Summary
piRNAs guide Piwi/Panoramix-dependent H3K9me3 chromatin modification and transposon silencing during Drosophila germline development. The THO RNA export complex is composed of Hpr1, Tho2, and Thoc5–7. Null thoc7 mutations, which displace Thoc5 and Thoc6 from a Tho2-Hpr1 sub-complex, reduce expression of a subset of germline piRNAs and increase transposon expression, suggesting that THO silences transposons by promoting piRNA biogenesis. Here we show that the thoc7 null mutant combination increases transposon transcription, but does not reduce anti-sense piRNAs targeting half of the transcriptionally activated transposon families. These mutations also fail to reduce piRNA-guided H3K9me3 chromatin modification or block Panoramix-dependent silencing of a reporter transgene, and unspliced transposon transcripts co-precipitate with THO through a Piwi- and Panoramix-independent mechanism. Mutations in piwi also dominantly enhance germline defects associated with thoc7 null alleles. THO thus functions in a piRNA-independent transposon silencing pathway, which acts cooperatively with Piwi to support germline development.
Introduction
Transposons can mobilize and trigger genome instability, but also appear to generate beneficial genetic diversity that contributes to adaptive evolution (Bourque et al., 2018; Hedges and Deininger, 2007; Huang et al., 2012; Khurana and Theurkauf, 2008; Parhad and Theurkauf, 2019). The PIWI-interacting RNA (piRNA) pathway silences transposons during germline development, controlling the balance between transposition and host genome maintenance (Aravin et al., 2007; Khurana and Theurkauf, 2008; Senti and Brennecke, 2010). The 23–30nt piRNAs are loaded into PIWI clade Argonaute proteins and guide sequence-specific transcriptional and post-transcriptional silencing (Czech et al., 2018; Ozata et al., 2019). In Drosophila ovaries, piRNA precursors are produced from dual-strand and uni-strand piRNA clusters composed of nested transposon fragments (Bergman et al., 2006; Brennecke et al., 2007), and from a subset of dispersed euchromatic transposon insertions that function as “mini-clusters” (Mohn et al., 2014; Parhad et al., 2020). Germline clusters are bound by the Rhino-Deadlock-Cuff complex, which promotes non-canonical transcription from both genomic strands, suppresses splicing and polyadenylation, and recruits UAP56 and the THO complex to piRNA cluster transcripts (Chen et al., 2016; Hur et al., 2016; Klattenhoff et al., 2009; Mohn et al., 2014; Zhang et al., 2012a; Zhang et al., 2018; Zhang et al., 2014). The resulting RNPs are delivered to the processing machinery in perinuclear nuage and at the surface of mitochondria by specialized nuclear export factors (Fabry et al., 2019; Kneuss et al., 2019; Murano et al., 2019). Germline piRNAs bind the PIWI proteins Aub, Ago3 and Piwi. Aub and Ago3 localize to perinuclear nuage and drive Ping-Pong amplification and post-transcriptional silencing. Ago3 bound piRNAs also guide cleavage of RNAs that undergo phased biogenesis at mitochondria, producing piRNAs that bind to Piwi. The resulting Piwi-piRNA complexes localize to the nucleus and function with Panoramix (Panx) to direct transcriptional silencing, which is associated with increased repressive H3K9me3 modification and LSD1 dependent reductions in H3K4me2/3 at promoters (Brennecke et al., 2007; Han et al., 2015; Malone et al., 2009; Mohn et al., 2015; Sienski et al., 2012; Wang et al., 2015; Yu et al., 2015).
The THO complex has a conserved function in co-transcriptional RNA processing and nuclear export (Reed and Cheng, 2005). In metazoans, this complex is composed of Hpr1, Tho2, and Thoc5–7. Tho2 and Hpr1 are conserved from yeast to humans, and null mutations in Drosophila tho2 and mouse Hpr1/Thoc1 are lethal, consistent with an essential function for the complex in RNA metabolism (Jagut et al., 2013; Wang et al., 2006). The Thoc5–7 subunits are specific to metazoans, and mutations in Drosophila thoc5 and thoc7 are viable and do not alter protein-coding gene expression, but disrupt transposon silencing, reduce germline piRNA expression, and lead to female sterility (Hur et al., 2016; Zhang et al., 2018). In null thoc7 mutant ovaries, Thoc5 and 6 are displaced from a stable dimer of the deeply conserved Hpr1 and Tho2 subunits, and hypomorphic thoc5 mutations displace Thoc6 and 7 from Hpr1-Tho2 (Zhang et al., 2018). These findings suggest that an Hpr1/Tho2 subcomplex supports essential functions for THO, while the heteropentamer is critical to germline development and transposon silencing.
To gain insight into the function of the intact THO heteropentamer in germline development, we performed a detailed analysis of thoc7 null mutants. These mutations increase steady state transposon expression (Hur et al., 2016; Zhang et al., 2018), and here we show that this is linked to increased transcription. The null thoc7 mutant combination significantly reduces piRNA expression from the major germline clusters (Hur et al., 2016; Zhang et al., 2018). However, we here show that loss of Thoc7 does not reduce anti-sense piRNAs targeting half of the transcriptionally activated transposon families, block piRNA-directed H3K9me3 chromatin modification of transposons, or disrupt Panx-dependent silencing of a reporter transgene. We also show that unspliced transposon transcripts co-precipitate with Hpr1, and that binding is disrupted by thoc7 mutations, but not by mutations in piwi or panx. Finally, we show that thoc7 and piwi mutations produce strong synergistic defects in oogenesis, and that double mutants lead to loss of germline tissue from adult females. The THO heteropentamer is therefore required for biogenesis of a subset of germline piRNAs, and functions in a piRNA-independent transposon silencing pathway that cooperates with piRNAs to maintain genome integrity and support germline development.
Results
piRNA-independent silencing by the THO complex
In the Drosophila germline, the HP1 homolog Rhino (Rhi) binds dual-strand clusters, anchoring a complex that drives transcription from both genomic strands, suppresses splicing, and promotes precursor transcript binding to the THO complex and UAP56 (Hur et al., 2016; Klattenhoff et al., 2009; Mohn et al., 2014; Zhang et al., 2018; Zhang et al., 2014). UAP56 and THO interact within the Transcription and Export (TREX) complex (Reed and Cheng, 2005). A point mutation that reduces UAP56 binding to THO, and mutations in thoc5 and thoc7 that block UAP56 co-precipitation with Hpr1, are viable and do not significantly alter protein-coding gene expression. However, mutant females are sterile, dual strand cluster piRNA expression is reduced, and silencing of a subset of transposon families is disrupted (Hur et al., 2016; Zhang et al., 2012a; Zhang et al., 2018). These findings suggest that the TREX functions downstream of Rhino in a pathway that leads from clusters to piRNA-guided transposon silencing.
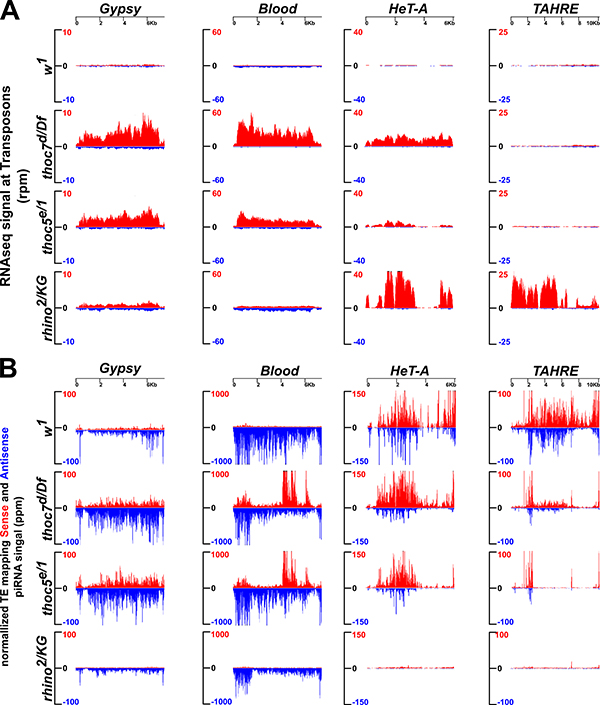
If THO functions downstream of Rhino in a linear pathway, thoc5, thoc7, and rhino (rhi) mutants should produce similar defects in transposon silencing and piRNA production. Figure 1 shows transcript and piRNA profiles of four transposon families, which represent the range of patterns observed across all transposon families in these genotypes. Mutations in rhi nearly eliminate piRNAs targeting the non-LTR retrotransposons HeT-A and TAHRE and lead to a modest decrease in Blood and Gypsy piRNAs. Consistent with these changes in piRNA expression, HeT-A and TAHRE are significantly over-expressed, while Blood and Gypsy show modest increases in expression. In thoc7 mutants, by contrast, Gypsy and Blood are significantly over-expressed, but antisense piRNAs targeting these elements show a modest increase relative to w1 controls. There is also a significant increase in sense strand piRNAs, which are produced through anti-sense directed transcript cleavage during the ping-pong cycle. The anti-sense piRNAs expressed in thoc7 mutants thus appear to function in ping-pong amplification, but are not sufficient to silence the target elements. HeT-A is over-expressed in thoc7, thoc5, and rhi mutants, and anti-sense piRNAs are reduced in all three genotypes.
Figure 1. Representative transposons show overexpression without piRNA defects in thoc7 and thoc5 mutants.
Long (A) and short (B) RNAs profiles mapping to Gypsy, Blood, HeT-A and TAHRE consensus sequences in w1 (control), thoc7d/Df, thoc5e/1, and rhino2/KG mutants. Sense reads are in red and antisense in blue.
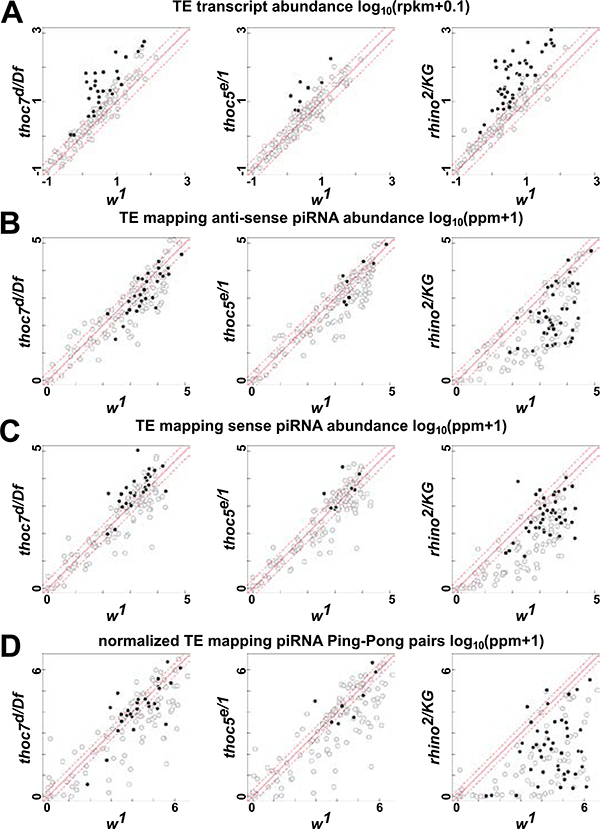
The scatter plots in Figure 2 compare mutant and wild type transcript expression, anti-sense piRNA abundance, sense piRNA abundance and normalized “ping-pong” pairs across all transposon families. Each scatter plot is an average of highly correlated deep sequencing replicates (Figure S1A–D). Using two-fold change as cutoff, these plots show that antisense piRNAs are not reduced for half of the transposon families that are over-expressed in thoc5 and thoc7 mutants (Figure 2A and B; black dots highlight overexpressed transposons with cutoff: Fold change>4 and FDR< 0.01). THO mutations also do not reduce the 10nt overlap between sense and anti-sense piRNAs, which is a hallmark of ping-pong biogenesis driven by catalytically active piRNA-Aub and -Ago3 complexes (Figure 2D). By contrast, rhino mutants reduce anti-sense piRNAs and the ping-pong signature for the majority of transposon families (Figure 2B and D). Thus, there is abundance sense piRNAs in thoc5 and thoc7 mutants, but not in rhino mutant (Figure 2C). The THO complex functions with UAP56 in the TREX complex, and thoc7 null and uap56 hypomorhic mutations reduce piRNA production from germline clusters (Zhang et al., 2012a; Zhang et al., 2018). However, hypomorphic uap56 mutant combination produces more significant reductions in anti-sense piRNA abundance than the thoc7 null (compare Figure 2B and Figure S2 second panel). We therefore speculate that UAP56, possibly within the TREX, has a primary function in Rhi- and piRNA-dependent transposon silencing. The THO heteropentamer, by contrast, appears to have an additional piRNA-independent role in transposon silencing.
Figure 2. Mutation in thoc7 and thoc5 lead to transposon overexpression without corresponding piRNA defects. See also Figure S1 and Figure S2.
Scatter plots compare transposon expression (A), transposon consensus mapping antisense piRNA abundance (B), sense piRNA abundance (C) and normalized ping-pong pairs (D) between thoc7d/Df, thoc5e/1, rhino2/KG and w1 control. Solid circles highlight overexpressed transposons in respective mutants over w1 (Fold change > 4 and FDR<0.01 see methods for details).
Thoc7 is required for transcriptional silencing
piRNAs guide transcriptional and post-transcriptional silencing (Czech et al., 2018; Ozata et al., 2019). Post-transcriptional silencing is driven by reciprocal RNA slicing by Aub and Ago3 during the ping-pong cycle (Brennecke et al., 2007). Ago3 bound piRNAs also initiate phased piRNA biogenesis, which produces Piwi-piRNA complexes that localize to the nucleus and guide transcriptional silencing (Wang et al., 2015). Mutations in thoc5 and thoc7 do not reduce the 10nt overlap between piRNAs from opposite strands, which is a signature of the ping-pong cycle (Figure 2D). However, thoc7 mutants disrupt Ago3 and Aub localization to nuage (Figure S1E), and reduce, but do not eliminate, Piwi localization to the nucleus (Figure S1E). These observations suggested that some piRNA-guided transcriptional and post-transcriptional silencing may persist in thoc7 mutants.
To directly examine the mechanism of transposon silencing by THO, we therefore quantified nascent transcript elongation using modified SLAM-seq pulse labeling (Herzog et al., 2017; Schofield et al., 2018). For these experiments, dissected ovaries were pulse labeled for 15 and 60 minutes with 4-thiouridine (4sU), which incorporates into nascent RNA during transcript elongation. RNA was then isolated and treated with iodoacetamide (IAA), generating carboxyamidomethyl modified 4sU, which is read as cytosine during reverse transcription (Herzog et al., 2017). 4sU incorporation into nascent transcripts was therefore estimated by the “T” to “C” conversion rate, measured by paired-end RNA-seq. The background “T” to “C” sequencing error rate, measured by sequencing unmodified RNA, was subtracted from the experimental signal (Figure S3A and B) (Herzog et al., 2017; Schofield et al., 2018).
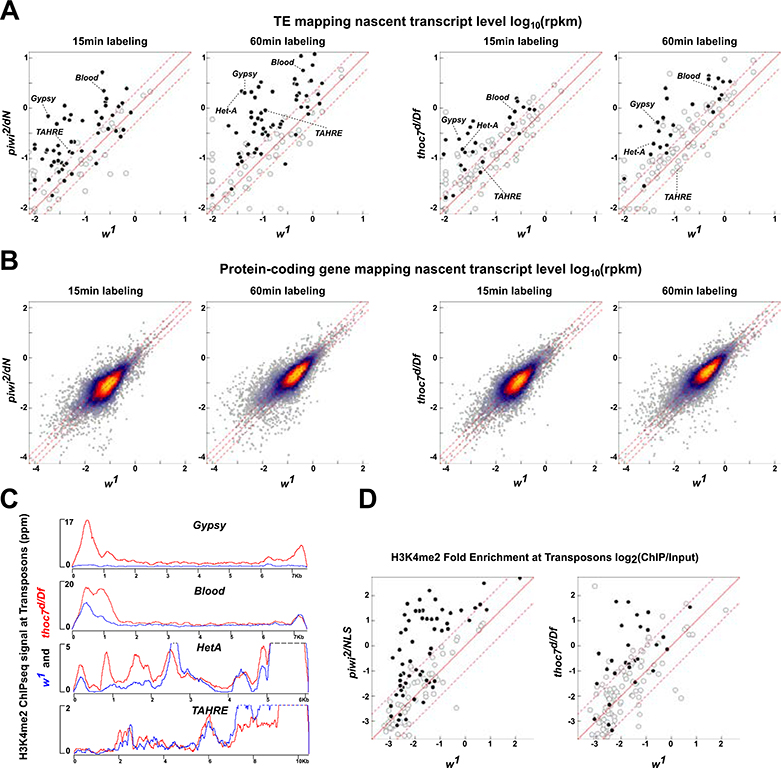
For these studies, and subsequent detailed molecular and genetic analyses, we focused on the null thoc7 allelic combination, which does not produce detectable RNA or protein. By contrast, the thoc5 allelic combination is hypomorphic, complicating interpretation of associated phenotypes. Piwi directs transcriptional silencing, and piwi nuclear localization mutants were therefore used as a positive control. The w1 strain was used a wild type control. Relative to w1, piwi and thoc7 mutations increased 4sU incorporation into transposon transcripts, but did not increase incorporation into protein-coding gene transcripts (Figure 3A and B). Mutations that disrupt THO thus appear to selectively increase transposon transcription.
Figure 3. Mutations in thoc7 lead to transposon transcriptional activation. See also Figure S3.
A. Scatter plots compare background subtracted nascent transposon transcripts signal, measured by SLAM-seq, between the indicated genotypes after 15 and 60 minutes of labeling. Solid circles indicate transposons that are overexpressed in respective mutants.
B. Heat scatter plots compare background subtracted nascent transcript signal for protein-coding genes between the indicated genotypes, after 15 and 60 minutes of labeling.
C. Gypsy, Blood, HeT-A and TAHRE consensus mapping H3K4me2 ChIP-seq signal in w1 (blue) and thoc7d/Df mutants (red).
D. Scatter plots comparing fold-enrichment of H3K4me2 ChIP-seq signal (ChIP/input) mapping to the first two kilo-bases of transposon consensus sequences, in piwi2/NLS, thoc7d/Df and w1. Solid circles are transposons overexpressed in respective mutants comparing to w1.
Transcriptionally active promoters are marked by H3K4me2/3, and piwi nuclear localization mutants increase H3K4me2/3 at transposons promoters (Klenov et al., 2014; Le Thomas et al., 2013; Sienski et al., 2012). We therefore assayed H3K4me2 in thoc7 mutants and w1 controls by ChIP-seq. Figure 3C shows H3K4me2 signal mapping across the consensus sequences of the four transposon families shown in Figure 1. Gypsy, Blood, HeT-A are overexpressed in thoc7 mutants, and all three show higher H3K4me2 signal at 5’ end of the consensus sequence, which carries promoter elements. TAHRE is not overexpress in thoc7 mutants, and does not show increased H3K4me2. The scatter plots in Figure 3D show H3K4me2 (ChIP/input) signal at the 5’ ends of all transposon consensus sequences in thoc7 and piwi mutants, relative to w1 controls. Both mutations lead to a global increase in H3K4me2 at transposon promoters, with higher H3K4me2 modification and transposon expression in piwi mutant (Figure S3C and D). By contrast, only 115 of the 5607 protein-coding gene promoters showed a two-fold or greater increase in H3K4me2 in thoc7 mutants (Figure S3E and see Experimental Procedures). RNA-seq revealed only 108 protein-coding genes with increased steady state expression in thoc7 mutants (Cutoff: Fold change>4 and FDR< 0.01), but the promoters for these genes did not show a corresponding increase in H3K4me2. The combined SLAM-seq and ChIP-seq data thus indicate that thoc7 mutations selectively increase transcription of transposons.
Thoc7 mutations do not reduce H3K9me3 at transposon insertions.
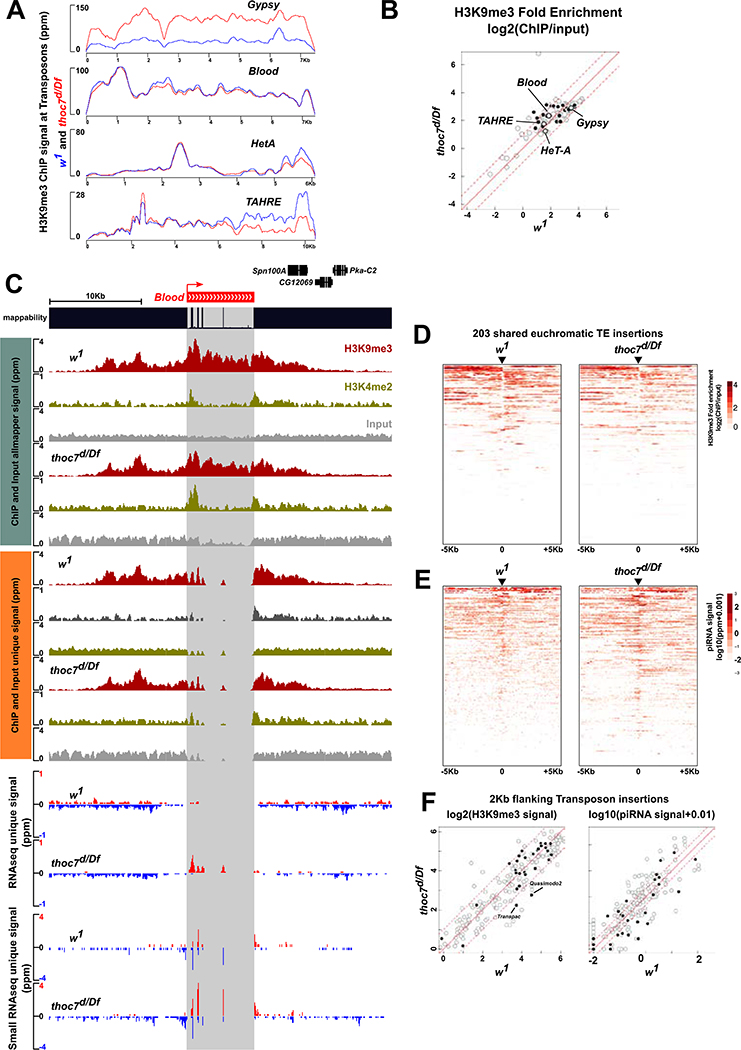
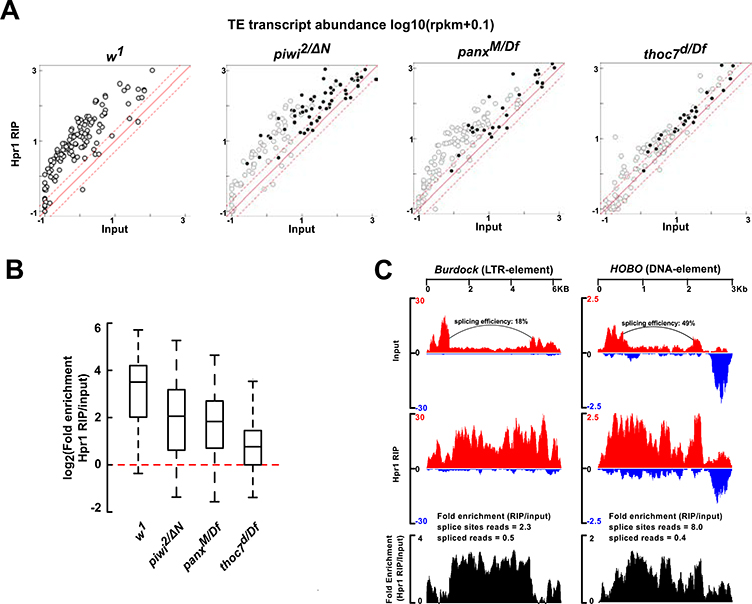
piRNAs bound to Piwi are proposed to base pair with nascent transcripts and recruit accessory factors, including Panx, which promotes inhibitory H3K9me3 chromatin modification over the body of transposon insertions (Sienski et al., 2015; Sienski et al., 2012; Yu et al., 2015). To determine if thoc7 mutations disrupt this repressive chromatin modification, we assayed H3K9me3 in thoc7 mutants by ChIP-seq. As shown in Figure 4A, thoc7 mutations do not alter ChIP-seq signal mapping to consensus sequences of four example transposons, and the scatter plot in Figure 4B shows that this extends to all transposon families.
Figure 4. Mutations in thoc7 do not affect piRNA mediated H3K9me3 deposition. See also Figure S4.
A. H3K9me3 ChIP-seq signal mapping to Gypsy, Blood, HeT-A and TAHRE transposon consensus sequences in w1 (blue) and thoc7d/Df (red).
B. Scatter plot compares fold-enrichment of transposon consensus mapping H3K9me3 ChIP-seq signals (ChIP/input) in thoc7d/Df and w1. Solid circles are transposons overexpressed in thoc7d/Df over w1, and four representative transposons in A are labeled.
C. Genome browser view of a Blood insertion on chromosome 3R that is present in thoc7d/df, w1, and the reference genome. Signals of H3K9me3 ChIP-seq, H3K4me2 Chip-seq and corresponding input, long RNA-seq and small RNA-seq are shown, for both unique and multi-mapping reads. The mappability track reflects the uniqueness of the sequences.
D. The heat maps show average H3K9me3 ChIP-seq signal of two biological replicates, for regions 5 kb up- and down-stream of transposon insertions present in thoc7d/Df and w1. Both heat maps were sorted based on decreasing H3K9me3 signal in the w1 control.
E. The heat maps show average small RNA signal (sum of both genomic strands) of two biological replicates flanking shared transposon insertions as in D. Both heat maps were sorted based on decreasing small RNA signal in the w1 control.
F. Scatter plots compare the total H3K9me3 (left) and small RNA signal (right) at one kilobase regions flanking individual transposon insertions from heat maps in D and E between thoc7d/Df and w1. Solid circles are transposons overexpressed in thoc7d/Df over w1. Two insertions of overexpressed transposon families with more than two folds reduction in H3K9me3 signal are labeled (left panel).
These findings suggested that THO is not required for piRNA-guided H3K9me3 modification, but most transposon insertions are truncated, transcriptionally inactive, and embedded in constitutive heterochromatin marked by H3K9me3. Signal from these “dead” elements could mask changes in H3K9me3 at active transposons. To determine if thoc7 mutants alter chromatin at active elements, we assayed H3K9me3 in unique regions flanking dispersed euchromatic insertions (Sienski et al., 2015; Sienski et al., 2012). For this analysis, we used input genomic sequence from our ChIP-seq experiments to define the locations of euchromatic insertions shared by thoc7 mutants and w1. We also found three insertions that were present in thoc7, w1, and the reference genome (Figure 4C, S4A and B). We used the reference genome sequence to identify unique internal polymorphisms within these insertions, and mapped short RNAseq, long RNA-seq, H3K9me3 and H3K4me2 ChIP-seq signal to the unique internal sites and to unique flaking regions. For all three insertions, thoc7 mutations increased transcript expression and promoter H3K4me2 modification, but did not reduce piRNAs or H3K9me3 mapping to internal regions or flanking sequences (Figure 4C and Figure S4A and B). We next analyzed H3K9me3 ChIP-seq signal mapping to unique sequences flanking all 203 euchromatic transposon insertions that are shared by thoc7 and w1. As shown in the heat maps in Figure 4D, thoc7 mutations do not significantly alter H3K9me3 signal flanking these insertions, with the vast majority showing less than a two-fold change in signal (Figure 4F left panel). The thoc7 null combination thus disrupts transposon silencing, but does not alter piRNA-directed H3K9me3 modification.
A subset of euchromatic transposon insertions function as “mini-clusters” that are bound by Rhi, transcribed from both strands, and produce piRNAs. These insertions can be identified by piRNAs mapping to unique flanking sequences (Mohn et al., 2014; Parhad et al., 2020). As shown in the heat map in Figure 4E and scatter plot in Figure 4F (right panel), thoc7 mutations do not reduce piRNA expression from these mini-clusters. This is in striking contrast to germline clusters in pericentric heterochromatin, which show significantly reduced piRNA expression in thoc5 and thoc7 mutants (Hur et al., 2016; Zhang et al., 2018). The intact THO complex thus promotes Rhi-dependent piRNA biogenesis from heterochromatic clusters, but is not required for Rhi-dependent piRNA production from euchromatic insertions.
Thoc7 is not required for Panoramix mediated silencing.
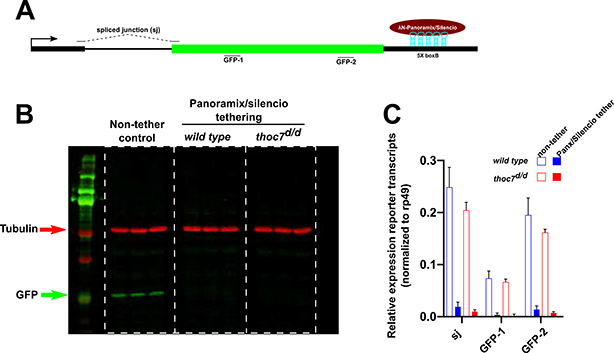
Panx binds Piwi/piRNA complexes, which associate with nascent transcripts and recruit histone-modifying enzymes, establishing H3K9me3 marks associated with transcriptional silencing (Sienski et al., 2015; Yu et al., 2015). To determine if Tho7/THO functions downstream of Panx binding during transcriptional silencing, we exploited a transgenic assay in which a λN-Panx fusion is tethered to the 3’UTR of a GFP reporter (Figure 5A) (Sienski et al., 2015). The reporter was crossed into control and thoc7 mutants, and expression was assayed in the presence and absence of the λN-Panx fusion, controlled by the constitutive Act5c->Gal4 driver, as diagrammed in Figure S5A (Sienski et al., 2015). In both w1 and thoc7 mutants, tethering λN-Panx silences GFP protein and RNA expression (Figure 5B and C). These findings, with our analysis of histone modifications at endogenous targets, indicate that THO is not required for Piwi/piRNA guided repressive chromatin modification.
Figure 5. THO is not required for Panoramix/Silencio mediated transgene silencing. See also Figure S5.
A. Schematic representation of the Panoramix/silencio RNA tethering reporter. Also shown are the positions of qPCR probes.
B. Western blot for GFP (green) upon tethering the indicated proteins to the transgenic reporter in the indicated genotypes. Three biological replicates are loaded for each experiment condition. Tubulin (red) serves as loading control.
C. Relative expression of reporter transcripts (2-ΔCt value normalized to rp49) under indicated experimental conditions. The bar graphs represent three biological replicates under each experimental condition.
Retrotransposon transcripts co-precipitate with intact THO.
THO associates with RNA Pol2 and is recruited to nascent gene transcripts co-transcriptionally (Jimeno and Aguilera, 2010; Zhang et al., 2018). An analysis of our published RIP-seq data showed that transposon transcripts co-precipitate with the core THO complex protein Hpr1 (Figure 6A and B) (Zhang et al., 2018). Splicing is co-transcriptional, and intron sequences thus provide an indirect marker for nascent transcripts. In w1 ovaries, 95 transposon families are expressed at greater than 1 rpkm, but only two of these families produce spliced mRNAs. For these two families, only the unspliced transcripts co-precipitate with Hpr1 (Figure 6C). THO thus appears to co-transcriptionally associate with unspliced transposon transcripts.
Figure 6. THO bind unspliced transposon transcripts.
A. Scatter plots comparing transposon transcript abundance between Hpr1 RIP and input in the indicated genotypes. Solid circles are transposons overexpressed in the respective mutants relative to w1.
B. The boxplot compares fold enrichment of transposon transcripts (Hpr RIP/input from A) among indicated genotypes.
C. RNA-seq profile from Hpr1 RIP with input and fold enrichment in Hpr1 RIP (Hpr1 RIP/input) mapping to consensus sequences of two transposon families that produce spliced transcripts.
Transposon transcripts show a modest reduction in Hpr1 binding in both piwi and panx mutants (Figure 6A and B). This could reflect a role for Piwi-Panx in promoting THO binding. However, these mutations disrupt transposon silencing and lead genome instability, indicating that transposon transcripts are exported from the nucleus and translated. Because THO is restricted to the nucleus, reduced Hpr1 binding may reflect inaccessibility of the cytoplasmic transcripts. By contrast, transposon transcript binding to Hpr1 is nearly eliminated by thoc7 mutations (Figure 6A and B). Transposon transcripts thus associate with the intact THO heteropentamer, through a process that does not require Piwi or Panoramix, although these factors could promote binding.
Synergistic genetic interactions between thoc7 and piwi.
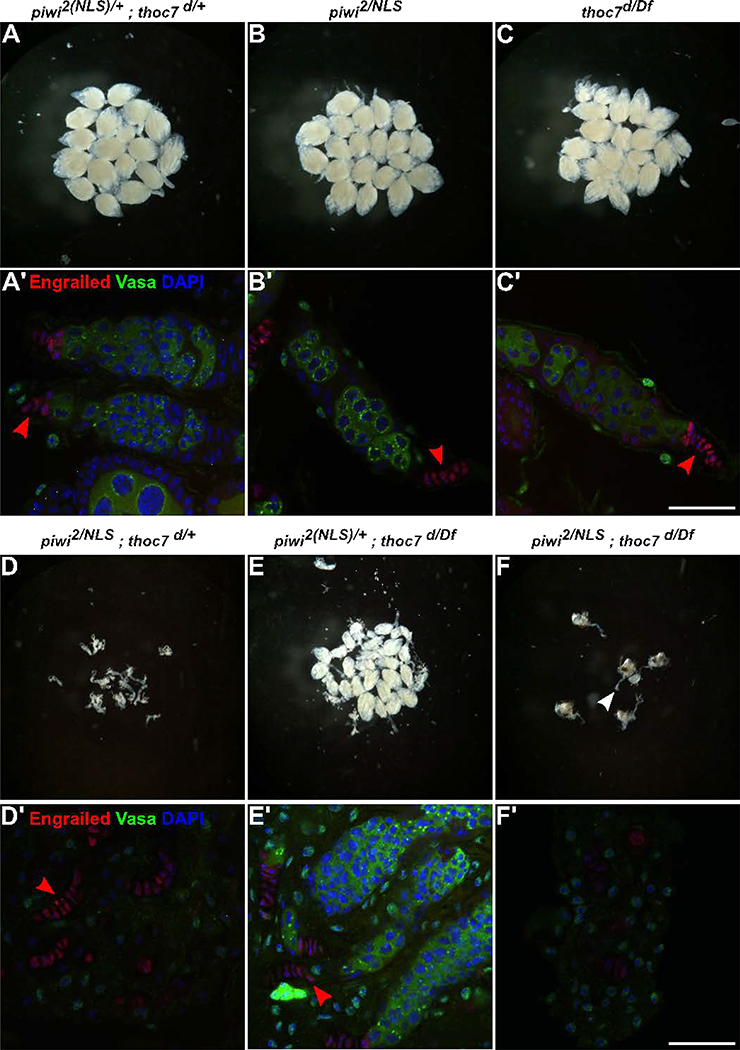
The studies described above strongly imply that Thoc7 functions in a Piwi independent transposon silencing system, predicting that thoc7 and piwi mutations will show synergistic genetic interactions. We therefore analyzed a series of thoc7 and piwi mutant combinations. For these studies, we used the null thoc7 allelic combination (thoc7d/Df), which is viable and does not alter gene expression, germline development or oogenesis, but disrupts transposon silencing (Zhang et al., 2018). Null piwi mutations, by contrast, lead to arrest during early oogenesis, which reflects a function for piwi in the somatic follicle cells (Cox et al., 1998; Cox et al., 2000; Jin et al., 2013). However, a piwi nuclear localization mutation (piwi-NLS), when combined with a null allele (piwi2), disrupts transcriptional silencing of transposons in the germline, but supports Piwi function in the soma and allows germline development and oogenesis (Klenov et al., 2011). To assay for genetic interactions between THO and Piwi in the germline, we therefore analyzed combination of piwi-NLS and thoc7 null mutants. Ovaries from thoc7d/Df, piwi-NLS/piwi2 and trans-heterozygous thoc7d/+; piwi-NLS/+ females are essentially identical to w1 controls (Figures 7A–C). By contrast, thoc7d/Df; piwi-NLS/+ females produce ovaries with reduced size (Figure 7E), and piwi-NLS/piwi2; thoc7d/+ females have rudimentary ovaries (Figure 7D). Strikingly, in thoc7d/Df; piwi-NLS/piwii2 double mutants females have somatic oviducts, but appear to lack ovarian tissue (Figure 7F).
Figure 7. Genetic interactions between thoc7 and piwi during ovary development. See also Figure S6.
A-F. Light microscope images (28X zoom) of Drosophila ovaries from the indicated genotypes. The heterozygous control and single mutant females carry morphologically similar ovaries (A-C). The thoc7 null allele dominantly enhances oogenesis defects associated with piwi mutations (D), and piwi mutations dominantly enhance the thoc7 null combination (E). There are no identifiable ovaries in piwi2/NLS; thoc7d/Df double mutants (F). White arrowhead marks the end of the oviduct, lacking attached ovaries. A’-F’. Immunofluorescence (IF) images of germaria from the indicated genotypes, stained with Engrailed (Red) and Vasa (Green). Engrailed marks the terminal filament cells (TF) and cap cells of the germline stem cell niche (red arrowheads in the images). Vasa marks the germline stem cells and developing germline cyst. Scale bar: 25μm.
To characterize these phenotypes in greater detail, we labeled the mutant combinations for Vasa and Engrailed. Vasa is a conserved DEAD box protein that marks the germline stem cells at the tip of the germarium, and developing germline cysts that bud from the germarium (Hay et al., 1990; Lasko and Ashburner, 1990). Engrailed is expressed in the somatic terminal filament (TF) cells and cap cells that surround the germline stem niche (Barton et al., 2016). In w1 controls and the single mutants, we consistently observed Vasa positive germline cells associated with Engrailed positive TF cells in the germarium, which were followed by Vasa positive egg chambers (Figures 7A’–C’ and E’). By contrast, the tip of piwi-NLS/piwii2; thoc7d/+ ovarioles frequently carried stacked Engrailed positive TF cells without neighboring Vasa positive cells, indicating that somatic ovary structures are present, but the germline is absent (Figure 7D’). In this mutant combination, each individual ovary contained 11.3 ± 3 TF arrays (nine ovaries scored), which is comparable to wild type (Bastock and St Johnston, 2008). However, only eight ovarioles in nine total ovaries had Vasa positive cells in association with the TF arrays (0.89 ± 1.36 per ovary lobe). In thoc7d/Df; piwi-NLS/piwii2 double mutants, we did not detect any Engrailed positive terminal filament arrays or Vasa positive cells (Figure 7F’). Nuclear localization mutations in piwi thus dominantly enhance the thoc7 null combination, leading to loss of somatic and germline cells of the ovary.
We also sought to determine if piwi and thoc7 mutants produce synergistic defects in transposon silencing. However, the piwi-NLS/piwii2; thoc7d/+ and thoc7d/Df; piwi-NLS/piwii2 mutant females have rudimentary ovaries that lack germline cells, and could not be assayed (Figure 7D and F). By contrast, thoc7d/Df; piwi-NLS/+ females produce ovaries, and thoc7d/+; piwi-NLS/+ double heterozygous females are fertile and have wild type ovaries. We therefore assayed for dominant enhancement of the thoc7 null transposon silencing defects by comparing Gypsy, Blood, HeT-A and TAHRE expression in the thoc7d/Df to thoc7d/Df; piwi-NLS/+. The fertile double heterozygous combination was used as a control. Both THARE and HeT-A showed significantly enhanced expression in thoc7d/Df; piwi-NLS/+ relative to thoc7d/Df (Figure S6A). By contrast, Gypsy and Blood showed only modest expression increases in the thoc7d/Df; piwi-NLS/+ combination, which were not statistically significant (Figure S6A). Reducing piwi dosage thus enhances both the developmental and transposon silencing defects associated with thoc7 mutations.
These findings are consistent with a piRNA-independent function for the THO complex in transposon silencing, but the thoc7 null mutant combination reduces piRNAs mapping to THARE and HeT-A (Figure 1), which also show the most significant increases in expression in thoc7d/Df; piwi-NLS/+ mutants (Figure S6A). These telomeric LINE elements are also over-expressed in piwi mutants. We therefore speculate that HeT-A and THARE are primarily silenced through piRNAs, which are partially dependent on THO. Reduced piRNA expression in thoc7 mutants thus sensitizes these elements to reduced Piwi dosage, which compromise piRNA silencing. By contrast, mutations in thoc7 and piwi disrupt transcriptional silencing of Blood and Gypsy (Figure 3A), and reducing piwi in the thoc7 mutant background does not enhance expression of these elements (Figure S6A). Mutations in thoc7 also fail to reduce anti-sense piRNAs targeting Blood and Gypsy (Figure 1), or reduce H3K9me3 modification of these elements (Figure 4A). We therefore speculate that a subset of elements, including Blood and Gypsy, are redundantly silenced by piRNAs and a piRNA-independent and THO-dependent mechanism (Figure S6B). How the transposon family-specific balance of these silencing mechanisms is determined remains to be explored.
Discussion
piRNA-independent transposon silencing by THO.
piRNAs guide an adaptive genome immune system that provides sequence specific transposon silencing during germline development. In this system, immune “memory” is carried in heterochromatic clusters and euchromatic “mini-cluster” insertions (Aravin et al., 2007; Mohn et al., 2014; Ophinni et al., 2019). Mutations in Drosophila thoc5 and thoc7 disrupt the heteropentameric THO complex, reduce expression of piRNAs from germline clusters, and increase expression of a subset of transposon families. Cluster transcripts also co-precipitate with Hpr1 and Thoc5, supporting a simple model in which THO binding to cluster transcripts promotes biogenesis of germline piRNA, which guide transposon silencing (Hur et al., 2016; Zhang et al., 2018). Here we show that thoc7 mutations increase transposon transcription (Figures 3), but do not reduce anti-sense piRNAs targeting half of the activated transposon families (Figures 1 and 2). In addition, piRNAs bound by Piwi function with Panx-dependent H3K9me3 modification of target elements, and thoc7 mutations do not block Panx-dependent silencing of a reporter transgene (Figure 5), or reduce H3K9me3 at transposon insertions (Figure 4). Significantly, even transposons families that show reduced piRNA targeting retain H3K9me3, implying that the piRNAs that are expressed in thoc7 mutants are sufficient to support Piwi/piRNA dependent chromatin modification. These observations, with the strong synergistic defects in germline development produced by thoc7/piwi mutant combinations (Figure 7), suggest that THO promotes biogenesis of a subset of germline piRNAs, but also functions in a piRNA-independent transposon silencing system.
These observations imply that the THO-dependent piRNAs produced from germline clusters are not essential to Piwi-mediated silencing. However, these piRNAs could function redundantly with piRNAs derived from dispersed insertions, which are produced by a THO-independent mechanism (Figure 4E). Alternatively, cluster piRNAs could primarily target older, inactive elements, and provide a genetic “memory” of previously active transposons. In this model, the clusters protect against genome invasion by transposons related to older elements, but are dispensable for silencing newer, active transposons. Both possibilities are consistent with the recent finding that deletion of several major piRNA clusters does not compromise transposon silencing or fertility (Gebert et al., Mol. Cell, in press).
Sequence-specific and sequence-independent genome immunity?
Adaptive immunity is acquired during pathogen exposure, specific to prior invaders, and carries immune memory (Marshall et al., 2018). Transposons that insert into heterochromatic clusters during genome invasion produce piRNAs that guide sequence-specific silencing and provide a hardwired genetic memory of the invading element (Andersen et al., 2017; Aravin et al., 2007; Brennecke et al., 2007). In addition, conversion of dispersed insertions into piRNA producing loci, through a mechanism that is not understood, appears to provide epigenetic memory of past invaders (Mohn et al., 2014). The established piRNA system thus provides sequence-specific “adaptive” genome immunity. Innate immune systems recognize sequence-independent Pathogen Associated Molecular Patterns (PAMPs), which are detected by pattern recognition receptors (Kumar et al., 2011). We speculate that THO functions in an “innate” genome immune system, which recognizes and silences target transposons through a sequence-independent mechanism (Figure S6B).
Distinguishing self from non-self is the basis of all immune systems, and thoc7 mutations disrupt silencing of transposons (non-self) but do not significantly alter gene expression (self). How are transposons distinguished from genes? Intriguingly, LTR retrotransposons, but not DNA transposable elements, are significantly over-expressed in thoc7 mutants. These endogenous retroviruses rely on very inefficient splicing to generate unspliced genomic transcripts, which encode the gag-pol polyprotein, and spliced transcripts encoding the Envelope protein. Analysis of RNAseq data from the control w1 strain indicates that 93 of 95 expressed transposon families produce only unspliced transcripts, and splicing is very inefficient at the remaining 2 families (Figure 6C). In striking contrast, we find that 13,961 protein-coding genes expressed at 1 RPKM or more, and all 11,651 with annotated introns are spliced at greater than 90% efficiency, and the remaining 2,265 are intronless and are never spliced. Significantly, intronless genes are not over-expressed in thoc7 mutants. Inefficient splicing thus appears to be specific to retrotransposons, and we speculate that this helps generate a sequence-independent PAMP that is recognize by THO, triggering transcriptional silencing (Figure S6B).
Observations in remarkably diverse systems implicate failed or defective splicing in small RNA biogenesis and transposons silencing. Pioneering studies in the pathogenic yeast Cryptococcus first showed that stalled splicing intermediates are processed into siRNAs, which silence transposons in trans (Dumesic et al., 2013). In addition, long unspliced transcripts co-precipitate with THO and are processed into piRNAs in flies and mice (Yu et al., 2021; Zhang et al., 2014). In addition, in C. elegans, genes producing unspliced transcripts are silenced through a piRNA-independent process. In this system, the unspliced transcripts are processed into siRNAs, which are loaded into Worm Argonauts (WAGOs) and trans-silence homologous spliced targets. Mutations that disarm the C. elegans WAGO pathway block trans-silencing, but do not prevent silencing of the “unspliced” source locus. In worms, expression of unspliced transcripts thus leads to small RNA-independent silencing in cis, and production of siRNAs that silence in trans (Makeyeva and Mello, personal communication). Finally, the KoRV-A retrovirus is invading the Koala genome, and unspliced KoRV-A transcripts, but not the more abundant spliced KoRV-A ENV mRNAs, are selectively processed into sense strand piRNAs (Yu et al., 2019). By consuming RNAs that encode the Gag-Pol fusion protein and the viral genome, processing of unspliced KoRV-A transcripts into piRNAs could provide an “innate” form of genome immunity (Yu et al., 2019). The splicing and small silencing RNA pathways also appear to be co-evolving (Tabach et al., 2013). Inefficient or failed splicing may therefore represent a deeply conserved non-self signal, which transcriptional or post-transcriptional silencing of targets through diverse downstream mechanisms.
Limitations of the study
The THO complex has been previously implicated in piRNA biogenesis, but here we show that mutations that disrupt this complex increase transcription of a subset of transposon families without reducing piRNAs or piRNA-directed inhibitory H3K9me3 modifications. These findings suggest that THO has a piRNA-independent function in transposon silencing, which may act redundantly with sequence specific silencing by the piRNA pathway. However, our studies do not define how transposon families that show piRNA-independent silencing are differentiated from families that are silenced primarily by piRNAs, or how THO drives piRNA-independent transcriptional silencing.
STAR Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, William E. Theurkauf (william.theurkauf@umassmed.edu).
Materials availability
Fly strains used in this study are all available on request.
Data and code availability
RNAseq, Small-RNAseq and ChIPseq data generated in this study have been deposited at the NCBI BioProject and Sequence Read Archive (SRA) as of the date of publication. Accession numbers for existing, publicly available data are listed in the Key Resources Table.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-Hpr1 | (Rehwinkel et al., 2004) | |
| Rabbit anti Piwi | (Li et al., 2009a) | |
| Rabbit anti Ago3 | (Li et al., 2009a) | |
| Rabbit anti Aub | (Li et al., 2009a) | |
| Rat IgM anti-Vasa | DSHB | |
| Mouse anti-Engrailed (4D9) | DSHB | |
| Rabbit anti H3K9me3 | Abcam | Cat# ab8898 |
| Rabbit anti H3K4me2 | EMDMillipore | Cat# 07–030 |
| Mouse Anti-α-Tubulin | Sigma Aldrich | Cat# T5168 |
| Rabbit anti-GFP | ThermoFisher Scientific | Cat# A11122 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-Thiouridine | MilliporeSigma | Cat# T4509 |
| Iodoacetamide | MilliporeSigma | Cat# I1149 |
| Superscript III | ThermoFisher Scientific | Cat# 18080–085 |
| RNase OUT | ThermoFisher Scientific | Cat# 10777–019 |
| TURBO DNase | ThermoFisher Scientific | Cat# AM2238 |
| RNaseH | ThermoFisher Scientific | Cat# 18021–071 |
| T4 RNA Ligase | ThermoFisher Scientific | Cat# AM2141 |
| AccuPrime™ Pfx DNA Polymerase | ThermoFisher Scientific | Cat# 12344024 |
| TRIzol™ Reagent | ThermoFisher Scientific | Cat# 15596026 |
| UltraPure™ Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v) | ThermoFisher Scientific | Cat# 15593031 |
| dNTP Set (100 mM) | ThermoFisher Scientific | Cat# 10297018 |
| dUTP Solution (100 mM) | ThermoFisher Scientific | Cat# R0133 |
| Grace's Insect Medium, unsupplemented | ThermoFisher Scientific | Cat# 11595030 |
| Hybridase | Lucigen | Cat# H39500 |
| dNTP mix | NEB | Cat# N0447L |
| DNA polymerase I | NEB | Cat# M0209S |
| T4 DNA polymerase | NEB | Cat# M0203L |
| Klenow DNA polymerase | NEB | Cat# M0210S |
| T4 PNK | NEB | Cat# M0201L |
| Klenow 3’ to 5’ exo | NEB | Cat# M0212L |
| UDG | NEB | Cat# M0280S |
| Phusion Polymerase | NEB | Cat# M0530S |
| T4 RNA Ligase 2, truncated | NEB | Cat# M0242L |
| 50% PEG8000 | NEB | Cat# B1004S |
| T4 DNA ligase | Enzymatics Inc. | Cat# L6030-HC-L |
| EDTA-free Protease Inhibitor Cocktail (Roche) | Sigma | Cat# 11873580001 |
| 16% formaldehyde | Ted Pella Inc | Cat# 18505 |
| Miracloth membrane (Calbiochem) | EMDMillipore | Cat# 475855 |
| Critical Commercial Assays | ||
| mirVANA™ miRNA isolation kit | ThermoFisher Scientific | Cat# AM1560 |
| RNeasy Mini Kit | Qiagen | Cat# 74104 |
| QuantiTect SYBR® Green PCR Kits | Qiagen | Cat# 204145 |
| Dynabeads® Protein G | ThermoFisher Scientific | Cat# 10004D |
| Dynabeads® Protein A | ThermoFisher Scientific | Cat# 10001D |
| RNA Clean & Concentrator-5 | Zymo Research | Cat# R1015 |
| In-Fusion® HD Cloning Plus | Takara | Cat# 638909 |
| Agencourt AMPure XP | Beckman Coulter | Cat# A63880 |
| Deposited Data | ||
| High throughput Sequencing | This study | PRJNA590287 |
| small RNAseq for thoc5[e00906]/thoc5[1]_rep1 | (Zhang et al., 2018) | SRR7686976 |
| small RNAseq for thoc5[e00906]/thoc5[1]_rep2 | (Zhang et al., 2018) | SRR7686977 |
| small RNAseq for w[1]_rep1 | (Zhang et al., 2018) | SRR7408119 |
| small RNAseq for thoc7[d05792]/Df(3L)BSC128 | (Zhang et al., 2018) | SRR7408136 |
| Small RNAseq of rhi mut ovary | (Parhad et al., 2017) | SRR5803097 |
| RNAseq for thoc5[e00906]/thoc5[1]_rep1 | (Zhang et al., 2018) | SRR7408102 |
| RNAseq for thoc7[d05792]/Df(3L)BSC128_rep2 | SRR7408139 | |
| input RNAseq from w[1]_rep1 | (Zhang et al., 2018) | SRR7408152 |
| input RNAseq from w[1]_rep2 | (Zhang et al., 2018) | SRR7408157 |
| Hpr1IP RNAseq from w[1]_rep1 | (Zhang et al., 2018) | SRR7408151 |
| Hpr1IPRNAseq from w[1]_rep2 | (Zhang et al., 2018) | SRR7408156 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster/thoc7d05792 | Harvard Exelixis stock collection | Stock # d05792 |
| D. melanogaster/Df(3L)BSC128 | Bloomington Drosophila Stock Center | Stock# 9293 |
| D. melanogaster/thoc5e00906 | Harvard Exelixis stock collection | Stock# e00906 |
| D. melanogaster/thoc51 | (Moon et al., 2011) | N.A. |
| D. melanogaster/rhino2 | (Klattenhoff et al., 2009) | N.A. |
| D. melanogaster/rhinoKG | (Klattenhoff et al., 2009) | N.A. |
| D. melanogaster/piwi2 | Bloomington Drosophila Stock Center | Stock# 43319 |
| D. melanogaster/piwiNLS | (Klenov et al., 2011) | N.A. |
| D. melanogaster/panoramixM4 | (Yu et al., 2015) | N.A. |
| D. melanogaster/Df(2R)BSC821 | (Yu et al., 2015) | Stock# 27582 |
| D. melanogaster/ pUASp>lambdaN-HA-CG9754 [attP40]/CyO; tub>EGFP_5xBoxB_SV40 [attP2]/TM3, Ser; | VDRC Stock Center | Stock# 313393 |
| D. melanogaster/w1 | William Theurkauf lab | N.A. |
| Oligonucleotides | ||
| Random primers | ThermoFisher Scientific | Cat# 48190011 |
| Primers for qPCR, see Table S1 | N.A. | |
| Software and Algorithms | ||
| Prism 7 | GraphPad Prism | https://www.graphpad.com/ |
| Image Studio™ Lite | LI-COR | https://www.licor.com/bio/products/software/image_studio_lite/ |
| RStudio | https://www.rstudio.com/ | |
| ImageJ | https://imagej.nih.gov/ij/ | |
| UCSC Genome Browser | (Kent et al., 2002) | https://genome.ucsc.edu/cgi-bin/hgGateway |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All genetic crosses were maintained at 25oC on standard cornmeal medium. All experiments were performed on ovaries from 2–4 days old female Drosophila melanogaster raised in the presence of yeast paste. The w1 strain served as general wild type in this study. The fly strains used in this study are listed in Key resource table.
METHOD DETAILS
Immunofluorescent staining and image acquisition
The antibody used for IF are listed in the Key resource table. Fixation and immuno-staining of Drosophila ovaries was performed with Buffer A protocol as described previously (Theurkauf, 1994). The images were acquired with Leica TCS SP8 confocal microscope.
Western blotting, RNA and Chromatin immune-precipitation and RT-qPCR
2–4 day old Drosophila ovaries were lysed in RIPA lysis buffer (Tris.HCl 25mM pH 7.6; NaCl 150mM; Na Deoxycholate 1%; SDS 1%) supplied with protease inhibitor (Roche). The concentration of the lysate was measured by BCA kit (Pierce). The same amount of protein lysates were resolved by 10% SDS-PAGE and transferred to Nylon membrane (Amersham Biosciences). The blot was blocked by Li-COR blocking buffer and sequentially probed by anti-GFP and anti-tubulin antibody and corresponding secondary antibodies using Li-COR protocol. The images were acquired by LI-COR Odyssey Infrared Imaging System.
RNA Immuno-precipitation (RIP) from ovary lysate was performed as described previously (Zhang et al., 2018). In brief, sixty flies’ ovaries were lysed in NP40 lysis buffer (HEPES 50mM pH 7.5, KCl 150mM, MgCl2 3.2mM, NP40 0.5%, PMSF 1mM, Proteinase Inhibitor (Roche) 1X) by homogenizing pestle and sonication. The lysate was cleared by centrifugation. The Rat anti Hpr1 antibody was first conjugated to Magnetic Dynabeads protein G (Invitrogen) in Citric phosphate buffer (7.10 g Na2HPO4, 11.5g Citric acid in 1 liter water, ph 5.6) for 2 hours at room temperature with rotation and washed with Citric phosphate buffer with 0.1%Tween 20 and lysis buffer. The antibody-conjugated beads were incubated overnight at 4oC with lysate and washed three times with lysis buffer. The washed beads were resuspended in RTL buffer from RNeasy mini kit to extract RNA (Qiagen) and the kit procedure was followed to purify RNA.
The Chromatin Immuno-precipitation (ChIP) was performed as previously described with sixty flies’ ovaries per ChIP with Anti-H3K9me3 (Abcam) and Anti-H3K4me2 (EMDMillipore/upstate) (Parhad et al., 2017; Zhang et al., 2014).
The qPCR was performed using the QuantiTect SYBR Green PCR Kit (Qiagen) with Step ONE plus real time PCR system (Applied Biosystem). PCR primer sequences are presented in Table S1. The results are graphed using Prism 7 (GraphPad).
Nascent transcript labeling
The protocol for labeling nascent transcript in Drosophila ovaries was adapted from SLAM-seq protocol (Herzog et al., 2017; Schofield et al., 2018). Around fifteen flies’ ovaries were teased apart in room temperature unsupplemented Grace’s Insect Medium (Invitrogen). The dissociated ovaries were incubated with 100 μM 4-Thiouridine (4SU) in unsupplemented Grace’s Insect Medium at room temperature for 15mins or 60mins and snap frozen in liquid nitrogen to stop labeling. The RNA was isolated with mirVana miRNA Isolation Kit (Ambion). The total RNA was treated by iodoacetamide as in described in SLAM-seq (10 to 15 μg total RNA; 5 μl of 100mM iodoacetamide; 50mM NaPO4 Ph8 and 50% DMSO to a final volume of 50 μl)(Herzog et al., 2017). Equal amount of RNA was mock treated with ethanol (solvent) under same condition as negative control (10 to 15 μg total RNA; 5 μl ethanol; 50mM NaPO4 Ph8; and 50% DMSO to a final volume of 50 μl). The reactions were stopped by adding 1μl 1M Dithiothreitol. The treated RNA is purified by ethanol precipitation with glycogen as carrier.
High-throughput sequencing
Strand specific RNA-seq libraries were constructed as described previously (Zhang et al., 2012b) with modification in the rRNA depletion procedure using enzymatic digestion of rRNA by Hybridase™ Thermostable RNase H (Epicenter) with a comprehensive mixture of antisense rRNA oligos (Fu et al., 2018). The small RNA-seq library is constructed as detailed previously (Li et al., 2009a) with 2S rRNA depletion as described in (Zhang et al., 2011). The ChIP-seq libraries were prepared as described previously (Zhang et al., 2014), RNA-seq and ChIP-seq libraries were paired-end sequenced, and small RNA-seq libraries were single-end sequenced on the Nextseq 500 platform (Illumina).
Bioinformatics Analysis
The bioinformatics analysis was performed as described previously (Yu et al., 2019). The Drosophila reference genome (dm6), gene annotations, rRNA sequences and hairpin sequences are obtained from Flybase (Version 6.13).
Transposon consensus and annotations:
The transposon consensus sequences were downloaded from Repbase (Bao et al., 2015). For transposons from Repbase, LTR transposons with both flanking LTRs and internal sequences were merged with LTR-int-LTR order. For RepeatMasker annotation, transposon names were fixed accord to fixed Repbase name and then the same transposon copies within 200 bps were merged. The merged transposon consensus sequences and genomic insertion annotations were used for downstream analysis.
RNA-seq:
The raw RNA-seq reads were first mapped to rRNA sequences using Bowtie2 (Version 2.2.5) with default setting (Langmead and Salzberg, 2012). The remaining reads were mapped to Drosophila genome (dm6) and transposon consensus sequences using STAR (Version 020201) and Hisat2 with default parameters (Dobin et al., 2013; Kim et al., 2015). The splicing junctions/introns are extracted from STAR mapping results of two replicates of w1 RNAseq. Introns with less than 1 unique mapped read and introns without canonical splice junction (GT/AG) and less than 2 unique mapped reads were filtered out. The remaining introns from two replicates are merged to generate 9047 introns present in ovary. The transcript abundance for each gene, intron and transposon (RPKM: Reads Per Kilobase per Million mapped reads) was counted by BEDTools (Version 2.27.1) (Quinlan and Hall, 2010) and normalized to total number of genome mapping reads, after excluding rRNA mapping reads. We performed differentially expression analysis of transposons and protein-coding genes together with DESeq2 (Love et al., 2014) and default parameters from two biological replicates.
Small RNA-seq:
After removing 3’ end adaptor via cutadapt (Version 1.15) (Martin, 2011), the raw small RNA-seq reads were sequentially mapped to rRNA, miRNA hairpin, snoRNA, snRNA and tRNA sequences using Bowtie (Version 1.1.0) (Langmead et al., 2009) by allowing 1 mismatches. The remaining reads were mapped to Drosophila genome (dm6) and transposon consensus sequences. The small RNA abundances across different libraries were normalized to the total hairpin mapping reads. For ping-pong analysis for piRNA reads, 5´ to 5´ overlaps between all pairs of piRNAs that mapped to the opposite genomic strands were calculated, and then the Z-score for the 10-nt overlap was calculated using the 1–9 nt and 11–30 nt overlaps as the background (Li et al., 2009a). The ping-pong pairs were normalized to quadratic of the total hairpin mapping reads.
ChIP-seq:
The raw ChIP-seq reads were mapped to Drosophila genome (dm6) and transposon consensus sequences using Bowtie2 (Version 2.2.5) with default parameters (Langmead and Salzberg, 2012). The ChIP-seq signal mapping to transposon consensus sequences is normalized to total number of genome mapping ChIP-seq reads. H3K4me2 peaks in w1 and thoc7d/Df ovaries are called by MACS2 (Version 2.1.1) (q<0.01) from two replicates (Zhang et al., 2008). After peak calling, peaks in w1 and thoc7d/Df ovaries were merged using BEDtools merge with default parameters (Quinlan and Hall, 2010), then w1 specific, thoc7d/Df specific and shared peaks and fold enrichment in these peaks were extracted.
Transposon insertion analysis:
To obtain comprehensive transposon insertion annotations specific to the studied genome types, we integrated insertions in the reference genome and new insertions detected by TEMP in our experimental strains (Zhuang et al., 2014). First, we extracted the full-length transposon insertions that are longer than 80% of their consensus sequences. Next, “insertion” and “absence” modules of TEMP was run with two replicates of ChIP input DNA sequencing for w1 and thoc7d/Df (together with at least fifty times genome coverage) to detect new transposon insertions and the absence of reference annotated full-length transposon insertions. The reference annotated full-length insertions and new insertions (supported by paired-end reads on both ends) with penetrance higher than 0.5 were merged and regarded as transposon insertions in the studied genome types. Then shared and genotype specific insertions were identified via BEDTools (Quinlan and Hall, 2010). We then defined heterochromatic regions in Drosophila genome (dm6) based on H3K9me3 ChIP-seq. The five kilobases regions flanking euchromatic transposon insertions were divided into 100 base pairs bins. The H3K9me3 ChIP-seq and input signal and small RNA signal were quantified for each bin to generate heatmaps in Figure 4D and 4E.
Nascent RNAseq:
Four-thiouridine (4sU) labeled nascent RNA-seq libraries were firstly mapped to rRNA and unmapped reads were then mapped to dm6 genome and transposon consensus sequences by STAR and Hisat2 respectively with 7 mismatches allowed. To remove false positive nucleotide conversion due to single nucleotide polymorphism, genome and transposon consensus mapping file (BAM format; sorted and duplication removed) from w1 RNA-seq used in this study were used for SNP calling by samtools pileup and bcftools with default parameters (Li, 2011; Li et al., 2009b). Nucleotide conversions in each read were calculated and further filtered if the nucleotide quality is less than 30 or overlapped with a SNP. After filtering, reads with more than one T>C conversions were considered as newly synthesized message RNA. Finally, newly synthesized mRNA abundance was calculated for each gene, piRNA cluster and transposon element and normalized to total sequencing depth and gene, piRNA cluster and transposon length.
Quantification and statistical analysis
The replicates and sample size are reported in the Figures and corresponding legends.
Supplementary Material
Acknowledgements
We would like to thank the members of Theurkauf and Weng labs for their insightful discussions and comments throughout the project; Elisa Izaurralde for anti Hpr1 and Tho2 antibodies; Yu Yang and Gregory Hannon for panxM4 and Df(2R)BSC821; Bloomington, Harvard Exelixis and VDRC stock centers for fly strains. Special thanks to Yekaterina Makeyev and Craig Mello for sharing unpublished data and their manuscript. This work was supported by NIH grants R01 HD049116 and P01 HD078253 to W.T. and Z. W.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Andersen PR, Tirian L, Vunjak M, and Brennecke J (2017). A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 549, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, and Brennecke J (2007). The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318, 761–764. [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, and Kohany O (2015). Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton LJ, Lovander KE, Pinto BS, and Geyer PK (2016). Drosophila male and female germline stem cell niches require the nuclear lamina protein Otefin. Dev Biol 415, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastock R, and St Johnston D (2008). Drosophila oogenesis. Curr Biol 18, R1082–1087. [DOI] [PubMed] [Google Scholar]
- Bergman CM, Quesneville H, Anxolabehere D, and Ashburner M (2006). Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol 7, R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, Imbeault M, Izsvak Z, Levin HL, Macfarlan TS, et al. (2018). Ten things you should know about transposable elements. Genome Biol 19, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, and Hannon GJ (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. [DOI] [PubMed] [Google Scholar]
- Chen YA, Stuwe E, Luo Y, Ninova M, Le Thomas A, Rozhavskaya E, Li S, Vempati S, Laver JD, Patel DJ, et al. (2016). Cutoff Suppresses RNA Polymerase II Termination to Ensure Expression of piRNA Precursors. Mol Cell 63, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, and Lin H (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12, 3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, and Lin H (2000). piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127, 503–514. [DOI] [PubMed] [Google Scholar]
- Czech B, Munafo M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, and Hannon GJ (2018). piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu Rev Genet 52, 131–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumesic PA, Natarajan P, Chen C, Drinnenberg IA, Schiller BJ, Thompson J, Moresco JJ, Yates JR 3rd, Bartel DP, and Madhani HD (2013). Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell 152, 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabry MH, Ciabrelli F, Munafo M, Eastwood EL, Kneuss E, Falciatori I, Falconio FA, Hannon GJ, and Czech B (2019). piRNA-guided co-transcriptional silencing coopts nuclear export factors. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Wu P-H, Beane T, Zamore PD, and Weng Z (2018). Elimination of PCR duplicates in RNA-seq and small RNA-seq using unique molecular identifiers. BMC genomics 19, p. 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, and Zamore PD (2015). Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348, 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B, Jan LY, and Jan YN (1990). Localization of vasa, a component of Drosophila polar granules, in maternal-effect mutants that alter embryonic anteroposterior polarity. Development 109, 425–433. [DOI] [PubMed] [Google Scholar]
- Hedges DJ, and Deininger PL (2007). Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res 616, 46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog VA, Reichholf B, Neumann T, Rescheneder P, Bhat P, Burkard TR, Wlotzka W, von Haeseler A, Zuber J, and Ameres SL (2017). Thiol-linked alkylation of RNA to assess expression dynamics. Nat Methods 14, 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CR, Burns KH, and Boeke JD (2012). Active transposition in genomes. Annu Rev Genet 46, 651–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Luo Y, Moon S, Ninova M, Marinov GK, Chung YD, and Aravin AA (2016). Splicing-independent loading of TREX on nascent RNA is required for efficient expression of dual-strand piRNA clusters in Drosophila. Genes Dev 30, 840–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagut M, Mihaila-Bodart L, Molla-Herman A, Alin MF, Lepesant JA, and Huynh JR (2013). A mosaic genetic screen for genes involved in the early steps of Drosophila oogenesis. G3 (Bethesda) 3, 409–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno S, and Aguilera A (2010). The THO complex as a key mRNP biogenesis factor in development and cell differentiation. J Biol 9, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Flynt AS, and Lai EC (2013). Drosophila piwi mutants exhibit germline stem cell tumors that are sustained by elevated Dpp signaling. Curr Biol 23, 1442–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002. ). The human genome browser at UCSC. Genome Res 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, and Theurkauf WE (2008). piRNA function in germline development. In StemBook (Cambridge (MA)). [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. (2009). The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138, 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Korbut AP, Stolyarenko AD, Yakushev EY, Reuter M, Pillai RS, and Gvozdev VA (2014). Impact of nuclear Piwi elimination on chromatin state in Drosophila melanogaster ovaries. Nucleic Acids Res 42, 6208–6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, and Gvozdev VA (2011). Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci U S A 108, 18760–18765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneuss E, Munafo M, Eastwood EL, Deumer US, Preall JB, Hannon GJ, and Czech B (2019). Specialization of the Drosophila nuclear export family protein Nxf3 for piRNA precursor export. Genes Dev 33, 1208–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, and Akira S (2011). Pathogen recognition by the innate immune system. Int Rev Immunol 30, 16–34. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko PF, and Ashburner M (1990). Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev 4, 905–921. [DOI] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, and Toth KF (2013). Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27, 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. (2009a). Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009b). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, and Hannon GJ (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JS, Warrington R, Watson W, and Kim HL (2018). An introduction to immunology and immunopathology. Allergy Asthma Clin Immunol 14, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal 17, 10–12. [Google Scholar]
- Mohn F, Handler D, and Brennecke J (2015). Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348, 812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, and Brennecke J (2014). The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157, 1364–1379. [DOI] [PubMed] [Google Scholar]
- Moon S, Cho B, Min SH, Lee D, and Chung YD (2011). The THO complex is required for nucleolar integrity in Drosophila spermatocytes. Development 138, 3835–3845. [DOI] [PubMed] [Google Scholar]
- Murano K, Iwasaki YW, Ishizu H, Mashiko A, Shibuya A, Kondo S, Adachi S, Suzuki S, Saito K, Natsume T, et al. (2019). Nuclear RNA export factor variant initiates piRNA-guided co-transcriptional silencing. EMBO J 38, e102870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophinni Y, Palatini U, Hayashi Y, and Parrish NF (2019). piRNA-Guided CRISPR-like Immunity in Eukaryotes. Trends Immunol 40, 998–1010. [DOI] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, and Zamore PD (2019). PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet 20, 89–108. [DOI] [PubMed] [Google Scholar]
- Parhad SS, and Theurkauf WE (2019). Rapid evolution and conserved function of the piRNA pathway. Open Biol 9, 180181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parhad SS, Tu S, Weng Z, and Theurkauf WE (2017). Adaptive Evolution Leads to Cross-Species Incompatibility in the piRNA Transposon Silencing Machinery. Dev Cell 43, 60–70 e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parhad SS, Yu T, Zhang G, Rice NP, Weng Z, and Theurkauf WE (2020). Adaptive Evolution Targets a piRNA Precursor Transcription Network. Cell Rep 30, 2672–2685 e2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, and Cheng H (2005). TREX, SR proteins and export of mRNA. Curr Opin Cell Biol 17, 269–273. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, Ciccarelli FL, Wilm M, and Izaurralde E (2004). Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 11, 558–566. [DOI] [PubMed] [Google Scholar]
- Schofield JA, Duffy EE, Kiefer L, Sullivan MC, and Simon MD (2018). TimeLapse-seq: adding a temporal dimension to RNA sequencing through nucleoside recoding. Nat Methods 15, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, and Brennecke J (2010). The piRNA pathway: a fly’s perspective on the guardian of the genome. Trends Genet 26, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Batki J, Senti KA, Donertas D, Tirian L, Meixner K, and Brennecke J (2015). Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery. Genes Dev 29, 2258–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Donertas D, and Brennecke J (2012). Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151, 964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y, Billi AC, Hayes GD, Newman MA, Zuk O, Gabel H, Kamath R, Yacoby K, Chapman B, Garcia SM, et al. (2013). Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature 493, 694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE (1994). Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis. Methods Cell Biol 44, 489–505. [DOI] [PubMed] [Google Scholar]
- Wang W, Han BW, Tipping C, Ge DT, Zhang Z, Weng Z, and Zamore PD (2015). Slicing and Binding by Ago3 or Aub Trigger Piwi-Bound piRNA Production by Distinct Mechanisms. Mol Cell 59, 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chang Y, Li Y, Zhang X, and Goodrich DW (2006). Thoc1/Hpr1/p84 is essential for early embryonic development in the mouse. Mol Cell Biol 26, 4362–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Fan K, Ozata DM, Zhang G, Fu Y, Theurkauf WE, Zamore PD, and Weng Z (2021). Long first exons and epigenetic marks distinguish conserved pachytene piRNA clusters from other mammalian genes. Nat Commun 12, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Koppetsch BS, Pagliarani S, Johnston S, Silverstein NJ, Luban J, Chappell K, Weng Z, and Theurkauf WE (2019). The piRNA Response to Retroviral Invasion of the Koala Genome. Cell 179, 632–643 e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Gu J, Jin Y, Luo Y, Preall JB, Ma J, Czech B, and Hannon GJ (2015). Panoramix enforces piRNA-dependent cotranscriptional silencing. Science 350, 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, et al. (2012a). UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Tu S, Yu T, Zhang XO, Parhad SS, Weng Z, and Theurkauf WE (2018). Co-dependent Assembly of Drosophila piRNA Precursor Complexes and piRNA Cluster Heterochromatin. Cell Rep 24, 3413–3422 e3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Theurkauf WE, Weng Z, and Zamore PD (2012b). Strand-specific libraries for high throughput RNA sequencing (RNA-Seq) prepared without poly(A) selection. Silence 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, Vreven T, Zamore PD, Weng Z, and Theurkauf WE (2014). The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 157, 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, and Zamore PD (2011). Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol Cell 44, 572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, Wang J, Theurkauf W, and Weng Z (2014). TEMP: a computational method for analyzing transposable element polymorphism in populations. Nucleic Acids Res 42, 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq, Small-RNAseq and ChIPseq data generated in this study have been deposited at the NCBI BioProject and Sequence Read Archive (SRA) as of the date of publication. Accession numbers for existing, publicly available data are listed in the Key Resources Table.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-Hpr1 | (Rehwinkel et al., 2004) | |
| Rabbit anti Piwi | (Li et al., 2009a) | |
| Rabbit anti Ago3 | (Li et al., 2009a) | |
| Rabbit anti Aub | (Li et al., 2009a) | |
| Rat IgM anti-Vasa | DSHB | |
| Mouse anti-Engrailed (4D9) | DSHB | |
| Rabbit anti H3K9me3 | Abcam | Cat# ab8898 |
| Rabbit anti H3K4me2 | EMDMillipore | Cat# 07–030 |
| Mouse Anti-α-Tubulin | Sigma Aldrich | Cat# T5168 |
| Rabbit anti-GFP | ThermoFisher Scientific | Cat# A11122 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-Thiouridine | MilliporeSigma | Cat# T4509 |
| Iodoacetamide | MilliporeSigma | Cat# I1149 |
| Superscript III | ThermoFisher Scientific | Cat# 18080–085 |
| RNase OUT | ThermoFisher Scientific | Cat# 10777–019 |
| TURBO DNase | ThermoFisher Scientific | Cat# AM2238 |
| RNaseH | ThermoFisher Scientific | Cat# 18021–071 |
| T4 RNA Ligase | ThermoFisher Scientific | Cat# AM2141 |
| AccuPrime™ Pfx DNA Polymerase | ThermoFisher Scientific | Cat# 12344024 |
| TRIzol™ Reagent | ThermoFisher Scientific | Cat# 15596026 |
| UltraPure™ Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v) | ThermoFisher Scientific | Cat# 15593031 |
| dNTP Set (100 mM) | ThermoFisher Scientific | Cat# 10297018 |
| dUTP Solution (100 mM) | ThermoFisher Scientific | Cat# R0133 |
| Grace's Insect Medium, unsupplemented | ThermoFisher Scientific | Cat# 11595030 |
| Hybridase | Lucigen | Cat# H39500 |
| dNTP mix | NEB | Cat# N0447L |
| DNA polymerase I | NEB | Cat# M0209S |
| T4 DNA polymerase | NEB | Cat# M0203L |
| Klenow DNA polymerase | NEB | Cat# M0210S |
| T4 PNK | NEB | Cat# M0201L |
| Klenow 3’ to 5’ exo | NEB | Cat# M0212L |
| UDG | NEB | Cat# M0280S |
| Phusion Polymerase | NEB | Cat# M0530S |
| T4 RNA Ligase 2, truncated | NEB | Cat# M0242L |
| 50% PEG8000 | NEB | Cat# B1004S |
| T4 DNA ligase | Enzymatics Inc. | Cat# L6030-HC-L |
| EDTA-free Protease Inhibitor Cocktail (Roche) | Sigma | Cat# 11873580001 |
| 16% formaldehyde | Ted Pella Inc | Cat# 18505 |
| Miracloth membrane (Calbiochem) | EMDMillipore | Cat# 475855 |
| Critical Commercial Assays | ||
| mirVANA™ miRNA isolation kit | ThermoFisher Scientific | Cat# AM1560 |
| RNeasy Mini Kit | Qiagen | Cat# 74104 |
| QuantiTect SYBR® Green PCR Kits | Qiagen | Cat# 204145 |
| Dynabeads® Protein G | ThermoFisher Scientific | Cat# 10004D |
| Dynabeads® Protein A | ThermoFisher Scientific | Cat# 10001D |
| RNA Clean & Concentrator-5 | Zymo Research | Cat# R1015 |
| In-Fusion® HD Cloning Plus | Takara | Cat# 638909 |
| Agencourt AMPure XP | Beckman Coulter | Cat# A63880 |
| Deposited Data | ||
| High throughput Sequencing | This study | PRJNA590287 |
| small RNAseq for thoc5[e00906]/thoc5[1]_rep1 | (Zhang et al., 2018) | SRR7686976 |
| small RNAseq for thoc5[e00906]/thoc5[1]_rep2 | (Zhang et al., 2018) | SRR7686977 |
| small RNAseq for w[1]_rep1 | (Zhang et al., 2018) | SRR7408119 |
| small RNAseq for thoc7[d05792]/Df(3L)BSC128 | (Zhang et al., 2018) | SRR7408136 |
| Small RNAseq of rhi mut ovary | (Parhad et al., 2017) | SRR5803097 |
| RNAseq for thoc5[e00906]/thoc5[1]_rep1 | (Zhang et al., 2018) | SRR7408102 |
| RNAseq for thoc7[d05792]/Df(3L)BSC128_rep2 | SRR7408139 | |
| input RNAseq from w[1]_rep1 | (Zhang et al., 2018) | SRR7408152 |
| input RNAseq from w[1]_rep2 | (Zhang et al., 2018) | SRR7408157 |
| Hpr1IP RNAseq from w[1]_rep1 | (Zhang et al., 2018) | SRR7408151 |
| Hpr1IPRNAseq from w[1]_rep2 | (Zhang et al., 2018) | SRR7408156 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster/thoc7d05792 | Harvard Exelixis stock collection | Stock # d05792 |
| D. melanogaster/Df(3L)BSC128 | Bloomington Drosophila Stock Center | Stock# 9293 |
| D. melanogaster/thoc5e00906 | Harvard Exelixis stock collection | Stock# e00906 |
| D. melanogaster/thoc51 | (Moon et al., 2011) | N.A. |
| D. melanogaster/rhino2 | (Klattenhoff et al., 2009) | N.A. |
| D. melanogaster/rhinoKG | (Klattenhoff et al., 2009) | N.A. |
| D. melanogaster/piwi2 | Bloomington Drosophila Stock Center | Stock# 43319 |
| D. melanogaster/piwiNLS | (Klenov et al., 2011) | N.A. |
| D. melanogaster/panoramixM4 | (Yu et al., 2015) | N.A. |
| D. melanogaster/Df(2R)BSC821 | (Yu et al., 2015) | Stock# 27582 |
| D. melanogaster/ pUASp>lambdaN-HA-CG9754 [attP40]/CyO; tub>EGFP_5xBoxB_SV40 [attP2]/TM3, Ser; | VDRC Stock Center | Stock# 313393 |
| D. melanogaster/w1 | William Theurkauf lab | N.A. |
| Oligonucleotides | ||
| Random primers | ThermoFisher Scientific | Cat# 48190011 |
| Primers for qPCR, see Table S1 | N.A. | |
| Software and Algorithms | ||
| Prism 7 | GraphPad Prism | https://www.graphpad.com/ |
| Image Studio™ Lite | LI-COR | https://www.licor.com/bio/products/software/image_studio_lite/ |
| RStudio | https://www.rstudio.com/ | |
| ImageJ | https://imagej.nih.gov/ij/ | |
| UCSC Genome Browser | (Kent et al., 2002) | https://genome.ucsc.edu/cgi-bin/hgGateway |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.