Abstract
Background
The coronavirus disease (COVID-19) effected a global health crisis in 2019, 2020, and beyond. Currently, methods such as temperature detection, clinical manifestations, and nucleic acid testing are used to comprehensively determine whether patients are infected with the severe acute respiratory syndrome coronavirus 2. However, during the peak period of COVID-19 outbreaks and in underdeveloped regions, medical staff and high-tech detection equipment were limited, resulting in the continued spread of the disease. Thus, a more portable, cost-effective, and automated auxiliary screening method is necessary.
Objective
We aim to apply a machine learning algorithm and non-contact monitoring system to automatically screen potential COVID-19 patients.
Methods
We used impulse-radio ultra-wideband radar to detect respiration, heart rate, body movement, sleep quality, and various other physiological indicators. We collected 140 radar monitoring data from 23 COVID-19 patients in Wuhan Tongji Hospital and compared them with 144 radar monitoring data from healthy controls. Then, the XGBoost and logistic regression (XGBoost + LR) algorithms were used to classify the data according to patients and healthy subjects.
Results
The XGBoost + LR algorithm demonstrated excellent discrimination (precision = 92.5%, recall rate = 96.8%, AUC = 98.0%), outperforming other single machine learning algorithms. Furthermore, the SHAP value indicates that the number of apneas during REM, mean heart rate, and some sleep parameters are important features for classification.
Conclusion
The XGBoost + LR-based screening system can accurately predict COVID-19 patients and can be applied in hotels, nursing homes, wards, and other crowded locations to effectively help medical staff.
Keywords: COVID-19, Screening system, Non-contact vital signs, XGBoost, Logistic regression
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in a large-scale global pandemic with its extreme contagiousness and high fatality rate [1]. From the onset of the outbreak in late 2019 to January 14, 2021, 25.86 million infected cases have been reported worldwide, which have resulted in 1,362,700 deaths. Some effective methods of controlling the spread of the virus include large-scale screening, patient isolated treatment, and symptom surveillance. Most of the current testing methods are based on the reverse transcription-polymerase chain reaction (RT-PCR). At the peak of the SARS-CoV-2 outbreak, there was a severe RT-PCR test kit shortage; thus, computed tomography (CT), clinical features, and temperature testing were proposed as alternative diagnostic methods in hospitals [2,3]. However, owing to the relative novelty of the disease, doctors and medical personnel encounter challenges in accurately identifying coronavirus disease (COVID-19) cases in underdeveloped regions. As a result, many suspected cases cannot be tested, treated, and quarantined in time; thus, the spread of the virus may continue [[4], [5], [6]]. Moreover, because SARS-CoV-2 is highly infectious, the risk of infection is three times higher in doctors and nurses than in the general population, as they are closer to patients during treatment [7]. Therefore, this study aims at finding a non-contact automated detection device and method for testing SARS-CoV-2 infections without assistance from professional physicians.
Infected patients experience symptoms such as fever, fatigue, dry cough, and dyspnea [8]. Some researchers have attempted to detect these symptoms using non-contact devices and machine learning algorithms [9]. They focused on two main issues: the features extracted from the subject and the algorithms used to recognize infected subjects.
Regarding the features, many infectious disease detection systems have focused on detecting abnormal changes in heart rate, respiration rate, and facial temperature. Yang et al. [10] designed a contactless dengue fever screening system with microwave sensors (detecting heart rate, respiration rate, and the standard deviation of heartbeat interval) while using a neural network and the SoftMax function to determine whether the disease is infectious as well as the probability of infection, achieving a 98% accuracy. However, they believed that facial temperature could be greatly influenced by the environment. Matsui et al. [18,19] used respiratory rate, heart rate, and facial thermal imaging to rapidly screen influenza based on linear discriminant analysis (LDA). Their results revealed that the system has higher accuracy than systems with only thermal imaging, achieving an 88.9% accuracy.
In terms of single classification methods, most studies applied a single machine learning classifier to detect infections; for example, logistic regression [10], support vector machine (SVM) [11,12], k-means [13], k-nearest neighbor (KNN) [14], decision trees [15], and random forest (RF) [16,17] were broadly studied in the infectious disease screening field. Yao et al. [14] compared the performance of six different single classification models, including LDA, quadratic discriminant analysis (QDA), SVM, KNN, logistic regression (LR), and the NaiveBeyesian classifier, among which SVM and QDA exhibited the best results. Their research also indicated that the classification performance was not affected when the respiratory rate was excluded. A possible explanation is that the respiratory rate is difficult to measure owing to the short observation period and large noise. Sumiyakhand et al. [15] used the random tree method to predict the influenza infection status in patients. The sensitivity and negative predictive value (NPV) were 96.2% and 96.0%, respectively; the random tree showed a high accuracy. Sun et al. [20] used an LR model and features such as facial temperature, respiratory rate, and heart rate to recognize infections, with a sensitivity and NPV of 87.5% and 91.7%, respectively. These results showed that the facial temperature and heart rate of influenza patients were higher than those of the healthy group, although there was no significant difference in the respiratory rate.
The abovementioned studies mainly discussed a few short-term features, meaning that these features only contain information about the subject over several seconds or minutes. Moreover, the short-term monitoring data within 20 s are easily disturbed and cannot reflect the long-term fluctuation of patients’ physical data. In addition, some studies have shown that the respiration rate over a short period of time rarely contributes to the screening of infectious diseases. In our study, we attempted to determine more long-term features to describe the status of the subject, especially at night—such as sleep quality—to screen for infection possibilities.
Another limitation concerning these studies is that they used a single feature classification model, which can result in over-fitting and a less robust performance, despite these models performing well on their datasets. Furthermore, because we studied more features than these studies did, it was necessary to propose a robust model that can handle high-dimensional features.
In this study, we used a biological radar to continuously monitor patients throughout the night and obtained 25 long-term and short-term features. A combined XGBoost and LR algorithm (XGBoost + LR) was applied to the radar monitoring data to screen COVID-19 patients, in which the XGBoost model was used to select effective features, and the results of the constructed trees of this model were treated as inputs to the LR model. In this way, we combined the advantage of decision trees—i.e., selecting suitable features—and that of LR, which in turn allows the model to deal with massive features.
The major contributions of this study are as follows:
-
●
Providing a long-term non-contact COVID-19 screening system for use in hotels, wards, and other places located in isolated regions.
-
●
Illustrating the efficiency of long-term features and the ensemble XGBoost + LR model for night monitoring and COVID-19 disease screening.
2. Materials and methods
2.1. Equipment and data
In this study, a non-contact vital sign monitoring system was used to monitor COVID-19 patients at Wuhan Tongji Hospital, Wuhan, China. A total of 140 radar monitoring data from 23 patients were collected from March 17, 2020, to April 4, 2020, and 144 sleep monitoring data of healthy controls were analyzed. In this experiment, patients ranged from mild to moderate cases and were all in the general isolation ward during data collection. The healthy subjects’ data were acquired in their own homes. Basic information such as age, sex, underlying diseases, and medication was recorded. The basic information from both groups displayed a scattered distribution with no significant difference. Therefore, these factors can be regarded as small interference items; thus, the effectiveness of this control experiment is validated.
Doctors collect patient data by placing the monitor at the patient's bedside. This data collection method does not affect the daily activities of patients and affords ease of operation to medical staff. This research was approved by the Peking University Third Hospital Medical Research Ethics Committee (No.2020(081-01)), and the patients (or their caregivers) provided written informed consent prior to participating in the experiment.
The monitoring system consists of a non-contact vital sign monitor [23] and remote data service platform. The monitor transmits radar signals and then filters the radar echo signals to separate the heartbeat, respiratory, and body motion signals, thus extracting the respiration rate, heart rate, body movement, and sleep apnea. The monitoring data were uploaded to the data platform in real-time via Wi-Fi. After one night's sleep, the system analyzes the sleep and apnea data of the entire night and obtains a sleep monitoring report.
The sleep monitoring report has 25 data points, as shown in Table 1 , which fully reflect the patient's nighttime breathing, heartbeat, sleep structure, body movement, apnea, and other aspects.
Table 1.
Monitoring data during sleep.
| Metric | Abbreviation | Metric | Abbreviation |
|---|---|---|---|
| Mean respiratory rate | meanRR | Mean heart rate | meanHR |
| Median respiratory rate | medRR | Median heart rate | medHR |
| Maximum respiratory rate | maxRR | Maximum heart rate | maxHR |
| Minimum respiratory rate | minRR | Minimum heart rate | minHR |
| Percentage of awake sleep | awakPrct | Percentage of rem sleep | REMSPrct |
| Percentage of light sleep | lightSPrct | Percentage of deep sleep | deepSPrct |
| Sleep latency | latnMin | Sleep duration | slepMin |
| Sleep efficiency | slepEffic | Sleep score | slepScore |
| Mean body dynamic density | meanNMD | Percentage of body movement per minute | movMinPrct |
| Wake up times | awakTims | Turn over times | timesTO |
| Number of apneas during sleep | slepATims | Apnea-hypopnea index | AHI |
| Number of apneas during rem | REMSATims | Number of apneas during light sleep | lightSATims |
| Number of apneas during deep sleep | deepSATims |
2.2. Data exploration
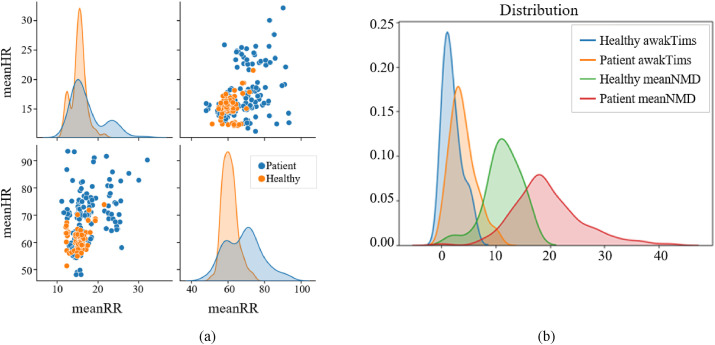
First, we verified whether differences existed in the nighttime respiration, heartbeat, body movement, and sleep quality between the COVID-19 patients and the healthy subjects. Fig. 1 shows that the heart and respiratory rates of the healthy subjects are relatively concentrated, whereas those of the patients are scattered and the values are relatively large. The mean body dynamic density and wake-up times of patients were approximately twice those of healthy subjects. The data distribution of patients was significantly different from that of healthy subjects. Therefore, we can classify patients and healthy subjects based on these features.
Fig. 1.
Distribution of a) heart rate and respiratory rate and b) mean body dynamic density and wake up times in healthy subjects and COVID-19 patients.
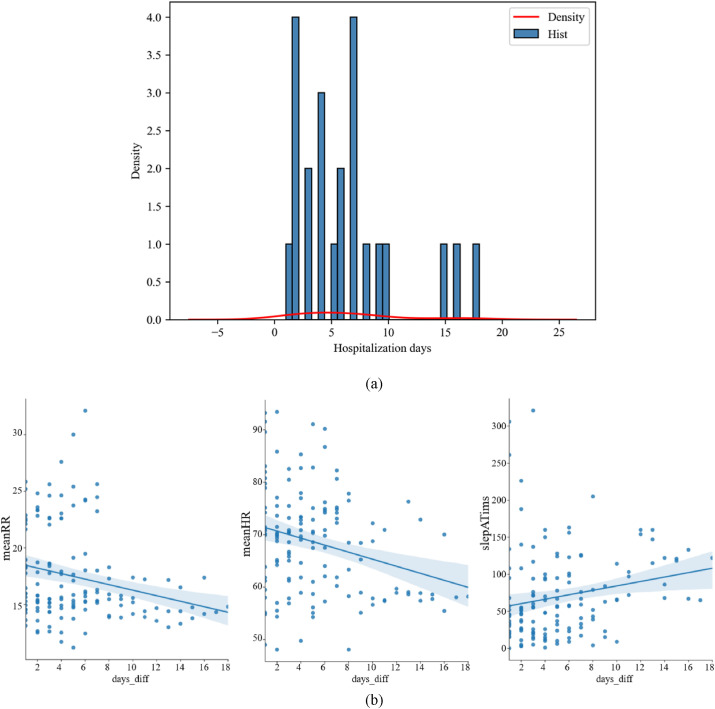
Fig. 2 shows the distribution of hospitalization days and the joint distribution of average heart rate, average respiratory rate, and apnea frequency during sleep and hospitalization days. The average heart rate and average respiratory rate decreased with an increase in hospitalization days, and the wake up time increased with an increase in hospitalization days.
Fig. 2.
a) Distribution of patients' hospitalization days. b) Fitting of average heart rate, average respiratory rate, apnea frequency during sleep, and hospitalization days. The shaded area represents the 95% confidence band.
2.3. Model building
XGBoost is an ensemble-learning algorithm based on gradient descent iterations. XGBoost uses decision tree as the base learner for integration. The algorithm continuously splits features to grow a tree. Each decision tree calculates the feature and threshold with the best branch effect and completes the split construction. Finally, the prediction results become consistent with the patient's diagnosis results, and the screening model is obtained.
| (1) |
| (2) |
where hypothesis is the output of the -th tree, is the current output of the model, and is the actual result. represents the number of decision trees, and represents the -th iteration, that is, every time we find an optimal model to add to the existing model to make the predicted value closer to the real value. We build the optimal model by minimizing the loss function. When the training dataset is small, it is easy to over-fit; therefore, it is generally necessary to add a regular term to reduce the complexity of the model.
| (3) |
where F is the hypothetical space and is the control of the complexity of the model.
Therefore, the objective function is given by the following equation:
| (4) |
The first part on the right side of equation (4) is the training error, and the middle part is the complexity of the penalty model (the sum of the complexity of all trees), which contains two parts: the number of leaf nodes and the value of each leaf node. The expression is given as
| (5) |
where T is the number of leaf nodes, ||w|| is the module of the leaf node vector, is the difficulty of node segmentation, and is the L2 regularization coefficient.
We expand the loss function using Taylor's approximation to the quadratic term and use the greedy algorithm to solve the model parameters.
LR is a basic binary classification model. Based on linear regression, the input function value is mapped to interval (0,1) by the sigmoid function to represent the probability of various types of discrimination. Let y be a secondary dependent variable, indicating whether the subject is a COVID-19 patient or not; y = 0 means the subject is healthy, y = 1 means he/she/they are a patient. denotes the probability that y belongs to category 1 under the condition of a given feature vector . Let ; can be expressed as
| (6) |
The above formula is called the LR model, where represents the coefficient corresponding to each feature. Parameters can be obtained using the gradient descent method.
The XGBoost + LR algorithm consists of two parts, where XGBoost is used to extract features from the training set as new training input data and LR is the classifier for the new training input data.
Using XGBoost to construct combined features means every leaf node of each decision tree is considered as a new feature. Therefore, the number of features constructed is the same as that of the XGBoost leaf nodes. For each decision tree, if the input sample falls into a leaf node, the value of the leaf node is 1; otherwise, it is 0.
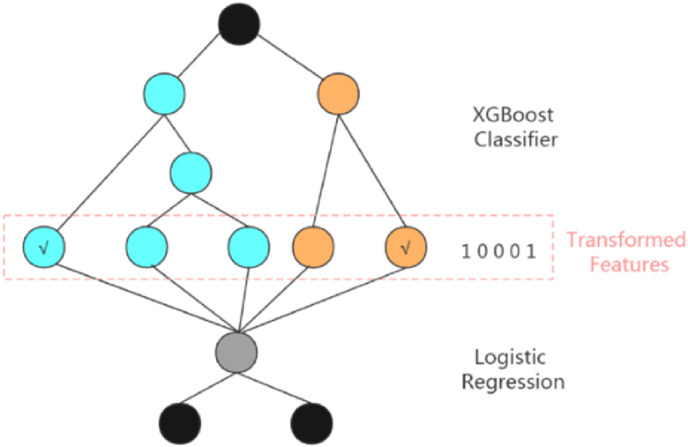
Fig. 3 shows how our proposed XGBoost + LR model processes a sample data. The XGBoost model is obtained by training the original sample data. The model consists of two decision trees. Tree 1 has three leaf nodes and Tree 2 has two leaf nodes. Each leaf node is regarded as a new feature. For example, if a sample falls into the first leaf node of Tree 1 through Tree 1 and falls in to the second leaf node of Tree 2 through Tree 2, then the newly constructed feature vector of this sample would be [1, 0, 0, 0, 1].
Fig. 3.
Structural features of the proposed XGBoost + LR model. First, the XGBoost classifier is used as a feature selection model, whose result would be one-hot coded. Second, using the results of the XGBoost model as the LR model input, the final result is predicted.
For model training, four steps are required to obtain the trained proposed ensemble XGBoost + LR model. First, we train the XGBoost model with the original training dataset to construct the combined features, where the grid searching method is employed to find the optimal parameters. Second, the training samples are input to the trained model from step 1, and the output of the leaf nodes constitutes new construction features. Third, we combine the new features constructed from step 2 and the original features. Lastly, the final LR model with the combined features is trained.
For model testing, first, the test sample is input into the trained XGBoost model to obtain new construction feature vectors. Then, the new construction feature vectors and the original feature vectors are combined into new feature vectors, which are input to the trained LR model, and the output is the classification result of the test sample.
2.4. Feature selection
Feature selection improves the speed of model building, enhances the generalization ability of the model, and reduces over-fitting problems. A good global feature importance metric must satisfy both consistency and accuracy. We use the SHAP value [24] to describe and evaluate the importance of the features.
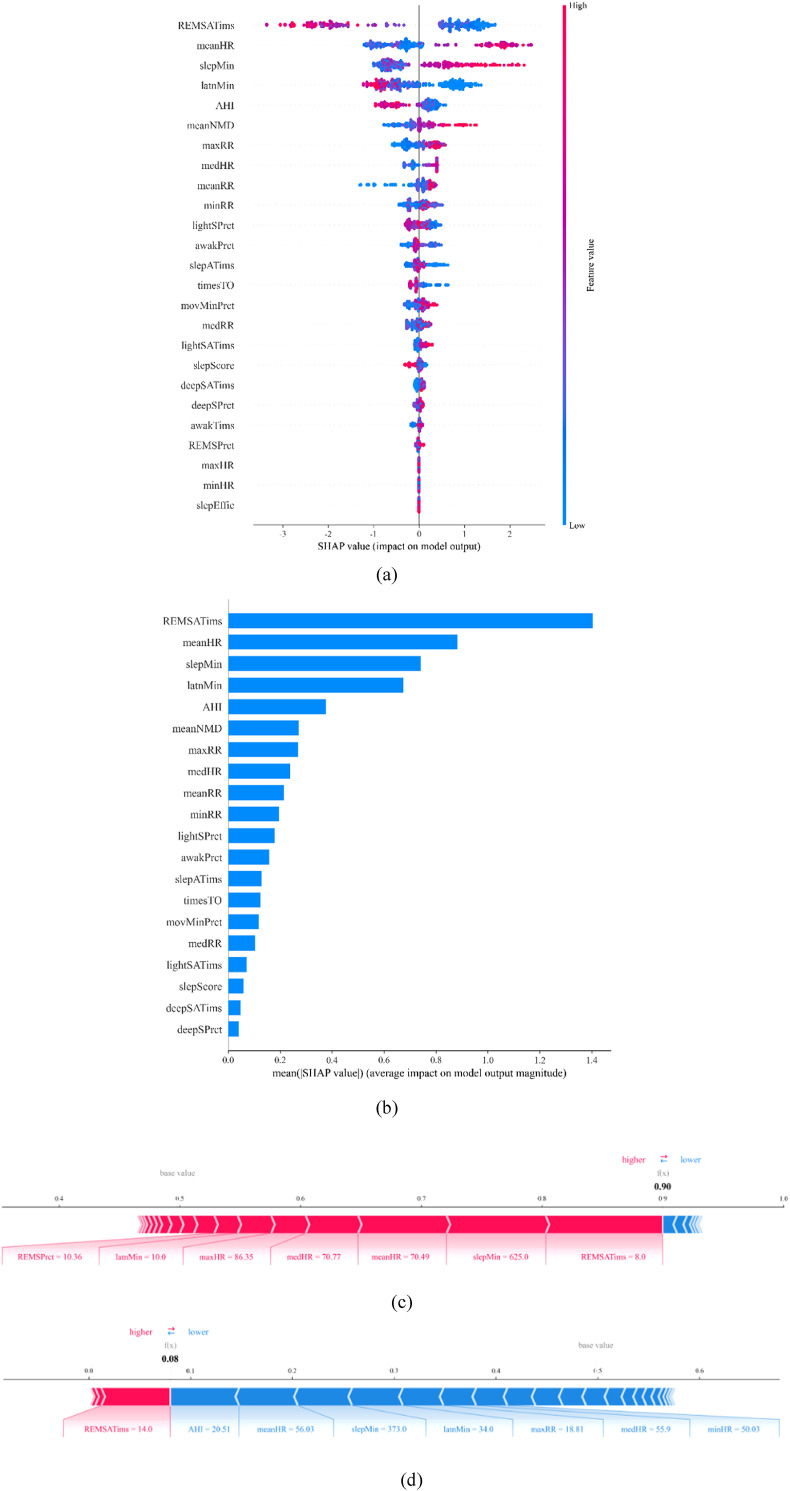
The SHAP value can visualize the features as a whole. Fig. 4 (a) shows the impact of the features on each sample. Each row represents a feature and the abscissa represents the SHAP value. A point represents a sample, and the color represents the feature value (red and blue represent high and low values, respectively). For example, Fig. 4(a) shows that a lower value of the “REMSATims” feature increases the risk of becoming a patient, whereas a higher value of the “meanHR” feature increases the risk.
Fig. 4.
a) Overall visualization of features by SHAP. b) Feature importance sorted by SHAP. c) SHAP value of a single patient sample. d) SHAP value of a single healthy subject sample.
The average absolute SHAP value of each feature represents a unified measure of its importance. A standard bar graph is shown in Fig. 4(b). These features are sorted by mean (|Tree SHAP|); revealing that the “REMSATims” feature is the strongest factor for detecting COVID-19 patients.
We randomly collected data from a group of patients and healthy subjects to compare and observe the impact of the features on the classification results. Fig. 4(c) and (d) show the SHAP values of patients and healthy subjects, respectively. These figures show that each feature has its contribution, thus nudging the model result from the base value to the model output. Red indicates that the contribution of the feature is positive. For example, the longest red bar in the patient graph is “REMSATims,” and the SHAP value increased by 0.09886. The longest blue bar is “lightSprct,” and the SHAP value decreased by 0.00747. The final SHAP value is 0.90.
Differences are observed in heart rate, respiratory rate, sleep quality, and apnea index between patients and healthy subjects. Specifically, patients exhibited a higher heart rate, longer sleep duration, shorter sleep latency, deeper sleep, and lower apnea index. As previously reported, heart rate is one of the most useful characteristics in the classification of infected patients, whereas the contribution of the respiratory rate is insignificant. Furthermore, we found that nighttime sleep quality and apnea index are important distinguishing indicators.
Based on the importance of the feature values, the top eight features, namely, “REMSATims,” “meanHR,” “slepMin,” “latnMin,” “AHI,” “meanNMD,” “maxRR,” and “medHR,” are selected for model training.
2.5. Evaluation method
We selected precision, recall, and the receiver operating characteristic (ROC) curves as the evaluation criteria [25]. The precision measures the accuracy of the model in terms of false positives, that is, the number of healthy subjects misdiagnosed with the disease. The lower the false positive rate, the higher the precision of the model. The recall rate provides information regarding false-negative cases, that is, the number of infected patients predicted to be healthy. In the context of disease screening, the missed detection of infected patients is considered more serious than that of healthy subjects. Therefore, in terms of evaluation models, recall is more significant than precision. The ROC curve draws a comparison chart of the true positive and false positive rates to further evaluate the performance of the model. AUC is defined as the area under the ROC curve; the larger its value, the better the effect of the corresponding classifier. The precision and recall are defined as follows, where TP, FP, and FN represent true positive, false positive, and false negative, respectively.
| (7) |
| (8) |
where TP = true positive, which is the number of correctly classified positive samples; FP = false positive, incorrectly classified negative samples; TN = true negative, correctly classified negative samples, and FN = false negative, incorrectly classified positive samples.
2.6. Comparison of the results
In this study, six machine learning algorithms, namely, LR, KNN, SVM, RF, XGBoost, and XGBoost + LR, were compared. The data of healthy subjects and COVID-19 patients were divided, with 75% of the data being randomly selected as the training set, while the remaining 25% were used as the test set. The training set was used to train the infection screening model, and the test set was used to test the screening performance of the model.
The 10-fold cross-validation method [26] was used to further divide the training set into 10 copies, one such copy was cyclically extracted as the validation set for the optimal parameters. Table 2 lists the final parameter settings for each algorithm.
Table 2.
Final parameter settings for the six algorithms used in this study.
| Logistic Regression |
kNN | SVM | RF | XGBoost | XGBoost + LR |
|---|---|---|---|---|---|
| C = 0.01 | k_neighbors = 5 | kernel = ‘rbf’ | n_estimators = 80 | learning_rate = 0.2 | learning_rate = 0.2 |
| penalty = ‘l2’ | p = 1 | C = 100 | max_depth = 5 | n_estimators = 80 | n_estimators = 80 |
| gamma = 0.001 | max_depth = 7 | max_depth = 7 | |||
| gamma = 0.001 | gamma = 0.001 |
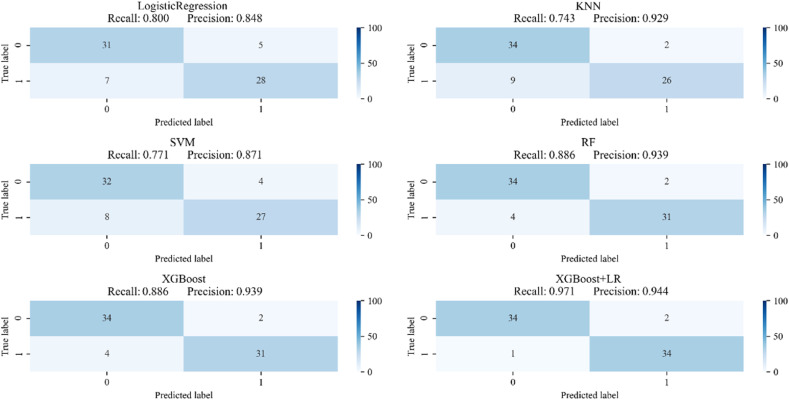
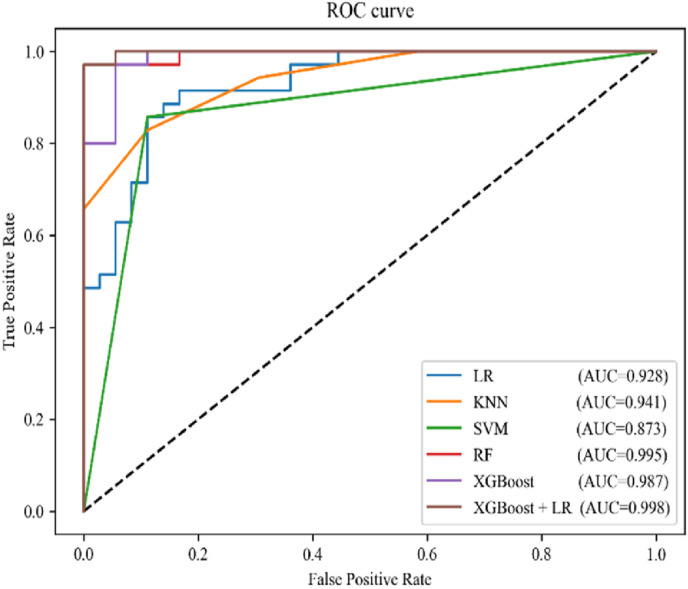
The confusion matrix of the six algorithms is shown in Fig. 5 . Here, it can be seen that the RF and XGBoost models have the best classification results among single algorithms, with both exhibiting a recall rate and precision of 88.6% and 93.9%, respectively. The XGBoost + LR model exhibits better results, with a recall rate and precision of 97.1% and 94.4%, respectively. The ROC curve of the six classification models is shown in Fig. 6 . The classification performance of XGBoost + LR is also better than that of other algorithms, with an AUC of 0.988.
Fig. 5.
Confusion matrix of the six models.
Fig. 6.
ROC curves of the six models.
To reduce the randomness of the results, the data extraction and modeling were repeated 1000 times. Table 3 shows the average performance for each algorithm. Based on these results, it is confirmed that the XGBoost + LR model outperforms the other models, with a recall, precision, and AUC of 96.8%, 92.5%, and 98.0%, respectively. Any model with such performance metrics should be considered suitable for clinical use and helpful to doctors.
Table 3.
Comparison of the classification results of the six models.
| Model | Recall (%) (95%CI) |
Precision (%) (95%CI) |
AUC (%) (95%CI) |
|---|---|---|---|
| LR | 85.1 (82.9, 87.3) | 87.3 (85.2, 89.4) | 92.6 (91.0, 94.2) |
| KNN | 77.6 (75.0, 80.2) | 90.5 (88.7, 92.3) | 91.3 (89.5, 93.1) |
| SVM | 80.6 (78.2, 83.0) | 91.5 (89.8, 93.2) | 92.8 (91.2, 94.4) |
| RF | 89.1 (87.2, 91.0) | 97.4 (96.4, 98.4) | 97.9 (97.0, 98.8) |
| XGBoost | 91.3 (89.6, 93.0) | 96.6 (95.5, 97.7) | 97.8 (96.9, 98.7) |
| XGBoost + LR | 96.8 (95.8 97.8) | 92.5 (90.9, 94.1) | 98.0 (97.1, 98.9) |
Compared with the SVM classifier, our model achieved a 16.2% improvement in terms of recall. This improvement is due to the creation of new features (OneHOT features) by using the XGBoost model. The features determine the upper limit of the effect of all algorithms, and different algorithms only differ in the distance from this upper limit. By creating new and effective features, our model achieved better performance.
Our proposed XGBoost + LR model combines the advantages of both models. The LR algorithm is a linear model, which is easy to parallelize, and can easily process hundreds of millions of data, although its learning ability is very limited because it requires a lot of feature engineering work for it to be improved. However, processing numerous features is time- and effort-consuming and might not necessarily improve the results. Therefore, the automatic identification of effective features and feature combinations to compensate for the lack of manual experience and shorten the cycle of feature engineering in LR models remains a major challenge.
Although the XGBoost and LR algorithms have also achieved better results when used alone, the integrated use of the XGBoost and LR algorithms can reduce the probability of misdiagnosing patients as healthy. In this study, the results of the XGBoost training were encoded by OneHOT, and these new results were treated as newly constructed features and fed into logistic classification, along with the original features. Herein, XGBoost functioned as a feature selection tool and created new effective features from the original features for the next layer of the logistic classification model. It first performs nonlinear changes in samples, derives some crossover features, and then uses the LR model for discrimination. Compared with single models, the integrated model has unique advantages in dealing with complex interactions and nonlinear relationships among variables.
3. Conclusion
This study had several advantages. First, more than 20 physiological indicators, such as heart rate and sleep quality, were innovatively measured to connect patients' nighttime sleep information with infectious disease prediction to increase the prediction reliability. Among these features, apneas during REM, mean heart rate, and sleep parameters are shown to be the key features. Second, the proposed XGBoost + LR-based classification algorithm was used to strengthen the feature selection and could measure the difference between patients and healthy subjects in different features. We innovatively used a XGBoost model to process the original data and created new and effective features, which were used to train an LR model. Furthermore, the proposed combined model exhibited a comparable performance to that of traditional machine learning algorithms, achieving a precision, recall rate, and AUC of 92.5%, 96.8%, and 98.0%, respectively. Finally, because infectious diseases have similar inflammatory symptoms, during the next pandemic, it could be used as a convenient, low-cost, automatic screening method without the guidance of professional doctors, especially in underdeveloped countries and regions without sufficient health facilities and experienced doctors.
However, this study also had some drawbacks. First, owing to the limitations of the conditions, this study included patients and healthy subjects with different experimental backgrounds. Environmental factors and other parameters could interfere with the controlled trials. In addition, the sample size of this study was small and did not include multiple age groups; therefore, in practice, the accuracy index may decline. Moreover, the proposed method and system can only monitor one patient at a time, which limits the monitoring efficiency in a real-world hospital setting. More specifically, the proposed system uses a continuous-wave radar, which limits the application to monitor multiple subjects simultaneously. In future work, a larger sample size should be used to cover a more comprehensive population. Furthermore, the algorithm and sensor should also be updated to support multi-person monitoring while using only one device. We collected data from patients at different stages of disease development. If we accumulate sufficient data, we can continue to distinguish the severity of patients and provide a reference for clinical triage and reasonable allocation of medical resources. In this study, we used physical sign data to predict potential infections; however, these prediction results cannot be directly used for medical diagnosis. Although our system has been sufficiently tested on COVID-19 patients, it may also incorrectly diagnose COVID-19 when the patient has a similar disease that causes the body's inflammatory response and the system is unable to distinguish them. Nevertheless, this prediction system can still be very effective in assisting screening during the outbreak of infectious diseases while saving cost and labor.
In summary, we demonstrated the relationship between the data obtained from non-contact sleep monitoring equipment and the prediction of COVID-19. These preliminary results provide a basis for further evaluation of the nocturnal cardiopulmonary status and sleep status as physiological signs of COVID-19 infection, which helps to identify COVID-19 and other infections in an economical and fast manner.
Funding
This work was supported by the Alliance of International Science Organization (No. ANSO-CR-SP-2020-04 (2020000147)).
Declaration of competing interest
None Declared.
References
- 1.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ring E.F., McEvoy H., Jung A., Zuber J., Machin G. New standards for devices used for the measurement of human body temperature. J. Med. Eng. Technol. 2010;34(4):249–253. doi: 10.3109/03091901003663836. [DOI] [PubMed] [Google Scholar]
- 3.Lahiri B.B., Bagavathiappan S., Jayakumar T., Philip J. Medical applications of infrared thermography: a review. Infrared Phys. Technol. 2012;55(4):221–235. doi: 10.1016/j.infrared.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aw J. The non-contact handheld cutaneous infra-red thermometer for fever screening during the COVID-19 global emergency. J. Hosp. Infect. 2020;104(4):451. doi: 10.1016/j.jhin.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow E.J., Schwartz N.G., Tobolowsky F.A., et al. Symptom screening at illness onset of health care personnel with SARS-CoV-2 infection in king county, Washington. JAMA J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yombi J.C., De Greef J., Marsin A.S., et al. Symptom-based screening for COVID-19 in health care workers: the importance of fever. J. Hosp. Infect. 2020 doi: 10.1016/j.jhin.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirten R.P., Danieletto M., Tomalin L., et al. Use of physiological data from a wearable device to identify SARS-CoV-2 infection and symptoms and predict COVID-19 diagnosis: observational study. J. Med. Internet Res. 2021;23(2) doi: 10.2196/2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen N., Zhou M., Dong X., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/s0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdonk C., Verdonk F., Dreyfus G. How machine learning could be used in clinical practice during an epidemic. Crit. Care. 2020 May 26;24(1):265. doi: 10.1186/s13054-020-02962-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X., Kumagai K., Sun G., et al. 2019 IEEE Sensors Applications Symposium (SAS); 2019 11-13 March 2019. 2019. Dengue fever screening using vital signs by contactless microwave radar and machine learning; pp. 1–6. [Google Scholar]
- 11.Cortes C., Vapnik V. Support-vector networks. Mach. Learn. 1995;20(3):273–297. doi: 10.1007/BF00994018. [DOI] [Google Scholar]
- 12.Cuong V., Nguyen, et al. A non-contact infection screening system using medical radar and Linux-embedded FPGA: implementation and preliminary validation. Inform. Med. Unlocked. 2019;16(C) doi: 10.1016/j.imu.2019.100225. 100225-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun G., Hakozaki Y., Abe S., Vinh N.Q., Matsui T. A novel infection screening method using a neural network and k-means clustering algorithm which can be applied for screening of unknown or unexpected infectious diseases. J. Infect. 2012;65(6):591–592. doi: 10.1016/j.jinf.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Yao Y., Sun G., Matsui T., Hakozaki Y., van Waasen S., Schiek M. Multiple vital-sign-based infection screening outperforms thermography independent of the classification algorithm. IEEE Trans. Biomed. Eng. 2016;63(5):1025–1033. doi: 10.1109/tbme.2015.2479716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sumiyakhand, Dagdanpurev, Shigeto, et al. A novel machine-learning-based infection screening system via 2013-2017 seasonal influenza patients' vital signs as training datasets. J. Infect. 2019 doi: 10.1016/j.jinf.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 16.Breiman L. Random forests. Mach. Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 17.Bishop C.M. Springer-Verlag New York, Inc.; 2006. Pattern Recognition and Machine Learning. [Google Scholar]
- 18.Matsui T., Hakozaki Y., Suzuki S., et al. A novel screening method for influenza patients using a newly developed non-contact screening system. J. Infect. 2010;60(4):271–277. doi: 10.1016/j.jinf.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X., Ishibashi K., Negishi T., et al. Springer; Singapore: 2017. Short Time and Contactless Virus Infection Screening System with Discriminate Function Using Doppler Radar. Bio-Inspired Computing: Theories and Applications. [Google Scholar]
- 20.Sun G., Nakayama Y., Dagdanpurev S., et al. Remote sensing of multiple vital signs using a CMOS camera-equipped infrared thermography system and its clinical application in rapidly screening patients with suspected infectious diseases. Int. J. Infect. Dis.: IJID: Off. Publ. Int. Soc. Infect. Dis. 2017;55:113–117. doi: 10.1016/j.ijid.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu W., Zhao Z., Wang Y., Zhang H., Lin F. Noncontact accurate measurement of cardiopulmonary activity using a compact quadrature Doppler radar sensor. IEEE Trans. Biomed. Eng. 2014;61(3):725–735. doi: 10.1109/tbme.2013.2288319. [DOI] [PubMed] [Google Scholar]
- 24.Lundberg S., Lee S.I. Nips; 2017. A Unified Approach to Interpreting Model Predictions. [Google Scholar]
- 25.Goutte C., Gaussier E. Advances in Information Retrieval. Springer Berlin Heidelberg; Berlin, Heidelberg: 2005. A probabilistic interpretation of precision, recall and F-score, with implication for evaluation; pp. 345–359. [Google Scholar]
- 26.Kohavi R. Proceedings of the 14th International Joint Conference on Artificial Intelligence. Morgan Kaufmann Publishers Inc.; Montreal, Quebec, Canada: 1995. A study of cross-validation and bootstrap for accuracy estimation and model selection; pp. 1137–1143. [Google Scholar]