Abstract
A key pathway for mRNA degradation in bacterial cells begins with conversion of the initial 5′-terminal triphosphate to a monophosphate, a modification that renders transcripts more vulnerable to attack by ribonucleases whose affinity for monophosphorylated 5′ ends potentiates their catalytic efficacy. In Escherichia coli, the only proteins known to be important for controlling degradation via this pathway are the RNA pyrophosphohydrolase RppH, its heteromeric partner DapF, and the 5′-monophosphate-assisted endonucleases RNase E and RNase G. We have now identified the metabolic enzyme cytidylate kinase as another protein that affects rates of 5′-end-dependent mRNA degradation in E. coli. It does so by utilizing two distinct mechanisms to influence the 5′-terminal phosphorylation state of RNA, each dependent on the catalytic activity of cytidylate kinase and not its mere presence in cells. First, this enzyme acts in conjunction with DapF to stimulate the conversion of 5′ triphosphates to monophosphates by RppH. In addition, it suppresses the direct synthesis of monophosphorylated transcripts that begin with cytidine by reducing the cellular concentration of cytidine monophosphate, thereby disfavoring the 5′-terminal incorporation of this nucleotide by RNA polymerase during transcription initiation. Together, these findings suggest dual signaling pathways by which nucleotide metabolism can impact mRNA degradation in bacteria.
INTRODUCTION
In all living organisms, messenger RNA is an indispensable intermediary that carries instructions for protein synthesis from genes to ribosomes. Critically important for determining the levels of synthesis of individual proteins is the cellular concentration of their respective mRNAs, which depends not only on the rate of mRNA synthesis but also on the rate of mRNA degradation. Decay rates can vary significantly between different mRNAs or for the same mRNA under different conditions. As a result, mRNA lifetimes in the same cell can differ from one another by as much as two orders of magnitude in both eukaryotic and prokaryotic organisms (1).
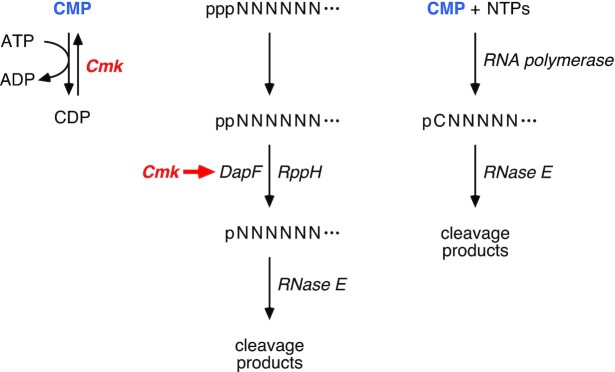
In Escherichia coli and countless other bacterial species, the half-lives of mRNAs are governed primarily by the rate at which they are cleaved by RNase E and, to a lesser degree, the low-abundance RNase E paralog RNase G (2). These low-specificity endonucleases cut RNA in single-stranded regions that are AU-rich (3–8). They are able to gain access to those sites either directly or by a 5′-end-dependent mechanism. The latter mechanism requires prior conversion of the 5′ triphosphate of the primary transcript to a monophosphate (9,10), which can stimulate RNase E and RNase G cleavage at downstream sites up to 100-fold by binding to an enzyme pocket that is distinct from the catalytic center (5,11–14). In E. coli, removal of the 5′-terminal γ and β phosphates generally occurs by the sequential action of two enzymes: an unidentified RNA triphosphatase that converts the 5′ triphosphate to a diphosphate and the RNA pyrophosphohydrolase RppH, which subsequently releases the β phosphate to generate a monophosphorylated 5′ end (10,15). At a slower rate, RppH is also capable of removing pyrophosphate from triphosphorylated 5′ termini to generate a 5′ monophosphate in a single step.
A member of the Nudix hydrolase family, which also includes eukaryotic mRNA decapping enzymes such as Dcp2, E. coli RppH is well conserved at the sequence level in most proteobacteria as well as in flowering plants (16). More distantly related homologs of this enzyme are present in other bacterial phyla (16–19). It can react with any diphosphorylated or triphosphorylated RNA bearing at least two and preferably three or more unpaired nucleotides at the 5′ terminus (15,16). Despite the pivotal role of RppH in converting 5′ ends to monophosphates, the rate of this process is not limited by the cellular concentration of RppH, whose overproduction does not alter levels of monophosphorylated 5′ termini in E. coli (20). Consistent with this observation, the specific activity of RppH is enhanced in E. coli by complex formation with the diaminopimelate epimerase DapF, a metabolic enzyme that catalyzes a step in lysine and peptidoglycan biosynthesis (21). Together these proteins assemble into a heterotetramer in which one RppH protomer is bound to each subunit of a DapF homodimer (22,23). The mechanism by which DapF binding stimulates RppH activity is not yet clear. Moreover, no systematic study has yet been undertaken to identify other proteins that may activate or inhibit RppH.
We have now conducted a screen for E. coli mutants that are impaired for 5′-end-dependent mRNA degradation. This genetic screen has implicated cytidylate kinase in the early events of that regulatory pathway, where it influences the generation of monophosphorylated mRNAs by two distinct mechanisms. Interestingly, the impact of cytidylate kinase on that process depends in part on the identity of the 5′-terminal RNA nucleotide.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions
All measurements of RNA half-lives, 5′ phosphorylation states, and intracellular protein levels were performed with the E. coli K-12 strain BW25113 (24) and its isogenic derivatives. Unless otherwise noted, cells were grown at 37°C in LB medium, which was supplemented with kanamycin (30 μg/ml), ampicillin (100 μg/ml), tetracycline (10 μg/ml) and/or chloramphenicol (20 μg/ml) when necessary. A list of the strains used in this study can be found in Supplementary Table S4.
Chromosomal mutations were introduced into various genetic backgrounds by P1 transduction, allelic exchange, or a combination of the two techniques. To knock out genes by creating in-frame deletions, bacteriophage P1 was used to introduce a kan-substituted gene deletion from the Keio collection (25) into the target strain, and the kan gene was then excised with the aid of plasmid pCP20 (24). P1 transduction was also used to move a kan-linked RNase III null allele (rnc-105) into BW25113.
To introduce point mutations, small deletions, or epitope tags into the chromosome, the mutation of interest flanked on each side by 750–1000 bp of chromosomal sequence was amplified by SOE-PCR (splicing by overlap extension PCR) and cloned between the XbaI and SacI sites of the allelic exchange vector pRE112 (26). The resulting plasmid was conjugated into the target strain by using E. coli strain S17-1λpir, and single recombinants were selected on MOPS-glucose agar containing chloramphenicol (20 μg/ml). Double recombinants were then selected on LB agar containing sucrose (10%) but no sodium chloride, and their genotype was verified by sequencing the chromosomal locus that had been modified.
The plasmids used in this study are described in Supplementary Table S5. Plasmid pYeiP-GFP encoded an in-frame translational fusion of E. coli yeiP to the gene for a fast-folding form of GFP (27). Plasmids pPlacHis6-Cmk and pPlacHis6-CmkR41E encoded active or catalytically inactive (R41E) E. coli cytidylate kinase bearing an amino-terminal hexahistidine tag. Plasmid pPM30 cmk encoded wild-type cytidylate kinase and was used for complementation. Plasmids pYeiP1, pYdfG1, pTrxB1, pYajQ1, pYfcZ1, and pEfp1 encoded the wild-type E. coli yeiP, ydfG, trxB, yajQ, yfcZ and efp transcripts, respectively, synthesized from their native promoters and were used to facilitate detection by Northern blotting. Plasmid pEfp1-C1A encoded a variant of efp mRNA in which the 5′-terminal nucleotide had been changed from C to A. Plasmid pR1.3-yeiP encoded a chimeric transcript comprising the T7 R1.3 stem loop (28) fused to the 5′ end yeiP mRNA.
Screening of the Keio collection
The reporter plasmid pYeiP-GFP, encoding a yeiP-GFP translational fusion, was transformed into each strain of the Keio collection by using TSS solution, as previously described (29). The resulting transformants were grown in microtiter plates alongside wild-type and ΔrppH controls at 37°C in modified LB medium (10 g bacto-tryptone, 10 g NaCl, and 0.25 g yeast extract per liter of deionized water) to reduce background fluorescence. At mid-log phase (OD620 ≈ 0.2), GFP fluorescence was measured by using a Tecan SpectraFluor Plus plate reader at an excitation wavelength of 485 nm and an emission wavelength of 535 nm.
RNA extraction and Northern blot analysis
Total cellular RNA was extracted from E. coli by the hot-phenol procedure (15). For Northern blotting, RNA was subjected to electrophoresis on a 6% polyacrylamide gel containing 7 M urea (PABLO, PACO, and rpsT P2 half-life measurements) or a 1.5% agarose gel containing 2.4% formaldehyde (other half-life measurements). Following electrophoresis, polyacrylamide gels were electroblotted onto a Hybond-XL membrane (GE Healthcare), whereas agarose gels were blotted onto Hybond-XL by downward capillary transfer (30). The membrane was UV-crosslinked and then probed with a 5′ radiolabeled oligodeoxynucleotide complementary to the transcript of interest. Bands were visualized with a Typhoon Trio imager (GE Healthcare) and quantified with ImageQuant TL software.
Measuring RNA lifetimes
E. coli cells grown to OD600 ≈ 0.3 were harvested at the indicated time intervals after treatment with rifampicin (0.2 mg/ml) to halt transcription. Following RNA extraction, 10 μg of total cellular RNA isolated at each time point was subjected to electrophoresis on a 1.5% agarose/2.4% formaldehyde gel or, in the case of the rpsT P2 transcript, on a 6% polyacrylamide/7 M urea gel to enable separation from the rpsT P1 transcript. The RNA was then transferred to a Hybond-XL membrane (GE Healthcare), crosslinked, and probed as outlined above. RNA half-lives and standard deviations were calculated by linear regression analysis of band intensities plotted semilogarithmically as a function of time after rifampicin addition.
5′ End mapping
The 5′-terminal nucleotide of the efp, efp-C1A and rpsT P2 transcripts was identified by rapid amplification of cDNA ends (5′ RACE) as previously described (20). Purified E. coli RppH (200 nM) (15) was substituted for tobacco acid pyrophosphatase (TAP) to convert all 5′ ends to monophosphates prior to ligation to oligonucleotide RACE-1, cDNA synthesis with primer efp rev or rpsTP2 rev, and PCR amplificaton with oligonucleotide- and transcript-specific primers (Supplementary Table S6).
Analyzing the 5′ phosphorylation state of RNA by PABLO and PACO
The percentage of monophosphorylated 5′ ends for individual RNAs was measured by PABLO analysis (31,32) with minor modifications. Briefly, total cellular RNA (10 μg) extracted from E. coli cells grown to OD600 ≈ 0.3 was combined with the appropriate 10–23 DNAzyme, oligonucleotide X, oligonucleotide Y, and T4 DNA ligase (see Supplementary Table S6 for oligonucleotide sequences) to achieve site-specific cleavage and transcript-specific ligation of monophosphorylated 5′ ends, as previously described, and the reaction products were visualized by Northern blotting and quantified to measure the ligation yield (32). The percentage of a cellular transcript that was monophosphorylated was determined from the ratio of its ligation yield before and after exhaustive treatment with purified RppH (15) to render it fully monophosphorylated. Measurements were performed in triplicate to allow the mean and standard deviation to be calculated. The detection of yeiP, trxB, yajQ, ydfG and yfcZ mRNA was aided by supplementing expression with a plasmid-borne copy of the gene. For PABLO analysis of efp and efp-C1A, plasmid-encoded transcripts were examined in a Δefp background.
The percentage of diphosphorylated 5′ ends for individual RNAs was measured by PACO analysis, as previously described (15). Briefly, total cellular RNA (5 μg) extracted from E. coli cells that had been grown to OD600 ≈ 0.3 was treated consecutively with Schizosaccharomyces pombe Pce1 + GTP, calf intestine alkaline phosphatase, and E. coli RppH. In parallel, monophosphorylated, diphosphorylated, and triphosphorylated forms of yeiP mRNA, synthesized by in vitro transcription, were added separately to total RNA (5 μg) from E. coli ΔyeiP cells and then treated with the same three enzymes. Finally, the monophosphorylated yeiP mRNA or R1.3-yeiP processing product generated in this manner was assayed by PABLO as described above, and the data for yeiP mRNA were analyzed by Monte Carlo simulation (15).
Purification of Cmk and Cmk-R41E bearing an amino-terminal hexahistidine tag
E. coli strain BL21(DE3) rne-131 Δrna (33) harboring either pPlacHis6-Cmk or pPlacHis6-CmkR41E was grown in 1.0 l of LB + ampicillin to OD600 = 0.8, at which time protein synthesis was induced with 1 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG) for 4 h at 30°C. Cells were harvested and resuspended in 10 ml of binding buffer (50 mM Tris–HCl pH 7.5, 200 mM NaCl, 2 mM DTT) containing 0.2 mM phenylmethane sulfonyl fluoride (PMSF) and 1 cOmplete mini EDTA-free protease inhibitor tablet (Roche). The cells were lysed by sonication, and the resulting lysate was centrifuged at 20 000 × g for 1 h to remove cellular debris. After incubating the lysate with 1 ml of BD TALON metal affinity resin (Clontech) for 1 h at 4°C with gentle rocking, the resin was washed with 5 ml of binding buffer containing 10 mM imidazole, and the protein was eluted with binding buffer containing 100–300 mM imidazole. Fractions containing pure Cmk or Cmk-R41E, as determined by SDS-PAGE and Coomassie Blue staining, were pooled and dialyzed into storage buffer (50 mM Tris–HCl pH 7.5, 200 mM NaCl, 2 mM DTT, 50% glycerol). Final protein concentrations were quantified by a Bradford assay.
To confirm the activity (or lack thereof) of the purified proteins, Cmk or Cmk-R41E was incubated with 1 mM CMP and 1 mM ATP in Cmk buffer (50 mM Tris–HCl pH 7.5, 0.5 mg/ml bovine serum albumin, 5 mM DTT, 2 mM MgCl2, 15 mM NaCl) for 1 h. The reaction was quenched with excess EDTA, and the products were analyzed by thin layer chromatography (see below).
Immunoblotting
To visualize intracellular protein levels by immunoblotting, whole cell extracts were separated by SDS-PAGE (15% polyacrylamide for RppH-FH and DapF-FLAG, 10% polyacrylamide for RNase E) and wet-transferred onto Hybond-P membranes (GE Healthcare). Proteins of interest were detected with monoclonal mouse anti-His antibodies (Clontech; for RppH-FH) or anti-FLAG M2 antibodies (Sigma; for DapF-FLAG) used at a dilution of 1:5000 or with polyclonal rabbit anti-RNase E antibodies (a gift from George Mackie, University of British Columbia) used at a dilution of 1:100 000. Bands were visualized with goat horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse antibodies (BioRad), used at a dilution of 1:100 000, in combination with the Supersignal West Pico Maximum Sensitivity Substrate (ThermoFisher Scientific).
Synthesis of RNA substrates and standards by in vitro transcription
Radiolabeled monophosphorylated CG(A)26 RNA was synthesized by in vitro transcription at 37°C in a 40-μl overnight reaction containing 40 mM Tris pH 7.9, 6 mM MgCl2, 20 mM DTT, 10 mM NaCl, 2 mM spermidine, 1 mM ATP, 7.5 mM CMP, 2.4 μM [α-32P] GTP (1.1 mCi/ml), 375 nM CGAA sense oligonucleotide, 375 nM CGAA antisense oligonucleotide, 1 unit of rRNasin, and 100 units of T7 RNA polymerase. Radiolabeled diphosphorylated CG(A)26 was synthesized under similar conditions except that the CMP was replaced with CDP (7.5 mM). In each case, the reaction products were separated by electrophoresis on a 16% polyacrylamide/7M urea gel, extracted from a gel fragment, and ethanol precipitated, and the 5′ phosphorylation state of the transcript was verified by alkaline hydrolysis and thin layer chromatography (see below).
Radiolabeled triphosphorylated AG(CU)13 was synthesized by in vitro transcription at 37°C in 40 mM Tris pH 7.9, 6 mM MgCl2, 20 mM DTT, 10 mM NaCl, 2 mM spermidine, 1.25 mM ATP, 1 mM CTP, 1 mM UTP, 2.4 μM [α-32P] GTP (1.1 mCi/ml), 375 nM AGCU sense oligonucleotide, 375 nM AGCU antisense oligonucleotide, 1 unit of rRNasin, and 100 units of T7 polymerase. Radiolabeled diphosphorylated AG(CU)13 was synthesized under similar conditions except that the ATP was replaced with ADP.
The triphosphorylated, diphosphorylated, and monophosphorylated yeiP-U2G standards for PACO were synthesized by in vitro transcription as previously described (15).
Analysis of RNA phosphorylation by alkaline hydrolysis and thin layer chromatography
To test for γ phosphate removal from triphosphorylated RNA by cytidylate kinase, radiolabeled triphosphorylated AG(CU)13 (1.9 μCi/ml, 0.4 nM) was incubated in Cmk buffer (50 mM Tris–HCl pH 7.5, 0.5 mg/ml bovine serum albumin, 5 mM DTT, 2 mM MgCl2, 15 mM NaCl) with 1 mM CMP and 200 nM Cmk or Cmk-R41E in a 20-μl reaction at 37°C for 1 h. To test for β phosphate addition to monophosphorylated RNA by cytidylate kinase, the reactions instead contained radiolabeled monophosphorylated CG(A)26 (1.9 μCi/ml, 0.4 nM) and 1 mM ATP as substrates. In each case, the reaction was stopped with 20 mM EDTA, after which 5 μl was subjected to electrophoresis on a 13.5% polyacrylamide/7M urea gel to confirm RNA integrity, and 10 μl was analyzed by alkaline hydrolysis and thin layer chromatography. Alkaline hydrolysis was accomplished by treatment with 100 mM NaOH for 15 min at 95°C, followed by neutralization with 1 M formic acid. The hydrolysis products (400 dpm) were spotted onto prewashed PEI Cellulose F TLC plates (Millipore) and developed in a glass chamber with 0.3 M KH2PO4 as the mobile phase. Radioactive spots on the TLC plates were visualized using a Typhoon Trio imager (GE Healthcare).
RESULTS
Screening for mutants impaired for 5′-end-dependent mRNA decay
Previous studies have demonstrated the importance of the RNA pyrophosphohydrolase RppH and its ancillary factor DapF in the 5′-end-dependent pathway for mRNA degradation in E. coli (10,21). To identify other proteins that might influence this decay pathway, we screened a library of E. coli mutants for defects in 5′-end-dependent degradation. The library chosen for this purpose was the Keio collection, which contains individual deletions of almost every non-essential gene in E. coli K-12 (25). The reporter used to screen this library was a plasmid-borne yeiP-GFP translational fusion, chosen because the rapid degradation of yeiP mRNA is controlled by its 5′ untranslated region and requires both RppH and RNases E and G (10,34,35). The 3,985 strains of the Keio collection were each transformed with the reporter plasmid, and the GFP fluorescence of the transformants was compared quantitatively in 96-well plates. In particular, we sought E. coli mutants in which yeiP-GFP expression was elevated, as this would be the expected result of abolishing the synthesis of proteins that facilitate RppH-dependent degradation and because other mechanisms by which gene knockouts could increase reporter expression were expected to be uncommon. Among the deleted genes with the desired phenotype was one that was unanticipated: cmk, which encodes the pyrimidine recycling enzyme cytidylate kinase (2.3-fold increase in GFP fluorescence per cell; Figure 1A). Other knockouts with a qualitatively similar phenotype were of genes (rppH and dapF) whose protein products were already known to be important for 5′-end-dependent mRNA degradation.
Figure 1.
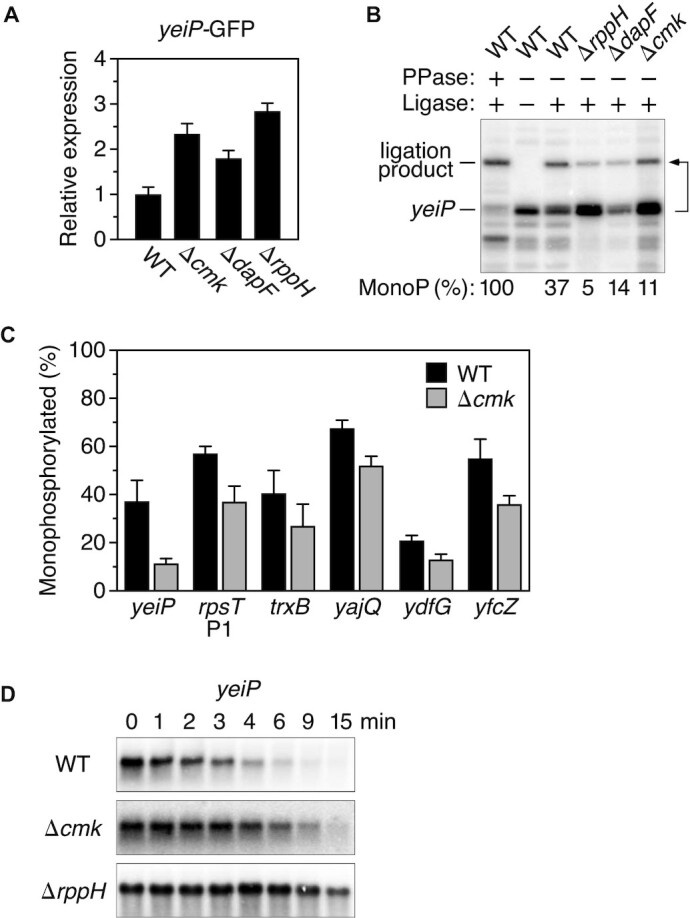
Influence of cytidylate kinase on the phosphorylation state and decay rate of A-initiated transcripts in E. coli. (A) Effect of gene deletions on the expression of a yeiP-GFP reporter. Reporter gene expression was calculated from the ratio of cellular GFP fluorescence to cell density and normalized to reporter expression in wild-type cells (WT). Each value is the average of three biological replicates. Error bars correspond to standard deviations. (B) Effect of gene deletions on the 5′ phosphorylation state of yeiP mRNA. The 5′ phosphorylation state of yeiP mRNA was analyzed by using a splinted ligation assay (PABLO) to determine the percentage of 5′ ends that were monophosphorylated (MonoP) in an isogenic set of E. coli strains. This percentage was calculated by normalizing the ligation yield to that of fully monophosphorylated yeiP mRNA generated by exhaustive pretreatment of cellular RNA with an excess of purified E. coli RppH (PPase), as the ligation of monophosphorylated 5′ ends generally is incomplete. The bent arrow beside the blot leads from the RNA substrate to its ligation product. A representative experiment is shown. (C) Effect of a cmk gene deletion on the 5′ phosphorylation state of A-initiated mRNAs in E. coli. The percentage of monophosphorylated 5′ ends for each cellular transcript was determined by PABLO, as in (B). Each value is the average of three biological replicates. Error bars correspond to standard deviations. (D) Effect of gene deletions on the decay rate of yeiP mRNA. The decay of yeiP mRNA in an isogenic set of E. coli strains was monitored as a function of time after inhibiting transcription with rifampicin. Representative experiments are shown.
Phenotype of the Δcmk mutant
To validate the influence of cmk on RppH-dependent mRNA decay, we transferred the Δcmk::kan allele from the Keio strain to E. coli strain BW25113 by P1 transduction and excised the kan marker to create an in-frame deletion of all but seven codons of the chromosomal cmk gene (25). We then examined the effect of cmk on the percentage of wild-type yeiP transcripts that bore a 5′-terminal monophosphate as a consequence of RppH-mediated phosphate removal. This was accomplished by PABLO (Phosphorylation Assay By Ligation of Oligonucleotides), a quantitative assay based on the ability of monophosphorylated but not triphosphorylated or diphosphorylated RNA 5′ termini to undergo splinted ligation to a DNA oligonucleotide (9,31). These measurements revealed that the steady-state percentage of yeiP 5′ ends that were monophosphorylated fell from 37 ± 9% in the cmk+ parent strain to 11 ± 2% in the isogenic Δcmk strain, a reduction similar in magnitude to that observed in the absence of DapF but smaller than that in cells lacking RppH (Figure 1B; Supplementary Table S1A). The 5′ phosphorylation state of other mRNAs targeted by RppH (10,20) was similarly affected (Figure 1C; Supplementary Table S1A).
In principle, the reduction in the steady-state percentage of yeiP transcripts that were monophosphorylated could result either from the slower formation or the more rapid breakdown of this decay intermediate in Δcmk cells. To distinguish between these possibilities, we determined the effect of this mutation on the decay rate of yeiP mRNA after treating cells with rifampicin to block further RNA synthesis (Figure 1D; Supplementary Table S2). In Δcmk cells, yeiP mRNA decayed half as fast as in wild-type cells (half-life of 2.9 ± 0.2 min in the Δcmk strain versus 1.4 ± 0.1 min in the isogenic cmk+ strain). The finding that the degradation of yeiP mRNA is impeded in the Δcmk mutant suggests that this genetic lesion diminishes the percentage of yeiP transcripts that are monophosphorylated by inhibiting the production of this intermediate rather than by hastening its destruction.
Origin of the Δcmk phenotype
The observed phenotype of the Δcmk mutant could have various possible origins. First, it might result from abolishing the catalytic activity of cytidylate kinase, which mediates the reaction of (deoxy)cytidine monophosphate ((d)CMP) with ATP to generate (d)CDP and ADP (36). Alternatively, it might be a consequence of eliminating the physical presence of cytidylate kinase if this protein structurally assists another enzyme important for converting 5′ ends to monophosphates, much as DapF is thought to do. Finally, it might be an indirect result of impairing translation by reducing expression of the ribosomal protein S1 gene rpsA, some promoters of which are located within the deleted cmk coding region (37).
To test these possibilities, two new mutations were created in the chromosomal cmk gene (Figure 2A). One was an in-frame deletion of codons 95–142, a deletion large enough to disrupt the structure and catalytic activity of cytidylate kinase but small enough to avoid deleting any of the known rpsA promoters. The other was a point mutation (R41E) in an active-site arginine residue that interacts with the α phosphate of both a substrate and a product of cytidylate kinase (CMP and CDP) (38,39). This residue is conserved among diverse nucleoside monophosphate kinases (36,38) and is required for catalytic activity (Supplementary Figure S1). Both of these mutations phenocopied the effect of the Δcmk lesion on the phosphorylation state and half-life of yeiP mRNA and could be complemented in trans by a wild-type cmk gene on a plasmid (Figure 2B, C; Supplementary Tables S1B and S2). These findings indicate that the catalytic function of cytidylate kinase is important for the conversion of 5′ triphosphates to monophosphates and that this protein does not merely play a structural role in this process. In addition, they rule out the alternative interpretation that was based on reduced rpsA expression. The qualitatively similar effect of the cmk-R41E mutation on the phosphorylation state and half-life of ydfG mRNA (Supplementary Figure S2; Supplementary Tables S1C and S2), another E. coli transcript known to be degraded by a mechanism dependent on RppH and RNases E and G (20), suggests that the impact of cytidylate kinase activity on this degradation pathway is likely to be widespread.
Figure 2.
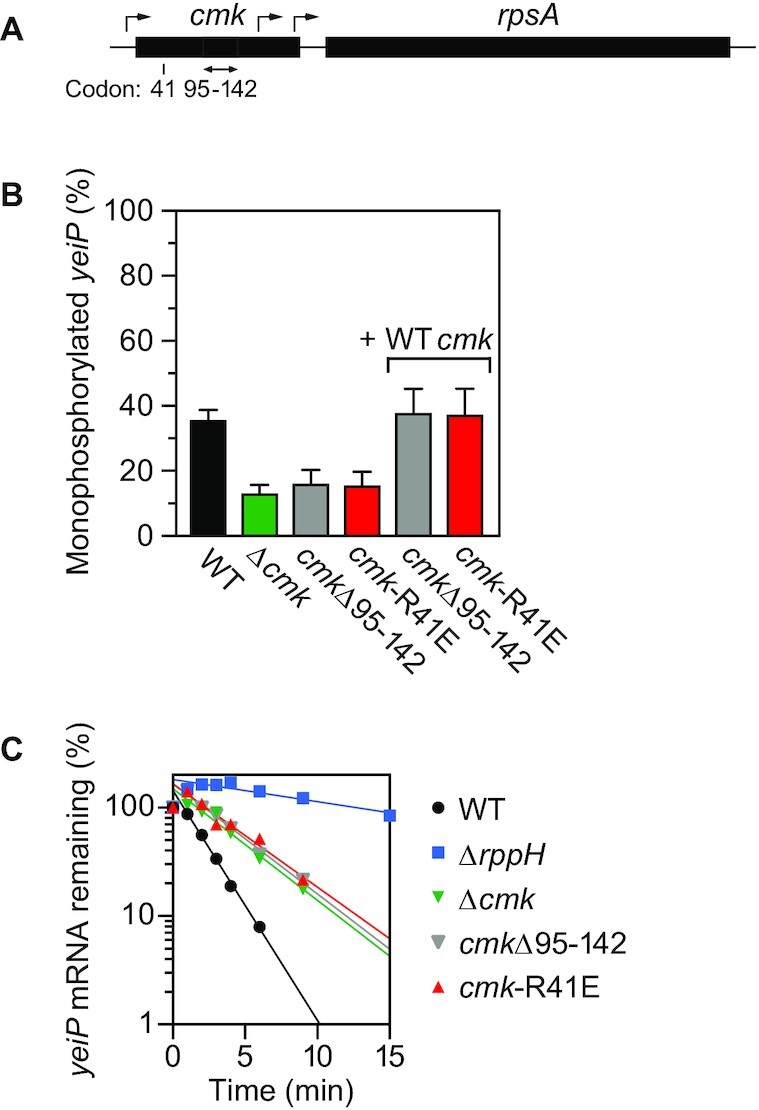
Importance of the catalytic activity of cytidylate kinase for its effect on mRNA degradation. (A) Chromosomal cmk-rpsA locus in E. coli. The location of the cmk-R41E codon substitution is indicated. Black rectangles, cmk and rpsA genes. Bent arrows, transcription initiation sites (37). Double-headed arrow, cmk gene segment spanning codons 95–142. (B) Effect of cmk mutations on the phosphorylation state of yeiP mRNA. The percentage of monophosphorylated yeiP 5′ ends was determined by PABLO, as in Figure 1. + WT cmk, complementation of cmkΔ95–142 and cmk-R41E with a plasmid-borne copy of the wild-type cmk gene. Each value is the average of three biological replicates. Error bars correspond to standard deviations. (C) Effect of cmk mutations on the decay rate of yeiP mRNA. The decay of yeiP mRNA in an isogenic set of E. coli strains was monitored as a function of time after inhibiting transcription with rifampicin, plotted semilogarithmically, and analyzed by linear regression. Representative experiments are shown.
Mechanism by which cytidylate kinase assists the generation of 5′ monophosphates
In its capacity as a pyrimidine recycling enzyme, cytidylate kinase catalyzes the production of (d)CDP by transferring the γ phosphate of ATP to (d)CMP, prompting us to ask how this activity might be related to the conversion of triphosphorylated RNA to monophosphorylated RNA. Recent evidence indicates that 5′ phosphate removal from primary transcripts occurs sequentially in E. coli, with the γ phosphate released by an unidentified triphosphatase to produce a diphosphorylated intermediate from which RppH removes the β phosphate, thereby generating a monophosphorylated RNA vulnerable to attack by RNase E (15). These observations raised the tantalizing possibility that cytidylate kinase might assist the production of monophosphorylated RNA by catalyzing the transfer of the γ phosphate of A-initiated transcripts, such as yeiP and ydfG, to (d)CMP so as to generate the diphosphorylated intermediate on which RppH acts.
To determine whether cytidylate kinase can react not only with ATP but also with the 5′-terminal nucleotide of A-initiated transcripts, we added this enzyme or a catalytically inactive mutant (R41E) to a mixture of CMP and triphosphorylated AG(CU)13 that had been radiolabeled site-specifically between the first two nucleotides. The 5′ phosphorylation state of the RNA reaction product was then examined by alkaline hydrolysis, thin layer chromatography, and autoradiography to detect the 5′-terminal nucleotide. Under these conditions, cytidylate kinase but not its catalytically inactive counterpart was able to convert a significant fraction of the RNA 5′ triphosphates to diphosphates in vitro (Supplementary Figure S3).
We then tested the importance of this enzyme for the production of diphosphorylated RNA 5′ ends in vivo. This was accomplished by PACO (Phosphorylation Assay by Capping Outcome), an assay that quantifies the percentage of 5′ termini that are diphosphorylated on the basis of the ability of the yeast guanylyltransferase Pce1 to preferentially cap such ends (15). However, instead of declining in cells lacking cytidylate kinase activity, the percentage of yeiP 5′ termini that were diphosphorylated increased from 58 ± 17% in cmk+ cells to 94 ± 27% in cmk-R41E cells (Figure 3A; Supplementary Table S3A–D). Thus, rather than enabling the production of diphosphorylated RNA 5′ ends in E. coli, cytidylate kinase appears to facilitate their subsequent conversion to 5′ monophosphates. As a result, the absence of this activity allows diphosphorylated 5′ ends to accumulate. Therefore, even though cytidylate kinase is capable of transferring the γ phosphate of A-initiated transcripts to CMP in vitro, it does not contribute appreciably to the generation of diphosphorylated 5′ ends in vivo, presumably because another cellular mechanism for creating such ends predominates.
Figure 3.
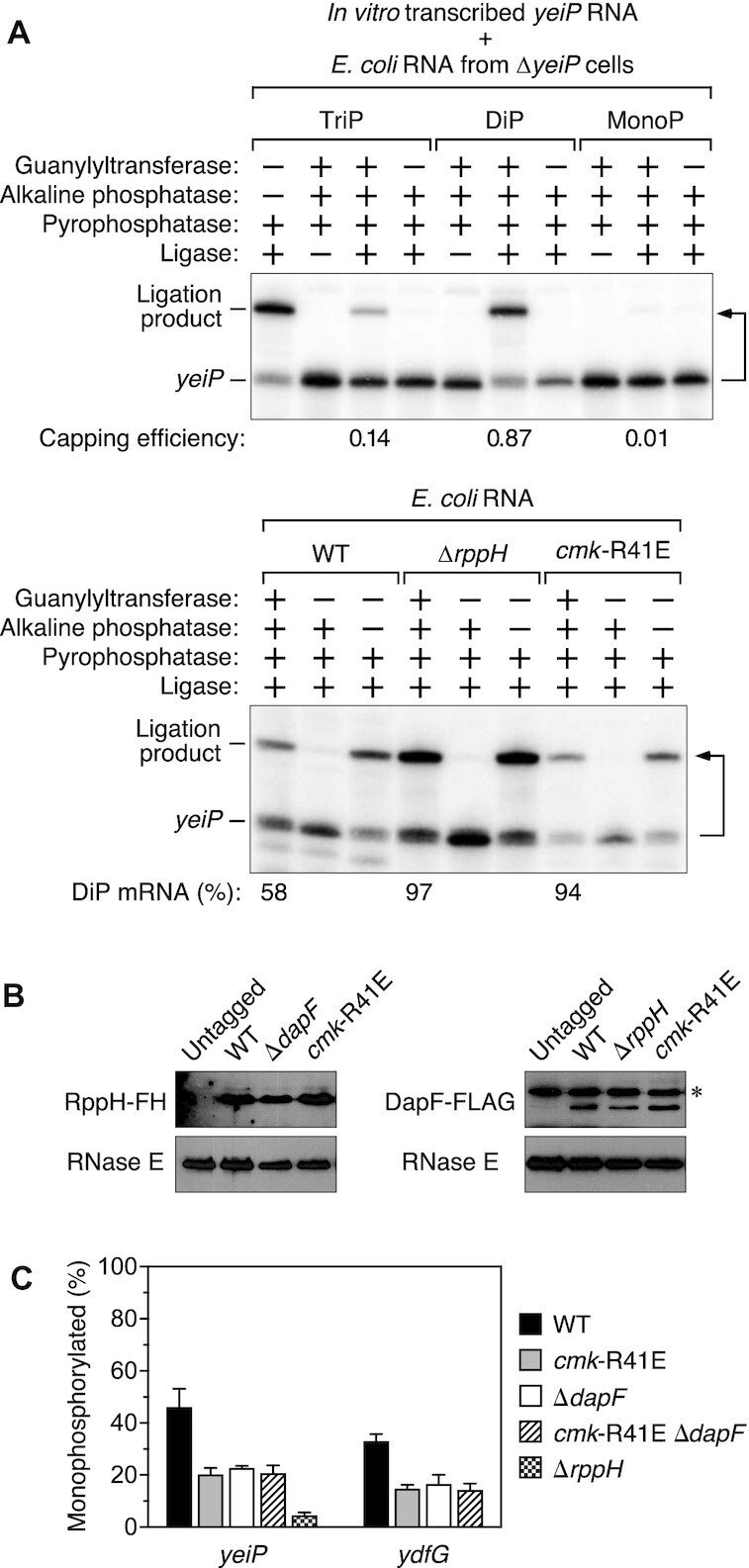
Mechanism by which cytidylate kinase influences the degradation of A-initiated transcripts in E. coli. (A) Increased percentage of yeiP 5′ ends that are diphosphorylated in cells lacking cytidylate kinase activity. The 5′ phosphorylation state of yeiP mRNA was analyzed by using a capping assay (PACO) to determine the percentage of yeiP 5′ ends that were diphosphorylated in an isogenic set of E. coli strains (bottom) versus a set of triphosphorylated (TriP), diphosphorylated (DiP) and monophosphorylated (MonoP) yeiP standards synthesized by in vitro transcription (top). Capping by the guanylyltransferase Pce1 protects the 5′-terminal phosphates of diphosphorylated RNA (and to a lesser extent triphosphorylated RNA) from removal by alkaline phosphatase; subsequent decapping by the pyrophosphatase RppH generates a monophosphorylated 5′ end able to undergo splinted ligation to an oligonucleotide (15). A bent arrow leads from each RNA to its ligation product. The calculated percentage of diphosphorylated (DiP) yeiP 5′ ends in each strain, as determined by comparison to the yeiP standards (Supplementary Tables S3A–D), is indicated below the lane in which the cellular RNA underwent a complete series of four consecutive enzyme treatments. Representative experiments are shown. (B) Invariant concentration of RppH and DapF in cells lacking cytidylate kinase activity. Equal amounts of total cellular protein extracted from an isogenic set of E. coli strains were analyzed by immunoblotting. RppH and DapF were detected via a chromosomally encoded epitope tag added to the carboxyl end of the protein (FLAG-hexahistidine (FH) or FLAG, respectively). RNase E served as an internal standard. ✻, untagged E. coli protein bound nonspecifically by the anti-FLAG antibodies. Representative experiments are shown. (C) Epistatic effect of cytidylate kinase and DapF on the phosphorylation state of yeiP and ydfG mRNA. The percentage of monophosphorylated yeiP and ydfG 5′ ends in an isogenic set of E. coli strains was determined by PABLO, as in Figure 1. Each value is the average of three biological replicates. Error bars correspond to standard deviations.
Subsequent immunoblotting assays showed no change in the cellular concentration of either RppH or its ancillary factor DapF in cmk-R41E cells versus cmk+ cells (Figure 3B), thus eliminating the possibility that cytidylate kinase stimulates the production of monophosphorylated 5′ ends by altering the concentration of these enzymes. However, the similar effects of cytidylate kinase and DapF on the conversion of diphosphorylated RNA to monophosphorylated RNA in E. coli led us to consider whether cytidylate kinase might influence the specific activity of RppH by acting directly or indirectly through DapF. If so, the effects of cmk and dapF mutations should be epistatic. To test this hypothesis, we compared the phosphorylation state of yeiP and ydfG mRNA in a cmk-R41E ΔdapF double mutant to their phosphorylation state in cells bearing a mutation in only one or the other of these genes. In each case, the percentage of 5′ ends that were monophosphorylated was no lower in the double mutant than in either single mutant (Figure 3C; Supplementary Table S1C). This epistasis suggests that cytidylate kinase and DapF somehow act together to stimulate the generation of monophosphorylated RNA 5′ ends by RppH.
Distinct effect of cytidylate kinase on C-initiated transcripts
All of the E. coli transcripts discussed so far begin with a 5′-terminal A. In every case, the absence of cytidylate kinase activity impaired conversion of the 5′ end to a monophosphate. Interestingly, we observed that this mutation had the opposite effect on two transcripts (efp and rpsT P2) that instead begin with C (40), as confirmed by rapid amplification of cDNA ends (5′ RACE). Rather than declining, the percentage of efp and rpsT P2 transcripts that were monophosphorylated increased substantially in cmk-R41E cells (Figure 4A; Supplementary Table S1D) even though both are degraded by a mechanism dependent on RppH (Figure 4B; Supplementary Table S2) and RNase E (10). Moreover, changing the 5′-terminal nucleotide of efp mRNA from C to A reversed the effect of the cmk-R41E mutation, enabling this genetic lesion to decrease rather than increase the percentage of monophosphorylated 5′ ends (Figure 4A; Supplementary Table S1D). Thus, the observed phenotypic disparity is governed by the identity of the 5′-terminal nucleotide. Nevertheless, the half-lives of the efp and rpsT P2 transcripts were slightly prolonged in cmk-R41E cells (Figure 4B; Supplementary Table S2), as had been observed for the A-initiated yeiP and ydfG transcripts in those cells.
Figure 4.
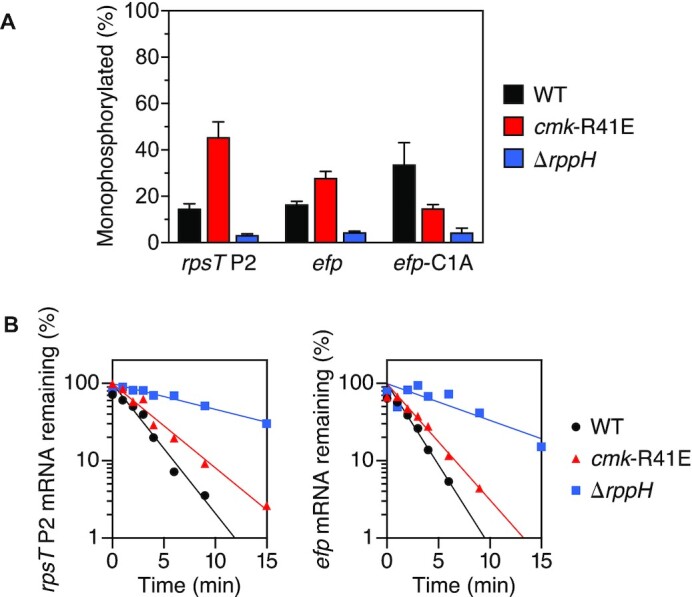
Influence of cytidylate kinase on the phosphorylation state and lifetime of C-initiated transcripts in E. coli. (A) Effect of cytidylate kinase on the phosphorylation state of rpsT P2, efp and efp-C1A mRNA. The percentage of rpsT P2, efp, and efp-C1A 5′ ends that were monophosphorylated in an isogenic set of E. coli strains was determined by PABLO, as in Figure 1. Each value is the average of three biological replicates. Error bars correspond to standard deviations. (B) Effect of cytidylate kinase on the decay rate of rpsT P2 and efp mRNA. The decay of rpsT P2 and efp mRNA in an isogenic set of E. coli strains was monitored as a function of time after inhibiting transcription with rifampicin, plotted semilogarithmically, and analyzed by linear regression. Representative experiments are shown.
Hypothetically, the distinct effect of cytidylate kinase on the phosphorylation state of C-initiated mRNAs could be explained if the enzyme was able to convert the 5′ end of those transcripts from a monophosphate to a diphosphate, much as it can convert CMP to CDP by transferring a phosphate from ATP. To determine whether E. coli cells contain an enzyme activity that can convert C-initiated RNA 5′ termini from a monophosphate to a diphosphate, we modified the 5′ untranslated region of yeiP mRNA to introduce a bacteriophage T7 stem-loop (R1.3) whose cleavage by RNase III is known to generate a 3′ product bearing a monophosphorylated C at the 5′ end (28) (Figure 5A). However, as determined by PABLO, the resulting 5′ ends remained fully monophosphorylated in both cmk+ and cmk-R41E cells, even when the rppH gene was deleted to stabilize any diphosphorylated intermediates that might have been formed (Figure 5B; Supplementary Table S1E). Consistent with this observation, no diphosphorylated 5′ ends could be detected on the RNase III-cleaved processing product by PACO (Figure 5C). The inability of cytidylate kinase to convert the 5′ monophosphate of a C-initiated RNA substrate to a diphosphate was confirmed in vitro by treating a mixture of monophosphorylated CG(A)26 and an excess of ATP with this enzyme. No diphosphorylated RNA product was generated (Supplementary Figure S4), whereas CDP was produced when the monophosphorylated RNA substrate was replaced with CMP (Supplementary Figure S1).
Figure 5.
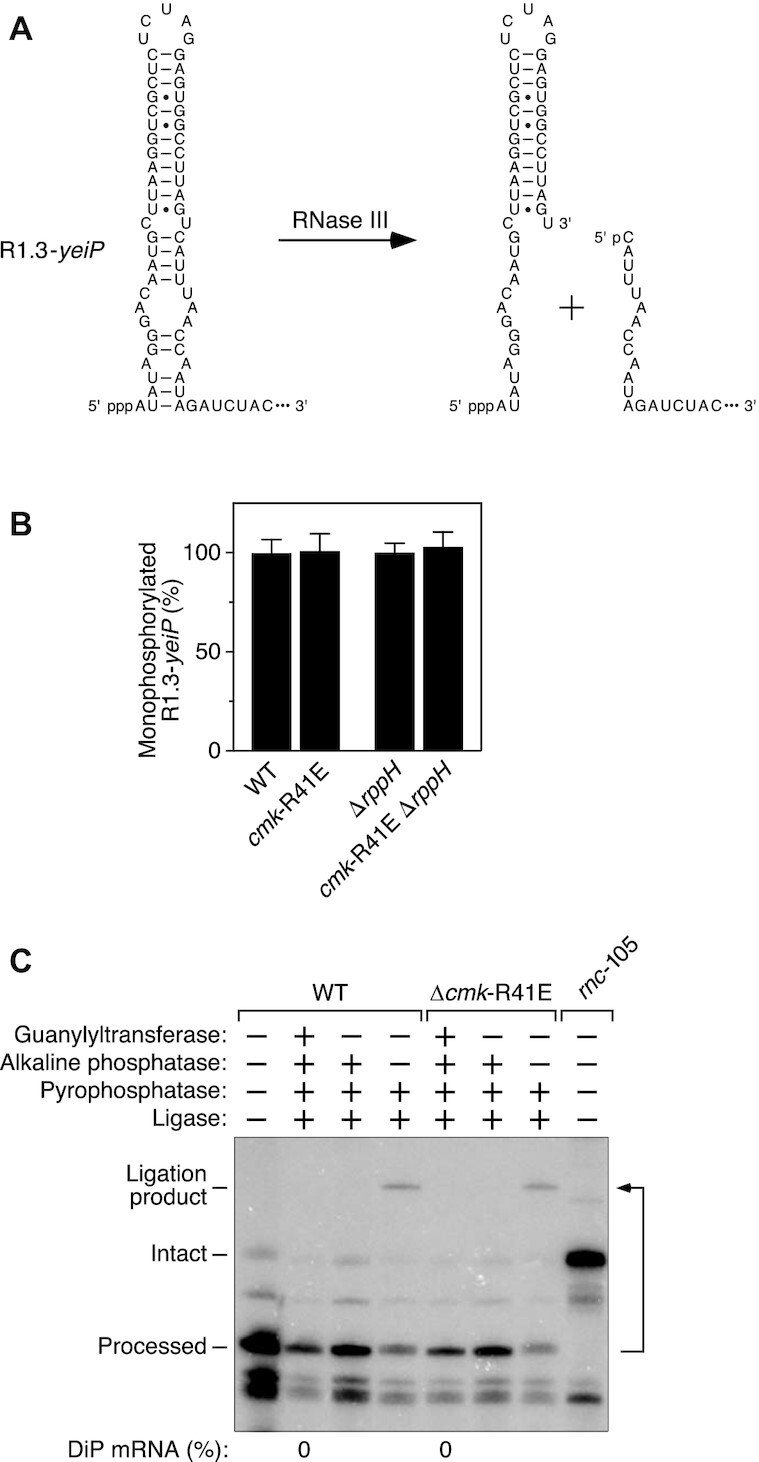
Inability of cytidylate kinase to phosphorylate a C-initiated E. coli RNA bearing a 5′ monophosphate. (A) RNase III cleavage within the 5′ untranslated region of R1.3-yeiP mRNA to generate a processed product that is monophosphorylated and begins with C. Base pairs are indicated by horizontal lines. The remainder of each RNA (not shown) is represented by an ellipsis. The natural 5′ untranslated region of wild-type yeiP mRNA begins eight nucleotides (ACACACAC) downstream of the sequence shown. p, phosphate. (B) Invariant percentage of processed R1.3-yeiP 5′ ends that are monophosphorylated in cells lacking cytidylate kinase activity. The phosphorylation state of the processed 5′ end of R1.3-yeiP RNA in an isogenic set of E. coli strains was analyzed by PABLO, as in Figure 1. Each value is the average of three biological replicates. Error bars correspond to standard deviations. (C) Absence of processed R1.3-yeiP 5′ ends that are diphosphorylated. The phosphorylation state of the processed 5′ end of R1.3-yeiP RNA in an isogenic pair of E. coli strains containing or lacking cytidylate kinase activity was analyzed by PACO, as in Figure 3. No processed 5′ ends capable of being capped by the guanylyltransferase Pce1 were detected in either strain, as evidenced by the absence of a splinted ligation product after consecutive treatment with all four enzymes (DiP mRNA: 0%). As expected, the R1.3-yeiP transcript did not undergo processing by RNase III in a mutant strain (rnc-105) lacking that endonuclease. A representative experiment is shown.
Another potential explanation for the distinct behavior of C-initiated transcripts is that the absence of cytidylate kinase activity might allow CMP to accumulate to such a high cellular concentration that it can outcompete CTP for incorporation by RNA polymerase at the 5′ end of nascent transcripts. Indeed, one known consequence of mutating the cmk gene is to elevate the level of CMP by impairing its conversion to CDP (37). To investigate this possibility, we examined whether the cmk-R41E mutation could increase the percentage of C-initiated transcripts that were monophosphorylated in ΔrppH cells, as cells lacking RppH would have no mechanism for generating such 5′ ends other than 5′-terminal incorporation of CMP into newly synthesized transcripts. Consistent with this interpretation, the percentage of efp 5′ ends that were monophosphorylated increased from 4.8 ± 1.4% in ΔrppH cells to 11.9 ± 0.9% in cmk-R41E ΔrppH cells, as determined by PABLO (Figure 6A; Supplementary Table S1F). By contrast, the same small fraction of efp-C1A 5′ ends were monophosphorylated in both strains (5.5 ± 0.4% versus 5.4 ± 0.1%), as expected in the absence of RppH for an enzyme mutation that does not significantly perturb AMP levels in E. coli. Moreover, by PABLO analysis of RNA extracted at time intervals after arresting transcription, the monophosphorylated efp transcripts were found to be lost from cmk-R41E ΔrppH cells much faster than their tri/diphosphorylated counterparts (measured half-lives of 1.1 ± 0.1 min versus 12.2 ± 2.3 min, respectively, for ligated and unligated efp mRNA; Figure 6B; Supplementary Table S2), as anticipated for two RNA pools that are metabolically isolated from one another in a strain lacking RppH, whose absence prevents resupply of the short-lived form from a long-lived precursor.
Figure 6.
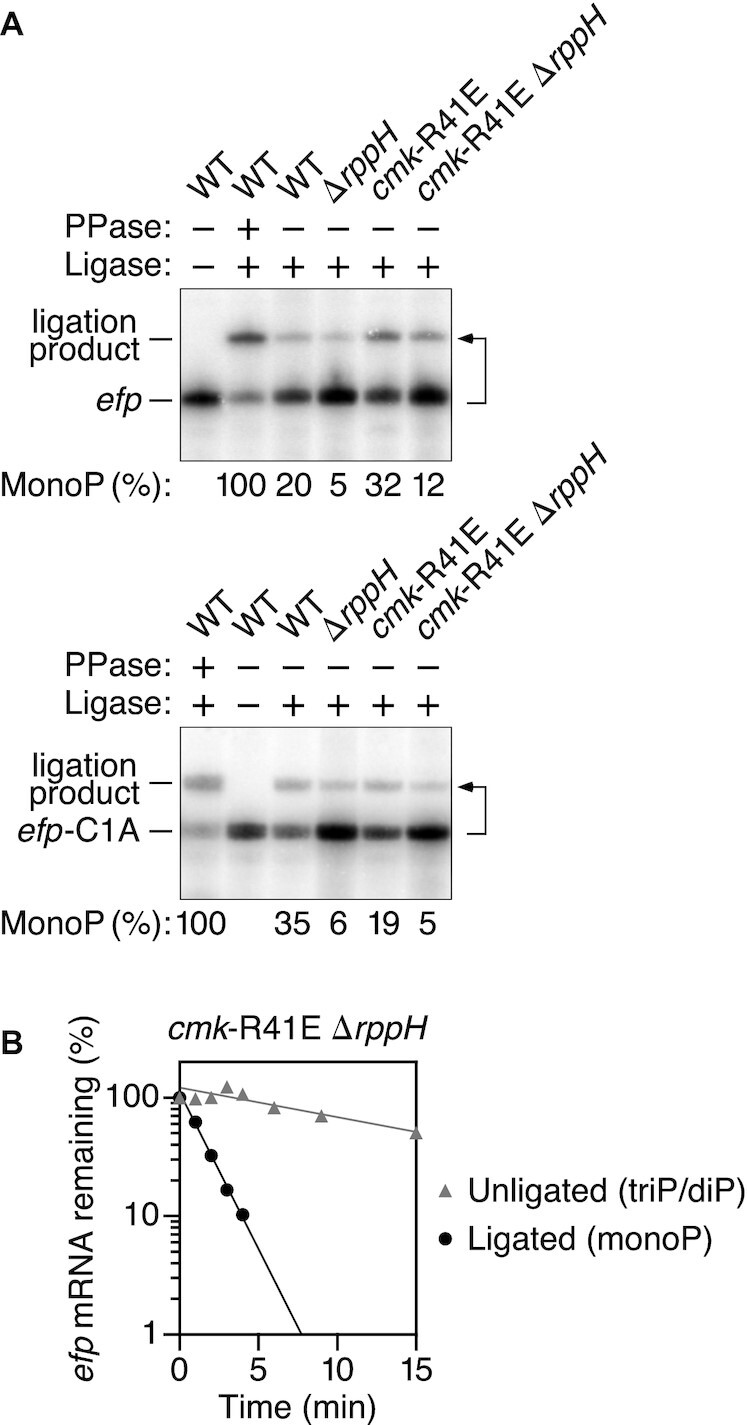
Increased synthesis of monophosphorylated C-initiated transcripts in E. coli cells lacking cytidylate kinase activity. (A) Effect of cytidylate kinase on the percentage of efp 5′ ends that were monophosphorylated in E. coli cells lacking RppH. The phosphorylation state of the 5′ end of efp mRNA (top) and efp-C1A mRNA (bottom) in an isogenic set of E. coli strains was analyzed by PABLO, as in Figure 1, to quantify 5′ termini that were monophosphorylated (MonoP). A bent arrow leads from each RNA to its ligation product. PPase, pyrophosphatase. Representative experiments are shown. (B) Difference in the decay rate of monophosphorylated versus diphosphoryated/triphosphoryated efp mRNA in E. coli cells lacking both cytidylate kinase activity and RppH. Total cellular RNA extracted at time intervals after inhibiting transcription with rifampicin was analyzed by PABLO, as in Figure 1, to differentiate between efp 5′ ends that were monophosphorylated (ligated) or diphosphoryated/triphosphoryated (unligated), and the resulting band intensities were plotted semilogarithmically and analyzed by linear regression. Representative experiments are shown.
DISCUSSION
Prior studies have established the importance of an RNA triphosphatase, the RNA pyrophosphohydrolase RppH, and the ancillary factor DapF in the stepwise conversion of triphosphorylated to monophosphorylated 5′ ends as a prelude to mRNA degradation in E. coli (10,15,21). However, the identity of the RNA triphosphatase has not been determined, and the possible contribution of other proteins to the generation of 5′ monophosphates has not previously been investigated. Our findings have now implicated cytidylate kinase in this critical regulatory process by showing that it functions in two distinct ways to influence the percentage of mRNA 5′ ends that are monophosphorylated in E. coli (Figure 7). For A-initiated transcripts and probably others, it accelerates the generation of monophosphorylated 5′ ends by acting in conjunction with DapF to stimulate β phosphate removal by RppH. For C-initiated mRNAs, cytidylate kinase appears to suppress the direct synthesis of monophosphorylated transcripts by minimizing the incorporation of cytidine monophosphate during transcription initiation by RNA polymerase. Key to these effects is the catalytic activity of cytidylate kinase and not the mere presence of the protein.
Figure 7.
Cellular functions of cytidylate kinase. (Left) Role of cytidylate kinase (Cmk) in the conversion of CMP to CDP. (Center) Importance of the catalytic activity of cytidylate kinase for enabling DapF to potentiate RppH-mediated 5′ phosphate removal from diphosphorylated RNA as a prelude to rapid endonucleolytic cleavage of the monophosphorylated RNA product by RNase E. Represented by a short red arrow, this stimulatory effect of cytidylate kinase presumably is a direct or indirect consequence of the depletion of one of its substrates or the accumulation of one of its products. (Right) Direct synthesis of monophosphorylated C-initiated transcripts by RNA polymerase when the CMP that accumulates in cells lacking cytidylate kinase activity is incorporated into RNA during transcription initiation. ppp, triphosphate; pp, diphosphate; p, monophosphate. N, RNA nucleotide; C, cytidine nucleoside. Ellipsis, 3′-terminal RNA segment.
In principle, cytidylate kinase could increase the percentage of A-initiated transcripts that are monophosphorylated either by functioning as an RNA triphosphatase that reacts with primary transcripts to generate diphosphorylated 5′ ends, by facilitating the conversion of the resulting 5′ diphosphates to monophosphates, or by inhibiting the subsequent degradation of monophosphorylated RNA by RNase E. The last of these possibilities is contradicted by the observation that the decay of both yeiP and ydfG mRNA is slower rather than faster in cmk-R41E cells. Interestingly, cytidylate kinase, which catalyzes phosphoryl transfer from ATP to (d)CMP, is also able to transfer the γ phosphate of A-initiated transcripts to CMP in vitro, indicating that it cannot readily distinguish between ATP and the 5′-terminal nucleotide of triphosphorylated A-initiated RNA as substrates. However, it evidently does not contribute significantly to generating diphosphorylated 5′ ends in E. coli, as such ends are more abundant, not less abundant, in cells lacking cytidylate kinase activity. Moreover, cytidylate kinase does not itself appear to possess RNA pyrophosphohydrolase activity, as its mutational inactivation does not reduce the low level of monophosphorylated 5′ ends in E. coli cells lacking RppH (Figure 6A; Supplementary Table S1F). Instead, this enzyme seems to potentiate the activity of RppH as an RNA pyrophosphohydrolase, thereby facilitating β phosphate removal from diphosphorylated RNA. The failure of the genetic screen to identify a physiologically consequential RNA triphosphatase suggests either that this triphosphatase is encoded by an essential gene not represented among the Keio mutants or that multiple enzymes contribute redundantly to γ phosphate removal from RNA in E. coli.
The mechanism by which cytidylate kinase accelerates β phosphate removal by RppH has been investigated by testing for epistasis. The non-additive effects of mutations in cmk and dapF suggest that these two proteins somehow work together to stimulate RppH activity. However, unlike DapF, which acts by forming a complex with RppH (21), the mere presence of the cytidylate kinase protein is not sufficient for it to hasten the conversion of 5′ diphosphates to monophosphates in E. coli, and there currently is no evidence for complex formation between cytidylate kinase and either DapF or RppH (41). Instead, the catalyic activity of this kinase is required for its effect on the phosphorylation state of cellular transcripts. Because cytidylate kinase does not appear to influence the cellular concentration of either RppH or DapF, we hypothesize that a nucleotide substrate or product of cytidylate kinase affects the ability of DapF to activate RppH, either directly or indirectly.
In contrast, cytidylate kinase has the opposite effect on the 5′ ends of C-initiated transcripts such as efp and rpsT P2, reducing their steady-state level of monophosphorylation by an entirely different mechanism that is RppH-independent. By catalyzing the conversion of CMP to CDP, cytidylate kinase shrinks the pool of CMP available to compete with CTP for 5′-terminal incorporation into RNA during transcription initiation by RNA polymerase. In the absence of this kinase, the molar ratio of CMP to CTP rises 40-fold in E. coli (37), greatly increasing the potential for CMP assimilation at the 5′ end of nascent C-initiated transcripts. In principle, other nucleoside monophosphate kinases—the products of the essential adk, gmk and pyrH genes—might similarly inhibit NMP incorporation at the 5′ end of A-, G-, and U-initiated transcripts, respectively. An alternative explanation, that cytidylate kinase may be able to convert the 5′ end of C-initiated RNAs from a monophosphate to a diphosphate, is excluded by evidence to the contrary both in vitro and in vivo. The inability of cytidylate kinase to react with C-initiated RNAs that are monophosphorylated is consistent with the structure of this enzyme bound to CMP, whose 3′ hydroxyl is buried deep in the active site (39), sterically precluding the binding of a CMP-initiated RNA polymer there. By contrast, the 3′ hydroxyl of ADP bound to cytidylate kinase is solvent-exposed (PDB ID code 3AKC), a finding congruous with the ability of this enzyme to react in vitro not only with ATP but also with the ATP-like 5′ end of triphosphorylated transcripts that begin with adenosine.
Despite the elevated level of monophosphorylated efp and rpsT P2 5′ ends in cmk-R41E cells, the lifetimes of these C-initiated transcripts are prolonged, much like the half-lives of A-initiated transcripts. Though seemingly incompatible, these two observations can be reconciled in light of the fact that RppH stimulates the rapid decay of both of these mRNAs and the finding that, even in a cmk mutant, fewer than half of the efp and rpsT P2 5′ termini are monophosphorylated at steady state. Therefore, it appears that most efp and rpsT P2 transcripts in those cells are initially synthesized with a 5′ triphosphate that must subsequently undergo γ and β phosphate removal to trigger RNA degradation by RNase E. Since there is no reason to expect that the influence of cytidylate kinase on 5′ phosphate removal is limited to A-initiated transcripts, a majority of the 5′ ends of these C-initiated RNAs should be similarly impacted by the impediment to RppH-mediated deprotection caused by cmk mutations, resulting in a modest increase in the aggregate lifetime of all forms of each C-initiated transcript in cells lacking cytidylate kinase activity. Consistent with this explanation, the aggregate half-life of efp mRNA in cmk-R41E cells (2.0 ± 0.1 min; Figure 4B) is almost twice the half-life of the monophosphorylated form of this transcript (1.1 ± 0.1 min; Figure 6B).
Three metabolic enzymes are now known to influence rates of mRNA decay in E. coli: cytidylate kinase, the diaminopimelate epimerase DapF (21), and the glycolytic enzyme enolase (42,43). Whereas the latter two proteins form heteromeric complexes with enzymes involved in RNA degradation (RppH and RNase E, respectively (21–23,44–46)), the effect of cytidylate kinase depends on its catalytic activity. The ability of cytidylate kinase activity to influence the phosphorylation state of mRNA by two distinct mechanisms reveals the multifaceted regulatory impact of crosstalk between nucleotide metabolism and mRNA degradation. While the deterrent effect of this enzyme on 5′-terminal CMP incorporation during transcription initiation can readily be explained, how CMP phosphorylation enhances the capacity of DapF to stimulate the deprotection of RNA 5′ ends by RppH remains to be elucidated. Whatever the details of this signaling mechanism may be, it could enable rates of mRNA decay to adjust to fluctuations in cellular nucleotide levels.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dan Luciano for his assistance with in vitro transcription and immunoblotting, Kevin Belasco for help with statistical analysis, Thomas Bernhardt and Andrew Darwin for plasmids and strains, Jane Hubbard and Victor Torres for their 96-pin replicators, and George Mackie for RNase E antibodies.
Author contributions: M.P.H. and J.G.B. designed the experiments, M.P.H. conducted the experiments, and M.P.H. and J.G.B. interpreted the data and wrote the paper.
Contributor Information
Monica P Hui, Skirball Institute of Biomolecular Medicine, New York University School of Medicine, 540 First Avenue, New York, NY 10016, USA; Department of Microbiology, New York University School of Medicine, 430 E. 29th Street, New York, NY 10016, USA.
Joel G Belasco, Skirball Institute of Biomolecular Medicine, New York University School of Medicine, 540 First Avenue, New York, NY 10016, USA; Department of Microbiology, New York University School of Medicine, 430 E. 29th Street, New York, NY 10016, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [F32GM101962 to M.P.H., R01GM035769 to J.G.B.]. Funding for open access charge: NIH [R01GM035769].
Conflict of interest statement. None declared.
REFERENCES
- 1. Belasco J.G. All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat. Rev. Mol. Cell Biol. 2010; 11:467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hui M.P., Foley P.L., Belasco J.G.. Messenger RNA degradation in bacterial cells. Annu. Rev. Genet. 2014; 48:537–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McDowall K.J., Lin-Chao S., Cohen S.N.. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 1994; 269:10790–10796. [PubMed] [Google Scholar]
- 4. Lin-Chao S., Wong T.-T., McDowall K.J., Cohen S.N.. Effects of nucleotide sequence on the specificity of rne-dependent and RNase E-mediated cleavages of RNA I encoded by the pBR322 plasmid. J. Biol. Chem. 1994; 269:10797–10803. [PubMed] [Google Scholar]
- 5. Tock M.R., Walsh A.P., Carroll G., McDowall K.J.. The CafA protein required for the 5'-maturation of 16 S rRNA is a 5'-end-dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem. 2000; 275:8726–8732. [DOI] [PubMed] [Google Scholar]
- 6. Kaberdin V.R. Probing the substrate specificity of Escherichia coli RNase E using a novel oligonucleotide-based assay. Nucleic. Acids. Res. 2003; 31:4710–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Del Campo C., Bartholomäus A., Fedyunin I., Ignatova Z.. Secondary structure across the bacterial transcriptome reveals versatile roles in mRNA regulation and function. PLoS Genet. 2015; 11:e1005613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chao Y., Li L., Girodat D., Förstner K.U., Said N., Corcoran C., Śmiga M., Papenfort K., Reinhardt R., Wieden H.J.et al.. In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell. 2017; 65:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Celesnik H., Deana A., Belasco J.G.. Initiation of RNA decay in Escherichia coli by 5' pyrophosphate removal. Mol. Cell. 2007; 27:79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deana A., Celesnik H., Belasco J.G.. The bacterial enzyme RppH triggers messenger RNA degradation by 5' pyrophosphate removal. Nature. 2008; 451:355–358. [DOI] [PubMed] [Google Scholar]
- 11. Mackie G.A. Ribonuclease E is a 5'-end-dependent endonuclease. Nature. 1998; 395:720–723. [DOI] [PubMed] [Google Scholar]
- 12. Jiang X., Diwa A., Belasco J.G.. Regions of RNase E important for 5’-end-dependent RNA cleavage and autoregulated synthesis. J. Bacteriol. 2000; 182:2468–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Callaghan A.J., Marcaida M.J., Stead J.A., McDowall K.J., Scott W.G., Luisi B.F.. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005; 437:1187–1191. [DOI] [PubMed] [Google Scholar]
- 14. Garrey S.M., Blech M., Riffell J.L., Hankins J.S., Stickney L.M., Diver M., Hsu Y.H., Kunanithy V., Mackie G.A.. Substrate binding and active site residues in RNases E and G: role of the 5'-sensor. J. Biol. Chem. 2009; 284:31843–31850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Luciano D.J., Vasilyev N., Richards J., Serganov A., Belasco J.G.. A novel RNA phosphorylation state enables 5′ end-dependent degradation in Escherichia coli. Mol. Cell. 2017; 67:44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Foley P.L., Hsieh P.K., Luciano D.J., Belasco J.G.. Specificity and evolutionary conservation of the Escherichia coli RNA pyrophosphohydrolase RppH. J. Biol. Chem. 2015; 290:9478–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Richards J., Liu Q., Pellegrini O., Celesnik H., Yao S., Bechhofer D.H., Condon C., Belasco J.G.. An RNA pyrophosphohydrolase triggers 5′-exonucleolytic degradation of mRNA in Bacillus subtilis. Mol. Cell. 2011; 43:940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsieh P.K., Richards J., Liu Q., Belasco J.G.. Specificity of RppH-dependent RNA degradation in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8864–8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Piton J., Larue V., Thillier Y., Dorleans A., Pellegrini O., Li de la Sierra-Gallay I., Vasseur J.J., Debart F., Tisne C., Condon C.. Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8858–8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luciano D.J., Hui M.P., Deana A., Foley P.L., Belasco K.J., Belasco J.G.. Differential control of the rate of 5′-end-dependent mRNA degradation in Escherichia coli. J. Bacteriol. 2012; 194:6233–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee C.R., Kim M., Park Y.H., Kim Y.R., Seok Y.J.. RppH-dependent pyrophosphohydrolysis of mRNAs is regulated by direct interaction with DapF in Escherichia coli. Nucleic Acids Res. 2014; 42:12746–12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao A., Vasilyev N., Luciano D.J., Levenson-Palmer R., Richards J., Marsiglia W.M., Traaseth N.J., Belasco J.G., Serganov A.. Structural and kinetic insights into stimulation of RppH-dependent RNA degradation by the metabolic enzyme DapF. Nucleic Acids Res. 2018; 46:6841–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Q., Zhang D., Guan Z., Li D., Pei K., Liu J., Zou T., Yin P.. DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia coli. Nucleic Acids Res. 2018; 46:6880–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006; 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Edwards R.A., Keller L.H., Schifferli D.M.. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998; 207:149–157. [DOI] [PubMed] [Google Scholar]
- 27. Dinh T., Bernhardt T.G.. Using superfolder green fluorescent protein for periplasmic protein localization studies. J. Bacteriol. 2011; 193:4984–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dunn J.J., Studier F.W.. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 1983; 166:477–535. [DOI] [PubMed] [Google Scholar]
- 29. Chung C.T., Niemela S.L., Miller R.H.. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:2172–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chomczynski P. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal. Biochem. 1992; 201:134–139. [DOI] [PubMed] [Google Scholar]
- 31. Celesnik H., Deana A., Belasco J.G.. PABLO analysis of RNA: 5'-phosphorylation state and 5'-end mapping. Methods Enzymol. 2008; 447:83–98. [DOI] [PubMed] [Google Scholar]
- 32. Luciano D.J., Belasco J.G.. Analysis of RNA 5′ ends: phosphate enumeration and cap characterization. Methods. 2019; 155:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luciano D.J., Levenson-Palmer R., Belasco J.G.. Stresses that raise Np4A levels induce protective nucleoside tetraphosphate capping of bacterial RNA. Mol. Cell. 2019; 75:957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Richards J., Luciano D.J., Belasco J.G.. Influence of translation on RppH-dependent mRNA degradation in Escherichia coli. Mol. Microbiol. 2012; 86:1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Richards J., Belasco J.G.. Distinct requirements for 5'-monophosphate-assisted RNA cleavage by Escherichia coli RNase E and RNase G. J. Biol. Chem. 2016; 291:5038–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bucurenci N., Sakamoto H., Briozzo P., Palibroda N., Serina L., Sarfati R.S., Labesse G., Briand G., Danchin A., Bărzu O.et al.. CMP kinase from Escherichia coli is structurally related to other nucleoside monophosphate kinases. J. Biol. Chem. 1996; 271:2856–2862. [DOI] [PubMed] [Google Scholar]
- 37. Fricke J., Neuhard J., Kelln R.A., Pedersen S.. The cmk gene encoding cytidine monophosphate kinase is located in the rpsA operon and is required for normal replication rate in Escherichia coli. J. Bacteriol. 1995; 177:517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Briozzo P., Golinelli-Pimpaneau B., Gilles A.M., Gaucher J.F., Burlacu-Miron S., Sakamoto H., Janin J., Bârzu O.. Structures of Escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity. Structure. 1998; 6:1517–1527. [DOI] [PubMed] [Google Scholar]
- 39. Bertrand T., Briozzo P., Assairi L., Ofiteru A., Bucurenci N., Munier-Lehmann H., Golinelli-Pimpaneau B., Bârzu O., Gilles A.M.. Sugar specificity of bacterial CMP kinases as revealed by crystal structures and mutagenesis of Escherichia coli enzyme. J. Mol. Biol. 2002; 315:1099–1110. [DOI] [PubMed] [Google Scholar]
- 40. Kim D., Hong J.S., Qiu Y., Nagarajan H., Seo J.H., Cho B.K., Tsai S.F., Palsson B.Ø.. Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS Genet. 2012; 8:e1002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rajagopala S.V., Sikorski P., Kumar A., Mosca R., Vlasblom J., Arnold R., Franca-Koh J., Pakala S.B., Phanse S., Ceol A.et al.. The binary protein-protein interaction landscape of Escherichia coli. Nat. Biotechnol. 2014; 32:285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morita T., Kawamoto H., Mizota T., Inada T., Aiba H.. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 2004; 54:1063–1075. [DOI] [PubMed] [Google Scholar]
- 43. Murashko O.N., Lin-Chao S.. Escherichia coli responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E8025–E8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Py B., Higgins C.F., Krisch H.M., Carpousis A.J.. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996; 381:169–172. [DOI] [PubMed] [Google Scholar]
- 45. Miczak A., Kaberdin V.R., Wei C.L., Lin-Chao S.. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:3865–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chandran V., Luisi B.F.. Recognition of enolase in the Escherichia coli RNA degradosome. J. Mol. Biol. 2006; 358:8–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.