Abstract
Orbiviruses are arboviruses with 10 double-stranded linear RNA segments, and some have been identified as pathogens of dramatic epizootics in both wild and domestic ruminants. Tibet orbivirus (TIBOV) is a new orbivirus isolated from hematophagous insects in recent decades, and, currently, most of the strains have been isolated from insects in PR China, except for two from Japan. In this study, we isolated a novel reassortment TIBOV strain, YN15-283-01, from Culicoides spp. To identify and understand more characteristics of YN15-283-01, electrophoresis profiles of the viral genome, electron microscopic observations, plaque assays, growth curves in various cell lines, and bioinformatic analysis were conducted. The results indicated that YN15-283-01 replicated efficiently in mosquito cells, rodent cells and several primate cells. Furthermore, the maximum likelihood phylogenetic trees and simplot analysis of the 10 segments indicated that YN15-283-01 is a natural reassortment isolate that had emerged mainly from XZ0906 and SX-2017a.
Keywords: Culicoides, reassortment, Tibet orbivirus, Yunnan
Introduction
Emerging and re-emerging arbovirus diseases remain one of the most serious threats to human and animal health, with potentially devastating social and economic consequences. For example, Zika virus (ZIKV) was unexpectedly associated with severe neurological disease (Guillain–Barré syndrome) in adults in French Polynesia [1], followed by clusters of microcephaly in Brazil [2], leading to the World Health Organization (WHO) declaring a public health emergency of international concern (PHEIC) in 2016. Dengue virus (DENV) is estimated to infect 390 million people annually, of which 96 million manifest clinically (with any severity of disease) [3]. Despite a risk of infection existing in 129 countries [4], 70% of the actual burden of DENV is in Asia. Furthermore, the emergence of the epidemic and epizootic West Nile virus (WNV) in the Americas (1999–2004), Rift Valley fever (RVF) in Africa (2009–2011) and the epidemics of chikungunya virus (CHIKV) (2004–2014) were truly black swan events (i.e., epidemics that are difficult to predict and that have an extreme effect), which had substantial public health and economic effects that took the world by surprise [5]. Since One Health is receiving attention for arbovirus infection prevention and control [6], scientists realized that globalization [7], climate change [8], increasing vector distribution [9] and urbanization [10] are all the reasons for rising incidence and geographical expansion of arboviruses.
Orbiviruses consist of 10 segments of linear double-stranded RNA and are members of the Reoviridae family with 14 other genera. Currently in the Orbivirus genus, there are 22 recognized species and at least 160 different serotypes worldwide [11]. Orbiviruses are distributed globally [12], and the majority are habitually found in tropical or subtropical zones, including Europe, Asia and Africa [13]. For a while, most attention has been given to four representatives of the genus Orbivirus that are known to cause economic burden and significant diseases of farm animals, bluetongue virus (BTV), epizootic haemorrhagic disease virus (EHDV) [14], African horse sickness virus (AHSV) [15] and equine encephalosis virus (EEV) [16].
Orbiviruses are transmitted between their vertebrate hosts, including domestic and wild ruminants [17], by a variety of vectors, such as mosquitoes, midges and ticks [18]. Among these vectors, members of the genus Culicoides [19] carry most orbiviruses and are considered a potent vector of important animal arbovirus diseases, which cause major economic losses in domestic animals [20, 21].
The first identified Tibet orbivirus (TIBOV, XZ0906) was isolated from Anopheles maculatus mosquito specimens collected in 2009 from Tibet, PR China. Molecular genetic analysis revealed that the isolate was a novel species within the genus Orbivirus [22]. Subsequently, several TIBOV strains were found in Culex fatigan mosquitoes and in Culicoides collected from Guangdong and Yunnan provinces (PR China) [23]. Additionally, almost all the TIBOVs have been found in PR China, except for two found in Japan [24]. Furthermore, serological evidence of TIBOV infection in livestock confirmed the long-term prevalence in the southwest border area of Yunnan, PR China, and that they potentially caused livestock disease outbreaks [25]. Due to the segmented structure of the virus, when coinfection occurs, genetic reassortment or exchange of segments between orbivirus species could transpire, which inevitably affects the host range, virulence, immune evasion and evolution of antiviral resistance [26–28].
In recent years, next-generation sequencing (NGS) and viral metagenomics have greatly increased the pace of new virus discovery from arthropod vectors such as mosquitoes, midges, ticks and sandflies, allowing nucleotide sequences, taxonomic assignments and phylogenetic and evolutionary relationships to be obtained without actual live virus isolates and with minimal biological data [29–31].
In this study, a Tibet orbivirus strain YN15-283-01 was isolated from specimens of members of the genus Culicoides collected in Yunnan in 2015, and genome sequencing revealed that the isolate is a natural reassortment virus. Cell lines derived from mosquitoes, humans, monkeys and hamsters are highly susceptible to infection by this isolate. Our findings expand the knowledge of diversity, evolutionary relationships and the characteristics of TIBOV.
Methods
Culicoides collection and sample preparation
A total of 7700 Culicoides were trapped in Lincang, Pu’er, Xishuangbanna and Honghe, Yunnan Province, PR China, between August and September 2015 and then assigned to 77 pools (100 midges per pool) based on their respective collection sites. Each Culicoides pool was triturated by the cryogenic grinding method at liquid nitrogen temperatures using sterile mortars and pestles. After sufficient grinding, 1 ml of Roswell Park Memorial Institute (RPMI) medium was added for homogenization [25], the samples were then clarified by centrifugation at 20000 g (4 °C for 30 min), filtered through a 0.22 µm membrane filter (Millipore) to remove cell debris and stored at −80 °C until further use.
Cell culture, virus isolation and Purification
In this study, invertebrate cell lines [Aedes albopictus RNAi-deficient (C6/36), Aedes aegypti (Aag2)] and vertebrate cell lines [African green monkey kidney (Vero E6), baby hamster kidney (BHK-21), Madin-Darby bovine kidney (MDBK), human hepatoma (Huh7) and human adrenal gland (SW13)] were used.
C6/36 cells were maintained at 28 °C (in the absence of CO2) in RPMI medium (Gibco) supplemented with 10% foetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin. Aag2 cells were passaged in Schneider’s Drosophila Medium (SDM; Gibco) containing 5% FBS, cultured at 28 °C in an incubator with 5% CO2. Vero E6, BHK-21, SW13 and MDBK cells were grown at 37 °C under a 5% CO2 atmosphere in Dulbecco’s minimal essential medium (DMEM; Gibco) (4.5 g l−1 d-glucose) supplemented with 10% FBS and 1% penicillin/streptomycin.
For virus isolation, the microfiltered supernatants (200 µl) from each pool of homogenized midges were inoculated into each well of 24-well plates with C6/36 cells as passage one. After 1 h of incubation at 28 °C, the inoculum was removed and replaced with RPMI (with 2% FBS) medium, and the cell plates were put into the incubator under 28 °C with 5 % CO2 for 5–7 days, followed by two passages in C6/36 cells. Then the supernatants (200 µl) from the third passage in C6/36 were seeded on BHK-21 cells plates (24-well plates) and incubated for 5–7 days to monitor the cytopathic effects (CPEs) associated with infection.
Virus purification was performed as described previously [32, 33]. Briefly, the supernatants from viral culture were harvested and clarified to remove cellular debris by centrifugation at 5000 g at 4°C for 30 min. Thereafter, ultracentrifugation was conducted by adding 2 ml of the harvested virus supernatant at the bottom of the ultracentrifuge tube (Type 70), followed by the careful addition of 4 ml of 20% (w/v) sucrose in phosphate-buffered saline (PBS) into the tube to remove impurities. Ultracentrifugation was performed at 40000 g at 4°C for 3.5 h in a Type 70 Ti rotor (Beckman Coulter Diagnostics). Upon completion, the supernatant was discarded. Then, 100 µl of PBS was added to the sediment at the bottom of the tube and the precipitate was resuspended. The purified virus was stored at −80°C.
Electron microscopy
Electron microscopy imaging was performed using the negative contrast method. A 20 µl sample of purified virus and an equal volume of PBS were mixed and then placed on a Formvar carbon-coated copper grid for 3 min. After staining with 2% phosphotungstic acid solution (pH 6.8) for 3 min, the excess liquid was absorbed and discarded. Finally, clear and well-contrasted electron microscope images were taken using a U8010 electron microscope (Hitachi).
dsRNA-polyacrylamide gel electrophoresis
Viral RNA was extracted from the culture supernatant of CPE-positive BHK-21 cells using a Direct-zol RNA MiniPrep kit (Zymo Research) according to the manufacturer’s requirements. The dsRNA was unpacked in a 65 °C -water bath for 10 min and then mixed with 5 µl of 6× loading buffer. The dsRNA was then separated on a standard discontinuous 10 % acrylamide slab gel, and electrophoresis was performed in an ice bath at 80 V for 30 min, followed by 100 V for 7 h. The virus dsRNA was visualized with silver nitrate staining.
Plaque assay
The virus was diluted 10-fold with DMEM to 10−9, and 100 µl virus diluent was added to the BHK-21, Vero E6 and SW13 cell monolayers in 24-well plates at 37 °C. After 1 h, the virus diluent was discarded, and 500 µl DMEM containing 1.5% methyl cellulose was added to the cells and the cells were cultured at 37 °C. Four days later, the cells were fixed overnight with 3.7% formaldehyde and stained with 2% crystal violet. The number of plaques was calculated, and the sizes of the plaques were measured.
RNA extraction and RT-qPCR
Viral RNA was extracted from the cell culture supernatant using a Direct-zol RNA MiniPrep kit (Zymo Research) according to the manufacturer’s instructions. RT-qPCR for the detection of viral RNA was performed using a Luna Universal Probe One-Step RT-qPCR Kit (New England Biolabs) according to the manufacturer’s recommendations with a thermocycler (BIO-RAD CFX96 Real-Time System).
The primers for RT-qPCR targeted the VP1 (Segment 1) of TIBOV, including TBV-VP1-F (5′–ATCACAATGGTCGTAATAAC −3′), TBV-VP1-R (5′–TCATCATTAACTGCTAATCTTG−3′) and TBV-VP1-Probe (5′FAM–CAGATCTAATAAGACGAACAAT-BHQ13′) [34]. Both oligoprimer DNAs were synthesized by TSINGKE (Wuhan Branch, PR China). Reaction mixtures (20 µl) containing 2 µl of viral RNA and 0.8 µl of each primer were incubated at 55 °C for 10 min and 95 °C for 1 min followed by 40 cycles of 95 °C for 10 s and 55 °C for 30 s.
Standard curve generation was initiated by a 10-fold serial dilution (from 2.7×107 p.f.u. ml−1 to 2.7×101 p.f.u. ml−1) from viral stock, and then RNA was extracted from 200 µl of each dilution and used as the template for RT-qPCR. Each assay was carried out in triplicate. The viral titres (p.f.u. ml−1) were automatically plotted against cycle threshold (Ct) values based on linear regression.
Growth characteristics of the viral isolate in the cell cultures
BHK-21, Vero E6, SW13, C6/36 and Aag2 cells grown in T25 flask plates were infected with the virus at MOI=1, 0.01 and 0.0001. Huh7 and MDBK cells grown in T25 flask plates were infected with the virus at MOI=10, 5, 1, 0.01 and 0.0001. After inoculation, 200 µl cell supernatant was collected every day (from day 0 to day 7) and then replenished with 200 µl fresh medium. Viral RNA was extracted and detected by RT-qPCR, and the virus concentration (p.f.u. ml−1) was calculated based on the standard curve according to the generated Ct values. The experiment was repeated three times.
Genome sequencing and PCR confirmation
Viral RNA was extracted from the passaged virus and sent to Nextomics Biosciences (Wuhan, PR China) for sequencing through the Illumina MiSeq System. The de novo assembly of viral sequences was performed using MEGAHIT (https://github.com/voutcn/megahit) and then further verified with PCR using multiple primers.
Bioinformatics analyses
The complete coding sequences (CDSs) of all ten segments of all seven isolates of TIBOV were used in the analysis (GenBank as of the first of June 2021). Background information of all the utilized strains is listed in Table 1. The nucleotide sequence identities of the CDSs of TIBOV strain YN15-283-01 and others were analysed using BioEdit v7.2.6. Alignment was conducted by the ClustalW function in MEGAv7.0.212. The GTR+I+G substitution model was selected as the best-fit nucleotide substitution model by jModelTest 23. Phylogenetic analyses of nucleotide sequences were performed by the maximum likelihood method using the general time-reversible model with 1000 bootstrap replicates in mega.
Table 1.
Background information on viruses used in bioinformatic analysis
|
Viruses |
Strains |
|||||||
|---|---|---|---|---|---|---|---|---|
|
YN15-283-01 |
XZ0906 |
SX-2017a |
DH13C120 |
D181/2008 |
KSB-8/C/09 |
KSB-8/C/10 |
||
|
Location |
Yunnan, PR China |
Tibet, PR China |
Yunnan, PR China |
Yunnan, PR China |
Guangdong, PR China |
Kagoshima, Japan |
Kagoshima, Japan |
|
|
Isolated source |
Culicoides midges |
Anopheles maculatus |
Culex tritaeniorhynchus |
Culicoides midges |
Culex pipiens fatigans |
Culicoides midges |
Culicoides midges |
|
|
Collection Date |
2015 |
2009 |
2007 |
2013 |
2008 |
2009 |
2010 |
|
|
Accession number |
Segment 1 |
|||||||
|
Segment 2 |
||||||||
|
Segment 3 |
||||||||
|
Segment 4 |
||||||||
|
Segment 5 |
||||||||
|
Segment 6 |
||||||||
|
Segment 7 |
||||||||
|
Segment 8 |
||||||||
|
Segment 9 |
||||||||
|
Segment 10 |
||||||||
Potential reassortment or recombination events in the complete genome of strain YN15-283-01 were identified using the Recombination Detection Programme (RDP 4 Version 4.33) through different algorithms (RDP, GENECONV, Bootscan, MaxChi, Chimaera, SiScan and 3Seq) [35]. The potential reassortment or recombination events were further verified by similarity plots (SimPlots) analysis in SimPlot version 3.5.1 [36].
Statistical analyses
All statistical data were analysed with R (version 4.0.5). One-way ANOVA was used to determine significant differences among groups of highest viral titres in different cell lines under the same MOI (MOI=1, 0.01 or 0.0001) and different MOIs of the same cell line (confidence interval 95%).
Results
Virus isolation and morphology
The supernatants from each pool of homogenized Culicoides were inoculated into C6/36 cells for three times and then into BHK-21 cells. After 120 h of infection, strong CPEs in BHK-21 cells were observed, with cells becoming round and dead under the microscope. The virus isolate was obtained from one of 77 pools and named as YN15-283-01.
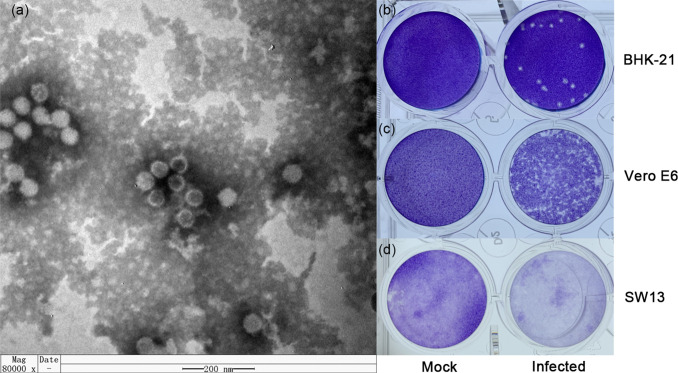
Viral purification was performed by sucrose density gradient centrifugation. Electron microscopic observation revealed that the virus particle showed a spherical morphology without an envelope and a diameter of 40–60 nm (Fig. 1a).
Fig. 1.
The viral morphology and viral plaques in cells of YN15-283-01. (a) Negative-stained ultracentrifuged virions. Viral plaques in (b) BHK-21 (10−5 dilution at four dpi), (c) Vero E6 (10−3 dilution at five dpi) and (d) SW13 (10−3 dilution at five dpi) cell monolayers. The wells were 16 mm in diameter.
In addition, in the plaque morphology test, YN15-283-01caused distinct, encircled plaques in BHK-21 cells (10−5 dilution) at day four after inoculation (Fig. 1b). However, no sharp plaques were produced (Fig. 1c) in Vero E6 cells (10−3 dilution) at day five after inoculation, and only detachment of the cell monolayer was observed in SW13 cells (10−3 dilution) on day five post-infection.
Identification of the dsRNA genome structure
Viral RNA was harvested from the culture supernatant of infected BHK-21 cells and analysed by PAGE, revealing a ten-segment double-stranded RNA genome with a migration pattern of 3-3-3-1. Within this pattern, segment 10 was very weak but still identifiable (Fig. 2).
Fig. 2.

Electrophoresis profile of the dsRNAs of YN15-283-01 using a 10% acrylamide slab gel.
Growth characteristics of the viral isolate in the cell cultures
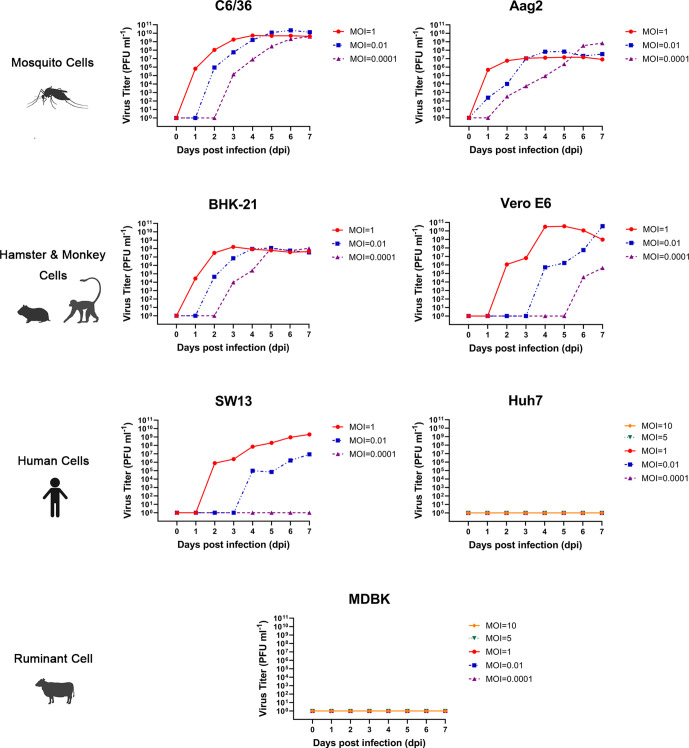
To understand viral growth characteristics in a variety of host cells, the virus was inoculated into five mammalian cell lines, BHK-21, Vero E6, MDBK, SW13 and Huh 7, and two mosquito cell lines, C6/36 and Aag2, with different MOIs. This virus effectively replicated and caused a typical CPE characterized by cell exfoliation, shrinkage and death in BHK-21, Vero E6 and C6/36, however much weaker CPE was observed in SW13 cells. No CPE was observed in Aag2, Huh7 and MDBK cells (Figures S1–S6, available in the online version of this article).
Viral growth kinetics analysis in the supernatant of the infected BHK-21 cells indicated that YN15-283-01 replicated efficiently and could reach peak titres of 108 p.f.u. ml−1 and 109 p.f.u. ml−1 when using MOIs of 1 and 0.01 or 0.0001, respectively, and then remained steady. Slight differences in Aag2 cells indicated that the virus titre peaked at 107 p.f.u. ml−1 when usingMOIs of 1 or 0.01 but reached 109 p.f.u. ml−1, with an MOI of 0.0001. The virus titres at an MOI of 1 reached the highest level on day four in Vero E6 cells and showed a downward trend, while the virus continued to multiply at MOIs of 0.01 and 0.0001. Viral replication was detected at an MOI of 1 or 0.01, but no viral growth was detected when using an MOI of 0.0001 in SW13 cells. In Huh7 and MDBK cell lines, the virus neither grew nor proliferated at either low or high MOIs from 0.0001 to 10. The results of statistical analysis indicated that there were no significant differences among the groups with the highest viral titres in different cell lines at the same MOI (MOI=1, 0.01 or 0.0001) (P>0.05). In addition, the differences were statistically significant in BHK-21 cells among different MOI virus treatments of the same cells (P<0.05), but there was no difference in other cell lines (P>0.05) (Fig. 3).
Fig. 3.
Growth curves of TIBOV YN15-283-01 with different MOIs in cells derived from mosquitoes and mammals. For C6/36, Aag2, BHK-21, Vero E6 and SW13 cells, the infected MOI=1, 0.01 and 0.0001; for Huh7 and MDBK cells, the infected MOI=10, 5, 1, 0.01 and 0.0001.
Viral genome, phylogenetic and reassortment analysis of YN15-283-01
De novo assembly of the next-generation sequenced data acquired 99.8% coverage of the complete TIBOV genome. The lengths for segments 1–10 were 3950, 2901, 2769, 1978, 1775, 1640, 1165, 1142, 1103 and 833 bp, respectively. The complete CDSs of S1 to S10 encoded 1304 (VP1), 950 (VP2), 899 (VP3), 643 (VP4), 554 (NS1), 526 (VP5), 349 (VP7), 359 (NS2), 347 (VP6) and 234 (NS3) amino acids, respectively. The full genome sequences (S1 to S10) have been deposited in GenBank (accession numbers: MT793636 to MT79345) (Table 1).
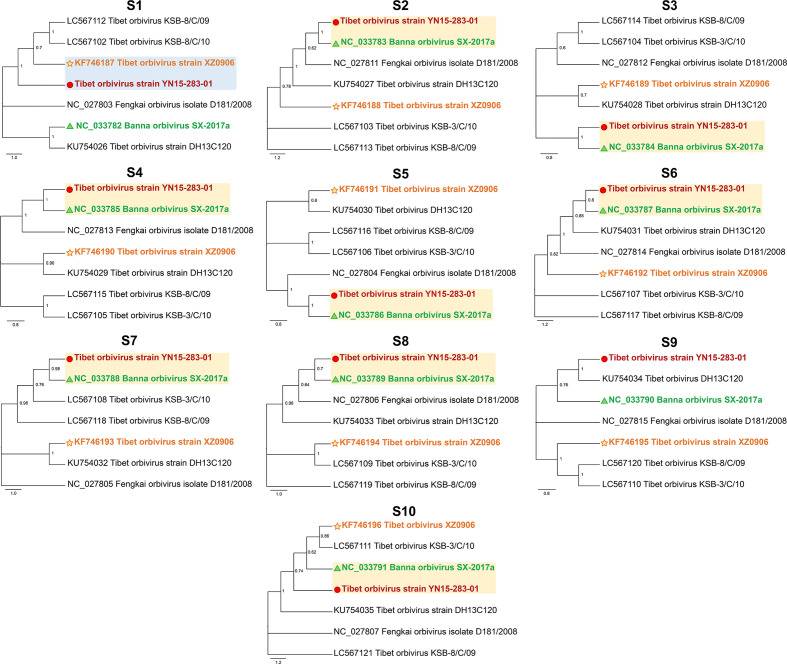
Nucleotide sequence identity analysis of all 10 segments was conducted among YN15-283-01 and the other six TIBOV isolates with full genome information (Table 2). The results indicated that the genomic sequences of eight segments (S2 to S9) in YN15-283-01 share the highest identity (from 94.2–99.6 %) with the corresponding segments of SX-2017a isolated from Culex tritaeniorhynchus from Yunnan, whereas S1 and S10 of YN15-283-01 show the most similarity with XZ0906 acquired from Anopheles maculatus from Tibet, indicating that YN15-283-01 may have undergone a reassorting event between SX-2017a and XZ0906.
Table 2.
Nucleotide sequence identity matrix for Tibet orbiviruses
|
Segments |
S1 |
S2 |
S3 |
S4 |
S5 |
S6 |
S7 |
S8 |
S9 |
S10 |
|---|---|---|---|---|---|---|---|---|---|---|
|
Genes |
VP1 |
VP2 |
VP3 |
VP4 |
NS1 |
VP5 |
VP7 |
NS2 |
VP6 |
NS3 |
|
TIBOV isolates |
YN15-283-01 |
|||||||||
|
XZ0906 |
0.974* |
0.544 |
0.802 |
0.956 |
0.929 |
0.702 |
0.951 |
0.883 |
0.970 |
0.984* |
|
SX-2017a |
0.910 |
0.993* |
0.942* |
0.994* |
0.996* |
0.981* |
0.996* |
0.977* |
0.984* |
0.979 |
|
DH13C120 |
0.911 |
0.967 |
0.802 |
0.959 |
0.930 |
0.959 |
0.955 |
0.971 |
0.980 |
0.979 |
|
D181/2008 |
0.934 |
0.970 |
0.797 |
0.973 |
0.962 |
0.965 |
0.928 |
0.973 |
0.974 |
0.849 |
|
KSB-8/C/09 |
0.970 |
0.436 |
0.801 |
0.934 |
0.932 |
0.645 |
0.969 |
0.777 |
0.964 |
0.721 |
|
KSB-3/C/10 |
0.973 |
0.552 |
0.798 |
0.939 |
0.929 |
0.695 |
0.984 |
0.882 |
0.966 |
0.980 |
*Bold type indicates the highest sequence identity value.
To further confirm the reassortment events, the phylogenetic tree was reconstructed based on each segment of all seven TIBOV strains. As shown in Fig. 4, the overall topology of the obtained phylogenetic trees with the S2 to S8 sequences were similar, as YN15-283-01 had the closest relationship with SX-2017a, while S1 of YN15-283-01 was clustered closely along with XZ0906.
Fig. 4.
Maximum likelihood phylogenetic trees of S1–S10 segments for the TIBOVs and related orbiviruses. The scale bar indicates the evolutionary distance in the number of substitutions per nucleotide substitution/site, and the principal bootstrap support levels are indicated.
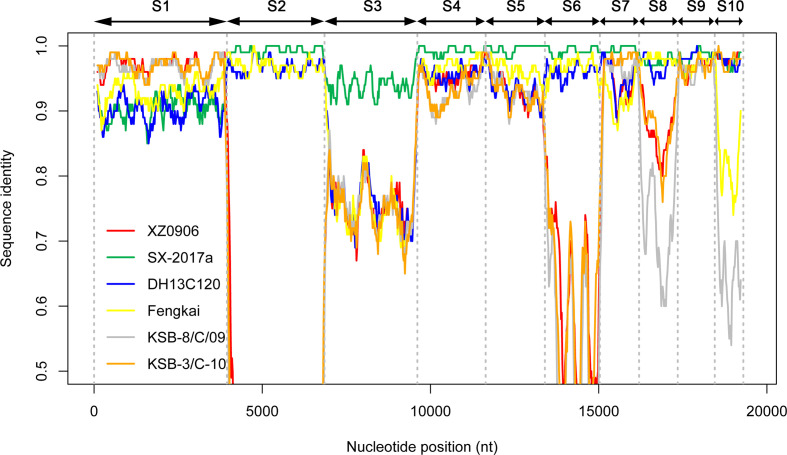
In addition, the Simplot (Fig. 5) and RDP4 analysis was used to detect the reassortment events in the genome of YN15-283-01, which were predicted using different algorithms (GENECONV, RDP, Bootscan, MaxChi, Chimaera, SiSscan and 3Seq) with a significance level set at P≤0.01. S1 of YN15-283-01 was potentially derived from XZ0906, S2, S4-S10 were derived from SX-2017a, whereas S3 was from an unknown source.
Fig. 5.
Simplot among seven TIBOV isolates. YN15-283-01 was used as the query, and the red, green, blue, yellow, grey and orange lines represent the strains XZ0906, SX-2017a, DH13C120, Fengkai, KSB-8/C/09 and KSB-3/C/10 respectively. The Y axis indicates the degree of sequence similarity, and the X axis indicates the position of genomic sequences of S1 to S10.
In summary, the results from similarity identity, phylogenetic and the Simplot/RDP4 reassortment analysis herein indicate that YN15-283-01 is a potential reassortment isolate that emerged from segment exchange mainly from XZ0906 and SX-2017a.
Discussion
In this study, we isolated a Tibet orbivirus YN15-283-01 from Culicoides collected from Yunnan Province (PR China) in 2015. The genomic dsRNA electrophoresis profile indicated that the ten segments of YN15-283-01 were similar to those of other TIBOVs with 3-3-3-1 migration pattern. TYN15-283-01 can cause CPE in C6/36 cells, which is similar to the reported effects of strain YN12246 and Fengkai virus, but in contrast to strain XZ0906 which did not show CPE [34].
The genetic reassortment of viruses with 10 segments in the orbivirus plays an important role in driving the diversity and evolution of this virus group [13]. Herein, we performed reassortment analyses among all reported TIBOVs with full genome information, based on the results of phylogenetic, similarity identity and Simplot/RDP4 reassortment analysis. The results supported the hypothesis that the major parent of YN15-283-01 was SX-2017a, and the minor parent was XZ0906. The S1 of YN15-283-01 had the closest relationship with XZ0906, while S2, and S4–S8 were derived from SX-2017a. However, for the S3, even though in the phylogenetic tree YN15-283-01 was clustered with SX-2017a, the nucleotide identity between them was only 94.2%, and the RDP4 analysis indicated that the parent strains for S3 is unknown. This could be because of the limitations of the database for TIBOVs currently available, thus increased release of genomes in the future will help to improve the reassortment analyses.
Vector is the most important factor affecting arbovirus transmission and outbreaks. The midge feeding model has been developed to study infection, replication and dissemination of BTV in vectors [37]. Results from another study have indicated that Culicoides sonorensis are susceptible to EHDV [38, 39]. Although our results showed that YN15-283-01 can replicate efficiency in C6/36 and Aag2 cells, the experimental infection ofvectors, such as mosquito or midges, should be conducted in the future to determine the vector competence to the TIBOV.
It has been reported that intraperitoneal injection of Yunnan orbivirus into naïve mice led to productive, non-lethal viral replication and viremia [40], and the results of animal challenge experiments with strain DH13C120 of TIBOV indicated that it can lead to lethal neurovirulence in suckling mice and fever illness in sheep. In addition, the positive prevalence of DH13C120 virus and YN12246 strain antibodies has been detected in cattle and pigs in Yunnan Province [25]. These studies implied that TIBOV could be a potential pathogen leading to livestock diseases. More animal infection models should be developed to help understand the pathogenicity of TIBOV.
Presently, the general concept of One Health is widely accepted, but we do not know when and which disease will become the next outbreak, and multisectoral cooperation in the surveillance and control of emerging infectious diseases is quite challenging to achieve due to the significant gap between the fields of animal and human health [29]. However, systematic surveillance of vector-borne viruses is still essential and necessary to screen for vector-borne diseases that are underestimated and neglected [41]. Thus, it is important to study the host range, transmission and pathogenicity of TIBOV, then further assess the risks it may pose to public, veterinary or environmental health.
Supplementary Data
Funding information
This work was supported by the Open Foundation of Key Laboratory of Tropical Translational Medicine of Ministry of Education, Hainan Medical University (2021TTM010) and the Wuhan Science and Technology Plan Project (2018201261638501).
Acknowledgements
We would like to thank Pei Zhang and An-Na Du from The Core Facility and Technical Support, Wuhan Institute of Virology, for their help with producing transmission electron micrographs.
Author contributions
Conceptualization: Z.Y., H.X., N.R., X.W., M.L. Methodology: H.X., N.R., X.W., M.L. Investigation: N.R., X.W., M.L. Software: H.X., X.W. Formal analysis: H.X., X.W. Resources: L.Z., D.H. Writing – original draft preparation: N.R., X.W., M.L. Writing – Review and Editing H.X., S.T., C.O., Q.X., N.R. Visualization: N.R. Supervision: H.X., Z.Y. Funding: Z.Y., H.X., Q.X., S.T. All authors agreed to the submission of this work to Journal of General Virology.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AHSV, African horse sickness virus; BTV, bluetongue virus; CDS, complete coding sequences; CPEs, cytopathic effects; DENV, dengue virus; dsRNA, double strands RNA; EHDV, epizootic hemorrhagic disease virus; MOI, multiplicity of infection; NGS, next-generation sequencing; RT-qPCR, quantitative reverse transcription PCR; RVF, Rift Valley fever; TIBOV, Tibet orbivirus; WNV, West Nile virus; ZIKV, Zika virus.
Six supplementary figures are available with the online version of this article.
References
- 1.Cao-Lormeau V-M, Blake A, Mons S, Lastère S, Roche C, et al. Guillain–Barré syndrome outbreak associated with Zika virus infection in French Polynesia: A case–control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wanderson K, Oliveira de, Eduardo H, Carmo CMH, Coelho G, et al. Zika virus infection and associated neurologic disorders in Brazil. N Engl J Med. 2017;376:1591–1593. doi: 10.1056/NEJMc1608612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, et al. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl Trop Dis. 2012;6:e1760. doi: 10.1371/journal.pntd.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musso D, Rodriguez-Morales AJ, Levi JE, Cao-Lormeau V-M, Gubler DJ. Unexpected outbreaks of arbovirus infections: Lessons learned from the Pacific and tropical America. Lancet Infect Dis. 2018;18:e355–e361. doi: 10.1016/S1473-3099(18)30269-X. [DOI] [PubMed] [Google Scholar]
- 6.Dente MG, Riccardo F, Van Bortel W, Marrama L, Mollet T, et al. Enhancing preparedness for arbovirus infections with a one health approach: the development and implementation of multisectoral risk assessment exercises. Biomed Res Int. 2020;2020:4832360. doi: 10.1155/2020/4832360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMichael AJ. Globalization, climate change, and human health. N Engl J Med. 2013;369:96. doi: 10.1056/NEJMc1305749. [DOI] [PubMed] [Google Scholar]
- 8.Zinsstag J, Crump L, Schelling E, Hattendorf J, Maidane YO, et al. Climate change and one health. FEMS Microbiol Lett. 2018;365 doi: 10.1093/femsle/fny085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelms BM, Macedo PA, Kothera L, Savage HM, Reisen WK. Overwintering biology of Culex (Diptera: Culicidae) mosquitoes in the Sacramento valley of California. jnl med entom. 2013;50:773–790. doi: 10.1603/ME12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowe R, Lee S, Martins Lana R, Torres Codeço C, Castro MC, et al. Emerging arboviruses in the urbanized Amazon Rainforest. BMJ. 2020:m4385. doi: 10.1136/bmj.m4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Attoui H, Mohd Jaafar F. Zoonotic and emerging orbivirus infections. Rev Sci Tech OIE. 2015;34:353–361. doi: 10.20506/rst.34.2.2362. [DOI] [PubMed] [Google Scholar]
- 12.Li C, Guo X, Deng Y, Xing D, Sun A, et al. Vector competence and transovarial transmission of two Aedes aegypti strains to Zika virus. Emerg Microbes Infect. 2019;6:1–7. doi: 10.1038/emi.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing S, Guo X, Zhang X, Zhao Q, Li L, et al. A novel mosquito-borne reassortant orbivirus isolated from Xishuangbanna, China. Virol Sin. 2016;32:159–162. doi: 10.1007/s12250-016-3886-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajko-Nenow P, Brown-Joseph T, Tennakoon C, Flannery J, Oura CAL, et al. Detection of a novel reassortant epizootic hemorrhagic disease virus serotype 6 in cattle in Trinidad, West Indies, containing nine RNA segments derived from exotic EHDV strains with an Australian origin. Infect Genet Evol. 2019;74:103931. doi: 10.1016/j.meegid.2019.103931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Gennip RGP, Drolet BS, Rozo Lopez P, Roost AJC, Boonstra J, et al. Vector competence is strongly affected by a small deletion or point mutations in bluetongue virus. Parasit Vectors. 2019;12:470. doi: 10.1186/s13071-019-3722-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhama K, Pawaiya RVS, Karthik K, Chakraborty S, Tiwari R. Equine encephalosis virus (EEV): A Review. Asian J Anim Vet Adv. 2014;9:123–133. [Google Scholar]
- 17.Drolet BS, van Rijn P, Howerth EW, Beer M, Mertens PP. A review of knowledge gaps and tools for Orbivirus research. Vector-Borne and Zoonotic Diseases. 2015;15:339–347. doi: 10.1089/vbz.2014.1701. [DOI] [PubMed] [Google Scholar]
- 18.Carpenter S, Wilson A, Mellor PS. Culicoides and the emergence of bluetongue virus in northern Europe. Trends in Microbiology. 2009;17:172–178. doi: 10.1016/j.tim.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Mellor PS, Boorman J, Baylis M. Culicoides biting midges their role as arbovirus vectors. Annu Rev Entomol. 2000;45:307–340. doi: 10.1146/annurev.ento.45.1.307. [DOI] [PubMed] [Google Scholar]
- 20.MacLachlan NJ, Guthrie AJ. Re-emergence of bluetongue, african horse sickness, and other orbivirus diseases. Vet Res. 2010;41:35. doi: 10.1051/vetres/2010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maclachlan NJ, Mayo CE. Potential strategies for control of bluetongue, a globally emerging, Culicoides-transmitted viral disease of ruminant livestock and wildlife. Antiviral Research. 2013;99:79–90. doi: 10.1016/j.antiviral.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Zheng Y, Zhao G, Fu S, Wang D, et al. Tibet Orbivirus, a novel Orbivirus species isolated from Anopheles maculatus mosquitoes in Tibet, China. PLoS ONE. 2014;9:e88738. doi: 10.1371/journal.pone.0088738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei W, Guo X, Fu S, Feng Y, Nie K, et al. Isolation of Tibet orbivirus, TIBOV, from Culicoides Collected in Yunnan, China. PLoS One. 2015;10:e0136257. doi: 10.1371/journal.pone.0136257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suda Y, Murota K, Shirafuji H, Yanase T. Genomic analysis of putative novel serotypes of Tibet orbivirus isolated in Japan. Arch Virol. 2021;166:1151–1156. doi: 10.1007/s00705-021-04966-7. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Li H, He Y, Zhou Y, Xin A, et al. Isolation of Tibet Orbivirus from Culicoides and associated infections in livestock in Yunnan, China. Virol J. 2017;14 doi: 10.1186/s12985-017-0774-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nora T, Charpentier C, Tenaillon O, Hoede C, Clavel F, et al. Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J Virol. 2007;81:7620–7628. doi: 10.1128/JVI.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon-Loriere E, Holmes EC. Why do RNA viruses recombine? Nat Rev Microbiol. 2011;9:617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald SM, Nelson MI, Turner PE, Patton JT. Reassortment in segmented RNA viruses: Mechanisms and outcomes. Nat Rev Microbiol. 2016;14:448–460. doi: 10.1038/nrmicro.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryu S, Kim BI, Lim JS, Tan CS, Chun BC. One health perspectives on emerging public health threats. J Prev Med Public Heal. 2017;50:411–414. doi: 10.3961/jpmph.17.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasilakis N, Tesh RB, Popov VL, Widen SG, Wood TG, et al. Exploiting the legacy of the arbovirus hunters. Viruses. 2019;11 doi: 10.3390/v11050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alonso C, Utrilla-Trigo S, Calvo-Pinilla E, Jiménez-Cabello L, Ortego J, et al. Inhibition of orbivirus replication by aurintricarboxylic acid. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21197294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coelho SVA, Neris RLS, Papa MP, Schnellrath LC, Meuren LM, et al. Development of standard methods for Zika virus propagation, titration, and purification. J Virol Methods. 2017;246:65–74. doi: 10.1016/j.jviromet.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Xia H, Liu H, Zhao L, Atoni E, Wang Y, et al. First isolation and characterization of a group C Banna virus (BAV) from Anopheles sinensis mosquitoes in Hubei, China. Viruses. 2018;10:8–10. doi: 10.3390/v10100555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu TQ, Zhang H, Huang P, Zhou H, Zhang X, et al. Genomic and biological features of a novel orbivirus isolated from mosquitoes, in China. Virus Res. 2020;285:197990. doi: 10.1016/j.virusres.2020.197990. [DOI] [PubMed] [Google Scholar]
- 35.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype c-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/JVI.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Rijn PA. Reference Module in Life Sciences. 2019. African Horse Sickness Virus. [Google Scholar]
- 38.Ruder MG, Stallknecht DE, Allison AB, Mead DG, Carter DL, et al. Host and potential vector susceptibility to an emerging orbivirus in the United States: Epizootic hemorrhagic disease virus serotype 6. Vet Pathol. 2016;53:574–584. doi: 10.1177/0300985815610387. [DOI] [PubMed] [Google Scholar]
- 39.Federici V, Ippoliti C, Goffredo M, Catalani M, Di Provvido A, et al. Epizootic haemorrhagic disease in Italy: vector competence of indigenous Culicoides species and spatial multicriteria evaluation of vulnerability. Vet Ital. 2016;52:271–279. doi: 10.12834/VetIt.894.4516.2. [DOI] [PubMed] [Google Scholar]
- 40.Attoui H, Jaafar FM, Belhouchet M, Aldrovandi N, Tao S, et al. Yunnan orbivirus, a new orbivirus species isolated from Culex tritaeniorhynchus mosquitoes in China. J Gen Virol. 2005;86:3409–3417. doi: 10.1099/vir.0.81258-0. [DOI] [PubMed] [Google Scholar]
- 41.Torii S, Orba Y, Hang’ombe BM, Mweene AS, Wada Y, et al. Discovery of Mwinilunga alphavirus: A novel alphavirus in Culex mosquitoes in Zambia. Virus Res. 2018;250:31–36. doi: 10.1016/j.virusres.2018.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.