Abstract
Asthma is a heterogeneous disease characterized by chronic airway inflammation with a genetic predisposition. Butyrophilin-like 2 (BTNL2) is a member of the immunoglobulin superfamily that plays an important role in regulating T cell activation and immune homeostasis. Here, we aimed to investigate the association of the genetic variants of BTNL2 with childhood asthma and asthma-related traits by utilizing extreme asthma phenotypes and employing a genome-wide association study. Our study included 243 children with well-defined moderate to severe atopic asthma and 134 healthy children with no history of allergic diseases and allergic sensitization. DNA from these subjects was genotyped using AxiomTM Genome-Wide Array Plates. Although no single nucleotide polymorphisms (SNPs) reached a genome-wide threshold of significance, 3 SNPs, rs3817971, rs41355746, and rs41441651, at BTNL2 were significantly associated with moderate to severe atopic asthma after performing Bonferroni correction. These SNPs were also associated with the risk of allergic sensitization toward house dust mites and the presence and degree of bronchial hyperresponsiveness. Thus, we identified that BTNL2 was associated with atopic moderate to severe persistent asthma in Korean children, and this may play an important role in disease development and susceptibility.
Keywords: allergic sensitization, asthma, bronchial hyperresponsiveness, children, genome-wide association study
1. Introduction
Asthma is a heterogenous and genetically complex respiratory disease that is a result of gene–environment interactions, especially early in life.[1] Gene discovery approaches in asthma were initially knowledge-based candidate gene association studies and family-based genome-wide linkage analyses. Recently, genome-wide association studies (GWASs) have dominated; these studies aid hypothesis-free discovery of novel risk loci, which in turn provide mechanistic insights into disease pathogenesis.[2–4]
GWASs of asthma have identified many risk loci, including 17q12-21 (ORMDL3, GSDMB), 6p21 (HLA region), 2q12 (IL1RL1/IL18R1), 5q22 (TSLP), and 9p24 (IL33) that provided novel insights into asthma biology.[4] In addition, GWASs have revealed significant genome-wide variants for asthma-related traits, such as bronchodilator response (BDR),[5] bronchial hyperresponsiveness (BHR),[6] blood eosinophils,[7] total serum immunoglobulin E (IgE) levels,[2,8] and allergic sensitizations.[9] Such studies are based on the assumptions that it is easy to detect genes influencing components of asthma by reducing asthma heterogeneity and that the same genes also contribute to asthma risk.
However, most previous GWASs of asthma comprise subjects of European ancestry; only a few studies have been conducted in ethnic minority populations. As populations vary with respect to allele frequencies, linkage disequilibrium (LD) patterns, and effect sizes of variants underlying disease risk,[10–12] the previous GWASs may have limited utility in other population groups. Furthermore, they have probably missed important risk variants in non-European populations.
In the Korean population, GWASs of asthma have focused on occupation-associated forms of asthma and aspirin-exacerbated respiratory disease in adults.[13–17] A recent GWAS in Korean subjects with asthma–chronic obstructive pulmonary disease overlap syndrome did not reveal any significant genome-wide hits.[18] Additionally, no significant genome-wide loci were reported in a GWAS based on total serum IgE levels in Korean subjects with asthma.[19] Such limited GWAS results in the Korean population are most likely because of the small sample size. Considering that GWAS adopts rigorous statistical control for false-discovery rate, which may miss several single nucleotide polymorphisms (SNPs) of true associations,[20] there is a need for alternative strategies to address such concerns.
Therefore, the aim of the present study was to identify asthma-related loci using cases of extreme phenotypes – that is, moderate-to-severe persistent atopic asthma – and healthy controls in a small-sized GWAS. In addition, we sought to determine the GWAS-based SNPs associated with asthma susceptibility traits, including BDR, BHR, and total or specific serum IgE levels.
2. Methods
2.1. Study subjects
For this case-control investigation, 243 subjects with moderate to severe persistent asthma with allergic sensitization, and 134 healthy children without a history of allergic disease or allergic sensitization were included in the study. All children were recruited from Severance Children's Hospital, Seoul, Korea during April, 2004 and May, 2010. The study was approved by the Institutional Review Board of Severance Hospital (IRB no. 4-2004-0036). All participants were unrelated, and either they or their parents provided written informed consent.
Subjects with asthma were confirmed based on consistent respiratory symptoms verified by physicians, presence of either a BDR of ≥12% increase in forced expiratory volume in 1 s (FEV1) or BHR of decrease in FEV1 by ≥20% upon inhalation of <16 mg/mL methacholine. Allergic sensitization was defined by specific serum IgE levels greater than 0.7 kUA/L for at least one of the following food or airborne allergens: egg white, milk, Dermatophagoides pteronyssinus (Der p), Dermatophagoides farina (Der f), Alternaria species, or Blattella germanica. Children with moderate to severe asthma according to the Global Initiative for Asthma guidelines were recruited.
Healthy controls included children who had visited the hospital for a general health checkup or vaccination, and had no history of allergic diseases based on interviews with their parents. Further, children who were not sensitized to the abovementioned 6 common allergens and had total serum IgE levels <100 kU/L were enrolled.
The total and specific IgE levels to selected allergens were measured from peripheral blood samples using the ImmunoCAP system (Pharmacia Diagnostics, Uppsala, Sweden). Complete blood count, including eosinophil count, was analyzed using the ADVIA 2120i hematology system with autoslide (Siemens Healthcare Diagnostics Inc., Deerfield, IL, USA).
2.2. Genotyping, quality control, and Butyrophilin-like 2 (BTNL2) region selection
Blood samples were collected from each subject to extract genomic DNA. The DNA was then genotyped using an Affymetrix Axiom array (Affymetrix Inc., Santa Clara, CA, USA). There was originally a total of 600,252 SNPs; after frequency and genotyping pruning using GRCh37/hg19, there were 574,420 autosomal SNPs. During quality control filtration, samples with an autosomal SNP call rate <95% were removed. SNPs with minor allele frequencies <5% or Hardy–Weinberg equilibrium P value <10−6 were also excluded. Finally, 423,461 SNPs with a genomic inflation factor (λ) of 1.0033 were used for the GWAS (Supplemental Figure S1). Among the finally refined SNPs, we then selected 46 SNPs located ± 500 base pairs around BTNL2 in order to further analyze the BTNL2 region.
2.3. Statistical analysis
Genome-wide associations between atopic asthma and variants were analyzed using PLINK (v.1.07).[21] Correlation coefficients were calculated and genome-wide statistical analyses were performed using PLINK. Genomic inflation factor (λ) was calculated based on median chi-square statistics. Manhattan plots and quantile–quantile plots were plotted using the “manhattanly” and “qqman” libraries, respectively, in R (v.3.3.3).[22] LD calculations were performed using PLINK, and loci, including variants significantly associated with asthma, were plotted using LocusZoom (v.0.4.8).[23] Association of genetic polymorphisms with BHR and levels of total and mite-specific IgE were evaluated by logistic and linear regression analyses using the software R. Age and sex were the adjustment factors for the analyses.
3. Results
3.1. Subject characteristics
Demographic characteristics of the study subjects are summarized in Table 1. The control and asthma groups did not differ in age. Males were predominant in the asthma group (n = 168, 69.1%). Blood eosinophil count and total serum IgE levels were significantly higher in the asthma group than in the control group (P value <.001 for both). Compared with healthy controls, children with asthma showed a lower percentage of FEV1/forced vital capacity ratio, percentage of predicted FEV1, percentage of predicted maximum mid-expiratory flow (FEF25-75), and peak expiratory flow (Table 1).
Table 1.
Clinical profiles of the study subjects (N = 377).
| Control (n = 134) | Atopic asthma (n = 243) | P value | |
| Sex, M (%) | 63 (47.0) | 168 (69.1) | <.001 |
| Age (yr) | 9.1 (7.3–11.6) | 8.4 (6.5–10.5) | .051 |
| Height (cm) | 133.6 (125.0–147.0) | 131.0 (119.2–142.0) | .034 |
| Weight (kg) | 31.0 (24.7–45.0) | 30.0 (23.0–40.0) | .096 |
| FVC (% pred) | 92.8 (84.0–102.1) | 91.9 (80.8–100.1) | .361 |
| FEV1 (% pred) | 98.3 ± 15.7 | 91.7 ± 15.6 | <.001 |
| Post-BD FEV1 (% pred) | 108.2 ± 11.9 | 101.4 ± 16.1 | <.001 |
| FEV1/FVC (%) | 92.0 (87.3–97.3) | 86.7 (79.3–93.2) | <.001 |
| PEF (% pred) | 94.5 (81.3–104.8) | 88.0 (77.0–99.0) | .002 |
| FEF25–75 (% pred) | 101.9 ± 23.6 | 80.9 ± 27.0 | <.001 |
| ΔFEV1 | 4.1 (1.1–6.1) | 9.6 (4.2–15.3) | <.001 |
| PC20 (mg/mL) | N/A | 3.7 (1.6–7.3) | N/A |
| PC20 category | <.001 | ||
| <1 | 0 (0.0) | 42 (17.6) | |
| 1–4 | 0 (0.0) | 85 (35.6) | |
| 4–16 | 0 (0.0) | 103 (43.1) | |
| ≧16 | 134 (100.0) | 9 (3.8) | |
| Blood eosinophils (/μL) | 140.0 (90.0–200.0) | 520.0 (300.0–740.0) | <.001 |
| Total serum IgE (kU/L) | 28.9 (15.8–53.0) | 584.0 (317.0–1035.0) | <.001 |
Data are given as number (%), mean (± standard deviation), or median (interquartile range).
ΔFEV1 = change in FEV1 after BD; BD = bronchodilator; FEF25–75 = maximum mid-expiratory flow; FEV1 = forced expiratory volume in 1 s; FVC = forced vital capacity; IgE = immunoglobulin E; N/A = not applicable; PC20 = provocative concentration of methacholine inducing a 20% decrease in FEV1; PEF = peak expiratory flow.
3.2. Genome-wide associations with moderate-to-severe persistent atopic asthma
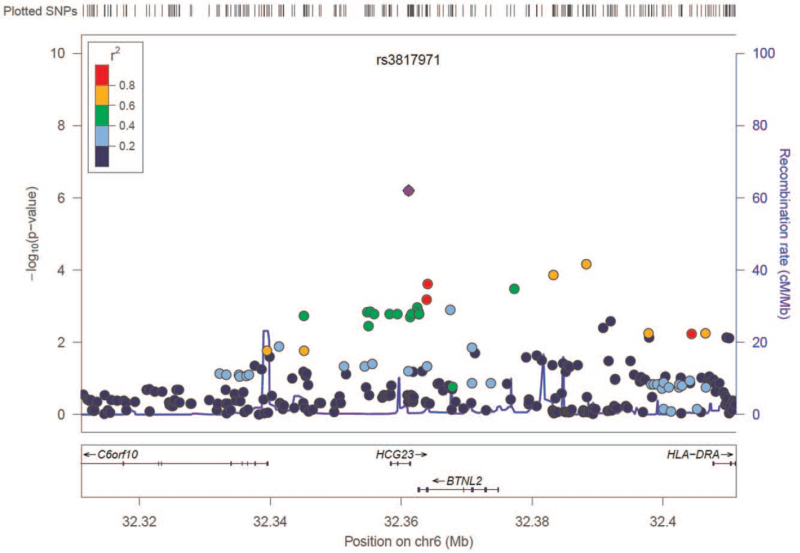
A total of 423,461 autosomal SNPs that passed quality control were analyzed for their association with asthma. GWAS of atopic asthma and healthy control subjects showed an excess of small P values compared with those expected by chance (Supplemental Figure S1). Although no SNPs reached a genome-wide threshold of significance (P < 1.18 × 10−7), several candidate loci were identified as top signals (Manhattan plot in Supplemental Figure S2); the most prominent SNP was located on chromosome 6p21.32. Figure 1 displays a regional association plot at the 6p21.32 locus for atopic asthma. The most highly associated SNP was rs3817971, which is located within BTNL2.
Figure 1.
Regional association plot of 6p21.3 locus for atopic moderate to severe persistent asthma. Statistical significance of each single nucleotide polymorphism (SNP) on the -log10 scale (left y-axis) according to its chromosomal position (x-axis) is shown. Relative location of the genes in each region and the direction of transcription is shown in the bottom panel. The blue line shows recombination rate across that region (right y-axis). The most associated SNP (rs3817971) is shown as a purple circle, and the remaining SNPs are color-coded according to the level of linkage disequilibrium (r2).
3.3. Association of genetic variations in BTNL2 with asthma
To analyze BTNL2 polymorphisms, we selected SNPs that were located ±500 base pairs around BTNL2. A total of 46 SNPs were reanalyzed, which yielded 3 SNPs (rs3817971, rs41355746, and rs41441651) with P values <.001087 (Bonferroni-corrected; Table 2 and Supplemental Table S1). Risk alleles of these SNPs were associated with an increased risk of asthma after age and sex adjustment (rs3817971: adjusted odds ratio [aOR] = 3.314, P < .001; rs41355746: aOR = 2.639, P = .001; rs41441651: aOR = 2.404, P = .002). These associations between genetic polymorphisms and asthma remained significant for both additive and dominant models (Supplemental Table S2).
Table 2.
Summary of GWASs of atopic moderate to severe persistent asthma at P value <1.087 × 10−3 (= 0.05/46) in the study subjects.
| Minor allele frequency | |||||||
| SNP | Position | Alleles (risk/alternative) | Asthma (n = 243) | Control (n = 134) | P value | OR | 95% CI |
| rs3817971 | 32361133 | A/G | 0.949 | 0.840 | 6.285 × 10−7 | 3.509 | 2.090–5.889 |
| rs41355746 | 32364052 | C/T | 0.951 | 0.877 | 2.456 × 10−4 | 2.703 | 1.562–4.679 |
| rs41441651 | 32363888 | C/T | 0.947 | 0.877 | 6.546 × 10−4 | 2.484 | 1.452–4.252 |
CI = confidence interval; GWAS = genome-wide association study; OR = odds ratio; SNP = single nucleotide polymorphism.
3.4. Associations with BDR and BHR
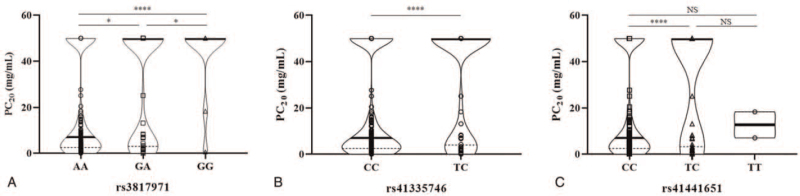
A case-control analysis adjusted for age and sex as covariates was performed for determining association with additional phenotypes. Table 3 represents the association results of the provocative concentration of methacholine that caused a 20% drop in FEV1 from baseline level (PC20). PC20 levels were further quadrichotomized as follows: <1, ≥1 to <4, ≥4 to <16, and ≥16. The 3 SNPs (rs3817971, rs41355746, and rs41441651) showed significant associations with the level and grade of PC20, as well as the presence of BHR. Association results between genetic polymorphisms and BHR are depicted in Supplemental Table S3. Differences in individual values of PC20 according to each genotype are presented in Figure 2.
Table 3.
Association results of 3 SNPs with the presence and degree of BHR.
| PC20; categorical | PC20; continuous | BHR† | ||||||||||
| SNP | Risk allele | <1 | 1–4 | 4–16 | >16 | P value | β | SE | P value | aOR (95% CI) | P value | |
| Number of cases | 79 | 160 | 197 | 240 | <.001 | −12.408 | 2.695 | <.001 | 3.599 (2.089–6.200) | <.001 | ||
| Proportion | 0.12 | 0.24 | 0.29 | 0.36 | ||||||||
| rs41355746 | C | Number of cases | 80 | 160 | 198 | 251 | .005 | −9.958 | 2.926 | .001 | 2.704 (1.532–4.775) | .001 |
| Proportion | 0.12 | 0.23 | 0.29 | 0.36 | ||||||||
| rs41441651 | C | Number of cases | 80 | 160 | 197 | 250 | .004 | −9.284 | 2.882 | .001 | 2.657 (1.520–4.645) | .001 |
| Proportion | 0.12 | 0.23 | 0.29 | 0.36 | ||||||||
Regression analyses were adjusted for age and sex.
β = β estimate; aOR = adjusted odds ratio; BHR = bronchial hyperresponsiveness; CI = confidence interval; PC20 = provocative concentration of methacholine inducing a 20% decrease in forced expiratory volume in 1 s; SE = standard errors; SNP = single nucleotide polymorphism.
BHR was defined as a decrease in forced expiratory volume in 1 s of ≥20% with inhalation of <16 mg/mL methacholine.
Figure 2.
Differences in PC20 according to genotype groups for each SNP. The y-axis represents the value of PC20 in mg/mL, and the x-axis represents the 3 genotyping groups for 3 SNPs of (A) rs3817971, (B) rs41335746, and (C) rs41441651. Violin plots depict the distribution of individual data in addition to the median (thick horizontal line) and interquartile range (dotted line). PC20 = provocative concentration of methacholine inducing a 20% decrease in forced expiratory volume in 1; SNP = single nucleotide polymorphism. NS, P > .05; ∗, P < .05; ∗∗∗∗, P < .0001.
For BDR, no significant associations were found with the 3 SNPs after comparisons between alleles.
3.5. Associations with allergic sensitization
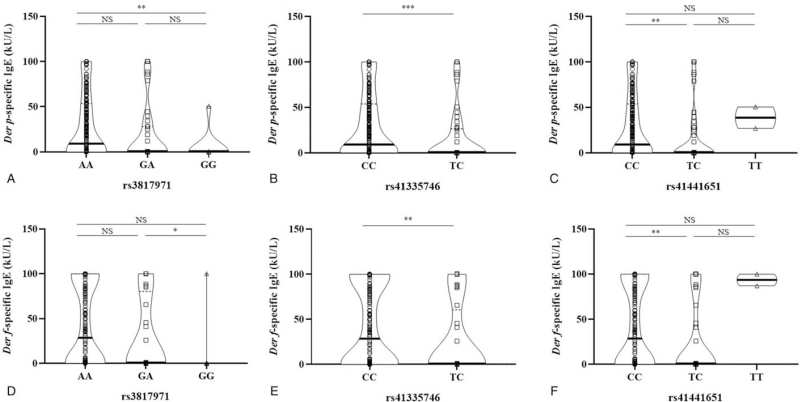
Linear regression analysis indicated that the 3 loci did not show significant associations with the serum level of total IgE. However, analyses with Der p- and Der f-specific IgE revealed significant associations (Table 4). rs3817971 showed a significant association with serum levels of Dep p- and Dep f-specific IgE under additive, dominant, and recessive models. Further, rs41441651 and rs41335746 showed a positive association under the additive and dominant models (Supplemental Table S4). Figure 3 depicts the level of mite-specific IgE by genotypes of each SNP, and reveals significant differences among genotypic variations.
Table 4.
Association results of 3 SNPs with the level of total and mite-specific serum IgE.
| Total IgE | Der p specific IgE | Der f specific IgE | ||||||||
| SNP | Risk allele | β | SE | P value | β | SE | P value | β | SE | P value |
| rs3817971 | A | 170.315 | 91.430 | .068 | 13.930 | 4.776 | .004 | 19.075 | 5.960 | .001 |
| rs41355746 | C | 149.837 | 98.680 | .129 | 11.511 | 5.196 | .027 | 14.958 | 6.501 | .022 |
| rs41441651 | C | 142.387 | 97.147 | .143 | 10.516 | 5.101 | .040 | 11.957 | 6.388 | .062 |
Analyses were adjusted for age and sex.
β = β estimate; Der f = Dermatophagoides farina; Der p = Dermatophagoides pteronyssinus; IgE = immunoglobulin E; SE = standard errors; SNP = single nucleotide polymorphism.
Figure 3.
Differences in mite-specific serum IgE levels according to genotyping groups for each SNP. Serum specific IgE levels towards Der p (A–C) and Der f (D–F) according to genotypes of each SNP are presented. Violin plots depict the distribution of individual data in addition to the median (thick horizontal line) and interquartile range (dotted line). IgE = immunoglobulin E; SNP = single nucleotide polymorphism; Der p = Dermatophagoides pteronyssinus; Der f = Dermatophagoides farina. NS, P > .05; ∗, P < .05; ∗∗, P < .01; ∗∗∗, P < .001.
4. Discussion
In this study, we conducted a GWAS of asthma in 243 subjects with moderate to severe persistent asthma and allergic sensitization and 134 controls. We observed that no SNP passed the genome-wide threshold of significance; however, genetic variations of BTNL2 harbored the top significant variant (rs3817971) and BTNL2 was associated with atopic moderate to severe persistent asthma. We also revealed that BTNL2 was associated with asthma-related traits such as allergic sensitization and BHR.
BTNL2 gene, a member of the immunoglobulin superfamily, is located at the junction of the HLA class II and class III regions on chromosome 6p21.[24] Owing to its structural homology to the CD80/CD86 family of costimulatory proteins, BTNL2 plays an important role in modulating costimulatory receptors involved in T cell activation during antigen presentation by antigen presenting cells.[25] Human BTNL2 inhibits the proliferation of T cells and reduces the levels of cytokines, such as interleukin-2 and interferon-γ, which are associated with T cell activation.[26–28]BTNL2 also induces the differentiation of regulatory T cells from mature naïve CD4+ T cells.[29,30] To date, there have been consistent reports on its strong genetic association with autoimmune and inflammatory diseases. For instance, sarcoidosis, a systemic inflammatory granulomatous disorder characterized by an exaggerated cellular immune response due to increased inflammatory activity of macrophages and CD4+ helper T cells,[31] is representative of diseases related to BTNL2.[32] The truncating mutation of rs2076530 and the concomitant loss of the IgC domain and transmembrane helix, which disrupt the protein's membrane localization, result in impaired T cell downregulatory function of BTNL2.[33] Moreover, BTNL2 reportedly plays a role in the genetic susceptibility to ulcerative colitis, which represents chronic relapsing inflammatory phenotypes of the atypical T helper 2 (Th2) immune responses affecting mucosal epithelial surfaces.[34,35]
Asthma is a chronic inflammatory disease of the conducting airways and involves both innate and adaptive immune mechanisms through a combination of Th1, Th2, and Th17 responses.[36] It is a heterogeneous disease with respect to its severity, presence of allergic sensitization, total or specific serum IgE levels, and prognosis as well as its difference in children and adults.[1] It is likely that the genetic architecture also differs between the phenotypic subtypes of asthma. Adoption of extreme phenotypes in our study effectively showed an association between genotypes and phenotypes. As we targeted moderate-to-severe atopic asthma in this study, BTNL2 could have been associated with asthma or allergic sensitization. All asthma cases had allergic sensitization; however, no controls had allergic sensitization. Here, we demonstrated that 3 SNPs of BTNL2 were associated with a risk of sensitization toward Der p and Der f. This finding is consistent with that of a previous report by Konno et al,[37] which suggests that BTNL2 is a candidate gene responsible for the pathogenesis of Der f-specific IgE responsiveness. Our results also support previous observations that suggest that non-HLA genes are important for regulating specific IgE-responsiveness to complex allergens such as Der p and Der f.[38,39] Unlike other low molecular weight allergens that show particular HLA class II-associated IgE responsiveness,[40] complex allergens expressing multiple epitopes and sequence motifs[41,42] are expected to be related to more complex immune mechanisms.[43]
BTNL2 polymorphisms were also significantly associated with the presence and degree of BHR in this study. As BHR is one of the important characteristics in asthma pathology, this novel finding also implicates the association between BTNL2 and asthma. Indeed, there have been a few reports from other populations that may reinforce the association of BTNL2 with asthma. Hirota et al[44] identified several susceptibility loci including rs3117098, which is located within the intron of BTNL2, from the GWAS of adult asthma cases in a Japanese cohort. Further, rs3117098 was found to be significantly associated with asthma susceptibility in adult Chinese Zhuang population.[45] Our 3 GWAS-supported variants, that is, these non-coding regions, may play important roles in the regulation of gene activity, thereby affecting disease development and susceptibility.[46]
The present study has several fundamental limitations. Our GWAS had a small sample size; inevitably, no significant variants were found. A GWAS using extreme phenotypes of both cases and controls cannot overcome the limitation of a small sample size.[47] However, we acknowledge that the statistical approach of using a Bonferroni-corrected P value to control the false positive rate and the stringent significance threshold would miss many true associations in a GWAS.[20] Thus, variants with small P values that do not meet genome-wide significance thresholds in GWASs remain to be further investigated as true associations. Additionally, our results should be interpreted with caution because we did not validate the identified variants in an independent cohort. Furthermore, considering BTNL2 is located on chromosome 6p21.3 near the HLA gene cluster, we cannot exclude the possibility that the identified BTNL2–asthma association was merely a reflection of LD with HLA genes. Moreover, this case-control study only provides a hypothesis of the epidemiological etiology. The potential functional mechanisms via which genetic polymorphisms may lead to asthma pathogenesis have not yet been elucidated and thus warrant further investigations.
In conclusion, we observed that BTNL2 was associated with atopic moderate to severe persistent asthma in Korean children. BTNL2 was also associated with asthma-related traits, including allergic sensitization to mite and BHR. To our knowledge, this is the first study in a Korean population that has demonstrated an association between the genetic variants of BTNL2 with childhood asthma and asthma-related traits using a GWAS. However, further studies are needed to clarify the immunogenetic basis of this association with childhood asthma.
Author contributions
Conceptualization, Soo Yeon Kim, Min Jung Kim, Yoon Hee Kim, Myung Hyun Sohn and Kyung Won Kim; Data curation, Ga Eun Kim, Min Jung Kim, Yoon Hee Kim and Kyung Won Kim; Formal analysis, Eun Gyul Kim, Mi Na Kim and Jung Yeon Hong; Funding acquisition, Kyung Won Kim; Investigation, Soo Yeon Kim, Jae Hwa Jung and Mireu Park; Methodology, Soo Yeon Kim, Eun Gyul Kim, Ga Eun Kim, Jae Hwa Jung and Mireu Park; Supervision, Myung Hyun Sohn and Kyung Won Kim; Writing – original draft, Soo Yeon Kim; Writing – review & editing, Kyung Won Kim. All authors have read and agreed to the published version of the manuscript.
Conceptualization: Soo Yeon Kim, Min Jung Kim, Yoon Hee Kim, Myung Hyun Sohn, Kyung Won Kim.
Data curation: Ga Eun Kim, Min Jung Kim, Yoon Hee Kim, Kyung Won Kim.
Formal analysis: Eun Gyul Kim, Mi Na Kim, Jung Yeon Hong.
Funding acquisition: Kyung Won Kim.
Investigation: Soo Yeon Kim, Jae Hwa Jung, Mireu Park.
Methodology: Soo Yeon Kim, Eun Gyul Kim, Ga Eun Kim, Jae Hwa Jung, Mireu Park.
Supervision: Myung Hyun Sohn, Kyung Won Kim.
Writing – original draft: Soo Yeon Kim.
Writing – review & editing: Kyung Won Kim.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: BDR = bronchodilator response, BHR = bronchial hyperresponsiveness, BTNL2 = Butyrophilin-like 2, Der f = Dermatophagoides farina, Der p = Dermatophagoides pteryonyssinus, FEV1 = forced expiratory volume in 1 s, GWAS = genome-wide association study, IgE = immunoglobulin E, LD = linkage disequilibrium, SNP = single nucleotide polymorphism.
How to cite this article: Kim SY, Kim EG, Kim MN, Hong JY, Kim GE, Jung JH, Park M, Kim MJ, Kim YH, Sohn MH, Kim KW. Genome-wide association study identifies BTNL2 associated with atopic asthma in children. Medicine. 2021;100:44(e27626).
This work was supported by a faculty research grant of Yonsei University College of Medicine (6-2012-0016) and the National Research Foundation Grant funded by the Korean Government (NRF-2019R1F1A1058910).
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of SEVERANCE HOSPITAL (IRB no. 4-2004-0036).
Informed consent was obtained from all subjects involved in the study.
The authors declare no conflict of interest.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplemental digital content is available for this article.
References
- [1].Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet 2008;372:1107–19. [DOI] [PubMed] [Google Scholar]
- [2].Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 2010;363:1211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet 2016;48:709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kim KW, Ober C. Lessons learned from GWAS of asthma. Allergy Asthma Immunol Res 2018;11:170–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Drake KA, Torgerson DG, Gignoux CR, et al. A genome-wide association study of bronchodilator response in Latinos implicates rare variants. J Allergy Clin Immunol 2014;133:370–8. e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nieuwenhuis M, Vonk J, Himes B, et al. PTTG 1 IP and MAML 3, novel genomewide association study genes for severity of hyperresponsiveness in adult asthma. Allergy 2017;72:792–801. [DOI] [PubMed] [Google Scholar]
- [7].Gudbjartsson DF, Bjornsdottir US, Halapi E, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet 2009;41:342–7. [DOI] [PubMed] [Google Scholar]
- [8].Weidinger S, Gieger C, Rodriguez E, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet 2008;4:e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bønnelykke K, Matheson MC, Pers TH, et al. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat Genet 2013;45:902–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science 2008;319:1100–4. [DOI] [PubMed] [Google Scholar]
- [11].Shriner D, Adeyemo A, Gerry NP, et al. Transferability and fine-mapping of genome-wide associated loci for adult height across human populations. PLoS One 2009;4:e8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Baye TM, Butsch Kovacic M, Biagini Myers JM, et al. Differences in candidate gene association between European ancestry and African American asthmatic children. PLoS One 2011;6:e16522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kim SH, Cho BY, Park CS, et al. Alpha-T-catenin (CTNNA3) gene was identified as a risk variant for toluene diisocyanate-induced asthma by genome-wide association analysis. Clin Exp Allergy 2009;39:203–12. [DOI] [PubMed] [Google Scholar]
- [14].Kim J-H, Park B-L, Cheong HS, et al. Genome-wide and follow-up studies identify CEP68 gene variants associated with risk of aspirin-intolerant asthma. PLoS One 2010;5:e13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shin SW, Park J, Kim YJ, et al. A highly sensitive and specific genetic marker to diagnose aspirin-exacerbated respiratory disease using a genome-wide association study. DNA Cell Biol 2012;31:1604–9. [DOI] [PubMed] [Google Scholar]
- [16].Park BL, Kim T-H, Kim J-H, et al. Genome-wide association study of aspirin-exacerbated respiratory disease in a Korean population. Human Genetics 2013;132:313–21. [DOI] [PubMed] [Google Scholar]
- [17].Kim S-H, Cho B-Y, Choi H, et al. The SNP rs3128965 of HLA-DPB1 as a genetic marker of the AERD phenotype. PLoS One 2014;9:e111220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Park S-Y, Jung H, Kim J-H, et al. Longitudinal analysis to better characterize Asthma-COPD overlap syndrome: findings from an adult asthma cohort in Korea (COREA). Clin Exp Allergy 2019; 10.1111/cea.13339 [DOI] [PubMed] [Google Scholar]
- [19].Kim J-H, Cheong HS, Park JS, et al. A genome-wide association study of total serum and mite-specific IgEs in asthma patients. PLoS One 2013;8:e71958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ober C. Asthma genetics in the post-GWAS era. Ann Am Thorac Soc 2016;13: (Suppl 1): S85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Turner SD. qqman: an R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv 2014;005165. [Google Scholar]
- [23].Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26:2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Traherne JA, Barcellos LF, Sawcer SJ, et al. Association of the truncating splice site mutation in BTNL2 with multiple sclerosis is secondary to HLA-DRB1∗15. Hum Mol Genet 2006;15:155–61. [DOI] [PubMed] [Google Scholar]
- [25].Arnett HA, Escobar SS, Viney JL. Regulation of costimulation in the era of butyrophilins. Cytokine 2009;46:370–5. [DOI] [PubMed] [Google Scholar]
- [26].Nguyen T, Liu XK, Zhang Y, Dong C. BTNL2, a butyrophilin-like molecule that functions to inhibit T cell activation. J Immunol 2006;176:7354–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Arnett HA, Escobar SS, Gonzalez-Suarez E, et al. BTNL2, a butyrophilin/B7-like molecule, is a negative costimulatory molecule modulated in intestinal inflammation. J Immunol 2007;178:1523–33. [DOI] [PubMed] [Google Scholar]
- [28].Yamazaki T, Goya I, Graf D, Craig S, Martin-Orozco N, Dong C. A butyrophilin family member critically inhibits T cell activation. J Immunol 2010;185:5907–14. [DOI] [PubMed] [Google Scholar]
- [29].Ammann JU, Cooke A, Trowsdale J. Butyrophilin Btn2a2 inhibits TCR activation and phosphatidylinositol 3-kinase/Akt pathway signaling and induces Foxp3 expression in T lymphocytes. J Immunol 2013;190:5030–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Swanson RM, Gavin MA, Escobar SS, et al. Butyrophilin-like 2 modulates B7 costimulation to induce Foxp3 expression and regulatory T cell development in mature T cells. J Immunol 2013;190:2027–35. [DOI] [PubMed] [Google Scholar]
- [31].Newman LS, Rose CS, Maier LA. Medical progress: sarcoidosis. N Engl J Med 1997;336:1224–34. [DOI] [PubMed] [Google Scholar]
- [32].Calender A, Weichhart T, Valeyre D, Pacheco Y. Current insights in genetics of sarcoidosis: functional and clinical impacts. J Clin Med 2020;9:2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Valentonyte R, Hampe J, Huse K, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet 2005;37:357–64. [DOI] [PubMed] [Google Scholar]
- [34].Mochida A, Kinouchi Y, Negoro K, et al. Butyrophilin-like 2 gene is associated with ulcerative colitis in the Japanese under strong linkage disequilibrium with HLA-DRB1∗ 1502. Tissue Antig 2007;70:128–35. [DOI] [PubMed] [Google Scholar]
- [35].Pathan S, Gowdy R, Cooney R, et al. Confirmation of the novel association at the BTNL2 locus with ulcerative colitis. Tissue Antig 2009;74:322–9. [DOI] [PubMed] [Google Scholar]
- [36].Lambrecht BN, Hammad H. The immunology of asthma. Nat Immunol 2015;16:45–56. [DOI] [PubMed] [Google Scholar]
- [37].Konno S, Takahashi D, Hizawa N, et al. Genetic impact of a butyrophilin-like 2 (BTNL2) gene variation on specific IgE responsiveness to Dermatophagoides farinae (Der f) in Japanese. Allergol Int 2009;58:29–35. [DOI] [PubMed] [Google Scholar]
- [38].Hizawa N, Freidhoff LR, Ehrlich E, et al. Genetic regulation of Dermatophagoides pteronyssinus-specific IgE responsiveness: a genome-wide multipoint linkage analysis in families recruited through 2 asthmatic sibs. J Allergy Clin Immunol 1998;102:436–42. [DOI] [PubMed] [Google Scholar]
- [39].Blumenthal M, Ober C, Beaty T, et al. Genome scan for loci linked to mite sensitivity: the Collaborative Study on the Genetics of Asthma (CSGA). Genes Immun 2004;5:226–31. [DOI] [PubMed] [Google Scholar]
- [40].Huang SK, Zwollo P, Marsh DG. Class II major histocompatibility complex restriction of human T cell responses to short ragweed allergen, Amb a V. Eur J Immunol 1991;21:1469–73. [DOI] [PubMed] [Google Scholar]
- [41].Yssel H, Johnson K, Schneider P, et al. T cell activation-inducing epitopes of the house dust mite allergen Der p I. Proliferation and lymphokine production patterns by Der p I-specific CD4+ T cell clones. J Immunol 1992;148:738–45. [PubMed] [Google Scholar]
- [42].van Neerven RJ, van t’Hof W, Ringrose J, et al. T cell epitopes of house dust mite major allergen Der p II. J Immunol 1993;151:2326–35. [PubMed] [Google Scholar]
- [43].Marsh DG, Bias WB, Ishizaka K. Genetic control of basal serum immunoglobulin E level and its effect on specific reaginic sensitivity. Proc Natl Acad Sci 1974;71:3588–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hirota T, Takahashi A, Kubo M, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet 2011;43:893–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liang SQ, Deng JM, Wei X, et al. Association of GWAS-supported noncoding area loci rs404860, rs3117098, and rs7775228 with asthma in Chinese Zhuang population. J Clin Lab Anal 2020;34:e23066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gloss BS, Dinger ME. Realizing the significance of noncoding functionality in clinical genomics. Exp Mol Med 2018;50:01–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kim KW, Myers RA, Lee JH, et al. Genome-wide association study of recalcitrant atopic dermatitis in Korean children. J Allergy Clin Immunol 2015;136:678–84. e674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.