Abstract
Background
Chromobacterium violaceum is an environmental opportunistic pathogen that causes rare but deadly infections in humans. The transcriptional regulators that C. violaceum uses to sense and respond to environmental cues remain largely unknown.
Results
Here, we described a novel transcriptional regulator in C. violaceum belonging to the MarR family that we named OsbR (oxidative stress response and biofilm formation regulator). Transcriptome profiling by DNA microarray using strains with deletion or overexpression of osbR showed that OsbR exerts a global regulatory role in C. violaceum, regulating genes involved in oxidative stress response, nitrate reduction, biofilm formation, and several metabolic pathways. EMSA assays showed that OsbR binds to the promoter regions of several OsbR-regulated genes, and the in vitro DNA binding activity was inhibited by oxidants. We demonstrated that the overexpression of osbR caused activation of ohrA even in the presence of the repressor OhrR, which resulted in improved growth under organic hydroperoxide treatment, as seem by growth curve assays. We showed that the proper regulation of the nar genes by OsbR ensures optimal growth of C. violaceum under anaerobic conditions by tuning the reduction of nitrate to nitrite. Finally, the osbR overexpressing strain showed a reduction in biofilm formation, and this phenotype correlated with the OsbR-mediated repression of two gene clusters encoding putative adhesins.
Conclusions
Together, our data indicated that OsbR is a MarR-type regulator that controls the expression of a large number of genes in C. violaceum, thereby contributing to oxidative stress defense (ohrA/ohrR), anaerobic respiration (narK1K2 and narGHJI), and biofilm formation (putative RTX adhesins).
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-021-02369-x.
Keywords: Chromobacterium violaceum, MarR family regulator, OsbR regulon, Transcriptome analysis, Oxidative stress, Biofilm formation, Nitrate respiration
Background
Chromobacterium violaceum is a Gram-negative betaproteobacterium found in tropical and subtropical ecosystems around the world, primarily in water and soil samples [1, 2]. C. violaceum strains produce several enzymes and secondary metabolites with biotechnological interest, including quitinases, hydrogen cyanide, siderophores, the antitumoral depsipeptide FR901228, and antibiotics, such as violacein. The purple pigment violacein has in vitro activity against eukaryotic cells and Gram-positive bacteria [1, 3]. From the environmental reservoir, C. violaceum can cause rare but deadly opportunistic infections in humans and other animals, entering the hosts mainly through skin lesions. The resulting disease manifests as fatal septicemia and abscesses in the lung and liver [2, 4]. Similar to other environmental pathogens, C. violaceum has complex regulatory systems and versatile metabolic capacities to survive under several stress conditions [5–7]. For example, genome sequence analyses indicate that C. violaceum possesses a nitrate reductase (Nar) to reduce nitrate to nitrite to obtain energy under anaerobic conditions [5, 6]. However, the transcriptional regulators involved in C. violaceum adaptation to diverse environments remain poorly investigated.
The MarR (multiple antibiotic resistance regulator) family of transcriptional regulators, widespread in bacteria and archaea, includes proteins involved in several bacterial processes, such as antibiotic resistance, virulence control, oxidative stress response, and catabolism of aromatic compounds [8–13]. Many MarR-type regulators act as transcriptional repressors that dissociate from DNA upon binding of small ligands or oxidation of conserved Cys residues [10–12]. One prototypical redox-sensing MarR regulator is OhrR, a Cys-based redox sensor that, upon oxidation by organic hydroperoxides, releases the transcription of ohrA, encoding a Cys-based peroxidase [14–17]. Other redox-sensing MarR regulators control large regulons and multiple bacterial responses in addition to antioxidant defense. For instance, OspR of Pseudomonas aeruginosa, an OhrR homolog, regulates pigment production, antibiotic resistance, virulence, and antioxidant enzymes [18, 19]; MgrA of Staphylococcus aureus is a global regulator of virulence, antibiotic resistance, biofilm, and clumping [20, 21]. Several homologs of OspR and MgrA have been characterized, including SarZ of S. aureus, MosR of Mycobacterium tuberculosis, AbfR of Staphylococcus epidermidis, BmoR of Bacteroides fragilis, and AsrR of Enterococcus faecium [22–26]. The homolog pairs OhrR/OspR in P. aeruginosa and MgrA/SarZ in S. aureus control distinct and overlapping functions [18, 22].
Based on the genome sequence, C. violaceum has more than two hundred predicted transcriptional regulators grouped in several families, including the MarR family with at least fifteen members [6]. However, only a few transcriptional regulators have been characterized in C. violaceum. The master regulator CilA activates most genes from the Chromobacterium pathogenicity islands 1 and 1a (Cpi-1/−1a) that encode a type III secretion system required for virulence [27, 28]. The iron-sensing regulator Fur controls several traits, including antioxidant defense, siderophore production, and virulence [29]. Recently, we described the regulons of the MarR-type regulators OhrR and EmrR in C. violaceum [30, 31]. We have shown that OhrR affects C. violaceum virulence and is involved in resistance to organic hydroperoxides by regulating ohrA [30, 32], while EmrR confers antibiotic resistance by regulating the MFS-type efflux pump EmrCAB [31]. In this work, we described a novel MarR family transcriptional regulator in C. violaceum, CV_3905, which we named OsbR (oxidative stress response and biofilm regulator). Transcriptome and phenotype analyses revealed that OsbR exerts a global regulatory role in C. violaceum, contributing to oxidative stress defense, anaerobic respiration, and biofilm formation. Our results also indicate a double regulation of OhrR and OsbR on the ohrR/ohrA system for fine-tuning the organic hydroperoxide response.
Results and discussion
Identification of C. violaceum OsbR (CV_3905) as an OspR/MgrA homolog
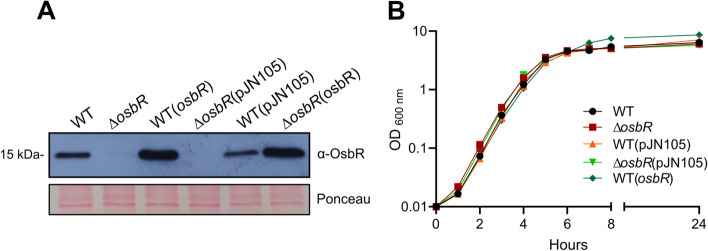
The MarR-type regulators OspR of P. aeruginosa, MgrA of S. aureus, and their homologs in other bacteria, such as AbfR of S. epidermidis and MosR of M. tuberculosis, control multiple and complex bacterial phenotypes [19, 21, 23, 25]. To identify the OspR/MgrA homolog in C. violaceum, we performed BLASTP analyses with OspR or MgrA against the proteome derived from the genome sequence of C. violaceum ATCC 12472, using default parameters. The search yielded a single hit in C. violaceum, OhrR, which showed 34 and 47% identity with MgrA and OspR, respectively. Considering that we have previously characterized OhrR in C. violaceum [30, 32], we performed a subsequent search using the Position-Specific Iteractive BLAST (PSI-BLAST), which uses a position-specific score matrix (PSSM) to found distantly related proteins. In addition to OhrR, this search found as a second hit CV_3905, which shared 30% identity with MgrA (E value 6e-08 and cover 58%) and 35% identity with OspR (E value 2e-07 and cover 52%). Based on transcriptome and phenotype characterization presented in this work, we named CV_3905 as OsbR. The OsbR protein harbors two cysteine residues located at positions 55 and 133. Multiple sequence alignment revealed that the Cys55 residue of OsbR is conserved among closely related unstudied proteins. Still, it does not align with the reactive conserved Cys residues of well-characterized redox-sensing OspR/MgrA homologs (Additional file 1, Fig. S1). To characterize OsbR in C. violaceum, we constructed a null mutant strain ∆osbR and an overexpressing strain WT(osbR). The absence of OsbR in ∆osbR and the high levels of OsbR in WT(osbR) were confirmed by western blot (Fig. 1a). All strains presented the same growth pattern as the WT strain in the LB medium (Fig. 1b).
Fig. 1.
Construction and analyses of C. violaceum strains with deletion (∆osbR) or overexpression (WT(osbR)) of osbR. a The levels of OsbR in the indicated strains were analyzed by western blot using a polyclonal anti-OsbR antiserum. Equal protein loading was confirmed by Ponceau staining. Full-length blot and Ponceau-stained membrane are presented in Additional file 3, Fig. S3. b Growth curves of the indicated strains in LB medium. All strains showed a similar growth profile
Transcriptome analyses reveal a large and diverse OsbR regulon
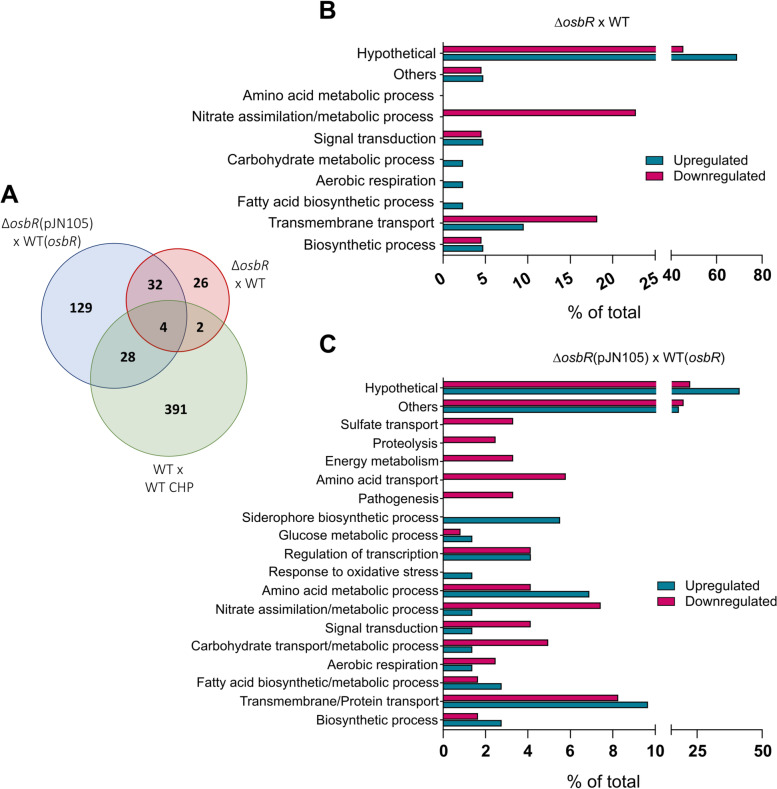
To identify the OsbR regulon in C. violaceum, we performed two sets of DNA microarray analyses using, in both cases, three biological replicates of mid-log phase bacteria. Comparison of the transcriptome profiles of WT with ∆osbR revealed 65 differentially expressed genes (43 downregulated and 21 upregulated genes in ∆osbR) (Additional file 2, Table S1). In comparison, the transcriptome profiles of WT(osbR) versus ∆osbR(pJN105) showed a higher number of differentially expressed genes (72 downregulated and 121 upregulated genes in ∆osbR) (Additional file 2, Table S2) and more robust differences in the fold-change, including in the genes common to the two datasets (Fig. 2a, Additional file 2, Tables S1 and S2). Interesting, a subset of these genes, including the ohrA/ohrR system (organic hydroperoxide detoxification), the narGHJI operon (nitrate reduction), and the cbaABECF operon (siderophore production) (Fig. 2a, Additional file 2, Table S3), were previously identified as members of the cumene hydroperoxide stimulon in C. violaceum [30], indicating a link between OsbR and oxidative stress (see below).
Fig. 2.
Microarray analyses revealed that ObsR regulates many genes involved in several biological processes. a Venn diagrams comparing the differentially expressed genes from the microarray analyses of this work (osbR strains) with published data (WT treated with the oxidant CHP). b, c Functional categorization by Gene Ontology (GO) of the differentially expressed genes from the comparison wild-type versus ∆osbR (b) and WT(osbR) versus ∆osbR(pJN105) (c)
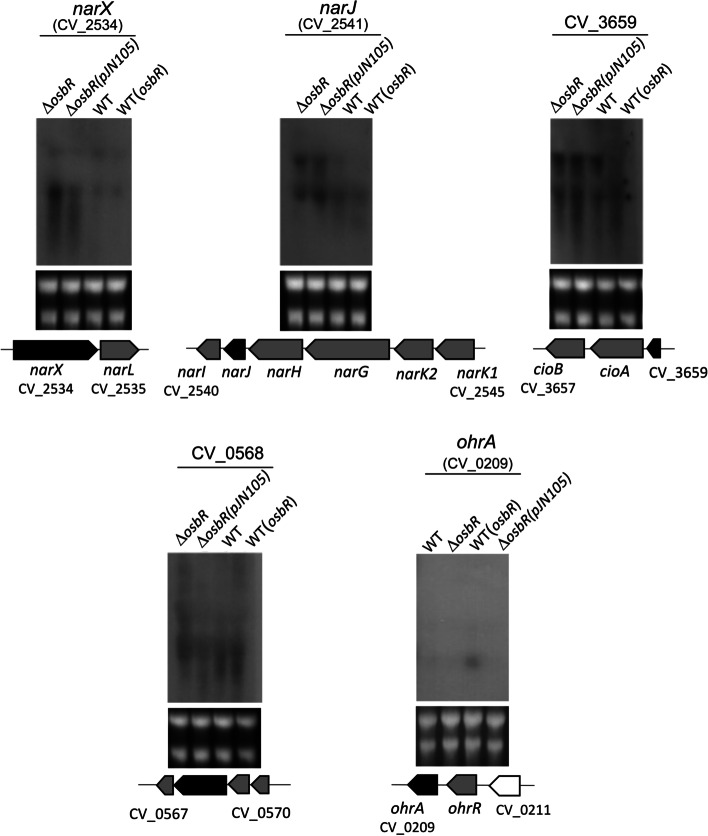
Functional categorization of the up- and downregulated genes identified in both microarray analyses reinforced that OsbR exerts a global transcriptional effect on the C. violaceum transcriptome with an increase in the number of genes and functional processes from the comparison WT versus ∆osbR (Fig. 2b) to the comparison WT(osbR) versus ∆osbR(pJN105) (Fig. 2c). Examples of biological processes affected by OsbR include the categories nitrogen metabolism, membrane transport, iron/siderophore, sulfur and amino acid metabolism, metabolic pathways, regulatory and signaling pathways, among others (Fig. 2b and c). To validate the microarray data, we performed a Northern blot assay probing the genes narX (narXL operon), narJ (narK1K2GHJI operon), CV_3659 (a MarR family regulator in an operon with cioAB), ohrA (ohrR/ohrA), and CV_0568 (anthranilate synthase in CV_0567-68-69-70 operon) (Fig. 3). The results from both microarrays and the Northern blot assay matched for all genes tested (narX, narJ, and CV_3659 were repressed by OsbR while CV_0558 and ohrA were activated by OsbR). The exception was CV_0568 in the comparison WT with ∆osbR since it was downregulated in ∆osbR in the microarray but not in the Northern blot assay.
Fig. 3.
Validation by Northern blot of selected genes regulated by OsbR. RNA samples from the indicated C. violaceum strains (the same strains used in the microarray analyses) were transferred to a membrane and hybridized with specific radiolabeled probes for the selected genes (indicated by black arrows in the gene maps). The selected genes were repressed (narX, narJ, and CV_3659) or activated (CV_0568 and ohrA) by OsbR. Grey arrows indicate neighbor genes also differentially expressed according to microarray analyses. The rRNA were used as a loading control (Bottom gels). Full-length blot and RNA loading gel are presented in Additional file 3, Fig. S4
OsbR acts directly repressing and activating genes of its regulon
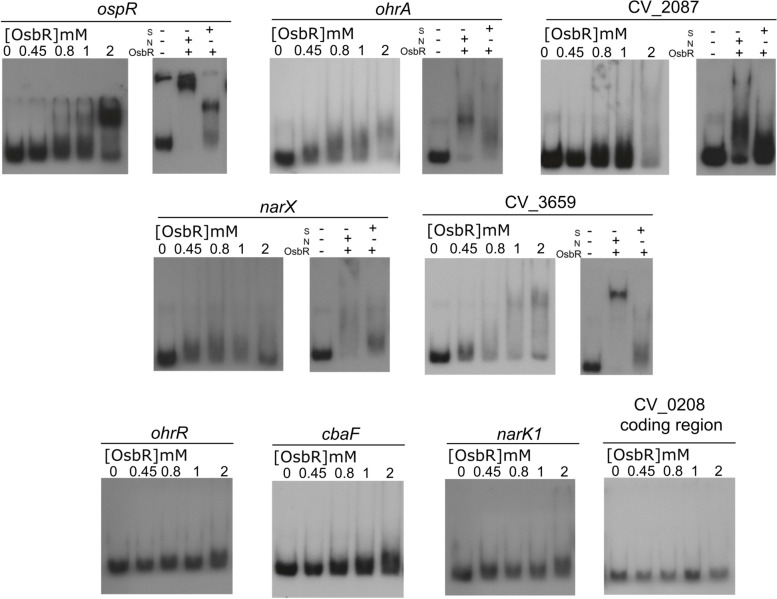
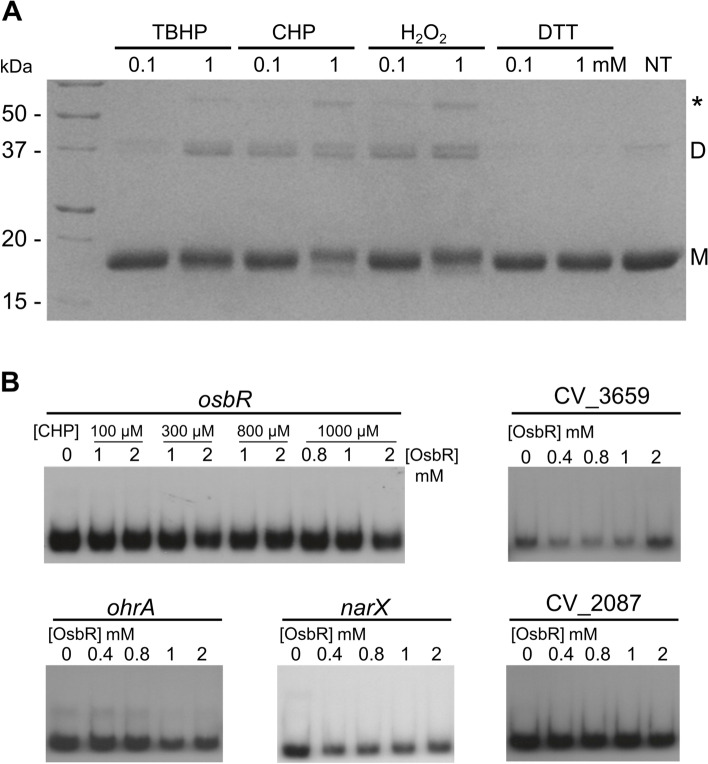
Several MarR family regulators act as repressors by directly binding in the promoter regions of their target genes [10, 11]. To verify if OsbR interacts in vitro with its own promoter and the promoter region of OsbR-regulated genes, we purified the His-OsbR protein (Additional file 1, Fig. S2). In the EMSA assays, an increasing amount of His-OsbR was incubated with labeled DNA fragments of the promoter regions (Fig. 4). The DNA binding assay showed that OsbR binds in different concentrations to the promoter regions of osbR, ohrA, CV_2087, narX, and CV_3659. The specificity of these interactions was confirmed by competition assays, adding in the reactions an unspecific (N) or a specific (S) cold probe (Fig. 4, right panels). Interestingly, OsbR binds to the promoter of both repressed (narX and CV_3659) and activated (ohrA, CV_2087) genes, suggesting a dual role of this regulator as repressor and activator. No band-shift was observed to the promoters of narK1, ohrR, cbaF and to the negative control (the coding region of CV_0208) (Fig. 4), indicating that these genes are indirectly regulated by OsbR. Considering that many transcriptional regulators are members of the OsbR regulon (Fig. 2c, Additional file 2, Table S2), it is tempting to propose that OsbR has its global regulatory role amplified by regulating other transcriptional regulators.
Fig. 4.
OsbR directly activates and represses genes from its regulon. EMSA assay was performed with radiolabeled probes corresponding to the promoter regions of the indicated genes incubated with increasing concentrations of OsbR. The coding region of CV_0208 was used as a negative control. Competition assays were performed with an excess of specific (S) or unspecified (N) unlabeled fragments
OsbR responds to oxidation and has an antioxidant role
Considering that OsbR has two cysteine residues (Additional file 1, Fig. S1) and when overexpressed activates the genes ohrA/ohrR (Fig. 3, Fig. 4, Additional file 2, Table S2), which encode an organic hydroperoxide antioxidant system in C. violaceum [30, 32], we investigated whether OsbR dimerizes upon oxidation by intermolecular disulfide bond formation, a feature of many Cys-based redox sensing regulators [16, 17, 19, 23, 32]. Indeed, the dimerization of monomeric purified His-OsbR was verified in non-reducing SDS-PAGE gel when this protein was incubated with the oxidants tert-butyl hydroperoxide (TBHP), cumene hydroperoxide (CHP), and hydrogen peroxide (H2O2) at 0.1 and 1 mM, but not with the reductant agent dithiothreitol (DTT) (Fig. 5a). Accordingly, when pretreated with CHP, the oxidized OsbR did not bind to the promoter regions of ohrA, CV_2087, CV_3659, narX, and osbR (Fig. 5b). Although further investigation is required to define the role of the OsbR Cys residues on oxidation sensing, our data indicated that the DNA binding activity of OsbR is inhibited by its oxidation.
Fig. 5.
OsbR oxidation causes dimerization and inhibition of its DNA binding activity. a Formation of covalent OsbR dimers under oxidative stress in vitro. After 2 hours of exposure to oxidants or DTT, the His-OsbR protein was analyzed by non-reducing SDS-PAGE. NT refers to nontreated protein. Monomeric and dimeric forms are indicated by M and D, respectively. Asterisk indicates a third unknown multimeric form. b EMSA assay with oxidized OsbR. Prior to incubation with the DNA probes, His-OsbR was oxidized with 1 mM CHP for 1 hour (for the osbR probe, OsbR was oxidized with the indicated CHP concentrations)
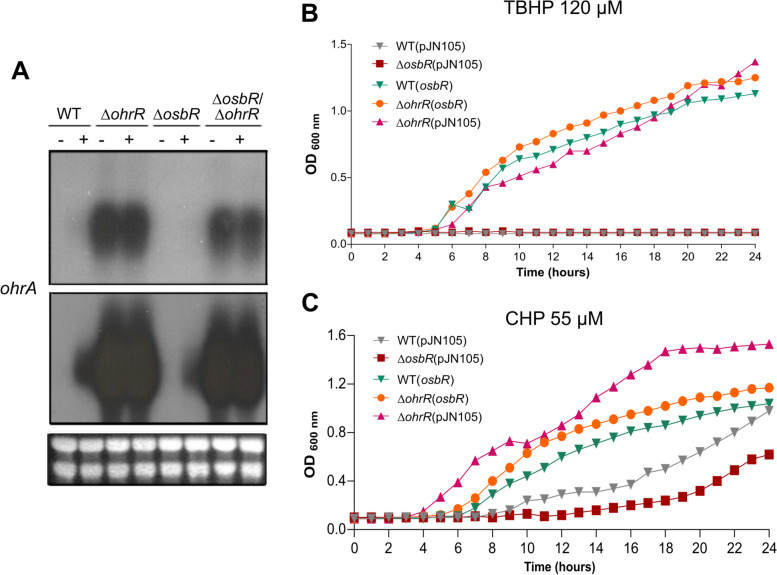
Next, we investigated the relationship between OsbR and the OhrA/OhrR system on the response to organic hydroperoxides by Northern blot and growth curve assays (Fig. 6). The ohrA expression was increased in the wild-type strain under CHP treatment and reached the maximum levels even without treatment in the ∆ohrR mutant (Fig. 6a), confirming our previous data that OhrR is a repressor of ohrA [30, 32]. Importantly, deletion of osbR in both the wild-type and ∆ohrR backgrounds decreased the ohrA expression under CHP treatment (Fig. 6a), suggesting that the maximum ohrA expression requires both OhrR derepression and OsbR activation. Indeed, overexpression of OsbR caused activation of ohrA even in the absence of oxidative stress (Fig. 3). Consistently with this regulatory model, the growth curves under treatment with the oxidants TBHP (Fig. 6b) and CHP (Fig. 6c) indicated that the strains with high ohrA levels (WT(osbR), ∆ohrR(osbR), and ∆ohrR) showed better growth than the wild-type and ∆ospR strains. Our results showing an interconnection of OhrR and OsbR on the regulation of ohrA in C. violaceum resemble the cross-regulation of OhrR and OspR on the ohr and gpx genes described in P. aeruginosa [18]. The OspR of P. aeruginosa is a homolog of OhrR that binds and represses ohr and can even functionally complement an ohrR mutant strain [18]. Despite OspR being a repressor of ohr and OsbR a putative activator of ohrA, both transcriptional regulators are responsive to organic hydroperoxides.
Fig. 6.
OsbR is required for fully ohrA expression and protection against oxidative stress. a Northern blot of ohrA using the indicated strains untreated (−) or treated (+) with 100 μM CHP for 10 min. The same film was exposed for 24 h (top) or 4 days (bottom). Ribosomal RNA loading is shown below. Full-length blot and RNA loading gel are presented in Additional file 3, Fig. S5. b, c Growth curves of different strains under oxidative stress. The indicated C. violaceum strains were grown in LB treated with either 120 μM TBHP (b) or 55 μM CHP (c)
Among the genes upregulated by CHP oxidative stress and activated by OsbR overexpression (Fig. 2a, Additional file 2, Table S3), we highlight the cbaABECF operon in addition to the ohrA and ohrR genes. Recently, we determined that this operon encodes enzymes required for the production of a catecholate-type siderophore in C. violaceum [33]. The E. coli catecholate-type siderophore enterobactin has recently been shown to be involved in protection against oxidative stress via an iron-independent radical scavenging activity [34]. The fact that the cbaABECF operon is activated by OsbR like ohrA in C. violaceum could suggest a dual OsbR-mediated mechanism for protection against oxidative stress.
OsbR regulates genes involved in anaerobic nitrate respiration in C. violaceum
Among the differentially expressed genes found in both microarray analyses and showing the highest upregulation in the ∆osbR mutant were those involved in nitrate respiration (Fig. 2, Additional file 2, Tables S1 and S2). These genes include the narXL operon encoding for a two-component system (NarXL) and the narK1K2GHJI operon encoding for nitrate/nitrite transporters (NarK1 and NarK2) and a respiratory nitrate reductase (NarGHJI). The OsbR-mediated repression of both operons was validated by Northern blot (Fig. 3). As the OsbR binding occurred in the promoter of the narXL operon but not in the promoter of narK1K2GHJI (Fig. 4), we propose that OsbR acts via NarXL to regulate the narK1K2GHJI operon in C. violaceum. Indeed, the NarXL is a well-characterized two-component system that senses the nitrate/nitrite levels to activate narGHJI and narK expression in Escherichia coli and Pseudomonas stutzeri [35, 36]. Consistently with our data, it has been demonstrated that other MgrA/OspR homologs, such as MosR in M. tuberculosis [25, 37] and RosR in Corynebacterium glutamicum [38] regulate nar genes associated with the nitrate reduction for growth under hypoxic and anaerobic conditions [39].
Based on genome prediction [5, 6] and experimental data [40], C. violaceum can undergo anaerobic respiration using nitrate as an electron acceptor. Considering the remarkable role of OsbR on repressing the nar genes, we checked the involvement of osbR in anaerobic respiration, growing the C. violaceum strains in LB medium in an anaerobic jar (Fig. 7). No bacterial growth occurred in LB unless this medium was supplemented with NaNO3, confirming that C. violaceum uses nitrate to obtain energy in anaerobic conditions. When compared with the WT strain, the ∆osbR strain had an impaired growth, while the WT(osbR) strain showed an improved growth (Fig. 7). These data indicate that the overexpression of the nar genes (in ∆osbR) was detrimental while their repression (in WT(osbR)) was beneficial under these conditions. Although C. violaceum possesses the enzymes nitrate reductase (Nar), nitrite reductase (Nir), and nitric oxide reductase (Nor) allowing denitrification until nitrous oxide (N2O) [5, 6, 40], OsbR affected the expression only of the nar genes. Therefore, the elevated expression of the nar genes could enhance the reduction of nitrate to nitrite in the osbR mutant, causing nitrite accumulation and toxicity. Nitrite accumulation is a challenge in bacteria that lack nitrite reductase [41, 42]. Otherwise, in P. aeruginosa, a strain overexpressing the response regulator AtvR had upregulation of all denitrification genes (nar, nir, nor, and nos operons) and improved growth coupled to anaerobic nitrate reduction [43].
Fig. 7.
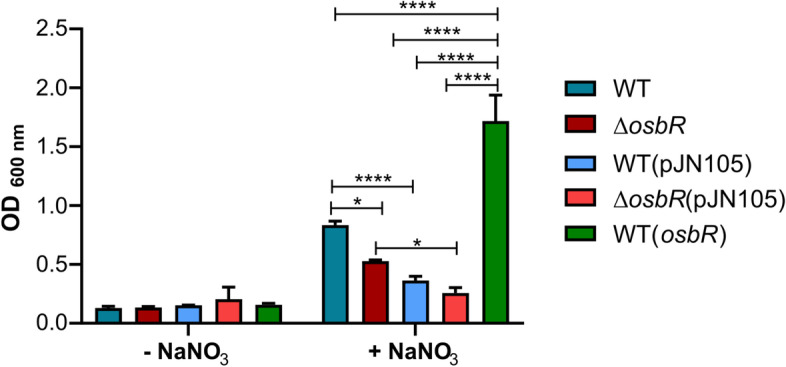
OsbR affects C. violaceum growth under anaerobic conditions. The indicated strains of C. violaceum were diluted to OD600 = 0.1 in LB medium or LB plus 0.5% NaNO3. The cultures were grown without agitation in an anaerobic jar and the OD600 was measured after 48 h. Statistical analysis was performed by Two-way ANOVA and P value < 0.05 was considered significant. P value < 0.05 = *; P value < 0.01 = **; P value < 0.001 = ***; P value < 0.0001 = ****
OsbR represses biofilm formation in C. violaceum
Several MarR-type regulators homologs to OspR/MgrA have been associated with biofilm formation, including MgrA and SarZ of S. aureus [21, 44, 45], AbfR of S. epidermidis [23], and AsrR of E. faecium [26]. To investigate whether OsbR affects biofilm formation in C. violaceum, we performed a biofilm assay with cultures grown statically in LB medium. Deletion of osbR had no effect, while overexpression of osbR caused a pronounced reduction in the biofilm formation (Fig. 8). Detailed inspection of our microarray data indicated that genes from two clusters encoding putative type I secretion systems and related RTX adhesins (CV_1734-35-36-37 and CV_0513-15-16) were upregulated in the absence of OsbR when comparing WT(osbR) with ∆osbR(pJN105) (Additional file 2, Table S2). As adhesins are important to surface attachment and biofilm development [46], we propose that the osbR overexpression caused a reduction in the biofilm formation owing to the OsbR-mediated repression of these two gene clusters. However, more work will be needed to prove this assumption and to define the role of RTX proteins in C. violaceum. In agreement with our data, in S. aureus and E. faecium, the regulators MgrA and AsrR negatively regulate biofilm production by repressing several surface adhesins [21, 26, 44].
Fig. 8.
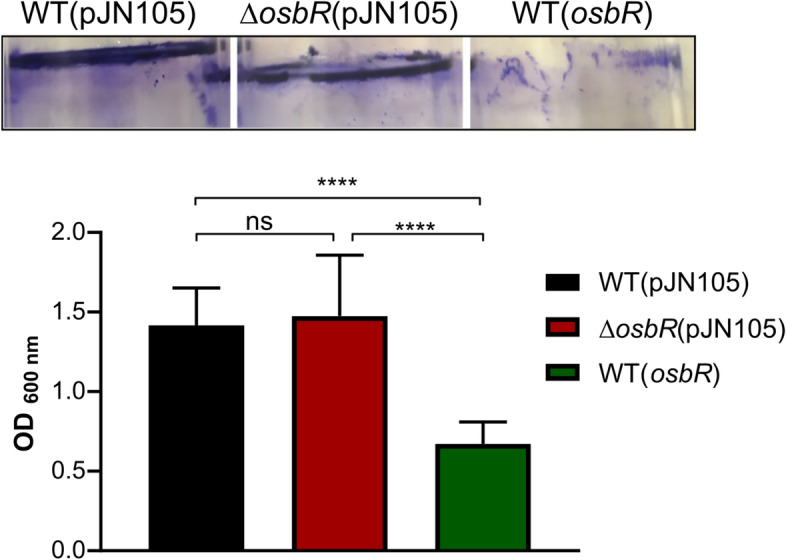
OsbR represses biofilm formation in C. violaceum. Strains were diluted to OD600 = 0.01 in LB medium, and the biofilm formation was measured after growth in static condition for 24 h. Upper: Representative test tubes for each strain after biofilm staining with violet crystal. Bottom: Quantification of biofilm formation by measurement of OD600. Statistical analysis was performed with One-way ANOVA with P value < 0.05 being considered significant. ****, P value < 0.0001
It will be of interest in future studies to address whether OsbR senses oxidants via its cysteine residues to regulate its target genes. Moreover, genetic epistasis analyses could provide a link of the phenotypes related to OsbR (oxidative stress, biofilm formation, and anaerobic growth) and the specific genes of the OsbR regulon.
Conclusions
In this work, we presented transcriptome and phenotypic characterization of OsbR, a yet unstudied MarR family transcriptional regulator of C. violaceum. We showed that OsbR regulates many genes involved in diverse cellular functions as its counterparts OspR/MgrA. Among the OsbR-regulated genes, we highlighted those encoding proteins associated with organic hydroperoxide detoxification (ohrA/ohrR), nitrate assimilation (narK1K2), anaerobic nitrate respiration (narGHJI), and biofilm formation (putative type I secretion systems and associated adhesins). Phenotypic assays using C. violaceum strains harboring null mutation or overexpression of osbR confirmed the importance of OsbR to organic hydroperoxide resistance (probably by OsbR activation of ohrA), anaerobe nitrate respiration (by strong repression of nar operons), and biofilm formation (by repression of adhesins). The EMSA assays revealed that reduced but not oxidized OsbR binds to the promoter regions of either repressed or activated genes. Together, the results outline the global function and importance of OsbR as a novel MarR-type regulator in C. violaceum.
Methods
Bacterial strains and growth conditions
All the bacterial strains and plasmids used in this work are listed in Table 1. The Chromobacterium violaceum wild-type (WT) strain ATCC 12472 and its derivative strains were grown at 37 °C in Luria-Bertani (LB) medium supplemented with gentamycin (40 μg/ml) or kanamycin (50 μg/ml), when necessary. Mutant strains harboring an in-frame deletion of osbR (ΔosbR) and of osbR and ohrR (ΔosbRΔohrR) were constructed by homologous recombination, using a previously described allelic exchange protocol based on sucrose selection [29, 31–33]. All mutant strains were confirmed by PCR and DNA sequencing (data not shown). To obtain an osbR overexpressing strain (WT(osbR)), the osbR gene with its own promoter was cloned into the pJN105 plasmid, and the resulting construct was introduced in the WT strain by conjugation. DNA cloning was performed by digestion with restriction enzymes of PCR-amplified DNA products (Table 2).
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | Strain for cloning | [47] |
| S17–1 | Strain for plasmid mobilization | [48] |
| BL21(DE3) | Strain for protein expression | Novagen |
| Chromobacterium violaceum strains | ||
| ATCC 12472 | Wild-type strain (Sequenced genome) | [6] |
| JF3905 | Wild-type strain with the CV_3905 gene deleted (∆osbR) | This work |
| JF0210 | Wild-type strain with the CV_0210 gene deleted (∆ohrR) | [32] |
| JF39050210 | Wild-type strain with the CV_3905 and CV_0210 genes deleted (ΔosbRΔohrR) | This work |
| WT(pJN105) | Wild-type strain containing the vector pJN105 empty | This work |
| WT(osbR) | Wild-type strain overexpressing osbR | This work |
| ∆osbR(pJN105) | Mutant osbR containing the vector pJN105 empty | This work |
| ∆osbR(osbR) | Mutant osbR strain overexpressing osbR | This work |
| Plasmids | ||
| pNPTS138 | Suicide vector containing oriT sacB; Kanamycin resistance | D. Alley |
| pJN105 | Broad-host-range vector; Gentamycin resistance | [49] |
| pET15b | His-tagged protein expression vector; Ampicillin resistance | Novagen |
| pNPTS138 ∆osbR | In frame null deletion of osbR | This work |
| pNPTS138 ∆ohrR | In frame null deletion of ohrR | [32] |
| pJN105 osbR | Overexpression of osbR | This work |
| pET15b osbR | Heterologous expression of the His-OsbR protein | This work |
Table 2.
Primers used in this work
| Primer | Sequence (5′ - > 3′) |
|---|---|
| Mutant and overexpressing strains | |
| CV3905-del1 | TAGGGCCCGAAACTGCGGCTCGAACGCC |
| CV3905-del2 | ATCAAGCTTCATCGAAAGAGTCCCCGTGC |
| CV3905-del3 | ATCAAGCTTGAAAACACGCCGGCCTGAGC |
| CV3905-del4 | TTGGATCCCTGGTGGCGGGCTTTGTTGC |
| CV_3905-Comp-Fw | GGTACCCTGCAGAGCCAGTCCGCATTAAGGCG |
| CV_3905-Sup-Rv | GGTTACGAGCTCGACGCTCAGCGTCAGAGCTG |
| Northern blot | |
| CV0209-NB-Fw | TGCAGGTGAAGTTGAGCACCCC |
| CV0209-NB-Rv | CCGATGAAGCAGGCGGAATAGCC |
| NB CV2534 -Fw | GATGGTGCTGATCGCGATGG |
| NB CV2534 -Rv | CCAGGGTCTCGATCAGCATC |
| NB CV2541-Fw | CCATTCTGTCCGCGCTGTTG |
| NB CV2541-Rv | GCATTTCGTCCATGTCGCGG |
| NB CV3659-Fw | CACCTCTGGTGCAGGCTTTC |
| NB CV3659-Rv | GACAGCGACTGGGTGAACTG |
| NB CV0568-Fw | ACGCCAAGCTGGGACTCAAC |
| NB CV0568-Rv | GCTCTACCTCCCGTTCGATG |
| EMSA | |
| pCV0209-Fw | TTGGATCCTGCCGGAGCGGAATGCAACT |
| pCV0209-Rv | ATTACTGCAGGCGGTGGCTTCGGCGGTATAC |
| pCV3905-Fw | GGTACCCTGCAGAGCCAGTCCGCATTAAGGCG |
| pCV3905-Rv | ATCAAGCTTCATCGAAAGAGTCCCCGT |
| pCV0210-Fw | CTCGCTGGCCGATCTGGGAC |
| pCV0210-Rv | GGCGGAATACAGCGCGAAGC |
| pCV1486Fw | GACCAAGTGATAGACGCCGG |
| CV1486-DEL2 | GGCCTAAAGCTTCTCGGTCATGAGGATGTCTG |
| pCV2087Fw | CCTCGTAAAAGCGGATGGCG |
| pCV2087Rv | CGTCGAAACCCTTCATCGCG |
| pCV2534Fw | GATCACCGACACCCTCAAGC |
| pCV2534Rv | GCGACAACACCAGCAGCTTG |
| pCV2545Fw | TGACTCTCCCCCATTGGGAG |
| pCV2545Rv | ATCGGGATGCCCAGCACCGC |
| pCV3659Fw | GACCGACAATGACGGCTACG |
| pCV3659Rv | GCCGAAATGCAGCACGAAAG |
| CV0208 Fw | GACCATACTAGACAGCTACGCC |
| CV0208Rv | CAGCGTGGTGATTTGCGGATAG |
Underlined letters indicate restriction enzyme recognition sites used for cloning purposes.
Growth curves
Growth curves of the C. violaceum strains were performed in LB medium at 37 °C and 250 rpm agitation in Erlenmeyer flasks. Overnight liquid cultures were diluted to an optical density at 600 nm (OD600) of 0.01. Samples were collected at the indicated time points to measure the OD600 in a BioPhotometer (Eppendorf). The growth curves under oxidant conditions were performed in 96-well plates. We added 120 μM tert-butyl hydroperoxide (TBHP) or 55 μM cumene hydroperoxide (CHP) to the C. violaceum LB cultures at OD600 of 0.01. The growth was determined by measuring for 24 h the OD600 at 15 min intervals in a SpectraMax i3 MiniMax Imaging Cytometer. Data were plotted in one-hour intervals for clarity.
Growth under anaerobic conditions
Overnight cultures of C. violaceum were diluted in 2 mL of LB to OD600nm of 0.1. Cultures were grown in LB or LB plus 5% NaNO3 without agitation at 37 °C in an anaerobic jar with a GasPak EZ Anaerobe Container System (catalog number 260001; Becton Dickinson) to establish anaerobic conditions. The growth was determined by measuring the OD600 after cultivation by 48 h.
Biofilm formation
Biofilm was quantified using a crystal violet method as previously described [29]. Briefly, C. violaceum liquid cultures at an OD600 of 0.01 were grown without agitation at 37 °C for 24 h in 1.5 mL LB in glass tubes. The biofilm was stained with crystal violet, solubilized with ethanol, and quantified by measuring the OD600.
RNA isolation
The C. violaceum strains in three biological replicates (for DNA microarray assays) were grown in LB medium at 37 °C and 250 rpm agitation until mid-log phase (OD600 of 0.8 to 1.2). The cultures were centrifugated, and the total RNA was extracted with the TRIzol reagent (Ambion) and purified with the illustra RNAspin Mini RNA isolation kit (GE Healthcare). The RNA integrity was evaluated by formaldehyde agarose gel. The absence of DNA contamination was confirmed by PCR. Quantification of total RNA was measured with a NanoDrop spectrophotometer (Thermo Scientific).
DNA microarray
The procedures for cRNA labeling, hybridization, and washing were performed according to the Agilent Two-Color Microarray-Based Exon Analysis protocol (Agilent Technologies). Briefly, 100 ng RNA of each sample were converted in cRNA and labeled with Cy3 and Cy5 using the Low Input Quick Amp WT Two-Color kit (Agilent Technologies). Equimolar amounts of oppositely labeled cRNA were hybridized at 65 °C by 17 h in a custom-designed oligonucleotide microarray slide [30]. The slides were scanned with an Agilent High-Resolution C Scanner. The data were extracted and normalized by using Agilent Feature Extraction Image Analysis Software (Version 10.7.3). Data analysis to determine the differentially expressed genes was performed as previously described [30, 31]. Briefly, the values for the relative expression of each gene were calculated as the average of the values of all probes corresponding to the same gene (ratio values of at least 27 probes, 9 from each biological replicate). Statistical significance was calculated according to the chi square p-value compositional method [50], as implemented in [51]. Briefly, the log-ratio p-values for each probe were obtained from Agilent Feature Extraction Software. The global significance level was controlled at 0.05 using the highly stringent Bonferroni multiple correction method. Considering that multiple hypothesis testing were performed (45,220*3 = 135,660, number of probes in biological triplicates), the effective cutoff is 0.05/135660 = 3.68*10− 7. Genes were considered differentially expressed if showed simultaneously (i) 2-fold change and (ii) statistical significance at 0.05 (< 3.68*10− 7).
Northern blot
For the microarray validation, we used 7.5 μg of the same RNA samples. For the expression of ohrA under oxidant condition, we used 5 μg of total RNA extracted from the WT, ΔosbR, ΔohrR, and ΔosbRΔohrR strains grown in LB untreated or treated with 100 μM of cumene hydroperoxide (CHP) for 10 min. After running in denaturing formaldehyde agarose gel, the RNA samples were transferred onto nylon Hybond-XL membranes (GE Healthcare). Specific probes for the indicated genes were generated by PCR (Table 2) and radiolabeled with [α 32P]-dCTP according to the DECAprime kit (Ambion) protocol. RNA transfer, membrane hybridization and washing, and signal detection were performed as described [30, 32]. Briefly, RNA was transferred to membranes by capillarity. Membranes were prehybridized and incubated with the labeled probes in 10 ml of ULTRAhyb (Ambion). Washing was performed according to the manufacturer’s instructions and as previously described [30].
Expression and purification of his-OsbR
The osbR gene (CV_3905) was PCR-amplified and cloned into the expression vector pET15b (Table 2). The E. coli BL21(DE3) harboring the plasmid pET15b-osbR was used to express the recombinant His-tagged OsbR protein after induction with 1 mM IPTG for 2 h at 37 °C. The His-OsbR protein was purified by affinity chromatography in a Ni-NTA superflow column (Qiagen) as previously described [32].
OsbR dimerization in vitro
The purified His-OsbR protein was incubated with the reductant agent dithiothreitol (DTT) or the oxidants cumene hydroperoxide (CHP), tert-butyl hydroperoxide (TBHP), and hydrogen peroxide (H2O2) at concentrations of 0.1 and 1 mM for 30 min at room temperature. The samples were alkylated with 100 mM N-ethylmaleimide (NEM) for 2 h. The presence of OsbR as monomers or dimers was analyzed after non-reducing SDS-PAGE and Coomassie brilliant blue staining.
Electrophoretic mobility shift assay (EMSA)
DNA probes corresponding to the promoter region of the indicated genes and the coding region of CV_0208 were amplified by PCR with specific primers (Table 2). These fragments were labeled with [γ32P]-ATP by T4 polynucleotide kinase (Thermo Scientific) and purified with the Wizard SV gel and PCR clean-up system (Promega). The interaction of reduced OsbR with the DNA probes was performed as already described [30, 32]. The EMSA with oxidized OsbR was performed by pretreatment of OsbR with 1 mM CHP by 1 h at room temperature.
Western blot
The C. violaceum strains were grown in LB until the mid-log phase. Samples were collected, centrifugated and the pellets were resuspended in an SDS sample buffer. Proteins were resolved by 15% SDS-PAGE and transferred onto a nitrocellulose membrane. The western blot was performed with the Protein Detector LumiGLO Western blot (KPL) kit, using an anti-OsbR polyclonal antiserum (1:1000) developed in mice by a subcutaneous injection of the His-OsbR protein. The six-week-old female BALB/c mice (n = 5) used in this study were obtained from the facility Biotério Geral (PUSP-RP), and maintained in the Animal Facilities of the Faculdade de Medicina de Ribeirão Preto (FMRP-USP). The animals were euthanized with an excessive dose of anesthetic (one dose of 200 mg/kg ketamine and 20 mg/kg xylazine).
Supplementary Information
Additional file 1: Figure S1 OsbR alignment. Figure S2 His-OsbR purification by affinity chromatography.
Additional file 2: Table S1 Comparison of the transcriptome profiles of WT with ΔosbR. Table S2 Comparison of the transcriptome profiles of WT(osbR) versus ΔosbR(pJN105). Table S3 List of genes shared among our microarray analyses and the CHP stimulon.
Additional file 3: Figure S3 Uncropped blot. Figure S4 Uncropped Northen blot. Figure S5 Uncropped Northern blot.
Acknowledgments
We are grateful to the Sergio L Salvador lab for assistance with the anaerobic growth assays and the Geraldo A Passos lab for assistance with the microarray scanning.
Authors’ contributions
JAA and JFSN planned the experiments and wrote the manuscript; JAA, MPM, and KCMM performed the experimental work; JAA, MPM, and JFSN analyzed and interpreted data; TK performed the statistical analysis of the microarray data; JFSN participated in study coordination and funding acquisition. All authors read and approved the final manuscript.
Funding
This research was supported by grants from the São Paulo Research Foundation (FAPESP; grants 2012/20435–9 and 2020/00259–8) and Fundação de Apoio ao Ensino, Pesquisa e Assistência do Hospital das Clínicas da FMRP-USP (FAEPA). During the course of this work, JAA (grant 2016/08728–1), MPM (grant 2013/25745–9), and KCMB (grant 2013/18797–2) were supported by FAPESP fellowships. The funders had no role in the design of the study, collection, analysis, and interpretation of data, and in writing the manuscript.
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its additional files. The microarray datasets are available in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE171860.
Declarations
Ethics approval and consent to participate
The experiments with mice (antibody production) were performed following the Ethical Principles in Animal Research adopted by the National Council for the Control of Animal Experimentation (CONCEA), and the protocol was approved by the Local Ethical Animal Committee (CEUA) from FMRP-USP (number 147/2014). The study was carried out in compliance with the ARRIVE guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Júlia A. Alves, Email: julia.aparecida.alves@usp.br
Maristela Previato-Mello, Email: maristelapreviato@usp.br.
Kelly C. M. Barroso, Email: kellybarroso@usp.br
Tie Koide, Email: tkoide@fmrp.usp.br.
José F. da Silva Neto, Email: jfsneto@usp.br
References
- 1.Durán N, Menck CFM. Chromobacterium violaceum: a review of pharmacological and industrial perspectives. Crit Rev Microbiol. 2001;27(3):201–222. doi: 10.1080/20014091096747. [DOI] [PubMed] [Google Scholar]
- 2.Batista JH, da Siva Neto JF. Chromobacterium violaceum pathogenicity: updates and insights from genome sequencing of novel Chromobacterium species. Front Microbiol. 2017;8:2213. doi: 10.3389/fmicb.2017.02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durán N, Justo GZ, Ferreira CV, Melo PS, Cordi L, Martins D. Violacein: properties and biological activities. Biotechnol Appl Biochem. 2007;48(Pt 3):127–133. doi: 10.1042/BA20070115. [DOI] [PubMed] [Google Scholar]
- 4.Yang CH, Li YH. Chromobacterium violaceum infection: a clinical review of an important but neglected infection. J Chinese Med Assoc. 2011;74(10):435–441. doi: 10.1016/j.jcma.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Creczynski-Pasa TB, Antônio RV. Energetic metabolism of Chromobacterium violaceum. Genet Mol Res. 2004;3(1):162–166. [PubMed] [Google Scholar]
- 6.Vasconcelos ATR, Almeida DF, Hungria M, Guimarães CT, Antônio RV, Almeida FC, et al. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc Natl Acad Sci U S A. 2003;100(20):11660–11665. doi: 10.1073/pnas.1832124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hungria M, Nicolás MF, Guimarães CT, Jardim SN, Gomes EA, De Vasconcelos ATR. Tolerance to stress and environmental adaptability of Chromobacterium violaceum. Genet Mol Res. 2004;3(1):102–116. [PubMed] [Google Scholar]
- 8.Cohen SP, Hachler H, Levy SB. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175(5):1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellison DW, Miller VL. Regulation of virulence by members of the MarR/SlyA family. Curr Opin Microbiol. 2006;9(2):153–159. doi: 10.1016/j.mib.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Wilkinson SP, Grove A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol. 2006;8(1):51–62. [PubMed] [Google Scholar]
- 11.Perera IC, Grove A. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J Mol Cell Biol. 2010;2(5):243–254. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- 12.Hao Z, Lou H, Zhu R, Zhu J, Zhang D, Zhao BS, et al. The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli. Nat Chem Biol. 2014;10(1):21–28. doi: 10.1038/nchembio.1380. [DOI] [PubMed] [Google Scholar]
- 13.Roy A, Ranjan A. HosA, a MarR family transcriptional regulator, represses nonoxidative Hydroxyarylic acid decarboxylase operon and is modulated by 4-Hydroxybenzoic acid. Biochemistry. 2016;55(7):1120–1134. doi: 10.1021/acs.biochem.5b01163. [DOI] [PubMed] [Google Scholar]
- 14.Fuangthong M, Helmann JD. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc Natl Acad Sci U S A. 2002;99(10):6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J Bacteriol. 2001;183(14):4134–4141. doi: 10.1128/JB.183.14.4134-4141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox Signal. 2011;14(6):1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Netto LES, Oliveira MA, Tairum CA, da Silva Neto JF. Conferring specificity in redox pathways by enzymatic thiol/disulfide exchange reactions. Free Radic Res. 2016;50(2):206–245. doi: 10.3109/10715762.2015.1120864. [DOI] [PubMed] [Google Scholar]
- 18.Atichartpongkul S, Vattanaviboon P, Wisitkamol R, Jaroensuk J, Mongkolsuk S, Fuangthong M. Regulation of organic hydroperoxide stress response by two OhrR homologs in Pseudomonas aeruginosa. PLoS One. 2016;11(8):e0161982. doi: 10.1371/journal.pone.0161982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan L, Murray TS, Kazmierczak BI, He C. Pseudomonas aeruginosa OspR is an oxidative stress sensing regulator that affects pigment production, antibiotic resistance and dissemination during infection. Mol Microbiol. 2010;75(1):76–91. doi: 10.1111/j.1365-2958.2009.06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luong TT, Dunman PM, Murphy E, Projan SJ, Lee CY. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J Bacteriol. 2006;188(5):1899–1910. doi: 10.1128/JB.188.5.1899-1910.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crosby HA, Schlievert PM, Merriman JA, King JM, Salgado-Pabón W, Horswill AR. The Staphylococcus aureus global regulator MgrA modulates clumping and virulence by controlling surface protein expression. PLoS Pathog. 2016;12(5):e1005604. doi: 10.1371/journal.ppat.1005604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen PR, Nishida S, Poor CB, Cheng A, Bae T, Kuechenmeister L, et al. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol Microbiol. 2009;71(1):198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Sun X, Wu Y, Xie C, Zhang W, Wang D, et al. Oxidation-sensing regulator AbfR regulates oxidative stress responses, bacterial aggregation, and biofilm formation in Staphylococcus epidermidis. J Biol Chem. 2013;288(6):3739–3752. doi: 10.1074/jbc.M112.426205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teixeira FL, Silva DN, Pauer H, Ferreira LQ, de Oliveira Ferreira E, Domingues RM, et al. The role of BmoR, a MarR family regulator, in the survival of Bacteroides fragilis during oxidative stress. Int J Med Microbiol. 2013;303(8):443–448. doi: 10.1016/j.ijmm.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Brugarolas P, Movahedzadeh F, Wang Y, Zhang N, Bartek IL, Gao YN, et al. The oxidation-sensing regulator (MosR) is a new redoxdependent transcription factor in mycobacterium tuberculosis. J Biol Chem. 2012;287(45):37703–37712. doi: 10.1074/jbc.M112.388611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebreton F, van Schaik W, Sanguinetti M, Posteraro B, Torelli R, Le Bras F, et al. AsrR is an oxidative stress sensing regulator modulating enterococcus faecium opportunistic traits, antimicrobial resistance, and pathogenicity. PLoS Pathog. 2012;8(8):e1002834. doi: 10.1371/journal.ppat.1002834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miki T, Iguchi M, Akiba K, Hosono M, Sobue T, Danbara H, et al. Chromobacterium pathogenicity island 1 type III secretion system is a major virulence determinant for Chromobacterium violaceum-induced cell death in hepatocytes. Mol Microbiol. 2010;77(4):855–872. doi: 10.1111/j.1365-2958.2010.07248.x. [DOI] [PubMed] [Google Scholar]
- 28.Miki T, Akiba K, Iguchi M, Danbara H, Okada N. The Chromobacterium violaceum type III effector CopE, a guanine nucleotide exchange factor for Rac1 and Cdc42, is involved in bacterial invasion of epithelial cells and pathogenesis. Mol Microbiol. 2011;80(5):1186–1203. doi: 10.1111/j.1365-2958.2011.07637.x. [DOI] [PubMed] [Google Scholar]
- 29.Santos RERS, Batista BB, da Silva Neto JF. Ferric uptake regulator Fur coordinates Siderophore production and defense against Iron toxicity and oxidative stress and contributes to virulence in Chromobacterium violaceum. Appl Environ Microbiol. 2020;86(21):e01620. doi: 10.1128/AEM.01620-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Previato-Mello M, de Abreu Meireles D, Netto LES, da Silva Neto JF. Global transcriptional response to organic hydroperoxide and the role of OhrR in the control of virulence traits in Chromobacterium violaceum. Infect Immun. 2017;85(8):e00017. doi: 10.1128/IAI.00017-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barroso KCM, Previato-Mello M, Batista BB, Batista JH, da Silva Neto JF. EmrR-dependent Upregulation of the efflux pump EmrCAB contributes to antibiotic resistance in Chromobacterium violaceum. Front Microbiol. 2018;9:2756. doi: 10.3389/fmicb.2018.02756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.da Silva Neto JF, Negretto CC, Netto LES. Analysis of the organic hydroperoxide response of Chromobacterium violaceum reveals that OhrR is a cys-based redox sensor regulated by thioredoxin. PLoS One. 2012;7(10):e47090. doi: 10.1371/journal.pone.0047090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Batista BB, De Souza Santos RER, Ricci-Azevedo R, Da Silva Neto JF. Production and uptake of distinct endogenous catecholate-type siderophores are required for iron acquisition and virulence in Chromobacterium violaceum. Infect Immun. 2019;87(12):e00577–e00519. doi: 10.1128/IAI.00577-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peralta DR, Adler C, Corbalán NS, Paz García EC, Pomares MF, Vincent PA. Enterobactin as part of the oxidative stress response repertoire. PLoS One. 2016;11(6):e0157799. doi: 10.1371/journal.pone.0157799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabin RS, Stewart V. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J Bacteriol. 1993;175(11):3259–3268. doi: 10.1128/jb.175.11.3259-3268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartig E, Schiek U, Vollack KU, Zumft WG. Nitrate and nitrite control of respiratory nitrate reduction in denitrifying pseudomonas stutzeri by a two-component regulatory system homologous to NarXL of Escherichia coli. J Bacteriol. 1999;181(12):3658–3665. doi: 10.1128/JB.181.12.3658-3665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abomoelak B, Hoye EA, Chi J, Marcus SA, Laval F, Bannantine JP, et al. mosR, a novel transcriptional regulator of hypoxia and virulence in mycobacterium tuberculosis. J Bacteriol. 2009;191(19):5941–5952. doi: 10.1128/JB.00778-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bussmann M, Baumgart M, Bott M. RosR (Cg1324), a hydrogen peroxide-sensitive MarR-type transcriptional regulator of Corynebacterium glutamicum. J Biol Chem. 2010;285(38):29305–29318. doi: 10.1074/jbc.M110.156372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.González PJ, Correia C, Moura I, Brondino CD, Moura JJG. Bacterial nitrate reductases: molecular and biological aspects of nitrate reduction. J Inorg Biochem. 2006;100(5–6):1015–1023. doi: 10.1016/j.jinorgbio.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 40.Bazylinski DA, Palome E, Blakemore NA, Blakemore RP. Denitrification by Chromobacterium violaceum. Appl Environ Microbiol. 1986;52(4):696–699. doi: 10.1128/aem.52.4.696-699.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bollag JM, Henninger NM. Effects of nitrite toxicity on soil bacteria under aerobic and anaerobic conditions. Soil Biol Biochem. 1978;10:377–381. doi: 10.1016/0038-0717(78)90061-5. [DOI] [Google Scholar]
- 42.Bueno E, Sit B, Waldor MK, Cava F. Anaerobic nitrate reduction divergently governs population expansion of the enteropathogen vibrio cholerae. Nat Microbiol. 2018;3(12):1346–1353. doi: 10.1038/s41564-018-0253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaihami GH, Breda LCD, Almeida JRF, Pereira TO, Nicastro GG, Boechat AL, et al. The atypical response regulator AtvR is a new player in Pseudomonas aeruginosa. Infect Immun. 2017;85(8):e00207–e00217. doi: 10.1128/IAI.00207-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang Q, Jin Z, Sun B. MgrA negatively regulates biofilm formation and detachment by repressing the expression of psm operons in Staphylococcus aureus. Appl Environ Microbiol. 2018;84(16):e01008. doi: 10.1128/AEM.01008-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamber S, Cheung AL. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect Immun. 2009;77(1):419–428. doi: 10.1128/IAI.00859-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berne C, Ducret A, Hardy GG, Brun YV. Adhesins involved in attachment to abiotic surfaces by negative bacteria. Microbiol Spectr. 2015;3(4):MB-0018-2015. doi: 10.1128/microbiolspec.MB-0018-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166(4):557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 48.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 49.Newman JR, Fuqua C. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 1999;227(2):197–203. doi: 10.1016/S0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 50.Fisher RA. Statistical Methods for Research Workers. 4th ed. London; 1932.
- 51.Cortez DA, Tonon AP, Colepicolo P, Vêncio RZN. Combining P values to improve classification of differential gene expression in the HTself software. Genet Mol Res. 2011;10(4):3586–3595. doi: 10.4238/2011.December.5.5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1 OsbR alignment. Figure S2 His-OsbR purification by affinity chromatography.
Additional file 2: Table S1 Comparison of the transcriptome profiles of WT with ΔosbR. Table S2 Comparison of the transcriptome profiles of WT(osbR) versus ΔosbR(pJN105). Table S3 List of genes shared among our microarray analyses and the CHP stimulon.
Additional file 3: Figure S3 Uncropped blot. Figure S4 Uncropped Northen blot. Figure S5 Uncropped Northern blot.
Data Availability Statement
The data supporting the conclusions of this article are included within the article and its additional files. The microarray datasets are available in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE171860.