Abstract
Tandem mass spectrometry (MS/MS) is an accurate tool to assess modified ribonucleosides and their dynamics in mammalian cells. However, MS/MS quantification of lowly abundant modifications in non-ribosomal RNAs is unreliable, and the dynamic features of various modifications are poorly understood. Here, we developed a 13C labeling approach, called 13C-dynamods, to quantify the turnover of base modifications in newly transcribed RNA. This turnover-based approach helped to resolve mRNA from ncRNA modifications in purified RNA or free ribonucleoside samples and showed the distinct kinetics of the N6-methyladenosine (m6A) versus 7-methylguanosine (m7G) modification in polyA+-purified RNA. We uncovered that N6,N6-dimethyladenosine (m62A) exhibits distinct turnover in small RNAs and free ribonucleosides when compared to known m62A-modified large rRNAs. Finally, combined measurements of turnover and abundance of these modifications informed on the transcriptional versus posttranscriptional sensitivity of modified ncRNAs and mRNAs, respectively, to stress conditions. Thus, 13C-dynamods enables studies of the origin of modified RNAs at steady-state and subsequent dynamics under nonstationary conditions. These results open new directions to probe the presence and biological regulation of modifications in particular RNAs.
Keywords: isotopic labeling; RNA modifications; RNA turnover; N6-methyladenosine; 7-methylguanosine; N6,N6-dimethyladenosine; mass spectrometry; metabolism; methylation dynamics; metabolic stress
Abbreviations: f6A, N6-formyladenosine; FBS, fetal bovine serum; hm6A, N6-hydroxymethyladenosine; m6A, N6-methyladenosine; m7G, 7-methylguanosine; m62A, N6,N6-dimethyladenosine; MS/MS, tandem mass spectrometry; NGS, next-generation sequencing; rRNA, ribosomal RNA; SAM, S-adenosylmethionine
RNA methylation modulates crucial RNA–protein interactions at various stages of RNA metabolism. Comprehensive studies of individual RNA modifications have been mostly advanced by applications of next-generation sequencing (NGS), which rely on chemical, enzymatic, and/or antibody-based detection of modified ribonucleosides (1, 2, 3). These methods have provided a wealth of information on the sites of modification across the transcriptome (1, 2, 3), which have uncovered the dynamic behavior of N6-methyladenosine (m6A) mRNA methylation in development (4, 5, 6, 7, 8). Notably, dysregulation of m6A levels has been recently linked to cancer, aging, and neurodegeneration (9, 10, 11, 12, 13, 14, 15), which can occur via metabolic inhibition of m6A demethylation (9, 10). Furthermore, various tRNA modifications have been reported to be posttranscriptionally sensitive to cellular stress (16, 17, 18, 19). A current caveat of NGS-based profiling is the lack of high-specificity reagents for every modification (1, 2, 3). Also, the effects of transcription, which often predominate in differential expression analyses (20), make it challenging to quantify general changes in methylation levels, with limited insight being obtained into biological associations between various modifications. As a consequence, the dynamic behavior of RNA methylation often remains poorly understood, and complementary approaches are needed to quantify multiple modifications across biological contexts and to assess their associations to transcriptional versus posttranscriptional events.
Tandem mass spectrometry (LC-MS/MS) is a highly accurate tool for analysis of modified RNAs, which has so far been applied primarily in two approaches to identify and quantify RNA modifications (21). The first approach employs LC-MS/MS of intact RNA oligonucleotides to detect multiple modifications with positional information in specific RNA sequences. This approach is chromatographically challenging (22) and requires advanced data mining for unambiguous identification of RNA fragments, and it has so far been applied for comprehensive characterization of RNA modifications in abundant or short RNAs (22), such as rRNA (23, 24, 25, 26), tRNAs (27, 28), and miRNAs (29). The second approach employs LC-MS/MS of ribonucleosides for sensitive quantification of RNA modifications (21). This approach, often combined with stable isotopes, can simultaneously quantify the abundance of multiple modifications in specific RNA classes of interest, being highly suitable to assess the presence of tRNA and rRNA modifications and their dynamics under various biological scenarios (19, 27, 30, 31, 32, 33, 34, 35). These methods have so far been applied mainly to study these abundant RNA species because MS detection of mRNA modifications is unreliable due to the heavily modified ncRNAs, which invariably contaminate mRNA pools purified using either polyA+-enrichment or rRNA depletion (36, 37). Therefore, MS approaches are needed that are capable to account for and quantitatively resolve the origin of multiple RNA modifications in purified RNA. Moreover, the dynamic behavior of RNA modifications is insufficiently explained solely by changes in their abundance (or levels), as these do not inform on the underlying pathways driving these changes. Since the deposition (and removal) of RNA modifications is linked to the life cycle of the RNAs themselves (38, 39, 40), the change in methylation levels can result from changes either in RNA (de)methylation rates or in the transcription or decay of methylated RNAs, From this lens, methods are needed that can assess transcriptional and posttranscriptional effects on methylated RNAs under nonstationary conditions.
Stable isotope labeling is a well-established method to quantify metabolic activity in cultured cells (41, 42, 43). Here, we developed a quantitative approach using [13C-methyl]-methionine labeling and mass spectrometry (MS) to assess the turnover of base modifications (13C-dynamods) in newly transcribed RNA. With 13C-dynamods, we trace the proportion of newly methylated ribonucleosides and their decay through time, i.e., methylation turnover, from digested polyadenylated RNA and ncRNA. We first showed that polyadenylated RNA and ncRNA were distinguished by the different turnover frequencies (in h−1) of modified ribonucleosides, which are inherently linked to the different half-lives of mRNA, rRNA, and tRNA (39, 44, 45). Examining the kinetics of methylation turnover at steady state within and across RNA classes as well as in free ribonucleosides enabled us to resolve the origin of RNA modifications in digested RNA and thereby uncover the presence of modifications in uncharacterized RNA classes, such as N6,N6-dimethyladenosine (m62A). We then applied 13C-dynamods in conjugation with abundance measurements of modified ribonucleosides in polyA+ and ncRNA, which resolved their transcriptional versus posttranscriptional sensitivity in response to actinomycin D and metabolic stresses. Thus, the quantitative nature of 13C-dynamods demonstrates its capacity for sensitive characterization of modified ribonucleosides, their origin and dynamics, in multiple RNA classes of interest.
Results
13C labeling of polyadenylated and ribosomal RNA modifications
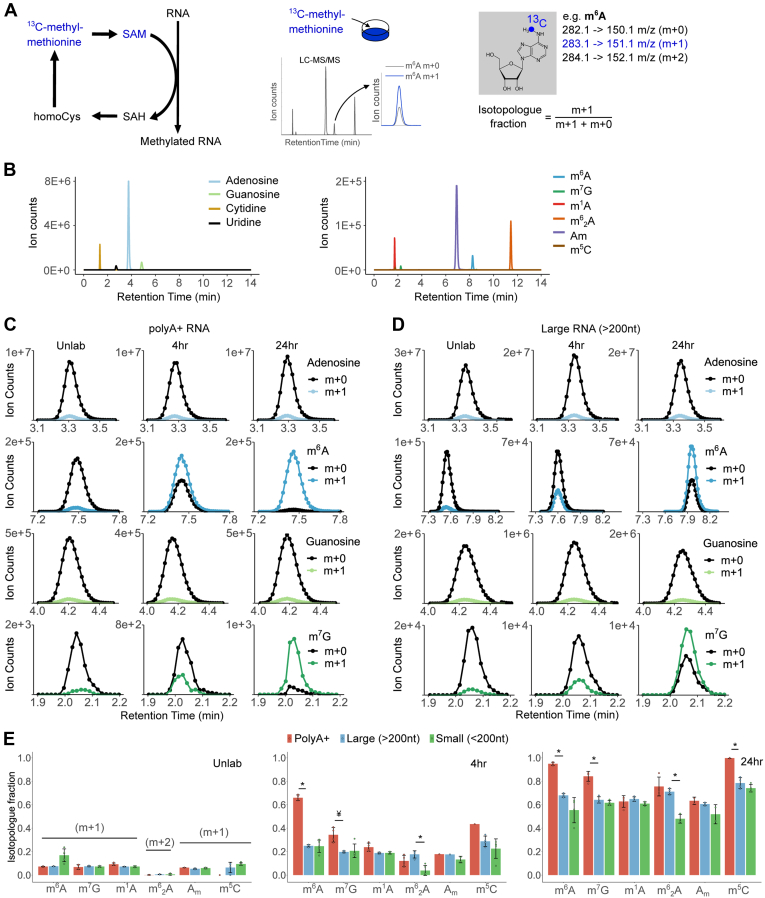
S-adenosylmethionine (SAM) is the direct substrate of RNA methylation reactions in eukaryotic cells (46). To trace the incorporation of SAM into RNA, we cultured 786O cells in methionine-free DMEM medium supplemented with either unlabeled methionine (“Unlab”) or [13C-methyl]-methionine and analyzed the isotopologues (m + 0, m + 1, m + 2) of modified and unmodified ribonucleosides by tandem mass spectrometry (LC-MS/MS) (Fig. 1, A and B). The m + 0 isotopologue (e.g., 150.1 m/z for m6A) represents the mass of the analyzed molecule where all atoms are present as the most common isotope, whereas the m + 1 isotopologue (e.g., 151.1 m/z for m6A) indicates the mass shift due to 13C incorporation from the 13C-labeled methionine tracer or from the natural abundance of 13C, 15N, 18O, and 2H stable isotopes. With 13C-dynamods, we measure the isotopologue fractions [m + 1/(m + 1 + m + 0)] of modified and unmodified ribonucleosides, which reflect the amount of 13C enrichment (m + 1) in the measured ribonucleoside relative to the total pool of that ribonucleoside (m + 1 + m + 0) (Fig. 1A). Thus, the isotopologue fractions of each ribonucleoside during [13C-methyl]-methionine labeling are internally controlled for the amount of preexisting ribonucleosides (m + 0 and naturally labeled m + 1) prior to 13C labeling. Here, we analyzed the isotopologues of N6-methyladenosine (m6A), 7-methylguanosine (m7G), 1-methyladenosine (m1A), N6,N6-dimethyladenosine (m62A), 2′-O-methyladensone (Am), 5-methylcytosine (m5C), and unmodified ribonucleosides from digested polyadenylated (polyA+), large (>200 nt) and small (<200 nt) RNA. We observed increased m + 1 and concomitant decreased m + 0 ion counts in modified ribonucleosides from polyA+ and large RNA after 4 and 24 h of [13C-methyl]-methionine labeling, while the m + 1 fractions (natural abundance) of the unmodified ribonucleosides was unaltered during the labeling period (Figs. 1, C and D and S1A; Supplementary Materials). We determined that the 13C enrichment of intracellular methionine and SAM reaches a 98 to 100% plateau within 30 min and remains constant thereafter (Fig. S1B). Thus, the change in the “heavy” (m + 1) isotopologue fraction relative to the unlabeled condition (natural abundance) indicates SAM-dependent RNA methylation of newly synthesized RNA, whose turnover we have examined within and across RNA classes. In contrast to singly methylated ribonucleosides, the m62A modification exhibited an enrichment mostly of the m + 2 isotopologue upon 13C labeling (Fig. S1A), and so the m + 2 fraction was used to assess m62A dynamics in subsequent analyses.
Figure 1.
13C labeling of RNA modifications across RNA classes.A, the 13C-dynamods workflow shows the tracing of RNA methylation: cells are cultured with [13C-methyl]-methionine, the RNA is isolated, digested to ribonucleosides and subjected to LC-MS/MS analysis. The isotopologues detected for m6A are shown. B, representative chromatogram of unmodified and modified ribonucleosides from total RNA. C and D, the m + 0 and m + 1 isotopologues of modified and unmodified ribonucleosides (representative chromatograms) in polyA+ (C) and large RNA (D). E, quantification of the isotopologue fractions of each ribonucleoside in polyA+, large and tRNA under unlabeled (‘Unlab’) conditions, after 4 and 24 h of culture with [13C-methyl]-methionine. Error bars represent standard deviation of 3 to 4 biological replicates, with the exception of m5C in polyA+-purified RNA (two replicates at 24 h, one replicate in the Unlab/4 h time points). In all cases, each replicate is the average of two technical replicas. ∗ denotes p < 0.005, ¥ denotes p < 0.05, of a two-sided Student’s t test comparing samples as indicated. Am, 2′-O-methyladenosine; homoCys, homocysteine; m1A, 1-methyladenosine; m1G, 1-methylguanosine; m2G, 2-methylguanosine; m5C, 5-methylcytidine; m6A, N6- methyladenosine; m62A, N6,N6-dimethyladenosine; m7G, 7-methylguanosine; SAH, sadenosylhomocysteine; SAM, s-adenosylmethionine.
Quantification of isotopologue fractions showed a faster kinetics for m6A, m7G, and m5C in polyA+-purified RNA when compared with large (>200 nt) and small (<200 nt) RNA (Fig. 1E), in accordance with the faster life cycle of mRNA in mammalian cells (45). In contrast, the kinetics of m1A, m62A, and Am methylation was similar between polyA+ and large RNA fractions (Fig. 1E), suggesting that a large portion of the signal for these modifications in the polyA+ fraction might derive from contaminating ribosomal RNA (rRNA). Of note, the isotopologues are analyzed on all ribonucleosides from the same polyA+ pool (Fig. S1C), so the purity of polyA+ enrichment is the same in all cases. This indicates that the relative abundance in mRNAs versus contaminating ncRNA is much higher for m6A, m7G, and m5C compared with m1A, m62A, and Am. Interestingly, we detected m62A in small RNAs from 786O cells, and its kinetics was different when compared with large RNA (Fig. 1E).
Methylation turnover to resolve the origin of RNA modifications and their presence in uncharacterized RNA classes
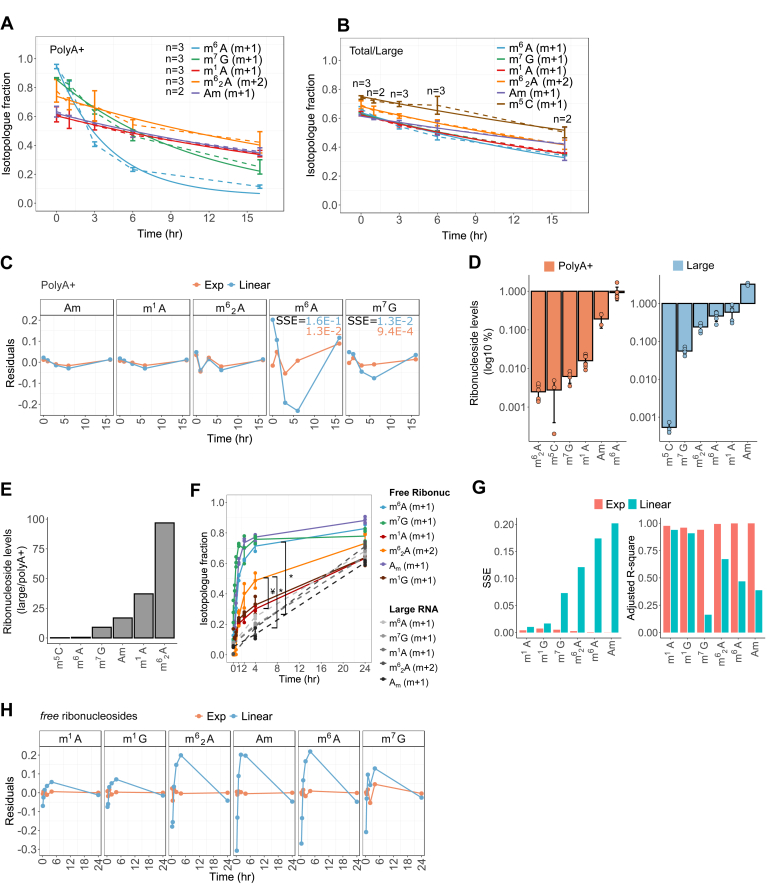
To examine the kinetics of methylation turnover, we cultured cells for 24 h with [13C-methyl]-methionine followed by replacement (chase) with unlabeled methionine and analyzed the isotopologues over time. In accordance with the preceding findings (Fig. 1E), we observed an exponential decay of the m + 1 isotopologue fraction for m6A/m7G but not for Am/m1A/m62A in polyA+ RNA (Fig. 2A). Conversely, the modifications of total/large RNAs exhibited uniformly slow turnover (Fig. 2B), consistent with the high stability of rRNA and tRNA in growing cells (44). To test the kinetic behavior underlying methylation turnover, we examined the goodness of fit of a linear versus exponential regression of the isotopologue fraction. Analysis of residual errors showed that the linear regression (m(t) = m(0) + kt) fits well the turnover of m1A/m62A/Am but not m6A/m7G in polyA+ RNA, in which the “U-shaped” curve supports a nonlinear model (m(t) = m(0) e(−kt)) (Figs. 2C and S2A). Conversely, the linear regression fitted well the turnover of ncRNA modifications (Fig. S2, B and C). The turnover frequency determined for Am/m1A/m62A in polyA+ was similar to ncRNA modifications (k = 0.031 h−1 on average) and significantly slower than m6A (k = 0.244 h−1) or m7G (k = 0.089 h−1) in polyA+ RNA (Fig. 2, A and B). Thus, the turnover of m1A/m62A/Am from polyA+ fractions is incompatible with the exponential turnover of mRNA modifications (44, 45), confirming that they originate from ncRNA contamination.
Figure 2.
Turnover of modified ribonucleosides in polyA+, ncRNA, and free ribonucleoside pool.A and B, isotopologue fractions during the “chase” of 13C-labeled modifications with naturally labeled methionine for 0, 1, 3, 6, and 16 h, in polyA+ (A) and total/large RNA (B); dashed lines connect data points; solid lines, exponential (A) or linear (B) fit of isotopologue fractions. C, residuals of a linear versus exponential regression of isotopologue fractions in polyA+ RNA in the chase experiment. D, normalized ion counts of modified ribonucleosides relative to the ion counts sum of all ribonucleosides, shown for polyA+ and large RNA. E, ratio of normalized ion counts between polyA+ and large RNA, as determined in (D). F, isotopologue fraction of free ribonucleosides analyzed from metabolic extracts; gray datapoints refer to large RNA modifications (Fig. 1E) for comparison. G and H, goodness of fit of a linear versus exponential fit of the isotopologue fractions in free modified ribonucleosides. Error bars represent 90% confidence intervals in (A and B), or standard deviation in (D and F) of at least three biological replicates with exception of Am (two replicates) and in (B) as indicated. In all cases, each replicate is the average of two technical replicas. ∗ denotes p < 0.005, ¥ denotes p < 0.05, of a two-sided Student’s t test comparing m62A and m6A in the free ribonucleoside pool (free ribonuc) to large RNA, or m62A to m1A in the free ribonuc pool. A, adenosine; Am, 2′-O-methyladenosine; G, guanosine; m1A, 1-methyladenosine; m1G, 1-methylguanosine; m5C, 5-methylcytidine; m6A, N6-methyladenosine; m62A, N6,N6-dimethyladenosine; m7G, 7-methylguanosine; SSE, sum of squared errors.
The normalized abundance (frequency) of m7G was in between that of m62A and m1A/Am in polyA+ RNA (Fig. 2D), and the sensitivity of MS detection was similar across all measured modifications except m5C, as determined from equimolar injections of pure compounds (Fig. S2D). Thus, the distinct turnover of RNA modifications in the polyA+-purified fraction was not explained by differences in normalized abundances alone or analytical sensitivity of ribonucleoside detection. Instead, the modified ribonucleosides were more abundant in large RNA relative to polyA+ RNA by a factor of 17/37/97/9 for Am/m1A/m62A/m7G, respectively, while m6A level was greater in polyA+ RNA by a factor of 2 (Fig. 2E). From the determined turnover frequencies (h−1) of ncRNA (Fig. 2B), we estimated that a 50% contribution of ncRNA signal to the ribonucleoside pool in polyA+-purified RNA would lower the detected turnover of the bona fide m6A mRNA modification, with a turnover frequency of 0.242 h−1, to that of m7G, with a turnover frequency of ∼0.09 h−1 (Fig. S2E). That is, reliable turnover-based detection of a hypothetical mRNA modification that is more abundant by a factor of 17 to 97 in ncRNA relative to mRNA is attainable with a maximal ncRNA contamination of 0.5% (50%/97) to 2.9% (50%/17), but even an optimal performance of available methods to purify polyA+ RNA commonly contains 2 to 3% of contaminating ncRNA (Fig. S2F) (36, 37). Thus, the ability to resolve bona fide mRNA modifications based on methylation turnover is limited by the depletion efficiency of highly abundant rRNA modifications.
As the turnover frequency of m6A was approximately eight times faster in polyA+ RNA than ncRNA (Fig. 2, A and B), we reasoned that free ribonucleosides derived from metabolic extracts would be derived mainly from endogenous degradation of mRNAs, rather than ncRNAs. Thus, we employed 13C-dynamods to examine the methylation turnover of free ribonucleosides, thereby enriching for mRNA-derived ribonucleosides and decreasing interference from rRNA-derived ribonucleosides. We found a fast kinetics following 13C labeling that was similar for m6A, m7G, and Am, indicating that they are primarily derived from the same RNA type (i.e., mRNAs) (Fig. 2, F–H). Interestingly, the m62A modification also exhibited a nonlinear, faster kinetics than m1A or 1-methylguanosine (m1G) (Fig. 2, F–H), which are known tRNA modifications and were present in the metabolic extracts at high levels (Fig. S2G) (47). We did not include the isotopologue analysis of m5C in the time-series experiments due to low abundance of its free ribonucleosides and due to low MS sensitivity for m5C (Figs. 2D and S2, D and G), which compromises the quantification under conditions of partial 13C labeling. Thus, while the turnover analysis of the polyA+ fraction could not be used to validate lowly abundant modifications due to rRNA contamination (Fig. 2E), the turnover of Am and m62A in the free pool suggests that these modifications indeed partly derive from rapidly decaying RNAs (Fig. 2F), which may include non-ribosomal RNAs. This was supported by the normalized levels of Am and m62A in the free pool being more similar to those of polyA+ than rRNA (Fig. S2H). These results confirm the presence of Am in mRNA (48) and suggest that m62A is more common in short-lived RNAs than m1A, which has already been studied in mRNAs (49, 50).
Sensitivity of RNA modifications to metabolic stresses
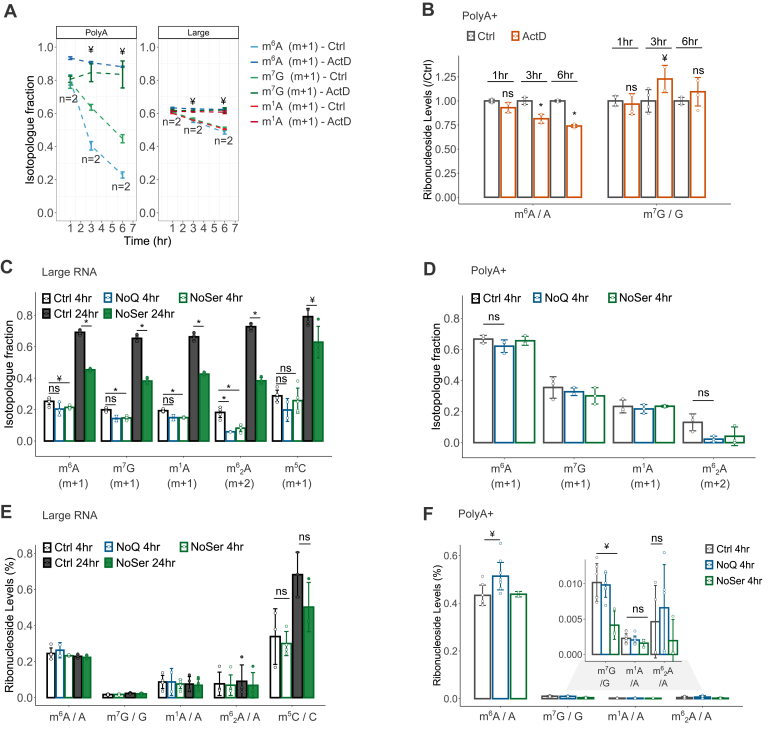
The maintenance of methylation reactions requires SAM and serine/glycine, which feed one-carbon metabolism and SAM synthesis (51). Conversely, RNA demethylation is catalyzed by α-ketoglutarate (α-kg)-dependent dioxygenases such as FTO and ALKBH5, which act on m6A, N6,2-O-dimethyladenosine (m6Am), and m1A (52, 53). We measured both the methylation turnover and abundance of modified ribonucleosides to obtain insights into the sensitivity of mRNA versus ncRNA to metabolic stresses linked to RNA (de)methylation. First, we confirmed that actinomycin (ActD), a pan inhibitor of eukaryotic transcription, completely inhibited the methylation turnover of polyA+ and large RNA (Fig. 3A), in accordance with the cotranscriptional deposition of mRNA and most rRNA modifications (54, 55, 56, 57). Treatment with ActD decreased the normalized levels of m6A (m6A/A) in polyA+ RNA (Fig. 3B), but not of m7G, which supports the role of m6A in global mRNA destabilization, as initially reported (40). Deprivation of serine or glutamine inhibited mainly the methylation turnover of large RNA (Fig. 3, C and D). In contrast, the abundance of modified ribonucleosides in large RNA was unaltered under these forms of stress (Fig. 3E), whereas glutamine deprivation led to increased m6A levels in polyA+ RNA (Fig. 3F), which was in contrast to ActD treatment (Fig. 3B). Thus, the reduced methylation turnover of large RNA simply results from an inhibited transcription of the rRNAs themselves, while the altered abundance of m6A without significant changes in its turnover indicates its posttranscriptional lability to stress conditions via altered (i) decay of m6A-enriched versus m6A-depleted mRNAs or (ii) dynamics of the m6A modification itself. These data show that measurements of methylation turnover and ribonucleoside abundances resolve transcriptional versus posttranscriptional effects on RNA modifications in nonstationary (stimulus-dependent) experiments.
Figure 3.
Dynamics of RNA modifications under stress conditions.A, isotopologue fractions of modified ribonucleosides in polyA+ and large RNA during a 13C-dynamods chase experiment in 786O cells cultured with complete DMEM medium (ctrl) or DMEM supplemented with actinomycin D (ActD, 10 μM); dashed lines connect data points. B, normalized levels of m6A (m6A/A) and m7G (m7G/G) in polyA+ RNA derived from ctrl and ActD-treated 786O cells, normalized relative to the ctrl data points in the chase experiment as indicated. C and D, isotopologue fractions of modified ribonucleosides in large RNA (C) and polyA+ RNA (D) following [13C-methyl]-methionine labeling under control (ctrl), glutamine-(NoQ), or serine-(NoSer) deprived conditions. E and F, normalized levels of modified ribonucleosides in large RNA (E) and polyA+ RNA (F) in 786O cells cultured with ctrl, NoQ or NoSer DMEM medium for the times indicated. Error bars represent standard deviation of three or more biological replicates, with exception of (A), the NoQ condition in (C and E) and m6A under NoSer (D and F) (two biological replicates). In all cases, each replicate is the average of two technical replicas. ∗ denotes p < 0.005, ¥ denotes p < 0.05, ns denotes not significant, of a two-sided Student’s t test comparing stress to ctrl samples, as indicated.
Discussion
Isotopic labeling of cultured cells is a well-established method to quantify metabolic activity, but its application to RNA modifications has been limited to ncRNAs (33, 58). Here, we demonstrate that quantification of methylation turnover with dynamic 13C labeling informs on the distinct dynamics of polyA+ and ncRNA modifications and their sensitivity to metabolic perturbations of mammalian cells. 13C-dynamods presents several advances in the application of MS approaches to study RNA turnover and dynamics. First, in contrast to approaches that rely on prior/in-parallel labeling with 15N/13C-enriched nucleosides to distinguish between preexisting and newly modified RNA (33, 58), the “heavy” isotopologue fraction derived from [13C-methyl]-methionine is internally controlled for the amount of unlabeled, preexisting ribonucleosides and is thus specific to modified ribonucleosides. Second, in contrast to approaches using multiplexing or spiking from isotopically labeled cultures (24, 25, 32, 58), the dynamic 13C labeling over time directly informs on the turnover frequency (in h−1) (41, 43) of various RNA modifications, as the direct substrate (SAM) of the targeted reaction (RNA methylation) is close to 100% labeled in less than 30 min in cell culture (Fig. S1B) (43). Moreover, SAM need not be fully labeled for comparisons of dynamic 13C labeling between modifications at early, single time points as its 13C enrichment is expected to be equal across SAM-dependent RNA modifications. Third, conventional RNA labeling approaches using ribonucleoside analogues or precursors rely on their incorporation into newly synthesized RNA through the salvage pathway (e.g., uridine and its analogs) (45, 59) or the de novo pathway (glucose, serine/glycine, or water tracers) of ribonucleotide (NTP) synthesis (60, 61, 62, 63, 64). In contrast, the methylation of newly transcribed RNA occurs downstream (cotranscriptionally) of NTP synthesis, making it a reliable readout of RNA turnover, particularly under nonstationary conditions that may alter the relative activity of NTP synthesis pathways (65, 66). Also, the nucleotide recycling through the salvage pathway may lead to ineffective “washout” with unlabeled nucleotides in chase experiments (59, 66), whereas a new SAM molecule is used in each (cotranscriptional) methylation cycle in 13C-dynamods, adding to the specificity and versatility of experimental designs (59). Finally, combined measurements of methylation turnover and ribonucleoside levels allow 13C-dynamods to also inform on transcriptional versus posttranscriptional sensitivity of RNA modifications in response to a stimulus.
Reliable study of mRNA modifications by MS analyses of ribonucleosides is challenging due to the high abundance of heavily modified ncRNAs that invariably contaminate the polyA+ fraction (36, 37). To address this challenge, we exploited the fact that the life cycle of methylated RNA follows the life cycle of the RNAs at metabolic steady state (38, 39, 40). The decay of mRNA is fast with a median half-life from 40 min to 9 h in mammalian cells (44, 67, 68), while rRNA and tRNA exhibit half-lives of 60 to 70 h in growing fibroblasts (44). By definition, the methylation turnover as determined by 13C incorporation into newly methylated RNA does not assess nonmodified RNAs and hence does not capture the removal of RNA modifications. That is, any RNA demethylation activity can only affect the abundances of modified ribonucleosides as a whole (sum of m + 1 and m + 1), not the m + 1 or m + 0 isotopologues individually, which would be required to cause a differential m + 1 fraction. Since RNA transcription and decay rates are constant at metabolic steady state, we propose that the methylation turnover is an accurate readout of RNA turnover, which allowed us to assess the contributions of short-lived (mRNAs) and long-lived (noncoding) RNAs to a pool of modified RNAs and free ribonucleosides. Indeed, this information could not be drawn if labeled ribonucleoside themselves or its precursors were used to trace ribonucleotide synthesis (via the salvage and de novo pathways) or transcription. Likewise, the source RNA cannot be pinpointed if the abundance (levels) of modified ribonucleosides is measured in isolation.
Using this approach, we could not reliably determine highly abundant rRNA modifications (Am/m1A/m62A) in polyA+-purified RNA as being derived from mRNAs. Based on methylation turnover, we estimated that these modifications in the polyA+ fraction could be detected with a maximal ncRNA contamination of 0.5 to 2.9%, since Am/m1A/m62A levels were found higher in ncRNAs by a factor of 17–37–97, respectively. In contrast, a ∼6% of ncRNA contamination would be required to fully account for the slower m7G turnover detected in polyA+ RNA, since m7G levels were found only approximately nine times higher in ncRNAs. Since the m7G turnover reflected that of a bona fide mRNA modification, its slower kinetics in polyA+-purified RNA suggests a temporal delay relative to m6A turnover. A further evidence for the distinct turnover of m7G relative to m6A in mRNAs is the response to metabolic stresses, where the m7G turnover changed in large RNA but not polyA+ RNA following 4 h of 13C labeling (Fig. 3, C and D). In this respect, while m6A is cotranscriptionally deposited in mRNA (39, 54, 56), m7G is an essential modification at the 5′ cap of mRNA that can be placed in both the nucleus and cytoplasm (69, 70). Moreover, m7G sites have been reported internally within mRNA, catalyzed by the cytosolic METTL1 methyltransferase (71). Taken together, the distinct kinetics of m7G and m6A in polyadenylated RNA suggest compartmentalization differences affecting their temporal deposition into newly synthesized mRNA.
Previous reports identified m5C and Am as mRNA modifications (48, 72) and more recently also m1A (49, 50). Our analysis of free ribonucleosides showed a similarly fast kinetics for the turnover of Am, m6A and m7G, consistent with these modifications being derived from the rapidly decaying mRNAs. This was supported by the normalized levels of free modified ribonucleosides being more similar to those of polyA+-purified than of ncRNAs. In contrast, the turnover of free m1A and m1G ribonucleosides was significantly slower, indicating that these modifications are predominantly derived from an ncRNA class, likely tRNAs, where they are present at high levels (47). This aligns with the recent conclusions that m1A might be restricted to a handful of mRNAs (3, 49). Finally, despite the low sensitivity of m5C detection in chase experiments, the quantification of m5C isotopologues at 24 h is consistent with it being derived from mRNA (72).
The m62A modification is thought to be present primarily in the 18S and 12S rRNA of mammalian transcriptomes (26, 73, 74). While m62A has been detected in bacterial tRNA (75), care must be taken with abundant m62A-modified rRNA fragments that copurify with tRNA (76), and are not resolved by standard MS quantifications. Here, we detected slower turnover of m62A in mammalian small RNAs relative to large RNA, raising the possibility that this modification is present in non-18S/12S RNA species, e.g., in small rRNA and/or tRNAs. Nevertheless, an unlikely possibility from 13C-dynamods measurements remains that m62A-containing 18S/12S rRNA fragments are turned over at a slower rate than m62A-containing intact rRNAs. Interestingly, the turnover of free m62A ribonucleosides exhibited a faster kinetics than free m1A/m1G (canonical ncRNA modifications) (47) and large RNA modifications, suggesting that m62A is also present in high turnover RNAs, which likely include short-lived ncRNAs and mRNAs. The presence and role of m62A beyond intact 18S/12S rRNAs thus merit further investigation. Of note, single methylation intermediates of m62A have been detected in vitro (77), but we did not detect m + 1 isotopologues for m62A above its natural abundance. As SAM was 98 to 100% 13C-labeled within 30 min, the time resolution of our experiments does not capture sequential methylation of adenosine into m62A. These various findings highlight the value examining the turnover of methylated RNA to uncover the presence of modifications in uncharacterized in RNA subclasses, which warrants investigation.
A nonstationary condition, e.g., a stress condition, may affect the afferent (i.e., methylation of newly transcribed RNA) or efferent (i.e., decay of methylated RNAs) turnover parts of methylated RNAs. Combined with abundance measurements, this allowed us to assess transcriptional versus nontranscriptional sensitivity of modified RNAs to stress conditions. This was evidenced by the reduced methylation turnover of ncRNAs following serine deprivation and ActD treatment, wherein the abundance of modifications was unaltered in the former (transcriptional effect) but m6A levels decreased in the latter (posttranscriptional effect). The observed inhibited turnover of methylated ncRNAs under serine deprivation is in line with the expected inhibition of mTOR activity and thereby of rRNA biogenesis (78). In contrast to ncRNAs, glutamine deprivation increased m6A levels in polyA+-purified RNA without significantly changing (not increasing) its turnover. These data suggest that m6A is posttranscriptionally sensitive to glutamine levels through either (i) an altered decay of m6A-enriched versus m6A-depleted RNAs or (ii) inhibition of m6A demethylation itself. The demethylation of m6A is a stepwise conversion into adenosine through the formation of N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (f6A) intermediates (79), whose presence or turnover was not assessed here. Thus, related developments are needed to address if posttranscriptional dynamics of m6A by metabolic stress conditions are due to RNA demethylation or differential decay of m6A-modified versus m6A-depleted RNAs. As glutamine is the main carbon source of α-ketoglutarate, a cosubstrate of RNA demethylases (52, 53), it is plausible that its depletion could inhibit RNA demethylation. This supports the notion that the reversibility of m6A in mRNA is likely context-dependent (80). With exception of ActD treatment, the stimuli examined did not lead to changes in both the turnover and abundances of modified ribonucleosides. In this respect, while altered methylation turnover (and unaltered abundance) indicates a transcriptional effect, it is plausible that a stimulus may alter methylation turnover at the posttranscriptional level, e.g., if the newly methylated (m + 1 fraction) and preexisting methylated (m + 0 fraction) RNAs differently are exposed differently to RNA degradation, e.g., through nuclear/cytosolic compartmentalization.
Our study demonstrates how quantification of methylation turnover and abundance can be used to examine the presence of RNA modifications in RNA classes, their temporal dynamics, and sensitivity to stress conditions. These insights open new directions to be further explored by MS and orthogonal approaches to obtain information on particular RNAs.
Experimental procedures
Cell culture and metabolic labeling
786O cells were obtained from the Crick Cell Services and cultured at 37 °C with 5% CO2 in high-glucose DMEM medium (Thermo Fisher Scientific, #61965026) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, #21875034). For 13C labeling experiments, 786O cells were grown in high-glucose DMEM medium without glutamine, methionine, and cystine (Thermo Fisher Scientific, #21013024) supplemented with 10% dialyzed FBS (Thermo Fisher Scientific, #26400044) to minimize contributions of unlabeled amino acids typically present in regular FBS, 2 mM glutamine, 0.1 mM cystine, and 0.2 mM [13C-methyl]-methionine (CK Isotopes Limited). Cells were maintained at 50 to 60% confluence (or 30–40% in chase experiments) and washed with PBS before switching to 13C-labeled medium (or regular medium in chase) for the indicated time periods.
RNA purification
At the conclusion of metabolic labeling, the medium was aspirated and extraction of total RNA was performed with the mirVana isolation kit according to manufacturer’s instructions (Thermo Fisher Scientific, #AM1560). Large (>200 nt) RNAs were purified by adding 1/3 volume of 100% ethanol to the aqueous phase recovered from the organic extraction before loading into the filter cartridge of the mirVana Kit. Small (<200 nt) RNAs were purified by collecting the total filtrate, addition of 2/3 volume of 100% ethanol, and loading into the filter cartridge. Polyadenylated RNA was purified from total/large RNA via two rounds of polyA tail hybridization with Oligo-dT magnetic Dynabeads (Thermo Fisher Scientific, #61002).
LC-MS/MS analysis of ribonucleosides
Purified RNA (100–250 ng) was digested into ribonucleosides using one unit of nuclease P1 (Sigma-Aldrich, #N8630-1VL) in 25 μl of buffer 25 mM NaCl, 2.5 mM ZnCl2, and 10 mM NaCH3COO pH 5.3 and incubated for 2 h at 37 °C. Subsequently, NH4HCO3 (100 mM) and 5 units of alkaline phosphatase (CIP) (NEB, #M0525S) were added and the sample incubated for 2 h (or 20 min, with Quick CIP) at 37 °C. Formic acid was added at 0.1% v/v in a final volume of 50 μl, samples were filtered (0.22 μm, Millipore) and 15 to 20 μl analyzed in duplicate by LC-MS. Ribonucleosides were resolved with a C18 reverse-phase column (100 × 2.1 mm, 3 μm particle size, Chromex Scientific, #F18-020503) and eluted with a gradient of 0.1% v/v formic acid (solvent A) and 80% acetonitrile in 0.1% formic acid (solvent B) at a flow rate of 0.2 ml/min and 40 °C: 100% solvent A for 3 min, 12% solvent B for 12 min, and 100% solvent B for 2 min after which the column was re-equilibrated with 100% solvent A for 3 min (20 min total run time). The ribonucleoside separation was performed using U3000 HPLC (Thermo Scientific) and the detection by a TSQ Quantiva Triple Quadrupole mass spectrometer (TSQ Quantiva, Thermo Scientific) controlled by the Xcalibur software version 4.0.27 (Thermo Scientific). The HPLC was coupled to the TSQ Quantiva using a HESI (heated electrospray) ion source (Thermo Scientific) operating in positive ionization mode with the following parameters: capillary voltage, 3500 V; sheath gas flow, 7.35 l/min; gas temperature, 325C. The first and third quadrupoles (Q1 and Q3) were stringently fixed to 0.2 units of resolution and set to detect the mass of the precursor ribonucleoside ion (Q1) and of the base and ribose product ions (Q3). The ribonucleosides were identified by comparison of the retention time and detected mass transitions to commercially available standards. The collision energies were experimentally defined based on the fragmentation pattern of each ribonucleoside standard and chosen based on the maximum intensity of the base product; the ribose ring was used only as a qualifying transition. The retention time, mass transitions (m/z), and collision energies of each ribonucleoside were: adenosine, ∼4.1 min, 268.1 -> 136.1 m/z, 20 V; guanosine, ∼5.4 min, 284.1 -> 135.0 m/z, 35.5 V; cytidine, ∼1.4 min, 244.2 -> 112.05 m/z, 12 V; m6A, ∼8.7 min, 282.1 -> 150.1 m/z, 20 V; m1A, ∼1.8 min, 282.1 -> 150.1 m/z, 20 V, m7G, ∼2.3 min, 298.05 -> 166.1 m/z, 20 V; m5C, ∼1.8 min, 258.2 -> 126.1, m/z, 13 V; m62A, ∼11.9 min, 296.2 -> 164.1 m/z, 22 V. Each mass transition above corresponded to the m + 0 isotopologue and increased by one (m + 1), two (m + 2), and three (m + 3) units for detection of the other isotopologues, e.g., m62A: 297.2 -> 165.1 (m + 1), 298.2 -> 166.1 (m + 2), 299.2 -> 167.1 (m + 3). The dwell time for each transition was 30 ms for a duty cycle of 930 ms (31 transitions), and 8 to 20 data points per chromatographic peak were obtained for “short” and “long” peaks, respectively. A mix of ribonucleoside standards containing 0.5, 1, 5, 10, 50, 100, 500 fmol, 1, 5, 10, 50, or 100 pmol of each ribonucleoside was run in parallel after the biological samples for absolute quantifications, a subset of which is shown in Fig. S2D. Data were recorded using the Xcalibur 3.0.63 software (Thermo Fisher Scientific) and analyzed using Skyline (version 19.1) (81) (Supplementary Materials).
Metabolite extraction and LC-MS analysis of SAM/free ribonucleosides
At the end of cell culture with [13C-methyl]-methionine, metabolic activity quenched by adding ice-cold PBS. Metabolites were extracted by addition of 600 μl ice-cold 1:1 (vol/vol) methanol/water to the cell pellets, samples were transferred to a chilled microcentrifuge tube containing 300 μl chloroform and 600 μl methanol (1500 μl total, in 3:1:1 vol/vol methanol/water/chloroform). Samples were sonicated in a water bath for 8 min at 4 °C and centrifuged (13,000 rpm) for 10 min at 4 °C. The supernatant containing the extract was transferred to a new tube for evaporation in a speed-vacuum centrifuge, resuspended in 3:3:1 (vol/vol/vol) methanol/water/chloroform (350 μl total) to phase separate polar metabolites (upper aqueous phase) from nonpolar metabolites (lower organic phase), and centrifuged. The aqueous phase was transferred to a new tube for evaporation in a speed-vacuum centrifuge and resuspended in 100 μl water for LC-MS acquisition. LC-MS analysis was performed using a Dionex UltiMate LC system (Thermo Fisher Scientific) with a ZIC-pHILIC column (150 mm × 4.6 mm, 5 μm particle, Merck Sequant), as described previously (82). A 15 min elution gradient of 80% Solvent A (20 mM ammonium carbonate in Optima HPLC grade water, Sigma Aldrich) to 20% Solvent B (acetonitrile Optima HPLC grade, Sigma Aldrich) was used, followed by a 5 min wash of 95:5 Solvent A to Solvent B and 5 min re-equilibration. Other parameters were as follows: flow rate, 300 μl/min; column temperature, 25 °C; injection volume, 10 μl; autosampler temperature, 4 °C. All metabolites were detected across a mass range of 70 to 1050 m/z using a Q Exactive Orbitrap instrument (Thermo Fisher Scientific) with heated electrospray ionization and polarity switching mode at a resolution of 70,000 (at 200 m/z). MS parameters were as follows: spray voltage 3.5 kV for positive mode and 3.2 kV for negative mode; probe temperature, 320 °C; sheath gas, 30 arbitrary units; auxiliary gas, five arbitrary units. Parallel reaction monitoring (PRM) was used at a resolution of 17,500 to confirm the identification of metabolites; collision energies were set individually in HCD (high-energy collisional dissociation) mode. Data were recorded using the Xcalibur 3.0.63 software and analyzed using Tracefinder 4.1 (ThermoFisher Scientific) according to the manufacturer’s workflows.
Quantification of methylation turnover
The isotopologue fractions were defined as the total ion counts of the m + 1 isotopologue (except for m + 2 in m62A) relative to the total ion counts of the m + 0 plus m + 1 isotopologues. The kinetics of isotopologue fractions and goodness of fit were determined using the Curve Fitting toolbox of Matlab R2020a (MathWorks) either by a linear regression [f(x) = p1∗x + p2] or an exponential fit: a one-term function in the chase experiments [f(x) = a∗exp(b∗x), Levenberg–Marquardt algorithm], and a two-term function to fit the isotopologue fractions of free ribonucleosides [f(x) = a∗exp(b∗x) + c∗exp(d∗x), Levenberg–Marquardt algorithm].
Data availability
The mass spectrometry.raw data and Skyline files pertaining Figure 1 and Fig. S1 are made available in the Supplementary Materials. All remaining mass spectrometry data can be obtained from the authors upon request.
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This project has received funding from Wellcome Trust (103760/Z/14/Z) and the European Research Council (ERC) under the European Union's Seventh Framework Program (FP7/2007-2013)/ERC grant agreement ID: 617837. The Francis Crick Institute receives its core funding from Cancer Research UK (grant no. FC001002), the UK Medical Research Council (grant no. FC001002), and the Wellcome Trust (grant no. FC001002).
Author contributions
P. A. G. conceptualization; P. A. G. data curation; P. A. G., V. E., and M. S. D. S. formal analysis; P. A. G., J. I. M., and J. U. funding acquisition; P. A. G. investigation; P. A. G. and V. E. methodology; P. A. G., J. I. M., and J. U. resources; J. U. supervision; P. A. G. visualization; P. A. G. and J. U. writing—original draft; P. A. G., J. I. M., and J. U. writing—review and editing.
Funding and additional information
P. A. G. was supported by a Marie Sklodowska-Curie individual fellowship (grant agreement ID: 701730).
Edited by Karin Musier-Forsyth
Supporting information
References
- 1.Capitanchik C., Toolan-Kerr P., Luscombe N.M., Ule J. How do you identify m6 A methylation in transcriptomes at high resolution? A comparison of recent datasets. Front. Genet. 2020;11:398. doi: 10.3389/fgene.2020.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helm M., Motorin Y. Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 2017;18:275–291. doi: 10.1038/nrg.2016.169. [DOI] [PubMed] [Google Scholar]
- 3.Grozhik A.V., Jaffrey S.R. Distinguishing RNA modifications from noise in epitranscriptome maps. Nat. Chem. Biol. 2018;14:215–225. doi: 10.1038/nchembio.2546. [DOI] [PubMed] [Google Scholar]
- 4.Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frye M., Harada B.T., Behm M., He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batista P.J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley D.M., Lujan E., Haddad B., Daneshvar K., Carter A.C., Flynn R.A., Zhou C., Lim K.-S., Dedon P. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H.-B., Tong J., Zhu S., Batista P.J., Duffy E.E., Zhao J., Bailis W., Cao G., Kroehling L., Chen Y., Wang G., Broughton J.P., Chen Y.G., Kluger Y., Simon M.D. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon K.J., Ringeling F.R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., Su Y., Kim N.S., Zhu Y., Zheng L., Kim S., Wang X., Doré L.C., Jin P., Regot S. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017;171:877–889.e17. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su R., Dong L., Li C., Nachtergaele S., Wunderlich M., Qing Y., Deng X., Wang Y., Weng X., Hu C., Yu M., Skibbe J., Dai Q., Zou D., Wu T. R-2HG exhibits anti-tumor activity by targeting FTO/m 6 A/MYC/CEBPA signaling. Cell. 2017;172:90–91.e23. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qing Y., Dong L., Gao L., Li C., Li Y., Han L., Prince E., Tan B., Deng X., Wetzel C., Shen C., Gao M., Chen Z., Li W., Zhang B. R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m6A/PFKP/LDHB axis. Mol. Cell. 2021;81:922–939.e9. doi: 10.1016/j.molcel.2020.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min K.W., Zealy R.W., Davila S., Fomin M., Cummings J.C., Makowsky D., Mcdowell C.H., Thigpen H., Hafner M., Kwon S.H., Georgescu C., Wren J.D., Yoon J.H. Profiling of m6A RNA modifications identified an age-associated regulation of AGO2 mRNA stability. Aging Cell. 2018;17 doi: 10.1111/acel.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z., Shi Y., Lu M., Song M., Yu Z., Wang J., Wang S., Ren J., Yang Y.G., Liu G.H., Zhang W., Ci W., Qu J. METTL3 counteracts premature aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids Res. 2020;48:11083–11096. doi: 10.1093/nar/gkaa816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livneh I., Moshitch-Moshkovitz S., Amariglio N., Rechavi G., Dominissini D. The m6A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020;21:36–51. doi: 10.1038/s41583-019-0244-z. [DOI] [PubMed] [Google Scholar]
- 14.Chen X., Yu C., Guo M., Zheng X., Ali S., Huang H., Zhang L., Wang S., Huang Y., Qie S., Wang J. Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem. Neurosci. 2019;10:2355–2363. doi: 10.1021/acschemneuro.8b00657. [DOI] [PubMed] [Google Scholar]
- 15.Han M., Liu Z., Xu Y., Liu X., Wang D., Li F., Wang Y., Bi J. Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. Front. Neurosci. 2020;14:98. doi: 10.3389/fnins.2020.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaefer M., Pollex T., Hanna K., Tuorto F., Meusburger M., Helm M., Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanco S., Dietmann S., Flores J.V., Hussain S., Kutter C., Humphreys P., Lukk M., Lombard P., Treps L., Popis M., Kellner S., Holter S.M., Garrett L., Wurst W., Becker L. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gkatza N.A., Castro C., Harvey R.F., Heiß M., Popis M.C., Blanco S., Bornelöv S., Sajini A.A., Gleeson J.G., Griffin J.L., West J.A., Kellner S., Willis A.E., Dietmann S., Frye M. Cytosine-5 RNA methylation links protein synthesis to cell metabolism. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu C., Begley T.J., Dedon P.C. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014;588:4287–4296. doi: 10.1016/j.febslet.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lovén J., Orlando D.A., Sigova A.A., Lin C.Y., Rahl P.B., Burge C.B., Levens D.L., Lee T.I., Young R.A. Revisiting global gene expression analysis. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wetzel C., Limbach P.A. Mass spectrometry of modified RNAs: Recent developments. Analyst. 2016;141:16–23. doi: 10.1039/c5an01797a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber C.G., Oberacher H. Analysis of nucleic acids by on-line liquid chromatography-mass spectrometry. Mass Spectrom. Rev. 2001;20:310–343. doi: 10.1002/mas.10011. [DOI] [PubMed] [Google Scholar]
- 23.Taoka M., Nobe Y., Hori M., Takeuchi A., Masaki S., Yamauchi Y., Nakayama H., Takahashi N., Isobe T. A mass spectrometry-based method for comprehensive quantitative determination of post-transcriptional RNA modifications: The complete chemical structure of Schizosaccharomyces pombe ribosomal RNAs. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waghmare S.P., Dickman M.J. Characterization and quantification of RNA post-transcriptional modifications using stable isotope labeling of RNA in conjunction with mass spectrometry analysis. Anal. Chem. 2011;83:4894–4901. doi: 10.1021/ac200547y. [DOI] [PubMed] [Google Scholar]
- 25.Popova A.M., Williamson J.R. Quantitative analysis of rRNA modifications using stable isotope labeling and mass spectrometry. J. Am. Chem. Soc. 2014;136:2058–2069. doi: 10.1021/ja412084b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taoka M., Nobe Y., Yamaki Y., Sato K., Ishikawa H., Izumikawa K., Yamauchi Y., Hirota K., Nakayama H., Takahashi N., Isobe T. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 2018;46:9289–9298. doi: 10.1093/nar/gky811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T., Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42:7346–7357. doi: 10.1093/nar/gku390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wein S., Andrews B., Sachsenberg T., Santos-Rosa H., Kohlbacher O., Kouzarides T., Garcia B.A., Weisser H. A computational platform for high-throughput analysis of RNA sequences and modifications by mass spectrometry. Nat. Commun. 2020;11:926. doi: 10.1038/s41467-020-14665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kullolli M., Knouf E., Arampatzidou M., Tewari M., Pitteri S.J. Intact microRNA analysis using high resolution mass spectrometry. J. Am. Soc. Mass Spectrom. 2014;25:80–87. doi: 10.1007/s13361-013-0759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan C.T.Y., Dyavaiah M., DeMott M.S., Taghizadeh K., Dedon P.C., Begley T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su D., Chan C.T.Y., Gu C., Lim K.S., Chionh Y.H., McBee M.E., Russell B.S., Babu I.R., Begley T.J., Dedon P.C. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 2014;9:828–841. doi: 10.1038/nprot.2014.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kellner S., Ochel A., Th K., Spenkuch F., Neumann J., Sharma S., Entian K., Schneider D., Helm M. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heiss M., Hagelskamp F., Marchand V., Motorin Y., Kellner S. Cell culture NAIL-MS allows insight into human tRNA and rRNA modification dynamics in vivo. Nat. Commun. 2021;12:389. doi: 10.1038/s41467-020-20576-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Endres L., Dedon P.C., Begley T.J. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 2015;12:603–614. doi: 10.1080/15476286.2015.1031947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki T., Suzuki T., Wada T., Saigo K., Watanabe K. Taurine as a constituent of mitochondrial tRNAs: New insights into the functions of taurine and human mitochondrial diseases. EMBO J. 2002;21:6581–6589. doi: 10.1093/emboj/cdf656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao W., He X., Hoadley K.A., Parker J.S., Hayes D.N., Perou C.M. Comparison of RNA-Seq by poly (A) capture, ribosomal RNA depletion, and DNA microarray for expression profiling. BMC Genomics. 2014;15:419. doi: 10.1186/1471-2164-15-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui P., Lin Q., Ding F., Xin C., Gong W., Zhang L., Geng J., Zhang B., Yu X., Yang J., Hu S., Yu J. A comparison between ribo-minus RNA-sequencing and polyA-selected RNA-sequencing. Genomics. 2010;96:259–265. doi: 10.1016/j.ygeno.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 38.Wei C.M., Gershowitz A., Moss B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 39.Chen-Kiang S., Nevins J.R., Darnell J.E. N-6-methyl-adenosine in adenovirus type 2 nuclear RNA is conserved in the formation of messenger RNA. J. Mol. Biol. 1979;135:733–752. doi: 10.1016/0022-2836(79)90174-8. [DOI] [PubMed] [Google Scholar]
- 40.Sommer S., Lavi U., Darnell J.E. The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol. 1978;124:487–499. doi: 10.1016/0022-2836(78)90183-3. [DOI] [PubMed] [Google Scholar]
- 41.Jang C., Chen L., Rabinowitz J.D. Metabolomics and isotope tracing. Cell. 2018;173:822–837. doi: 10.1016/j.cell.2018.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelleher J.K. Flux estimation using isotopic tracers: Common ground for metabolic physiology and metabolic engineering. Metab. Eng. 2001;3:100–110. doi: 10.1006/mben.2001.0185. [DOI] [PubMed] [Google Scholar]
- 43.Buescher J.M., Antoniewicz M.R., Boros L.G., Burgess S.C., Brunengraber H., Clish C.B., DeBerardinis R.J., Feron O., Frezza C., Ghesquiere B., Gottlieb E., Hiller K., Jones R.G., Kamphorst J.J., Kibbey R.G. A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abelson H.T., Johnson L.F., Penman S., Green H. Changes in RNA in relation to growth of fibroblast.2. Lifetime of messenger-RNA, ribosomal-RNA, and transfer-RNA in resting and growing cells. Cell. 1974;1:161–165. [Google Scholar]
- 45.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rana A.K., Ankri S. Reviving the RNA world: An insight into the appearance of RNA methyltransferases. Front. Genet. 2016;7:99. doi: 10.3389/fgene.2016.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boccaletto P., Machnicka M.A., Purta E., Piątkowski P., Bagiński B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A., Helm M., Bujnicki J.M. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keith J.M., Ensinger M.J., Moss B. HeLa cell RNA(2’-O-methyladenosine-N6-)-methyltransferase specific for the capped 5’-end of messenger RNA. J. Biol. Chem. 1978;253:5033–5039. [PubMed] [Google Scholar]
- 49.Safra M., Sas-Chen A., Nir R., Winkler R., Nachshon A., Bar-Yaacov D., Erlacher M., Rossmanith W., Stern-Ginossar N., Schwartz S. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017;551:251–255. doi: 10.1038/nature24456. [DOI] [PubMed] [Google Scholar]
- 50.Li X., Xiong X., Zhang M., Wang K., Chen Y. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell. 2017;68:993–1005.e9. doi: 10.1016/j.molcel.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kinnaird A., Zhao S., Wellen K.E., Michelakis E.D. Metabolic control of epigenetics in cancer. Nat. Rev. Cancer. 2016;16:694–707. doi: 10.1038/nrc.2016.82. [DOI] [PubMed] [Google Scholar]
- 52.Gerken T., Girard C.A., Tung Y.-C.L., Webby C.J., Saudek V., Hewitson K.S., Yeo G.S.H., McDonough M.A., Cunliffe S., McNeill L.A., Galvanovskis J., Rorsman P., Robins P., Prieur X., Coll A.P. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M.M., Li C.J., Vågbø C.B., Shi Y., Wang W.-L.L., Song S.-H.H., Lu Z., Bosmans R.P.G., Dai Q., Hao Y.-J.J., Yang X. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Slobodin B., Han R., Calderone V., Vrielink J.A.F.O., Loayza-puch F., Elkon R., Agami R., Methylation N. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine. Cell. 2017;169:326–337.e12. doi: 10.1016/j.cell.2017.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bertero A., Brown S., Madrigal P., Osnato A., Ortmann D., Yiangou L., Kadiwala J., Hubner N.C., De Los Mozos I.R., Sadée C., Lenaerts A.S., Nakanoh S., Grandy R., Farnell E., Ule J. The SMAD2/3 interactome reveals that TGFβ controls m 6 A mRNA methylation in pluripotency. Nature. 2018;555:256–259. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang H., Weng H., Zhou K., Wu T., Zhao B.S., Sun M., Chen Z., Deng X., Xiao G., Auer F., Klemm L., Wu H., Zuo Z., Qin X., Dong Y. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature. 2019;567:414–419. doi: 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koš M., Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heiss M., Reichle V.F., Kellner S. Observing the fate of tRNA and its modifications by nucleic acid isotope labeling mass spectrometry: NAIL-MS. RNA Biol. 2017;14:1260–1268. doi: 10.1080/15476286.2017.1325063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolfe M.B., Goldstrohm A.C., Freddolino P.L. Global analysis of RNA metabolism using bio-orthogonal labeling coupled with next-generation RNA sequencing. Methods. 2019;155:88–103. doi: 10.1016/j.ymeth.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macallan D.C., Fullerton C.A., Neese R.A., Haddock K., Park S.S., Hellerstein M.K. Measurement of cell proliferation by labeling of DNA with stable isotope-labeled glucose: Studies in vitro, in animals, and in humans. Proc. Natl. Acad. Sci. U. S. A. 1998;95:708–713. doi: 10.1073/pnas.95.2.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Defoiche J., Zhang Y., Lagneaux L., Pettengell R., Hegedus A., Willems L., Macallan D.C. Measurement of ribosomal RNA turnover in vivo by use of deuterium-labeled glucose. Clin. Chem. 2009;55:1824–1833. doi: 10.1373/clinchem.2008.119446. [DOI] [PubMed] [Google Scholar]
- 62.Busch R., Neese R.A., Awada M., Hayes G.M., Hellerstein M.K. Measurement of cell proliferation by heavy water labeling. Nat. Protoc. 2007;2:3045–3057. doi: 10.1038/nprot.2007.420. [DOI] [PubMed] [Google Scholar]
- 63.Maddocks O.D.K., Labuschagne C.F., Adams P.D., Vousden K.H. Serine metabolism supports the methionine cycle and DNA/RNA methylation through de novo ATP synthesis in cancer cells. Mol. Cell. 2016;61:210–221. doi: 10.1016/j.molcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perez J.F., Reeds P.J. A new stable isotope method enables the simultaneous measurement of nucleic acid and protein synthesis in vivo in mice. J. Nutr. 1998;128:1562–1569. doi: 10.1093/jn/128.9.1562. [DOI] [PubMed] [Google Scholar]
- 65.Fairbanks L.D., Bofill M., Ruckemann K., Simmonds H.A. Importance of ribonucleotide availability to proliferating T-lymphocytes from healthy humans: Disproportionate expansion of pyrimidine pools and contrasting effects of de novo synthesis inhibitors. J. Biol. Chem. 1995;270:29682–29689. [PubMed] [Google Scholar]
- 66.Nikolov E.N., Dabeva M.D. Re-utilization of pyrimidine nucleotides during rat liver regeneration. Biochem. J. 1985;228:27–33. doi: 10.1042/bj2280027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 68.Tani H., Mizutani R., Salam K.A., Tano K., Ijiri K., Wakamatsu A., Isogai T., Suzuki Y., Akimitsu N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:1382. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramanathan A., Robb G.B., Chan S.H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Galloway A., Cowling V.H. mRNA cap regulation in mammalian cell function and fate. Biochim. Biophys. Acta Gene Regul. Mech. 2019;1862:270–279. doi: 10.1016/j.bbagrm.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang L.S., Liu C., Ma H., Dai Q., Sun H.L., Luo G., Zhang Z., Zhang L., Hu L., Dong X., He C. Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol. Cell. 2019;74:1304–1316.e8. doi: 10.1016/j.molcel.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Squires J.E., Patel H.R., Nousch M., Sibbritt T., Humphreys D.T., Parker B.J., Suter C.M., Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lafontaine D., Delcour J., Glasser A.L., Desgrès J., Vandenhaute J. The DIM1 gene responsible for the conserved m26Am26A dimethylation in the 3′-terminal loop of 18 S rRNA is essential in yeast. J. Mol. Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- 74.O’Sullivan M., Rutland P., Lucas D., Ashton E., Hendricks S., Rahman S., Bitner-Glindzicz M. Mitochondrial m.1584A 12S m62A rRNA methylation in families with m.1555A>G associated hearing loss. Hum. Mol. Genet. 2015;24:1036–1044. doi: 10.1093/hmg/ddu518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chan C.T.Y., Chionh Y.H., Ho C.H., Lim K.S., Babu I.R., Ang E., Wenwei L., Alonso S., Dedon P.C. Identification of N6,N6-dimethyladenosine in transfer RNA from Mycobacterium bovis bacille calmette-guerin. Molecules. 2011;16:5168–5181. doi: 10.3390/molecules16065168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCloskey J. a, Graham D.E., Zhou S., Crain P.F., Ibba M., Konisky J., Söll D., Olsen G.J. Post-transcriptional modification in archaeal tRNAs: Identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic methanococcales. Nucleic Acids Res. 2001;29:4699–4706. doi: 10.1093/nar/29.22.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ensfelder T.T., Kurz M.Q., Iwan K., Geiger S., Matheisl S., Müller M., Beckmann R., Carell T. ALKBH5-induced demethylation of mono- and dimethylated adenosine. Chem. Commun. (Camb.) 2018;54:8591–8593. doi: 10.1039/c8cc03980a. [DOI] [PubMed] [Google Scholar]
- 78.Goberdhan D.C.I., Wilson C., Harris A.L. Amino acid sensing by mTORC1: Intracellular transporters mark the spot. Cell Metab. 2016;23:580–589. doi: 10.1016/j.cmet.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fu Y., Jia G., Pang X., Wang R.N., Wang X., Li C.J., Smemo S., Dai Q., Bailey K.A., Nobrega M.A., Han K.-L., Cui Q., He C. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 2013;4:1798. doi: 10.1038/ncomms2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mauer J., Jaffrey S.R. FTO, m6Am , and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Lett. 2018;592:2012–2022. doi: 10.1002/1873-3468.13092. [DOI] [PubMed] [Google Scholar]
- 81.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fets L., Driscoll P.C., Grimm F., Jain A., Nunes P.M., Gounis M., Doglioni G., Papageorgiou G., Ragan T.J., Campos S., Silva Dos Santos M., MacRae J.I., O’Reilly N., Wright A.J., Benes C.H. MCT2 mediates concentration-dependent inhibition of glutamine metabolism by MOG. Nat. Chem. Biol. 2018;14:1032–1042. doi: 10.1038/s41589-018-0136-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry.raw data and Skyline files pertaining Figure 1 and Fig. S1 are made available in the Supplementary Materials. All remaining mass spectrometry data can be obtained from the authors upon request.