated with Tri-reagent (Molecular Research Center, Inc.) by the manufacturer's protocol. The full-length human E2F4 cDNA was labeled and used as probe under high-stringency conditions. Murine cDNA probes were obtained from Tim Kowalik (p19ARF) and Julie DeLoia (cyclin E).
For E2F4 Western blot analysis, whole-cell protein extract was made from primary keratinocytes as described previously (19). Antibody specific for E2F4 (C-20; catalog number sc-1082) was purchased from Santa Cruz Biotechnology, Inc.
Immunohistochemistry.
Frozen tissue sections were fixed in acetone for 5 min, air dried, and placed in methanol containing 0.3% H2O2 for 20 min. Tissue sections were then rinsed with phosphate-buffered saline (PBS) three times for 5 min each before addition of primary E2F4 antiserum (catalog number sc-866, 1:100 dilution; Santa Cruz Biotechnology, Inc.). Sections were incubated for 30 min, rinsed with PBS, and then incubated with biotinylated anti-rabbit immunoglobulin G (Vector) for 30 min. Slides were rinsed with PBS and developed with a streptavidin-horseradish peroxidase conjugate (Vectastain kit; Vector) and a diaminobenzidine substrate. Tissue sections were rinsed again with H2O and counterstained before being mounted.
EMSA.
E2F electrophoretic mobility shift assays (EMSA) used extract from primary keratinocytes as previously described (20). A fragment derived from the adenovirus E2 gene containing two E2F sites was used as a probe. Antisera used in supershift studies were purchased from Santa Cruz Biotechnology (E2F4, catalog number sc-1082; p107, catalog number sc-250; p130, catalog number sc-317) and Oncogene Research Products (Rb, AB-2).
BrdU incorporation and TUNEL assays.
Mice were injected with bromodeoxyuridine (BrdU), and skin samples were immunostained using antibody specific for BrdU (Becton Dickinson) as previously described (19). For determining percent incorporation, interfollicular basal keratinocytes were examined and the numbers of unstained and stained cells were determined. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays were performed using formalin-fixed, paraffin-embedded skin sections and an ApopTag in situ apoptosis detection kit (Oncor). TUNEL-positive epidermal keratinocytes were visualized by peroxidase-diaminobenzidine staining, and the average number of positive cells per 10 mm of linear skin was determined.
Mouse skin two-stage carcinogenesis assay.
Tumors were initiated by topical application of 10 nmol of DMBA (9,10-dimethyl-1,2-benzathracene) in 200 μl of acetone to previously shaved dorsal skin. Tumors were promoted twice weekly by topical application of 1.0 μg of 12-O-tetradecanoylphorbol-13-acetate (TPA) in 200 μl of acetone beginning 2 weeks after initiation. Mice were scored for papillomas weekly. For short-term TPA treatments, the shaved dorsal skin of mice was treated with 2.0 μg of TPA twice weekly for 2 weeks.
RESULTS
Expression and localization of E2F4 in K5 E2F4 transgenic mice.
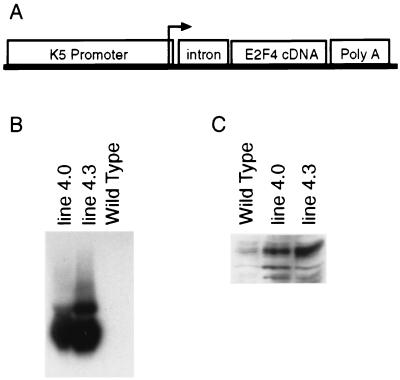
The K5 E2F4 transgene was generated by subcloning the human E2F4 cDNA into a vector containing the bovine K5 promoter, the rabbit β-globin intron 2, and the simian virus 40 polyadenylation signal (Fig. 1A). The bovine K5 promoter fragment has been shown to direct expression to the basal cell layer of the epidermis, the hair follicles, and other stratified squamous epithelia in transgenic mice (26). Four founders containing the K5 E2F4 transgene were originally identified by PCR analysis of genomic DNA. One of the founders did not pass the transgene, while mice from another line did not express the transgene in skin keratinocytes. Transgenic mouse lines that overexpressed E2F4 in the epidermis and in primary keratinocytes were established from the two remaining founders. By Northern blot analysis, line 4.0 and line 4.3 expressed the human E2F4 transgene to similar extents in transgenic keratinocytes (Fig. 1B). Under the high-stringency conditions used for this Northern blot analysis, endogenous murine E2F4 was not detected. Western blot analysis using antiserum that recognizes both mouse and human E2F4 confirmed that transgene overexpression resulted in a five- to sevenfold increase in E2F4 protein levels in each line (Fig. 1C).
FIG. 1.
Generation of K5 E2F4 transgenic lines. (A) Schematic representation of the K5 E2F4 transgene. Primary keratinocytes were isolated and cultured from the epidermis of newborn nontransgenic (wild-type) and K5 E2F4 transgenic mice (lines 4.0 and 4.3). (B) Total RNA (20 μg per lane) from primary keratinocytes was subjected to Northern blot analysis using the human E2F4 cDNA as a probe. (C) Whole-cell protein lysate (20 μg per lane) from primary-keratinocyte cultures was subjected to Western blot analysis using polyclonal antiserum specific for E2F4.
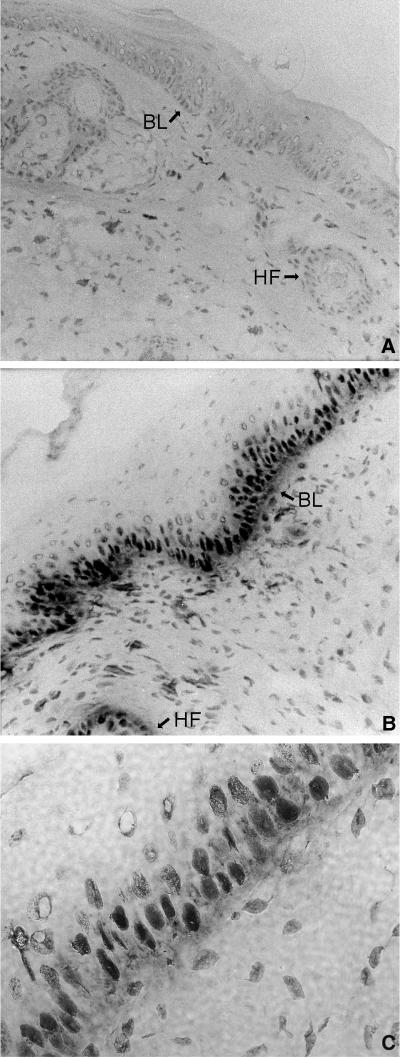
Overexpression of E2F4 in transgenic tissues could also be detected by immunohistochemistry. Previous experiments have demonstrated that E2F4 lacks a nuclear localization signal and that when it is overexpressed in immortalized fibroblasts or human cancer cell lines, it is localized primarily to the cytoplasm (12, 14, 17, 33). In contrast to these findings, immunostaining revealed that the overexpressed E2F4 protein localized to the nuclei of transgenic keratinocytes in the basal cell layer of the epidermis and in the hair follicles (Fig. 2). Our immunohistochemistry assay did not detect endogenous E2F4 in nontransgenic tissues.
FIG. 2.
Ectopically expressed E2F4 localizes to the nucleus in transgenic tissue. Tail sections were taken from nontransgenic (A) or K5 E2F4 line 4.0 (B) mice and immunohistochemically stained with antiserum specific for E2F4. BL, basal layer; HF, hair follicle. (C) Higher magnification of K5 E2F4 line 4.0 tissue demonstrating the nuclear localization of E2F4.
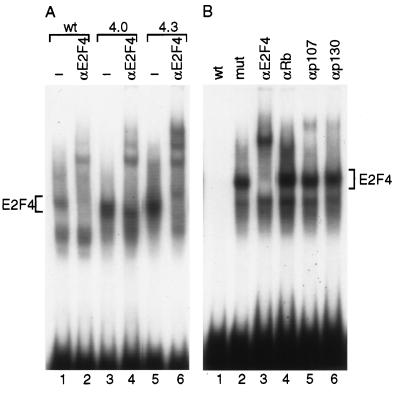
To examine the resultant change in E2F DNA-binding activity associated with expression of the E2F4 transgene, whole-cell extract was prepared from primary keratinocytes isolated from newborn transgenic mice. Extracts from each of the transgenic lines and from nontransgenic mice were used in an E2F EMSA. In wild-type keratinocytes, two prominent E2F complexes were observed (Fig. 3A). One of these bands corresponded to a complex containing E2F4, as was evidenced by a supershift with E2F4 antiserum. The faster-migrating complex likely contained one or more of the other E2F family members since it also was specifically competed by excess unlabeled oligonucleotide containing a wild-type E2F site but not a mutated E2F site. Neither complex appeared to contain an Rb family member, as was evidenced by a lack of supershift with antibody specific for Rb, p107, or p130 (Fig. 3B). Minor complexes above the two more-prominent complexes were supershifted by the p107 and p130 antisera. In keratinocytes from K5 E2F4 line 4.0 or 4.3, the intensity of the E2F4 DNA-binding complex was modestly increased between 1.8- and 1.6-fold as measured by densitometry. These findings are consistent with previous findings from K5 E2F1 transgenic mice demonstrating that expression of the E2F1 transgene results in a relatively small increase in E2F DNA-binding activity (20).
FIG. 3.
E2F DNA-binding activity from K5 E2F4 transgenic keratinocytes. (A) An E2F EMSA was performed using whole-cell extracts (10 μg) from primary keratinocytes isolated from nontransgenic (lanes 1 and 2), line 4.0 (lanes 3 and 4), or line 4.3 mice (lanes 5 and 6). Antiserum specific for E2F4 was added (lanes 2, 4, and 6) to identify complexes containing E2F4. (B) An E2F EMSA was performed using whole-cell extract (10 μg) from primary keratinocytes isolated from line 4.0 mice. Excess double-stranded oligonucleotide (20 ng) containing either wild-type (wt) E2F sites (lane 1) or mutated (mut) E2F sites (lane 2) was added to the binding reaction mixtures to distinguish specific E2F complexes. Antisera specific for E2F4 (lane 3), Rb (lane 4), p107 (lane 5), and p130 (lane 6) were added to identify proteins in specific complexes.
Growth-regulatory activities of E2F4 in transgenic epidermis.
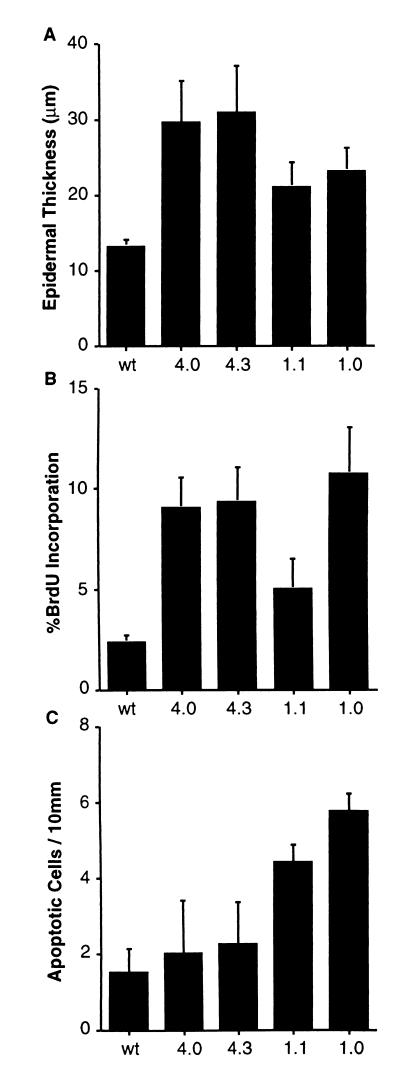
To examine the effect of deregulated E2F4 expression on cell growth control in vivo, skin samples from adult K5 E2F4 transgenic mice were analyzed. Epidermis from both K5 E2F4 transgenic lines was found to be hyperplastic (Fig. 4 and 5A). The thickness of the epidermis of nontransgenic sibling controls averaged 13 μm, while the average thickness of the epidermis from line 4.0 and 4.3 mice was approximately 30 μm. K5 E2F4 epidermis was more hyperplastic than epidermis from either K5 E2F1 transgenic mouse line. The proliferation index of interfollicular basal keratinocytes was determined for transgenic mice and nontransgenic siblings by measuring BrdU incorporation. Two to three percent of basal keratinocytes were in S phase in wild-type mice, while this is increased to 9.0 and 9.4 percent in line 4.0 and 4.3 transgenic mice, respectively (Fig. 5B). These levels of epidermal hyperproliferation in K5 E2F4 mice were between those found in the two K5 E2F1 transgenic lines.
FIG. 4.
Histological appearance of K5 E2F4 transgenic skin. Photomicrographs of skin samples from a nontransgenic mouse (A) and K5 E2F4 transgenic line 4.0 (B) and line 4.3 (C) mice stained with hematoxylin and eosin.
FIG. 5.
Hyperplasia, proliferation, and apoptosis in K5 E2F1 and K5 E2F4 transgenic epidermis. (A) Epidermal thickness was measured from skin samples taken from nontransgenic (wild-type [wt]), K5 E2F4 line 4.0 (4.0), K5 E2F4 line 4.3 (4.3), K5 E2F1 line 1.1 (1.1), and K5 E2F1 line 1.0 (1.0) mice. Samples were taken from five different mice in each group, and 100 measurements were taken for each sample to calculate the average thickness. (B) The percentage of interfollicular basal keratinocytes in S phase was calculated for the same mice as those used to obtain the results in panel A by measuring BrdU incorporation. Mice were injected with BrdU 30 min prior to sacrifice, and antibody specific for BrdU was used to immunostain skin samples. Five hundred cells were counted per sample, and the average percentages of positive cells from five mice in each group are presented. (C) The TUNEL assay was used to examine apoptosis in skin sections from the same mice. At least 40 measurements were taken from each sample (five mice per group) to calculate the average number of TUNEL-positive epidermal cells per 10 mm of linear skin.
The effect of transgene expression on apoptosis in the epidermis was also measured by performing the TUNEL assay on skin sections. K5 E2F4 transgenic mice were found to have only a slight increase in the number of apoptotic cells over background levels (Fig. 5C). As shown previously (20), K5 E2F1 transgenic mice had a three- to fourfold increase in the number of TUNEL-positive cells in the epidermis compared to the number for nontransgenic mice (Fig. 5C). Thus, the K5 E2F4 transgene induces proliferation, but not apoptosis, as efficiently as the K5 E2F1 transgene. The increased survival of cells in K5 E2F4 epidermis may explain why the level of hyperplasia, as measured by epidermal thickness, is greater in K5 E2F4 mice than in K5 E2F1 mice.
Expression of p19ARF and cyclin E in K5 E2F1 and K5 E2F4 primary keratinocytes.
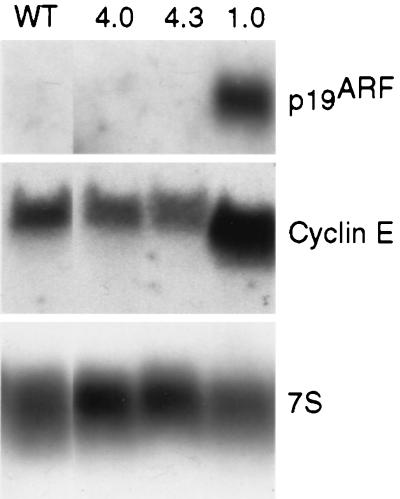
It has been suggested that the ability of E2F1 to induce apoptosis is related to its ability to transcriptionally activate the p19ARF tumor suppressor gene (1, 31). The p19ARF protein is encoded at the INK4a locus in an overlapping, alternative reading frame from the cyclin-dependent kinase inhibitor p16INK4a (24). The p19ARF protein activates the p53 tumor suppressor by inhibiting the activity of mdm2 (23, 34, 37). The p19ARF gene promoter contains a consensus E2F DNA-binding site and is transcriptionally activated by overexpression of E2F1 (1, 3). To determine if differential regulation of p19ARF contributes to the differential abilities of E2F1 and E2F4 to induce apoptosis in the skin of transgenic mice, Northern blot analysis was performed on RNAs isolated from primary keratinocytes derived from K5 E2F1, K5 E2F4, and nontransgenic mice. In nontransgenic and K5 E2F4 transgenic keratinocytes, p19ARF expression was virtually undetectable (Fig. 6). In contrast, p19ARF expression was significantly induced in keratinocytes from K5 E2F1 transgenic mice. Expression of another E2F target, cyclin E, was also found to be upregulated in keratinocytes from K5 E2F1 transgenic mice but not from K5 E2F4 transgenic mice (Fig. 6). After normalization with 7S RNA expression, the level of cyclin E expression in K5 E2F1 keratinocytes was found to be twice the level found in nontransgenic keratinocytes while a slight decrease in cyclin E expression was observed in K5 E2F4 keratinocytes.
FIG. 6.
Induction of p19ARF and cyclin E expression in K5 E2F1 but not K5 E2F4 keratinocytes. Total RNA (20 μg per lane) was isolated from primary keratinocytes derived from nontransgenic (wild-type [wt]), K5 E2F4 line 4.0 (4.0), K5 E2F4 line 4.3 (4.3), and K5 E2F1 line 1.0 (1.0) transgenic mice. Northern blot analysis was performed on the same filter using probes for murine p19ARF, cyclin E, and 7S.
Analysis of E2F4 in multistage carcinogenesis.
Previously we found that the expression of several E2F family members, including E2F4, is increased during premalignant progression in the mouse skin model of multistage carcinogenesis (27). This model has been extensively used to study the molecular events that occur during the multistep process of cancer development. In one of the most common protocols, initiation of carcinogenesis is carried out by a single application of DMBA, which induces an activating mutation at codon 61 of the c-Ha-ras gene (25). Initiated cells are expanded during the promotion stage by repetitive treatments with TPA, which results in the outgrowth of exophytic papillomas. A subset of these benign papillomas can progress to malignant skin carcinomas.
To examine the role of increased E2F4 activity in tumor development, K5 E2F4 transgenic mice were used in the mouse skin model of multistage carcinogenesis. First, K5 E2F4 transgenic mice and nontransgenic sibling controls were treated with TPA twice weekly for 2 weeks to examine their responses to TPA. K5 E2F4 transgenic mice responded to TPA treatment in a manner similar to that of nontransgenic control mice (Fig. 7). Compared to results with acetone vehicle-treated controls, TPA treatment increased epidermal thickness 1.6-fold in nontransgenic mice and 2.0-fold in K5 E2F4 transgenic mice. TPA treatment induced proliferation in the epidermis as measured by BrdU incorporation 3.2-fold in nontransgenic mice and 3.4-fold in K5 E2F4 transgenic mice. The hyperplastic and hyperproliferative responses to TPA observed in K5 E2F4 transgenic mice were similar to what was seen in K5 E2F1 transgenic mice (19).
FIG. 7.
Response of K5 E2F4 transgenic mice to TPA. K5 E2F4 line 4.3 transgenic mice (E2F4.3) and nontransgenic siblings (wild type) were treated with 2 μg of TPA or acetone vehicle control (ACE) twice weekly for 2 weeks. Mice were sacrificed 24 h following the last TPA treatment and 30 min following BrdU injection. (A) The average percentage of basal keratinocytes in S phase was determined for anti-BrdU immunostained skin sections from three mice in each group. (B) The average epidermal thickness was determined from hematoxylin- and eosin-stained skin sections from three mice in each group.
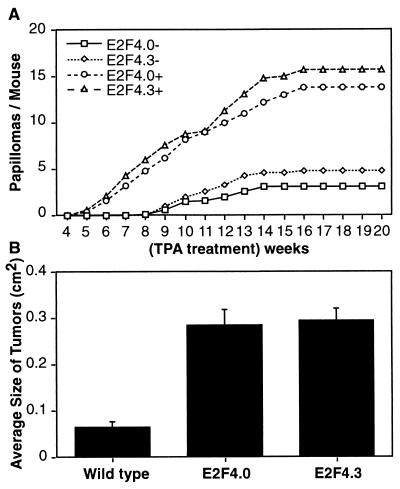
K5 E2F4 transgenic mice and nontransgenic siblings were also used in DMBA-TPA two-stage carcinogenesis experiments. Tumor development was initiated in 6- to 8-week-old transgenic and wild-type sibling mice by topical application of DMBA. Two weeks following initiation, tumors were promoted twice weekly with TPA for a total of 20 weeks. K5 E2F4 transgenic mice from both lines developed tumors sooner after the beginning of promotion and in greater numbers than did control mice (Fig. 8A). Both lines of K5 E2F4 transgenic mice had approximately three times as many tumors as their nontransgenic control siblings. This is in sharp contrast to the lack of tumor development observed in K5 E2F1 transgenic mice following the DMBA-TPA two-stage protocol (21). The size of each tumor was measured in nontransgenic and K5 E2F4 transgenic mice at 18 weeks of promotion. At this time, tumor size was relatively stable, with only a few tumors enlarging or regressing. The average size of the tumors from K5 E2F4 transgenic mice was four times greater than that from nontransgenic mice (Fig. 8B).
FIG. 8.
Enhanced tumor development in K5 E2F4 transgenic mice following two-stage carcinogenesis. (A) Carcinogenesis was initiated in mice from both K5 E2F4 transgenic lines (E2F4.0+ and E2F4.3+) and nontransgenic siblings (E2F4.0− and E2F4.3−) by topical application of 10 nmol of DMBA to shaved dorsal skin. Two weeks following initiation, tumors were promoted by twice weekly applications of TPA (1 μg) to dorsal skin for 20 weeks. The average numbers of palpable tumors per mouse at each week are presented. The number of mice in each group was five for line 4.0+, 8 for line 4.0−, six for line 4.3+, and three for line 4.3−. (B) The size of each tumor in the study was calculated at 18 weeks of promotion by multiplying tumor length by tumor width. The average sizes of tumors from nontransgenic (39 tumors total), line 4.0 (61 tumors total), and line 4.3 (89 tumors total) mice are presented.
DISCUSSION
To directly examine the effect of E2F4 upregulation on cell growth control and tumorigenesis, we generated transgenic mouse lines expressing E2F4 under the control of a K5 promoter. K5 E2F4 transgenic mice are the first animal model for studying the in vivo properties of this E2F family member. A caveat of this study is that E2F4 is overexpressed, and so it may not accurately reflect the normal role of E2F4. It can be argued, however, that overexpression of E2F4 in some ways mimics the activation of E2F4 that occurs when Rb function is lost. Loss of Rb function results in deregulated proliferation, increased apoptosis, and tumorigenesis. The downstream factors regulated by Rb that are responsible for this phenotype are not entirely clear, although E2F family members (E2F1 to -4) may play an important role. The data presented here suggest that E2F4 contributes to the hyperproliferative and tumorigenic effects of Rb inactivation but not to the apoptotic effects.
In transgenic tissue, exogenous E2F4 protein is detected in the nucleus. This is in contrast to results of previous in vitro studies that found that overexpressed E2F4 was localized primarily to the cytoplasm because it lacks a nuclear localization signal (12, 14, 17, 33). In immortalized cultured fibroblasts, the subcellular localization of endogenous E2F4 is regulated in response to the growth state of the cell. In quiescence, endogenous E2F4, in association with Rb family members, is found to be predominately nuclear. When these cultured cells are cycling, however, the majority of E2F4, both in the free form and complexed with p107, is in the cytoplasm. In K5 E2F4 transgenic mice, exogenous E2F4 is localized to the nuclei of keratinocytes in the basal layer of the epidermis and the outer root sheath of hair follicles despite the fact that the majority of these cells are cycling (4). In vitro, coexpression of a DP2 splice variant that contains a nuclear localization signal was found to promote nuclear localization of overexpressed E2F4 through heterodimerization (12, 14, 33). Coexpression of p107 or p130 may also promote nuclear localization of E2F4 (12, 14). According to EMSA and supershift experiments using extract from E2F4 transgenic keratinocytes, the majority of E2F4 DNA-binding activity does not appear to be in complex with p107 or p130 and is found in association with DP1, not DP2 (data not shown). Thus, the factor(s) that promotes E2F4 nuclear localization in K5 E2F4 transgenic tissue is unclear.
Previous in vitro studies also demonstrated that E2F4 is a weaker inducer of proliferation than E2F1. When transiently overexpressed in serum-starved rat fibroblast cells, E2F4 either was unable to induce S-phase entry (13) or was less than half as effective as E2F1 at inducing S phase (3). In the K5 transgenic mouse model, overexpression of E2F4 induces proliferation, as was indicated by an increase in BrdU-incorporating cells in the basal layer of the epidermis. The levels of hyperproliferation observed in the epidermis of both K5 E2F4 transgenic lines are between those found in the epidermis of the two K5 E2F1 transgenic lines. The increases in E2F DNA-binding activity as a result of transgene expression are also similar between K5 E2F4 and K5 E2F1 transgenic mice (20). This finding suggests that E2F4 is as effective at inducing proliferation as E2F1 in this model system. The apparent discrepancy between the in vitro and in vivo data regarding E2F4's ability to induce proliferation may be related to the difference in the subcellular localizations of E2F4 in the two systems. When a nuclear localization signal is fused to E2F4, it can induce S-phase entry as efficiently as E2F1 in vitro (13). Since E2F4 is localized to the nuclei of transgenic cells through another mechanism, it may not need a nuclear localization signal to efficiently induce proliferation.
In contrast to the comparable abilities of E2F4 and E2F1 to induce proliferation, E2F4 is less efficient than E2F1 at inducing apoptosis when it is overexpressed in the epidermis of transgenic mice. In vitro studies have also found that E2F4 lacks E2F1's potent apoptosis-promoting activity (3). The finding that K5 E2F4 transgenic epidermis has only a minor increase in TUNEL-positive cells over background levels is consistent with the gross phenotype of K5 E2F4 transgenic mice. K5 E2F1 transgenic mice have alopecia due to aberrant, p53-dependent apoptosis in the hair follicles (20). In contrast, K5 E2F4 transgenic mice have a normal hair coat. The increased survival of K5 E2F4 transgenic keratinocytes compared to that of K5 E2F1 transgenic keratinocytes may explain why the skin of K5 E2F4 mice is more hyperplastic than the skin of K5 E2F1 mice as measured by epidermal thickness.
The abilities of E2F1 and E2F4 to induce apoptosis correlates with their differential abilities to stimulate expression from the p19ARF gene. The p19ARF protein interacts with mdm2 and sequesters it in the nucleolus (34, 37). This prevents the negative feedback of mdm2 on p53 and results in the accumulation of active p53 in the nucleoplasm. The p19ARF gene promoter contains a consensus E2F-binding site (1), but this site appears to respond specifically to E2F1 and not to E2F4. While p19ARF expression was significantly induced in primary keratinocytes from K5 E2F1 transgenic mice, p19ARF expression was virtually undetectable in K5 E2F4 and nontransgenic keratinocytes. The same differential expression patterns for p19ARF were observed when Northern blot analysis was performed on RNAs isolated directly from the epidermis of transgenic and nontransgenic mice (data not shown). The idea that p19ARF induction participates in E2F1-mediated apoptosis in our transgenic model is supported by the finding that apoptosis in K5 E2F1 epidermis is largely p53 dependent (20). In a p53 null background, apoptosis in K5 E2F1 transgenic mice is reduced to near background levels.
Expression of the cyclin E gene was also found to be upregulated only in primary keratinocytes from K5 E2F1 transgenic mice and not from K5 E2F4 transgenic mice. This finding is in contrast to a previous report suggesting that E2F4 could stimulate cyclin E expression to a similar extent as E2F1 in immortalized rat fibroblast cells infected with recombinant adenoviruses (3). We have also found that cdk2 gene expression is upregulated only in E2F1 transgenic cells and not in E2F4 transgenic cells (data not shown). Despite the lack of cyclin E or cdk2 upregulation, K5 E2F4 transgenic epidermis is as hyperproliferative as K5 E2F1 transgenic epidermis. At present, it is unclear which E2F target genes, if any, are upregulated by E2F4 to mediate hyperproliferation in K5 E2F4 transgenic mice.
E2F1 and E2F4 have both been shown to behave as oncogenes in cell culture-based transformation assays (2, 7, 11, 36). However, a direct comparison between the oncogenic capacities of E2F1 and E2F4 has not been performed. In the K5 transgenic model, E2F4 does not appear to be as oncogenic as E2F1. We have yet to observe spontaneous tumors in K5 E2F4 transgenic mice. The K5 E2F4 transgene also does not appear to cooperate with p53 deficiency to induce skin carcinomas, as does the K5 E2F1 transgene (D. Wang and D. G. Johnson, unpublished data). An oncogenic activity for E2F4 can be revealed when K5 E2F4 mice are used in the two-stage mouse skin carcinogenesis model. K5 E2F4 transgenic mice develop tumors sooner and in greater numbers following DMBA-TPA treatment than nontransgenic mice. Tumors from K5 E2F4 mice are also larger than those from nontransgenic mice. These results are in sharp contrast to the resistance to two-stage carcinogenesis observed for K5 E2F1 transgenic mice (21). This difference in responses to chemical carcinogenesis may be related to the differential abilities of E2F1 and E2F4 to stimulate genes such as p19ARF and induce apoptosis. These findings demonstrate that E2F4 has an oncogenic activity but suggest that it, unlike E2F1, lacks a tumor-suppressive activity.
ACKNOWLEDGMENTS
We are very grateful to Claudio Conti, Robin Schneider-Broussard, and Aijin Wang for advice and assistance during this work. We thank Shawnda Sanders and Michelle Gardiner for preparation of the manuscript, Dale Weiss and coworkers for animal care, Judy Ing and Chris Yone for artwork, and Jennifer Smith and Jennifer Philhower for expert technical assistance.
This work was supported by grants from the National Institutes of Health (GM56144 to D.G.J., CA 79648 to D.G.J., NIEHS Center grant ES007784, and CA16672).
REFERENCES
- 1.Bates S, Phillips A C, Clark P A, Stott F, Peters G, Ludwig R L, Vousden K H. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 2.Beijersbergen R L, Kerkhoven R M, Zhu L, Carlee L, Voorhoeve P M, Bernards R. E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 1994;8:2680–2690. doi: 10.1101/gad.8.22.2680. [DOI] [PubMed] [Google Scholar]
- 3.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dover R, Wright N A. The cell proliferation kinetics of the epidermis. In: Goldsworth L A, editor. Physiology, biochemistry, and molecular biology of the skin. Vol. 1. New York, N.Y: Oxford University Press; 1991. pp. 239–265. [Google Scholar]
- 5.Dynlacht B D, Moberg K, Lees J A, Harlow E, Zhu L. Specific regulation of E2F family members by cyclin-dependent kinases. Mol Cell Biol. 1997;17:3867–3875. doi: 10.1128/mcb.17.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 7.Ginsberg D, Vairo G, Chittenden T, Xiao Z X, Xu G, Wydner K L, DeCaprio J A, Lawrence J B, Livingston D M. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 1994;8:2665–2679. doi: 10.1101/gad.8.22.2665. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda M, Orimo H, Moriyama H, Nakajima E, Matsubara N, Mibu R, Tanaka N, Shimada T, Kimura A, Shimizu K. Close correlation between mutations of E2F4 and hMSH3 genes in colorectal cancers with microsatellite instability. Cancer Res. 1998;58:594–598. [PubMed] [Google Scholar]
- 9.Ikeda M A, Jakoi L, Nevins J R. A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation. Proc Natl Acad Sci USA. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson D G. Regulation of E2F-1 gene expression by p130(Rb2) and D-type cyclin kinase activity. Oncogene. 1995;11:1685–1692. [PubMed] [Google Scholar]
- 11.Johnson D G, Cress W D, Jakoi L, Nevins J R. Oncogenic capacity of the E2F1 gene. Proc Natl Acad Sci USA. 1994;91:12823–12827. doi: 10.1073/pnas.91.26.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindeman G J, Gaubatz S, Livingston D M, Ginsberg D. The subcellular localization of E2F-4 is cell-cycle dependent. Proc Natl Acad Sci USA. 1997;94:5095–5100. doi: 10.1073/pnas.94.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukas J, Petersen B O, Holm K, Bartek J, Helin K. Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol Cell Biol. 1996;16:1047–1057. doi: 10.1128/mcb.16.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magae J, Wu C L, Illenye S, Harlow E, Heintz N H. Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J Cell Sci. 1996;109:1717–1726. doi: 10.1242/jcs.109.7.1717. [DOI] [PubMed] [Google Scholar]
- 15.Moberg K, Starz M A, Lees J A. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morkel M, Wenkel J, Bannister A J, Kouzaridesand T, Hagemeier C. An E2F-like repressor of transcription. Nature. 1997;390:567–568. doi: 10.1038/37507. [DOI] [PubMed] [Google Scholar]
- 17.Muller H, Moroni M C, Vigo E, Petersen B O, Bartek J, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nevins J R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 19.Pierce A M, Fischer S M, Conti C J, Johnson D G. Deregulated expression of E2F1 induces hyperplasia and cooperates with ras in skin tumor development. Oncogene. 1998;16:1267–1276. doi: 10.1038/sj.onc.1201666. [DOI] [PubMed] [Google Scholar]
- 20.Pierce A M, Gimenez-Conti I B, Schneider-Broussard R, Martinez L A, Conti C J, Johnson D G. Increased E2F1 activity induces skin tumors in mice heterozygous and nullizygous for p53. Proc Natl Acad Sci USA. 1998;95:8858–8863. doi: 10.1073/pnas.95.15.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierce A M, Schneider-Broussard R, Gimenez-Conti I B, Russell J L, Conti C J, Johnson D J. E2F1 has both oncogenic and tumor-suppressive properties in a transgenic model. Mol Cell Biol. 1999;19:6408–6414. doi: 10.1128/mcb.19.9.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierce A M, Schneider-Broussard R, Philhower J L, Johnson D G. Differential activities of E2F family members suggest unique functions in regulating transcription. Mol Carcinog. 1998;22:190–198. doi: 10.1002/(sici)1098-2744(199807)22:3<190::aid-mc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 23.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 24.Quelle D E, Zindy F, Ashman R A, Sherr C J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 25.Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez A, Bravo A, Jorcano J L, Vida M. Sequences 5′ of the bovine keratin 5 gene direct tissue-and-cell-type-specific expression of a lacZ gene in the adult and during development. Differentiation. 1994;58:53–64. doi: 10.1046/j.1432-0436.1994.5810053.x. [DOI] [PubMed] [Google Scholar]
- 27.Rodriquez-Puebla M L, LaCava M, Gimenez-Conti I B, Johnson D J, Conti C J. Deregulated expression of cell-cycle proteins during premalignant progression in SENCAR mouse skin. Oncogene. 1998;17:2251–2258. doi: 10.1038/sj.onc.1202131. [DOI] [PubMed] [Google Scholar]
- 28.Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, Weinberg R A. E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc Natl Acad Sci USA. 1995;92:2403–2407. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sellers W R, Rodgers J W, Kaelin W G, Livinston D M. A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 31.Sherr C J. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 32.Trimarchi J M, Fairchild B, Verona R, Moberg K, Andon N, Lees J A. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc Natl Acad Sci USA. 1998;95:2850–2855. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verona R, Moberg K, Estes S, Starz M, Vernon J P, Lees J A. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol Cell Biol. 1997;17:7268–7282. doi: 10.1128/mcb.17.12.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber J D, Taylor L J, Roussel M F, Sherr C J, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 35.Weintraub S J, Chow K N, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 36.Xu G, Livingston D M, Krek W. Multiple members of the E2F transcription factor family are the products of oncogenes. Proc Natl Acad Sci USA. 1995;92:1357–1361. doi: 10.1073/pnas.92.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]