Summary
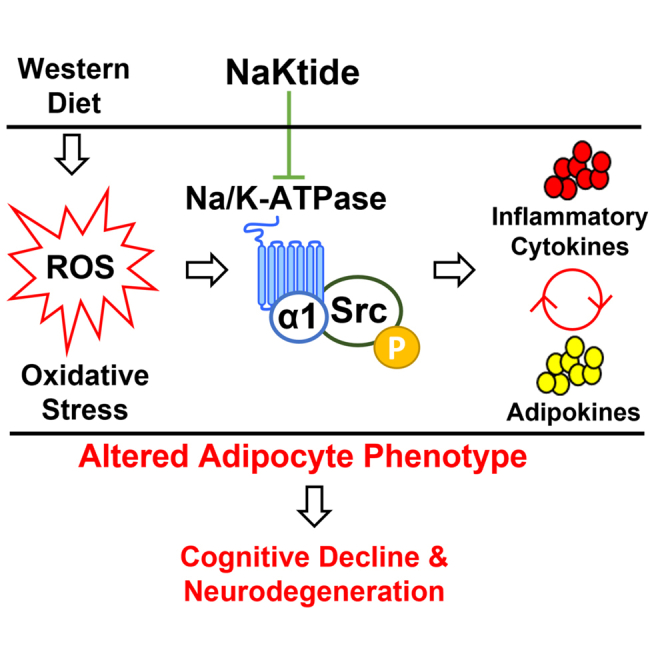
Recent studies suggest that a western diet may contribute to clinical neurodegeneration and dementia. Adipocyte-specific expression of the Na,K-ATPase signaling antagonist, NaKtide, ameliorates the pathophysiological consequences of murine experimental obesity and renal failure. In this study, we found that a western diet produced systemic oxidant stress along with evidence of activation of Na,K-ATPase signaling within both murine brain and peripheral tissues. We also noted this diet caused increases in circulating inflammatory cytokines as well as behavioral, and brain biochemical changes consistent with neurodegeneration. Adipocyte specific NaKtide affected by a doxycycline on/off expression system ameliorated all of these diet effects. These data suggest that a western diet produces cognitive decline and neurodegeneration through augmented Na,K-ATPase signaling and that antagonism of this pathway in adipocytes ameliorates the pathophysiology. If this observation is confirmed in humans, the adipocyte Na,K-ATPase may serve as a clinical target in the therapy of neurodegenerative disorders.
Subject areas: Molecular physiology, Neuroscience, Cell biology
Graphical abstract
Highlights
-
•
Adipocytes induce phenotypical changes in neurodegeneration through adipocyte NKAL
-
•
Adipocyte specific NaKtide inhibited NKAL and improved systemic homeostasis
-
•
NaKtide improved adipocyte phenotype and improved markers of neurodegeneration
-
•
NaKtide improved genomic profile of hippocampus and adipose tissue in WD fed mice
Molecular physiology; Neuroscience; Cell biology
Introduction
A number of clinical conditions are characterized by impairment in brain functions, specifically memory and cognitive ability (Moya-Alvarado et al., 2016; Sonkusare et al., 2005). Alzheimer’s disease is the most common and best studied. The dementias associated with these pathophysiological consequences have been grouped under the broad category of neurodegeneration. Recent studies have demonstrated that obesity exacerbates the progression of these neurodegenerative disorders (Ashrafian et al., 2013; Kothari et al., 2017; Miller and Spencer, 2014). Several mechanisms associated with obesity have been implicated in the neurodegeneration, including altered lipid and fatty acid metabolism (Chew et al., 2020; Uranga and Keller, 2019), ceramide dysregulation causing insulin resistance and impaired cognitive function (de la Monte et al., 2010), as well as mechanisms associated with oxidant stress. One might infer that the excessive production of reactive oxygen species (ROS) and inflammatory cytokines associated with obesity might either cross or alter the blood brain barrier (BBB), and through effects on central neuronal tissues, contribute to neurodegeneration (Bartolome et al., 2017; Duffy et al., 2019; Kothari et al., 2017; Pratt et al., 2019). Previous studies suggest that the ability of cytokines to alter or cross BBB is primarily achieved through either active transport across BBB or passive transport via circumventricular organs specifically through areas in the brain where the BBB is incomplete and cytokines may cross by simple diffusion (Banks, 2005; Benveniste, 1992; Rothwell and Hopkins, 1995; Yarlagadda et al., 2009). Studies have also shown that cytokines may also damage the BBB through activation or destruction of tight junctions of microvascular endothelial cells that form the BBB, hence increasing its permeability without entering the brain (Yarlagadda et al., 2009). It is well known that some cytokines, especially interleukin-6 (IL-6) (Banks et al., 1994, 1995), can cross the BBB and induce neuroinflammation (Banks, 2005; Tarro et al., 2014). Although there are certainly distinctions amongst the mechanisms by which different neurodegenerative diseases progress, there is some overlap between these disease pathways and those underlying obesity (Mattson et al., 1999; Perl et al., 1998) such as those related to oxidative stress, mitochondrial dysfunction, and increased inflammation.
Neurons have been shown to express receptors for various adipokines, indicating that factors released from adipose tissue have the potential to communicate directly with the brain (Xu et al., 2003). Metabolic changes and increased inflammatory conditions associated with obesity can cause damage to the central nervous system (CNS), which can lead to neural death and altered synaptic plasticity of neurons; this metabolic dysfunction increases the risk of neurodegenerative disorders (Jiang et al., 2019; Mazon et al., 2017; Saltiel and Olefsky, 2017). Although a significantly altered glucose tolerance, seen in obesity and related comorbidities, can have a severe impact on brain function, insulin in the brain regulates ligand-gated ion channels, modulates synaptic plasticity and plays a critical role in the development and maintenance of excitatory synapses (Jiang et al., 2019; Kothari et al., 2017; Miller and Spencer, 2014). The cumulative evidence suggests that a western diet (WD) can increase plasma inflammatory cytokine levels, which in turn can directly induce neuroinflammation (Banks, 2005; Ferreira et al., 2014).
In light of understanding molecular mechanisms operant in this diseased phenotype, recent investigations from our lab have demonstrated oxidant stress within adipocytes attributable to Na,K-ATPase signaling, in a manner distinct from its previously well-known ion pumping activity of this complex (Liang et al., 2007; Liu et al., 2012; Yan et al., 2012). The Na,K-ATPase signaling is either directly stimulated by cardiotonic steroids which bind to the Na,K-ATPase analogous to a ligand-receptor interaction or by ROS which appear to act through direct carbonylation of the Na,K-ATPase α1 subunit. Signaling through this Na,K-ATPase signaling pathway requires the phosphorylation of Src, the transactivation of the epithelial growth factor receptor and a series of steps which include the generation of more ROS, activation of phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) and extracellular signal-regulated kinase (ERK) (Tian et al., 2006; Wang et al., 2014; Yan et al., 2012, 2013, 2016). Because the Na,K-ATPase is stimulated by oxidant stress and results in further generation of ROS, we have characterized this pathway as the Na,K-ATPase oxidant amplification loop (NKAL). Work from our laboratories has demonstrated the involvement of NKAL in the pathogenesis of several conditions such as diet-induced metabolic syndrome, nonalcoholic steatohepatitis and renal failure associated cardiomyopathy (Liu et al., 2016; Sodhi et al., 2015, 2017). Our studies have also extensively demonstrated that a synthetic peptide, NaKtide, derived from the segment of Src kinase-binding domain of the α1 subunit, abrogates NKAL by preventing the phosphorylation of Src (Li et al., 2009). Although the systemic administration of pNaKtide (a cell permeant version of NaKtide) in various murine models effectively attenuates oxidative stress, inflammation and the ameliorated diseased phenotype, we have more recently shown that adipocyte-specific NaKtide expression is effective in ameliorating experimental obesity induced by a WD and uremic cardiomyopathy induced by partial nephrectomy (Pratt et al., 2019; Sodhi et al., 2020b). Some of these data in the WD model suggested effects of the WD ameliorated by adipocyte specific NaKtide expression on whole brain biochemistry (Pratt et al., 2019).
Although previous studies have established a causal relationship for crosstalk between adipose tissue and brain, a direct effect of adipocytes on different neuronal regions of the brain remains elusive. Hence, based on the evidence present in literature and our previous studies, this study aims to demonstrate a central role of adipocytes, specifically through adipocyte NKAL, in neurodegeneration. We believe that understanding the role of the adipocyte Na/K-ATPase signaling in the pathogenesis of neurodegeneration could potentially lead to the development of new therapeutic strategies in addition to NaKtide, to fight neurodegenerative disorders.
Results
Doxycycline inducible adipocyte-specific NaKtide improves adiposity, metabolic balance, locomotion, and systemic inflammation in mice fed a WD
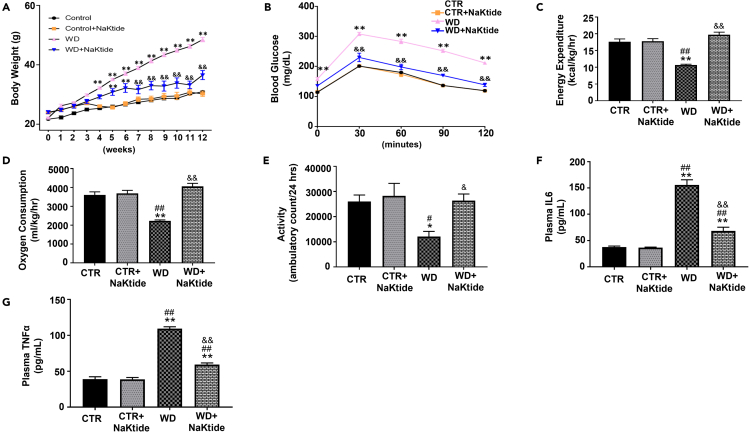
Transgenic mice expressing NaKtide in a tetracycline (TET) dependent manner (Tet-On), specifically in adipocytes, under the control of adiponectin promoter were used in this study. Because these mice used a TET dependent regulatory on/off system, the ingestion of doxycycline activated the adiponectin promoter and expressed NaKtide specifically in adipocytes. Mice were either fed a normal chow or WD with or without doxycycline for 12 weeks to demonstrate whether adipocyte-specific NaKtide can ameliorate diet induced metabolic changes. Mice fed a WD showed a significant increase in the body weight over the period of 12-weeks as compared to mice fed a normal chow (Figure 1A). The increase in the body weight of mice fed WD was significantly attenuated (Figure 1A) by doxycycline induced adipocyte specific NaKtide expression. Our results also showed significant insulin resistance as noted by impaired glucose tolerance in mice fed a WD, which was prevented by doxycycline induced adipocyte specific NaKtide expression (Figure 1B). Further, our results showed marked reduction in energy expenditure, oxygen consumption, and locomotion in the mice fed a WD as compared to mice fed a normal chow (Figures 1C–1E). This WD induced metabolic imbalance was attenuated by doxycycline induced adipocyte specific NaKtide expression (Figures 1C–1E). Our results also showed significantly increased levels of plasma inflammatory cytokines, IL-6 and tumor necrosis factor alpha (TNFα), in mice fed a WD (Figures 1F and 1G). The doxycycline induced adipocyte specific NaKtide expression significantly attenuated the increases in these levels of these inflammatory cytokines induced by a WD (Figures 1F and 1G).
Figure 1.
Doxycycline induced adipocyte-specific NaKtide expression improves adiposity, metabolic imbalance, locomotion and systemic inflammation in mice fed a WD
(A and B) (A) Body weight over the period of 12 weeks. Statistical analysis by two-way ANOVA, where (B) Glucose tolerance test.
(C–G) (C) Energy Expenditure, (D) Oxygen Consumption and (E) Locomotion determined by 48 h CLAMS assessment. Plasma levels of inflammatory markers (F) IL-6 and (G) TNFα. Results are expressed as means ± SEM. N = 6–10/group. Statistical analysis by two-way ANOVA for Panel (A and B), one-way ANOVA for Panel (C – G), multiple comparison using Tukey's test, where ∗p < 0.05 vs. CTR, ∗∗p < 0.01 vs. CTR, #p < 0.05 vs. CTR + NaKtide, ##p < 0.01 vs. CTR + NaKtide, &p < 0.05 vs. WD, &&p < 0.01 vs. WD. Groups: CTR (normal chow diet); CTR + NaKtide (normal chow diet with doxycycline inducible adipocyte specific NaKtide); WD (Western diet); WD + NaKtide (western diet with doxycycline inducible adipocyte specific NaKtide).
Doxycycline induced adipocyte-specific NaKtide improves neurodegenerative phenotype through inhibition of Na,K-ATPase signaling in visceral adipose tissue of mice fed a WD
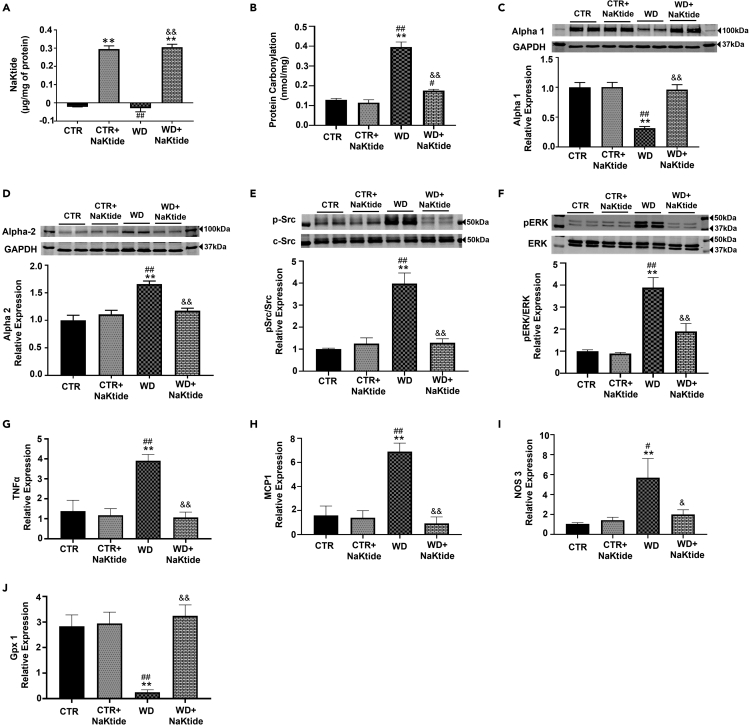
To test the efficacy and specificity of the doxycycline induced NaKtide expression in adipocytes, immunofluorescence staining was performed in visceral, subcutaneous, heart and brain tissues. Our results showed GFP (green) and NaKtide (red) expression in the visceral and subcutaneous adipose tissue of mice fed doxycycline (with chow or WD) (Figures S1A and S1B), while no detectable expression of GFP and NaKtide was noted in the heart and brain tissues of these mice (Figures S1C and S1D). We further confirmed the presence of doxycycline induced NaKtide expression in visceral adipose tissues by determining the concentration of NaKtide using a competitive ELISA (Sodhi et al., 2020b). Specifically, we found significant levels of NaKtide in visceral fat of mice fed doxycycline (added to chow or WD), whereas NaKtide concentrations were undetectable in mice groups without doxycycline (Figure 2A). Of note, the adipose tissue levels of NaKtide achieved with this Tet-On system were approximately 50% of that which we previously reported with the lentivirus adiponectin-NaKtide transduction (Sodhi et al., 2020b). We have previously shown that ROS initiates protein carbonylation of the Na,K-ATPase α1 subunit, followed by phosphorylation of Src and activation of downstream signaling cascades with modulation of extracellular signal-regulated kinase 1/2 (ERK 1/2) (Sodhi et al., 2015, 2017). Protein carbonylation in visceral adipose tissue showed significant increases in WD fed mice; this was attenuated by doxycycline induced NaKtide expression (Figure 2B). Further, our results showed significantly downregulated expression of α1 subunit of the Na,K-ATPase in WD fed mice which was increased by doxycycline induced NaKtide expression (Figure 2C). Conversely, the expression of the α2 subunit of the Na/K-ATPase was significantly upregulated with WD, as compared to control. This increase was significantly reverted by doxycycline induced NaKtide expression (Figure 2D). Adipose expression of phospho-Src was increased by the WD and attenuated by doxycycline induced NaKtide expression (Figure 2E). Our results also showed significant upregulation in the protein expression of ERK 1/2, in visceral adipose tissue of WD fed mice, which was attenuated by doxycycline induced NaKtide expression (Figure 2F). The mRNA expression of inflammatory markers, TNFα, monocyte chemoattractant protein 1 (MCP1) and nitric oxide synthase 3 (Nos3) were increased in mice fed a WD (Figures 2G–2I), and these changes were also attenuated by doxycycline induced NaKtide expression. Conversely, mice fed a WD alone had significantly downregulated mRNA expression of the antioxidant, glutathione peroxidase 1 (GPx1), which was also reverted by doxycycline induced NaKtide expression (Figure 2J).
Figure 2.
Doxycycline induced adipocyte-specific NaKtide expression improves oxidative stress in visceral adipose tissue of mice fed a WD
(A) NaKtide concentration by competitive ELISA.
(B–D)(B) Protein carbonylation assay. Immunoblot analysis of (C) α-1 subunit and (D) α-2 subunit with data shown as mean band density normalized to GAPDH.
(E) pSrc immunoblot analysis with data shown as mean band density normalized to c-Src.
(F–J)(F) Immunoblot analysis of pERK 1/2 with mean band density normalized to total ERK. mRNA expression of (G) TNFα, (H) MCP1, (I) Nos3 and (J) GPx1. Results are expressed as means ± SEM. N = 4–6/group. Statistical analysis by one-way ANOVA, multiple comparison using Tukey's test, where ∗p < 0.05 vs. CTR, ∗∗p < 0.01 vs. CTR, #p < 0.05 vs. CTR + NaKtide, ##p < 0.01 vs. CTR + NaKtide, &p < 0.05 vs. WD, &&p < 0.01 vs. WD.
Doxycycline induced adipocyte-specific NaKtide improves memory and markers of cognitive function in the hippocampus of mice fed a WD
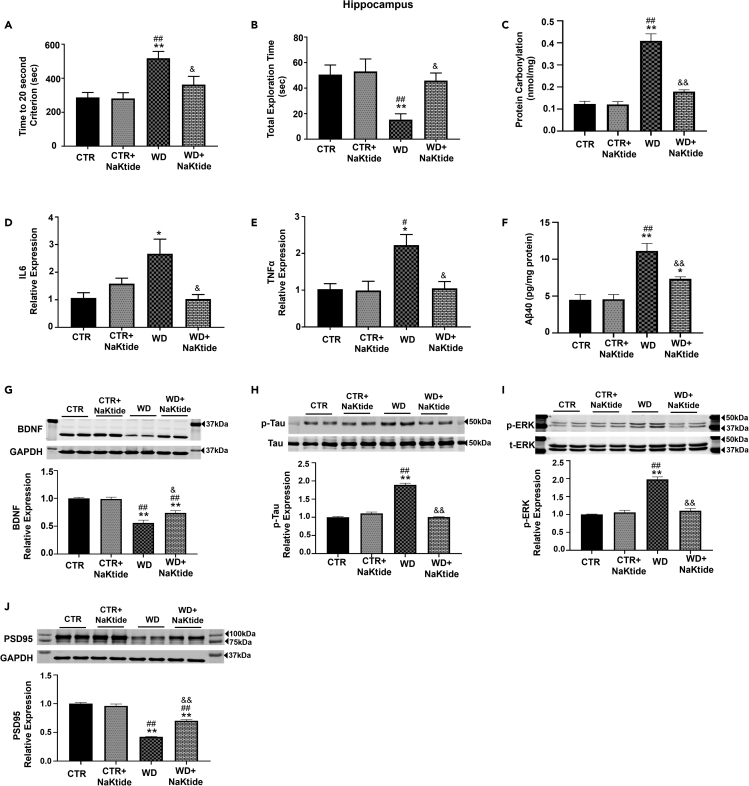
To determine the extent of cognitive decline in WD fed mice, we evaluated their learning and memory with several procedures. First, mice were subjected to an open field test, which demonstrated no significant differences among the experimental groups. The mice in all groups spent most of their time on the edge of the open arena followed by the intermediate zone with the least amount of time spent in the center zone of the open arena (data not shown). Using a novel object recognition test, mice fed a WD required significantly more time to reach the 20 s criterion, which was associated with significantly decreased exploratory times in these mice (Figures 3A and 3B). Doxycycline induced NaKtide expression reverted these changes. However, our results from novel object recognition test showed no significant change in the distance traveled by WD fed mice (7.34m ± 0.79) and WD fed mice with doxycycline induced NaKtide expression (7.45m ± 1.1), as compared to control mice fed normal chow (7.65m ± 0.93) and normal chow fed mice with doxycycline induced NaKtide expression (6.56m ± 1.54). Similarly, there was no significant change noted in the speed measurement of WD fed mice (0.024m/s ± 0.003) and WD fed mice with doxycycline induced NaKtide expression (0.025m/s ± 0.004), as compared to control mice fed normal chow (0.025m/s ± 0.003) and normal chow fed mice with doxycycline induced NaKtide expression (0.022m/s ± 0.005).
Figure 3.
Doxycycline induced adipocyte-specific NaKtide expression improves behavioral function, neuroinflammation and markers of cognitive function in the hippocampus of mice fed a WD
(A and B) Novel object recognition test analysis for (A) Time to 20 s criterion and (B) Total Exploration time.
(C–E) (C) Protein carbonylation assay in hippocampus homogenates. mRNA expression of (D) IL-6 and (E) TNFα.
(F) Plasma Aβ40 concentration. Immunoblot analysis for (A) BDNF (n = 6), (B) pTau (n = 4), (C) pERK (n = 6) and (D) PSD95 (n = 6). Results are expressed as means ± SEM. N = 4–14/group. Statistical analysis by one-way ANOVA, multiple comparison using Tukey's test, where ∗p < 0.05 vs. CTR, ∗∗p < 0.01 vs. CTR, #p < 0.05 vs. CTR + NaKtide, ##p < 0.01 vs. CTR + NaKtide, &p < 0.05 vs. WD, &&p < 0.01 vs. WD.
Protein carbonylation levels in the hippocampus were significantly increased in WD fed mice, which was attenuated by doxycycline induced NaKtide expression (Figure 3C). To assess Na/K-ATPase hydrolytic capacity in the hippocampus, we measured ouabain-sensitive Na/K-ATPase activity in crude membrane preparations obtained from mouse hippocampus. Our results showed that the maximal Na/K-ATPase activity in WD fed mouse hippocampus was unchanged (71.86 μmol Pi/h/mg protein ±1.87) compared with controls (77.85 μmol Pi/h/mg protein ±1.63). Doxycycline induce adipocyte specific NaKtide expression in WD fed mice (71.67 μmol Pi/h/mg protein ±1.63) or normal chow mice (71.33 μmol Pi/h/mg protein ±2.35) also did not show any significant change in the Na/K-ATPase activity, as compared to either control or WD alone. Next, we demonstrated changes in the isoforms of Na,K-ATPase in the hippocampus of these mice. Our results showed no significant changes in the protein expression of the α1, α2, and α3 subunits of Na,K-ATPase in the hippocampus (Figures S2A–S2C). Furthermore, our results showed significantly increased mRNA expression of inflammatory cytokines, IL-6, and TNFα, in the hippocampus of WD fed mice (Figures 3D and 3E), as compared to control. Doxycycline induced adipocyte specific NaKtide expression in WD fed mice significantly attenuated the expression of IL-6 and TNFα in the hippocampus (Figures 3D and 3E). Because microglial activation occurs in neuroinflammation, we performed immunohistochemical analysis of the hippocampal sections (Ito et al., 1998; Yanez Lopez et al., 2019; Zhang et al., 2021). Our results showed significant increase in the expression of ionized calcium-binding adaptor protein-1 (Iba1), a marker of microglial activation, in WD fed mice as compared to mice fed normal chow (Figure S3). Doxycycline induced adipocyte specific NaKtide expression in these WD fed mice significantly attenuated the Iba1-positive microglia in the hippocampus (Figure S3). These findings were consistent with the plasma levels of amyloid beta-40 (Aβ-40), a marker of cognitive decline (Jakel et al., 2020; Rosu et al., 2020; Tsai et al., 2021; Watt et al., 2020), which showed significant increases in the WD fed mice (Figure 3F). The increase in Aβ-40 levels was significantly attenuated by doxycycline induced NaKtide expression (Figure 3F). Brain derived neurotrophic factor (BDNF), a key marker of synaptic plasticity associated with learning and memory (Miranda et al., 2019), was decreased in mice fed a WD and restored by doxycycline induced NaKtide expression (Figure 3G). Mice fed a WD showed significant increases in phosphorylated Tau expression in the hippocampus, which is a marker of neuronal damage and its hyperphosphorylation, occurring at early stages of tauopathies, is correlated with cellular dysfunction (Bousiges et al., 2016; Duquette et al., 2020; Fossati et al., 2019; Kothari et al., 2017; Majd et al., 2017; Utrera et al., 2010) (Figure 3H). This was significantly attenuated by doxycycline induced NaKtide expression (Figure 3H). Our results also showed that the WD resulted in significant upregulation of phosphorylated ERK in the hippocampus, a marker of neuronal plasticity and neurodegeneration (Albert-Gasco et al., 2020) as well as downstream marker of Na,K-ATPase signaling; this was also attenuated by doxycycline induced NaKtide expression (Figure 3I). Finally, the WD also induced decreases in the expression of postsynaptic density 95 (PSD95), a marker of synaptic plasticity (Kothari et al., 2017), in the hippocampus of WD fed mice. Doxycycline induced NaKtide expression significantly upregulated the expression of PSD95 in WD fed mice (Figure 3J). Further, our results showed that doxycycline induced adipocyte specific NaKtide expression decreased protein carbonylation (Figure S4A), as well as restored expression of BDNF (Figure S4B), phosphorylated Tau (Figure S4C), phosphorylated ERK (Figure S4D) and PSD95 (Figure S4E) in the motor cortex of WD fed mice.
Doxycycline induced adipocyte-specific NaKtide improves cellular transcriptomic profile and biological pathways in the hippocampus of mice fed a WD
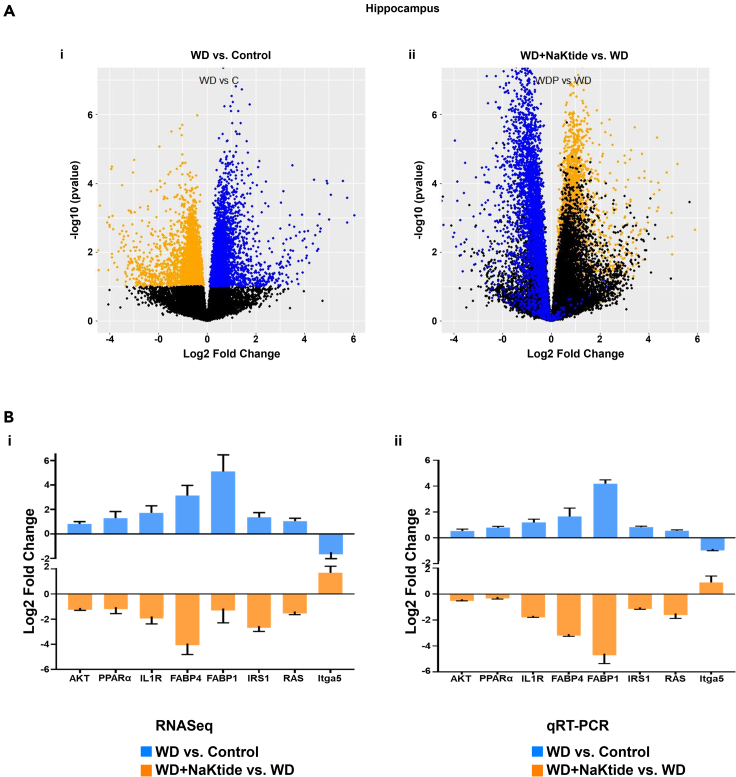
To characterize the effects of WD on brain transcriptome, RNA sequencing (RNAseq) was performed in the hippocampus of mice. WD induced marked changes in gene expression, all of which were largely reversed by doxycycline induced adipocyte specific NaKtide expression. These results are shown as volcano plots comparing mice fed WD and Control (left panel; Figure 4Ai) and comparing mice fed WD with doxycycline induced NaKtide vs. mice fed WD alone (right panel; Figure 4Aii). Very similar patterns were observed looking at the transcriptome of cerebellum, visceral and subcutaneous fat, and liver tissues (Figures S5A–S5D). The differential expression of some of the key genes from our transcriptomic analysis was validated by performing real time reverse transcriptase polymerase chain reaction (qRT-PCR) in the hippocampus. Specifically, the expression levels of genes including, AKT, peroxisome proliferator activated receptor alpha (PPARα), interleukin 1 receptor (IL1R), insulin receptor substrate 1 (IRS1), RAS, integrin alpha 5 (Itga5), fatty acid binding protein (FABP) −1 and 4, were evaluated in the hippocampus of mice fed WD with or without doxycycline induced NaKtide. Our results from qRT-PCR (Figure 4Bi) were in concordance with the findings from our transcriptomic analysis (Figure 4Bii) in the hippocampus, showing similar log2 fold change of these genes in WD fed mice as compared to control, which was reverted by doxycycline induced NaKtide.
Figure 4.
Doxycycline induced adipocyte-specific NaKtide expression improves cellular transcriptomic profile and biological pathways in the hippocampus of mice fed a WD
(A) Volcano plots of gene expression in WD vs. Control (left panel) and WD + NaKtide vs. WD (right panel) plotting antilog of unadjusted p value on y axis vs. log2 Fold Change on x axis. Genes downregulated (unadjusted p value of <0.1) by WD colored orange and genes upregulated (unadjusted p value of <0.1) by WD colored blue. We note that doxycycline induced NaKtide expression “moved” genes dysregulated with a high fat diet in the opposite direction.
(B) Validation of differential expression of selected genes from RNAseq analysis by qRT-PCR in the hippocampus. Genes validated by qRT-PCR includes akt3, ppara, il1r, fabp4, fabp1, irs1, rasa1 and itga5. Results expressed as log2 values of the fold change (±SEM) show similar up- or down regulation of these genes by qRT-PCR as compared to the gene expression by RNAseq in mice fed WD with or without doxycycline induced NaKtide. N = 6/group.
When we performed over-representation analysis (ORA) on the hippocampus mRNA with an over/under expression adjusted p value of 0.1, we found that there were 53 pathways which were differentially expressed with this methodology using a false discovery rate (FDR) cutoff of 0.1 (R package WebgestaltR using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and the mouse genome as reported previously (Luo and Brouwer, 2013; Wang et al., 2020). These pathways are summarized in Table S1. Notable pathways which were differentially expressed included those associated with the endocytosis, ubiquitin mediated proteolysis, dopaminergic synapse, mechanistic target of rapamycin (mTOR) signaling pathway, cyclic adenosine monophosphate (cAMP) signaling pathway, GABAergic synapse, and phospholipase D signaling pathway (all FDR<0.05).
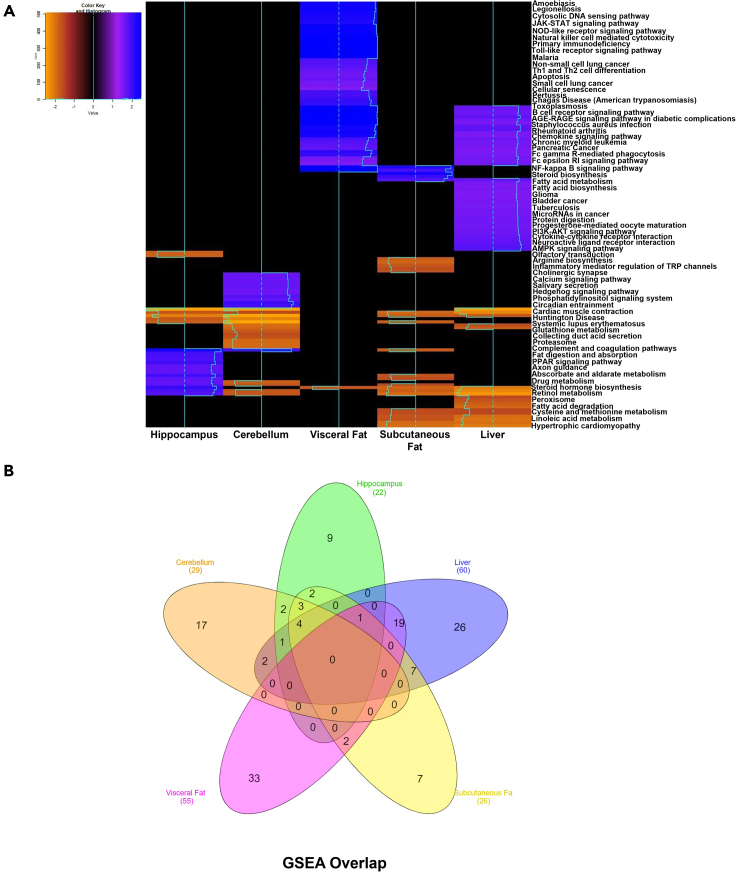
ORA performed on the cerebellum transcriptome yielded 11 pathways including pathways associated with Huntington disease, Parkinson’s disease, oxidative phosphorylation, thermogenesis and Alzheimer disease (all FDR <0.01). ORA performed on visceral fat yielded 93 pathways including those associated with the oxidative phosphorylation, parkinson’s disease, TNF signaling, NF-κB signaling, Alzheimer disease, Huntington disease, apoptosis, thermogenesis, and mitogen-activated protein kinases (MAPK) signaling (all FDR <0.05). ORA with subcutaneous fat resulted in 39 differentially expressed pathways including PPAR signaling, thermogenesis, oxidative phosphorylation, AMPK signaling, Parkinson’s, peroxisome, mTOR signaling and FoxO signaling (all FDR <0.05). Liver ORA demonstrated 59 differentially expressed pathways including PPAR signaling, peroxisome, glutathione metabolism, AMPK signaling, and thermogenesis, (all FDR <0.05). Overlap between these differentially expressed pathways detected with ORA is summarized in a Venn diagram (Figure S6A). Notably, the differential expression of pathway for mitochondrial thermogenesis was common among all 5 tissues.
Gene set enrichment analysis (GSEA) can also be used as an alternative method to examine differential gene expression. Unlike ORA which uses thresholds to identify differentially expressed genes, GSEA ranks the whole gene set and compares this ranking to random sorting (Subramanian et al., 2005). Although the pathways identified with GSEA often overlap with those detected by ORA, it is considered to be a complementary methodology which advantageously allows detection of both over- and under-expression of pathways (Liu and Ruan, 2013) using the normalized enrichment score (NES) which can be compared between gene sets. Heatmaps comparing the NES of significantly over- and under-expressed pathways among all tissues is shown in Figure 5A. Using GSEA, 22 pathways were noted to be over- or under-expressed in hippocampus (Figure S6B), including those associated with the ribosome, oxidative phosphorylation, PPAR signaling, transforming growth factor β (TGFβ) signaling and Parkinson' disease (all FDR <0.05). A GSEA rank plot was generated for the oxidative phosphorylation, Parkinson’s disease, Alzheimer’s disease pathway, and one wiki pathway of oxidative stress in the hippocampus (not shown). Specifically, we found differential gene expression in the hippocampus of WD fed mice as compared to control, for various genes involved in oxidative phosphorylation (Figure S7Ai), oxidative stress pathway (Figure S7Bi), Parkinson’s disease (Figure S8Ai), and Alzheimer’s disease (Figure S8Bi). Remarkably, most of the genes differentially expressed among these pathways were reversed with doxycycline induced NaKtide expression (Figures S7Aii, S7Bii, S8Aii, and S8Bii).
Figure 5.
Enriched biological pathways, using GSEA, among tissues in mice fed a WD
(A) Heatmap showing upregulation or downregulation of pathways enriched by GSEA, in each tissue of mice fed a WD.
(B) Venn diagram from pathway enrichment analysis performed using GSEA, depicting the overlap of all enriched pathways among cerebellum, hippocampus, liver, subcutaneous and visceral adipose tissue. N = 6/group.
GSEA performed on gene expression from cerebellum yielded 29 differentially expressed pathways including pathways associated with oxidative phosphorylation, Parkinson’s disease, ribosome, proteasome, thermogenesis, and Alzheimer disease (all FDR <0.05) (Figure S6C). Subcutaneous fat GSEA revealed 26 pathways including those associated with NF-κB, oxidative phosphorylation and Parkinson’s disease (all FDR<0.05) (Figure S6D). GSEA performed on visceral fat showed 55 pathways including those associated with NF-κB, TNF, JAK-STAT, apoptosis, and cellular senescence signaling (all FDR <0.05) (Figure S6E). GSEA performed on liver tissue mRNA showed 60 pathways including those associated with ribosome, Alzheimer’s disease, oxidative phosphorylation, and peroxisome (all FDR <0.05) (Figure S6F). Heatmaps comparing the NES of differentially regulated pathways among tissues in WD fed mice showed that doxycycline induced adipocyte specific NaKtide expression mostly reverted these changes (Figures S6B–S6F). The overlap of GSEA pathways among all tissues is shown with a Venn diagram (Figure 5B).
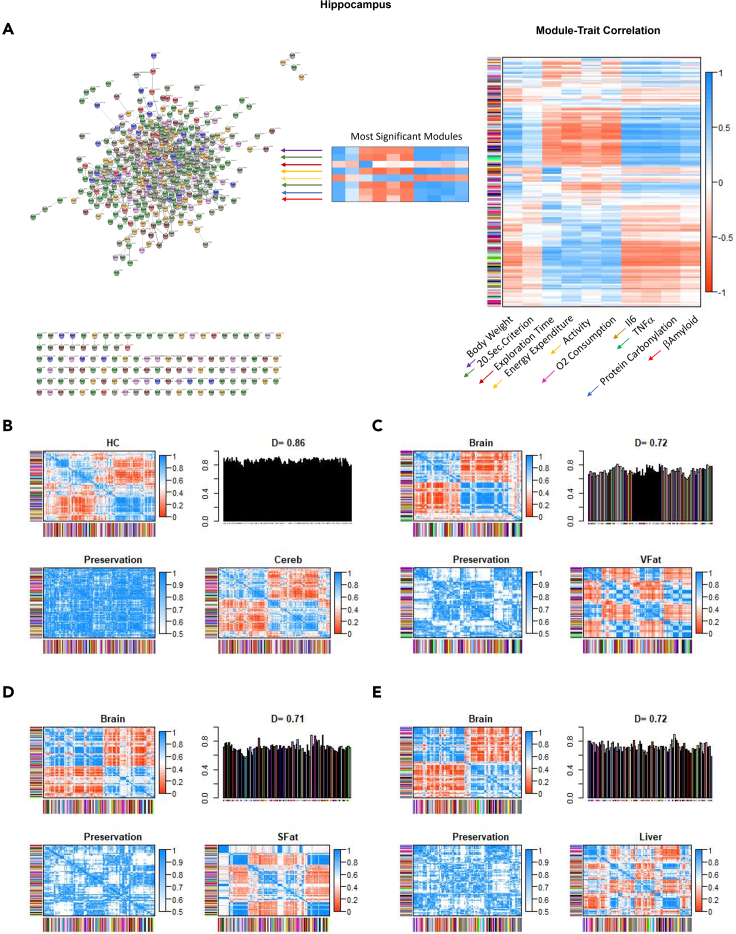
We next employed weighted gene correlation network analysis (WGCNA) to better understand the topology of gene expression with the distance metric based on the Pearson correlation coefficient (Langfelder and Horvath, 2008). Using this approach, we first confirmed that the phenotypical features indeed corresponded to the experimental groups, and that topological features of the gene expression network required a soft threshold of approximately 14 for optimal separation (data not shown). We next identified modules that had different degrees of correlation with 10 phenotypical features with a dendrogram and module assignments. Specifically, the groups of genes that exhibited the highest absolute correlation with the selected phenotypic features (body weight, time to 20 s criterion, exploration time, locomotion, energy expenditure, oxygen consumption, plasma IL-6, plasma TNFα, protein carbonylation, and Aβ40) were then mapped using protein-protein interactions (STRING database (Szklarczyk et al., 2017)) (Figure 6A), and heat maps detailing selected gene expression in these different groups were produced. The heatmaps for each color module which correlated best with one or more phenotypic features showed that the doxycycline induced NaKtide expression reversed most of the differential gene expression noted in the hippocampus that was induced by the WD (data not shown). We next applied gene sequence expression analysis to these modules again by using the KEGG pathways. Although each module had some association with a number of KEGG pathways with this approach, the FDR values were not significant except for the dark olive green module (best correlation with TNFα levels) which had an FDR value <0.05 for the Ras signaling pathway (other ORA data not shown). Perhaps of greater interest, we note on Figure 6A that a number of genes which were identified as members of these modules did not have known connections through the STRING protein-protein database.
Figure 6.
Differential eigengene network analysis and module-trait correlation of datasets from tissues of mice
(A–E)(A) Module-trait correlations. Each row in the table (right panel) corresponds to different gene groupings, and each column to selected phenotypical features. Based on the highest correlations with these phenotypic features (body weight, time to 20 s criterion, exploration time, locomotion, energy expenditure, oxygen consumption, plasma IL-6, plasma TNFα, protein carbonylation, and Aβ40). Modules which correlated best with the 10 phenotypic traits (there were 7 such modules as some correlated best with more than one trait) were placed into a cytoscape network plot using the STRING protein-protein interaction dataset (left panel). We note that although most of these module genes are assigned to such interactions in this dataset, a significant minority were not. Summary plot of consensus eigengene networks and their differential analysis from (B) hippocampus and cerebellum datasets, (C) brain and visceral fat datasets, (D) brain and subcutaneous fat datasets, (E) brain and liver datasets. Heat maps show high (red) and low (or negative, green) adjacency. Preservation heatmap is the 1-absolute difference of the eigengene networks in the two sets. Bar plot shows mean preservation of adjacency for each eigengene to other eigengenes with a D value calculated as the arithmetic mean of these measurements. N = 6/group.
When we performed comparisons of the gene expression networks in the different tissues using WGCNA, we noted extremely strong similarities between networks produced by the gene expression patterns of the different tissues studied. Specifically, comparison of the eigengene expressions in hippocampus and cerebellum yielded high similarity (D = 0.86; Figure 6B). When we combined the hippocampus and cerebellum under the category brain, we also noted high similarity with eigengene expression compared with visceral fat (D = 0.72, Figure 6C), subcutaneous fat (D = 0.71; Figure 6D) and liver (D = 0.72, Figure 6E). We also noted high similarity between networks produced by visceral and subcutaneous fat (D = 0.77, data not shown). Ergo, although different tissues had differences in differential gene and network expressions induced by the experimental maneuvers employed, there were even more notable similarities identified by the WGCNA strategy.
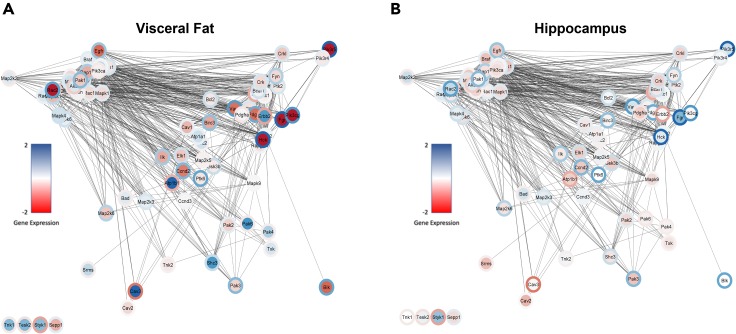
Our results from RNASeq analysis showed that genes, associated with Na,K-ATPase signaling, in mouse visceral fat and hippocampus demonstrated differences in expression with a WD compared to control. These differences were opposed or reversed by concomitant doxycycline induced NaKtide expression in the adipocytes (Figures 7A and 7B).
Figure 7.
Heatmap for protein-protein interaction of genes associated with the activation of Na,K-ATPase signaling
(A and B)WD gene expression shown as a heatmap in (A) visceral fat and (B) hippocampus. Protein interaction strength derived from the STRING database is shown as line thickness and inverse length using Cytoscape. Colors of WD vs Control (core) and WD + NaKtide vs WD (rim) range from log2FC of −2 to +2. Colors of the core and rim are reciprocal, indicating that NaKtide expression opposed or reversed the WD diet changes. N = 6/group.
Discussion
We previously found that oxidative stress within adipocytes attributable to the NKAL plays a key role in diet-induced obesity, neurodegeneration, and uremic cardiomyopathy, suggesting that NKAL may be a common factor behind these obesity-induced changes (Pratt et al., 2019; Sodhi et al., 2017, 2020b). In this study, we demonstrated a central role of the adipocyte NKAL in WD induced alterations in specific regions of the brain, which to the best of our knowledge has never been explored, most notably in the hippocampus, which is critical to memory (Dere et al., 2005). We also performed behavioral tests to assess the functional implications of the morphological and biochemical alterations. Studies in mice models of AD have shown that the novel object recognition test can evaluate an association between memory processing and hippocampal synaptic efficacy (Clarke et al., 2010; Taglialatela et al., 2009). Our findings showing reduced exploration time and increased time to criterion in WD-fed mice are in concordance with these findings. Our findings that doxycycline-induced adipocyte-specific NaKtide expression reversed these changes as well as reversed the decline in cognitive memory and restored BDNF, PSD95, and markers of neurodegenerative changes in hippocampus of WD fed mice indicate involvement of NKAL. Our findings are consistent with other reports of metabolic dysfunction (Bittencourt et al., 2020) and obesity-associated cognitive decline (Cope et al., 2018) producing Alzheimer disease-like pathology (Kacirova et al., 2020).
The transcriptional changes in the hippocampus demonstrated that a WD induced significant effects on genes specific for neurodegenerative disorders and synaptic signaling. Loss of neuronal function and synapses in the hippocampus, a characteristic of neurodegenerative disorders (Canchi et al., 2019), strongly correlates with cognitive impairment and high levels of Aβ production (Sheng et al., 2012). Other investigators have found evidence of the involvement of many of these pathways, including pathways for axon guidance (Lee et al., 2019) and histidine metabolism (Hunsberger et al., 2020; Smith et al., 2010). Neuroinflammation contributes to increased oxidative stress, phosphorylation of tau, reduced Aβ clearance, synaptic degeneration, and neuronal death (Chen et al., 2016; Kempuraj et al., 2017). Owing to the contribution of mitochondria in producing oxidant stress, apoptosis, and inflammation, impaired mitochondrial function may also be an important factor in neurodegeneration (Hroudova et al., 2014). Our RNA-seq results support previous suggestions that changes in oxidative phosphorylation (Federico et al., 2012), the PPAR signaling pathway (Jamwal et al., 2020; Wojtowicz et al., 2020) and the TGF-β signaling pathway (Tesseur and Wyss-Coray, 2006) may contribute to neurodegenerative disorders such as AD. The doxycycline-mediated expression of NaKtide in adipocytes antagonized all of these changes, indicating a possible contribution of obesity to mitochondrial dysfunction in the brain.
In the cerebellum, pathways associated with loss of neuronal function and synapses were also dysregulated, including those associated with glutamatergic synapses (Lewerenz and Maher, 2015), cholinergic synapses (Ferreira-Vieira et al., 2016), and calcium signaling (Ramanan et al., 2012). Pathways associated with mitochondrial function were also dysregulated in cerebellum, including oxidative phosphorylation, thermogenesis (Yang and Ruan, 2015), and glutathione metabolism (Gegg et al., 2003). Our results showed that adipocyte specific NaKtide expression reversed most of these transcriptomic changes. The similarity of transcriptomic changes in cerebellum to those in the hippocampus raises the question why obesity more strongly affects hippocampal function rather than cerebellar functions such as balance or motor learning. One possibility is that the hippocampus is more sensitive to mitochondrial and synaptic changes induced by a WD (Jacka et al., 2015). Further research will be required to answer this question.
Our findings clearly suggest a central role for the peripheral adipocytes in the pathogenesis of the WD induced changes in brain function and biochemistry. It is well-known that WD promotes excessive energy intake and body weight gain inducing inflammation in murine models. However, recent studies suggest that WD can lead to alteration in hippocampal dependent spatial learning and memory function, further causing impaired glucoregulation, reduced level of neurotrophins, neuroinflammation and alterations in the structural integrity of BBB (Kanoski and Davidson, 2011). WD-fed animals express impaired glucose tolerance and energy homeostasis, reduced locomotion, and increases in circulating inflammatory cytokines. Work by our group and others (Le Lay et al., 2014; Pratt et al., 2019; Sodhi et al., 2015) ascribes these changes to oxidant stress within adipocytes (Manna and Jain, 2015; Sodhi et al., 2020a). In the current study, the WD severely altered the transcriptomic profile of important pathways in both visceral and subcutaneous adipose tissues, most notably those associated with oxidative damage, inflammation, oxidative phosphorylation, and mitochondrial dysfunction. In addition, the NF-κB, TNF, IL-17, JAK-STAT, chemokine, and toll-like receptor signaling pathways were also dysregulated. Oxidative stress and subsequent release of inflammatory cytokines associated with these pathways in adipocytes can alter neuronal and synaptic plasticity (Cacci et al., 2005; Monje et al., 2003), impair learning and memory processes (Butler et al., 2004), and inhibit neurogenesis by inducing neuroinflammation (Parimisetty et al., 2016; Qin et al., 2016). As excessive fat accumulation and adipocyte dysfunction in response to a WD can also induce severe hepatic lipid accumulation and inflammation (Geisler and Renquist, 2017), it is not surprising that we observed a host of transcriptomic changes in liver tissue as well as adipose tissue. It is certainly possible that altered hepatic functions, either in the production or clearance of toxins and neuroactive molecules, could play a role in the neurodegenerative phenotype produced by the WD (Estrada et al., 2019; Geisler and Renquist, 2017; Nho et al., 2019; Pacholko et al., 2019). However, the large-scale improvement observed in specific regions of the brain as well as other tissues after targeting NKAL suggests that these transcriptomic changes in the liver are secondary to alterations in peripheral adipocytes.
Adipose tissue dysfunction can dramatically influence brain function and biochemistry, leading to neurodegeneration (Letra et al., 2014; Parimisetty et al., 2016). Our data demonstrate that the adipocyte NKAL is central to these changes. Although the exact contribution of adipocytes and cells regulated by adipocytes (e.g., macrophages) is not yet clear, it is possible that many, if not most, of the observed changes in brain function and metabolism could be because of the neurotoxic effects of inflammatory cytokines (Butler et al., 2004; Cacci et al., 2005; Monje et al., 2003; Pickering et al., 2005; Selmaj and Raine, 1988), which can be transported across the BBB (Letra et al., 2014; Parimisetty et al., 2016). In our study, both IL-6 and TNFα plasma levels were substantially elevated by the WD and normalized by adipocyte-specific NaKtide expression. Other factors related to the adipocyte redox state could also be involved. Studies have shown that high fat diet increases the production of ceramides, which can also enter circulation and cross BBB, similar to cytokines, and induce brain insulin resistance, oxidative stress, and inflammation, leading to neurodegeneration (Holland et al., 2011; Sripetchwandee et al., 2018). These stress related agonists can contribute to pro-inflammatory phenotype by directly activating toll-like receptor 4 (TLR4) and NF-κB signaling pathways in the brain (Holland et al., 2011; Valdearcos et al., 2014). The present study and evidence from the literature supports that WD may alter peripheral immune profile; however, it is unclear whether inhibition of these peripheral immune responses would completely inhibit neurodegenerative changes in the brain (Baumgarner et al., 2014).
In summary, our study demonstrates that diet induced stimulation of the adipocyte NKAL leads to the development of a neurodegenerative phenotype which is ameliorated by cell-specific inhibition of oxidant production by the NKAL with NaKtide. It is clear that peripheral adipocytes play a central role in WD induced brain dysfunction. Our results suggest that strategies aimed at targeting the adipocyte NKAL may potentially have clinical utility in treating or preventing neurodegeneration.
Limitations of the study
Our study has certain limitations which should be understood by the reader. First and foremost, we have seen a number of systemic effects derived from adipocyte specific NaKtide expression which may be responsible for some (or many) of the behavioral and brain biochemical changes in the WD setting. Simply ameliorating obesity might have a substantial effect, however this might be achieved. Second, but also important, we infer that the NaKtide expression specifically involves the aforementioned NKAL. While we have published data supporting this concept (Lai et al., 2013; Li et al., 2009; Pratt et al., 2019; Sodhi et al., 2020a, 2020b), it is possible that this peptide interacts with other cellular targets yet unidentified. Finally, while we infer that the improvement in adipocyte redox state achieved by NaKtide expression results in attenuation of inflammation and systemic oxidant stress, it is not yet clear how this occurs. Future work will hopefully clarify these important limitations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal BDNF antibody | Cell Signaling Technology | Cat# 47808S: RRID:AB_2894709 |
| Mouse monoclonal Anti-Glyceraldehyde-3-Phosphate Dehydrogenase Antibody, clone 6C5 | Millipore Sigma | Cat# MAB374; RRID:AB_2107445 |
| Rabbit monoclonal PSD95 antibody | Cell Signaling Technology | Cat# 3450S: RRID:AB_2292883 |
| Rabbit monoclonal Phospho-Tau (Ser404) (D2Z4G) Antibody | Cell Signaling Technology | Cat# 20194S; RRID:AB_2798837 |
| Mouse monoclonal Tau (Tau46) Antibody | Cell Signaling Technology | Cat# 4019S; RRID:AB_10695394 |
| Rabbit polyclonal phospho-SRC (Tyr419) Antibody | Thermo Fisher Scientific | Cat# 44-660G; RRID:AB_2533714 |
| Mouse monoclonal Anti-c-Src/Src Antibody (B-12) | Santa Cruz Biotechnology Inc. | Cat# sc-8056; RRID:AB_627306 |
| Rabbit polyclonal NaKtide Primary Antibody | Covance Inc. | |
| Goat Anti-Rabbit IgG H&L (Alexa Fluor 647) | Abcam | Cat# Ab150079; RRID:AB_2722623 |
| Goat Anti-Rabbit IgG H&L (Alexa Fluor 488) | Abcam | Cat# Ab150077; RRID:AB_2630356 |
| Goat Anti-rabbit IgG H&L HRP | Invitrogen | Cat# 31460; RRID:AB_228341 |
| Mouse monoclonal Anti-Na+/K+ ATPase a-1 Antibody, clone C464.6 | Millipore Sigma | Cat# 05-369; RRID:AB_309699 |
| Rabbit polyclonal Anti-Na+K+ ATPase α-2 Antibody | Millipore Sigma | Cat# 07-674; RRID:AB_390164 |
| Rabbit polyclonal Anti-Na+/K+ ATPase alpha-3 antibody | Millipore Sigma | Cat# 06-172-I; RRID:AB_310066 |
| Rabbit monoclonal Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) Antibody | Cell Signaling Technology | Cat# 4370; RRID:AB_2315112 |
| Rabbit polyclonal p44/42 MAPK (Erk1/2) Antibody | Cell Signaling Technology | Cat# 9102; RRID:AB_330744 |
| Chemicals, peptides, and recombinant proteins | ||
| VECTASHIELD Antifade Mounting Medium With DAPI | Vector Laboratories | Cat# H1200; RRID:AB_2336790 |
| 1 Step Ultra TMB-ELISA Substrate Solution | ThermoFisher Scientific | Cat# 34028 |
| BIOMOL Green | Enzo Life Science | Cat# BML-AK111-0250 |
| Critical commercial assays | ||
| Amyloid beta 40 mouse ELISA Kit | ThermoFisher Scientific | Cat# KMB3481 |
| Mouse IL-6 ELISA Kit | Abcam | Cat# Ab100712 |
| Mouse TNFa ELISA Kit | Abcam | Cat# Ab46105 |
| Protein Carbonyl ELISA Assay Kit | BioCell Corporation | Cat# BPCK01 |
| RNeasy Protect Mini Kit | Qiagen | Cat# 74124 |
| RevertAid First Strand cDNA Synthesis Kit | ThermoFisher Scientific | Cat# K1621 |
| Deposited data | ||
| Gene Expression Omnibus Archive | NCBI | GSE164295 |
| Experimental models: Organisms/strains | ||
| Transgenic Mice – Tet-On/off dependent adipocyte specific NaKtide (C57BL6 background) | Cyagen Biosciences Inc. | TGMB-161024-AAJ |
| Software and algorithms | ||
| R 4.0.3 | R Core Team, 2016 | https://cran.r-project.org/ |
| RStudio Package: gplots | Warnes et al., 2020 | https://cran.r-project.org/web/packages/gplots/index.html |
| RStudio Package: WebgestaltR | Wang et al., 2013 | https://cran.r-project.org/web/packages/WebGestaltR/index.html |
| RStudio Package: pathview | Luo and Brouwer, 2013 | https://doi.org/10.1093/bioinformatics/btt285 |
| RStudio Package: DESeq2 | Love et al., 2014 | https://doi.org/10.1186/s13059-014-0550-8 |
| RStudio Package: WGCNA | Langfelder and Horvath, 2008, 2012 |
https://doi.org/10.1186/1471-2105-9-559; https://pubmed.ncbi.nlm.nih.gov/23050260/ |
| RStudio Package: fgsea | Sergushichev, 2016 | https://doi.org/10.1101/060012 |
| Cytoscape: Wikipathways App | Kutmon et al., 2014 | https://doi.org/10.12688/f1000research.4254.2 |
| ANY-Maze Behavioral Tracking Software | AnyMaze | N/A |
| Prism 8.0 | Graphpad | N/A |
| Comprehensive Lab Animal Monitoring System | CLAMS | N/A |
| ImageJ | National Institute of Health | N/A |
| Other | ||
| Normal mouse chow diet | LabDiet | Cat# 5001 |
| Western Diet | ENVIGO | Cat# TD.88137 |
| Normal Chow Diet + 300 Doxycycline | ENVIGO | Cat# TD.180829 |
| Western Diet + 300 Doxycycline | ENVIGO | Cat# TD.180830 |
| Glucose Test Strips | One Touch Ultra | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Joseph I Shapiro (shapiroj@marshall.edu).
Materials availability
-
•
Transgenic mice used in this study were generated by Cyagen Biosciences Inc (Santa Clara, CA).
-
•
This study did not generate any new unique reagents.
Experimental model and subject details
All animal studies were approved by the Marshall University Animal Care and Use Committee in accordance with the National Institutes of Health (NIH) Guide for Care and Use of Laboratory Animals. Transgenic mice (C57BL6 background) expressing NaKtide in a tetracycline (TET) dependent manner (Tet-On), specifically in adipocytes, under the control of adiponectin promoter were generated utilizing a custom plasmid vector generated by Cyagen Biosciences Inc (Santa Clara, CA). Briefly, a plasmid vector was constructed utilizing the TET response element (TRE) to drive expression of an open reading frame (ORF) encoding NaKtide. TET-suppressor and TET-activator ORFs were included under the control of the mouse adiponectin promoter, resulting in TET-dependent expression of NaKtide exclusively within adipocytes. Following confirmation of sequence integrity and orientation via restriction digestion and sequencing, the plasmid was provided to Cyagen Biosciences for pro-nuclear injection. Following plasmid purification and linearization of the plasmid, pro-nuclear injection was performed. Positive clones were identified by polymerase chain reaction (PCR) based genotyping, using a primer specific for the adiponectin promoter and a primer specific for NaKtide. We generated adipocyte-specific NaKtide transgenic founders that had undergone breeding to generate offspring of these mice and then they were subjected to PCR and Southern blot analysis with genomic DNA prepared from tail biopsies to check the transgene expression. All mice were placed in cages at the Robert C. Byrd Biotechnology Science Center Animal Resource Facility (ARF).
Since these mice used TET dependent regulatory on/off system, the ingestion of doxycycline automatically activated the adiponectin promoter and expressed NaKtide specifically in adipocytes without substantial off target effects in these mice (Cawthorne et al., 2007; Das et al., 2016; Gengenbacher et al., 2020; Ingusci et al., 2019; Mansuy and Bujard, 2000; Redelsperger et al., 2016). Doxycycline is preferable to tetracycline as an inducer in tetracycline (tet)-dependent regulatory systems due to doxycycline's high potency, superior tissue penetration, and its widespread availability (Cawthorne et al., 2007; Redelsperger et al., 2016). In addition to that, doxycycline in mouse chow offers advantages such as improved doxycycline stability without any signs of animal discomfort.
All experimental male mice were 10-12 weeks old and were housed in a pathogen-free animal facility in designated rooms equipped with cages that supplied purified air under a 12-hour light/dark cycle. Mice were placed on a normal chow diet containing 11% fat, 62% carbohydrate, and 27.0% protein with total calories of 12.6 KJ/g and had free access to water or the mice were placed on WD containing 42% fat, 42.7 % carbohydrate, and 15.2% protein yielding 4.5 KJ/g and had free access to water (Pratt et al., 2019). To induce NaKtide expression in these mice, custom normal chow or WD (Envigo RMS, Inc.-Teklad Diets) with doxycycline, at a concentration of 300 mg/kg (Cawthorne et al., 2007; Redelsperger et al., 2016), was used (Normal chow with doxycycline: TD.180829; WD with Doxycycline: TD.180830). Animals were randomly divided into four primary groups (n=8-10/group) as follows: 1. Control (normal chow); 2. Control+NaKtide (Normal chow+Doxycycline); 3. WD (WD) and 4. WD+NaKtide (WD+Doxycycline). All mice groups were placed on their respective diets on Day 0 for a total of 12 weeks. Body weights were measured weekly over the course of 12 weeks. At the time of euthanization, weights of total body, visceral fat, subcutaneous fat, heart, liver and kidneys were determined. Blood samples were collected for the determination of plasma inflammatory cytokines and Aβ-40. Whole brain was individually dissected to obtain hippocampus, motor cortex and cerebellum. All brain parts and other tissues were flash-frozen in liquid nitrogen and maintained at -80οC, preserved in optimal cutting temperature (OCT) compound for sectioning or placed in paraformaldehyde for paraffin embedding.
Method details
Indirect calorimetry and locomotor activity
At the end of 12-week period prior to euthanization, mice underwent metabolic cages for the measurement of energy expenditure, oxygen consumption and locomotor activity using the Comprehensive Lab Animal Monitoring System (CLAMS) for the simultaneous assessment of these metabolic parameters as described previously (Sodhi et al., 2017).
Glucose tolerance test
At the end of 12-week period, mice were fasted for 8 hours. After the fasting period, a 10% glucose solution (2g/kg body weight) was injected intraperitoneally. Samples were taken from the tail vein at 0, 30, 60, 90 and 120 minutes after glucose injection. Blood Glucose was measured using glucometer as reported previously (Sodhi et al., 2017).
Measurement of plasma cytokines, Aβ-40 and protein carbonylation
Plasma IL-6 and TNFα were measured using an Enzyme Linked Immunosorbent Assay (ELISA) kit according to manufacturer’s protocol (Abcam, Cambridge, MA), as reported previously (Sodhi et al., 2017). Plasma Aβ-40 was also measure using ELISA Kit according to manufacturer’s protocol (Thermo Fisher Scientific, Waltham, MA). Whole cell lysates from visceral adipose tissue, hippocampus and motor cortex were prepared with RIPA buffer and protein carbonylation was measured using Protein Carbonyl ELISA Kit (BioCell Corporation, Auckland, New Zealand), according to manufacturer’s protocol (Sodhi et al., 2020b).
Measurement of NaKtide concentration in visceral fat by Competitive ELISA
To confirm the presence of NaKtide in the mice fed Doxycycline added normal chow or WD, visceral adipose tissues were homogenized in RIPA buffer and protein quantification was performed normalized to the final concentration of 1mg/ml. We also performed deproteinization with perchloric acid, followed by neutralization with 1M NaOH and extraction with chloroform. Polystyrene 96-well microtiter plates were coated with 100uL/well of NaKtide antigen in 1% DMSO at a concentration of 50ng/mL and the plates were incubated overnight at 4οC. ELISA was performed on the visceral adipose tissue homogenates using primary NaKtide antibody, as described previously (Sodhi et al., 2020b).
Immunofluorescence studies in adipose tissues
Adipose tissue and heart tissues were frozen in OCT compound, cut into 6μm sections and mounted on slides. The sections were fixed with 4% PFA for 15 minutes, washed once with TBS, probed with 1:50 primary NaKtide antibody dilution and 1:1000 secondary antibody (Alexa Fluor 647 Red), and then mounted with DAPI solution and coverslips as described previously (Pratt et al., 2019; Sodhi et al., 2020b). Expression of GFP and NaKtide was determined using a GFP and RFP filter respectively on a Nikon Eclipse 80i microscope equipped with a Nikon camera head DS-Fi1 (Nikon, Japan).
Open field maze and novel object recognition
Open field test was performed in the mice to assess exploratory behavior, anxiety, general activity level and gross locomotor activity, as described previously (Seibenhener and Wooten, 2015). Prior to this behavioral assessment, mice were habituated to the experimental room for 1 hour prior to testing. Following 1 hour of habituation, mice were individually placed in a square plexiglass box. The mice were allowed to freely move in an empty arena for about 10 minutes while being recorded by an overhead camera. The sensorimotor overhead camera is connected to a video recording software (ANY-maze) and the lenses are adjusted to fit the four corners of the square plexiglass box. The automatic tracking system in the software recorded the footage of the mice movement. Hence, following 10 minutes of free movement of the mice, the footage was analyzed for the following parameters: distance moved, velocity and time spent in the predefined zones (edge, intermediate and center). This test also serves as a habituation period to the experimental room for the novel object recognition test the following day.
The novel object recognition test was performed to evaluate cognition, particularly recognition memory, as described previously (Lueptow, 2017). Briefly, mice were habituated to the experimental room for 1 hour prior to testing. Following habituation, mice underwent training where mice were exposed to two identical objects, in the square plexiglass box (used in open field maze above). The two identical objects were placed at an equal distance of approximately 10 cm apart in the activity cage, with normal bedding. Mice were placed in the cage with the objects. Video Recording Software (ANY-maze) was used to record each instance of the mouse investigating each object. Two hours following the training, one of the objects was replaced with a new object and the mice were allowed to explore the open field in the presence of the familiar object and a novel object to test recognition memory. The video recording software was used to record each instance of mice investigating either the novel object or the familiar object. A day without testing occurred between each test. The footage was analyzed to assess the time spent exploring each object and the discrimination index percentage, which allows for the determination of impaired cognitive ability.
Ouabain-sensitive ATPase (Na/K-ATPase) in mouse hippocampus
The crude membrane preparation and Na/K-ATPase activity was assayed according to the protocol previously described (Gamaro et al., 2003; Liang et al., 2006; Xie et al., 1989) with modification. Mouse hippocampus (8-12mg) were homogenized in (1ml) ice-cold Skou C buffer (30mM histidine, 250mM sucrose, 1mM EDTA, pH 7.4) and briefly sonicated. After centrifugation (800 x g for 10 min), the supernatants were further centrifuged (45,000 X g for 45 min) with BECKMAN TL-100 Ultracentrifuge and BECKMAN TLA-45 rotors. The membrane-enriched pellets (crude membrane preparation) were resuspended in Skou C buffer. To allow access of substrates and inhibitor to their respective binding sites on the catalytic subunit of NKA in closed membrane vesicles, the crude preparations were treated with alamethicin (0.3 mg/mg of protein, which was the optimal concentration revealed by a calibration curve in the conditions of this assay) for 10 min at room temperature. The preparation was then incubated in a buffer containing 20mM Tris (pH 7.2), 1mM EGTA, 1mM MgCl2, 20mM KCl, 5mM NaN3, and 100mM NaCl. After 15min of preincubation at 37°C, ATP/Mg2+ was added to a final concentration of 2mM to start the reaction. The reaction was stopped after 15 min. by addition of 8% ice-cold trichloroacetic acid. The inorganic phosphate generated during the ATP hydrolysis was measured by a colorimetric assay using the BIOMOL GREEN Reagent (Enzo Life Science, NY, USA) and a microplate reader (SpectraMax M3). Ouabain-sensitive Na/K-ATPase activities were calculated as the difference between inorganic phosphate released in the absence and in the presence of ouabain 1mM.
Immunohistochemistry in hippocampus tissue
Immunohistochemistry experiment was performed by Wax-it Histology Services Inc. Canada. Briefly, brain was fixed overnight in 4% paraformaldehyde solution at 4°C. After paraffin embedding, the materials were sectioned at 25 μm using cryostat microtome throughout the hippocampus area. Following melting and dewaxing, the antigen retrieval was performed in a steamer at pH 6. After washing and blocking of endogenous peroxidase using Waxit Inc proprietary blocking solution, the slides were incubated overnight at 4°C with primary antibody for the microglial cell marker Iba1. Secondary antibody treatment and detection was performed using Polymer HRP detection system followed by incubation with DAB (3,3′-Diaminobenzidine) until desired staining is achieved. After counter staining and proper rinsing with water, the stained slides were imaged with Aperio AT2 whole slide imaging system at 20X. Using Positive Pixel Count V9 algorithm (Leica Imagescope software), positive stained pixels for Iba-1 were counted based on staining intensities and total pixels in the selected area of the image. The normalized values of positivity were calculated and represented by total positive pixels/total pixels.
Western blot analysis
Hippocampus, motor cortex, and visceral adipose tissue were pulverized with liquid nitrogen and placed in RIPA homogenization buffer. Homogenates were centrifuged, the supernatant was isolated, and immunoblotting performed (Pratt et al., 2019). Hippocampus and motor cortex were used for expression of BDNF, phosphorylated Tau, phosphorylated ERK and PSD95. All antibodies were obtained from Cell Signaling (Danvers, MA). Activation of c-Src was determined in visceral adipose tissue as described previously (Pratt et al., 2019; Yan et al., 2013). Polyclonal anti-Src [pY419] phospho-Src specific antibody was from Thermo Fisher Scientific (Waltham, MA). Monoclonal antibody against total c-Src was from Santa Cruz (Santa Cruz, CA). After immunoblotting for phospho-Src, the same membrane was stripped and blotted for total c-Src. Activation of c-Src was expressed as the ratio of phospho-Src/c-Src with measurements normalized to 1 for control samples. Immunoblot analysis was also performed for expression of Na/K-ATPase α1 subunit, α2 subunit and α3 subunit in hippocampus using mouse monoclonal anti-Na+/K+ ATPase α1 antibody (Sigma Aldrich, MO), rabbit polyclonal anti-Na+/K+ ATPase α2 antibody (Sigma Aldrich, MO) and rabbit polyclonal anti-Na+/K+ ATPase α3 antibody (Sigma Aldrich, MO), respectively. Similarly, immunoblot analysis was also performed in visceral fat for expression of Na/K-ATPase α1 subunit and α2 subunit.
RNA extraction and real-time PCR
Total RNA was extracted from visceral adipose tissue using RNeasy Protect Mini Kit (QIAGEN, Germantown, MD) as described previously (Sodhi et al., 2018). Total RNA was analyzed by a qRT-PCR. Each reaction was done in triplicate. All experimental samples were normalized using GAPDH. Specific predesigned primers (IDT DNA Technologies) were used for NOS (gene symbol: Nos3), MCP1 (gene symbol: Ccl2), GPx1 (gene symbol: Gpx1), TNFα (gene symbol: tnfa) (Table S2). In order to validate the high throughput RNAseq data, total RNA was also extracted from hippocampus as described, followed by preparation of cDNA libraries. qRT-pCR reactions were performed as described previously (Sodhi et al., 2020a). The relative mRNA expression levels of eight differentially expressed genes was determined. These genes included AKT (gene symbol: Akt3), PPARα (gene symbol: Ppara), IL1R (gene symbol: il1r), FABP4 (gene symbol: fabp4), FABP1 (gene symbol: fabp1), IRS1 (gene symbol: irs1), RAS (gene symbol: rasa1), Itga5 (gene symbol: itga5) (Table S2). The fold changes were calculated from relative expression for WD versus Control and WD + NaKtide versus WD, followed by estimation of log2 Fold Change values per candidate gene to compare RNAseq and qRT-PCR findings. Specific predesigned primers (IDT DNA Technologies) were also used for IL-6 (gene symbol: il6) and TNFα (gene symbol: tnfa) for their relative mRNA expression in hippocampus (Table S2).
RNA-seq and data analysis
Total RNA was extracted from visceral adipose tissue, subcutaneous adipose tissue, liver, hippocampus, and cerebellum using RNeasy Protect Mini Kit (Qiagen, Germantown, MD) as described previously (Sodhi et al., 2018, 2020b). With two exceptions, 5 replicates were studied in the 4 experimental in vivo groups for the different tissues. In the case of hippocampus tissues, there were 6 control and 6 WD + NaKtide expression samples. All samples had RNA integrity numbers greater than 8. A complete RNA-Seq analysis was performed by Marshall University Genomics Core, as described previously (Sodhi et al., 2020b). Differential gene expression was established with a false discovery rate (FDR) threshold of 0.1 using the DESeq2 package (Love et al., 2014). ORA and GSEA analyses were performed using the WebgestaltR package (Wang et al., 2020). All GSEA plots were produced with fgsea package (Sergushichev, 2016). Selected KEGG pathway analyses were displayed utilizing the pathview (Luo and Brouwer, 2013) R package. Wikipathway map was produced using Cytoscape program utilizing Wikipathway software (Kutmon et al., 2014). The differential gene expression from various tissues were also examined in concert with several in vivo parameters using the WCGNA package (Langfelder and Horvath, 2008, 2012) in a similar fashion to that described previously (Sodhi et al., 2020b). Parameters used for the WCGNA analyses are listed in the legend to the appropriate figure.
Raw sequencing reads have been deposited at the Gene Expression Omnibus at the National Center for Biotechnology Information and can be accessed using the accession number GSE164295.
Quantification and statistical analysis
Data was analyzed using GraphPad Prism 8.0. All data were tested for normality and then subjected to parametric analysis. When more than two groups were compared, one-way ANOVA was performed prior to comparison of individual groups, and the post-hoc t-tests were adjusted for multiple comparisons using the Tukey-Kramer correction (Sodhi et al., 2017). All data comparisons are presented at the NS, p<0.05 and p<0.01 levels. Each bar represent values as means ± standard error of mean (SEM), where ∗p < 0.05 vs. CTR, ∗∗p < 0.01 vs. CTR, #p < 0.05 vs. CTR+NaKtide, ##p < 0.01 vs. CTR+NaKtide, &p < 0.05 vs. WD, &&p < 0.01 vs. WD, unless specified otherwise.
Data from RNA-Seq experiments are presented in “volcano” plots as the anti-log10 of the raw p-value against the base 2 log of the fold change (“log2 fold change”). A Benjamini–Hochberg adjusted p-value of 0.1 was used as a threshold for the construction of these volcano plots (Love et al., 2014). The significance values for the overrepresentation of different pathways with either ORA or GSEA were calculated with the R package WebgestaltR (Wang et al., 2013) employing the more conservative FDR value and only examining those FDR values < 0.10 (Love et al., 2014). The gmt file "c2.cp.kegg.v7.2.entrez.gmt" obtained at the Molecular Signatures Database (website https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp) was used for reference. Heatmaps were constructed using the heatmap.2 command within the gplots package (Warnes et al., 2020).
Acknowledgments
We acknowledge the Genomics and Bioinformatics Core (GABC) of Marshall University for performing the RNA-Seq experiments, providing primary and secondary data analysis, and assisting with the interpretation. This work was supported by the National Institutes of Health Grants 1R15HL150721 (to K.S.), NIH Bench-to-Bedside award made possible by the Office of Research on Women's Health (ORWH) 736214 (to K.S. and J.I.S.), and by the BrickStreet Foundation (to J.I.S.) and by the Huntington Foundation, Inc. (J.I.S.). The GABC is supported by WV-INBRE (NIH/NIGMS P20GM103434), the COBRE ACCORD (1P20GM121299), and the WV-CTSI (2U54GM104942).
Author contributions
Conceptualization, J.I.S; Investigation, R.P., X.W., H.V.L, S.S.P, M.Z., J.W., L.G., B.H., and J.L.; Data Curation, J.D. and J.I.S.; Software, J.I.S.; Writing – Original Draft, K.S., and J.I.S.; Writing – Review & Editing, L.G., B.H., J.D., J.L. and T.N.; Supervision, K.S.; Project Administration, K.S. and J.I.S.; Funding Acquisition, K.S. and J.I.S.
Declaration of interests
The authors declare no competing interests.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103262.
Supplemental information
Datasets from hippocampus, cerebellum, liver, subcutaneous and visceral adipose tissue were queried using the WebGestalt toolkit including “KEGG” pathways and ORA was performed using R packages. ORA showed dysregulated KEGG pathways in these tissues. The table illustrates pathways dysregulated in each tissue with the p-value of <0.05 and FDR of <0.1.
This table lists the key RT-PCR primers employed in the study for evaluation of relative expression of genes in visceral adipose tissue and hippocampus.
Data and code availability
-
•
Raw sequencing reads have been deposited at the Gene Expression Omnibus at the National Center for Biotechnology Information and can be accessed using the accession number GSE164295. This data is publicly available as of the date of publication. The accession number is also listed in the key resources table.
-
•
All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplemental information. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
References
- Albert-Gasco H., Ros-Bernal F., Castillo-Gomez E., Olucha-Bordonau F.E. MAP/ERK signaling in developing cognitive and emotional function and its effect on pathological and neurodegenerative processes. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21124471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafian H., Harling L., Darzi A., Athanasiou T. Neurodegenerative disease and obesity: what is the role of weight loss and bariatric interventions? Metab. Brain Dis. 2013;28:341–353. doi: 10.1007/s11011-013-9412-4. [DOI] [PubMed] [Google Scholar]
- Banks W.A. Blood-brain barrier transport of cytokines: a mechanism for neuropathology. Curr. Pharm. Des. 2005;11:973–984. doi: 10.2174/1381612053381684. [DOI] [PubMed] [Google Scholar]
- Banks W.A., Kastin A.J., Broadwell R.D. Passage of cytokines across the blood-brain barrier. Neuroimmunomodulation. 1995;2:241–248. doi: 10.1159/000097202. [DOI] [PubMed] [Google Scholar]
- Banks W.A., Kastin A.J., Gutierrez E.G. Penetration of interleukin-6 across the murine blood-brain barrier. Neurosci. Lett. 1994;179:53–56. doi: 10.1016/0304-3940(94)90933-4. [DOI] [PubMed] [Google Scholar]
- Bartolome F., Antequera D., Tavares E., Pascual C., Maldonado R., Camins A., Carro E. Obesity and neuroinflammatory phenotype in mice lacking endothelial megalin. J. Neuroinflammation. 2017;14:26. doi: 10.1186/s12974-017-0800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarner K.M., Setti S., Diaz C., Littlefield A., Jones A., Kohman R.A. Diet-induced obesity attenuates cytokine production following an immune challenge. Behav. Brain Res. 2014;267:33–41. doi: 10.1016/j.bbr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste E.N. Inflammatory cytokines within the central nervous system: sources, function, and mechanism of action. Am. J. Physiol. 1992;263:C1–C16. doi: 10.1152/ajpcell.1992.263.1.C1. [DOI] [PubMed] [Google Scholar]
- Bittencourt A., Brum P.O., Ribeiro C.T., Gasparotto J., Bortolin R.C., de Vargas A.R., Heimfarth L., de Almeida R.F., Moreira J.C.F., de Oliveira J., Gelain D.P. High fat diet-induced obesity causes a reduction in brain tyrosine hydroxylase levels and non-motor features in rats through metabolic dysfunction, neuroinflammation and oxidative stress. Nutr. Neurosci. 2020:1–15. doi: 10.1080/1028415X.2020.1831261. [DOI] [PubMed] [Google Scholar]
- Bousiges O., Cretin B., Lavaux T., Philippi N., Jung B., Hezard S., Heitz C., Demuynck C., Gabel A., Martin-Hunyadi C., Blanc F. Diagnostic value of cerebrospinal fluid biomarkers (Phospho-Tau181, total-tau, Abeta42, and Abeta40) in prodromal stage of Alzheimer's disease and dementia with Lewy bodies. J. Alzheimers Dis. 2016;51:1069–1083. doi: 10.3233/JAD-150731. [DOI] [PubMed] [Google Scholar]
- Butler M.P., O'Connor J.J., Moynagh P.N. Dissection of tumor-necrosis factor-alpha inhibition of long-term potentiation (LTP) reveals a p38 mitogen-activated protein kinase-dependent mechanism which maps to early-but not late-phase LTP. Neuroscience. 2004;124:319–326. doi: 10.1016/j.neuroscience.2003.11.040. [DOI] [PubMed] [Google Scholar]
- Cacci E., Claasen J.H., Kokaia Z. Microglia-derived tumor necrosis factor-alpha exaggerates death of newborn hippocampal progenitor cells in vitro. J. Neurosci. Res. 2005;80:789–797. doi: 10.1002/jnr.20531. [DOI] [PubMed] [Google Scholar]
- Canchi S., Raao B., Masliah D., Rosenthal S.B., Sasik R., Fisch K.M., De Jager P.L., Bennett D.A., Rissman R.A. Integrating gene and protein expression reveals perturbed functional networks in Alzheimer's disease. Cell Rep. 2019;28:1103–1116.e4. doi: 10.1016/j.celrep.2019.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthorne C., Swindell R., Stratford I.J., Dive C., Welman A. Comparison of doxycycline delivery methods for Tet-inducible gene expression in a subcutaneous xenograft model. J. Biomol. Tech. 2007;18:120–123. [PMC free article] [PubMed] [Google Scholar]
- Chen W.W., Zhang X., Huang W.J. Role of neuroinflammation in neurodegenerative diseases (Review) Mol. Med. Rep. 2016;13:3391–3396. doi: 10.3892/mmr.2016.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew H., Solomon V.A., Fonteh A.N. Involvement of lipids in Alzheimer's disease pathology and potential therapies. Front. Physiol. 2020;11:598. doi: 10.3389/fphys.2020.00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J.R., Cammarota M., Gruart A., Izquierdo I., Delgado-Garcia J.M. Plastic modifications induced by object recognition memory processing. Proc. Natl. Acad. Sci. U S A. 2010;107:2652–2657. doi: 10.1073/pnas.0915059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope E.C., LaMarca E.A., Monari P.K., Olson L.B., Martinez S., Zych A.D., Katchur N.J., Gould E. Microglia play an active role in obesity-associated cognitive decline. J. Neurosci. 2018;38:8889–8904. doi: 10.1523/JNEUROSCI.0789-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A.T., Tenenbaum L., Berkhout B. Tet-on systems for doxycycline-inducible gene expression. Curr. Gene Ther. 2016;16:156–167. doi: 10.2174/1566523216666160524144041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Monte S.M., Tong M., Nguyen V., Setshedi M., Longato L., Wands J.R. Ceramide-mediated insulin resistance and impairment of cognitive-motor functions. J. Alzheimers Dis. 2010;21:967–984. doi: 10.3233/JAD-2010-091726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E., Huston J.P., De Souza Silva M.A. Integrated memory for objects, places, and temporal order: evidence for episodic-like memory in mice. Neurobiol. Learn Mem. 2005;84:214–221. doi: 10.1016/j.nlm.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Duffy C.M., Hofmeister J.J., Nixon J.P., Butterick T.A. High fat diet increases cognitive decline and neuroinflammation in a model of orexin loss. Neurobiol. Learn Mem. 2019;157:41–47. doi: 10.1016/j.nlm.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette A., Pernegre C., Veilleux Carpentier A., Leclerc N. Similarities and differences in the pattern of tau hyperphosphorylation in physiological and pathological conditions: Impacts on the elaboration of therapies to prevent tau pathology. Front. Neurol. 2020;11:607680. doi: 10.3389/fneur.2020.607680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada L.D., Ahumada P., Cabrera D., Arab J.P. Liver dysfunction as a novel player in Alzheimer's progression: looking outside the brain. Front. Aging Neurosci. 2019;11:174. doi: 10.3389/fnagi.2019.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federico A., Cardaioli E., Da Pozzo P., Formichi P., Gallus G.N., Radi E. Mitochondria, oxidative stress and neurodegeneration. J. Neurol. Sci. 2012;322:254–262. doi: 10.1016/j.jns.2012.05.030. [DOI] [PubMed] [Google Scholar]
- Ferreira-Vieira T.H., Guimaraes I.M., Silva F.R., Ribeiro F.M. Alzheimer's disease: targeting the cholinergic system. Curr. Neuropharmacol. 2016;14:101–115. doi: 10.2174/1570159x13666150716165726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira S.T., Clarke J.R., Bomfim T.R., De Felice F.G. Inflammation, defective insulin signaling, and neuronal dysfunction in Alzheimer's disease. Alzheimers Dement. (Amst) 2014;10:S76–S83. doi: 10.1016/j.jalz.2013.12.010. [DOI] [PubMed] [Google Scholar]
- Fossati S., Ramos Cejudo J., Debure L., Pirraglia E., Sone J.Y., Li Y., Chen J., Butler T., Zetterberg H., Blennow K., de Leon M.J. Plasma tau complements CSF tau and P-tau in the diagnosis of Alzheimer's disease. Alzheimers Dement. (Amst) 2019;11:483–492. doi: 10.1016/j.dadm.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamaro G.D., Streck E.L., Matté C., Prediger M.E., Wyse A.T., Dalmaz C. Reduction of hippocampal Na+, K+-ATPase activity in rats subjected to an experimental model of depression. Neurochem. Res. 2003;28:1339–1344. doi: 10.1023/a:1024988113978. [DOI] [PubMed] [Google Scholar]
- Gegg M.E., Beltran B., Salas-Pino S., Bolanos J.P., Clark J.B., Moncada S., Heales S.J. Differential effect of nitric oxide on glutathione metabolism and mitochondrial function in astrocytes and neurones: implications for neuroprotection/neurodegeneration? J. Neurochem. 2003;86:228–237. doi: 10.1046/j.1471-4159.2003.01821.x. [DOI] [PubMed] [Google Scholar]
- Geisler C.E., Renquist B.J. Hepatic lipid accumulation: cause and consequence of dysregulated glucoregulatory hormones. J. Endocrinol. 2017;234:R1–R21. doi: 10.1530/JOE-16-0513. [DOI] [PubMed] [Google Scholar]
- Gengenbacher M., Zimmerman M.D., Sarathy J.P., Kaya F., Wang H., Mina M., Carter C., Hossen M.A., Su H., Trujillo C. Tissue distribution of doxycycline in animal models of tuberculosis. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.02479-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland W.L., Bikman B.T., Wang L.P., Yuguang G., Sargent K.M., Bulchand S., Knotts T.A., Shui G., Clegg D.J., Wenk M.R. Lipid-induced insulin resistance mediated by the proinflammatory receptor TLR4 requires saturated fatty acid-induced ceramide biosynthesis in mice. J. Clin. Invest. 2011;121:1858–1870. doi: 10.1172/JCI43378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hroudova J., Singh N., Fisar Z. Mitochondrial dysfunctions in neurodegenerative diseases: relevance to Alzheimer's disease. Biomed. Res. Int. 2014;2014:175062. doi: 10.1155/2014/175062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunsberger H.C., Greenwood B.P., Tolstikov V., Narain N.R., Kiebish M.A., Denny C.A. Divergence in the metabolome between natural aging and Alzheimer's disease. Sci. Rep. 2020;10:12171. doi: 10.1038/s41598-020-68739-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingusci S., Verlengia G., Soukupova M., Zucchini S., Simonato M. Gene therapy tools for brain diseases. Front. Pharmacol. 2019;10:724. doi: 10.3389/fphar.2019.00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D., Imai Y., Ohsawa K., Nakajima K., Fukuuchi Y., Kohsaka S. Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res. Mol. Brain Res. 1998;57:1–9. doi: 10.1016/s0169-328x(98)00040-0. [DOI] [PubMed] [Google Scholar]
- Jacka F.N., Cherbuin N., Anstey K.J., Sachdev P., Butterworth P. Western diet is associated with a smaller hippocampus: a longitudinal investigation. BMC Med. 2015;13:215. doi: 10.1186/s12916-015-0461-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel L., Biemans E., Klijn C.J.M., Kuiperij H.B., Verbeek M.M. Reduced influence of apoE on Abeta43 Aggregation and reduced vascular Abeta43 toxicity as compared with Abeta40 and Abeta42. Mol. Neurobiol. 2020;57:2131–2141. doi: 10.1007/s12035-020-01873-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamwal S., Blackburn J.K., Elsworth J.D. PPARgamma/PGC1alpha signaling as a potential therapeutic target for mitochondrial biogenesis in neurodegenerative disorders. Pharmacol. Ther. 2020:107705. doi: 10.1016/j.pharmthera.2020.107705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Wang Y., Su L., Ren H., Wang C., Chen J., Fu X. Donepezil attenuates obesity-associated oxidative stress and central inflammation and improves memory deficit in mice fed a high-fat diet. Dement Geriatr. Cogn. Disord. 2019;48:154–163. doi: 10.1159/000504800. [DOI] [PubMed] [Google Scholar]
- Kacirova M., Zmeskalova A., Korinkova L., Zelezna B., Kunes J., Maletinska L. Inflammation: major denominator of obesity, Type 2 diabetes and Alzheimer's disease-like pathology? Clin. Sci. (Lond) 2020;134:547–570. doi: 10.1042/CS20191313. [DOI] [PubMed] [Google Scholar]
- Kanoski S.E., Davidson T.L. Western diet consumption and cognitive impairment: links to hippocampal dysfunction and obesity. Physiol. Behav. 2011;103:59–68. doi: 10.1016/j.physbeh.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempuraj D., Thangavel R., Selvakumar G.P., Zaheer S., Ahmed M.E., Raikwar S.P., Zahoor H., Saeed D., Natteru P.A., Iyer S., Zaheer A. Brain and peripheral Atypical inflammatory mediators potentiate neuroinflammation and neurodegeneration. Front. Cell. Neurosci. 2017;11:216. doi: 10.3389/fncel.2017.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothari V., Luo Y., Tornabene T., O'Neill A.M., Greene M.W., Geetha T., Babu J.R. High fat diet induces brain insulin resistance and cognitive impairment in mice. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:499–508. doi: 10.1016/j.bbadis.2016.10.006. [DOI] [PubMed] [Google Scholar]
- Kutmon M., Lotia S., Evelo C.T., Pico A.R. WikiPathways App for Cytoscape: making biological pathways amenable to network analysis and visualization. F1000Res. 2014;3:152. doi: 10.12688/f1000research.4254.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F., Madan N., Ye Q., Duan Q., Li Z., Wang S., Si S., Xie Z. Identification of a mutant alpha1 Na/K-ATPase that pumps but is defective in signal transduction. J. Biol. Chem. 2013;288:13295–13304. doi: 10.1074/jbc.M113.467381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 2012;46 [PMC free article] [PubMed] [Google Scholar]
- Le Lay S., Simard G., Martinez M.C., Andriantsitohaina R. Oxidative stress and metabolic pathologies: from an adipocentric point of view. Oxid Med. Cell Longev. 2014;2014:908539. doi: 10.1155/2014/908539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.S., Lee W.H., Bae Y.C., Suk K. Axon guidance molecules guiding neuroinflammation. Exp. Neurobiol. 2019;28:311–319. doi: 10.5607/en.2019.28.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letra L., Santana I., Seica R. Obesity as a risk factor for Alzheimer's disease: the role of adipocytokines. Metab. Brain Dis. 2014;29:563–568. doi: 10.1007/s11011-014-9501-z. [DOI] [PubMed] [Google Scholar]
- Lewerenz J., Maher P. Chronic glutamate toxicity in neurodegenerative diseases-what is the evidence? Front. Neurosci. 2015;9:469. doi: 10.3389/fnins.2015.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Cai T., Tian J., Xie J.X., Zhao X., Liu L., Shapiro J.I., Xie Z. NaKtide, a Na/K-ATPase-derived peptide Src inhibitor, antagonizes ouabain-activated signal transduction in cultured cells. J. Biol. Chem. 2009;284:21066–21076. doi: 10.1074/jbc.M109.013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Cai T., Tian J., Qu W., Xie Z.J. Functional characterization of Src-interacting Na/K-ATPase using RNA interference assay. J. Biol. Chem. 2006;281:19709–19719. doi: 10.1074/jbc.M512240200. [DOI] [PubMed] [Google Scholar]
- Liang M., Tian J., Liu L., Pierre S., Liu J., Shapiro J., Xie Z.J. Identification of a pool of non-pumping Na/K-ATPase. J. Biol. Chem. 2007;282:10585–10593. doi: 10.1074/jbc.M609181200. [DOI] [PubMed] [Google Scholar]
- Liu J., Kennedy D.J., Yan Y., Shapiro J.I. Reactive oxygen species modulation of Na/K-ATPase regulates fibrosis and renal proximal tubular Sodium handling. Int. J. Nephrol. 2012;2012:381320. doi: 10.1155/2012/381320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Tian J., Chaudhry M., Maxwell K., Yan Y., Wang X., Shah P.T., Khawaja A.A., Martin R., Robinette T.J. Attenuation of Na/K-ATPase mediated oxidant amplification with pNaKtide ameliorates experimental uremic cardiomyopathy. Sci. Rep. 2016;6:34592. doi: 10.1038/srep34592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Ruan J. Network-based pathway enrichment analysis. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2013:218–221. doi: 10.1109/BIBM.2013.6732493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueptow L.M. Novel object recognition test for the investigation of learning and memory in mice. J. Vis. Exp. 2017 doi: 10.3791/55718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majd S., Power J.H.T., Koblar S.A., Grantham H.J.M. The impact of tau hyperphosphorylation at Ser(262) on memory and learning after global brain ischaemia in a rat model of reversible cardiac arrest. IBRO Rep. 2017;2:1–13. doi: 10.1016/j.ibror.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna P., Jain S.K. Obesity, oxidative stress, adipose tissue dysfunction, and the associated Health risks: causes and therapeutic strategies. Metab. Syndr. Relat. Disord. 2015;13:423–444. doi: 10.1089/met.2015.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansuy I.M., Bujard H. Tetracycline-regulated gene expression in the brain. Curr. Opin. Neurobiol. 2000;10:593–596. doi: 10.1016/s0959-4388(00)00127-6. [DOI] [PubMed] [Google Scholar]
- Mattson M.P., Pedersen W.A., Duan W., Culmsee C., Camandola S. Cellular and molecular mechanisms underlying perturbed energy metabolism and neuronal degeneration in Alzheimer's and Parkinson's diseases. Ann. N. Y. Acad. Sci. 1999;893:154–175. doi: 10.1111/j.1749-6632.1999.tb07824.x. [DOI] [PubMed] [Google Scholar]
- Mazon J.N., de Mello A.H., Ferreira G.K., Rezin G.T. The impact of obesity on neurodegenerative diseases. Life Sci. 2017;182:22–28. doi: 10.1016/j.lfs.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Miller A.A., Spencer S.J. Obesity and neuroinflammation: a pathway to cognitive impairment. Brain Behav. Immun. 2014;42:10–21. doi: 10.1016/j.bbi.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Miranda M., Morici J.F., Zanoni M.B., Bekinschtein P. Brain-derived neurotrophic factor: a key molecule for memory in the healthy and the pathological brain. Front. Cell. Neurosci. 2019;13:363. doi: 10.3389/fncel.2019.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje M.L., Toda H., Palmer T.D. Inflammatory blockade restores adult hippocampal neurogenesis. Science. 2003;302:1760–1765. doi: 10.1126/science.1088417. [DOI] [PubMed] [Google Scholar]
- Moya-Alvarado G., Gershoni-Emek N., Perlson E., Bronfman F.C. Neurodegeneration and Alzheimer's disease (AD). What can proteomics tell us about the Alzheimer's brain? Mol. Cell Proteomics. 2016;15:409–425. doi: 10.1074/mcp.R115.053330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nho K., Kueider-Paisley A., Ahmad S., MahmoudianDehkordi S., Arnold M., Risacher S.L., Louie G., Blach C., Baillie R., Han X. Association of altered liver enzymes with Alzheimer disease diagnosis, cognition, neuroimaging measures, and cerebrospinal fluid biomarkers. JAMA Netw. Open. 2019;2:e197978. doi: 10.1001/jamanetworkopen.2019.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholko A.G., Wotton C.A., Bekar L.K. Poor diet, stress, and Inactivity converge to form a "perfect Storm" that drives Alzheimer's disease pathogenesis. Neurodegener. Dis. 2019;19:60–77. doi: 10.1159/000503451. [DOI] [PubMed] [Google Scholar]
- Parimisetty A., Dorsemans A.C., Awada R., Ravanan P., Diotel N., Lefebvre d'Hellencourt C. Secret talk between adipose tissue and central nervous system via secreted factors-an emerging frontier in the neurodegenerative research. J. Neuroinflammation. 2016;13:67. doi: 10.1186/s12974-016-0530-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl D.P., Olanow C.W., Calne D. Alzheimer's disease and Parkinson's disease: distinct entities or extremes of a spectrum of neurodegeneration? Ann. Neurol. 1998;44:S19–S31. doi: 10.1002/ana.410440705. [DOI] [PubMed] [Google Scholar]
- Pickering M., Cumiskey D., O'Connor J.J. Actions of TNF-alpha on glutamatergic synaptic transmission in the central nervous system. Exp. Physiol. 2005;90:663–670. doi: 10.1113/expphysiol.2005.030734. [DOI] [PubMed] [Google Scholar]
- Pratt R.D., Brickman C., Nawab A., Cottrill C., Snoad B., Lakhani H.V., Jelcick A., Henderson B., Bhardwaj N.N., Sanabria J.R. The adipocyte Na/K-ATPase oxidant amplification loop is the central regulator of western diet-induced obesity and associated comorbidities. Sci. Rep. 2019;9:7927. doi: 10.1038/s41598-019-44350-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Qin H., Buckley J.A., Li X., Liu Y., Fox T.H., 3rd, Meares G.P., Yu H., Yan Z., Harms A.S., Li Y. Inhibition of the JAK/STAT pathway protects against alpha-Synuclein-induced neuroinflammation and dopaminergic neurodegeneration. J. Neurosci. 2016;36:5144–5159. doi: 10.1523/JNEUROSCI.4658-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . Vienna; Austria: 2016. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
- Ramanan V.K., Kim S., Holohan K., Shen L., Nho K., Risacher S.L., Foroud T.M., Mukherjee S., Crane P.K., Aisen P.S. Genome-wide pathway analysis of memory impairment in the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort implicates gene candidates, canonical pathways, and networks. Brain Imaging Behav. 2012;6:634–648. doi: 10.1007/s11682-012-9196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redelsperger I.M., Taldone T., Riedel E.R., Lepherd M.L., Lipman N.S., Wolf F.R. Stability of doxycycline in feed and water and minimal effective doses in tetracycline-inducible systems. J. Am. Assoc. Lab. Anim. Sci. 2016;55:467–474. [PMC free article] [PubMed] [Google Scholar]
- Rosu G.C., Catalin B., Balseanu T.A., Laurentiu M., Claudiu M., Kumar-Singh S., Daniel P. Inhibition of Aquaporin 4 decreases amyloid Abeta40 drainage Around cerebral vessels. Mol. Neurobiol. 2020;57:4720–4734. doi: 10.1007/s12035-020-02044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell N.J., Hopkins S.J. Cytokines and the nervous system II: Actions and mechanisms of action. Trends Neurosci. 1995;18:130–136. doi: 10.1016/0166-2236(95)93890-a. [DOI] [PubMed] [Google Scholar]
- Saltiel A.R., Olefsky J.M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Invest. 2017;127:1–4. doi: 10.1172/JCI92035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener M.L., Wooten M.C. Use of the Open Field Maze to measure locomotor and anxiety-like behavior in mice. J. Vis. Exp. 2015:e52434. doi: 10.3791/52434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmaj K., Raine C.S. Tumor necrosis factor mediates myelin damage in organotypic cultures of nervous tissue. Ann. N. Y. Acad. Sci. 1988;540:568–570. doi: 10.1111/j.1749-6632.1988.tb27175.x. [DOI] [PubMed] [Google Scholar]
- Sergushichev A.A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. 2016 doi: 10.1101/060012. [DOI] [Google Scholar]
- Sheng M., Sabatini B.L., Sudhof T.C. Synapses and Alzheimer's disease. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.G., Ciccotosto G.D., Tew D.J., Perez K., Curtain C.C., Boas J.F., Masters C.L., Cappai R., Barnham K.J. Histidine 14 modulates membrane binding and neurotoxicity of the Alzheimer's disease amyloid-beta peptide. J. Alzheimers Dis. 2010;19:1387–1400. doi: 10.3233/JAD-2010-1334. [DOI] [PubMed] [Google Scholar]
- Sodhi K., Denvir J., Liu J., Sanabria J.R., Chen Y., Silverstein R., Xie Z., Abraham N.G., Shapiro J.I. Oxidant-induced alterations in the adipocyte transcriptome: role of the Na,K-ATPase oxidant amplification loop. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21165923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi K., Maxwell K., Yan Y., Liu J., Chaudhry M.A., Getty M., Xie Z., Abraham N.G., Shapiro J.I. pNaKtide inhibits Na/K-ATPase reactive oxygen species amplification and attenuates adipogenesis. Sci. Adv. 2015;1:e1500781. doi: 10.1126/sciadv.1500781. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sodhi K., Nichols A., Mallick A., Klug R.L., Liu J., Wang X., Srikanthan K., Goguet-Rubio P., Nawab A., Pratt R. The Na/K-ATPase oxidant amplification loop regulates aging. Sci. Rep. 2018;8:9721. doi: 10.1038/s41598-018-26768-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sodhi K., Srikanthan K., Goguet-Rubio P., Nichols A., Mallick A., Nawab A., Martin R., Shah P.T., Chaudhry M., Sigdel S. pNaKtide attenuates steatohepatitis and Atherosclerosis by blocking Na/K-ATPase/ROS amplification in C57Bl6 and ApoE knockout mice fed a western diet. Sci. Rep. 2017;7:193. doi: 10.1038/s41598-017-00306-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sodhi K., Wang X., Chaudhry M.A., Lakhani H.V., Zehra M., Pratt R., Nawab A., Cottrill C.L., Snoad B., Bai F. Central role for adipocyte Na,K-ATPase oxidant amplification loop in the pathogenesis of experimental uremic cardiomyopathy. J. Am. Soc. Nephrol. 2020;31:1746–1760. doi: 10.1681/ASN.2019101070. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sonkusare S.K., Kaul C.L., Ramarao P. Dementia of Alzheimer's disease and other neurodegenerative disorders--memantine, a new hope. Pharmacol. Res. 2005;51:1–17. doi: 10.1016/j.phrs.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Sripetchwandee J., Chattipakorn N., Chattipakorn S.C. Links between obesity-induced brain insulin resistance, brain mitochondrial dysfunction, and dementia. Front. Endocrinol. (Lausanne) 2018;9:496. doi: 10.3389/fendo.2018.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela G., Hogan D., Zhang W.R., Dineley K.T. Intermediate- and long-term recognition memory deficits in Tg2576 mice are reversed with acute calcineurin inhibition. Behav. Brain Res. 2009;200:95–99. doi: 10.1016/j.bbr.2008.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarro L., Llaurado E., Albaladejo R., Morina D., Arija V., Sola R., Giralt M. A primary-school-based study to reduce the prevalence of childhood obesity--the EdAl (Educacio en Alimentacio) study: a randomized controlled trial. Trials. 2014;15:58. doi: 10.1186/1745-6215-15-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesseur I., Wyss-Coray T. A role for TGF-beta signaling in neurodegeneration: evidence from genetically engineered models. Curr. Alzheimer Res. 2006;3:505–513. doi: 10.2174/156720506779025297. [DOI] [PubMed] [Google Scholar]
- Tian J., Cai T., Yuan Z., Wang H., Liu L., Haas M., Maksimova E., Huang X.Y., Xie Z.J. Binding of Src to Na+/K+-ATPase forms a functional signaling complex. Mol. Biol. Cell. 2006;17:317–326. doi: 10.1091/mbc.e05-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.H., Tsai L.K., Lo Y.L., Lin C.H. Amyloid related cerebral microbleed and plasma Abeta40 are associated with cognitive decline in Parkinson's disease. Sci. Rep. 2021;11:7115. doi: 10.1038/s41598-021-86617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uranga R.M., Keller J.N. The complex interactions between obesity, metabolism and the brain. Front. Neurosci. 2019;13:513. doi: 10.3389/fnins.2019.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utrera J., Junyent F., de Lemos L., Pallas M., Camins A., Romero R., Auladell C. Tau hyperphosphorylation and axonal damage induced by N,N-diethyldithiocarbamate (DEDTC) treatment along late postnatal development is followed by a rescue during adulthood. J. Neurosci. Res. 2010;88:1083–1093. doi: 10.1002/jnr.22284. [DOI] [PubMed] [Google Scholar]
- Valdearcos M., Robblee M.M., Benjamin D.I., Nomura D.K., Xu A.W., Koliwad S.K. Microglia dictate the impact of saturated fat consumption on hypothalamic inflammation and neuronal function. Cell Rep. 2014;9:2124–2138. doi: 10.1016/j.celrep.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Duncan D., Shi Z., Zhang B. WEB-based GEne SeT AnaLysis toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Liao Y., Jaehnig E., Shi Z., Sheng Q. CRAN; 2020. WebGestaltR: Gene Set Analysis Toolkit WebGestaltR. [Google Scholar]
- Wang Y., Ye Q., Liu C., Xie J.X., Yan Y., Lai F., Duan Q., Li X., Tian J., Xie Z. Involvement of Na/K-ATPase in hydrogen peroxide-induced activation of the Src/ERK pathway in LLC-PK1 cells. Free Radic. Biol. Med. 2014;71:415–426. doi: 10.1016/j.freeradbiomed.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., Moeller S. CRAN; 2020. Gplots: Various R Programming Tools for Plotting Data. [Google Scholar]
- Watt G., Shang K., Zieba J., Olaya J., Li H., Garner B., Karl T. Chronic treatment with 50 mg/kg cannabidiol improves cognition and moderately reduces Abeta40 levels in 12-month-old male AbetaPPswe/PS1DeltaE9 transgenic mice. J. Alzheimers Dis. 2020;74:937–950. doi: 10.3233/JAD-191242. [DOI] [PubMed] [Google Scholar]
- Wojtowicz S., Strosznajder A.K., Jezyna M., Strosznajder J.B. The novel role of PPAR alpha in the brain: promising target in therapy of Alzheimer's disease and other neurodegenerative disorders. Neurochem. Res. 2020;45:972–988. doi: 10.1007/s11064-020-02993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z.J., Wang Y.H., Ganjeizadeh M., McGee R., Jr., Askari A. Determination of total (Na+ + K+)-ATPase activity of isolated or cultured cells. Anal Biochem. 1989;183:215–219. doi: 10.1016/0003-2697(89)90470-3. [DOI] [PubMed] [Google Scholar]
- Xu H., Barnes G.T., Yang Q., Tan G., Yang D., Chou C.J., Sole J., Nichols A., Ross J.S., Tartaglia L.A., Chen H. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Haller S., Shapiro A., Malhotra N., Tian J., Xie Z., Malhotra D., Shapiro J.I., Liu J. Ouabain-stimulated trafficking regulation of the Na/K-ATPase and NHE3 in renal proximal tubule cells. Mol. Cell. Biochem. 2012;367:175–183. doi: 10.1007/s11010-012-1331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Shapiro A.P., Haller S., Katragadda V., Liu L., Tian J., Basrur V., Malhotra D., Xie Z.J., Abraham N.G. Involvement of reactive oxygen species in a feed-forward mechanism of Na/K-ATPase-mediated signaling transduction. J. Biol. Chem. 2013;288:34249–34258. doi: 10.1074/jbc.M113.461020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Shapiro A.P., Mopidevi B.R., Chaudhry M.A., Maxwell K., Haller S.T., Drummond C.A., Kennedy D.J., Tian J., Malhotra D. Protein carbonylation of an amino acid residue of the Na/K-ATPase alpha1 subunit determines Na/K-ATPase signaling and Sodium transport in renal proximal tubular cells. J. Am. Heart Assoc. 2016;5 doi: 10.1161/JAHA.116.003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez Lopez M., Pardon M.C., Baiker K., Prior M., Yuchun D., Agostini A., Bai L., Auer D.P., Faas H.M. Myoinositol CEST signal in animals with increased Iba-1 levels in response to an inflammatory challenge-Preliminary findings. PLoS One. 2019;14:e0212002. doi: 10.1371/journal.pone.0212002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Ruan H.B. Neuronal control of adaptive thermogenesis. Front. Endocrinol. (Lausanne) 2015;6:149. doi: 10.3389/fendo.2015.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarlagadda A., Alfson E., Clayton A.H. The blood brain barrier and the role of cytokines in neuropsychiatry. Psychiatry (Edgmont) 2009;6:18–22. [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Boska M., Zheng Y., Liu J., Fox H.S., Xiong H. Minocycline attenuation of rat corpus callosum abnormality mediated by low-dose lipopolysaccharide-induced microglia activation. J. Neuroinflammation. 2021;18:100. doi: 10.1186/s12974-021-02142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Datasets from hippocampus, cerebellum, liver, subcutaneous and visceral adipose tissue were queried using the WebGestalt toolkit including “KEGG” pathways and ORA was performed using R packages. ORA showed dysregulated KEGG pathways in these tissues. The table illustrates pathways dysregulated in each tissue with the p-value of <0.05 and FDR of <0.1.
This table lists the key RT-PCR primers employed in the study for evaluation of relative expression of genes in visceral adipose tissue and hippocampus.
Data Availability Statement
-
•
Raw sequencing reads have been deposited at the Gene Expression Omnibus at the National Center for Biotechnology Information and can be accessed using the accession number GSE164295. This data is publicly available as of the date of publication. The accession number is also listed in the key resources table.
-
•
All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplemental information. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.