Abstract
The architecture of flowering plants exhibits both phenotypic diversity and plasticity, determined, in part, by the number and activity of axillary meristems and, in part, by the growth characteristics of the branches that develop from the axillary buds. The plasticity of shoot branching results from a combination of various intrinsic and genetic elements, such as number and position of nodes and type of growth phase, as well as environmental signals such as nutrient availability, light characteristics, and temperature (Napoli et al., 1998; Bennett and Leyser, 2006; Janssen et al., 2014; Teichmann and Muhr, 2015; Ueda and Yanagisawa, 2019). Axillary meristem initiation and axillary bud outgrowth are controlled by a complex and interconnected regulatory network. Although many of the genes and hormones that modulate branching patterns have been discovered and characterized through genetic and biochemical studies, there are still many gaps in our understanding of the control mechanisms at play. In this review, we will summarize our current knowledge of the control of axillary meristem initiation and outgrowth into a branch.
The key regulatory genes and the role of multiple plant hormones coordinate the process of axillary meristem initiation and subsequent growth into a branch.
Axillary meristem initiation
The axillary meristem is initiated from the boundary between the shoot apex and leaf primordia
Axillary meristem initiation occurs in the meristematic cell niche at the adaxial side of the leaf axils and requires the establishment of a boundary between the main shoot and the leaf primordium. Boundary formation is critical for plant organogenesis, separating the pluripotent meristematic cell population that remains indeterminate from the developing organs. The cells in boundaries have reduced cell division, down-regulated cell-cycle-related genes, and relatively stiff cell walls (reviewed in Wang et al., 2016; Richardson and Hake 2019). In addition to physically separating neighboring tissues, boundaries contribute to various developmental processes, including axillary meristem initiation, leaf shape, fruit dehiscence, and organ abscission (reviewed in (Hepworth and Pautot, 2015)). The low rate of cell division within the boundary is controlled by a regulatory network in which regulatory genes such as CUC (CUP-SHAPED COTYLEDON), STM (SHOOT MERISTEMLESS), and LOB (LATERAL ORGAN BOUNDARIES) suppress cell division and differentiation and modulate the spatial distribution of growth promoting hormones, such as auxin, gibberellin (GA), and brassinosteroids (BR; Jasinski et al., 2005; Borghi et al., 2007; Takeda et al., 2011; Bell et al., 2012; Wang et al., 2016; Richardson and Hake, 2019). Several studies indicate that the boundary-expressed genes described above are required for axillary meristem formation, with loss-of-function or down-regulation of these genes resulting in organ fusion and axillary meristem defects, while overexpression of these genes led to ectopic axillary meristem formation (Greb et al., 2003; Vroemen et al., 2003; Hibara et al., 2006; Gómez-Mena and Sablowski, 2008; Raman et al., 2008; Lee et al., 2009; Spinelli et al., 2011).
Advances
Recent work has shown that axillary meristem initiation and outgrowth is controlled by a complex network involving interactions between multiple hormones, miRNAs and transcription factors that integrate environmental information to optimize growth.
BRC1 is a key hub protein that is involved in the control of branching. However, there is also BRC1 independent regulation of branching involving SL, auxin, CK, and T6P.
The SL signaling repressors SMXL6,7,8 can act as TFs, and SMXL6 is able to bind directly to the SMXL6,7,8 and BRC1 promoters to suppress their expression.
Both photosynthetic and signaling sugars have been increasingly recognized as important regulators of branching.
Organ boundary and meristem identity genes form linked regulatory networks for axillary meristem initiation
The maintenance of meristematic competence and subsequent axillary meristem initiation is regulated by a set of transcription factors (TFs), which appear to be conserved between species. Some of these TFs are important for multiple developmental processes; however, in this section, particular attention is given to their roles in axillary meristem initiation (Figure 1). Axillary meristem initiation involves protein movements, transcriptional regulation, protein–protein interactions, and feedback regulation in multiple pathways. Such complexity can be illustrated by the regulation of STM, a meristematic marker, by at least two interconnected yet independent pathways, while at the same time reporting recent data on the wider network.
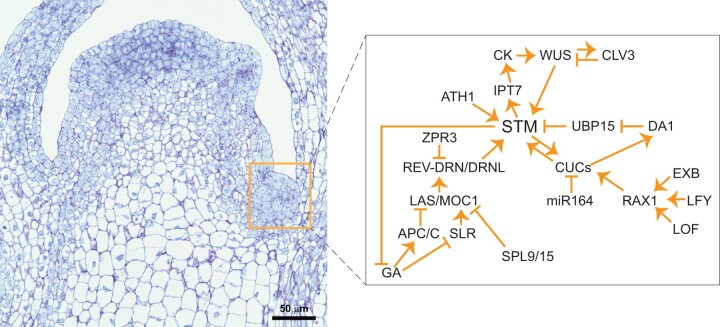
Figure 1.
Interacting regulatory pathways for axillary meristem initiation at the boundary between shoot and leaf primordium. The photograph shows a longitudinal section of a petunia SAM and leaf primordium (scale bar is 50 µm). The inserted box denotes the region where the axillary meristem initiates. The expanded box shows the proposed regulatory networks. STM, a meristematic marker, promotes axillary meristem initiation through CK and the WUS-CLVs pathway. STM is regulated by both LAS/MOC1 through the REV-DRN/DRNL pathway and RAXs through CUCs and/or the DA1 pathway. These two pathways are interconnected by CUCs. LAS is degraded by the APC/C complex that involves GA signaling, while RAX1 is able to interact with a number of TFs such as EXB, LFY, and LOF. The arrows and blunt lines represent positive and negative regulation, respectively.
STM, like its homologs from other species (ORYZA SATIVA HOMEOBOX1 (OSH1) in rice (Oryza sativa) and knotted1 (kn1) in maize, Zea mays), is required for establishing and maintaining apical meristems as well as axillary meristem identity (Long et al., 1996; Shi et al., 2016). STM expression changes coinciding with axillary meristem initiation and is controlled by a number of regulators (Greb et al., 2003; Bolduc et al., 2012; Shi et al., 2016; Wang et al., 2017; Scofield et al., 2018; Zhang et al., 2018; Cao et al., 2020; Xue et al., 2020). Initially, STM expression in the leaf axils is low yet appears to be sufficient to maintain meristematic cell competence (Shi et al., 2016; Zhang et al., 2018; Cao et al., 2020). An ATH1 (ARABIDOPSIS THALIANA HOMEOBOX GENE1)-STM heterodimer can bind to the STM gene and maintain its expression (Cao et al., 2020). In addition, REVOLUTA (REV) can interact with DORNRÖSCHEN (DRN)/DORNRÖSCHEN-LIKE (DRNL) to promote STM expression. Early in development, LITTLE ZIPPER3 (ZPR3) prevents the interaction of REV-DRN/DRNL, resulting in low levels of STM. Later in development, ZPR3 expression is decreased allowing REV-DRN/DRNL to interact, promoting STM expression (Shi et al., 2016; Zhang et al., 2018).
STM can promote axillary meristem initiation by upregulating the expression of cytokinin (CK) biosynthesis genes (e.g. ISOPENTENYLTRANSFERASE7 (IPT7)) and CK-response regulators (Jasinski 2005; Yanai et al., 2005). After STM-induced meristematic activity, activation of WUSCHEL (WUS) by CK signaling promotes axillary meristem initiation in the leaf axils (Shi et al., 2016; Wang et al., 2017). Interestingly, a recent study showed that WUS proteins are able to activate STM expression and bind to STM protein directly to promote CLAVATA3 (CLV3) expression (Su et al., 2020). These results suggest that STM plays a crucial role in maintaining the shoot–leaf boundary during axillary meristem initiation and regulating hormone content during axillary meristem initiation.
LATERAL SUPPRESSOR (LAS) of Arabidopsis thaliana and its homologs, Ls of tomato (Solanum lycopersicum), MONOCULM 1 (MOC1) of rice, ERAMOSA (ERA) of snapdragon (Antirrhinum majus), and AaLAS of Arabis alpina are boundary-specific GRAS family TFs that control meristem initiation through regulation of STM (Schumacher et al., 1999; Otsuga et al., 2001; Greb et al., 2003; Li et al., 2003; Mizzotti et al., 2017; Ponraj and Theres, 2020). LAS regulates STM expression through REV; however, mutation of LAS affects vegetative axillary meristems, while mutation of STM affects both shoot and floral meristem function (Endrizzi et al., 1996; Greb et al., 2003; Shi et al., 2016). Rice moc1 mutants affect the expression of OSH1 and OsTB1 (a homolog of maize teosinte branched1 (tb1)) in leaf axils only, whereas the overexpression of MOC1 in rice resulted in tiller outgrowth from higher order tiller buds (Li et al., 2003), implying that MOC1 may be a key regulator for both axillary meristem initiation and outgrowth.
Although the LAS function was conserved between perennial A. alpina and annual Arabidopsis and tomato (Ponraj and Theres, 2020), legumes appear to have lost a LAS-like TF during evolution (Mizzotti et al., 2017). The regulatory mechanism of MOC1 in rice has been studied in some detail: Lin et al. (2012) and Xu et al. (2012) both reported that the anaphase promoting complex/cyclosome (APC/C) E3 ubiquitin ligase complex mediates the degradation of MOC1 in leaf axils, resulting in down-regulation of OSH1 and repression of axillary meristem initiation. Xu et al. (2012) also showed that MOC1 degradation is cell-cycle dependent; when the degradation was blocked, the accumulation of MOC1 resulted in an increase in tiller numbers. Based on these results, it is possible that the APC/C–MOC1 complex activates during the G1-phase to maintain MOC1 at a relatively low level in axillary buds (Xu et al., 2012); however, whether MOC1 directly regulates cell division is still unclear and whether OSH1 or OsTB1 are the transcriptional targets of MOC1 is yet to be confirmed. Liao et al. (2019) have shown that the DELLA protein SLENDER RICE 1 can prevent the degradation of MOC1. High levels of GAs are also able to activate the APC/C complex and promote degradation of MOC1 (Lin et al., 2020).
In Arabidopsis, a number of TFs have been shown to affect LAS expression, such as SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE9 (SPL9) and SPL15 and CUC2 (Tian et al., 2014). Although evidence suggests that CUC2 proteins and probably CUC1 bind to the LAS promoter to enhance the expression of LAS (Hibara et al., 2006; Tian et al., 2014), an additive effect of axillary meristem defects was found in cuc2 las and cuc3 las double mutants (Hibara et al., 2006), suggesting LAS works at least partially independently of CUC2/CUC3. These lines of evidence suggest that LAS/MOC1 is a major regulator that integrates a number of signals and contributes to the STM-WUS axillary meristem initiation mechanism.
Studies of double mutants of LAS and REGULATOR OF AXILLARY MERISTEMS1 (RAX1)/Blind reveal additional complexities in the regulation of axillary meristems through STM involving RAX, CUCs, and microRNA164 (miR164; Schmitz et al., 2002; Müller et al., 2006). In the Arabidopsis rax1 rax2 rax3 triple mutant, almost no axillary meristems were able to initiate and the STM mRNA was undetectable in the leaf axils, while the expression of STM in the shoot apical meristem (SAM) was unaffected (Müller et al., 2006). The axillary meristem defect in the rax mutants could be restored by exogenous application of CK or overproduction of CK in leaf axils (Wang et al., 2014). The expression of RAX genes is regulated by a number of TFs, such as EXB1 (EXCESSIVE BRANCHES, Guo et al., 2015), LFY (LEAFY, Chahtane et al., 2013), and LOF1 (LATERAL ORGAN FUSION1, Lee et al., 2009). RAXs modulate the expression of CUC2 (Keller et al., 2006), which, as mentioned above, promotes LAS expression.
CUC1 and STM regulate each other’s expression through a positive feedback loop, which also involves miR164 (Raman et al., 2008; Spinelli et al., 2011; Scofield et al., 2018). This interaction requires the movement of STM protein from the SAM to the boundary, as a nonmobile version of STM failed to rescue the stm mutant and caused down-regulation of CUC1–CUC3 expression in inflorescence meristems, resulting in phenotypes similar to those in the cuc2 cuc3 double mutant (Balkunde et al., 2017). Recently, Li et al. (2020) found that CUC2 and CUC3 proteins also regulate the expression of DA1 and the DA1 substrate UBIQUITIN-SPECIFIC PROTEASE15 (UBP15). When DA1 is mutated, STM expression is lost and axillary meristems fail to initiate in Arabidopsis (Li et al., 2020), although a direct link between UBP15 or DA1 and STM has yet to be identified.
Axillary meristem outgrowth
Bud outgrowth from axillary meristems leads to the development of branches and is triggered by the perception of developmental and environmental cues, such as meristem age and location, nutrients, light, and temperature (Janssen et al., 2014). The decision to grow into a branch also takes into account information from throughout the plant, such as the presence of additional growing shoots. Both internal and external factors are integrated to determine where and when a bud will grow to form a branch. Much of our current knowledge of the control of branch growth centers on the roles of plant hormones, particularly of auxin, CK, and strigolactones (SL; for in-depth reviews of the roles of auxin and CK in particular, see Muller and Leyser, 2011; Harrison, 2017; Barbier et al., 2019). Here, we will summarize advances that help clarify the role of SLs in branching, along with key recent results for other plant hormones.
SL is a core component of signaling and regulatory networks for branching
SL biosynthesis genes, such as CAROTENOID CLEAVAGE DIOXYGENASE7 (CCD7), CCD8, and MORE AXILLARY GROWTH1 (MAX1), and signaling perception genes, such as DECREASED APICAL DOMINANCE (DAD2)/DWARF14 (D14)/AtD14/RAMOSUS3 (RMS3) and MAX2/D3/RMS4/PhMAX2A, have been identified mainly through analysis of highly branched mutants from a number of model species (Beveridge et al., 1994, 1997, 2000; Booker et al., 2005; Snowden et al., 2005; Zou et al., 2006; Arite et al., 2007; Simons et al., 2007; Stirnberg et al., 2007; Arite et al., 2009; Xie et al., 2010; Hamiaux et al., 2012; Janssen and Snowden, 2012; Jiang et al., 2013). For axillary buds on these plants, reduced or abolished SL production or signaling resulted in alteration of their outgrowth potential from dormant to active growth, for instance, petunia (Petunia hybrida) dad1 mutant plants produced branches at the leaf axils of the cotyledons and the first two basal nodes, where normally no branches develop in wild-type (WT; Snowden et al., 2005). The highly branched phenotype of SL-deficient mutants can be rescued by treatment with a synthetic SL analogue, GR24, or by grafting the mutant scion onto WT rootstocks, whereas these treatments cannot rescue the branched phenotype from the SL-insensitive mutants (Foo et al., 2001; Sorefan et al., 2003; Simons et al., 2007; Arite et al., 2009; Lin et al., 2009; Hamiaux et al., 2012; Waters et al., 2012). CCD7 and CCD8 have also been isolated in some woody perennial plants, such as poplar, kiwifruit, apple, and grape, and evidence suggests they have comparable roles to those observed in model systems, particularly in regulating branching in the shoot (Ledger et al., 2010; Vogel et al., 2010; Kohlen et al., 2012; Muhr et al., 2016; Pan et al., 2016; Foster et al., 2017; Gao et al., 2018; Wu et al., 2019; Ren et al., 2020). Collectively, these studies reveal a conserved function and regulatory pathway of SL signaling in controlling axillary bud outgrowth (Delaux et al., 2012; Ruyter-Spira et al., 2013; Janssen et al., 2014; Machin et al., 2019; Walker et al., 2019).
Grafting studies using SL mutants and corresponding WT plants from several species suggest that SL made in either the root or stem of plants can influence branching in the shoot system, and that SL moves acropetally through the plant. The mechanism of SL transport from the root to the shoot and into axillary buds is not completely resolved. Currently, PLEIOTROPIC DRUG RESISTANCE1 (PDR1) from petunia is the only characterized SL transporter, and pdr1 mutants have reduced SL exudation from roots and produce more branches with vigorous outgrowth (Kretzschmar et al., 2012). PDR1 appears to be involved in local short-distance transport, such as unloading SLs from roots into soil and from the shoot into axillary buds (Sasse et al., 2015; Shiratake et al., 2019). PDR1 might also contribute to the long-distance root-to-shoot transport of SL; however, grafting experiments indicate that there must also be a PDR1 independent pathway, which might involve the vasculature and/or other transporters that target SL precursors (Sasse et al., 2015; Shiratake et al., 2019). It is unclear whether SL is transported through the xylem (Kohlen et al., 2011; Xie et al., 2015). Xie et al. (2015) could not detect SLs or carlactone, an SL precursor, from xylem sap of Arabidopsis, rice, tomato, cucumber (Cucumis sativus), sorghum (Sorghum bicolor), or tobacco (Nicotiana tabacum), contrasting with the results from Kohlen et al. (2011) who did report detection of SLs (orobanchol and orobanchyl acetate) from Arabidopsis and tomato xylem sap.
The effects of SL on shoot branching are mediated through a complex containing an α/β fold hydrolase SL receptor, AtD14/DAD2/RMS3/D14, and an F-box protein, MAX2/PhMAX2A/RMS4/D3, targeting SMXLs/PhD53A/D53 for ubiquitination and subsequent degradation, and downstream transcriptional targets include TFs such as BRANCHED1 (BRC1) and IDEAL PLANT ARCHITECTURE1 (IPA1; Ishikawa et al., 2005; Stirnberg et al., 2007; Hamiaux et al., 2012; Janssen and Snowden, 2012; Nakamura et al., 2013; Bennett et al., 2016; Figure 2). The SL receptor contains a canonical hydrolase catalytic triad in its cavity and has slow hydrolytic activity toward SLs, resulting in covalent attachment of a fragment of SL to the receptor protein (Hamiaux et al., 2012; de Saint Germain et al., 2016; Yao et al., 2016). This hydrolytic activity might occur after the perception of SL and signal transduction, as AtD14D218A, a mutant for one of the catalytic triad residues, was able to complement the atd14 mutant despite lacking hydrolytic activity (Seto et al., 2019).
Figure 2.
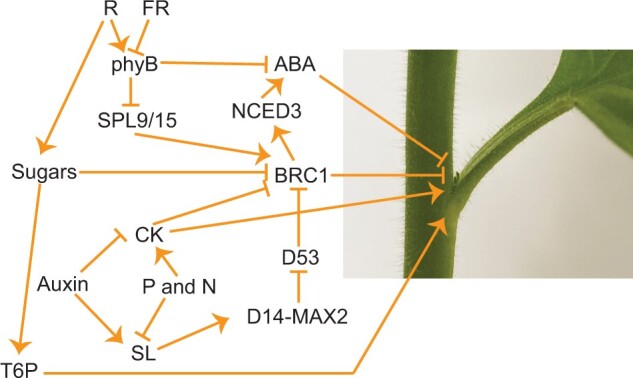
Selected components of axillary bud outgrowth regulation. Red light (R) promotes branching through phyB and SPL9/15 that regulate BRC1 (note FR is far-red light). Light quality also reduces ABA level, and increases sugar content, which suppresses BRC1 expression. Nutrient availability positively regulates CK content but negatively regulates SL production. SLs are transported at least partly by PDR1 and are perceived by the DAD2-MAX2-D53 complex. The degradation of D53 triggers BRC1 expression that inhibits axillary bud outgrowth. There are likely BRC1 independent pathways that involve T6P and SL and CK signaling. The arrows and blunt lines represent positive and negative regulation respectively.
D53 in rice and its homologs, SUPPRESSOR OF MORE AXILLARY GROWTH1-LIKE6,7, and 8 (SMXL6,7,8) in Arabidopsis are nuclear-localized SL-signaling repressors and are the primary targets of the SL receptor complex (Jiang et al., 2013; Zhou et al., 2013; Wang et al., 2015; Bennett et al., 2016). Phenotypes in single, double, and triple mutants of SMXL6, SMXL7, and SMXL8 in Col-0 and the max3 mutant background suggested that they function redundantly in promoting axillary bud outgrowth in Arabidopsis; however, there may be differences in the functions of the different SMXLs. For instance, SMXL6 can interact with MAX2 and AtD14 without GR24, while SMXL7 can interact with AtD14 only in the presence of GR24 (Wang et al., 2015; Liang et al., 2016). A recent study showed that SMXLs function as TFs that repress their own transcription through direct binding to the promoters (Wang et al., 2020); meanwhile, SMXL6 is also able to interact with other corepressor/partner proteins, such as TOPLESS-RELATED PROTEIN (TRP) and SPL9 and SPL15 to repress the expression of BRC1 (Wang et al., 2015, 2020; Xie et al., 2020). Similarly, it has been shown that D53 directly interacts with OsSPL14/IPA1 and TRP in rice and TaSPL17 in wheat (Liu et al., 2017; Ma et al., 2017; Song et al., 2017). D53 and SMXLs are able to regulate a number of targets (Wang et al., 2020); however, TFs such as BRC1/FC1/TB1 and SPLs also play a major role in the branching regulatory network.
As mentioned above, auxin and CK also play important roles in the branching regulatory network (reviewed in Muller and Leyser, 2011; Harrison, 2017; Barbier et al., 2019). In addition, it is clear that there is a role for abscisic acid (ABA) in regulating bud dormancy (Luo et al., 2019). Although BRs and GAs might not be considered to be part of the core hormone signaling pathway for controlling shoot branching (Bennett et al., 2016), recent evidence suggests they are part of the complex regulatory networks linking pathways and affect branching indirectly or as supplementary mechanisms (Ito et al., 2017; Fang et al., 2020; Zhang et al., 2020). In many plant species, further development of axillary buds into branches is suppressed by the influence of the primary shoot. The suppression mechanism of auxin on axillary bud outgrowth is yet to be fully elucidated, especially when auxin seems to act indirectly as it moves basipetally within the main stem and does not move into axillary buds (Muller and Leyser, 2011). Recently, a study indicated that at the initial stage, decapitation and CK induced bud outgrowth is independent of auxin flow from the bud (Chabikwa et al., 2019). Thus, auxin is likely to indirectly regulate branching by modulating other hormones such as CKs and SLs (Rameau et al., 2014; Barbier et al., 2019). Auxin positively regulates the expression of SL biosynthesis genes CCD7 and CCD8 in Arabidopsis, pea (Pisum sativum), and rice (Foo et al., 2005; Arite et al., 2007; Hayward et al., 2009) and, in turn, SL signaling modulates polar auxin transport and auxin biosynthesis (Hayward et al., 2009; Crawford et al., 2010; Shinohara et al., 2013; Ligerot et al., 2017).
There are several studies that suggest CK promotes branching in model species as well as woody perennials (Dun et al., 2012; Tan et al., 2019; and reviewed in Barbier et al., 2019). However, in some species, for instance Rosa hybrida, suppressing CK biosynthesis and signaling does not repress axillary bud outgrowth (Barbier et al., 2015). CK signaling results in a large number of transcriptional and other changes in relation to axillary bud outgrowth including changes in other hormone signaling pathways (Ni et al., 2017; Li et al., 2018; Waldie and Leyser, 2018). SL and CK regulatory pathways interact on a number of levels; first, at the transcriptional target level, both SLs and CK regulate the expression of BRC1, at least in Arabidopsis and pea (Dun et al., 2013); second, at the hormone production and degradation level, SLs have been shown to reduce CK levels through the transcriptional activation of CYTOKININ OXIDASE/DEHYDROGENASE 9 in rice (Duan et al., 2019). In pea SL mutants (rms1 and rms4), the expression of one of the CK biosynthesis genes, ISOPENTYLTRANSFERASE1 (IPT1) was increased significantly compared with WT in node 3 of pea plants (Dun et al., 2012). These studies further support the cross talk among hormones during bud outgrowth.
The transcription factor BRC1 is a key regulator of branching in axillary buds
The most well-characterized transcriptional regulator in branching regulatory pathways is BRC1 and its homologs in the TCP (TB1, CYCLOIDIA, and PROLIFERATING CELL FACTOR) TF family, including tb1 of maize, FINE CULM1 (FC1)/OsTB1 of rice, BRC1 and BRC2 from Arabidopsis, and PhTCP3 of petunia and PsBRC1 of pea. These genes are expressed in axillary buds or in the leaf axils where axillary buds develop, where they can repress bud outgrowth without altering the number of axillary buds (Doebley et al., 1997; Takeda et al., 2003; Aguilar-Martínez et al., 2007; Finlayson, 2007; Minakuchi et al., 2010; Braun et al., 2012; Drummond et al., 2015; Nicolas and Cubas, 2016). Mutation and knock down of BRC1 resulted in increased rosette branching in Arabidopsis (Aguilar-Martínez et al., 2007; Finlayson, 2007), strong basal branching in pea (Braun et al., 2012), and enhanced lateral tillering in rice (Takeda et al., 2003). BRC1 expression is significantly higher in basal buds than apical buds and hence corresponded with branching (Finlayson, 2007; Drummond et al., 2015). The expression of BRC1 is regulated by several pathways, such as SL and CK signaling, decapitation/auxin treatment, as well as sucrose treatment, nutrient availability, and light quality, and it is thought to integrate many of these shoot-branching signals in flowering plants (reviewed in Teichmann and Muhr, 2015; Barbier et al., 2019; Wang et al., 2019).
It is important to note that BRC1 is not the only mechanism that plants employ to inhibit branching (Figure 2). Not all nodes produce a branch in Arabidopsis brc1 brc2 double mutants or pea psbrc1 mutants, although they have twice as many or more branches than WT (Braun et al., 2012; Seale et al., 2017). The brc1 bcr2 double mutant Arabidopsis plants are still sensitive to low N-induced and SL-induced branch suppression (Seale et al., 2017); however, this is in contrast to the psbrc1 mutants in pea and fc1 mutants in rice that are insensitive to SL treatment (Minakuchi et al., 2010; Braun et al., 2012). Moreover, CK can increase branch length in psbrc1 mutants, suggesting CK has a BRC1-independent effect on sustaining bud outgrowth (Braun et al., 2012). The phenotypic difference between SL mutants and brc1 mutants suggests that BRC1 is not the sole regulator for branching. The pea rms1 mutant produced more branches at upper nodes, whereas psbrc1 mutant plants had more branches at the basal nodes. In addition, the rms1 psbrc1 double mutant had more branches than either rms1 or psbrc1 mutants at most of the nodes with the exception of node 2 (Braun et al., 2012). Similarly, in Arabidopsis, the max4 or d14 mutants had more branches than the brc1 brc2 double mutant and the triple mutant had an additive phenotype (Chevalier et al., 2014; Seale et al., 2017). On the other hand, fc1 d17 (an SL-deficient mutant) double mutant rice had a similar phenotype to d17 single mutants and GR24 application had no significant effect on the expression of FC1 (Minakuchi et al., 2010). A recent study suggested that ABCB19, an auxin transporter, might respond to GR24 but acts on a separate pathway from BRC1 (van Rongen et al., 2019). These observations suggest that BRC1/FC1/TB1 acts downstream of, but not exclusive to, the SL-signaling pathway, and that BRC1-independent branching regulation is likely, but yet to be identified and characterized.
Despite recent progress, our knowledge of the mechanism of action for BRC1 inhibition of bud growth remains incomplete. There are a number of possible routes downstream of BRC1 leading to bud growth inhibition. Recently, Shen et al. (2019) found that CsBRC1 in cucumber directly binds to the CsPIN3 promoter to repress the transcription of PIN-FORMED3 (PIN3), an auxin efflux carrier, possibly leading to reduced auxin export from axillary buds and subsequently suppression of branch outgrowth. BRC1 proteins are also able to bind to three HD-ZIP encoding genes, HB21, HB40, and HB53 to up-regulate their expression, resulting in up-regulation of 9-CIS-EPOXICAROTENOID DIOXIGENASE 3 (NCED3) and accumulation of ABA, causing inhibition of branch outgrowth (González-Grandío et al., 2017). BRC1 expression was not affected by ABA application, further supporting the idea that ABA signaling in branching regulation is downstream of BRC1 (Yao and Finlayson, 2015). More recently, Luo et al. (2019) provide further evidence of the role of ABA in the downstream regulation of bud dormancy, and this action is expected to be BRC1 dependent in some species (Wang et al., 2020).
Axillary branching is highly plastic in response to environmental signals
Our understanding of how the environment, especially nutrient concentrations and light quality, influence branching has advanced rapidly thanks to powerful genetic screening and transcriptomic tools (Figure 2).
Phosphate (P) and nitrogen (N) are the two macronutrients that limit plant growth and yield potential, and limited supply of N and/or P modulate root architecture, shoot branching, and flowering time through regulation of hormones and transcriptional changes (Péret et al., 2014; Ham et al., 2018; Luo et al., 2020). Hormones, especially SLs, are affected by nutrient availability (Umehara et al., 2008; Mayzlish-Gati et al., 2012; Al-Babili and Bouwmeester, 2015; Luo et al., 2020). Limited availability of P, N, and sometimes sulfur promotes SL production in, and exudation from, the roots, upregulates expression of SL biosynthetic genes, and inhibits branching in a number of species (Yoneyama et al., 2007, 2012; Drummond et al., 2015; Abuauf et al., 2018; Shindo et al., 2018). Conversely, P and N fertilization suppresses the expression of SL biosynthesis genes and subsequent SL exudation and increases branching (Umehara et al., 2008; Yoneyama et al., 2013, 2020). However, SL content is not the sole regulator of branching in response to low-nutrient conditions, as branch numbers were still reduced in SL-deficient and insensitive mutants of petunia under low-P or low-N conditions (Drummond et al., 2015). Indeed, there are reports indicating that low levels of P and N reduce CK production and signaling (reviewed in Wang et al., 2019). Low-N levels also reduced the branch angle in petunia (Drummond et al., 2015); however, the underlying mechanism is largely unknown.
Light quality is another environmental input that can greatly alter plant branching. When the red-to-far-red light (R:FR) ratio decreases, phytochrome (phy) proteins sense the change and trigger the shade-avoidance syndrome (Franklin, 2008; Franklin and Quail, 2010), suppressing axillary bud outgrowth in several plant species, similar to the phyB mutant phenotype in Arabidopsis and sorghum (Kebrom et al., 2006; Finlayson et al., 2010; Whipple et al., 2011; Reddy et al., 2013; Drummond et al., 2015). At least part of the branching response to changes in light is mediated by the SL pathway. MAX2, an F-box protein that is part of the SL receptor complex, is involved in photo-morphogenesis regulatory pathways in Arabidopsis, and, in petunia, SL pathway mutants have increased branching in altered light conditions (day length, and R-FR changes as well as crowding (Snowden and Napoli, 2003; Shen et al., 2007; Drummond et al., 2015)). In addition to regulating branch outgrowth, light appears to have a role in regulating branch angle in petunia (Snowden and Napoli, 2003; Drummond et al., 2015), in Arabidopsis through promoting the expression of TILLER ANGLE CONTROL (TAC1; Waite and Dardick, 2018), and in maize through supressing the expression of ZmLAZY1 (ZmLA1, Dong et al., 2013).
The mode of action for nutrient and light regulation of branching in axillary buds is largely mediated through BRC1 and its homologs. FR light increased the expression of DAD2 and PhTCP3 expression in petunia axillary buds (Drummond et al., 2015) as well as the expression of BRC1 and BRC2 in Arabidopsis and sorghum axillary buds (Kebrom et al., 2006; Finlayson et al., 2010; Gonzalez-Grandio et al., 2013; Xie et al., 2020). CsBRC1 transcript was enhanced after shade treatment in the inbred less-branched cucumber line R1461, whereas shading had little effect on the highly branched wild ancestor cucumber variety (Shen et al., 2019). In perennial crops, day length and temperature also contribute to shoot branching. Short days and low temperatures suppressed branching in Populus by up-regulating the expression of BRC1 and TERMINAL FLOWER 1/CENTRORADIALIS 1 (TFL1/CEN1, Maurya et al., 2020). There is also some evidence to support the control of BRC1 by nutrients like P and N; for instance, supply of nitrate enhanced the expression of OsMADS57, a MADS-box TF, which interacts with OsTB1 and targets D14 (a SL receptor) to regulate tiller outgrowth in rice (Guo et al., 2013; Huang et al., 2019). The nutrient regulation of BRC1 might also be mediated through hormone biosynthesis and signaling, including SLs, CK, and auxin.
Sugars have been shown to promote axillary bud outgrowth as observed in rice, pea, Arabidopsis, and chrysanthemum, with increasing evidence for the role of trehalose 6-phosphate (T6P) in this process (Mason et al., 2014; Fichtner et al., 2017, 2020; Barbier et al., 2019; Liu et al., 2020; Wang et al., 2020). Sugar availability is tightly linked to the carbon-fixation process that is heavily dependent on light quality. For example, removing leaves suppressed bud outgrowth in sorghum and pea, and girdling above the bud had the same effect on preventing branching in pea (Mason et al., 2014; Kebrom and Mullet, 2015). Sugars can act through CIRCADIAN CLOCK ASSOCIATED1 (OsCCA1) to regulate branching via the SL pathway and OsTB1 in rice (Wang et al., 2020). However, at least in Arabidopsis, branching changes due to alterations of T6P are additive with the brc1 mutation, indicating that sugars probably affect multiple pathways that control branching (Fichtner et al., 2020).
Concluding remarks
The growth of an axillary meristem to form a branch is a costly exercise that must balance the resources available with the current environmental conditions. A dynamic environment, especially one with changing nutrients and light quality, can lead to a range of shoot branching outcomes, from a high degree of active axillary meristem outgrowth to strong suppression of branching. In this context, different axillary meristems in a single plant can have dramatically different growth outcomes, ultimately affecting the overall shape of a plant. Over the last three decades, our knowledge of individual processes controlling axillary meristem initiation and branch growth has become increasingly detailed. However, the goal of integrating all our observations into the complex network of regulation is still incomplete (see Outstanding questions). Using model species to answer these questions has been extremely informative, but one particular challenge remains, given increasing stresses on the world’s food supply, how to control precisely the growth of specific branches in cultivated crops and perennial species, with the goal of enabling greater sustainable food production.
Acknowledgments
We wish to thank Ria Rebstock for the axillary meristem image, and Jo Putterill, Revel Drummond, Toshi Foster, Ed Morgan, Cath Kingston, and Anne Gunson for helpful discussions and feedback on this manuscript. We apologize to colleagues whose work could not be included due to space limitations.
Outstanding questions
How much regulation of axillary meristem initiation is conserved between monocots and eudicots?
What determines the different growth outcome of axillary buds that are located at different positions on the main shoot?
Is there a developmental trigger that makes a meristem competent for outgrowth?
How are all the different signals involved in meristem growth integrated?
How do axillary meristem cells initiate the outgrowth process?
How do we get the diversity of form observed in plants, given the evolutionary conservation of the signal transduction pathways discussed here?
How can developmental plasticity be used to improve crop yield?
Funding
The authors were supported by the Growing Futures Fund from Plant & Food Research.
Conflict of interest statement. None declared.
Z.L. and K.C.S. conceived the structure and content, and prepared the figures; all authors contributed to the writing. K.C.S. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Kimberley C. Snowden (kimberley.snowden@plantandfood.co.nz)
References
- Abuauf H, Haider I, Jia K-P, Ablazov A, Mi J, Blilou I, Al-Babili S (2018) The Arabidopsis DWARF27 gene encodes an all-trans-/9-cis-β-carotene isomerase and is induced by auxin, abscisic acid and phosphate deficiency. Plant Sci 277: 33–42 [DOI] [PubMed] [Google Scholar]
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Babili S, Bouwmeester HJ (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol 66: 161–186 [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a strigolactone–insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50: 1416–1424 [DOI] [PubMed] [Google Scholar]
- Balkunde R, Kitagawa M, Xu XM, Wang J, Jackson D (2017) SHOOT MERISTEMLESS trafficking controls axillary meristem formation, meristem size and organ boundaries in Arabidopsis. Plant J 90: 435–446 [DOI] [PubMed] [Google Scholar]
- Barbier F, Péron T, Lecerf M, Perez-Garcia M-D, Barrière Q, Rolčík J, Boutet-Mercey S, Citerne S, Lemoine R, Porcheron B, et al. (2015) Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J Exp Bot 66: 2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier FF, Dun EA, Kerr SC, Chabikwa TG, Beveridge CA (2019) An update on the signals controlling shoot branching. Trends Plant Sci 24: 220–236 [DOI] [PubMed] [Google Scholar]
- Bell EM, Lin W-c, Husbands AY, Yu L, Jaganatha V, Jablonska B, Mangeon A, Neff MM, Girke T, Springer PS (2012) Arabidopsis LATERAL ORGAN BOUNDARIES negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc Natl Acad Sci 109: 21146–21151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T, Leyser O (2006) Something on the side: axillary meristems and plant development. Plant Mol Biol 60: 843–854 [DOI] [PubMed] [Google Scholar]
- Bennett T, Liang Y, Seale M, Ward S, Müller D, Leyser O (2016) Strigolactone regulates shoot development through a core signaling pathway. Biol Open 5: 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge C, Ross J, Murfet I (1994) Branching mutant rms-2 in Pisum sativum (grafting studies and endogenous indole-3-acetic acid levels). Plant Physiol 104: 953–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge C, Symons G, Murfet I, Ross J, Rameau C (1997) The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s). Plant Physiol 115: 1251–1258 [Google Scholar]
- Beveridge CA (2000) Long-distance signaling and a mutational analysis of branching in pea. Plant Growth Regul 32: 193–203 [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O'Connor D, Grotewold E, Hake S (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell 8: 443–449 [DOI] [PubMed] [Google Scholar]
- Borghi L, Bureau M, Simon R (2007) Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell 19: 1795–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot J-P, Boutet–Mercey S, Dalmais M, Antoniadi I, Li X, Maia-Grondard A, Le Signor C, Bouteiller N, et al. (2012) The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol 158: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Wang J, Xiong Y, Yang H, Yang M, Ye P, Bencivenga S, Sablowski R, Jiao Y (2020) A self-activation loop maintains meristematic cell fate for branching. Curr Biol 30: 1893–1904.e1894 [DOI] [PubMed] [Google Scholar]
- Chabikwa TG, Brewer PB, Beveridge CA (2019) Initial bud outgrowth occurs independent of auxin flow from out of buds. Plant Physiol 179: 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahtane H, Vachon G, Le Masson M, Thévenon E, Périgon S, Mihajlovic N, Kalinina A, Michard R, Moyroud E, Monniaux M, et al. (2013) A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J 74: 678–689 [DOI] [PubMed] [Google Scholar]
- Chevalier F, Nieminen K, Sanchez-Ferrero JC, Rodriguez ML, Chagoyen M, Hardtke CS, Cubas P (2014) Strigolactone promotes degradation of DWARF14, an alpha/beta hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26: 1134–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Müller D, Domagalska MA, Leyser O (2010) Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137: 2905–2913 [DOI] [PubMed] [Google Scholar]
- de Saint Germain A, Clave G, Badet-Denisot M-A, Pillot J-P, Cornu D, Le Caer J-P, Burger M, Pelissier F, Retailleau P, Turnbull C, et al. (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol 12: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux P-M, Xie X, Timme RE, Puech-Pages V, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Bécard G, Séjalon-Delmas N (2012) Origin of strigolactones in the green lineage. New Phytol 195: 857–871 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Dong Z, Jiang C, Chen X, Zhang T, Ding L, Song W, Luo H, Lai J, Chen H, Liu R, Zhang X, Jin W (2013) Maize LAZY1 mediates shoot gravitropism and inflorescence development through regulating auxin transport, auxin signaling, and light response. Plant Physiol 163: 1306–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RS, Janssen BJ, Luo Z, Oplaat C, Ledger SE, Wohlers MW, Snowden KC (2015) Environmental control of branching in petunia. Plant Physiol 168: 735–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Yu H, Yuan K, Liao Z, Meng X, Jing Y, Liu G, Chu J, Li J (2019) Strigolactone promotes cytokinin degradation through transcriptional activation of CYTOKININ OXIDASE/DEHYDROGENASE 9 in rice. Proc Natl Acad Sci 116: 14319–14324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA (2012) Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA (2013) Dynamics of strigolactone function and shoot branching responses in Pisum sativum. Mol Plant 6: 128–140 [DOI] [PubMed] [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T (1996) The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J 10: 967–979 [DOI] [PubMed] [Google Scholar]
- Fang Z, Ji Y, Hu J, Guo R, Sun S, Wang X (2020) Strigolactones and brassinosteroids antagonistically regulate the stability of the D53–OsBZR1 complex to determine FC1 expression in rice tillering. Mol Plant 13: 586–597 [DOI] [PubMed] [Google Scholar]
- Fichtner F, Barbier FF, Annunziata MG, Feil R, Olas JJ, Mueller-Roeber B, Stitt M, Beveridge CA, Lunn JE (2020) Regulation of shoot branching in Arabidopsis by trehalose 6-phosphate. New Phytol 229: 2135–2151 [DOI] [PubMed] [Google Scholar]
- Fichtner F, Barbier FF,, Feil R, Watanabe M, Annunziata MG, Chabikwa TG, Höfgen R, Stitt M, Beveridge CA, Lunn JE (2017) Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). Plant J 92: 611–623 [DOI] [PubMed] [Google Scholar]
- Finlayson SA (2007) Arabidopsis TEOSINTE BRANCHED1-LIKE 1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant Cell Physiol 48: 667–677 [DOI] [PubMed] [Google Scholar]
- Finlayson SA, Krishnareddy SR, Kebrom TH, Casal JJ (2010) Phytochrome regulation of branching in arabidopsis. Plant Physiol 152: 1914–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA (2005) The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17: 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Turnbull CGN, Beveridge CA (2001) Long-distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiol 126: 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TM, Ledger SE, Janssen BJ, Luo Z, Drummond RSM, Tomes S, Karunairetnam S, Waite CN, Funnell KA, van Hooijdonk BM, et al. (2017) Expression of MdCCD7 in the scion determines the extent of sylleptic branching and the primary shoot growth rate of apple trees. J Exp Bot 69: 2379–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA (2008) Shade avoidance. New Phytol 179: 930–944 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Zhang T, Xu B, Jia L, Xiao B, Liu H, Liu L, Yan H, Xia Q (2018) CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 8 (CCD8) in tobacco affects shoot and root architecture. Int J Mol Sci 19: 1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Mena C, Sablowski R (2008) ARABIDOPSIS THALIANA HOMEOBOX GENE1 establishes the basal boundaries of shoot organs and controls stem growth. Plant Cell 20: 2059–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Grandío E, Pajoro A, Franco-Zorrilla JM, Tarancón C, Immink RGH, Cubas P (2017) Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc Natl Acad Sci 114: E245–E254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Grandio E, Poza-Carrion C, Sorzano CO, Cubas P (2013) BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25: 834–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Clarenz O, Schäfer E, Müller D, Herrero R, Schmitz G, Theres K (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Zhang J, Wang X, Han X, Wei B, Wang J, Li B, Yu H, Huang Q, Gu H, et al. (2015) The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. Plant Cell 27: 3112–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun 4: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham B-K, Chen J, Yan Y, Lucas WJ (2018) Insights into plant phosphate sensing and signaling. Curr Opin Biotechnol 49: 1–9 [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22: 2032–2036 [DOI] [PubMed] [Google Scholar]
- Harrison CJ (2017) Auxin transport in the evolution of branching forms. New Phytol 215: 545–551 [DOI] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O (2009) Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, , Pautot VA (2015) Beyond the Divide: Boundaries for Patterning and Stem Cell Regulation in Plants. Front Plant Sci 6: 1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara K-i, Karim MR, Takada S, Taoka K-i, Furutani M, Aida M, Tasaka M (2006) Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Liang Z, Chen S, Sun H, Fan X, Wang C, Xu G, Zhang Y (2019) A transcription factor, OsMADS57, regulates long-distance nitrate transport and root elongation. Plant Physiol 180: 882–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46: 79–86 [DOI] [PubMed] [Google Scholar]
- Ito S, Yamagami D, Umehara M, Hanada A, Yoshida S, Sasaki Y, Yajima S, Kyozuka J, Ueguchi-Tanaka M, Matsuoka M, et al. (2017) Regulation of strigolactone biosynthesis by gibberellin signaling. Plant Physiol 174: 1250–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen B, Snowden K (2012) Strigolactone and karrikin signal perception: receptors, enzymes, or both? Front Plant Sci 3: 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BJ, Drummond RSM, Snowden KC (2014) Regulation of axillary shoot development. Curr Opin Plant Biol 17: 28–35 [DOI] [PubMed] [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M (2005) KNOX action in arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, et al. (2013) DWARF 53 acts as a repressor of strigolactone signaling in rice. Nature 504: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom TH, Burson BL, Finlayson SA (2006) Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol 140: 1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom TH, Mullet JE (2015) Photosynthetic leaf area modulates tiller bud outgrowth in sorghum. Plant Cell Environ 38: 1471–1478 [DOI] [PubMed] [Google Scholar]
- Keller T, Abbott J, Moritz T, Doerner P (2006) Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18: 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Lammers M, Pollina T, Toth P, Haider I, Pozo MJ, de Maagd RA, Ruyter-Spira C, Bouwmeester HJ, et al. (2012) The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol 196: 535–547 [DOI] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska MA, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C (2011) Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol 155: 974–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J,, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signaling and branching. Nature 483: 341–344 [DOI] [PubMed] [Google Scholar]
- Ledger SE, Janssen BJ, Karunairetnam S, Wang T, Snowden KC (2010) Modified CAROTENOID CLEAVAGE DIOXYGENASE8 expression correlates with altered branching in kiwifruit (Actinidia chinensis). New Phytol 188: 803–813 [DOI] [PubMed] [Google Scholar]
- Lee D-K, Geisler M, Springer PS (2009) LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 136: 2423–2432 [DOI] [PubMed] [Google Scholar]
- Li G, Tan M, Cheng F, Liu X, Qi S, Chen H, Zhang D, Zhao C, Han M, Ma J (2018) Molecular role of cytokinin in bud activation and outgrowth in apple branching based on transcriptomic analysis. Plant Mol Biol 98: 261–274 [DOI] [PubMed] [Google Scholar]
- Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D, Wang X, Liu X, Teng S, Hiroshi F, et al. (2003) Control of tillering in rice. Nature 422: 618–621 [DOI] [PubMed] [Google Scholar]
- Li Y, Xia T, Gao F, Li Y (2020) Control of plant branching by the CUC2/CUC3-DA1-UBP15 regulatory module. Plant Cell 32: 1919–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ward S, Li P, Bennett T, Leyser O (2016) SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 28: 1581–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Z, Yu H, Duan J, Yuan K, Yu C, Meng X, Kou L, Chen M, Jing Y, Liu G, et al. (2019) SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat Commun 10: 2738–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligerot Y, de Saint Germain A, Waldie T, Troadec C, Citerne S, Kadakia N, Pillot J-P, Prigge M, Aubert G, Bendahmane A, et al. (2017) The pea branching RMS2 gene encodes the PsAFB4/5 auxin receptor and is involved in an auxin-strigolactone regulation loop. PLOS Genet 13: e1007089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z, Yan C, Jiang B, Su Z, Li J, et al. (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Wang D, Dong H, Gu S, Cheng Z, Gong J, Qin R, Jiang L, Li G, Wang JL, et al. (2012) Rice APC/CTE controls tillering by mediating the degradation of MONOCULM 1. Nat Commun 3: 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Zhang Z, Wu F, Feng M, Sun Y, Chen W, Cheng Z, Zhang X, Ren Y, Lei C, et al. (2020) The APC/CTE E3 ubiquitin ligase complex mediates the antagonistic regulation of root growth and tillering by ABA and GA. Plant Cell 32: 1973–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cheng X, Liu P, Sun J (2017) miR156-targeted SBP-box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol 174: 1931–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Peng B, Song A, Jiang J, Chen F (2020) Sugar transporter, CmSWEET17, promotes bud outgrowth in chrysanthemum morifolium. Genes 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Luo L, Takahashi M, Kameoka H, Qin R, Shiga T, Kanno Y, Seo M, Ito M, Xu G, Kyozuka J (2019) Developmental analysis of the early steps in strigolactone-mediated axillary bud dormancy in rice. Plant J 97: 1006–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Zhang Y, Xu G (2020) How does nitrogen shape plant architecture? J Exp Bot 71: 4415–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Duan J, Ke J, He Y, Gu X, Xu T-H, Yu H, Wang Y, Brunzelle JS, Jiang Y, et al. (2017) A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex. Sci Adv 3: e1601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin DC, Hamon-Josse M, Bennett T (2019) Fellowship of the rings: a saga of strigolactones and other small signals. New Phytol [DOI] [PubMed] [Google Scholar]
- Mason MG, Ross JJ, Babst BA, Wienclaw BN, Beveridge CA (2014) Sugar demand, not auxin, is the initial regulator of apical dominance. Proc Natl Acad Sci 111: 6092–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya JP, Miskolczi PC, Mishra S, Singh RK, Bhalerao RP (2020) A genetic framework for regulation and seasonal adaptation of shoot architecture in hybrid aspen. Proc Natl Acad Sci 117: 11523–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayzlish-Gati E, De-Cuyper C, Goormachtig S, Beeckman T, Vuylsteke M, Brewer PB, Beveridge CA, Yermiyahu U, Kaplan Y, Enzer Y, et al. (2012) Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiol 160: 1329–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N, Umehara M, Luo L, Kobayashi K, Hanada A, Ueno K, Asami T, Yamaguchi S, et al. (2010) FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol 51: 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzotti C, Galliani BM, Dreni L, Sommer H, Bombarely A, Masiero S (2017) ERAMOSA controls lateral branching in snapdragon. Sci Rep 7: 41319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhr M, Prüfer N, Paulat M, Teichmann T (2016) Knockdown of strigolactone biosynthesis genes in populus affects BRANCHED1 expression and shoot architecture. New Phytol 212: 613–626 [DOI] [PubMed] [Google Scholar]
- Muller D, Leyser O (2011) Auxin, cytokinin and the control of shoot branching. Ann Bot 107: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D, Schmitz G, Theres K (2006) Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Xue YL, Miyakawa T, Hou F, Qin HM, Fukui K, Shi X, Ito E, Ito S, Park SH, et al. (2013) Molecular mechanism of strigolactone perception by DWARF14. Nat Commun 4: 2613. [DOI] [PubMed] [Google Scholar]
- Napoli CA, Beveridge CA, Snowden KC (1998) Reevaluating concepts of apical dominance and the control of axillary bud outgrowth. InPedersen RA, Schatten GP, eds, Current Topics in Developmental Biology, Vol 44. Academic Press, Cambridge, pp 127–169 [DOI] [PubMed] [Google Scholar]
- Ni J, Zhao M-L, Chen M-S, Pan B-Z, Tao Y-B, Xu Z-F (2017) Comparative transcriptome analysis of axillary buds in response to the shoot branching regulators gibberellin A3 and 6–benzyladenine in Jatropha curcas. Sci Rep 7: 11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M, Cubas P (2016) TCP factors: new kids on the signaling block. Curr Opin Plant Biol 33: 33–41 [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE (2001) REVOLUTA regulates meristem initiation at lateral positions. Plant J 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Pan X, Zheng H, Zhao J, Xu Y, Li X (2016) ZmCCD7/ZpCCD7 encodes a carotenoid cleavage dioxygenase mediating shoot branching. Planta 243: 1407–1418 [DOI] [PubMed] [Google Scholar]
- Péret B, Desnos T, Jost R, Kanno S, Berkowitz O, Nussaume L (2014) Root architecture responses: in search of phosphate. Plant Physiol 166: 1713–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponraj U, Theres K (2020) Keep a distance to be different: axillary buds initiating at a distance from the shoot apical meristem are crucial for the perennial lifestyle of Arabis alpina. New Phytol 227: 116–131 [DOI] [PubMed] [Google Scholar]
- Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K (2008) Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55: 65–76 [DOI] [PubMed] [Google Scholar]
- Rameau C, Bertheloot J, Leduc N, Andrieu B, Foucher, S (2015) Multiple pathways regulate shoot branching. Front Plant Sci 5: 741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SK, Holalu SV, Casal JJ, Finlayson SA (2013) Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol 163: 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Guo Y, Kong J, Lecourieux F, Dai Z, Li S, Liang Z (2020) Knockout of VvCCD8 gene in grapevine affects shoot branching. BMC Plant Biol 20: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AE, Hake S (2019) Drawing a line: grasses and boundaries. Plants 8: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Al-Babili S, van der Krol S, Bouwmeester H (2013) The biology of strigolactones. Trends Plant Sci 18: 72–83 [DOI] [PubMed] [Google Scholar]
- Sasse J, Simon S, Gübeli C, Liu G-W, Cheng X, Friml J, Bouwmeester H, Martinoia E, Borghi L (2015) Asymmetric localizations of the ABC transporter PaPDR1 trace paths of directional strigolactone transport. Curr Biol 25: 647–655 [DOI] [PubMed] [Google Scholar]
- Schmitz G, Tillmann E, Carriero F, Fiore C, Cellini F, Theres K (2002) The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc Natl Acad Sci USA 99: 1064–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci 96: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S, Murison A, Jones A, Fozard J, Aida M, Band LR, Bennett M, Murray JAH (2018) Coordination of meristem and boundary functions by transcription factors in the SHOOT MERISTEMLESS regulatory network. Development 145: dev157081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale M, Bennett T, Leyser O (2017) BRC1 expression regulates bud activation potential but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development 144: 1661–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y, Yasui R, Kameoka H, Tamiru M, Cao M, Terauchi R, Sakurada A, Hirano R, Kisugi T, Hanada A, et al. (2019) Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat Commun 10: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Luong P, Huq E (2007) The F-Box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol 145: 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zhang Y, Ge D, Wang Z, Song W, Gu R, Che G, Cheng Z, Liu R, Zhang X (2019) CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. Proc Natl Acad Sci 116: 17105–17114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Zhang C, Tian C, Wang J, Wang Q, Xu T, Xu Y, Ohno C, Sablowski R, Heisler MG, et al. (2016) Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis. PLOS Genet 12: e1006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo M, Shimomura K, Yamaguchi S, Umehara M (2018) Upregulation of DWARF27 is associated with increased strigolactone levels under sulfur deficiency in rice. Plant Direct 2: e00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O (2013) Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 11: e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratake K, Notaguchi M, Makino H, Sawai Y, Borghi L (2019) Petunia PLEIOTROPIC DRUG RESISTANCE 1 is a strigolactone short-distance transporter with long-distance outcomes. Plant Cell Physiol 60: 1722–1733 [DOI] [PubMed] [Google Scholar]
- Simons JL, Napoli CA, Janssen BJ, Plummer KM, Snowden KC (2007) Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiol 143: 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden K, Napoli C (2003) A quantitative study of lateral branching in petunia. Funct Plant Biol 30: 987–994 [DOI] [PubMed] [Google Scholar]
- Snowden K, Simkin A, Janssen B, Templeton K, Loucas H, Simons J, Karunairetnam S, Gleave A, Clark D, Klee H (2005) The decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17: 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Lu Z, Yu H, Shao G, Xiong J, Meng X, Jing Y, Liu G, Xiong G, Duan J, et al. (2017) IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res 27: 1128–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogne K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, et al. (2003) MAX4 and RMS1 are orthologous dioxygenase–like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17: 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli SV, Martin AP, Viola IL, Gonzalez DH, Palatnik JF (2011) A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol 156: 1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Leyser HMO (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J 50:80–94 [DOI] [PubMed] [Google Scholar]
- Su YH, Zhou C, Li YJ, Yu Y, Tang LP, Zhang WJ, Yao WJ, Huang R, Laux T, Zhang XS (2020) Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc Natl Acad Sci 117: 22561–22571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Hanano K, Kariya A, Shimizu S, Zhao L, Matsui M, Tasaka M, Aida M (2011) CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J 66: 1066–1077 [DOI] [PubMed] [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J 33: 513–520 [DOI] [PubMed] [Google Scholar]
- Tan M, Li G, Chen X, Xing L, Ma J, Zhang D, Ge H, Han M, Sha G, An N (2019) Role of cytokinin, strigolactone, and auxin export on outgrowth of axillary buds in apple. Front Plant Sci 10: 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann T, Muhr M (2015) Shaping plant architecture. Front Plant Sci 6: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Zhang X, He J, Yu H, Wang Y, Shi B, Han Y, Wang G, Feng X, Zhang C, et al. (2014) An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol Syst Biol 10: 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Yanagisawa S (2019) Perception, transduction, and integration of nitrogen and phosphorus nutritional signals in the transcriptional regulatory network in plants. J Exp Bot 70: 3709–3717 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- van Rongen M, Bennett T, Ticchiarelli F, Leyser O (2019) Connective auxin transport contributes to strigolactone-mediated shoot branching control independent of the transcription factor BRC1. PLOS Genet 15: e1008023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Walter MH, Giavalisco P, Lytovchenko A, Kohlen W, Charnikhova T, Simkin AJ, Goulet C, Strack D, Bouwmeester HJ, et al. (2010) SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J 61: 300–311 [DOI] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MACJ, de Vries SC (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite JM, Dardick C (2018) TILLER ANGLE CONTROL 1 modulates plant architecture in response to photosynthetic signals. J Exp Bot 69: 4935–4944 [DOI] [PubMed] [Google Scholar]
- Waldie T, Leyser O (2018) Cytokinin targets auxin transport to promote shoot branching. Plant Physiol 177: 803–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CH, Siu-Ting K, Taylor A, O’Connell MJ, Bennett T (2019) Strigolactone synthesis is ancestral in land plants, but canonical strigolactone signaling is a flowering plant innovation. BMC Biol 17: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Han T, Song Q, Ye W, Song X, Chu J, Li J, Chen ZJ (2020) The rice circadian clock regulates tiller growth and panicle development through strigolactone signaling and sugar sensing. Plant Cell 32: 3124–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian C, Zhang C, Shi B, Cao X, Zhang T-Q, Zhao Z, Wang J-W, Jiao Y (2017) Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell 29: 1373–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J (2015) Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27: 3128–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang B, Yu H, Guo H, Lin T, Kou L, Wang A, Shao N, Ma H, Xiong G, et al. (2020) Transcriptional regulation of strigolactone signaling in Arabidopsis. Nature 583: 277–281 [DOI] [PubMed] [Google Scholar]
- Wang M, Le Moigne M-A, Bertheloot J, Crespel L, Perez-Garcia M-D, Ogé L, Demotes-Mainard S, Hamama L, Davière J-M, Sakr S (2019) BRANCHED1: a key hub of shoot branching. Front Plant Sci 10: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hasson A, Rossmann S, Theres K (2016) Divide et impera: boundaries shape the plant body and initiate new meristems. New Phytol 209: 485–498 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang J, Shi B, Yu T, Qi J, Meyerowitz EM, Jiao Y (2014) The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26: 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA (2012) The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol 159: 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP (2011) Grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci USA 108: E506–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Li H, Chen H, Qi Q, Ding Q, Xue J, Ding J, Jiang X, Hou X, Li Y (2019) Identification and expression analysis of strigolactone biosynthetic and signaling genes reveal strigolactones are involved in fruit development of the woodland strawberry (Fragaria vesca). BMC Plant Biol 19: 73–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Kisugi T, Nomura T, Akiyama K, Asami T, Yoneyama K (2015) Strigolactones are transported from roots to shoots, although not through the xylem. J Pestic Sci 40: 214–216 [Google Scholar]
- Xie X, Yoneyama K, Yoneyama K (2010) The strigolactone story. Annu Rev Phytopathol 48: 93–117 [DOI] [PubMed] [Google Scholar]
- Xie Y, Liu Y, Ma M, Zhou Q, Zhao Y, Zhao B, Wang B, Wei H, Wang H (2020) Arabidopsis FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nat Commun 11: 1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang Y, Yu Y, Duan J, Liao Z, Xiong G, Meng X, Liu G, Qian Q, Li J (2012) Degradation of MONOCULM 1 by APC/C(TAD1) regulates rice tillering. Nat Commun 3: 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Liu L, Zhang C (2020) Regulation of shoot apical meristem and axillary meristem development in plants. Int J Mol Sci 21: 2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, Ori N (2005) . Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Current Biol: CB 15(17): 1566–71 [DOI] [PubMed] [Google Scholar]
- Yao C, Finlayson SA (2015) Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol 169: 611–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L, et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536: 469–473 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Akiyama K, Brewer PB, Mori N, Kawano-Kawada M, Haruta S, Nishiwaki H, Yamauchi S, Xie X, Umehara M, et al. (2020) Hydroxyl carlactone derivatives are predominant strigolactones in Arabidopsis. Plant Direct 4: e00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kim HI, Kisugi T, Nomura T, Sekimoto H, Yokota T, Yoneyama K (2012) How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 235: 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kisugi T, Nomura T, Yoneyama K (2013) Nitrogen and phosphorus fertilization negatively affects strigolactone production and exudation in sorghum. Planta 238: 885–894 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kusumoto D, Sekimoto H, Sugimoto Y, Takeuchi Y, Yoneyama K (2007) Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 227: 125–132 [DOI] [PubMed] [Google Scholar]
- Zhang C, Wang J,, Wenkel S, Chandler JW, Werr W, Jiao Y (2018) Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development 145: dev158352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q-Q, Wang J-G, Wang L-Y, Wang J-F, Wang Q, Yu P, Bai M-Y, Fan M (2020) Gibberellin repression of axillary bud formation in Arabidopsis by modulation of DELLA-SPL9 complex activity. J Integr Plant Biol 62: 421–432 [DOI] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al. (2013) D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L (2006) The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J 48: 687–698 [DOI] [PubMed] [Google Scholar]