ABSTRACT
Intestinal microbiota, dominated by bacteria, plays an important role in the occurrence and the development of alcohol-associated liver disease (ALD), which is one of the most common liver diseases around the world. With sufficient studies focusing on the gut bacterial community, chronic alcohol consumption is now known as a key factor that alters the composition of gut bacterial community, increases intestinal permeability, causes intestinal dysfunction, induces bacterial translocation, and exacerbates the process of ALD via gut-liver axis. However, gut non-bacterial communities including fungi, viruses, and archaea, which may also participate in the disease, has received little attention relative to the gut bacterial community. This paper will systematically collect the latest literatures reporting non-bacterial communities in mammalian health and disease, and review their mechanisms in promoting the development of ALD including CLEC7A pathway, Candidalysin (a peptide toxin secreted by Candida albicans), metabolites, and other chemical substances secreted or regulated by gut commensal mycobiome, virome, and archaeome, hoping to bring novel insights on our current knowledge of ALD.
KEYWORDS: Alcohol-associated liver disease, gut-liver axis, gut barrier, liver sinusoid endothelial cells, mycobiome, CLEC7A, virome, archaeome, immune response
Graphical abstract
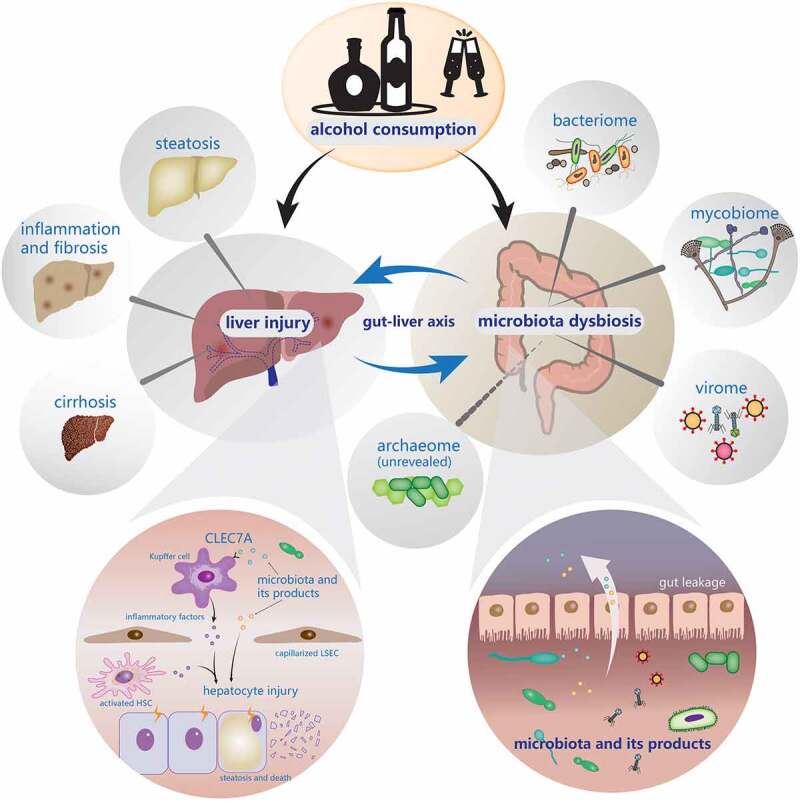
Introduction
Alcohol-associated liver disease (ALD) is a common disease caused by alcohol use disorder (AUD), ranging from asymptomatic liver steatosis to alcohol-associated hepatitis (AH), cirrhosis, and potentially hepatocellular carcinoma (HCC). ALD is the leading indication for liver transplantation in the United States.1,2 Globally, about 2 million people die of liver diseases each year. In Western Europe, one-third of liver cirrhosis can be attributed to alcohol.3 Moreover, it has been estimated that 60–80% of liver-related deaths can be attributed to alcohol consumption.4 Currently, the pathogenetic mechanisms have not been fully elucidated, but they might be related to oxidative stress, acetaldehyde-induced toxicity, cytokine, and chemokine-induced inflammation.5 There is no effective therapeutic method for ALD till now except for liver transplantation. A great number of studies have reported that gut microbiota has an intimate relationship with ALD, which provides broader insights and opportunities for understanding and treating this disease.6–14 However, most studies put emphasis upon the interactions between gut bacteria and ALD, while the roles of non-bacterial communities remain unclear. Hence, this review mainly focuses on addressing the relationships between ALD and the non-bacterial communities (fungi, viruses, and archaea) in the gut, and the possible pathogenetic mechanisms mediated by those microorganisms.
The gut-liver axis
The term gut-liver axis was first described by Volta et al in 1987, which pointed out the interaction between intestine and liver.15 During the embryonic stage, both the liver and intestine originate from the same foregut, and the two organs are anatomically connected by the portal vein system after reaching their maturity.16 About 70 ~ 80% of the blood from the portal vein flows to the liver,17 transmitting a variety of signals generated by dietary, genetic, and environmental factors.(Figure 1)18 As a virtual metabolic organ, the gut-liver axis achieves close-knit functional collaboration and forms a sophisticated network structure through substance metabolism, immune regulation, and interaction with the neuroendocrine system.
Figure 1.
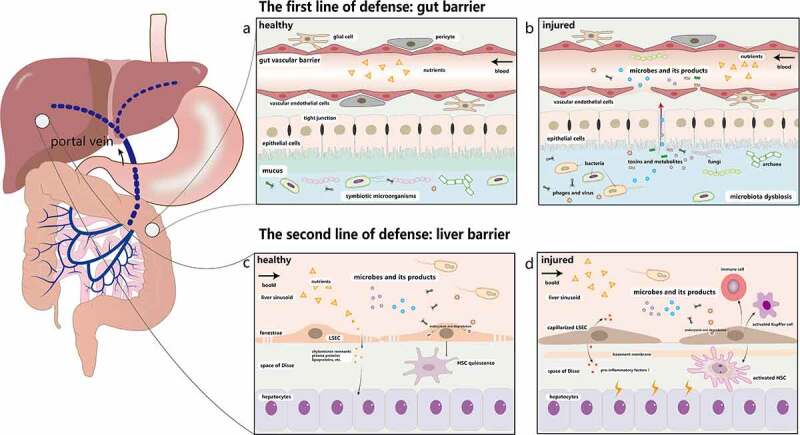
The double line of defense in the gut-liver axis. a. The first line of defense is the gut barrier, which is composed of the mucus barrier, the intestinal epithelial barrier, and the GVB. The mucus layer consists of a microbiota-colonized outer layer and an almost sterile inner layer. The epithelial barrier below the mucus layer is formed by epithelial cells. The deepest protective layer is the GVB. Under physiological circumstances, the healthy intestine barrier allows the absorption of nutrients while prevents the penetration of most toxic and harmful substances. b. Once it is damaged, intestinal microorganisms and their metabolites will move into the liver through the portal vein. c. LSECs are highly fenestrated cells, which can remove recycled waste products by endocytosis and degradation to protect liver against microorganisms and toxicants. They can also induce hepatic immune tolerance and keep the quiescence of HSCs and Kupffer cells. d. Injured LSECs become capillarized and the main characteristics of capillarization are formation of basement membrane, reduction of fenestrae and increased expression of pro-inflammatory mediators. They recruit immune cells and activate HSCs and Kupffer cells to cause liver inflammation. GVB: gut vascular barrier; LSECs: liver sinusoid endothelial cells. HSCs: hepatic stellate cells
Normally, the healthy intestinal barrier is composed of the mucus barrier, the intestinal epithelial barrier, and the gut vascular barrier (GVB),18–21 which together protects the body against external insults (Figure 1(a)). In the colon, the mucus layer usually consists of a microbiota-colonized outer layer and an almost sterile inner layer. These inner and outer layers work together to provide the needed nutrients for microorganisms survival and unique niches for microorganisms inhabitation.22,23 Below the mucus barrier is the epithelial barrier, which is a single layer formed by the epithelial cells including enterocytes, goblet cells, tuft cells, and enterochromaffin cells.24 The neighboring epithelial cells also remain in close contact through tight junctions.25–27 Epithelial barrier’s main functions are regulating the interactions between gut microbiota and the host immune system, and selectively absorbing water, electrolytes, and nutrients.28,29 The GVB identified in 2015 is comprised of vascular endothelial cells, pericytes, and glial cells, which prevents the invasion of microbiota.30,31 As the deepest protective layer and the last defense line of the gut barrier, once the GVB is injured, the intestinal pathogens will go into the blood stream and reach other organs (Figure 1(b)). Hence, in order to fight against exogenous substances in the gut lumen, these three layers summarized above constitute the body’s first line of defense.
Once the intestinal barrier collapses, the liver becomes the first organ to encounter intestinal products, which makes it susceptible to pathological changes.32,33 Liver sinusoidal endothelial cells (LSECs), a major member of the hepatic barrier, provide a second line of defense against gut-derived antigens and inflammatory factors.34 LSECs are highly fenestrated cells (Figure 1(c)). The diameters of transcellular pores on LSECs are 50 ~ 200 nm and these LSECs usually act as a dynamic filter under physiological conditions.35,36 For example, small molecules, such as chylomicron remnants, plasma proteins, and lipoproteins, can cross fenestrae and reach the space of Disse for uptake and utilization by hepatocytes and HSCs.37 Furthermore, LSECs can remove recycled waste products and toxicants, including enteric viruses,38,39 bacteriophages,40 lipopolysaccharides (LPS),41 and immune complexes.42 LSECs can also maintain the quiescent state of hepatic stellate cells, induce hepatic immune tolerance and exert an anti-inflammatory effect.43,44 Due to the existence of the second line of defense, the sole injury of the intestinal barrier is unlikely to lead to significant liver damage, which has been confirmed experimentally in the previous studies.32,33
However, when LSECs are impaired by intestinal pathogenic factors, all the above functions are repressed. LSECs become capillarized45 and their phenotype is transformed into a pro-inflammatory pattern, with the formation of the basement membrane, a significant reduction of fenestrae, and increased expression of pro-inflammatory mediators such as tumor necrosis factor α (TNF-α), macrophage inflammatory protein 1 alpha (MIP-1α), monocyte chemoattractant protein-1 (MCP-1), and chemokine (C-C motif) ligand 5 (CCL5).46 At the same time, LSECs recruit immune cells and activate HSCs and Kupffer cells to induce liver inflammation.47 (Figure 1(d)). Inflammatory factors and metabolites secreted by the injured liver will further aggravate dysfunction of the intestinal barrier, creating a vicious circle eventually. In the future, understanding the mechanism of barrier damage and repair, finding damage markers that are easily detectable, developing drugs to repair the barrier are important steps for the treatment of liver and intestine-related diseases, and achieving the transformation from basic research to clinical implementations.
A complete gut-liver axis relies on not only the intact intestinal barrier and normal liver function but also the healthy gut microbiota. Gut microbiota is composed of 500 to 1000 different species belonging to more than 70 genera.48,49 The number of bacteria residing in the human body is close to the number of human cells.50,51 Not merely can they participate in digestion, absorption and metabolism of food but can also influence the intestinal structure and immune function both directly and indirectly. Therefore, clarifying the changes and roles of gut microbiota in ALD is of great importance for us to understand its pathogenesis and to develop effective treatment strategies. Whereas an immense number of reviews have covered gut bacteria in detail,52–56 thus we center our discussion on the gut mycobiome, virome, and archaeome in human and animal models.
Gut mycobiome
Fungi are ubiquitous in the natural environment and are important parts of the earth’s ecosystem.57 The fungal community is considered as a fundamental component of the human microbiota, coexisting and interacting with other microorganisms in the gut.58 However, they are usually neglected in studies owing to their relatively low abundance in the human body. For instance, there have been almost 100 times more research reports on microbiota than on mycobiota from 2008 to 2018.59 With the increase of related research data and the development of methodology, scientists begin to realize its indispensable role in metabolism, immune regulation, and pathophysiology of the host.
Composition and colonization
Fungi occupy a relatively small proportion of the human microbiota. Fungi comprise approximately 0.03% of the fecal microbiome.60,61 However, this does not mean that it is meaningless to study fungi. Conversely, some scientists contended that previous efforts might underestimate the proportion of fungi in gut microbiota owing to limited annotated reference sequences.62 Besides, fungal cells are generally larger than bacterial cells, indicating that fungi contain much more biomass and metabolites, which cannot be reflected by simple genome-counting.62 Thus, more data and further studies are required for accurately estimating the amount of living gut fungi and completing the annotated fungal genome.
The gut fungi in adults are mainly composed of three phyla, Ascomycetes, Basidiomycetes, and Zygomycetes,63 which are regulated by many factors such as environment,64 diet,65–67 host immunity,68 etc. Both human and animal studies have confirmed that the composition of the intestinal mycobiome is dynamic over time and much more mutable than the component of gut bacteria.69,70 A previous study indicated that intestinal fungi have colonized in human 10 days after birth.71 But recently, fungi were found in the first-pass meconium, suggesting that colonization of gut fungi may exist earlier than expected.72 In the gut of 10-day to 3-month-old infants, the fungal species that had the highest abundance are Debaryomyces hansenii and Rhodotorula mucilaginosa, while S. cerevisiae becomes the most abundant fungal species in the gut of babies 1–2 years after birth.71 From that timepoint and on, the gut fungi develops and gets closer and closer to the intestinal fungal components of the adult. Therefore, given the variability of fungal components, some scholars advocate that fungi do not colonize the gastrointestinal tract in healthy adults, and they may originate from the fungi already present in the mouth or diet, and persistent fungal colonization may be a symptom of disease.67 In their study, when a healthy adult volunteer increased the frequency of cleaning teeth, the abundance of C. albicans in stool was lowered 10-fold to 100-fold, suggesting that the oral cavity may be the primary source of C. albicans detected in the stool of healthy people.67 Indeed, existing research rarely consider the influence of oral and dietary fungi, especially in the studies of exploring changes in the gut fungi compositions. It is difficult to determine whether differences in stool fungi are due to increased transient fungi or due to real intestinal colonization. Overall, the colonization and composition of intestinal fungi are still in their infancy. In the future, further studies with a larger sample size should be done in this field, and sequencing methods with higher sensitivity and accuracy need to be invented to detect intestinal fungi at various time points of embryonic development.
Intestinal fungi and alcohol-associated liver disease
The studies on the correlation between gut mycobiota and ALD are limited and mostly focus on the changes in fungal composition and exploration of potential pathogenic mechanisms. Previous studies reported a decrease in the diversity of mycobiota in the feces of alcohol consumers, with a significant overgrowth of Candida, and a decrease in Epicoccum, unclassified fungi, Galactomyces, and Debaryomyces.73,74 Similarly, recent research reported mycobiota dysbiosis with an overgrowth of Candida as well.75–77 Although there were differences in the severity of alcohol-related liver disease, there were no significant differences in the intestinal mycobiota among patients with non-progressive alcohol-associated liver disease, alcohol-associated hepatitis, and alcoholic cirrhosis.73
As for the role of gut fungi in the pathogenesis of ALD, there remain two main pathways at present: one is dependent on the C-type lectin domain family 7 member A (CLEC7A) pathway of Kupffer cells in liver and the other is independent of that pathway (Figure 2(a)).
Figure 2.
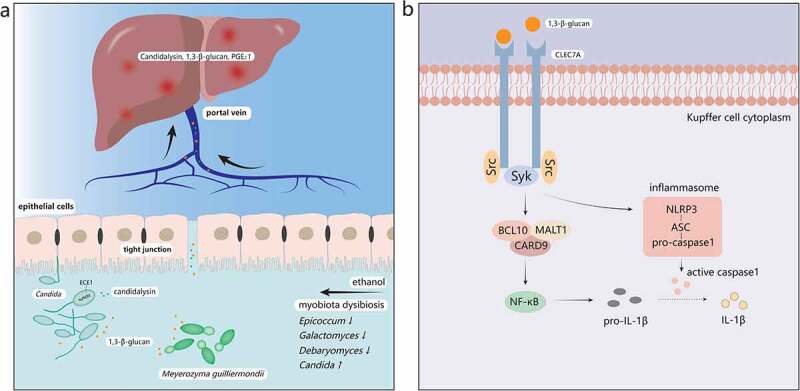
The mechanisms of ALD induced by the gut mycobiota. a. Ethanol abuse leads to disordered gut mycobiota. Epicoccum, Galactomyces, and Debaryomyces decrease, while Candida increases significantly in the intestine. 1,3-β-glucan, a main component of fungal cell walls, and Candidalysin, a fungal metabolite, translocate into the systemic circulation through the injured intestinal barrier and reach the liver first. b. 1,3-β-glucan binds to dectin-1 of Kupffer cells. Src kinases phosphorylate tyrosine residue of dectin-1 and recruit Syk, which can activate the CARD9/BCL10/MALT1 complex. Then NF-κB will be activated by the complex and produce pro-IL-1β. Syk also promotes the secretion of caspase-1 through the activated NLRP3/ASC/pro-caspase-1 complex. Caspase-1 then cleaves pro-IL-1β into mature IL-1β, mediating liver inflammation and damage. Syk, spleen tyrosine kinase; CARD9, caspase recruitment domain 9; BCL10, B-cell lymphoma 10; MALT1, mucosa-associated lymphoid tissue lymphoma-translocation gene 1; NF-κB, nuclear factor kappa B; NLRP3, NOD-like receptor family pyrin domain containing 3; ASC, apoptosis-associated speck-like protein containing a CARD
CLEC7A (also called dectin-1) is a pattern recognition receptor primarily located on the surface of Kupffer cells and macrophages in the liver.73 CLEC7A is capable of recognizing 1,3-β-glucan,78 which is widely found in fungal cell walls, and plays an important role in resisting fungi and fungal products.(Figure 2(b))79 Upon ligand binding, tyrosine residue of dectin-1 is phosphorylated by Src kinases, which creates a docking site for spleen tyrosine kinase (Syk), then Syk activates the caspase recruitment domain 9 (CARD9)/B-cell lymphoma 10 (BCL10)/mucosa-associated lymphoid tissue lymphoma-translocation gene 1 (MALT1) complex, which leads to the activation of nuclear factor kappa B (NF-κB) and promotes cytokines expression including pro-interleukin-1β (pro-IL-1β).80,81 Furthermore, Syk can activate NOD-like receptor family pyrin domain containing 3 (NLRP3) inflammasome, which usually forms a complex with apoptosis-associated speck-like protein containing a CARD (ASC) and pro-caspase-1. ASC can activate pro-caspase-1 into caspase-1, which will cleave pro-IL-1β to produce mature IL-1β and lead to liver damage by activating downstream cascade.82,83 An animal study reported that the development of ALD required IL-1β which was activated via a caspase-1-dependent process, and that IL-1β signaling was essential for alcohol-induced inflammation, steatosis, liver damage, and fibrosis.84 A subsequent study73 also revealed that chronic alcohol intake could increase the number of fungi in mice intestine, and that overgrowth of fungi could produce more fungal products such as 1,3-β-glucan, which can be translocated into the liver through already dysfunctional enteric tight junctions, bind to the Dectin-1 of Kupffer cells, and finally lead to the secretion of IL-1β to promote liver inflammation. However, one study indicated that water-insoluble polysaccharides (WIP), a 1,3-β-glucan from Wolfporia cocos, alleviated the hepatic inflammatory injury and steatosis effectively in mice with alcoholic hepatic steatosis (AHS), which suggested the protective effect of 1,3-β-glucan in liver diseases but the mechanism needs to be further studied.85
Another fungus-related mechanism is mainly associated with its metabolites. As an opportunistic pathogen in the intestine, Candida albicans has attracted extensive attention because of its overgrowth in ALD patients.73 Recent studies have proven that Candida albicans could secrete a type of peptide toxin called Candidalysin, which directly damages epithelial membranes and activates host immune responses.86 One previous study found an interesting phenomenon that the number of Candidalysin positive C. albicans increased in patients with AH and the expression of the extent of cell elongation 1 (ECE1) gene encoding Candidalysin also increased significantly.87 In addition, clinical studies suggested that Candidalysin was positively associated with the severity and mortality of AH.87 With regard to the underlying pathogenic mechanisms of Candidalysin, however, it does not affect the intestinal permeability nor interacts with the glucan receptor CLEC7A, but it increases the expression of IL-1β, CXCL1, and CXCL2 in mice liver, which contributes to recruit immune cells and induce hepatocyte death. In brief, the specific molecular mechanisms need further research.
A recent research also found the contribution of fungi-induced metabolite – prostaglandin E2 (PGE2) to the development of ALD.85 They observed an overwhelming increase of the commensal fungus Meyerozyma guilliermondii and hepatic PGE2 in mice with alcoholic hepatic steatosis (AHS). By introducing antifungal agents, the level of liver PGE2 was significantly reduced. Interestingly, it is still unclear whether PGE2 is produced directly by the gut fungi or indirectly by the fungi-stimulated liver. To sum up, the concrete mechanisms of gut fungi-induced PGE2 in the development of ALD awaits further study.
In addition, the host immune response plays a key role in the process of ALD as well. A recent study74 revealed a novel finding that the level of serum anti-Saccharomyces cerevisiae antibodies (ASCA) was significantly higher in patients with AH compared with alcohol use disorder and nonalcoholic controls. As a marker of the host immune response to fungus and fungal products, ASCA was positively associated with mortality in AH patients.
In summary, it is not difficult to find that a few potential noninvasive indicators, such as the level of serum ASCA, serum Candidalysin, serum 1,3-β-glucan, and the expression of ECE1 in stool, can help us determine the progress or prognosis of ALD. In addition, inhibiting the increase of alcohol-related gut fungi, especially some specific antifungal drugs against harmful mycobiome, may be a promising therapeutic method for ALD, but its clinical application has yet to be confirmed in large-scale cohort studies. Also, further compelling investigations should explore the causal relationship between the specific fungus strain isolated from the gut and the pathogenesis of ALD.
Probiotic fungi
In 2014, the International Scientific Association of Probiotics and Prebiotics (ISAPP) defined probiotics as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host”.88 There are many types of probiotics, such as Bifidobacterium bifidus, Lactobacillus, and Saccharomyces boulardii, of which S. boulardii is probably the most commonly used probiotic fungi. Animal studies have confirmed that S. boulardii could change gut microbiota and attenuate liver injury, inflammation,89–91 and fibrosis,92 potentially indicating its protective and therapeutic role in liver diseases. A recent study revealed that most strains of Saccharomyces and non-Saccharomyces yeasts evaluated in their work are safe microorganisms, and those could be regarded as a valid alternative to the widely available probiotic yeast S. boulardii.93 However, some side effects of S. boulardii, mainly presented as fungemia, were reported in clinical application, which are usually found in the elderly, immunosuppressed, and patients with broad-spectrum antibiotics.94–97 Furthermore, macrofungi with large sporocarps or fruiting bodies, also known as mushrooms, particularly showed their medicinal properties.98 Some types of macrofungi including Agaricus bisporus99–101 and Pleurotus ostreatus102 also possess hepato-protective activity. In conclusion, supplementing probiotic fungi to prevent or treat liver diseases can be expected in the future.
Gut virome
Composition and distribution
Viruses have their own important roles in the intestine. As the most numerous biological entities in nature,103,104 viruses are composed of endogenous retroviruses, eukaryotic viruses, and bacteriophages. Most viruses in the human body reside in the gastrointestinal tract.
The gut viruses, even called virus fingerprints, are unique to each person and mainly consist of bacteriophages.105 However, identifying viral sequences in large and mixed gut microbiota is extremely challenging because viruses lack a universal viral marker.106 Thus, an overwhelming number of gut bacteriophages remain uncultured and unclassified till now, and their specific hosts and infection strategies have not been clarified either. Scientists also named gut viruses “the dark matter” in the intestine, which showed that relevant research on them were far from enough.107–109
Fortunately, the Gut Virome Database (GVD)110 and the Gut Phage Database111 have been established and are continuously updating, which will advance research of gut virome considerably. According to the GVD,110 97.7% of viral populations are phages, 2.1% are eukaryotic viruses, and 0.1% are archaeal viruses. About 1015 bacteriophages settle in a healthy human intestine, which is 10 times the number of symbiotic bacteria.112 The main phages come from the order Caudovirales including Siphoviridae, Myoviridae, and Podoviridae.113,114 Some phages are highly specific to certain bacterial strains, while others have broader ranges of host cells.115 After infection of host by phages, highly virulent phages often cause cell lysis (lytic cycle) while mild phages either lyse the host cell or keep the host cell alive and reproduce normally (lysogenic cycle).116,117
In summary, large-scale efforts are still required urgently in order to culture and catalog gut phages, which is much like the process of building collections and genome databases of bacterial strains in the human microbiome.118
Is it pathogens?
Phages are regarded as potential human pathogens.119 This was probably first suggested with the genome of lambdoid phages that could encode Shiga toxins (Stx),120 which poses a severe and lethal threat to human health. Moreover, animal models revealed that phages may act as pathogens by affecting the gut bacterial communities and increasing the gut permeability.121 Besides, An increasing number of evidence indicates that human diseases, such as inflammatory bowel disease,109,122 diabetes,123 acquired immune deficiency syndrome,124 and colorectal cancer,125 are closely related to the gut viruses. Nevertheless, it is difficult to determine whether the changes in the gut virome are a cause or a result of those diseases. And the boundary between ‘normal’ and pathogenic virome is blurred because the same virus can be either a symbiont or a pathogen depending on the conditions such as the health status and development stage of the host. On the whole, the current knowledge of symbiotic viruses lags far behind that of pathogenic viruses.126
Gut virome and alcohol-associated liver disease
The exact role of gut virome in the etiology of ALD is still unclear, as not only the complex pathogenesis of ALD but also our fragmented understanding of the gut viruses and limited relevant studies. In 2020, scientists systematically described an intestinal virome signature in AH patients for the first time. They observed an increased viral diversity in the stools of patients with ALD. In AH patients, Escherichia-, Enterobacteria-, and Enterococcus phages were over-represented and mammalian viruses such as Parvoviridae and Herpesviridae significantly increased. Staphylococcus phages and Herpesviridae were associated with severity and mortality of ALD.127,128 Above findings were consistent with previous research. In fecal samples, most detected Herpesviridae such as herpesvirus‐6128,129 and herpesvirus‐8130,131 are associated with the severity of ALD, and they could be classified into Epstein-Barr virus (EBV). EBV infection often causes liver inflammation, but the pathogenesis remains unknown.129 A retrospective study of patients with liver cirrhosis found that EBV-positive patients had higher Child-Pugh scores, more severe liver injury and a higher rate of chronic acute liver failure,132 which suggested the reactivation of EBV might contribute to the development of alcoholic hepatitis.
Gut virome may regulate the pathogenesis of ALD through multiple interactions with symbiotic bacteria.133 Phages, as the main component of the gut virome, are able to mediate bacterial cells lysis and regulate the abundance of bacteria. A recent study confirmed that the interaction between bacteria and phages in patients with cirrhosis and hepatic encephalopathy centered on Streptococcus species.134 Moreover, gut bacteria have access to additional genomes (such as antibiotic-resistance genes or bacterial virulence factors) transferred by phages, which will adjust bacterial virulence and adaptability and modify the composition of intestine microbiota.135
Furthermore, gut virome can interact with the host immune system directly. The intestinal immune system is mainly comprised of intestinal intraepithelial lymphocytes (IELs), lamina propria lymphocytes (LPLs), and Peyer’s patches (PPs),136 which could be important in the pathogenesis of ALD. Among them, commensal viruses can be recognized by retinoic acid-inducible gene 1 (RIG-1) signaling in antigen-presenting cells (APCs), and RIG-1 signaling can promote IL-15 production, prevent inflammation and tissue damage as well as maintain the homeostasis of IELs.137 Moreover, scientists found that the immune cells including CD4+ and CD8+ T cells increased in the germ-free mice treated with bacteriophages, and the increased abundance of bacteriophages can exacerbate intestinal colitis through toll-like receptor 9 (TLR9) and interferon-gamma (IFN-γ), indicating that phages can alter mucosal immunity to impact mammalian health.138 But, the mechanism of the phage recognition by the mucosal immune system needs more studies in animal models. Moreover, some studies on pathogenic enteroviruses, such as norovirus and rotavirus, have revealed several signaling pathways of intestinal recognition of viral nucleic acids.139,140 Thus, the gut virome either commensal or pathogenic viruses is a hot topic in host immune response.
To sum up, gut virome may promote the progression of ALD through interventions with the symbiotic bacteria and immune system141 . However, it is still unclear for patients with ALD what specific impacts of increased gut viruses are on the other microorganisms or the host body. We do not know yet whether liver inflammation or damage is caused by the virus itself, or its metabolites, or changes of other microbes induced by the altered gut virome. The studies of gut virome are just starting and more studies are needed to help us understand the correlations between the gut virus, the health of the host and liver diseases.
Phage therapy of ALD
Recent animal studies creatively revealed the importance of phages in the treatment of ALD. According to a study in 2019,142 Enterococcus faecalis was significantly increased in fecal samples and the cytolysin-positive (cytolytic) E. faecalis correlated with the severity and mortality of AH patients. Then, researchers screened out bacteriophages that can lyse the cytolytic E. faecalis to treat mice transplanted with the gut microbiota of AH patients, which significantly reduced the levels of cytolysin in mice liver and attenuated alcohol-induced liver inflammation.
Editing intestinal microbes to treat or improve diseases related to microbial dysbiosis seems to be a quite promising direction. After all, antibiotics are not suitable for precise editing due to their broad spectrum, instead, some bacteriophages are highly specific to bacteria, suggesting a unique advantage of manipulating microbiota.143,144Certainly, the clinical application of phage therapy still needs the support of large-scale human data. In addition, the limitation of phage therapy may be due to its high specificity. Different patients need to deploy different phages in order to better efficacy, so its mass production-like antibiotics may be unrealistic in some extent. In the future, it is necessary to construct a huge phage bank, which will efficiently help us screen out concrete phages and make a personalized treatment within a short period of time.
Gut archaeome
Composition and distribution
Archaea were originally discovered and isolated from ecosystems with extreme conditions, including environments with high temperature, strong acid or base, and high ion concentration. However, with the continuous advancing of detection techniques, archaea had also been found in some mild environments such as the ocean ecosystems.145–149 Archaea are similar to bacteria in terms of shape, size, and genetic information expression including DNA replication, RNA transcription, and protein synthesis. Beside these similarities, there are also some obvious differences between archaea and bacteria. For instance, the archaeal cell walls do not contain peptidoglycans,150 and their cell membranes are composed of L-glycerol-ether/isoprenoid lipids, which are more stable and rigid than bacterial.151 Moreover, due to its special metabolic patterns, archaea can use sunlight, inorganic or organic substances as energy sources.152
The most abundant archaea in the human gastrointestinal tract are Methanobacteriales and Methanomassiliicoccales.153 The former, mainly consisting of Methanobrevibacter smithii and Methanosphaera stadtmanae,154 is identified as a keystone species that has a noticeable impact on the composition and function of other gut microbiome.155,156 The species of archaea are usually affected by various factors such as diet, environment, age, genetics, etc., and they are also related to non-archaea members of the host microbiota. Interestingly, the abundance of M. smithii was recently found to be stable over time, even after major changes in diet.157 In addition, the archaea present highly specific adaptability in the gastrointestinal tract, and they often evolve unique features such as modifications of cell surface (adhesin-like proteins (ALPs), glycans, bile salt hydrolases, and biofilm formation), which are not found in wide archaea, to escape the host-defense mechanism.158
Generally speaking, as an important part of the gastrointestinal microbiota of humans and animals, the role of archaea may be far more underestimated owing to the methodological shortcomings.159 For instance, most of the so-called “universal” 16S rRNA primers fail to picture the diversity of archaeal signatures and thus are unable to detect certain archaeal lineages in specific sample types.160,161 The archaea research is still in its infancy and the academics of microbiology lack relevant knowledge as well.
Archaea and human diseases
Whether archaea are pathogens or not has always been a controversial topic.158 Some studies verified the change of archaeome in certain human diseases. For example, M. smithii was found in urinary tract infections.161 M. oralis was reported in periodontitis, and its quantity increased as the illness severity increased, but it was no longer present after healing, which highlighted the close association of M. oralis with the inflamed site.162 The abundance of M. stadtmanae increased three-fold in inflammatory bowel disease (IBD) patients when compared to healthy individuals, indicating this archaeon might be involved in pathologic process within the human gut.163
Metabolites produced by archaea such as methane can also affect the health status of the host. In dog models, methane can weaken gastrointestinal motility.164 In a population with irritable bowel syndrome (IBS), the subjects with the constipation-dominant disease (IBS-C) were shown to have a higher proportion of methane producers than individuals with the diarrhea-dominant disease (IBS-D).165
Furthermore, people have also found the interaction between archaea and the human immune system. When stimulating the immune system with M. stadtmanae,154 high levels of proinflammatory cytokines including interleukin and interferon were released.166,167 Only recently was it demonstrated that RNA from M. stadtmanae was a potent immune stimulator, and toll-like receptor 7 (TLR7) and TLR8 were identified as the involved pattern recognition receptors. Moreover, this molecular interaction induced TLR8-dependent NLRP3 inflammasome activation.168 The proinflammatory ability of archaea also varies from species to species. For example, M. stadtmanae is capable of inducing a stronger immune response than both M. smithii and M. luminyensis.166,169 There are some speculations attempting to explain the mild response induced by M. smithii stating that M. smithii has the capacity to produce glycans which is similar to those found in the gut,170 and thus may help M. smithii escape from the host immune system. In conclusion, understanding the molecular mechanisms of how archaea induce inflammation in the body is thus an important step in uncovering how such diseases might develop.
The impact of ALD on the composition of archaea is rarely reported. According to a recent research, the proportion of archaea among the healthy control group, the alcohol use disorder group, and the alcoholic hepatitis group was 0.6%. 0.3%, and 0.0% respectively, which indicated that the abundance of archaea was reduced as the disease progresses.171 However, it is not clear which archaea have modified, nor is it clear whether these changes are a result of or a cause for the disease. But considering the proinflammatory ability, it is worthwhile to study the interspecies communication between archaea and other gut microbes, the potential role of archaeal metabolites, and the relationship of archaea and chronic liver diseases.
Conclusion
Although we have generally devoted significant efforts on studying the gut bacteriota, it is worth noting that non-bacterial communities also have potential functions. All these microbes will interact with the host and also with each other, in combination or alone, to influence the health and disease of the host. Eventually, we will unveil the role of non-bacterial microbiota in human with the remarkable development of technologies for the detection of fungi, viruses, and archaea. Furthermore, human microbiota investigations have now reached a critical inflection point. In the future, we need to move rapidly from mere description or correlation to causation, and ultimately to translation.172
To accomplish such a great mission, much work remains ahead of us. The first is the improvement of methodology. At present, there are three kinds of methods commonly used in studying intestinal microbiota: culture-dependent methods, culture-independent methods, and high-throughput sequencing (HTS). There are also several excellent literatures evaluating these methods in gut mycobiome,173 virome,174 and archaeome.160 Nevertheless, most research tools still have some defects. Culture methods are time-consuming and also susceptible to environmental factors. Scientists might misestimate the abundance of certain gut microorganisms owing to the incompleteness of annotated gene databases or improper primers. In addition, the amount of relic DNA in human gut remains unknown currently, and a few fungi DNA can even be detected after years of organism decomposition. Thus, we need to be more prudent in face of the results of DNA-based sequencing. The study of gut non-bacterial microbiota will be more efficient through establishing a uniform, accurate, and highly operable detection system for non-bacteriota. And ensuring that the microbiome from different regions, races, and species can be compared or referenced is highly expected in future work.
Secondly, it is necessary to expand the genomics, transcriptomics, and metabolomics data of fungi, viruses, and archaea in the existing database, so that future research can accurately and quickly identify the corresponding microbiota in multiple dimensions. Moreover, the study of microbiome cannot simply stop at the macroscopic appearance of the gut community composition changes, it should be conducted from bacterial species to specific strains, from cells to subcellular organs, from metabolites to molecular regulation. Study in such detail will provide a fresh and fundamental perspective for manipulating and editing the gut microbiota logically and efficiently in the future. Finally, the commensal microbiota in the intestine is an emerging and promising field that has aroused great interest from scientists, physicians and patients. With a deeper understanding and transformation of the gut-liver axis, the investments may be rewarded and the management of ALD patients will also be improved.
Supplementary Material
Funding Statement
This project was supported by National Natural Science Foundation of China (grant number 81770582 to JY and 82000561 to HKC).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Authors’ contributions
WKG contributed to the conception and drafted the manuscript. YXZ polished the language and grammar and revised the manuscript. JY checked and improved the manuscript. HKC contributed to the conception and critically revised the manuscript. All authors read and approved the final version of the manuscript.
Supplementary material
supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Cholankeril G, Wong RJ, Hu M, Perumpail RB, Yoo ER, Puri P, Younossi ZM, Harrison SA, Ahmed A.. 2017. Liver transplantation for nonalcoholic steatohepatitis in the US: temporal trends and outcomes. Dig Dis Sci. 62(10):2915–18. doi: 10.1007/s10620-017-4684-x. PMID: 28744836 [DOI] [PubMed] [Google Scholar]
- 2.Lee BP, Vittinghoff E, Dodge JL, Cullaro G, Terrault NA. 2019. National trends and long-term outcomes of liver transplant for alcohol-Associated liver disease in the United States. JAMA Intern Med. 179(3):340–348. doi: 10.1001/jamainternmed.2018.6536. PMID: 30667468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mokdad AA, Lopez AD, Shahraz S, Lozano R, Mokdad AH, Stanaway J, Murray CJ, Naghavi M. 2014. Liver cirrhosis mortality in 187 countries between 1980 and 2010: a systematic analysis. BMC Med. 12(1):145. doi: 10.1186/s12916-014-0145-y. PMID: 25242656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheron N. 2016. Alcohol and liver disease in Europe–Simple measures have the potential to prevent tens of thousands of premature deaths. J Hepatol. 64(4):957–967. doi: 10.1016/j.jhep.2015.11.006. PMID: 26592352 [DOI] [PubMed] [Google Scholar]
- 5.Seitz HK, Bataller R, Cortez-Pinto H, Gao B, Gual A, Lackner C, Mathurin P, Mueller S, Szabo G, Tsukamoto H, et al. 2018. Alcoholic liver disease. Nat Rev Dis Primers. 4(1):16. PMID: 30115921. doi: 10.1038/s41572-018-0014-7. [DOI] [PubMed] [Google Scholar]
- 6.Rivera Ca, Bradford BU, Seabra V, Thurman RG. 1998. Role of endotoxin in the hypermetabolic state after acute ethanol exposure. Am J Physiol. 275:G1252–8. PMID: 9843760. doi: 10.1152/ajpgi.1998.275.6.G1252. [DOI] [PubMed] [Google Scholar]
- 7.Yan AW, Fouts DE, Brandl J, Starkel P, Torralba M, Schott E, Tsukamoto H, E. Nelson K, A. Brenner D, Schnabl B, et al. 2011. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 53(1):96–105. PMID: 21254165. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR, et al. 2014. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 60(5):940–947. PMID: 24374295. doi: 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Starkel P, Windey K, Tremaroli V, Bäckhed F, Verbeke K, et al. 2014. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci U S A. 111(42):E4485–93. PMID: 25288760. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen P, Miyamoto Y, Mazagova M, Lee KC, Eckmann L, Schnabl B. 2015. Microbiota protects mice against acute alcohol-induced liver injury. Alcohol Clin Exp Res. 39(12):2313–2323. doi: 10.1111/acer.12900. PMID: 26556636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llopis M, Cassard AM, Wrzosek L, Boschat L, Bruneau A, Ferrere G, Puchois V, Martin JC, Lepage P, Le Roy T, et al. 2016. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut.65(5):830–839. PMID: 26642859. doi: 10.1136/gutjnl-2015-310585. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Fouts DE, Starkel P, Hartmann P, Chen P, Llorente C, DePew J, Moncera K, Ho S, Brenner D, et al. 2016. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing Mucosa-Associated microbiota and preventing bacterial translocation. Cell Host Microbe. 19(2):227–239. PMID: 26867181. doi: 10.1016/j.chom.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llorente C, Jepsen P, Inamine T, Wang L, Bluemel S, Wang HJ, Loomba R, Bajaj JS, Schubert ML, Sikaroodi M, et al. 2017. Gastric acid suppression promotes alcoholic liver disease by inducing overgrowth of intestinal enterococcus. Nat Commun. 8(1):837. PMID: 29038503. doi: 10.1038/s41467-017-00796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smirnova E, Puri P, Muthiah MD, Daitya K, Brown R, Chalasani N, Liangpunsakul S, Shah VH, Gelow K, Siddiqui MS, et al. 2020. Fecal microbiome distinguishes alcohol consumption from alcoholic hepatitis but does not discriminate disease severity. Hepatology. 72(1):271–286. PMID: 32056227. doi: 10.1002/hep.31178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volta U, Bonazzi C, Bianchi FB, Baldoni AM, Zoli M, Pisi E. 1987. IgA antibodies to dietary antigens in liver cirrhosis. Ric Clin Lab. 17:235–242. PMID: 3671996. doi: 10.1007/BF02912537. [DOI] [PubMed] [Google Scholar]
- 16.Abdel-Misih SR, Bloomston M. 2010. Liver anatomy. Surg Clin North Am. 90(4):643–653. doi: 10.1016/j.suc.2010.04.017. PMID: 20637938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ger R. 1989. Surgical anatomy of the liver. Surg Clin North Am. 69(2):179–192. doi: 10.1016/s0039-6109(16)44780-8. PMID: 2928899 [DOI] [PubMed] [Google Scholar]
- 18.Albillos A, de Gottardi A, Rescigno M. 2020. The gut-liver axis in liver disease: pathophysiological basis for therapy. J Hepatol. 72(3):558–577. doi: 10.1016/j.jhep.2019.10.003. PMID: 31622696 [DOI] [PubMed] [Google Scholar]
- 19.Brescia P, Rescigno M. 2021. The gut vascular barrier: a new player in the gut-liver-brain axis. Trends Mol Med. 27(9):844–855. doi: 10.1016/j.molmed.2021.06.007. PMID: 34229973 [DOI] [PubMed] [Google Scholar]
- 20.Ghosh S, Whitley CS, Haribabu B, Jala VR. 2021. Regulation of intestinal barrier function by microbial metabolites. Cell Mol Gastroenterol Hepatol. 11(5):1463–1482. doi: 10.1016/j.jcmgh.2021.02.007. PMID: 33610769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chopyk DM, Grakoui A. 2020. Contribution of the Intestinal microbiome and gut barrier to hepatic disorders. Gastroenterology. 159(3):849–863. doi: 10.1053/j.gastro.2020.04.077. PMID: 32569766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC. 2013. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastrointest Liver Physiol. 305(5):G341–7. doi: 10.1152/ajpgi.00046.2013. PMID: 23832518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, Brugiroux S, Keller I, Macpherson JA, Rupp S, et al. 2015. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun. 6(1):8292. PMID: 26392213. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurashima Y, Kiyono H. 2017. Mucosal ecological network of epithelium and immune cells for gut homeostasis and tissue healing. Annu Rev Immunol. 35(1):119–147. doi: 10.1146/annurev-immunol-051116-052424. PMID: 28125357 [DOI] [PubMed] [Google Scholar]
- 25.Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. 1986. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 103(3):755–766. doi: 10.1083/jcb.103.3.755. PMID: 3528172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. 1993. Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol. 123(6):1777–1788. PMID: 8276896. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. 1998. Claudin-1 and −2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 141(7):1539–1550. doi: 10.1083/jcb.141.7.1539. PMID: 9647647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odenwald MA, Turner JR. 2017. The intestinal epithelial barrier: a therapeutic target? Nat Rev Gastroenterol Hepatol. 14(1):9–21. doi: 10.1038/nrgastro.2016.169. PMID: 27848962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rescigno M. 2011. The intestinal epithelial barrier in the control of homeostasis and immunity. Trends Immunol. 32(6):256–264. doi: 10.1016/j.it.2011.04.003. PMID: 21565554 [DOI] [PubMed] [Google Scholar]
- 30.Spadoni I, Zagato E, Bertocchi A, Paolinelli R, Hot E, Di Sabatino A, Caprioli F, Bottiglieri L, Oldani A, Viale G, et al. 2015. A gut-vascular barrier controls the systemic dissemination of bacteria. Science. 350(6262):830–834. PMID: 26564856. doi: 10.1126/science.aad0135. [DOI] [PubMed] [Google Scholar]
- 31.Bouziat R, Jabri B. 2015. IMMUNOLOGY. Breaching the gut-vascular barrier. Science. 350(6262):742–743. doi: 10.1126/science.aad6768. PMID: 26564835 [DOI] [PubMed] [Google Scholar]
- 32.Gabele E, Dostert K, Hofmann C, Wiest R, Scholmerich J, Hellerbrand C, Obermeier F. 2011. DSS induced colitis increases portal LPS levels and enhances hepatic inflammation and fibrogenesis in experimental NASH. J Hepatol. 55(6):1391–1399. doi: 10.1016/j.jhep.2011.02.035. PMID: 21703208 [DOI] [PubMed] [Google Scholar]
- 33.El Kasmi KC, Anderson AL, Devereaux MW, Fillon SA, Harris JK, Lovell MA, Finegold MJ, Sokol RJ. 2012. Toll-like receptor 4-dependent Kupffer cell activation and liver injury in a novel mouse model of parenteral nutrition and intestinal injury. Hepatology. 55(5):1518–1528. doi: 10.1002/hep.25500. PMID: 22120983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Liu Y. 2021. Gut-liver-axis: barrier function of liver sinusoidal endothelial cell (LSEC). J Gastroenterol Hepatol. PMID: 33811372 10.1111/jgh.15512 [DOI] [PubMed] [Google Scholar]
- 35.Wisse E. 1970. An electron microscopic study of the fenestrated endothelial lining of rat liver sinusoids. J Ultrastruct Res. 31(1–2):125–150. doi: 10.1016/s0022-5320(70)90150-4. PMID: 5442603 [DOI] [PubMed] [Google Scholar]
- 36.Monkemoller V, Oie C, Hubner W, Huser T, McCourt P. 2015. Multimodal super-resolution optical microscopy visualizes the close connection between membrane and the cytoskeleton in liver sinusoidal endothelial cell fenestrations. Sci Rep. 5(1):16279. doi: 10.1038/srep16279. PMID: 26549018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poisson J, Lemoinne S, Boulanger C, Durand F, Moreau R, Valla D, Rautou P-E. 2017. Liver sinusoidal endothelial cells: physiology and role in liver diseases. J Hepatol. 66(1):212–227. doi: 10.1016/j.jhep.2016.07.009. PMID: 27423426 [DOI] [PubMed] [Google Scholar]
- 38.Ganesan LP, Mohanty S, Kim J, Clark KR, Robinson JM, Anderson CL, Desrosiers RC. 2011. Rapid and efficient clearance of blood-borne virus by liver sinusoidal endothelium. PLoS Pathog. 7(9):e1002281. doi: 10.1371/journal.ppat.1002281. PMID: 21980295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon-Santamaria J, Rinaldo CH, Kardas P, Li R, Malovic I, Elvevold K,McCourt P, Smedsrod B, Hirsch HH, Sorensen KK. 2014. Efficient uptake of blood-borne BK and JC polyomavirus-like particles in endothelial cells of liver sinusoids and renal vasa recta. PLoS One. 9(11):e111762. PMID: 25375646. doi: 10.1371/journal.pone.0111762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oie CI, Wolfson DL, Yasunori T, Dumitriu G, Sorensen KK, McCourt PA,Ahluwalia BS, Smedsrod B. 2020. Liver sinusoidal endothelial cells contribute to the uptake and degradation of entero bacterial viruses. Sci Rep. 10(1):898. PMID: 31965000. doi: 10.1038/s41598-020-57652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao Z, Mates JM, Cheplowitz AM, Hammer LP, Maiseyeu A, Phillips GS, Wewers MD, Rajaram MVS, Robinson JM, Anderson CL, et al. 2016. Blood-Borne lipopolysaccharide is rapidly eliminated by liver sinusoidal endothelial cells via high-density lipoprotein. J Immunol. 197(6):2390–2399. PMID: 27534554. doi: 10.4049/jimmunol.1600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ganesan LP, Kim J, Wu Y, Mohanty S, Phillips GS, Birmingham DJ,Robinson JM, Anderson CL. 2012. FcgammaRIIb on liver sinusoidal endothelium clears small immune complexes. J Immunol. 189(10):4981–4988. PMID: 23053513. doi: 10.4049/jimmunol.1202017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deleve LD, Wang X, Guo Y. 2008. Sinusoidal endothelial cells prevent rat stellate cell activation and promote reversion to quiescence. Hepatology. 48(3):920–930. doi: 10.1002/hep.22351. PMID: 18613151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie G, Wang X, Wang L, Wang L, Atkinson RD, Kanel GC, Gaarde WA, DeLeve LD. 2012. Role of differentiation of liver sinusoidal endothelial cells in progression and regression of hepatic fibrosis in rats. Gastroenterology. 142(4):918–27e6. doi: 10.1053/j.gastro.2011.12.017. PMID: 22178212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaffner F, Poper H. 1963. Capillarization of hepatic sinusoids in man. Gastroenterology. 44(3):239–242. PMID: 13976646. doi: 10.1016/S0016-5085(63)80130-4. [DOI] [PubMed] [Google Scholar]
- 46.Connolly MK, Bedrosian AS, Malhotra A, Henning JR, Ibrahim J, Vera V,Cieza-Rubio NE, Hassan BU, Pachter HL, Cohen S, et al. 2010. In hepatic fibrosis, liver sinusoidal endothelial cells acquire enhanced immunogenicity. J Immunol. 185(4):920–930. PMID: 20639479. doi: 10.1002/hep.22351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyao M, Kotani H, Ishida T, Kawai C, Manabe S, Abiru H, Tamaki K. 2015. Pivotal role of liver sinusoidal endothelial cells in NAFLD/NASH progression. Lab Invest. 95(10):1130–1144. doi: 10.1038/labinvest.2015.95. PMID: 26214582 [DOI] [PubMed] [Google Scholar]
- 48.Human Microbiome PC. 2012. Structure, function and diversity of the healthy human microbiome. Nature. 486:207–214. PMID: 22699609. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Almeida A, Nayfach S, Boland M, Strozzi F, Beracochea M, Shi ZJ, Pollard KS, Sakharova E, Parks DH, Hugenholtz P, et al. 2021. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol. 39(1):105–114. PMID: 32690973. doi: 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sender R, Fuchs S, Milo R. 2016. Are we really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell. 164(3):337–340. doi: 10.1016/j.cell.2016.01.013. PMID: 26824647 [DOI] [PubMed] [Google Scholar]
- 51.Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14(8):e1002533. doi: 10.1371/journal.pbio.1002533. PMID: 27541692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bajaj JS. 2019. Alcohol, liver disease and the gut microbiota. Nat Rev Gastroenterol Hepatol. 16(4):235–246. doi: 10.1038/s41575-018-0099-1. PMID: 30643227 [DOI] [PubMed] [Google Scholar]
- 53.Lang S, Schnabl B. 2020. Microbiota and fatty liver disease-the known, the unknown, and the future. Cell Host Microbe. 28(2):233–244. doi: 10.1016/j.chom.2020.07.007. PMID: 32791115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szabo G. 2015. Gut-liver axis in alcoholic liver disease. Gastroenterology. 148(1):30–36. doi: 10.1053/j.gastro.2014.10.042. PMID: 25447847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Betrapally NS, Gillevet PM, Bajaj JS. 2016. Changes in the intestinal microbiome and alcoholic and nonalcoholic liver diseases: causes or effects? Gastroenterology. 150(8):1745–55e3. doi: 10.1053/j.gastro.2016.02.073. PMID: 26948887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adolph TE, Grander C, Moschen AR, Tilg H. 2018. Liver-microbiome axis in health and disease. Trends Immunol. 39(9):712–723. doi: 10.1016/j.it.2018.05.002. PMID: 29843959 [DOI] [PubMed] [Google Scholar]
- 57.Peay KG, Kennedy PG, Talbot JM. 2016. Dimensions of biodiversity in the earth mycobiome. Nat Rev Microbiol. 14(7):434–447. doi: 10.1038/nrmicro.2016.59. PMID: 27296482 [DOI] [PubMed] [Google Scholar]
- 58.Seed PC. 2014. The human mycobiome. Cold Spring Harb Perspect Med. 5(5):a019810. doi: 10.1101/cshperspect.a019810. PMID: 25384764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forbes JD, Bernstein CN, Tremlett H, Van Domselaar G, Knox NC. 2018. A fungal world: could the gut mycobiome be involved in neurological disease? Front Microbiol. 9:3249. PMID: 30687254. doi: 10.3389/fmicb.2018.03249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ott SJ, Kuhbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S, et al. 2008. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol. 43(7):831–841. PMID: 18584522. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 61.Hillman ET, Lu H, Yao T, Nakatsu CH. 2017. Microbial ecology along the gastrointestinal tract. Microbes Environ. 32(4):300–313. doi: 10.1264/jsme2.ME17017. PMID: 29129876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Underhill DM, Iliev ID. 2014. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 14(6):405–416. doi: 10.1038/nri3684. PMID: 24854590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. 2013. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 8(6):e66019. doi: 10.1371/journal.pone.0066019. PMID: 23799070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeung F, Chen YH, Lin JD, Leung JM, McCauley C, Devlin JC, Hansen C, Cronkite A, Stephens Z, Drake-Dunn C, et al. 2020. Altered immunity of laboratory mice in the natural environment is associated with fungal colonization. Cell Host Microbe. 27(5):809–22e6. PMID: 32209432. doi: 10.1016/j.chom.2020.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ukhanova M, Wang X, Baer DJ, Novotny JA, Fredborg M, Mai V. 2014. Effects of almond and pistachio consumption on gut microbiota composition in a randomised cross-over human feeding study. Br J Nutr. 111(12):2146–2152. doi: 10.1017/S0007114514000385. PMID: 24642201 [DOI] [PubMed] [Google Scholar]
- 66.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 505(7484):559–563. PMID: 24336217. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Auchtung TA, Fofanova TY, Stewart CJ, Nash AK, Wong MC, Gesell JR, Auchtung JM, Ajami NJ, Petrosino JF. 2018. Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere. 3(2). PMID: 29600282. doi: 10.1128/mSphere.00092-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. 2012. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 336(6086):1314–1317. PMID: 22674328. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hallen-Adams HE, Kachman SD, Kim J, Legge RM, Martinez I. 2015. Fungi inhabiting the healthy human gastrointestinal tract: a diverse and dynamic community. Fungal Ecol. 15:9–17. PMID: WOS: 000356109500002. doi: 10.1016/j.funeco.2015.01.006. [DOI] [Google Scholar]
- 70.Dollive S, Chen YY, Grunberg S, Bittinger K, Hoffmann C, Vandivier L, Cuff C, Lewis JD, Wu GD, Bushman FD, et al. 2013. Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment. PLoS One. 8(8):e71806. PMID: 23977147. doi: 10.1371/journal.pone.0071806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schei K, Avershina E, Oien T, Rudi K, Follestad T, Salamati S, Ødegård RA. 2017. Early gut mycobiota and mother-offspring transfer. Microbiome. 5(1):107. doi: 10.1186/s40168-017-0319-x. PMID: 28837002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Willis KA, Purvis JH, Myers ED, Aziz MM, Karabayir I, Gomes CK, Peters BM, Akbilgic O, Talati AJ, Pierre JF, et al. 2019. Fungi form interkingdom microbial communities in the primordial human gut that develop with gestational age. FASEB J. 33(11):12825–12837. PMID: 31480903. doi: 10.1096/fj.201901436RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang AM, Inamine T, Hochrath K, Chen P, Wang L, Llorente C,Bluemel S, Hartmann P, Xu J, Koyama Y, et al. 2017. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest. 127(7):2829–2841. PMID: 28530644. doi: 10.1172/JCI90562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lang S, Duan Y, Liu J, Torralba MG, Kuelbs C, Ventura‐Cots M, Abraldes JG, Bosques‐Padilla F, Verna EC, Brown RS, et al. 2020. Intestinal fungal dysbiosis and systemic immune response to fungi in patients with alcoholic hepatitis. Hepatology. 71(2):522–538. PMID: 31228214. doi: 10.1002/hep.30832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bajaj JS, Liu EJ, Kheradman R, Fagan A, Heuman DM, White M, Gavis EA, Hylemon P, Sikaroodi M, Gillevet PM, et al. 2018. Fungal dysbiosis in cirrhosis. Gut. 67(6):1146–1154. PMID: 28578302. doi: 10.1136/gutjnl-2016-313170. [DOI] [PubMed] [Google Scholar]
- 76.Hartmann P, Lang S, Zeng S, Duan Y, Zhang X, Wang Y, Bondareva M, Kruglov A, Fouts DE, Stärkel P, et al. 2021. Dynamic changes of the fungal microbiome in alcohol use disorder. Front Physiol. 12:699253. PMID: 34349667. doi: 10.3389/fphys.2021.699253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu J, Wu D, Ma K, Wang T, Shi G, Shao J, Wang C, Yan G. 2020. Paeonol ameliorates murine alcohol liver disease via mycobiota-mediated Dectin-1/IL-1beta signaling pathway. J Leukoc Biol. 108(1):199–214. doi: 10.1002/JLB.3MA0120-325RR. PMID: 32129526 [DOI] [PubMed] [Google Scholar]
- 78.Brown GD, Gordon S. 2001. Immune recognition. A new receptor for beta-glucans. Nature. 413(6851):36–37. doi: 10.1038/35092620. PMID: 11544516 [DOI] [PubMed] [Google Scholar]
- 79.Kimberg M, Brown GD. 2008. Dectin-1 and its role in antifungal immunity. Med Mycol. 46(7):631–636. doi: 10.1080/13693780802140907. PMID: 18608924 [DOI] [PubMed] [Google Scholar]
- 80.Rogers NC, Slack EC, Edwards AD, Nolte MA, Schulz O, Schweighoffer E, Williams DL, Gordon S, Tybulewicz VL, Brown GD, et al. 2005. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 22(4):507–517. PMID: 15845454. doi: 10.1016/j.immuni.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 81.Gross O, Gewies A, Finger K, Schafer M, Sparwasser T, Peschel C, Förster I, Ruland J. 2006. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 442(7103):651–656. doi: 10.1038/nature04926. PMID: 16862125 [DOI] [PubMed] [Google Scholar]
- 82.Kankkunen P, Teirila L, Rintahaka J, Alenius H, Wolff H, Matikainen S. 2010. (1,3)-beta-glucans activate both dectin-1 and NLRP3 inflammasome in human macrophages. J Immunol. 184(11):6335–6342. doi: 10.4049/jimmunol.0903019. PMID: 20421639 [DOI] [PubMed] [Google Scholar]
- 83.Gross O, Poeck H, Bscheider M, Dostert C, Hannesschlager N, Endres S, Hartmann G, Tardivel A, Schweighoffer E, Tybulewicz V, et al. 2009. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 459(7245):433–436. PMID: 19339971. doi: 10.1038/nature07965. [DOI] [PubMed] [Google Scholar]
- 84.Petrasek J, Bala S, Csak T, Lippai D, Kodys K, Menashy V, Barrieau M, Min S-Y, Kurt-Jones EA, Szabo G, et al. 2012. IL-1 receptor antagonist ameliorates inflammasome-dependent alcoholic steatohepatitis in mice. J Clin Invest. 122(10):3476–3489. PMID: 22945633. doi: 10.1172/JCI60777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun S, Wang K, Sun L, Cheng B, Qiao S, Dai H, Shi W, Ma J, Liu H. 2020. Therapeutic manipulation of gut microbiota by polysaccharides of Wolfiporia cocos reveals the contribution of the gut fungi-induced PGE2 to alcoholic hepatic steatosis. Gut Microbes. 12(1):1830693. doi: 10.1080/19490976.2020.1830693. PMID: 33106075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Höfs S, Gratacap RL, Robbins J, Runglall M, et al. 2016. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature. 532(7597):64–68. PMID: 27027296. doi: 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chu H, Duan Y, Lang S, Jiang L, Wang Y, Llorente C, Liu J, Mogavero S, Bosques-Padilla F, Abraldes JG, et al. 2020. The Candida albicans exotoxin candidalysin promotes alcohol-associated liver disease. J Hepatol. 72(3):391–400. PMID: 31606552. doi: 10.1016/j.jhep.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. 2014. Expert consensus document. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 11(8):506–514. PMID: 24912386. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 89.Barssotti L, Abreu I, Brandao ABP, Albuquerque R, Ferreira FG, Salgado MAC, Dias DDS, De Angelis K, Yokota R, Casarini DE, et al. 2021. Saccharomyces boulardii modulates oxidative stress and renin angiotensin system attenuating diabetes-induced liver injury in mice. Sci Rep. 11(1):9189. PMID: 33911129. doi: 10.1038/s41598-021-88497-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Albuquerque R, Brandao ABP, De Abreu I, Ferreira FG, Santos LB, Moreira LN, Taddei CR, Aimbire F, Cunha TS. 2019. Saccharomyces boulardii Tht 500101 changes gut microbiota and ameliorates hyperglycaemia, dyslipidaemia, and liver inflammation in streptozotocin-diabetic mice. Benef Microbes. 10(8):901–912. doi: 10.3920/BM2019.0056. PMID: 31965836 [DOI] [PubMed] [Google Scholar]
- 91.Yu L, Zhao XK, Cheng ML, Yang GZ, Wang B, Liu HJ, Hu YX, Zhu LL, Zhang S, Xiao ZW, et al. 2017. Saccharomyces boulardii administration changes gut microbiota and attenuates D-Galactosamine-induced liver injury. Sci Rep. 7(1):1359. PMID: 28465509. doi: 10.1038/s41598-017-01271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li M, Zhu L, Xie A, Yuan J. 2015. Oral administration of Saccharomyces boulardii ameliorates carbon tetrachloride-induced liver fibrosis in rats via reducing intestinal permeability and modulating gut microbial composition. Inflammation. 38(1):170–179. doi: 10.1007/s10753-014-0019-7. PMID: 25227279 [DOI] [PubMed] [Google Scholar]
- 93.Fernandez-Pacheco P, Ramos Monge IM, Fernandez-Gonzalez M, Poveda Colado JM, Safety A-VM. 2021. Evaluation of yeasts with probiotic potential. Front Nutr. 8:659328. PMID: 34095190. doi: 10.3389/fnut.2021.659328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Santino I, Alari A, Bono S, Teti E, Marangi M, Bernardini A, Magrini L, Di Somma S, Teggi A. 2014. Saccharomyces cerevisiae fungemia, a possible consequence of the treatment of clostridium difficile colitis with a probioticum. Int J Immunopathol Pharmacol. 27(1):143–146. doi: 10.1177/039463201402700120. PMID: 24674691 [DOI] [PubMed] [Google Scholar]
- 95.Cherifi S, Robberecht J, Miendje Y. 2004. Saccharomyces cerevisiae fungemia in an elderly patient with Clostridium difficile colitis. Acta Clin Belg. 59(4):223–224. doi: 10.1179/acb.2004.033. PMID: 15597730 [DOI] [PubMed] [Google Scholar]
- 96.Henry S, D’Hondt L, Andre M, Holemans X, Canon JL. 2004. Saccharomyces cerevisiae fungemia in a head and neck cancer patient: a case report and review of the literature. Acta Clin Belg. 59(4):220–222. doi: 10.1179/acb.2004.032. PMID: 15597729 [DOI] [PubMed] [Google Scholar]
- 97.Cesaro S, Chinello P, Rossi L, Zanesco L. 2000. Saccharomyces cerevisiae fungemia in a neutropenic patient treated with Saccharomyces boulardii. Support Care Cancer. 8(6):504–505. doi: 10.1007/s005200000123. PMID: 11094997 [DOI] [PubMed] [Google Scholar]
- 98.Lu H, Lou H, Hu J, Liu Z, Chen Q. 2020. Macrofungi: a review of cultivation strategies, bioactivity, and application of mushrooms. Compr Rev Food Sci Food Saf. 19(5):2333–2356. doi: 10.1111/1541-4337.12602. PMID: 33336985 [DOI] [PubMed] [Google Scholar]
- 99.Huang J, Ou Y, Yew TW, Liu J, Leng B, Lin Z,Su Y, Zhuang Y, Lin J, Li X, et al. 2016. Hepatoprotective effects of polysaccharide isolated from Agaricus bisporus industrial wastewater against CCl(4)-induced hepatic injury in mice. Int J Biol Macromol. 82:678–686. PMID: 26454111. doi: 10.1016/j.ijbiomac.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 100.Li S, Li J, Zhang J, Wang W, Wang X, Jing H, Ren Z, Gao Z, Song X, Gong Z, et al. 2017. The antioxidative, antiaging, and hepatoprotective effects of Alkali-Extractable polysaccharides by agaricus bisporus. Evid Based Complement Alternat Med. 2017:7298683. PMID: 29104605. doi: 10.1155/2017/7298683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu Y, Zheng D, Su L, Wang Q, Li Y. 2018. Protective effect of polysaccharide from Agaricus bisporus in Tibet area of China against tetrachloride-induced acute liver injury in mice. Int J Biol Macromol. 118:1488–1493. PMID: 29969638. doi: 10.1016/j.ijbiomac.2018.06.179. [DOI] [PubMed] [Google Scholar]
- 102.Duan Z, Zhang Y, Zhu C, Wu Y, Du B, Ji H. 2020. Structural characterization of phosphorylated Pleurotus ostreatus polysaccharide and its hepatoprotective effect on carbon tetrachloride-induced liver injury in mice. Int J Biol Macromol. 162:533–547. PMID: 32565302. doi: 10.1016/j.ijbiomac.2020.06.107. [DOI] [PubMed] [Google Scholar]
- 103.Bergh O, Borsheim KY, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340(6233):467–468. doi: 10.1038/340467a0. PMID: 2755508 [DOI] [PubMed] [Google Scholar]
- 104.Cobian Guemes AG, Youle M, Cantu VA, Felts B, Nulton J, Rohwer F. 2016. Viruses as winners in the game of life. Annu Rev Virol. 3(1):197–214. doi: 10.1146/annurev-virology-100114-054952. PMID: 27741409 [DOI] [PubMed] [Google Scholar]
- 105.Shkoporov AN, Clooney AG, Sutton TDS, Ryan FJ, Daly KM, Nolan JA, McDonnell SA, Khokhlova EV, Draper LA, Forde A, et al. 2019. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe. 26(4):527–41e5. PMID: 31600503. doi: 10.1016/j.chom.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 106.Shkoporov AN, Hill C. 2019. Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe. 25(2):195–209. doi: 10.1016/j.chom.2019.01.017. PMID: 30763534 [DOI] [PubMed] [Google Scholar]
- 107.Roux S, Hallam SJ, Woyke T, Sullivan MB. 2015. Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. Elife. 4 PMID: 26200428. doi: 10.7554/eLife.08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krishnamurthy SR, Wang D. 2017. Origins and challenges of viral dark matter. Virus Res. 239:136–142. PMID: 28192164. doi: 10.1016/j.virusres.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 109.Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, O’Regan O, Ryan FJ, Draper LA, Plevy SE, Ross RP, et al. 2019. Whole-Virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe. 26(6):764–78e5. PMID: 31757768. doi: 10.1016/j.chom.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 110.Gregory AC, Zablocki O, Zayed AA, Howell A, Bolduc B, Sullivan MB. 2020. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe. 28(5):724–40e8. doi: 10.1016/j.chom.2020.08.003. PMID: 32841606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Camarillo-Guerrero LF, Almeida A, Rangel-Pineros G, Finn RD, Lawley TD. 2021. Massive expansion of human gut bacteriophage diversity. Cell. 184(4):1098–109e9. doi: 10.1016/j.cell.2021.01.029. PMID: 33606979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wommack KE, Colwell RR. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 64:69–114. PMID: 10704475. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dalmasso M, Hill C, Ross RP. 2014. Exploiting gut bacteriophages for human health. Trends Microbiol. 22(7):399–405. doi: 10.1016/j.tim.2014.02.010. PMID: 24656964 [DOI] [PubMed] [Google Scholar]
- 114.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. 2016. Healthy human gut phageome. Proc Natl Acad Sci U S A. 113(37):10400–10405. doi: 10.1073/pnas.1601060113. PMID: 27573828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ross A, Ward S, Hyman P. 2016. More is better: selecting for broad host range bacteriophages. Front Microbiol. 7:1352. PMID: 27660623. doi: 10.3389/fmicb.2016.01352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cahill J, Young R. 2019. Phage lysis: multiple genes for multiple barriers. Adv Virus Res. 103:33–70. PMID: 30635077. doi: 10.1016/bs.aivir.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. 2017. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 11(7):1511–1520. doi: 10.1038/ismej.2017.16. PMID: 28291233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Human Microbiome Jumpstart Reference Strains C. KE N, GM W, SK H, KC W, HH C, Wortman JR, Rusch DB, Mitreva M, Sodergren E, Chinwalla AT, et al. 2010. A catalog of reference genomes from the human microbiome. Science. 3285981: 994–999. 10.1126/science.1183605PMID: 20489017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lengeling A, Mahajan A, Gally DL. 2013. Bacteriophages as pathogens and immune modulators?. mBio. 4(6):e00868–13. doi: 10.1128/mBio.00868-13. PMID: 24222490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Scotland SM, Smith HR, Willshaw GA, Rowe B. 1983. Vero cytotoxin production in strain of Escherichia coli is determined by genes carried on bacteriophage. Lancet. 2(8343):216. doi: 10.1016/s0140-6736(83)90192-7. PMID: 6135046 [DOI] [PubMed] [Google Scholar]
- 121.Tetz GV, Ruggles KV, Zhou H, Heguy A, Tsirigos A, Tetz V. 2017. Bacteriophages as potential new mammalian pathogens. Sci Rep. 7(1):7043. doi: 10.1038/s41598-017-07278-6. PMID: 28765534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco C, Zhao G, Fleshner P, et al. 2015. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 160(3):447–460. PMID: 25619688. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ma Y, You X, Mai G, Tokuyasu T, Liu C. 2018. A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome. 6(1):24. doi: 10.1186/s40168-018-0410-y. PMID: 29391057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, Lankowski A, Baldridge M, Wilen C, Flagg M, et al. 2016. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe. 19(3):311–322. PMID: 26962942. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, Li X, Szeto C-H, Sugimura N, Lam TYT, et al. 2018. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology. 155(2):529–41e5. PMID: 29689266. doi: 10.1053/j.gastro.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 126.Koonin EV, Dolja VV, Krupovic M. 2021. The healthy human virome: from virus-host symbiosis to disease. Curr Opin Virol. 47:86–94. PMID: 33652230. doi: 10.1016/j.coviro.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 127.Jiang L, Lang S, Duan Y, Zhang X, Gao B, Chopyk J, Schwanemann LK, Ventura‐Cots M, Bataller R, Bosques‐Padilla F, et al. 2020. Intestinal virome in patients with alcoholic hepatitis. Hepatology. 72(6):2182–2196. PMID: 32654263. doi: 10.1002/hep.31459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rojo J, Simoes P, Krueger GR, Humberto CO, Ramon AM. 2003. Human herpesvirus-6 has no apparent influence on course of HCV hepatitis, but may complicate HBV hepatitis and alcoholic liver disease. A pilot study. Vivo. 17:29–33. PMID: 12655786. [PubMed] [Google Scholar]
- 129.Yurlov KI, Masalova OV, Kisteneva LB, Khlopova IN, Samokhvalov EI, Malinovskaya VV,Parfyonov VV, Shuvalov AN, Kushch AA. 2021. Human herpesviruses increase the severity of hepatitis. Biology (Basel). 10. PMID: 34072365. doi: 10.3390/biology10060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tsai JP, Tseng KC, Lin MN, Su CC. 2019. A high seroprevalence of human herpesvirus type 8 already present in patients with chronic hepatitis before the development of cirrhosis. Pathology. 51(1):86–90. doi: 10.1016/j.pathol.2018.10.005. PMID: 30497802 [DOI] [PubMed] [Google Scholar]
- 131.Chou AL, Huang WW, Tsao SM, Li CT, Su CC. 2008. Human herpesvirus type 8 in patients with cirrhosis: correlation with sex, alcoholism, hepatitis B virus, disease severity, and thrombocytopenia. Am J Clin Pathol. 130(2):231–237. doi: 10.1309/4FMBN316N4792UQ4. PMID: 18628092 [DOI] [PubMed] [Google Scholar]
- 132.Hu J, Zhang X, Yu G, Cai H, Gu J, Hu M,Xiang D, Lian J, Yu L, Jia H, et al. 2019. Epstein-Barr virus infection is associated with a higher Child-Pugh score and may predict poor prognoses for patients with liver cirrhosis. BMC Gastroenterol. 19(1):94. PMID: 31215410. doi: 10.1186/s12876-019-1021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Keen EC, Dantas G. 2018. Close encounters of three kinds: bacteriophages, commensal bacteria, and host immunity. Trends Microbiol. 26(11):943–954. doi: 10.1016/j.tim.2018.05.009. PMID: 29909042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bajaj JS, Sikaroodi M, Shamsaddini A, Henseler Z, Santiago-Rodriguez T, Acharya C, Fagan A, Hylemon PB, Fuchs M, Gavis E, et al. 2021. ssInteraction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut. 70(6):1162–1173. PMID: 32998876. doi: 10.1136/gutjnl-2020-322470. [DOI] [PubMed] [Google Scholar]
- 135.Pirnay JP, Blasdel BG, Bretaudeau L, Buckling A, Chanishvili N, Clark JR,Corte-Real S, Debarbieux L, Dublanchet A, De Vos D, et al. 2015. Quality and safety requirements for sustainable phage therapy products. Pharm Res. 32(7):2173–2179. PMID: 25585954. doi: 10.1007/s11095-014-1617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cheroutre H, Madakamutil L. 2004. Acquired and natural memory T cells join forces at the mucosal front line. Nat Rev Immunol. 4(4):290–300. doi: 10.1038/nri1333. PMID: 15057787 [DOI] [PubMed] [Google Scholar]
- 137.Liu L, Gong T, Tao W, Lin B, Li C, Zheng X, Zhu S, Jiang W, Zhou R. 2019. Commensal viruses maintain intestinal intraepithelial lymphocytes via noncanonical RIG-I signaling. Nat Immunol. 20(12):1681–1691. doi: 10.1038/s41590-019-0513-z. PMID: 31636462 [DOI] [PubMed] [Google Scholar]
- 138.Gogokhia L, Buhrke K, Bell R, Hoffman B, Brown DG, Hanke-Gogokhia C, Ajami NJ, Wong MC, Ghazaryan A, Valentine JF, et al. 2019. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe. 25(2):285–99e8. PMID: 30763538. doi: 10.1016/j.chom.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Metzger RN, Krug AB, Eisenacher K. 2018. Enteric virome sensing-its role in intestinal homeostasis and immunity. Viruses. 10(4):146. doi: 10.3390/v10040146. PMID: 29570694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Broquet AH, Hirata Y, McAllister CS, MF K. 2011. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol. 186(3):1618–1626. doi: 10.4049/jimmunol.1002862. PMID: 21187438 [DOI] [PubMed] [Google Scholar]
- 141.Norman JM, Handley SA, Virgin HW. 2014. Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology. 146(6):1459–1469. doi: 10.1053/j.gastro.2014.02.001. PMID: 24508599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, White RC, Clarke TH, Nguyen K, Torralba M, et al. 2019. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. 575(7783):505–511. PMID: 31723265. doi: 10.1038/s41586-019-1742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kortright KE, Chan BK, Koff JL, Turner PE. 2019. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe. 25(2):219–232. doi: 10.1016/j.chom.2019.01.014. PMID: 30763536 [DOI] [PubMed] [Google Scholar]
- 144.Loc-Carrillo C, Abedon ST. 2011. Pros and cons of phage therapy. Bacteriophage. 1(2):111–114. doi: 10.4161/bact.1.2.14590. PMID: 22334867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.DeLong EF. 1992. Archaea in coastal marine environments. Proc Natl Acad Sci U S A. 89(12):5685–5689. doi: 10.1073/pnas.89.12.5685. PMID: 1608980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schleper C, Jurgens G, Jonuscheit M. 2005. Genomic studies of uncultivated archaea. Nat Rev Microbiol. 3(6):479–488. doi: 10.1038/nrmicro1159. PMID: 15931166 [DOI] [PubMed] [Google Scholar]
- 147.Auguet JC, Barberan A, Casamayor EO. 2010. Global ecological patterns in uncultured Archaea. ISME J. 4(2):182–190. doi: 10.1038/ismej.2009.109. PMID: 19847207 [DOI] [PubMed] [Google Scholar]
- 148.Pereira O, Hochart C, Auguet JC, Debroas D, Galand PE. 2019. Genomic ecology of Marine Group II, the most common marine planktonic Archaea across the surface ocean. Microbiologyopen. 8(9):e00852. doi: 10.1002/mbo3.852. PMID: 31264806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Santoro AE, Richter RA, Dupont CL. 2019. Planktonic marine archaea. Ann Rev Mar Sci. 11(1):131–158. doi: 10.1146/annurev-marine-121916-063141. PMID: 30212260 [DOI] [PubMed] [Google Scholar]
- 150.Albers SV, Meyer BH. 2011. The archaeal cell envelope. Nat Rev Microbiol. 9(6):414–426. doi: 10.1038/nrmicro2576. PMID: 21572458 [DOI] [PubMed] [Google Scholar]
- 151.Koga Y, Morii H. 2007. Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol Mol Biol Rev. 71(1):97–120. doi: 10.1128/MMBR.00033-06. PMID: 17347520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Valentine DL. 2007. Adaptations to energy stress dictate the ecology and evolution of the archaea. Nat Rev Microbiol. 5(4):316–323. doi: 10.1038/nrmicro1619. PMID: 17334387 [DOI] [PubMed] [Google Scholar]
- 153.Borrel G, McCann A, Deane J, Neto MC, Lynch DB, Brugere JF,O'Toole PW. 2017. Genomics and metagenomics of trimethylamine-utilizing archaea in the human gut microbiome. ISME J. 11(9):2059–2074. PMID: 28585938. doi: 10.1038/ismej.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dridi B, Henry M, El Khechine A, Raoult D, Drancourt M, Dobrindt U. 2009. High prevalence of methanobrevibacter smithii and methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS One. 4(9):e7063. doi: 10.1371/journal.pone.0007063. PMID: 19759898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Horz HP, Conrads G. 2010. The discussion goes on: what is the role of Euryarchaeota in humans? Archaea. 2010:967271. PMID: 21253553. doi: 10.1155/2010/967271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hajishengallis G, Darveau RP, Curtis MA. 2012. The keystone-pathogen hypothesis. Nat Rev Microbiol. 10(10):717–725. doi: 10.1038/nrmicro2873. PMID: 22941505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gaci N, Borrel G, Tottey W, O’Toole PW, Brugere JF. 2014. Archaea and the human gut: new beginning of an old story. World J Gastroenterol. 20(43):16062–16078. doi: 10.3748/wjg.v20.i43.16062. PMID: 25473158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Borrel G, Brugere JF, Gribaldo S, Schmitz RA, Moissl-Eichinger C. 2020. The host-associated archaeome. Nat Rev Microbiol. 18(11):622–636. doi: 10.1038/s41579-020-0407-y. PMID: 32690877 [DOI] [PubMed] [Google Scholar]
- 159.Bang C, Schmitz RA, Robinson NP. 2018. Archaea: forgotten players in the microbiome. Emerg Top Life Sci. 2(4):459–468. doi: 10.1042/ETLS20180035. PMID: 33525830 [DOI] [PubMed] [Google Scholar]
- 160.Pausan MR, Csorba C, Singer G, Till H, Schopf V, Santigli E, Klug B, Högenauer C, Blohs M, Moissl-Eichinger C, et al. 2019. Exploring the archaeome: detection of archaeal signatures in the human body. Front Microbiol. 10:2796. PMID: 31866971. doi: 10.3389/fmicb.2019.02796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Grine G, Lotte R, Chirio D, Chevalier A, Raoult D, Drancourt M, Ruimy R. 2019. Co-culture of Methanobrevibacter smithii with enterobacteria during urinary infection. EBioMedicine. 43:333–337. PMID: 31072770. doi: 10.1016/j.ebiom.2019.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. 2004. Methanogenic archaea and human periodontal disease. Proc Natl Acad Sci U S A. 101(16):6176–6181. doi: 10.1073/pnas.0308766101. PMID: 15067114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Blais Lecours P, Marsolais D, Cormier Y, Berberi M, Hache C, Bourdages R, Duchaine C. 2014. Increased prevalence of methanosphaera stadtmanae in inflammatory bowel diseases. PLoS One. 9(2):e87734. doi: 10.1371/journal.pone.0087734. PMID: 24498365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pimentel M, Lin HC, Enayati P, van den Burg B, Lee HR, Chen JH, Park S, Kong Y, Conklin J. 2006. Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am J Physiol Gastrointest Liver Physiol. 290(6):G1089–95. doi: 10.1152/ajpgi.00574.2004. PMID: 16293652 [DOI] [PubMed] [Google Scholar]
- 165.Triantafyllou K, Chang C, Pimentel M. 2014. Methanogens, methane and gastrointestinal motility. J Neurogastroenterol Motil. 20(1):31–40. doi: 10.5056/jnm.2014.20.1.31. PMID: 24466443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bang C, Weidenbach K, Gutsmann T, Heine H, Schmitz RA, Foligne B. 2014. The intestinal archaea methanosphaera stadtmanae and methanobrevibacter smithii activate human dendritic cells. PLoS One. 9(6):e99411. doi: 10.1371/journal.pone.0099411. PMID: 24915454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Blais Lecours P, Duchaine C, Taillefer M, Tremblay C, Veillette M, Cormier Y, Marsolais D. 2011. Immunogenic properties of archaeal species found in bioaerosols. PLoS One. 6(8):e23326. doi: 10.1371/journal.pone.0023326. PMID: 21858070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Vierbuchen T, Bang C, Rosigkeit H, Schmitz RA, Heine H. 2017. The human-associated archaeon methanosphaera stadtmanae is recognized through its RNA and induces TLR8-Dependent NLRP3 inflammasome activation. Front Immunol. 8:1535. PMID: 29181003. doi: 10.3389/fimmu.2017.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bang C, Vierbuchen T, Gutsmann T, Heine H, Schmitz RA, Proost P. 2017. Immunogenic properties of the human gut-associated archaeon methanomassiliicoccus luminyensis and its susceptibility to antimicrobial peptides. PLoS One. 12(10):e0185919. doi: 10.1371/journal.pone.0185919. PMID: 28982164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, Latreille P, Kim K, Wilson RK, Gordon JI, et al. 2007. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci U S A. 104(25):10643–10648. PMID: 17563350. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gao B, Zhang X, Schnabl B. 2021. Fungi-bacteria correlation in alcoholic hepatitis patients. Toxins (Basel). 13(2):143. doi: 10.3390/toxins13020143. PMID: 33672887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Jansson JK, Dorrestein PC, Knight R. 2016. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 535(7610):94–103. doi: 10.1038/nature18850. PMID: 27383984 [DOI] [PubMed] [Google Scholar]
- 173.Wu X, Xia Y, He F, Zhu C, Ren W. 2021. Intestinal mycobiota in health and diseases: from a disrupted equilibrium to clinical opportunities. Microbiome. 9(1):60. doi: 10.1186/s40168-021-01024-x. PMID: 33715629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Townsend EM, Kelly L, Muscatt G, Box JD, Hargraves N, Lilley D, Jameson E. 2021. The human gut phageome: origins and roles in the human gut microbiome. Front Cell Infect Microbiol. 11:643214. PMID: 34150671. doi: 10.3389/fcimb.2021.643214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


