Abstract
The pocket domain of pRB is required for pRB to arrest the cell cycle. This domain was originally defined as the region of the protein that is necessary and sufficient for pRB's interaction with adenovirus E1A and simian virus s40 large T antigen. These oncoproteins, and other pRB-binding proteins that are encoded by a variety of plant and animal viruses, use a conserved LXCXE motif to interact with pRB. Similar sequences have been identified in multiple cellular pRB-binding proteins, suggesting that the viruses have evolved to target a highly conserved binding site of pRB that is critical for its function. Here we have constructed a panel of pRB mutants in which conserved amino acids that are predicted to make close contacts with an LXCXE peptide were altered. Despite the conservation of the LXCXE binding site throughout evolution, pRB mutants that lack this site are able to induce a cell cycle arrest in a pRB-deficient tumor cell line. This G1 arrest is overcome by cyclin D-cdk4 complexes but is resistant to inactivation by E7. Consequently, mutants lacking the LXCXE binding site were able to induce a G1 arrest in HeLa cells despite the expression of HPV-18 E7. pRB mutants lacking the LXCXE binding site are defective in binding to adenovirus E1A and human papillomavirus type 16 E7 protein but exhibit wild-type binding to E2F or DP, and they retain the ability to interact with CtIP and HDAC1, two transcriptional corepressors that contain LXCXE-like sequences. Consistent with these observations, the pRB mutants are able to actively repress transcription. These observations suggest that viral oncoproteins depend on the LXCXE-binding site of pRB for interaction to a far greater extent than cellular proteins that are critical for cell cycle arrest or transcriptional repression. Mutation of this binding site allows pRB to function as a cell cycle regulator while being resistant to inactivation by viral oncoproteins.
One of the best-known properties of the retinoblastoma tumor suppressor protein (pRB) is its ability to interact with proteins that contain an LXCXE peptide sequence. The LXCXE motif was first identified in proteins encoded by small DNA tumor viruses (28, 70, 78). Subsequently, LXCXE sequences have been found to be critical both for the transformation properties of adenovirus 5 E1A protein, human papillomavirus type 16 (HPV16) E7, and simian virus 40 large T antigen and for the ability of these proteins to bind to pRB (14, 22, 24, 63, 92, 93). These LXCXE motifs also allow the viral proteins to associate with pRB-related proteins p107 and p130 (20), two proteins that share many properties with pRB, pRB, p107, and p130 possess synergistic or overlapping functions as negative regulators of cell proliferation (62) and the viral proteins appear to use the LXCXE motif to target, and inactivate, all three family members (23).
Several observations have served to emphasize the significance of the LXCXE-pRB interaction. Not only are the pRB-binding activities provided by the LXCXE motifs required for the oncogenic properties of E1A, E7, and large T antigen, but these sequences are highly conserved among adenoviruses, papillomaviruses, and polyomaviruses independent of their transforming activities (13, 19, 63, 73). In addition to the small DNA tumor viruses, rubella virus encodes a pRB-binding protein, NSP90, that also uses an LXCXE motif to interact with pRB (2). The conservation of LXCXE sequences among distinct groups of viruses suggests that they contribute functions that are advantageous for the biology of the virus. The maintenance of this structure is further underscored by the observation that plant viruses (see above; bean yellow dwarf virus and wheat dwarf virus) encode LXCXE-containing proteins that utilize this motif to target pRB family members (55, 96). Since the inactivation of pRB activates E2F, allowing the expression of genes that are required for DNA synthesis, it has been suggested that the inactivation of pRB family proteins provides a cellular environment that promotes efficient viral DNA replication (67, 81).
Consistent with the selection for the LXCXE motif during viral evolution, the LXCXE-binding site is one of the most highly conserved features of the pRB structure. The ability to bind to LXCXE sequences is a feature shared by pRB-homologues in species as diverse as maize and humans (1). Moreover, cocrystallization of the pRB pocket with an E7-derived peptide has identified which amino acids of pRB contact the LXCXE peptide (52). These residues are noncontiguous in the linear sequence but are conserved between pRB, p107, and p130 and the pRB-related proteins found in Xenopus laevis, Drosophila melanogaster, and maize when these sequences are aligned using the crystal structure as a guide (52).
The maintenance of the LXCXE-binding site during evolution suggests that this structure is critical to the normal function of pRB (52). A simple explanation for this conservation might be provided if cellular pRB-binding proteins use this site to interact with pRB. At least 84 cellular proteins have been reported to bind to pRB. At least 19 of these contain an LXCXE sequence or a related sequence that may contribute to the pRB interaction (AhR, Bog, BRG1, hBrm, CtIP, cyclin D1, cyclin D2, cyclin D3, Elf-1, HBP1, histone deacetylase 1 [HDAC1], HDAC2, HEC1, hsp75, retinoblastoma binding protein 1 [RBP1], RBP2, Rim, RIZ, and UBF) (4, 6, 7, 9, 12, 15–17, 25, 29, 30, 46, 57, 58, 60, 76, 80, 85, 94). This diverse list includes pRB-binding proteins that have been proposed to play important roles in pRB-mediated activation and repression of transcription.
The regulation of E2F-dependent transcription is thought to be a key component of pRB's properties as a cell cycle regulator (18). Previous studies have shown that the repression of E2F target genes is sufficient to induce a cell cycle arrest (75). Moreover, the active repression of E2F target genes by pRB-family members is necessary for several types of cell cycle arrest (97). E2F proteins do not contain an LXCXE motif and are thought to bind to a distinct but poorly characterized site in the viral oncoprotein-binding or “pocket” domain of pRB (26, 52). However, four of the pRB-binding proteins that contain LXCXE-like sequences, HDAC1, HDAC2, RBP1, and CtIP, have been implicated in the active repression of E2F-dependent transcription (4, 57, 58, 60). This suggests a model in which pRB is recruited to the promoters of various S-phase-specific genes via E2F. Once tethered to the promoter by E2F it uses its LXCXE-binding site to interact with transcriptional repressors which in turn block transcription until such time in late G1 when pRB is inactivated and this repressor complex is disassembled to allow transcription.
In this study we have investigated the consequences of mutating the LXCXE-binding site of human pRB. The results show that the LXCXE-binding site can be eliminated without affecting pRB's ability to bind to E2F. Surprisingly, mutants lacking the LXCXE-binding site retain the ability to actively repress the transcription of E2F-responsive promoters, and they efficiently arrest Rb-deficient cells in G1. As a consequence, the LXCXE mutants generate a pRB arrest that is resistant to the inactivating effects of viral oncoproteins like E7.
MATERIALS AND METHODS
Plasmid construction.
Site-directed mutations were constructed by PCR as described by Chen and Przybyla (8) or as outlined by Ausubel et al. (3). Briefly, substitutions in the coding region of the B half of the pocket domain were constructed in a 0.7-kb NheI-BsmI fragment. All subclones of PCR products were sequenced to ensure that they contained only the desired substitutions. The resulting mutants were then ligated into the full-length RB cDNA already present in the pCMV-neo-Bam expression plasmid (71). The Gal4-RB9 fusion was constructed by swapping a 2-kb NheI fragment in the pM2-RB (amino acids 300 to 928) plasmid obtained from D. Dean (88). Other expression constructs used in this study are directed by the viral cytomegalovirus (CMV) promoter and have been described previously (4, 26, 53, 84, 95). The E2F4B-Luc reporter was constructed from a 0.2-kb fragment of the E2F4B-CAT plasmid (37) and contains four consensus E2F-binding sites and the E1B TATA ligated into a blunted NheI and BamHI site in the luciferase reporter pGL3 (Promega). The dihydrofolate reductase (DHFR)-Luc reporters accompanying were a kind gift of N. Heintz (89). Likewise, the b-Myb-Luc and E2F mutant were obtained from R. Watson (51), and the E2F1-Luc reporters were supplied by W. Kaelin (66). The G5-MLP-CAT and G5-SV-CAT reporters were provided by D. Dean (57).
Cell culture and transfections.
Cell lines Saos-2, C33A, and HeLa were obtained from the American Tissue Culture Collection. All tissue culture was carried out in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 2 mM l-glutamine, penicillin (50 U/ml), and streptomycin (50 μg/ml). Saos-2 and C33A cells were transfected with Fugene 6 (Boehringer-Mannheim) as recommended by the manufacturer or by calcium phosphate precipitation (3). HeLa cells were transfected by calcium phosphate. Precipitates were left on cells for 16 h before refeeding the cells with fresh growth medium.
Immunoprecipitations and Western blotting.
C33A cells were transfected in 10-cm-diameter tissue culture plates with a total of 8 μg of CMV expression vector DNA. At 48 h posttransfection, cells were washed in phosphate-buffered saline and lysed in 0.3 ml of ELB containing 400 mM NaCl (36). Lysates were diluted with an equal volume of ELB containing no NaCl and then mixed with 100 μl of monoclonal antibody culture supernatant. Immune complexes were precipitated with protein A-Sepharose beads (35) and resolved by electrophoresis on a sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis (SDS–8% PAGE) gel (48). Proteins were transferred to polyvinylidene difluoride (PVDF) membranes by standard techniques (82) and probed for pRB using a 1:5 dilution of tissue culture supernatant from the C36 hybridoma line (91). E1A was detected and immunoprecipitated with antibody M73 (34), hemagglutinin (HA)-tagged E2F3 and DP1 were both recognized on Western blots and immunoprecipitated with 12CA5 culture supernatant. Flag-tagged human HDAC1 was immunodetected and -precipitated using anti-Flag monoclonal antibody M2 (Sigma).
GST pulldown binding experiments.
Glutathione S-transferase (GST) fusion proteins were expressed and purified according to the manufacturer's recommended protocol. GST-E1A (amino acids 1 to 139) has been described previously (26); the GST-E7 (full-length) expression plasmid was a kind gift of K. Munger (Harvard Medical School). Saos-2 cells were plated at 6 × 106 cells per 15-cm-diameter plate and transfected with 50 μg of CMV-RB or mutant expression plasmid per plate. Extracts were prepared as described above in 2 ml of ELB per plate. Two hundred microliters of extract was mixed with 1 μg of GST-E7 protein and incubated on ice for 30 min. GST-E7 complexes were precipitated by mixing with 100 μl of a 10% slurry of glutathione-Sepharose beads. Bead-containing solutions were mixed by gently rocking tubes at 4°C for 1 h. Beads were washed twice with ELB and resuspended in 1× SDS-PAGE sample buffer. Samples were analyzed by SDS-PAGE and Western blotting for pRB.
Transcriptional reporter assays.
Saos-2 or C33A cells were plated at 5 × 105 cells per well of a six-well plate. Each transfection contained 100 ng of transcriptional reporter and 100 ng of a CMV-LacZ reporter to normalize transfection efficiencies. Up to 100 ng of CMV-RB or 200 ng or Gal4-RB expression vector was used along with sufficient CMV-neo-Bam or SV40-Gal4(1-147) vector DNA to normalize expression plasmid content in each transfection. In the case of E2F4B-Luc reporter assays, 50 ng each of CMV-HA-E2F2 and CMV-HA-DP1 was included. Carrier DNA (pBluescript) was also included to make the final concentration of DNA up to 1.2 μg. Trichostatin A (TSA) was added 16 h after transfection to a final concentration of 100 nM. Extracts were prepared from cells 36 to 48 h posttransfection by lysing cells in Luciferase assay buffer (Promega) or chloramphenicol acetyltransferase (CAT) assay buffer (Boehringer-Mannheim) according to the manufacturers' directions. Luciferase activity was measured on an EG&G Berthold Microlumat luminometer. CAT expression levels were measured by quantitative enzyme-linked immunosorbent assay as directed by the manufacturer. β-Galactosidase activity was quantitated by standard methods (61). All data points presented are the average measurement of three independent transfections; each experiment (consisting of three transfections) was repeated at least twice.
Flow cytometry of Saos-2 and HeLa cell transfectants.
One million Saos-2 cells were transfected in 6-cm-diameter dishes with 1 μg of CMV-CD20, 0.5 μg of CMV-RB (or RB mutant), and 4 μg of CMV-E7 or 2 μg each of CMV-HA-cdk4 and CMV-cyclin D1. Cells were replated on 10-cm-diameter plates 16 to 24 h posttransfection and harvested 48 h later. HeLa cells were transfected with 15 μg of CMV-RB expression vector and 5 μg of CMV-CD20. Cells were refed 16 to 24 h posttransfection and harvested 24 h later. Cells were harvested and stained for CD20 and DNA content as described previously (84). Subsequently, cells were analyzed on a Becton Dickinson FACScan, and the resulting data were processed using CellQuest and Modfit LT software.
RESULTS
Mutation of the LXCXE-binding site of pRB.
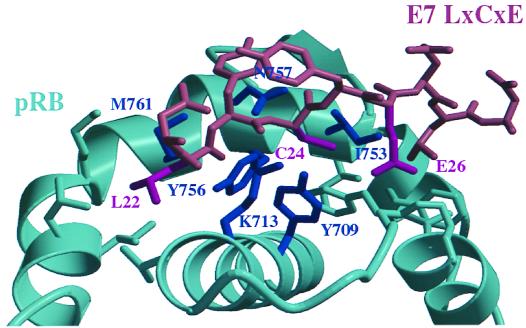
The three-dimensional structure of the small pocket domain of pRB bound to a 9-amino-acid LXCXE-containing peptide reveals that the LXCXE peptide binds into a cleft in the B half of the pocket (52). The side chains of the conserved leucine and cystine residues of the LXCXE motif fit tightly into hydrophobic pockets, while the carboxylate group of the glutamic acid is predicted to make two hydrogen bonds with two backbone amide groups of the α15 helix of pRB. Crystallographic data suggest that the peptide backbone is held in place by four hydrogen bonds with amino acids Tyr 709, Tyr 756, Asn 757, and Lys 713, respectively. These amino acid residues are highly conserved in pRB homologues (1, 56).
Several different mutants were prepared to disrupt the LXCXE-binding site. The mutants used in this study are listed in Table 1, and the position of the relevant amino acids is illustrated in Fig. 1. We mutated pRB amino acids Tyr 709, Lys 713, Ile 753, Tyr 756, Asn 757, and Met 761, since these residues are involved in the formation of the cleft and/or the formation of hydrogen bonds with the LXCXE motif. The mutations made in RB5 (Y709F and K713A) and RB6 (Y756A and N757A) were designed simply to eliminate hydrogen bonds between the side chains of these residues and the peptide backbone. The other mutants described in this study were constructed with the goal of disrupting the hydrophobic pockets that are occupied by the leucine and cystine residues of the LXCXE sequence. Alleles RB9 (I753A, N757A, and M761A) and RB10 (Y709A and K713A) remove the sides of this hydrophobic cleft and are predicted to make the leucine and cystine residues a poor fit for this hydrophobic groove. All mutations substitute phenylalanine or alanine for the wild-type amino acid. In this way partially buried side chains such as that of Tyr 756 can be altered in a minimally disruptive way. Alanine substitutions were chosen for amino acids that are mostly solvent exposed since they effectively truncate side chains but are not predicted to change the overall structure of the protein. In RB5, Tyr 756 was only changed to phenylalanine as only the hydroxyl group of the side chain is predicted to be solvent exposed.
TABLE 1.
Mutant alleles of RB used in this studya
| Allele no. | Substitutions |
|---|---|
| RB5 | Y709F, K713A |
| RB6 | Y756F, N757A |
| RB9 | I753A, N757A, M761A |
| RB10 | Y709A, K713A |
Designated mutant allele numbers are listed, accompanied by the corresponding changes that have been incorporated into pRB.
FIG. 1.
Diagram of an LXCXE peptide derived from E7 bound to pRB. Amino acids in the LXCXE-containing peptide are colored brown, with the side chains of Leu 22, Cys 24, and Glu 26 highlighted in maroon. The B half of the pocket domain is colored teal. The side chains of Tyr 709, Lys 713, Ile 753, Tyr 756, Asn 757, and Met 761 which have been mutated in this study are labeled and displayed in navy blue. The side chains of E7 amino acids shown in maroon have been shown to make extensive interactions with the pRB amino acids colored navy blue.
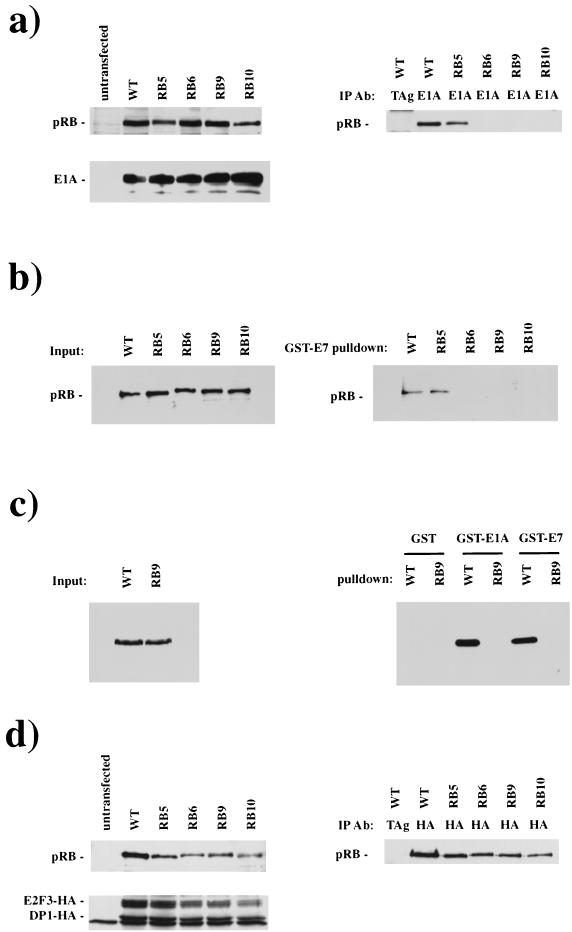
The binding properties of these pRB mutants were determined following cotransfection of RB and E1A expression vectors into C33A cells, a cervical carcinoma-derived cell line that does not express pRB (72) and is not arrested in G1 by ectopic pRB expression (98). Expression levels of pRB and E1A were determined by Western blotting of whole-cell extracts and the interaction between pRB and E1A was assessed by the immunoprecipitation of E1A and the detection of coprecipitated pRB (Fig. 2a). By this method RB6, RB9, and RB10 show dramatically reduced binding to E1A, whereas RB5 showed an intermediate level of binding. E1A CR1 binds to pRB with an approximately 100-fold-lower affinity than the LXCXE-containing CR2 (21, 91). Consistent with this, we detect a very low level of residual binding activity with the LXCXE-binding site mutants on long exposures of the Western blots. Similar results were obtained using in vitro binding assays in which cell lysates were prepared from Saos-2 cells following transfection with pRB or mutant pRB expression vectors and incubated with purified GST-E7 or GST-E1A proteins (Fig. 2b and c). As displayed in Fig. 2b, an input of equivalent quantities of pRB to the binding reactions results only in detectable interactions between E7 and wild-type pRB or RB5. Identical results were seen using purified GST-E7 or GST-E1A proteins (Fig. 2c). We conclude that the mutations introduced into the LXCXE-binding cleft in RB6, RB9, and RB10 eliminate the ability of pRB to bind to E1A or E7 in a stable manner, even when these proteins are expressed at high levels.
FIG. 2.
RB6, RB9, and RB10 are unable to bind to LXCXE-containing proteins, E1A and E7. Plasmids directing the expression of RB or mutant alleles 5, 6, 9, and 10 were transfected into C33A cells along with plasmids expressing E1A (a) or E2F3-HA and DP1-HA (d). Extracts were prepared and analyzed for expression of the transfected genes by Western blotting. The ability of wild-type or mutant forms of pRB to interact with E1A or E2F complexes was assessed by coimmunoprecipitation with E1A or HA antibodies. The quantity of pRB present in these complexes was determined by Western blotting. E7 and E1A interactions with RB were determined by transfecting cells with wild-type or mutant RB plasmids. Extracts were prepared, and pRB was precipitated through its interaction with glutathione-Sepharose-bound E7 or E1A. The quantity of pRB present in extracts and GST-E7 or -E1A pulldowns was determined by Western blotting (b and c). Abbreviations: IP Ab, immunoprecipitated antibody; TAg, T antigen; WT, wild type.
Binding experiments similar to those described above were used to test the ability of pRB mutants to interact with an E2F-DP complex. Peptide competition experiments have indicated E2F proteins bind to a site in the pRB-pocket that is distinct from the LXCXE-binding site (26, 52). Thus, subtle mutations in the LXCXE binding site that do not disrupt the overall conformation of the pRB pocket are not expected to affect the ability of pRB to bind to E2F. Figure 2d shows the results of an experiment in which wild-type or mutant pRB proteins were coexpressed in C33A cells with HA-tagged E2F-3 and DP-1, and complex formation was detected following immunoprecipitation with the HA tag. The ability of RB5, RB6, RB9, and RB10 to bind to E2F-3–DP-1 was indistinguishable from wild-type pRB, suggesting that mutation of the LXCXE binding cleft does not disrupt the overall structure of the pocket.
The LXCXE-binding site is not needed for pRB to repress E2F-containing reporters.
Studies of E2F have shown that pRB does not simply neutralize E2F by binding to its transcriptional activation domain; instead, pRB is an active inhibitor of transcription when recruited to DNA (32, 75, 87, 88), and active repression of E2F is required for several types of G1 arrest (97). Recent studies have found that pRB acts, at least at some E2F-regulated promoters, by recruiting HDACs to promote a chromatin structure that hinders transcription (4, 57, 58). Consistent with this model, TSA, a global inhibitor of deacetylases, derepresses several E2F-RB-regulated promoters (57). However, pRB-mediated repression of other promoters is insensitive to TSA, and pRB is thought to use other mechanisms to repress at these sites (57). Candidates for these alternative repressors include CtIP (60) and HBP1 (80), transcription repressors that have been shown to bind to pRB. In addition, two other pRB-binding proteins, hBrm and RBP1, have been found to cooperate with pRB to repress E2F-dependent transcription (49, 83). Intriguingly, HDAC1, HDAC2, CtIP, HBP1, hBrm, and RBP1 have all been suggested to bind to pRB through LXCXE or IXCXE motifs (27, 58, 60, 79, 80). We therefore tested whether pRB mutants lacking the LXCXE-binding site could repress E2F-dependent transcription.
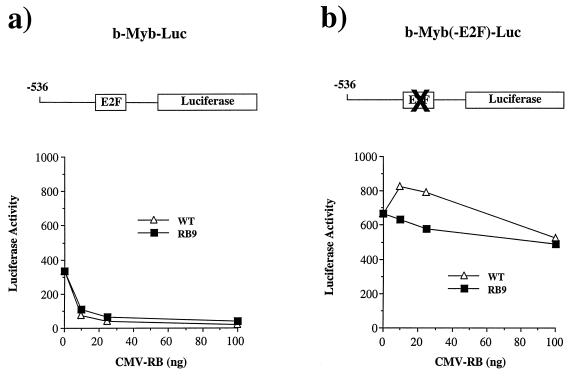
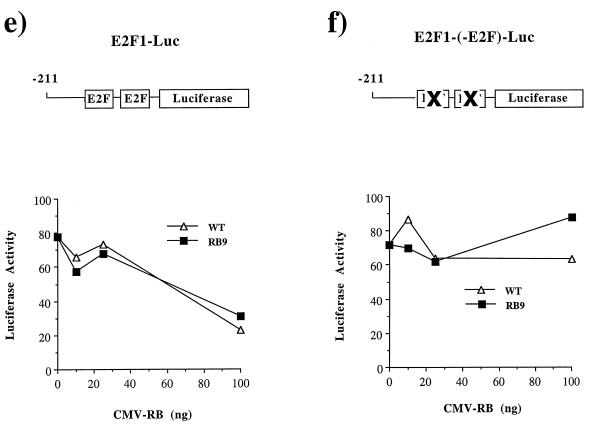
The E2F responsiveness of the b-Myb, DHFR, and E2F1 promoters has been well documented (39, 43, 50, 51, 66, 89, 90). Additionally, there is extensive literature indicating that pRB negatively regulates these promoters (5, 42, 50, 54, 66, 74, 77). Reporter constructs containing the wild-type promoters or sequences in which the E2F-binding sites are mutated were transfected into Saos-2 cells together with expression constructs encoding either wild-type RB or RB9, a mutant of pRB that fails to bind to E1A or E7. Dose-dependent repression of these promoters was observed using either wild-type pRB or RB9 (Fig. 3a, c, and e), although the RB9 repression is slightly weaker. In each case, pRB-mediated repression was completely dependent on the E2F-binding sites in these promoters (Fig. 3b, d, and f). Similar results were obtained following transfection of these reporters into C33A cells and from transfection of DHFR-luciferase into Rb−/− mouse embryo fibroblasts (data not shown).
FIG. 3.
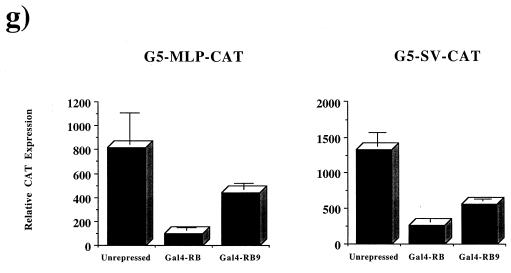
RB9 is capable of active repression of E2F site-containing reporters. Saos-2 cells were transiently transfected with reporter constructs for known E2F-regulated genes along with wild-type RB (WT) or RB9. The resultant activity of DHFR and b-Myb promoters regulating luciferase expression were plotted against increasing amounts of CMV-RB or -RB9 vector in the transfections. (a) Response of the murine b-Myb promoter to increasing quantities of RB or RB9. (b) An identical reporter containing a mutated E2F site was also tested for RB-mediated repression. (c and d) RB-induced repression of the DHFR promoter (c) or a DHFR promoter containing mutations in its overlapping E2F sites (d). (e and f) Similarly, the repressive effects of pRB on an E2F1 promoter and its E2F mutant are shown. Active repression by RB and RB9 was measured by expressing these forms of pRB fused to the Gal4 DNA-binding domain along with Gal4 site-containing reporters in C33A cells. (g) Normalized levels of CAT expression when reporters were transfected with Gal4 alone or with Gal4-RB or -RB9. Error bars indicate the standard deviation of each value.
Mutation of the E2F binding sites in the b-Myb, DHFR, and E2F1 promoters either increases or has no effect on the transcriptional activity of these reporters, indicating that the E2F binding sites are acting primarily as repressor elements. As pRB expression reduces the activity of the reporters in a dose-dependent and E2F site-dependent manner (compare luciferase activities shown in Fig. 3a to 3b, 3c to 3d, and 3e to 3f), these experiments suggest that pRB is actively repressing transcription from these E2F-regulated promoters. To extend this further we investigated whether the LXCXE-binding site mutants could repress transcription when recruited to a heterologous promoter. The large pocket domain of RB9 (amino acids 300 to 928) was fused to the DNA-binding domain of Gal4 and assayed for repression of two different reporter constructs, each containing five tandem Gal4 sites upstream of a viral promoter and a CAT reporter gene (Fig. 3g). Transcription from both constructs was repressed by Gal4-RB9, although the magnitude of repression seen with Gal4-RB9 was slightly reduced when compared with Gal4-RB. A similar trend was also observed on the E2F-responsive promoters shown in Fig. 3. We conclude that the LXCXE-binding cleft of pRB is not required for pRB to actively repress transcription.
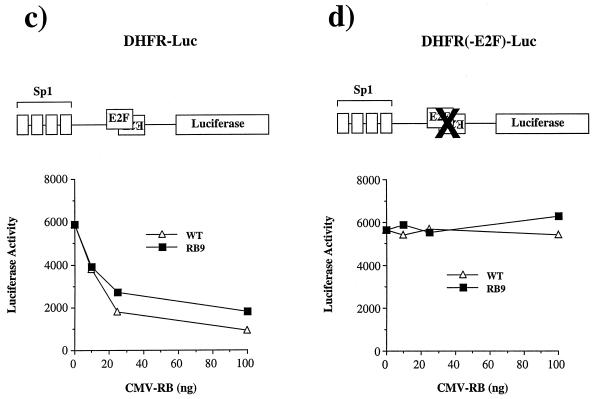
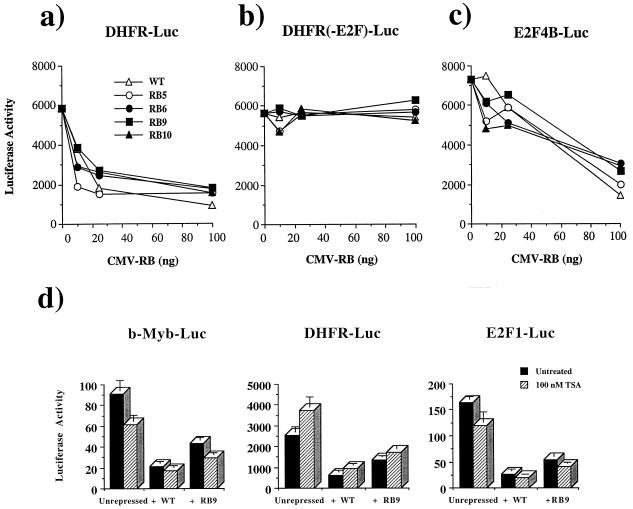
The properties of RB9 were surprising, since many of the pRB-binding proteins that have been linked to transcriptional repression contain LXCXE (or IXCXE) motifs and have been suggested to use these to interact with pRB. To rule out the possibility that these results were unique to RB9 and not shared by other LXCXE-binding site mutants, the other three mutant alleles were tested for E2F repression. As shown in Fig. 4a, each of the pRB mutants tested repress the DHFR-Luc reporter in a dose-dependent manner. This effect is dependent on the E2F-binding site (Fig. 4b). Similarly, a synthetic promoter construct containing four tandem E2F consensus sites and the adenovirus E1B TATA box, when cotransfected with E2F-2 and DP-1 expression vectors, is deactivated by all mutant and wild-type forms of pRB tested (Fig. 4c).
FIG. 4.
RB mutants 5, 6, 9, and 10 are all capable of repressing E2F site-containing reporters and do so in a TSA-insensitive manner. Saos-2 cells were transiently transfected with various amounts of CMV-RB or mutant expression plasmids along with DHFR-Luc reporters. The luciferase activity of these reporter constructs is plotted against increasing amounts of RB or mutant expression plasmids. The wild-type DHFR promoter activity is displayed in panel a; DHFR-containing mutations in known E2F sites is shown in panel b. The activity of a synthetic promoter containing four tandem E2F sites followed by the adenovirus E1B TATA is plotted in panel c. This reporter is cotransfected with CMV-E2F2 and CMV-DP1 to activate it. Wild-type RB (WT) and mutant RB are then titrated in to examine the deactivation of this reporter. The involvement of HDAC in repressing the luciferase activity of each E2F-responsive reporter was determined with or without RB or RB9 expression vectors when treated with 100 nM TSA for 24 h prior to harvesting (d). Error bars indicate the standard deviation of each value.
Although Gal4-RB repression of the G5-MLP-CAT promoter (Fig. 3g) is known to be TSA sensitive (57), treatment of transfected cells with TSA to inhibit deacetylases demonstrated that pRB repression of the E2F-1, b-Myb, and DHFR reporter constructs was primarily through a TSA-independent mechanism (Fig. 4d). TSA failed to reverse repression of either the E2F1 or b-Myb reporters. Although TSA partially reversed repression of DHFR-Luc by RB and RB9, we note that transcription from the DHFR reporter is stimulated by TSA. TSA also elevates the activity of the DHFR promoter construct lacking E2F-binding sites (data not shown), raising the possibility that the TSA effect on this promoter may not be mediated through pRB.
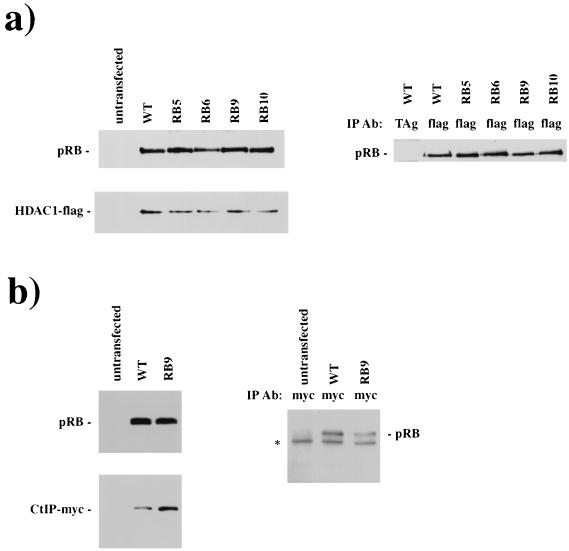
The ability of the LXCXE-binding site mutants to repress transcription raises two possibilities: either LXCXE-containing proteins are not needed for E2F repression, or such proteins do not depend on their LXCXE motifs to interact with pRB. We examined these possibilities for HDAC1 and CtIP, two proteins that have been reported to interact with pRB via an LXCXE (or related) sequence and implicated in pRB-mediated repression (58, 60). CMV-HDAC1 or CtIP plasmids were transfected into C33A cells together with plasmids directing the expression of wild-type or mutant pRB, and the physical interaction was scored by coimmunoprecipitation and Western blot analysis (Fig. 5a and b). Western blots of these extracts demonstrate that pRB and the cotransfected repressor expression levels were similar in all cell extracts and that similar amounts of wild-type or mutant pRB were coprecipitated with HDAC1 or CtIP. These results demonstrate that these repressor molecules must contain pRB-binding sequences that are independent of their LXCXE motif, providing a potential explanation for the repressor activity of the RB mutants. We note that these experiments do not exclude the possibility that the LXCXE-binding site contributes to pRB's recruitment of HDAC1 or CtIP at some promoters or that its interaction with other cellular proteins may be affected differently. Other extraction and immunoprecipitation conditions may reveal a defect in pRB-HDAC1 or CtIP-pRB interactions. However, under these experimental conditions, the mutation of the LXCXE-binding site has a dramatic effect on pRB's interaction with viral oncoproteins without greatly affecting its interactions with these cellular partners.
FIG. 5.
RB mutants bind to transcriptional repressors HDAC1 and CtIP. C33A cells were cotransfected with RB- and HDAC1-Flag-expressing plasmids. Extracts were prepared, and pRB and HDAC1-Flag expression levels were quantitated by Western blotting (a, left panels). The ability of pRB to bind to HDAC1 was determined by coimmunoprecipitation and subsequent Western blotting for pRB (a). (b) The ability of RB9 to interact with CtIP is similarly shown; a nonspecific immunoglobulin G band is marked by an asterisk. Abbreviations: IP Ab, immunoprecipitated antibody; TAg, T antigen; WT, wild type.
RB mutants lacking the LXCXE-binding site induce a cell cycle arrest that is insensitive to inactivation by E7.
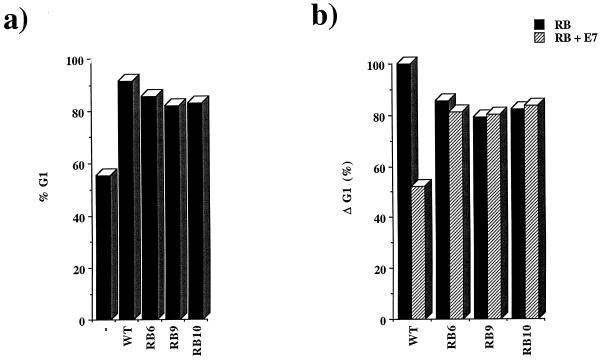
E2F regulation is only one aspect of pRB function, and additional activities may be required for pRB to regulate cell cycle progression. To test whether the LXCXE-binding site is required for pRB-mediated cell cycle arrest, the pRB mutants were expressed in the RB-deficient osteosarcoma cell line Saos-2, a cell line that is readily arrested in G1 by the reintroduction of wild-type pRB (38). The cell cycle arrest was monitored by the use of a CD20 cell surface marker to identify the transfected cells, as described previously (84). Cells were analyzed for CD20 and DNA content by flow cytometry. In these experiments, approximately 55% of cells transfected with CMV-CD20 alone displayed a G1 DNA content. The transient expression of wild-type pRB caused 90% of cells to accumulate in G1. Despite lacking an intact LXCXE-binding site, RB6, RB9, and RB10 each gave a robust cell cycle arrest that was similar to the arrest induced by wild-type pRB (Fig. 6a).
FIG. 6.
RB mutants arrest Saos-2 cells in G1 and are resistant to inactivation by E7. Saos-2 cells were transfected with CMV-RB (WT) or mutant RB, along with a CMV-CD20 expression plasmid. Cells were fixed and stained for CD20 with fluorescein-conjugated antibodies and for DNA content with propidium iodide. (a) Percentage of cells in G1 after being transfected with RB, an irrelevant CMV expression plasmid (−), or one of the mutants. (b) Change in G1 content of cell populations transfected with wild-type or mutant RB (black bars) or an RB expression plasmid and CMV-E7 (hatched bars). In this analysis the G1 content of pRB-arrested cells has been assigned a value of 100% and the G1 content of unarrested cells is set to 0.
These results suggest that the major binding site for E1A or E7 proteins on pRB is separable from the regions of pRB that are needed for pRB to impose a cell cycle arrest. In addition to the above conclusion, it is also formally possibly that the viral proteins do not need to bind to pRB in a stable manner in order to inactivate it. In this case, mutation of the LXCXE-binding cleft on pRB might prevent coprecipitation of these proteins without impairing the ability of viral proteins to overcome pRB function. To test this we investigated whether the cell cycle arrest caused by RB6, RB9, and RB10 could be reversed by E7. Since the ability of E7 to antagonize pRB depends on the relative levels of these proteins, the pRB and E7 expression plasmids were titrated to a level where E7 expression rescued 50% of the pRB-induced accumulation of G1 phase cells. Unlike the arrest induced by wild-type pRB, the cell cycle arrest caused by RB6, RB9, and RB10 was resistant to the expression of E7 (Fig. 6b), indicating that mutation of the LXCXE-binding site prevents E7 from functionally inactivating pRB.
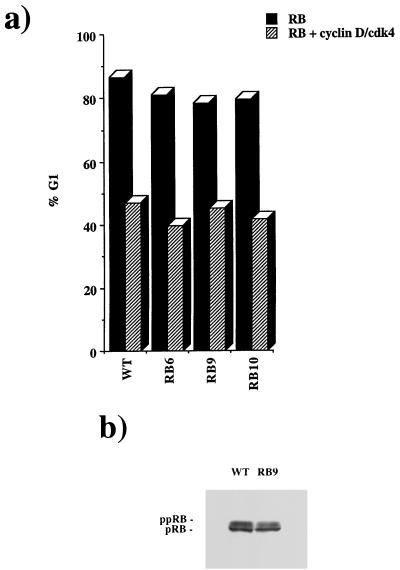
It has been proposed that the D-type cyclins use their LXCXE motif to recognize pRB as a substrate for phosphorylation. Given that these RB mutants may not be recognized by cyclin D, it was formally possible that the failure of E7 to overcome cell cycle arrest by RB6, RB9, or RB10 was due in part to a defect in RB phosphorylation. In this case the arrest might be similar to that caused by the mutation of phosphorylation sites of pRB (47). A number of observations show that this is not the case. First, coexpression of cyclin D1-cdk4 was readily able to overcome the cell cycle arrest induced by RB6, RB9, or RB10 (Fig. 7a). Second, cyclin D1-cdk4 rescue of the cell cycle arrest caused by these mutants produced the same pattern of hyperphosphorylated proteins that were seen with wild-type pRB (Fig. 7b). We conclude that mutation of the LXCXE-binding site does not interfere with the regulation of pRB by phosphorylation in these cells and that the arrest seen in the presence of E7 does not represent a gain-of-function property of these mutants.
FIG. 7.
RB-mediated cell cycle arrest is relieved by cyclin D-cdk4. Cell cycle arrest experiments were performed as in Fig. 5. The percentage of cells in G1 when transfected by RB or an RB mutant is shown by black bars. Cotransfection of wild-type (WT) or mutant RB with cyclin D-cdk4 (hatched bars) is shown in comparison (a). RB was transfected into C33A cells followed by immunoprecipitation and Western blotting. These samples were then analyzed by SDS–8% PAGE and Western blotting to display hyperphosphorylated forms of pRB (b).
As a rigorous test of the ability of the LXCXE-binding site mutants to escape E7 inactivation, we tested whether the expression of these pRB mutants is sufficient to cause a cell cycle arrest in HPV-transformed cells. Previous work has demonstrated that HeLa cells, despite their numerous passages in tissue culture, require the continued expression of HPV18 E6 and E7 proteins in order to proliferate (31, 41, 64). Wild-type or mutant pRB expression constructs were cotransfected with a CD20 marker into HeLa cells, and the effects on cell cycle distribution were measured by flow cytometry (Table 2). As expected from the ability of E7 to bind and inactivate pRB, the expression of wild-type pRB had no significant effect on the cell cycle profile of these cells, compared to the empty vector control. In contrast, a significant increase in the proportion of cells with 2N content was observed when each of the pRB mutants was expressed. Consistent with their inability to bind E7 or E1A (Fig. 2), RB6, RB9, and RB10 gave a strong cell cycle arrest. RB5, which appears to be weaker in binding in Fig. 2a, has an intermediate effect on cell cycle distribution. Thus, in these cells, the ability of the pRB mutants to arrest the cell cycle was dominant over E7-induced proliferation.
TABLE 2.
RB mutants 6, 9, and 10 arrest HeLa cells in G1a
| Transfected genes | % of cells (SD) in phase:
|
||
|---|---|---|---|
| G1 | S | G2/M | |
| None | 54.4 (5.5) | 25.9 (5.4) | 19.7 (0.1) |
| RB | 52.5 (0.6) | 23.0 (4.5) | 24.4 (3.9) |
| RB5 | 61.3 (1.0) | 23.1 (1.4) | 15.6 (2.4) |
| RB6 | 79.6 (2.8) | 8.0 (2.0) | 12.4 (4.8) |
| RB9 | 81.1 (0.4) | 6.6 (1.6) | 12.2 (2.0) |
| RB10 | 79.9 (4.2) | 8.1 (0.5) | 12.6 (4.7) |
CMV-RB constructs along with CMV-CD20 were transfected into HeLa cells. Cells were fixed and stained for CD20 and propidium iodide to quantitate the DNA content of transfected cells. The percentage of cells in G1, S, or G2/M was determined by the ModFit LT software program.
DISCUSSION
The retinoblastoma tumor suppressor protein (pRB) has an important impact on cell proliferation, cell differentiation, and cell survival (18, 68, 86). A detailed structure-function analysis of pRB has been hampered by the fact that many of its functional properties require the pocket domain, a structure that has proven difficult to analyze by systematic mutagenesis. The recently published crystal structure of the pRB pocket shows why this domain has been so difficult to study. Formation of the proper structure for this domain depends on an extensive interface between the A and B halves of the pocket (52). Most tumor-derived mutations of pRB generate large deletions or truncations that would remove this part of the molecule (33, 40). Similarly many tumor-derived point mutations in pRB affect amino acids that are buried in this interface and cause changes that are likely to destabilize the entire structure (52). Moreover, many of the amino acids that are highly conserved between pRB family members, which may have appeared to be attractive sites for mutagenesis, are buried within the structure and unlikely to be directly involved in intermolecular interactions (52).
Using the crystal structure as a guide, it is finally possible to target specific surfaces of the pRB pocket for mutation. In this study we have used this information to specifically eliminate the cleft in the B half of the pocket that allows pRB to interact with LXCXE peptides. A panel of mutants was prepared that perturbs the interaction between LXCXE and pRB in several different ways. These mutants have similar properties in cell cycle and transcription assays, and the effect of combining mutations does not appear to change their potency. We found that combining the RB9 and RB10 alleles creates a protein capable of arresting cells in G1 and repressing transcription as effectively as the RB6, RB9, or RB10 mutants alone, and is similarly rescued by cyclin D-cdk4 (data not shown). This indicates that RB6, RB9, and RB10 each can effectively eliminate the activity of this structure. Analysis of single amino acid substitutions reveals that Tyr 709 is a particularly important residue, as mutations of this site alone severely reduce binding to E1A. Taken together, these results confirm that the cleft identified in the crystal structure is essential for stable interaction between pRB and E7, even though short peptides containing the LXCXE motif bind to pRB with only 1/20 of the affinity of the full length E7 protein (45, 52).
The results described here show that mutation of the LXCXE-binding site prevents E7 from targeting pRB, yet the pRB mutant retains its ability to induce a cell cycle arrest and to repress transcription at E2F containing promoters, two activities that are thought to be central to pRB function. As a result, the expression of these mutants in cells that are transformed by HPV18 E6 and E7 restores a pRB-mediated cell cycle arrest. The properties of these mutants show that in principle, it is possible to prevent E7 from binding to pRB without inactivating normal functions of pRB. This study provides strong support for the idea that small compounds that are able to bind to the LXCXE-binding cleft might prevent viral proteins from inactivating pRB.
There is a great deal of circumstantial evidence indicating that the LXCXE-binding site is likely to be important for pRB function. One aspect of this argument is that the amino acids that form this groove are highly conserved between pRB homologues of different species (1, 52, 56). Divergent families of viruses have evolved proteins, typically expressed early during viral infection, which use an LXCXE motif to inactivate pRB. Multiple cellular pRB-binding proteins also contain LXCXE sequences. The simplest model is that pRB uses the LXCXE binding site to interact with an essential target, and viruses have evolved an LXCXE motif to mimic this interaction. How then does one explain that pRB can arrest the cell cycle, or repress transcription, without the LXCXE-binding cleft? We envision several possibilities.
First, it is possible that pRB can arrest the cell cycle in several different ways and that the interactions between pRB and cellular LXCXE-containing proteins contribute to this process without being essential. The relative importance of these interactions may vary between cell types or with types of arrest, and these results do not exclude the possibility that the pRB LXCXE-binding cleft is important for cell cycle arrest at a specific developmental stage or in response to a particular type of stimulus. Similarly, pRB appears to be able to interact with several different transcriptional repressors (4, 57, 58, 60, 80, 83), and redundancy between these mechanisms may allow the LXCXE-binding site mutants to repress E2F-dependent transcription in these assays, even though the repression of some E2F target genes may be mediated by LXCXE-binding proteins. In addition to the possibilities discussed above, it cannot be ruled out that some loss of binding between cellular LXCXE-containing proteins and our mutants occurs; however, this is masked by the overexpressed levels of pRB in these experiments. We observed a slight but consistent decrease in the ability of the RB mutants to repress transcription, particularly when fused to Gal4 and recruited to a heterologous promoter (Fig. 3g). However, the functional significance of this is unclear.
Another explanation for why the LXCXE-binding cleft is so well conserved but is dispensable in these experiments is that it may contribute to a specific pRB function that is distinct from cell cycle or E2F regulation. The pocket domain is required for a variety of pRB activities, including cell differentiation (10, 68), and the activation of transcription (10, 11, 65, 68, 76). Sellers et al. have identified RB mutants which are unable to regulate cell proliferation yet retain the ability of pRB to induce differentiation and activate transcription (74). These mutations are distant from the LXCXE-binding cleft and thus are not expected to affect binding to that site. Furthermore, expression of viral oncoproteins like E7 has been shown to block cellular differentiation (44, 59, 69). While proteins like E7 are able to use their LXCXE sequence to overcome pRB's functions in cell cycle regulation, there is no evidence that the cellular proteins that use this site are primarily involved in the cell cycle aspect of pRB function.
A third possible explanation stems from the idea that viral proteins may depend on the LXCXE motif to interact with pRB to a far greater extent than cellular pRB-binding proteins. This situation might arise if most cellular pRB proteins have a more extensive binding site on the surface of pRB or if they are components of complexes that have multiple contacts with pRB. It is clear that HDAC1 and CtIP, for example, have to bind to pRB in a manner independent of the LXCXE motif, since they are able to interact with mutants lacking the LXCXE-binding cleft. At first glance it might appear paradoxical for this cleft to remain so well conserved if it is not essential. Protein-protein interactions that require rigid conservation are usually ones in which the means of interaction is critical to their collective function, and thus alterations are selected against. However, interactions that are shared by numerous different proteins and a common binding site are also well conserved, since the simultaneous coevolution of a new interface between these molecules would be improbable. In this way the LXCXE-binding cleft might be maintained, because it contributes in some degree to pRB's interaction with many cellular partners.
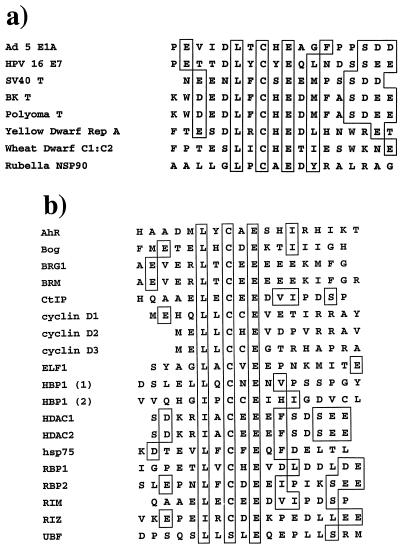
In support of the idea that viral and cellular LXCXE proteins might bind differently to pRB, we note that few of the cellular LXCXE-containing proteins contain all of the features that are conserved between viral RB-binding proteins (Fig. 8). The homology between E1A, E7, and T-antigen sequences includes an additional acidic residue 3 or 4 amino acids before the leucine (45), the presence of a hydrophobic side chain two or three residues after the glutamate (45, 52), and a series of acidic amino acids found C terminal to the LXCXE motif (45). Peptide competition assays have shown that the N-terminal acidic amino acid and the C-terminal hydrophobic residue have an important effect on the affinity of the interaction (45). Additionally, the importance of the hydrophobic amino acid just after the glutamate is also predicted by crystallographic data and supported by peptide competition experiments (45, 52). The conserved acidic sequence 5 or 6 amino acids C terminal to the LXCXE might also form ionic interactions with conserved lysine residues that surround the LXCXE binding cleft on pRB (52), adding to the strength of the interaction.
FIG. 8.
Sequence alignment of LXCXE-containing proteins. Amino acid sequences spanning the LXCXE motif of 8 viral proteins (a) and 18 cellular proteins (b) proposed to contain this motif. Conserved acid residues N terminal to the LXCXE motif, conserved hydrophobic and acid residues C terminal to the motif, and the LXCXE motif are boxed. Conserved residues surrounding the LXCXE motif of cellular proteins are likewise highlighted by a box. Abbreviations: Ad, adenovirus.
These results are consistent with the idea that viral and cellular pRB-binding proteins evolve under very different selective pressures. pRB's interaction with its cellular partners is regulated, and these interactions need to be reversible. In contrast, viral proteins bind to pRB in order to inactivate it, a process that is likely to be favored by high-affinity interactions. As a result, viral proteins may evolve a high-affinity pRB-binding site that only loosely mirrors the cellular proteins on which it is based. This study demonstrates that pRB's interaction with E7 is distinct from its interaction with any cellular protein that is essential to mediating cell cycle arrest within the context of the assays used here. However, further studies will be needed to determine whether such mutants can provide the full range of pRB functions that are needed for animal development.
ACKNOWLEDGMENTS
We thank past and present members of the Laboratory of Molecular Oncology—S. Boulton, E. Harrington, M. Classon, and S. Salama—for experimental suggestions or comments on the manuscript. We are particularly indebted to B. Schulman for advice early in this work. Plasmids were kindly provided by T. Kouzarides, N. Heintz, R. Watson, K. Munger, P. Sicinski, D. Dean, and B. Kaelin. Special thanks go to N. Pavletich, J.-O. Lee, and B. Schulman for providing the image used in Fig. 1. We also thank D. Dean and J. Wang for communicating their results prior to publication.
F.D. is a Fellow of the Leukemia Society of America. This work was supported by NIH grant CA64402 to N.D.
REFERENCES
- 1.Ach R A, Durfee T, Miller A B, Taranto P, Hanley-Bowdoin L, Zambryski P C, Gruissem W. RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol Cell Biol. 1997;17:5077–5086. doi: 10.1128/mcb.17.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atreya C D, Lee N S, Forng R-Y, Hofmann J, Washington G, Marti G, Nakhasi H L. The rubella virus putative replicase interacts with the retinoblastoma tumor suppressor protein. Virus Genes. 1998;16:177–183. doi: 10.1023/a:1007998023047. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates & Wiley-Interscience; 1988. [Google Scholar]
- 4.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 5.Buchmann A M, Swaminathan S, Thimmapaya B. Regulation of cellular genes in a chromosomal context by the retinoblastoma tumor suppressor protein. Mol Cell Biol. 1998;18:4565–4576. doi: 10.1128/mcb.18.8.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buyse I M, Shao G, Huang S. The retinoblastoma protein binds to RIZ, a zinc-finger protein that shares an epitope with the adenovirus E1A protein. Proc Natl Acad Sci USA. 1995;92:4467–4471. doi: 10.1073/pnas.92.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen B, Przybyla A E. An efficient site-directed mutagenesis method based on PCR. BioTechniques. 1994;17:657–659. [PubMed] [Google Scholar]
- 9.Chen C-F, Chen Y, Dai K, Chen P-L, Riley D J, Lee W-H. A new member of the hsp90 family of molecular chaperones interacts with the retinoblastoma protein during mitosis and after heat shock. Mol Cell Biol. 1996;16:4691–4699. doi: 10.1128/mcb.16.9.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen P-L, Riley D J, Chen Y, Lee W-H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 11.Chen P L, Riley D J, Chen-Kiang S, Lee W H. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Riley D J, Chen P-L, Lee W-H. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciccolini F, Di Pasquale G, Carlotti F, Crawford L, Tommasino M. Functional studies of E7 proteins from different HPV types. Oncogene. 1994;9:2633–2638. [PubMed] [Google Scholar]
- 14.DeCaprio J A, Ludlow J W, Figge J, Shew J W, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 15.Defeo-Jones D, Huang P S, Jones R E, Haskell K M, Vuocolo G A, Hanobik M G, Huber H E, Oliff A. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature. 1991;352:251–254. doi: 10.1038/352251a0. [DOI] [PubMed] [Google Scholar]
- 16.Dowdy S F, Hinds P W, Louie K, Reed S I, Arnold A, Weinberg R A. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- 17.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 18.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 19.Dyson N, Bernards R, Friend S H, Gooding L R, Hassell J A, Major E O, Pipas J M, Vandyke T, Harlow E. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J Virol. 1990;64:1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyson N, Buchkovich K, Whyte P, Harlow E. The cellular 107K protein that binds to adenovirus E1A also associates with the large T antigens of SV40 and JC virus. Cell. 1989;58:249–255. doi: 10.1016/0092-8674(89)90839-8. [DOI] [PubMed] [Google Scholar]
- 21.Dyson N, Guida P, McCall C, Harlow E. Adenovirus E1A makes two distinct contacts with the retinoblastoma protein. J Virol. 1992;66:4604–4611. doi: 10.1128/jvi.66.7.4606-4611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyson N, Guida P, Munger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dyson N, Harlow E, editors. Adenovirus E1A targets key regulators of cell proliferation. London, United Kingdom: Imperial Cancer Research Fund; 1992. [PubMed] [Google Scholar]
- 24.Ewen M E, Ludlow J W, Marsilio E, DeCaprio J A, Millikan R C, Cheng S H, Paucha E, Livingston D M. An N-terminal transformation-governing sequence of SV40 large T antigen contributes to the binding of both p110Rb and a second cellular protein, p120. Cell. 1989;58:257–267. doi: 10.1016/0092-8674(89)90840-4. [DOI] [PubMed] [Google Scholar]
- 25.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J, Livingston D M. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 26.Fattaey A R, Harlow E, Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol Cell Biol. 1993;13:7267–7277. doi: 10.1128/mcb.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fattaey A R, Helin K, Dembski M S, Dyson N, Harlow E, Vuocolo G A, Hanobik M G, Haskell K M, Oliff A, Defeo-Jones D, et al. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene. 1993;8:3149–3156. [PubMed] [Google Scholar]
- 28.Figge J, Webster T, Smith T F, Paucha E. Prediction of similar transforming regions in simian virus 40 large T, adenovirus E1A, and myc oncoproteins. J Virol. 1988;62:1814–1818. doi: 10.1128/jvi.62.5.1814-1818.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fusco C, Reymond A, Zervos A S. Molecular cloning and characterization of a novel retinoblastoma-binding protein. Genomics. 1998;51:351–358. doi: 10.1006/geno.1998.5368. [DOI] [PubMed] [Google Scholar]
- 30.Ge N L, Elferink C J. A direct interaction between the aryl hydrocarbon receptor and retinoblastoma protein. Linking dioxin signaling to the cell cycle. J Biol Chem. 1998;273:22708–22713. doi: 10.1074/jbc.273.35.22708. [DOI] [PubMed] [Google Scholar]
- 31.Goodwin E C, Naeger L K, Breiding D E, Androphy E J, DiMaio D. Transactivation-competent papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J Virol. 1998;72:3925–3934. doi: 10.1128/jvi.72.5.3925-3934.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamel P A, Gill R M, Phillips R A, Gallie B L. Transcriptional repression of the E2-containing promoters EIIaE, c-myc, and RB1 by the product of the RB1 gene. Mol Cell Biol. 1992;12:3431–3438. doi: 10.1128/mcb.12.8.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harbour J W. Overview of RB gene mutations in patients with retinoblastoma. Ophthalmology. 1998;105:1442–1447. doi: 10.1016/S0161-6420(98)98025-3. [DOI] [PubMed] [Google Scholar]
- 34.Harlow E, Franza B J, Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1988. [Google Scholar]
- 36.Harlow E, Whyte P, Franza B J, Schley C. Association of adenovirus early-region 1A proteins with cellular polypeptides. Mol Cell Biol. 1986;6:1579–1589. doi: 10.1128/mcb.6.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helin K, Wu C-L, Fattaey A, Lees J, Dynlacht B, Ngwu C, Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative transactivation. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 38.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 39.Hsiao K-M, McMahon S L, Farnham P J. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 40.Hu Q J, Dyson N, Harlow E. The regions of the retinoblastoma protein needed for binding to adenovirus E1A or SV40 large T antigen are common sites for mutations. EMBO J. 1990;9:1147–1155. doi: 10.1002/j.1460-2075.1990.tb08221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hwang E S, de Riese D J, Settleman J, Nilson L A, Honig J, Flynn S, DiMaio D. Inhibition of cervical carcinoma cell line proliferation by the introduction of a bovine papillomavirus regulatory gene. J Virol. 1993;67:3720–3729. doi: 10.1128/jvi.67.7.3720-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen D E, Black A R, Swick A G, Azizkhan J C. Distinct roles for Sp1 and E2F sites in the growth/cell cycle regulation of the DHFR promoter. J Cell Biochem. 1997;67:24–31. doi: 10.1002/(sici)1097-4644(19971001)67:1<24::aid-jcb3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 43.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F-1 expression in response to positive and negative regulators of cell cycle expression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 44.Jones D L, Alani R M, Munger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones R E, Wegrzyn R J, Patrick D R, Balishin N L, Vuocolo G A, Riemen M W, Defeo J D, Garsky V M, Heimbrook D C, Oliff A. Identification of HPV-16 E7 peptides that are potent antagonists of E7 binding to the retinoblastoma suppressor protein. J Biol Chem. 1990;265:12782–12785. [PubMed] [Google Scholar]
- 46.Kato J-Y, Matsushime H, Hiebert S W, Ewen M E, Sherr C J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 47.Knudsen E S, Buckmaster C, Chen T-T, Feramisco J R, Wang J Y J. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 1998;12:2278–2292. doi: 10.1101/gad.12.15.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 49.Lai A, Macellus R C, Corbeil H B, Branton P E. RBP1 induces growth arrest by repression of E2F-dependent transcription. Oncogene. 1999;18:2091–2100. doi: 10.1038/sj.onc.1202520. [DOI] [PubMed] [Google Scholar]
- 50.Lam E W, Bennett J D, Watson R J. Cell-cycle regulation of human B-myb transcription. Gene. 1995;160:277–281. doi: 10.1016/0378-1119(95)00184-8. [DOI] [PubMed] [Google Scholar]
- 51.Lam E W-F, Watson R J. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J-O, Russo A A, Pavletich N P. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 53.Lees J A, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13:7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J M, Hu P P, Shen X, Yu Y, Wang X F. E2F4-RB and E2F4-p107 complexes suppress gene expression by transforming growth factor beta through E2F binding sites. Proc Natl Acad Sci USA. 1997;94:4948–4953. doi: 10.1073/pnas.94.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu L, Saunders K, Thomas C L, Davies J W, Stanley J. Bean yellow dwarf virus RepA, but not rep, binds to maize retinoblastoma protein, and the virus tolerates mutations in the binding motif. Virology. 1999;256:270–279. doi: 10.1006/viro.1999.9616. [DOI] [PubMed] [Google Scholar]
- 56.Lu X, Horvitz H R. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–991. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- 57.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 58.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–604. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 59.McCaffrey J, Yamasaki L, Dyson N J, Harlow E, Griep A E. Disruption of retinoblastoma protein family function by human papillomavirus type 16 E7 oncoprotein inhibits lens development in part through E2F-1. Mol Cell Biol. 1999;19:6458–6468. doi: 10.1128/mcb.19.9.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meloni A R, Smith E J, Nevins J R. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc Natl Acad Sci USA. 1999;96:9574–9579. doi: 10.1073/pnas.96.17.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 62.Mulligan G, Jacks J. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- 63.Munger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naeger L K, Goodwin E C, Hwang E S, DeFilippis R A, Zhang H, DiMaio D. Bovine papillomavirus E2 protein activates a complex growth-inhibitory program in p53 negative HT-3 cervical carcinoma cells that includes repression of cyclin A and cdc25A phosphatase genes and accumulation of hypophosphorylated retinoblastoma protein. Cell Growth Differ. 1999;10:413–422. [PubMed] [Google Scholar]
- 65.Nead M A, Baglia L A, Antinore M J, Ludlow J W, McCance D L. Rb binds c-Jun and activates transcription. EMBO J. 1998;17:3242–3252. doi: 10.1093/emboj/17.8.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neuman E, Flemington E K, Sellers W R, Kaelin W G. Transcription of the E2F1 gene is rendered cell cycle-dependent by E2F DNA binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 68.Novitch B G, Mulligan G J, Jacks T, Lassar A B. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pan H, Gripe A E. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- 70.Phelps W C, Yee C L, Munger K, Howley P M. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell. 1988;53:539–547. doi: 10.1016/0092-8674(88)90570-3. [DOI] [PubMed] [Google Scholar]
- 71.Qin X Q, Chittenden T, Livingston D M, Kaelin W G., Jr Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 72.Scheffner M, Munger K, Byrne J C, Howley P M. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Natl Acad Sci USA. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmitt A, Harry J B, Rapp B, Wettstein F O, Iftner T. Comparison of the properties of the E6 and E7 genes of low- and high-risk cutaneous papillomaviruses reveals strongly transforming and high Rb-binding activity for the E7 protein of the low-risk human papillomavirus type 1. J Virol. 1994;68:7051–7059. doi: 10.1128/jvi.68.11.7051-7059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G., Jr Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sellers W R, Rodgers J W, Kaelin W G., Jr A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Singh P, Coe J, Hong W. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 77.Slansky J E, Farnham P J. Transcriptional regulation of the dihydrofolate reductase gene. Bioessays. 1996;18:55–62. doi: 10.1002/bies.950180111. [DOI] [PubMed] [Google Scholar]
- 78.Stabel S, Argos P, Philipson K. The release of growth arrest by microinjection of adenovirus E1A DNA. EMBO J. 1985;4:2329–2336. doi: 10.1002/j.1460-2075.1985.tb03934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strober B E, Dunaief J L, Guha S, Goff S P. Functional interactions between the hBrm/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996;16:1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tevosian S G, Shih H H, Mendelson K G, Sheppard K-A, Paulson K E, Yee A S. HBP1: a HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes Dev. 1997;11:383–396. doi: 10.1101/gad.11.3.383. [DOI] [PubMed] [Google Scholar]
- 81.Thomas J T, Hubert W G, Ruesch M N, Laimins L A. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci USA. 1999;96:8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm co-operate to repress the activation functions of E2F-1. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 85.Wang C Y, Petryniak B, Thompson C B, Kaelin W G, Leiden J M. Regulation of the Ets-related transcription factor Elf-1 by binding to the retinoblastoma protein. Science. 1993;260:1330–1335. doi: 10.1126/science.8493578. [DOI] [PubMed] [Google Scholar]
- 86.Wang J Y J. Retinoblastoma protein in growth suppression and death protection. Curr Opin Genet Dev. 1997;7:39–45. doi: 10.1016/s0959-437x(97)80107-4. [DOI] [PubMed] [Google Scholar]
- 87.Weintraub S J, Chow K N B, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 88.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 89.Wells J, Held P, Illenye S, Heintz N H. Protein-DNA interactions at the major and minor promoters of the divergently transcribed dhfr and rep3 genes during the chinese hamster ovary cell cycle. Mol Cell Biol. 1996;16:634–647. doi: 10.1128/mcb.16.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wells J M, Illenye S, Magae J, Wu C-L, Heintz N H. Accumulation of E2F-4/DP-1 DNA binding complexes correlates with induction of dhfr gene expression during the G1 to S phase transition. J Biol Chem. 1997;272:4483–4492. doi: 10.1074/jbc.272.7.4483. [DOI] [PubMed] [Google Scholar]
- 91.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 92.Whyte P, Ruley H E, Harlow E. Two regions of the adenovirus early region 1A proteins are required for transformation. J Virol. 1988;62:257–265. doi: 10.1128/jvi.62.1.257-265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Whyte P, Williamson N M, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 94.Woitach J T, Zhang M, Niu C-H, Thorgeirsson S S. A retinoblastoma-binding protein that affects cell-cycle control and confers transforming ability. Nat Genet. 1998;19:371–374. doi: 10.1038/1258. [DOI] [PubMed] [Google Scholar]
- 95.Wu C-L, Classon M, Dyson N, Harlow E. Expression of dominant-negative mutant DP-1 blocks cell cycle progression in G1. Mol Cell Biol. 1996;16:3698–3706. doi: 10.1128/mcb.16.7.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xie Q, Suarez-Lopez P, Gutierrez C. Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: requirement for efficient viral DNA replication. EMBO J. 1995;14:4073–4082. doi: 10.1002/j.1460-2075.1995.tb00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang H S, Postigo A A, Dean D C. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16, TGFb, and contact inhibition. Cell. 1999;97:53–61. doi: 10.1016/s0092-8674(00)80714-x. [DOI] [PubMed] [Google Scholar]
- 98.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]